Details of the Target
General Information of Target
| Target ID | LDTP06948 | |||||
|---|---|---|---|---|---|---|
| Target Name | Protein YIF1B (YIF1B) | |||||
| Gene Name | YIF1B | |||||
| Gene ID | 90522 | |||||
| Synonyms |
Protein YIF1B; YIP1-interacting factor homolog B |
|||||
| 3D Structure | ||||||
| Sequence |
MHPAGLAAAAAGTPRLRKWPSKRRIPVSQPGMADPHQLFDDTSSAQSRGYGAQRAPGGLS
YPAASPTPHAAFLADPVSNMAMAYGSSLAAQGKELVDKNIDRFIPITKLKYYFAVDTMYV GRKLGLLFFPYLHQDWEVQYQQDTPVAPRFDVNAPDLYIPAMAFITYVLVAGLALGTQDR FSPDLLGLQASSALAWLTLEVLAILLSLYLVTVNTDLTTIDLVAFLGYKYVGMIGGVLMG LLFGKIGYYLVLGWCCVAIFVFMIRTLRLKILADAAAEGVPVRGARNQLRMYLTMAVAAA QPMLMYWLTFHLVR |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
YIF1 family
|
|||||
| Subcellular location |
Endoplasmic reticulum membrane
|
|||||
| Function |
Functions in endoplasmic reticulum to Golgi vesicle-mediated transport and regulates the proper organization of the endoplasmic reticulum and the Golgi. Plays a key role in targeting to neuronal dendrites receptors such as HTR1A. Plays also a role in primary cilium and sperm flagellum assembly probably through protein transport to these compartments.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K108(9.64) | LDD0277 | [1] | |
|
Jackson_14 Probe Info |
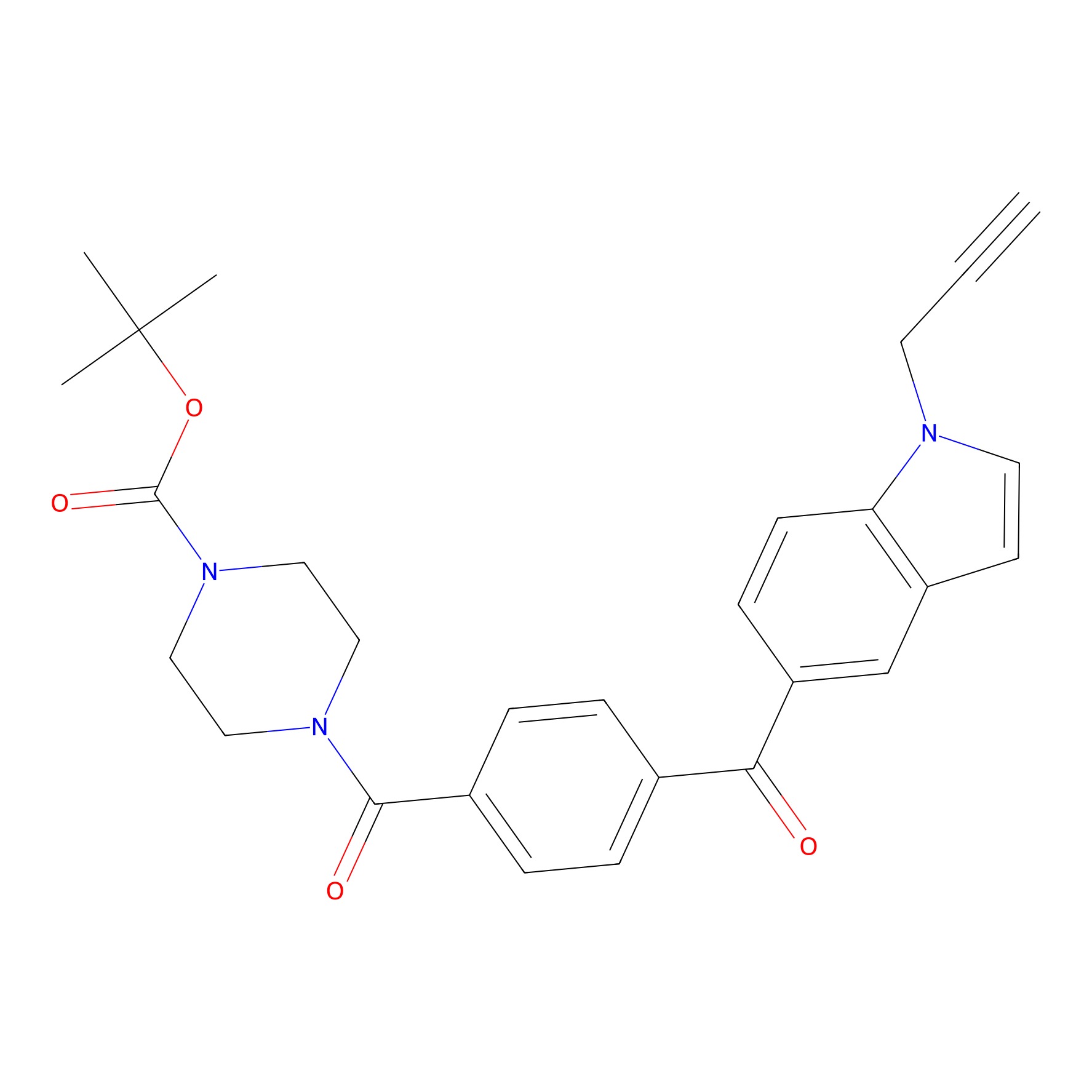 |
2.27 | LDD0123 | [2] | |
|
m-APA Probe Info |
 |
N.A. | LDD2231 | [3] | |
|
AOyne Probe Info |
 |
9.50 | LDD0443 | [4] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
DR-1 Probe Info |
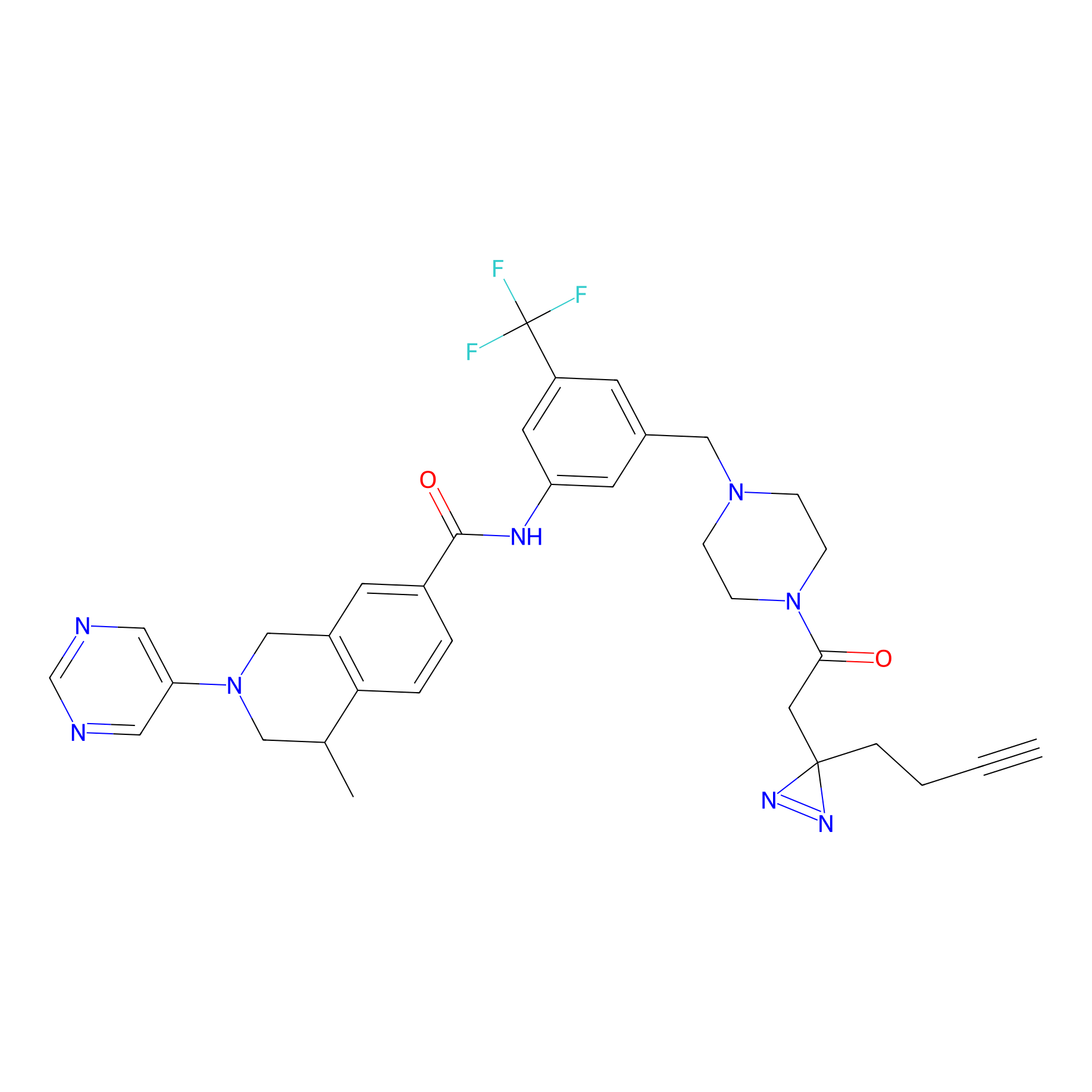 |
2.76 | LDD0398 | [5] | |
|
C017 Probe Info |
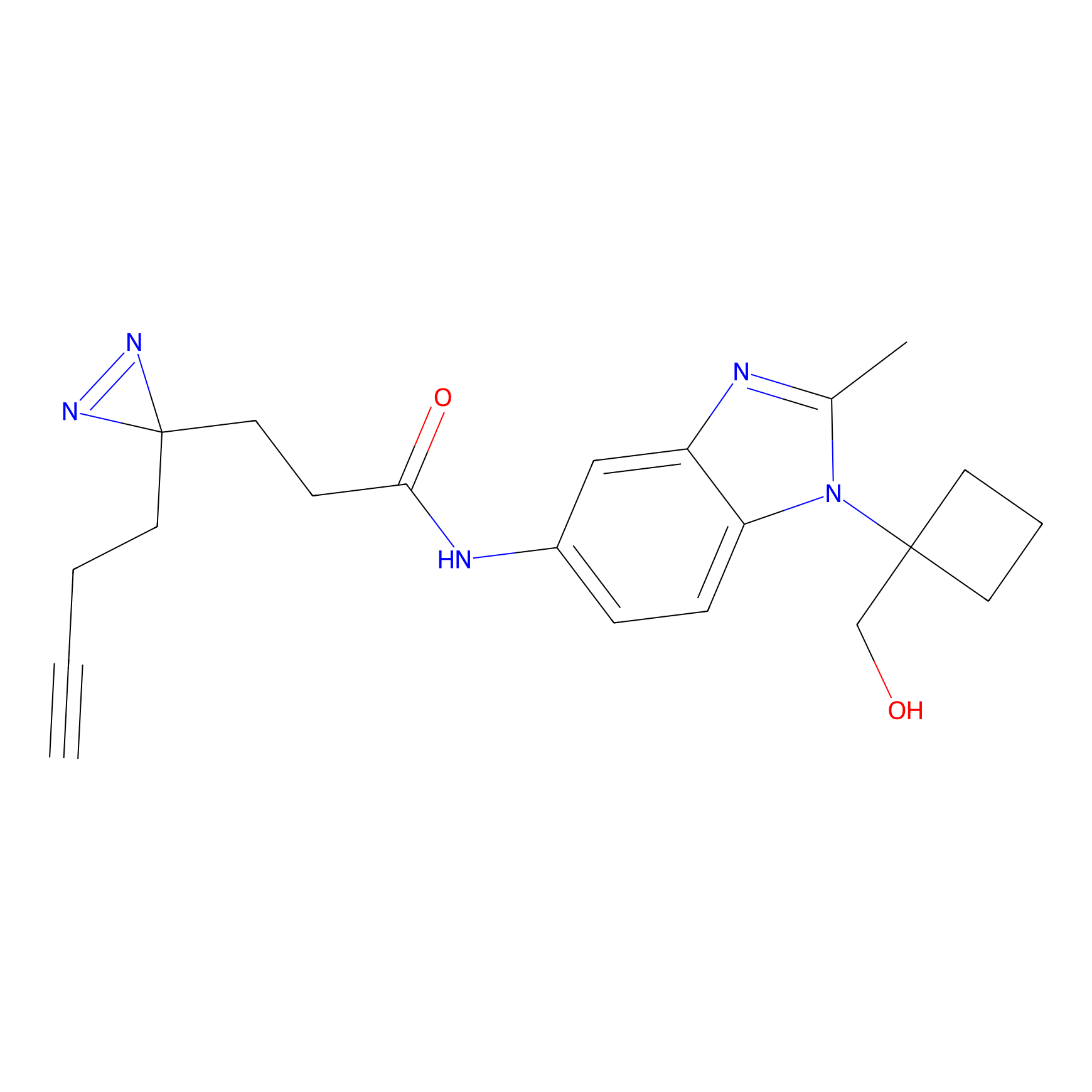 |
6.59 | LDD1725 | [6] | |
|
C055 Probe Info |
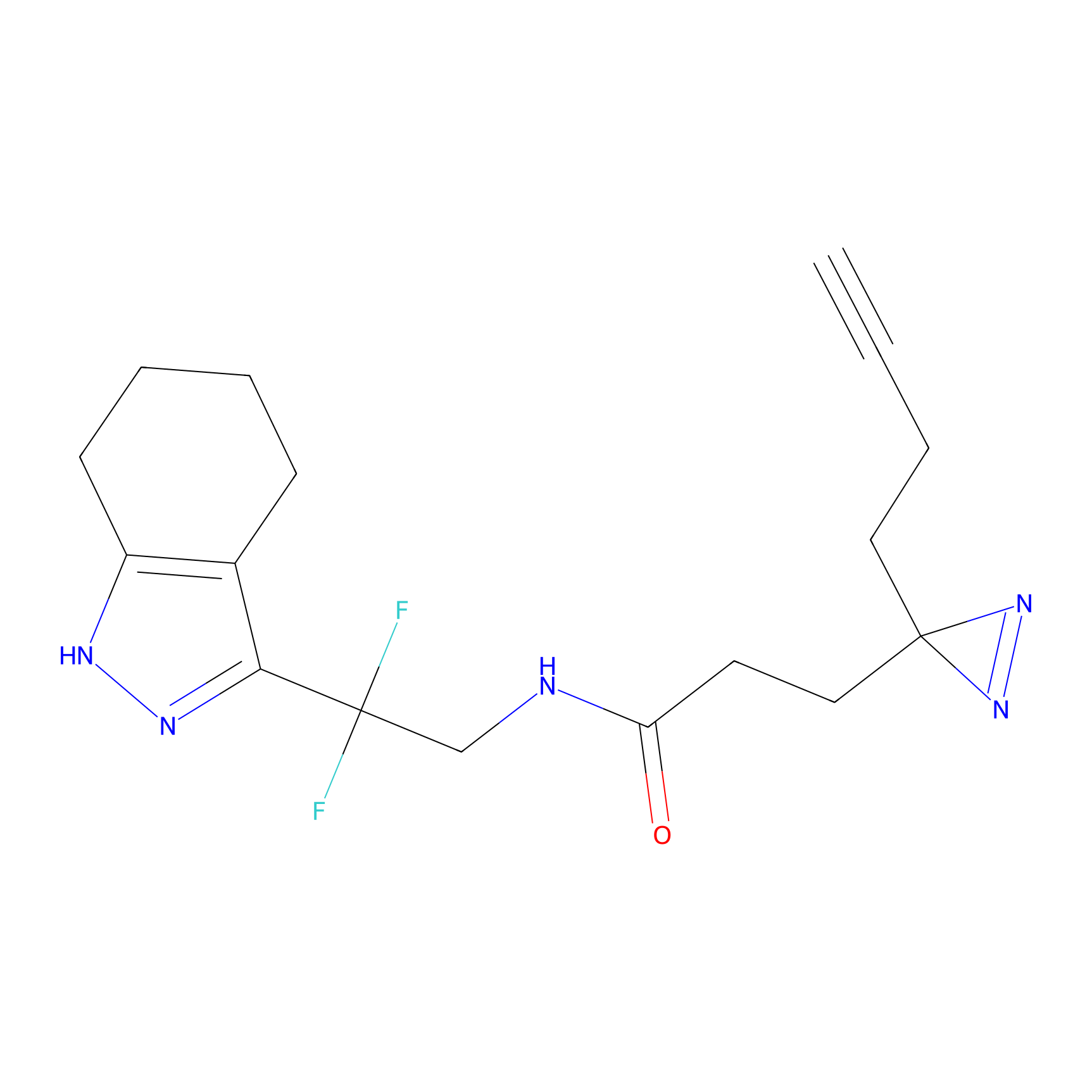 |
26.35 | LDD1752 | [6] | |
|
C056 Probe Info |
 |
57.28 | LDD1753 | [6] | |
|
C070 Probe Info |
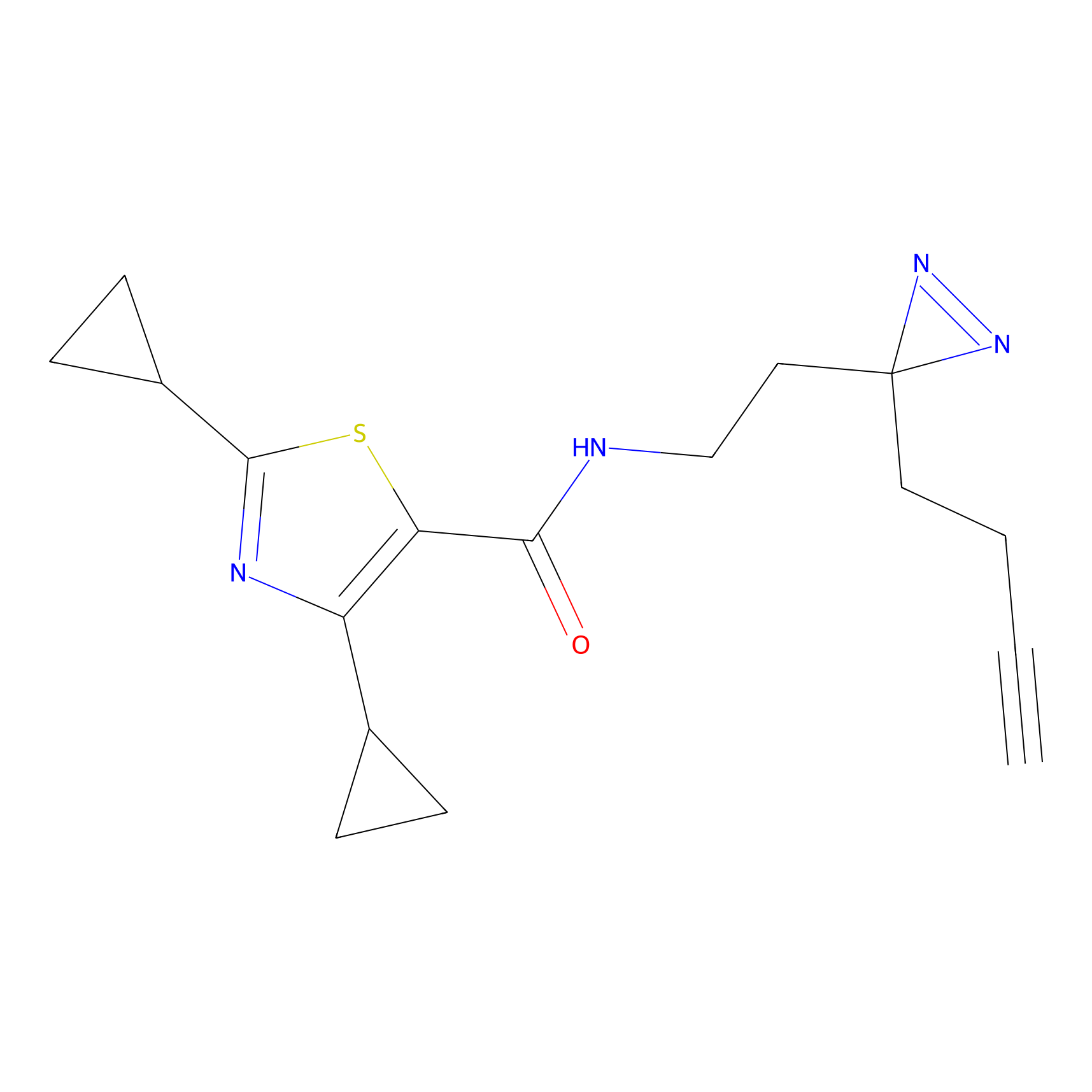 |
9.99 | LDD1766 | [6] | |
|
C072 Probe Info |
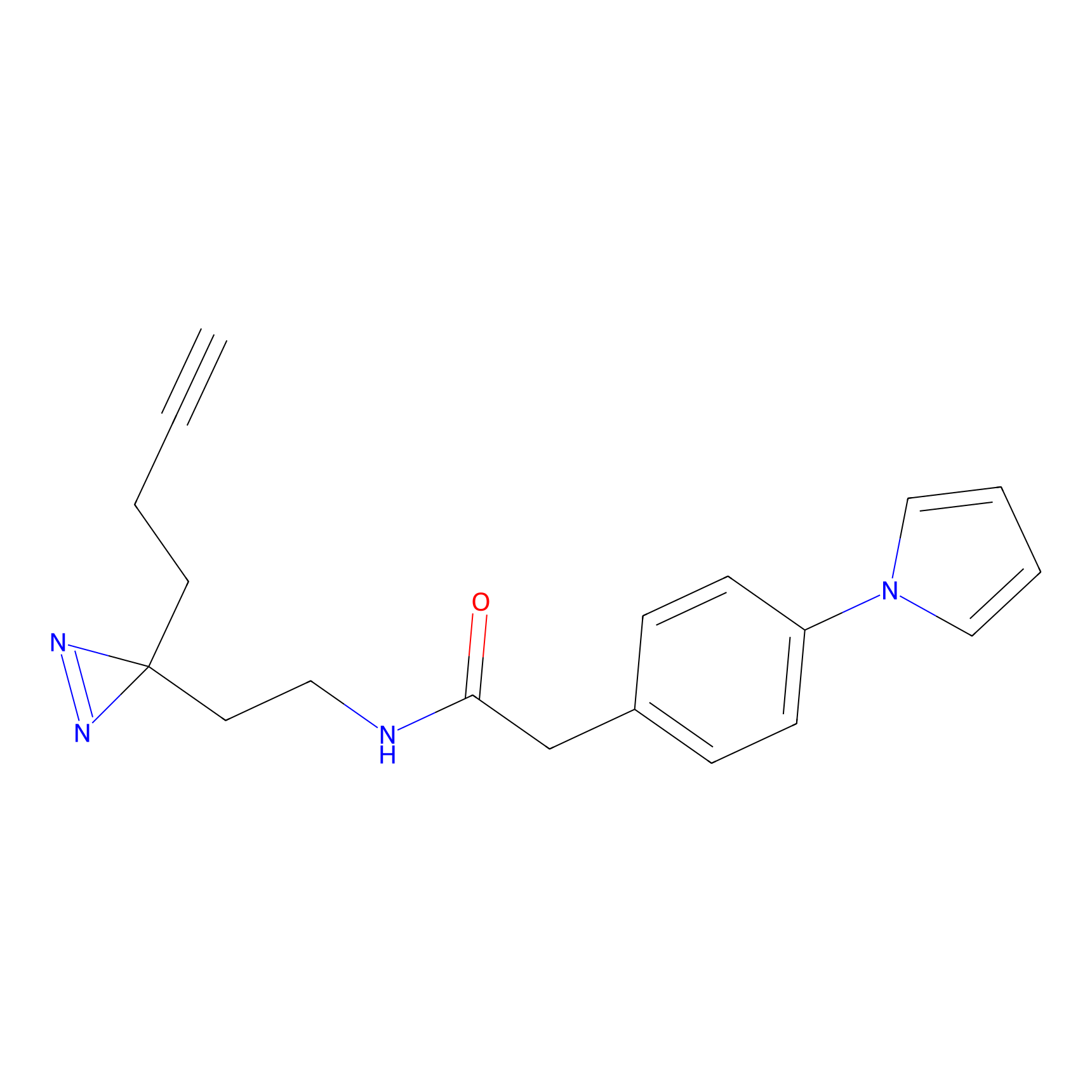 |
7.67 | LDD1768 | [6] | |
|
C112 Probe Info |
 |
37.79 | LDD1799 | [6] | |
|
C134 Probe Info |
 |
34.54 | LDD1816 | [6] | |
|
C135 Probe Info |
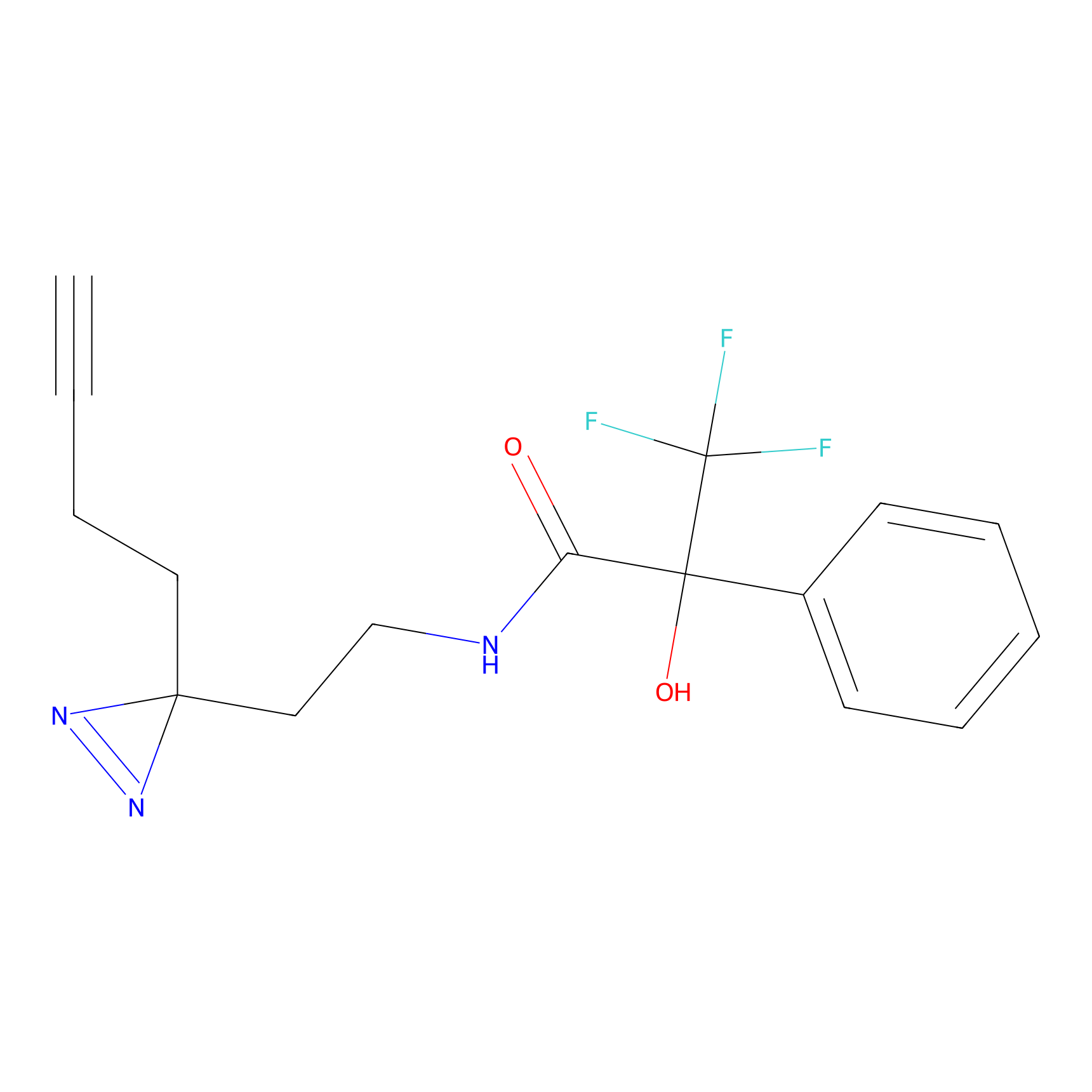 |
11.55 | LDD1817 | [6] | |
|
C177 Probe Info |
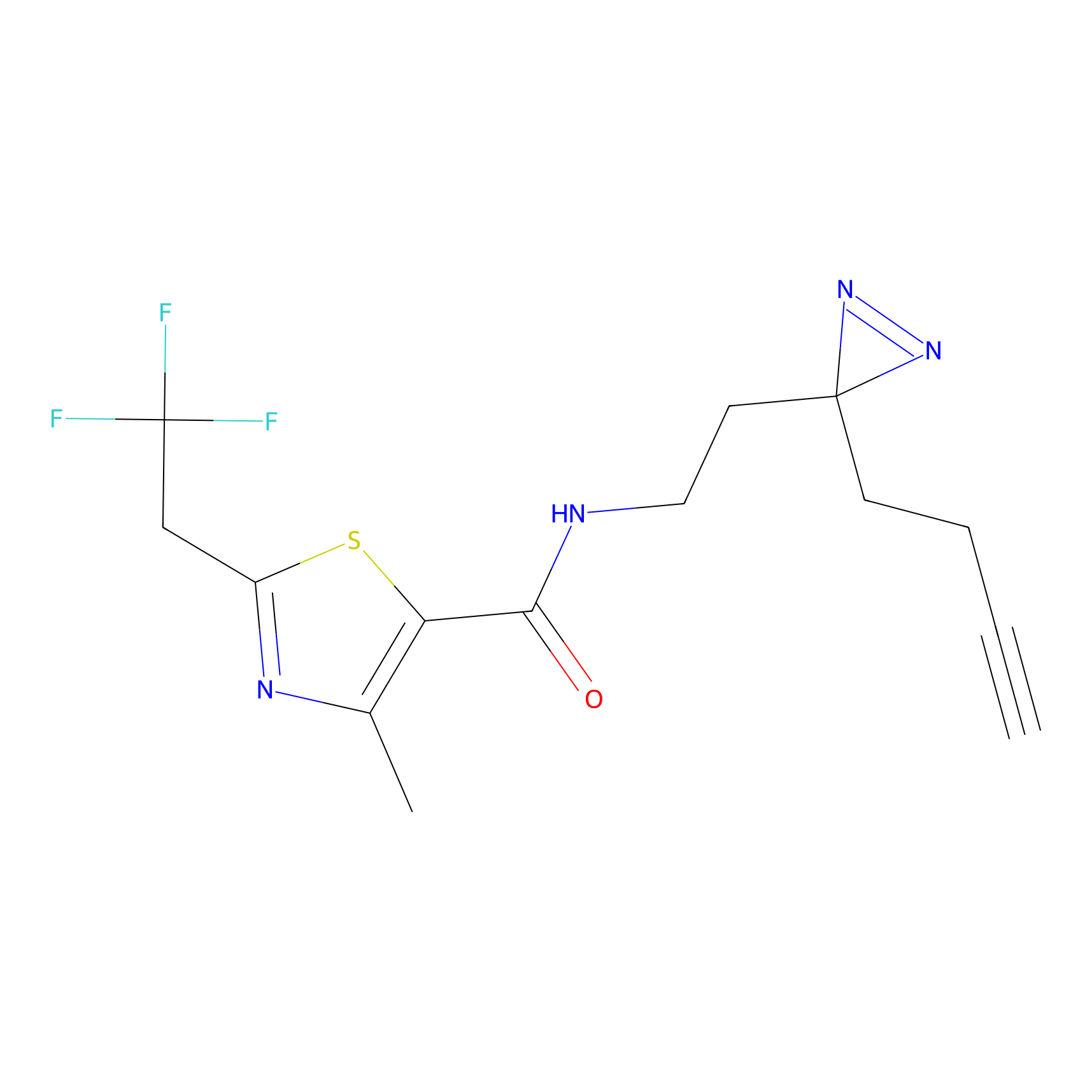 |
7.73 | LDD1856 | [6] | |
|
C178 Probe Info |
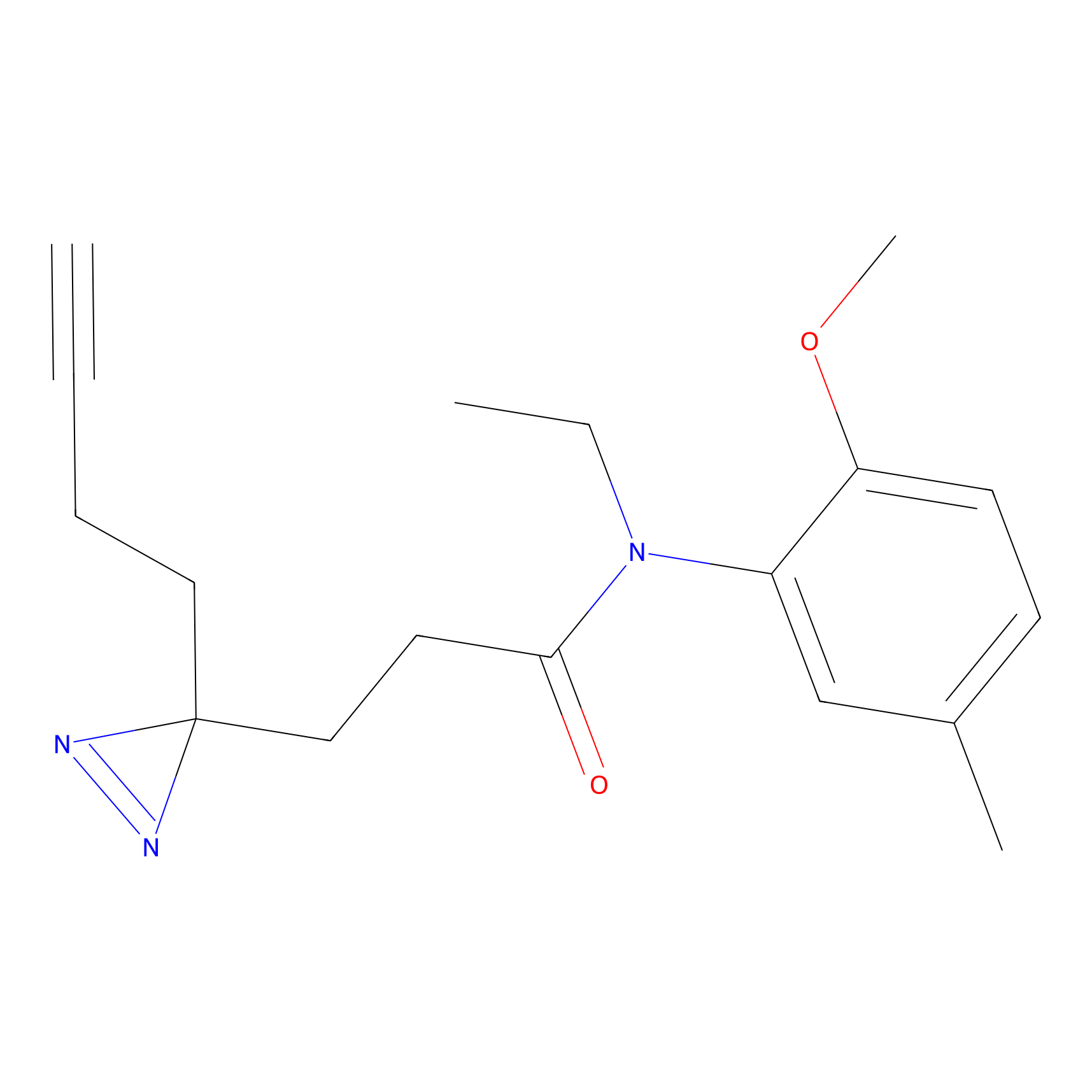 |
20.39 | LDD1857 | [6] | |
|
C246 Probe Info |
 |
15.56 | LDD1919 | [6] | |
|
C285 Probe Info |
 |
34.78 | LDD1955 | [6] | |
|
C286 Probe Info |
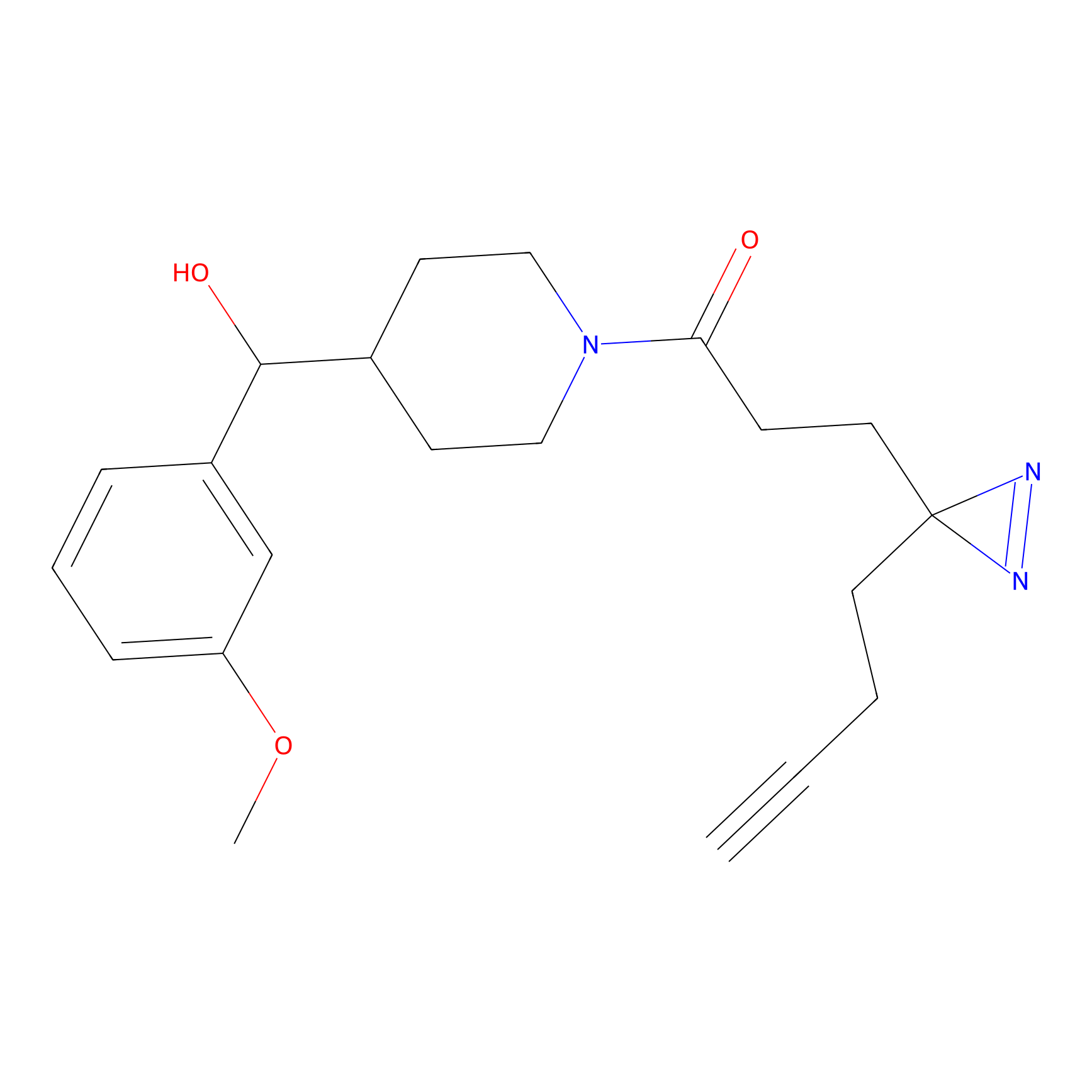 |
4.99 | LDD1956 | [6] | |
|
C287 Probe Info |
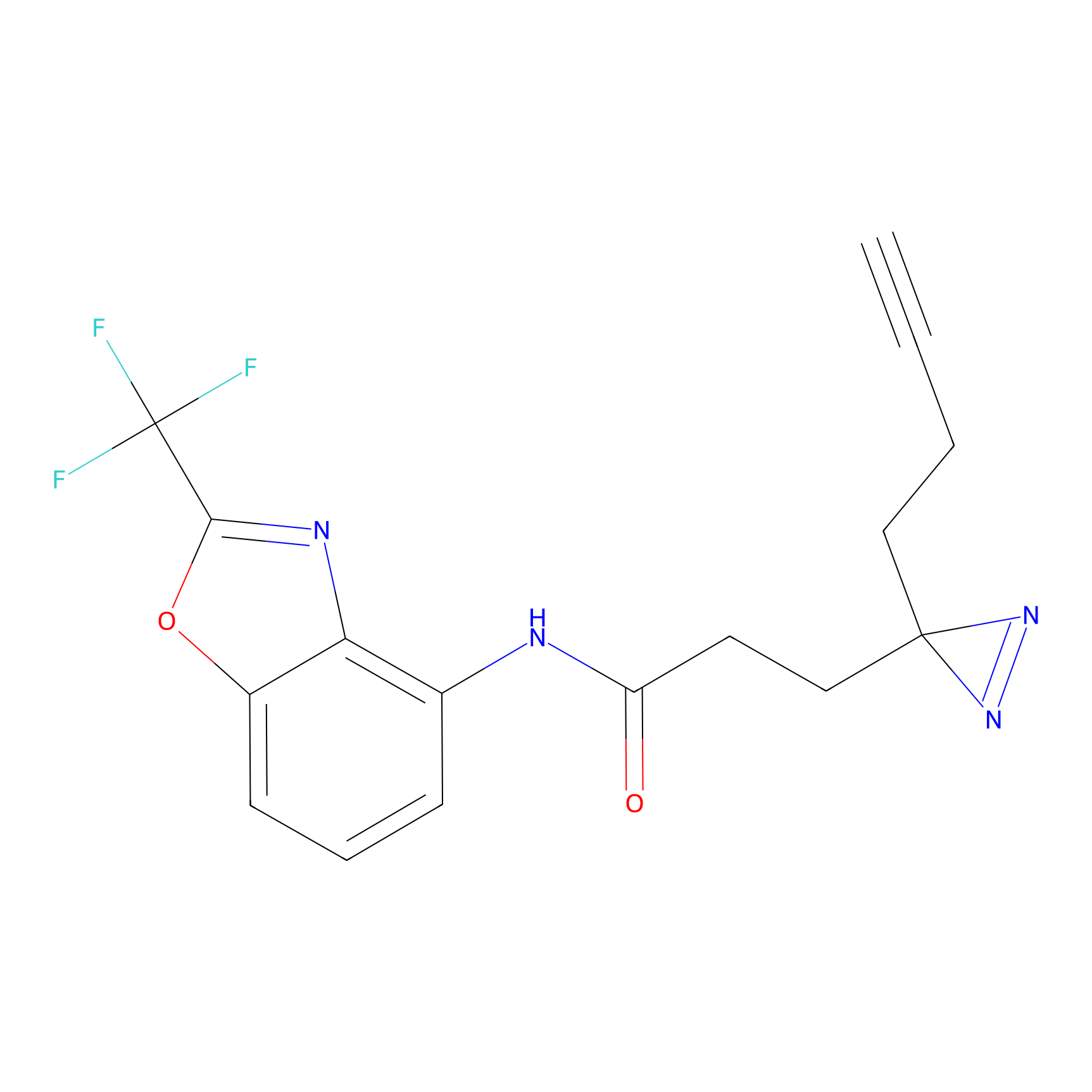 |
16.91 | LDD1957 | [6] | |
|
C288 Probe Info |
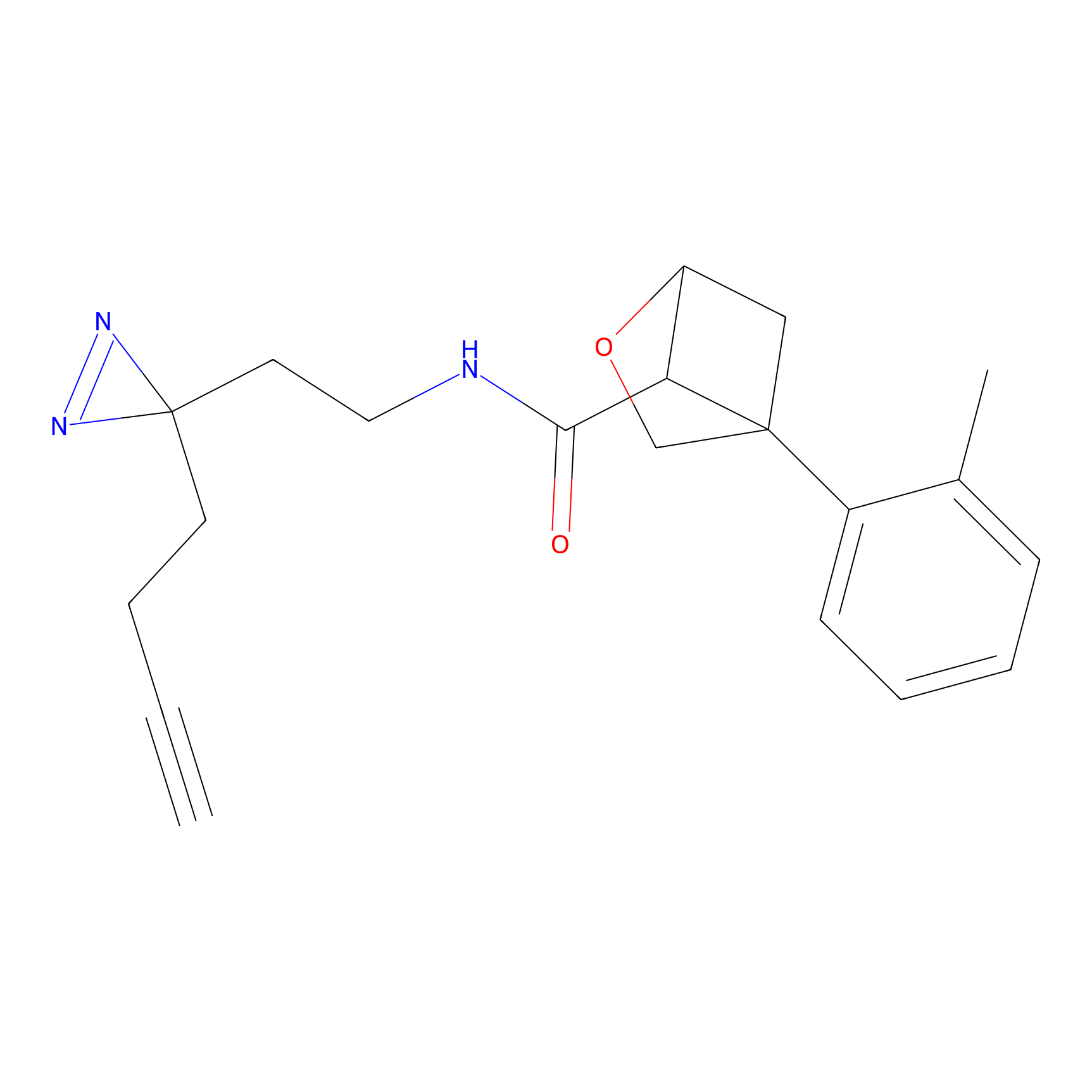 |
10.78 | LDD1958 | [6] | |
|
C289 Probe Info |
 |
83.29 | LDD1959 | [6] | |
|
C290 Probe Info |
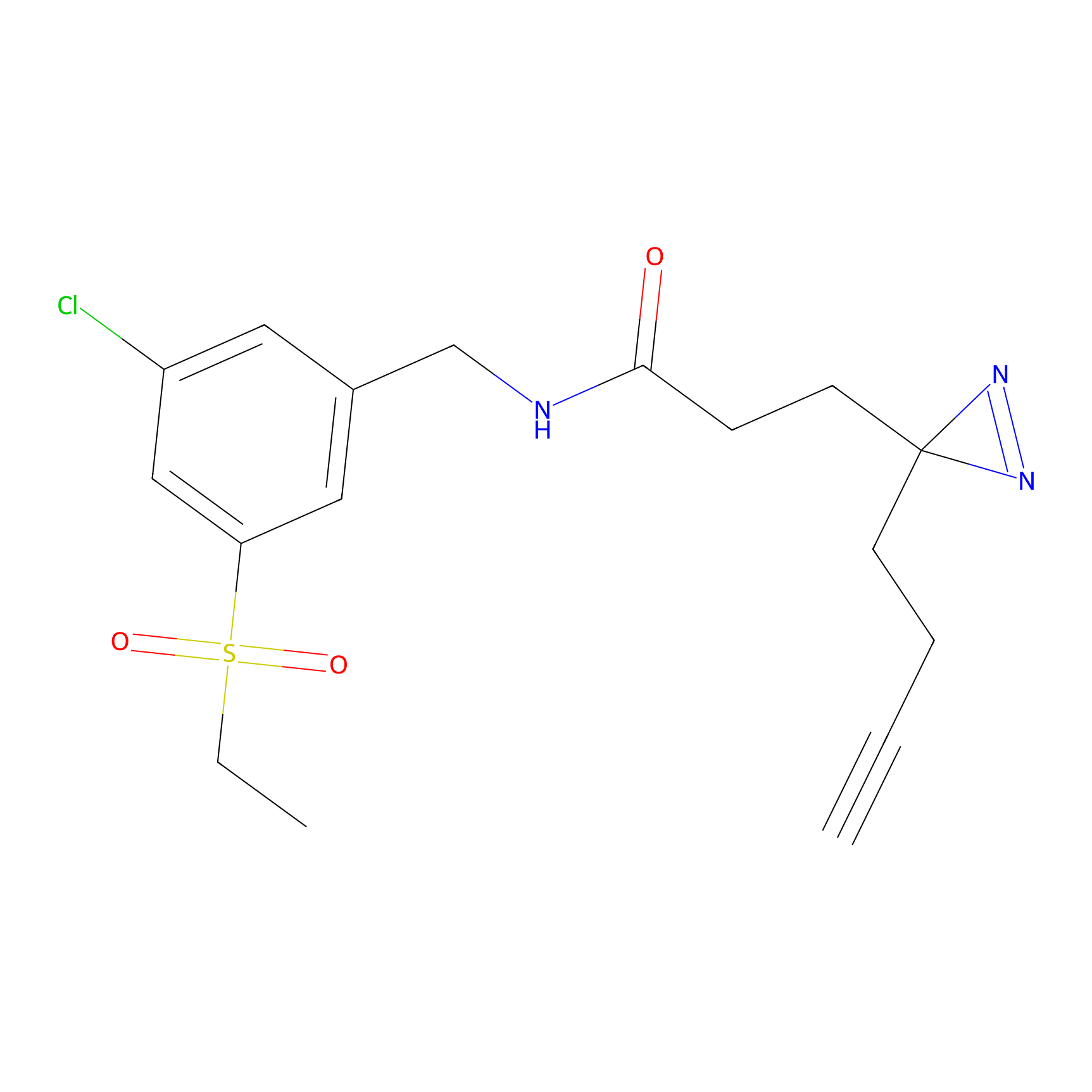 |
6.19 | LDD1960 | [6] | |
|
C362 Probe Info |
 |
26.91 | LDD2023 | [6] | |
|
C383 Probe Info |
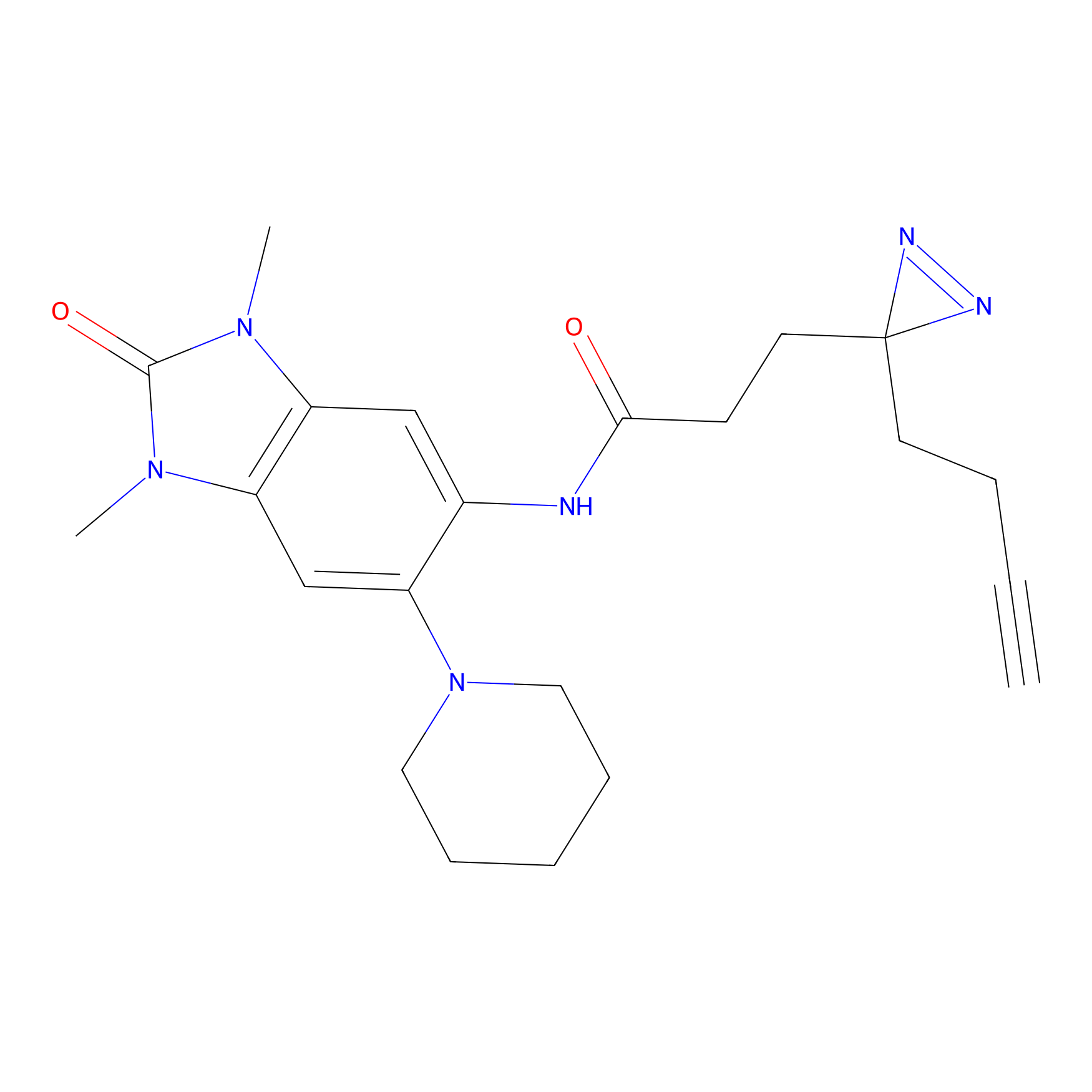 |
10.70 | LDD2042 | [6] | |
|
C388 Probe Info |
 |
99.73 | LDD2047 | [6] | |
|
C390 Probe Info |
 |
34.54 | LDD2049 | [6] | |
|
C391 Probe Info |
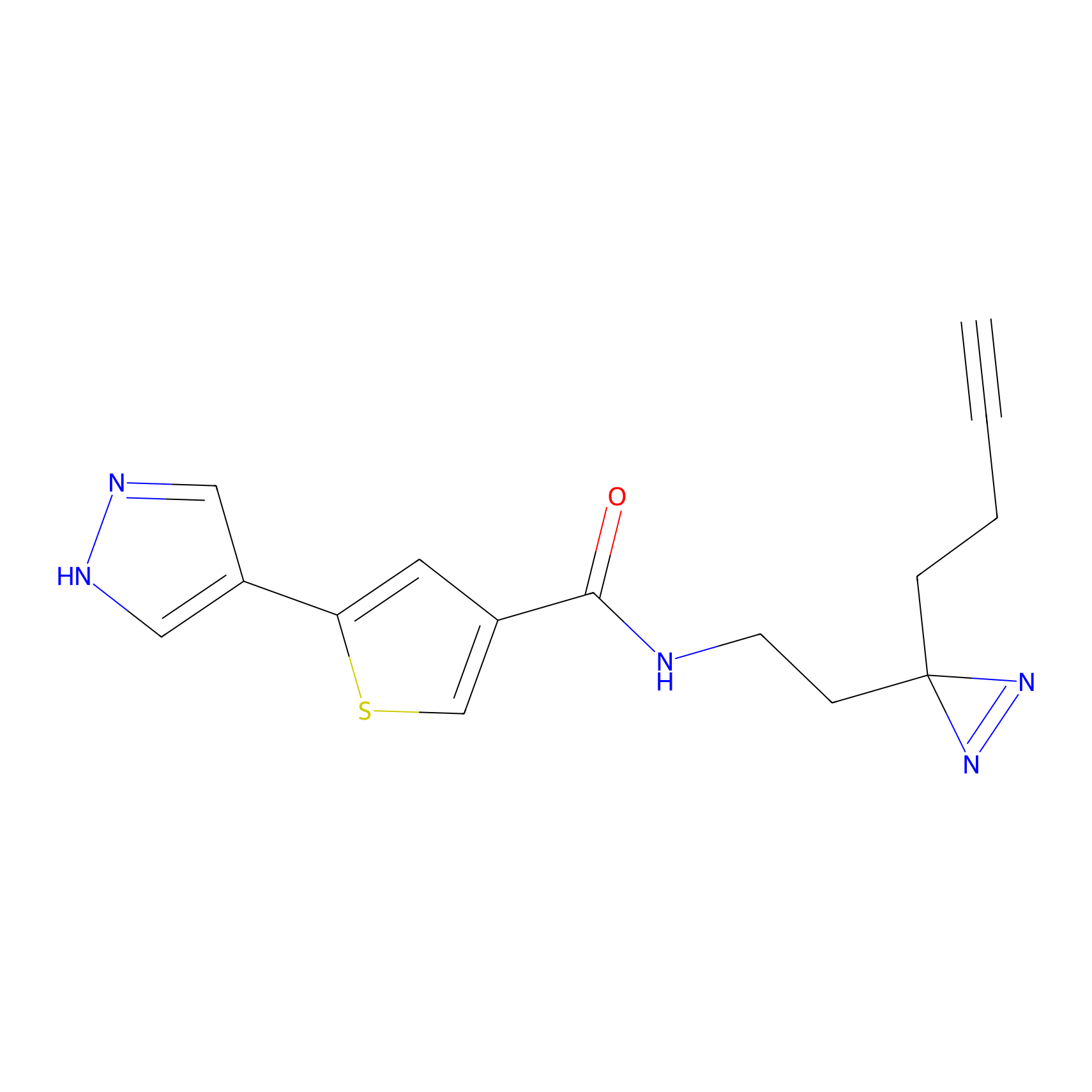 |
24.42 | LDD2050 | [6] | |
|
FFF probe11 Probe Info |
 |
20.00 | LDD0471 | [7] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [7] | |
|
FFF probe14 Probe Info |
 |
13.96 | LDD0477 | [7] | |
|
FFF probe2 Probe Info |
 |
18.84 | LDD0463 | [7] | |
|
A-DA Probe Info |
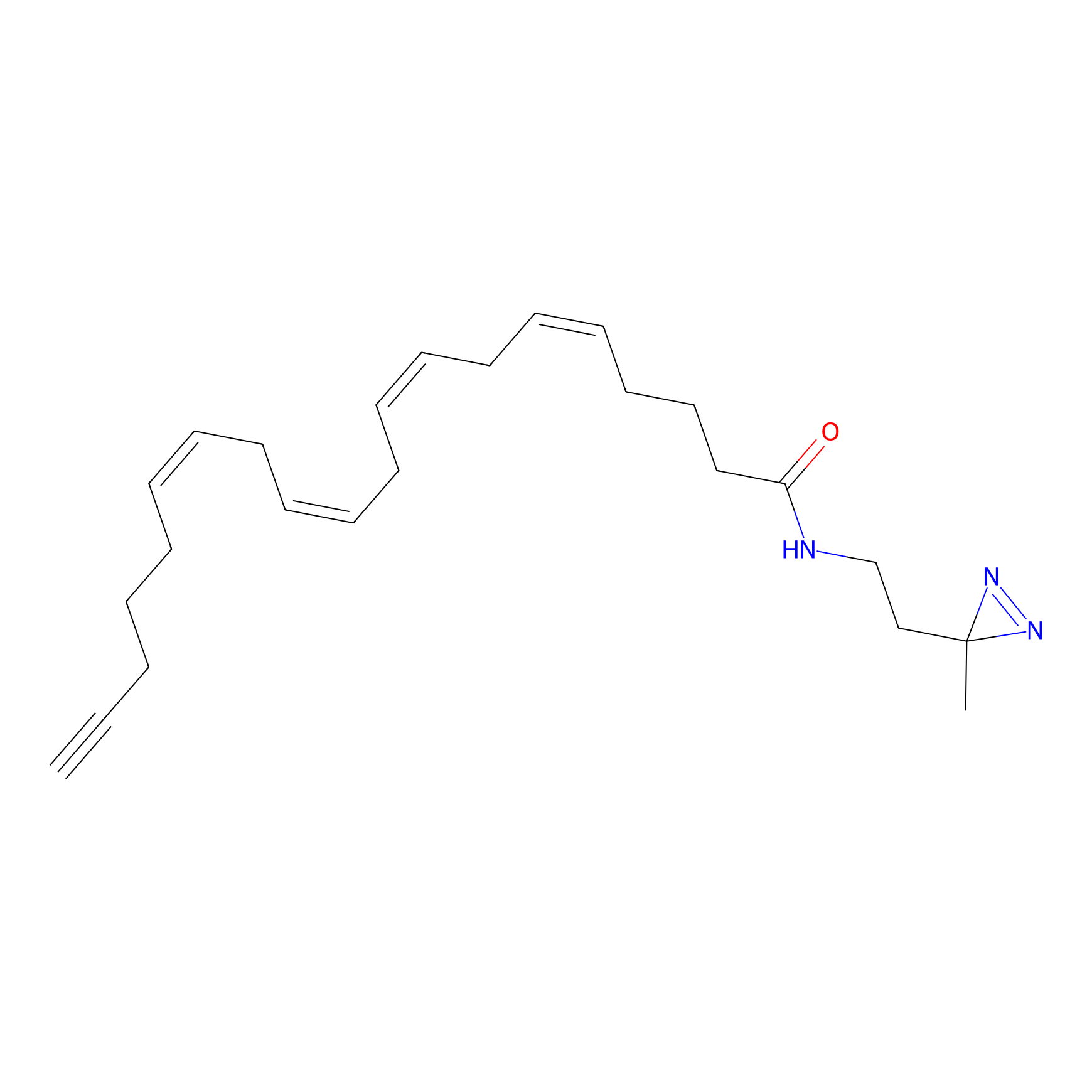 |
2.09 | LDD0145 | [8] | |
|
OEA-DA Probe Info |
 |
16.86 | LDD0046 | [9] | |
Competitor(s) Related to This Target
The Interaction Atlas With This Target
References
