Details of the Target
General Information of Target
| Target ID | LDTP07591 | |||||
|---|---|---|---|---|---|---|
| Target Name | Neutral cholesterol ester hydrolase 1 (NCEH1) | |||||
| Gene Name | NCEH1 | |||||
| Gene ID | 57552 | |||||
| Synonyms |
AADACL1; KIAA1363; Neutral cholesterol ester hydrolase 1; NCEH; EC 3.1.1.-; Acetylalkylglycerol acetylhydrolase; 2-acetyl MAGE hydrolase; EC 3.1.1.71; Arylacetamide deacetylase-like 1 |
|||||
| 3D Structure | ||||||
| Sequence |
MRSSCVLLTALVALAAYYVYIPLPGSVSDPWKLMLLDATFRGAQQVSNLIHYLGLSHHLL
ALNFIIVSFGKKSAWSSAQVKVTDTDFDGVEVRVFEGPPKPEEPLKRSVVYIHGGGWALA SAKIRYYDELCTAMAEELNAVIVSIEYRLVPKVYFPEQIHDVVRATKYFLKPEVLQKYMV DPGRICISGDSAGGNLAAALGQQFTQDASLKNKLKLQALIYPVLQALDFNTPSYQQNVNT PILPRYVMVKYWVDYFKGNYDFVQAMIVNNHTSLDVEEAAAVRARLNWTSLLPASFTKNY KPVVQTTGNARIVQELPQLLDARSAPLIADQAVLQLLPKTYILTCEHDVLRDDGIMYAKR LESAGVEVTLDHFEDGFHGCMIFTSWPTNFSVGIRTRNSYIKWLDQNL |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
'GDXG' lipolytic enzyme family
|
|||||
| Subcellular location |
Cell membrane
|
|||||
| Function |
Hydrolyzes 2-acetyl monoalkylglycerol ether (1-O-alkyl-2-acetyl-sn-glycerol), the penultimate precursor of the pathway for de novo synthesis of platelet-activating factor. May be responsible for the hydrolysis of cholesterol esters (such as cholesteryl (9Z-octadecenoate)) in macrophages. Also involved in organ detoxification by hydrolyzing exogenous organophosphorus compounds. May contribute to cancer pathogenesis by promoting tumor cell migration.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| HEC1 | SNV: p.Q331H | . | |||
| HEC1B | SNV: p.Q331H | . | |||
| LNCaP clone FGC | SNV: p.G89S | . | |||
| MDAMB231 | SNV: p.S108N | . | |||
| OSRC2 | SNV: p.R107H | DBIA Probe Info | |||
| PANC1 | SNV: p.G258D | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
HDSF-alk Probe Info |
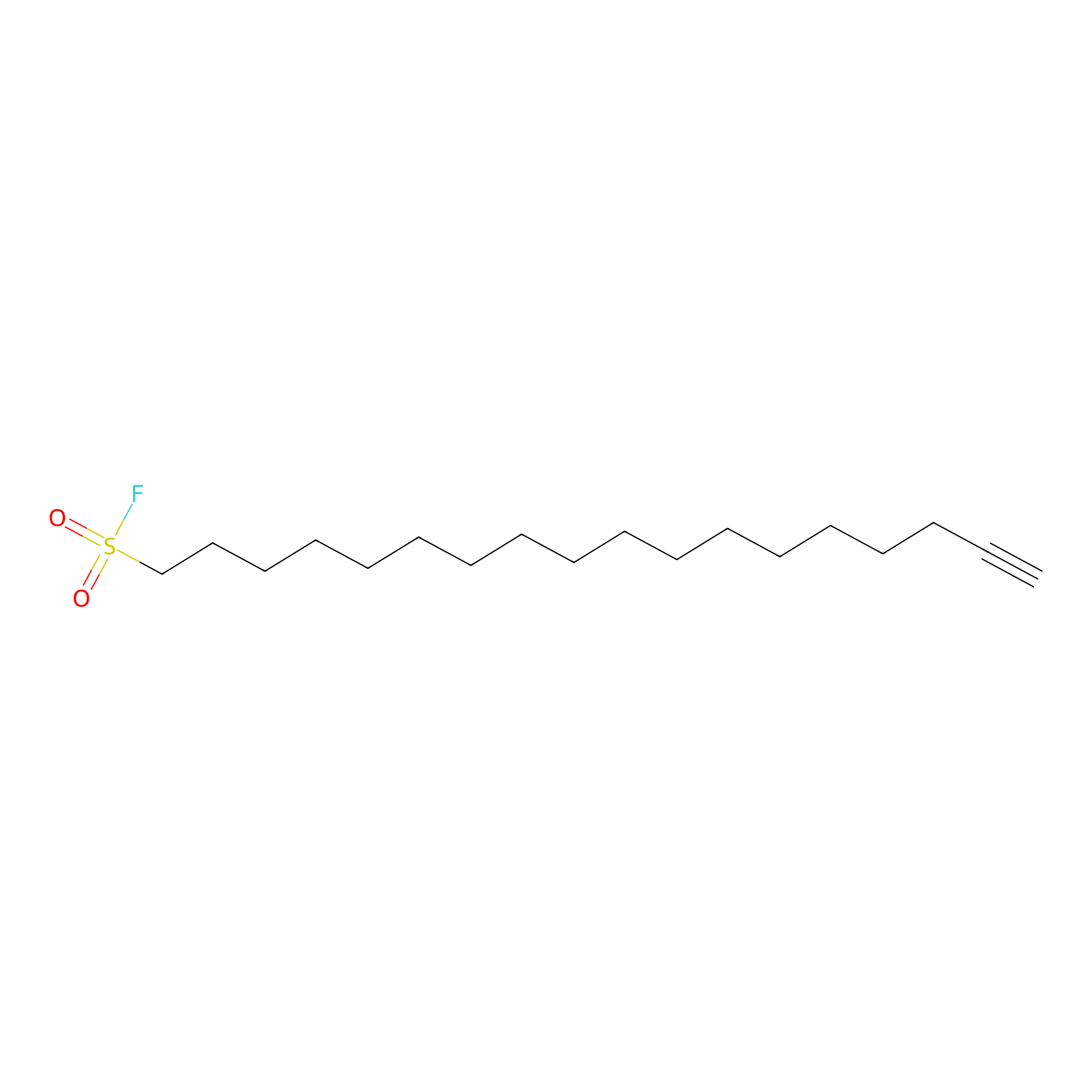 |
2.07 | LDD0197 | [1] | |
|
EN219-alkyne Probe Info |
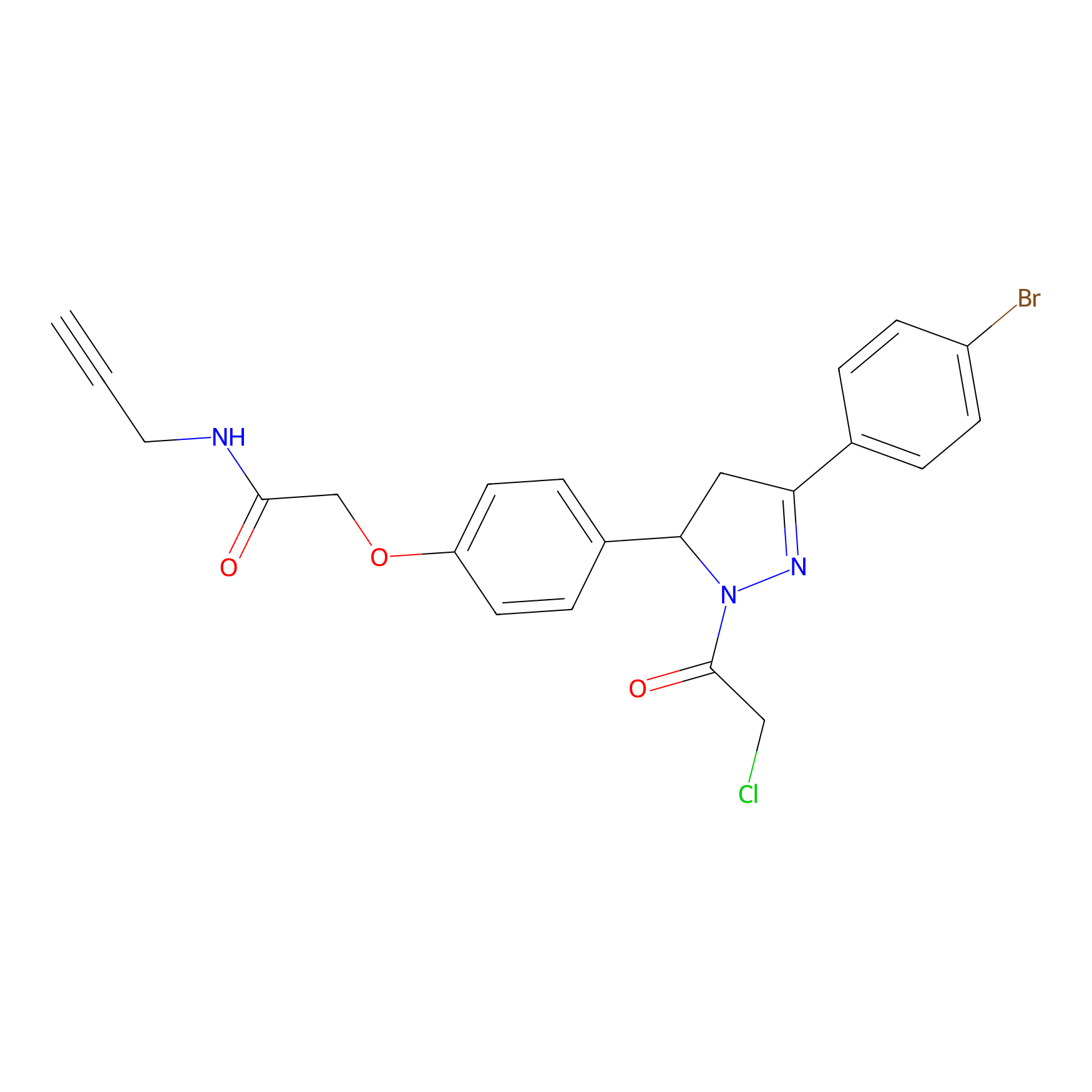 |
3.94 | LDD0297 | [2] | |
|
CHEMBL5175495 Probe Info |
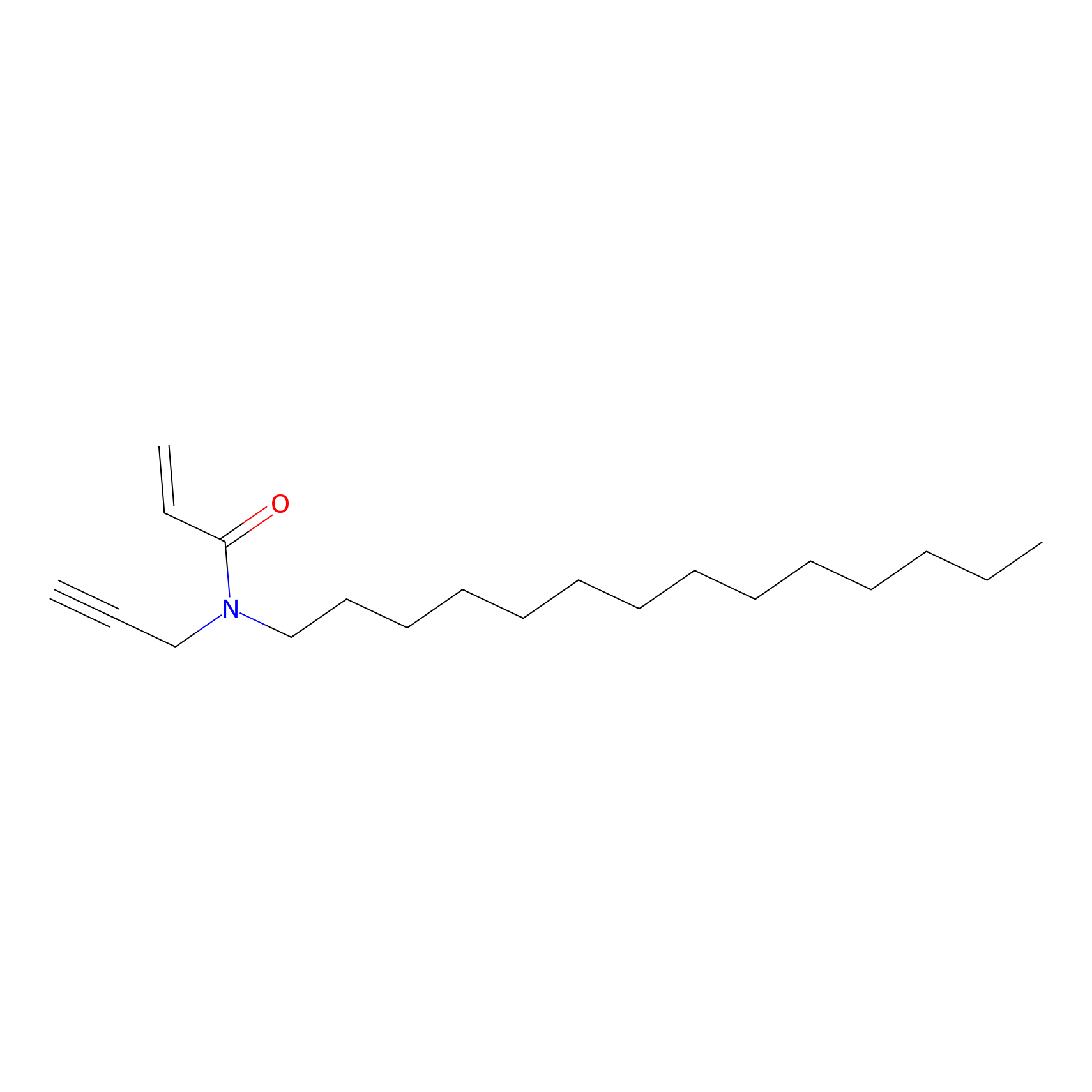 |
18.97 | LDD0196 | [3] | |
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [4] | |
|
CY4 Probe Info |
 |
100.00 | LDD0244 | [4] | |
|
N1 Probe Info |
 |
100.00 | LDD0242 | [4] | |
|
FBP2 Probe Info |
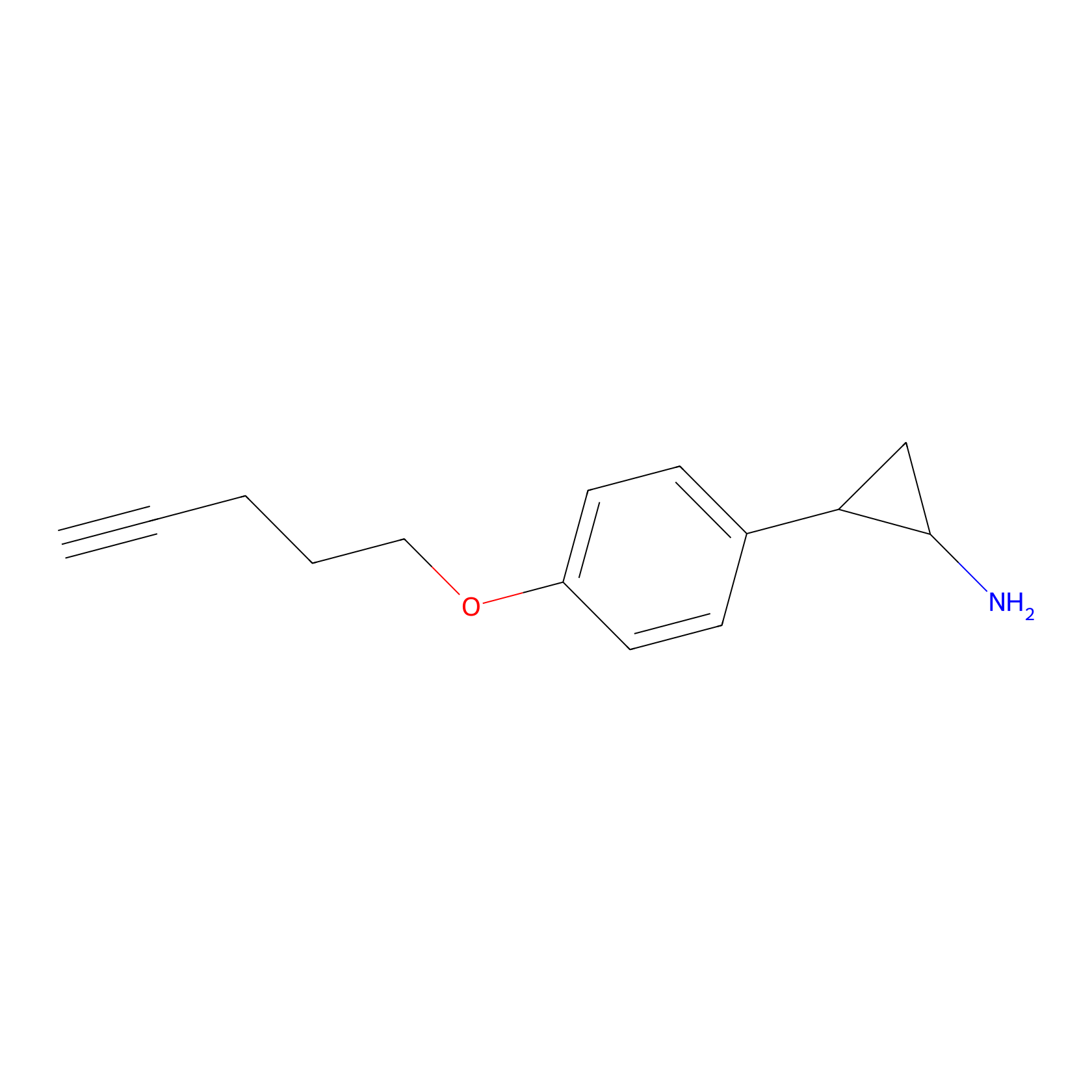 |
2.03 | LDD0323 | [5] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [6] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [6] | |
|
STPyne Probe Info |
 |
K152(8.51); K167(6.60); K301(10.00) | LDD0277 | [7] | |
|
OPA-S-S-alkyne Probe Info |
 |
K359(1.48) | LDD3494 | [8] | |
|
m-APA Probe Info |
 |
11.51 | LDD0403 | [9] | |
|
DBIA Probe Info |
 |
C353(1.97) | LDD3316 | [10] | |
|
THZ1-DTB Probe Info |
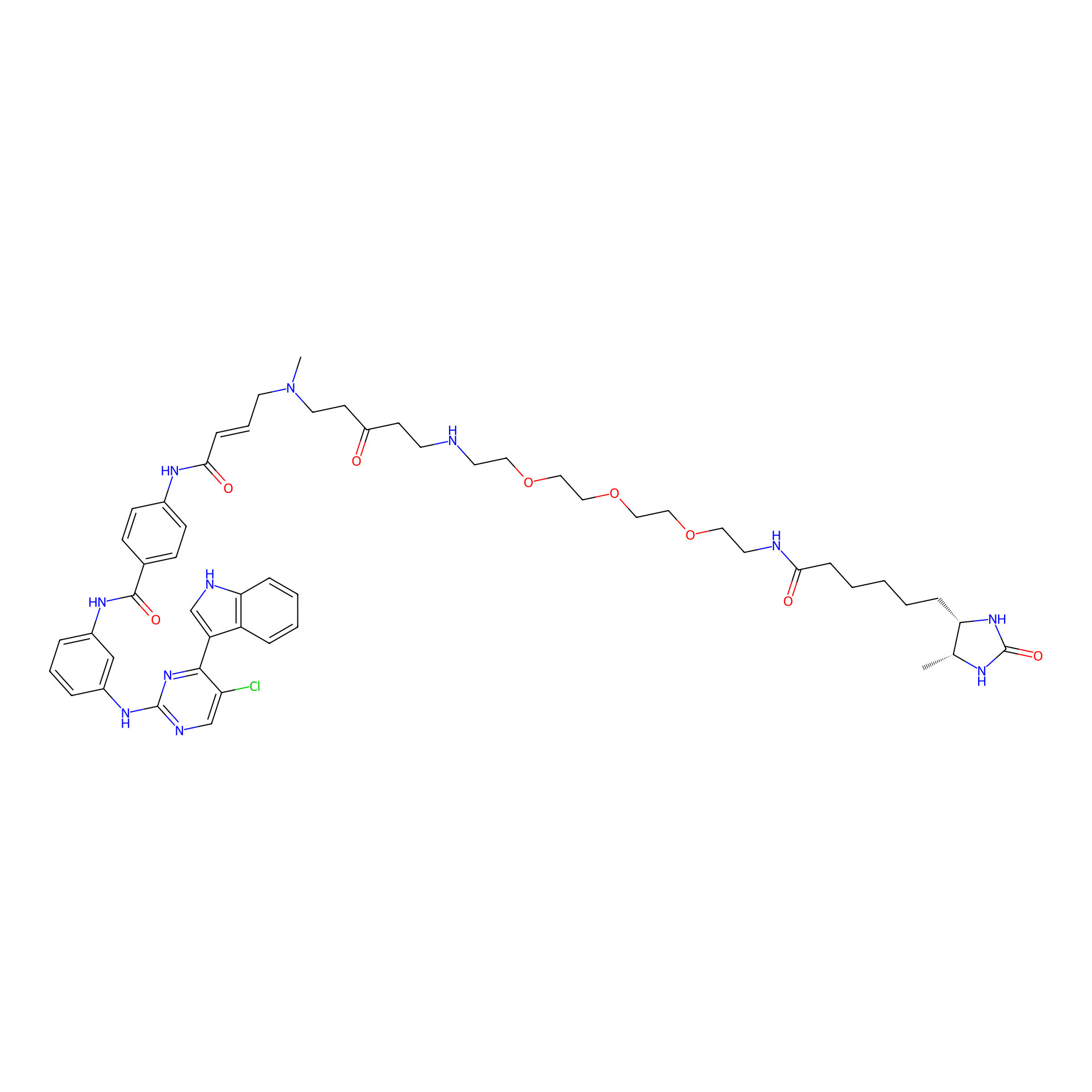 |
C186(2.47) | LDD0461 | [11] | |
|
BTD Probe Info |
 |
C345(0.41) | LDD2115 | [12] | |
|
Curcusone 37 Probe Info |
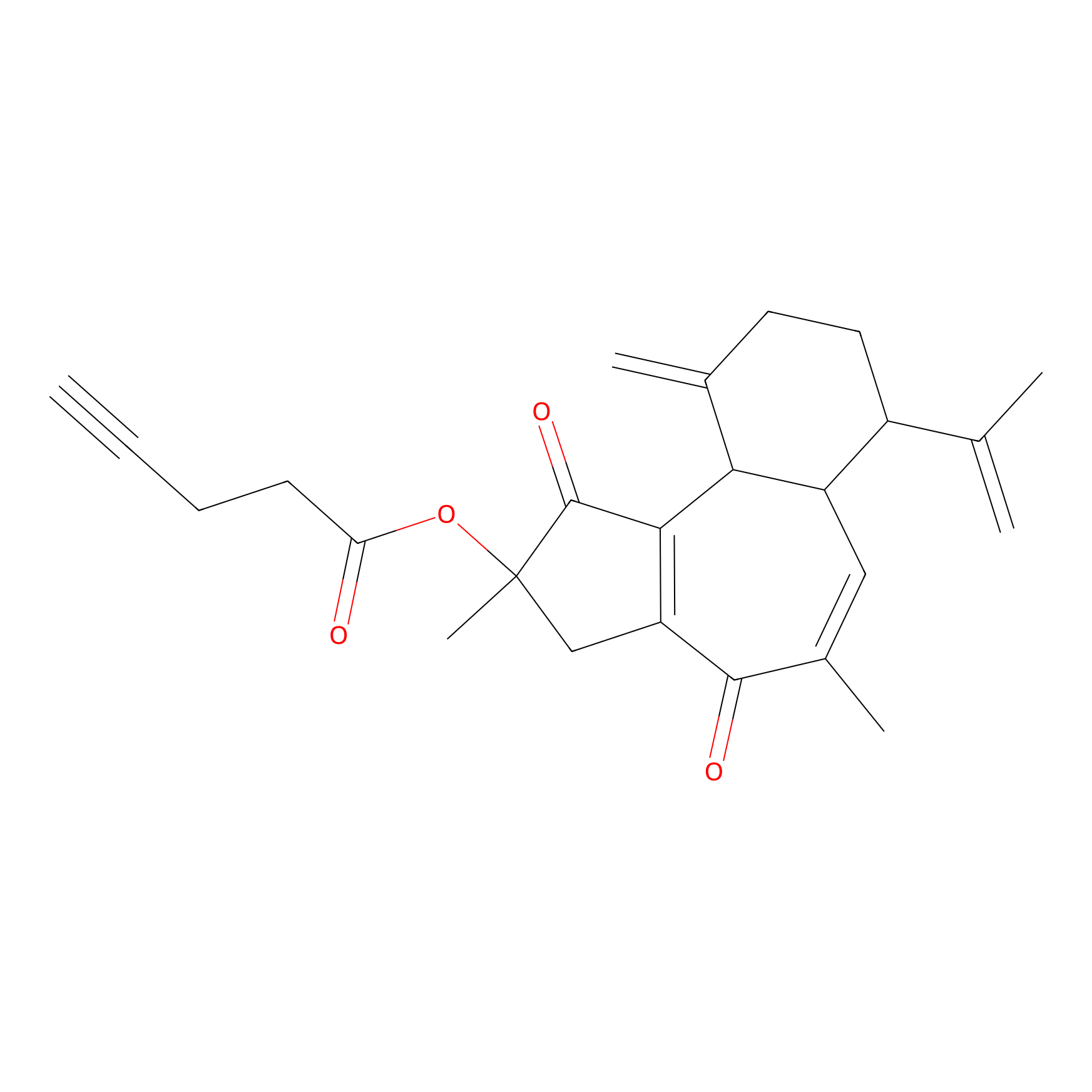 |
9.03 | LDD0188 | [13] | |
|
Jackson_14 Probe Info |
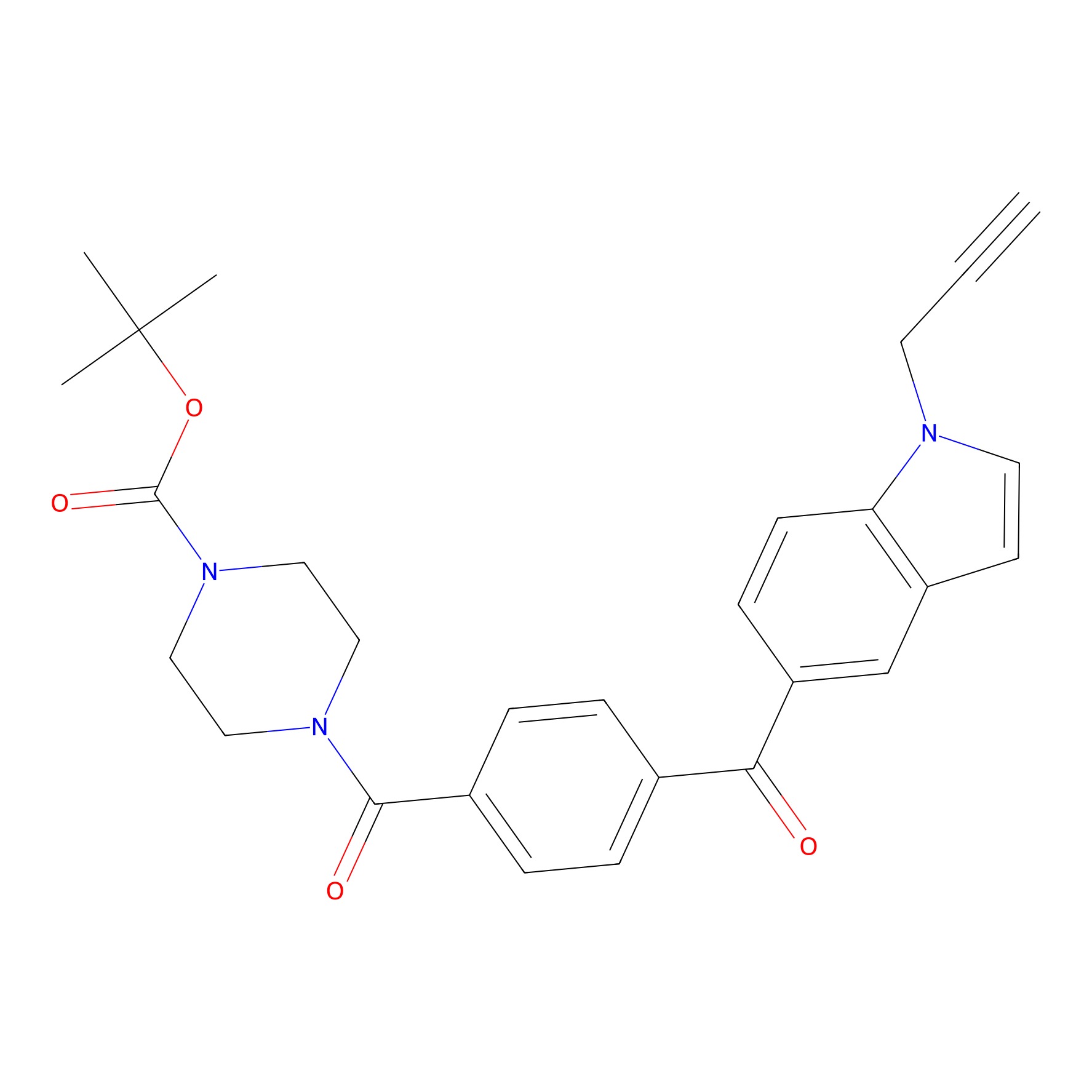 |
2.17 | LDD0123 | [14] | |
|
Johansson_61 Probe Info |
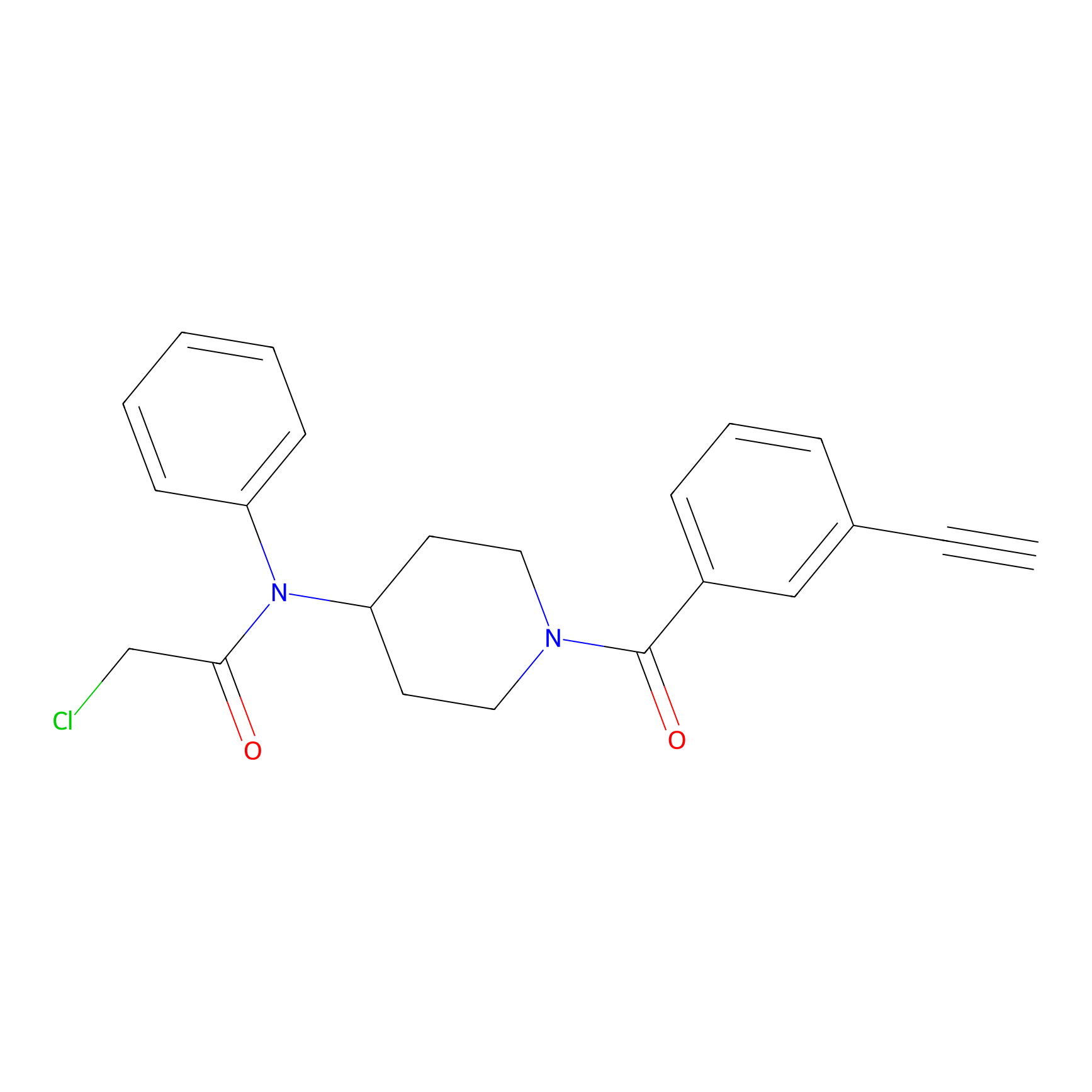 |
_(11.87) | LDD1485 | [15] | |
|
YY4-yne Probe Info |
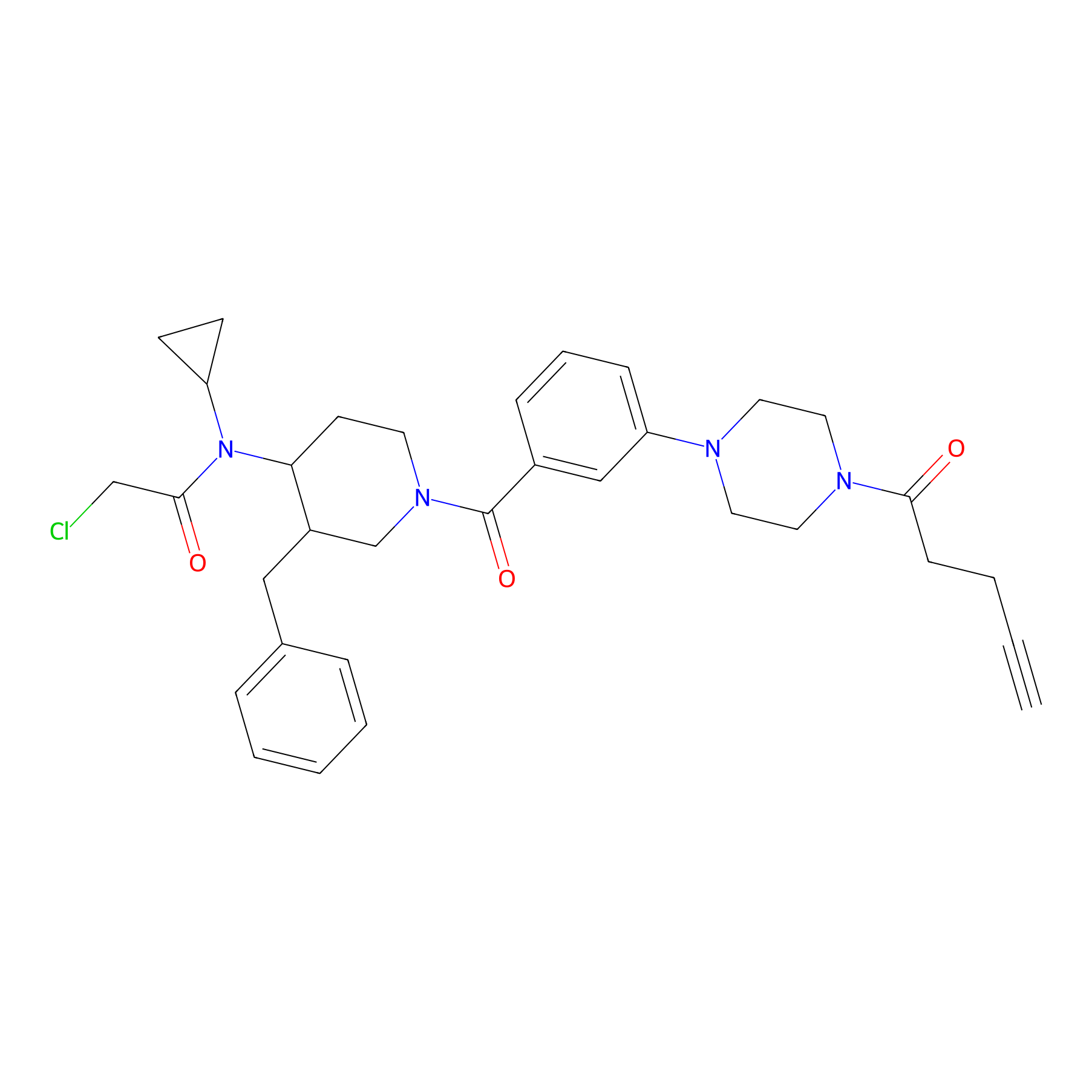 |
3.75 | LDD0400 | [16] | |
|
HPAP Probe Info |
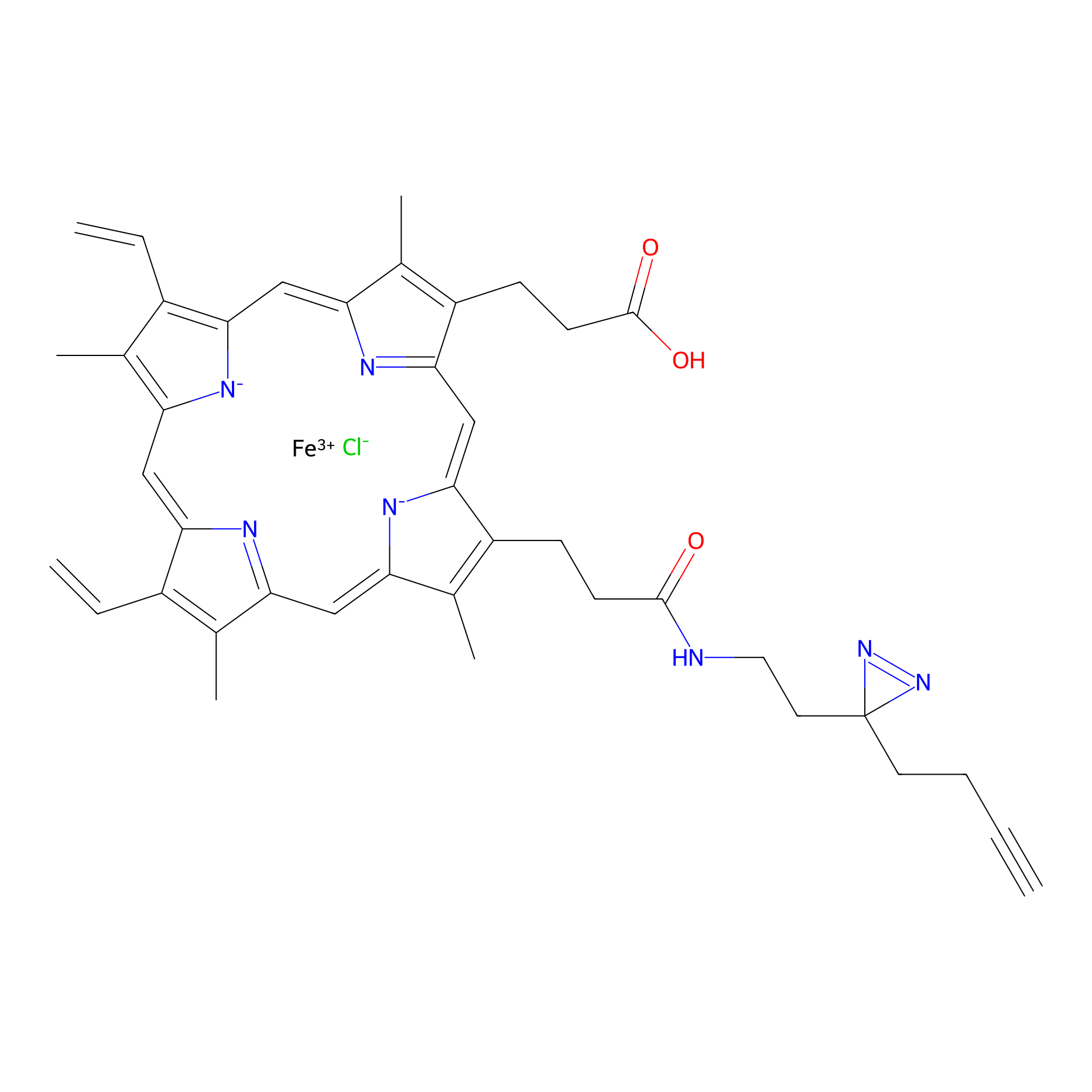 |
3.55 | LDD0062 | [17] | |
|
IA-alkyne Probe Info |
 |
C186(0.00); C345(0.00) | LDD0162 | [18] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [19] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [20] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
DR-1 Probe Info |
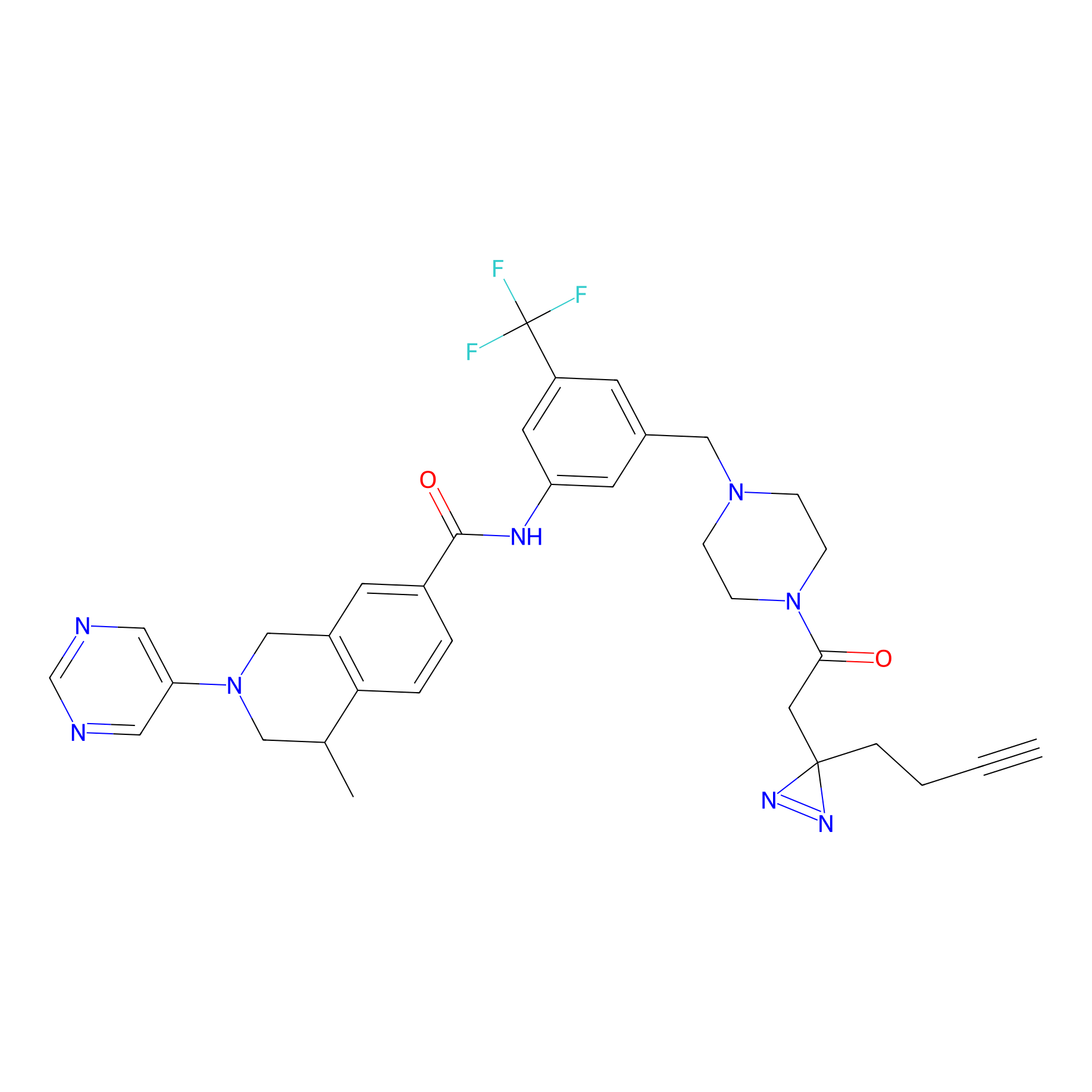 |
2.03 | LDD0398 | [21] | |
|
C027 Probe Info |
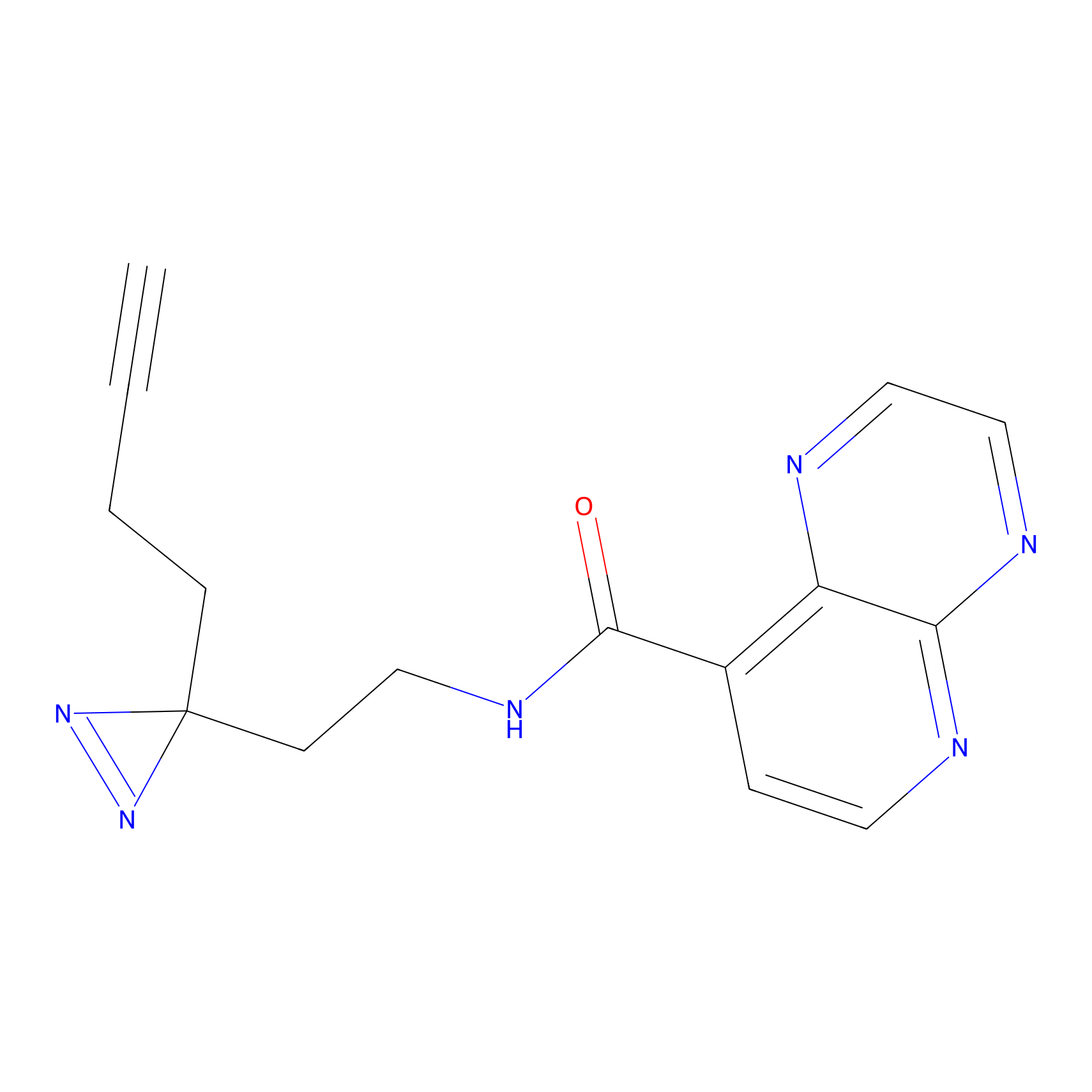 |
6.23 | LDD1733 | [22] | |
|
C056 Probe Info |
 |
17.39 | LDD1753 | [22] | |
|
C091 Probe Info |
 |
12.38 | LDD1782 | [22] | |
|
C094 Probe Info |
 |
34.06 | LDD1785 | [22] | |
|
C112 Probe Info |
 |
18.00 | LDD1799 | [22] | |
|
C178 Probe Info |
 |
15.78 | LDD1857 | [22] | |
|
C246 Probe Info |
 |
13.18 | LDD1919 | [22] | |
|
C251 Probe Info |
 |
37.01 | LDD1924 | [22] | |
|
C299 Probe Info |
 |
7.31 | LDD1968 | [22] | |
|
FFF probe11 Probe Info |
 |
20.00 | LDD0471 | [23] | |
|
FFF probe3 Probe Info |
 |
20.00 | LDD0464 | [23] | |
|
STS-1 Probe Info |
 |
3.57 | LDD0136 | [24] | |
|
STS-2 Probe Info |
 |
2.57 | LDD0138 | [24] | |
|
A-DA Probe Info |
 |
2.78 | LDD0145 | [25] | |
|
OEA-DA Probe Info |
 |
5.07 | LDD0046 | [26] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C345(0.43) | LDD2142 | [12] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C345(1.74) | LDD2117 | [12] |
| LDCM0156 | Aniline | NCI-H1299 | 11.51 | LDD0403 | [9] |
| LDCM0033 | Curcusone1d | MCF-7 | 9.03 | LDD0188 | [13] |
| LDCM0100 | EN219 | 231MFP | 4.55 | LDD0298 | [2] |
| LDCM0616 | Fragment61 | Jurkat | _(20.00) | LDD1489 | [15] |
| LDCM0615 | Fragment63-R | Jurkat | _(20.00) | LDD1487 | [15] |
| LDCM0617 | Fragment63-S | Jurkat | _(20.00) | LDD1490 | [15] |
| LDCM0569 | Fragment7 | Jurkat | _(11.87) | LDD1485 | [15] |
| LDCM0022 | KB02 | 42-MG-BA | C53(1.08) | LDD2244 | [10] |
| LDCM0023 | KB03 | 42-MG-BA | C53(1.38) | LDD2661 | [10] |
| LDCM0024 | KB05 | MEL167 | C353(1.97) | LDD3316 | [10] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C345(0.41) | LDD2115 | [12] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C345(1.52) | LDD2123 | [12] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C345(0.73) | LDD2141 | [12] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C345(0.59) | LDD2148 | [12] |
| LDCM0557 | Nucleophilic fragment 8b | MDA-MB-231 | C345(4.80) | LDD2151 | [12] |
| LDCM0014 | Panhematin | HEK-293T | 3.55 | LDD0062 | [17] |
| LDCM0016 | Ranjitkar_cp1 | MDA-MB-231 | 2.17 | LDD0123 | [14] |
| LDCM0084 | Ro 48-8071 | A-549 | 2.78 | LDD0145 | [25] |
| LDCM0178 | THZ531 | HeLa S3 | C186(2.47) | LDD0461 | [11] |
| LDCM0154 | YY4 | T cell | 3.75 | LDD0400 | [16] |
References
