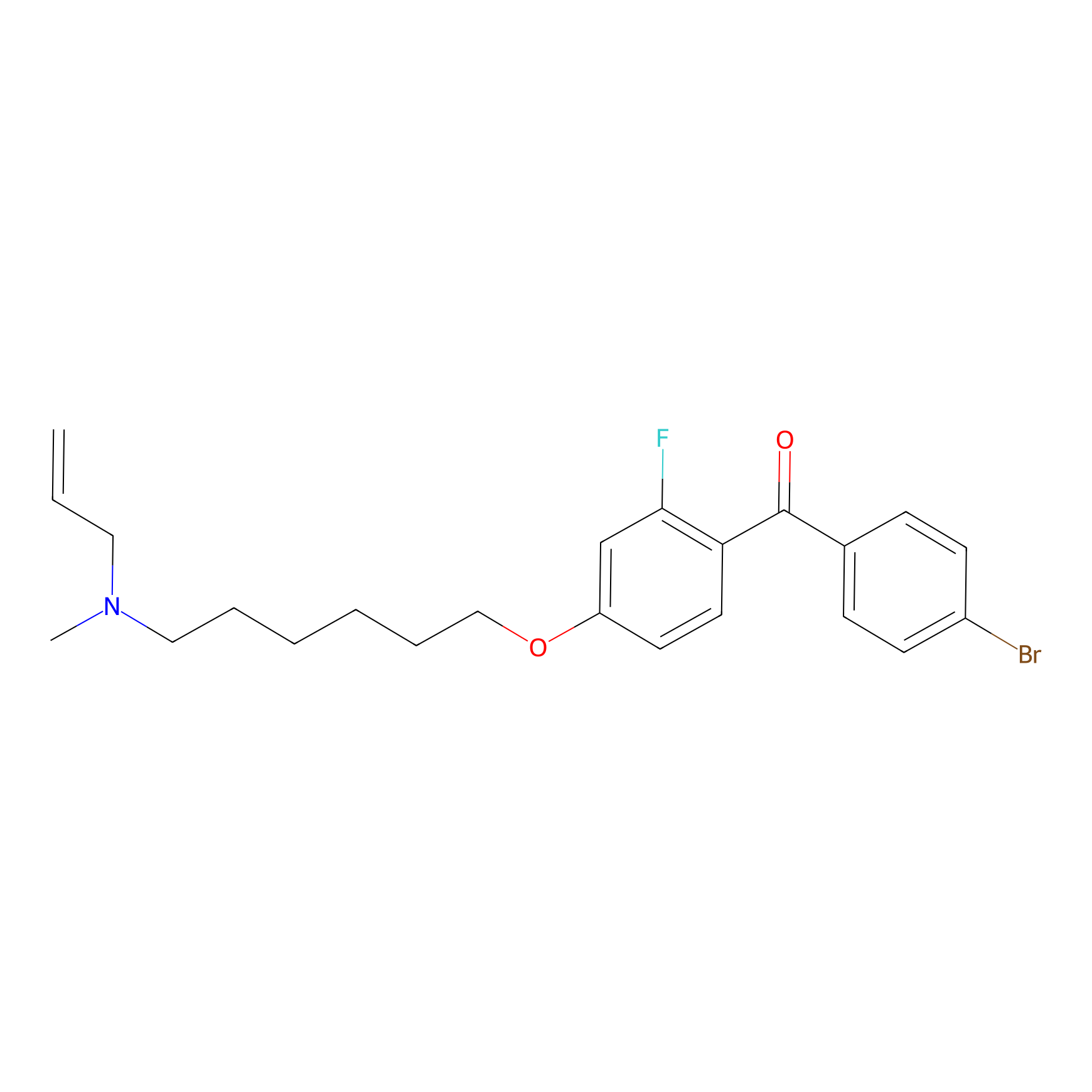
Details of the Competitor
General Information of Competitor
The Competitor Interaction Atlas
Probe(s) Related This Competitor
PAL-AfBPP Probe
| Probe Name | Structure | Concentration | Cell-system | Ref | |
|---|---|---|---|---|---|
|
A-DA Probe Info |
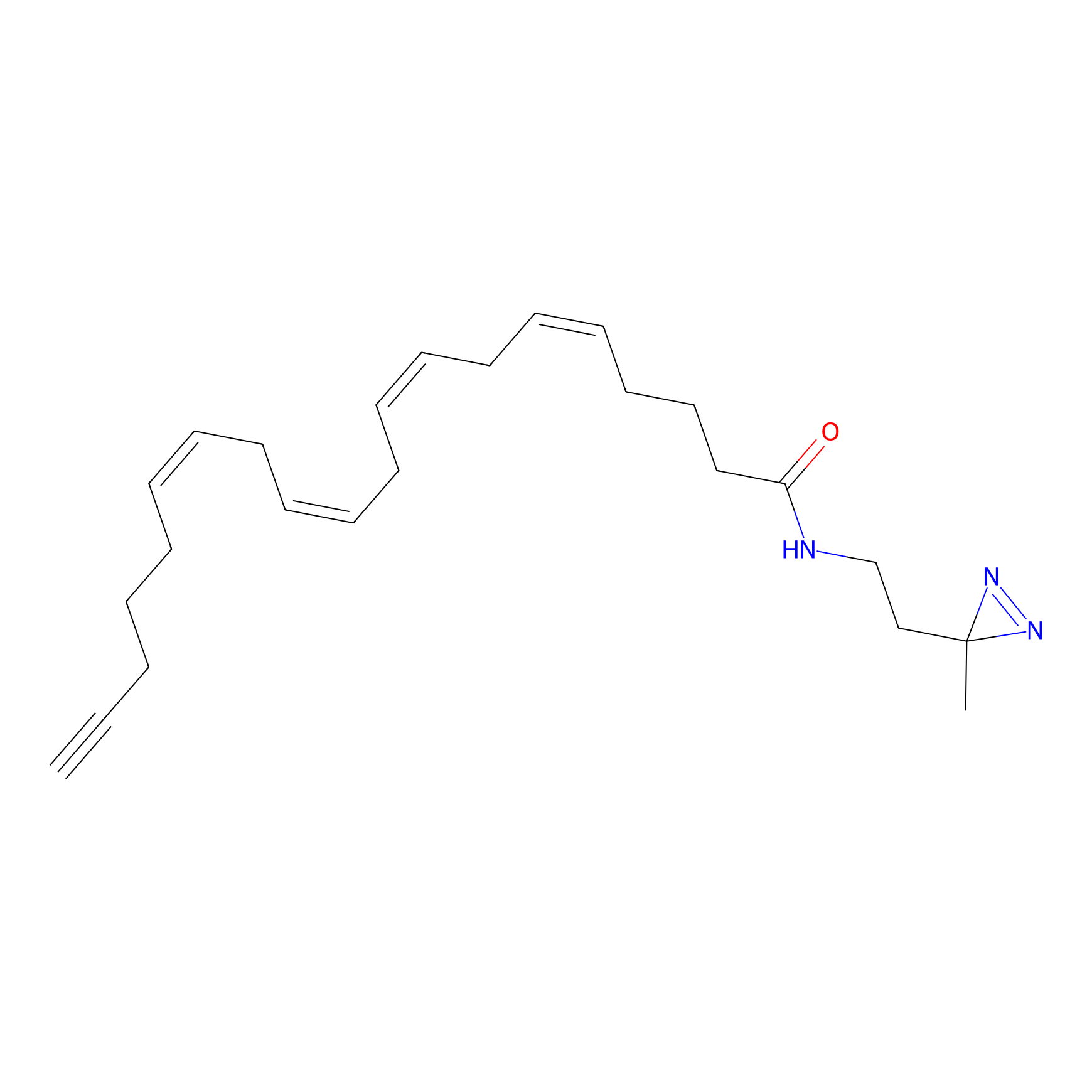 |
5 uM | Human lung adenocarcinoma cells (A-549) | [1] | |
Target(s) List of this Competitor
|
74 Enzyme Competed by This
Competitor
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
30 Transporter and channel Competed by This
Competitor
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
1 Transcription factor Competed by This
Competitor
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
2 Immunoglobulin Competed by This
Competitor
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
20 Other Competed by This
Competitor
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Full Information of The Labelling Profiles of This Competitor

A-DA
Quantification: Probe vs (Probe+Competitor)
|
Experiment 1 Reporting the Labelling Profiles of This Probe
|
||||
| Probe concentration | ||||
| Quantitative Method | ||||
| Competitor Name | ||||
| Competitor Concentration | ||||
| In Vitro Experiment Model |

|
|||

|
||||
| Lung adenocarcinoma [ICD-11:2C25] | ||||
| Human lung adenocarcinoma cells (A-549) | ||||
| Interaction Atlas ID |
 Download The Altas
Download The Altas
|
|||
|
Experiment 2 Reporting the Labelling Profiles of This Probe
|
||||
| Probe concentration | ||||
| Quantitative Method | ||||
| Competitor Name | ||||
| Competitor Concentration | ||||
| In Vitro Experiment Model |

|
|||

|
||||
| Lung adenocarcinoma [ICD-11:2C25] | ||||
| Human lung adenocarcinoma cells (A-549) | ||||
| Interaction Atlas ID |
 Download The Altas
Download The Altas
|
|||