Details of the Target
General Information of Target
| Target ID | LDTP12720 | |||||
|---|---|---|---|---|---|---|
| Target Name | Peptidyl-prolyl cis-trans isomerase FKBP14 (FKBP14) | |||||
| Gene Name | FKBP14 | |||||
| Gene ID | 55033 | |||||
| Synonyms |
FKBP22; Peptidyl-prolyl cis-trans isomerase FKBP14; PPIase FKBP14; EC 5.2.1.8; 22 kDa FK506-binding protein; 22 kDa FKBP; FKBP-22; FK506-binding protein 14; FKBP-14; Rotamase |
|||||
| 3D Structure | ||||||
| Sequence |
MQSCESSGDSADDPLSRGLRRRGQPRVVVIGAGLAGLAAAKALLEQGFTDVTVLEASSHI
GGRVQSVKLGHATFELGATWIHGSHGNPIYHLAEANGLLEETTDGERSVGRISLYSKNGV ACYLTNHGRRIPKDVVEEFSDLYNEVYNLTQEFFRHDKPVNAESQNSVGVFTREEVRNRI RNDPDDPEATKRLKLAMIQQYLKVESCESSSHSMDEVSLSAFGEWTEIPGAHHIIPSGFM RVVELLAEGIPAHVIQLGKPVRCIHWDQASARPRGPEIEPRGEGDHNHDTGEGGQGGEEP RGGRWDEDEQWSVVVECEDCELIPADHVIVTVSLGVLKRQYTSFFRPGLPTEKVAAIHRL GIGTTDKIFLEFEEPFWGPECNSLQFVWEDEAESHTLTYPPELWYRKICGFDVLYPPERY GHVLSGWICGEEALVMEKCDDEAVAEICTEMLRQFTGNPNIPKPRRILRSAWGSNPYFRG SYSYTQVGSSGADVEKLAKPLPYTESSKTAPMQVLFSGEATHRKYYSTTHGALLSGQREA ARLIEMYRDLFQQGT |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Subcellular location |
Endoplasmic reticulum lumen
|
|||||
| Function |
PPIase which accelerates the folding of proteins during protein synthesis. Has a preference for substrates containing 4-hydroxylproline modifications, including type III collagen. May also target type VI and type X collagens.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
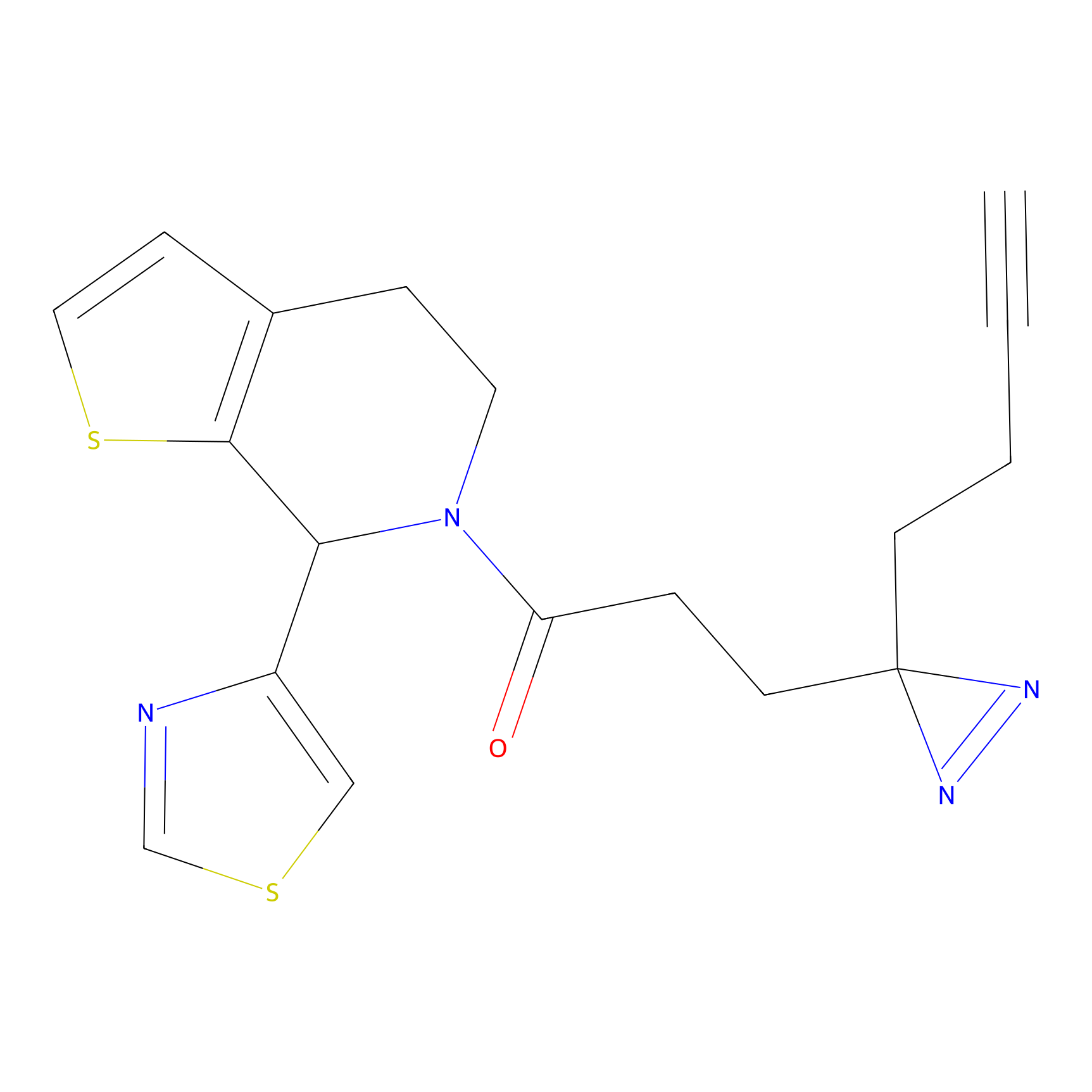
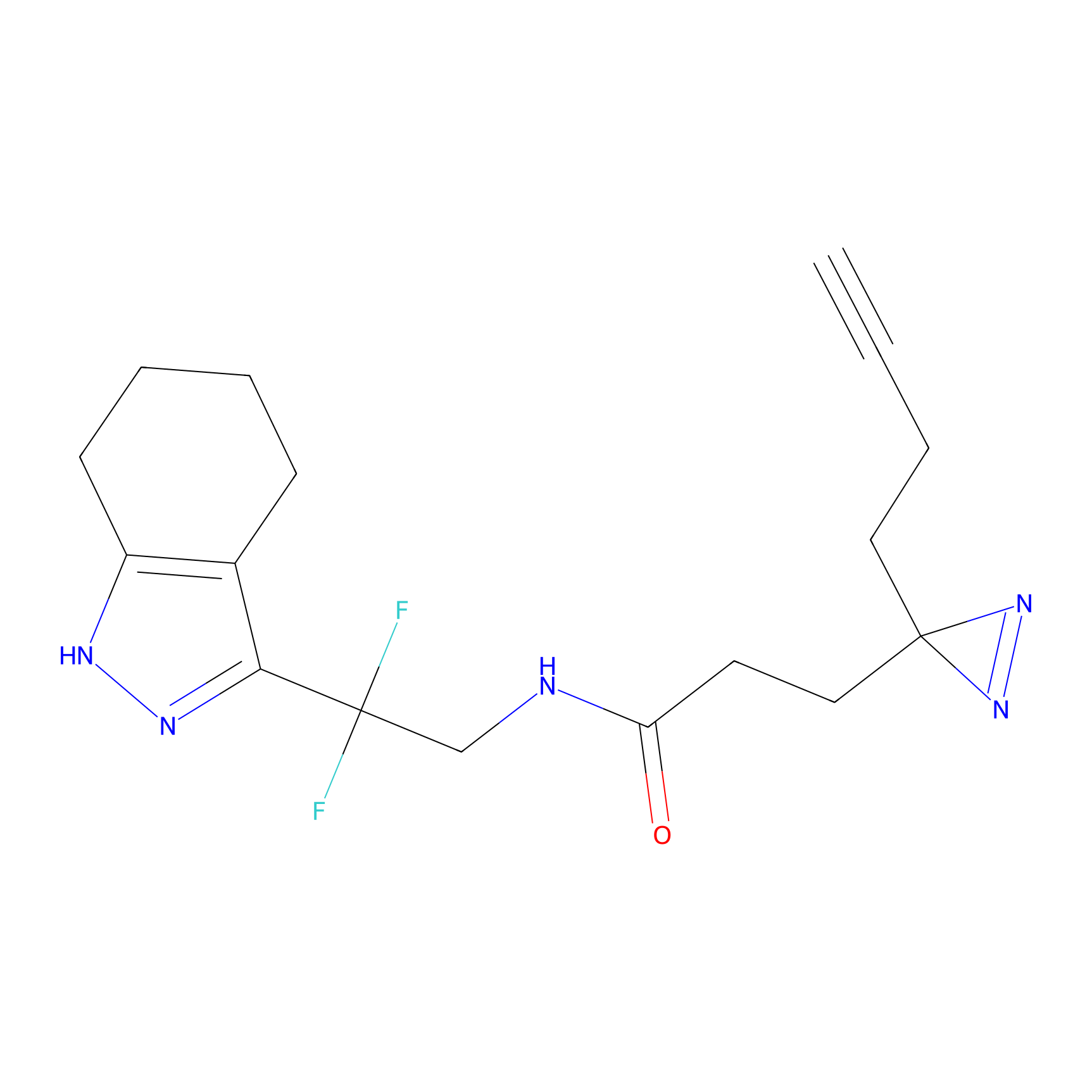
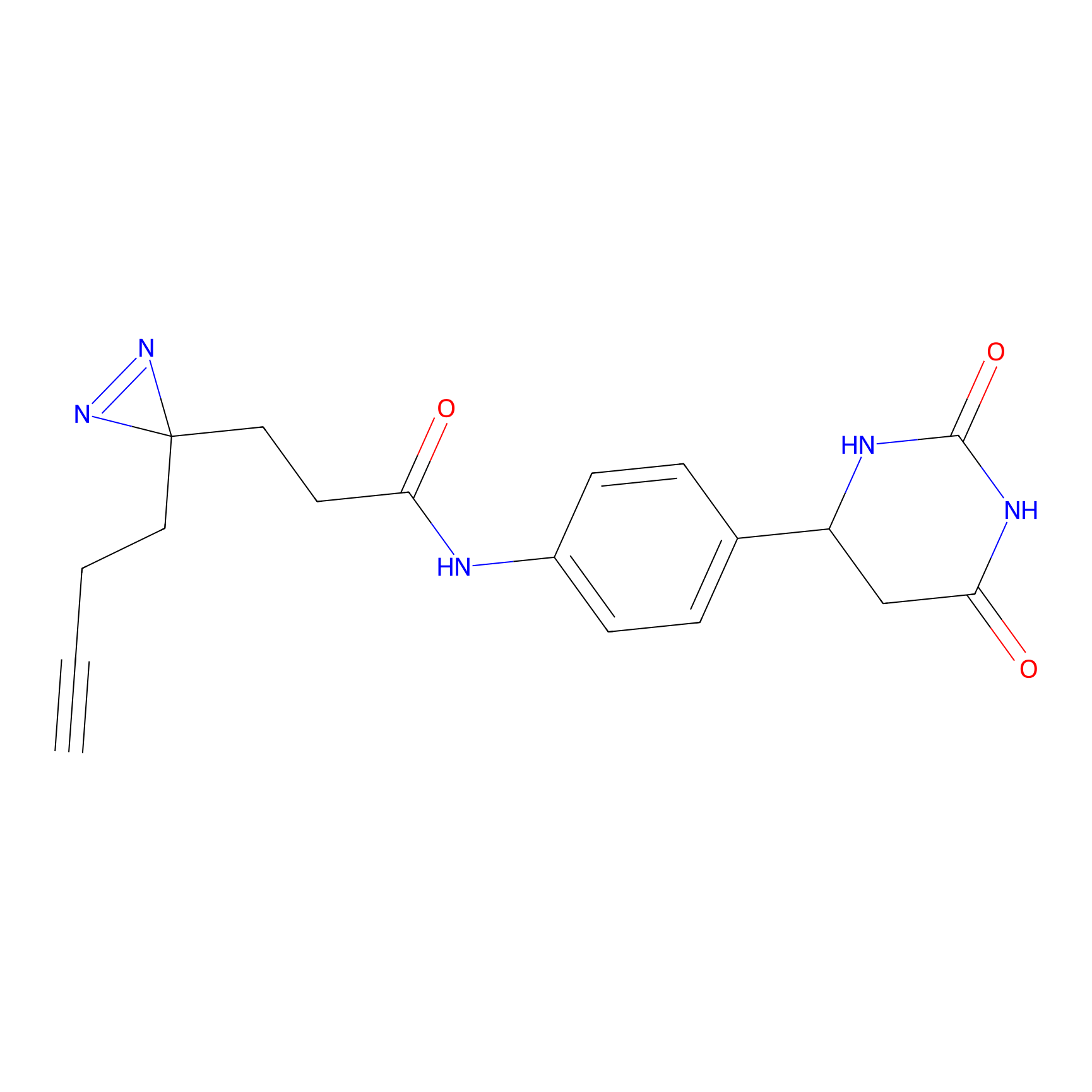
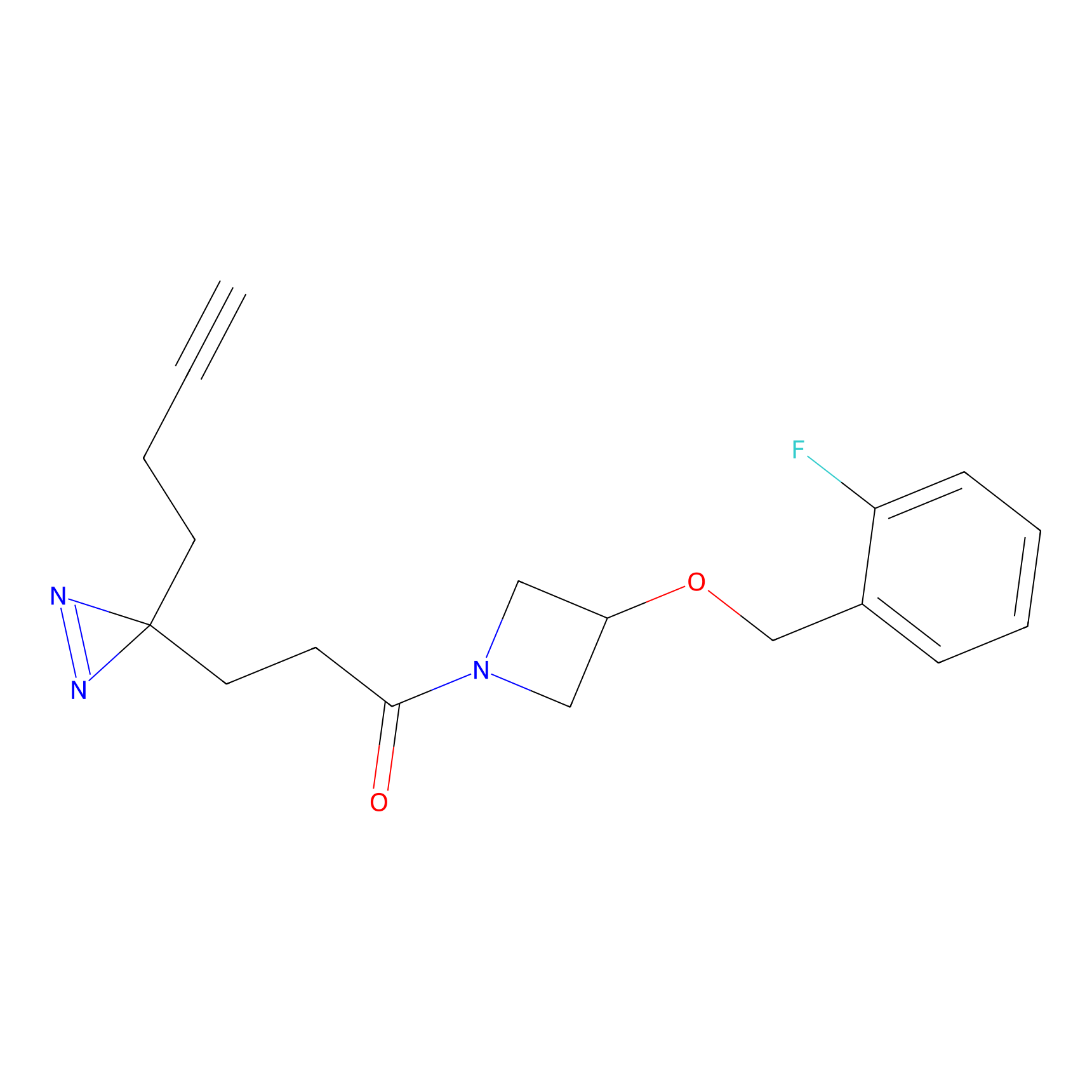
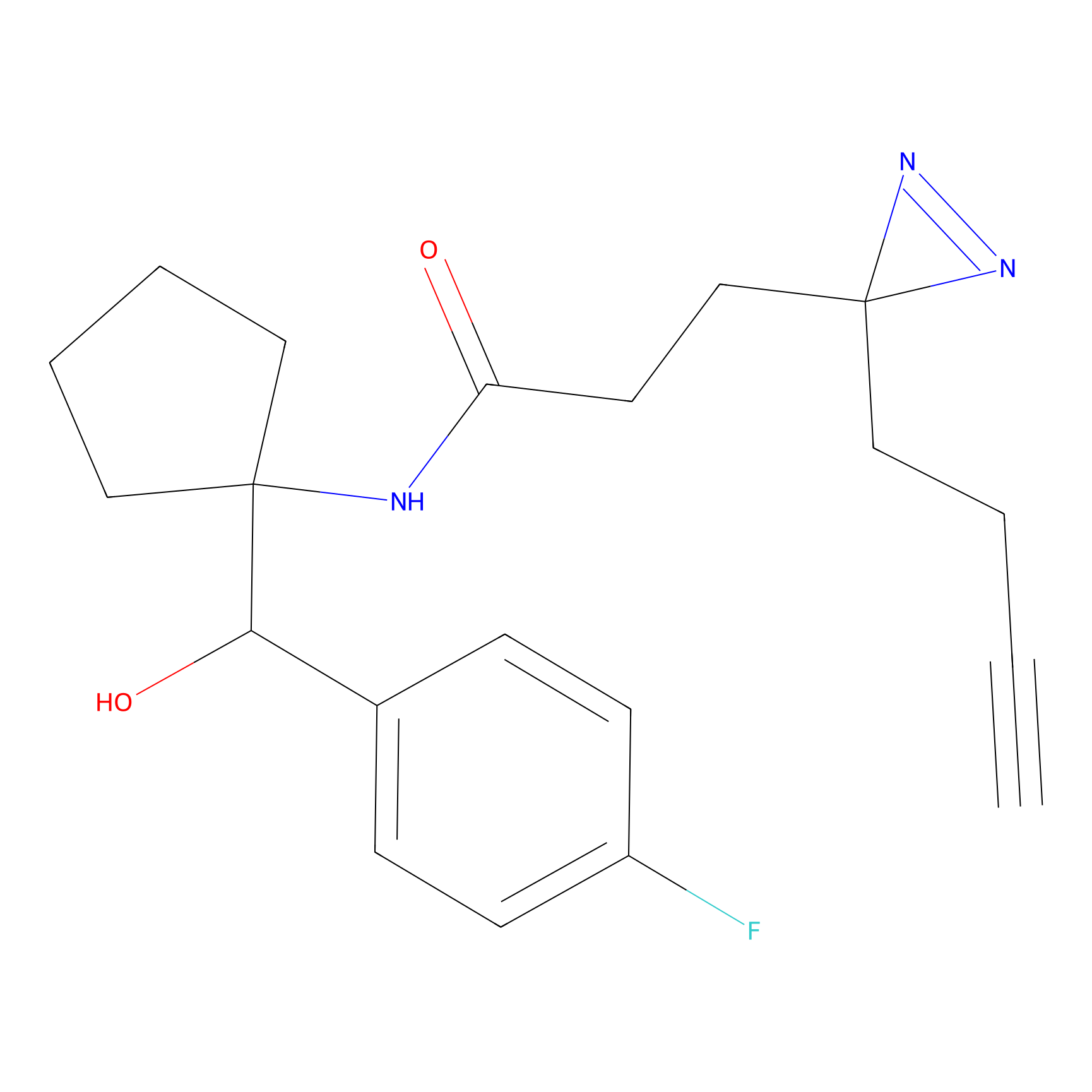
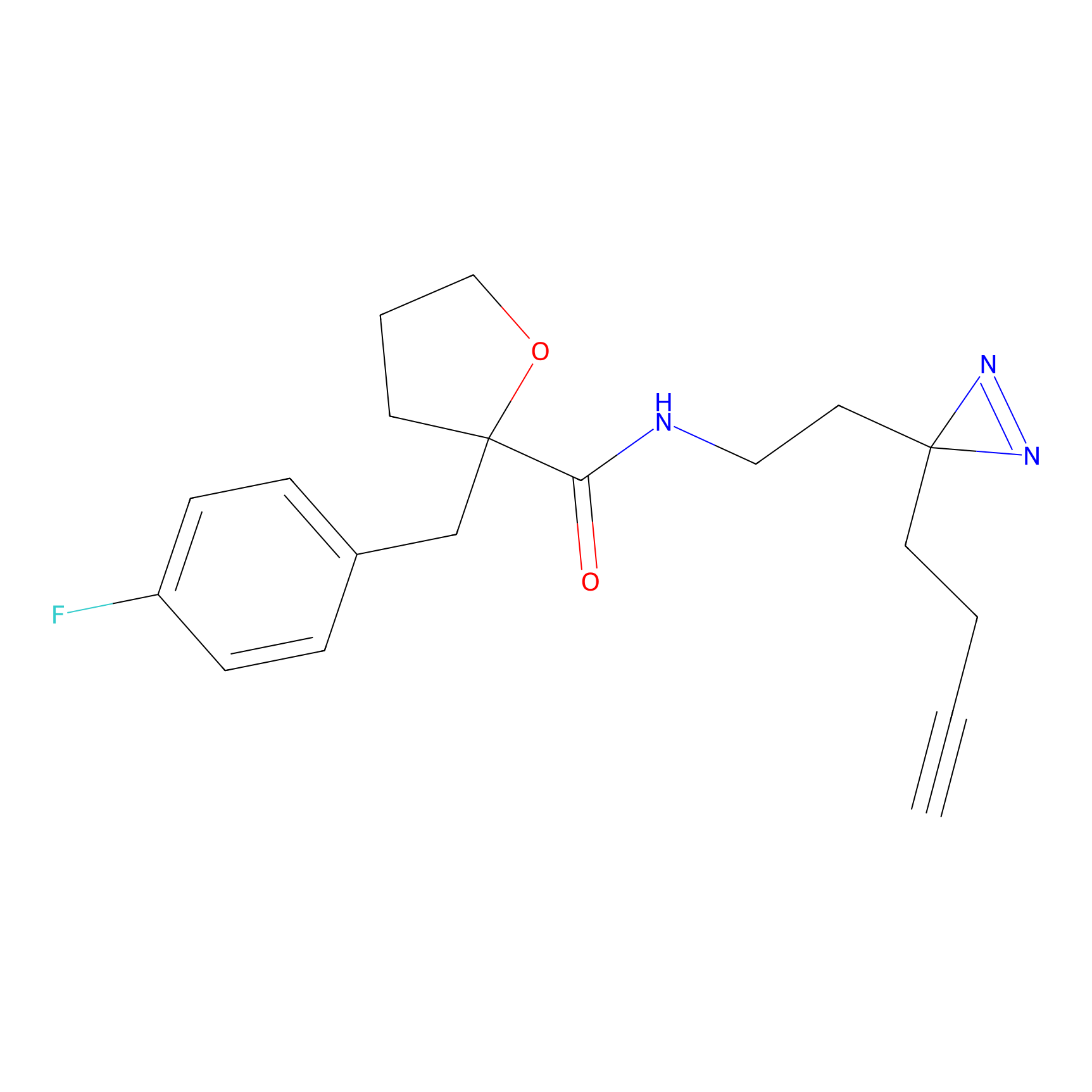
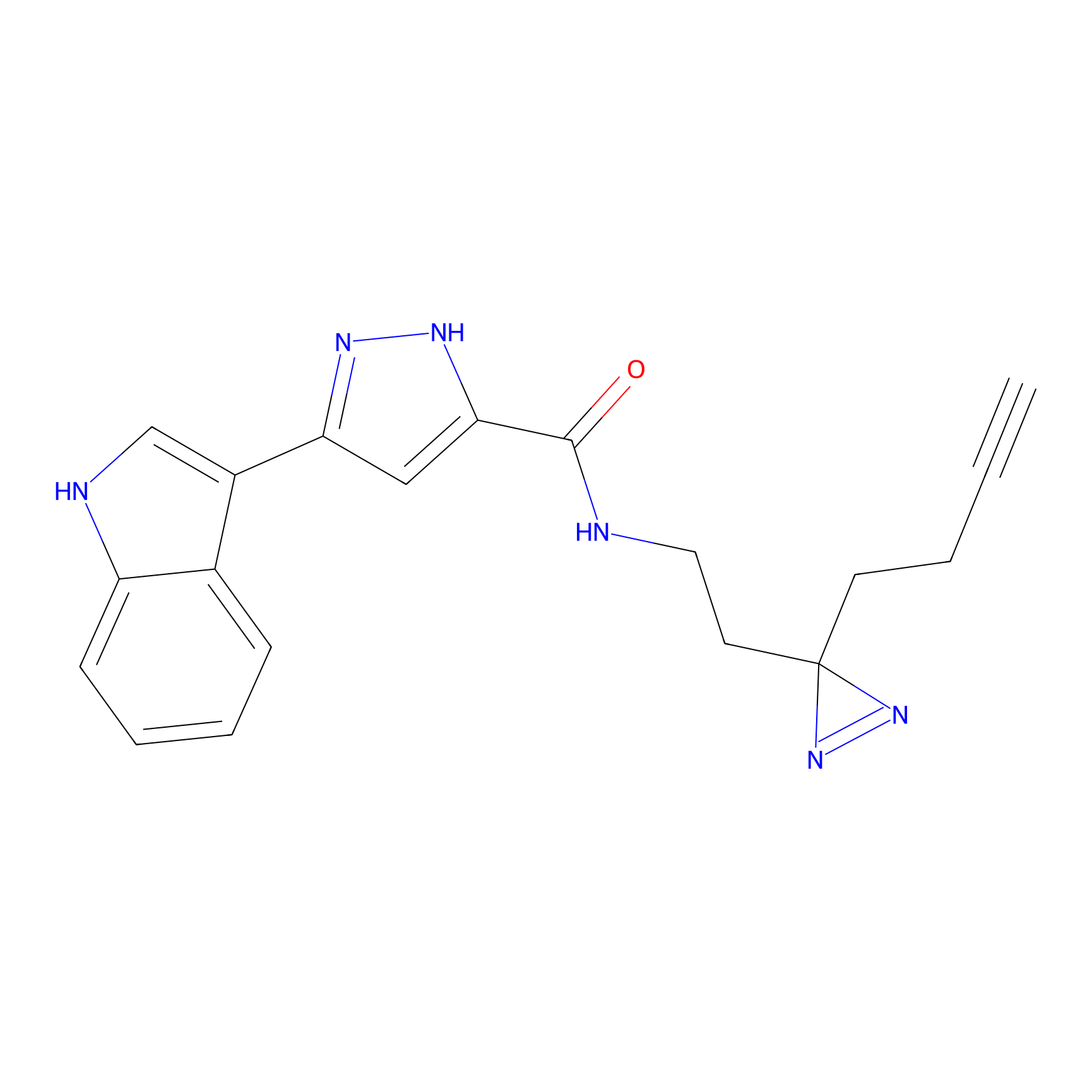
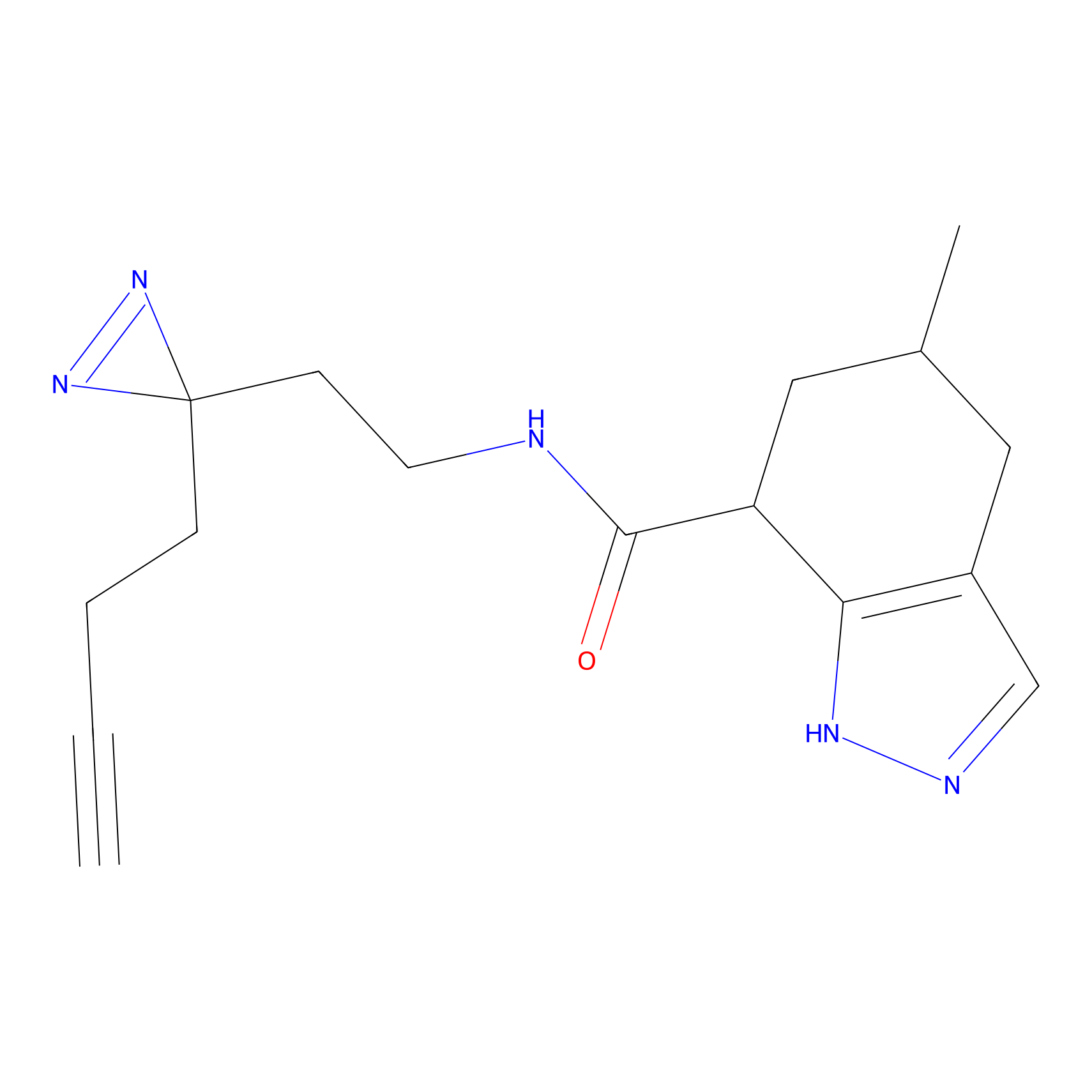
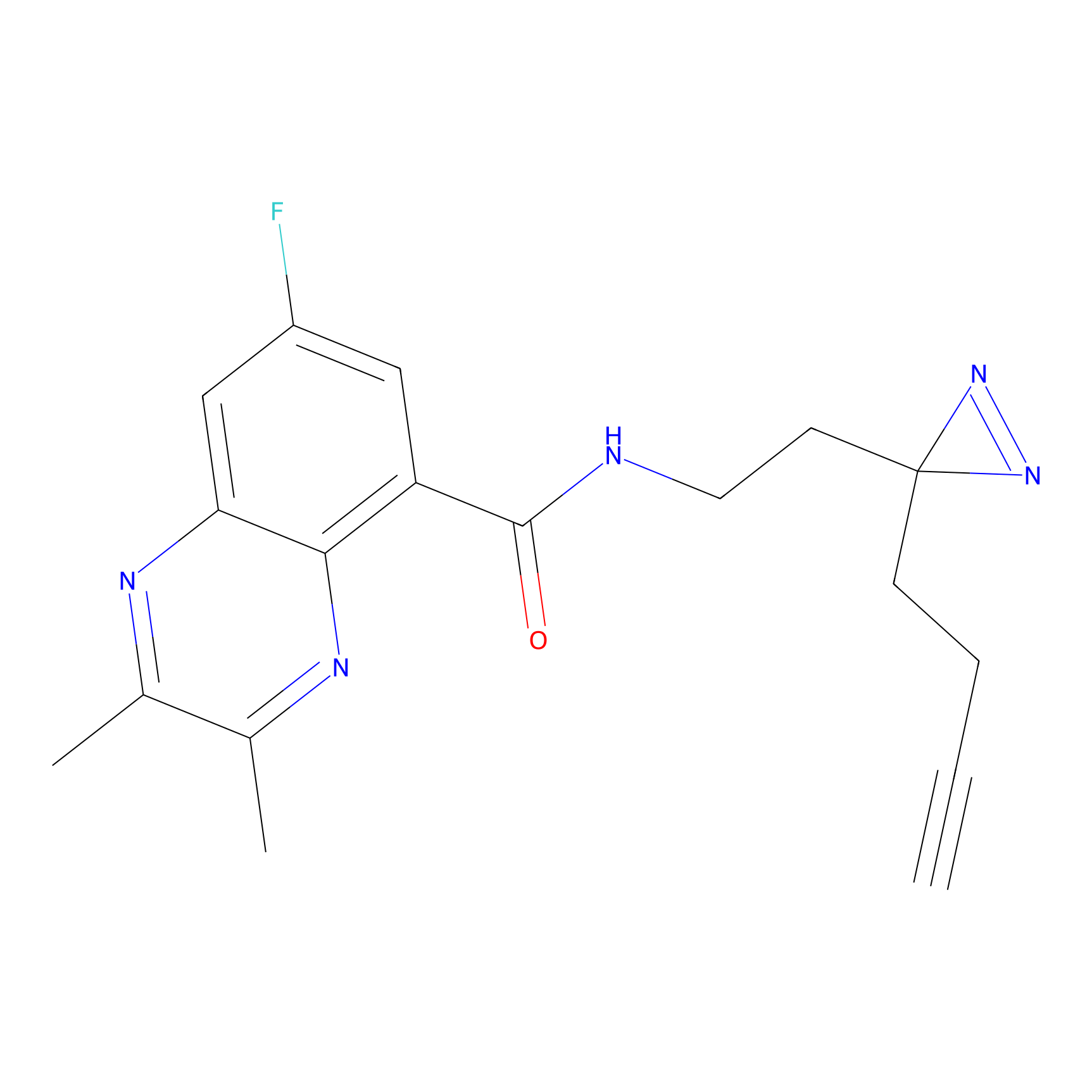
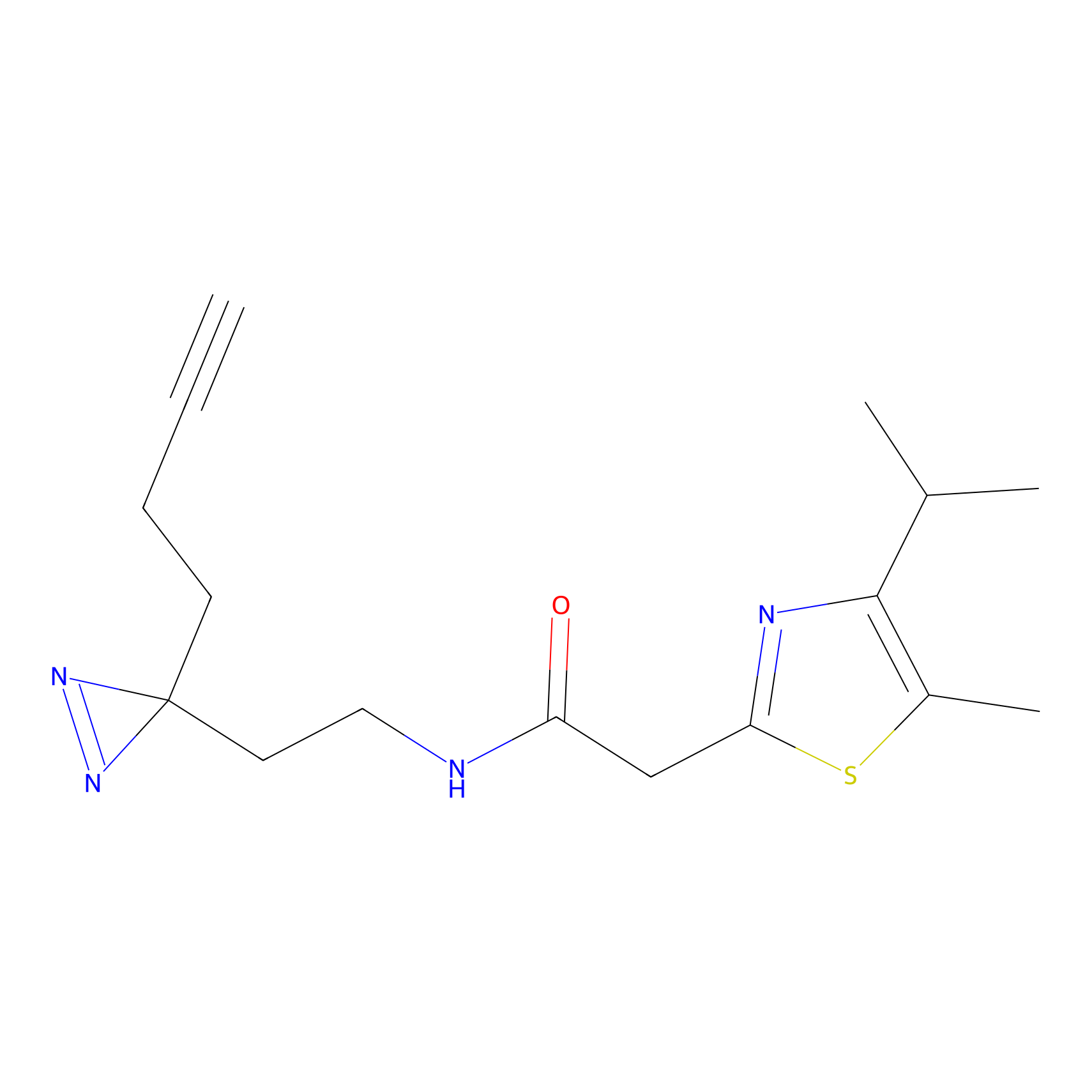
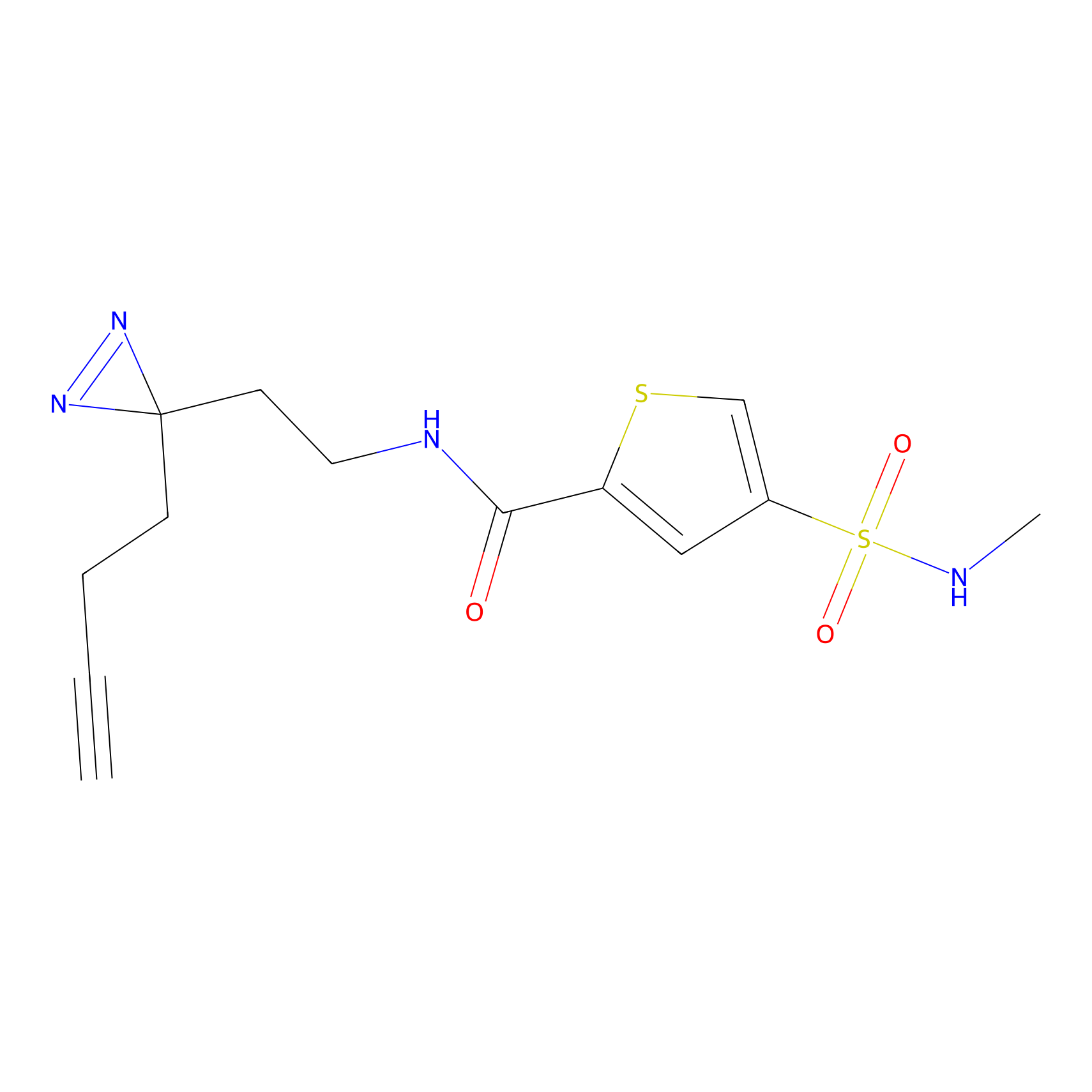
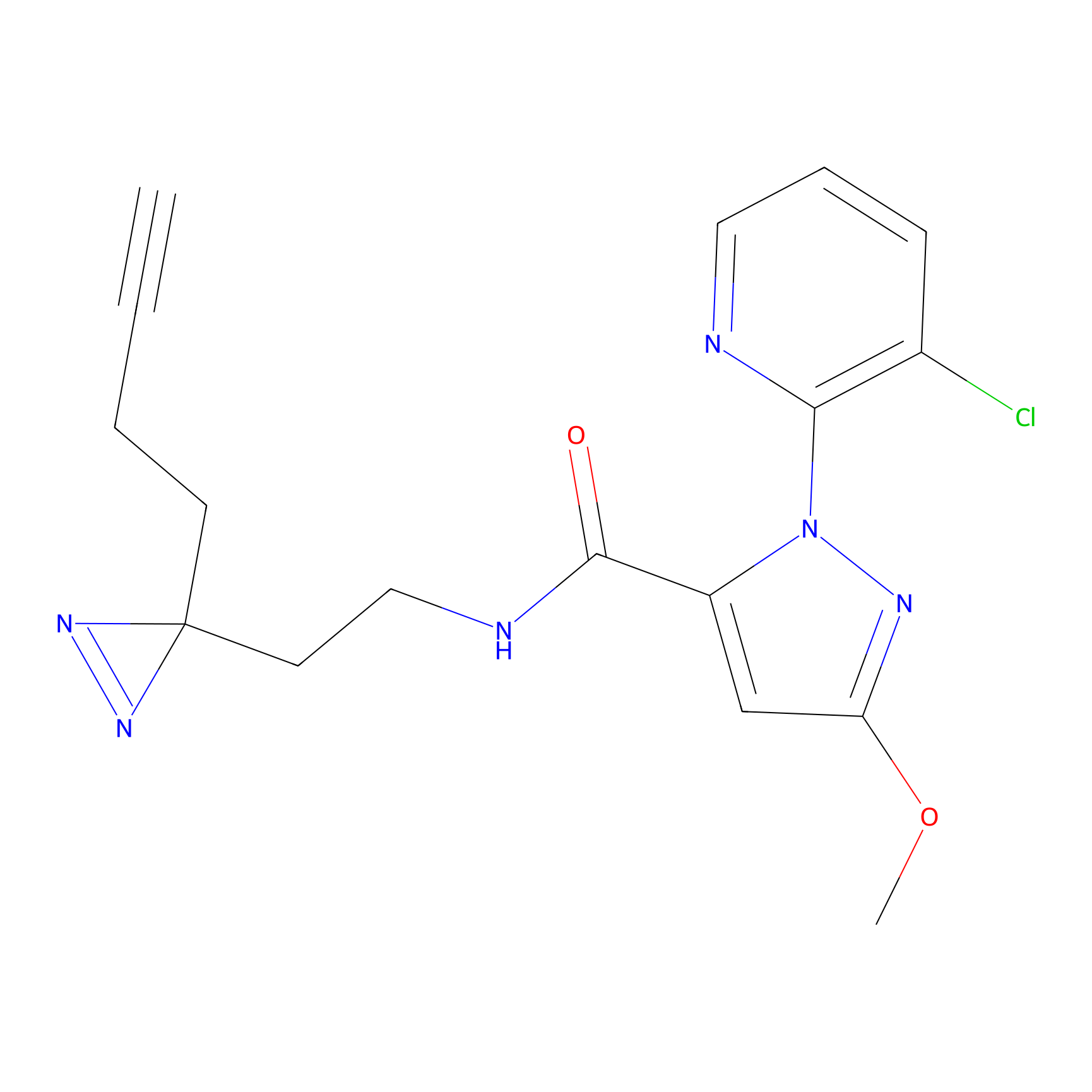
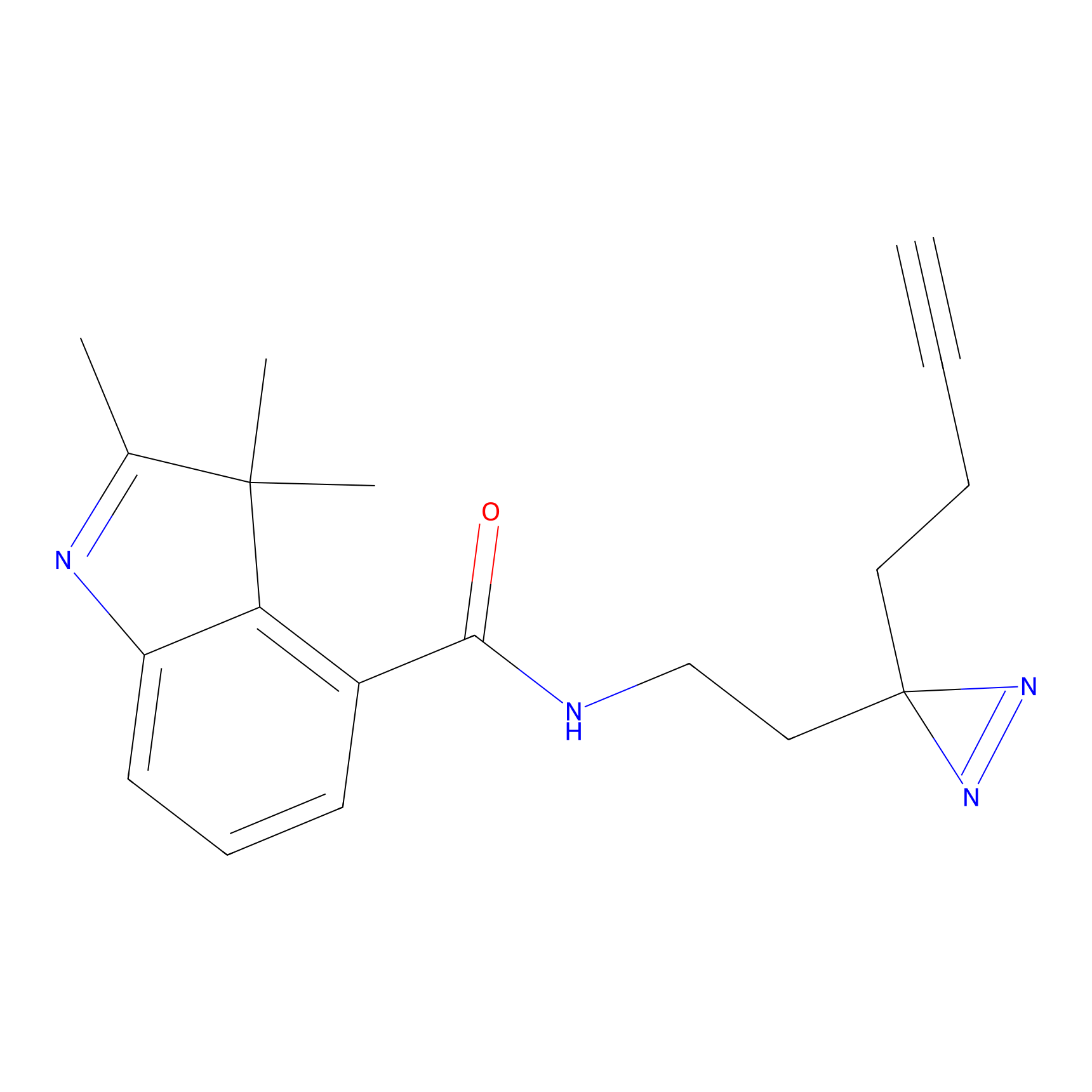
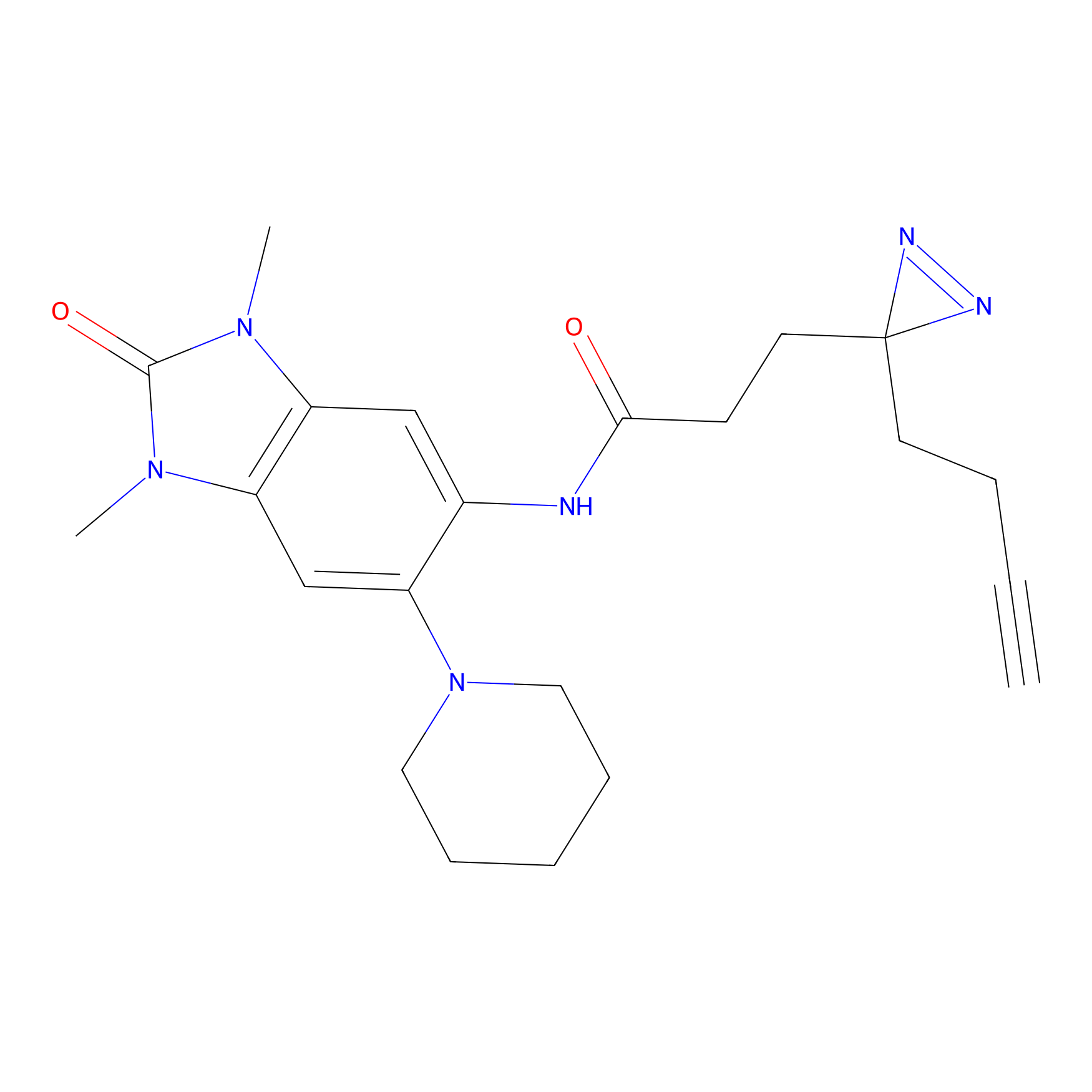
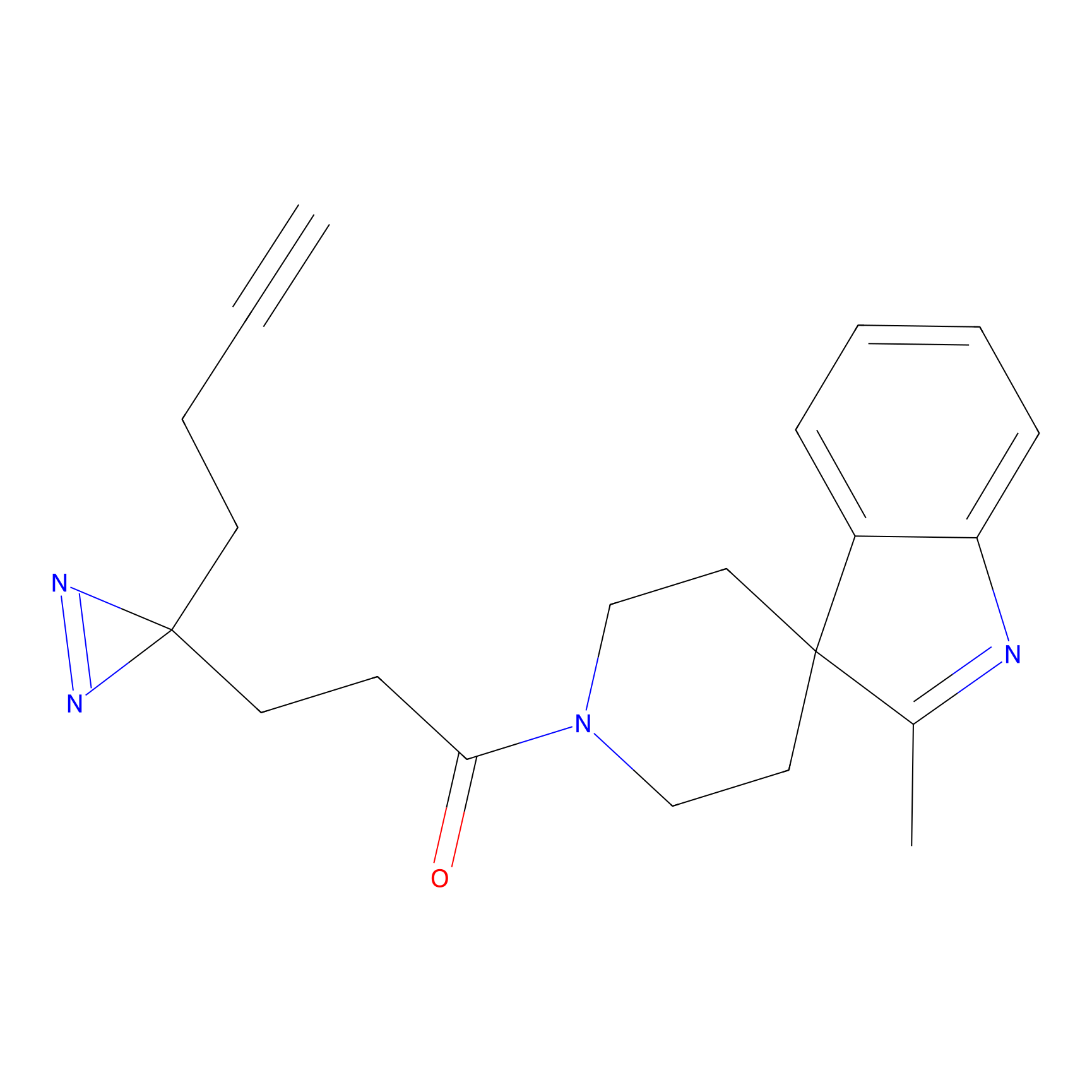
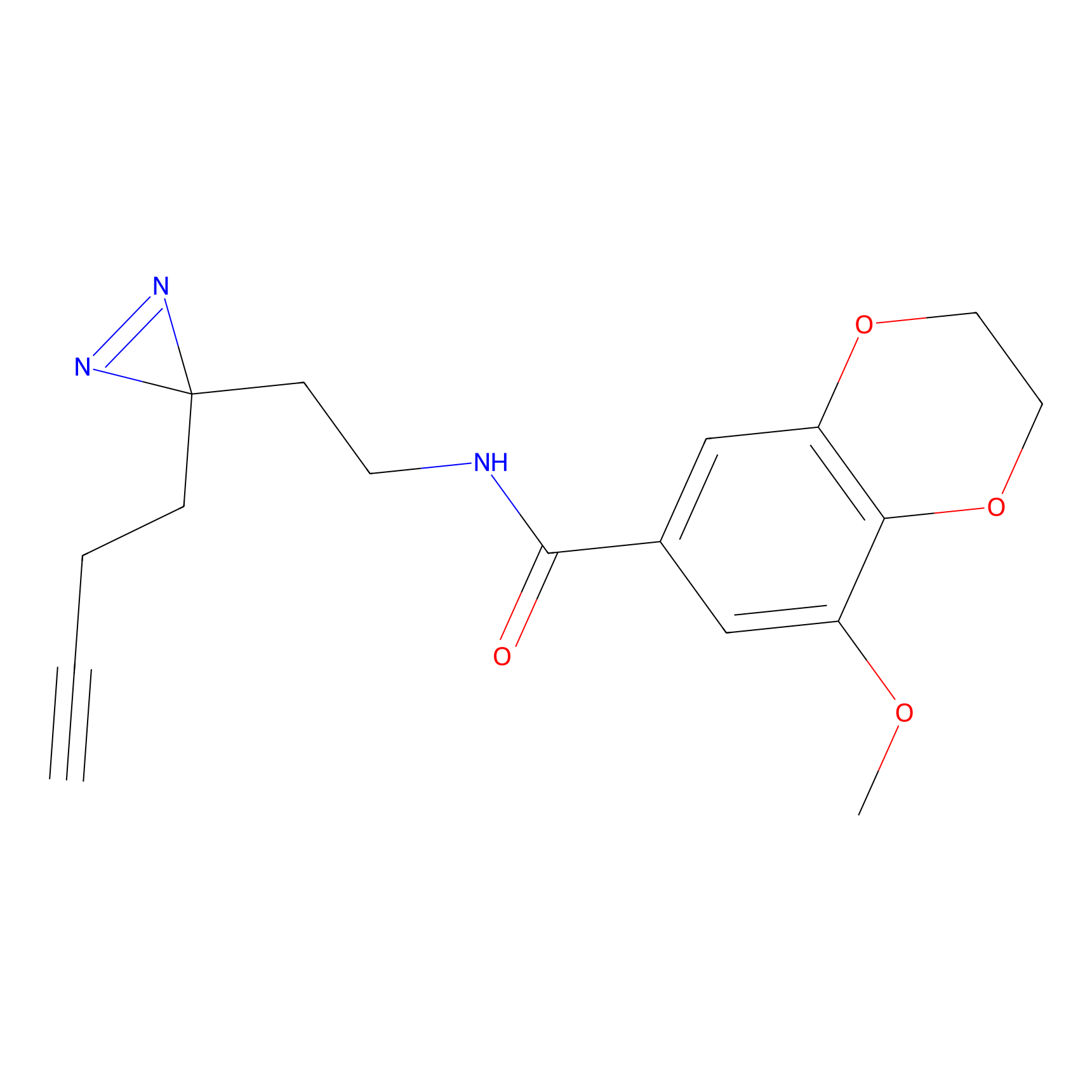
HHS-475 Probe Info |
 |
Y52(0.69) | LDD0264 | [1] | |
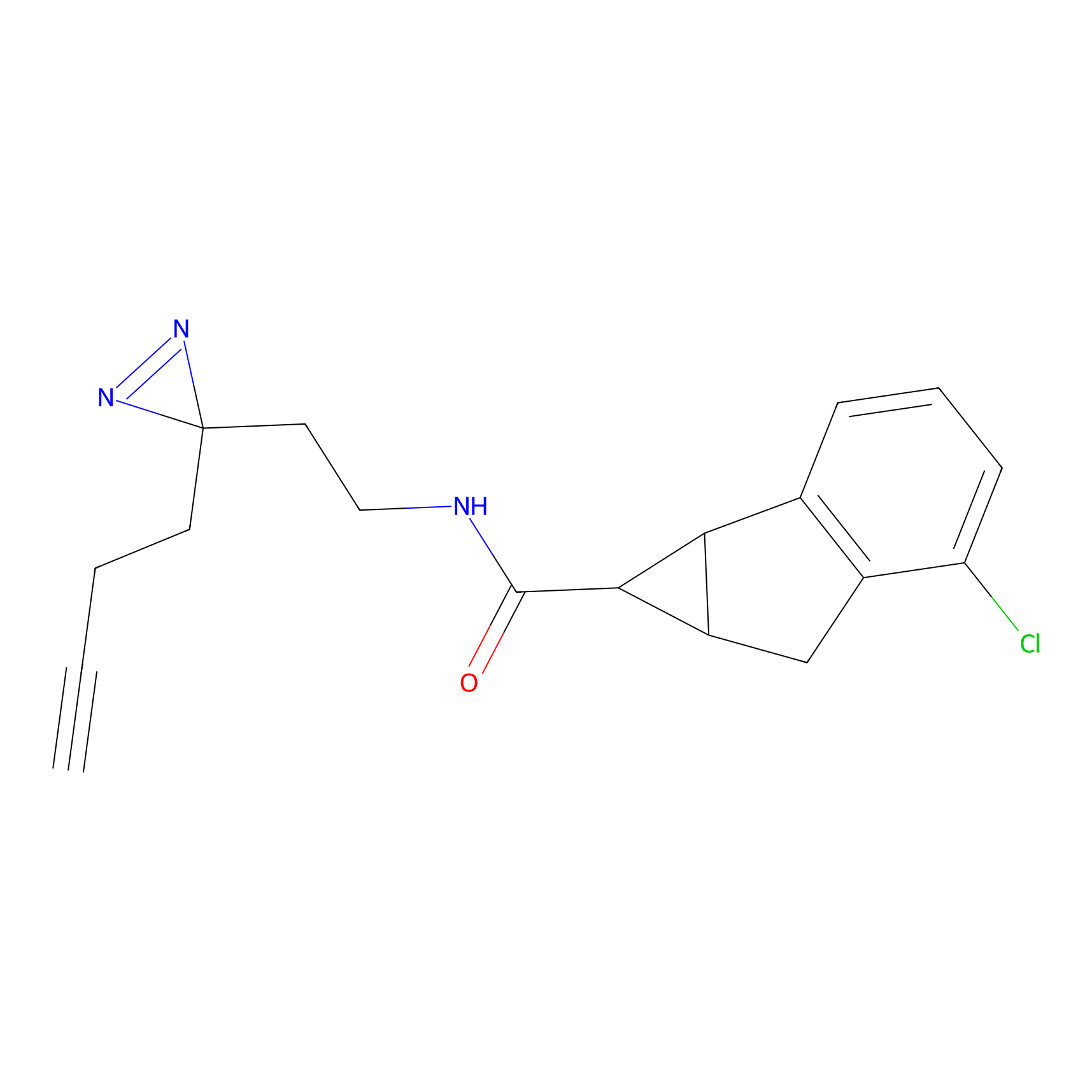
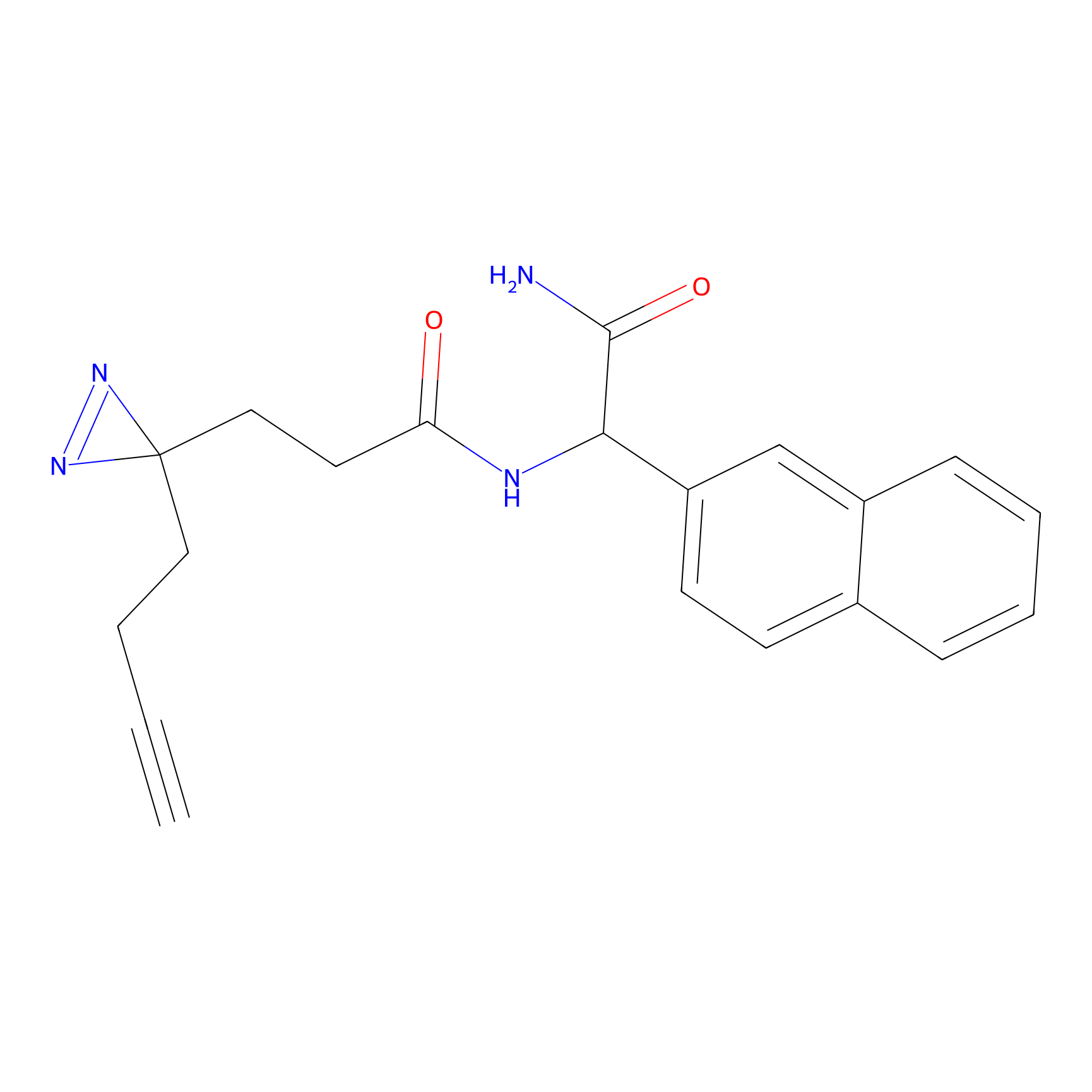
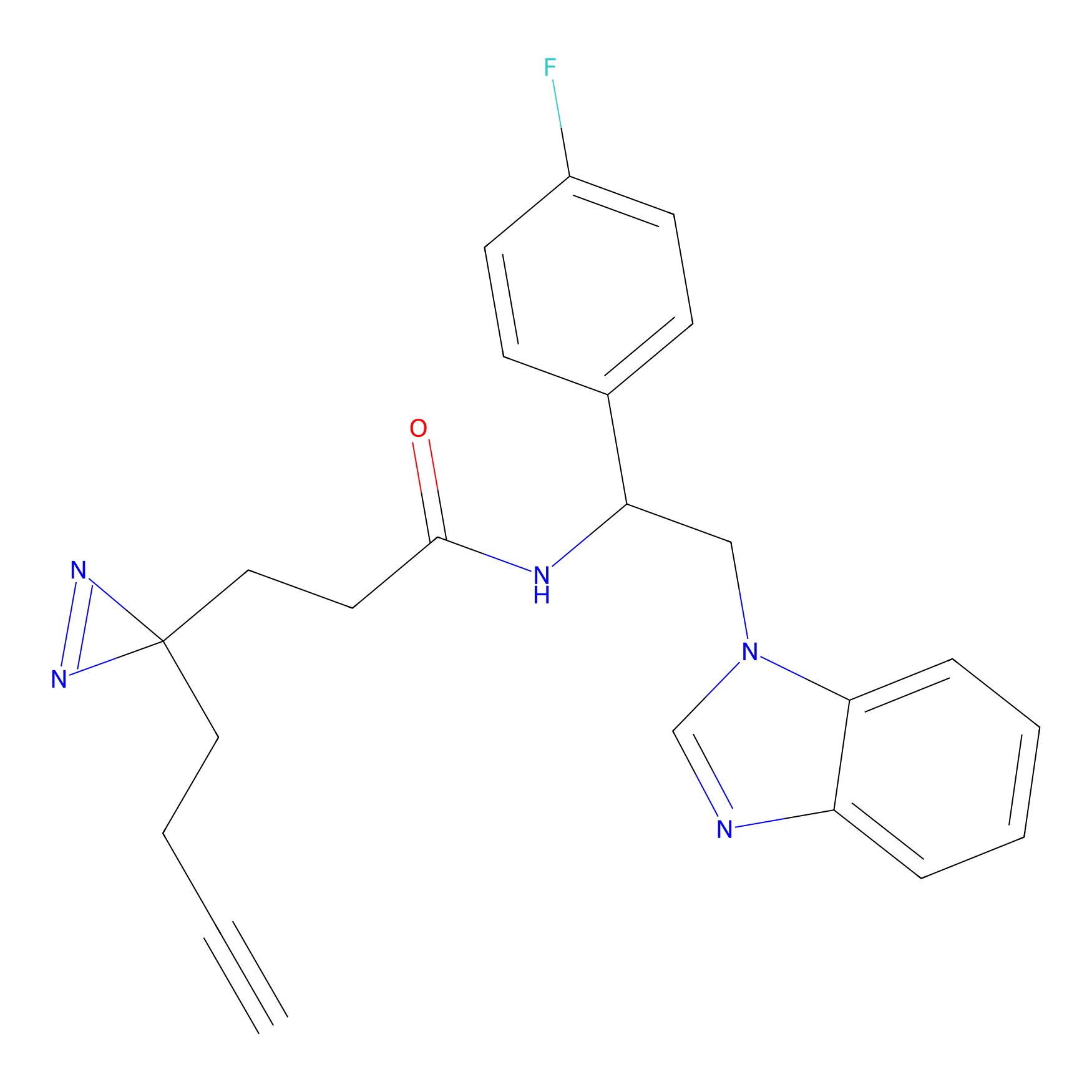
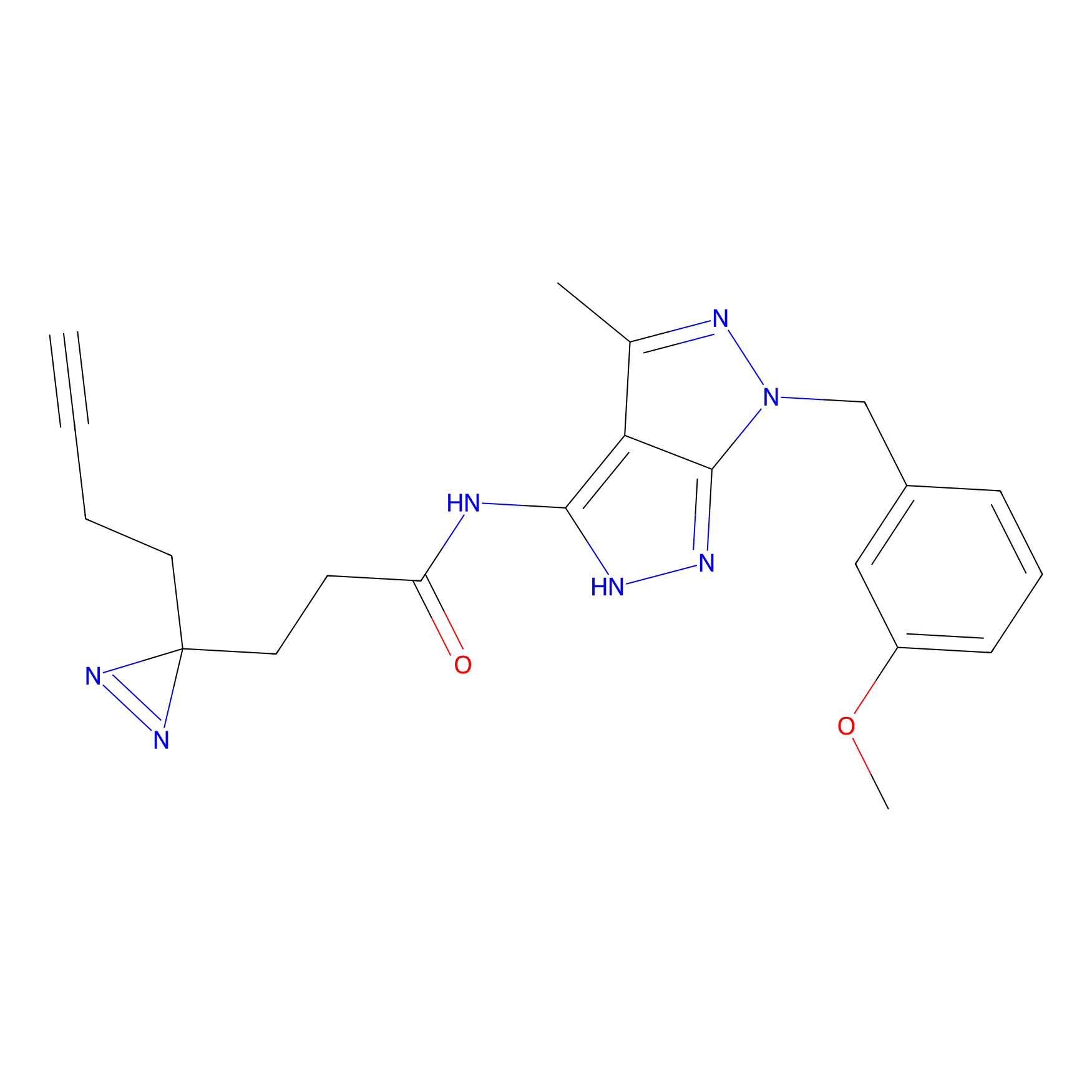
|
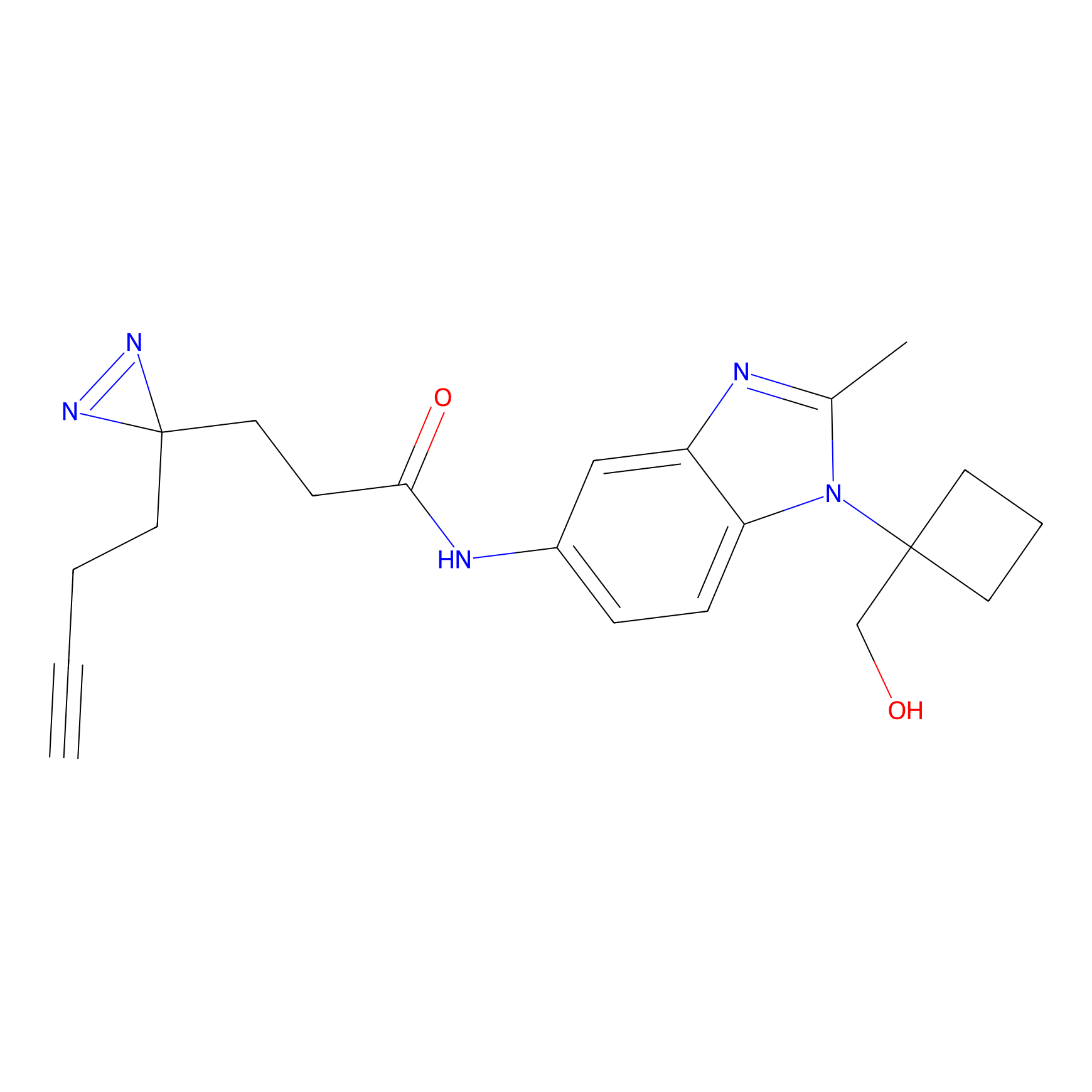
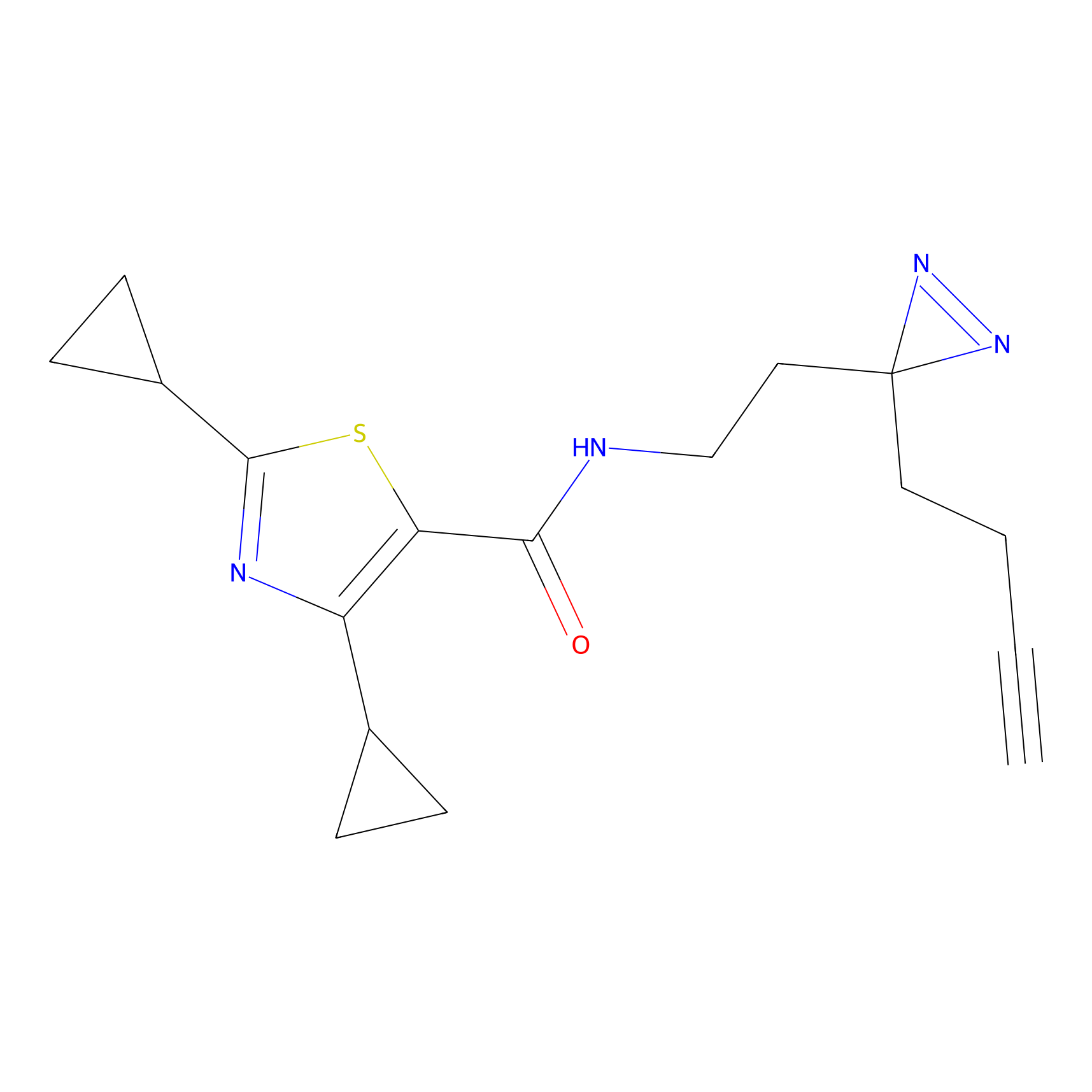
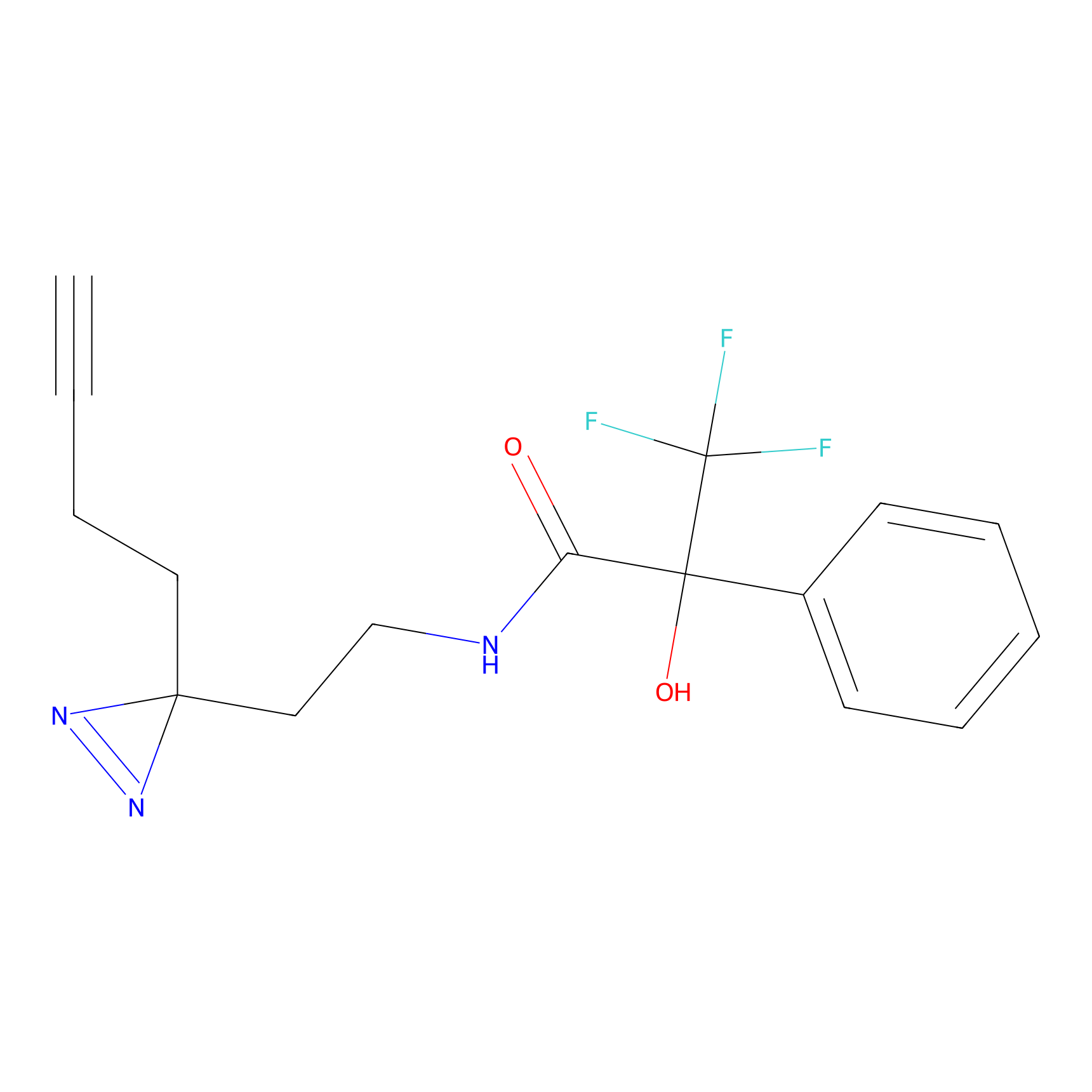
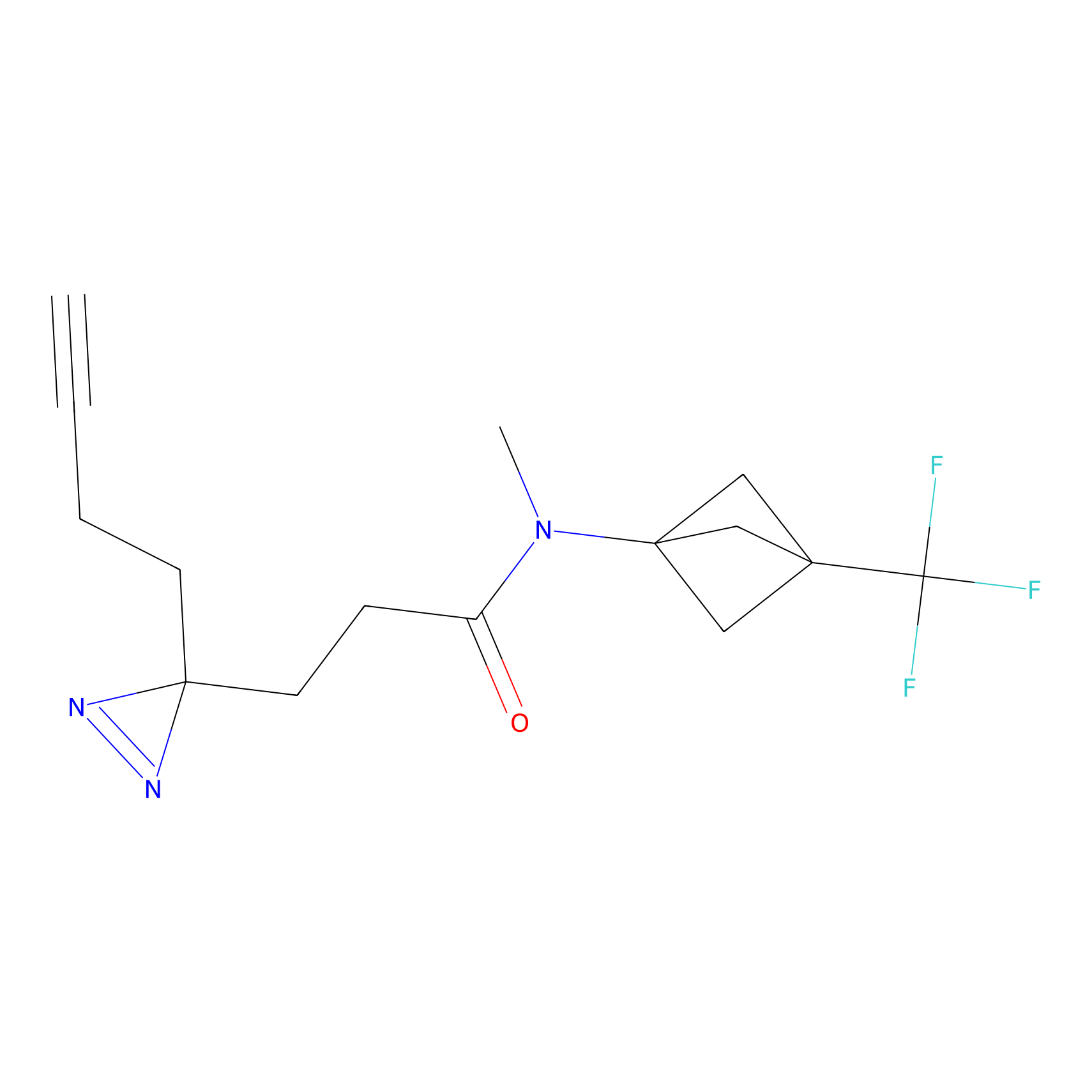
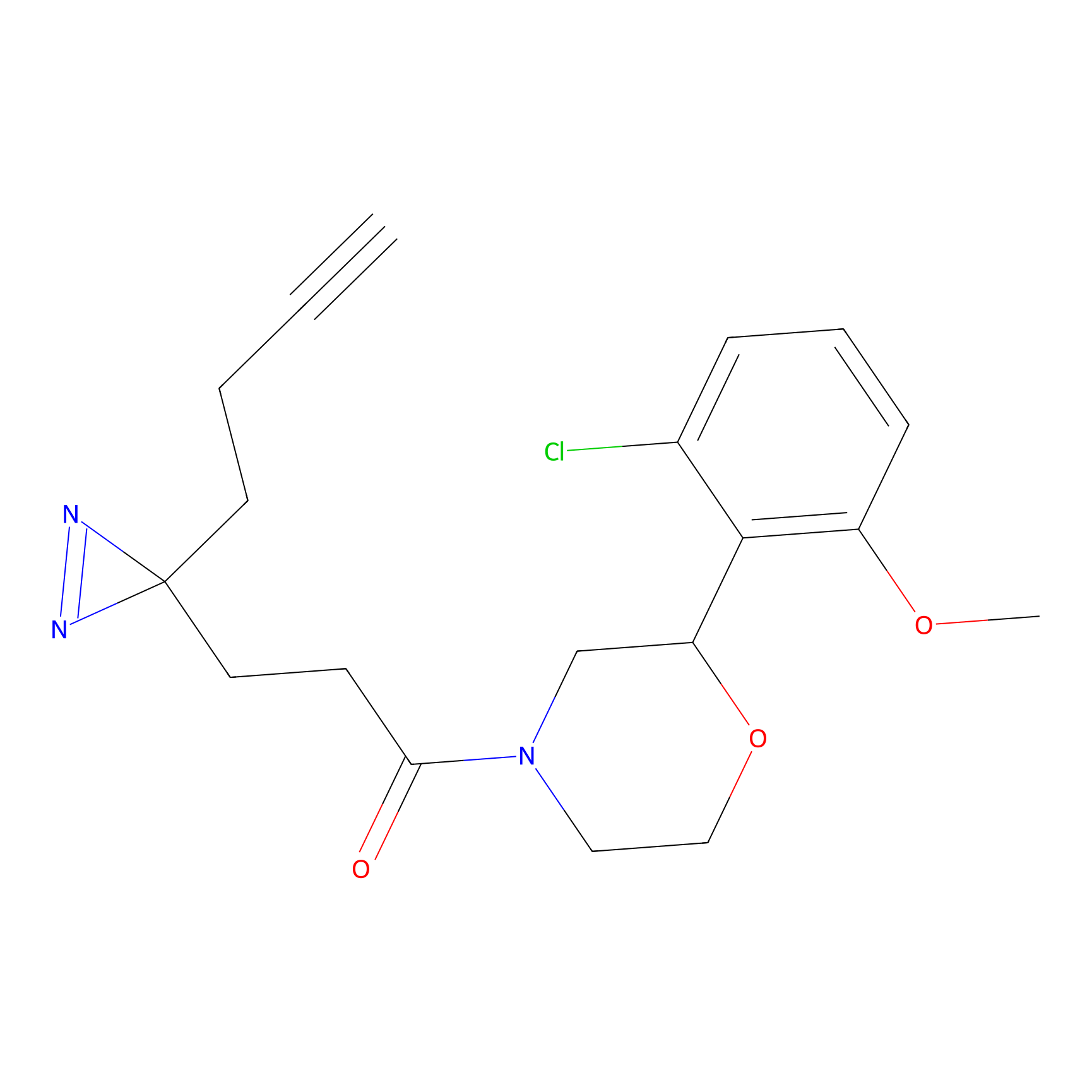
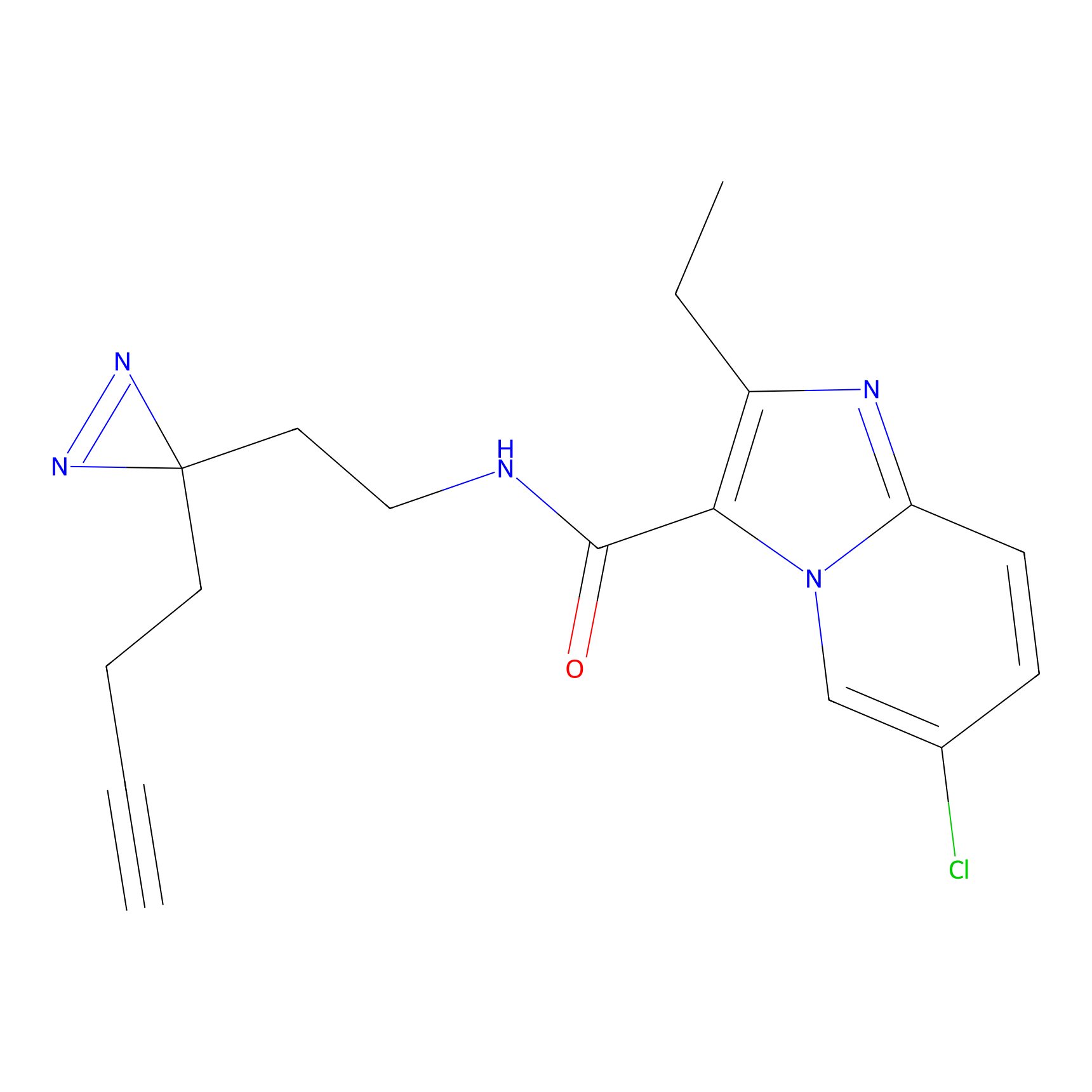
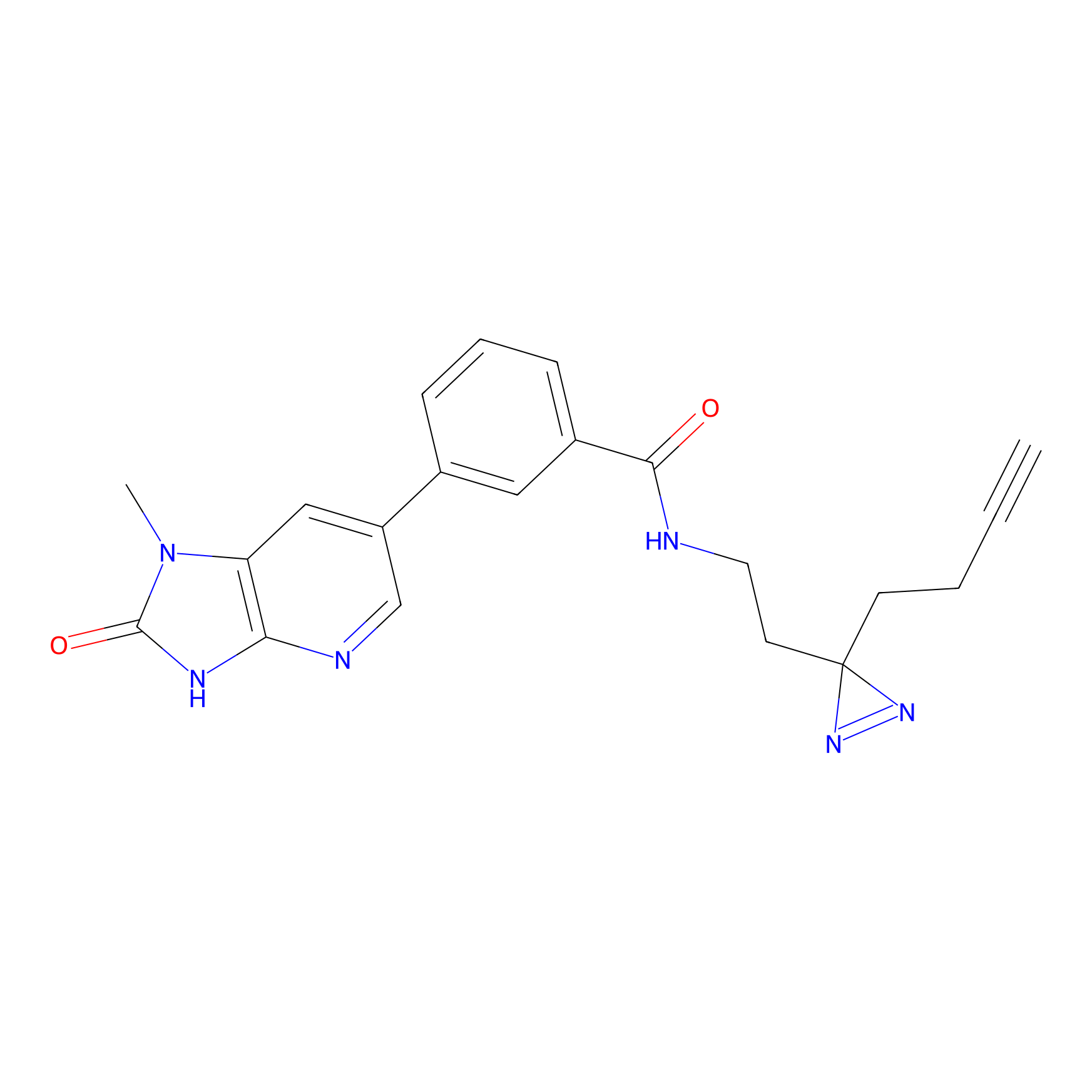
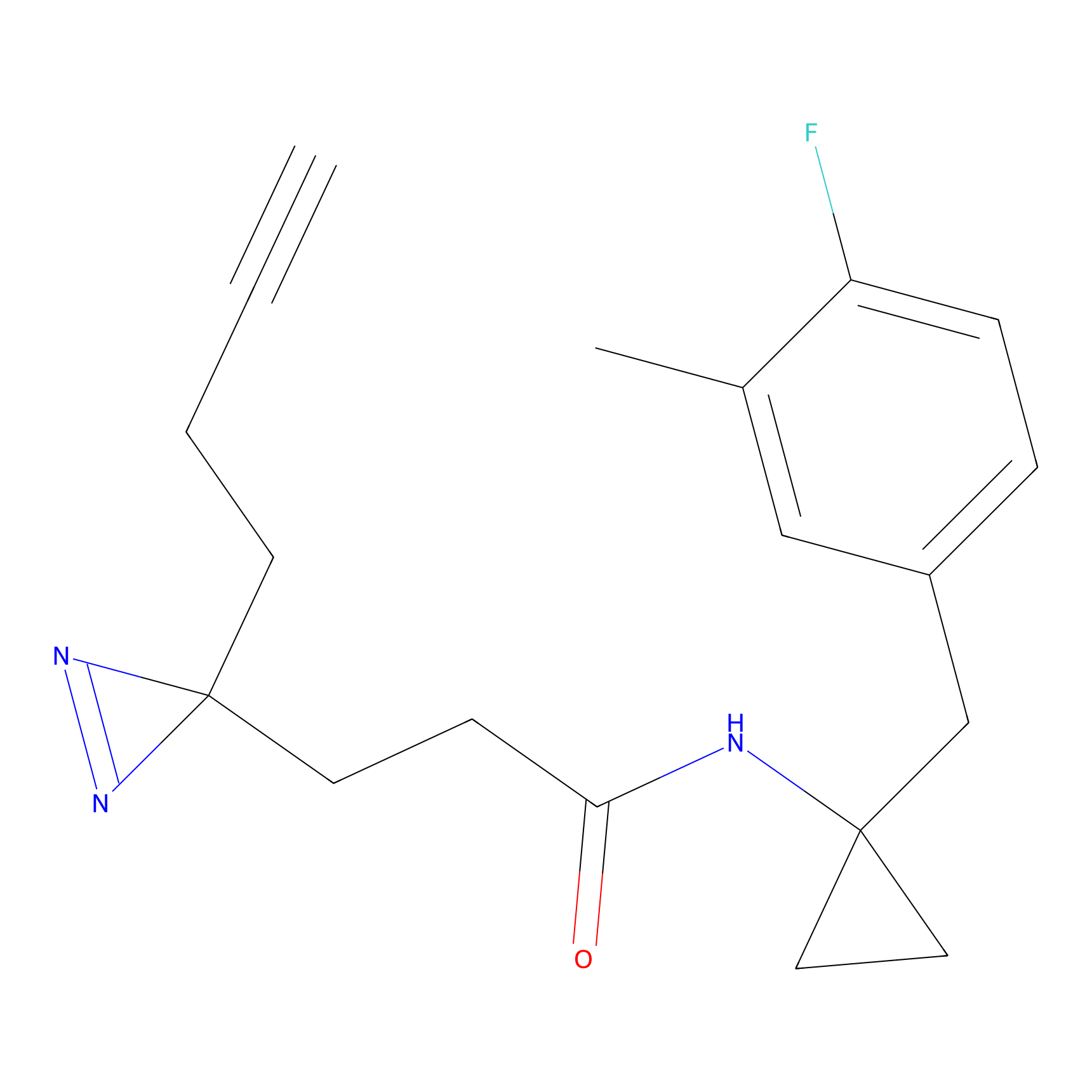
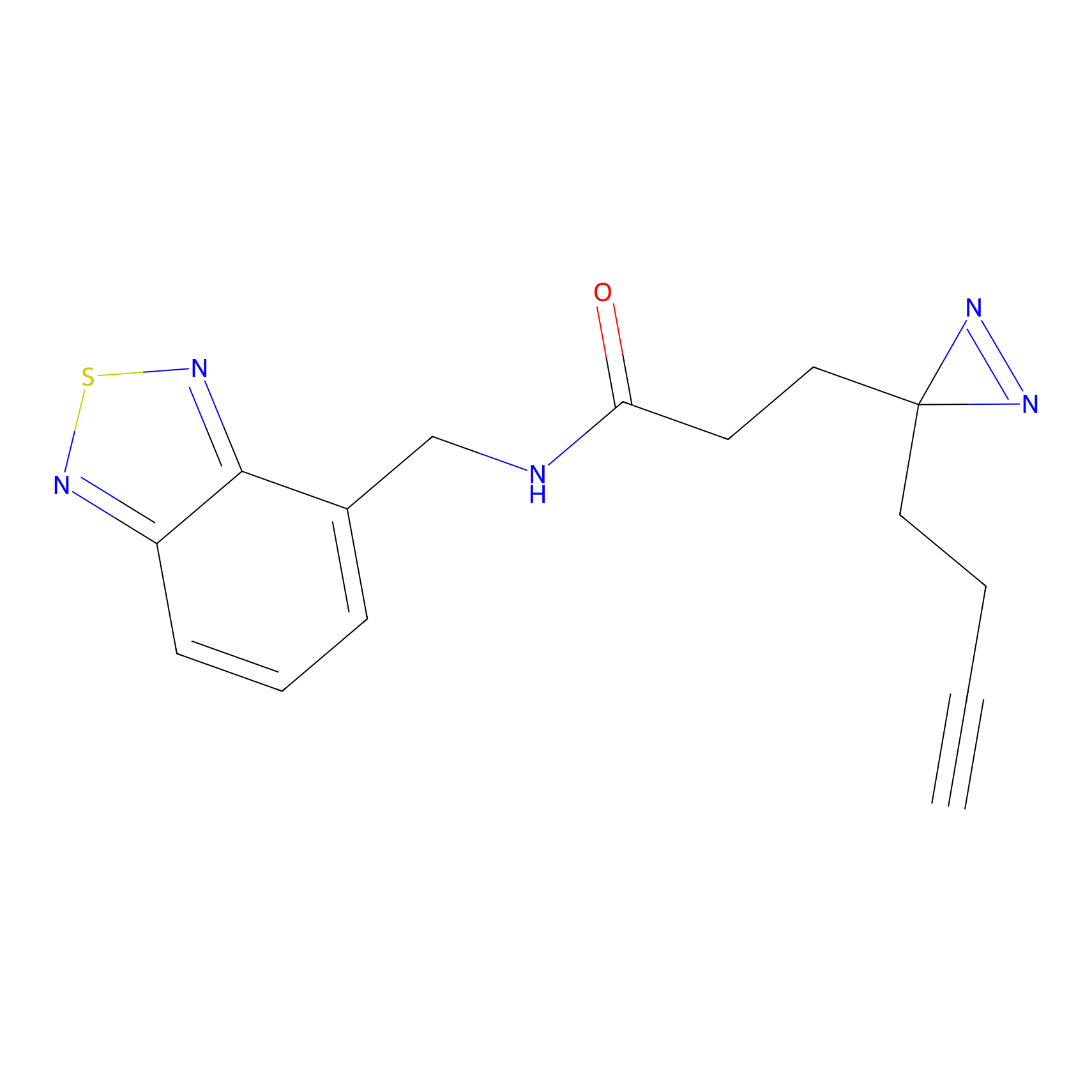
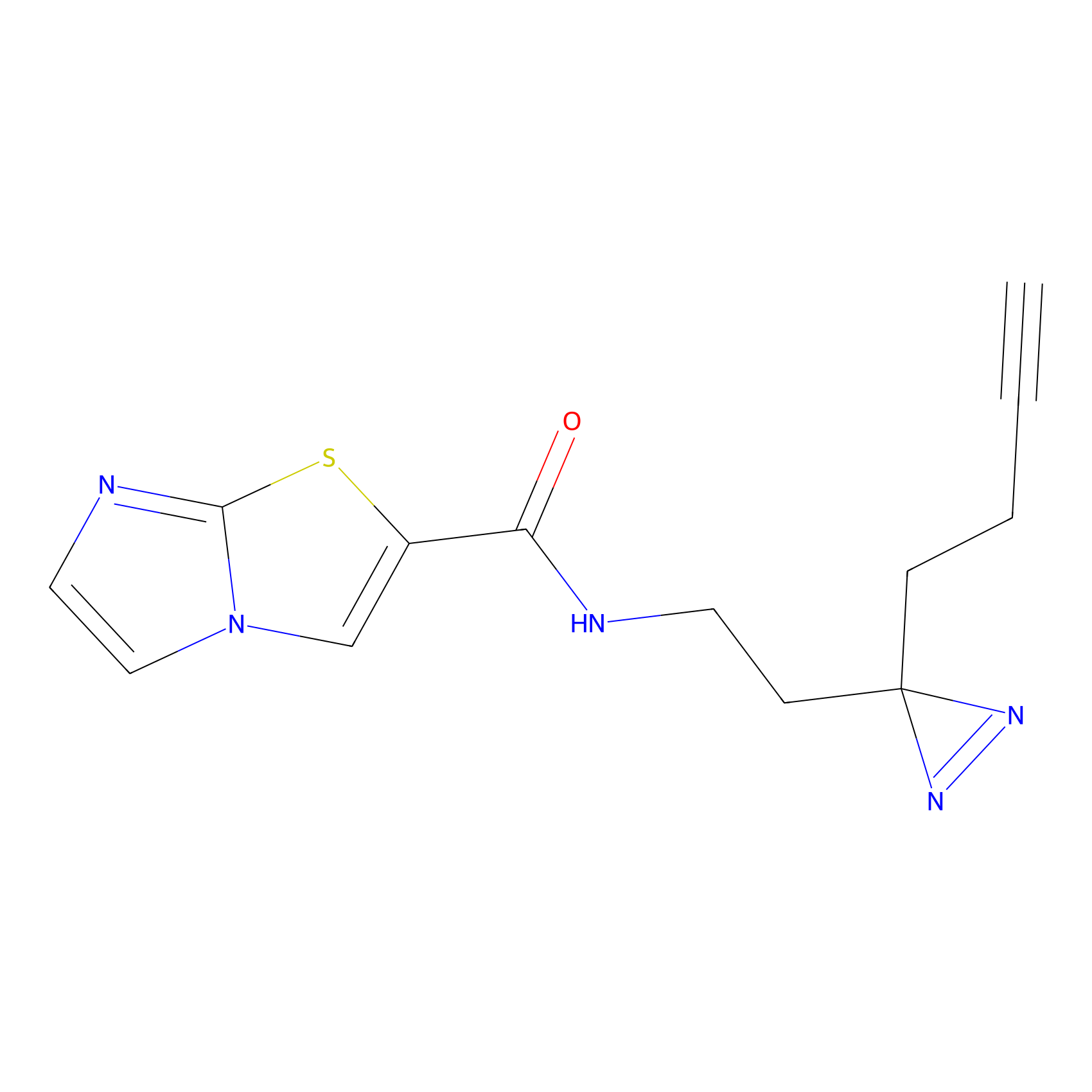
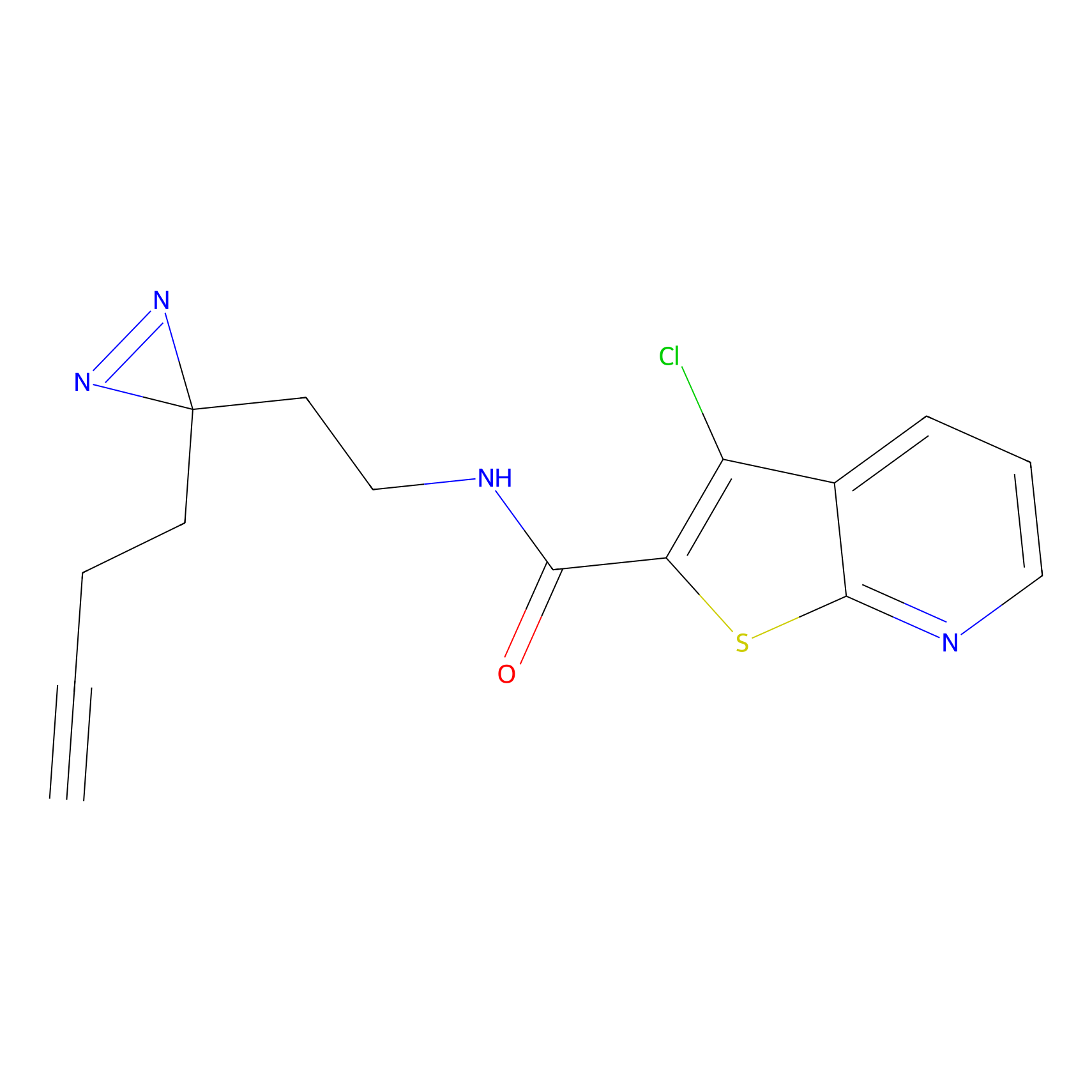
IA-alkyne Probe Info |
 |
N.A. | LDD0165 | [2] | |
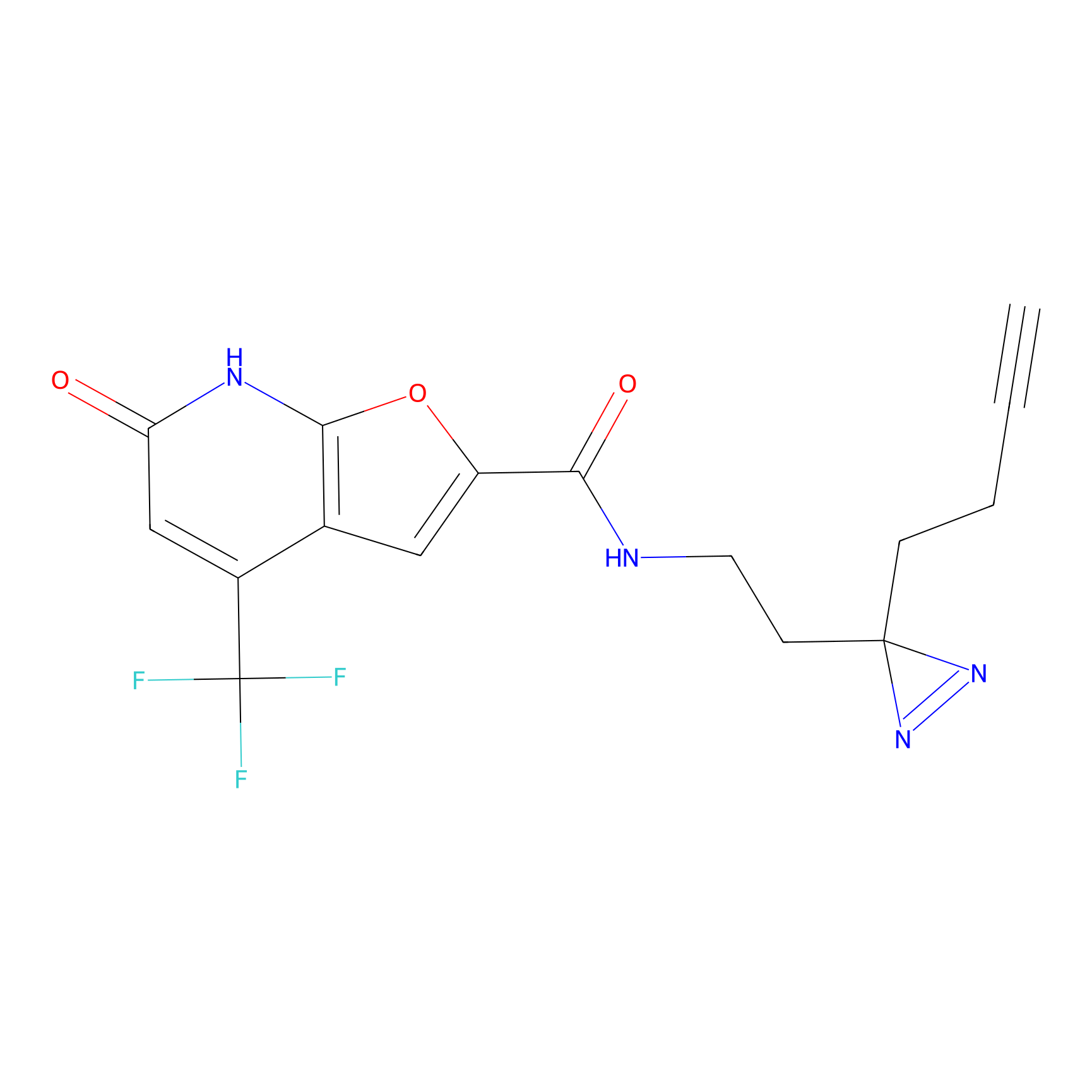
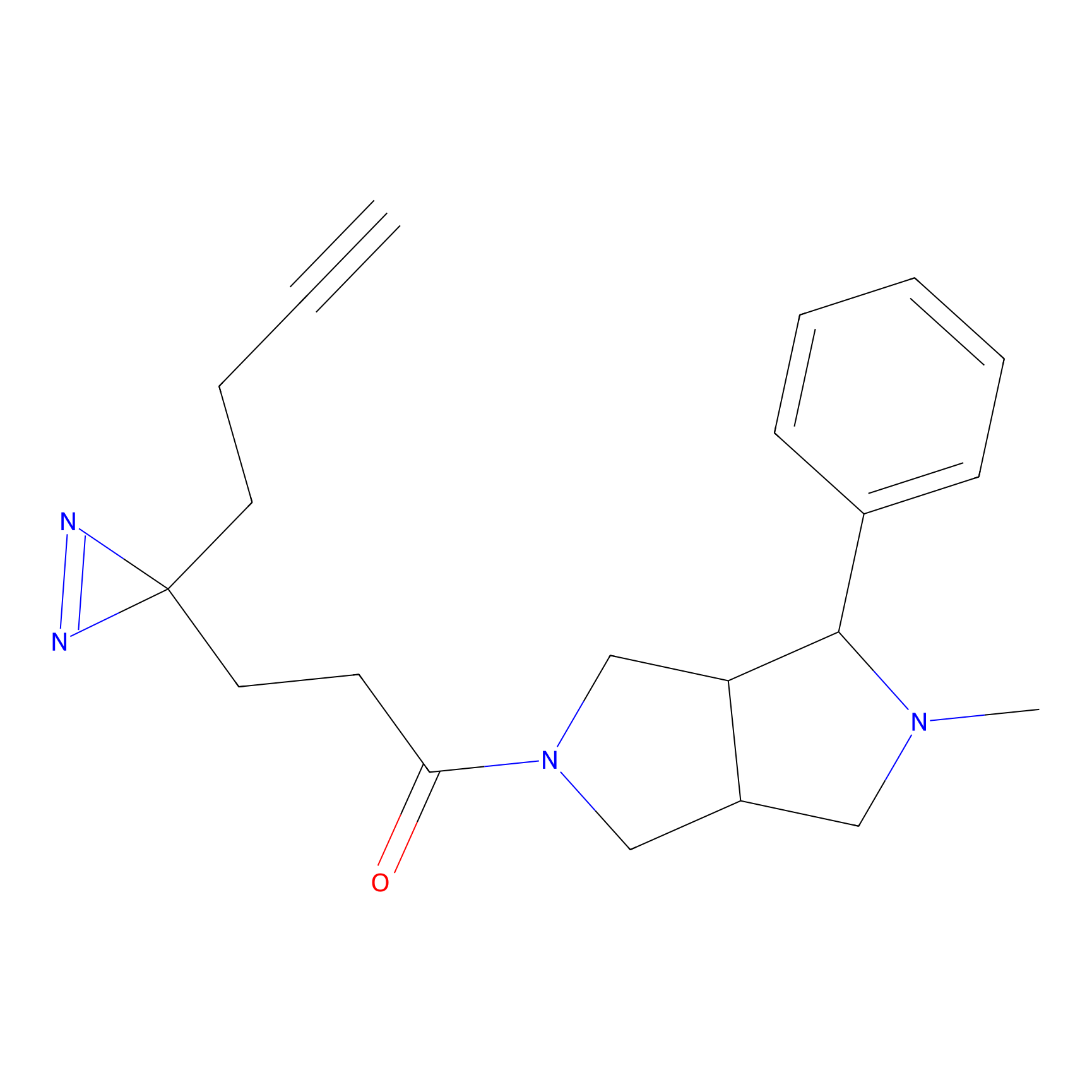
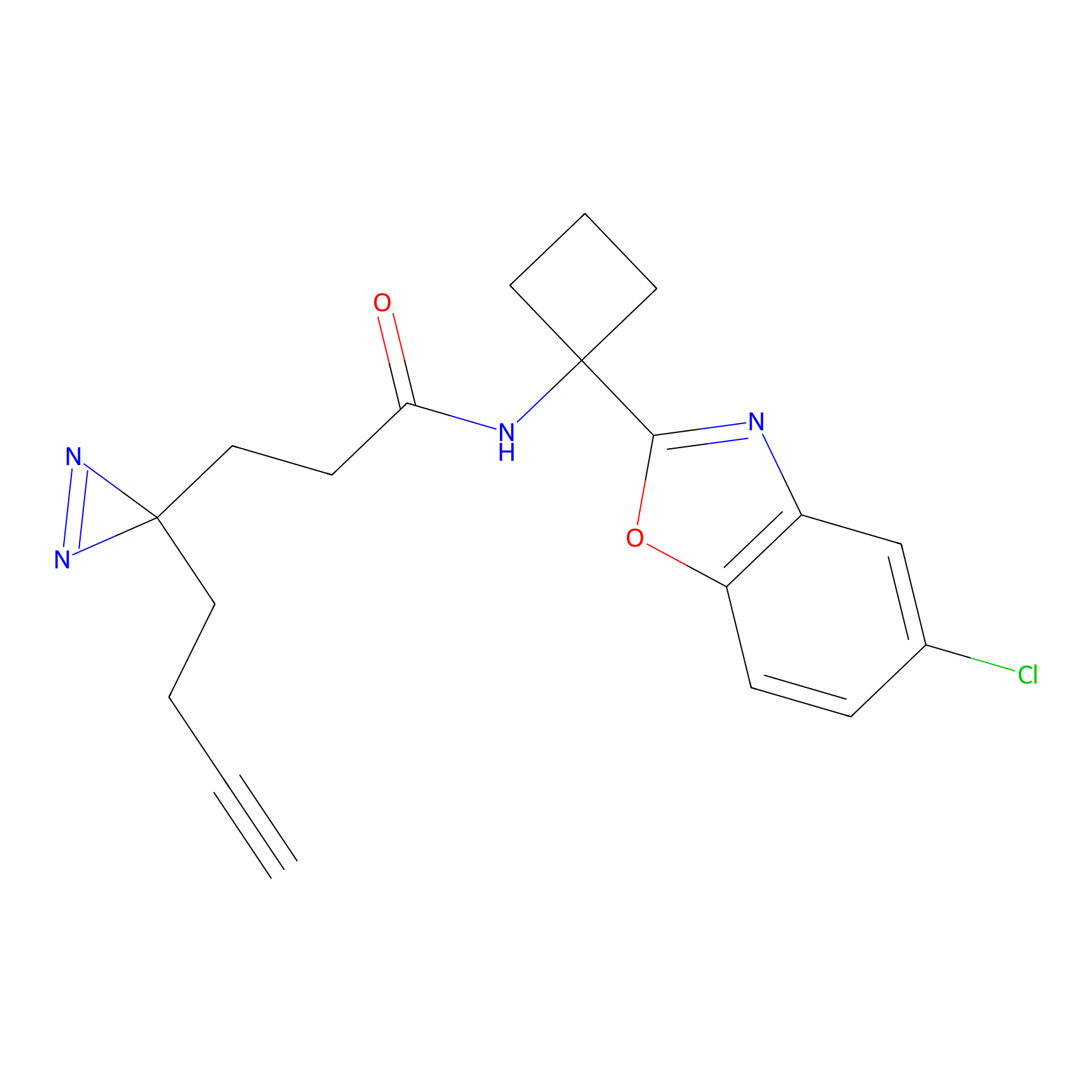
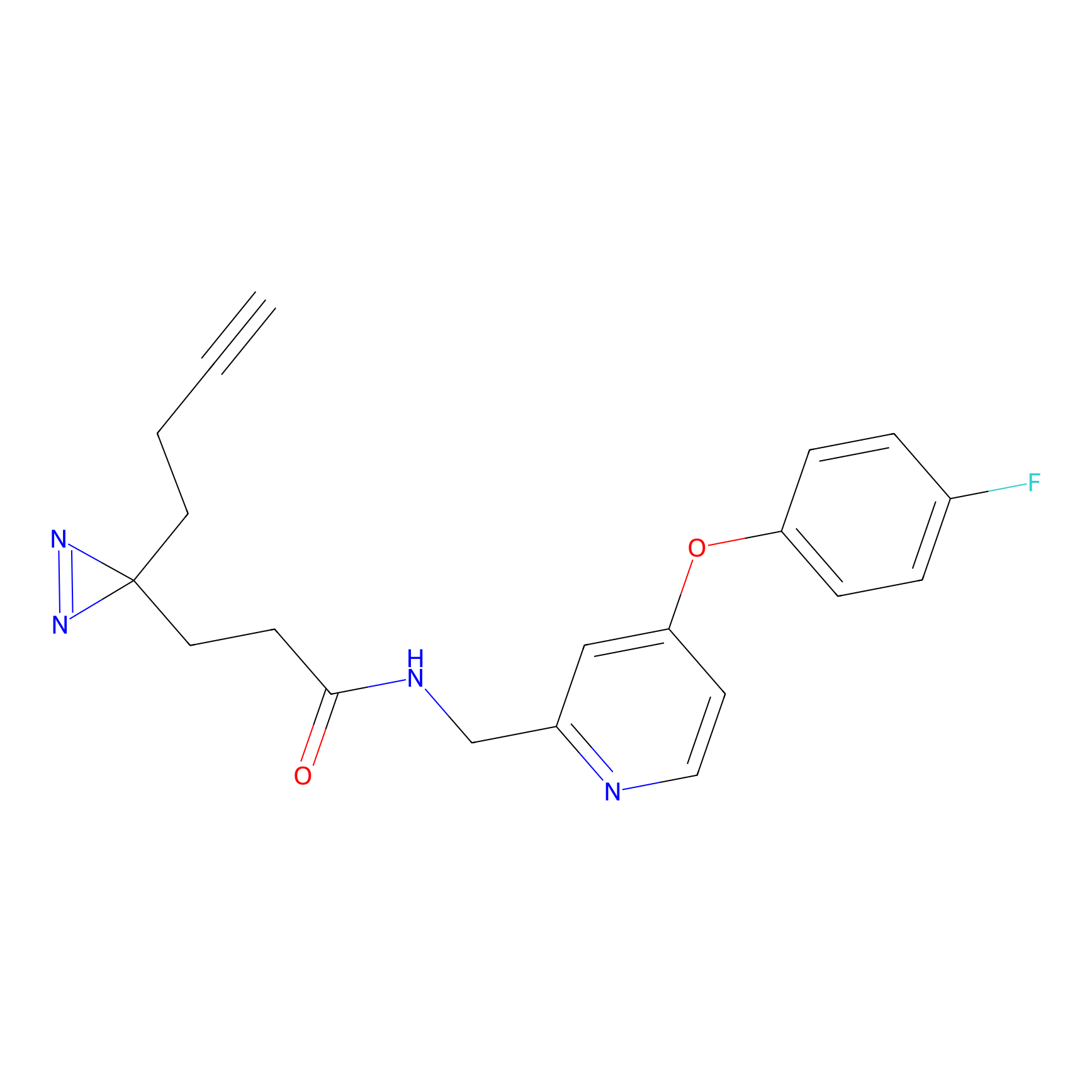
|
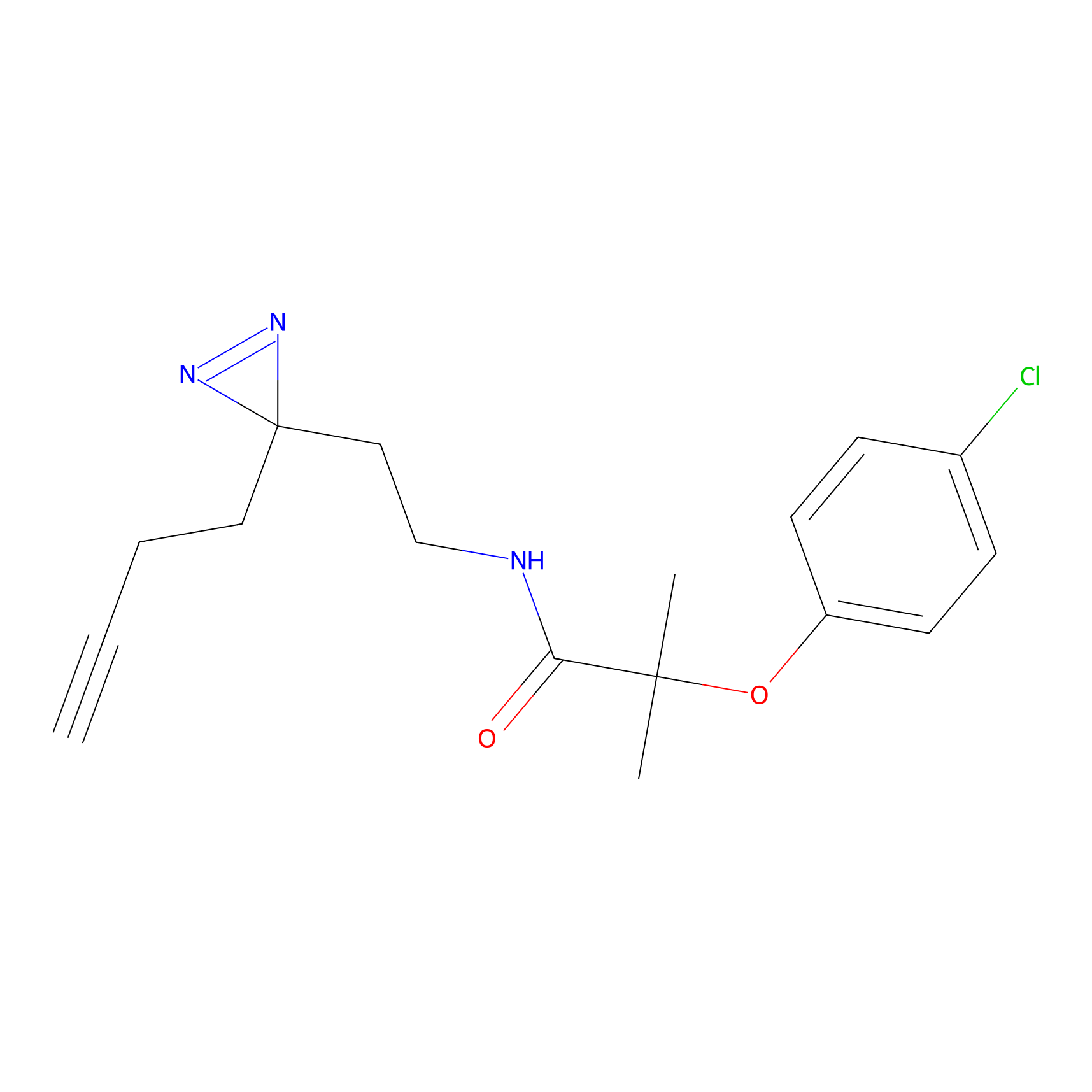
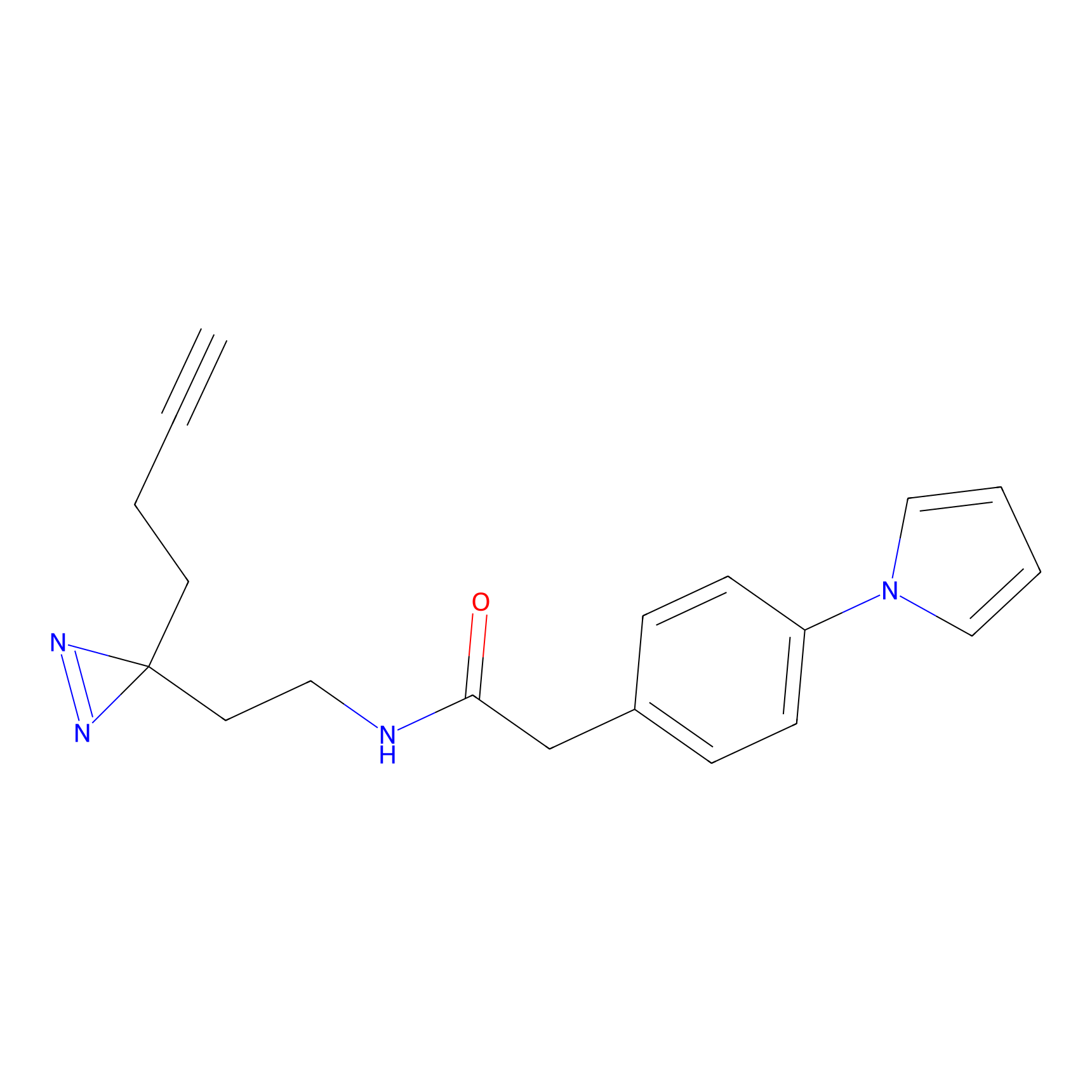
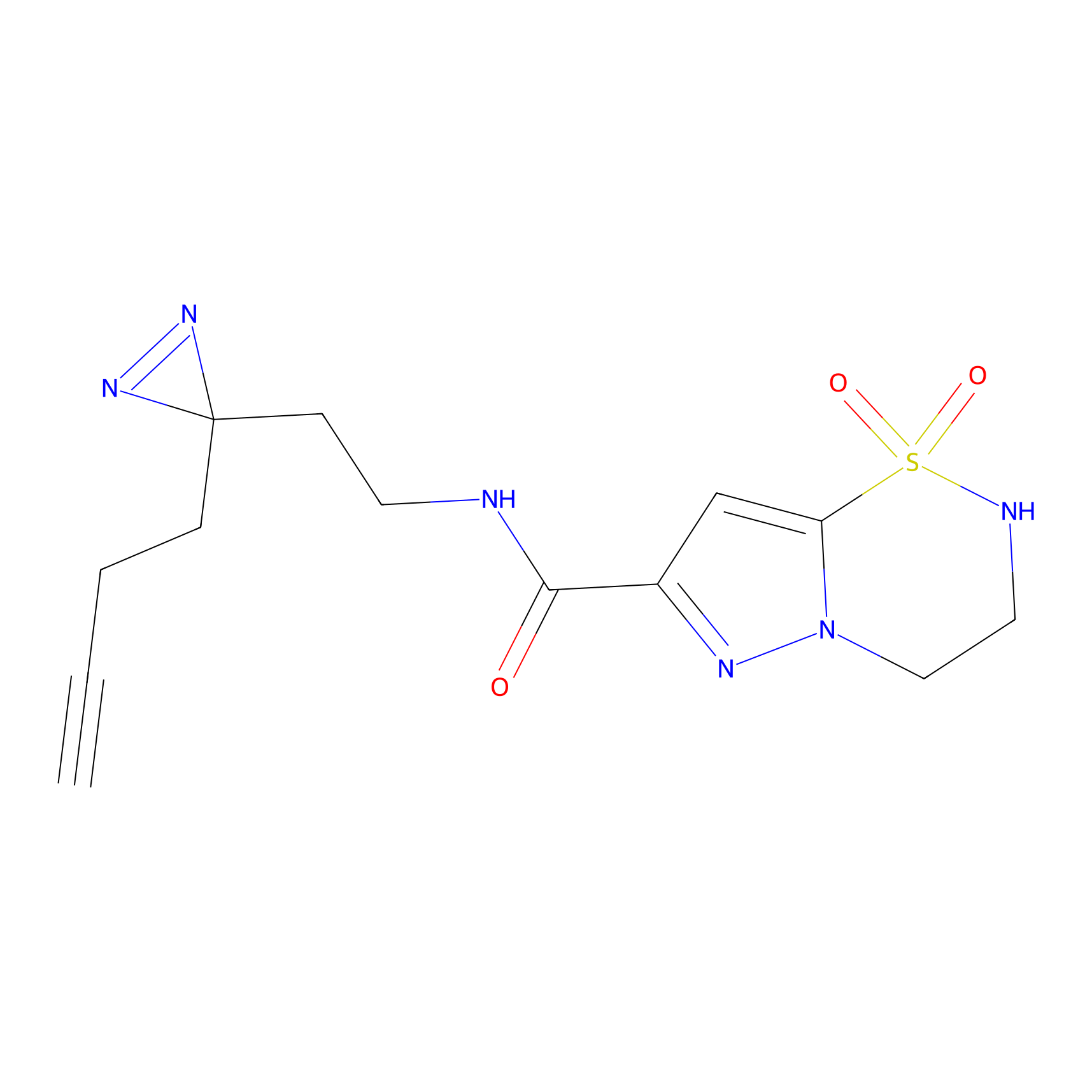
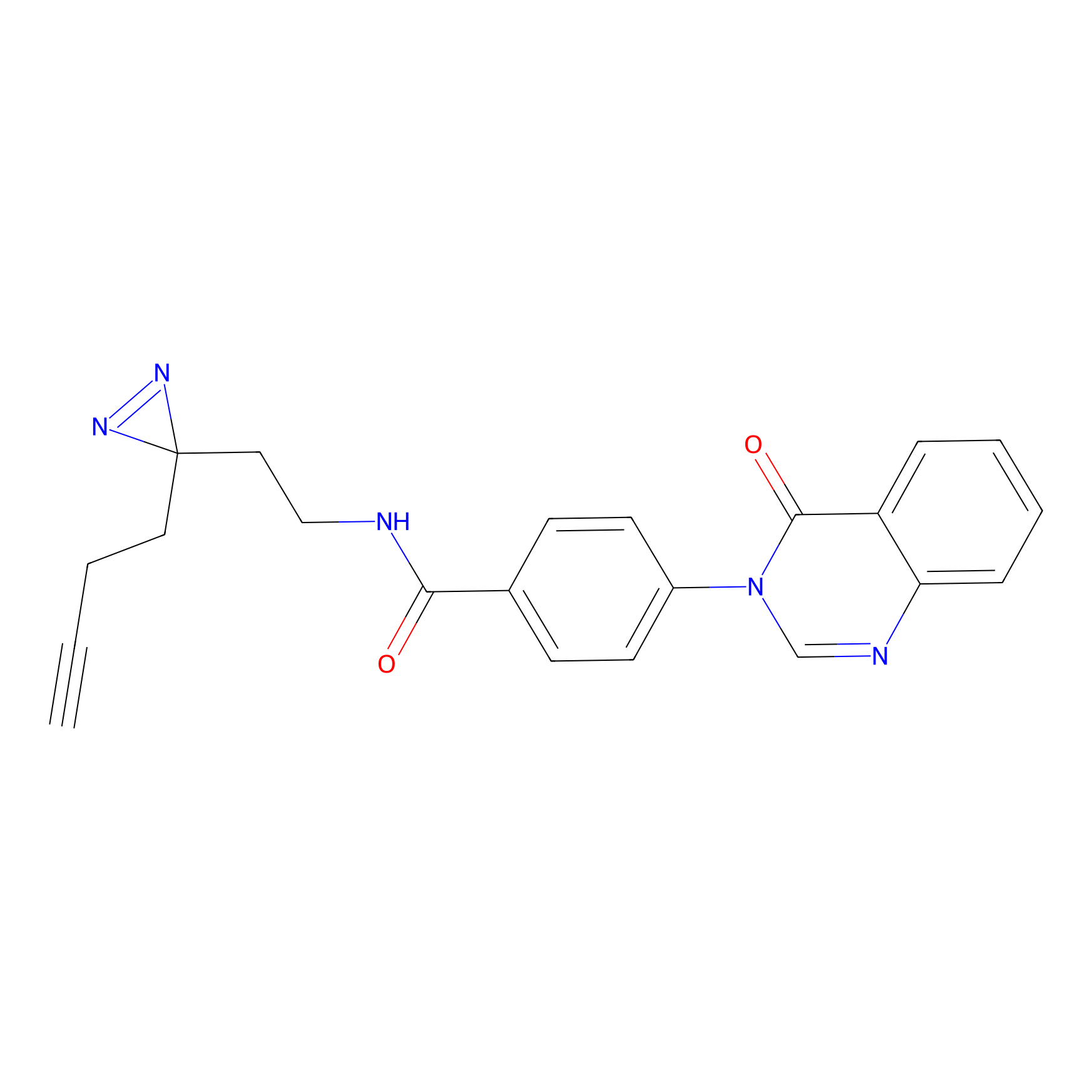
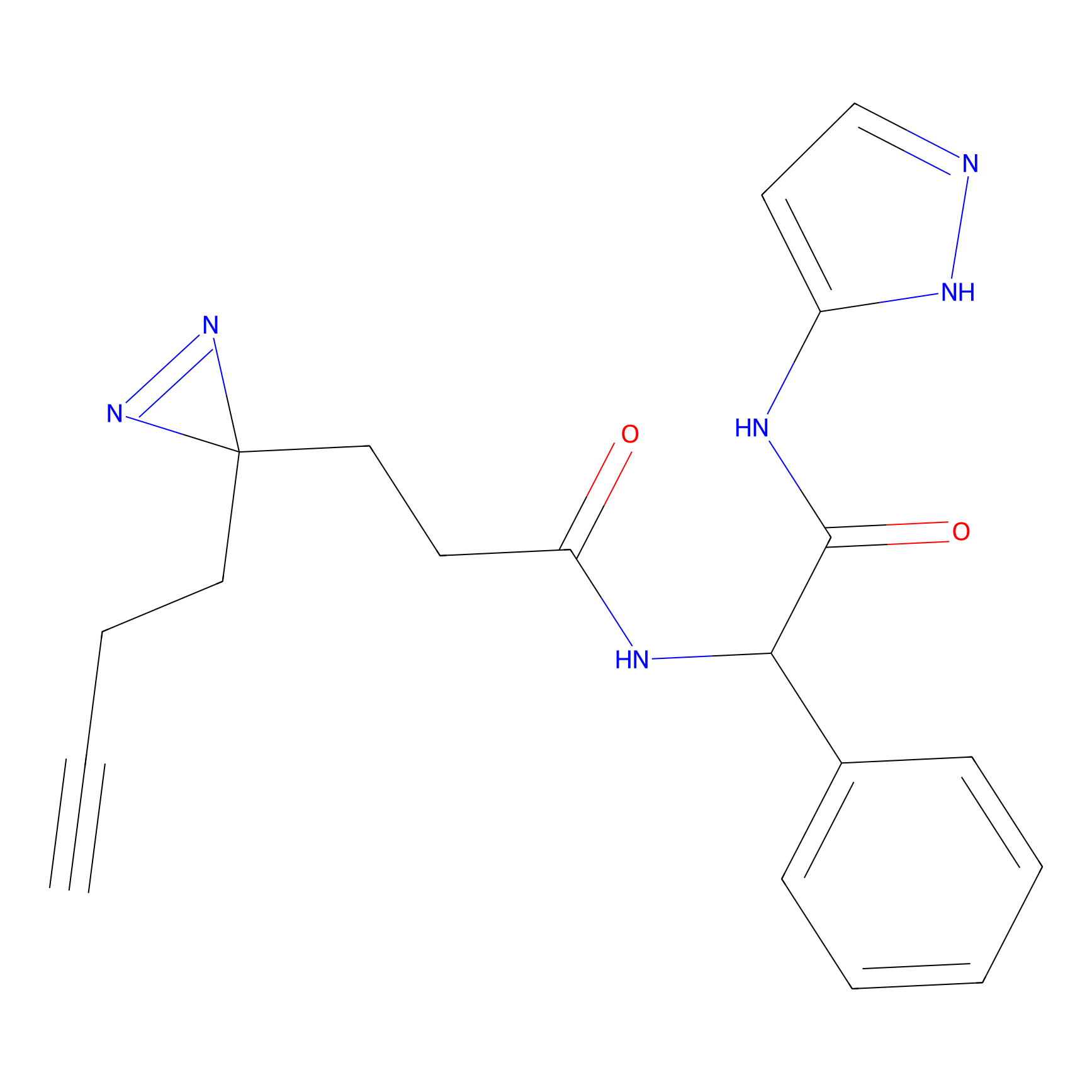
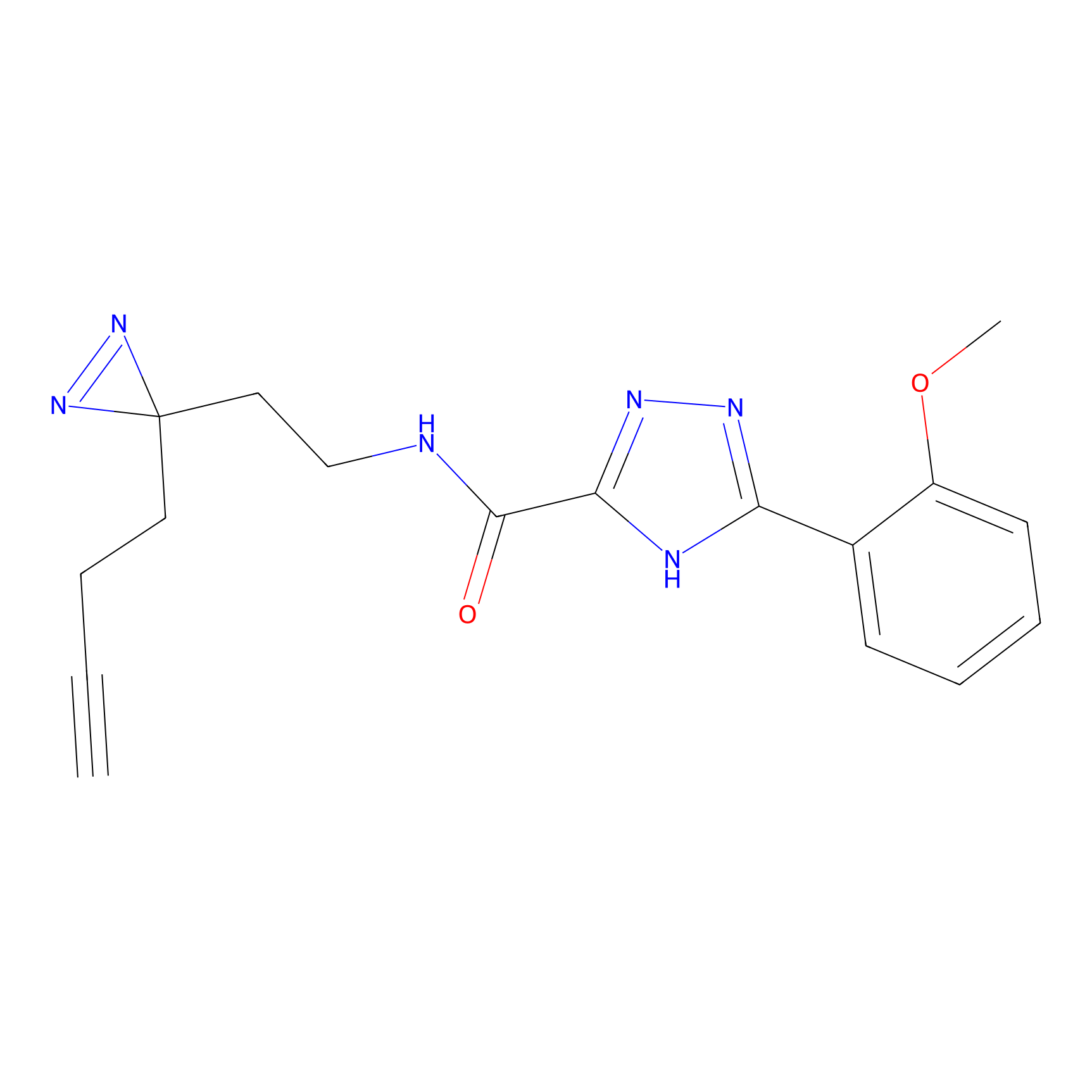
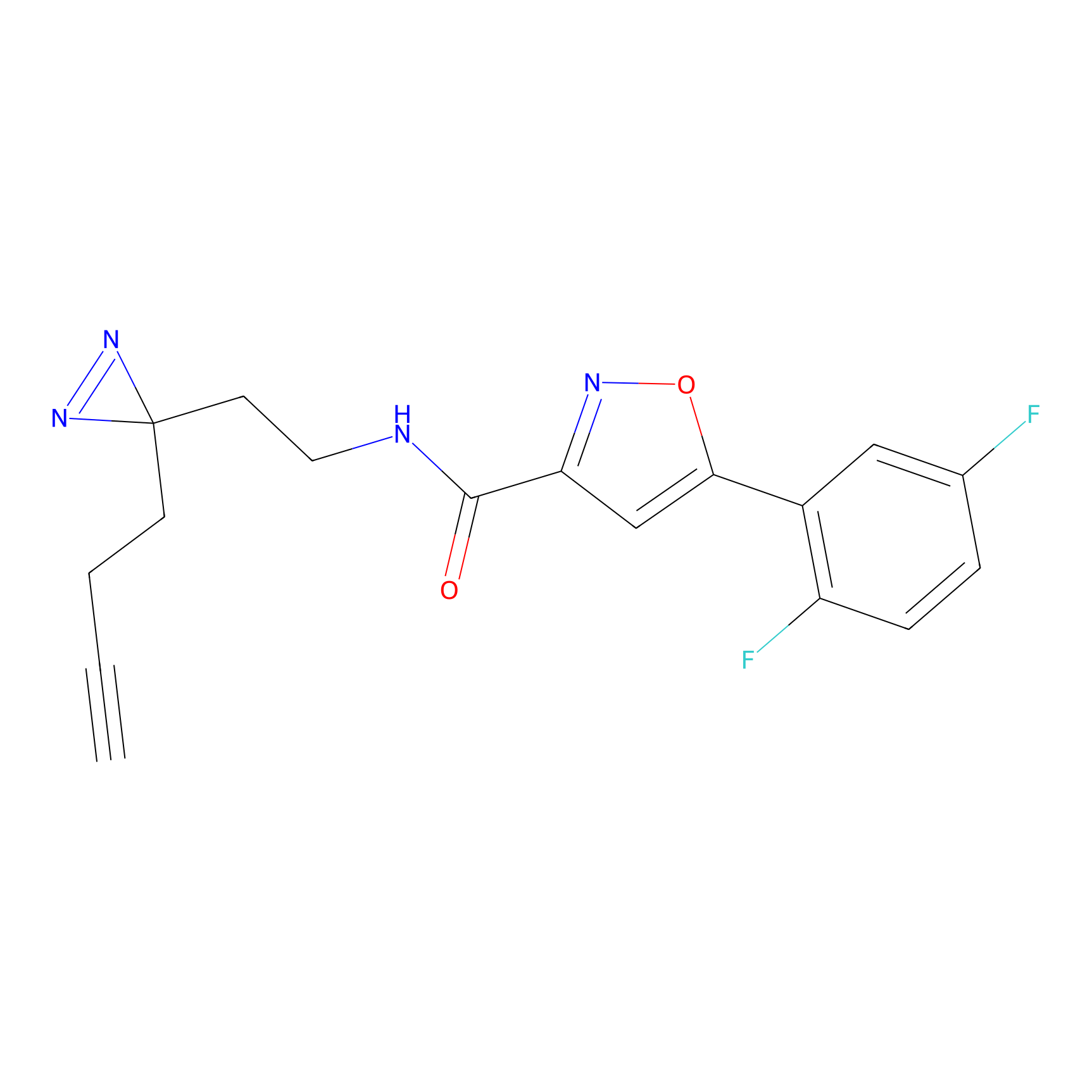
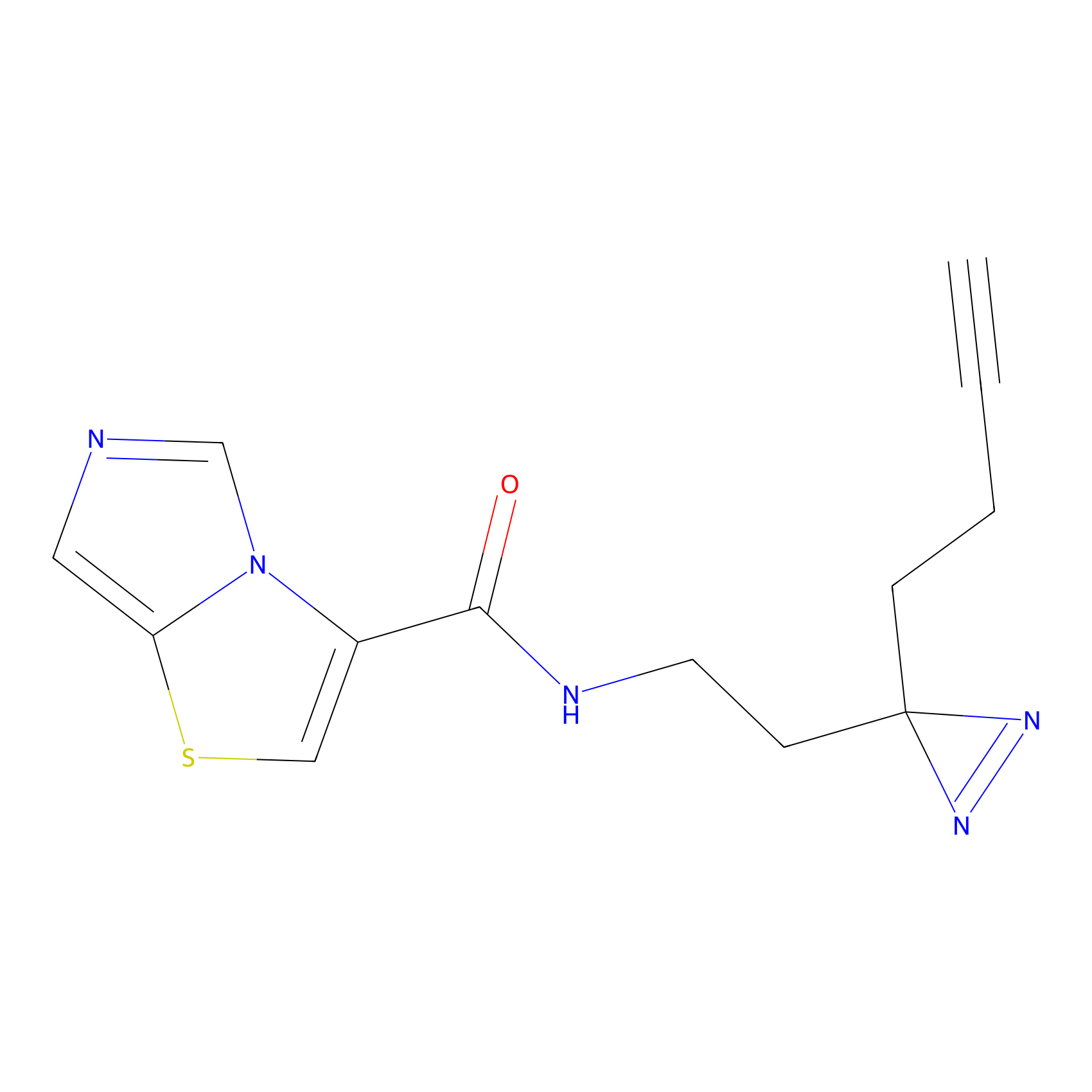
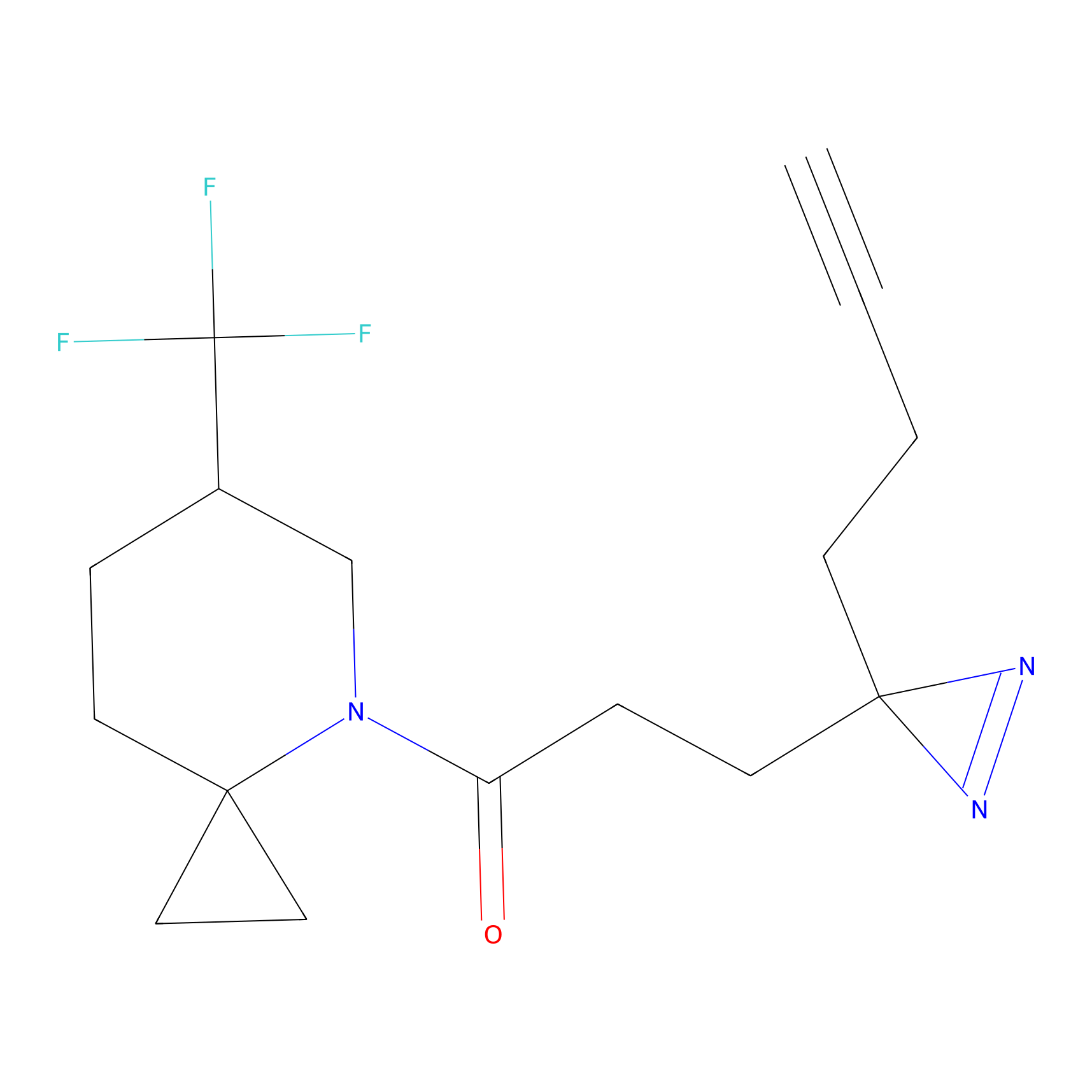
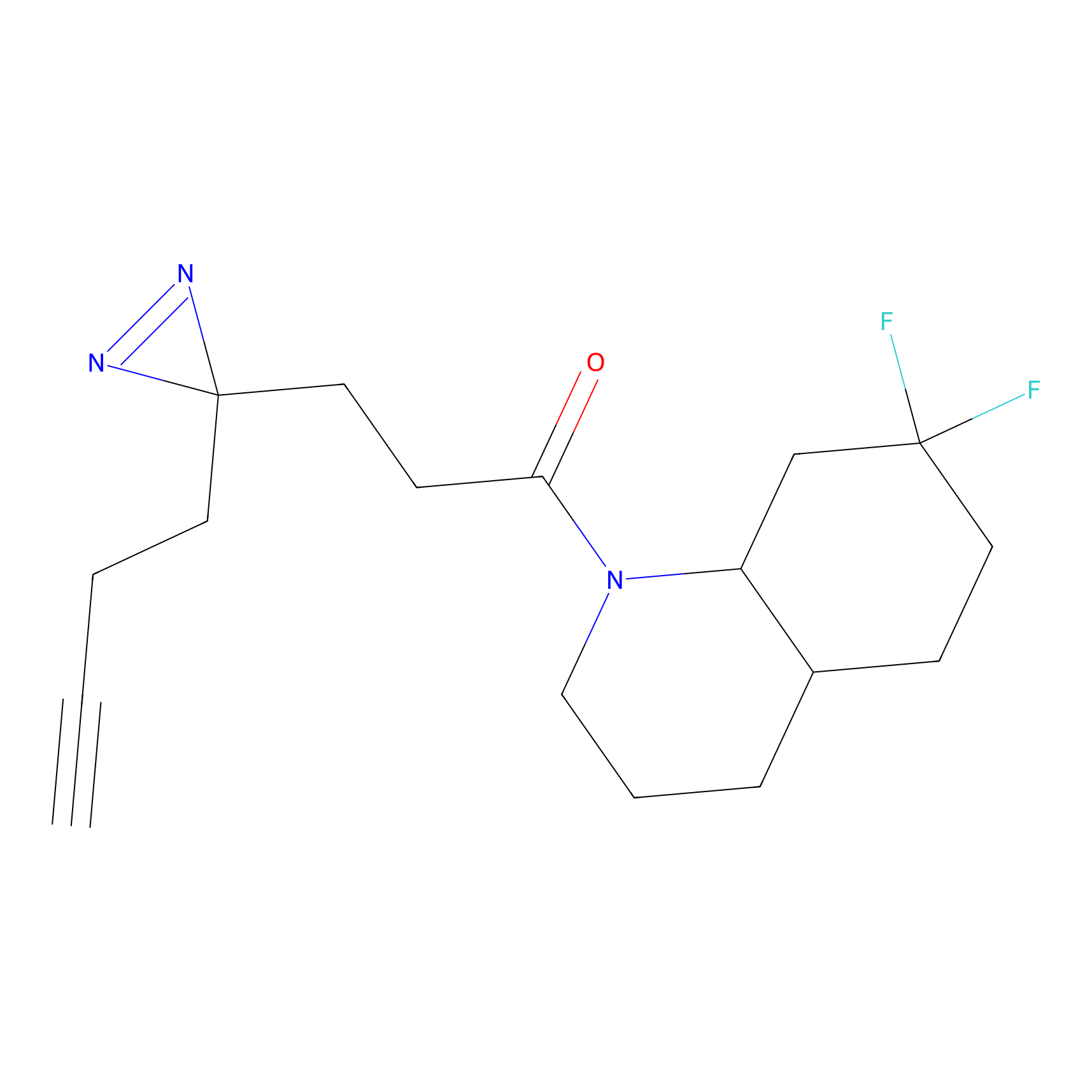
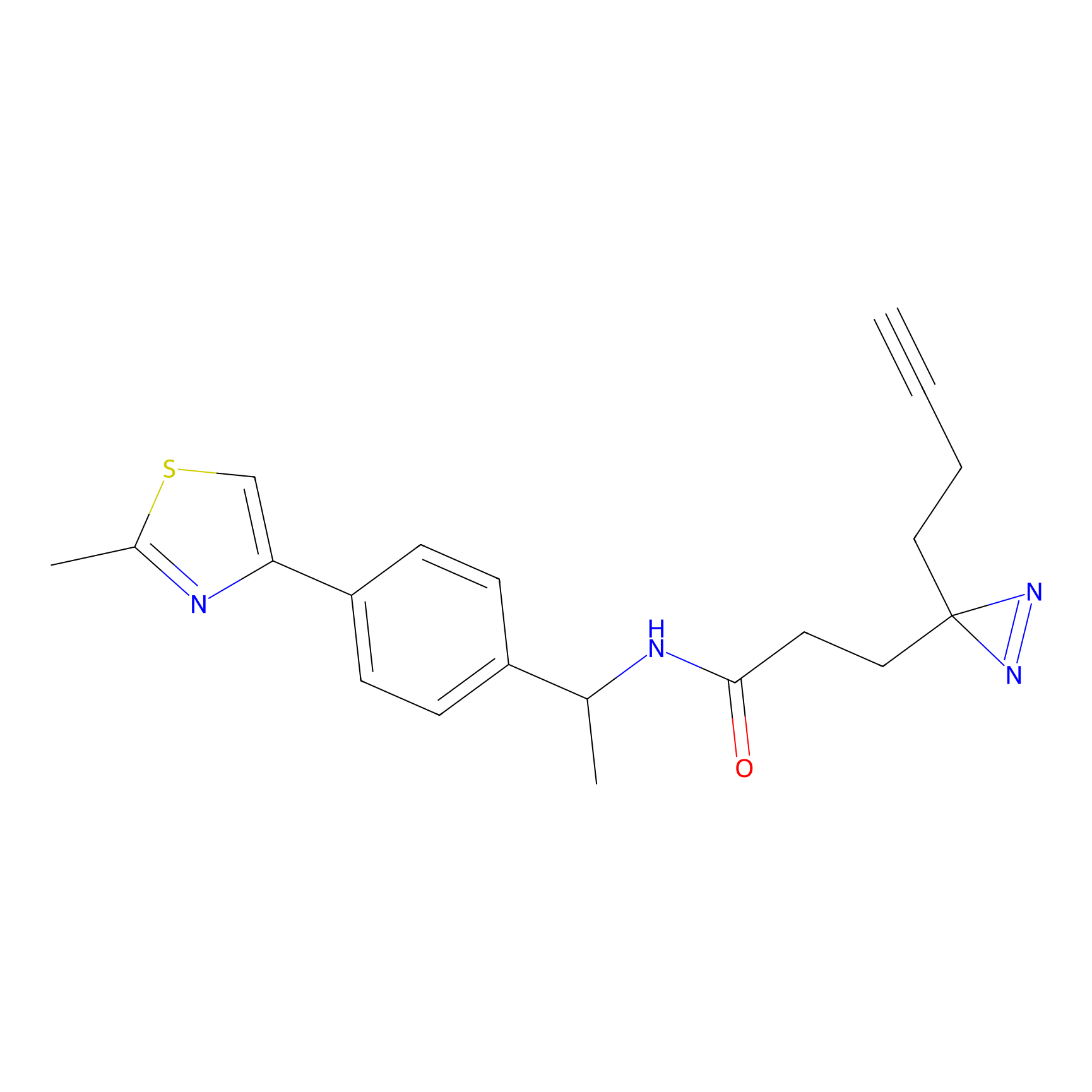
SF Probe Info |
 |
N.A. | LDD0028 | [3] | |
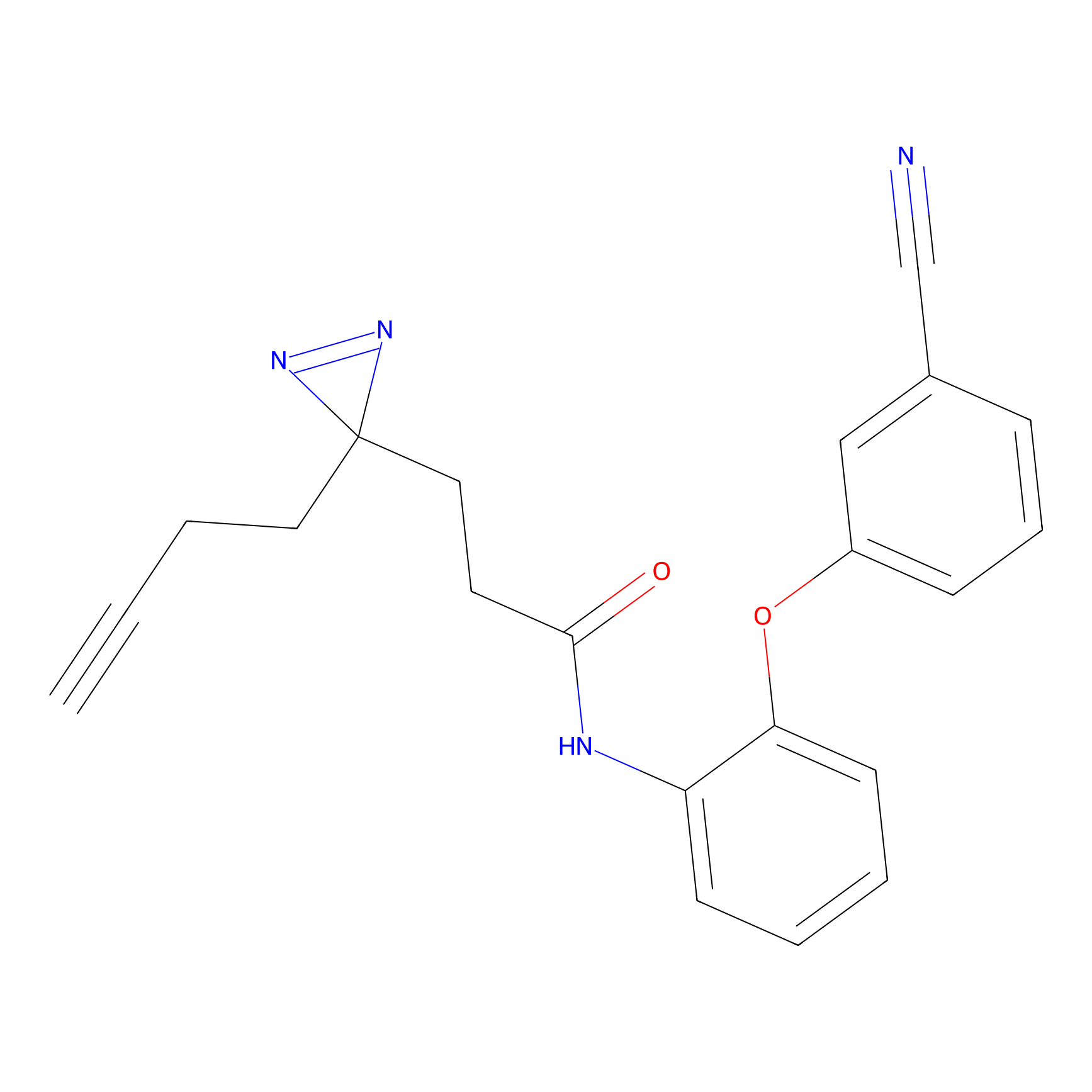
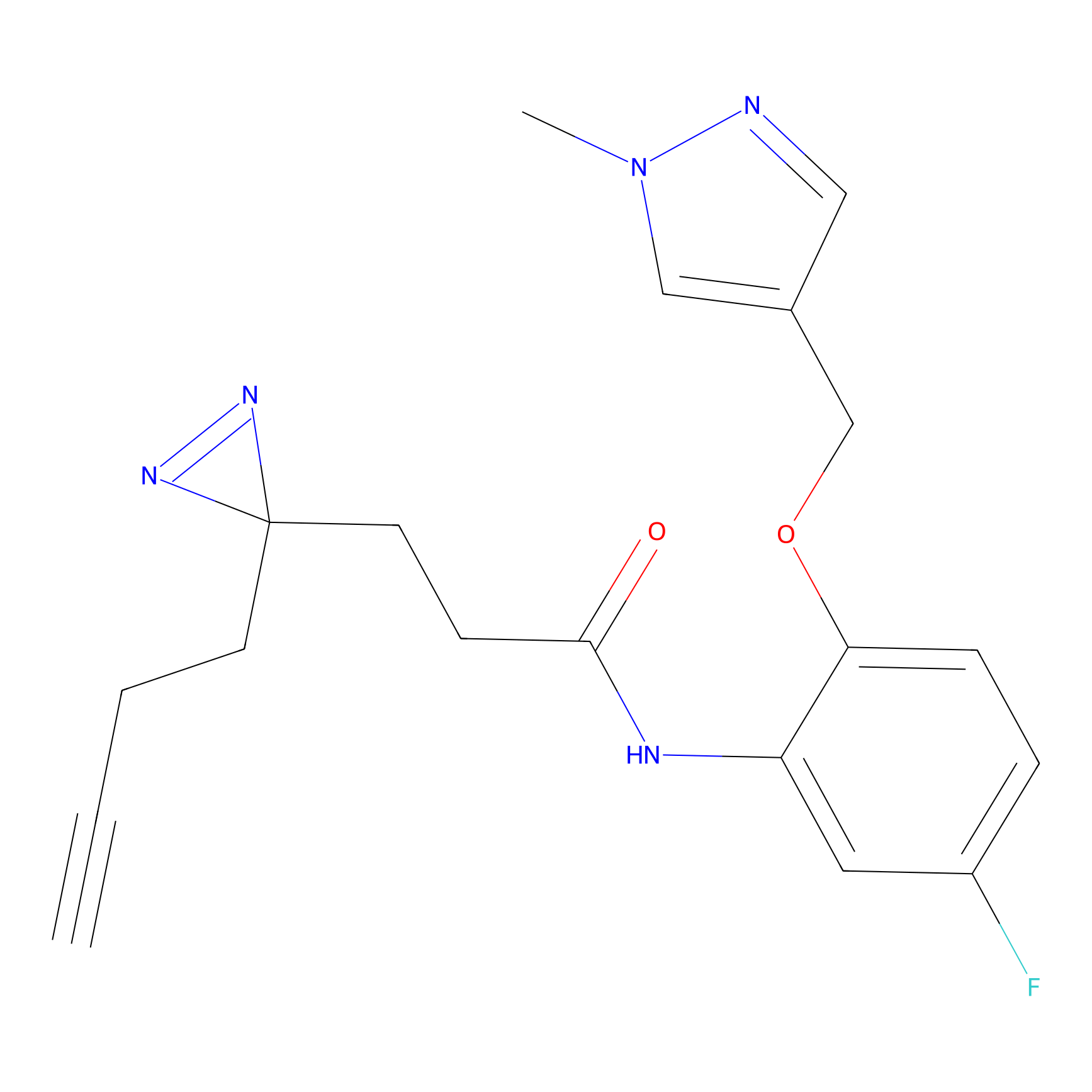
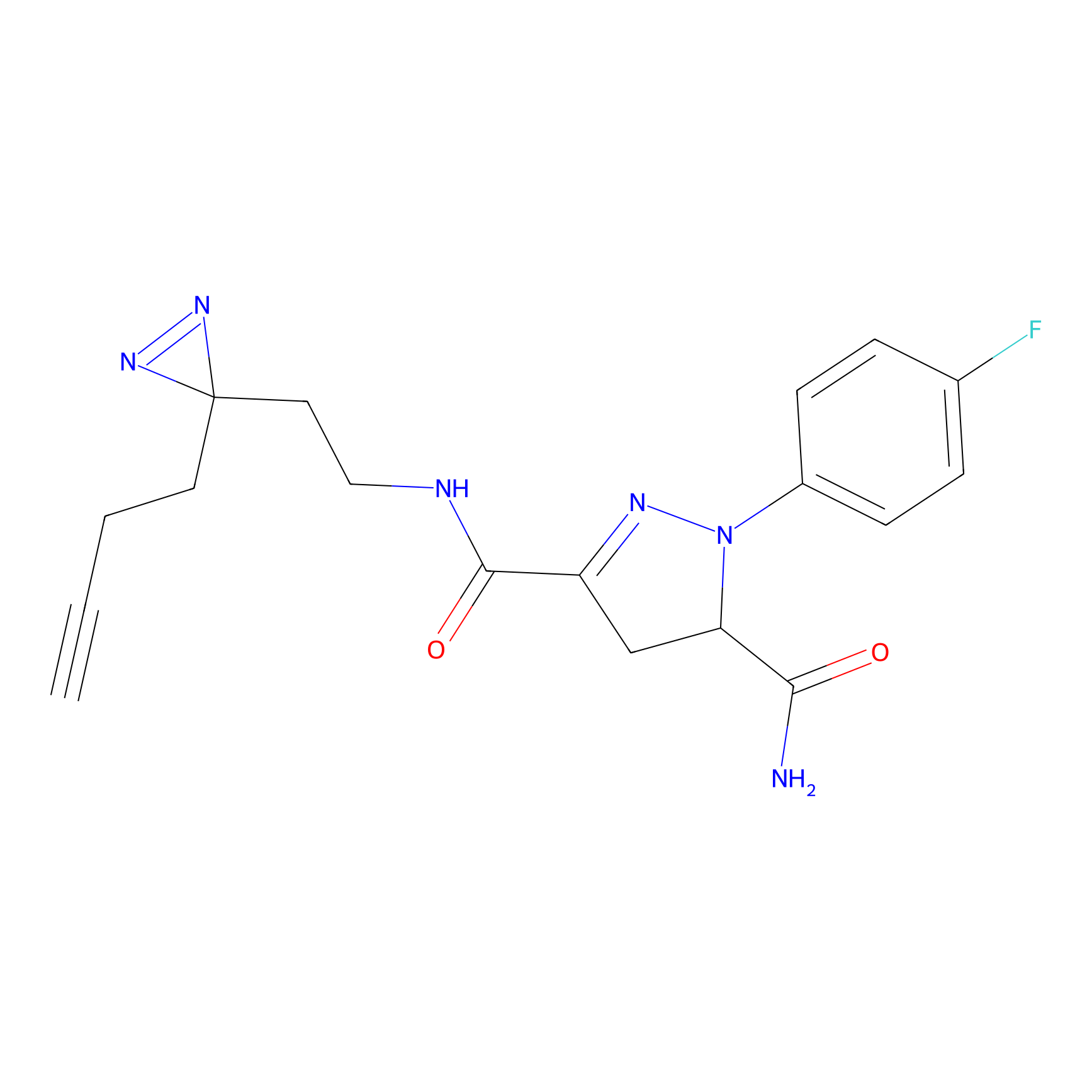
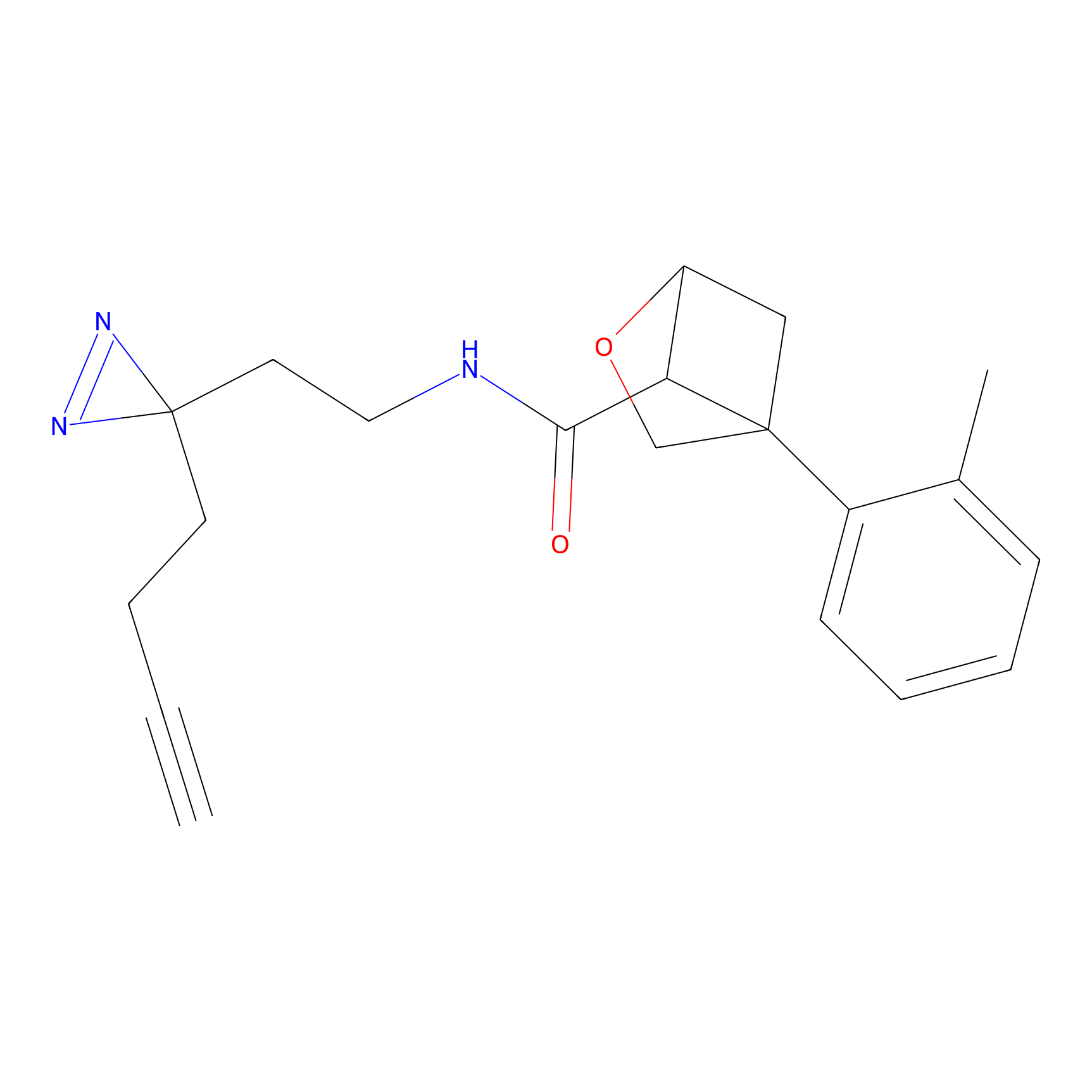
|
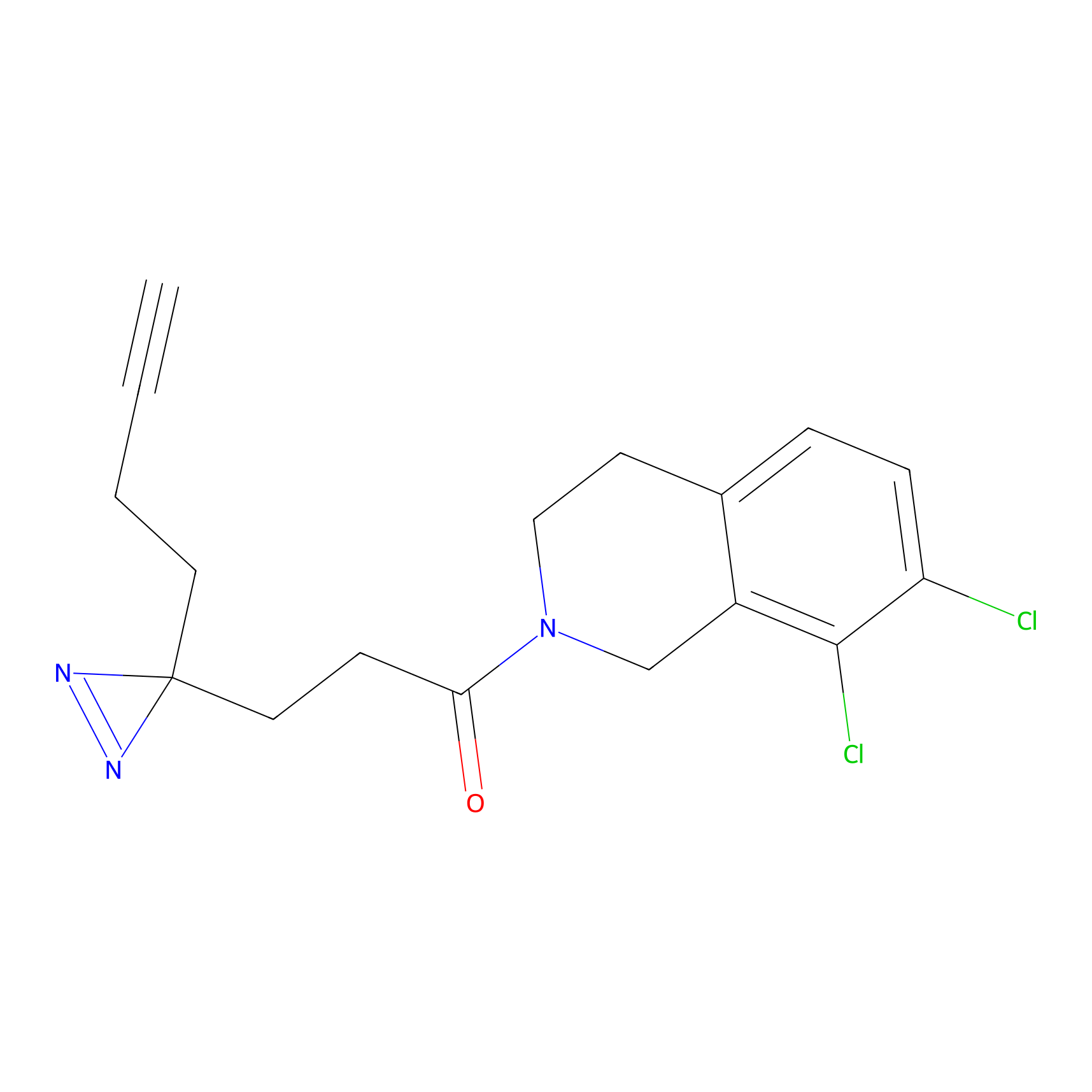
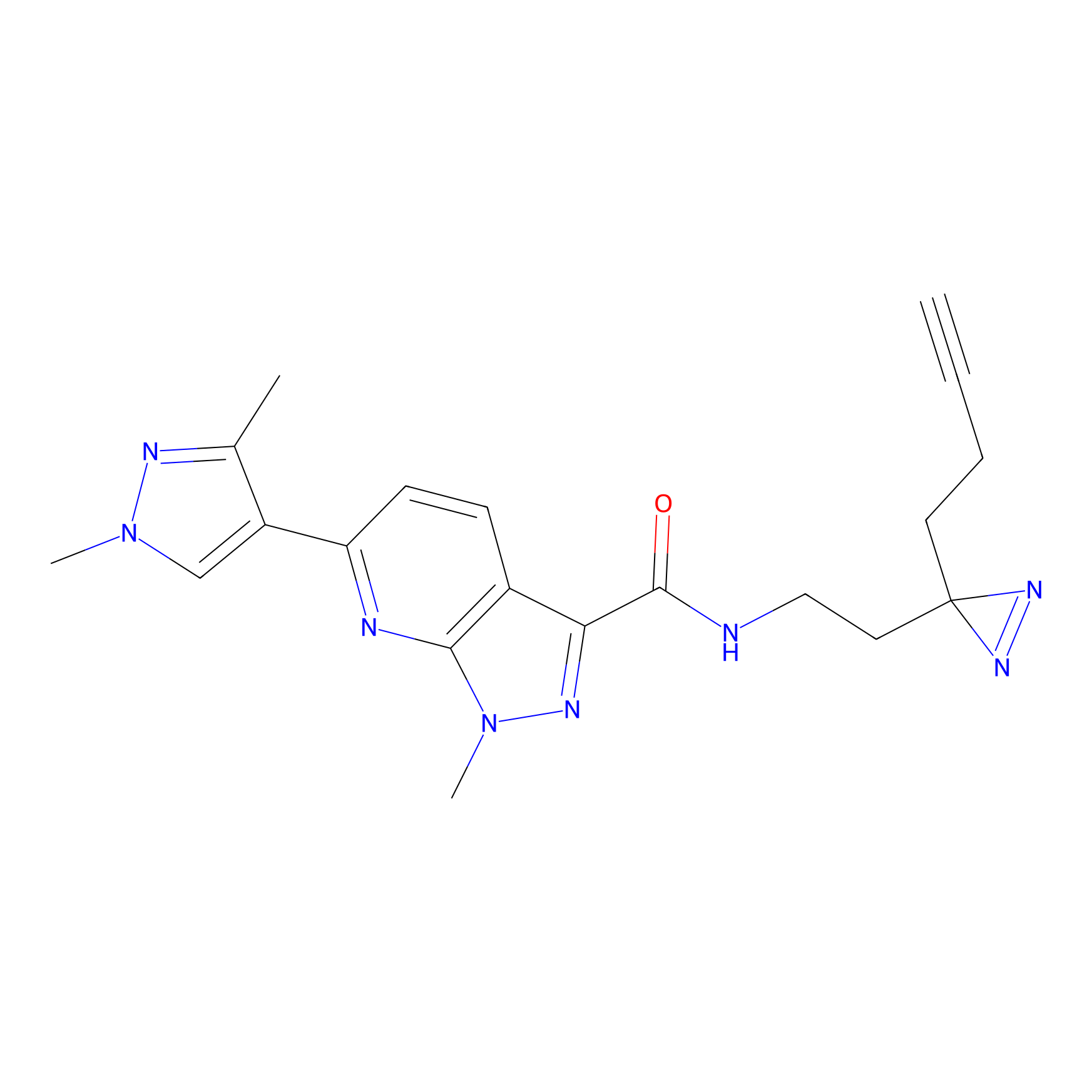
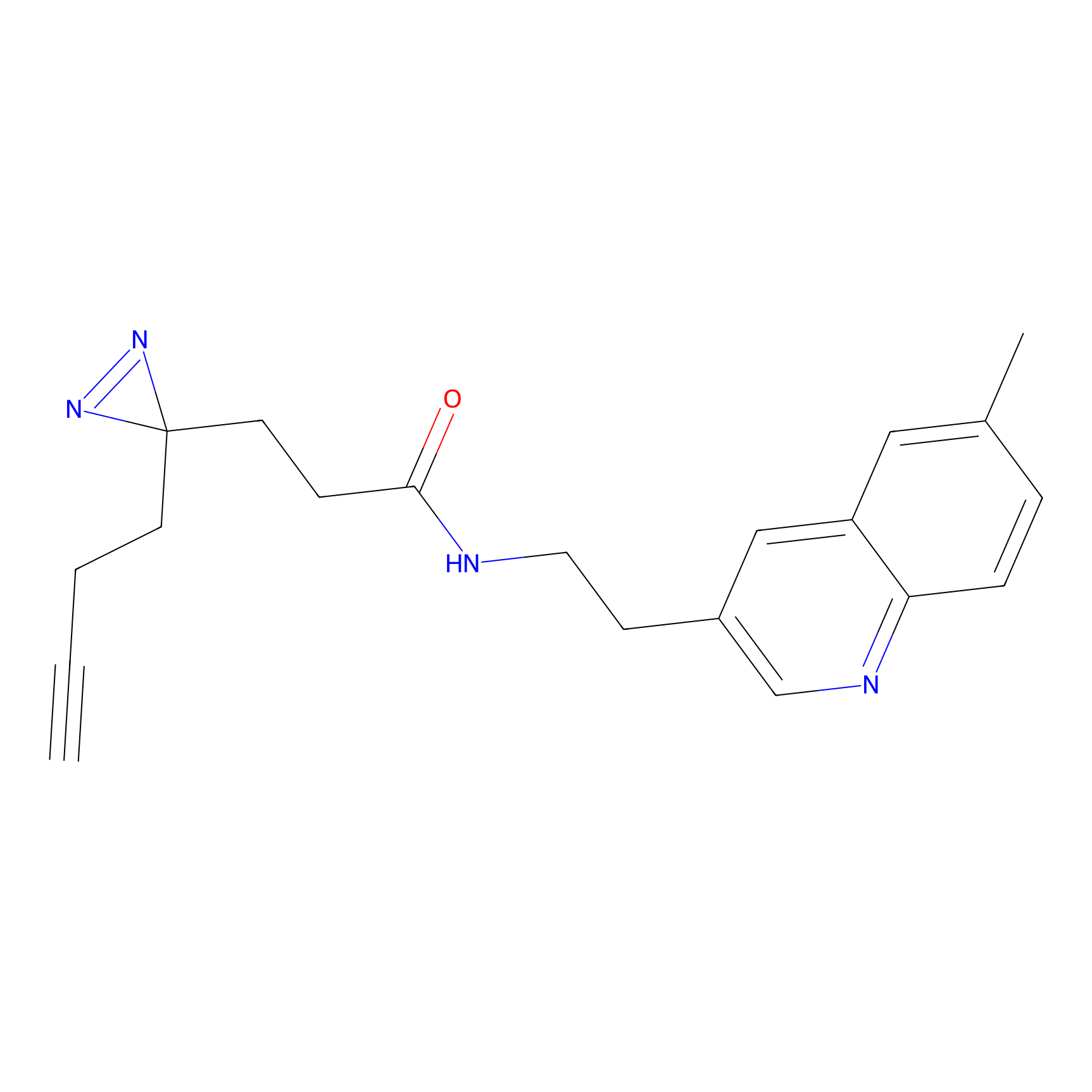
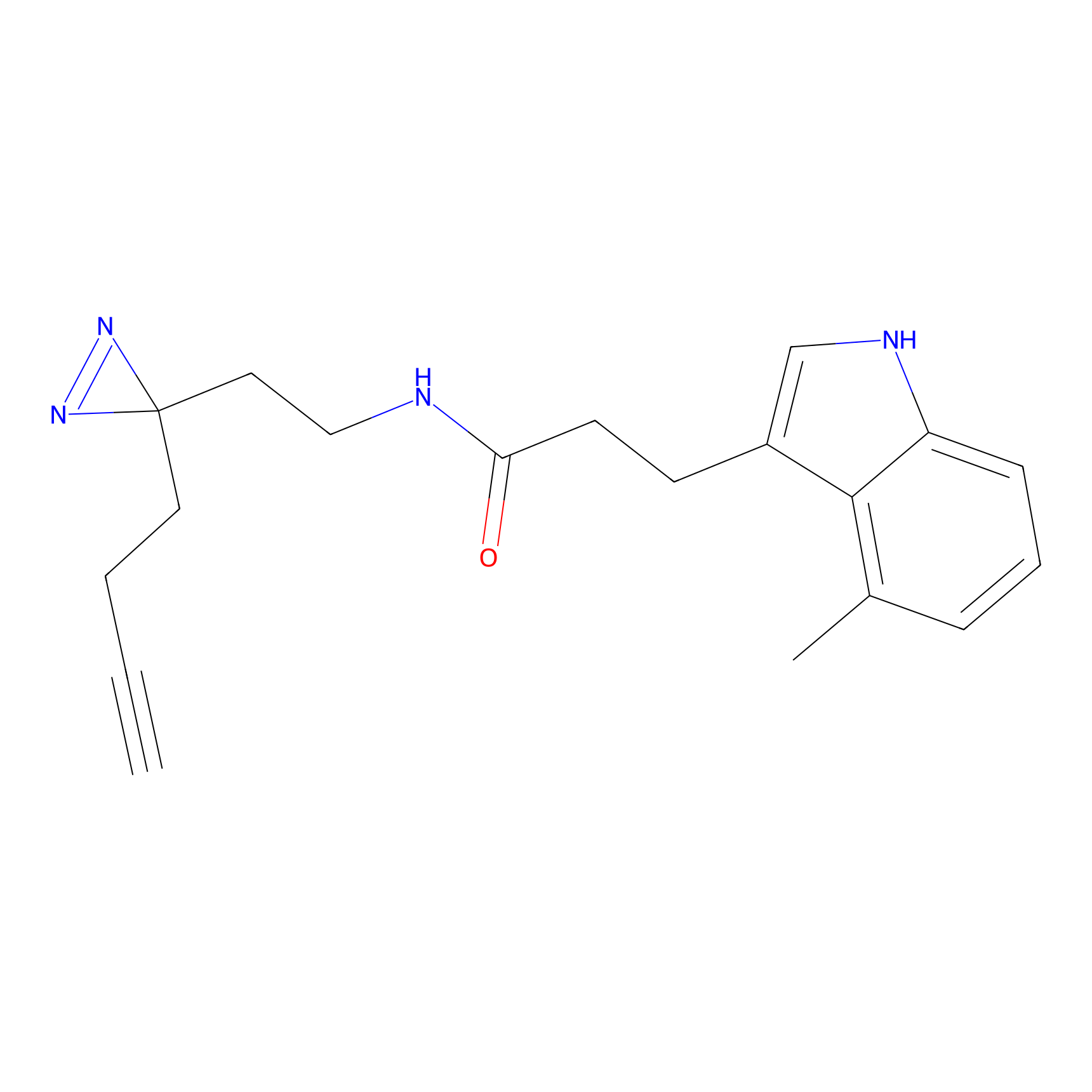
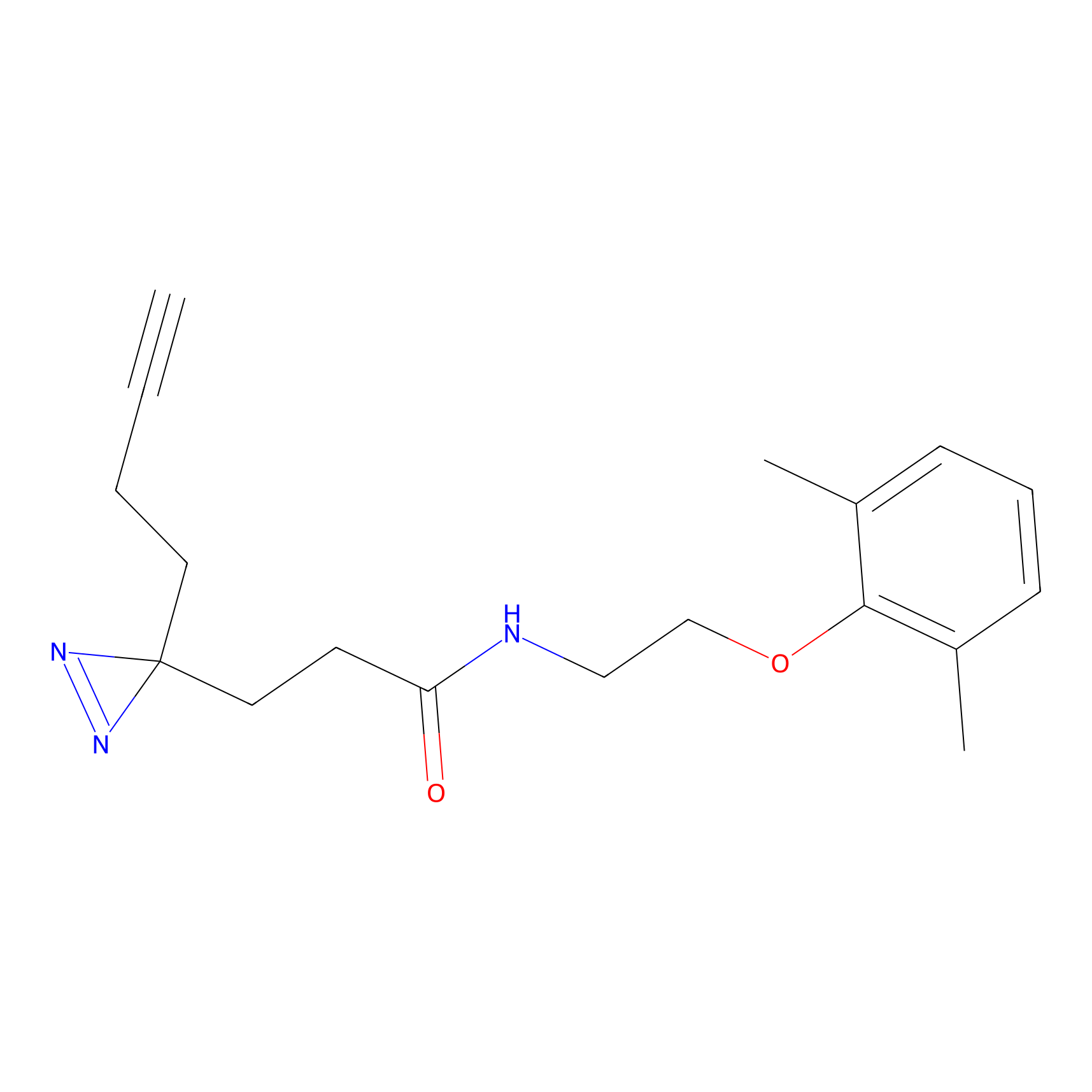
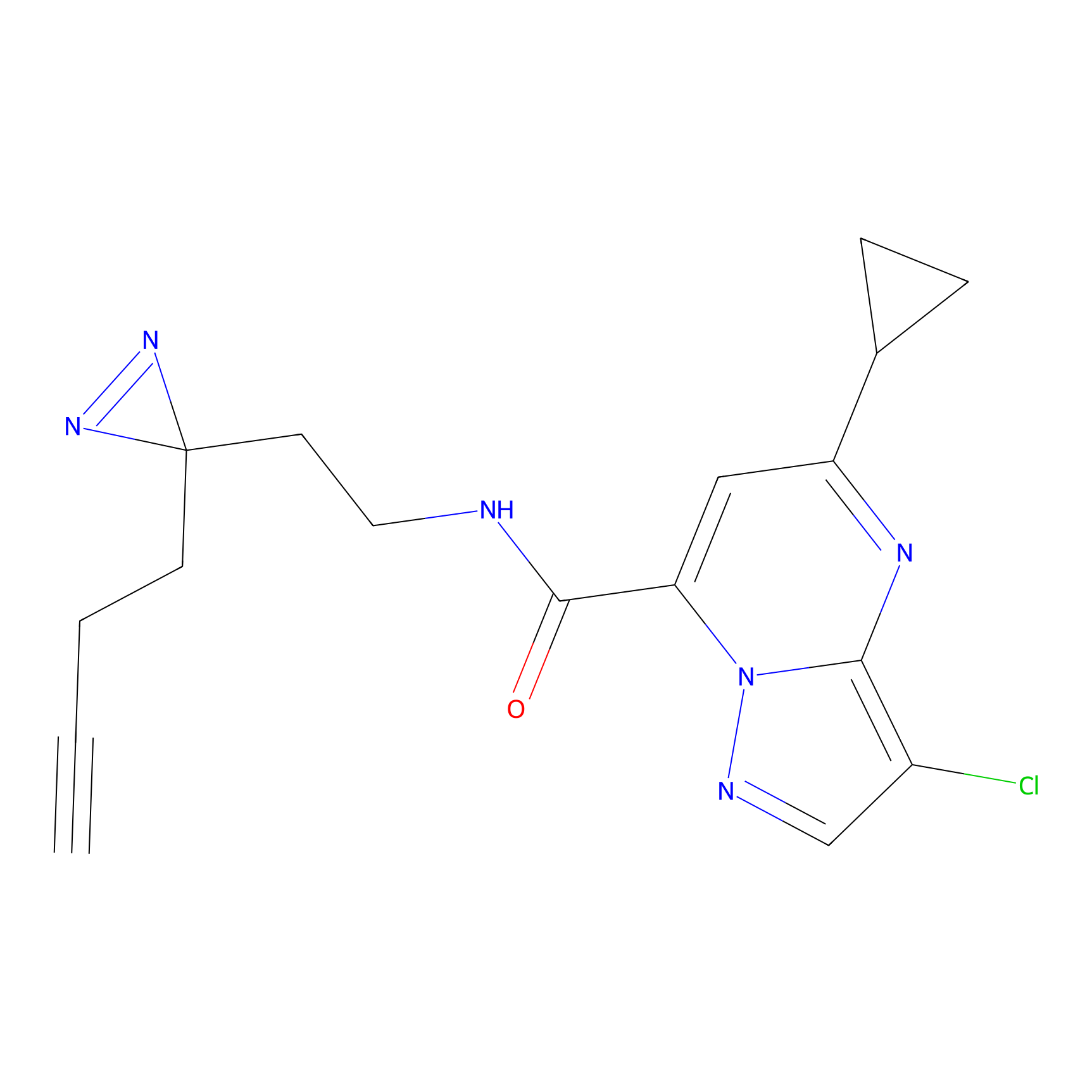
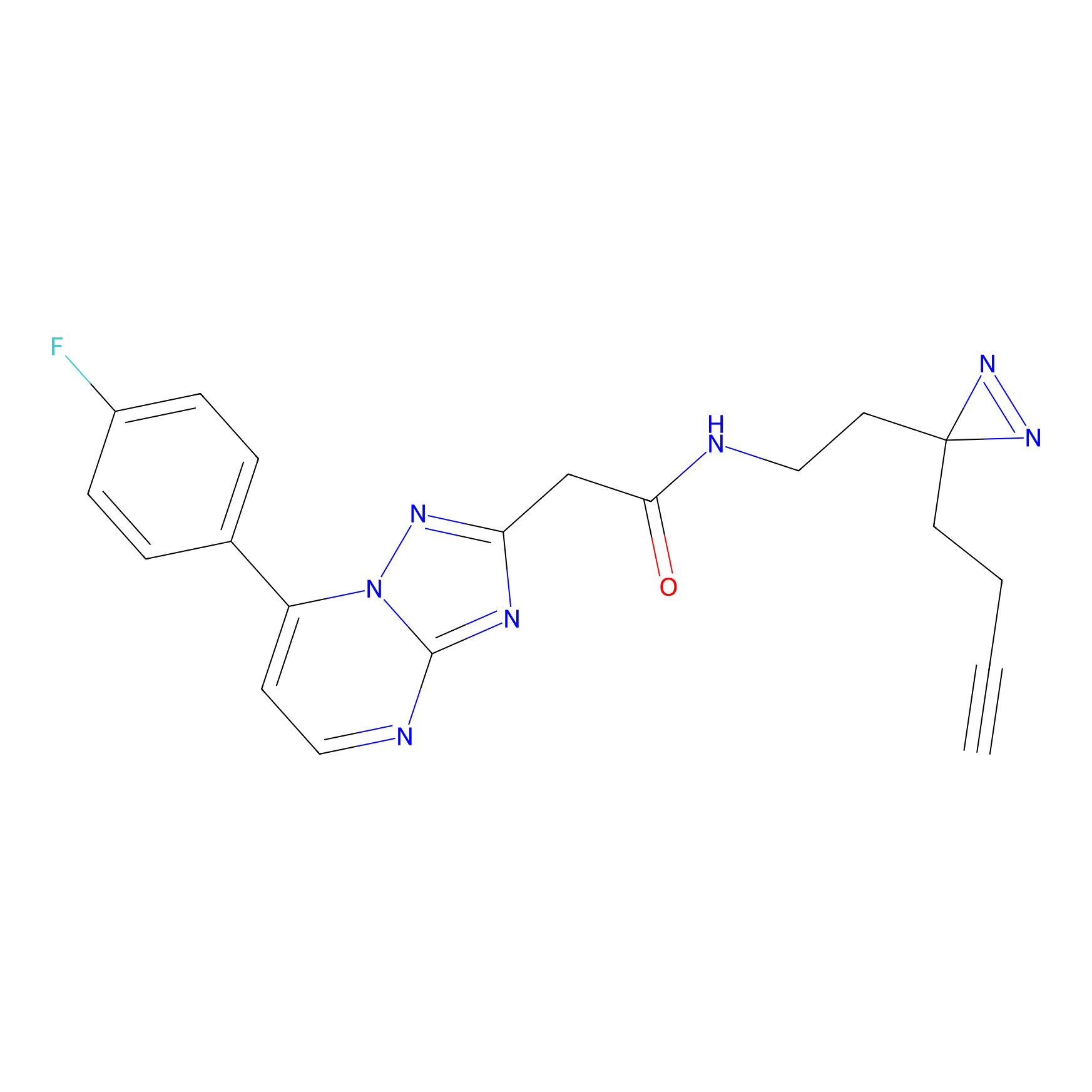
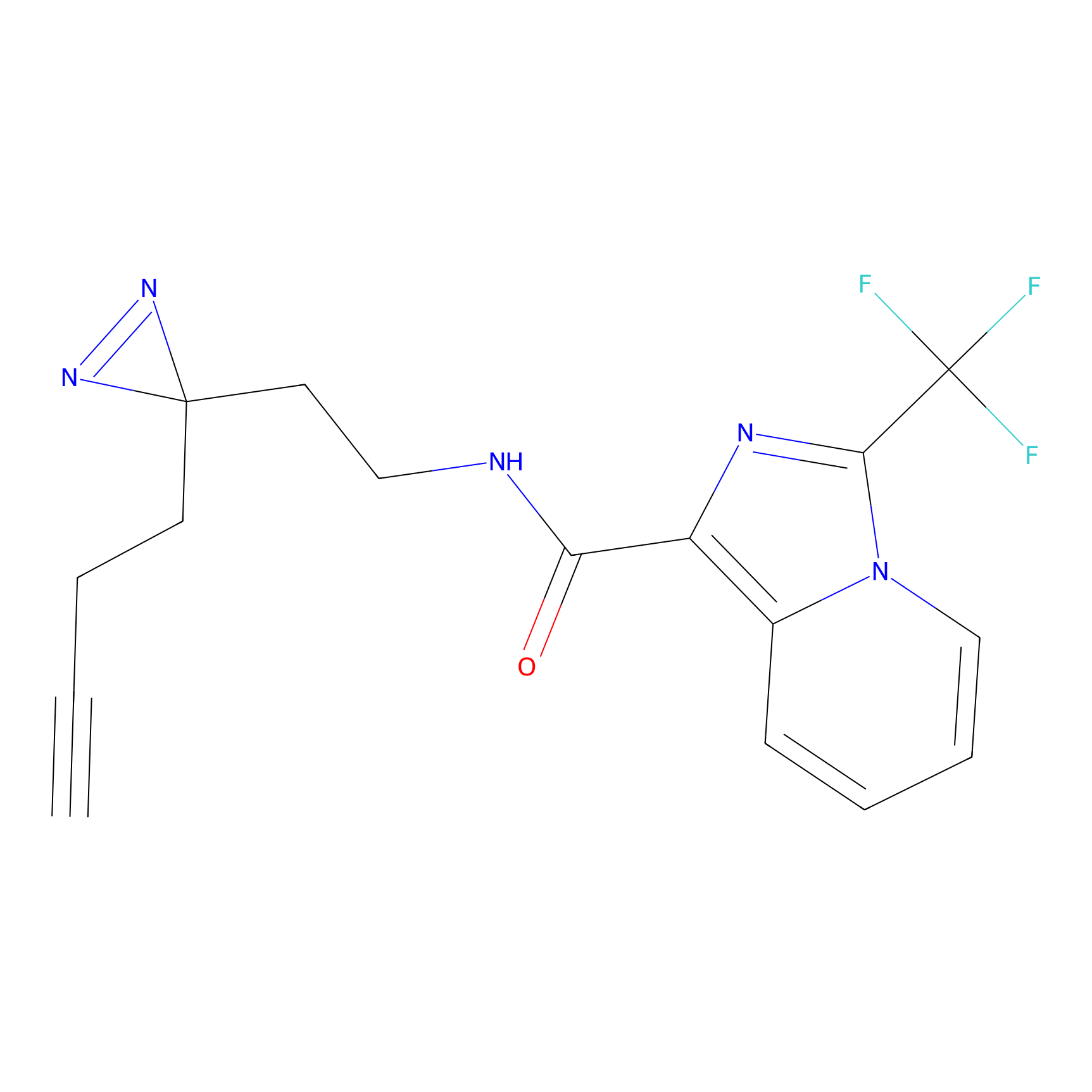
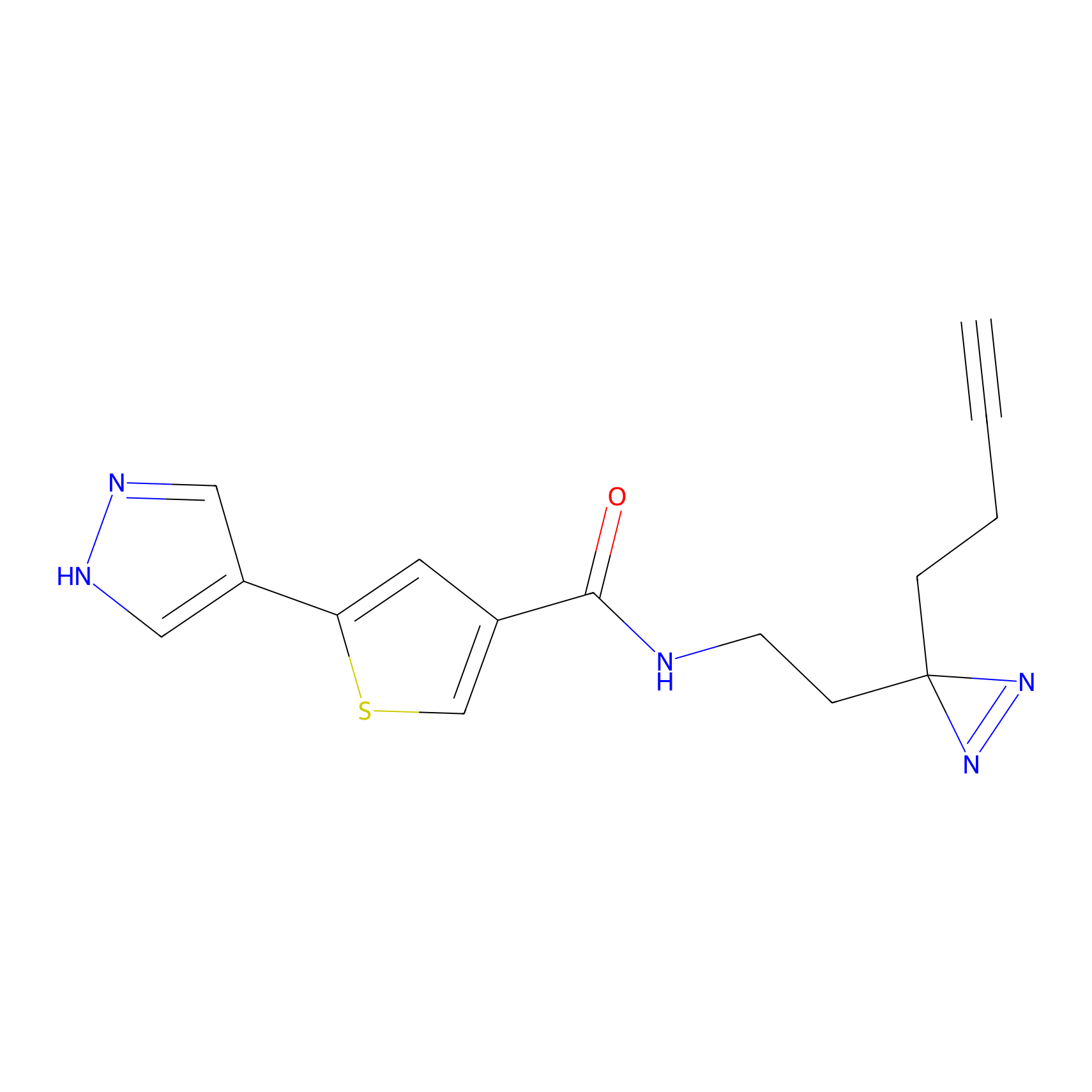
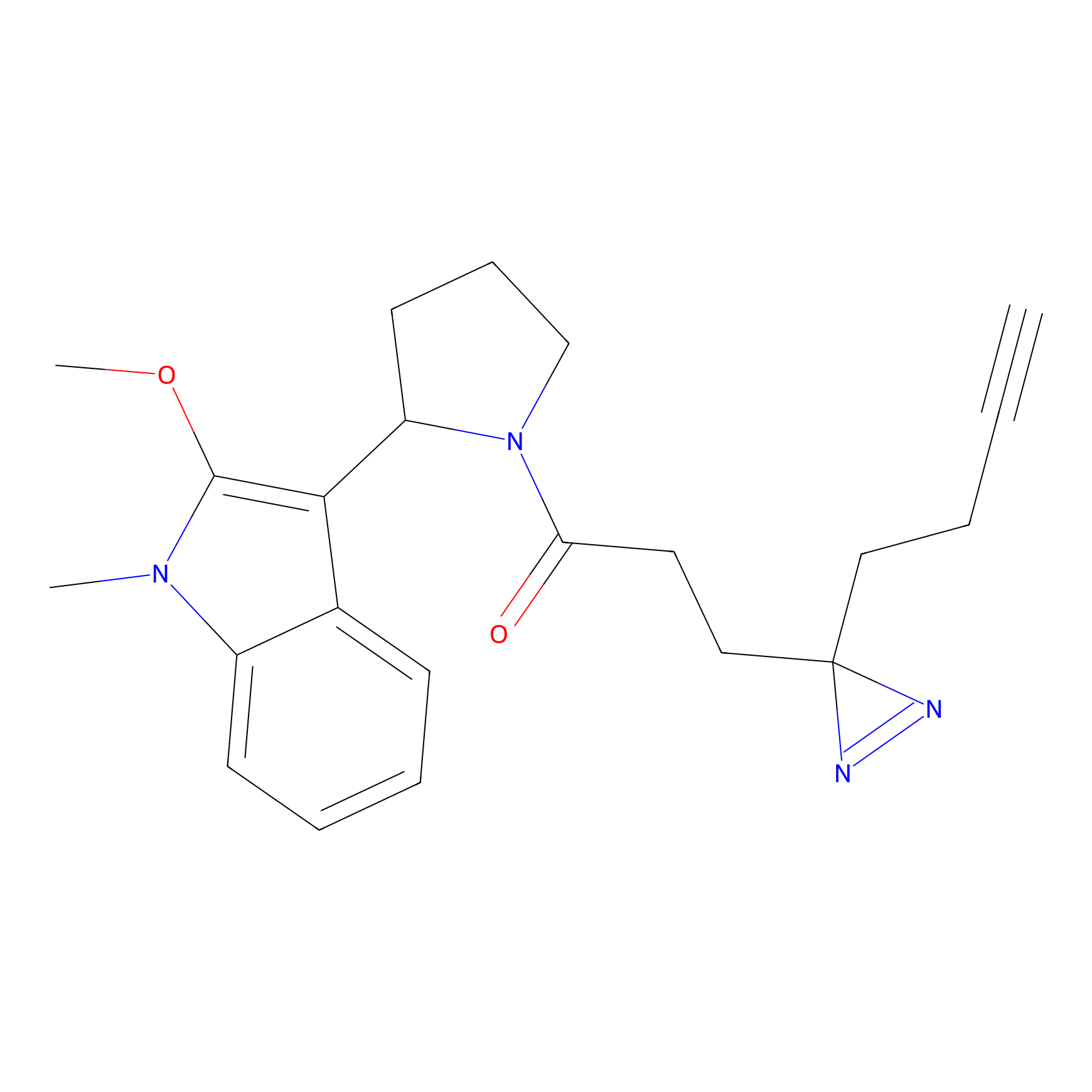
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [4] | |
|
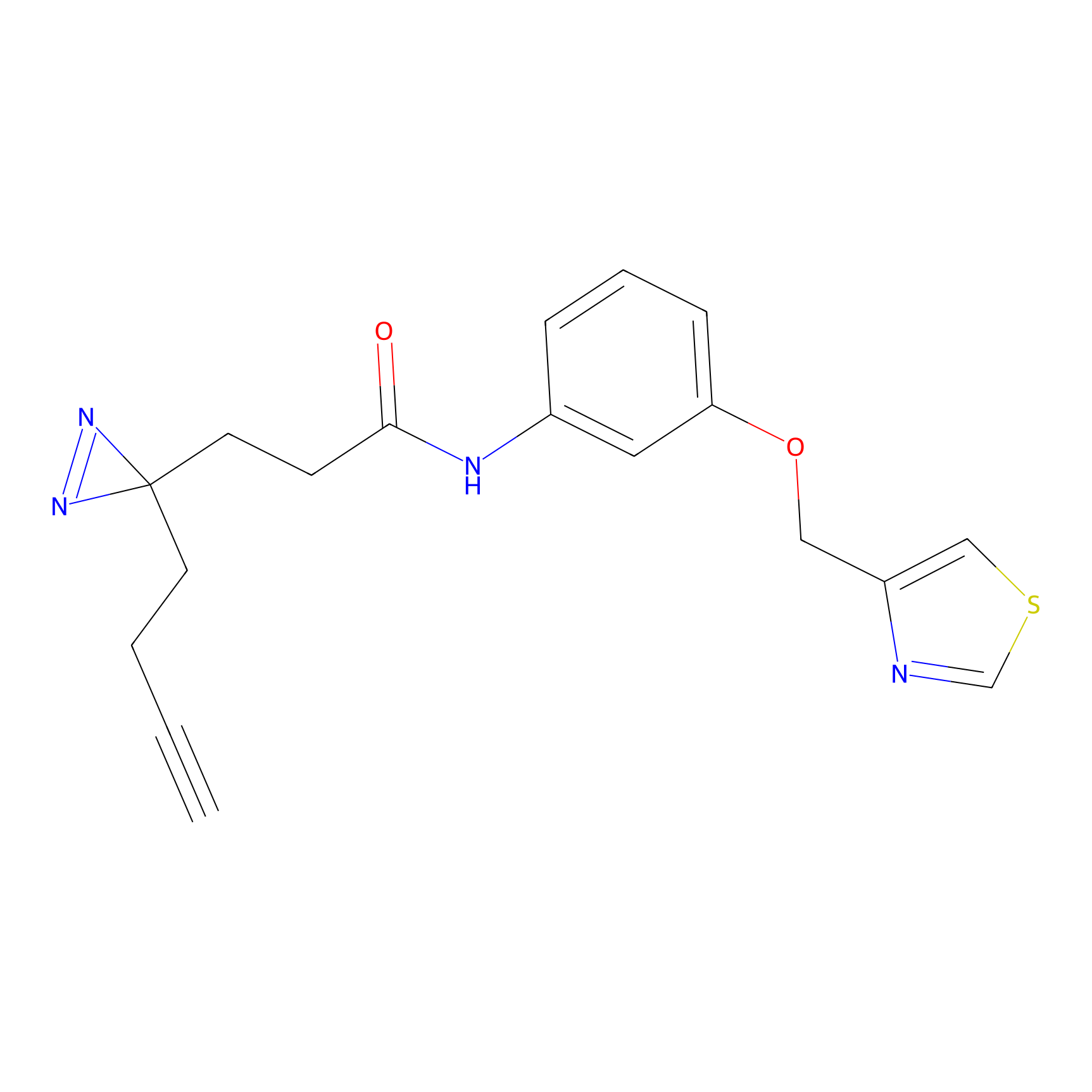
HHS-465 Probe Info |
 |
K113(0.00); Y52(0.00); Y111(0.00) | LDD2240 | [5] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
References