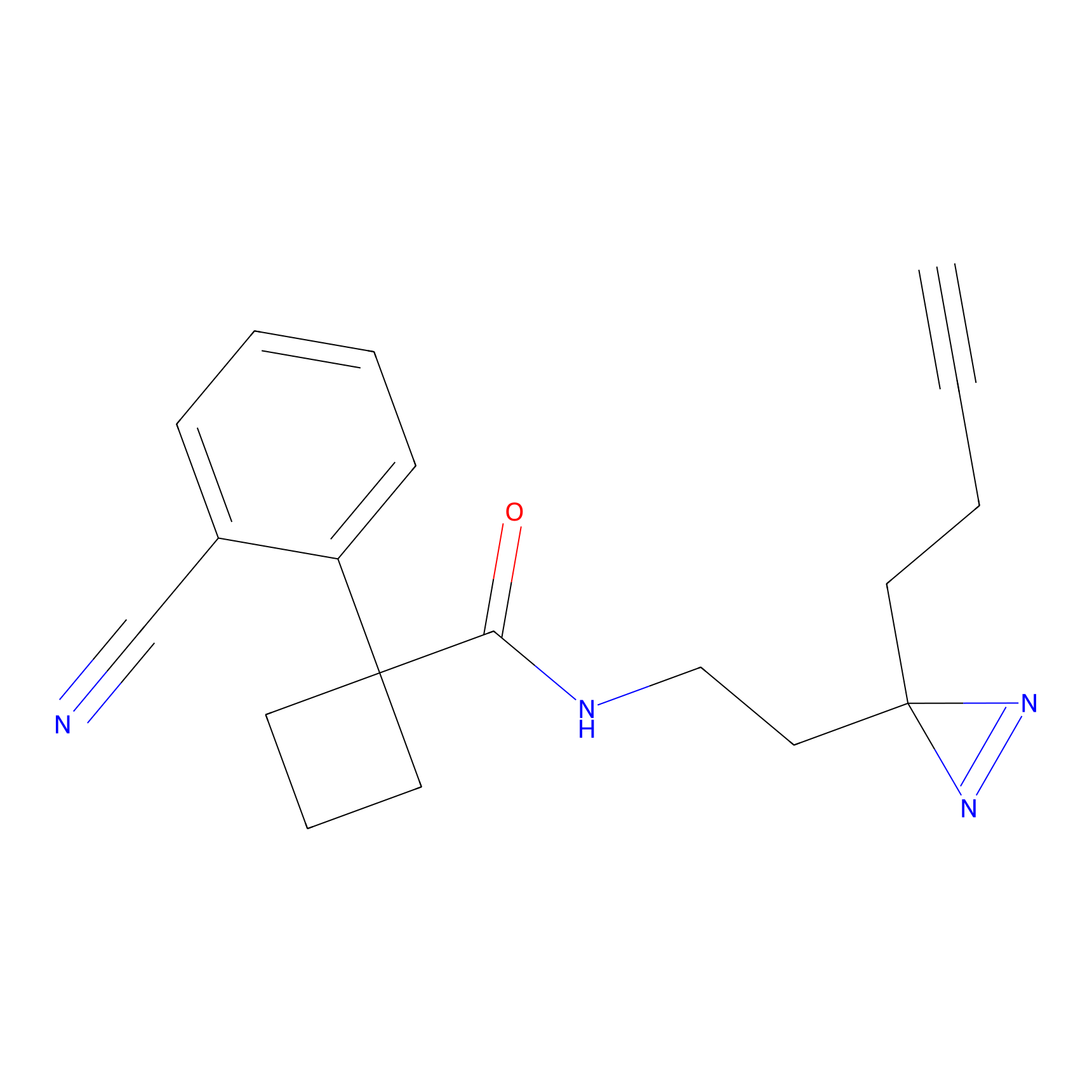
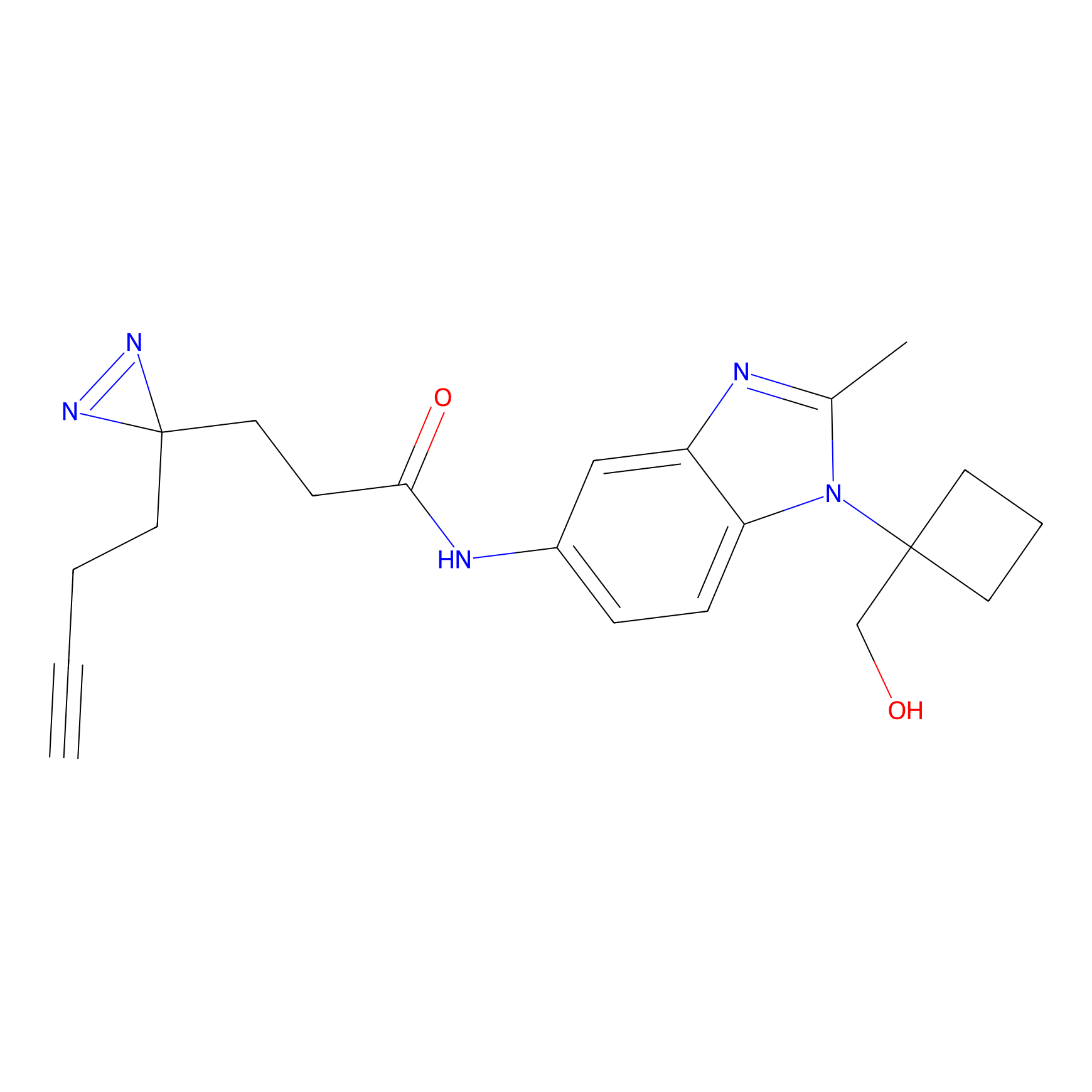
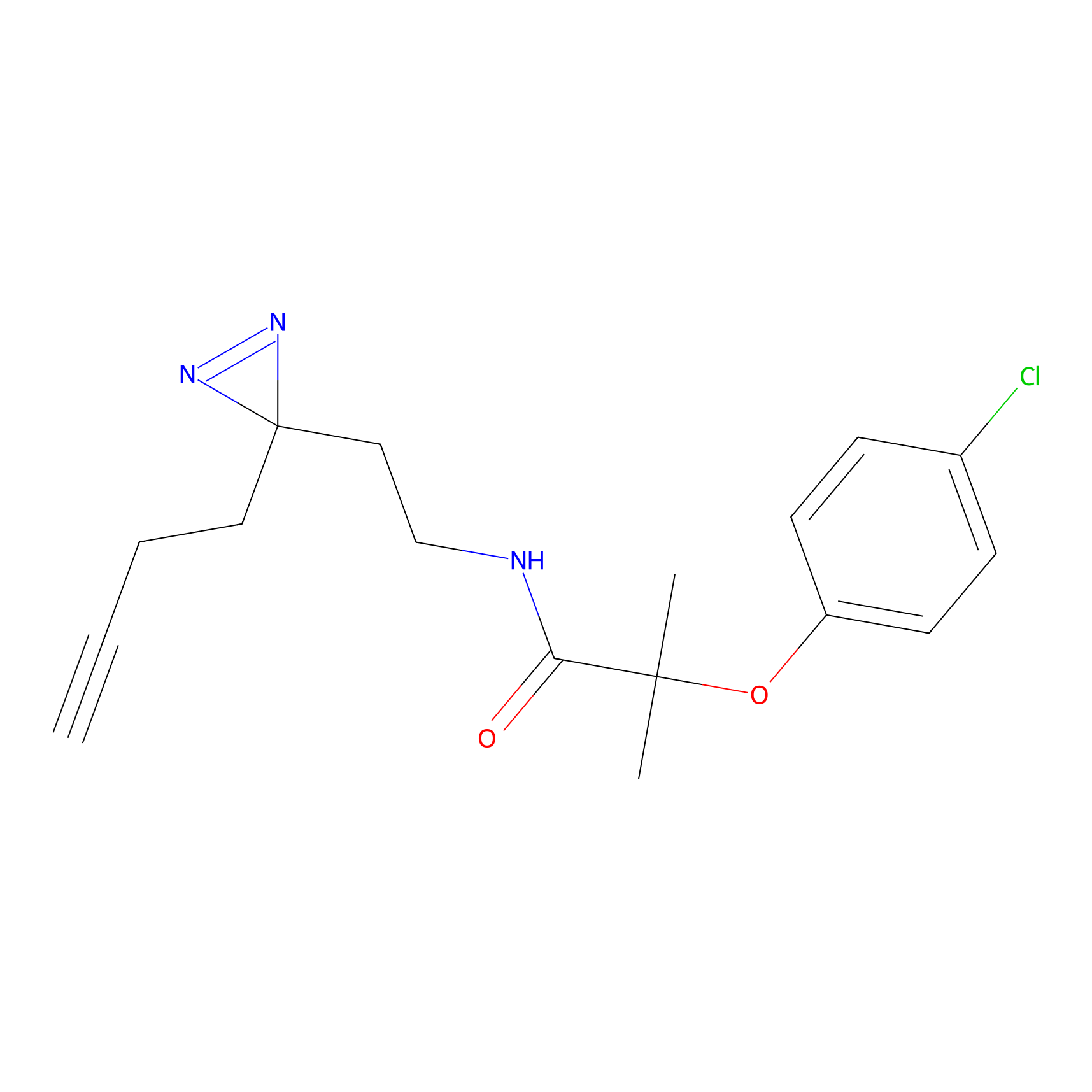
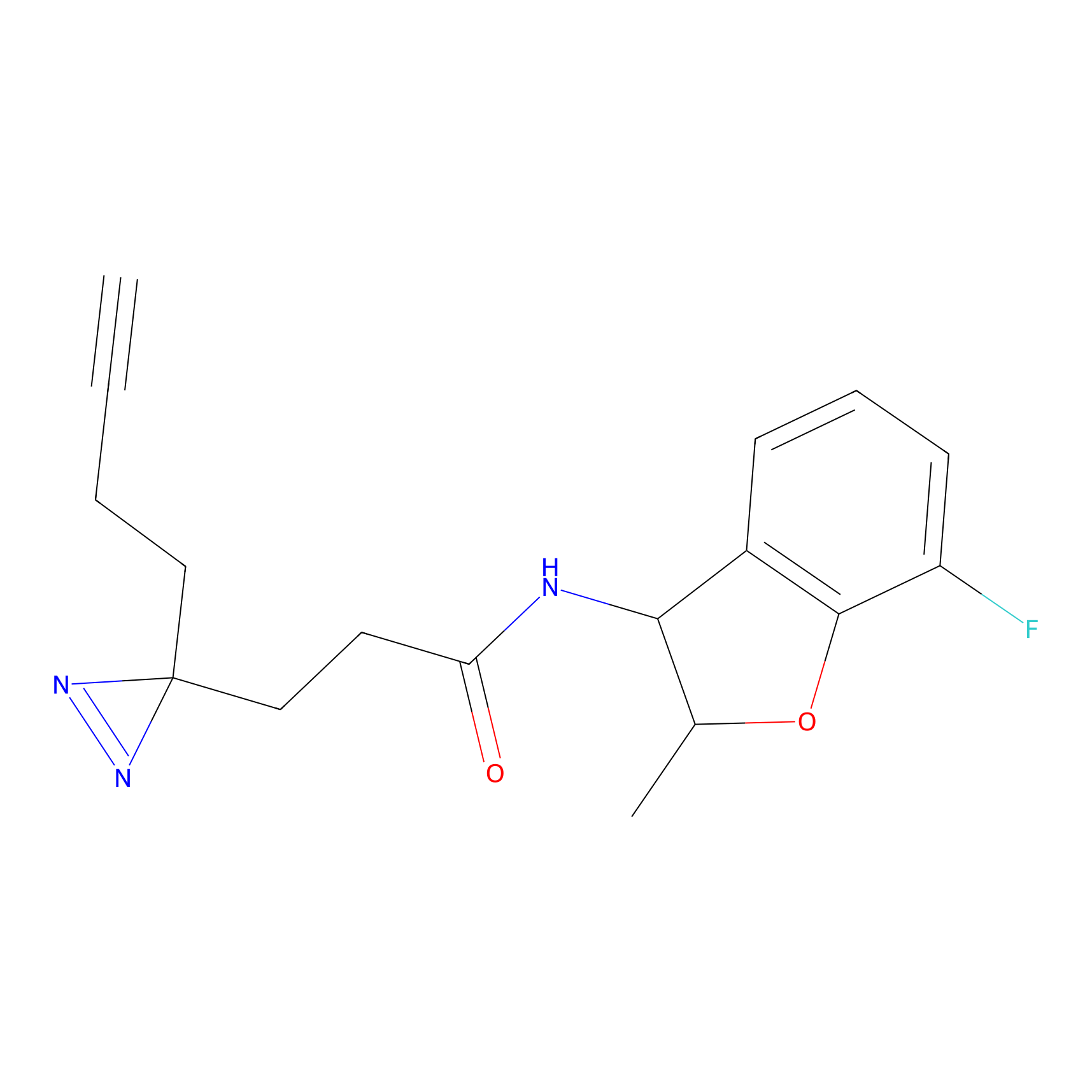
Details of the Target
General Information of Target
| Target ID | LDTP00281 | |||||
|---|---|---|---|---|---|---|
| Target Name | Golgi integral membrane protein 4 (GOLIM4) | |||||
| Gene Name | GOLIM4 | |||||
| Gene ID | 27333 | |||||
| Synonyms |
GIMPC; GOLPH4; GPP130; Golgi integral membrane protein 4; Golgi integral membrane protein, cis; GIMPc; Golgi phosphoprotein 4; Golgi-localized phosphoprotein of 130 kDa; Golgi phosphoprotein of 130 kDa
|
|||||
| 3D Structure | ||||||
| Sequence |
MGNGMCSRKQKRIFQTLLLLTVVFGFLYGAMLYYELQTQLRKAEAVALKYQQHQESLSAQ
LQVVYEHRSRLEKSLQKERLEHKKAKEDFLVYKLEAQETLNKGRQDSNSRYSALNVQHQM LKSQHEELKKQHSDLEEEHRKQGEDFSRTFNDHKQKYLQLQQEKEQELSKLKETVYNLRE ENRQLRKAHQDIHTQLQDVKQQHKNLLSEHEQLVVTLEDHKSALAAAQTQVAEYKQLKDT LNRIPSLRKPDPAEQQNVTQVAHSPQGYNTAREKPTREVQEVSRNNDVWQNHEAVPGRAE DTKLYAPTHKEAEFQAPPEPIQQEVERREPEEHQVEEEHRKALEEEEMEQVGQAEHLEEE HDPSPEEQDREWKEQHEQREAANLLEGHARAEVYPSAKPMIKFQSPYEEQLEQQRLAVQQ VEEAQQLREHQEALHQQRLQGHLLRQQEQQQQQVAREMALQRQAELEEGRPQHQEQLRQQ AHYDAMDNDIVQGAEDQGIQGEEGAYERDNQHQDEAEGDPGNRHEPREQGPREADPESEA DRAAVEDINPADDPNNQGEDEFEEAEQVREENLPDENEEQKQSNQKQENTEVEEHLVMAG NPDQQEDNVDEQYQEEAEEEVQEDLTEEKKRELEHNAEETYGENDENTDDKNNDGEEQEV RDDNRPKGREEHYEEEEEEEEDGAAVAEKSHRRAEM |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
GOLIM4 family
|
|||||
| Subcellular location |
Golgi apparatus, Golgi stack membrane
|
|||||
| Function | Plays a role in endosome to Golgi protein trafficking; mediates protein transport along the late endosome-bypass pathway from the early endosome to the Golgi. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
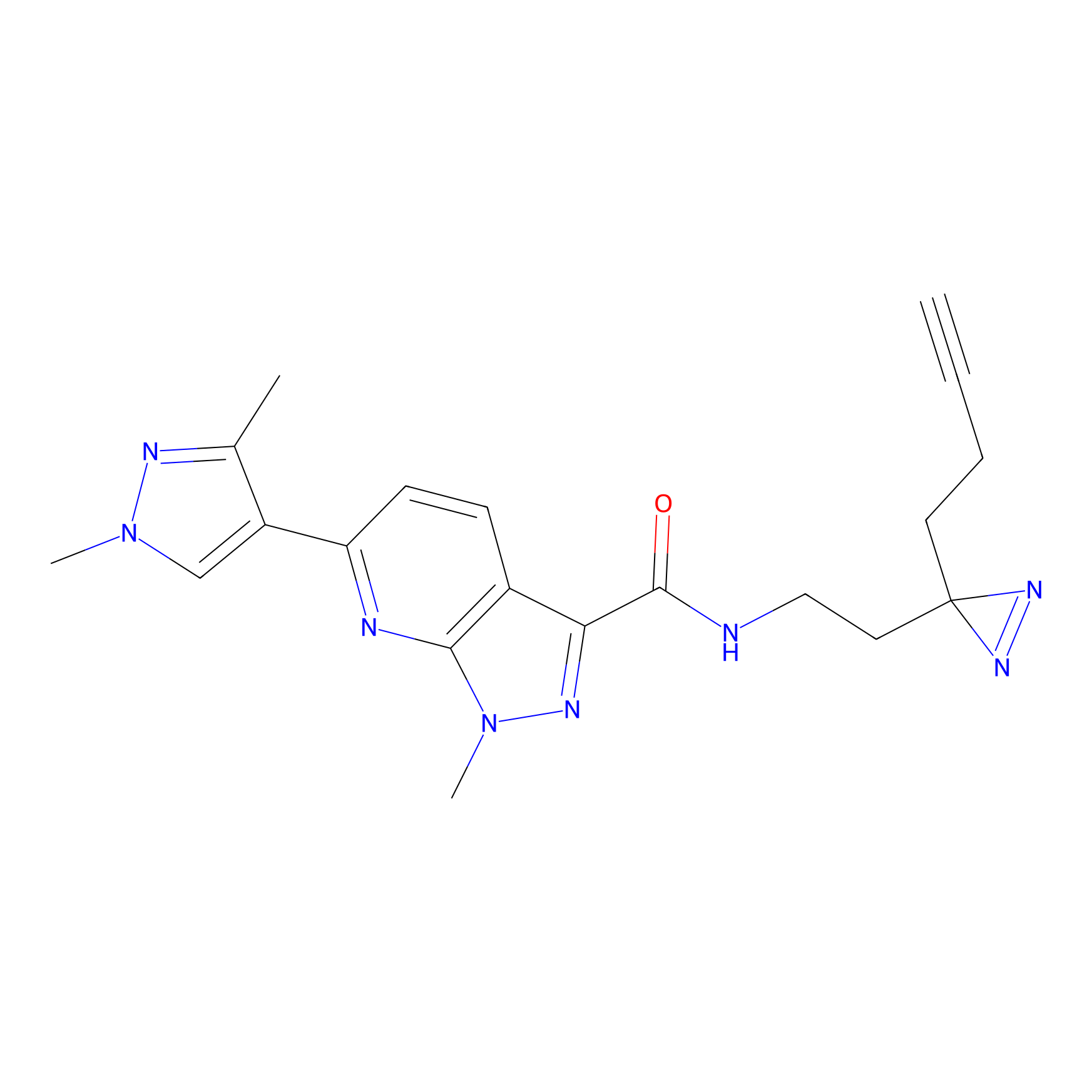
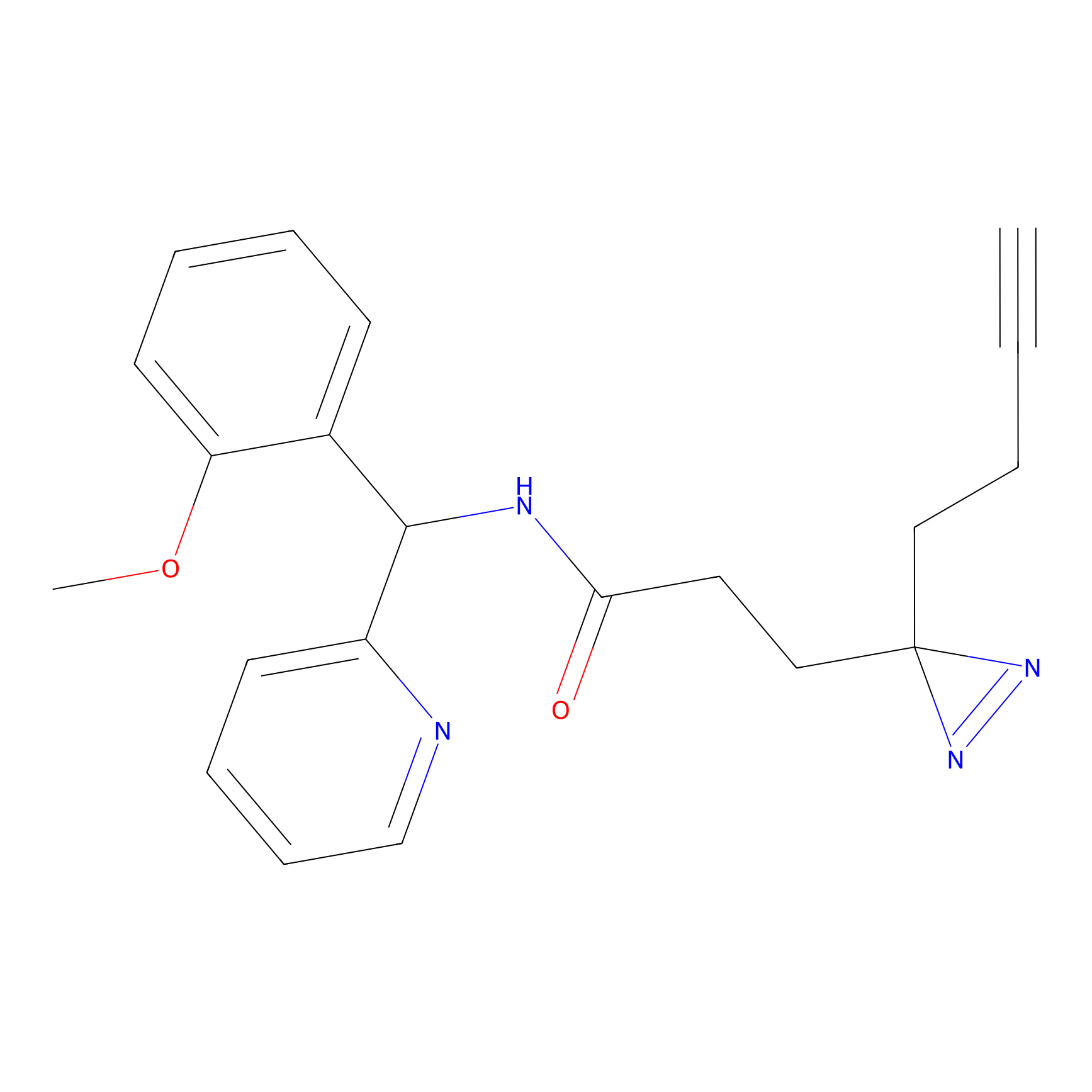
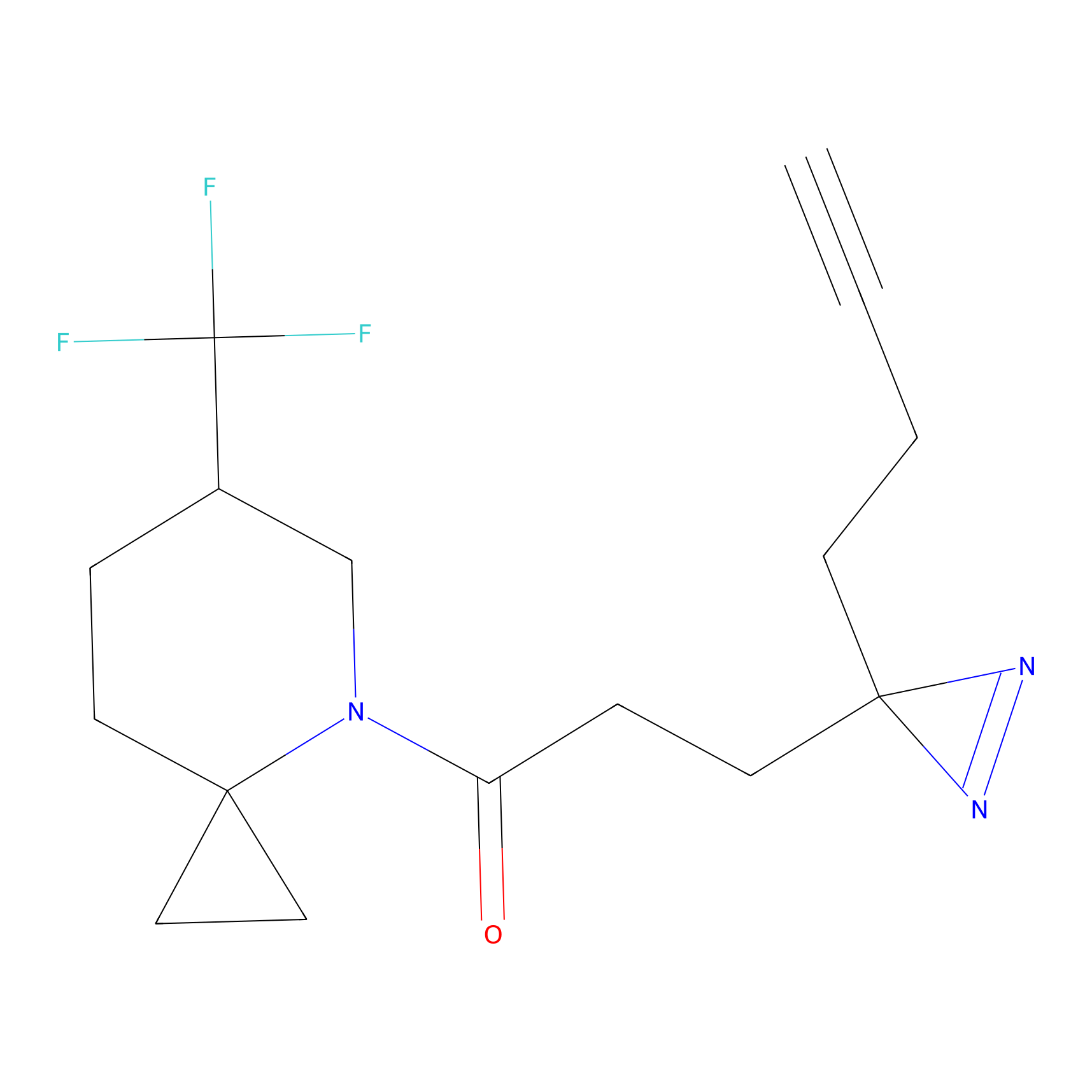
|
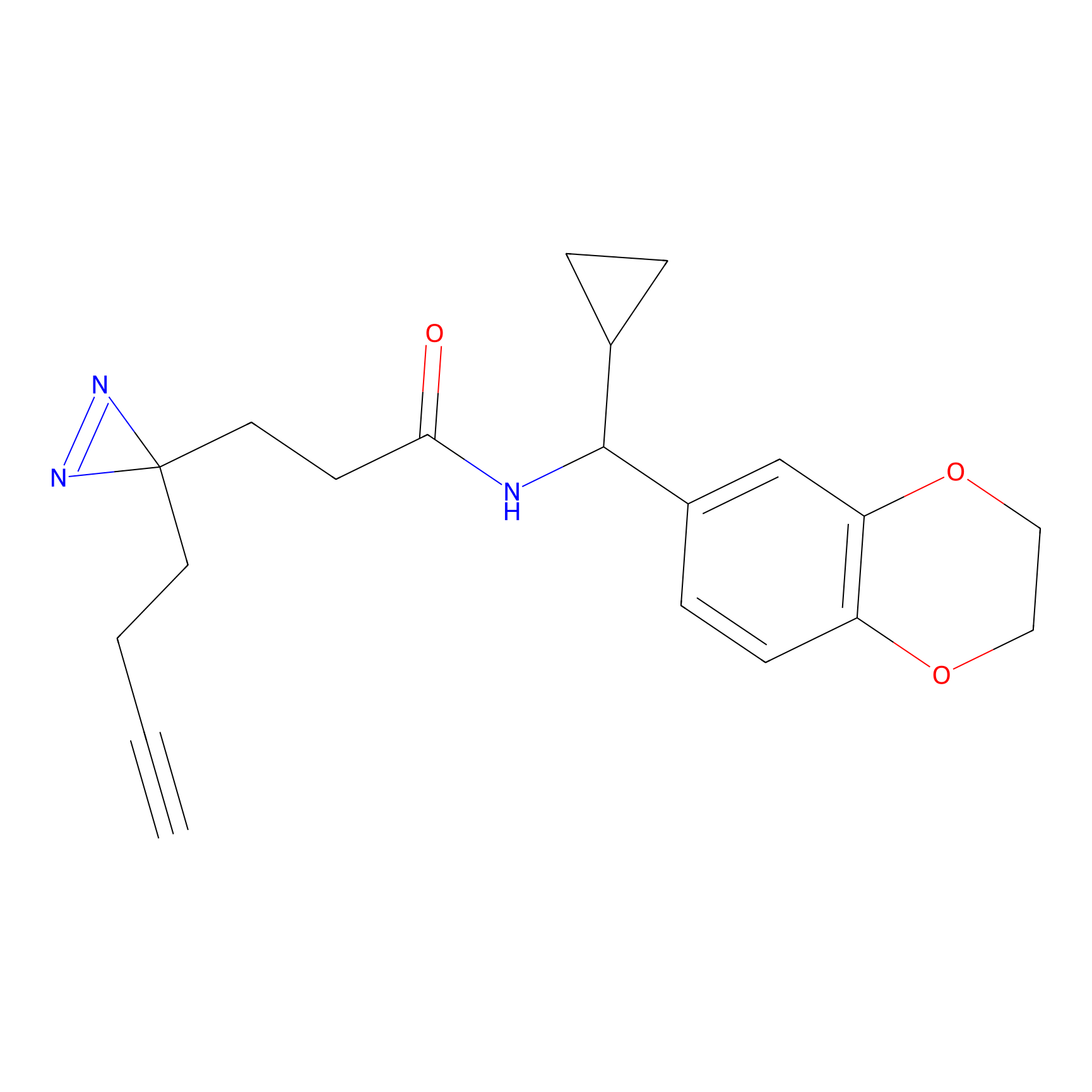
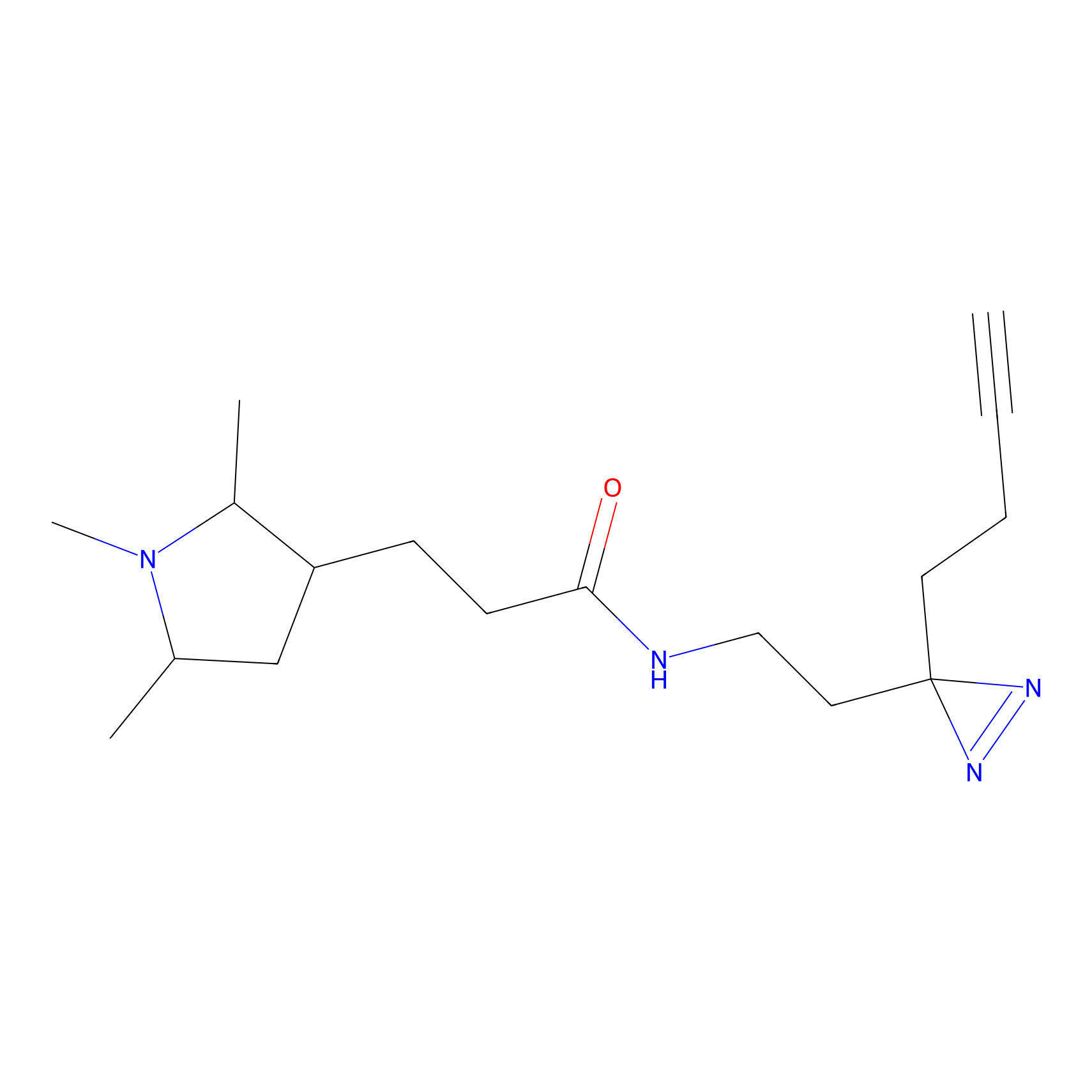
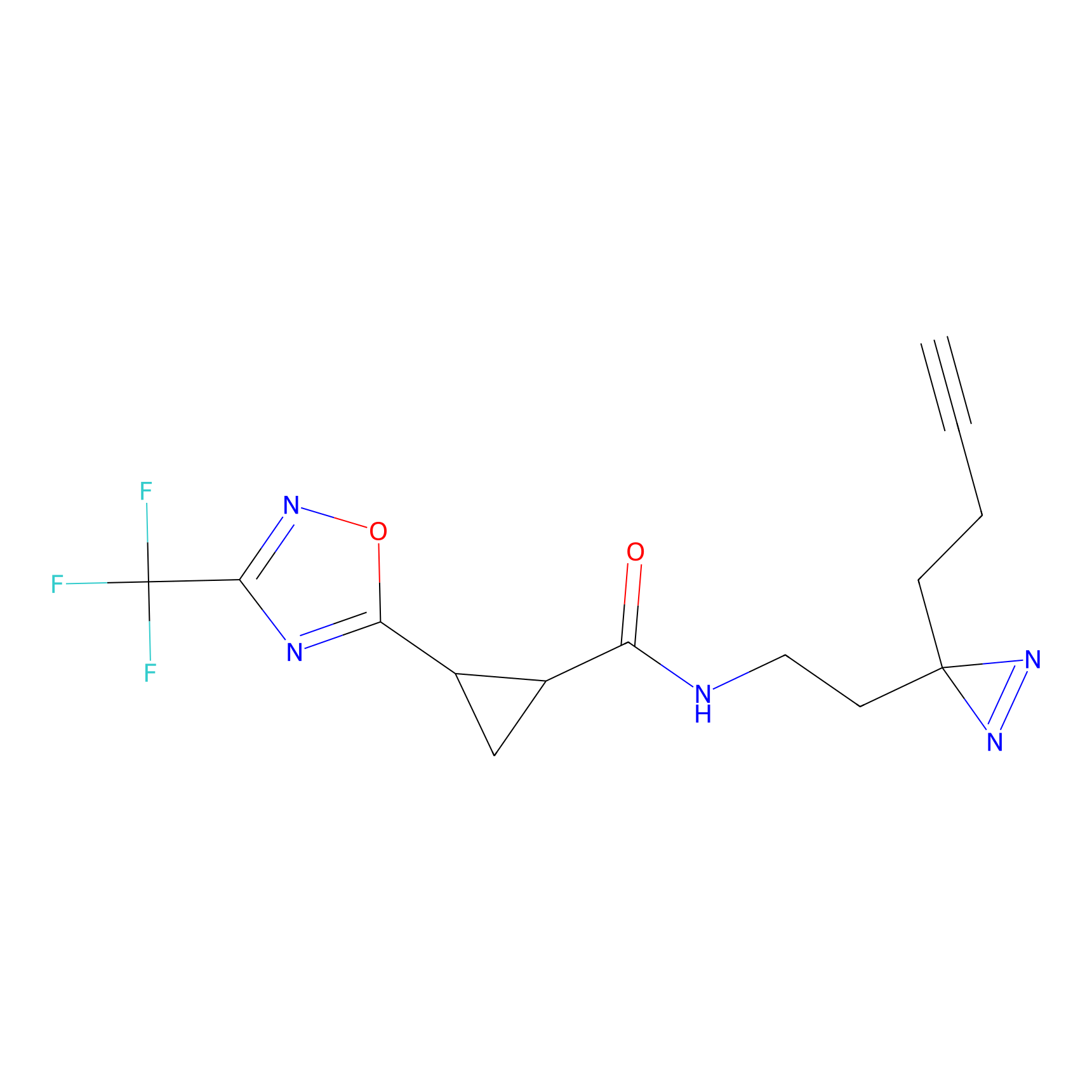
FBPP2 Probe Info |
 |
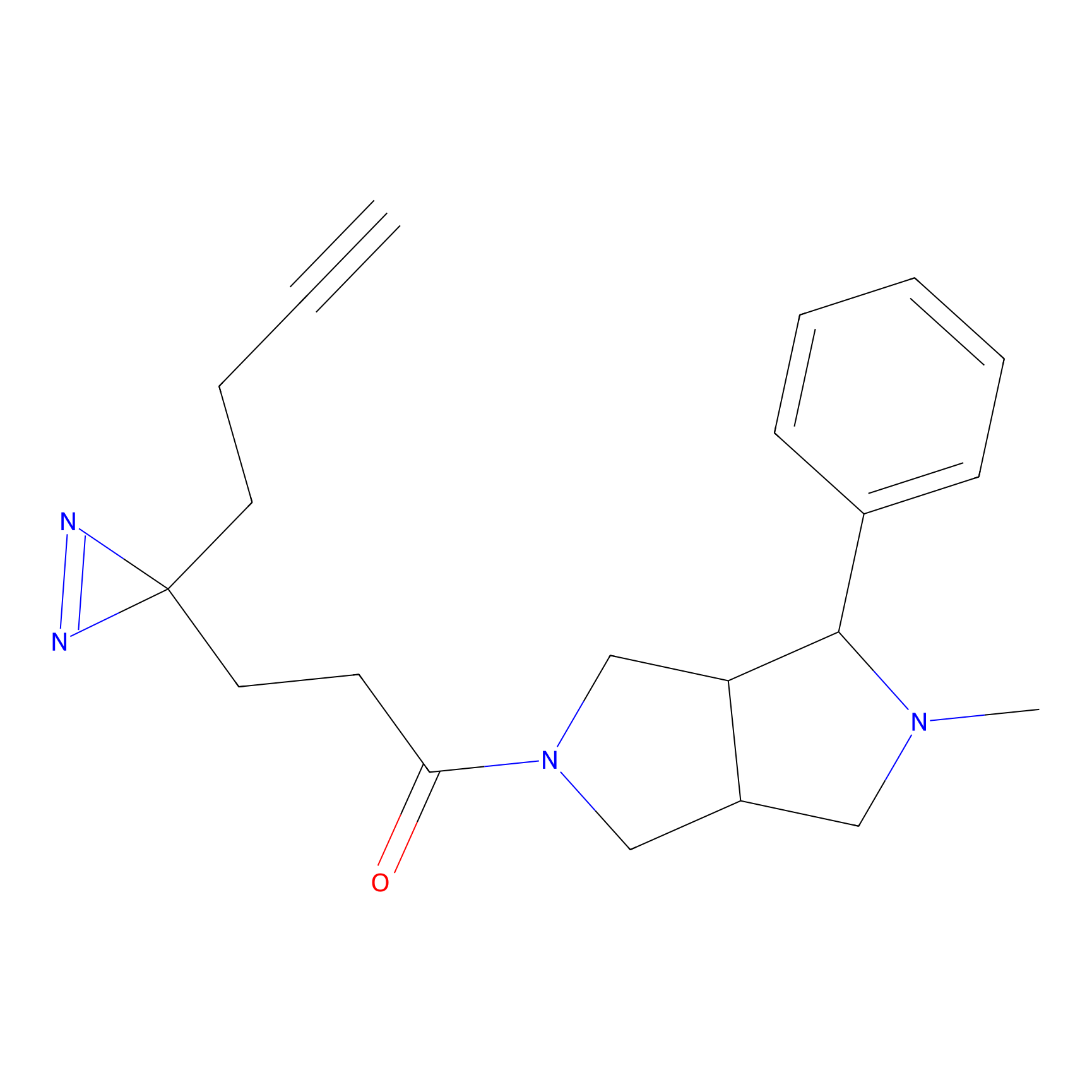
71.99 | LDD0318 | [1] | |
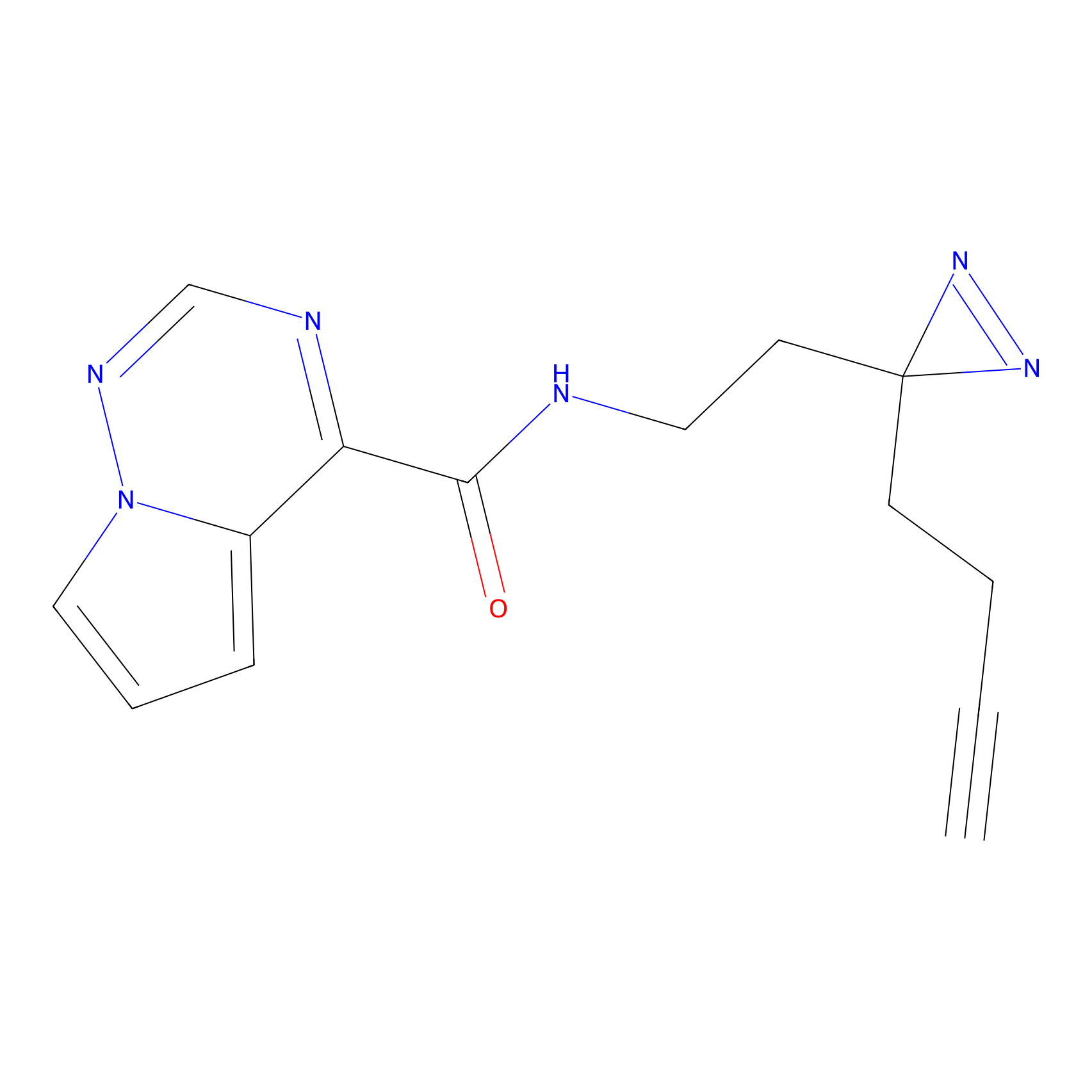
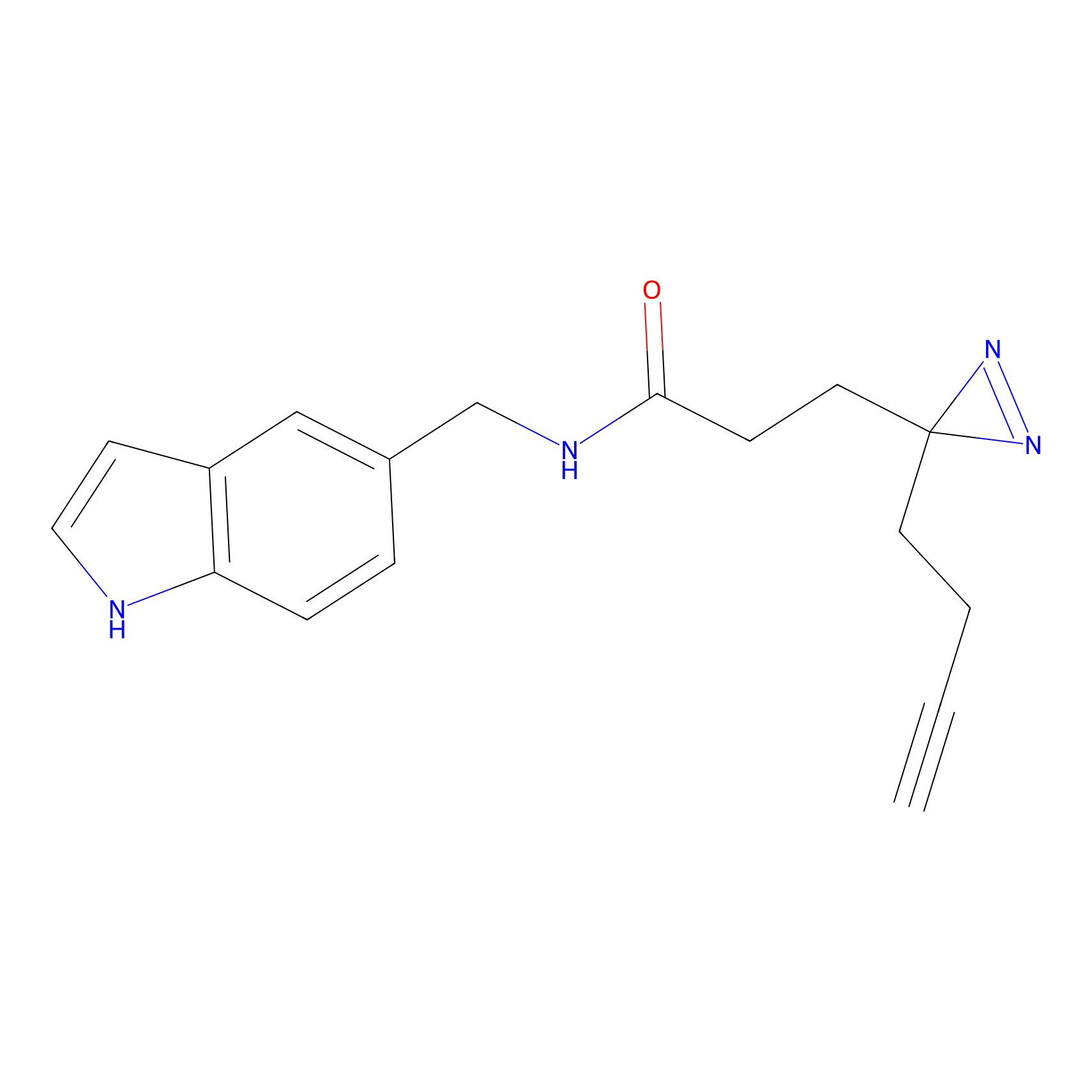
|
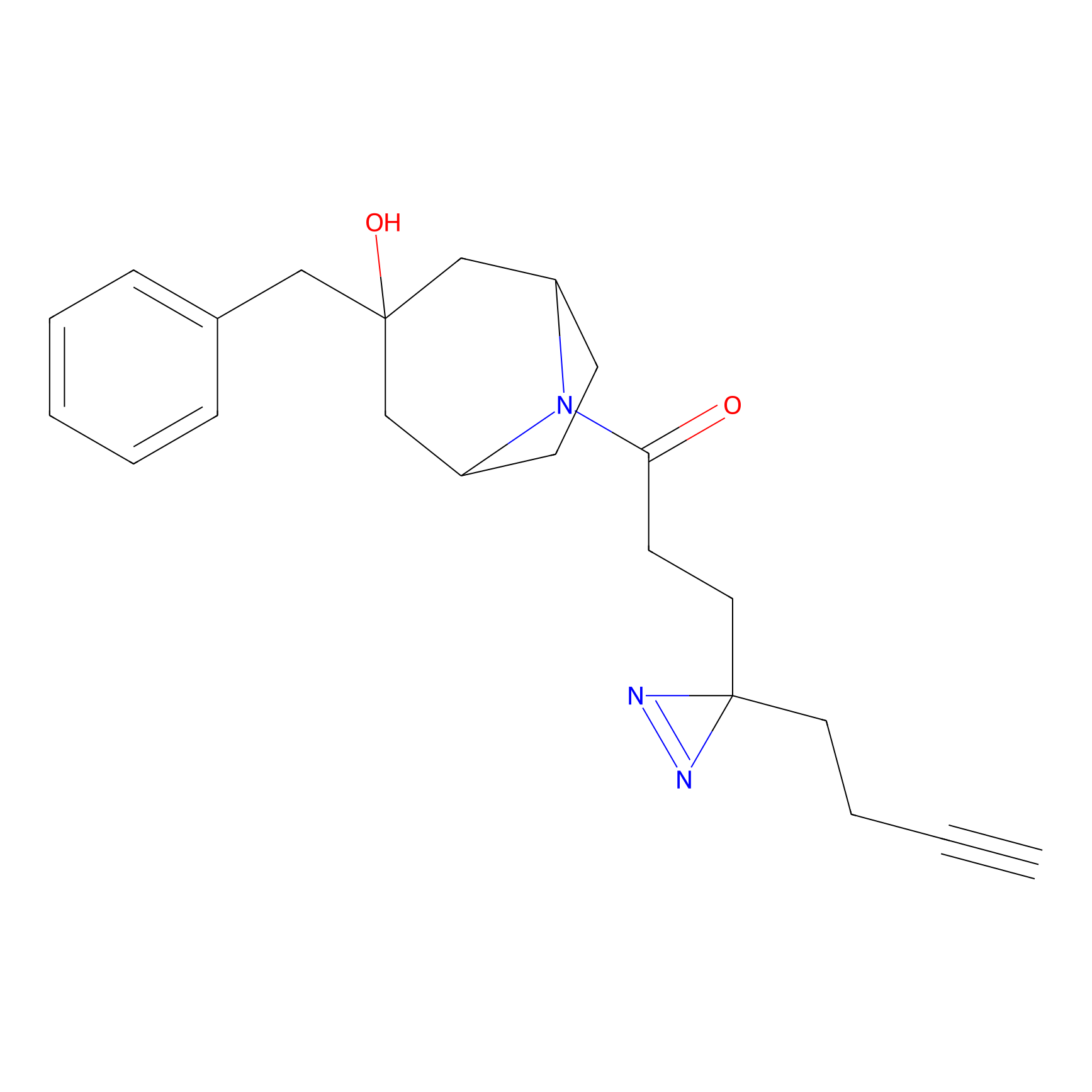
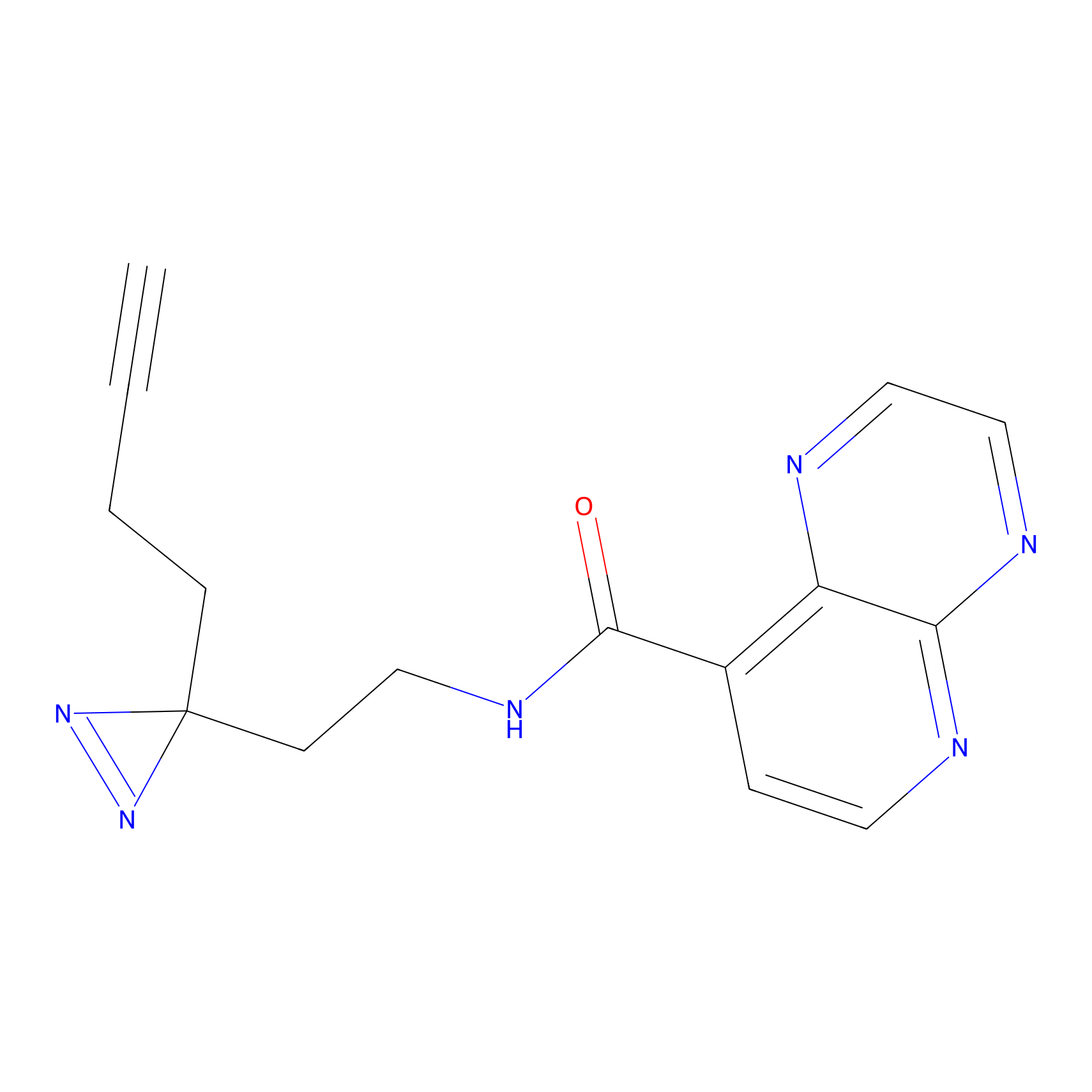
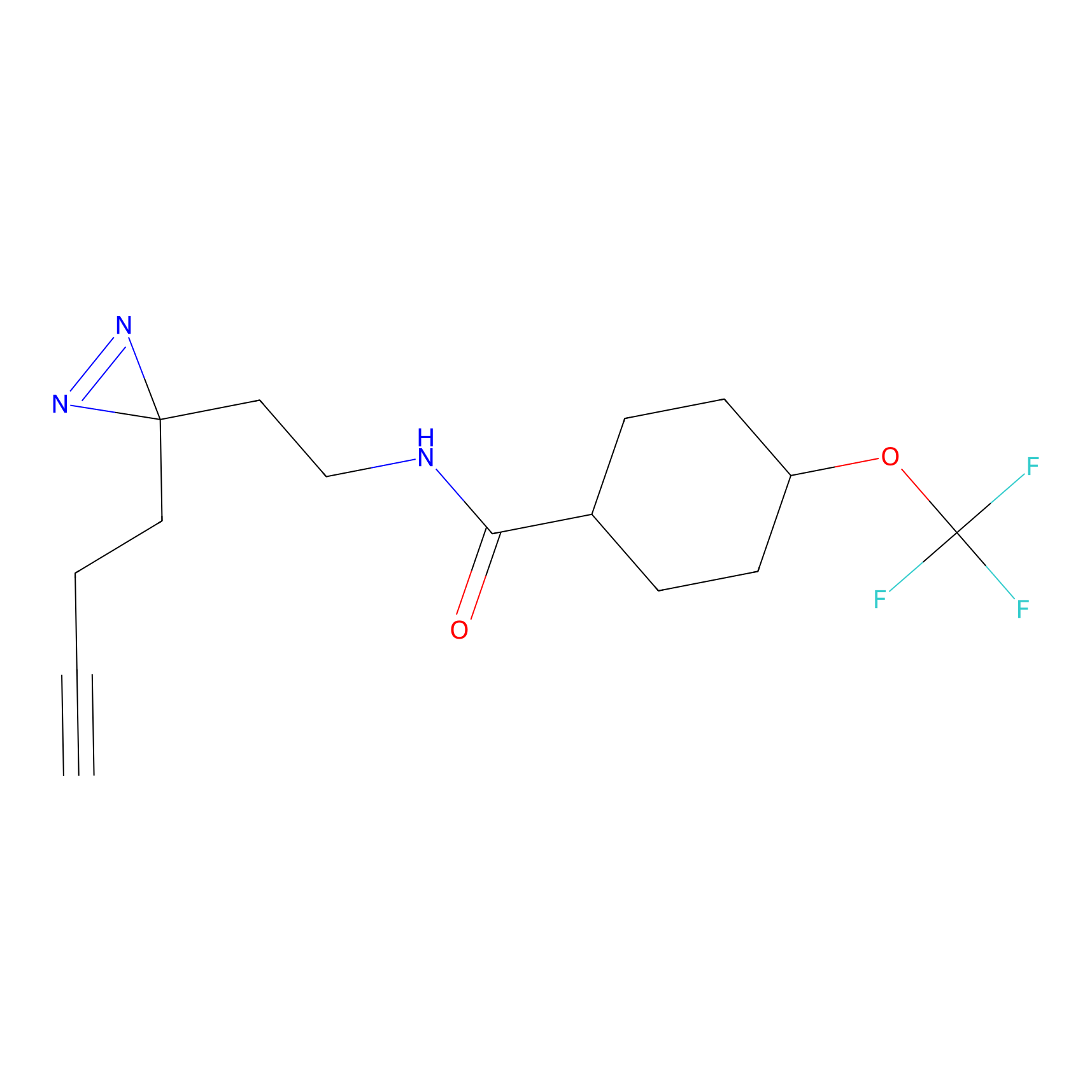
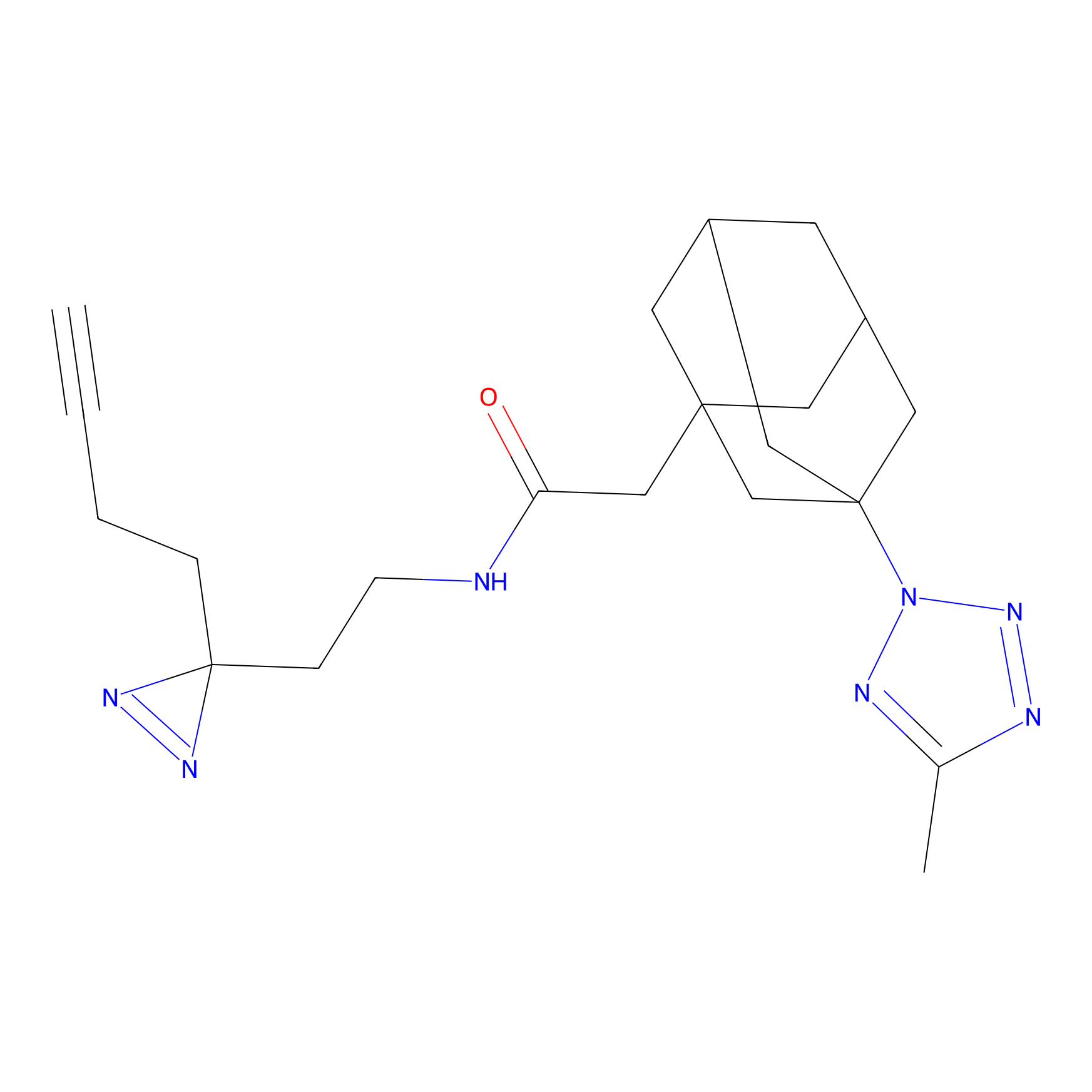
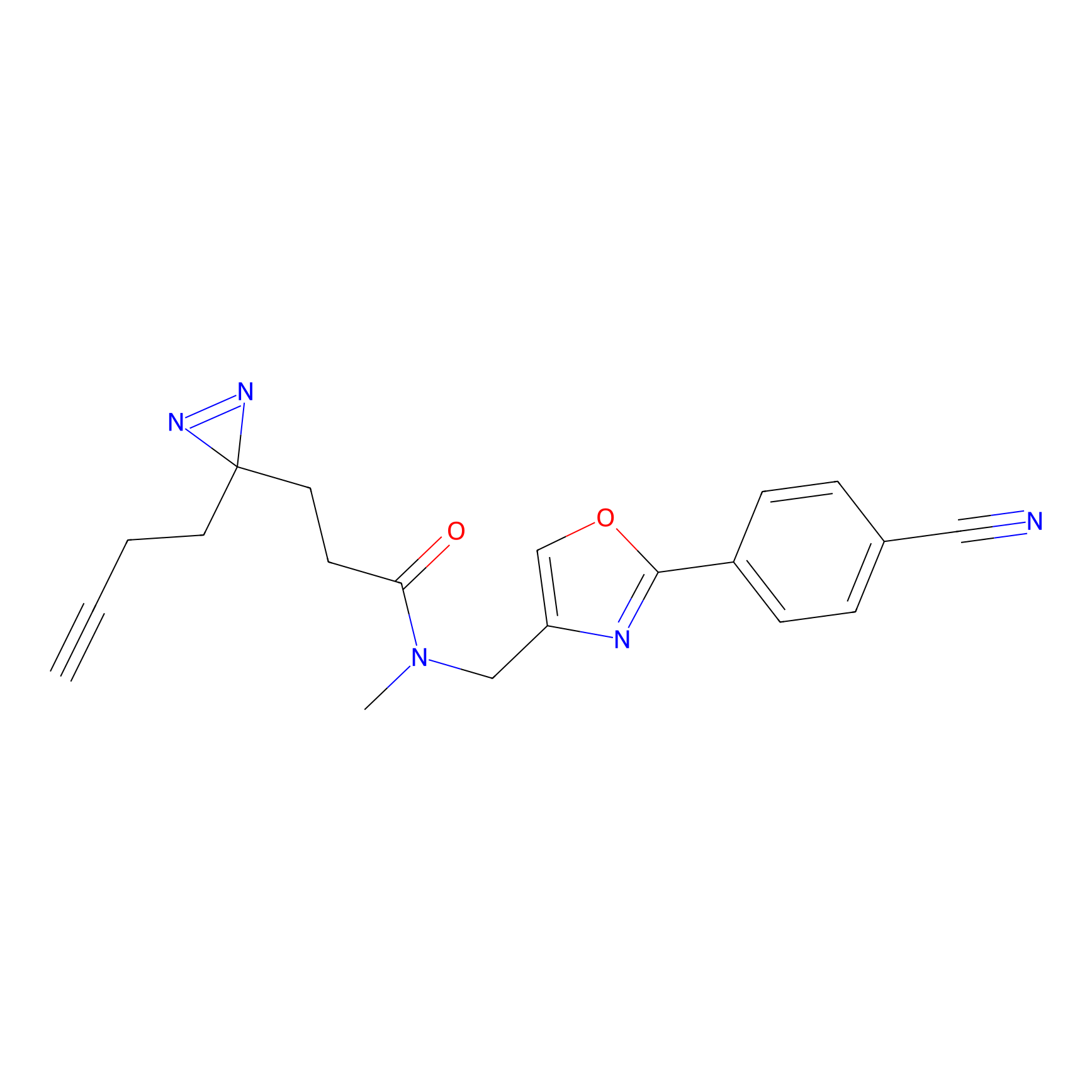
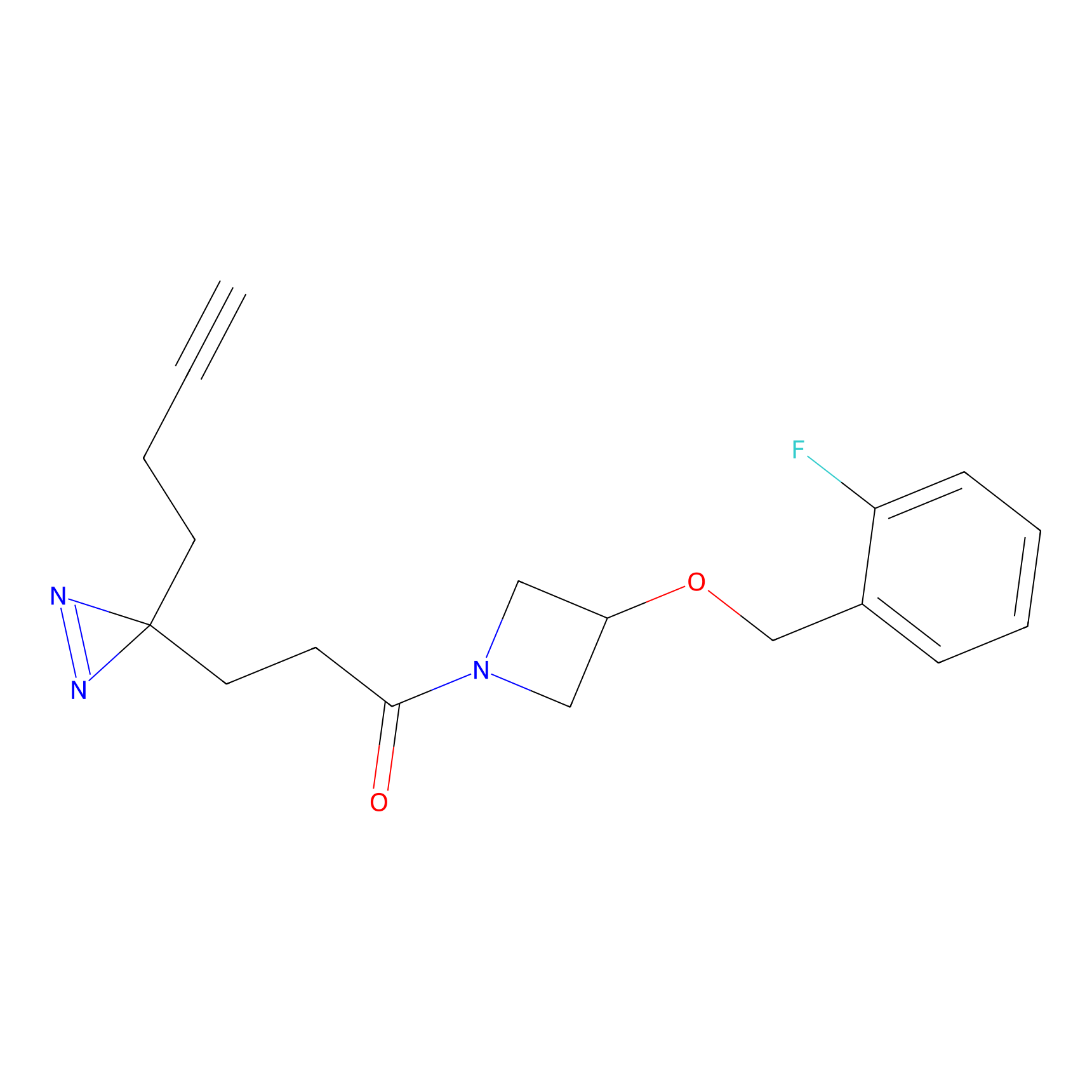
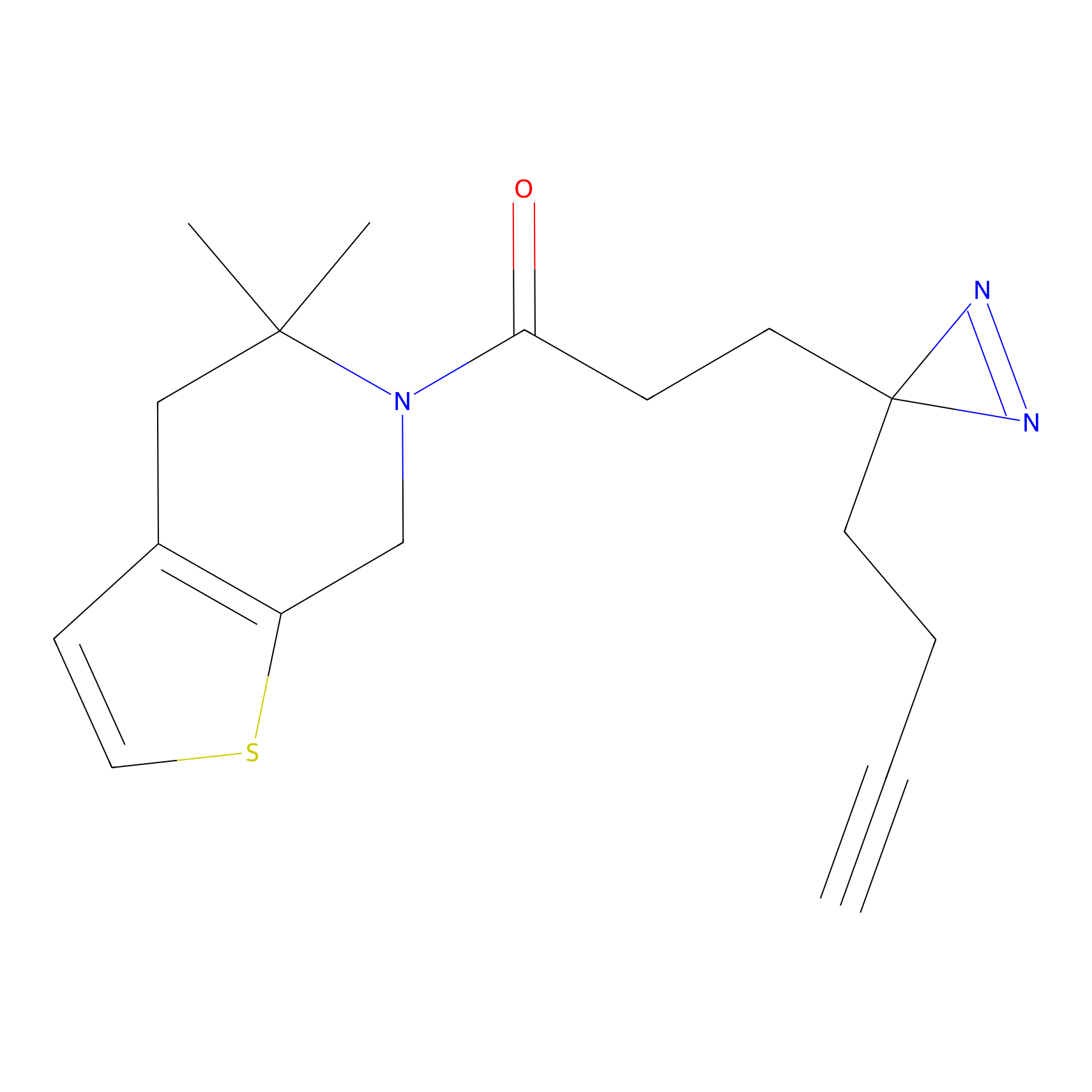
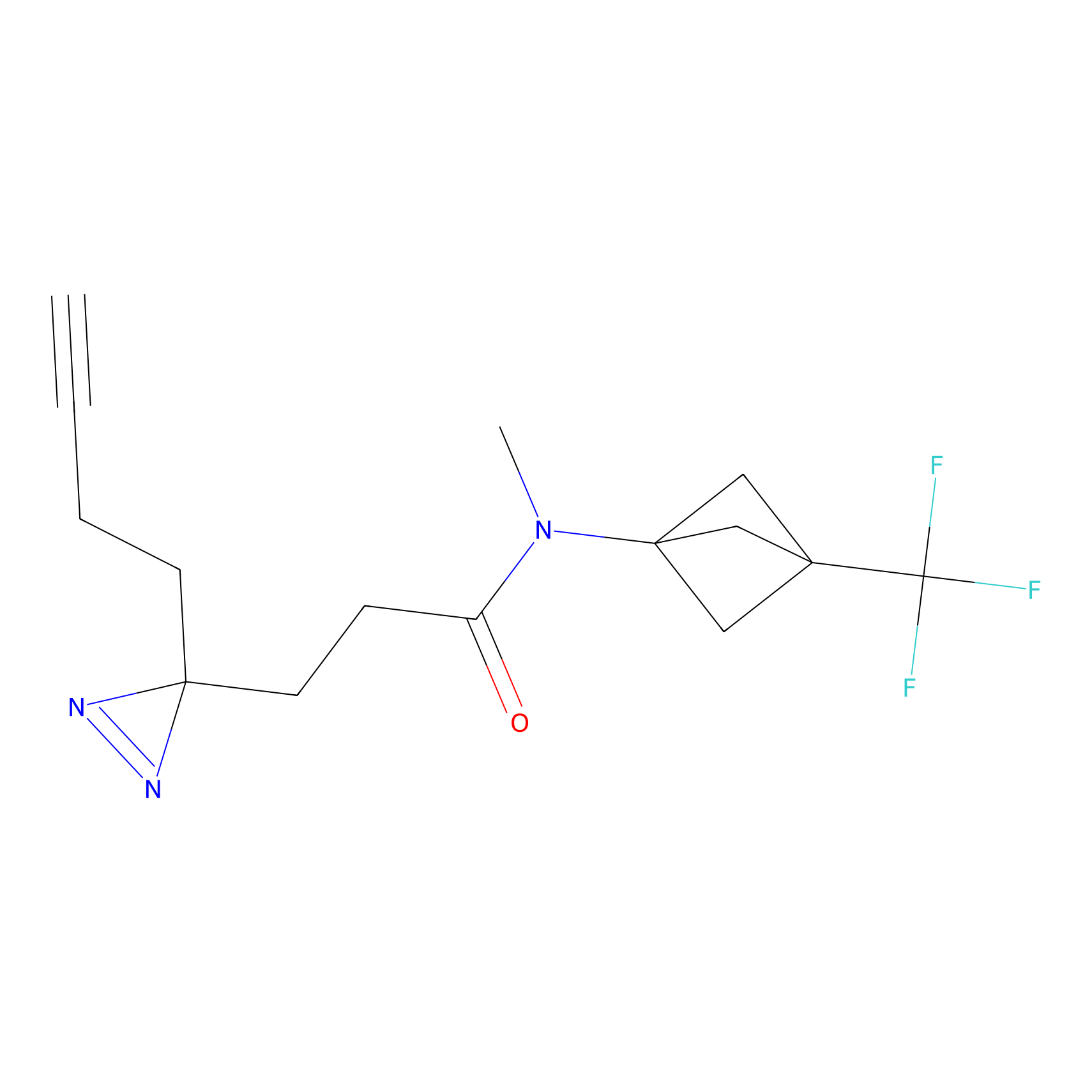
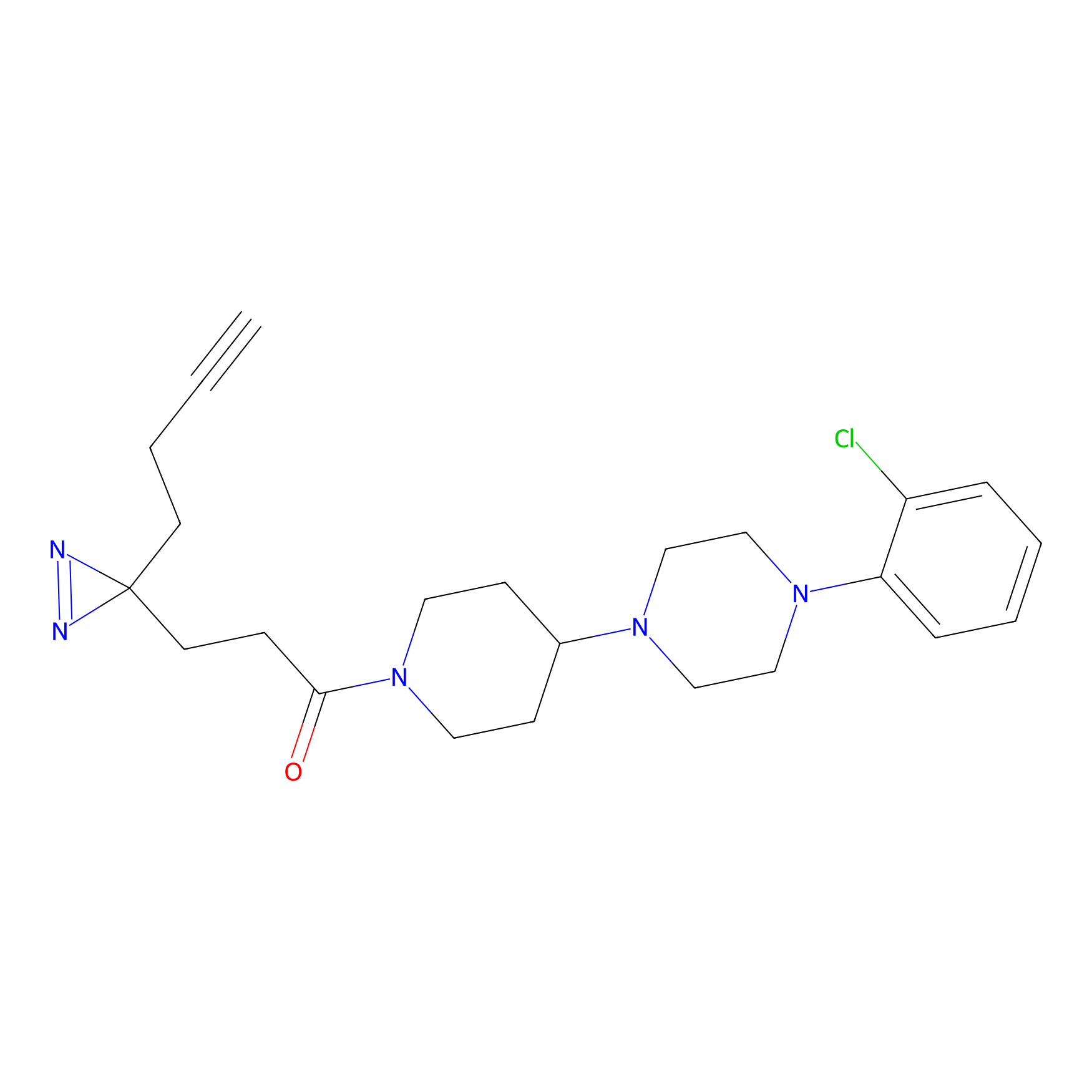
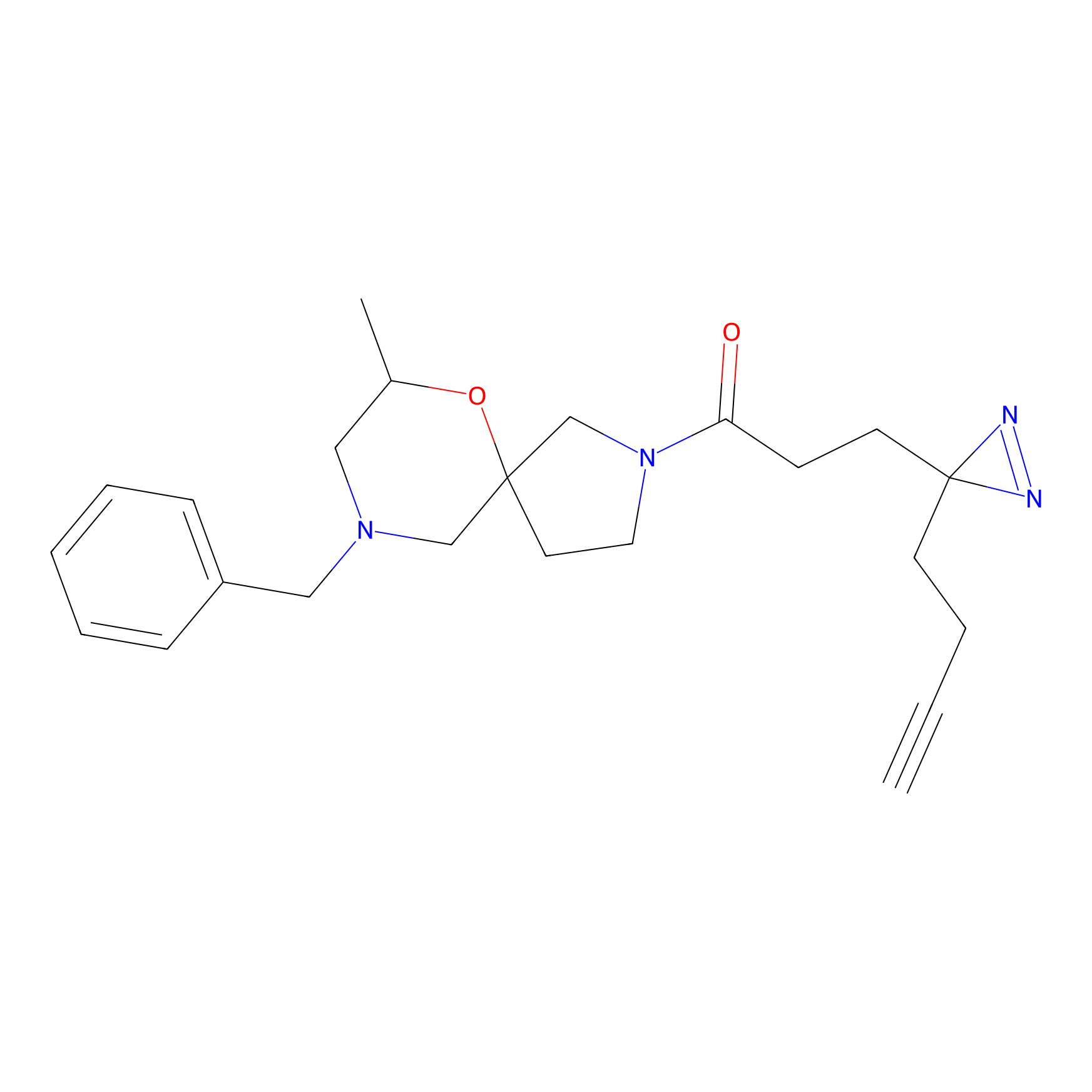
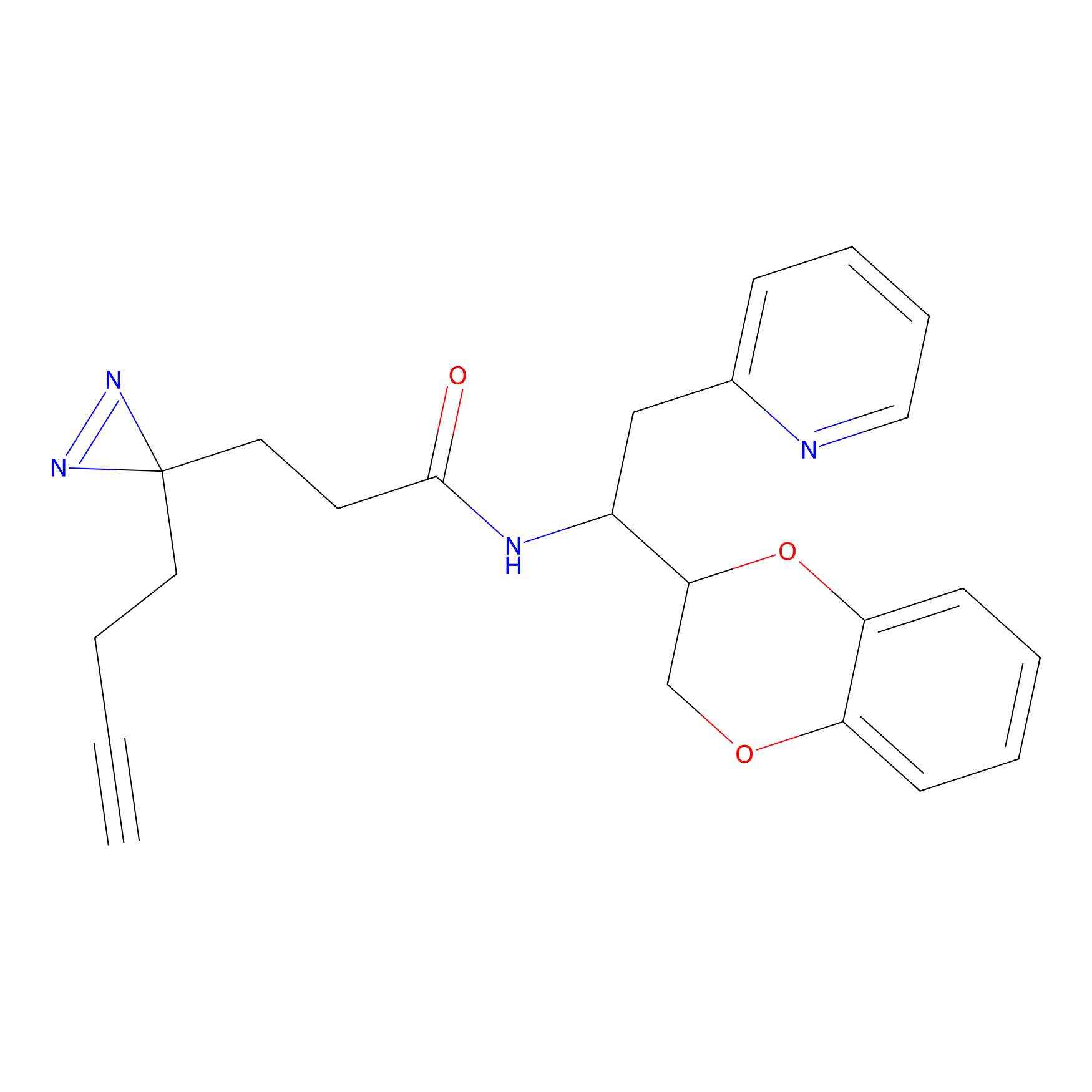
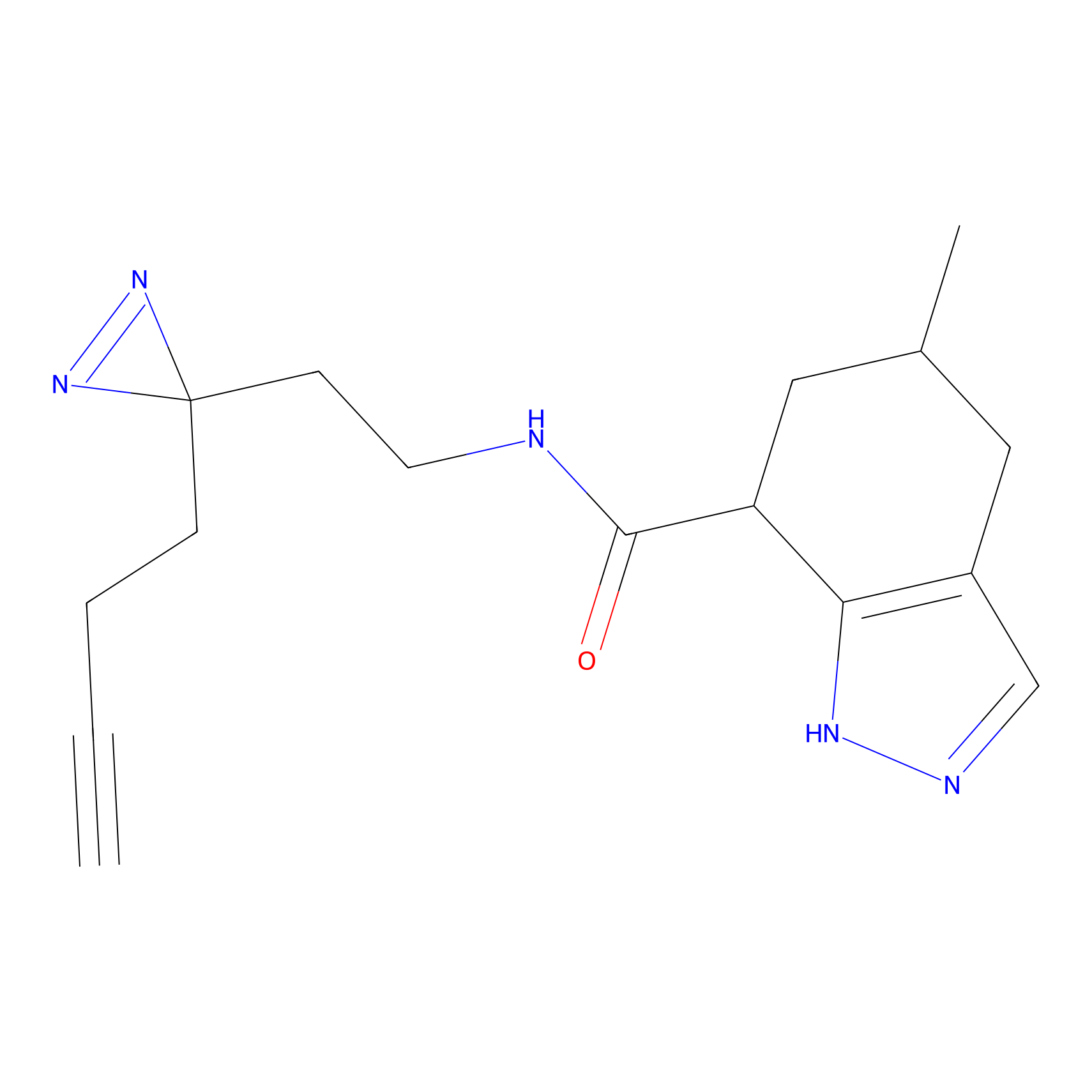
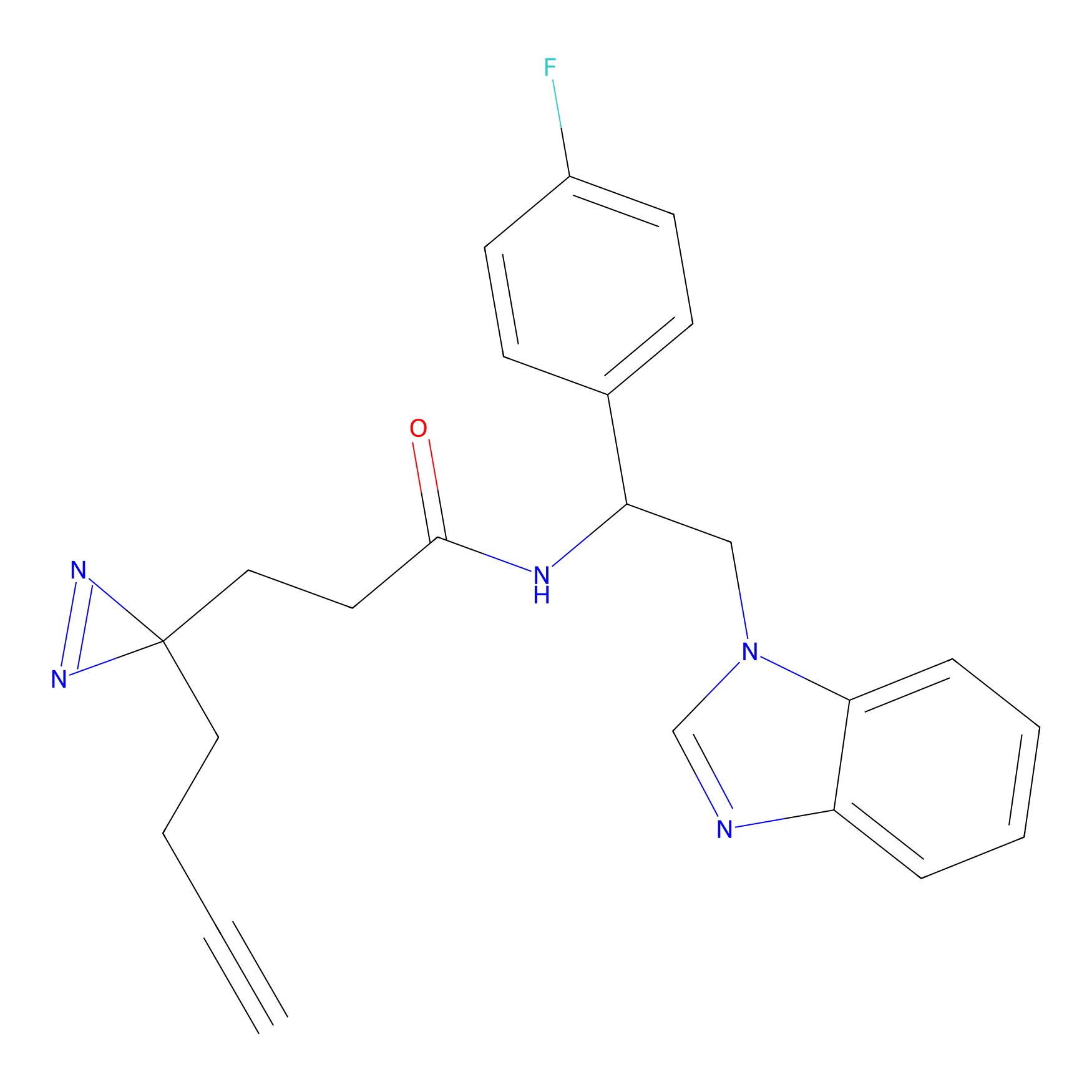
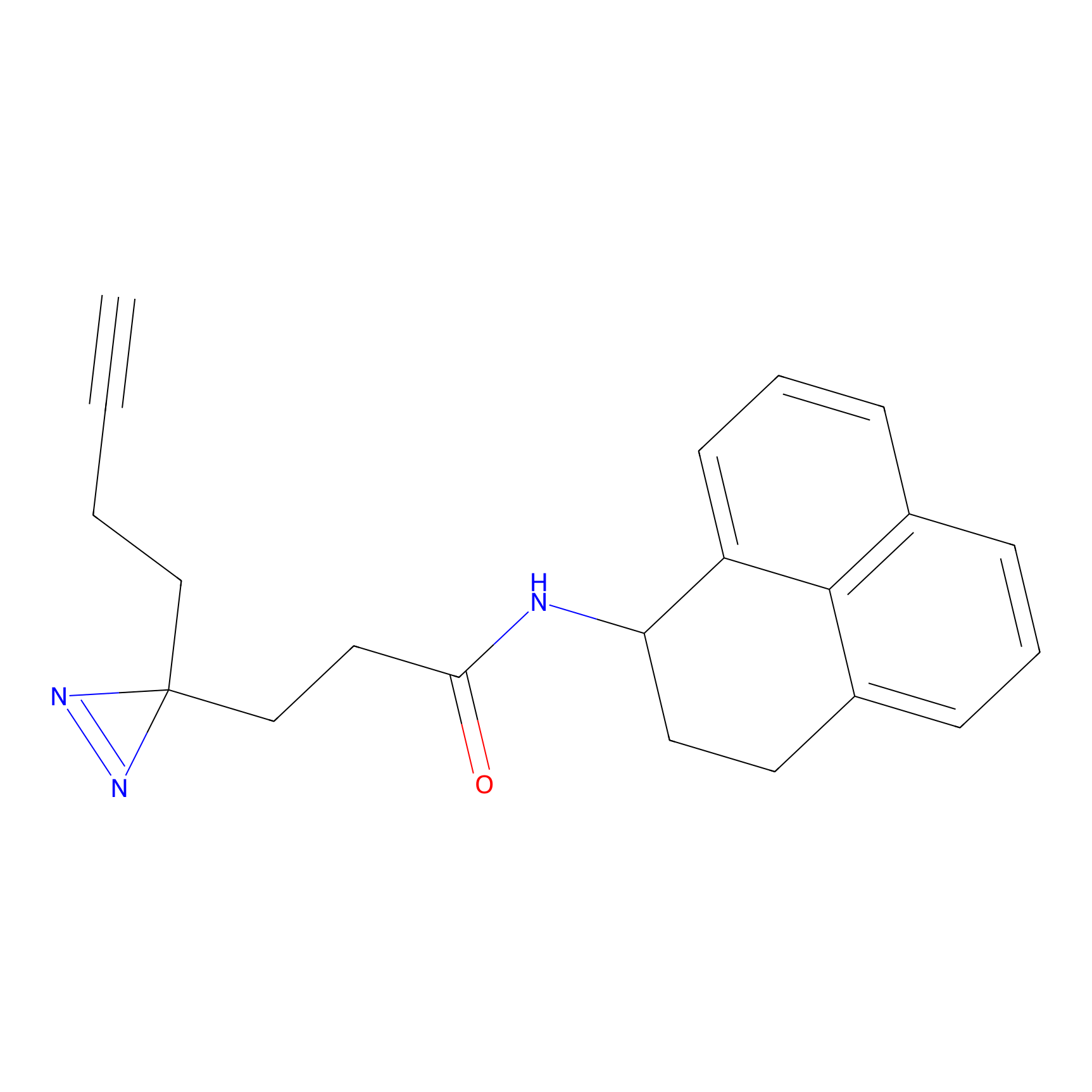
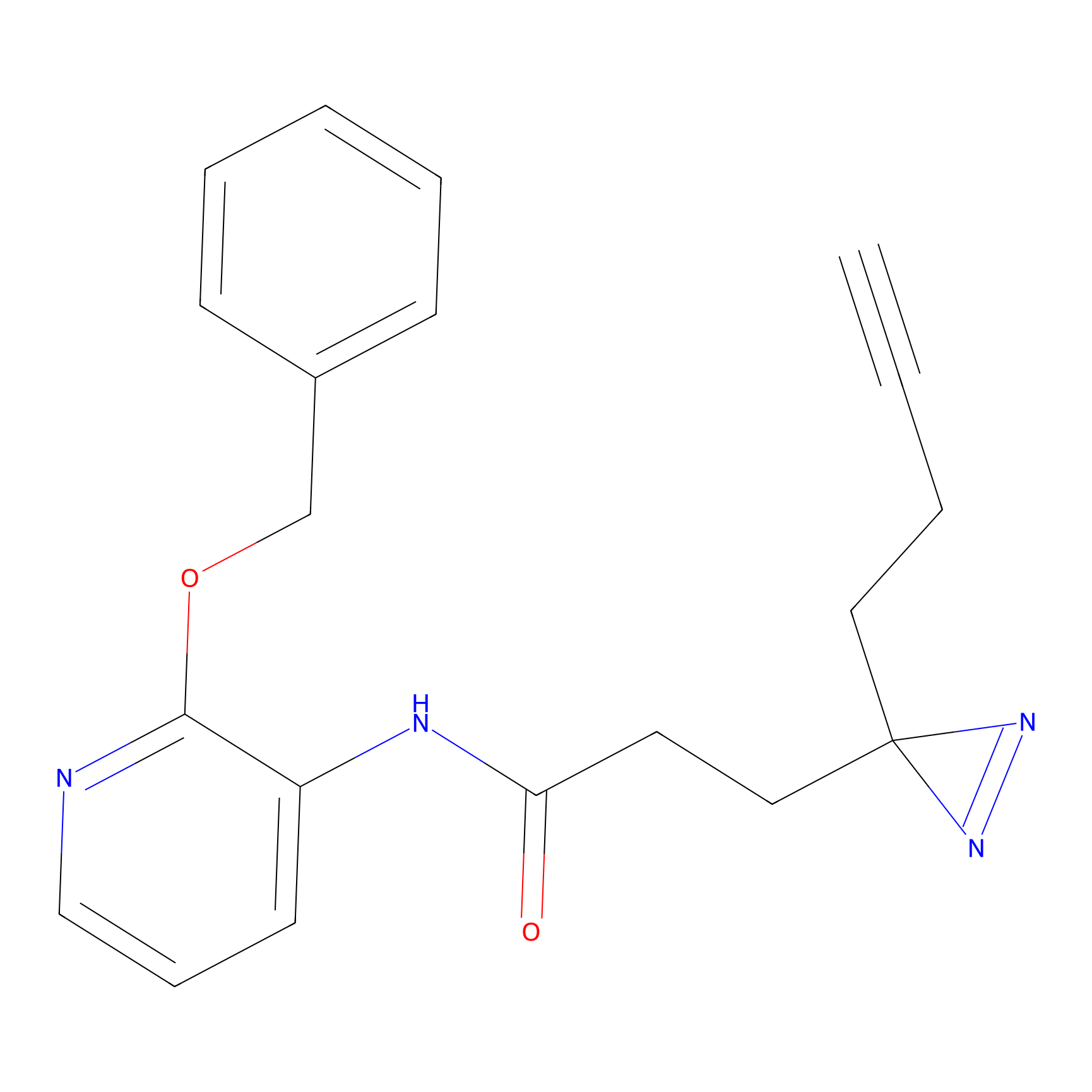
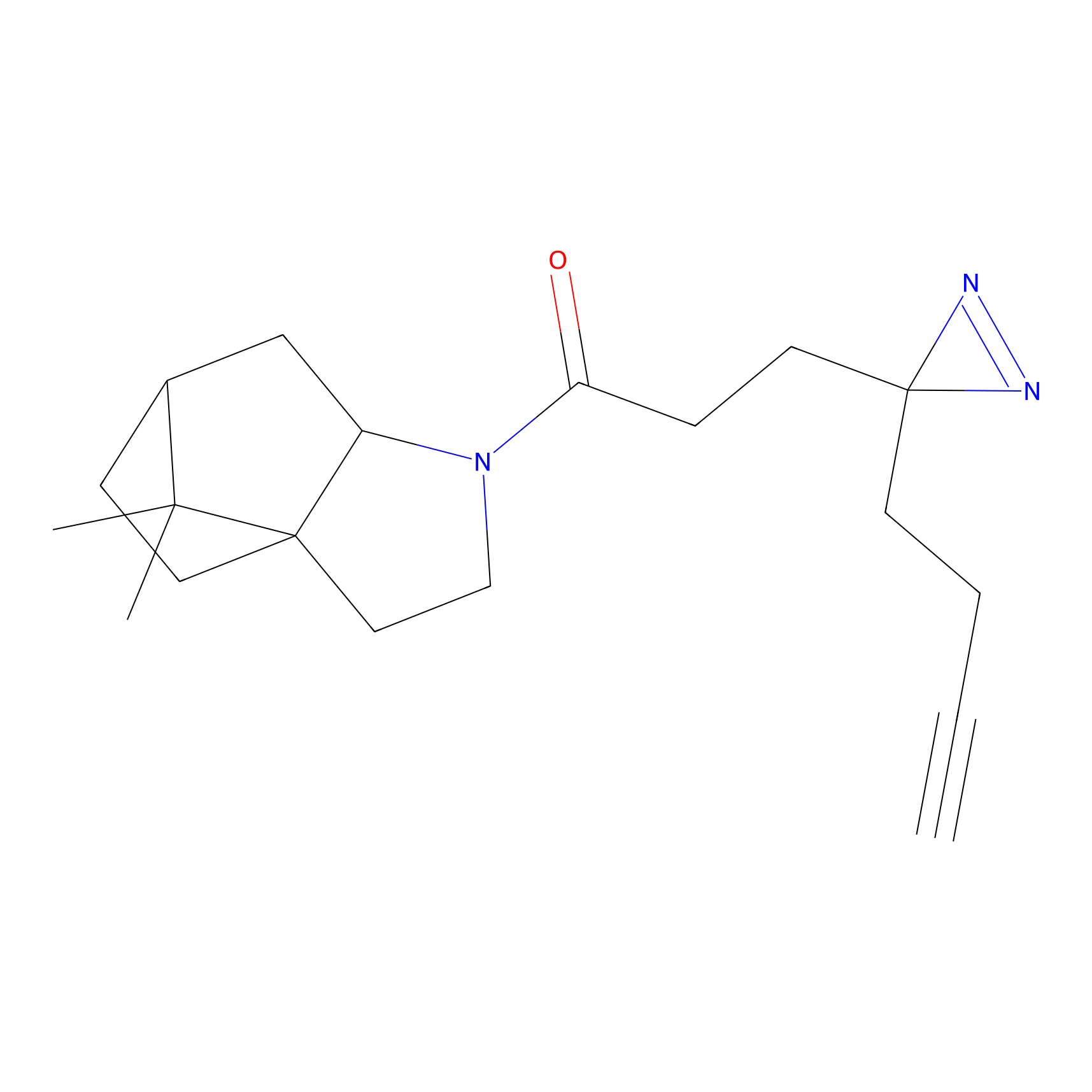
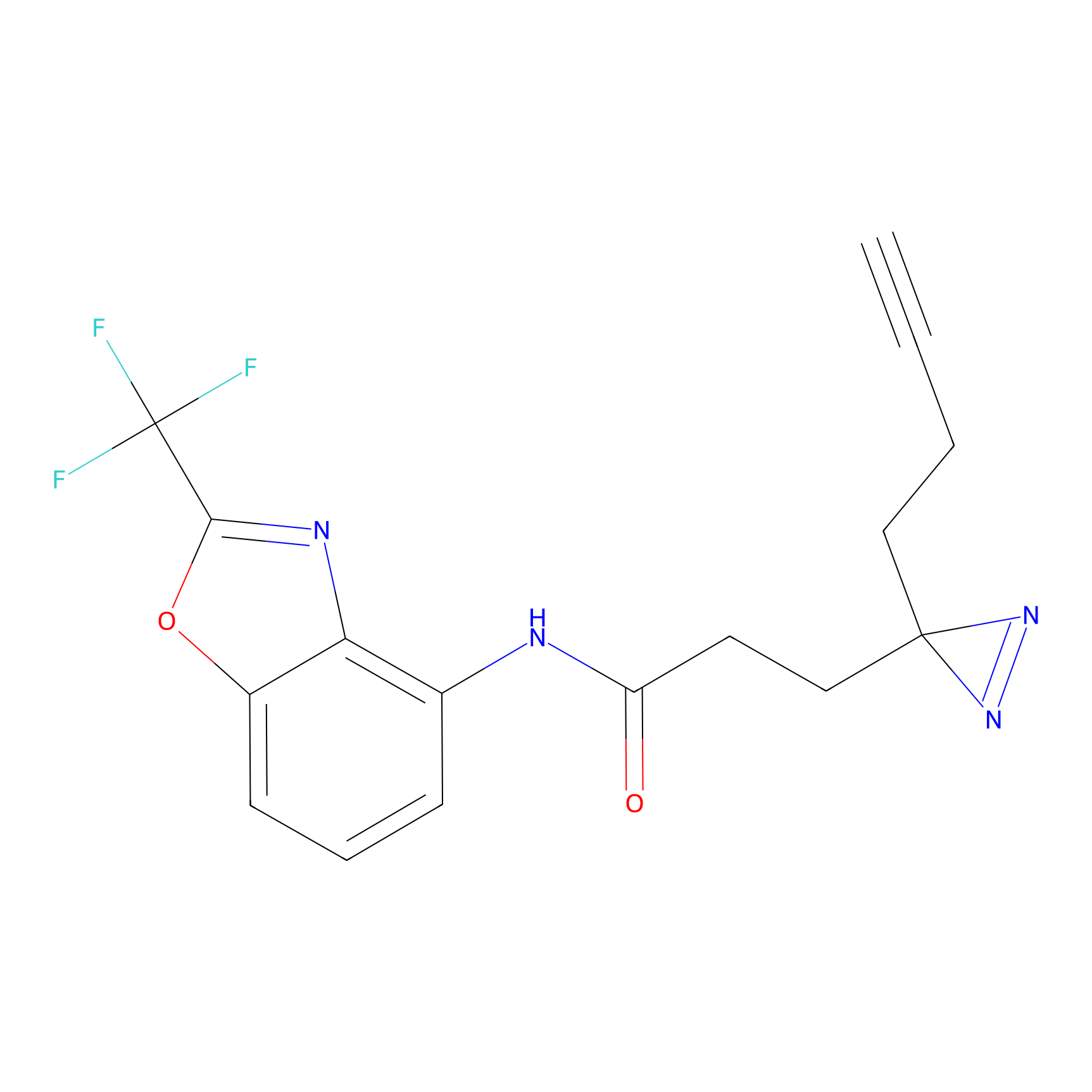
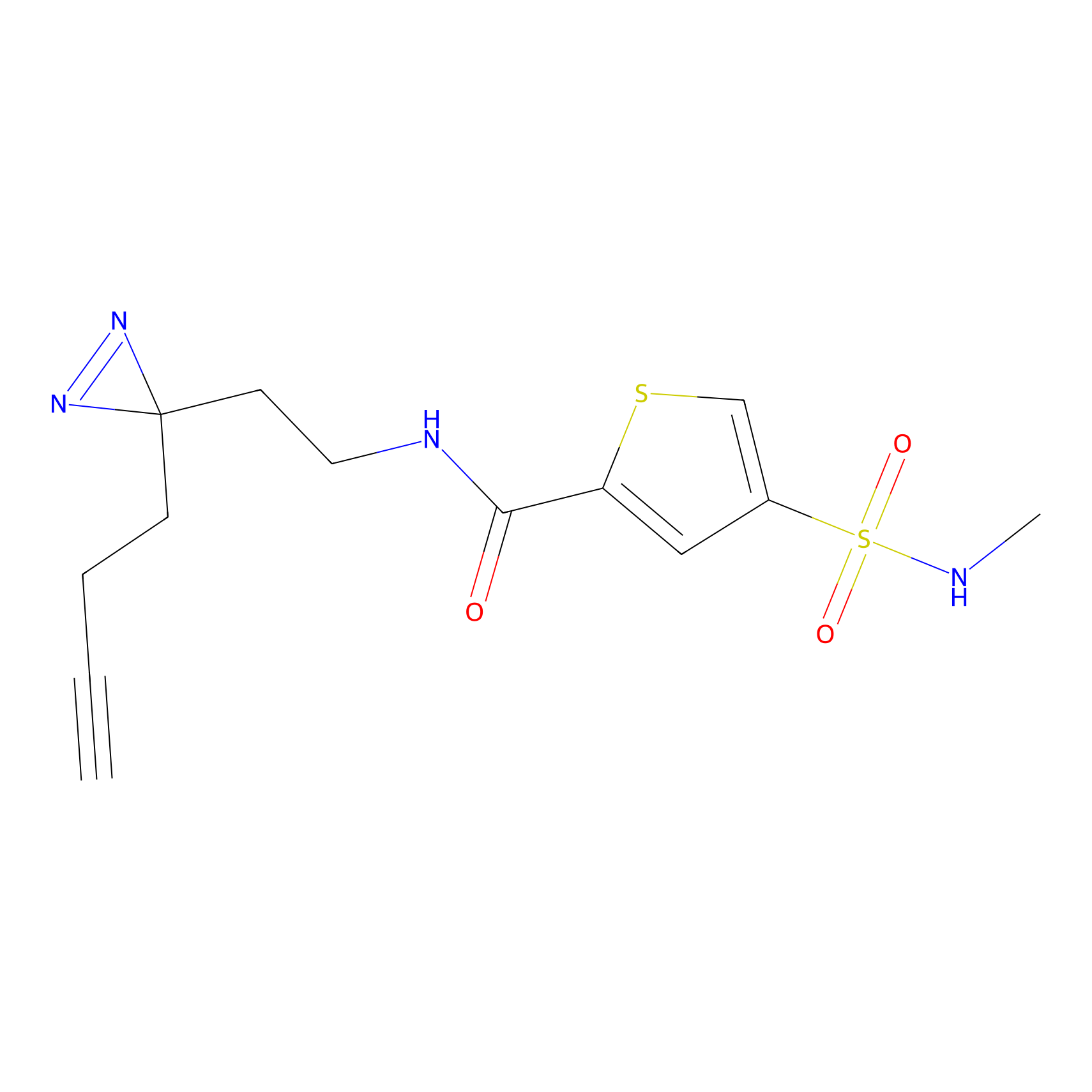
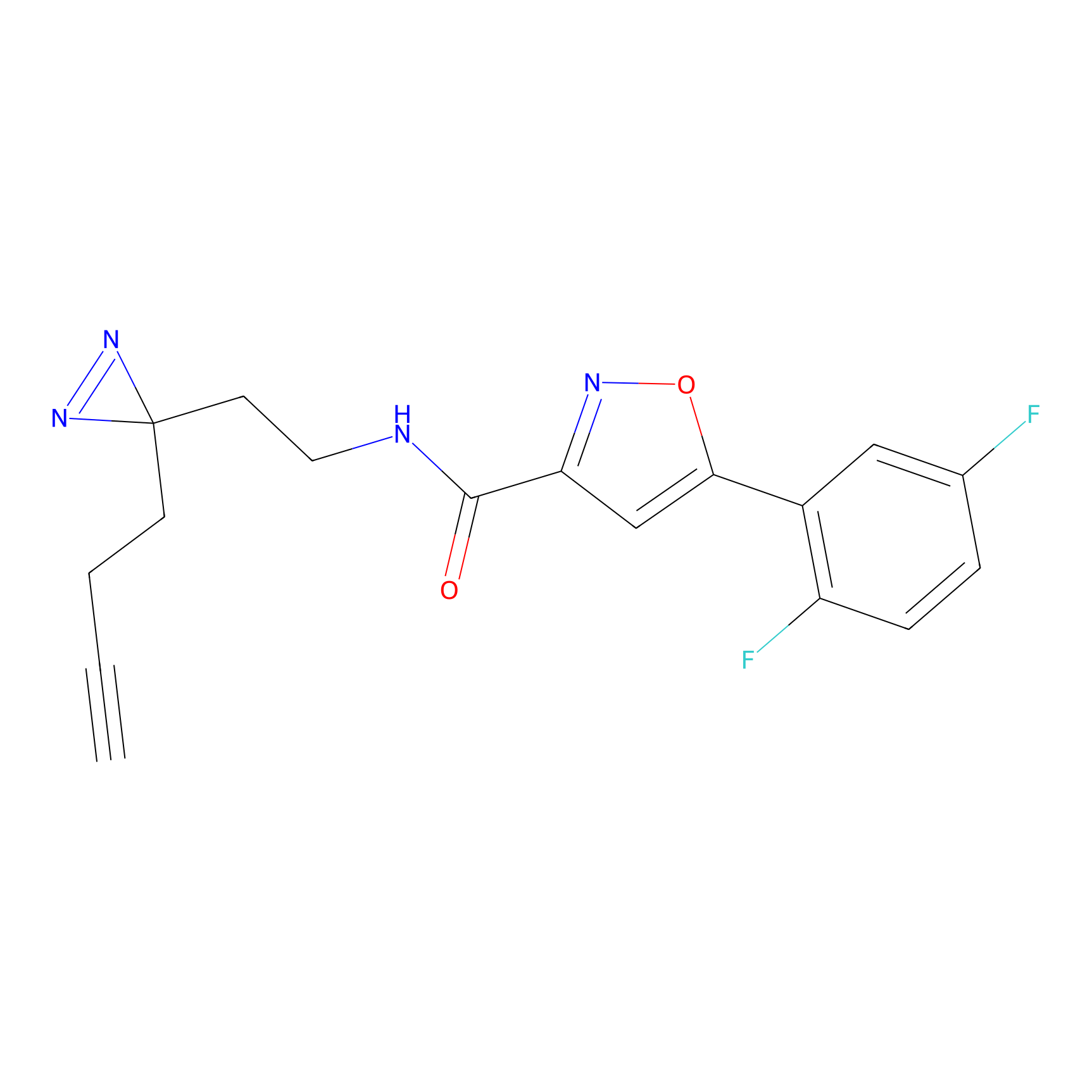
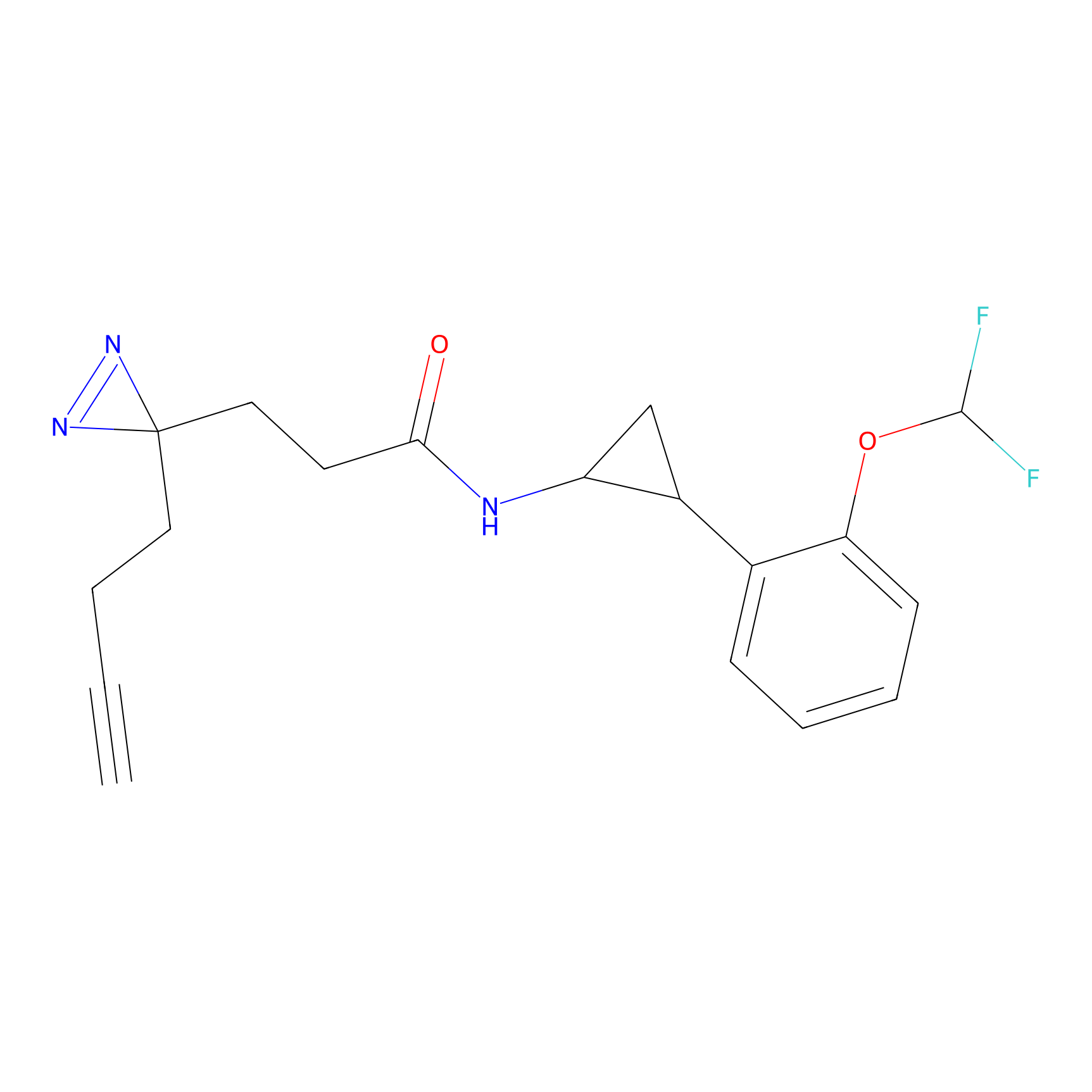
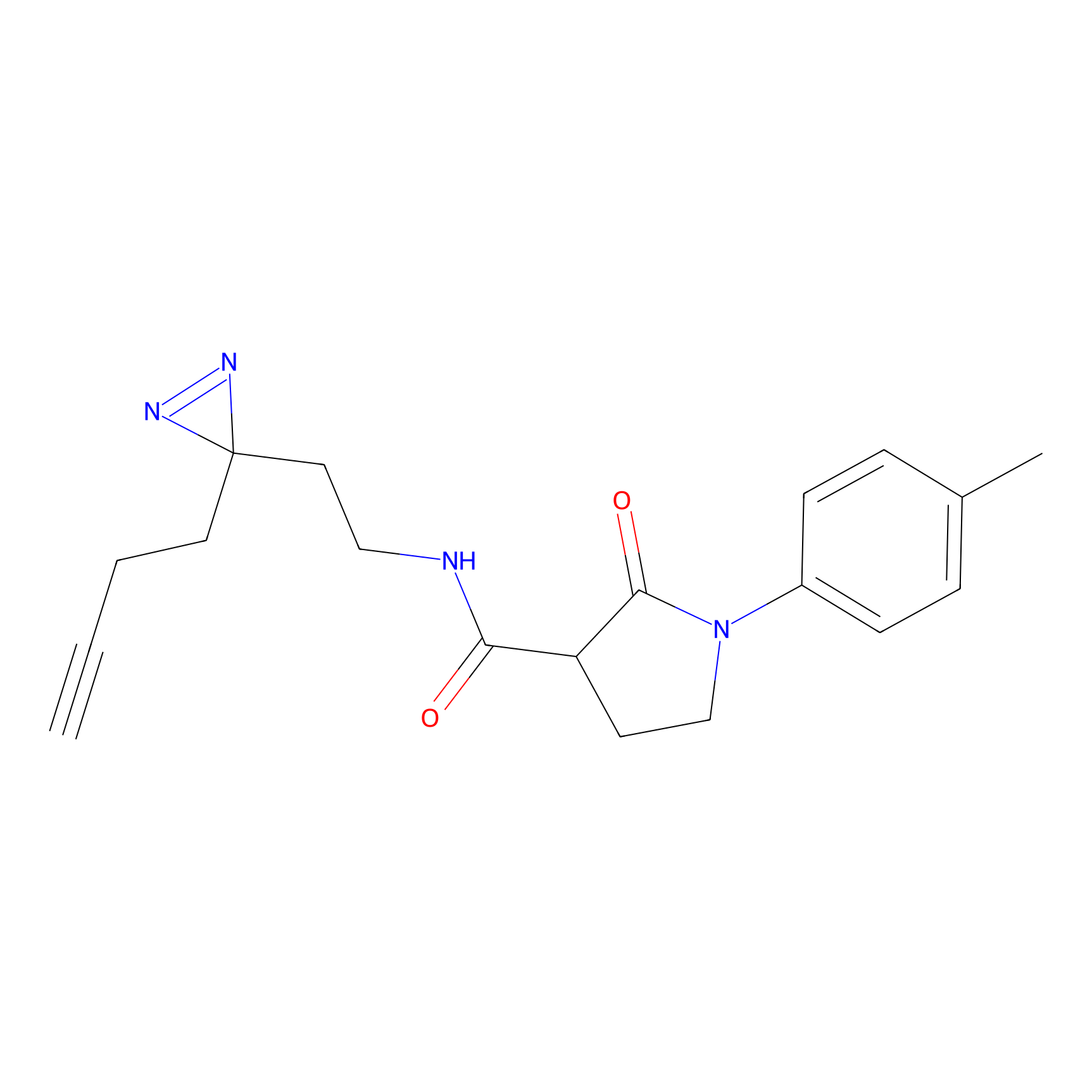
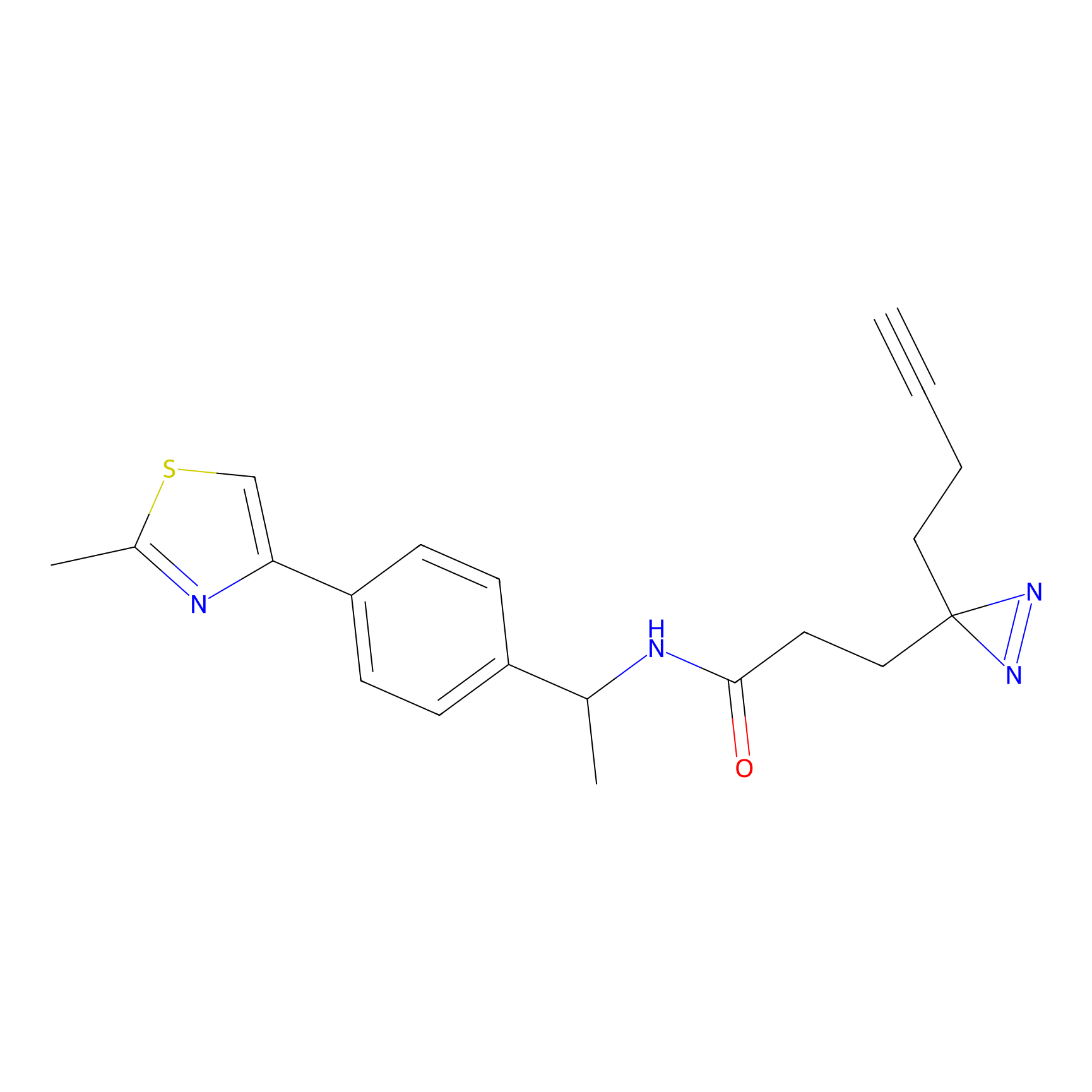
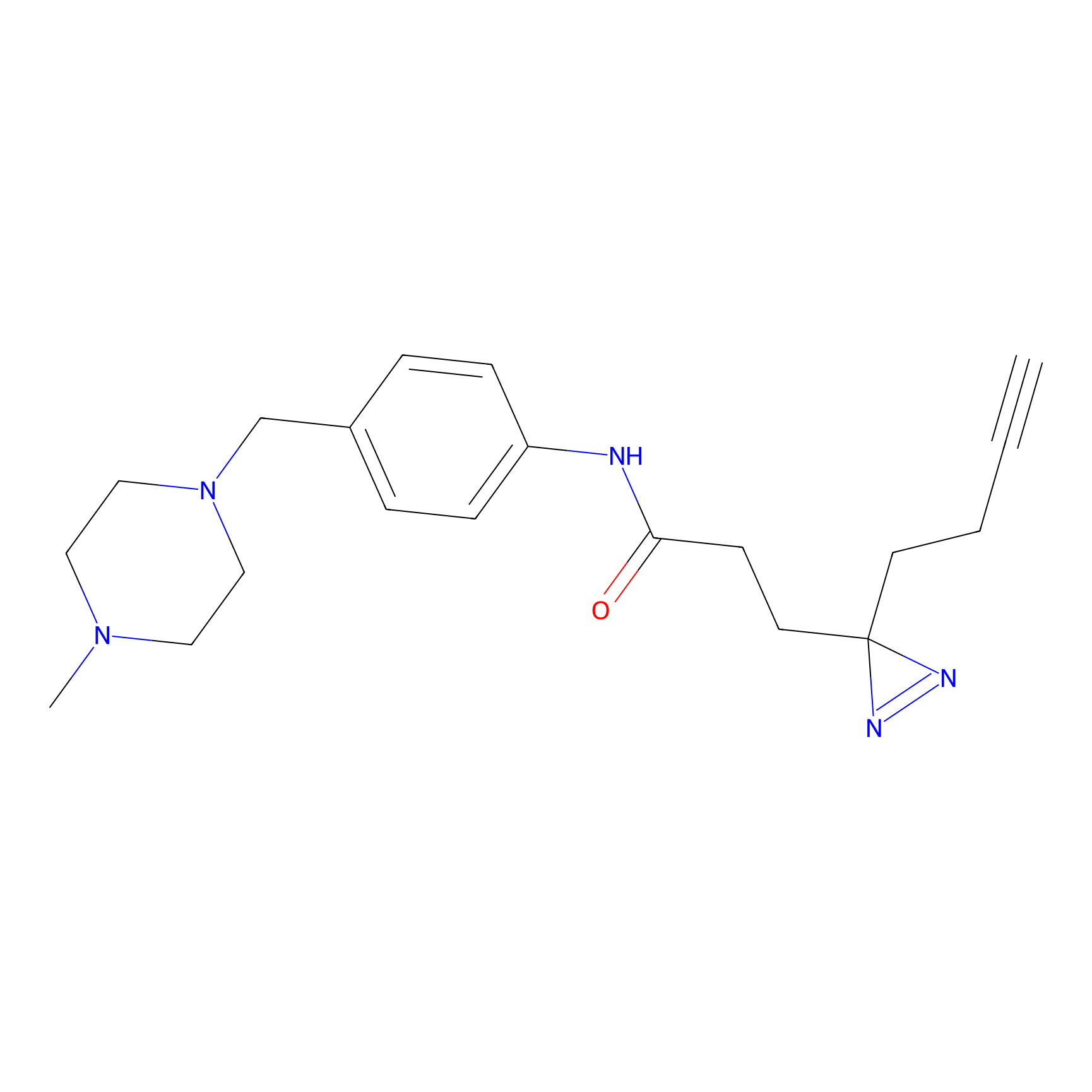
Probe 1 Probe Info |
 |
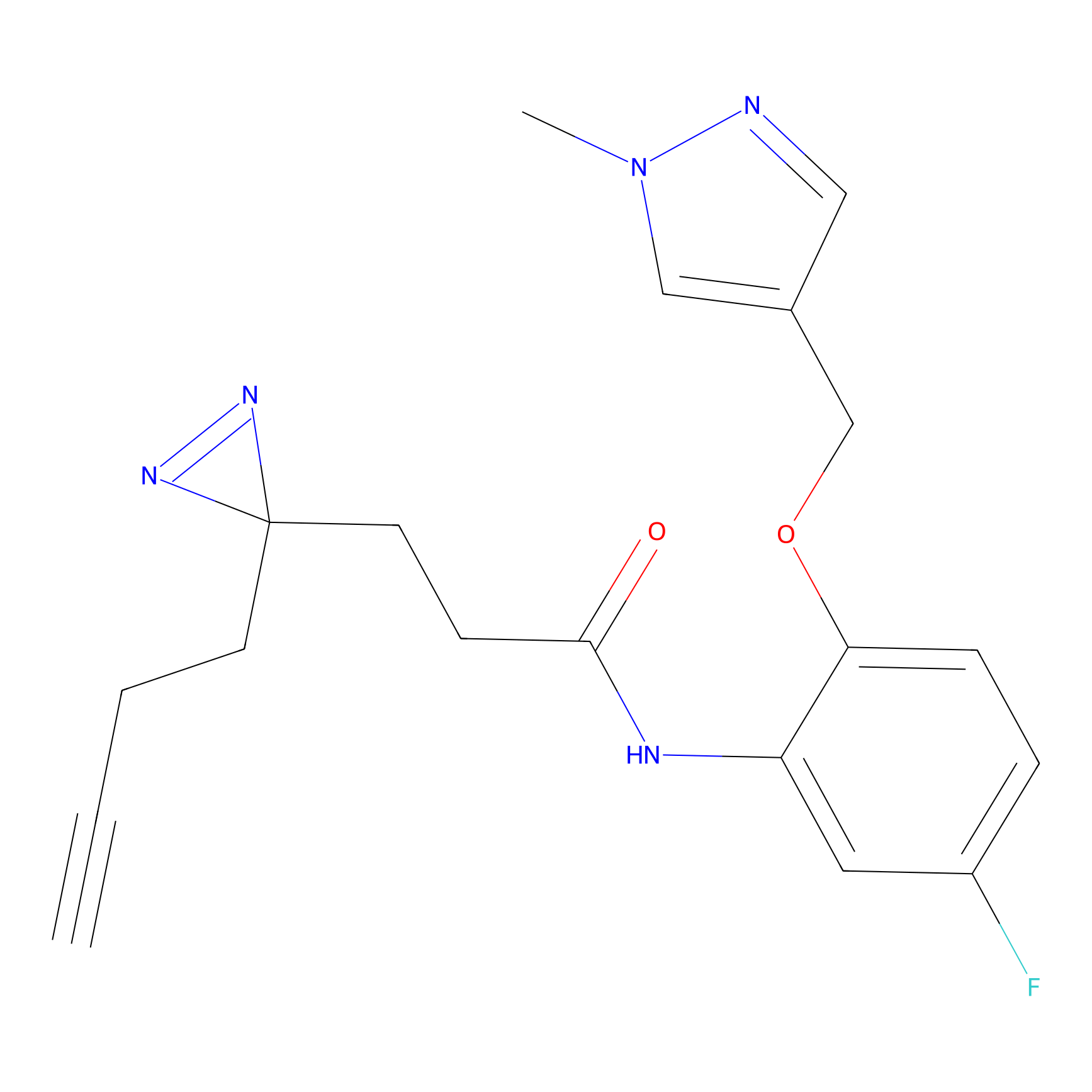
Y157(9.37) | LDD3495 | [2] | |
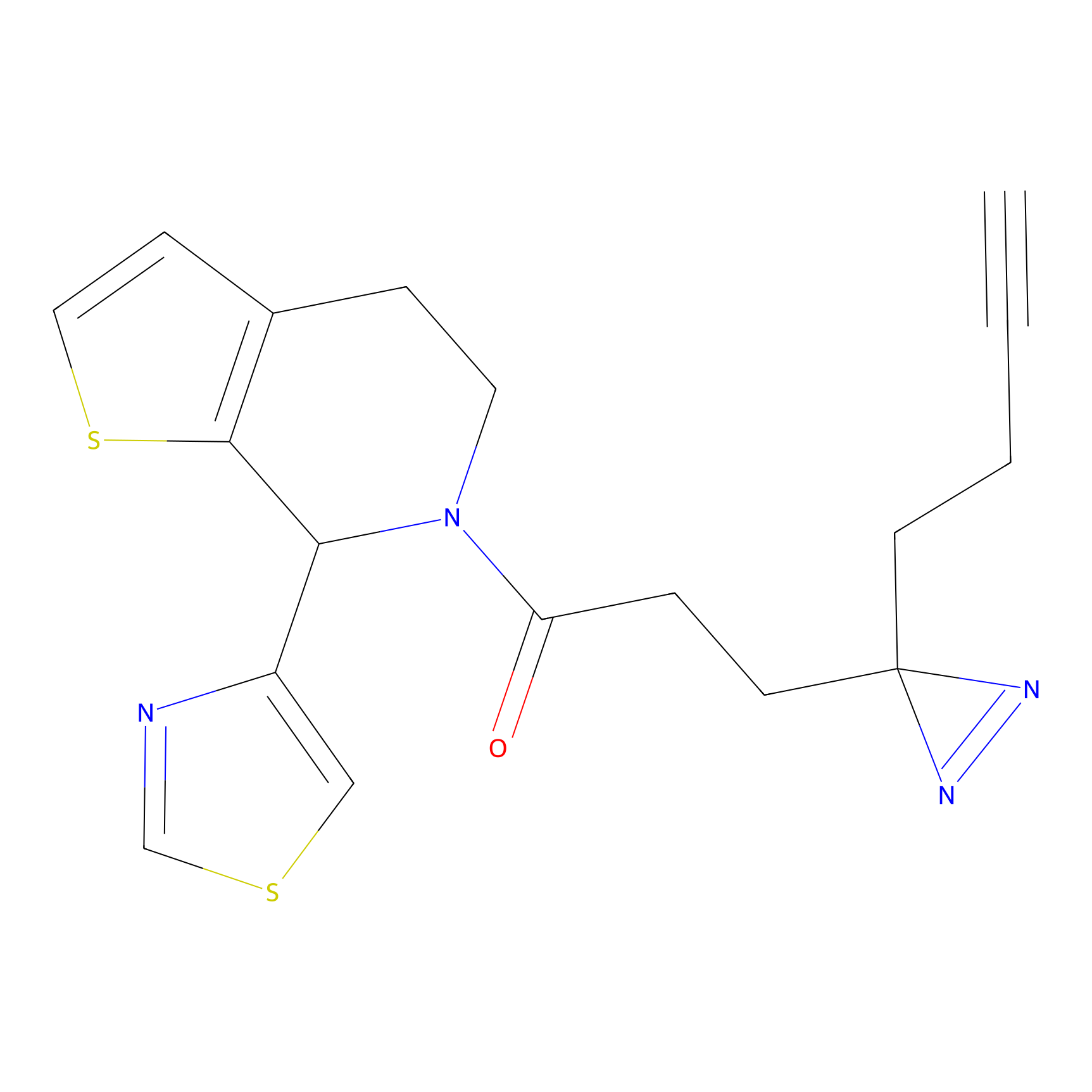
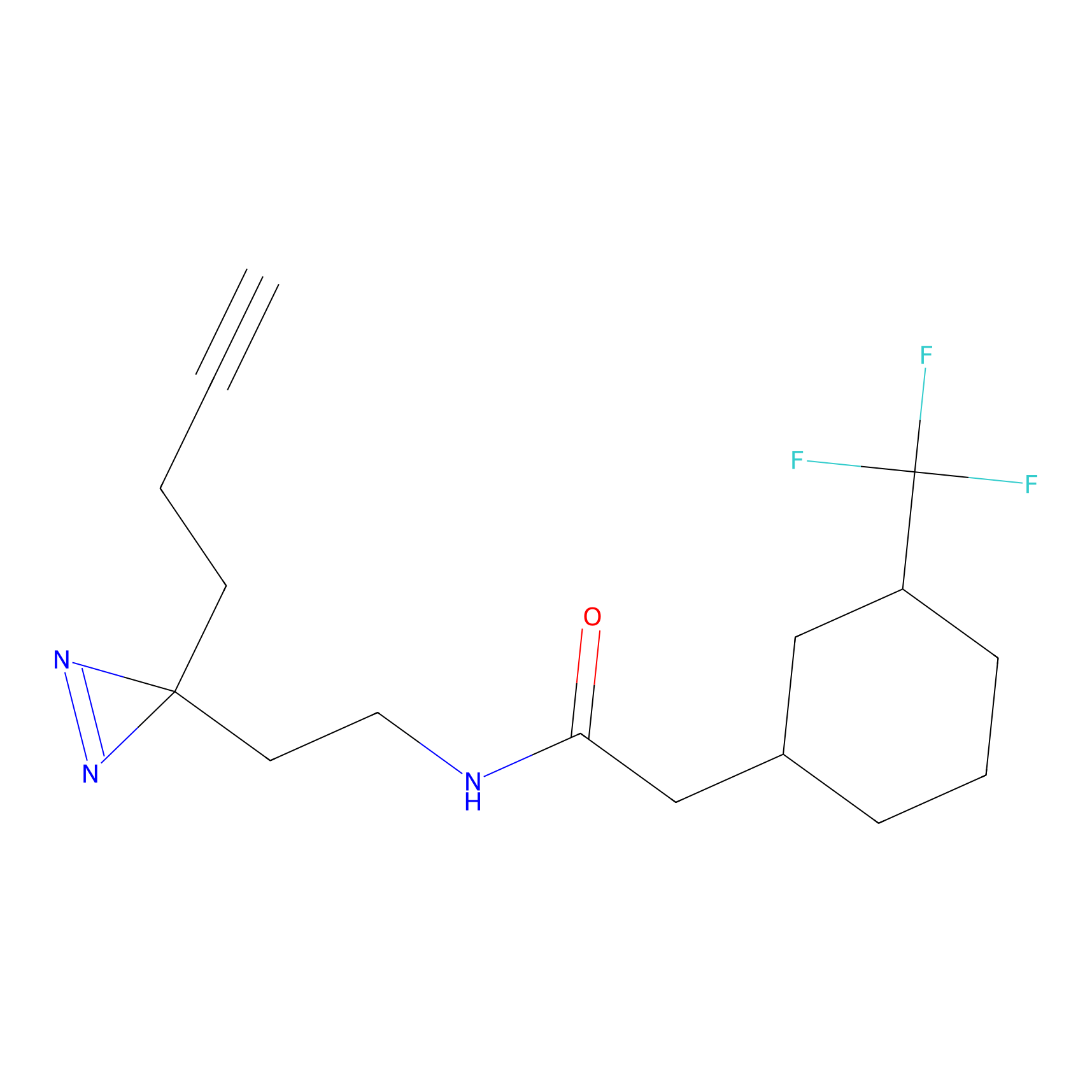
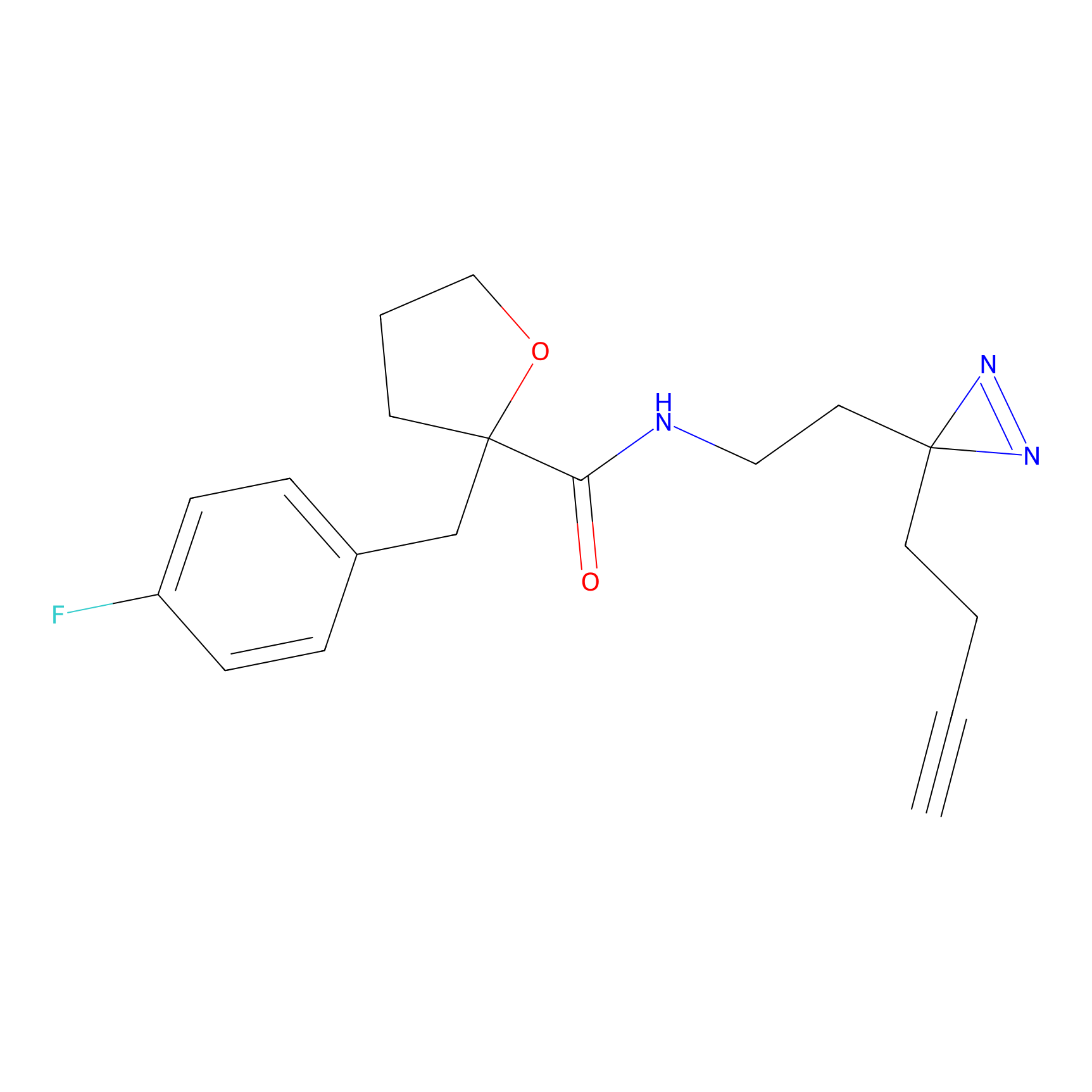
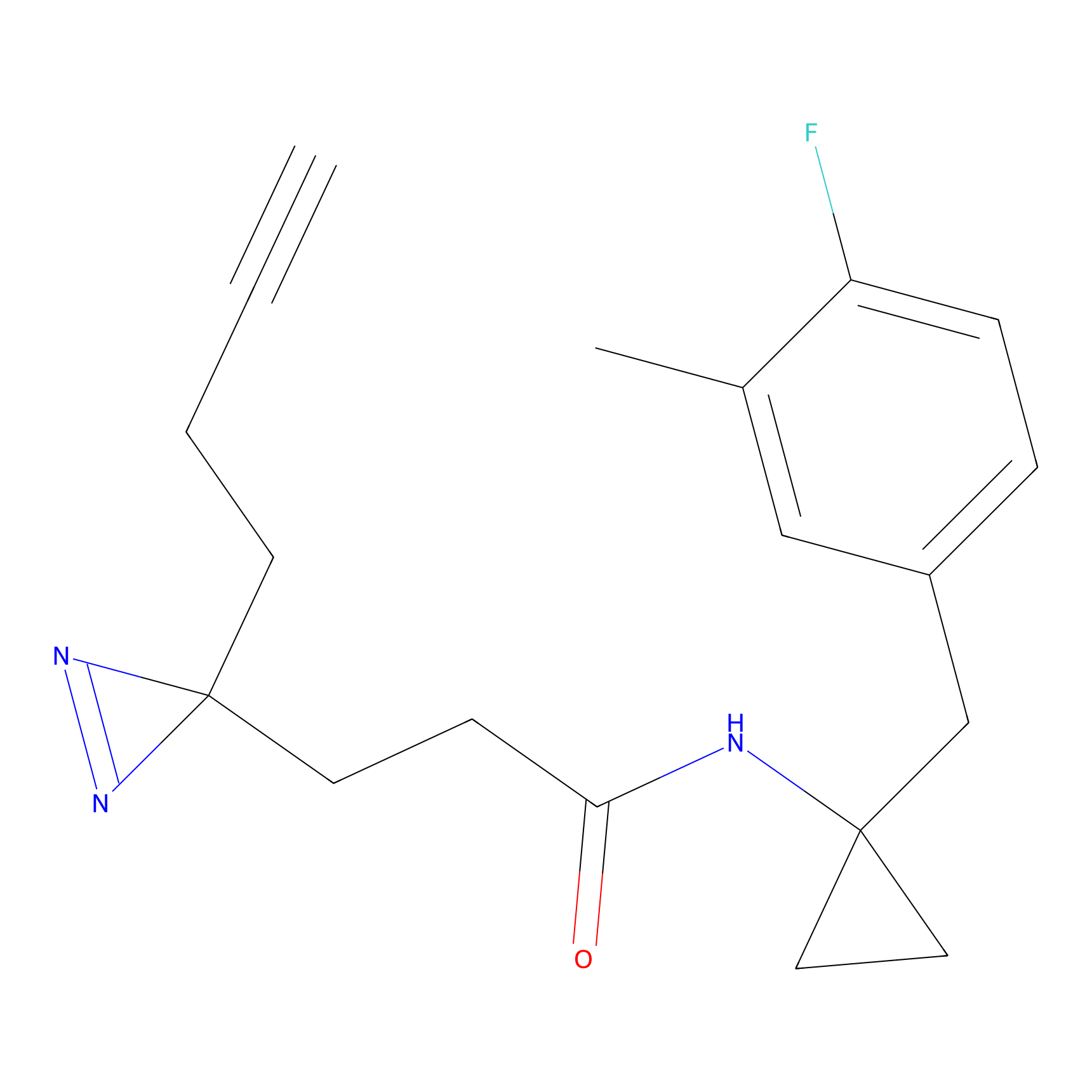
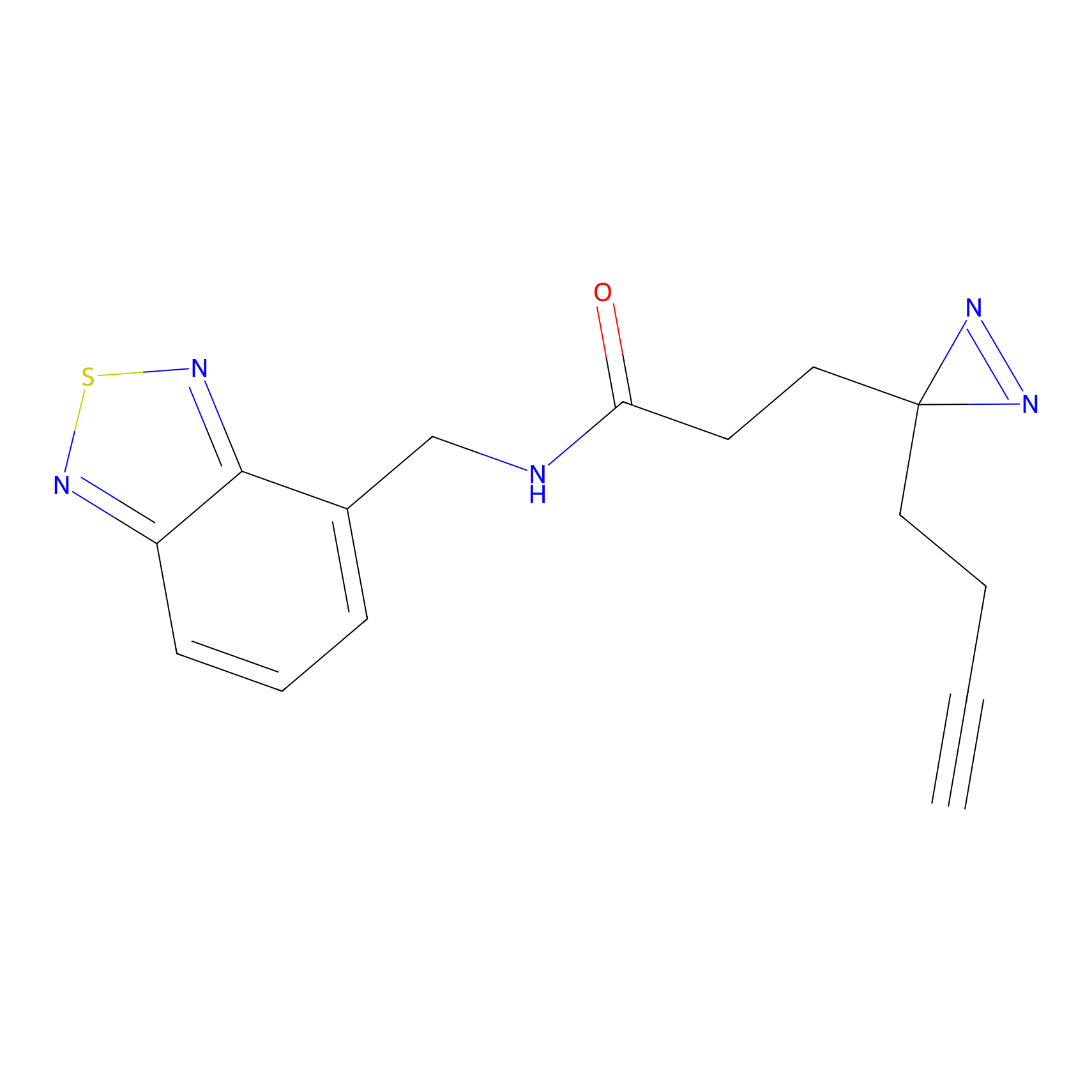
|
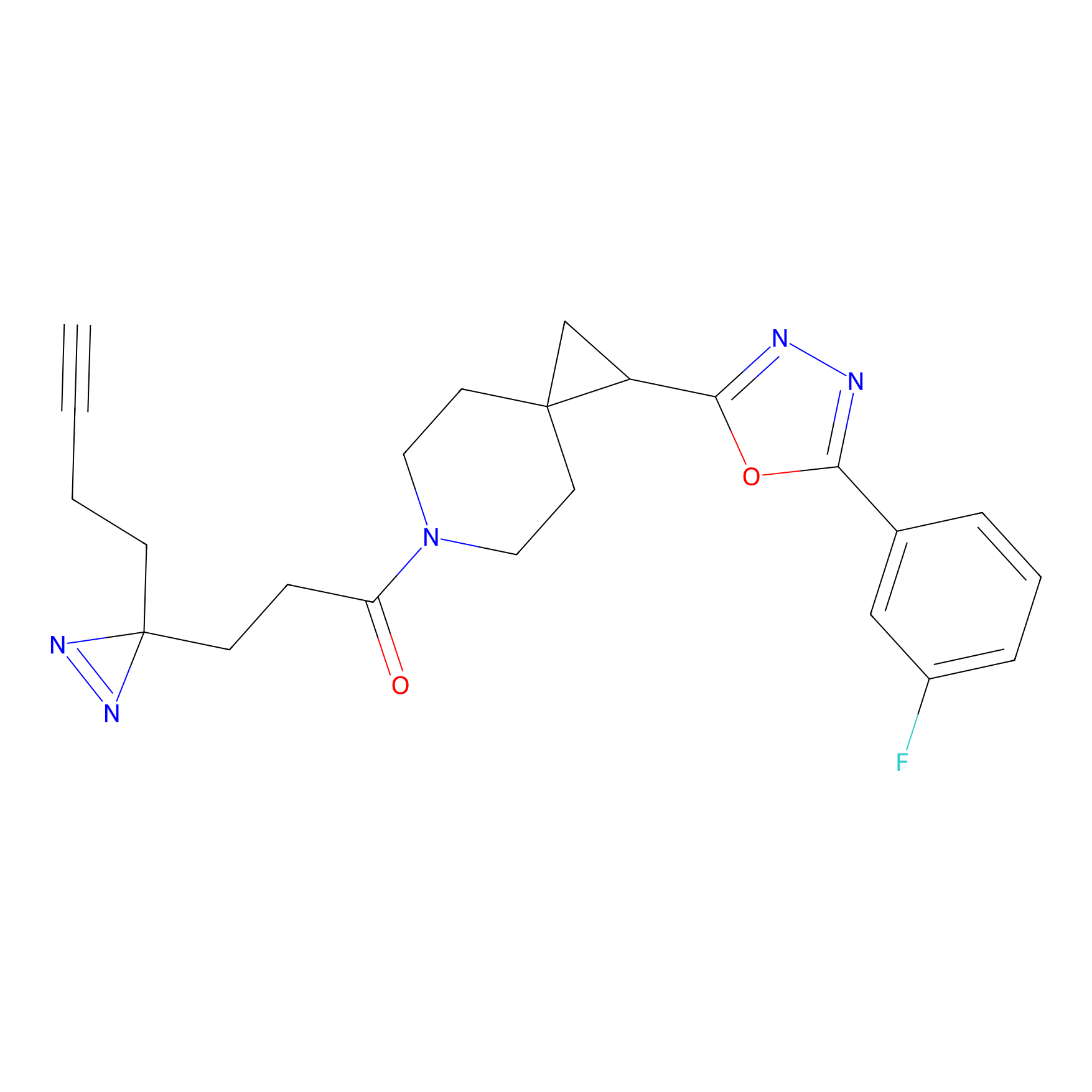
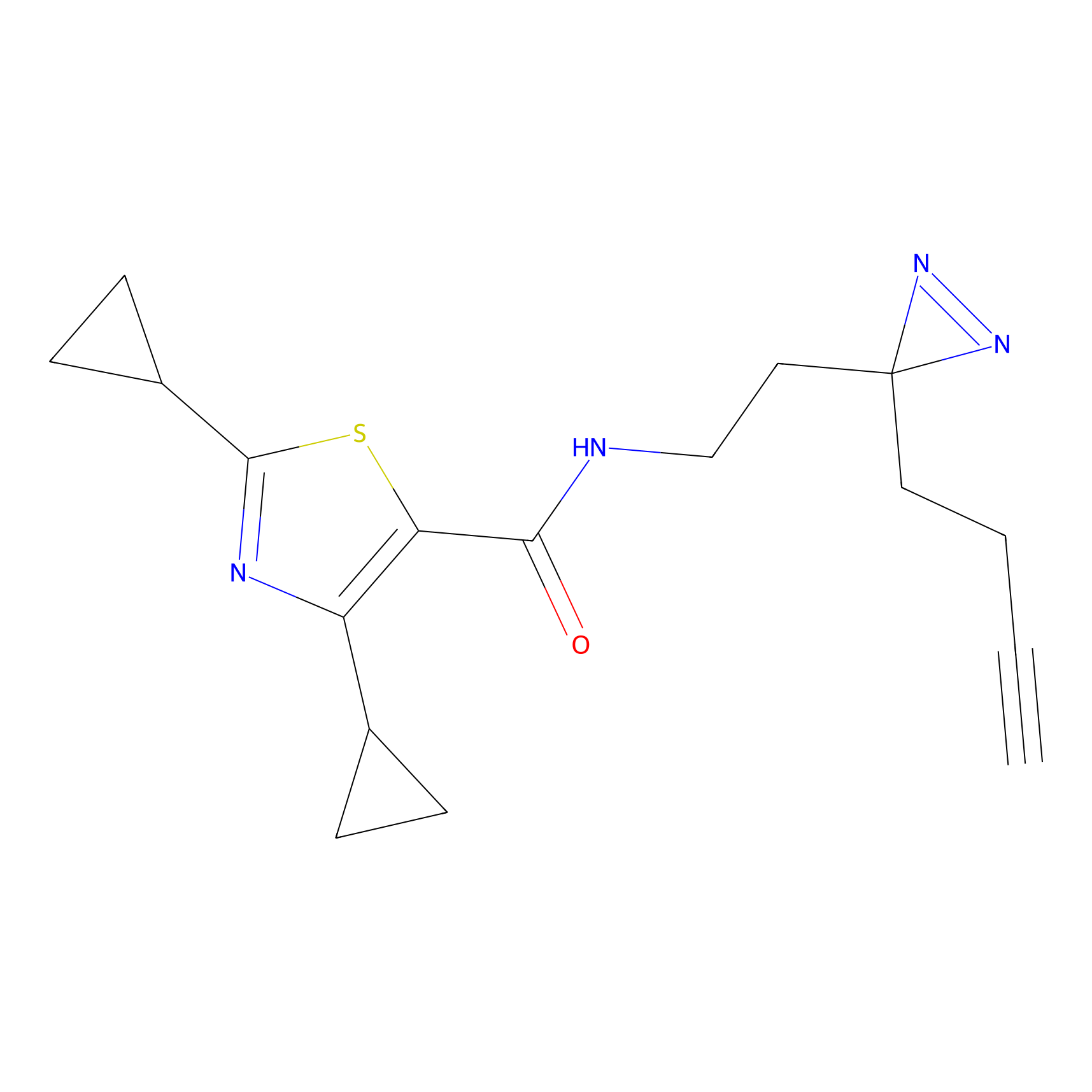
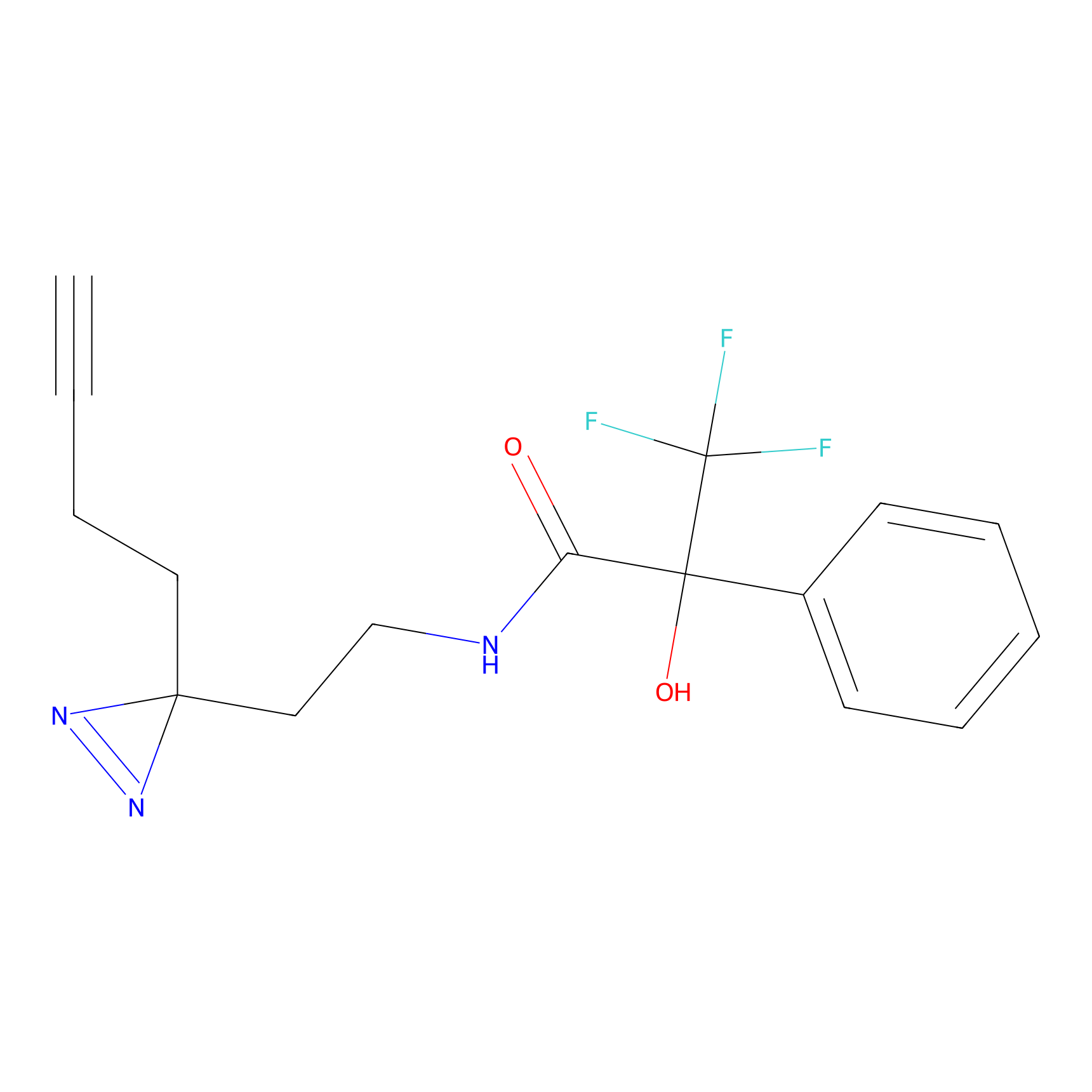
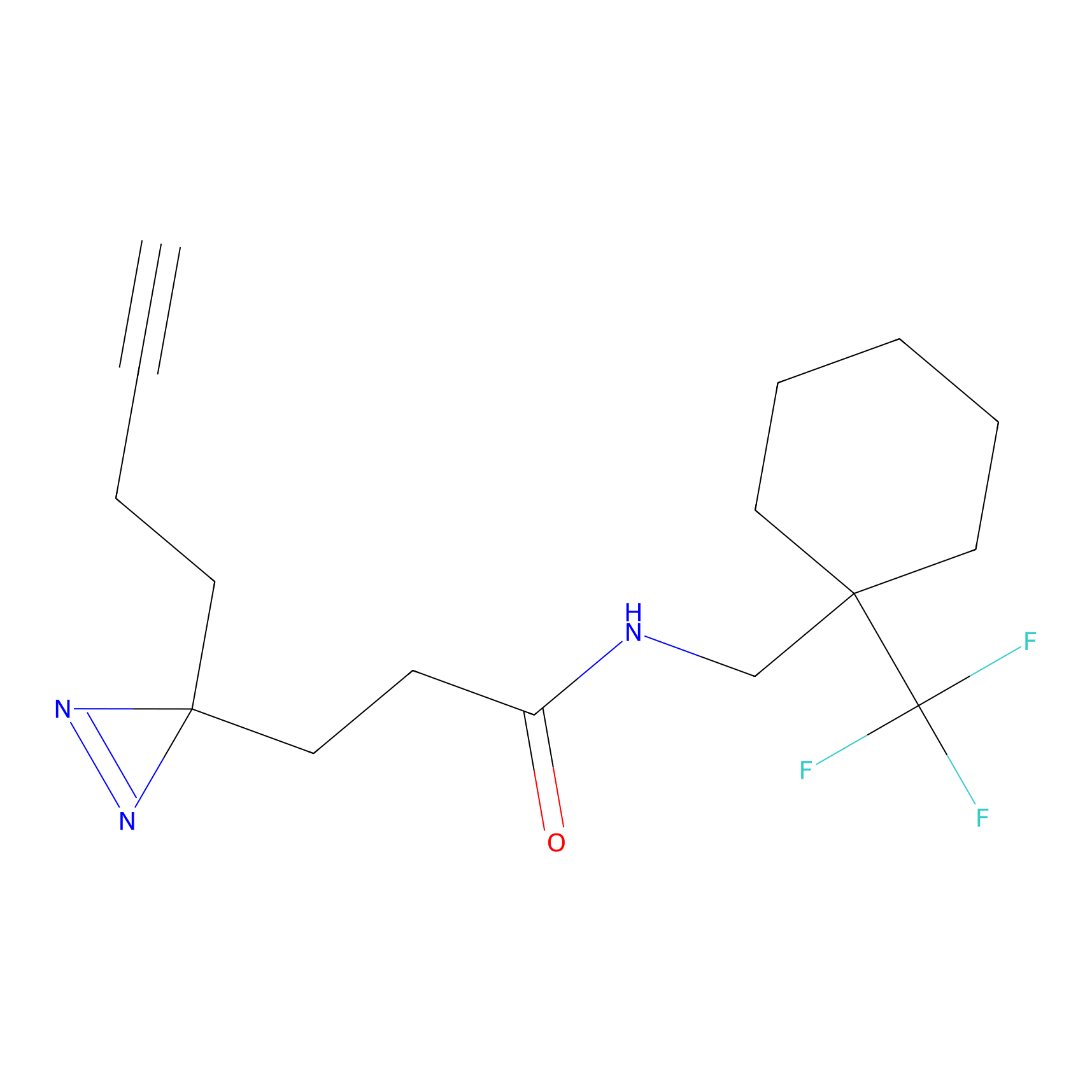
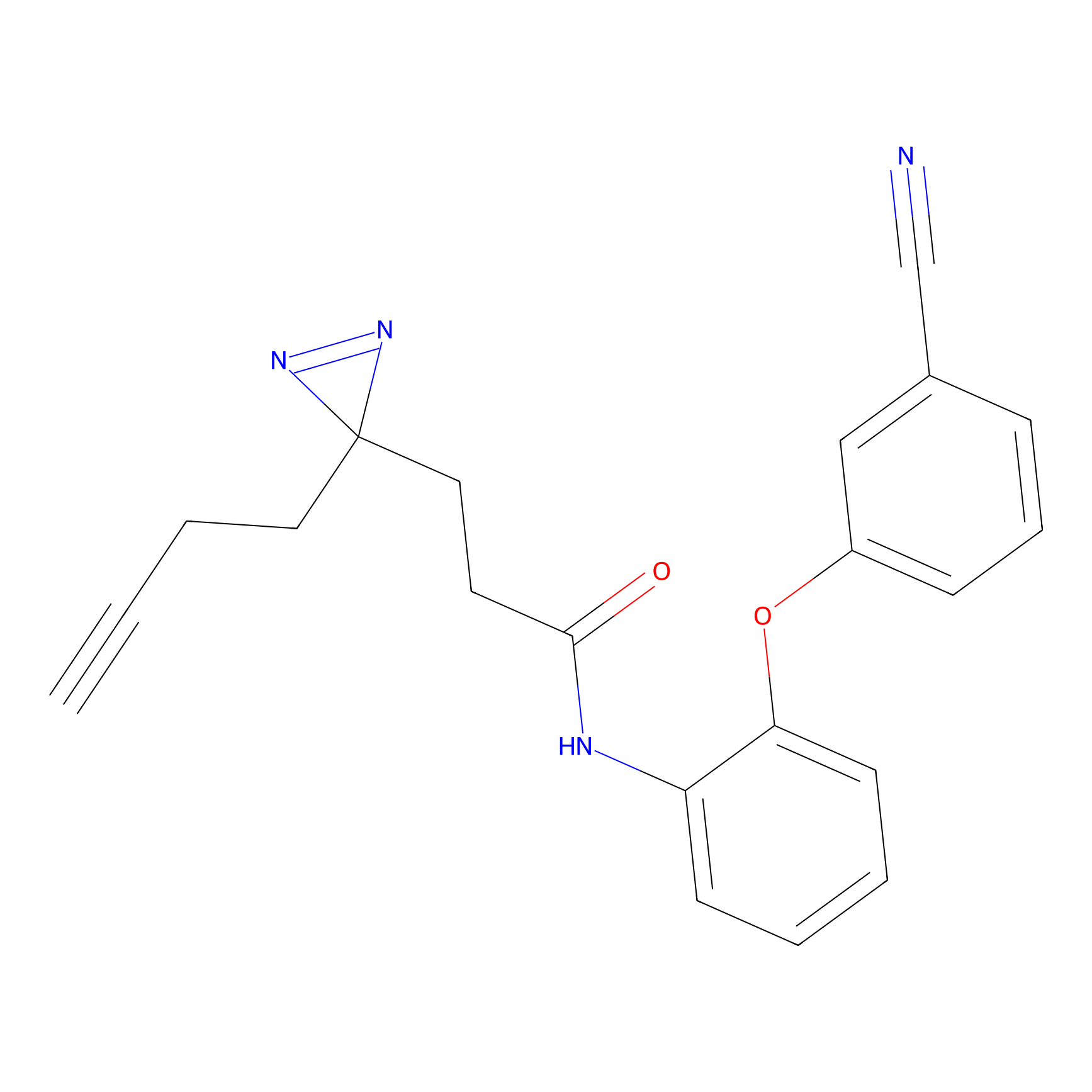
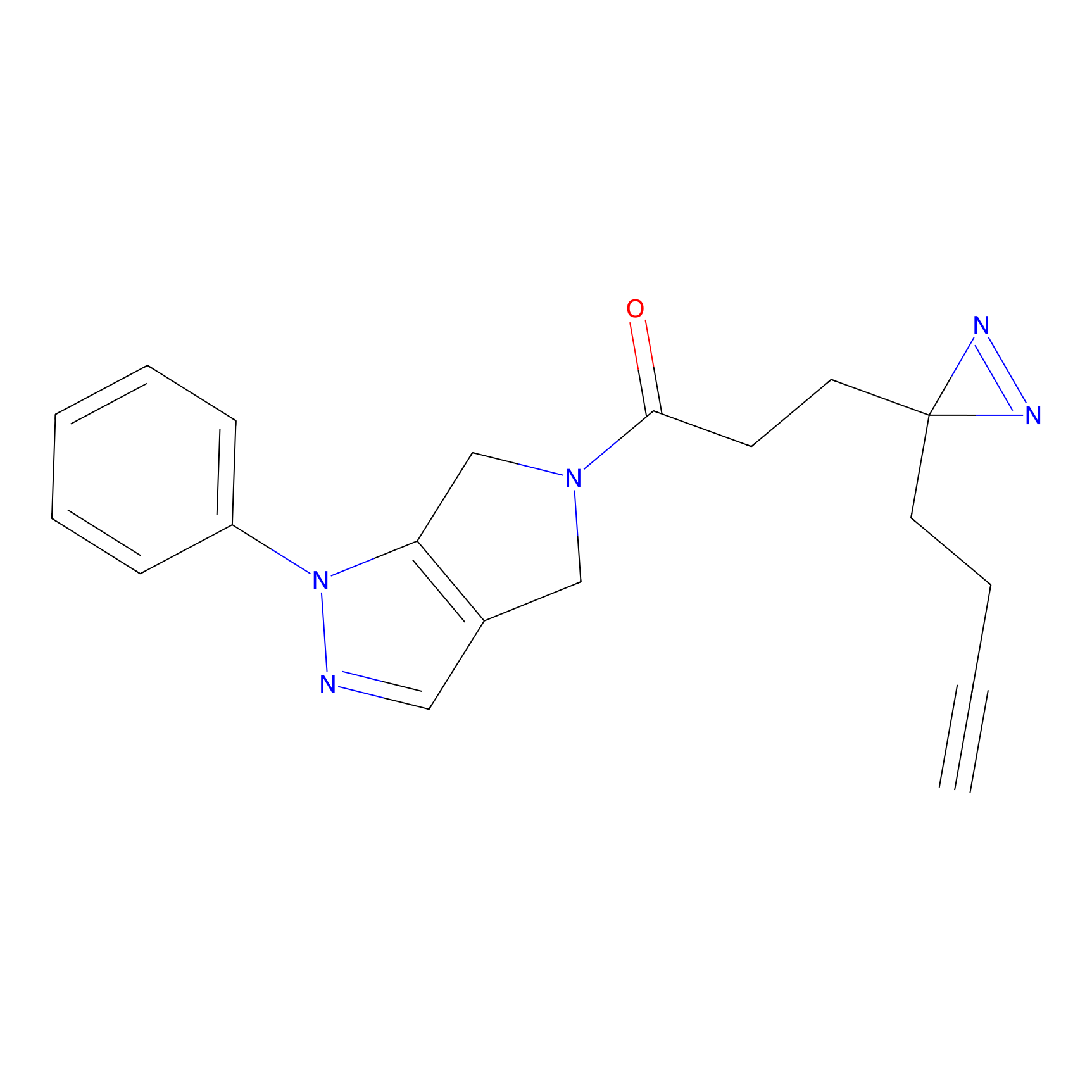
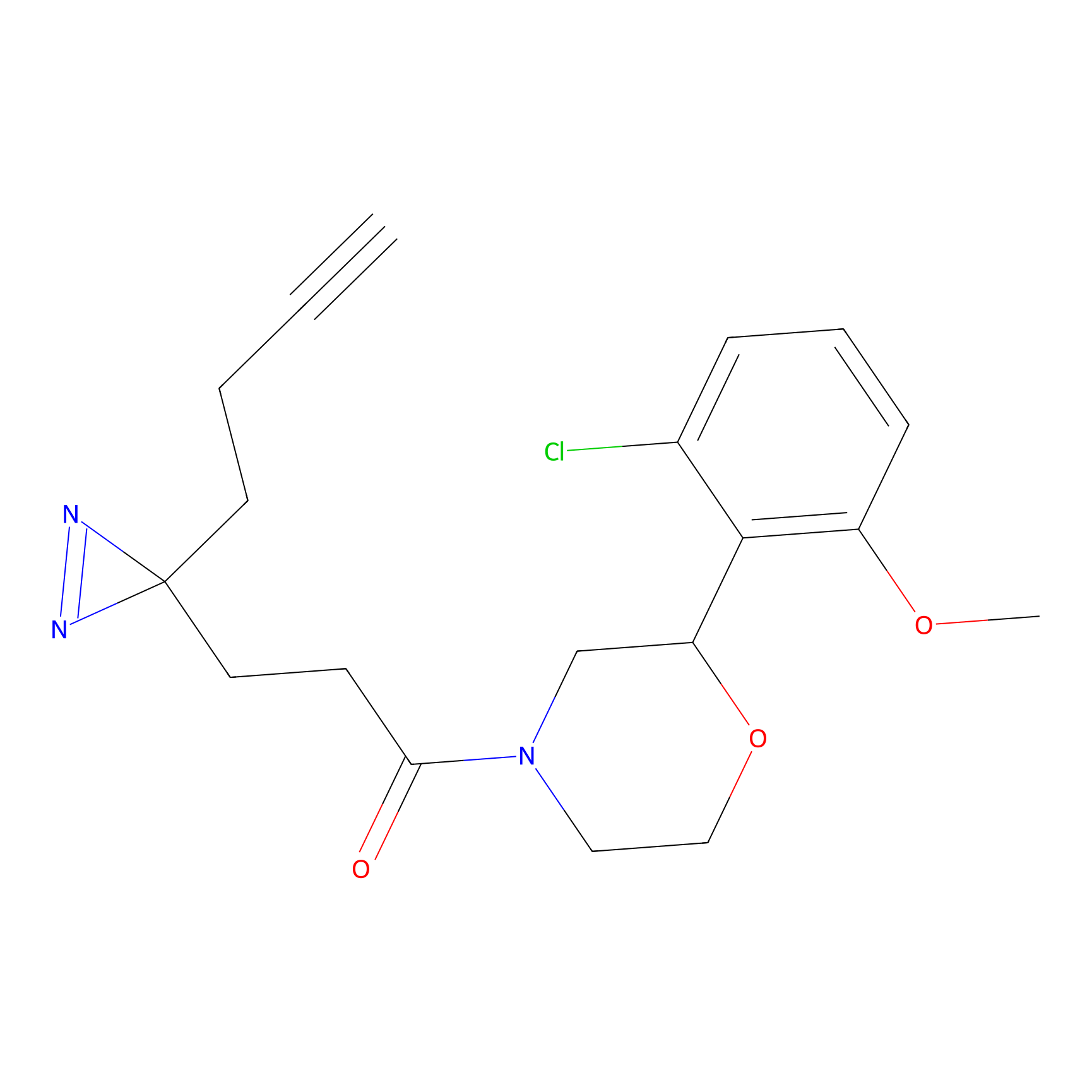
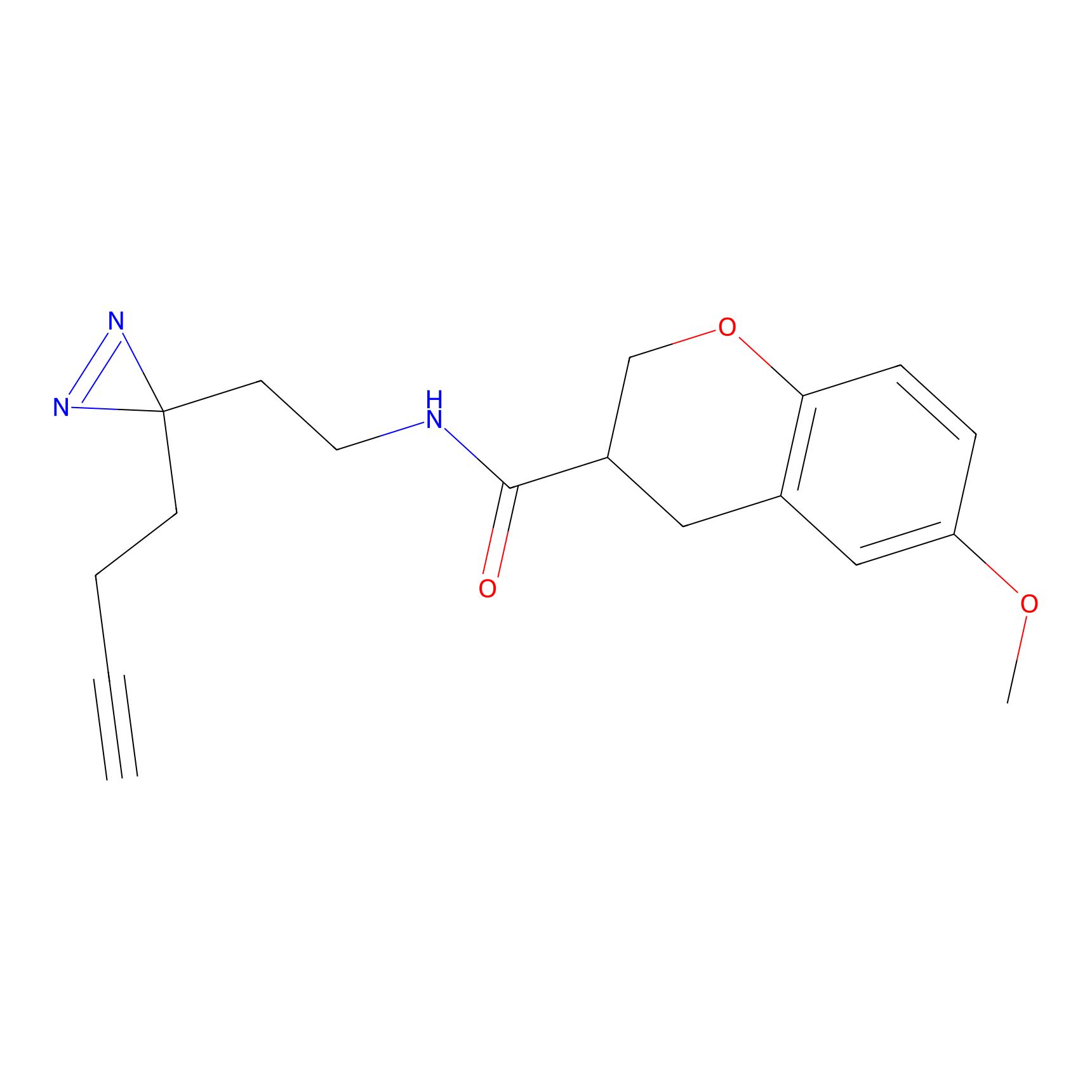
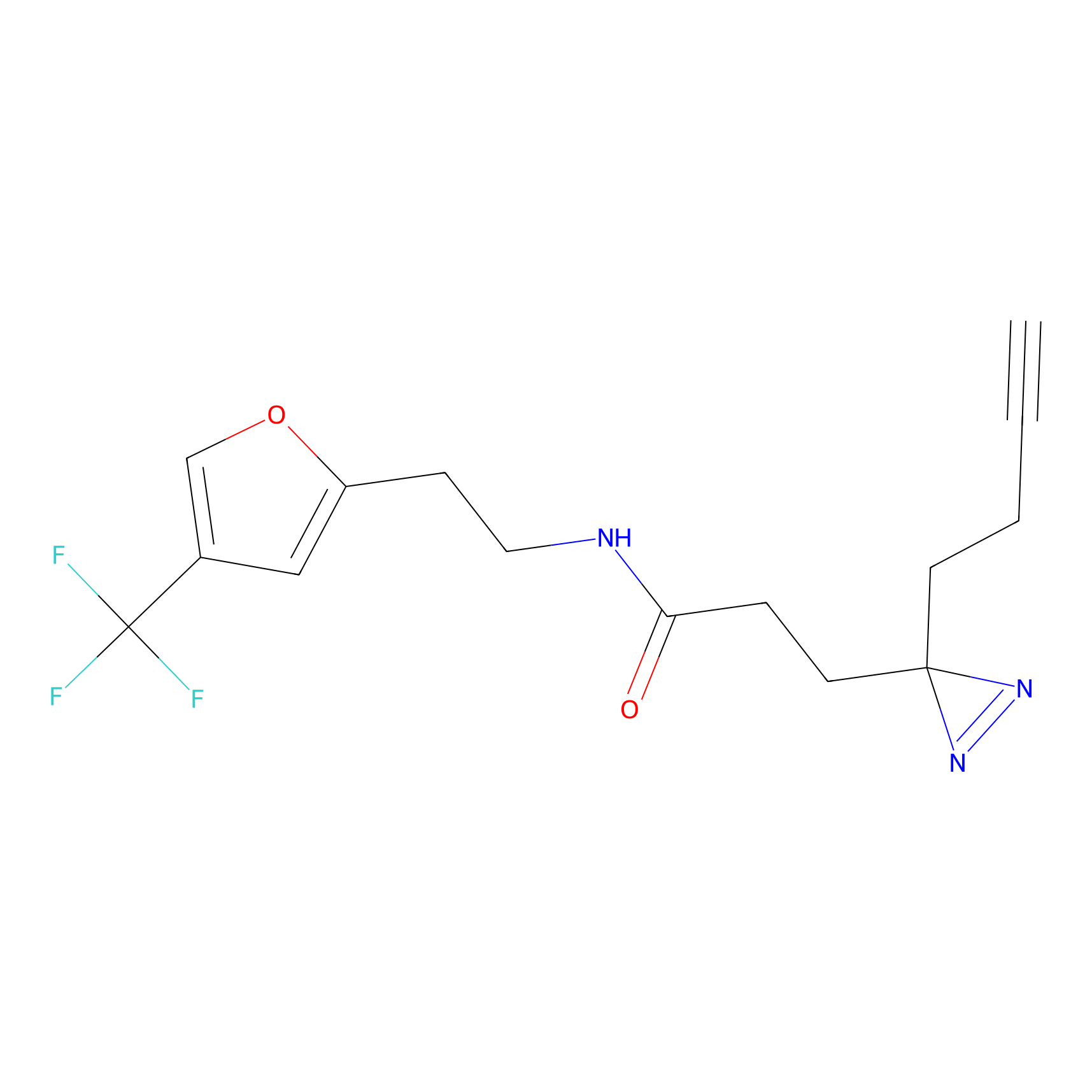
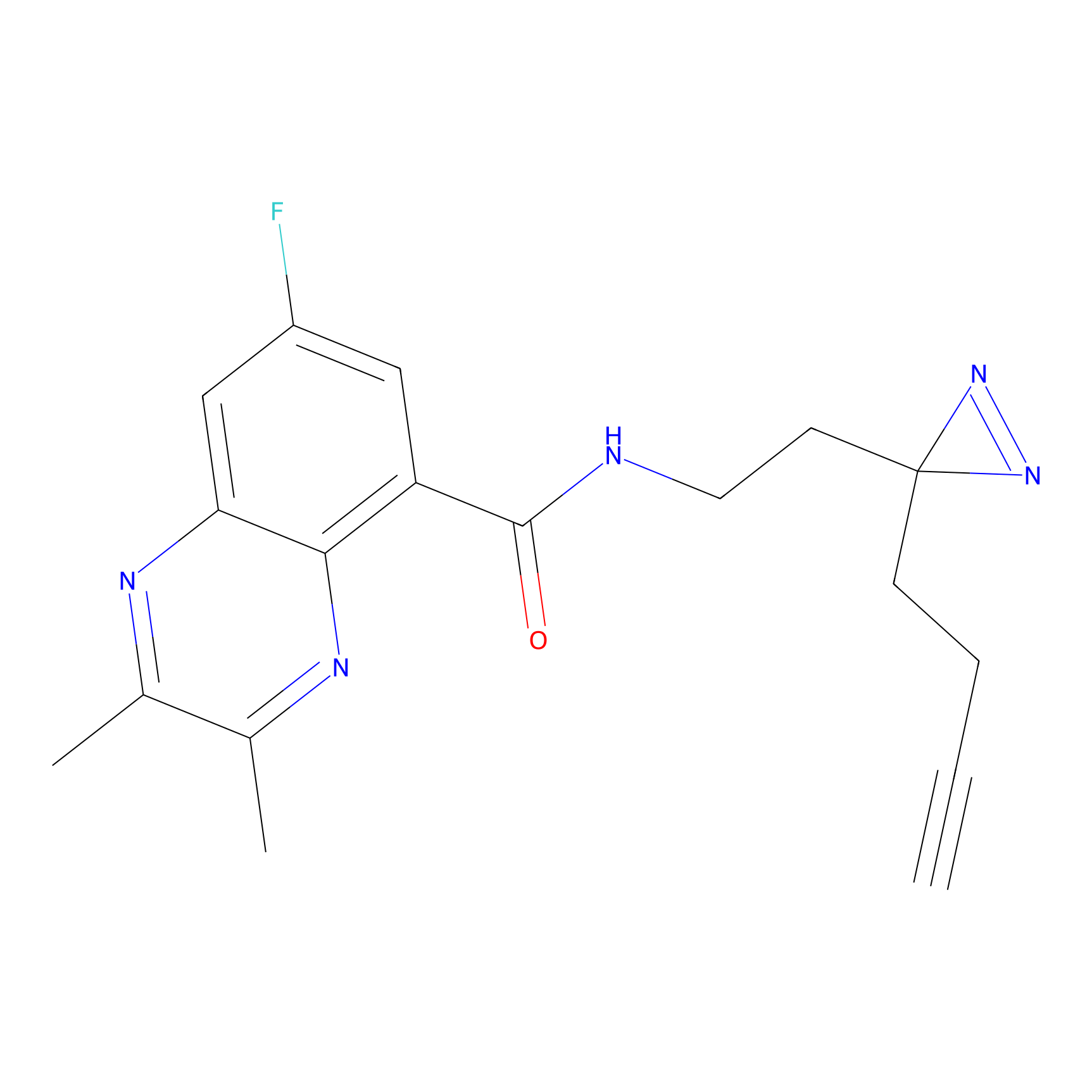
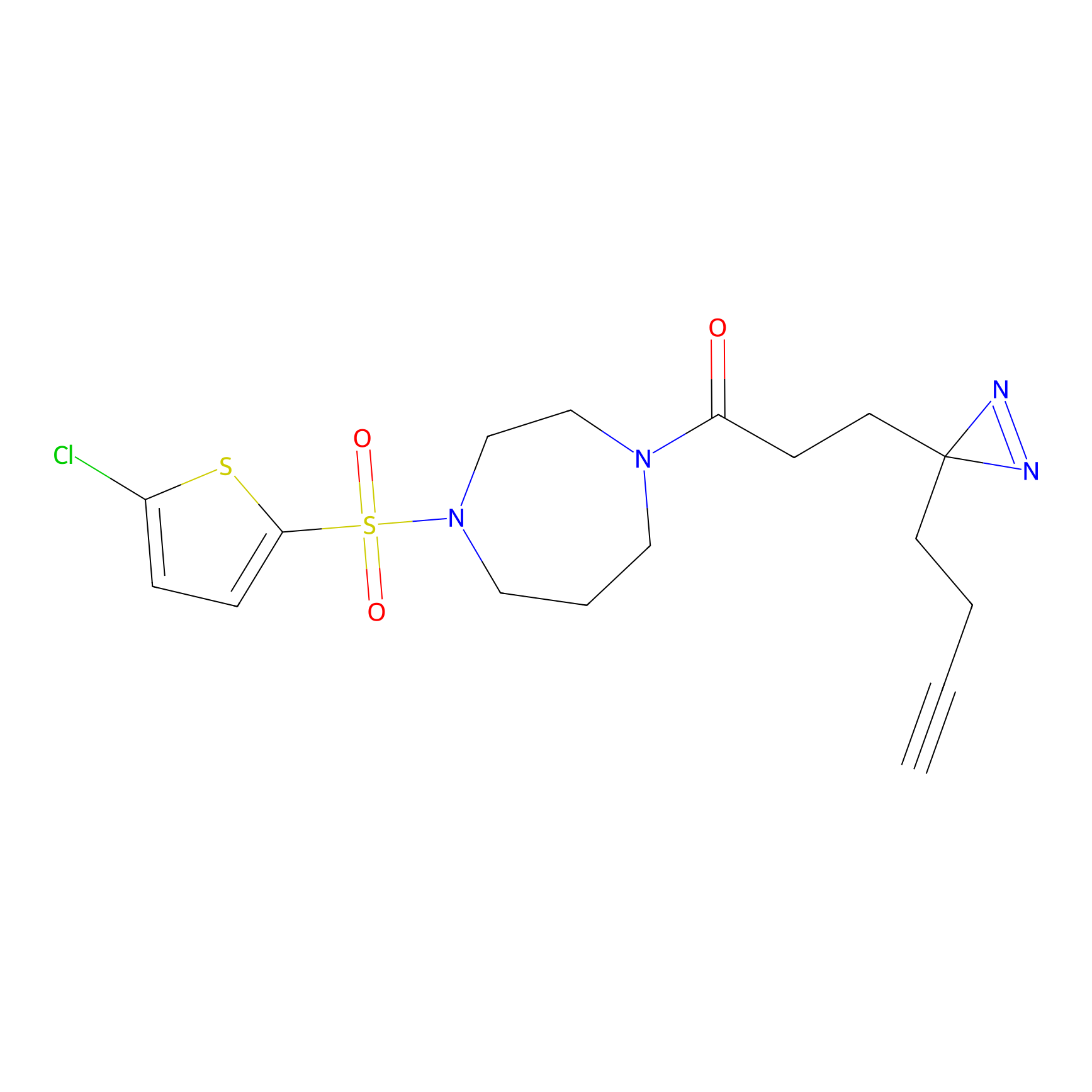
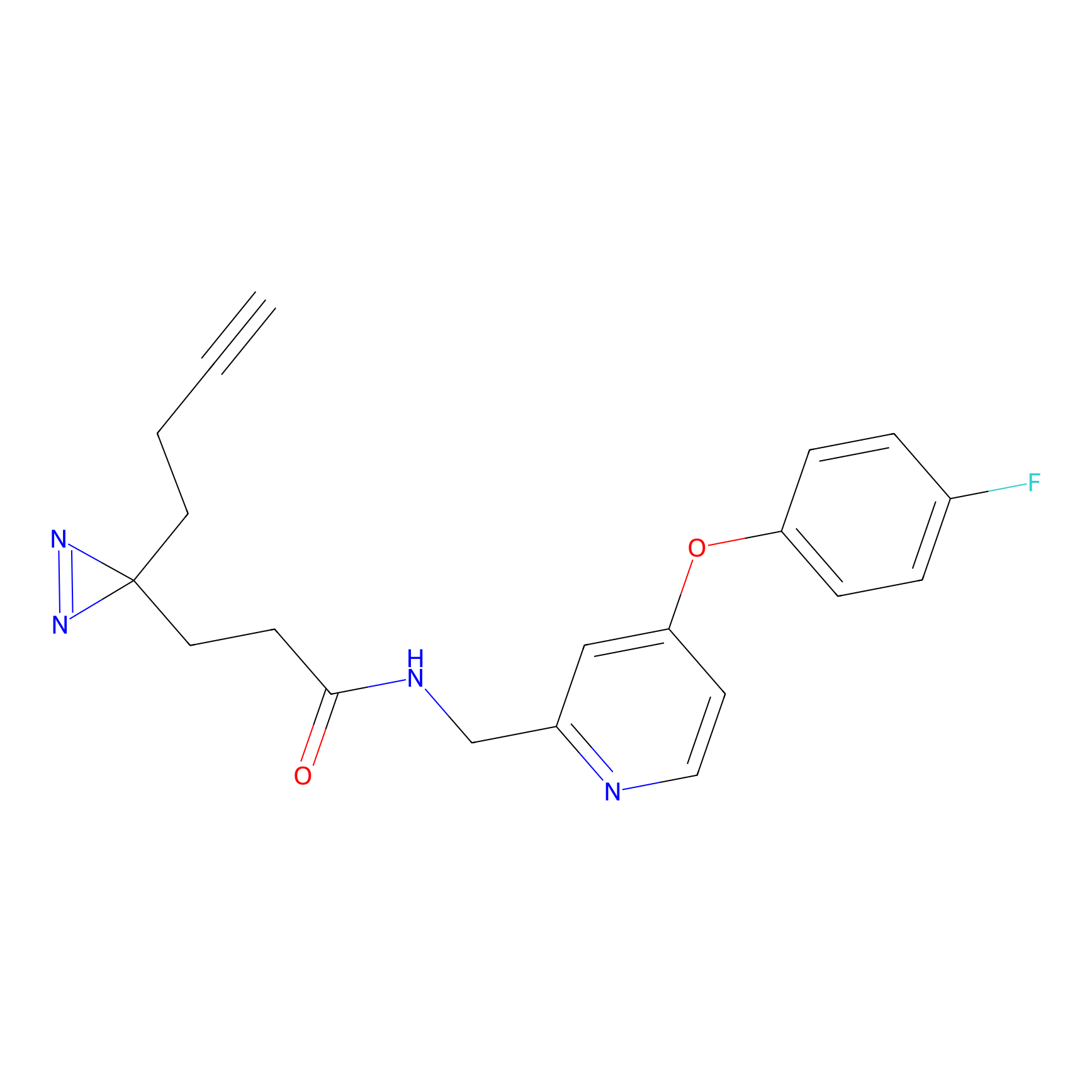
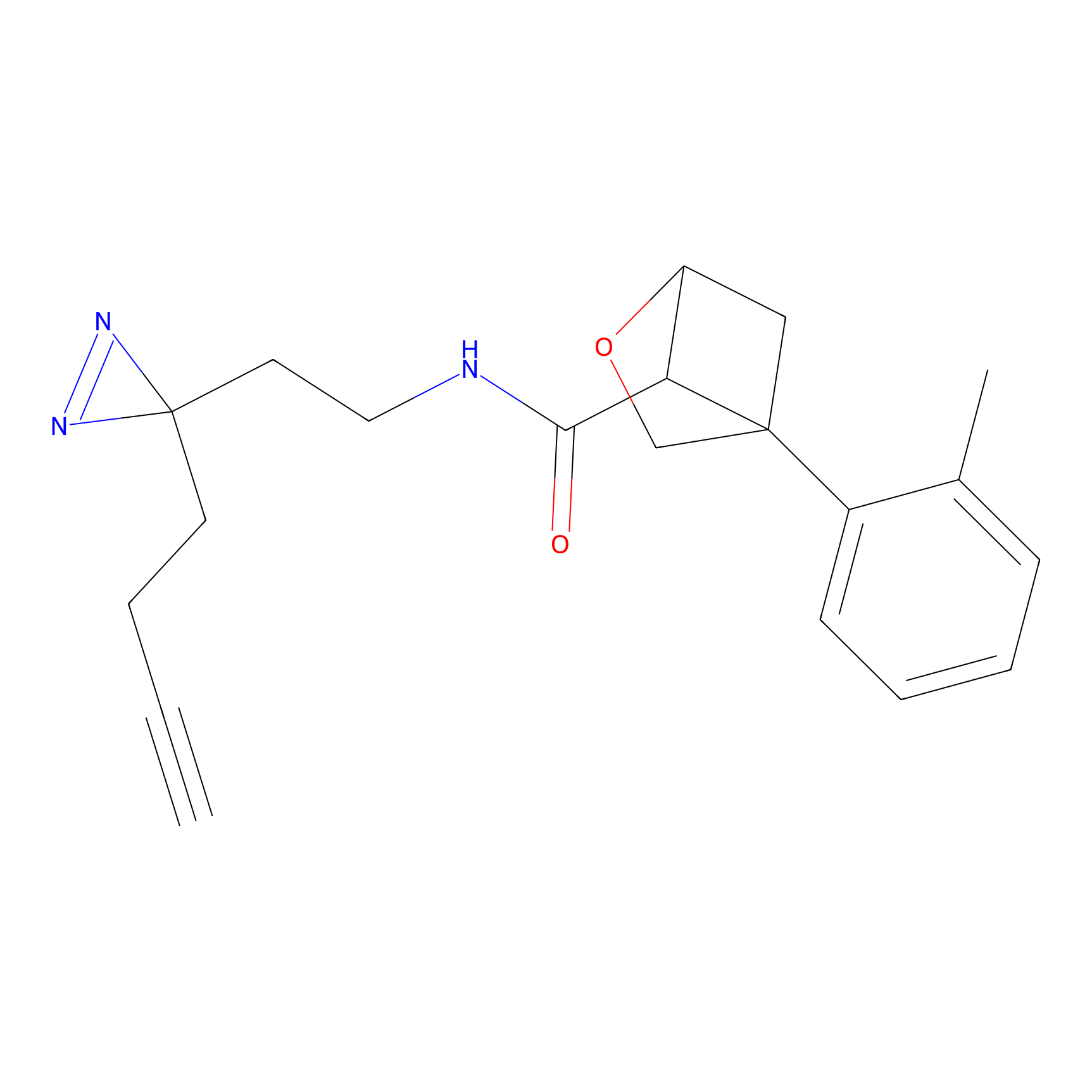
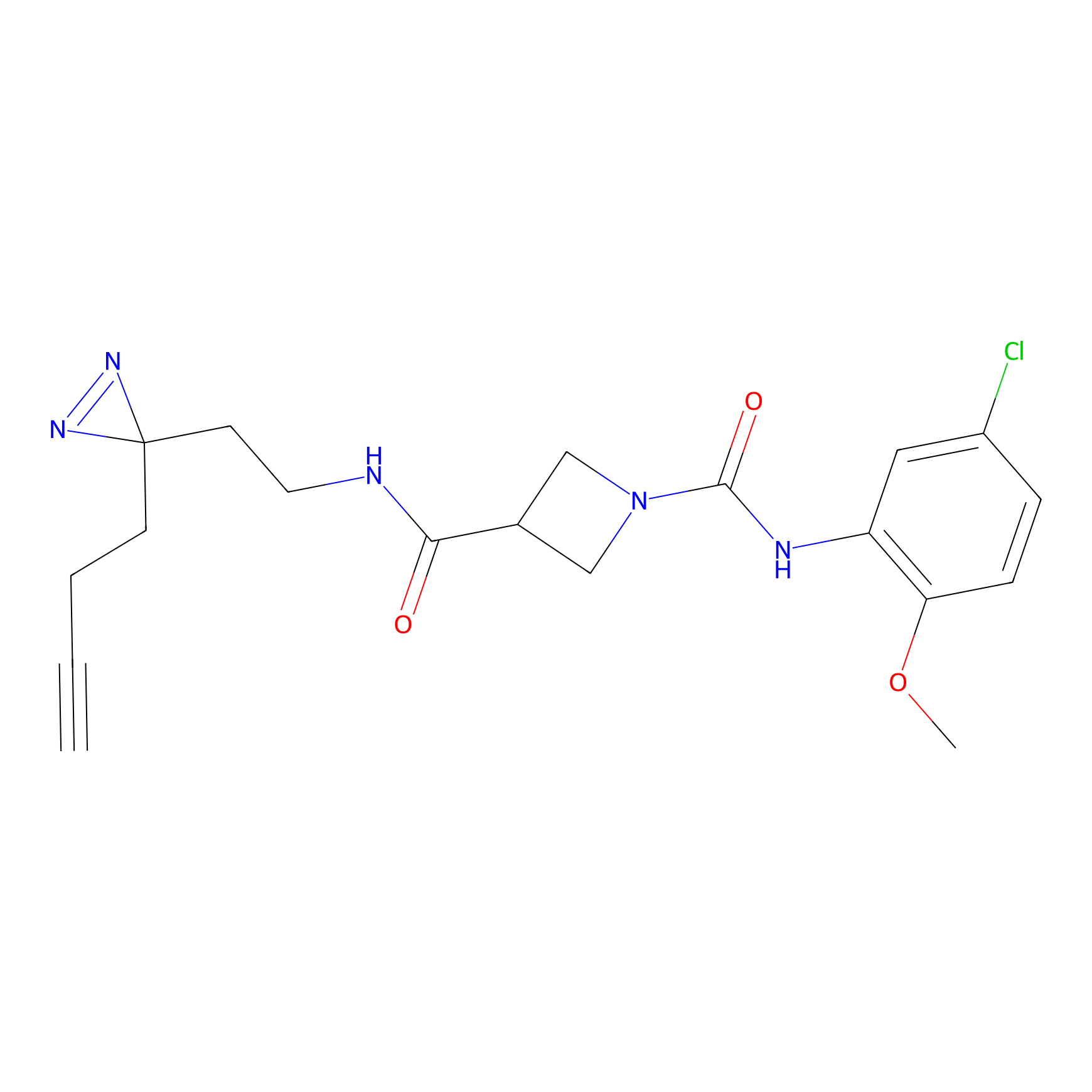
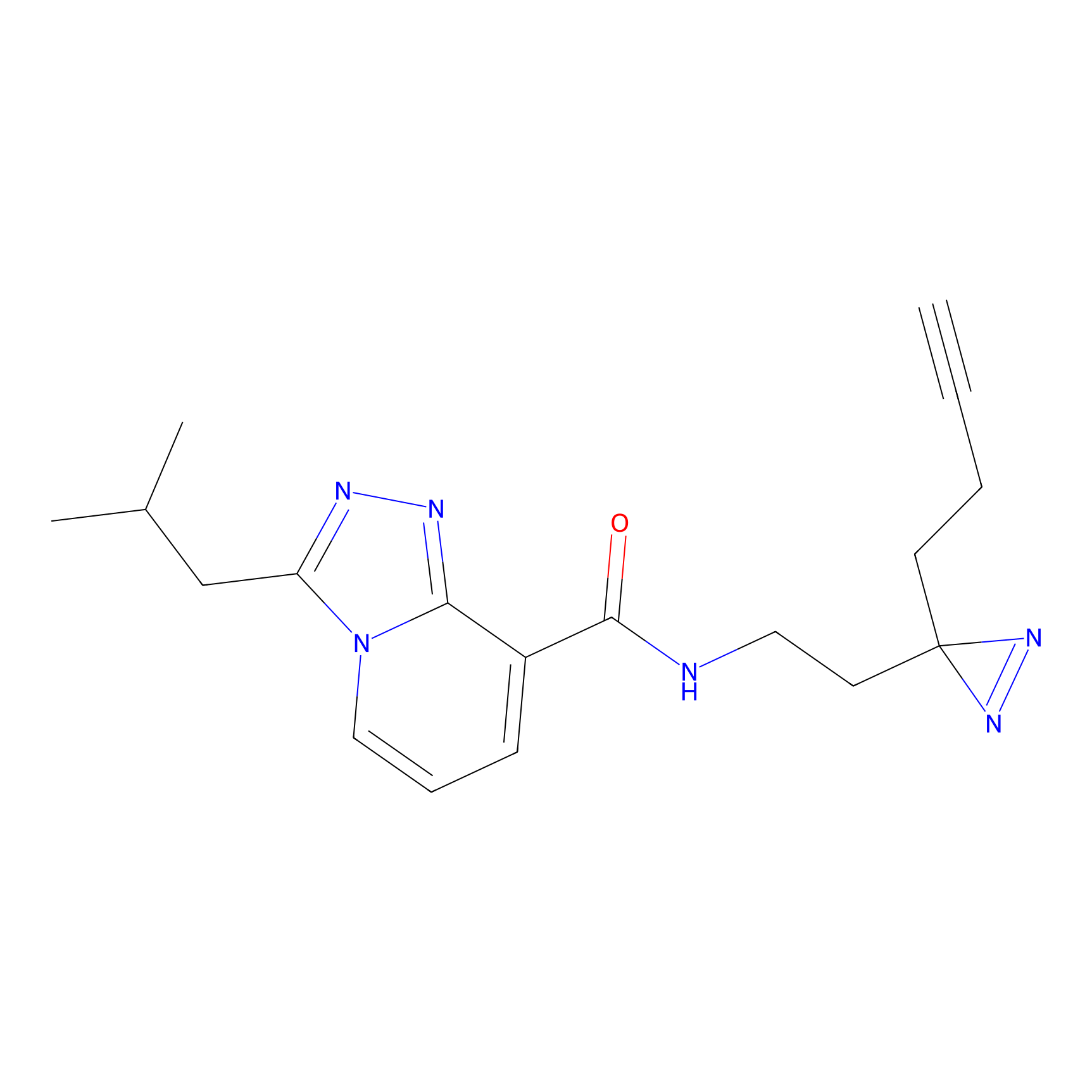
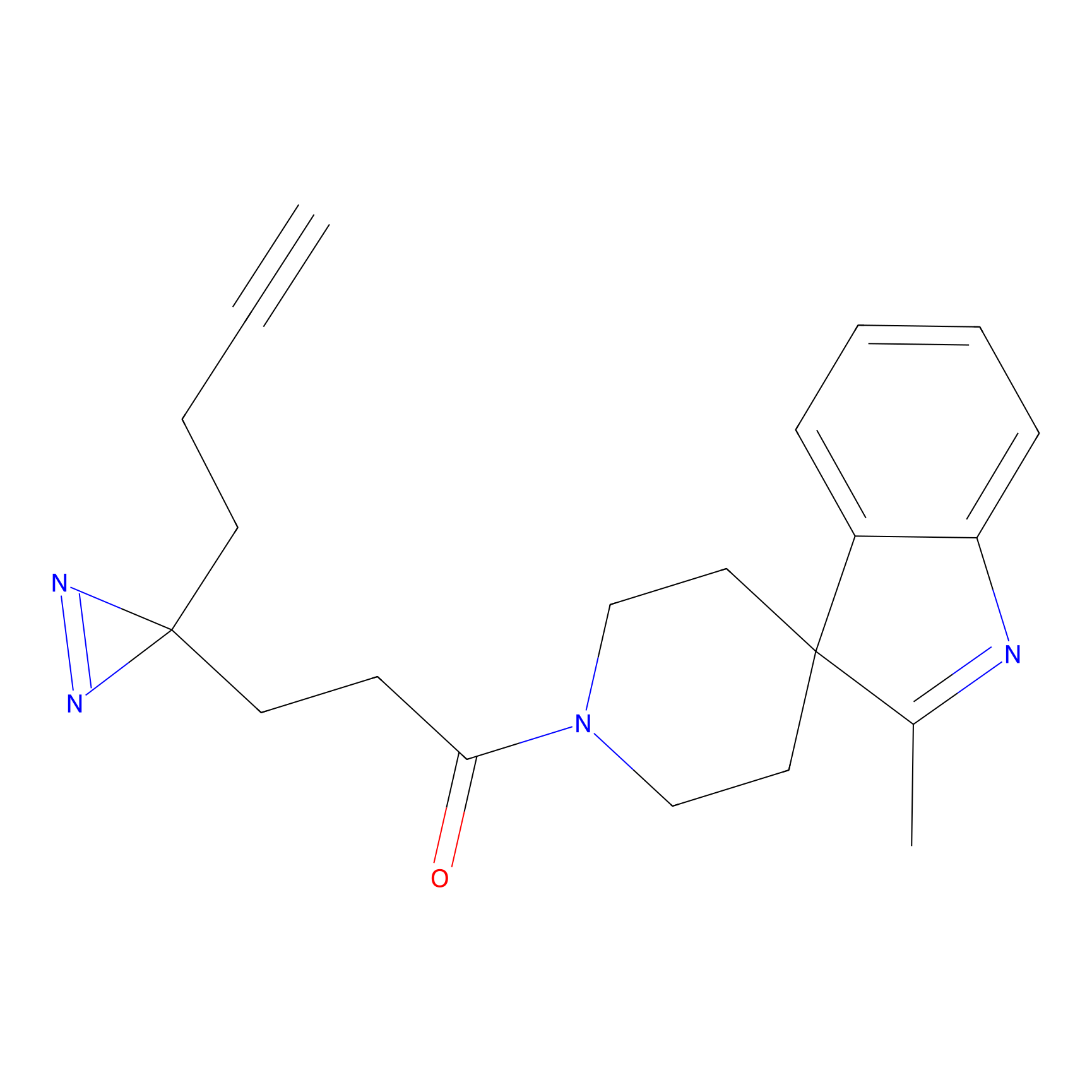
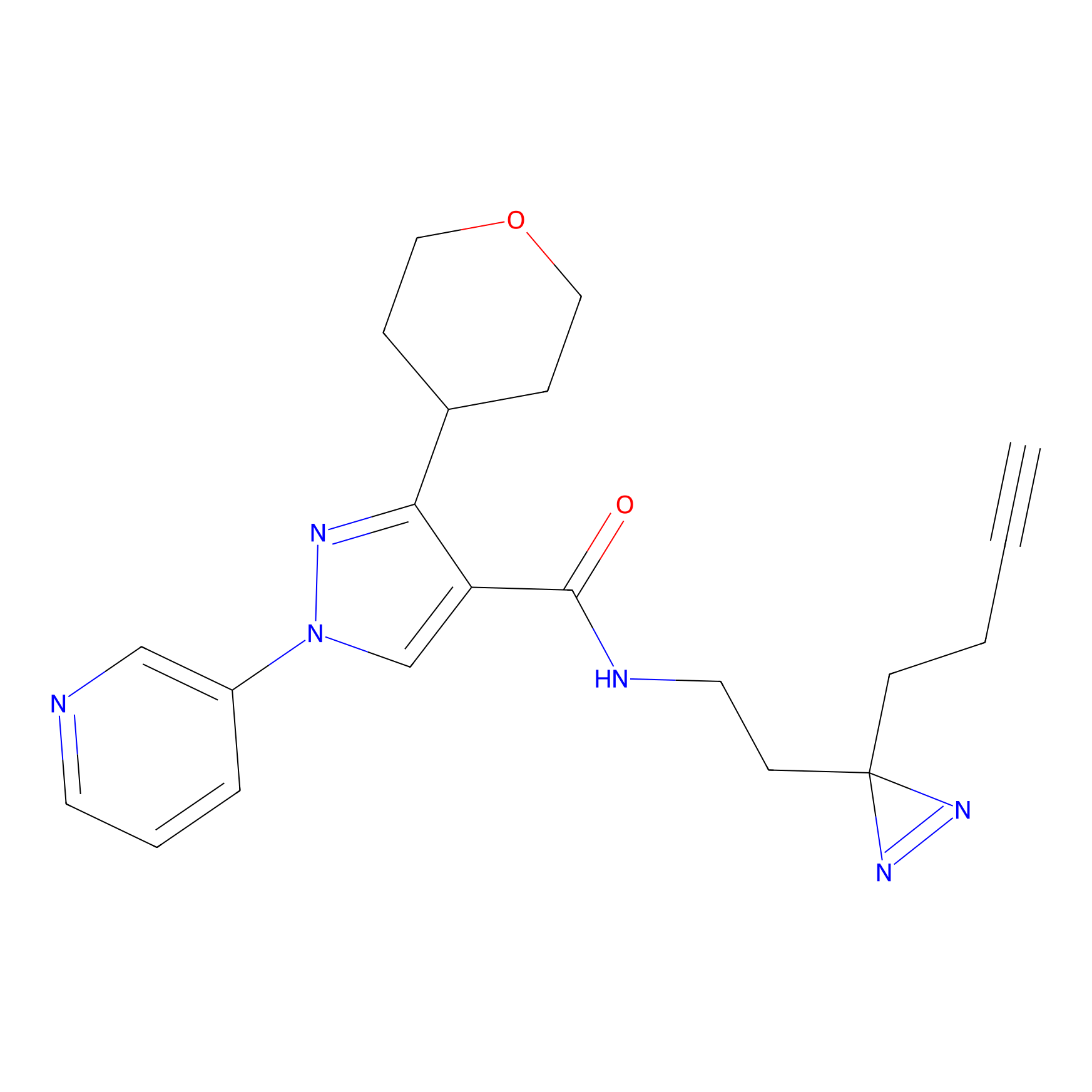
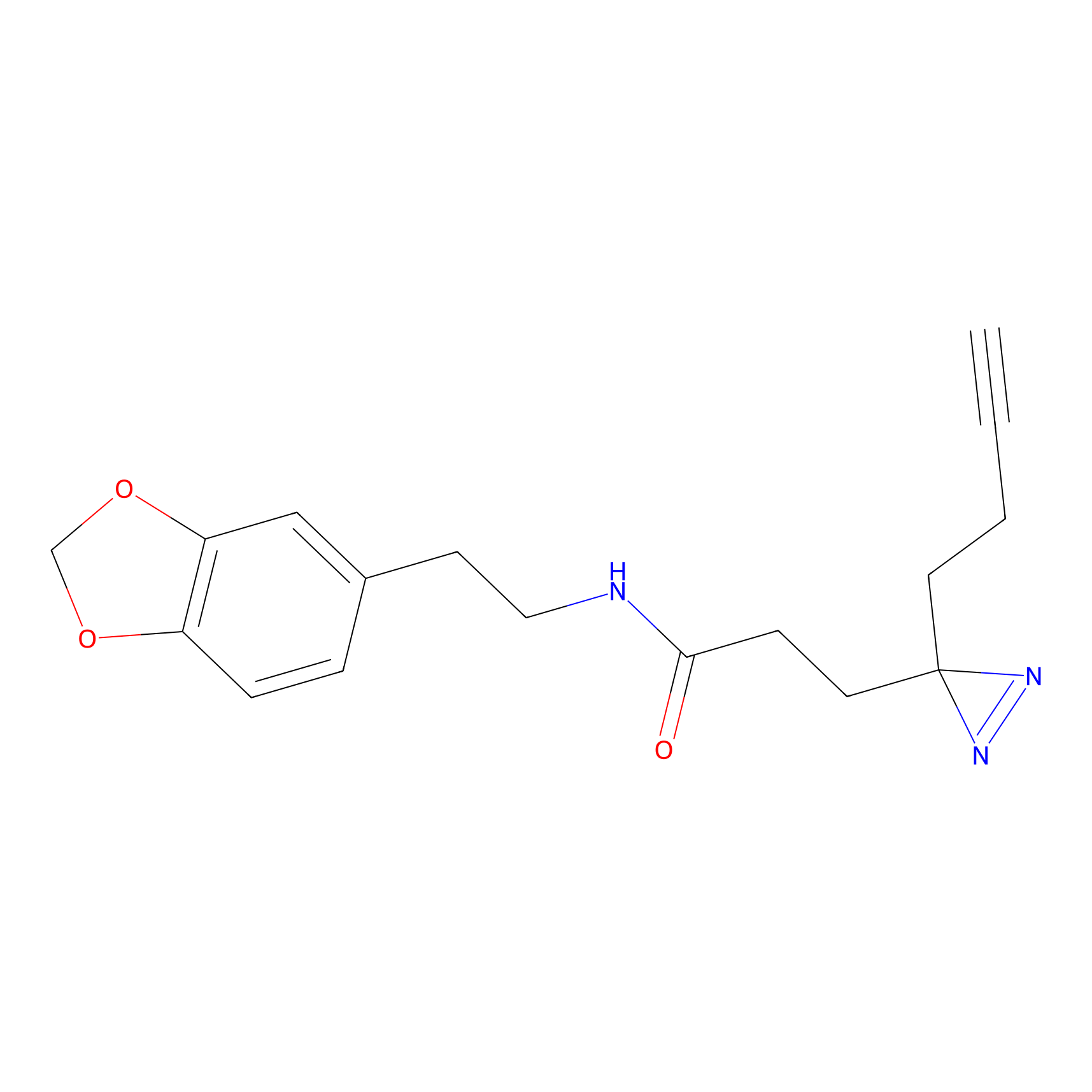
IPM Probe Info |
 |
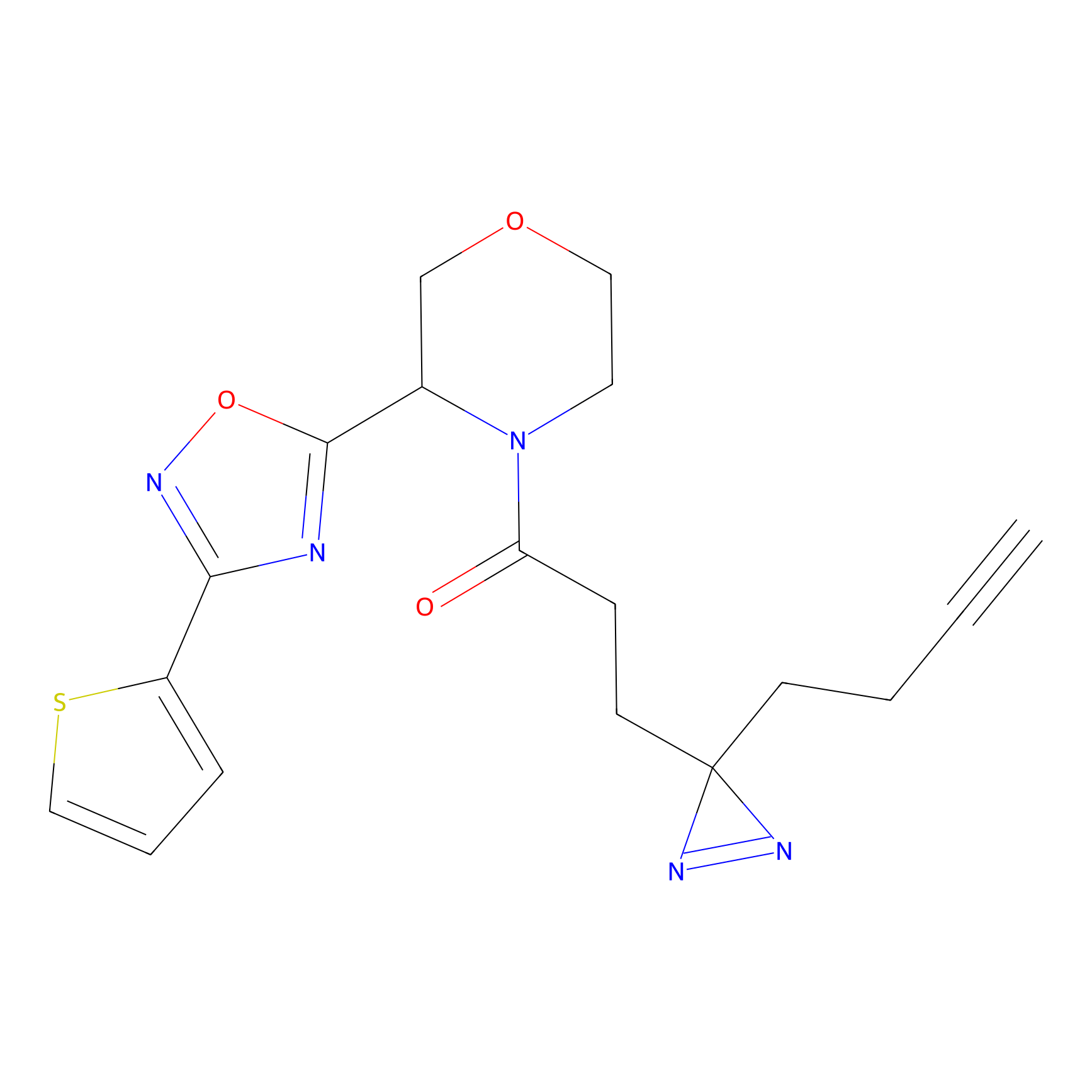
C6(1.79) | LDD1702 | [3] | |
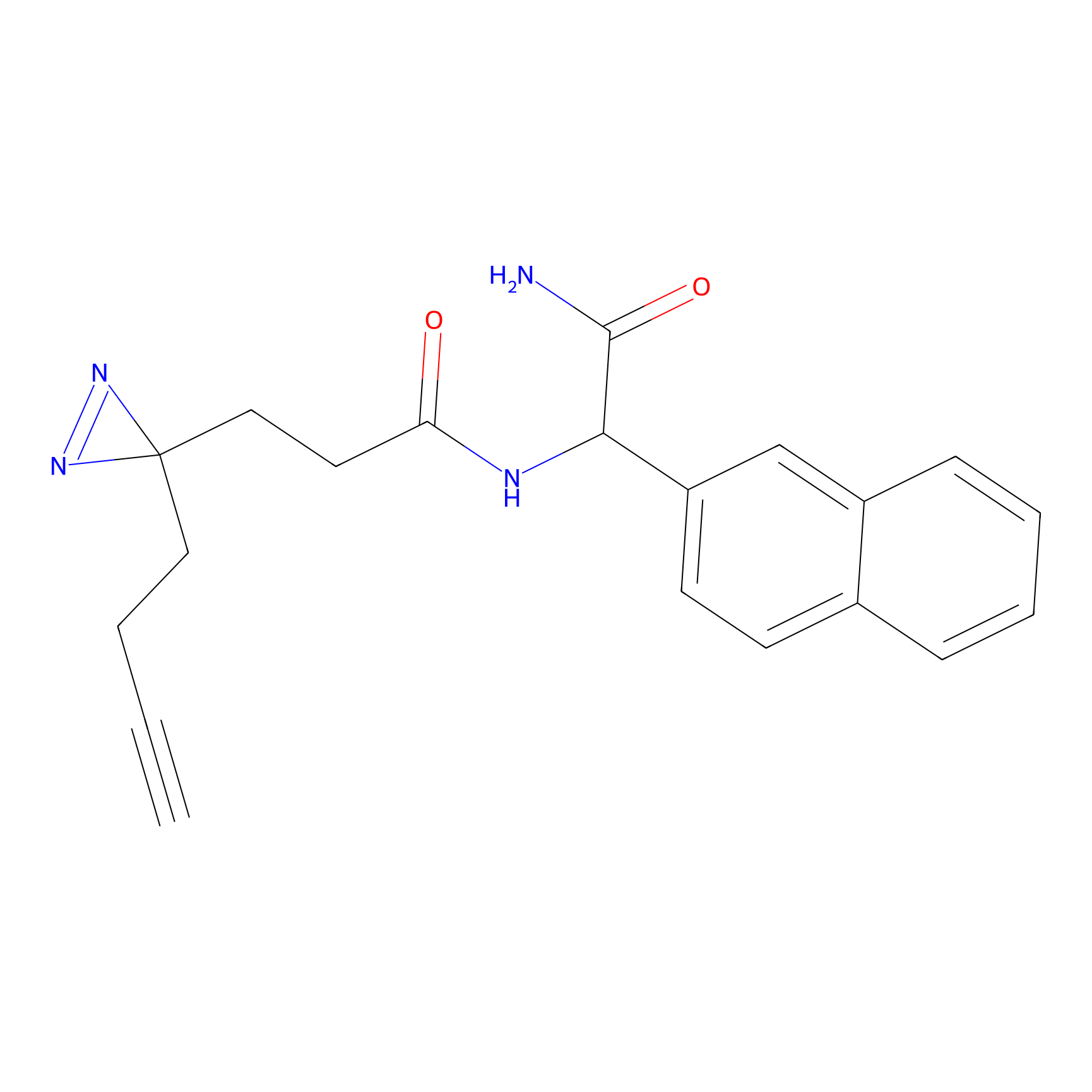
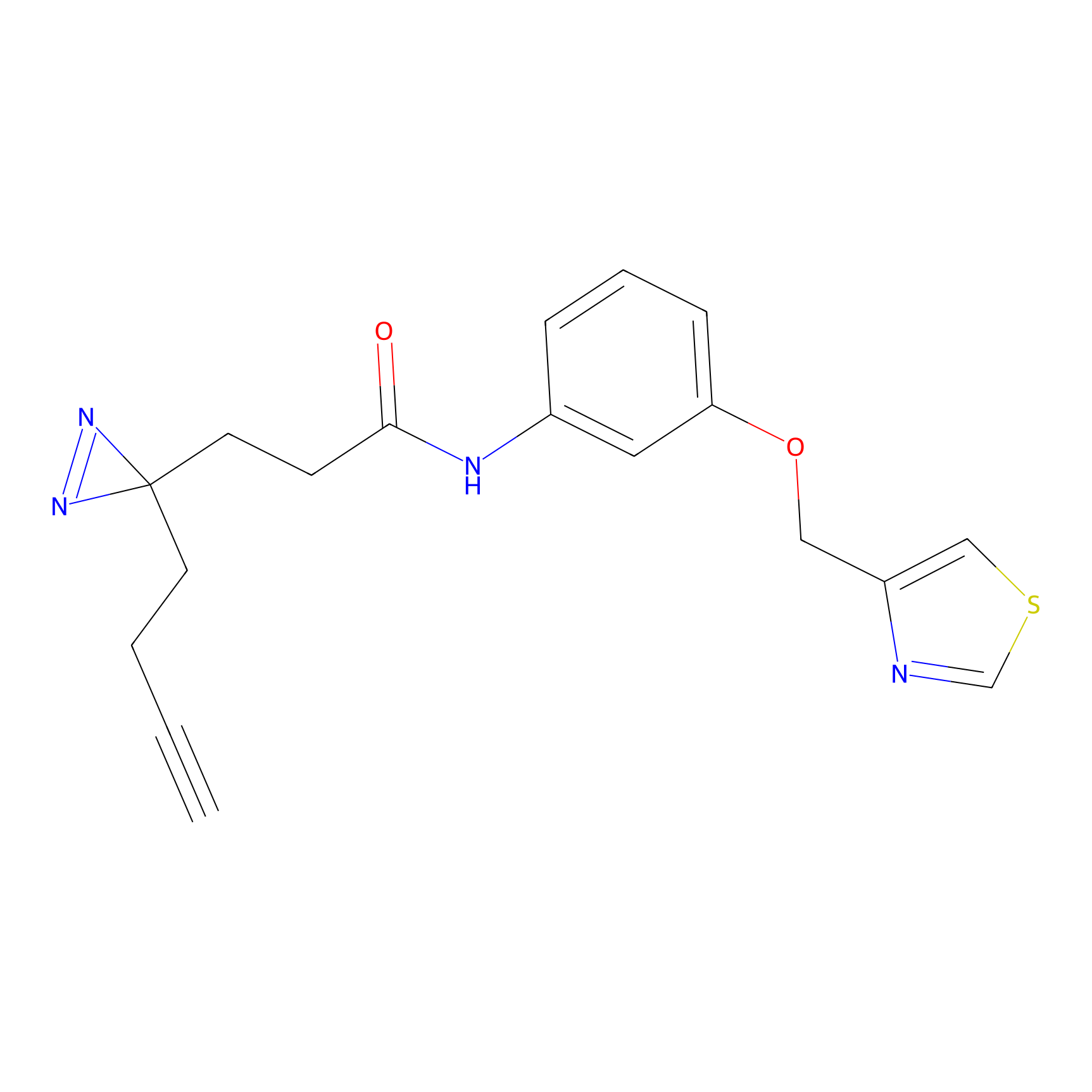
|
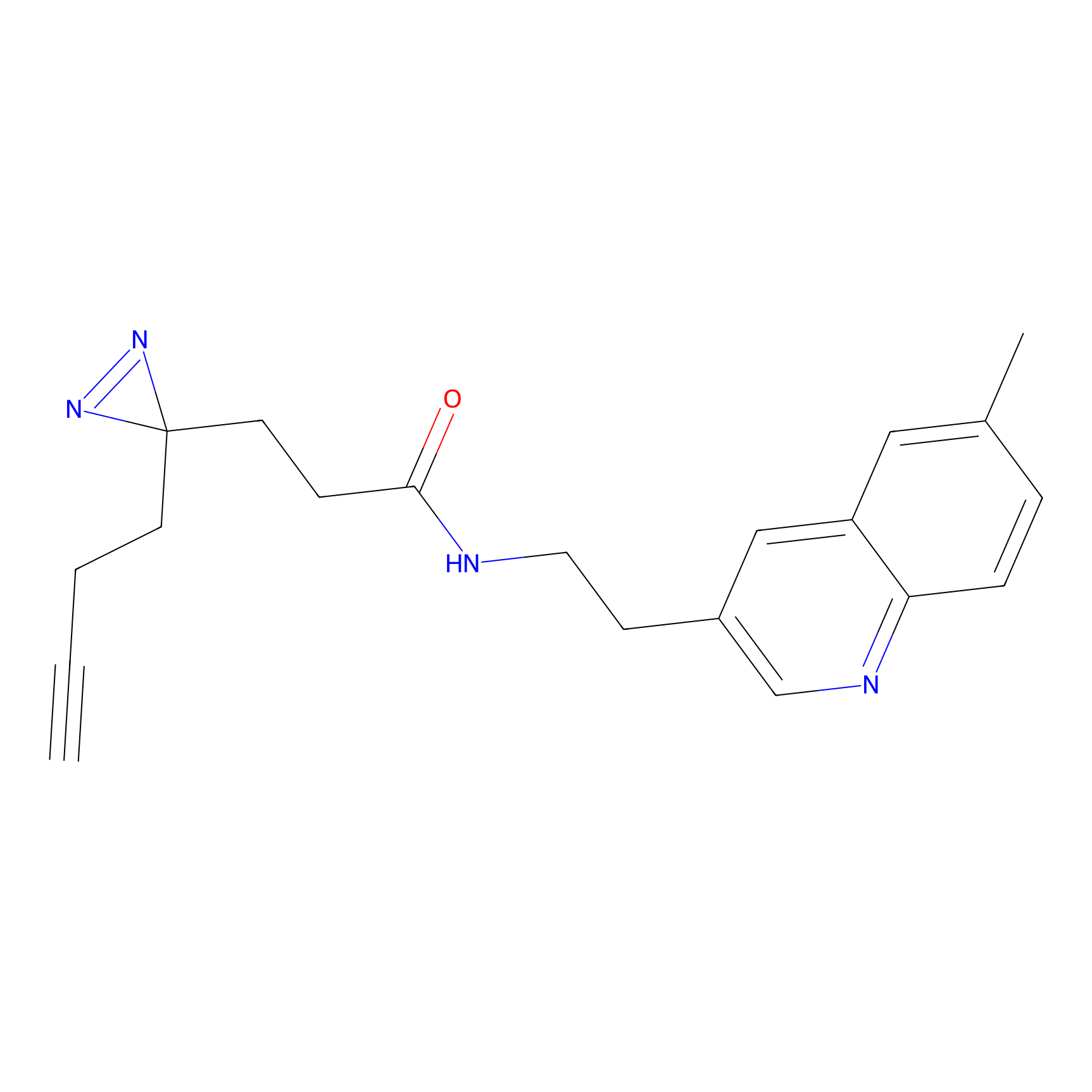
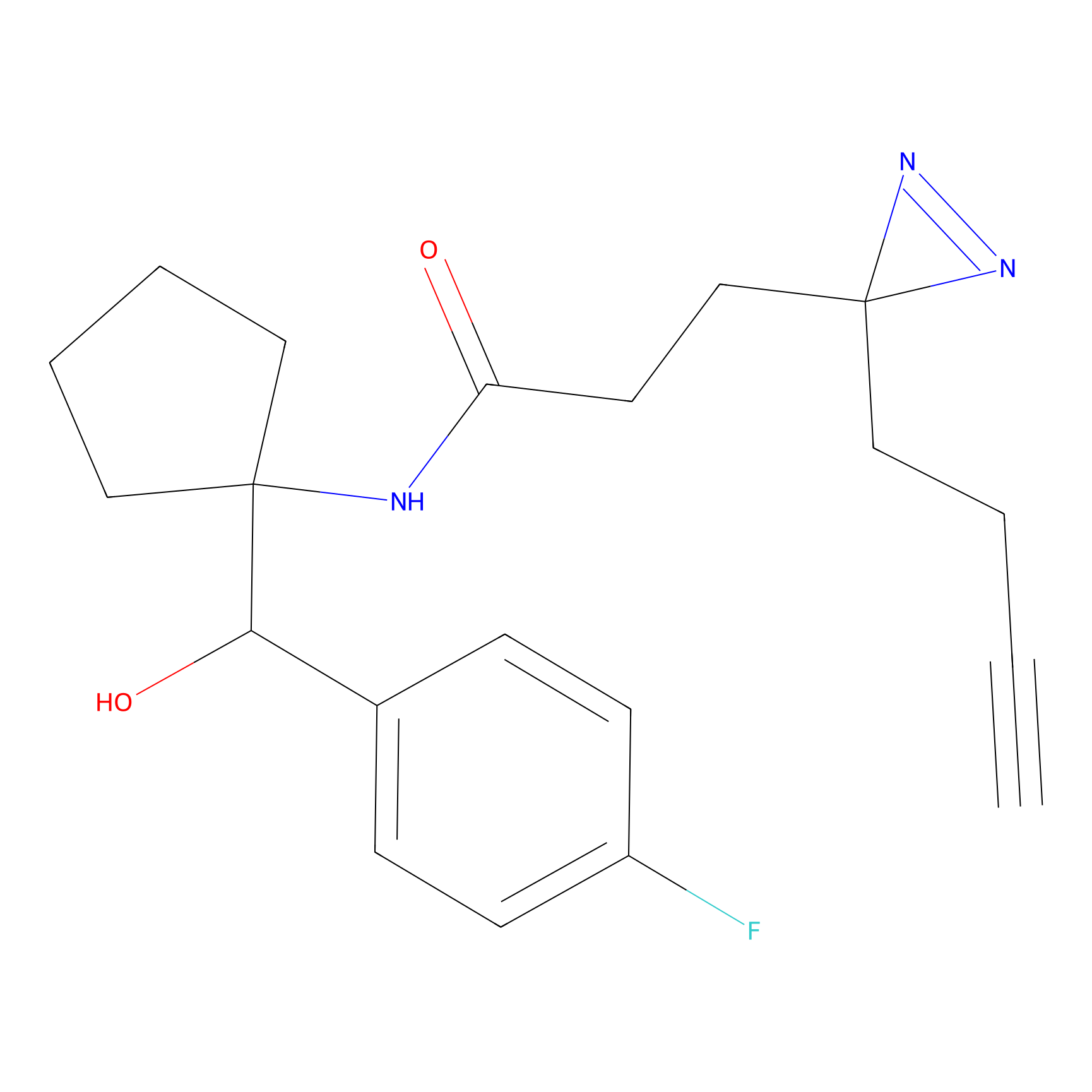
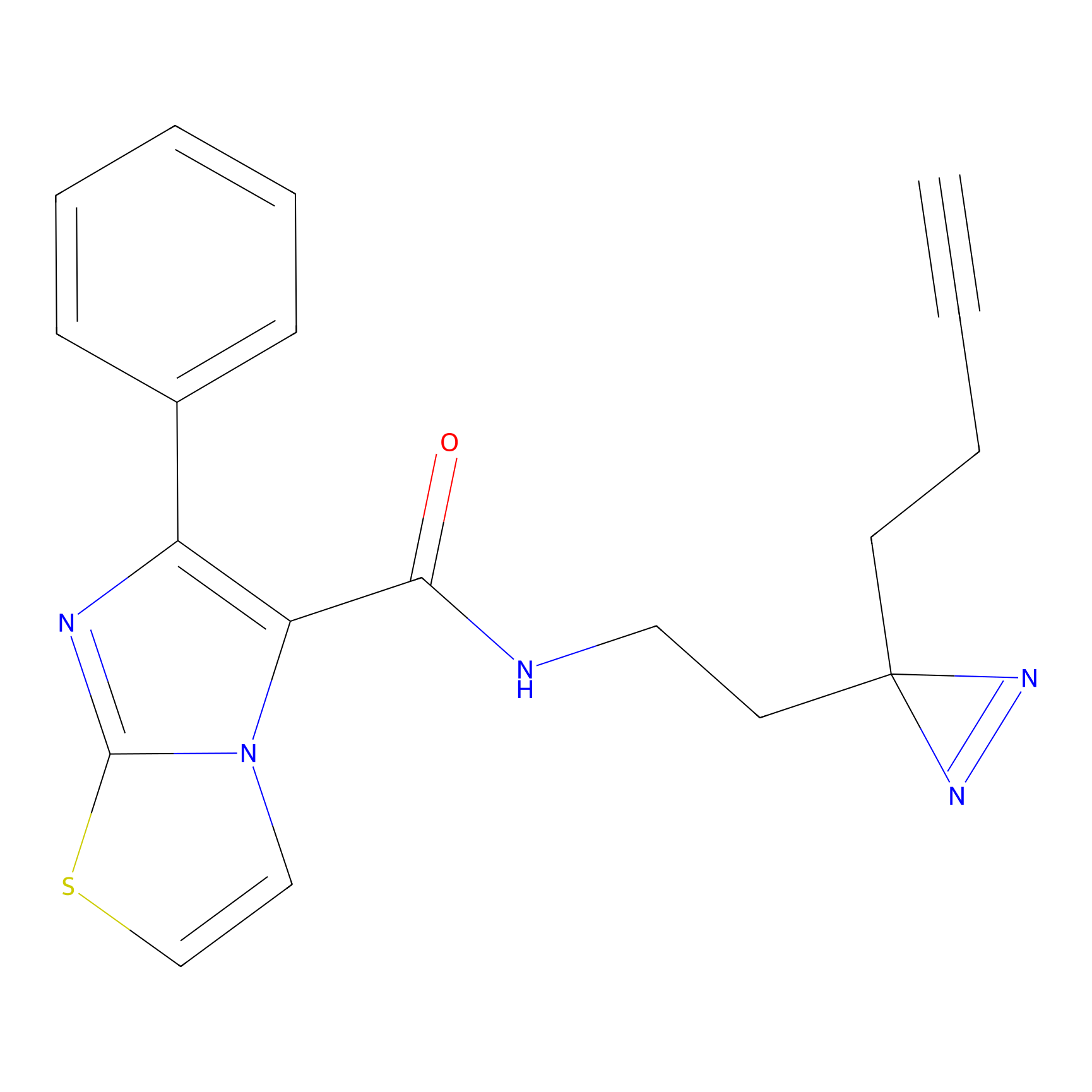
Acrolein Probe Info |
 |
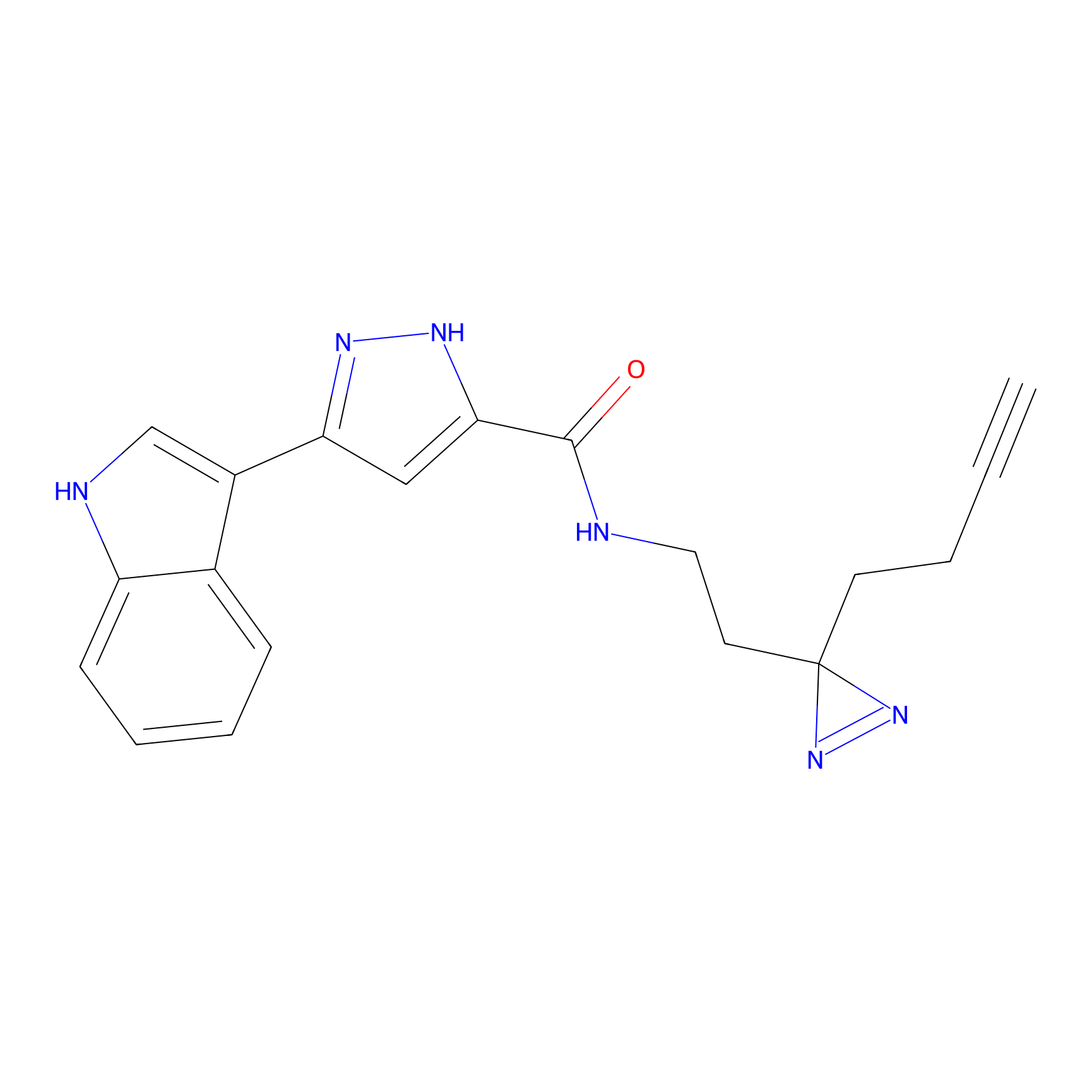
N.A. | LDD0221 | [4] | |
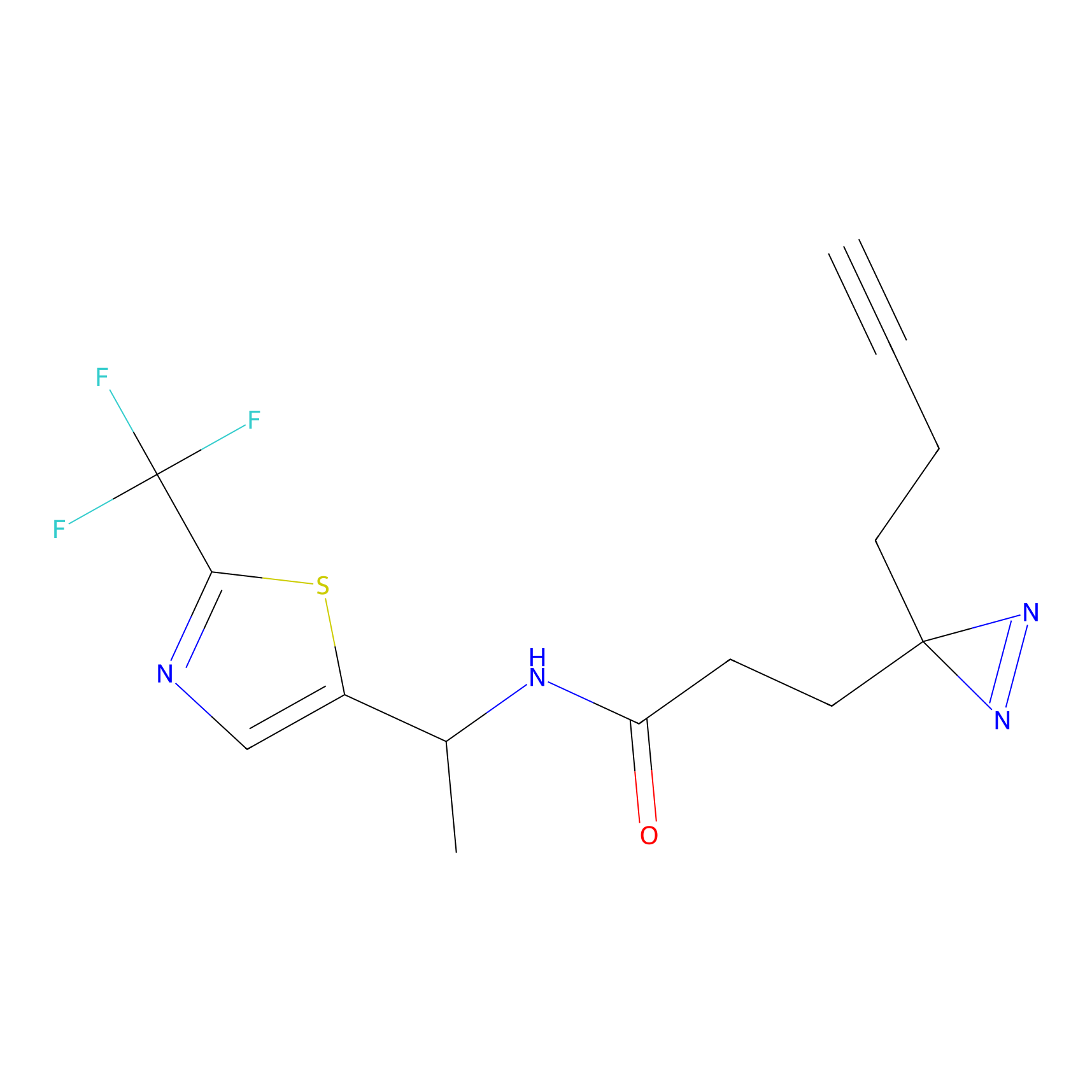
|
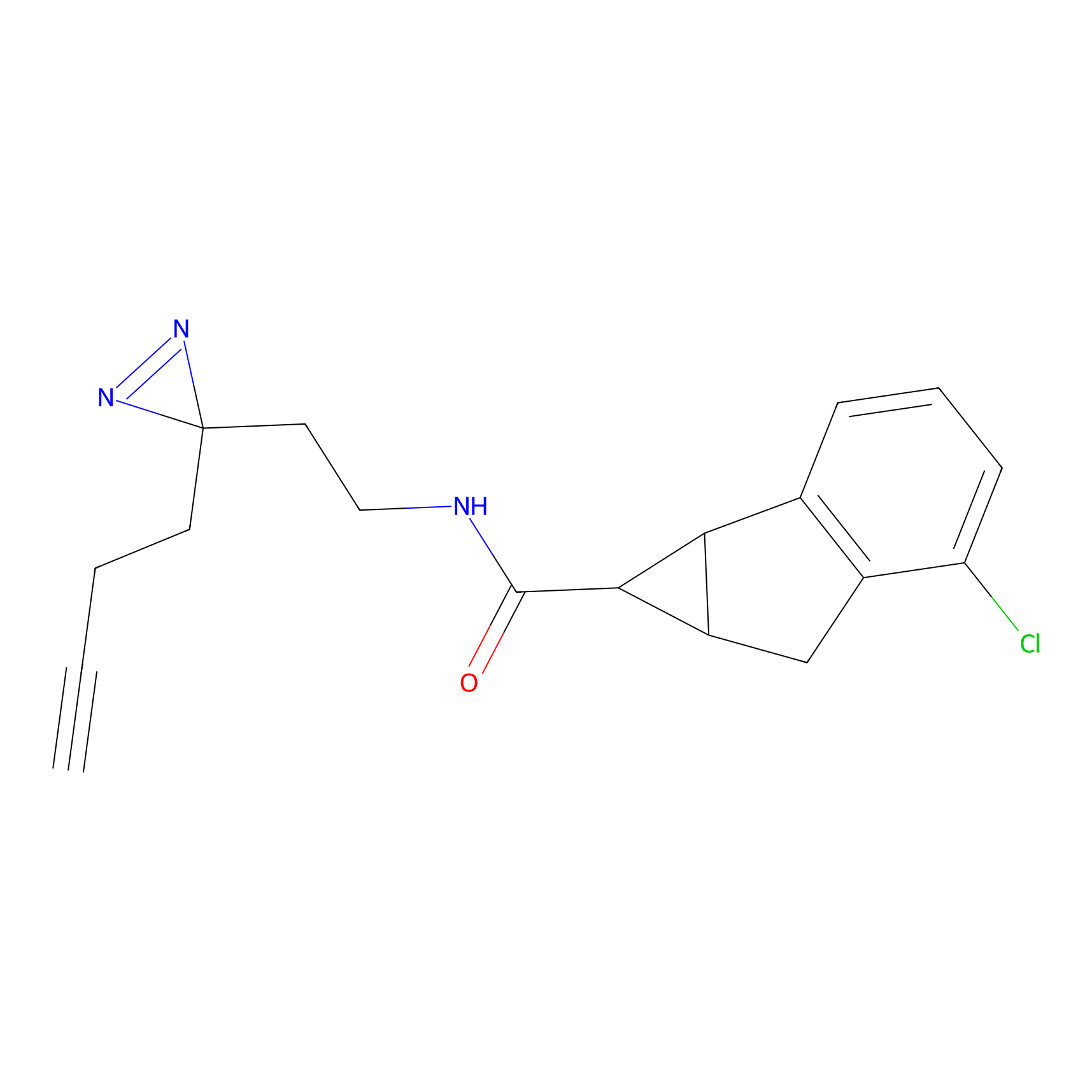
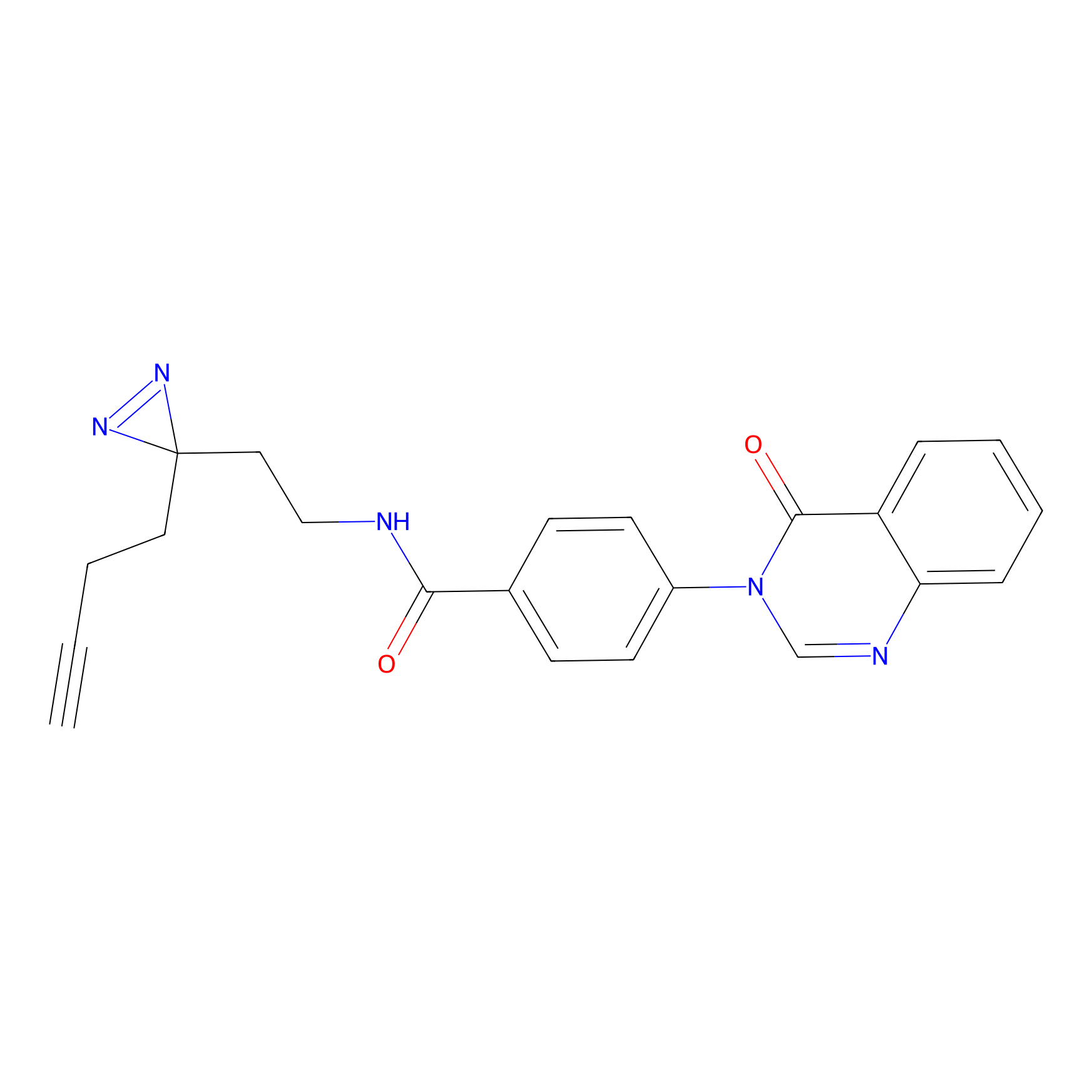
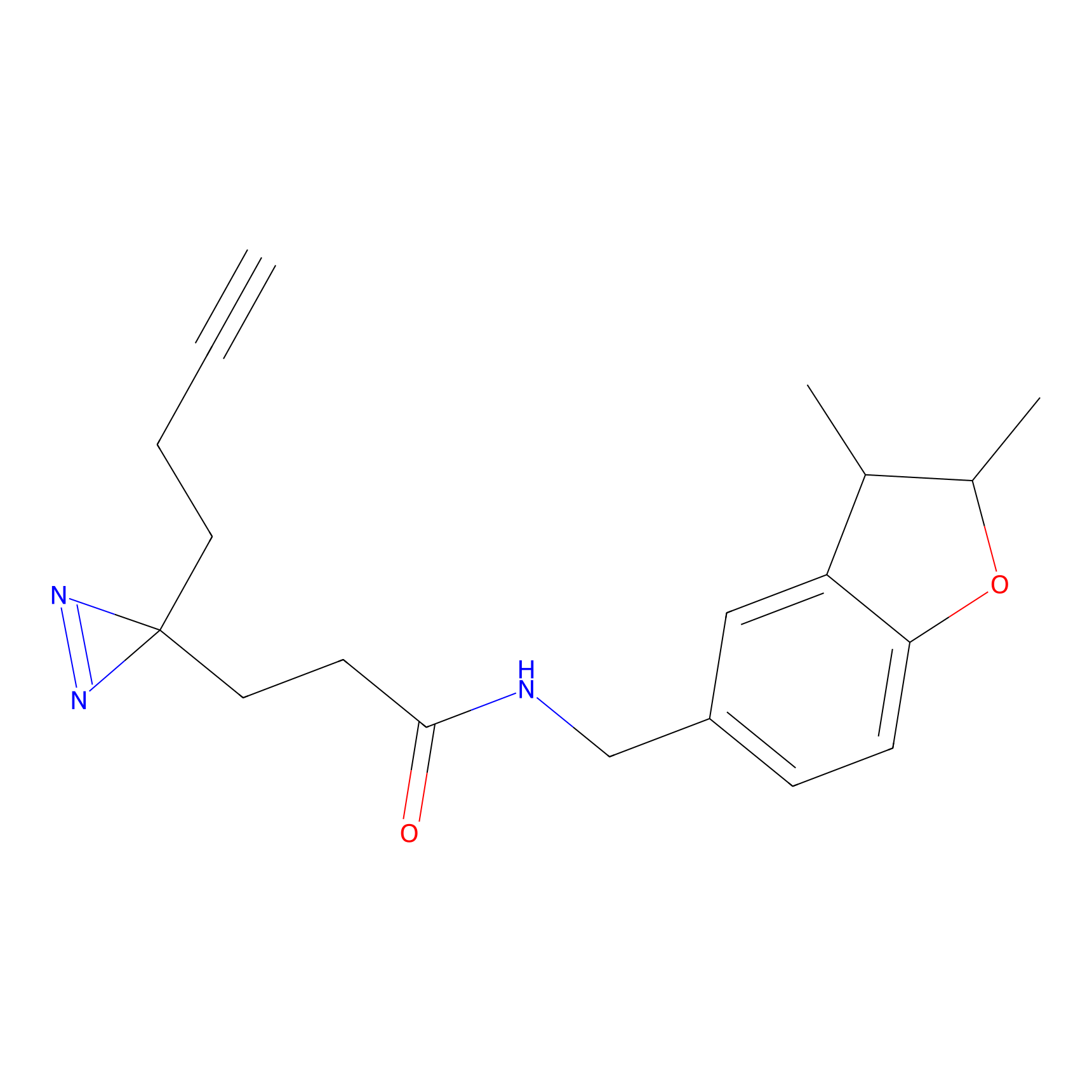
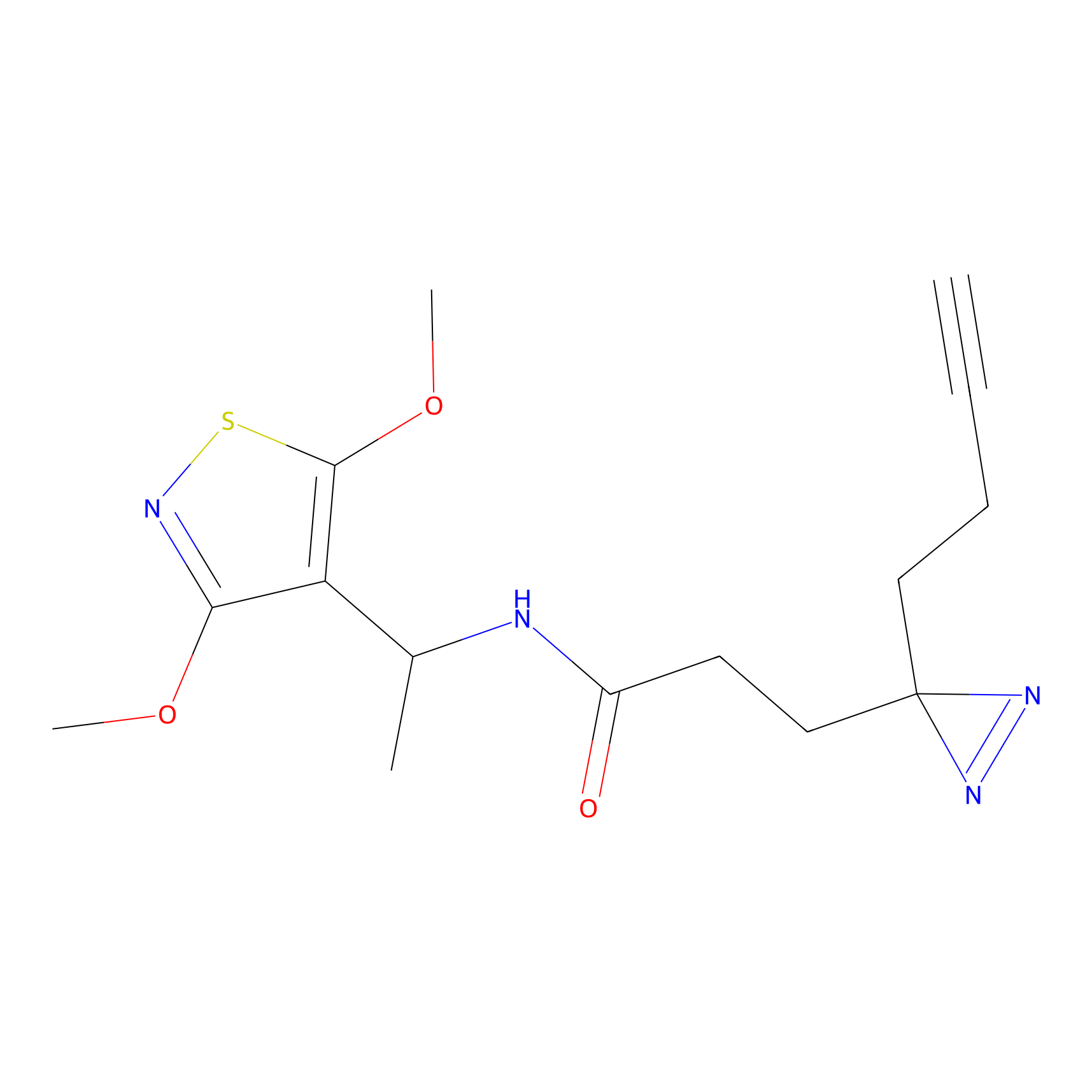
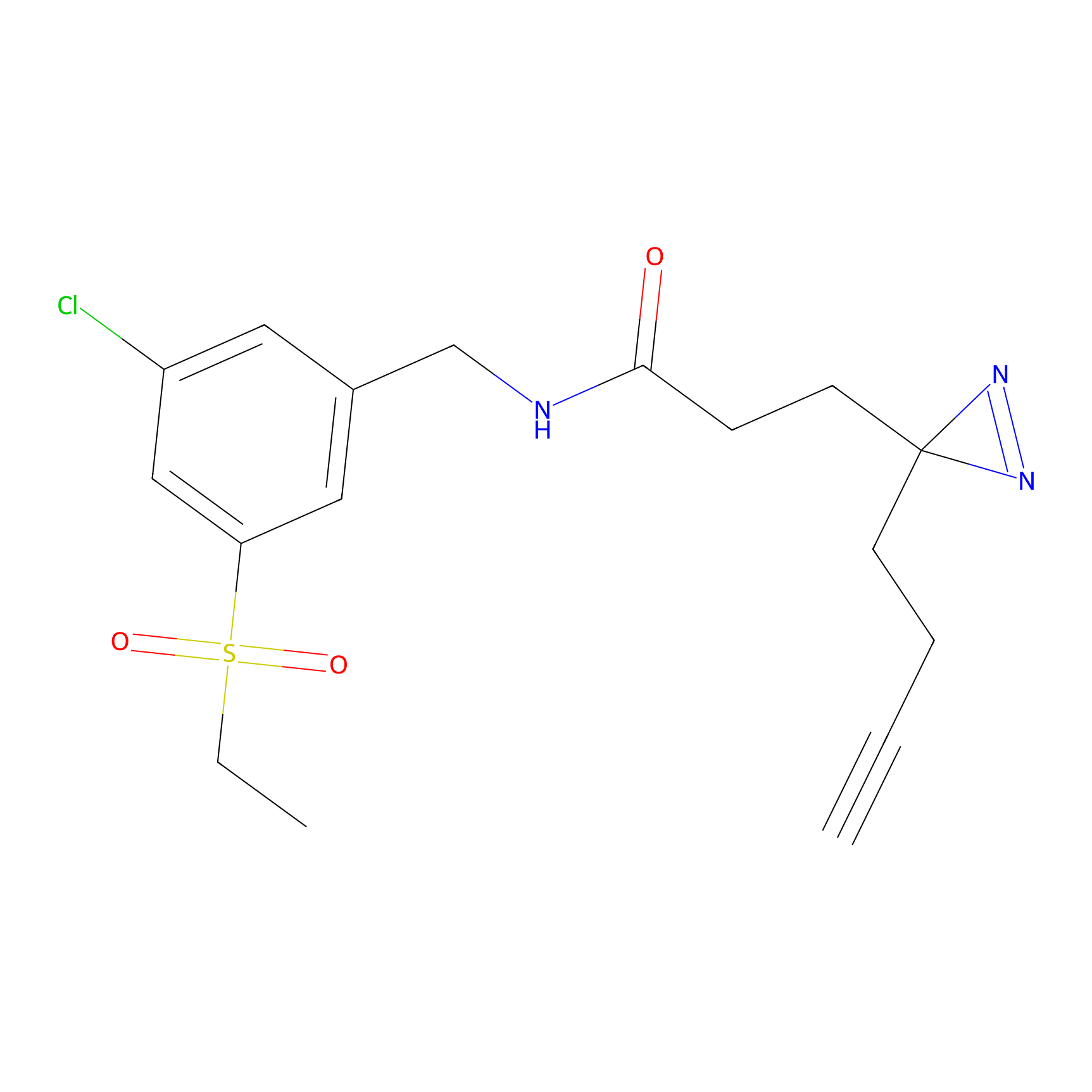
DBIA Probe Info |
 |
C6(0.98) | LDD1509 | [5] | |
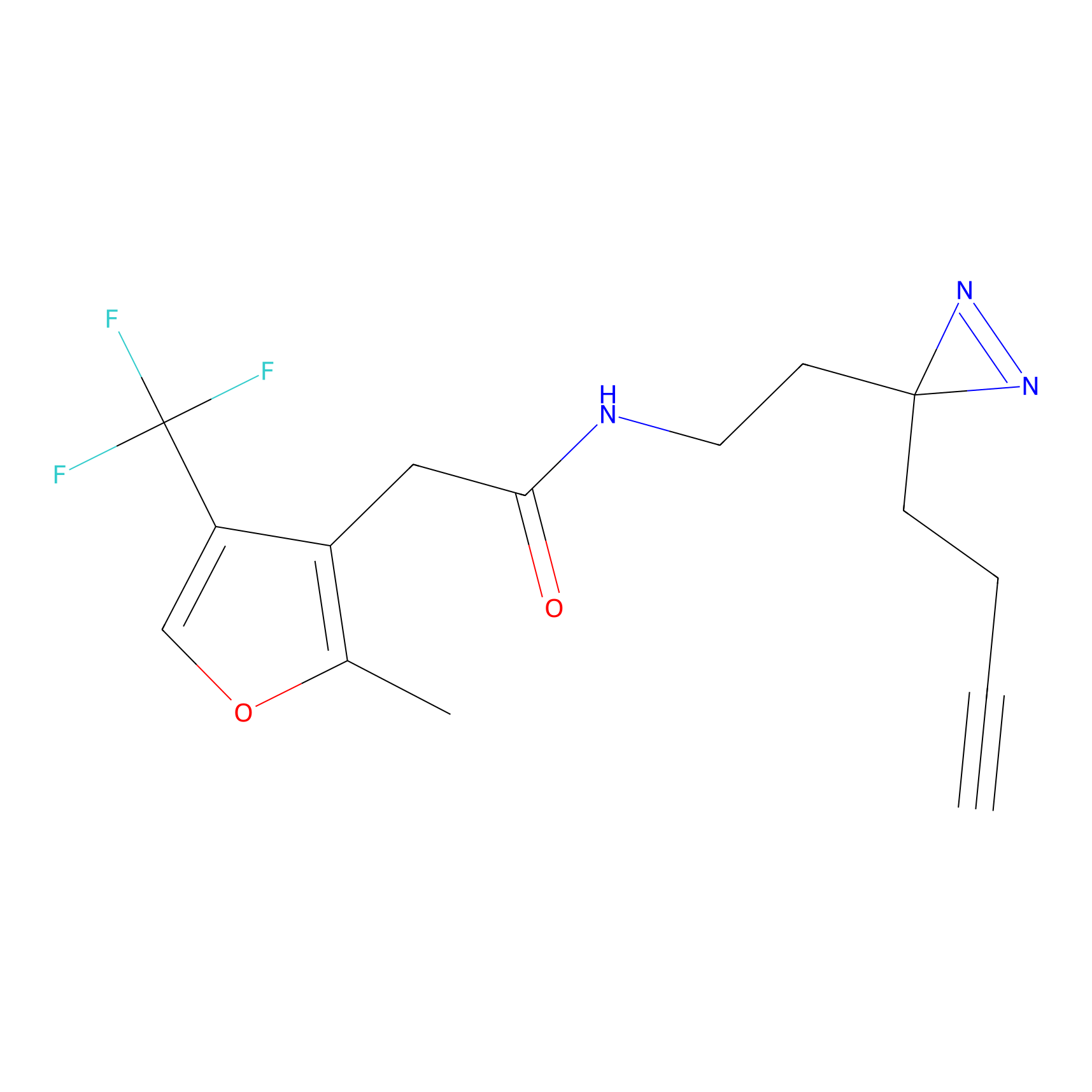
|
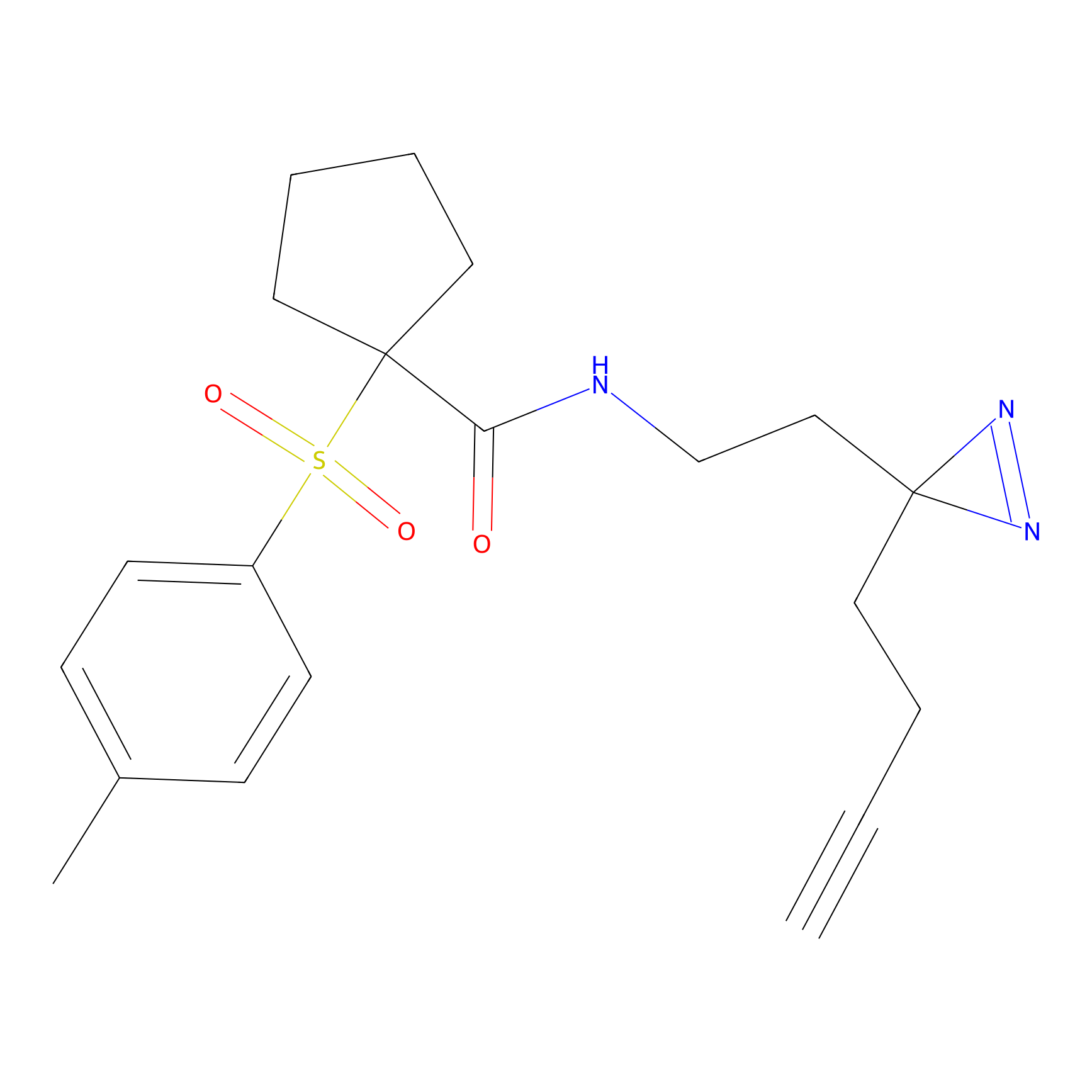
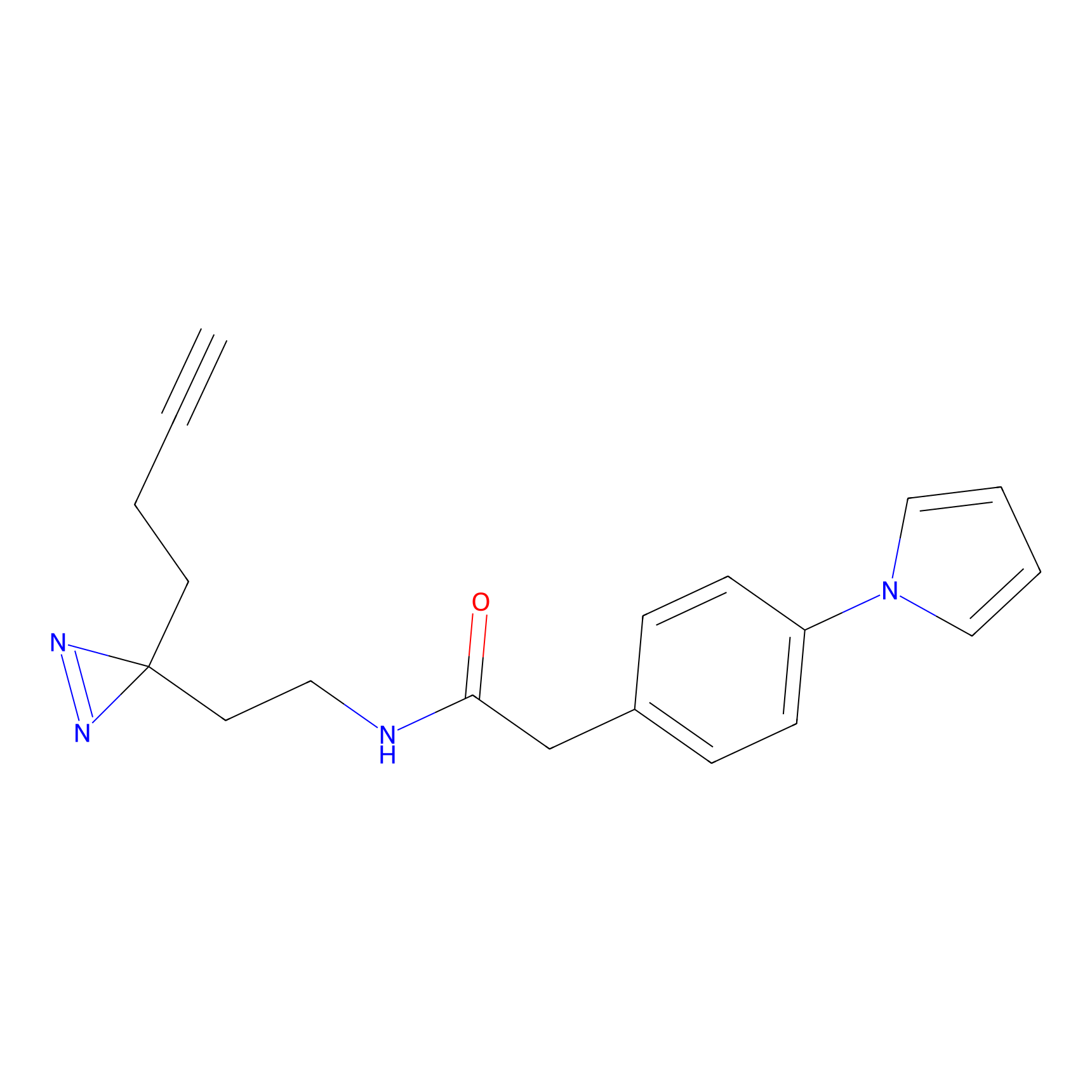
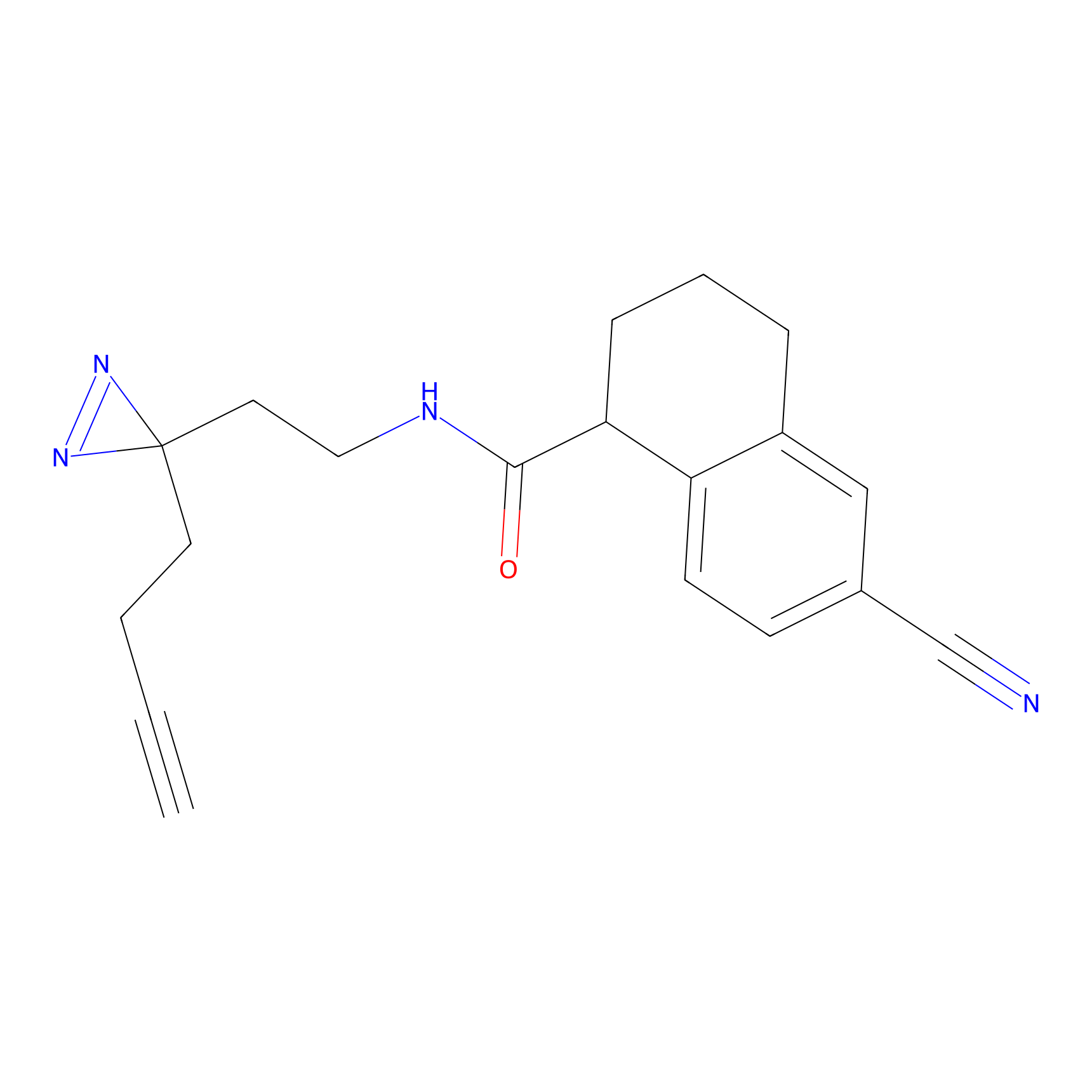
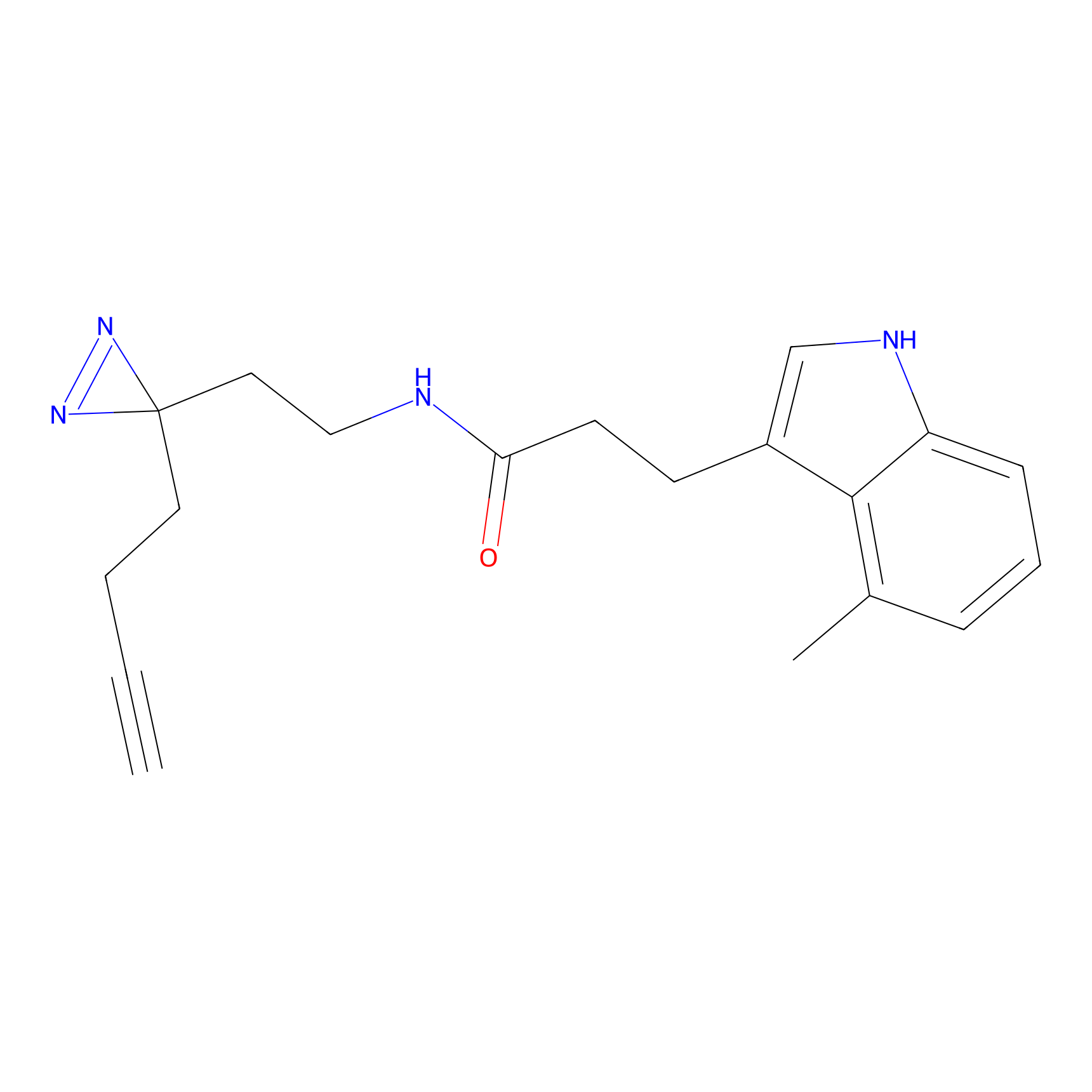
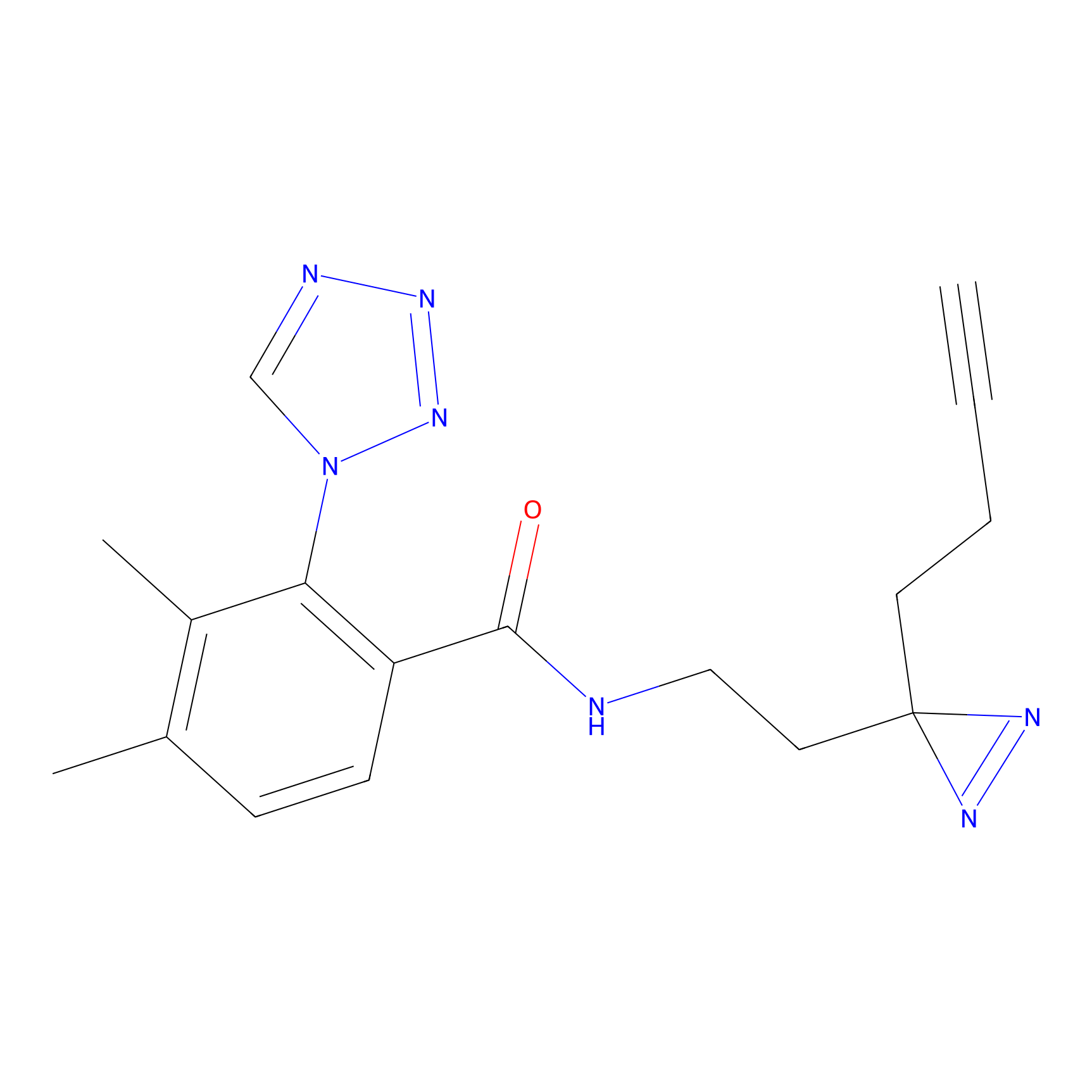
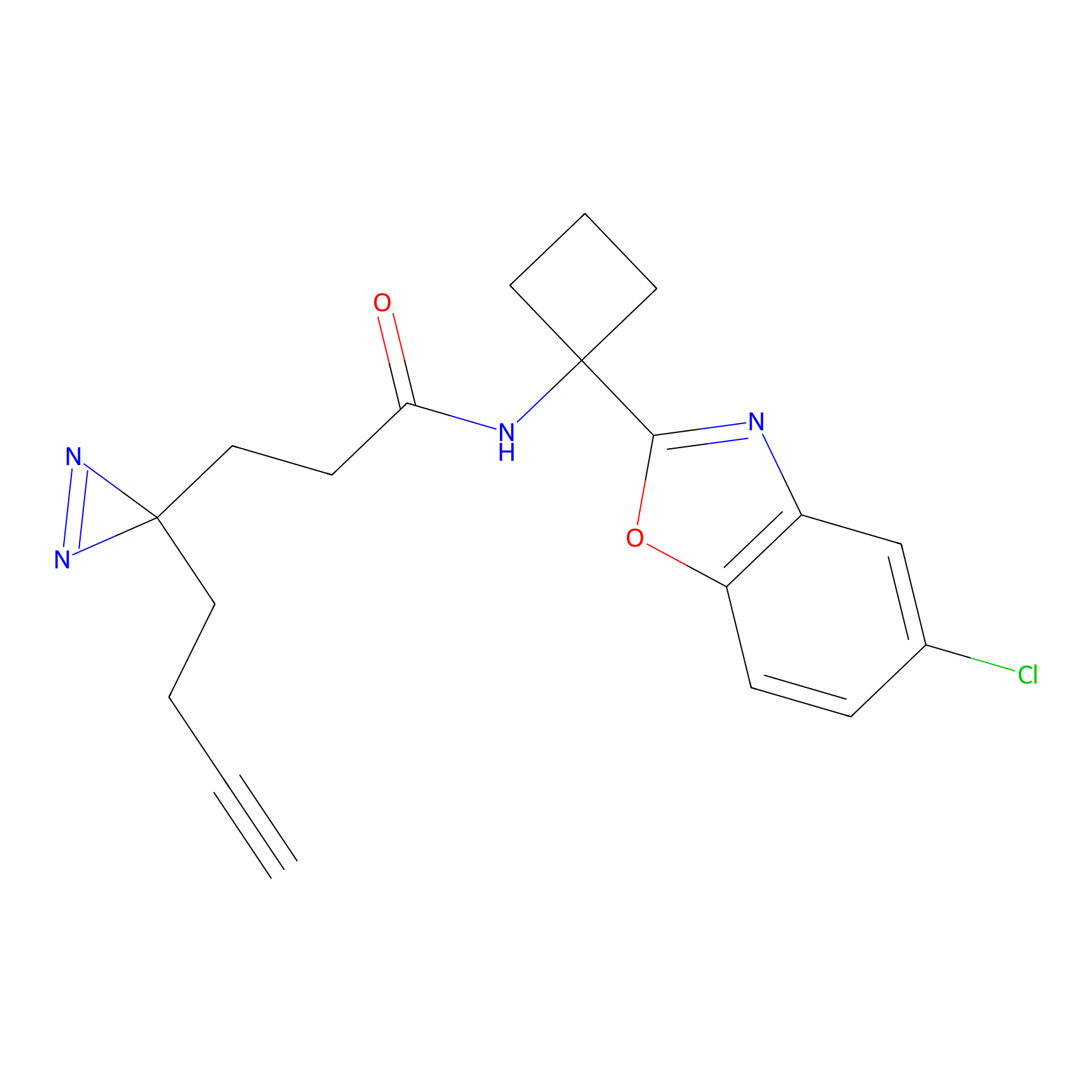
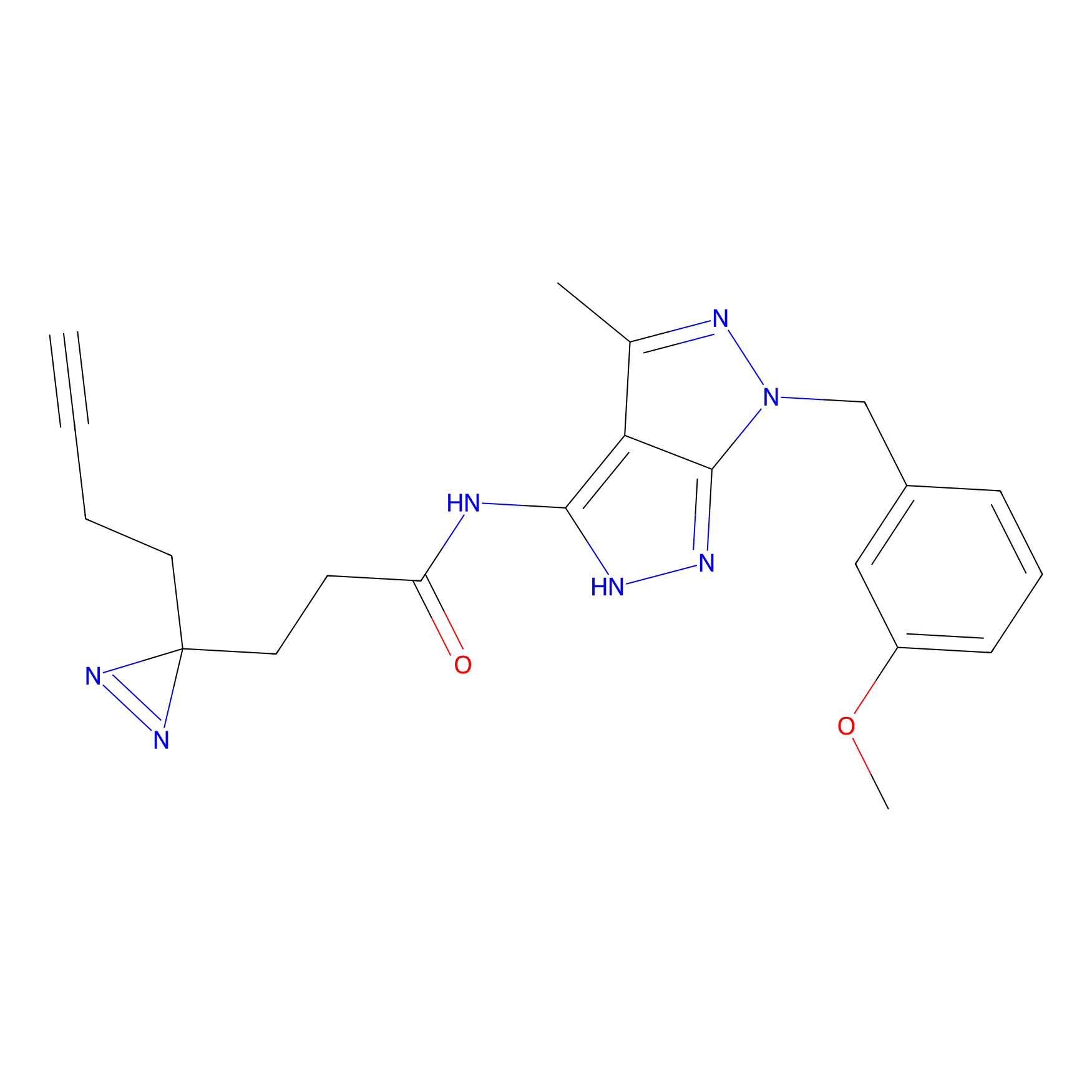
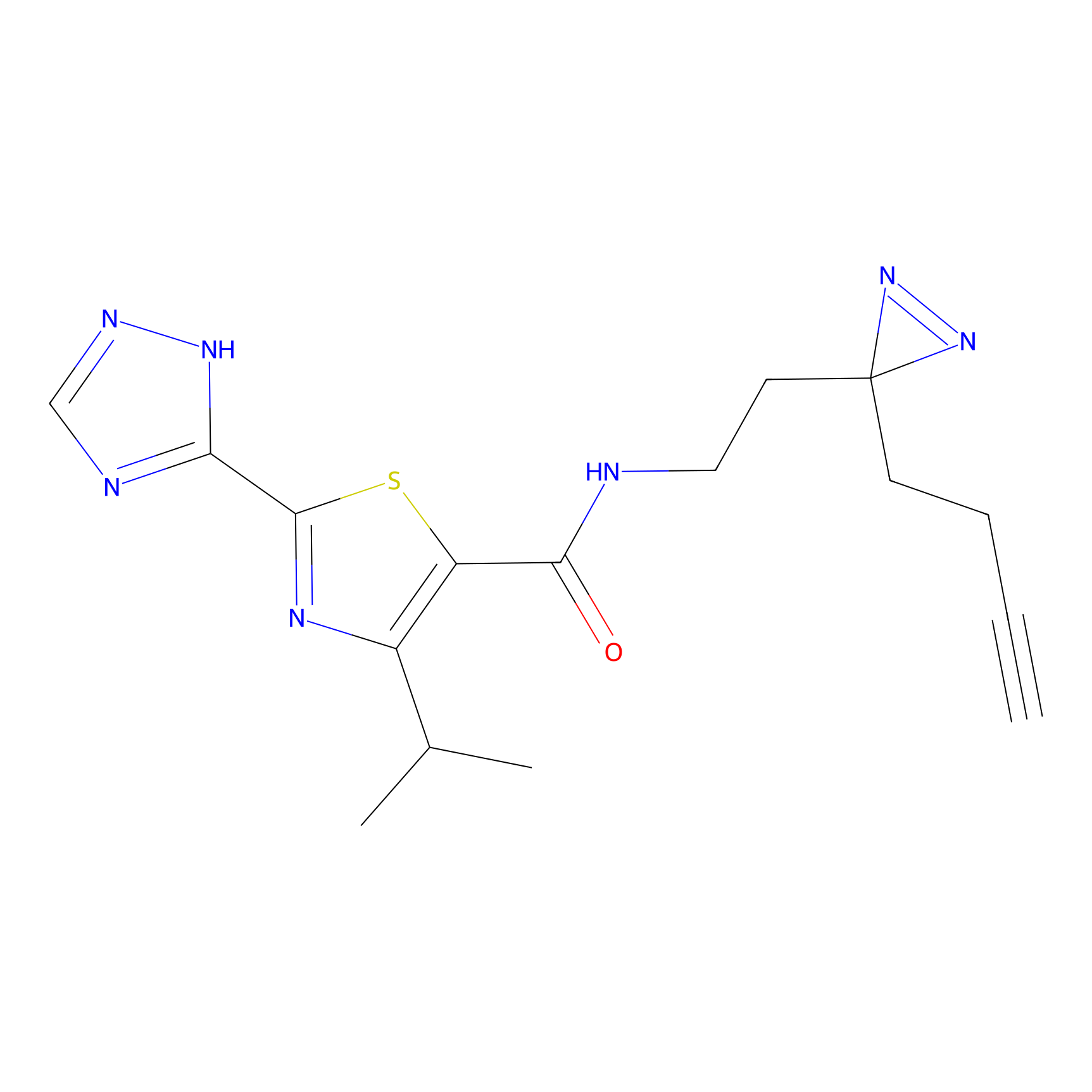
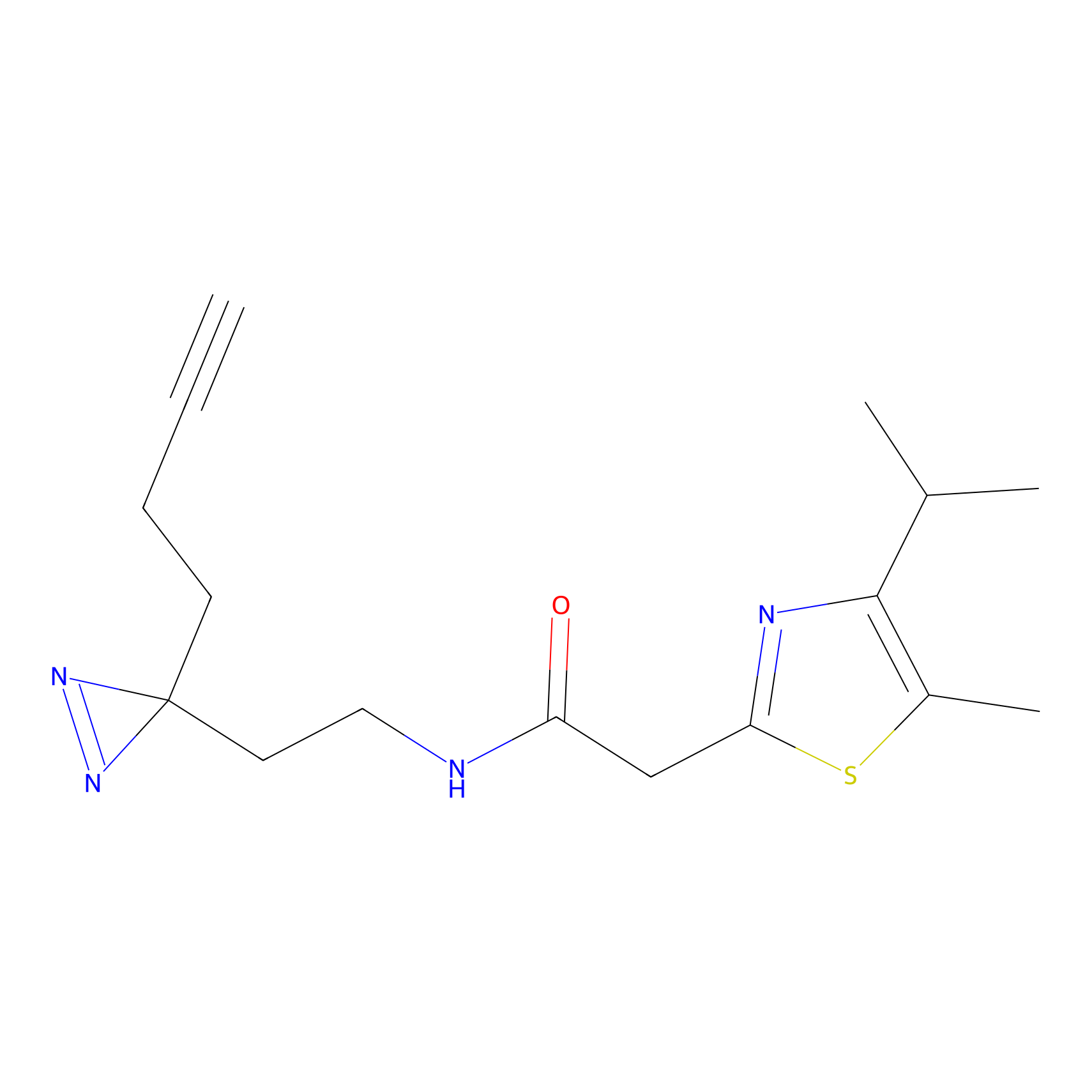
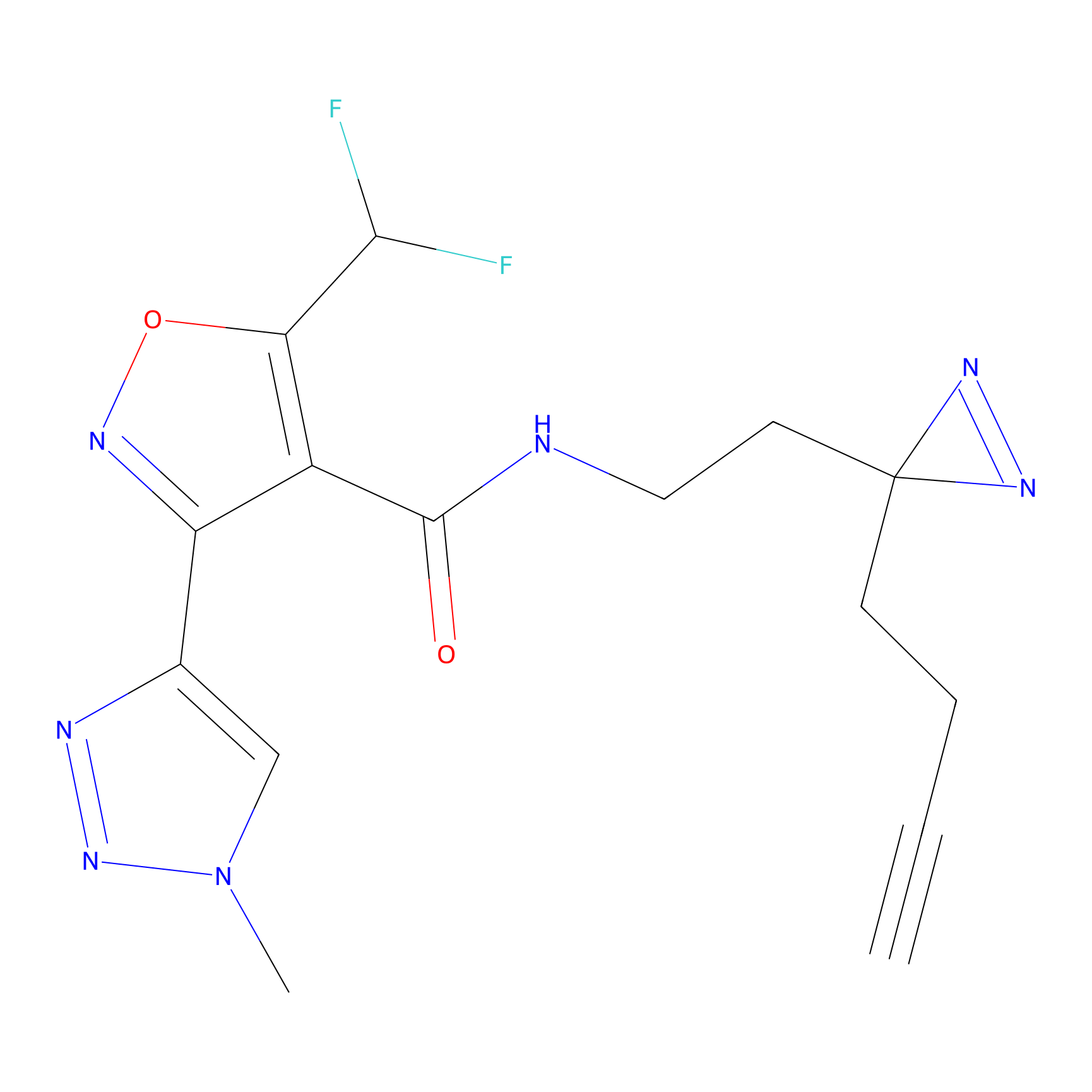
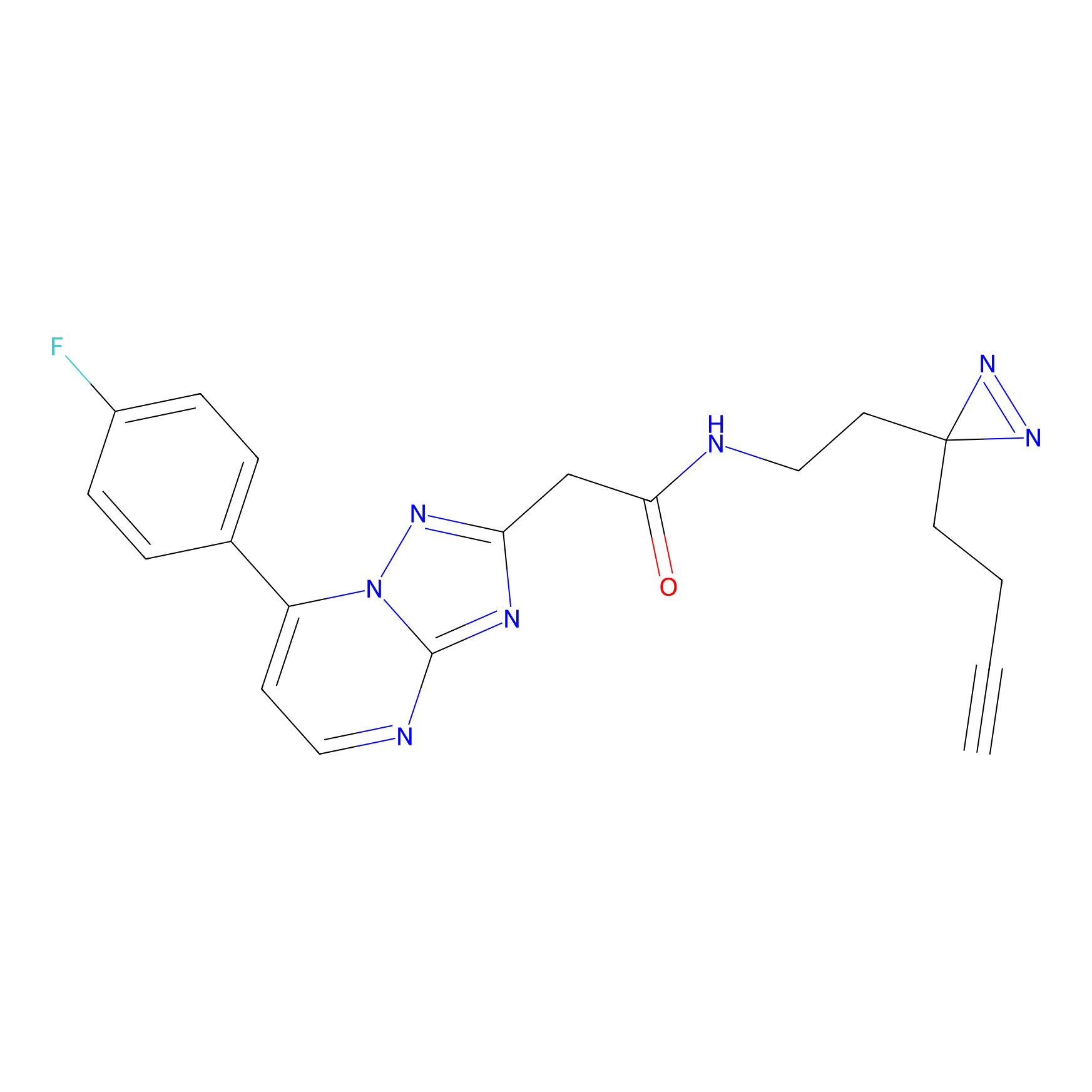
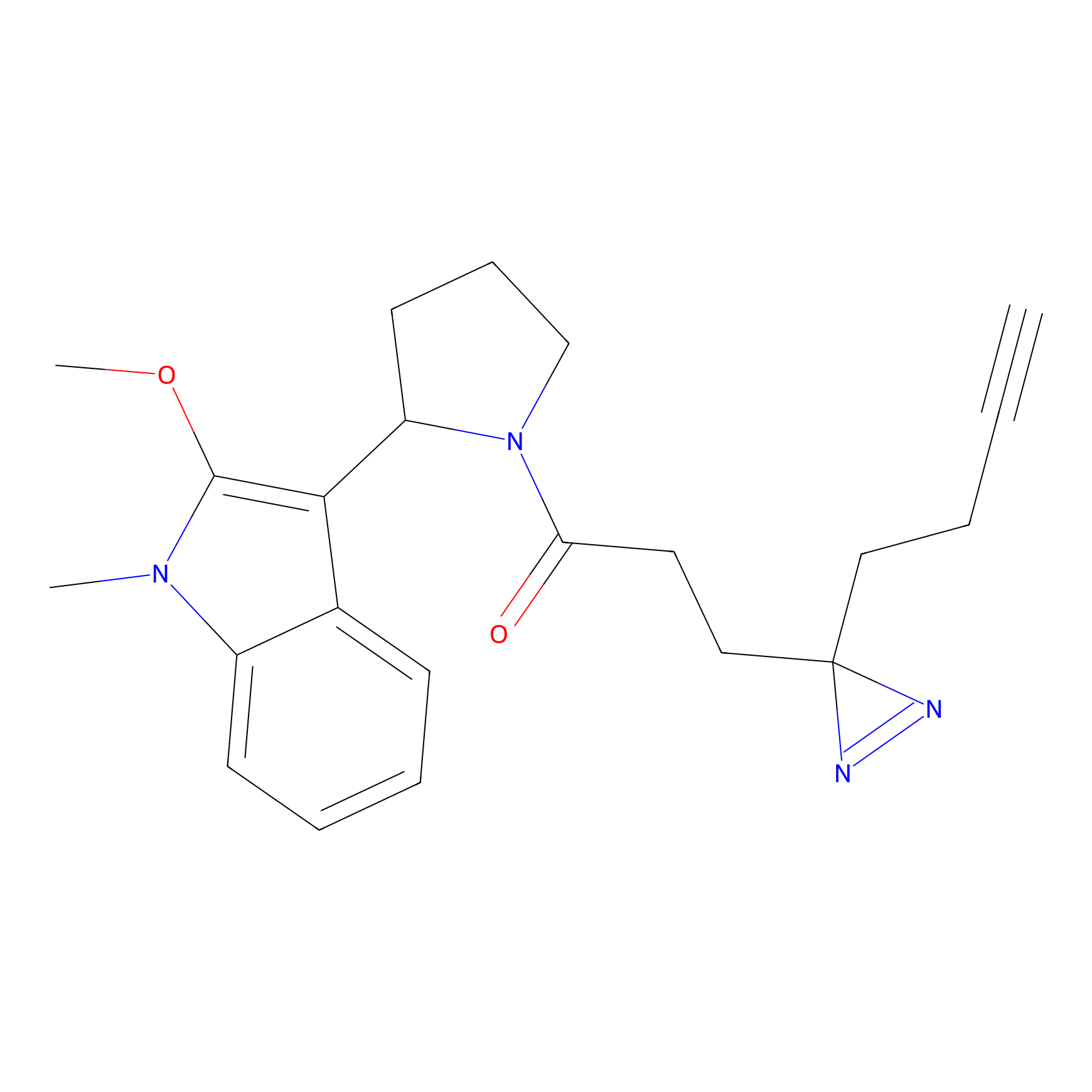
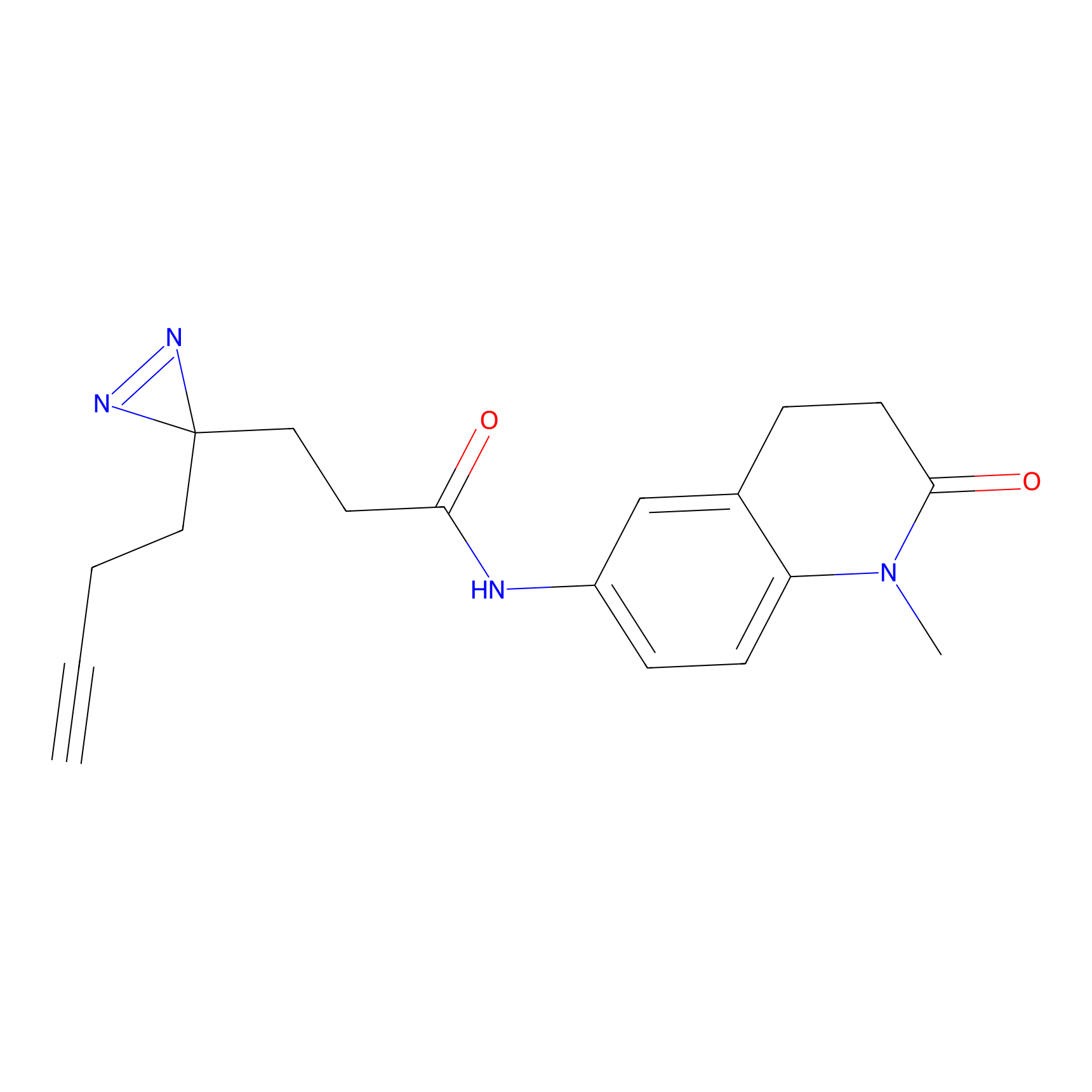
m-APA Probe Info |
 |
N.A. | LDD2231 | [6] | |
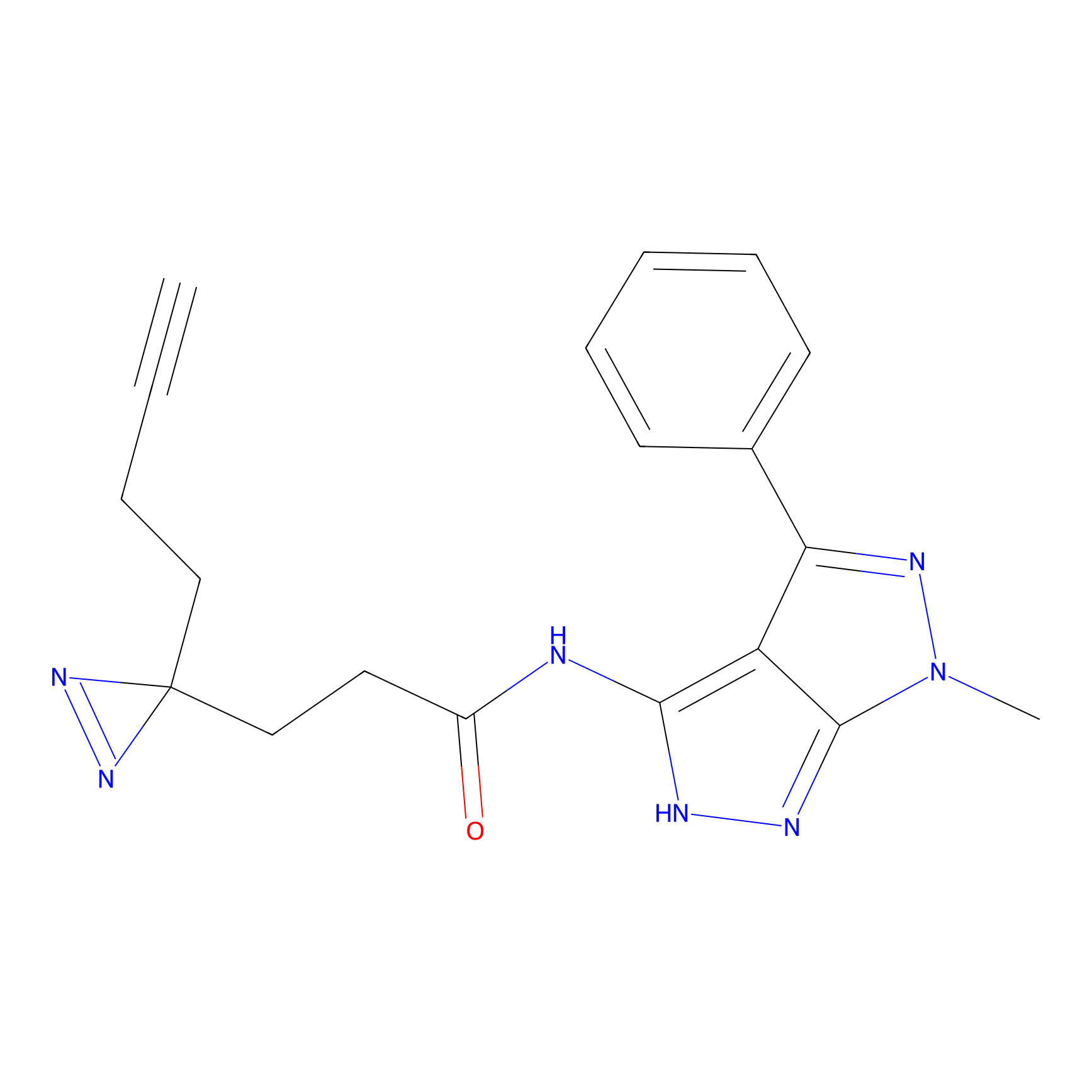
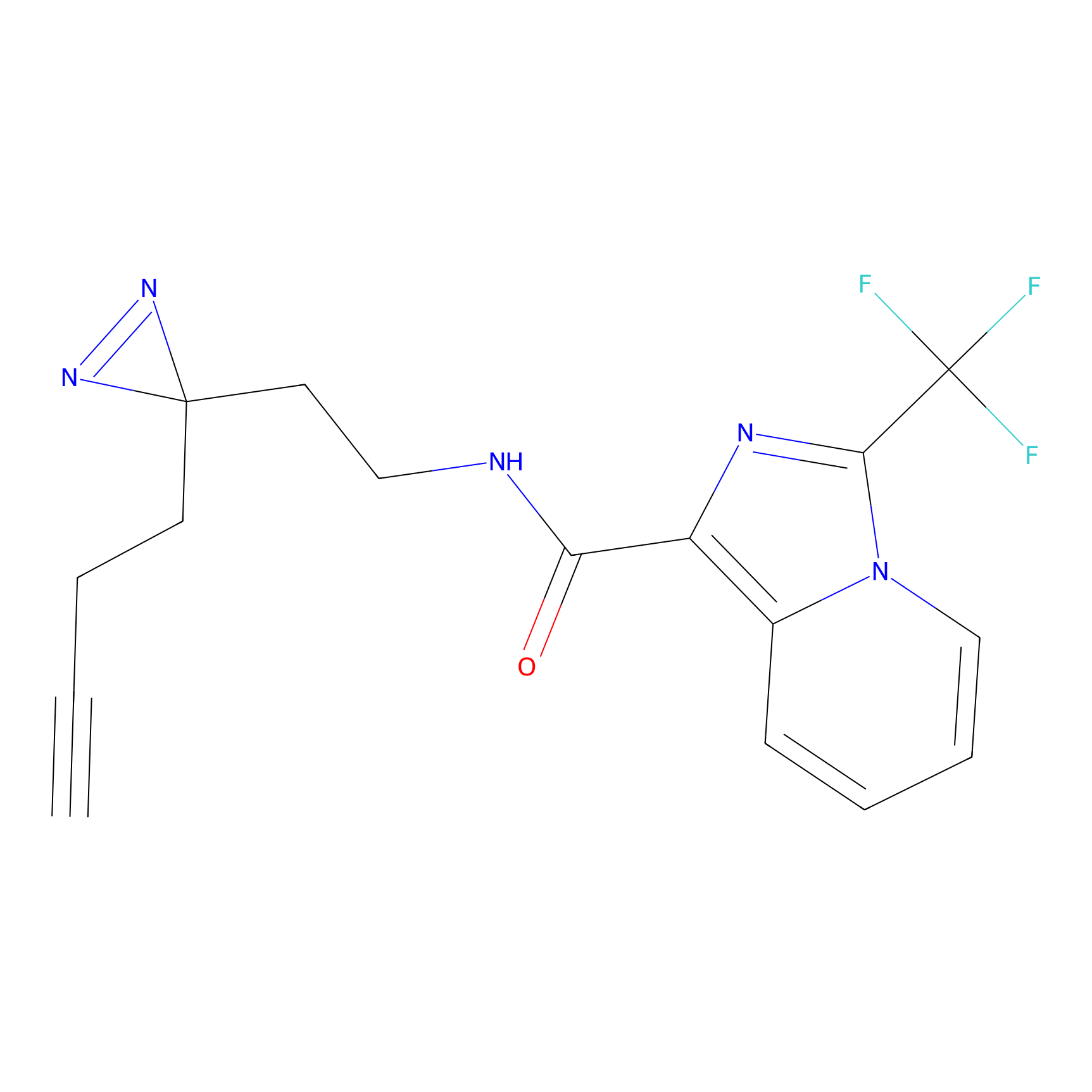
|
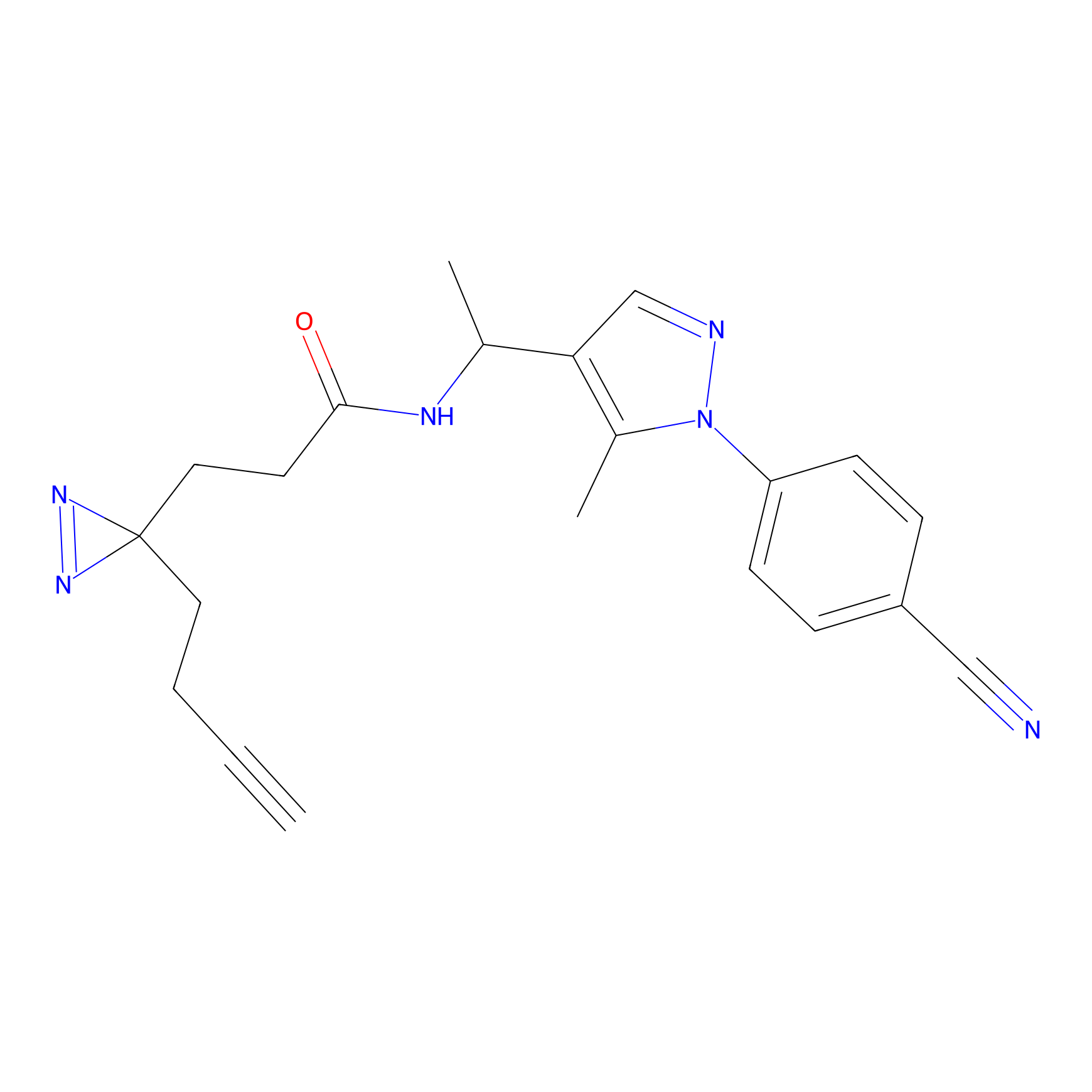
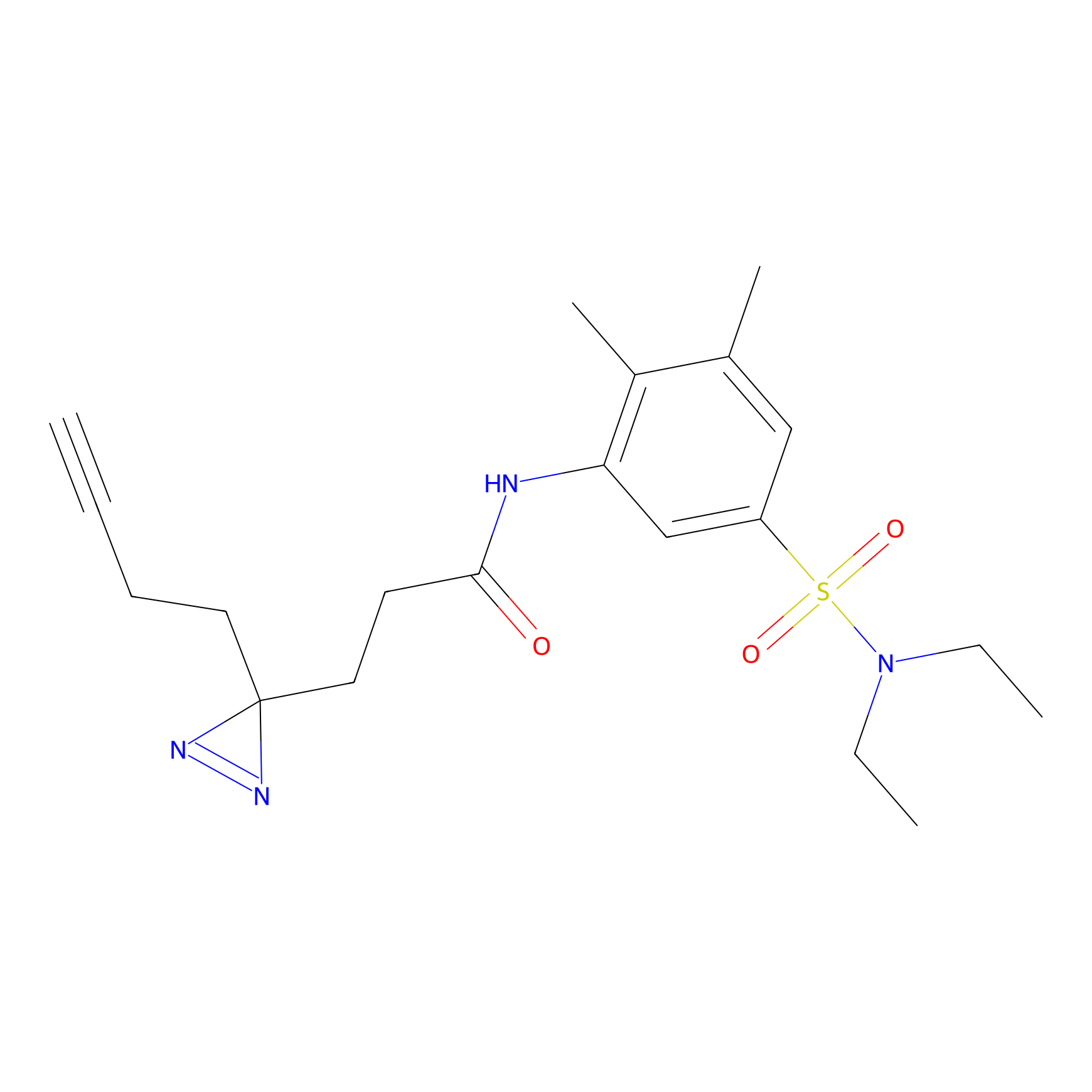
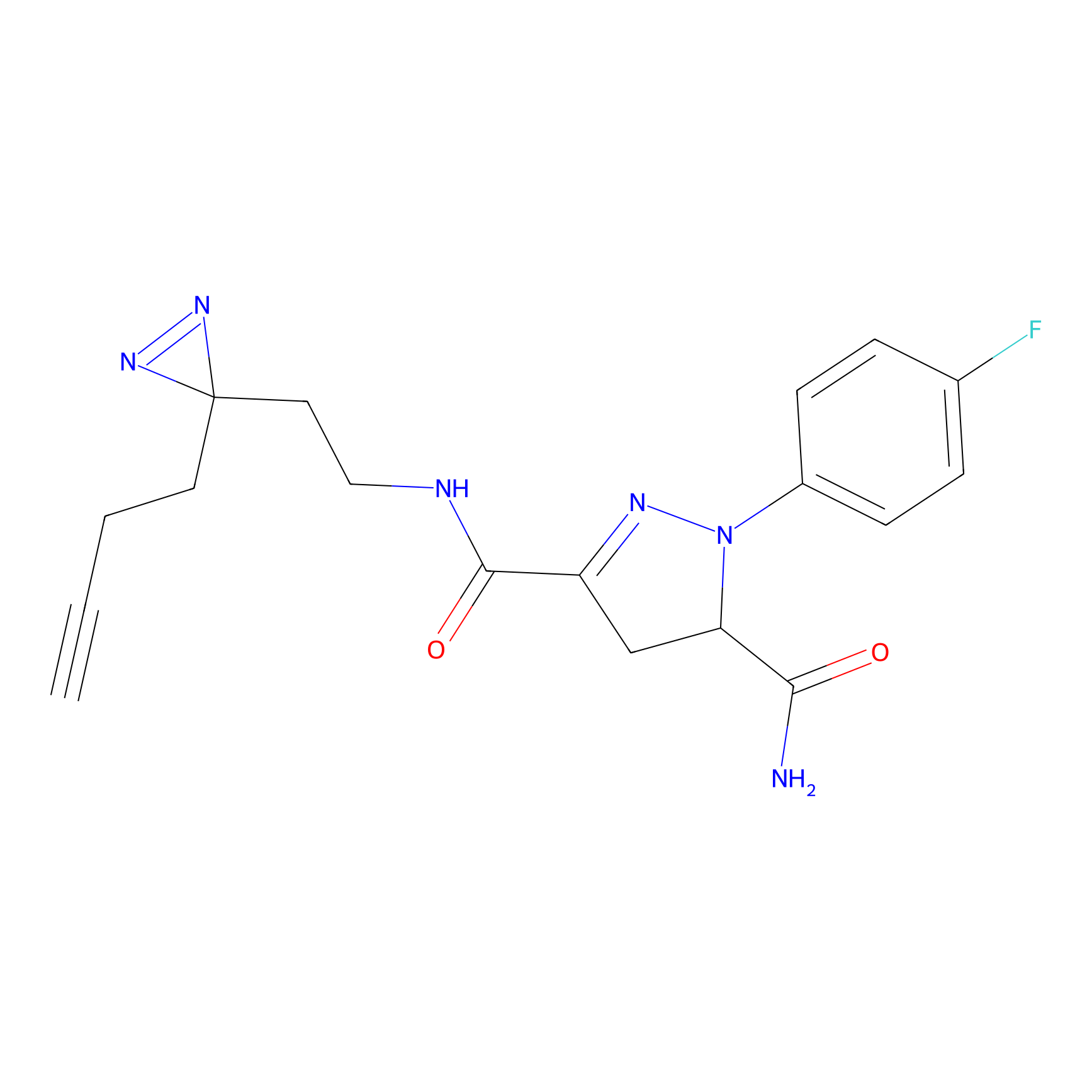
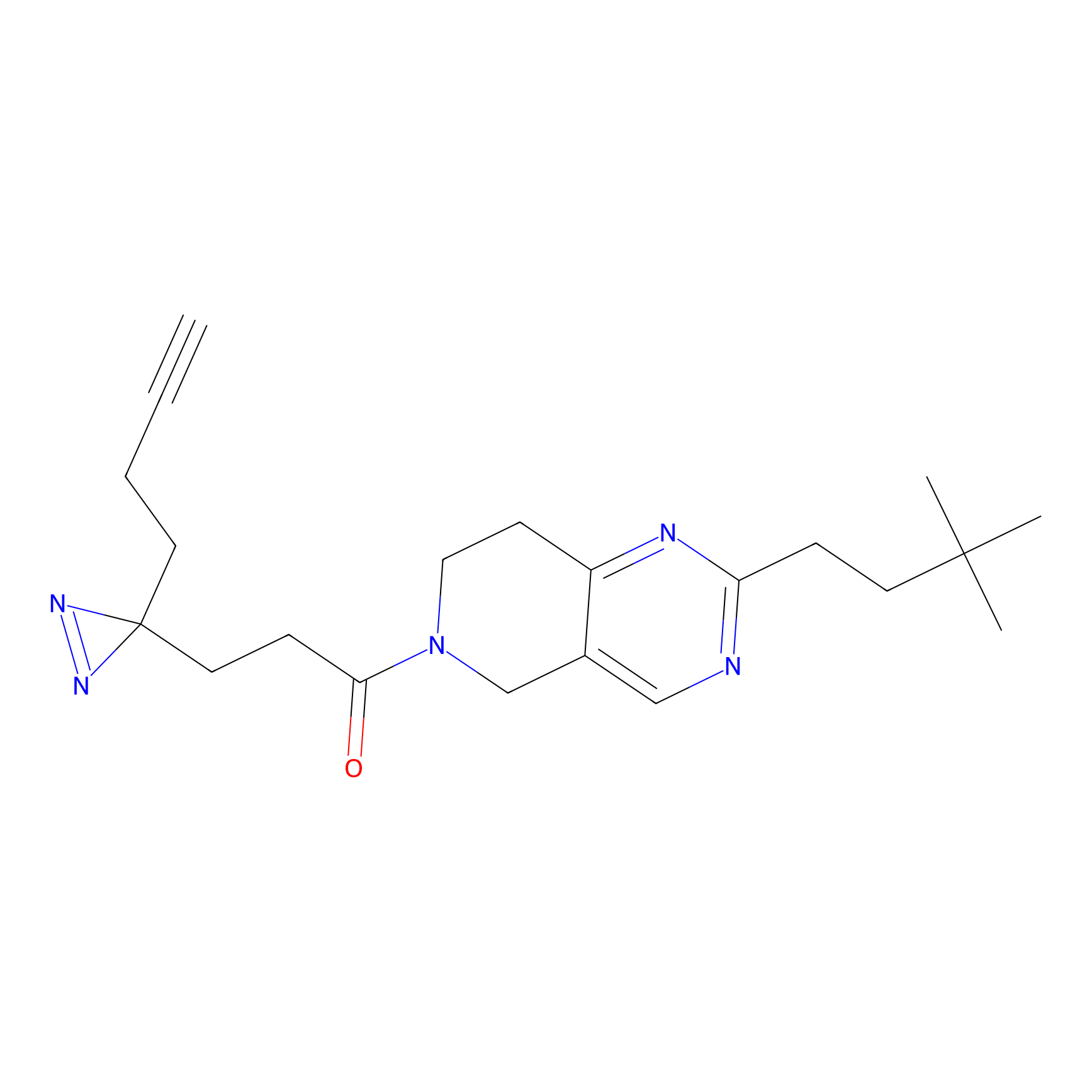
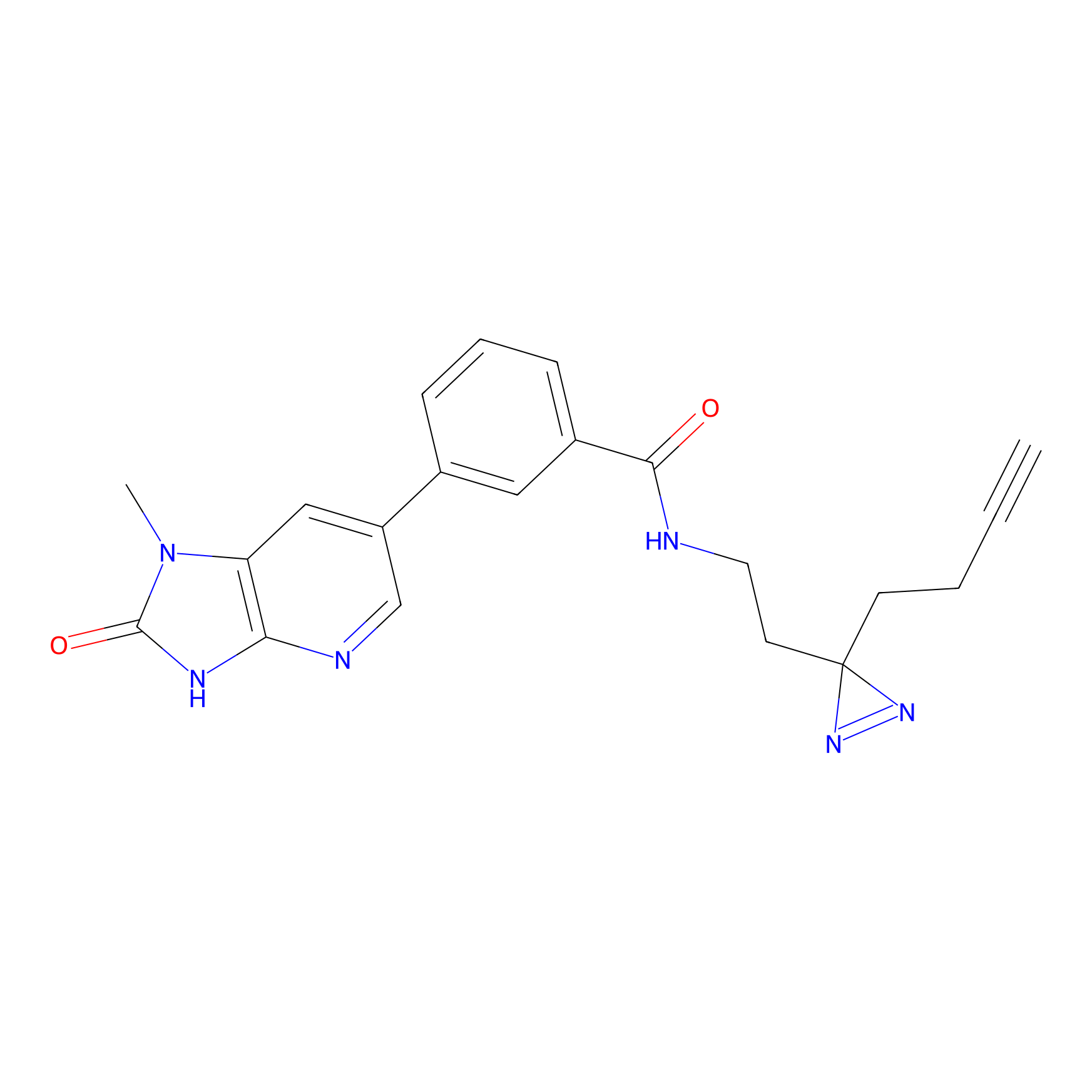
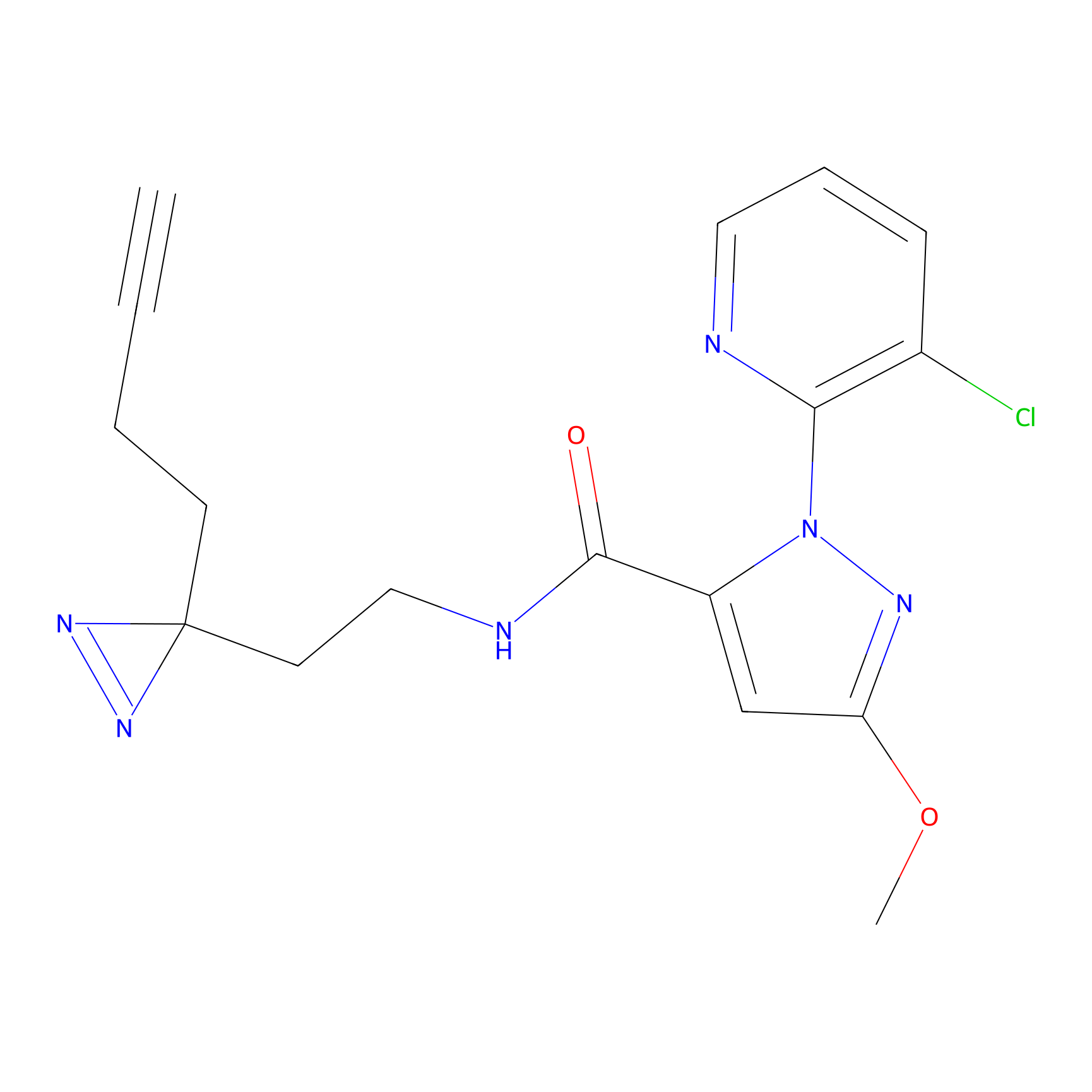
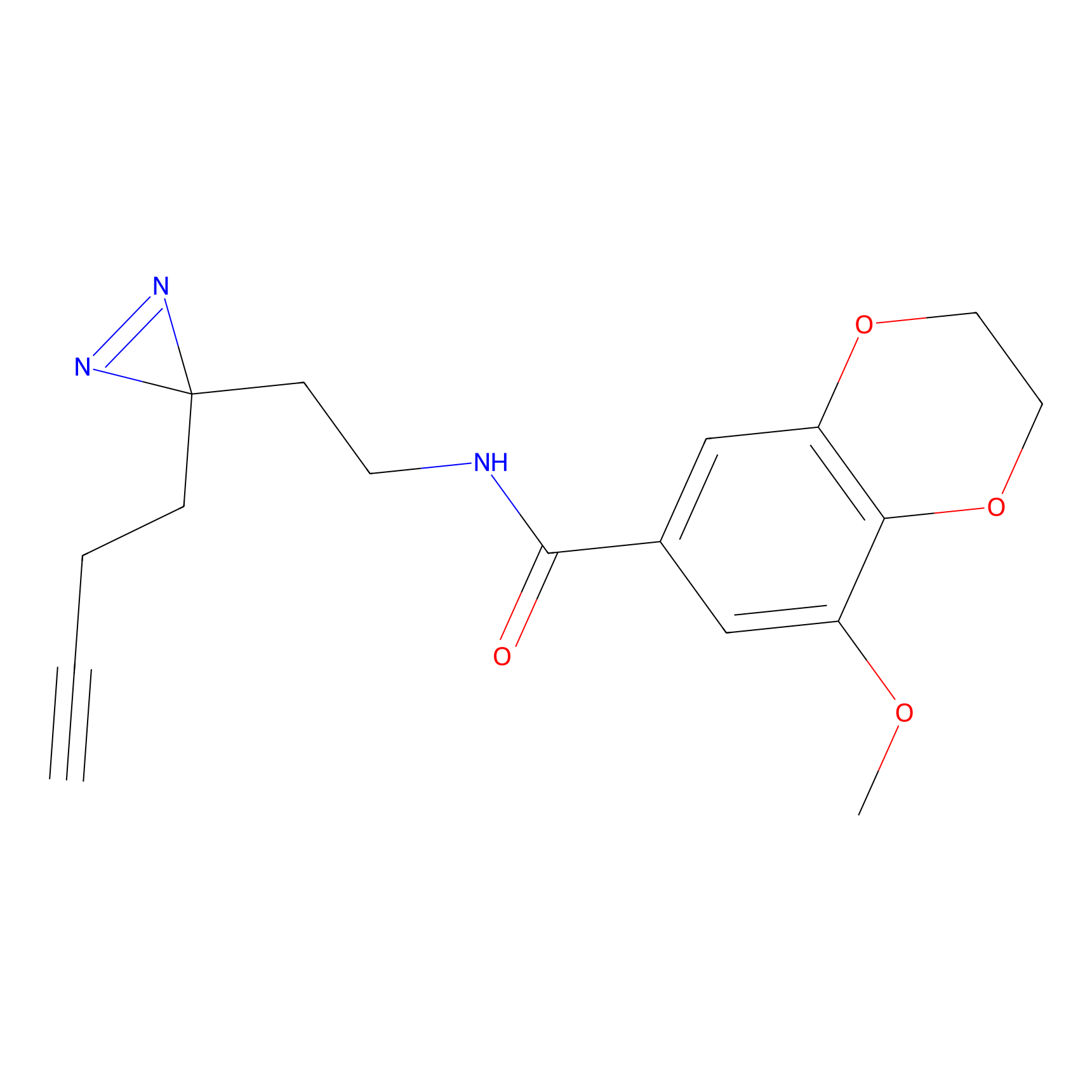
CY4 Probe Info |
 |
Q413(0.00); Q414(0.00) | LDD0247 | [7] | |
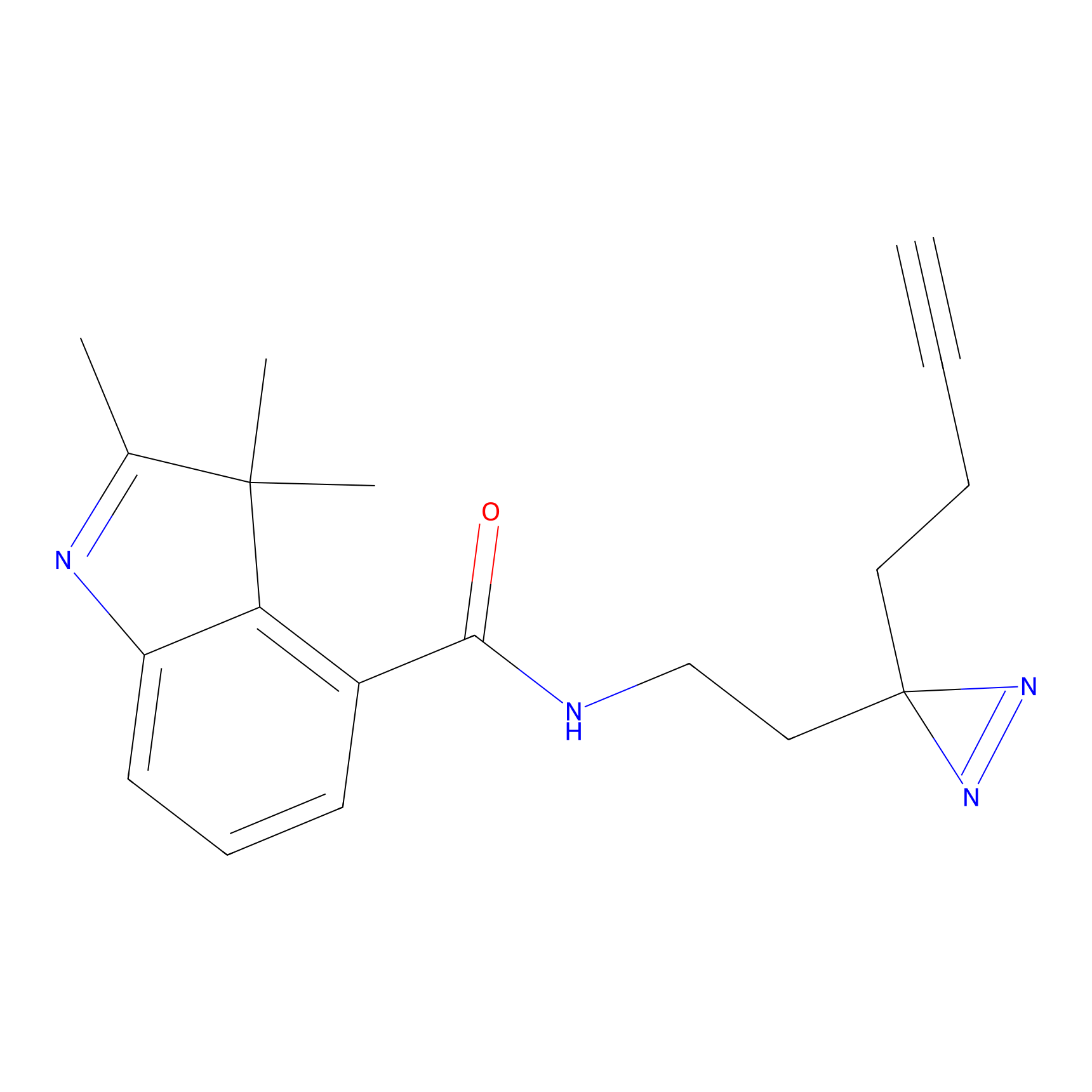
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [8] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [4] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0226 | AC11 | HEK-293T | C6(0.98) | LDD1509 | [5] |
| LDCM0278 | AC19 | HEK-293T | C6(1.19) | LDD1517 | [5] |
| LDCM0287 | AC27 | HEK-293T | C6(0.95) | LDD1526 | [5] |
| LDCM0290 | AC3 | HEK-293T | C6(0.98) | LDD1529 | [5] |
| LDCM0296 | AC35 | HEK-293T | C6(1.13) | LDD1535 | [5] |
| LDCM0305 | AC43 | HEK-293T | C6(1.06) | LDD1544 | [5] |
| LDCM0314 | AC51 | HEK-293T | C6(0.94) | LDD1553 | [5] |
| LDCM0322 | AC59 | HEK-293T | C6(1.00) | LDD1561 | [5] |
| LDCM0108 | Chloroacetamide | HeLa | H435(0.00); H339(0.00); H430(0.00) | LDD0222 | [4] |
| LDCM0406 | CL19 | HEK-293T | C6(1.03) | LDD1610 | [5] |
| LDCM0420 | CL31 | HEK-293T | C6(1.05) | LDD1624 | [5] |
| LDCM0433 | CL43 | HEK-293T | C6(1.06) | LDD1637 | [5] |
| LDCM0446 | CL55 | HEK-293T | C6(1.09) | LDD1649 | [5] |
| LDCM0459 | CL67 | HEK-293T | C6(1.02) | LDD1662 | [5] |
| LDCM0462 | CL7 | HEK-293T | C6(1.09) | LDD1665 | [5] |
| LDCM0472 | CL79 | HEK-293T | C6(1.01) | LDD1675 | [5] |
| LDCM0486 | CL91 | HEK-293T | C6(1.03) | LDD1689 | [5] |
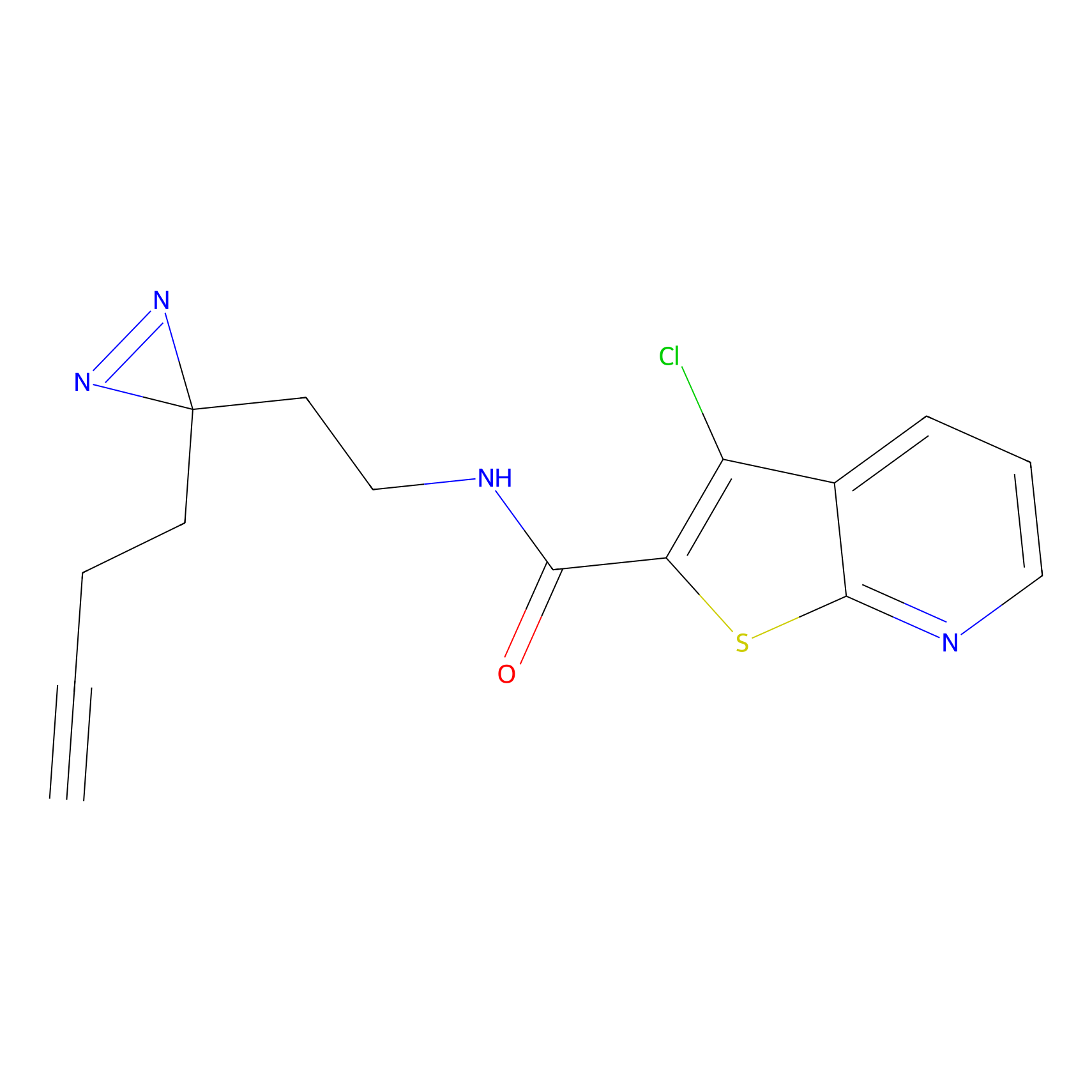
| LDCM0189 | Compound 16 | HEK-293T | 33.33 | LDD0492 | [10] |
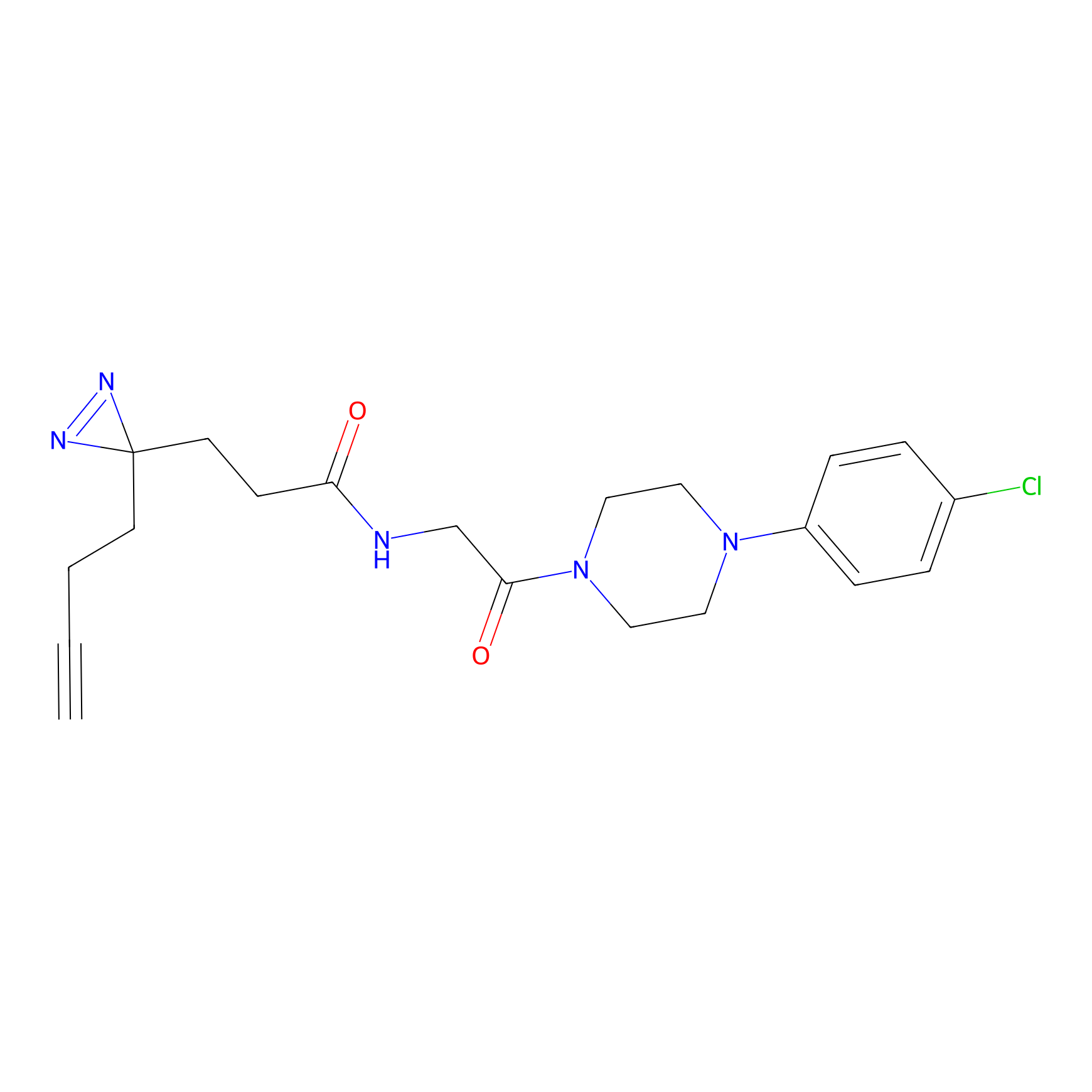
| LDCM0185 | Compound 17 | HEK-293T | 3.55 | LDD0512 | [10] |
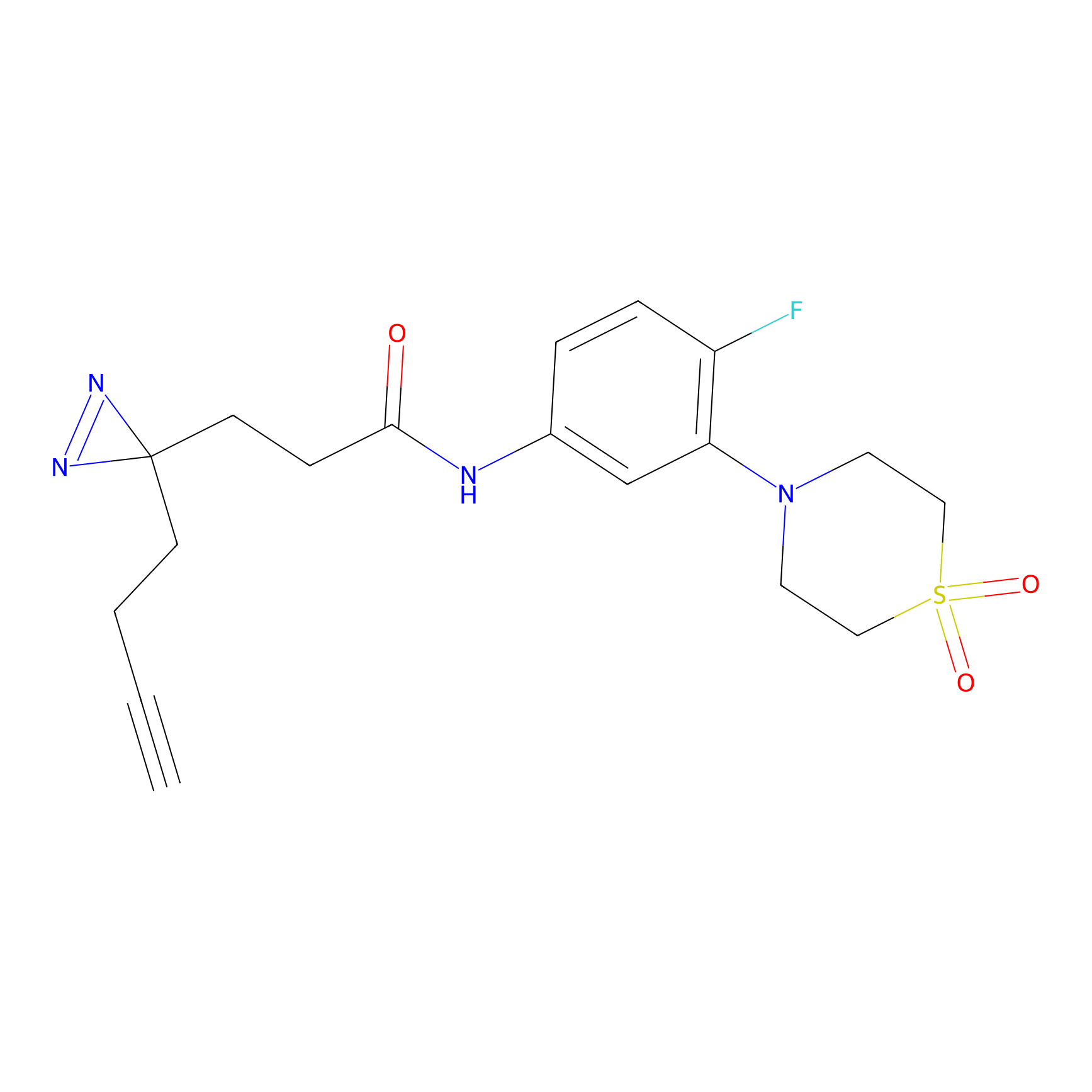
| LDCM0184 | Compound 20 | HEK-293T | 11.44 | LDD0503 | [10] |
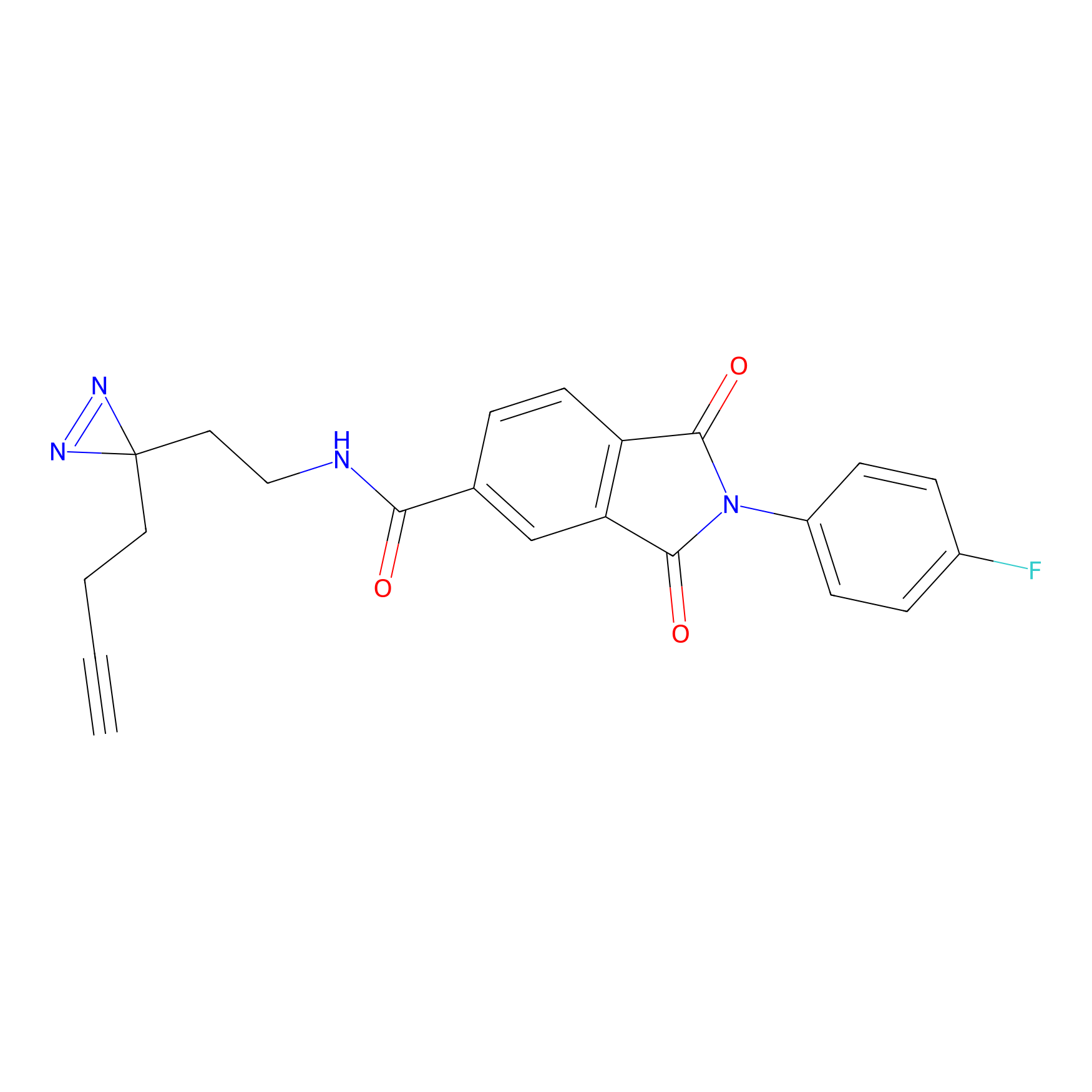
| LDCM0191 | Compound 21 | HEK-293T | 5.97 | LDD0493 | [10] |
| LDCM0190 | Compound 34 | HEK-293T | 30.30 | LDD0497 | [10] |
| LDCM0192 | Compound 35 | HEK-293T | 9.26 | LDD0491 | [10] |
| LDCM0193 | Compound 36 | HEK-293T | 13.71 | LDD0511 | [10] |
| LDCM0194 | Compound 37 | HEK-293T | 6.13 | LDD0498 | [10] |
| LDCM0195 | Compound 38 | HEK-293T | 15.50 | LDD0499 | [10] |
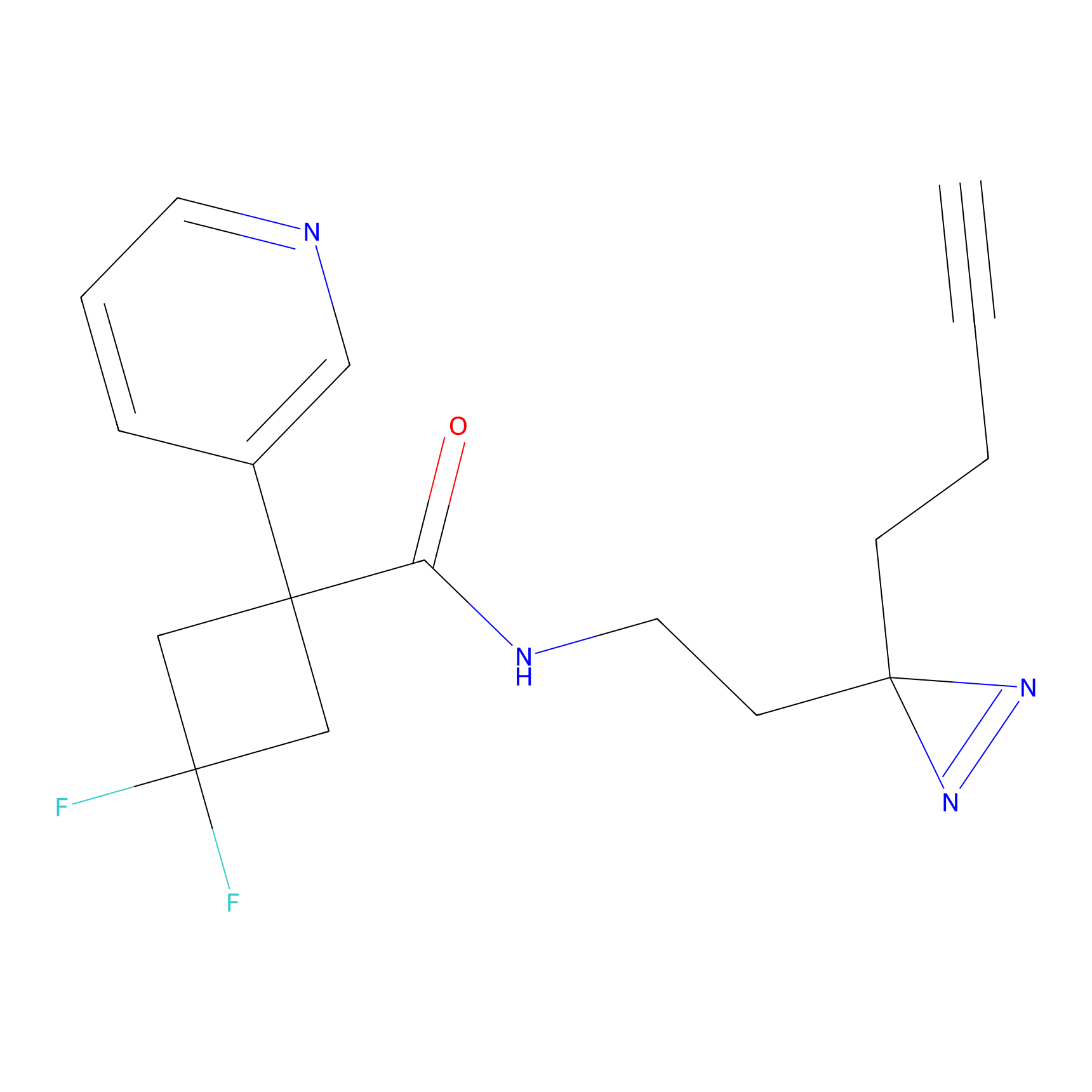
| LDCM0197 | Compound 40 | HEK-293T | 14.29 | LDD0495 | [10] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C6(1.79) | LDD1702 | [3] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [4] |
| LDCM0022 | KB02 | ICC18 | C6(1.95) | LDD2385 | [12] |
| LDCM0023 | KB03 | ICC3 | C6(7.20) | LDD2808 | [12] |
| LDCM0024 | KB05 | ICC26 | C6(1.96) | LDD3224 | [12] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0224 | [4] |
References