Details of the Target
General Information of Target
| Target ID | LDTP13606 | |||||
|---|---|---|---|---|---|---|
| Target Name | Mammalian ependymin-related protein 1 (EPDR1) | |||||
| Gene Name | EPDR1 | |||||
| Gene ID | 54749 | |||||
| Synonyms |
MERP1; UCC1; Mammalian ependymin-related protein 1; MERP-1; Upregulated in colorectal cancer gene 1 protein |
|||||
| 3D Structure | ||||||
| Sequence |
MRLRNGTVATALAFITSFLTLSWYTTWQNGKEKLIAYQREFLALKERLRIAEHRISQRSS
ELNTIVQQFKRVGAETNGSKDALNKFSDNTLKLLKELTSKKSLQVPSIYYHLPHLLKNEG SLQPAVQIGNGRTGVSIVMGIPTVKREVKSYLIETLHSLIDNLYPEEKLDCVIVVFIGET DIDYVHGVVANLEKEFSKEISSGLVEVISPPESYYPDLTNLKETFGDSKERVRWRTKQNL DYCFLMMYAQEKGIYYIQLEDDIIVKQNYFNTIKNFALQLSSEEWMILEFSQLGFIGKMF QAPDLTLIVEFIFMFYKEKPIDWLLDHILWVKVCNPEKDAKHCDRQKANLRIRFRPSLFQ HVGLHSSLSGKIQKLTDKDYMKPLLLKIHVNPPAEVSTSLKVYQGHTLEKTYMGEDFFWA ITPIAGDYILFKFDKPVNVESYLFHSGNQEHPGDILLNTTVEVLPFKSEGLEISKETKDK RLEDGYFRIGKFENGVAEGMVDPSLNPISAFRLSVIQNSAVWAILNEIHIKKATN |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Ependymin family
|
|||||
| Subcellular location |
Lysosome lumen
|
|||||
| Function | Binds anionic lipids and gangliosides at acidic pH. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
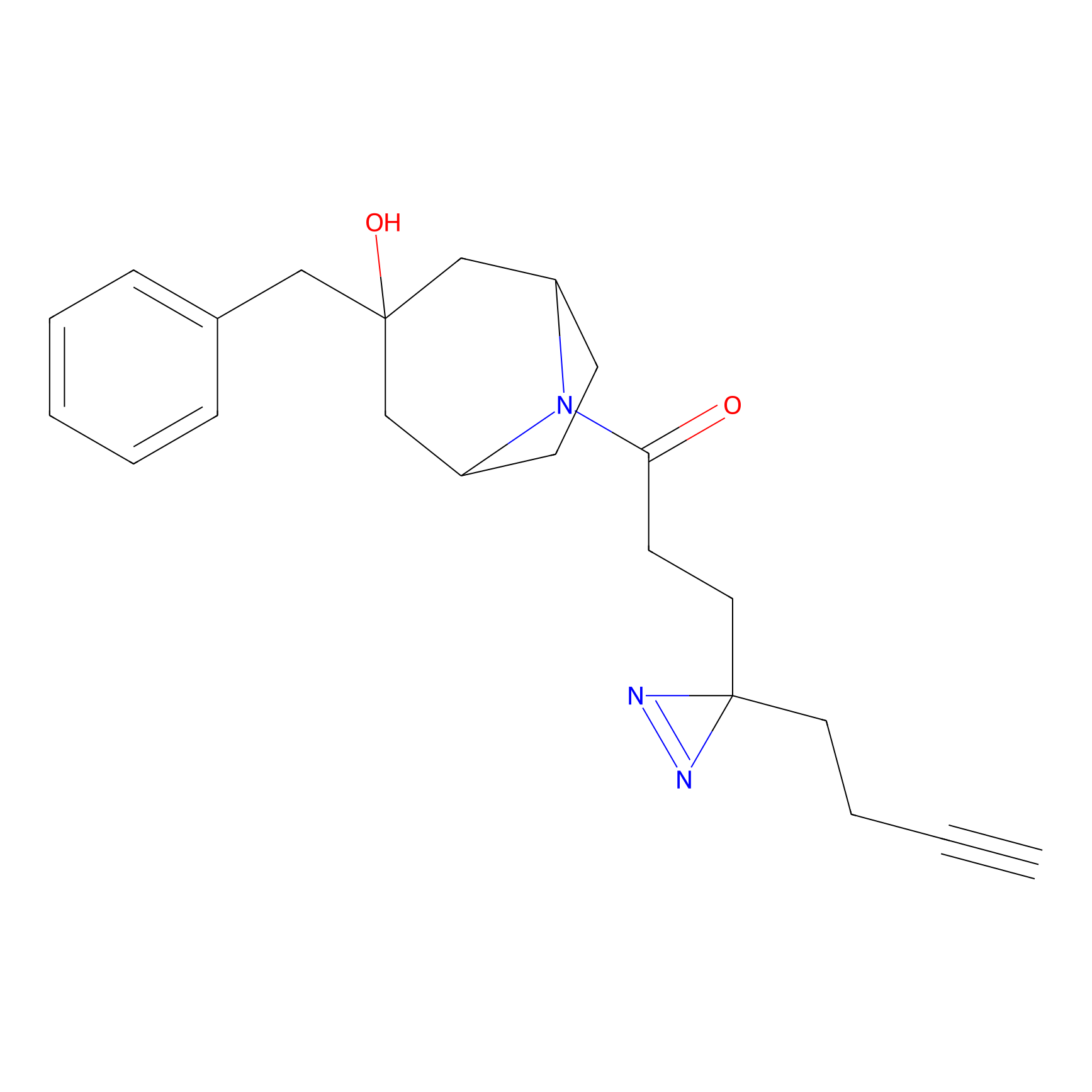
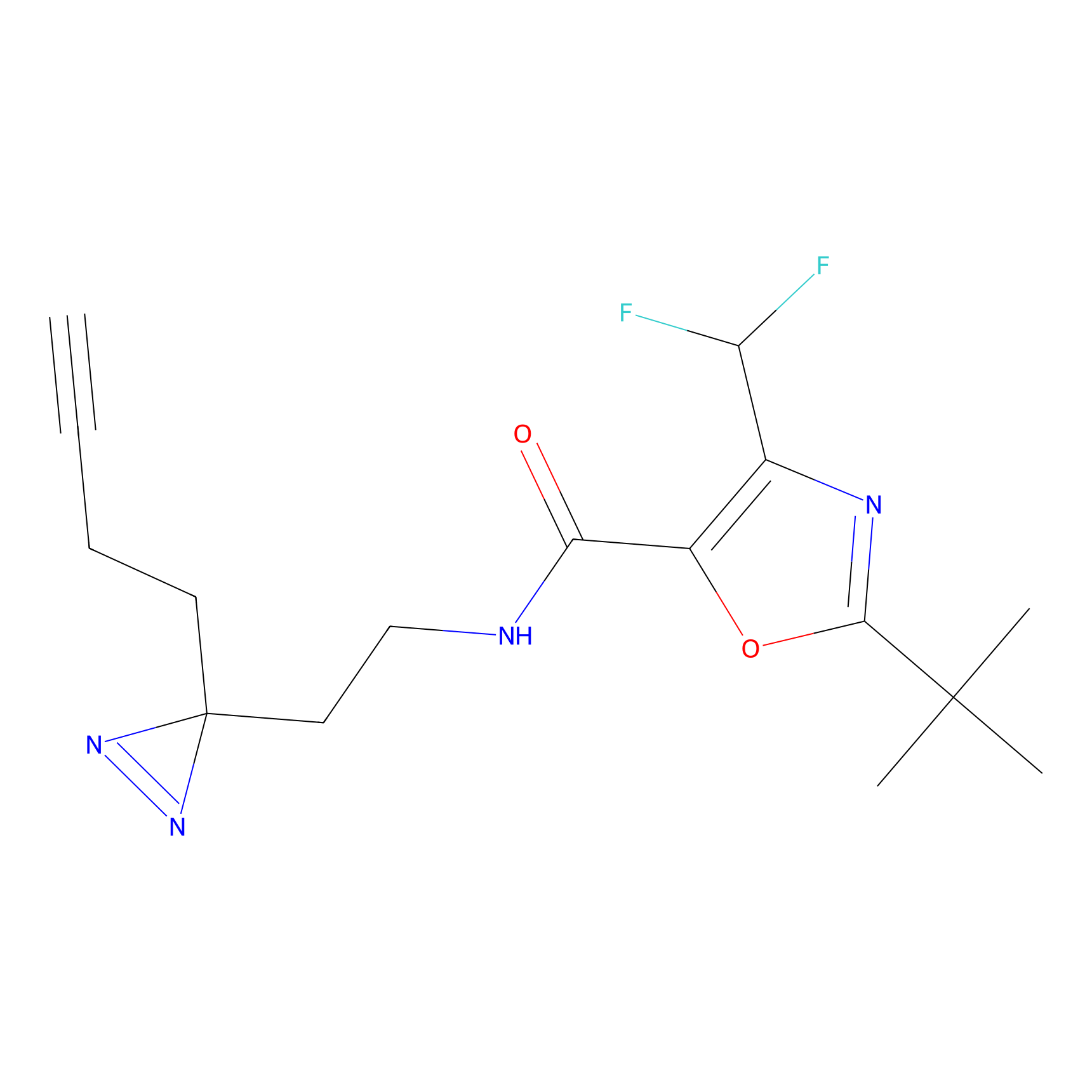
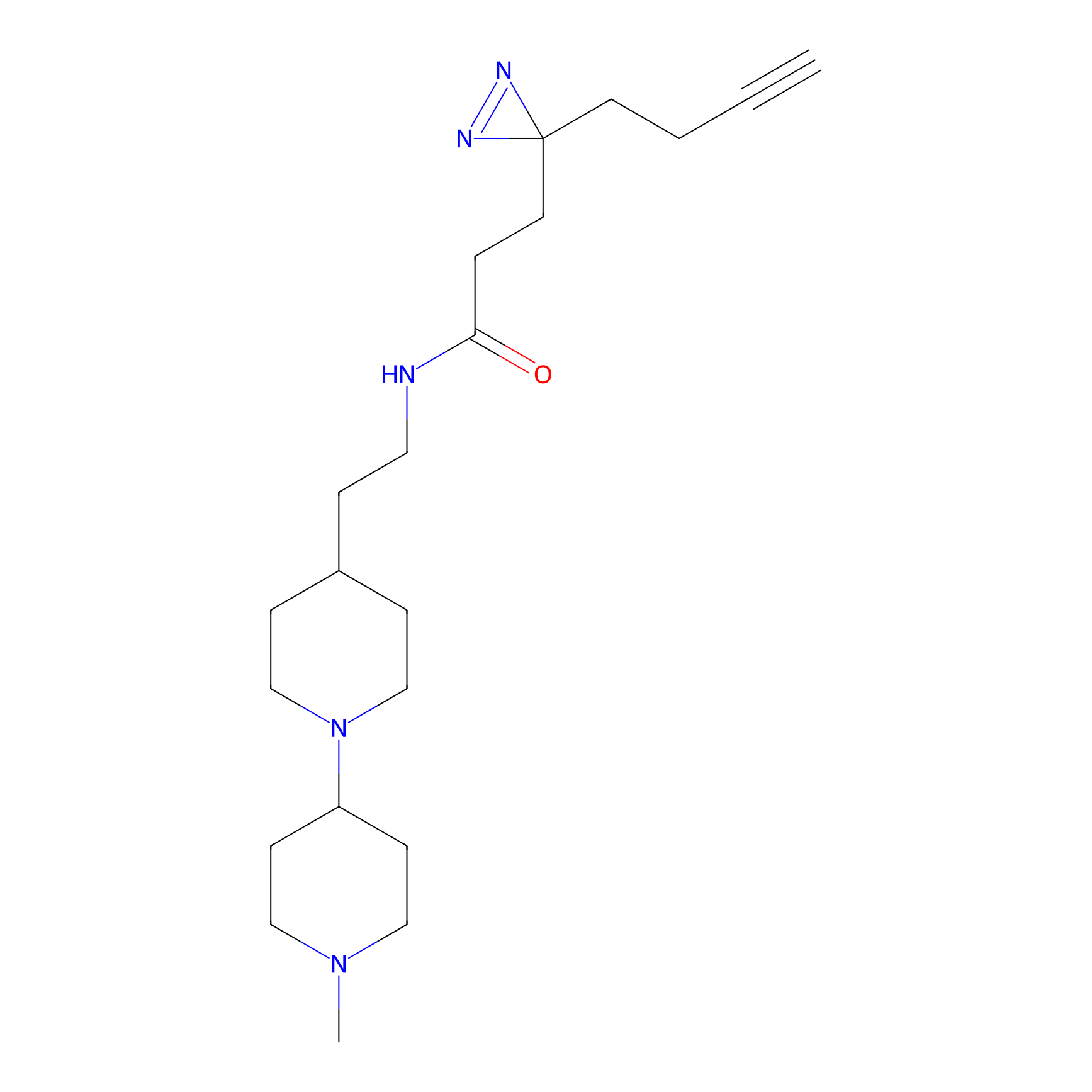
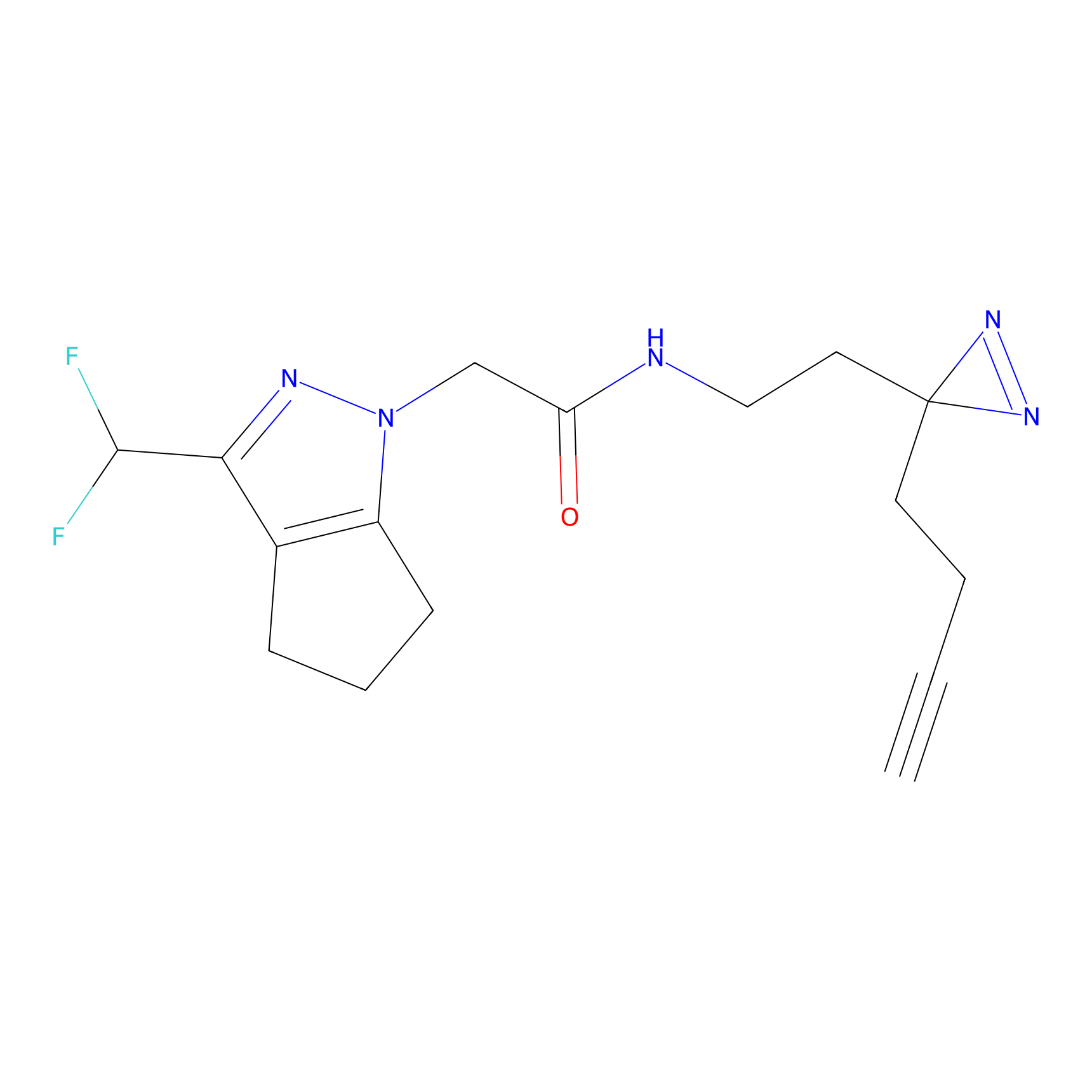
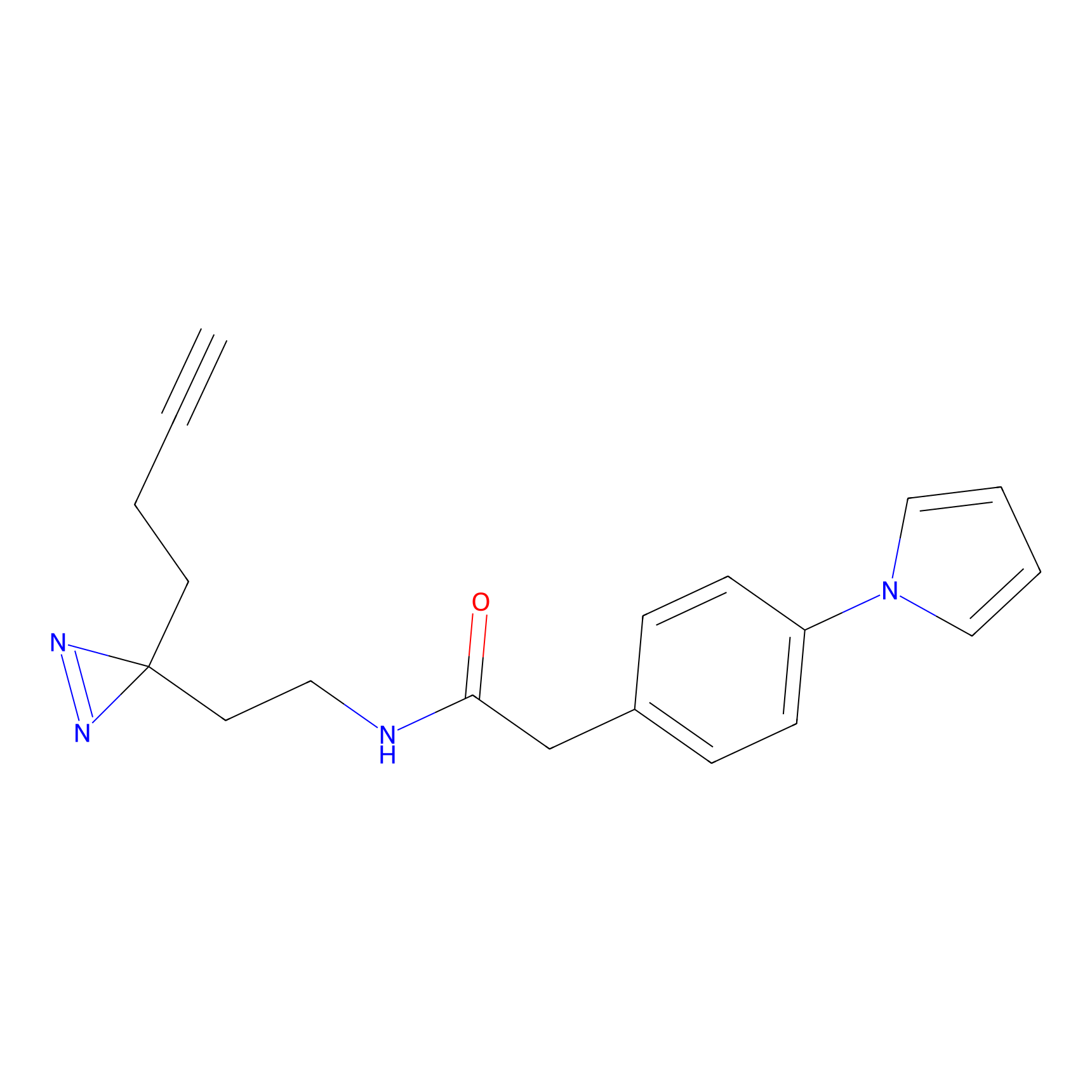
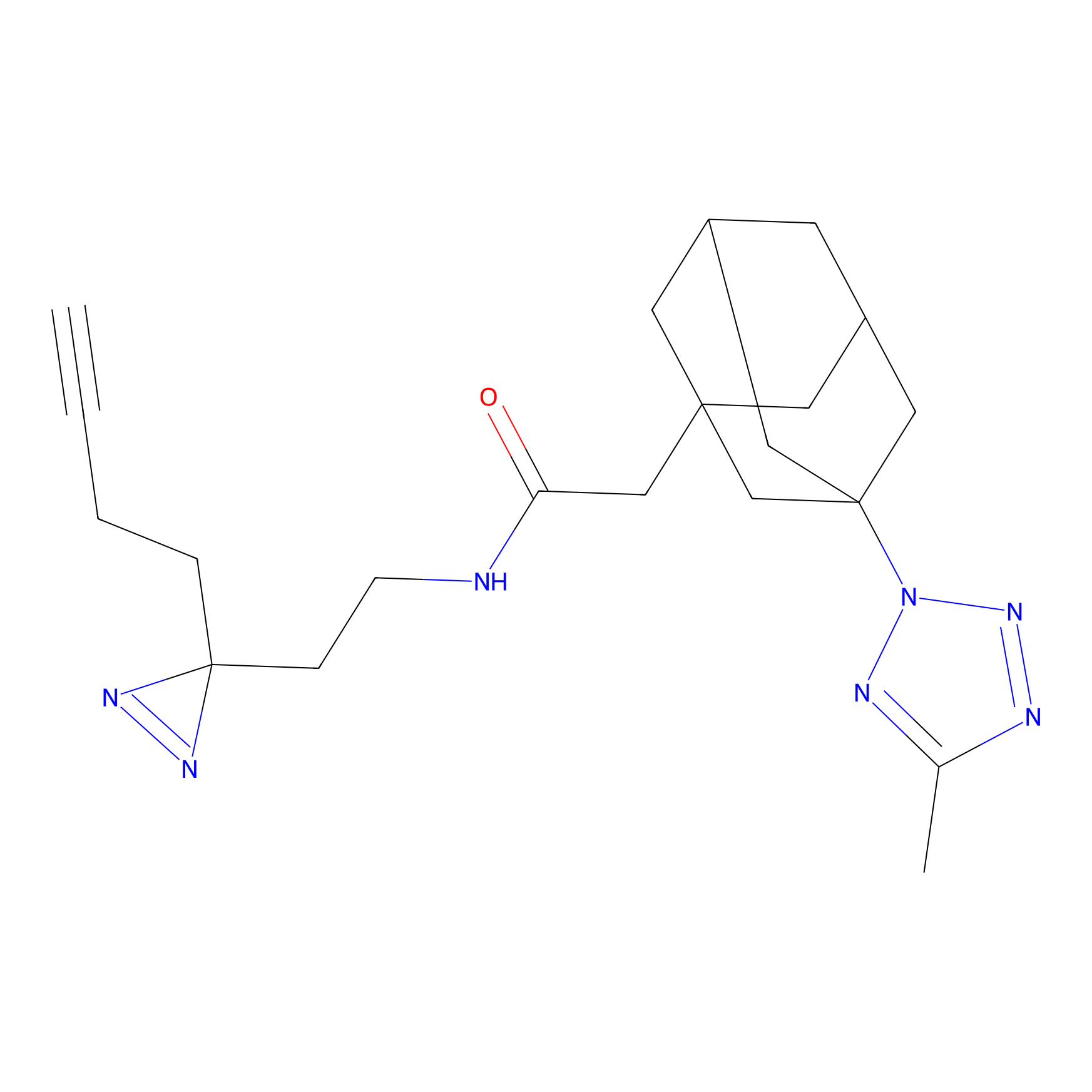
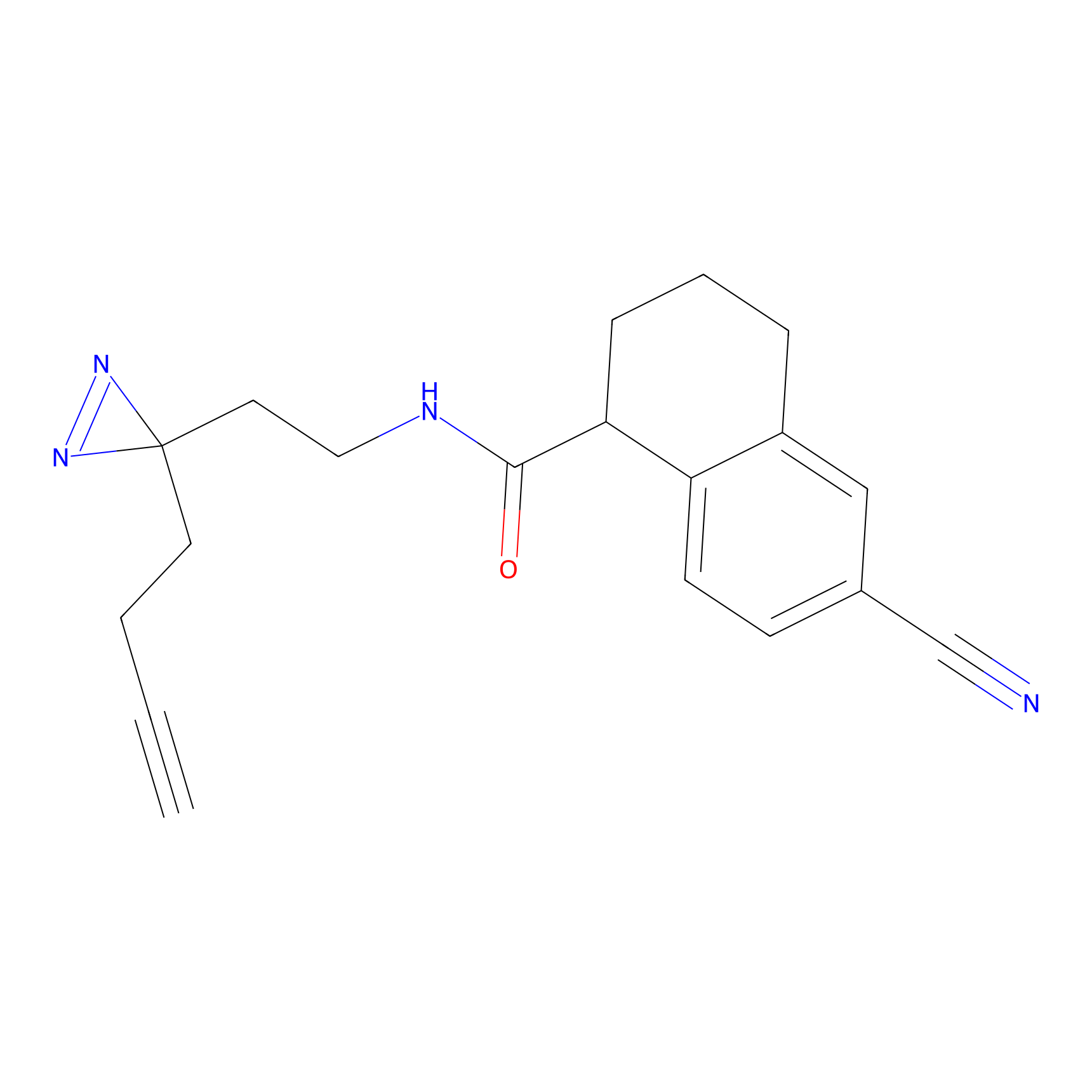
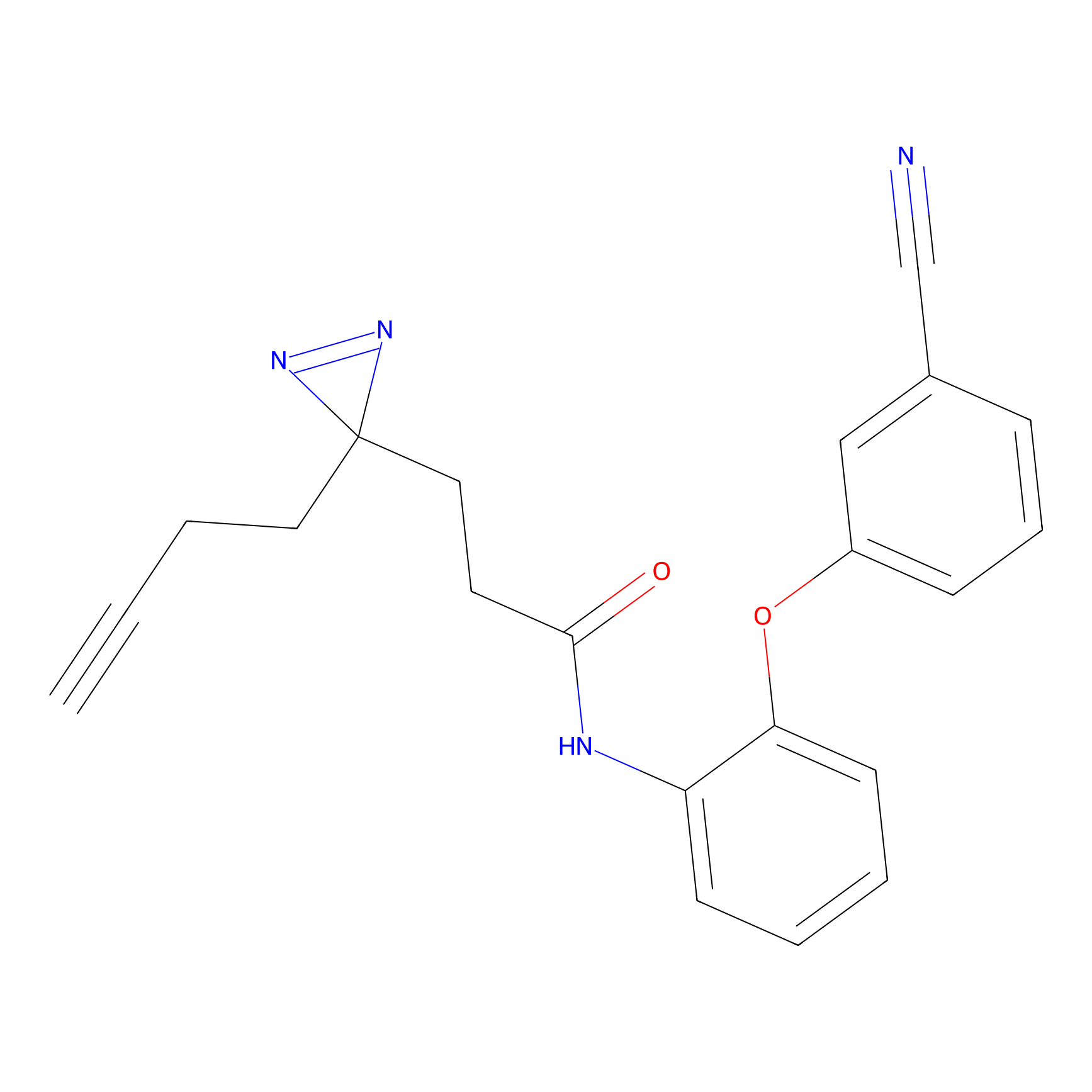
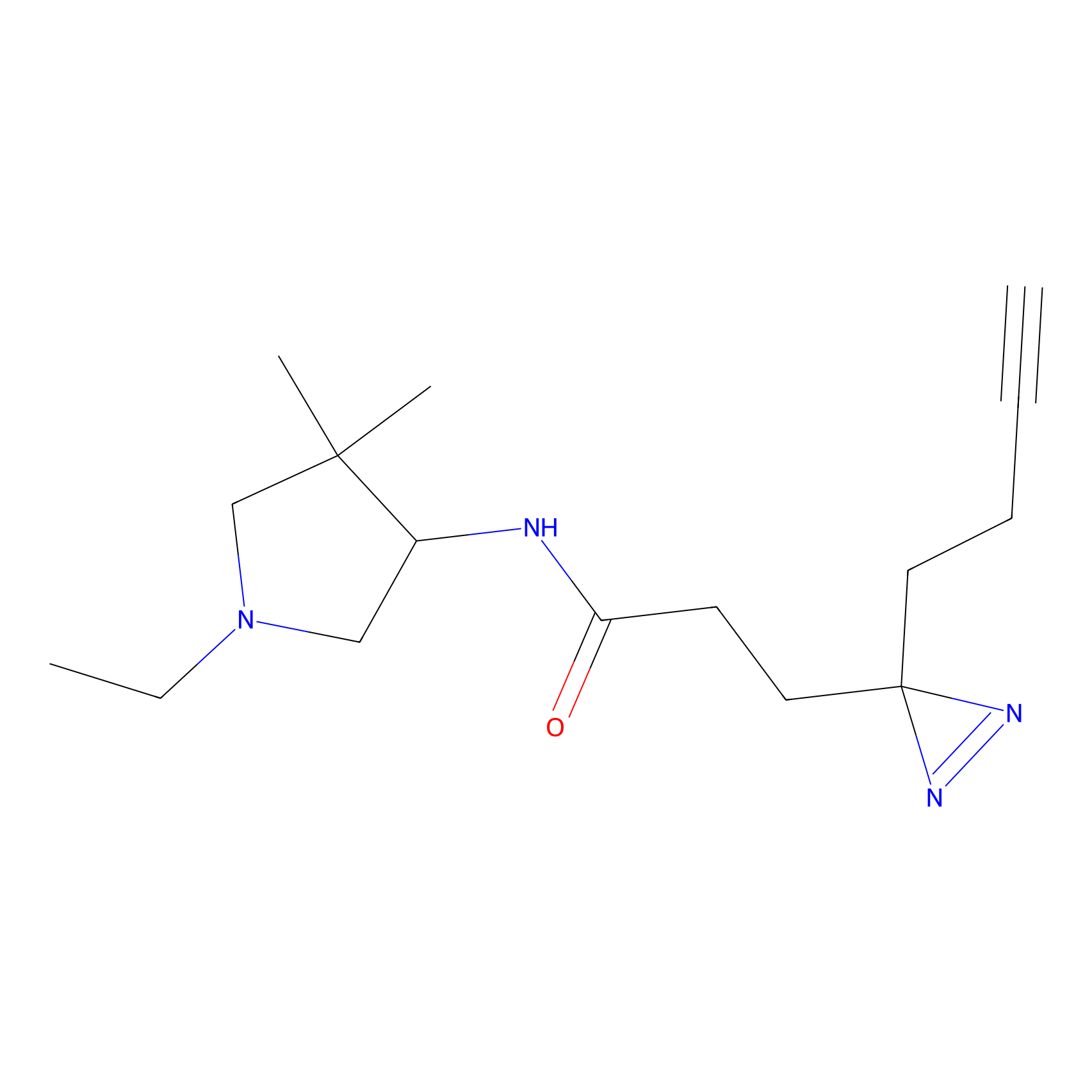
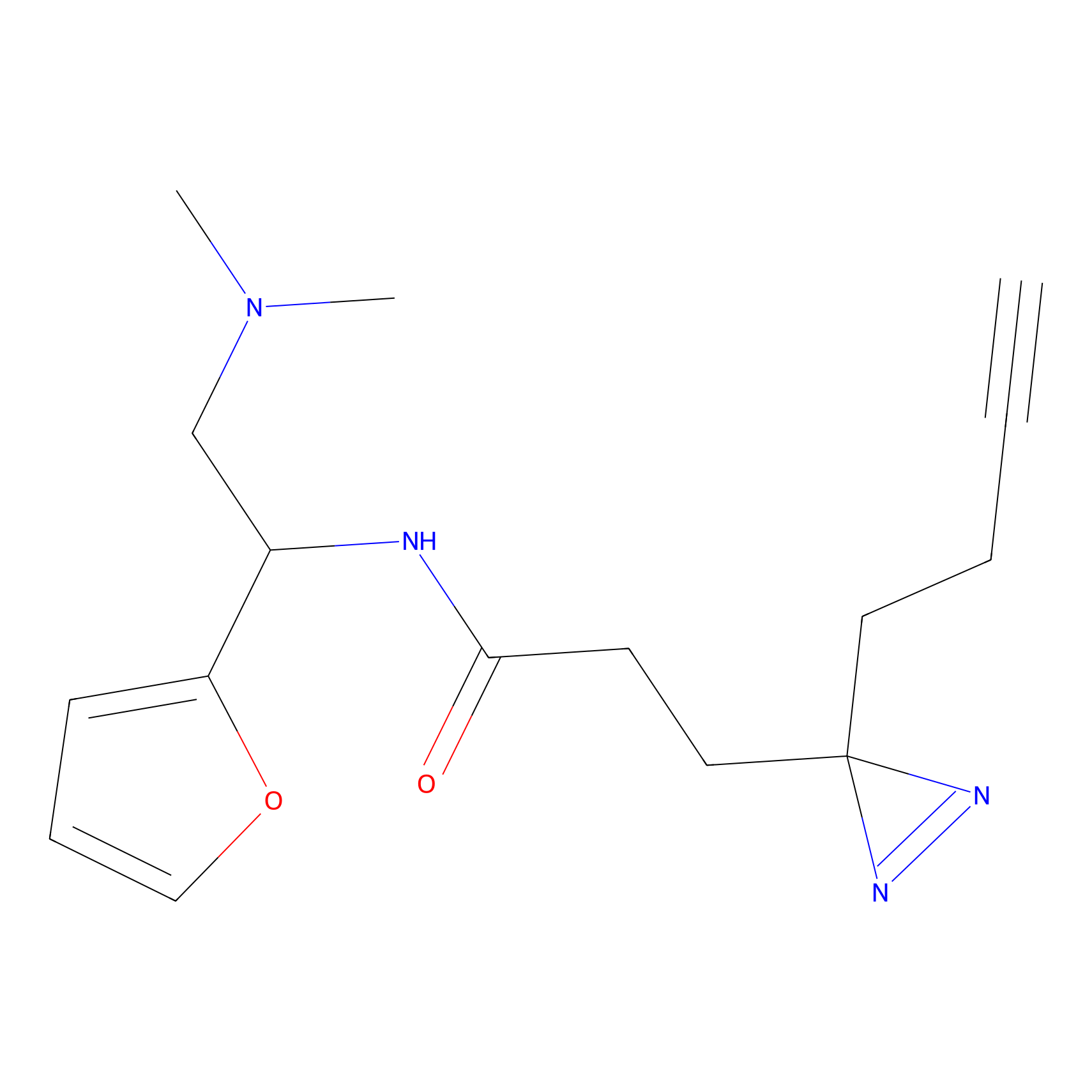
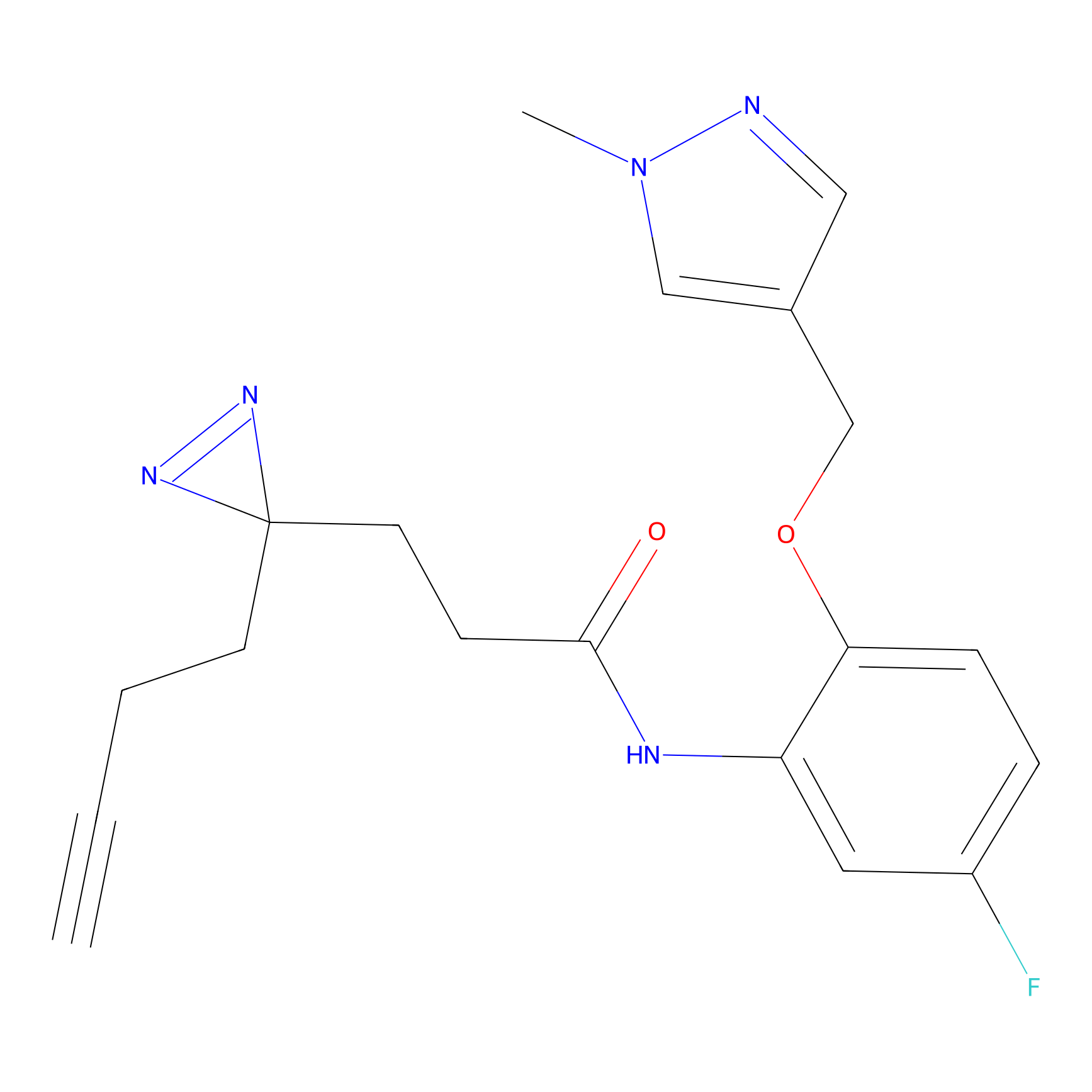
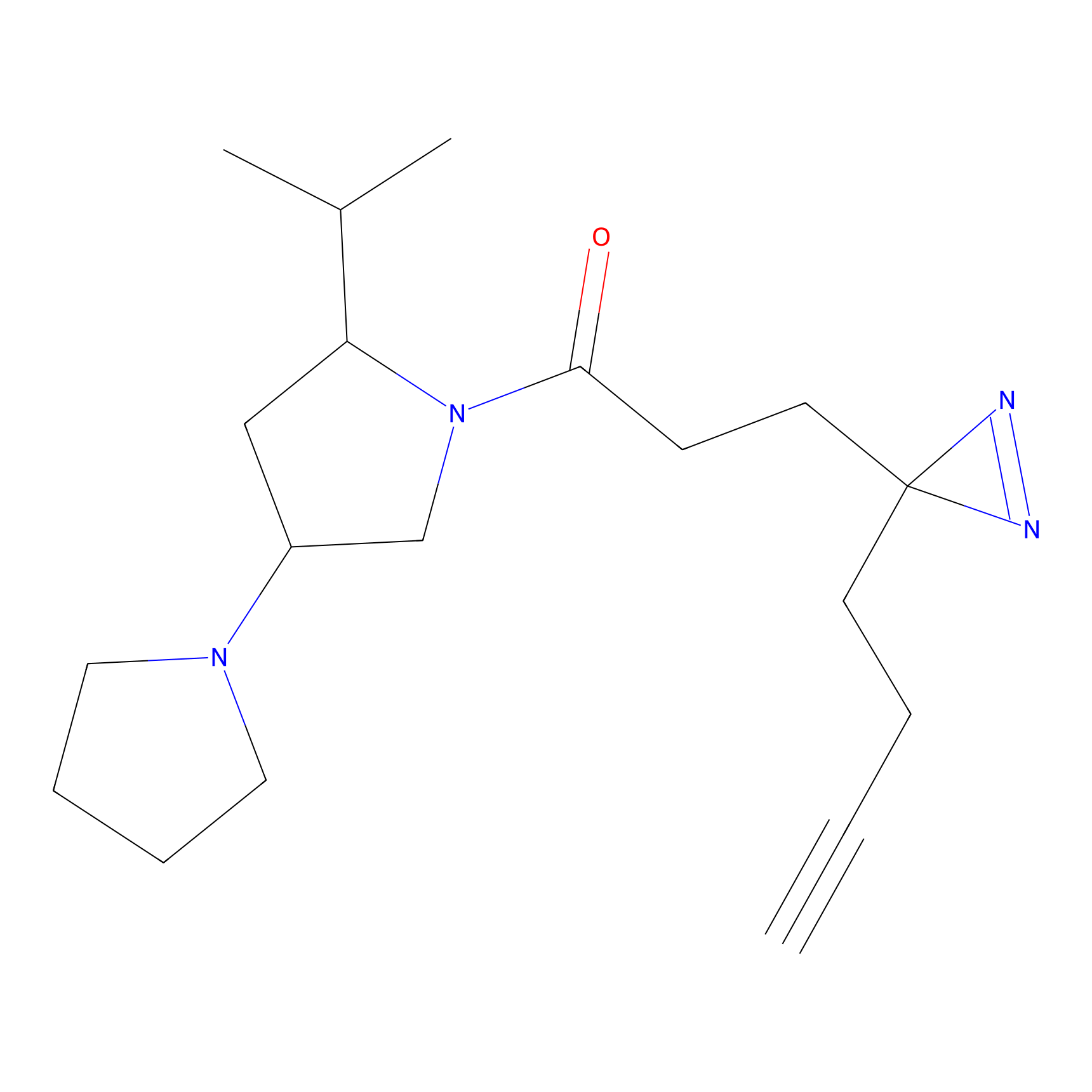
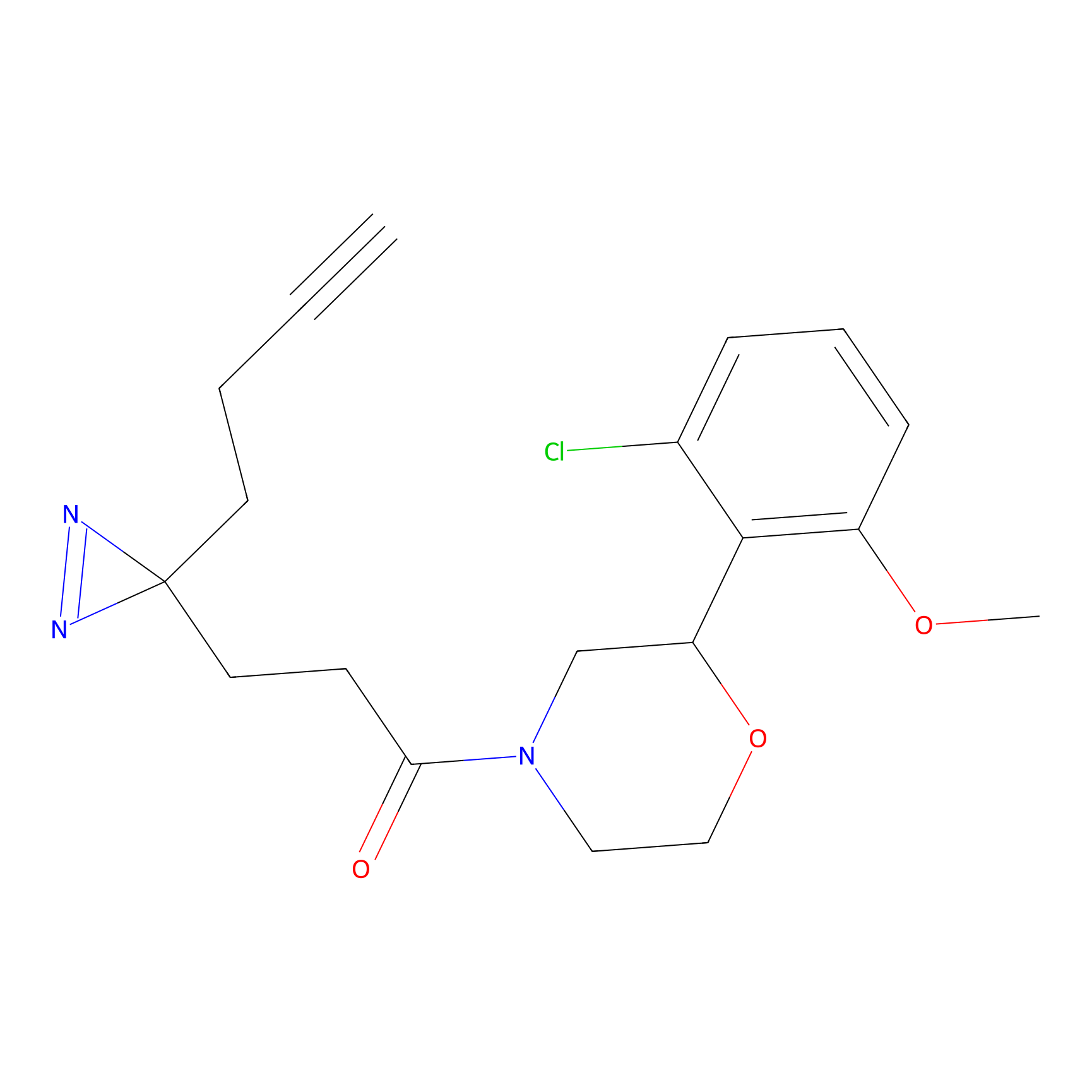
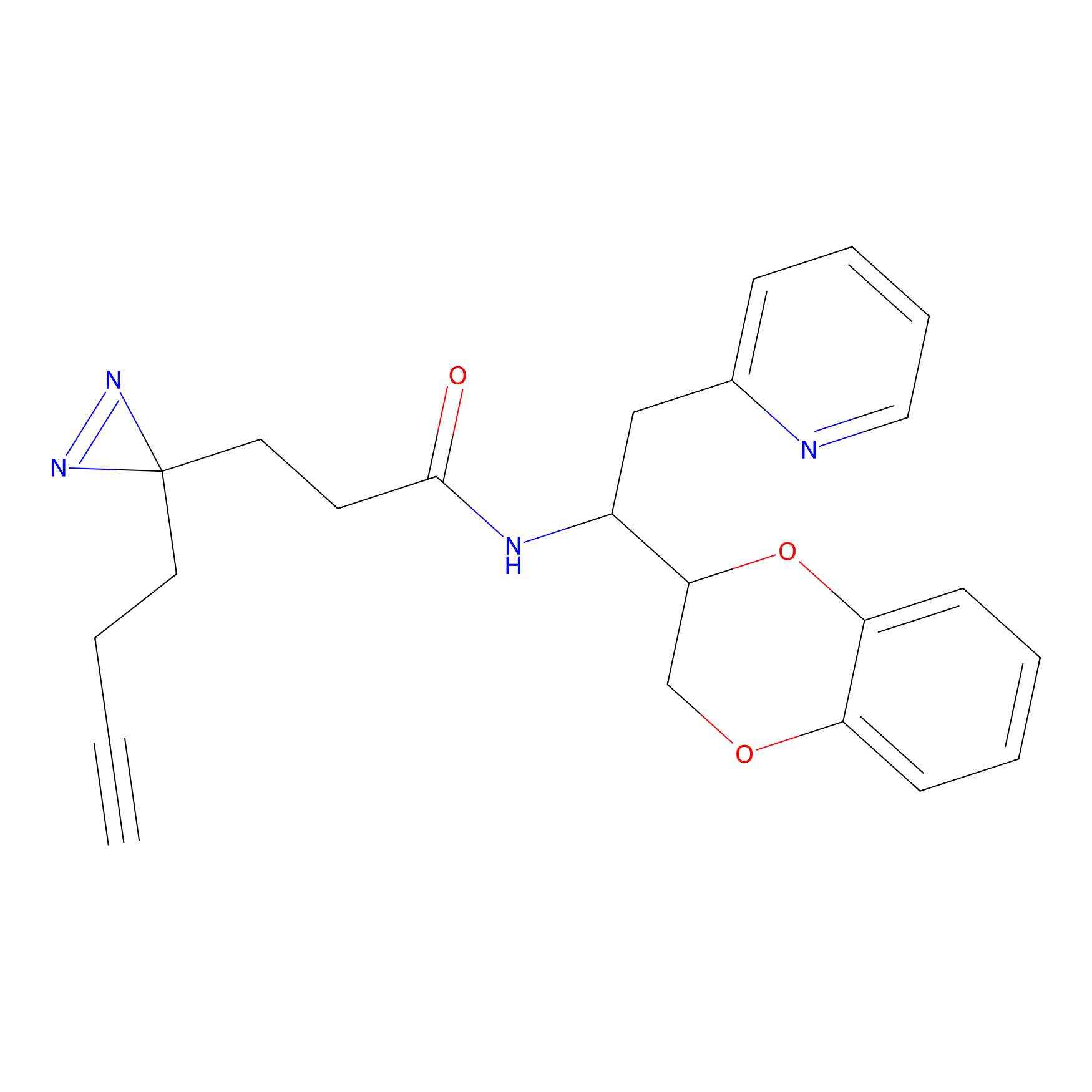
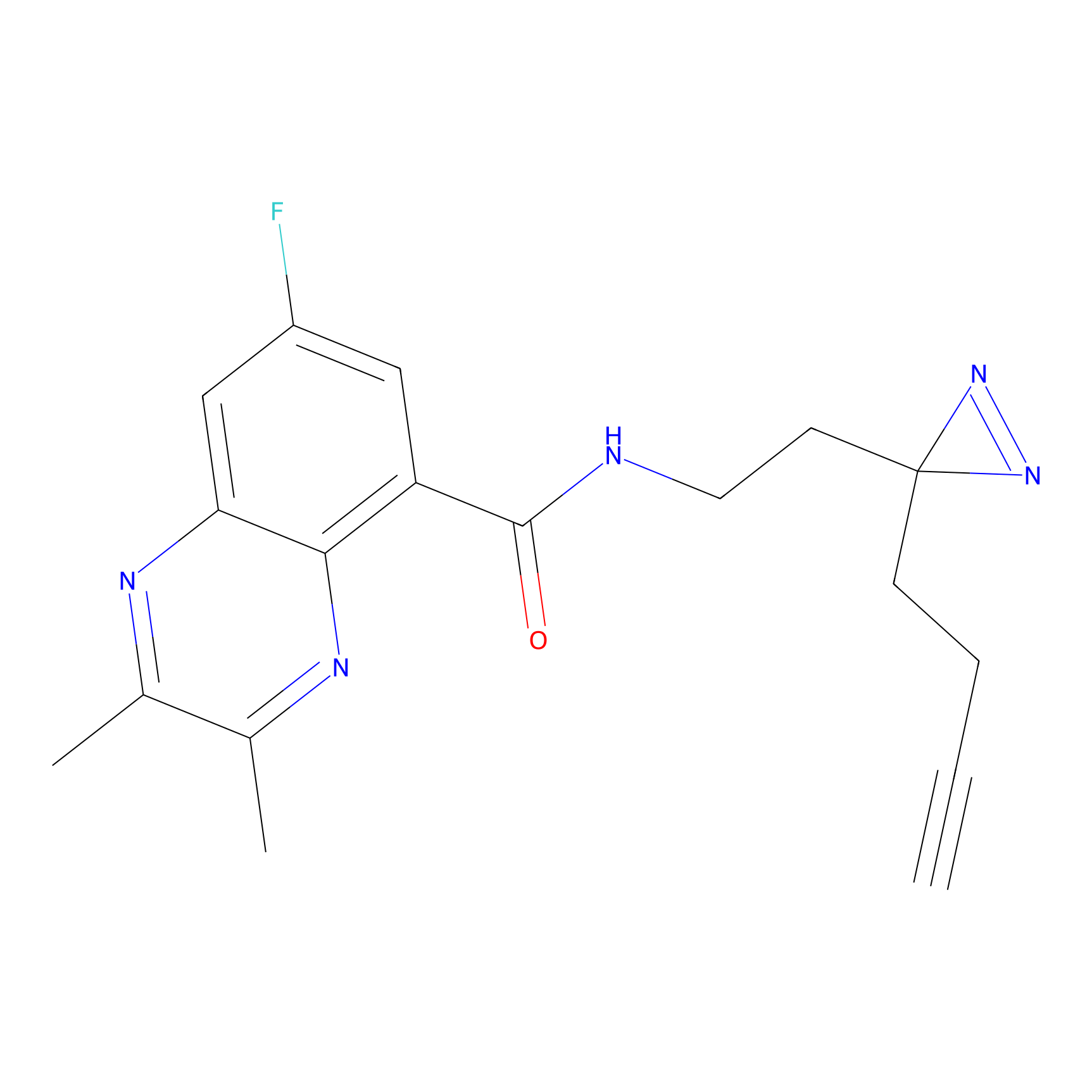
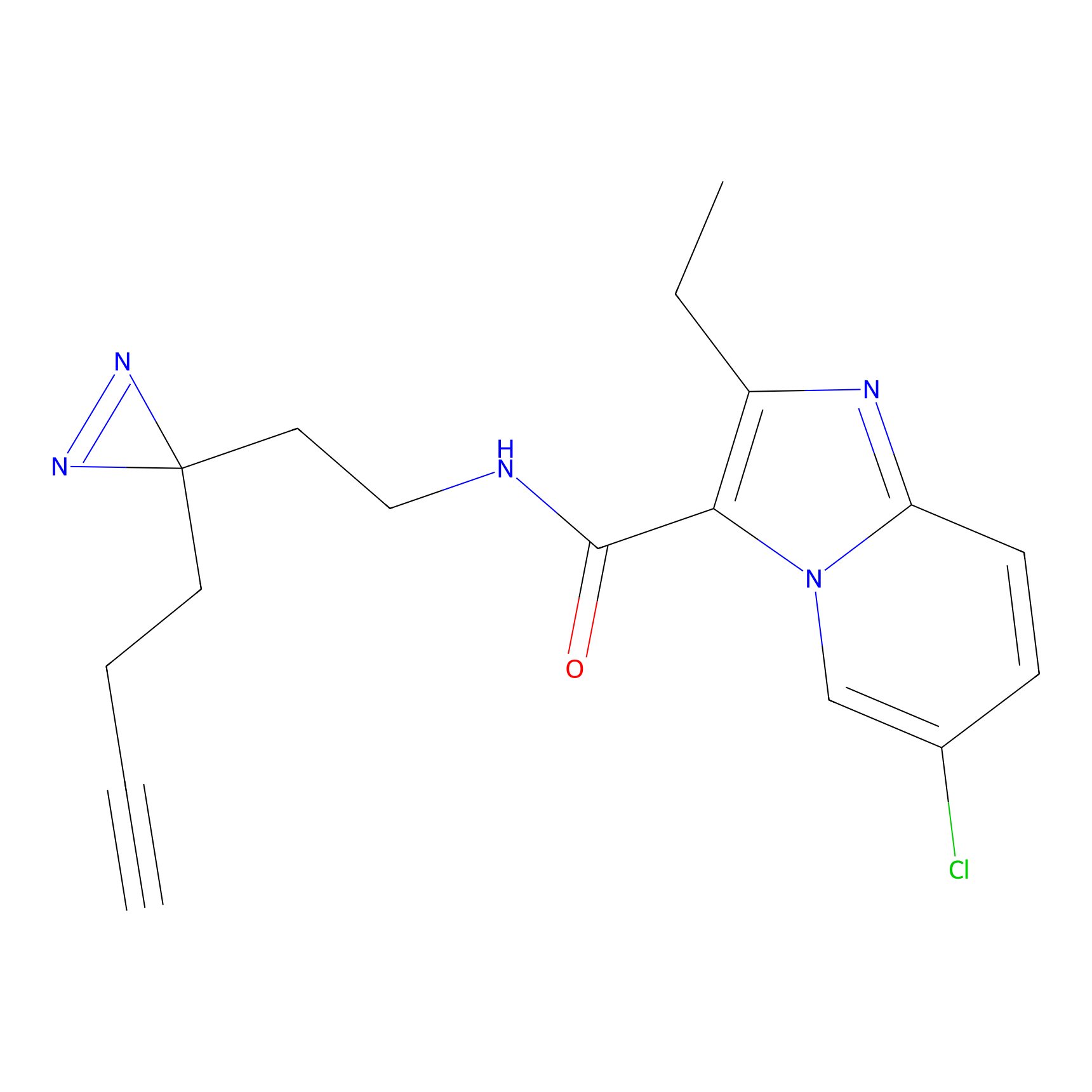
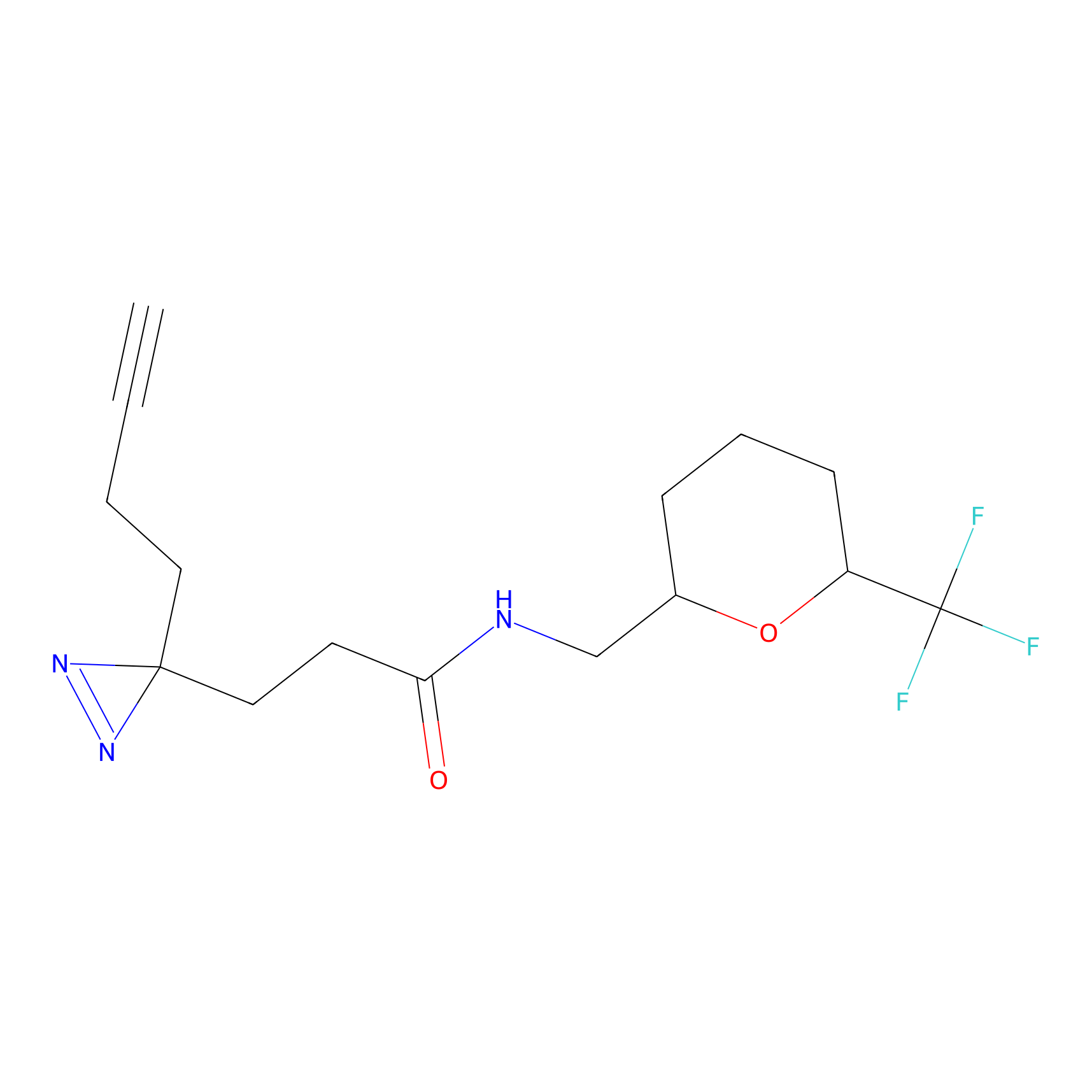
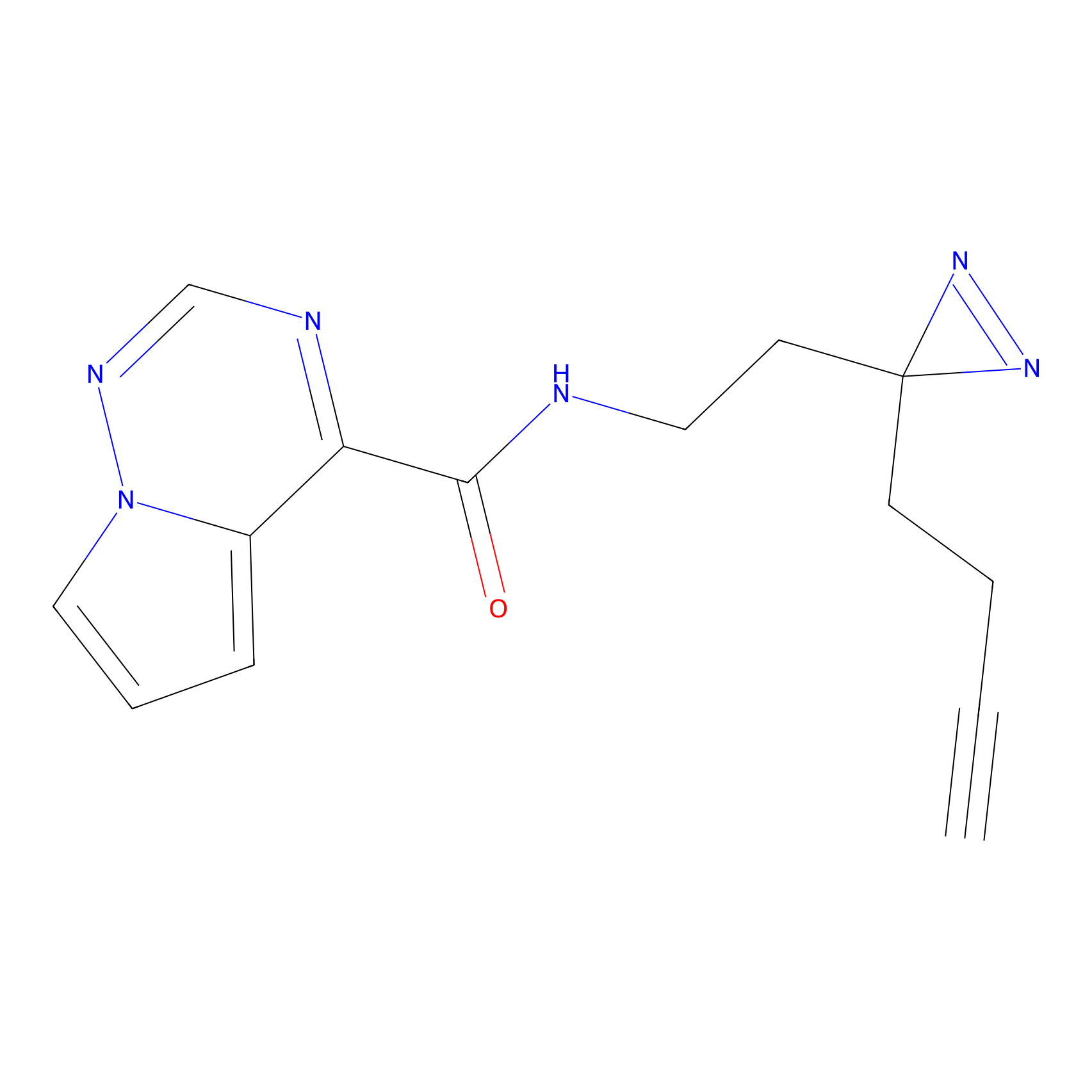
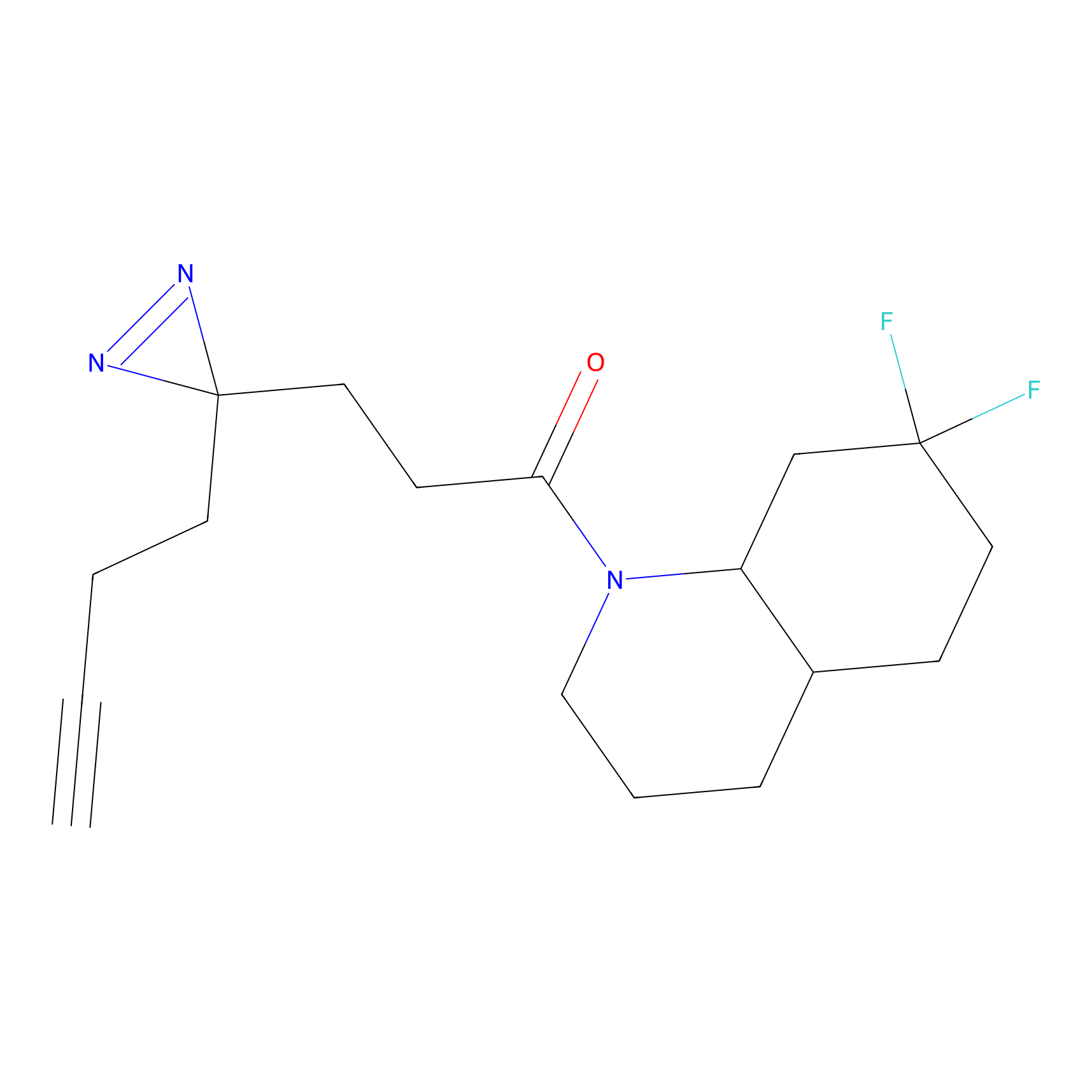
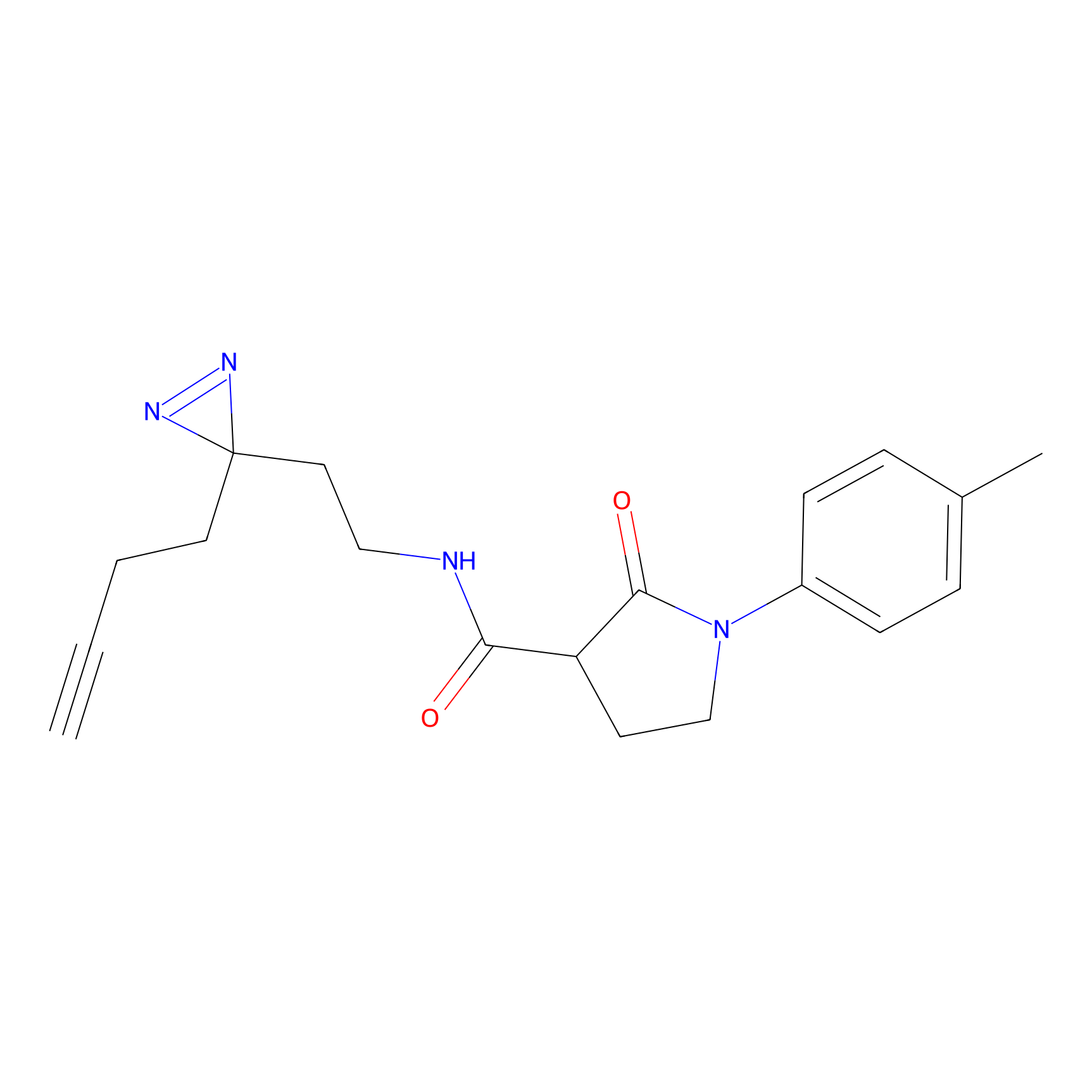
Alkylaryl probe 1 Probe Info |
 |
20.00 | LDD0387 | [1] | |
|
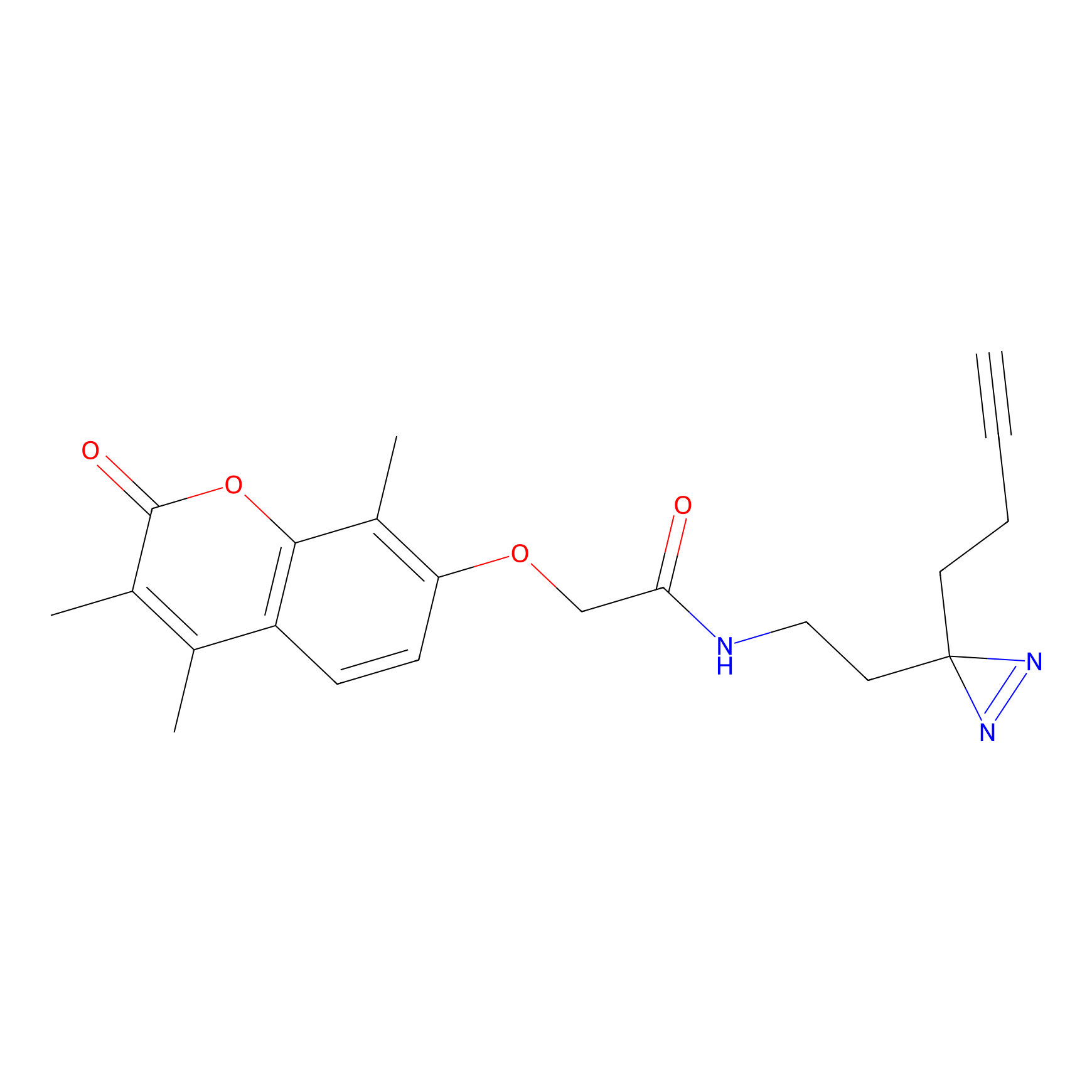
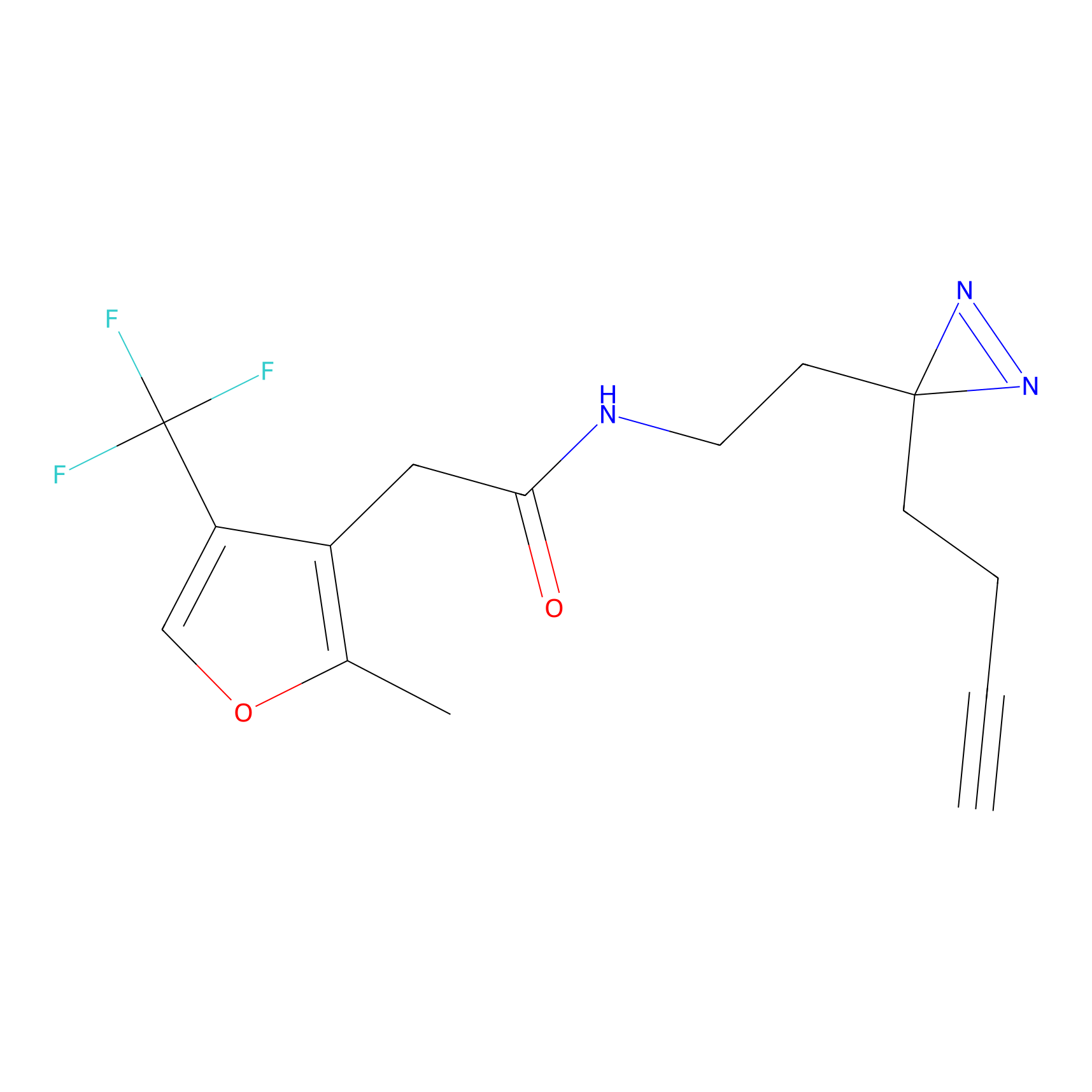
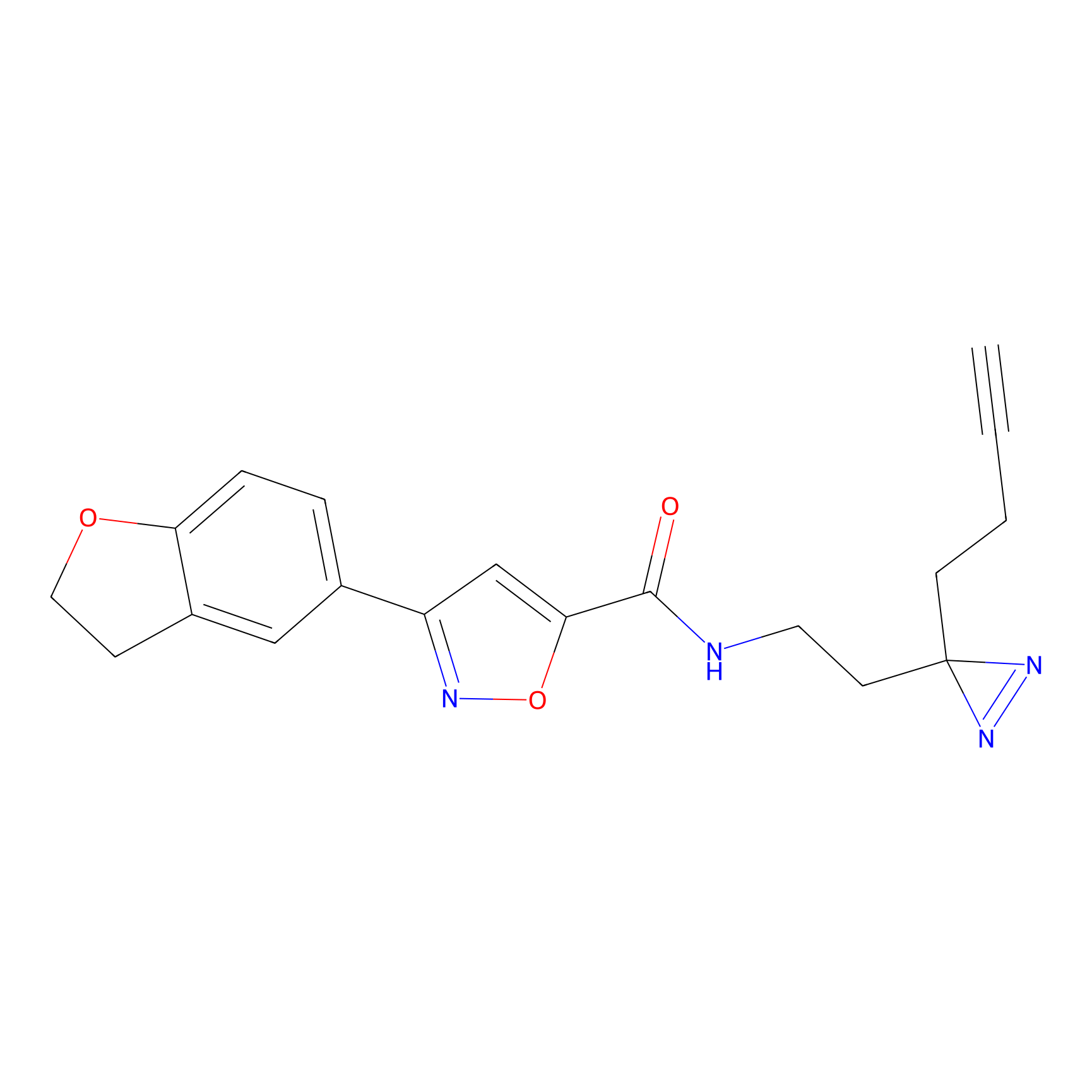
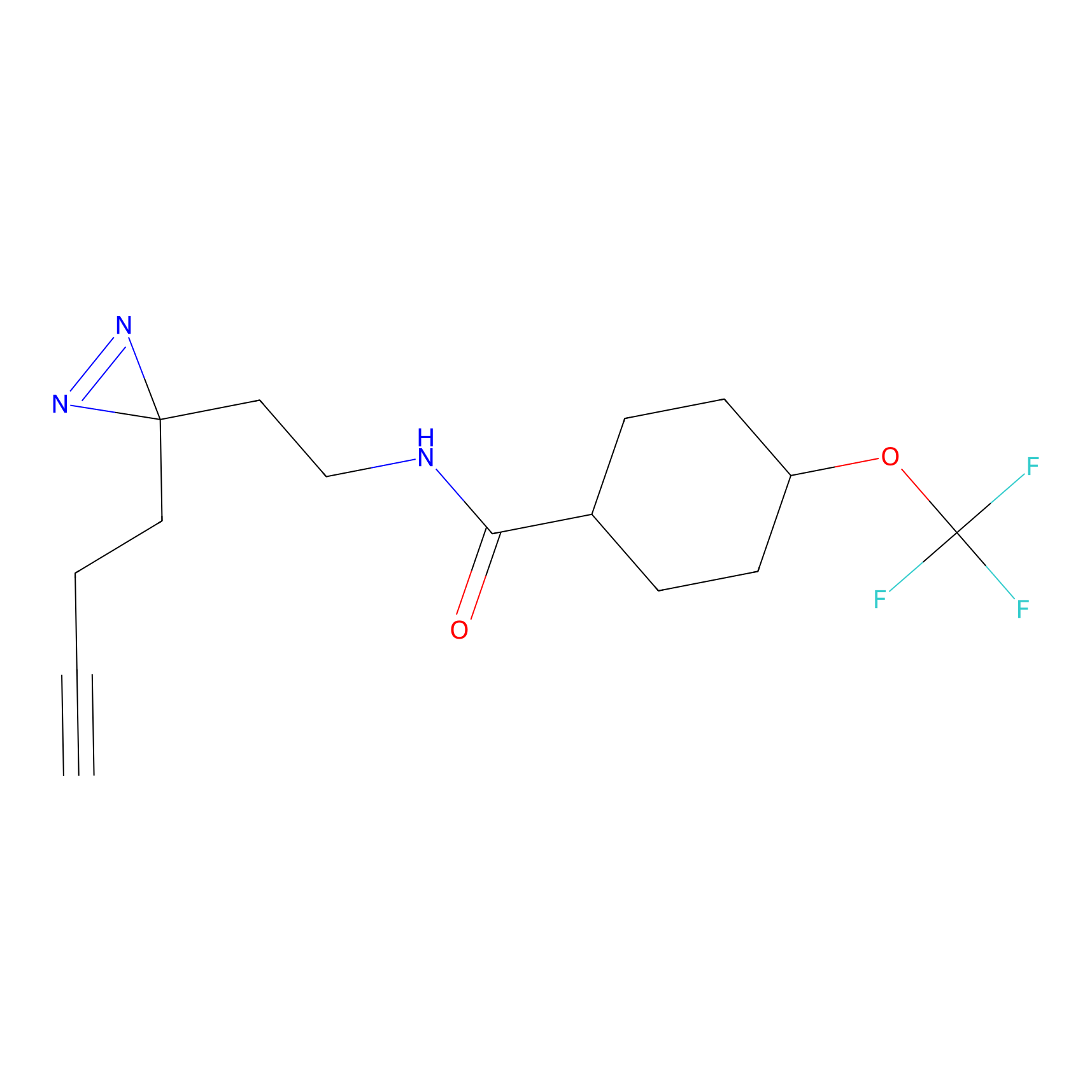
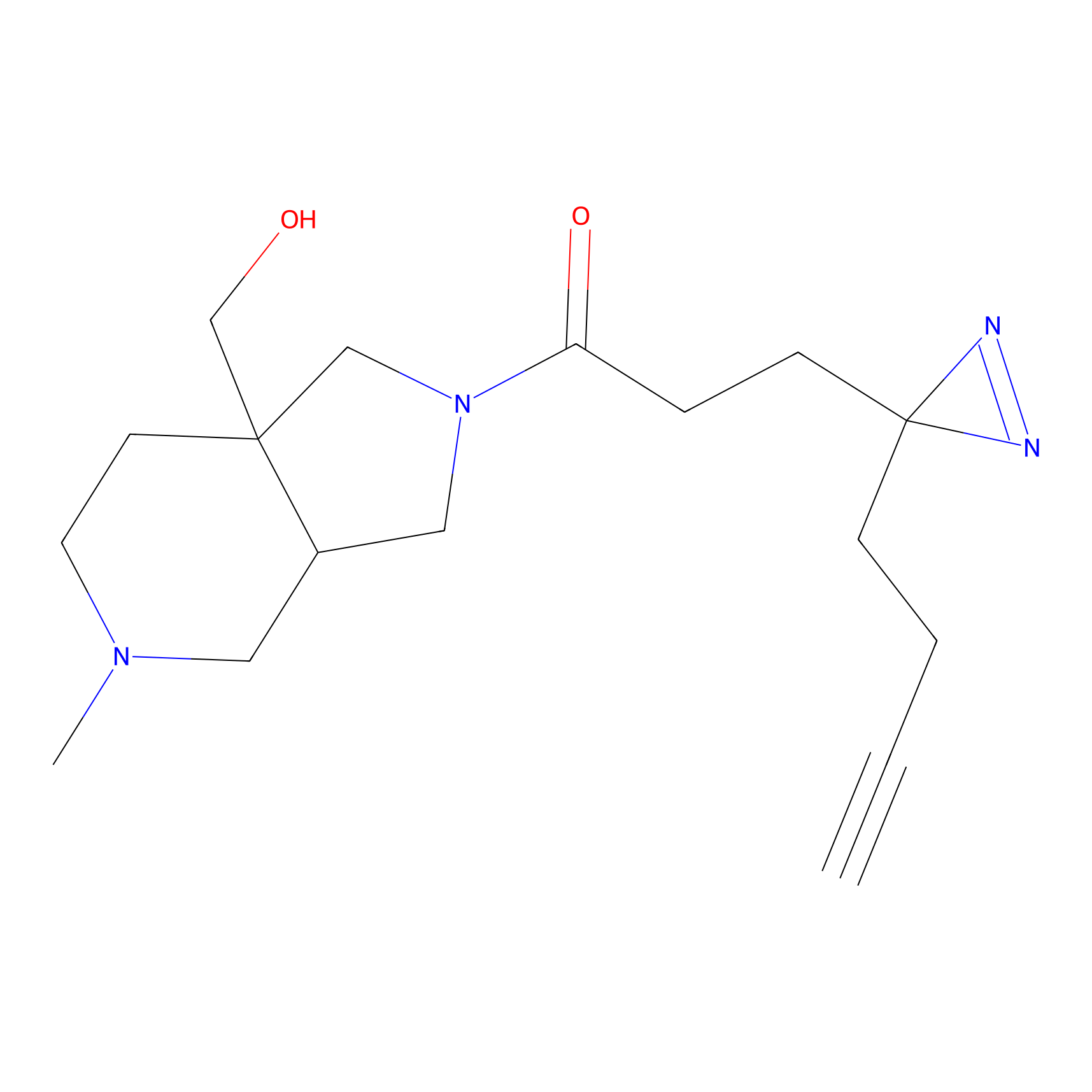
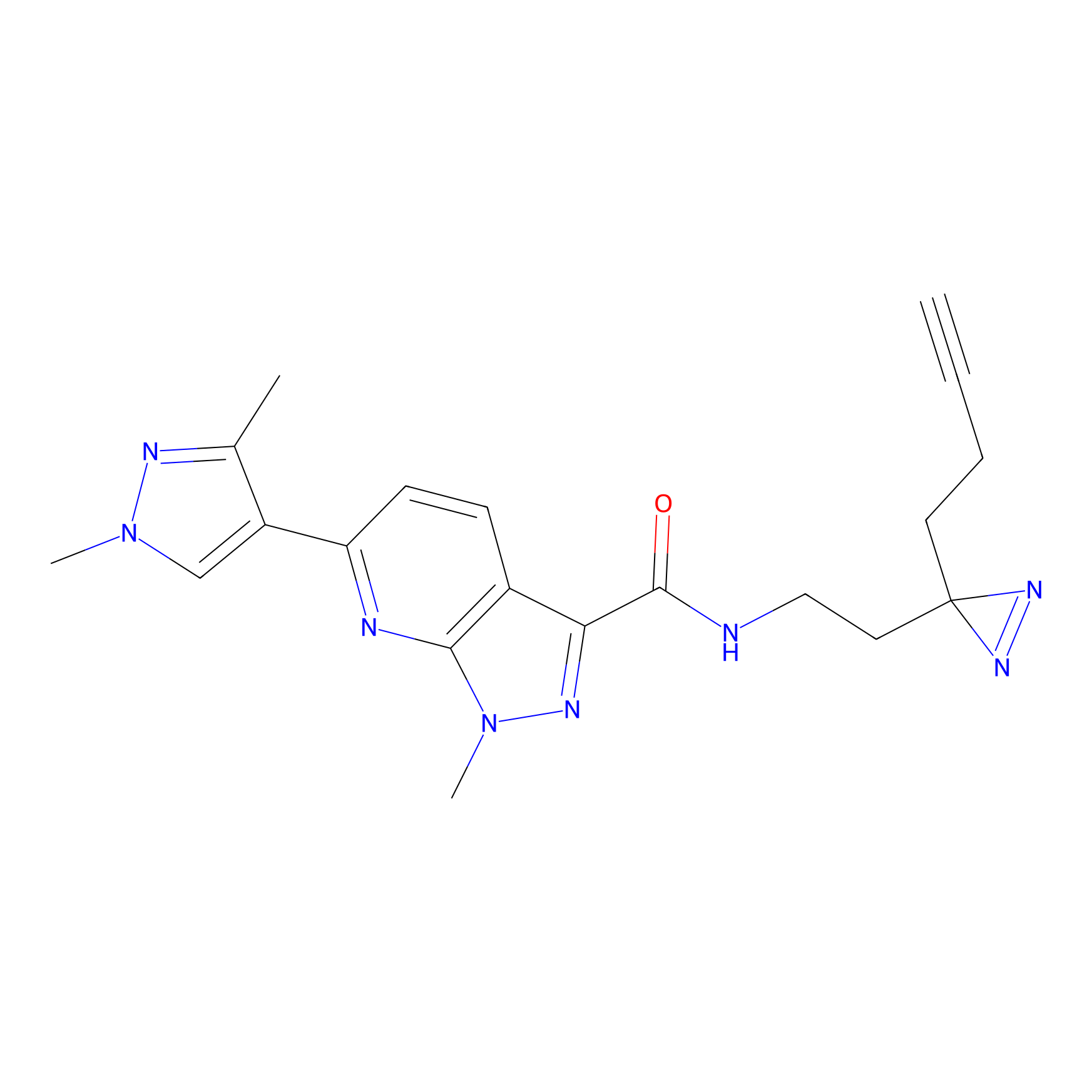
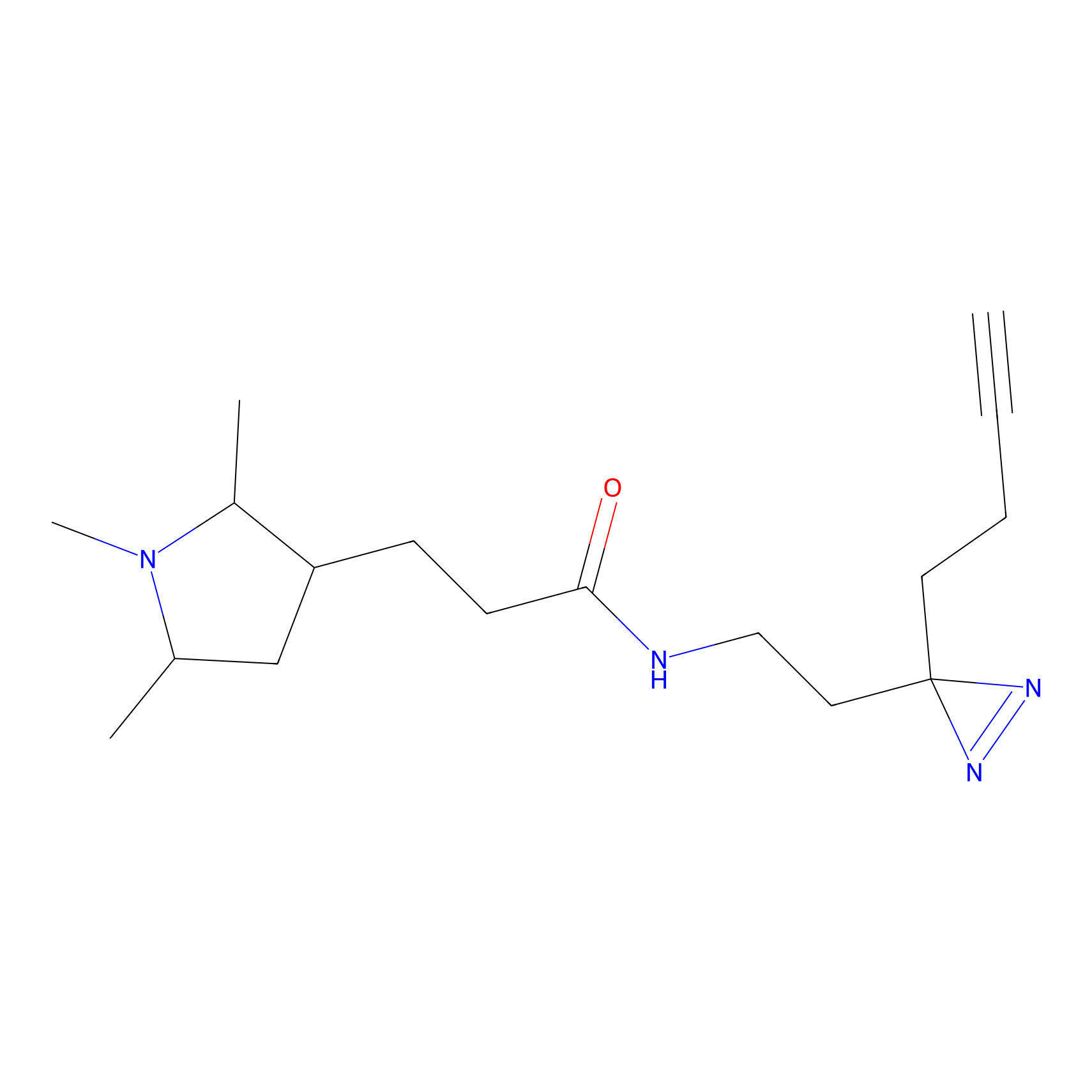
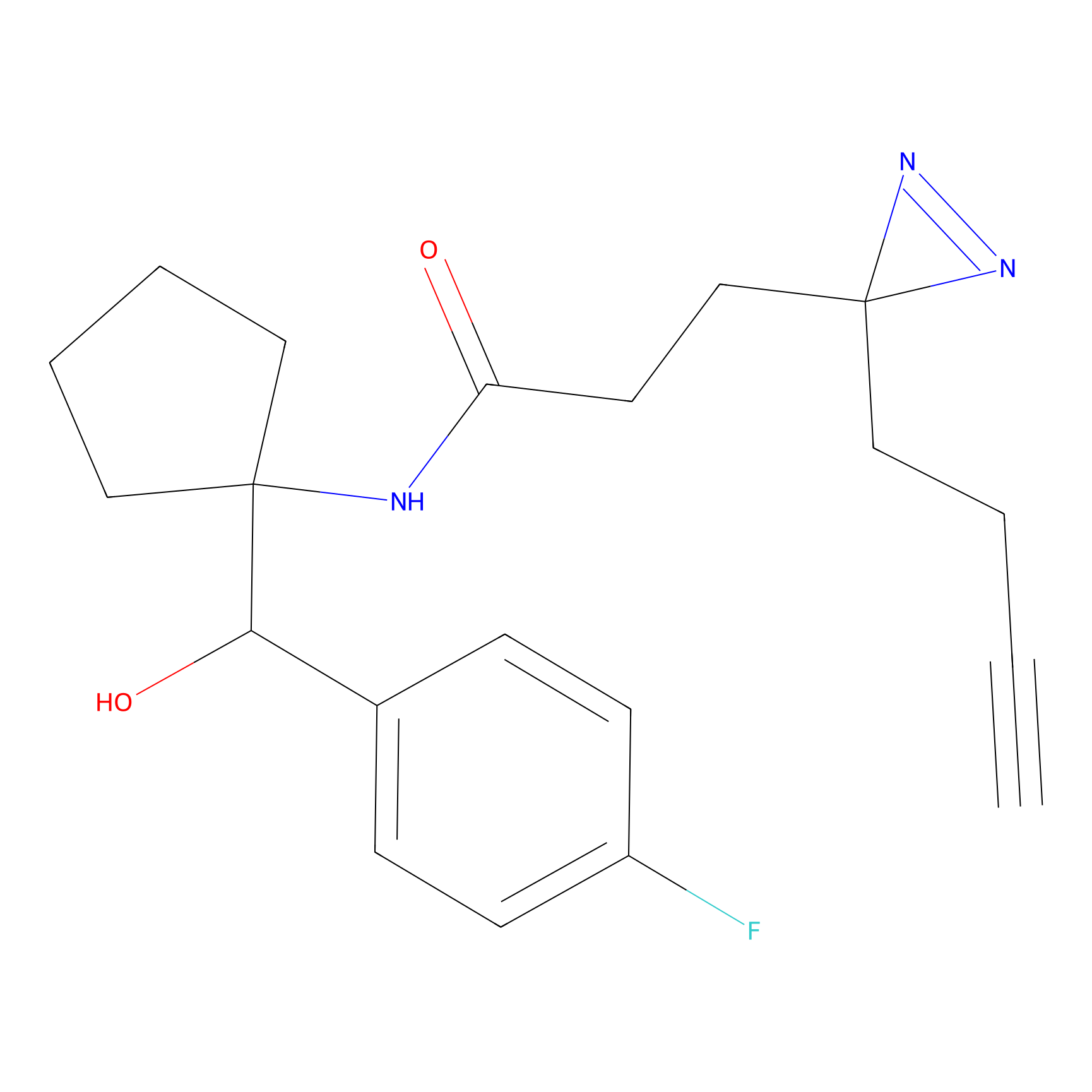
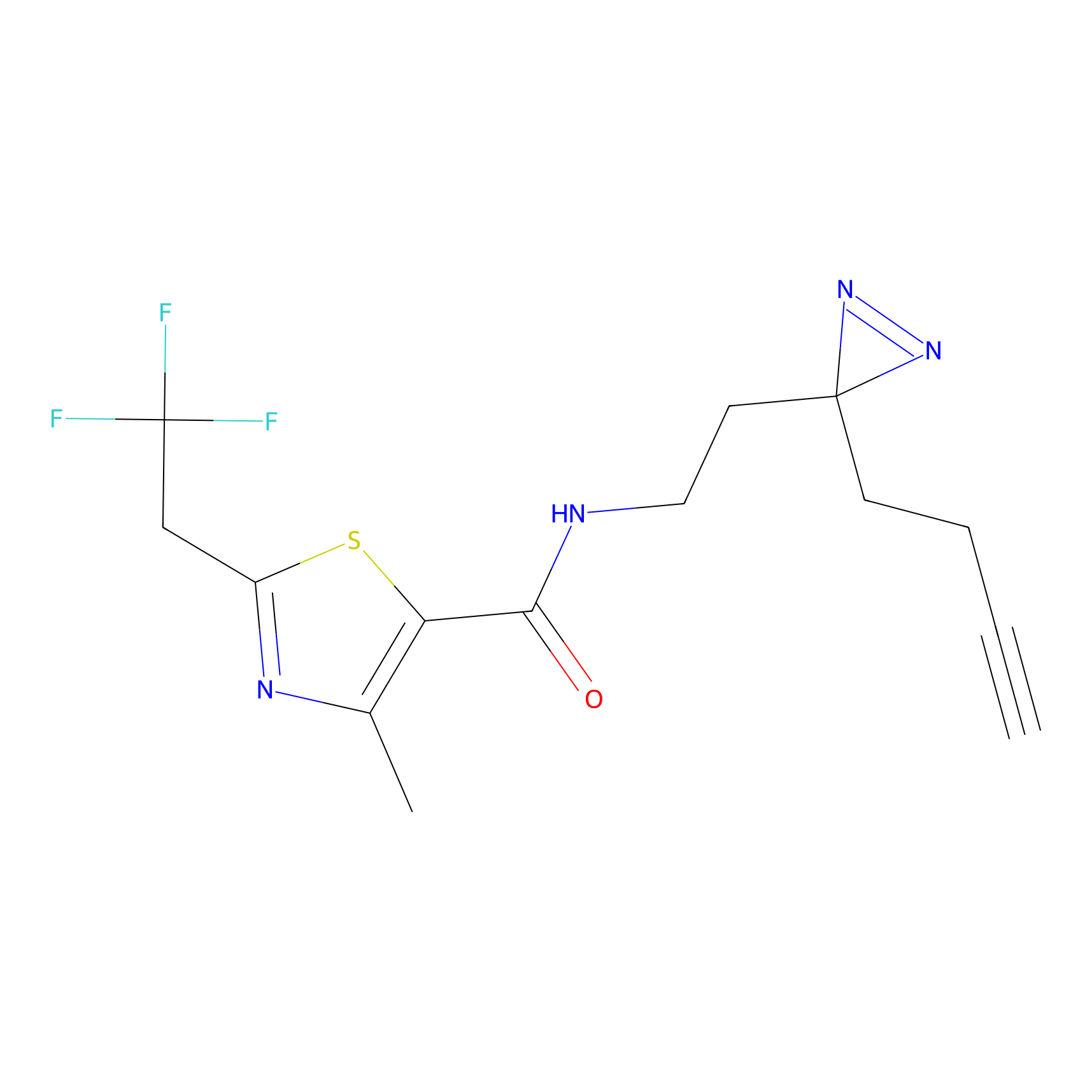
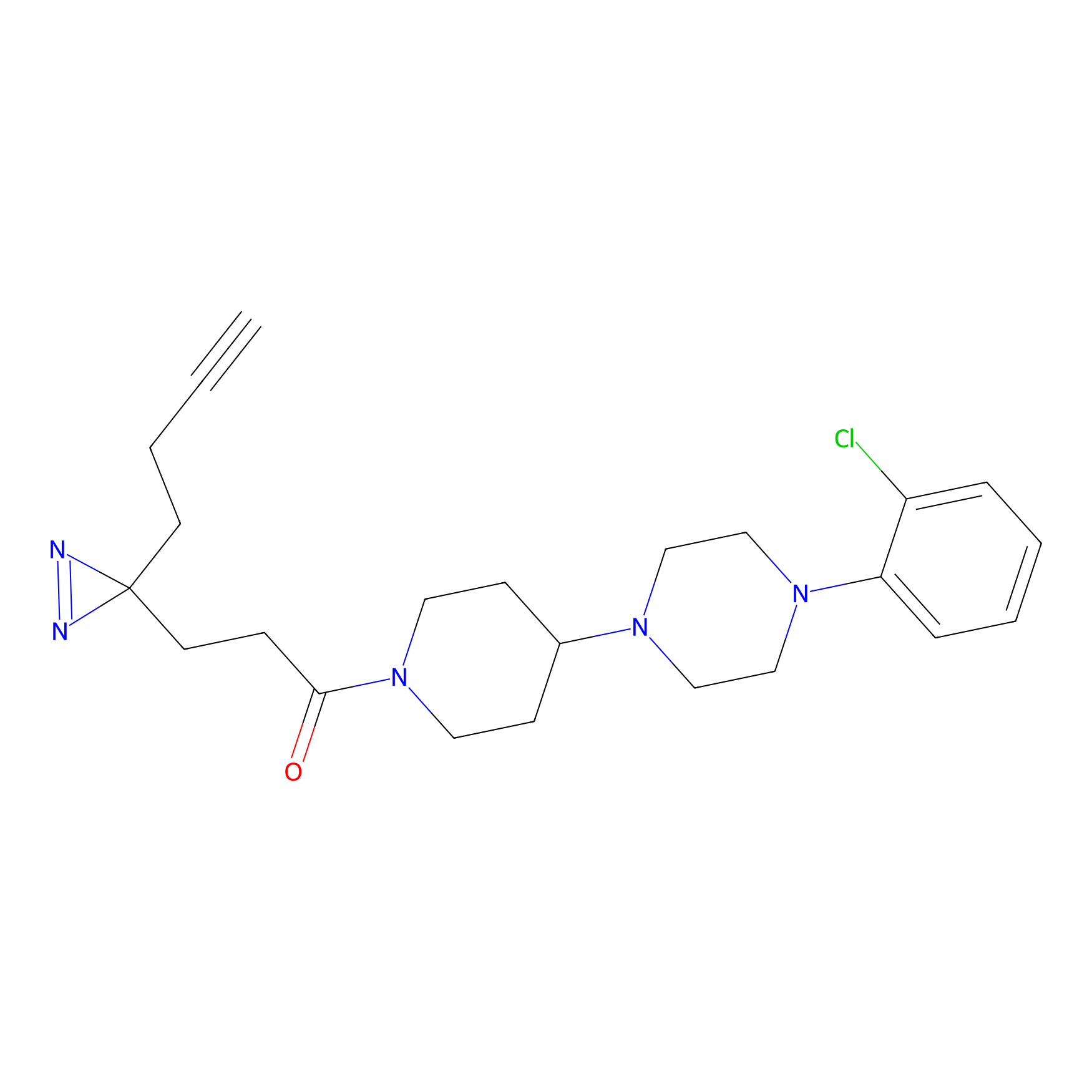
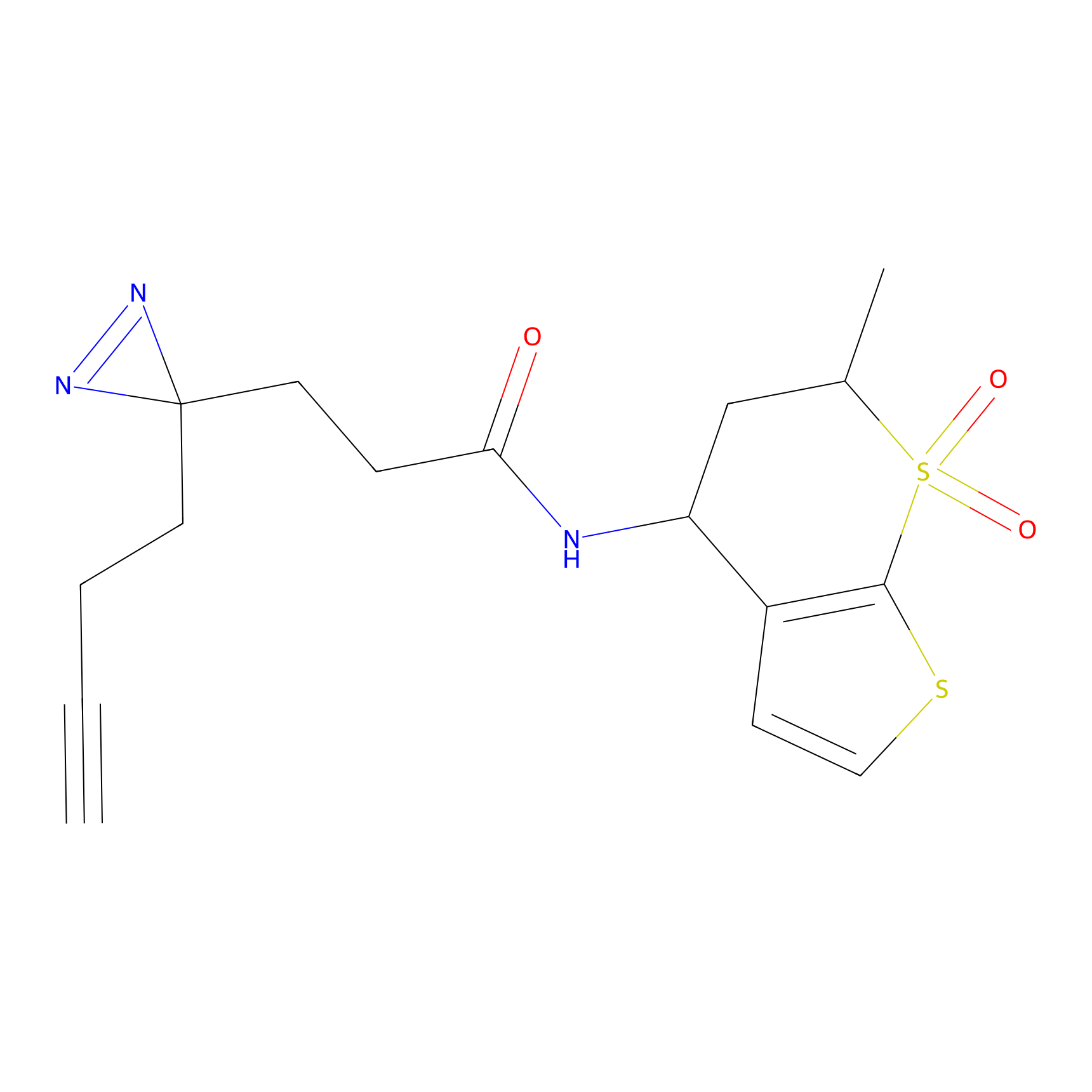
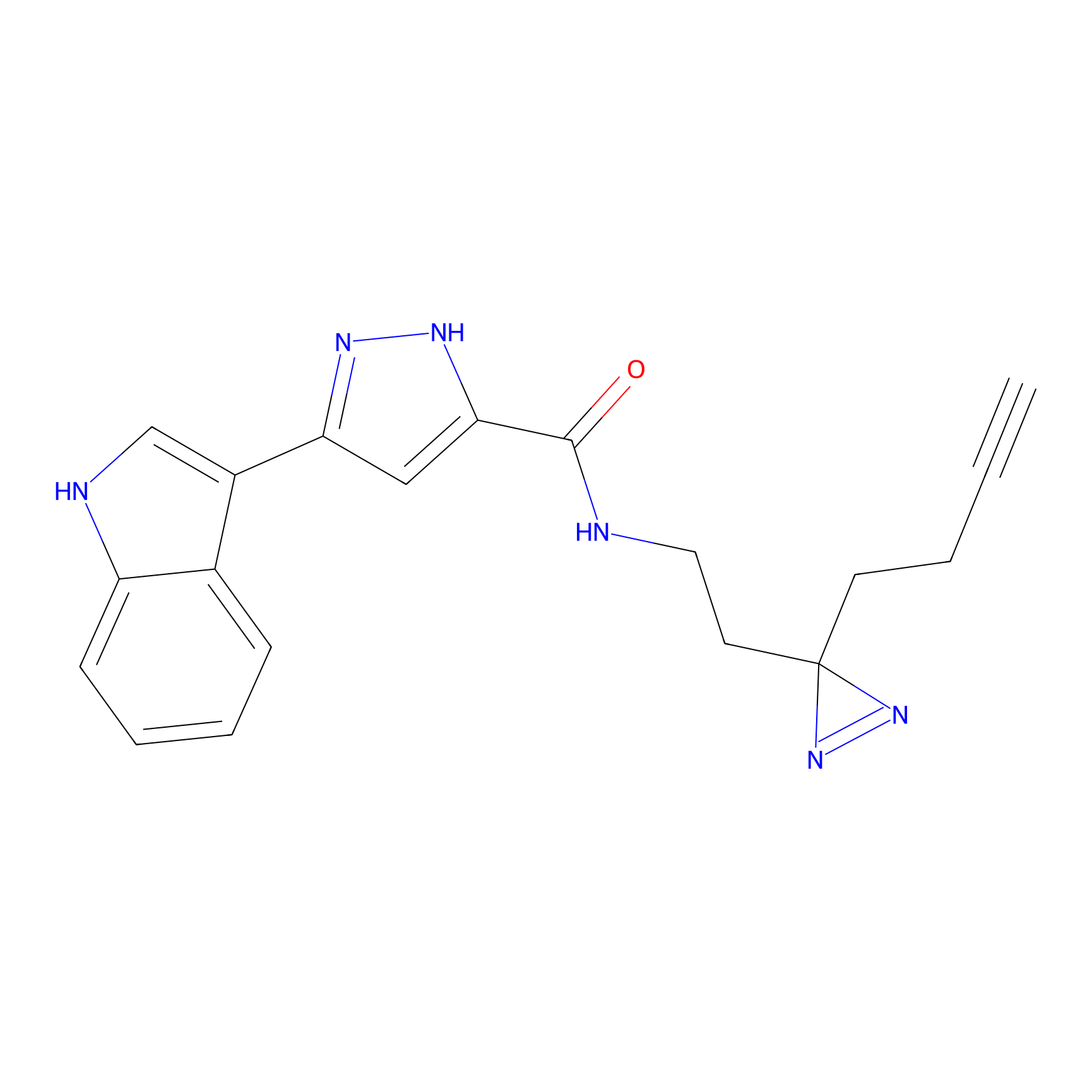
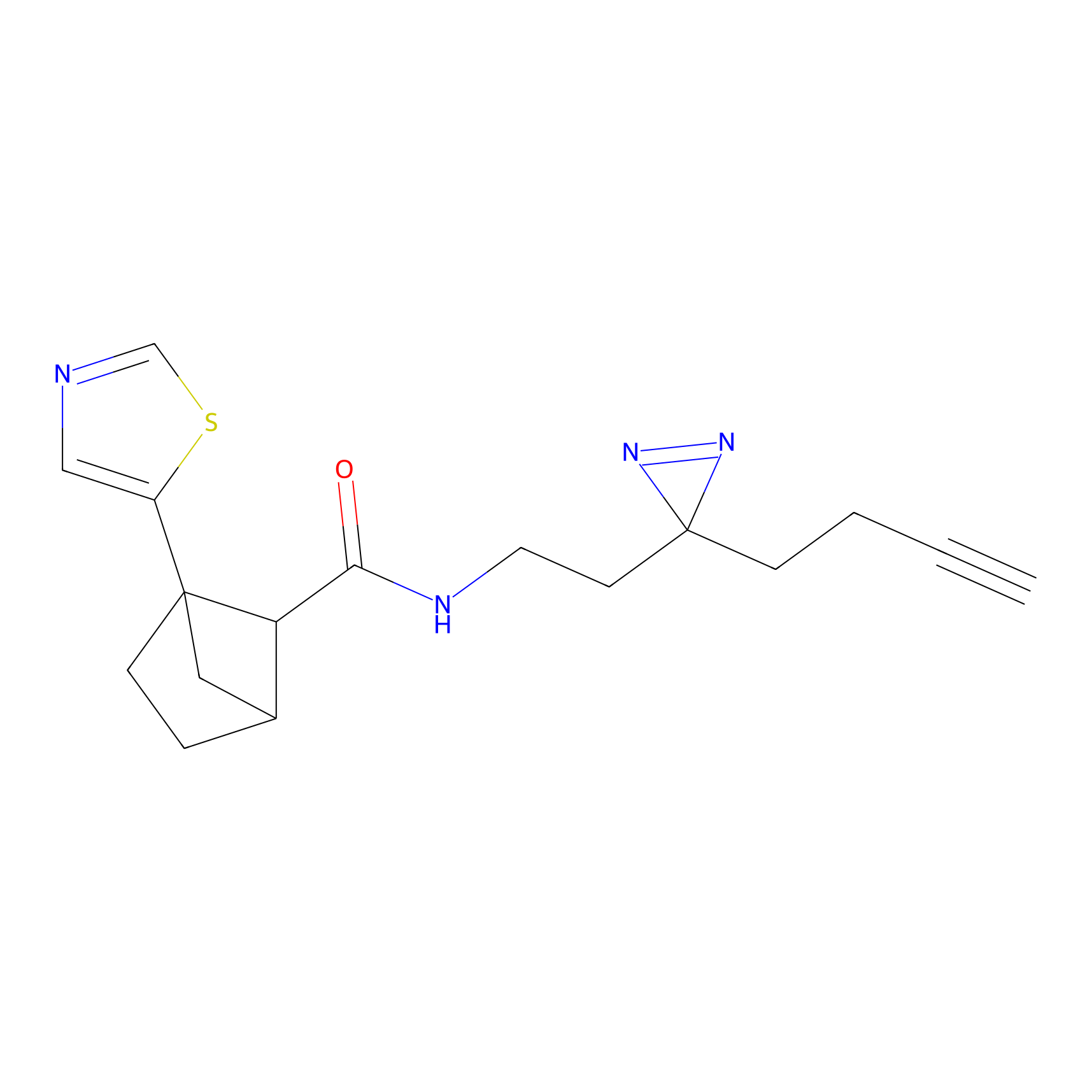
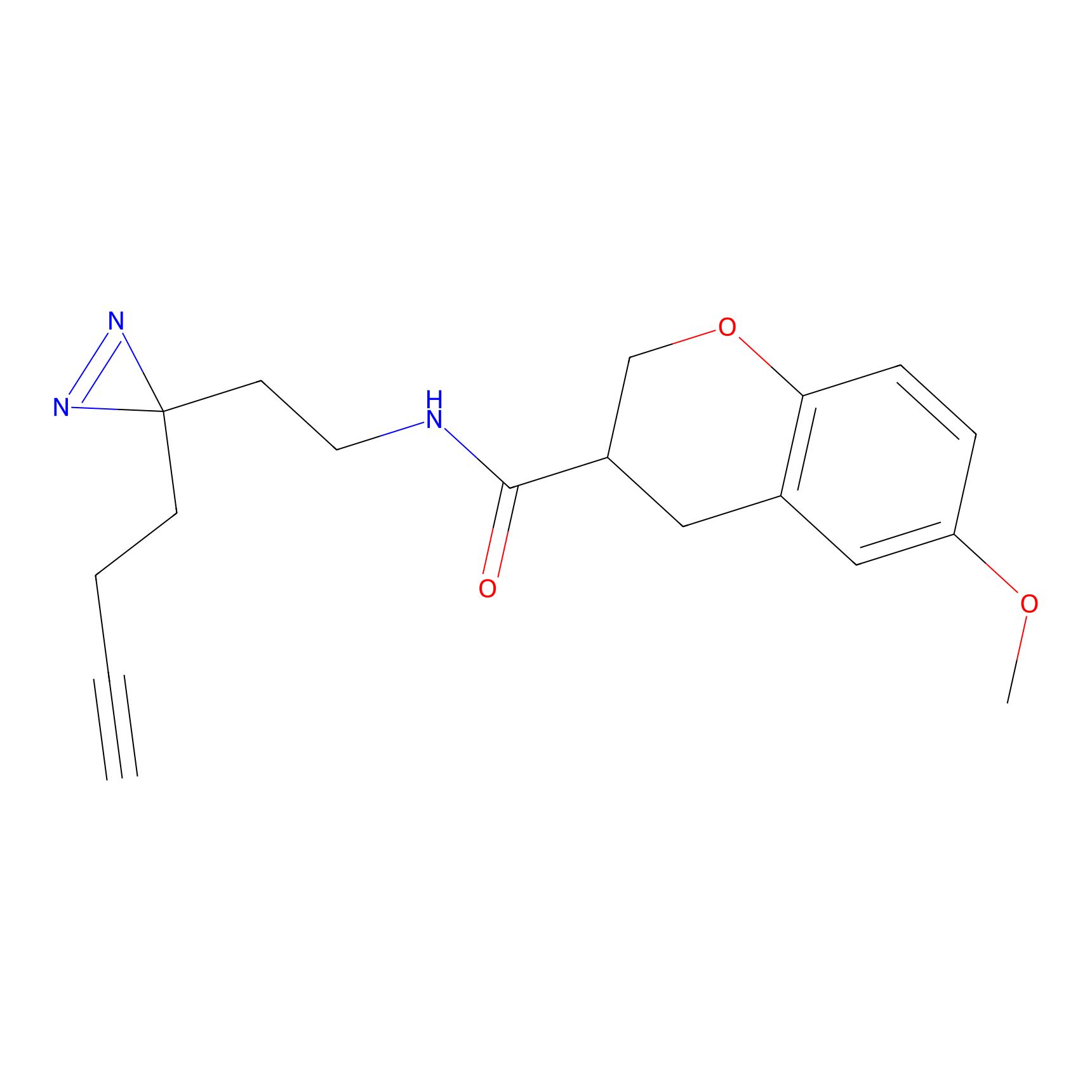
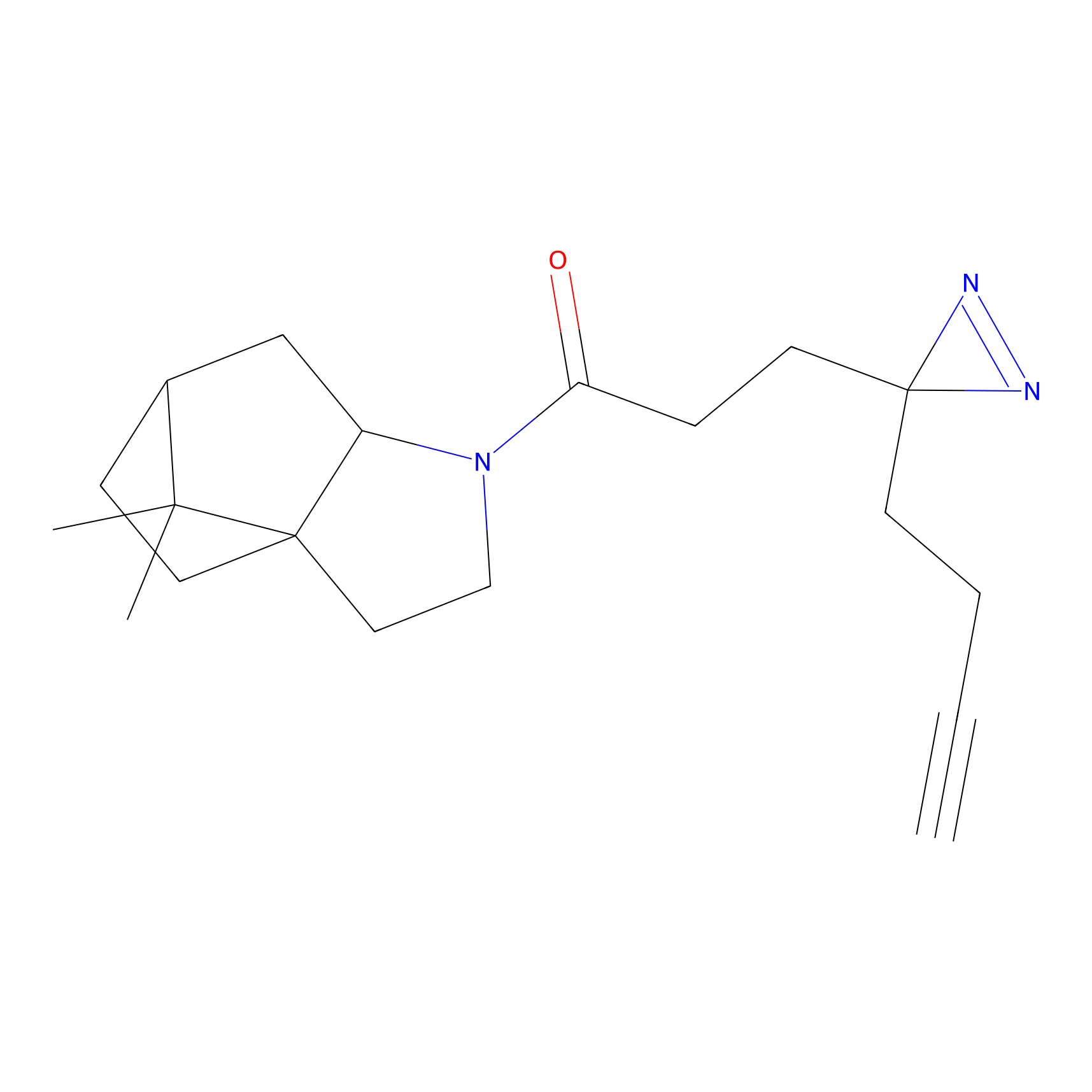
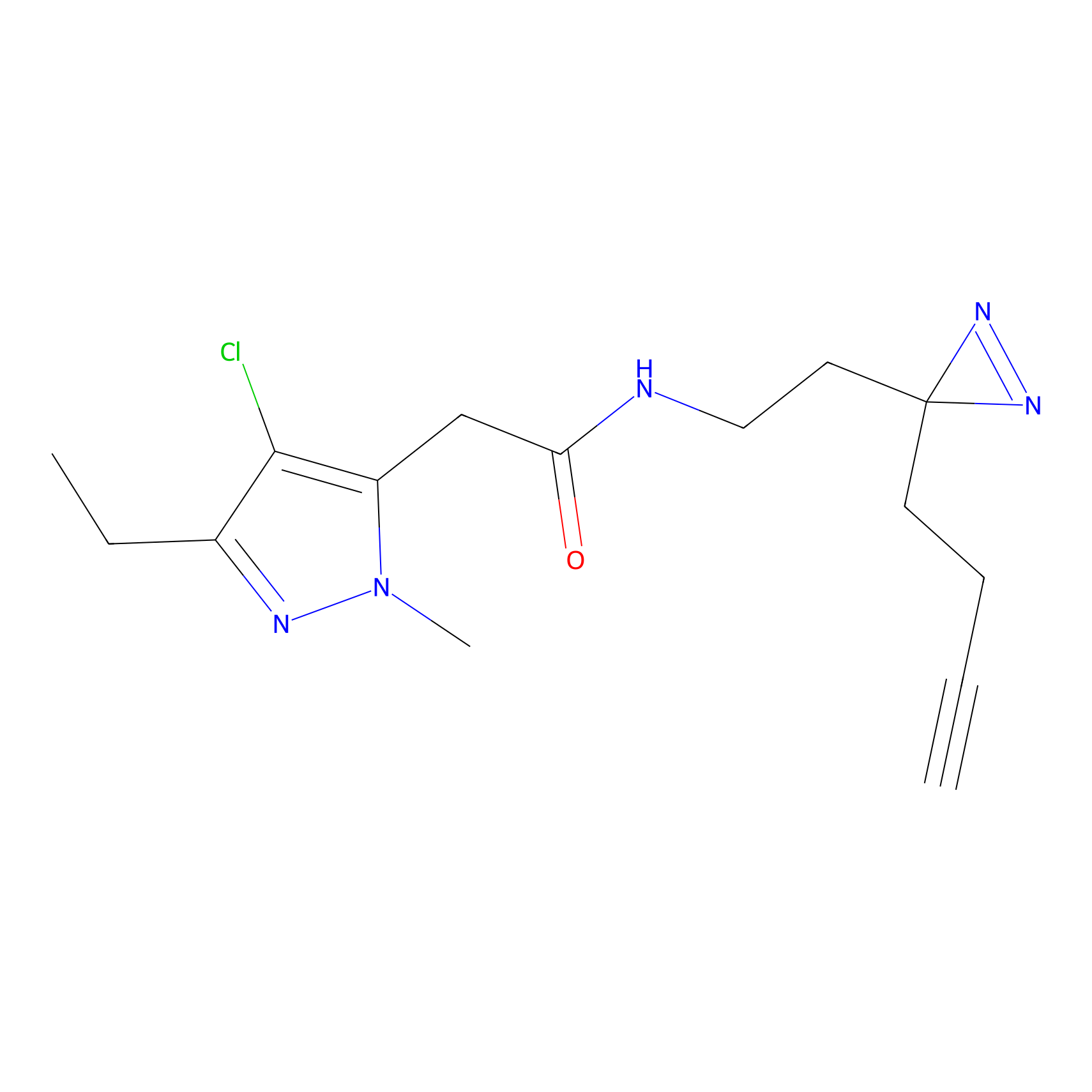
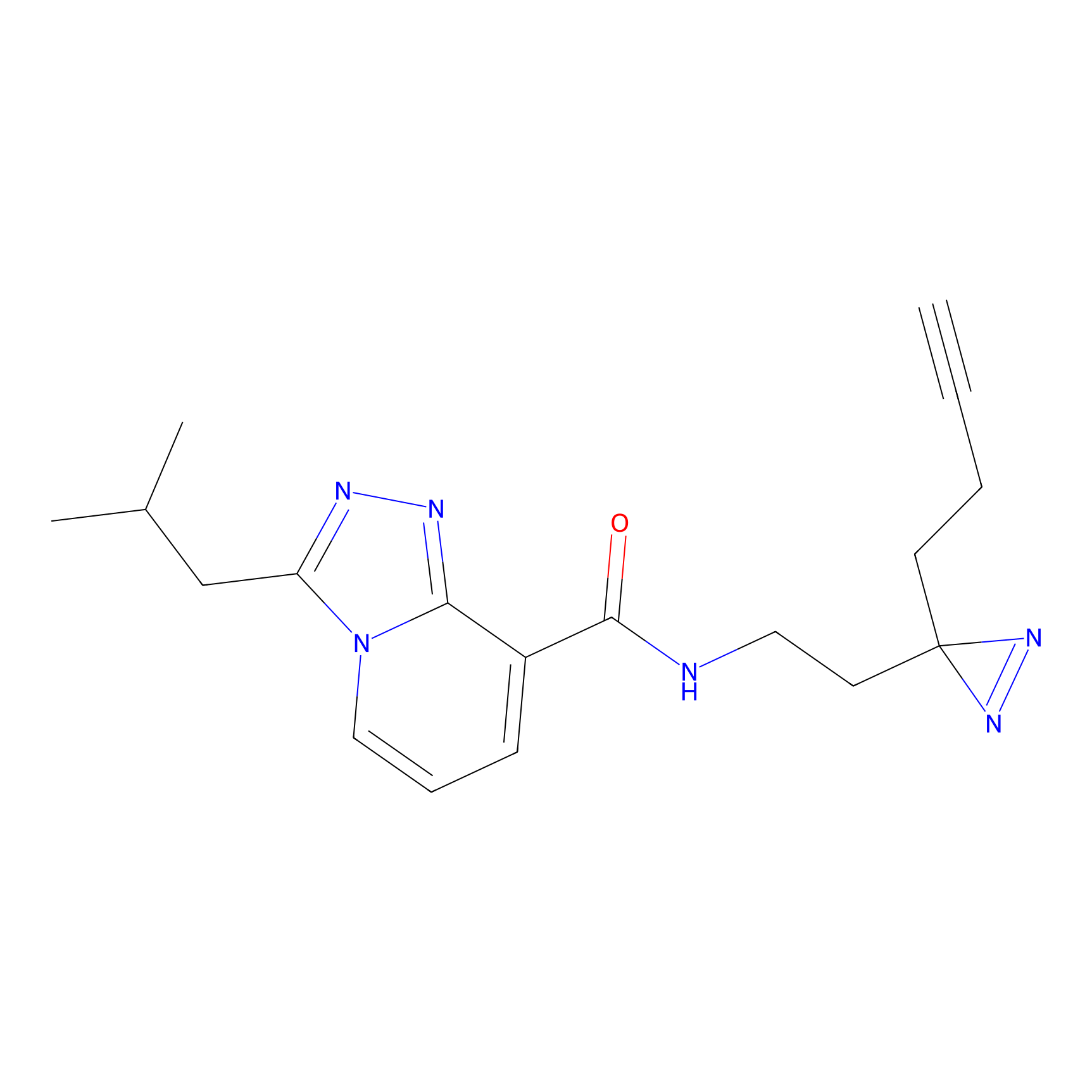
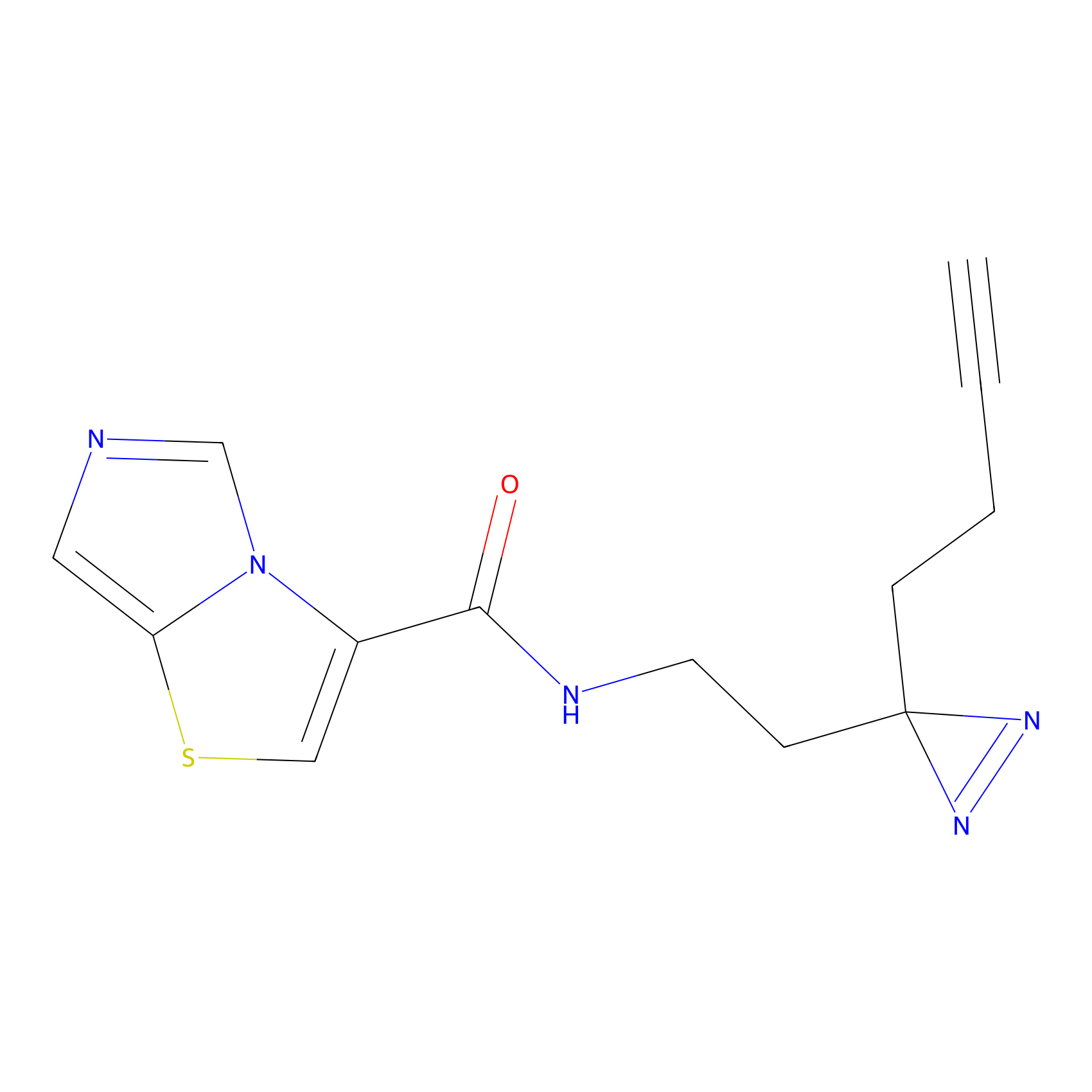
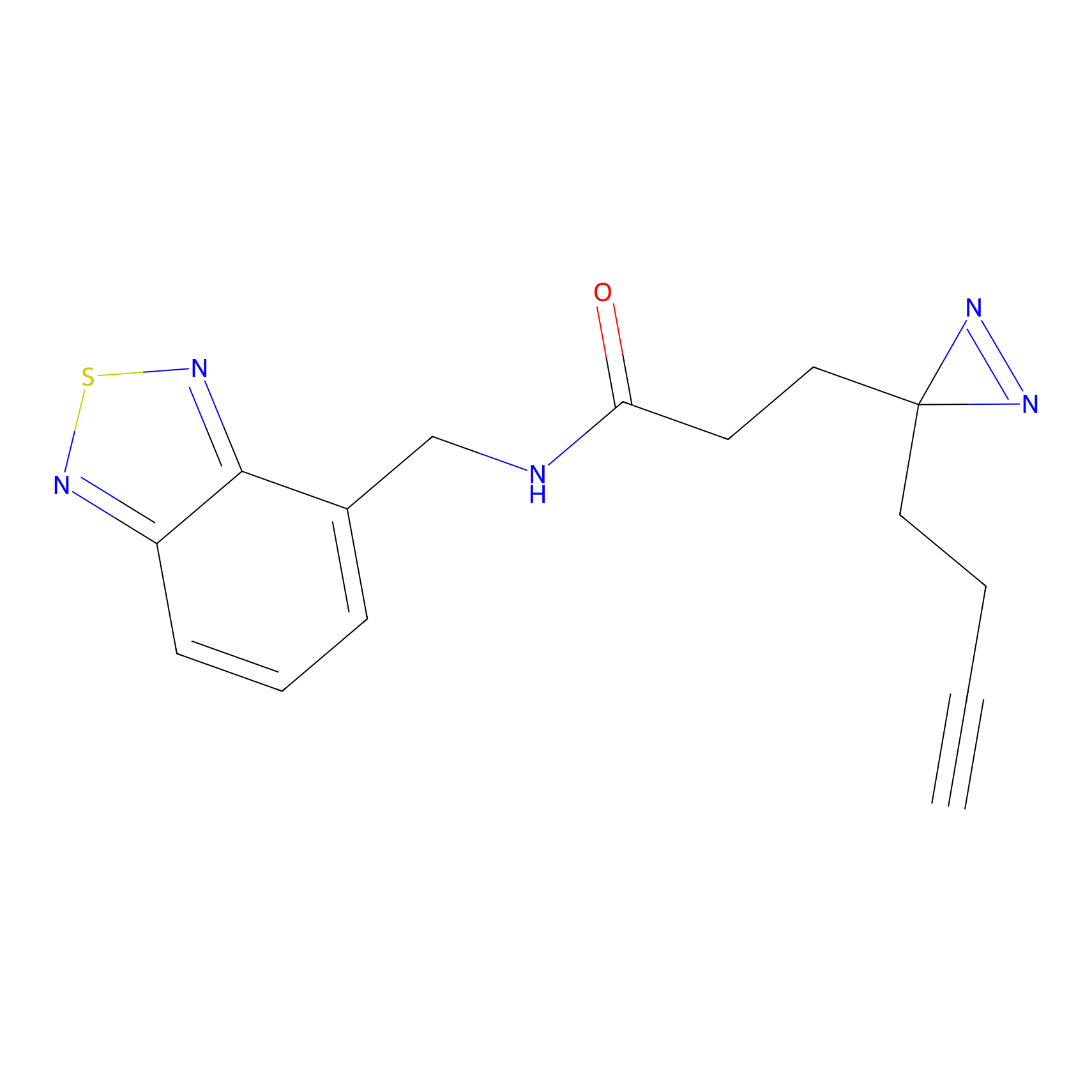
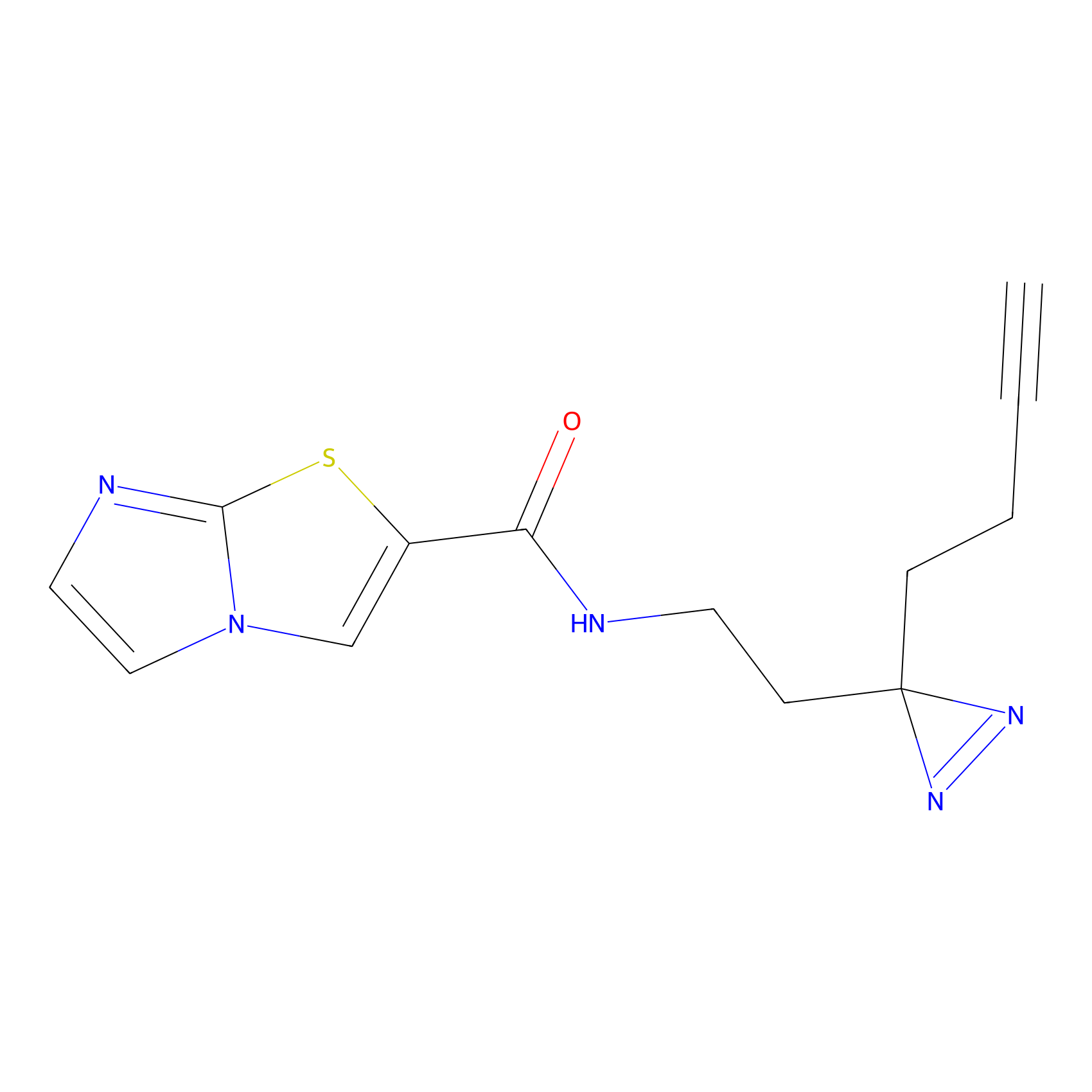
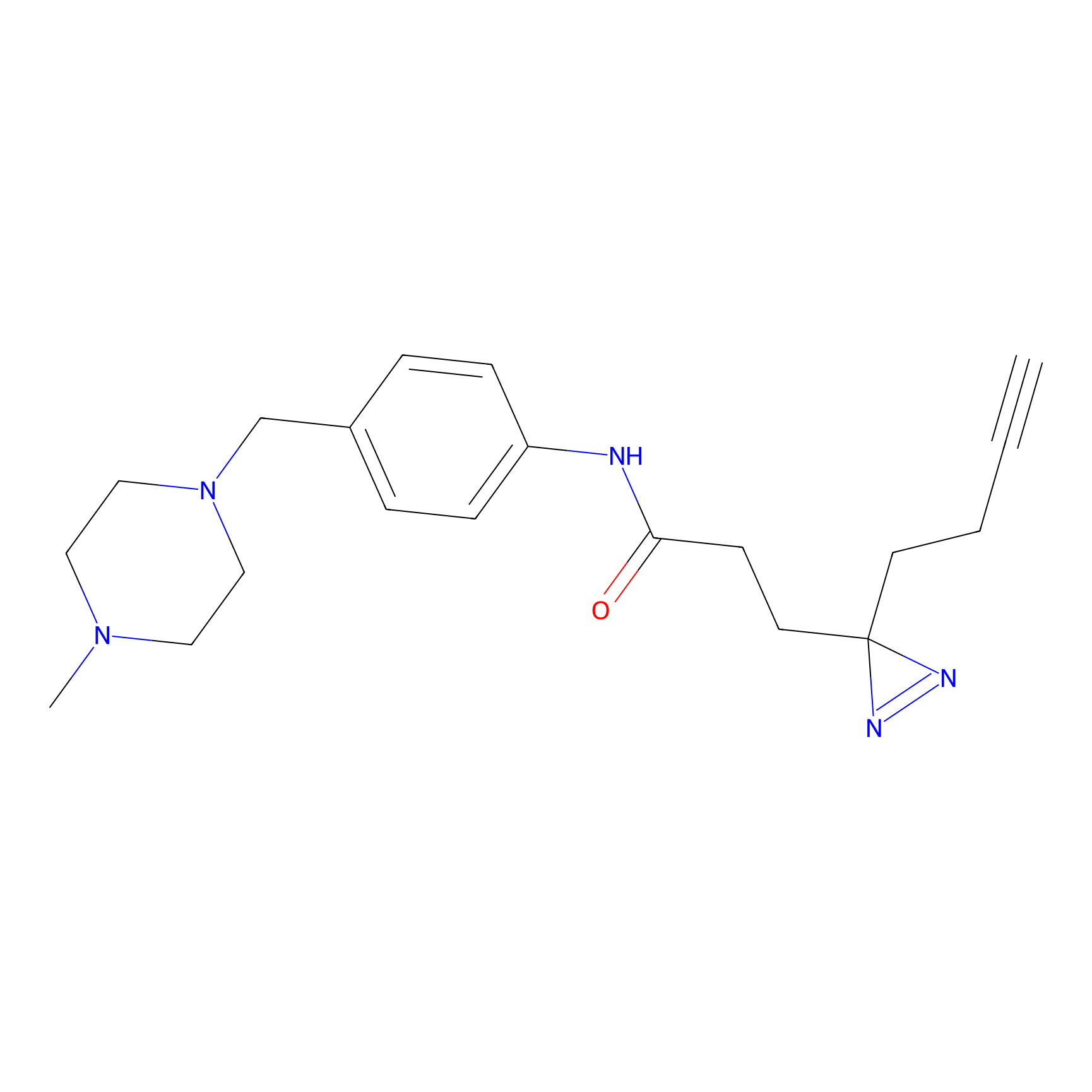
Alkylaryl probe 2 Probe Info |
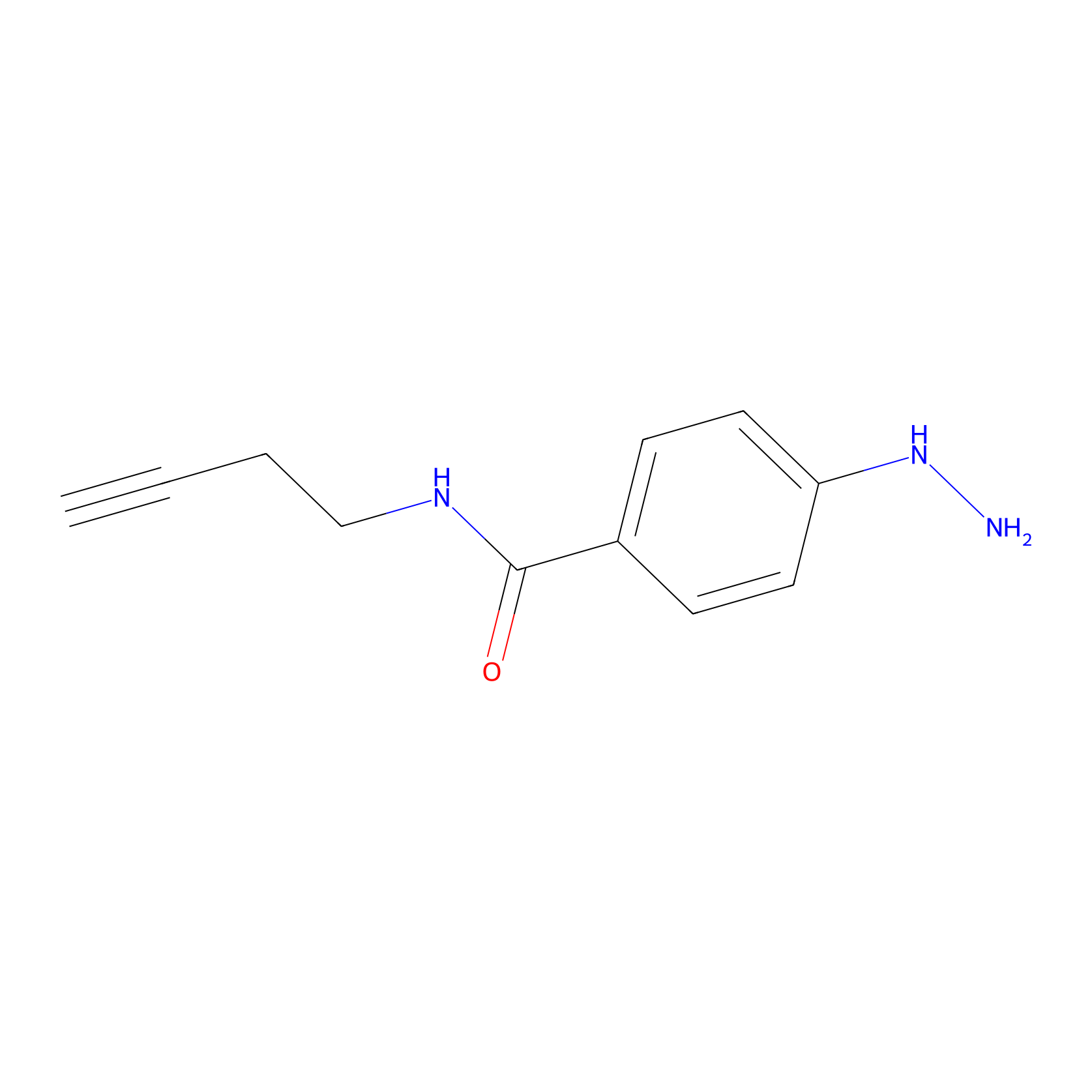 |
20.00 | LDD0391 | [1] | |
|
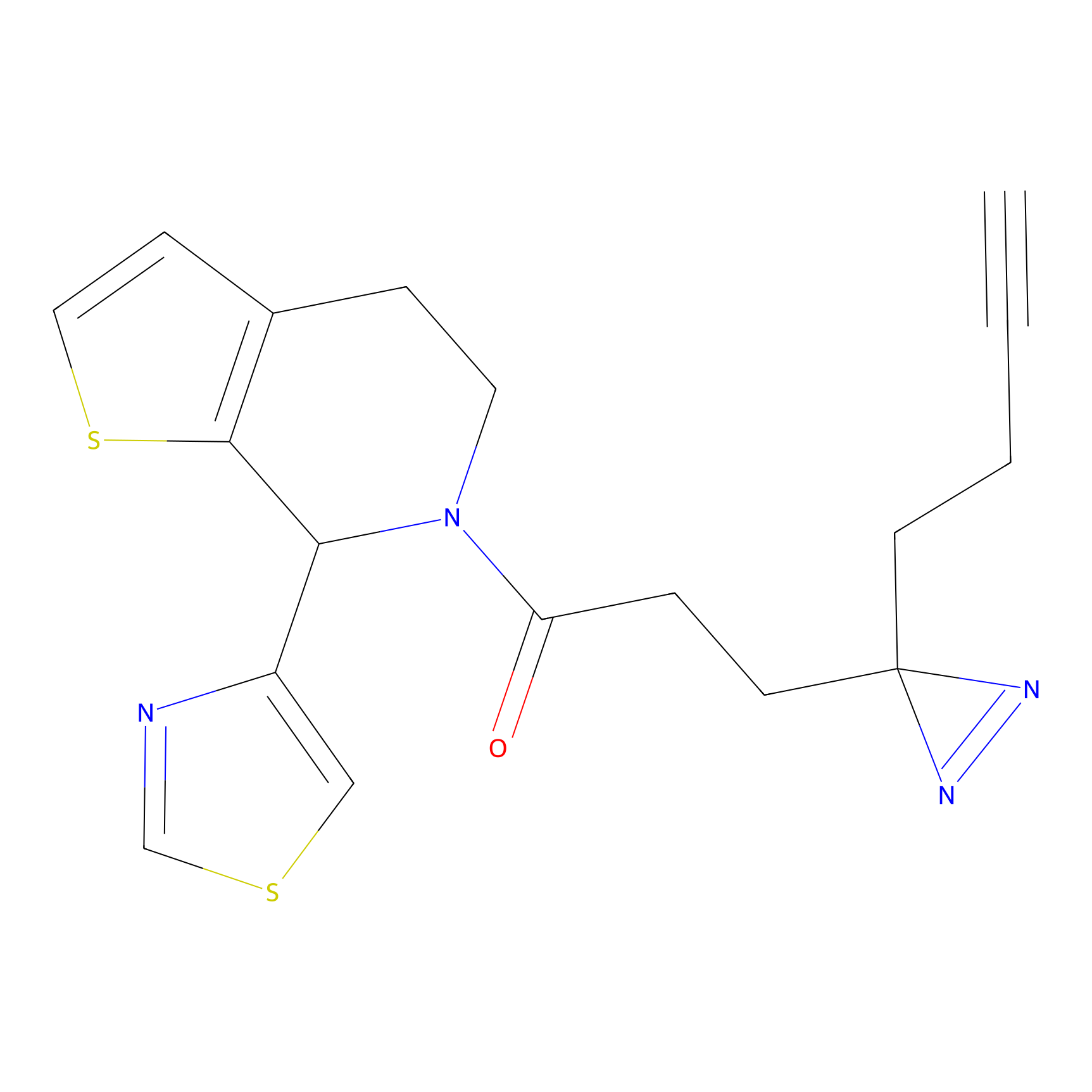
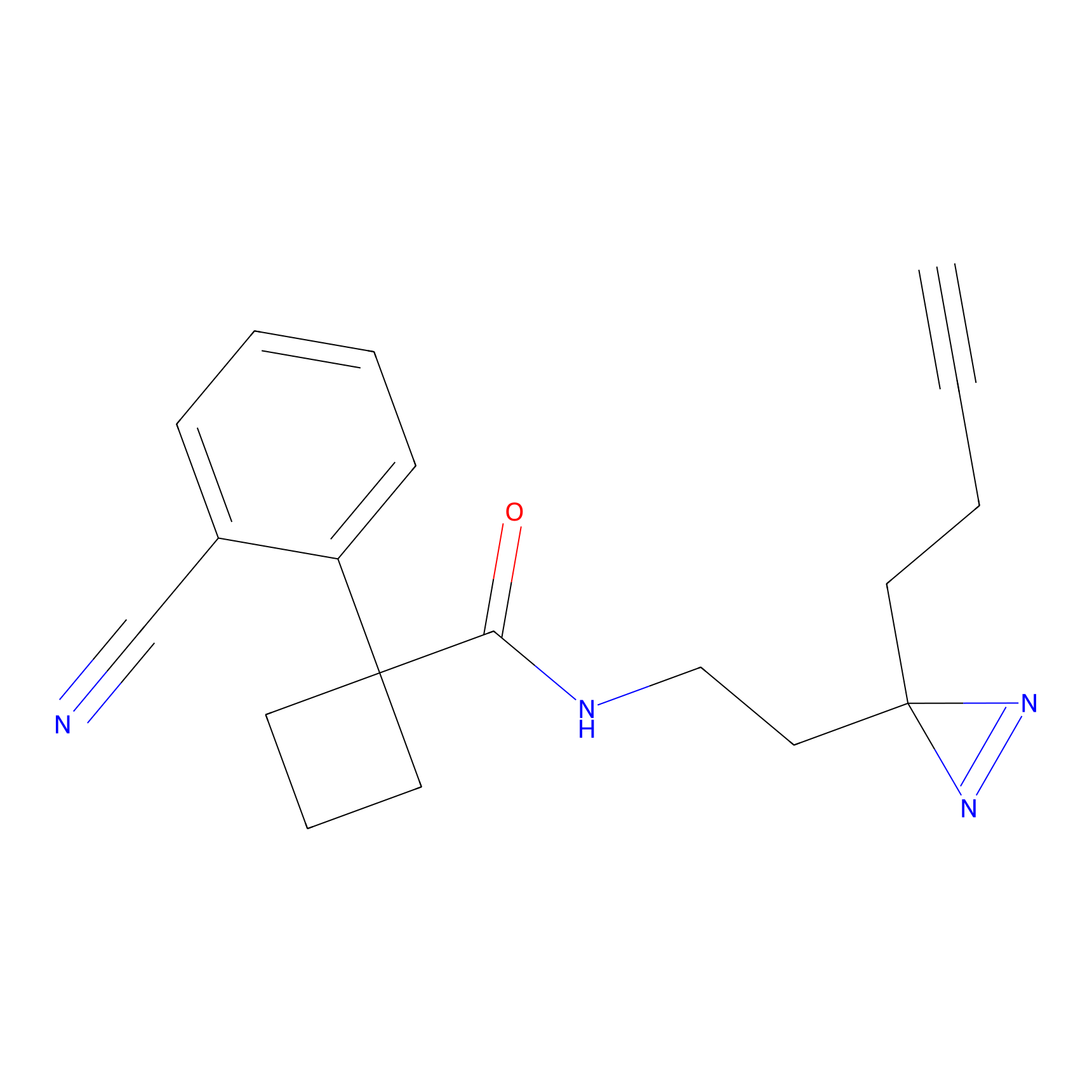
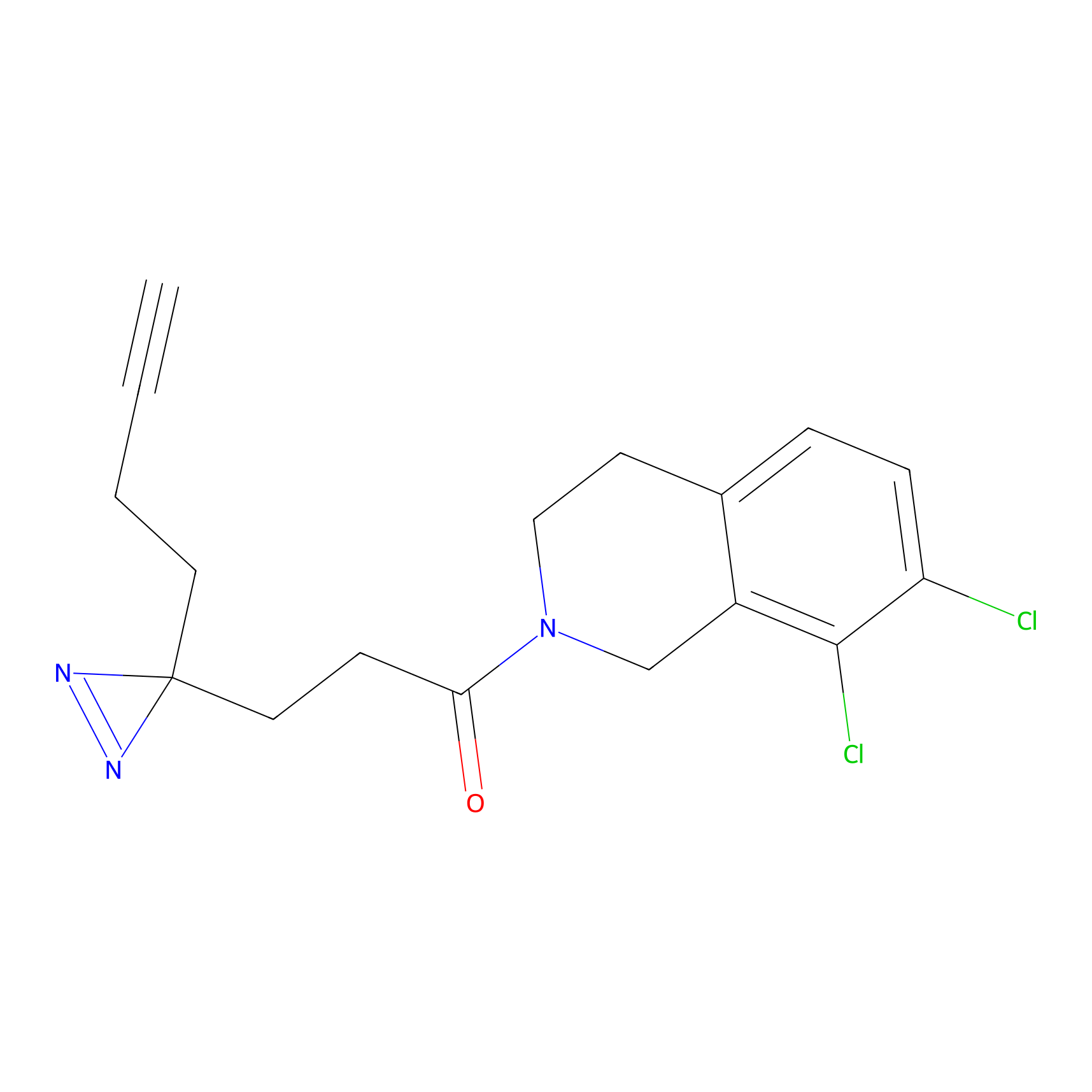
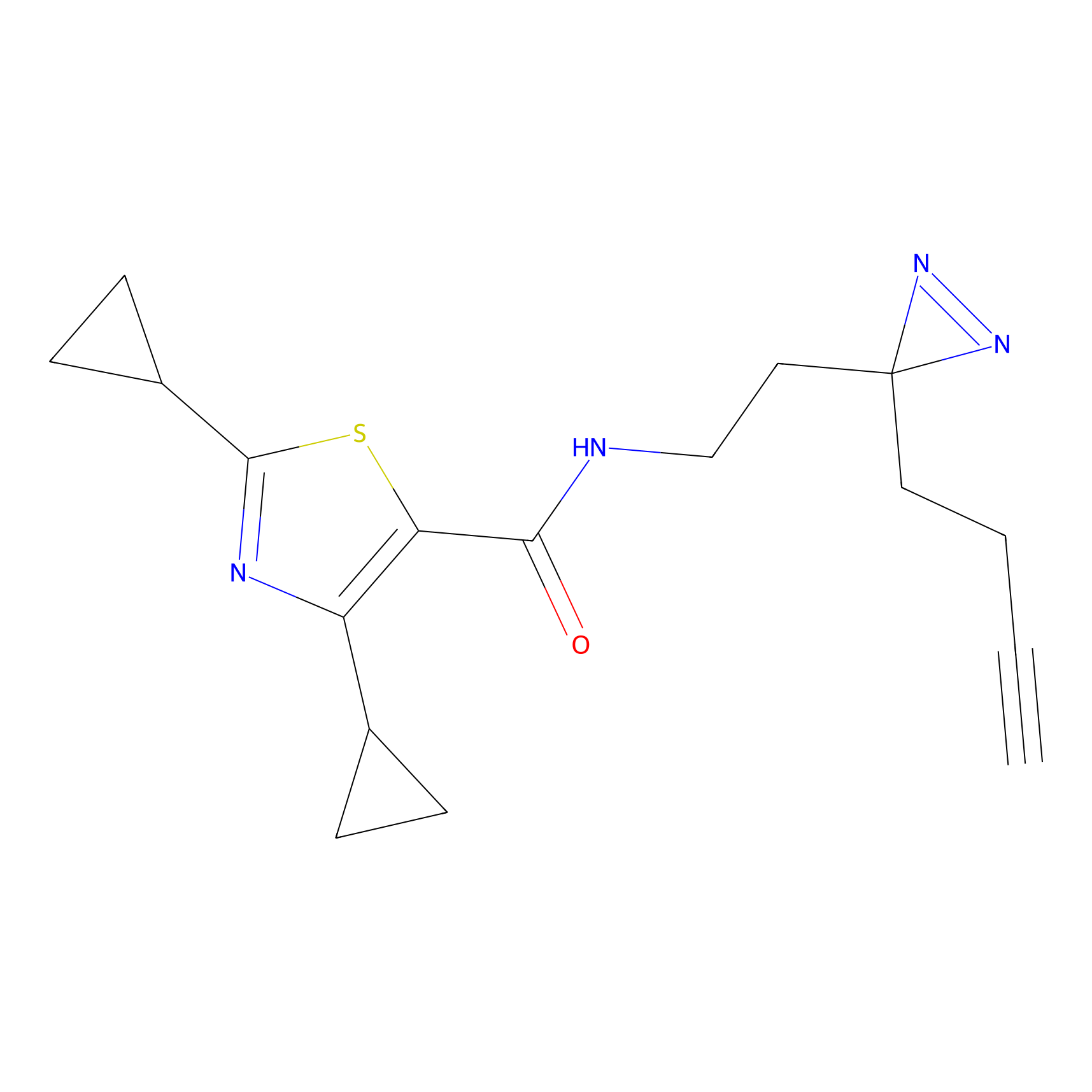
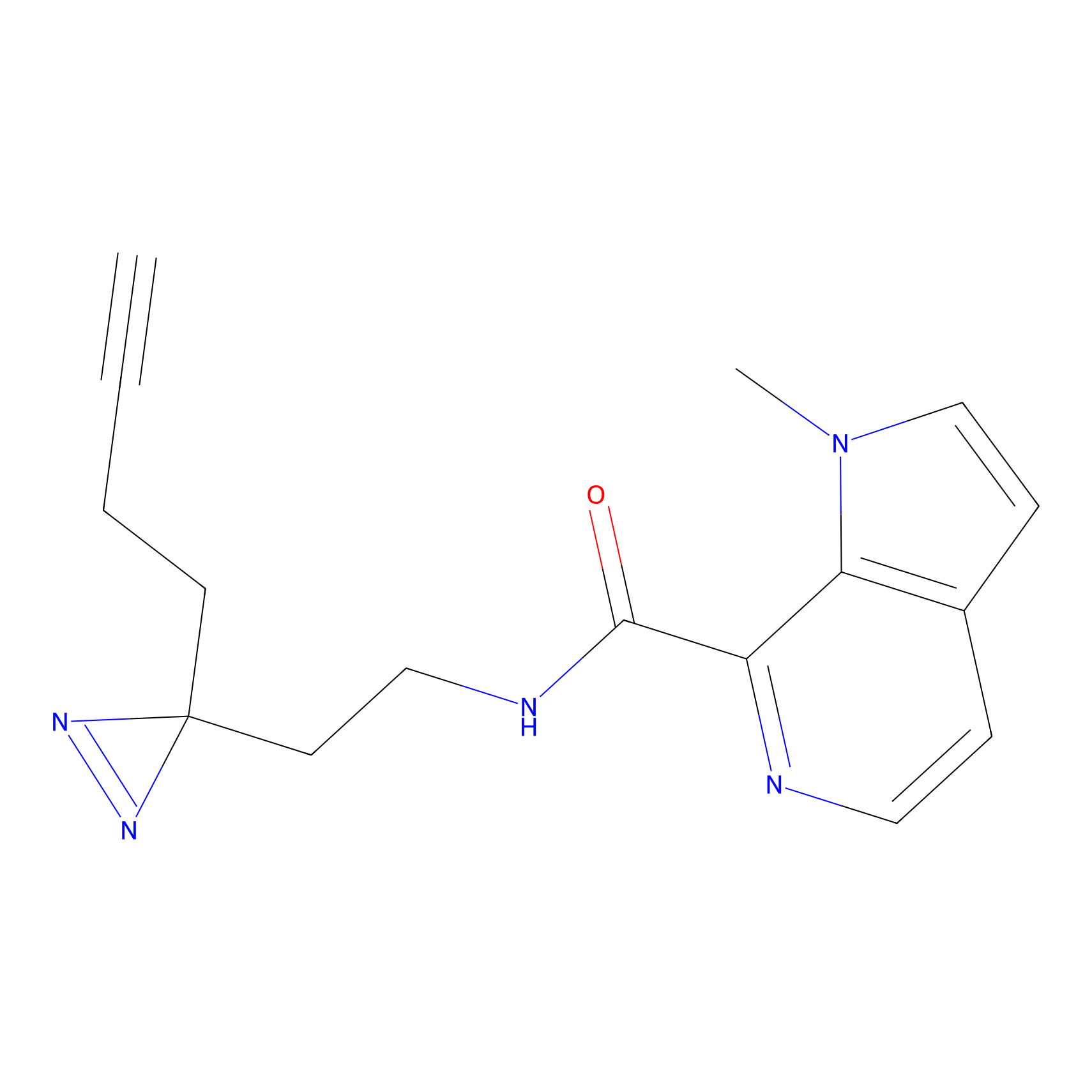
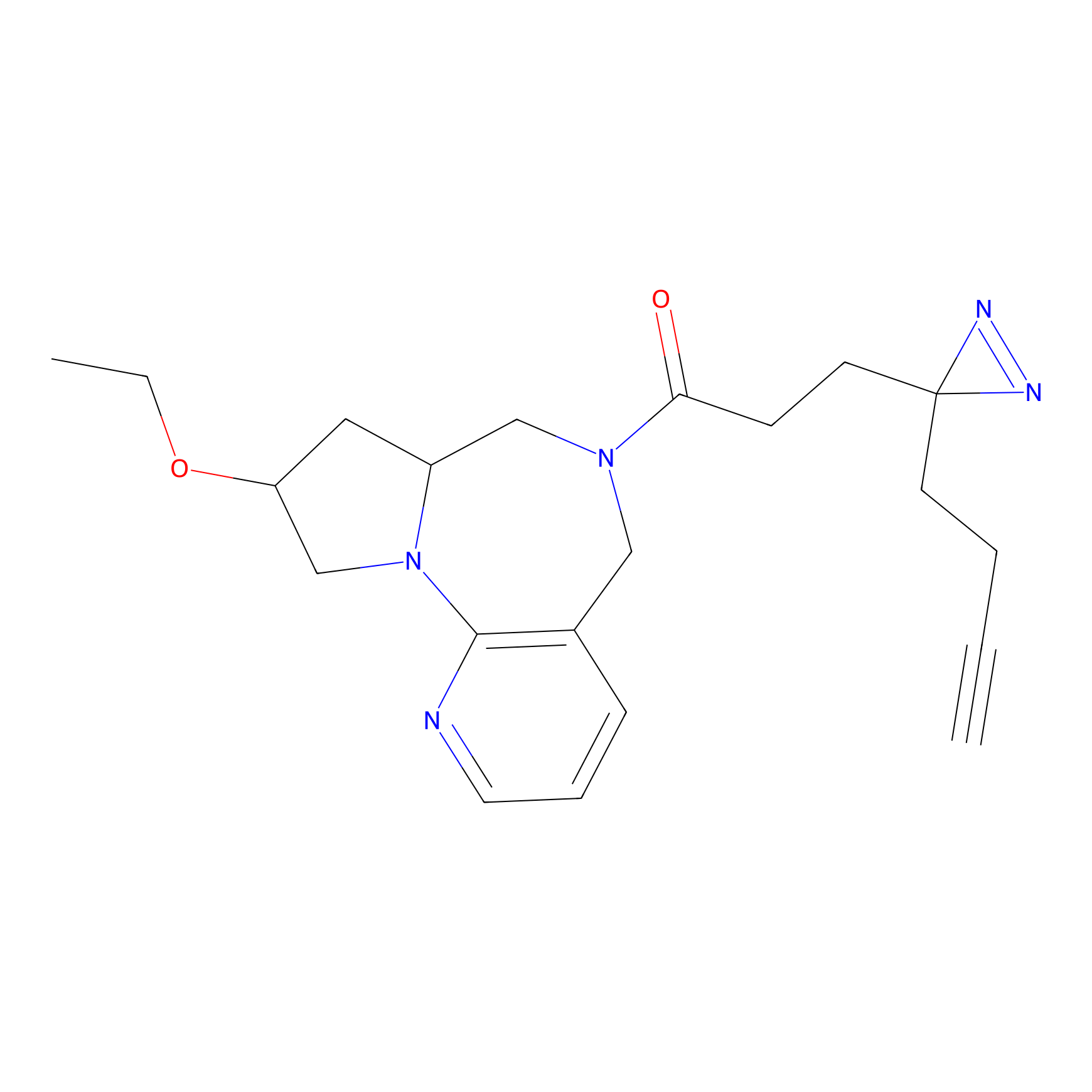
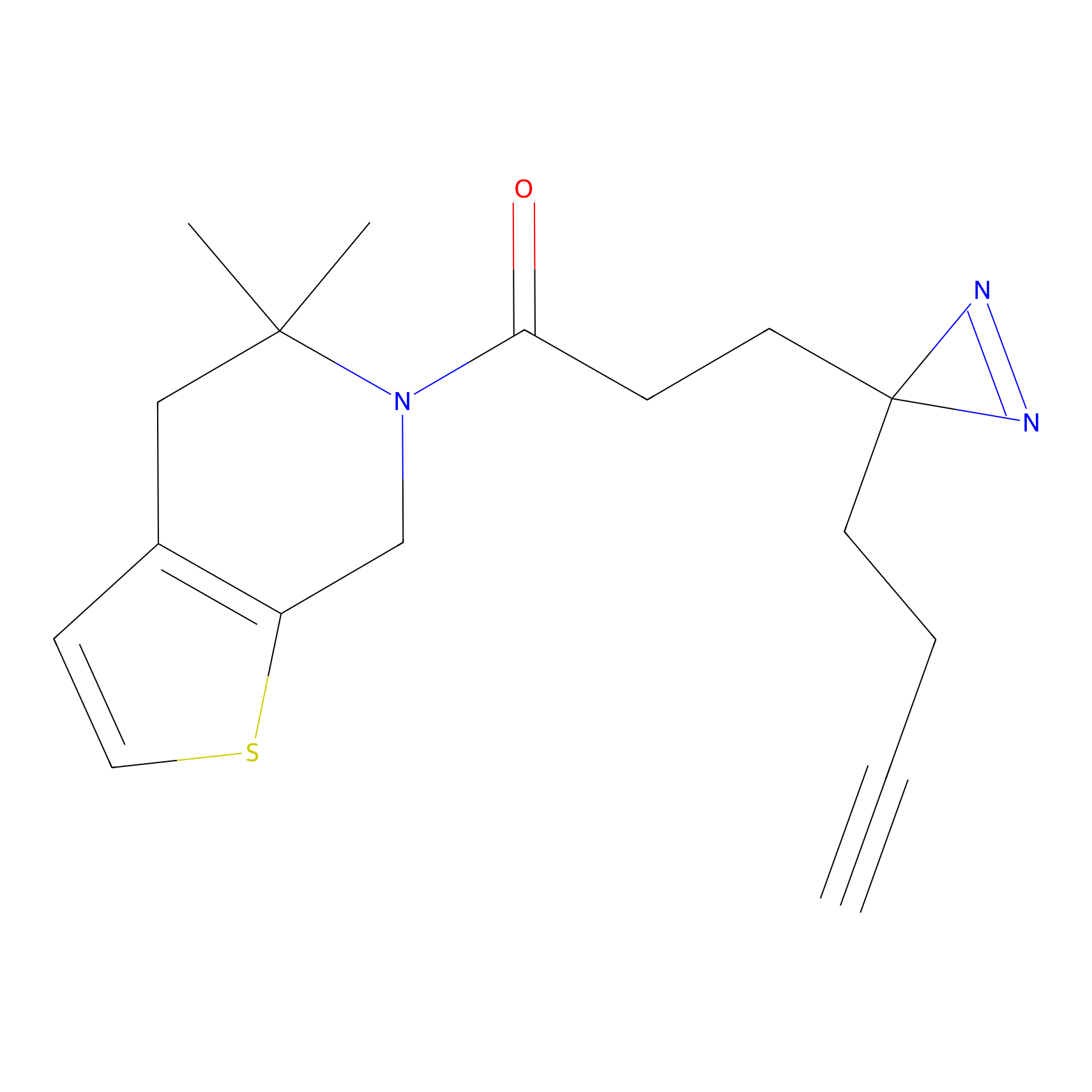
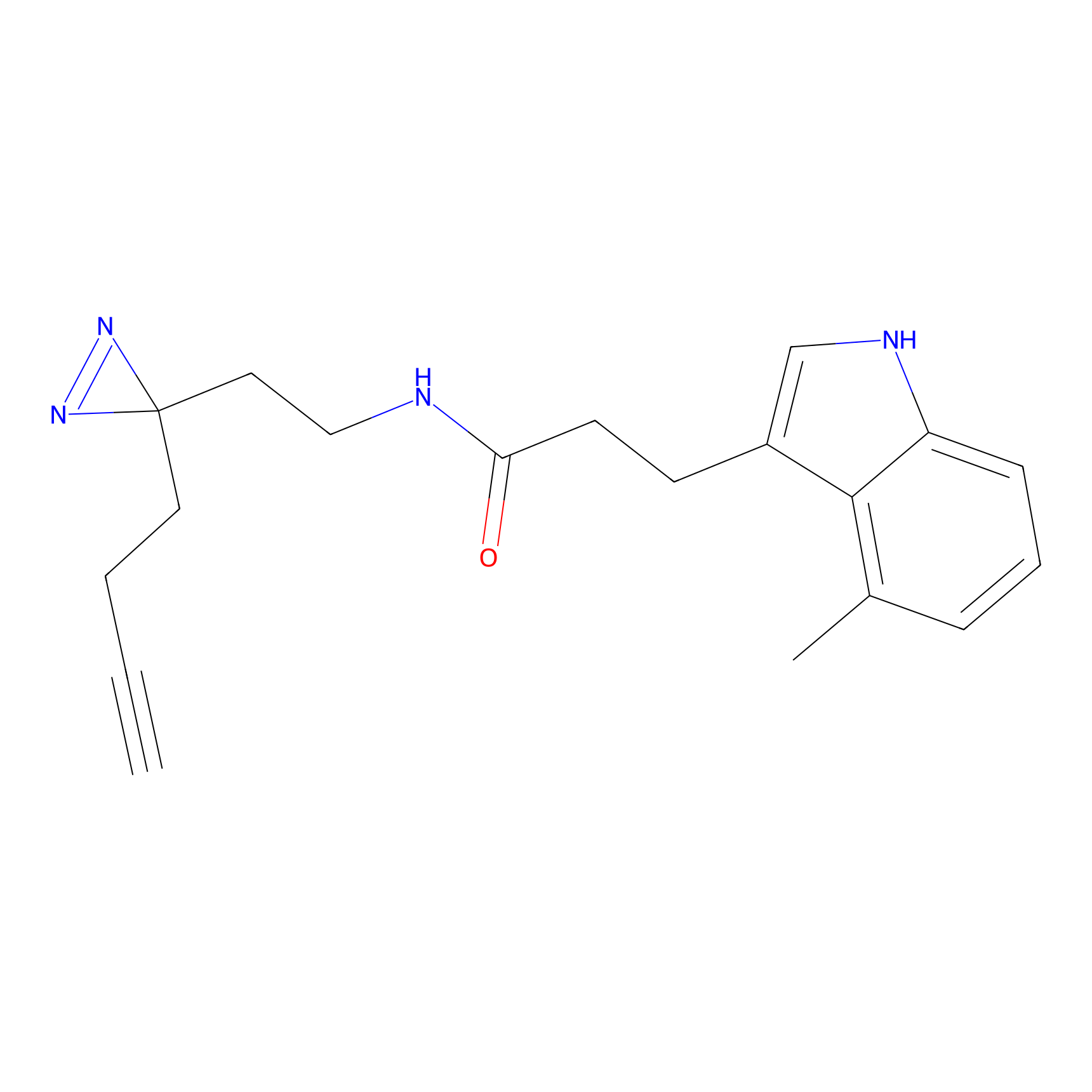
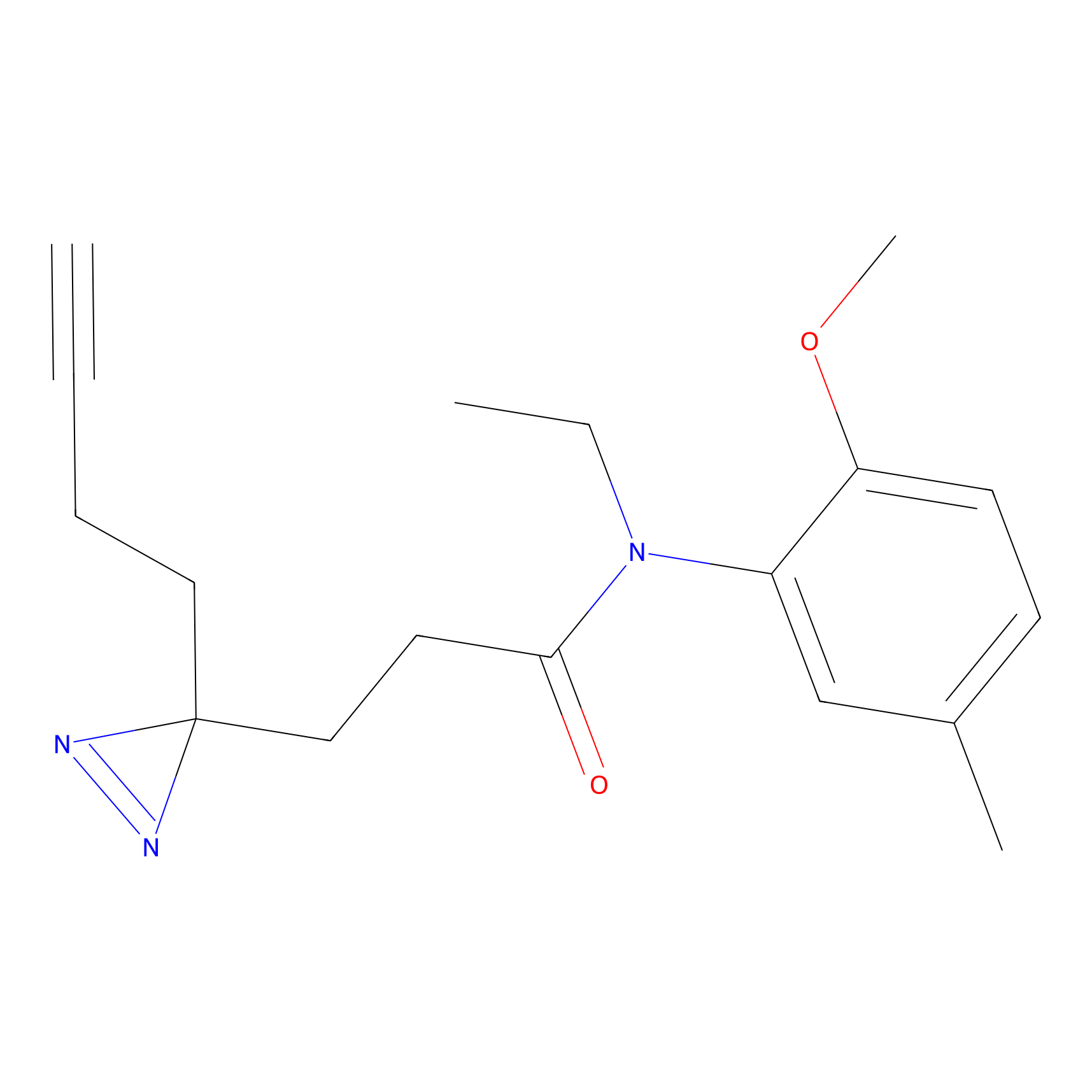
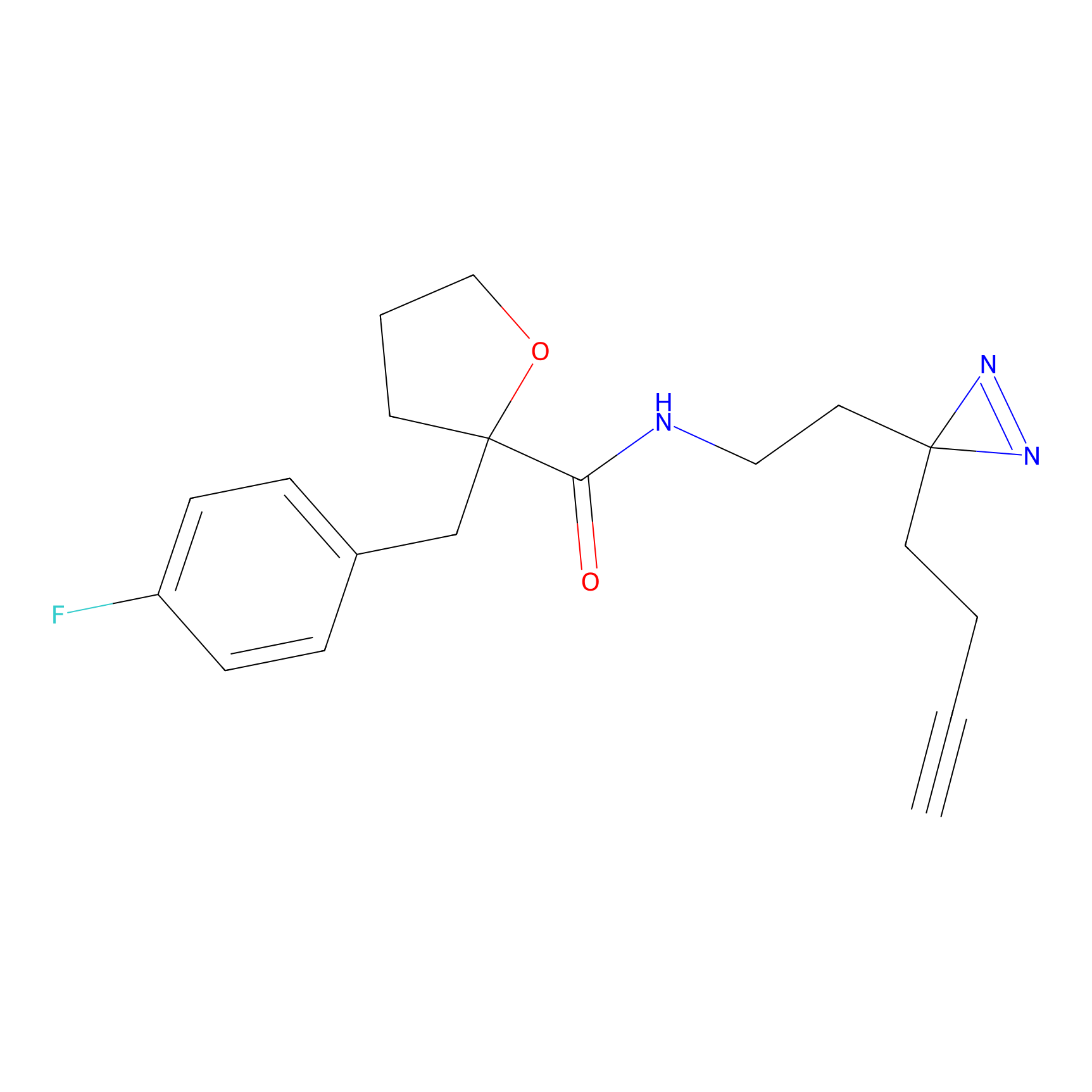
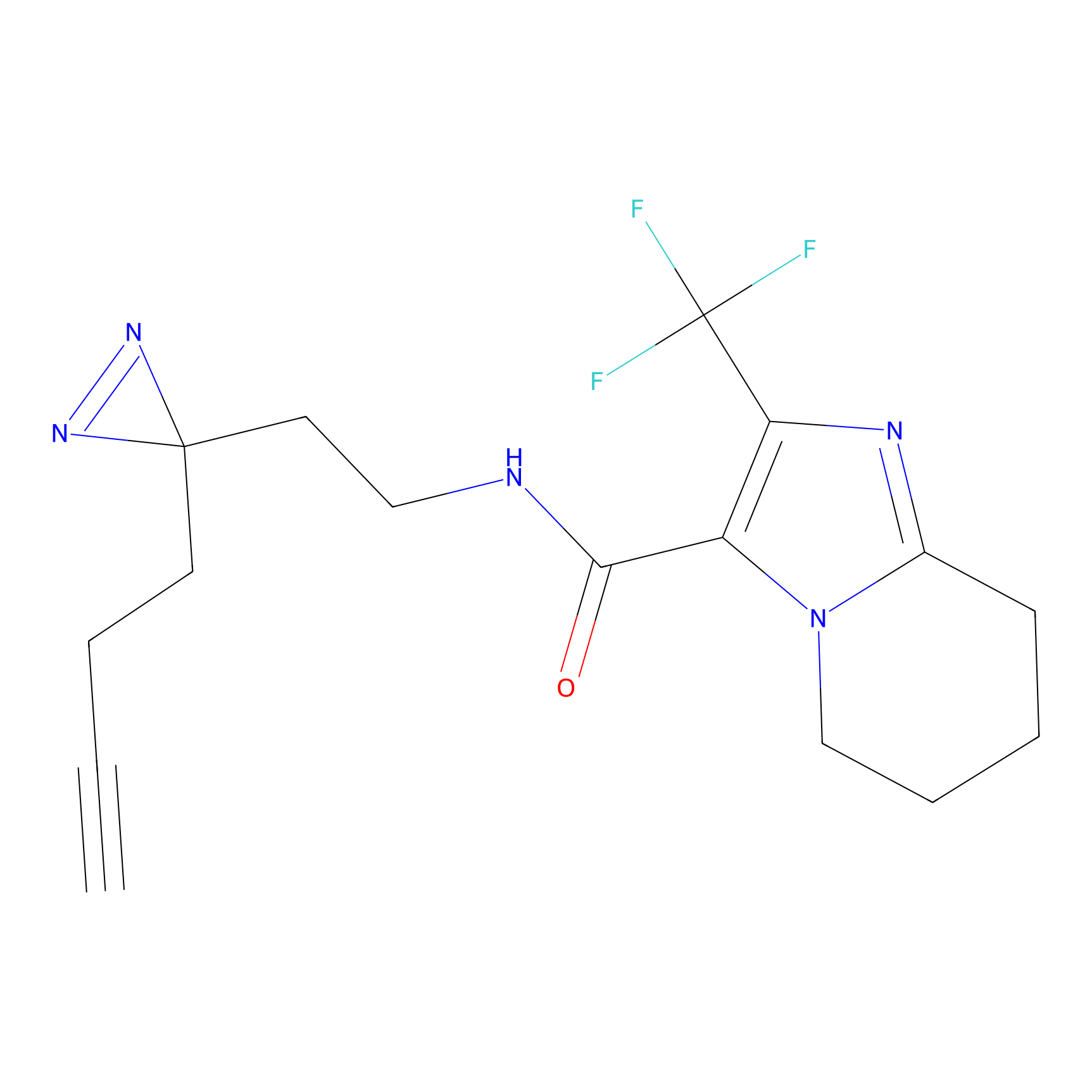
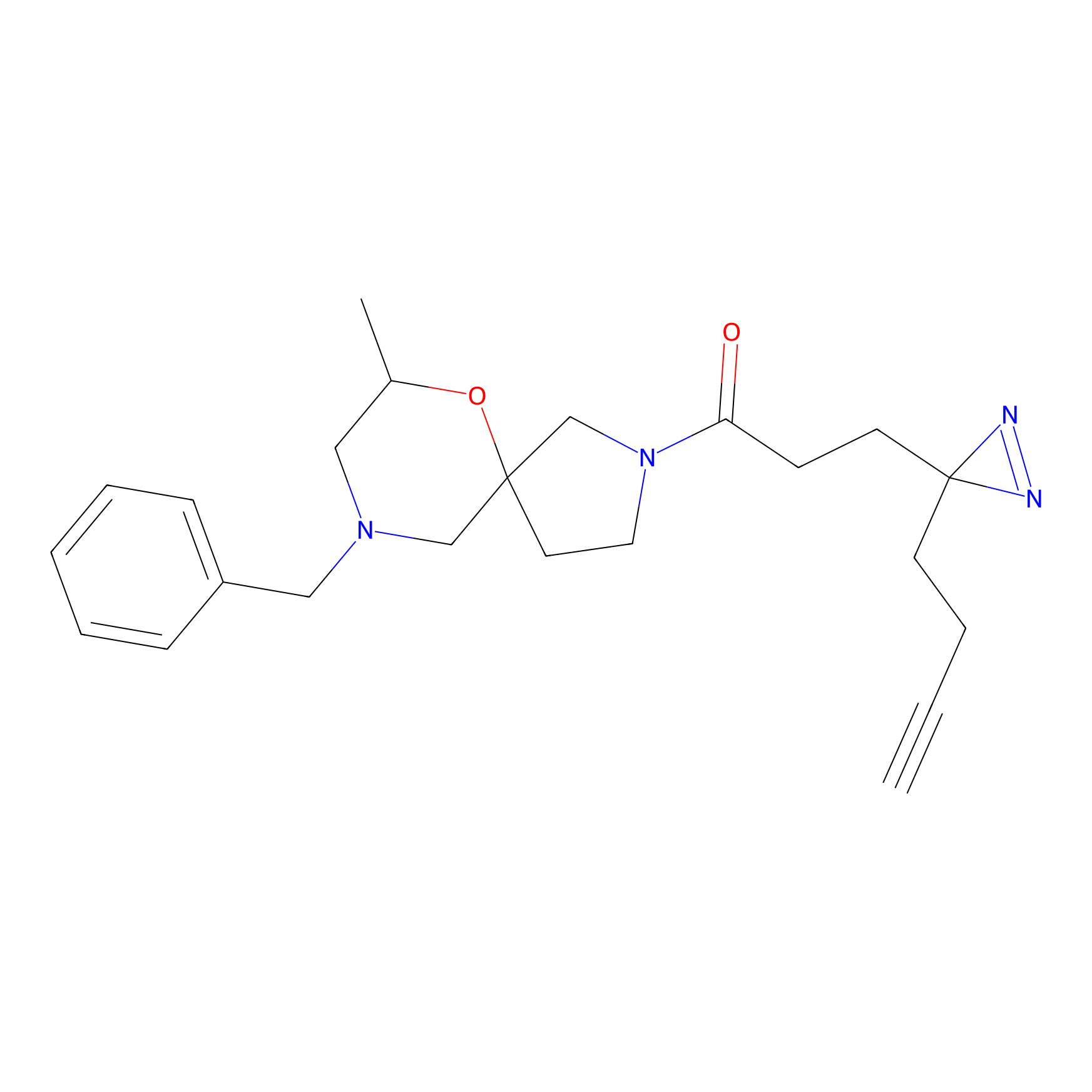
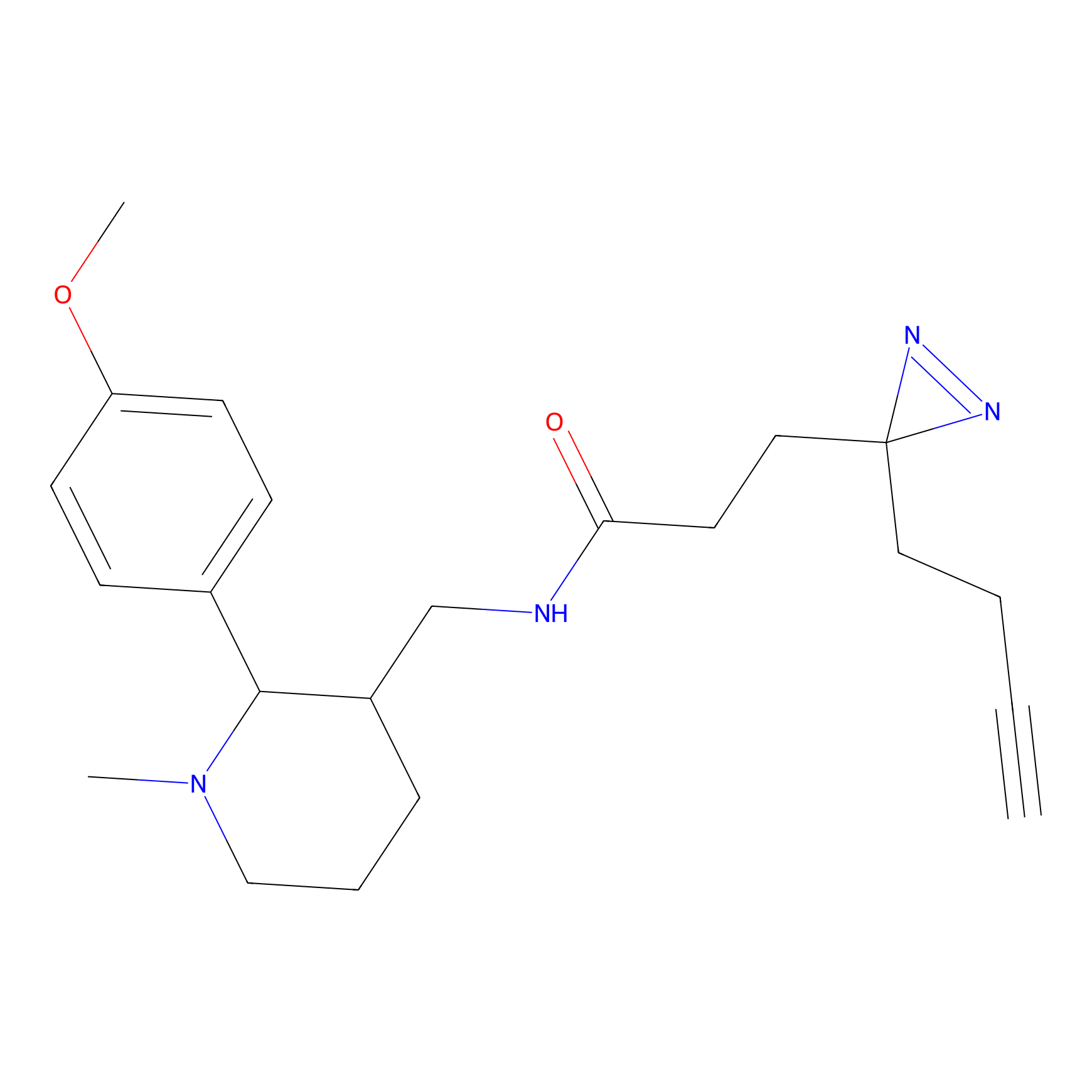
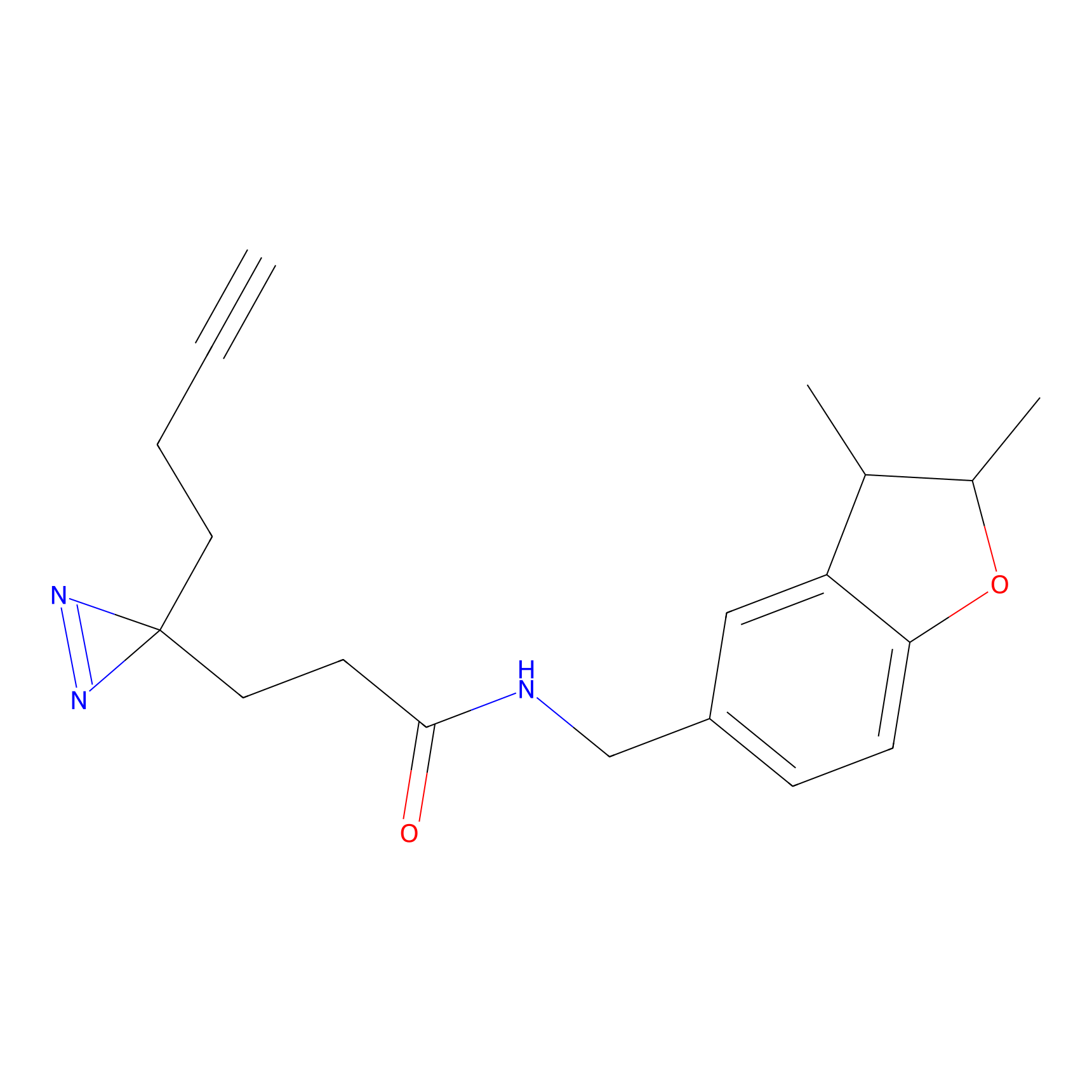
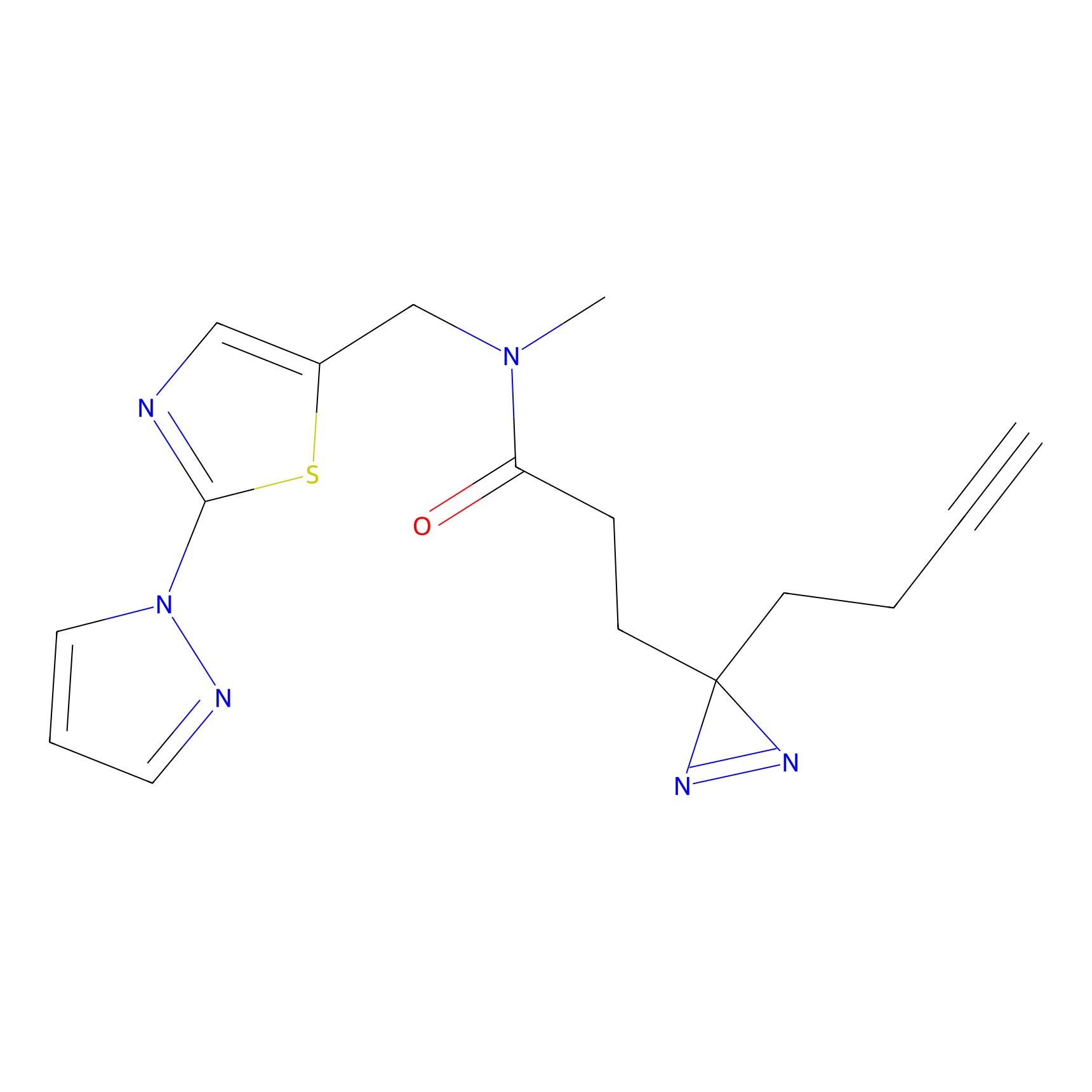
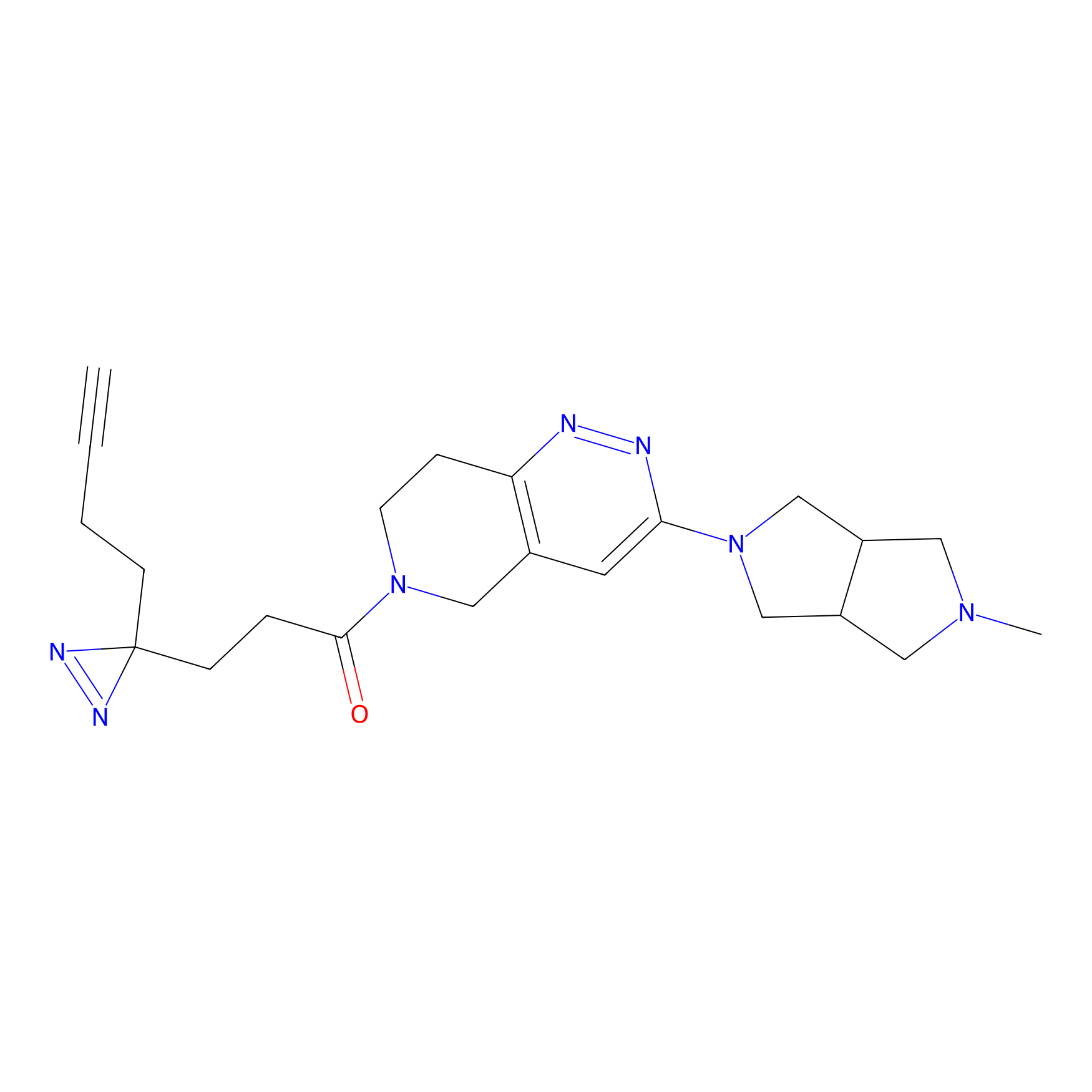
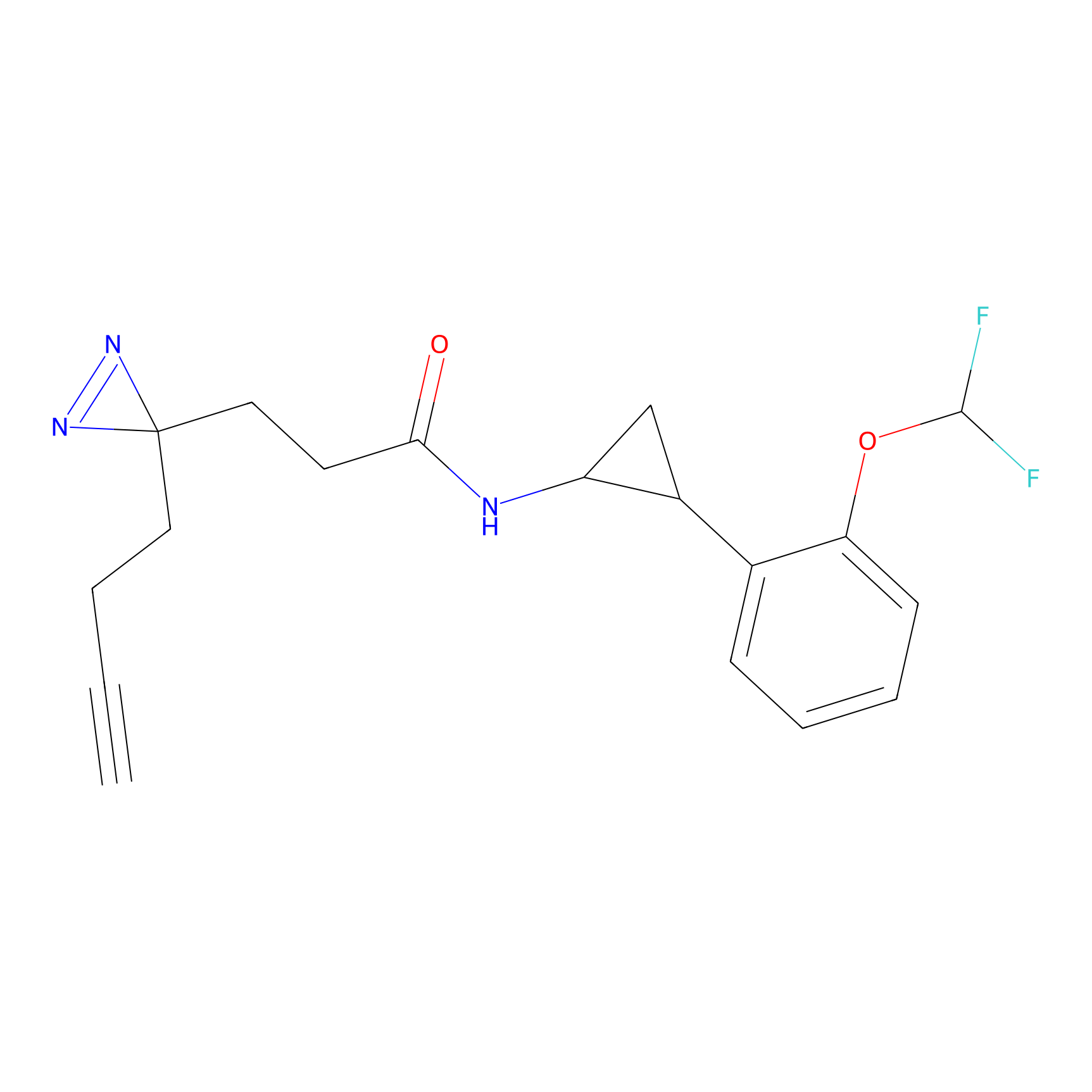
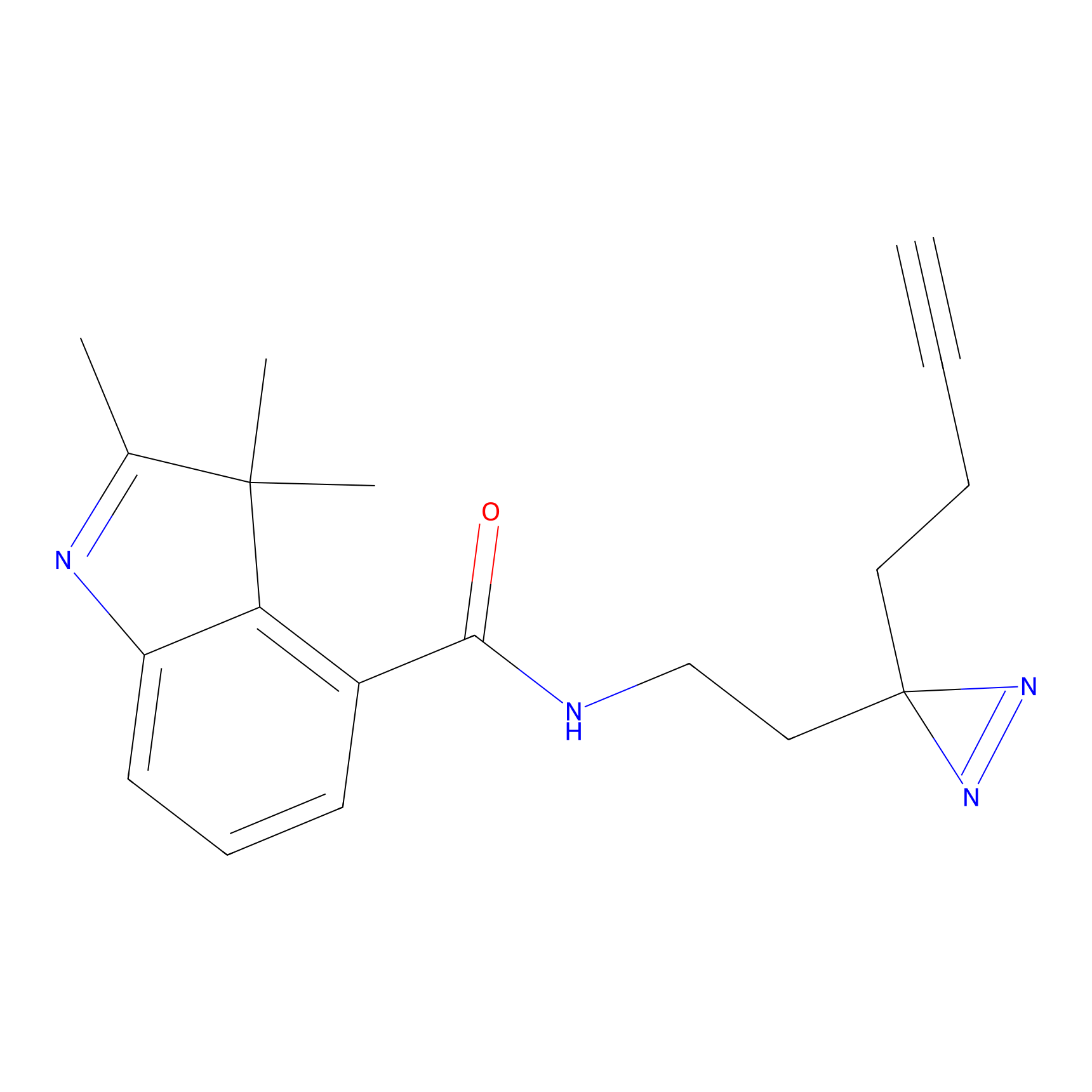
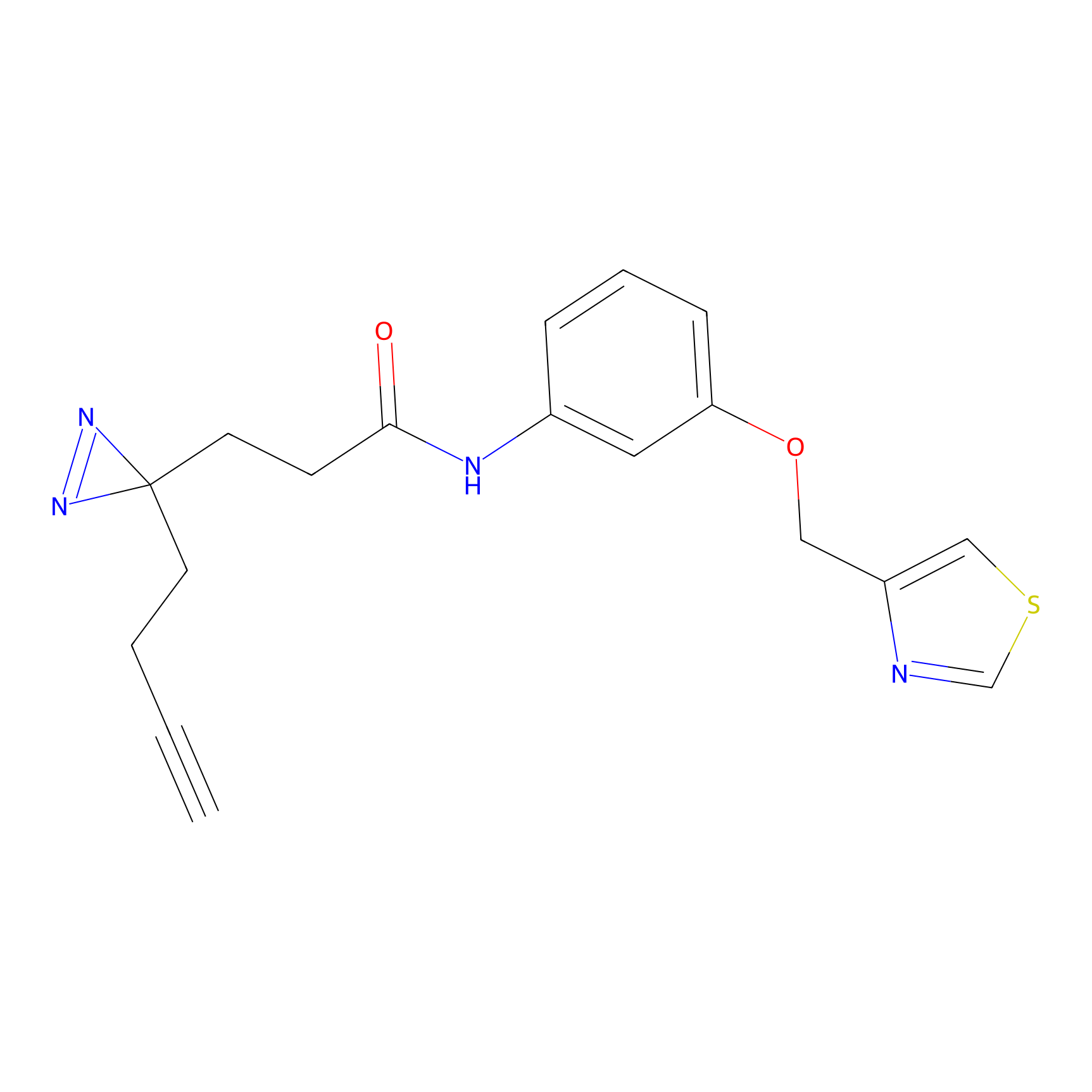
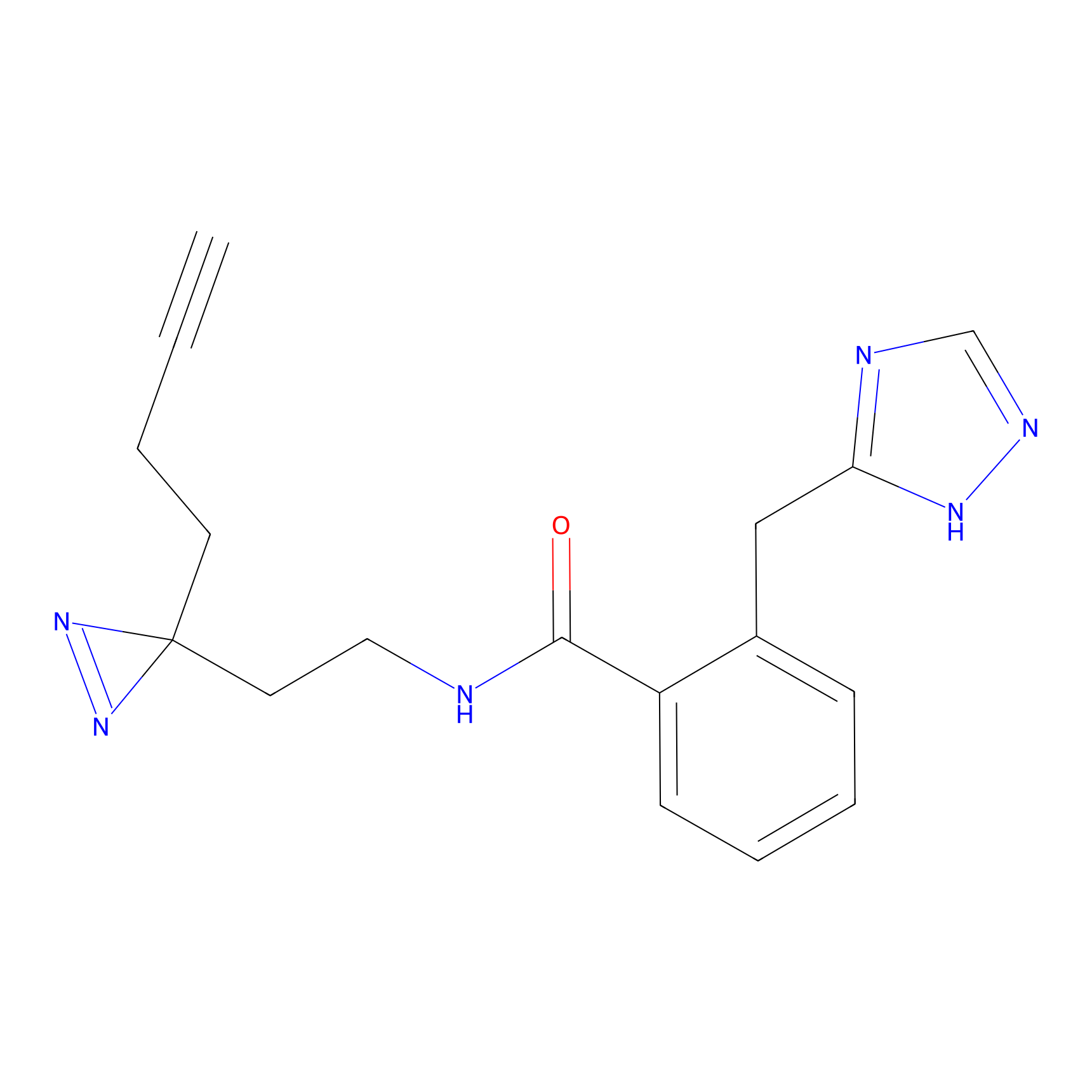
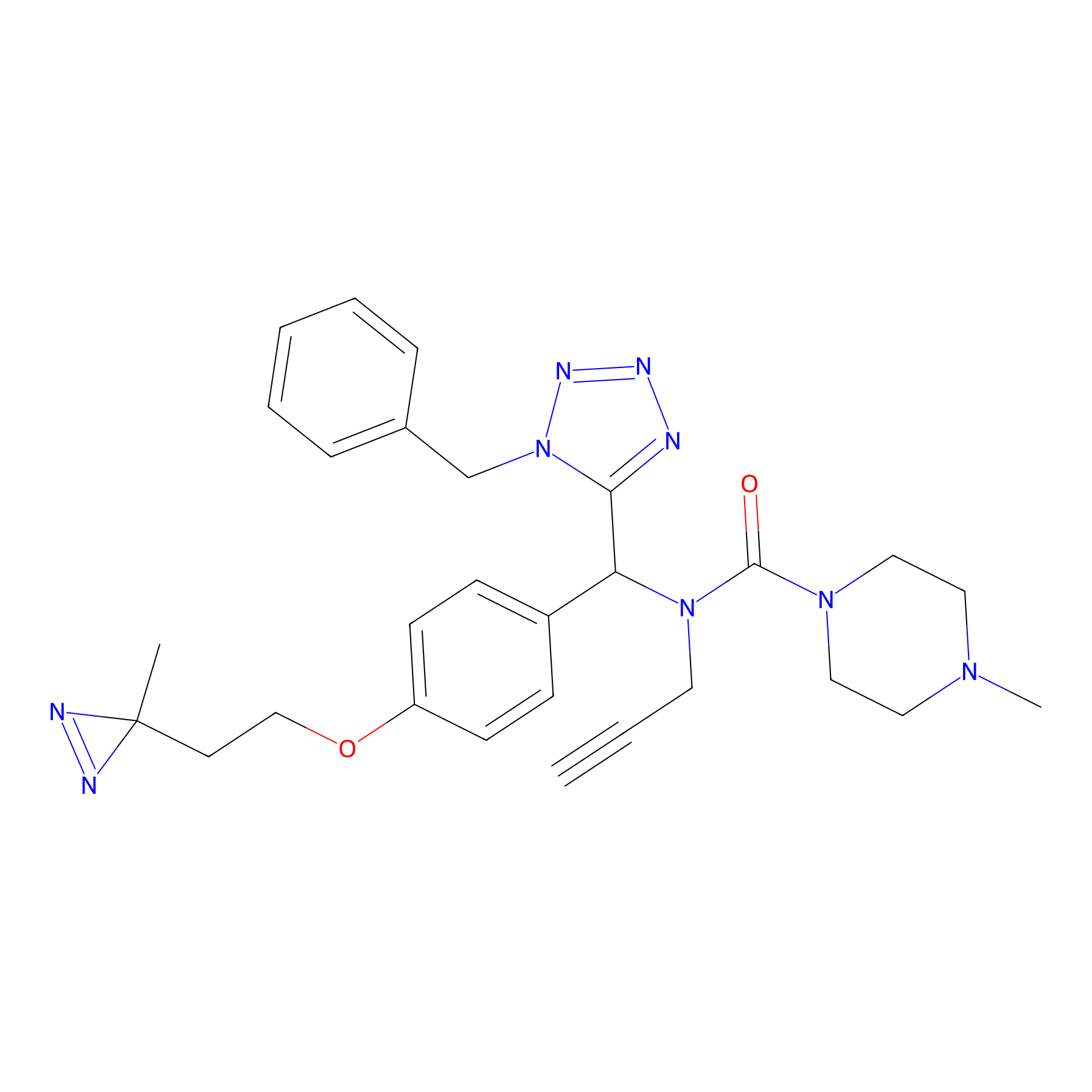
FBPP2 Probe Info |
 |
49.53 | LDD0318 | [2] | |
|
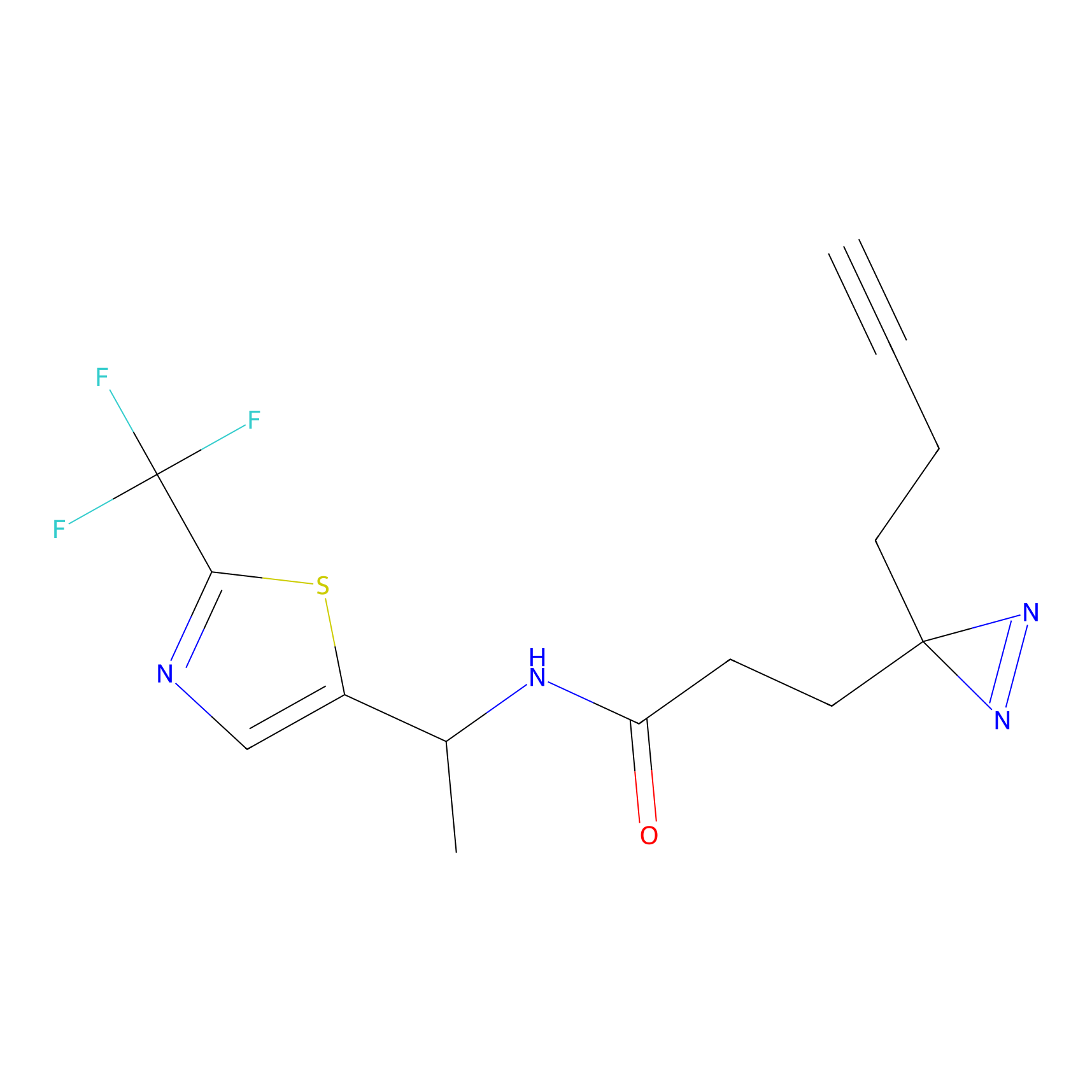
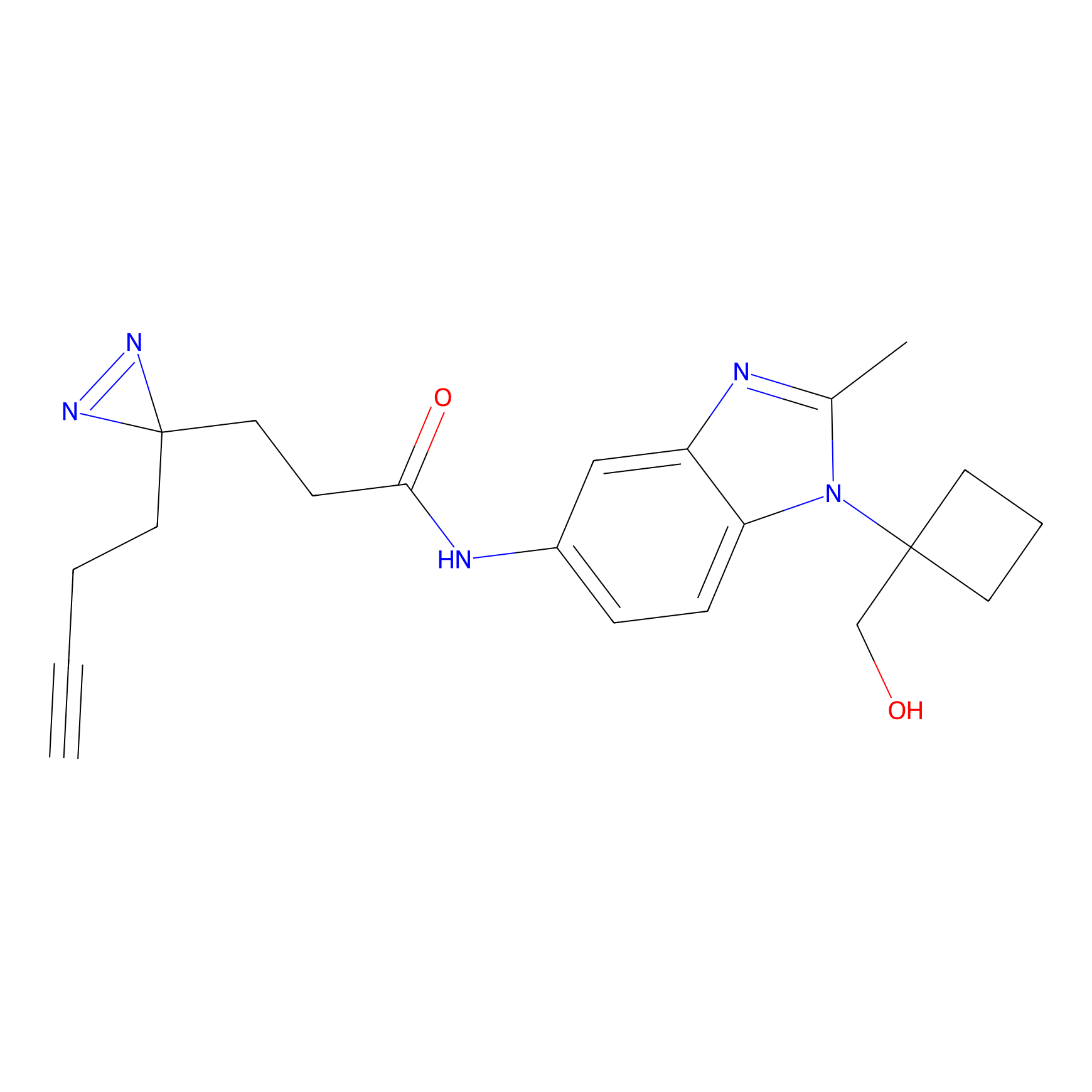
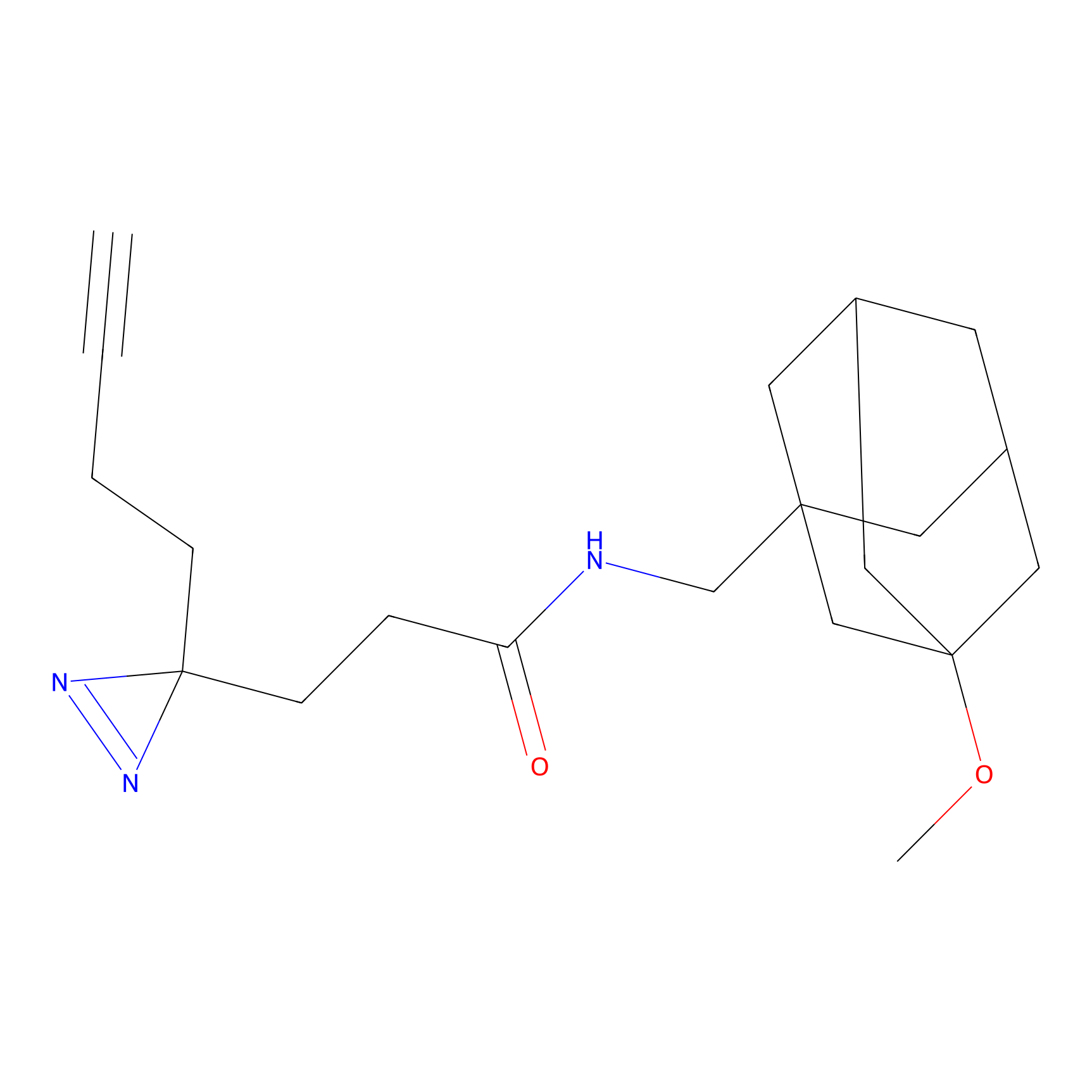
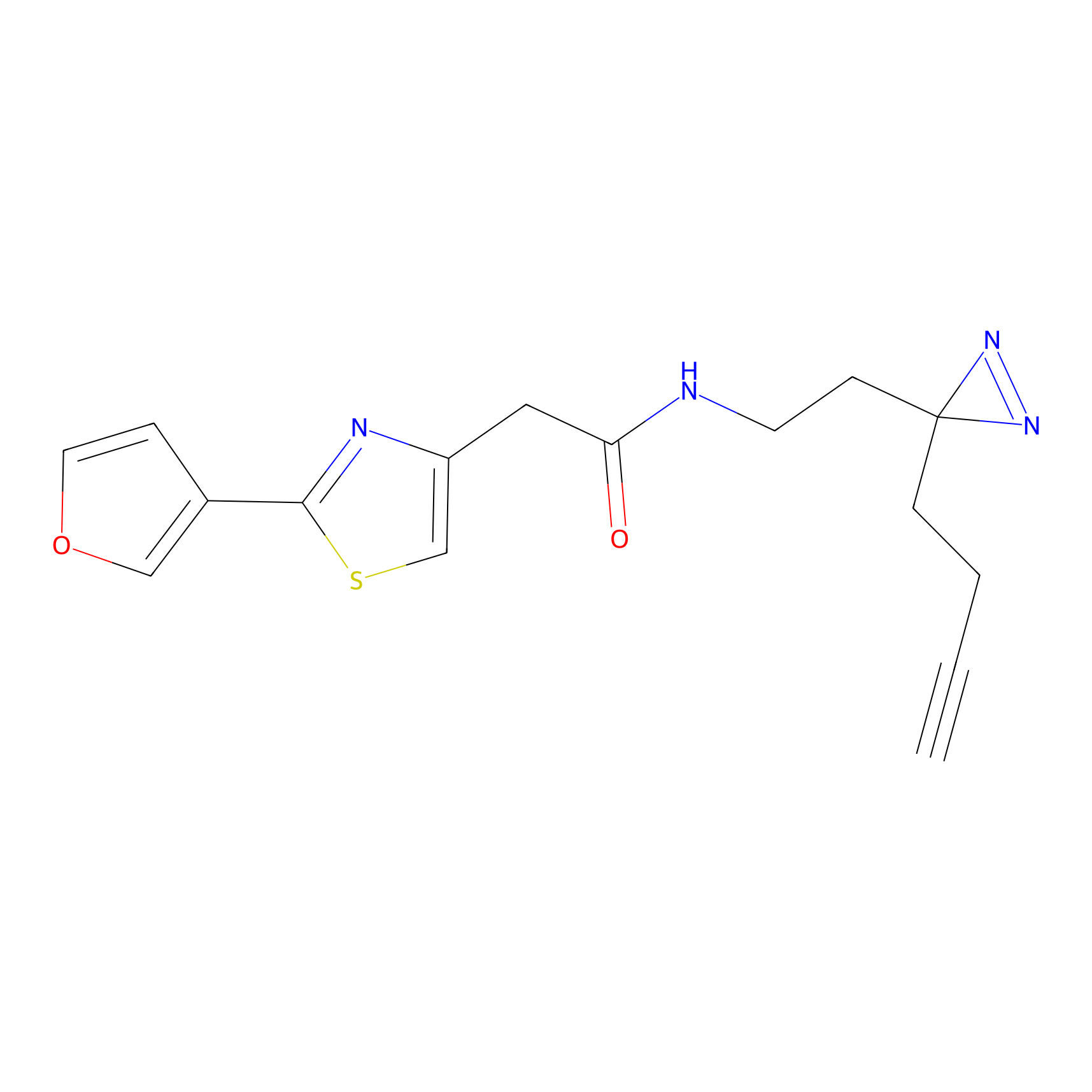
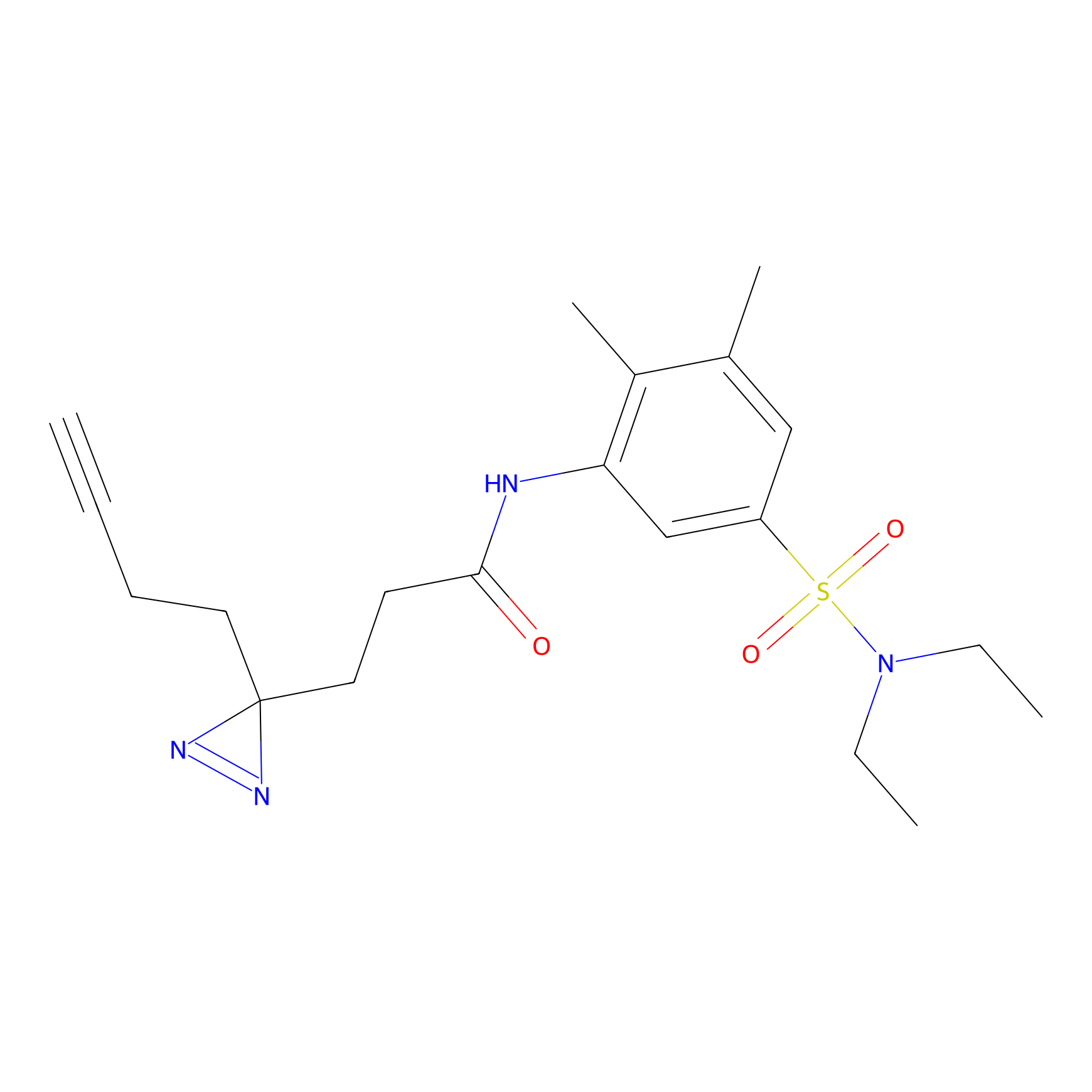
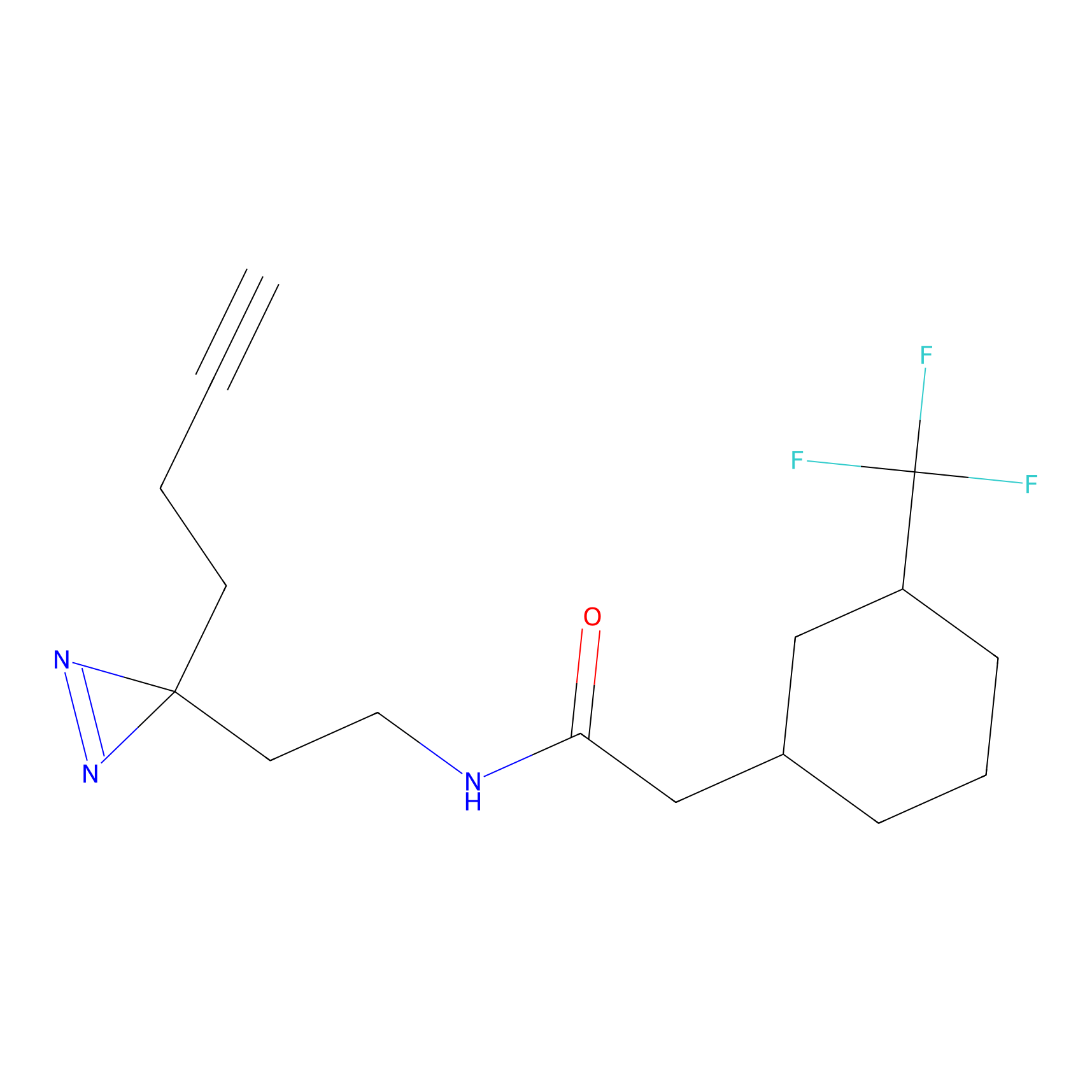
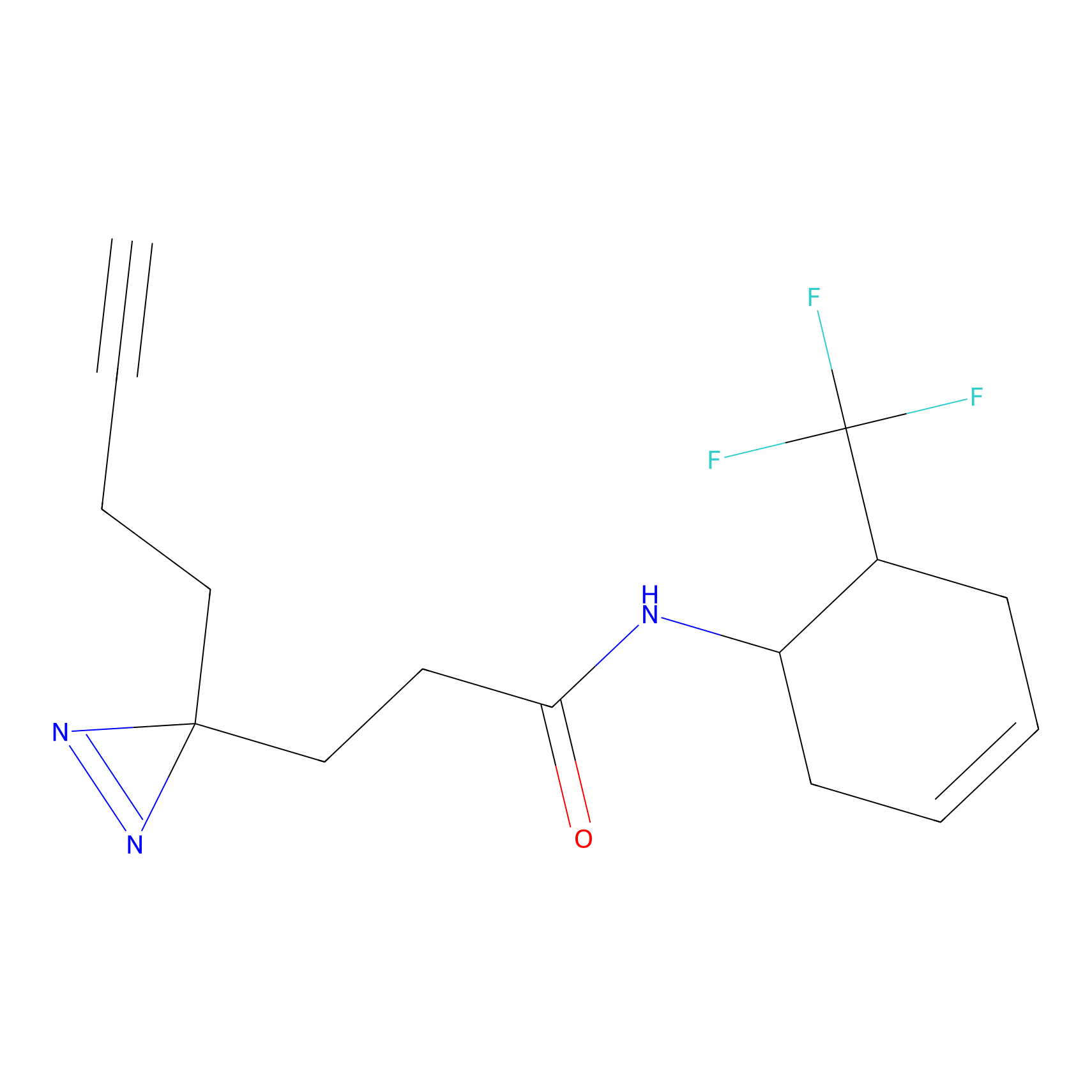
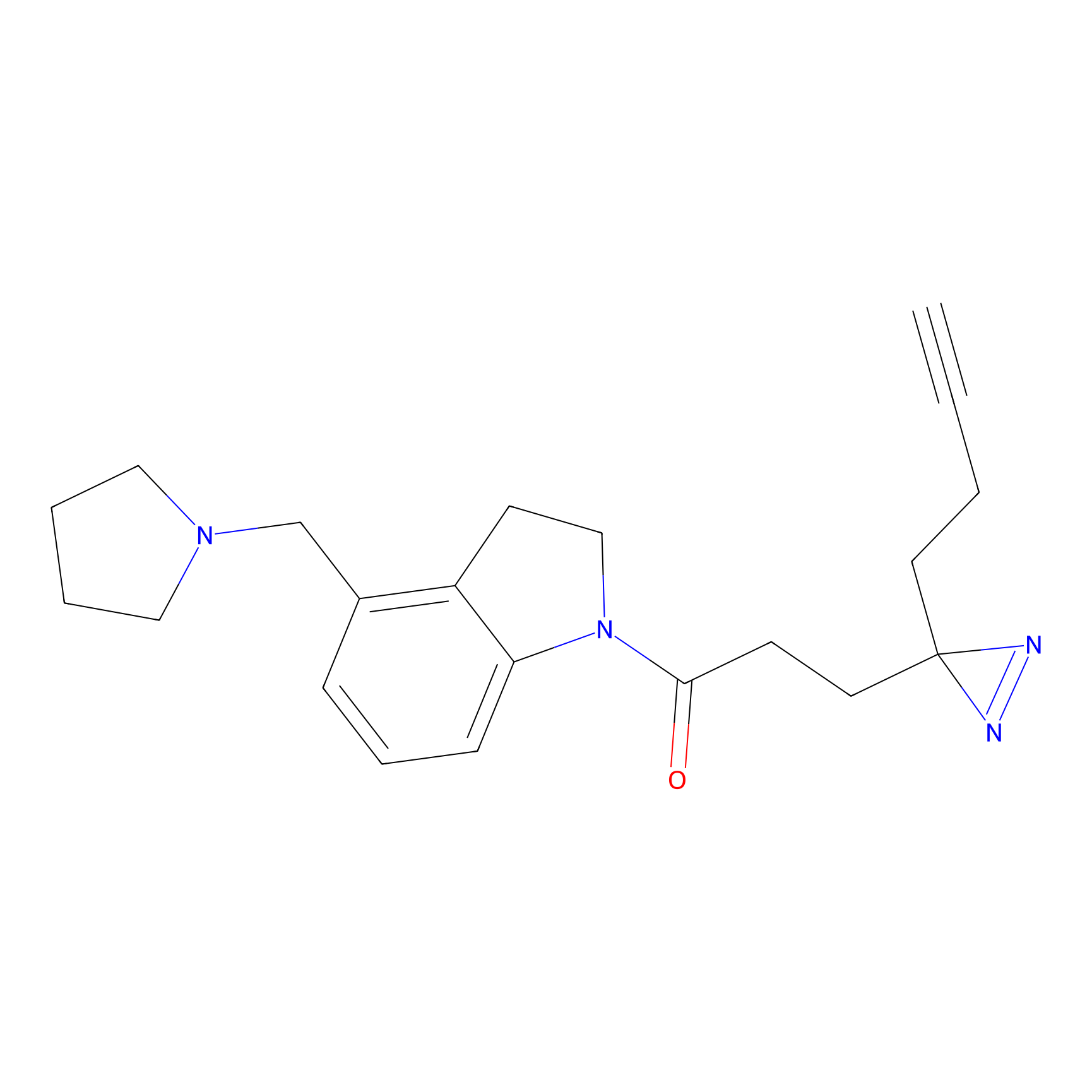
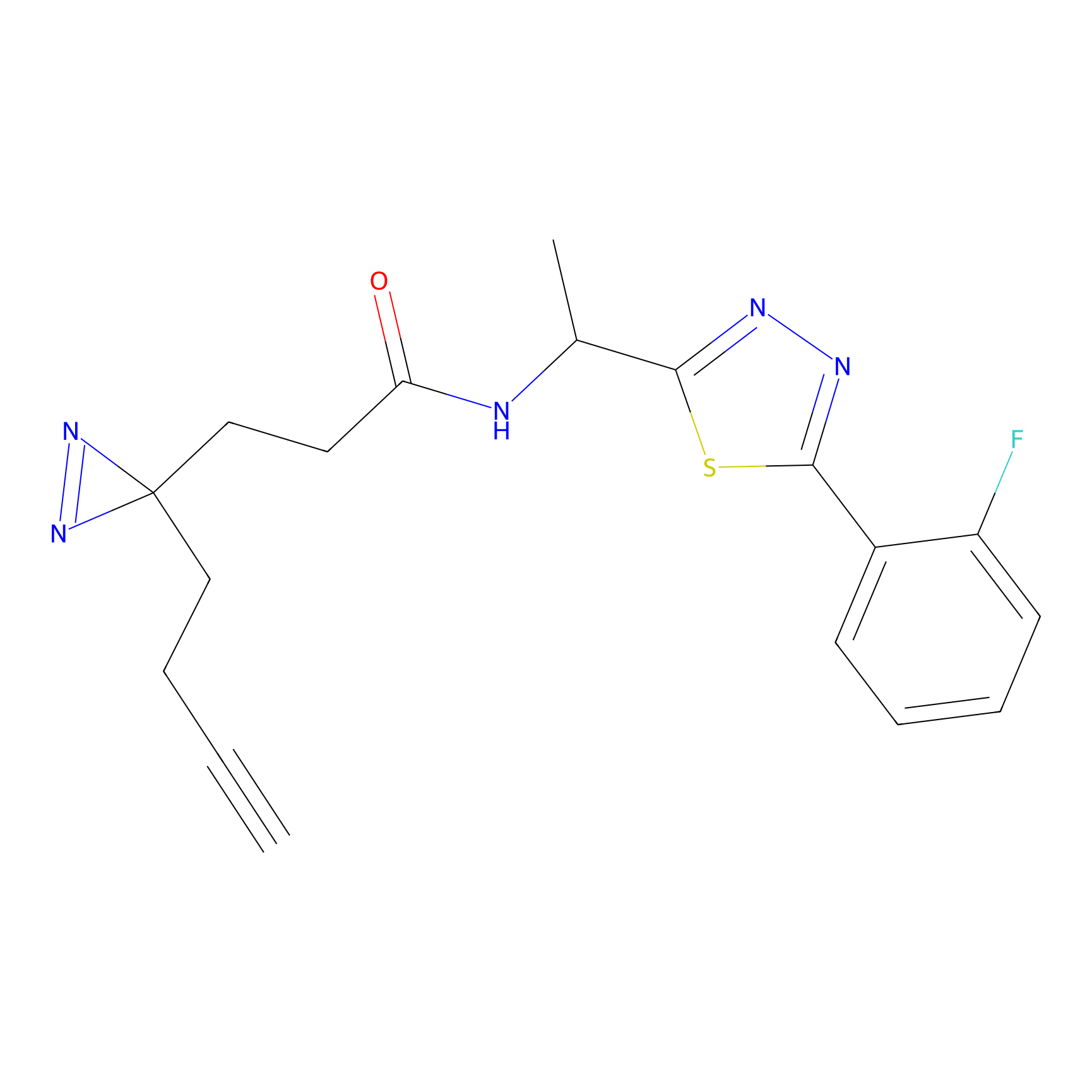
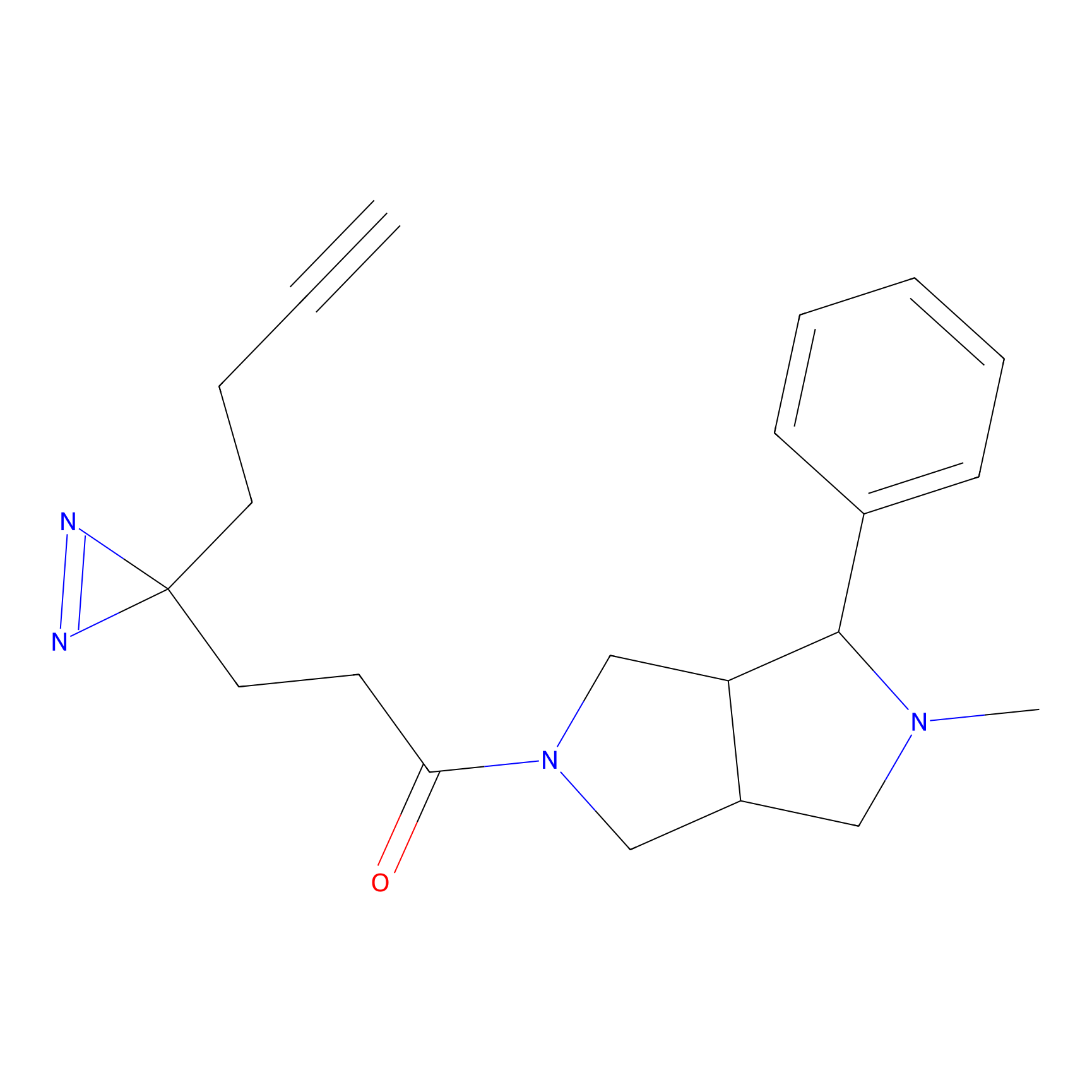
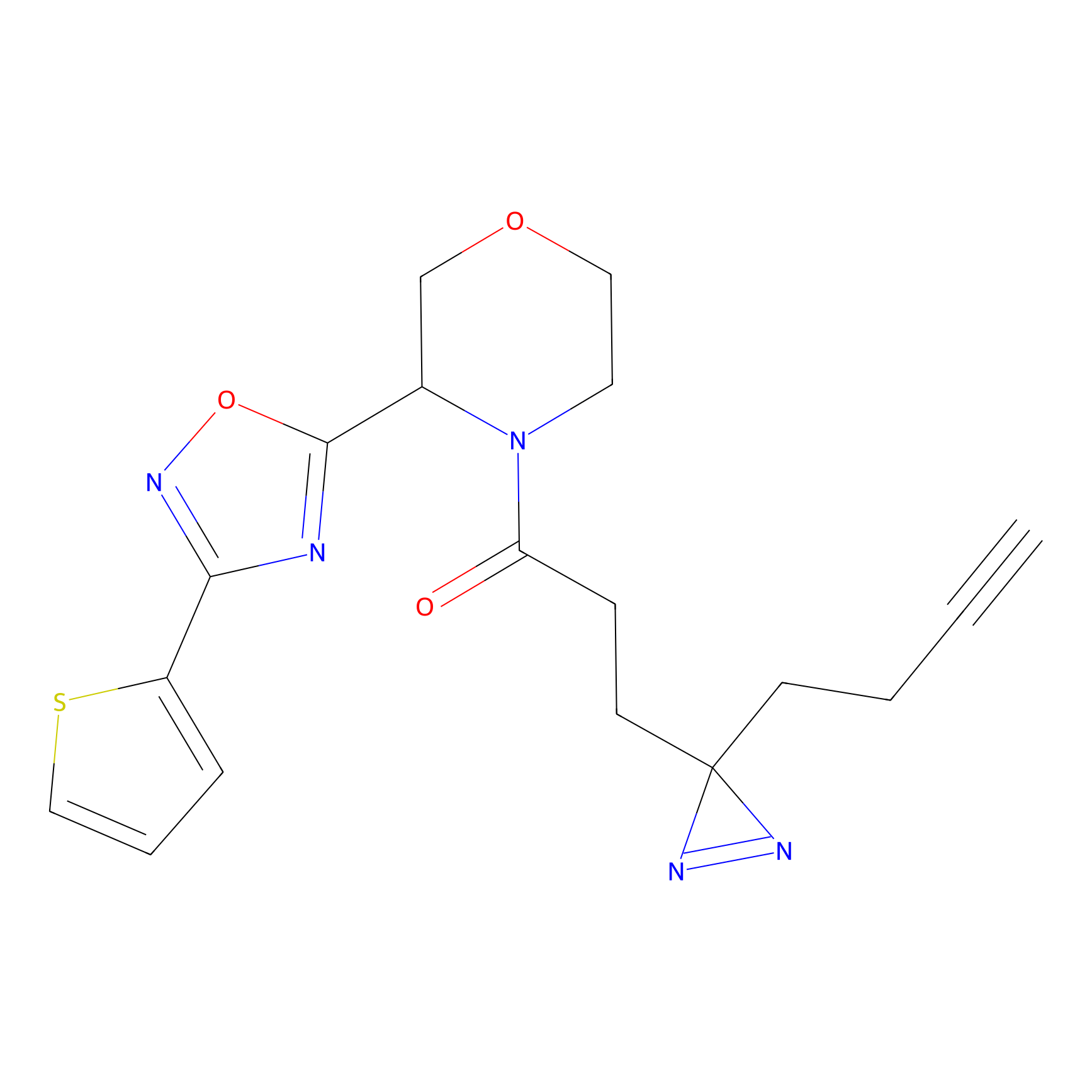
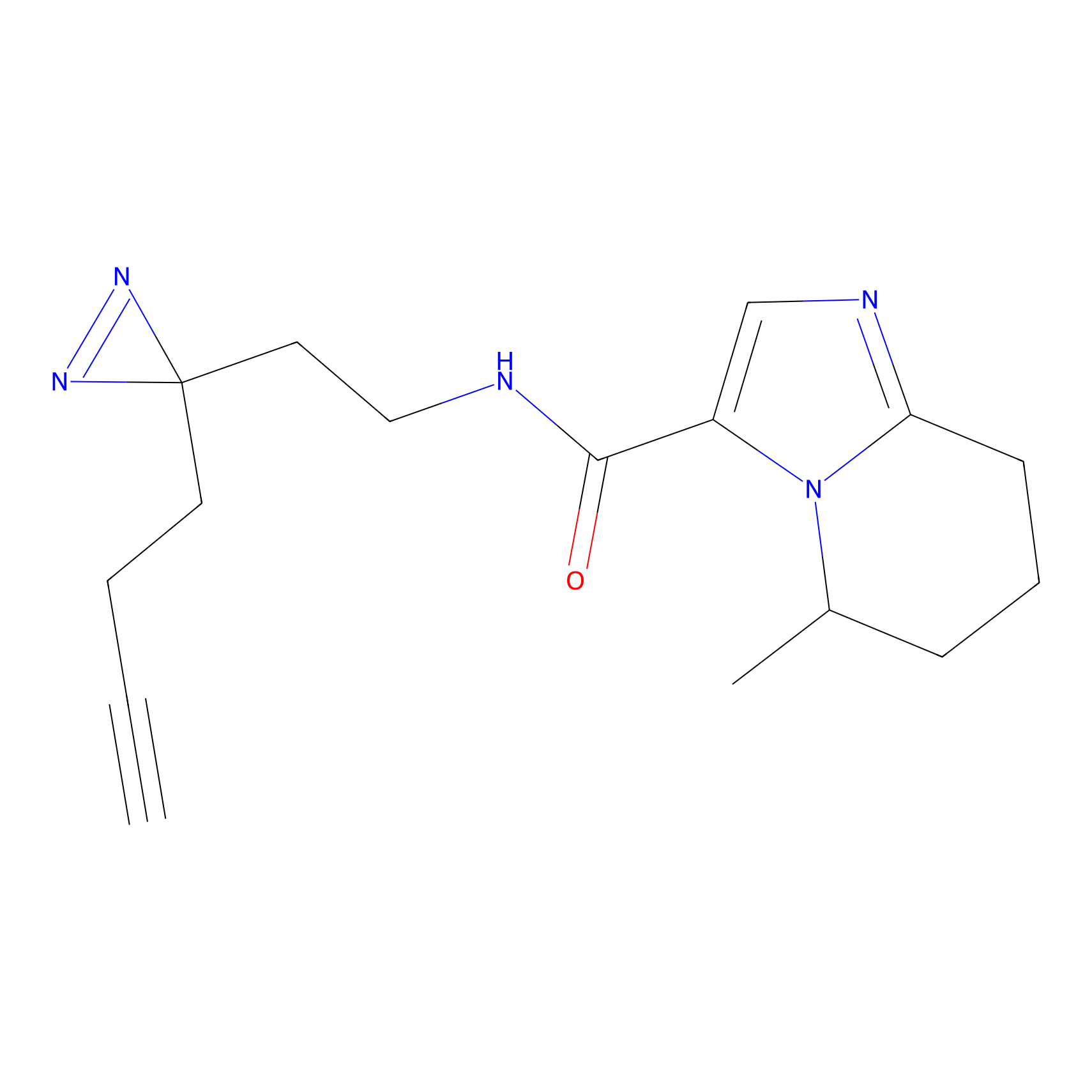
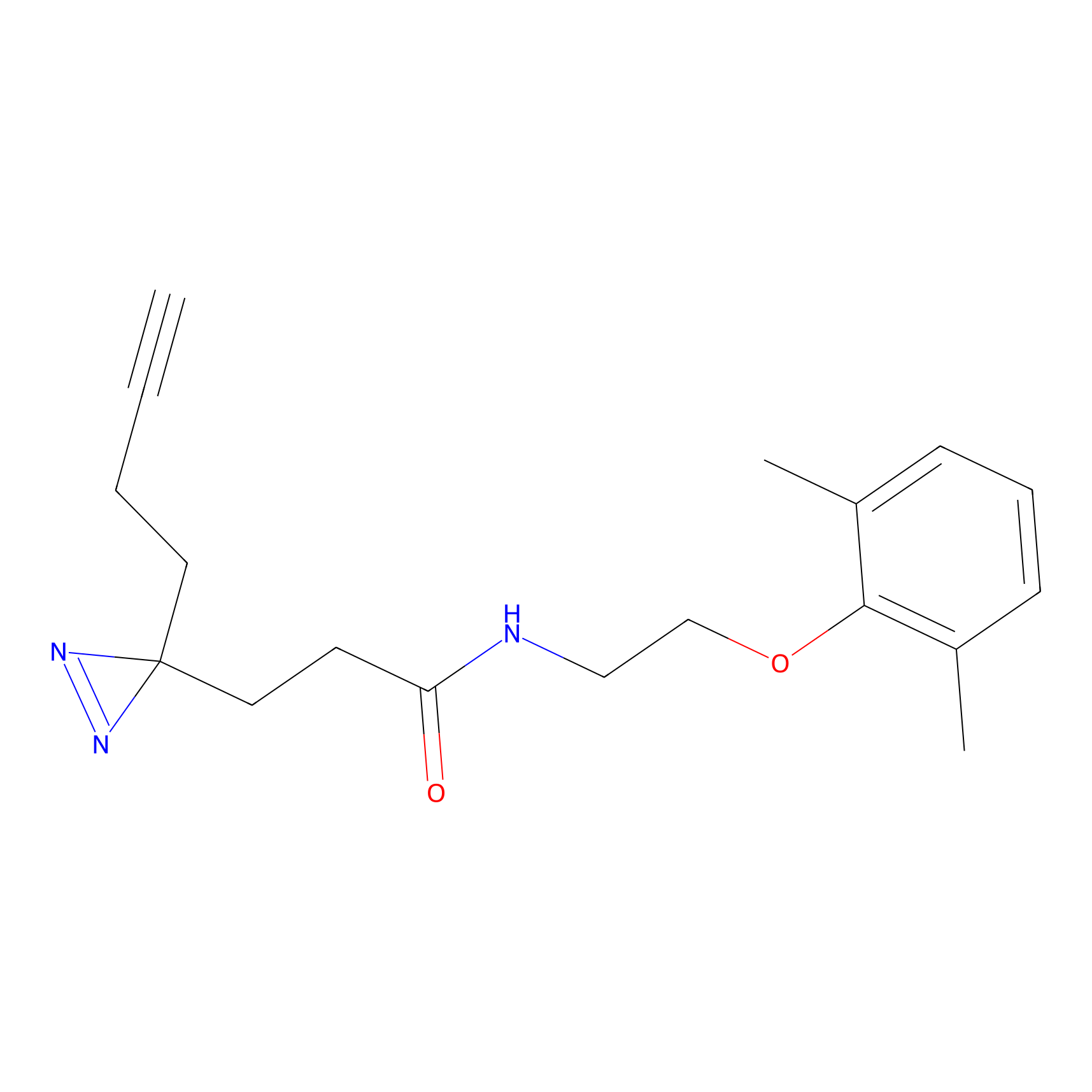
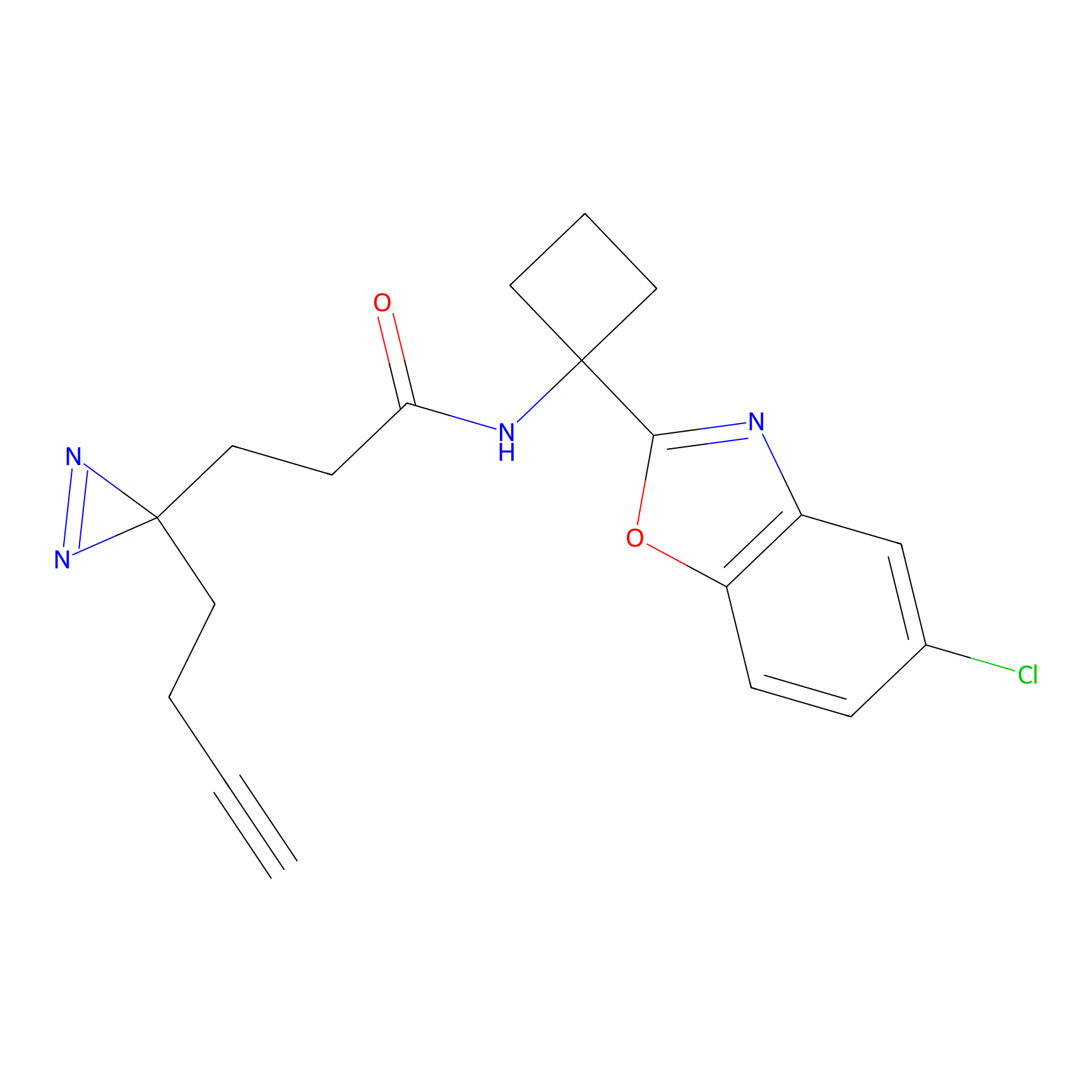
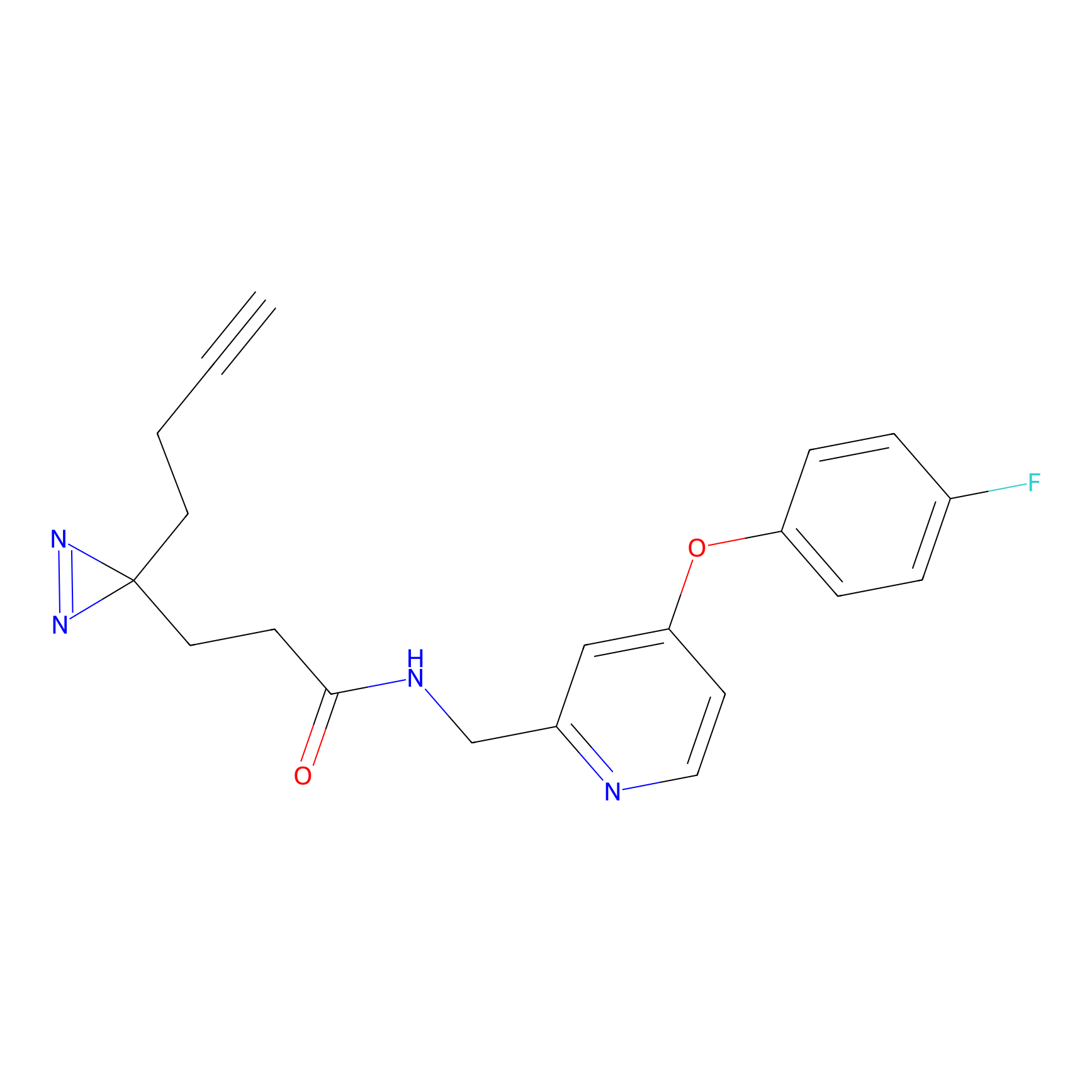
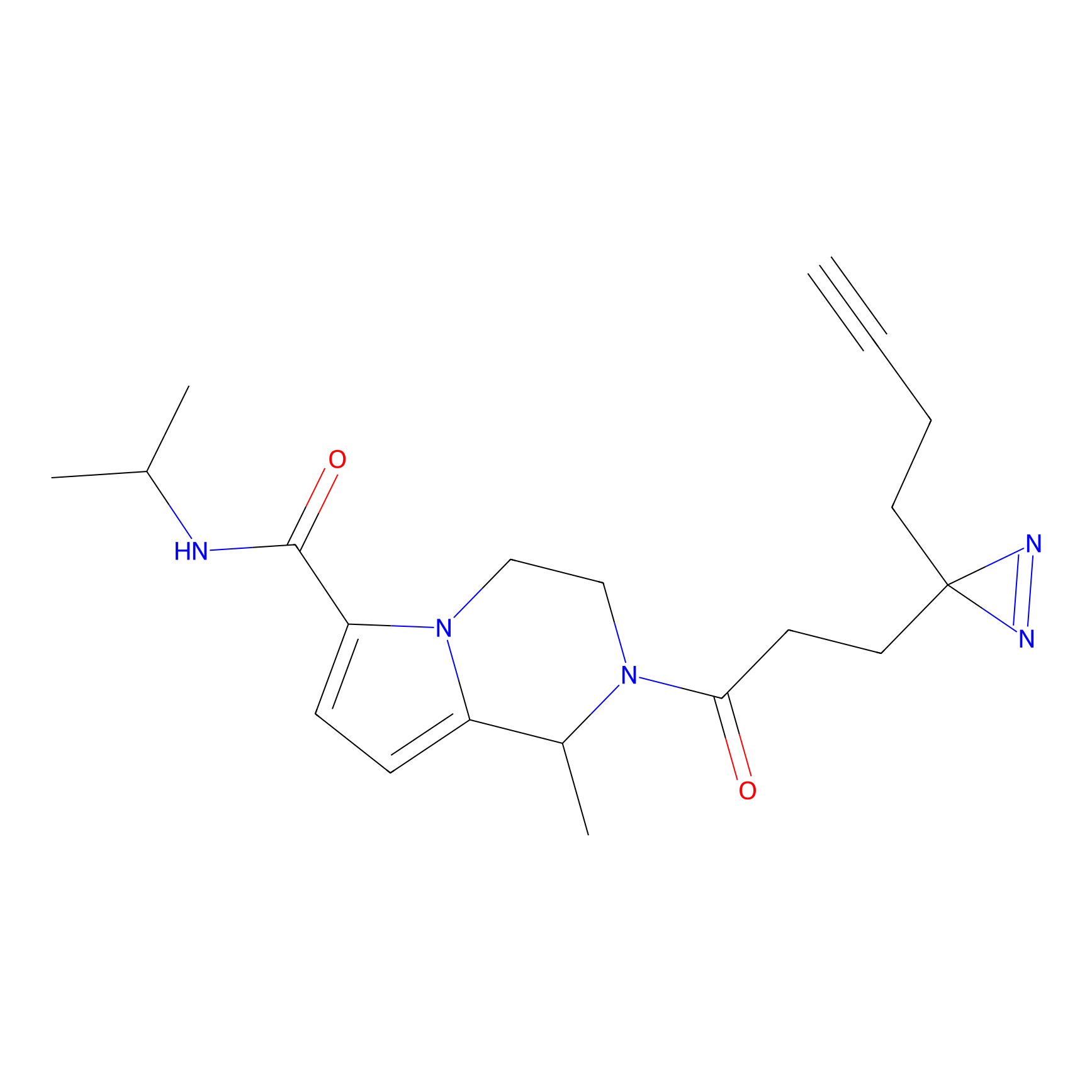
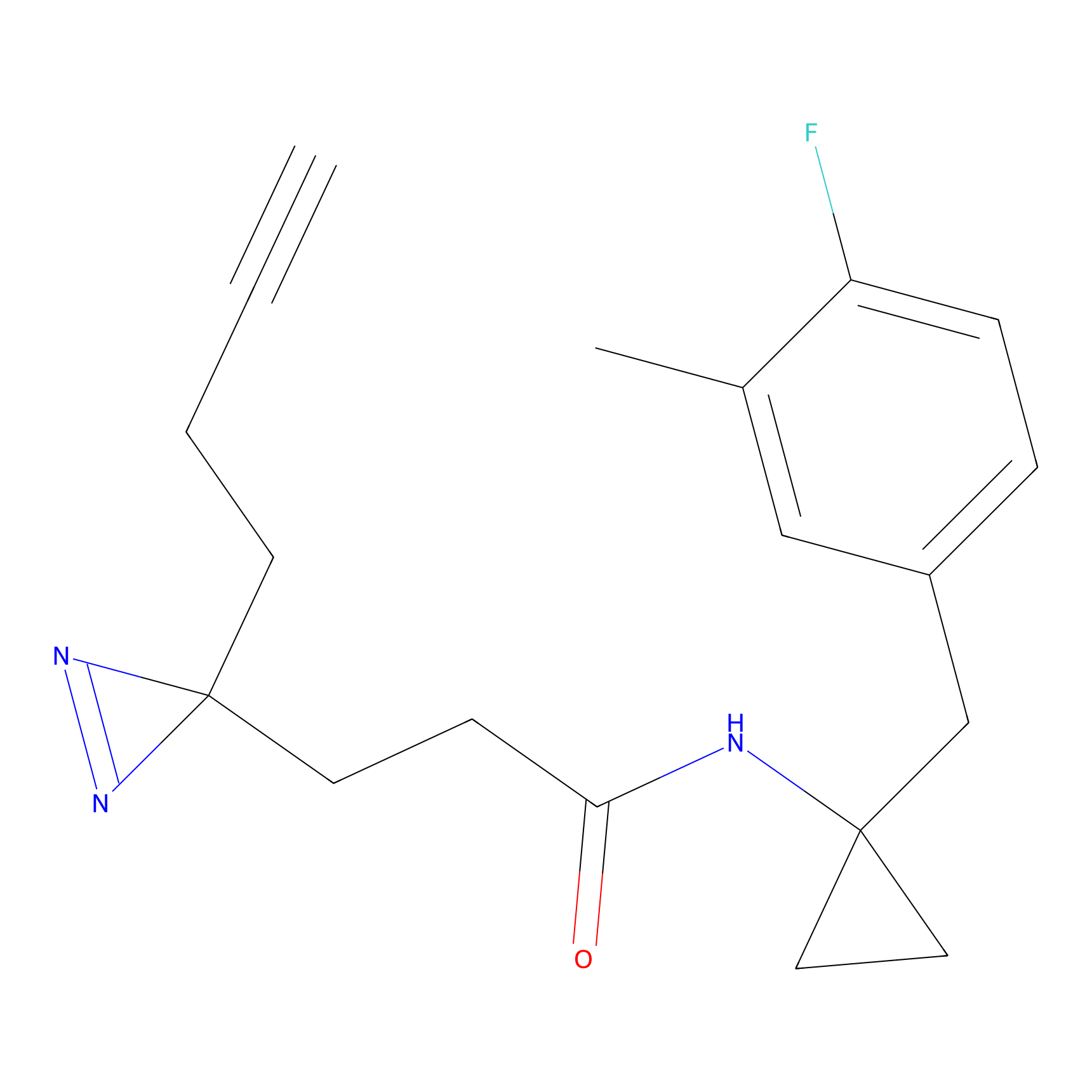
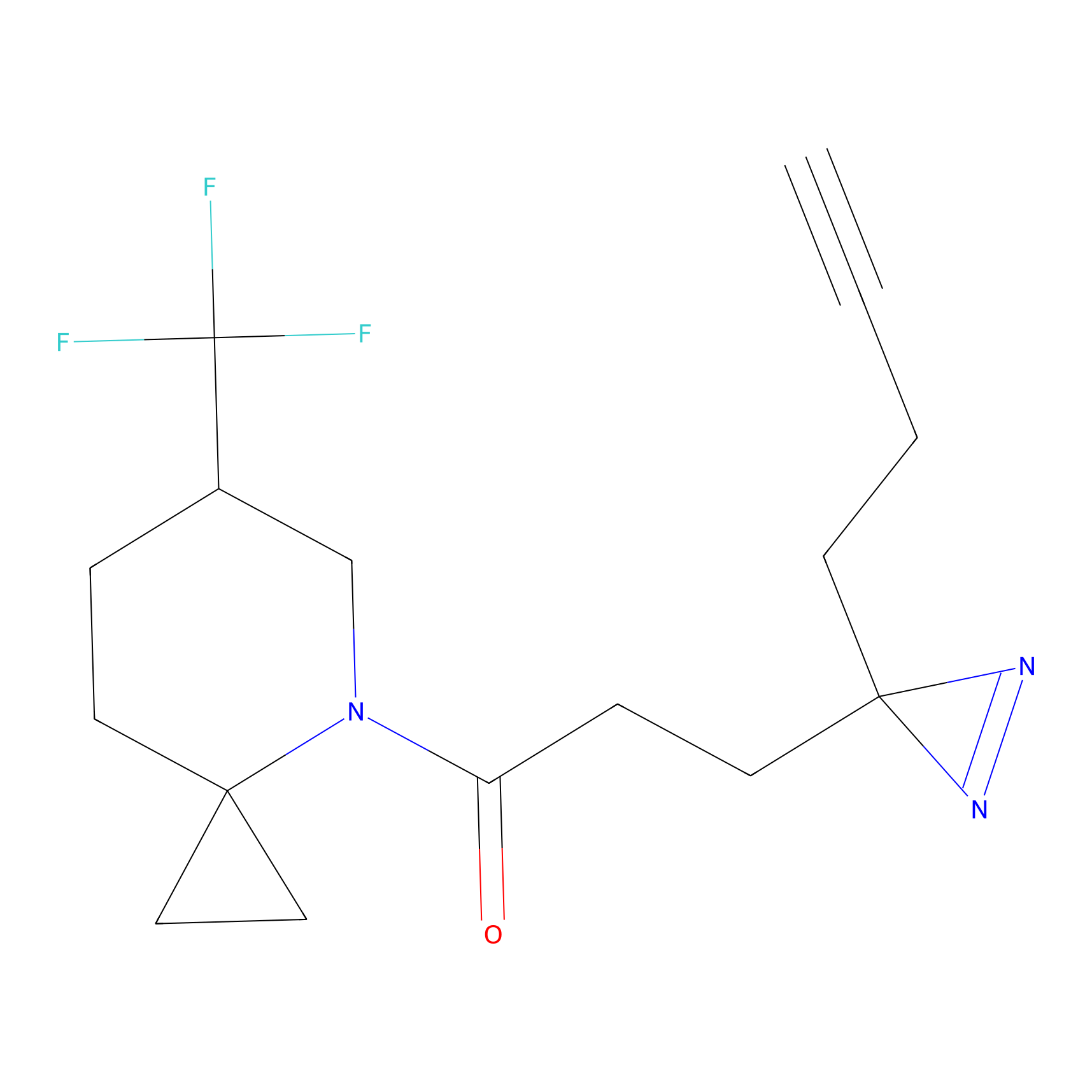
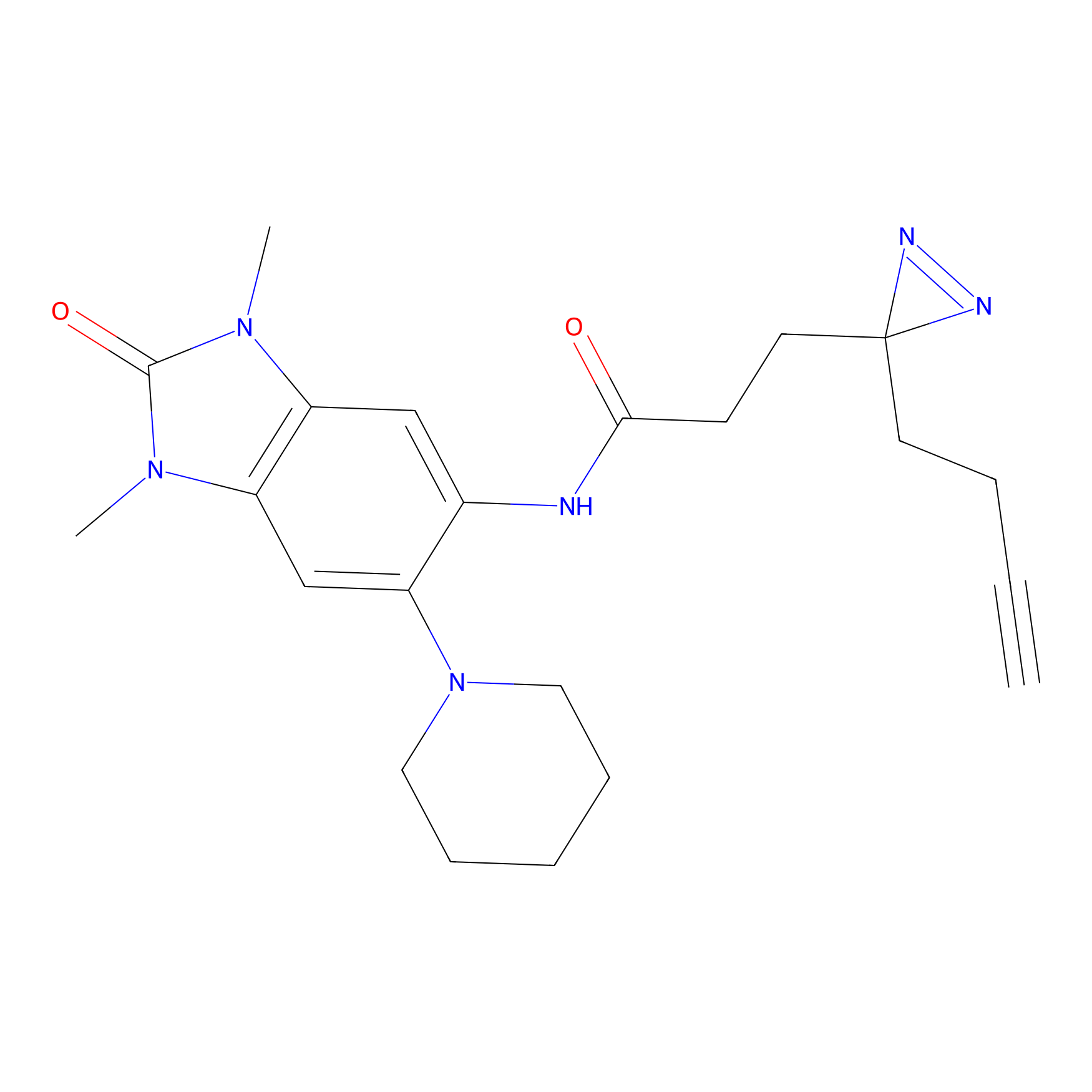
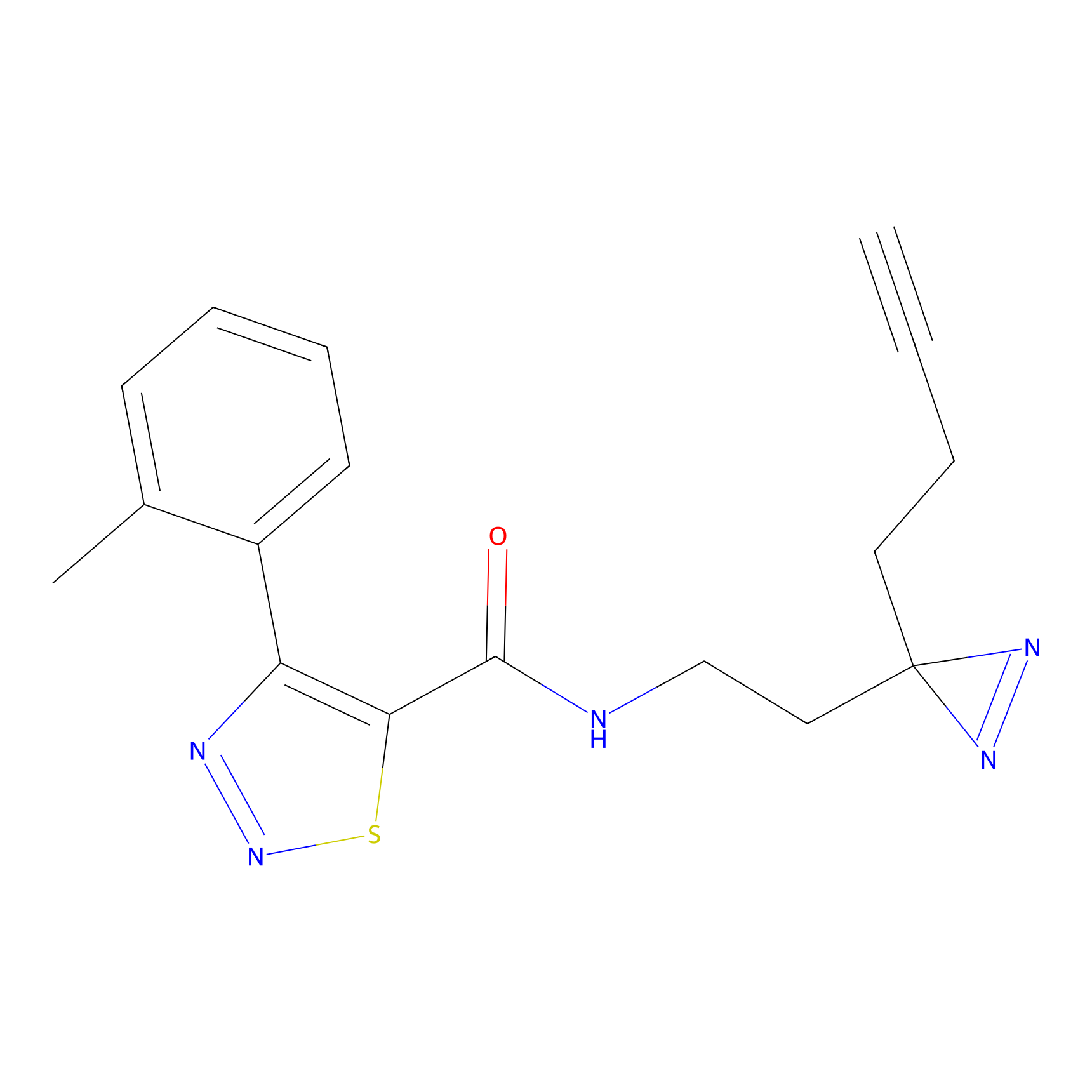
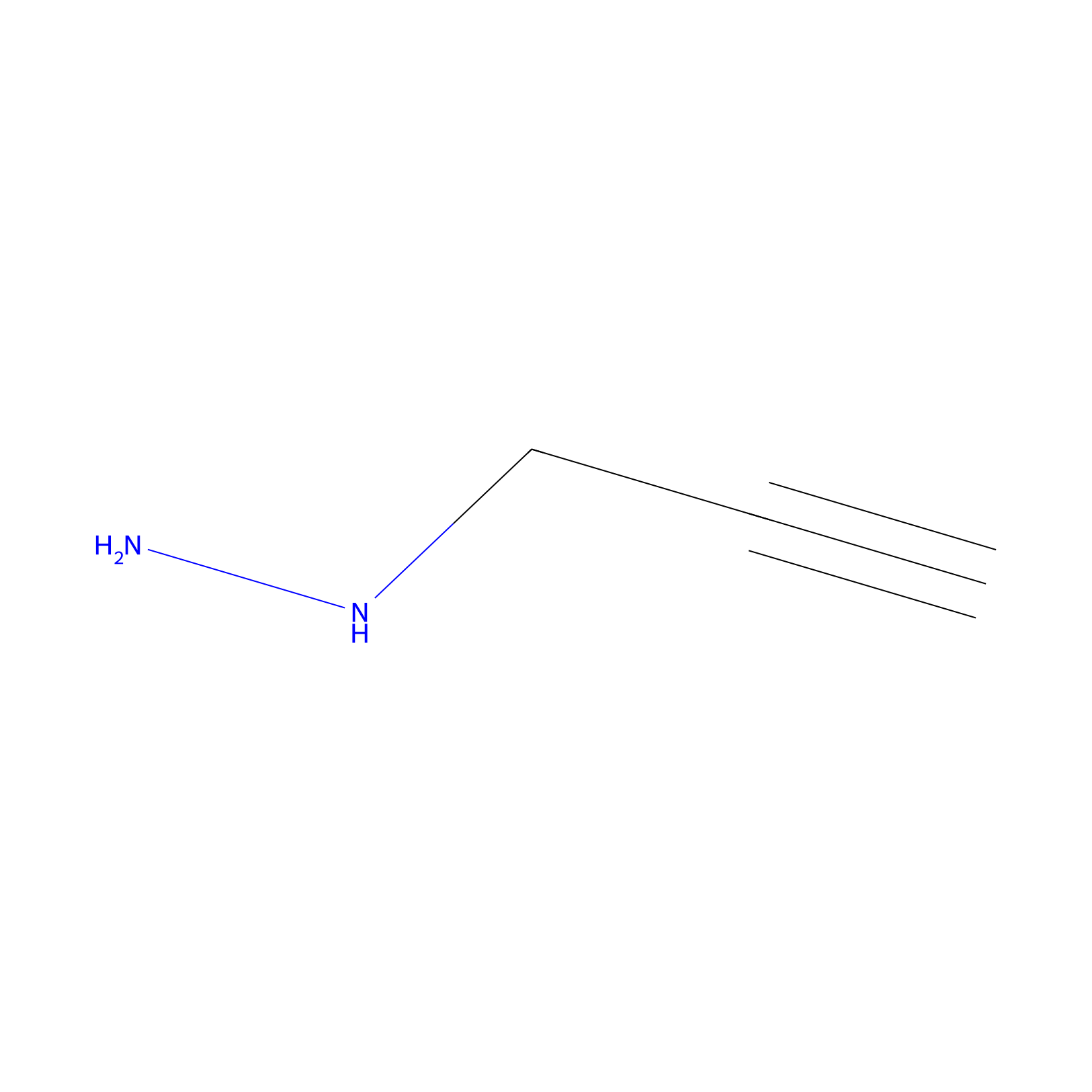
YN-1 Probe Info |
 |
100.00 | LDD0444 | [3] | |
|
DBIA Probe Info |
 |
C88(3.21) | LDD3314 | [4] | |
|
BTD Probe Info |
 |
C88(1.35) | LDD2093 | [5] | |
|
IPM Probe Info |
 |
C88(1.33) | LDD1701 | [5] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [6] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [6] | |
|
AOyne Probe Info |
 |
11.70 | LDD0443 | [7] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C88(1.27) | LDD2152 | [5] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C88(1.05) | LDD1702 | [5] |
| LDCM0073 | Kambe_cp3 | PC-3 | 5.44 | LDD0128 | [10] |
| LDCM0022 | KB02 | 22RV1 | C88(2.51) | LDD2243 | [4] |
| LDCM0023 | KB03 | MDA-MB-231 | C88(1.33) | LDD1701 | [5] |
| LDCM0024 | KB05 | IGR37 | C88(3.21) | LDD3314 | [4] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C88(1.35) | LDD2093 | [5] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C88(0.88) | LDD2099 | [5] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C88(0.88) | LDD2109 | [5] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C88(1.00) | LDD2125 | [5] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C88(1.39) | LDD2136 | [5] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C88(1.03) | LDD2137 | [5] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C88(1.35) | LDD2140 | [5] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C88(1.20) | LDD2146 | [5] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C88(0.18) | LDD2150 | [5] |
| LDCM0099 | Phenelzine | MDA-MB-231 | 20.00 | LDD0388 | [1] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Homeobox protein Hox-A1 (HOXA1) | Antp homeobox family | P49639 | |||
Other
References