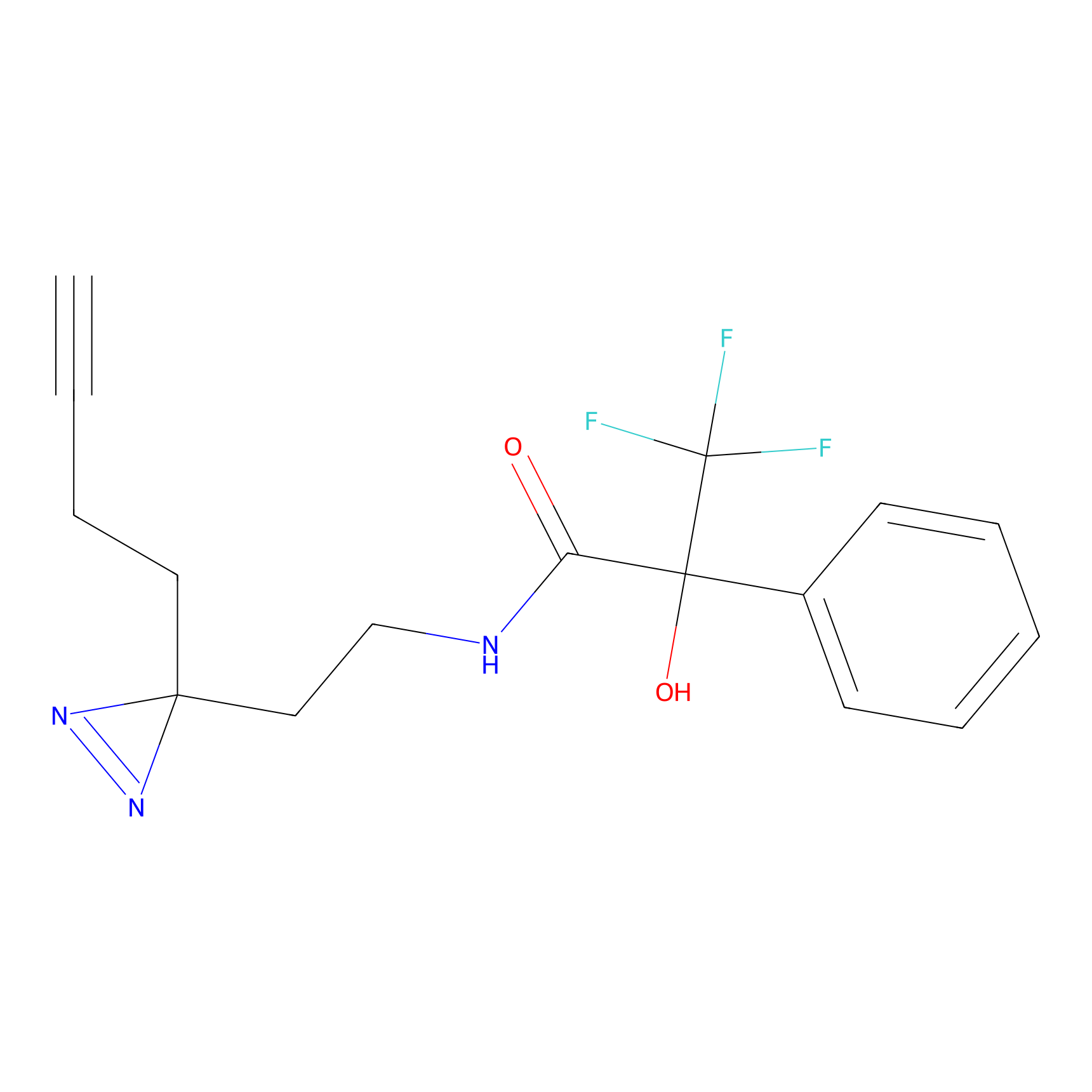
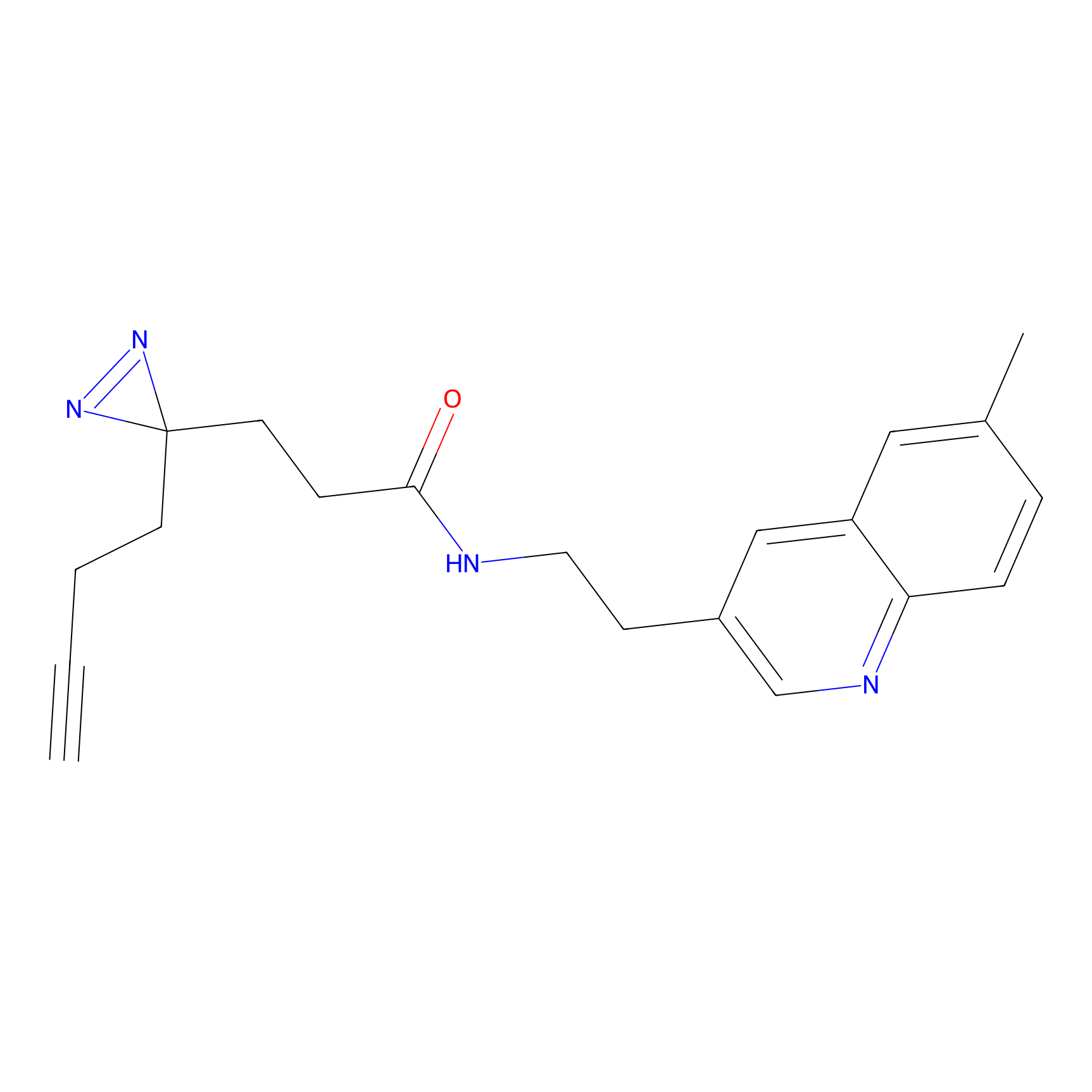
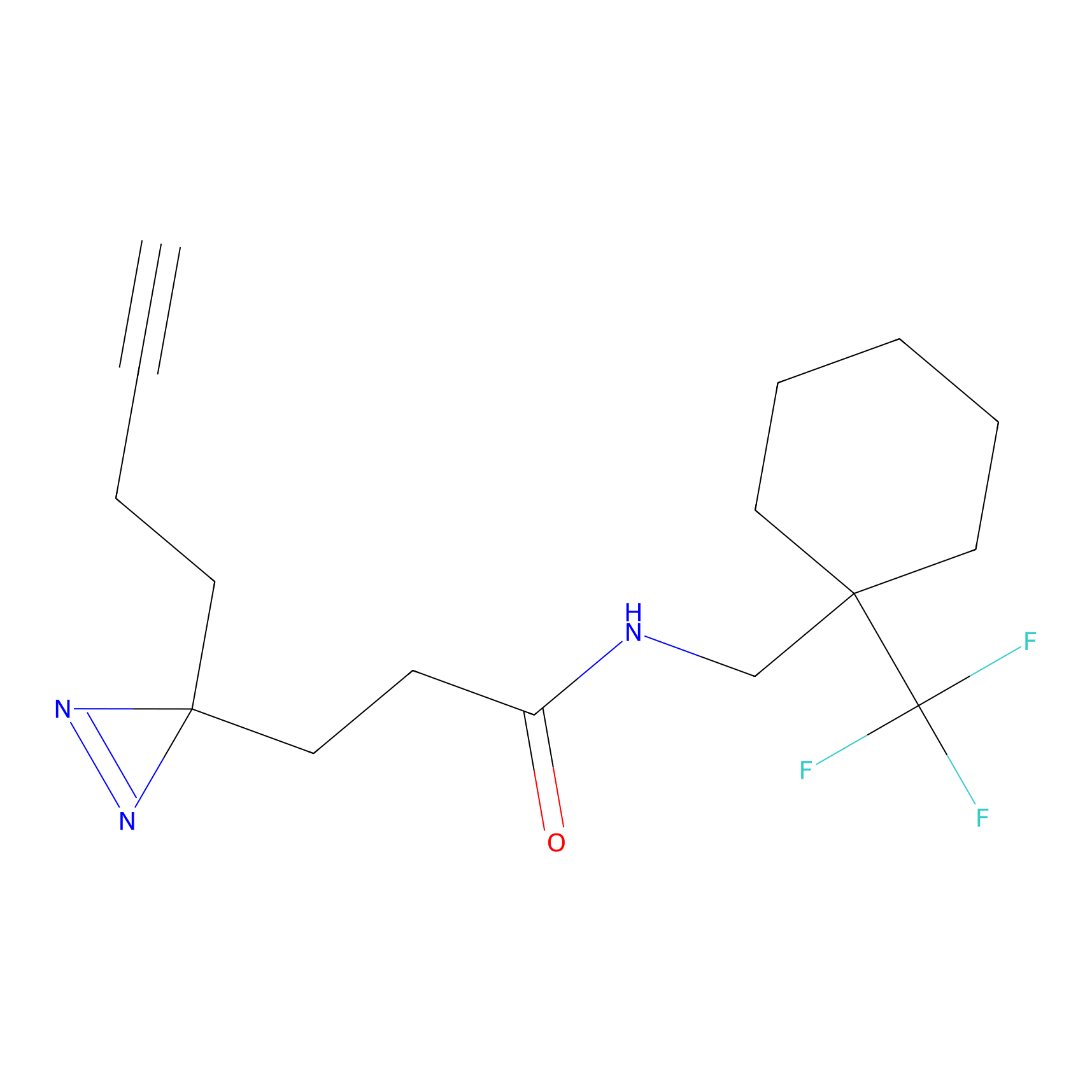
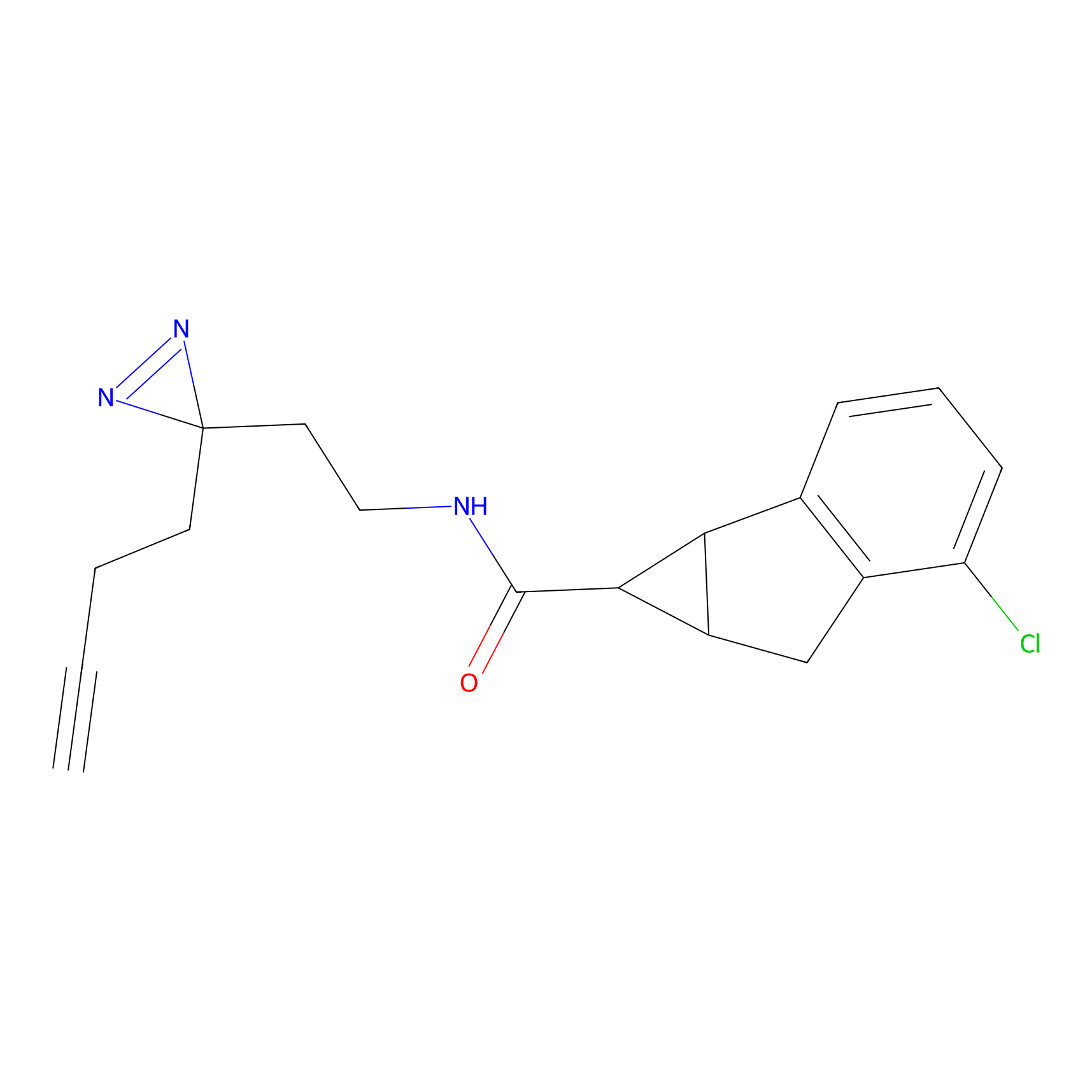
Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
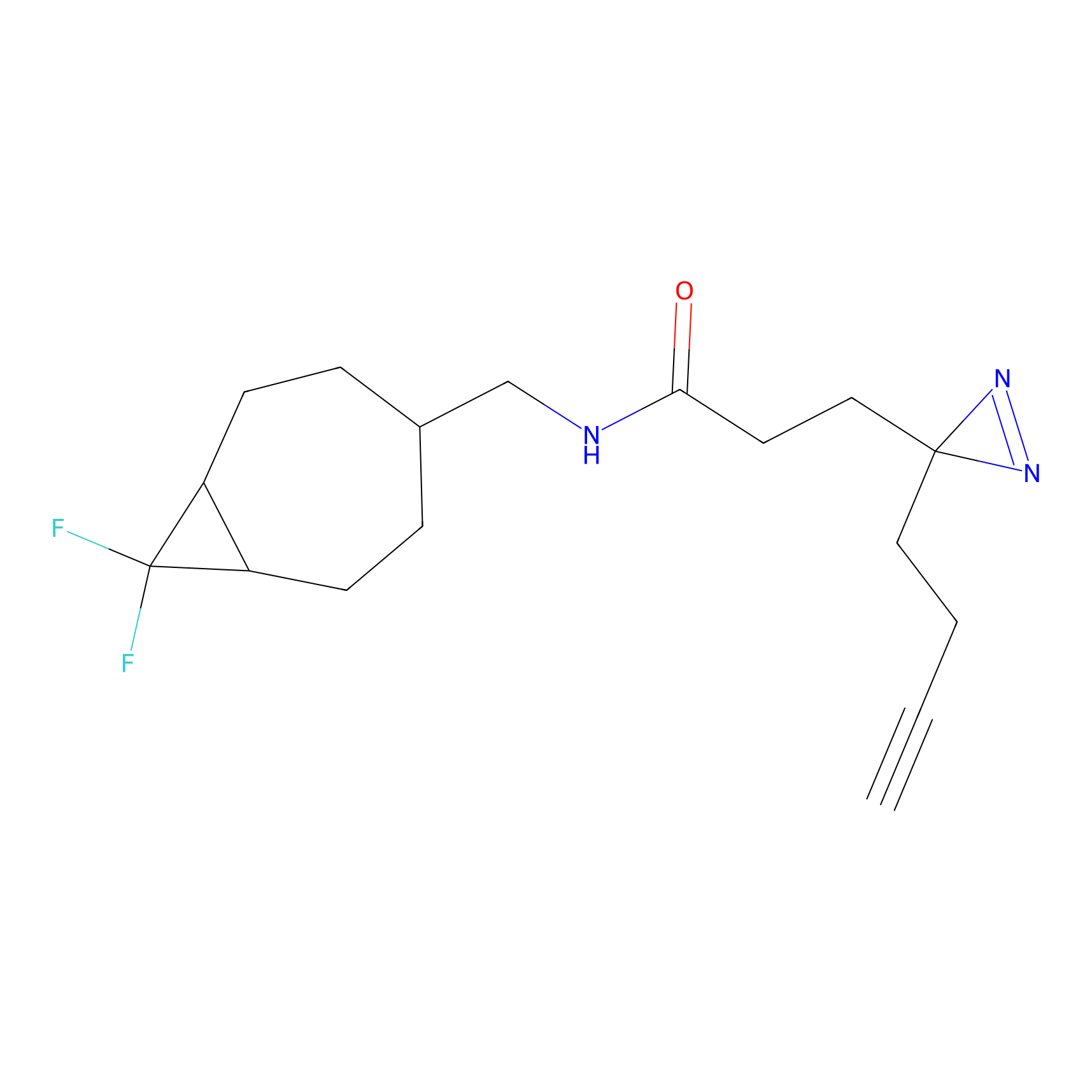
|
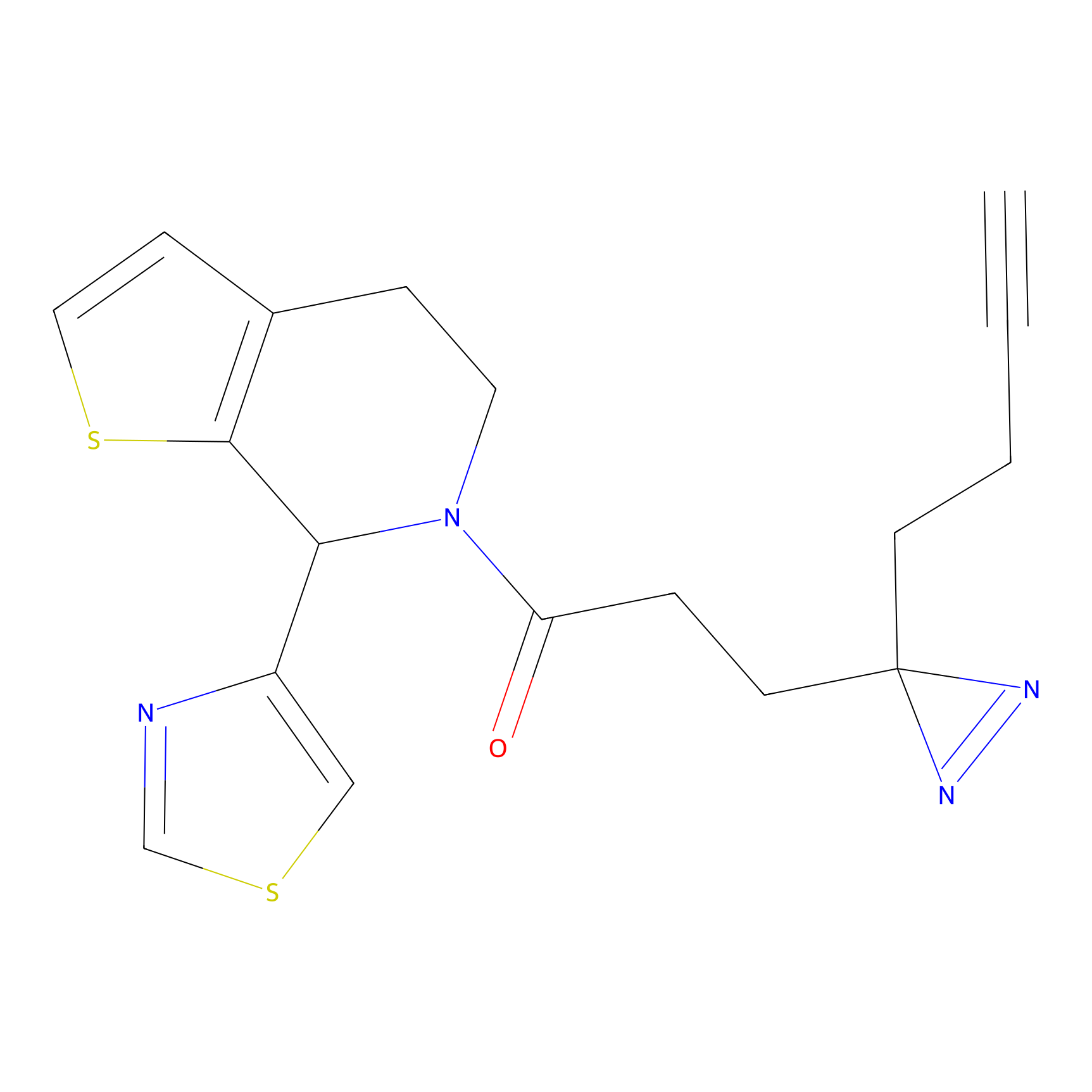
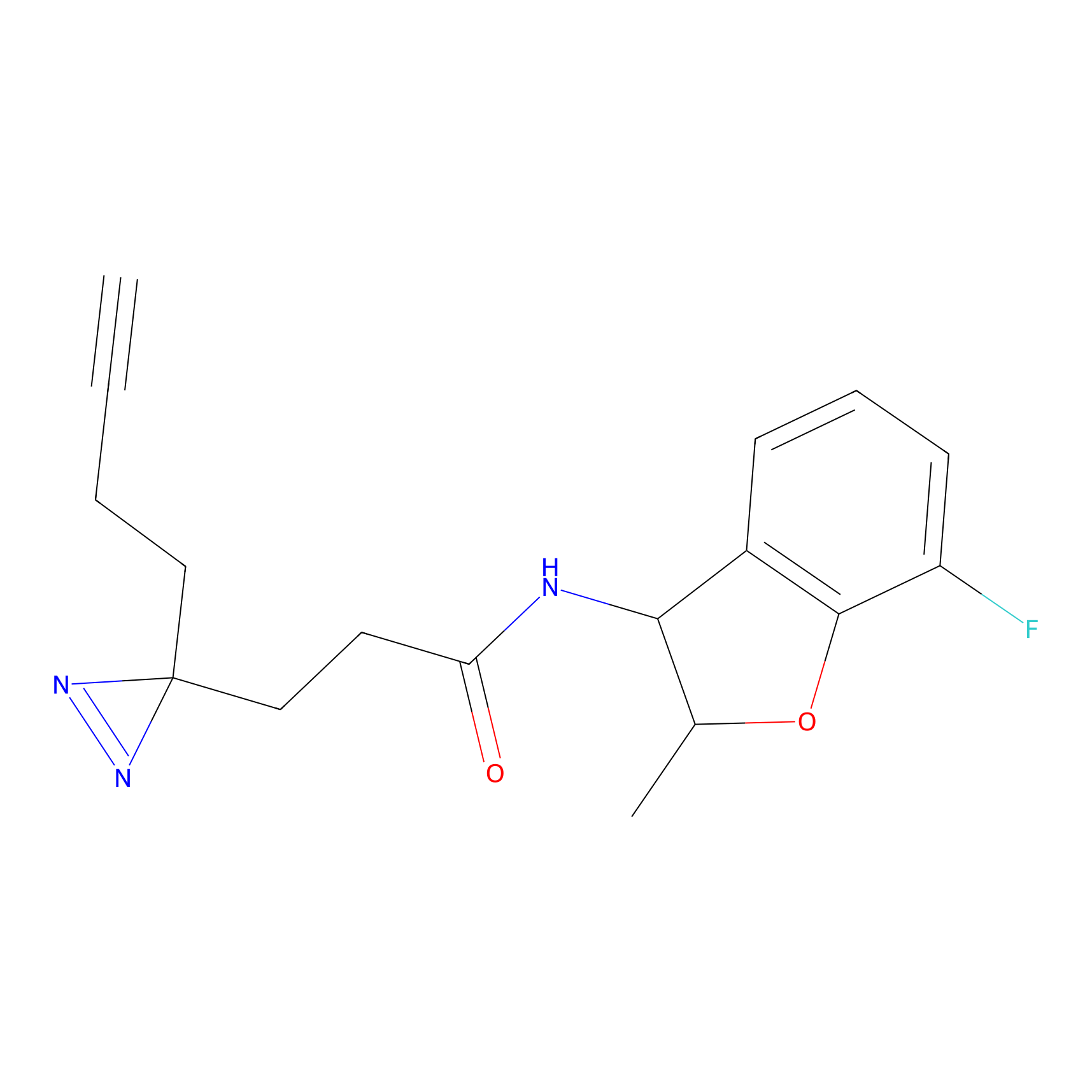
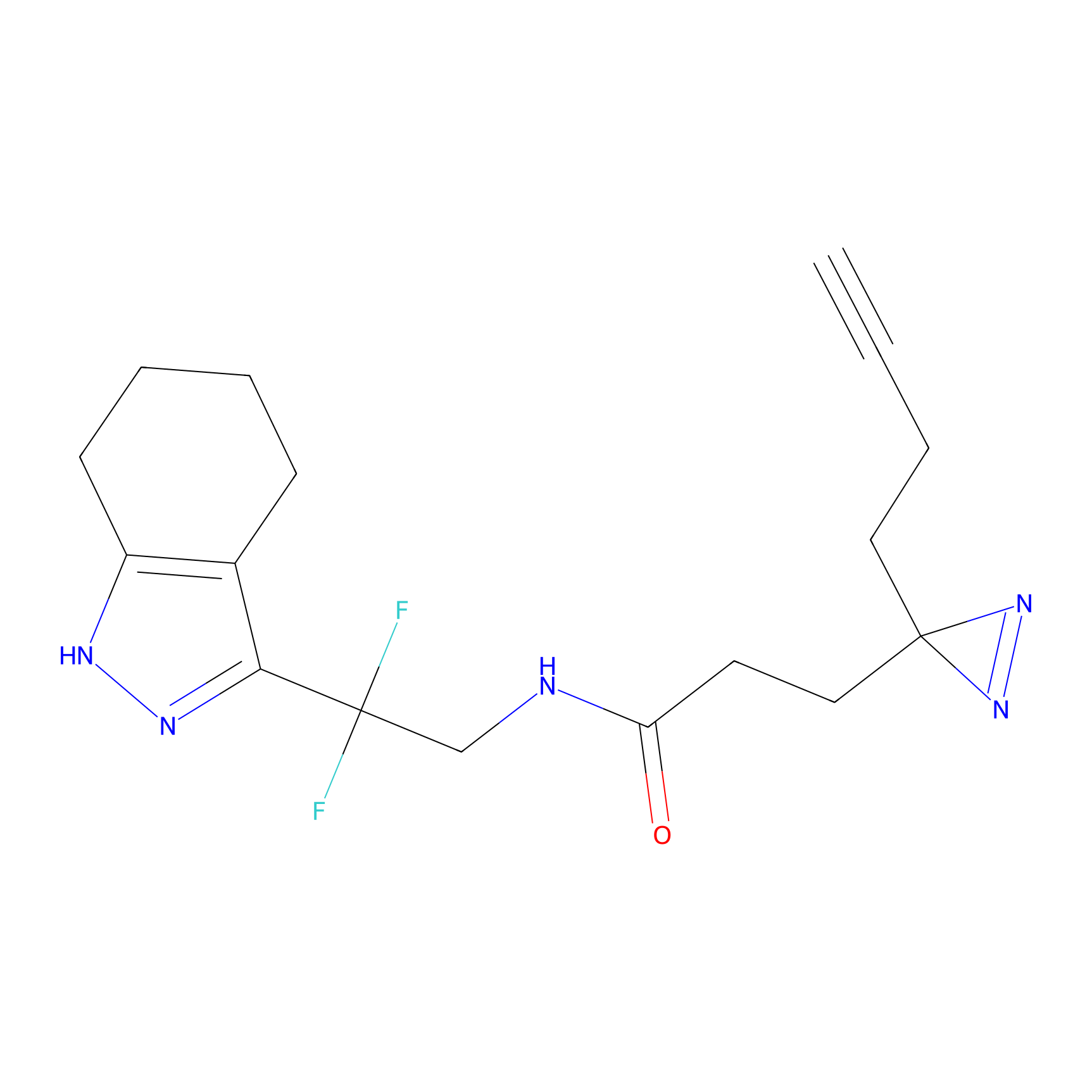
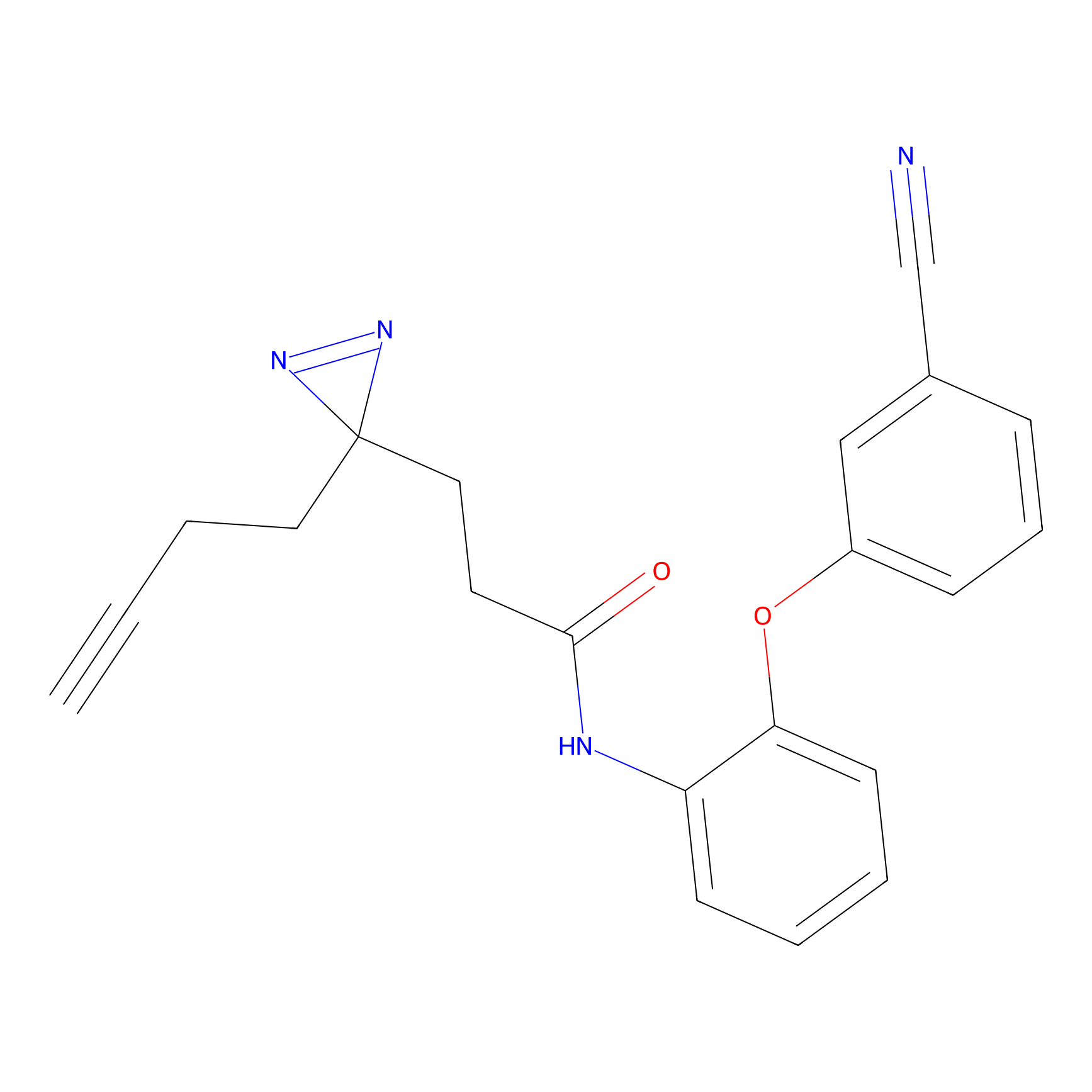
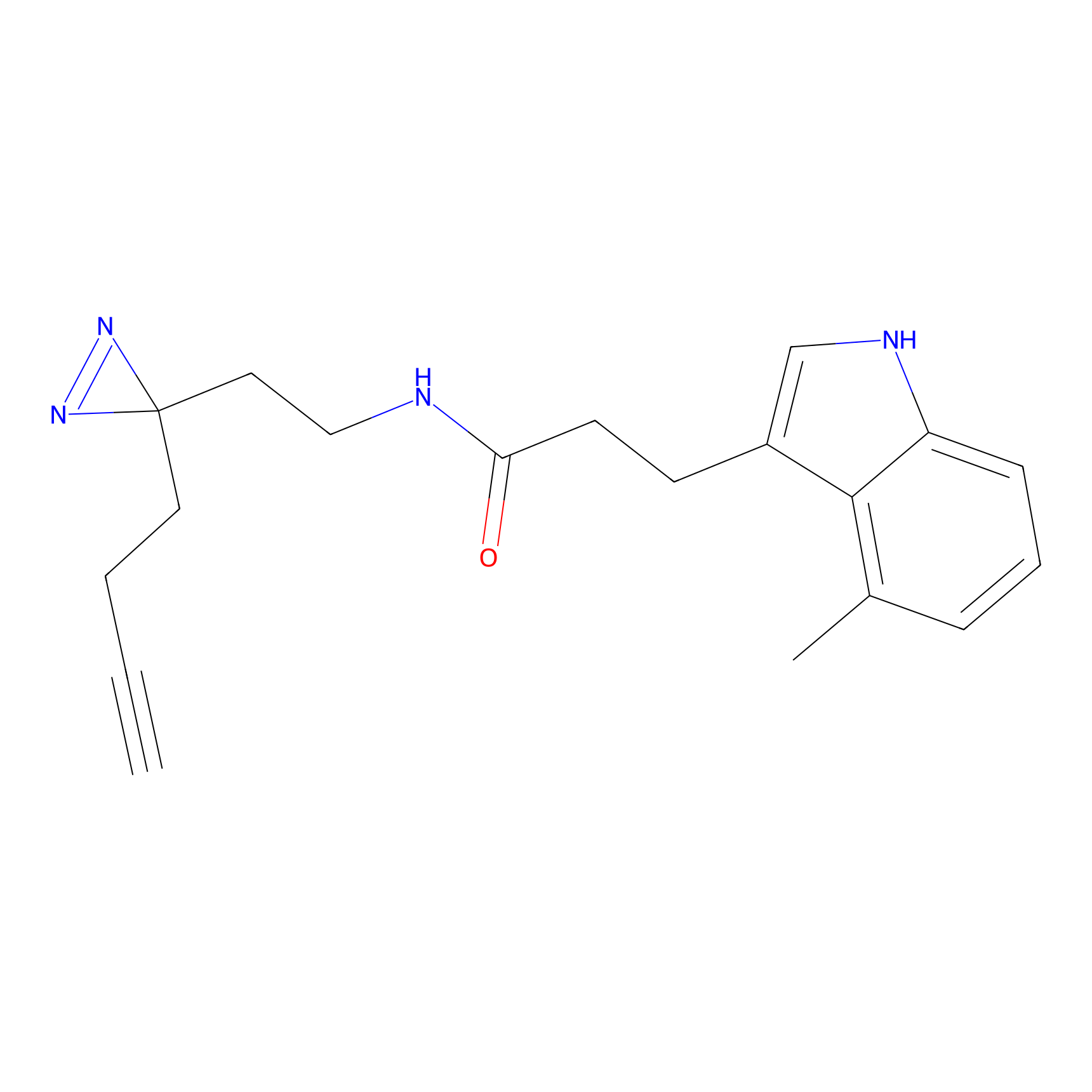
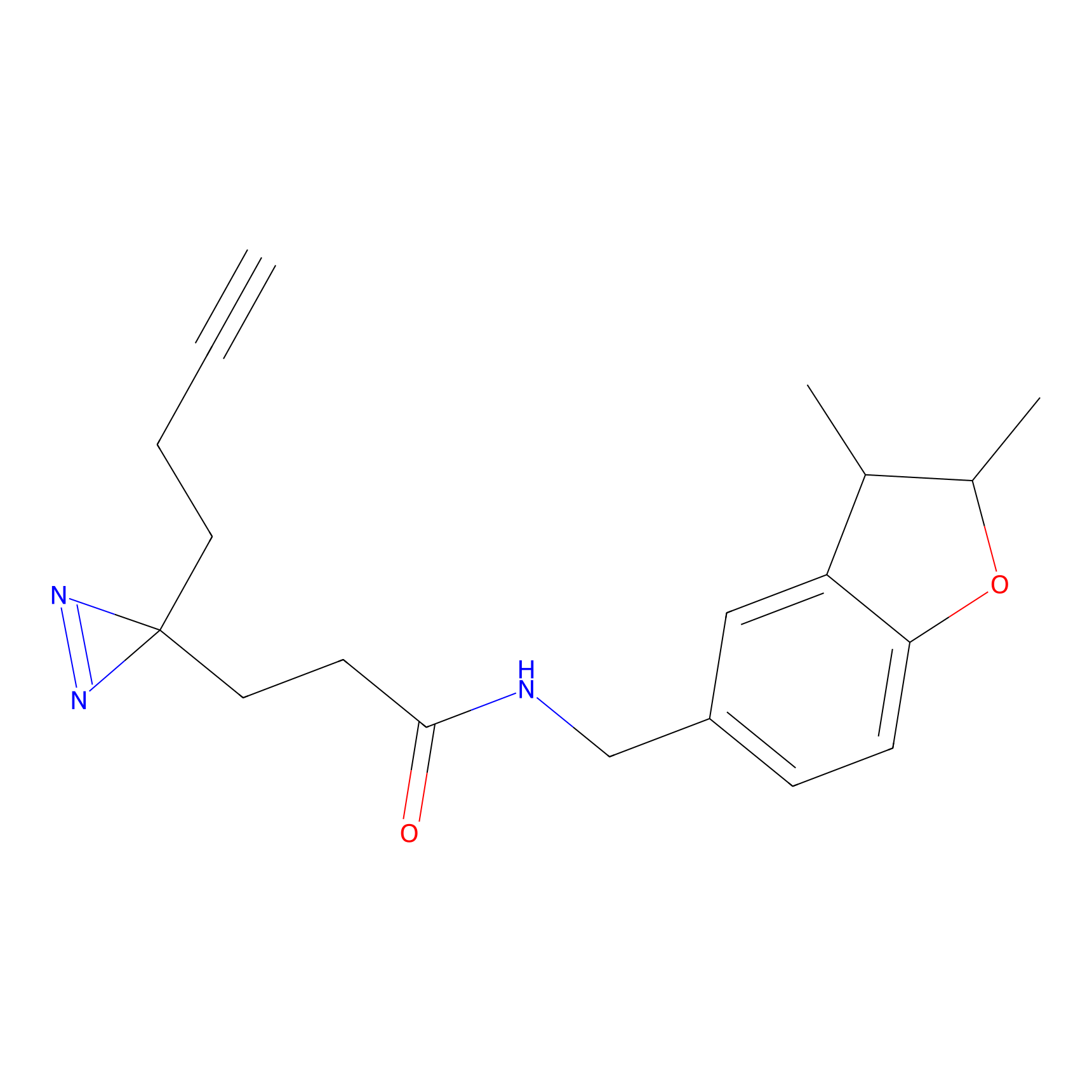
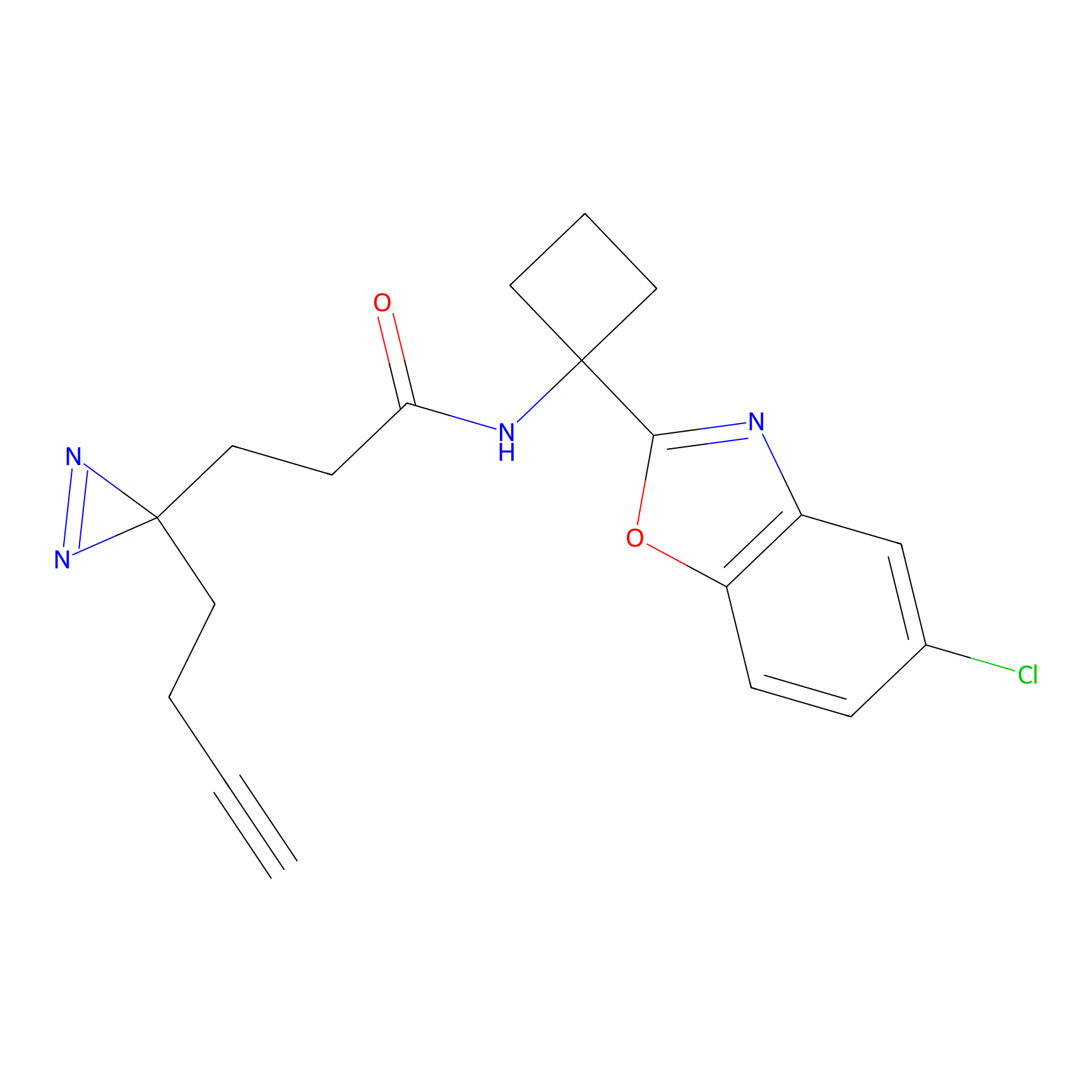
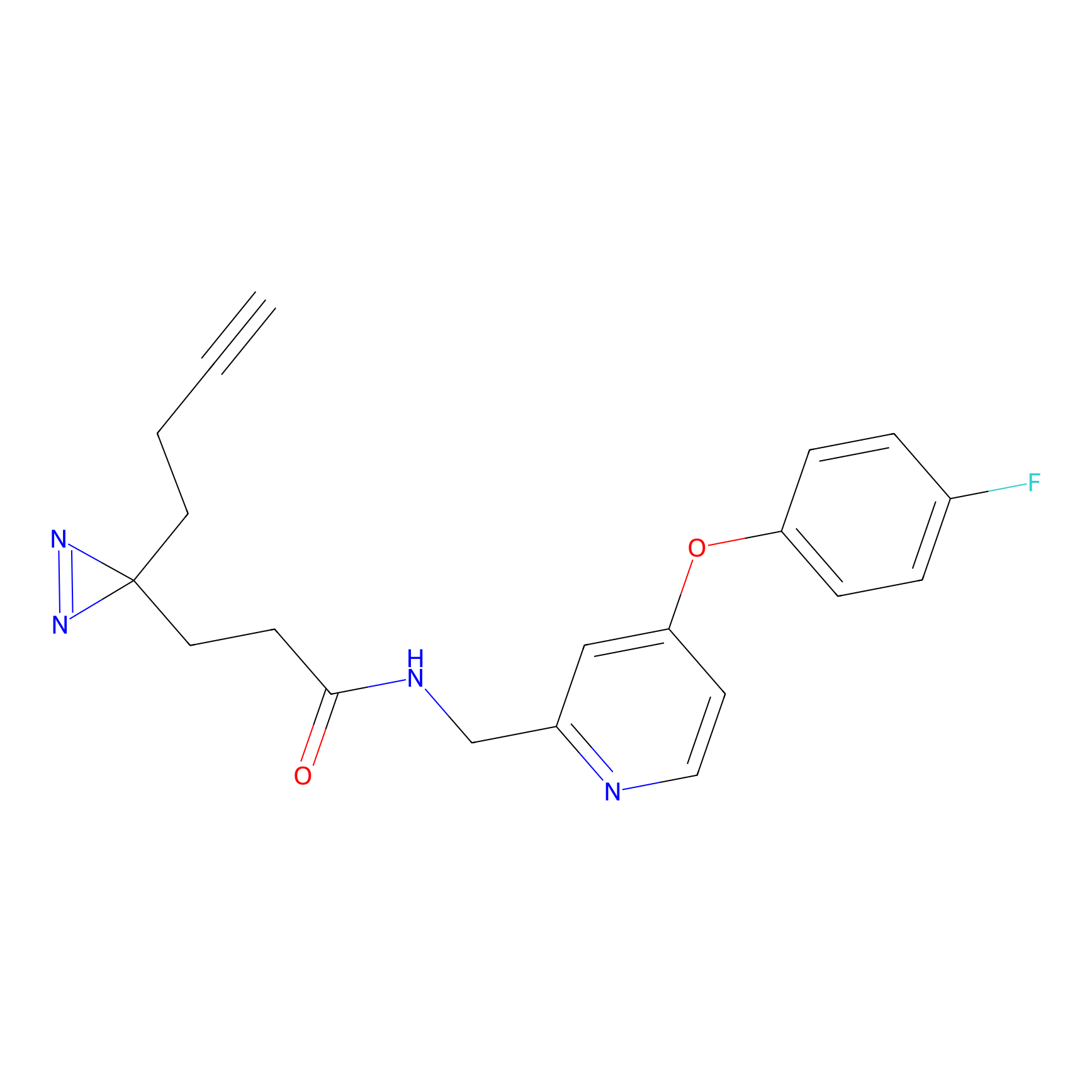
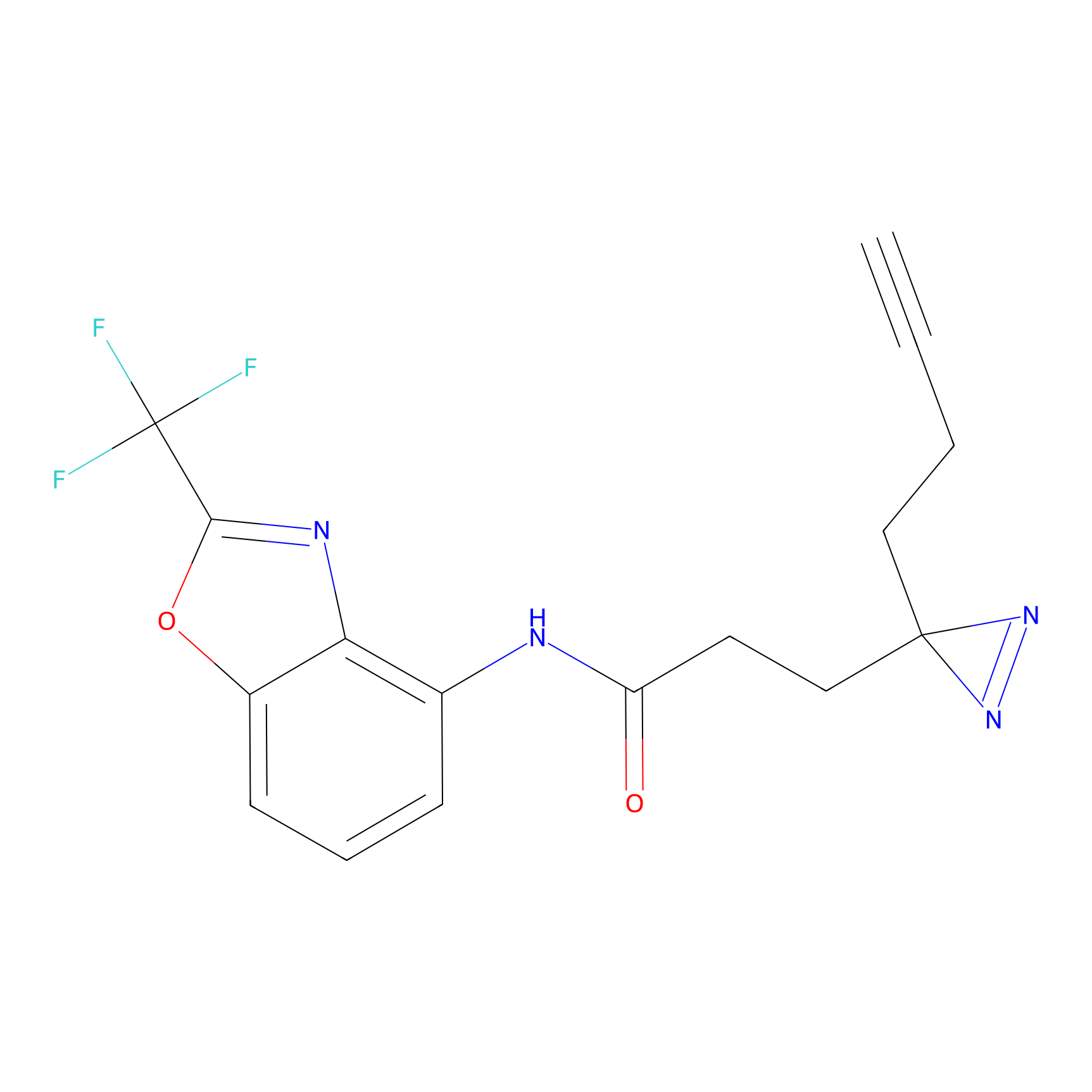
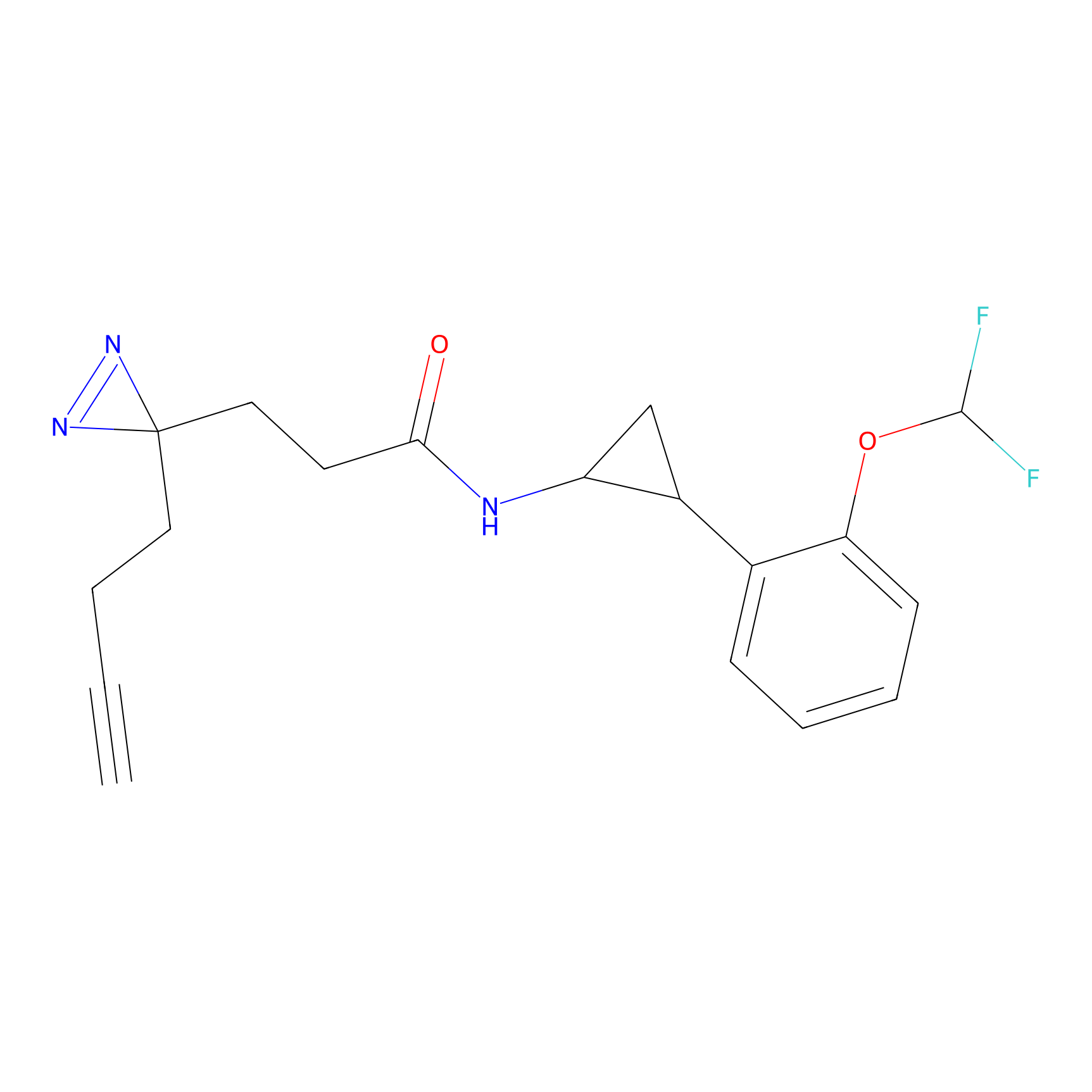
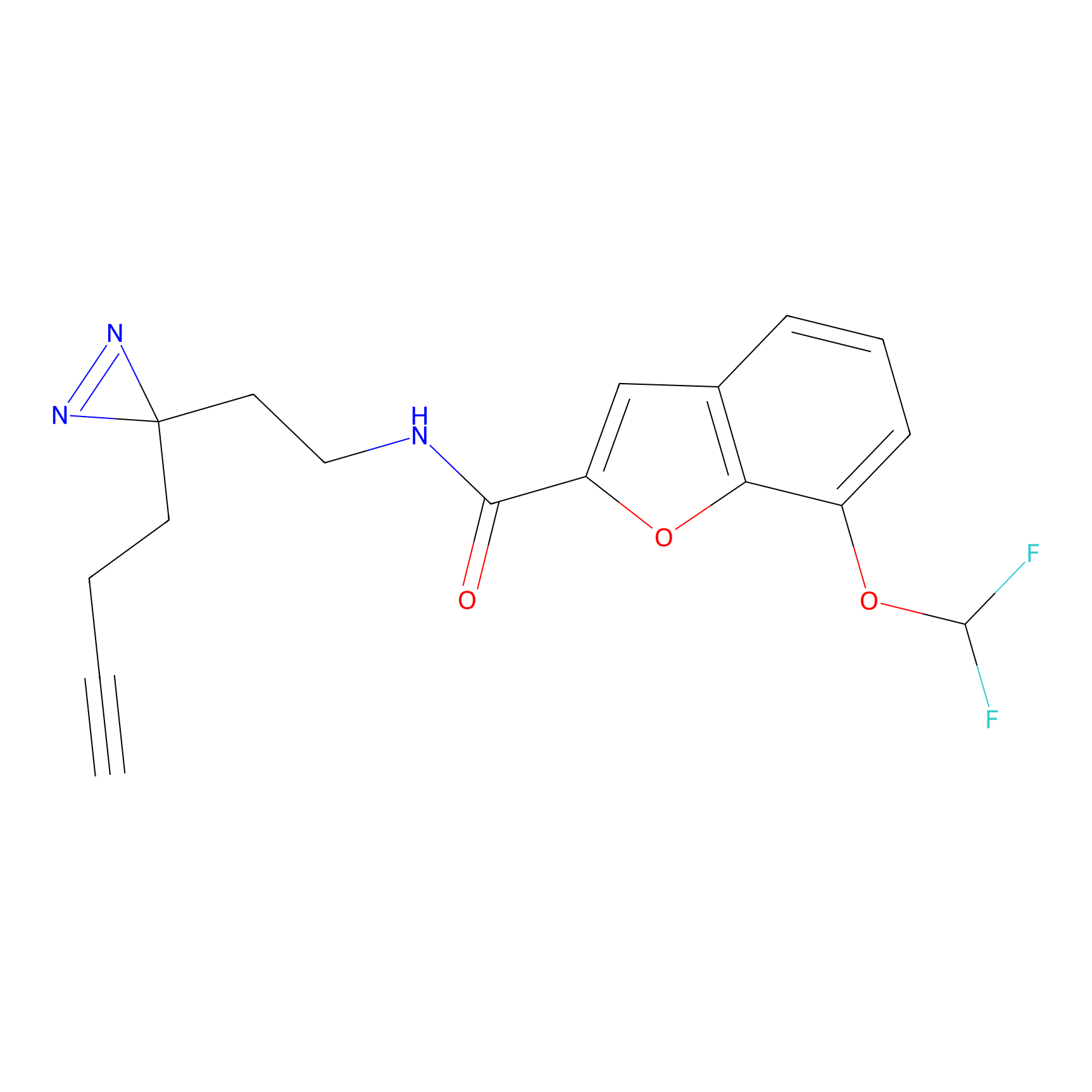
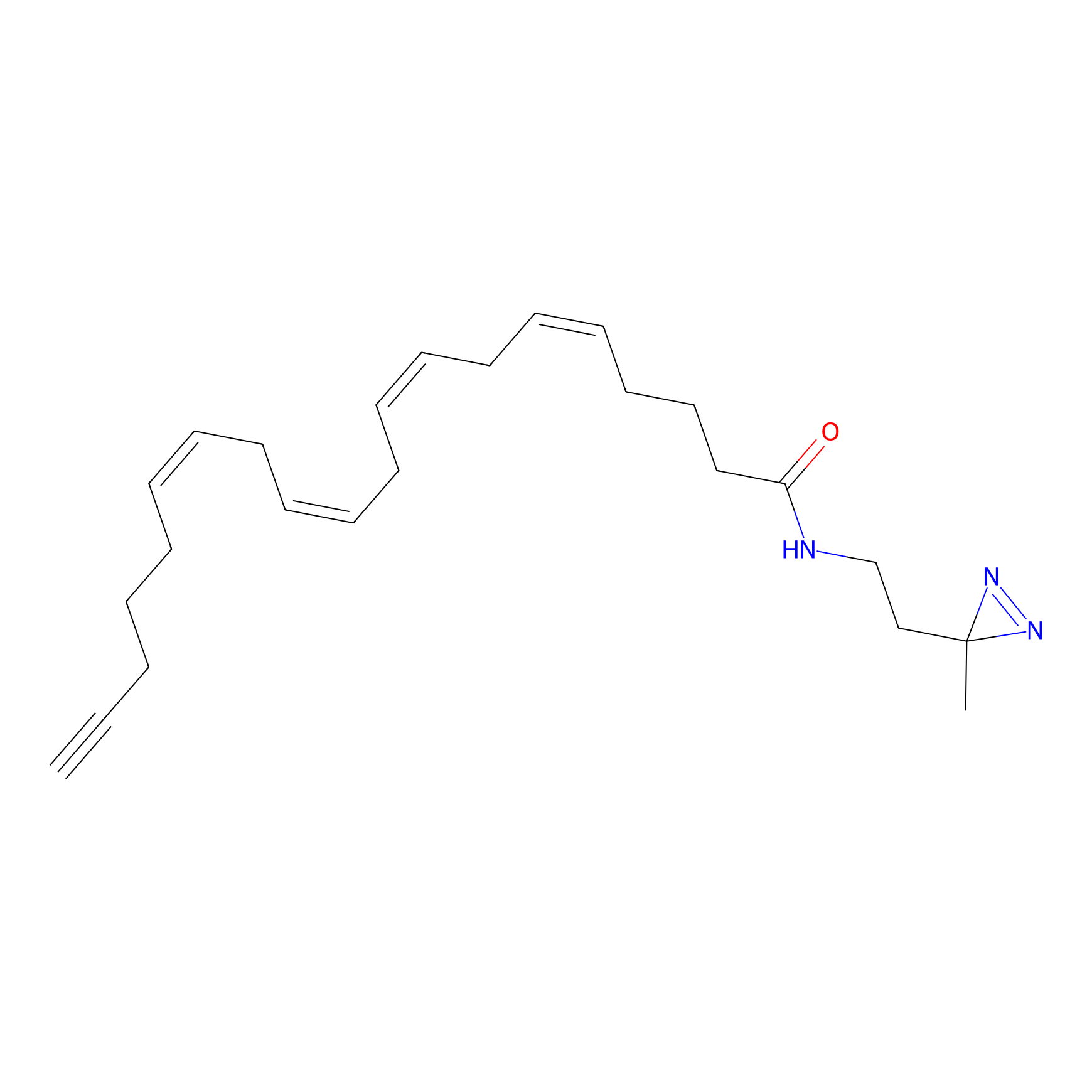
A-EBA Probe Info |
 |
3.99 | LDD0215 | [2] | |
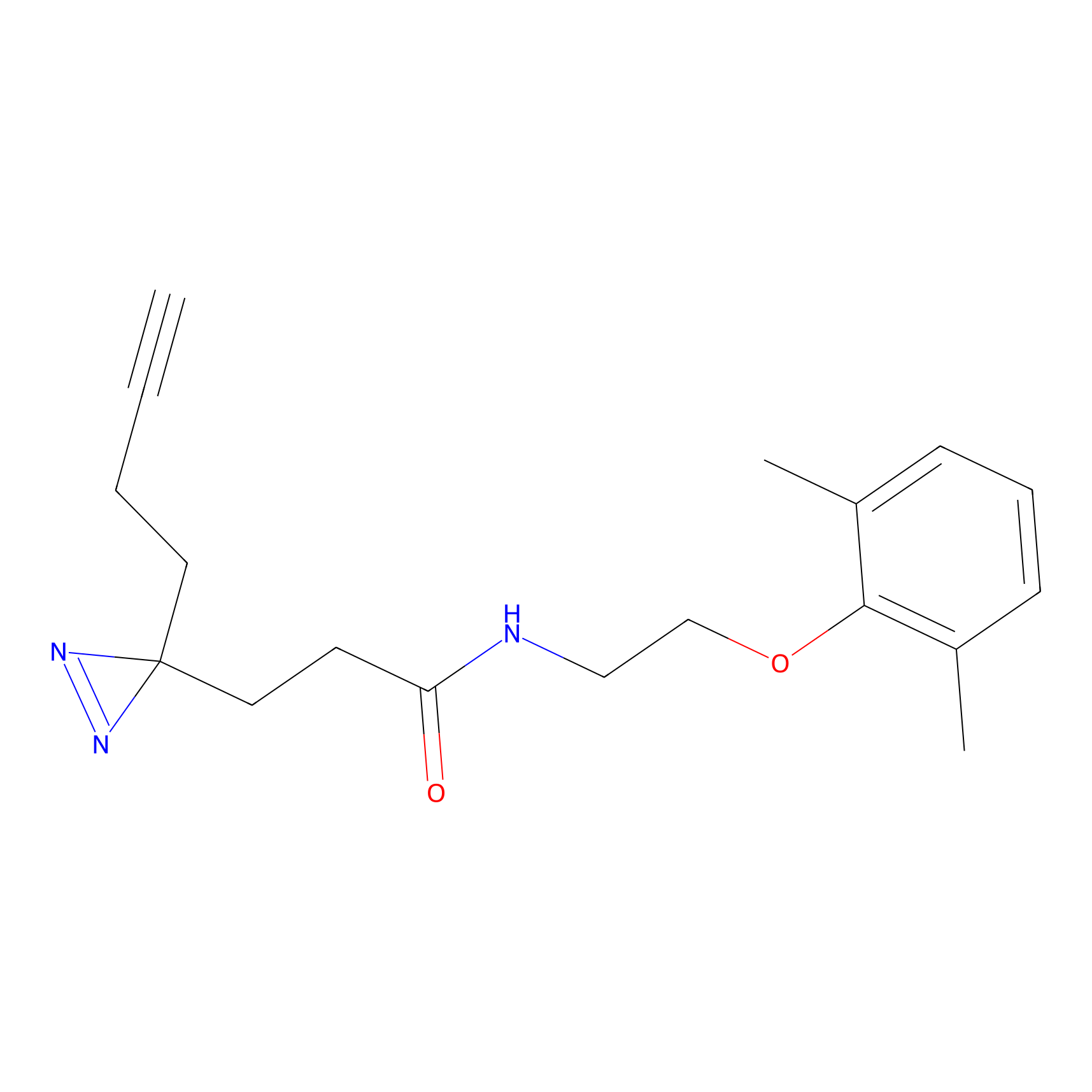
|
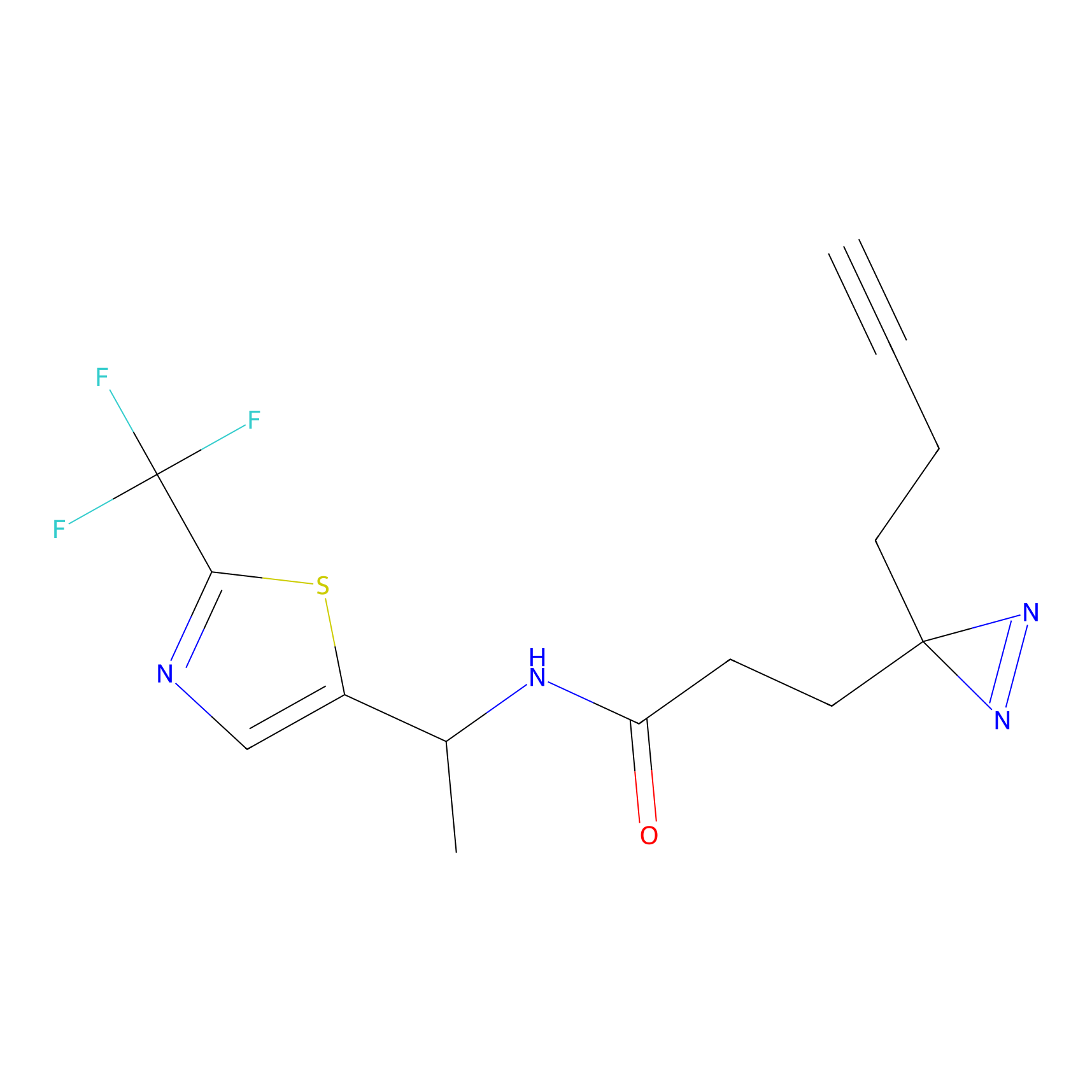
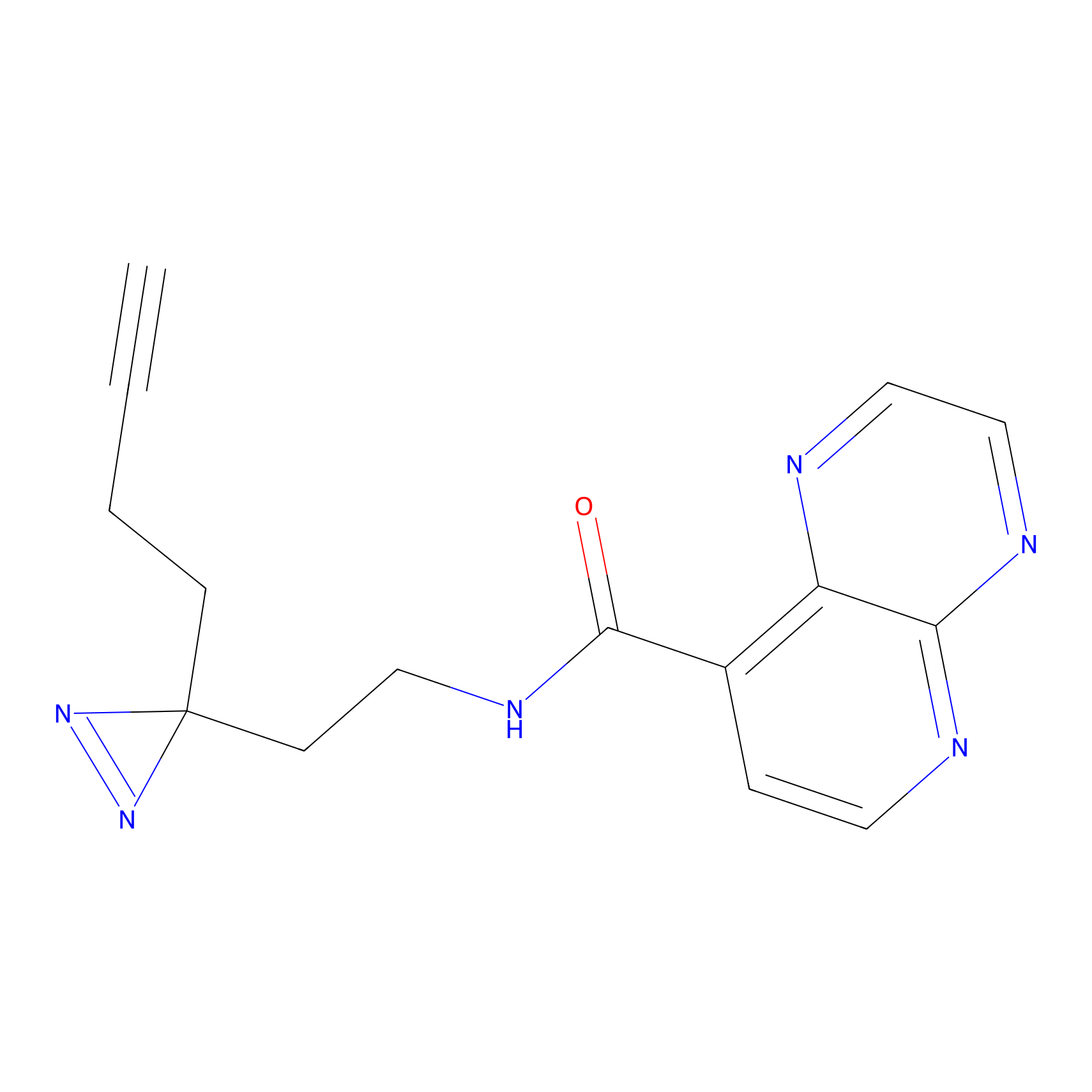
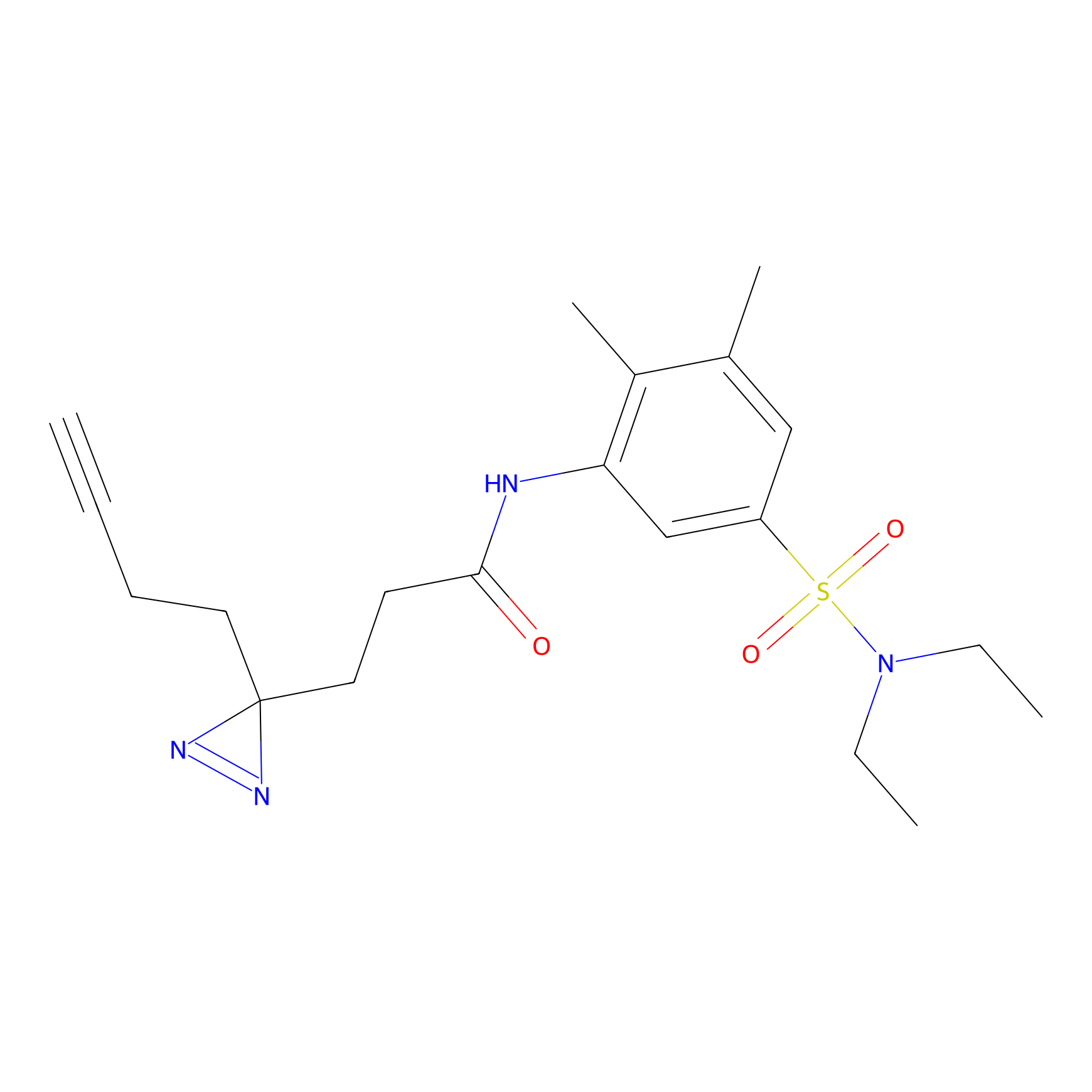
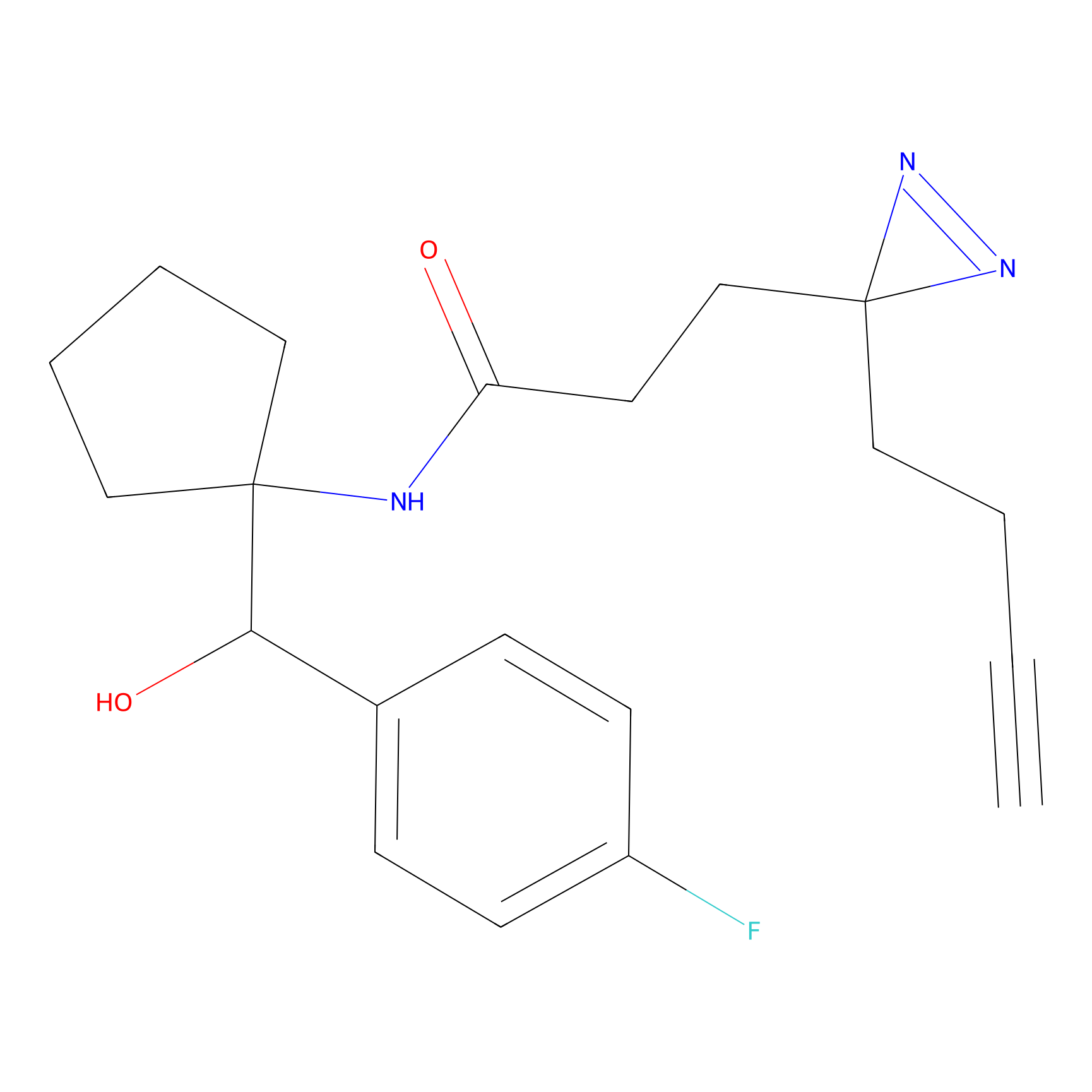
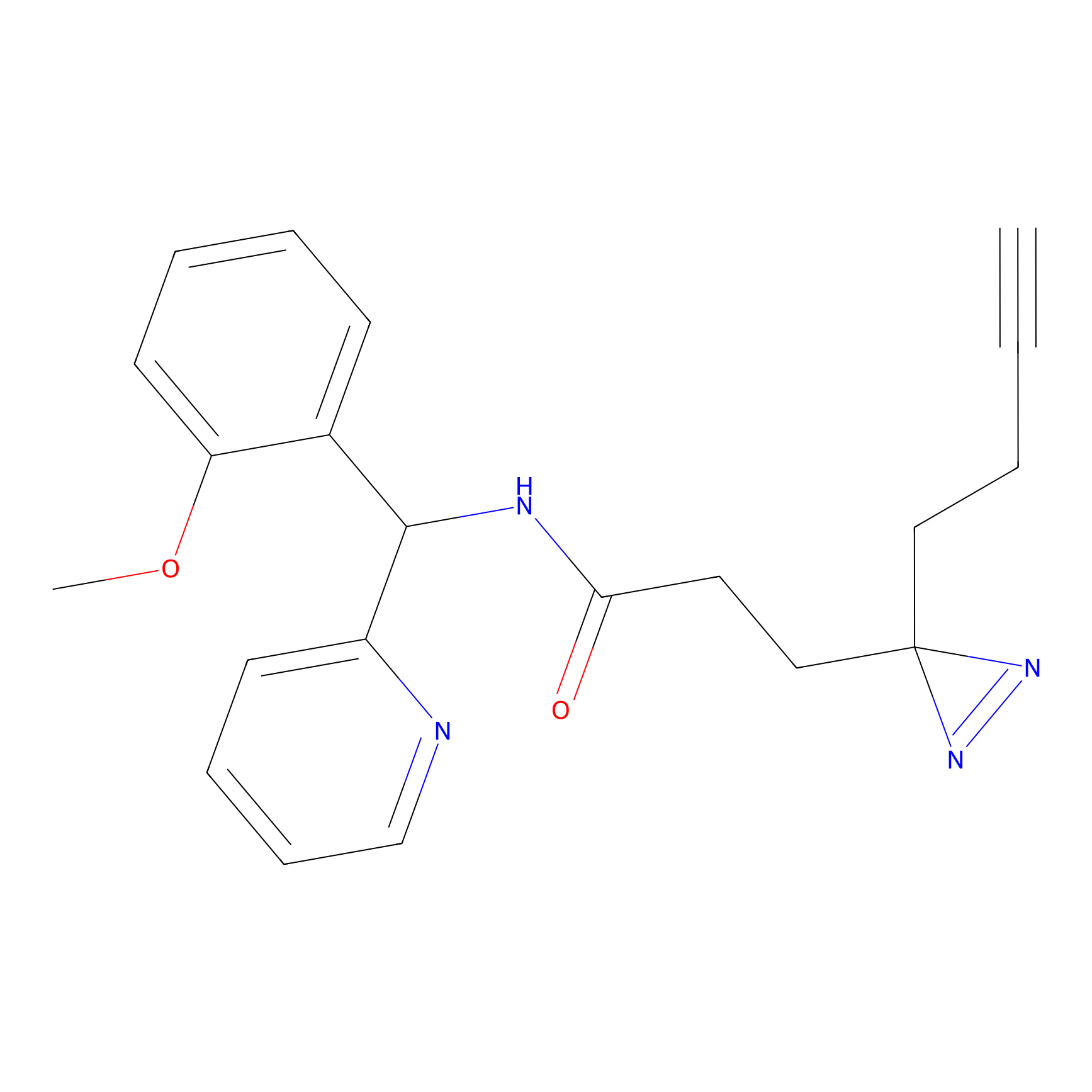
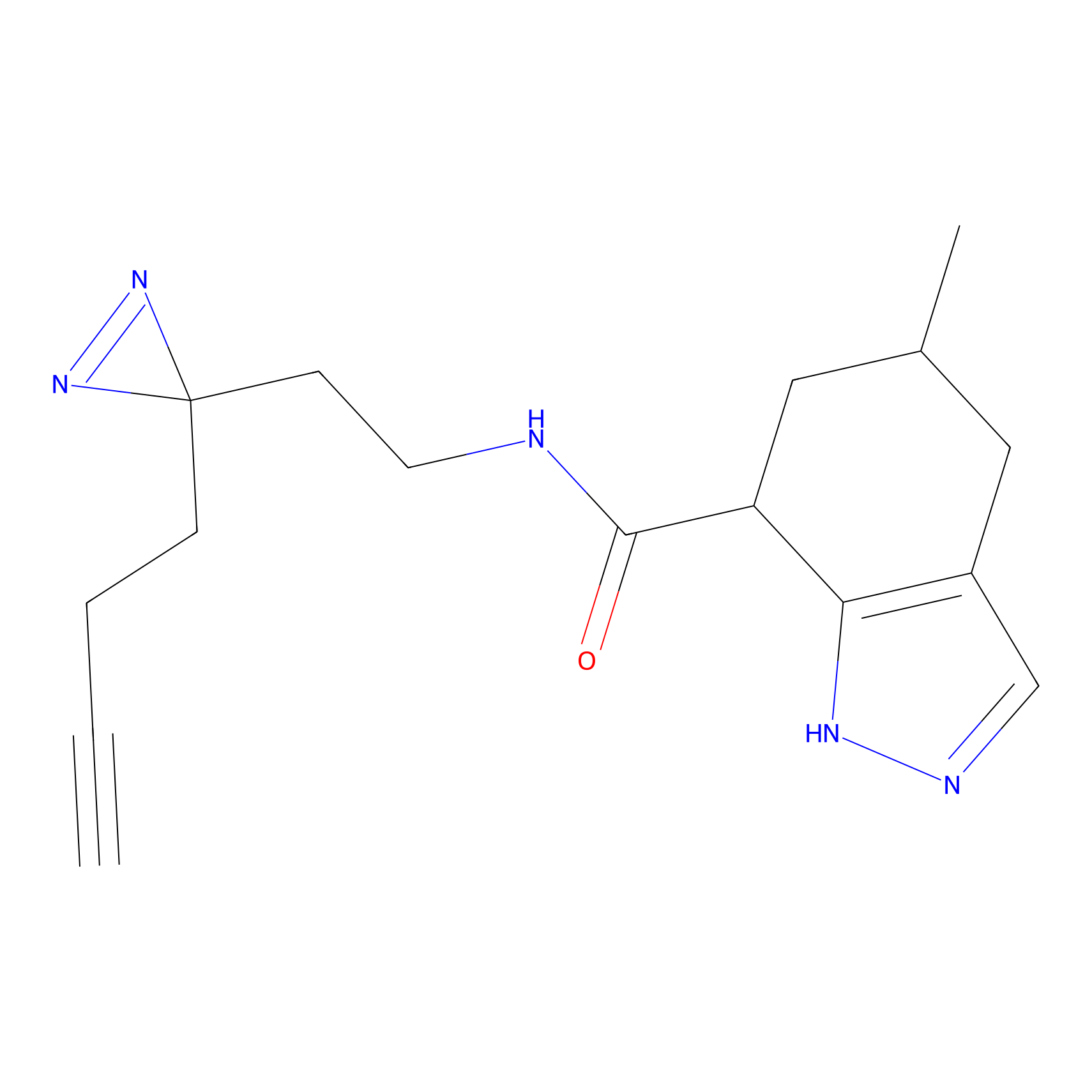
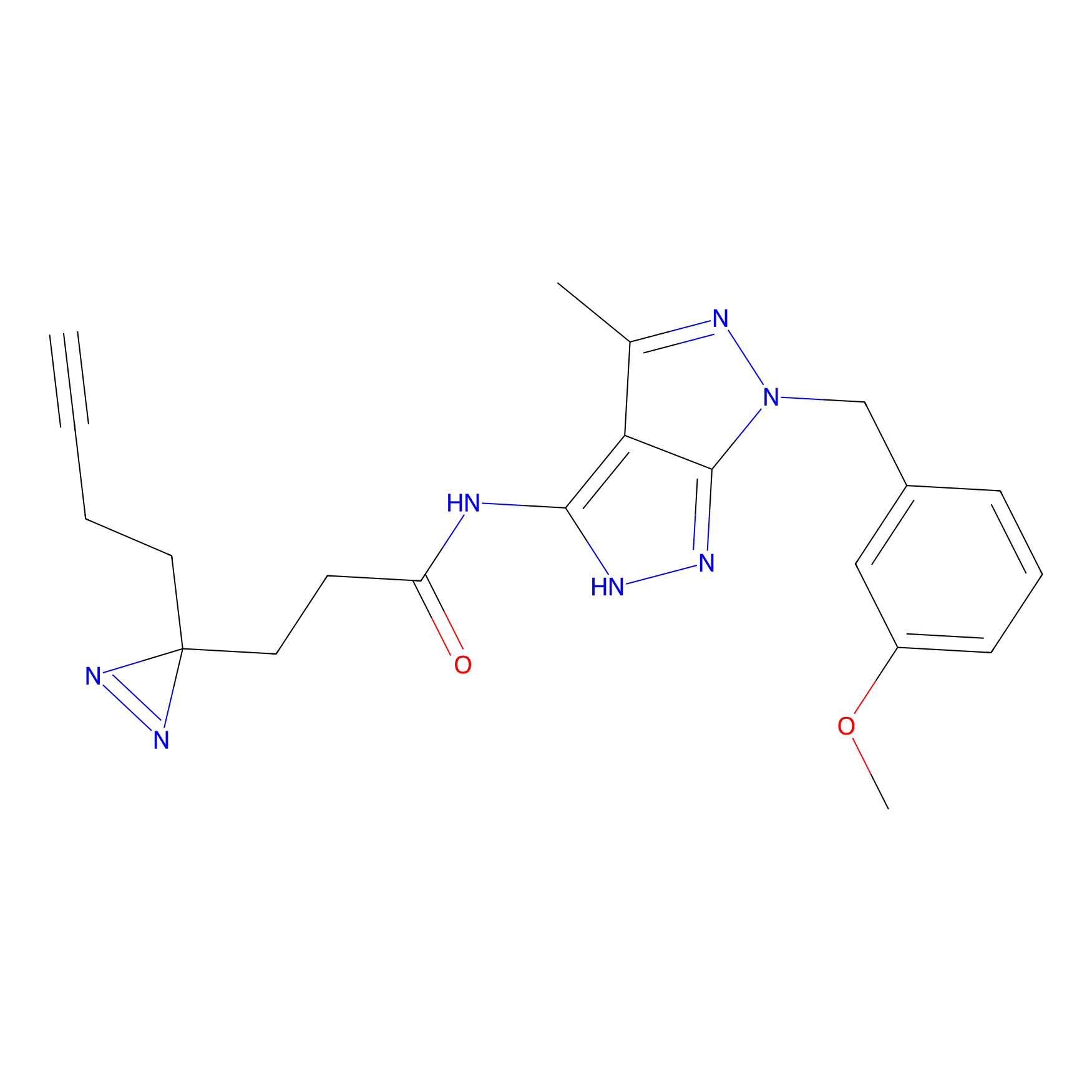
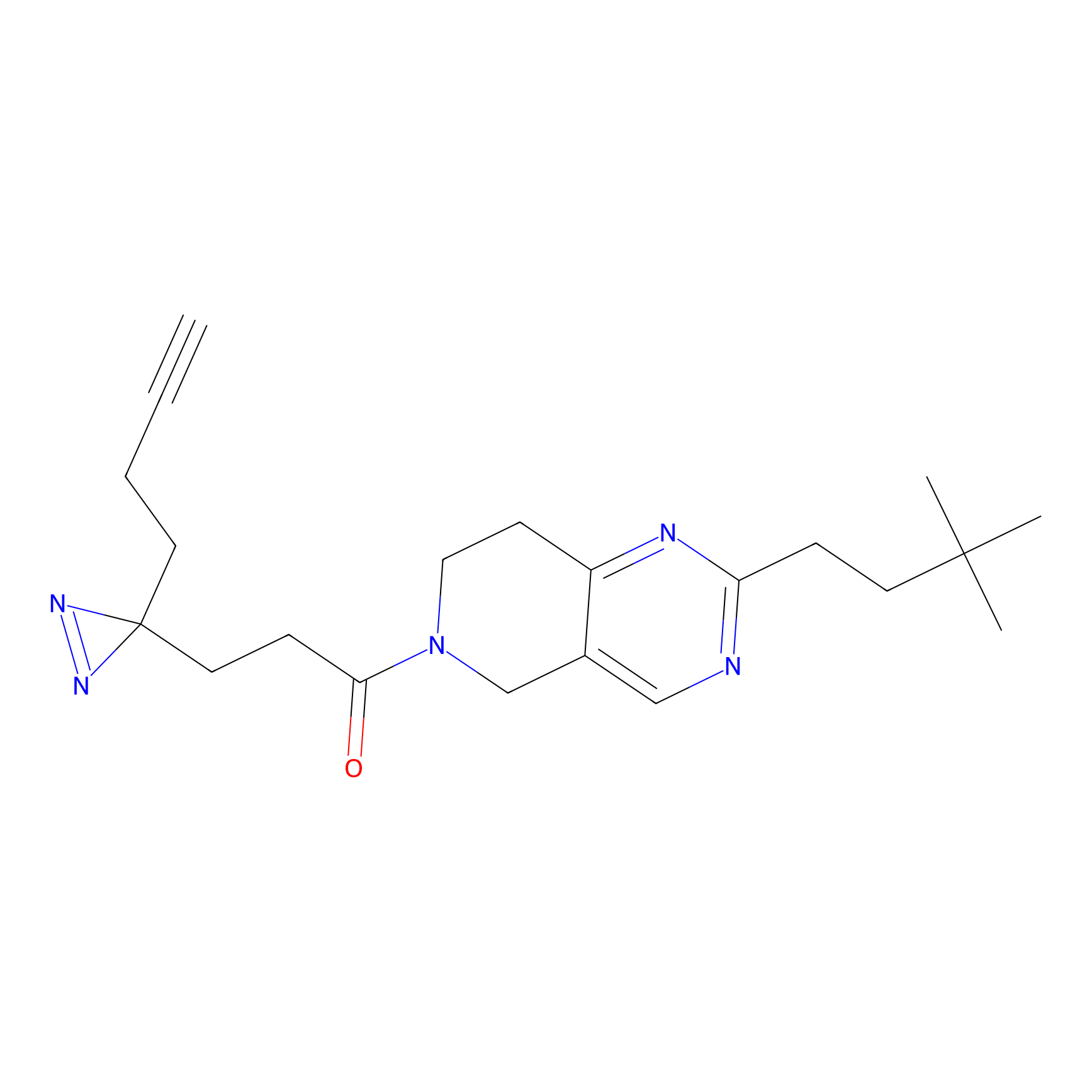
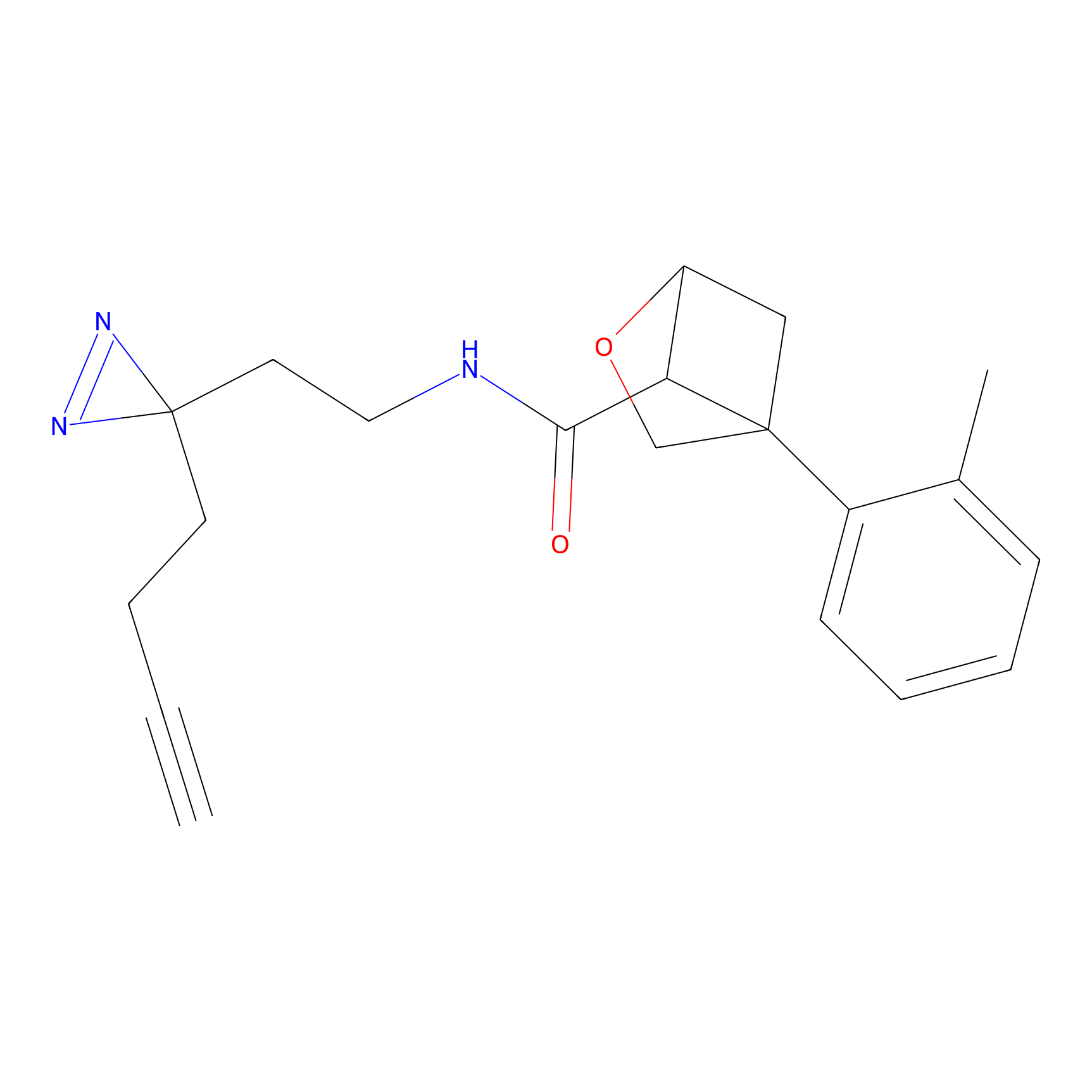
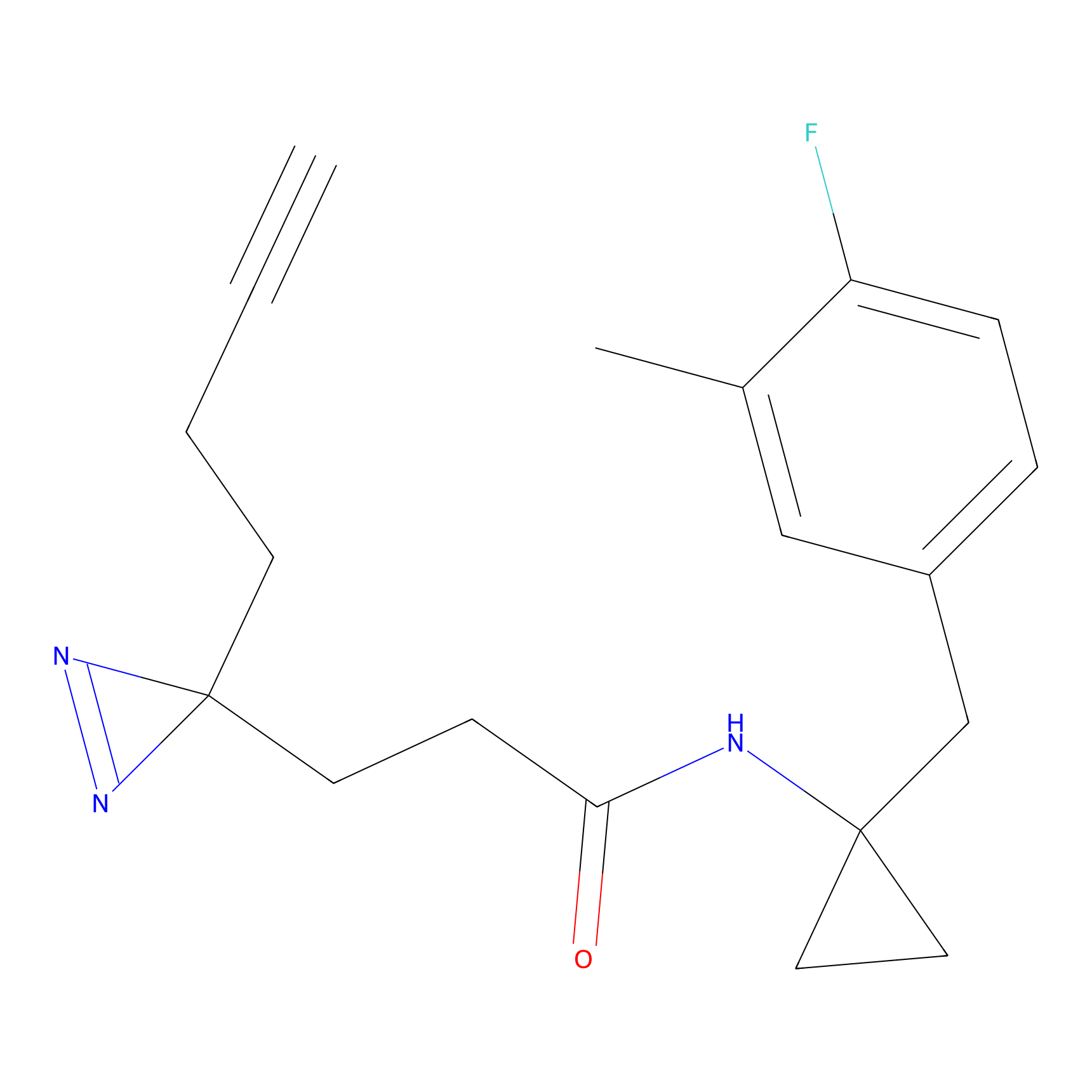
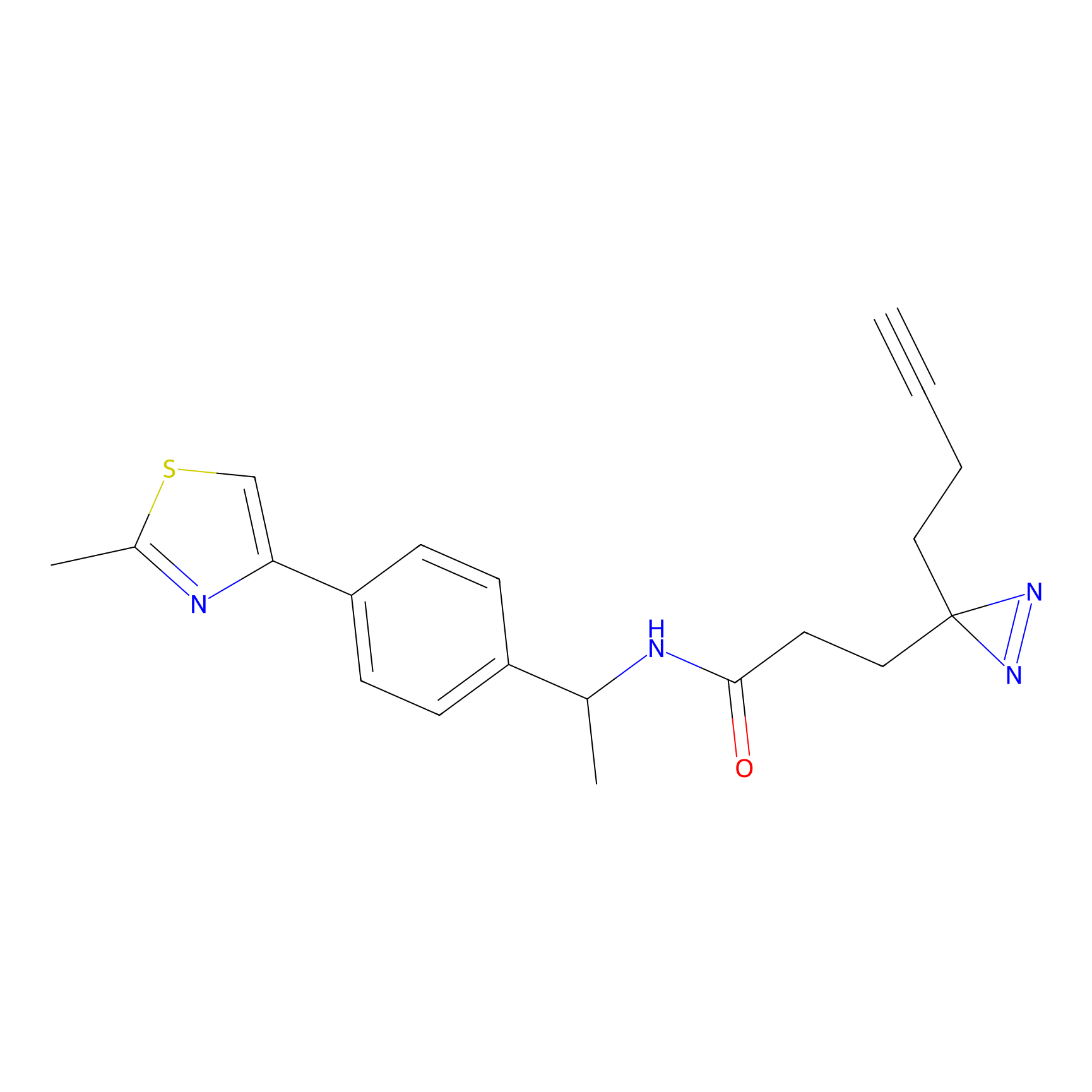
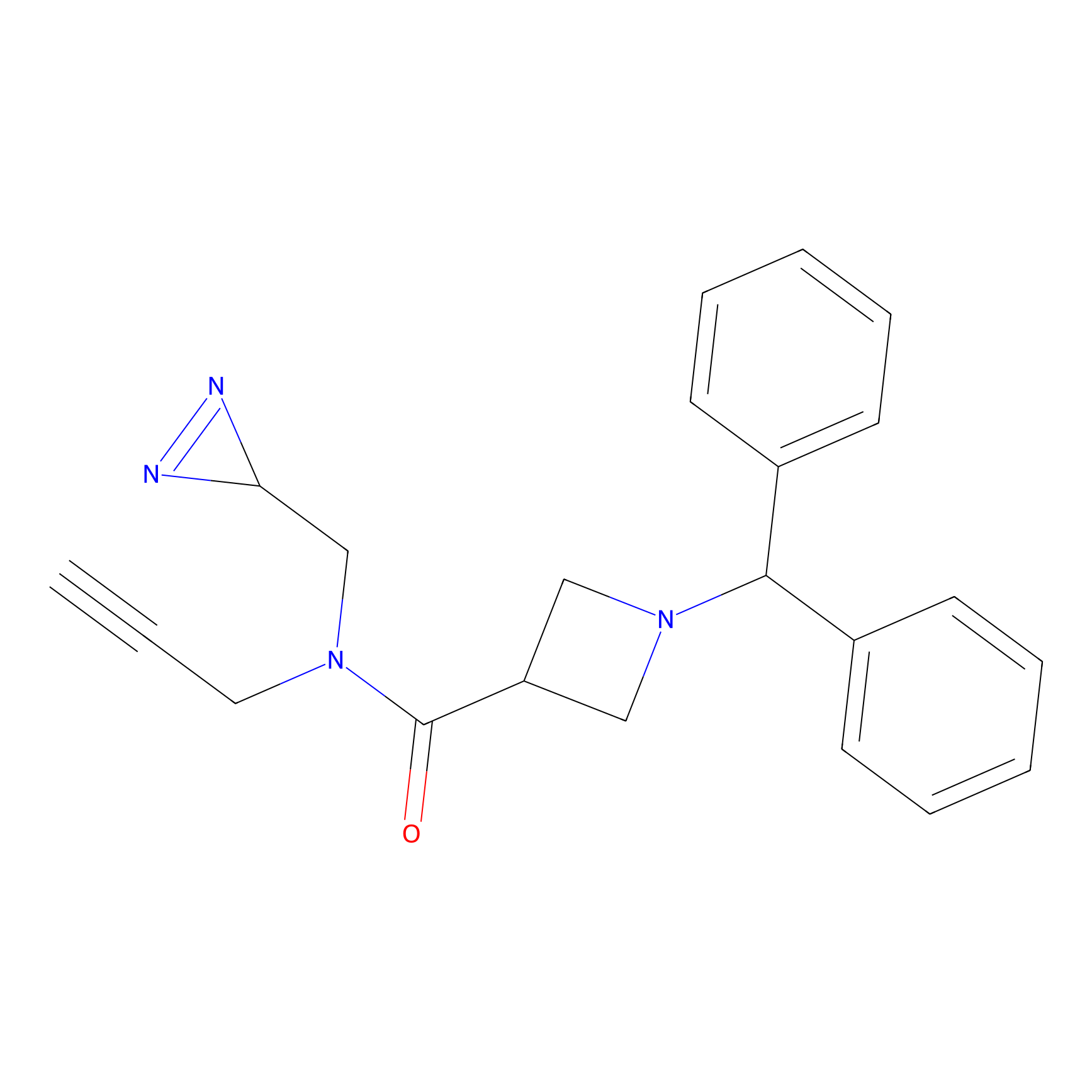
FBPP2 Probe Info |
 |
2.02 | LDD0318 | [3] | |
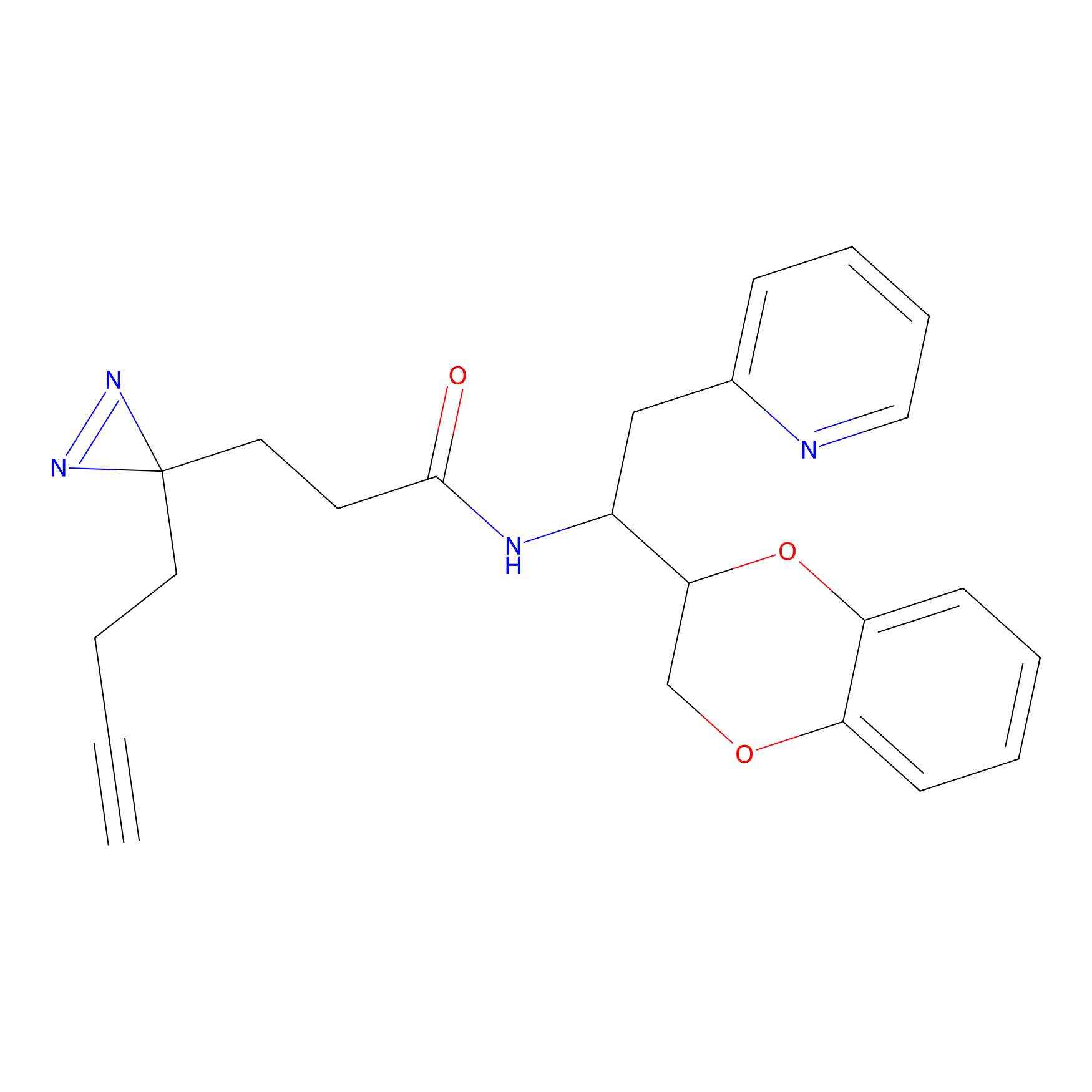
|
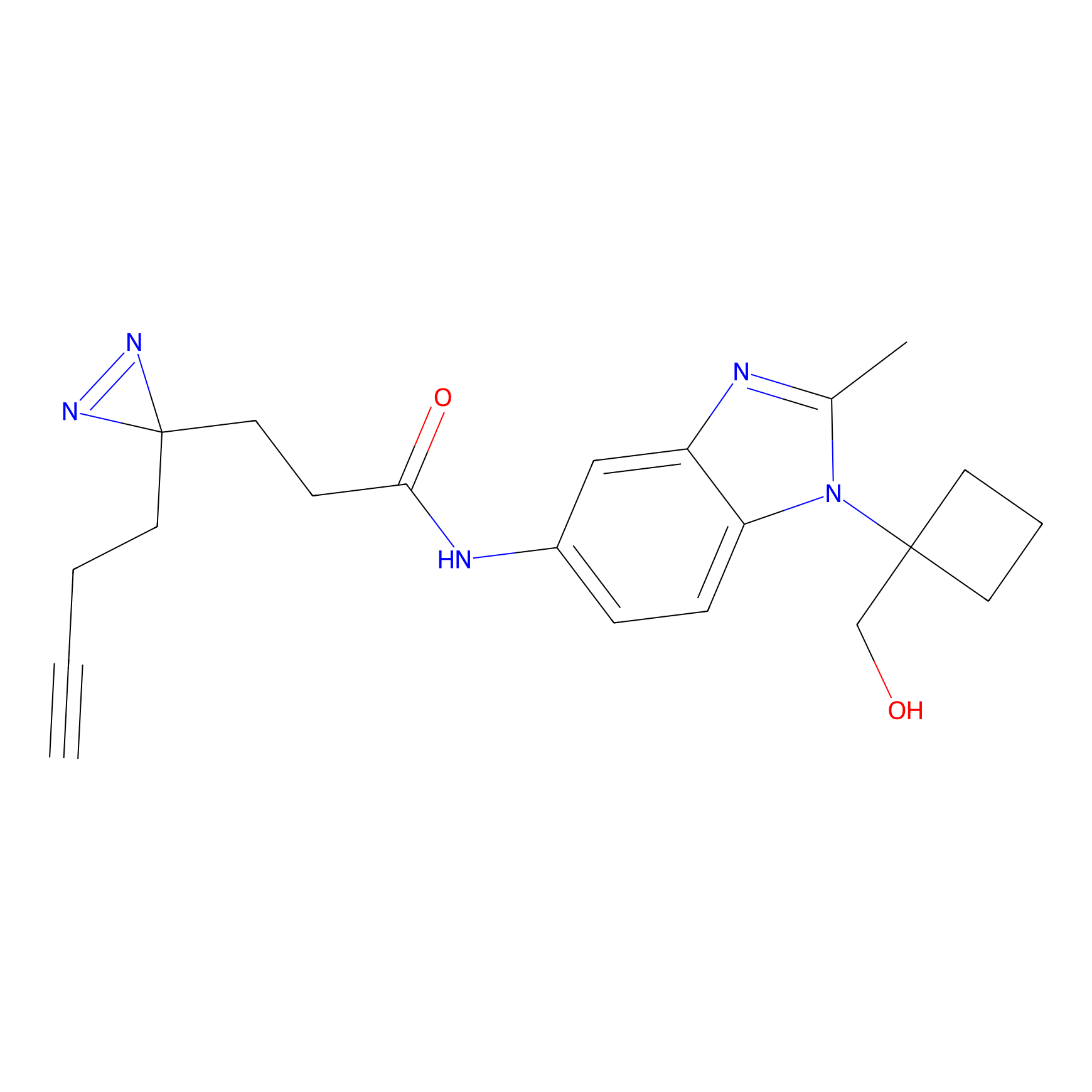
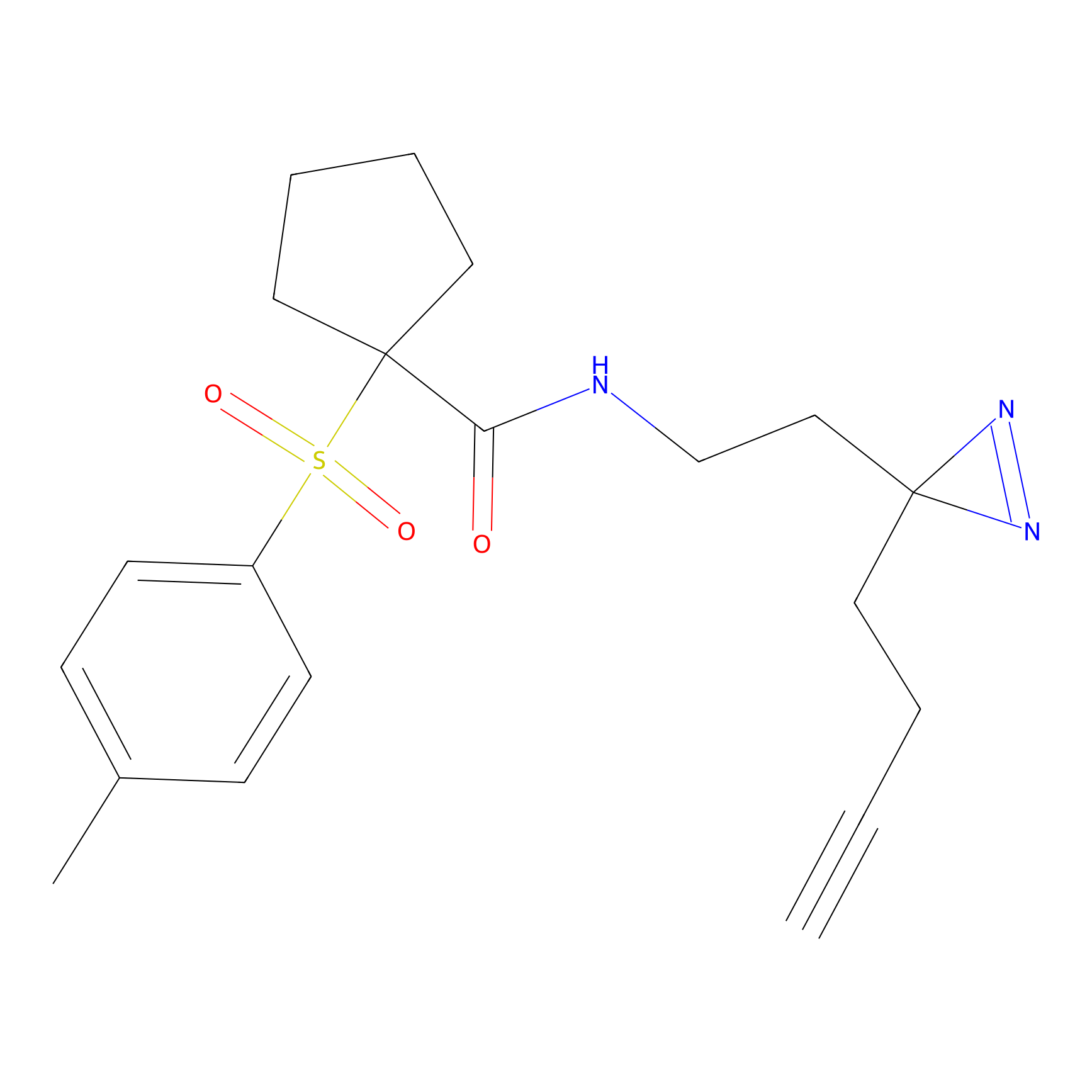
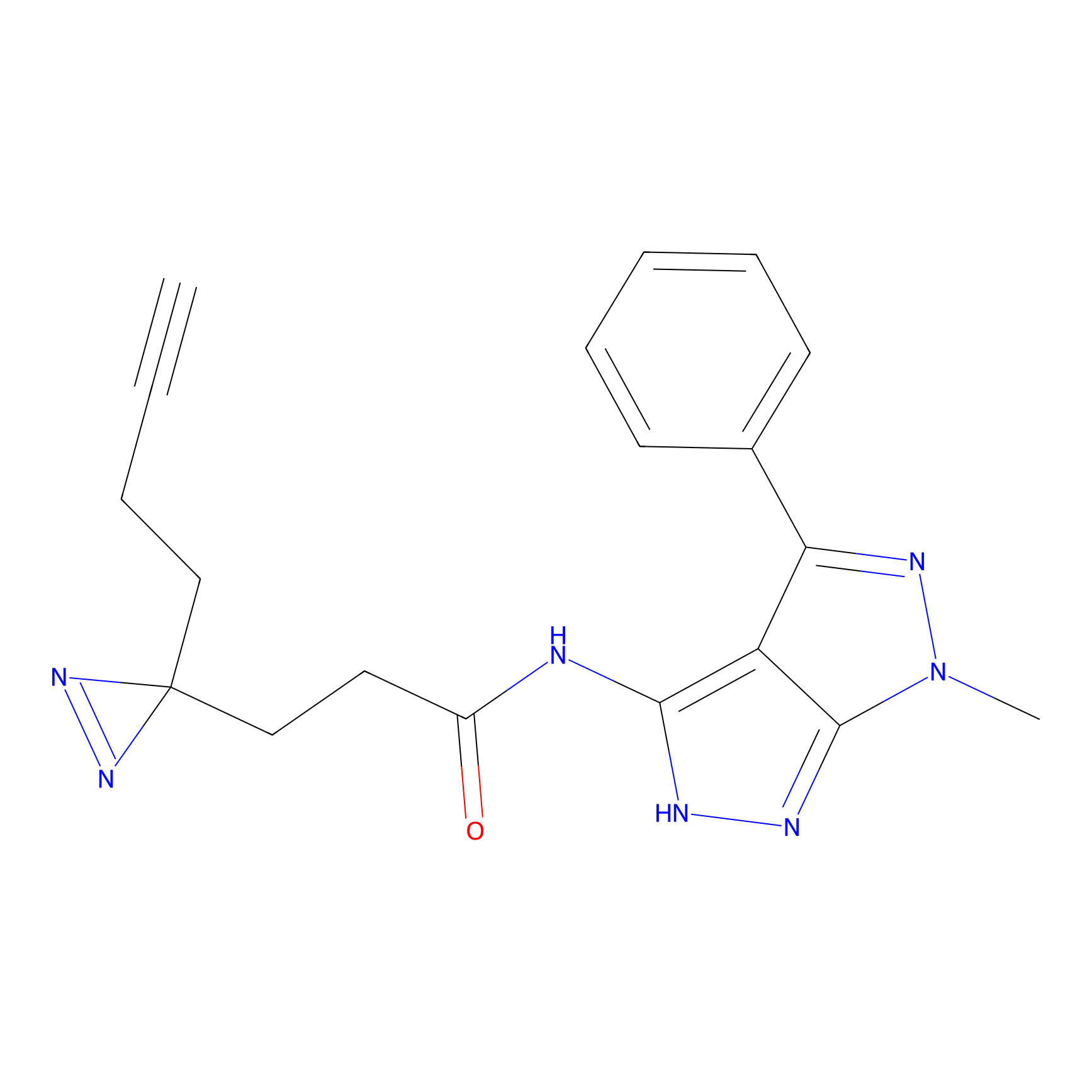
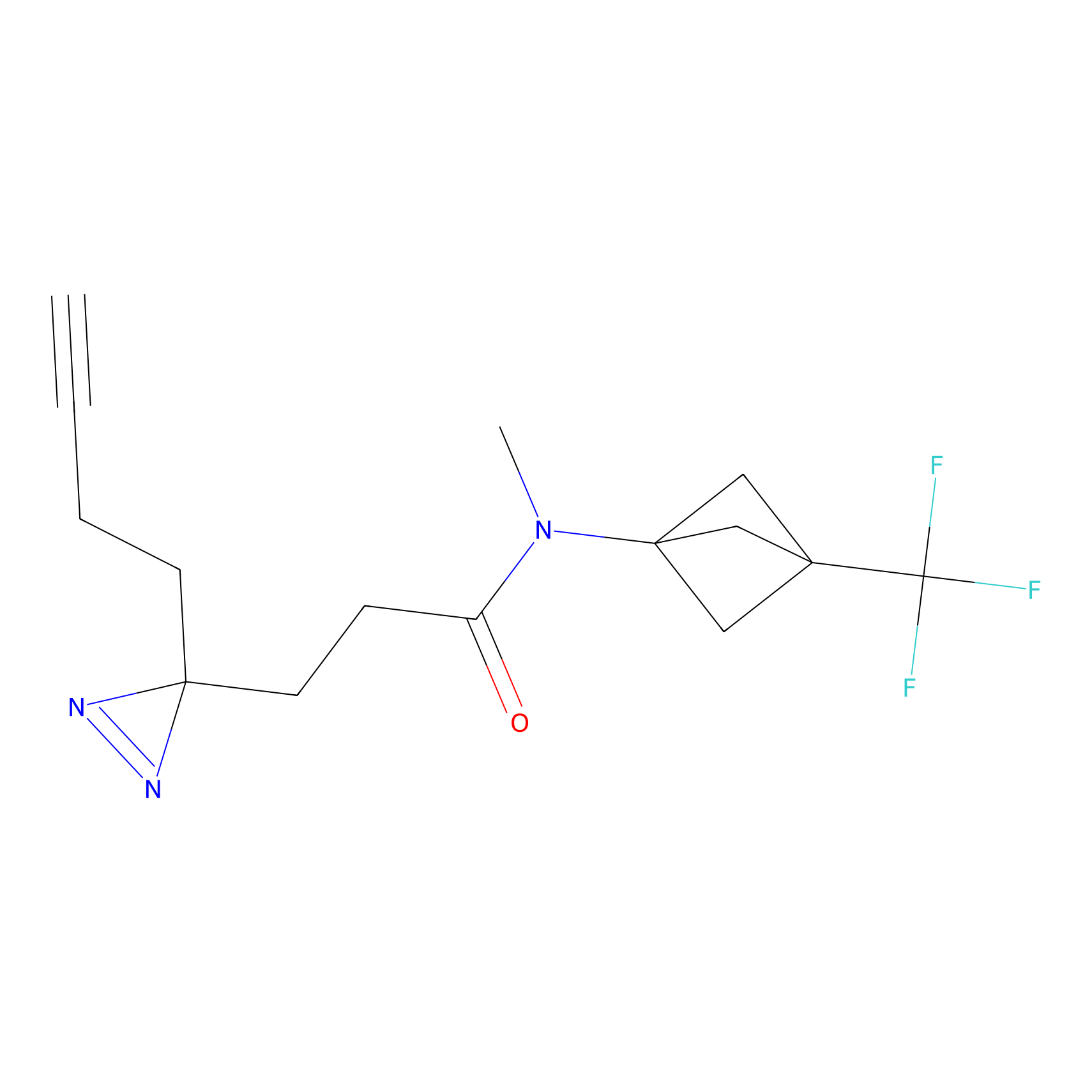
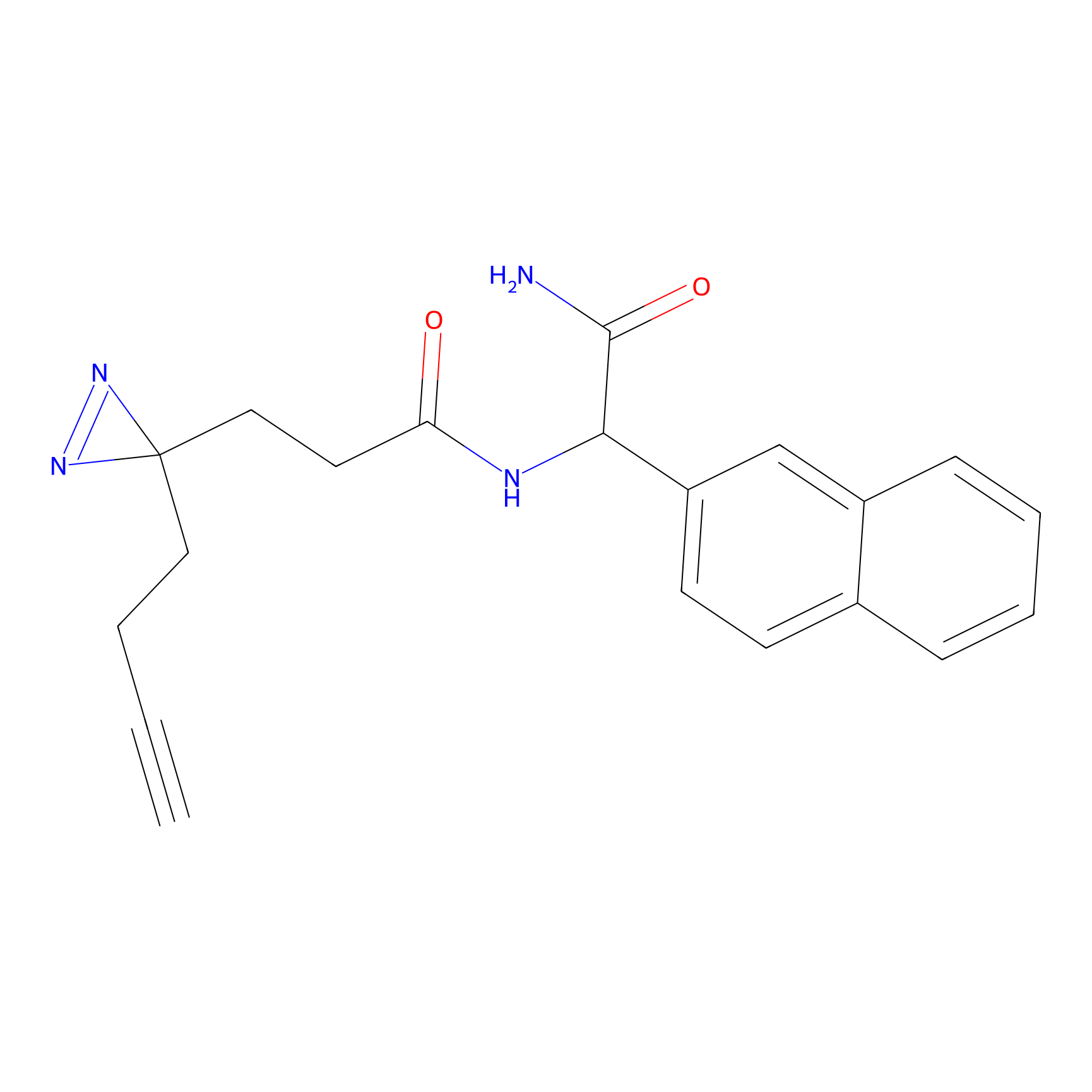
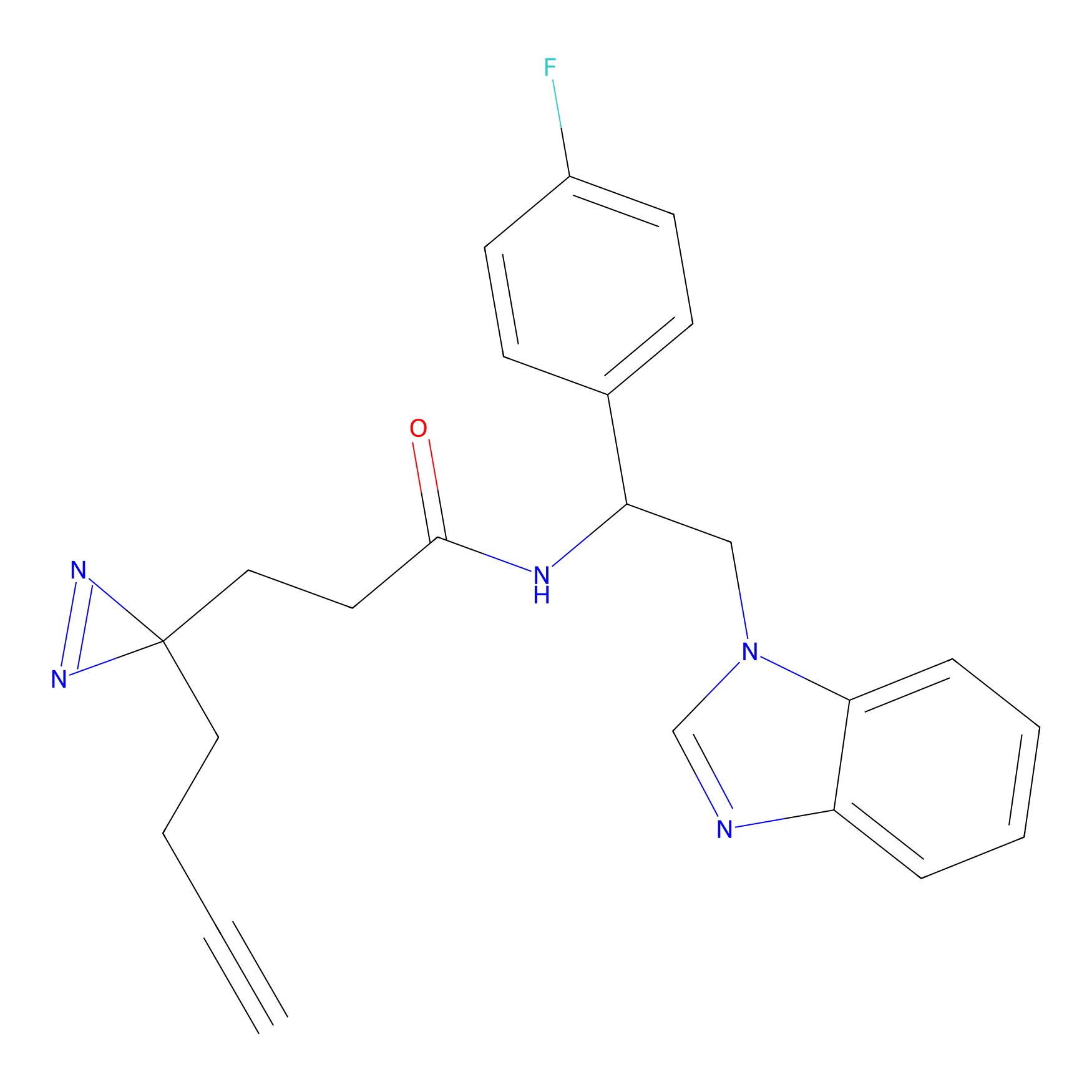
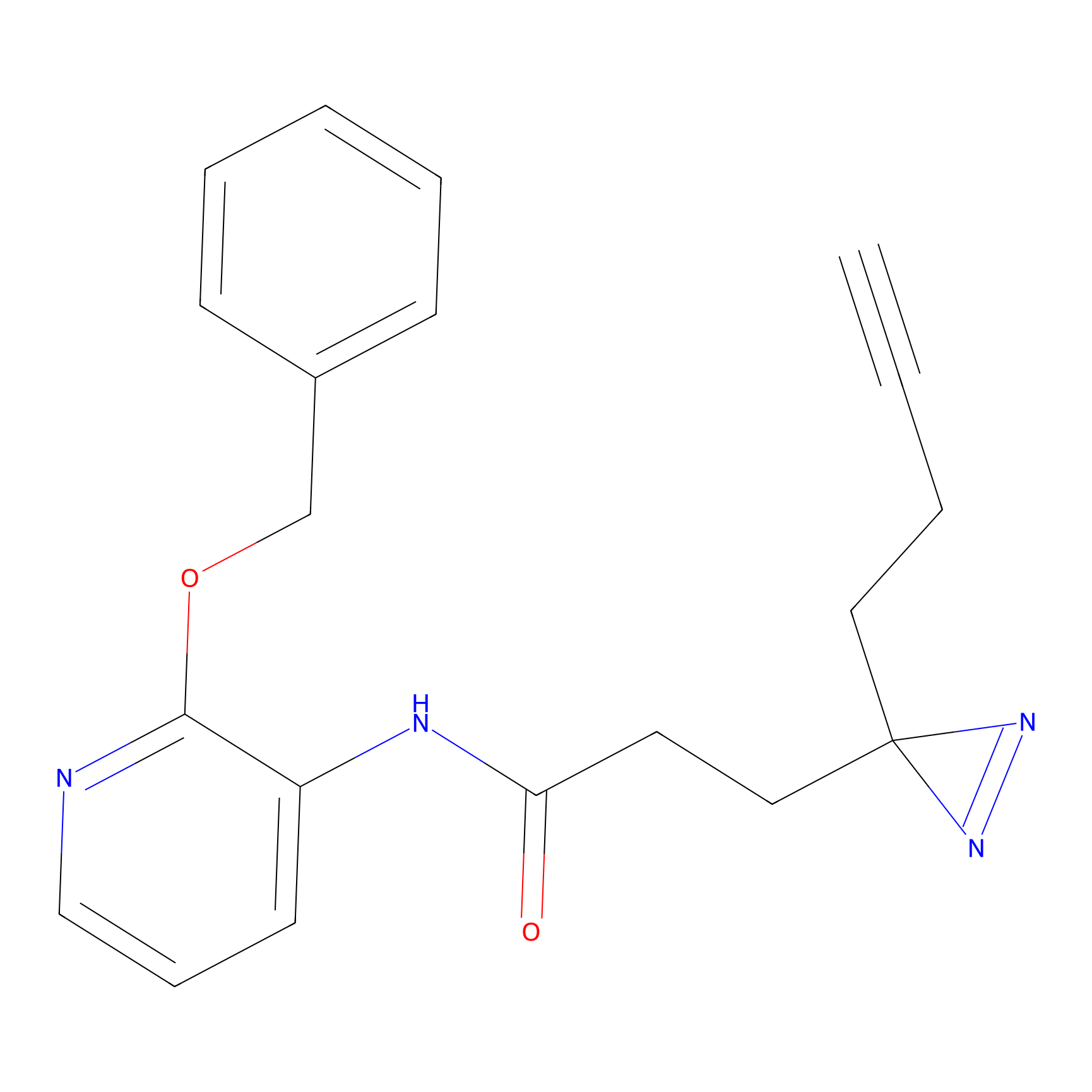
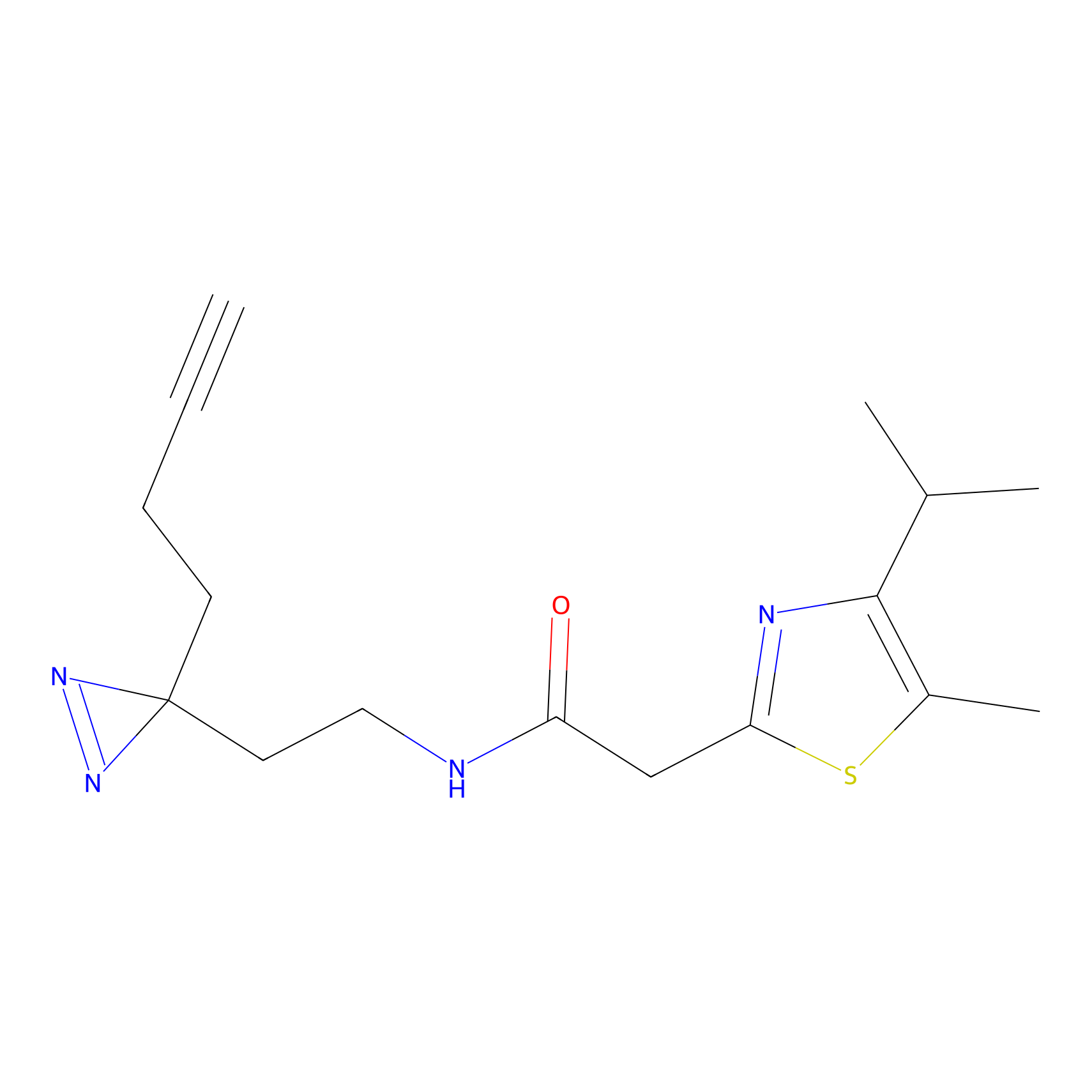
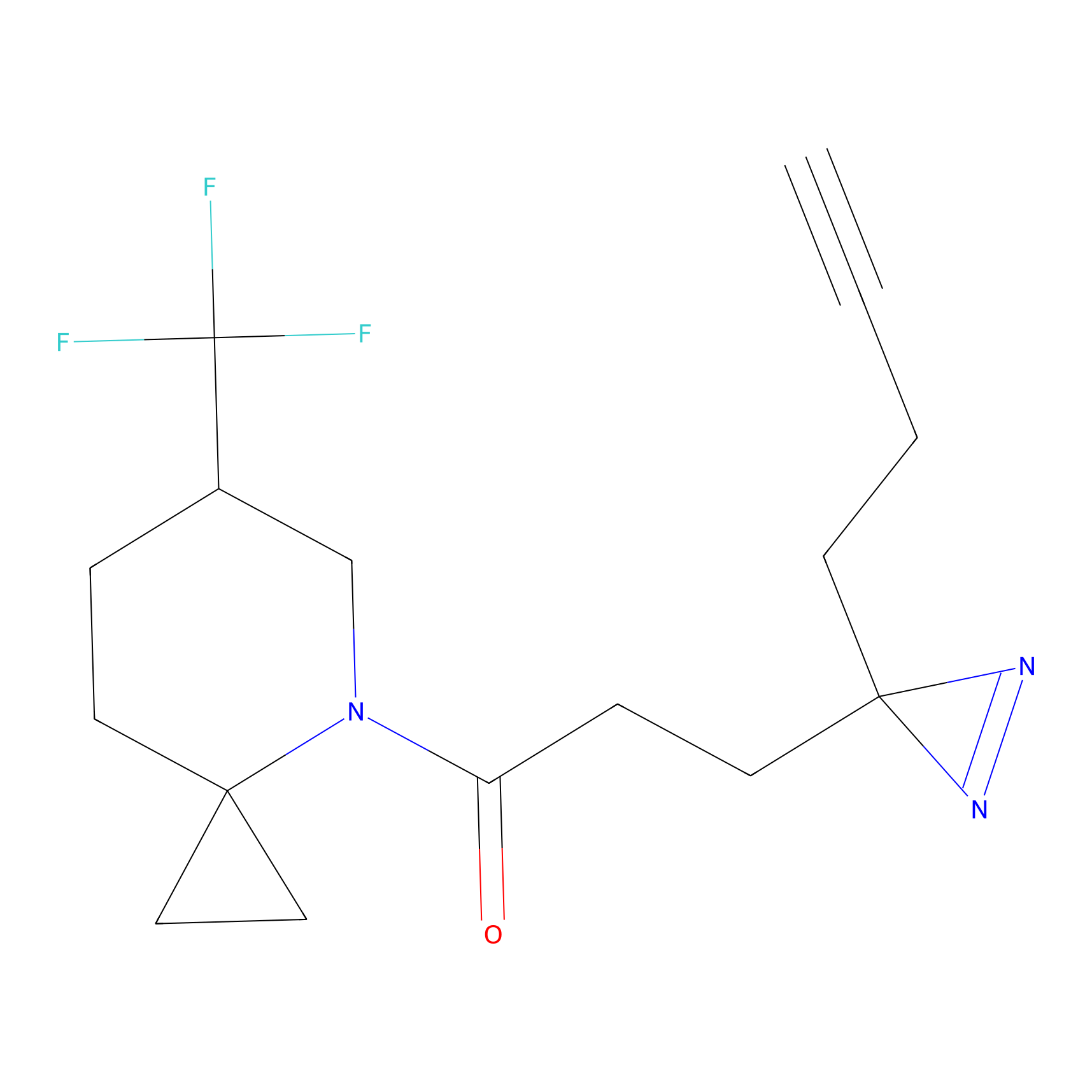
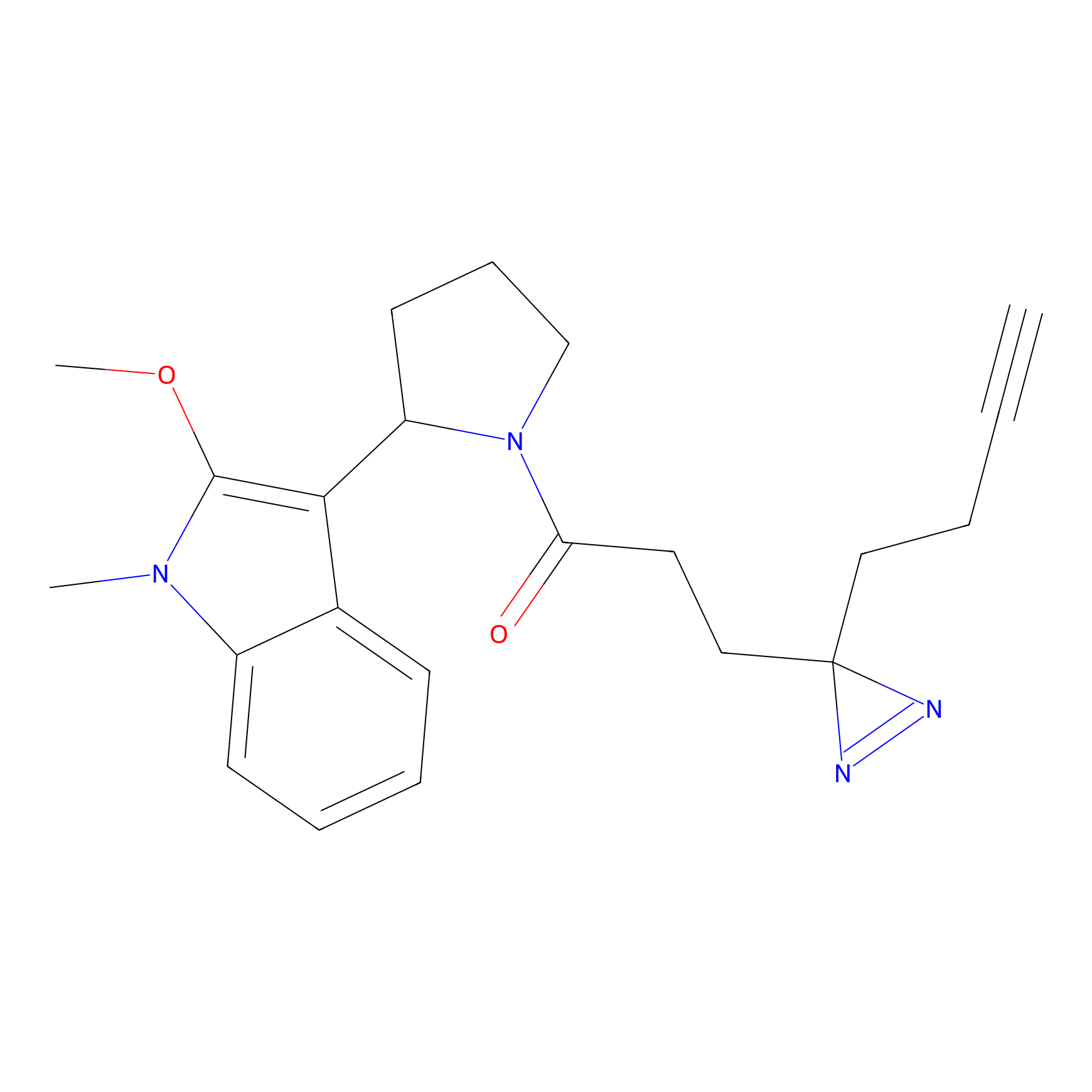
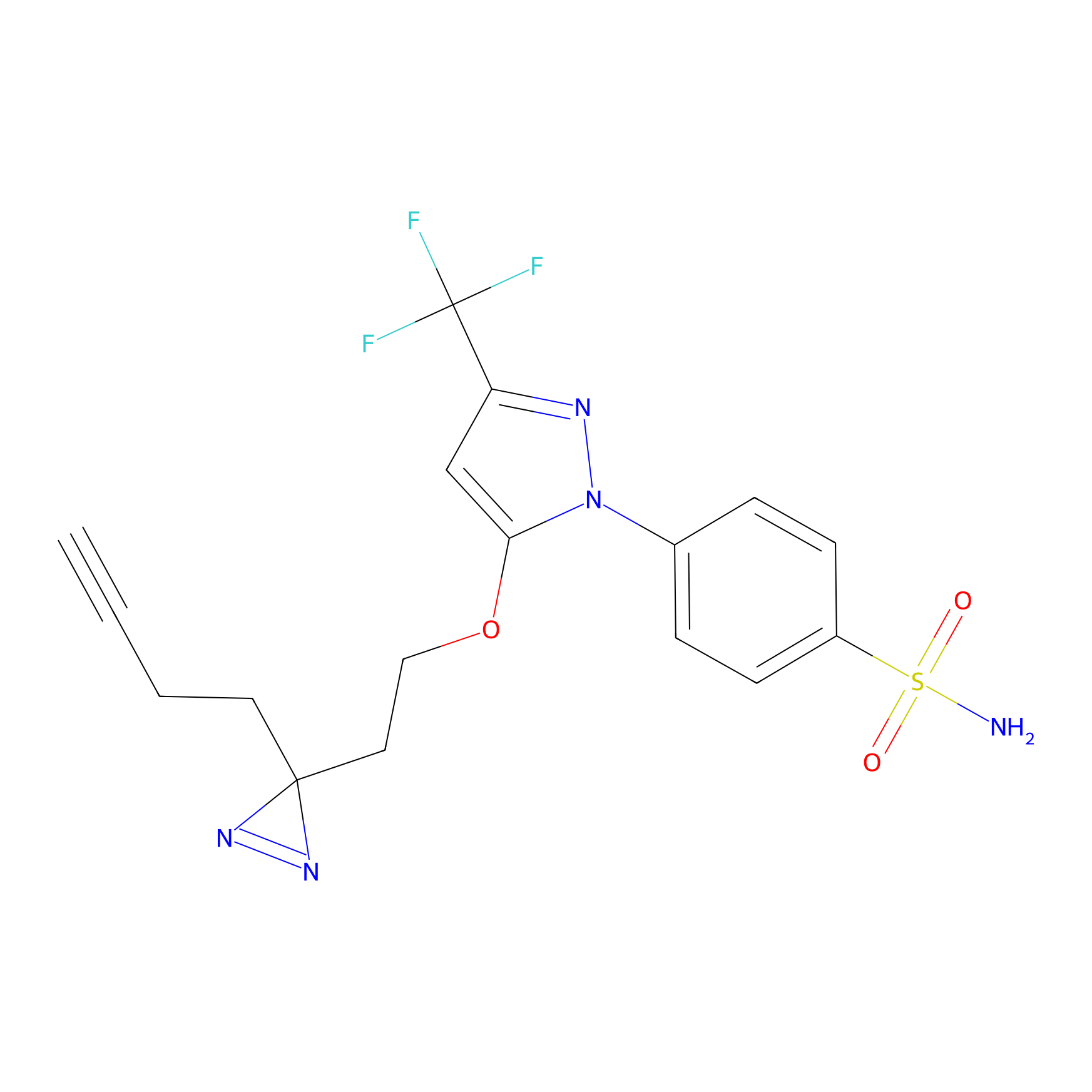
TG42 Probe Info |
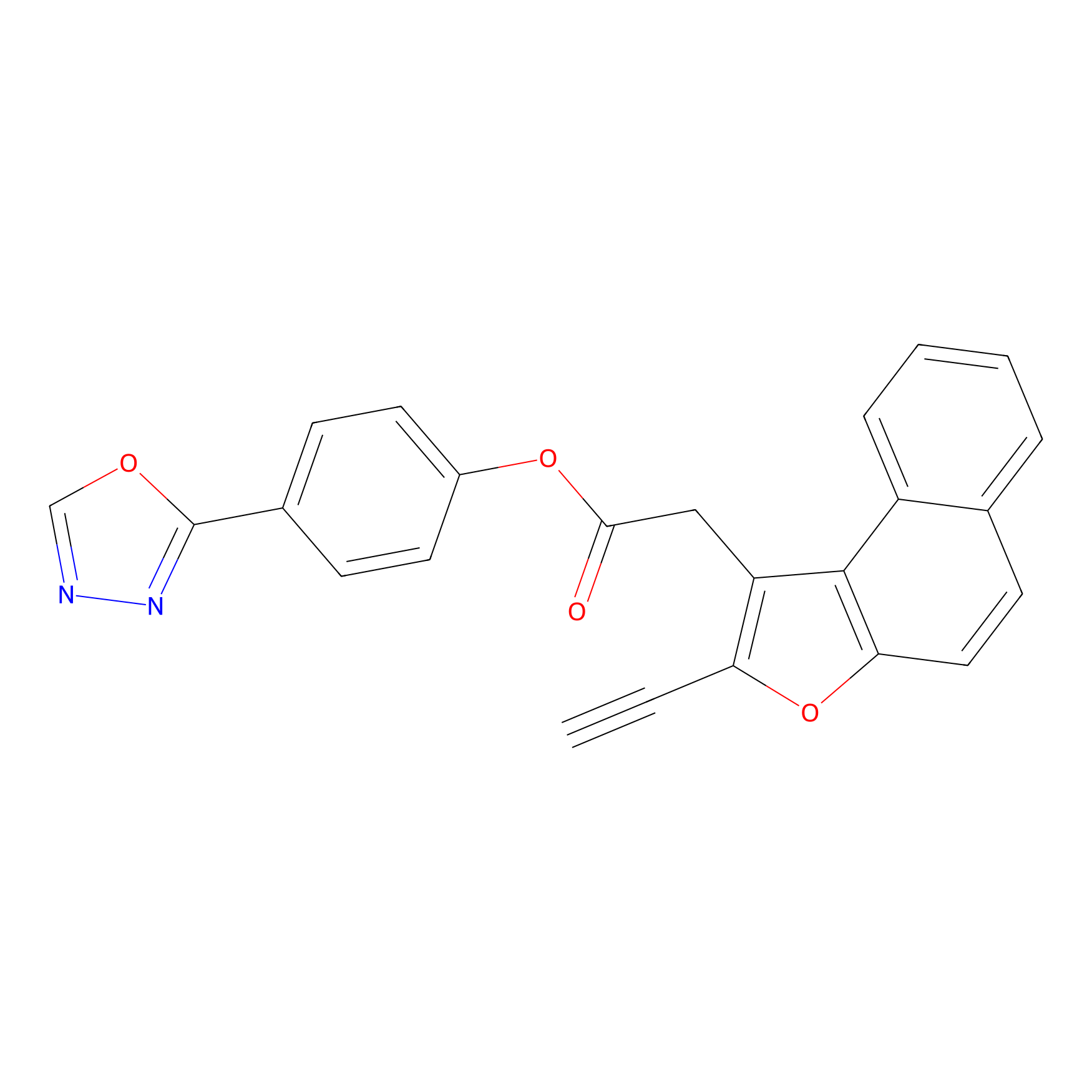 |
23.89 | LDD0326 | [4] | |
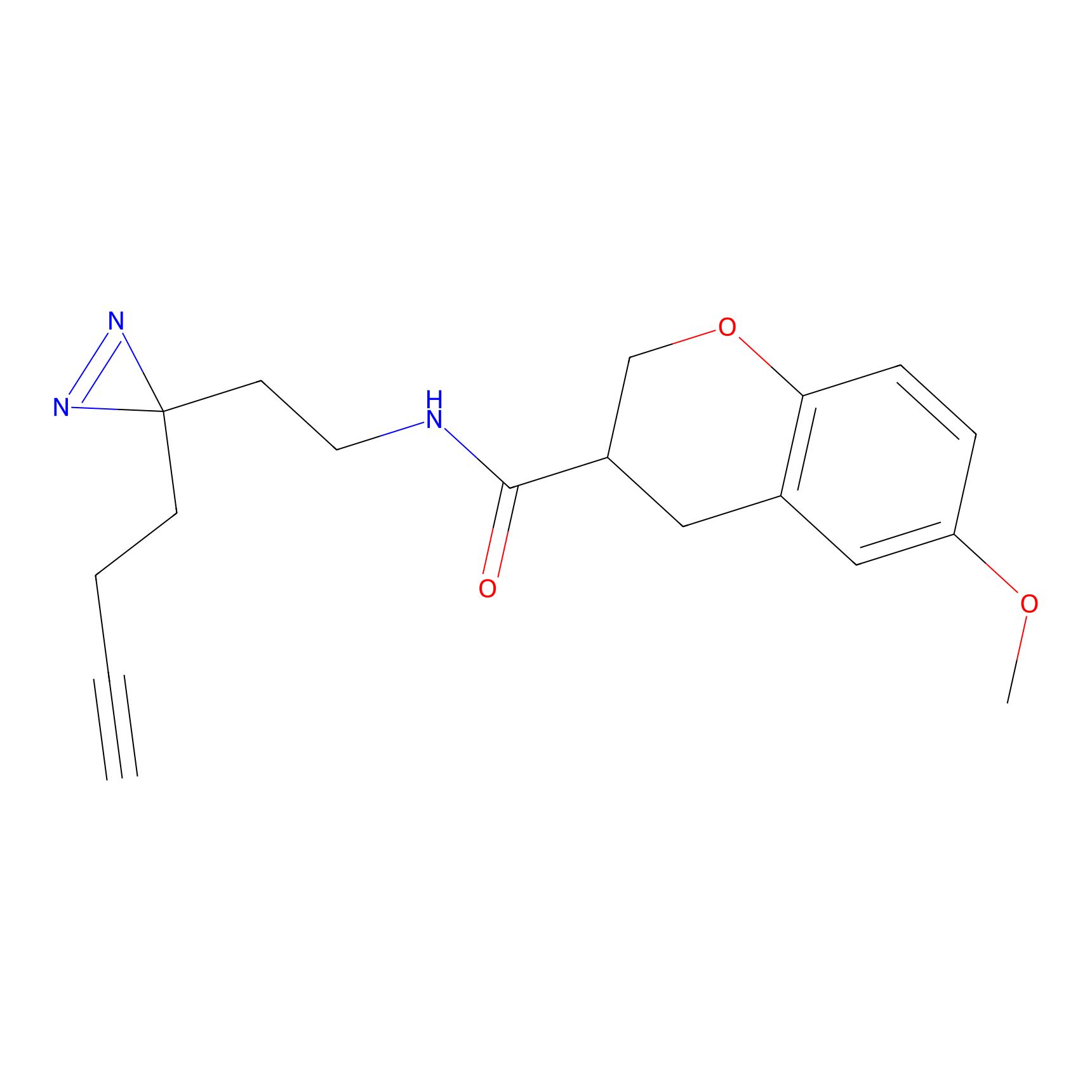
|
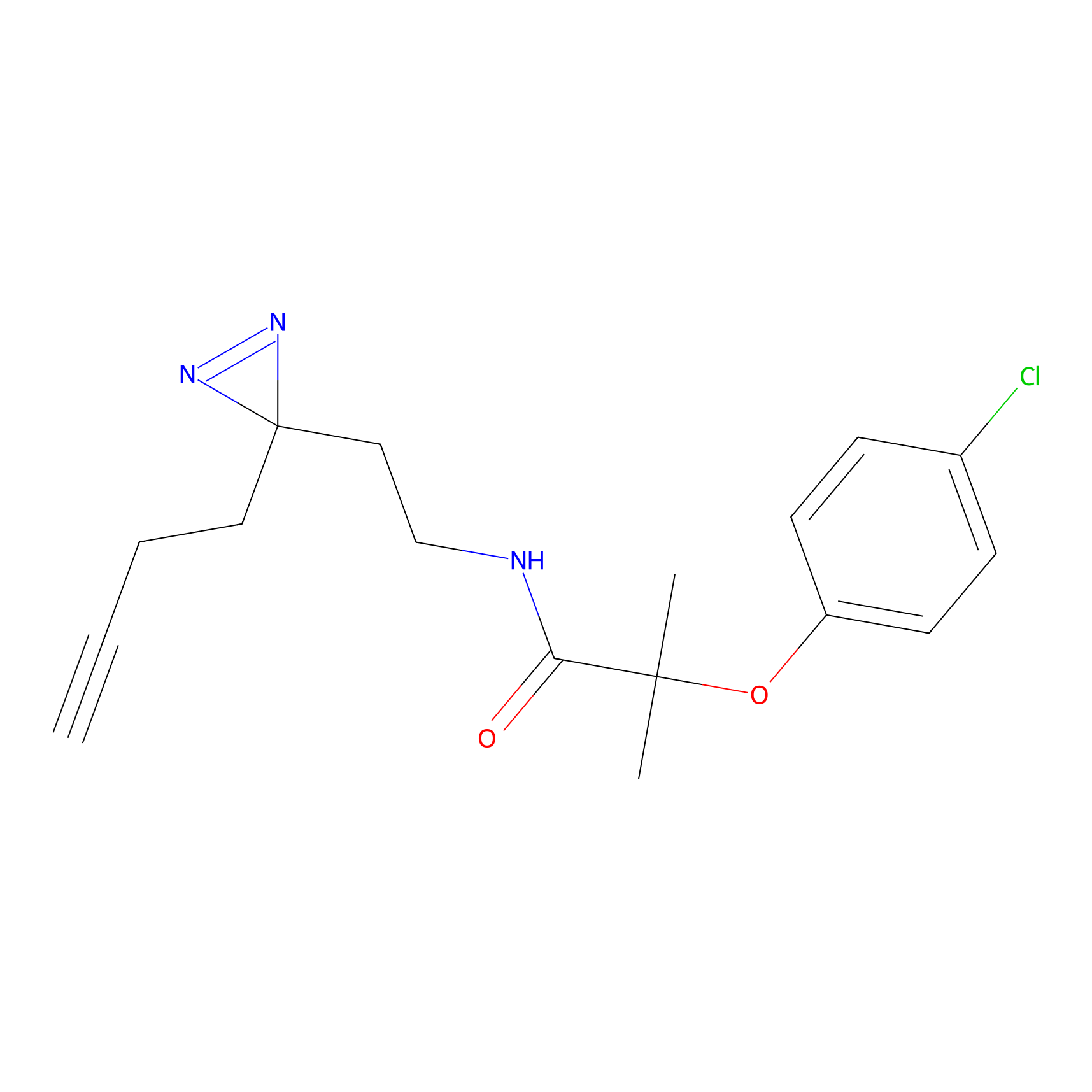
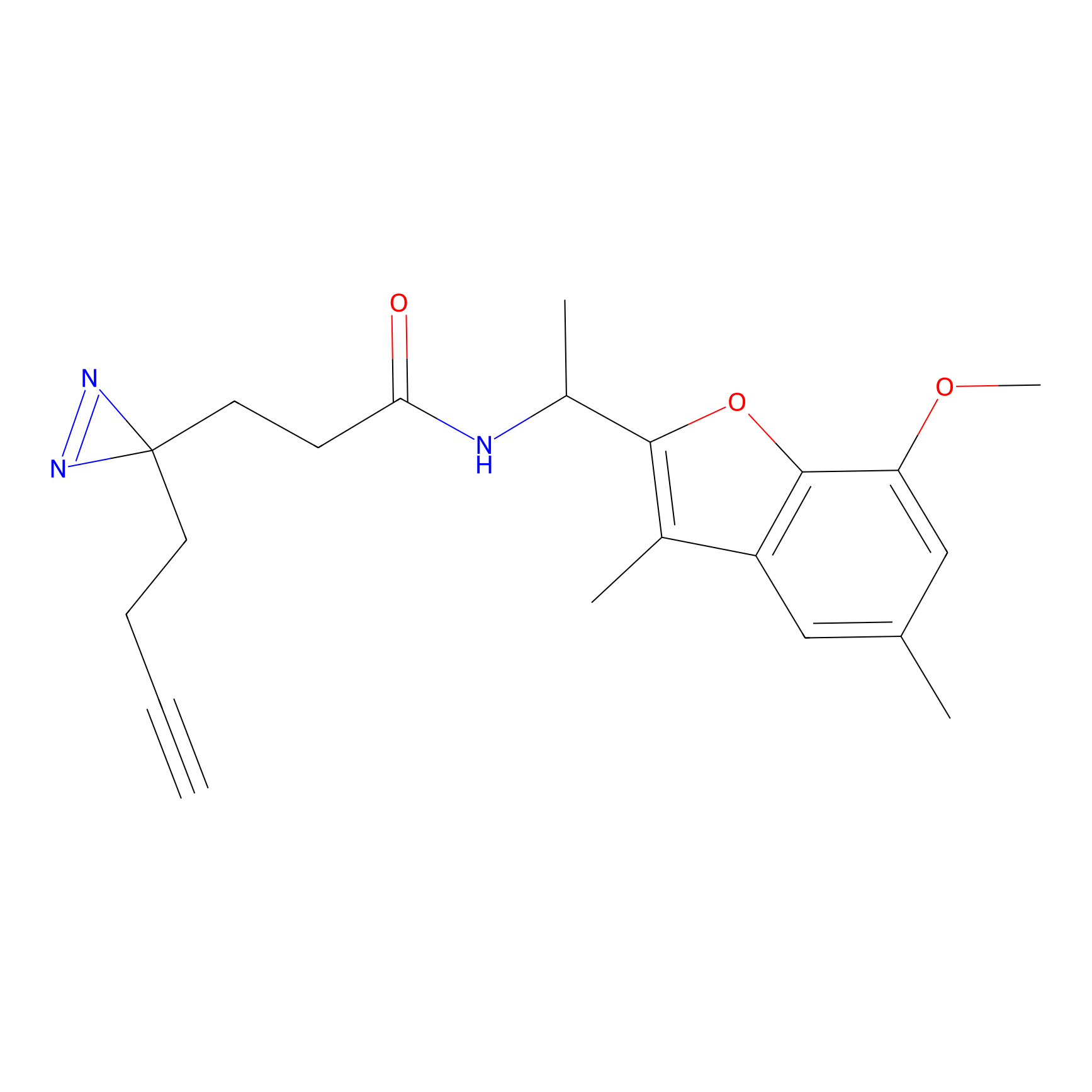
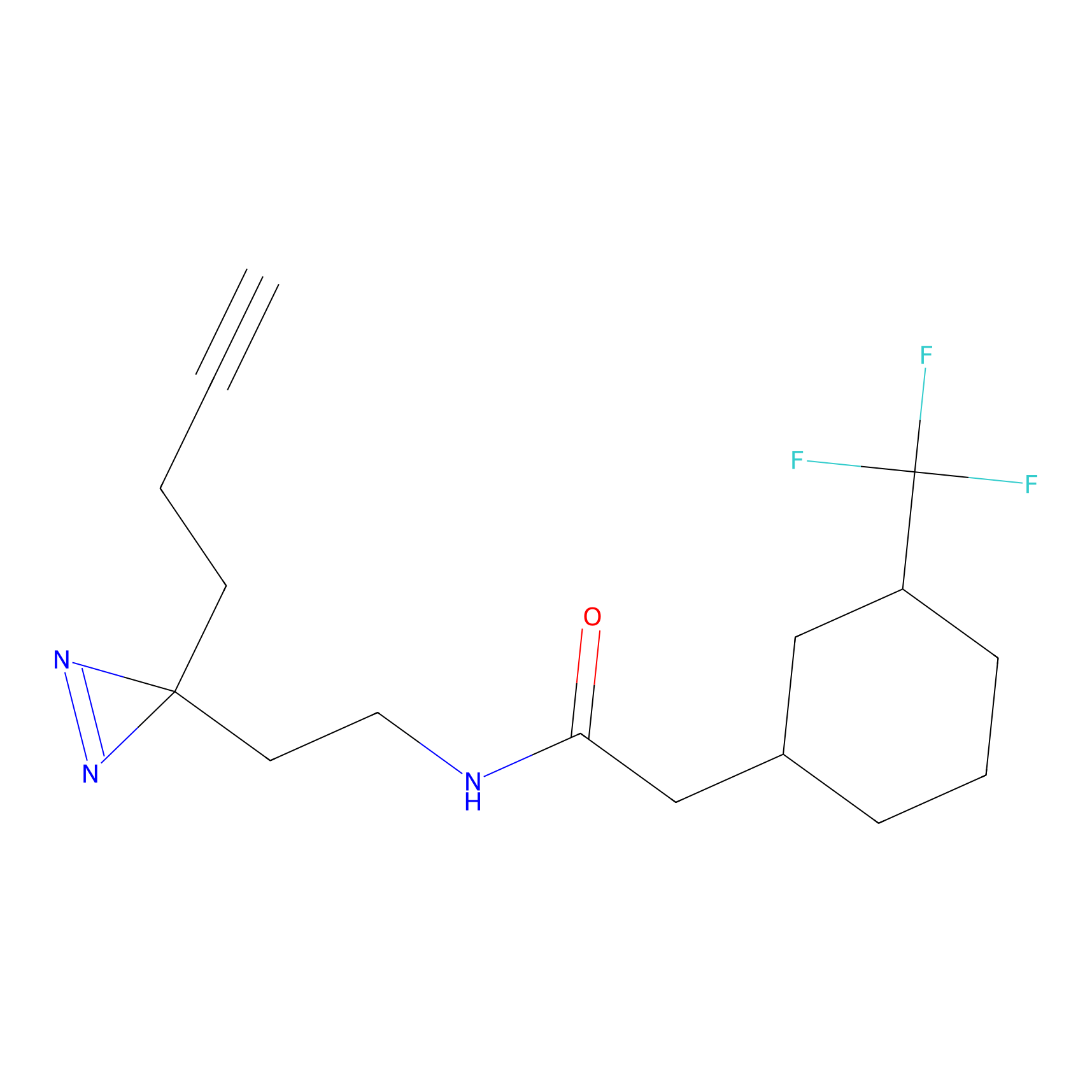
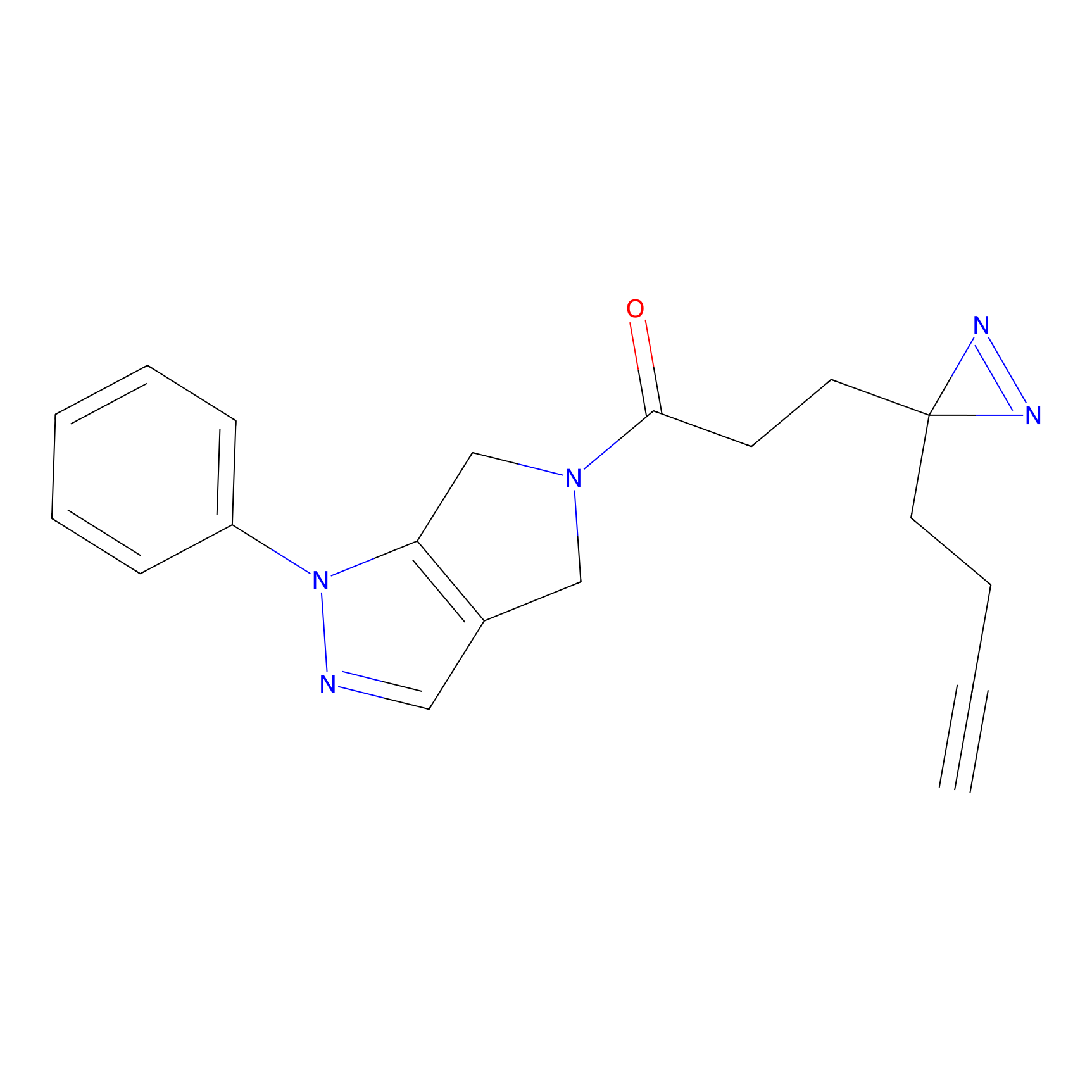
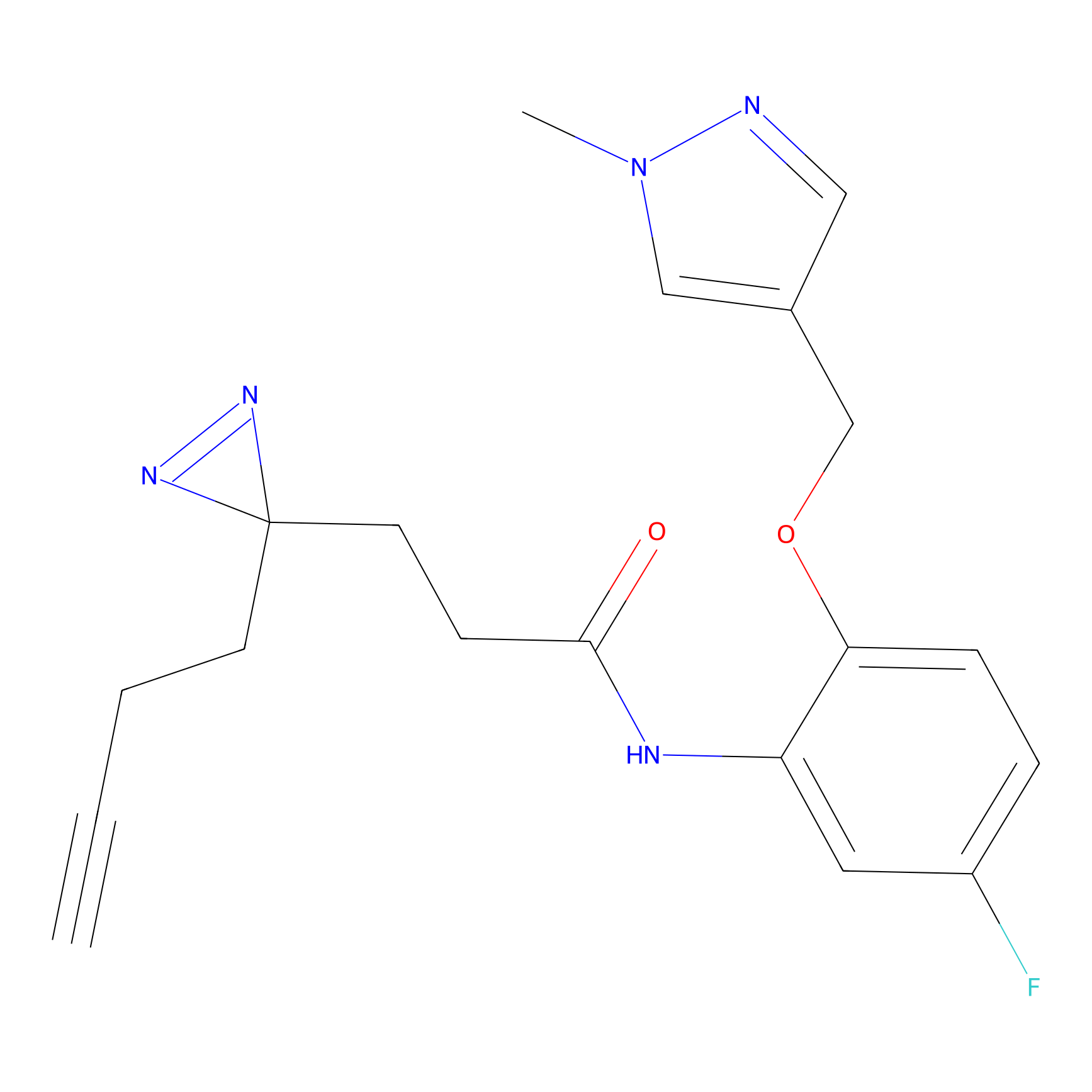
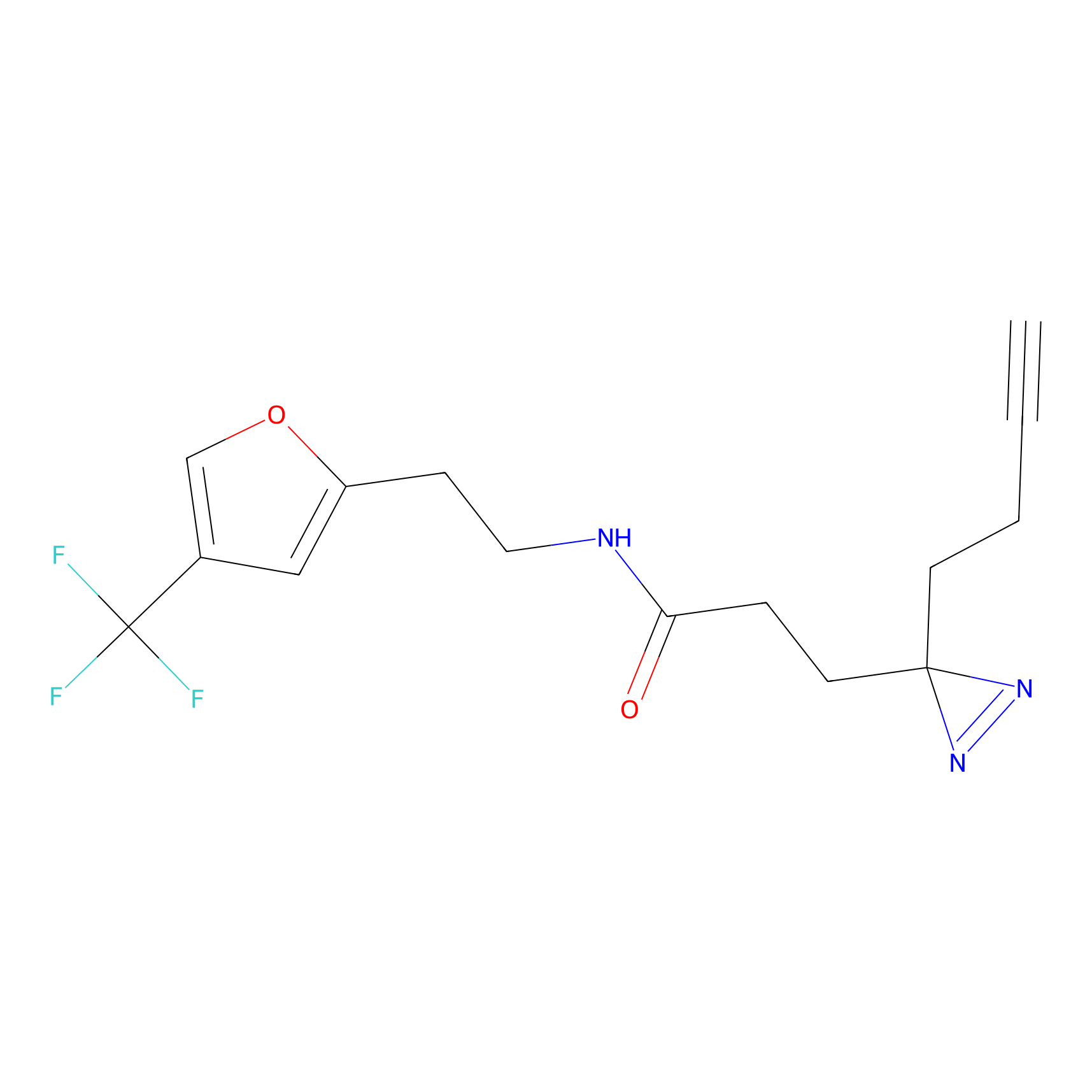
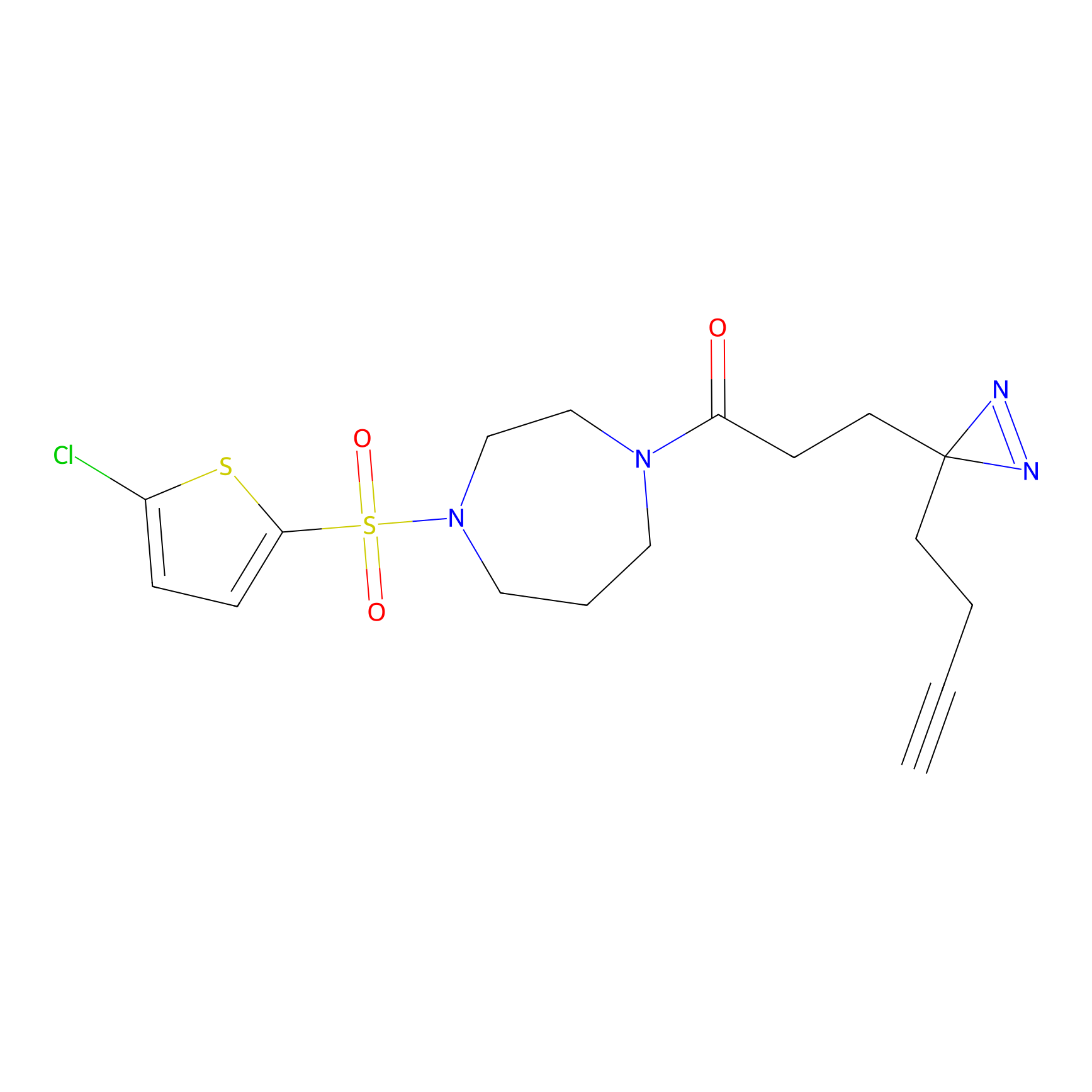
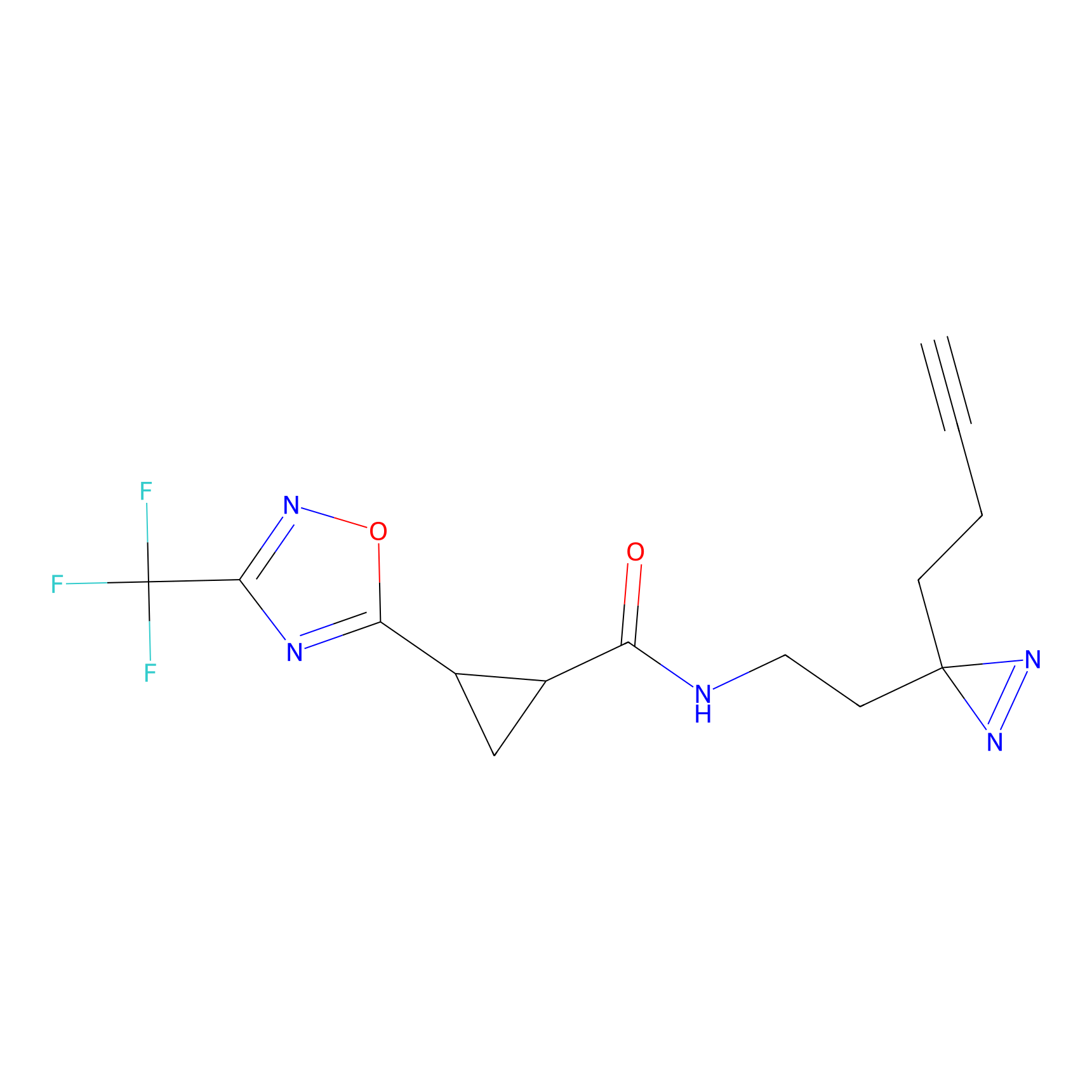
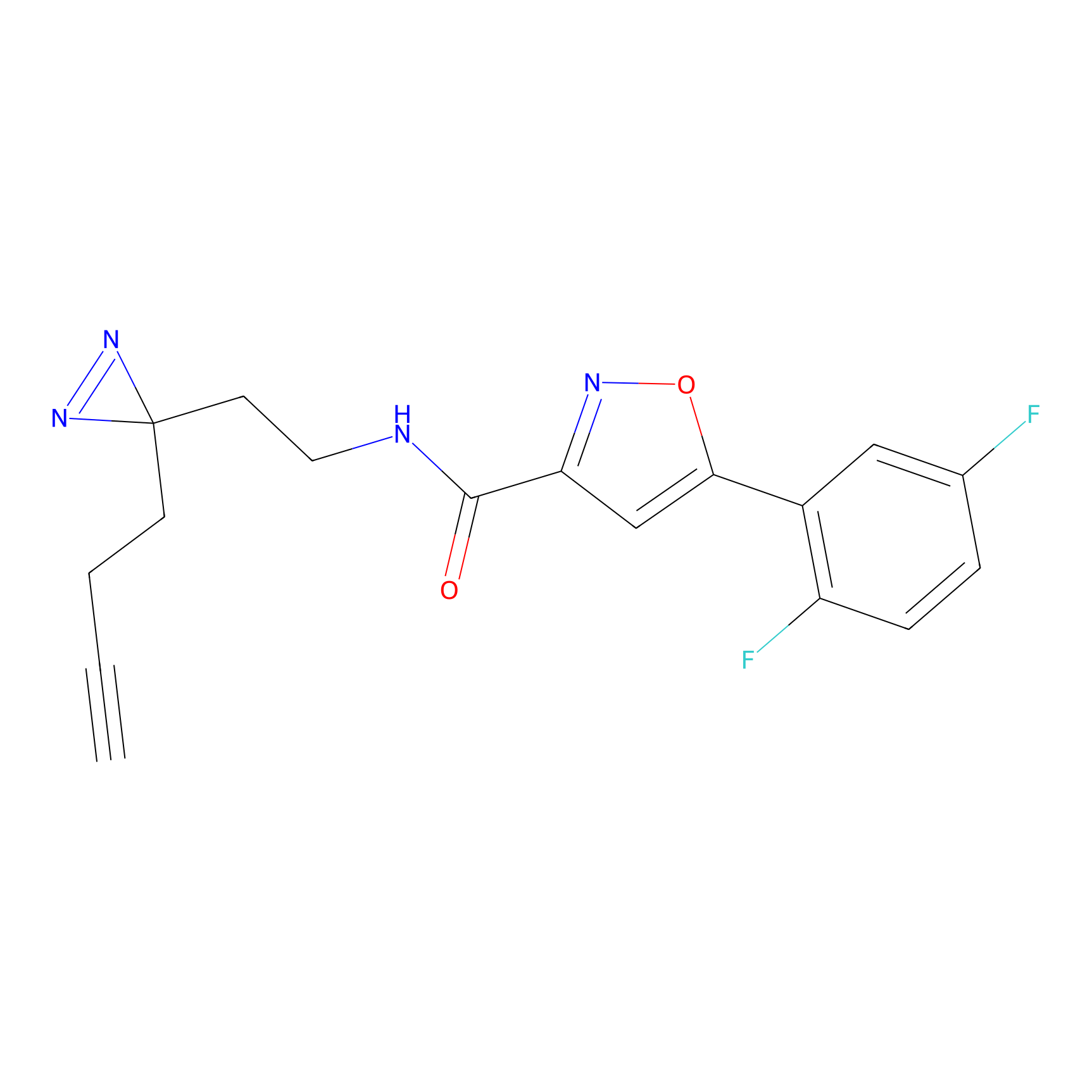
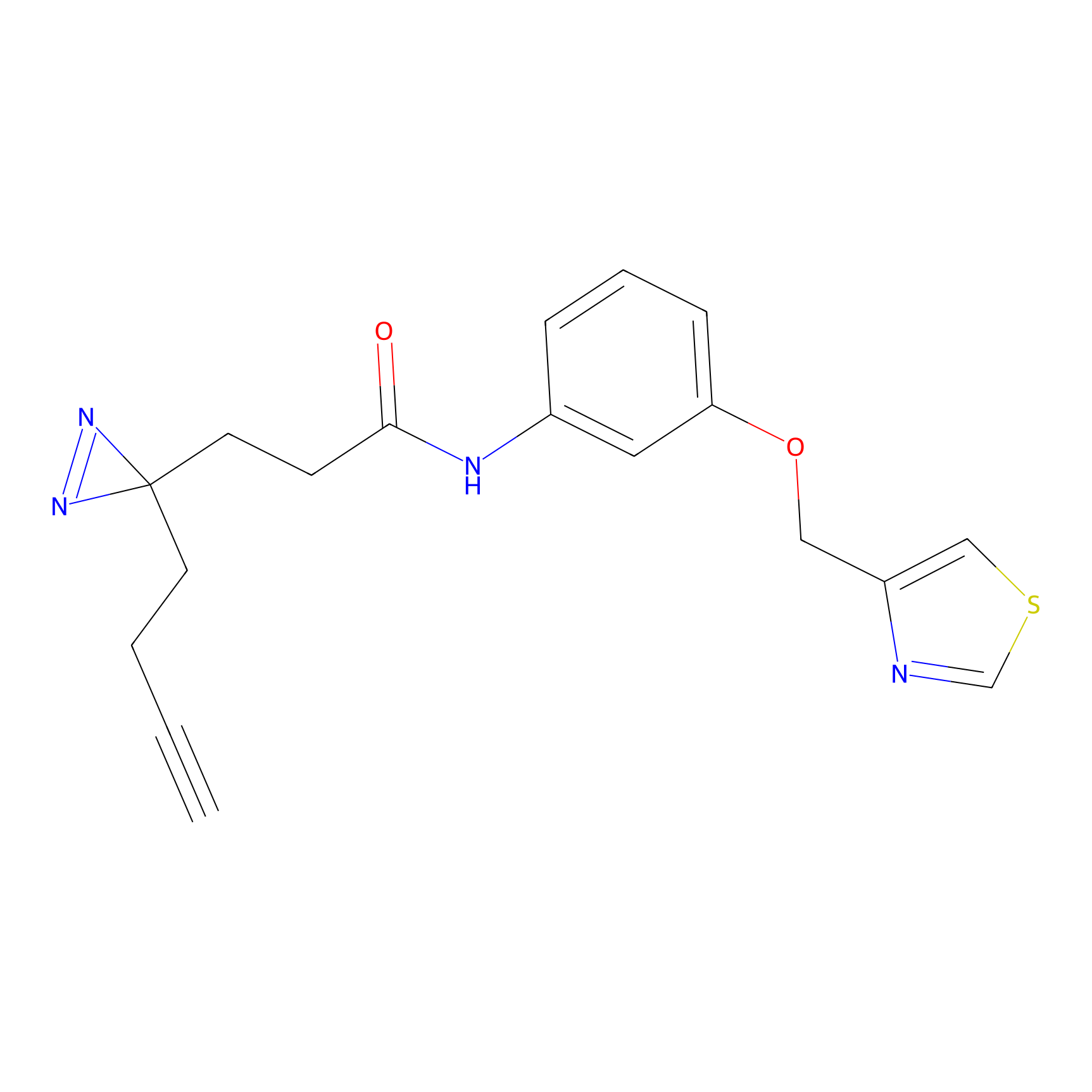
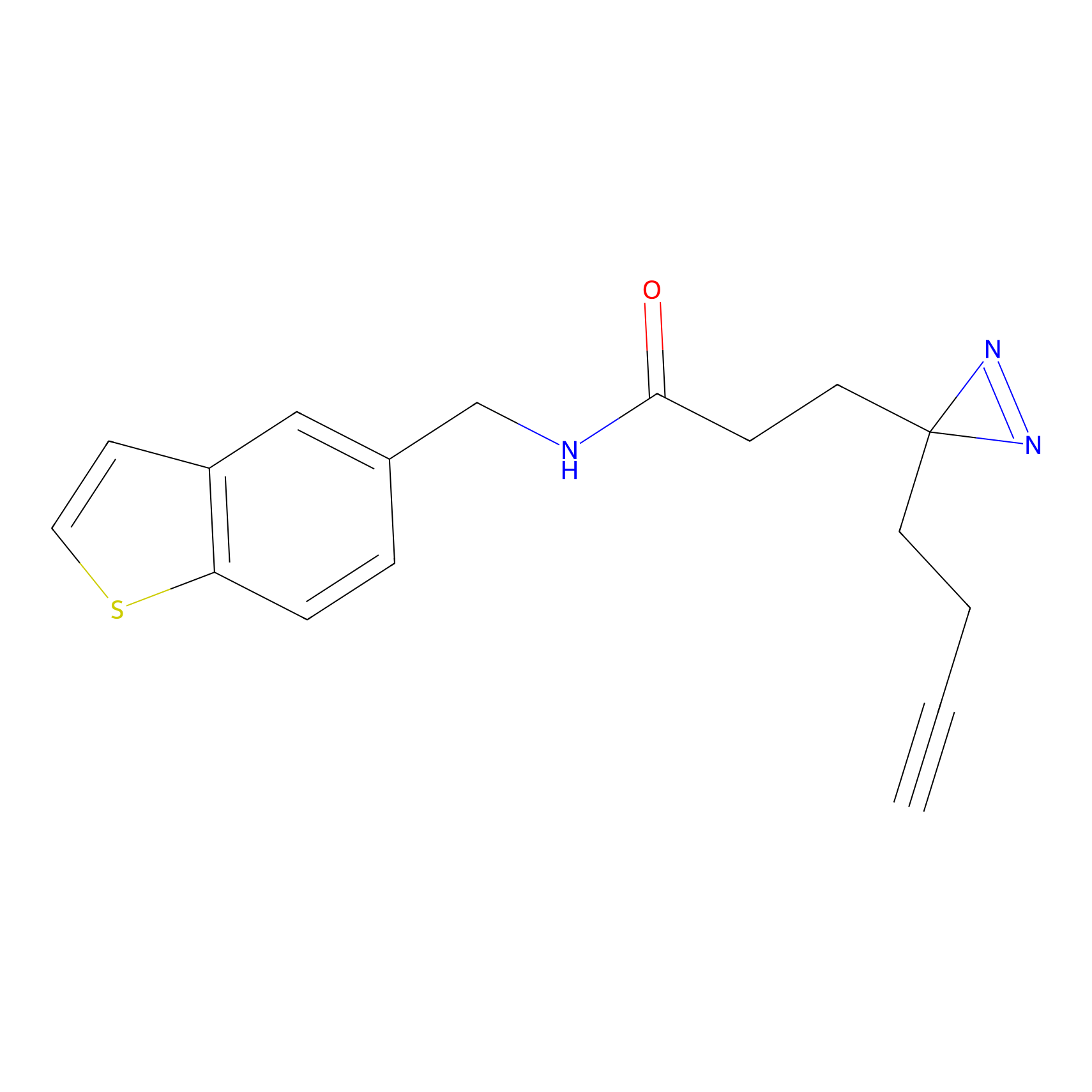
C-Sul Probe Info |
 |
2.12 | LDD0066 | [5] | |
|
STPyne Probe Info |
 |
K107(8.45); K174(10.00); K248(10.00); K486(2.13) | LDD0277 | [6] | |
|
OPA-S-S-alkyne Probe Info |
 |
K195(1.36) | LDD3494 | [7] | |
|
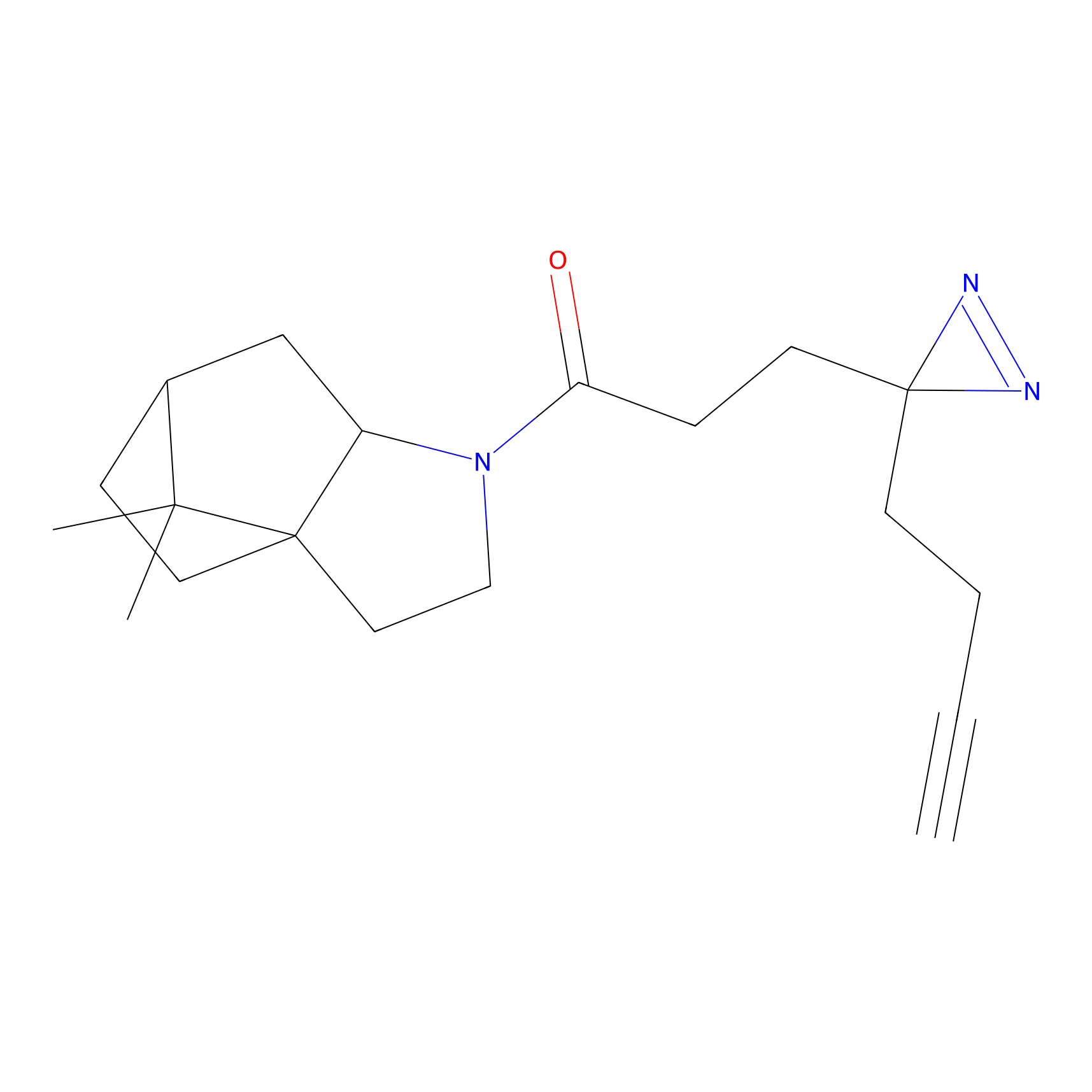
m-APA Probe Info |
 |
10.90 | LDD0403 | [8] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0221 | [9] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [10] | |
|
ATP probe Probe Info |
 |
K248(0.00); K314(0.00); K185(0.00) | LDD0199 | [11] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [12] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [13] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [14] | |
|
AOyne Probe Info |
 |
4.70 | LDD0443 | [15] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0156 | Aniline | NCI-H1299 | 10.90 | LDD0403 | [8] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [9] |
| LDCM0189 | Compound 16 | HEK-293T | 18.69 | LDD0492 | [17] |
| LDCM0185 | Compound 17 | HEK-293T | 9.52 | LDD0512 | [17] |
| LDCM0191 | Compound 21 | HEK-293T | 7.63 | LDD0493 | [17] |
| LDCM0190 | Compound 34 | HEK-293T | 16.53 | LDD0497 | [17] |
| LDCM0192 | Compound 35 | HEK-293T | 10.00 | LDD0491 | [17] |
| LDCM0193 | Compound 36 | HEK-293T | 6.56 | LDD0494 | [17] |
| LDCM0195 | Compound 38 | HEK-293T | 10.93 | LDD0499 | [17] |
| LDCM0196 | Compound 39 | HEK-293T | 4.15 | LDD0496 | [17] |
| LDCM0197 | Compound 40 | HEK-293T | 8.20 | LDD0495 | [17] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [9] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0227 | [9] |
| LDCM0084 | Ro 48-8071 | A-549 | 2.06 | LDD0145 | [19] |
The Interaction Atlas With This Target
References