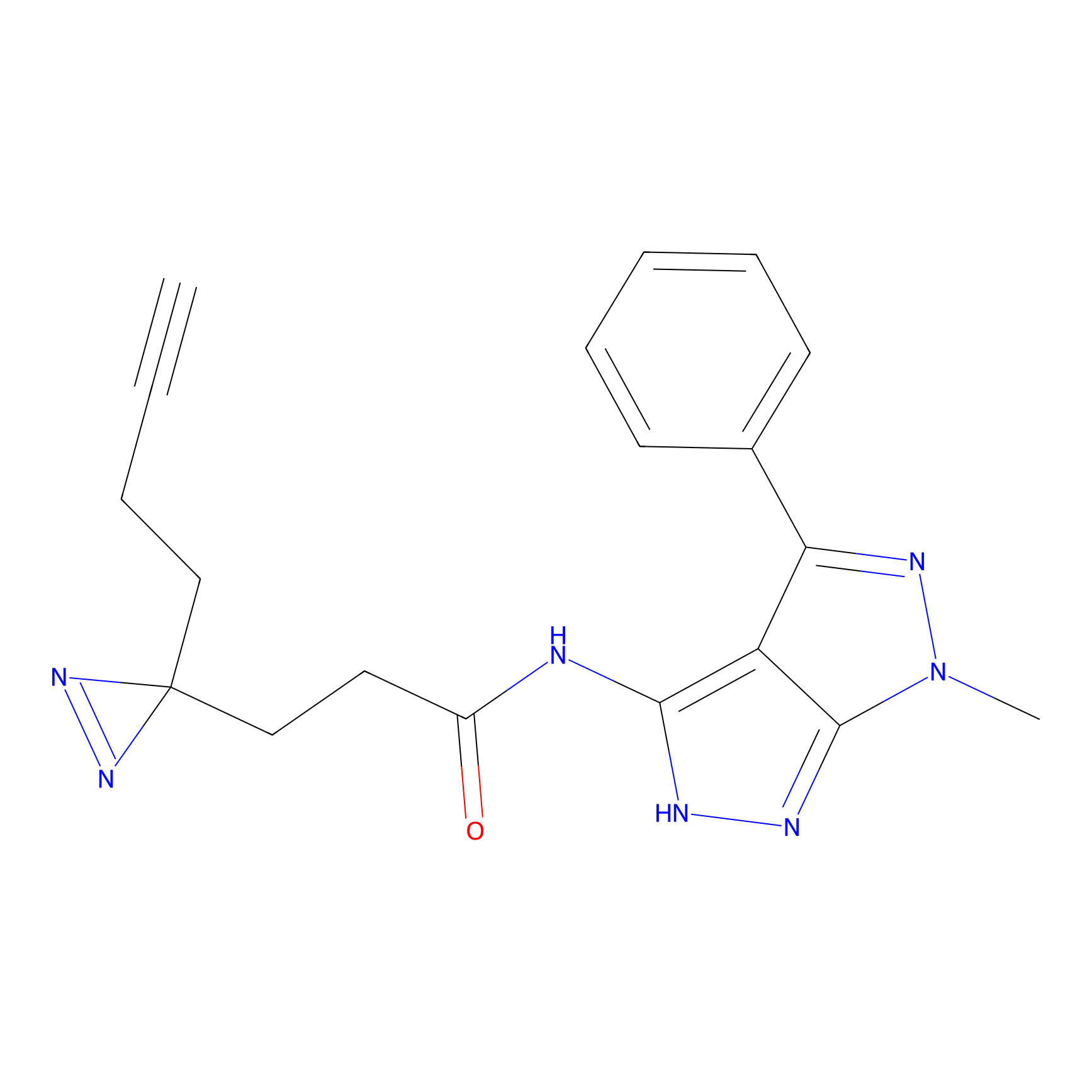
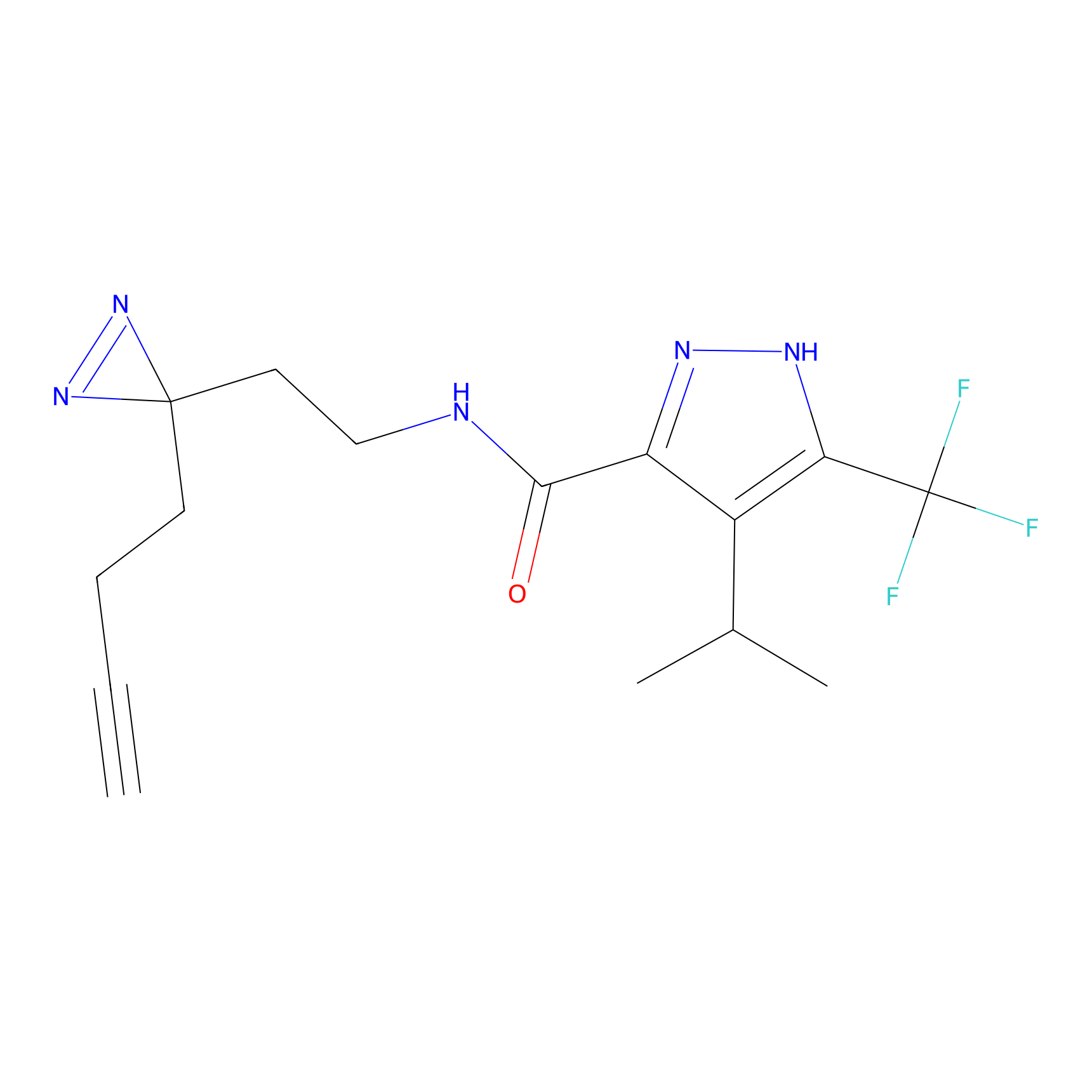
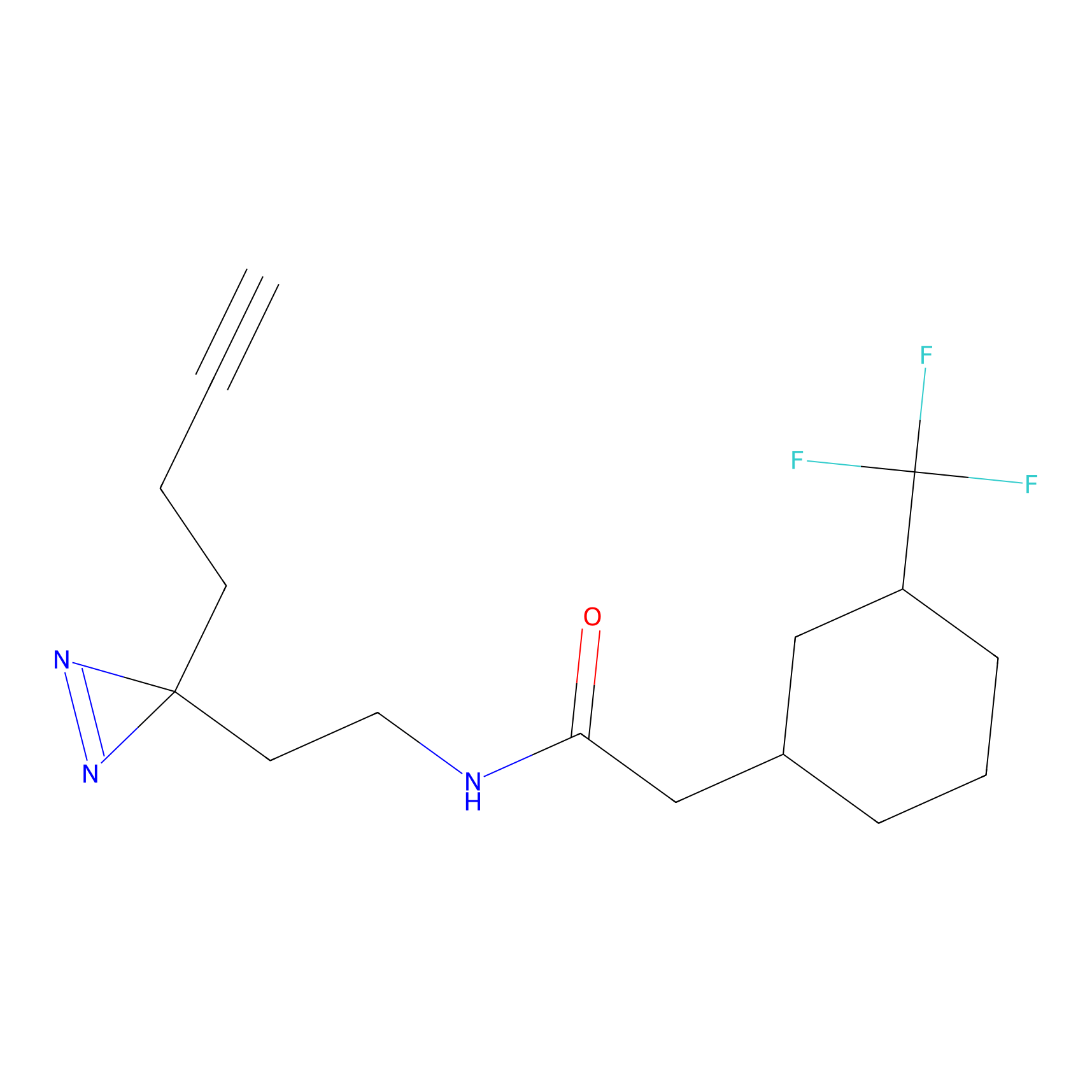
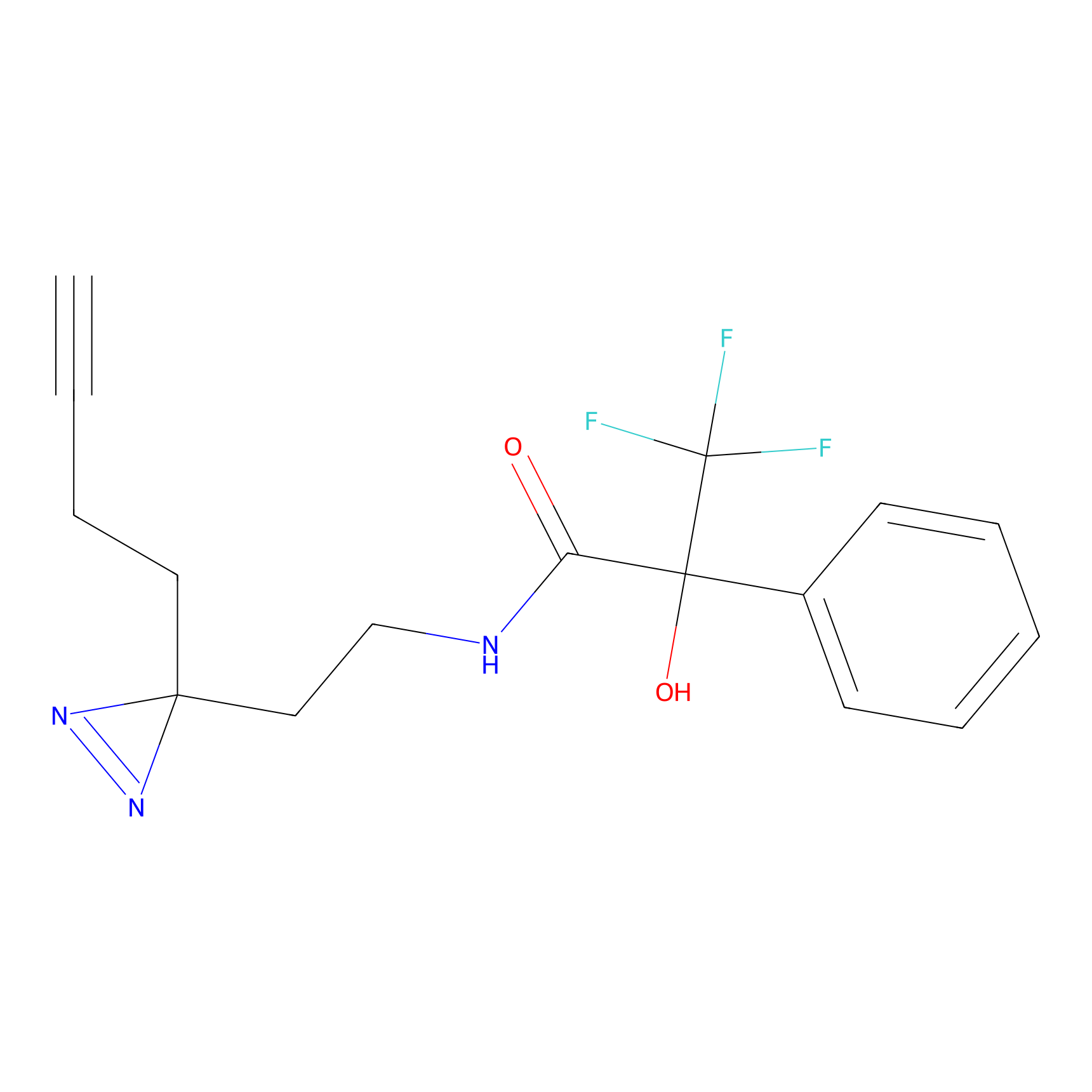
Details of the Target
General Information of Target
| Target ID | LDTP00477 | |||||
|---|---|---|---|---|---|---|
| Target Name | Mitochondrial import inner membrane translocase subunit Tim23 (TIMM23) | |||||
| Gene Name | TIMM23 | |||||
| Gene ID | 100287932 | |||||
| Synonyms |
TIM23; Mitochondrial import inner membrane translocase subunit Tim23 |
|||||
| 3D Structure | ||||||
| Sequence |
MEGGGGSGNKTTGGLAGFFGAGGAGYSHADLAGVPLTGMNPLSPYLNVDPRYLVQDTDEF
ILPTGANKTRGRFELAFFTIGGCCMTGAAFGAMNGLRLGLKETQNMAWSKPRNVQILNMV TRQGALWANTLGSLALLYSAFGVIIEKTRGAEDDLNTVAAGTMTGMLYKCTGGLRGIARG GLTGLTLTSLYALYNNWEHMKGSLLQQSL |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
Tim17/Tim22/Tim23 family
|
|||||
| Subcellular location |
Mitochondrion inner membrane
|
|||||
| Function | Essential component of the TIM23 complex, a complex that mediates the translocation of transit peptide-containing proteins across the mitochondrial inner membrane. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
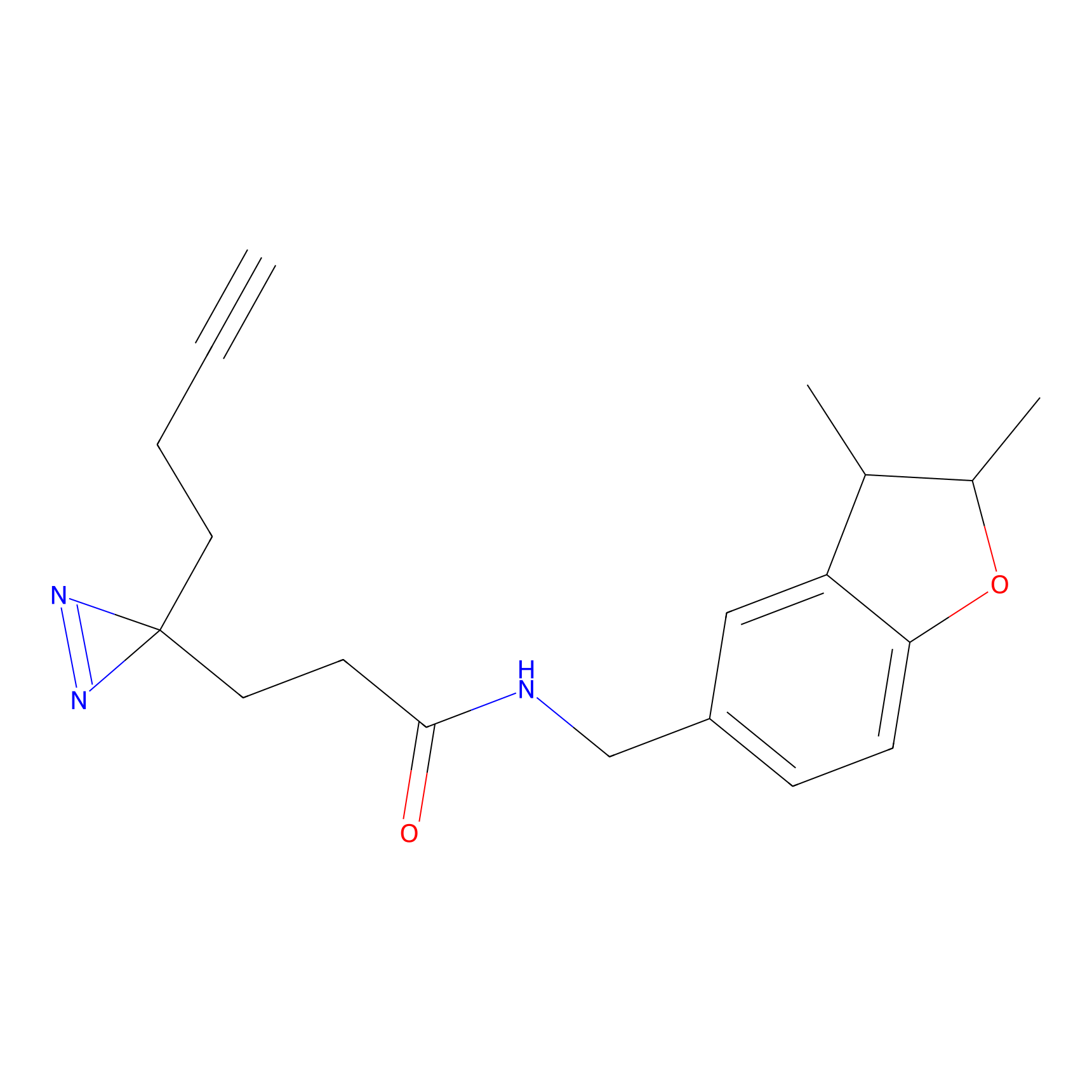
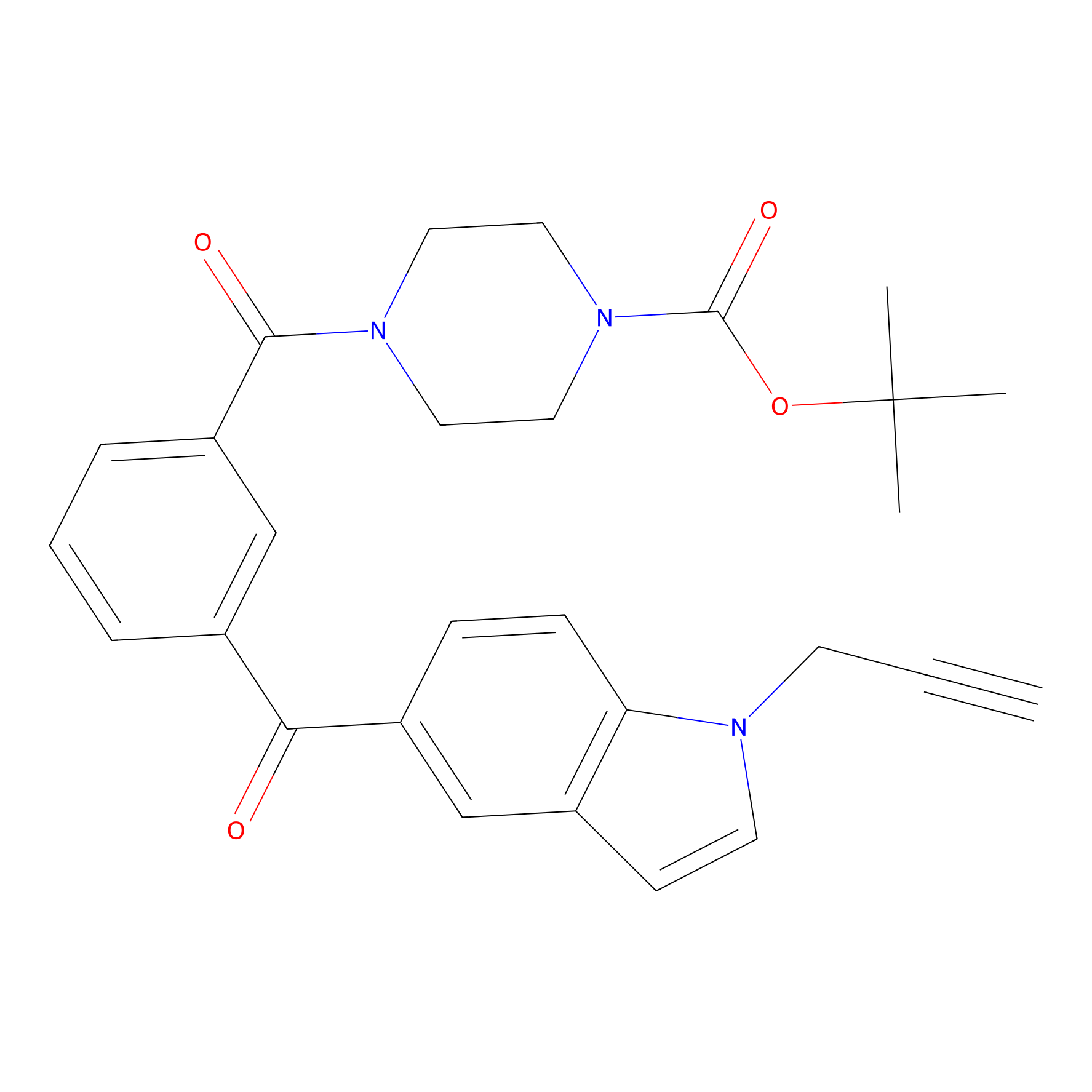
|
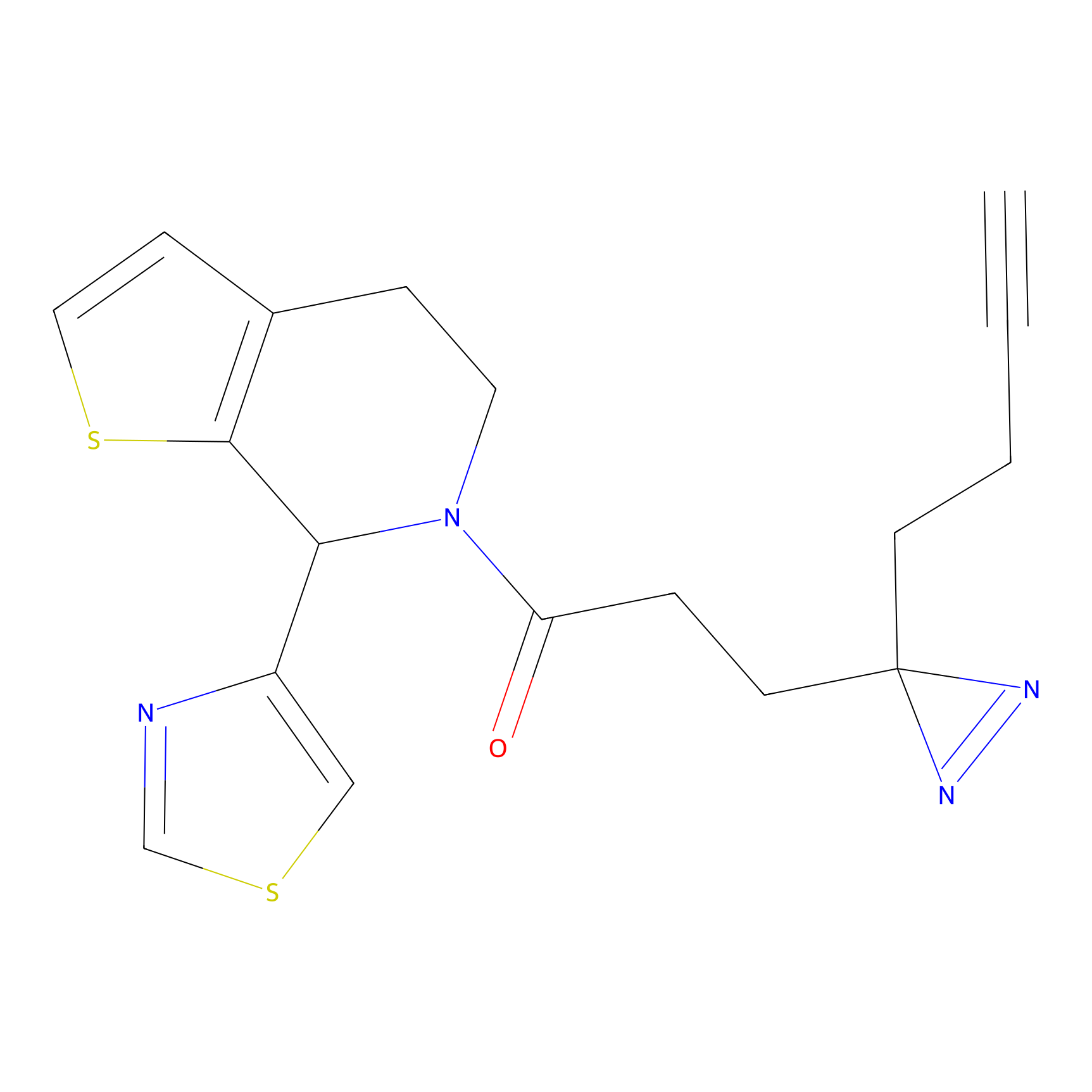
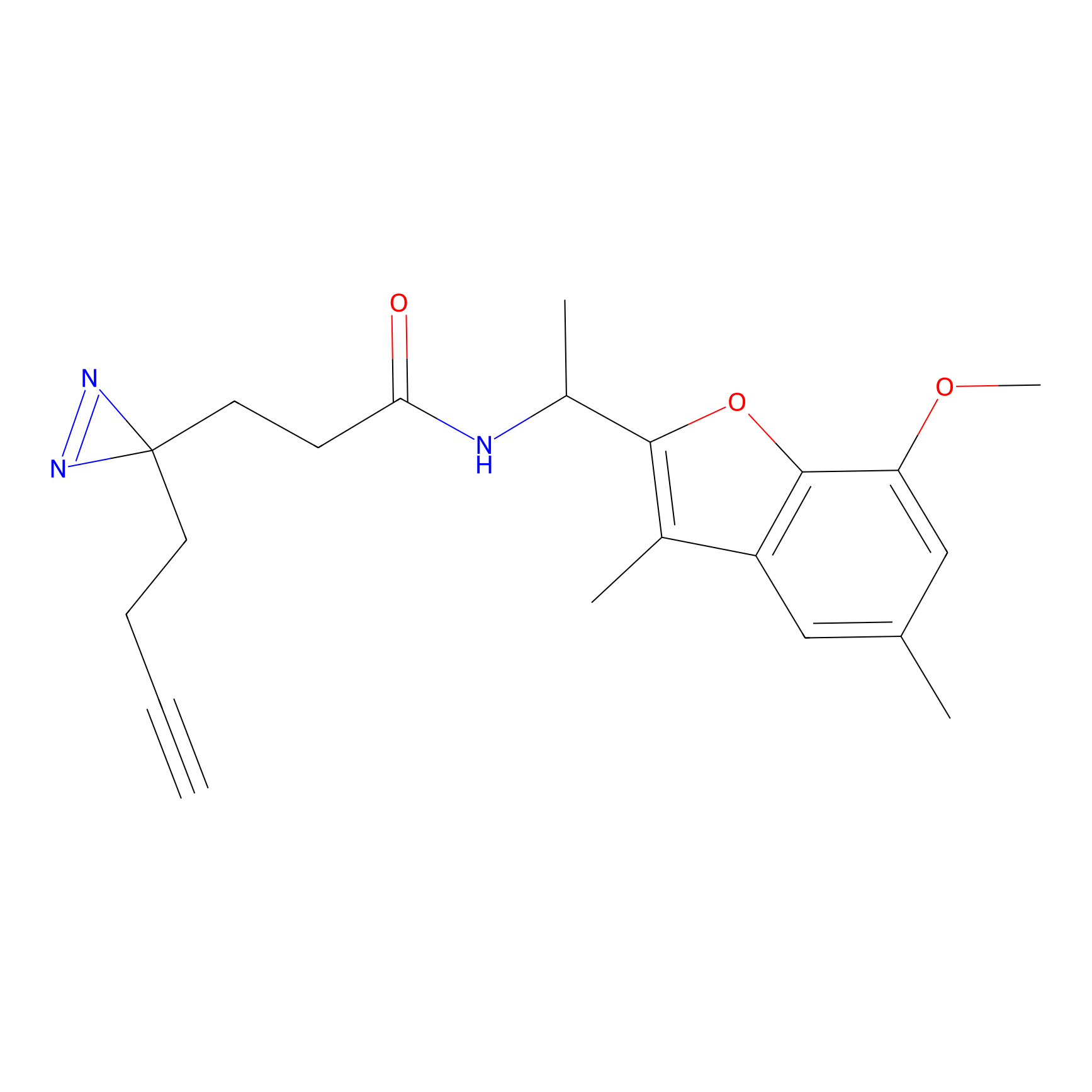
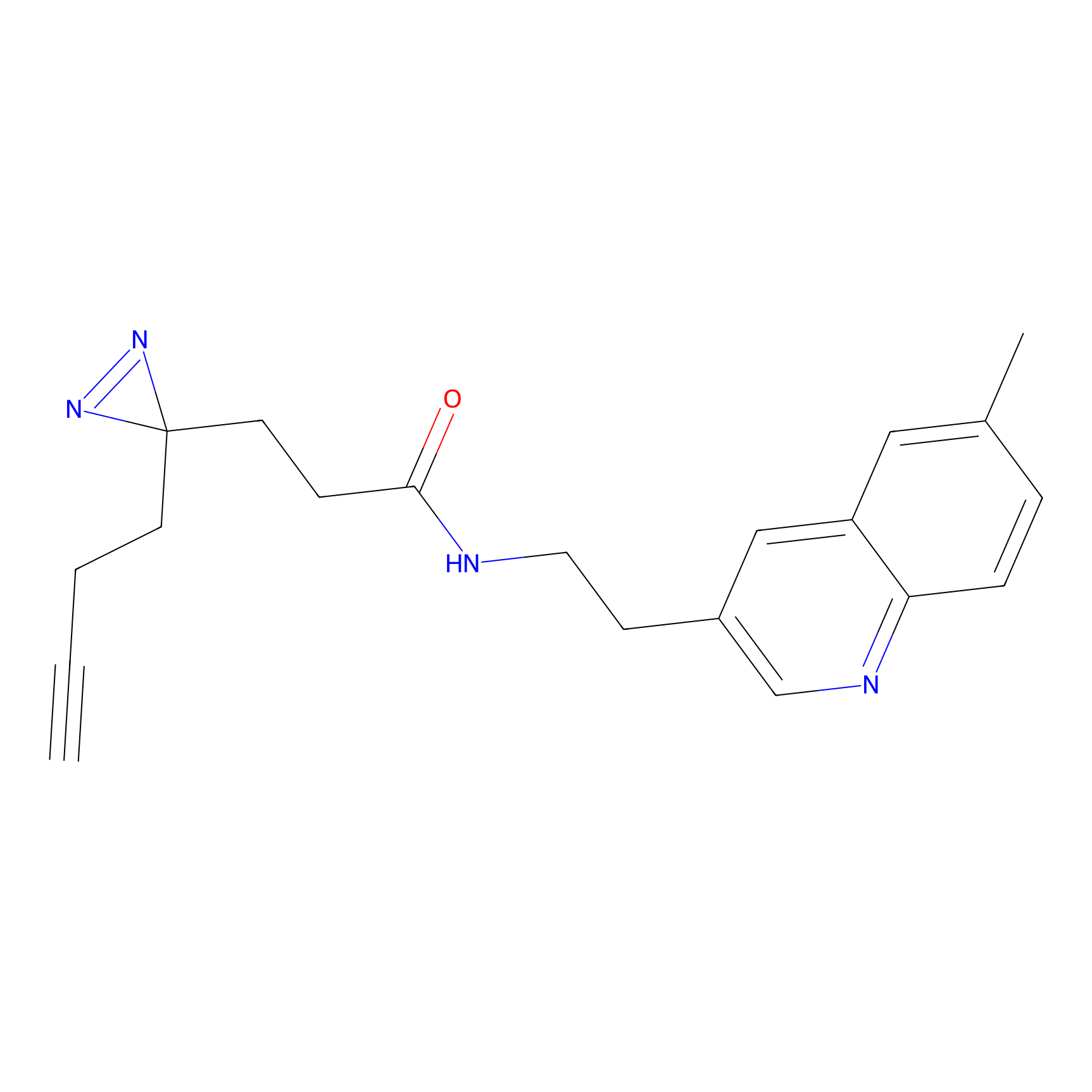
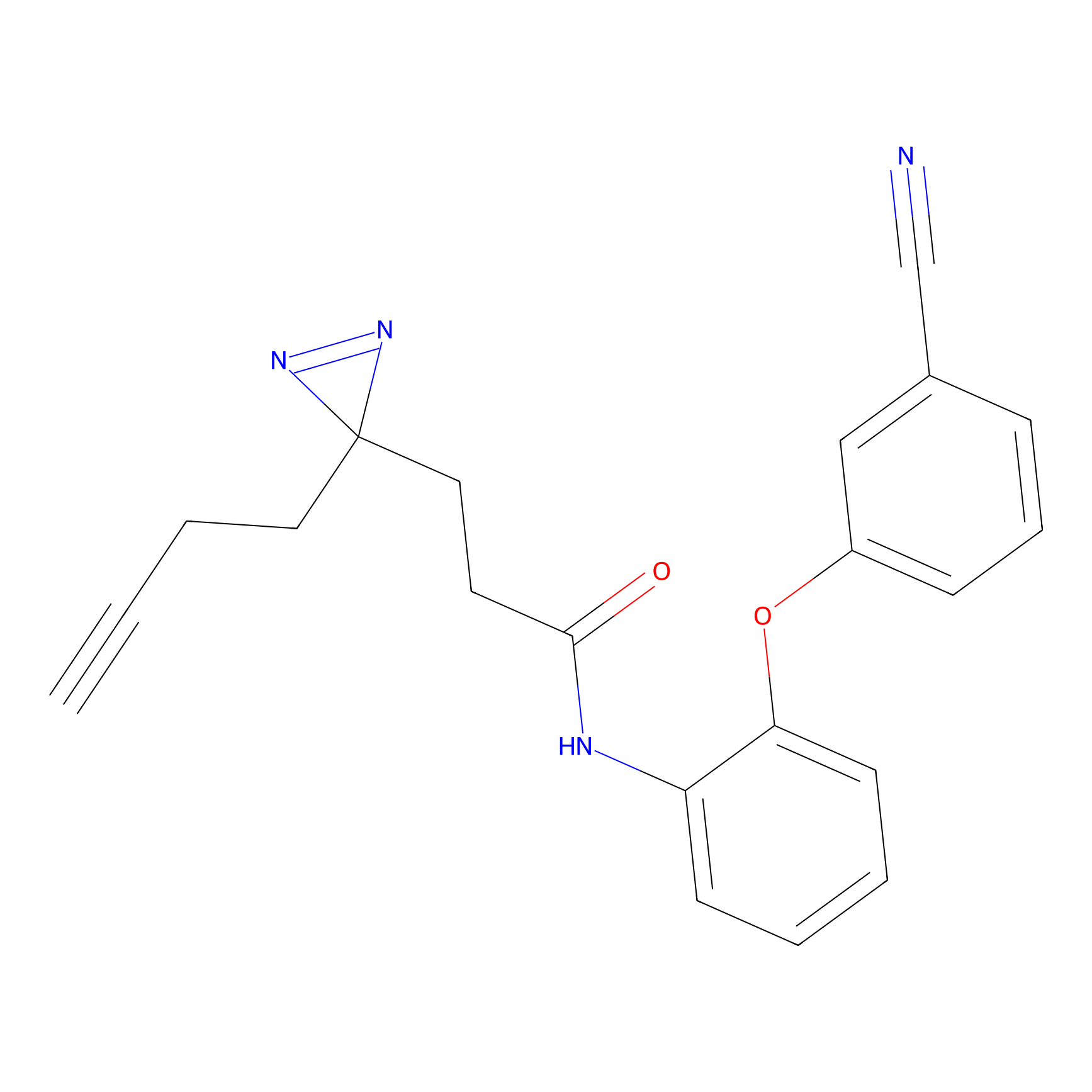
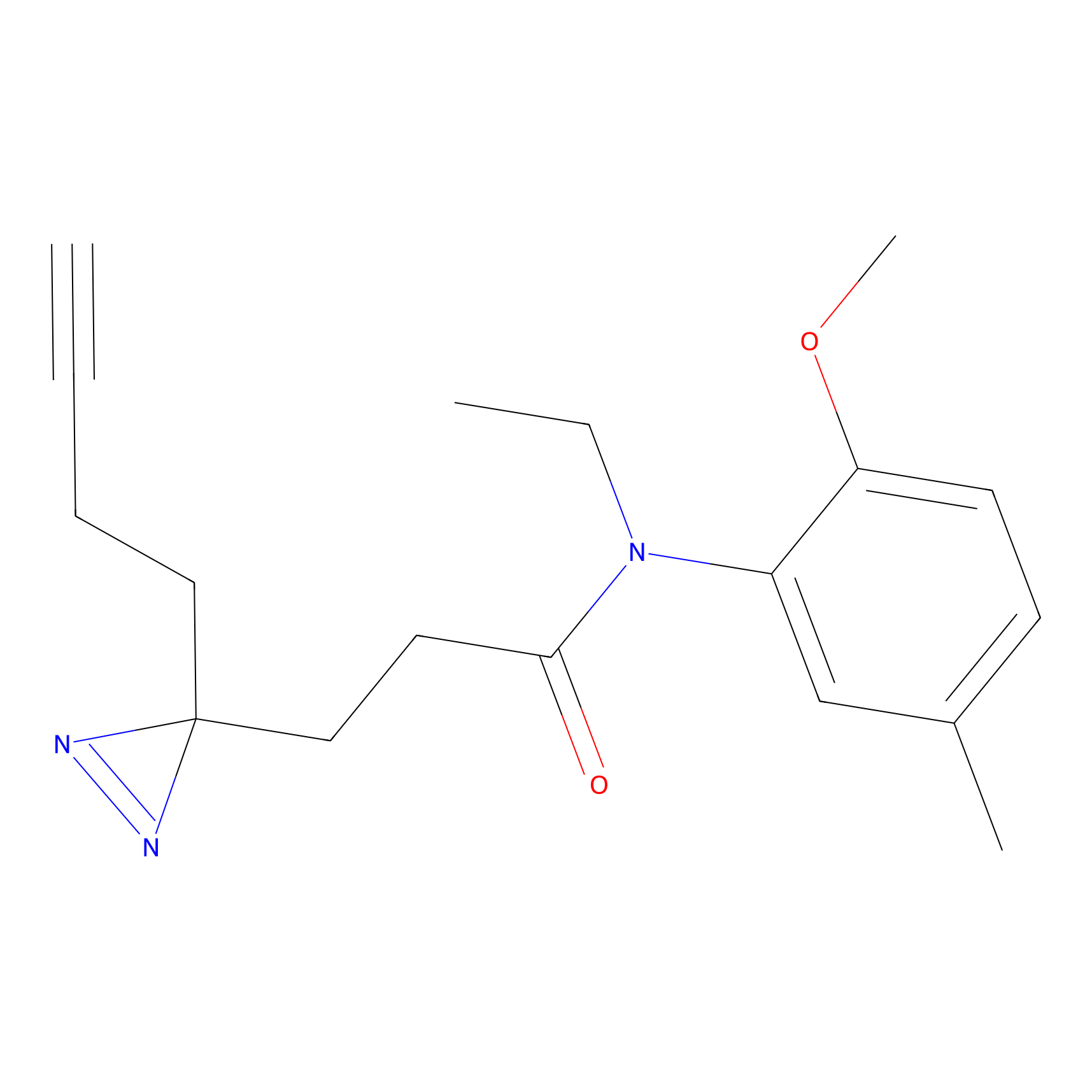
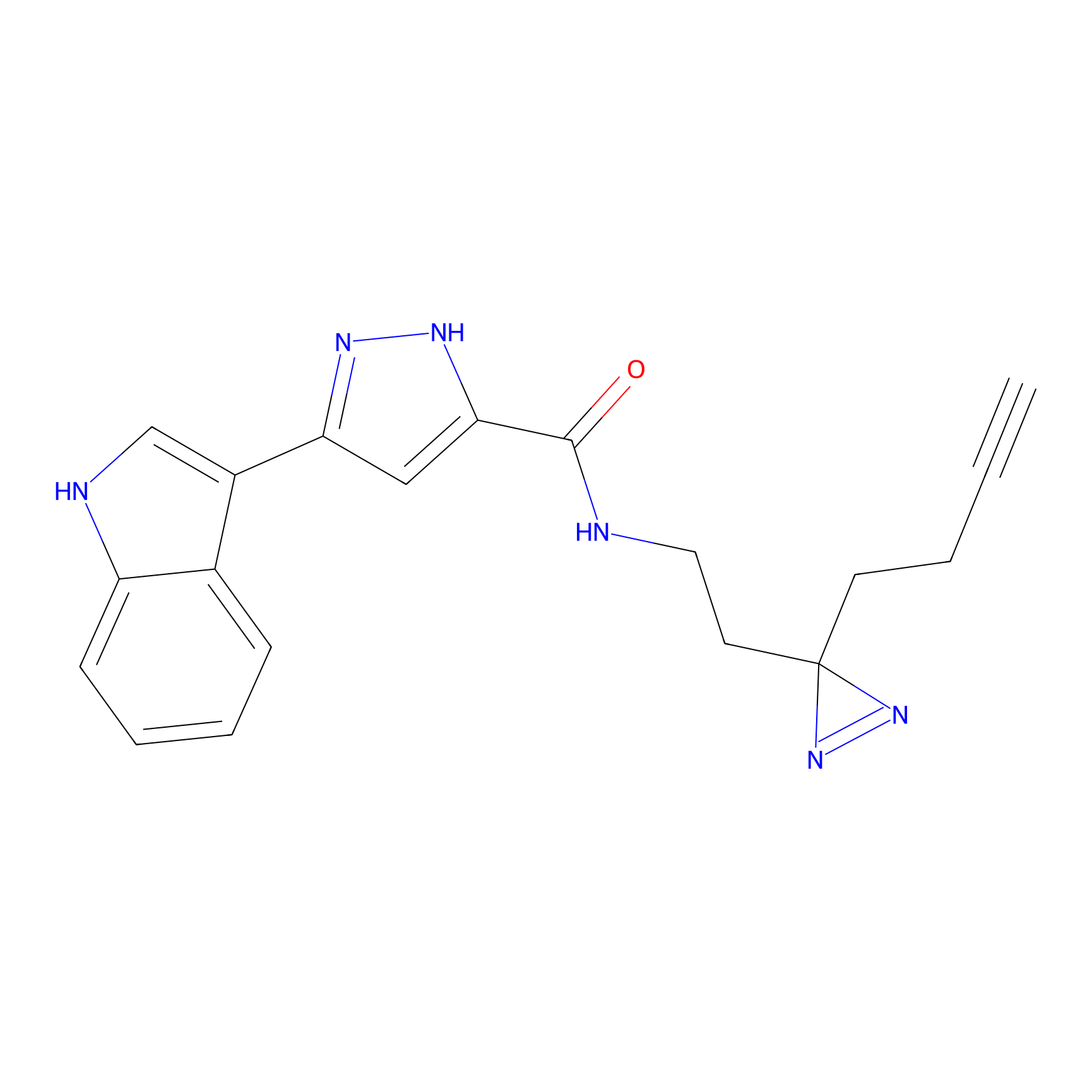
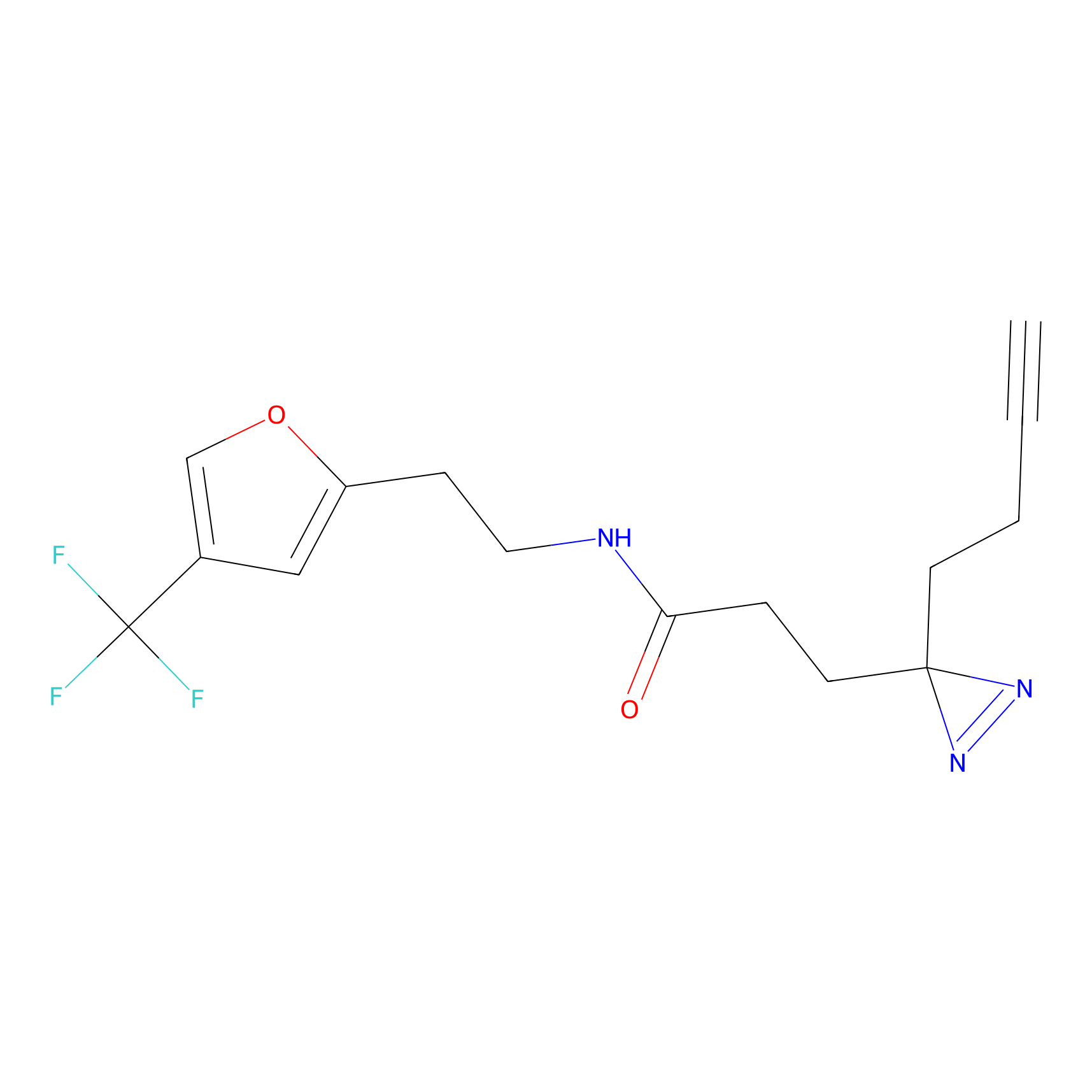
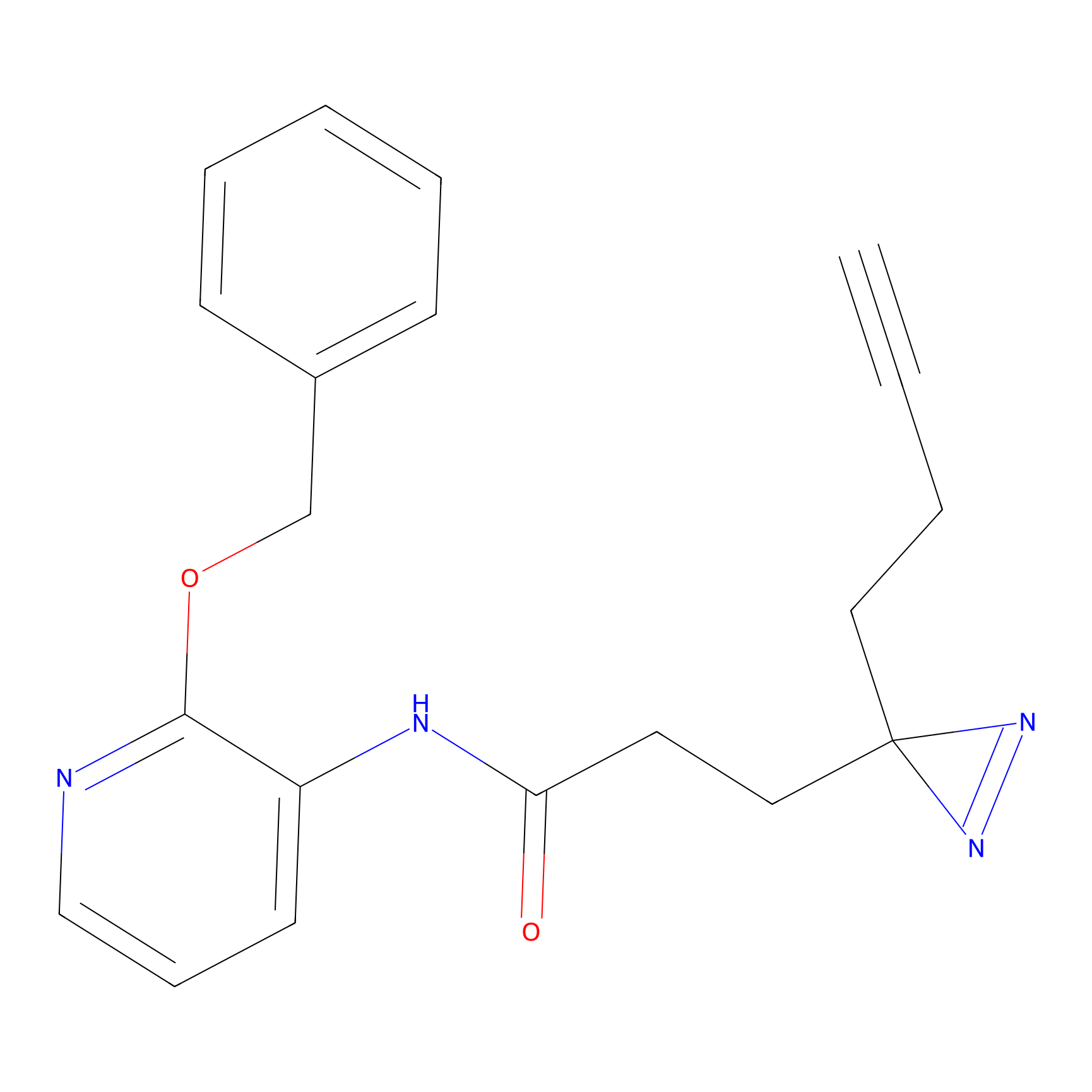
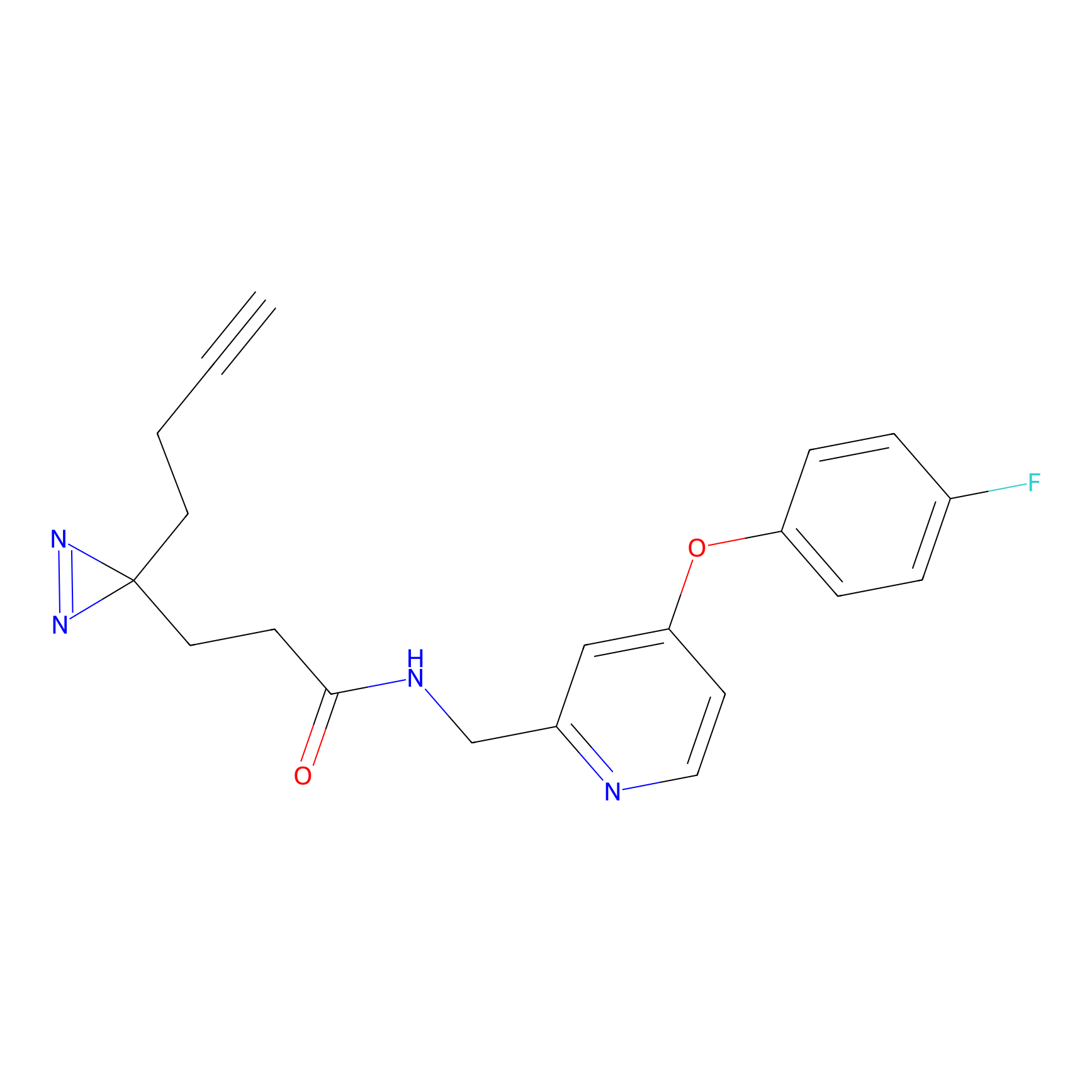
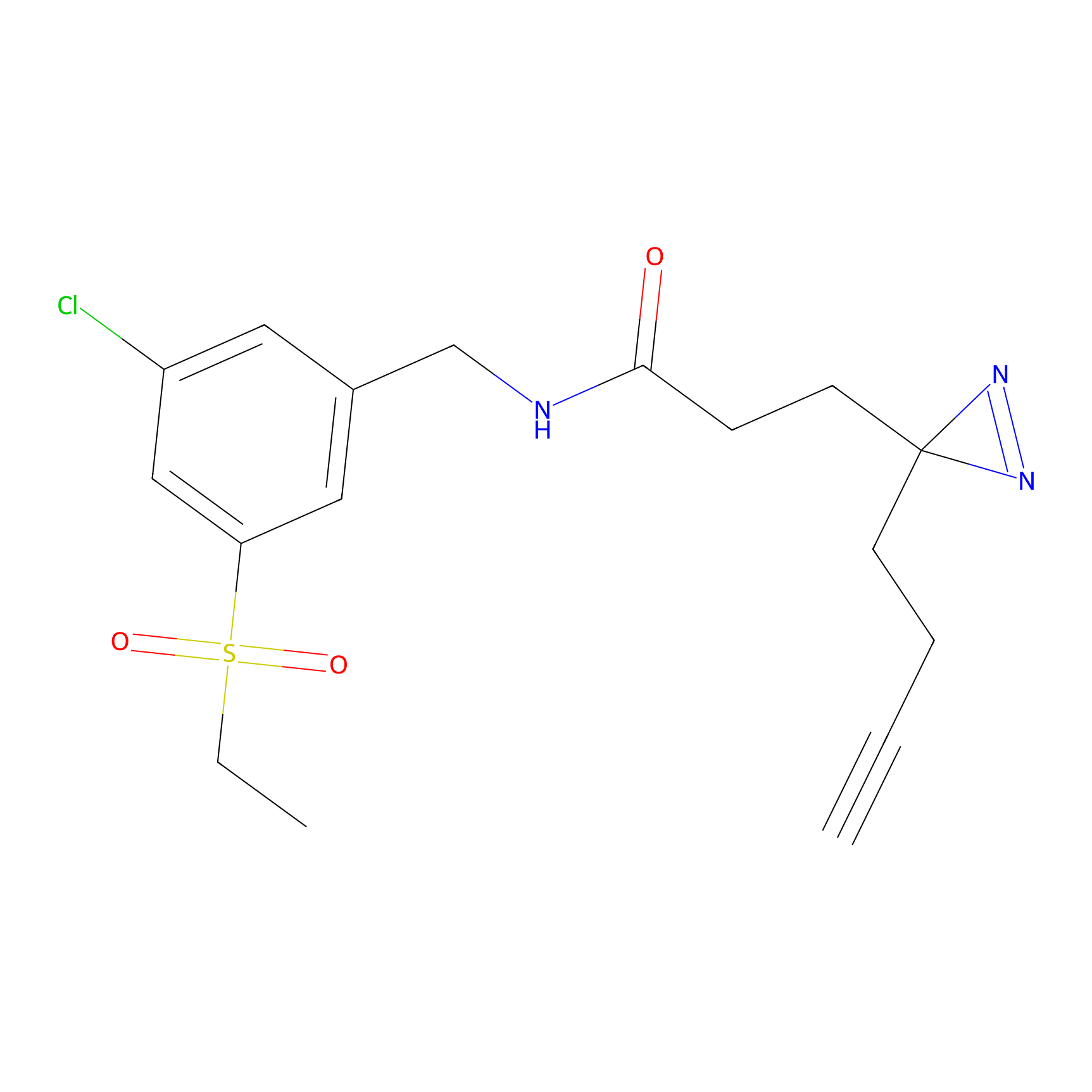
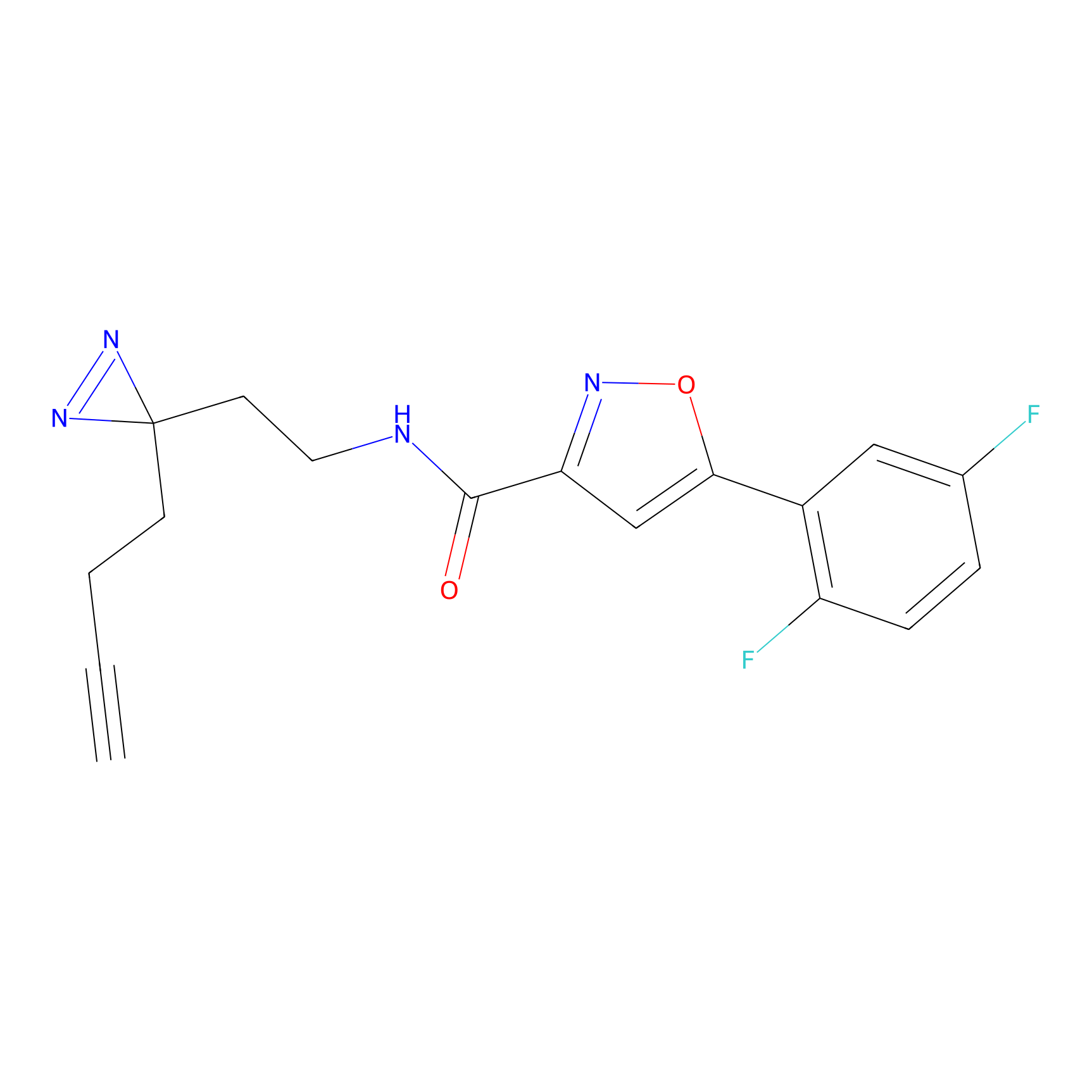
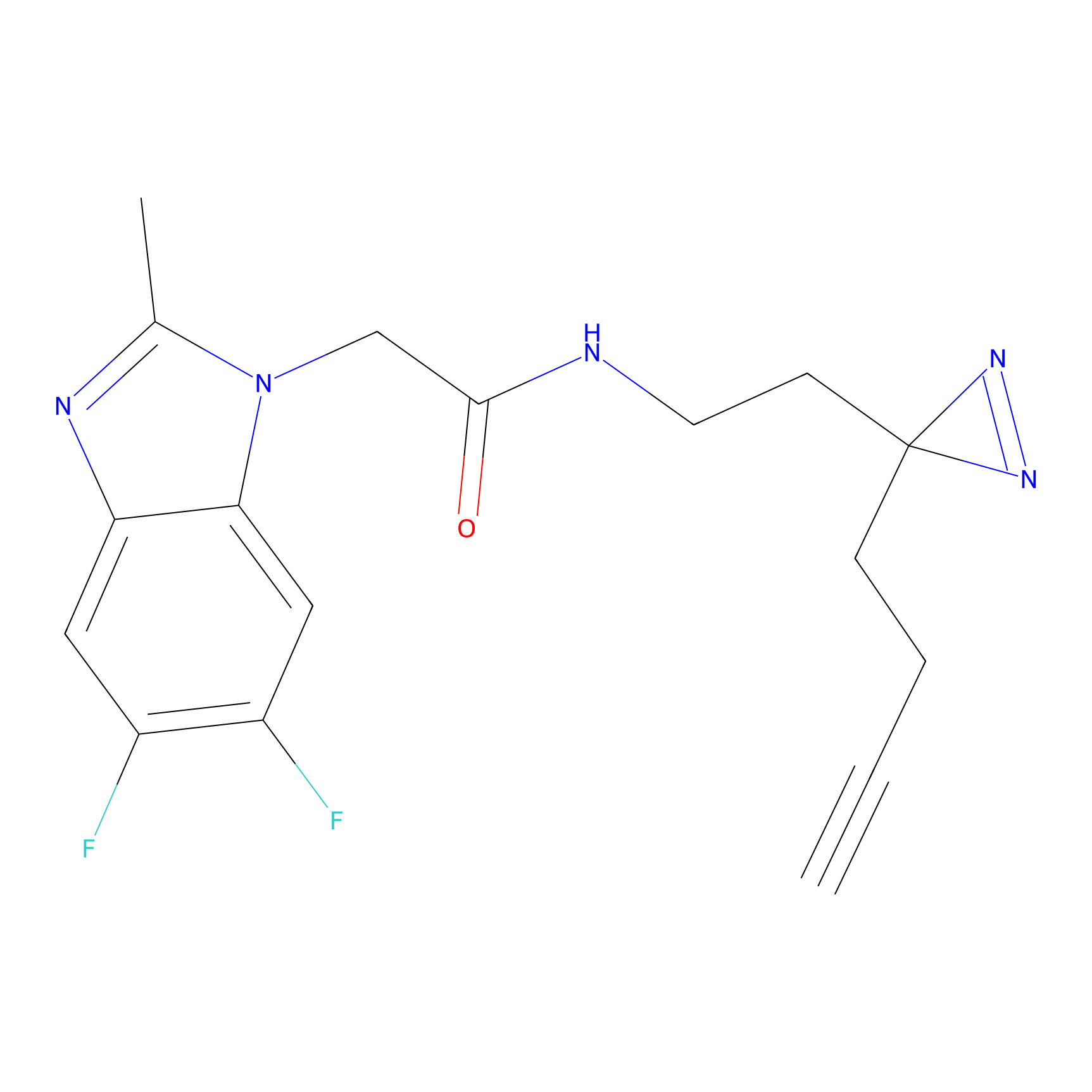
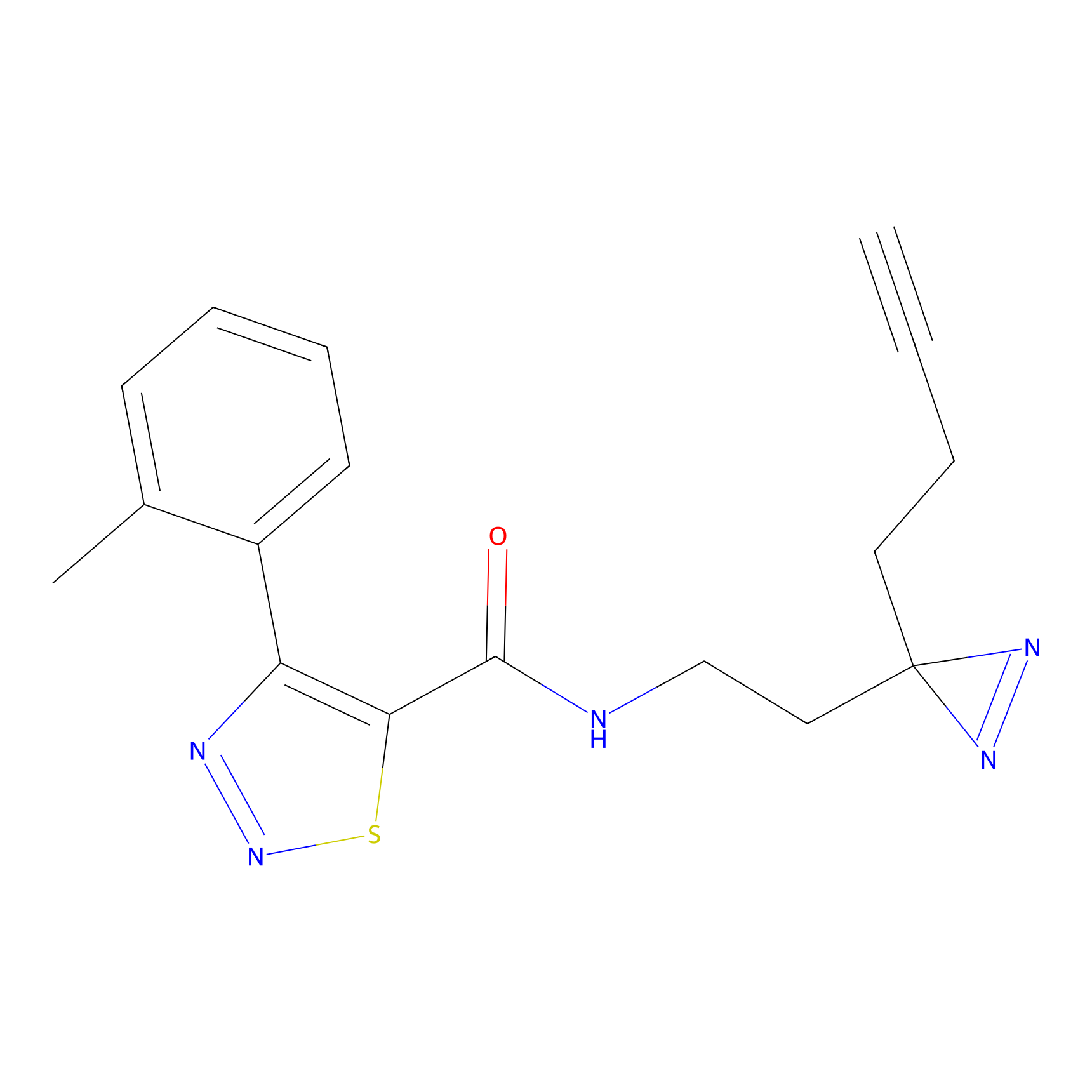
m-APA Probe Info |
 |
14.04 | LDD0402 | [1] | |
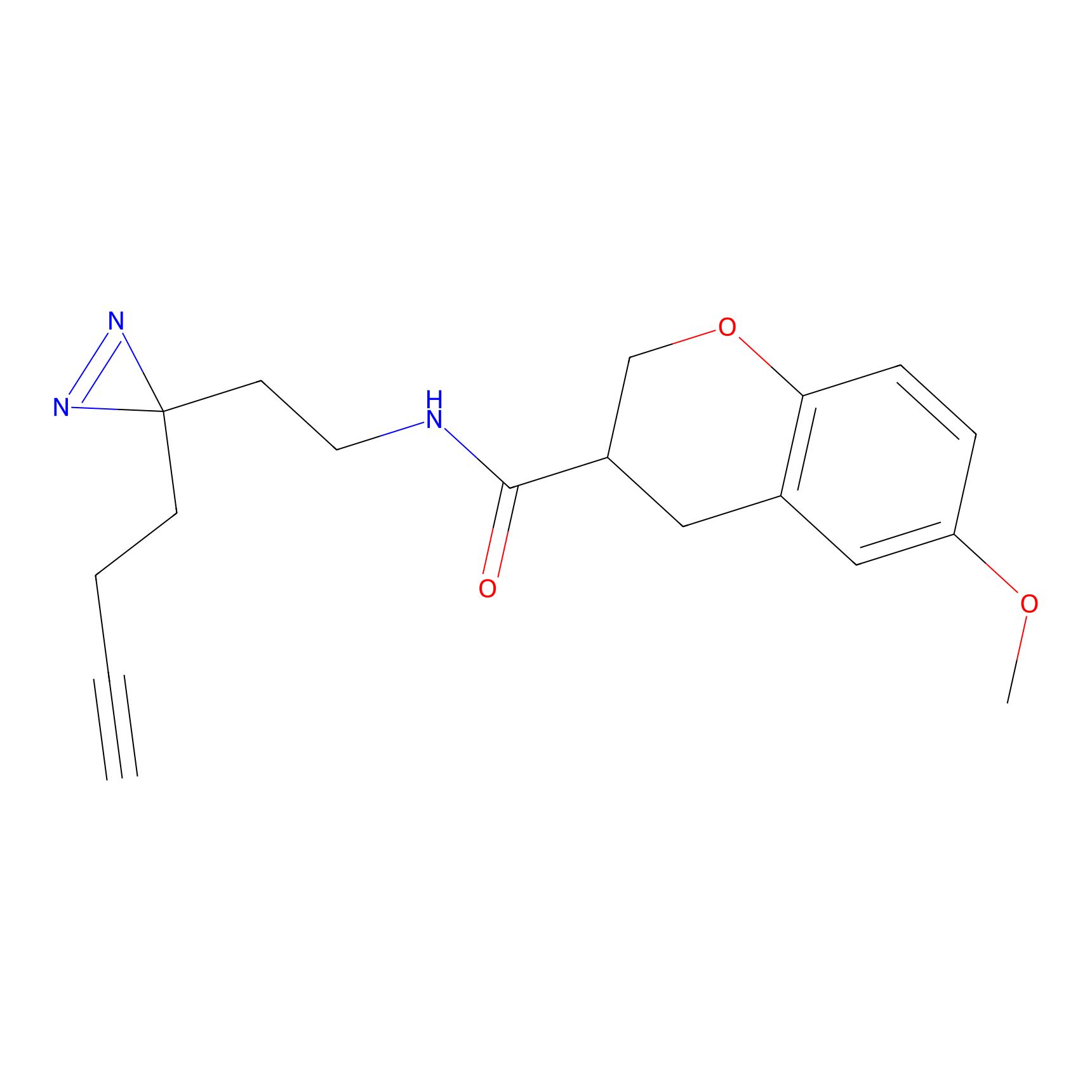
|
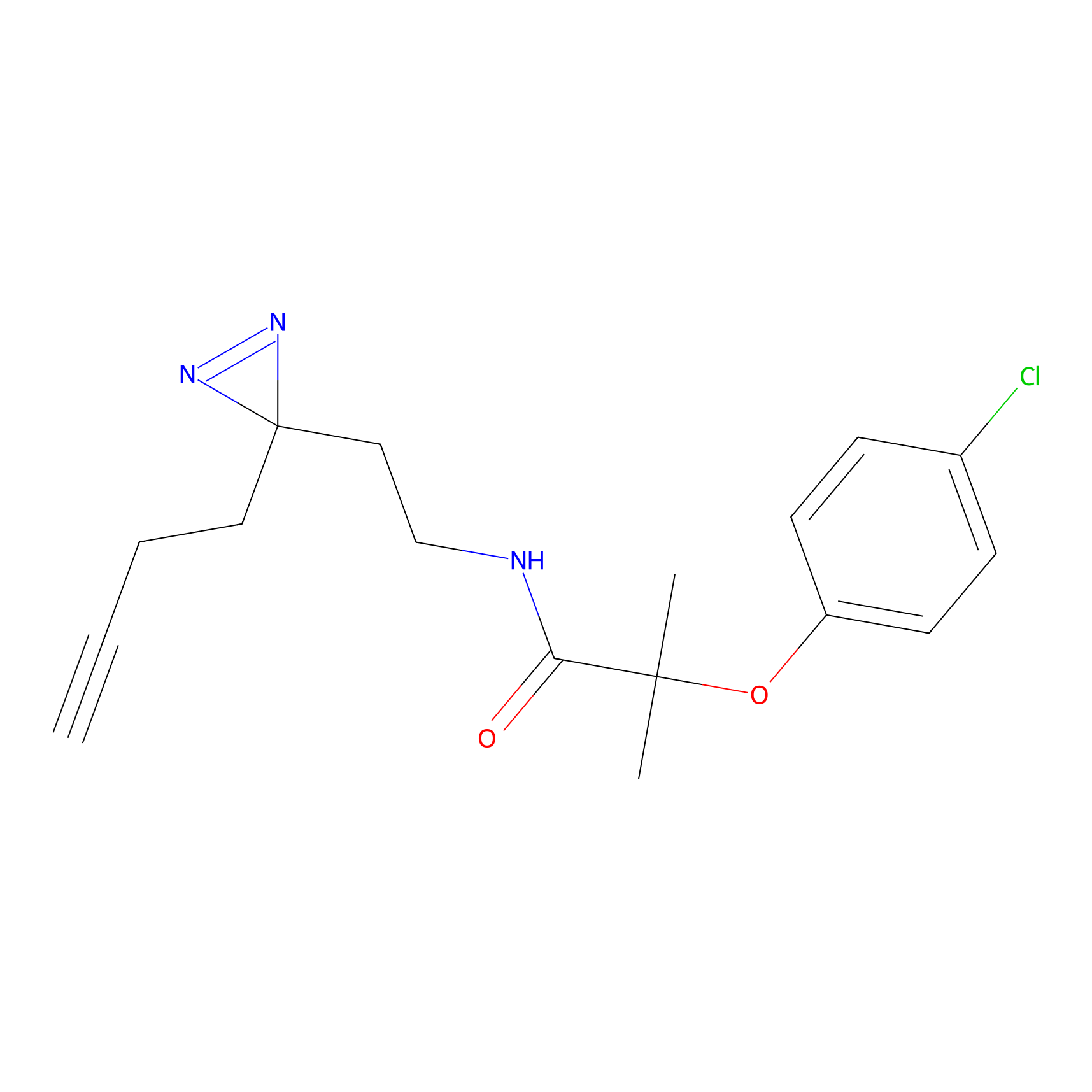
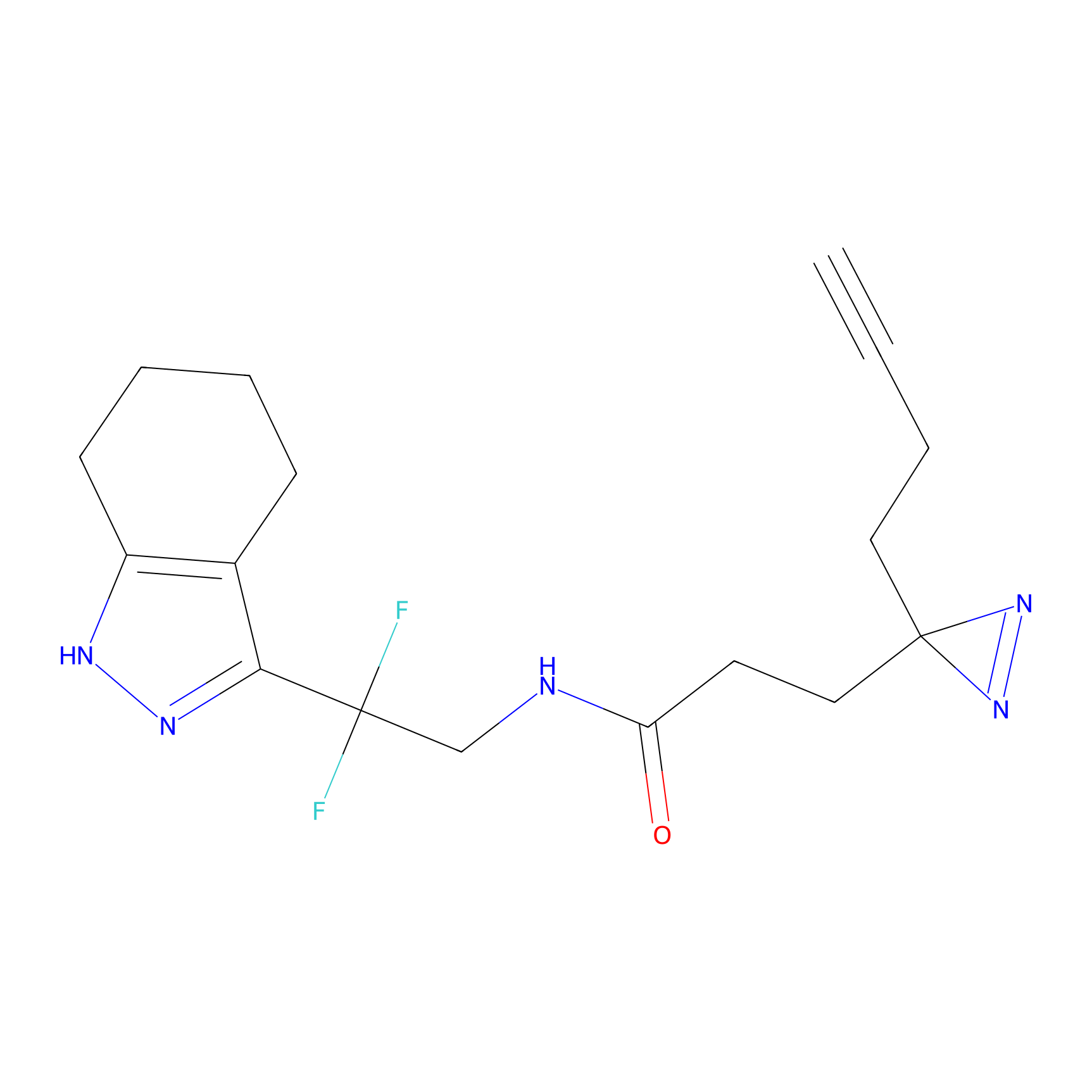
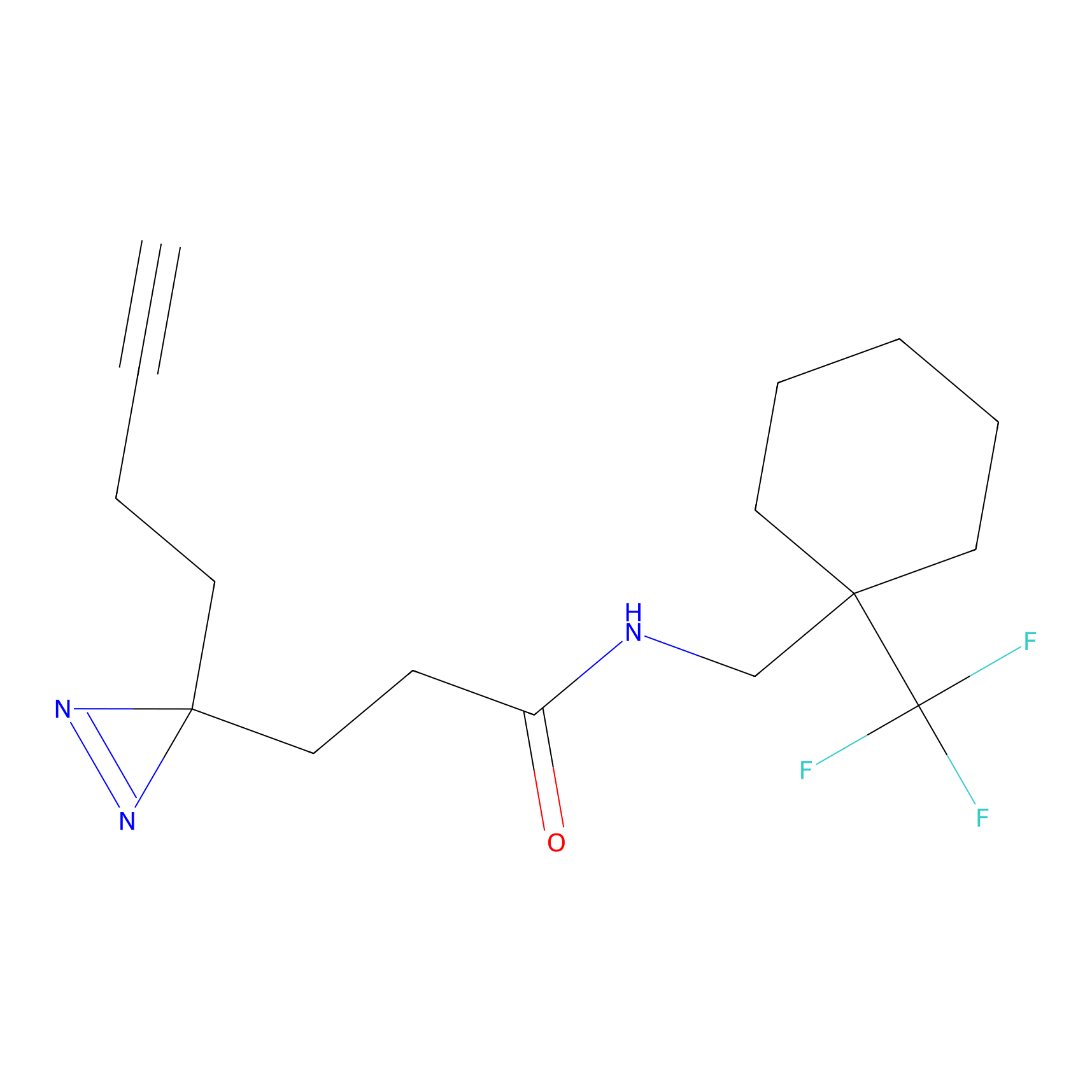
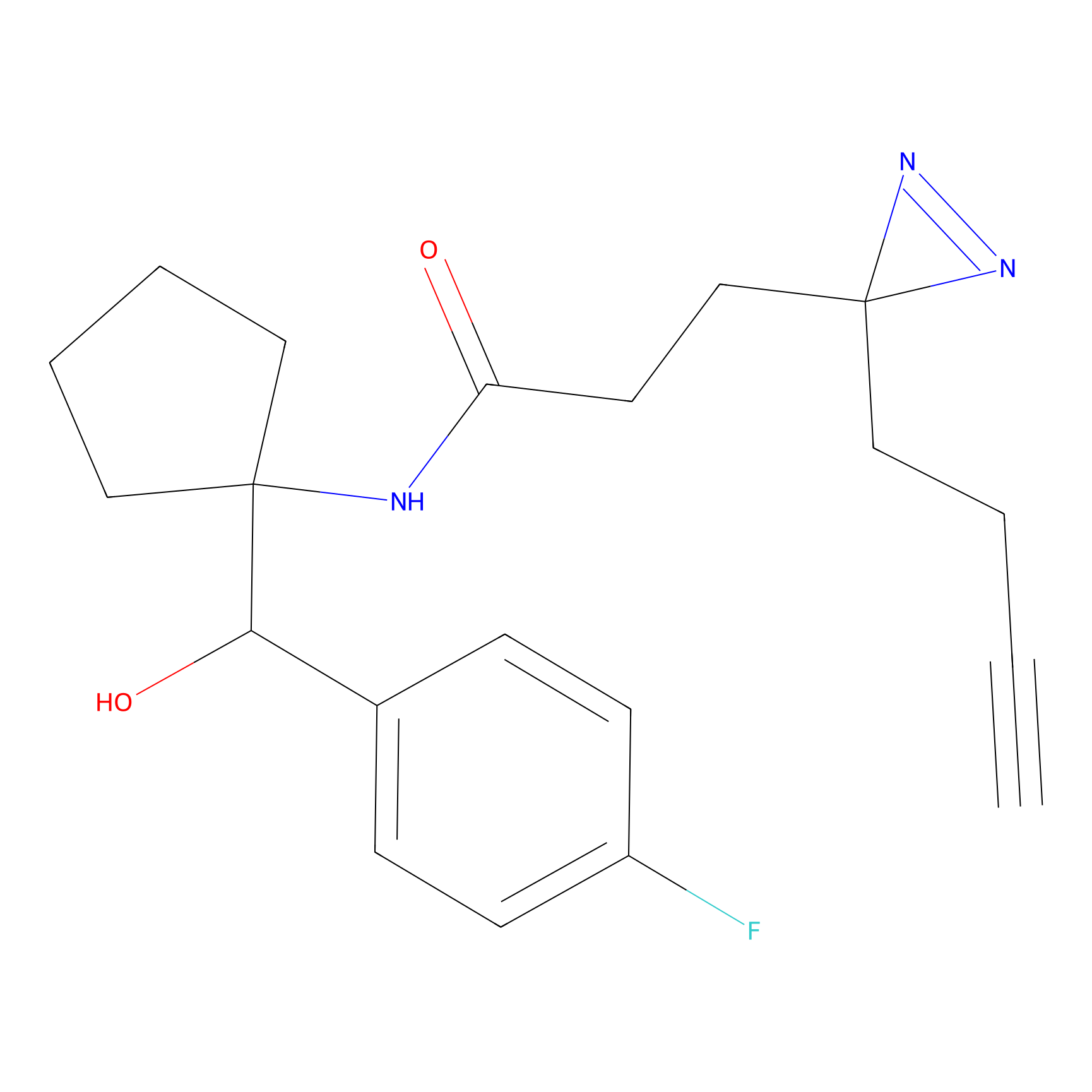
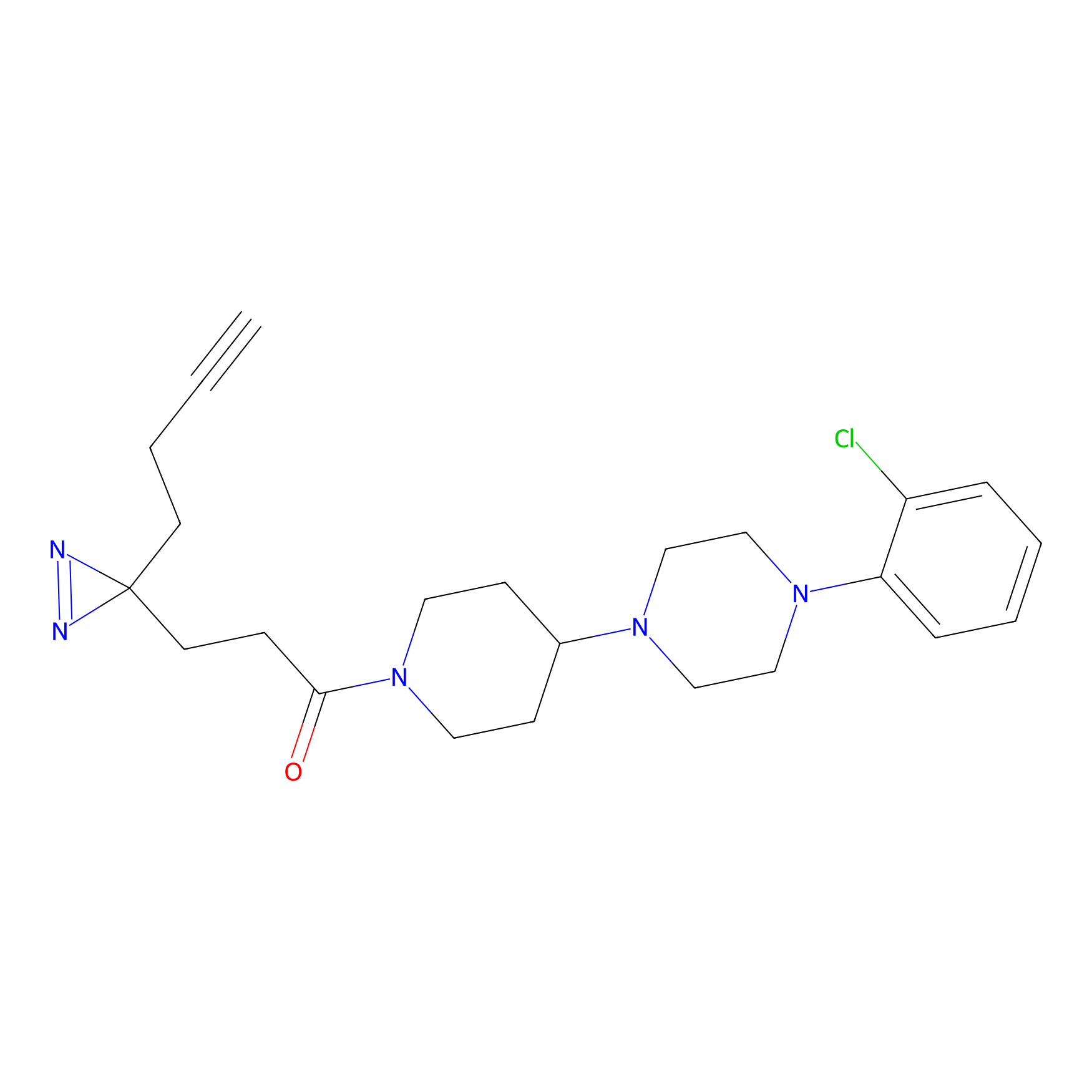
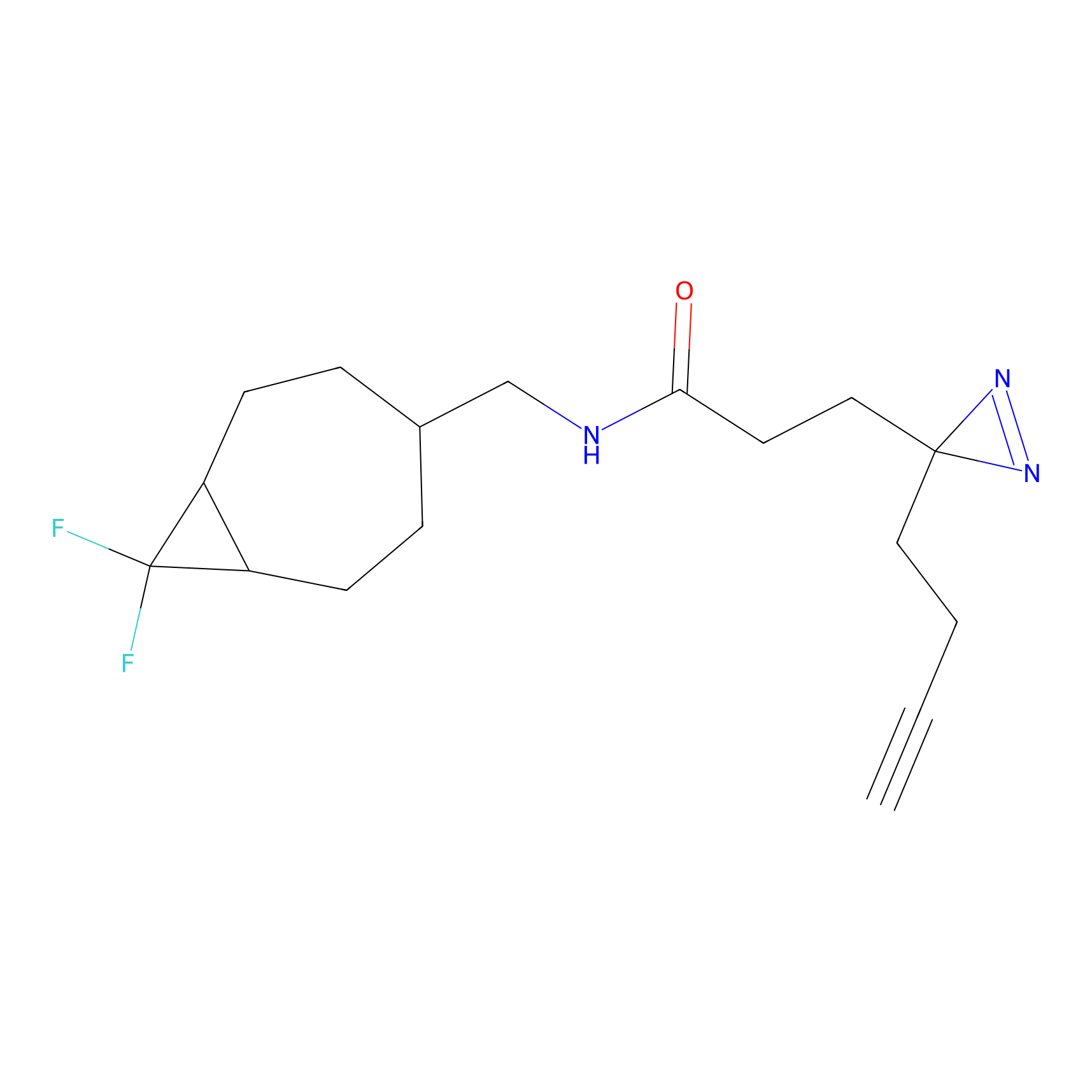
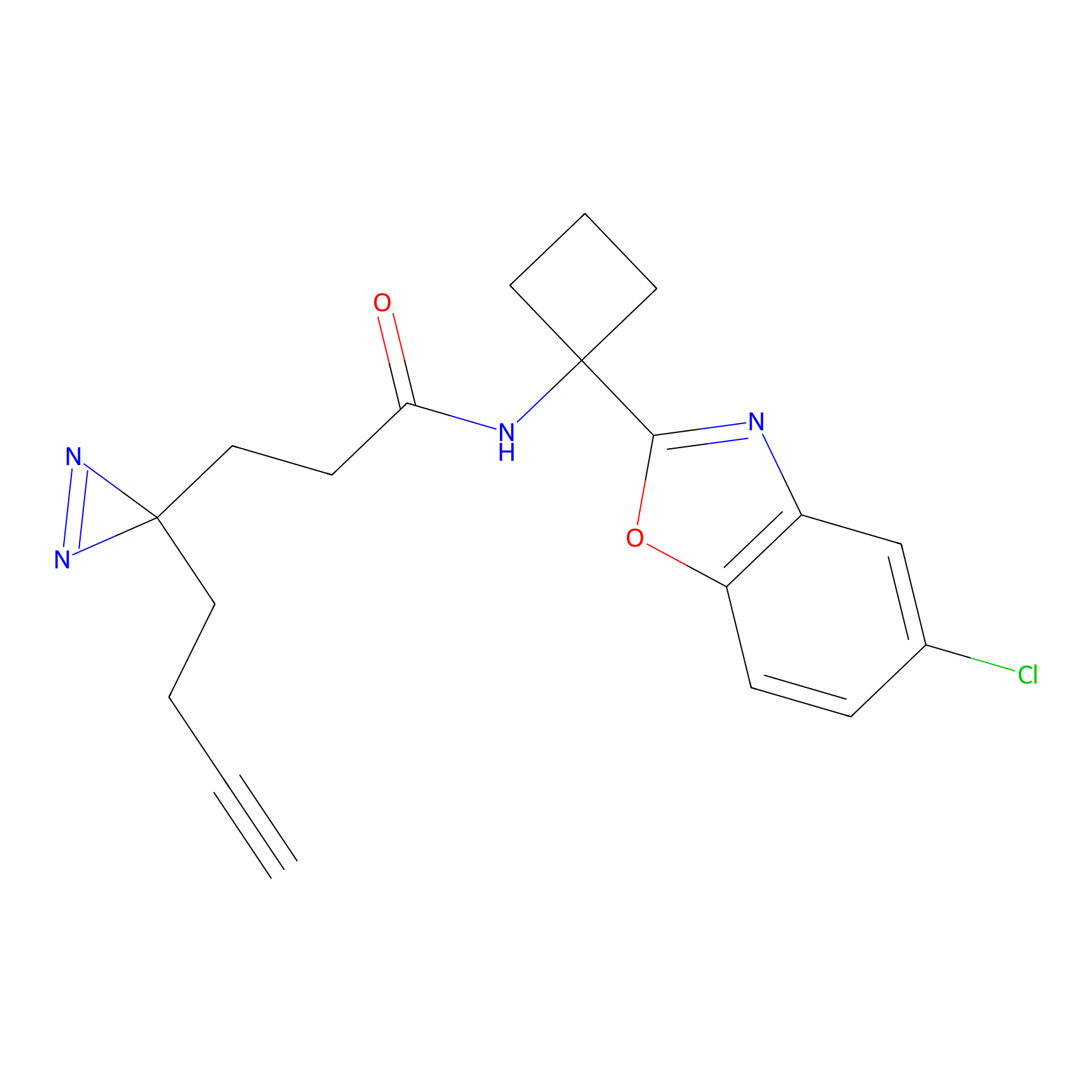
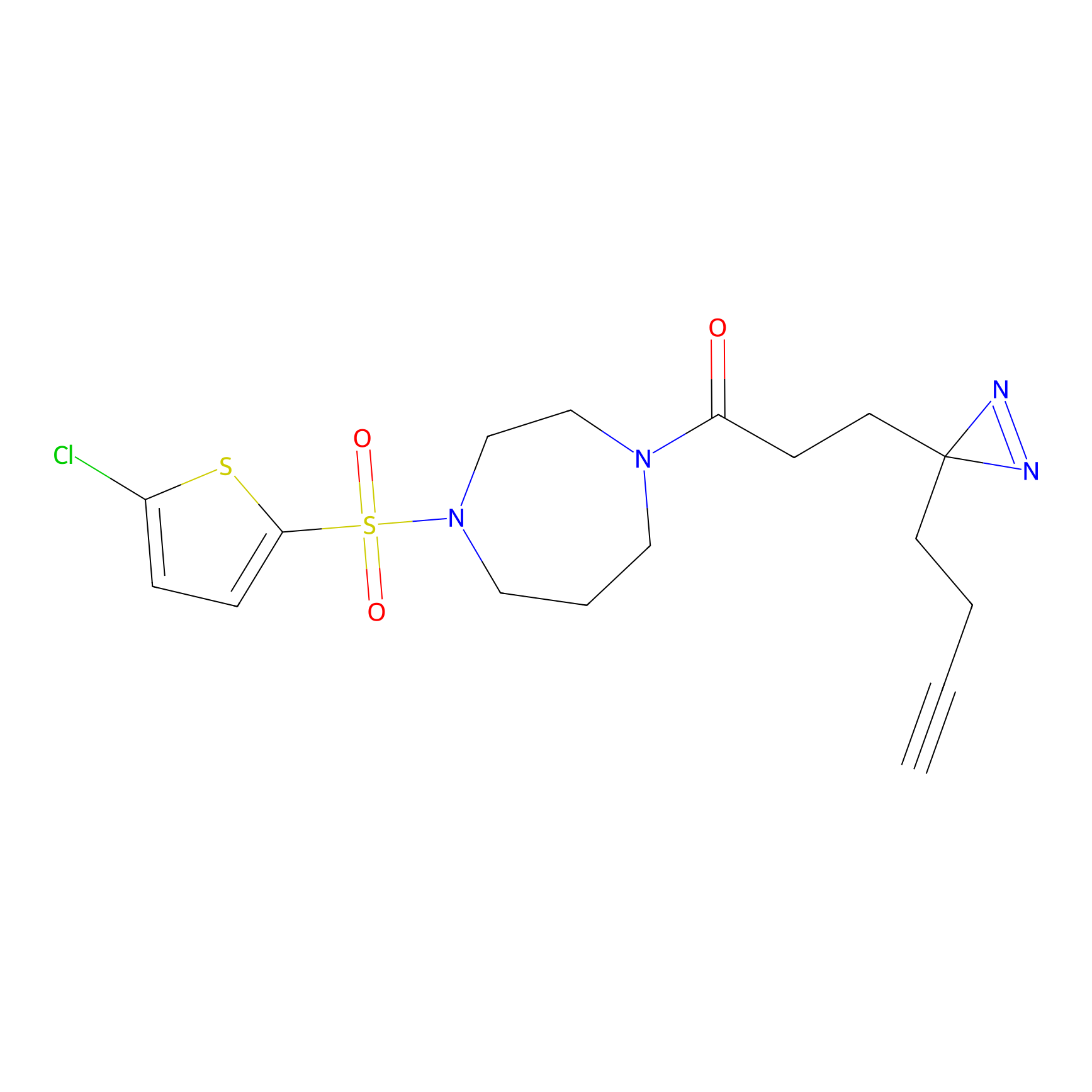
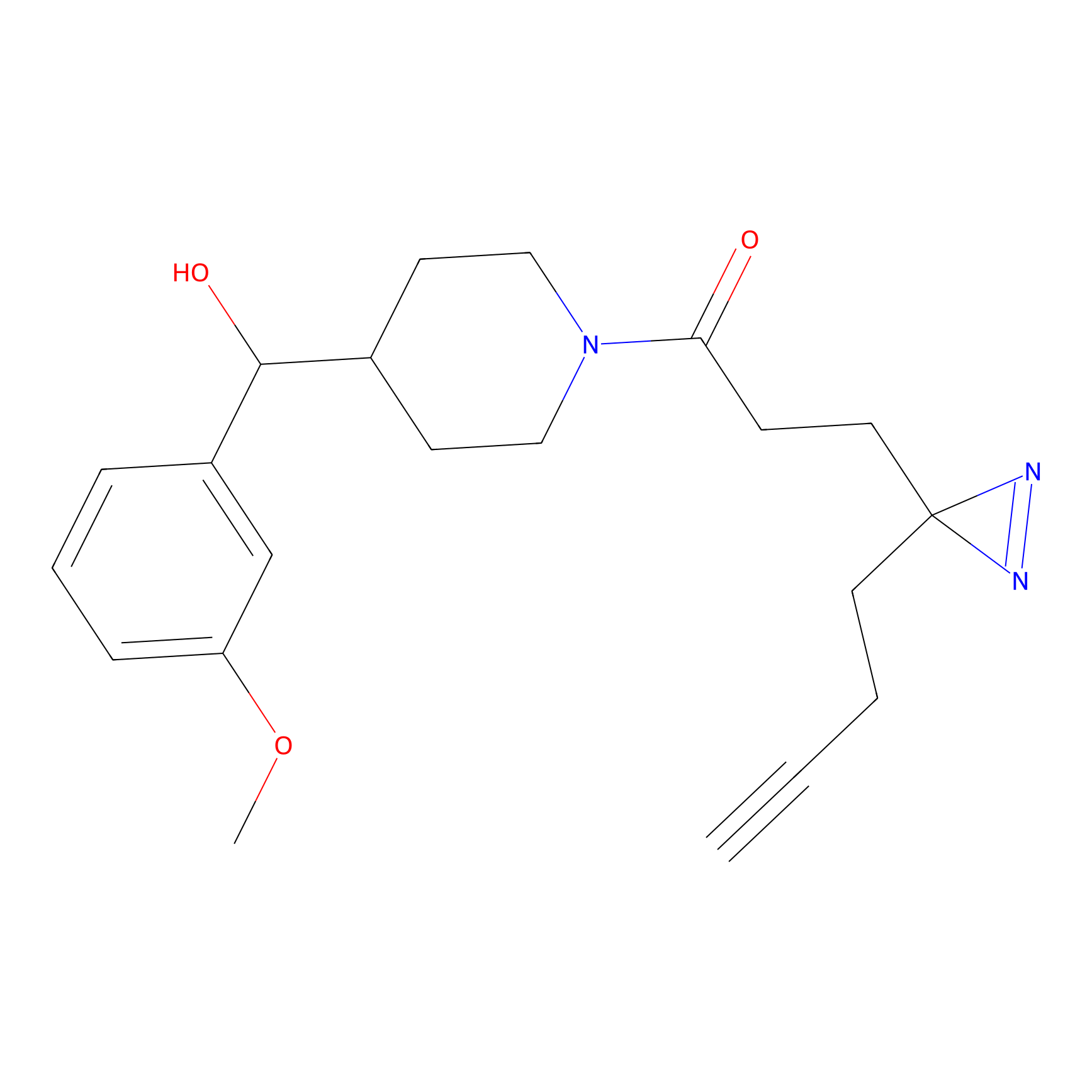
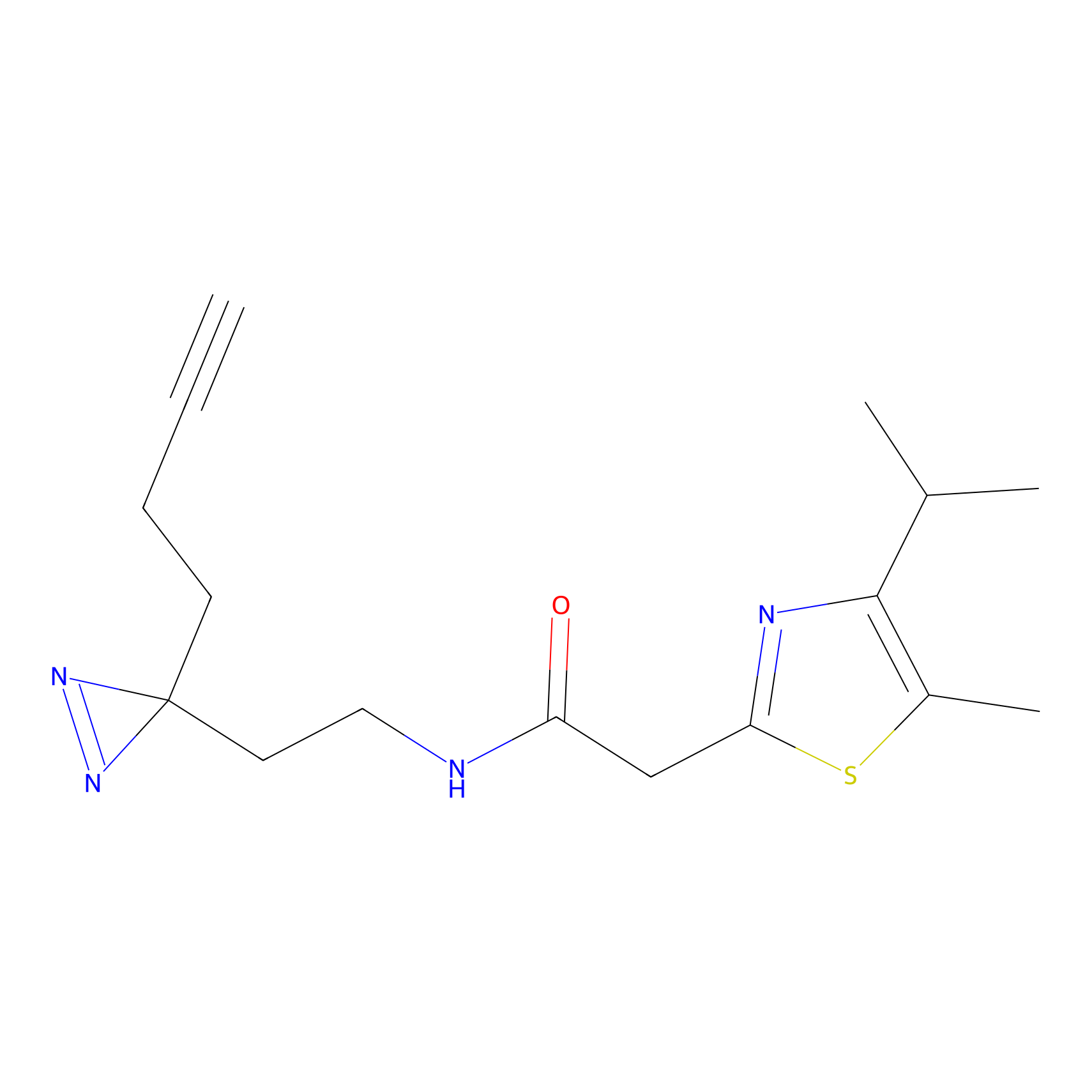
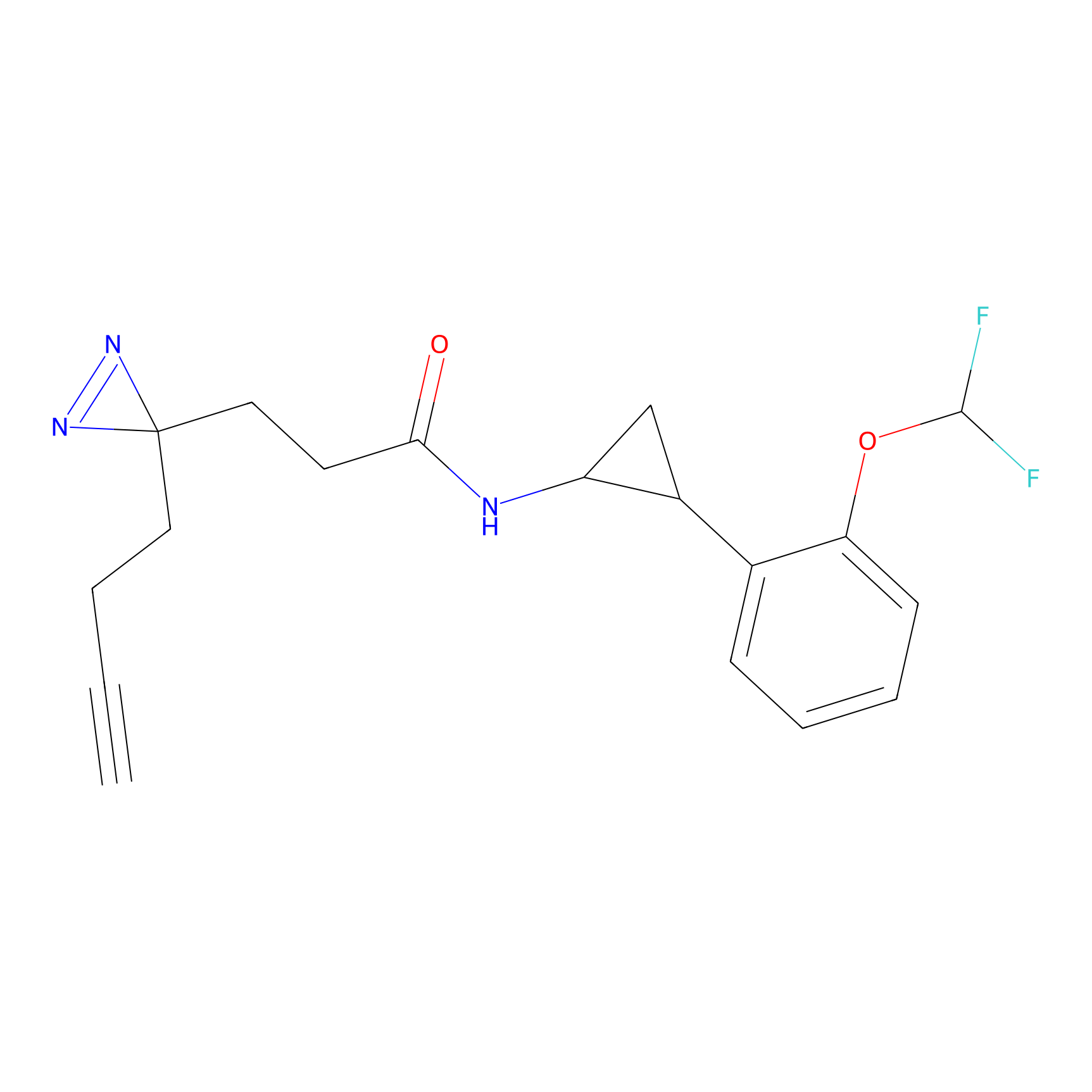
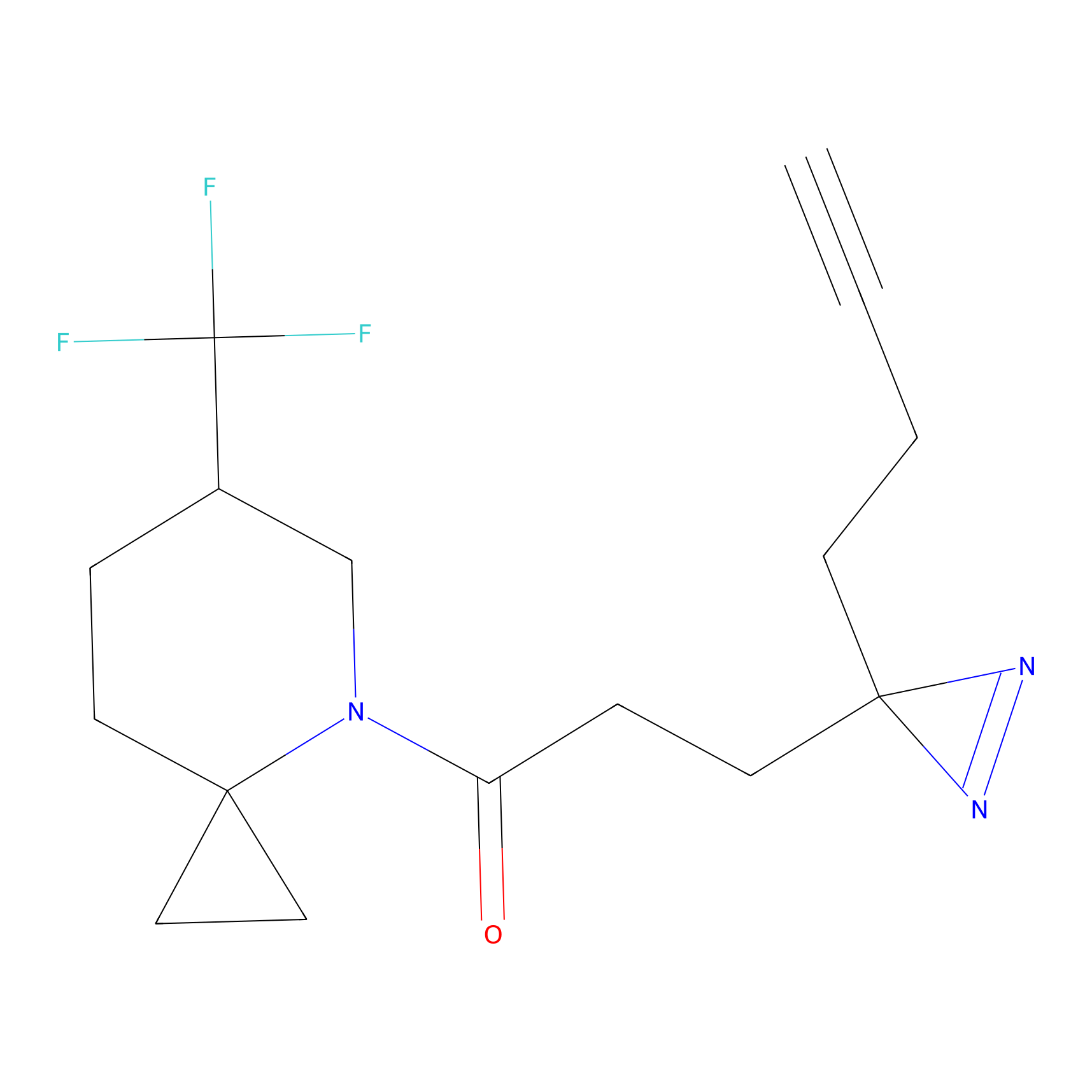
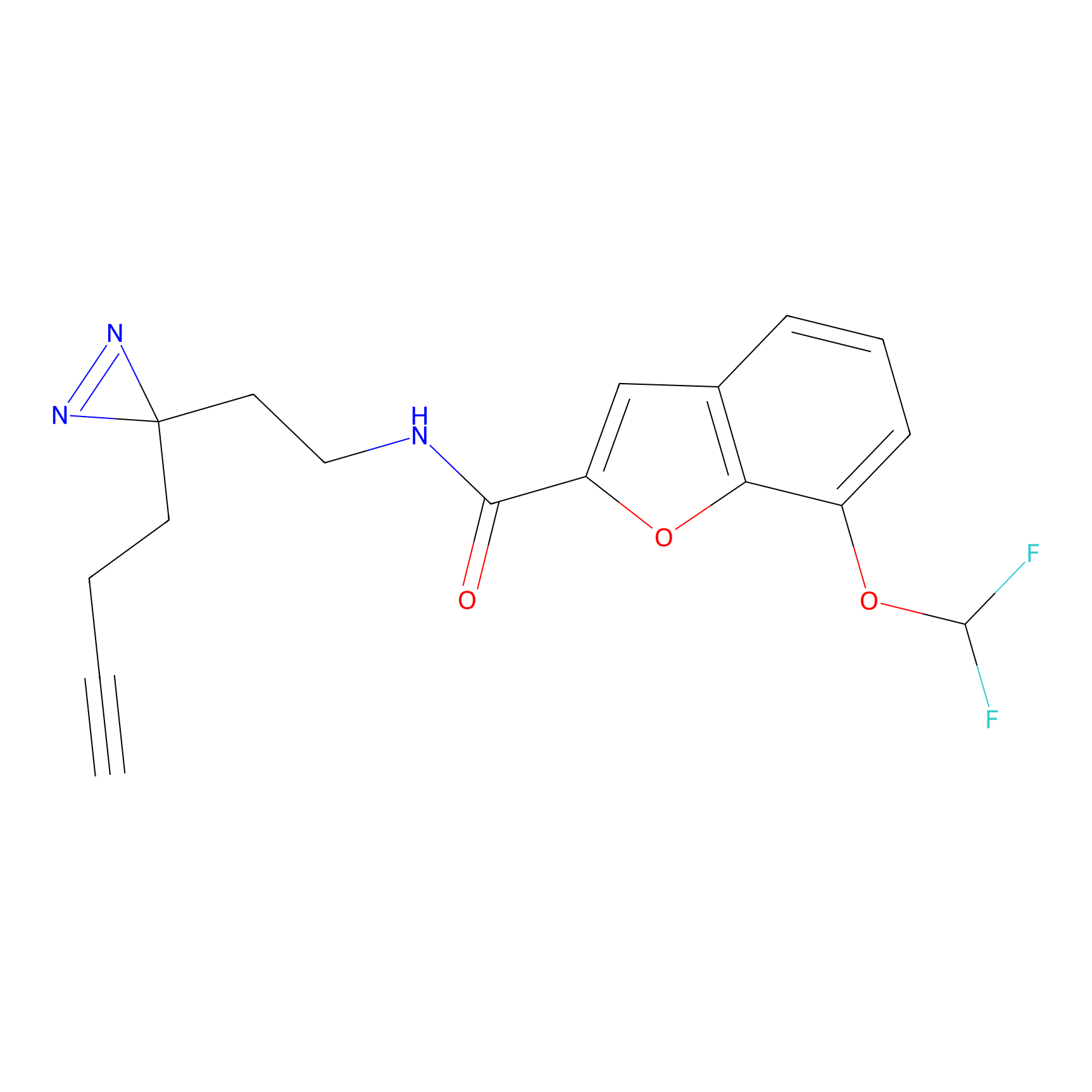
YN-1 Probe Info |
 |
100.00 | LDD0444 | [2] | |
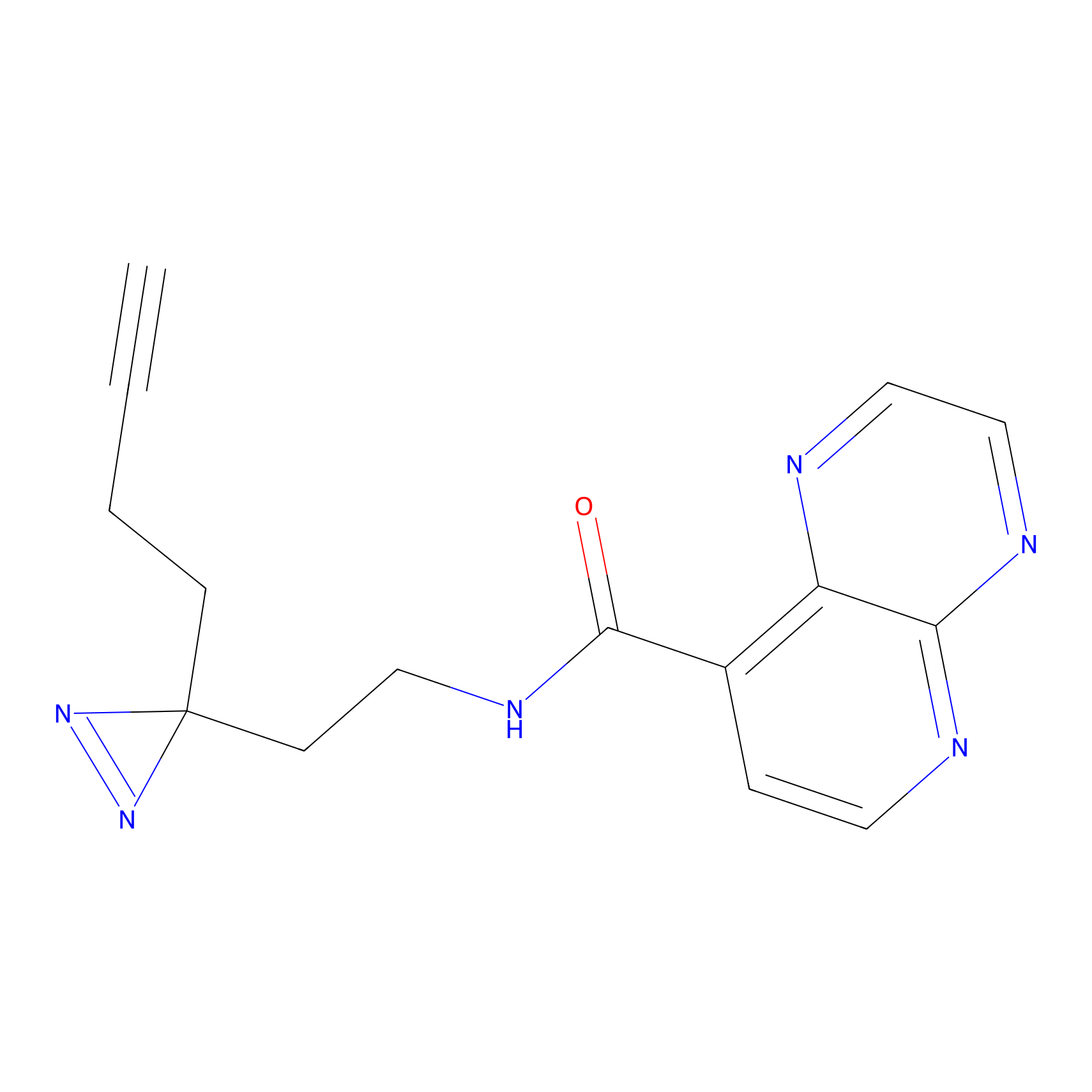
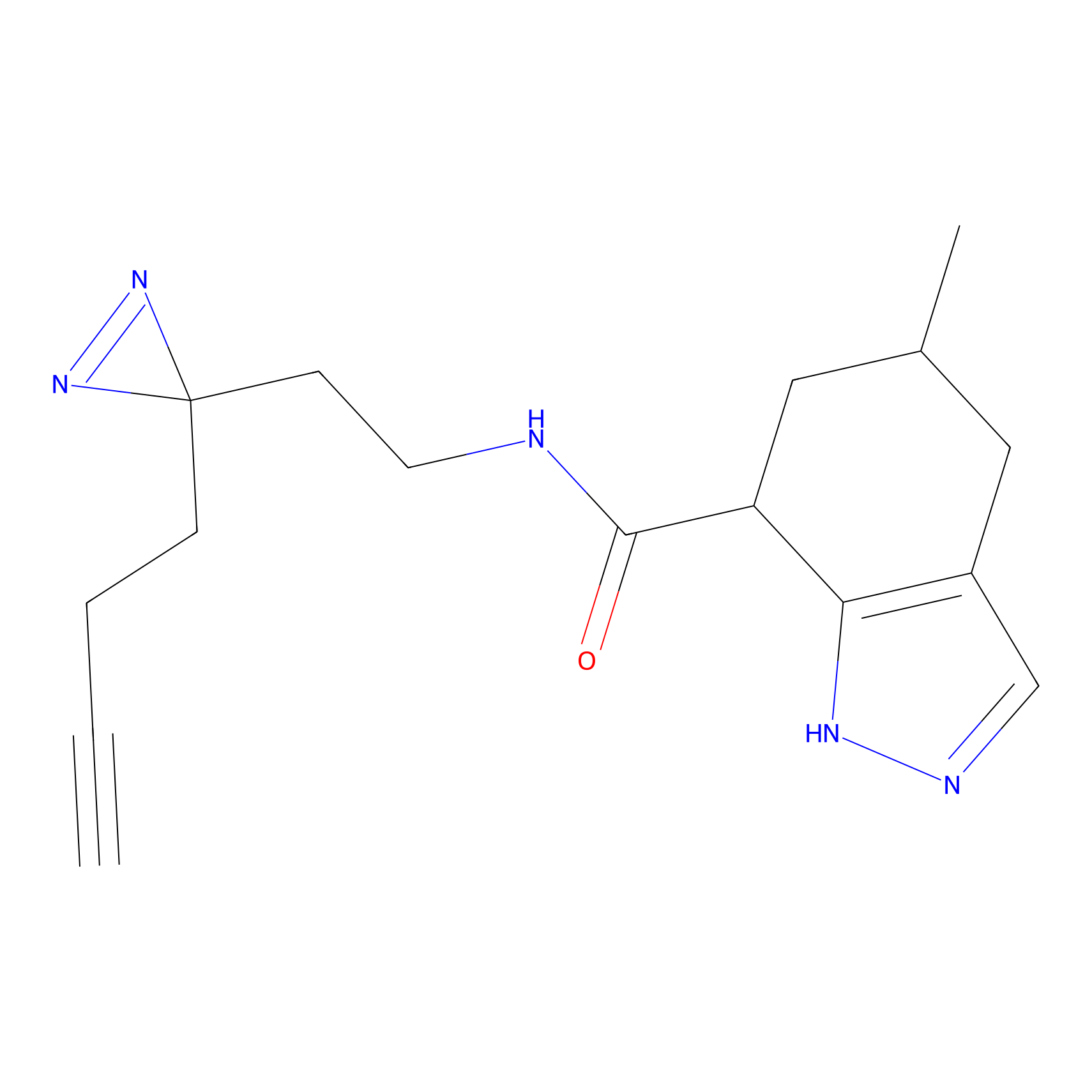
|
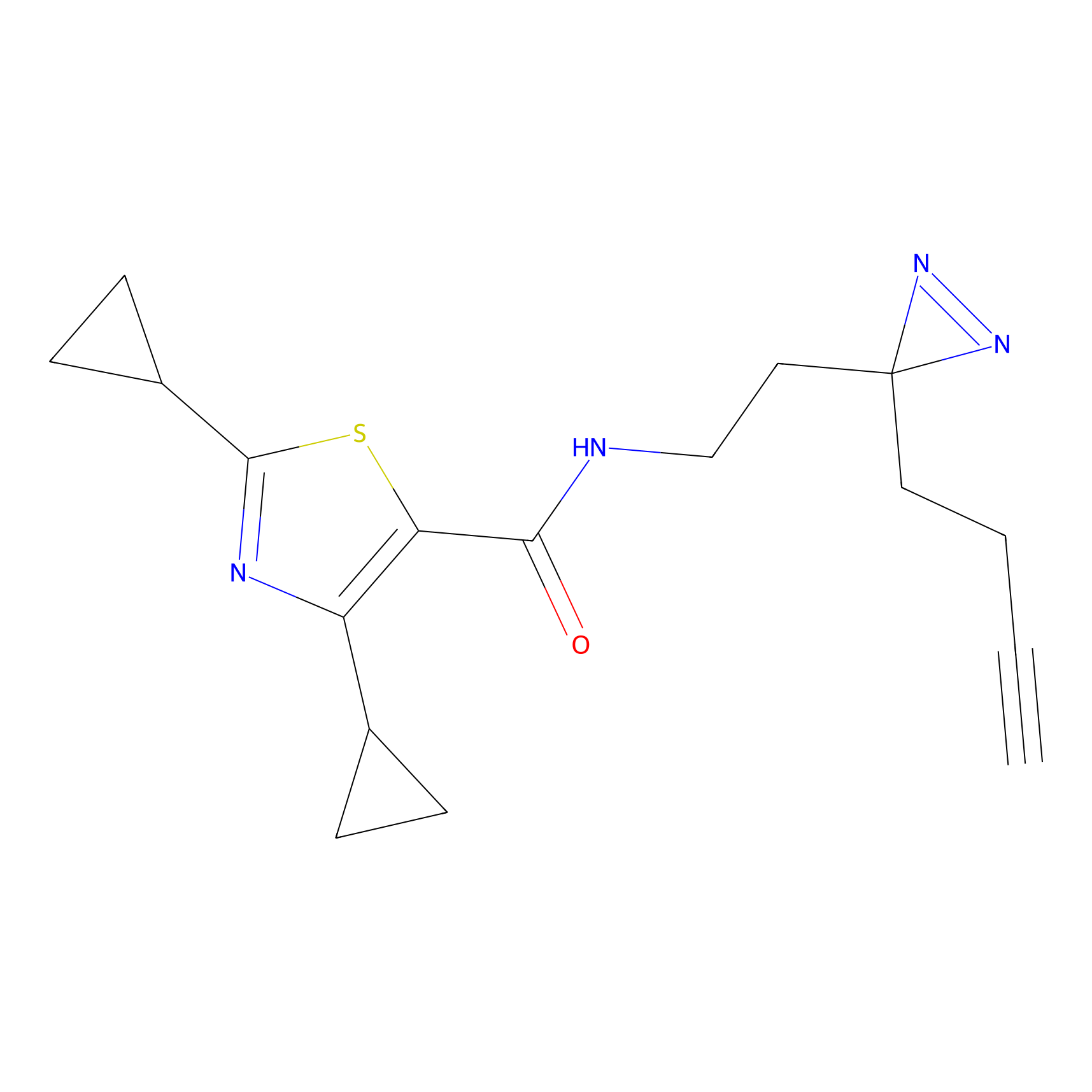
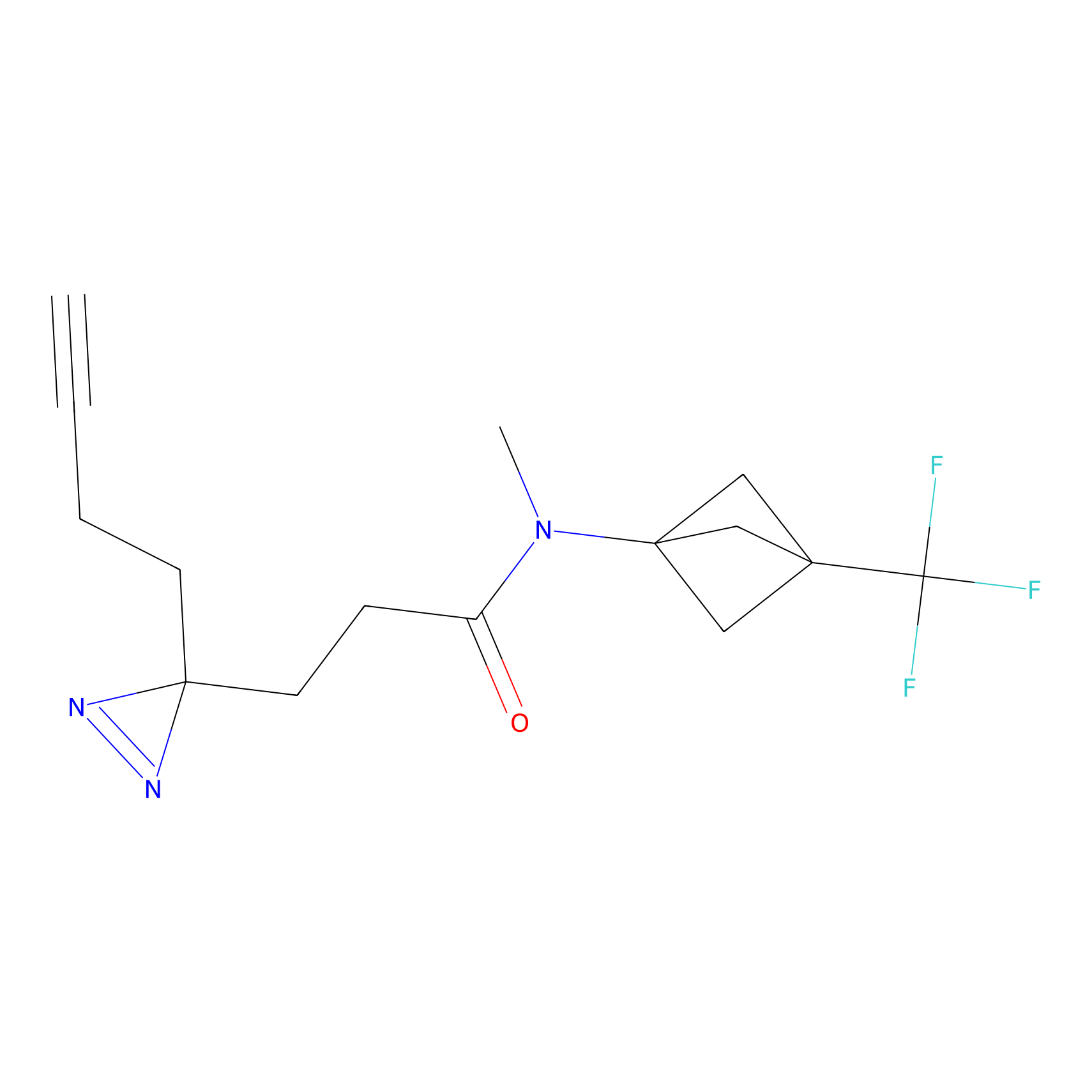
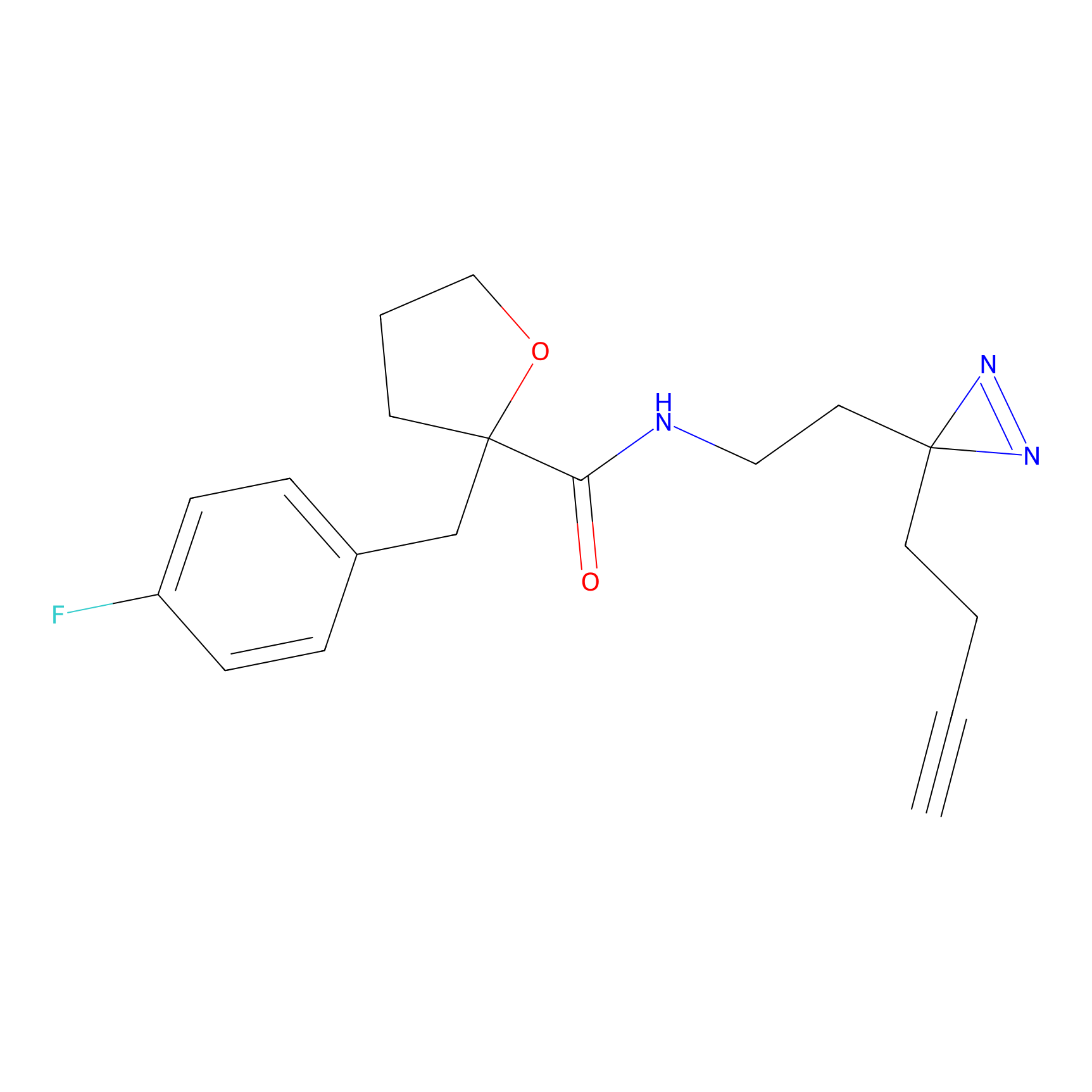
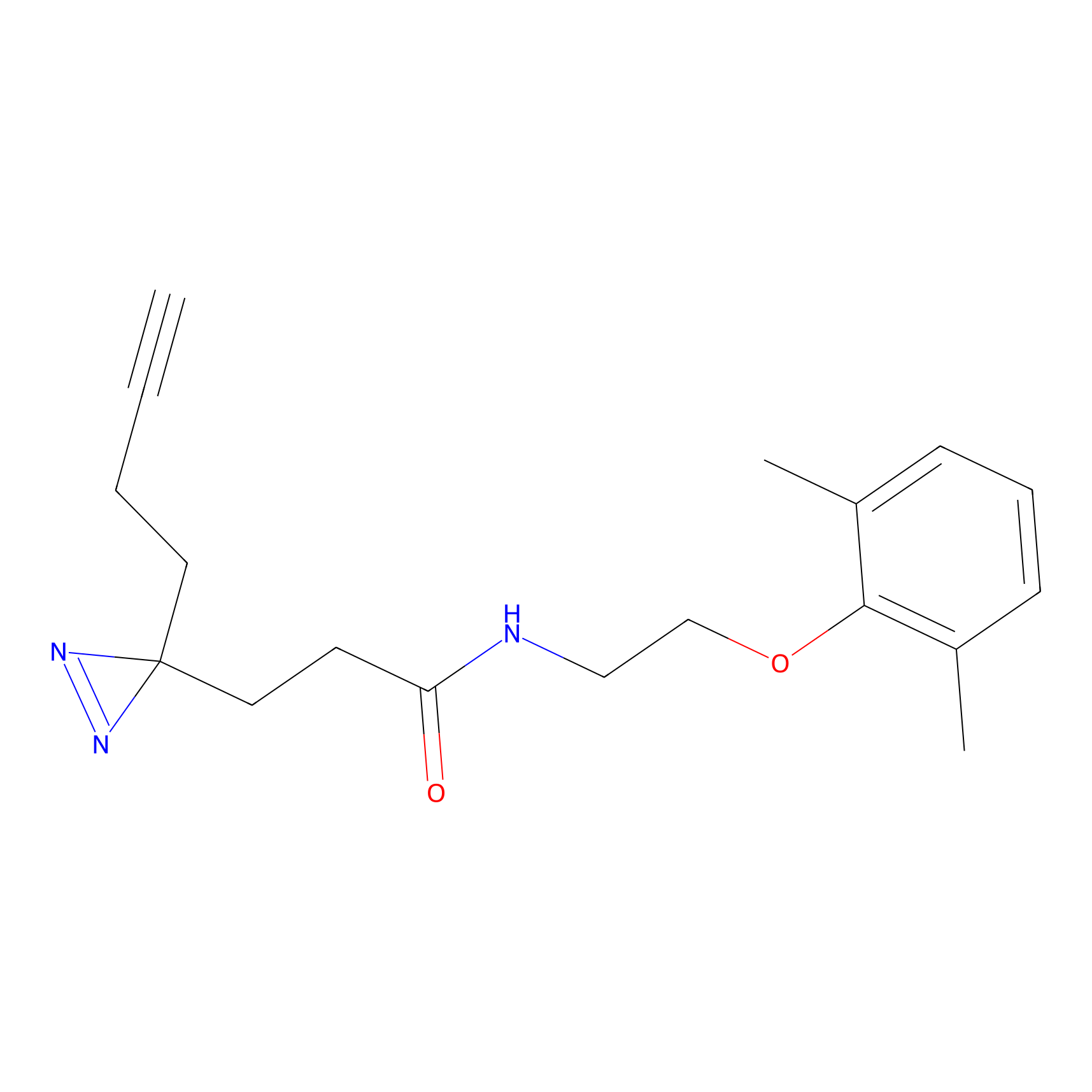
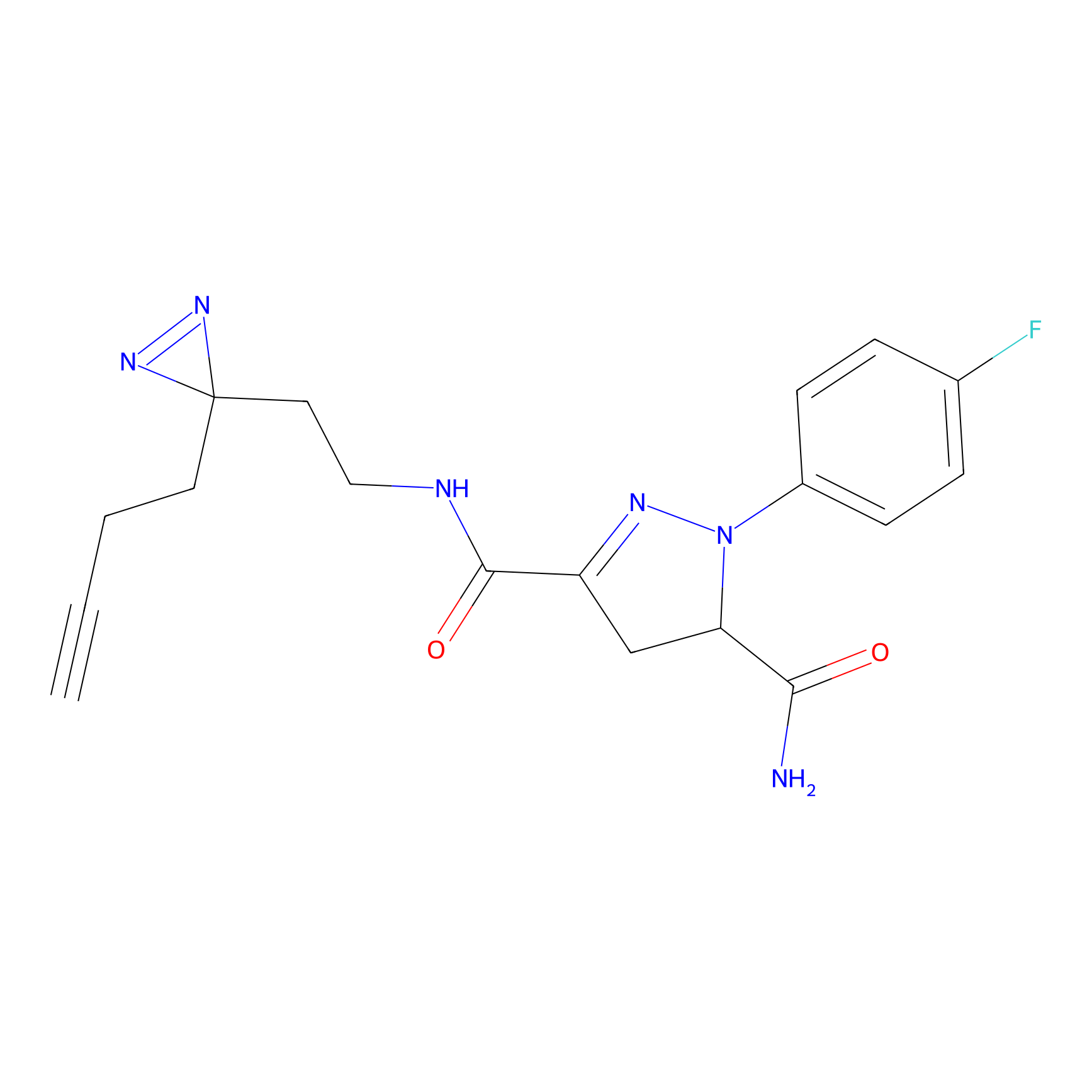
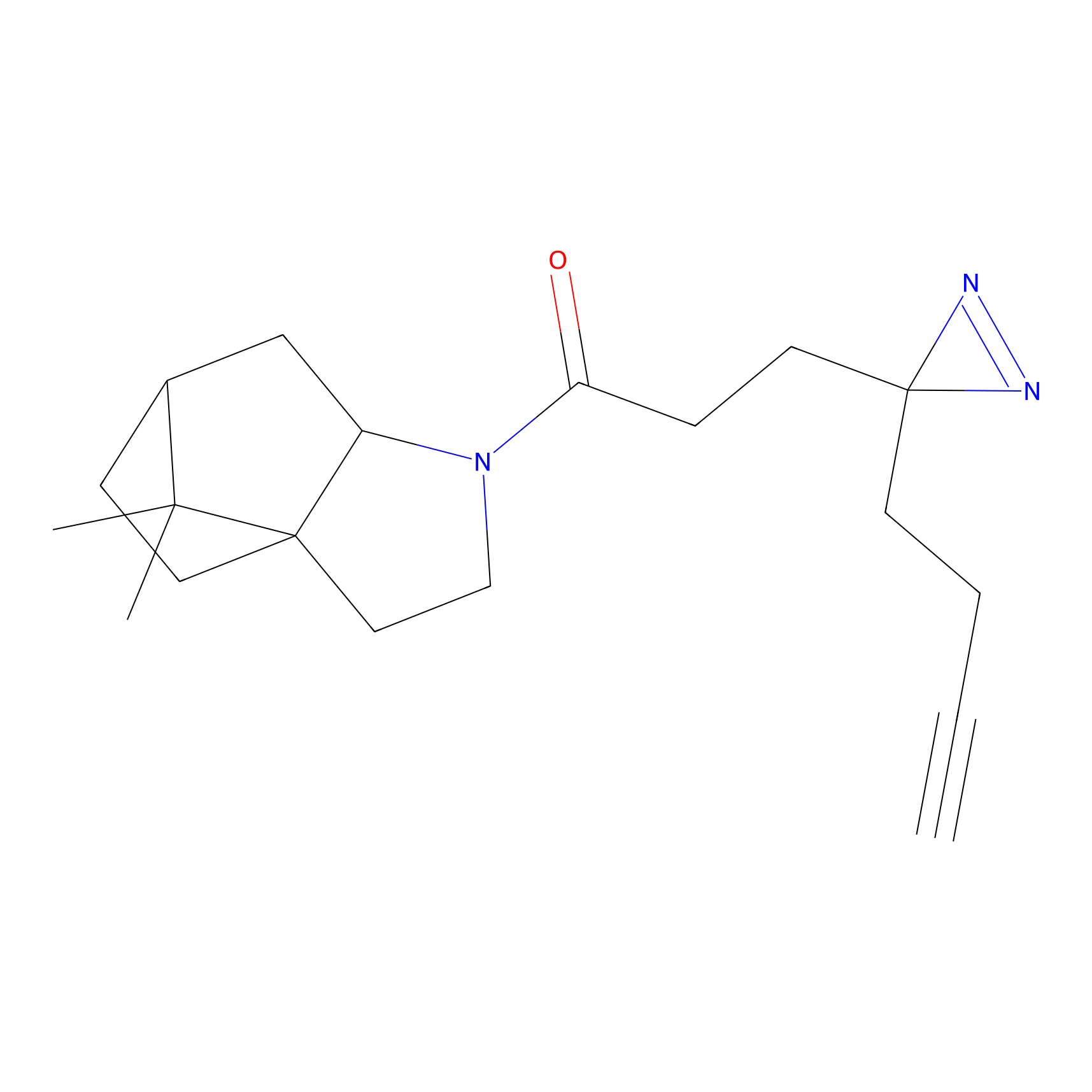
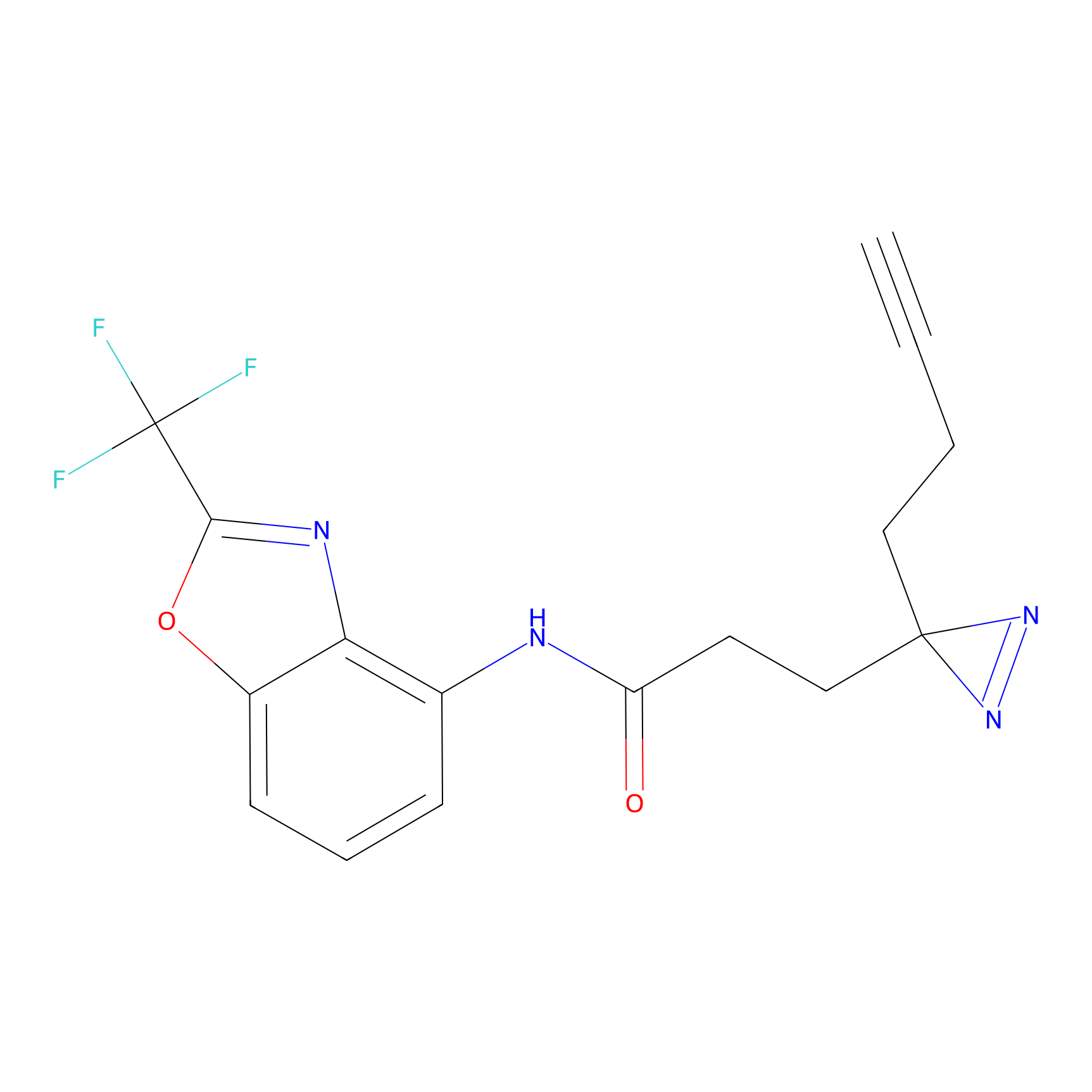
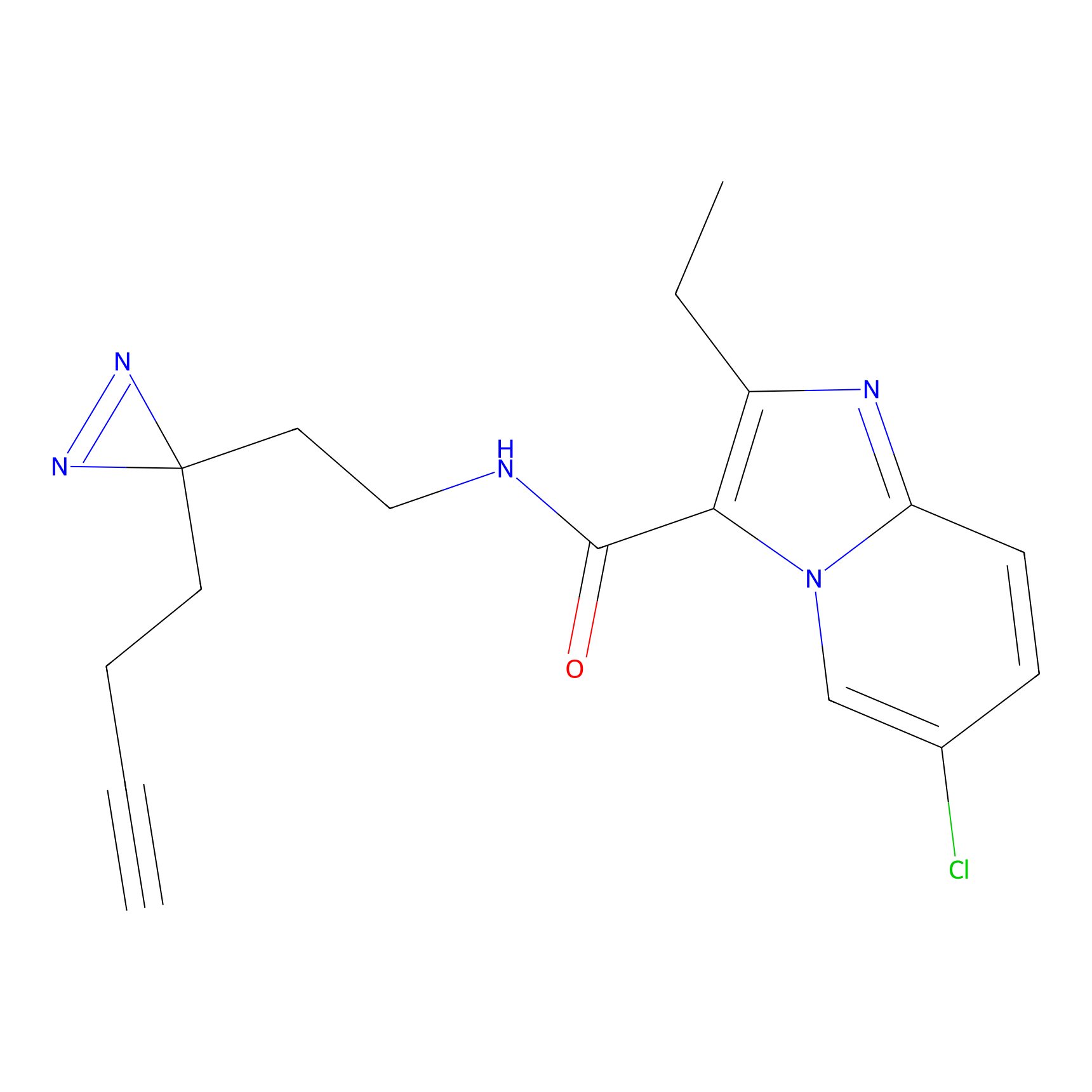
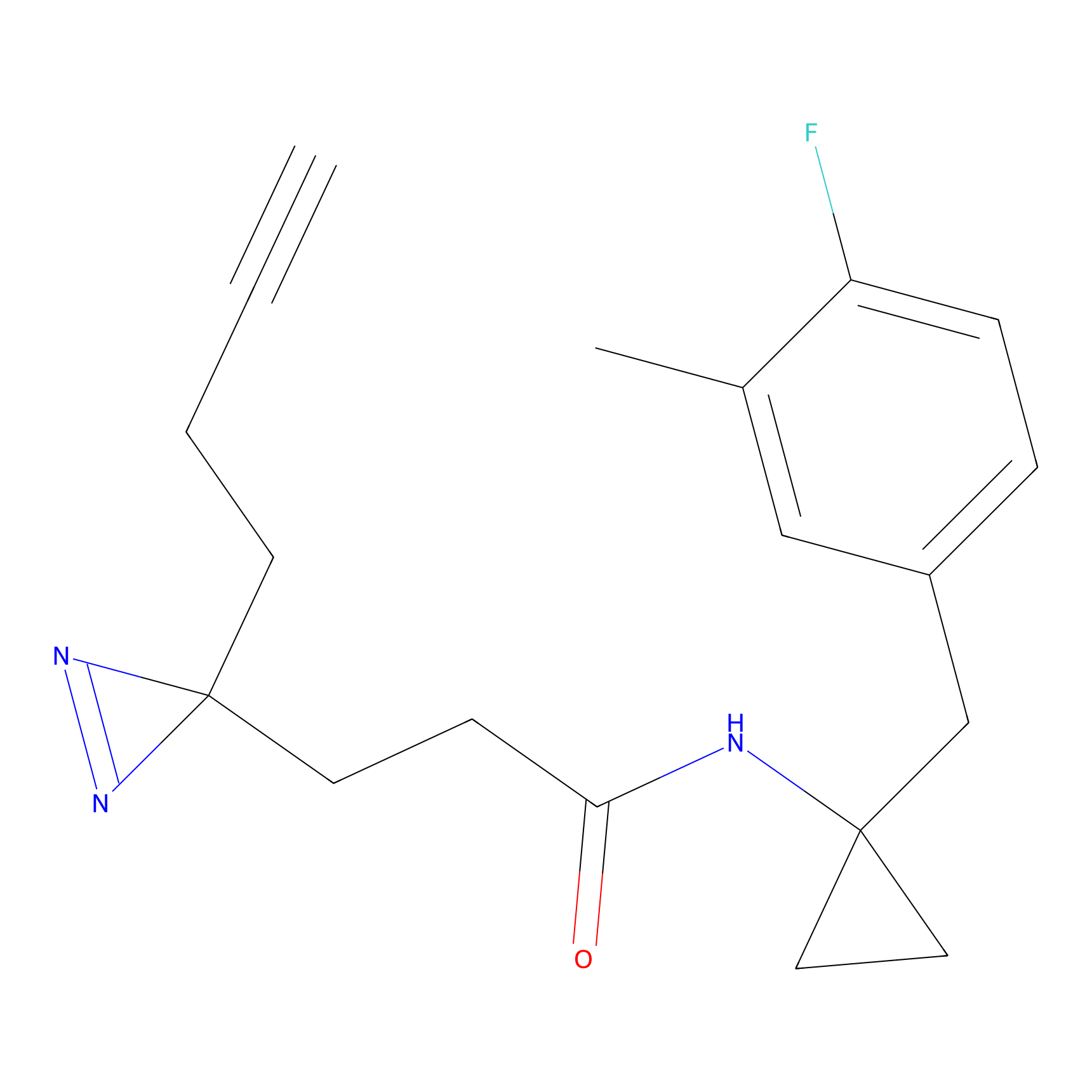
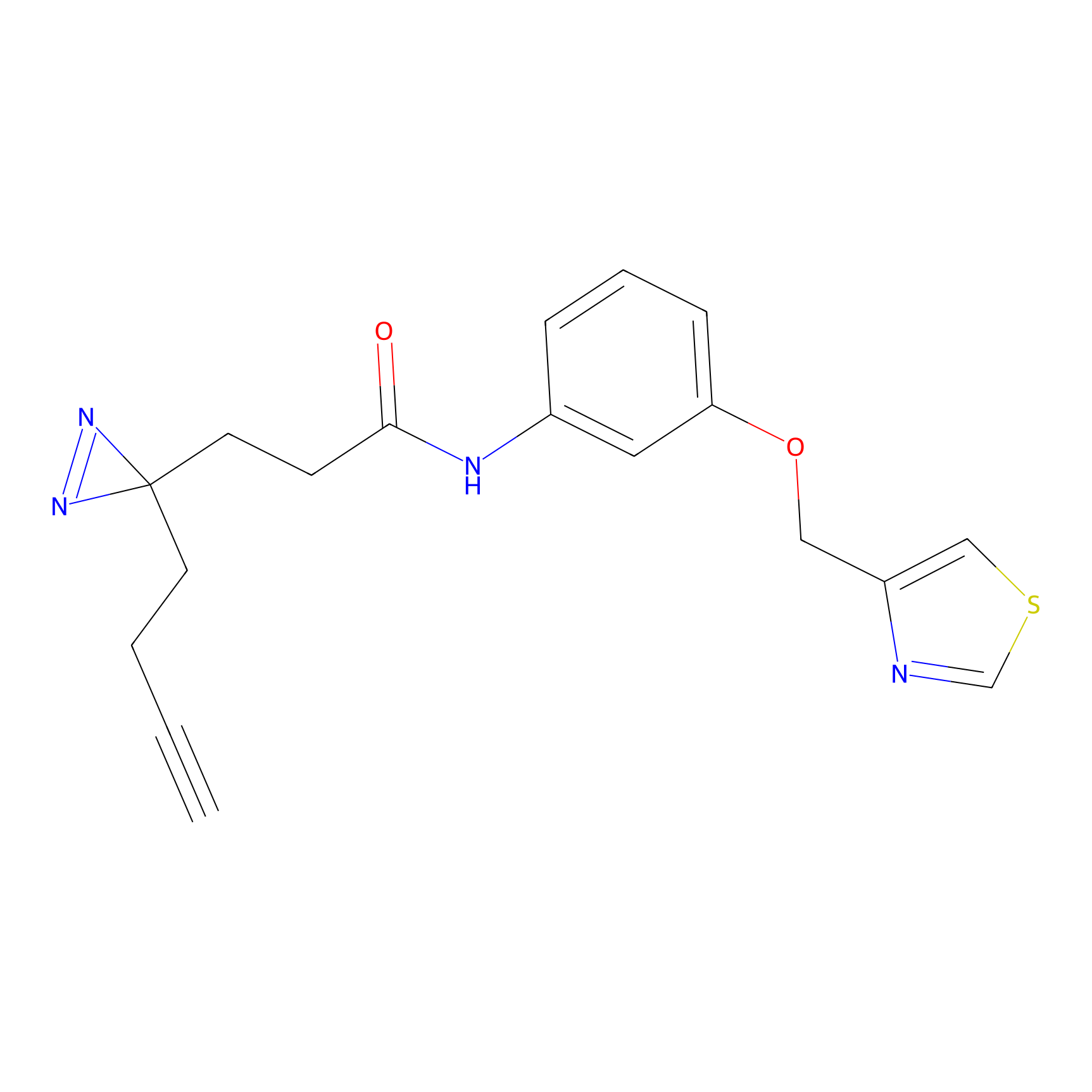
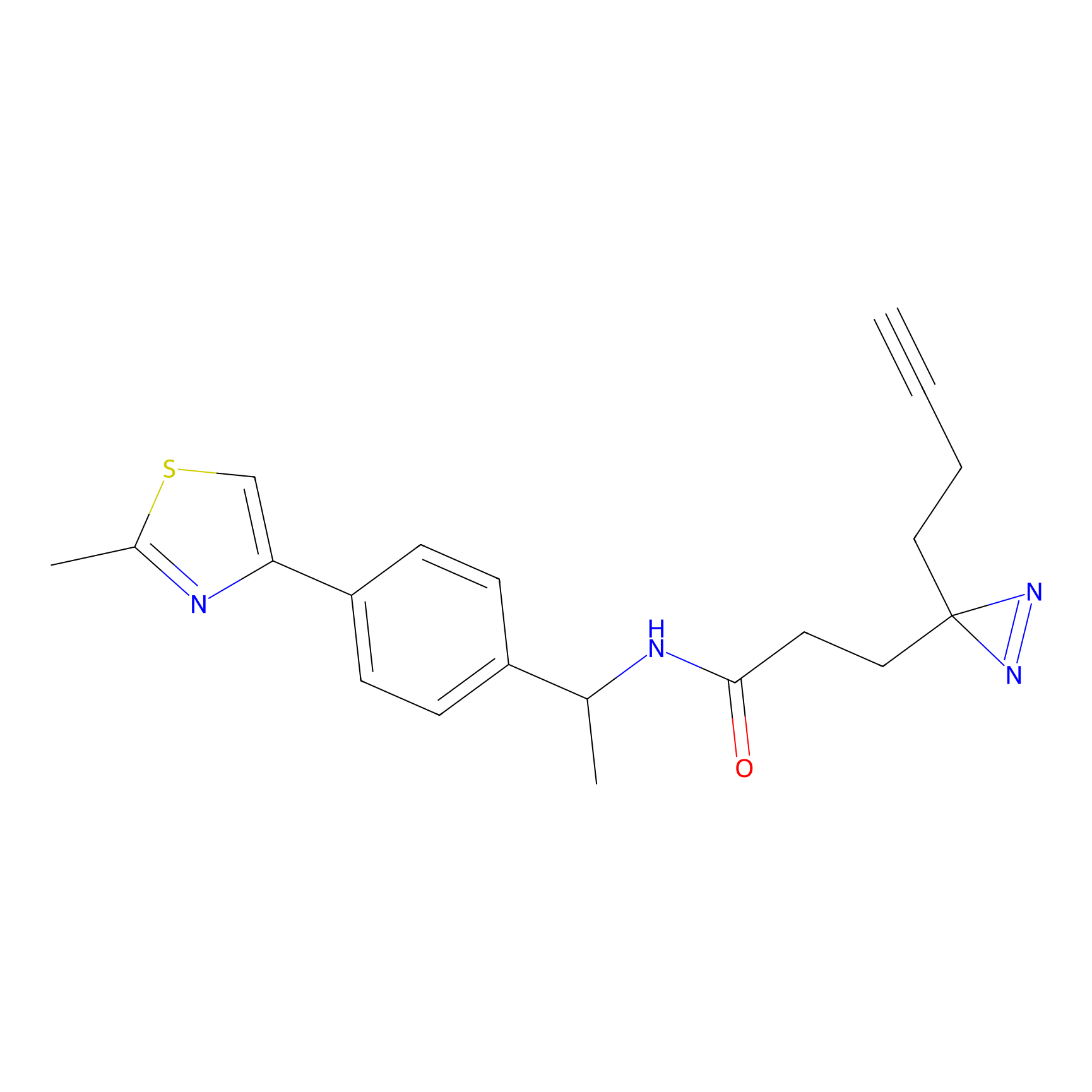
STPyne Probe Info |
 |
K110(0.57) | LDD0277 | [3] | |
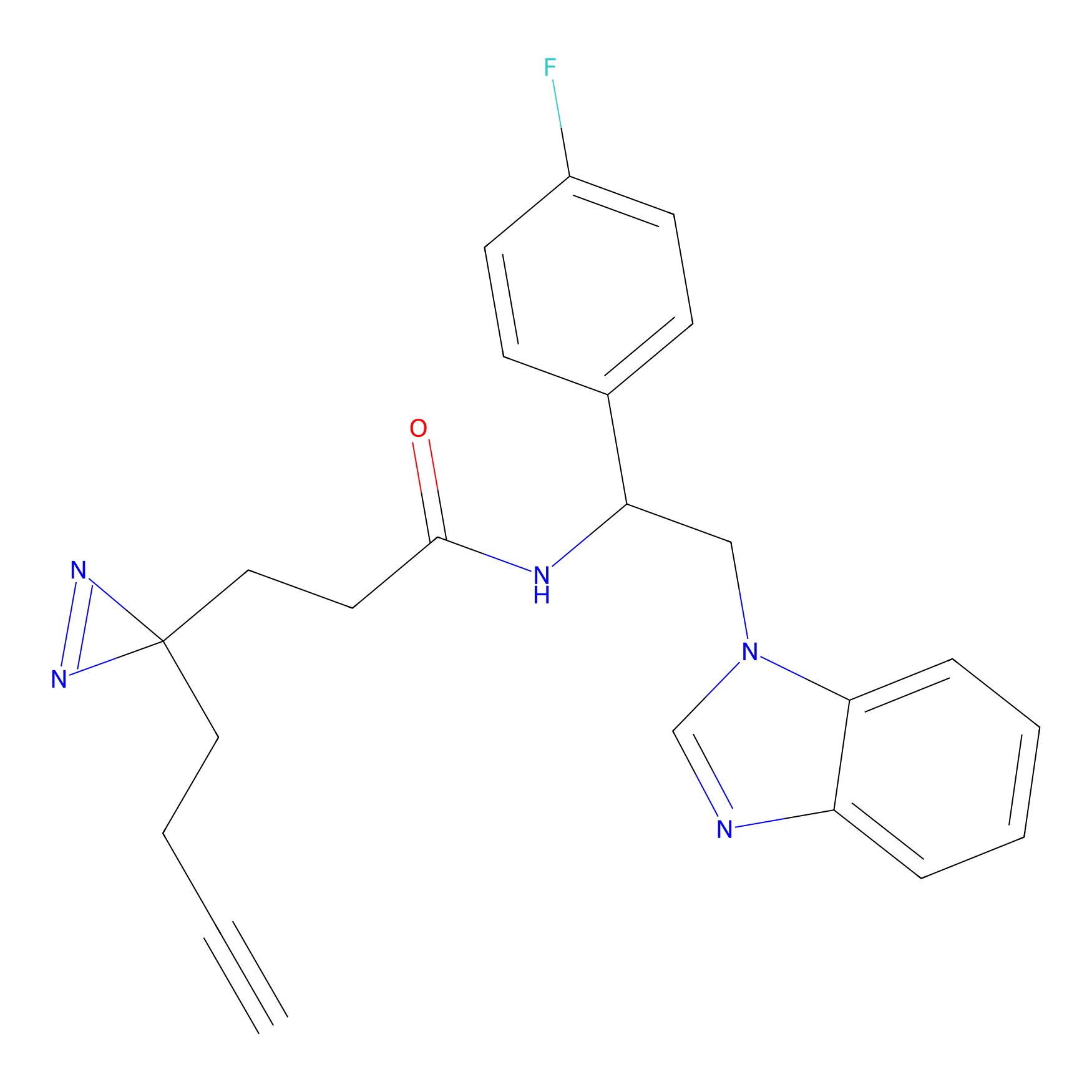
|
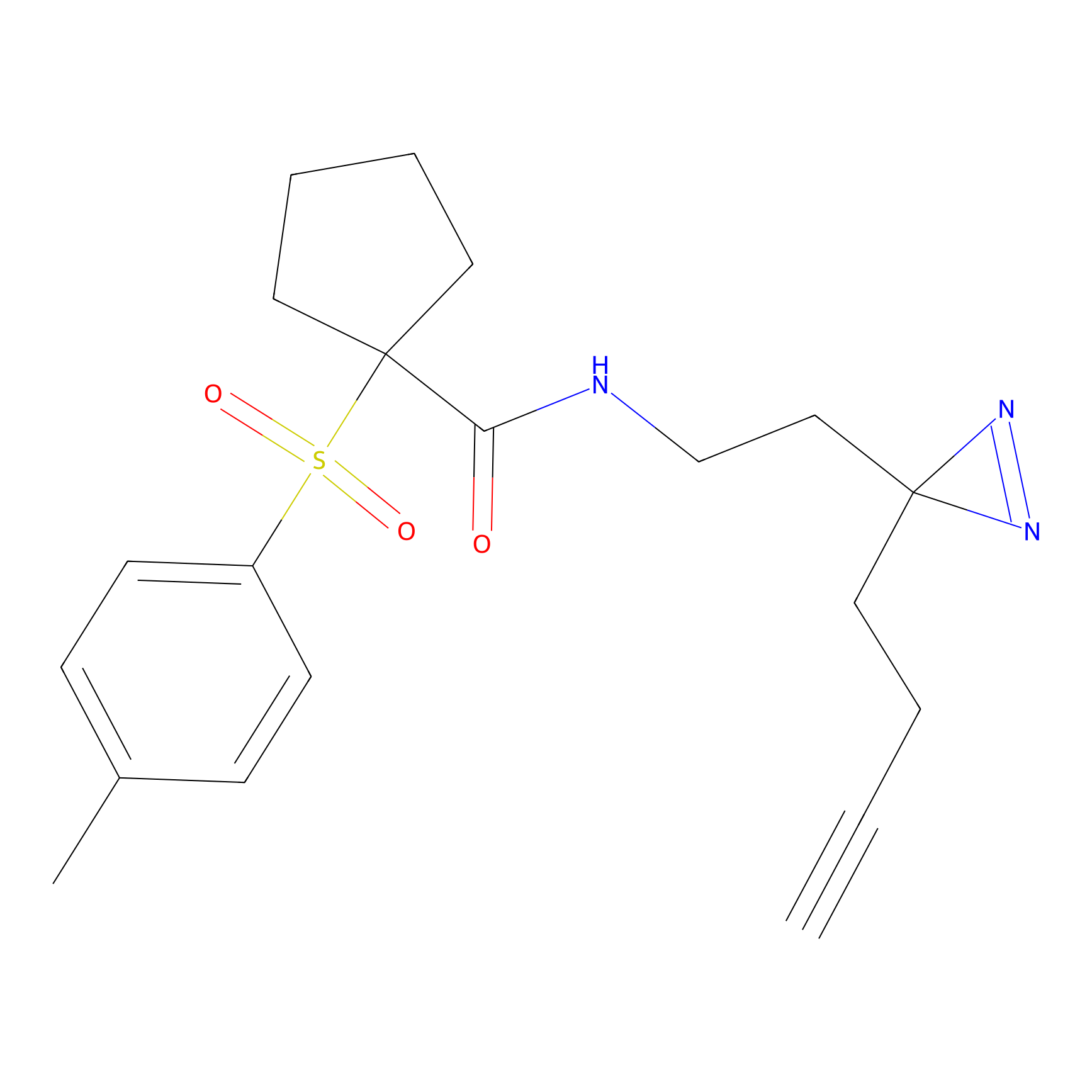
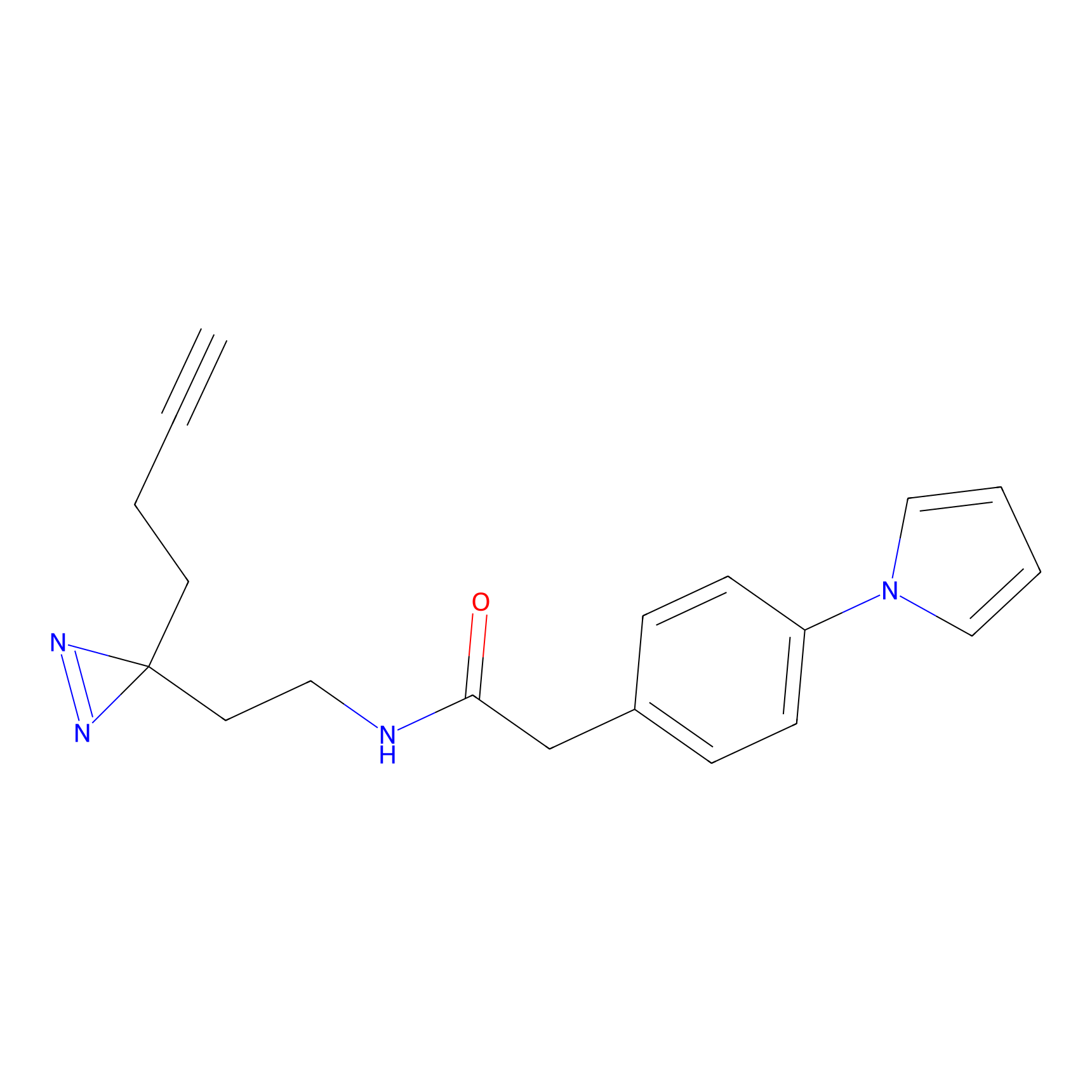
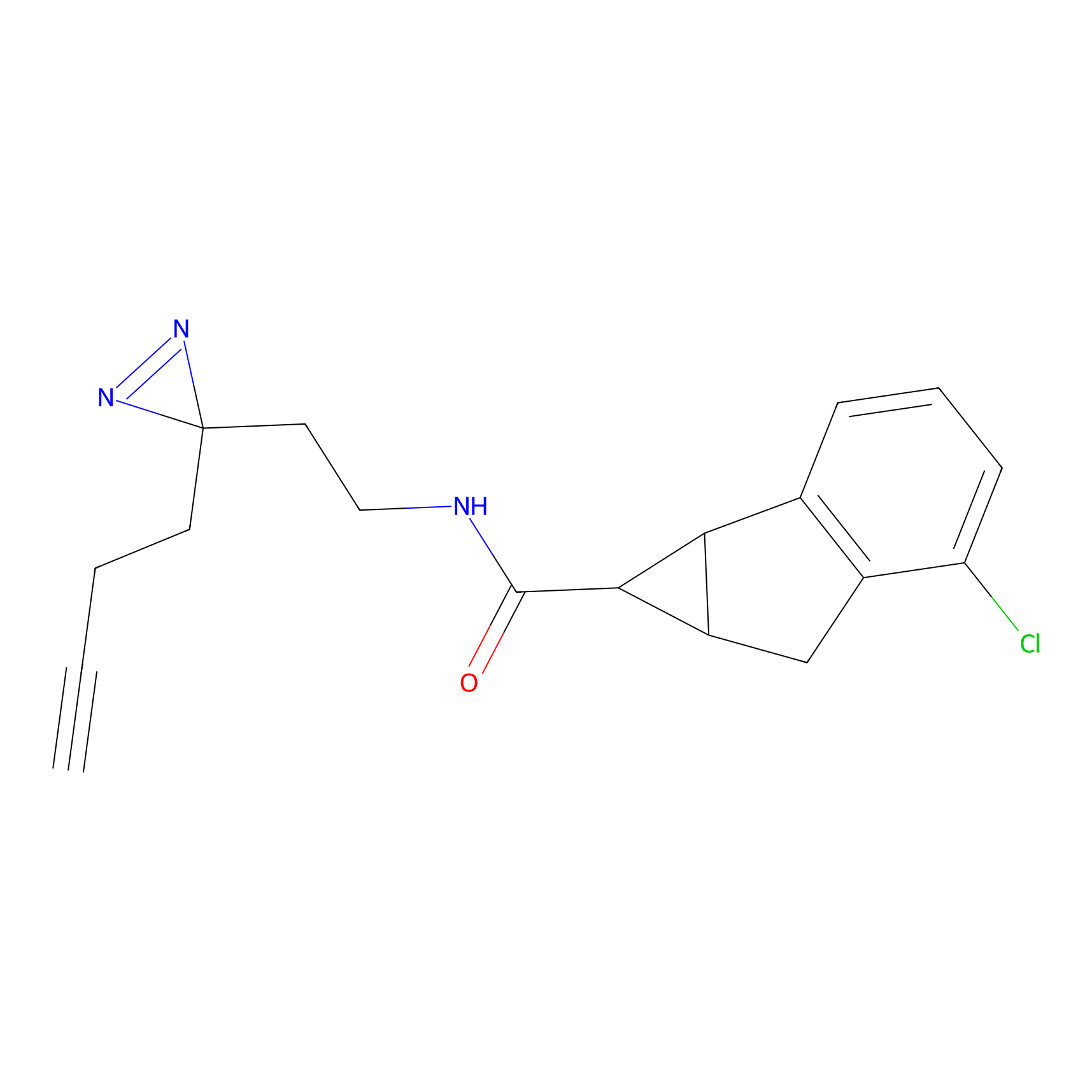
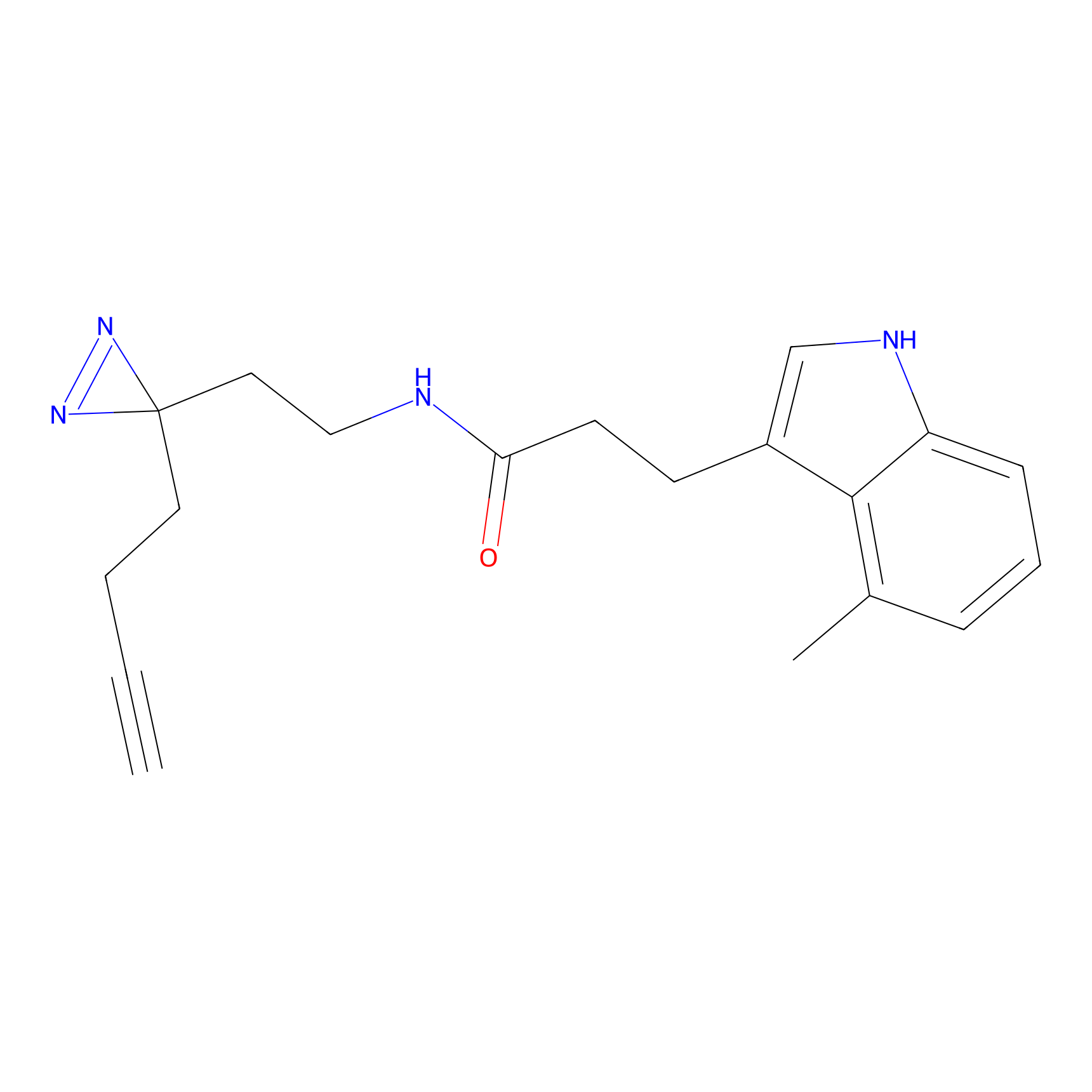
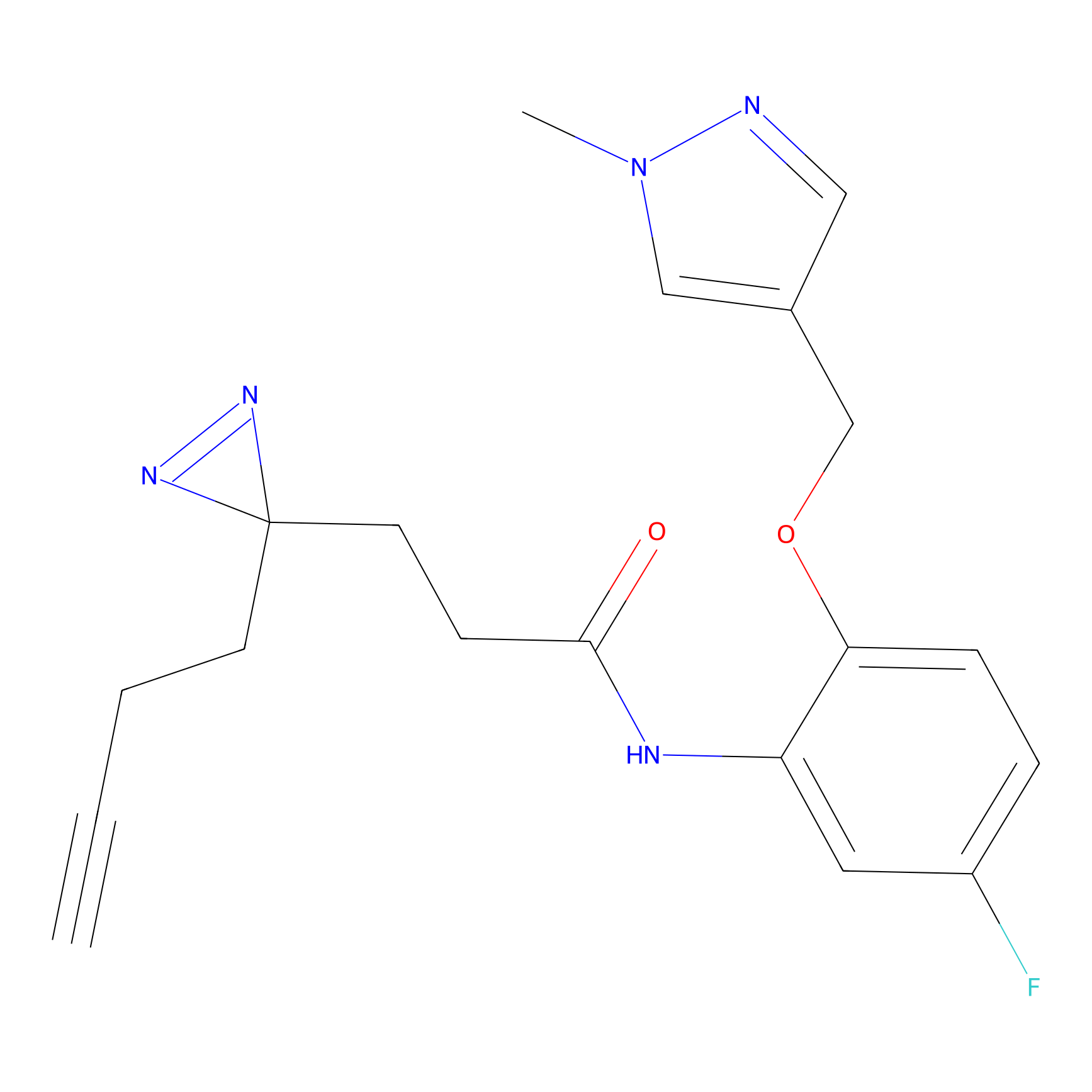
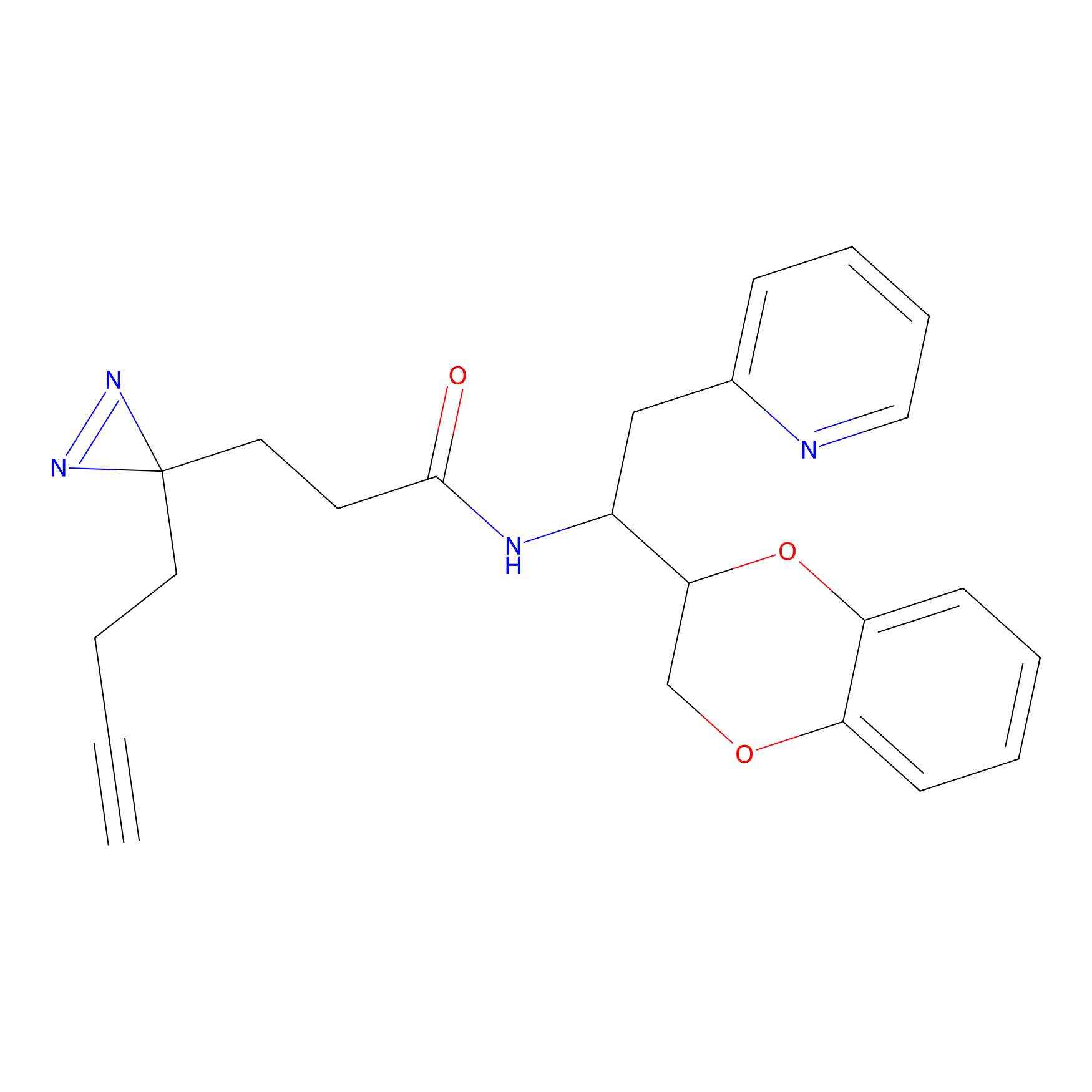
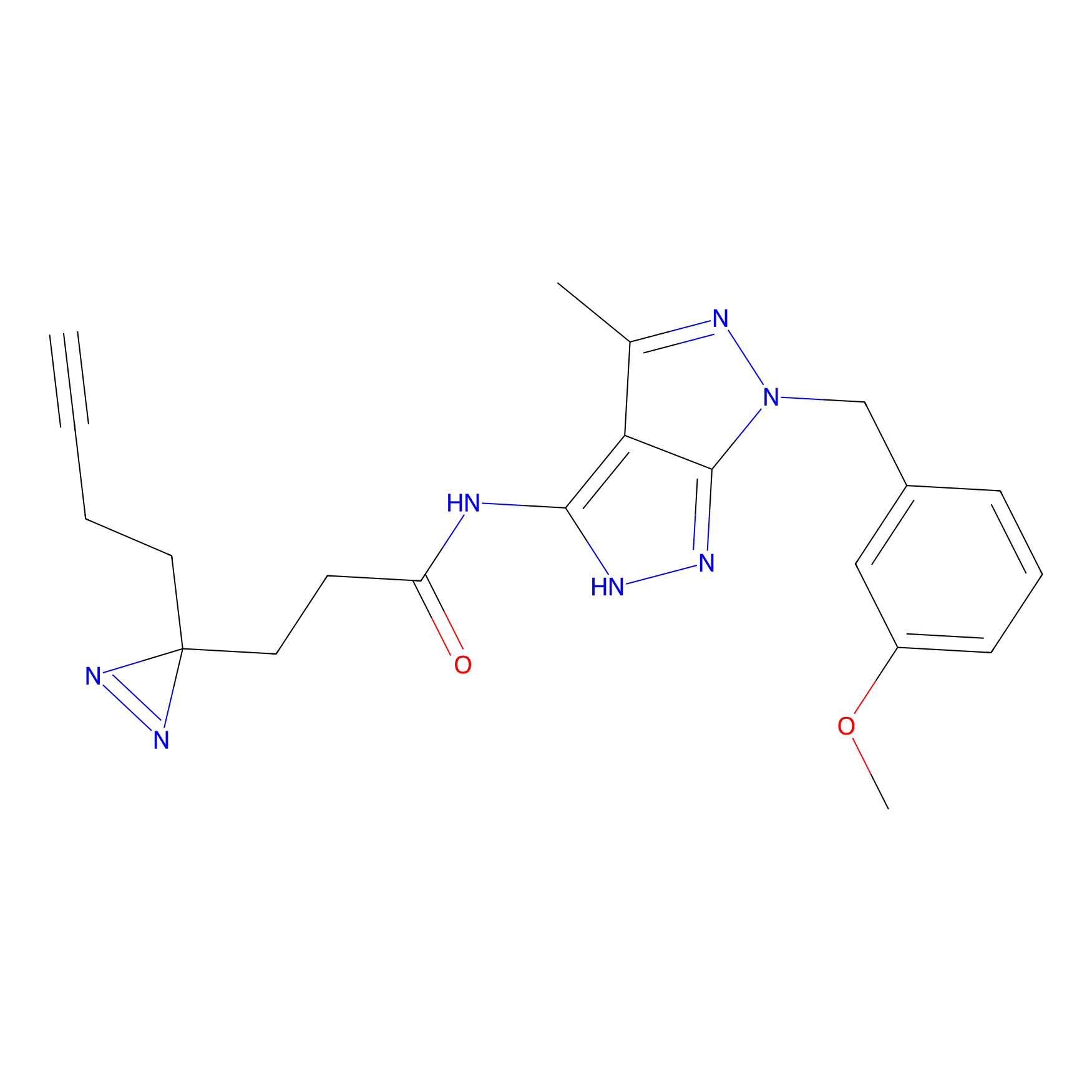
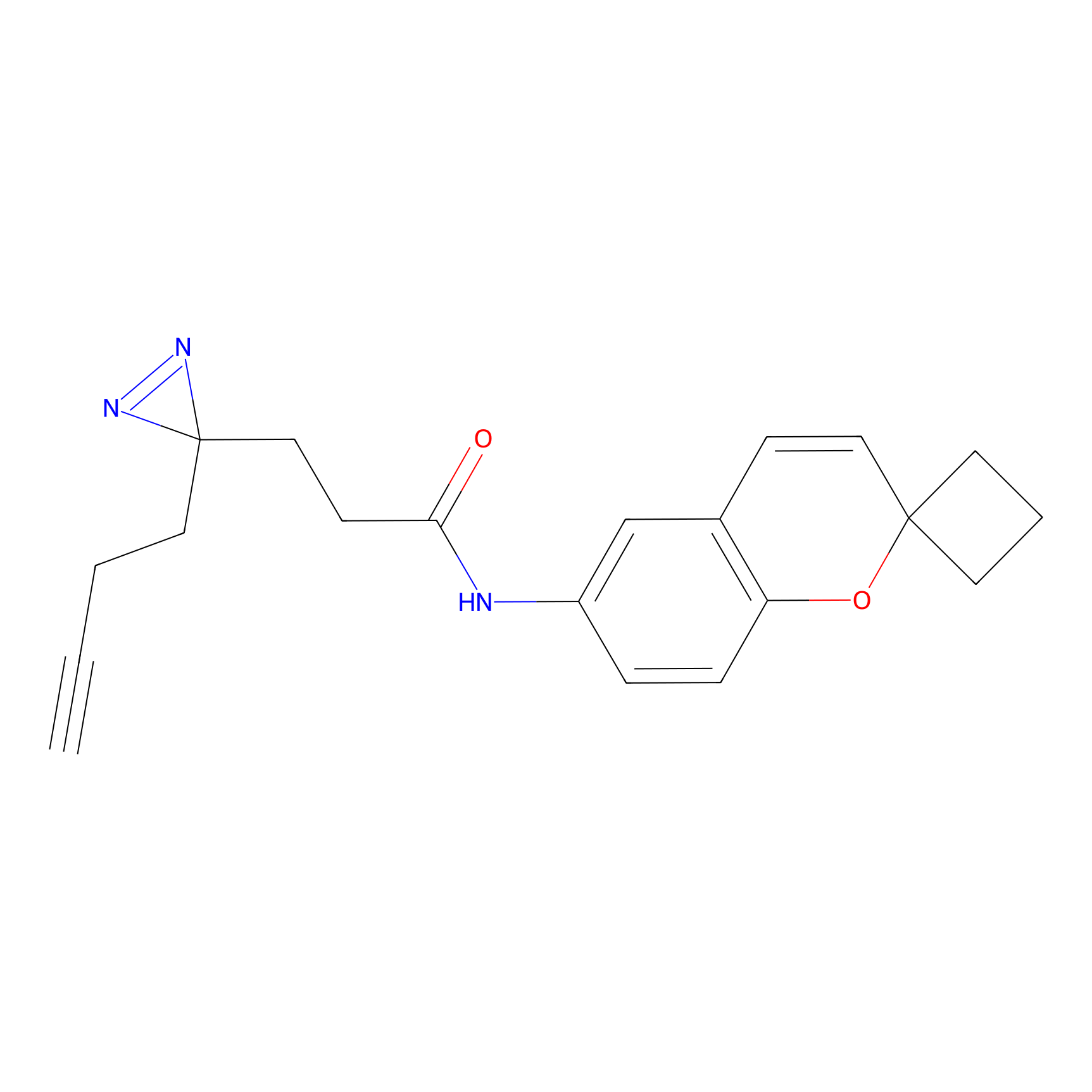
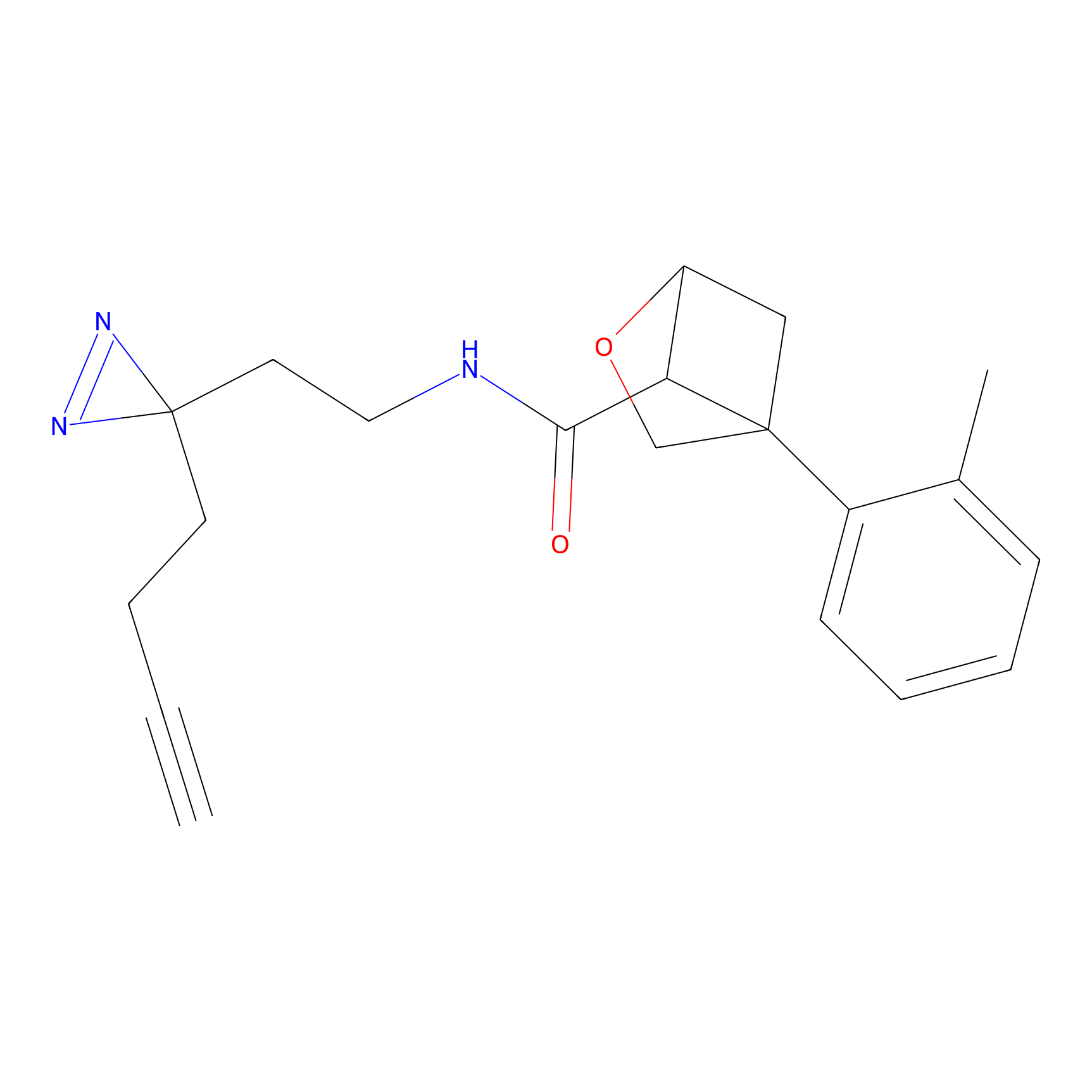
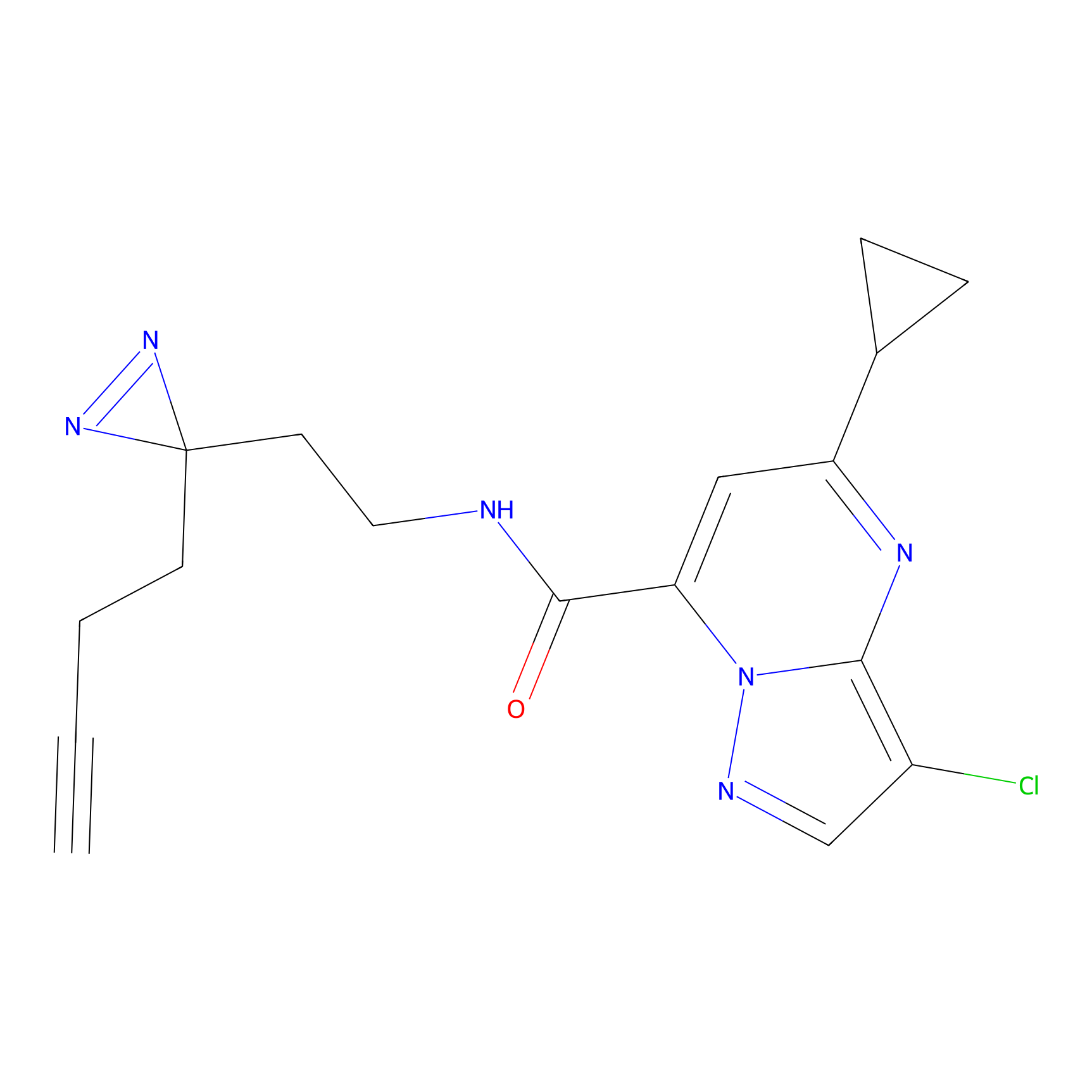
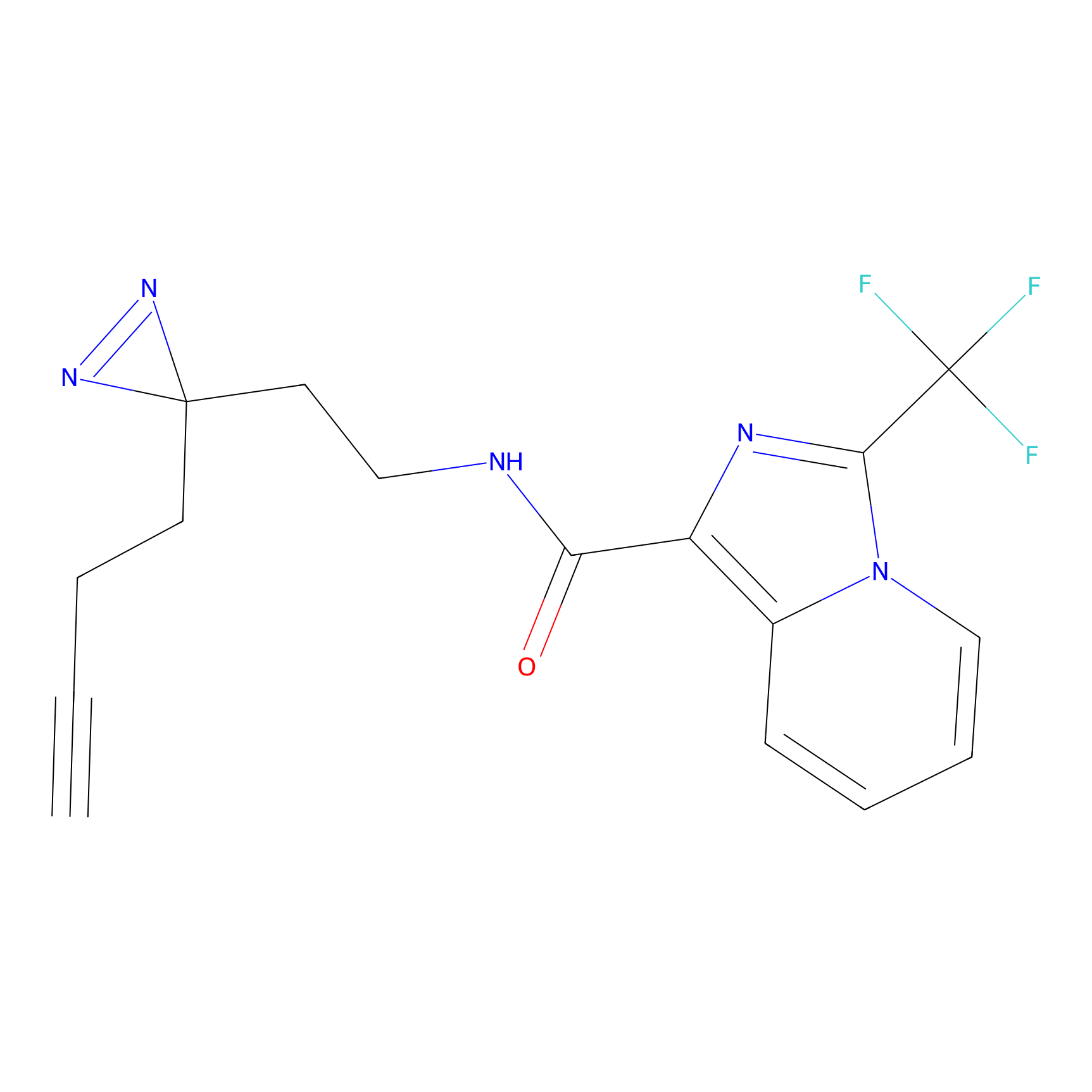
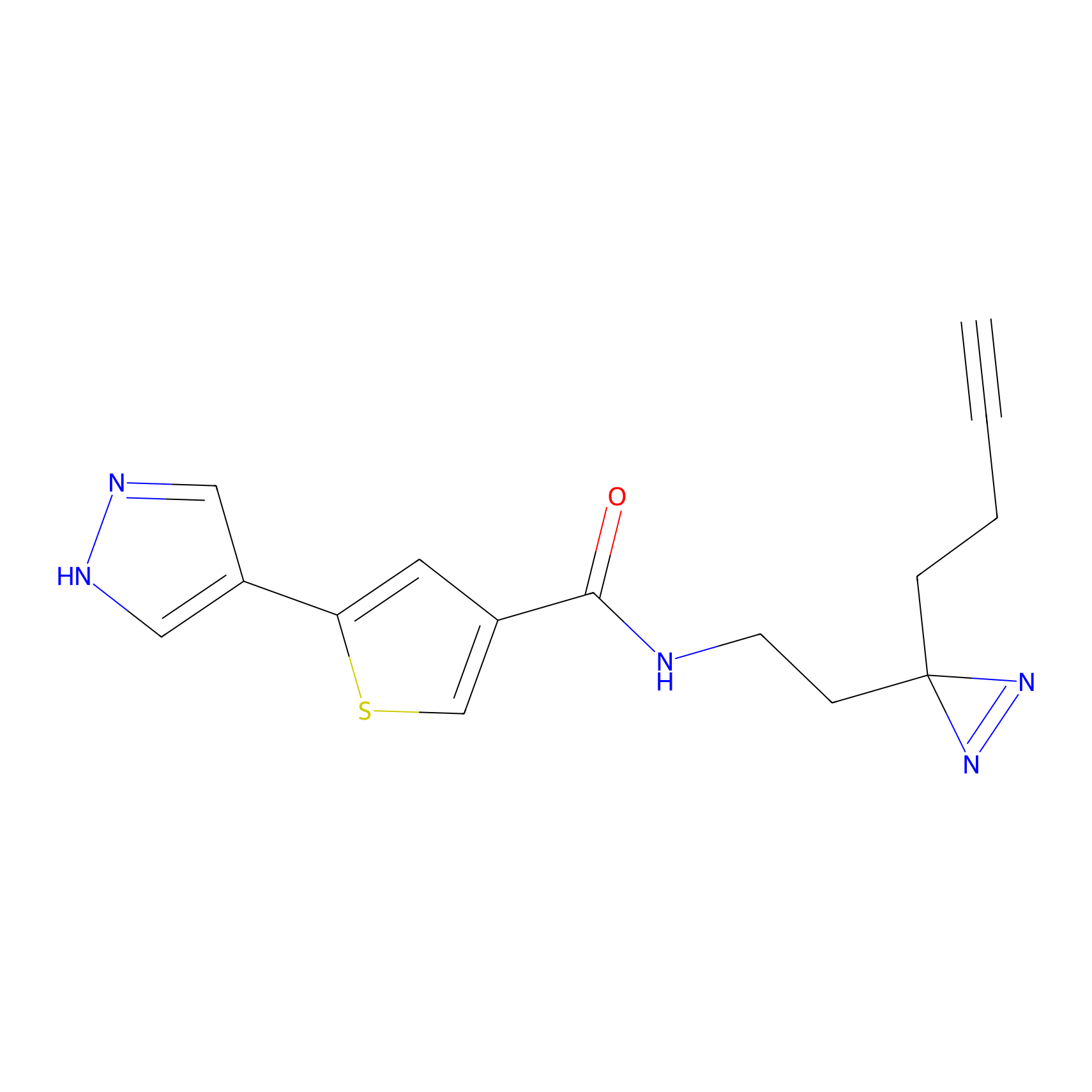
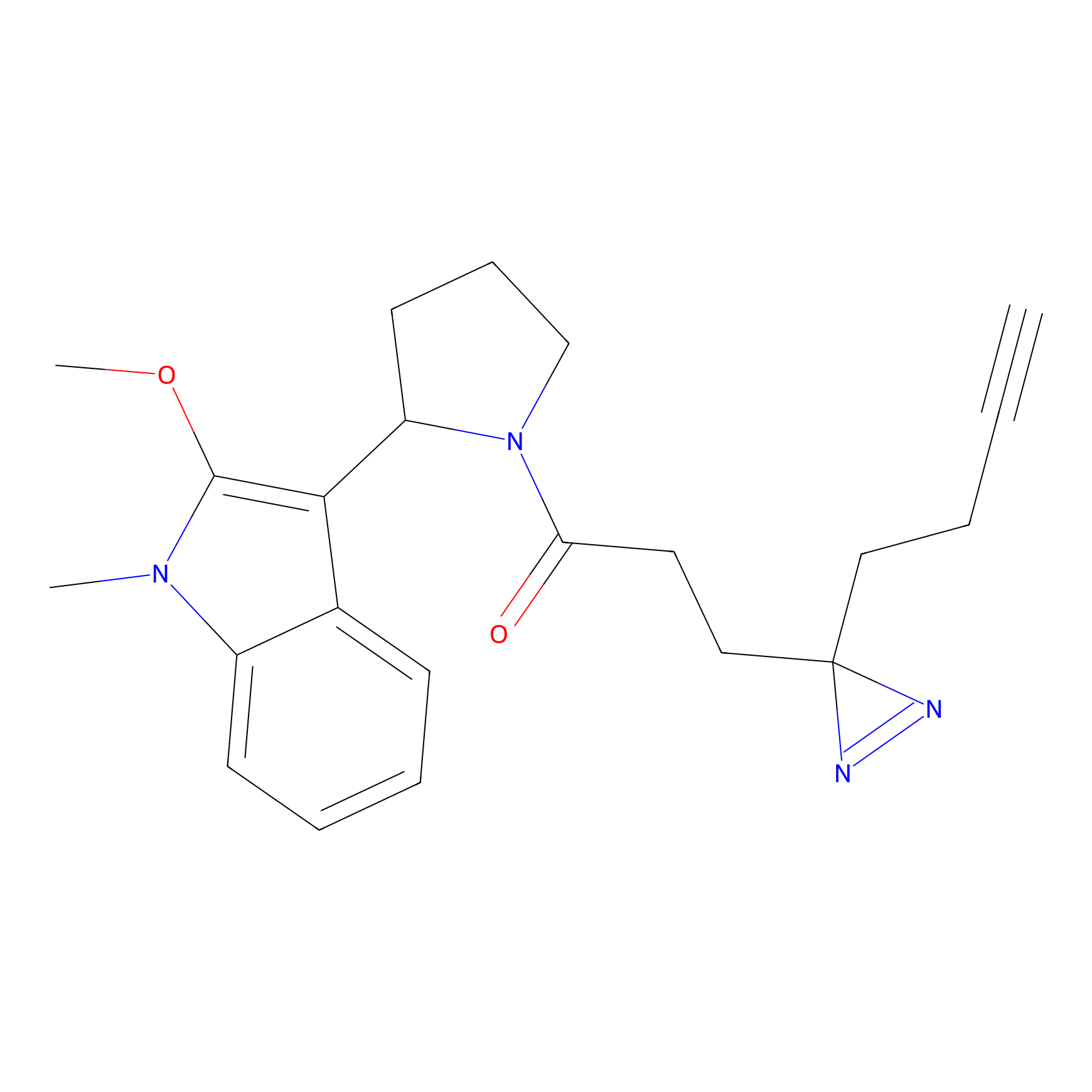
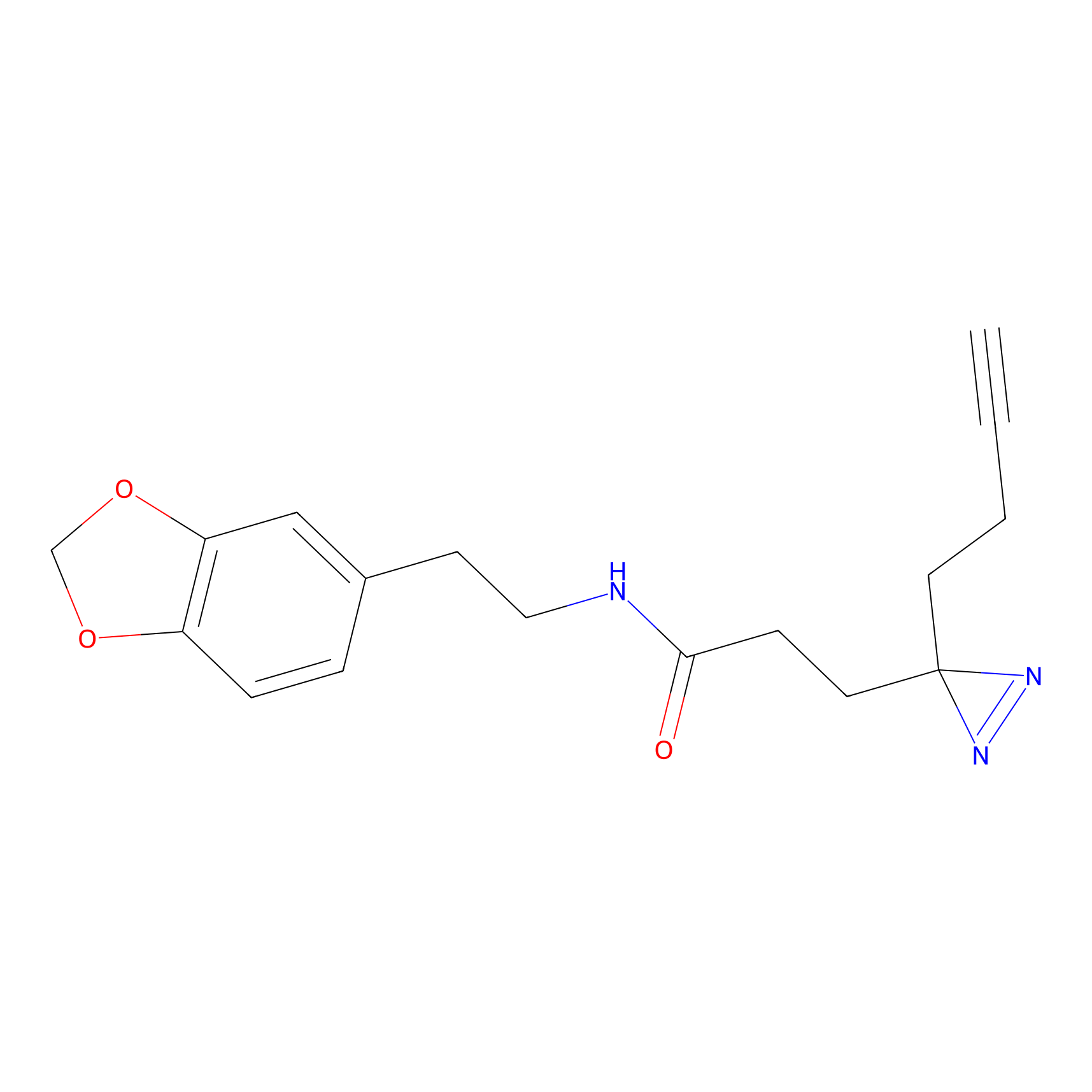
Jackson_1 Probe Info |
 |
20.00 | LDD0120 | [4] | |
|
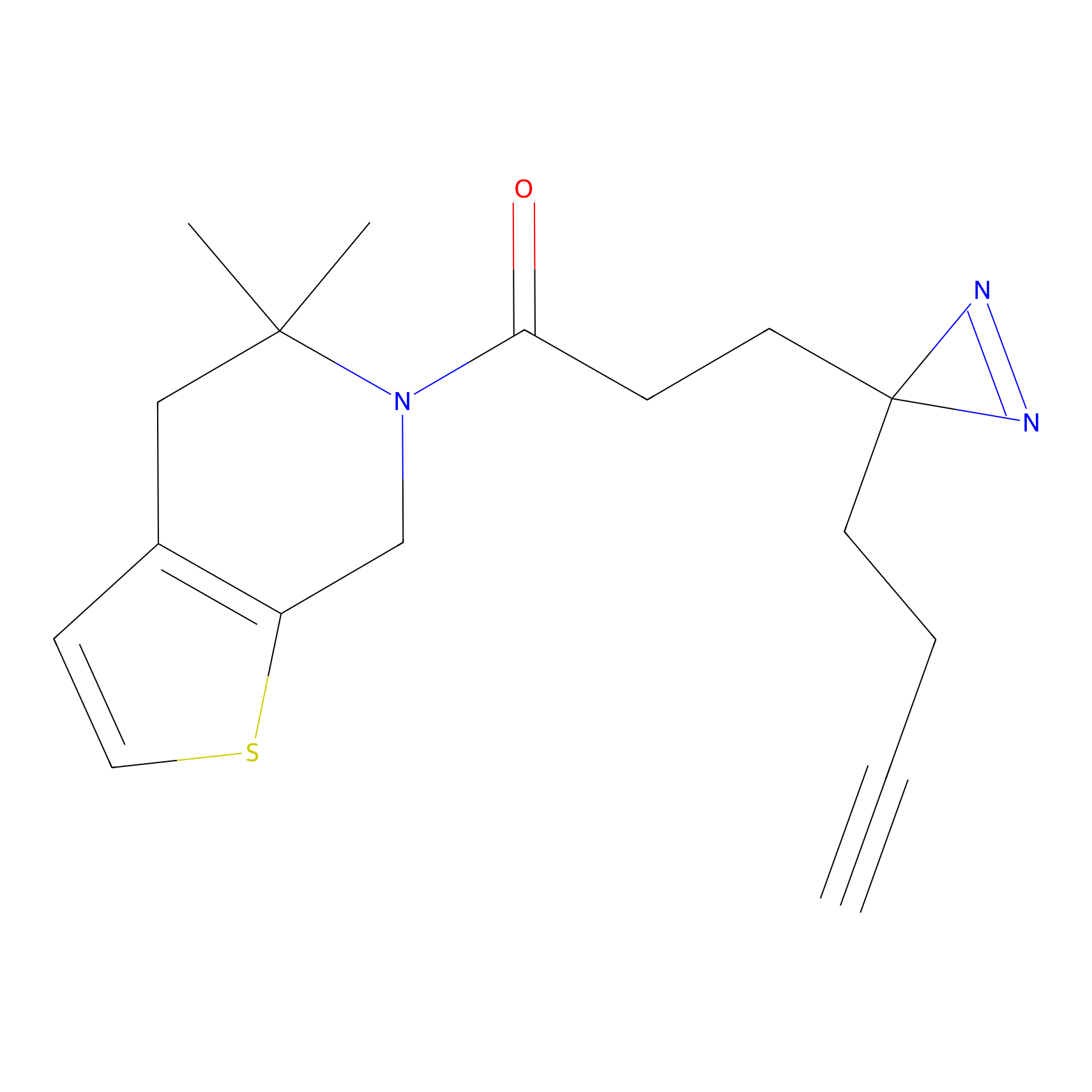
EA-probe Probe Info |
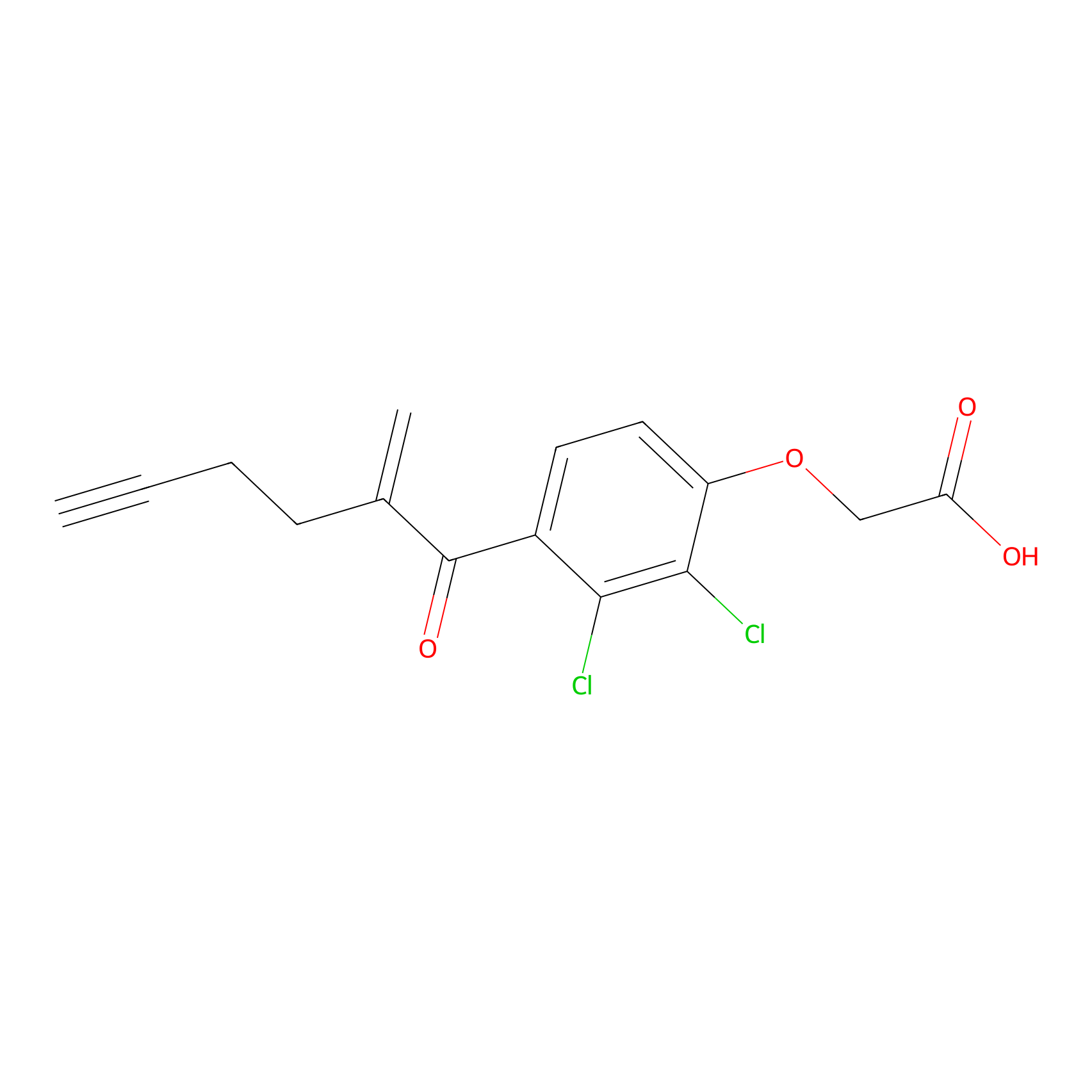 |
N.A. | LDD0440 | [5] | |
|
IA-alkyne Probe Info |
 |
C170(0.98) | LDD2206 | [6] | |
|
DBIA Probe Info |
 |
C170(0.96) | LDD1507 | [7] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [8] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [9] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [10] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HEK-293T | C170(0.96) | LDD1507 | [7] |
| LDCM0276 | AC17 | HEK-293T | C170(1.05) | LDD1515 | [7] |
| LDCM0285 | AC25 | HEK-293T | C170(1.14) | LDD1524 | [7] |
| LDCM0294 | AC33 | HEK-293T | C170(0.99) | LDD1533 | [7] |
| LDCM0303 | AC41 | HEK-293T | C170(1.01) | LDD1542 | [7] |
| LDCM0311 | AC49 | HEK-293T | C170(1.02) | LDD1550 | [7] |
| LDCM0320 | AC57 | HEK-293T | C170(1.02) | LDD1559 | [7] |
| LDCM0356 | AKOS034007680 | HEK-293T | C170(1.05) | LDD1570 | [7] |
| LDCM0156 | Aniline | NCI-H1299 | 14.20 | LDD0403 | [1] |
| LDCM0071 | Cisar_cp14 | MDA-MB-231 | 20.00 | LDD0121 | [4] |
| LDCM0070 | Cisar_cp37 | MDA-MB-231 | 20.00 | LDD0120 | [4] |
| LDCM0404 | CL17 | HEK-293T | C170(1.14) | LDD1608 | [7] |
| LDCM0417 | CL29 | HEK-293T | C170(1.05) | LDD1621 | [7] |
| LDCM0431 | CL41 | HEK-293T | C170(1.02) | LDD1635 | [7] |
| LDCM0440 | CL5 | HEK-293T | C170(1.10) | LDD1644 | [7] |
| LDCM0444 | CL53 | HEK-293T | C170(1.13) | LDD1647 | [7] |
| LDCM0457 | CL65 | HEK-293T | C170(1.10) | LDD1660 | [7] |
| LDCM0470 | CL77 | HEK-293T | C170(1.04) | LDD1673 | [7] |
| LDCM0483 | CL89 | HEK-293T | C170(1.01) | LDD1686 | [7] |
| LDCM0175 | Ethacrynic acid | HeLa | N.A. | LDD0440 | [5] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C170(0.98) | LDD2206 | [6] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C170(0.82) | LDD2207 | [6] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Cyclic AMP-responsive element-binding protein 3-like protein 1 (CREB3L1) | BZIP family | Q96BA8 | |||
Other
References