Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
9.87 | LDD0402 | [2] | |
|
Alkylaryl probe 1 Probe Info |
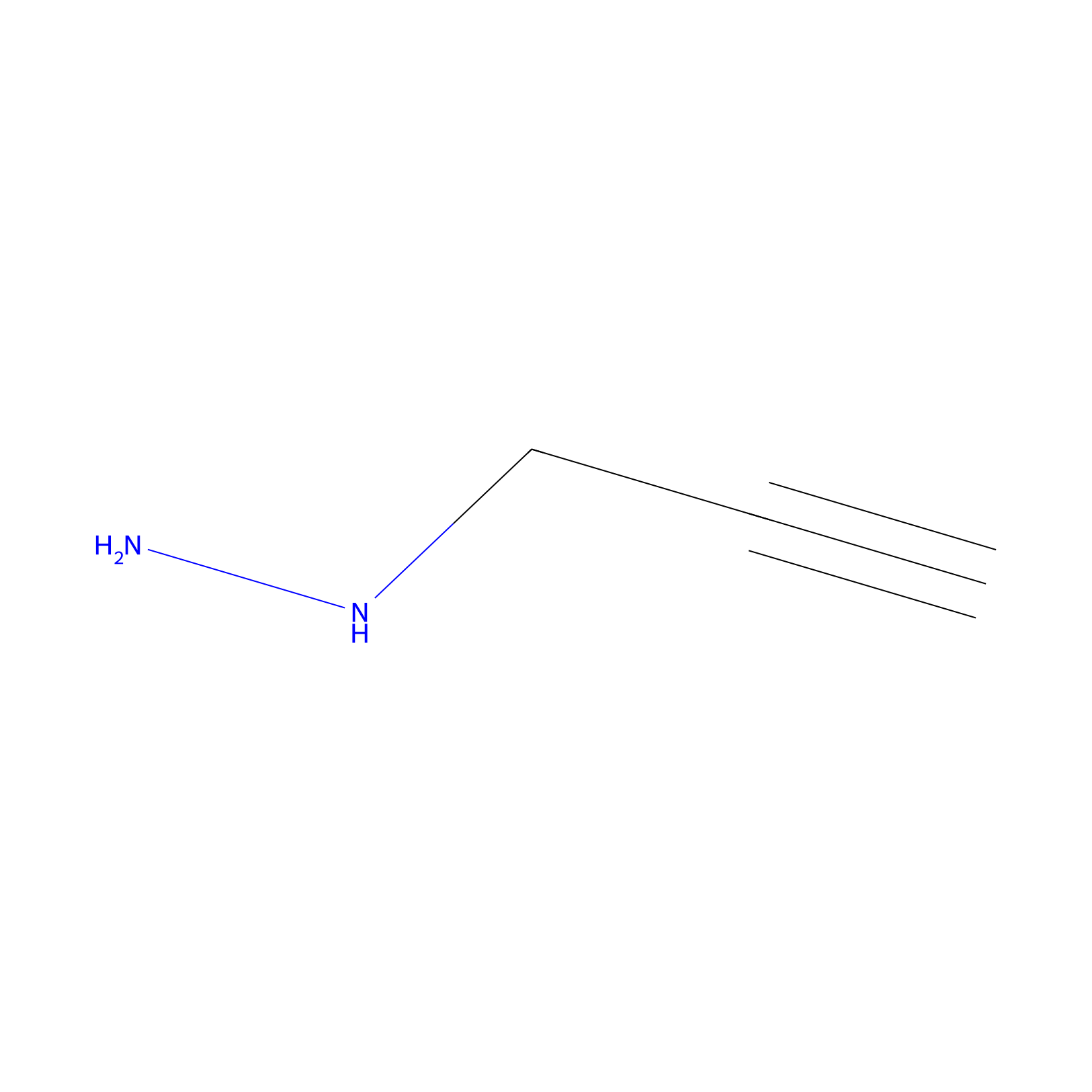 |
13.00 | LDD0385 | [3] | |
|
Alkylaryl probe 2 Probe Info |
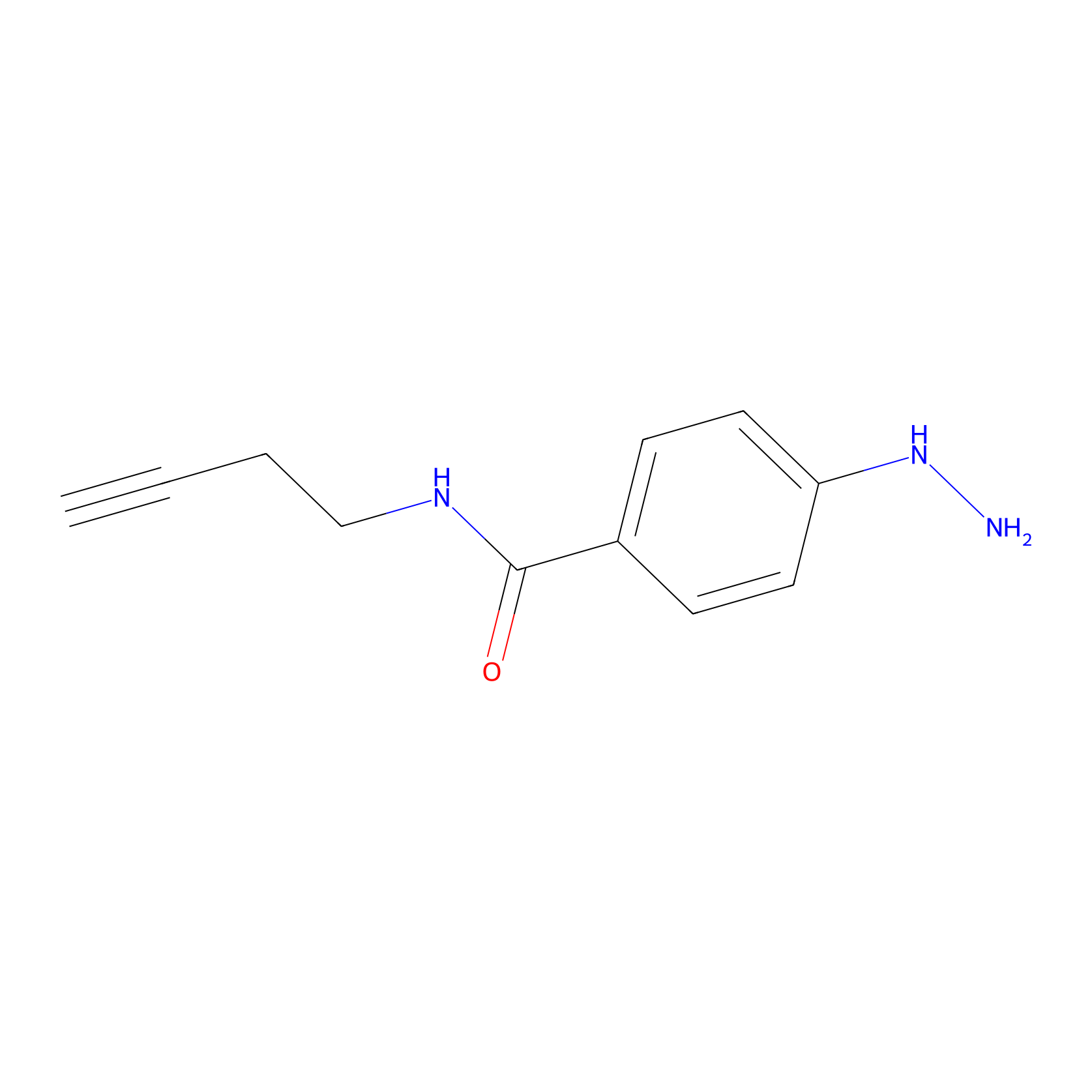 |
16.00 | LDD0391 | [3] | |
|
ILS-2 Probe Info |
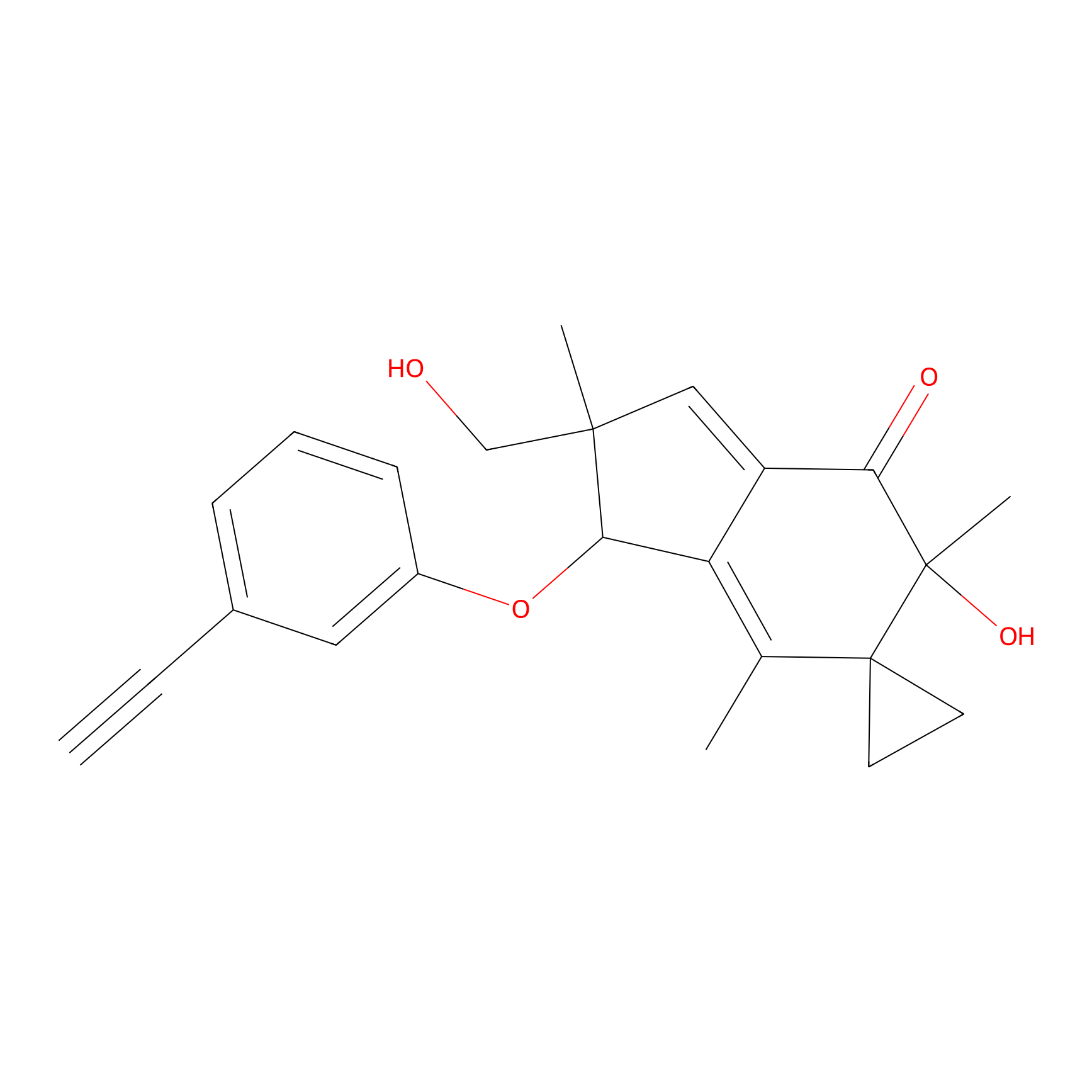 |
3.69 | LDD0416 | [4] | |
|
FBPP2 Probe Info |
 |
131.86 | LDD0318 | [5] | |
|
C-Sul Probe Info |
 |
4.31 | LDD0066 | [6] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [7] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [7] | |
|
ONAyne Probe Info |
 |
K345(0.63); K357(0.69) | LDD0274 | [8] | |
|
STPyne Probe Info |
 |
K122(8.68); K127(7.54); K184(6.99); K28(5.57) | LDD0277 | [8] | |
|
P11 Probe Info |
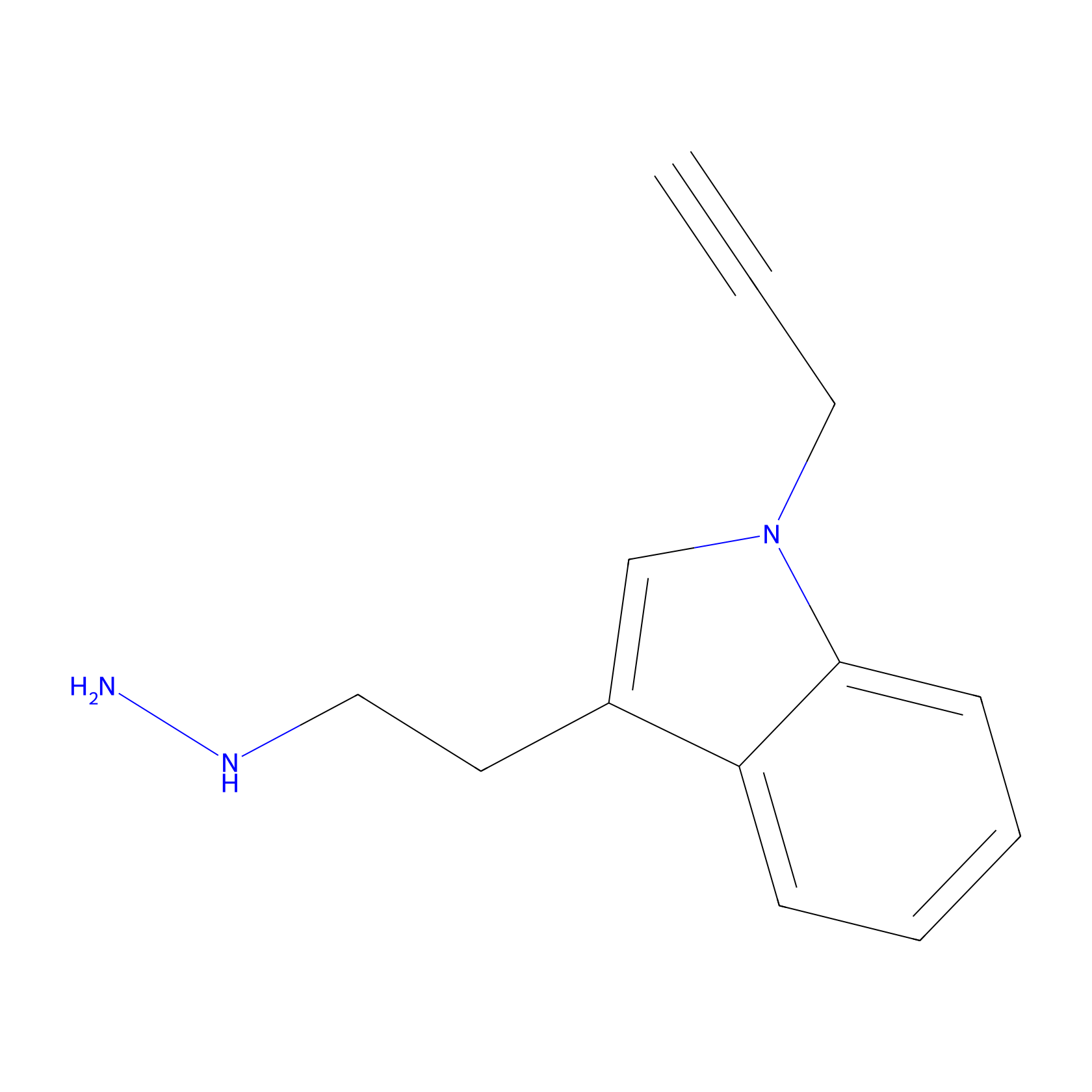 |
20.00 | LDD0201 | [9] | |
|
Alkylaryl probe 3 Probe Info |
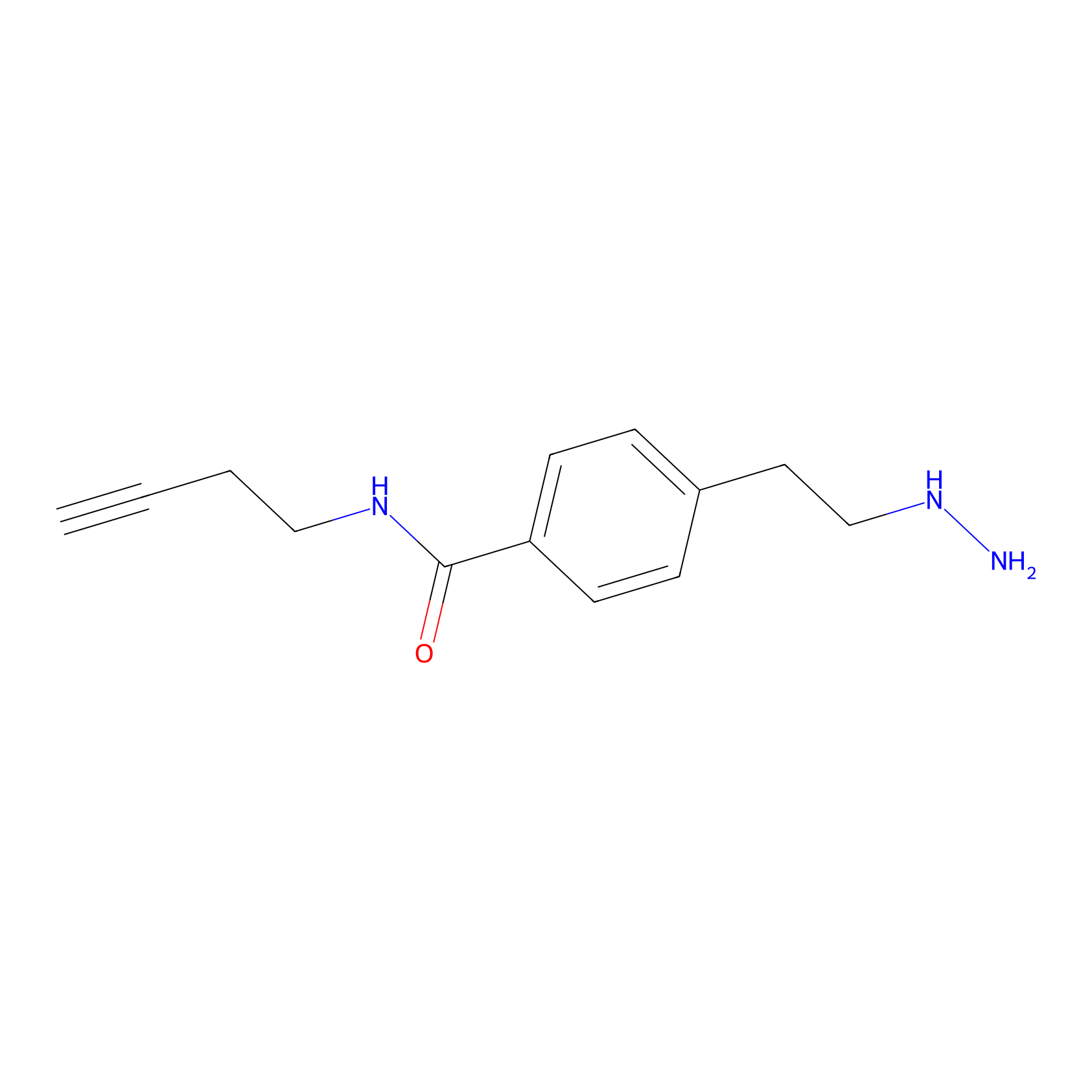 |
10.30 | LDD0282 | [10] | |
|
DBIA Probe Info |
 |
C286(1.53) | LDD3311 | [11] | |
|
THZ1-DTB Probe Info |
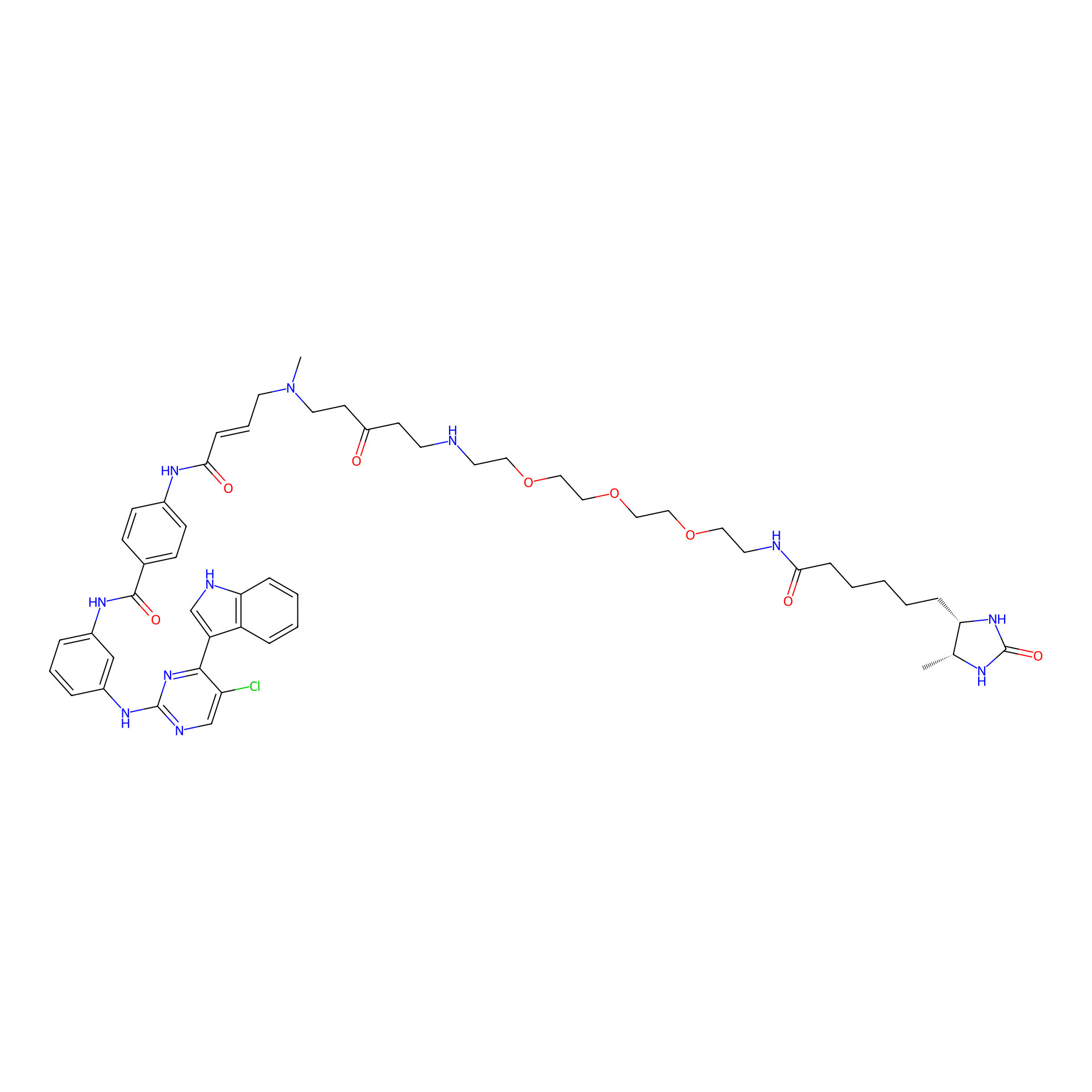 |
C329(1.06) | LDD0460 | [12] | |
|
BTD Probe Info |
 |
C329(1.06) | LDD2089 | [13] | |
|
AHL-Pu-1 Probe Info |
 |
C329(2.11) | LDD0171 | [14] | |
|
EA-probe Probe Info |
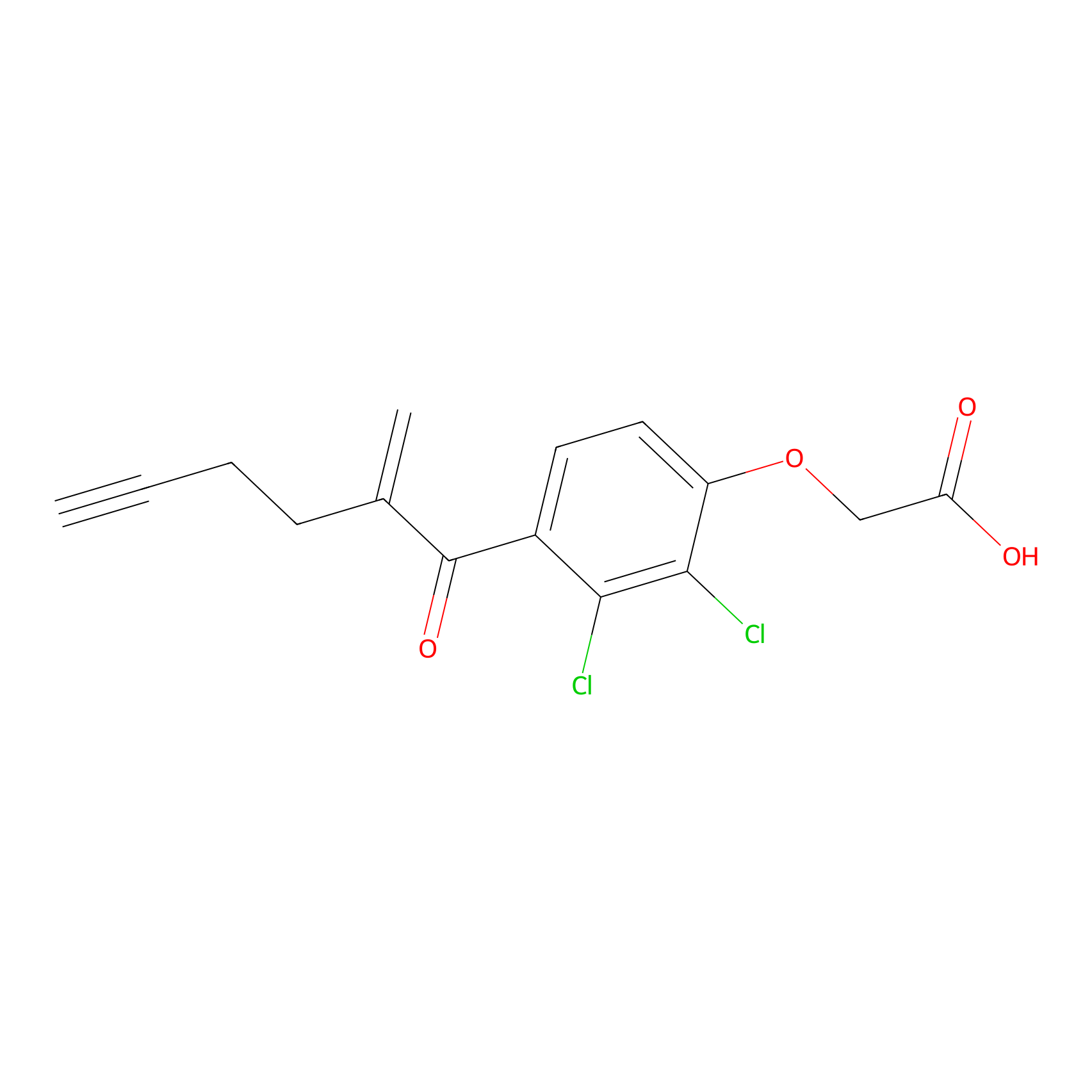 |
N.A. | LDD0440 | [15] | |
|
HHS-482 Probe Info |
 |
Y269(1.40); Y325(0.87) | LDD0285 | [16] | |
|
FBP2 Probe Info |
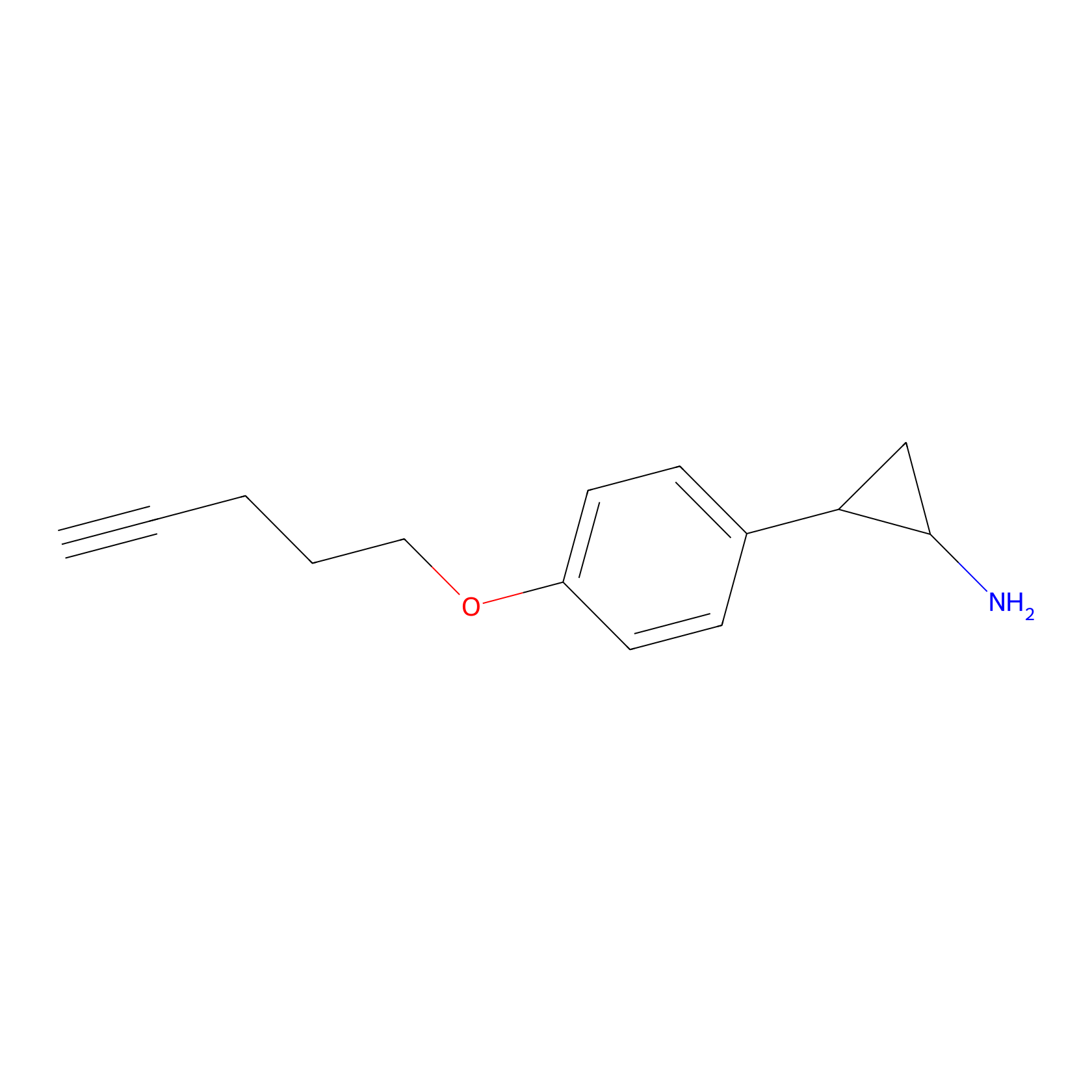 |
2.44 | LDD0053 | [5] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C329(0.00); C286(0.00) | LDD0038 | [17] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [18] | |
|
Lodoacetamide azide Probe Info |
 |
C329(0.00); C286(0.00); C290(0.00); C117(0.00) | LDD0037 | [17] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [19] | |
|
IPM Probe Info |
 |
N.A. | LDD0025 | [20] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [21] | |
|
NHS Probe Info |
 |
K127(0.00); K357(0.00) | LDD0010 | [21] | |
|
PF-06672131 Probe Info |
 |
N.A. | LDD0017 | [22] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [23] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [24] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [25] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [26] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [27] | |
|
MPP-AC Probe Info |
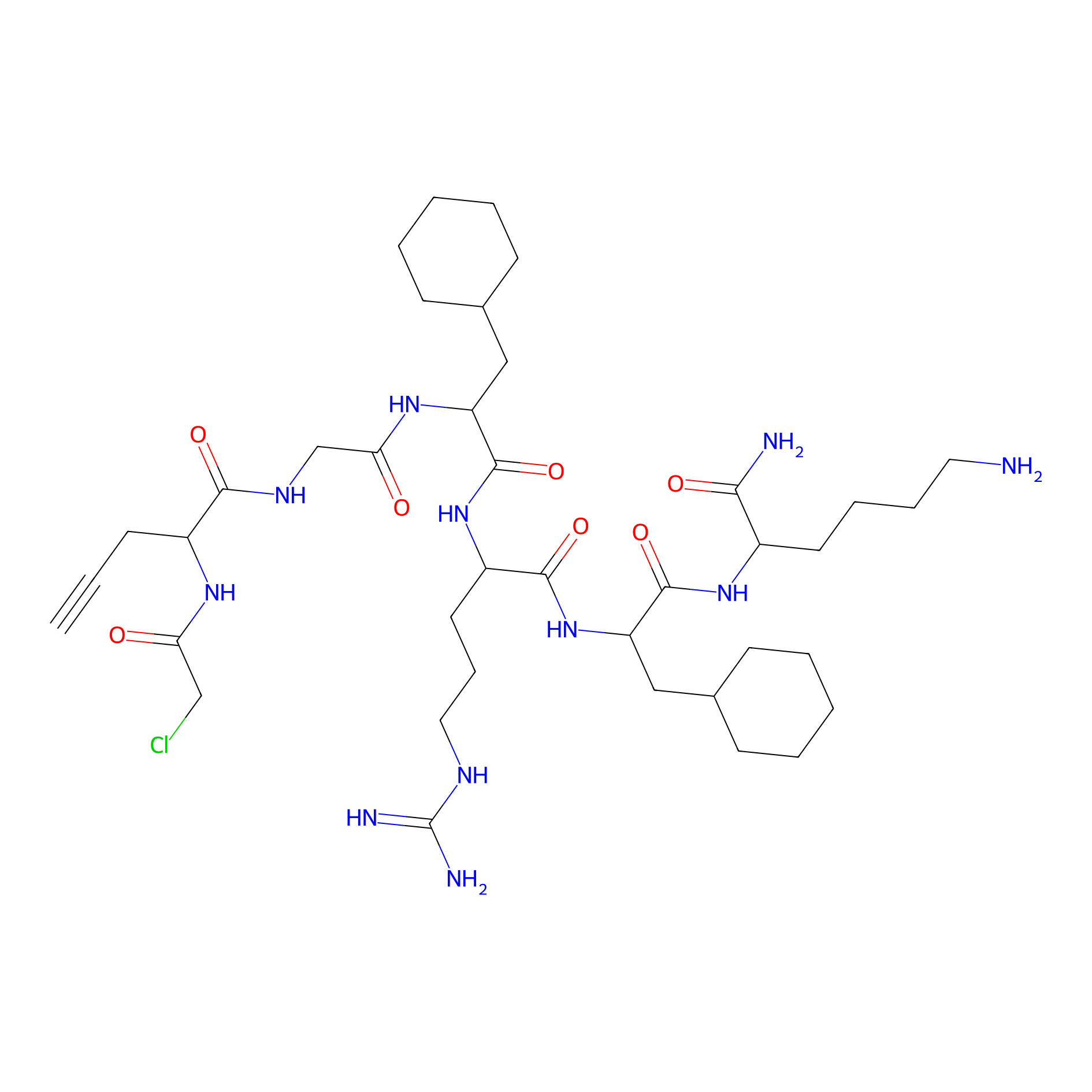 |
N.A. | LDD0428 | [28] | |
|
NAIA_5 Probe Info |
 |
C329(0.00); C286(0.00); C117(0.00); C366(0.00) | LDD2223 | [29] | |
|
TER-AC Probe Info |
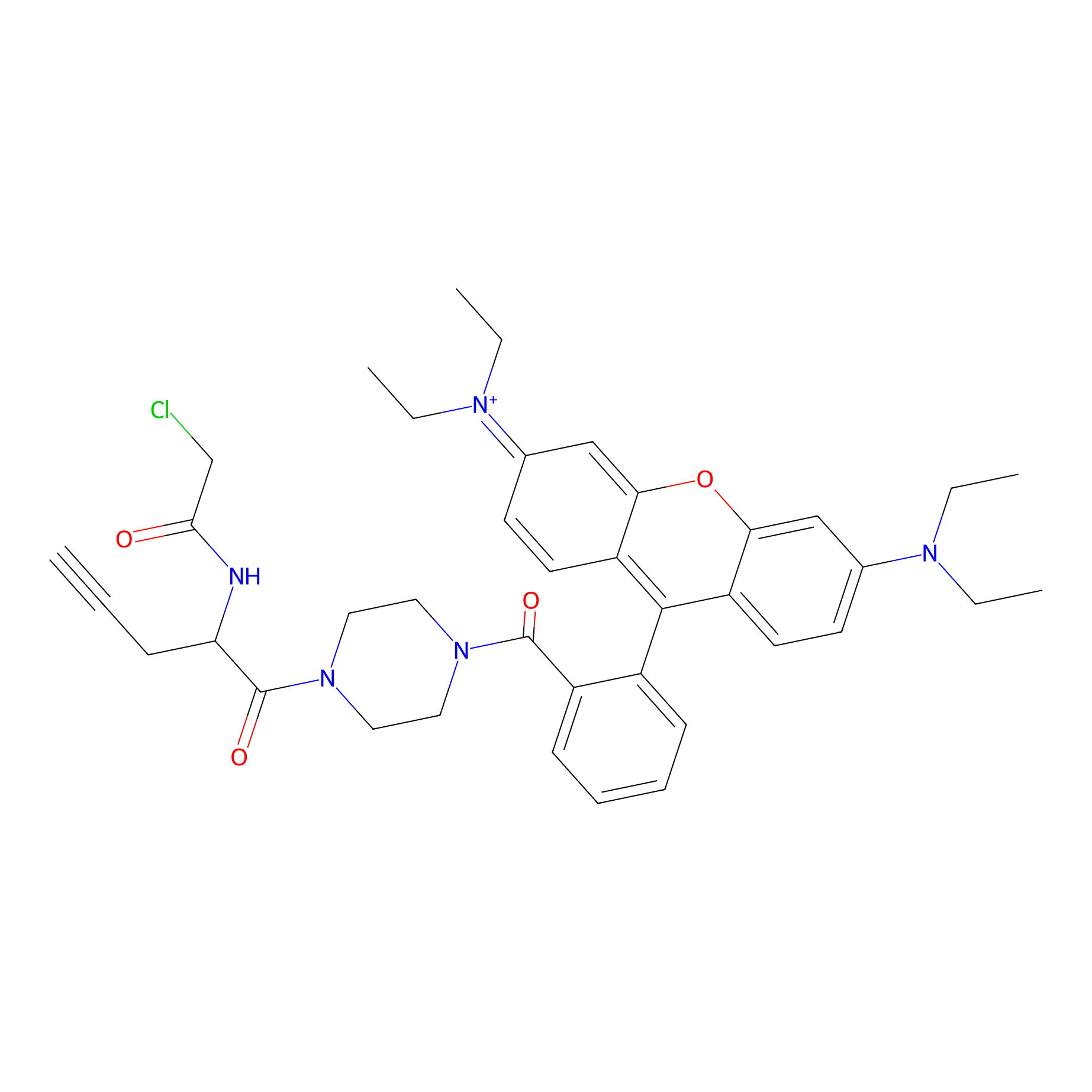 |
N.A. | LDD0426 | [28] | |
|
TPP-AC Probe Info |
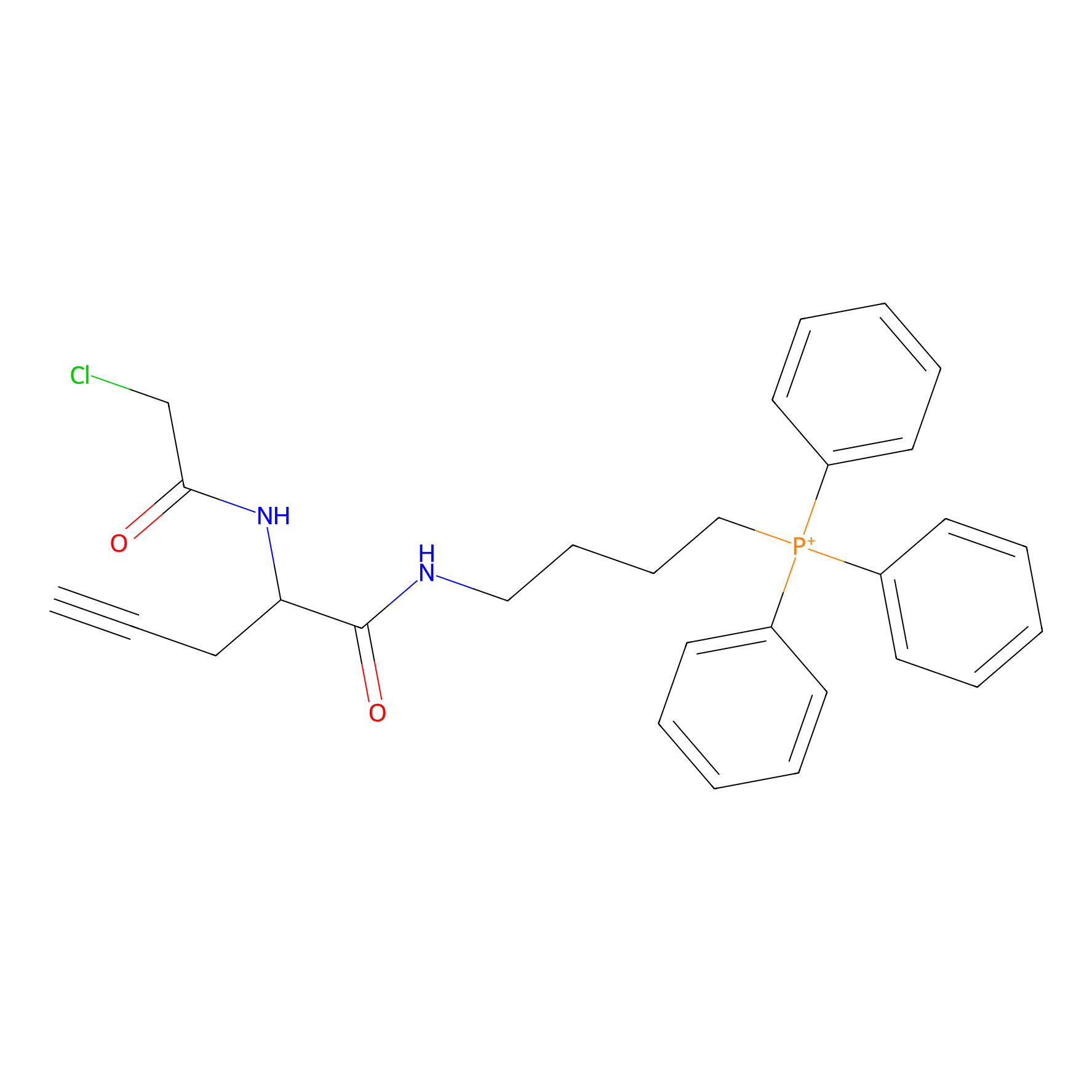 |
N.A. | LDD0427 | [28] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
PARPYnD Probe Info |
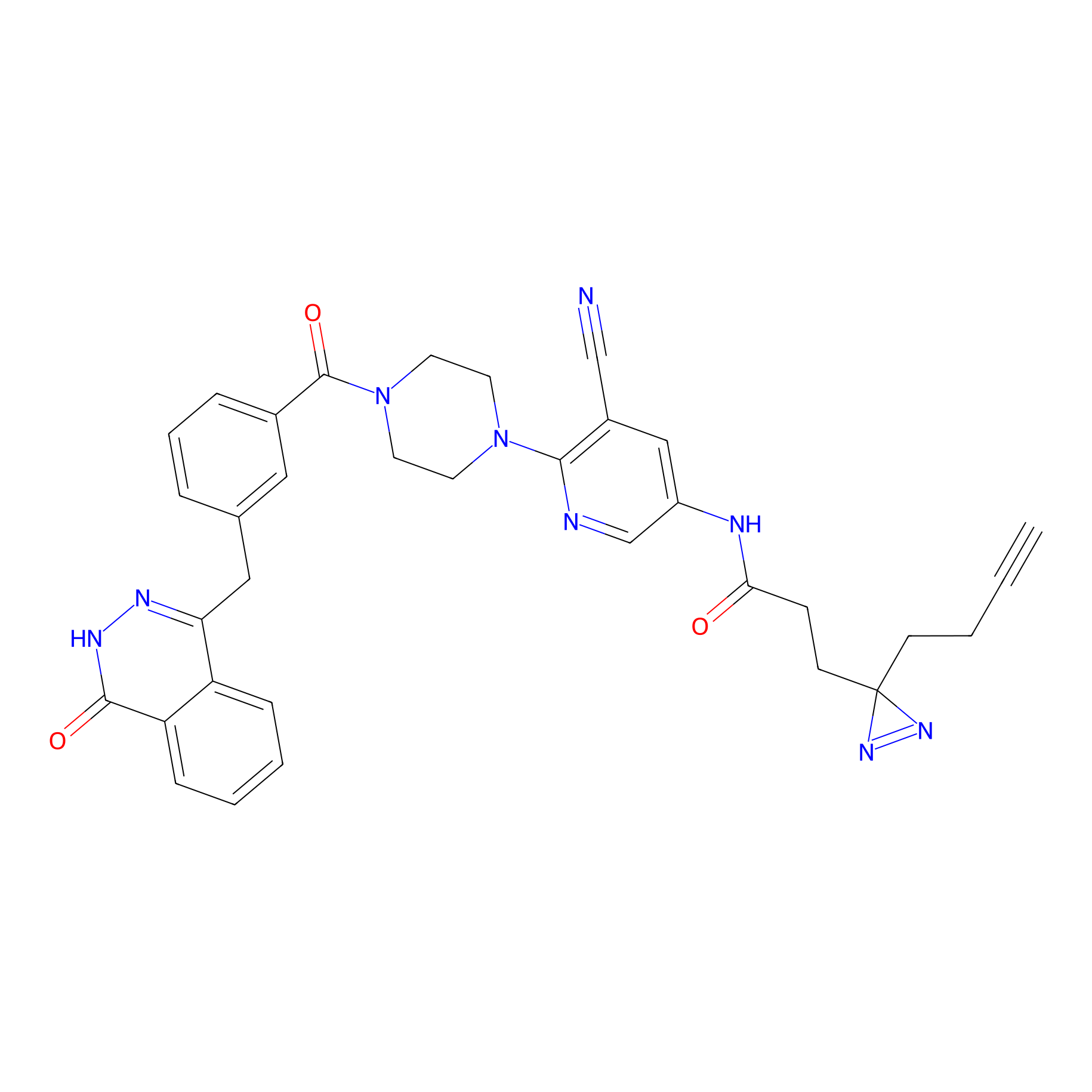 |
1.38 | LDD0374 | [30] | |
|
ILS-1 PP Probe Info |
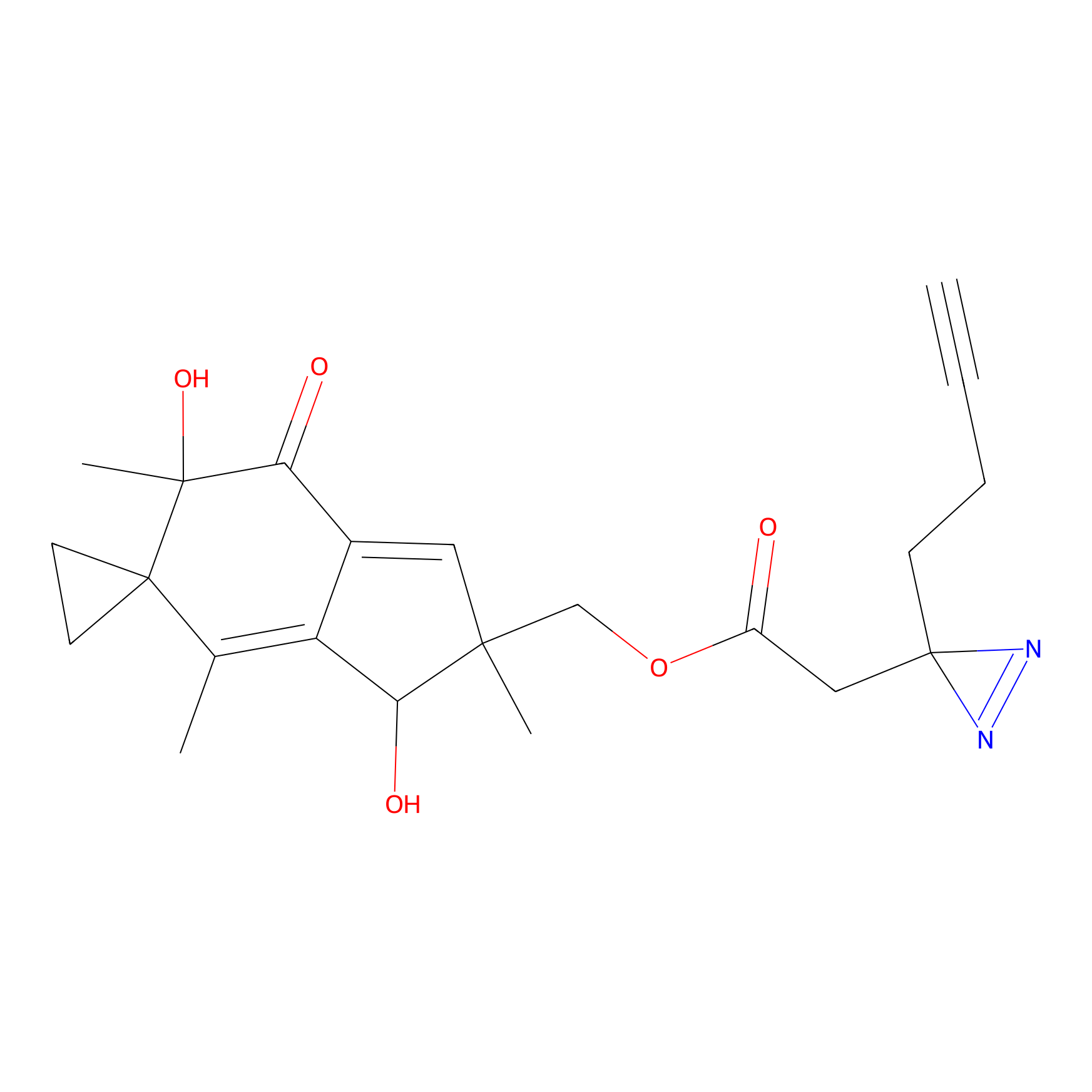 |
8.40 | LDD0417 | [4] | |
|
IMP2070 Probe Info |
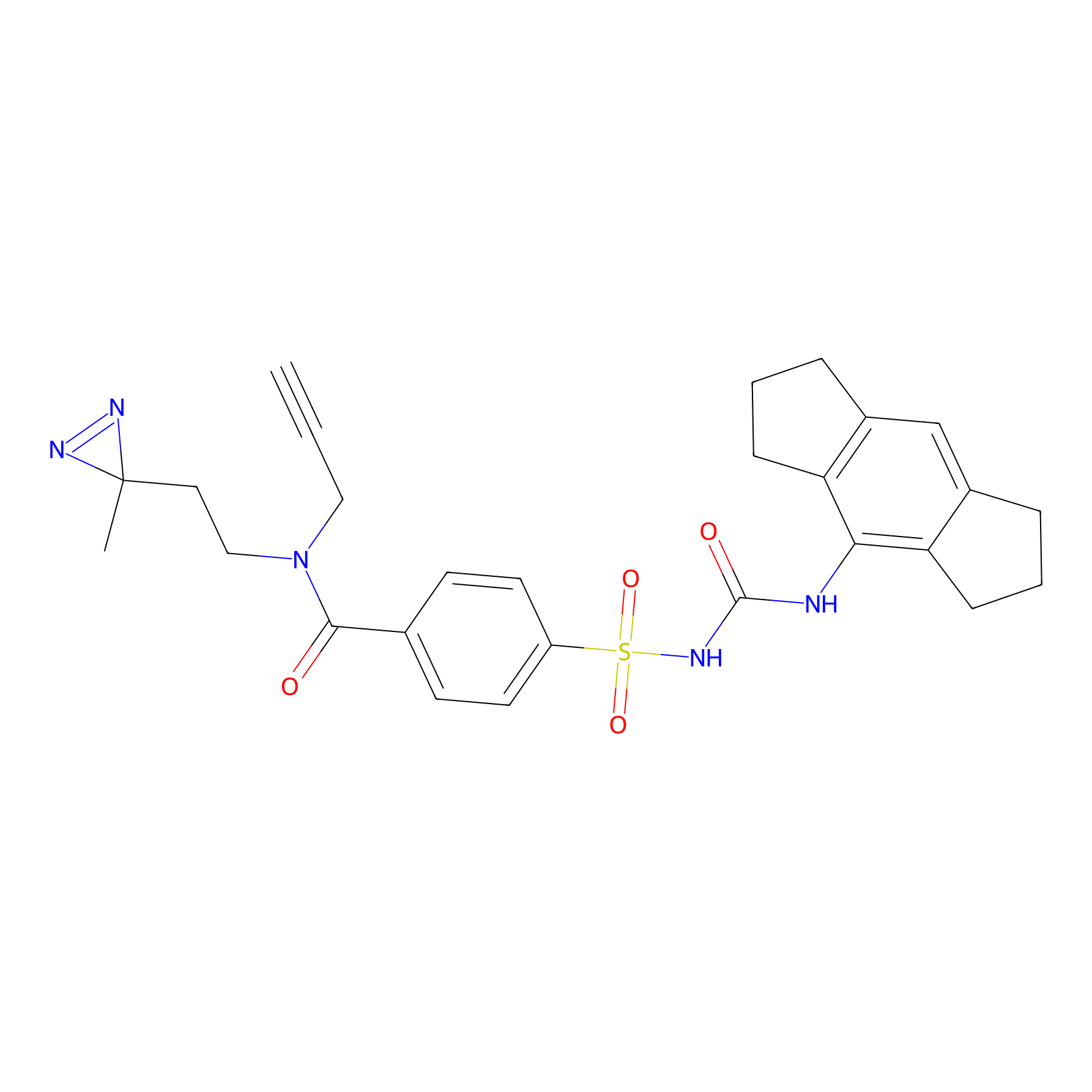 |
11.49 | LDD0367 | [31] | |
|
FFF probe11 Probe Info |
 |
18.81 | LDD0471 | [32] | |
|
FFF probe12 Probe Info |
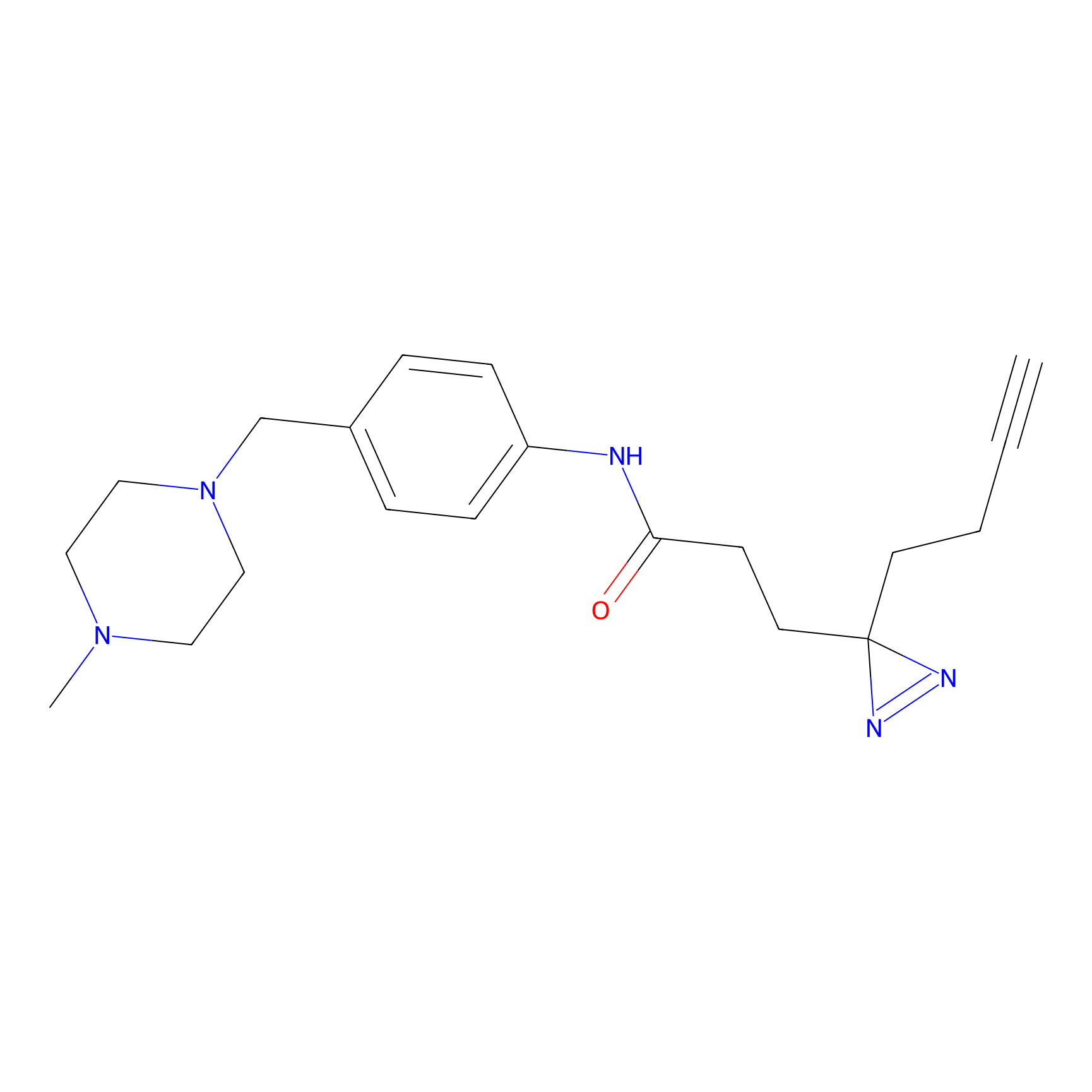 |
17.41 | LDD0473 | [32] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [32] | |
|
FFF probe14 Probe Info |
 |
19.81 | LDD0477 | [32] | |
|
FFF probe15 Probe Info |
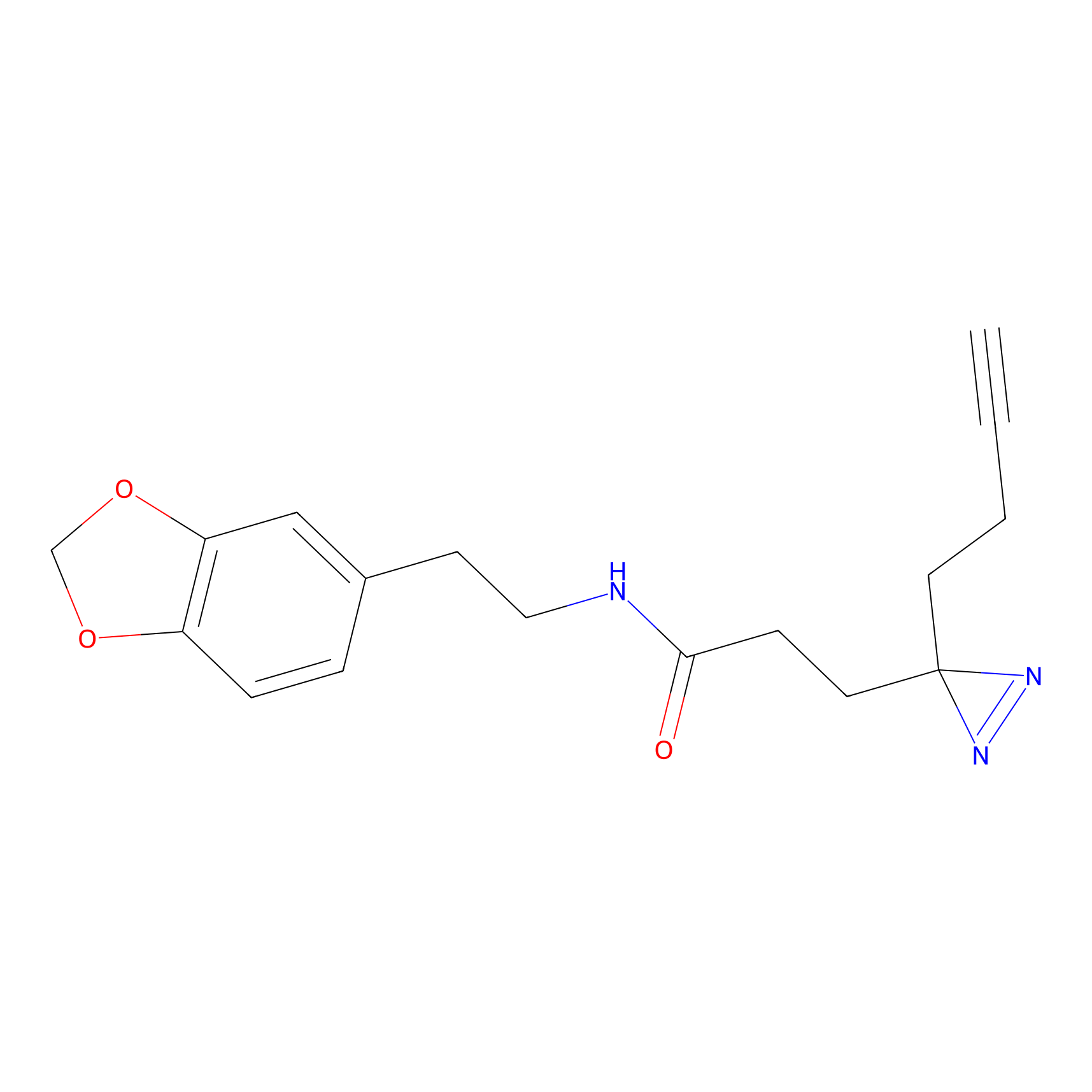 |
17.62 | LDD0478 | [32] | |
|
FFF probe2 Probe Info |
 |
18.03 | LDD0463 | [32] | |
|
FFF probe3 Probe Info |
 |
19.15 | LDD0464 | [32] | |
|
FFF probe4 Probe Info |
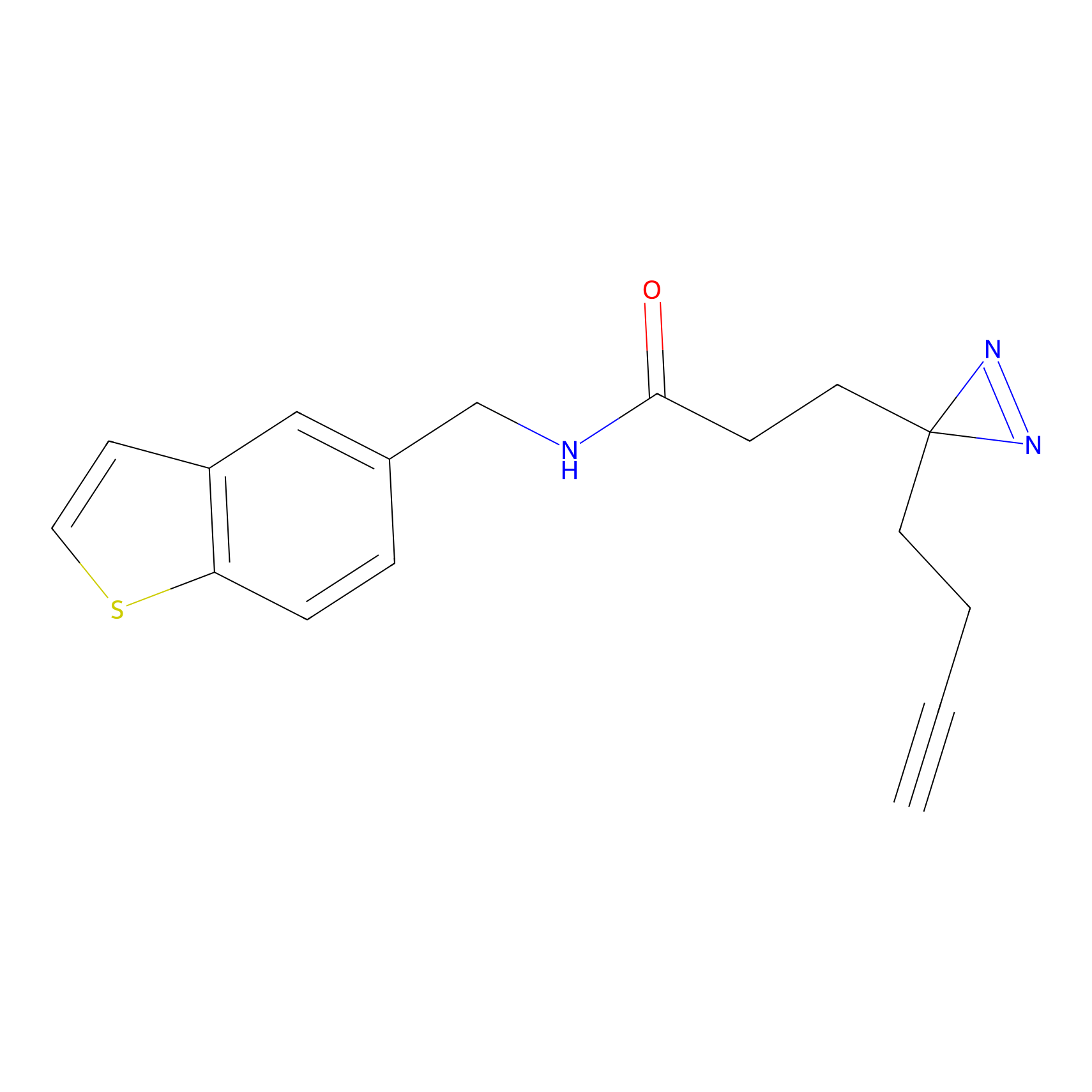 |
16.31 | LDD0466 | [32] | |
|
FFF probe6 Probe Info |
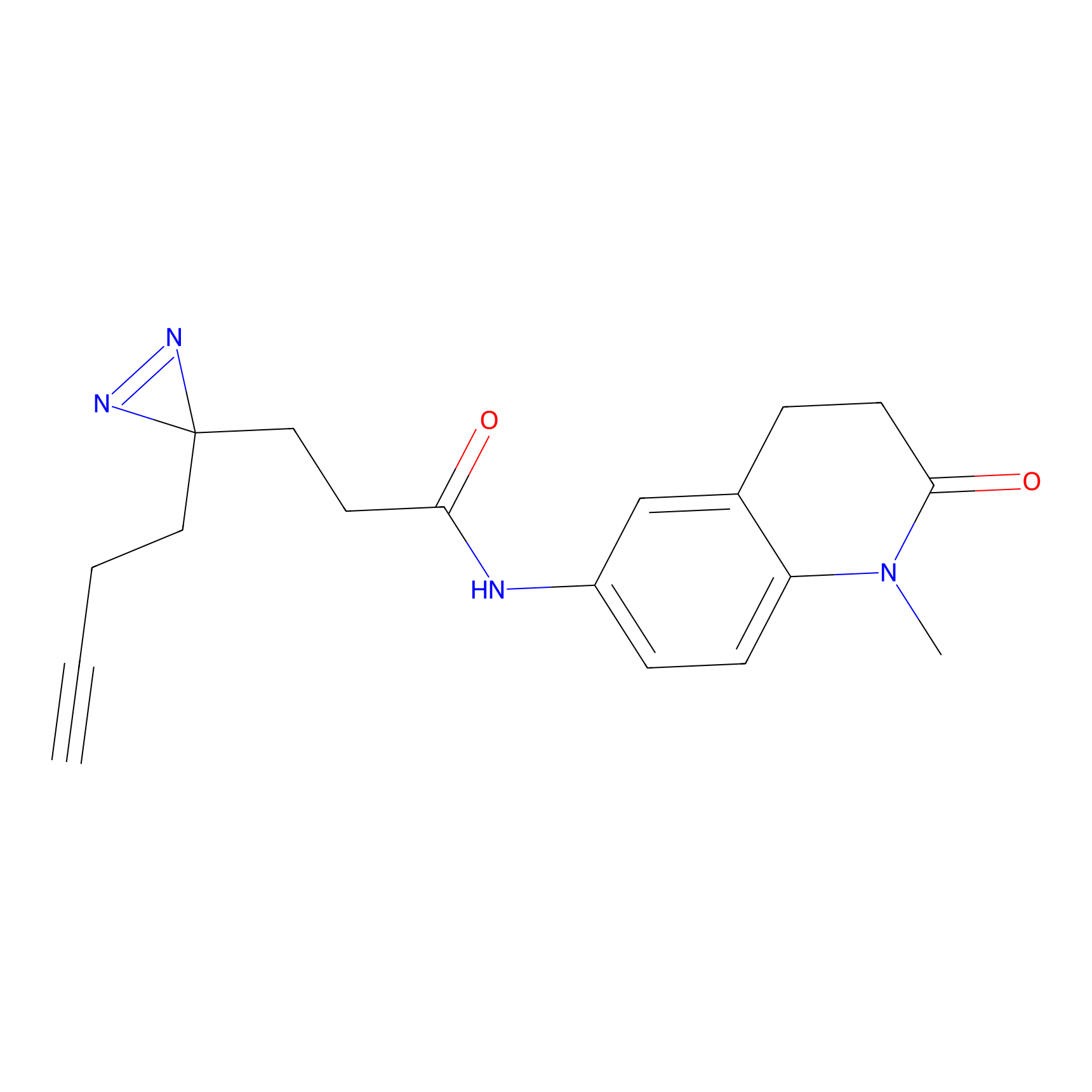 |
6.13 | LDD0467 | [32] | |
|
FFF probe7 Probe Info |
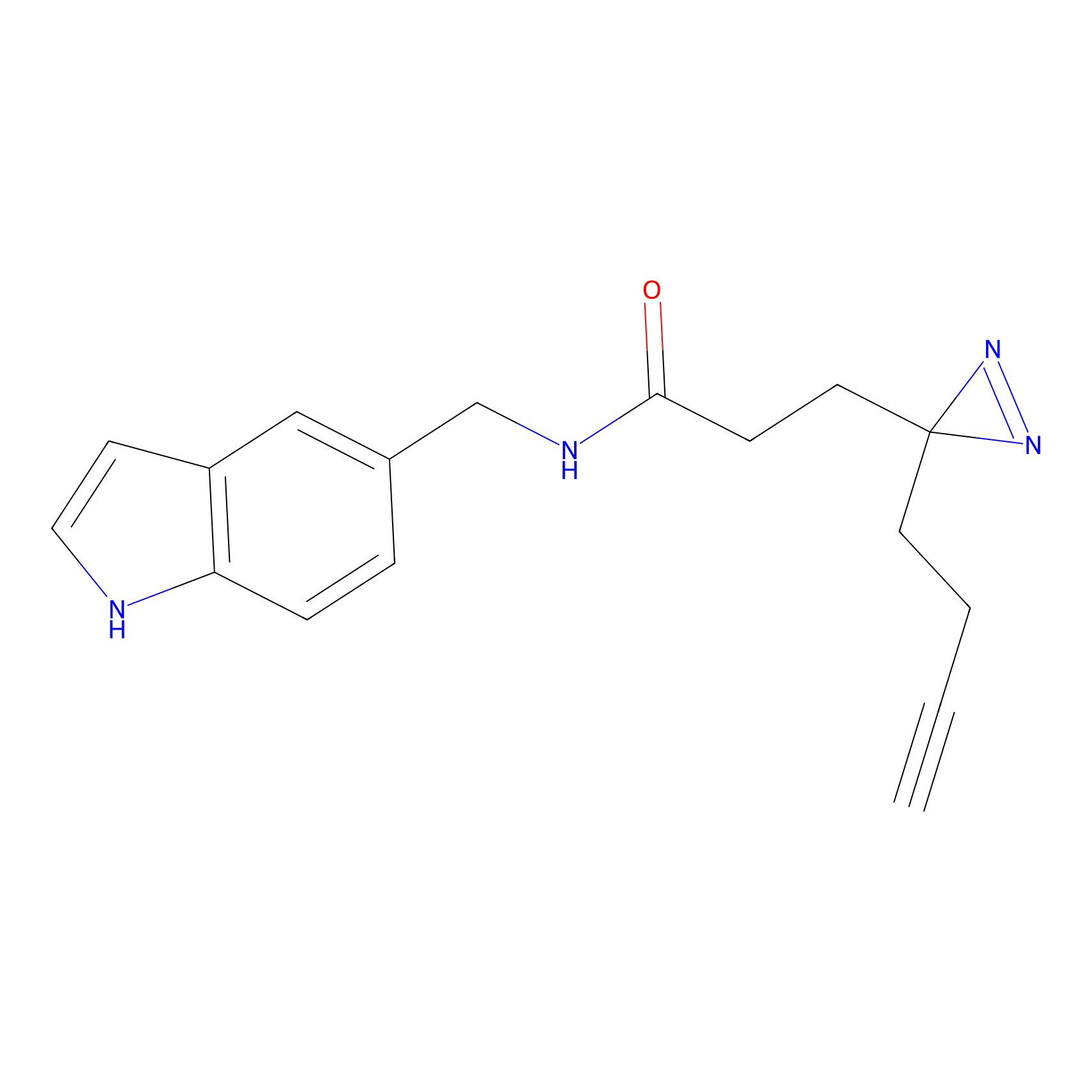 |
20.00 | LDD0483 | [32] | |
|
FFF probe9 Probe Info |
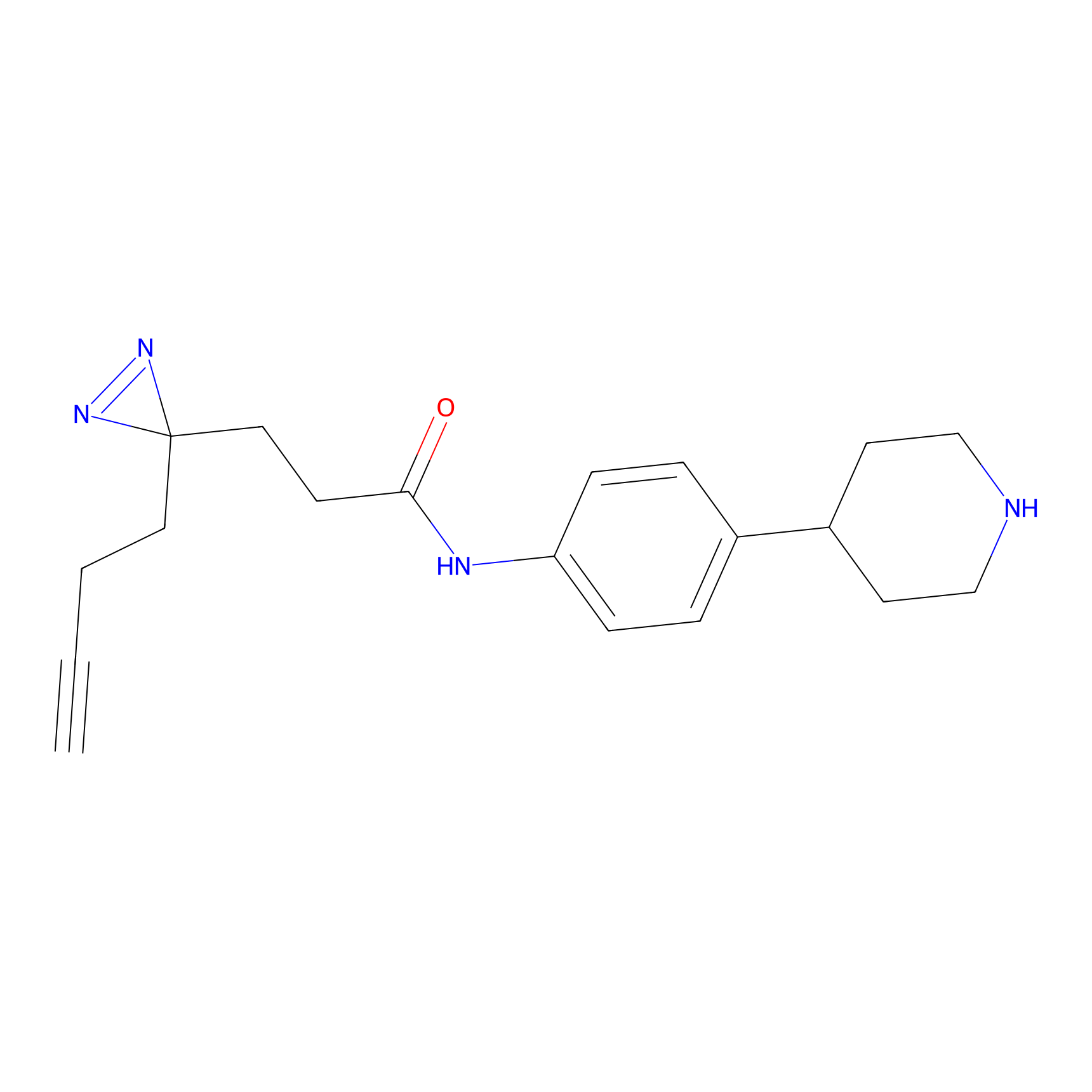 |
20.00 | LDD0470 | [32] | |
|
JN0003 Probe Info |
 |
20.00 | LDD0469 | [32] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [33] | |
|
DR-1 Probe Info |
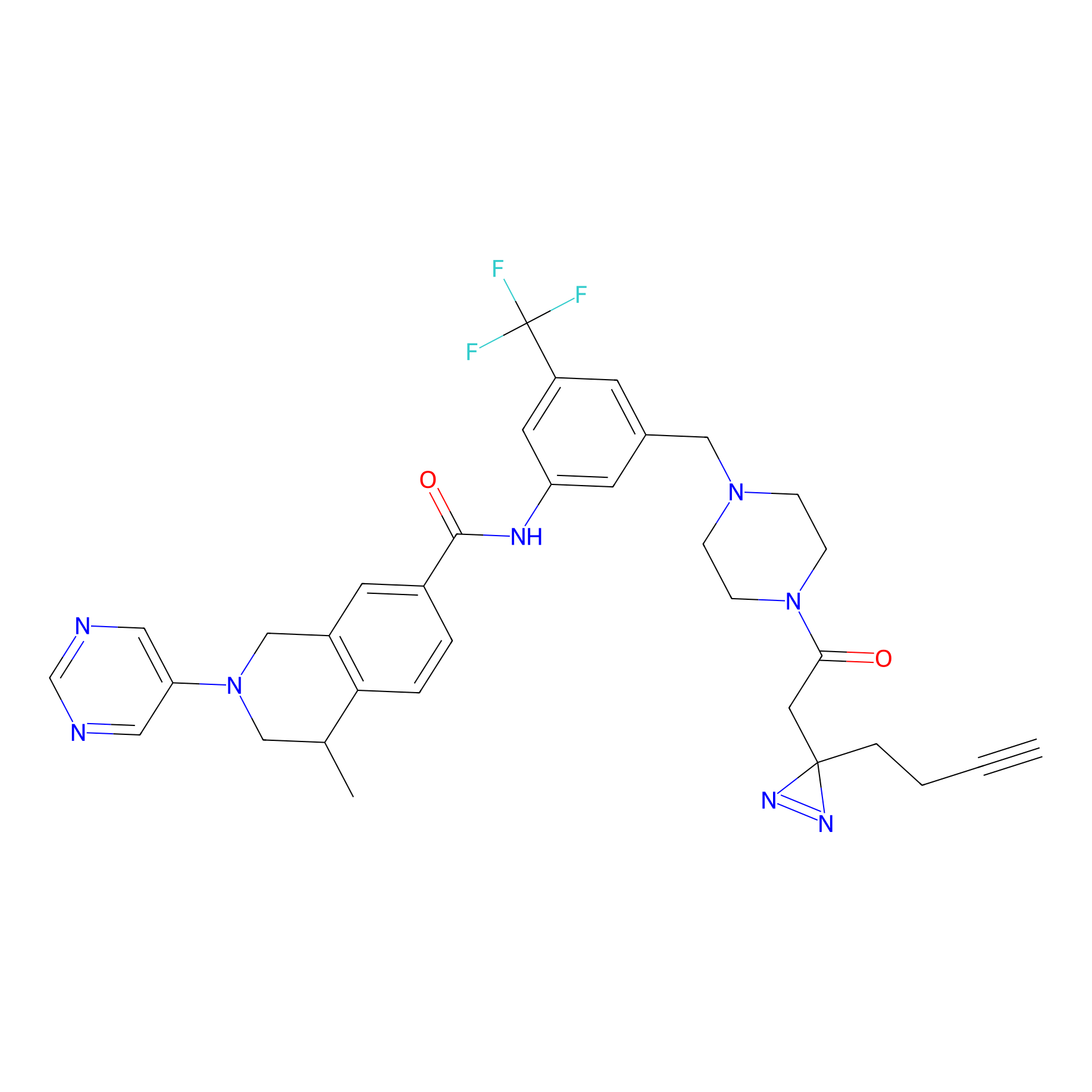 |
3.34 | LDD0397 | [34] | |
|
A-DA Probe Info |
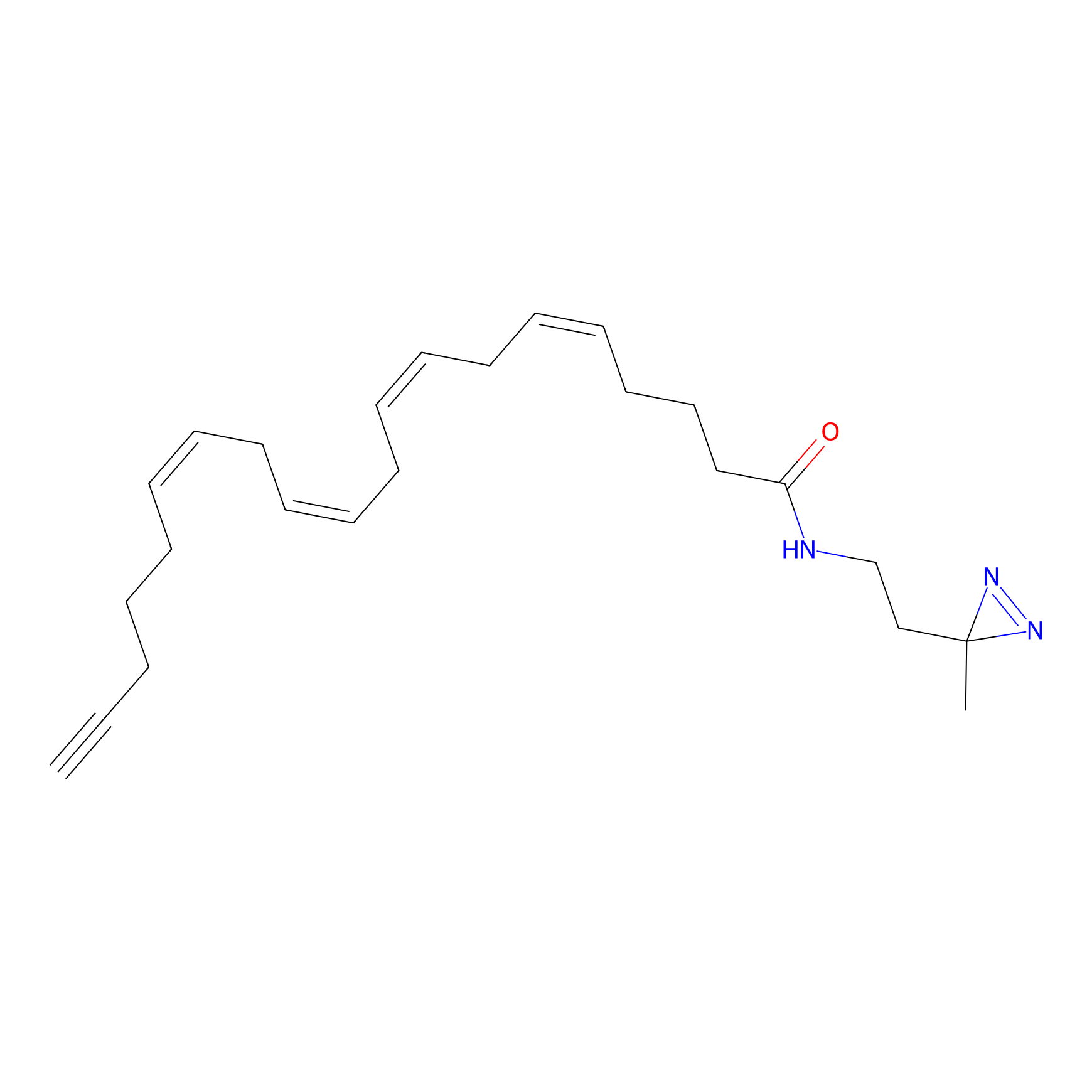 |
2.05 | LDD0145 | [35] | |
|
Kambe_31 Probe Info |
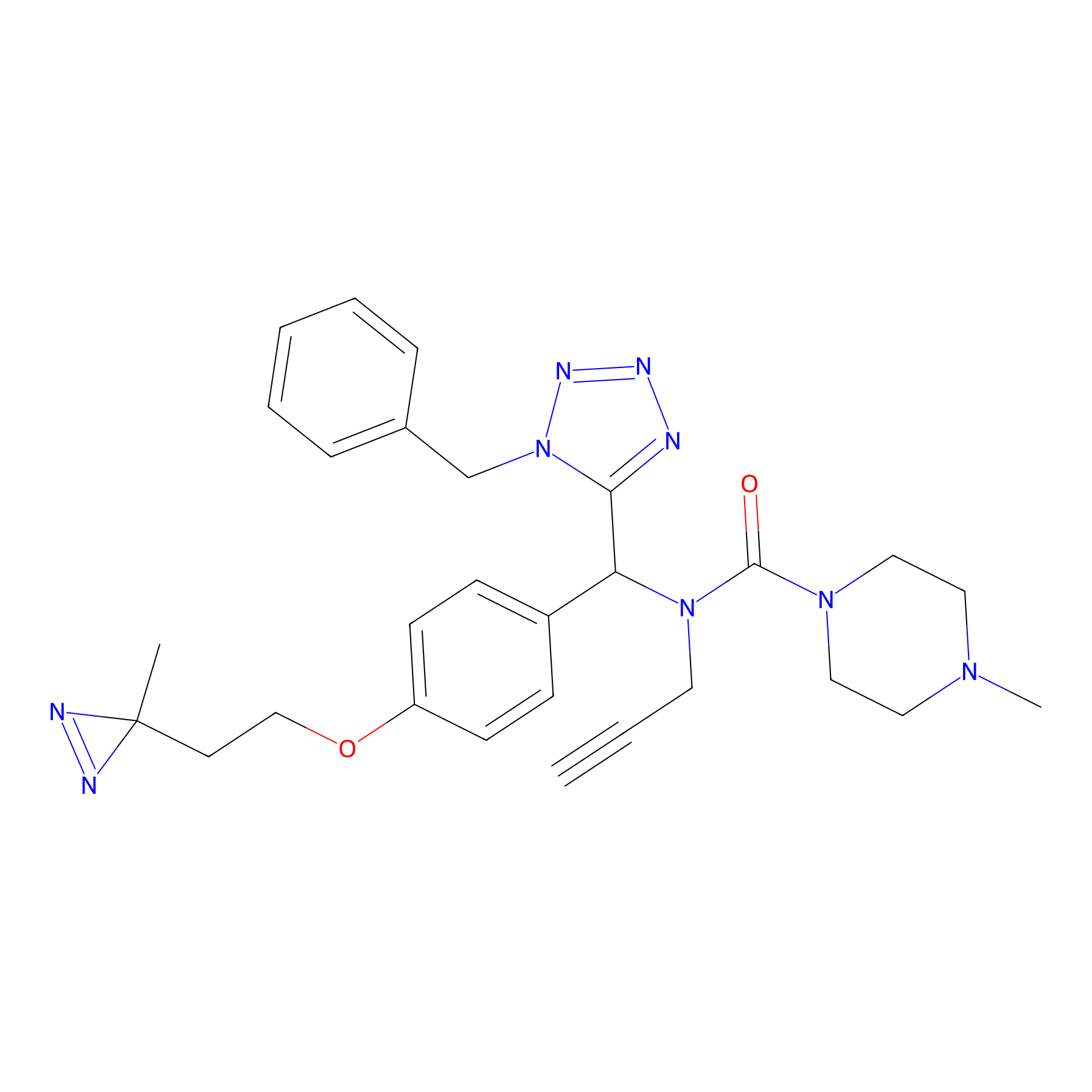 |
3.56 | LDD0128 | [36] | |
|
BD-F Probe Info |
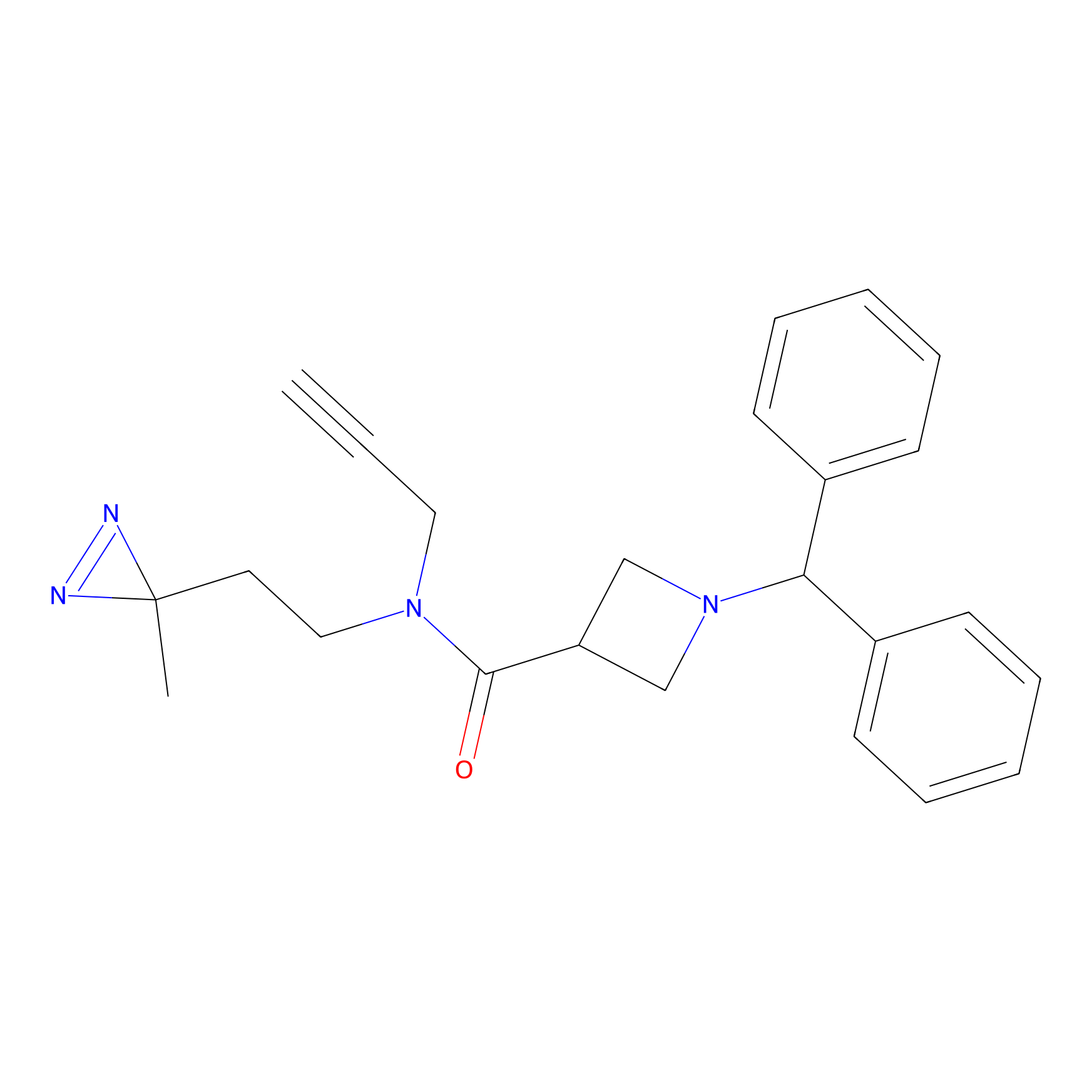 |
G323(0.00); Y325(0.00); E324(0.00) | LDD0024 | [37] | |
|
LD-F Probe Info |
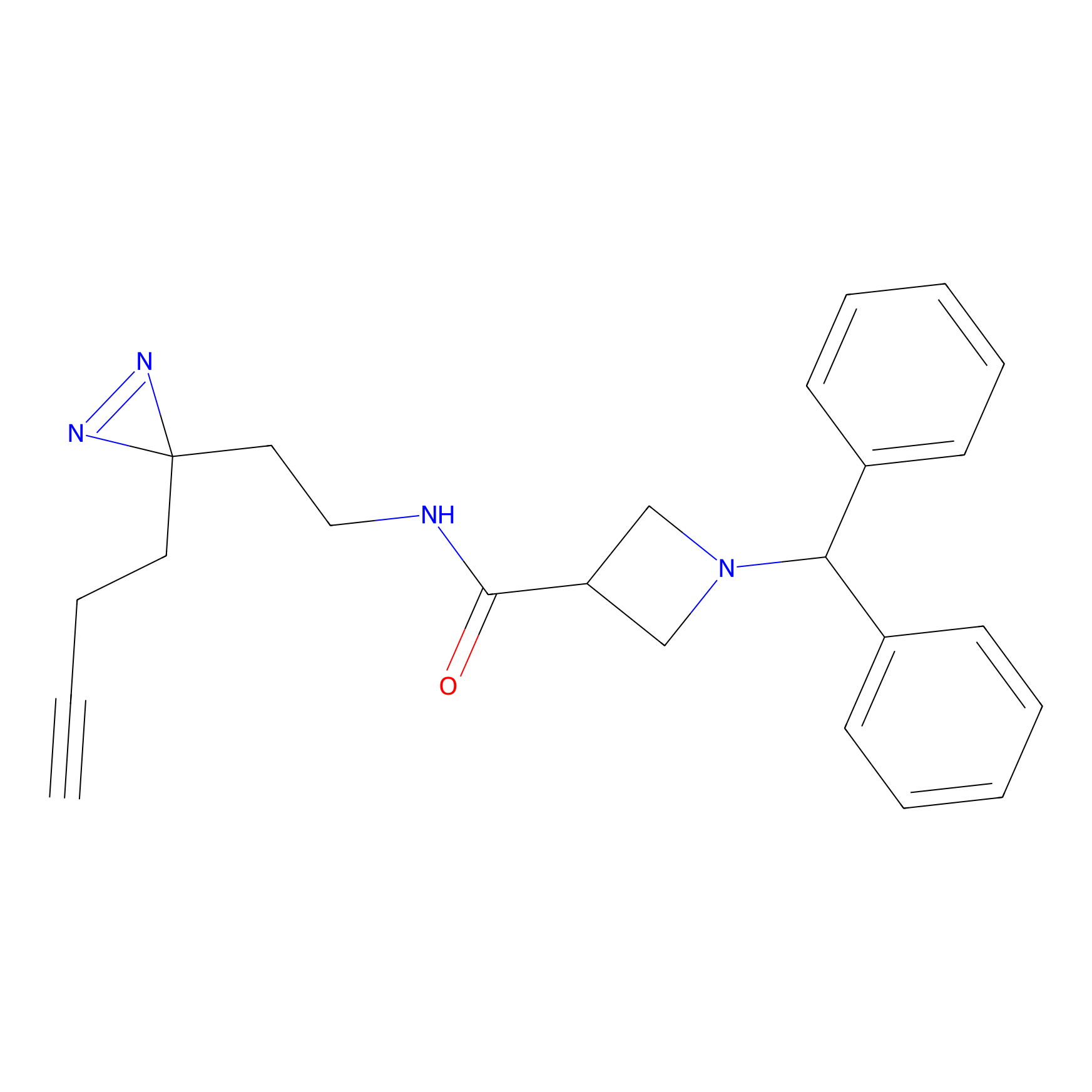 |
Q240(0.00); E324(0.00); G323(0.00); P237(0.00) | LDD0015 | [37] | |
|
Photocelecoxib Probe Info |
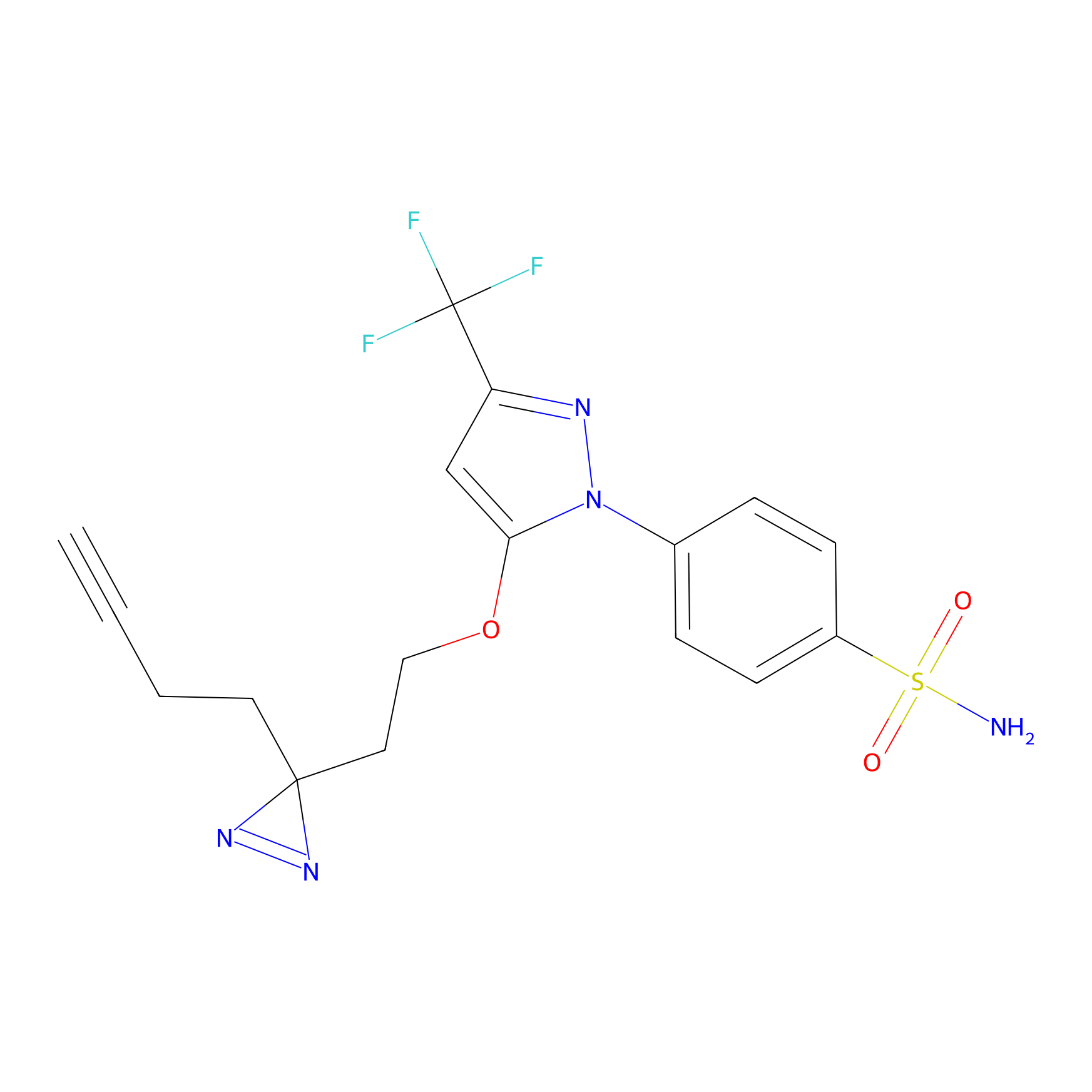 |
I321(0.00); G323(0.00) | LDD0019 | [38] | |
|
Photoindomethacin Probe Info |
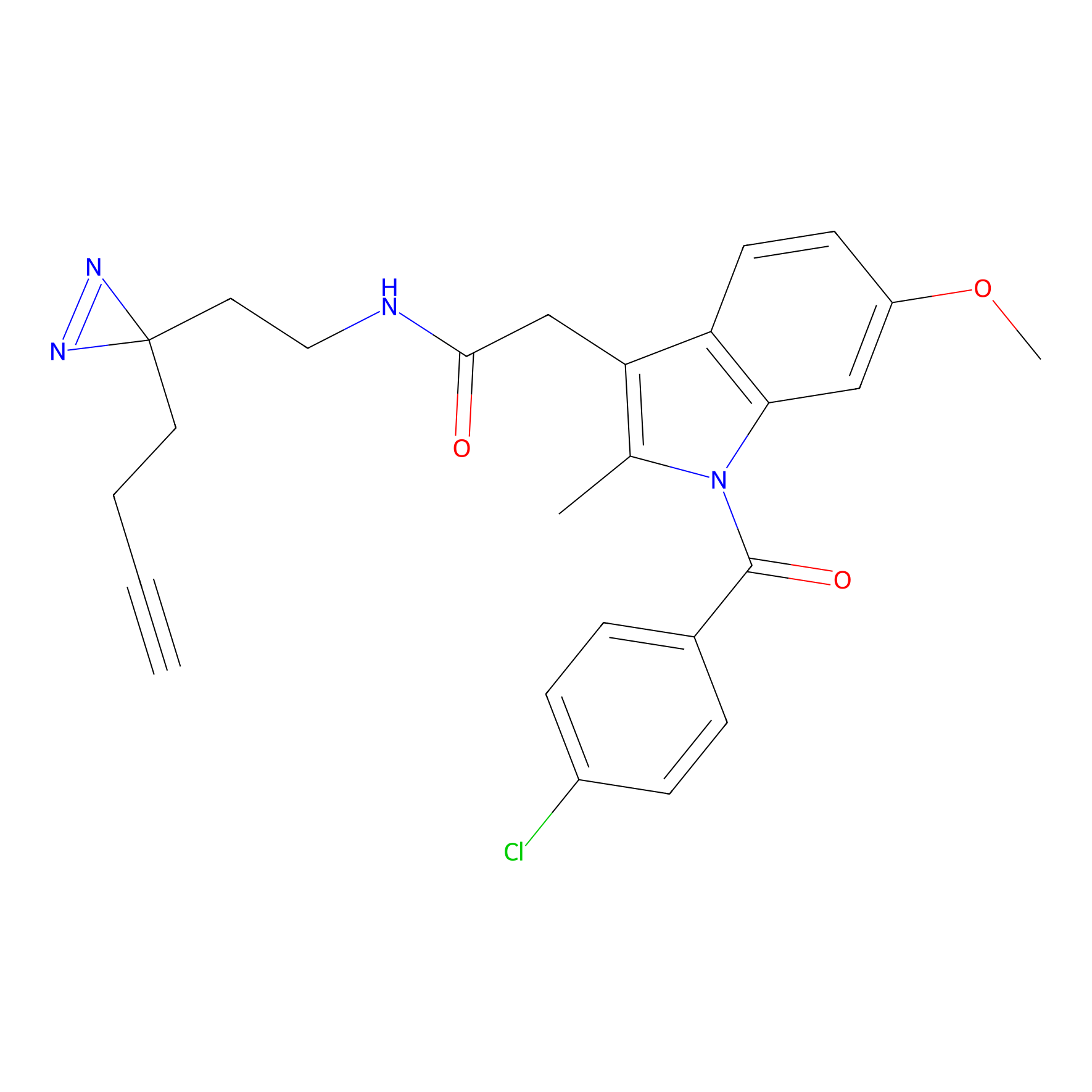 |
P319(0.00); G323(0.00); Y325(0.00) | LDD0154 | [38] | |
|
Photonaproxen Probe Info |
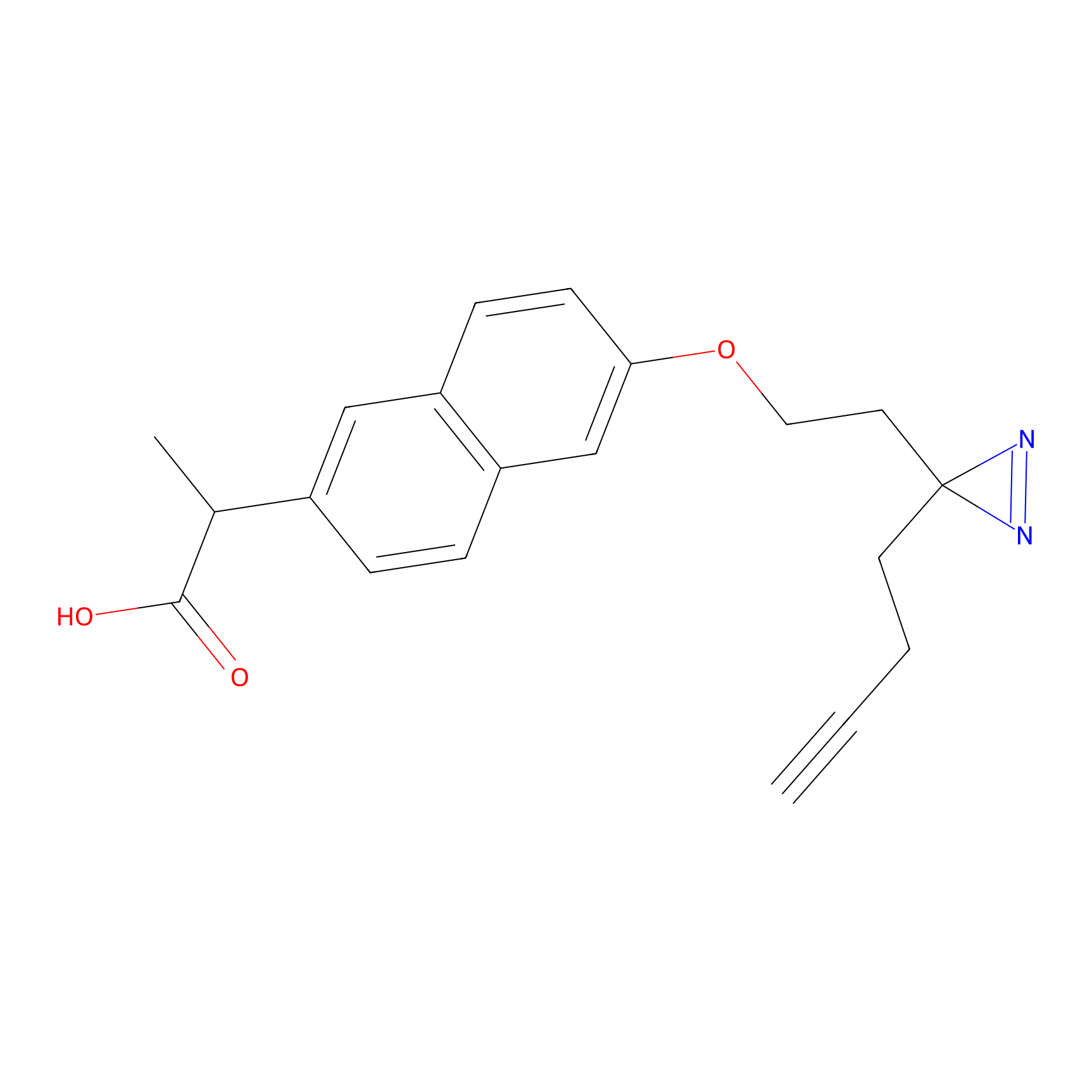 |
N.A. | LDD0156 | [38] | |
|
OEA-DA Probe Info |
 |
20.00 | LDD0046 | [39] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C329(0.76) | LDD2142 | [13] |
| LDCM0537 | 2-Cyano-N,N-dimethylacetamide | MDA-MB-231 | C329(0.96) | LDD2130 | [13] |
| LDCM0026 | 4SU-RNA+native RNA | DM93 | C329(2.11) | LDD0171 | [14] |
| LDCM0088 | C45 | HEK-293T | 20.00 | LDD0201 | [9] |
| LDCM0108 | Chloroacetamide | HeLa | C290(0.00); C329(0.00) | LDD0222 | [26] |
| LDCM0632 | CL-Sc | Hep-G2 | C329(1.63); C117(0.75); C117(0.67) | LDD2227 | [29] |
| LDCM0189 | Compound 16 | HEK-293T | 24.10 | LDD0492 | [32] |
| LDCM0185 | Compound 17 | HEK-293T | 8.89 | LDD0504 | [32] |
| LDCM0182 | Compound 18 | HEK-293T | 11.90 | LDD0501 | [32] |
| LDCM0184 | Compound 20 | HEK-293T | 17.24 | LDD0503 | [32] |
| LDCM0191 | Compound 21 | HEK-293T | 15.38 | LDD0493 | [32] |
| LDCM0186 | Compound 31 | HEK-293T | 13.99 | LDD0506 | [32] |
| LDCM0187 | Compound 32 | HEK-293T | 25.64 | LDD0507 | [32] |
| LDCM0188 | Compound 33 | HEK-293T | 10.20 | LDD0505 | [32] |
| LDCM0190 | Compound 34 | HEK-293T | 18.52 | LDD0497 | [32] |
| LDCM0192 | Compound 35 | HEK-293T | 15.87 | LDD0491 | [32] |
| LDCM0193 | Compound 36 | HEK-293T | 12.90 | LDD0494 | [32] |
| LDCM0194 | Compound 37 | HEK-293T | 18.87 | LDD0498 | [32] |
| LDCM0195 | Compound 38 | HEK-293T | 28.17 | LDD0499 | [32] |
| LDCM0196 | Compound 39 | HEK-293T | 11.30 | LDD0496 | [32] |
| LDCM0197 | Compound 40 | HEK-293T | 16.53 | LDD0495 | [32] |
| LDCM0181 | Compound 41 | HEK-293T | 10.64 | LDD0502 | [32] |
| LDCM0183 | Compound 42 | HEK-293T | 4.78 | LDD0500 | [32] |
| LDCM0152 | DR | NCI-H1299 | 3.34 | LDD0397 | [34] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C329(1.26) | LDD1702 | [13] |
| LDCM0175 | Ethacrynic acid | HeLa | N.A. | LDD0440 | [15] |
| LDCM0573 | Fragment11 | MDA-MB-231 | C117(6.60) | LDD1391 | [40] |
| LDCM0576 | Fragment14 | MDA-MB-231 | C117(0.66) | LDD1397 | [40] |
| LDCM0577 | Fragment15 | MDA-MB-231 | C117(1.21) | LDD1399 | [40] |
| LDCM0579 | Fragment20 | MDA-MB-231 | C117(12.51) | LDD1402 | [40] |
| LDCM0582 | Fragment23 | MDA-MB-231 | C117(3.10) | LDD1408 | [40] |
| LDCM0584 | Fragment25 | MDA-MB-231 | C117(0.97) | LDD1411 | [40] |
| LDCM0578 | Fragment27 | MDA-MB-231 | C117(0.57) | LDD1401 | [40] |
| LDCM0587 | Fragment29 | MDA-MB-231 | C117(1.42) | LDD1417 | [40] |
| LDCM0589 | Fragment31 | MDA-MB-231 | C117(1.32) | LDD1421 | [40] |
| LDCM0590 | Fragment32 | MDA-MB-231 | C117(2.87) | LDD1423 | [40] |
| LDCM0596 | Fragment38 | MDA-MB-231 | C117(1.23) | LDD1433 | [40] |
| LDCM0597 | Fragment39 | MDA-MB-231 | C117(1.29) | LDD1435 | [40] |
| LDCM0566 | Fragment4 | Ramos | C117(1.54) | LDD2184 | [41] |
| LDCM0598 | Fragment40 | MDA-MB-231 | C117(1.02) | LDD1436 | [40] |
| LDCM0599 | Fragment41 | MDA-MB-231 | C117(1.32) | LDD1438 | [40] |
| LDCM0601 | Fragment43 | MDA-MB-231 | C117(3.12) | LDD1441 | [40] |
| LDCM0603 | Fragment45 | MDA-MB-231 | C117(20.00) | LDD1444 | [40] |
| LDCM0604 | Fragment46 | MDA-MB-231 | C117(1.21) | LDD1445 | [40] |
| LDCM0605 | Fragment47 | MDA-MB-231 | C117(1.92) | LDD1446 | [40] |
| LDCM0608 | Fragment50 | MDA-MB-231 | C117(6.19) | LDD1449 | [40] |
| LDCM0610 | Fragment52 | MDA-MB-231 | C117(1.10) | LDD1452 | [40] |
| LDCM0614 | Fragment56 | MDA-MB-231 | C117(1.63) | LDD1458 | [40] |
| LDCM0568 | Fragment6 | MDA-MB-231 | C117(1.35) | LDD1382 | [40] |
| LDCM0569 | Fragment7 | MDA-MB-231 | C117(3.54) | LDD1383 | [40] |
| LDCM0570 | Fragment8 | MDA-MB-231 | C117(6.19) | LDD1385 | [40] |
| LDCM0571 | Fragment9 | MDA-MB-231 | C117(2.13) | LDD1387 | [40] |
| LDCM0123 | JWB131 | DM93 | Y269(1.40); Y325(0.87) | LDD0285 | [16] |
| LDCM0124 | JWB142 | DM93 | Y269(1.30); Y325(1.26) | LDD0286 | [16] |
| LDCM0125 | JWB146 | DM93 | Y269(1.37); Y325(1.17) | LDD0287 | [16] |
| LDCM0126 | JWB150 | DM93 | Y269(2.14); Y325(3.17) | LDD0288 | [16] |
| LDCM0127 | JWB152 | DM93 | Y269(1.56); Y325(1.67) | LDD0289 | [16] |
| LDCM0128 | JWB198 | DM93 | Y269(0.57) | LDD0290 | [16] |
| LDCM0129 | JWB202 | DM93 | Y269(0.74); Y325(0.92) | LDD0291 | [16] |
| LDCM0130 | JWB211 | DM93 | Y269(1.11); Y325(1.12) | LDD0292 | [16] |
| LDCM0073 | Kambe_cp3 | PC-3 | 3.56 | LDD0128 | [36] |
| LDCM0079 | kambe_cp64 | PC-3 | 2.30 | LDD0135 | [36] |
| LDCM0022 | KB02 | MDA-MB-231 | C117(20.00) | LDD1374 | [40] |
| LDCM0023 | KB03 | MDA-MB-231 | C117(20.00) | LDD1376 | [40] |
| LDCM0024 | KB05 | G361 | C286(1.53) | LDD3311 | [11] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C329(1.06) | LDD2089 | [13] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C329(0.83) | LDD2106 | [13] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C329(0.76) | LDD2125 | [13] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C329(1.30) | LDD2136 | [13] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C329(0.77) | LDD2150 | [13] |
| LDCM0099 | Phenelzine | HEK-293T | 10.30 | LDD0282 | [10] |
| LDCM0084 | Ro 48-8071 | A-549 | 2.05 | LDD0145 | [35] |
| LDCM0021 | THZ1 | HeLa S3 | C329(1.06) | LDD0460 | [12] |
| LDCM0008 | Tranylcypromine | SH-SY5Y | 2.44 | LDD0053 | [5] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Amyloid-beta precursor protein (APP) | APP family | P05067 | |||
| Voltage-dependent anion-selective channel protein 2 (VDAC2) | Eukaryotic mitochondrial porin family | P45880 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Zinc finger protein 497 (ZNF497) | Krueppel C2H2-type zinc-finger protein family | Q6ZNH5 | |||
GPCR
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Probable G-protein coupled receptor 141 (GPR141) | G-protein coupled receptor 1 family | Q7Z602 | |||
Other
The Drug(s) Related To This Target
Clinical trial
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Pmid10498202c1 | Small molecular drug | D0FE7H | |||
Investigative
Discontinued
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Ci-992 | Small molecular drug | D04TSV | |||
References
