Details of the Target
General Information of Target
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| HKGZCC | SNV: p.F19V | . | |||
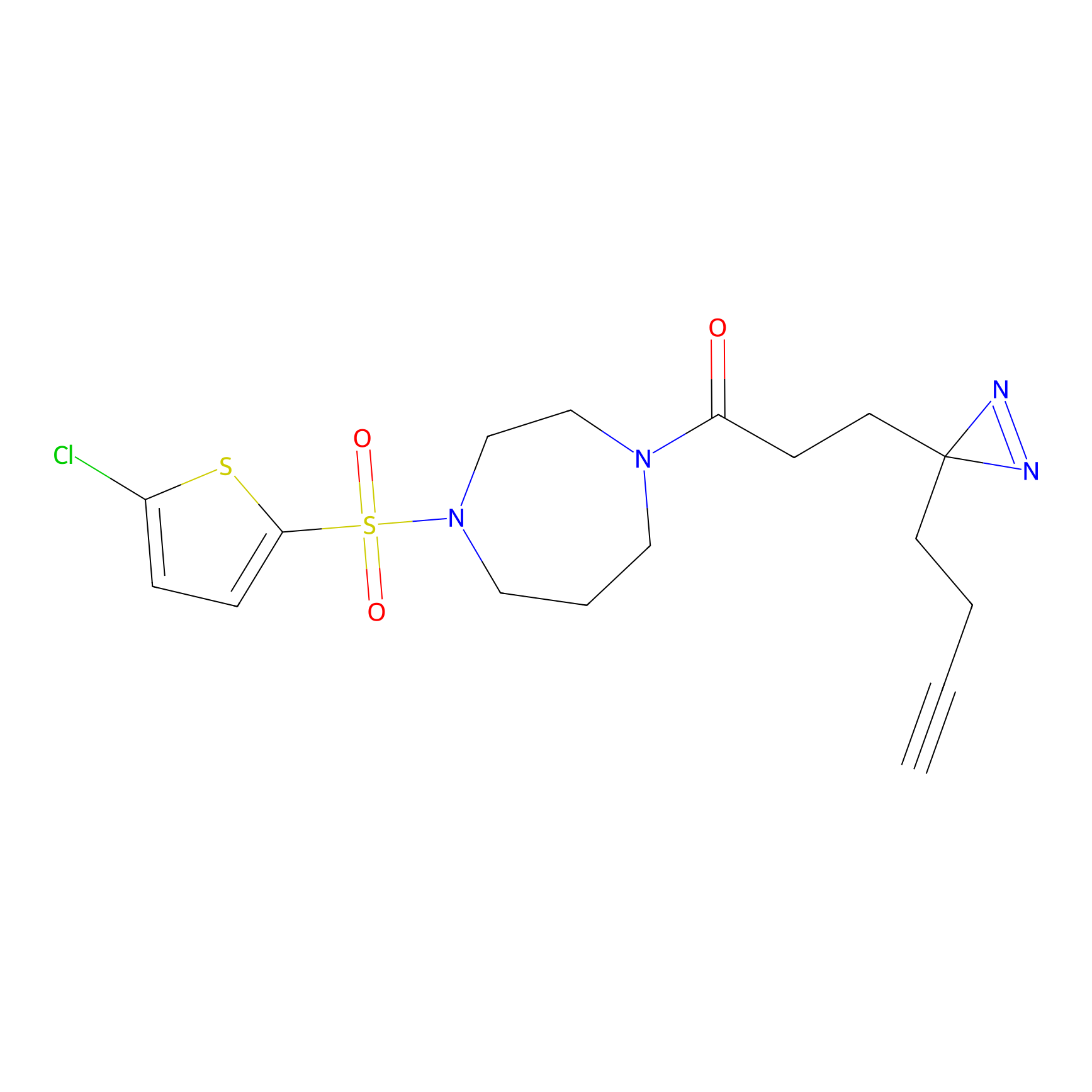
| NCIH1155 | SNV: p.A80T | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
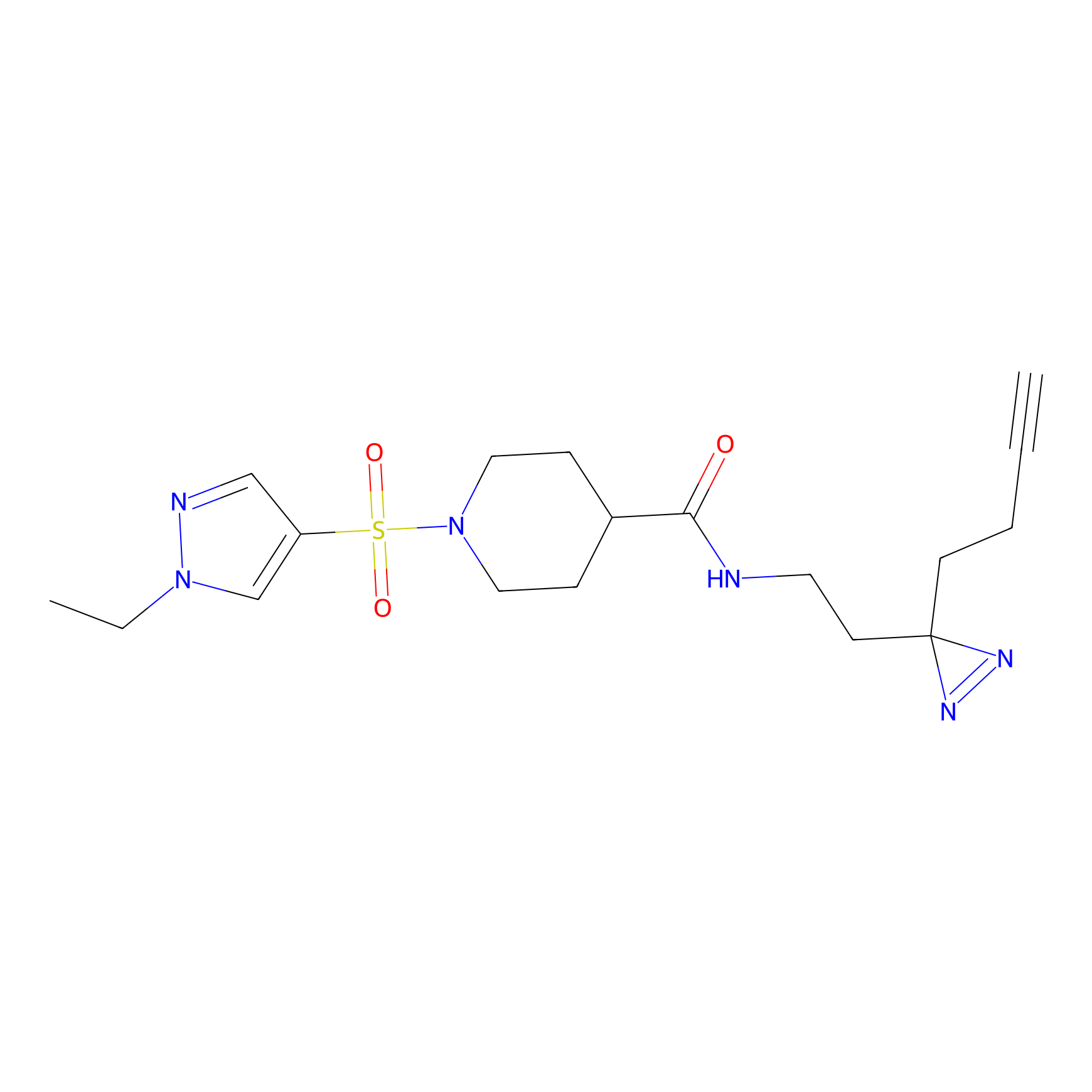
DBIA Probe Info |
 |
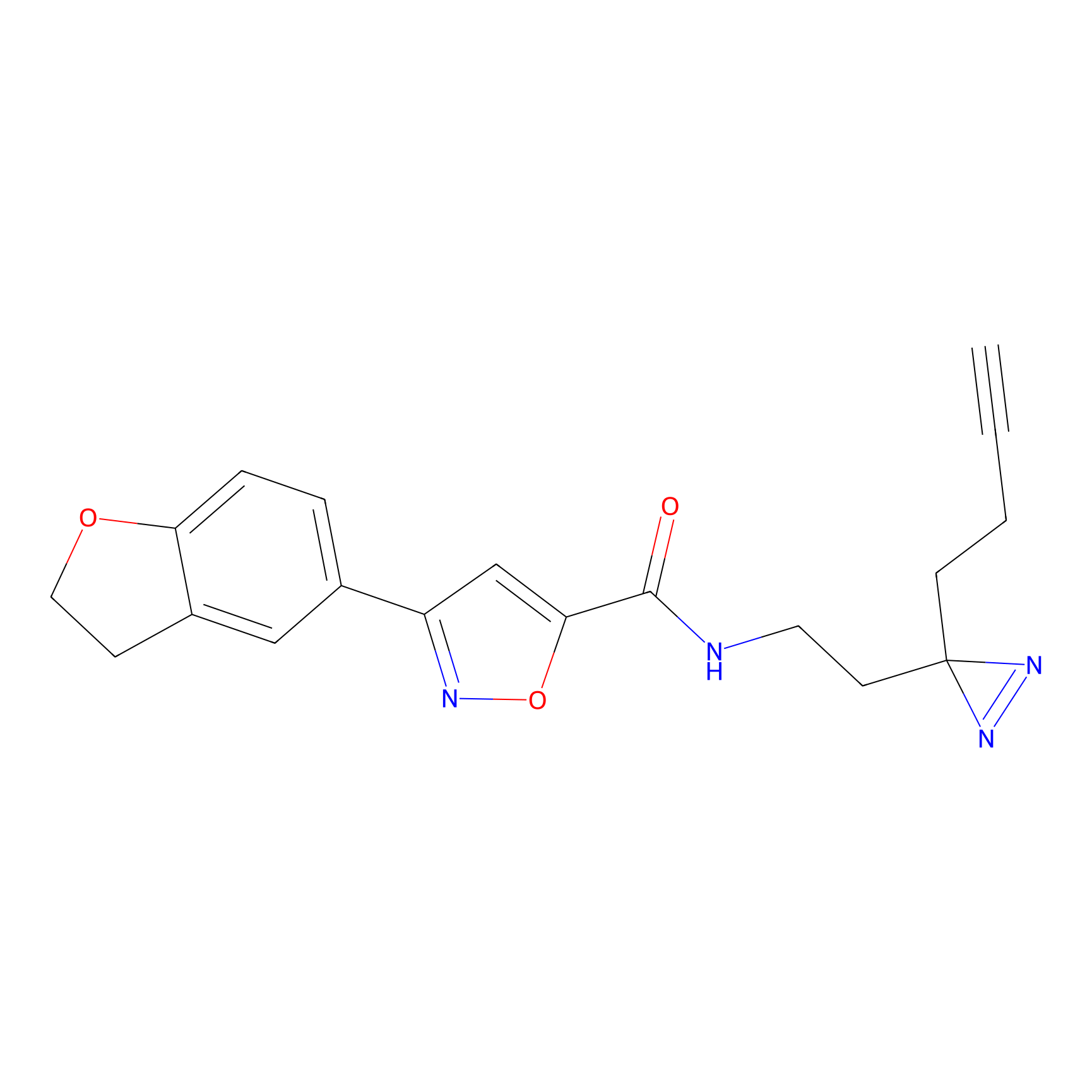
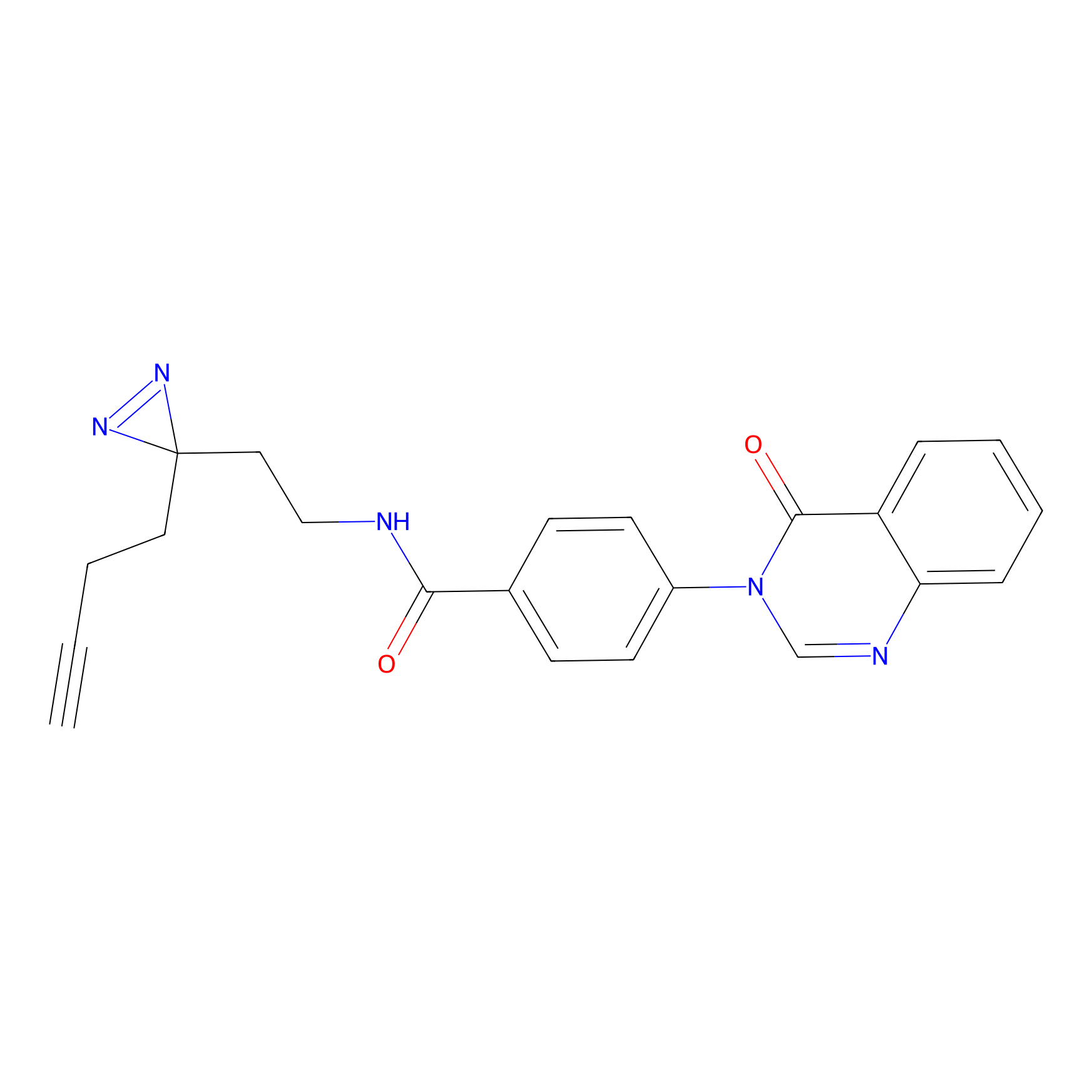
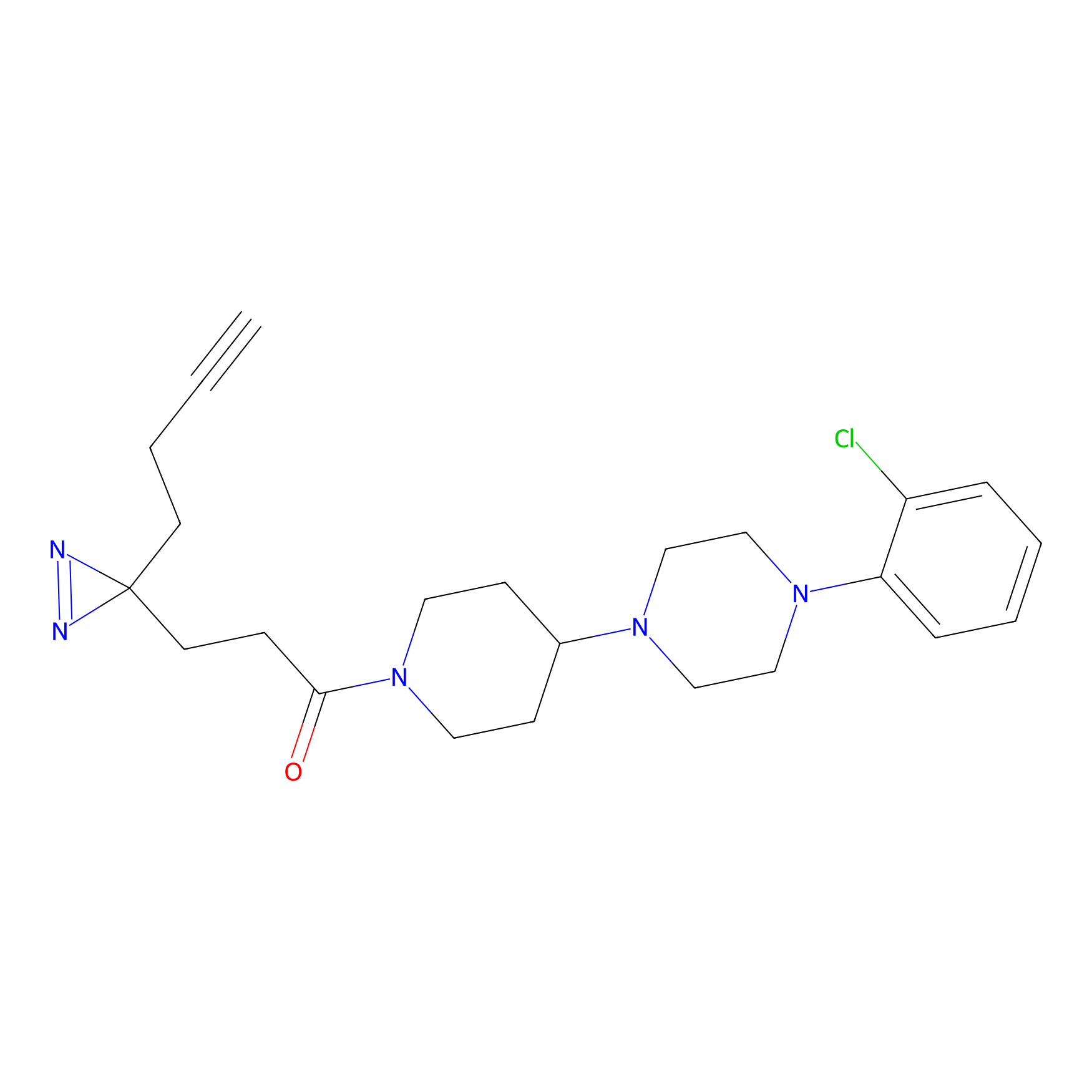
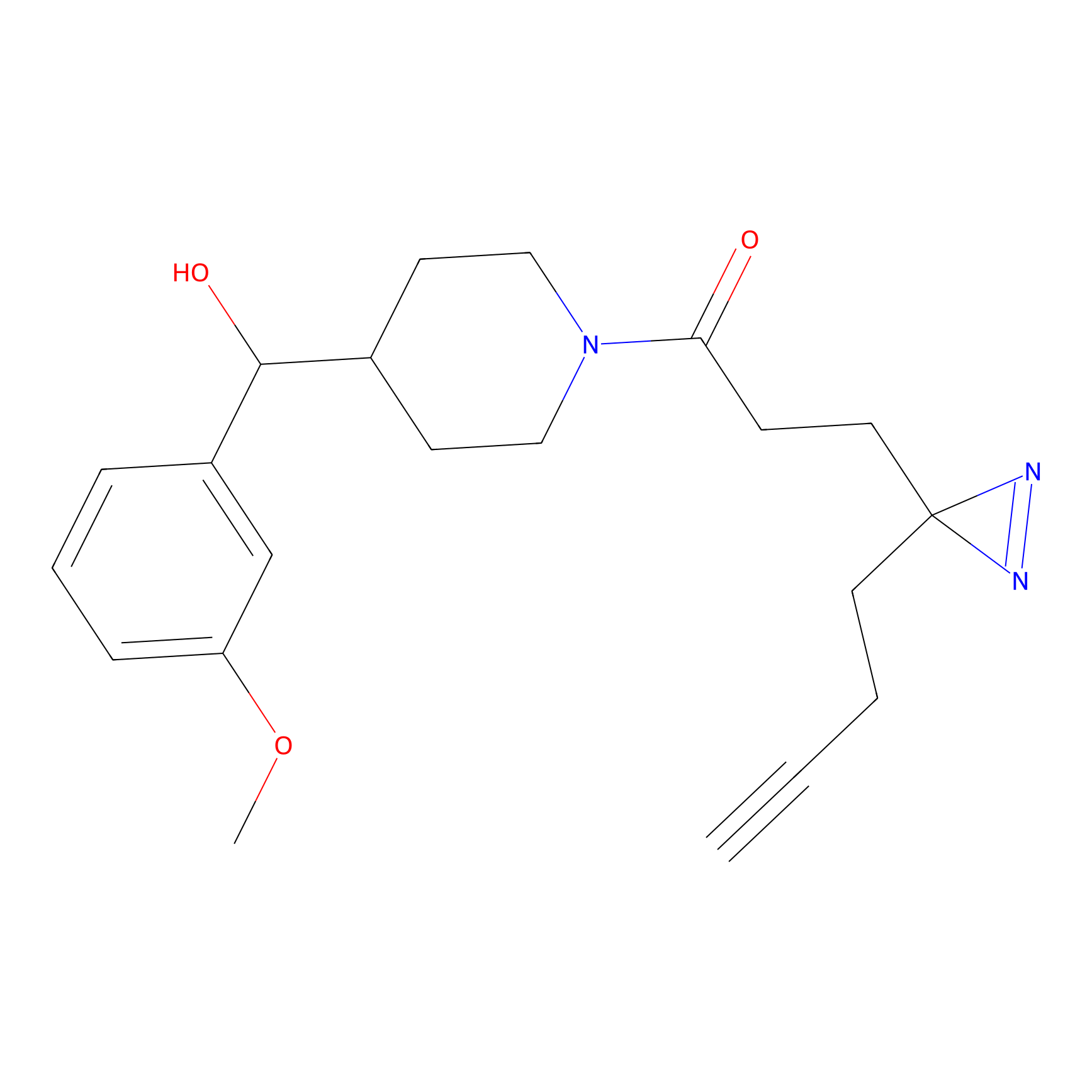
C349(1.34) | LDD3312 | [1] | |
|
BTD Probe Info |
 |
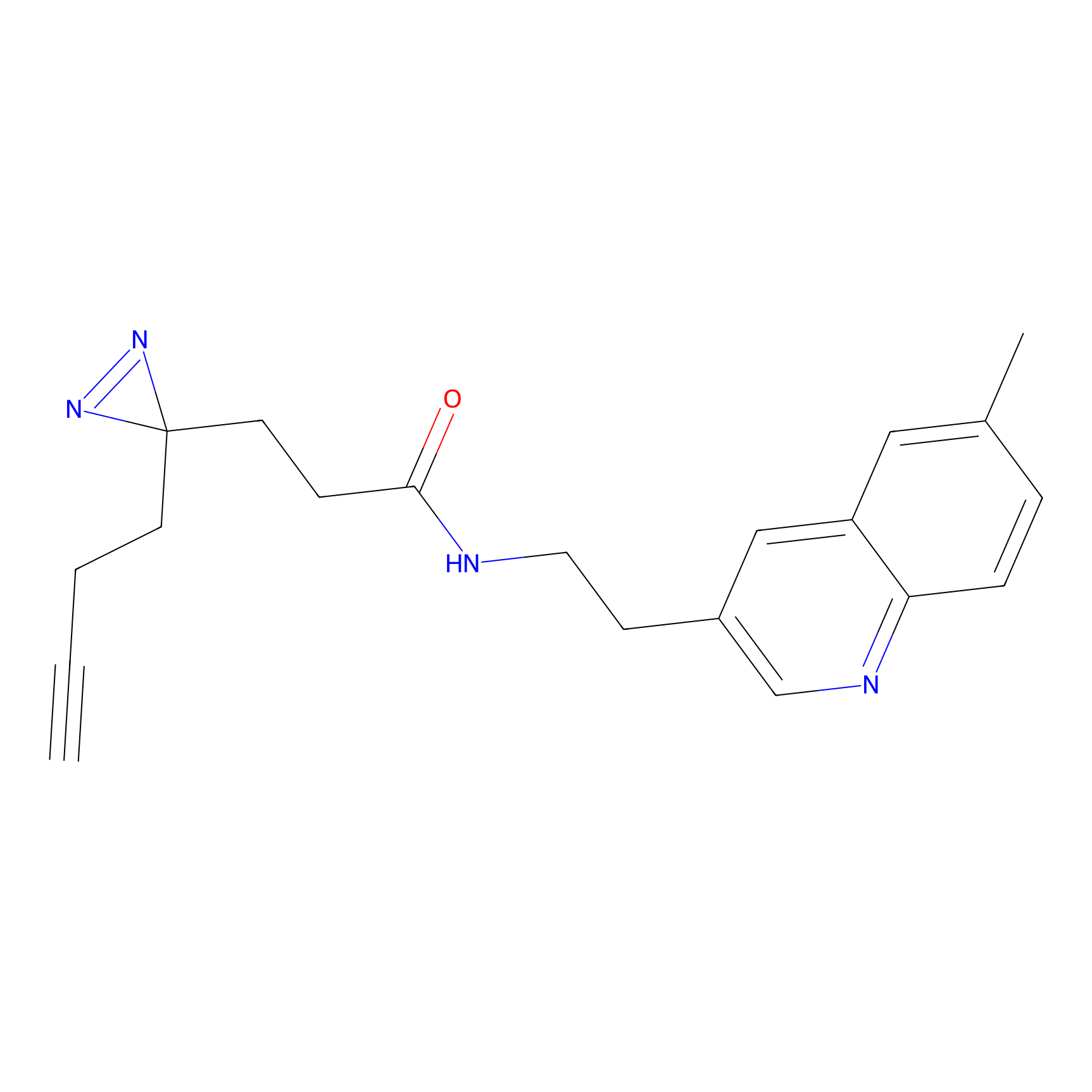
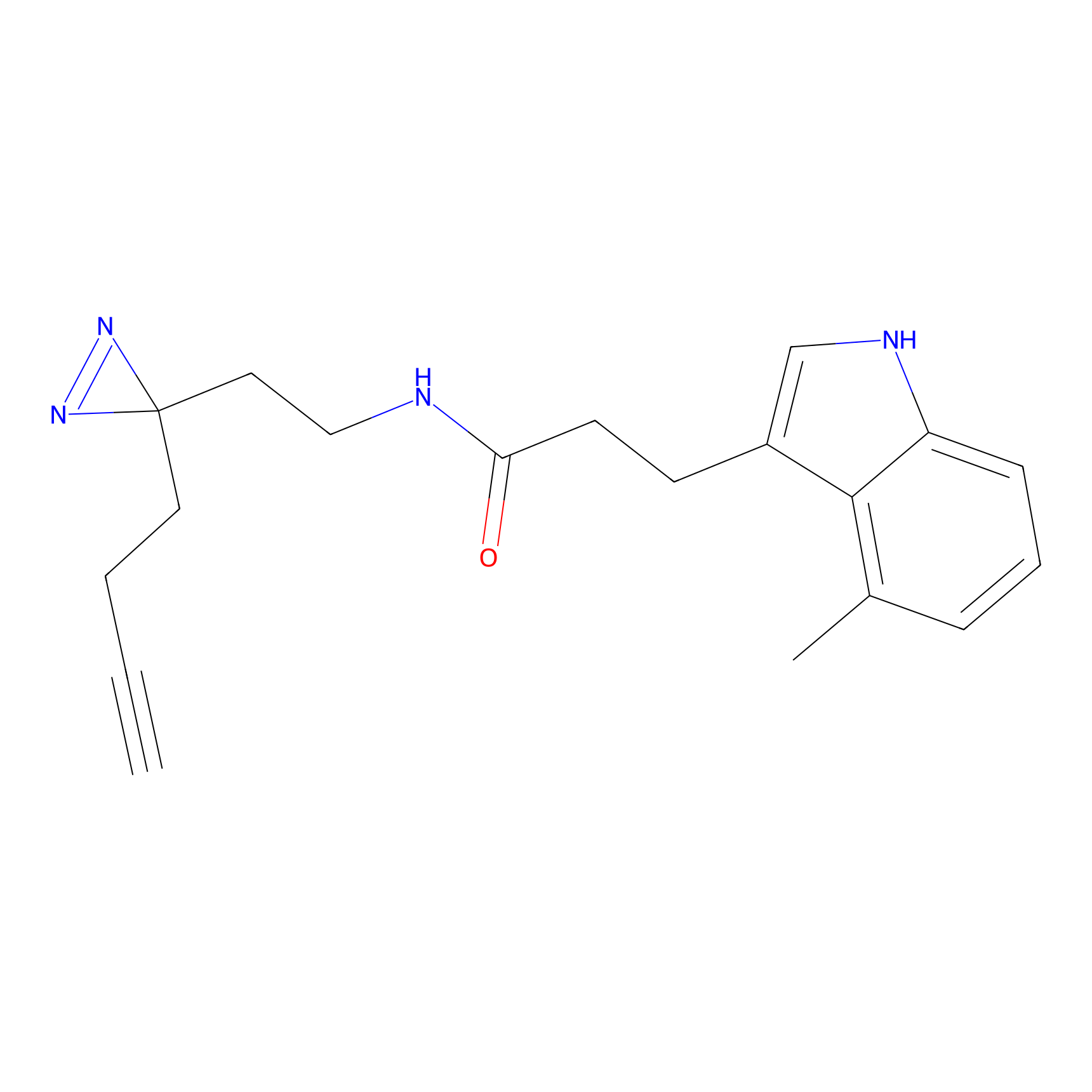
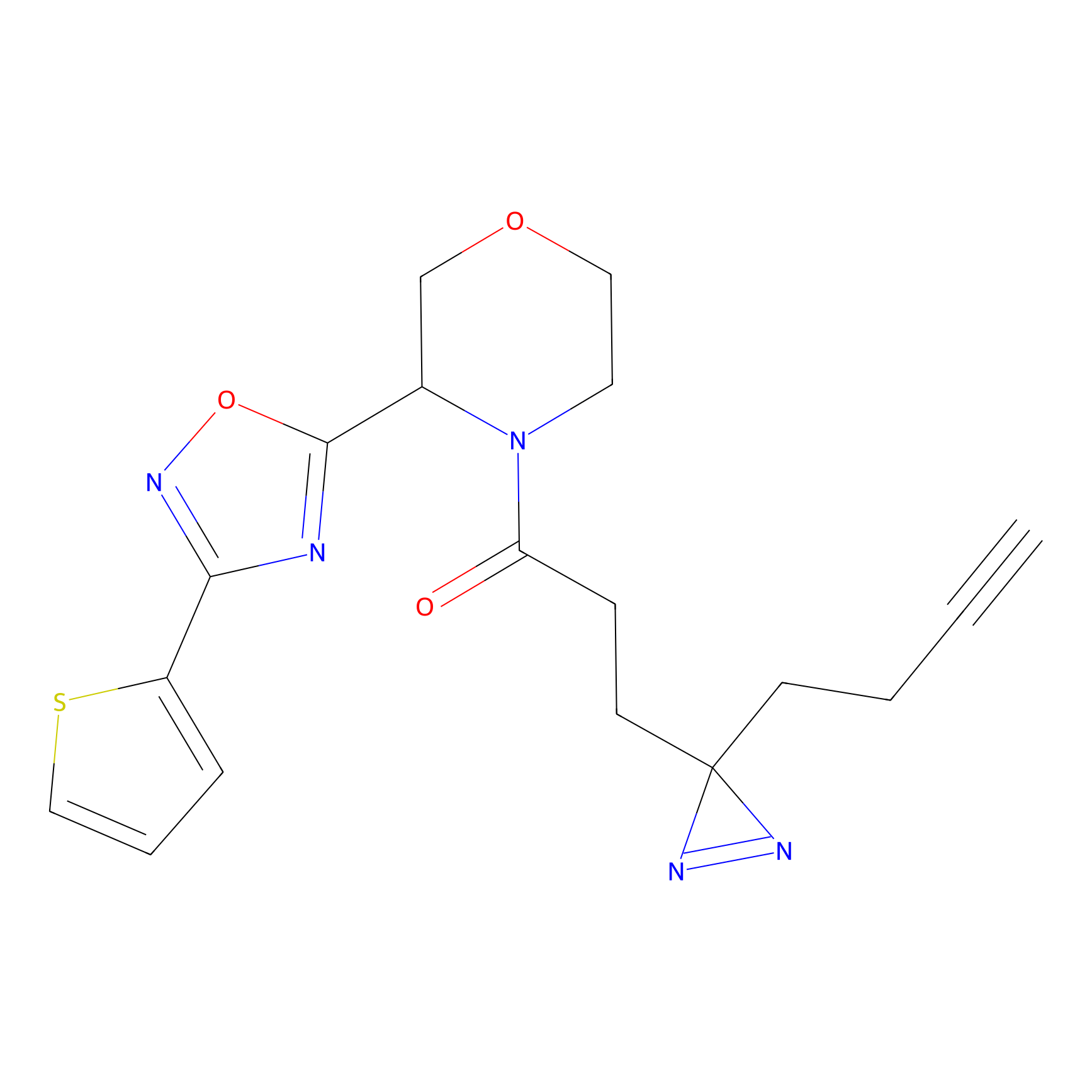
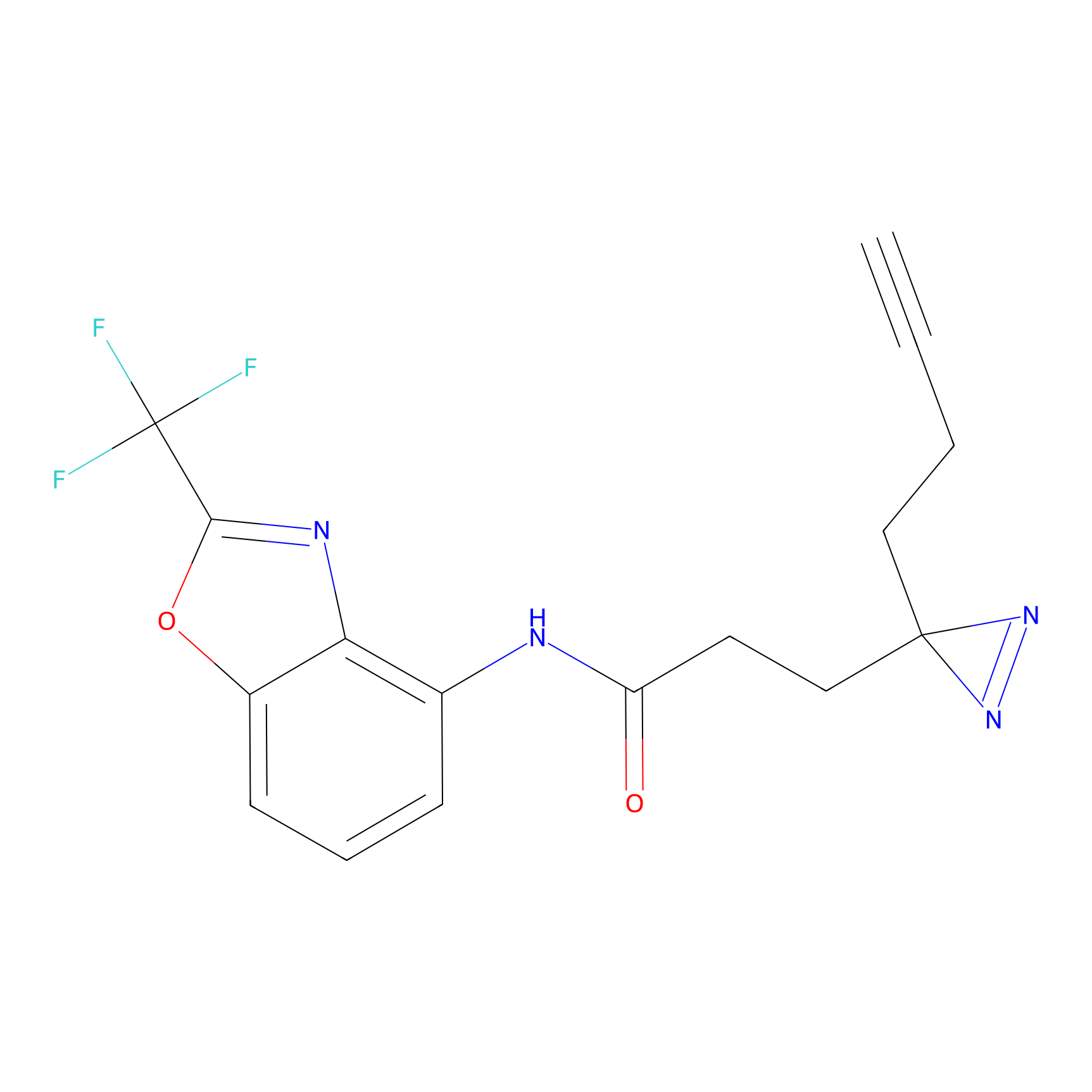
C203(0.78) | LDD2099 | [2] | |
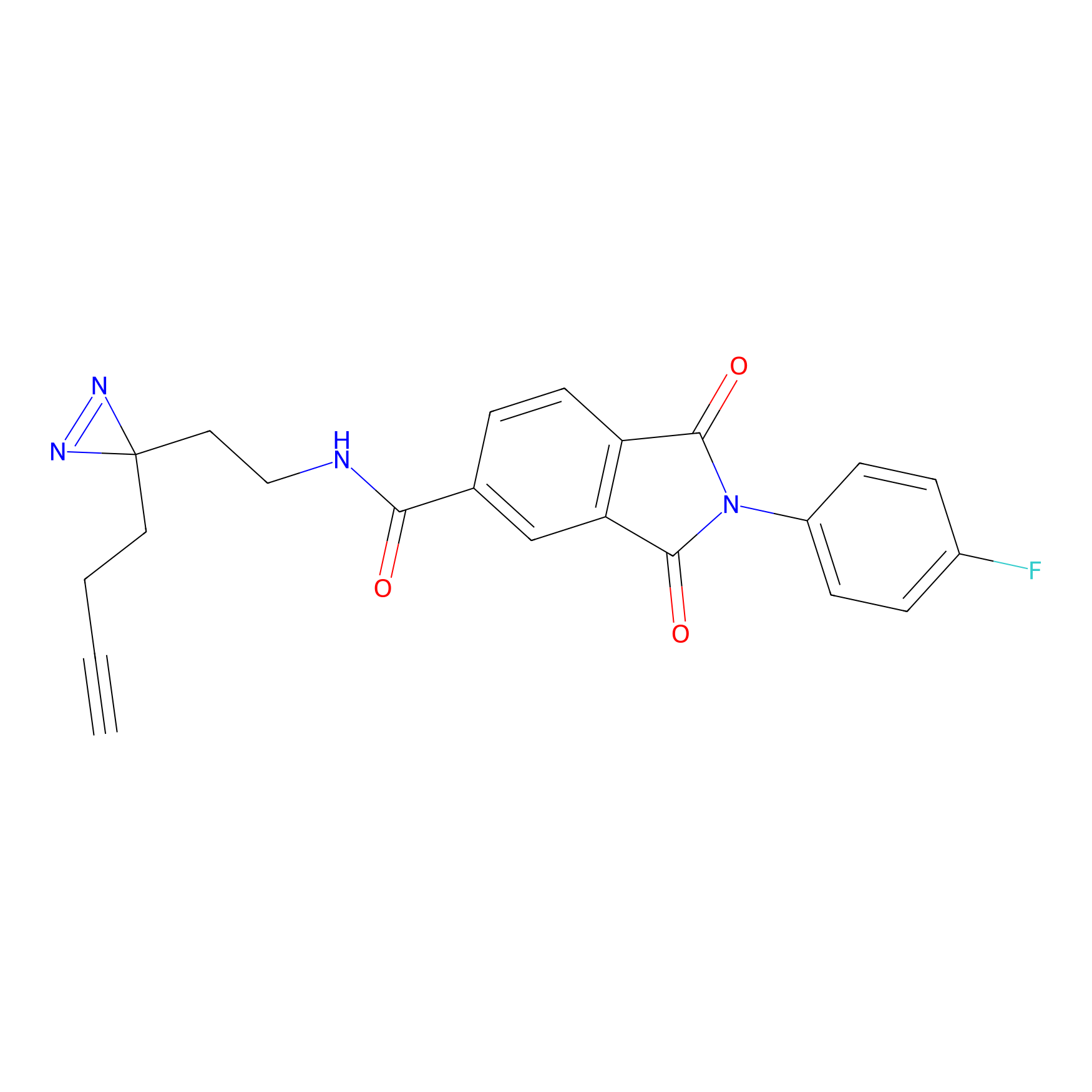
|
IA-alkyne Probe Info |
 |
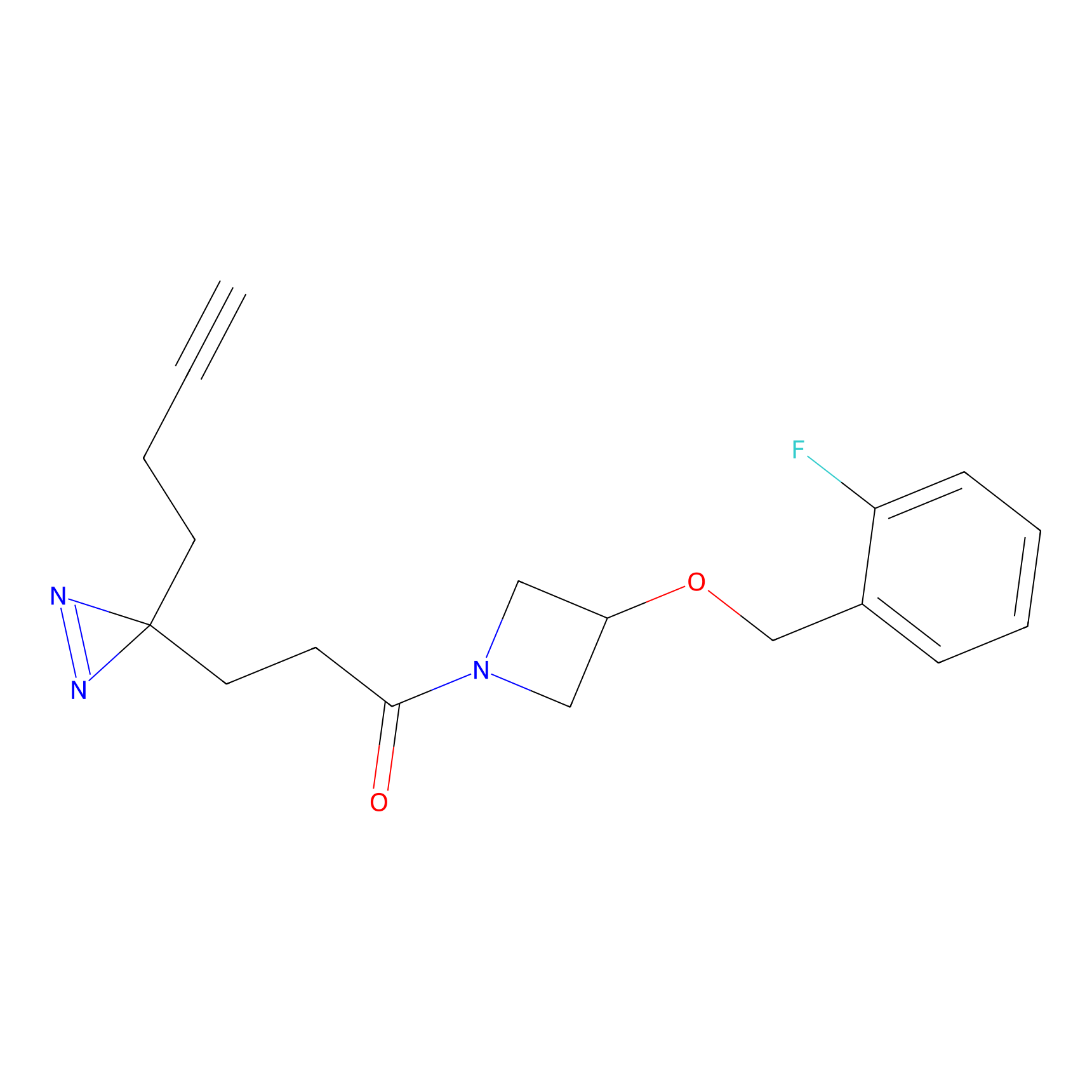
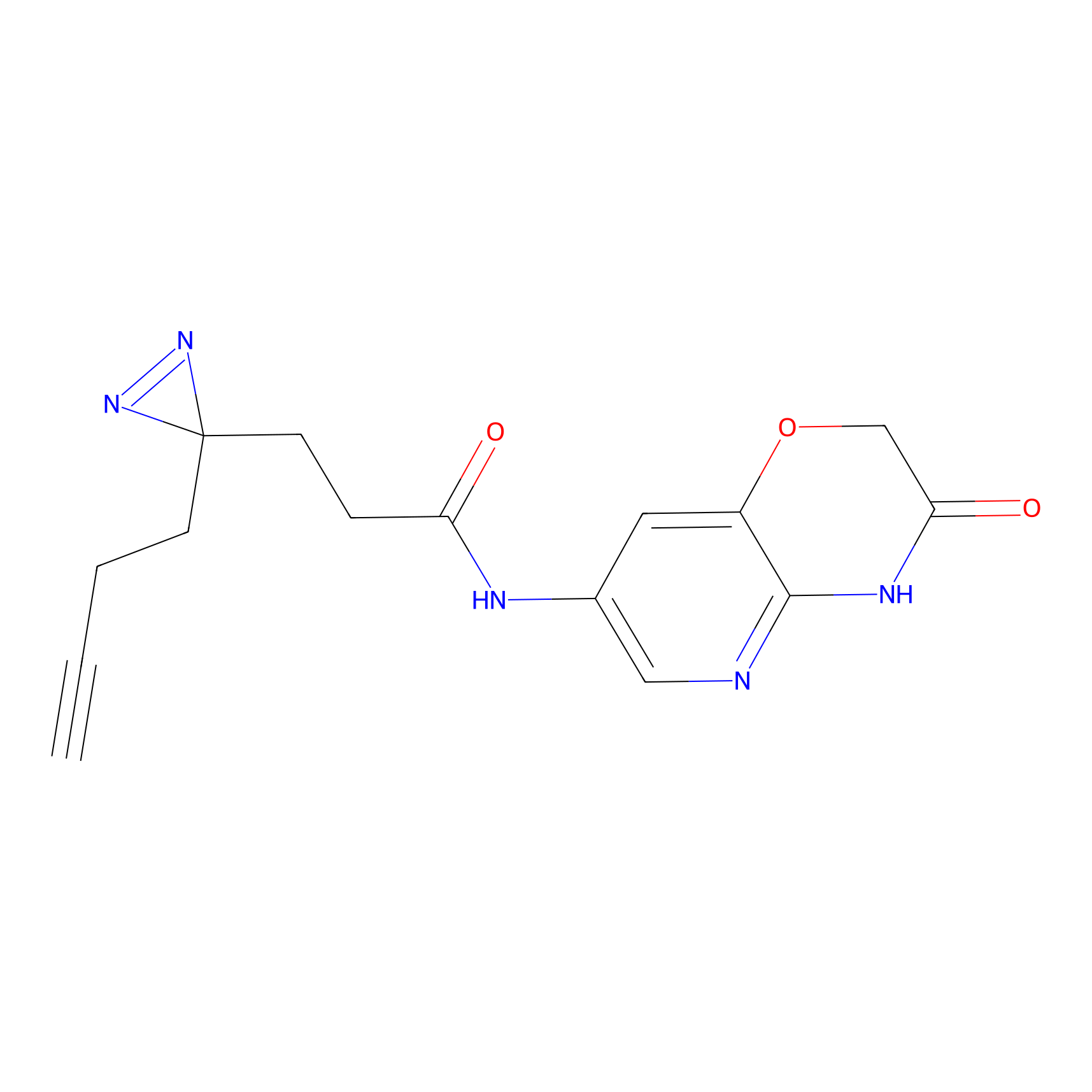
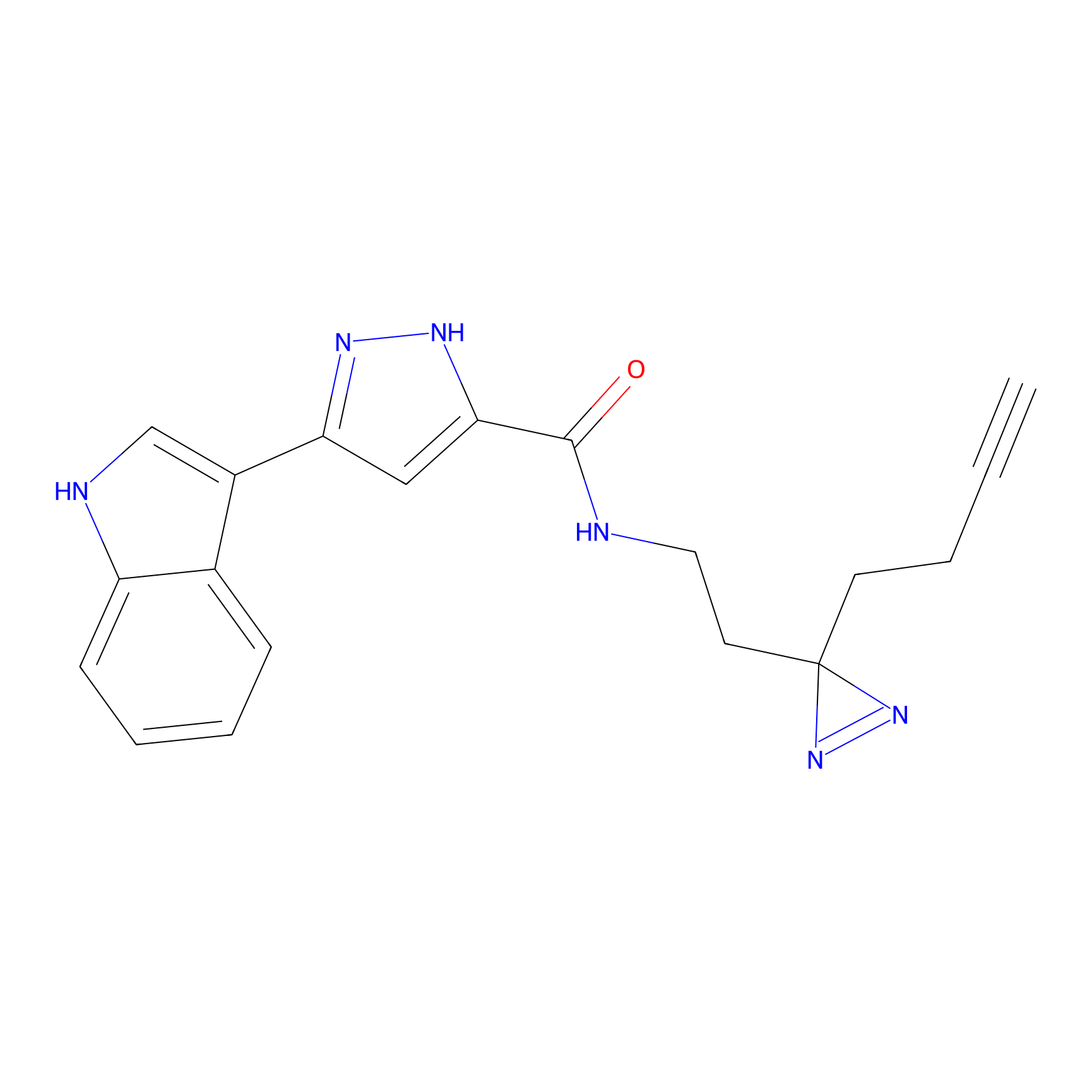
N.A. | LDD0036 | [3] | |
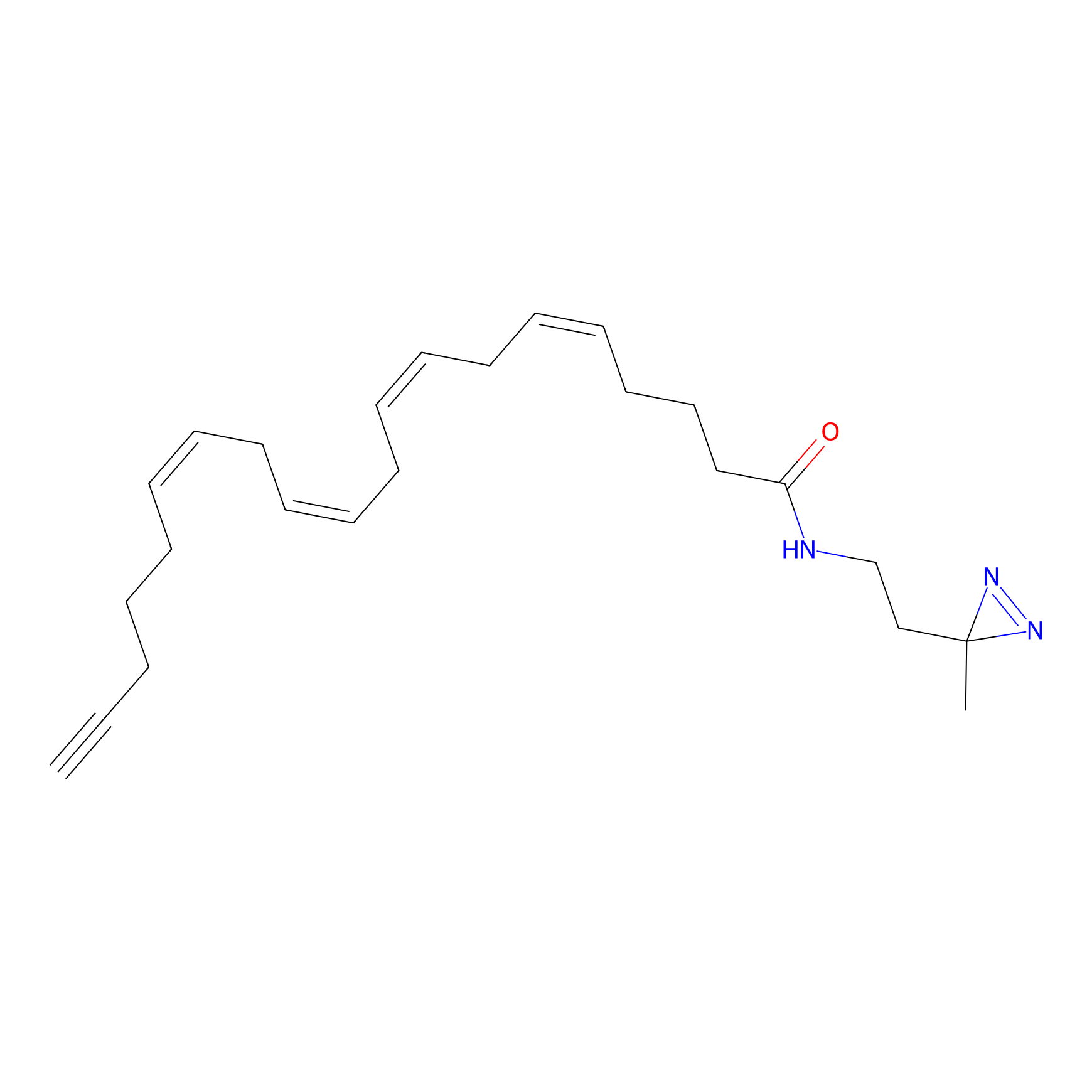
|
Lodoacetamide azide Probe Info |
 |
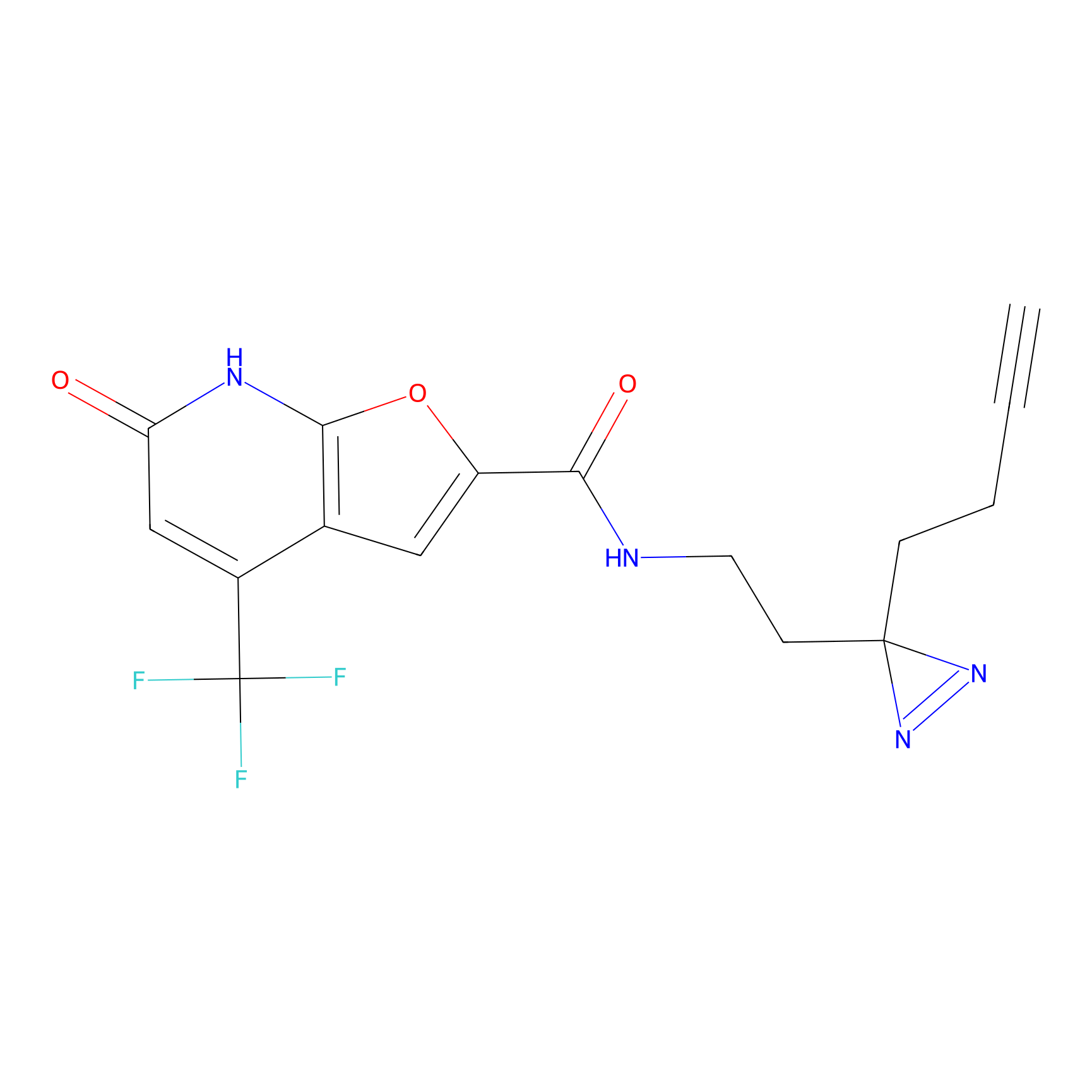
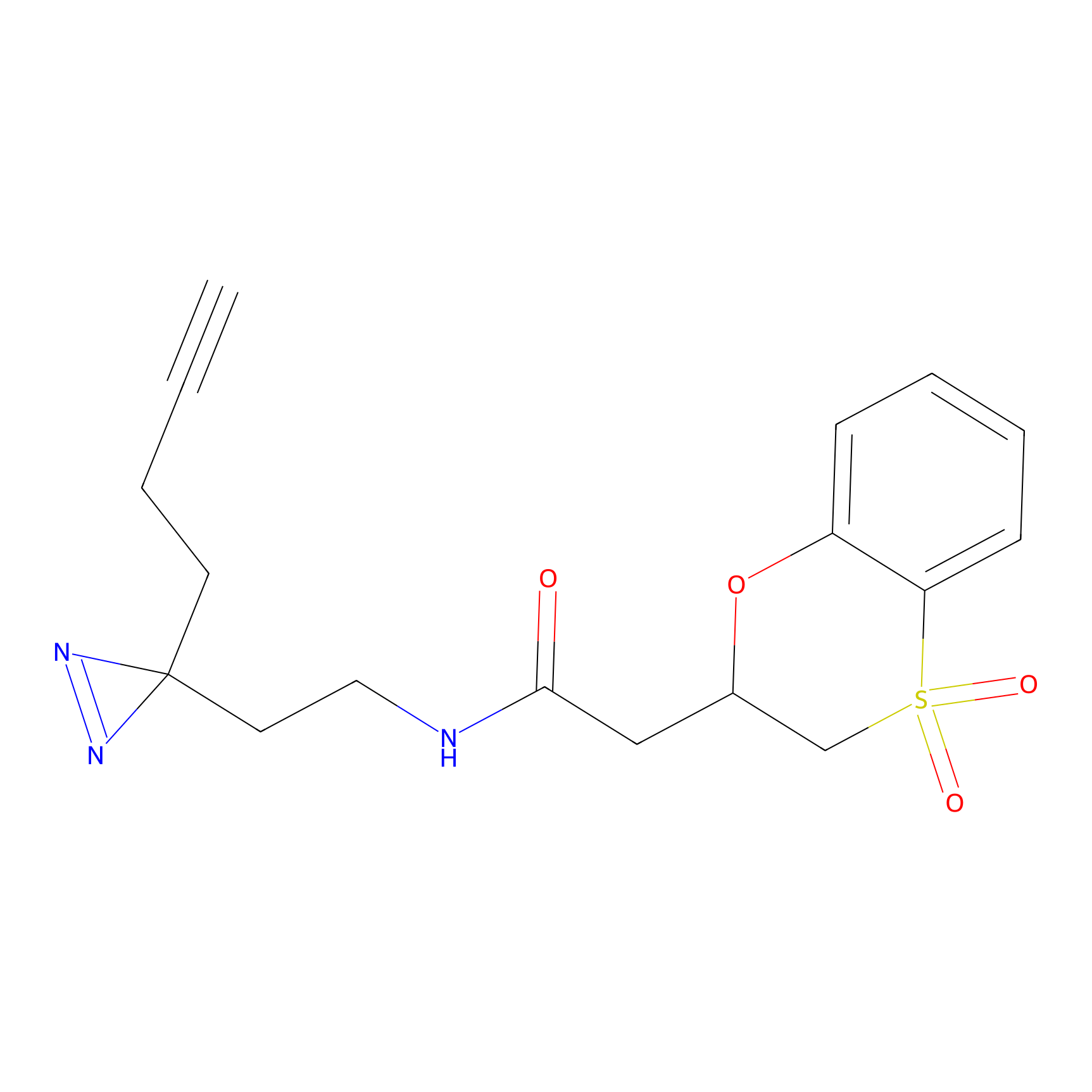
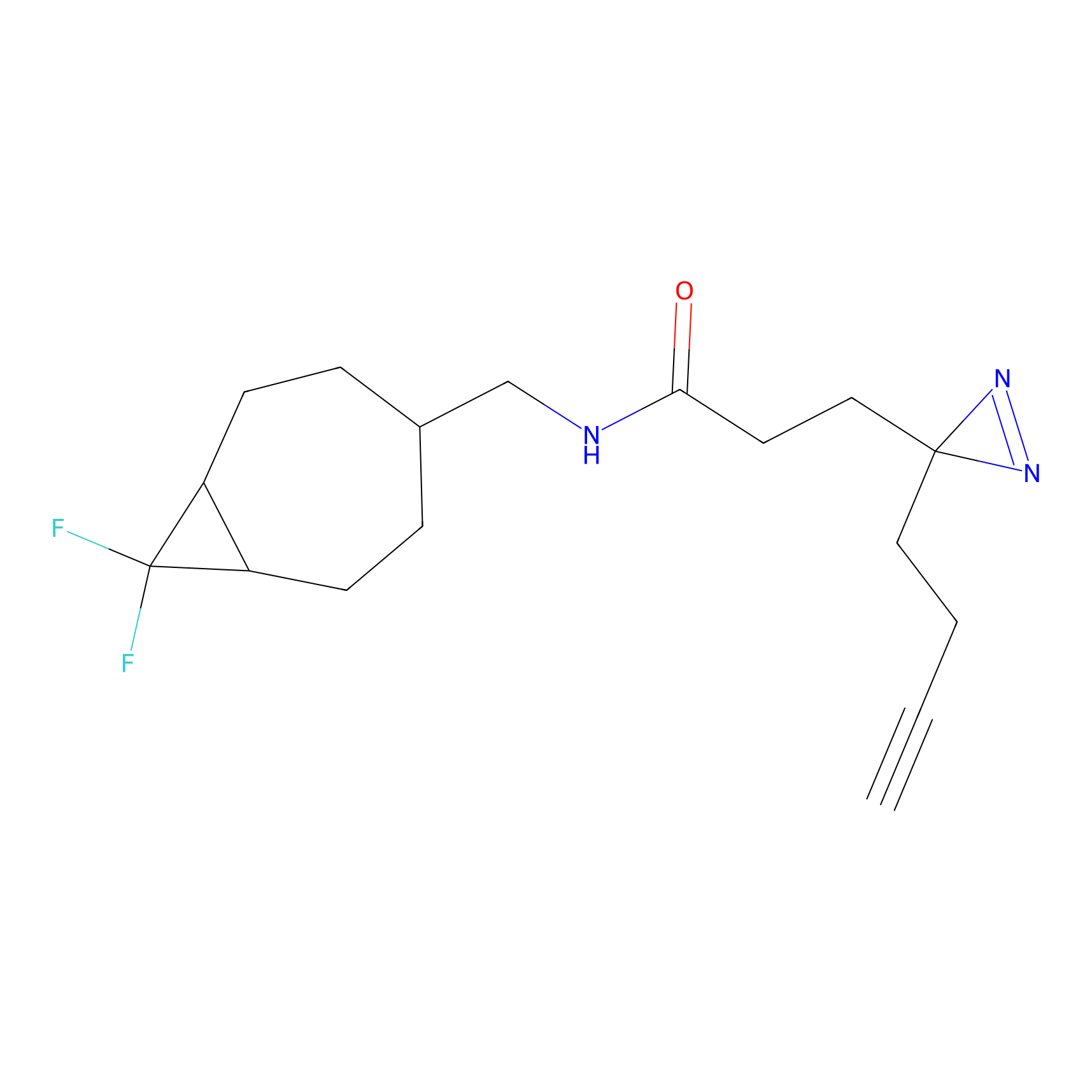
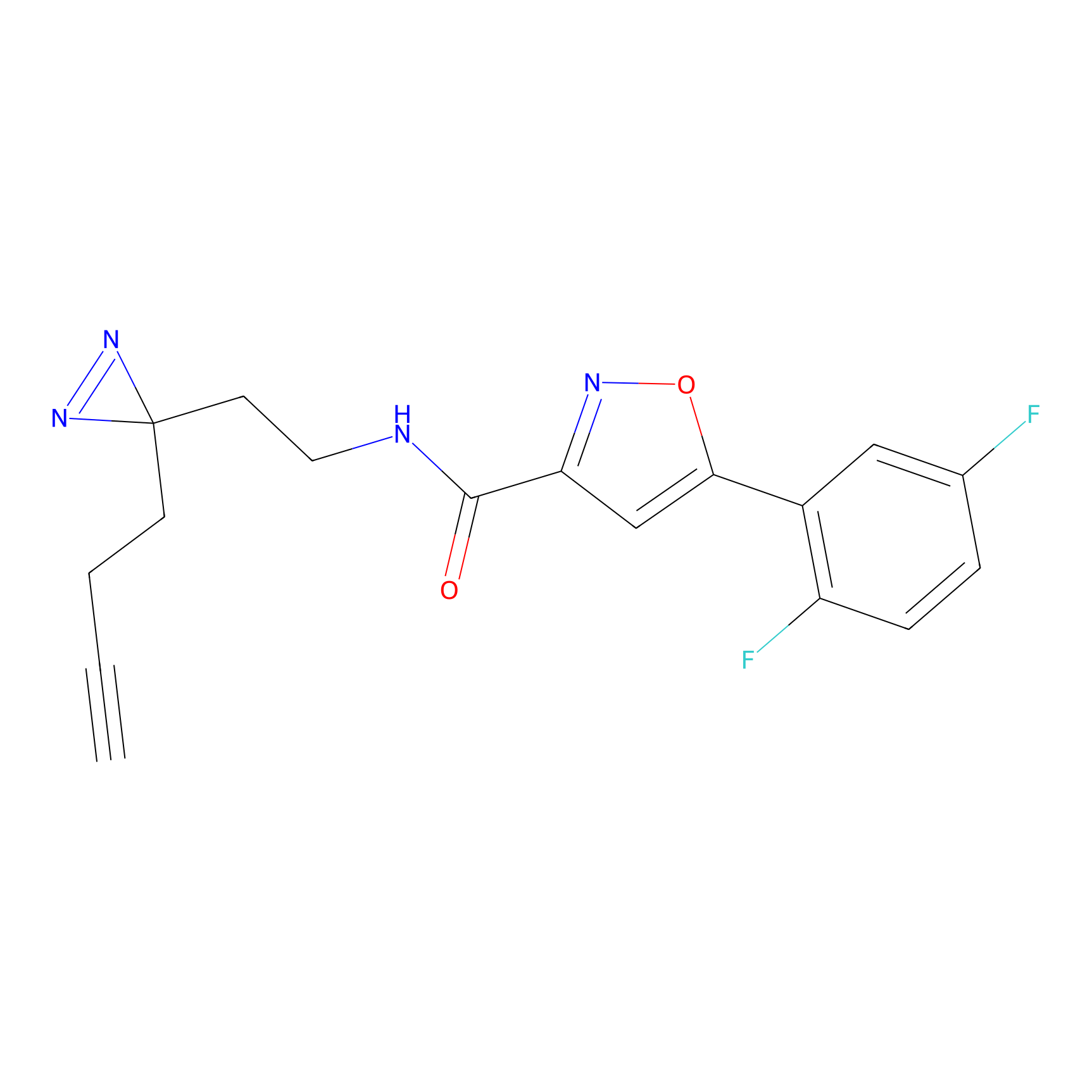
C236(0.00); C349(0.00) | LDD0037 | [3] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [4] | |
|
NAIA_5 Probe Info |
 |
C130(0.00); C203(0.00) | LDD2223 | [5] | |
|
HHS-465 Probe Info |
 |
N.A. | LDD2240 | [6] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HEK-293T | C349(1.13) | LDD1507 | [10] |
| LDCM0259 | AC14 | HEK-293T | C349(0.93) | LDD1512 | [10] |
| LDCM0276 | AC17 | HEK-293T | C349(1.45) | LDD1515 | [10] |
| LDCM0281 | AC21 | HEK-293T | C349(0.92) | LDD1520 | [10] |
| LDCM0282 | AC22 | HEK-293T | C349(0.94) | LDD1521 | [10] |
| LDCM0285 | AC25 | HEK-293T | C349(1.33) | LDD1524 | [10] |
| LDCM0289 | AC29 | HEK-293T | C349(1.00) | LDD1528 | [10] |
| LDCM0291 | AC30 | HEK-293T | C349(1.05) | LDD1530 | [10] |
| LDCM0294 | AC33 | HEK-293T | C349(1.34) | LDD1533 | [10] |
| LDCM0298 | AC37 | HEK-293T | C349(1.04) | LDD1537 | [10] |
| LDCM0299 | AC38 | HEK-293T | C349(1.01) | LDD1538 | [10] |
| LDCM0303 | AC41 | HEK-293T | C349(1.06) | LDD1542 | [10] |
| LDCM0307 | AC45 | HEK-293T | C349(0.93) | LDD1546 | [10] |
| LDCM0308 | AC46 | HEK-293T | C349(0.97) | LDD1547 | [10] |
| LDCM0311 | AC49 | HEK-293T | C349(1.20) | LDD1550 | [10] |
| LDCM0312 | AC5 | HEK-293T | C349(0.92) | LDD1551 | [10] |
| LDCM0316 | AC53 | HEK-293T | C349(0.93) | LDD1555 | [10] |
| LDCM0317 | AC54 | HEK-293T | C349(1.02) | LDD1556 | [10] |
| LDCM0320 | AC57 | HEK-293T | C349(1.14) | LDD1559 | [10] |
| LDCM0323 | AC6 | HEK-293T | C349(0.94) | LDD1562 | [10] |
| LDCM0325 | AC61 | HEK-293T | C349(0.96) | LDD1564 | [10] |
| LDCM0326 | AC62 | HEK-293T | C349(1.05) | LDD1565 | [10] |
| LDCM0248 | AKOS034007472 | HEK-293T | C349(1.00) | LDD1511 | [10] |
| LDCM0356 | AKOS034007680 | HEK-293T | C349(1.33) | LDD1570 | [10] |
| LDCM0083 | Avasimibe | A-549 | 7.68 | LDD0143 | [9] |
| LDCM0368 | CL10 | HEK-293T | C349(0.81) | LDD1572 | [10] |
| LDCM0404 | CL17 | HEK-293T | C349(1.04) | LDD1608 | [10] |
| LDCM0409 | CL21 | HEK-293T | C349(0.85) | LDD1613 | [10] |
| LDCM0410 | CL22 | HEK-293T | C349(1.08) | LDD1614 | [10] |
| LDCM0417 | CL29 | HEK-293T | C349(1.14) | LDD1621 | [10] |
| LDCM0422 | CL33 | HEK-293T | C349(0.76) | LDD1626 | [10] |
| LDCM0423 | CL34 | HEK-293T | C349(0.85) | LDD1627 | [10] |
| LDCM0431 | CL41 | HEK-293T | C349(1.15) | LDD1635 | [10] |
| LDCM0435 | CL45 | HEK-293T | C349(0.91) | LDD1639 | [10] |
| LDCM0436 | CL46 | HEK-293T | C349(0.96) | LDD1640 | [10] |
| LDCM0440 | CL5 | HEK-293T | C349(1.14) | LDD1644 | [10] |
| LDCM0444 | CL53 | HEK-293T | C349(1.07) | LDD1647 | [10] |
| LDCM0448 | CL57 | HEK-293T | C349(0.82) | LDD1651 | [10] |
| LDCM0449 | CL58 | HEK-293T | C349(1.02) | LDD1652 | [10] |
| LDCM0457 | CL65 | HEK-293T | C349(1.16) | LDD1660 | [10] |
| LDCM0461 | CL69 | HEK-293T | C349(1.01) | LDD1664 | [10] |
| LDCM0463 | CL70 | HEK-293T | C349(0.94) | LDD1666 | [10] |
| LDCM0470 | CL77 | HEK-293T | C349(0.93) | LDD1673 | [10] |
| LDCM0475 | CL81 | HEK-293T | C349(0.92) | LDD1678 | [10] |
| LDCM0476 | CL82 | HEK-293T | C349(0.97) | LDD1679 | [10] |
| LDCM0483 | CL89 | HEK-293T | C349(1.22) | LDD1686 | [10] |
| LDCM0484 | CL9 | HEK-293T | C349(0.98) | LDD1687 | [10] |
| LDCM0488 | CL93 | HEK-293T | C349(0.89) | LDD1691 | [10] |
| LDCM0489 | CL94 | HEK-293T | C349(0.98) | LDD1692 | [10] |
| LDCM0185 | Compound 17 | HEK-293T | 4.26 | LDD0512 | [8] |
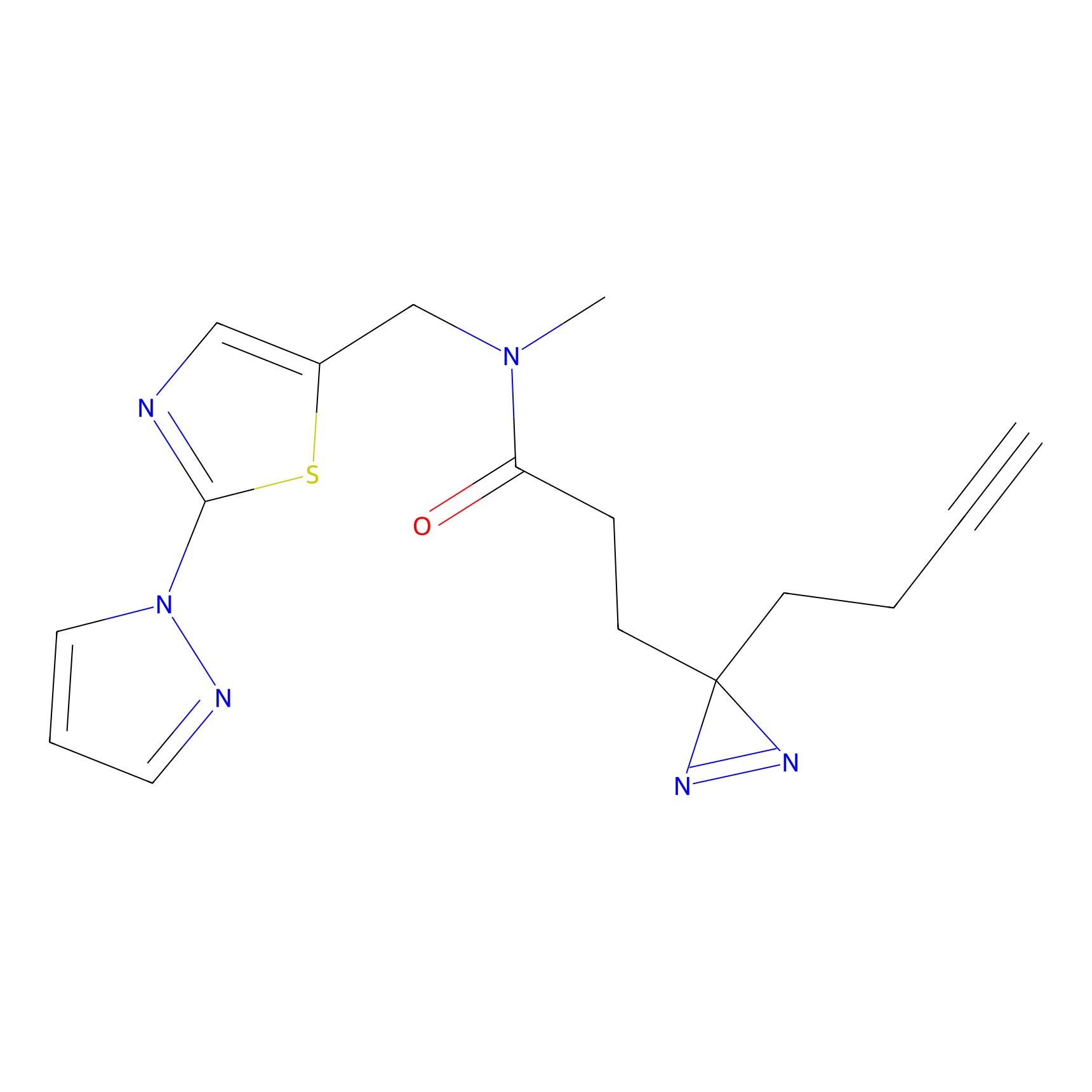
| LDCM0625 | F8 | Ramos | C203(4.67) | LDD2187 | [11] |
| LDCM0082 | FK866 | A-549 | 2.14 | LDD0142 | [9] |
| LDCM0572 | Fragment10 | Ramos | C203(1.66) | LDD2189 | [11] |
| LDCM0573 | Fragment11 | Ramos | C203(6.23) | LDD2190 | [11] |
| LDCM0574 | Fragment12 | Ramos | C203(2.17) | LDD2191 | [11] |
| LDCM0579 | Fragment20 | Ramos | C203(1.57) | LDD2194 | [11] |
| LDCM0580 | Fragment21 | Ramos | C203(0.91) | LDD2195 | [11] |
| LDCM0586 | Fragment28 | Ramos | C203(0.61) | LDD2198 | [11] |
| LDCM0588 | Fragment30 | Ramos | C203(1.81) | LDD2199 | [11] |
| LDCM0590 | Fragment32 | Ramos | C203(1.72) | LDD2201 | [11] |
| LDCM0610 | Fragment52 | Ramos | C203(1.42) | LDD2204 | [11] |
| LDCM0614 | Fragment56 | Ramos | C203(2.83) | LDD2205 | [11] |
| LDCM0569 | Fragment7 | Ramos | C203(2.19) | LDD2186 | [11] |
| LDCM0571 | Fragment9 | Ramos | C203(1.60) | LDD2188 | [11] |
| LDCM0022 | KB02 | Ramos | C203(1.66) | LDD2182 | [11] |
| LDCM0023 | KB03 | Ramos | C203(2.27) | LDD2183 | [11] |
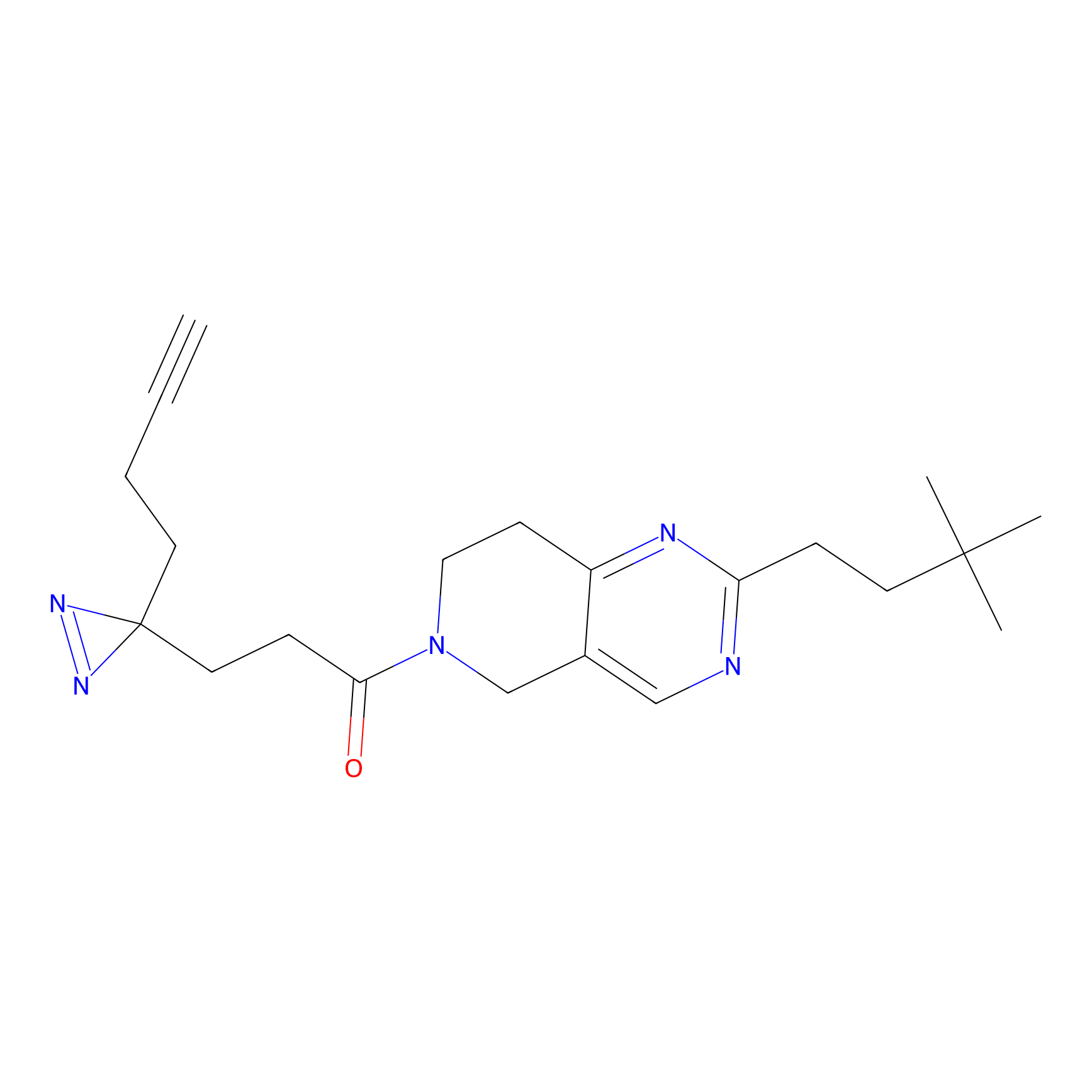
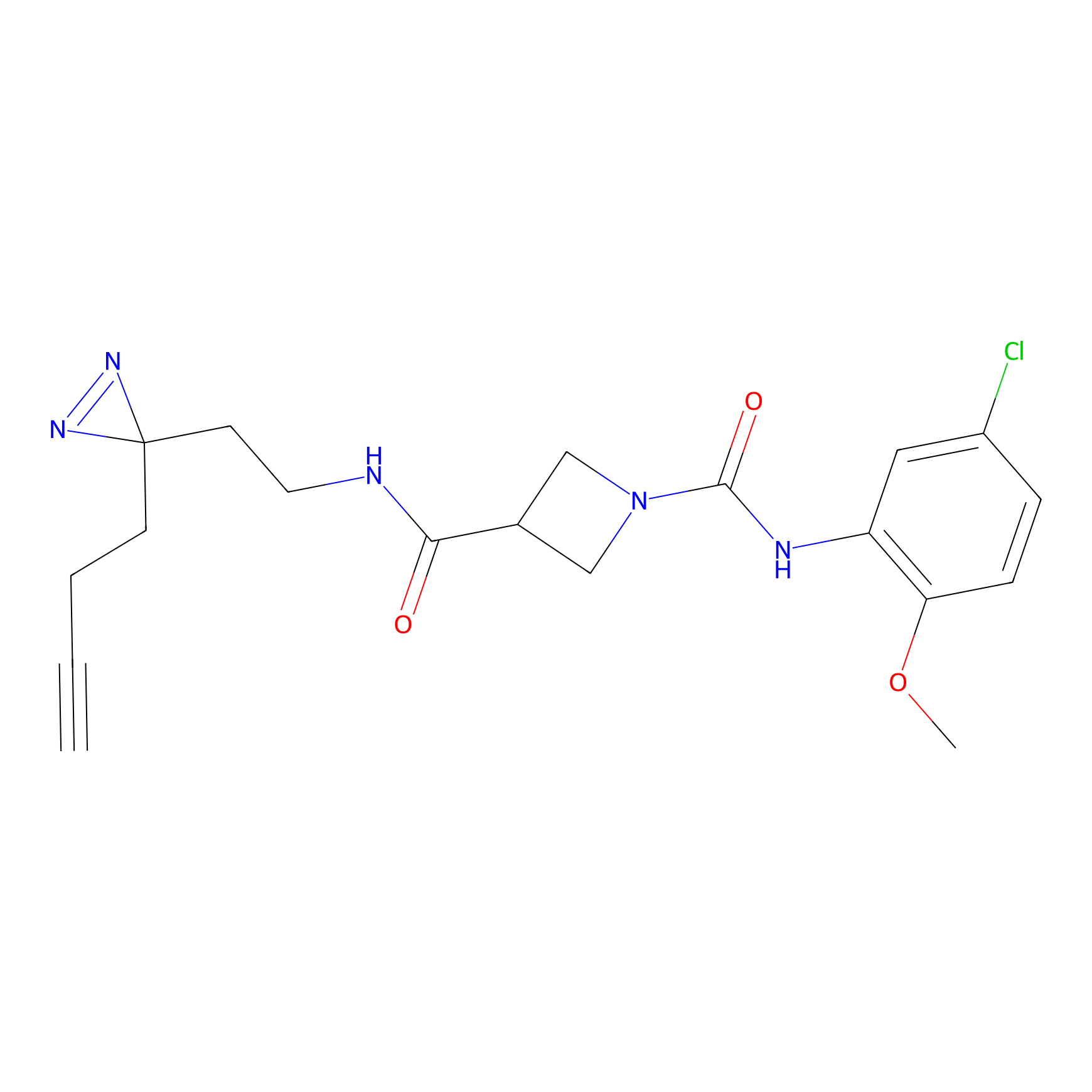
| LDCM0024 | KB05 | HMCB | C349(1.34) | LDD3312 | [1] |
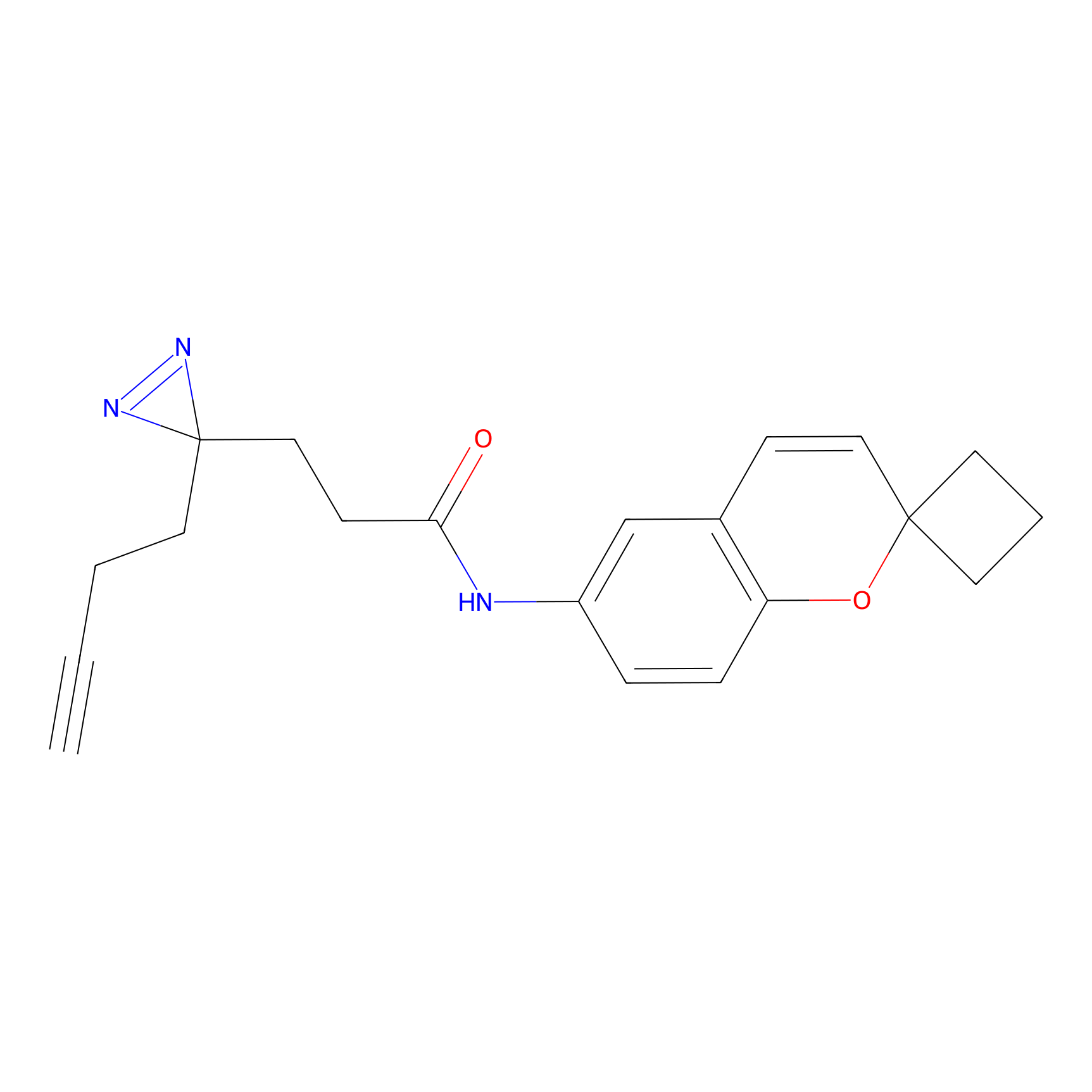
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C203(0.78) | LDD2099 | [2] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C203(0.72) | LDD2107 | [2] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C203(0.96) | LDD2146 | [2] |
| LDCM0084 | Ro 48-8071 | A-549 | 2.42 | LDD0145 | [9] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| E3 ubiquitin-protein ligase MARCHF5 (MARCHF5) | . | Q9NX47 | |||
| E3 ubiquitin-protein ligase SMURF2 (SMURF2) | . | Q9HAU4 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Splicing factor, arginine/serine-rich 19 (SCAF1) | Splicing factor SR family | Q9H7N4 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Highly divergent homeobox (HDX) | . | Q7Z353 | |||
Other
References