Details of the Target
General Information of Target
| Target ID | LDTP11906 | |||||
|---|---|---|---|---|---|---|
| Target Name | Phosphatidylinositol glycan anchor biosynthesis class U protein (PIGU) | |||||
| Gene Name | PIGU | |||||
| Gene ID | 128869 | |||||
| Synonyms |
CDC91L1; Phosphatidylinositol glycan anchor biosynthesis class U protein; Cell division cycle protein 91-like 1; Protein CDC91-like 1; GPI transamidase component PIG-U |
|||||
| 3D Structure | ||||||
| Sequence |
MGAAAWARPLSVSFLLLLLPLPGMPAGSWDPAGYLLYCPCMGRFGNQADHFLGSLAFAKL
LNRTLAVPPWIEYQHHKPPFTNLHVSYQKYFKLEPLQAYHRVISLEDFMEKLAPTHWPPE KRVAYCFEVAAQRSPDKKTCPMKEGNPFGPFWDQFHVSFNKSELFTGISFSASYREQWSQ RFSPKEHPVLALPGAPAQFPVLEEHRPLQKYMVWSDEMVKTGEAQIHAHLVRPYVGIHLR IGSDWKNACAMLKDGTAGSHFMASPQCVGYSRSTAAPLTMTMCLPDLKEIQRAVKLWVRS LDAQSVYVATDSESYVPELQQLFKGKVKVVSLKPEVAQVDLYILGQADHFIGNCVSSFTA FVKRERDLQGRPSSFFGMDRPPKLRDEF |
|||||
| Target Type |
Literature-reported
|
|||||
| Target Bioclass |
Other
|
|||||
| Family |
PIGU family
|
|||||
| Subcellular location |
Endoplasmic reticulum membrane
|
|||||
| Function | Component of the GPI transamidase complex, necessary for transfer of GPI to proteins. May be involved in the recognition of either the GPI attachment signal or the lipid portion of GPI. | |||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
HDSF-alk Probe Info |
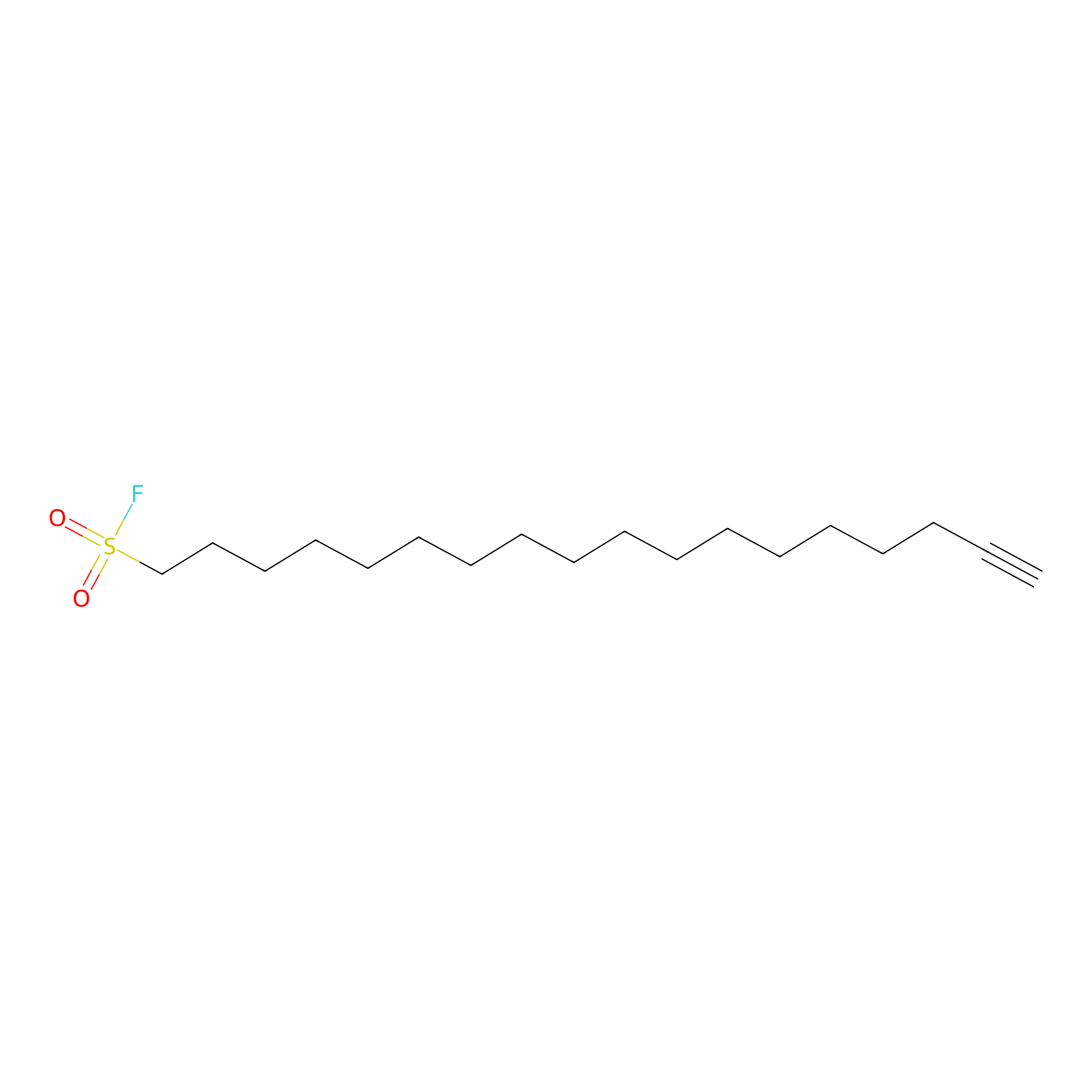 |
2.90 | LDD0197 | [1] | |
|
Jackson_1 Probe Info |
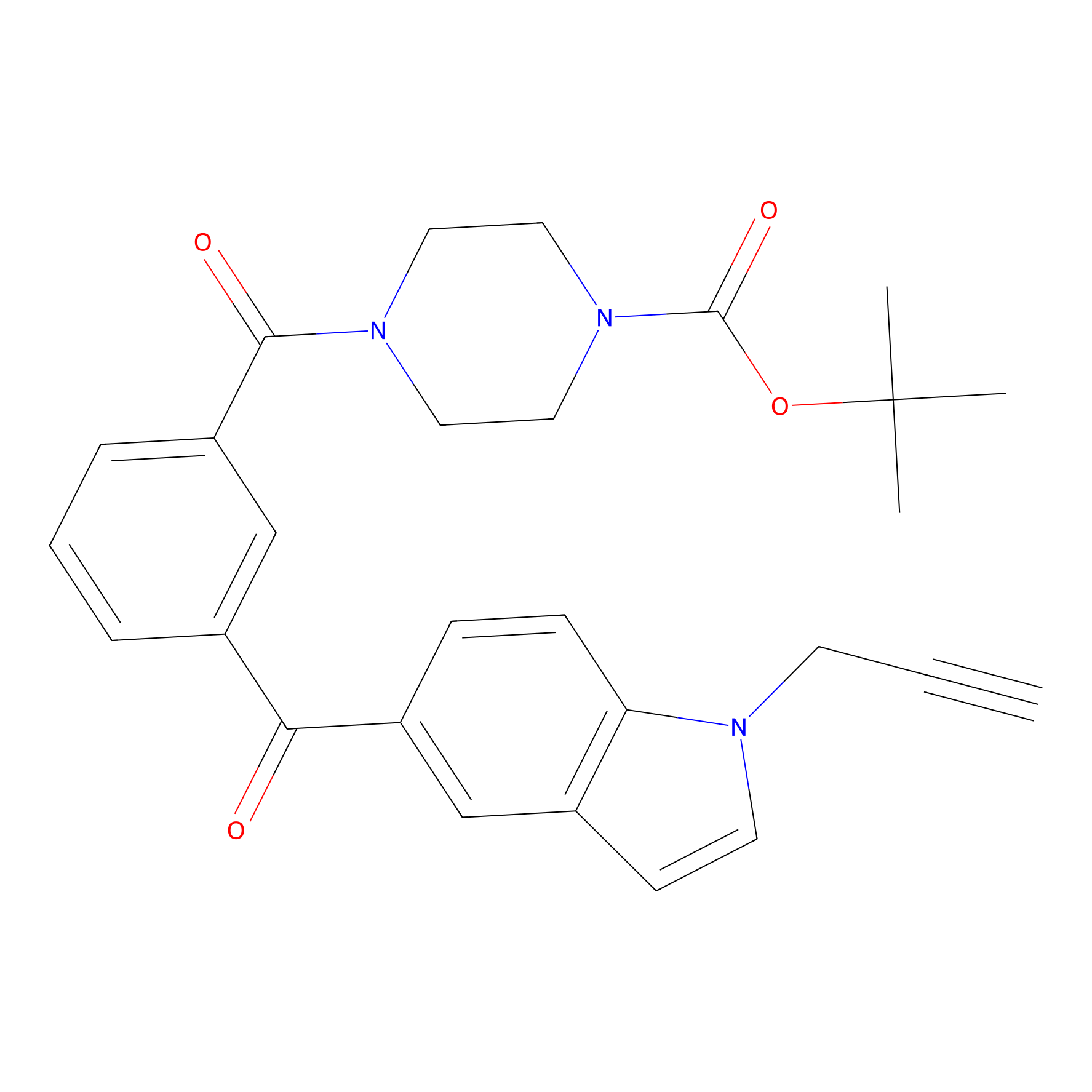 |
20.00 | LDD0122 | [2] | |
|
TH211 Probe Info |
 |
Y119(20.00) | LDD0260 | [3] | |
|
m-APA Probe Info |
 |
12.15 | LDD0403 | [4] | |
|
DBIA Probe Info |
 |
C154(1.48) | LDD3365 | [5] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C040 Probe Info |
 |
10.63 | LDD1740 | [6] | |
|
C091 Probe Info |
 |
17.15 | LDD1782 | [6] | |
|
C092 Probe Info |
 |
26.17 | LDD1783 | [6] | |
|
C094 Probe Info |
 |
42.22 | LDD1785 | [6] | |
|
C153 Probe Info |
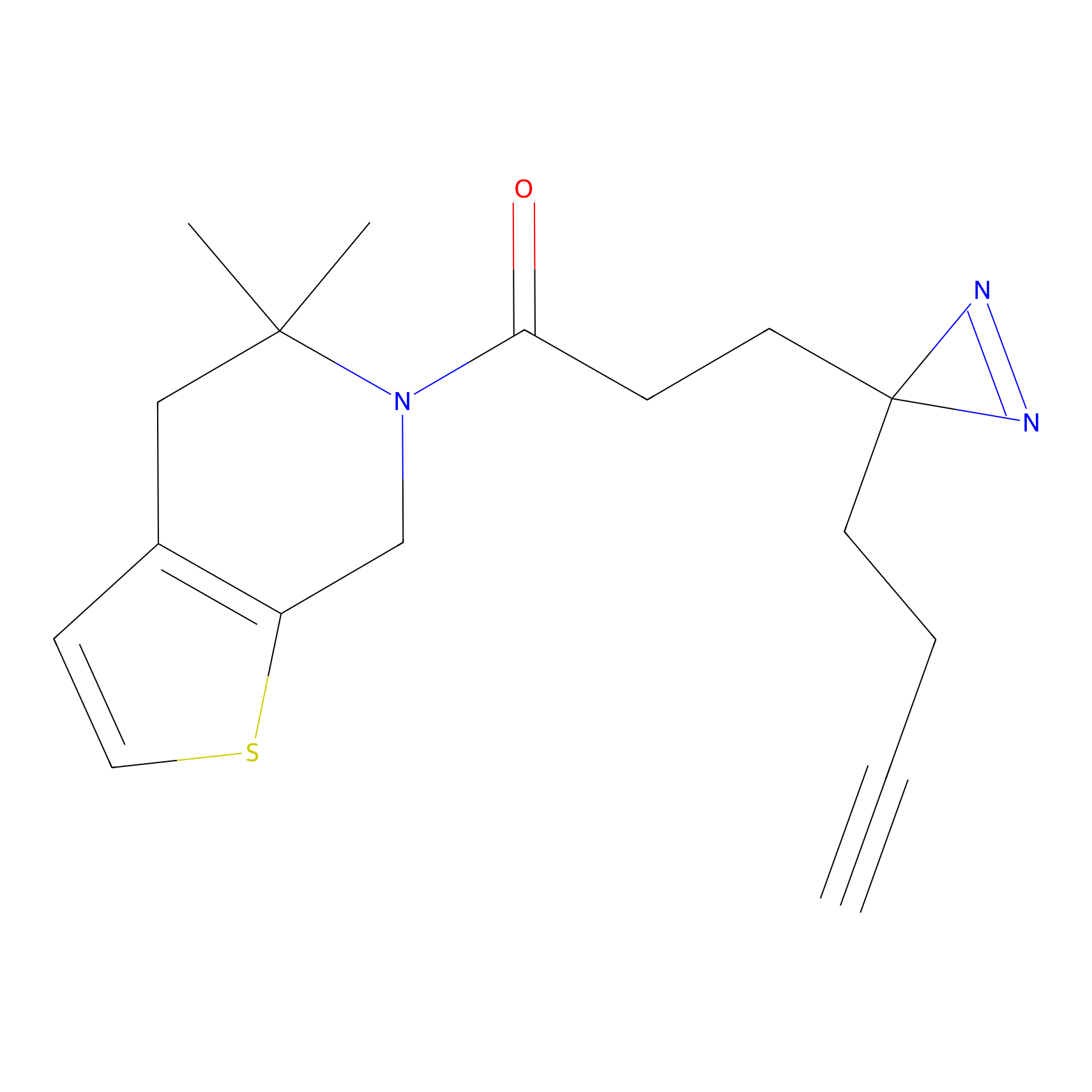 |
15.67 | LDD1834 | [6] | |
|
C161 Probe Info |
 |
10.78 | LDD1841 | [6] | |
|
C187 Probe Info |
 |
20.11 | LDD1865 | [6] | |
|
C218 Probe Info |
 |
18.64 | LDD1892 | [6] | |
|
C219 Probe Info |
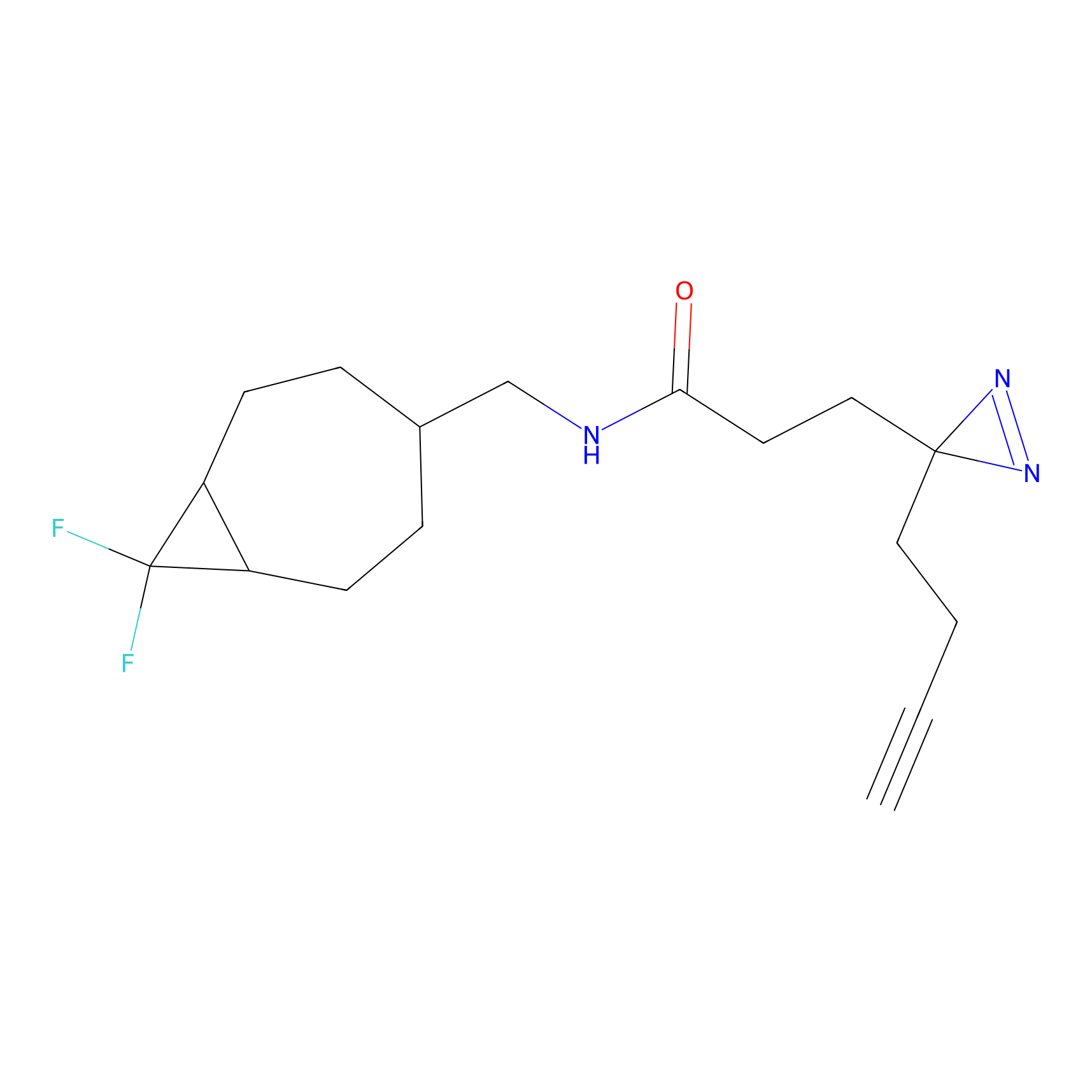 |
6.92 | LDD1893 | [6] | |
|
C220 Probe Info |
 |
22.94 | LDD1894 | [6] | |
|
C228 Probe Info |
 |
16.22 | LDD1901 | [6] | |
|
C231 Probe Info |
 |
24.59 | LDD1904 | [6] | |
|
C233 Probe Info |
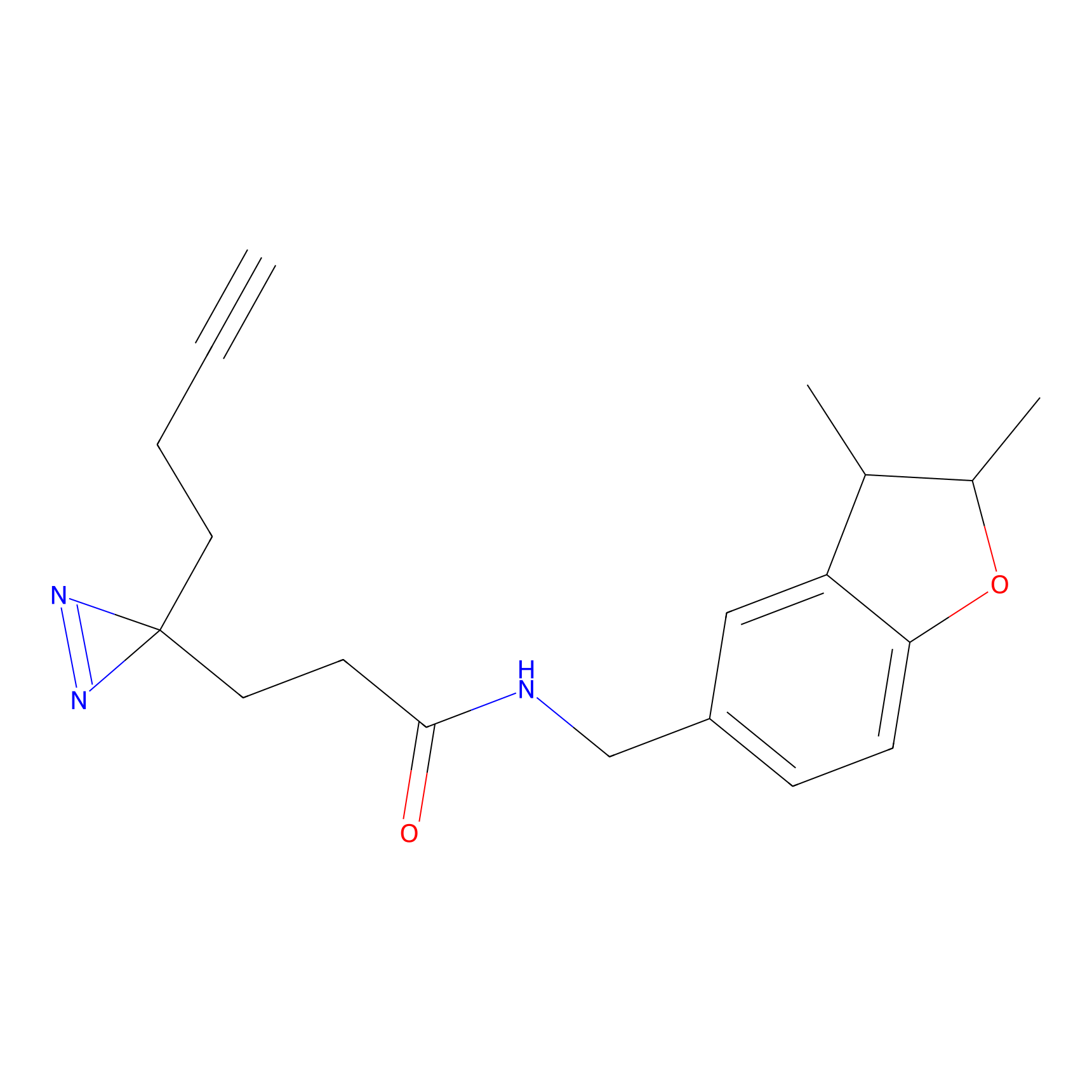 |
10.27 | LDD1906 | [6] | |
|
C235 Probe Info |
 |
25.28 | LDD1908 | [6] | |
|
C246 Probe Info |
 |
16.22 | LDD1919 | [6] | |
|
C278 Probe Info |
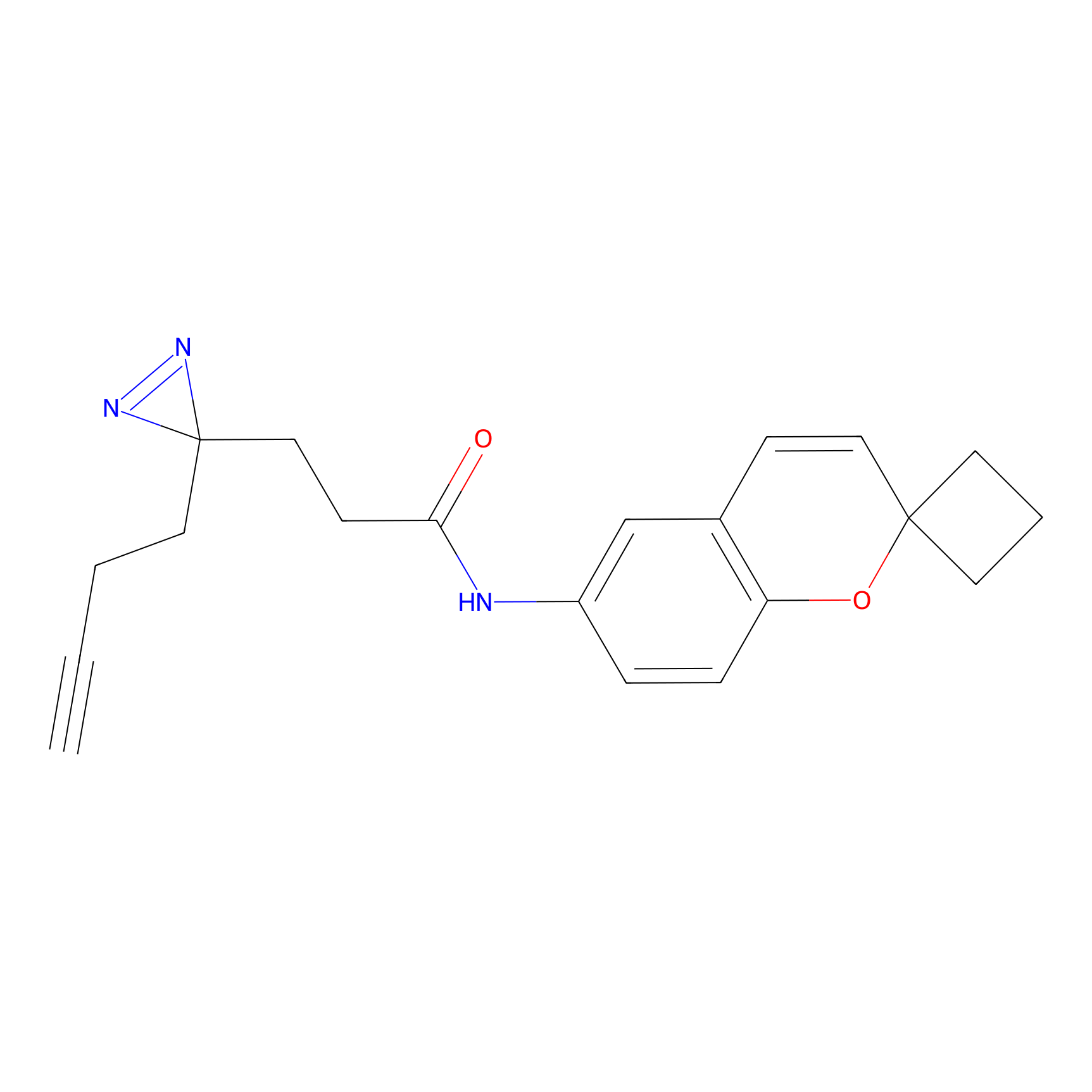 |
54.95 | LDD1948 | [6] | |
|
C282 Probe Info |
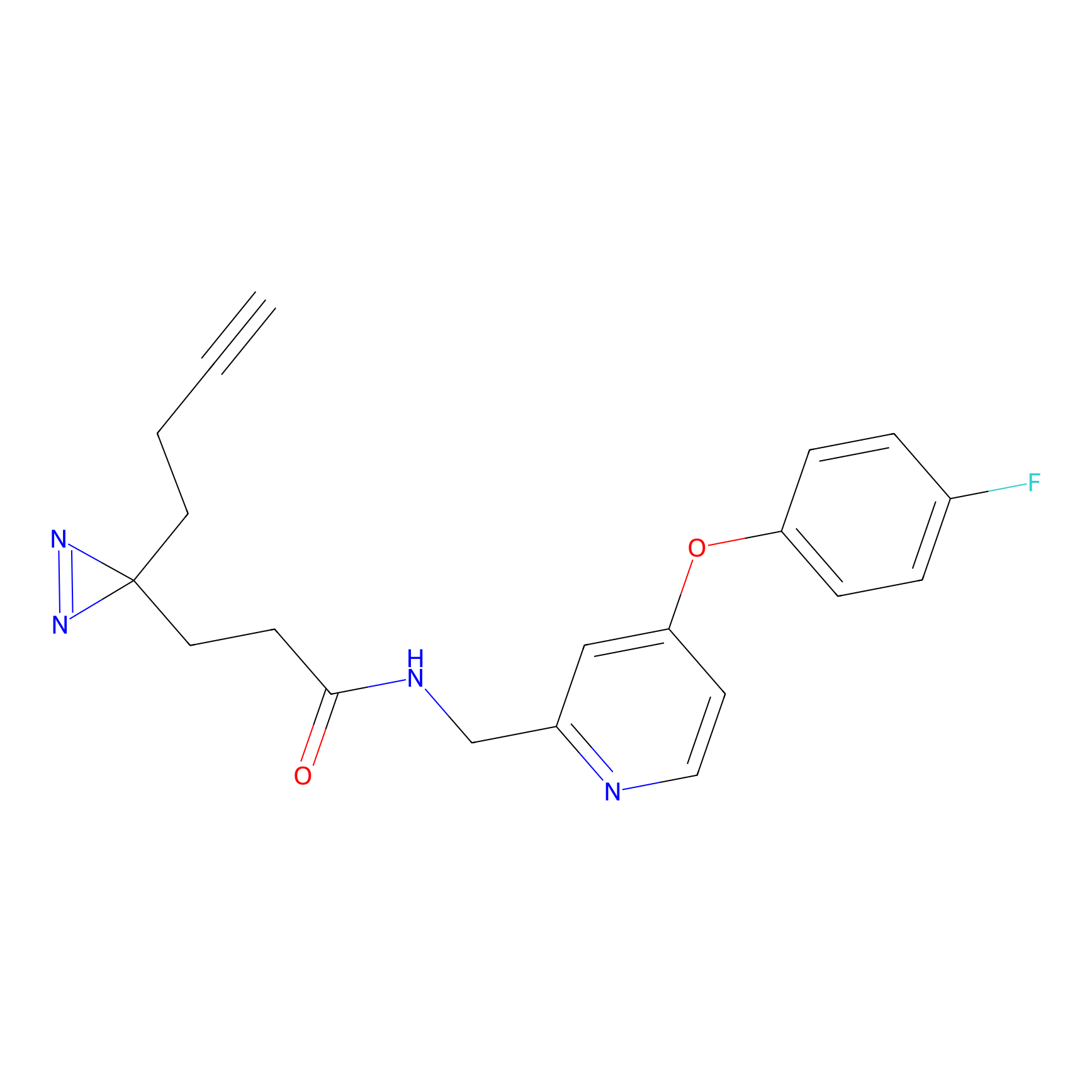 |
16.45 | LDD1952 | [6] | |
|
C285 Probe Info |
 |
19.16 | LDD1955 | [6] | |
|
C287 Probe Info |
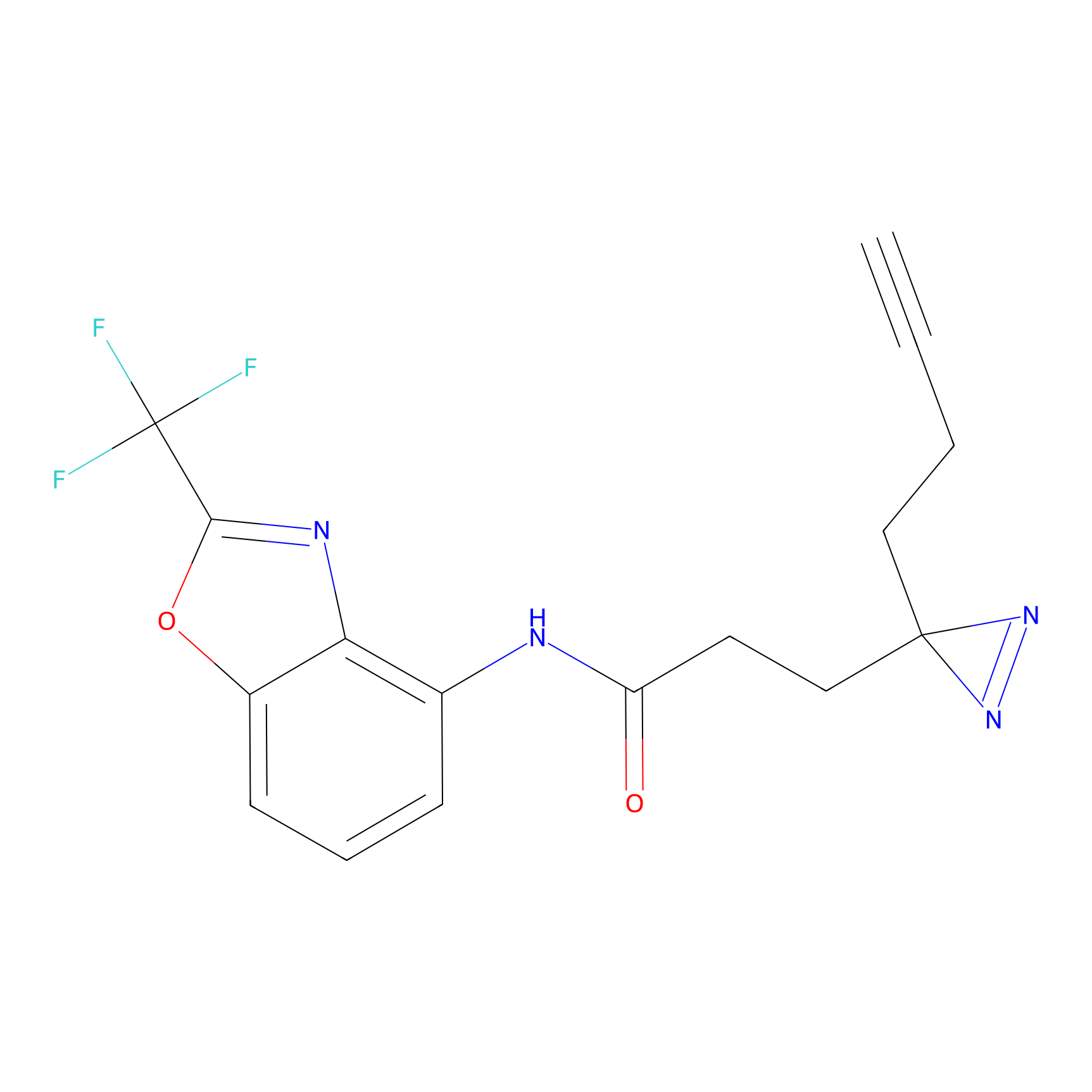 |
9.38 | LDD1957 | [6] | |
|
C289 Probe Info |
 |
33.59 | LDD1959 | [6] | |
|
C307 Probe Info |
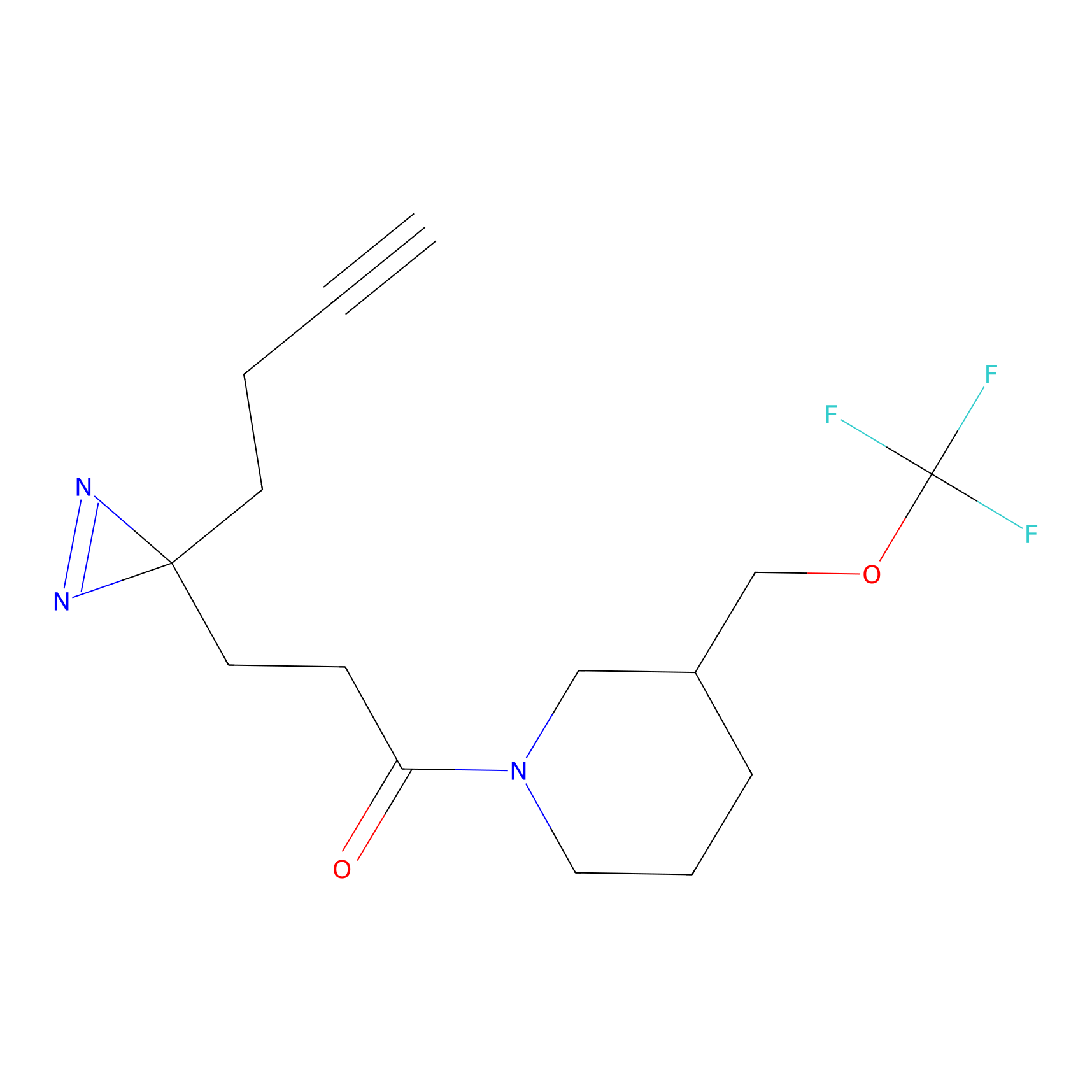 |
5.54 | LDD1975 | [6] | |
|
C310 Probe Info |
 |
13.83 | LDD1977 | [6] | |
|
C348 Probe Info |
 |
10.27 | LDD2009 | [6] | |
|
C349 Probe Info |
 |
9.25 | LDD2010 | [6] | |
|
C350 Probe Info |
 |
41.07 | LDD2011 | [6] | |
|
C388 Probe Info |
 |
37.01 | LDD2047 | [6] | |
|
FFF probe11 Probe Info |
 |
20.00 | LDD0471 | [7] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [7] | |
|
FFF probe3 Probe Info |
 |
19.99 | LDD0464 | [7] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [8] | |
|
A-DA Probe Info |
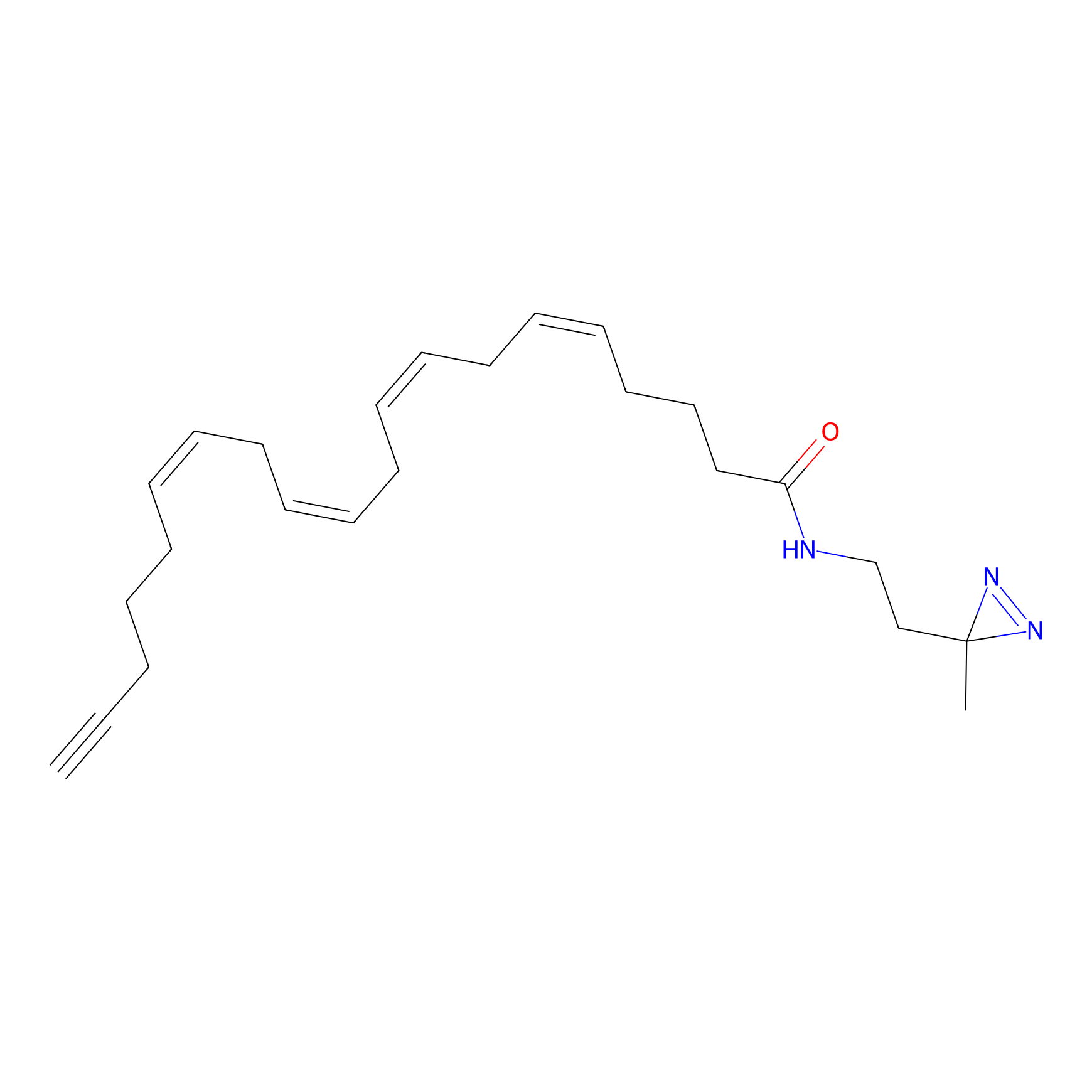 |
2.19 | LDD0145 | [9] | |
|
OEA-DA Probe Info |
 |
9.85 | LDD0046 | [10] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HEK-293T | C154(0.97) | LDD1507 | [11] |
| LDCM0237 | AC12 | HEK-293T | C154(1.22) | LDD1510 | [11] |
| LDCM0259 | AC14 | HEK-293T | C154(0.97) | LDD1512 | [11] |
| LDCM0276 | AC17 | HEK-293T | C154(0.94) | LDD1515 | [11] |
| LDCM0280 | AC20 | HEK-293T | C154(0.97) | LDD1519 | [11] |
| LDCM0281 | AC21 | HEK-293T | C154(0.89) | LDD1520 | [11] |
| LDCM0282 | AC22 | HEK-293T | C154(0.99) | LDD1521 | [11] |
| LDCM0285 | AC25 | HEK-293T | C154(1.12) | LDD1524 | [11] |
| LDCM0288 | AC28 | HEK-293T | C154(0.98) | LDD1527 | [11] |
| LDCM0289 | AC29 | HEK-293T | C154(0.91) | LDD1528 | [11] |
| LDCM0291 | AC30 | HEK-293T | C154(1.03) | LDD1530 | [11] |
| LDCM0294 | AC33 | HEK-293T | C154(0.97) | LDD1533 | [11] |
| LDCM0297 | AC36 | HEK-293T | C154(0.94) | LDD1536 | [11] |
| LDCM0298 | AC37 | HEK-293T | C154(0.80) | LDD1537 | [11] |
| LDCM0299 | AC38 | HEK-293T | C154(1.02) | LDD1538 | [11] |
| LDCM0301 | AC4 | HEK-293T | C154(1.16) | LDD1540 | [11] |
| LDCM0303 | AC41 | HEK-293T | C154(1.08) | LDD1542 | [11] |
| LDCM0306 | AC44 | HEK-293T | C154(1.16) | LDD1545 | [11] |
| LDCM0307 | AC45 | HEK-293T | C154(0.91) | LDD1546 | [11] |
| LDCM0308 | AC46 | HEK-293T | C154(0.97) | LDD1547 | [11] |
| LDCM0311 | AC49 | HEK-293T | C154(0.99) | LDD1550 | [11] |
| LDCM0312 | AC5 | HEK-293T | C154(0.85) | LDD1551 | [11] |
| LDCM0315 | AC52 | HEK-293T | C154(1.10) | LDD1554 | [11] |
| LDCM0316 | AC53 | HEK-293T | C154(0.87) | LDD1555 | [11] |
| LDCM0317 | AC54 | HEK-293T | C154(0.99) | LDD1556 | [11] |
| LDCM0320 | AC57 | HEK-293T | C154(0.98) | LDD1559 | [11] |
| LDCM0323 | AC6 | HEK-293T | C154(1.02) | LDD1562 | [11] |
| LDCM0324 | AC60 | HEK-293T | C154(1.18) | LDD1563 | [11] |
| LDCM0325 | AC61 | HEK-293T | C154(0.99) | LDD1564 | [11] |
| LDCM0326 | AC62 | HEK-293T | C154(1.24) | LDD1565 | [11] |
| LDCM0248 | AKOS034007472 | HEK-293T | C154(0.86) | LDD1511 | [11] |
| LDCM0356 | AKOS034007680 | HEK-293T | C154(1.00) | LDD1570 | [11] |
| LDCM0156 | Aniline | NCI-H1299 | 12.15 | LDD0403 | [4] |
| LDCM0367 | CL1 | HEK-293T | C154(1.24) | LDD1571 | [11] |
| LDCM0368 | CL10 | HEK-293T | C154(2.15) | LDD1572 | [11] |
| LDCM0370 | CL101 | HEK-293T | C154(0.99) | LDD1574 | [11] |
| LDCM0372 | CL103 | HEK-293T | C154(1.12) | LDD1576 | [11] |
| LDCM0374 | CL105 | HEK-293T | C154(1.40) | LDD1578 | [11] |
| LDCM0376 | CL107 | HEK-293T | C154(1.07) | LDD1580 | [11] |
| LDCM0378 | CL109 | HEK-293T | C154(1.16) | LDD1582 | [11] |
| LDCM0381 | CL111 | HEK-293T | C154(1.35) | LDD1585 | [11] |
| LDCM0383 | CL113 | HEK-293T | C154(1.18) | LDD1587 | [11] |
| LDCM0385 | CL115 | HEK-293T | C154(1.02) | LDD1589 | [11] |
| LDCM0387 | CL117 | HEK-293T | C154(0.87) | LDD1591 | [11] |
| LDCM0389 | CL119 | HEK-293T | C154(1.00) | LDD1593 | [11] |
| LDCM0392 | CL121 | HEK-293T | C154(0.94) | LDD1596 | [11] |
| LDCM0394 | CL123 | HEK-293T | C154(1.62) | LDD1598 | [11] |
| LDCM0396 | CL125 | HEK-293T | C154(1.21) | LDD1600 | [11] |
| LDCM0398 | CL127 | HEK-293T | C154(1.25) | LDD1602 | [11] |
| LDCM0400 | CL13 | HEK-293T | C154(1.04) | LDD1604 | [11] |
| LDCM0402 | CL15 | HEK-293T | C154(2.16) | LDD1606 | [11] |
| LDCM0404 | CL17 | HEK-293T | C154(1.93) | LDD1608 | [11] |
| LDCM0408 | CL20 | HEK-293T | C154(1.07) | LDD1612 | [11] |
| LDCM0409 | CL21 | HEK-293T | C154(1.12) | LDD1613 | [11] |
| LDCM0410 | CL22 | HEK-293T | C154(1.11) | LDD1614 | [11] |
| LDCM0413 | CL25 | HEK-293T | C154(1.36) | LDD1617 | [11] |
| LDCM0415 | CL27 | HEK-293T | C154(1.09) | LDD1619 | [11] |
| LDCM0417 | CL29 | HEK-293T | C154(1.00) | LDD1621 | [11] |
| LDCM0418 | CL3 | HEK-293T | C154(1.59) | LDD1622 | [11] |
| LDCM0421 | CL32 | HEK-293T | C154(1.15) | LDD1625 | [11] |
| LDCM0422 | CL33 | HEK-293T | C154(1.59) | LDD1626 | [11] |
| LDCM0423 | CL34 | HEK-293T | C154(1.21) | LDD1627 | [11] |
| LDCM0426 | CL37 | HEK-293T | C154(1.24) | LDD1630 | [11] |
| LDCM0428 | CL39 | HEK-293T | C154(1.14) | LDD1632 | [11] |
| LDCM0431 | CL41 | HEK-293T | C154(1.27) | LDD1635 | [11] |
| LDCM0434 | CL44 | HEK-293T | C154(0.99) | LDD1638 | [11] |
| LDCM0435 | CL45 | HEK-293T | C154(0.90) | LDD1639 | [11] |
| LDCM0436 | CL46 | HEK-293T | C154(0.95) | LDD1640 | [11] |
| LDCM0439 | CL49 | HEK-293T | C154(1.46) | LDD1643 | [11] |
| LDCM0440 | CL5 | HEK-293T | C154(1.19) | LDD1644 | [11] |
| LDCM0444 | CL53 | HEK-293T | C154(1.39) | LDD1647 | [11] |
| LDCM0447 | CL56 | HEK-293T | C154(1.14) | LDD1650 | [11] |
| LDCM0448 | CL57 | HEK-293T | C154(1.12) | LDD1651 | [11] |
| LDCM0449 | CL58 | HEK-293T | C154(1.17) | LDD1652 | [11] |
| LDCM0453 | CL61 | HEK-293T | C154(1.52) | LDD1656 | [11] |
| LDCM0455 | CL63 | HEK-293T | C154(0.96) | LDD1658 | [11] |
| LDCM0457 | CL65 | HEK-293T | C154(1.13) | LDD1660 | [11] |
| LDCM0460 | CL68 | HEK-293T | C154(1.17) | LDD1663 | [11] |
| LDCM0461 | CL69 | HEK-293T | C154(1.07) | LDD1664 | [11] |
| LDCM0463 | CL70 | HEK-293T | C154(1.09) | LDD1666 | [11] |
| LDCM0466 | CL73 | HEK-293T | C154(1.42) | LDD1669 | [11] |
| LDCM0470 | CL77 | HEK-293T | C154(1.27) | LDD1673 | [11] |
| LDCM0473 | CL8 | HEK-293T | C154(3.30) | LDD1676 | [11] |
| LDCM0474 | CL80 | HEK-293T | C154(1.04) | LDD1677 | [11] |
| LDCM0475 | CL81 | HEK-293T | C154(0.92) | LDD1678 | [11] |
| LDCM0476 | CL82 | HEK-293T | C154(0.98) | LDD1679 | [11] |
| LDCM0479 | CL85 | HEK-293T | C154(1.43) | LDD1682 | [11] |
| LDCM0481 | CL87 | HEK-293T | C154(1.29) | LDD1684 | [11] |
| LDCM0483 | CL89 | HEK-293T | C154(1.18) | LDD1686 | [11] |
| LDCM0484 | CL9 | HEK-293T | C154(1.07) | LDD1687 | [11] |
| LDCM0487 | CL92 | HEK-293T | C154(1.46) | LDD1690 | [11] |
| LDCM0488 | CL93 | HEK-293T | C154(1.24) | LDD1691 | [11] |
| LDCM0489 | CL94 | HEK-293T | C154(1.17) | LDD1692 | [11] |
| LDCM0492 | CL97 | HEK-293T | C154(1.89) | LDD1695 | [11] |
| LDCM0494 | CL99 | HEK-293T | C154(1.01) | LDD1697 | [11] |
| LDCM0495 | E2913 | HEK-293T | C154(1.01) | LDD1698 | [11] |
| LDCM0468 | Fragment33 | HEK-293T | C154(0.96) | LDD1671 | [11] |
| LDCM0022 | KB02 | HEK-293T | C154(0.94) | LDD1492 | [11] |
| LDCM0023 | KB03 | HEK-293T | C154(0.95) | LDD1497 | [11] |
| LDCM0024 | KB05 | OVCAR-3 | C154(1.48) | LDD3365 | [5] |
| LDCM0084 | Ro 48-8071 | A-549 | 2.19 | LDD0145 | [9] |
The Interaction Atlas With This Target
References
