Details of the Target
General Information of Target
| Target ID | LDTP11245 | |||||
|---|---|---|---|---|---|---|
| Target Name | Derlin-1 (DERL1) | |||||
| Gene Name | DERL1 | |||||
| Gene ID | 79139 | |||||
| Synonyms |
DER1; Derlin-1; Degradation in endoplasmic reticulum protein 1; DERtrin-1; Der1-like protein 1 |
|||||
| 3D Structure | ||||||
| Sequence |
MDDKKKKRSPKPCLAQPAQAPGTLRRVPVPTSHSGSLALGLPHLPSPKQRAKFKRVGKEK
CRPVLAGGGSGSAGTPLQHSFLTEVTDVYEMEGGLLNLLNDFHSGRLQAFGKECSFEQLE HVREMQEKLARLHFSLDVCGEEEDDEEEEDGVTEGLPEEQKKTMADRNLDQLLSNLEDLS NSIQKLHLAENAEPEEQSAA |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
Derlin family
|
|||||
| Subcellular location |
Endoplasmic reticulum membrane
|
|||||
| Function |
Functional component of endoplasmic reticulum-associated degradation (ERAD) for misfolded lumenal proteins. Forms homotetramers which encircle a large channel traversing the endoplasmic reticulum (ER) membrane. This allows the retrotranslocation of misfolded proteins from the ER into the cytosol where they are ubiquitinated and degraded by the proteasome. The channel has a lateral gate within the membrane which provides direct access to membrane proteins with no need to reenter the ER lumen first. May mediate the interaction between VCP and the misfolded protein. Also involved in endoplasmic reticulum stress-induced pre-emptive quality control, a mechanism that selectively attenuates the translocation of newly synthesized proteins into the endoplasmic reticulum and reroutes them to the cytosol for proteasomal degradation. By controlling the steady-state expression of the IGF1R receptor, indirectly regulates the insulin-like growth factor receptor signaling pathway.; (Microbial infection) In case of infection by cytomegaloviruses, it plays a central role in the export from the ER and subsequent degradation of MHC class I heavy chains via its interaction with US11 viral protein, which recognizes and associates with MHC class I heavy chains. Also participates in the degradation process of misfolded cytomegalovirus US2 protein.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
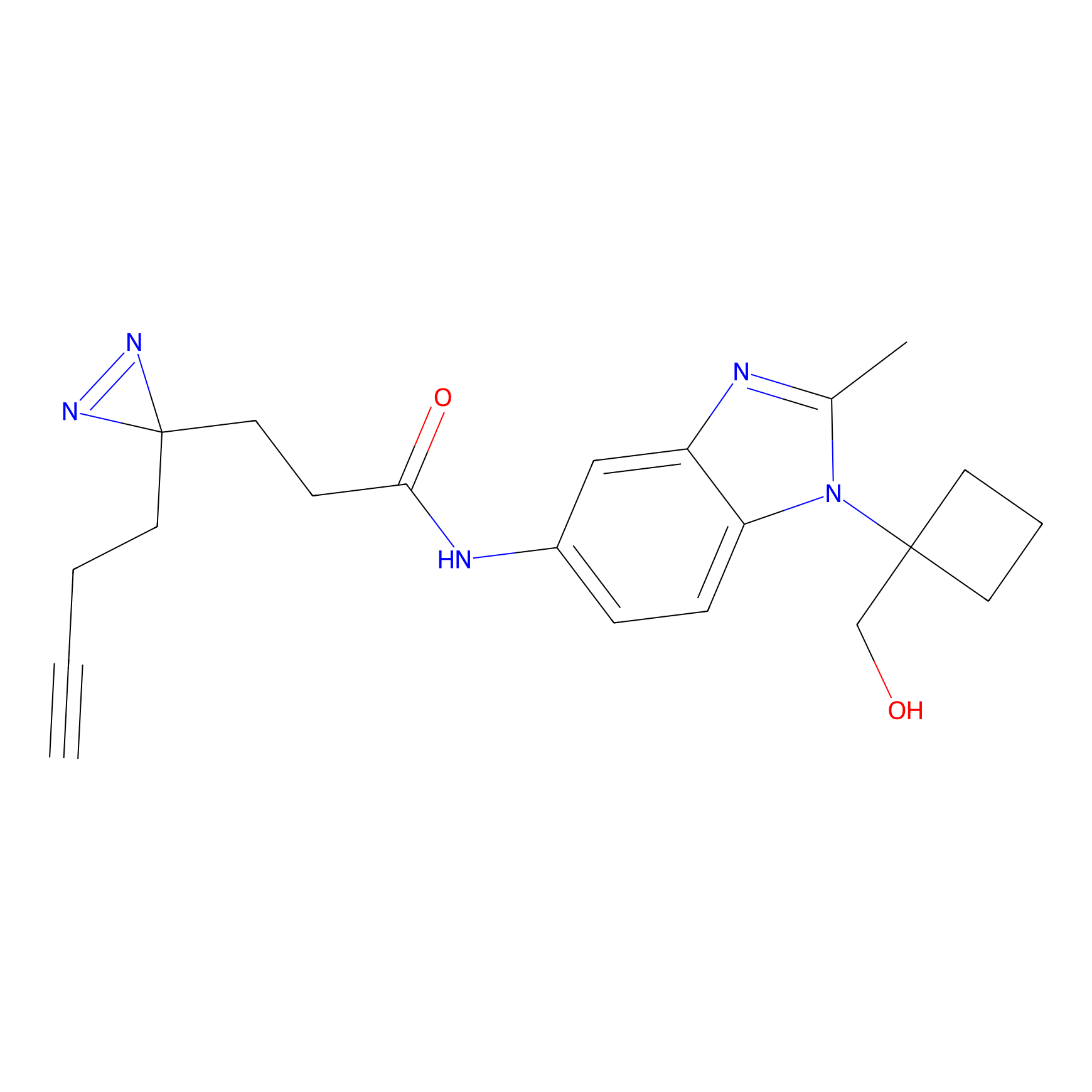
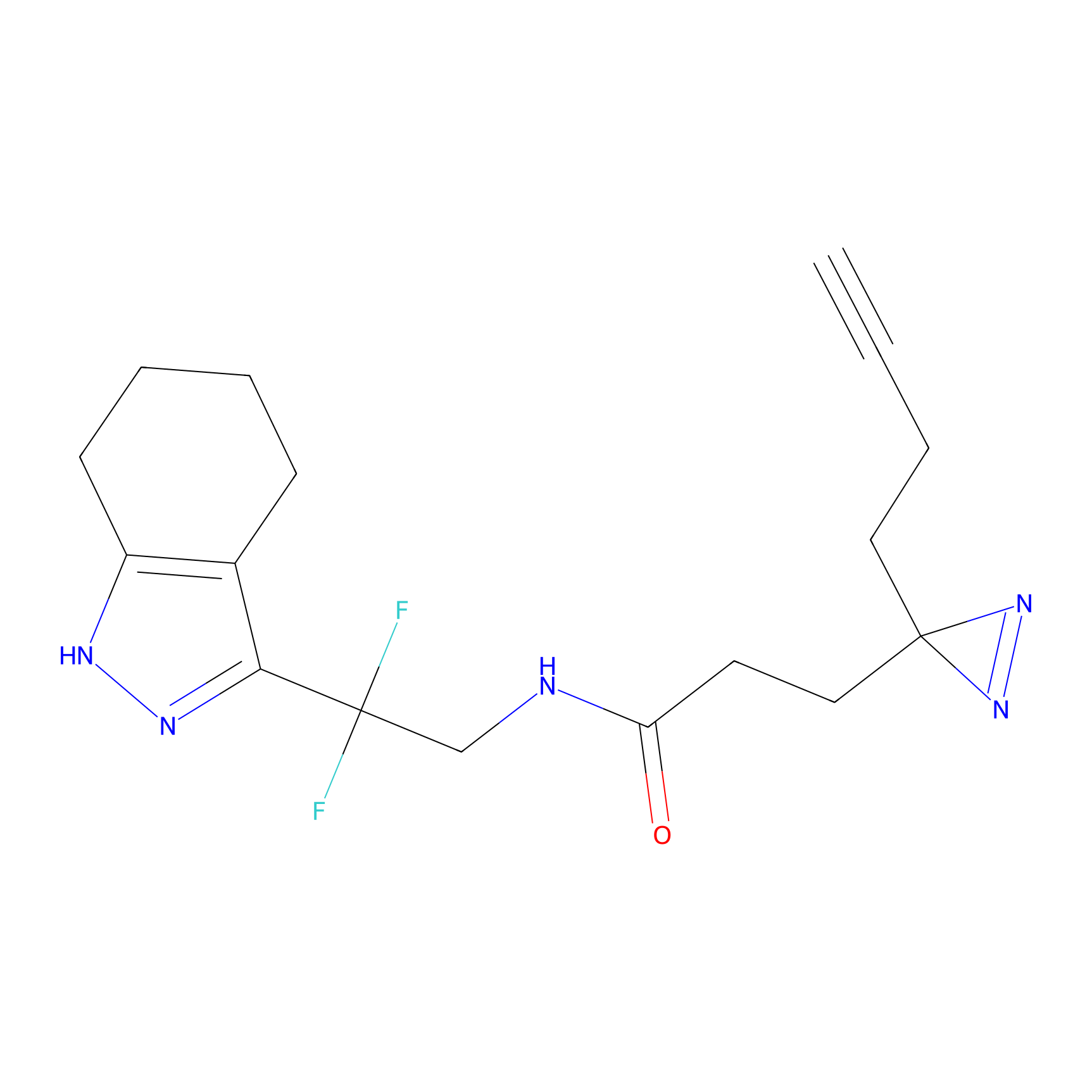
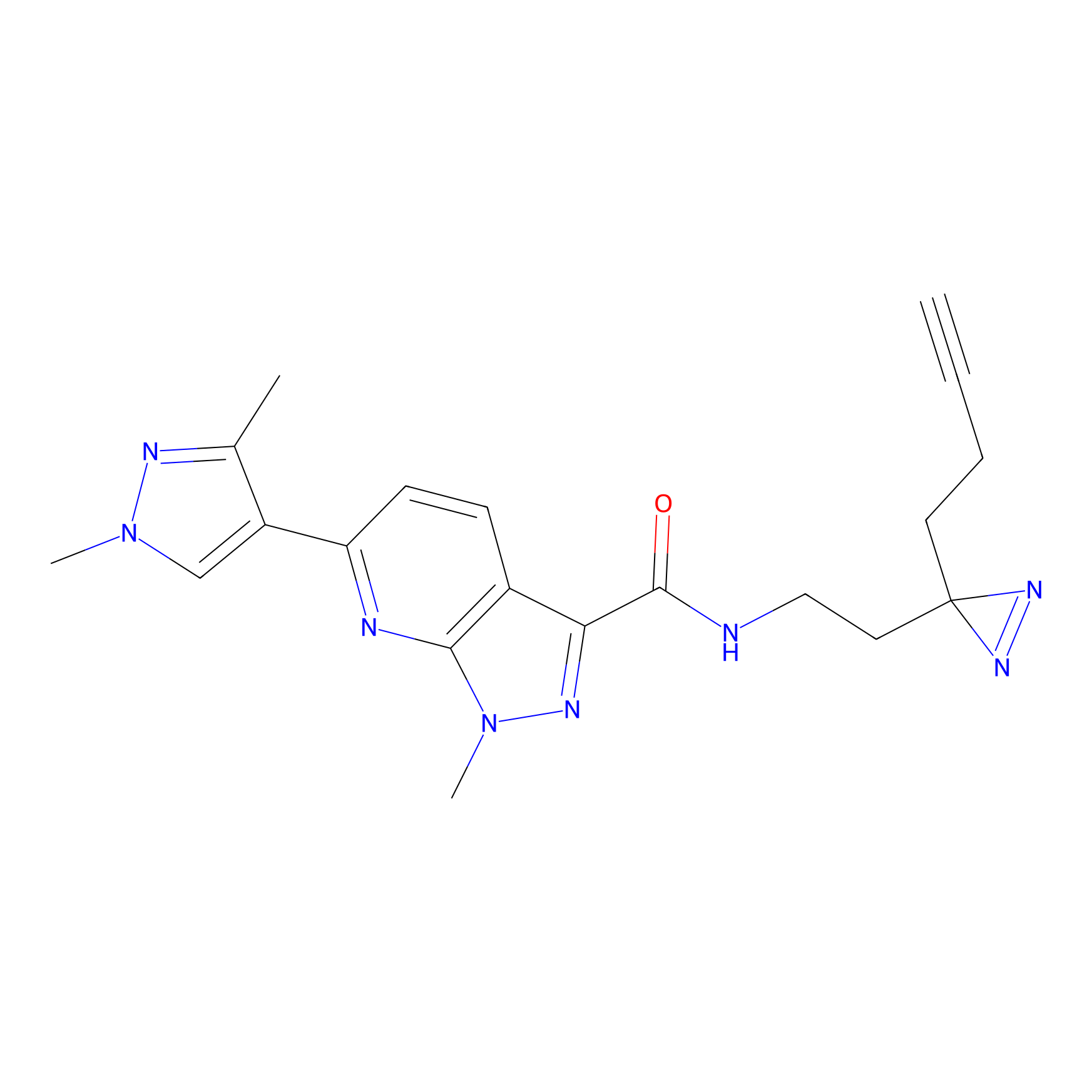
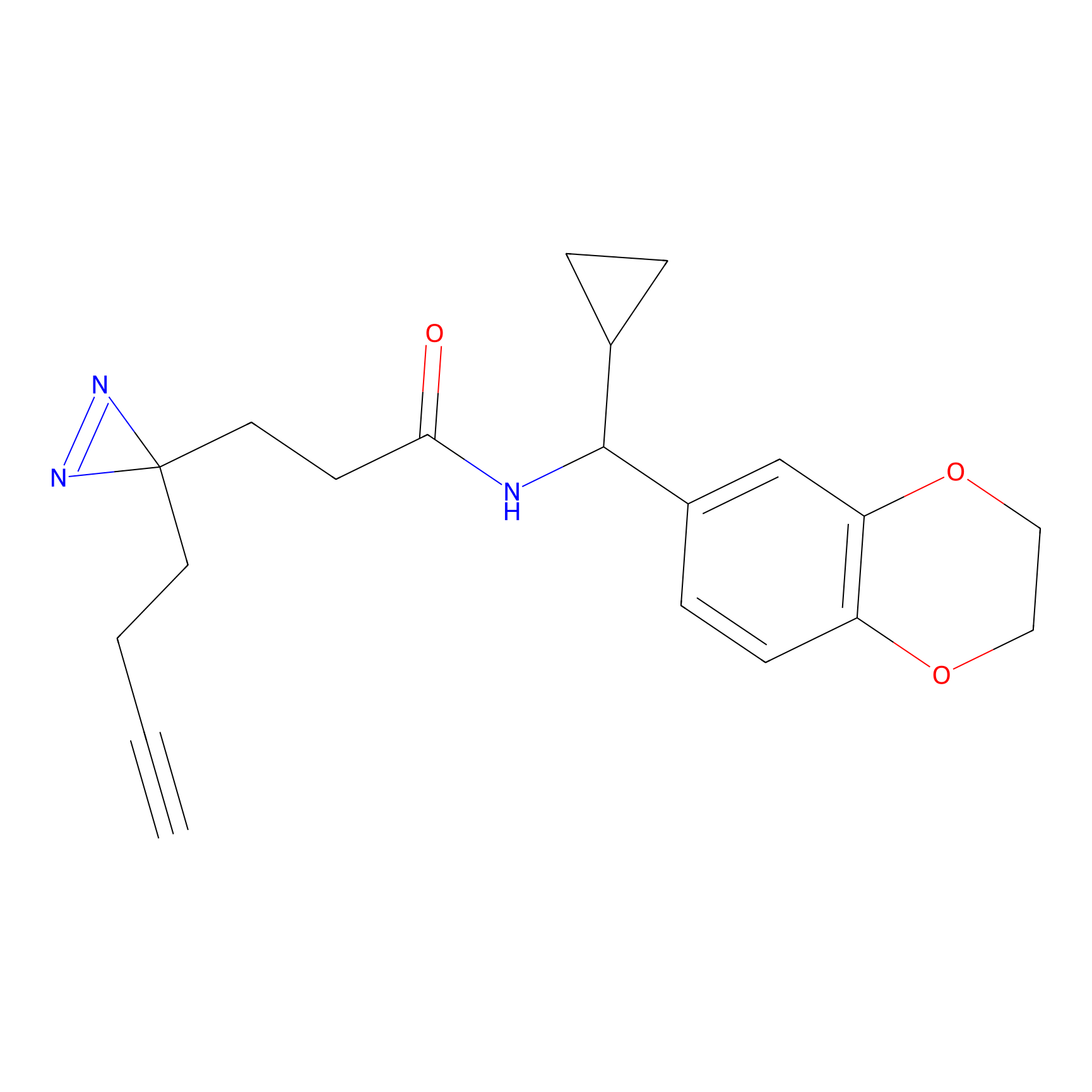
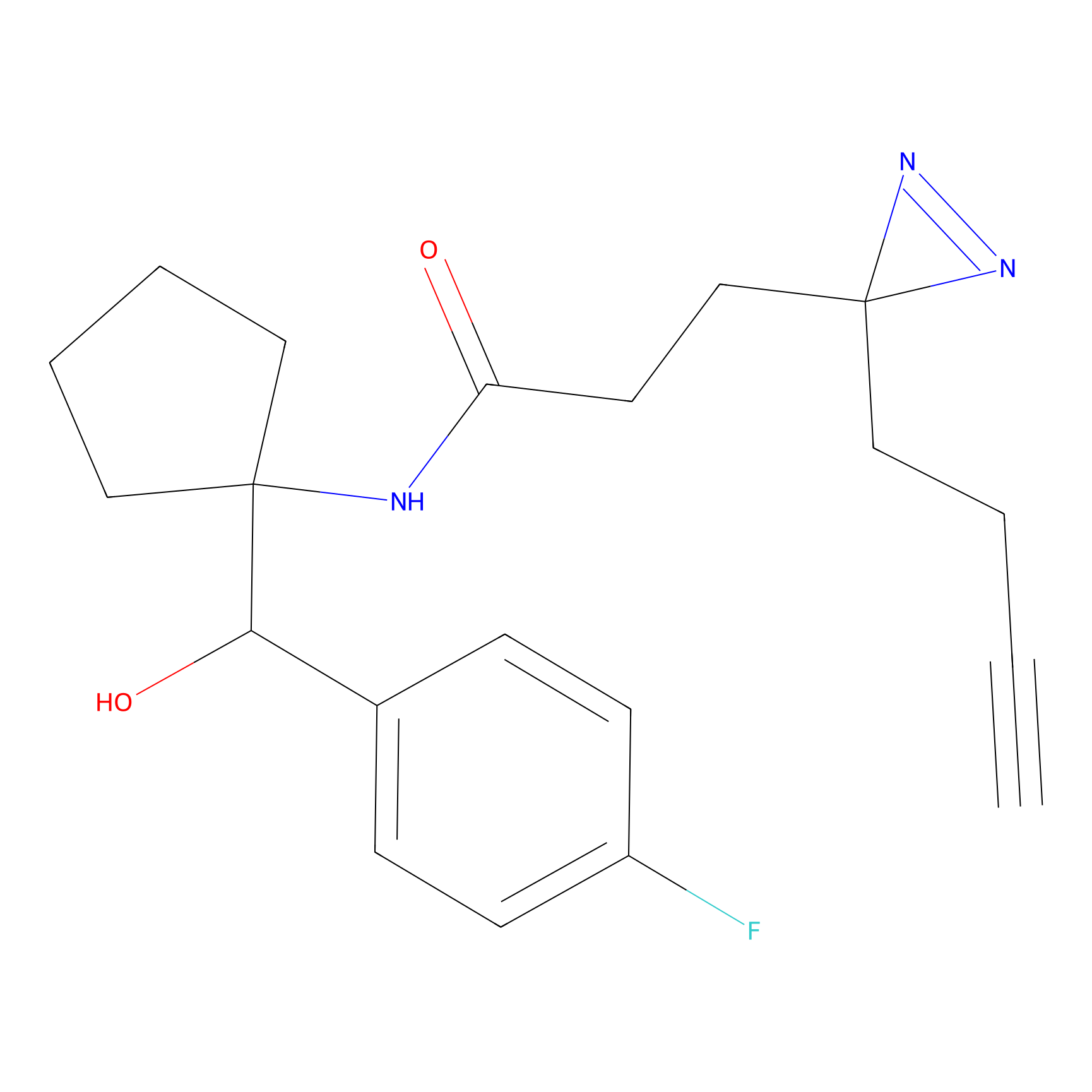
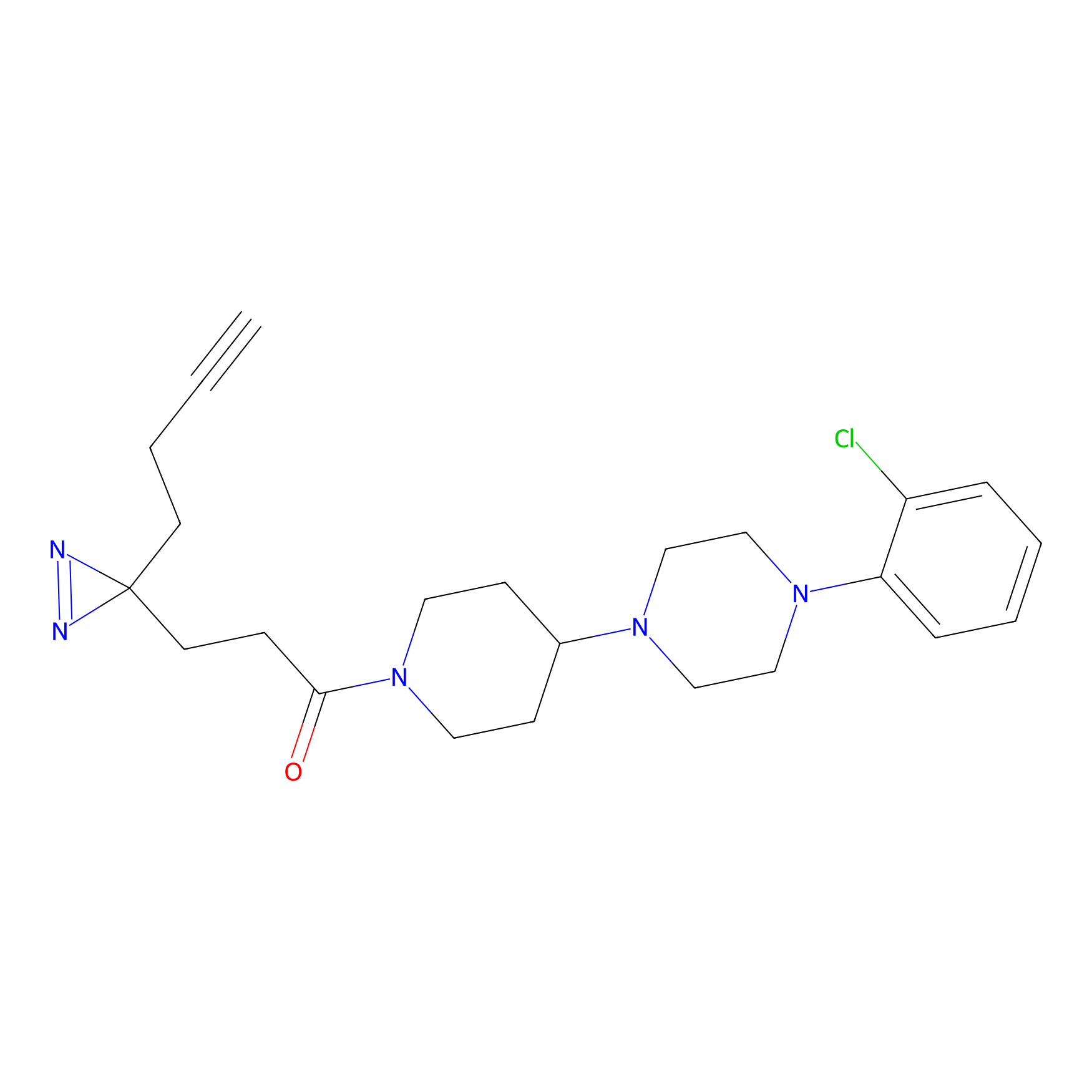
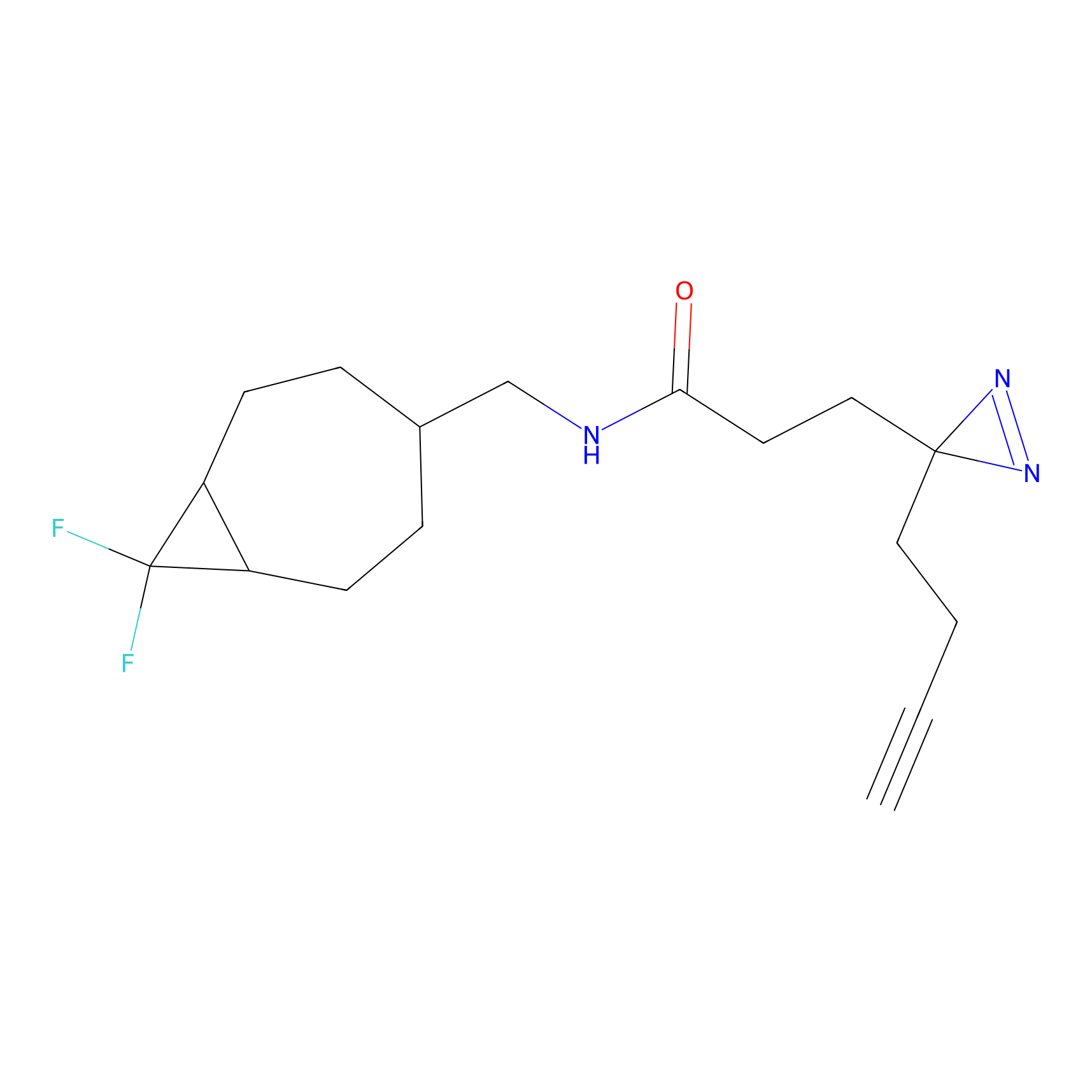
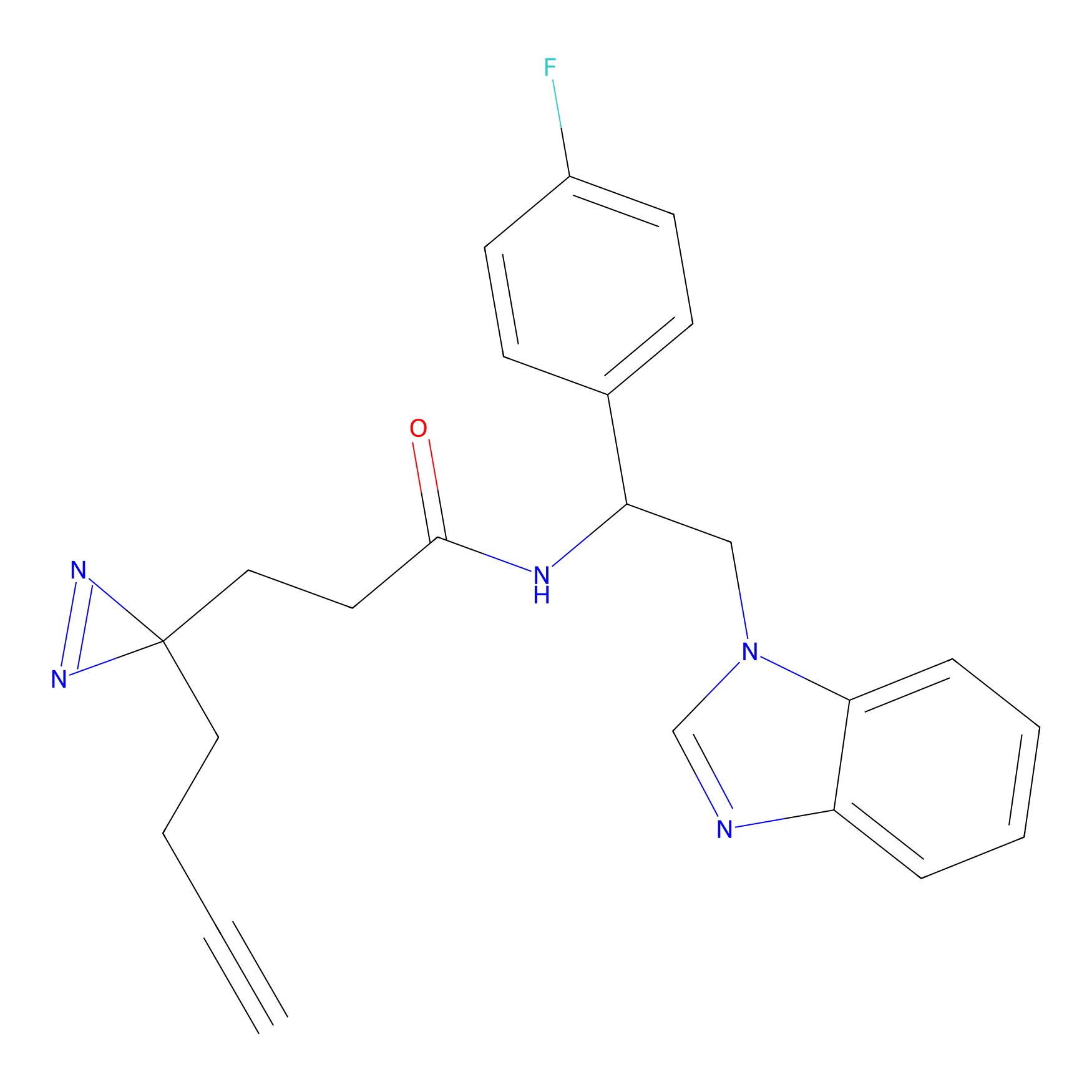
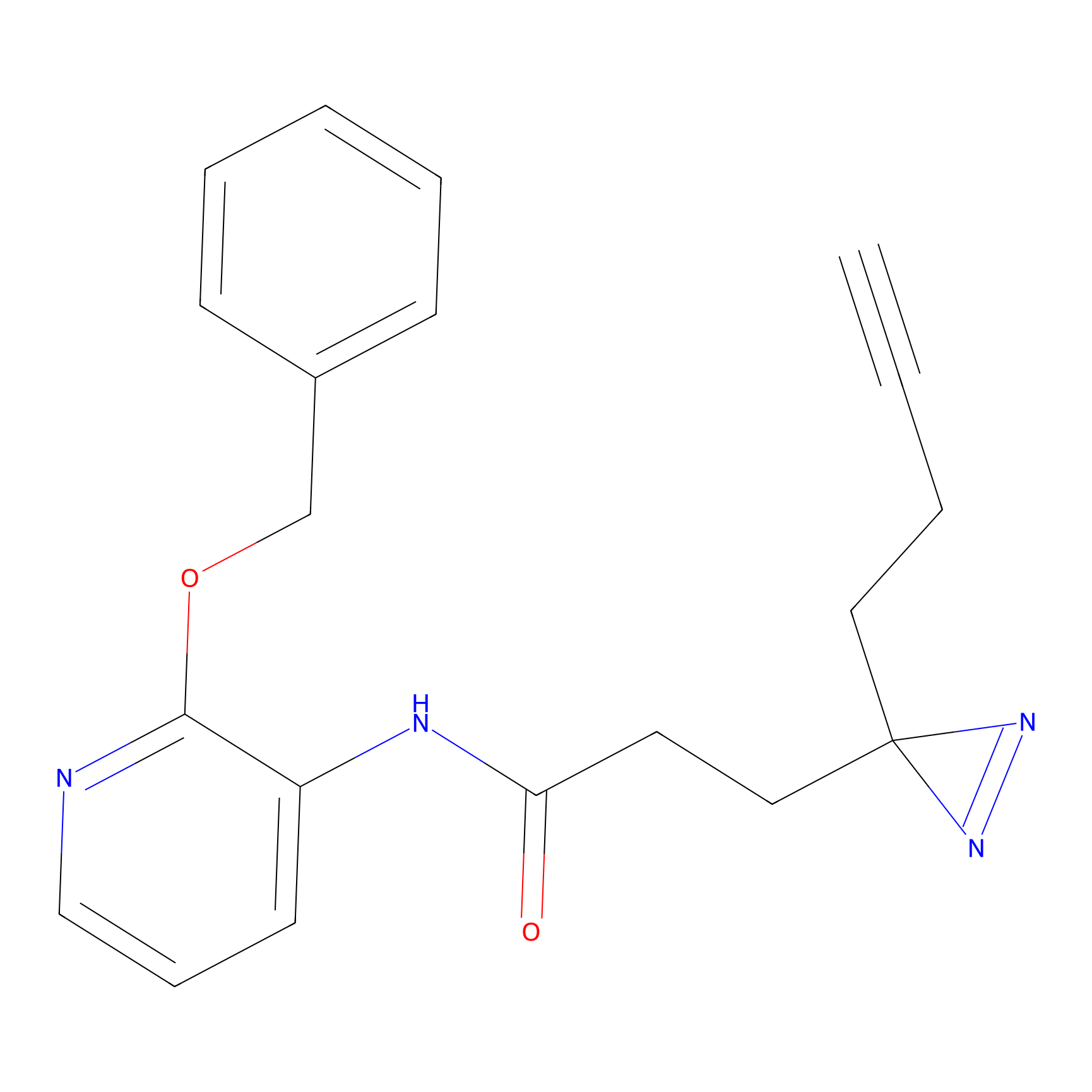
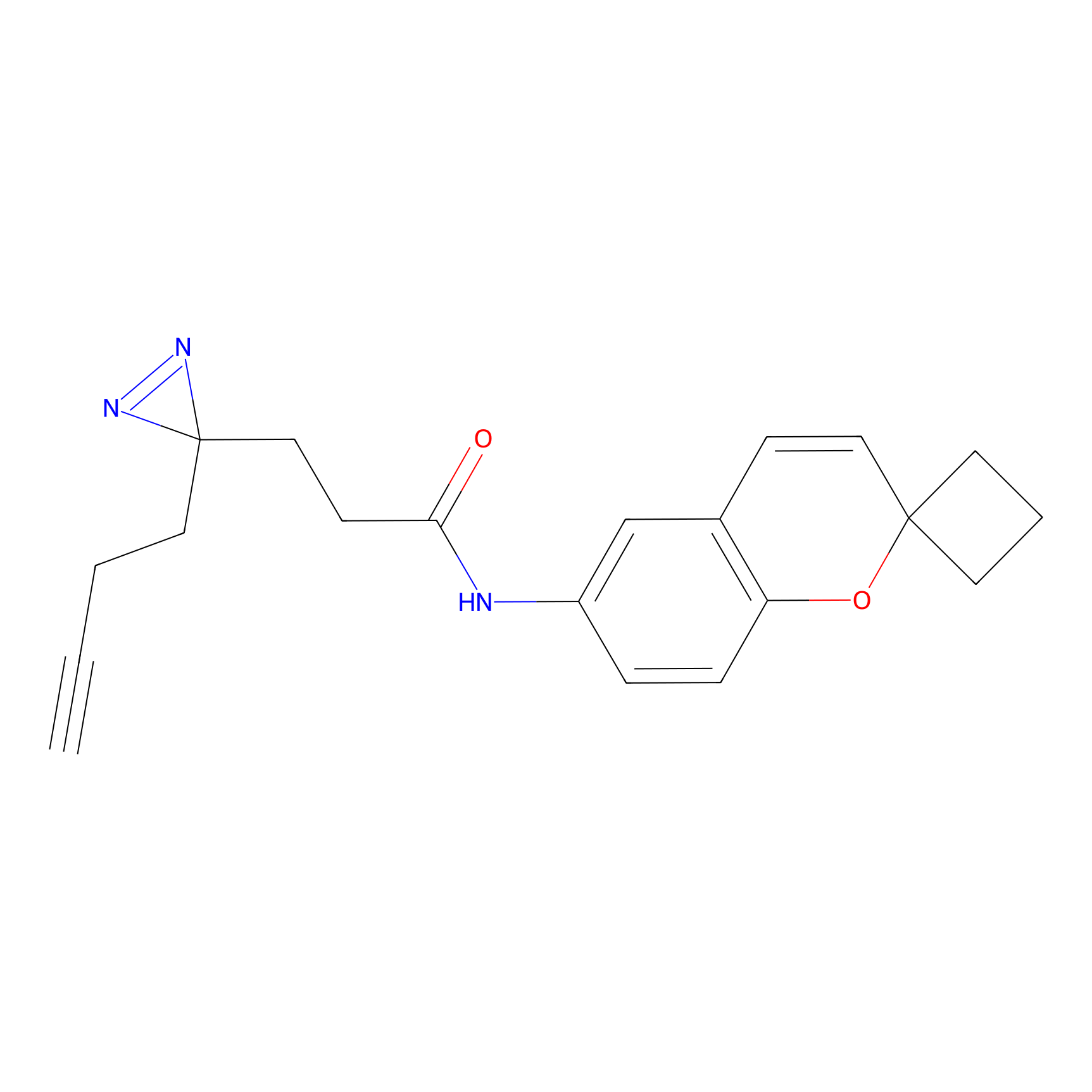
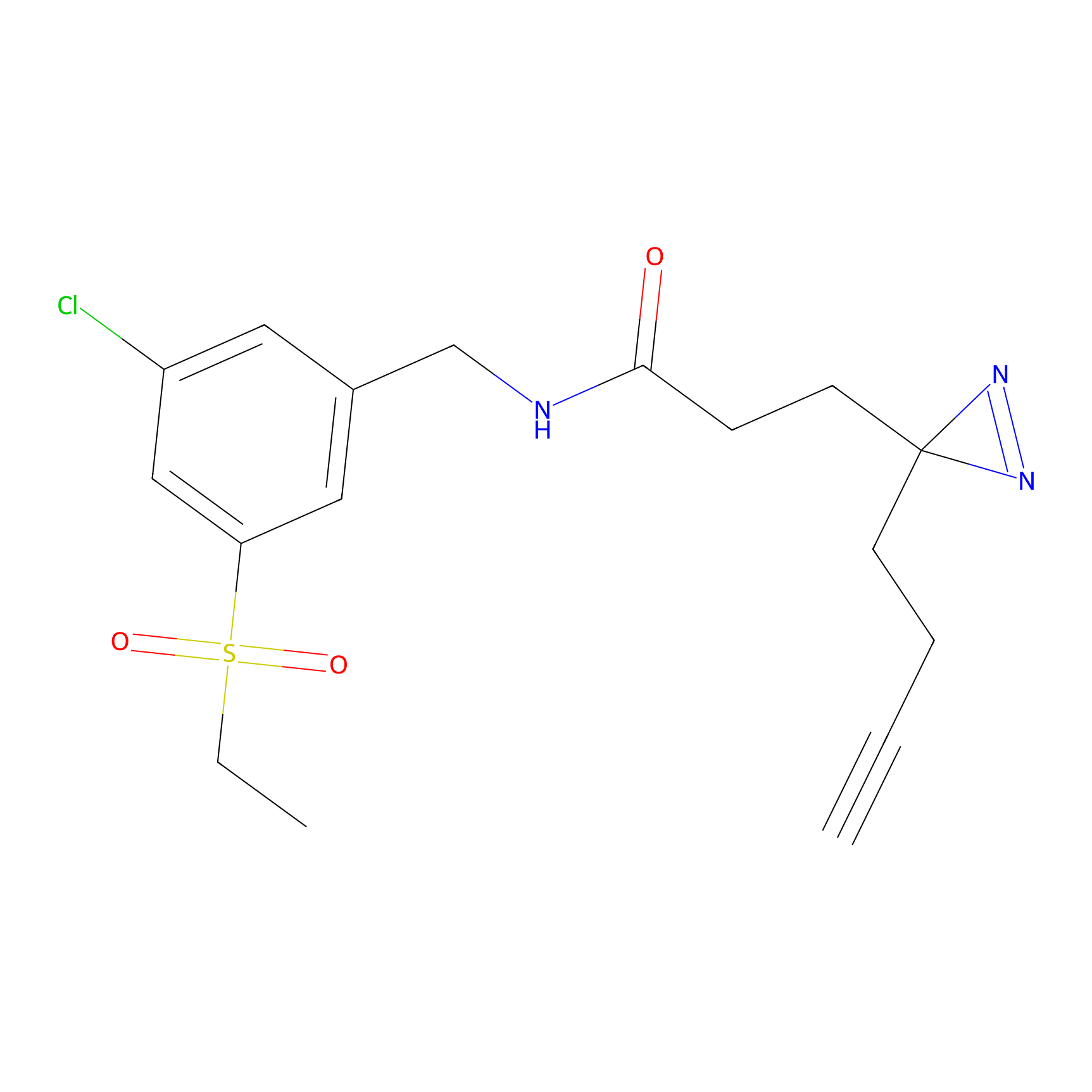
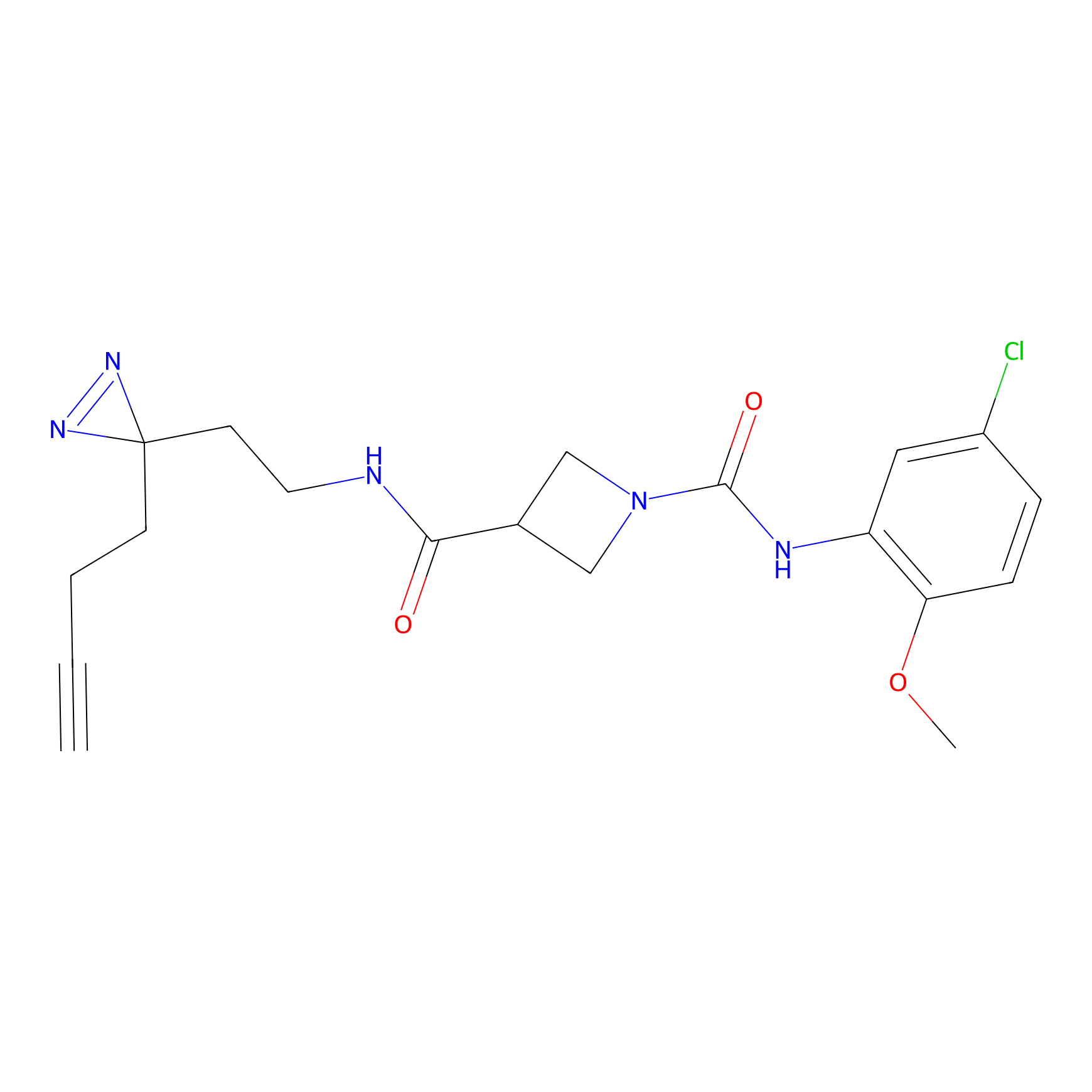
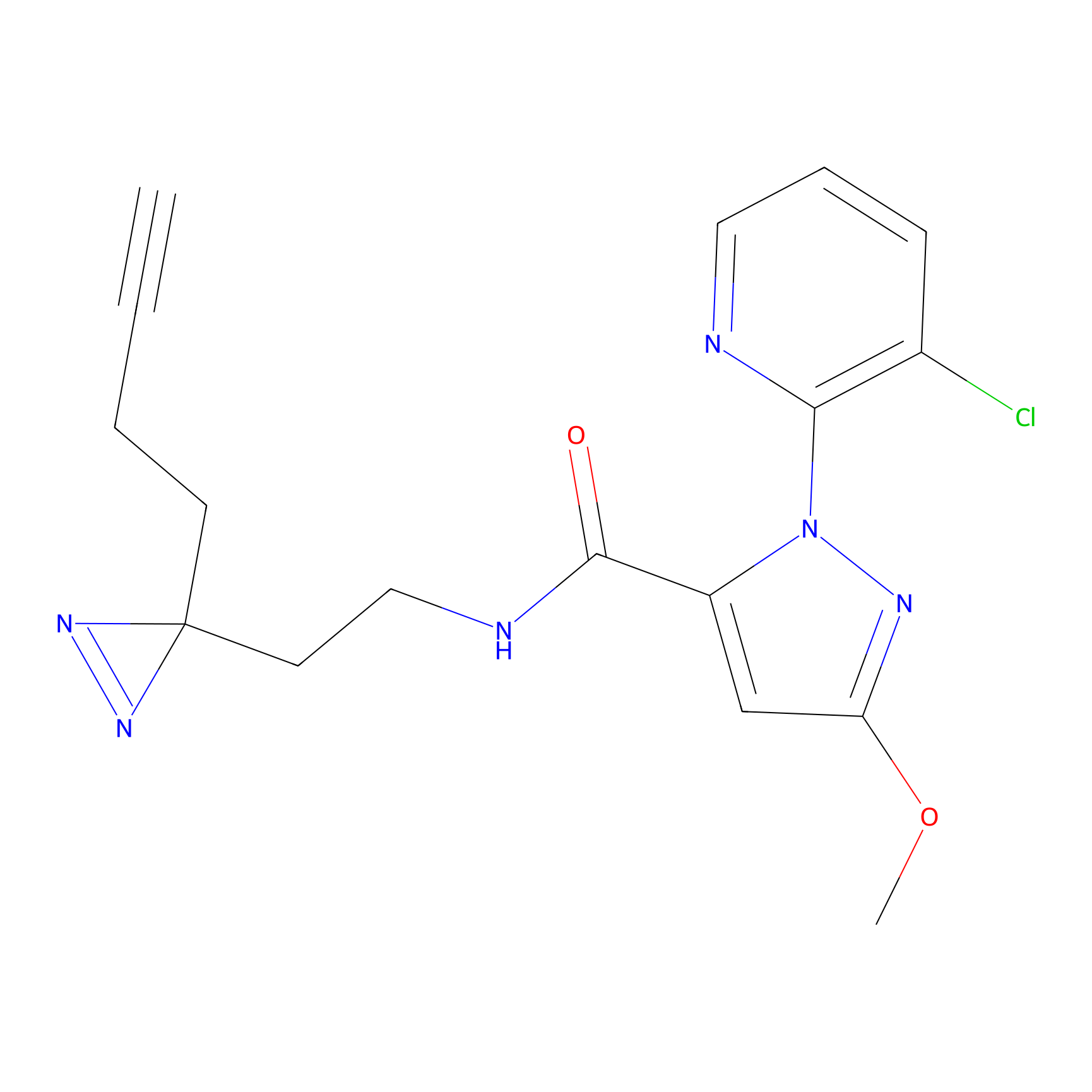
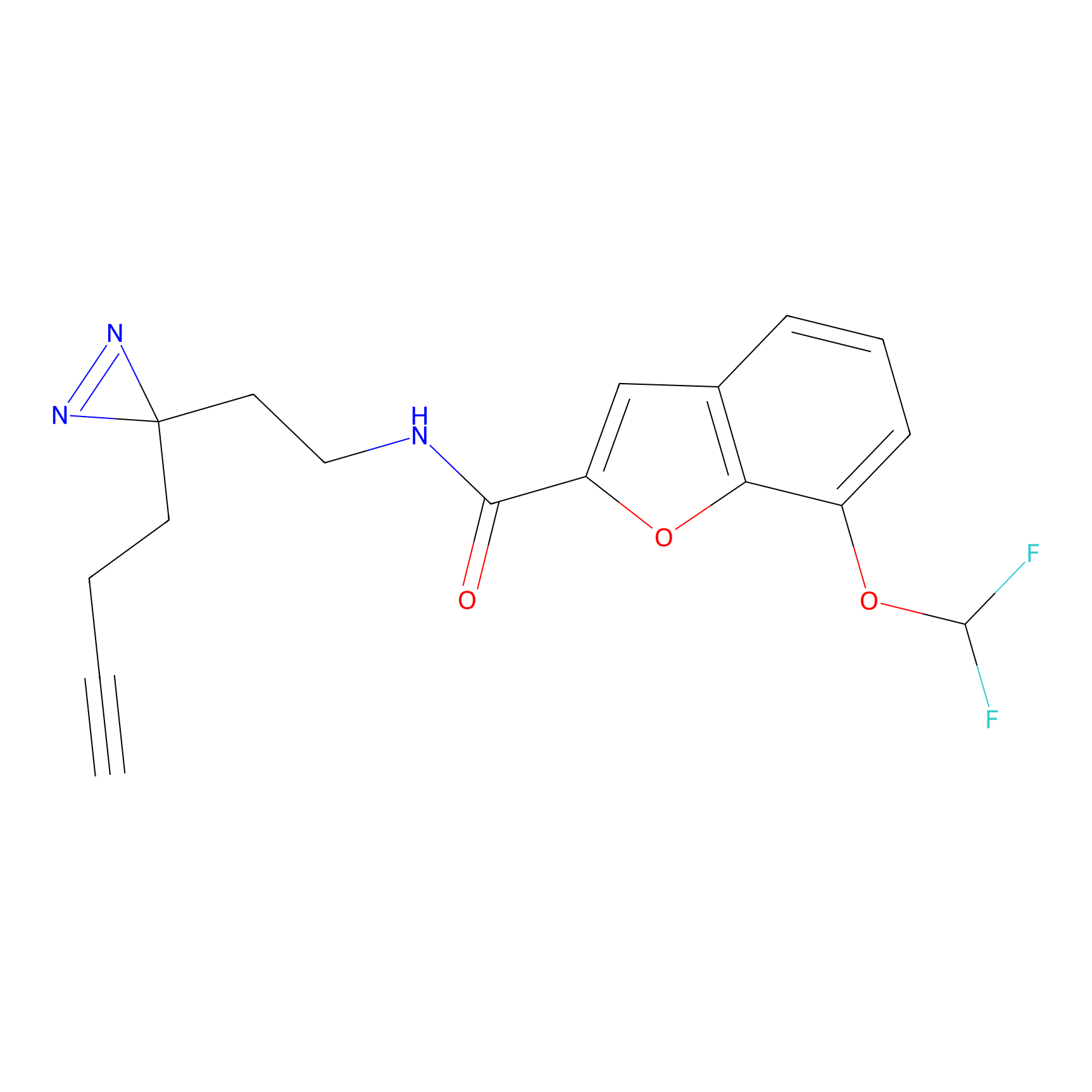
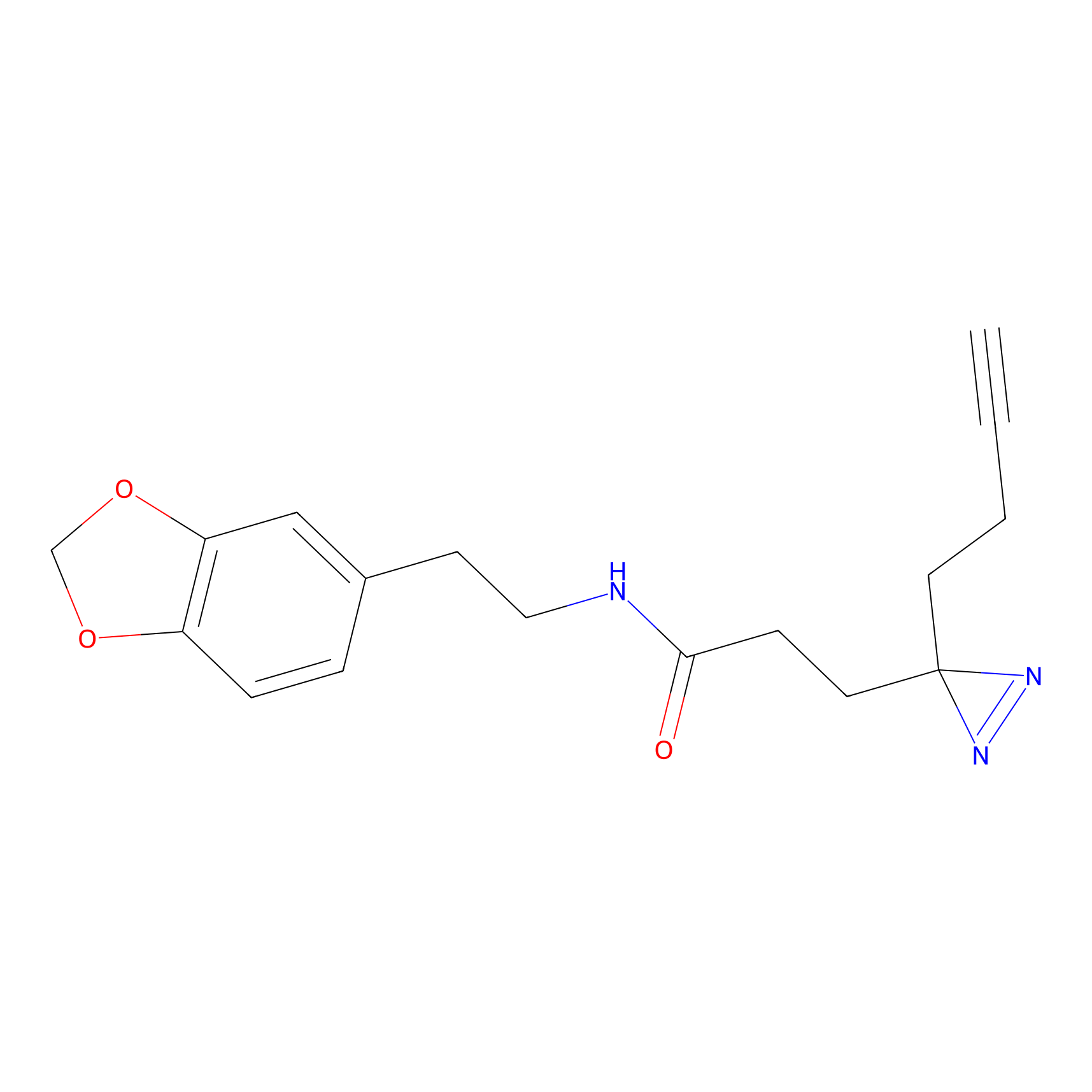
m-APA Probe Info |
 |
14.00 | LDD0402 | [1] | |
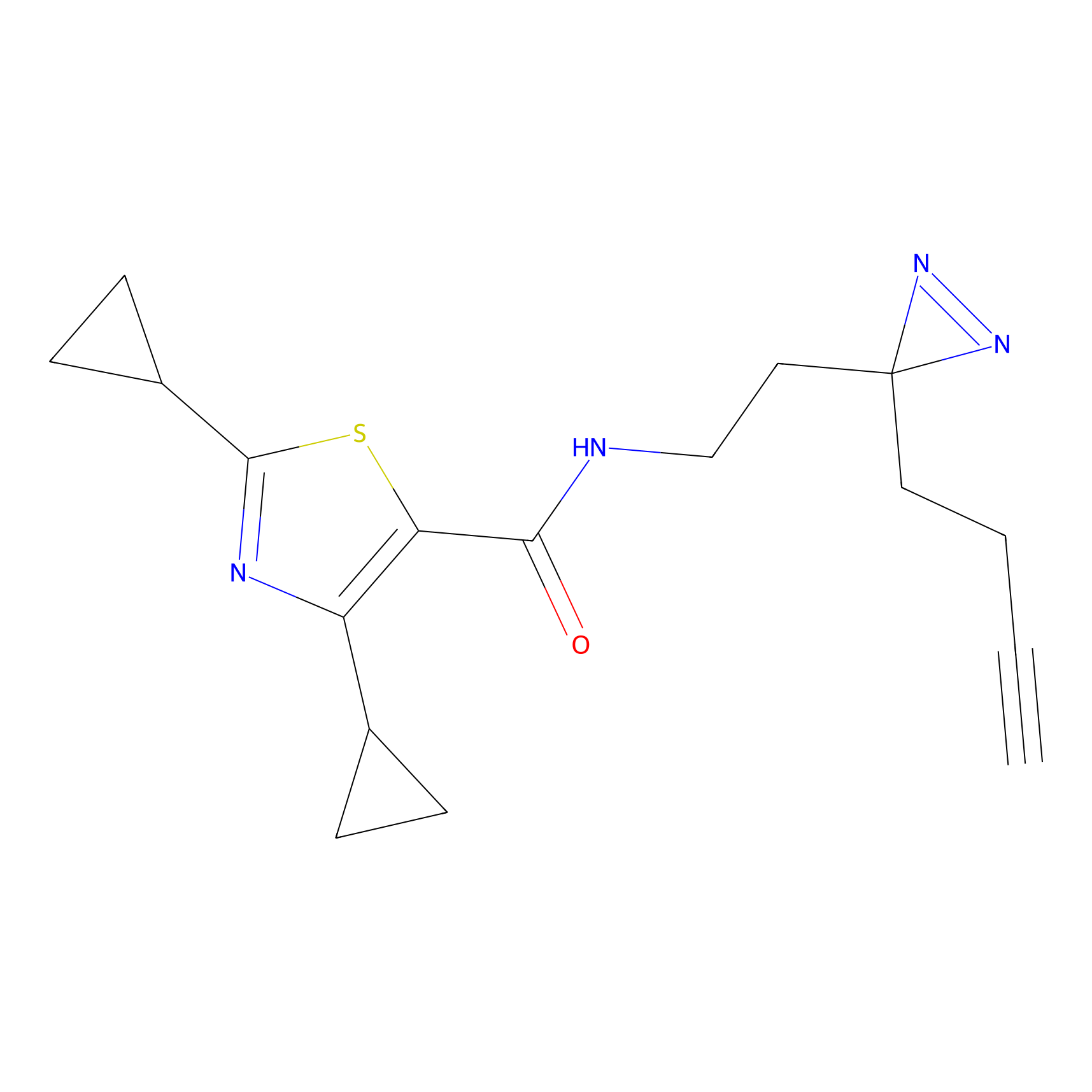
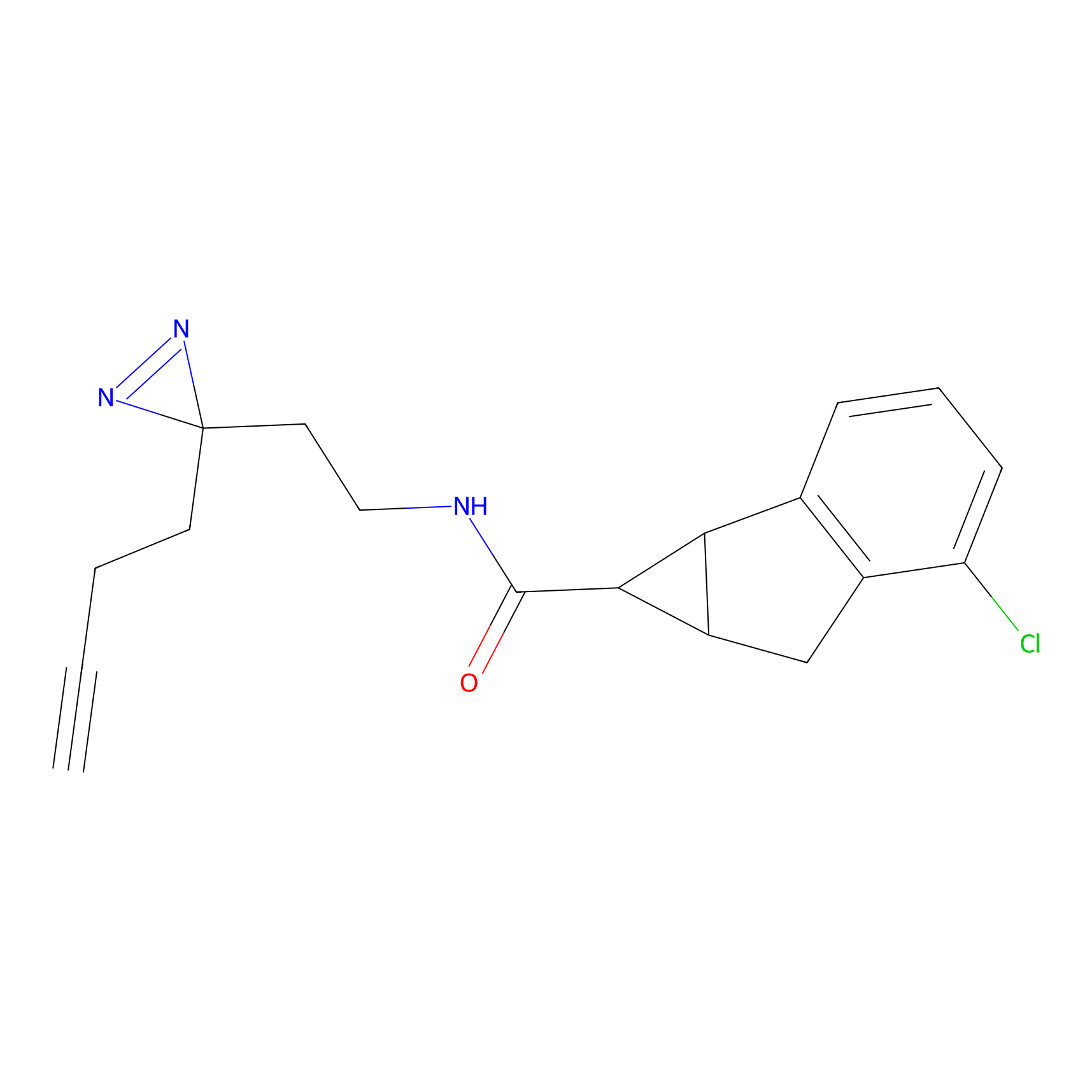
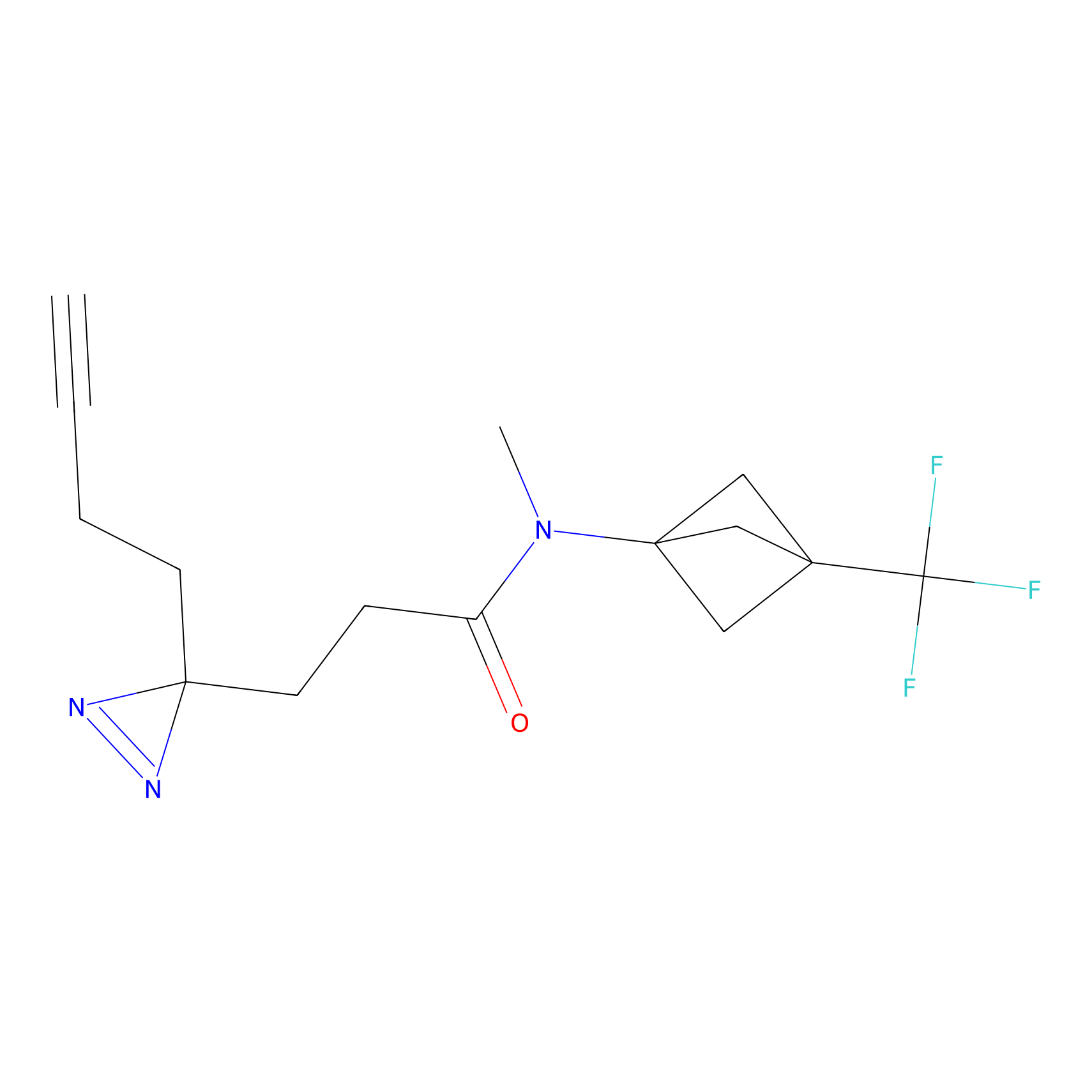
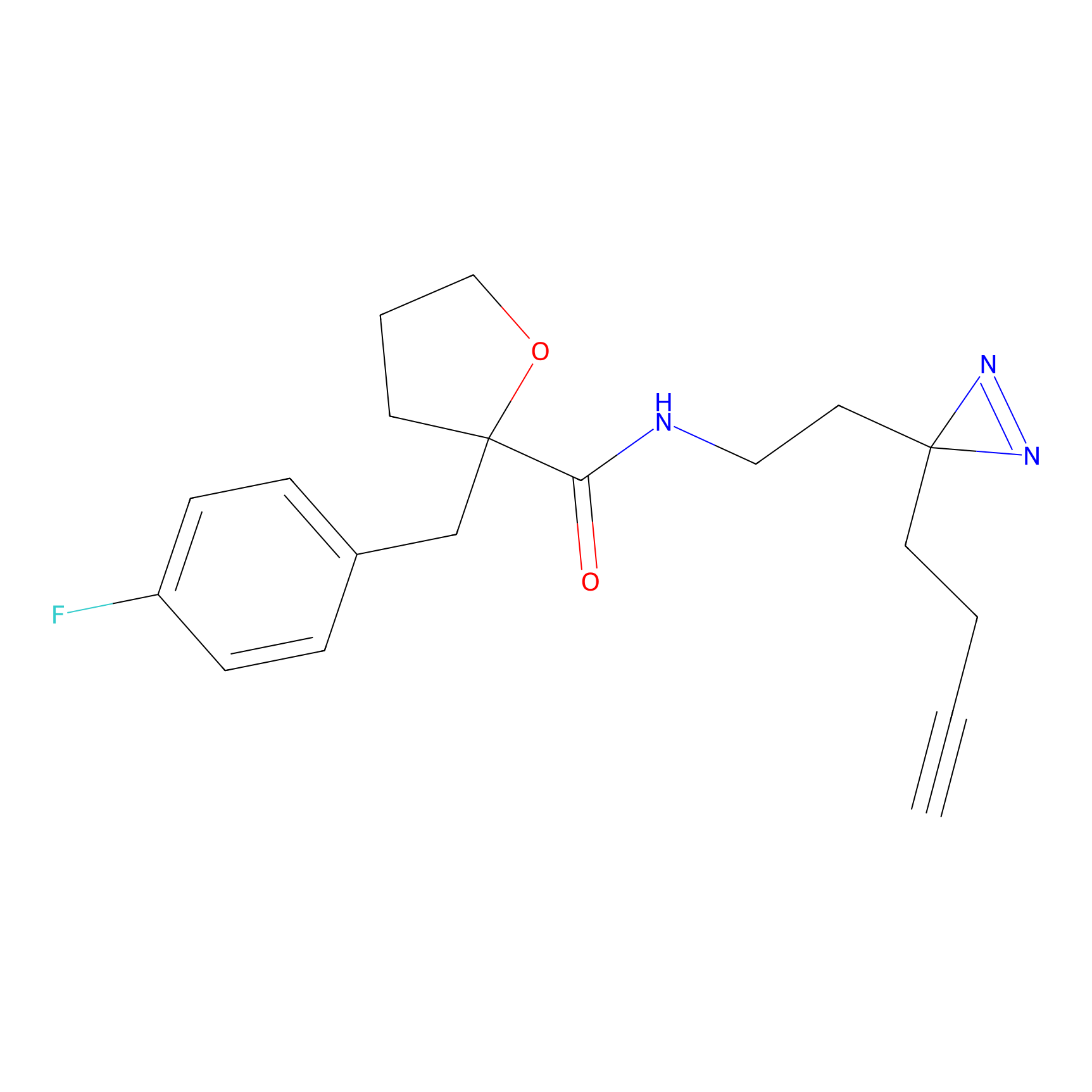
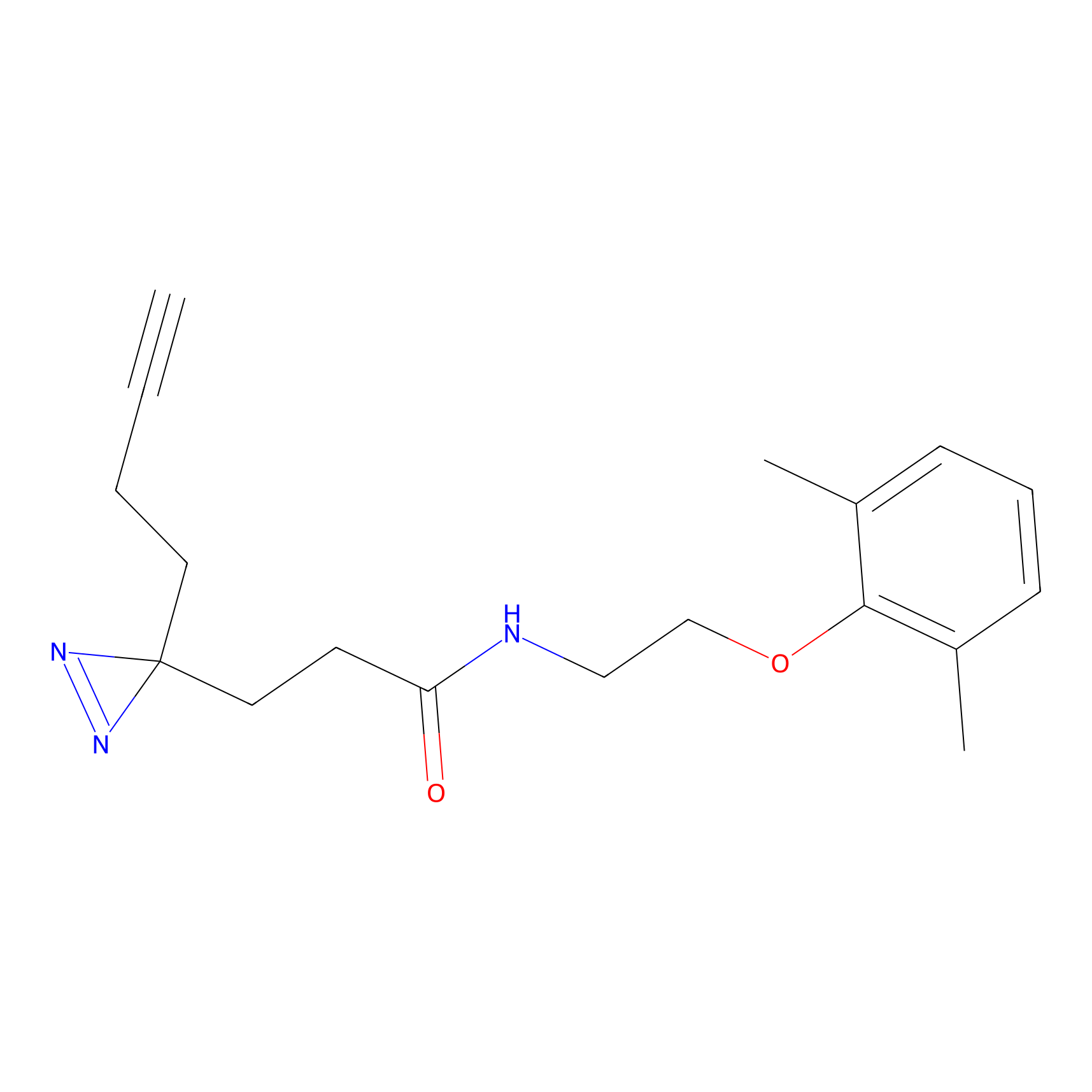
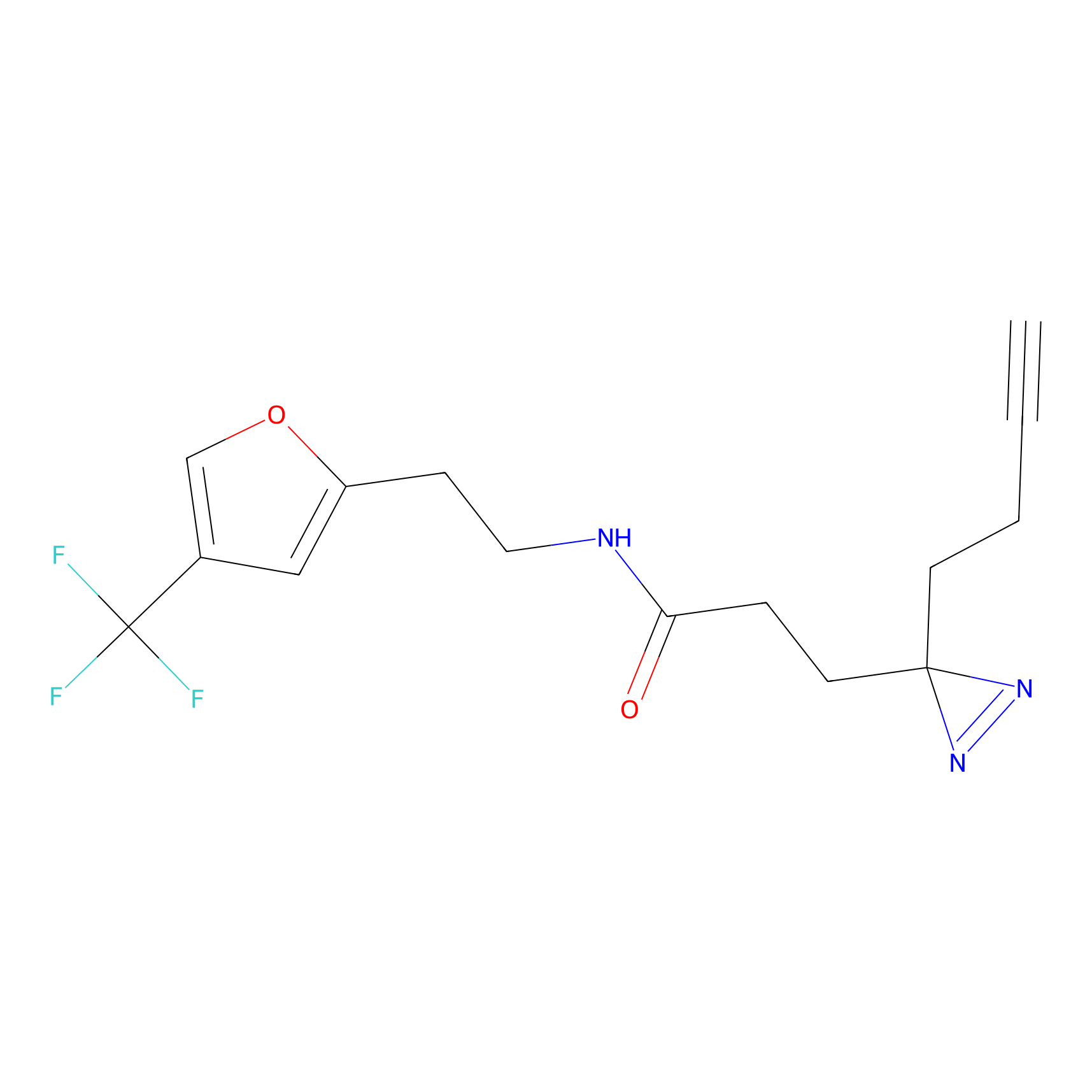
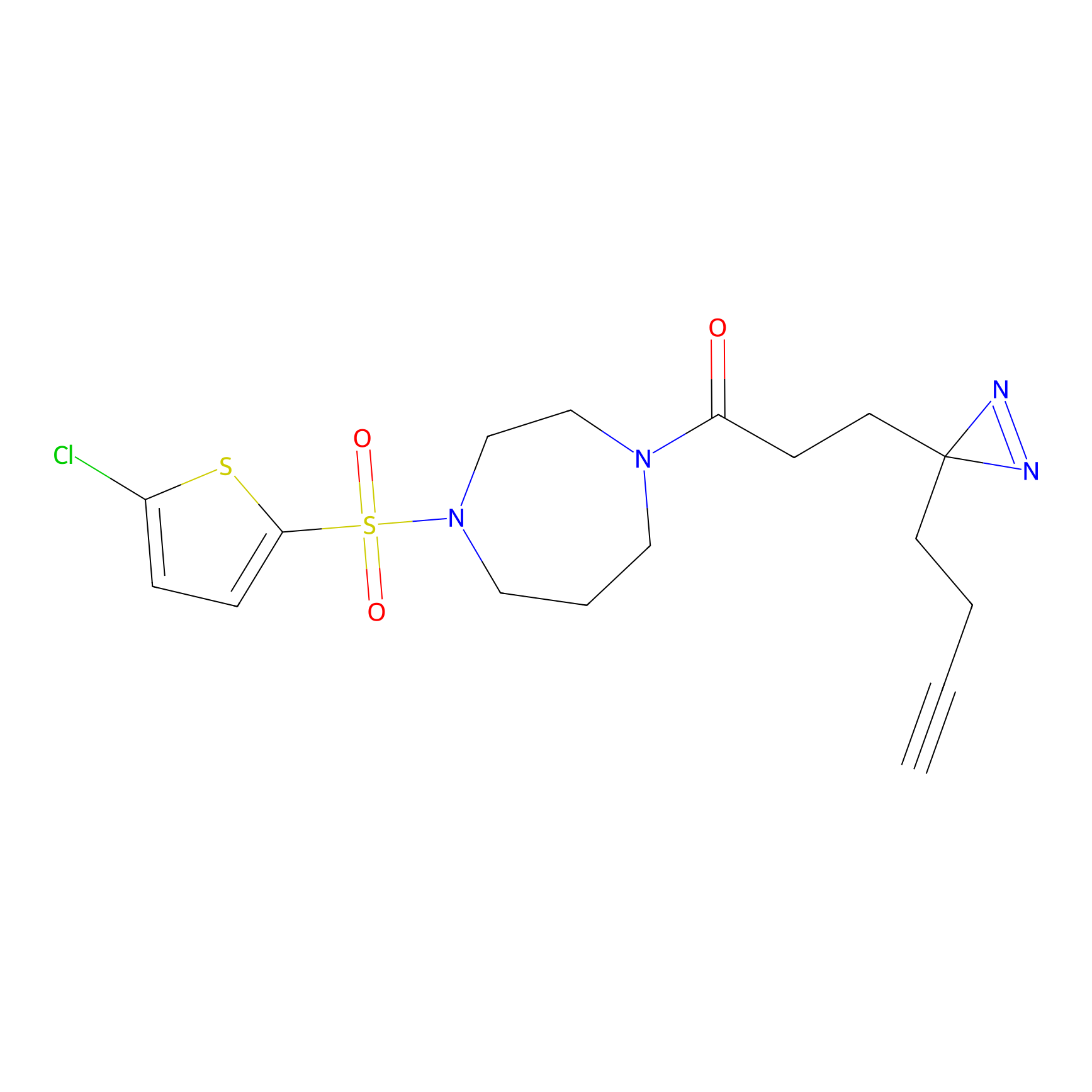
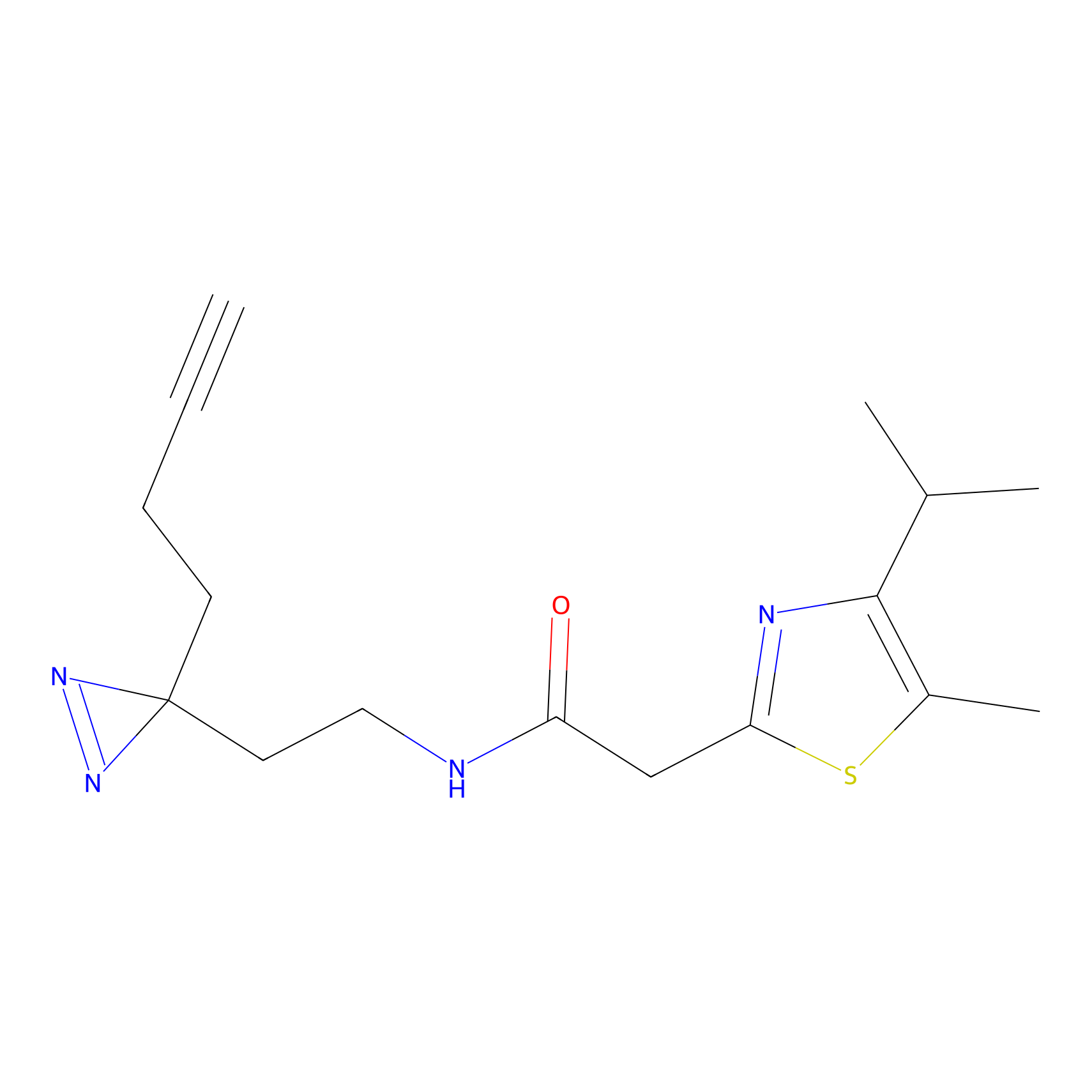
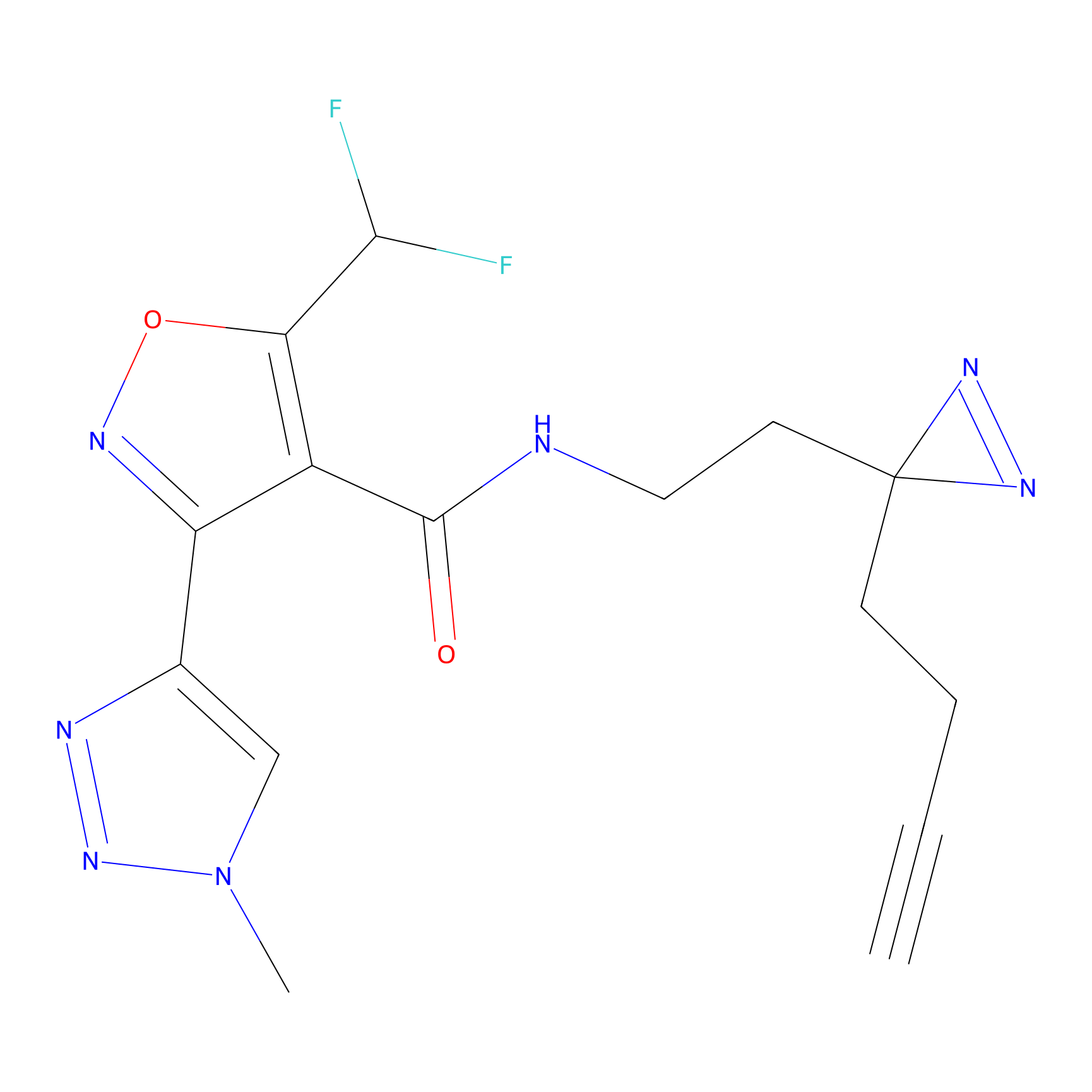
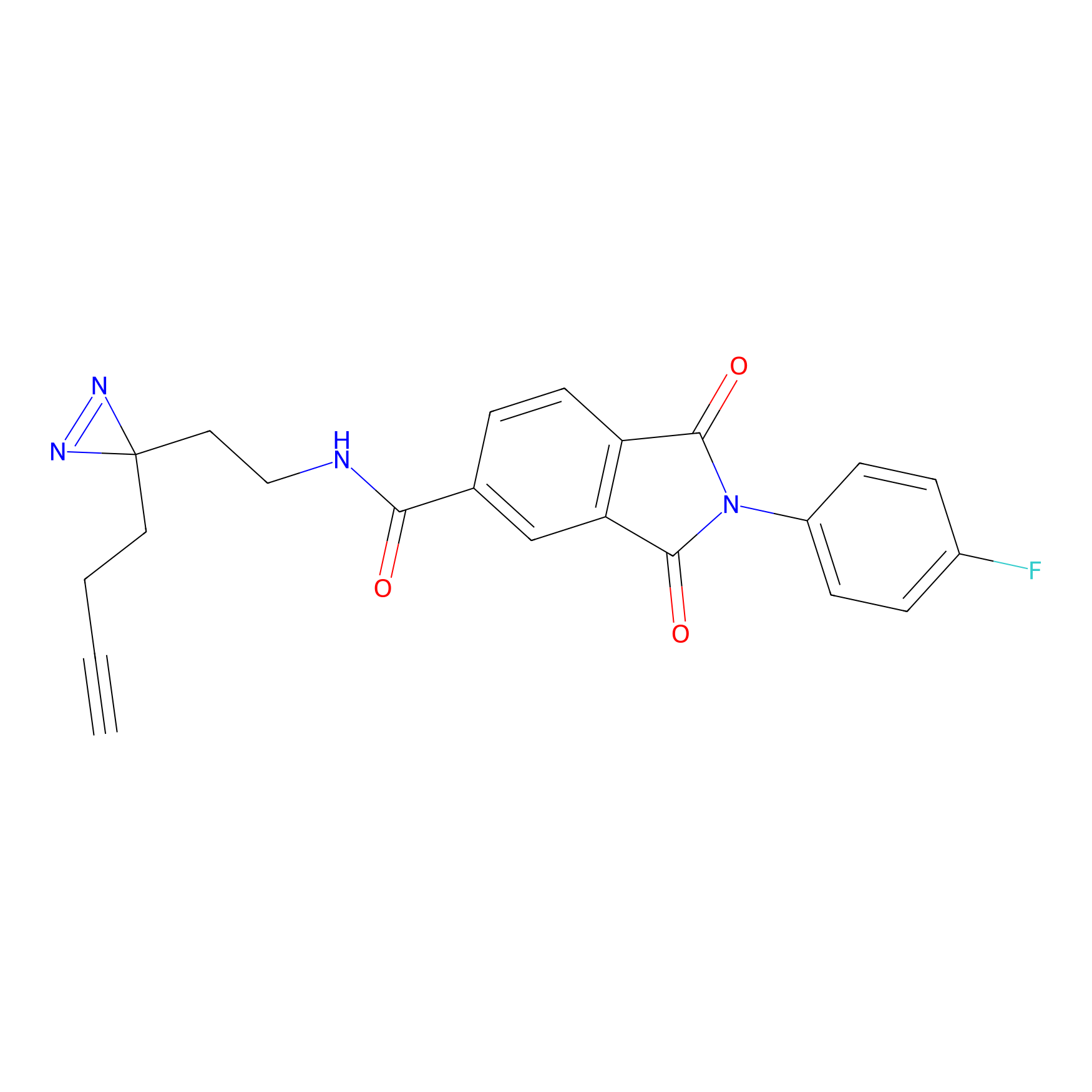
|
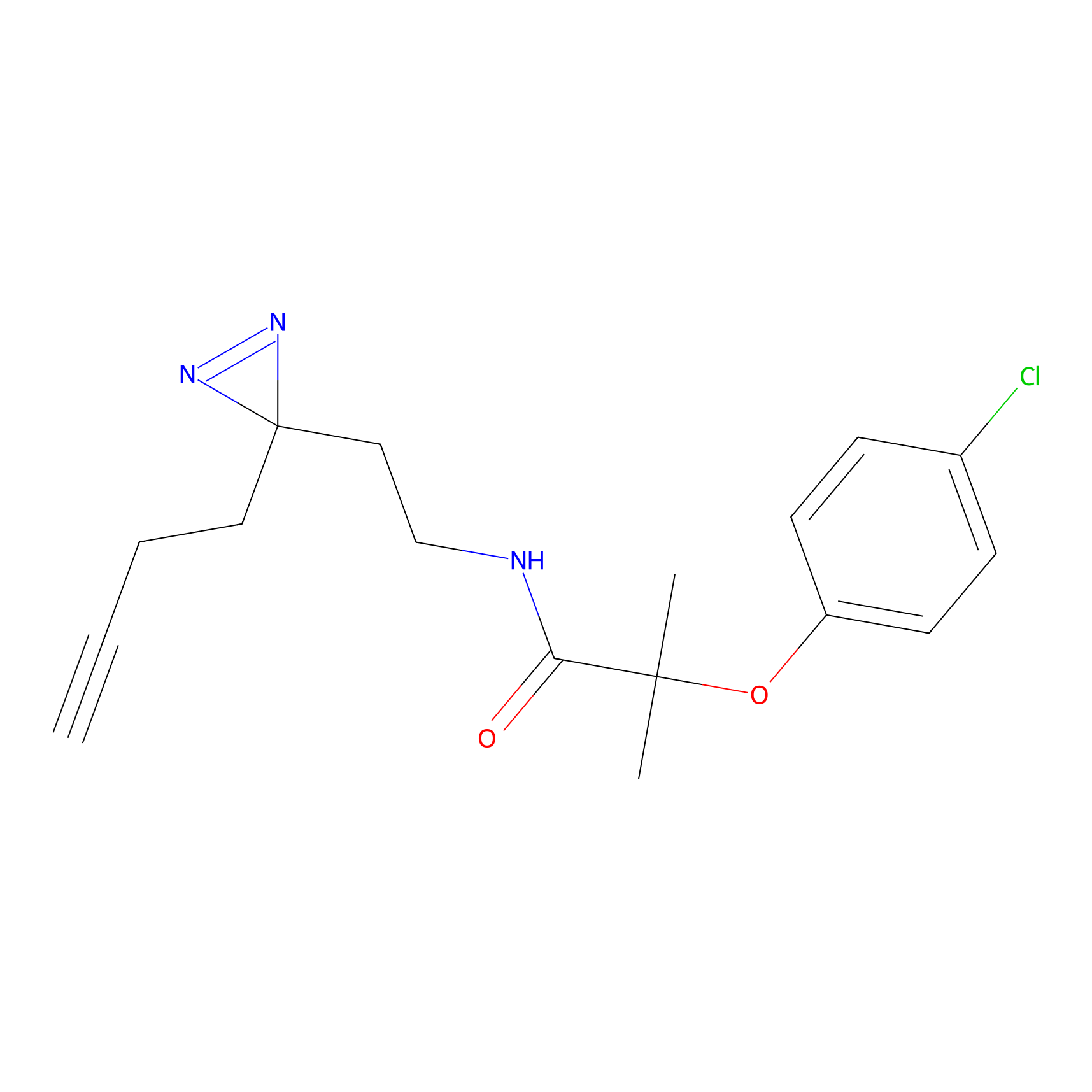
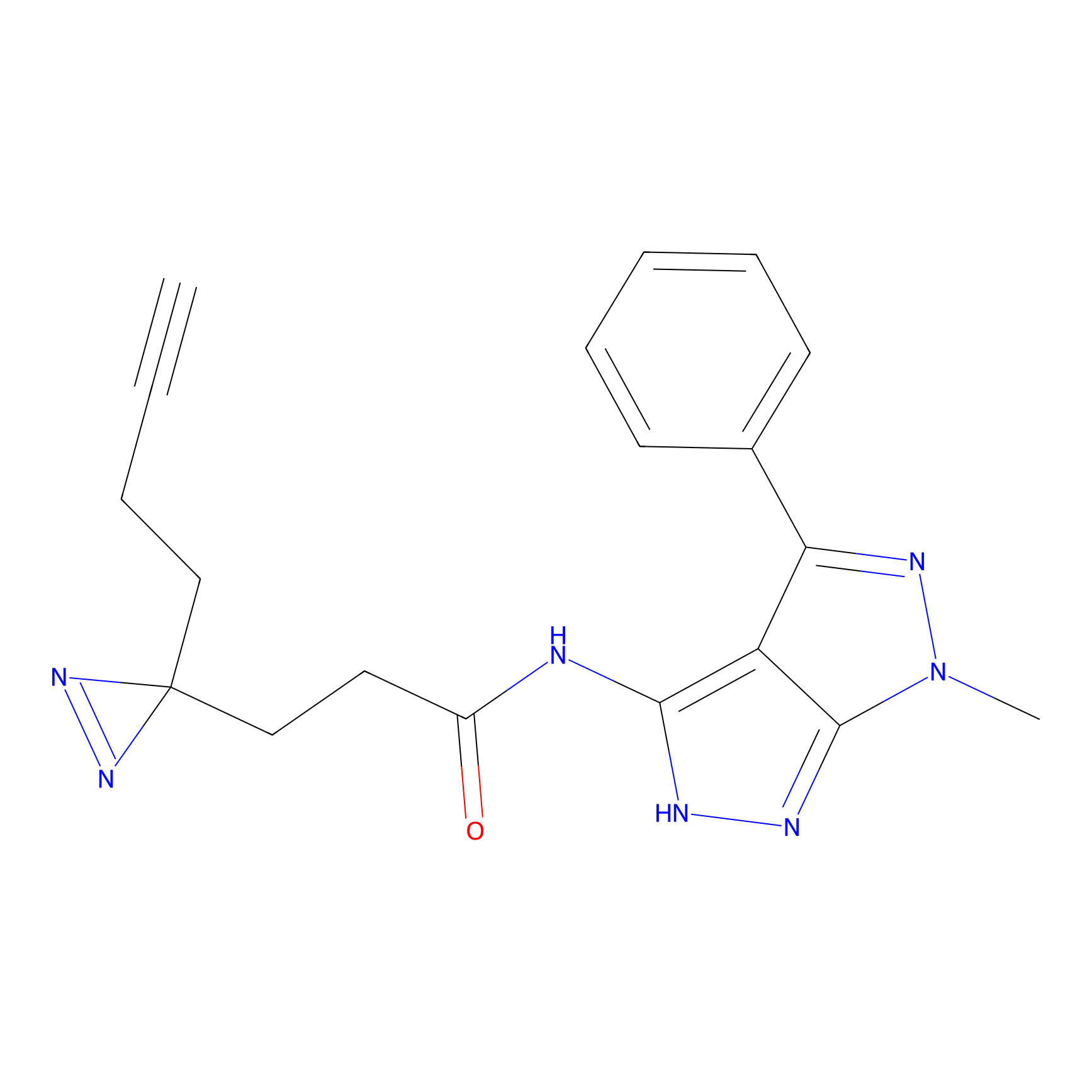
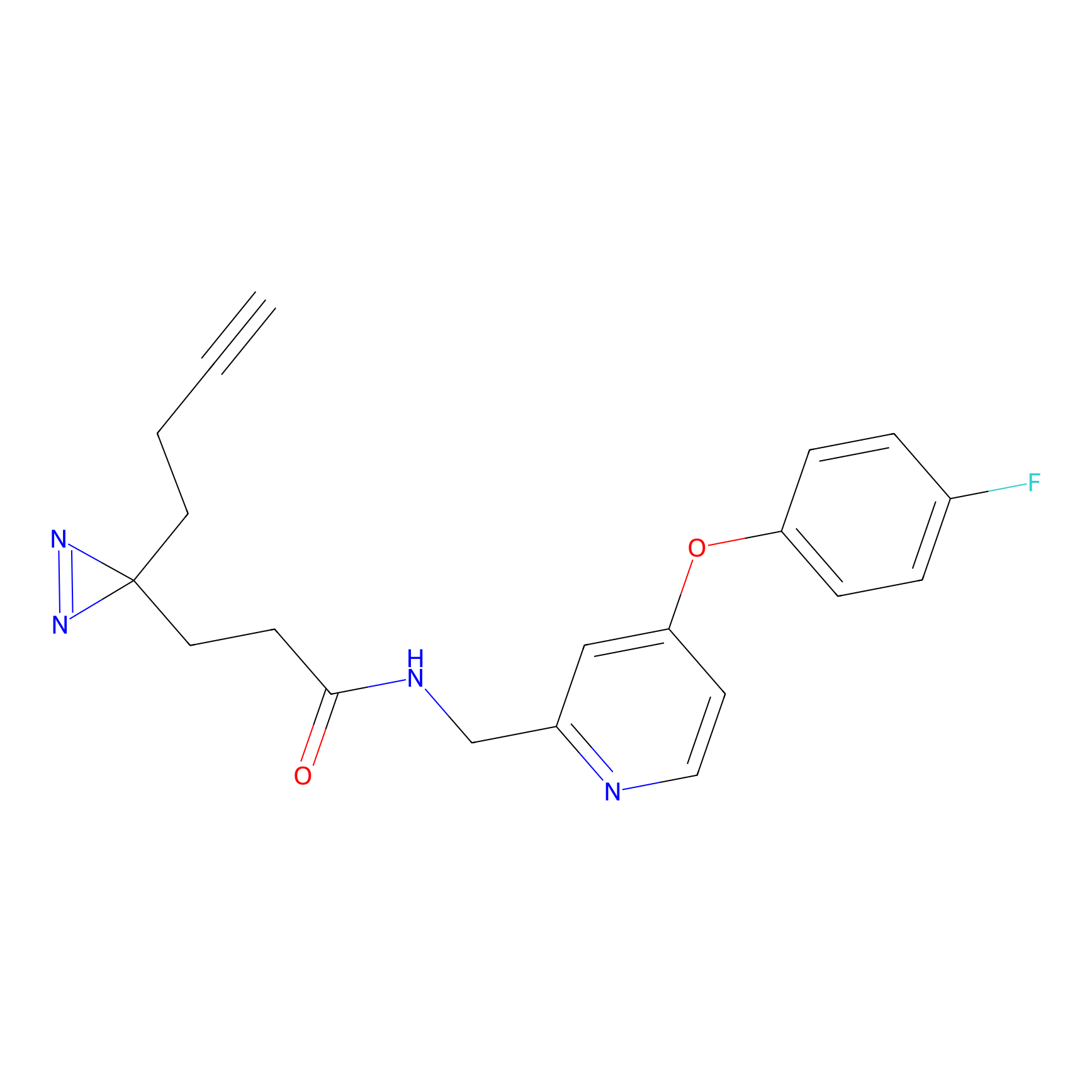
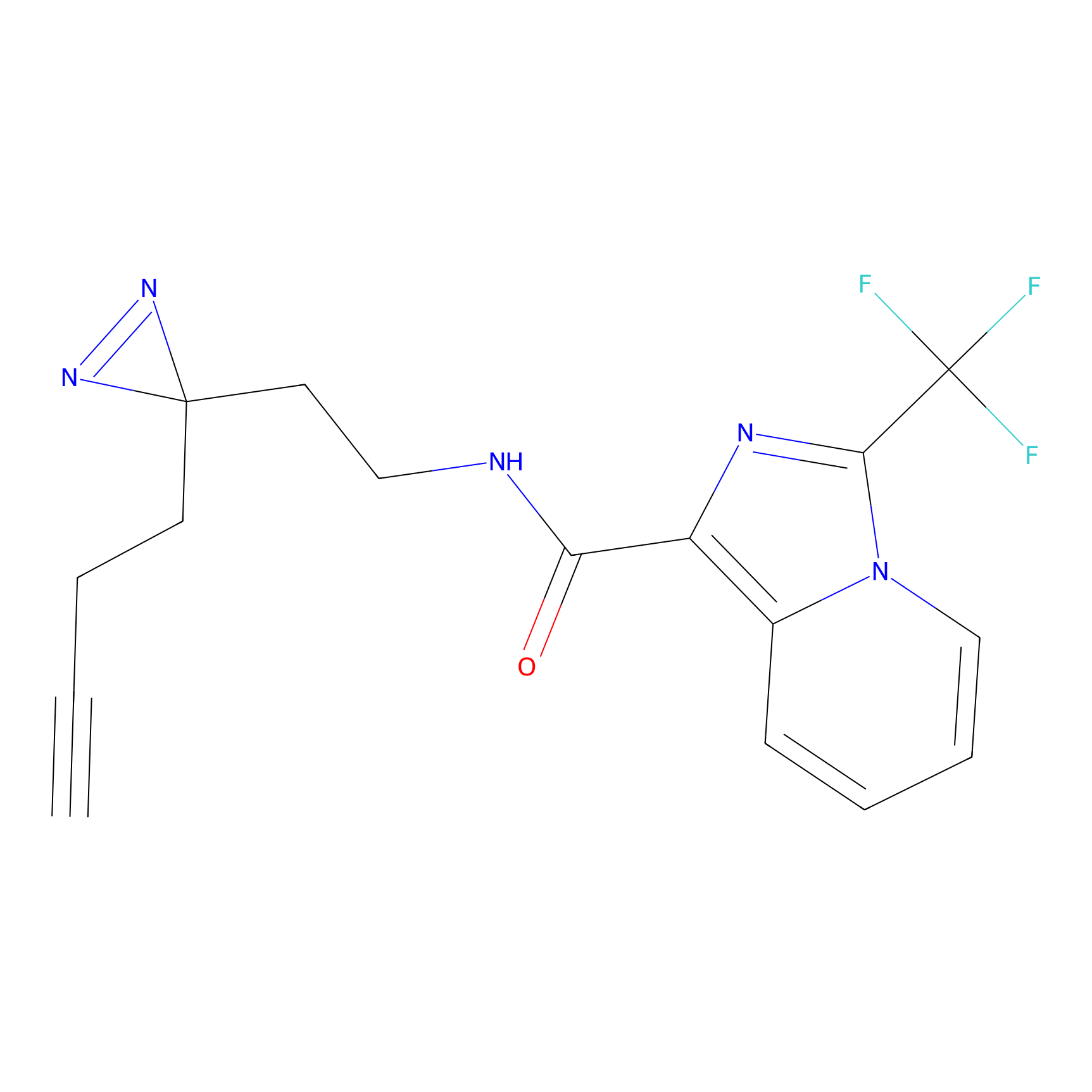
CHEMBL5175495 Probe Info |
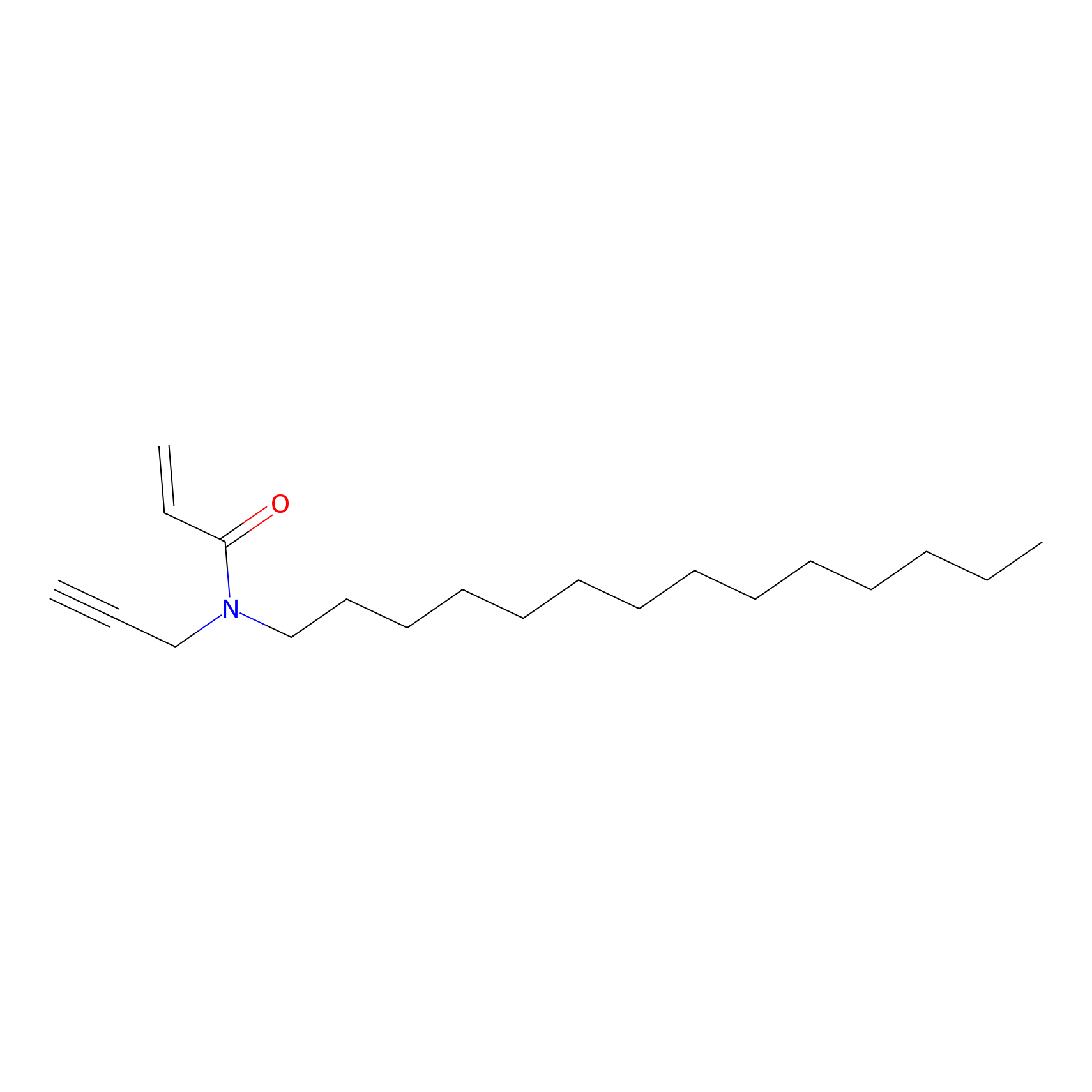 |
6.35 | LDD0196 | [2] | |
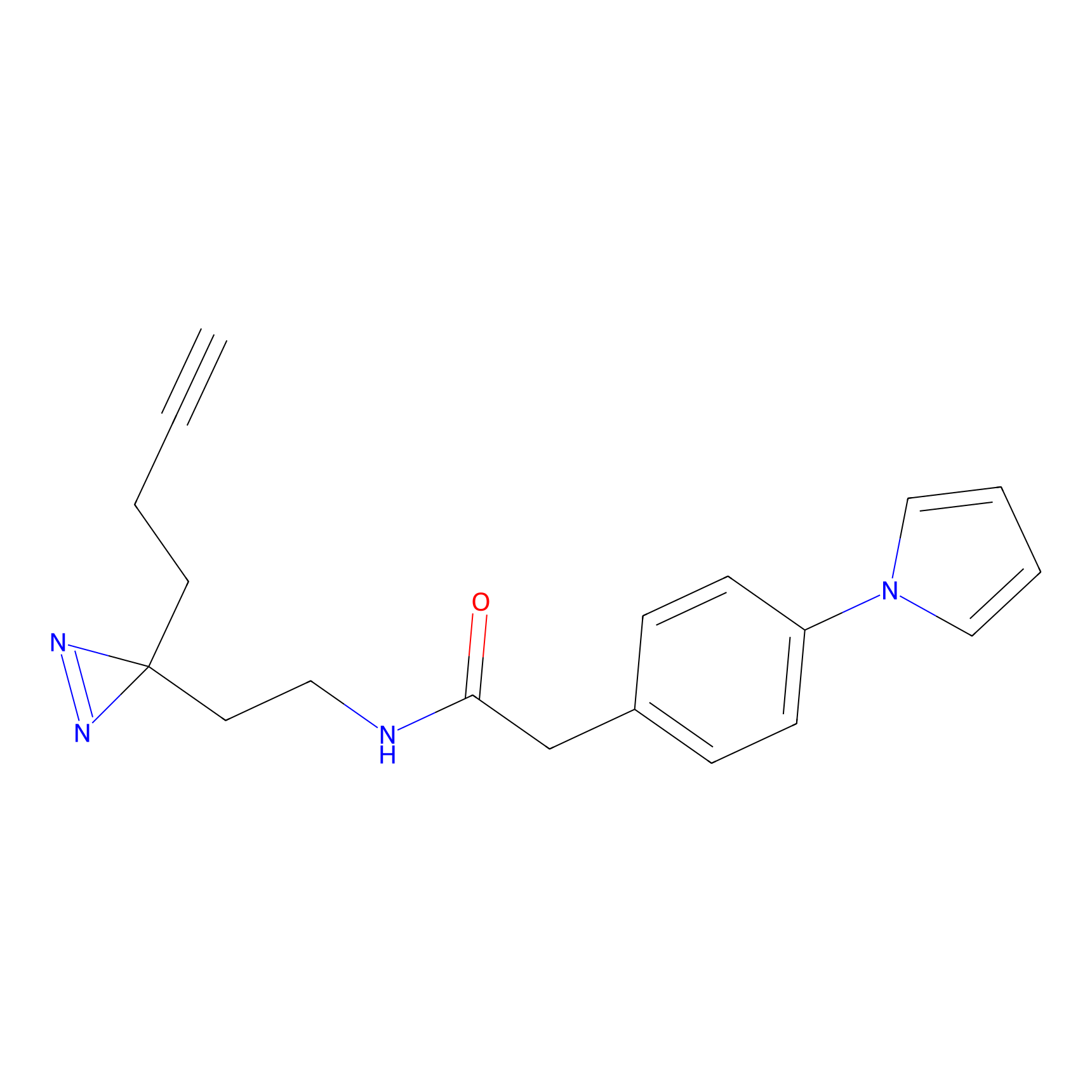
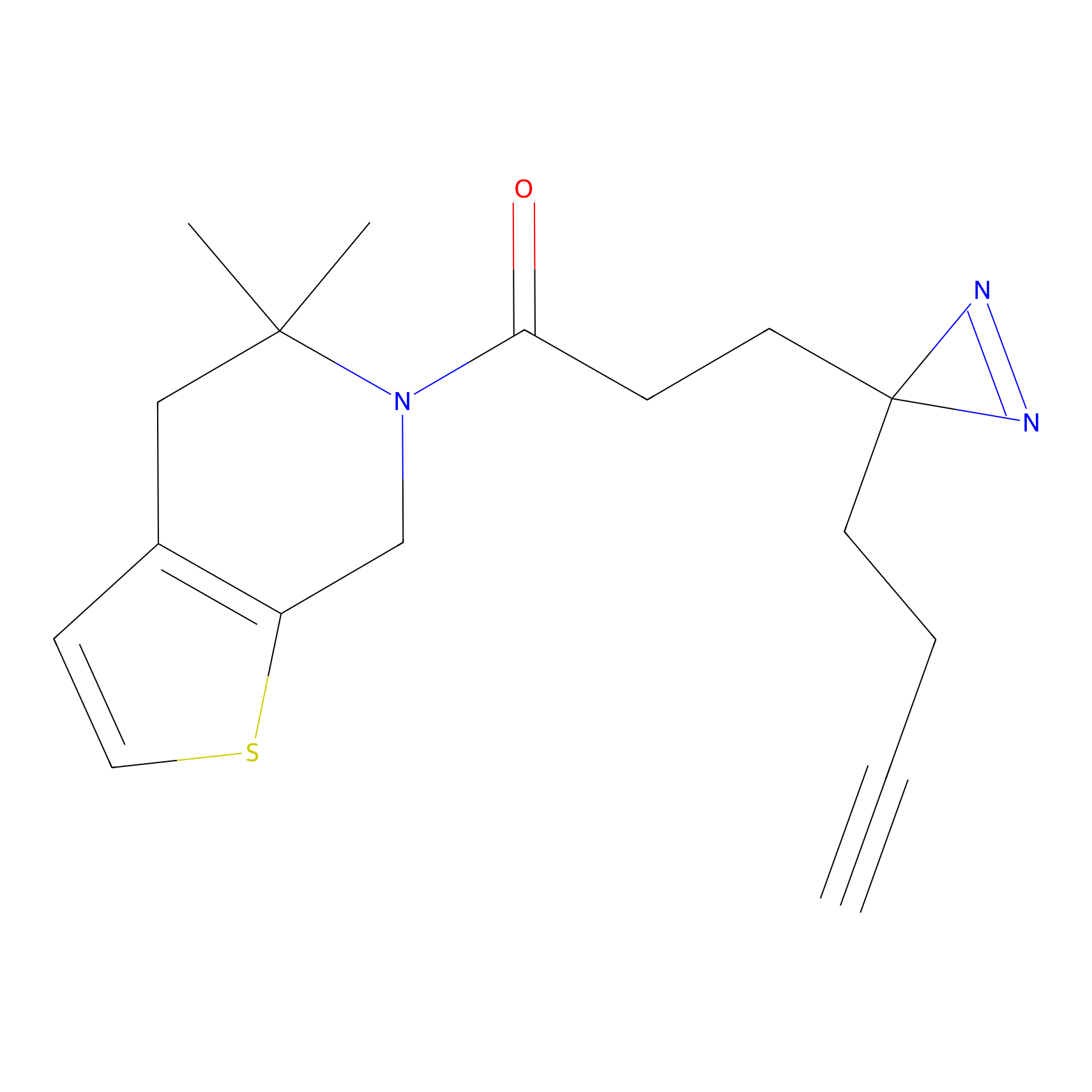
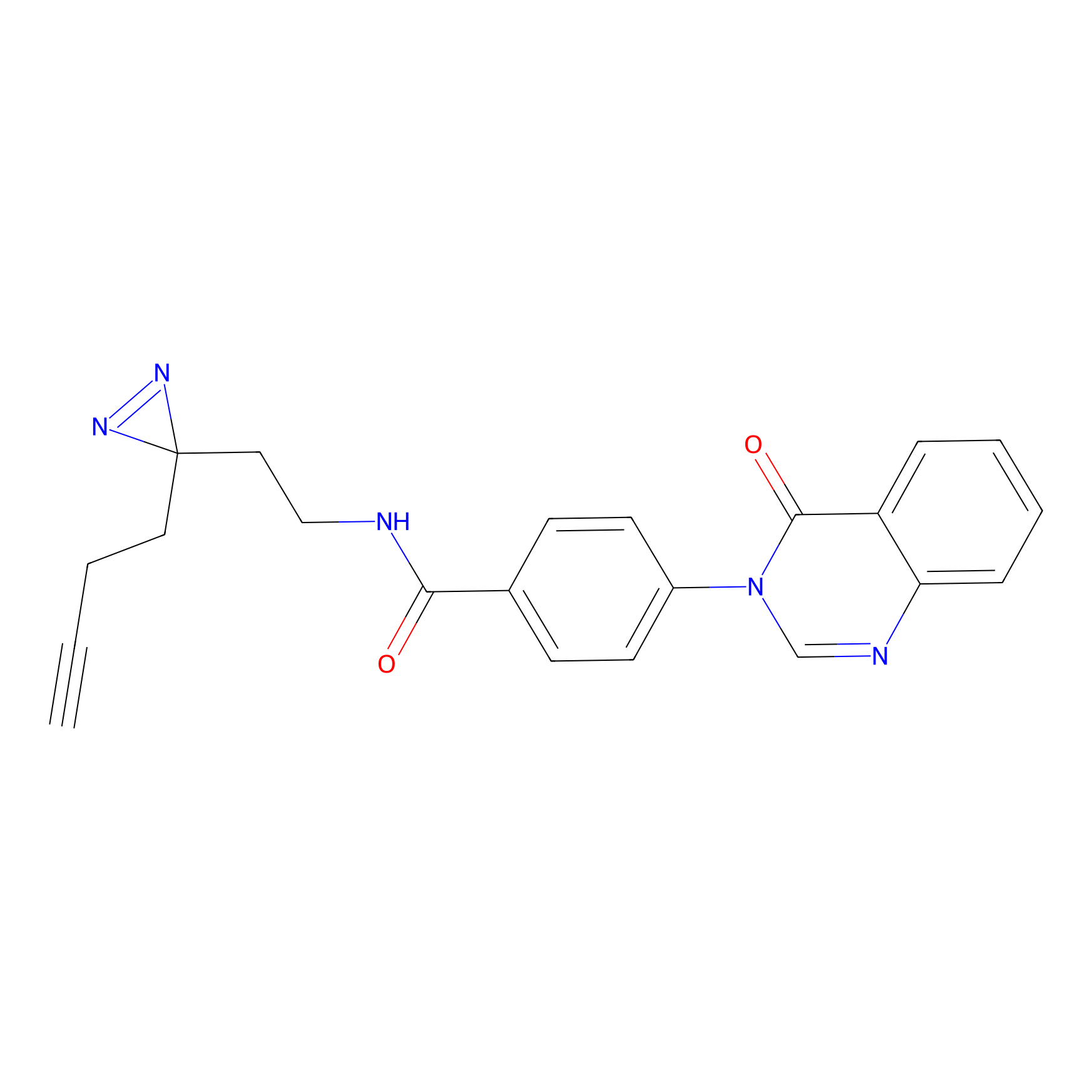
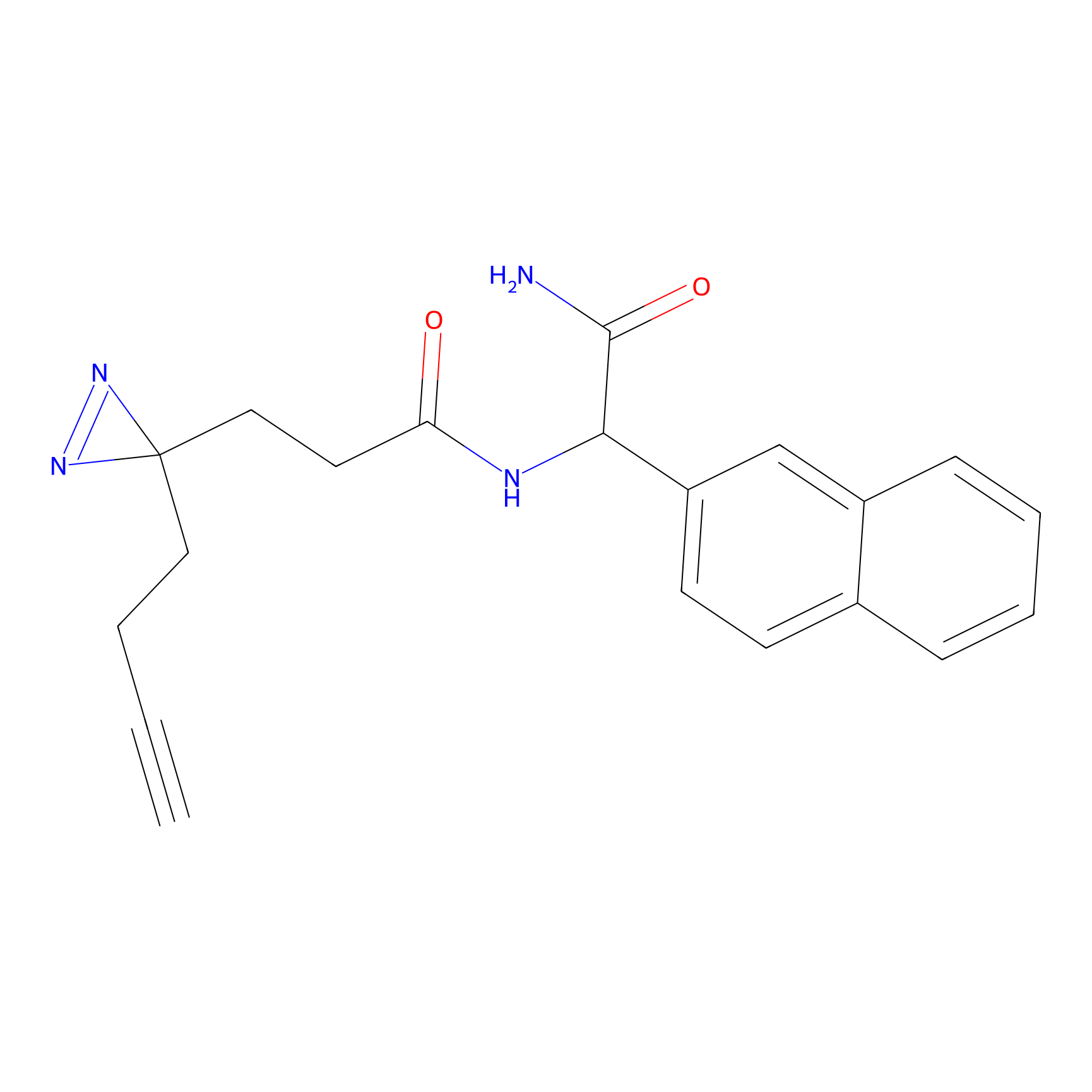
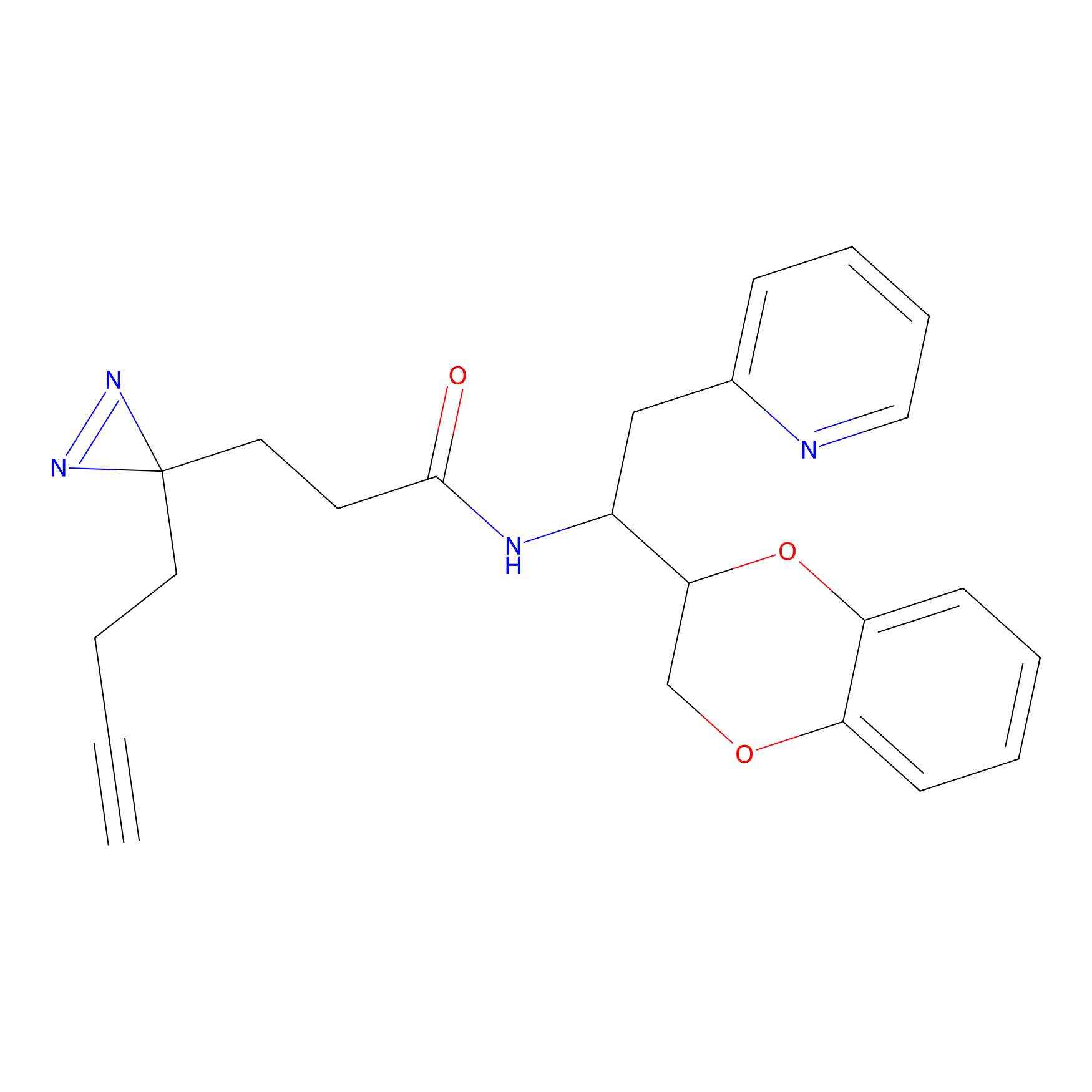
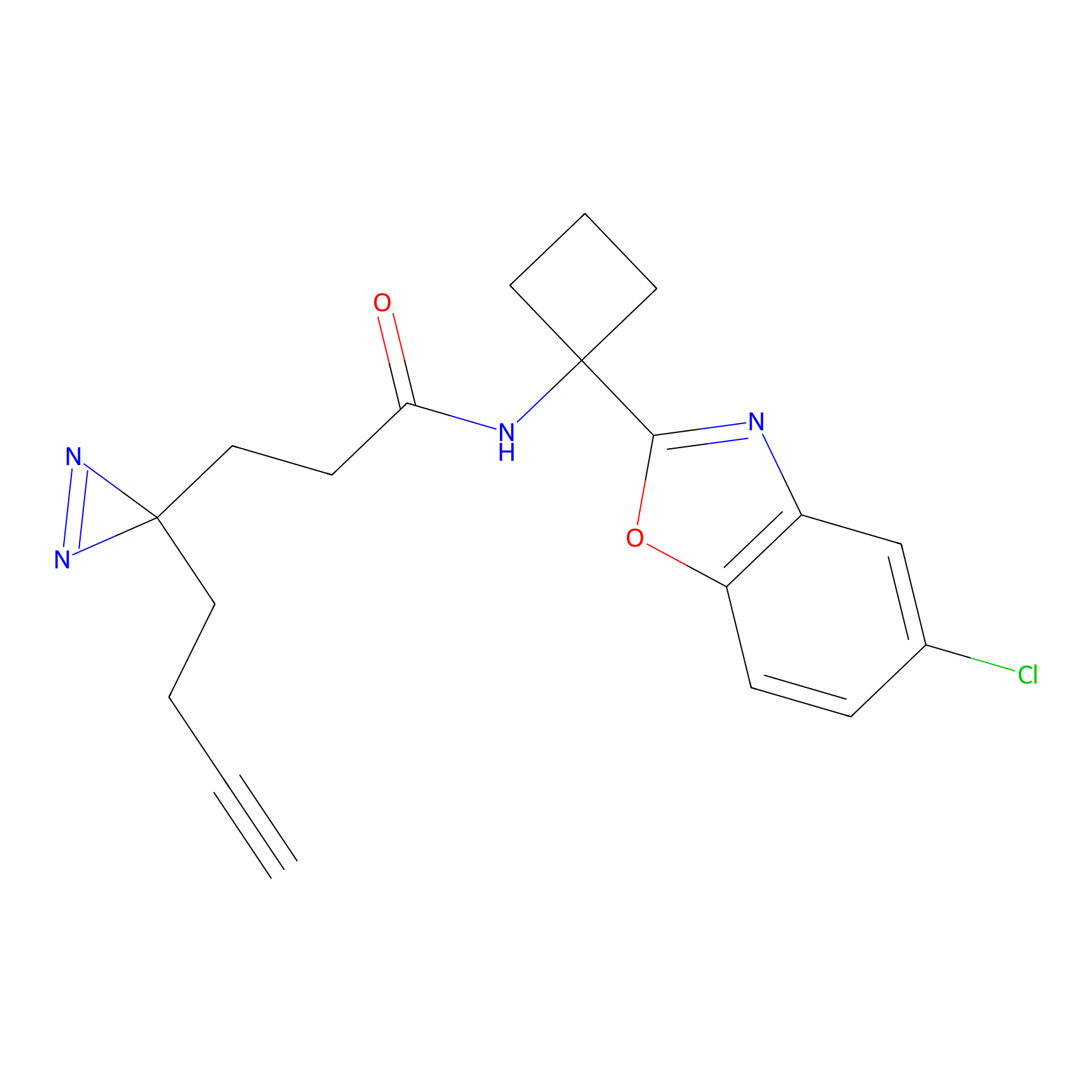
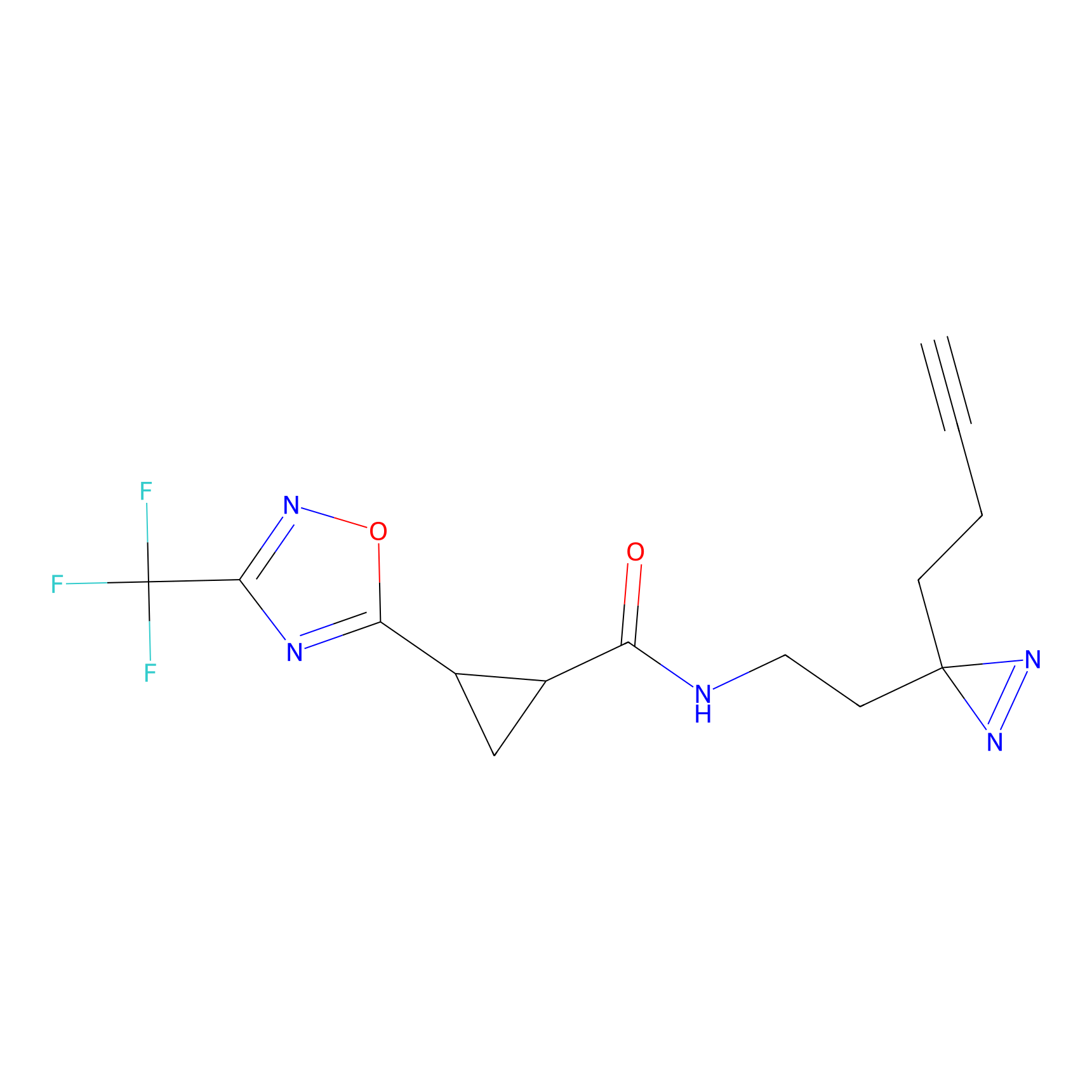
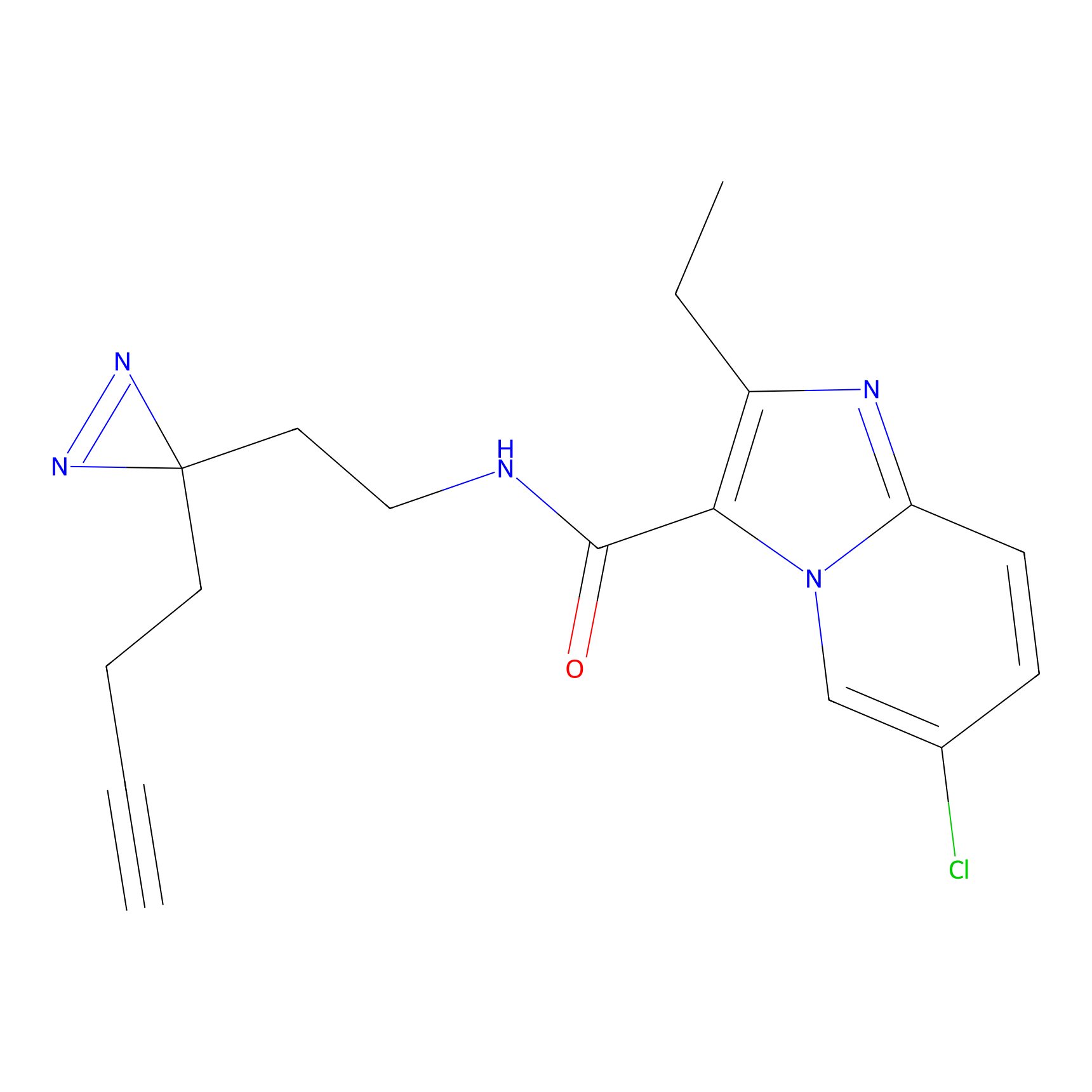
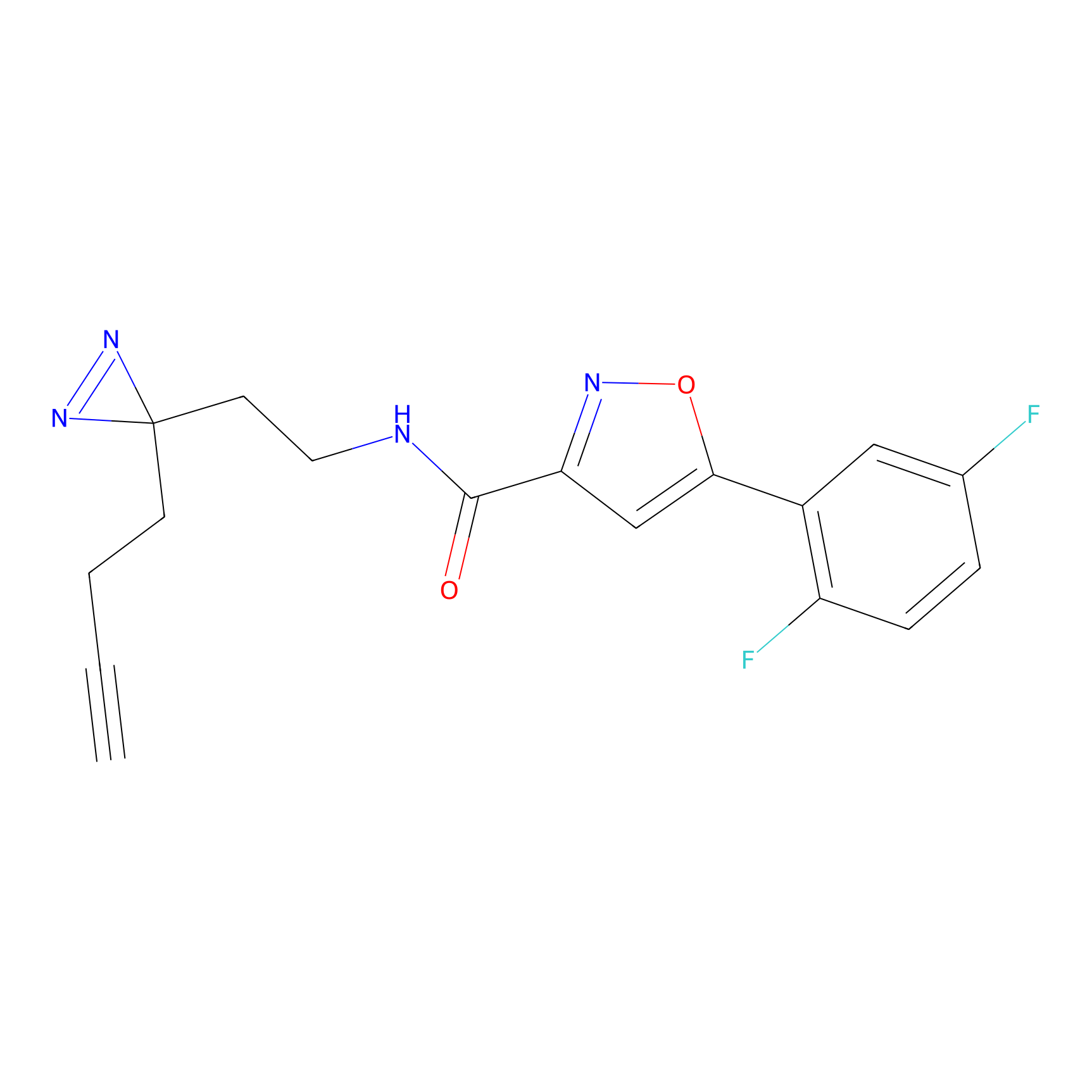
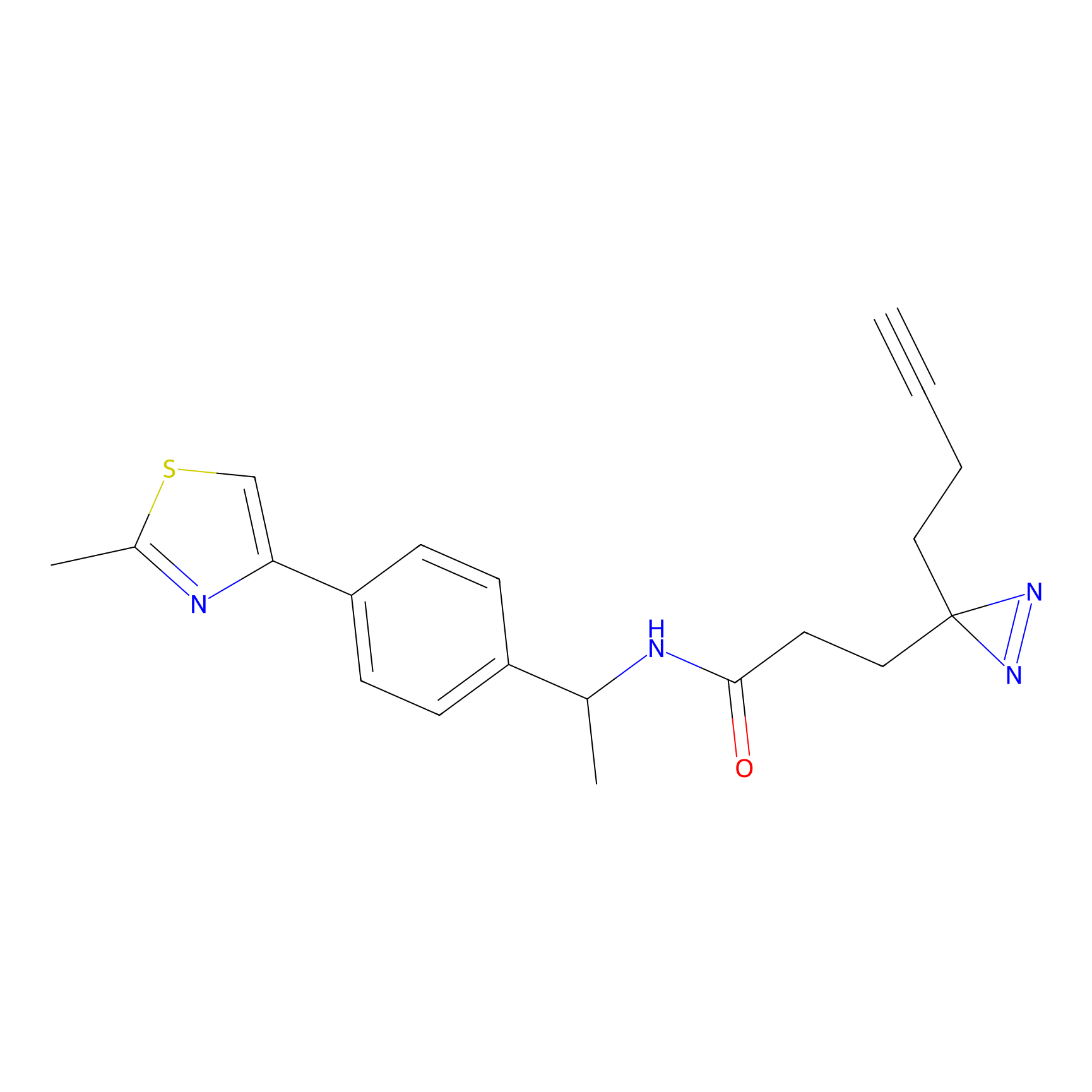
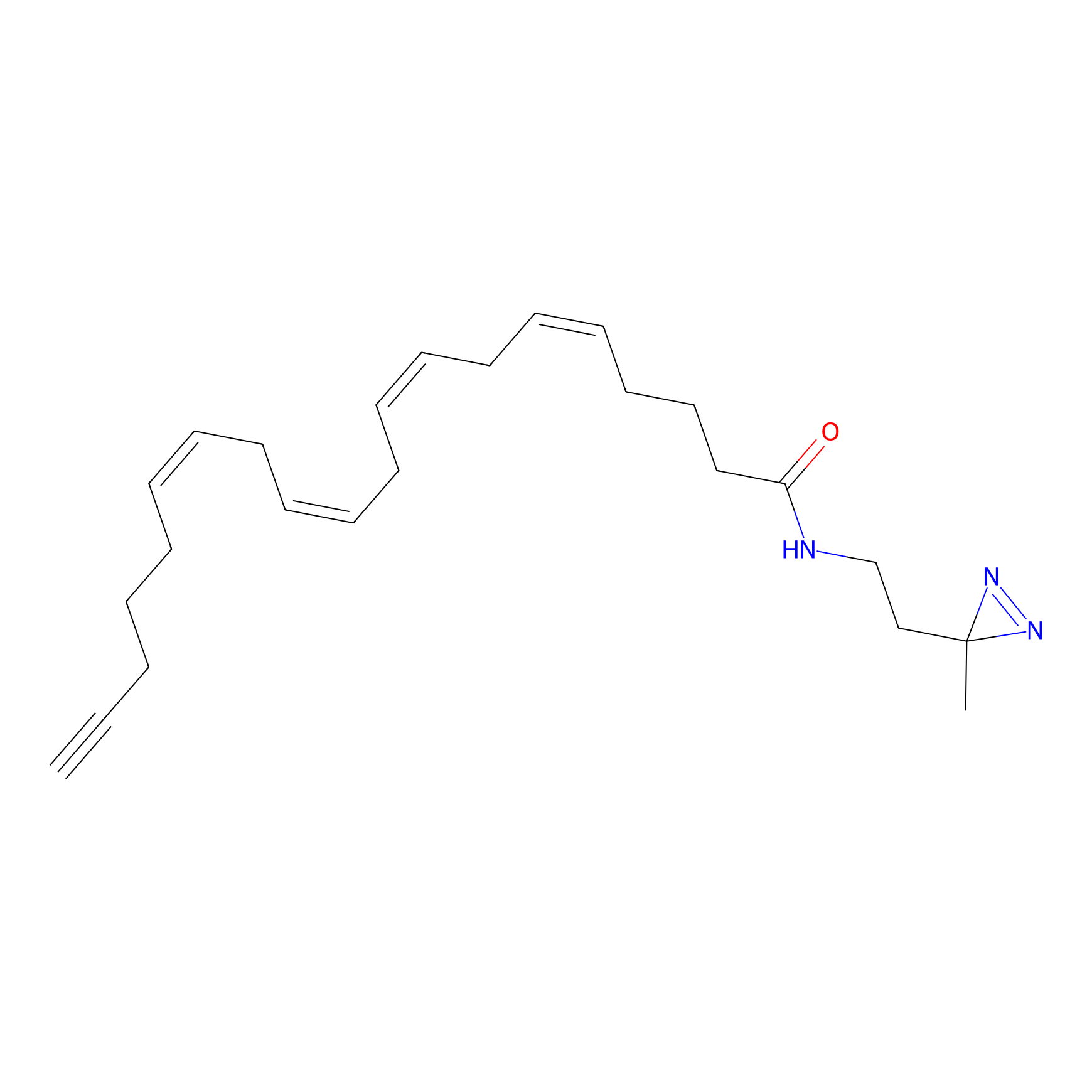
|
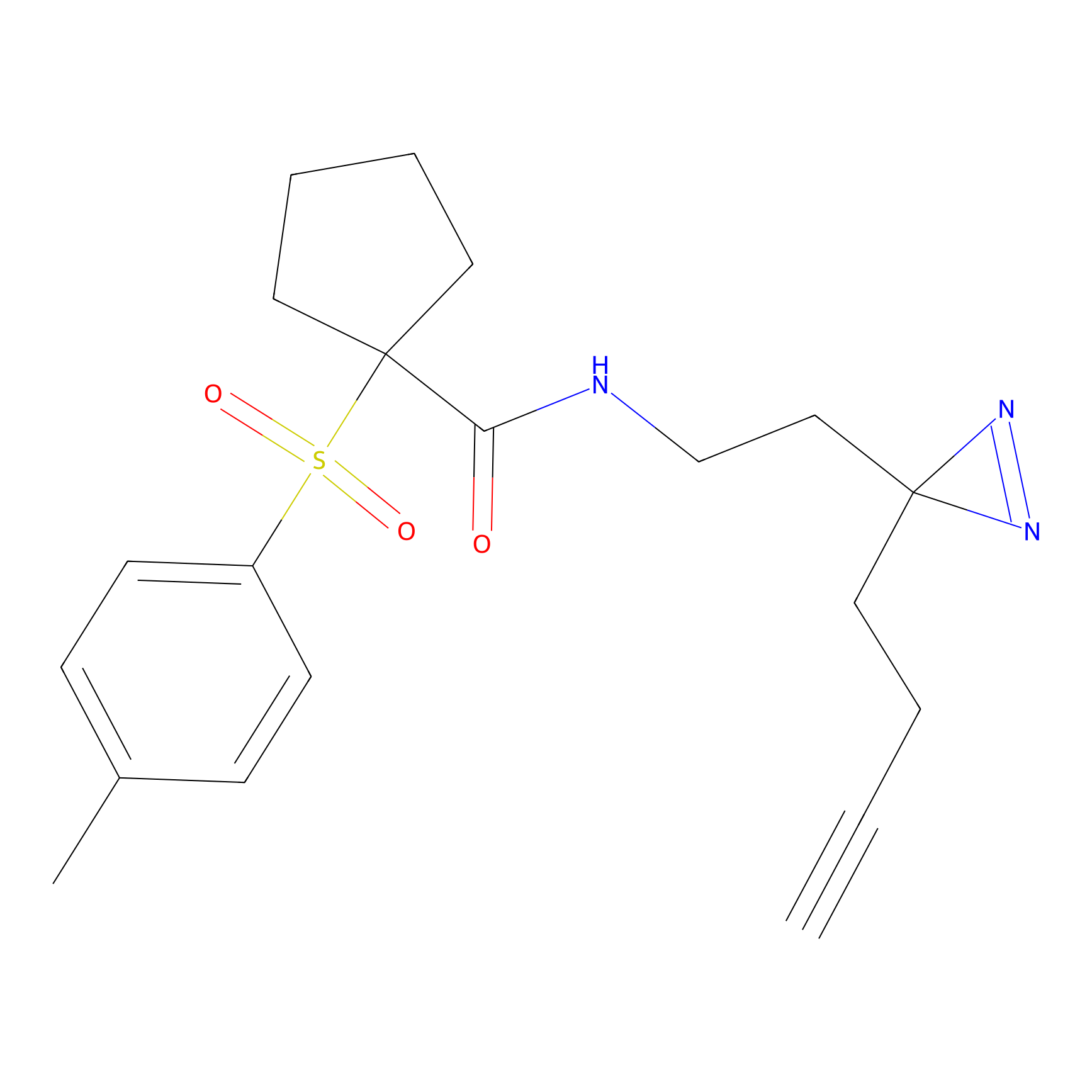
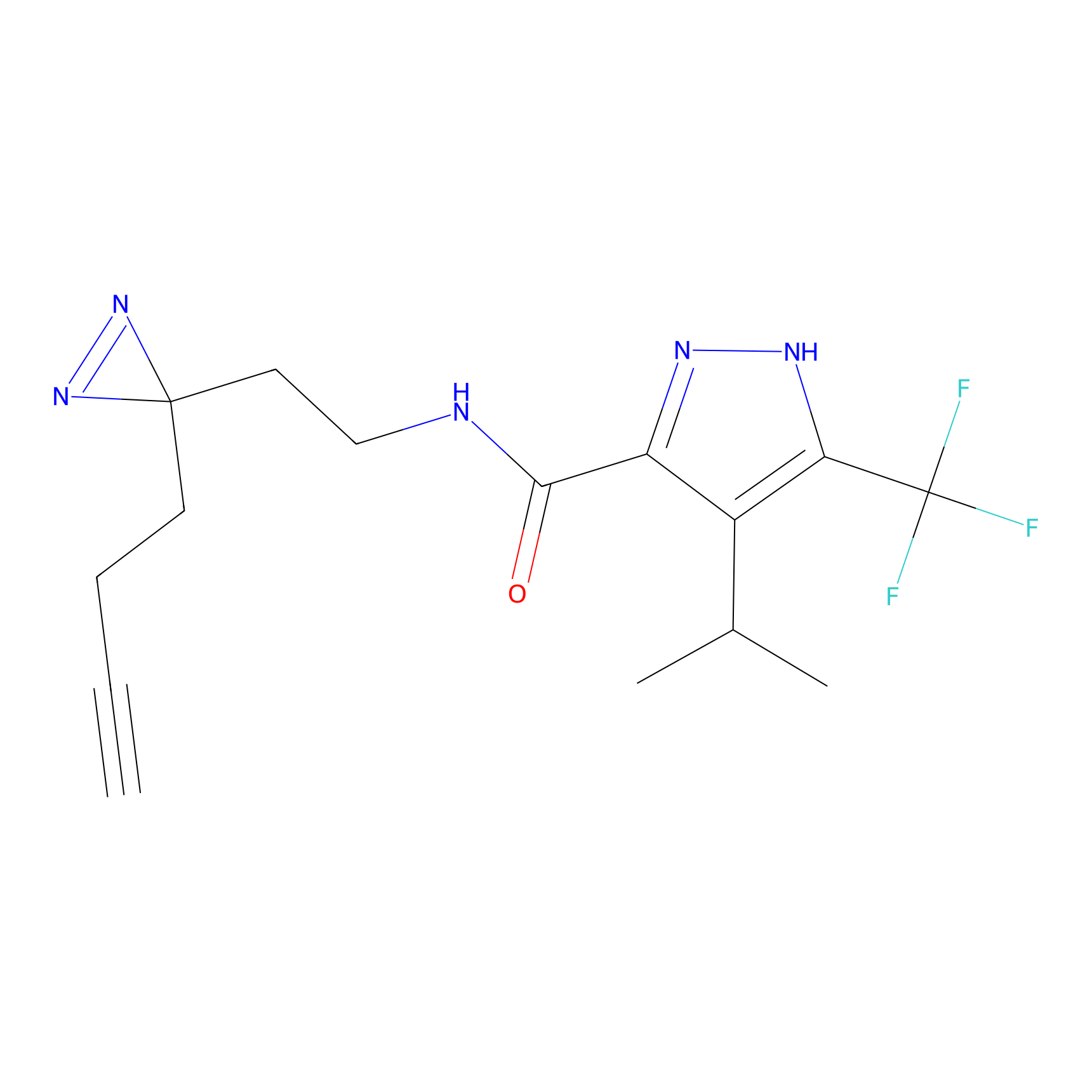
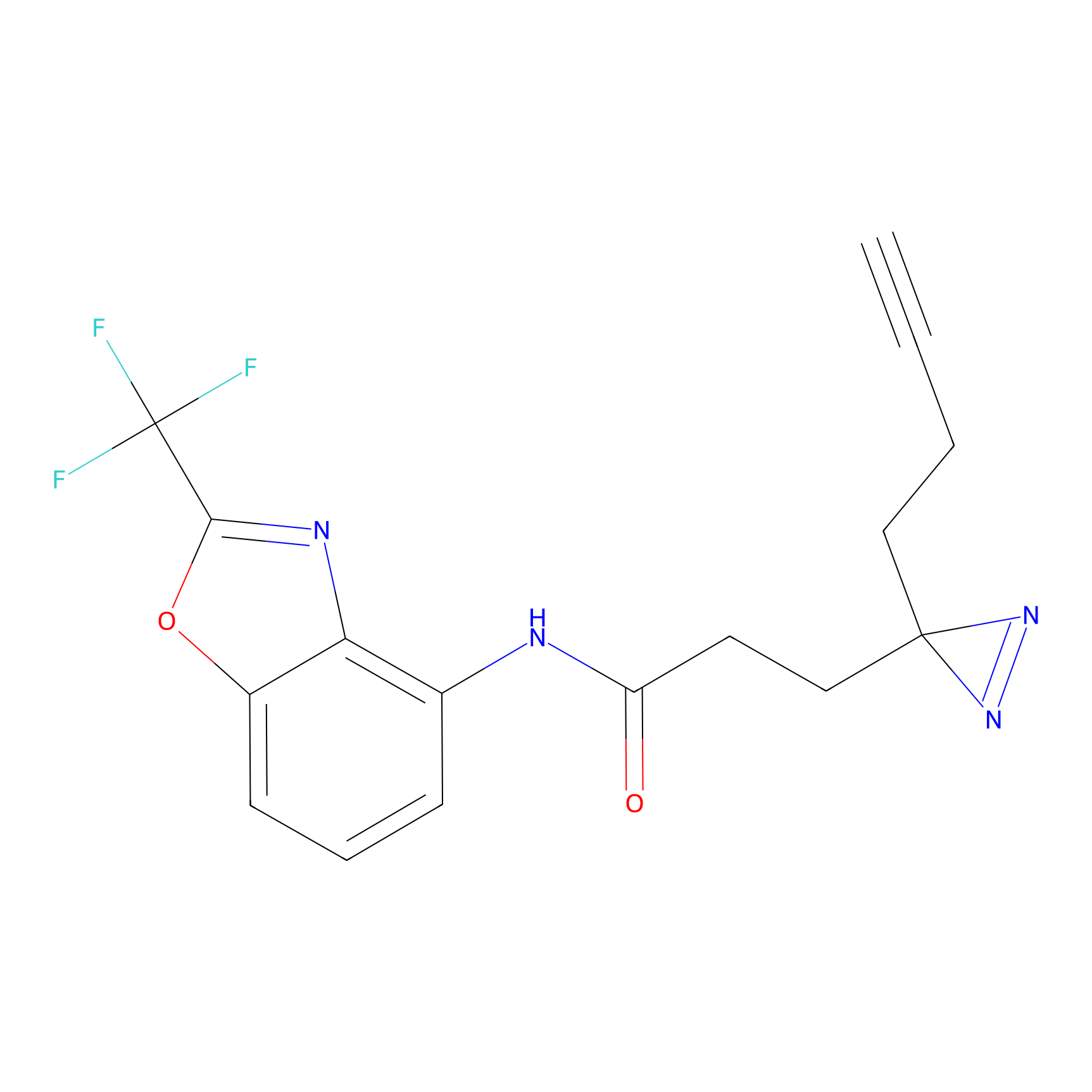
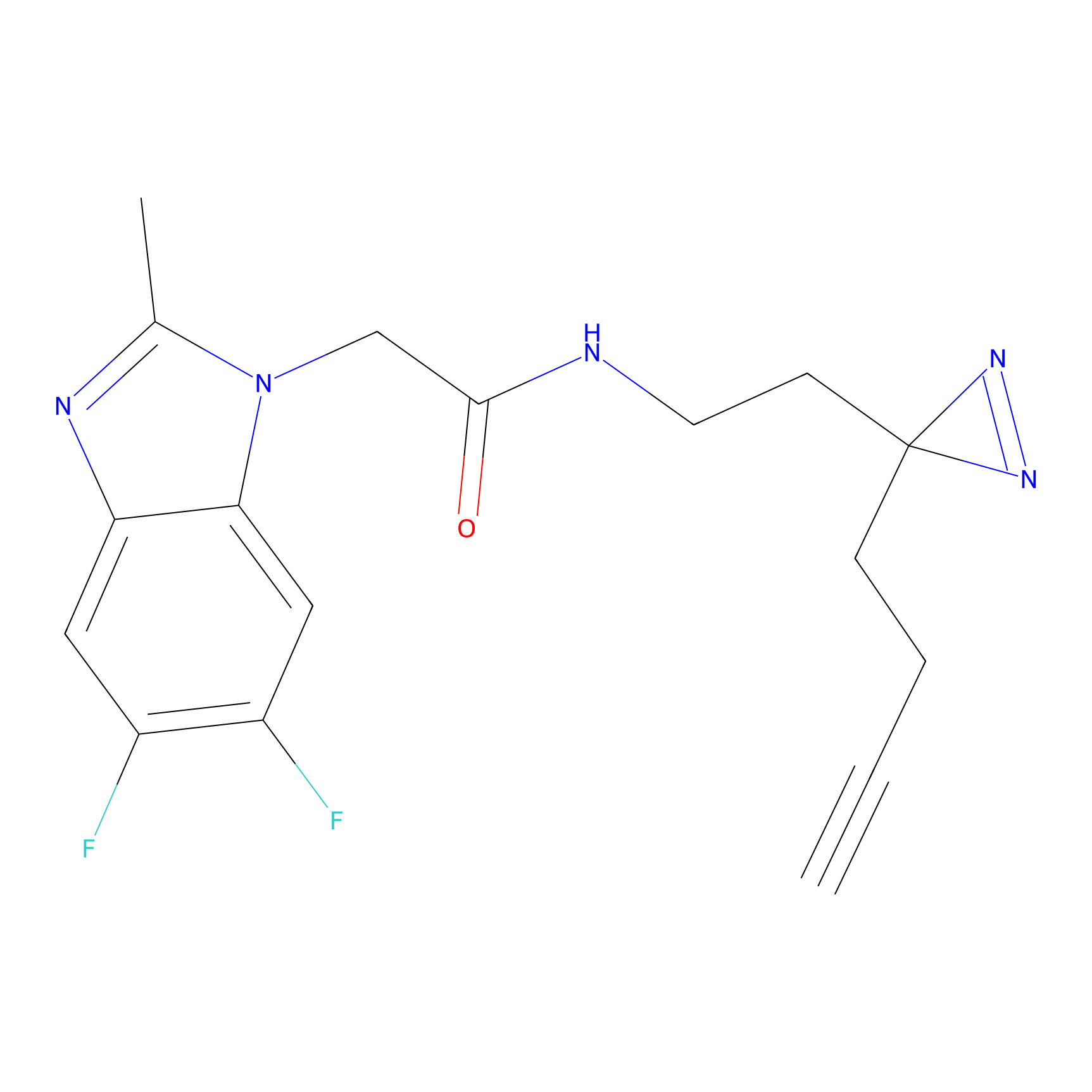
TH211 Probe Info |
 |
Y207(20.00) | LDD0260 | [3] | |
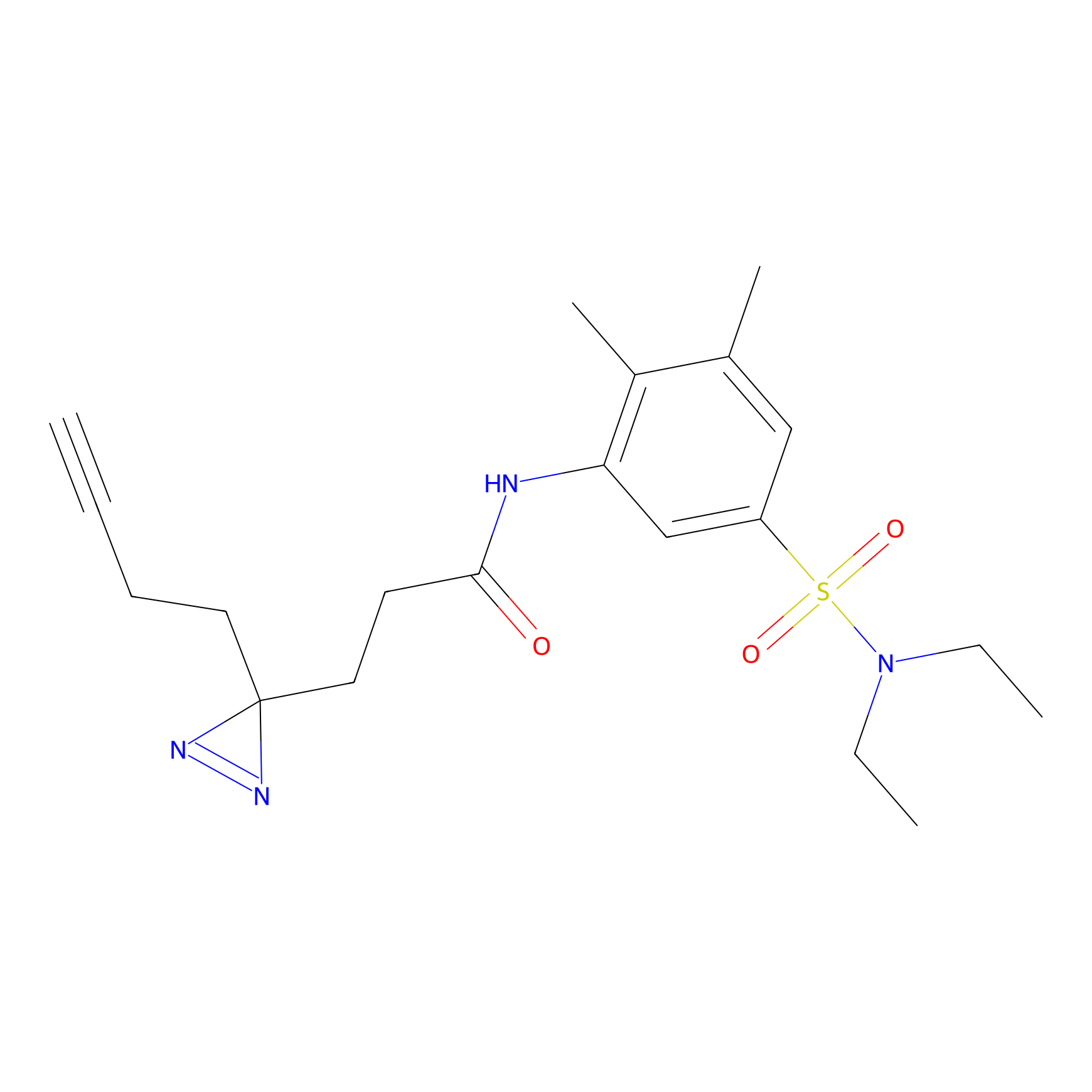
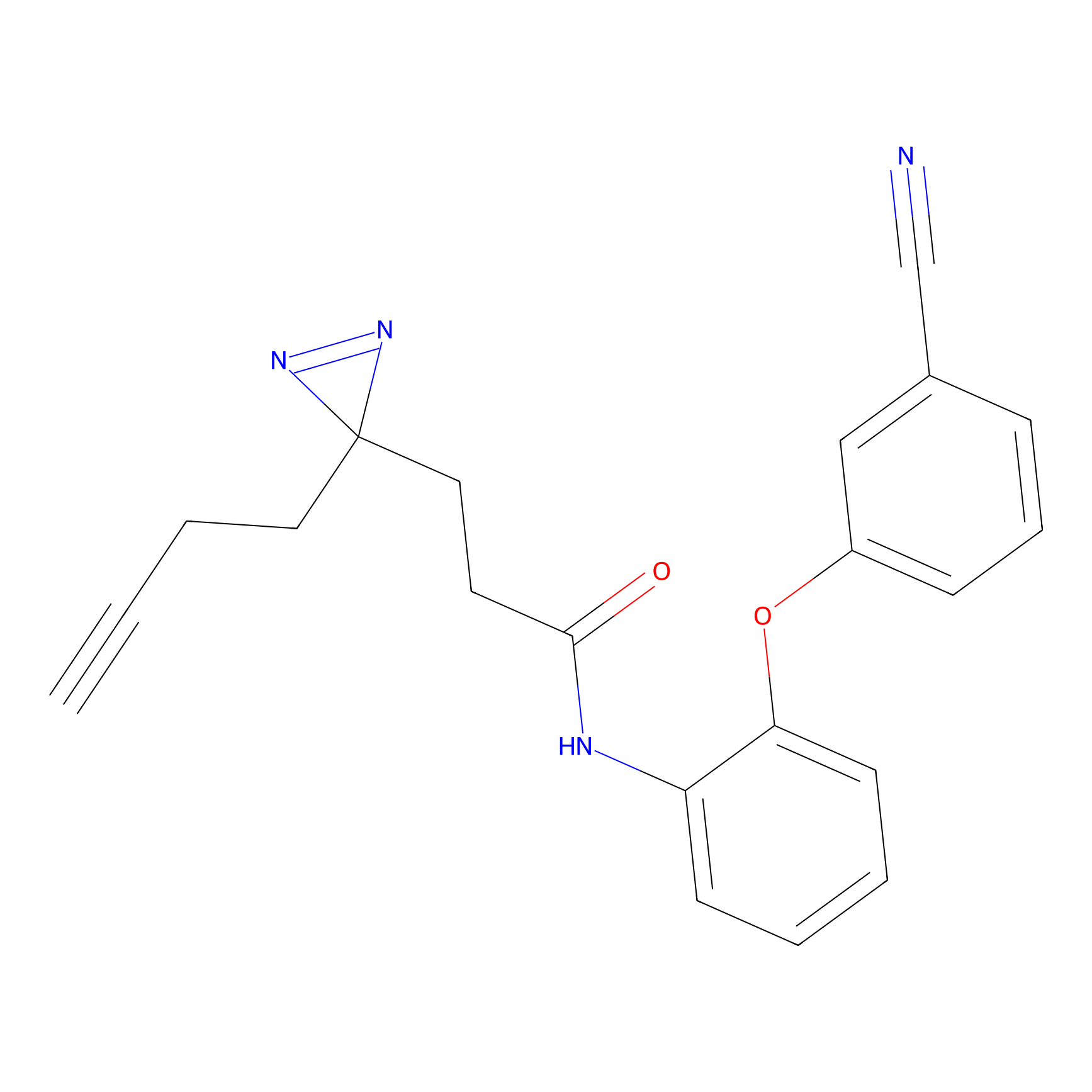
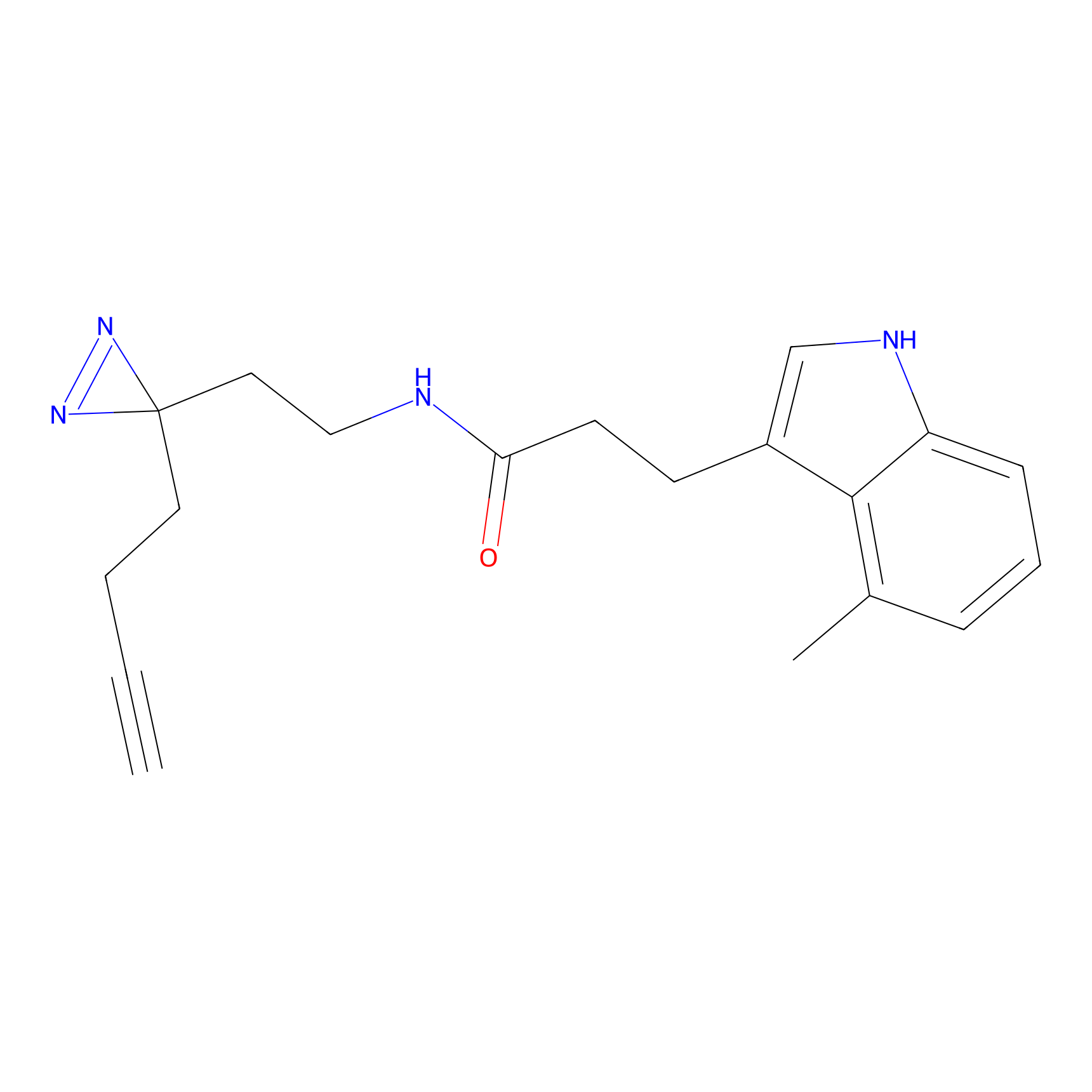
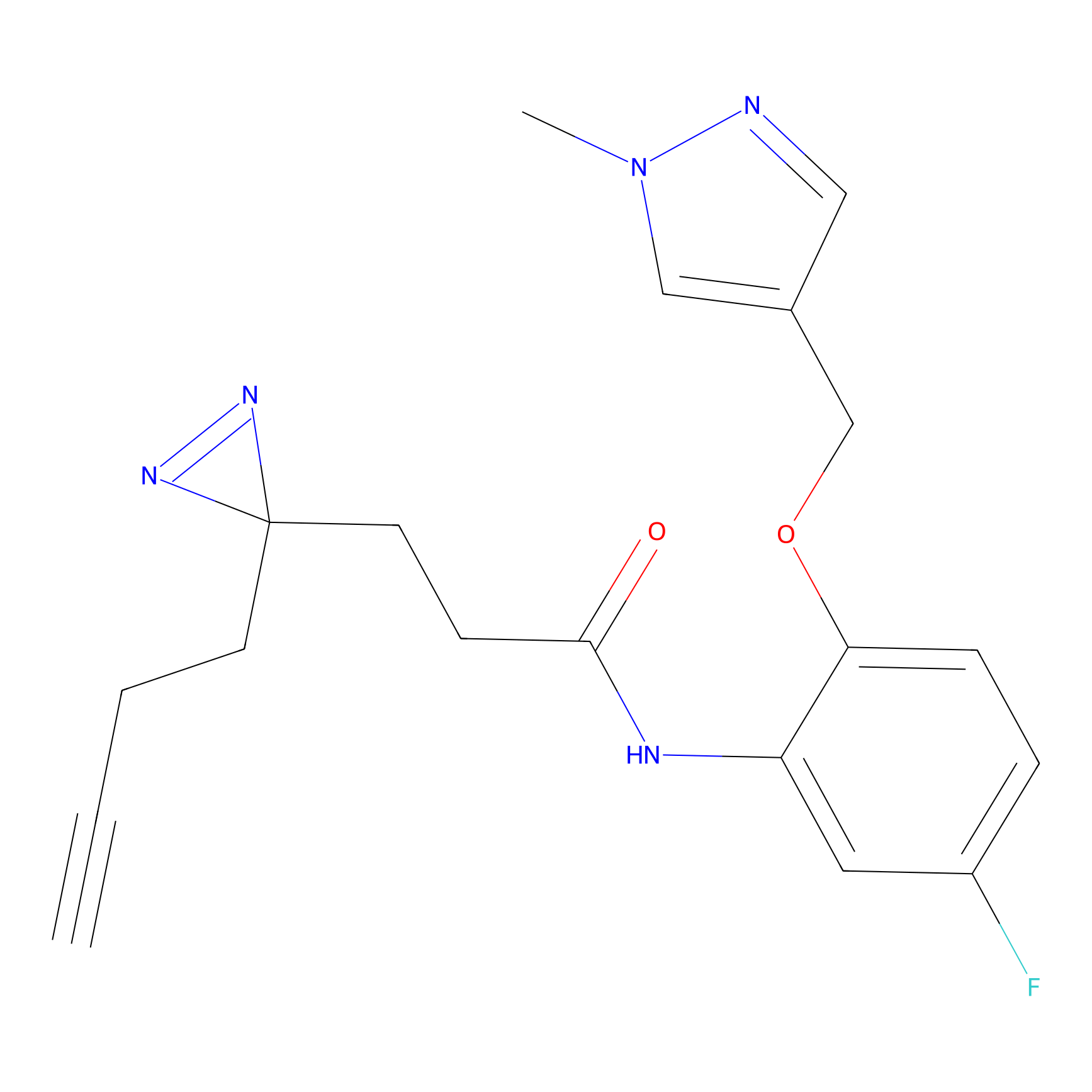
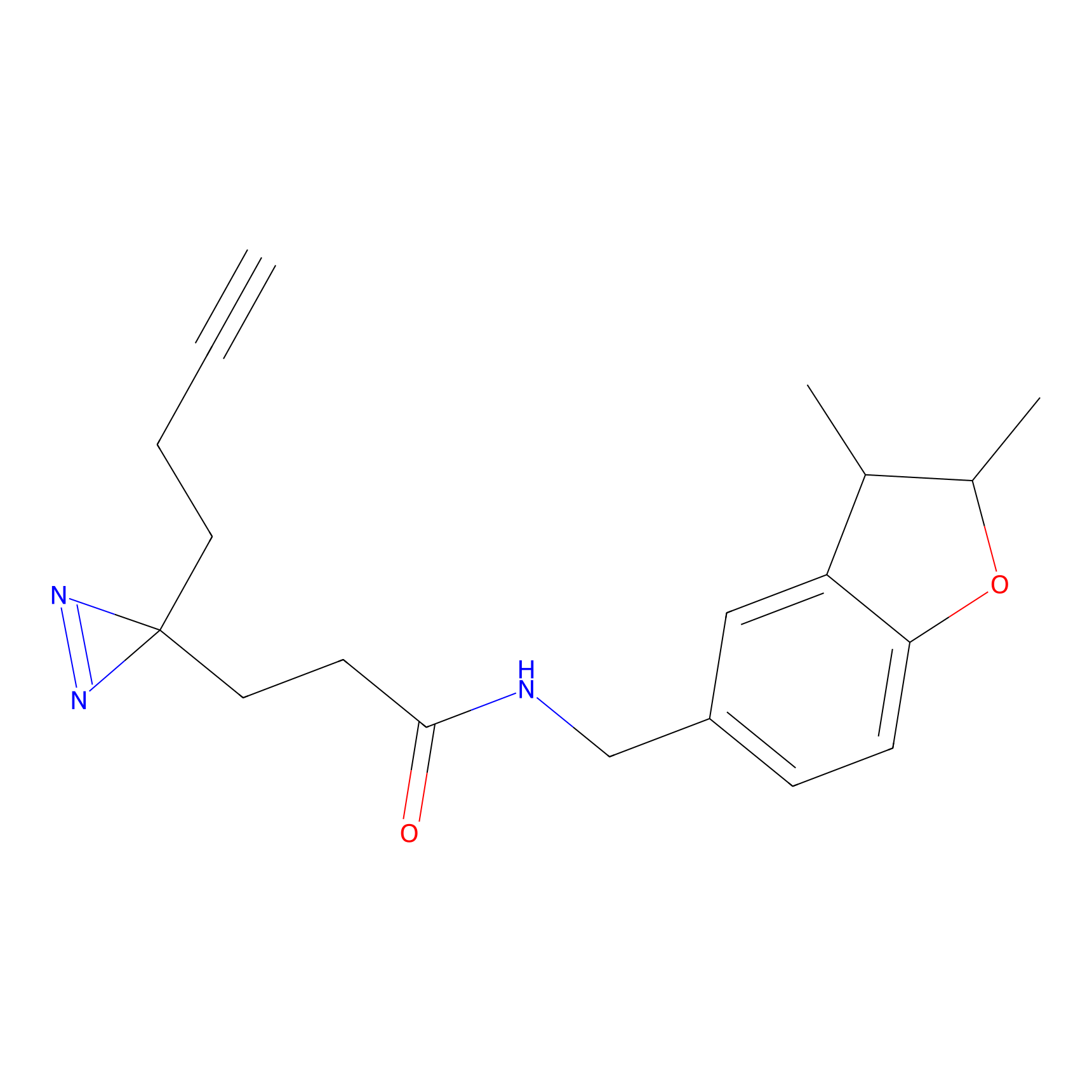
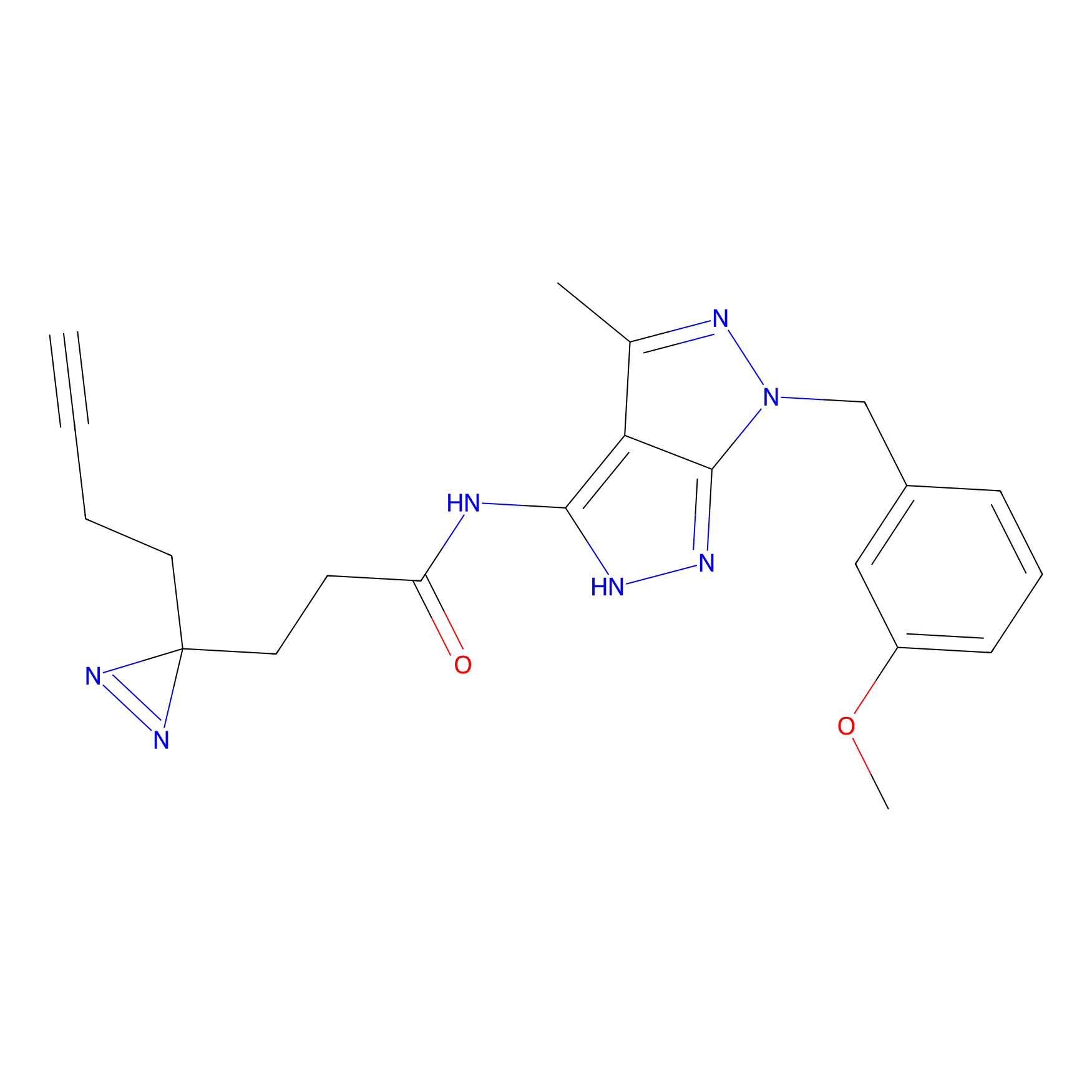
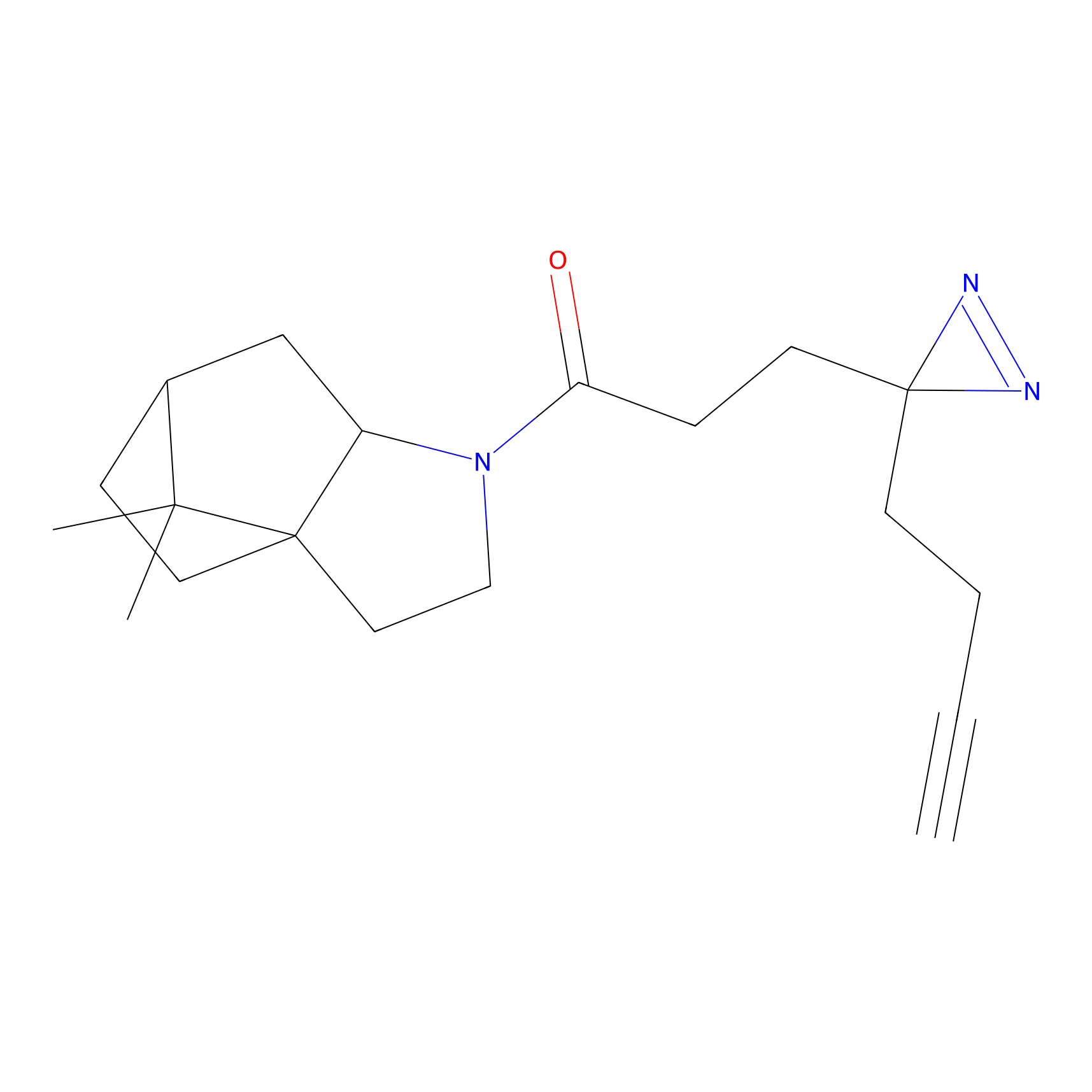
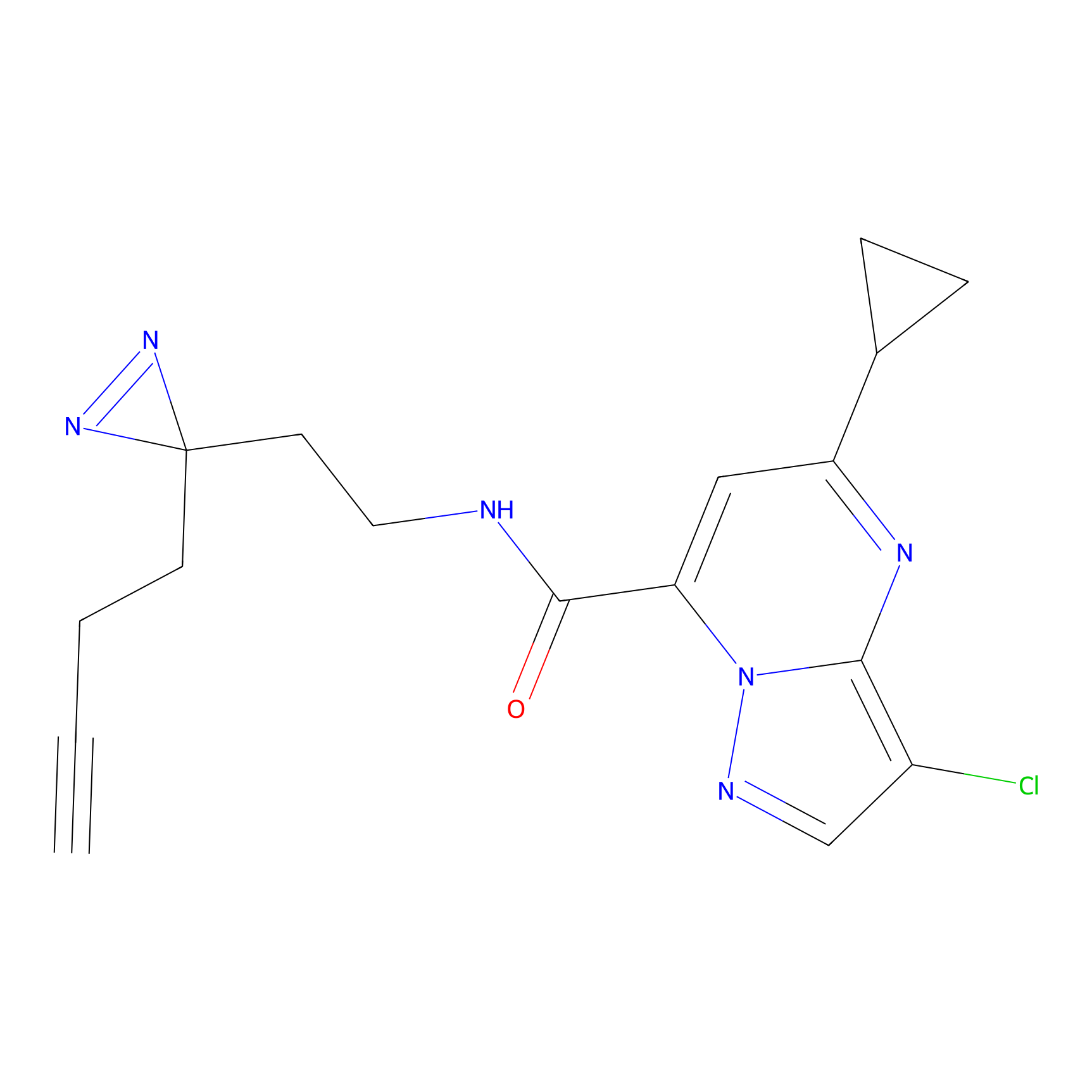
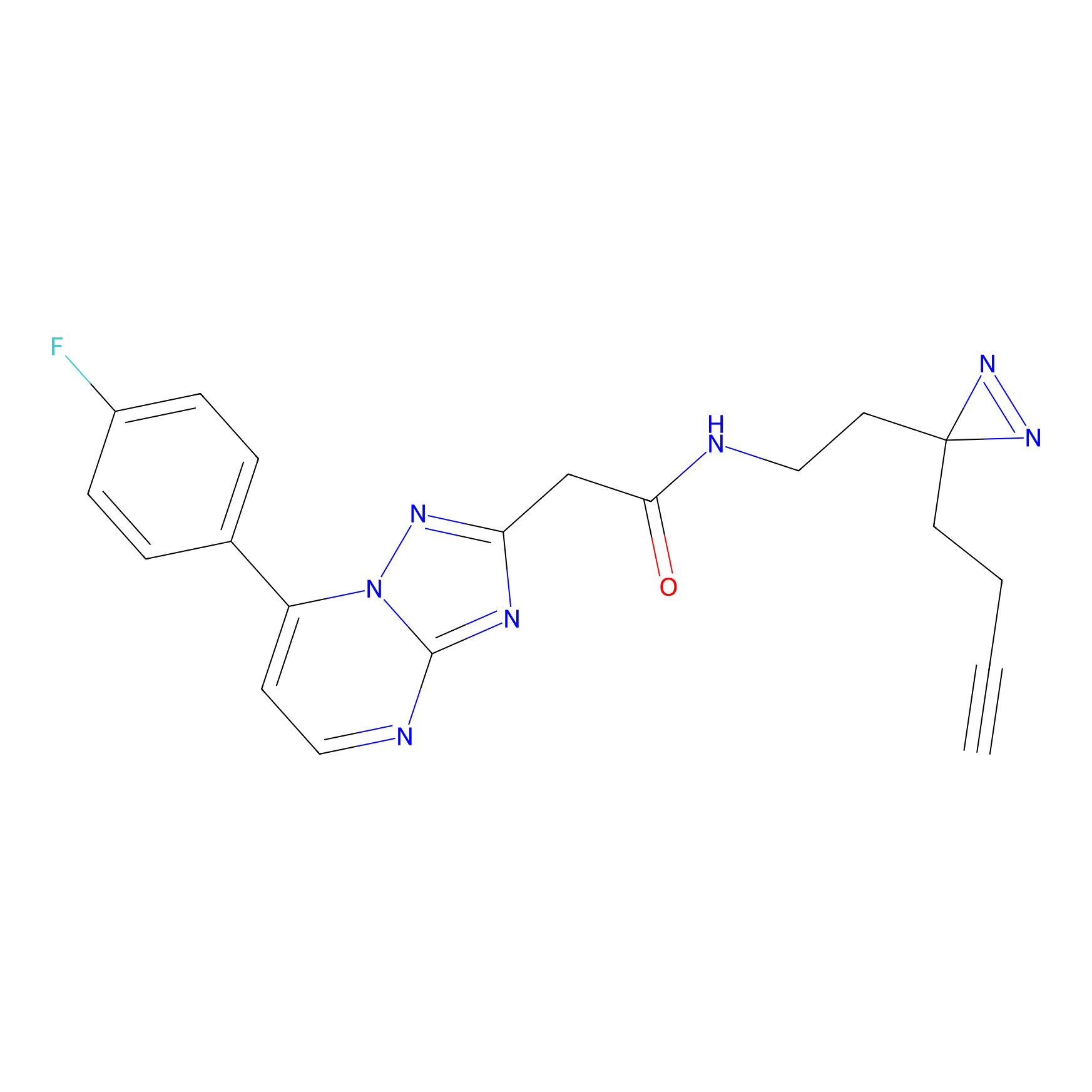
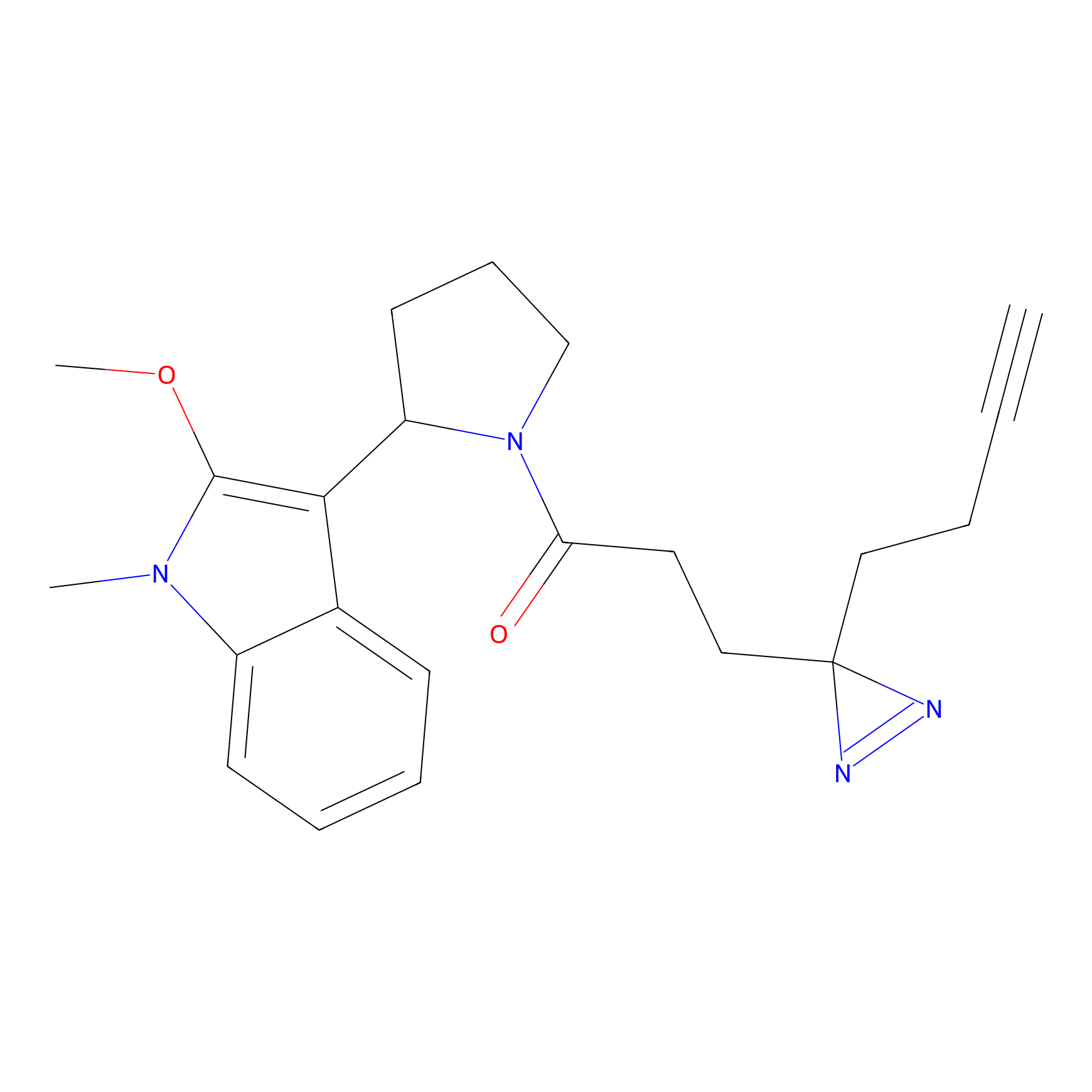
|
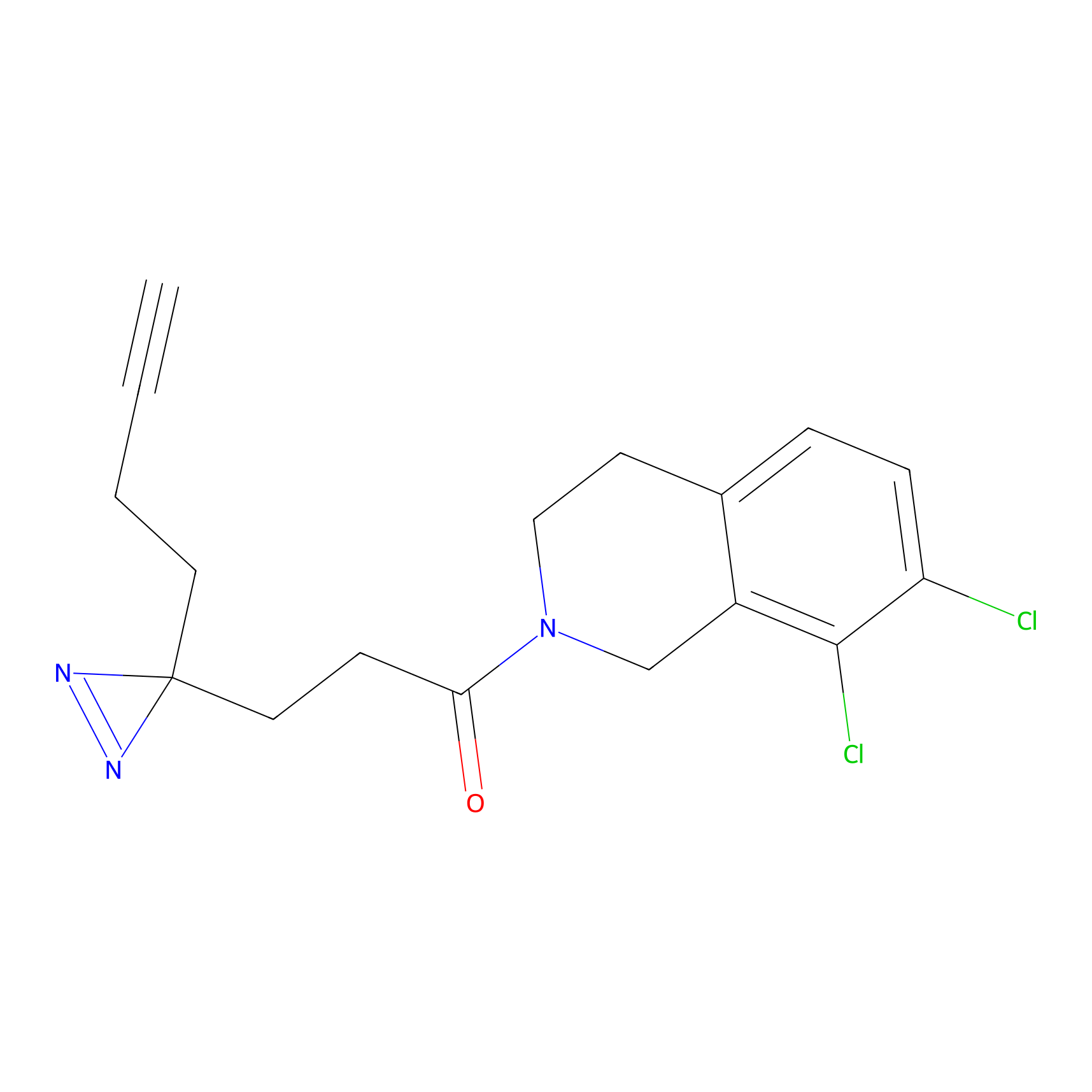
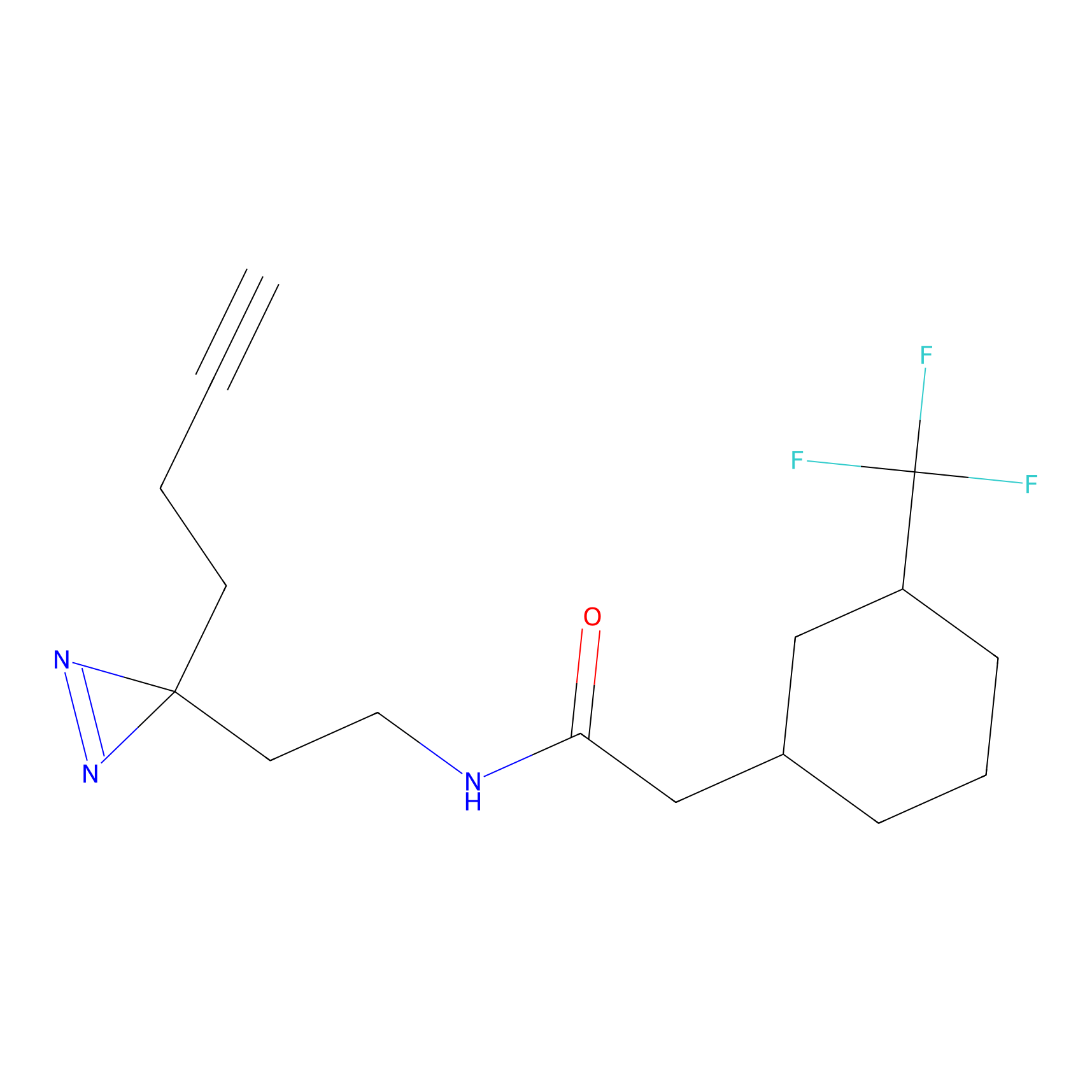
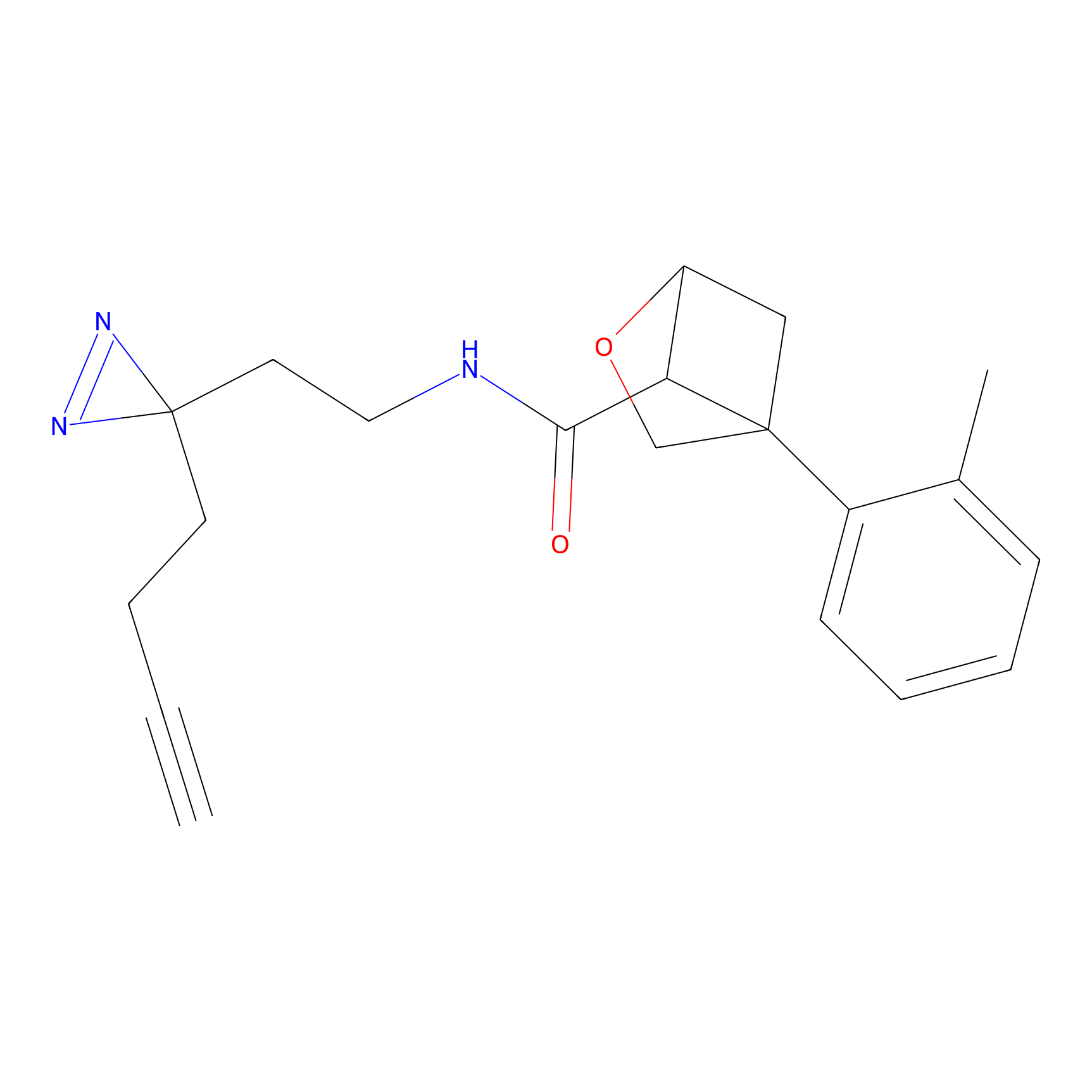
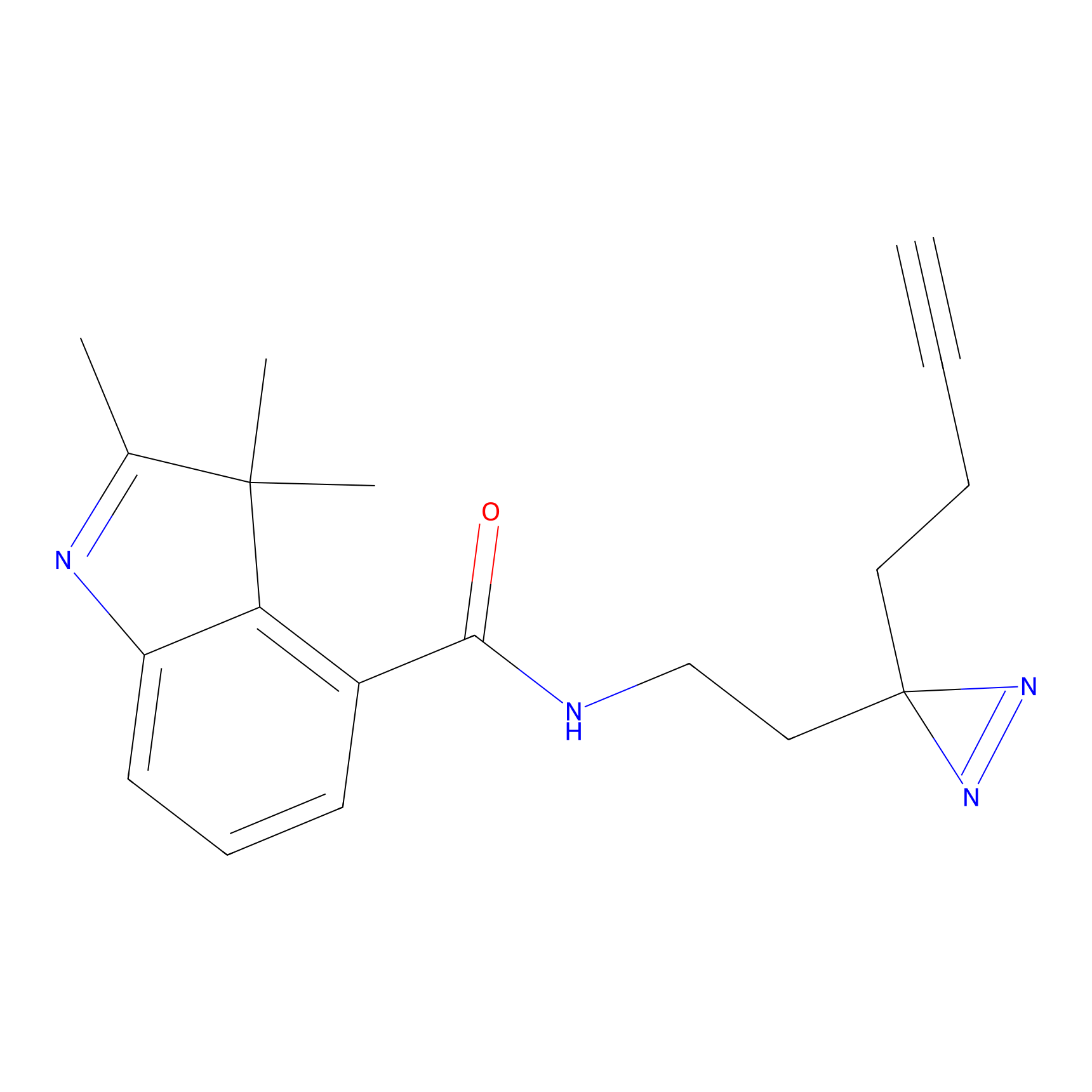
C-Sul Probe Info |
 |
2.69 | LDD0066 | [4] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0223 | [5] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [5] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Ribosome quality control complex subunit TCF25 (TCF25) | TCF25 family | Q9BQ70 | |||
GPCR
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Free fatty acid receptor 2 (FFAR2) | G-protein coupled receptor 1 family | O15552 | |||
Immunoglobulin
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Fc receptor-like protein 4 (FCRL4) | . | Q96PJ5 | |||
Other
References