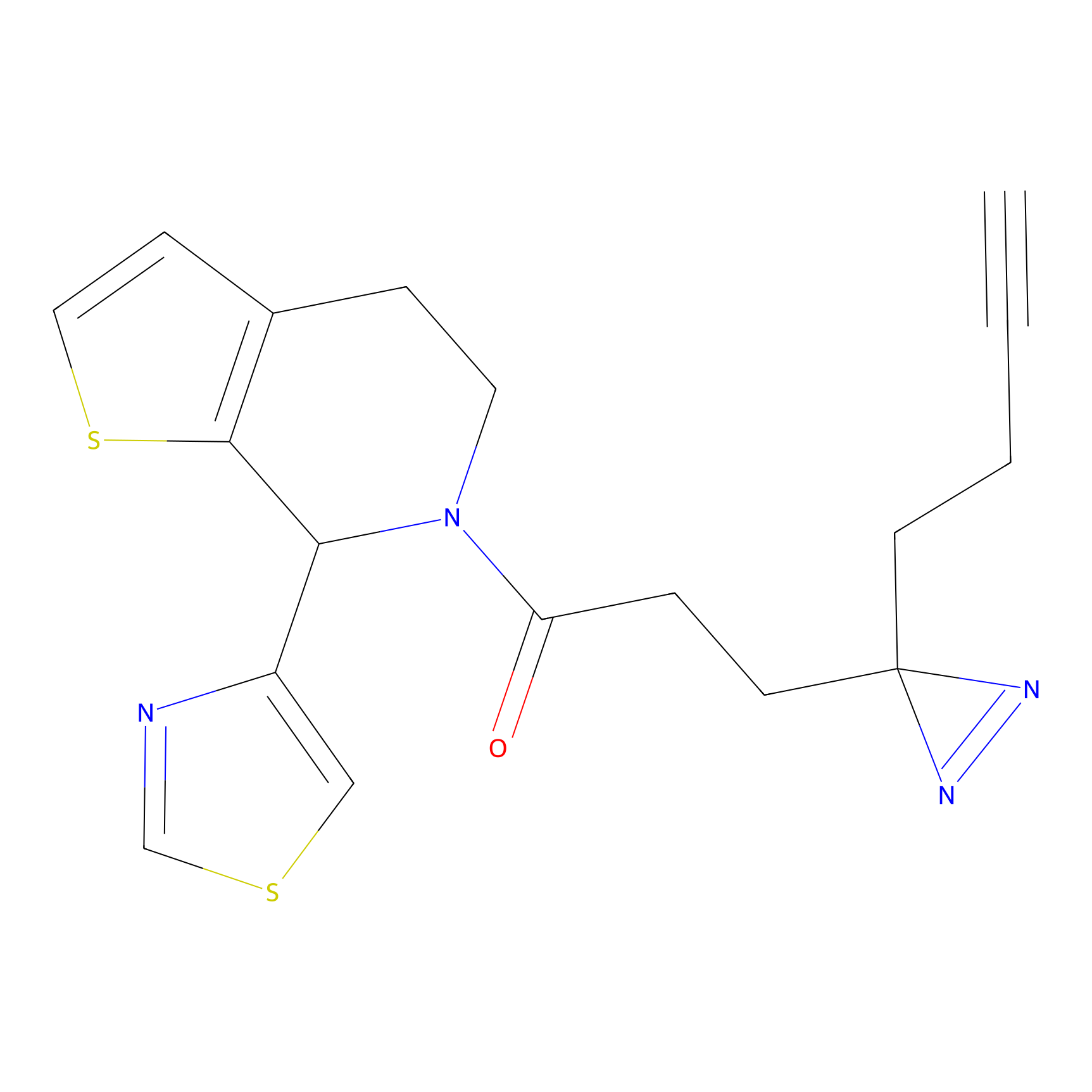
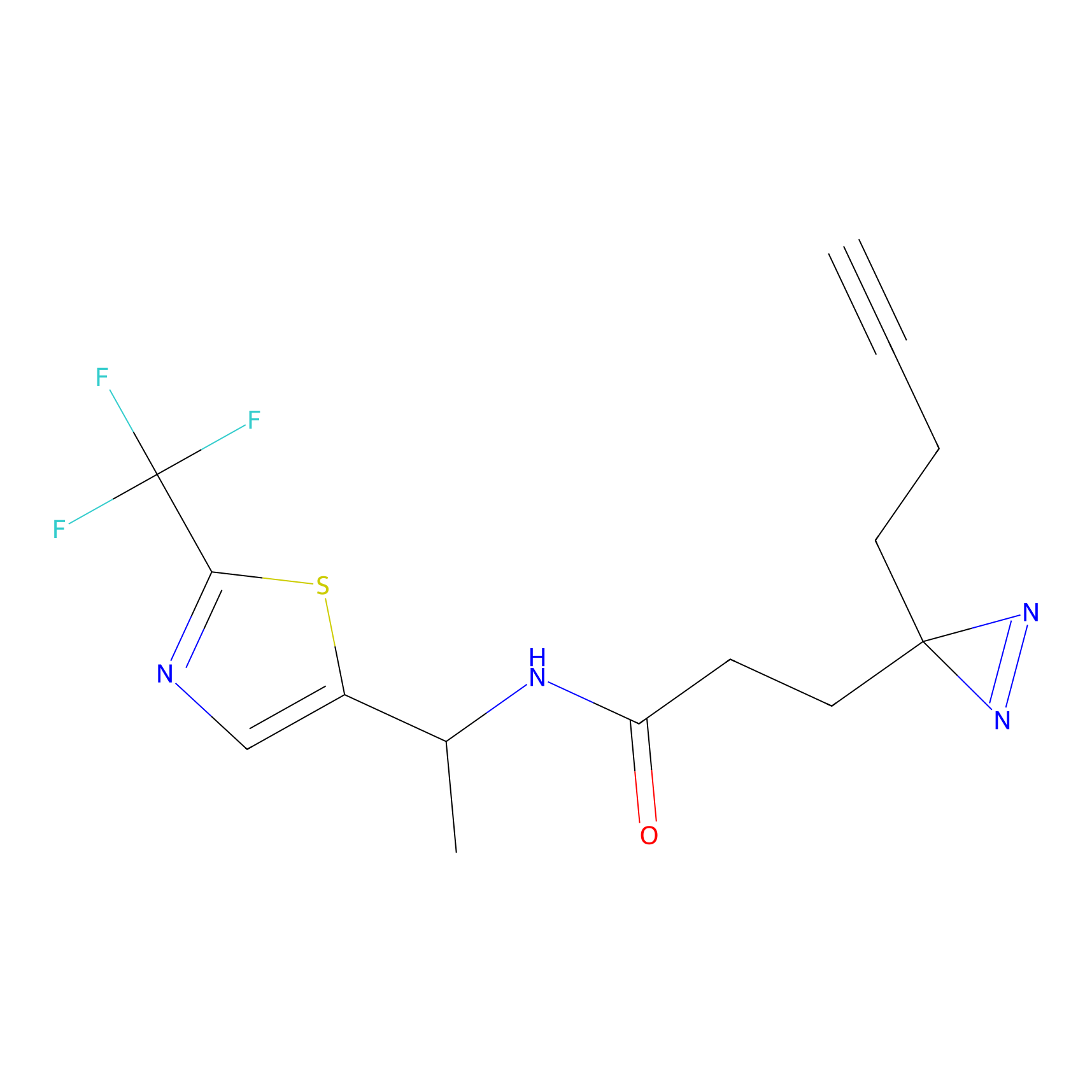
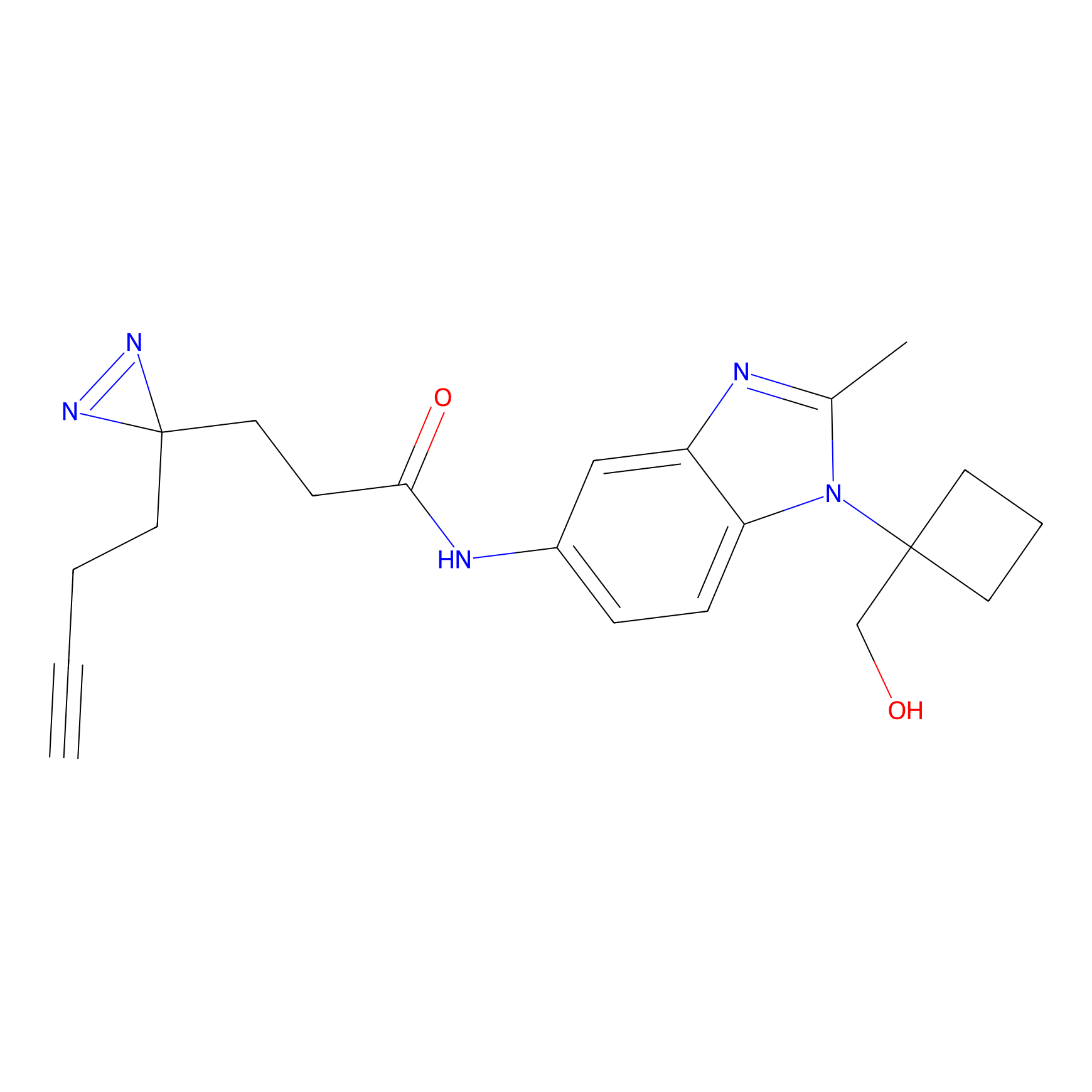
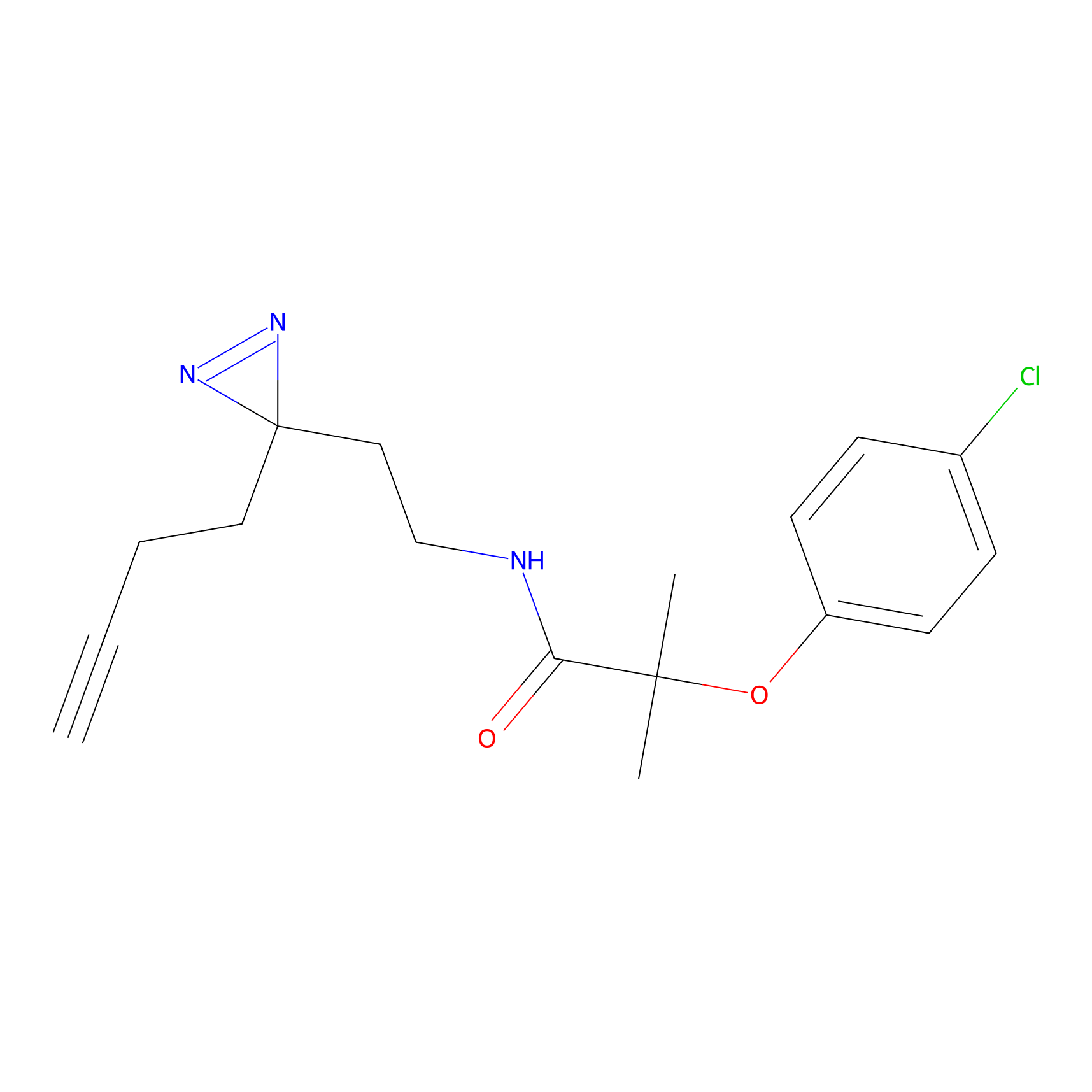
Details of the Target
General Information of Target
| Target ID | LDTP10056 | |||||
|---|---|---|---|---|---|---|
| Target Name | Protein FAM162A (FAM162A) | |||||
| Gene Name | FAM162A | |||||
| Gene ID | 26355 | |||||
| Synonyms |
C3orf28; E2IG5; Protein FAM162A; E2-induced gene 5 protein; Growth and transformation-dependent protein; HGTD-P |
|||||
| 3D Structure | ||||||
| Sequence |
MGKTFSQLGSWREDENKSILSSKPAIGSKAVNYSSTGSSKSFCSCVPCEGTADASFVTCP
TCQGSGKIPQELEKQLVALIPYGDQRLKPKHTKLFVFLAVLICLVTSSFIVFFLFPRSVI VQPAGLNSSTVAFDEADIYLNITNILNISNGNYYPIMVTQLTLEVLHLSLVVGQVSNNLL LHIGPLASEQMFYAVATKIRDENTYKICTWLEIKVHHVLLHIQGTLTCSYLSHSEQLVFQ SYEYVDCRGNASVPHQLTPHPP |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
UPF0389 family
|
|||||
| Subcellular location |
Mitochondrion membrane
|
|||||
| Function |
Proposed to be involved in regulation of apoptosis; the exact mechanism may differ between cell types/tissues. May be involved in hypoxia-induced cell death of transformed cells implicating cytochrome C release and caspase activation (such as CASP9) and inducing mitochondrial permeability transition. May be involved in hypoxia-induced cell death of neuronal cells probably by promoting release of AIFM1 from mitochondria to cytoplasm and its translocation to the nucleus; however, the involvement of caspases has been reported conflictingly.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
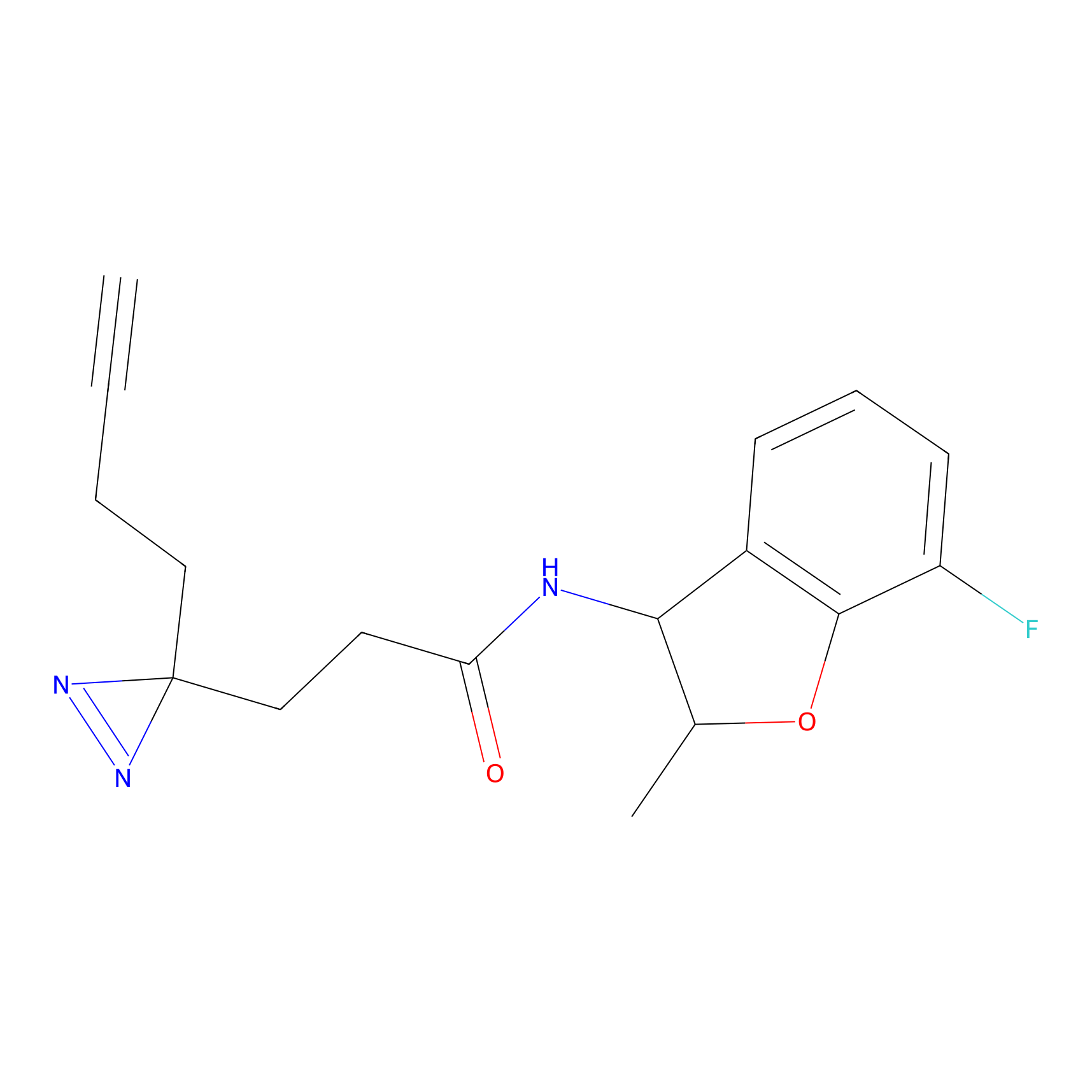
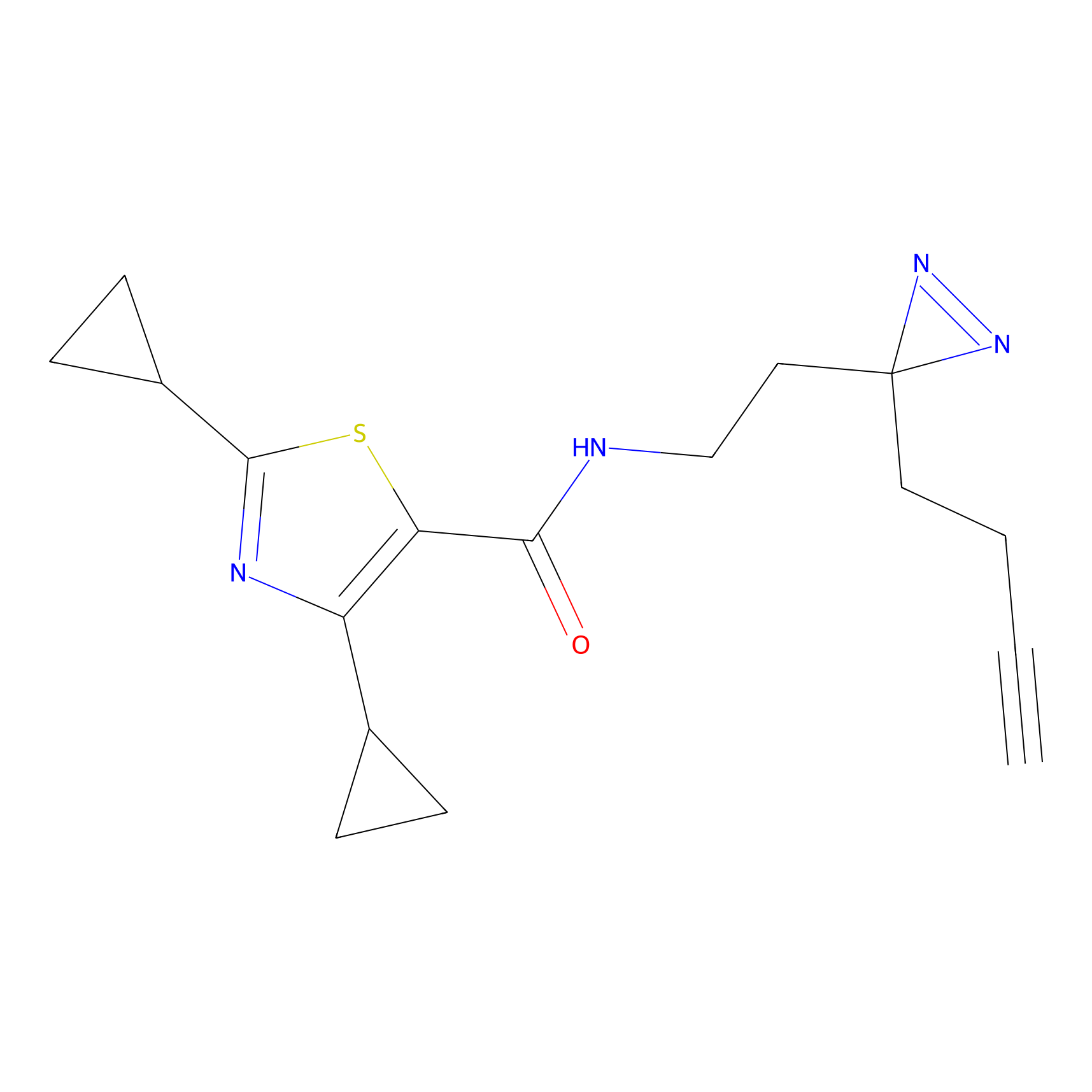
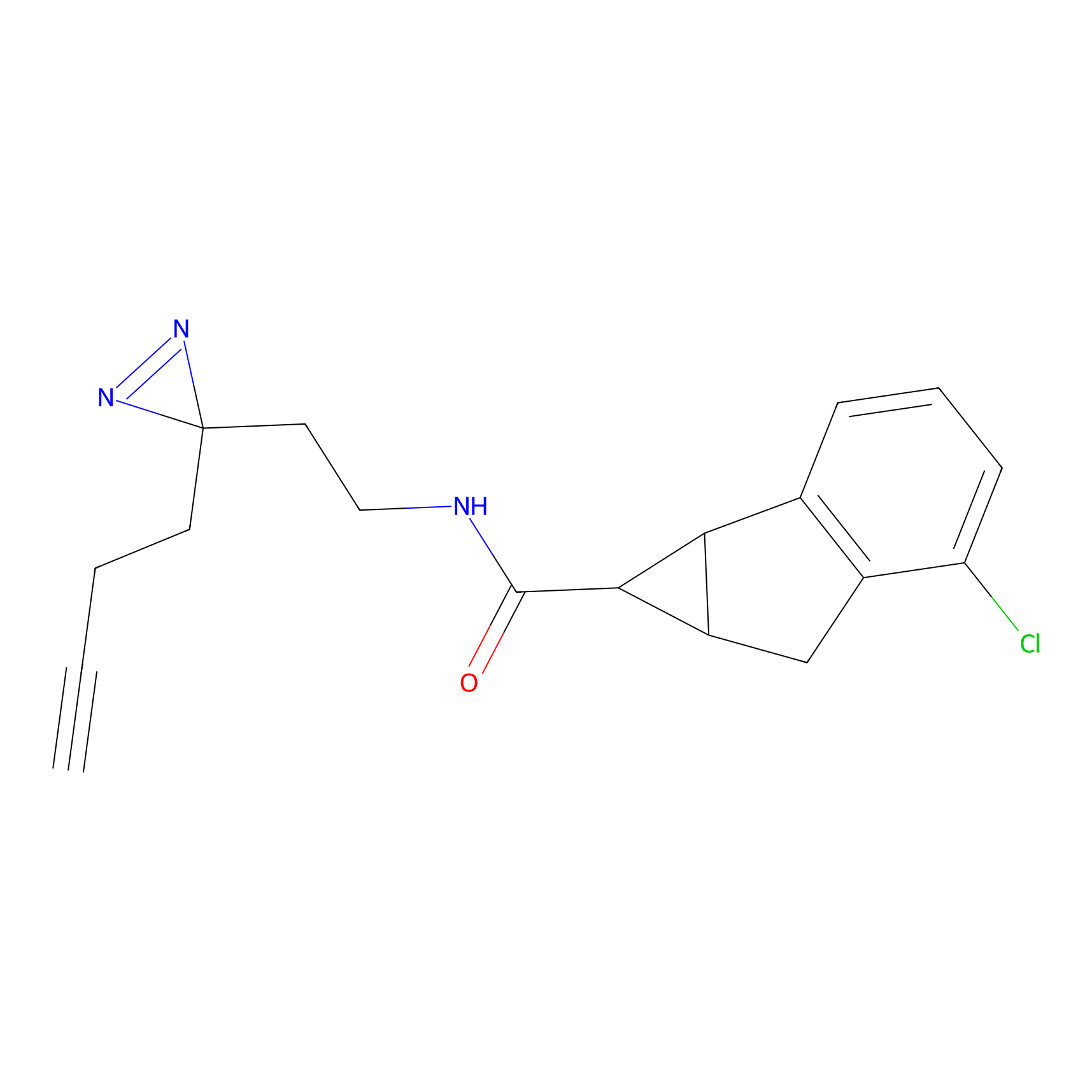
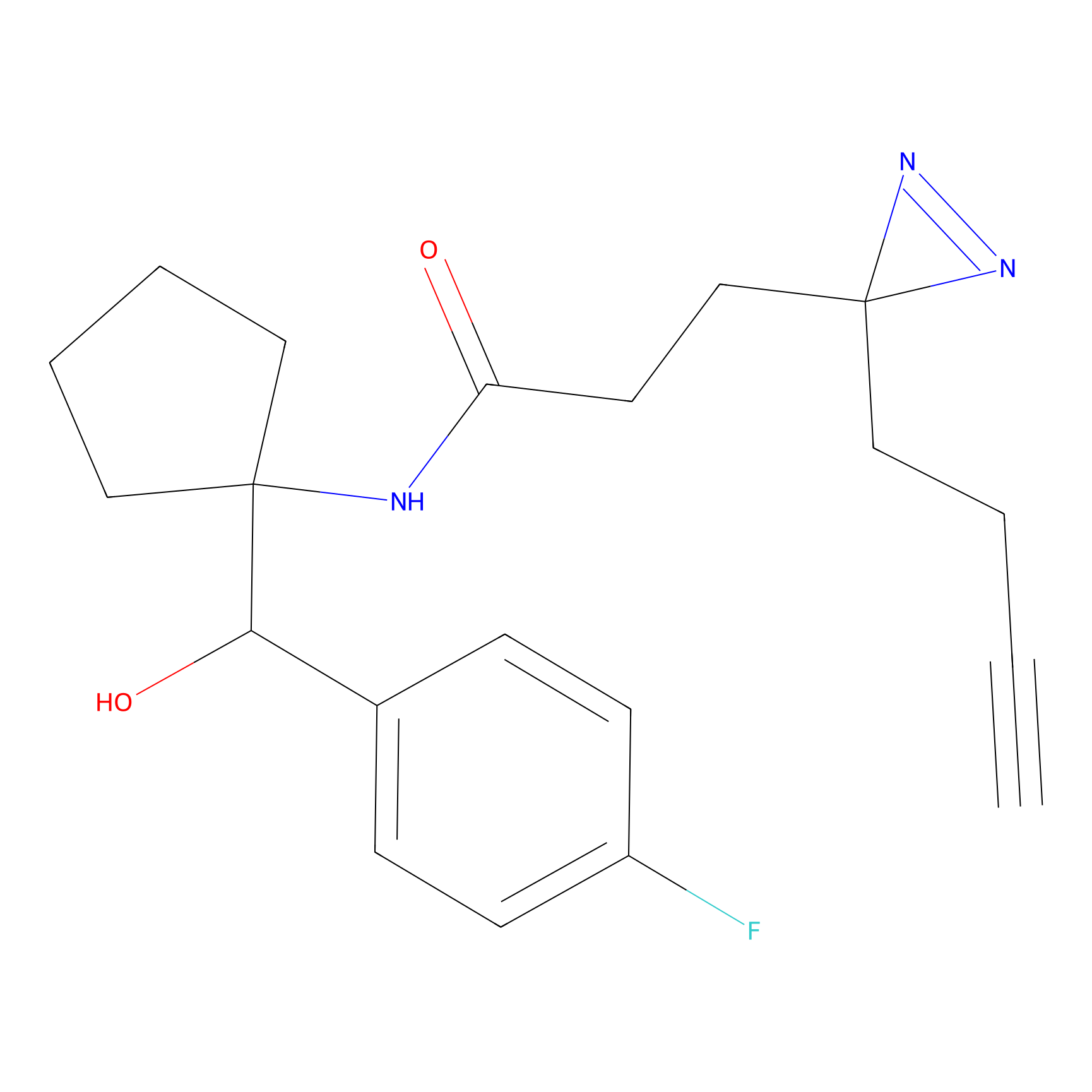
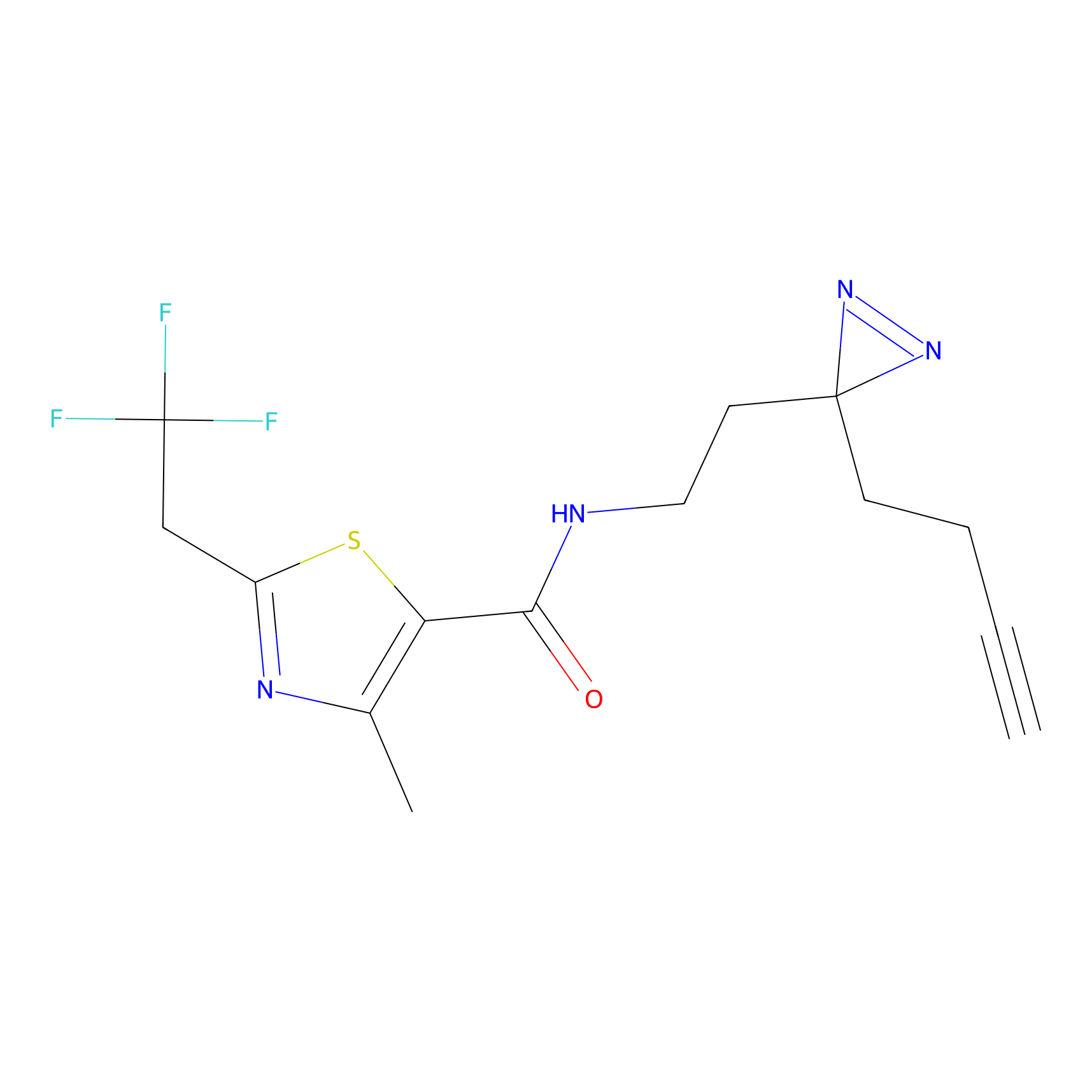
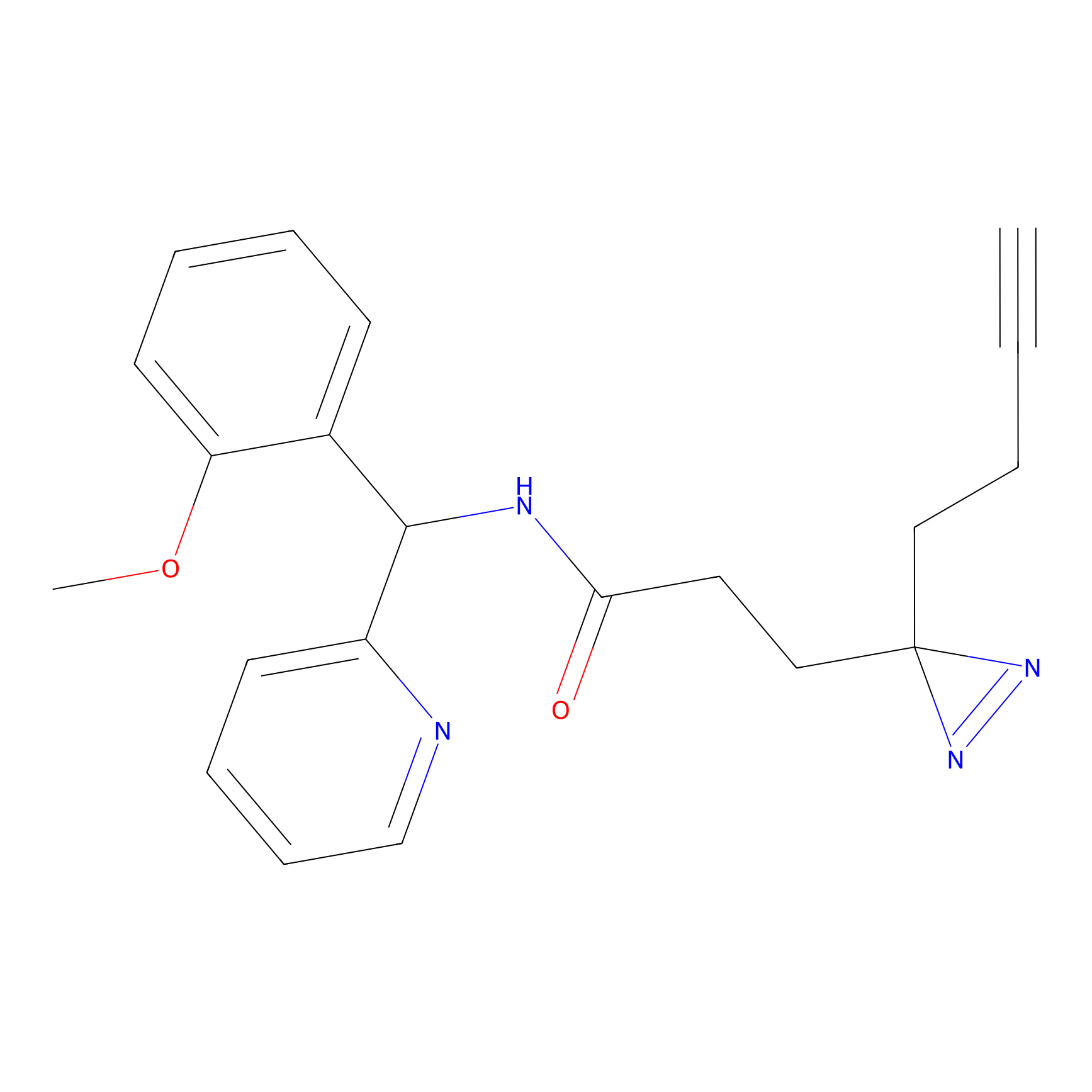
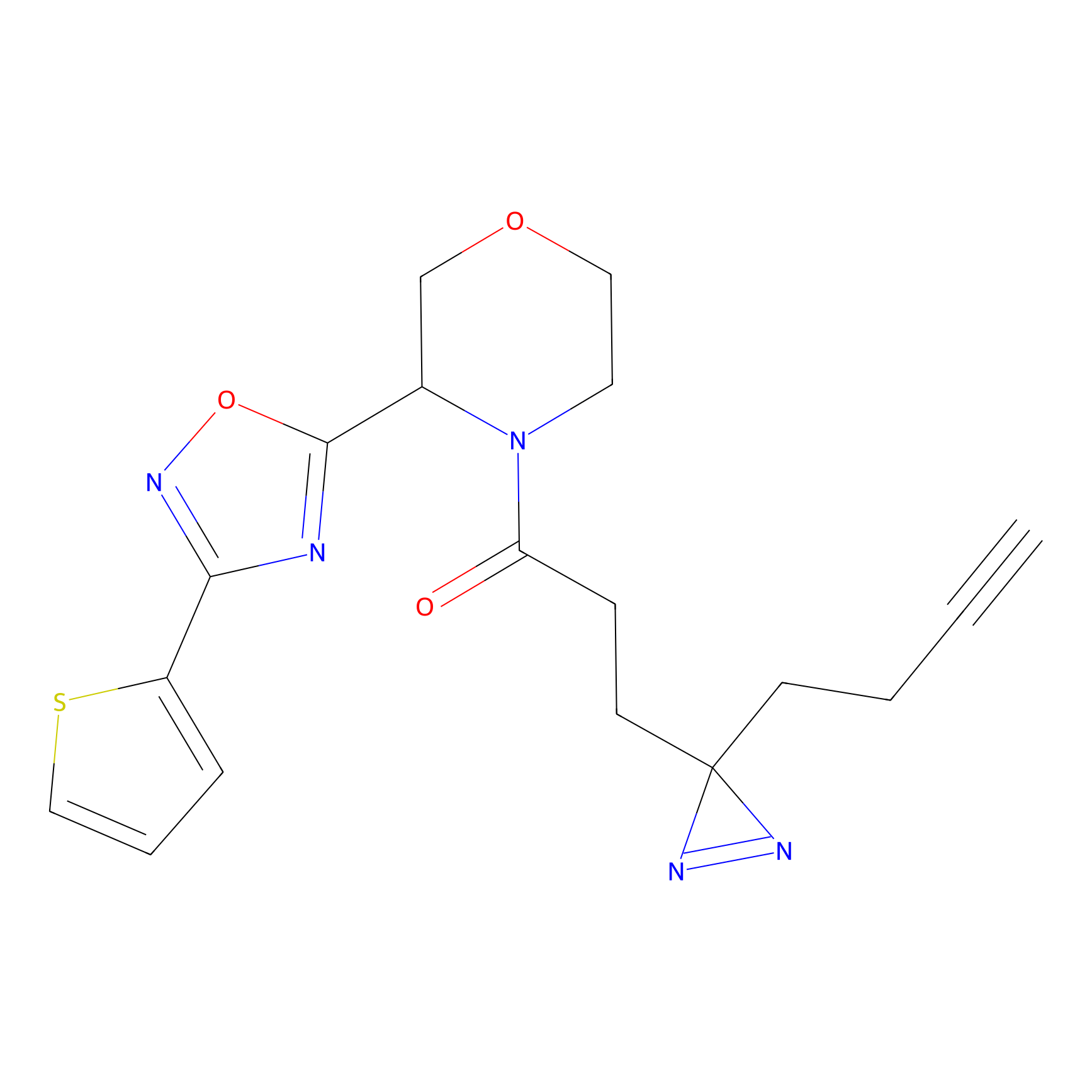
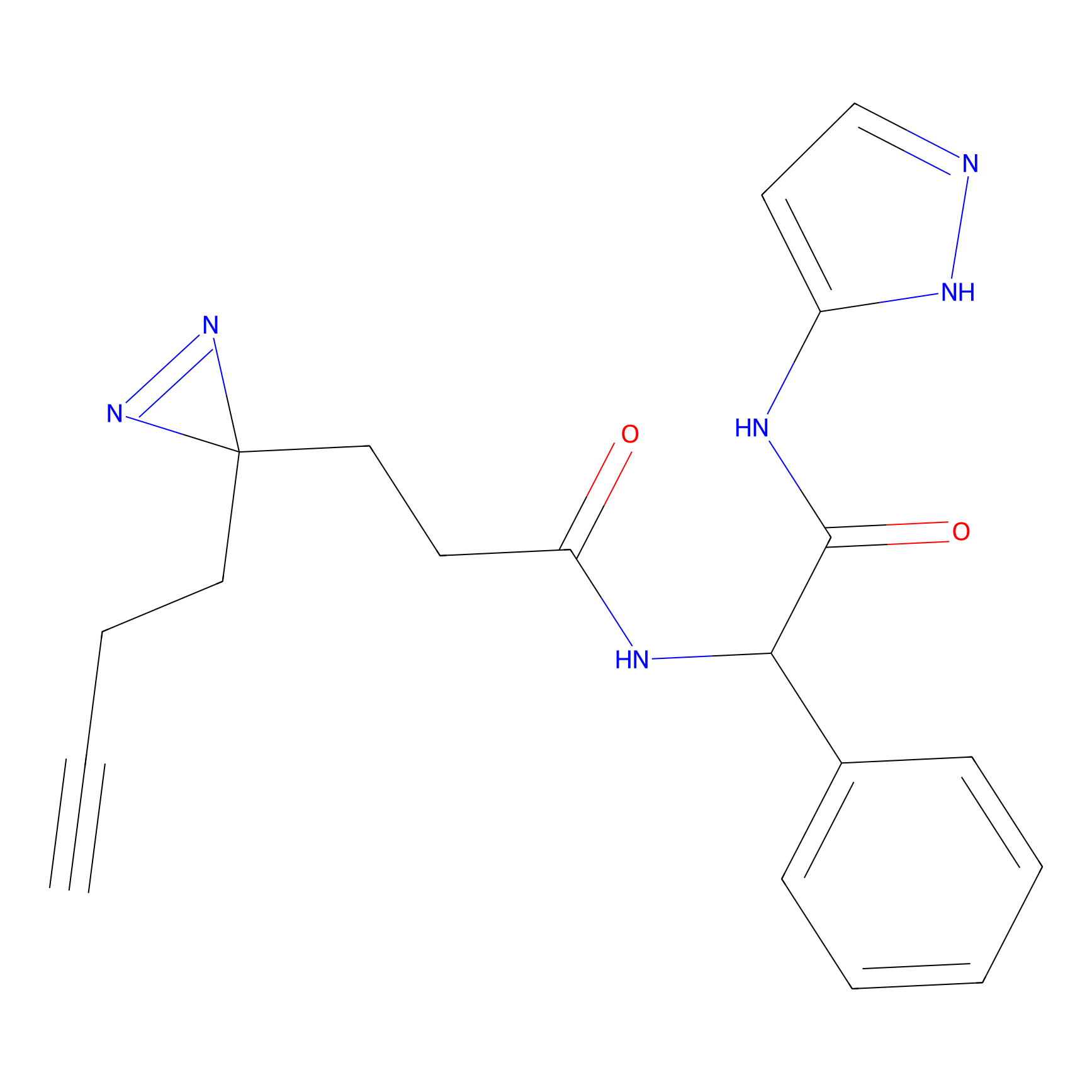
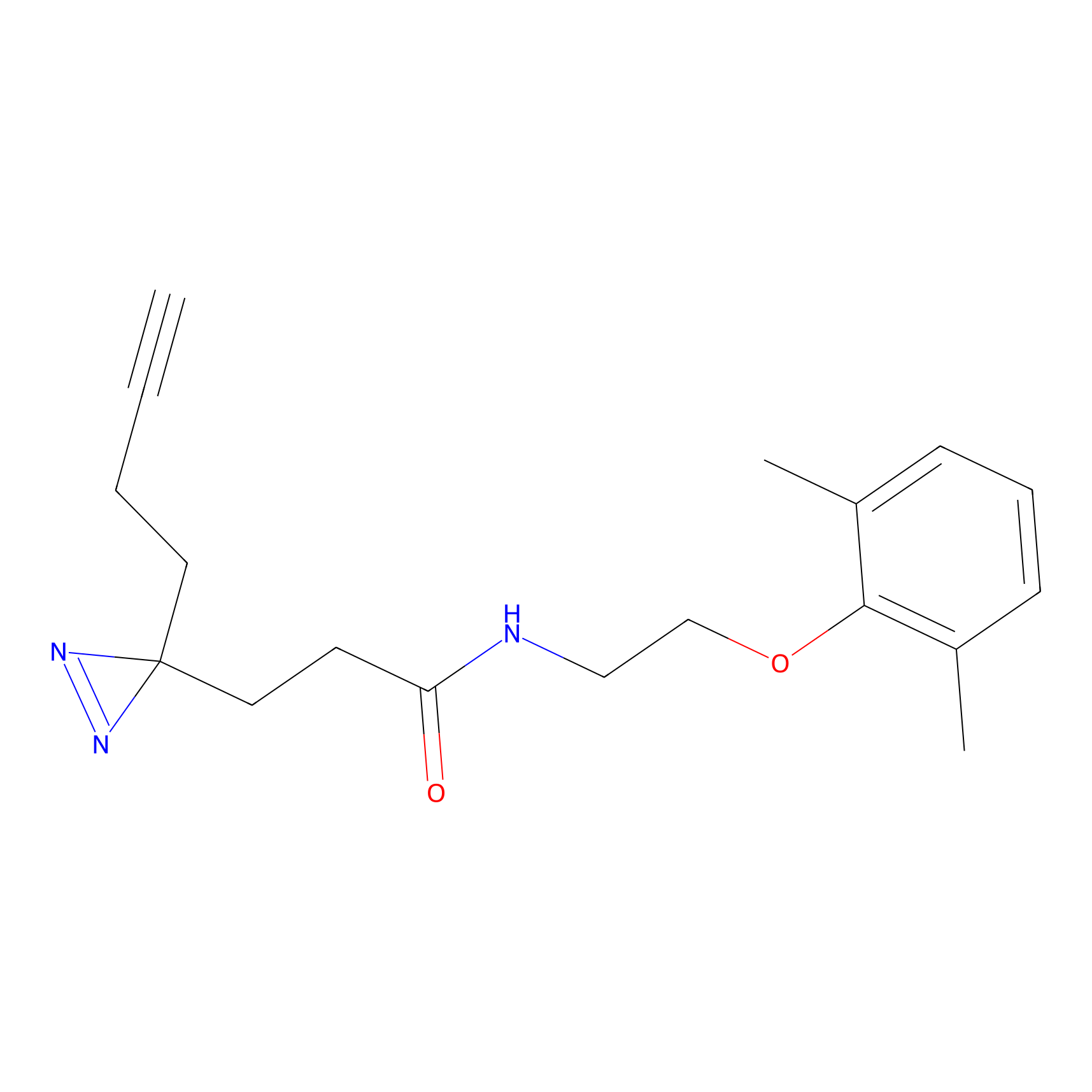
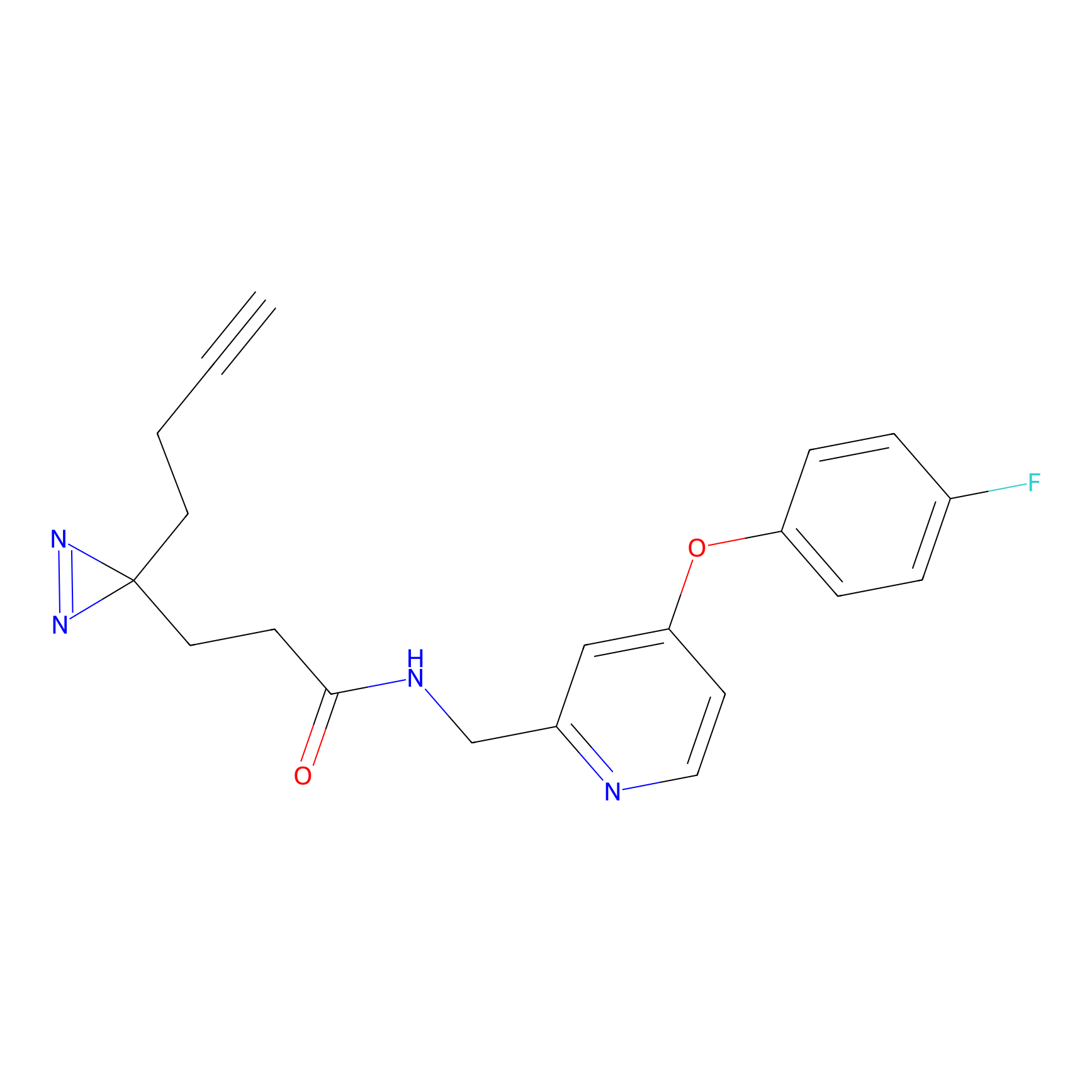
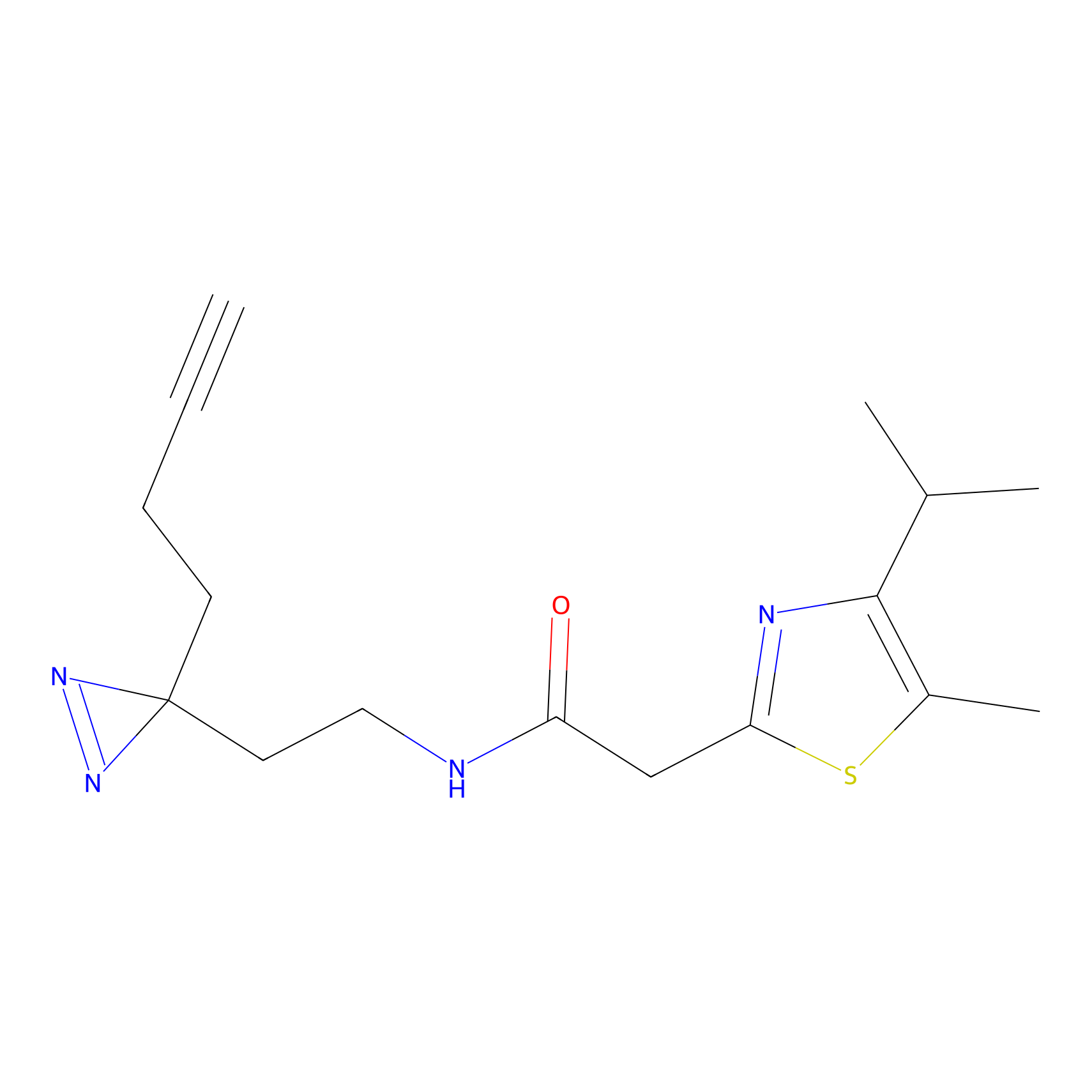
|
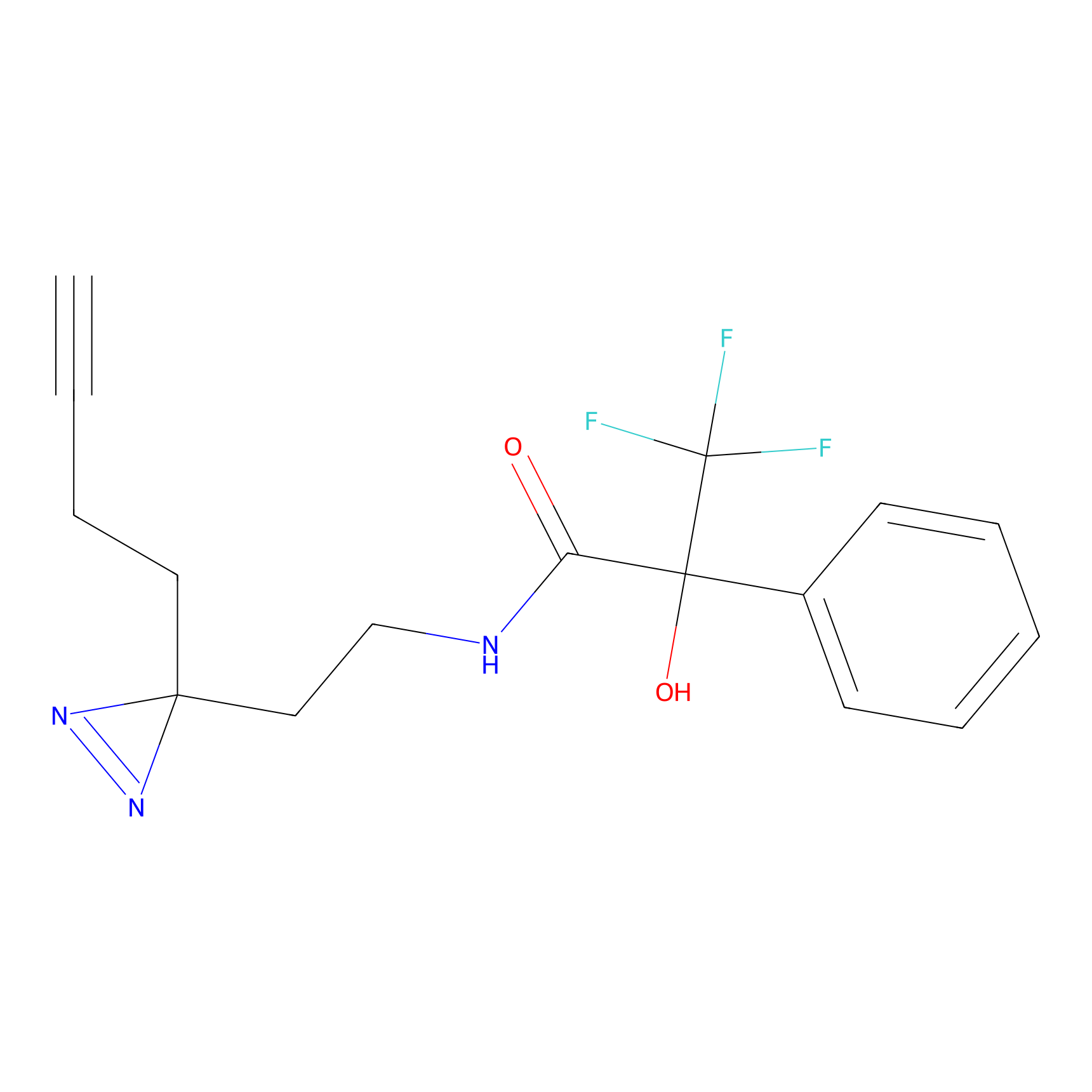
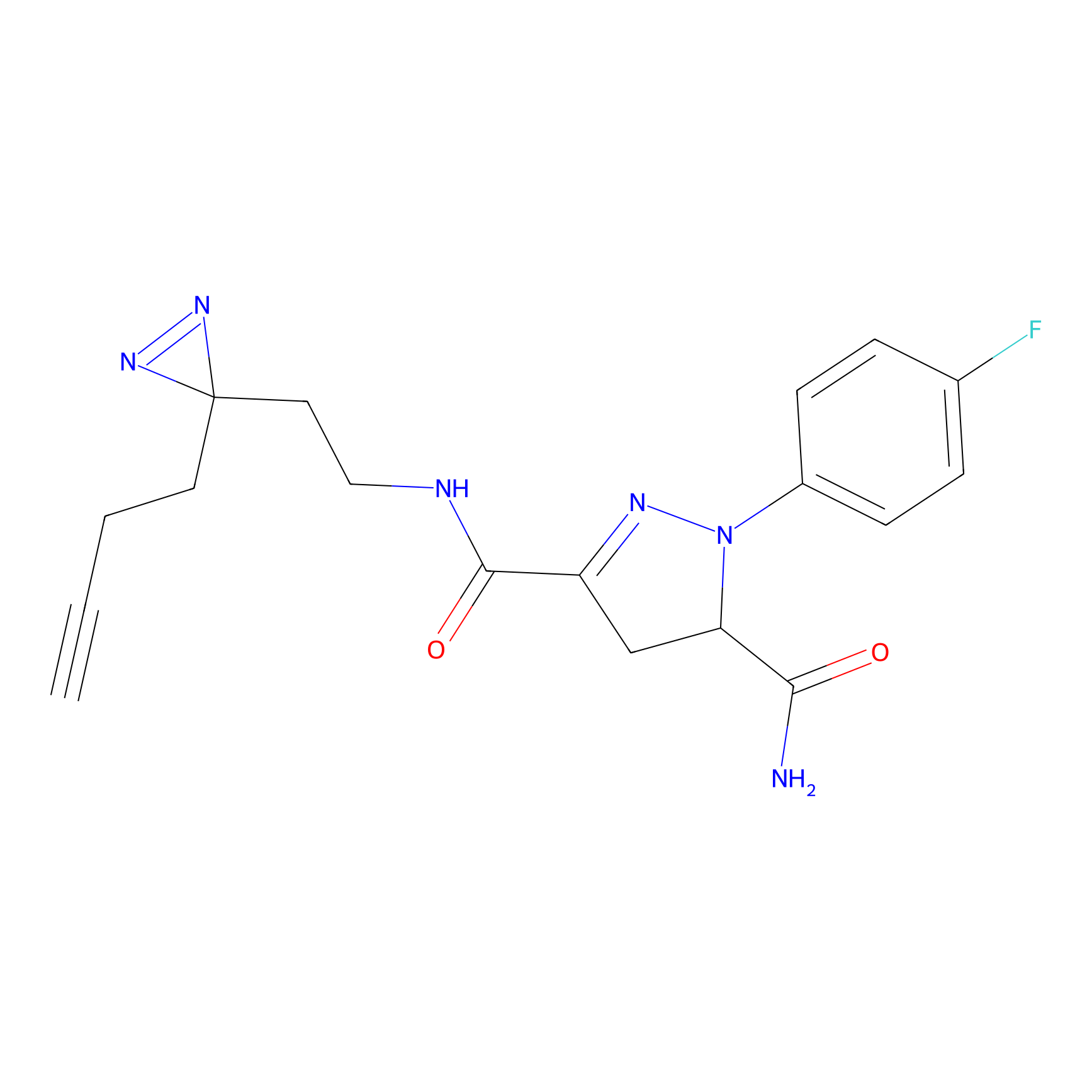
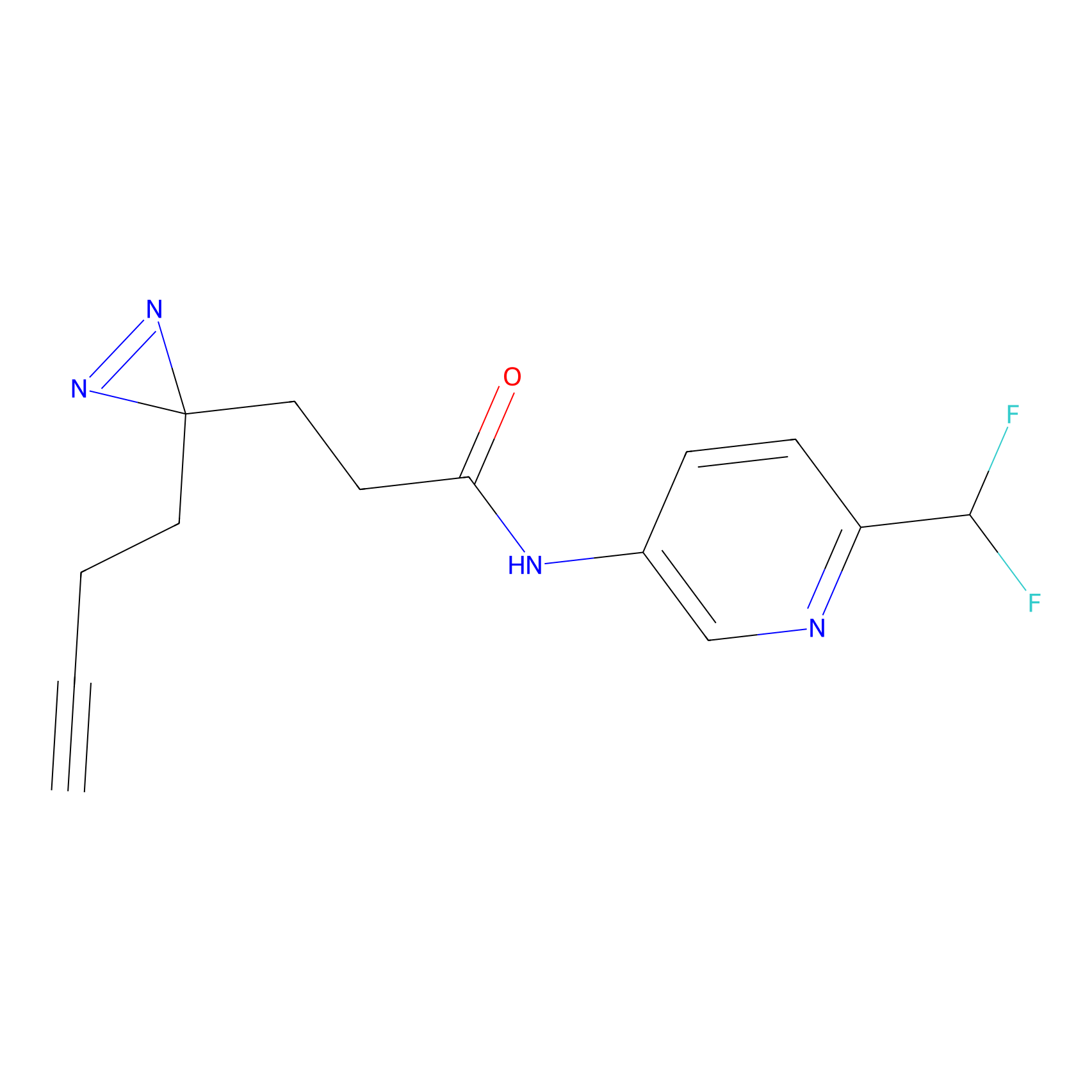
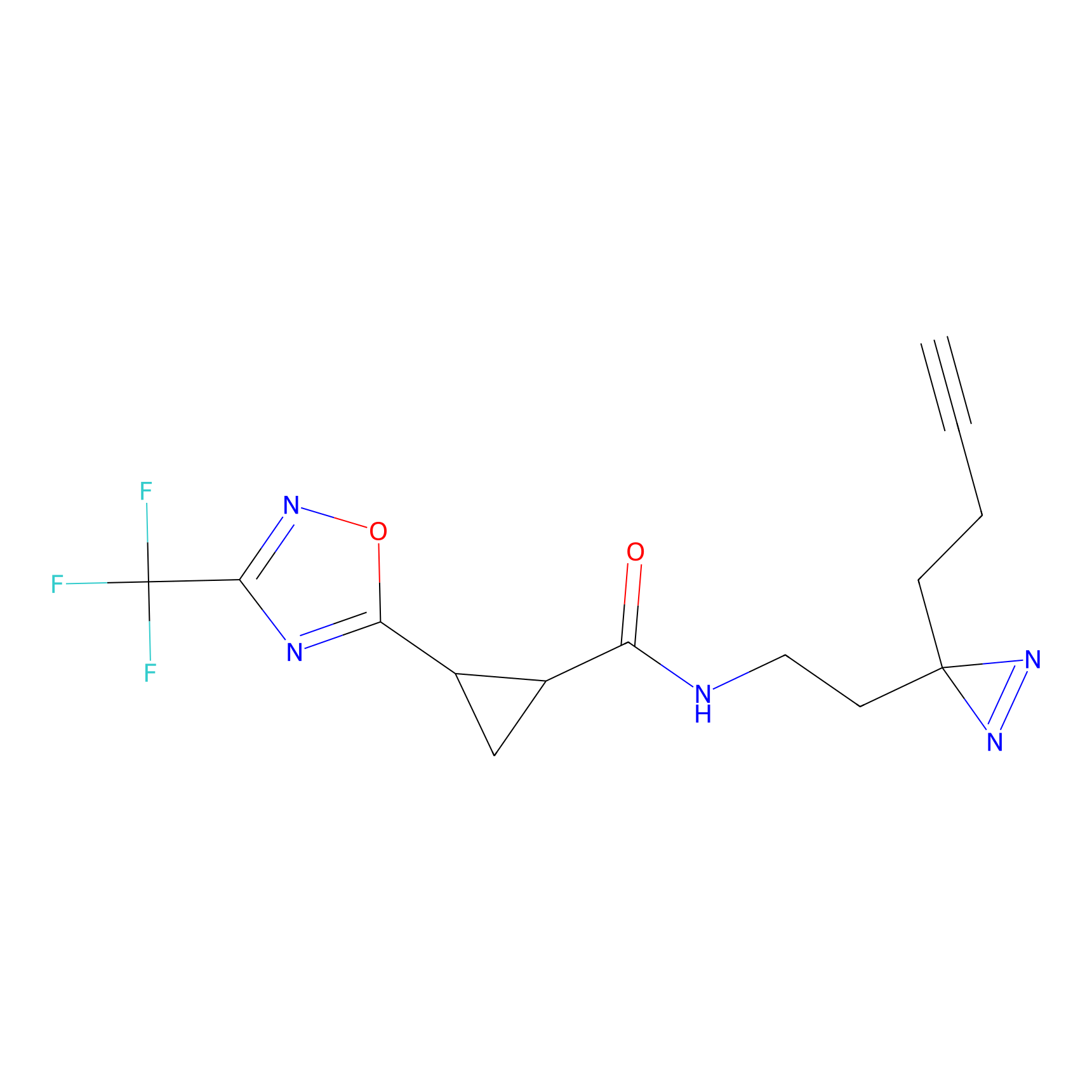
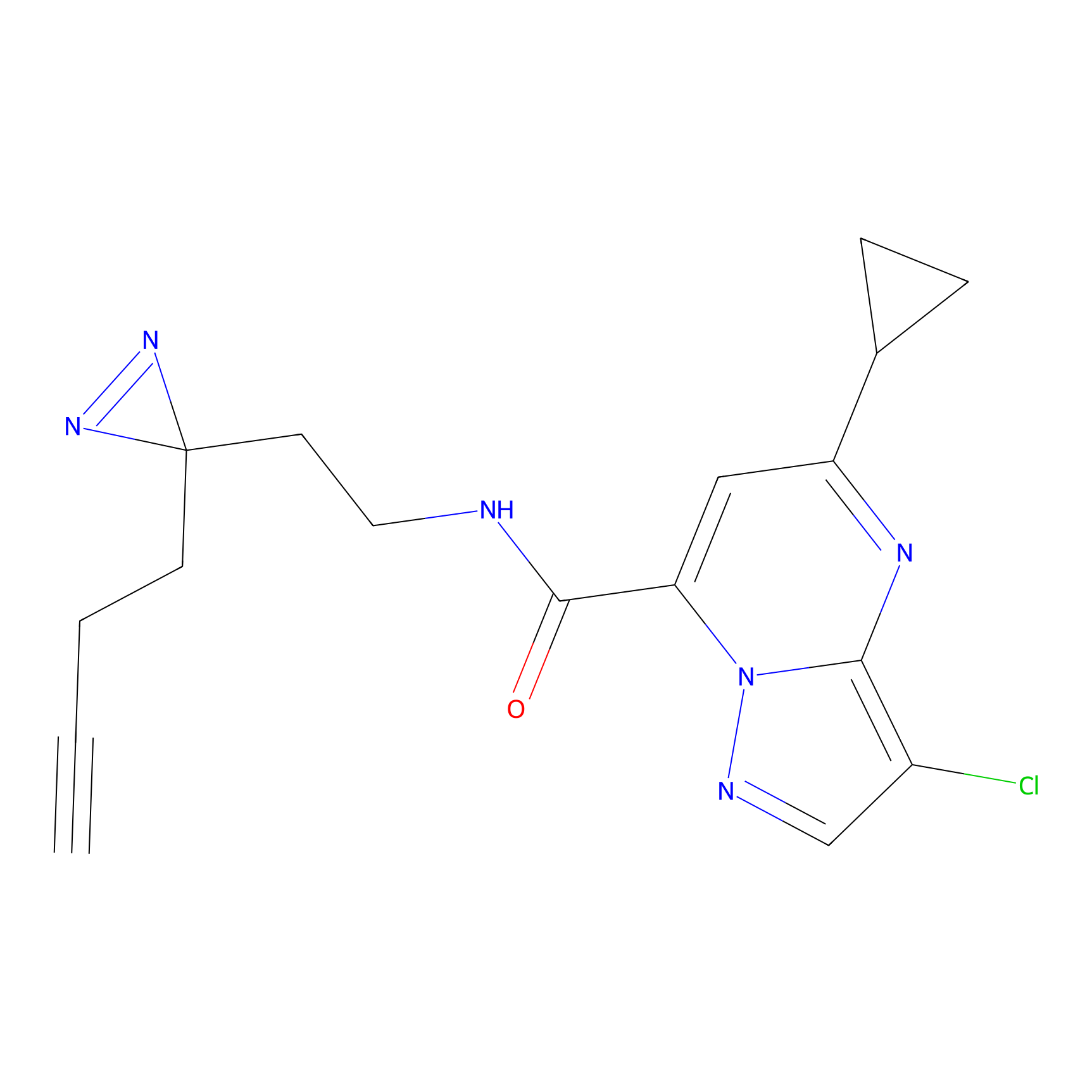
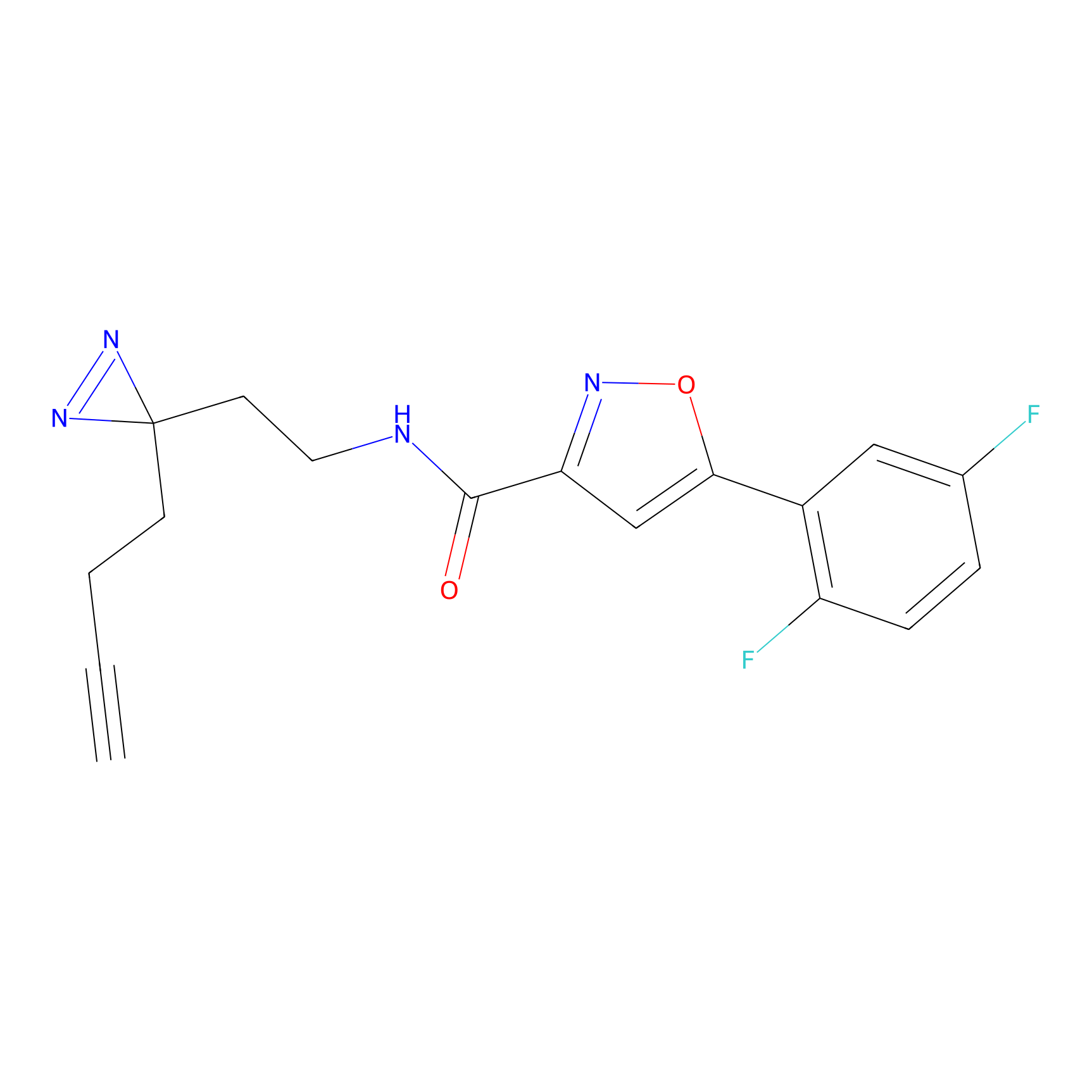
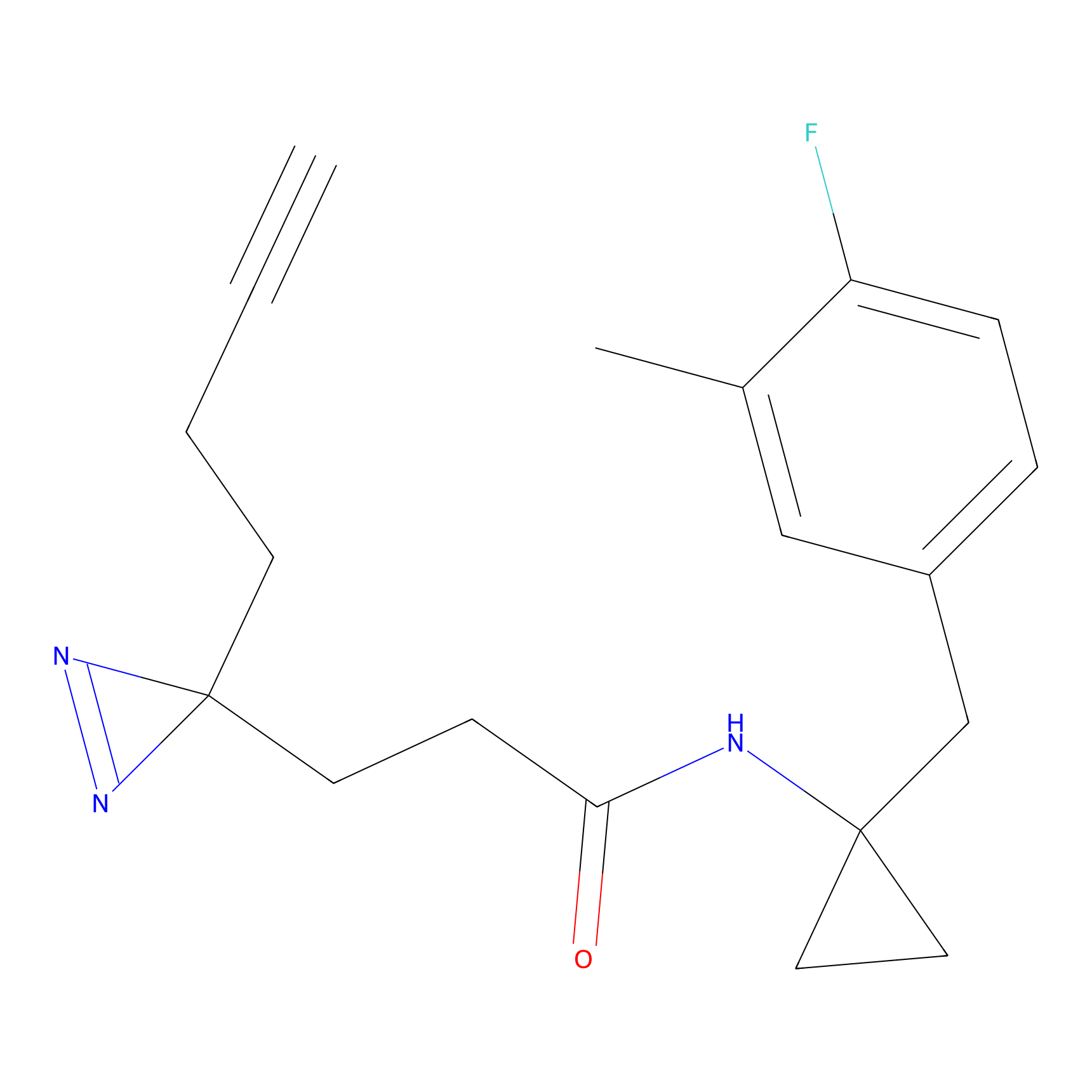
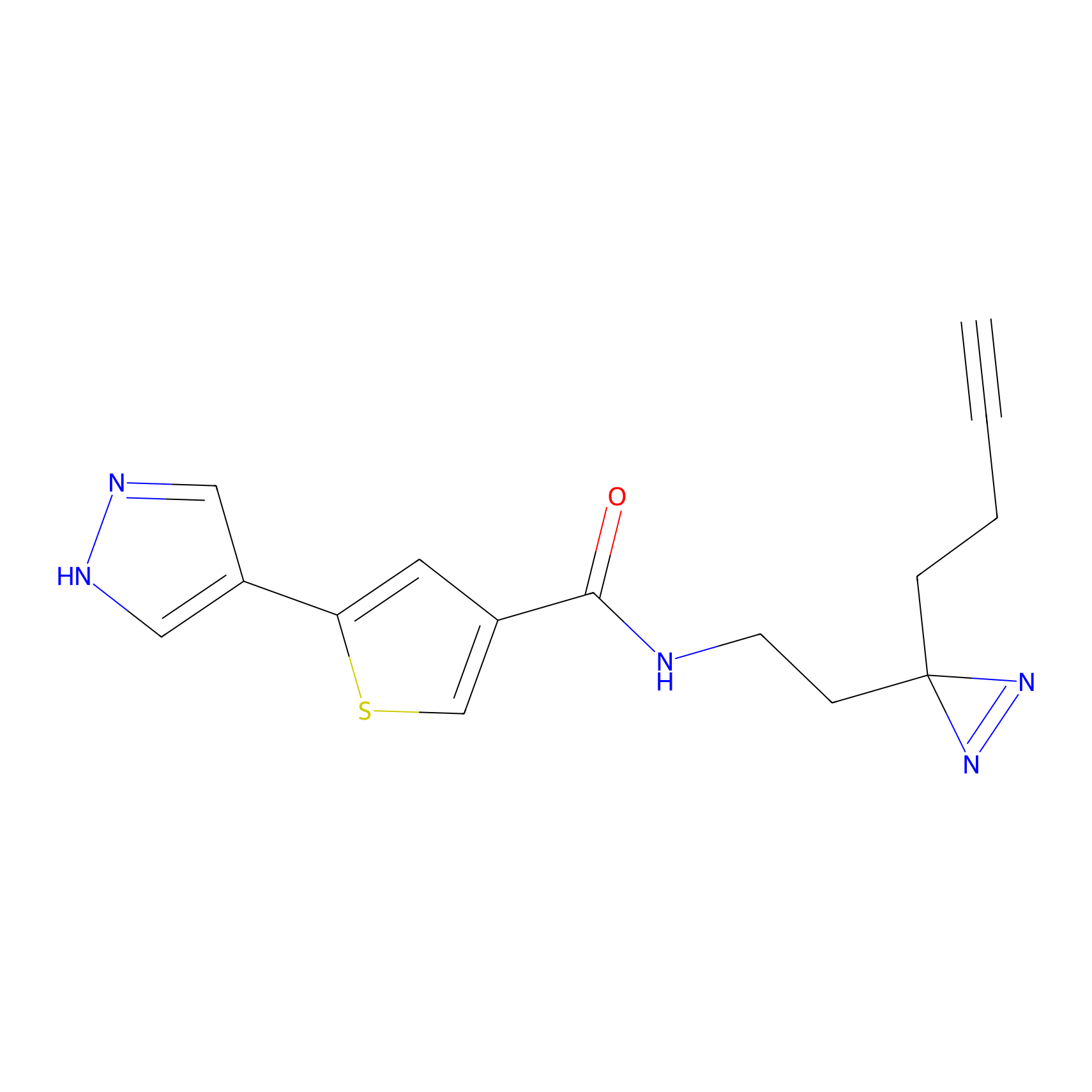
FBPP2 Probe Info |
 |
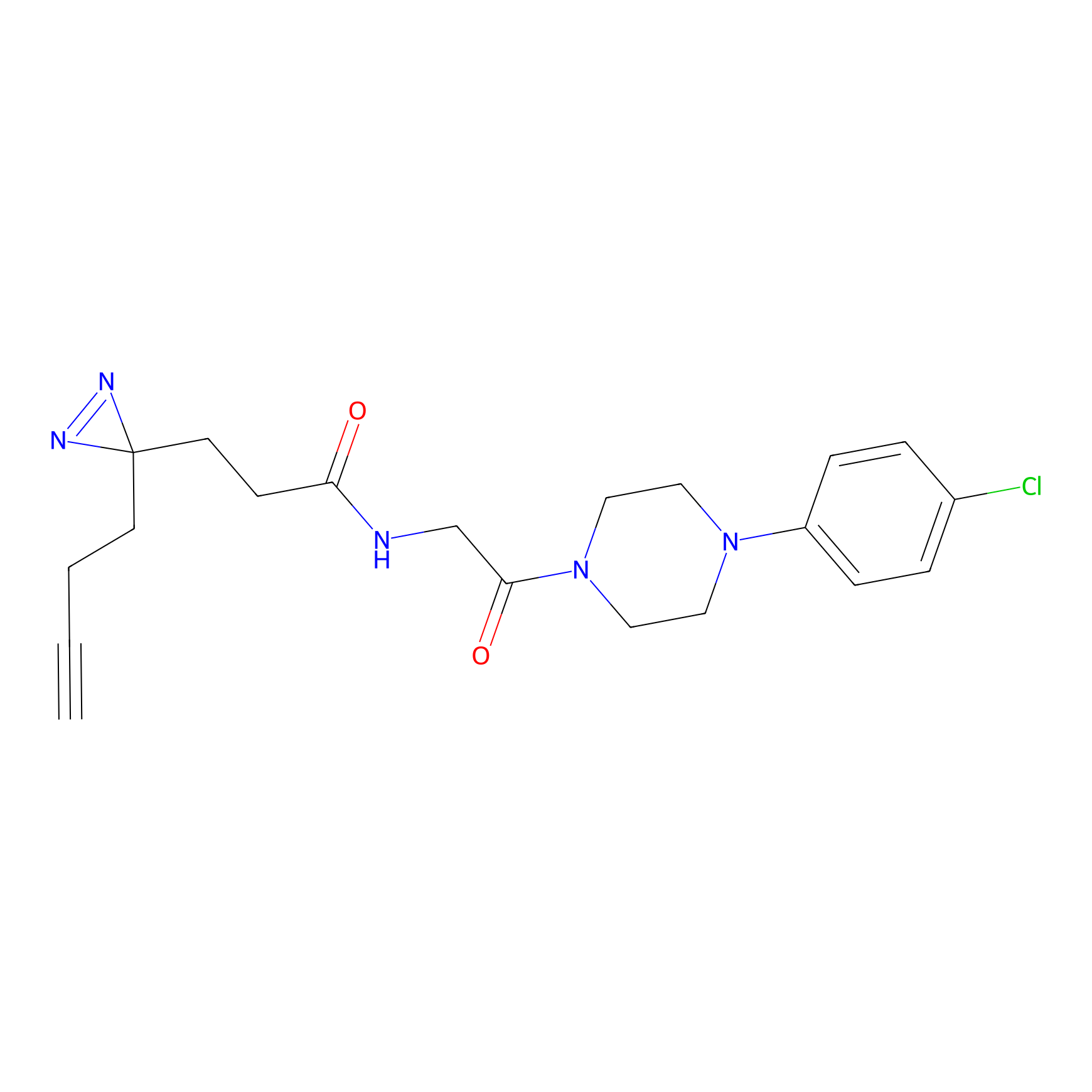
12.56 | LDD0318 | [1] | |
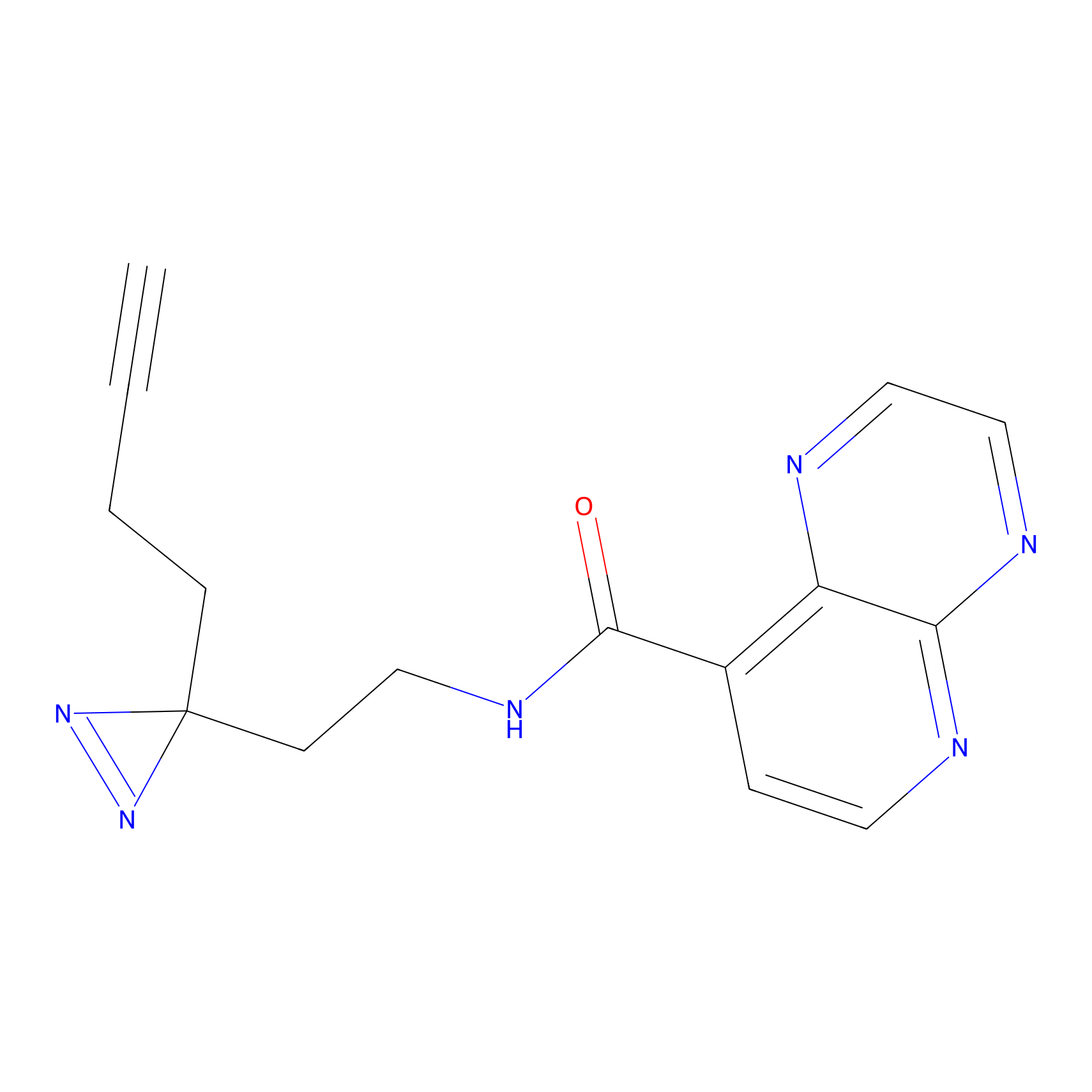
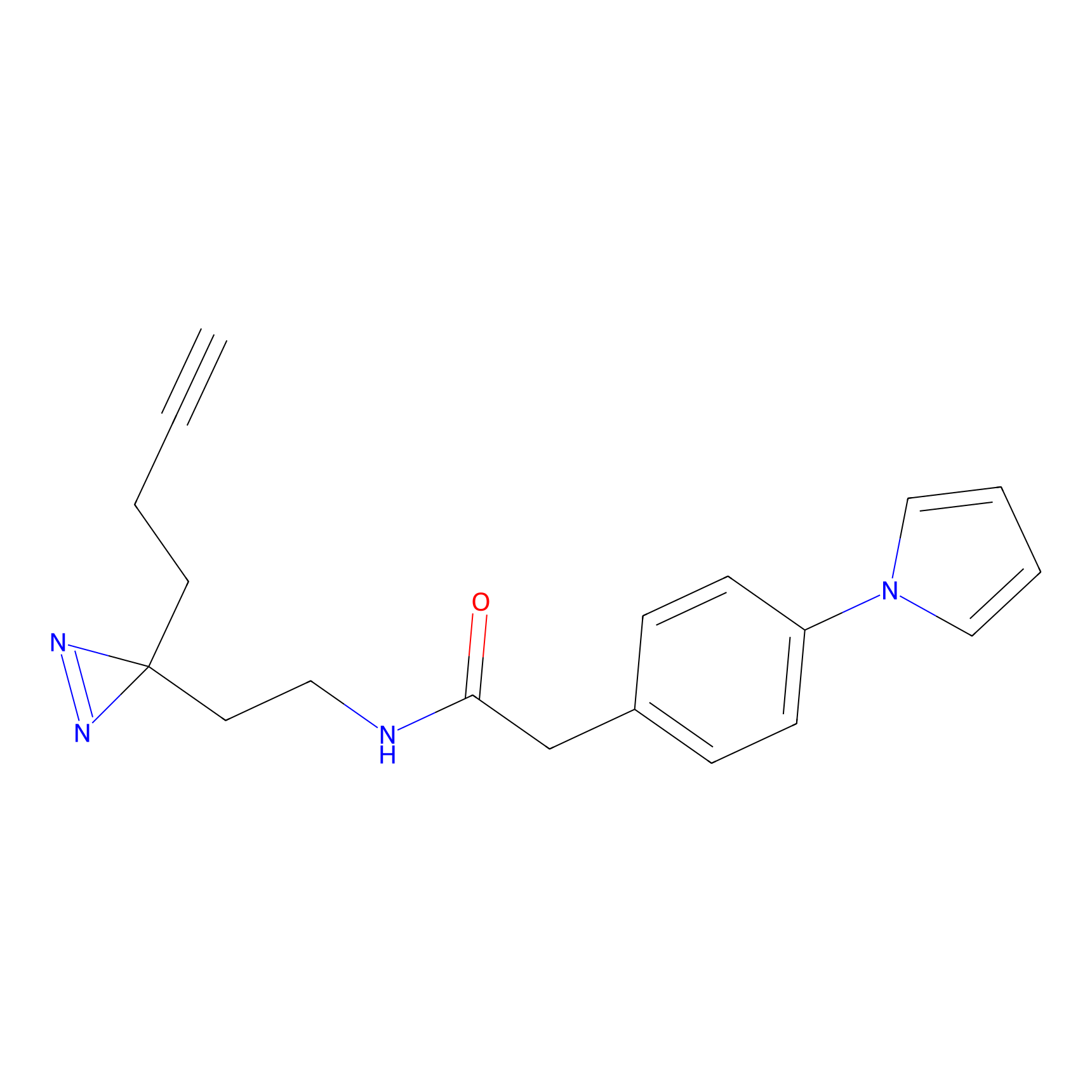
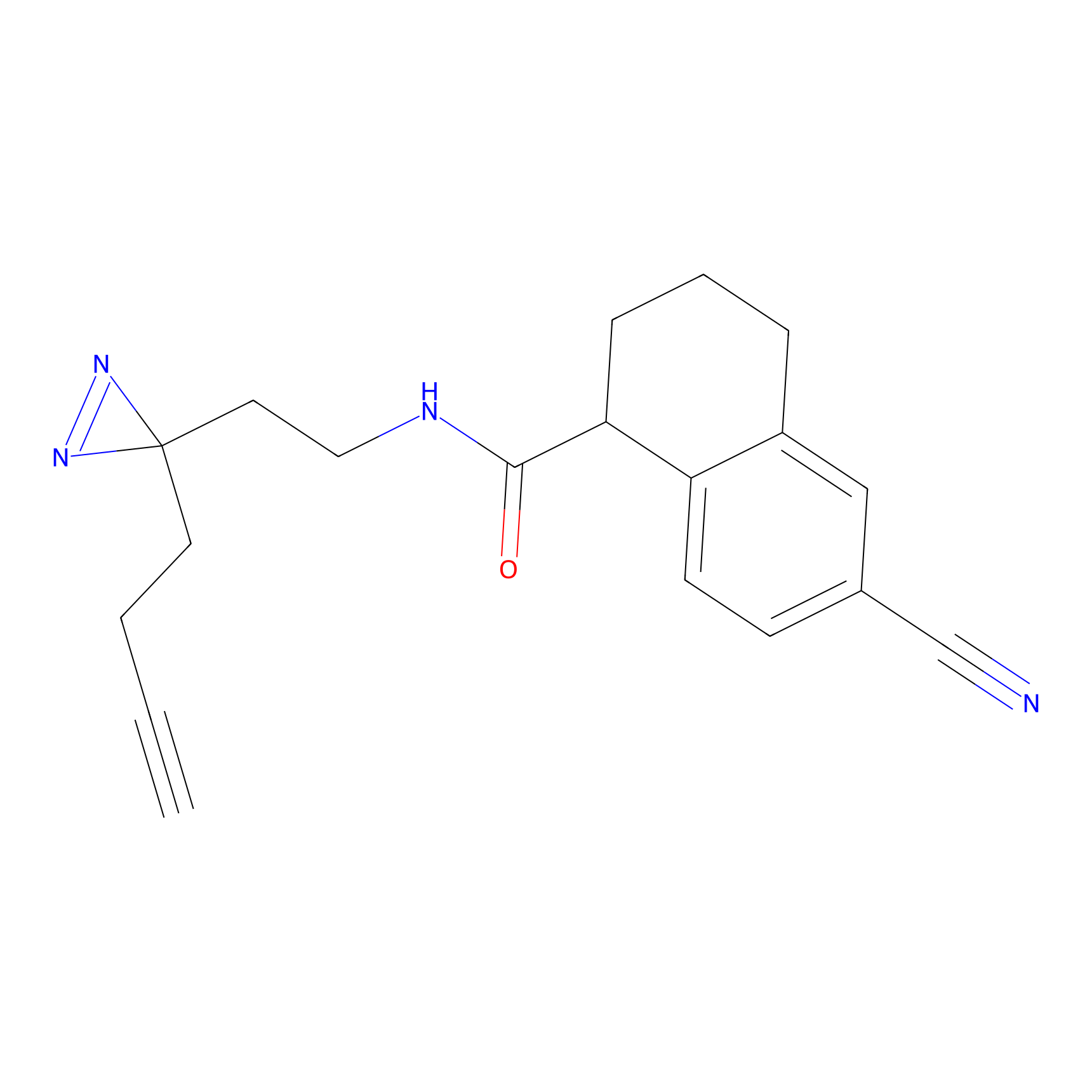
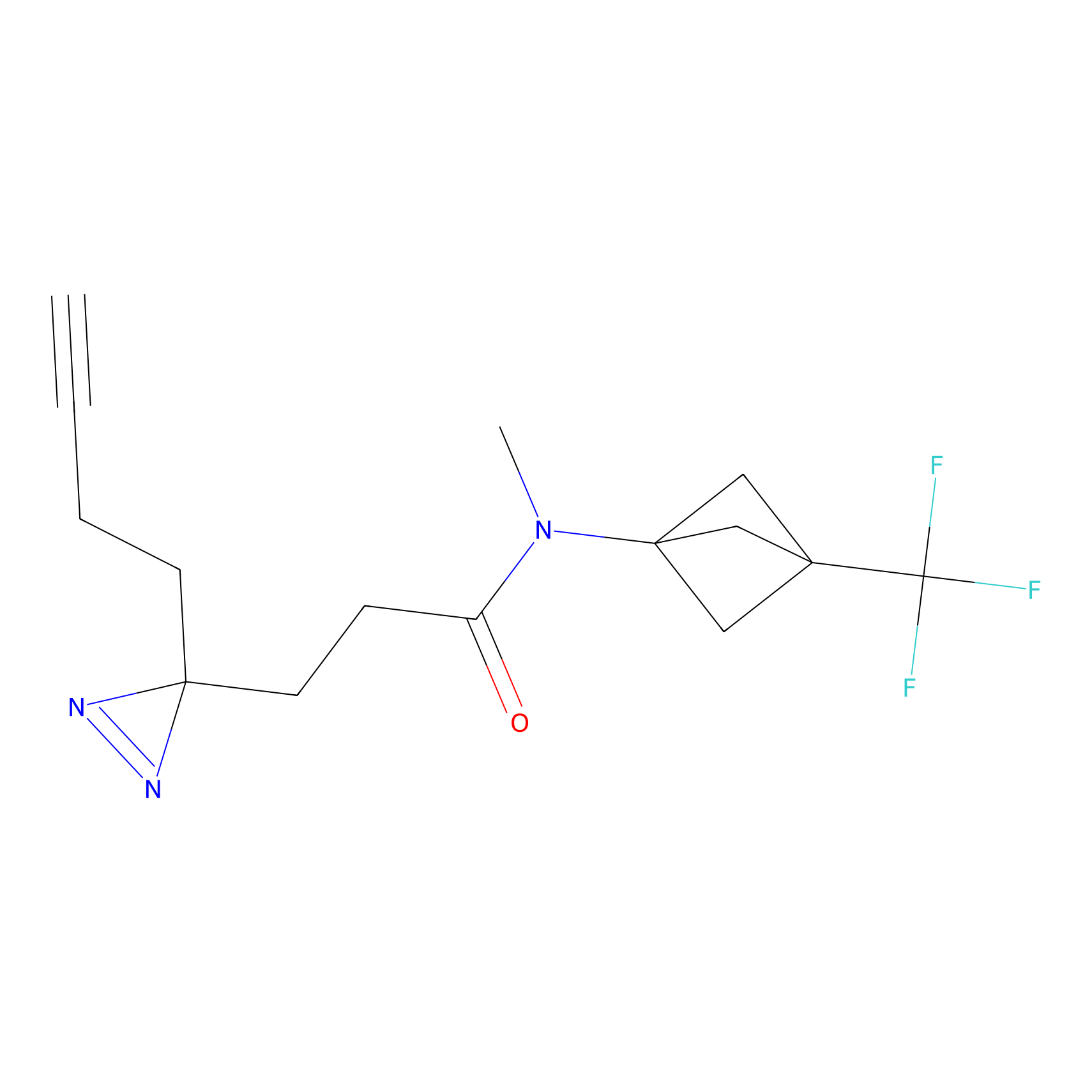
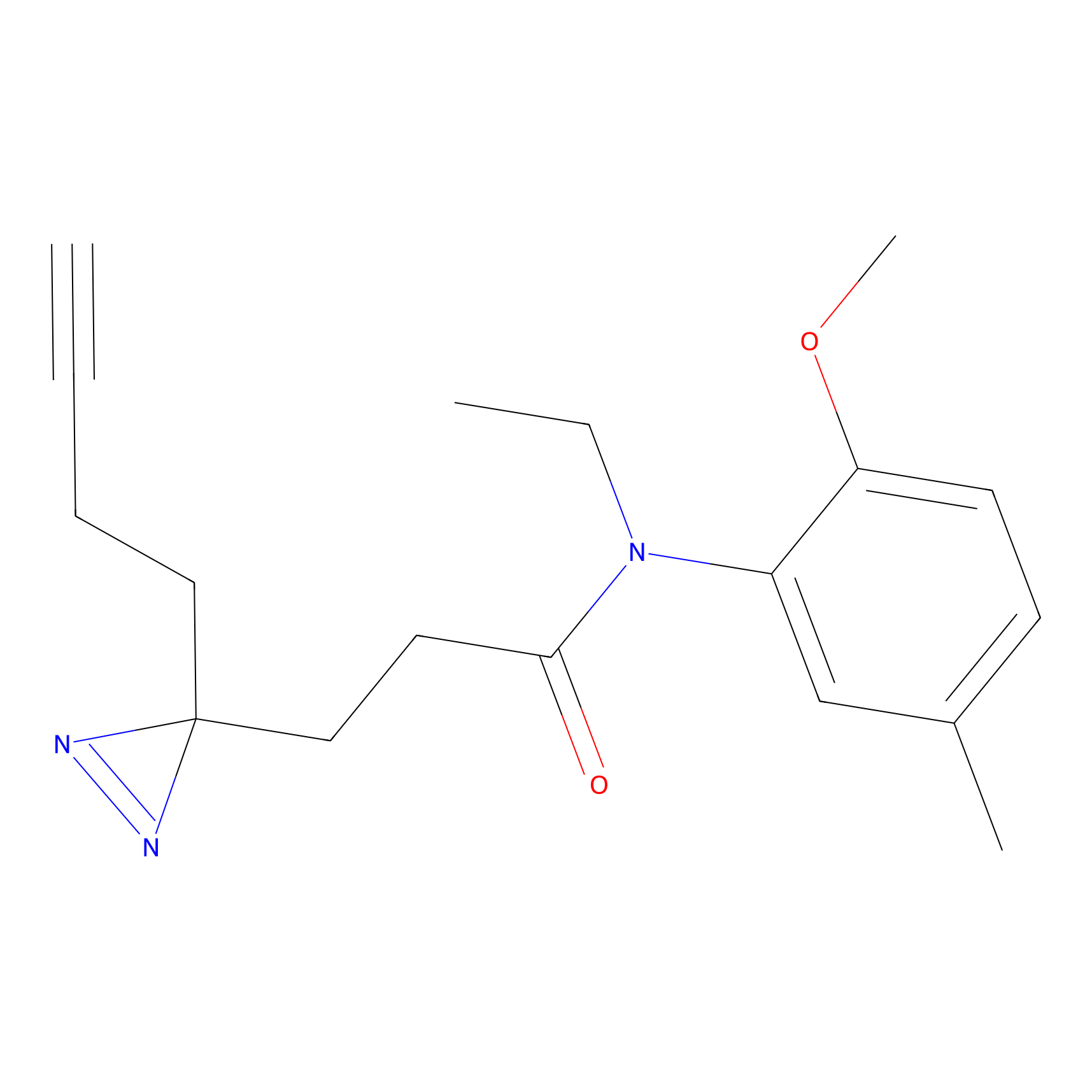
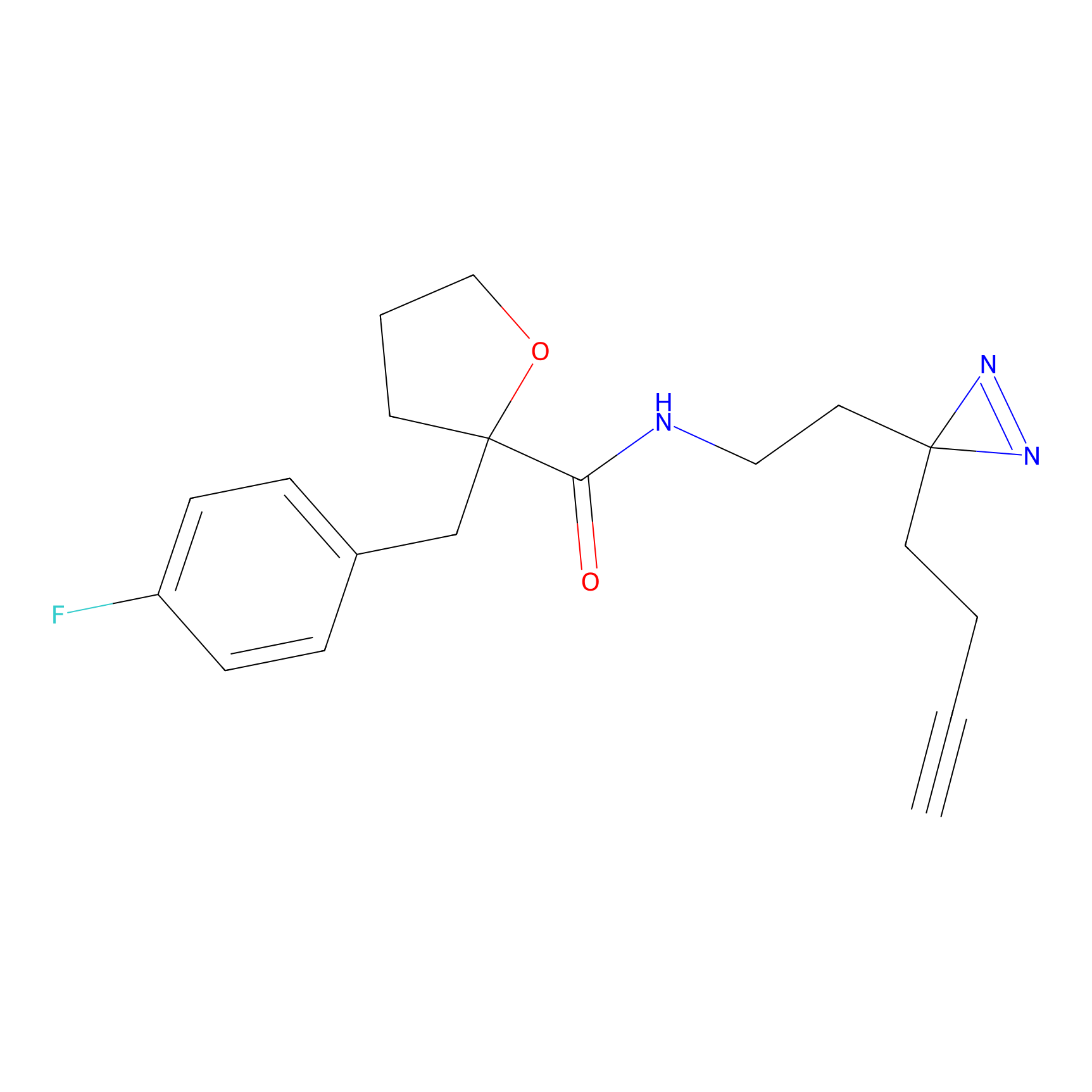
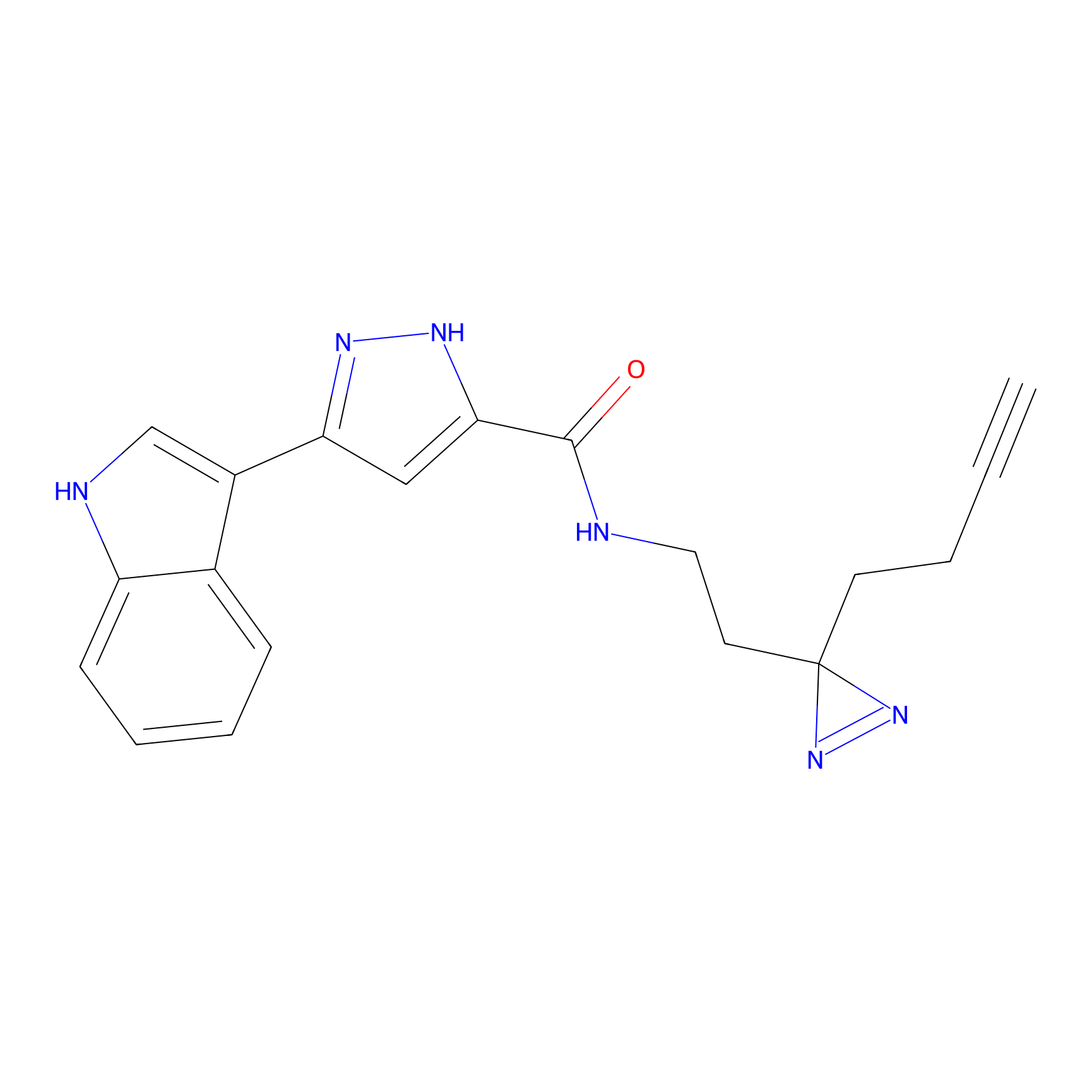
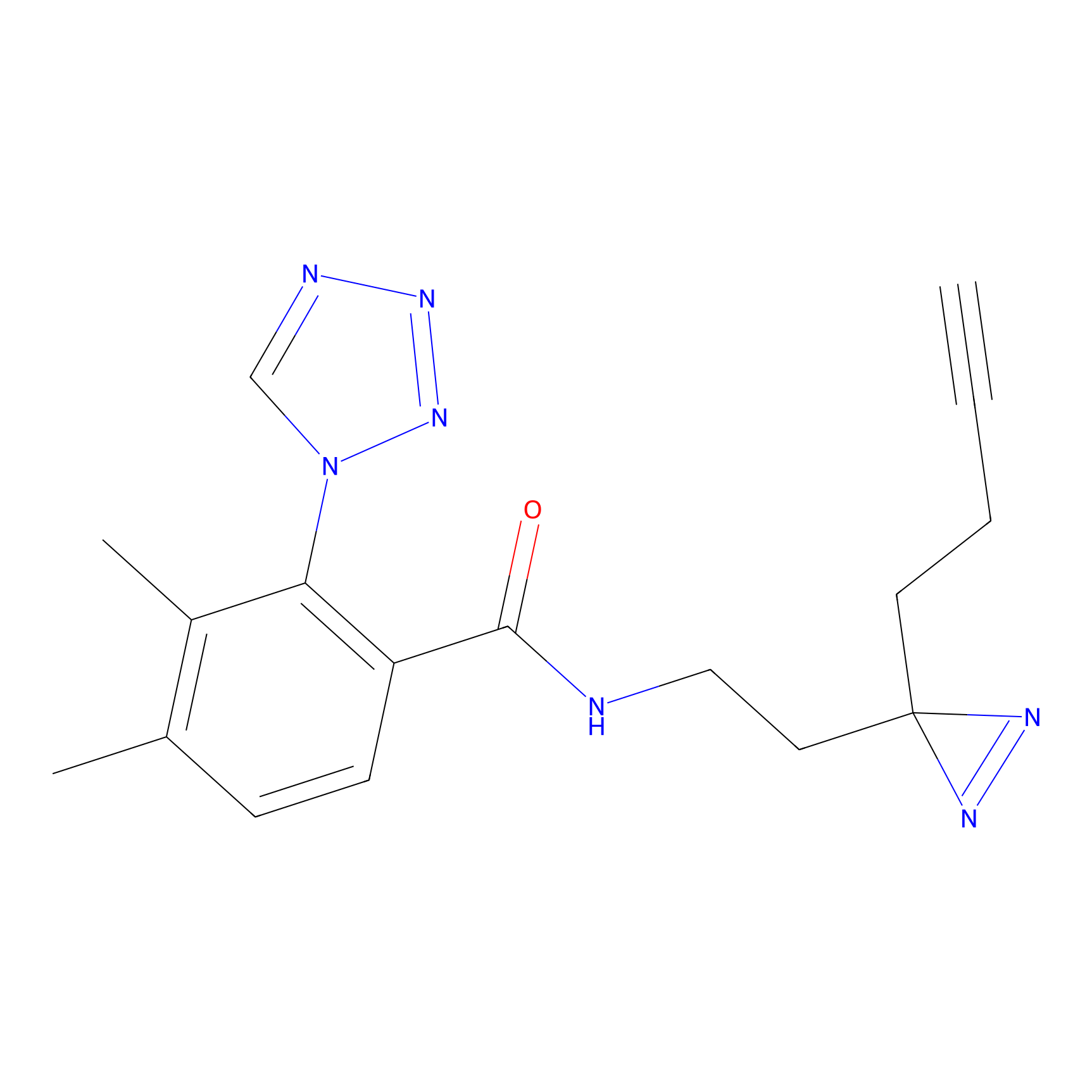
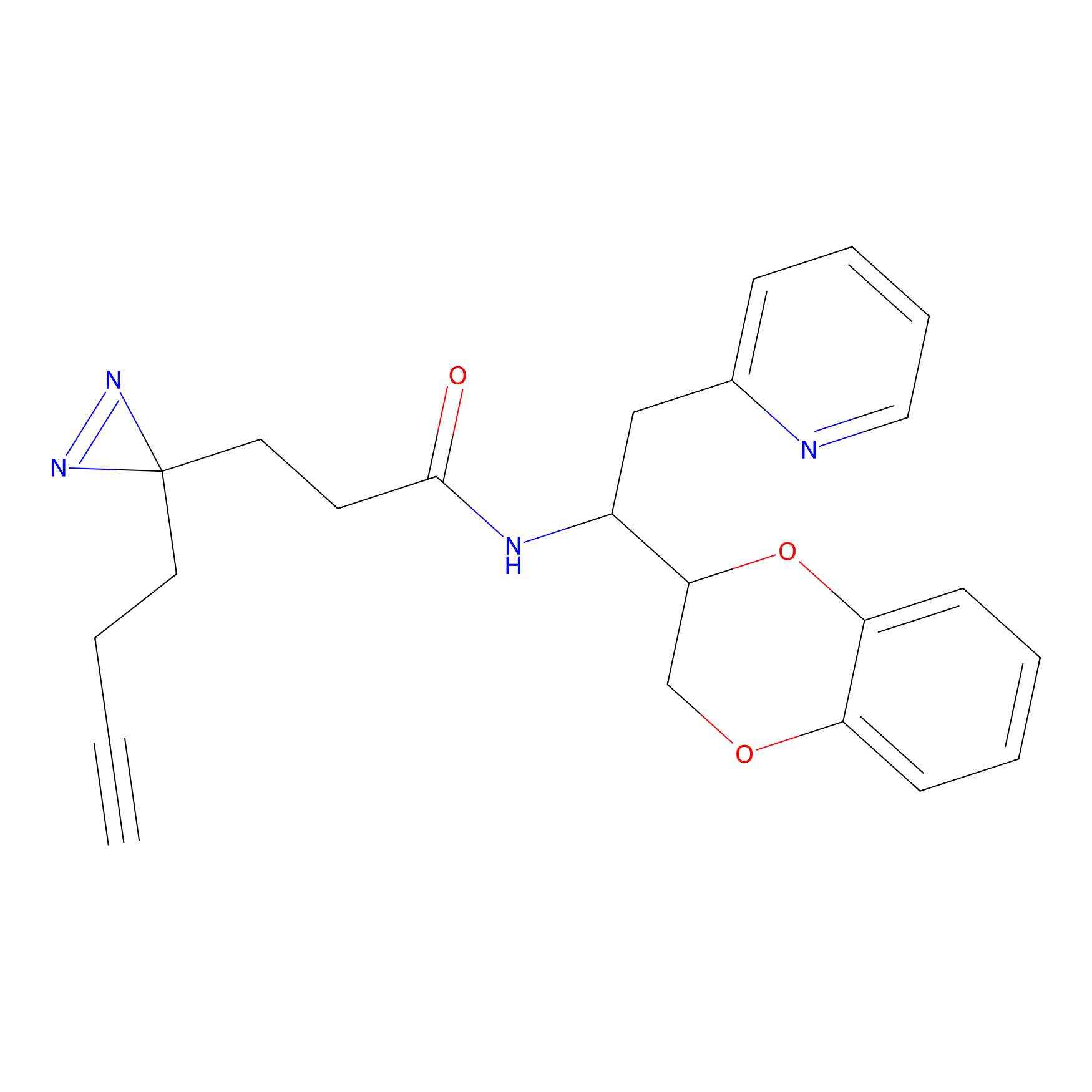
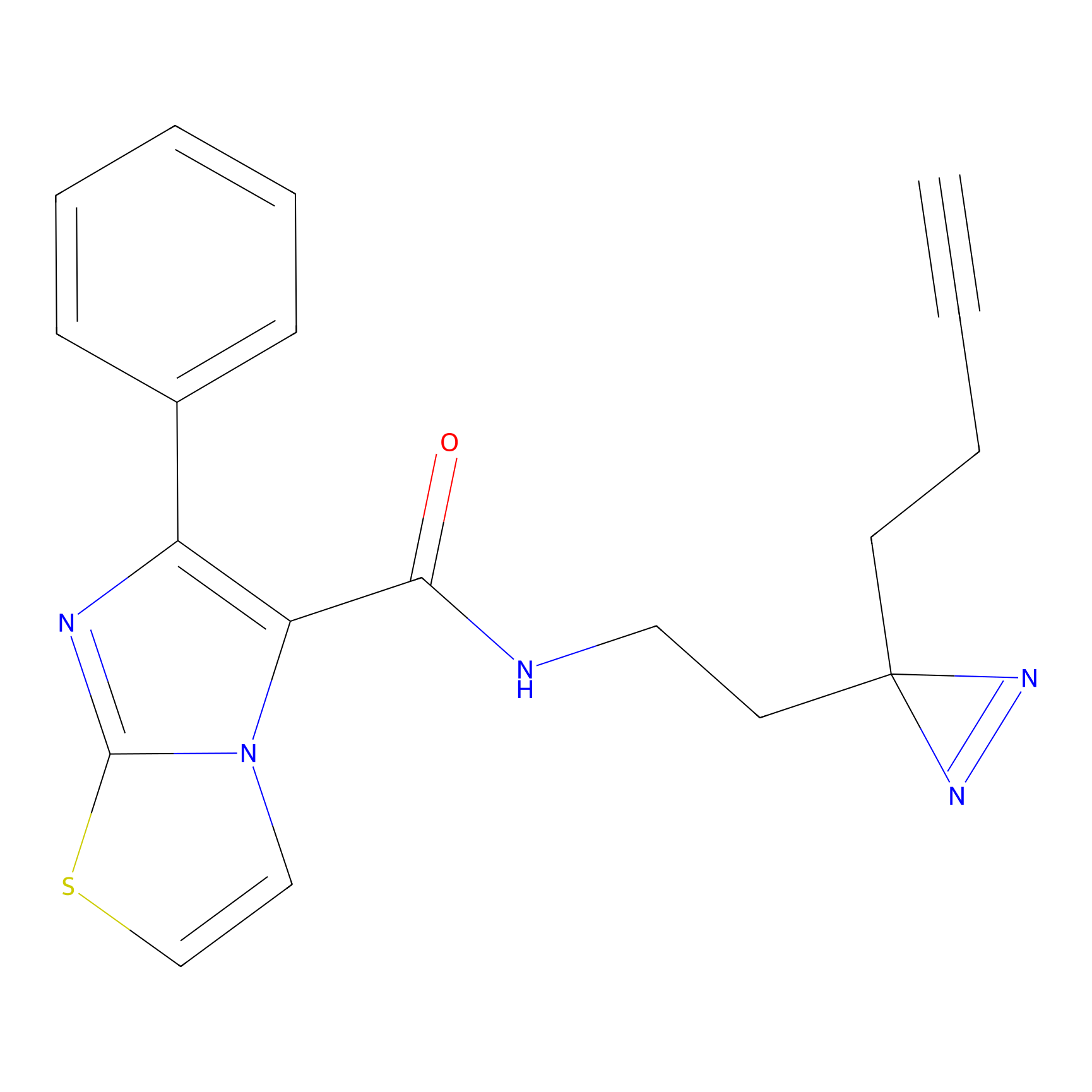
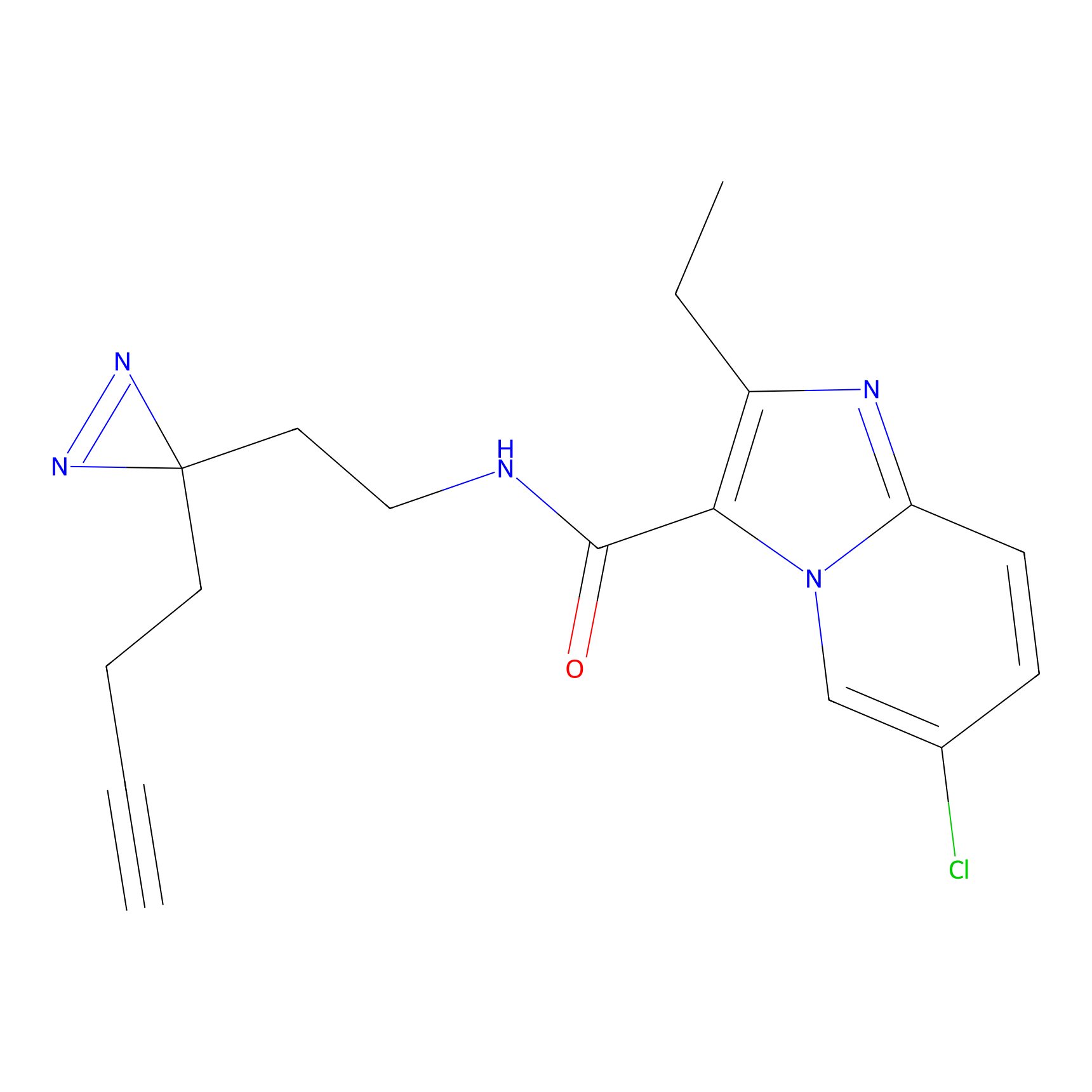
|
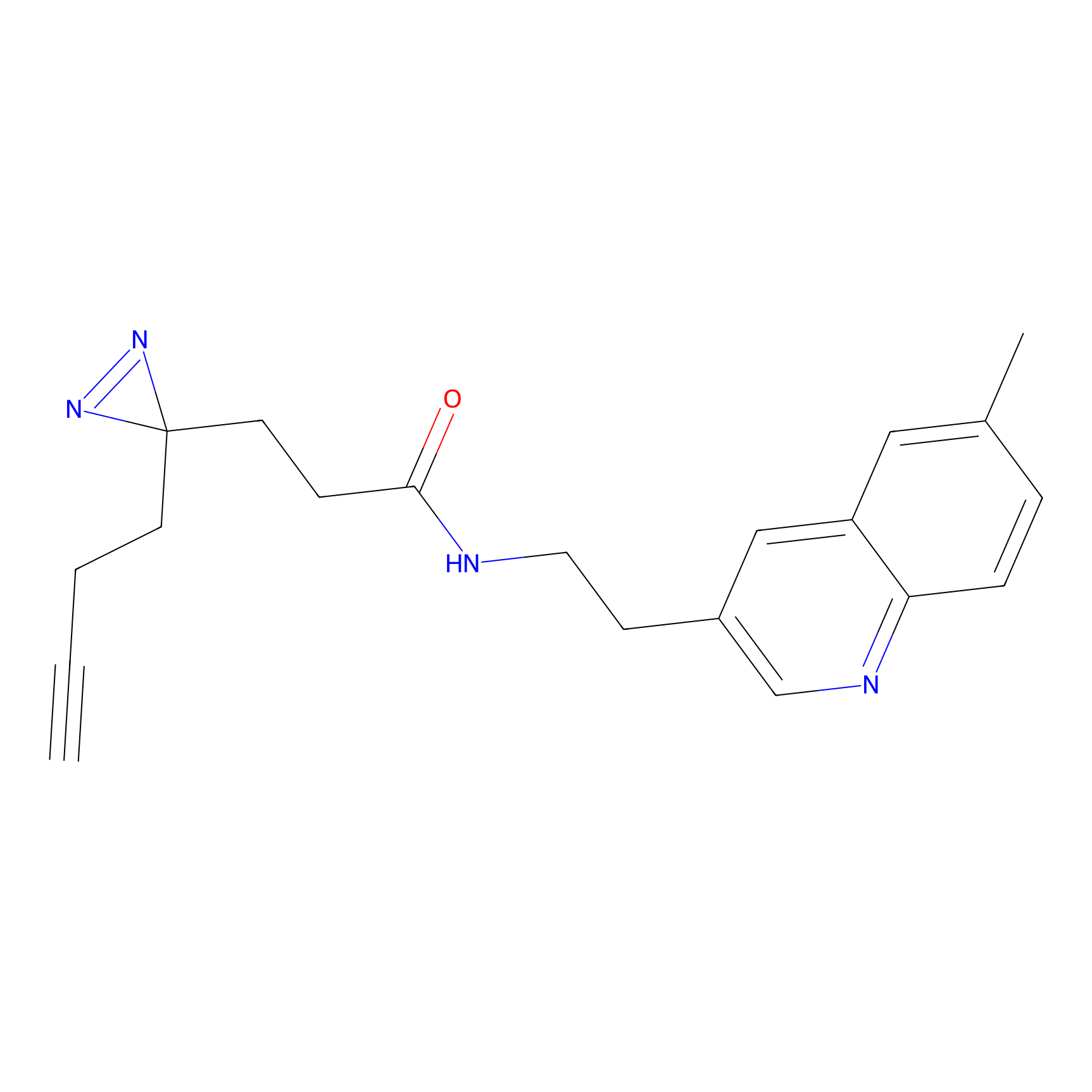
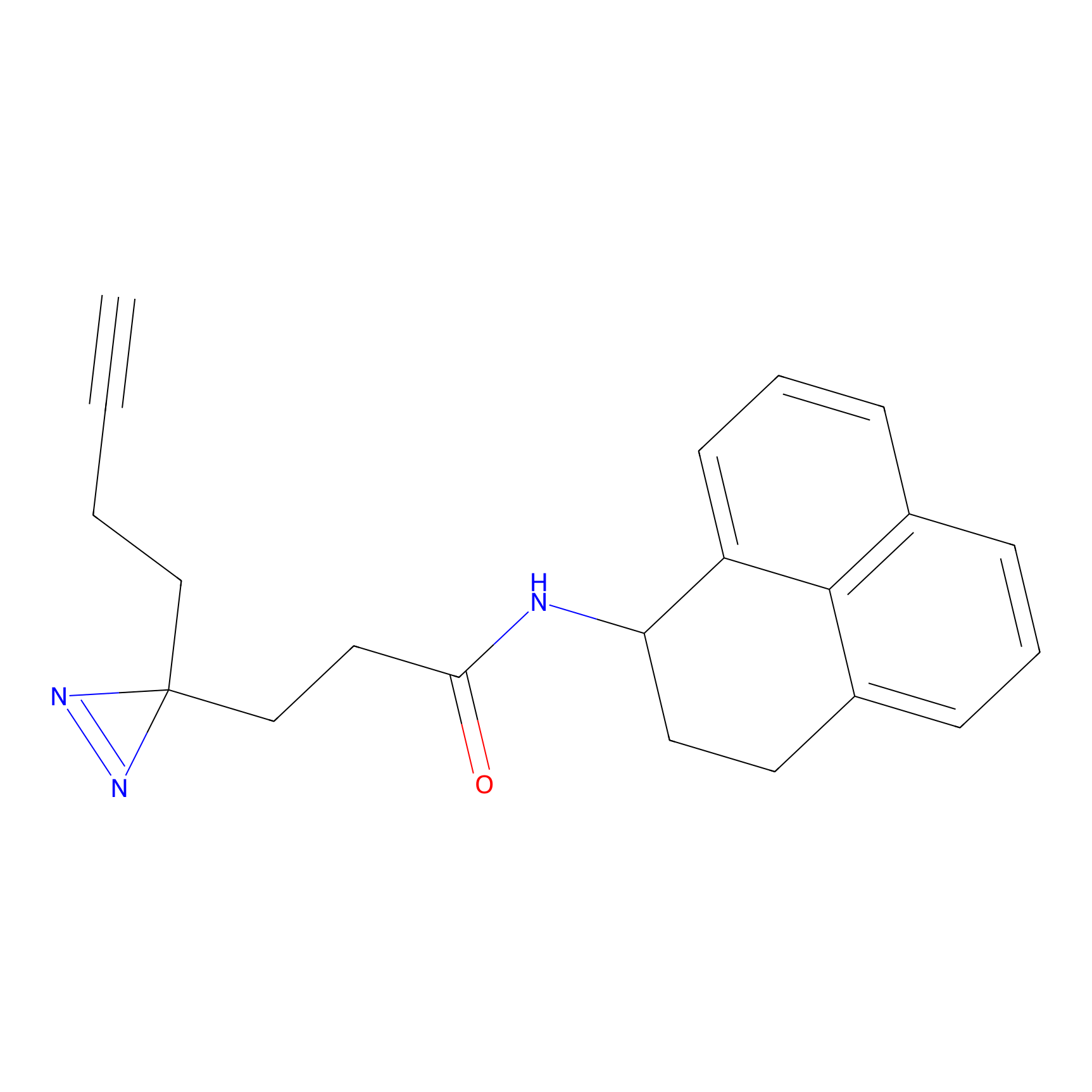
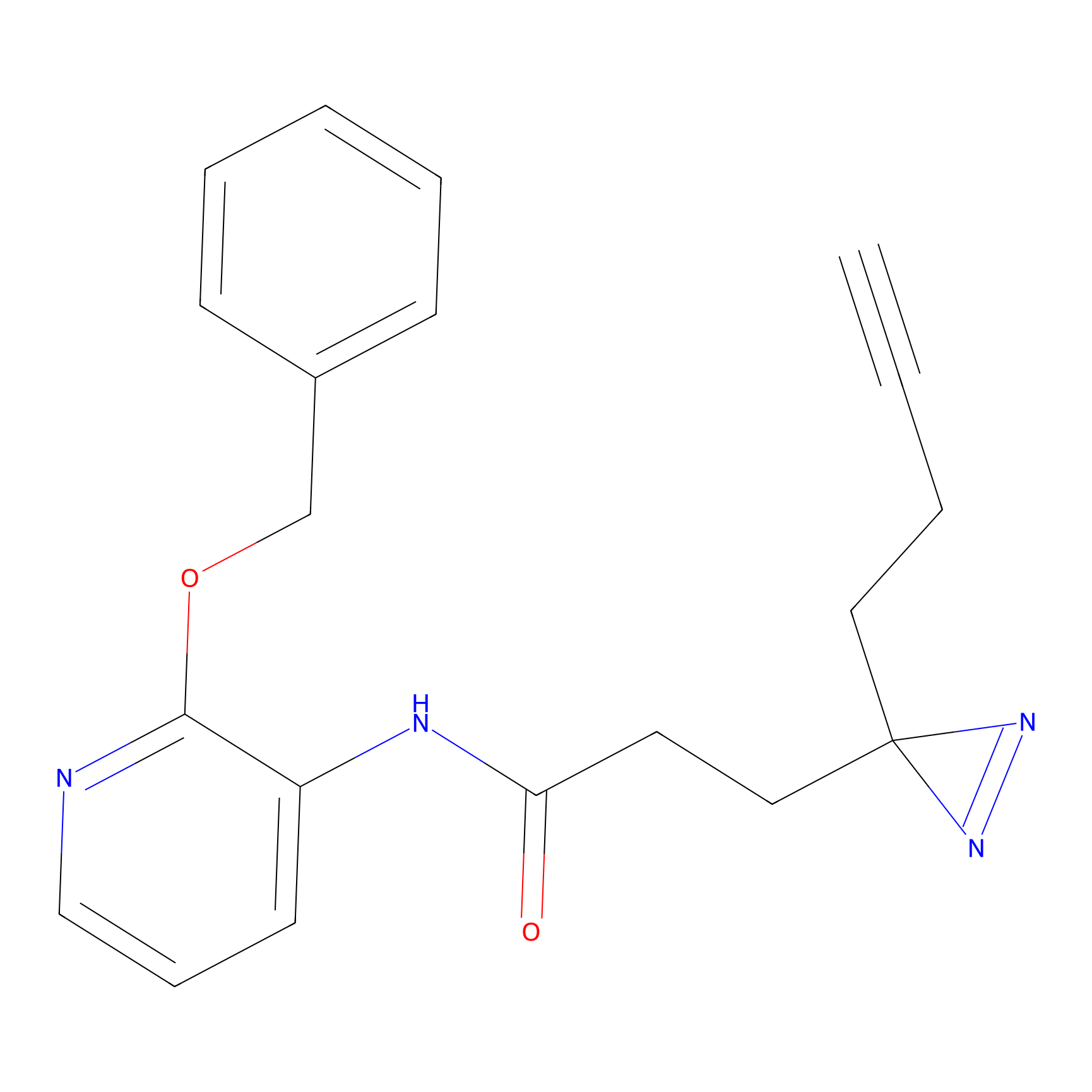
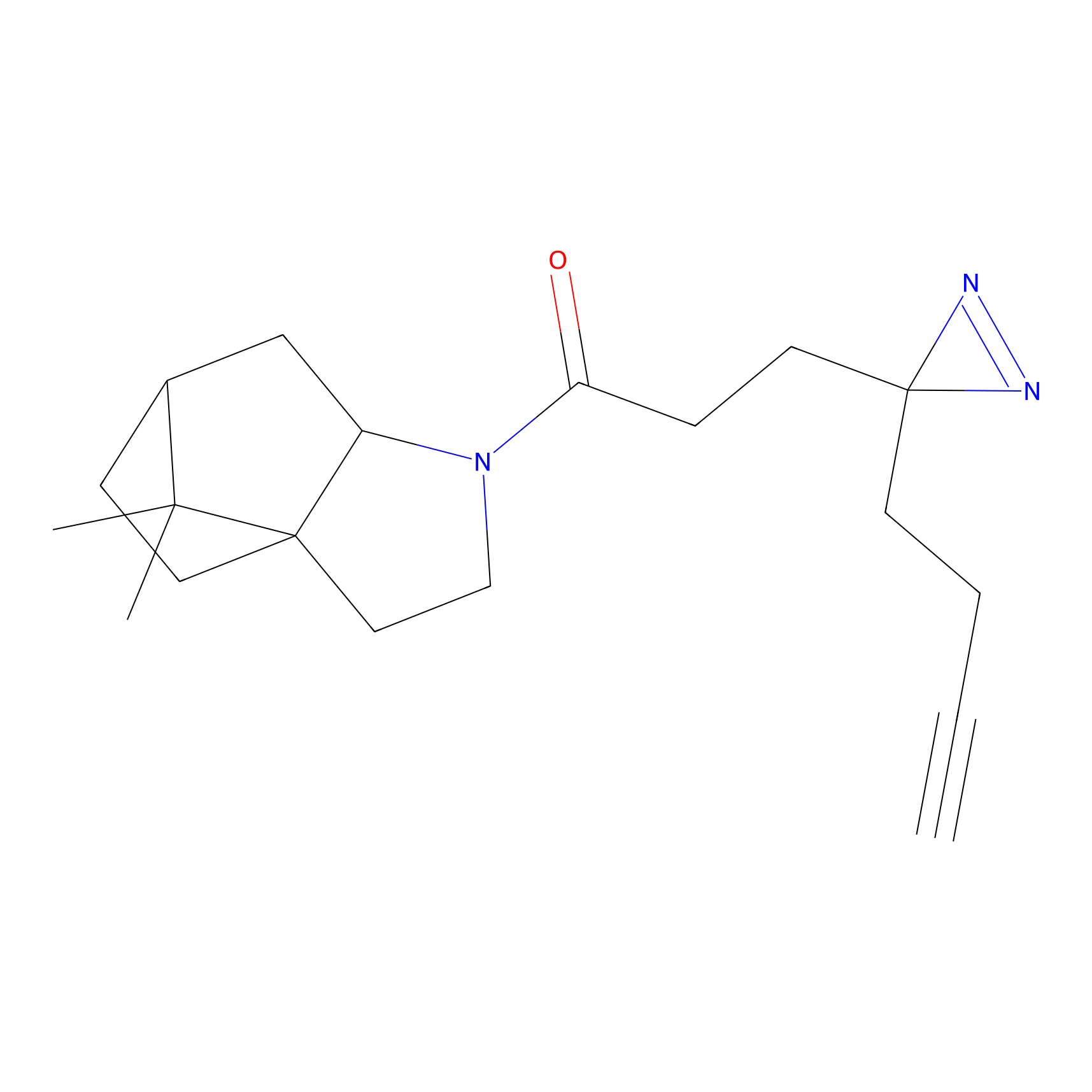
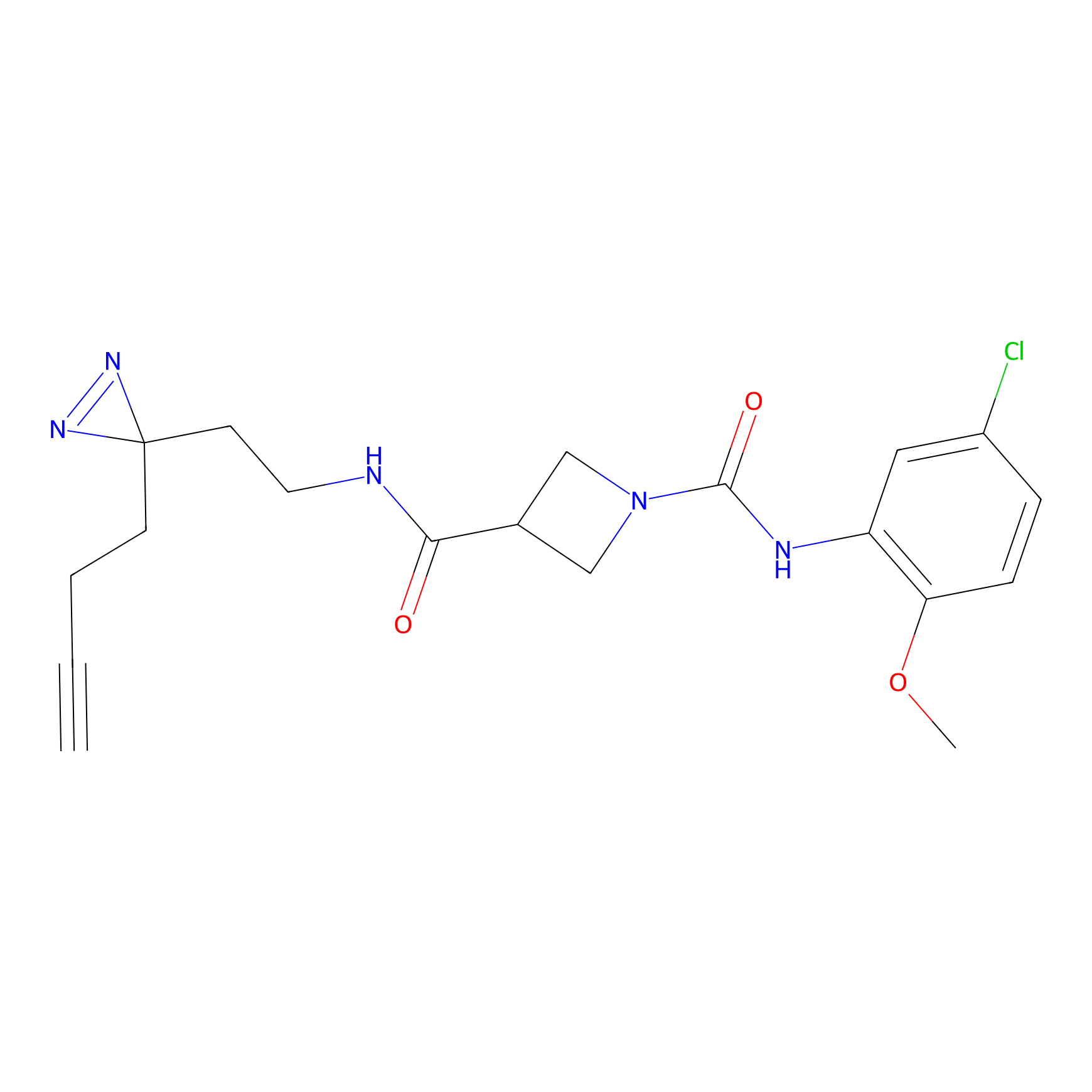
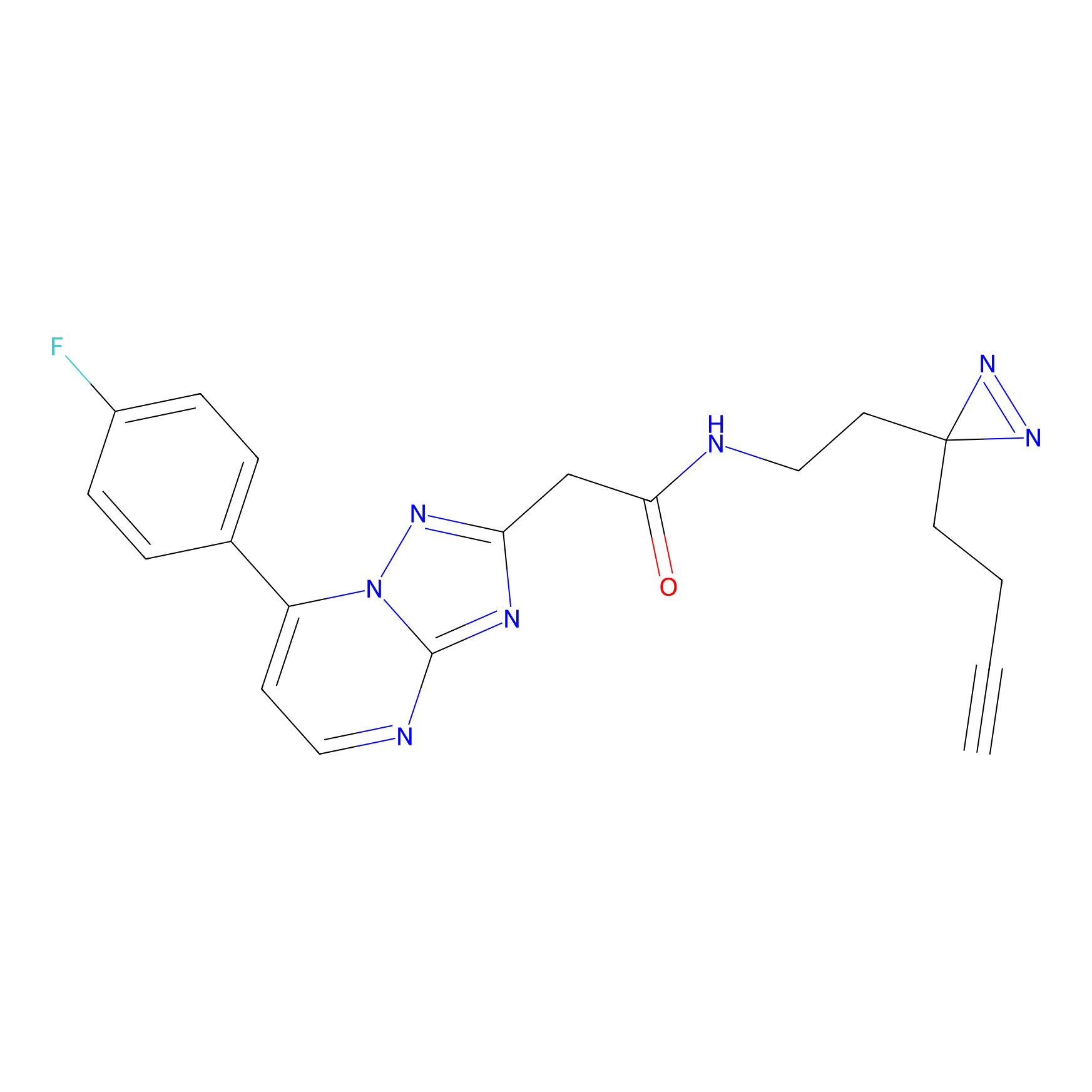
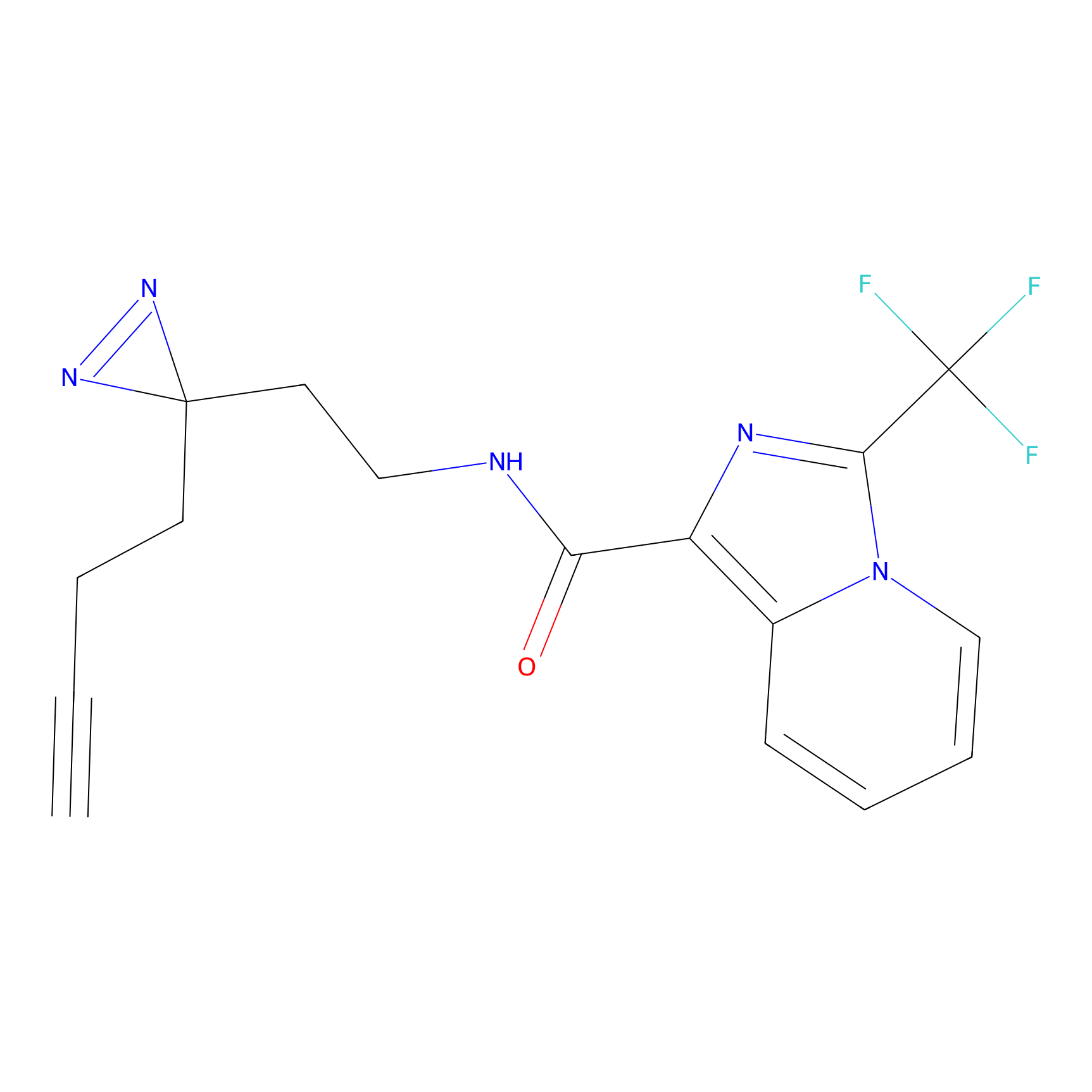
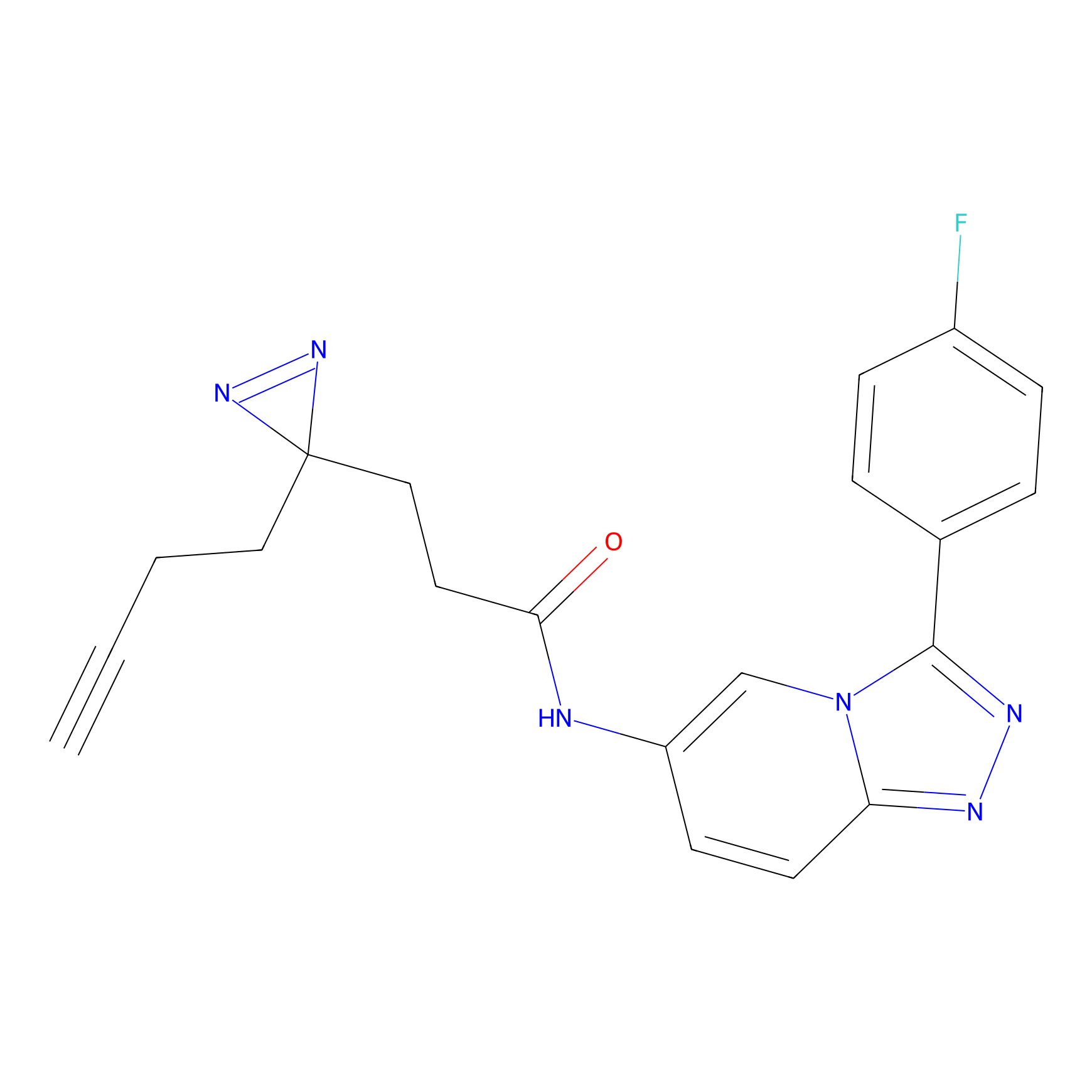
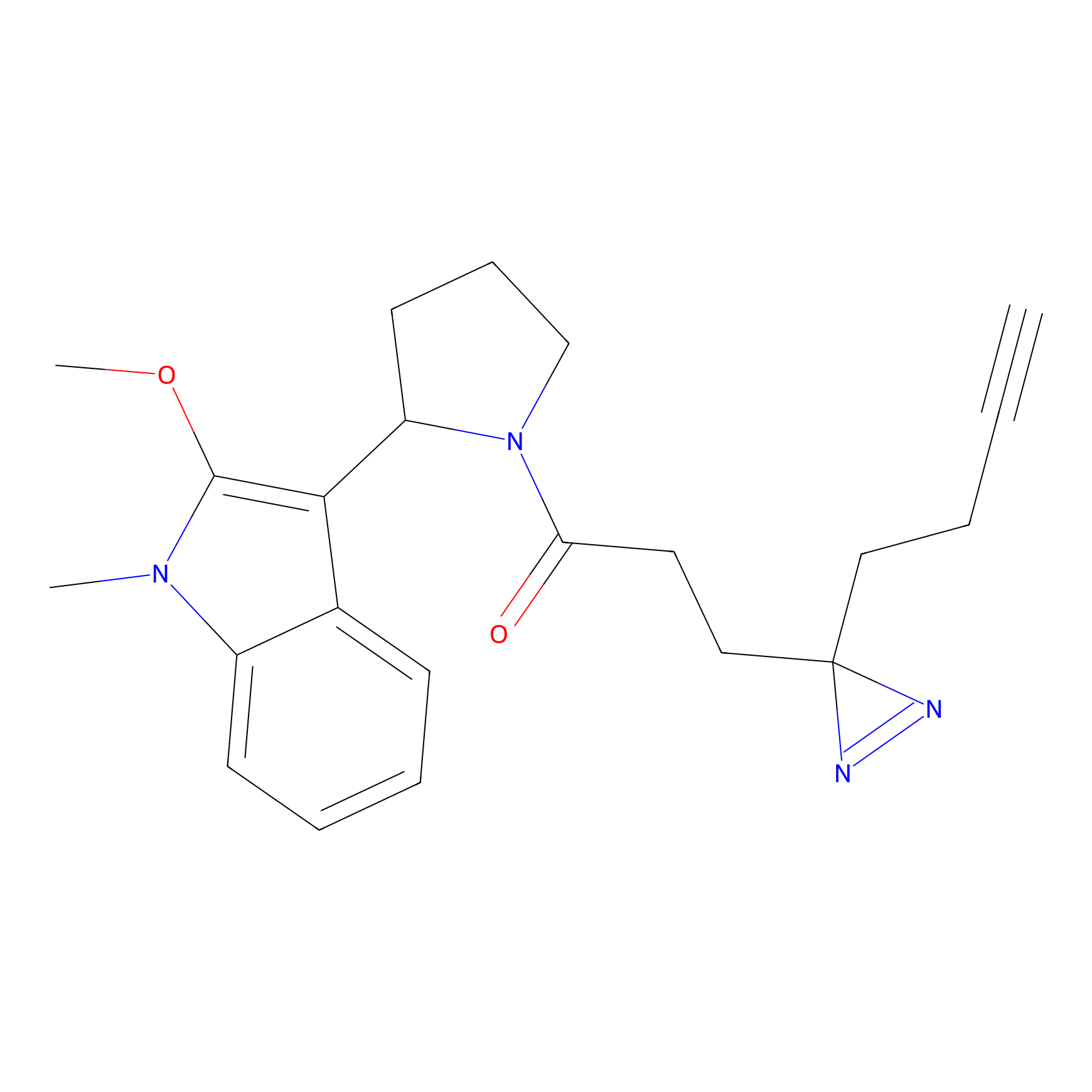
CHEMBL5175495 Probe Info |
 |
5.77 | LDD0196 | [2] | |
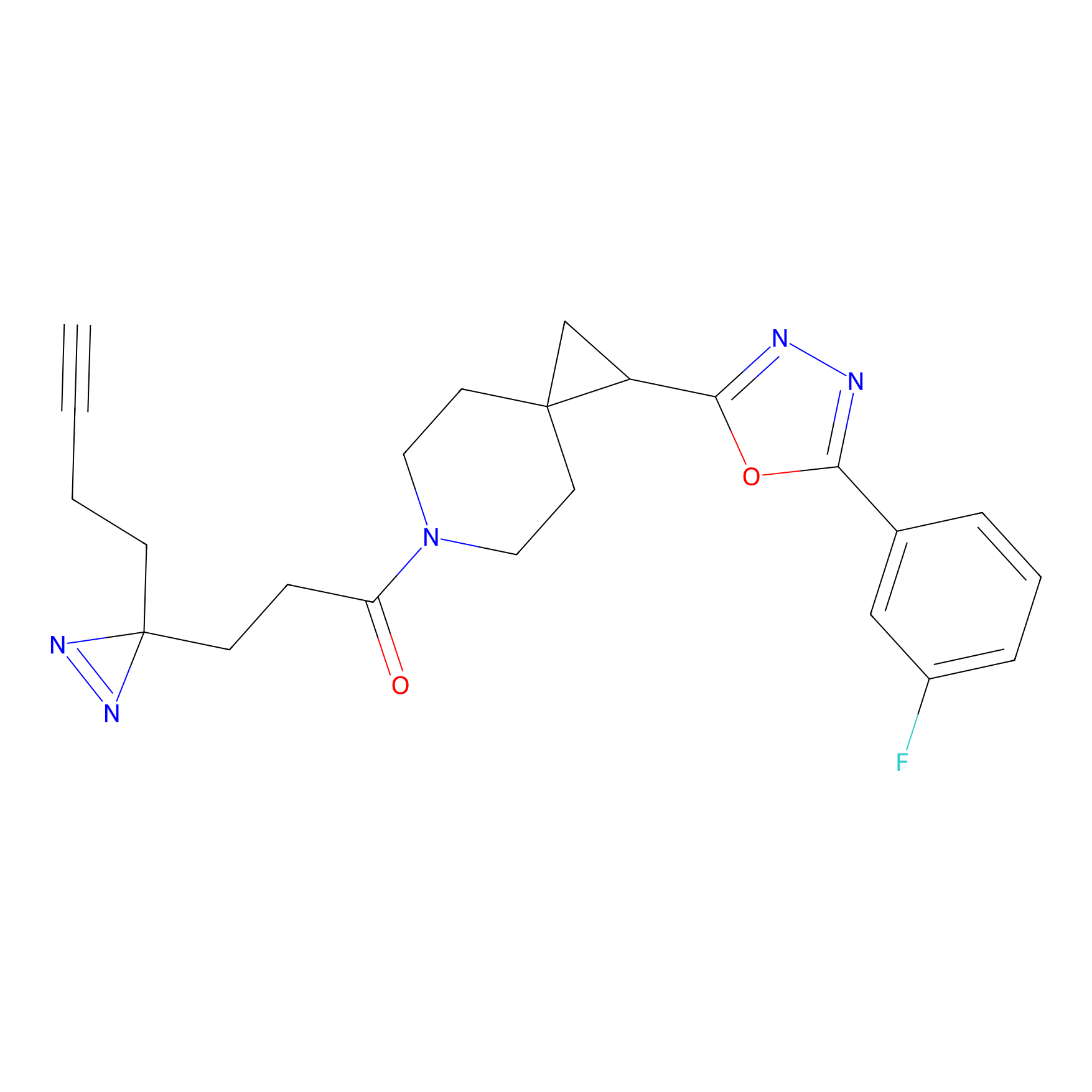
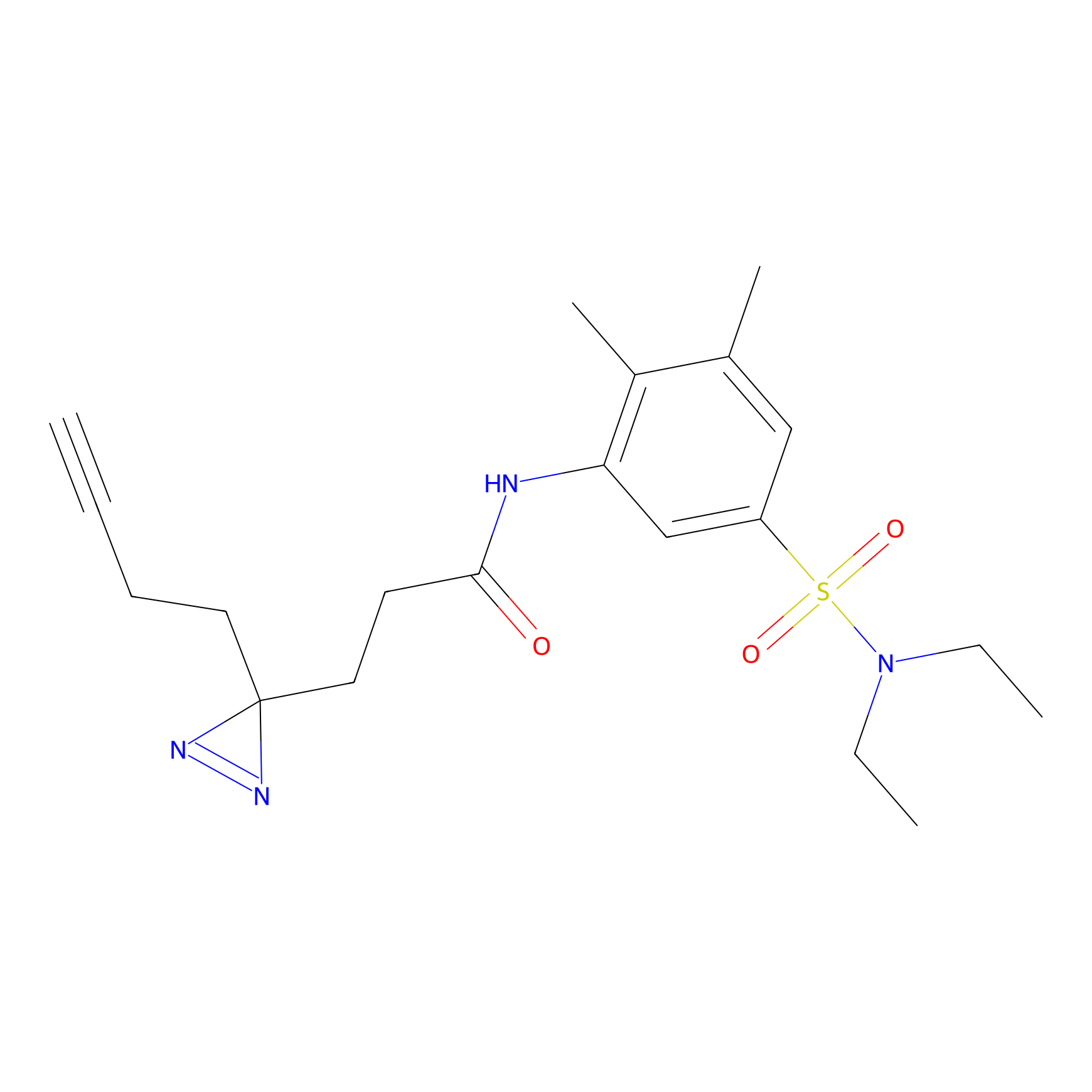
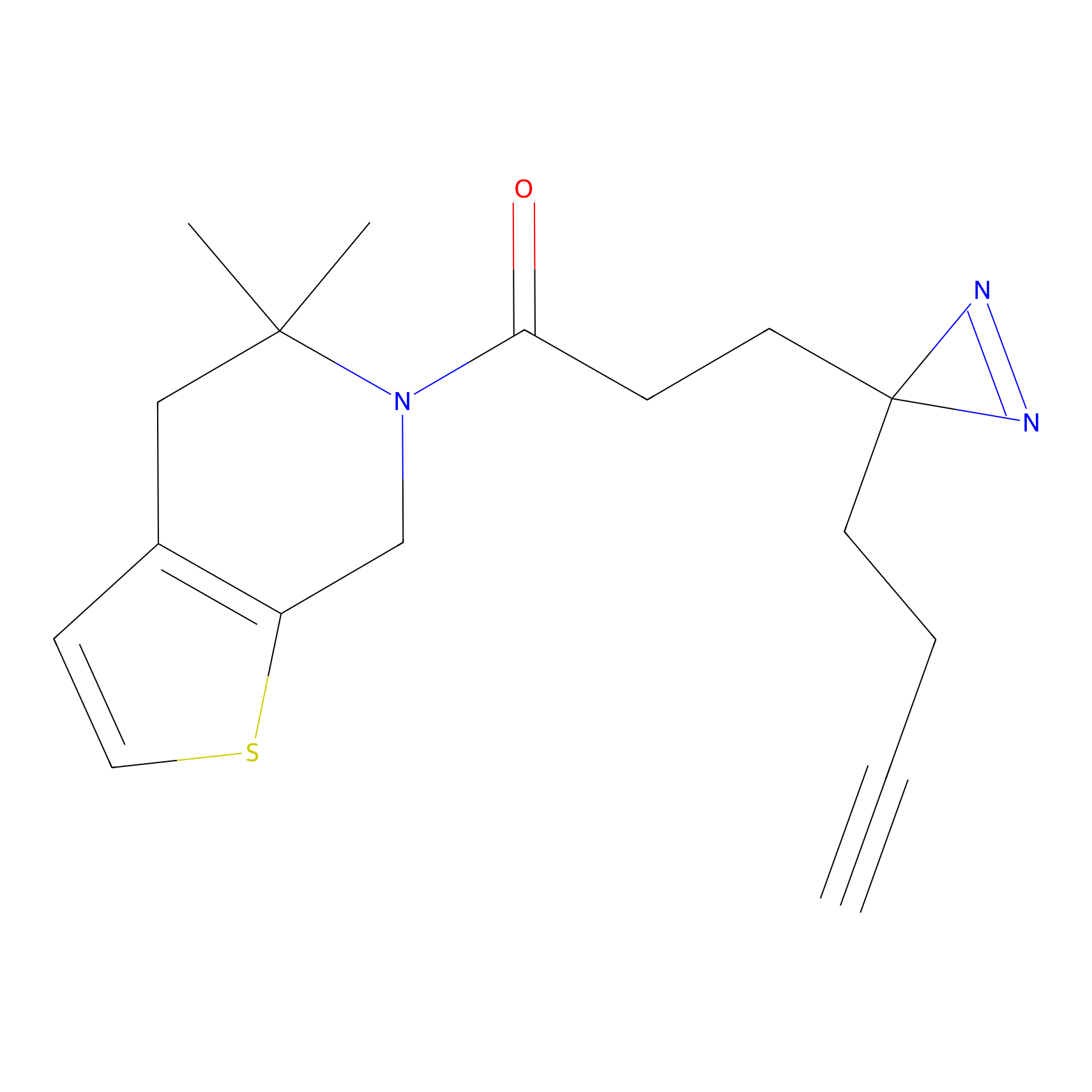
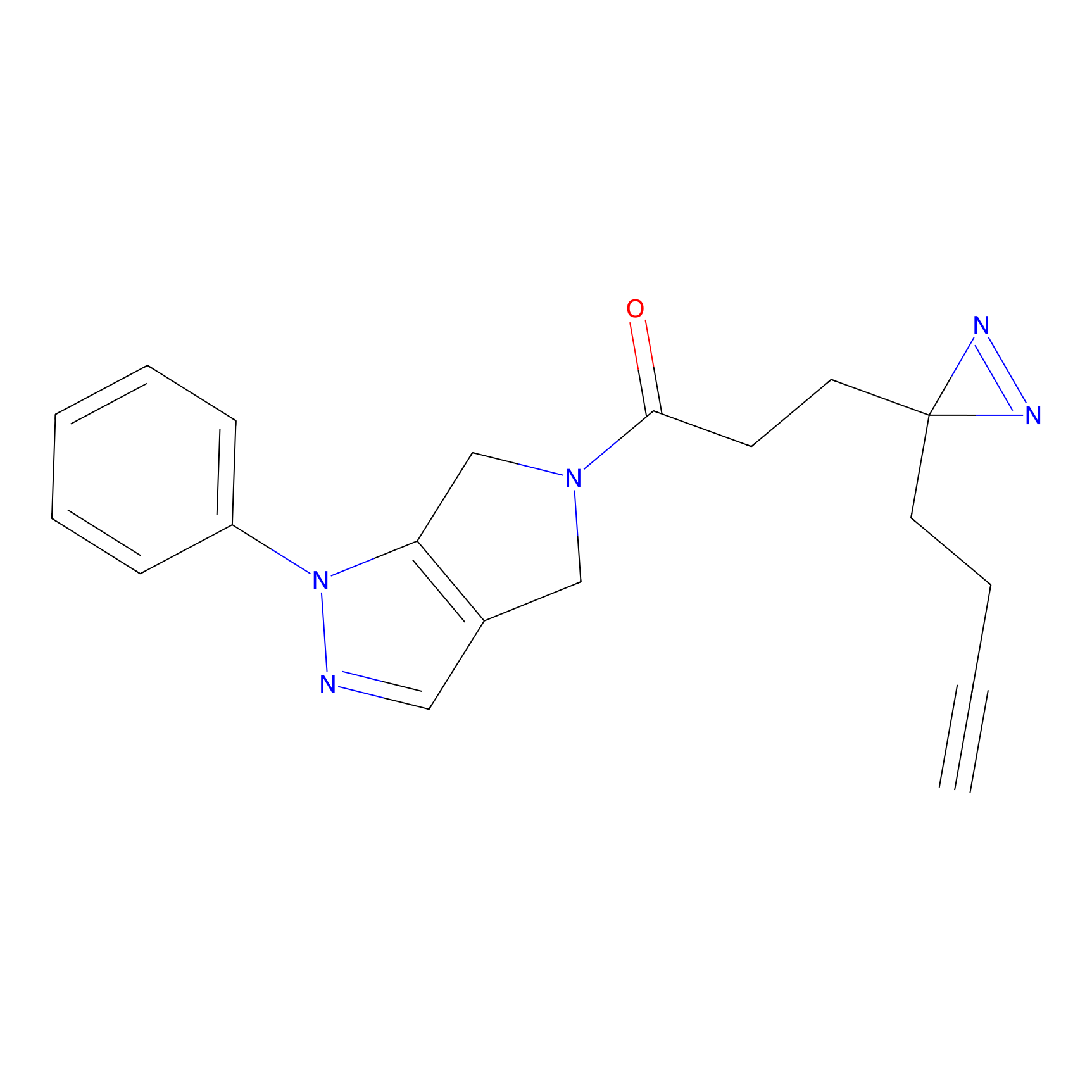
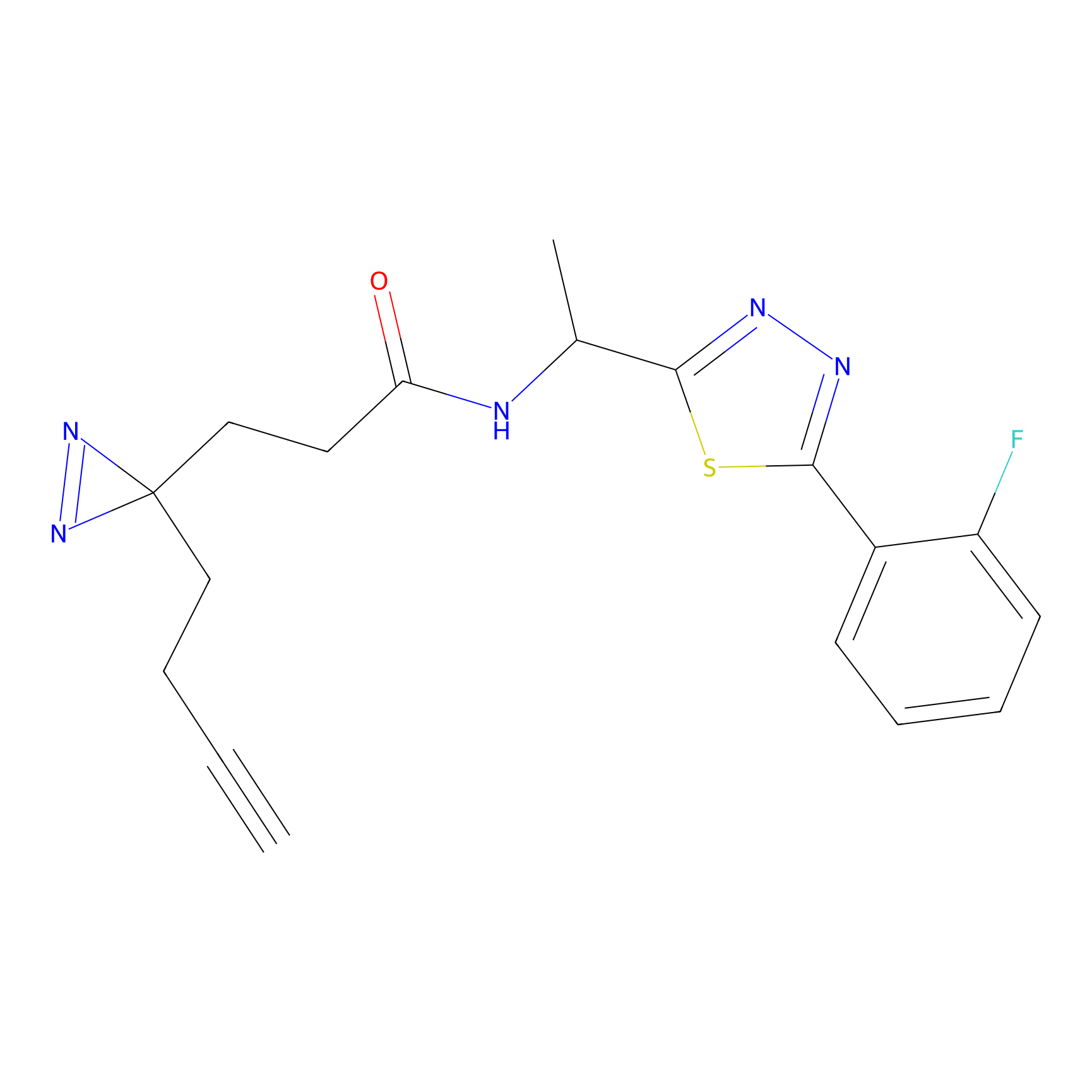
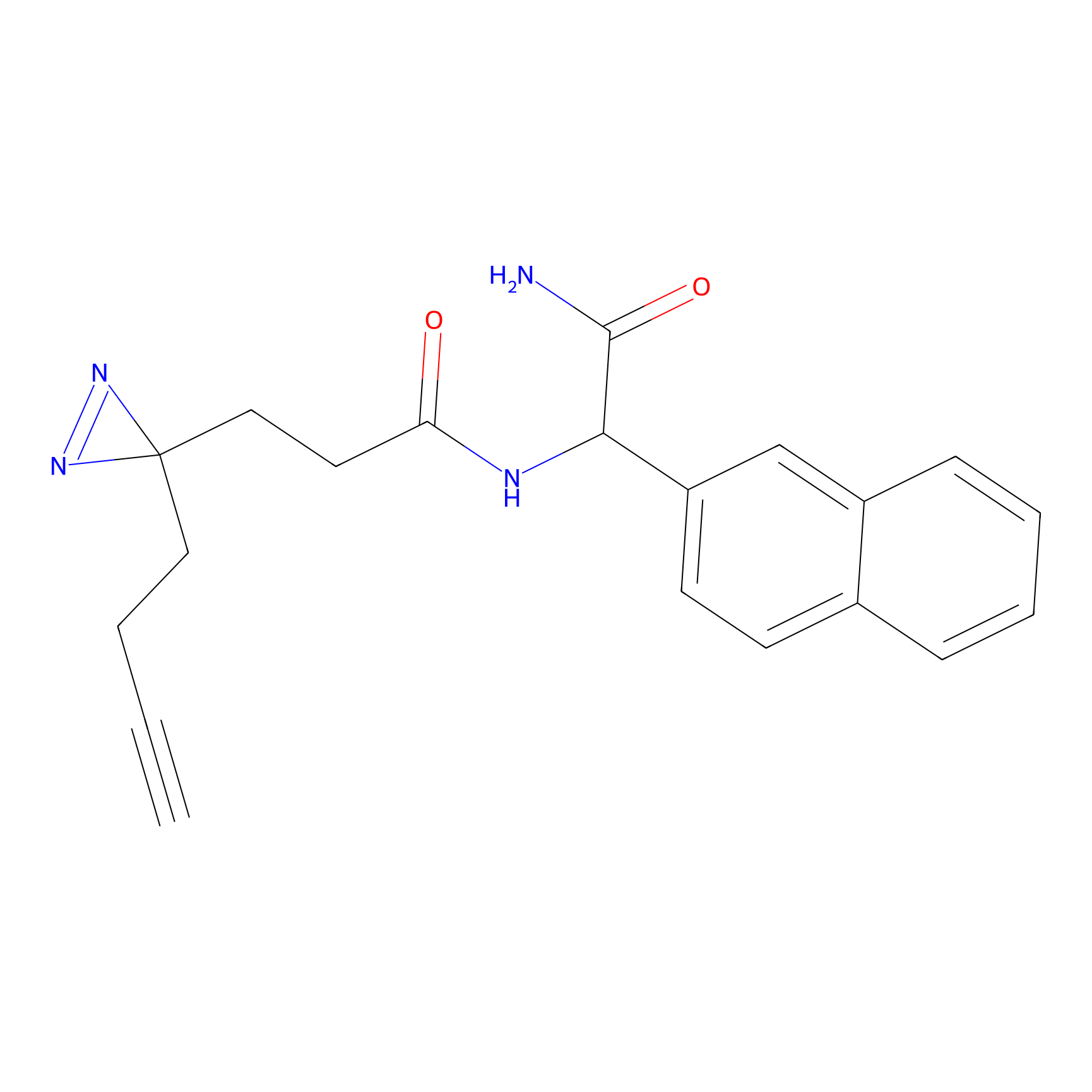
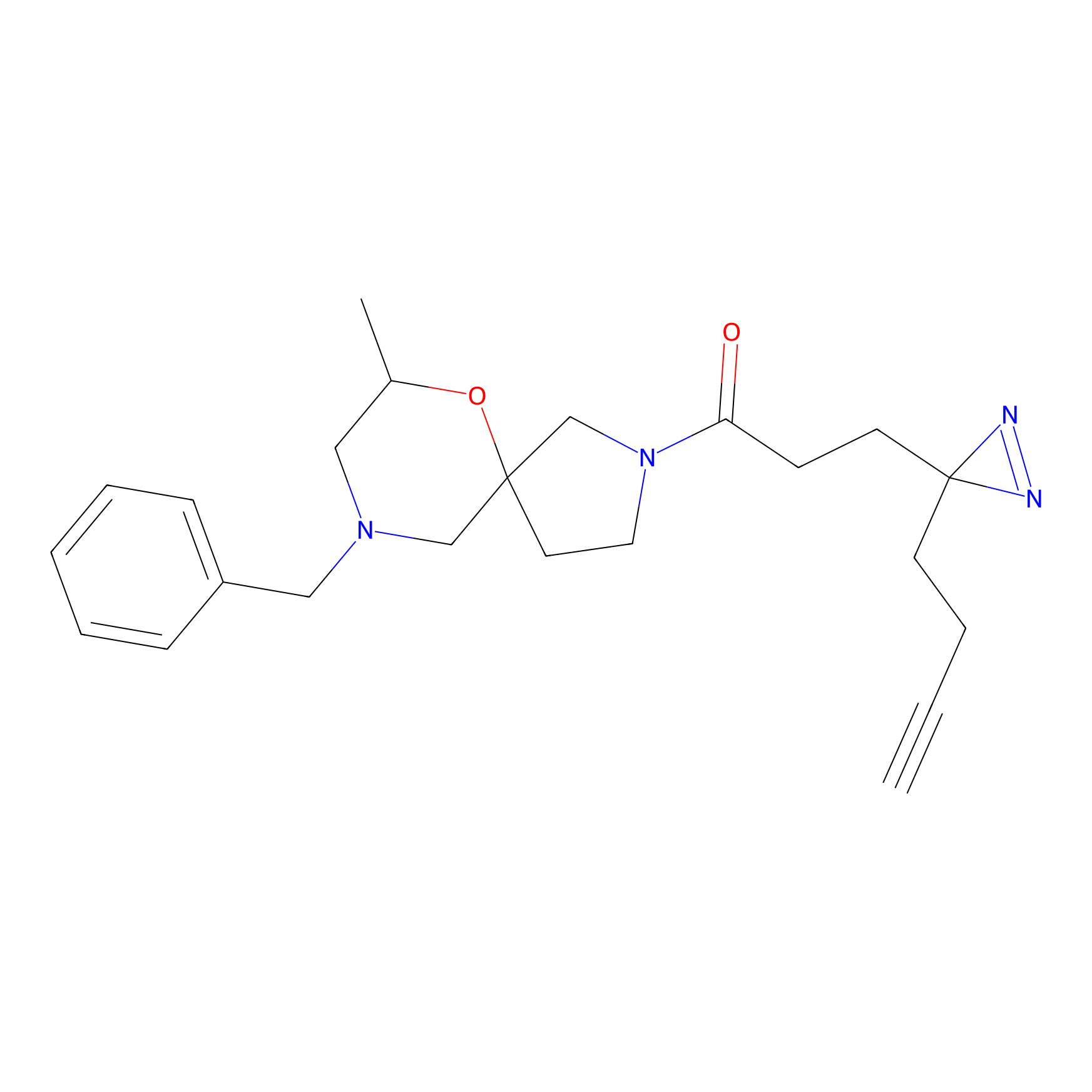
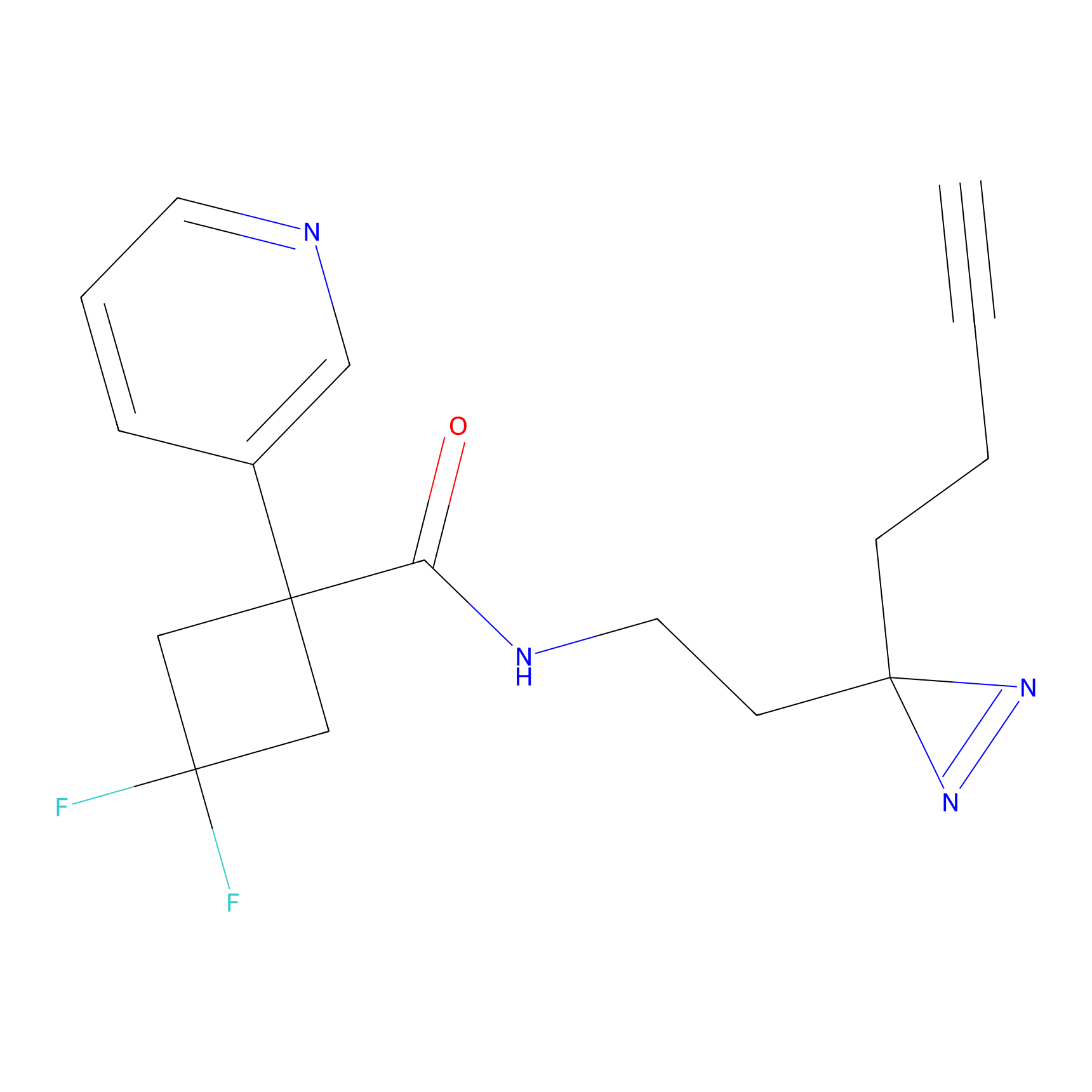
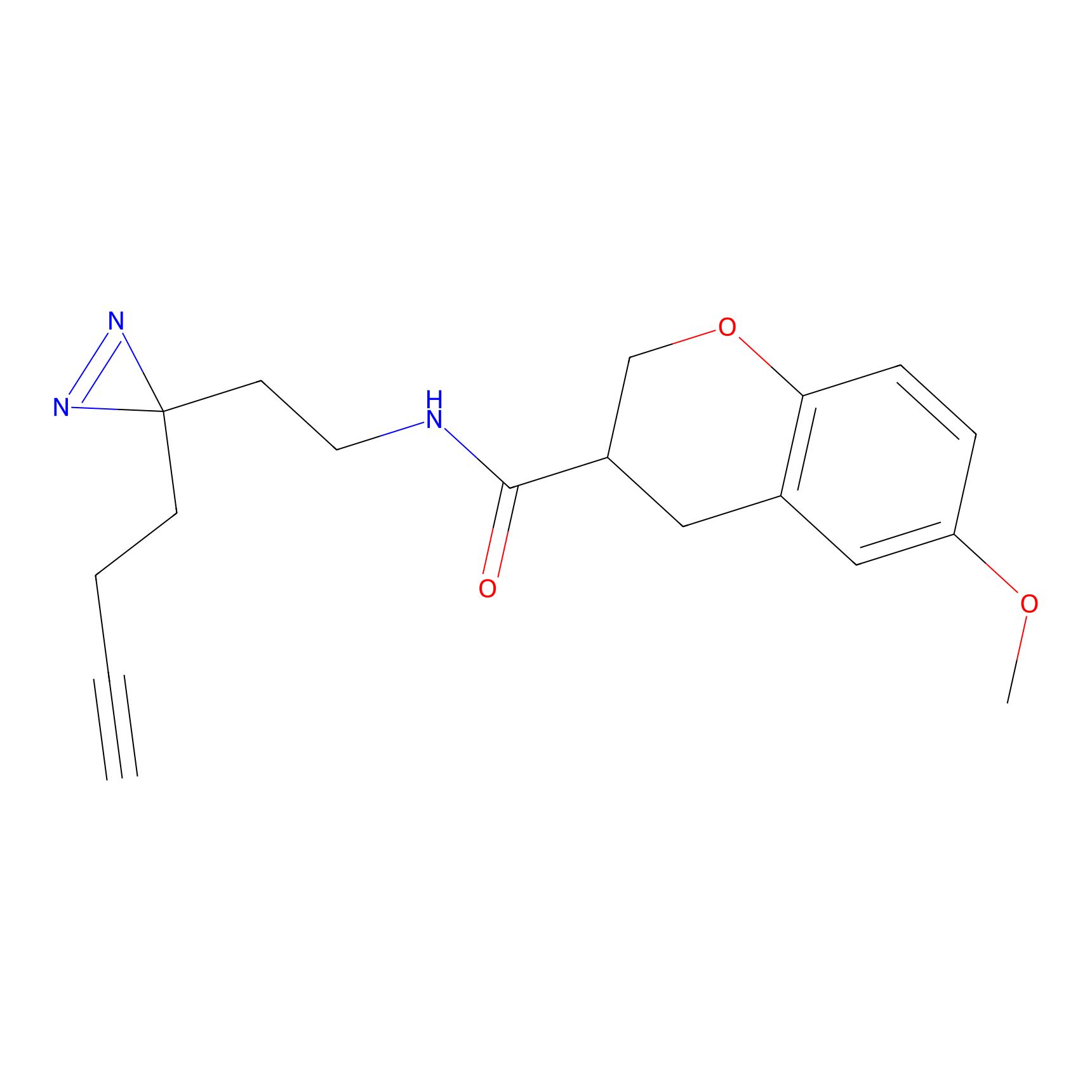
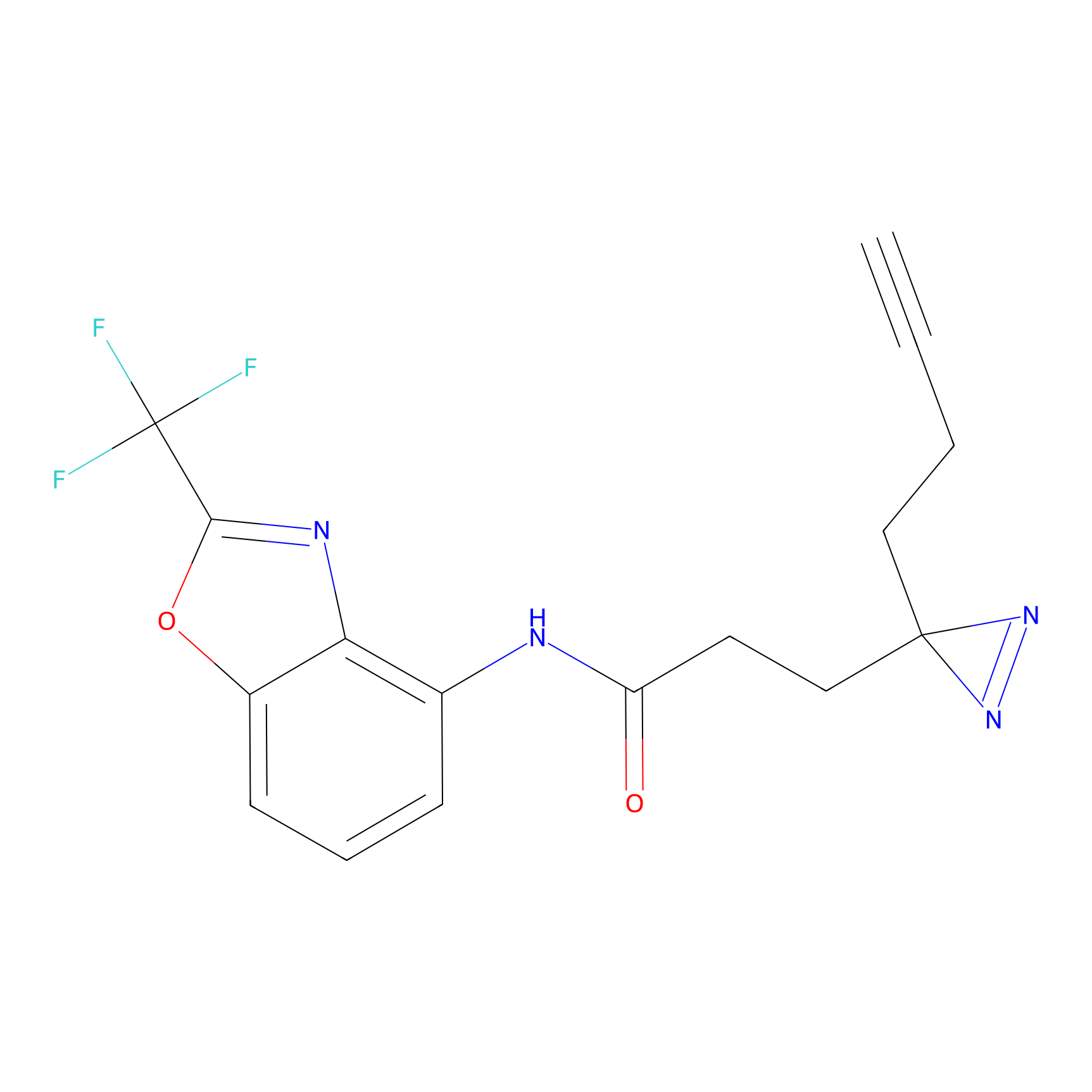
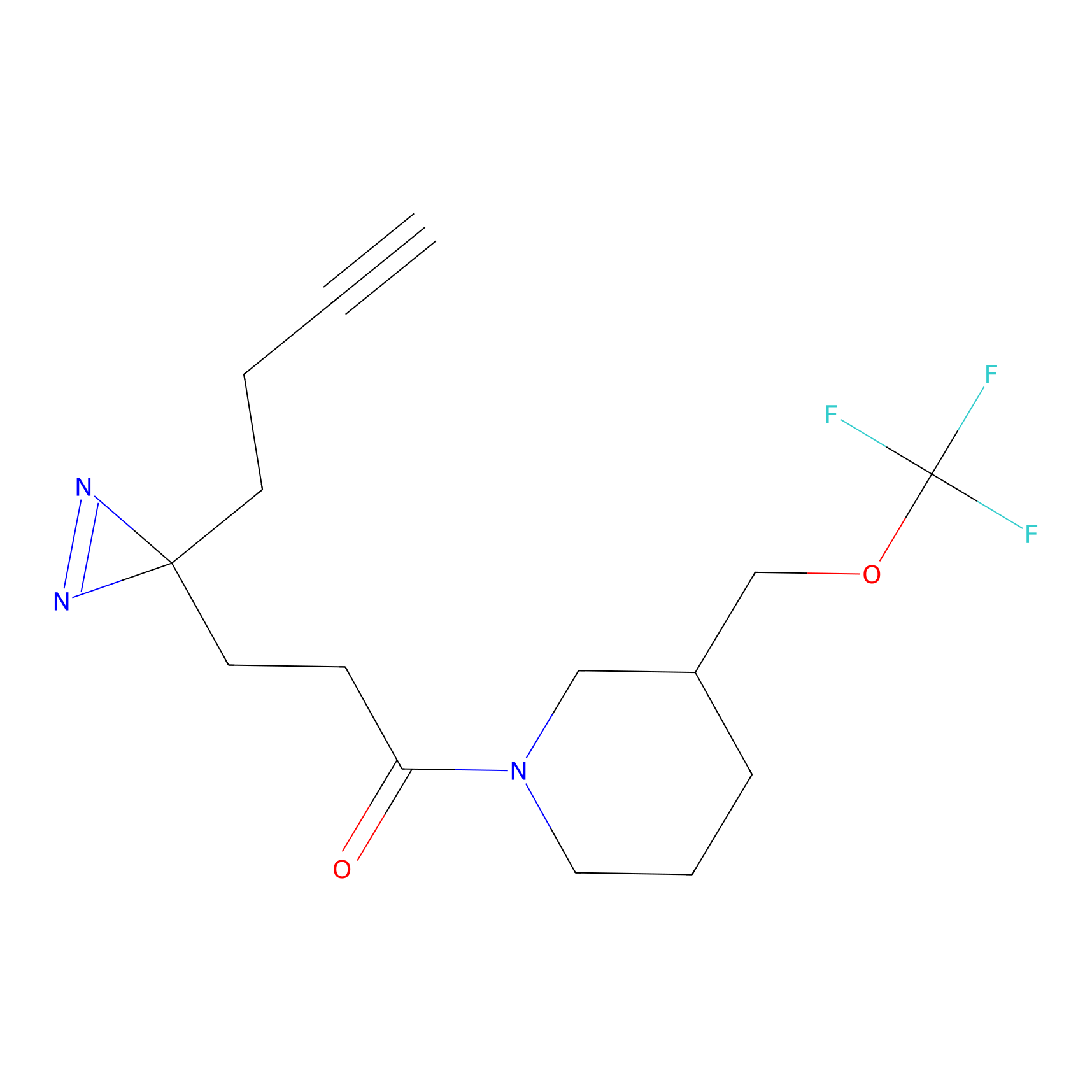
|
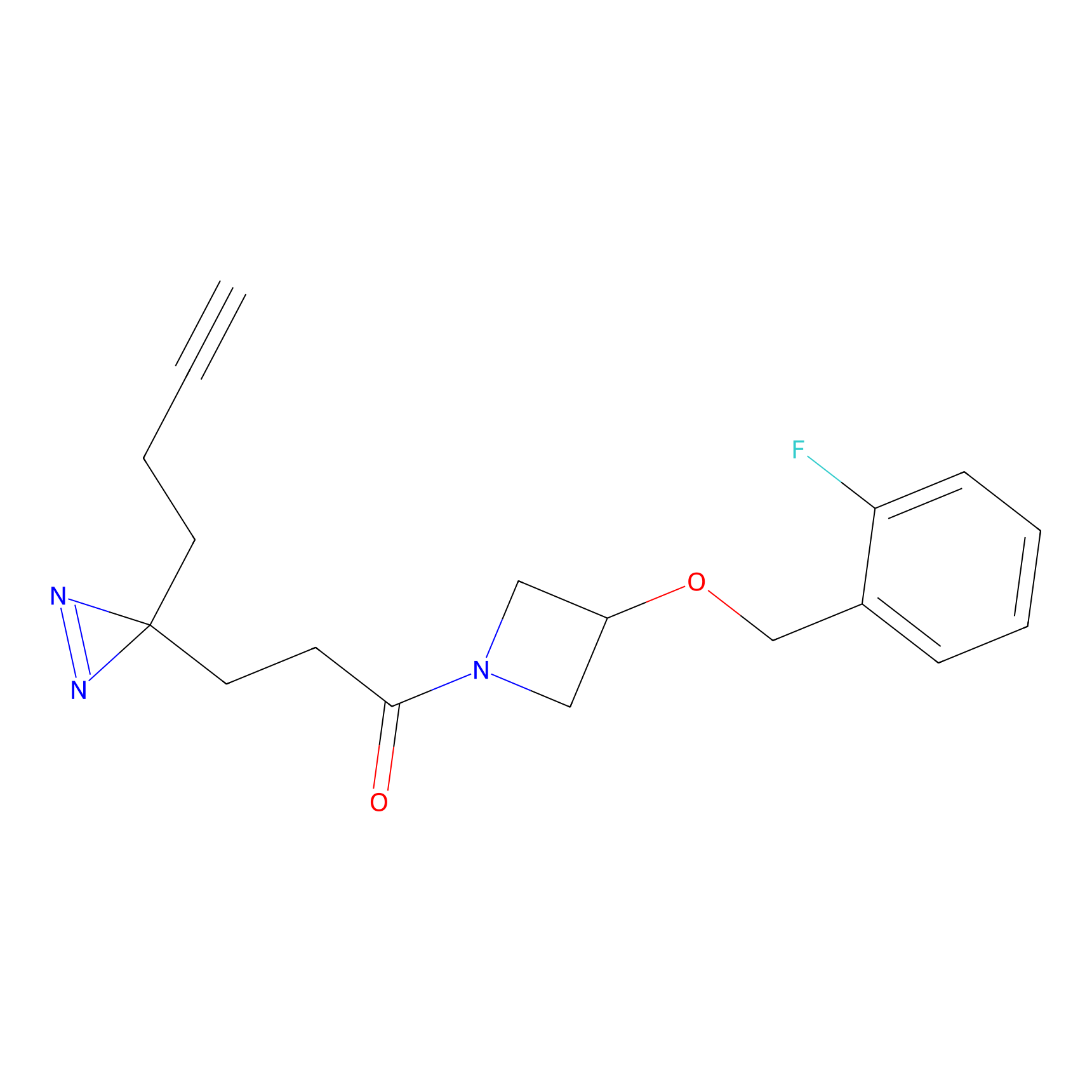
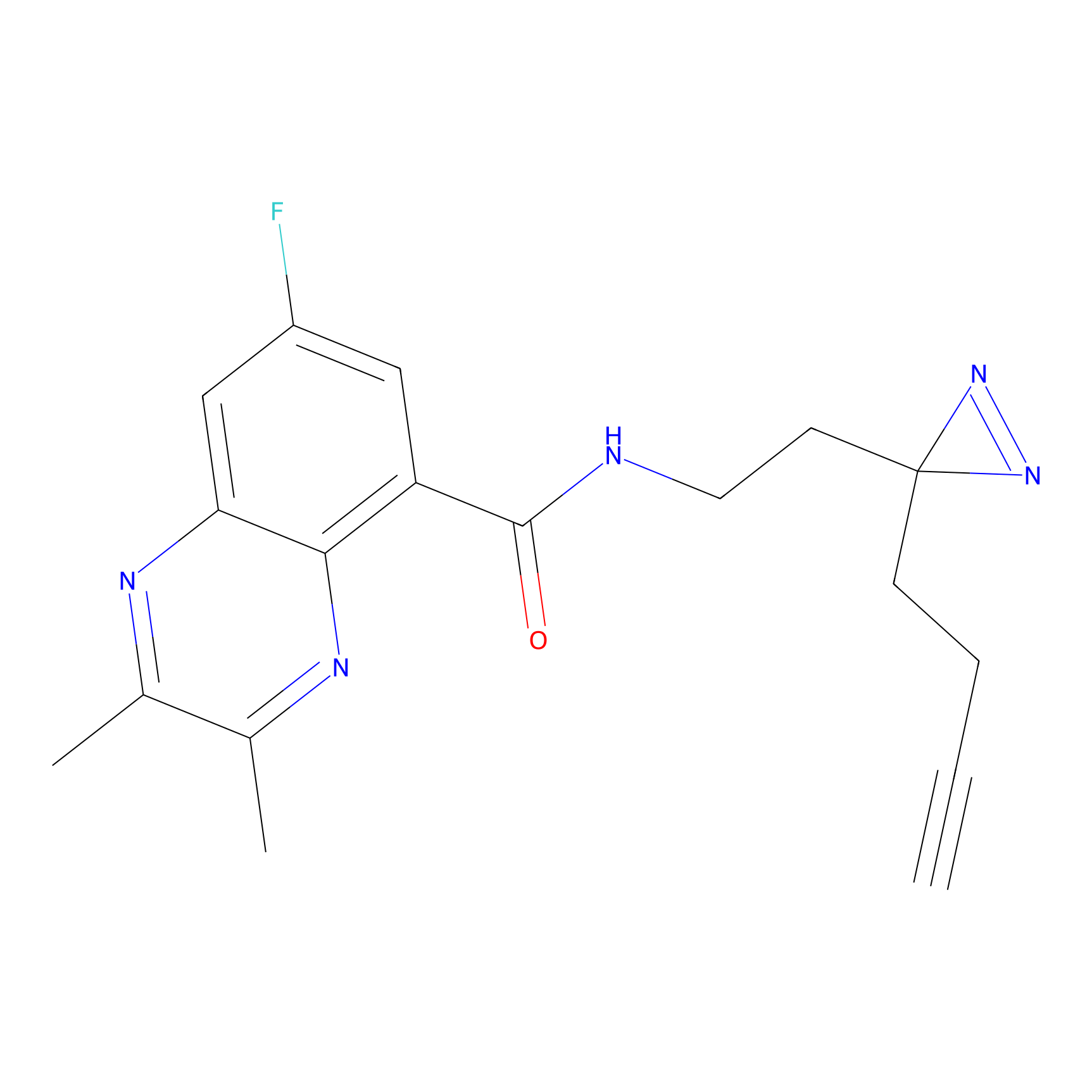
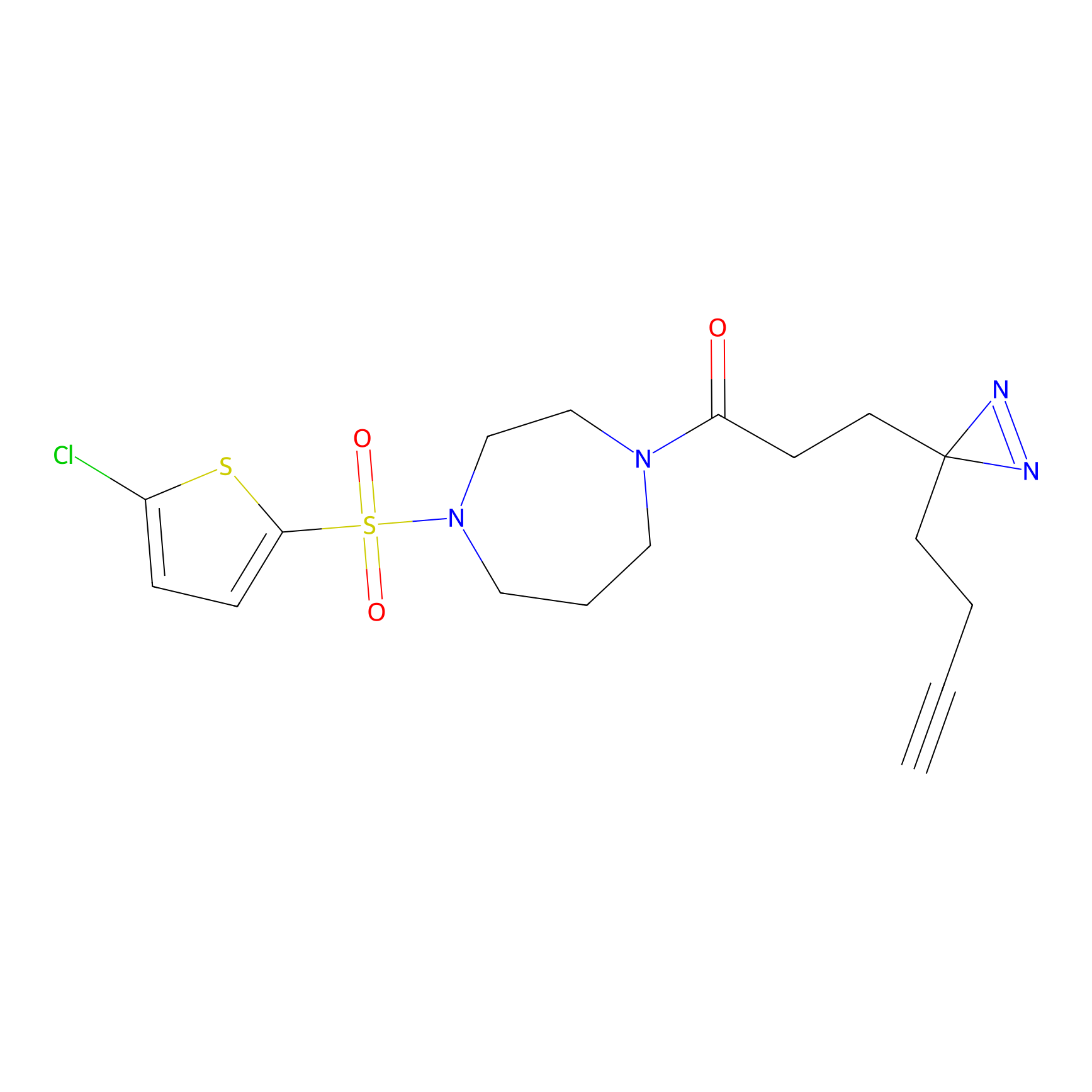
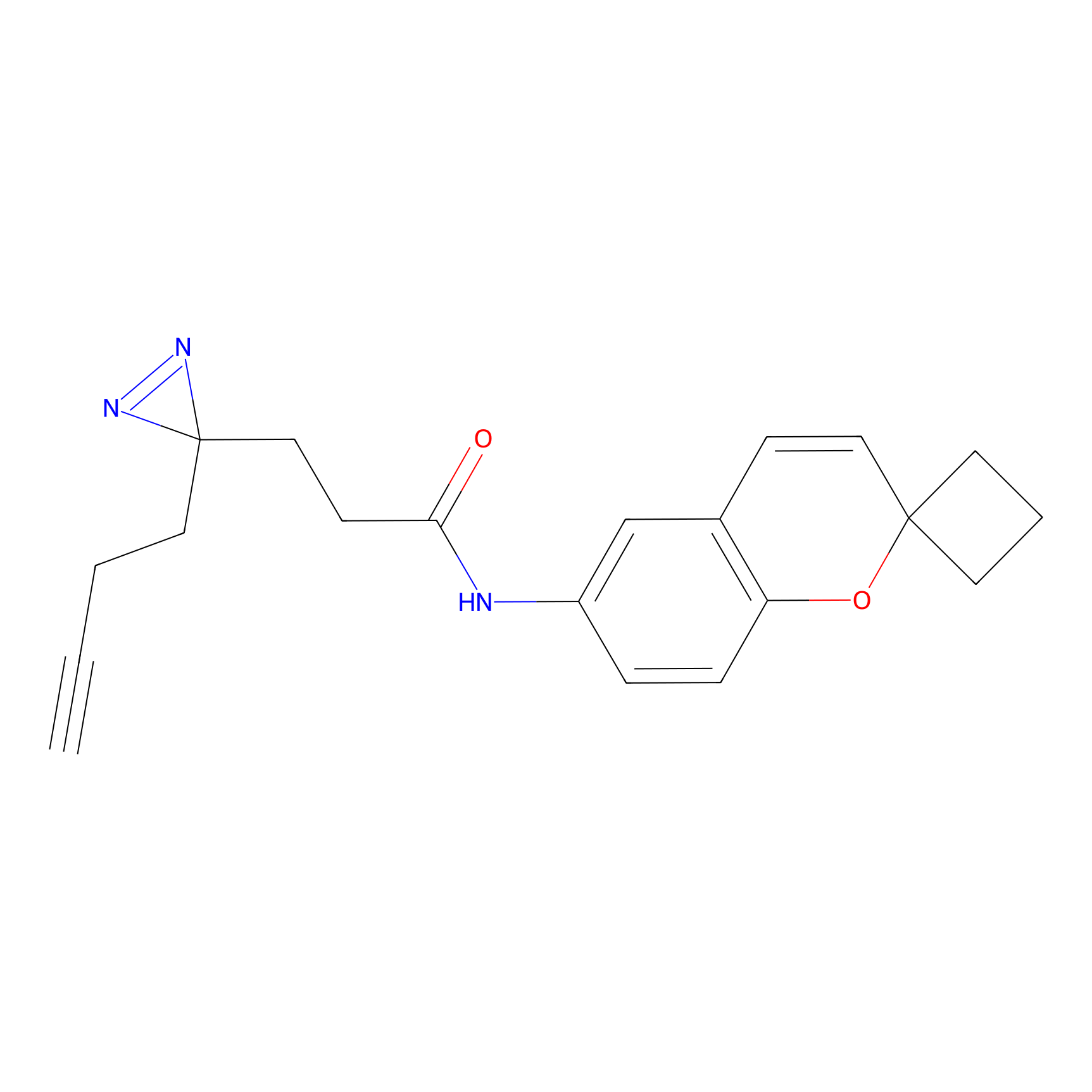
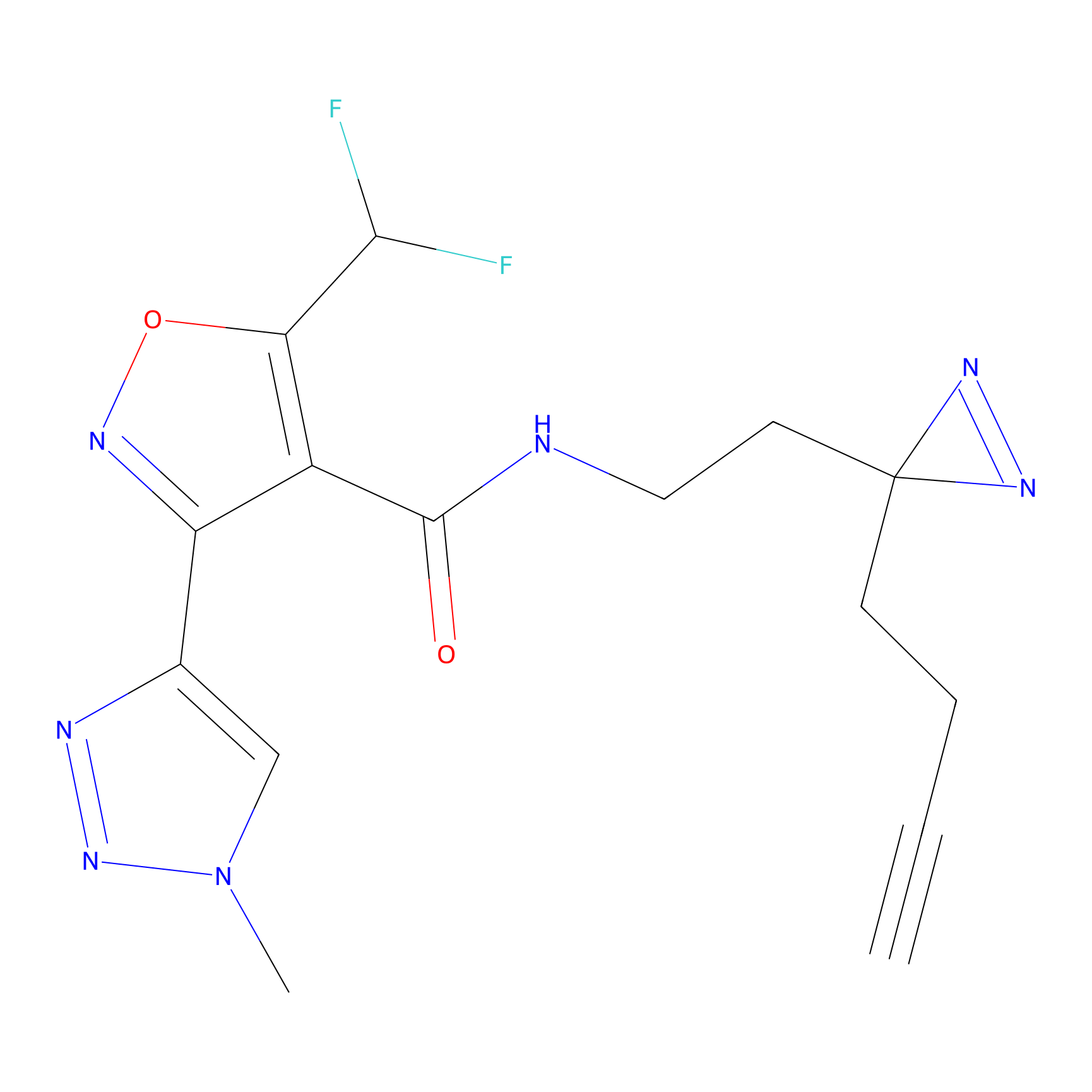
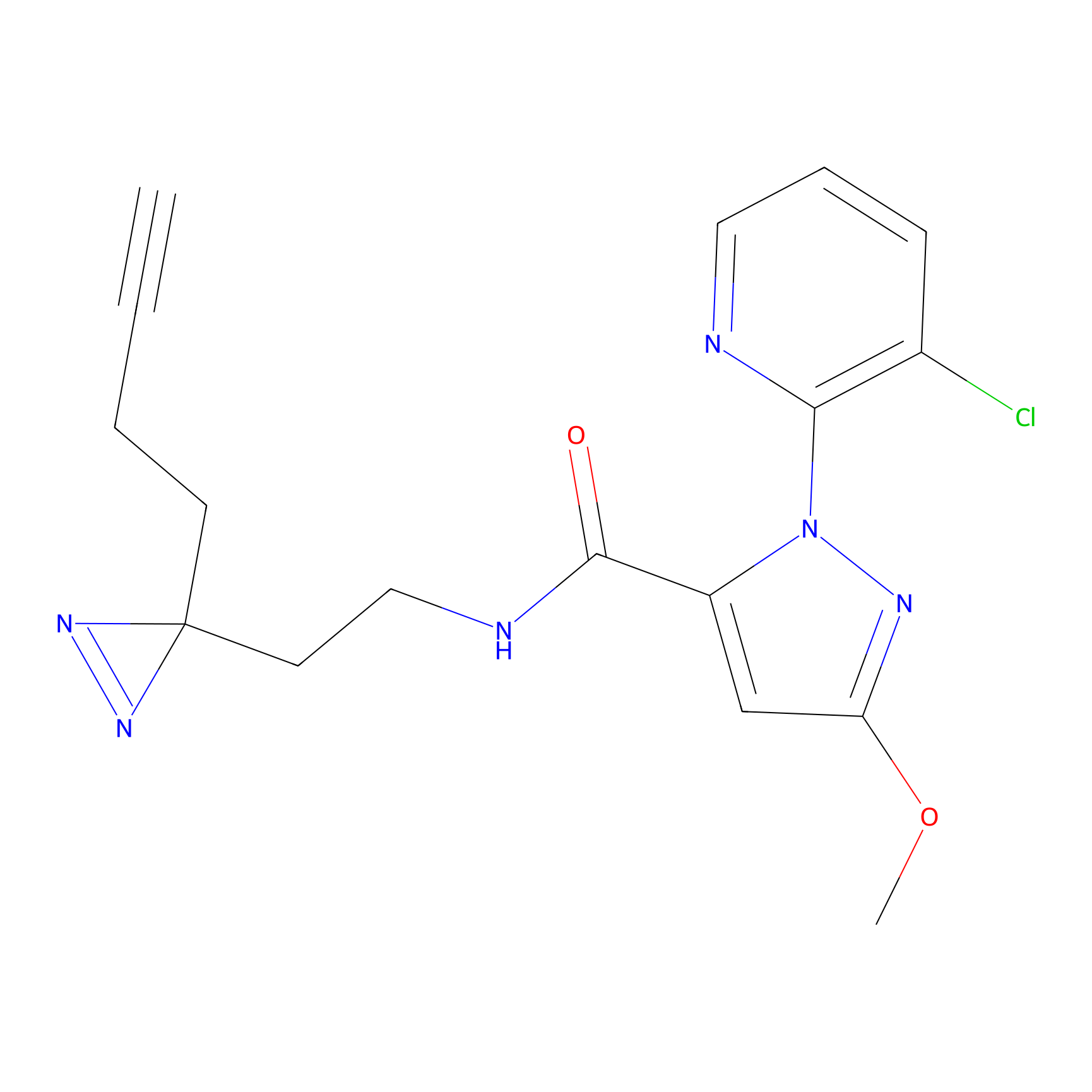
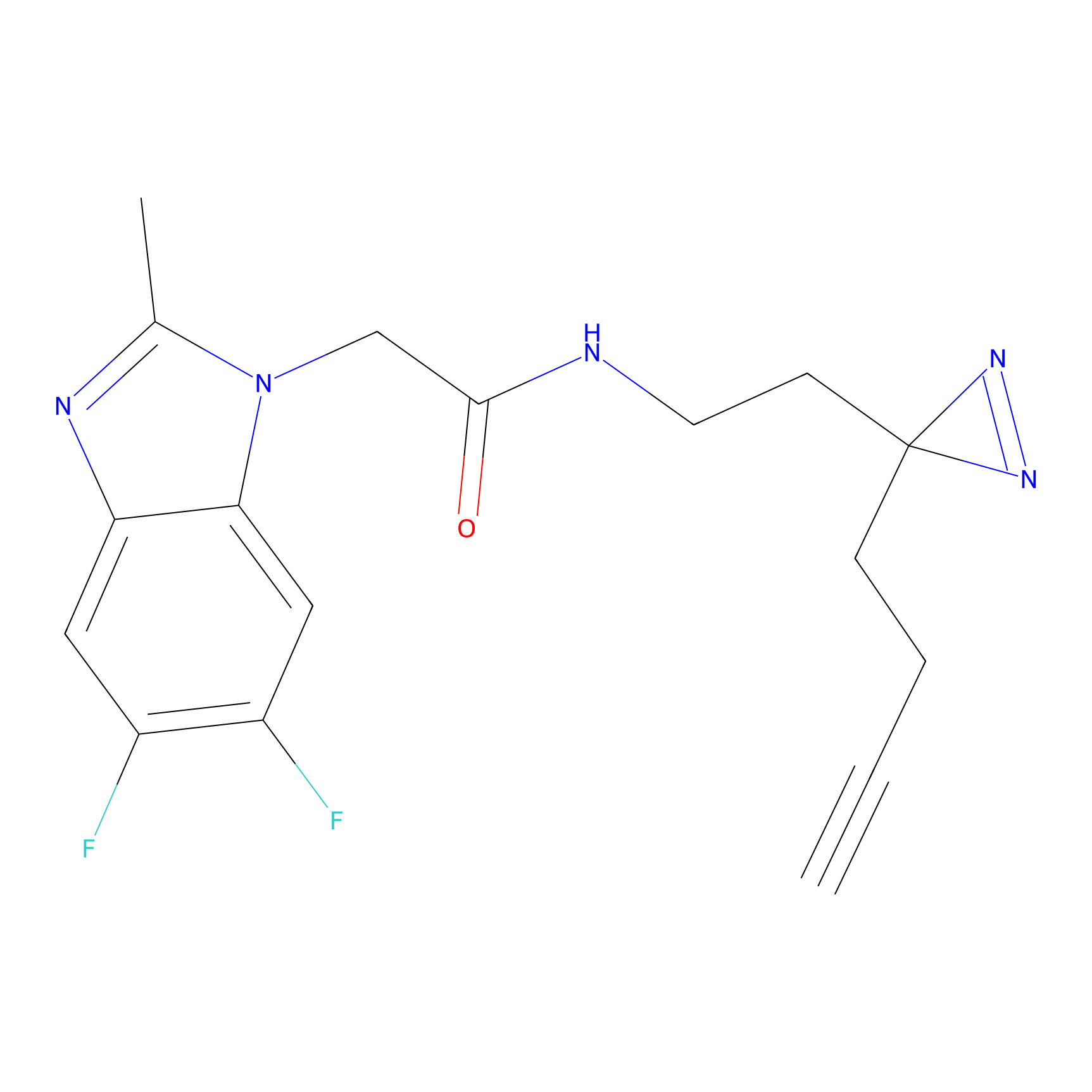
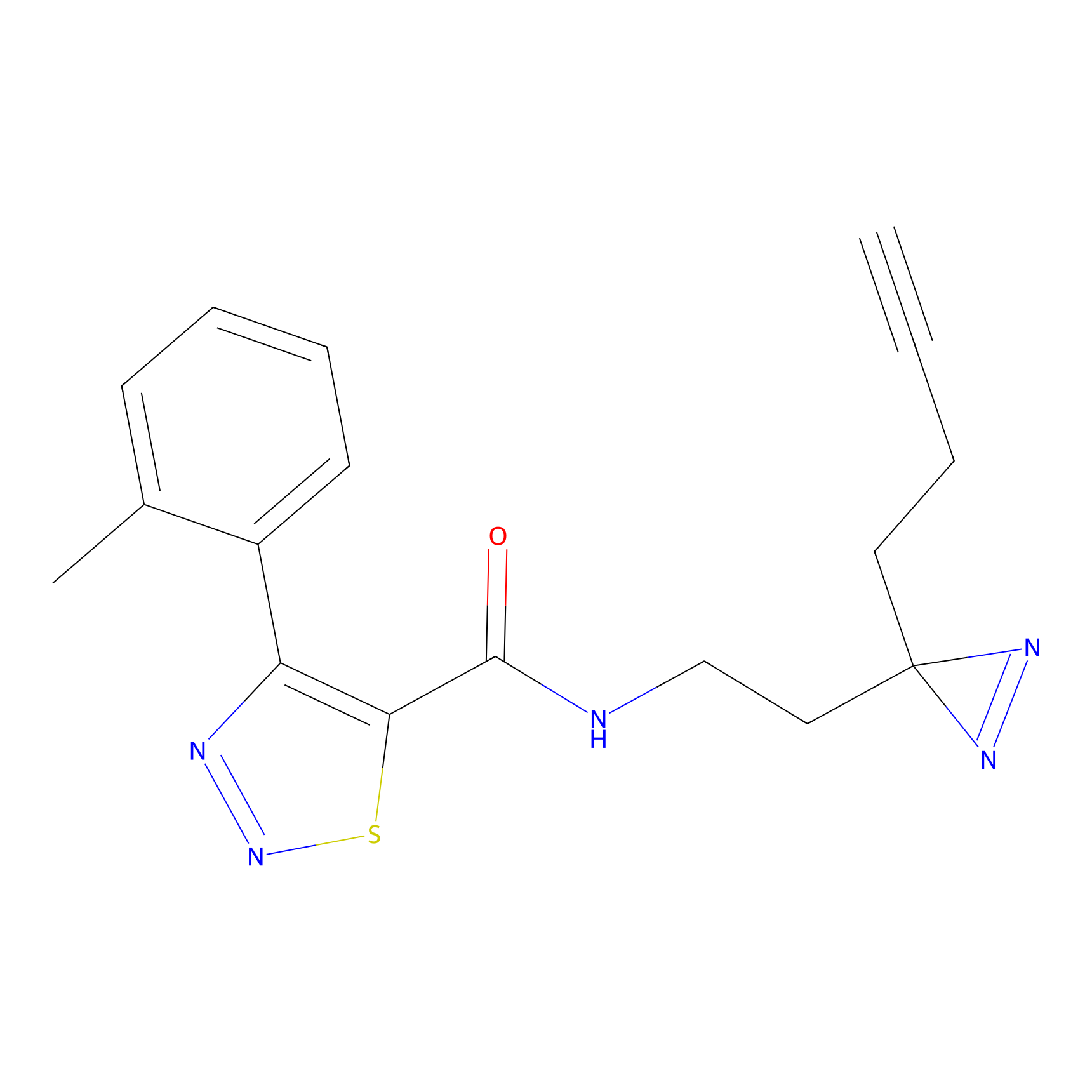
N1 Probe Info |
 |
100.00 | LDD0242 | [3] | |
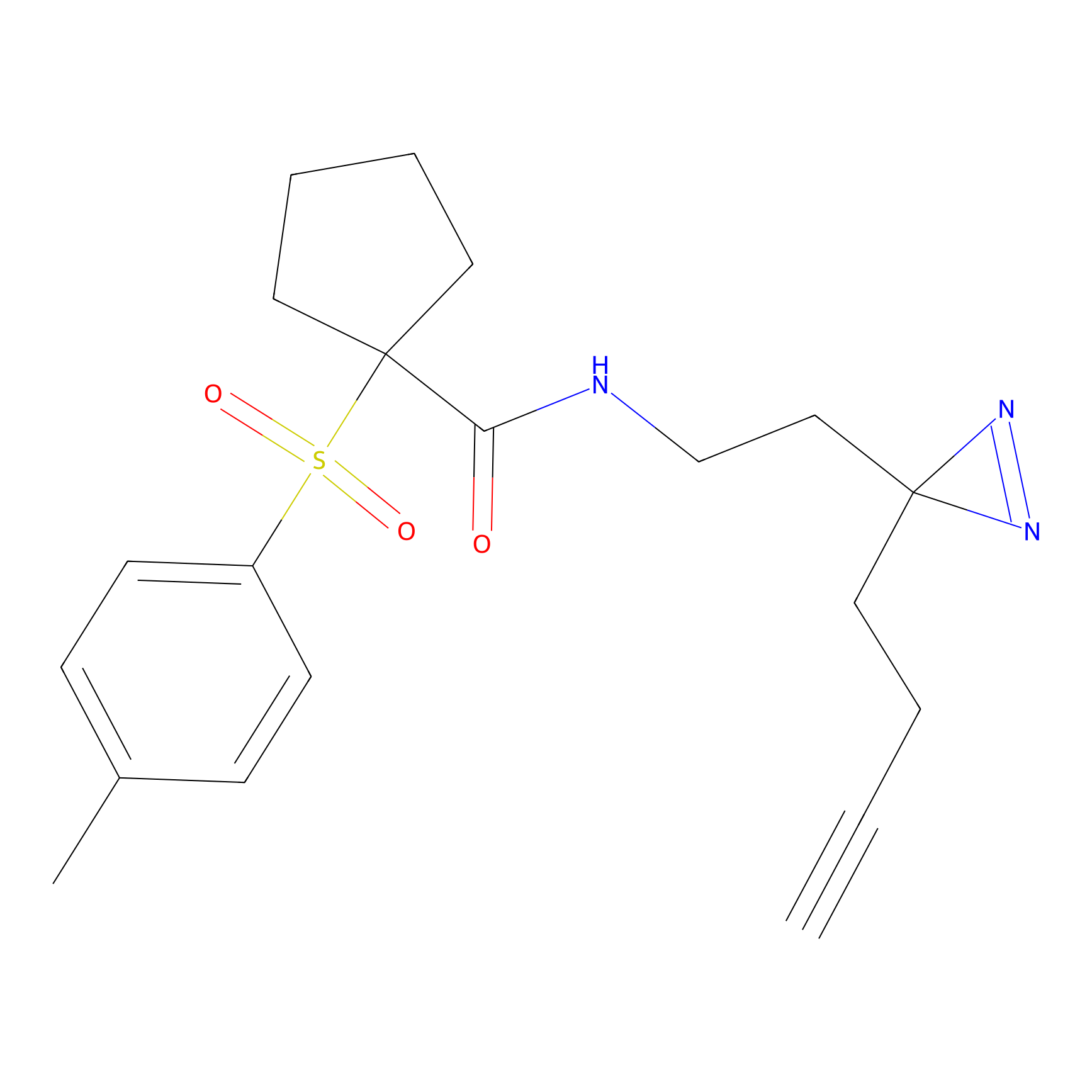
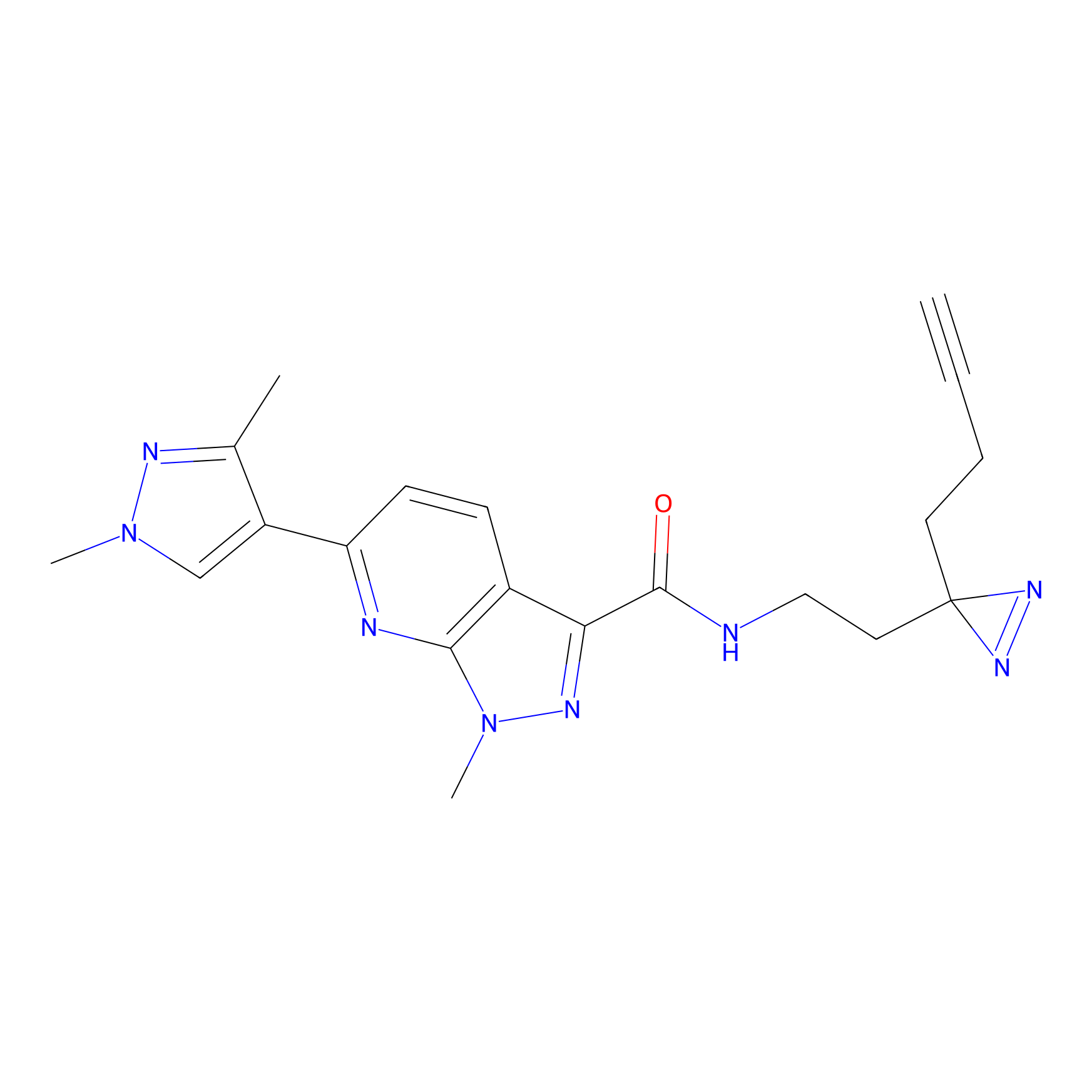
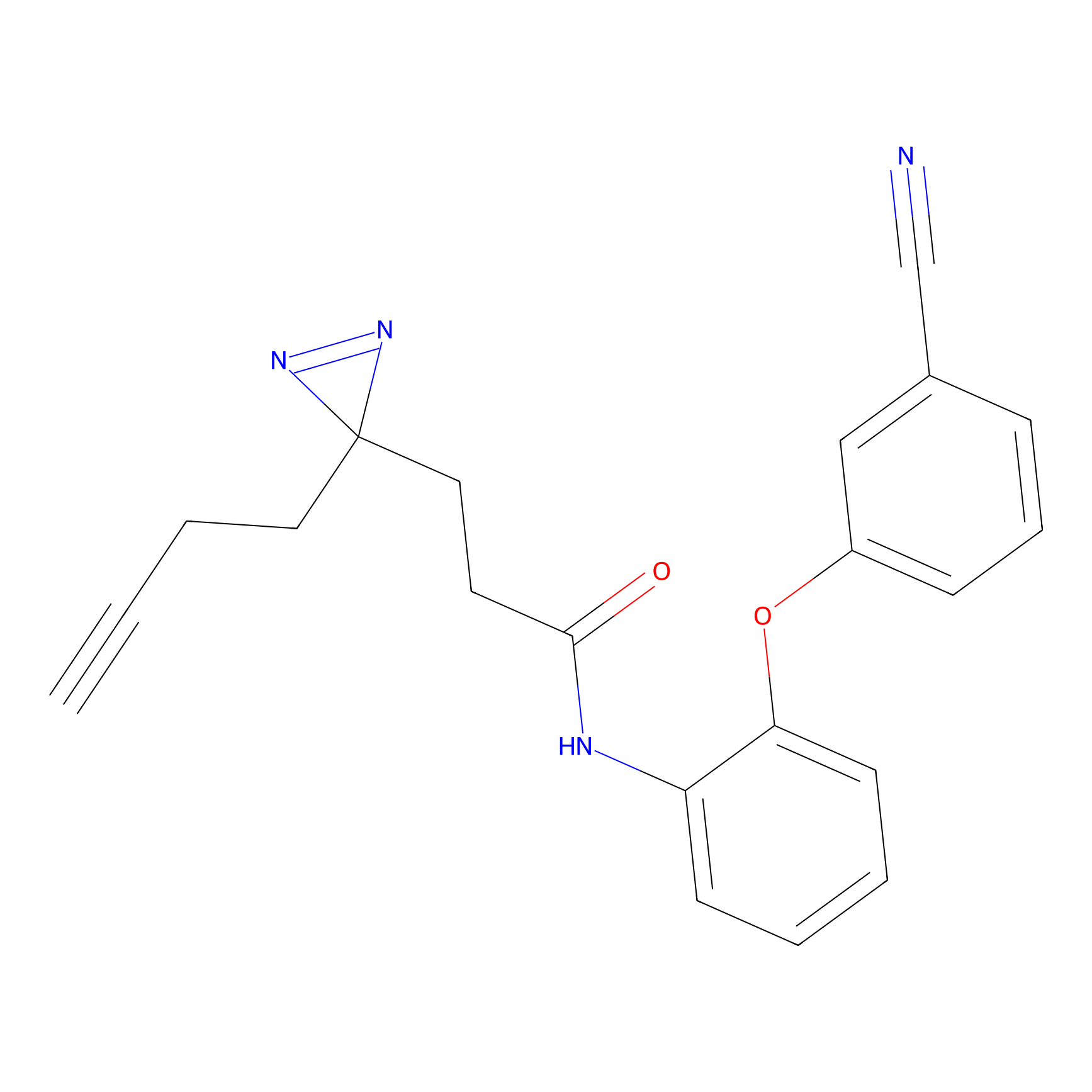
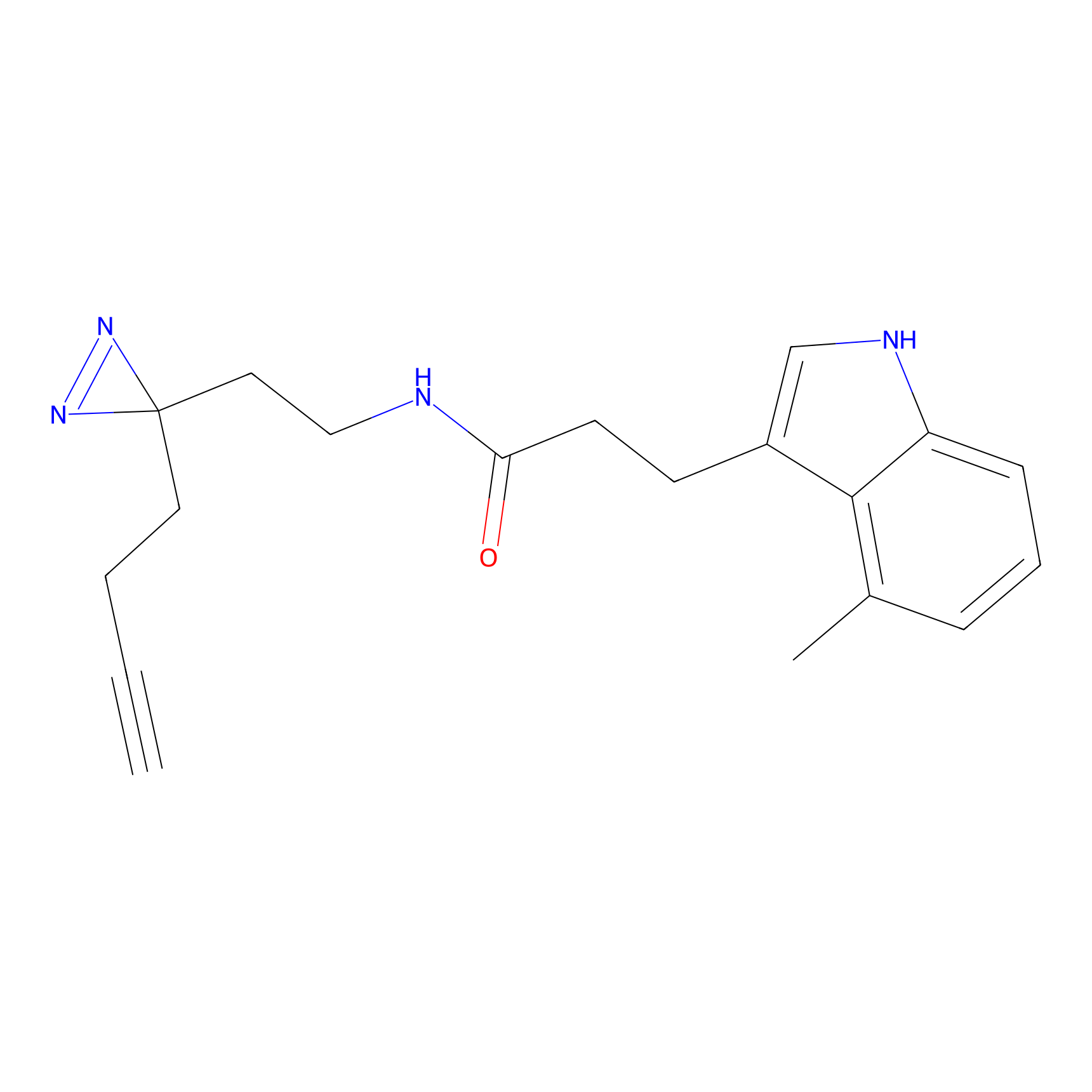
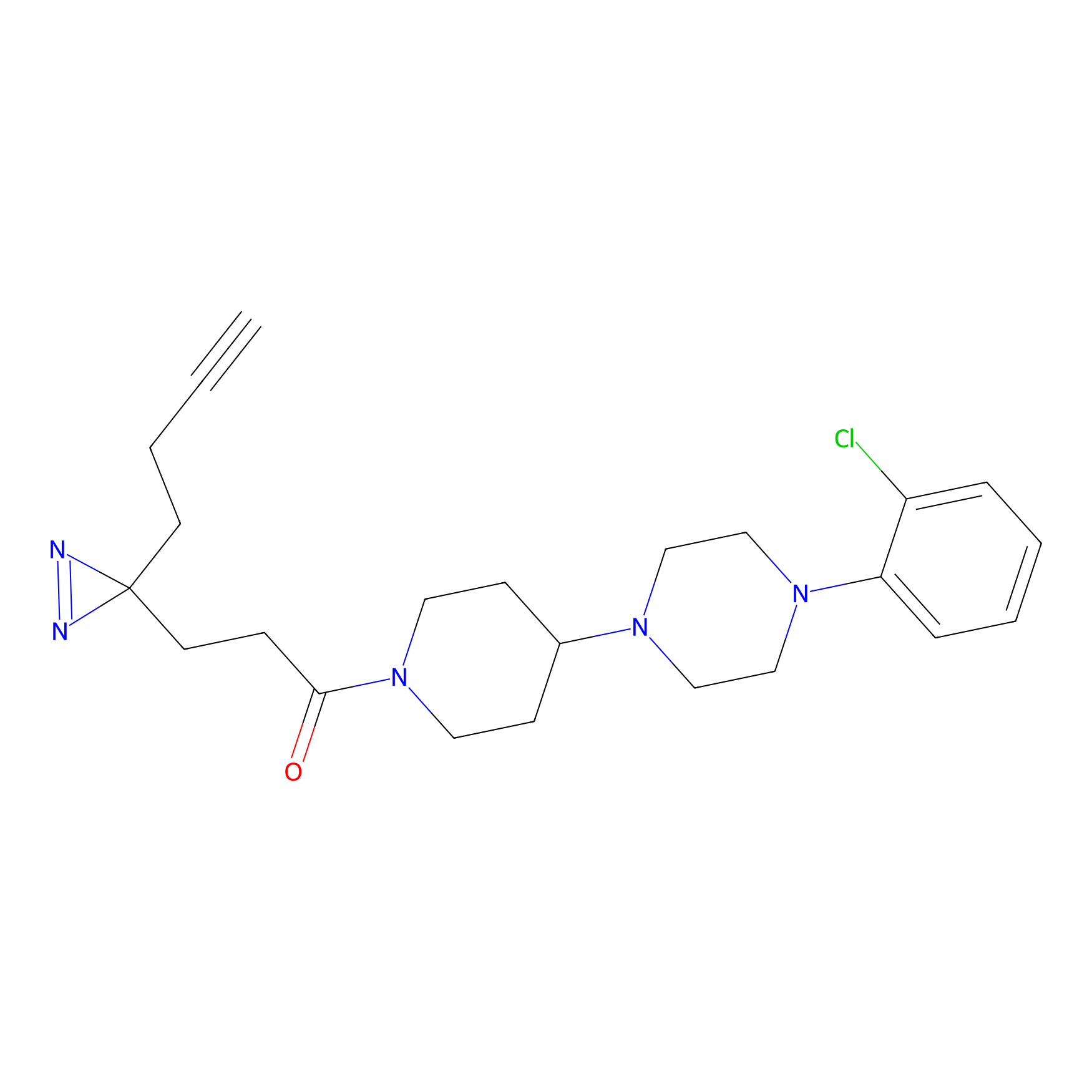
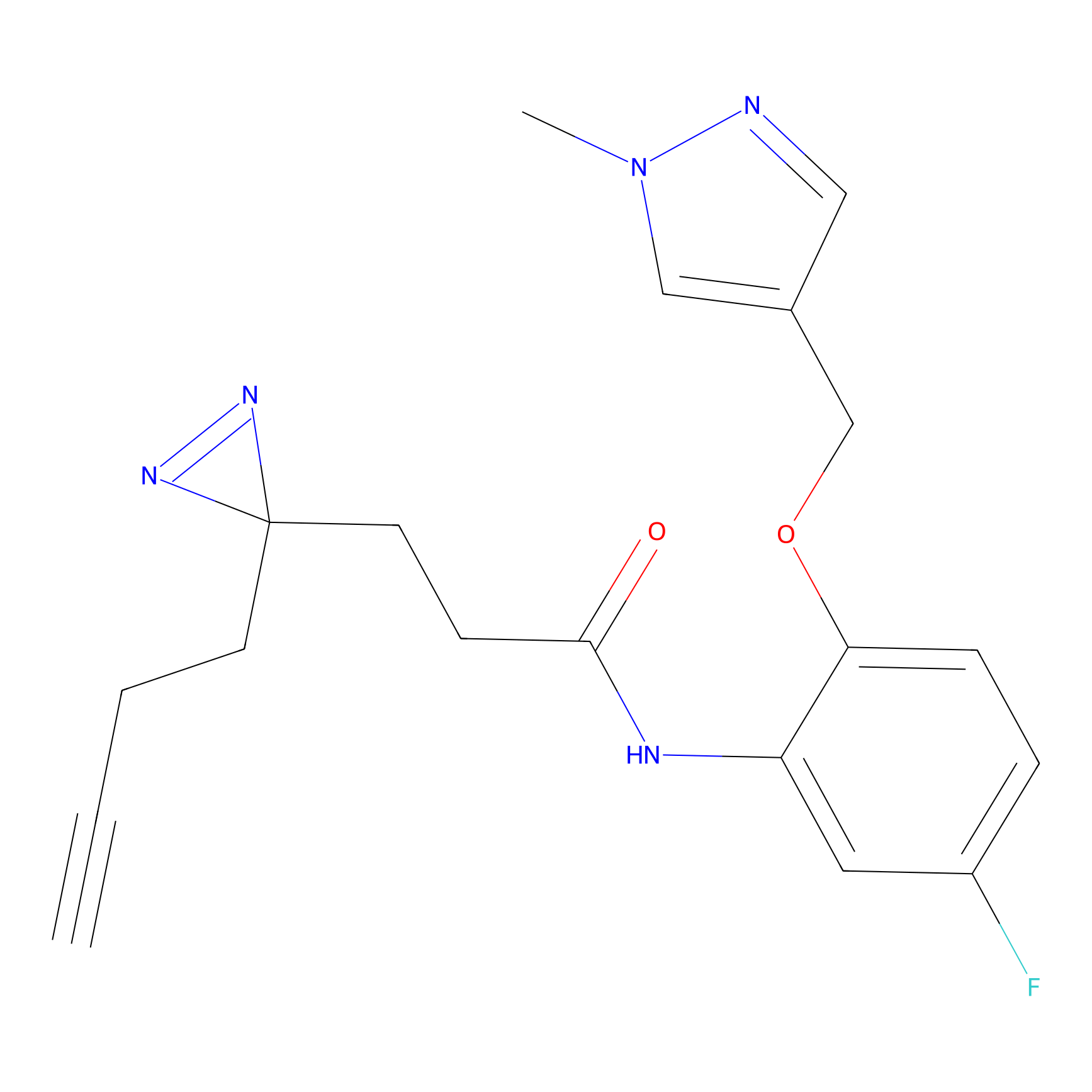
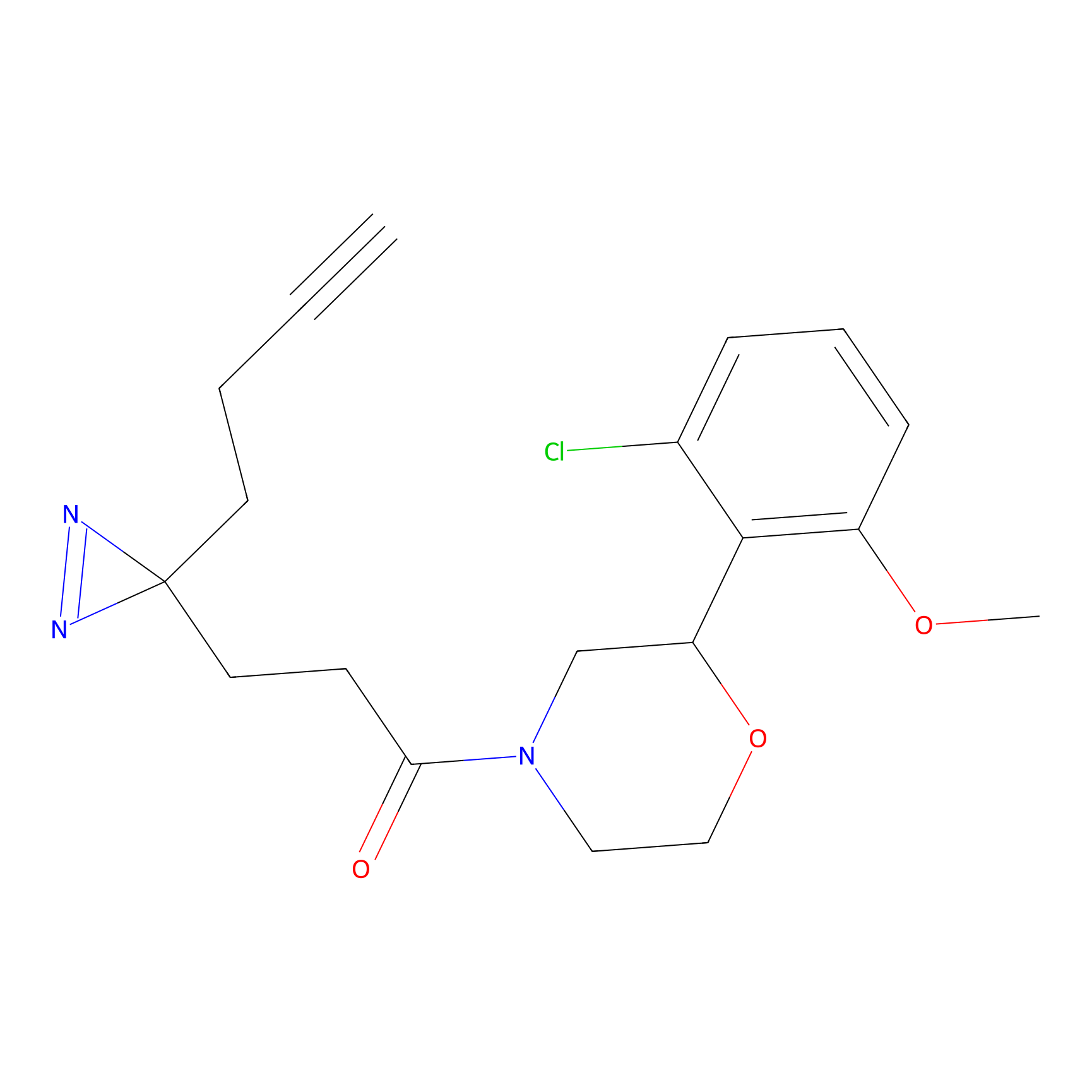
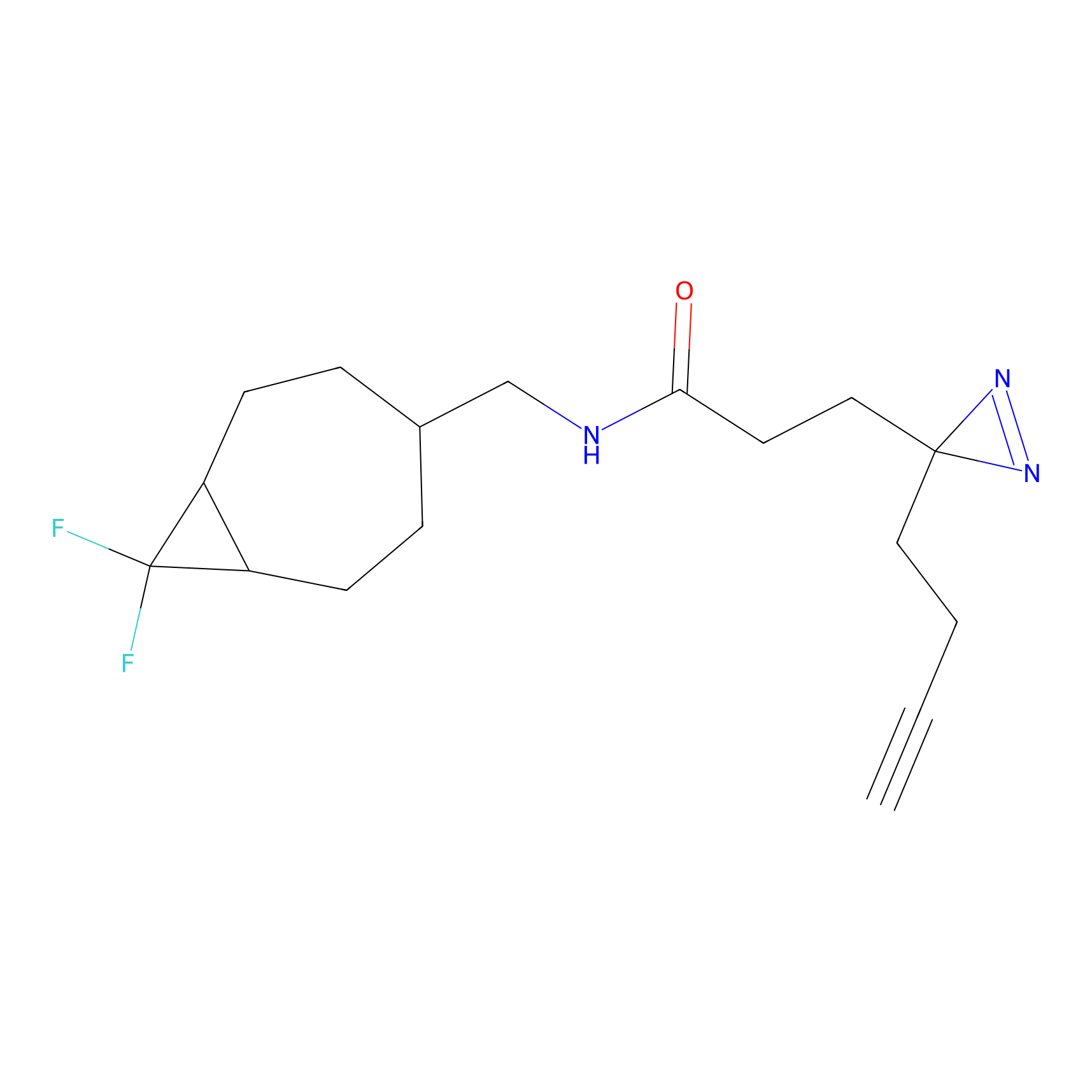
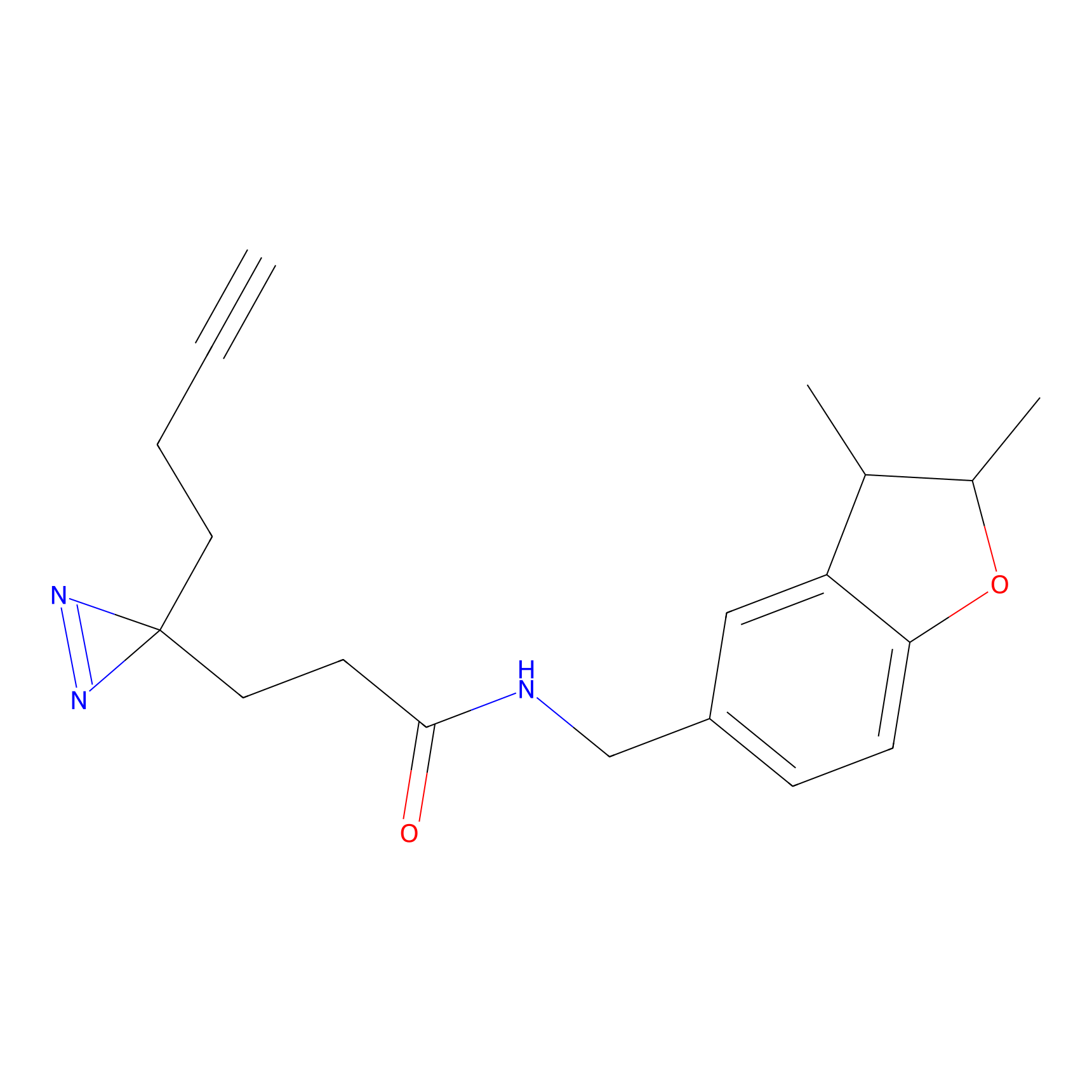
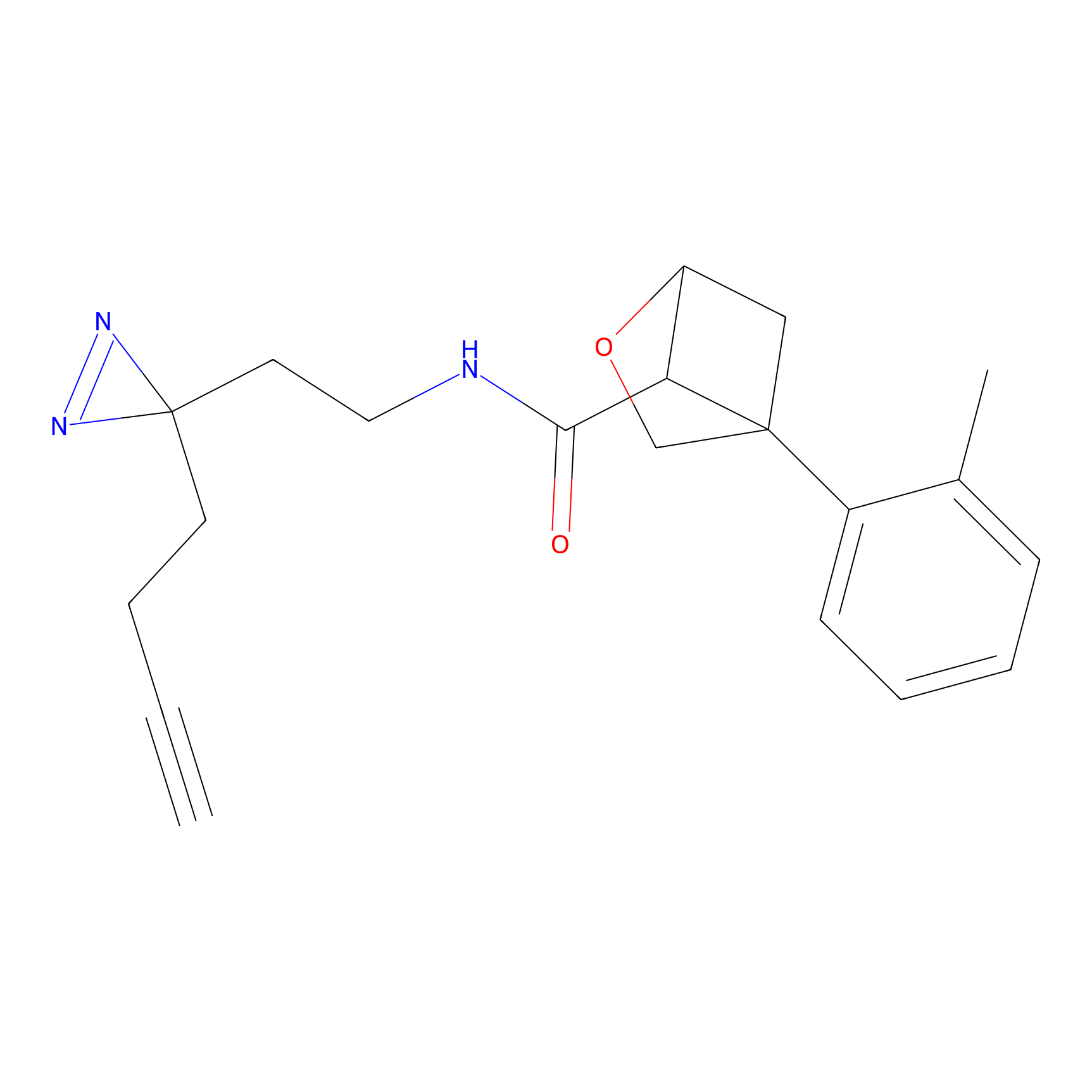
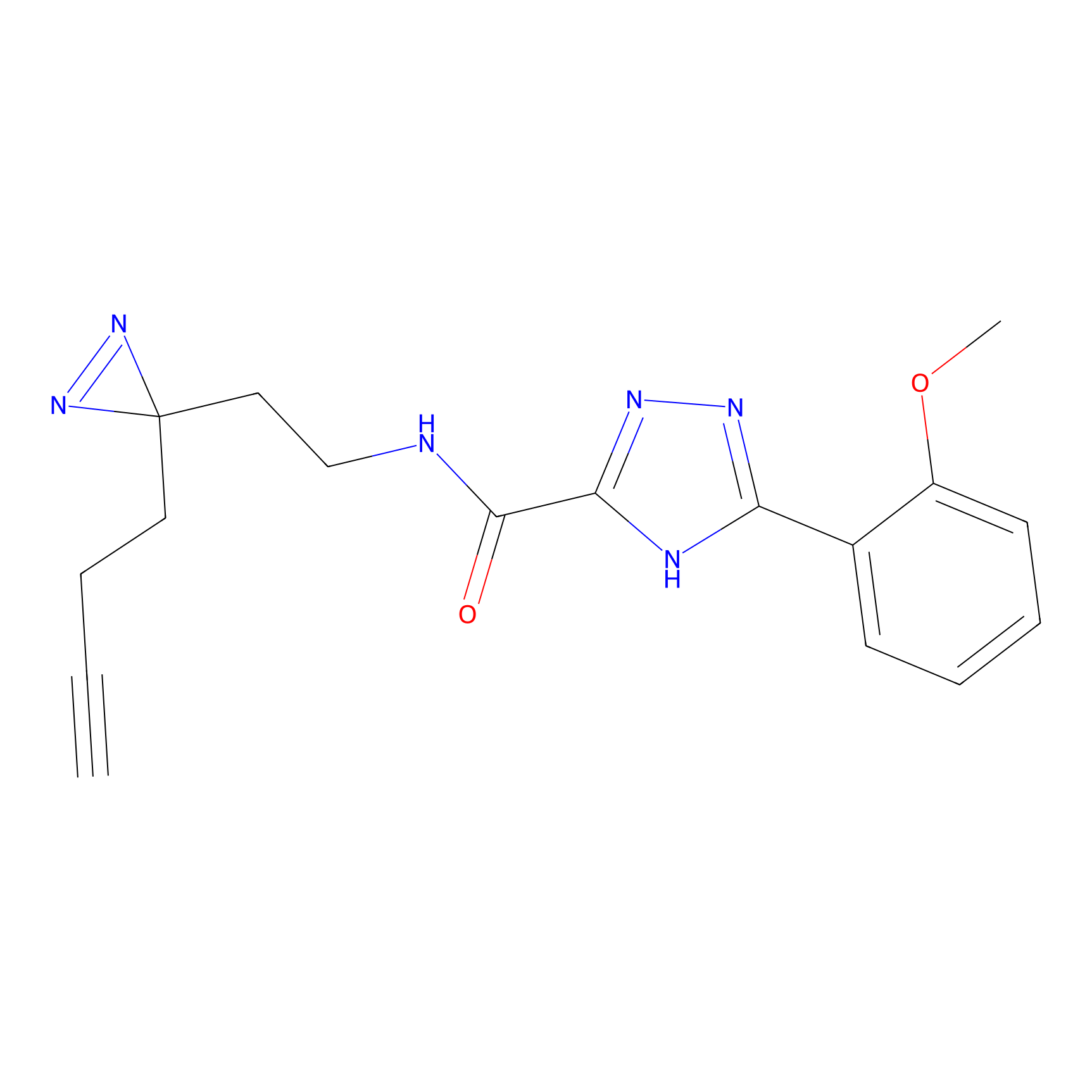
|
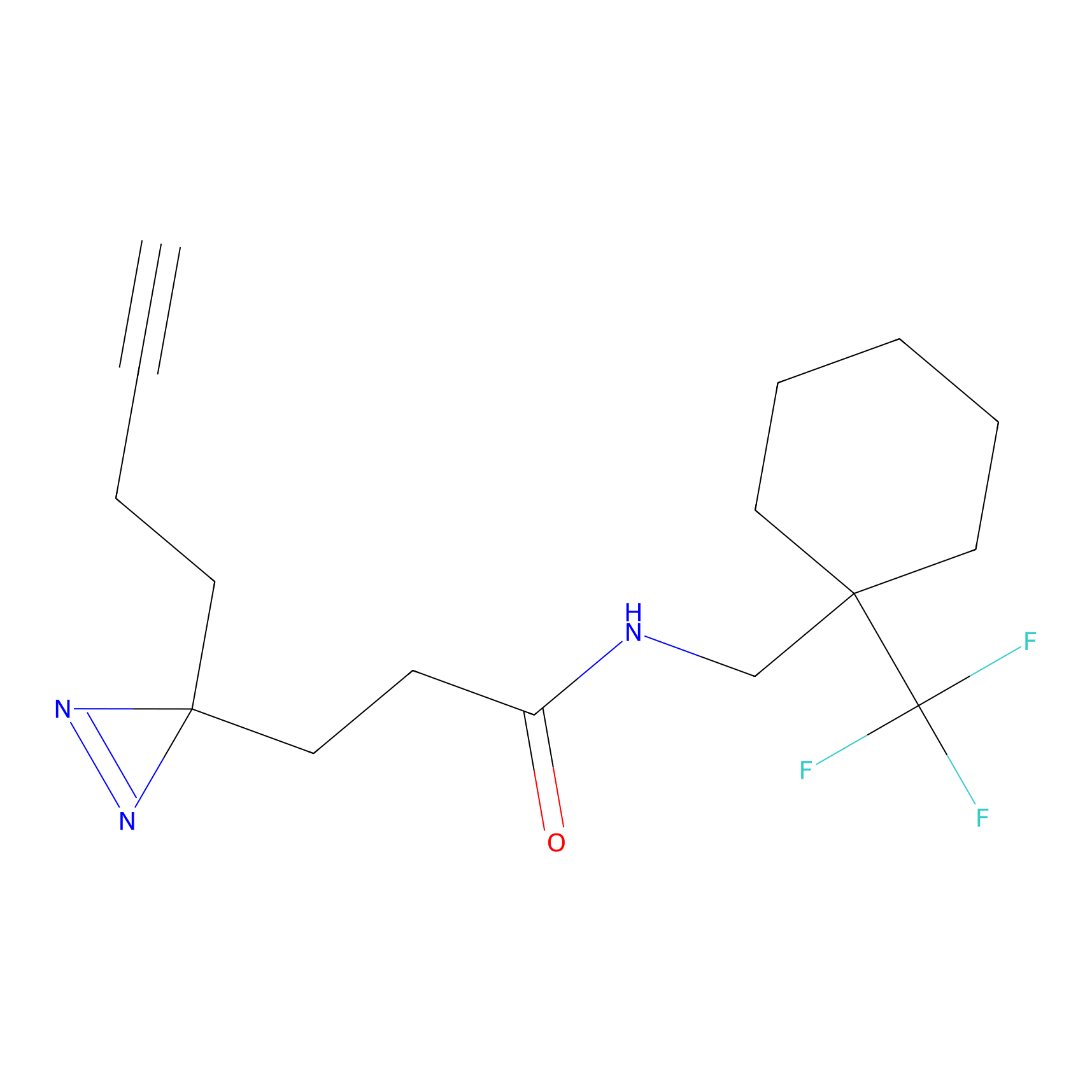
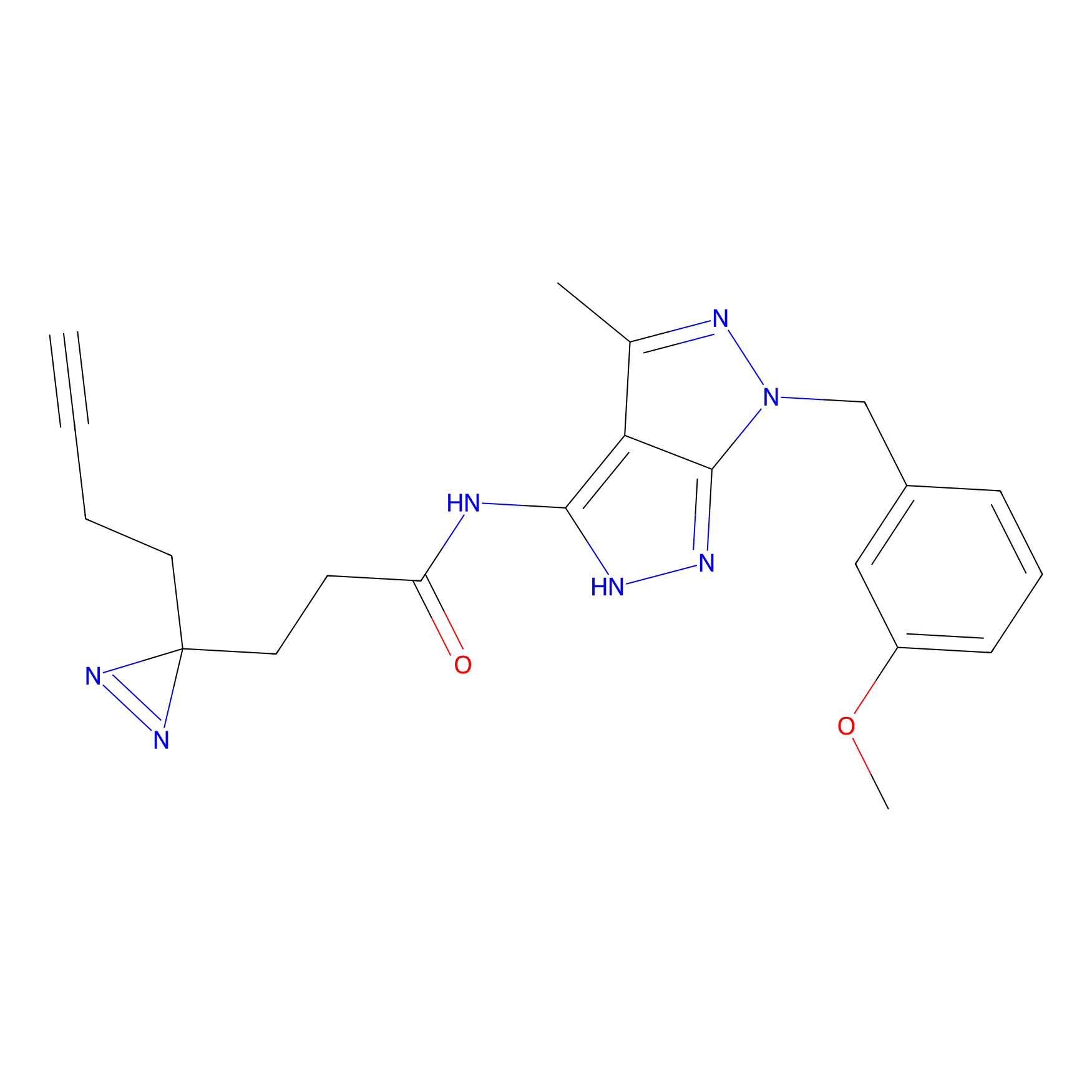
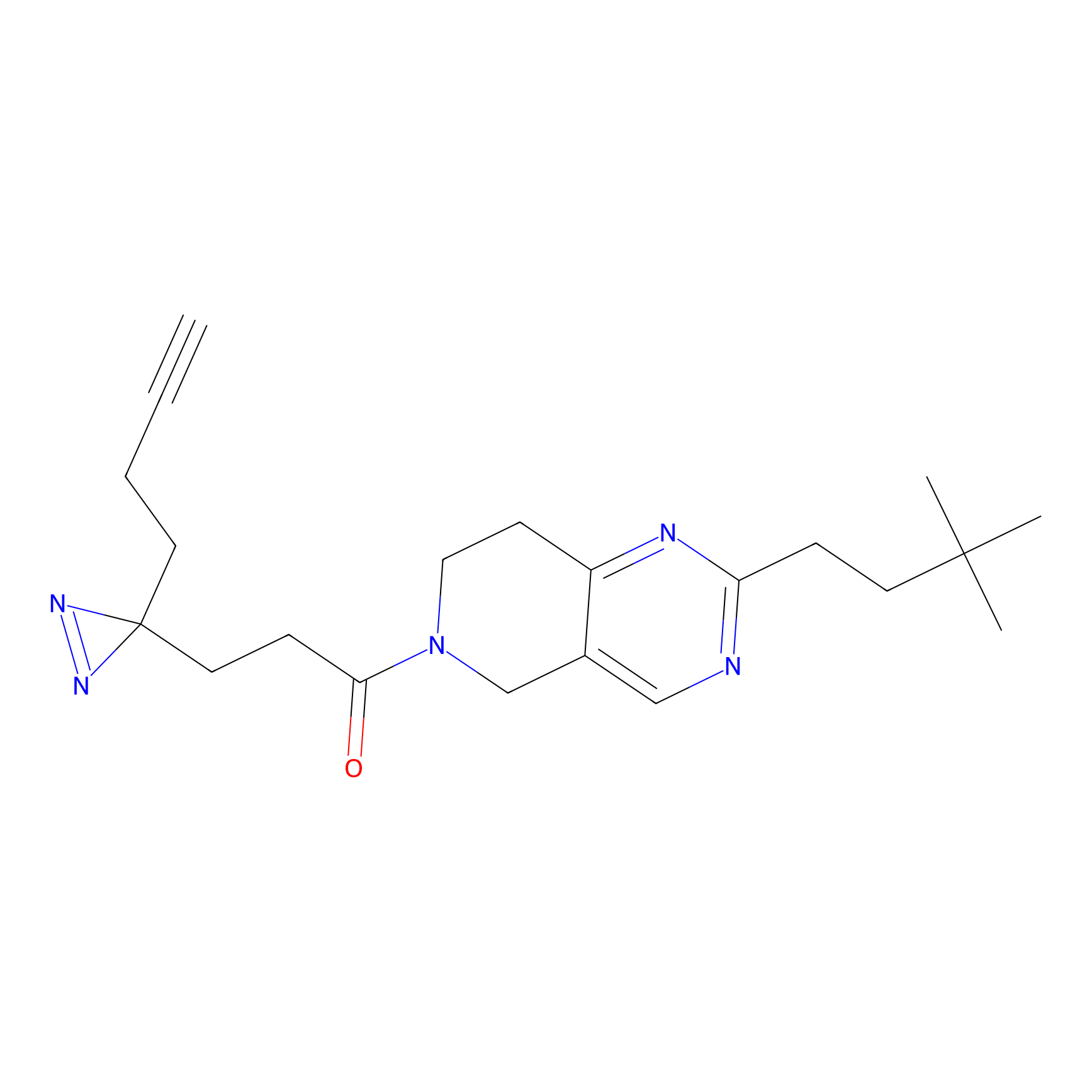
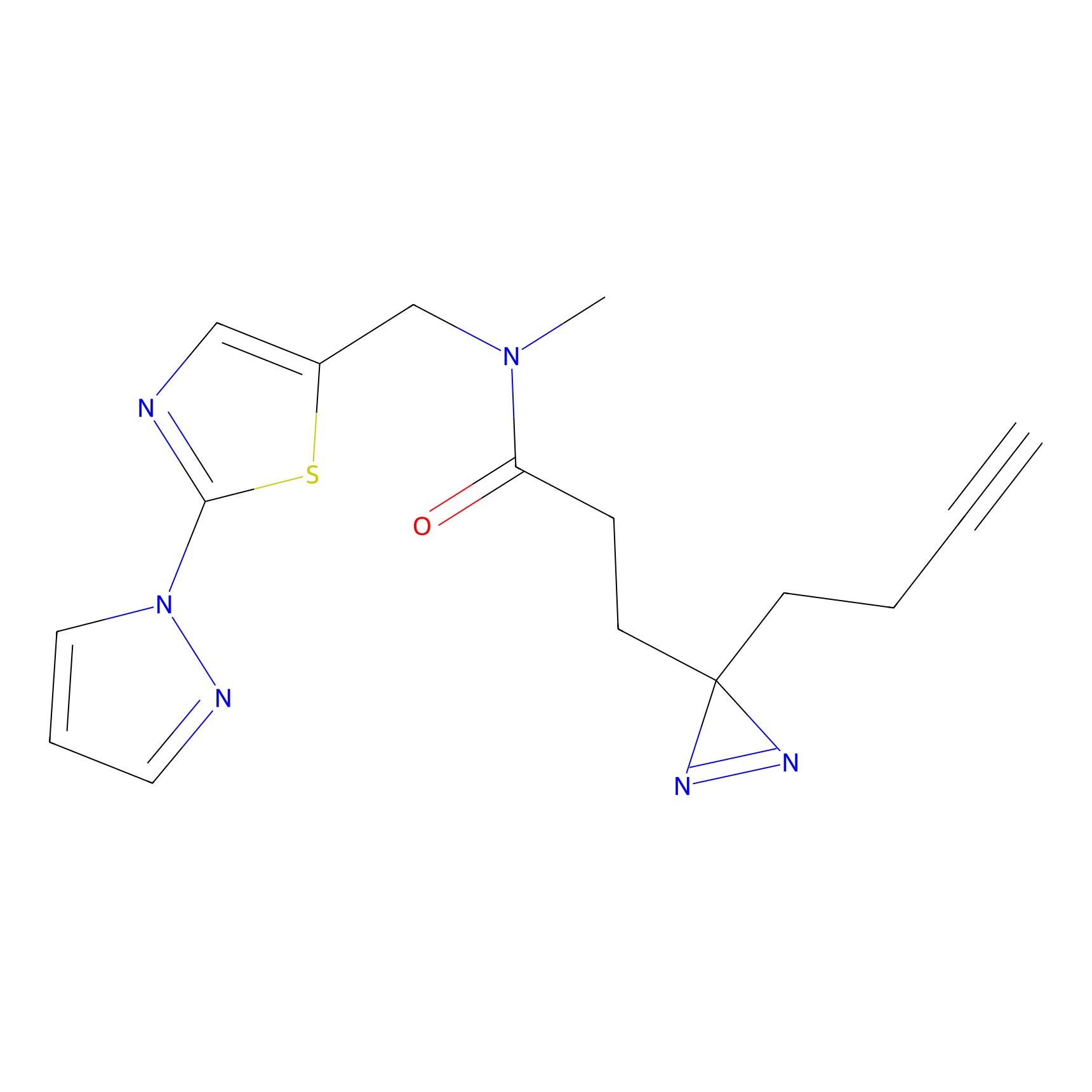
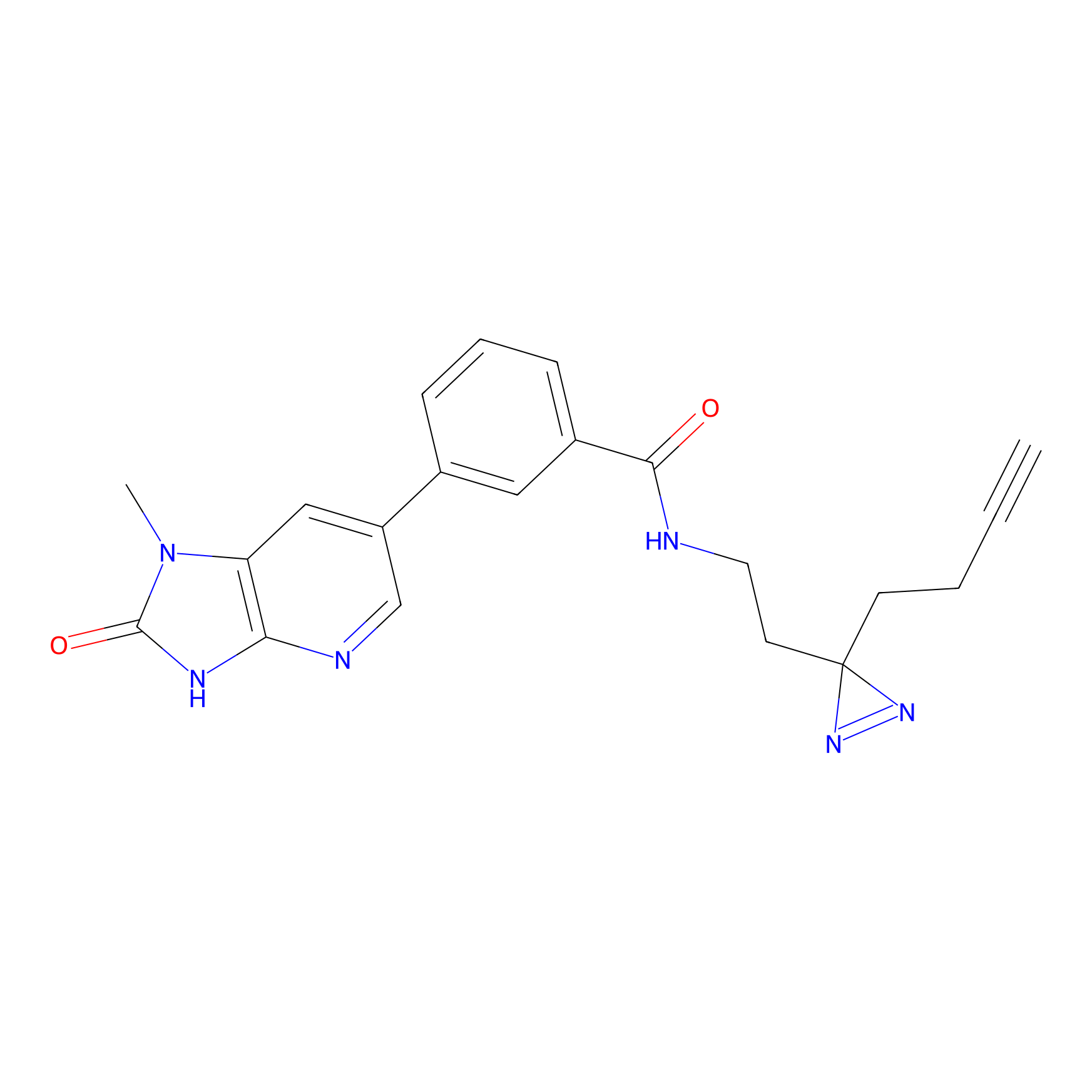
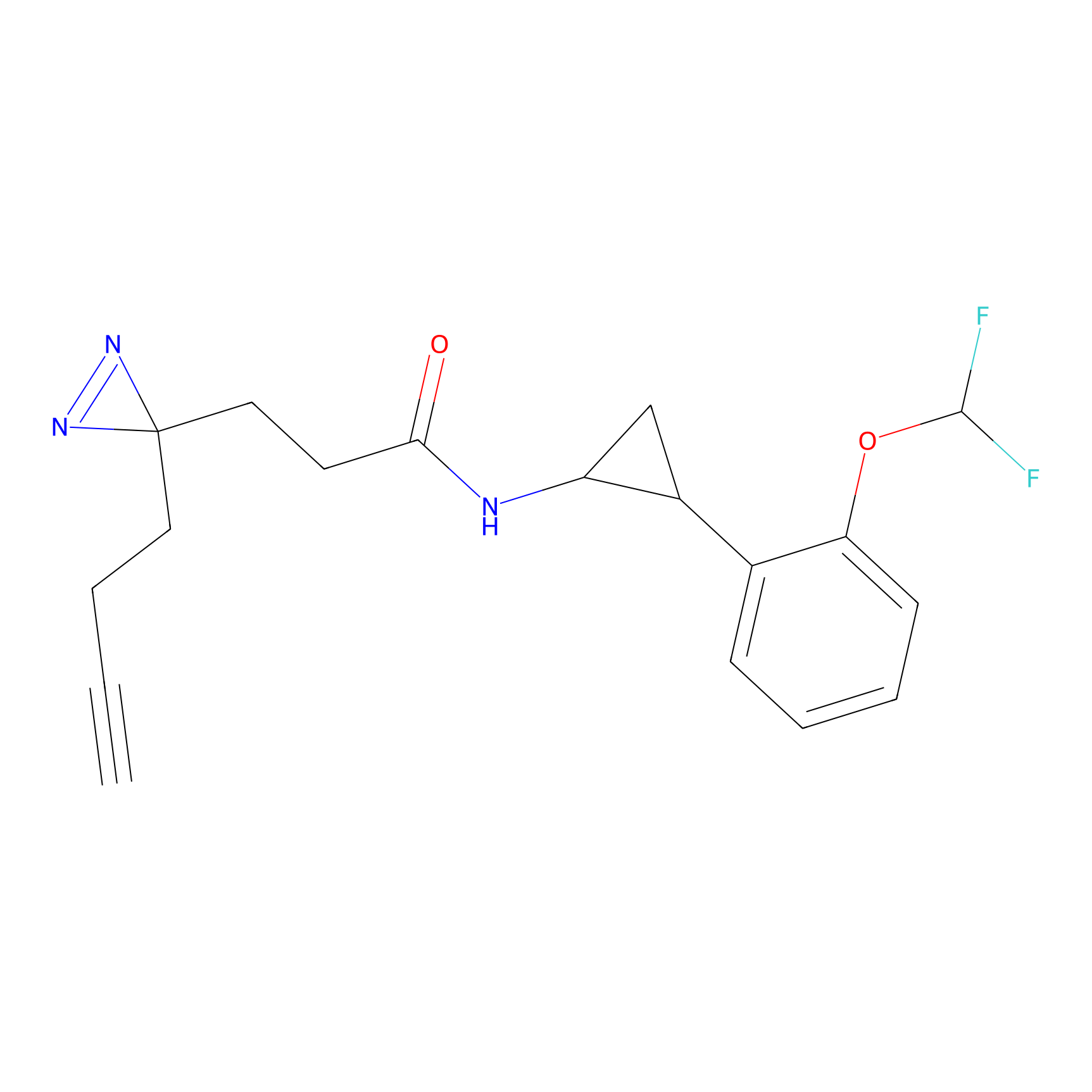
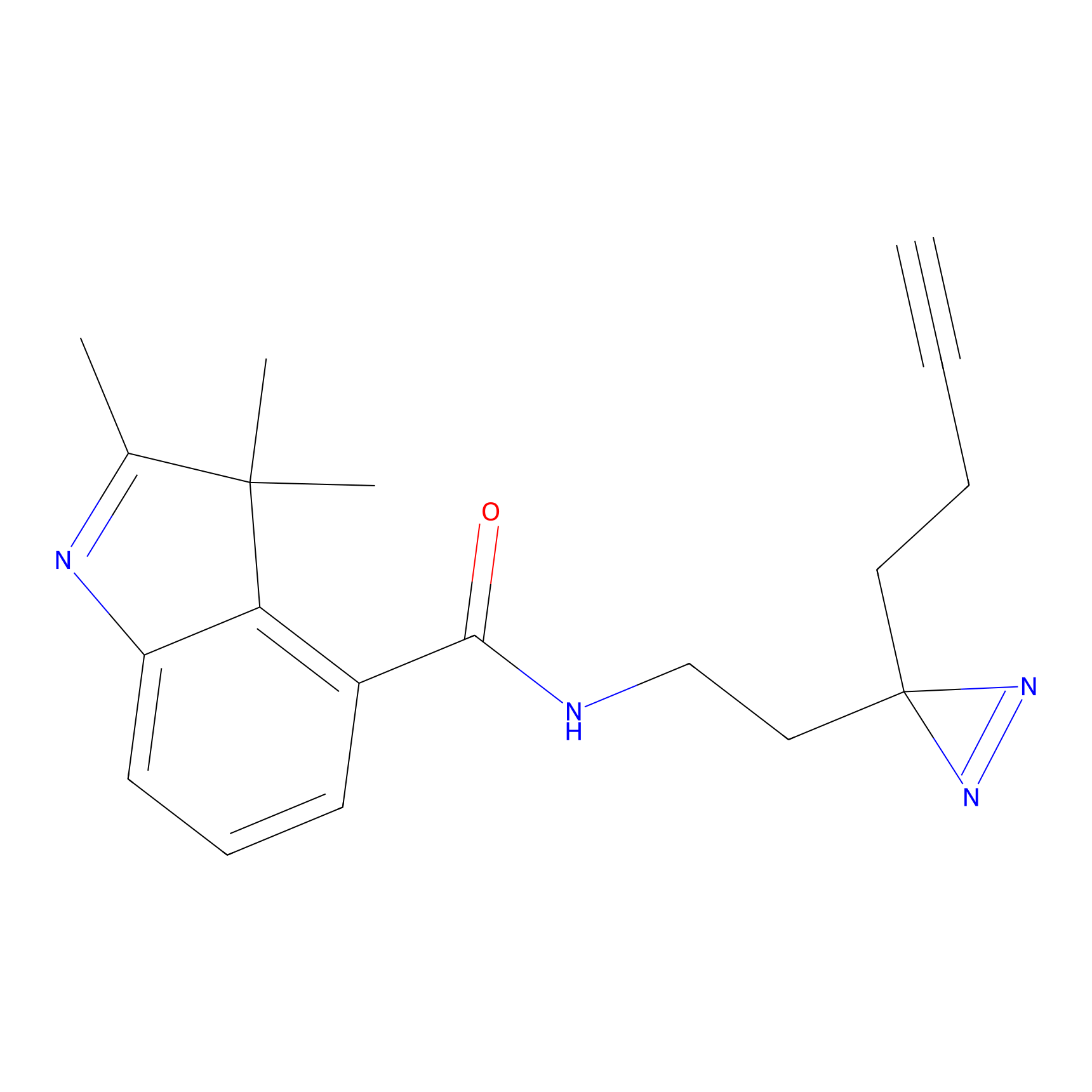
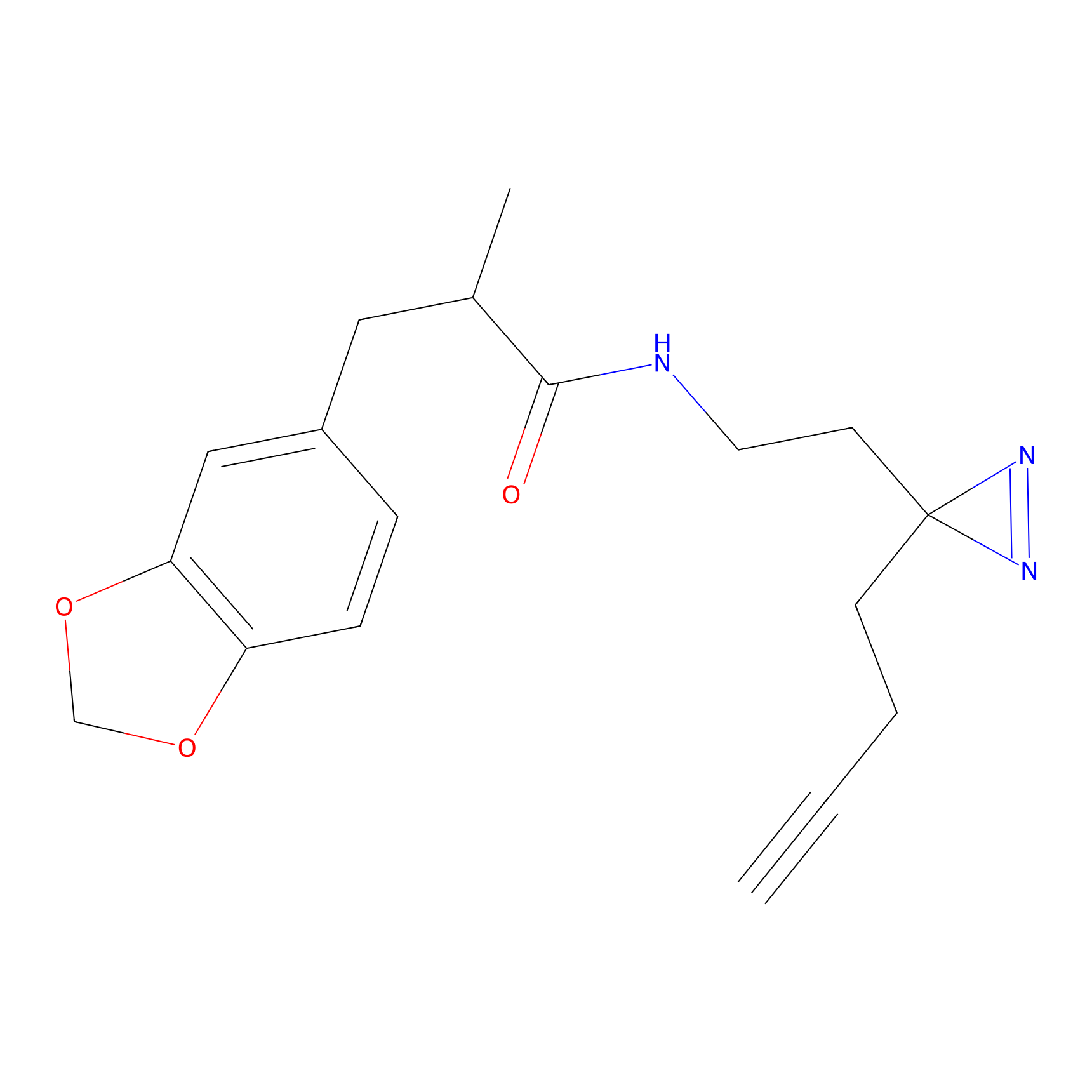
TH211 Probe Info |
 |
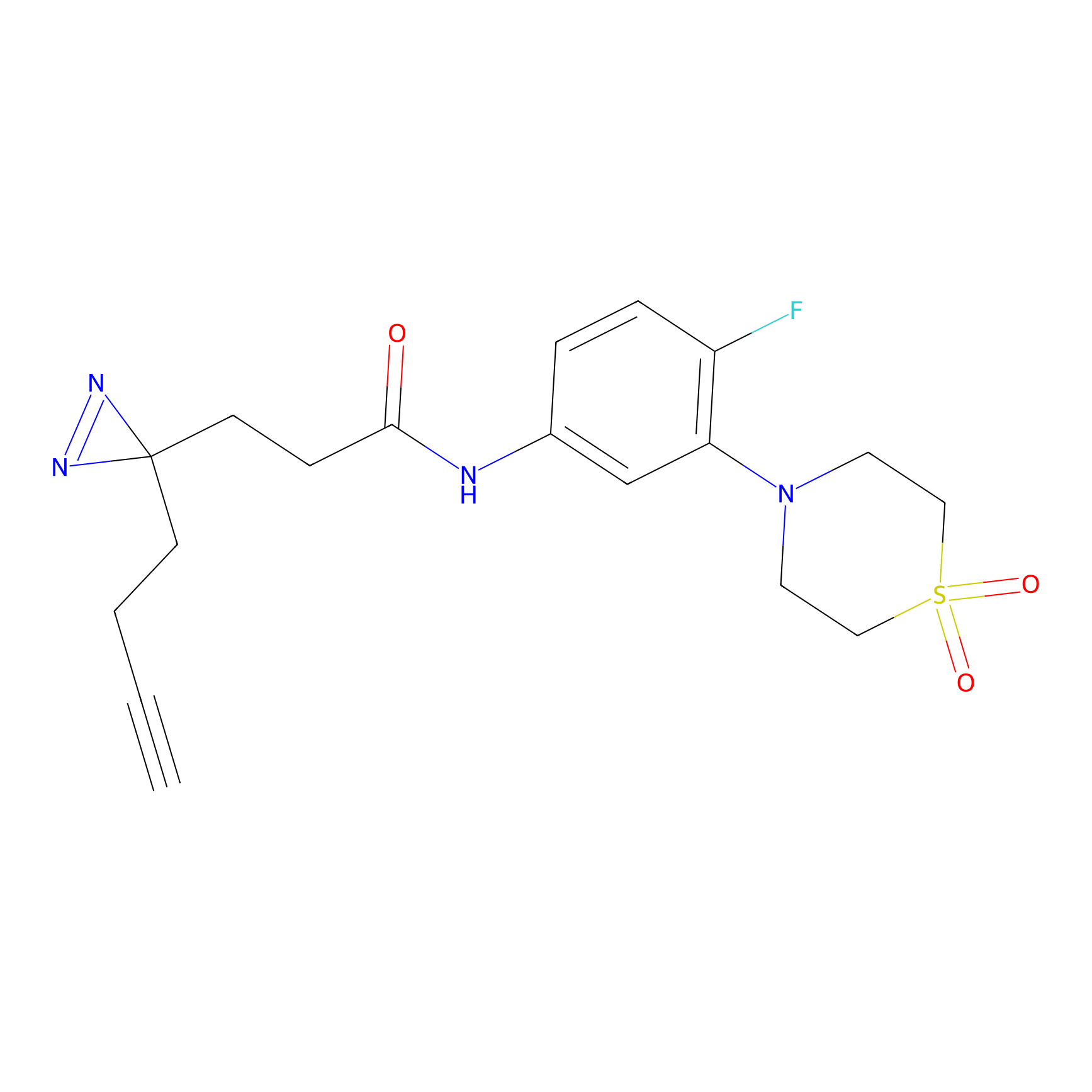
Y55(20.00) | LDD0260 | [4] | |
|
YN-1 Probe Info |
 |
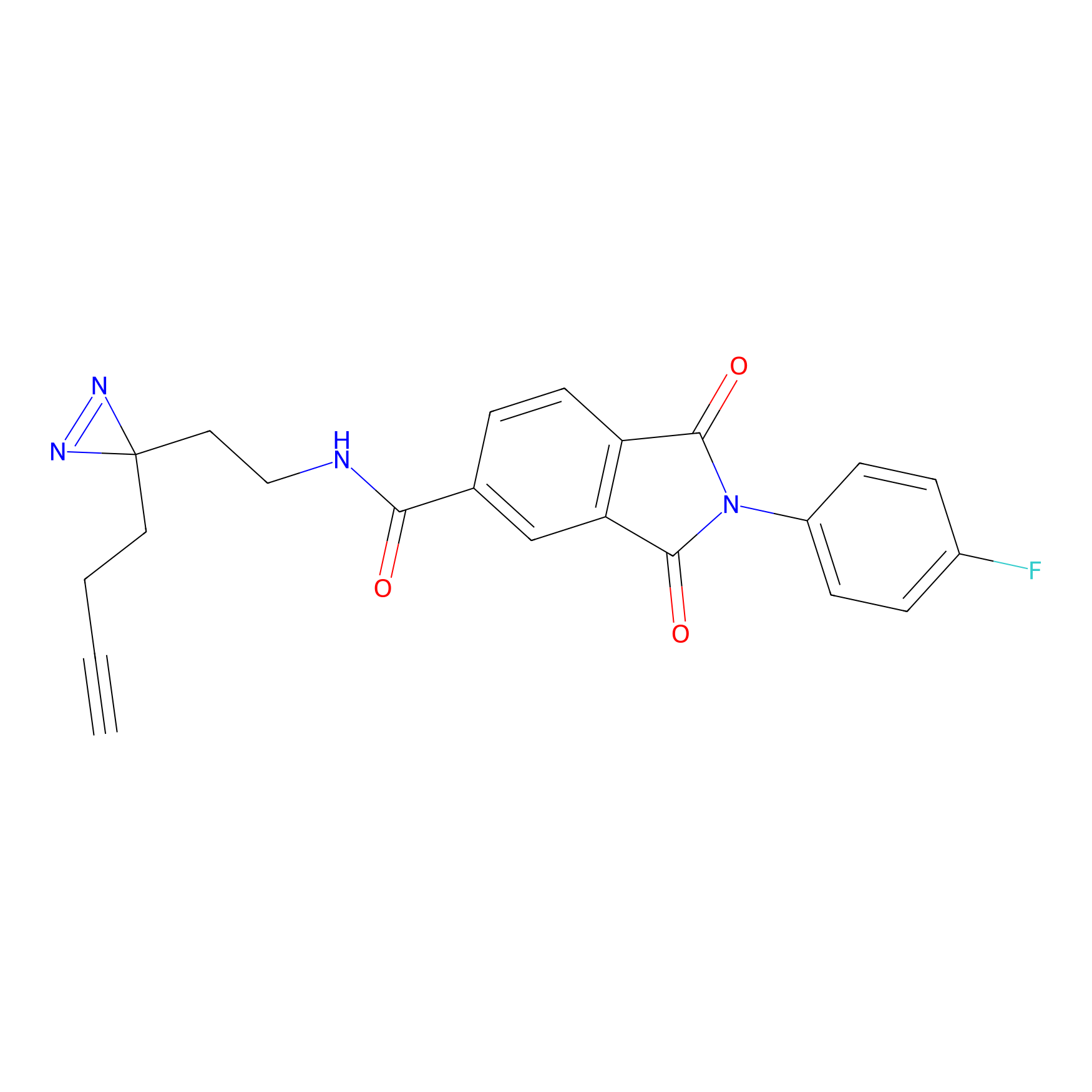
100.00 | LDD0444 | [5] | |
|
Jackson_14 Probe Info |
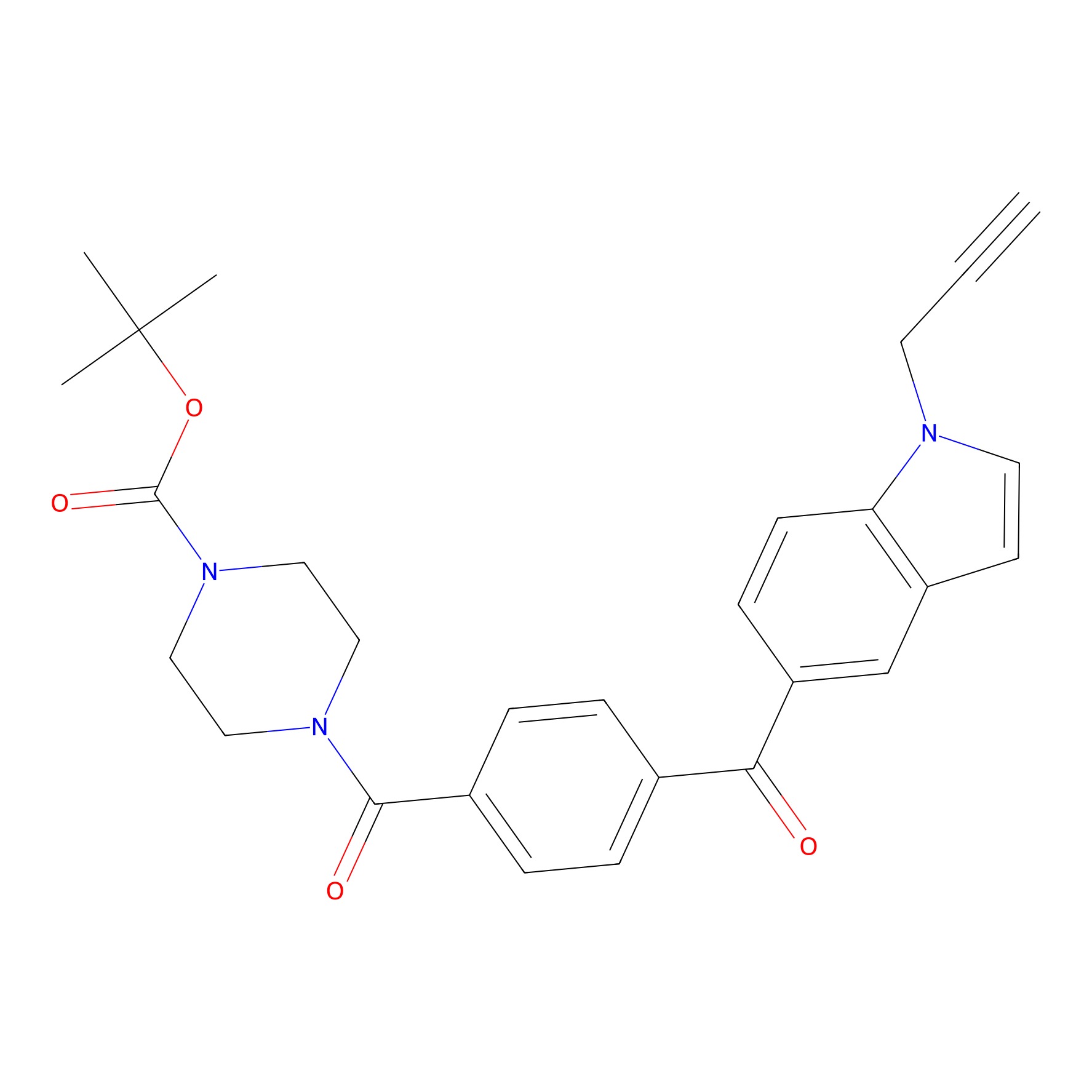 |
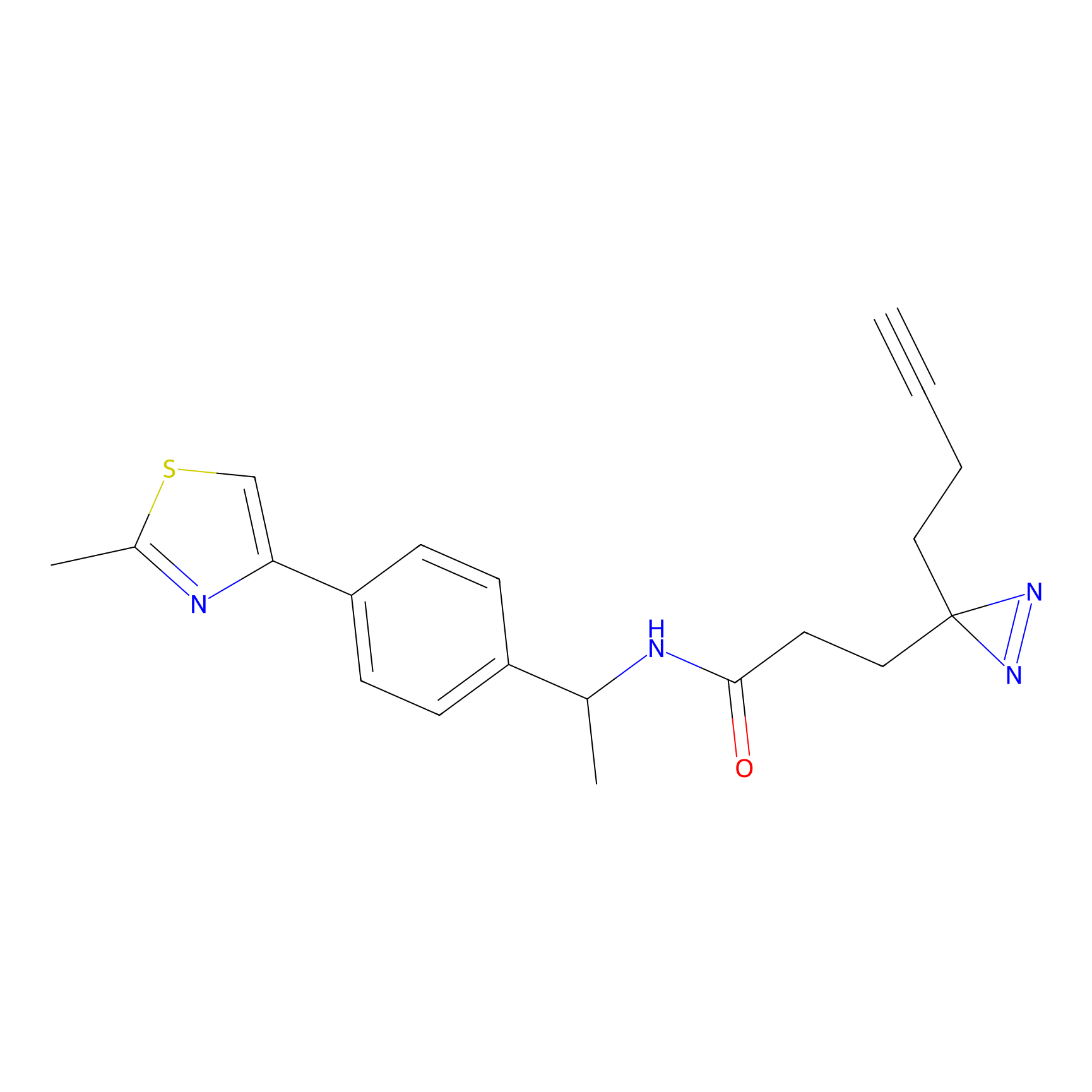
2.56 | LDD0123 | [6] | |
|
5E-2FA Probe Info |
 |
H129(0.00); H61(0.00) | LDD2235 | [7] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [8] | |
|
m-APA Probe Info |
 |
H129(0.00); H61(0.00) | LDD2231 | [7] | |
|
CY-1 Probe Info |
 |
K150(0.00); K152(0.00) | LDD0246 | [3] | |
|
IPM Probe Info |
 |
N.A. | LDD2156 | [9] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [10] | |
|
STPyne Probe Info |
 |
K98(0.00); K96(0.00) | LDD0009 | [10] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [11] | |
|
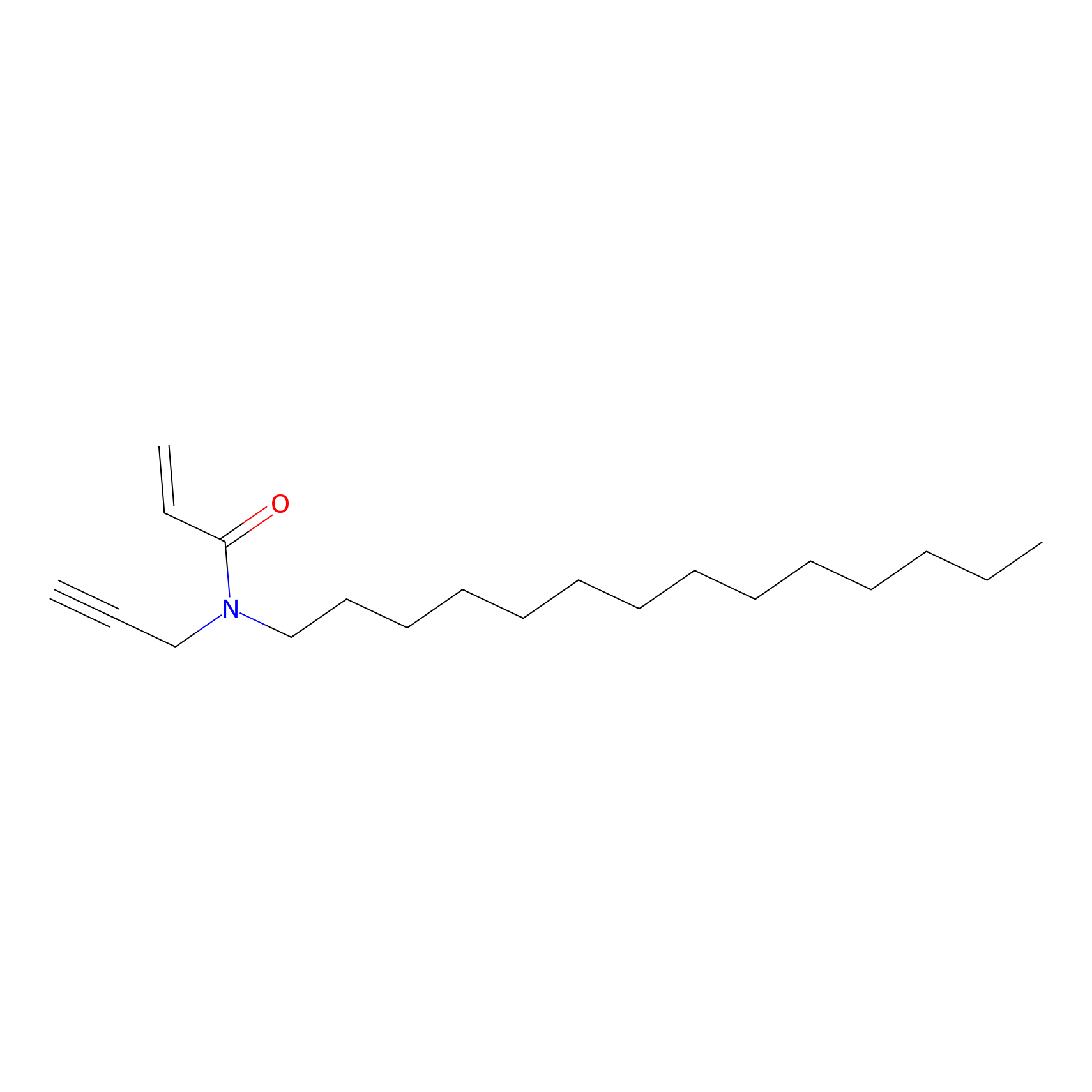
Acrolein Probe Info |
 |
H129(0.00); H61(0.00) | LDD0217 | [12] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [13] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
The Interaction Atlas With This Target
References