Details of the Target
General Information of Target
| Target ID | LDTP11124 | |||||
|---|---|---|---|---|---|---|
| Target Name | Ferroptosis suppressor protein 1 (AIFM2) | |||||
| Gene Name | AIFM2 | |||||
| Gene ID | 84883 | |||||
| Synonyms |
AMID; PRG3; Ferroptosis suppressor protein 1; FSP1; EC 1.6.5.-; Apoptosis-inducing factor homologous mitochondrion-associated inducer of death; AMID; p53-responsive gene 3 protein |
|||||
| 3D Structure | ||||||
| Sequence |
MGSTESSEGRRVSFGVDEEERVRVLQGVRLSENVVNRMKEPSSPPPAPTSSTFGLQDGNL
RAPHKESTLPRSGSSGGQQPSGMKEGVKRYEQEHAAIQDKLFQVAKREREAATKHSKASL PTGEGSISHEEQKSVRLARELESREAELRRRDTFYKEQLERIERKNAEMYKLSSEQFHEA ASKMESTIKPRRVEPVCSGLQAQILHCYRDRPHEVLLCSDLVKAYQRCVSAAHKG |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
FAD-dependent oxidoreductase family
|
|||||
| Subcellular location |
Lipid droplet
|
|||||
| Function |
A NAD(P)H-dependent oxidoreductase that acts as a key inhibitor of ferroptosis. At the plasma membrane, catalyzes reduction of coenzyme Q/ubiquinone-10 to ubiquinol-10, a lipophilic radical-trapping antioxidant that prevents lipid oxidative damage and consequently ferroptosis. Acts in parallel to GPX4 to suppress phospholipid peroxidation and ferroptosis. This anti-ferroptotic function is independent of cellular glutathione levels. Also acts as a potent radical-trapping antioxidant by mediating warfarin-resistant vitamin K reduction in the canonical vitamin K cycle: catalyzes NAD(P)H-dependent reduction of vitamin K (phylloquinone, menaquinone-4 and menadione) to hydroquinone forms. Hydroquinones act as potent radical-trapping antioxidants inhibitor of phospholipid peroxidation and ferroptosis. May play a role in mitochondrial stress signaling. Upon oxidative stress, associates with the lipid peroxidation end product 4-hydroxy-2-nonenal (HNE) forming a lipid adduct devoid of oxidoreductase activity, which then translocates from mitochondria into the nucleus triggering DNA damage and cell death. Capable of DNA binding in a non-sequence specific way.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| MCC13 | SNV: p.R267I | DBIA Probe Info | |||
| NCIH716 | SNV: p.A261D | . | |||
| SKOV3 | SNV: p.G337D | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FBP2 Probe Info |
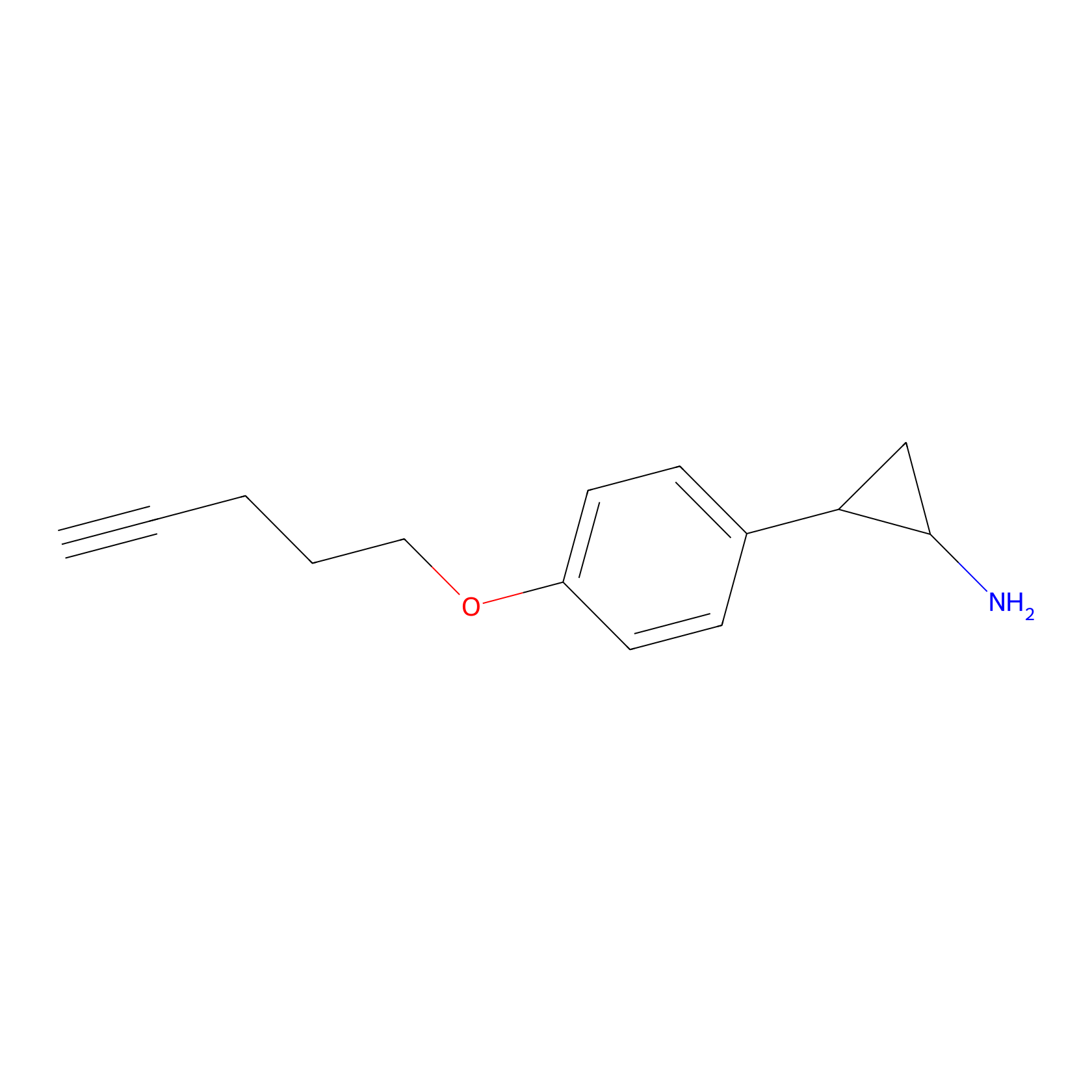 |
2.80 | LDD0323 | [1] | |
|
STPyne Probe Info |
 |
K254(7.30) | LDD0277 | [2] | |
|
m-APA Probe Info |
 |
9.03 | LDD0403 | [3] | |
|
DBIA Probe Info |
 |
C242(2.64) | LDD3314 | [4] | |
|
IPM Probe Info |
 |
C286(2.27) | LDD1701 | [5] | |
|
NAIA_4 Probe Info |
 |
C242(0.00); C286(0.00) | LDD2226 | [6] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
A-DA Probe Info |
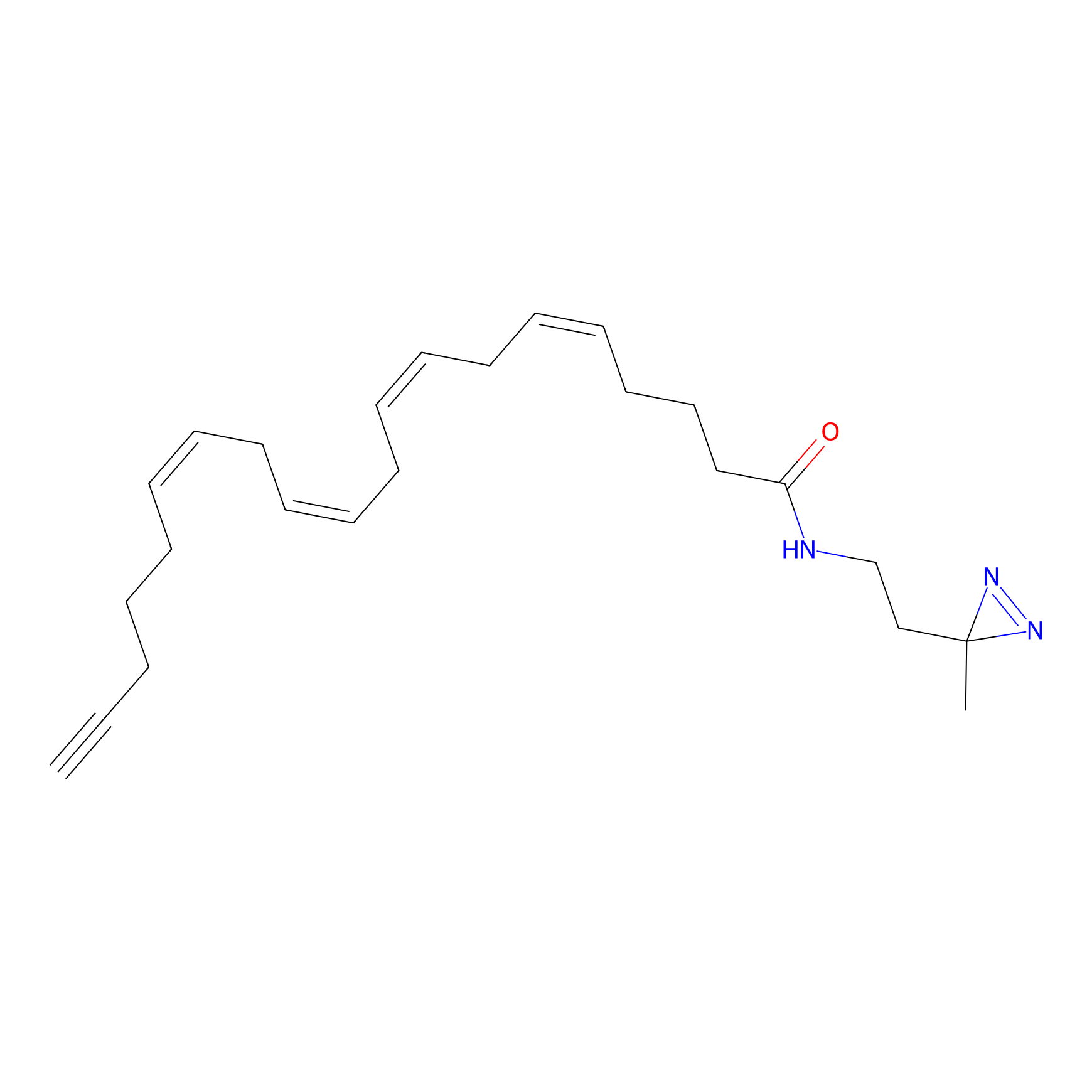 |
16.14 | LDD0143 | [7] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0156 | Aniline | NCI-H1299 | 9.03 | LDD0403 | [3] |
| LDCM0083 | Avasimibe | A-549 | 16.14 | LDD0143 | [7] |
| LDCM0022 | KB02 | 42-MG-BA | C242(1.97) | LDD2244 | [4] |
| LDCM0023 | KB03 | MDA-MB-231 | C286(2.27) | LDD1701 | [5] |
| LDCM0024 | KB05 | IGR37 | C242(2.64) | LDD3314 | [4] |
| LDCM0084 | Ro 48-8071 | A-549 | 2.20 | LDD0144 | [7] |
The Interaction Atlas With This Target
References
