Details of the Target
General Information of Target
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
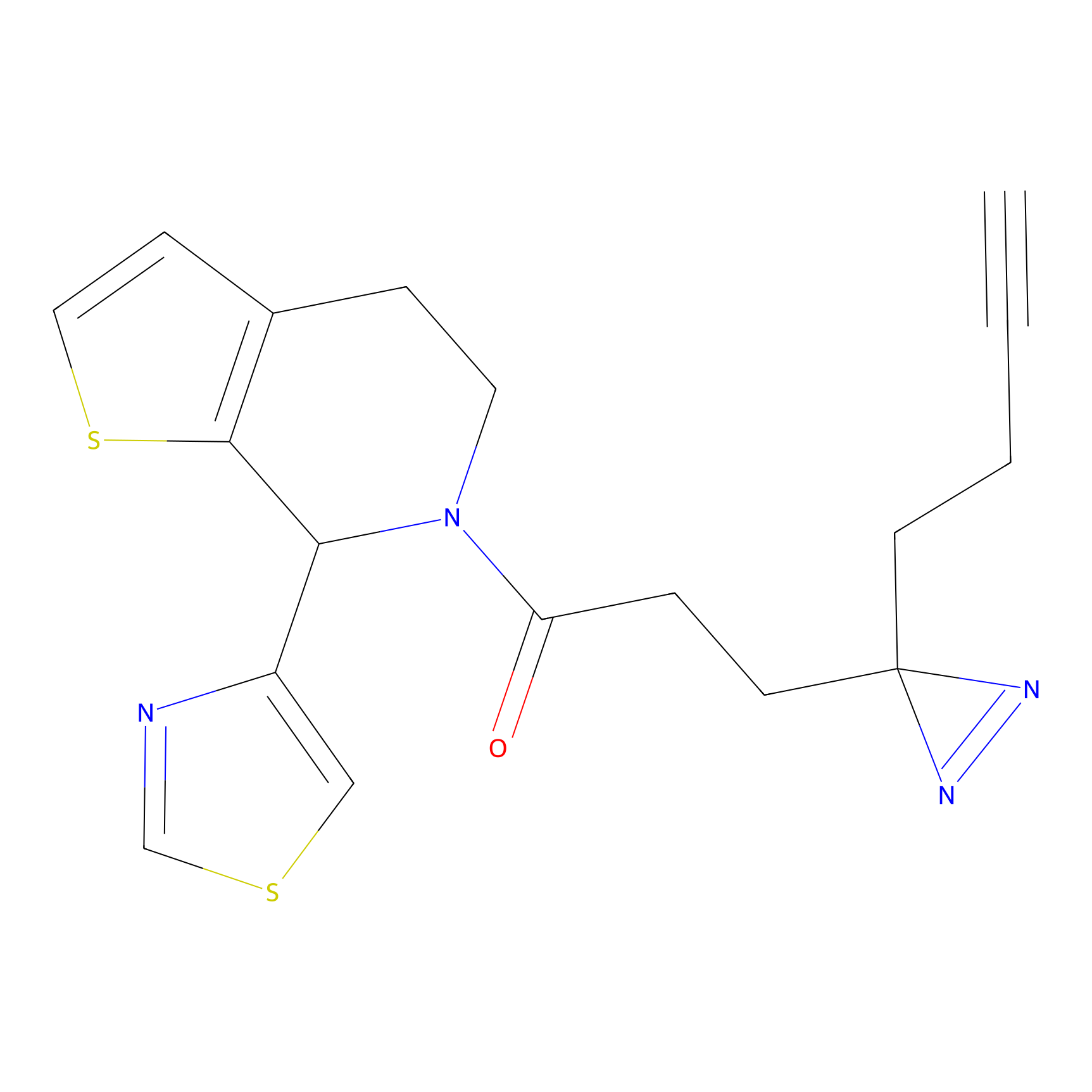
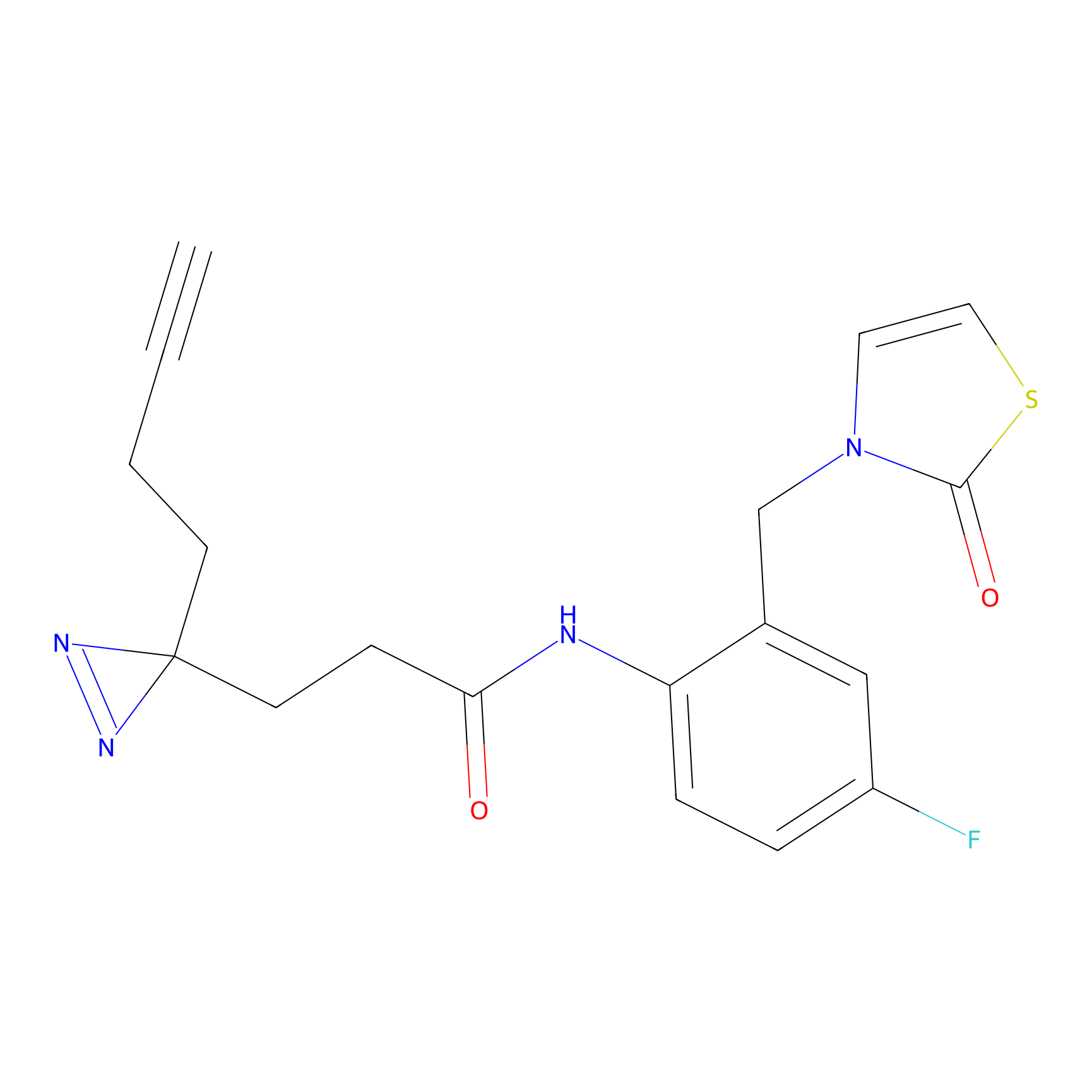
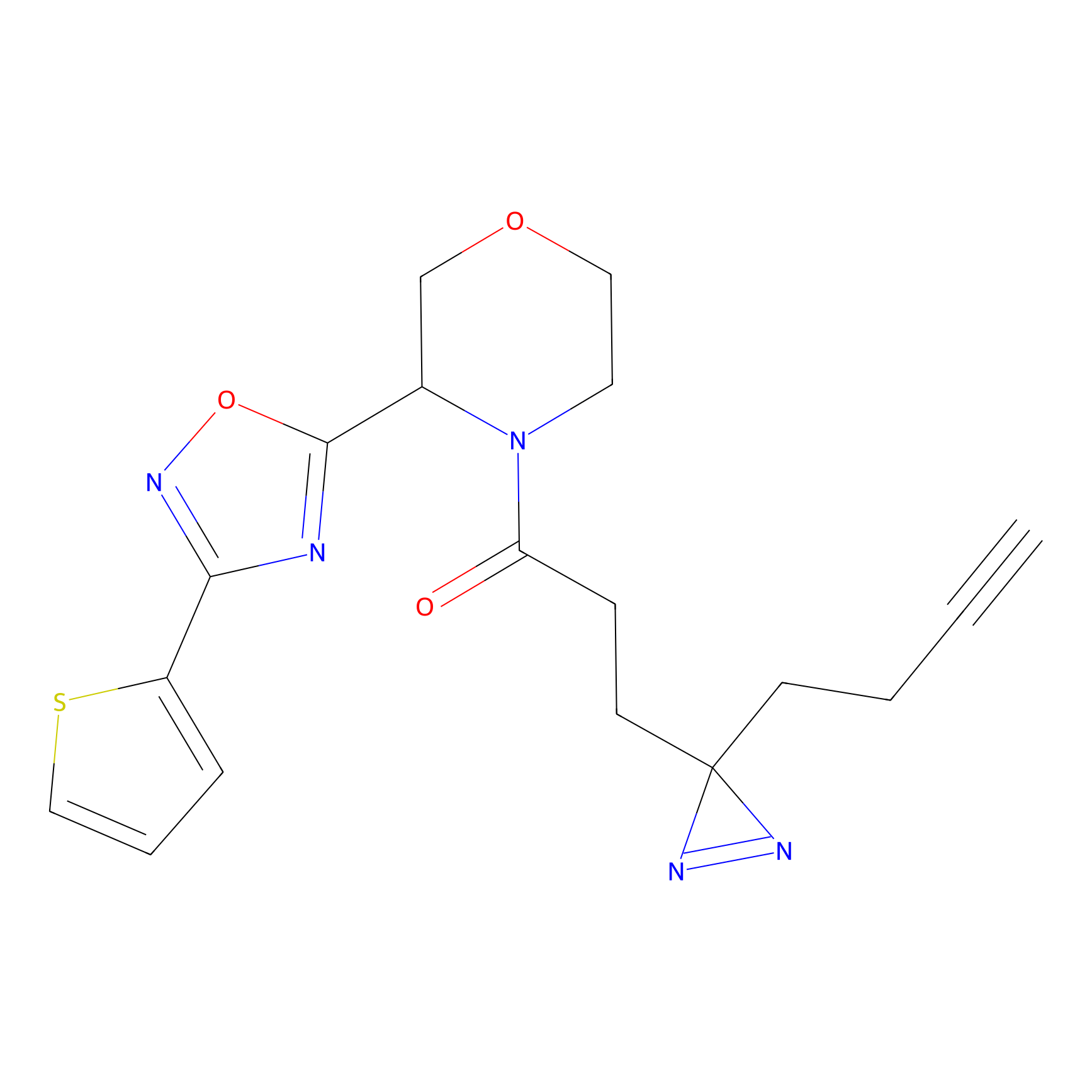
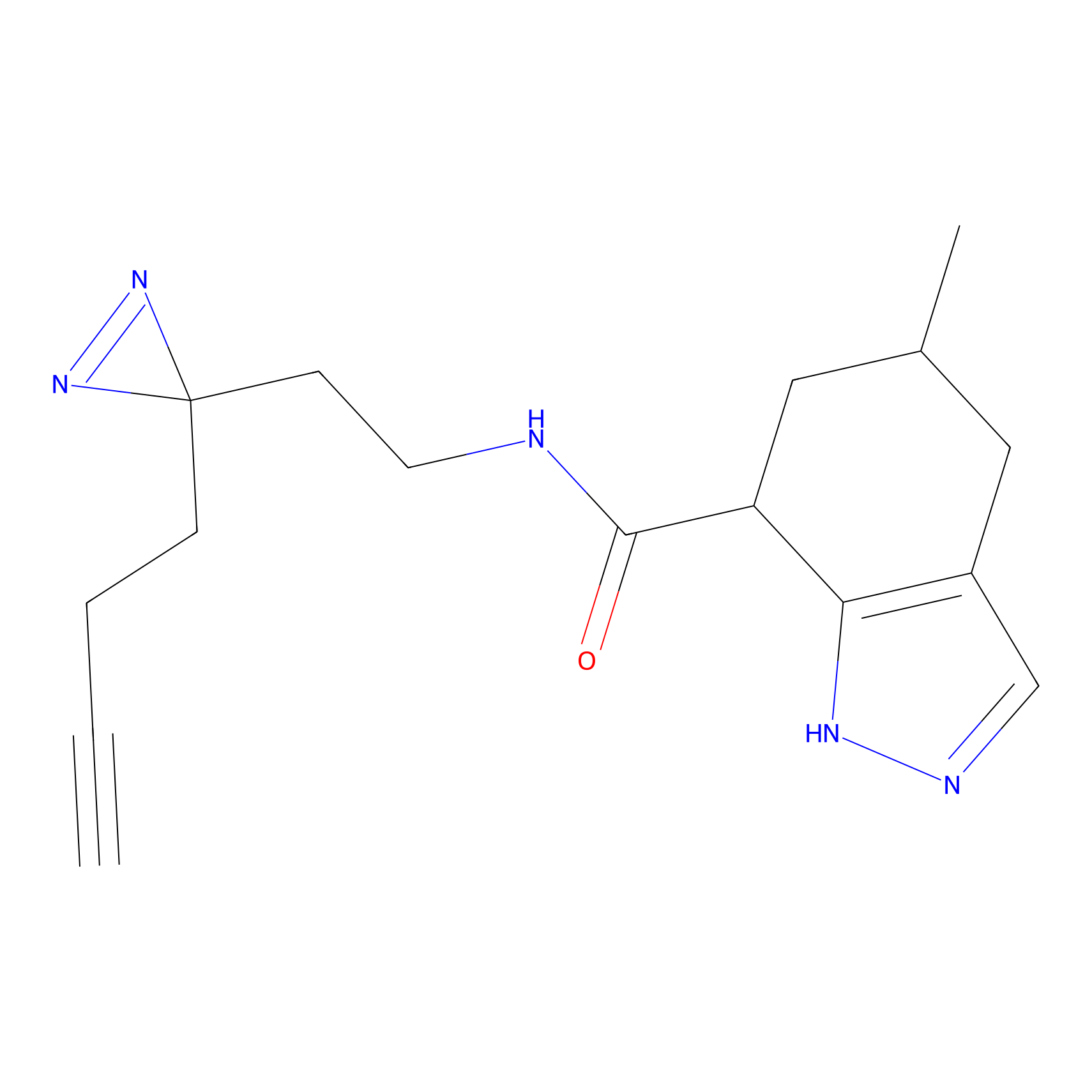
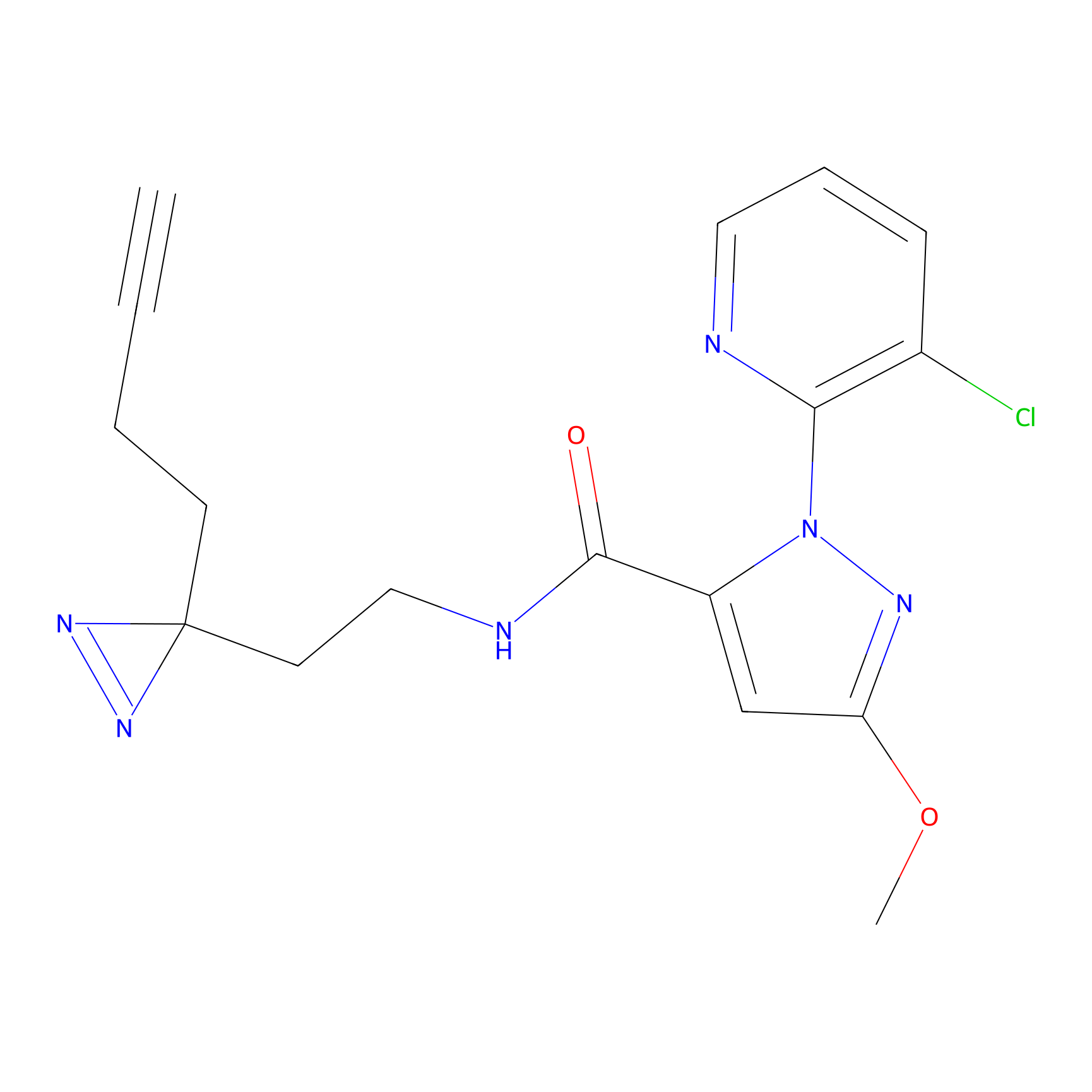
|
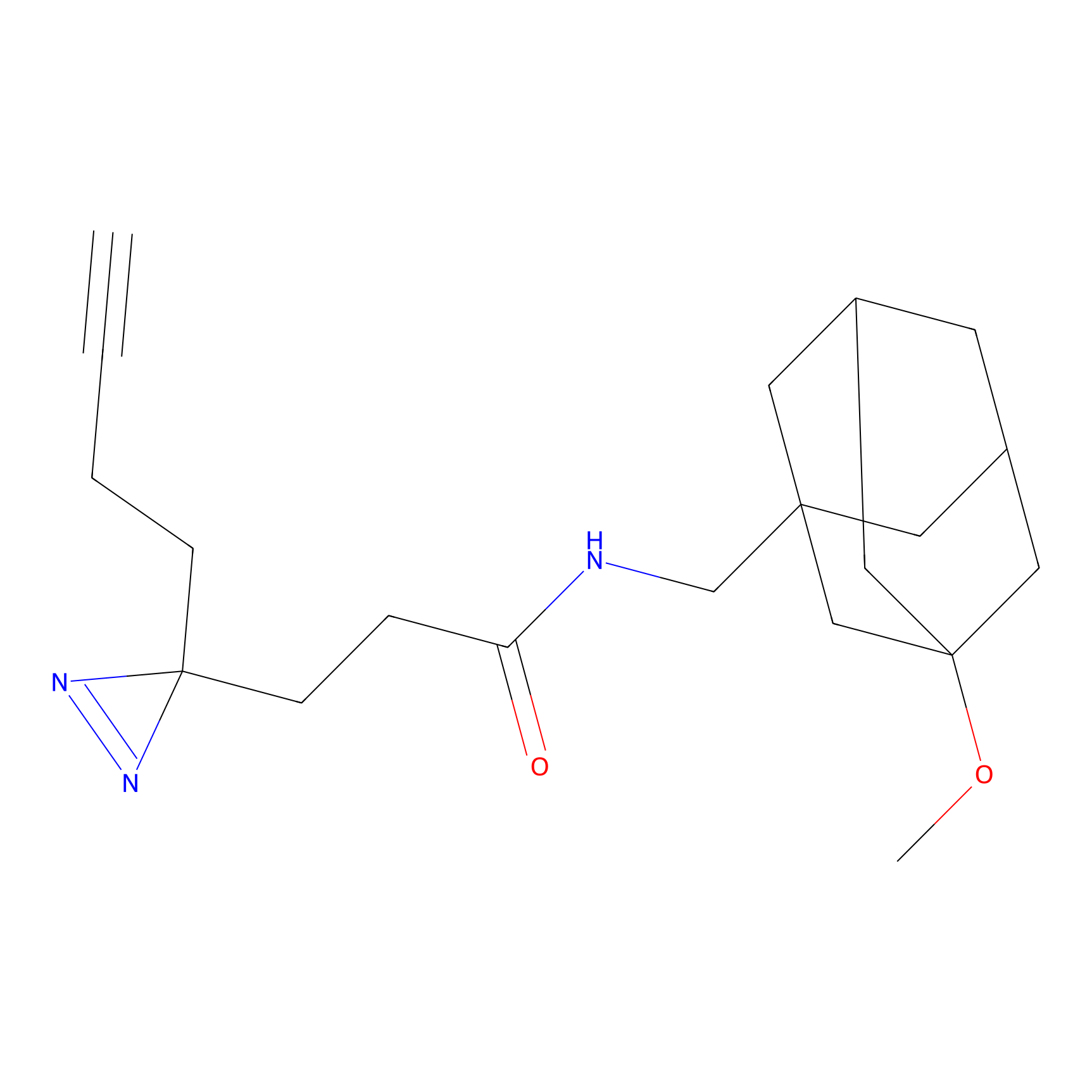
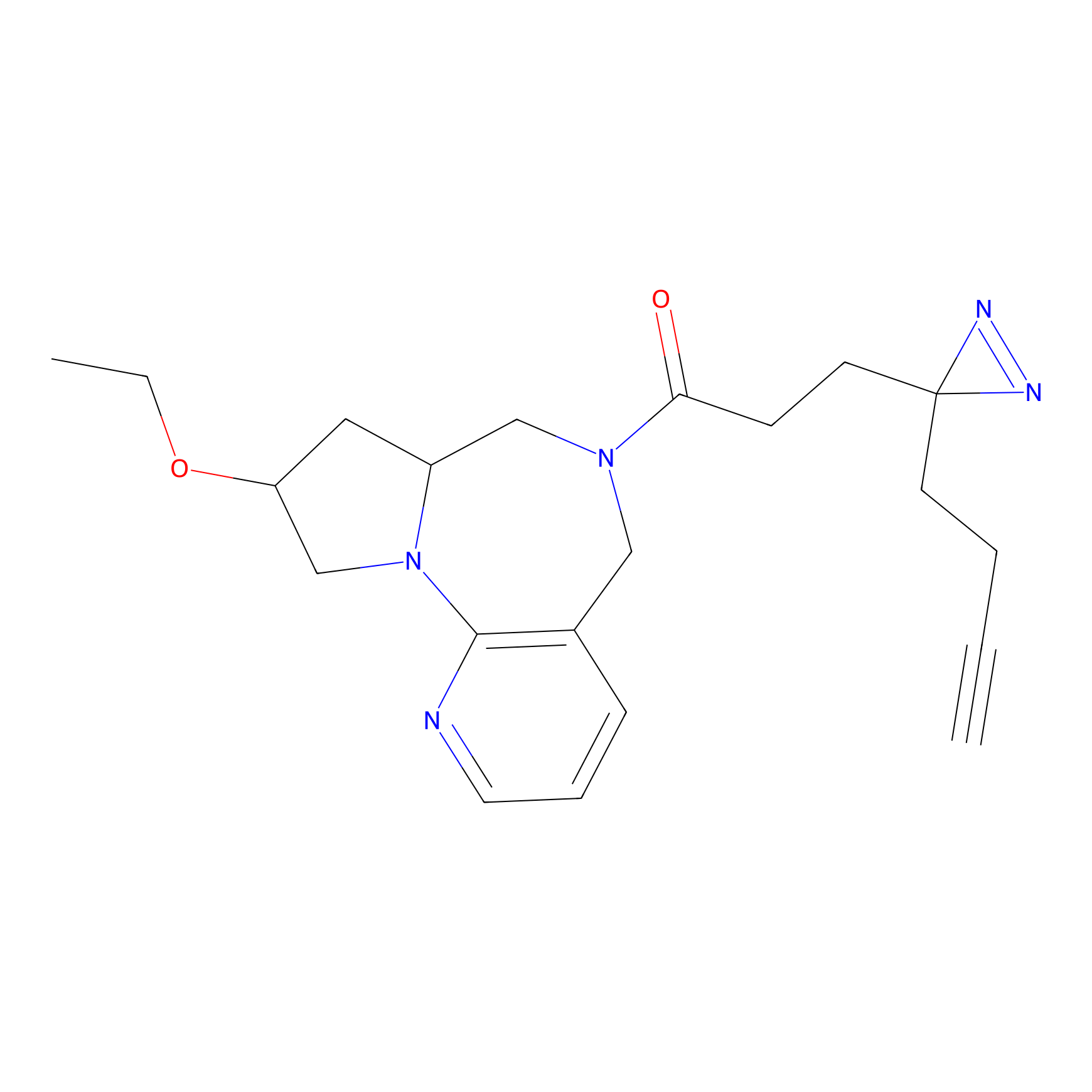
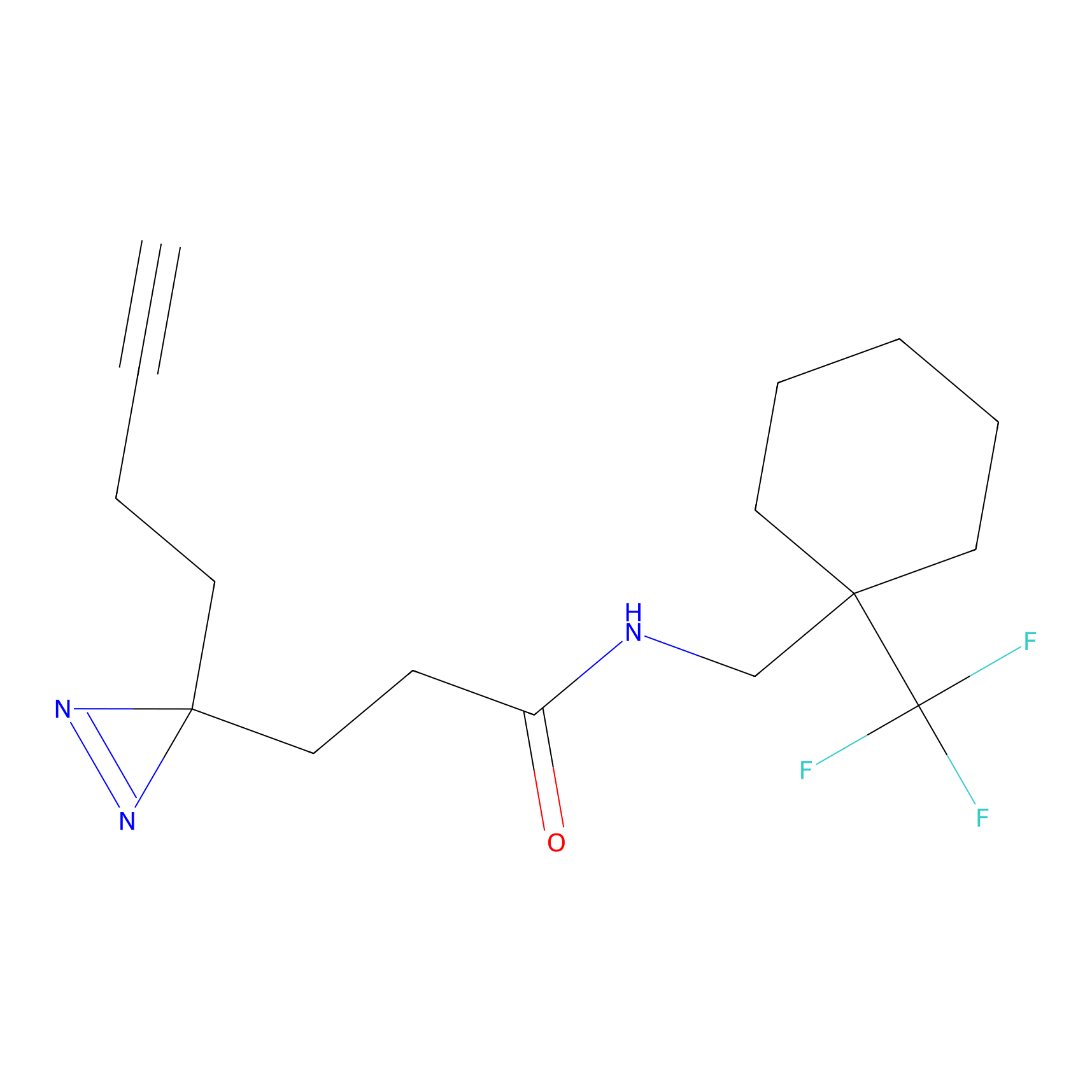
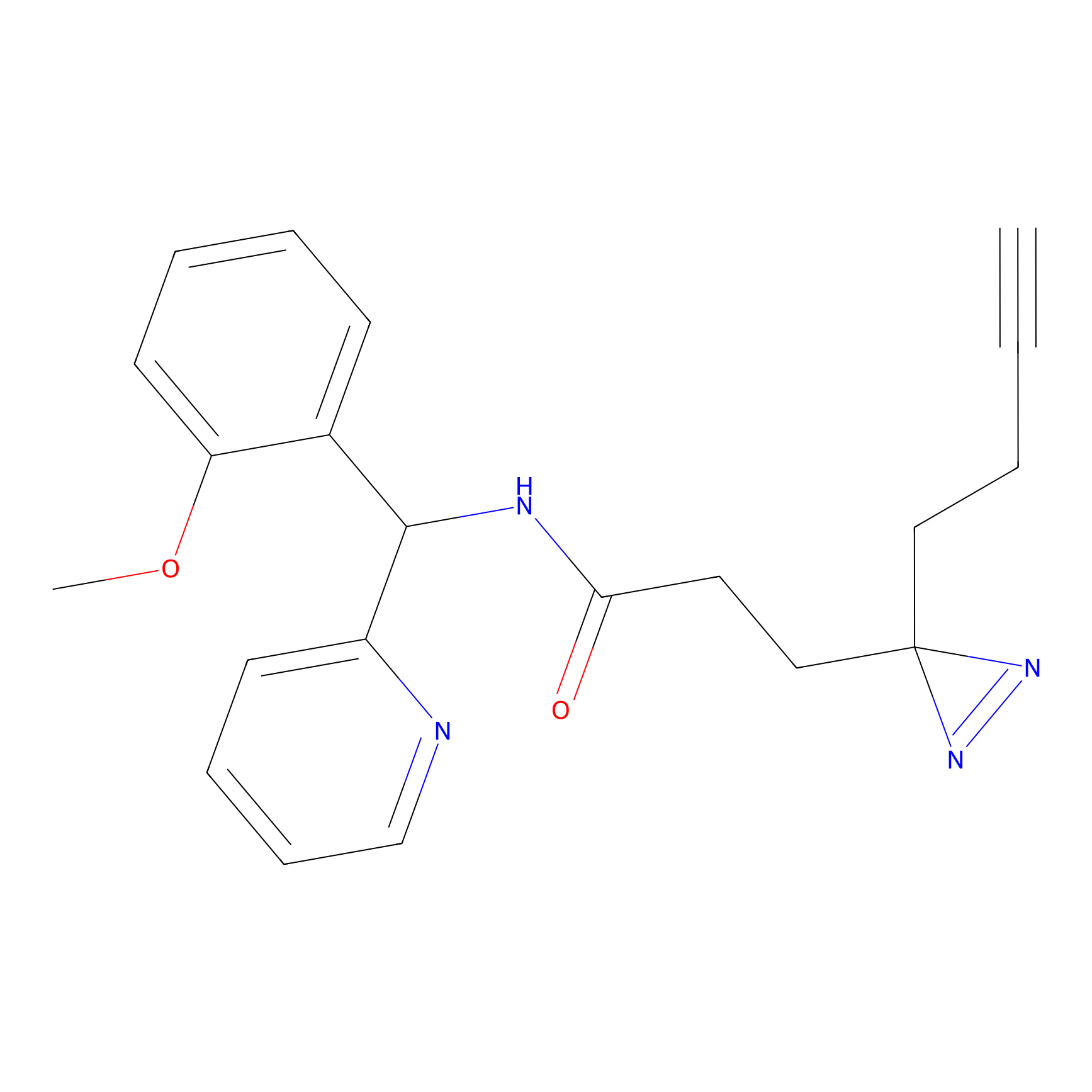
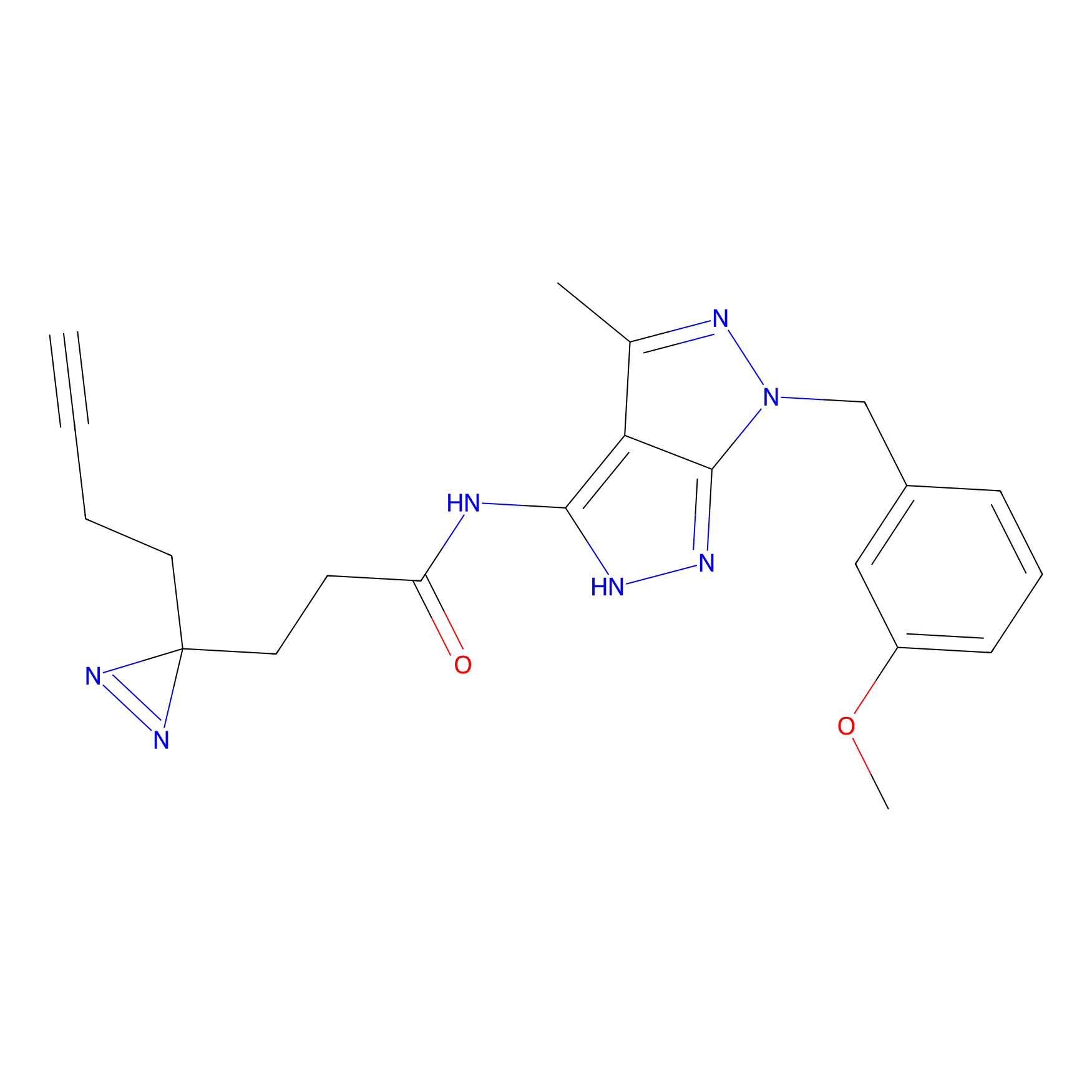
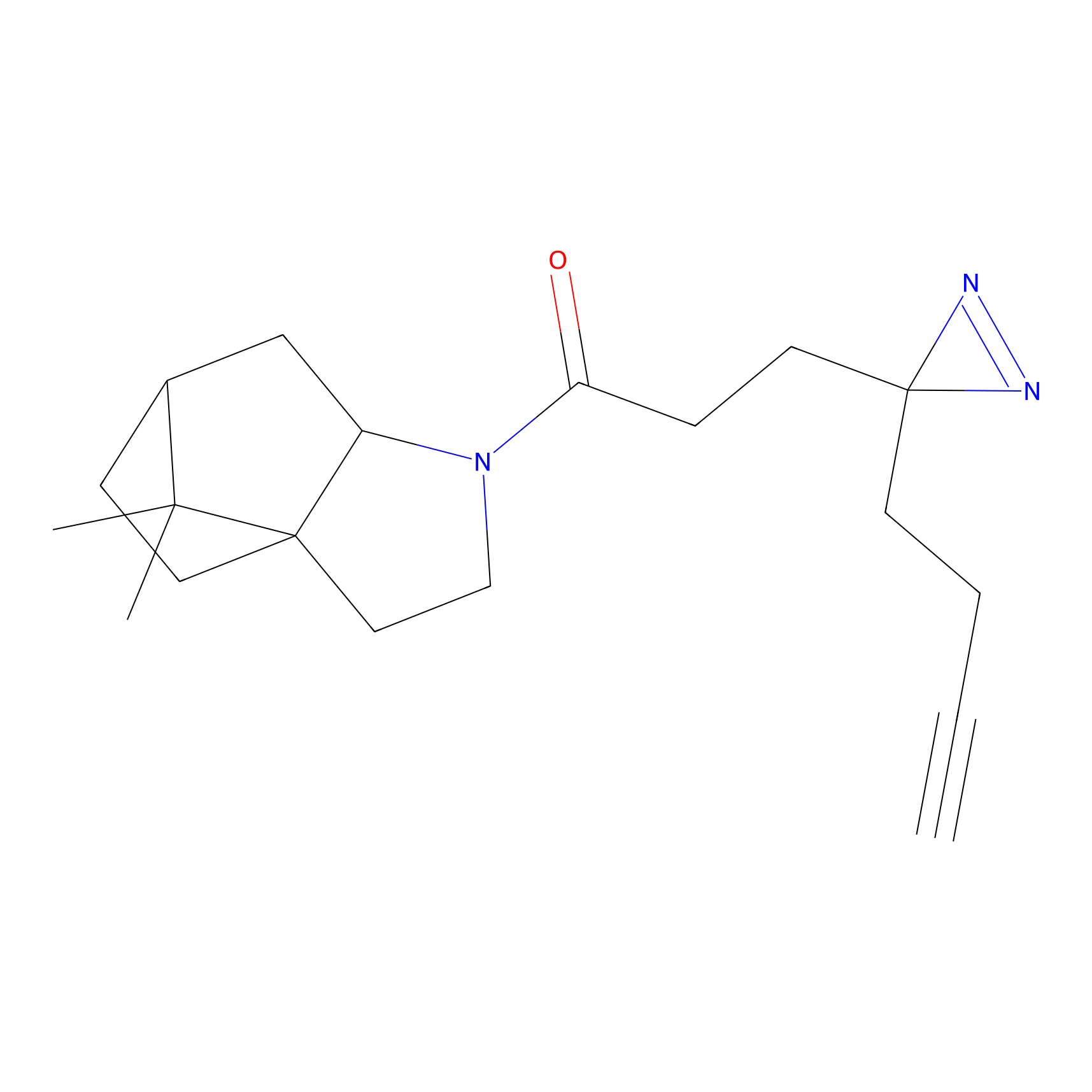
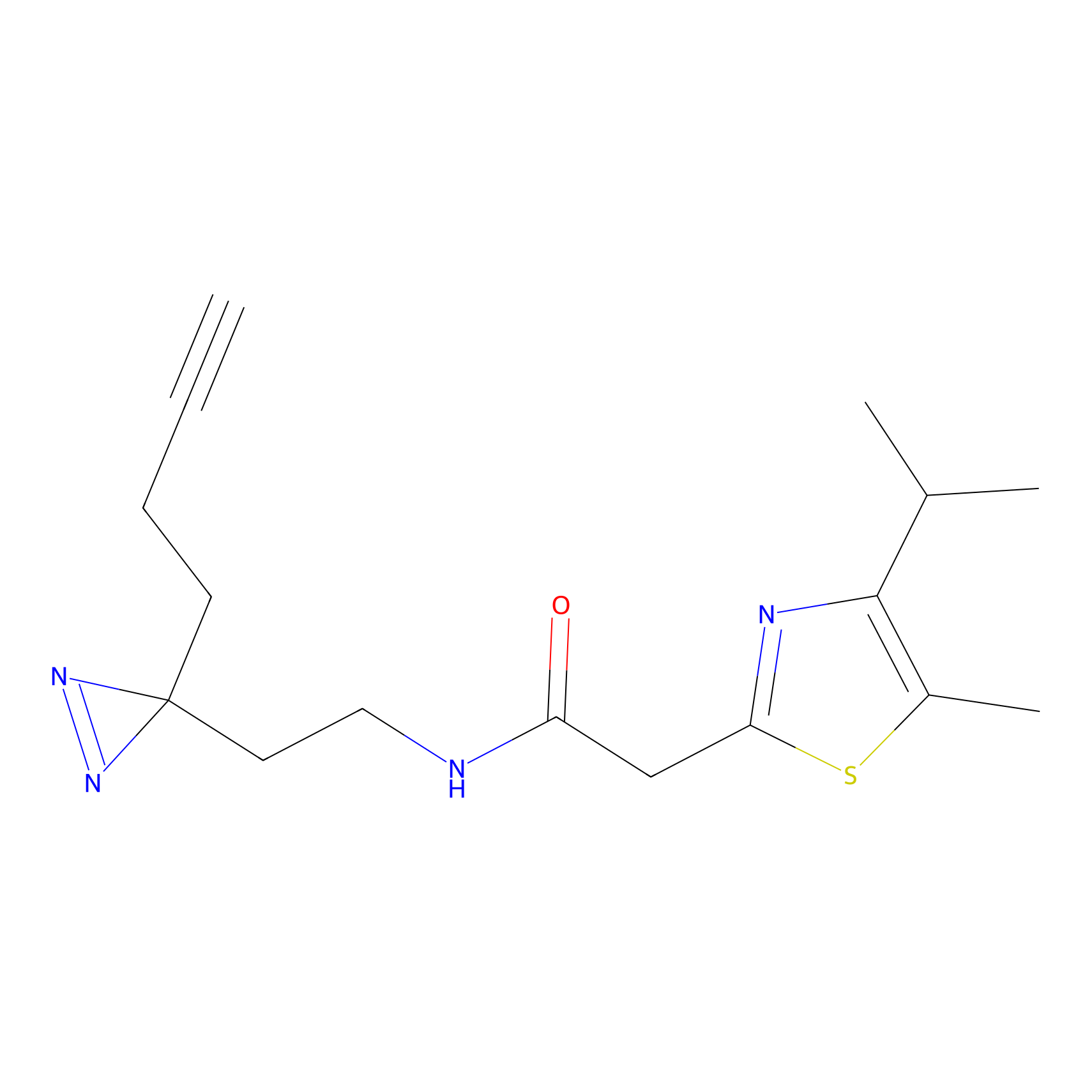
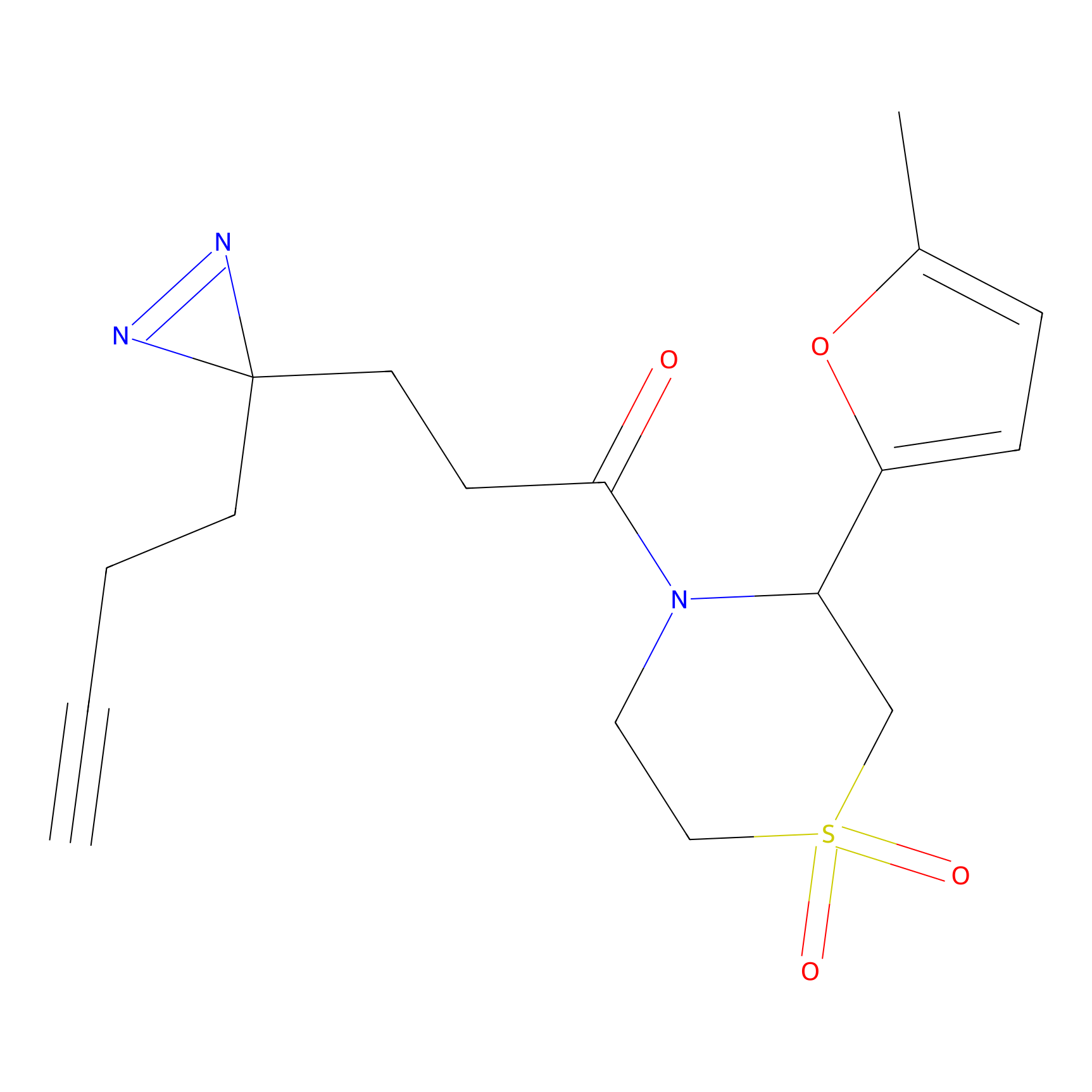
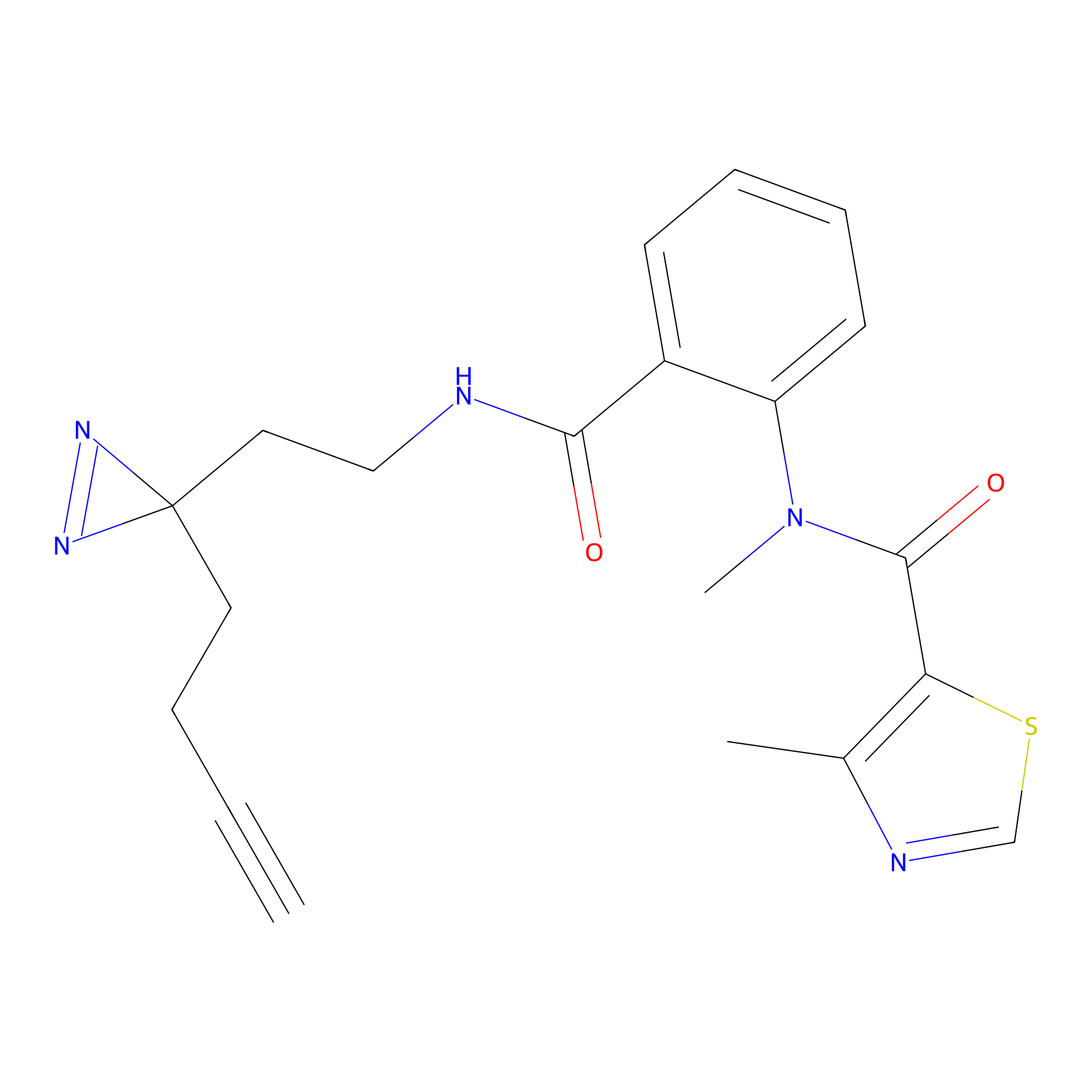
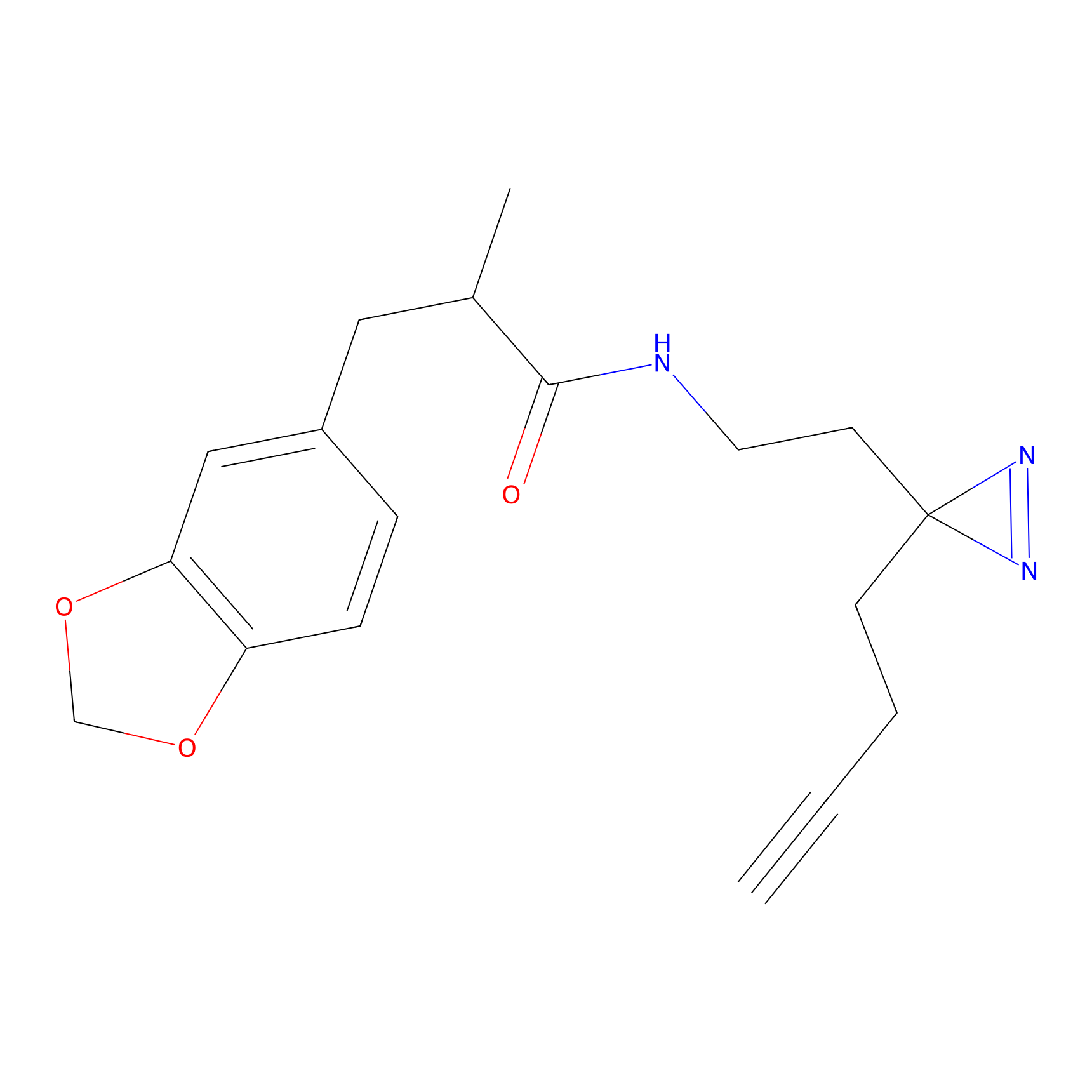
FBPP2 Probe Info |
 |
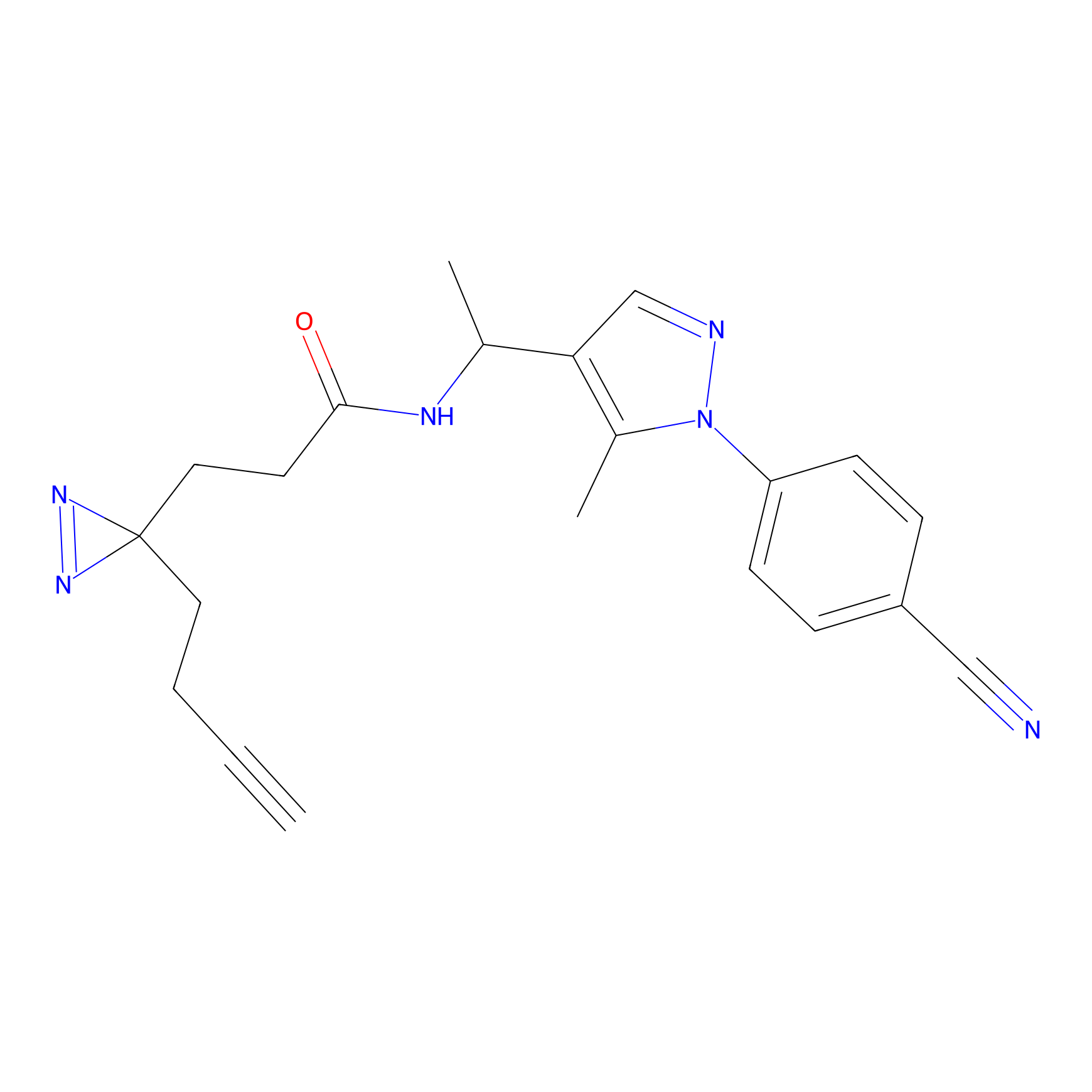
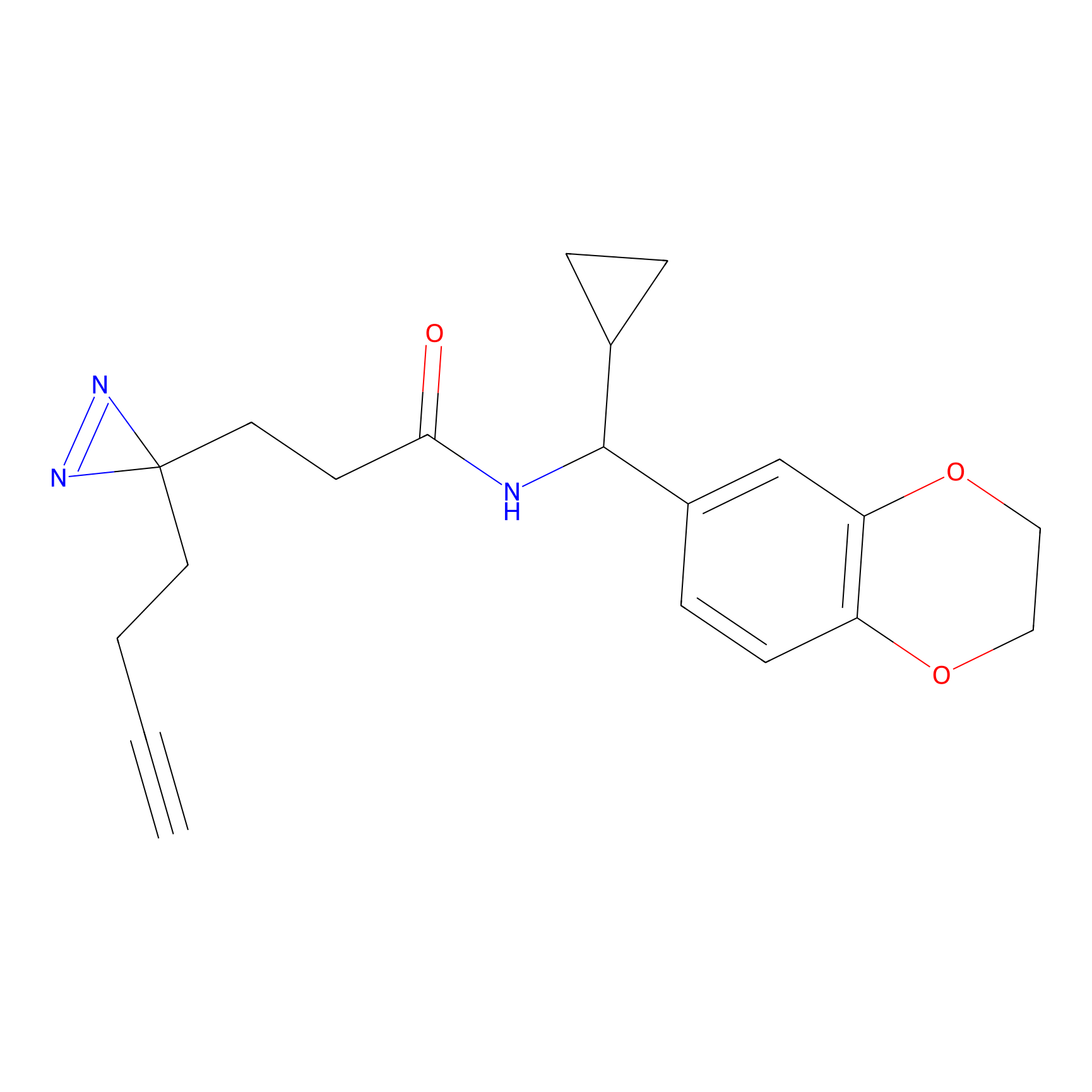
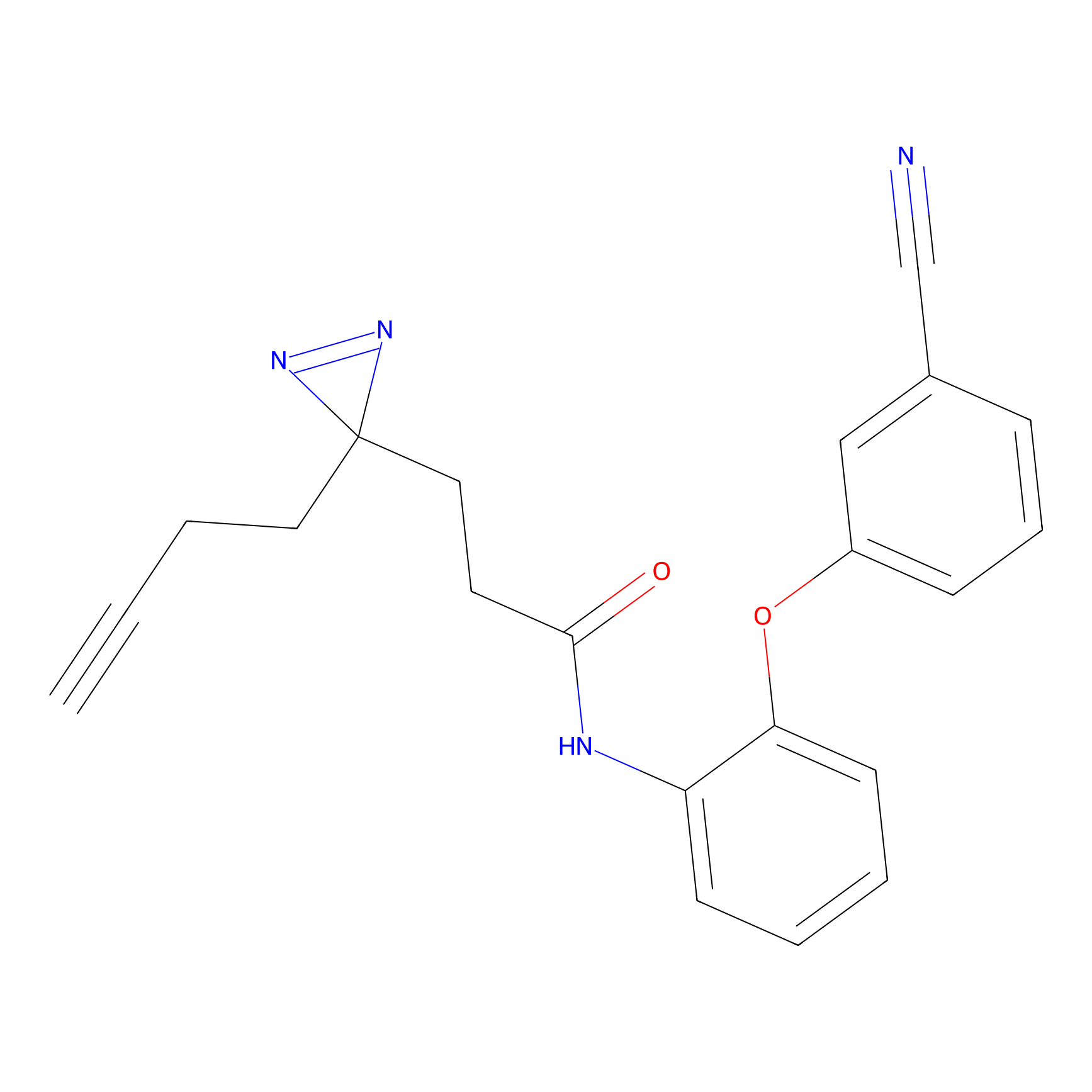
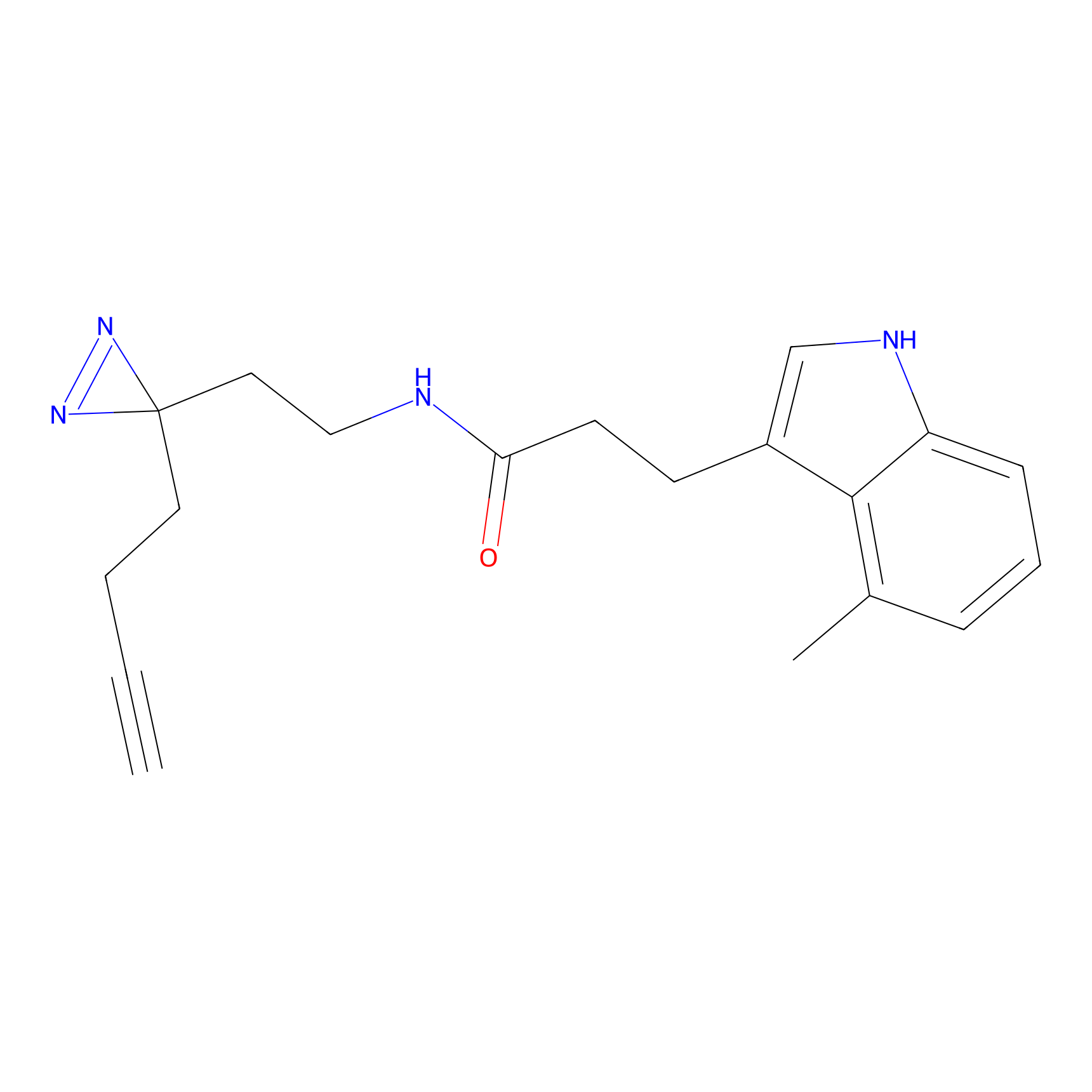
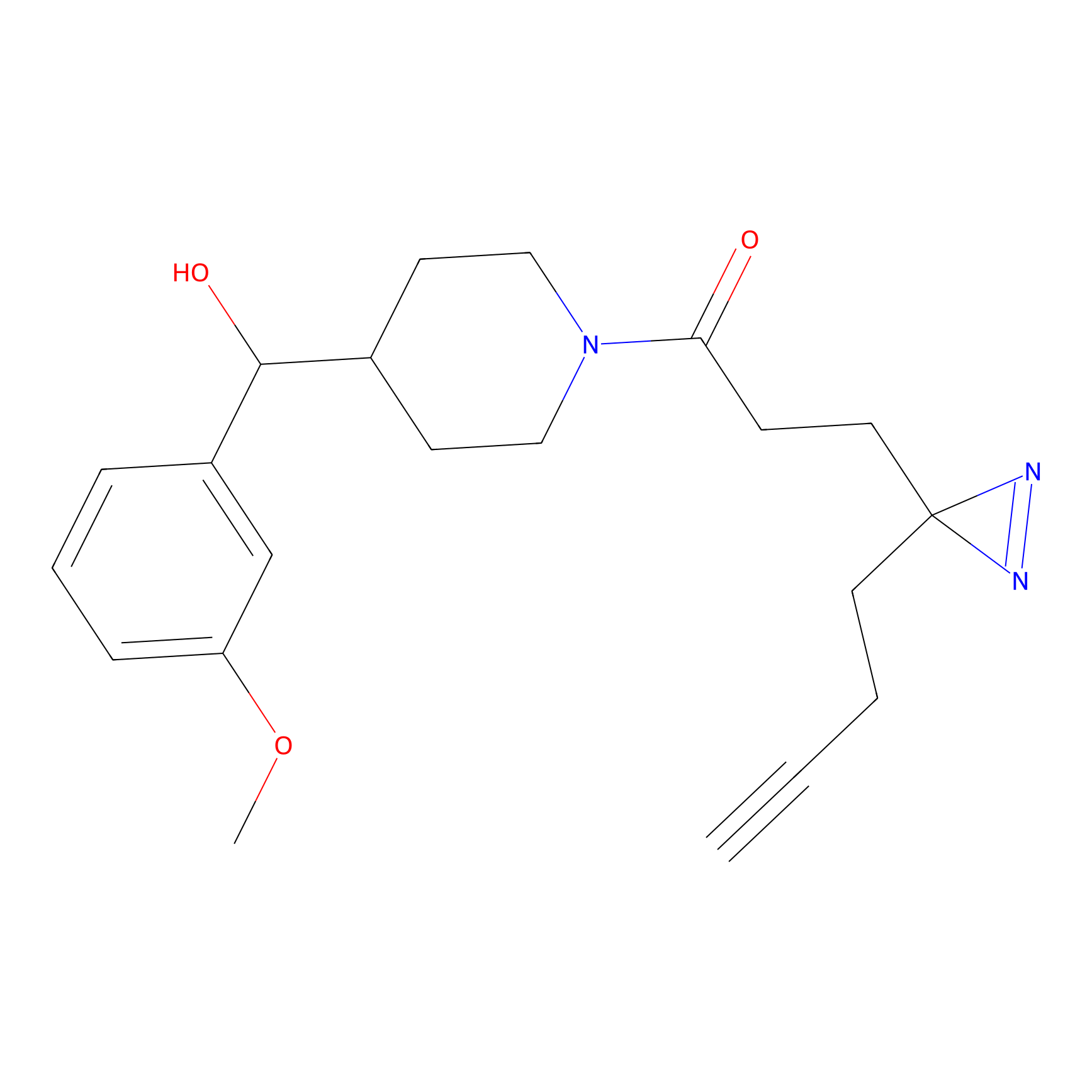
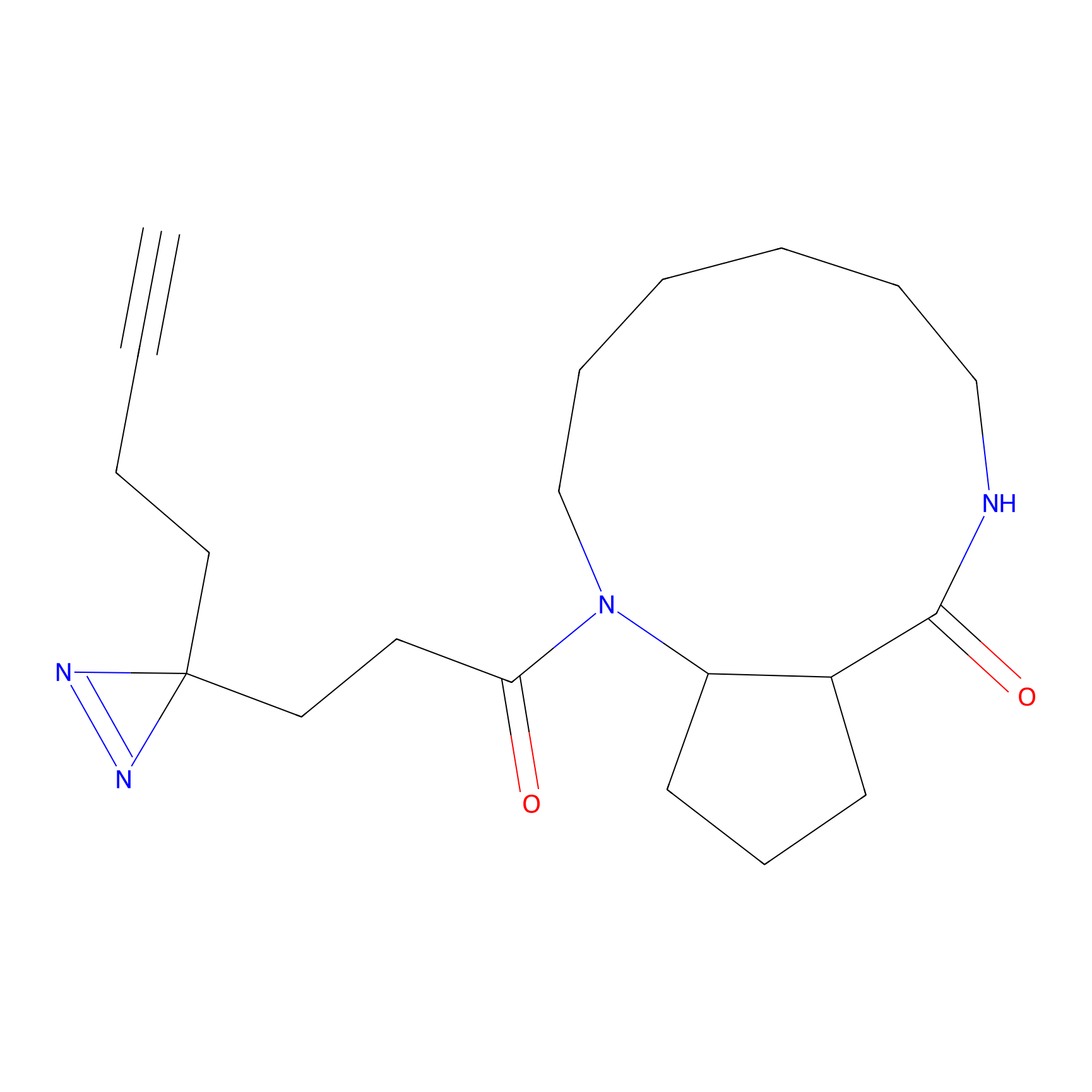
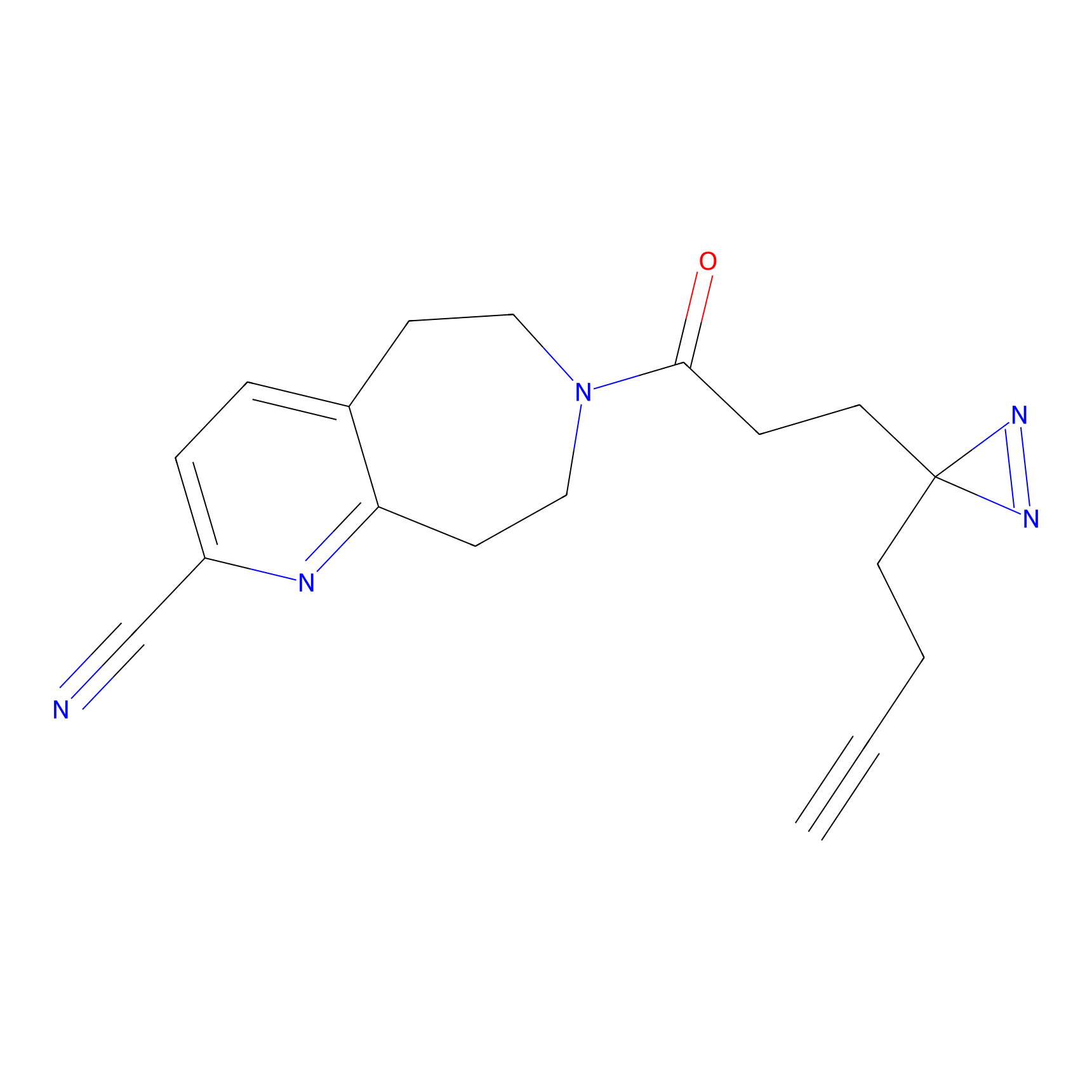
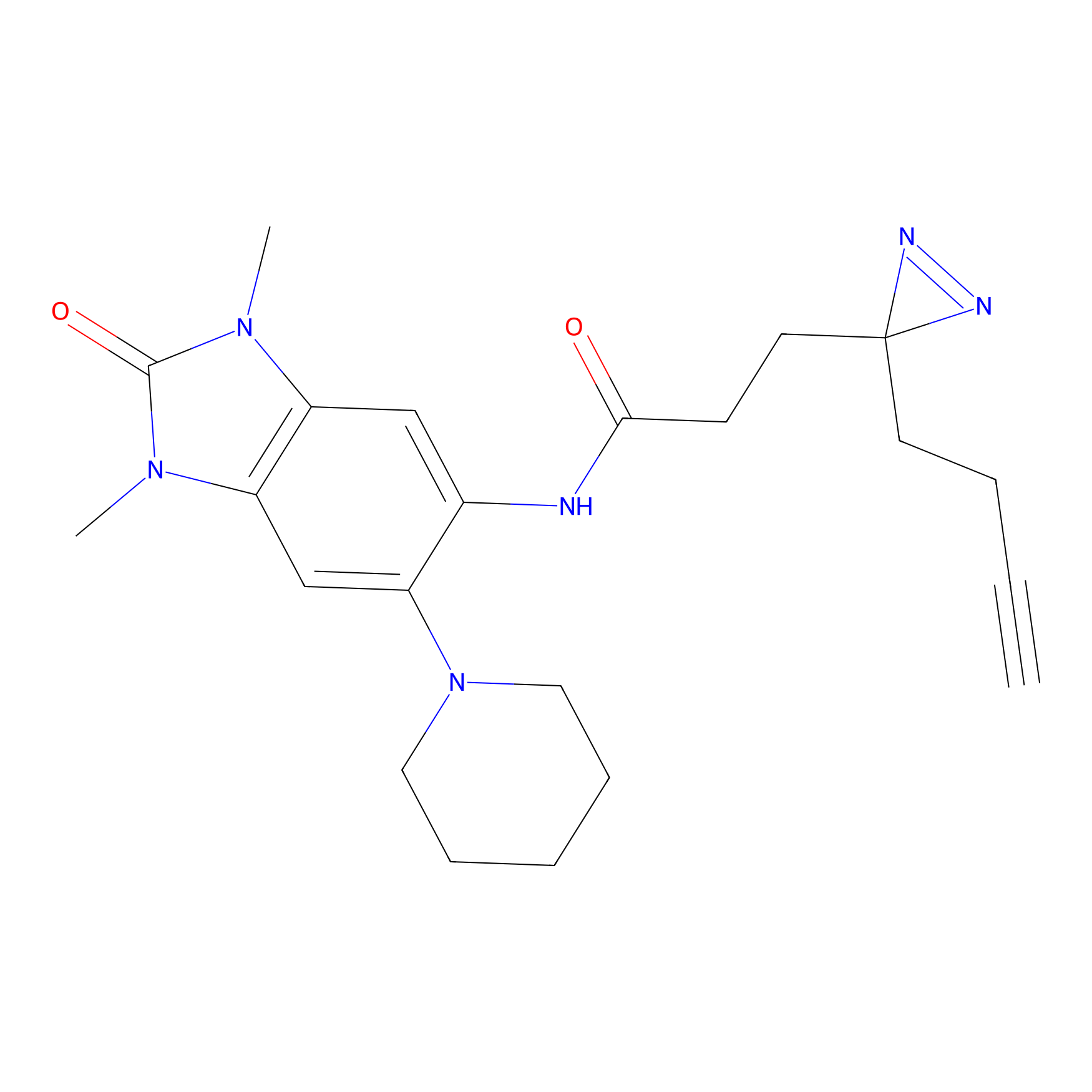
6.68 | LDD0318 | [1] | |
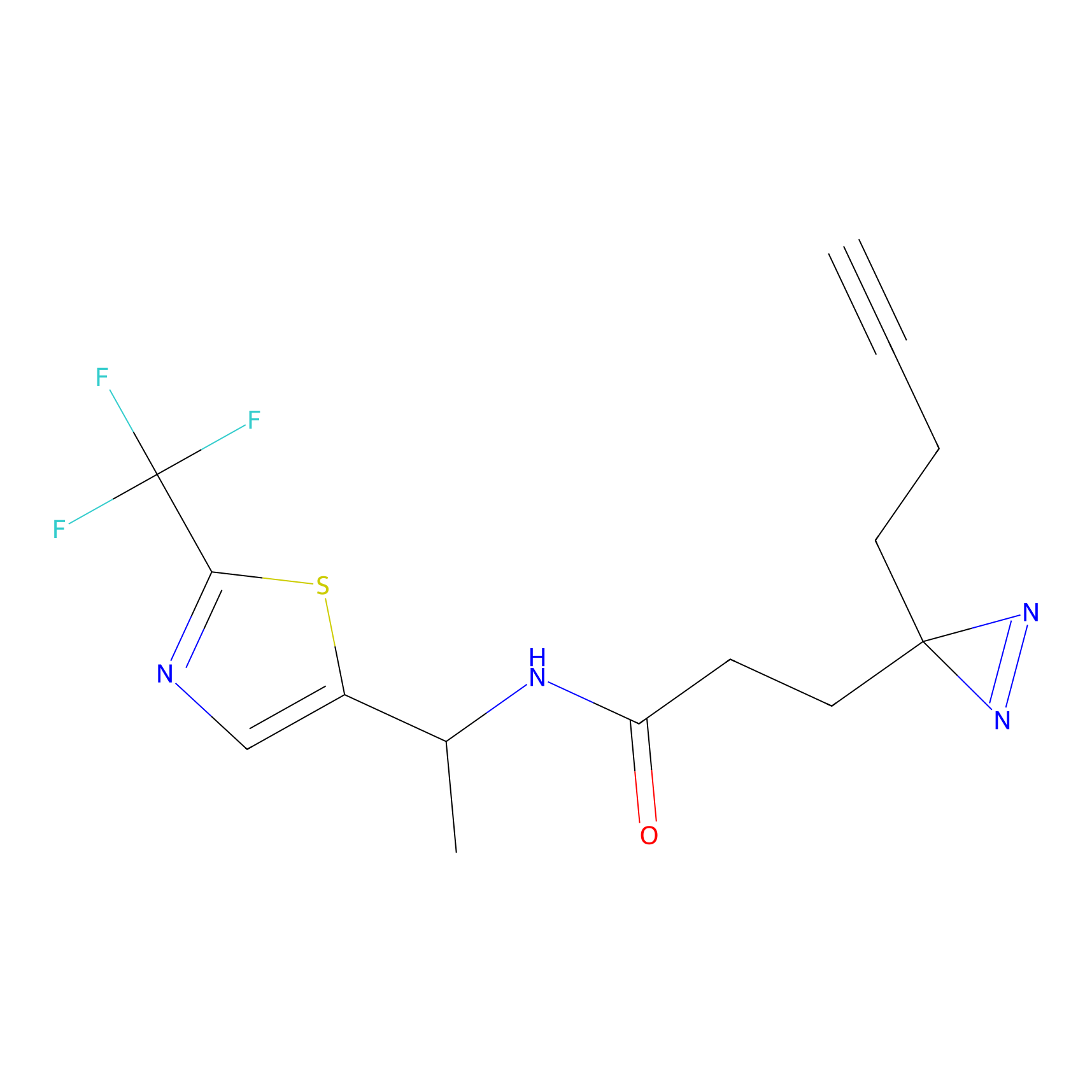
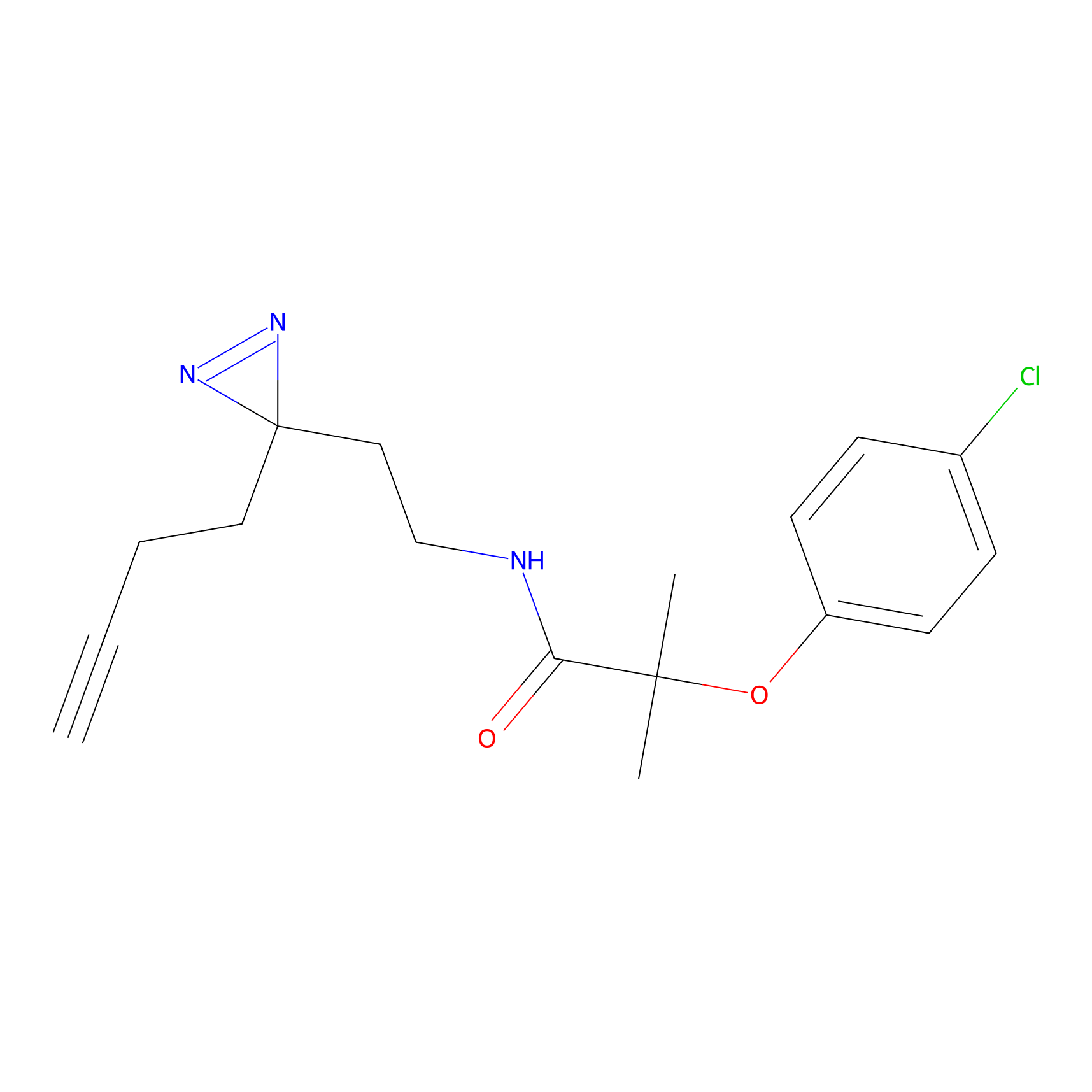
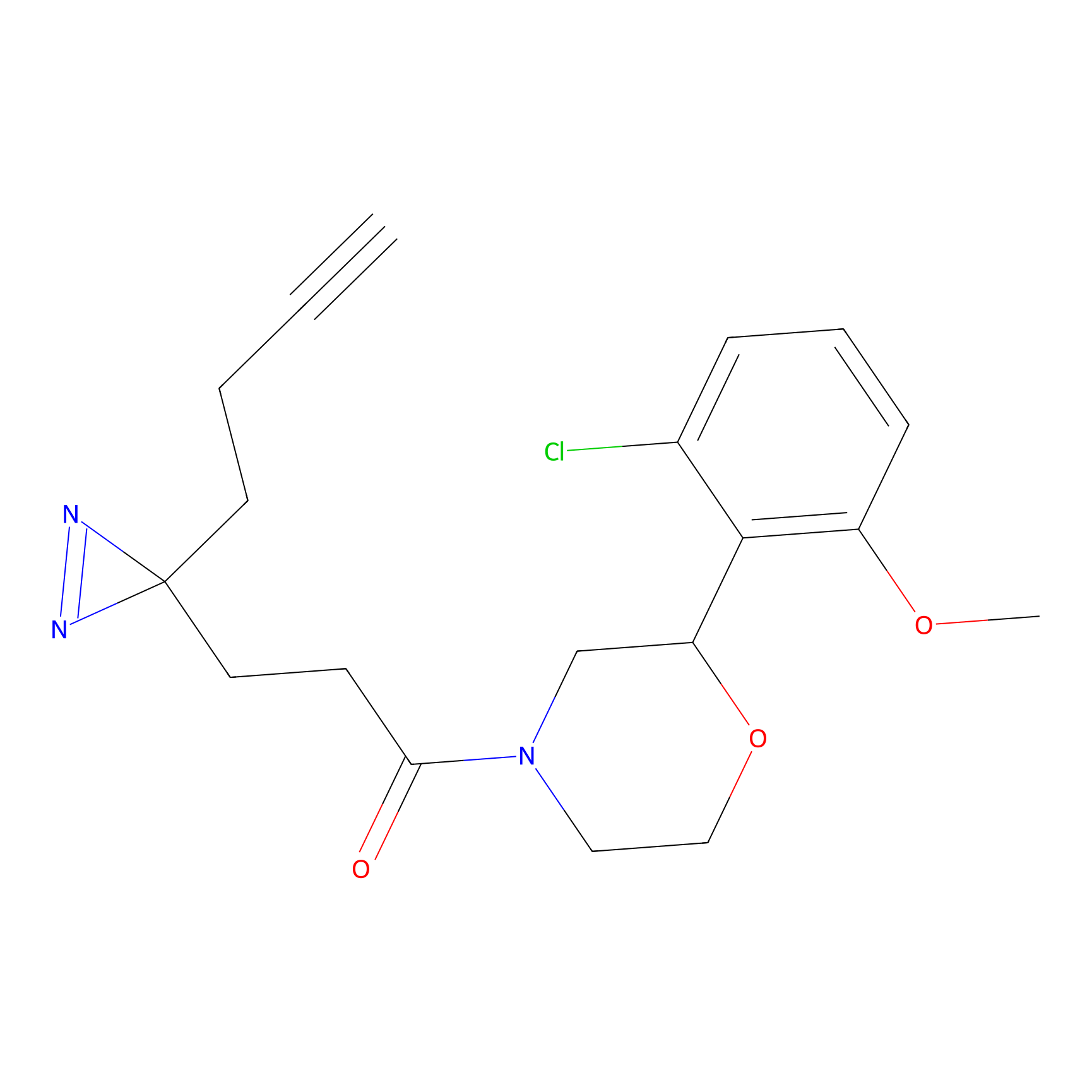
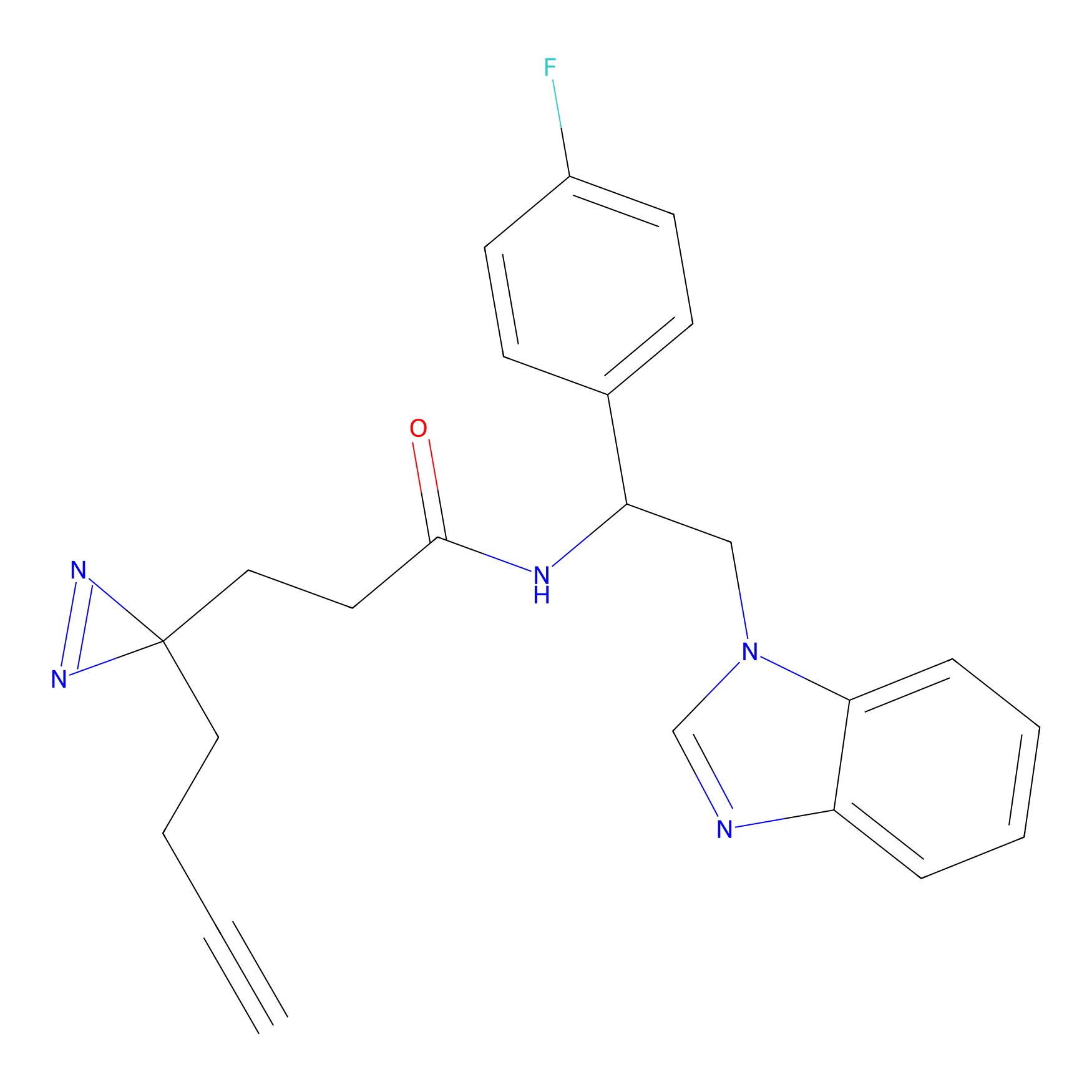
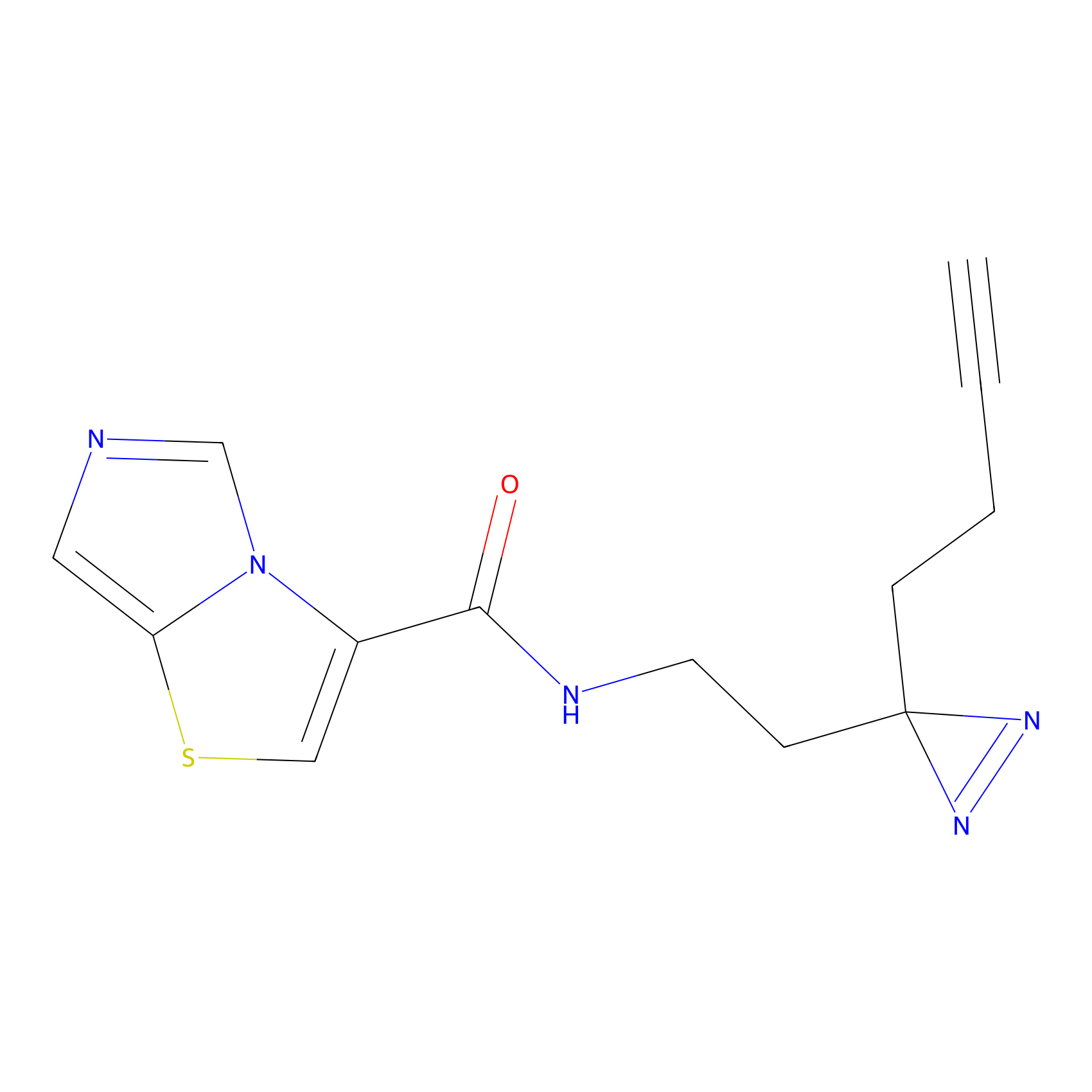
|
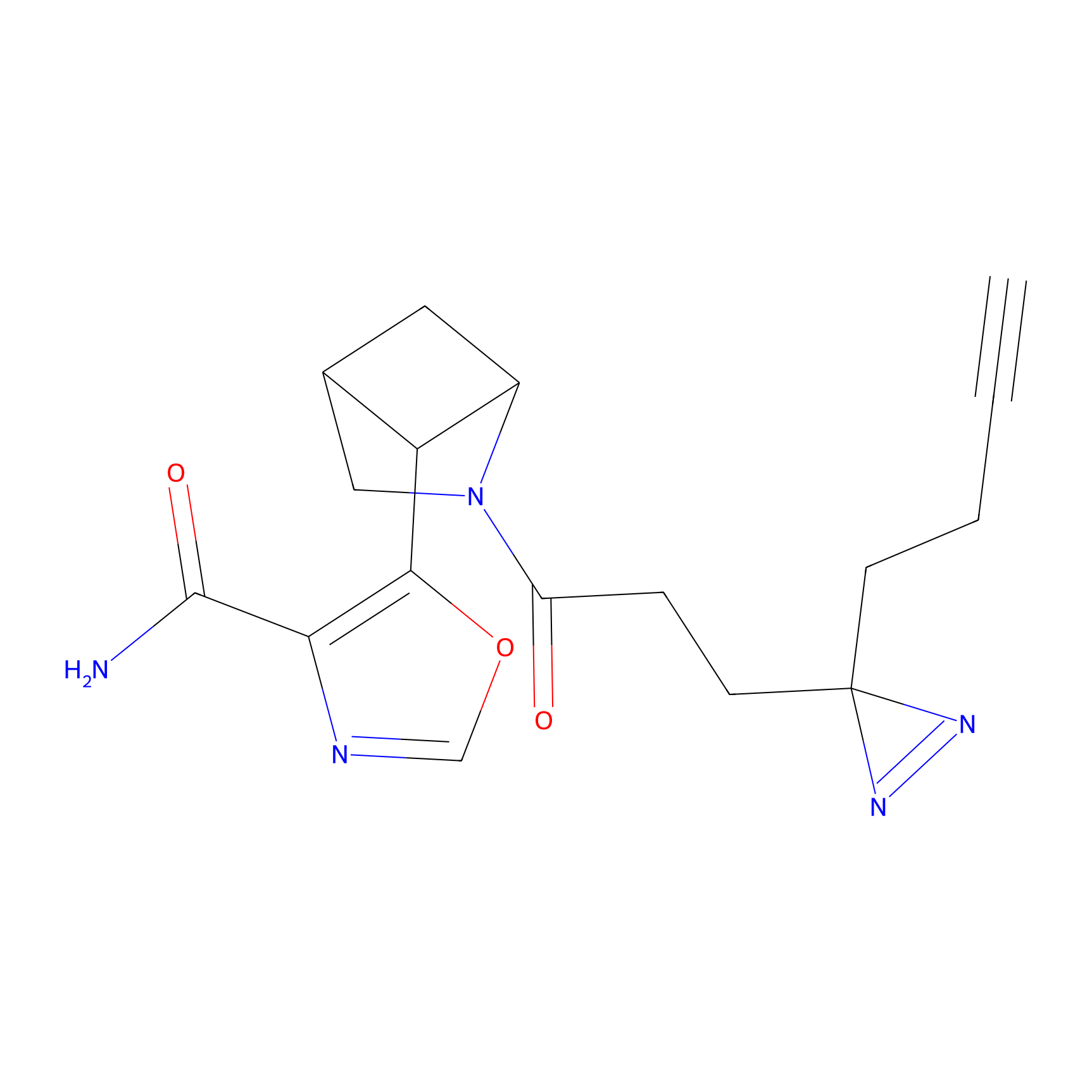
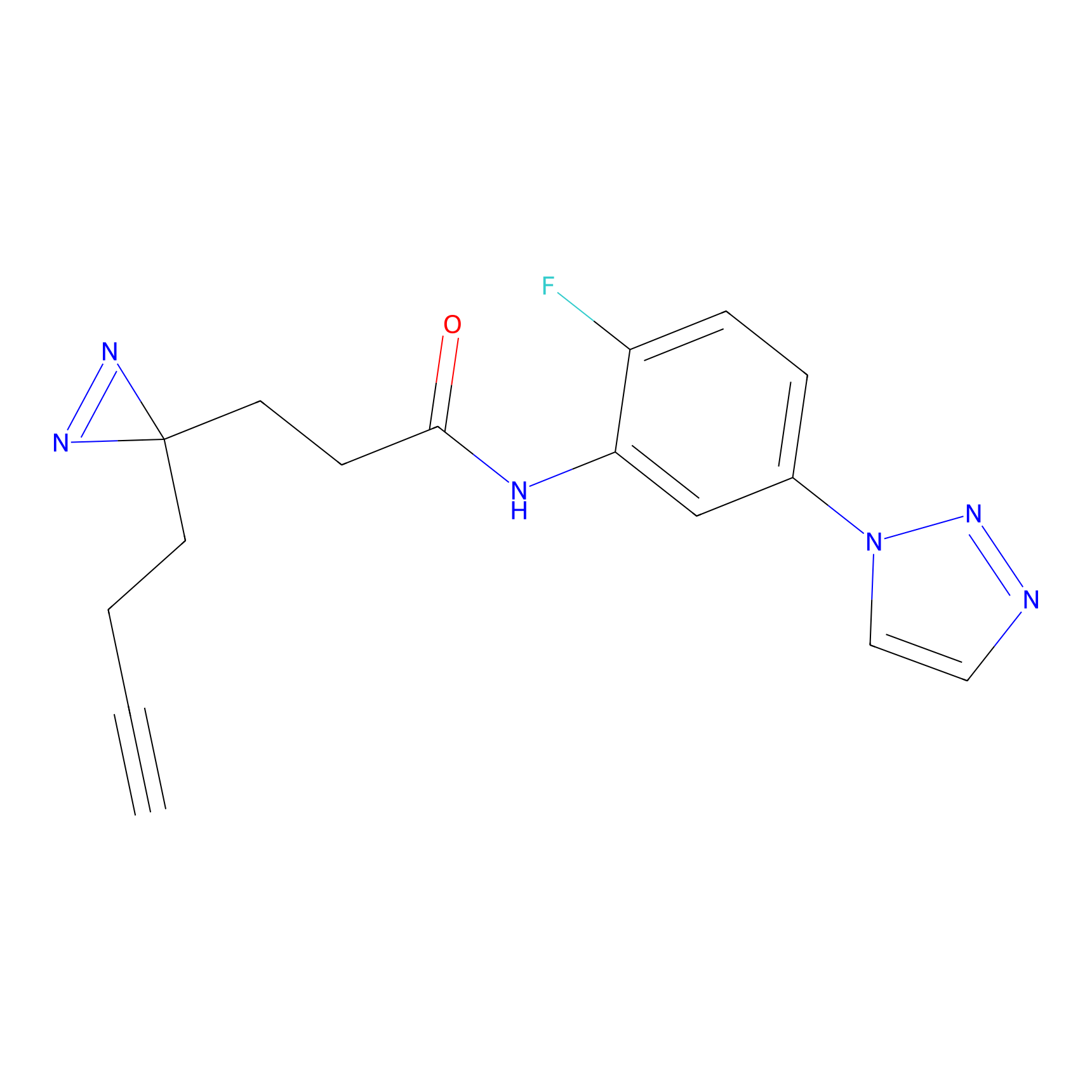
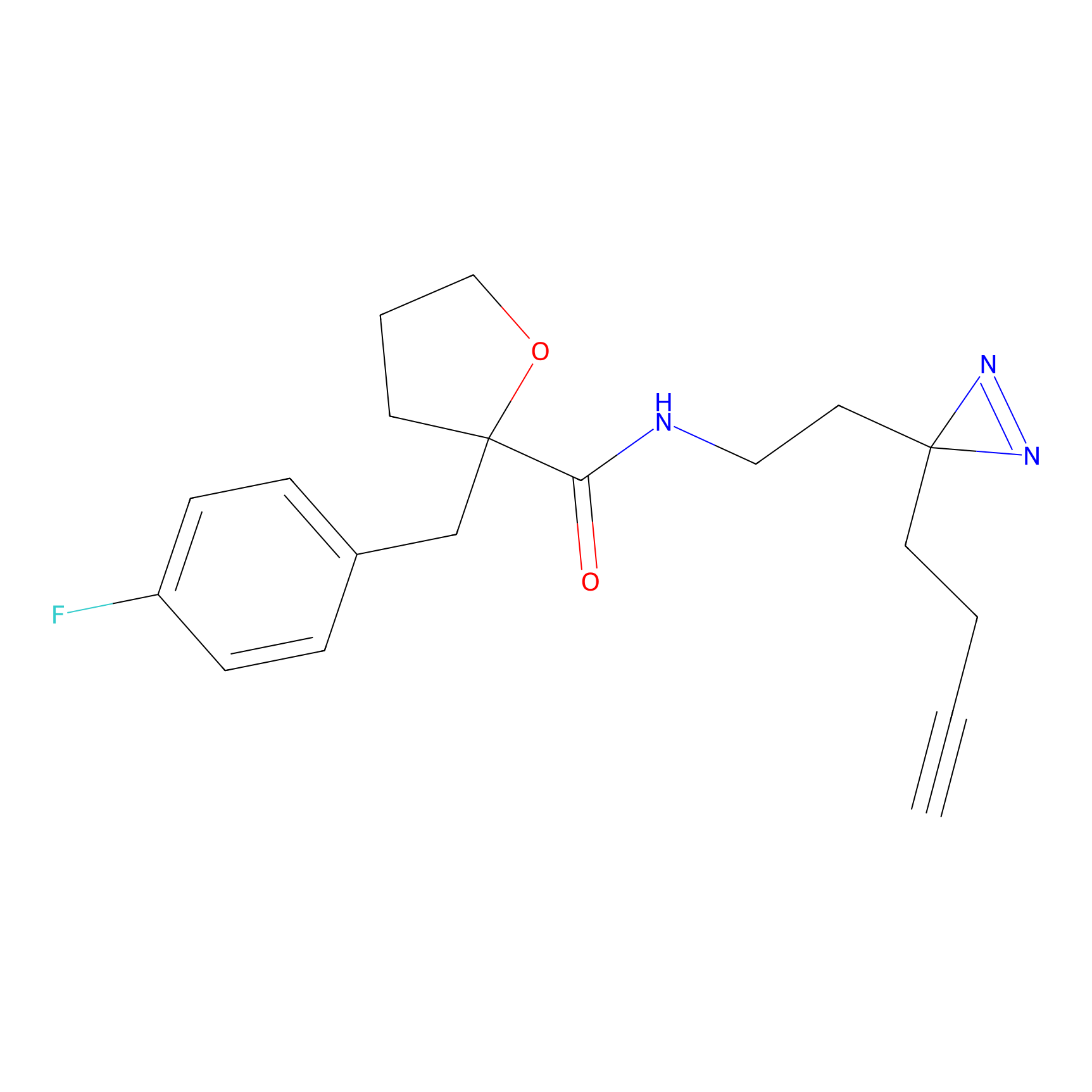
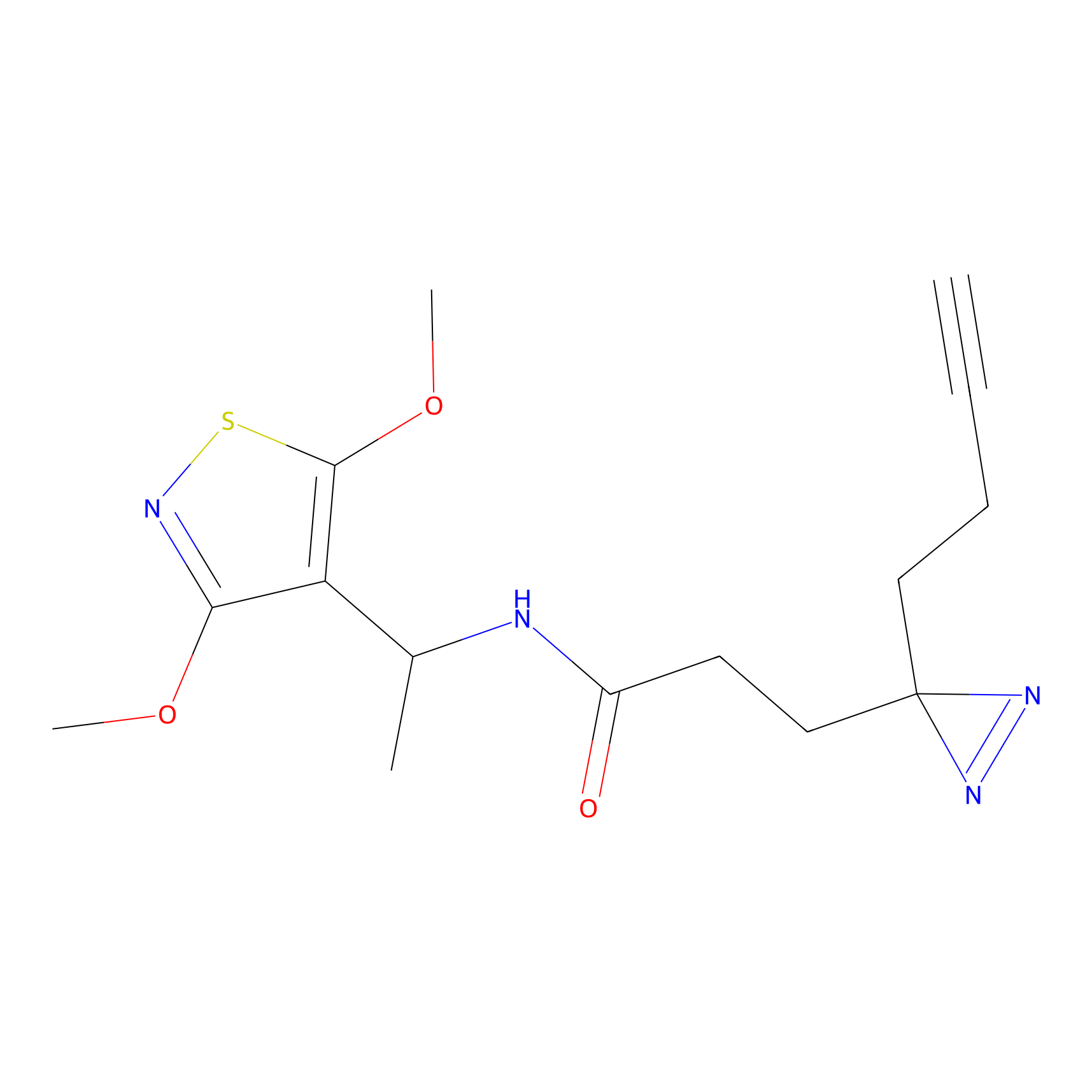
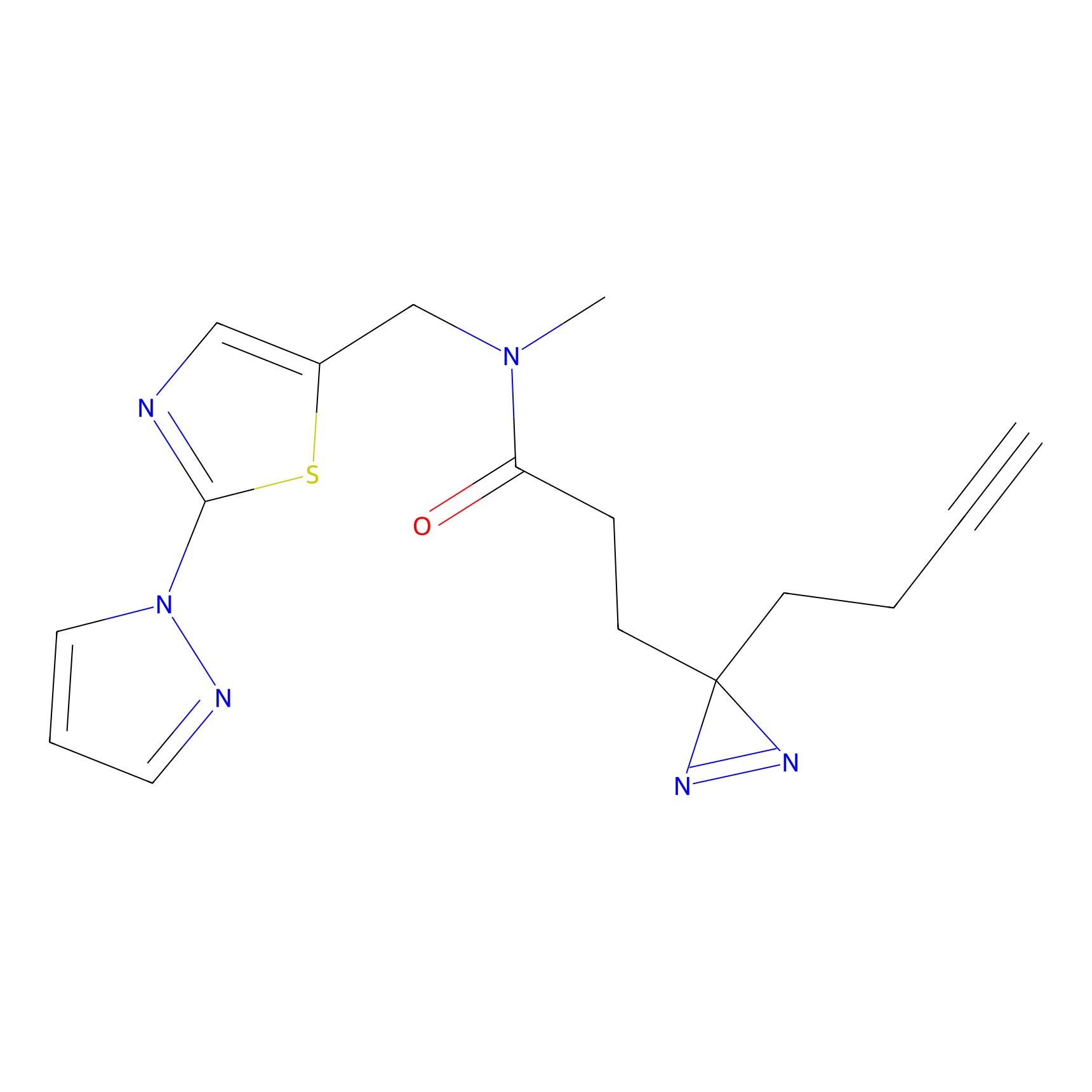
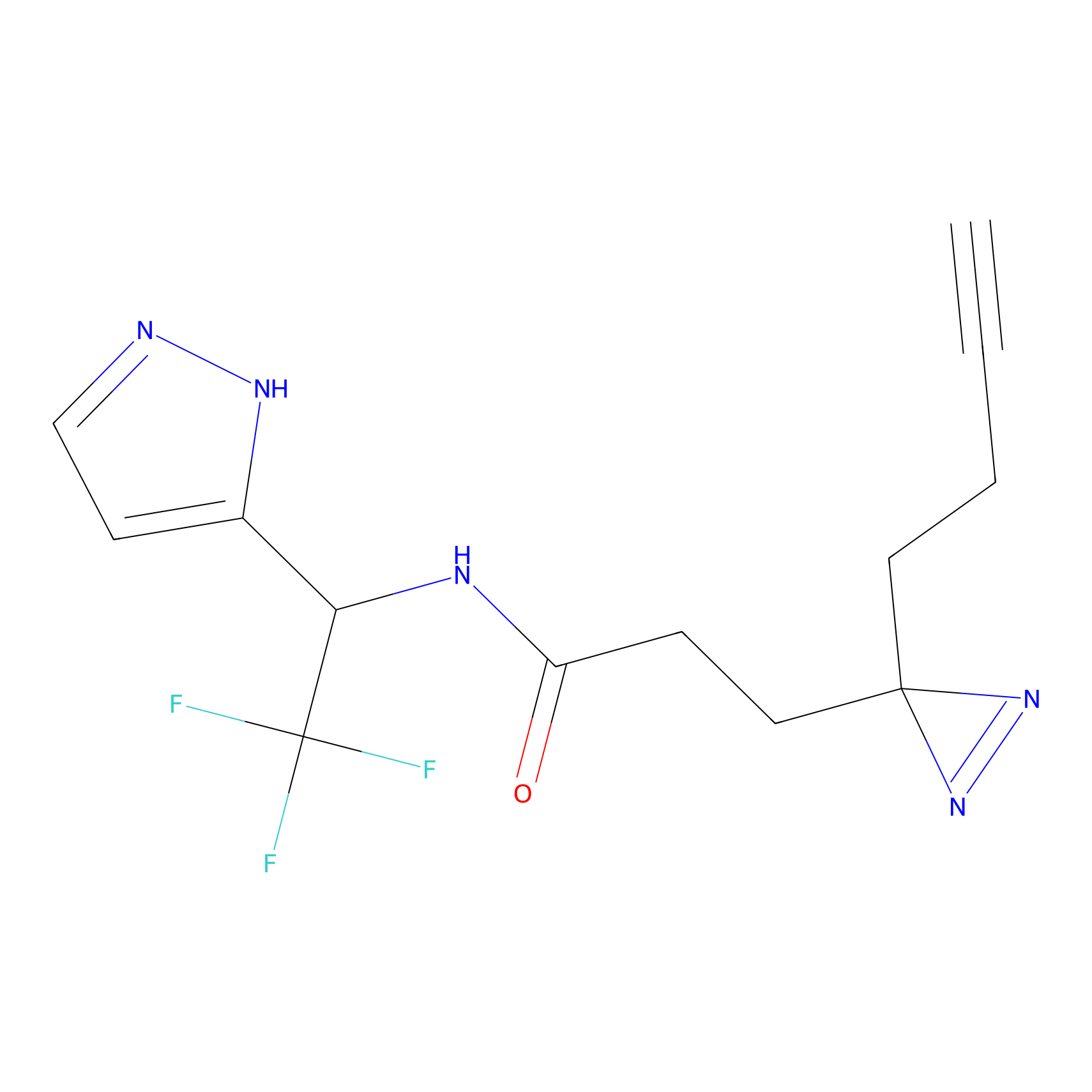
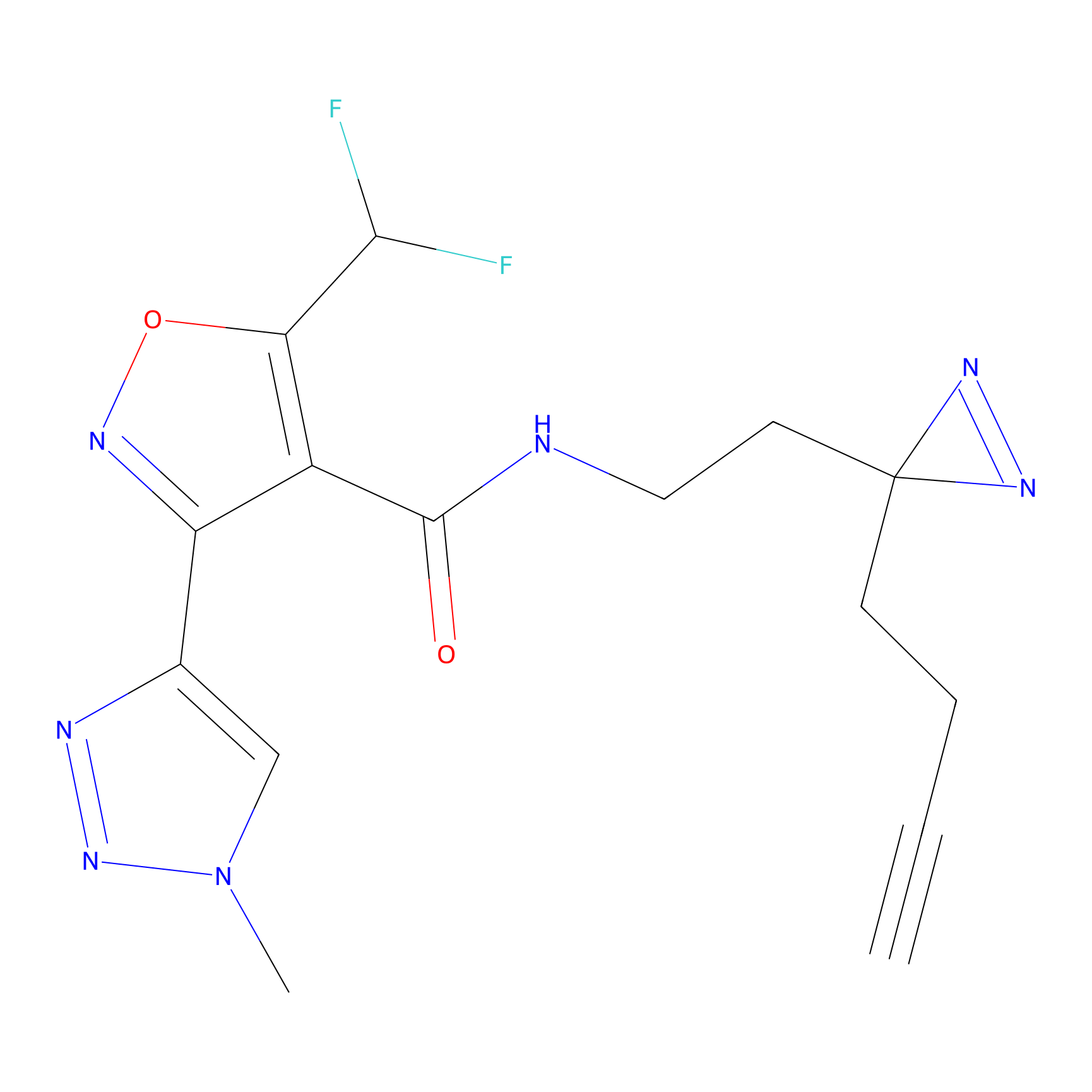
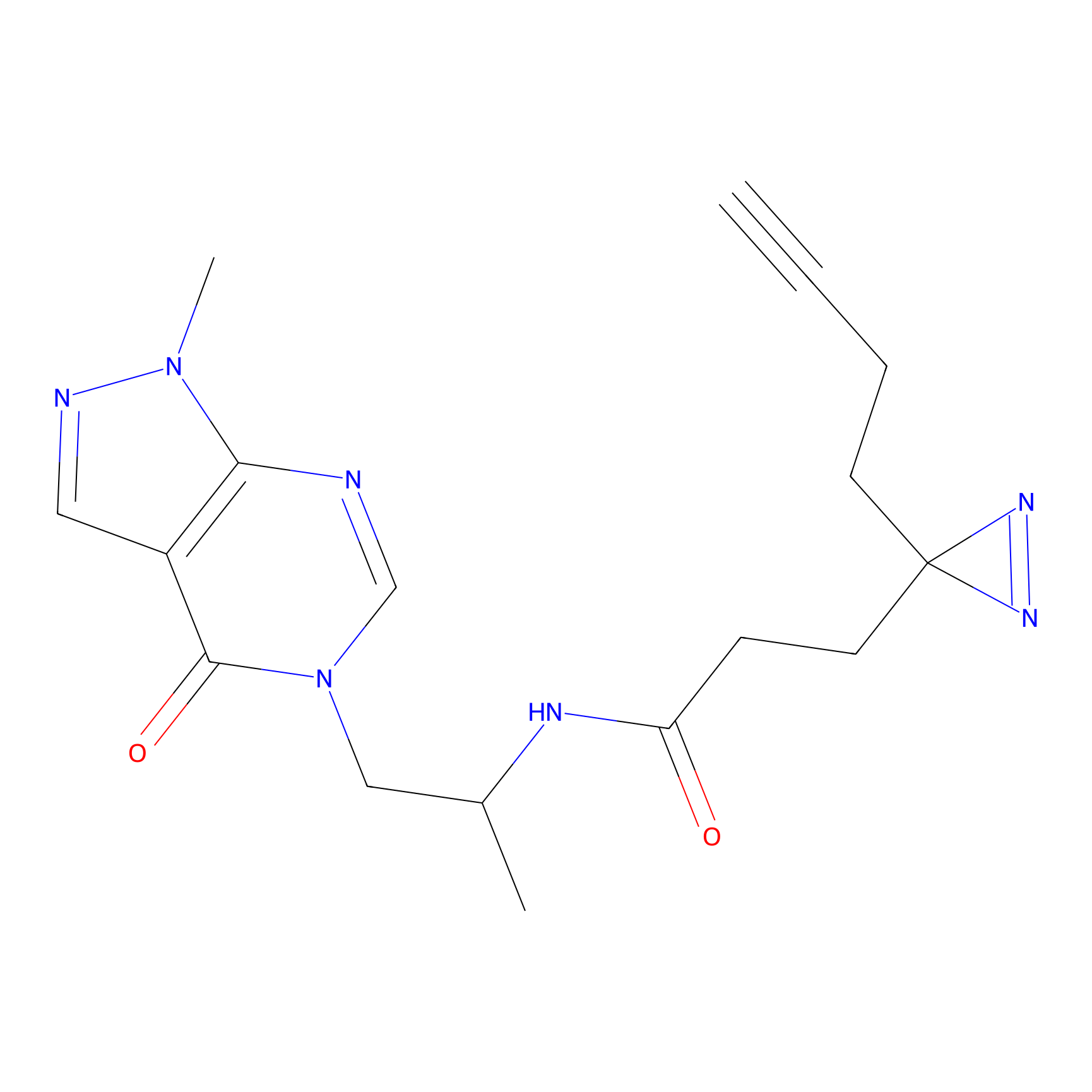
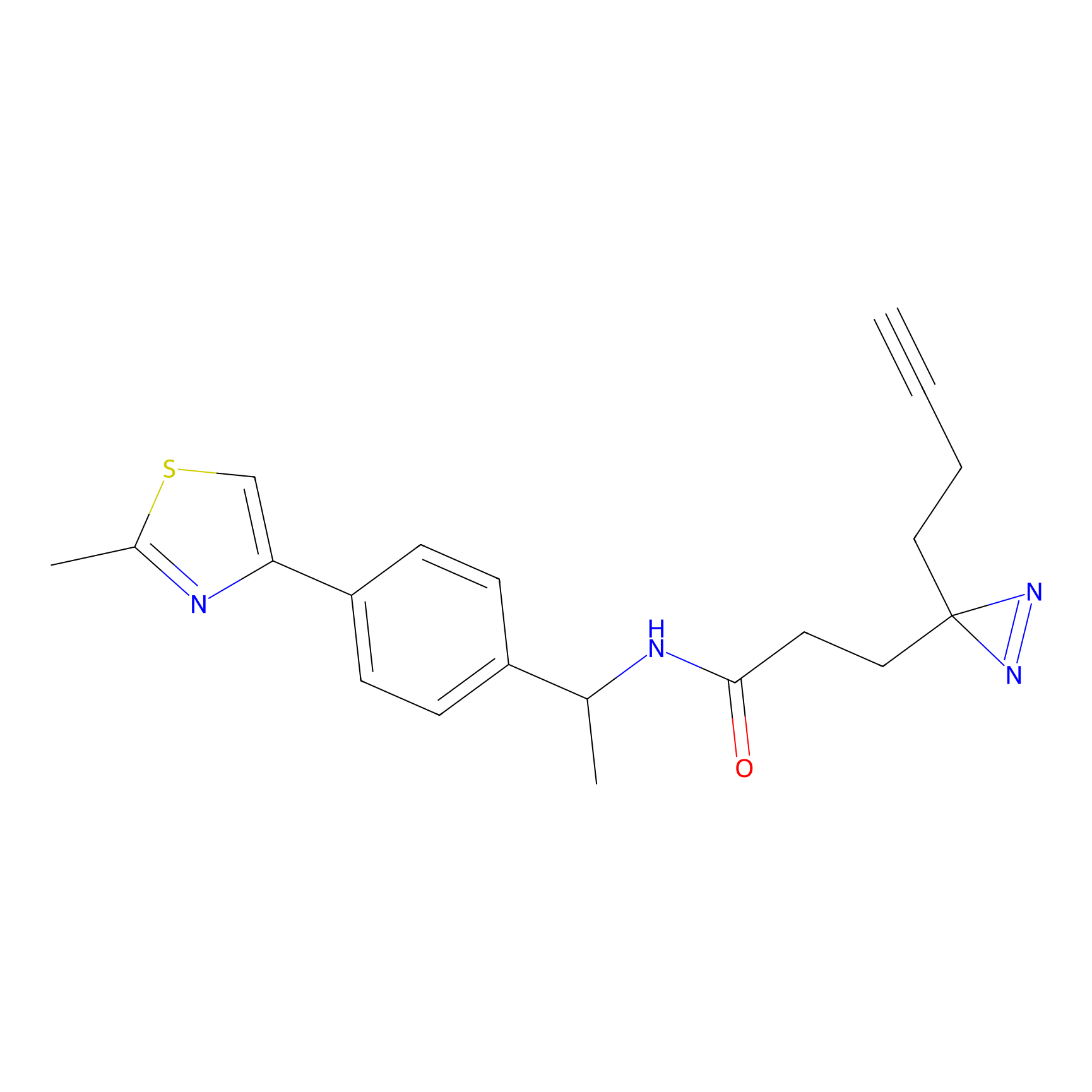
TH211 Probe Info |
 |
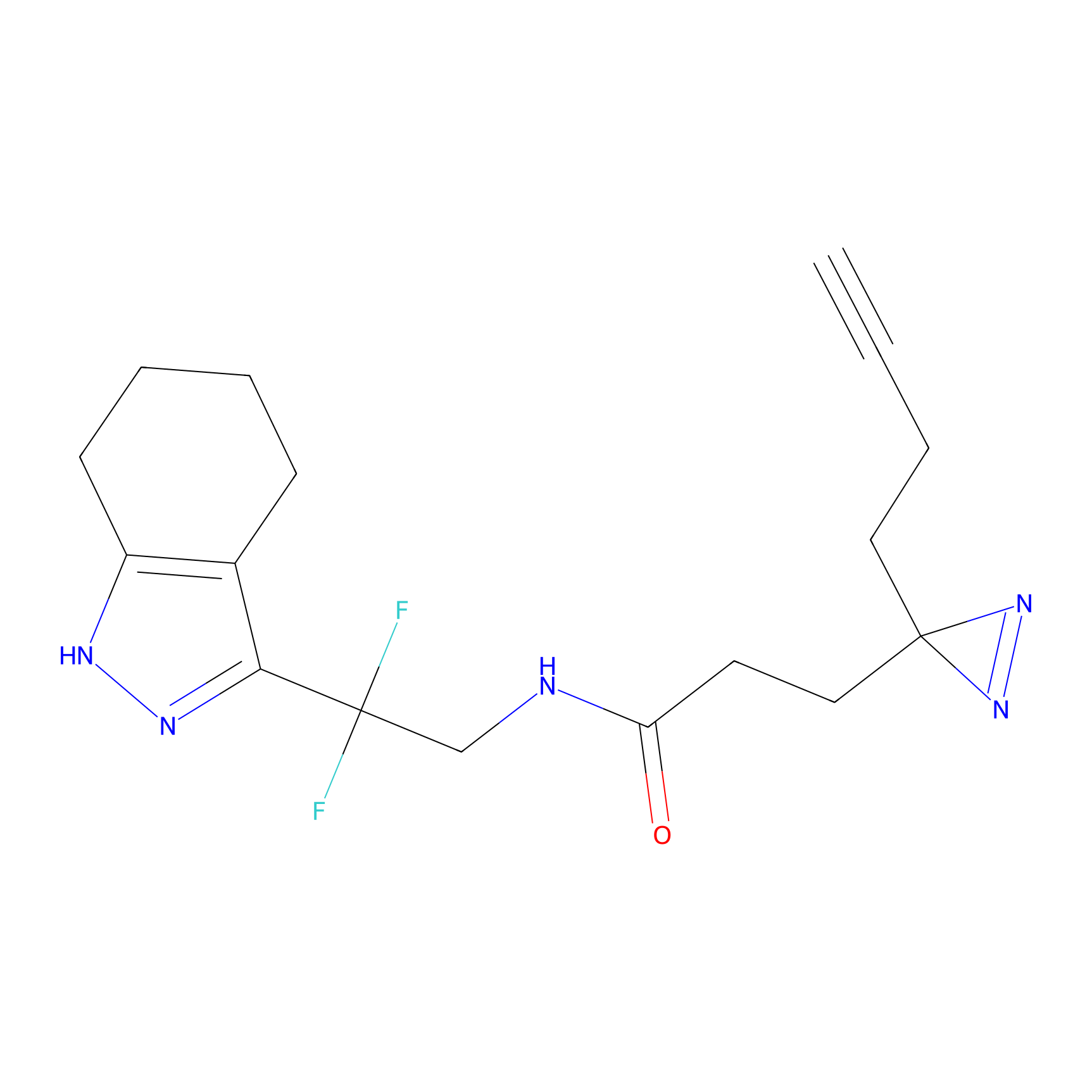
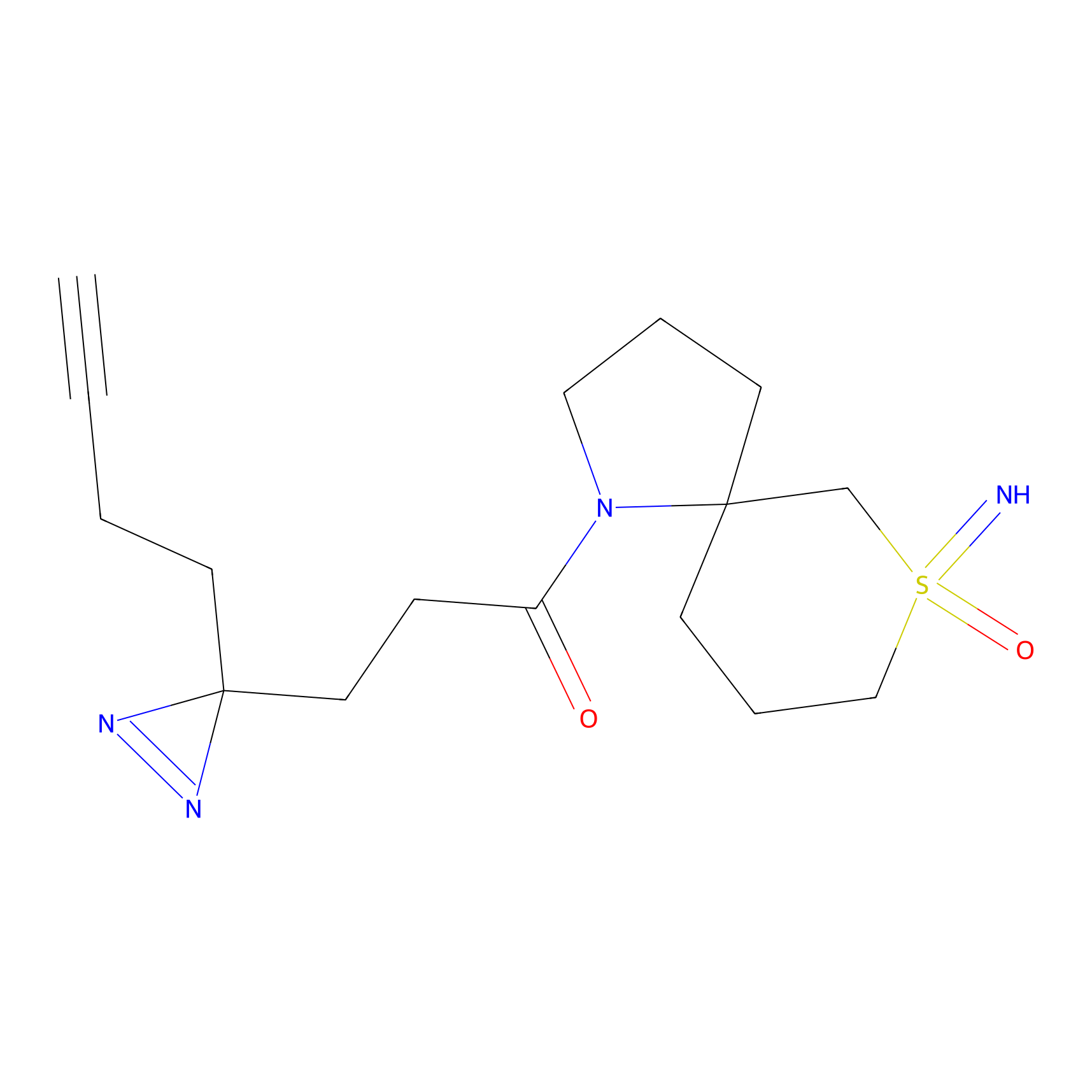
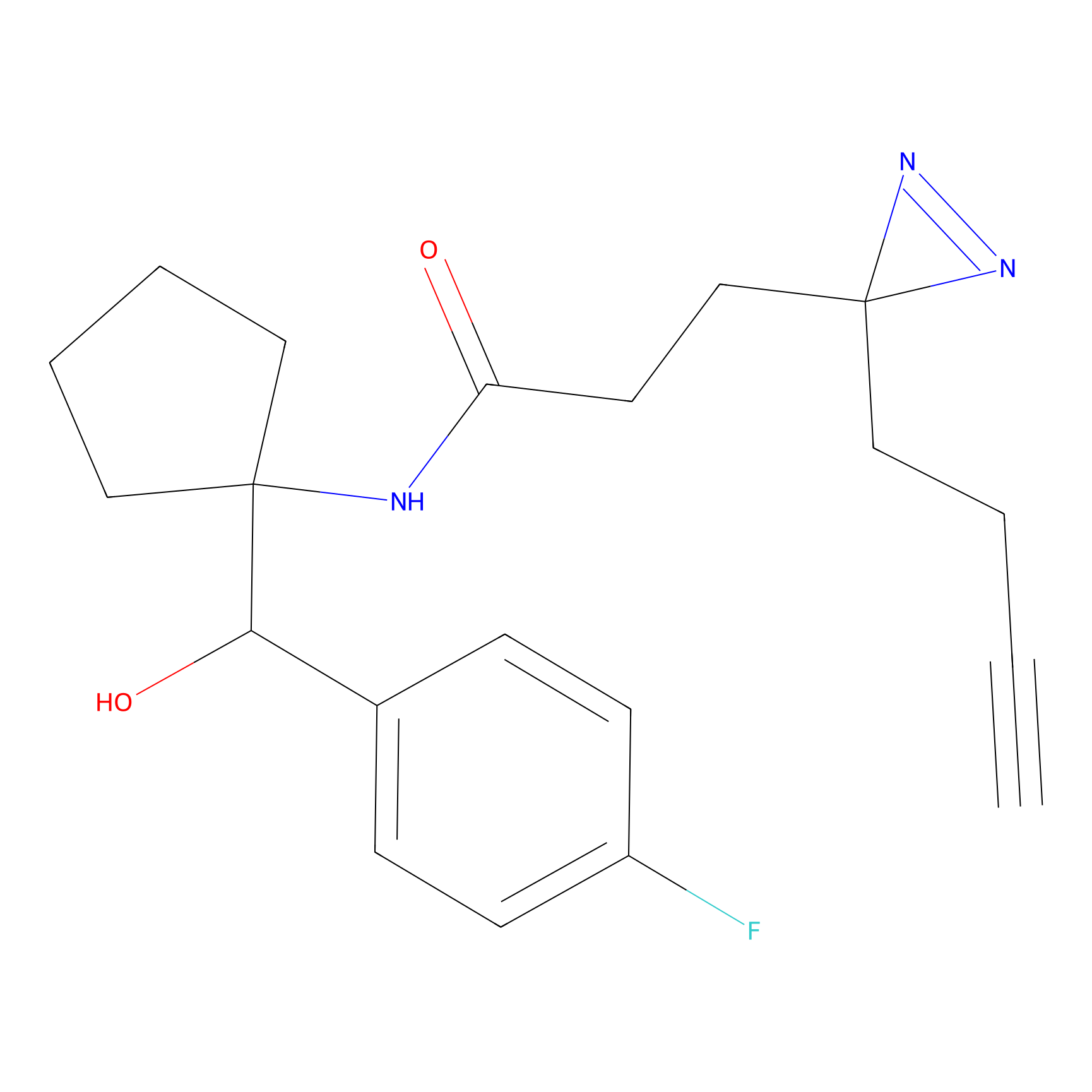
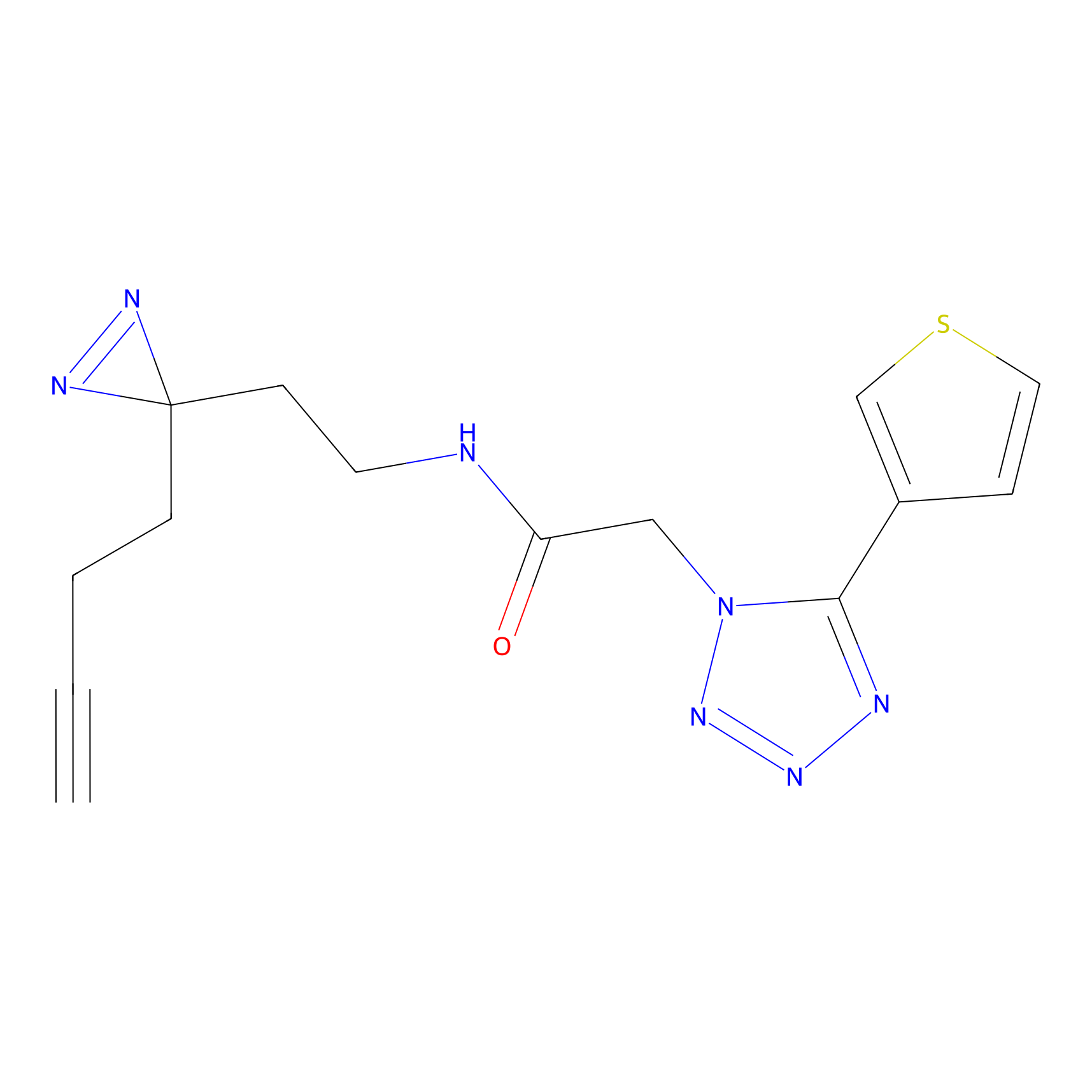
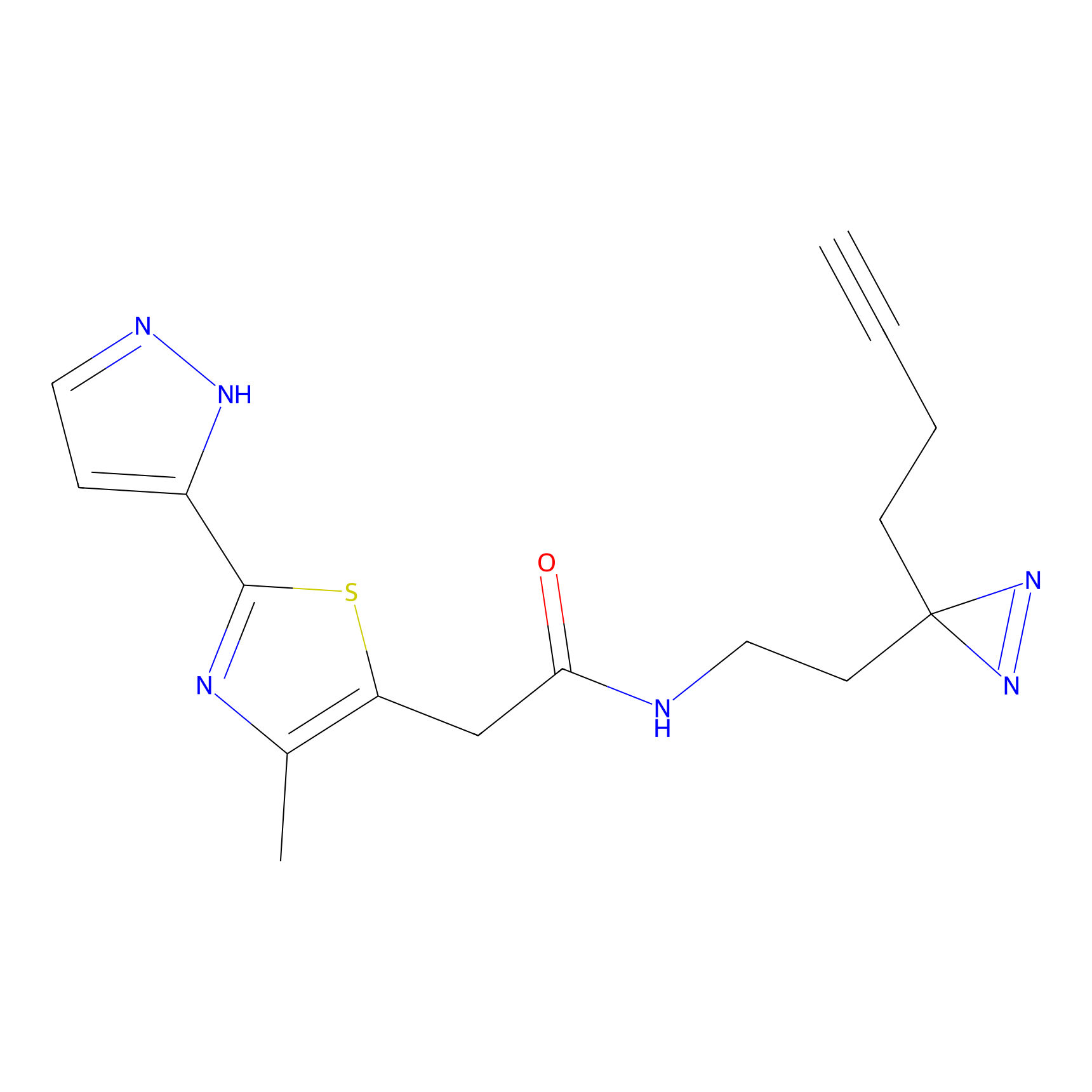
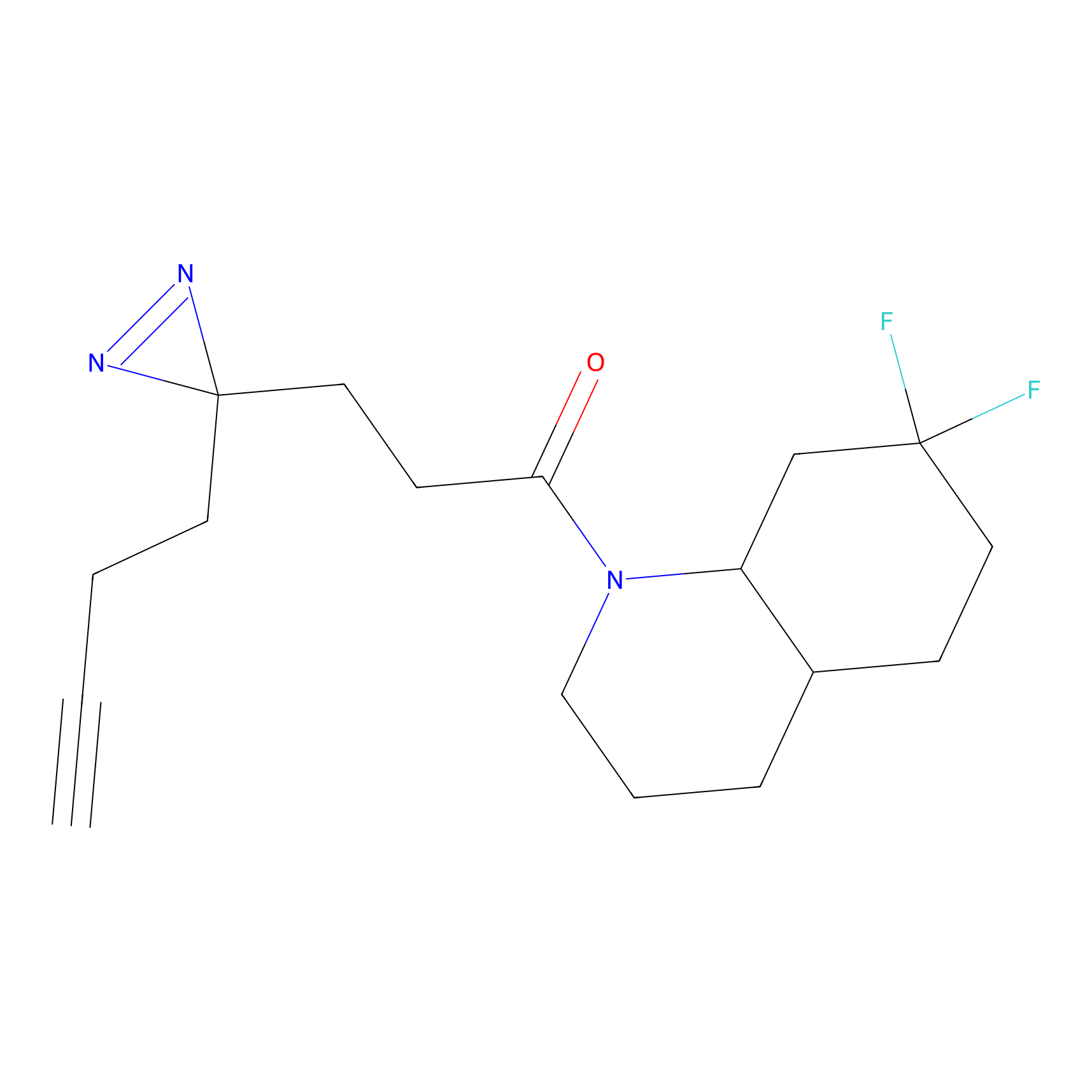
Y157(13.12); Y163(5.47) | LDD0257 | [2] | |
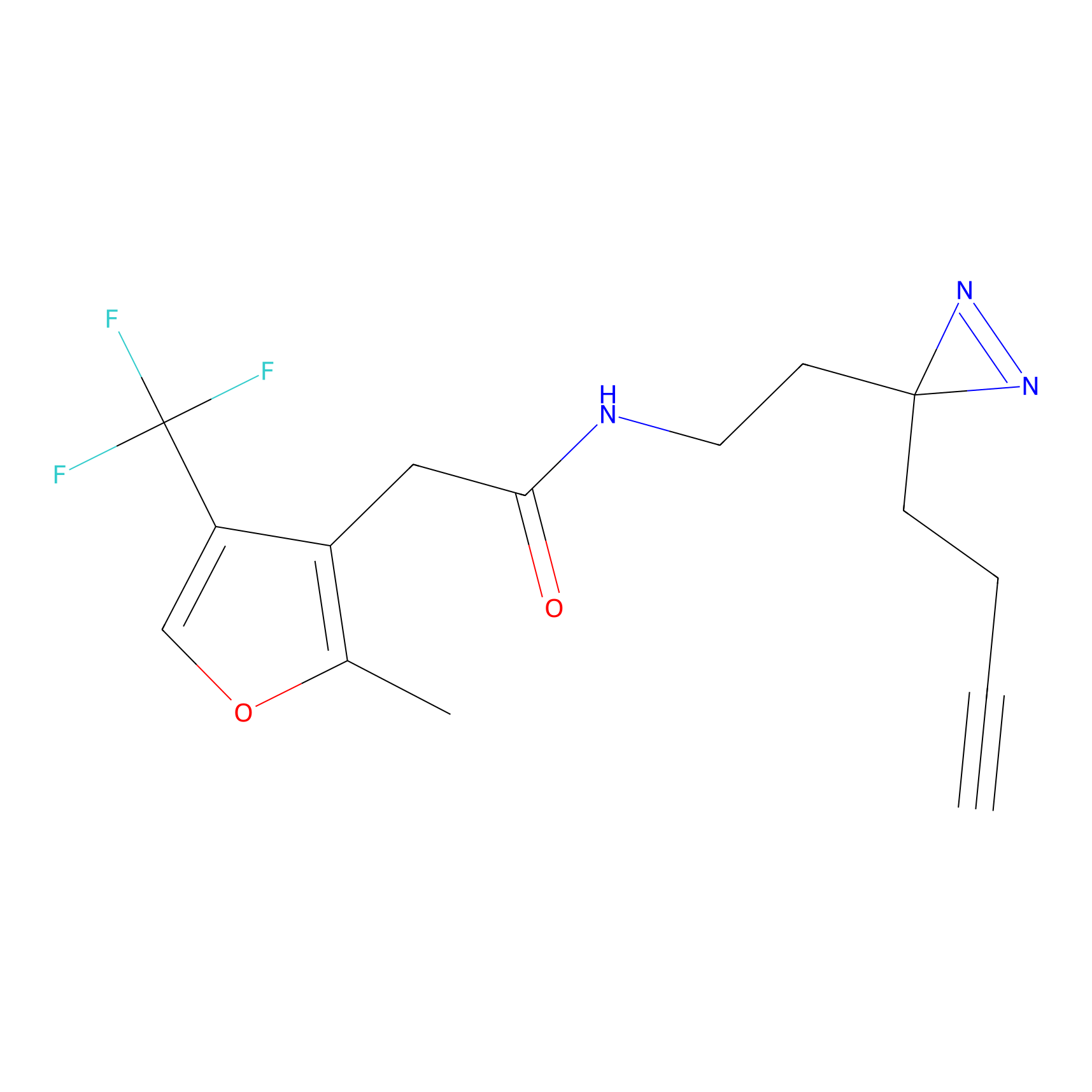
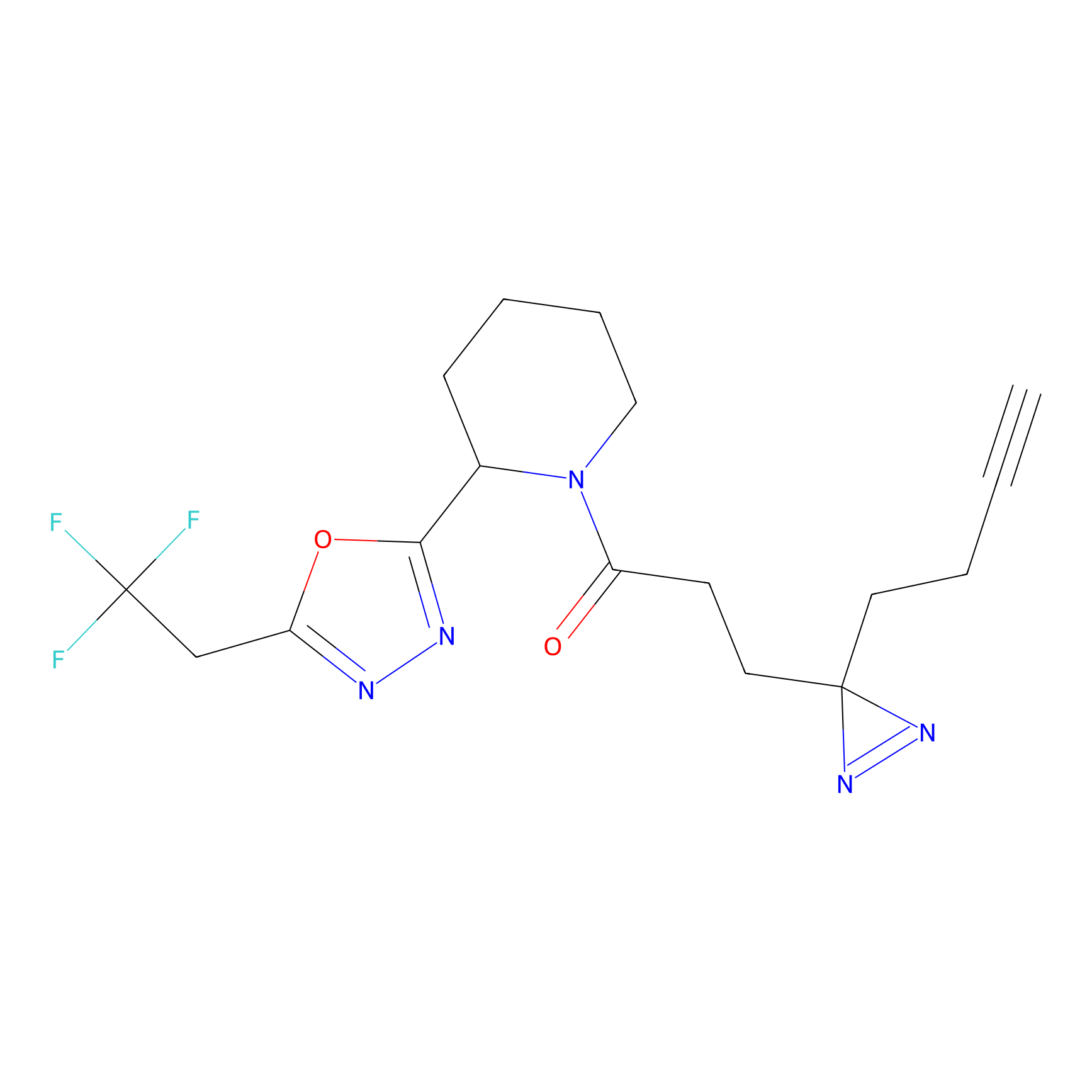
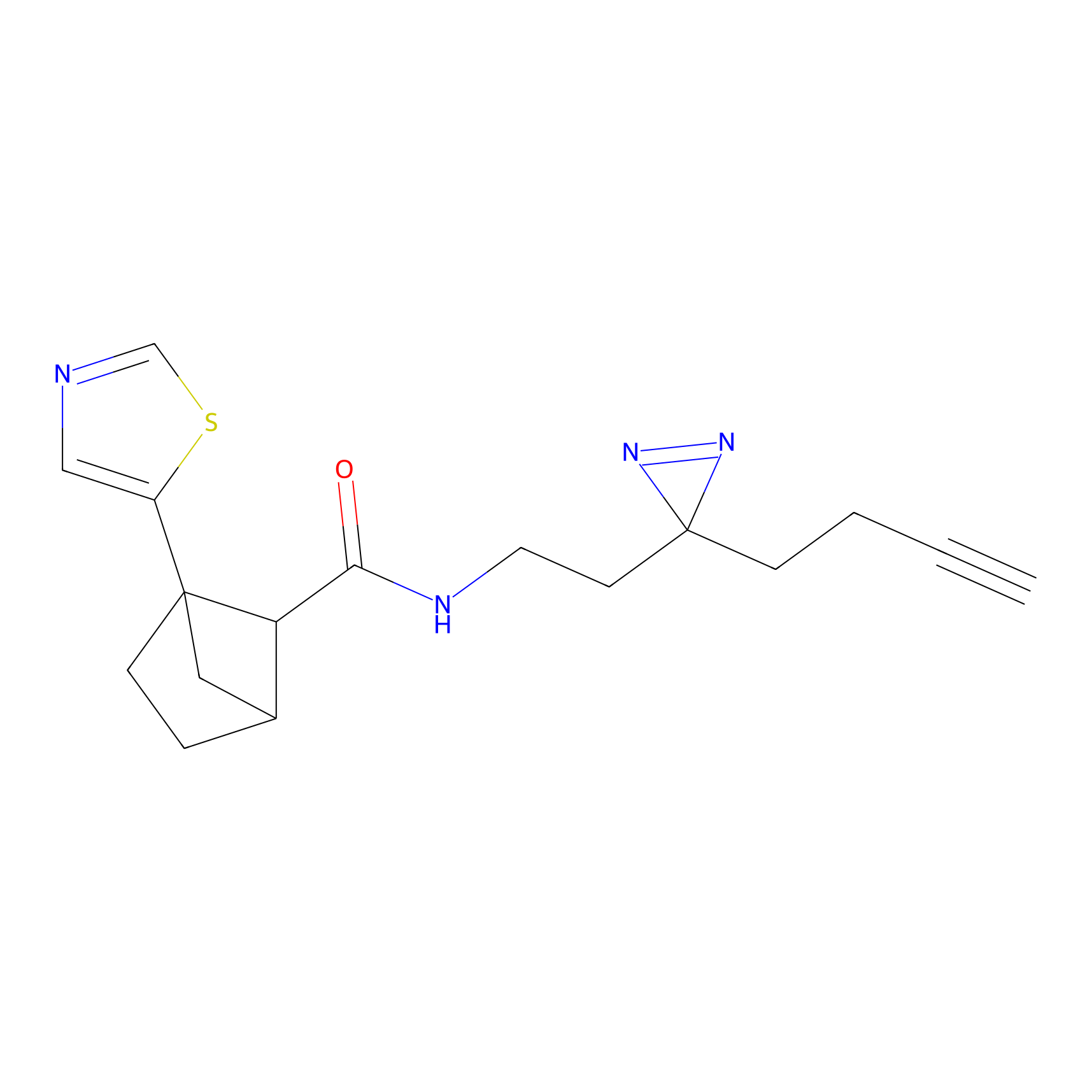
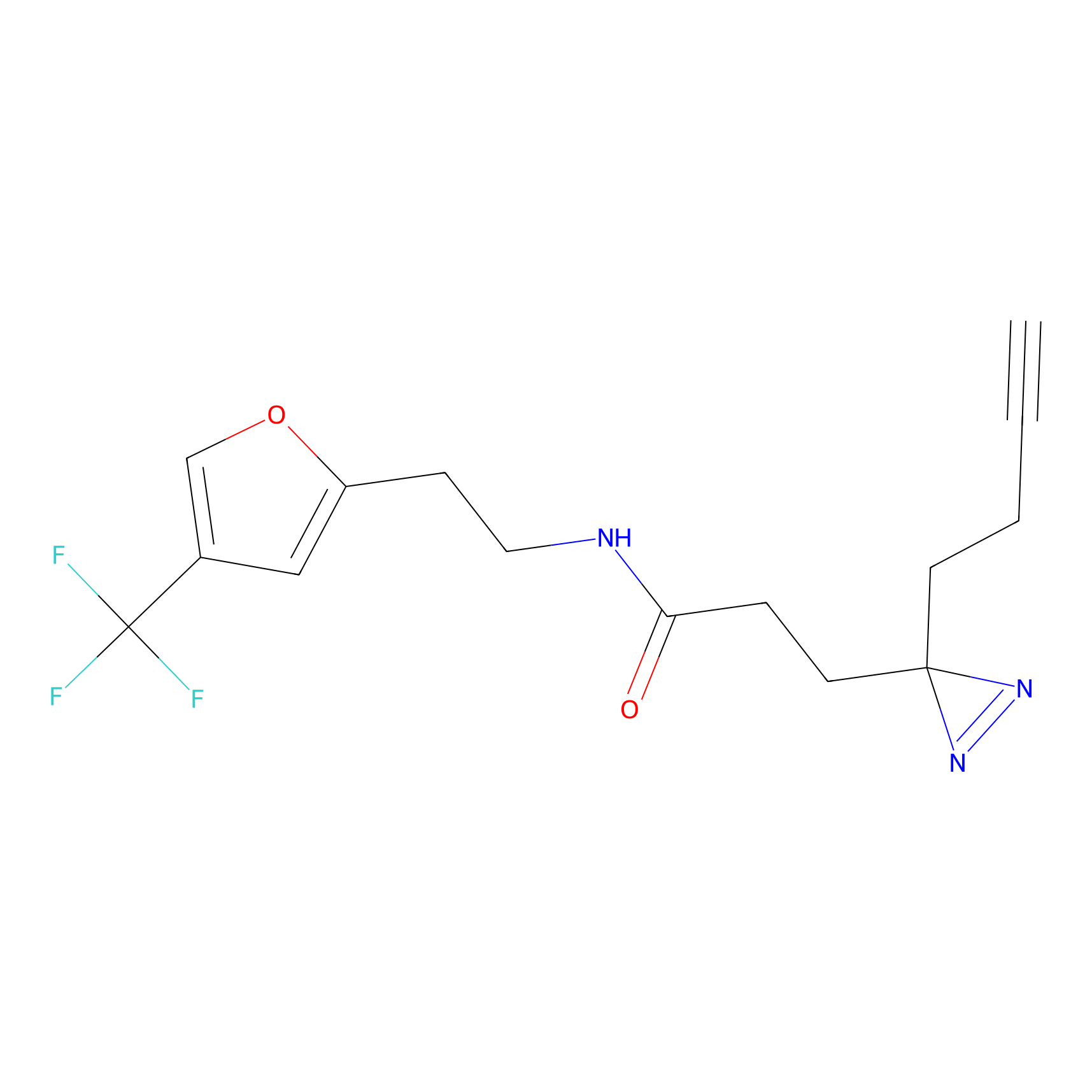
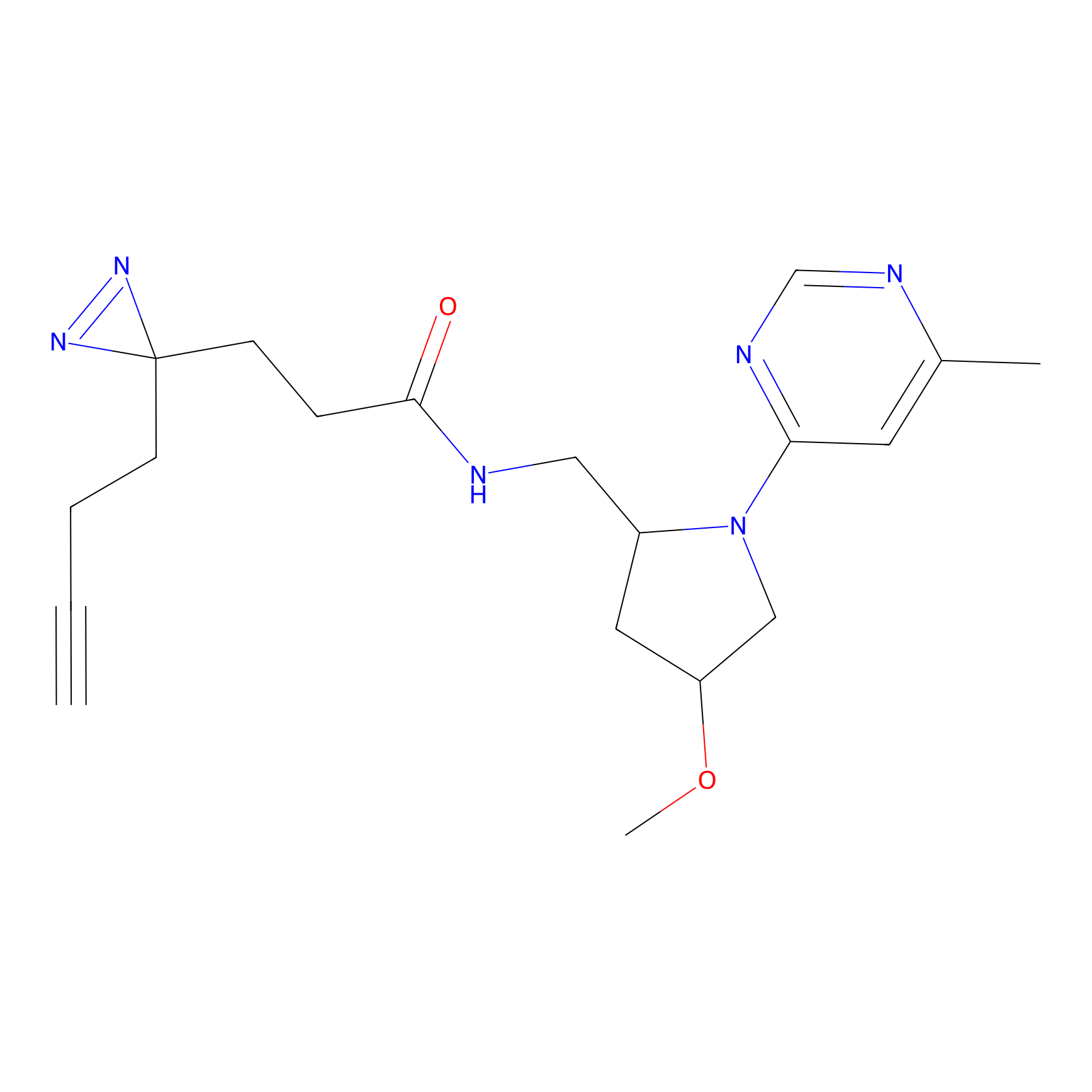
|
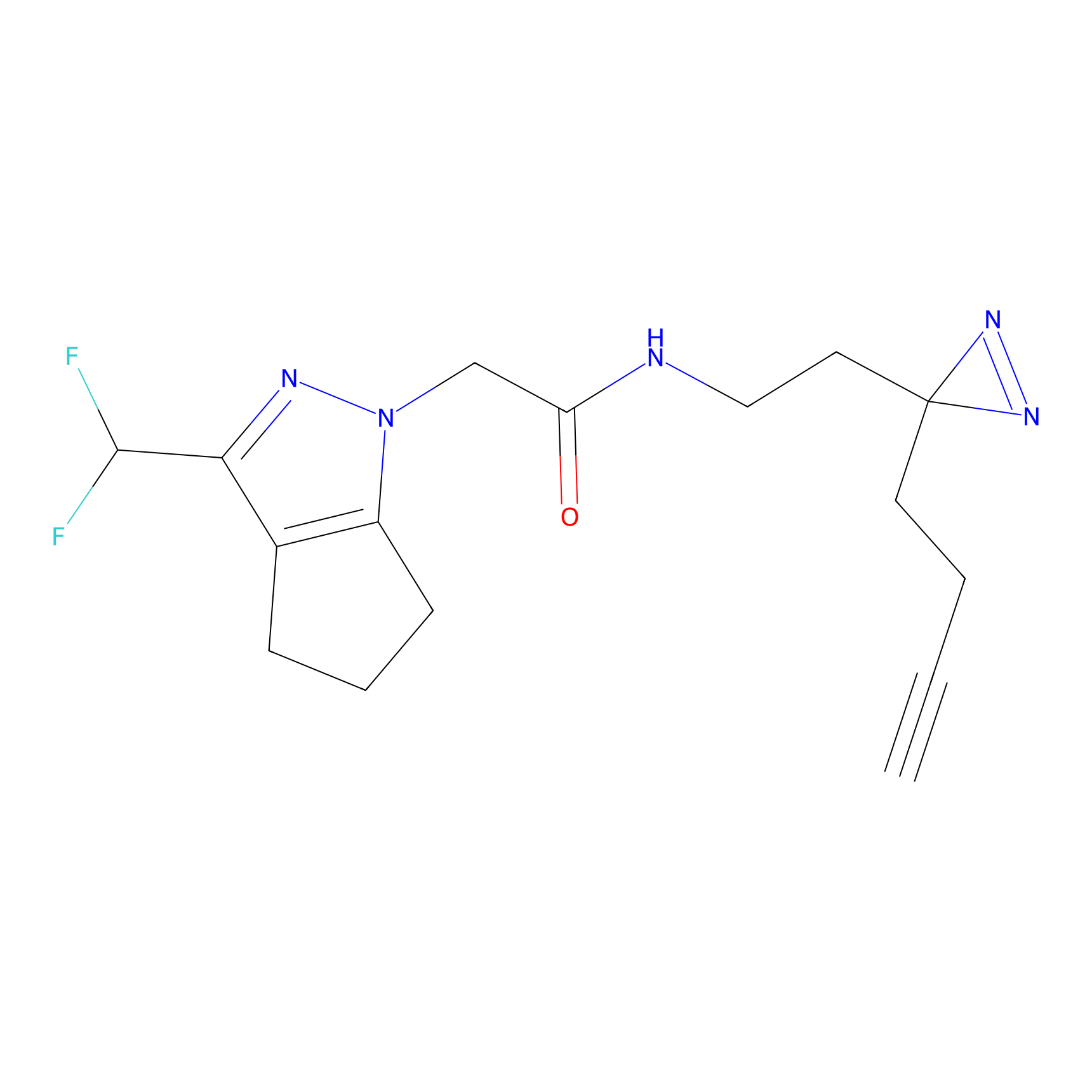
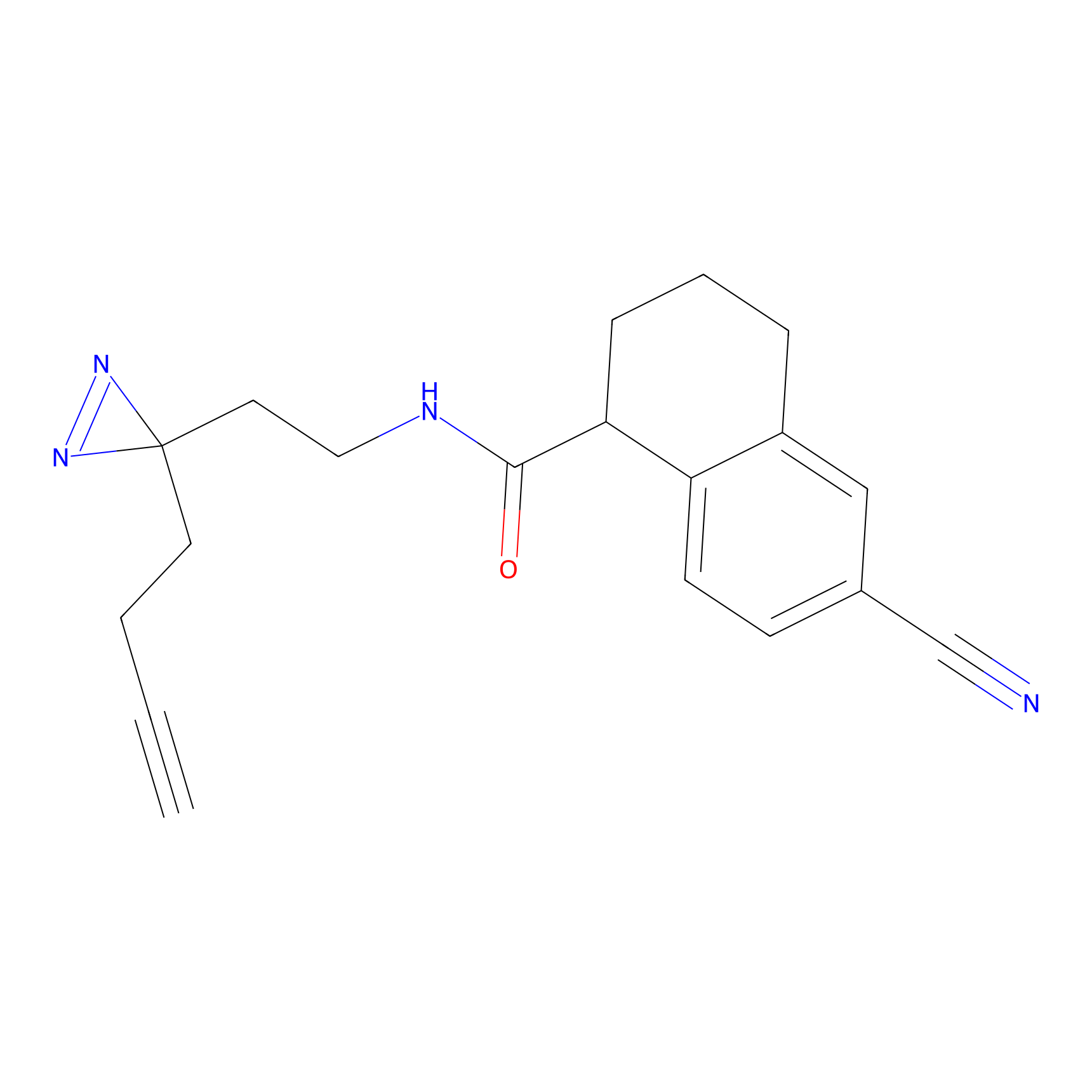
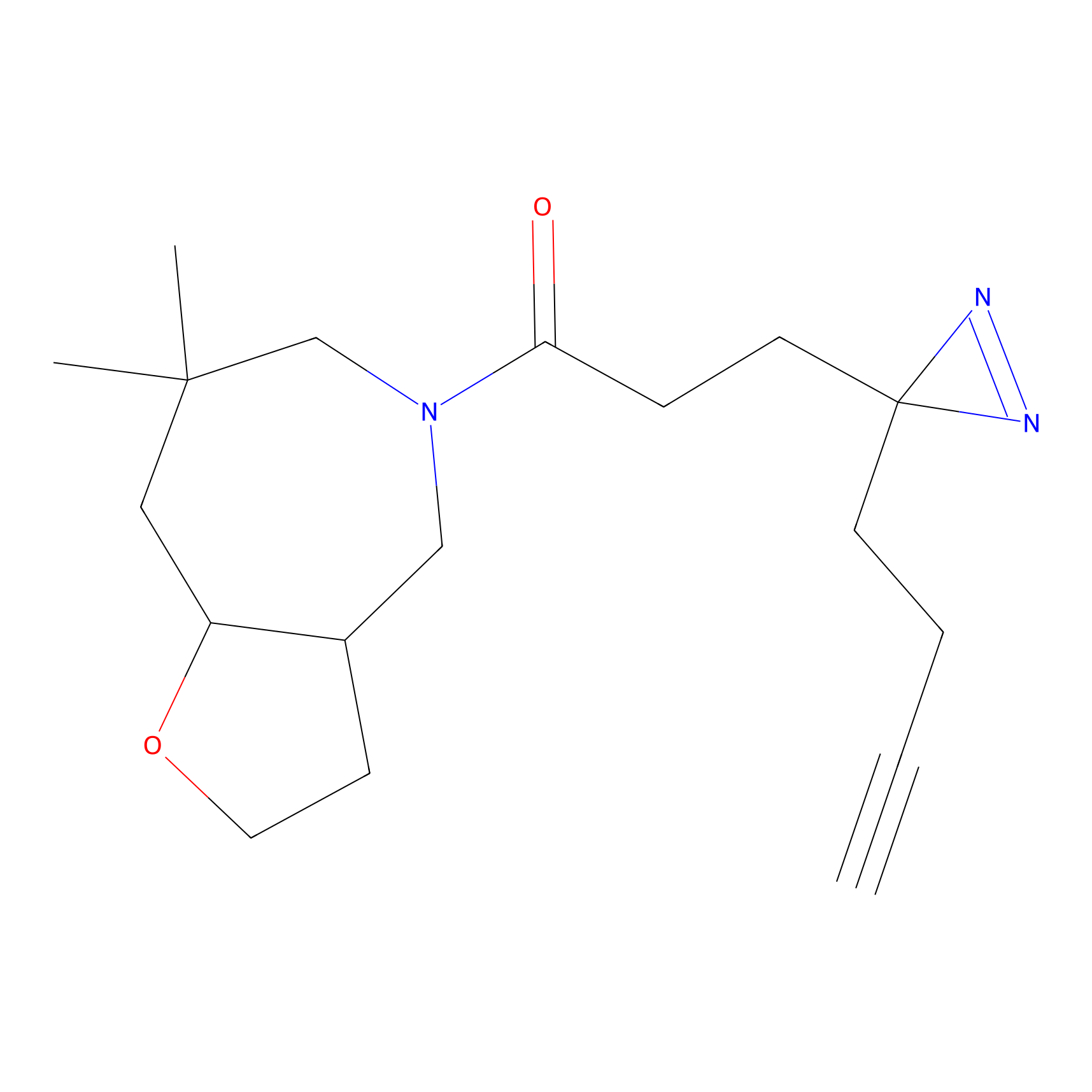
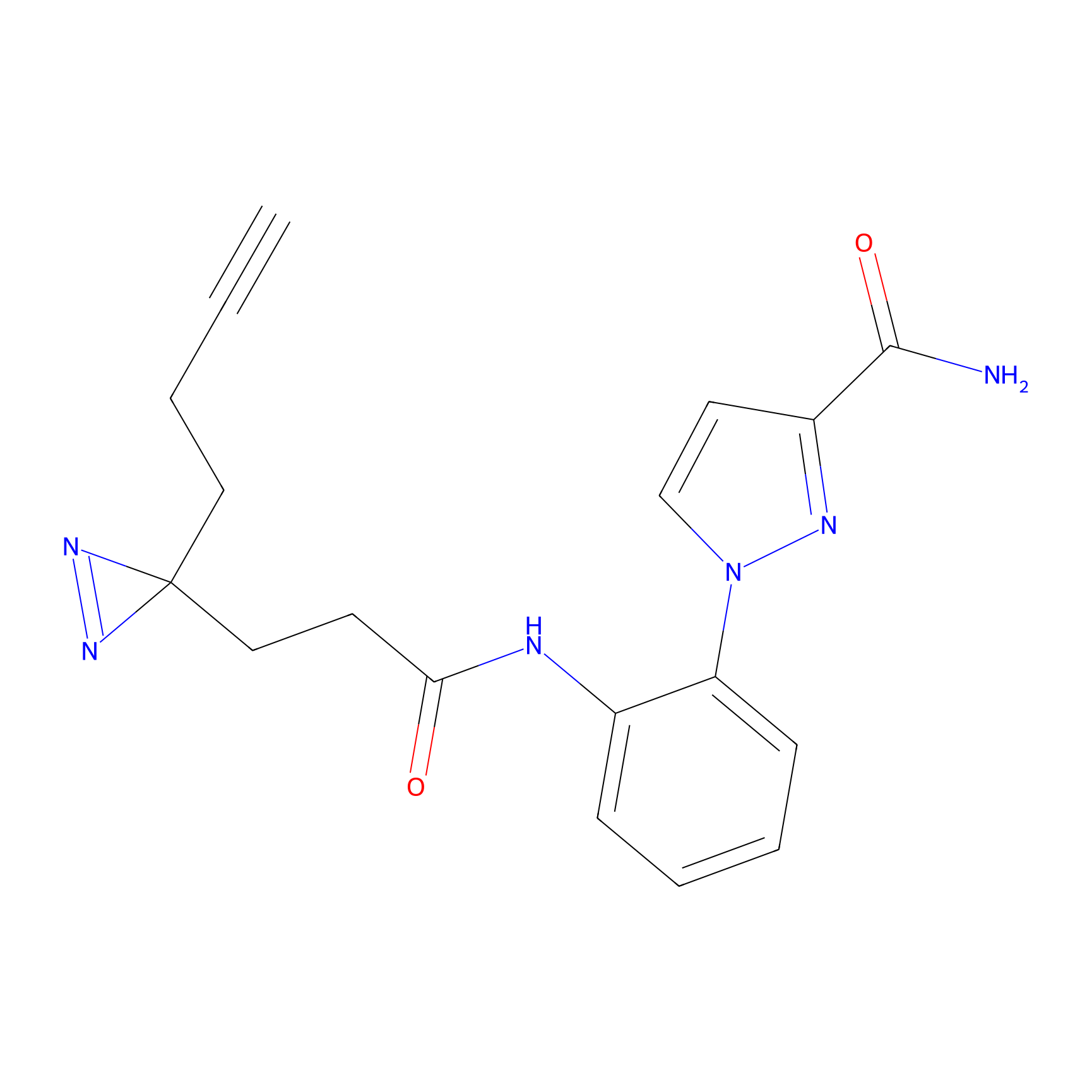
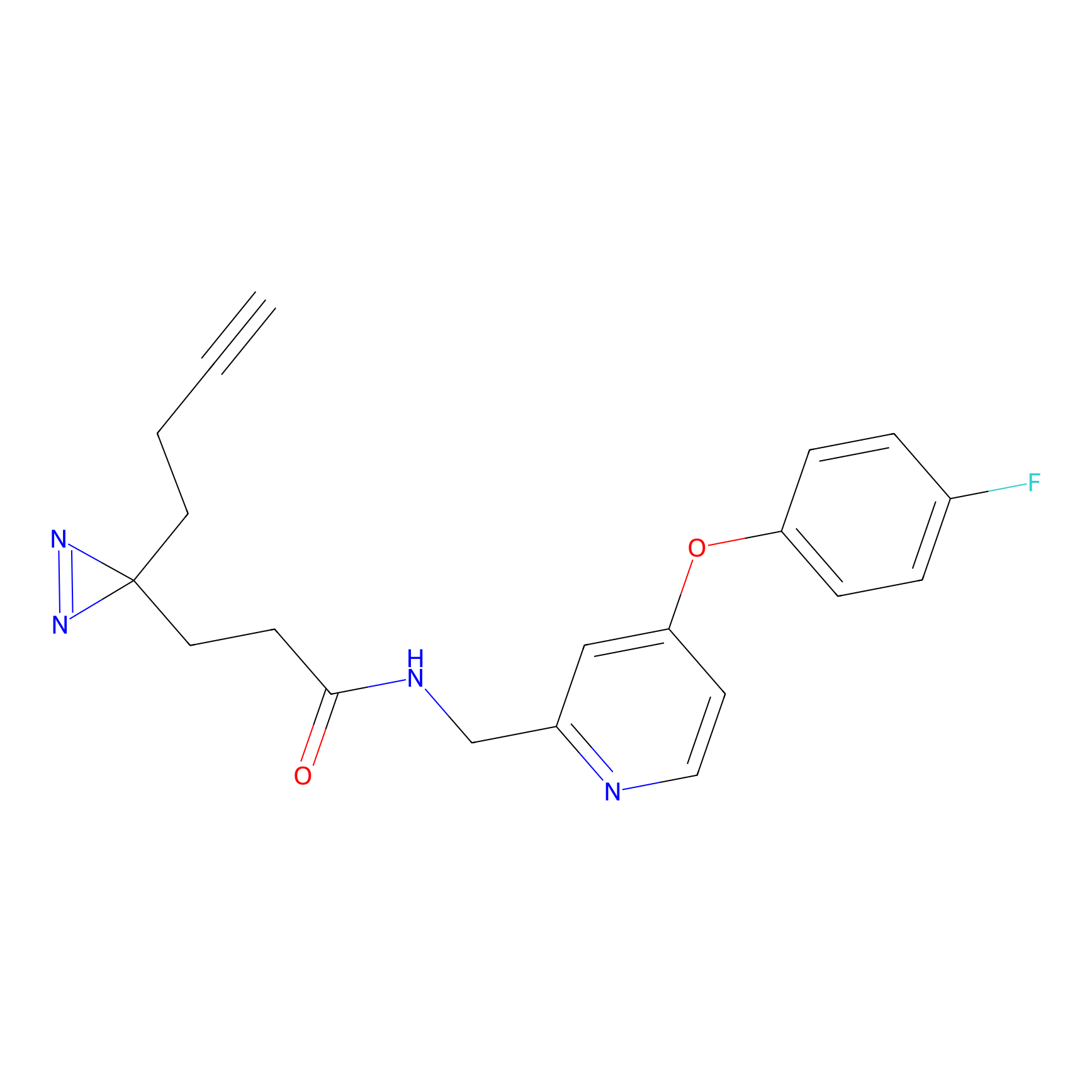
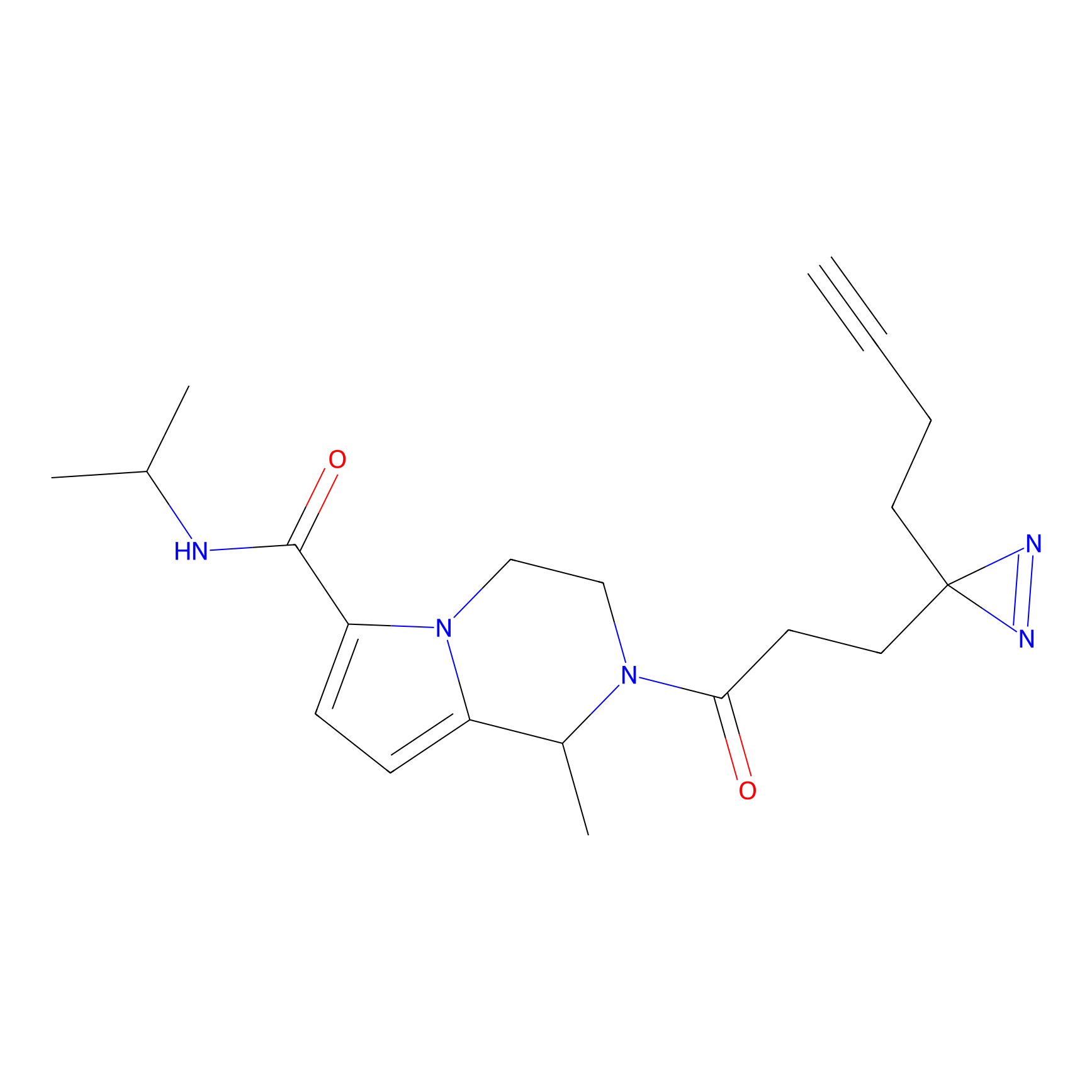
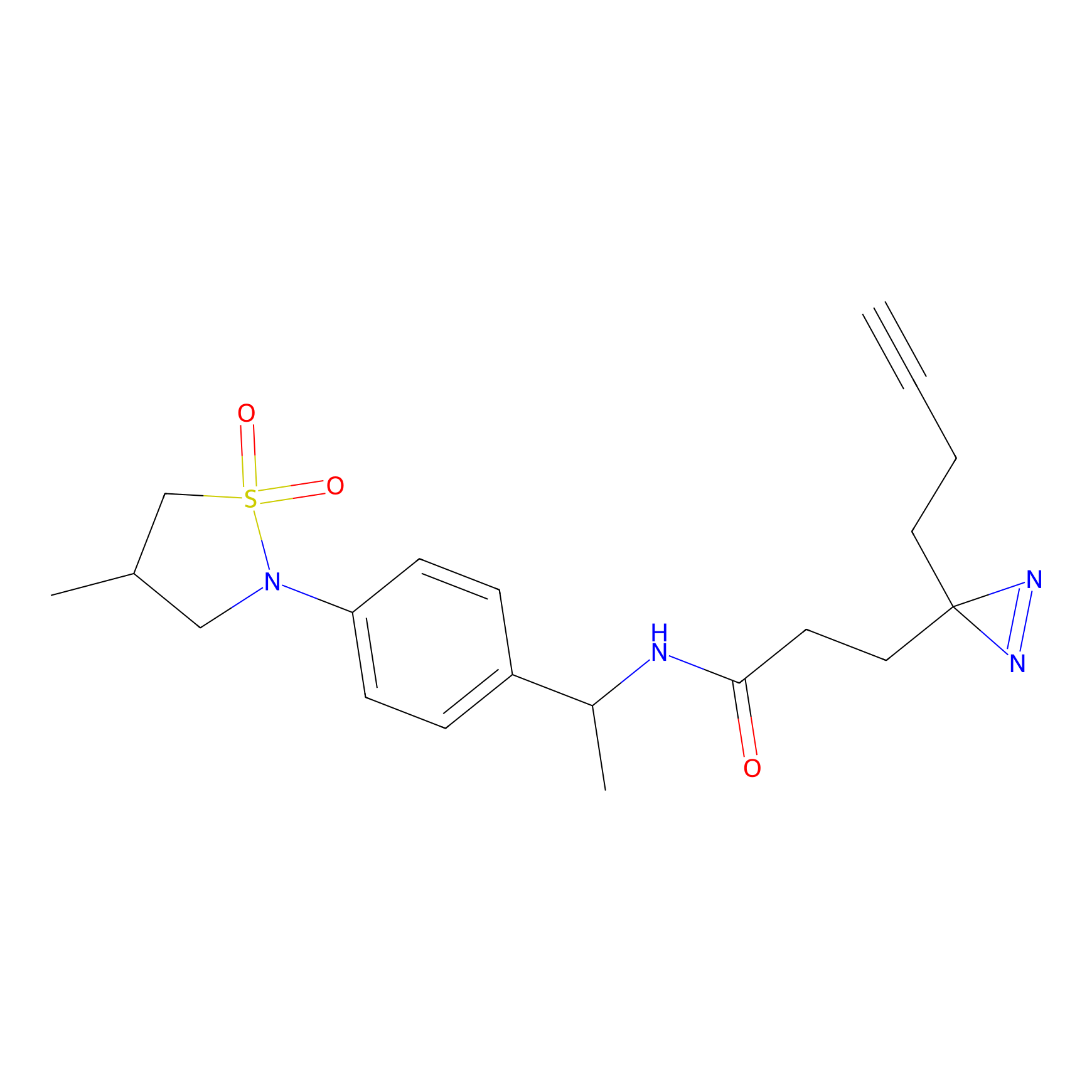
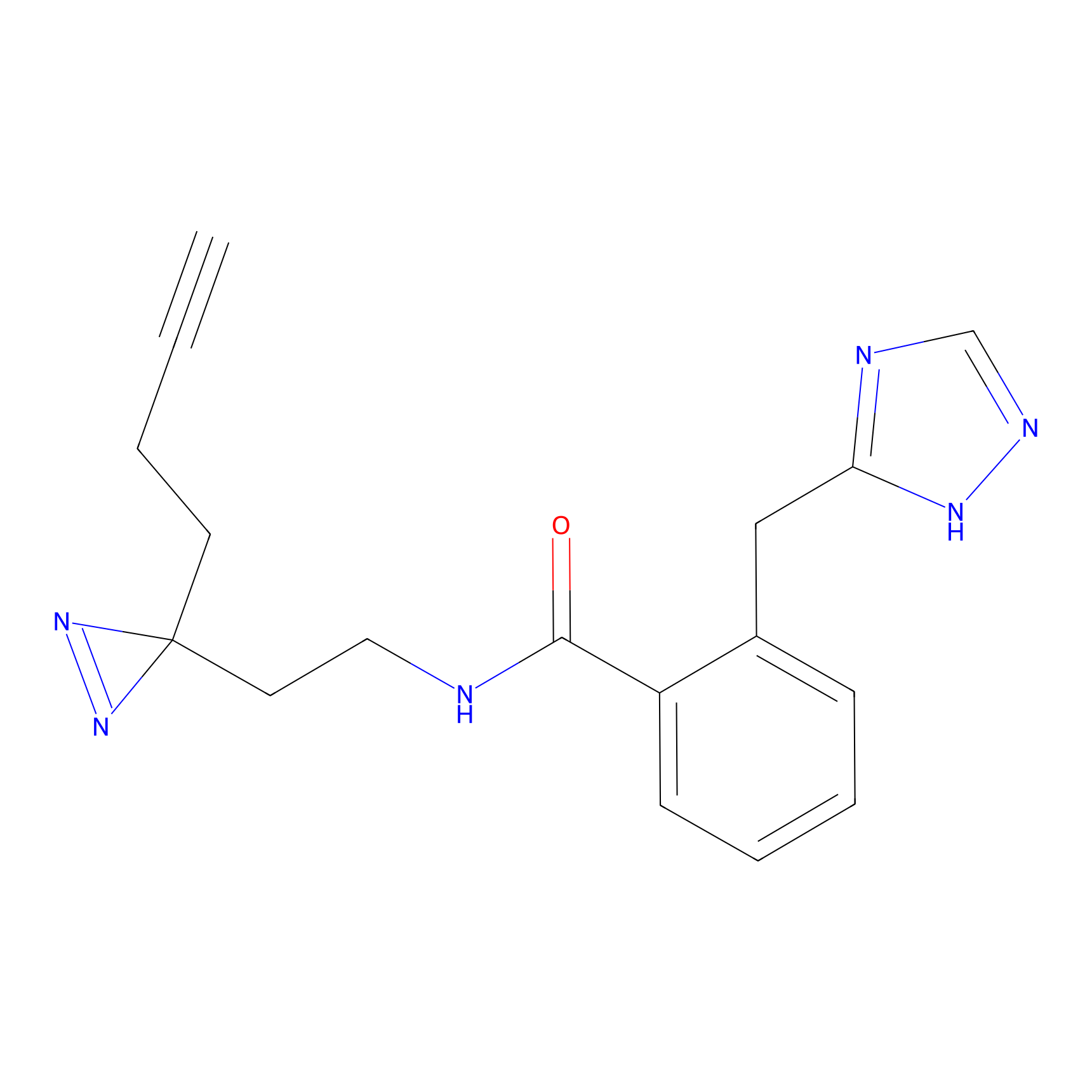
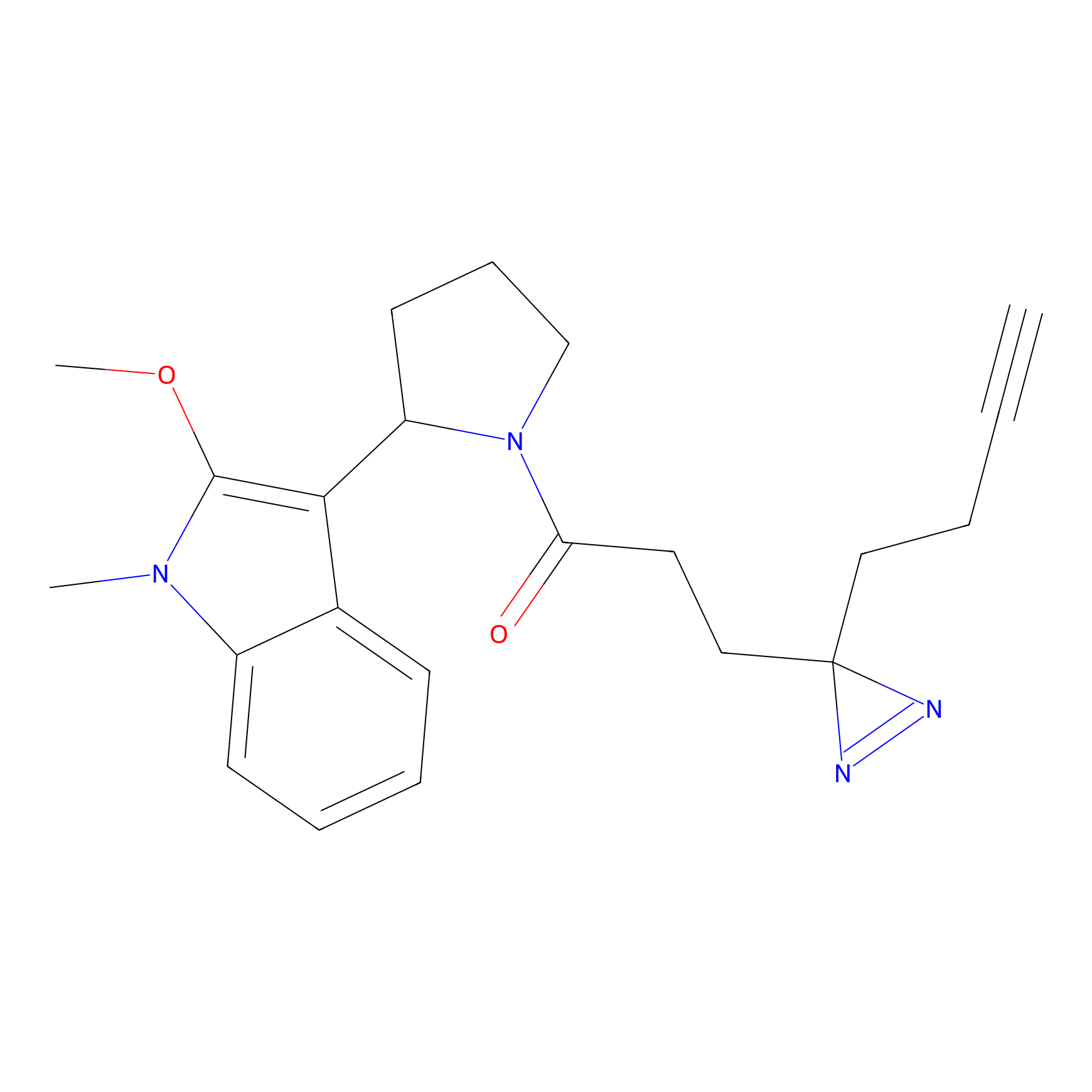
TH216 Probe Info |
 |
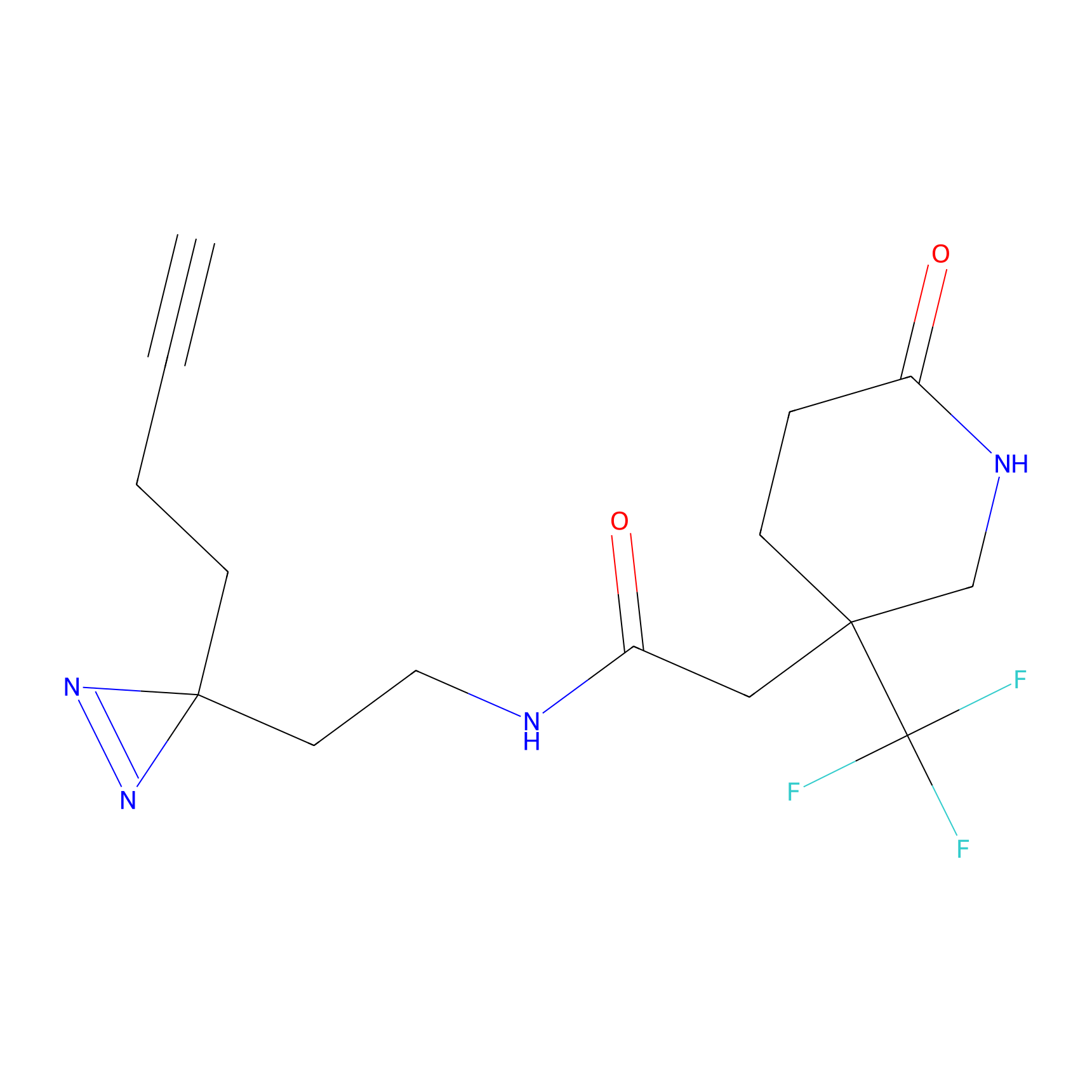
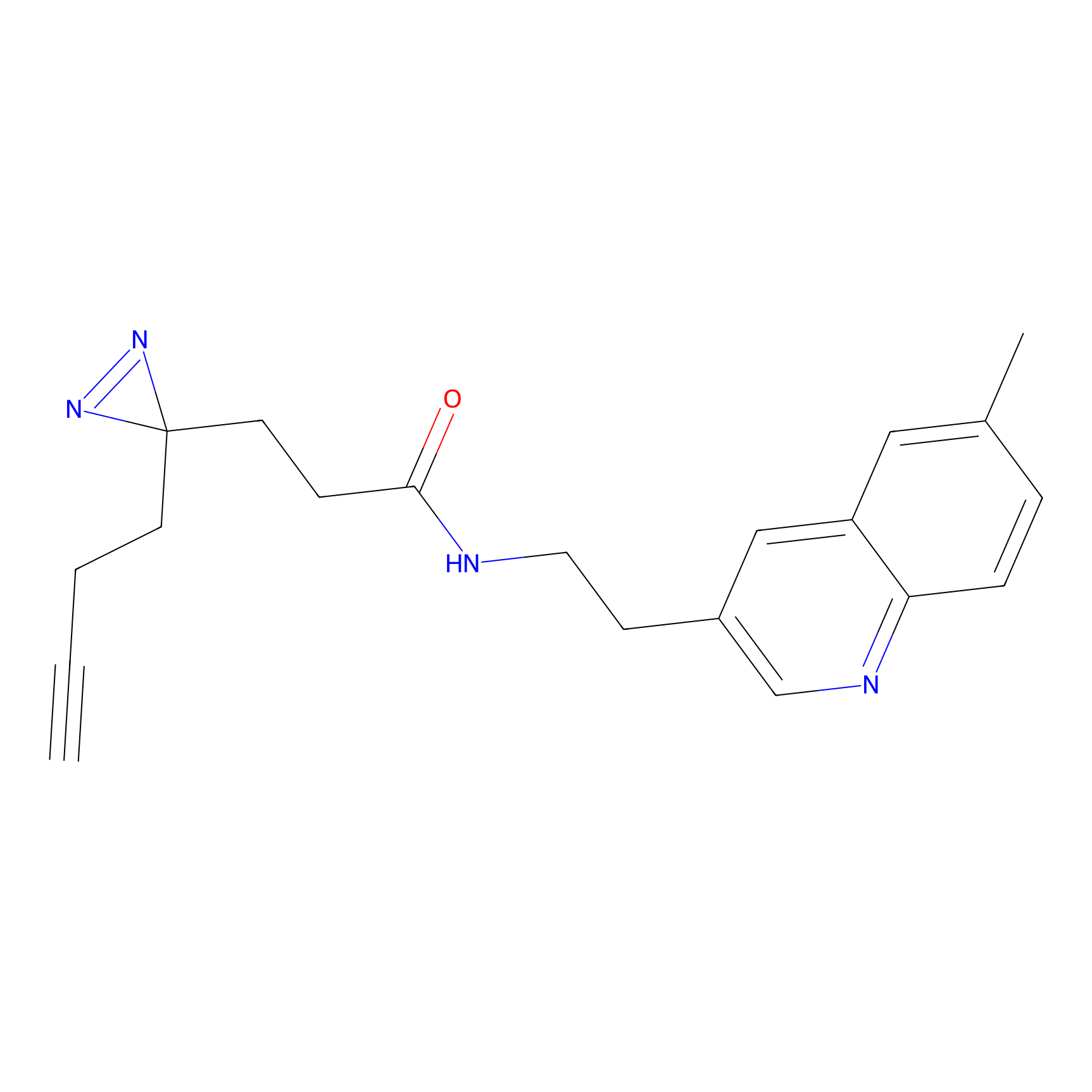
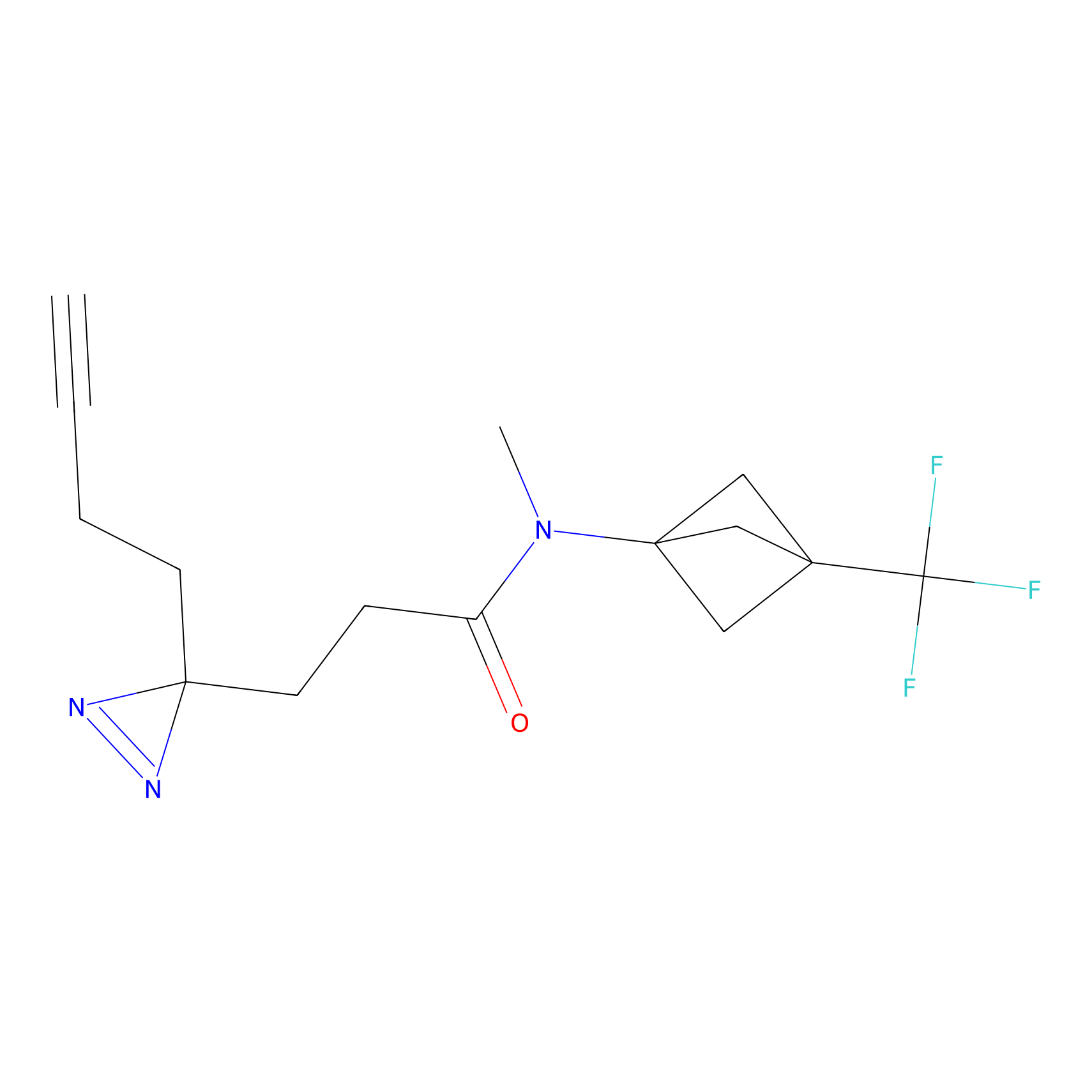
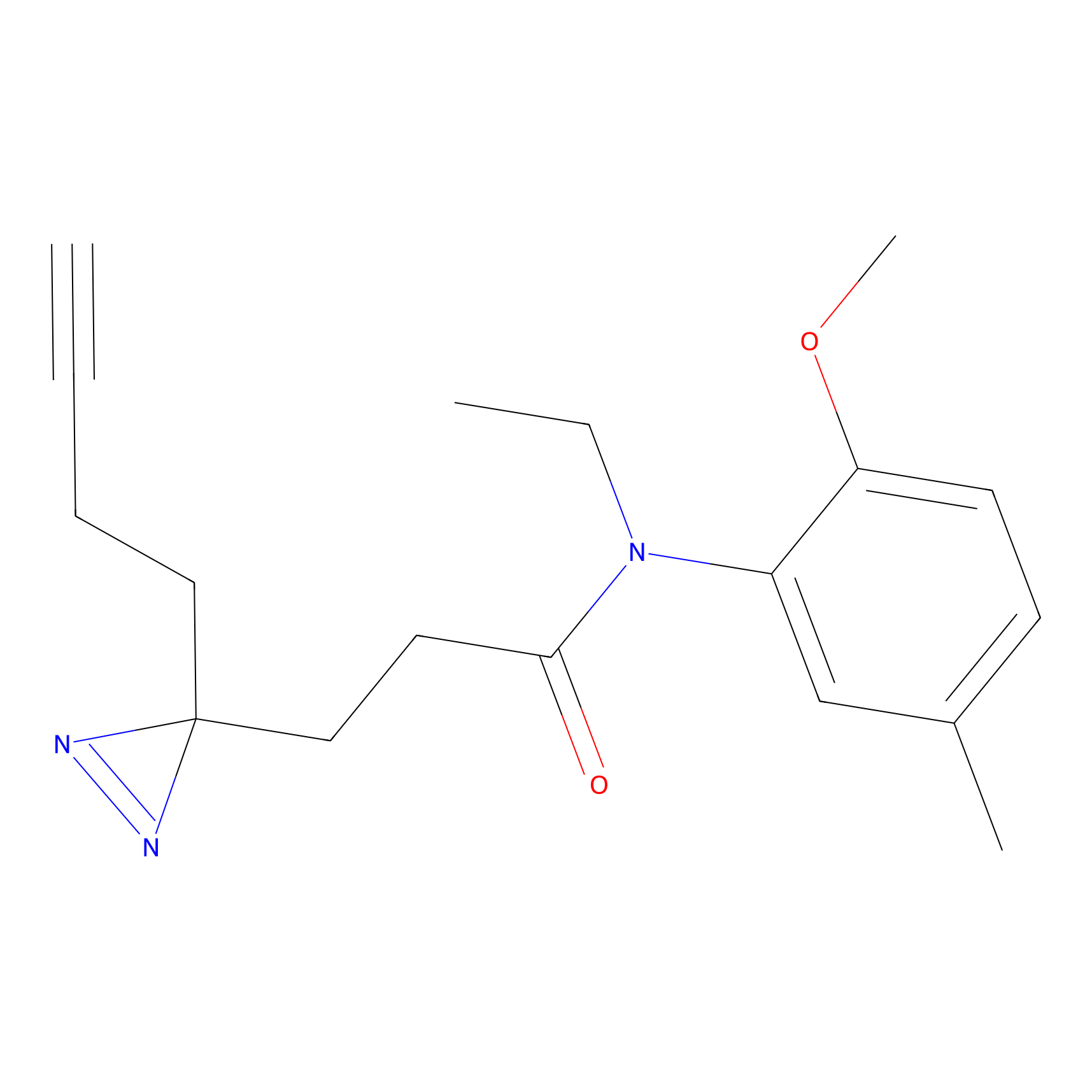
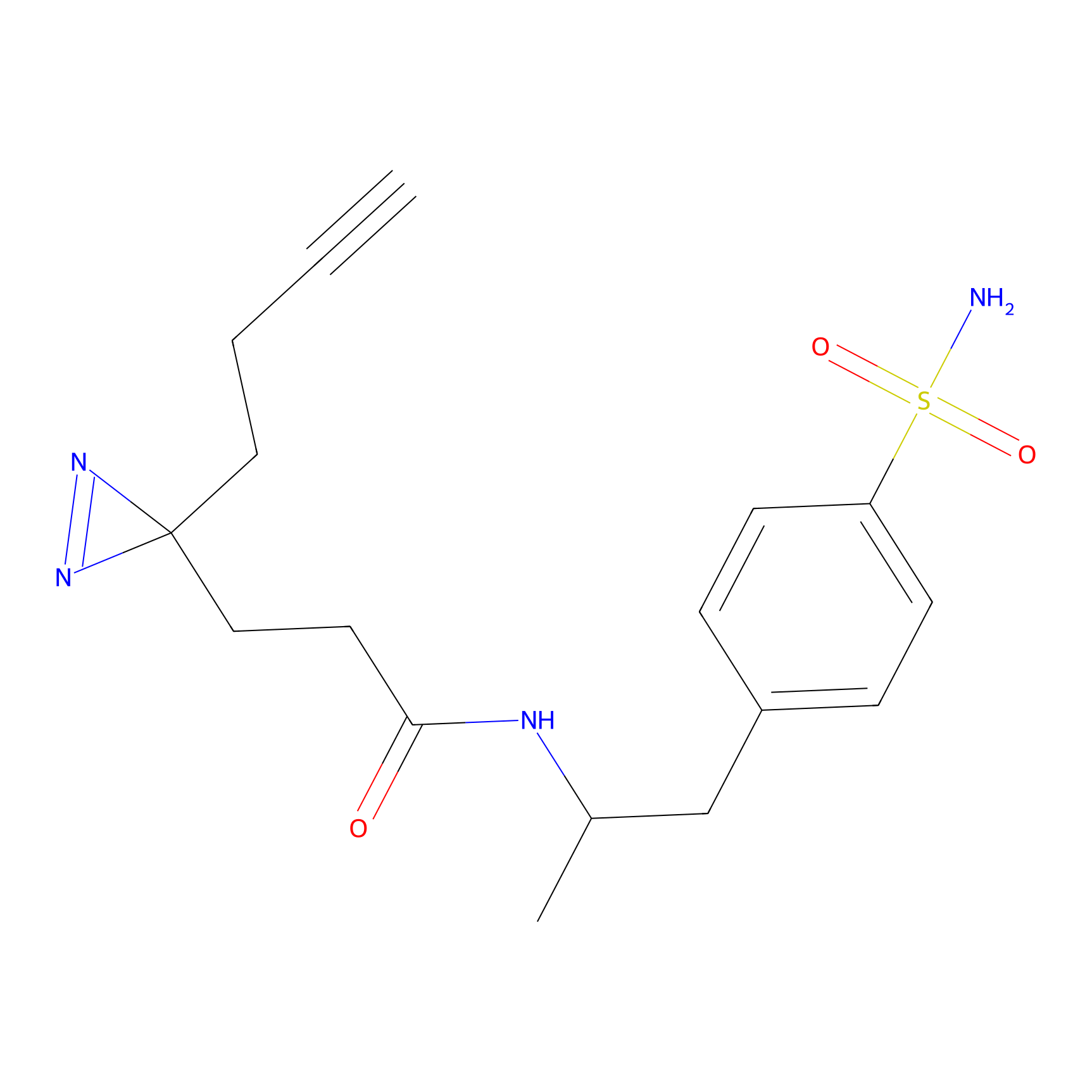
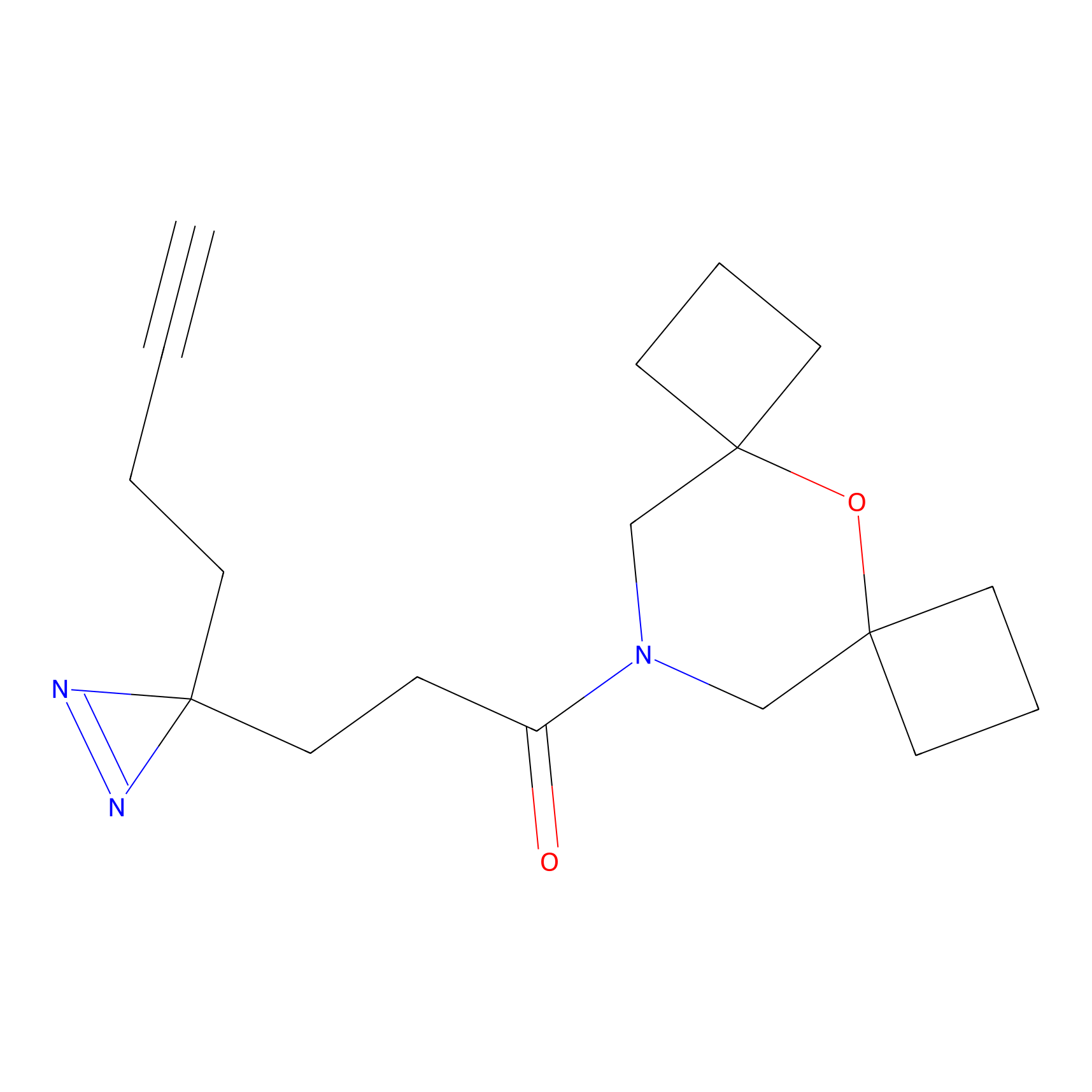
Y122(17.99); Y157(16.91); Y163(6.94) | LDD0259 | [2] | |
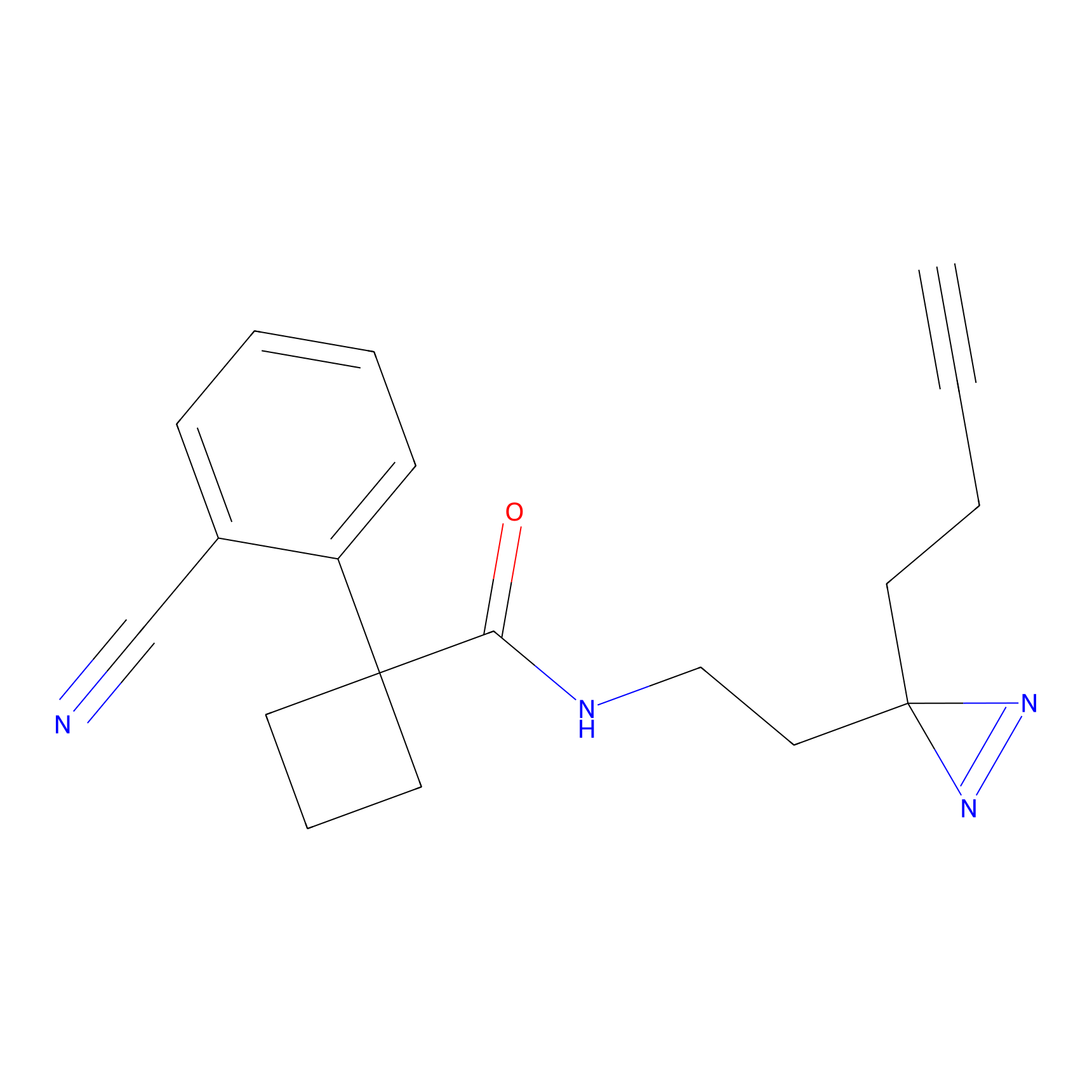
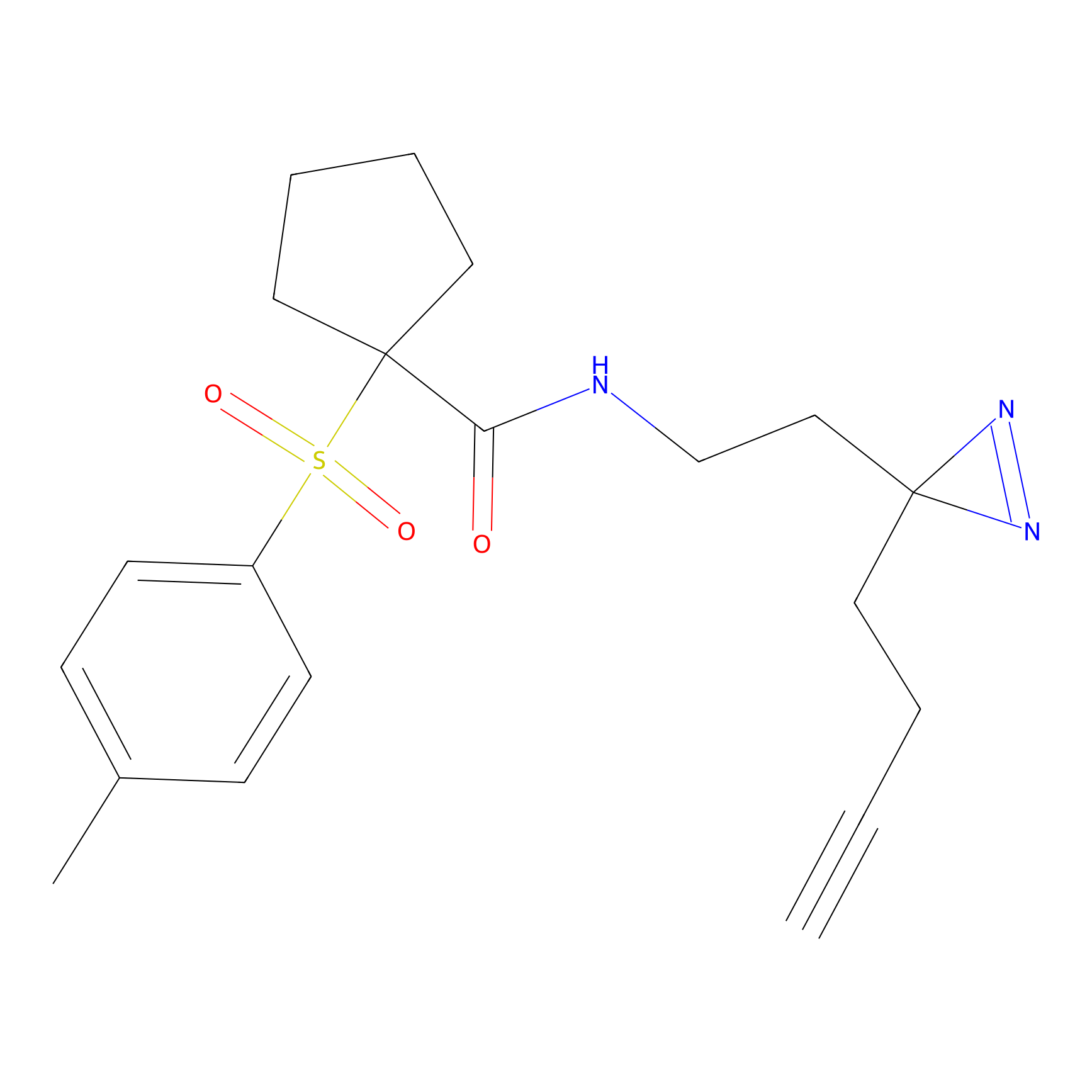
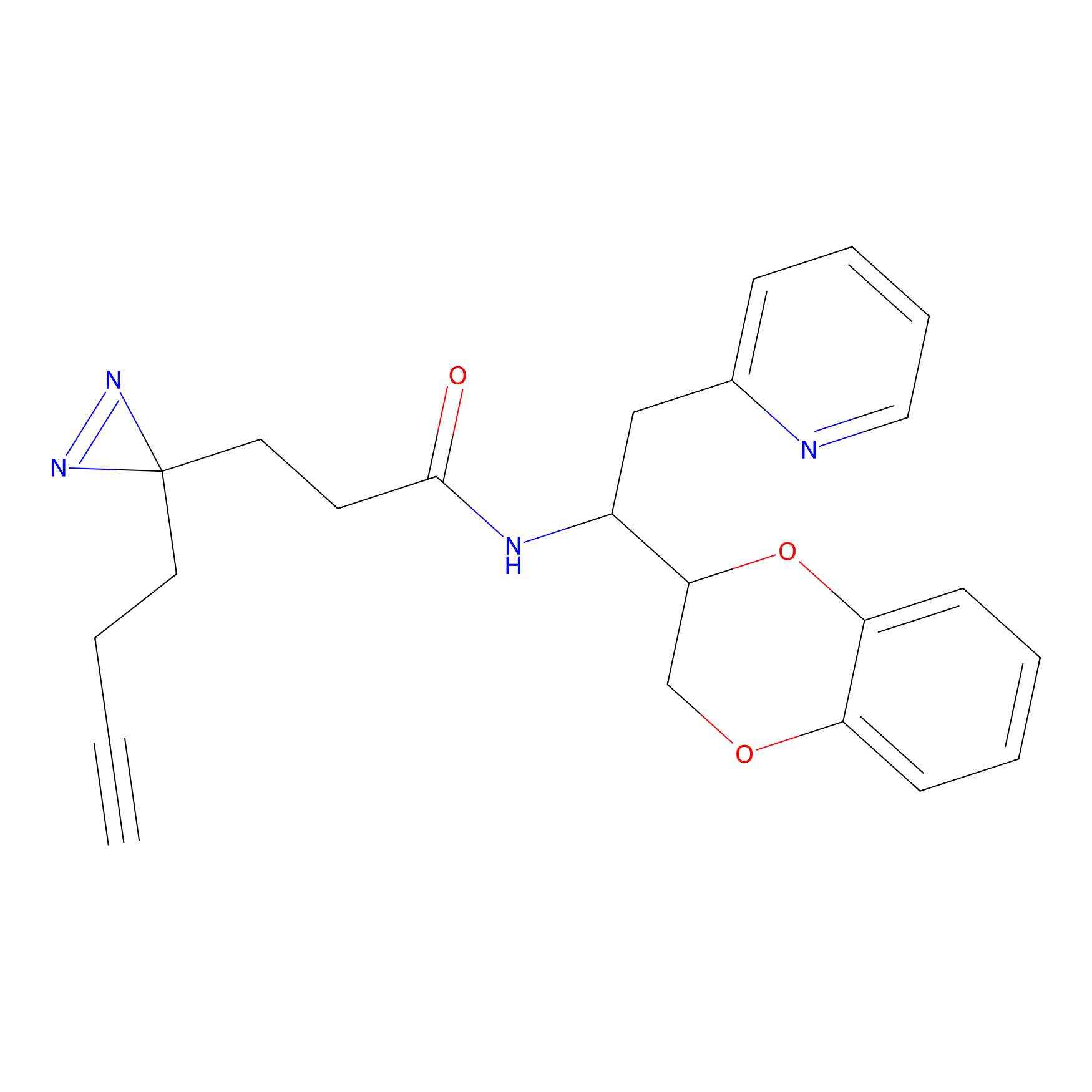
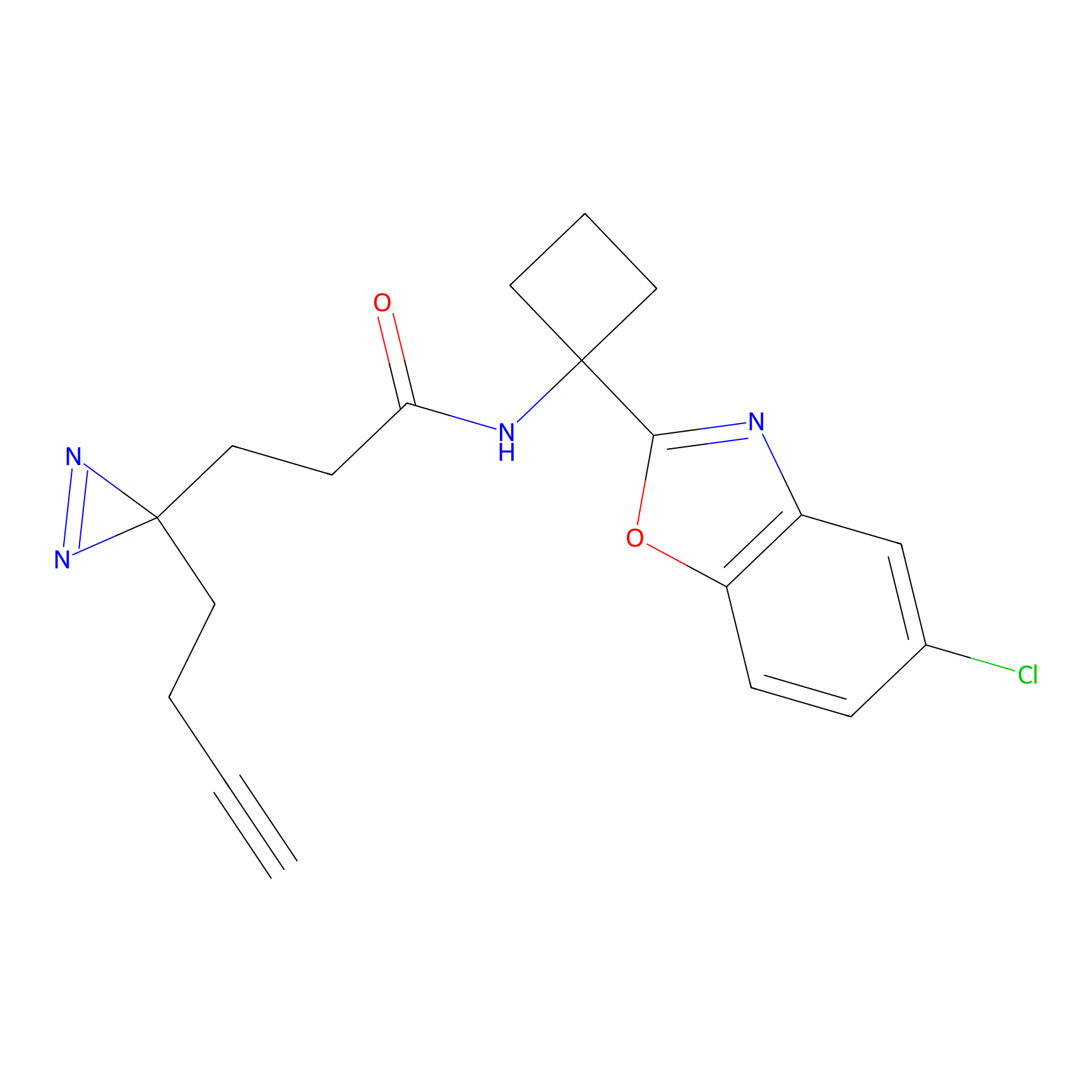
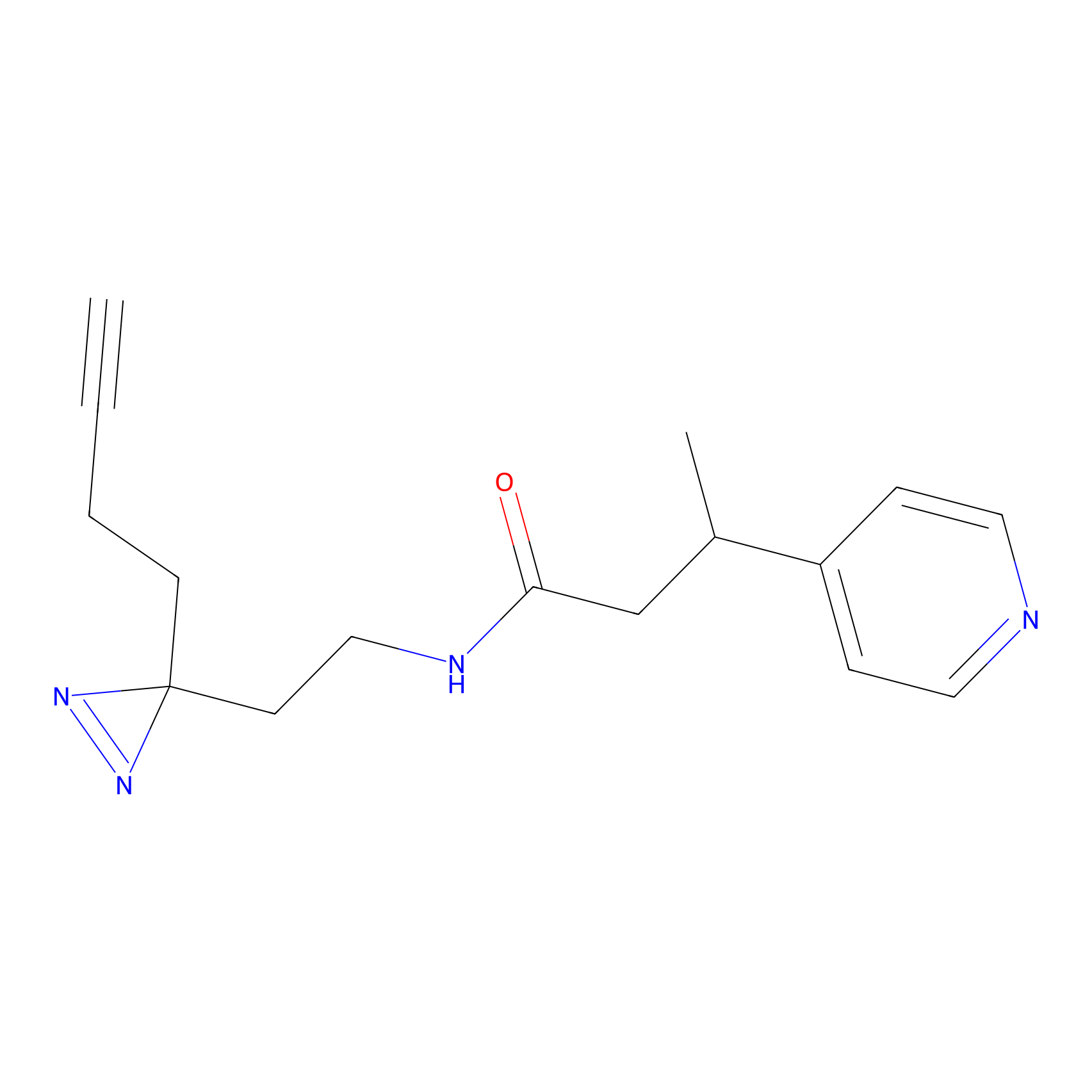
|
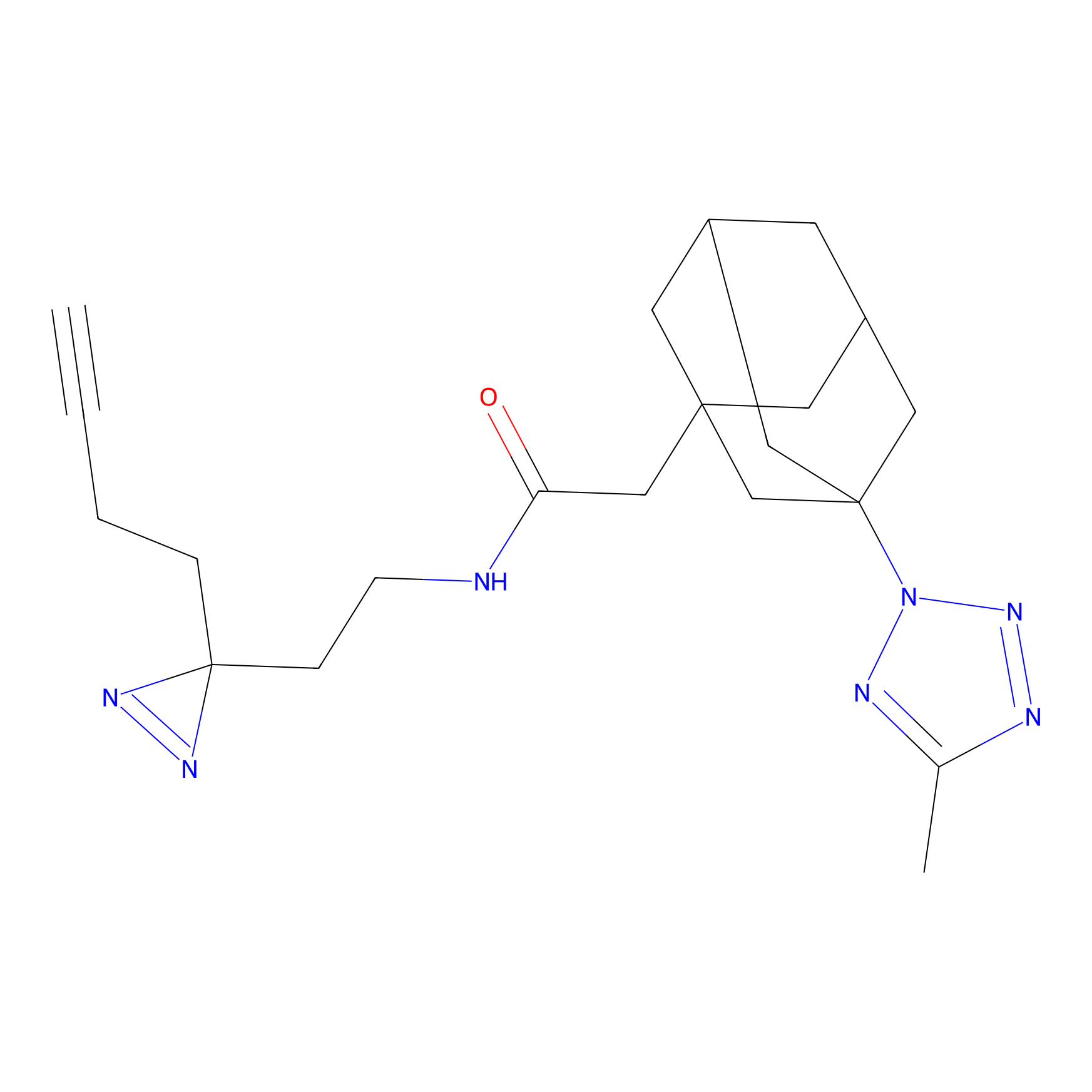
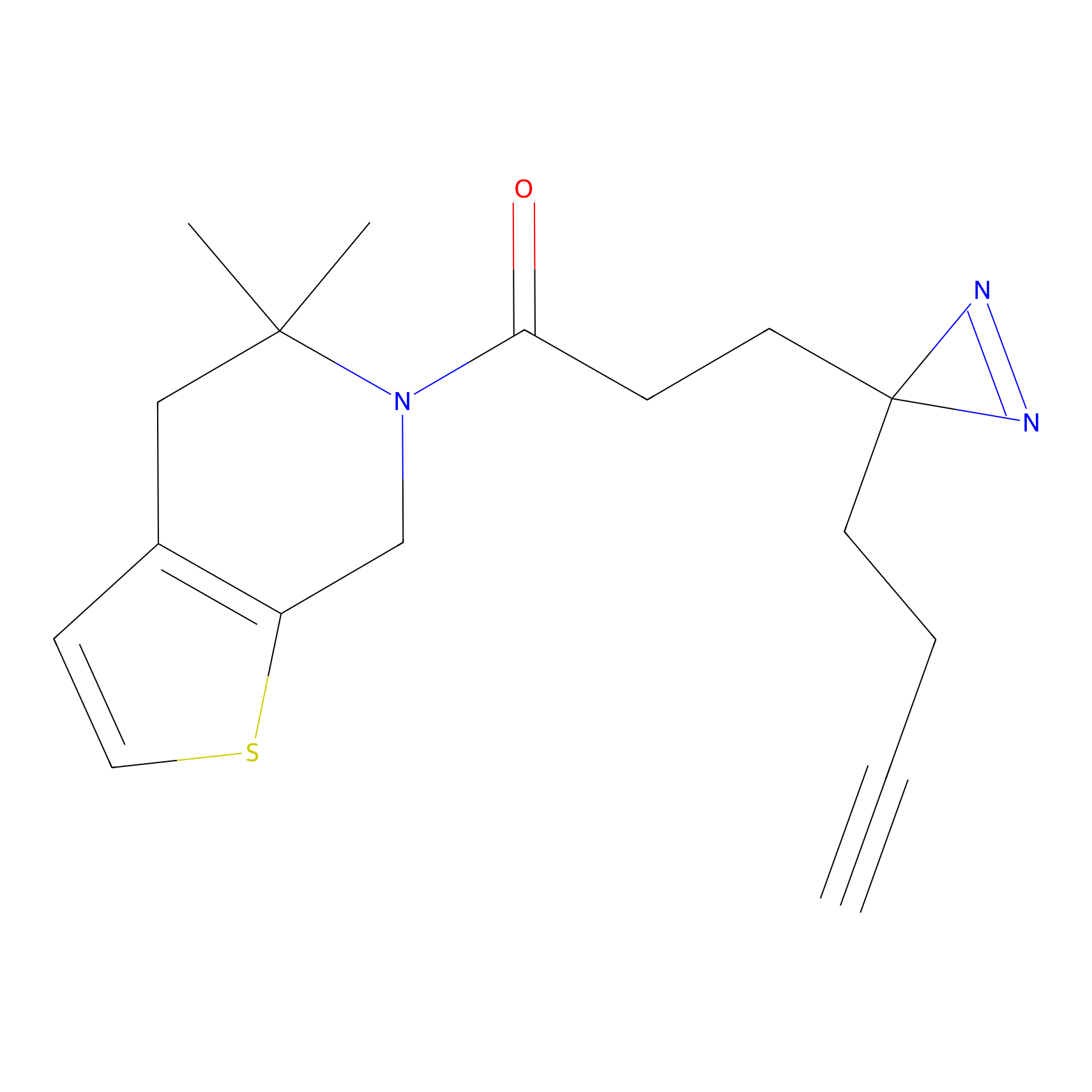
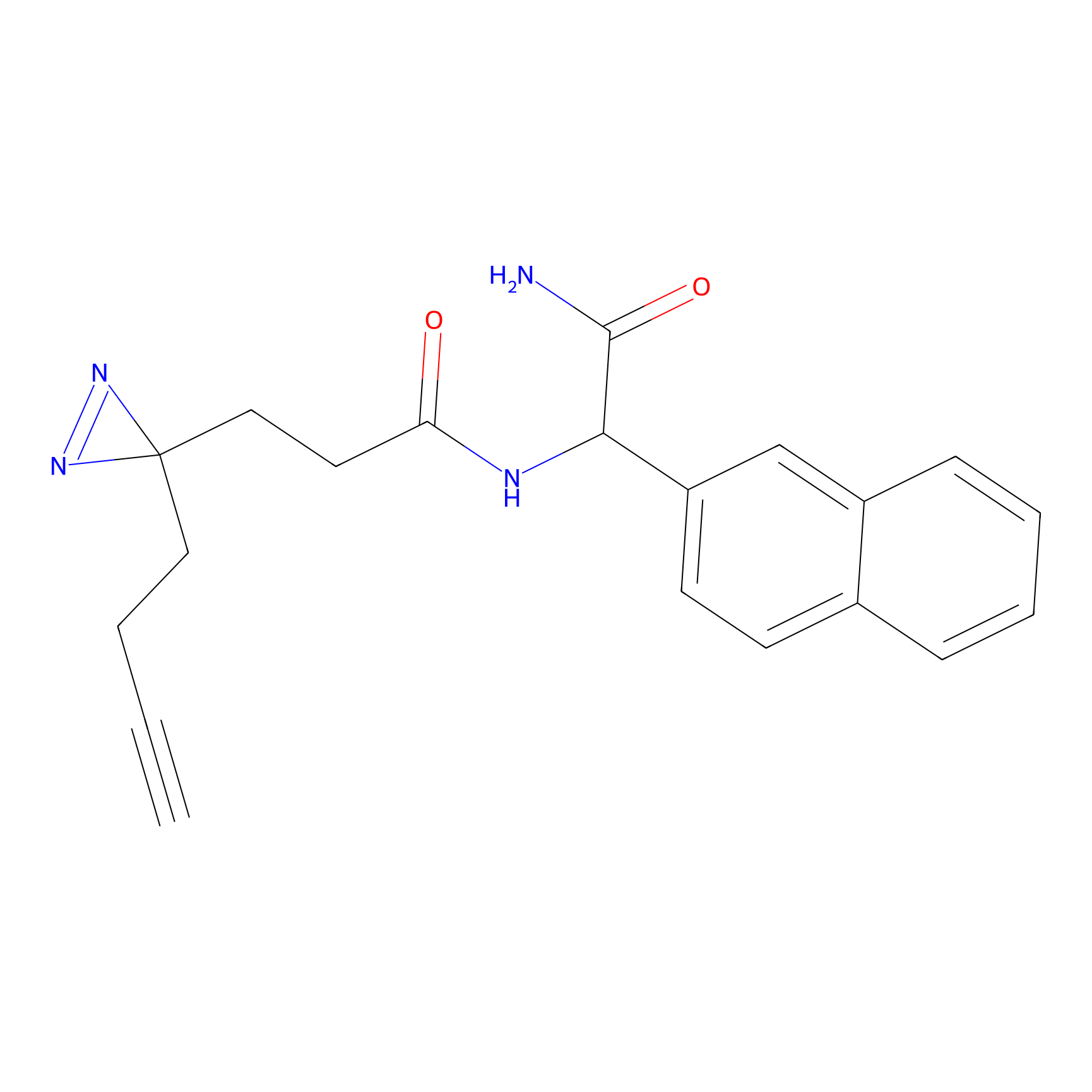
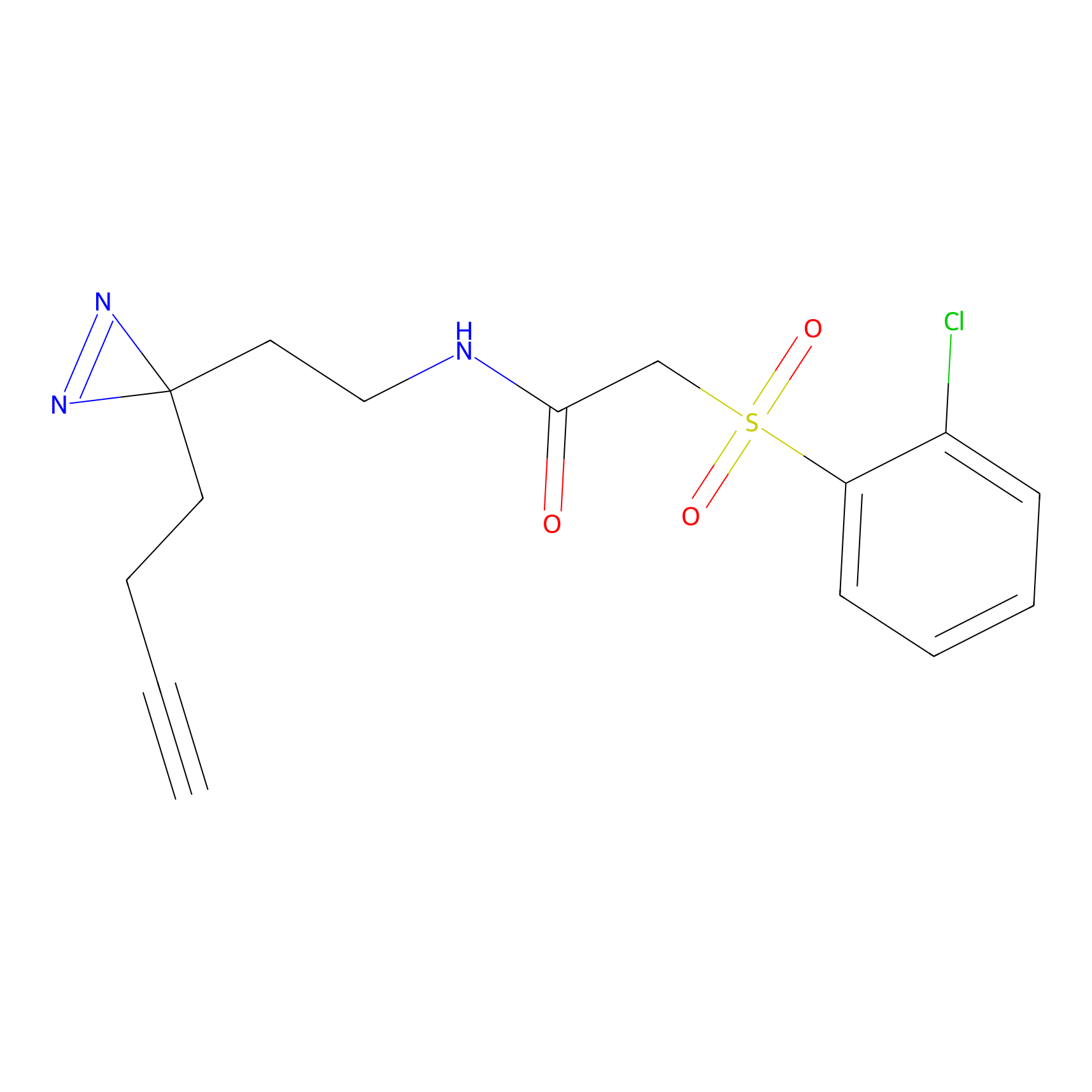
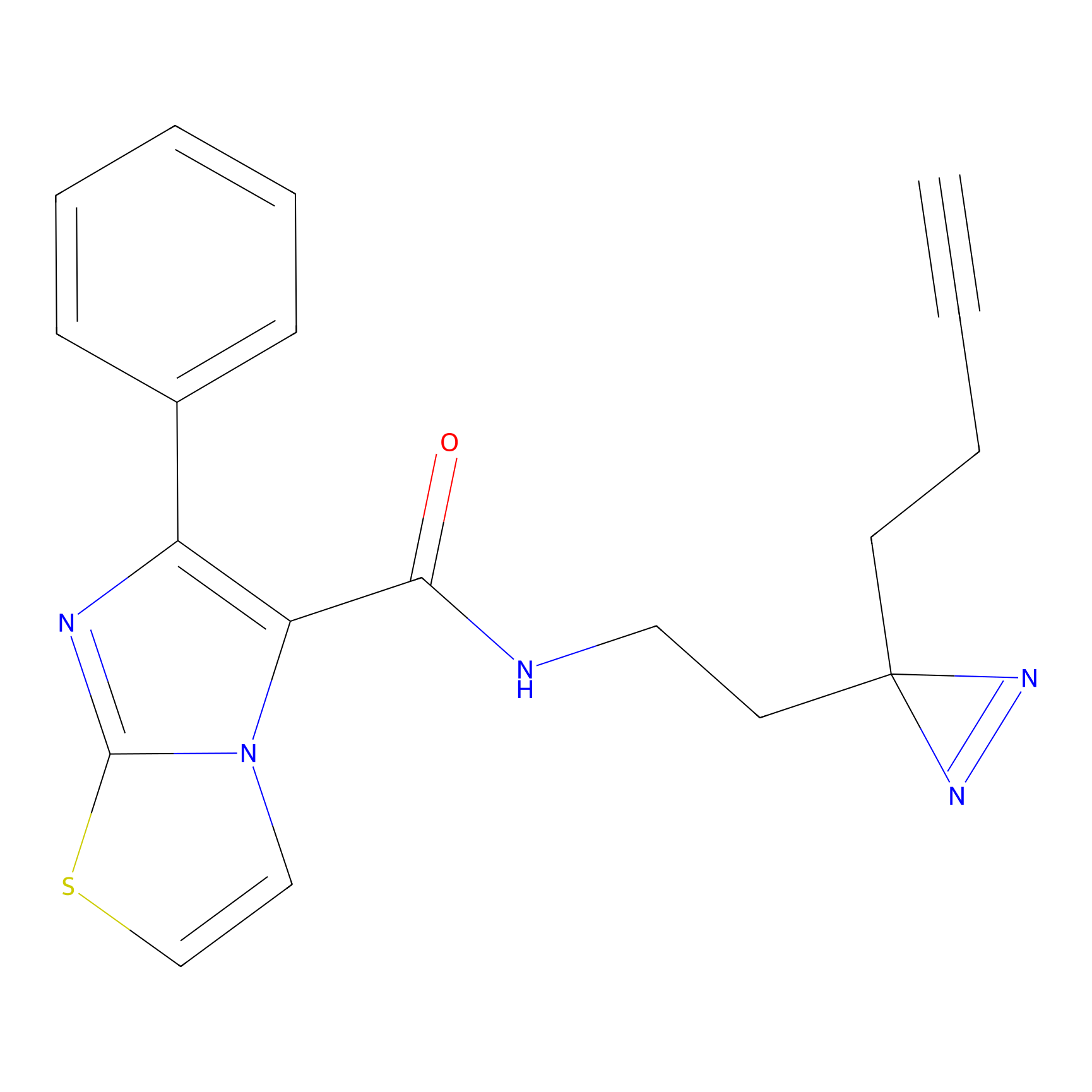
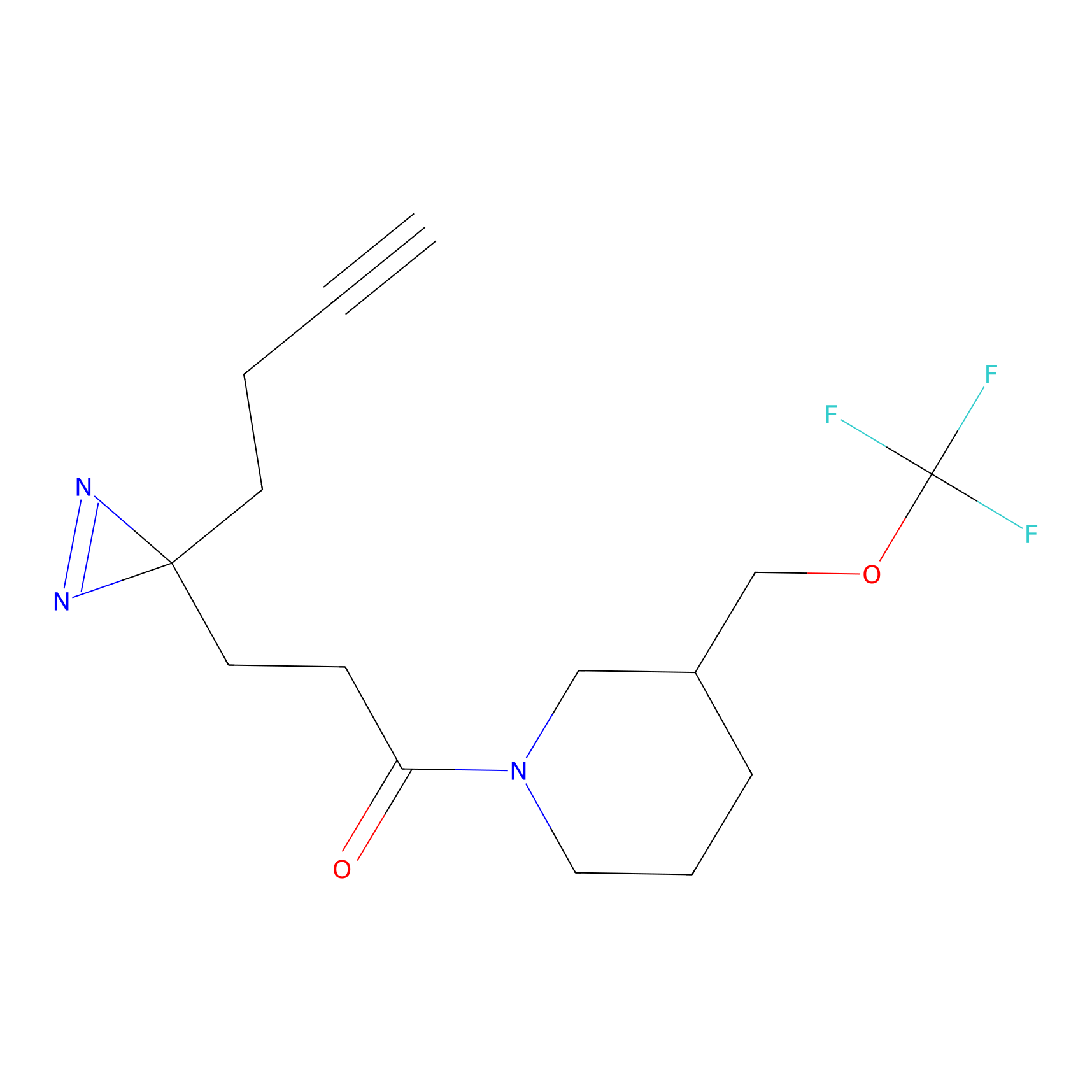
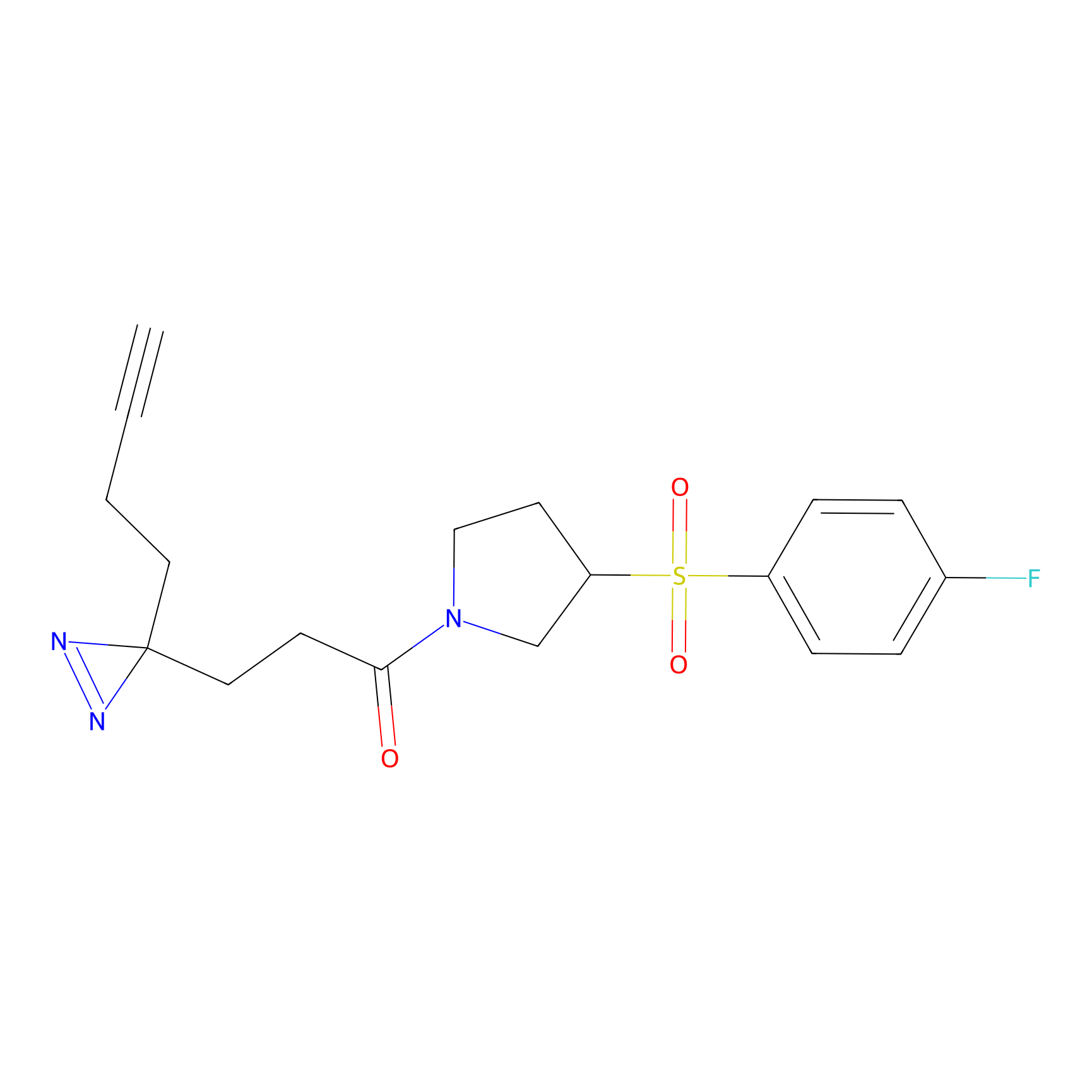
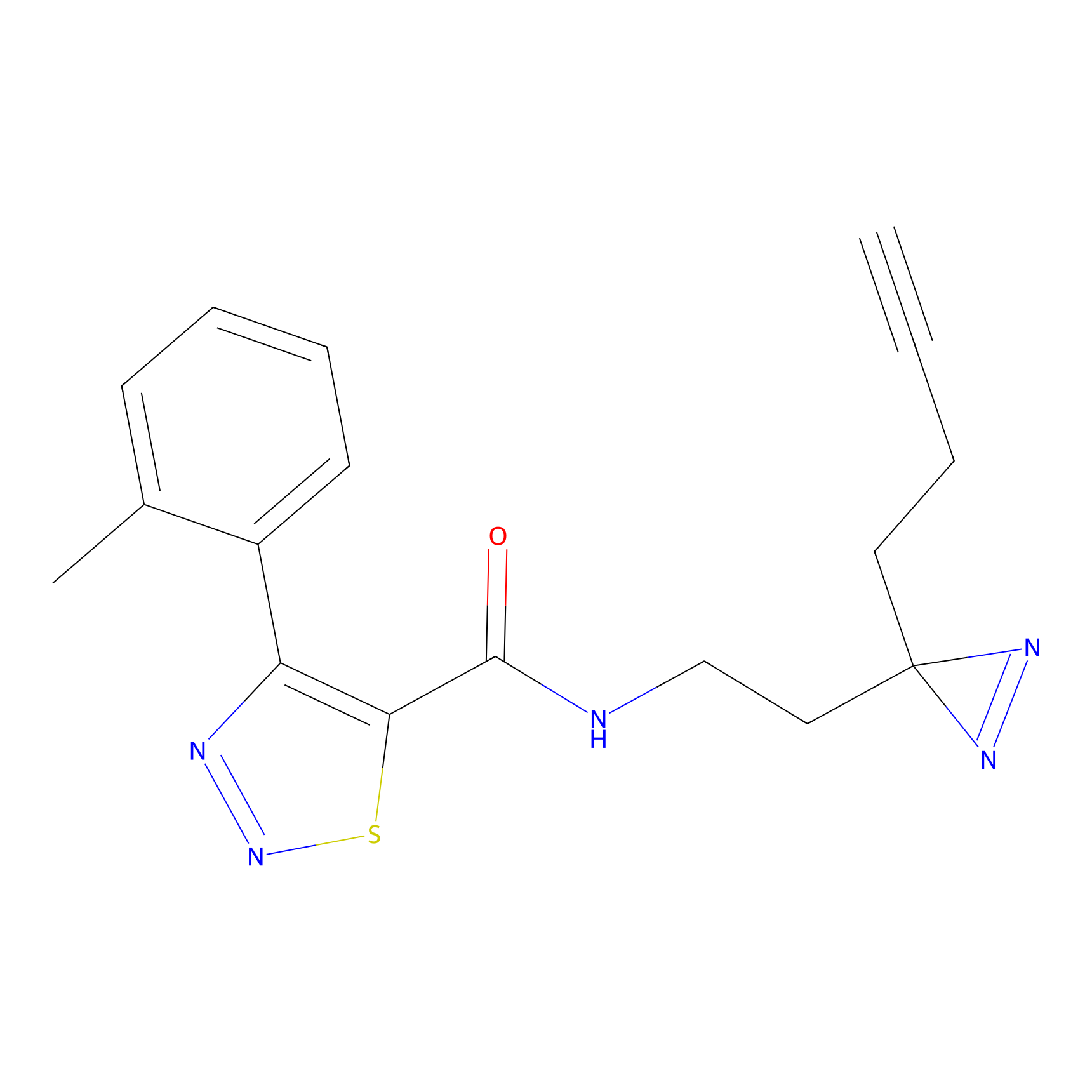
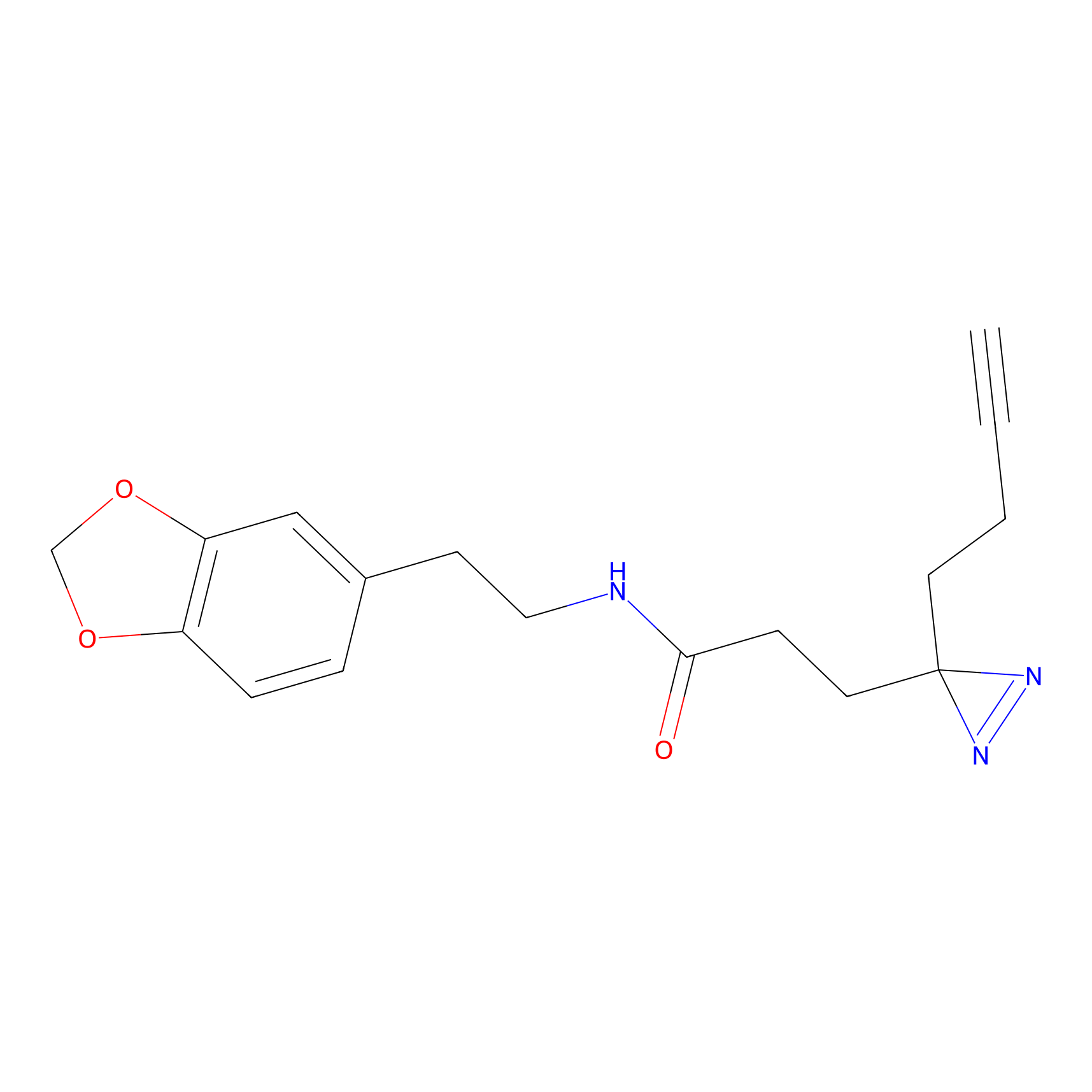
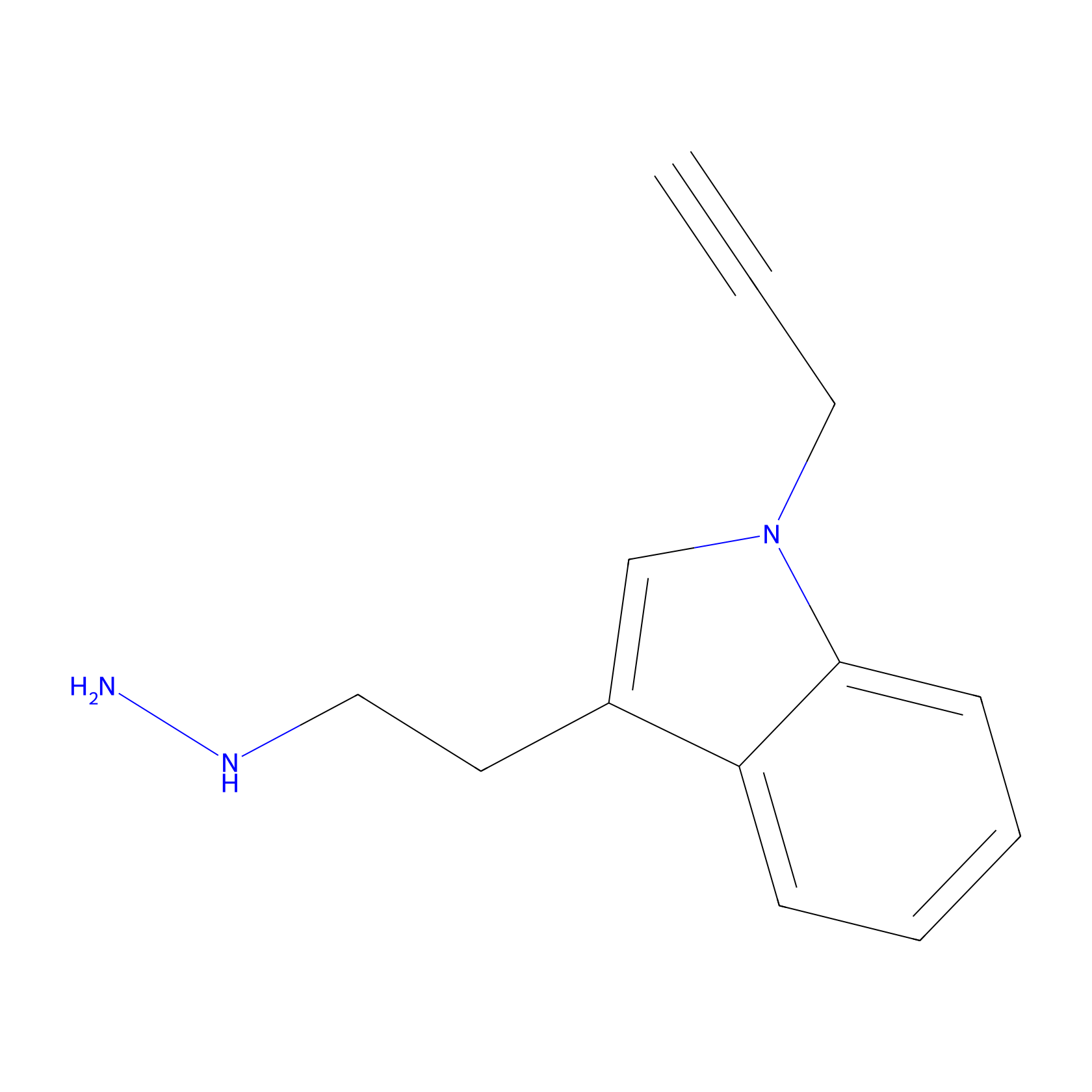
STPyne Probe Info |
 |
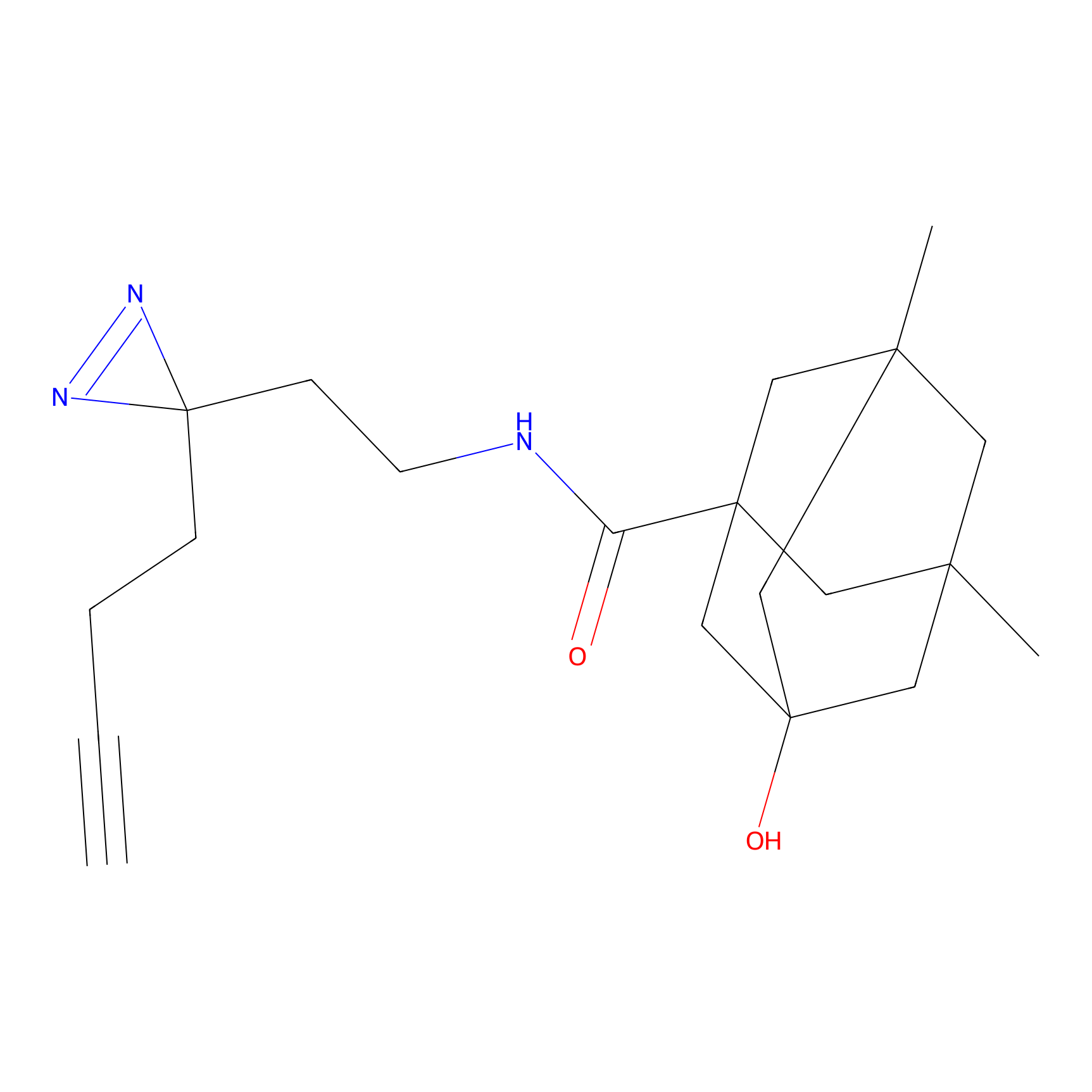
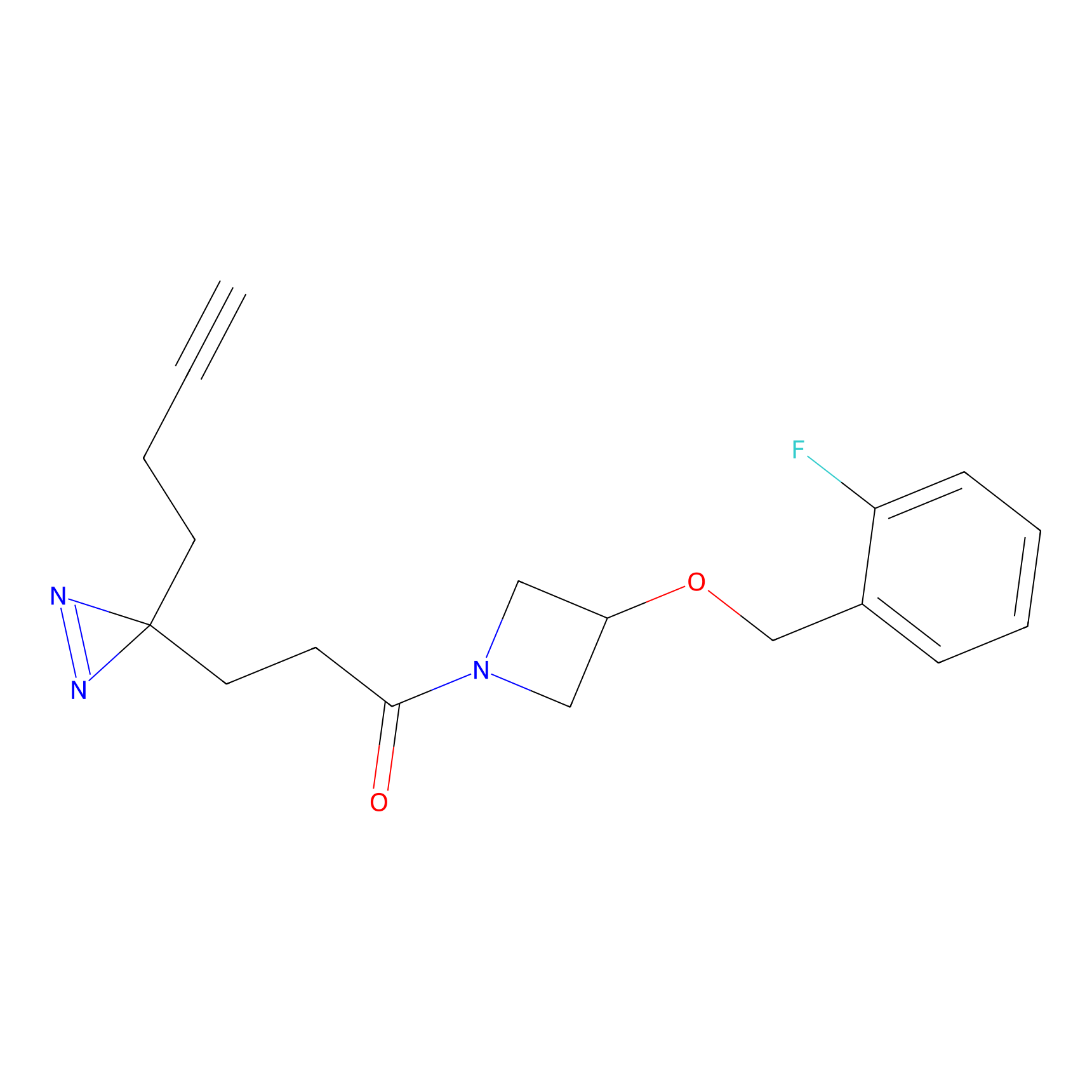
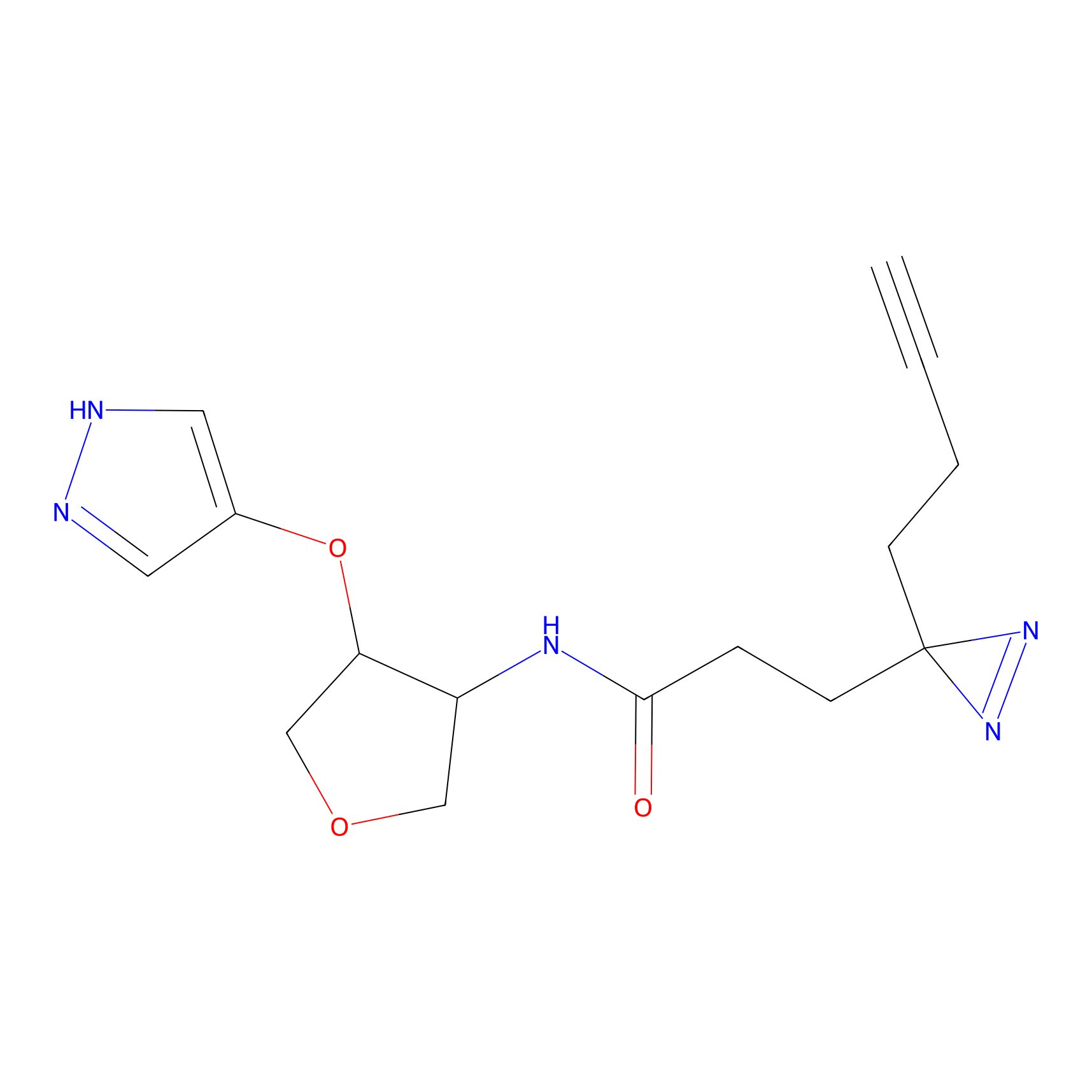
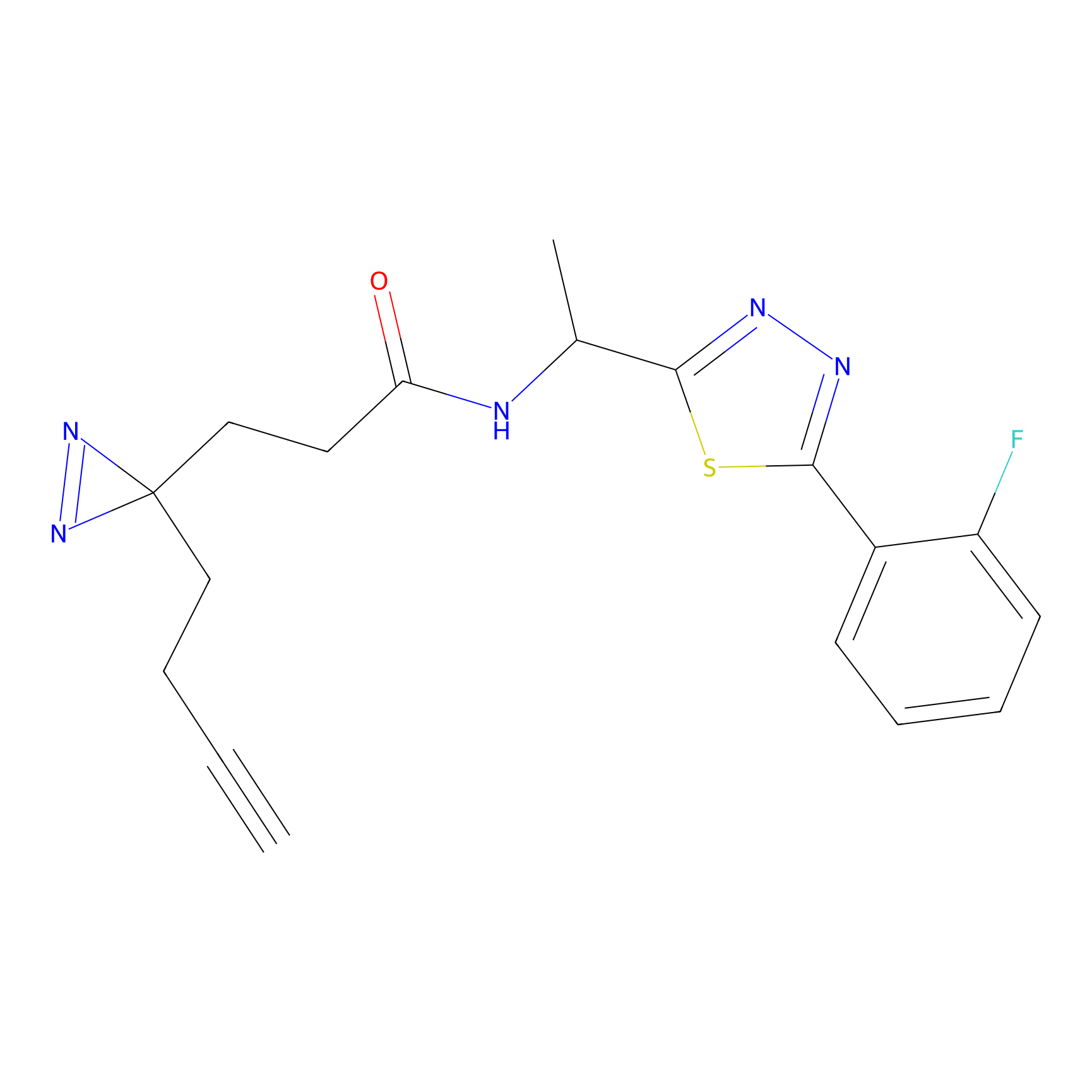
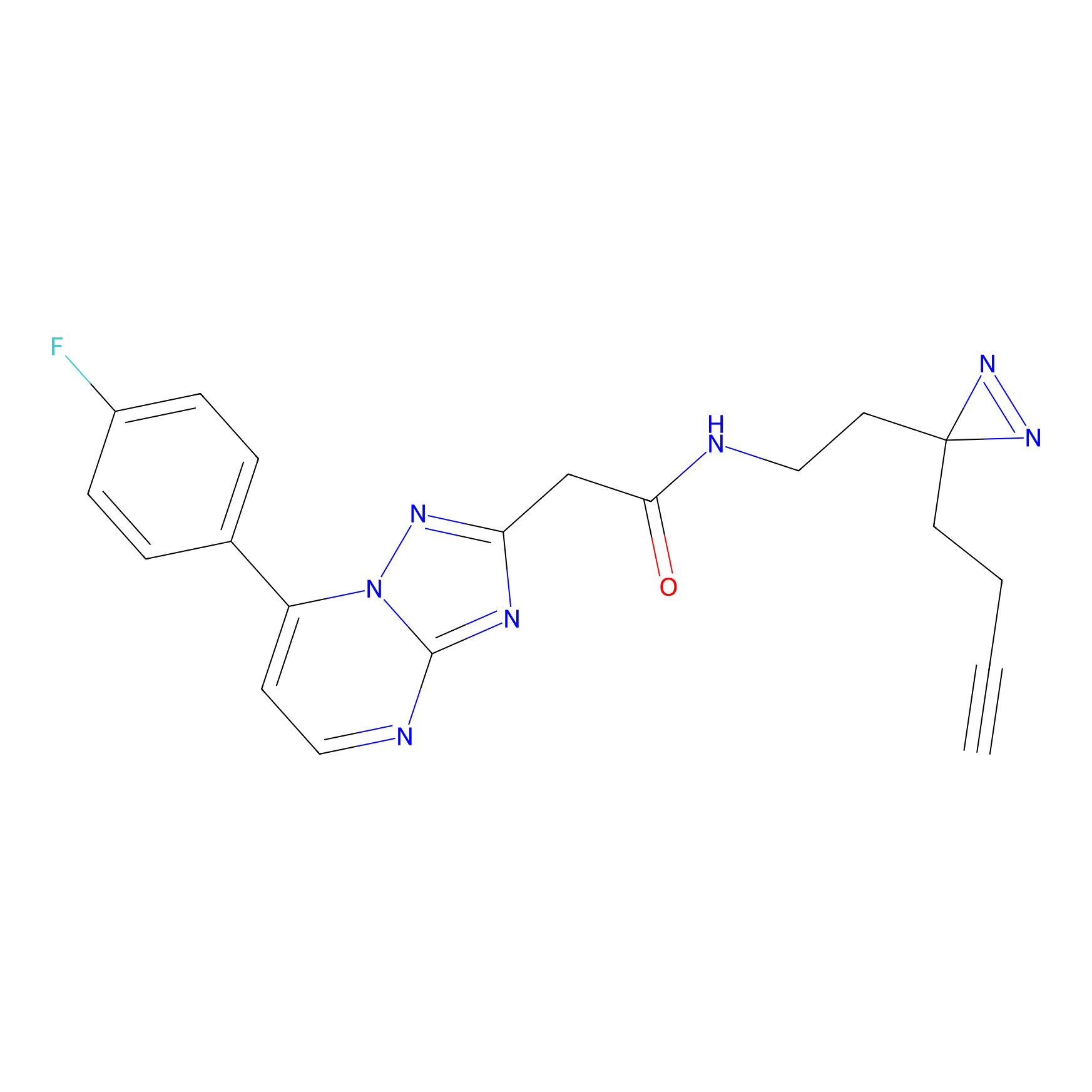
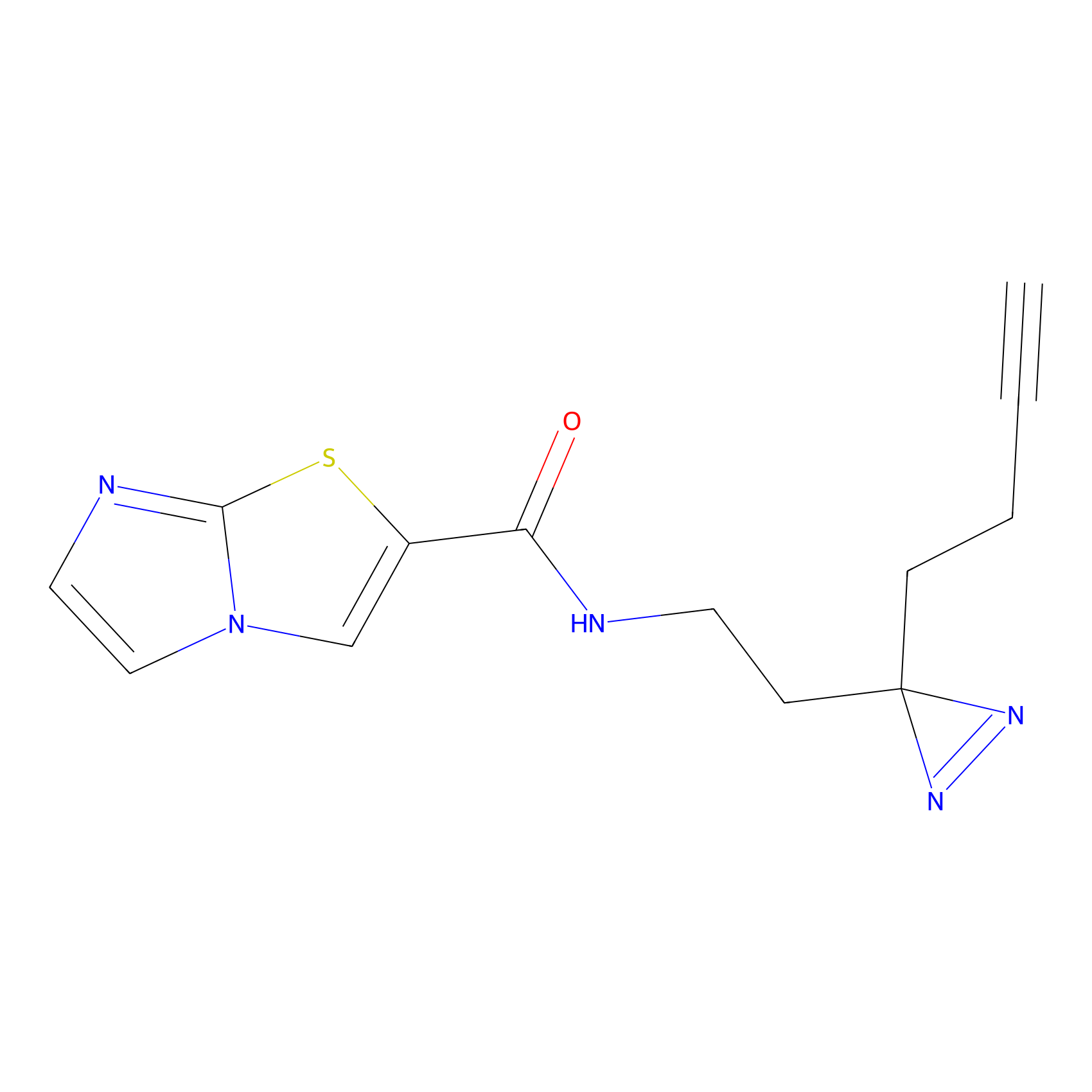
K174(8.06) | LDD0277 | [3] | |
|
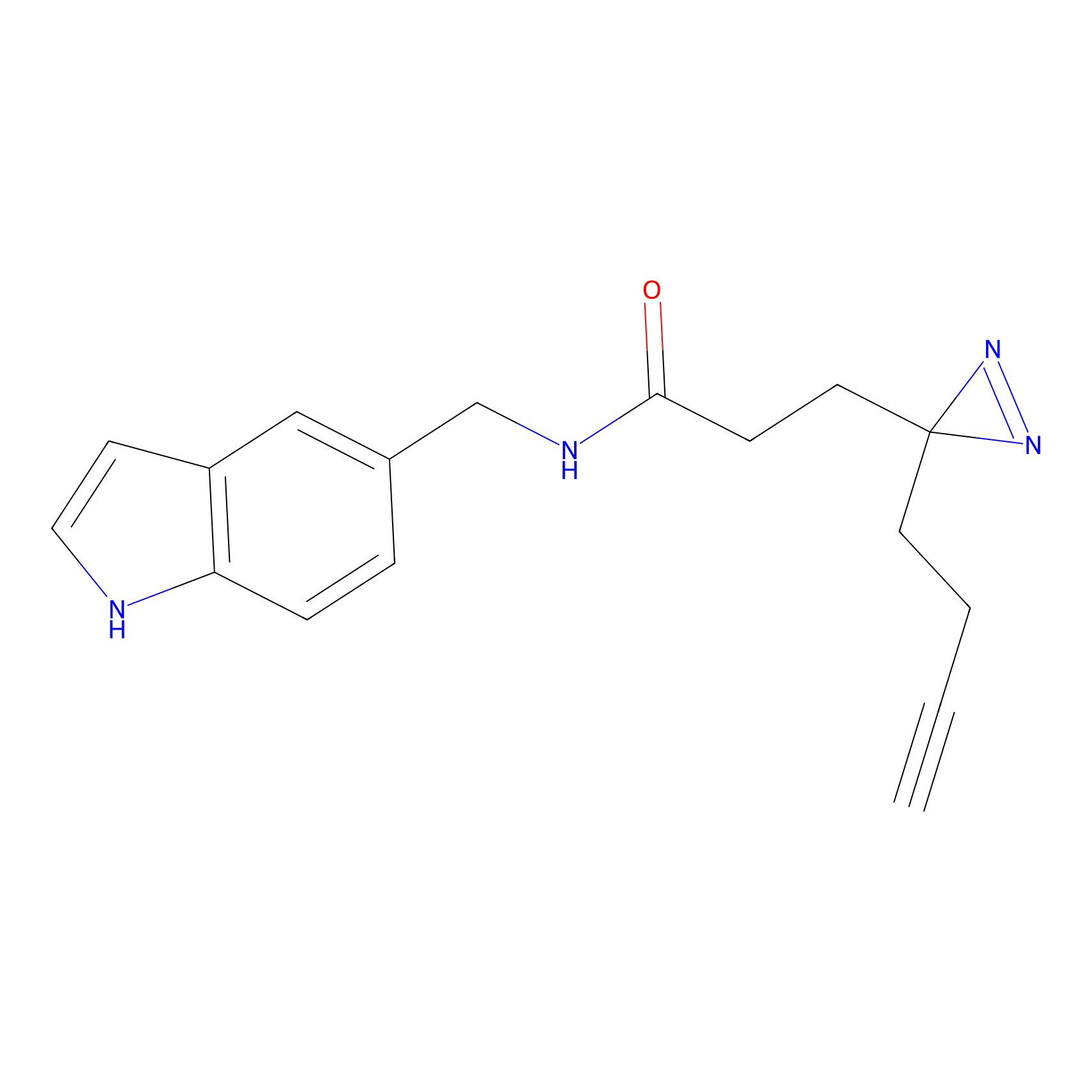
P11 Probe Info |
 |
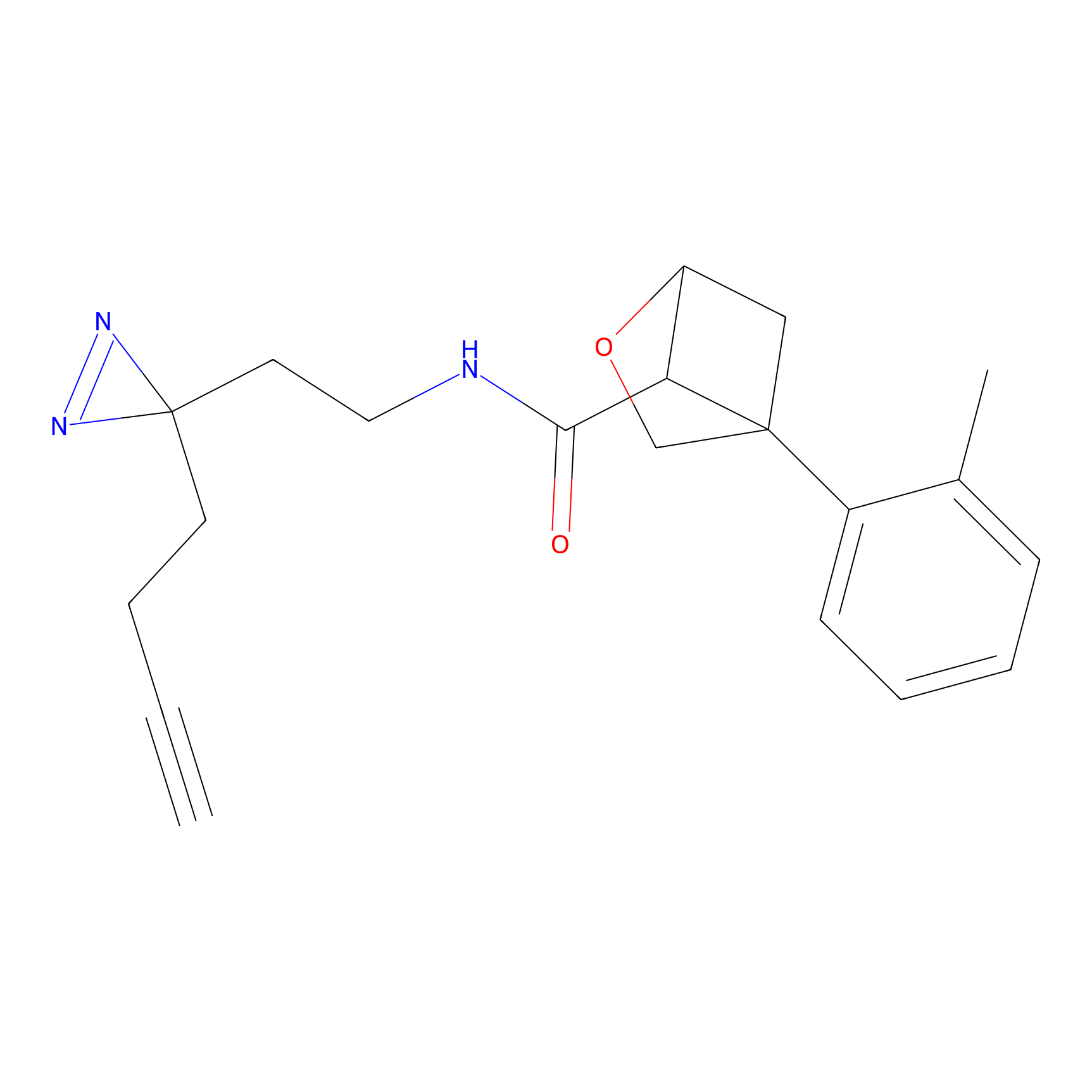
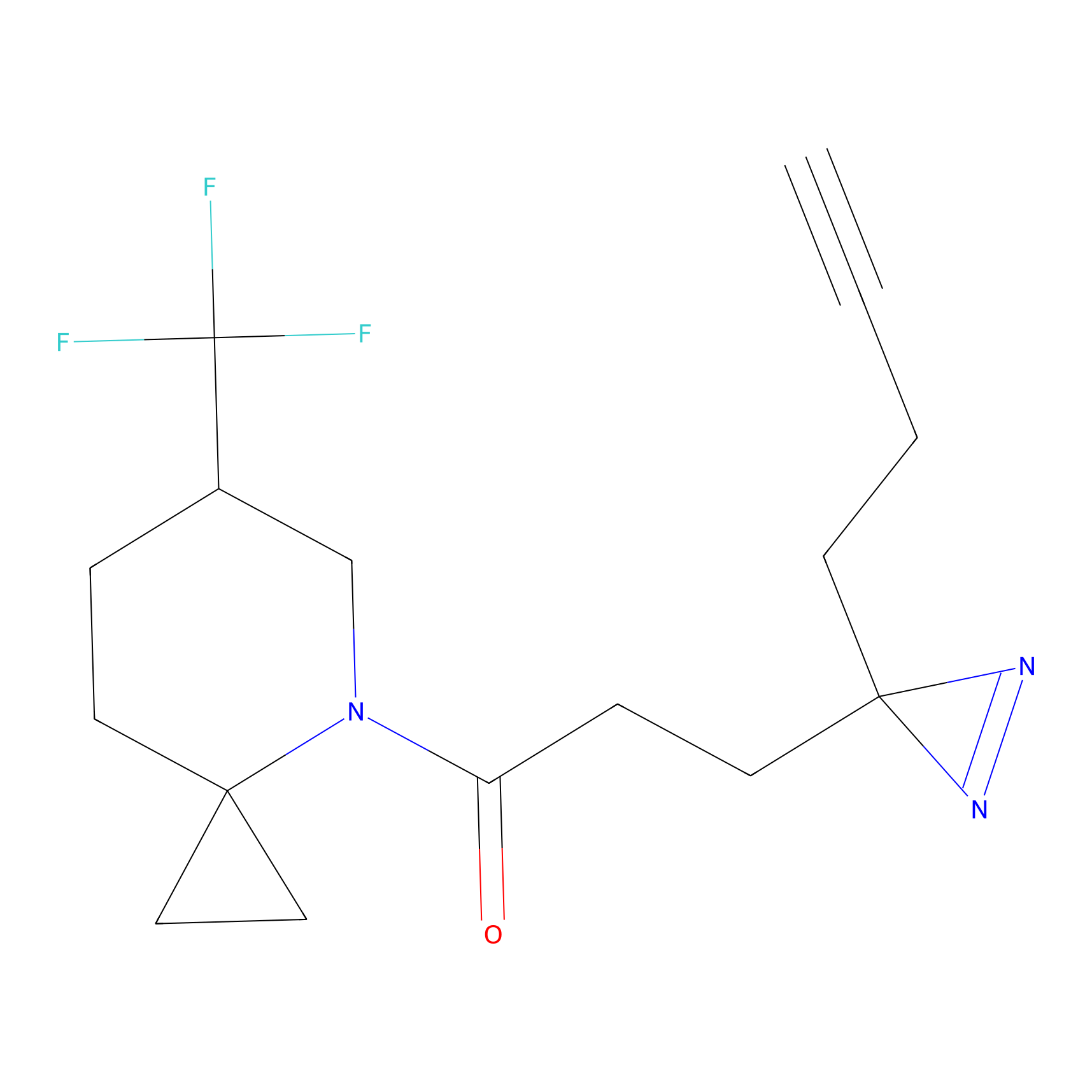
20.00 | LDD0201 | [4] | |
|
DBIA Probe Info |
 |
C7(1.42) | LDD2836 | [5] | |
|
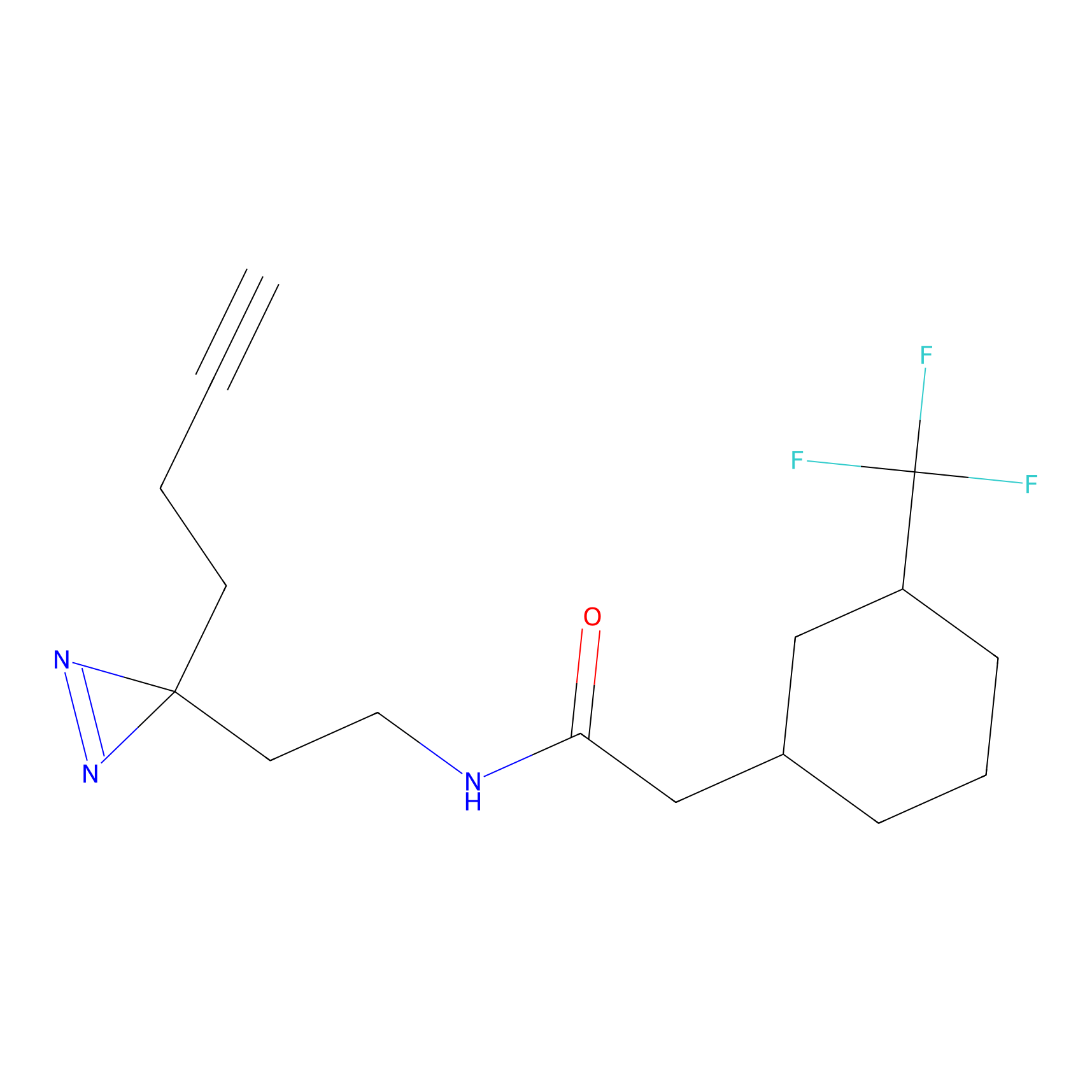
MCL-14 Probe Info |
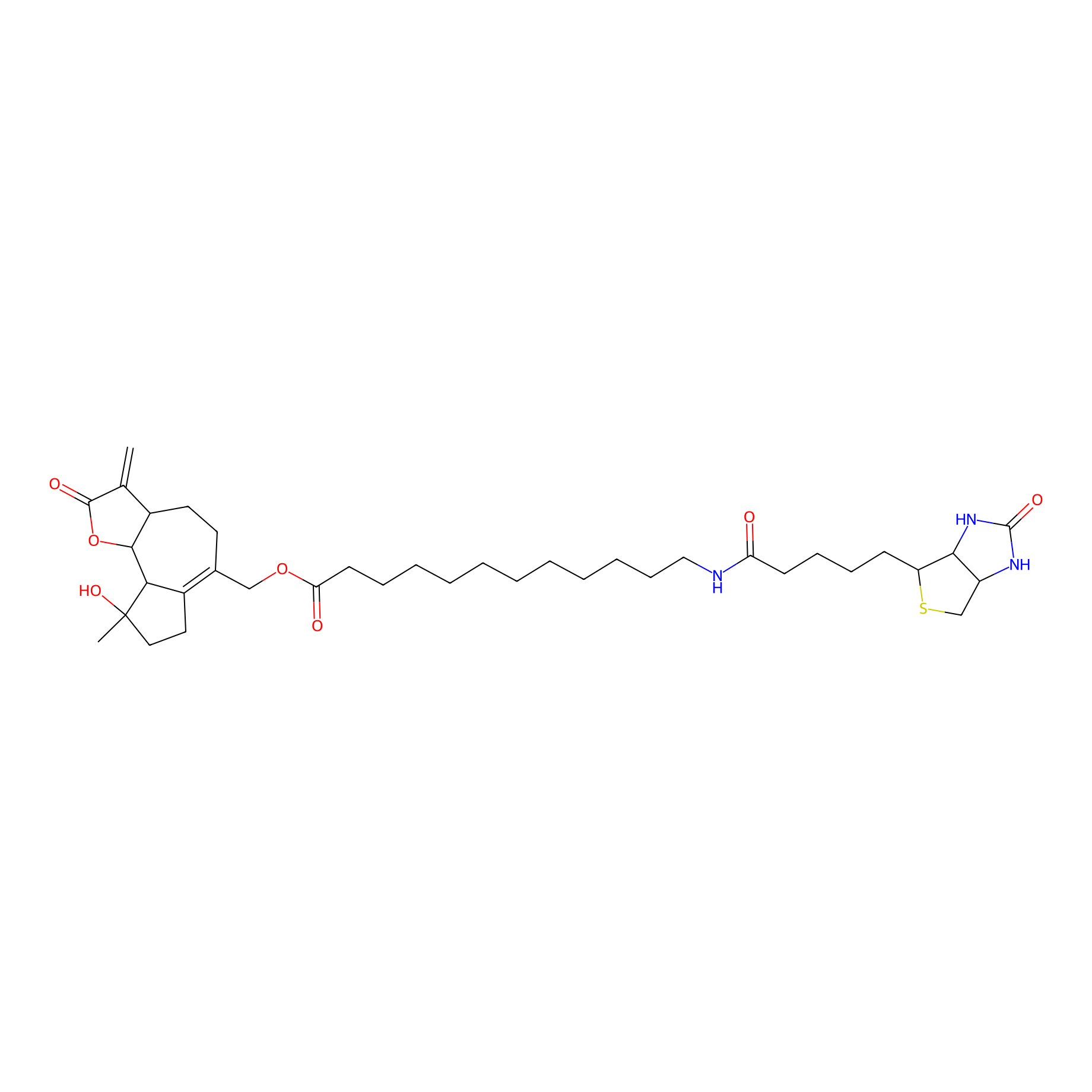 |
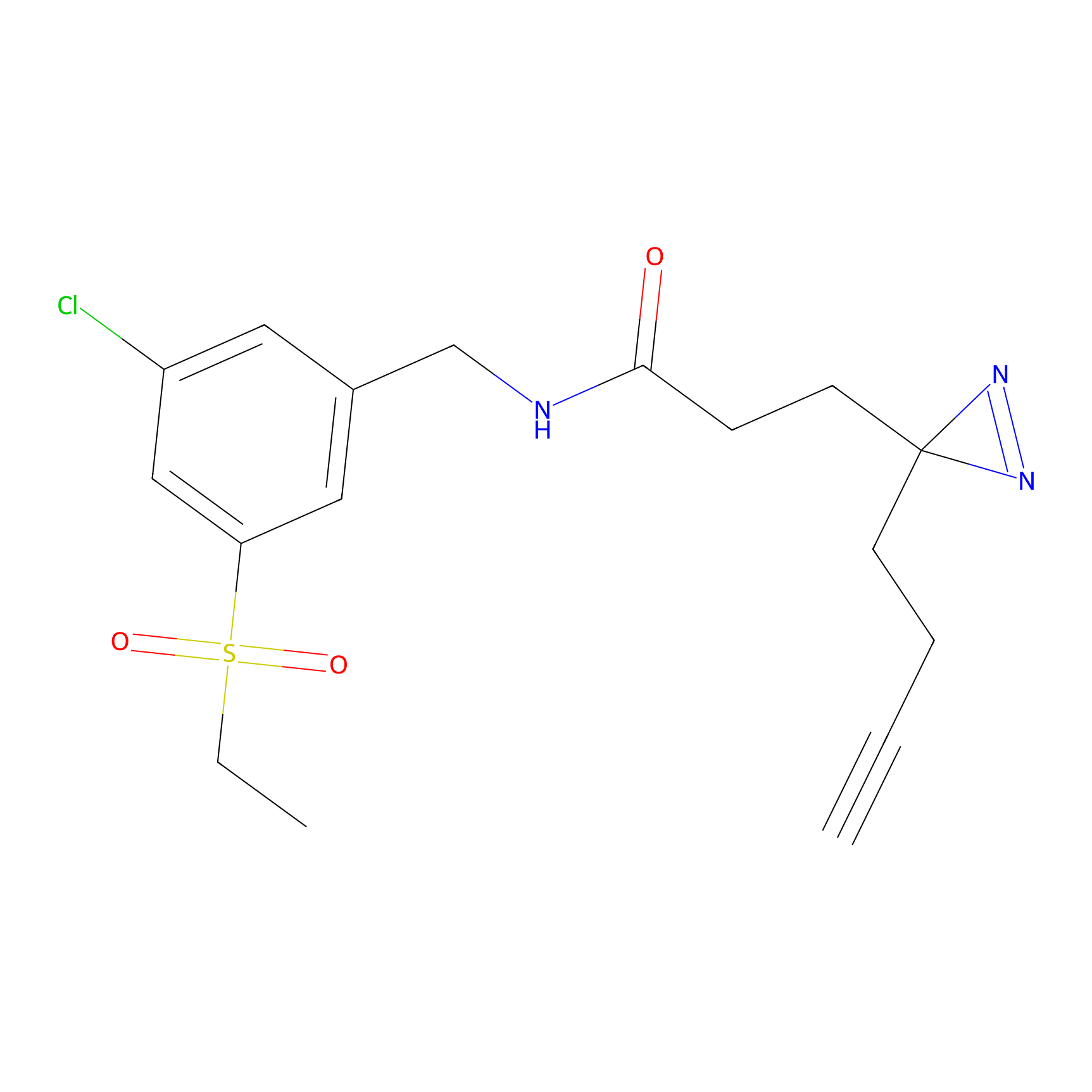
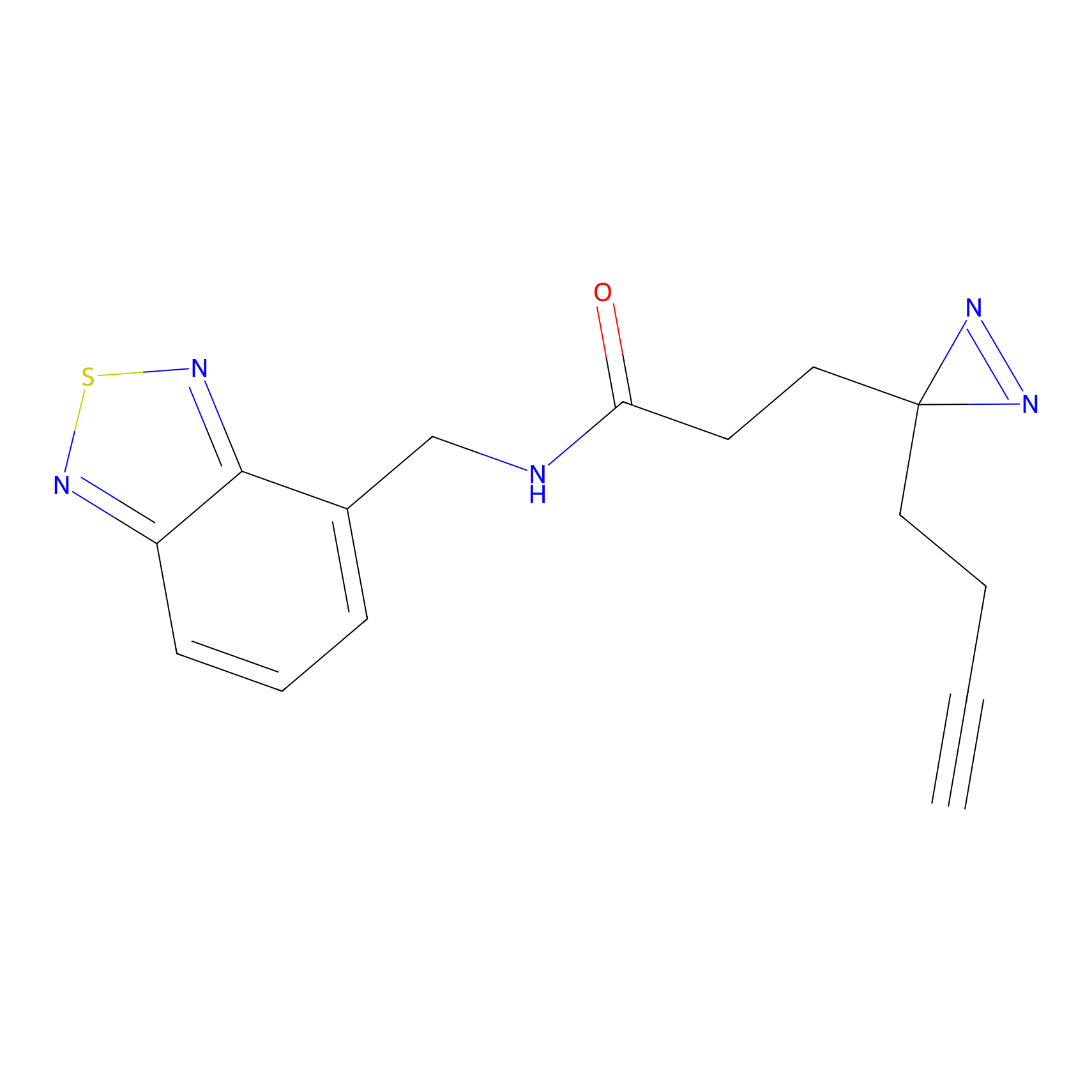
3.00 | LDD0050 | [6] | |
|
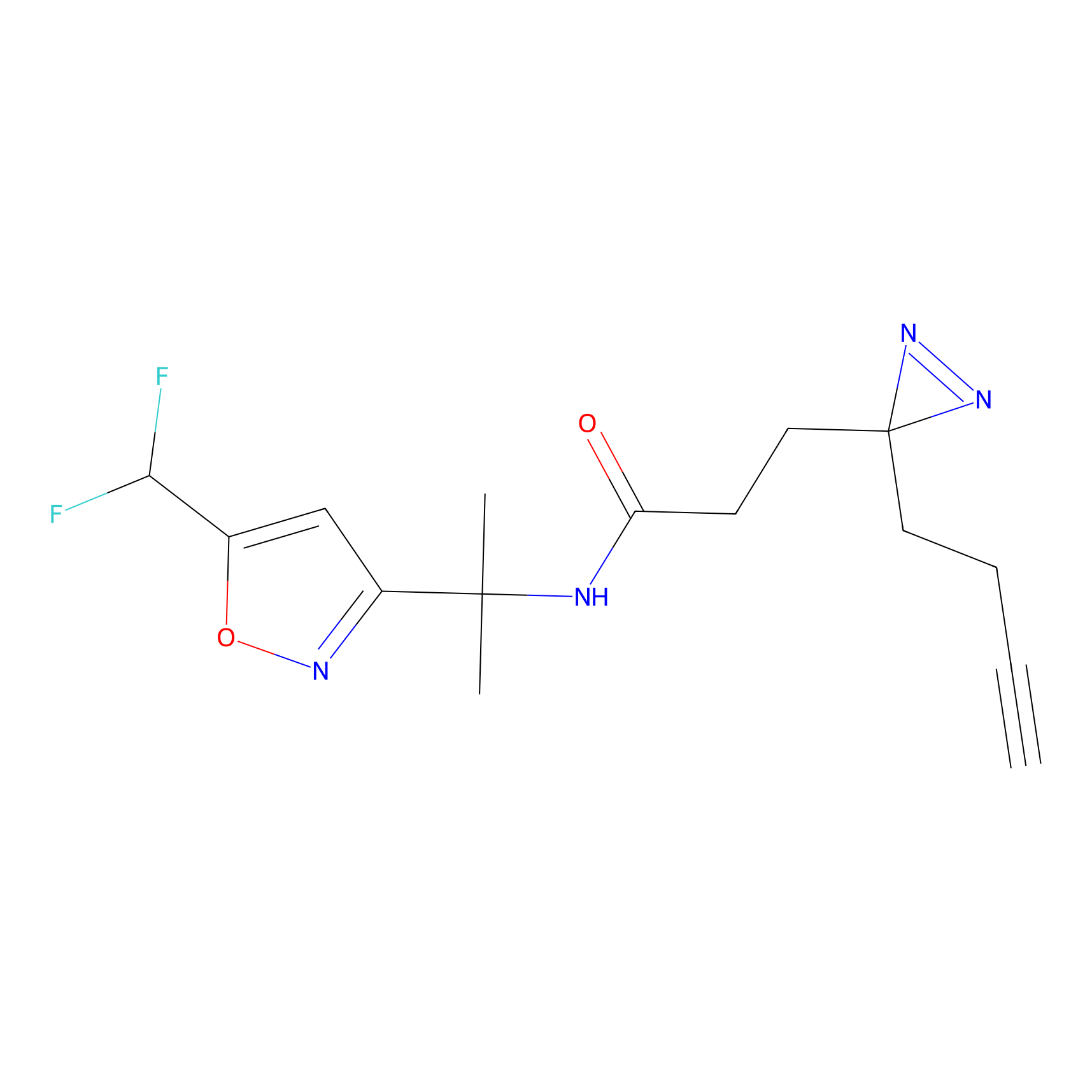
HHS-475 Probe Info |
 |
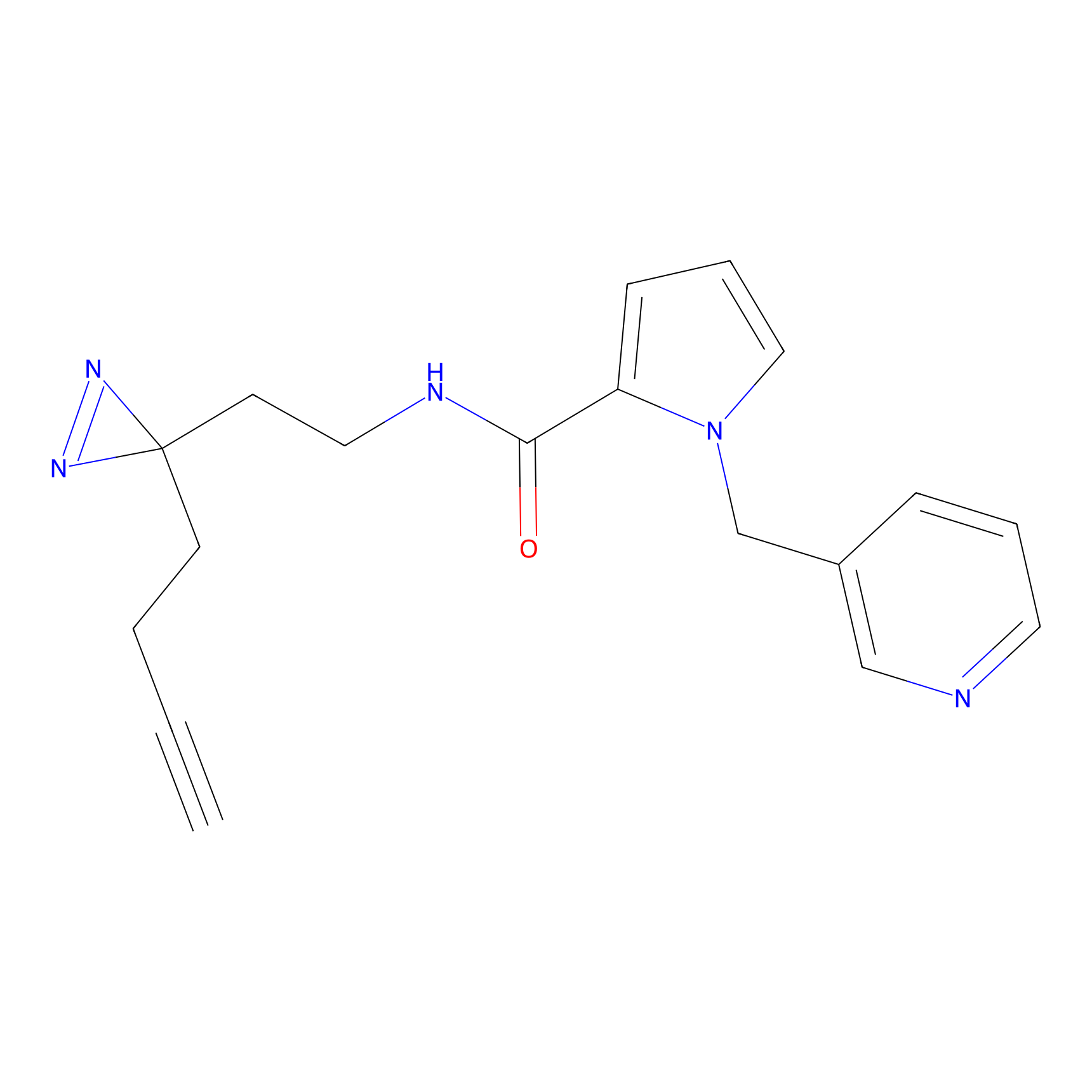
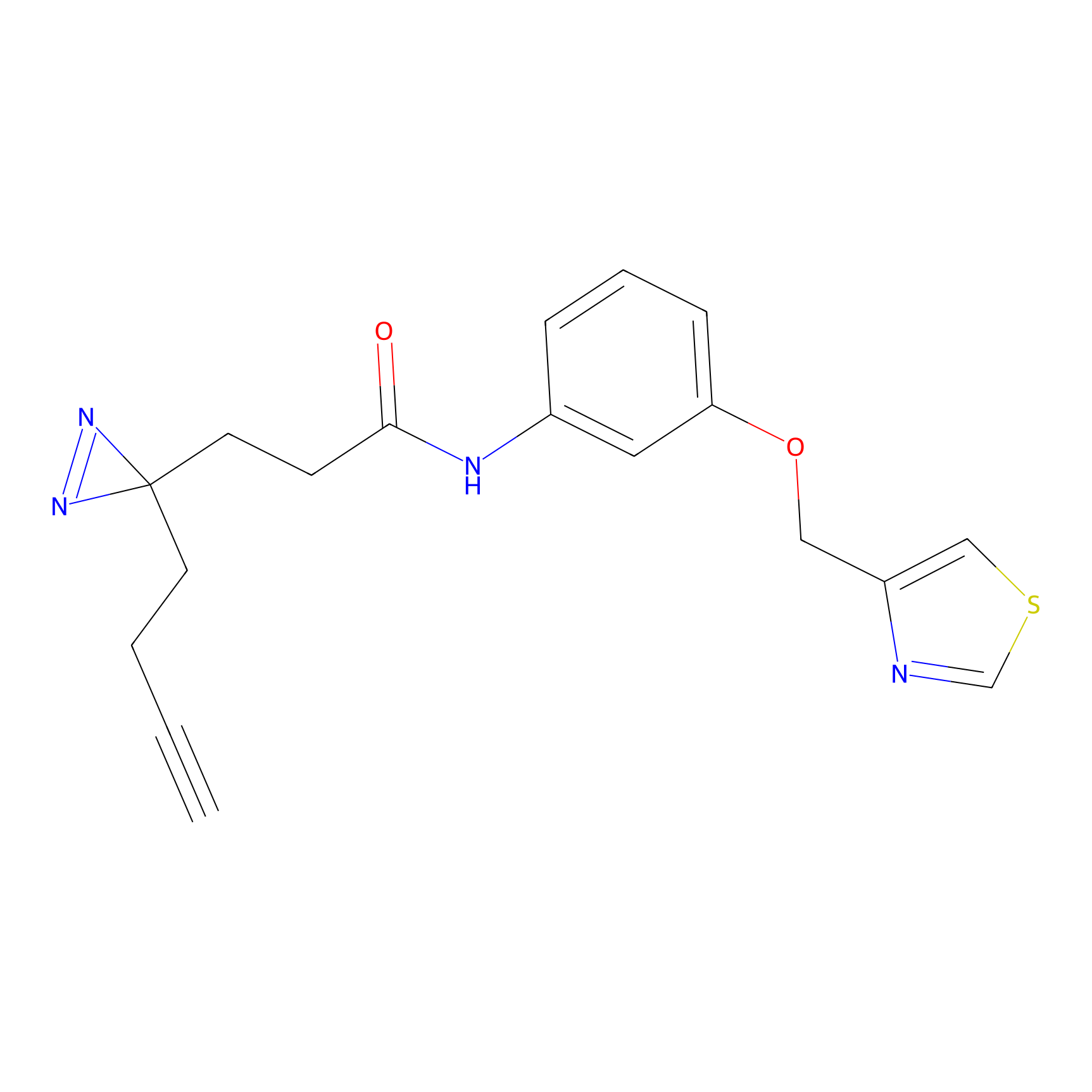
Y163(0.82) | LDD0264 | [7] | |
|
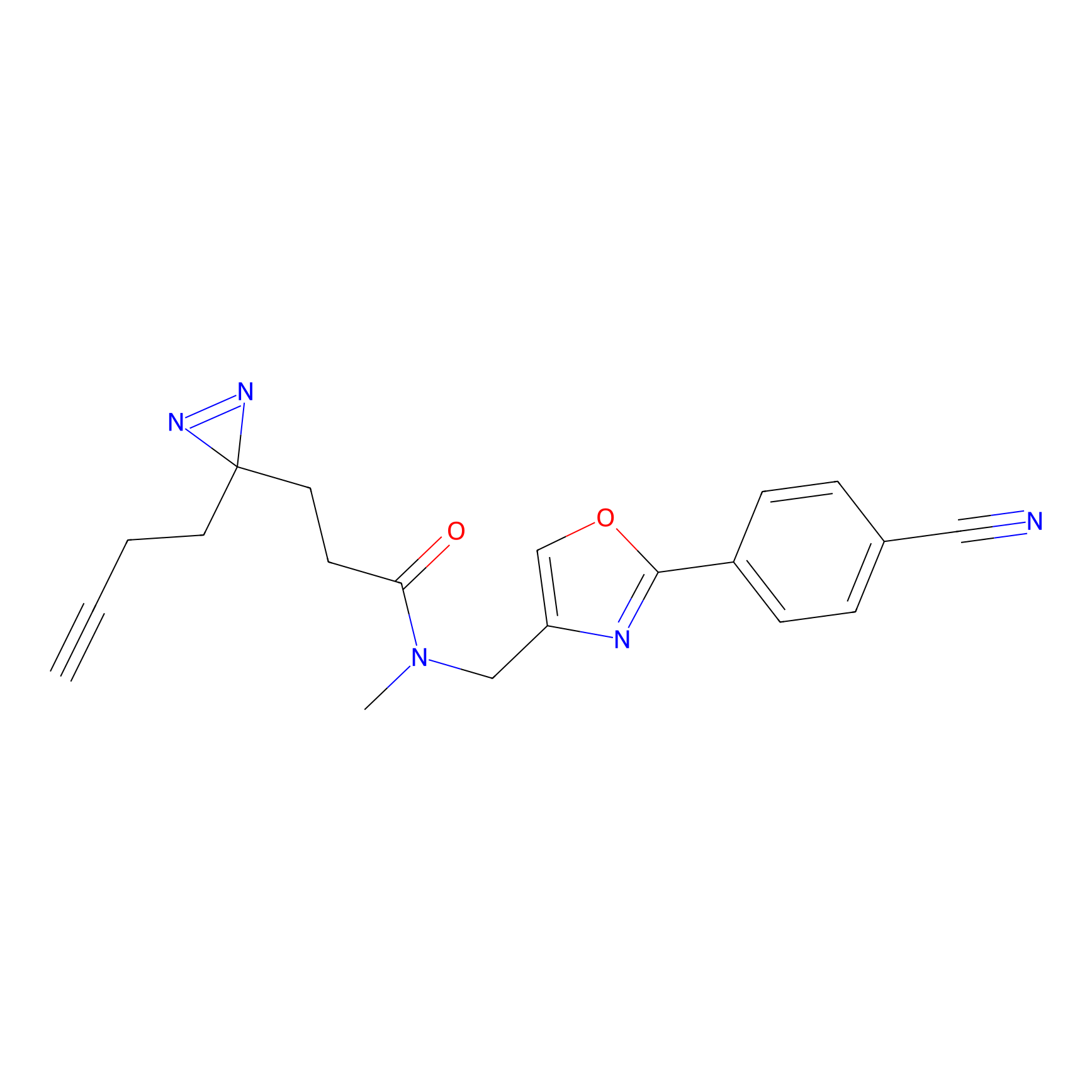
HHS-465 Probe Info |
 |
Y163(5.12) | LDD2237 | [8] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [9] | |
|
ATP probe Probe Info |
 |
K174(0.00); K177(0.00) | LDD0199 | [10] | |
|
m-APA Probe Info |
 |
H128(0.00); H88(0.00) | LDD2231 | [9] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [11] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [11] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [11] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [12] | |
|
IPM Probe Info |
 |
N.A. | LDD0025 | [13] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [13] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [13] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [14] | |
|
SF Probe Info |
 |
Y178(0.00); Y163(0.00) | LDD0028 | [15] | |
|
1c-yne Probe Info |
 |
K81(0.00); K109(0.00) | LDD0228 | [16] | |
|
AOyne Probe Info |
 |
11.60 | LDD0443 | [17] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [18] | |
|
TER-AC Probe Info |
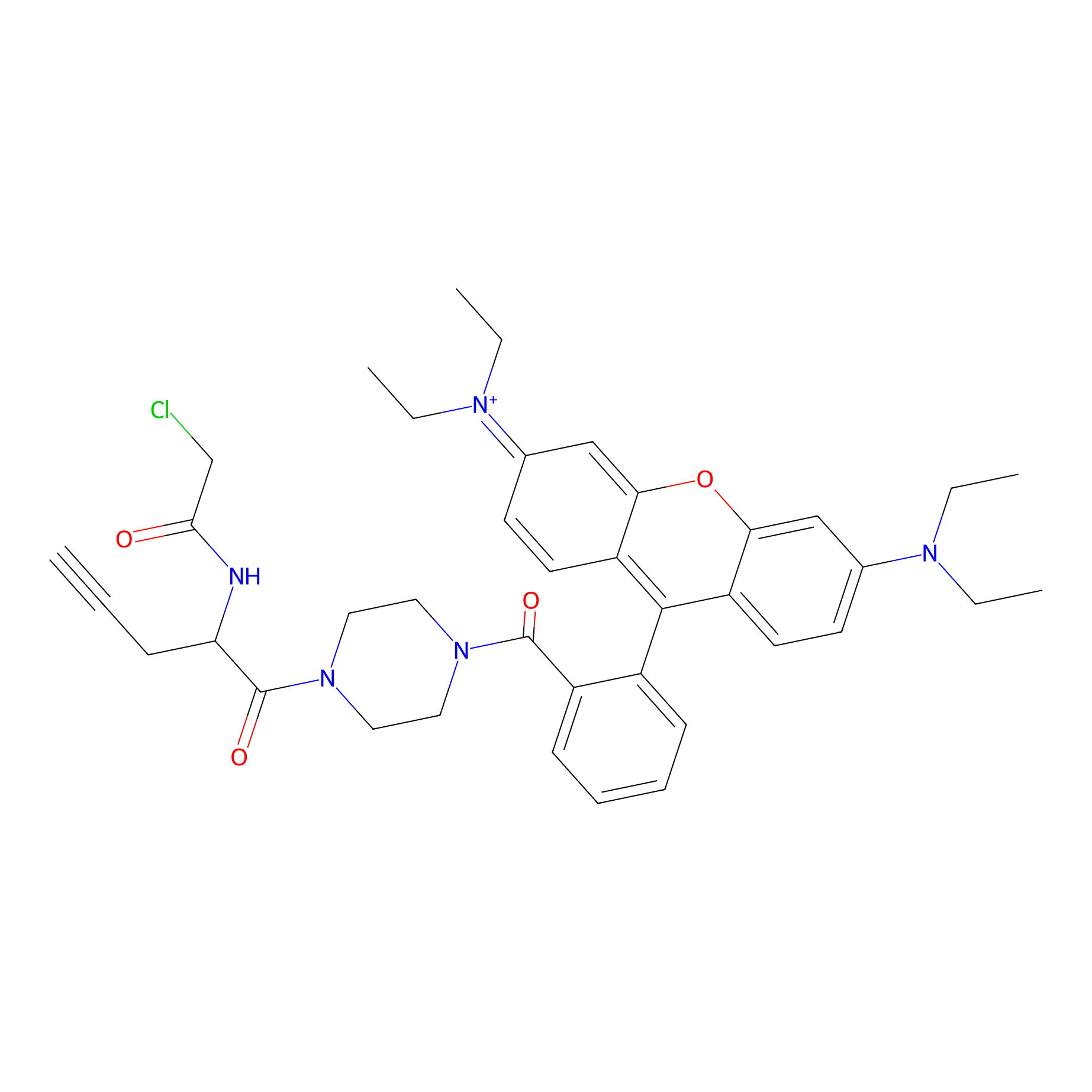 |
N.A. | LDD0426 | [19] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0088 | C45 | HEK-293T | 20.00 | LDD0201 | [4] |
| LDCM0632 | CL-Sc | Hep-G2 | C7(0.94) | LDD2227 | [18] |
| LDCM0625 | F8 | Ramos | C7(0.47) | LDD2187 | [23] |
| LDCM0572 | Fragment10 | Ramos | C7(0.46) | LDD2189 | [23] |
| LDCM0573 | Fragment11 | Ramos | C7(0.07) | LDD2190 | [23] |
| LDCM0574 | Fragment12 | Ramos | C7(0.78) | LDD2191 | [23] |
| LDCM0575 | Fragment13 | Ramos | C7(0.68) | LDD2192 | [23] |
| LDCM0576 | Fragment14 | Ramos | C7(0.84) | LDD2193 | [23] |
| LDCM0579 | Fragment20 | Ramos | C7(0.53) | LDD2194 | [23] |
| LDCM0580 | Fragment21 | Ramos | C7(0.72) | LDD2195 | [23] |
| LDCM0582 | Fragment23 | Ramos | C7(0.60) | LDD2196 | [23] |
| LDCM0578 | Fragment27 | Ramos | C7(0.62) | LDD2197 | [23] |
| LDCM0586 | Fragment28 | Ramos | C7(1.01) | LDD2198 | [23] |
| LDCM0588 | Fragment30 | Ramos | C7(0.48) | LDD2199 | [23] |
| LDCM0589 | Fragment31 | Ramos | C7(0.86) | LDD2200 | [23] |
| LDCM0590 | Fragment32 | Ramos | C7(0.56) | LDD2201 | [23] |
| LDCM0468 | Fragment33 | Ramos | C7(0.54) | LDD2202 | [23] |
| LDCM0596 | Fragment38 | Ramos | C7(0.86) | LDD2203 | [23] |
| LDCM0566 | Fragment4 | Ramos | C7(0.64) | LDD2184 | [23] |
| LDCM0610 | Fragment52 | Ramos | C7(0.50) | LDD2204 | [23] |
| LDCM0614 | Fragment56 | Ramos | C7(0.43) | LDD2205 | [23] |
| LDCM0569 | Fragment7 | Ramos | C7(0.48) | LDD2186 | [23] |
| LDCM0571 | Fragment9 | Ramos | C7(0.61) | LDD2188 | [23] |
| LDCM0116 | HHS-0101 | DM93 | Y163(0.82) | LDD0264 | [7] |
| LDCM0117 | HHS-0201 | DM93 | Y163(0.69) | LDD0265 | [7] |
| LDCM0118 | HHS-0301 | DM93 | Y163(0.72) | LDD0266 | [7] |
| LDCM0119 | HHS-0401 | DM93 | Y163(0.80) | LDD0267 | [7] |
| LDCM0120 | HHS-0701 | DM93 | Y163(0.92) | LDD0268 | [7] |
| LDCM0022 | KB02 | Ramos | C7(0.41) | LDD2182 | [23] |
| LDCM0023 | KB03 | Ramos | C7(0.69) | LDD2183 | [23] |
| LDCM0024 | KB05 | Ramos | C7(0.38) | LDD2185 | [23] |
| LDCM0006 | Micheliolide | M9-ENL1 | 3.00 | LDD0050 | [6] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C7(0.27) | LDD2207 | [24] |
The Interaction Atlas With This Target
The Drug(s) Related To This Target
Approved
Phase 3
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Iclaprim | Small molecular drug | D04LBC | |||
Phase 2
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Piritrexim | Small molecular drug | D09AYQ | |||
| Ch-4051 | . | D02OTK | |||
Phase 1
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| L-mdam | Small molecular drug | D0S9OH | |||
| Vyr 006 | Small molecular drug | D9ZQ7T | |||
| Mdam (Y-methylene-10-deazaaminopterin) | . | D0DO5J | |||
Preclinical
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| 1954u89 | . | D05HCO | |||
Investigative
Discontinued
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Tnp-351 | Small molecular drug | D00CQW | |||
References