Details of the Target
General Information of Target
| Target ID | LDTP08878 | |||||
|---|---|---|---|---|---|---|
| Target Name | ER membrane protein complex subunit 5 (MMGT1) | |||||
| Gene Name | MMGT1 | |||||
| Gene ID | 93380 | |||||
| Synonyms |
EMC5; TMEM32; ER membrane protein complex subunit 5; Membrane magnesium transporter 1; Transmembrane protein 32 |
|||||
| 3D Structure | ||||||
| Sequence |
MAPSLWKGLVGIGLFALAHAAFSAAQHRSYMRLTEKEDESLPIDIVLQTLLAFAVTCYGI
VHIAGEFKDMDATSELKNKTFDTLRNHPSFYVFNHRGRVLFRPSDTANSSNQDALSSNTS LKLRKLESLRR |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
Membrane magnesium transporter (TC 1.A.67) family
|
|||||
| Subcellular location |
Endoplasmic reticulum membrane
|
|||||
| Function |
Part of the endoplasmic reticulum membrane protein complex (EMC) that enables the energy-independent insertion into endoplasmic reticulum membranes of newly synthesized membrane proteins. Preferentially accommodates proteins with transmembrane domains that are weakly hydrophobic or contain destabilizing features such as charged and aromatic residues. Involved in the cotranslational insertion of multi-pass membrane proteins in which stop-transfer membrane-anchor sequences become ER membrane spanning helices. It is also required for the post-translational insertion of tail-anchored/TA proteins in endoplasmic reticulum membranes. By mediating the proper cotranslational insertion of N-terminal transmembrane domains in an N-exo topology, with translocated N-terminus in the lumen of the ER, controls the topology of multi-pass membrane proteins like the G protein-coupled receptors. By regulating the insertion of various proteins in membranes, it is indirectly involved in many cellular processes. May be involved in Mg(2+) transport.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
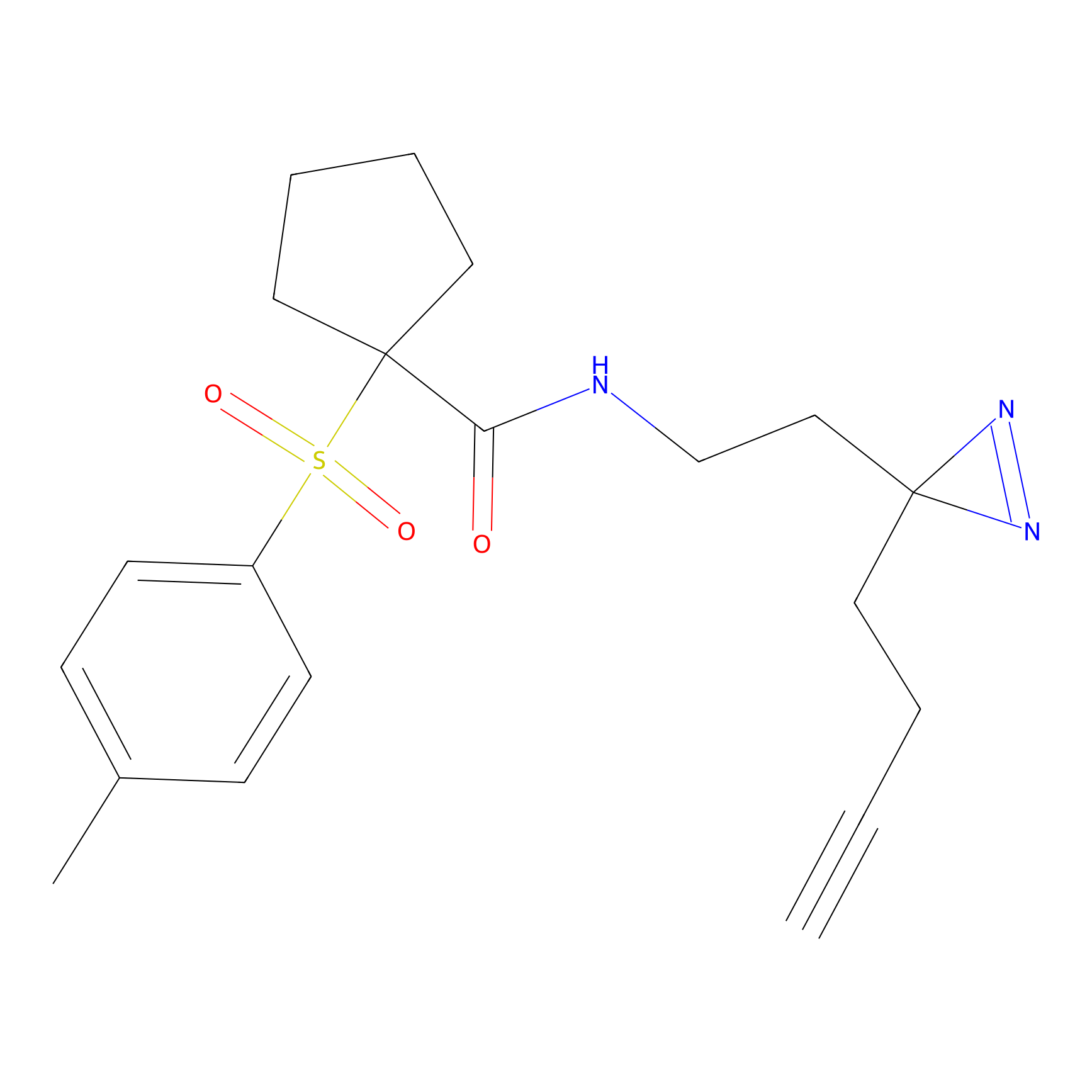
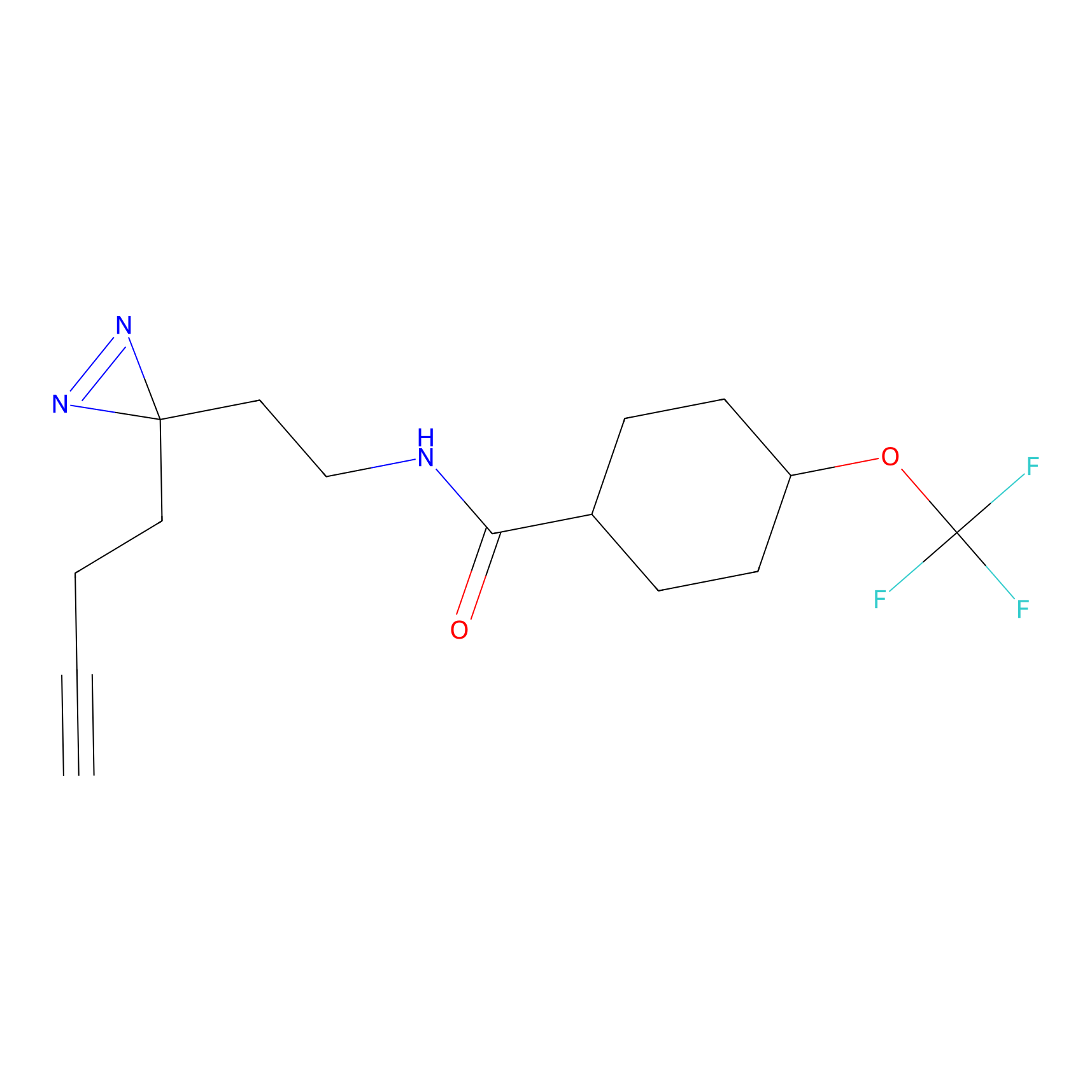
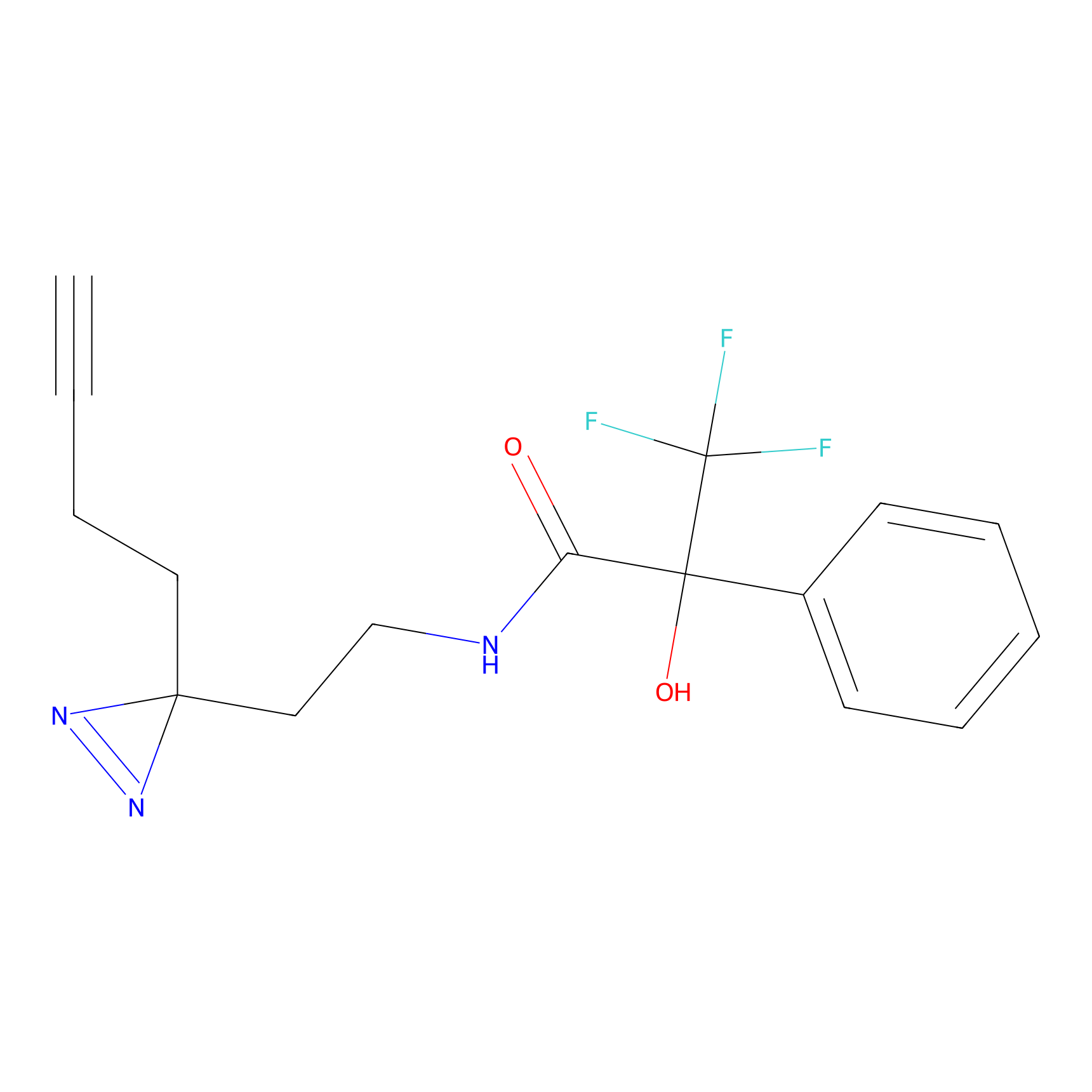
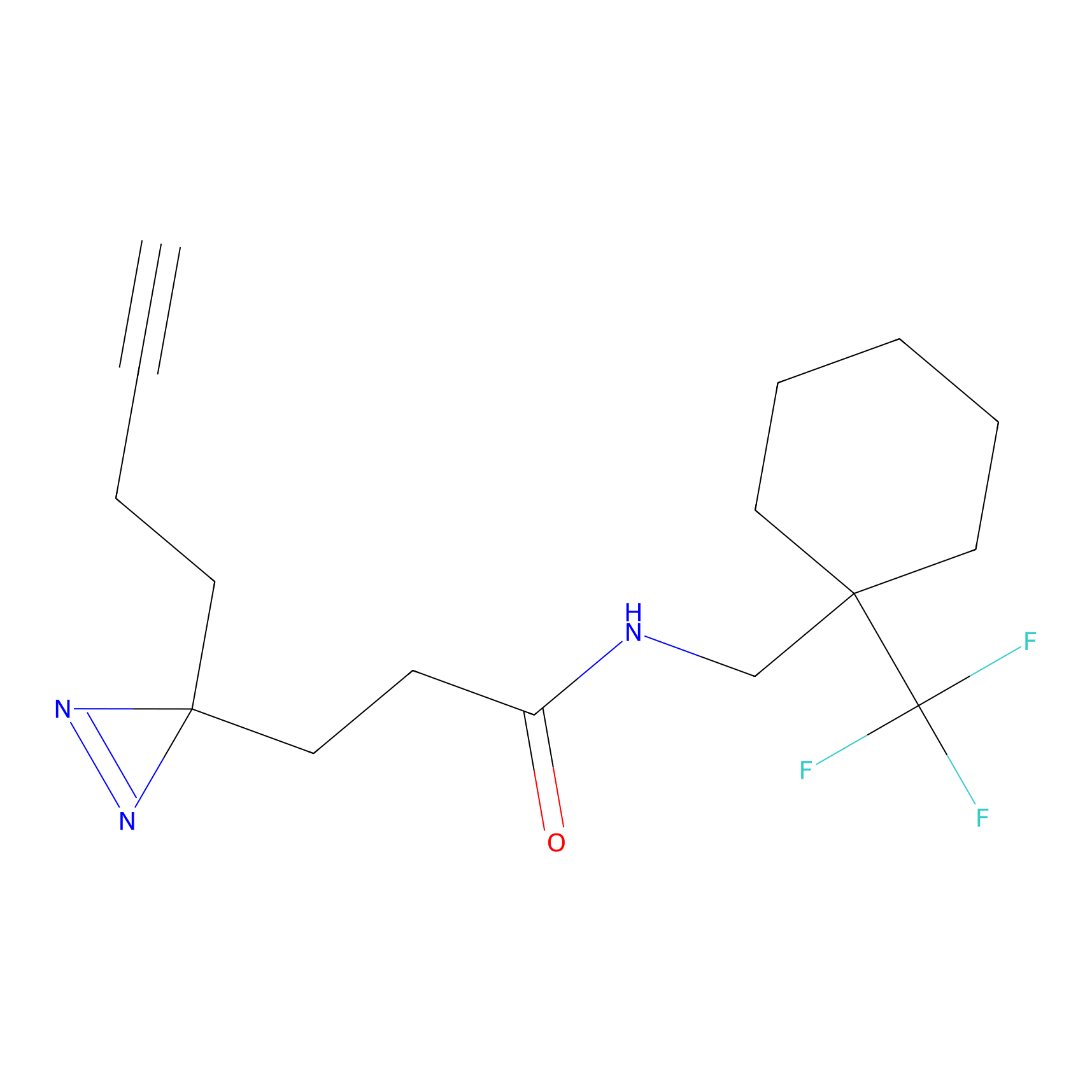
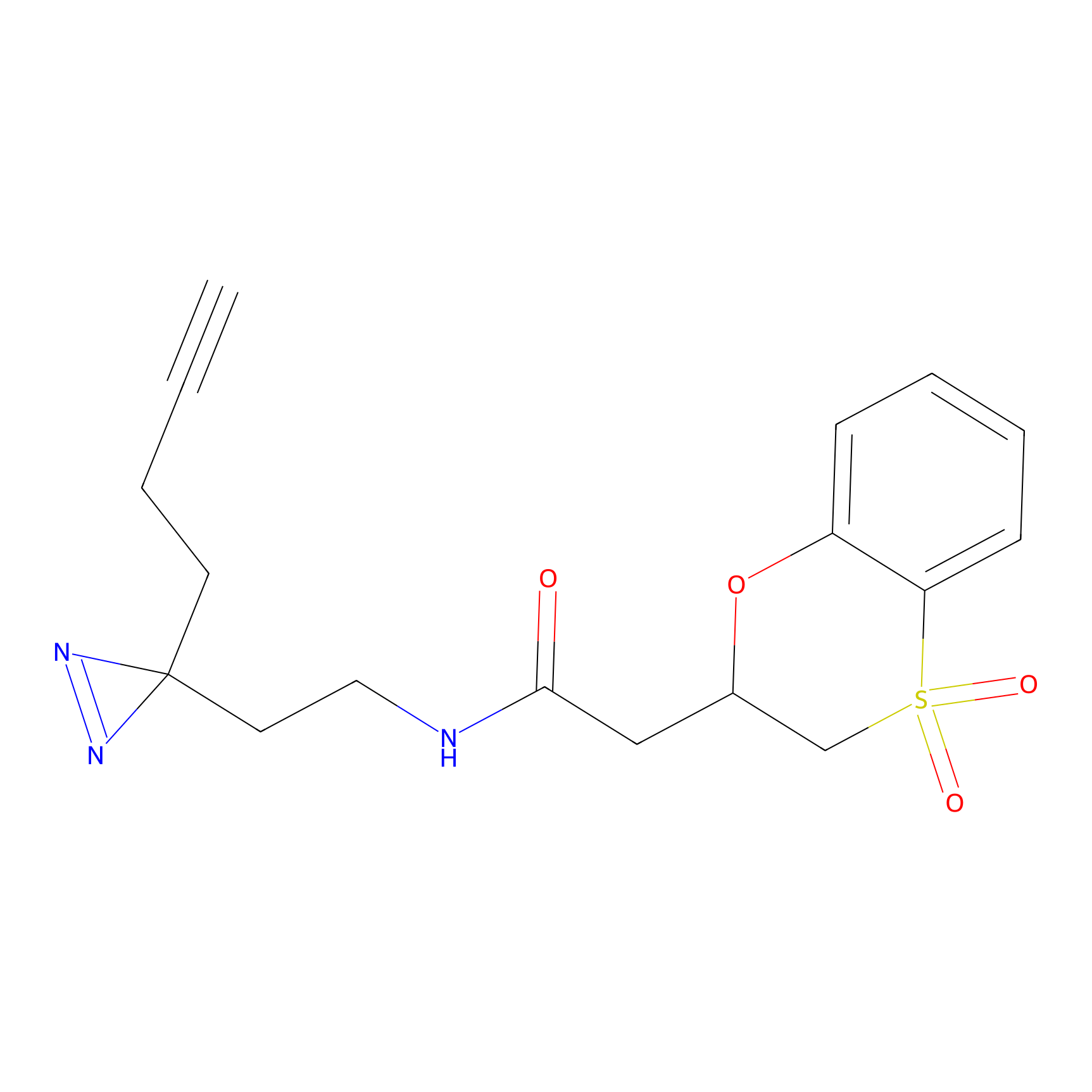
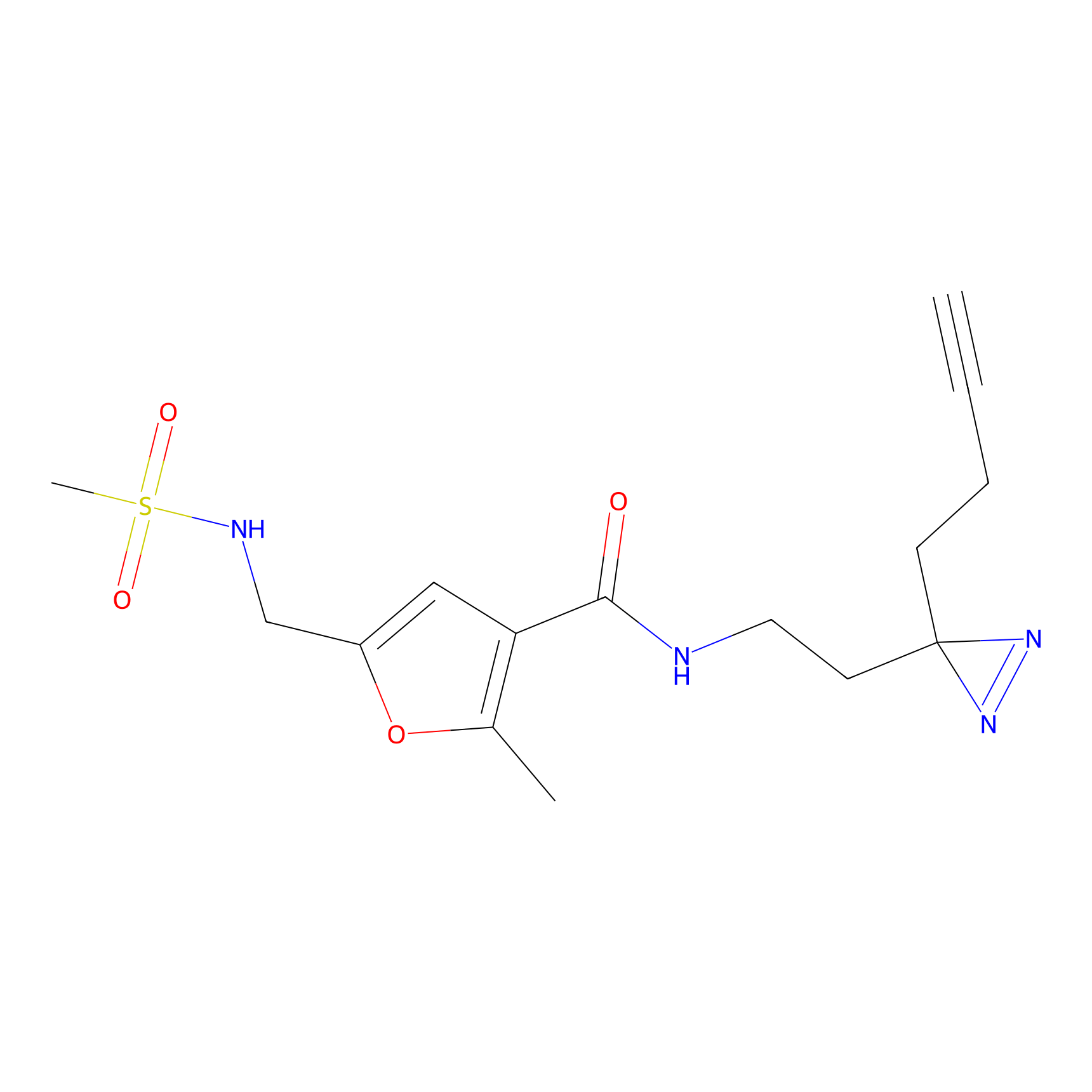
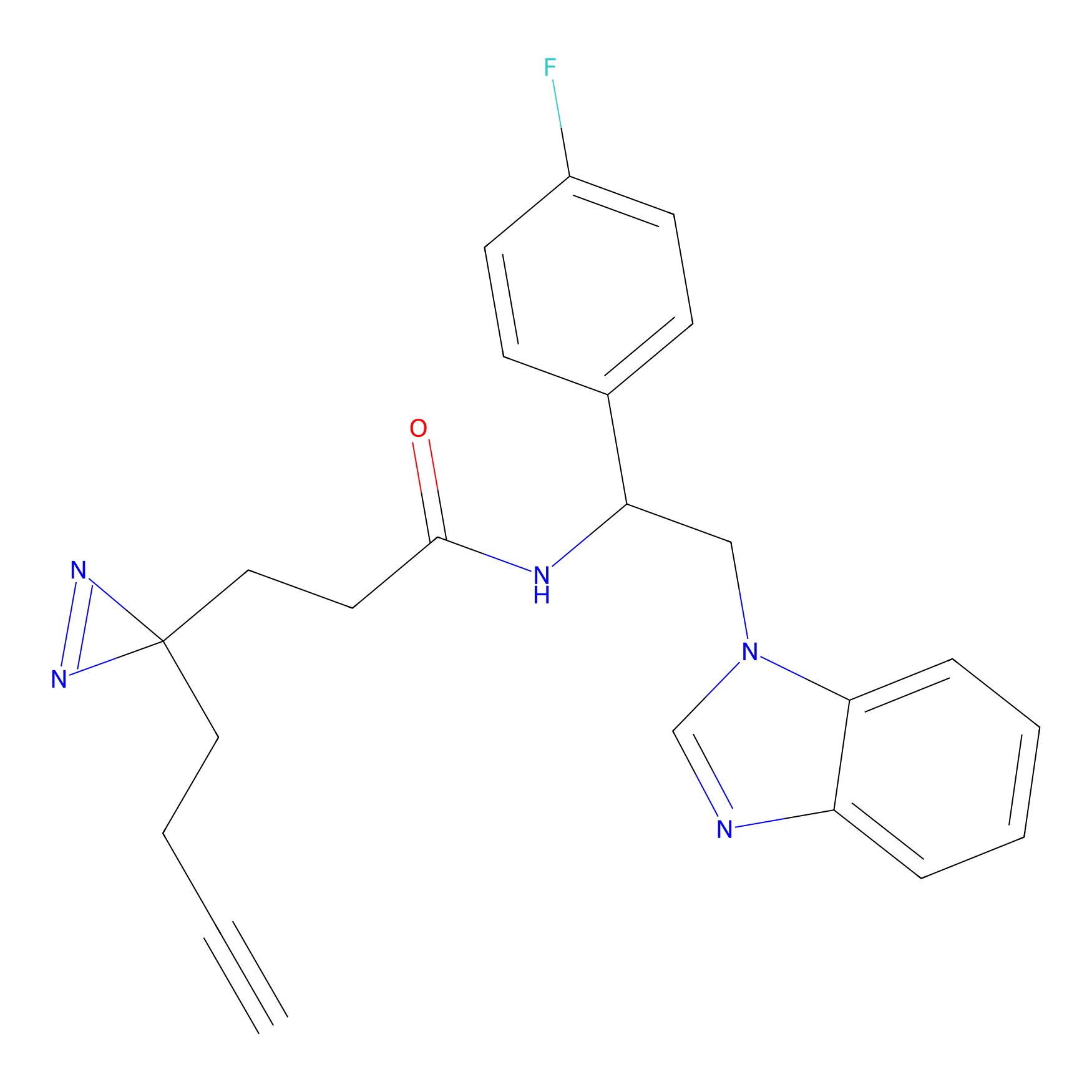
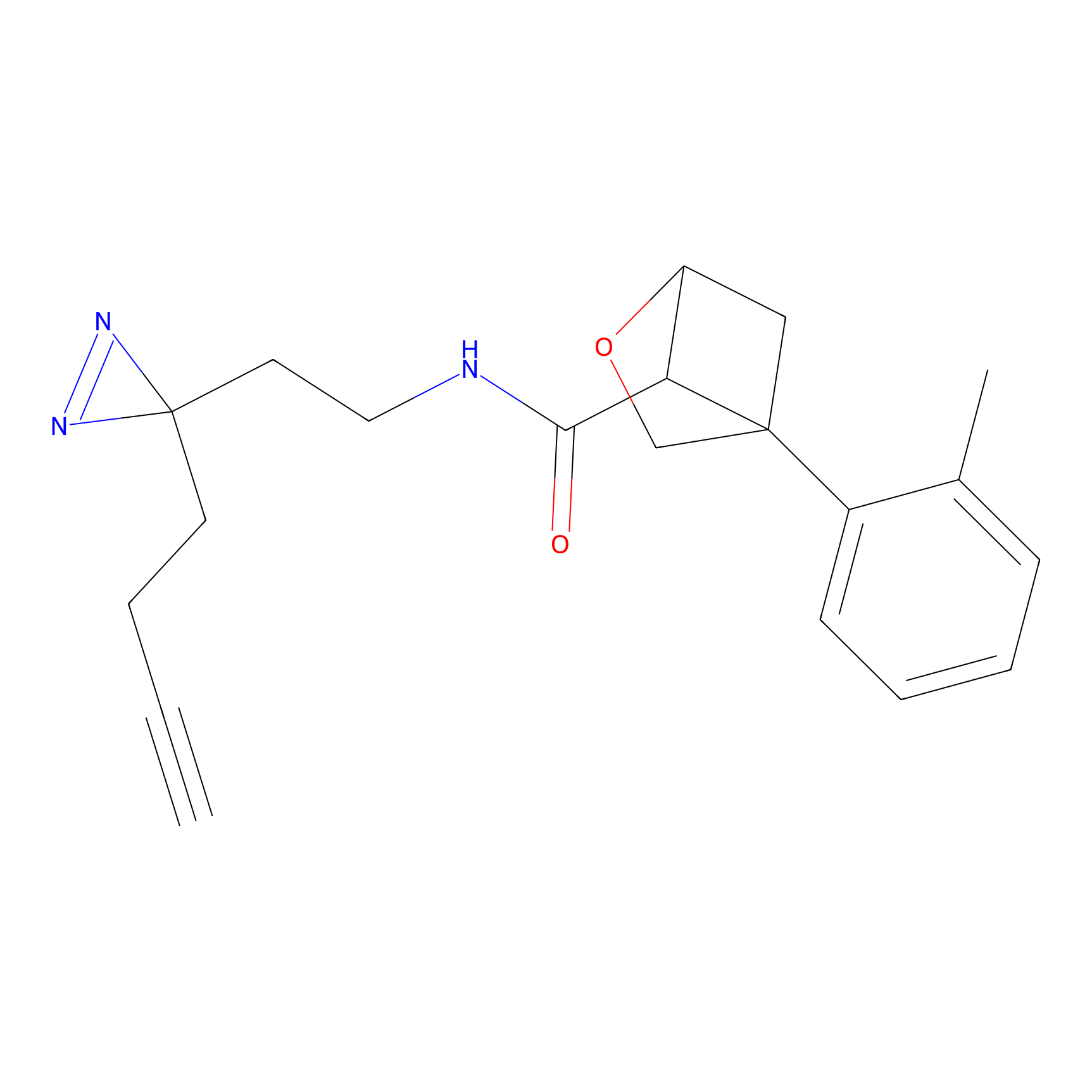
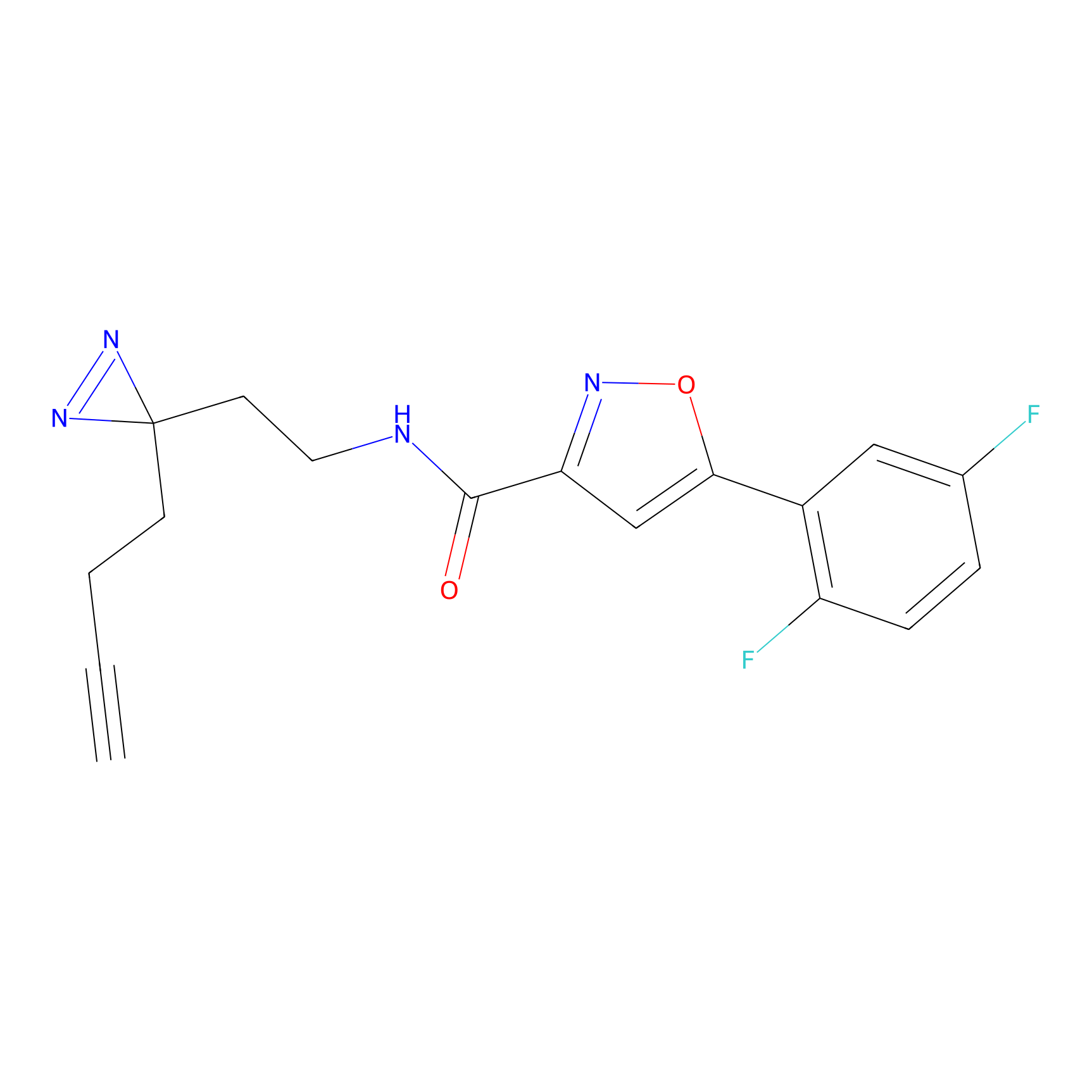
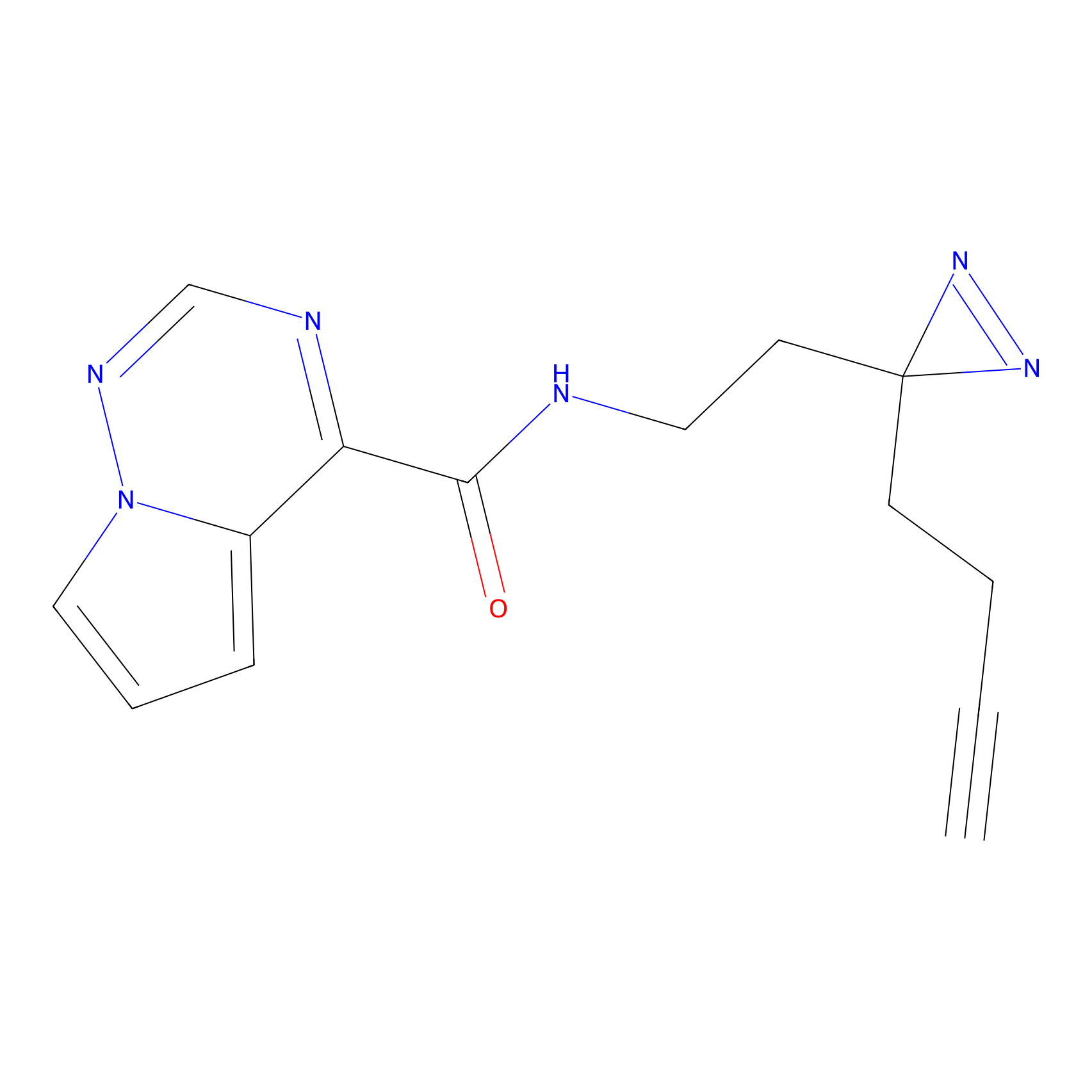
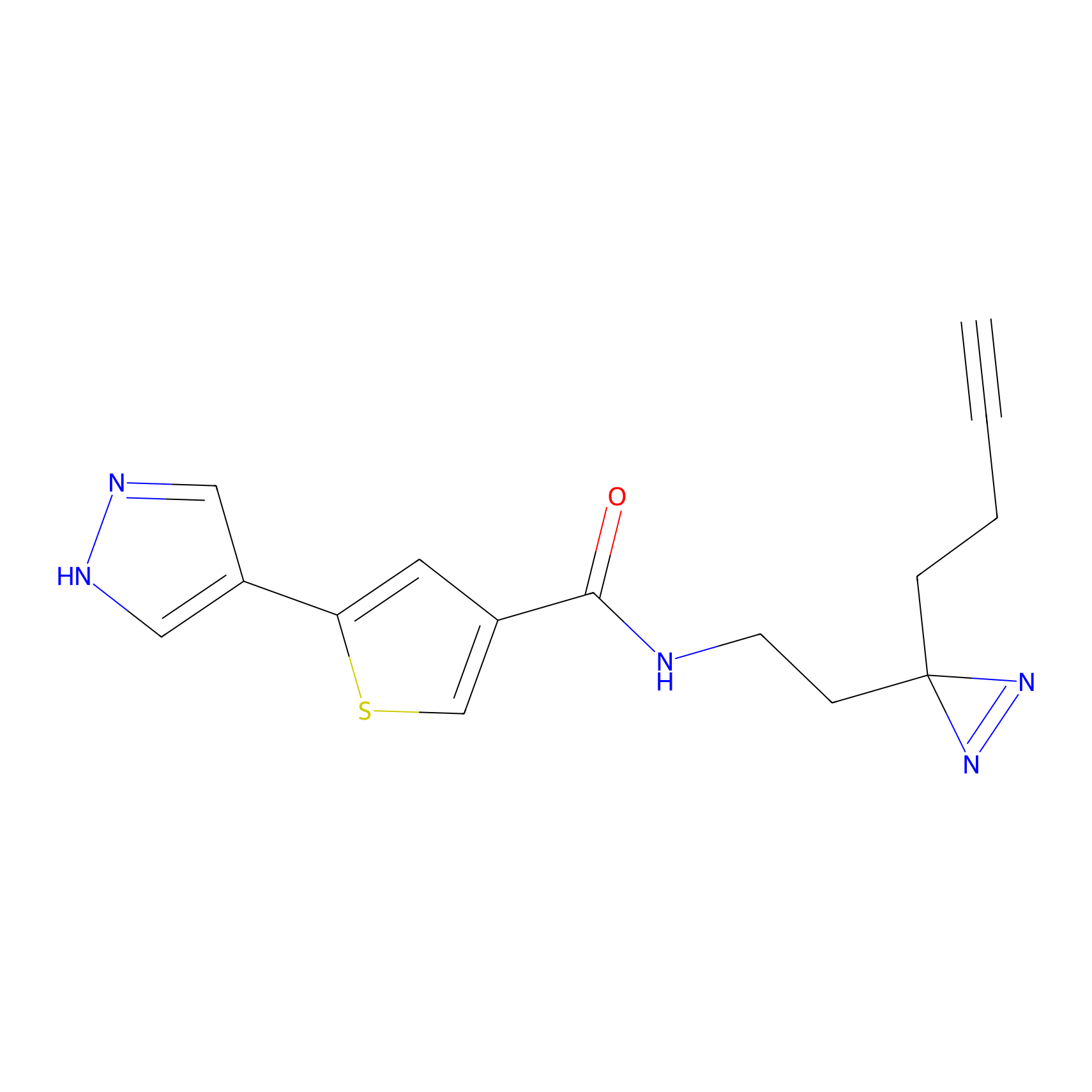
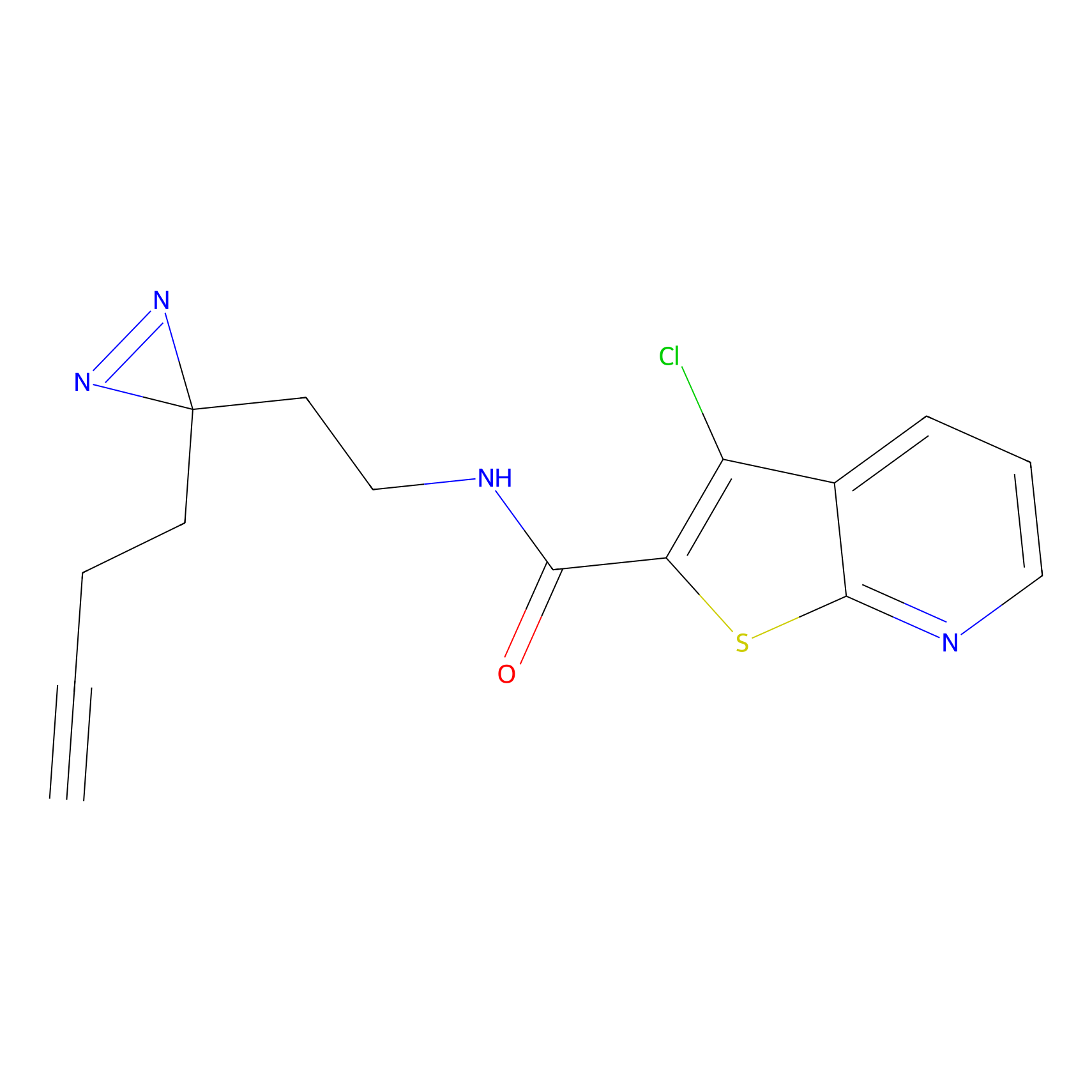
HDSF-alk Probe Info |
 |
2.18 | LDD0197 | [1] | |
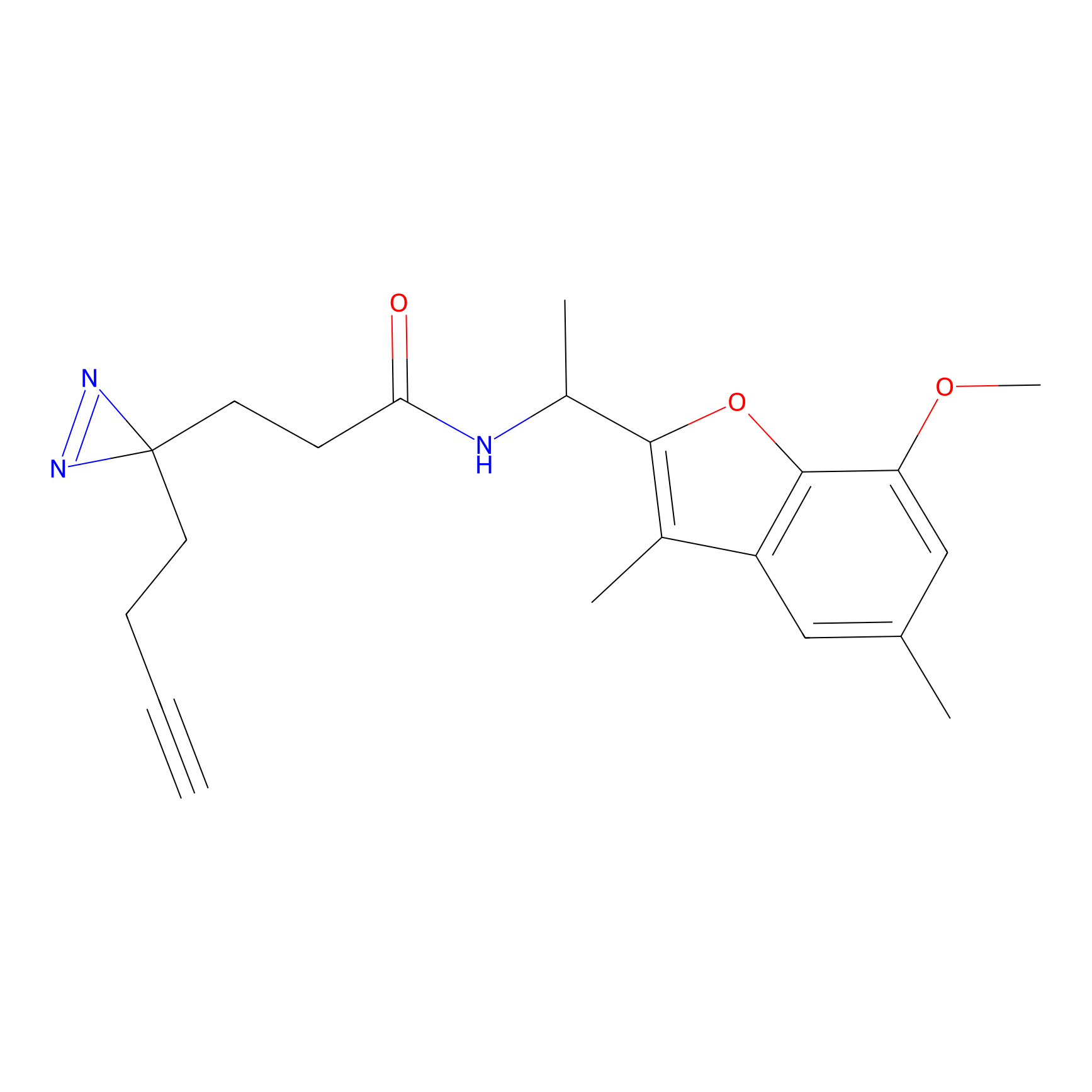
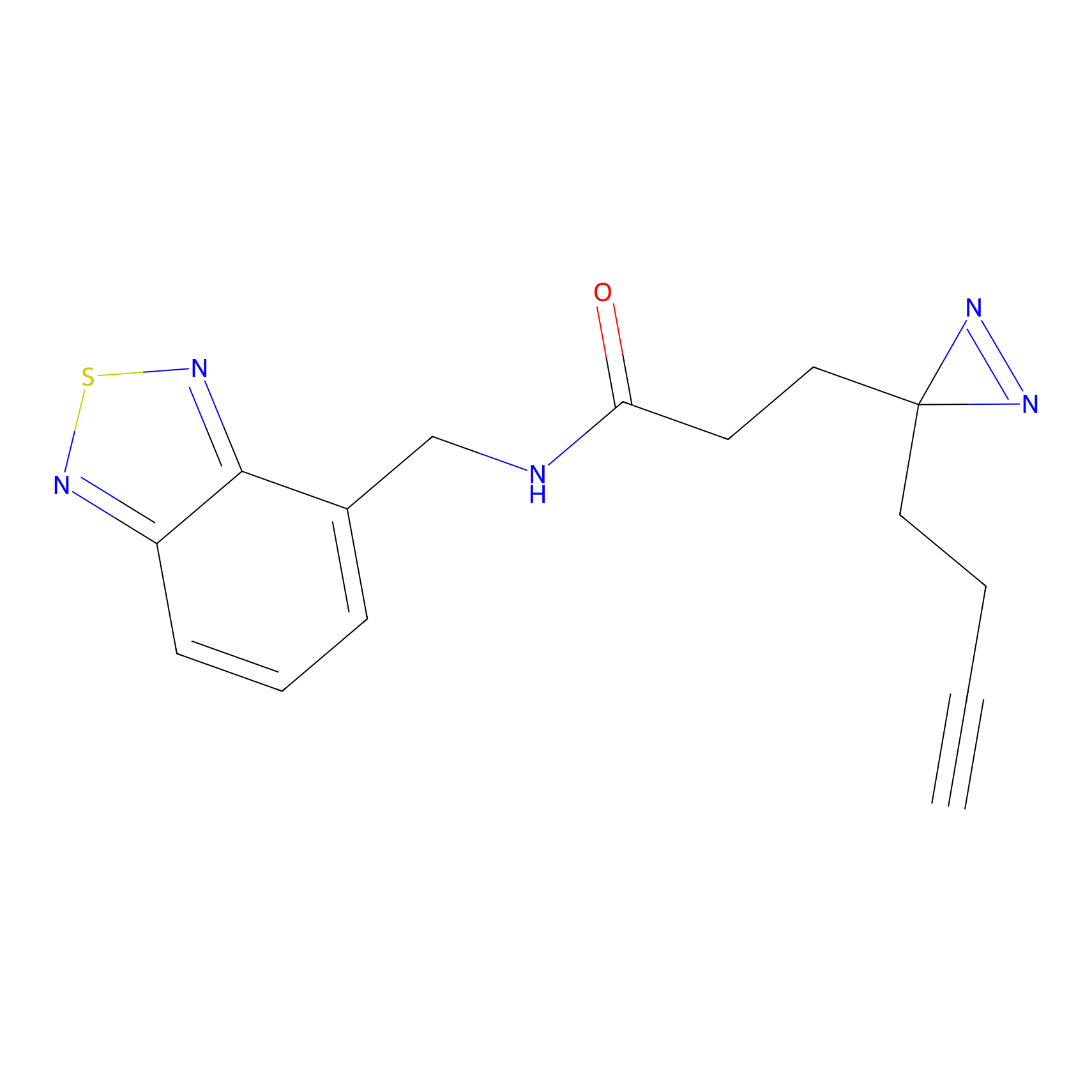
|
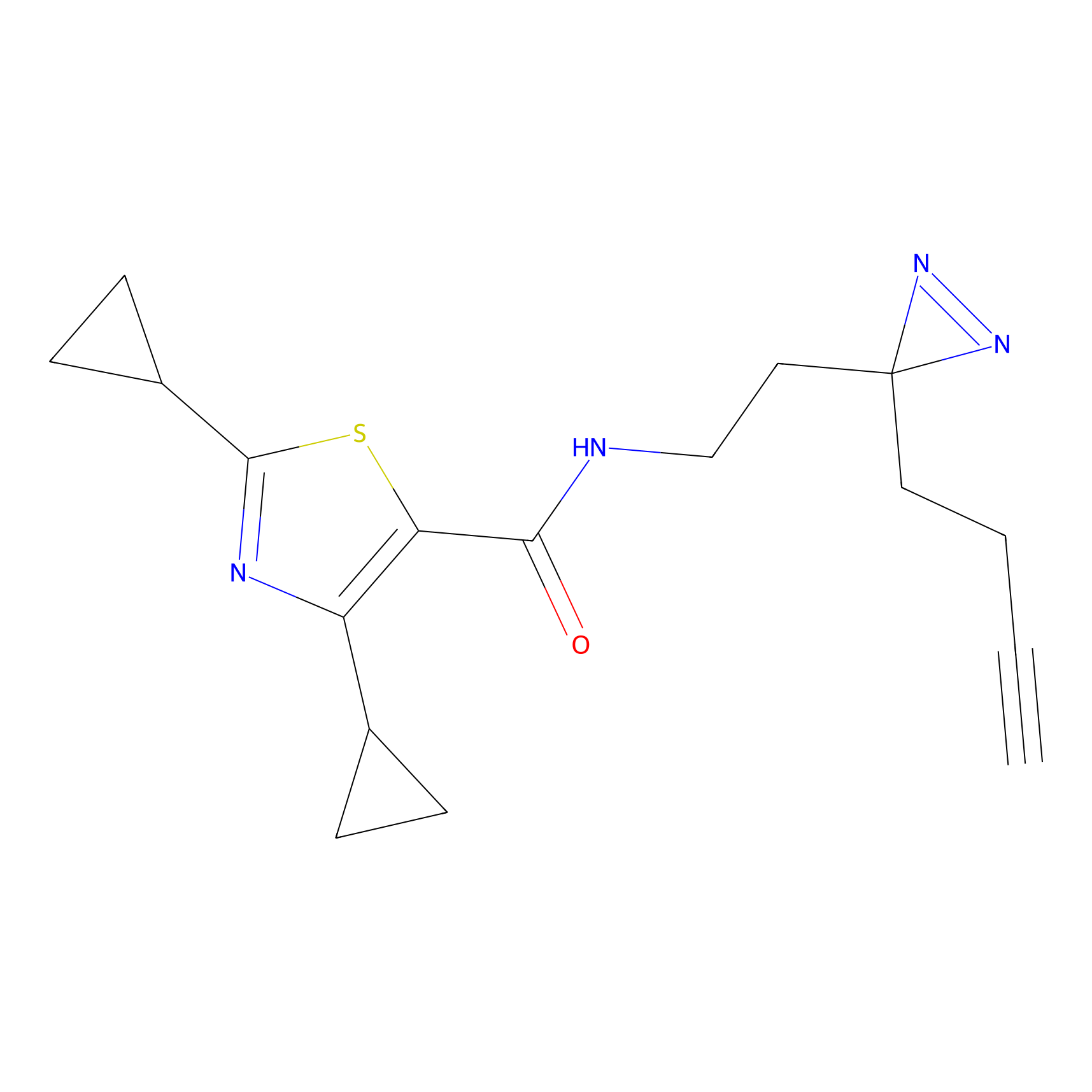
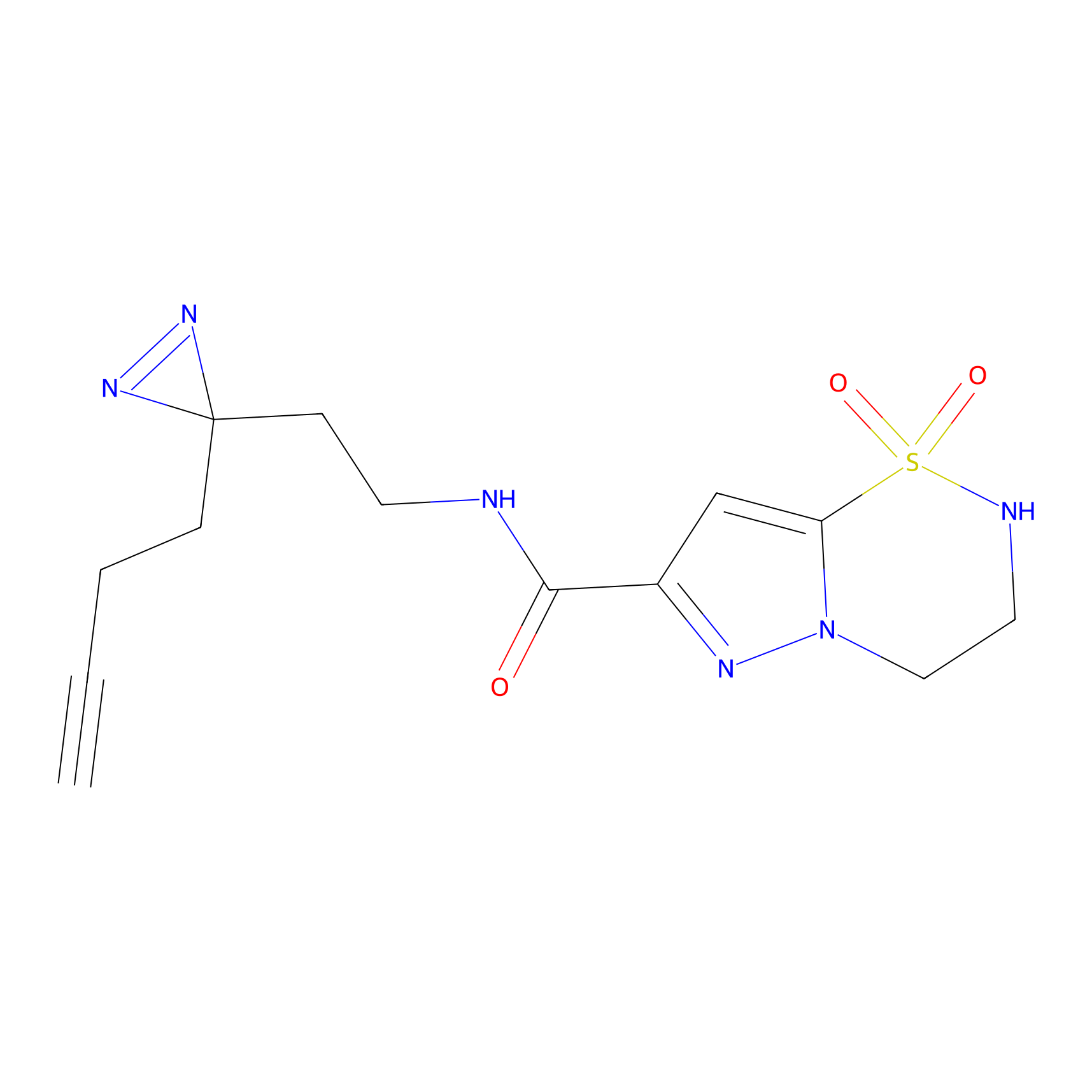
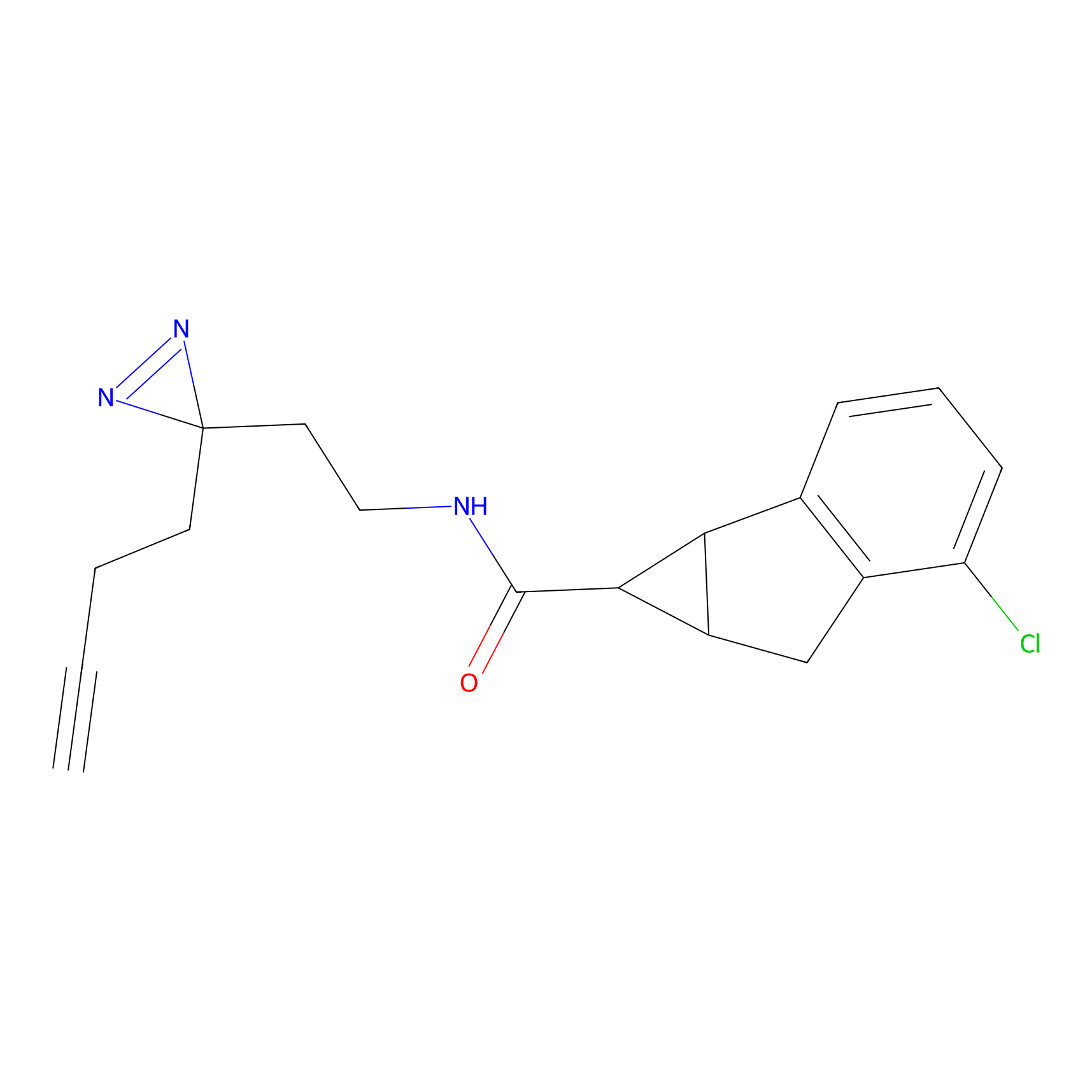
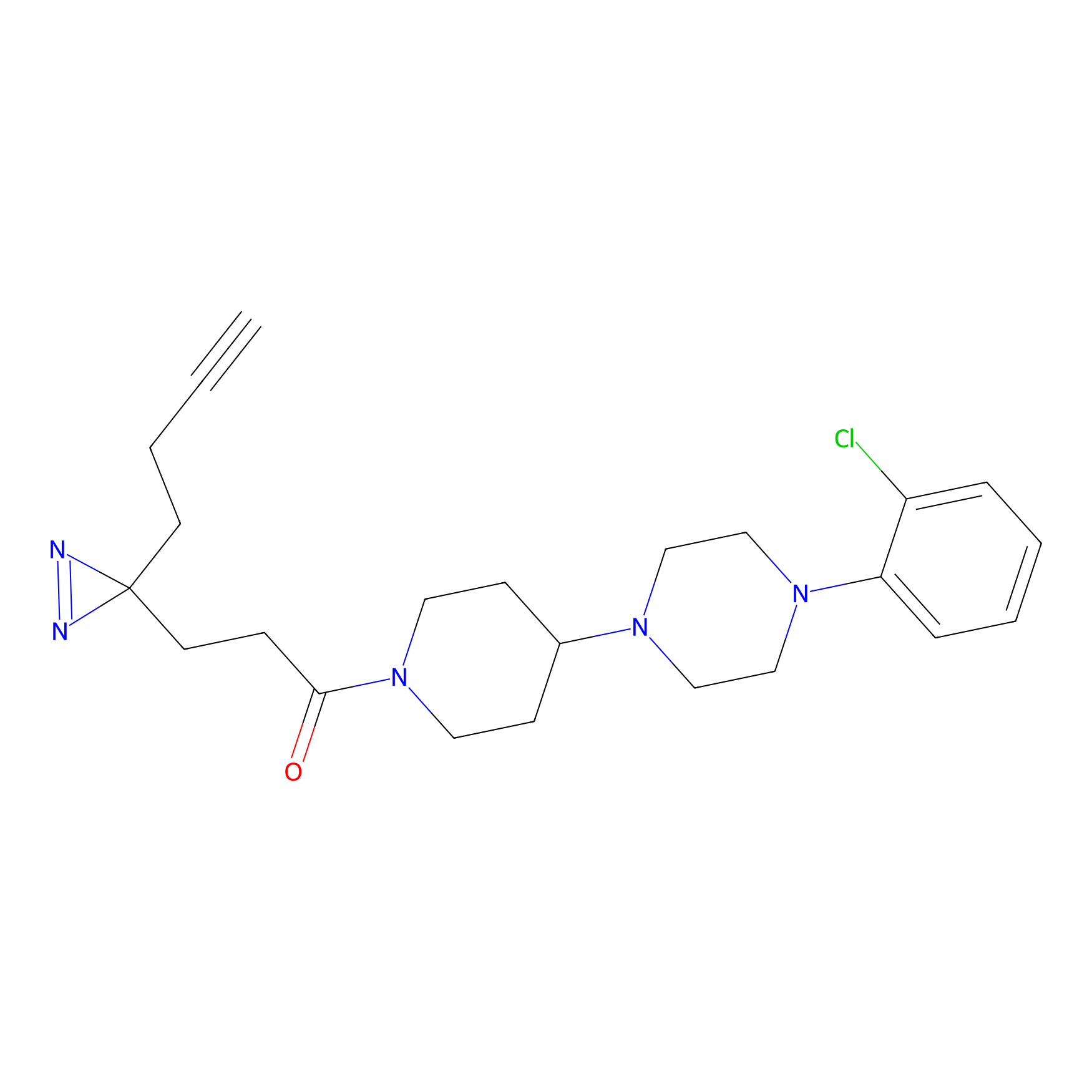
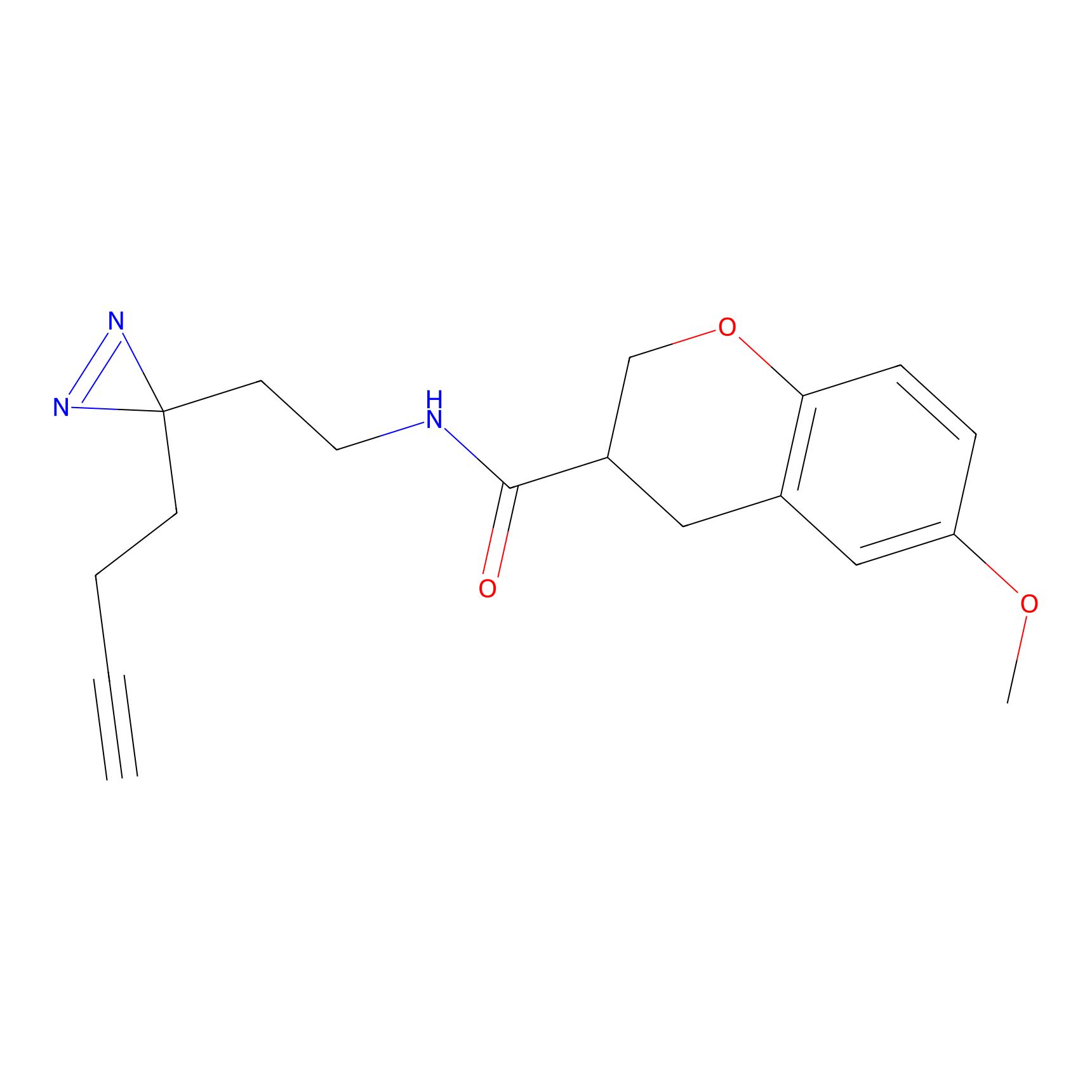
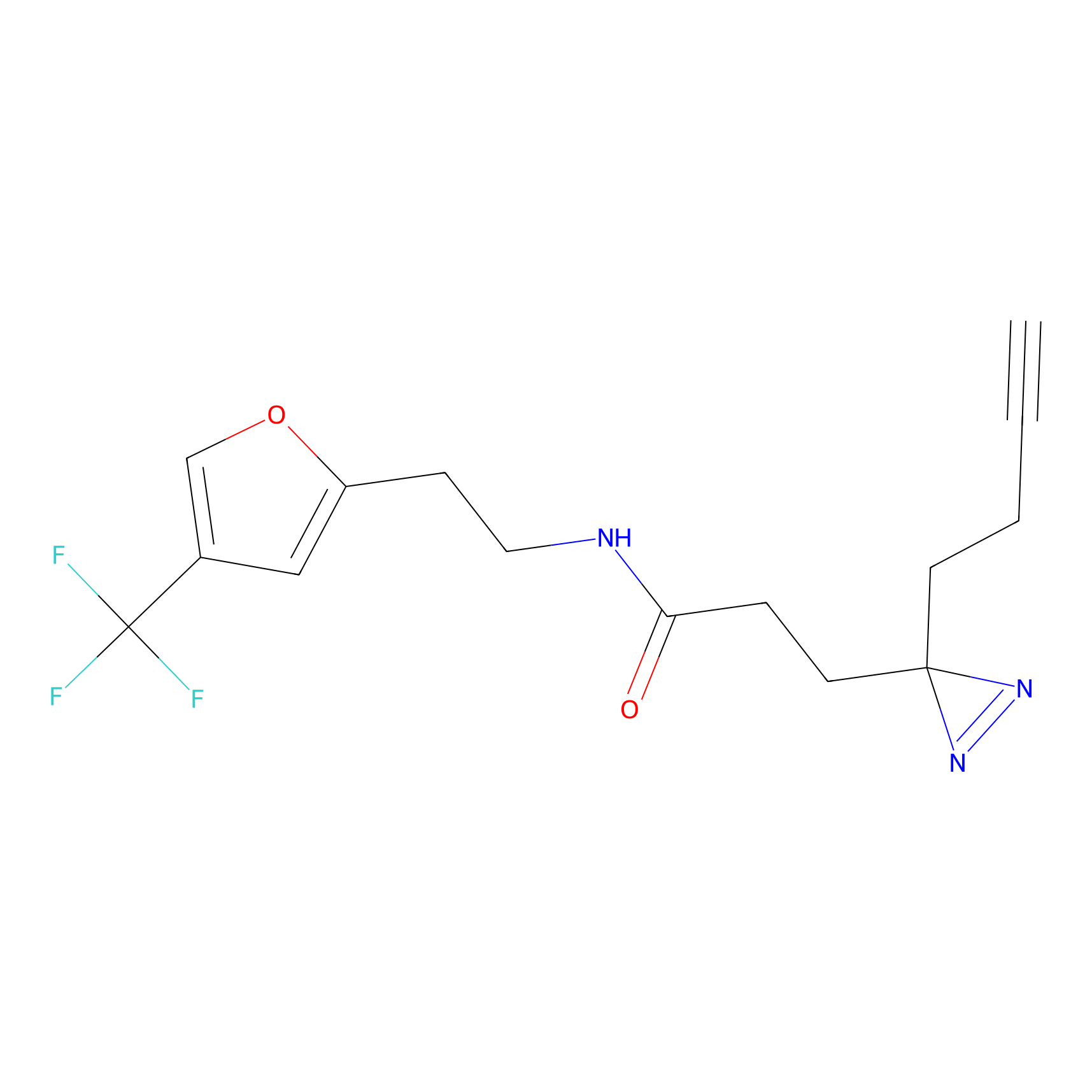
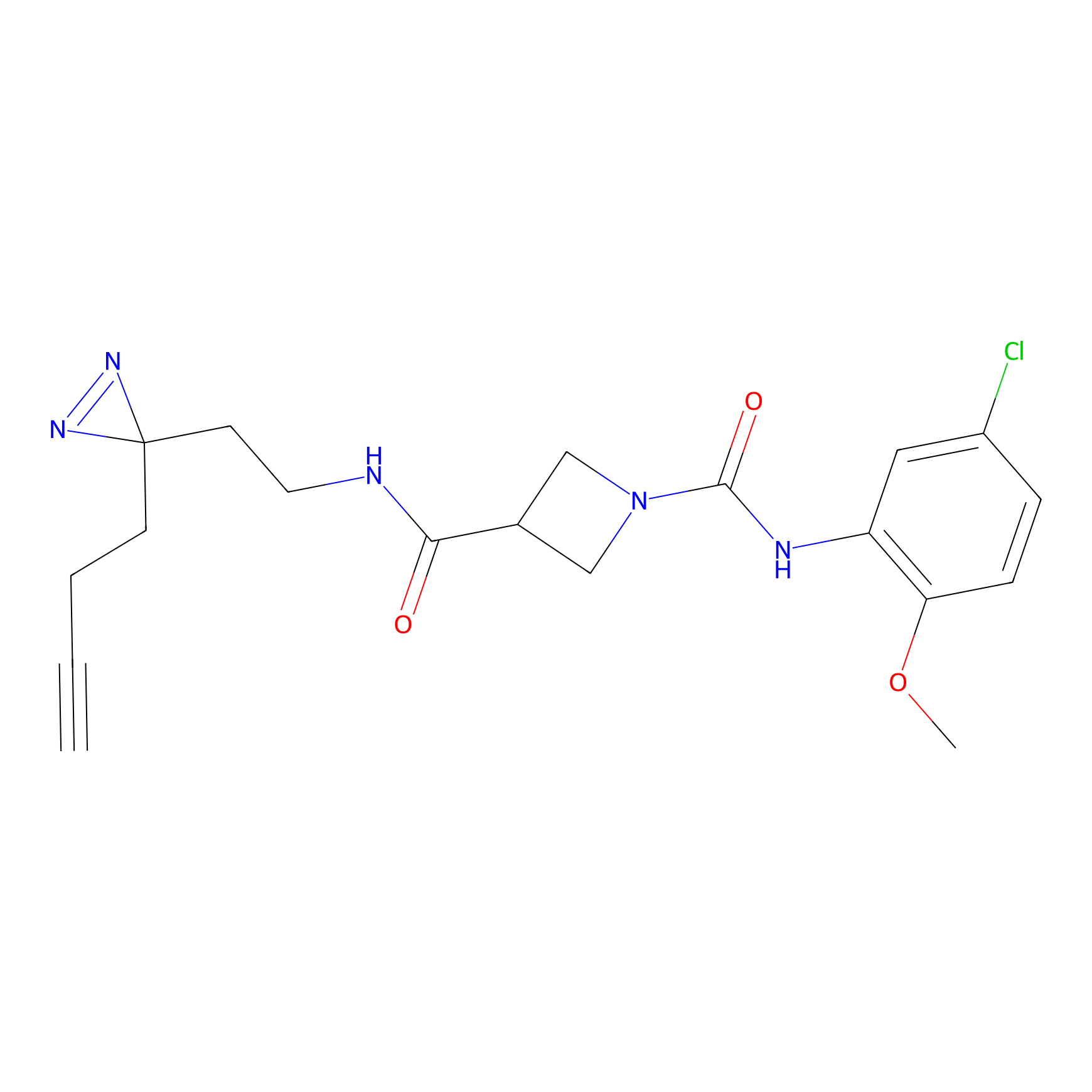
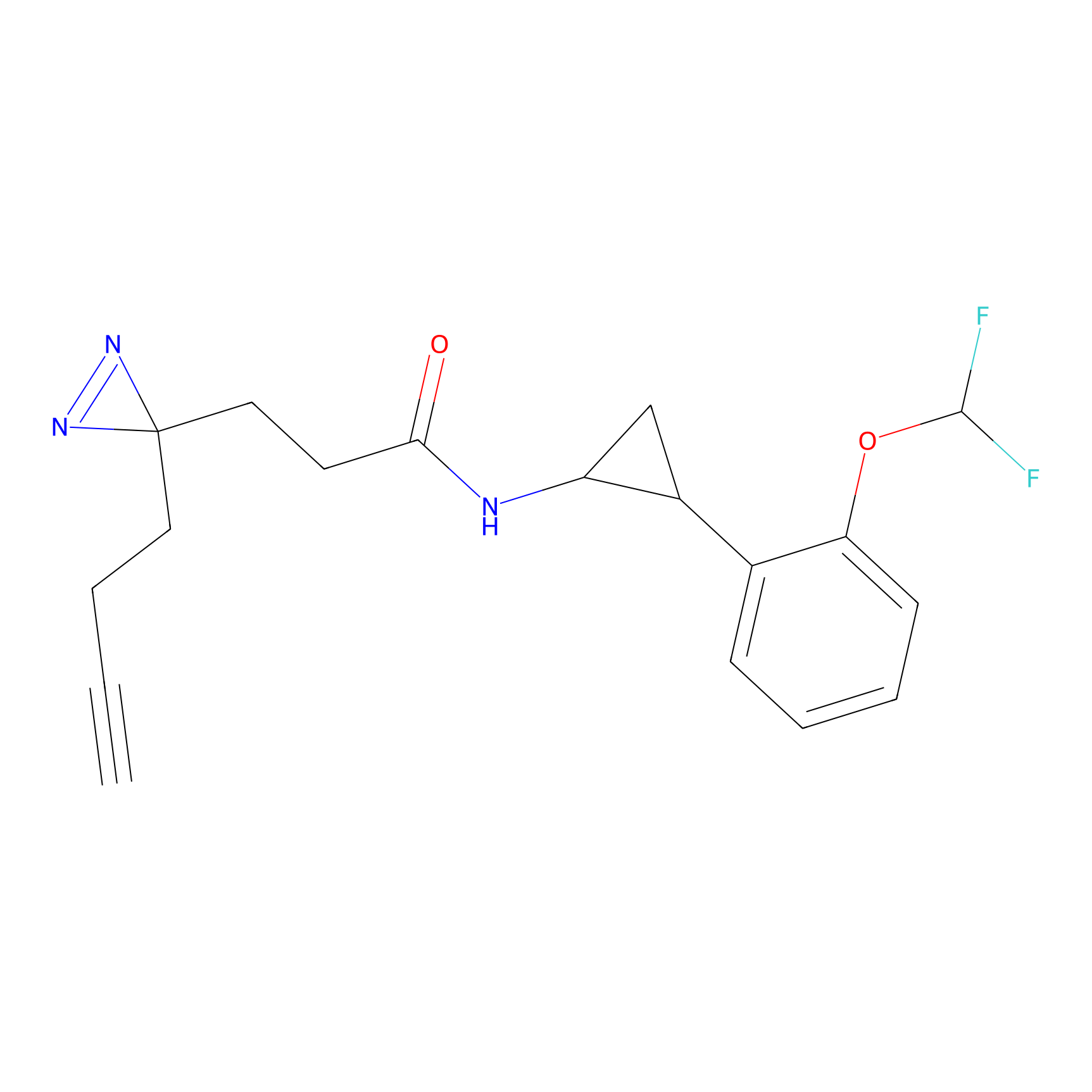
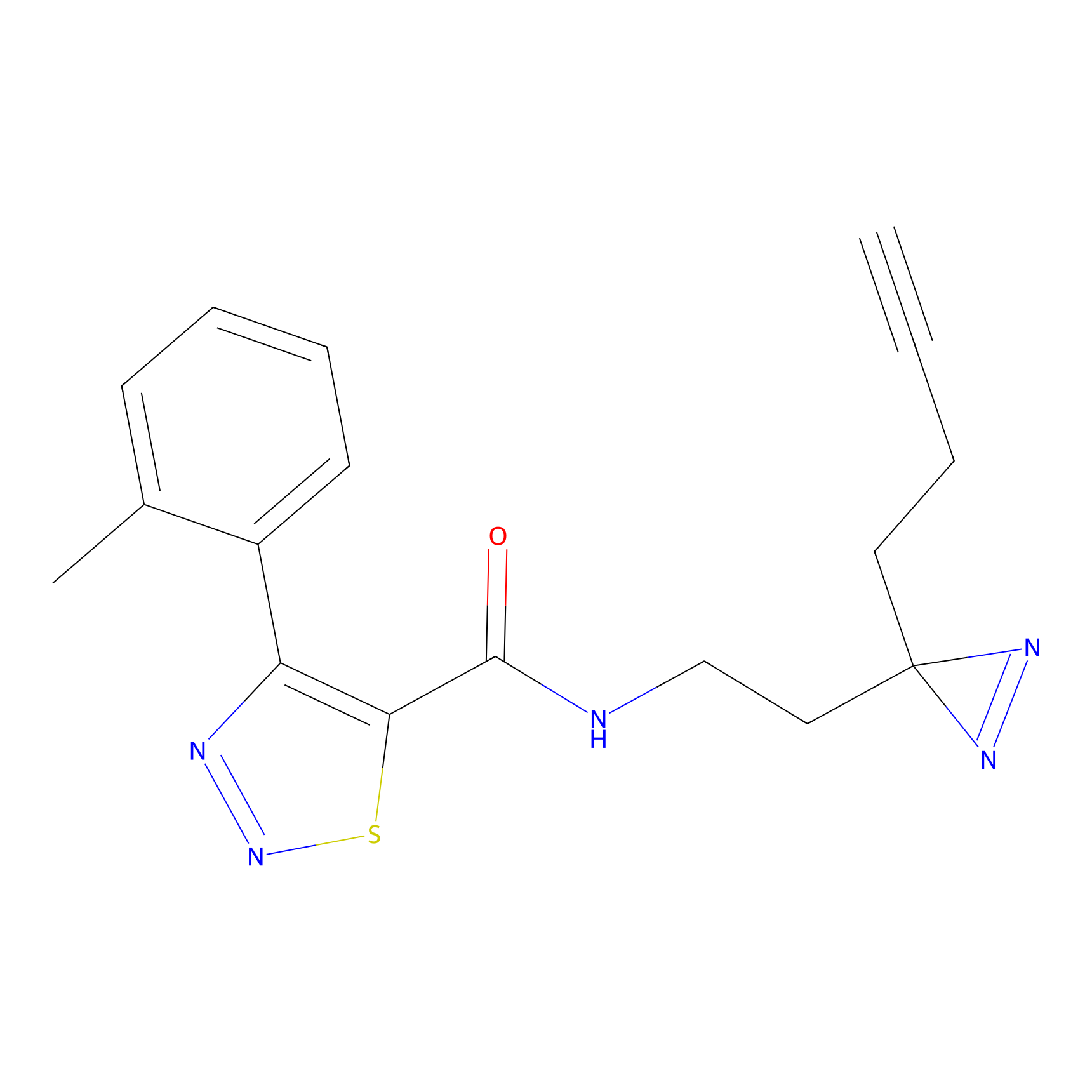
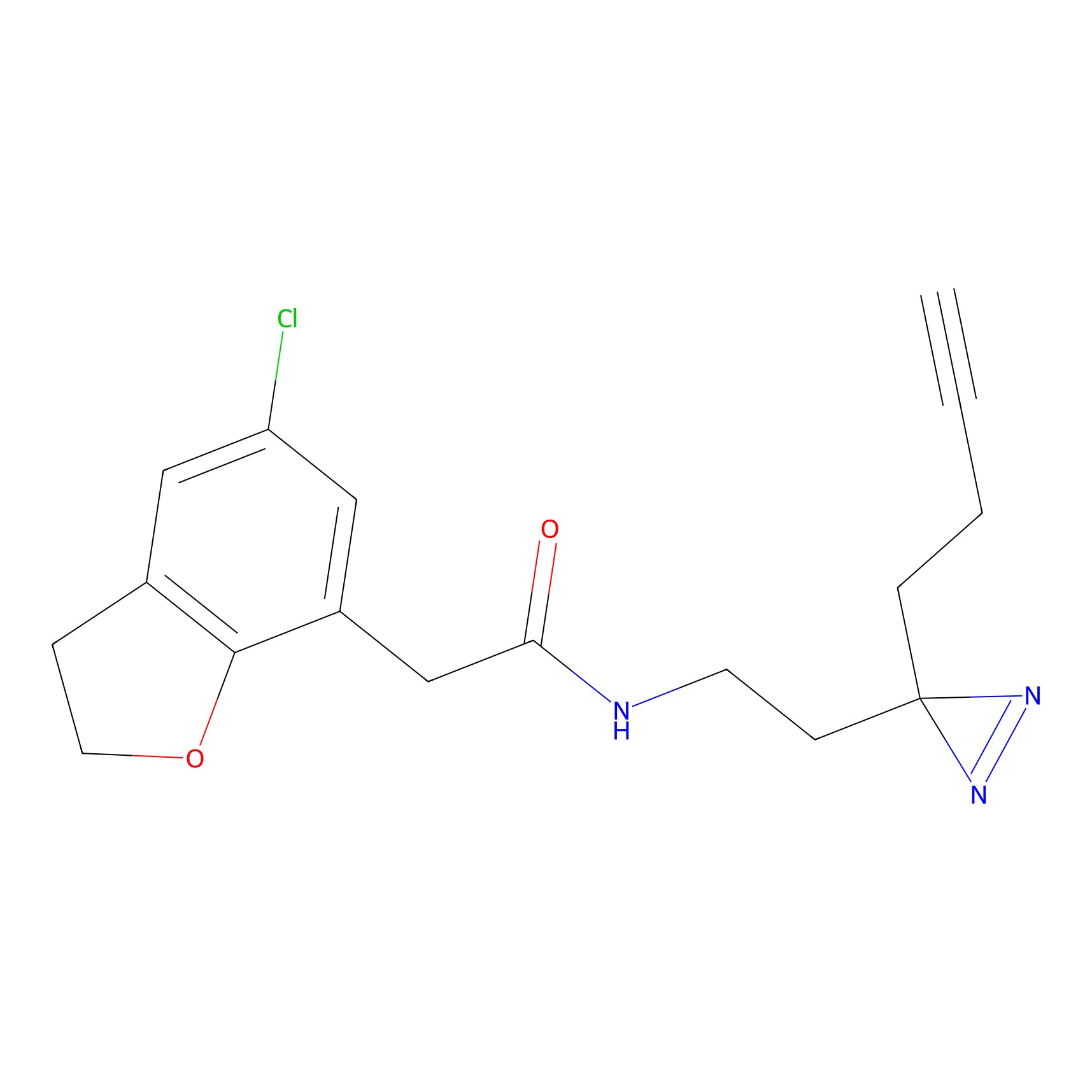
STPyne Probe Info |
 |
K79(9.09) | LDD0277 | [2] | |
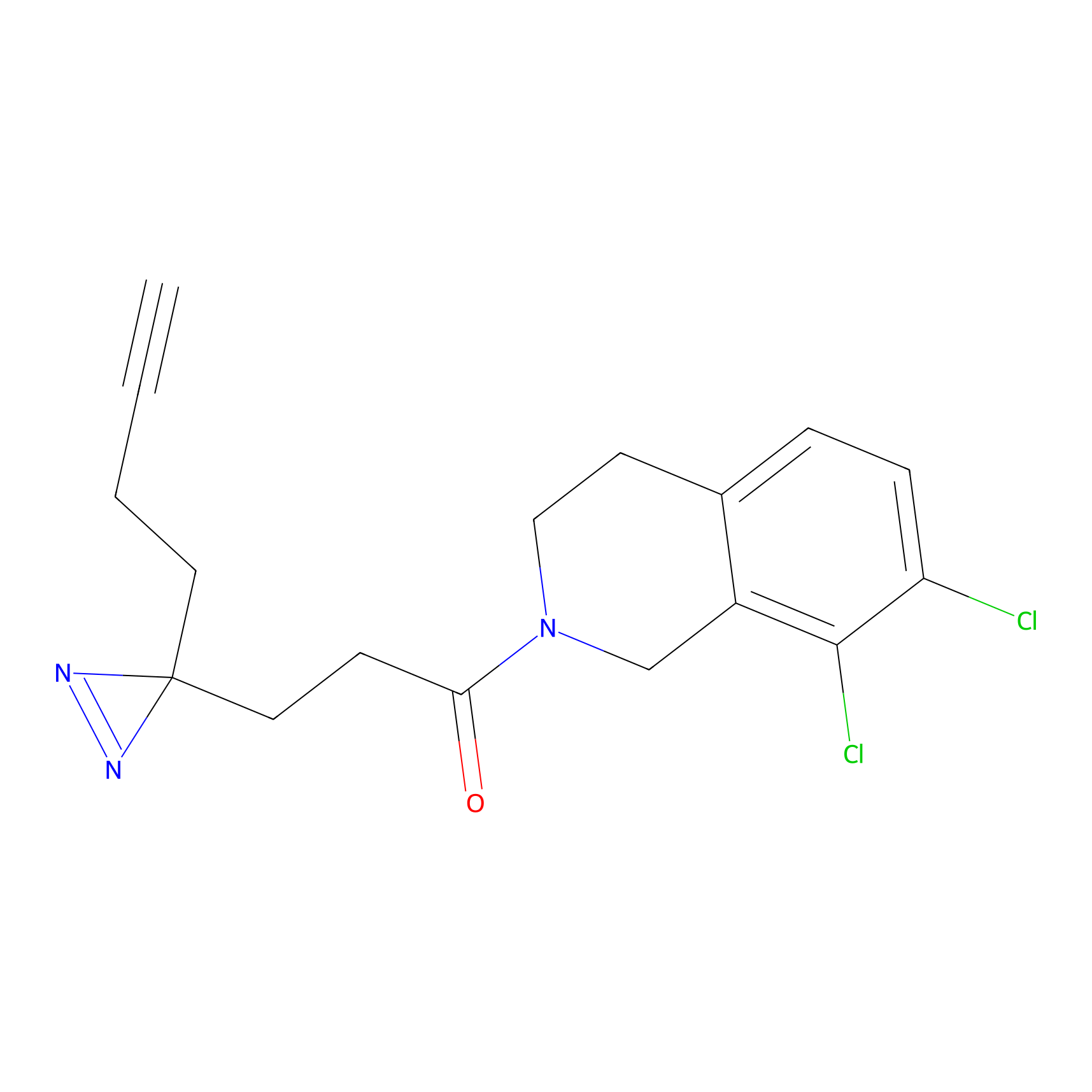
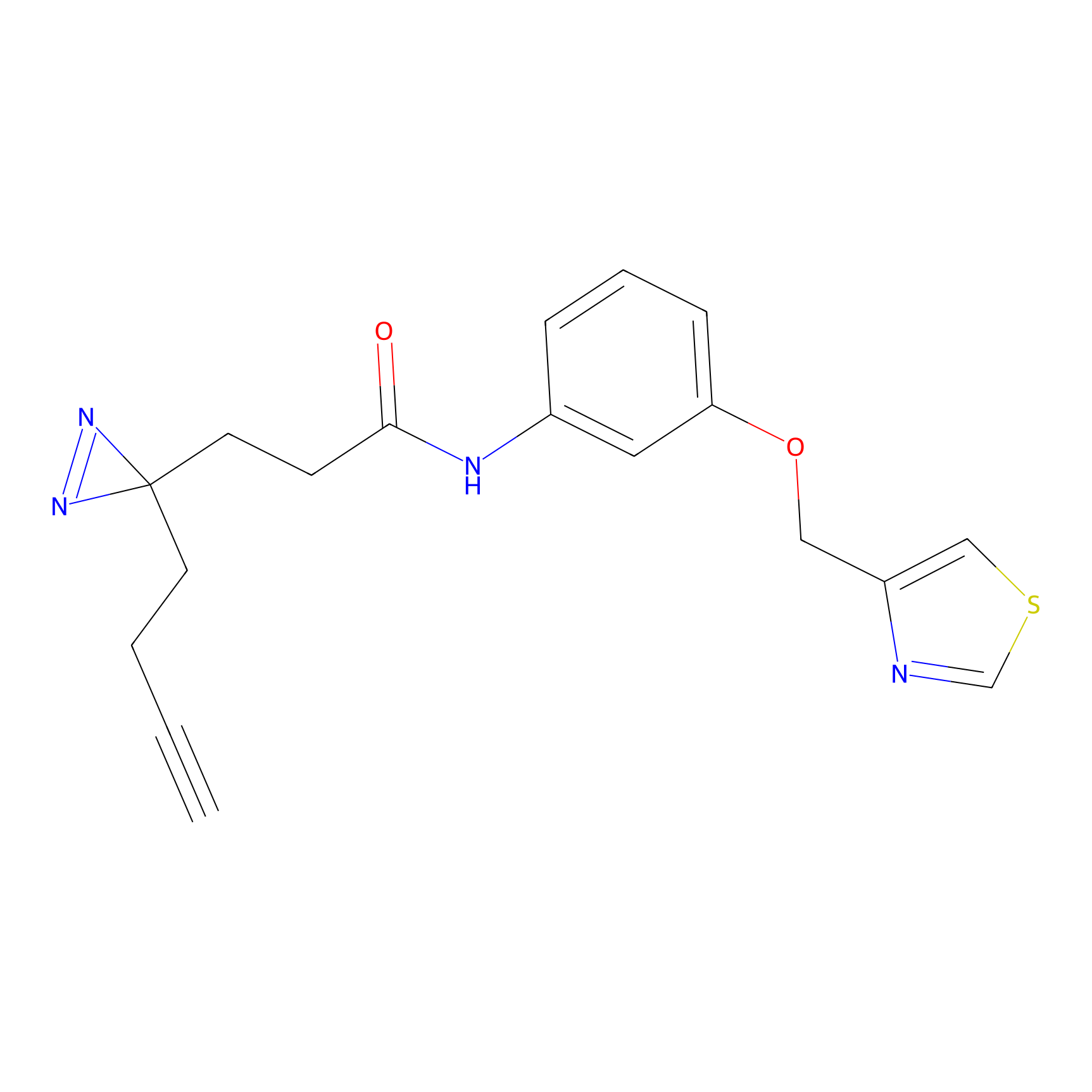
|
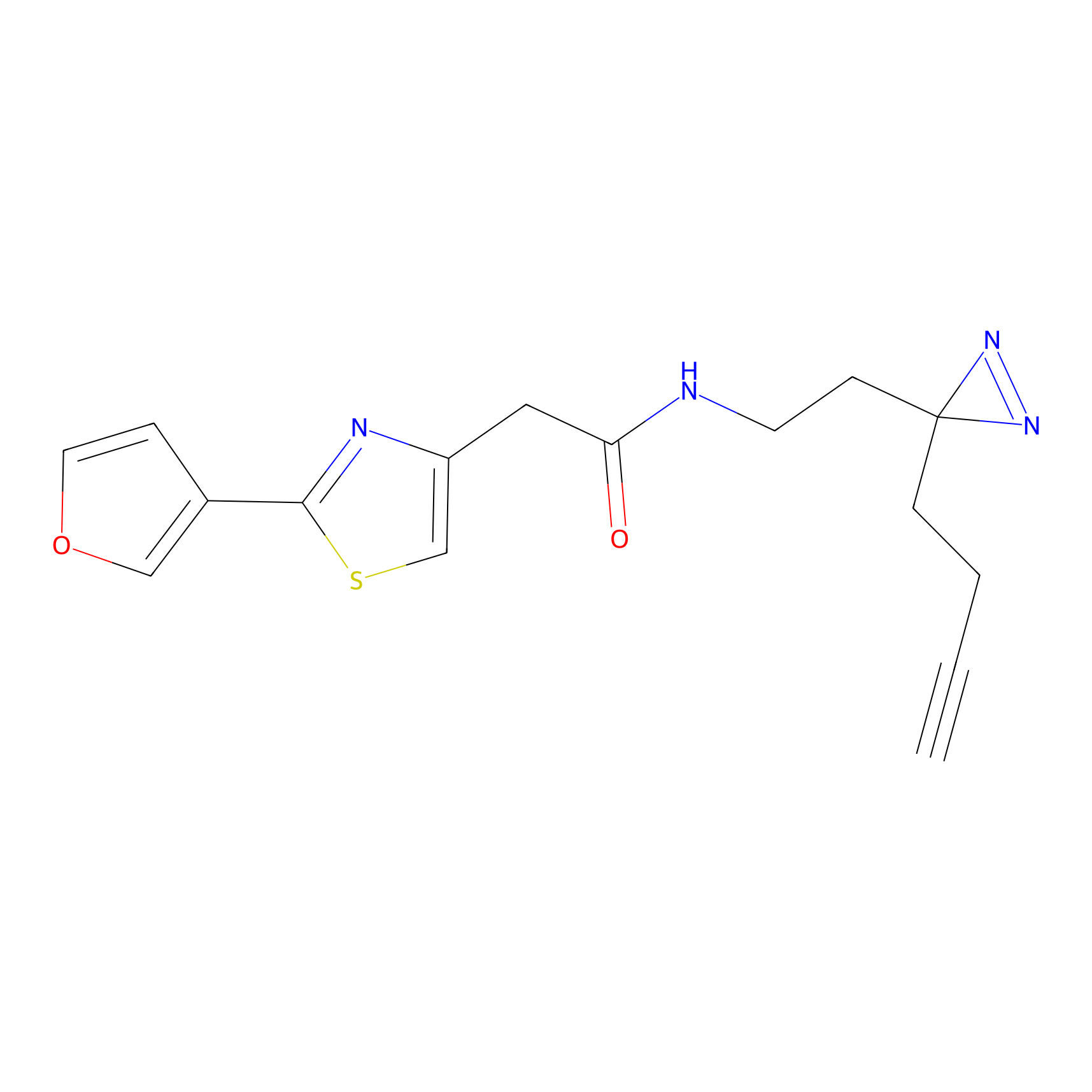
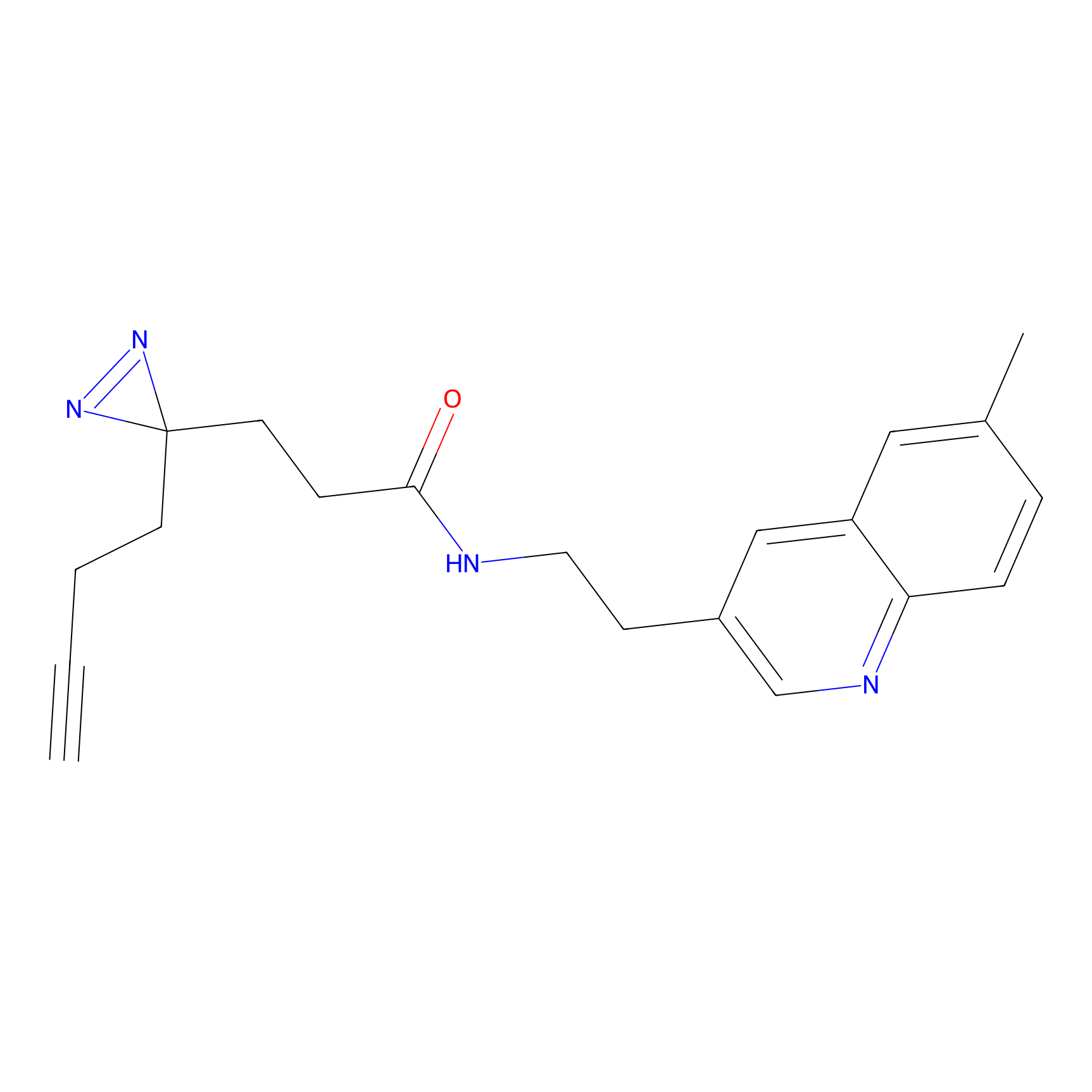
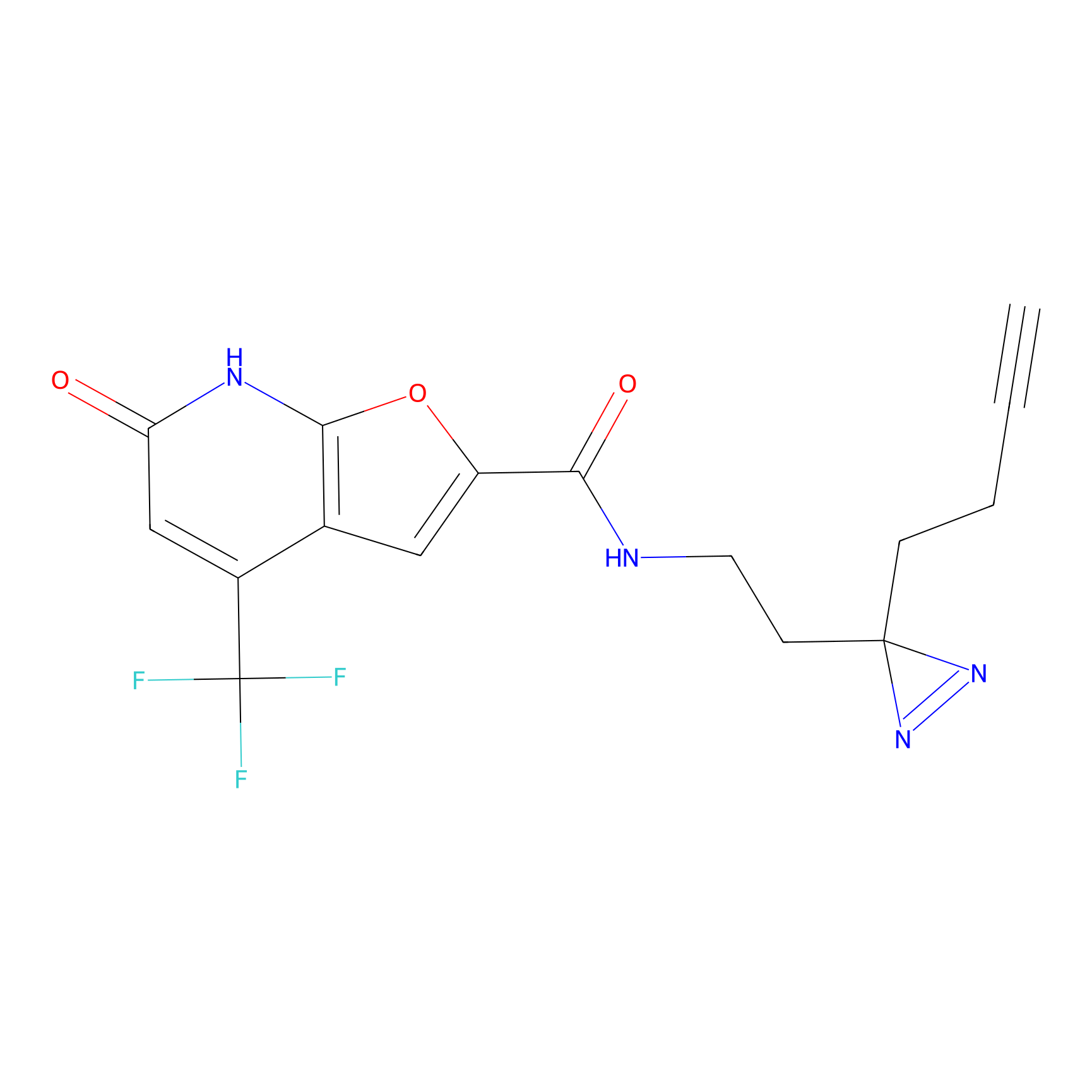
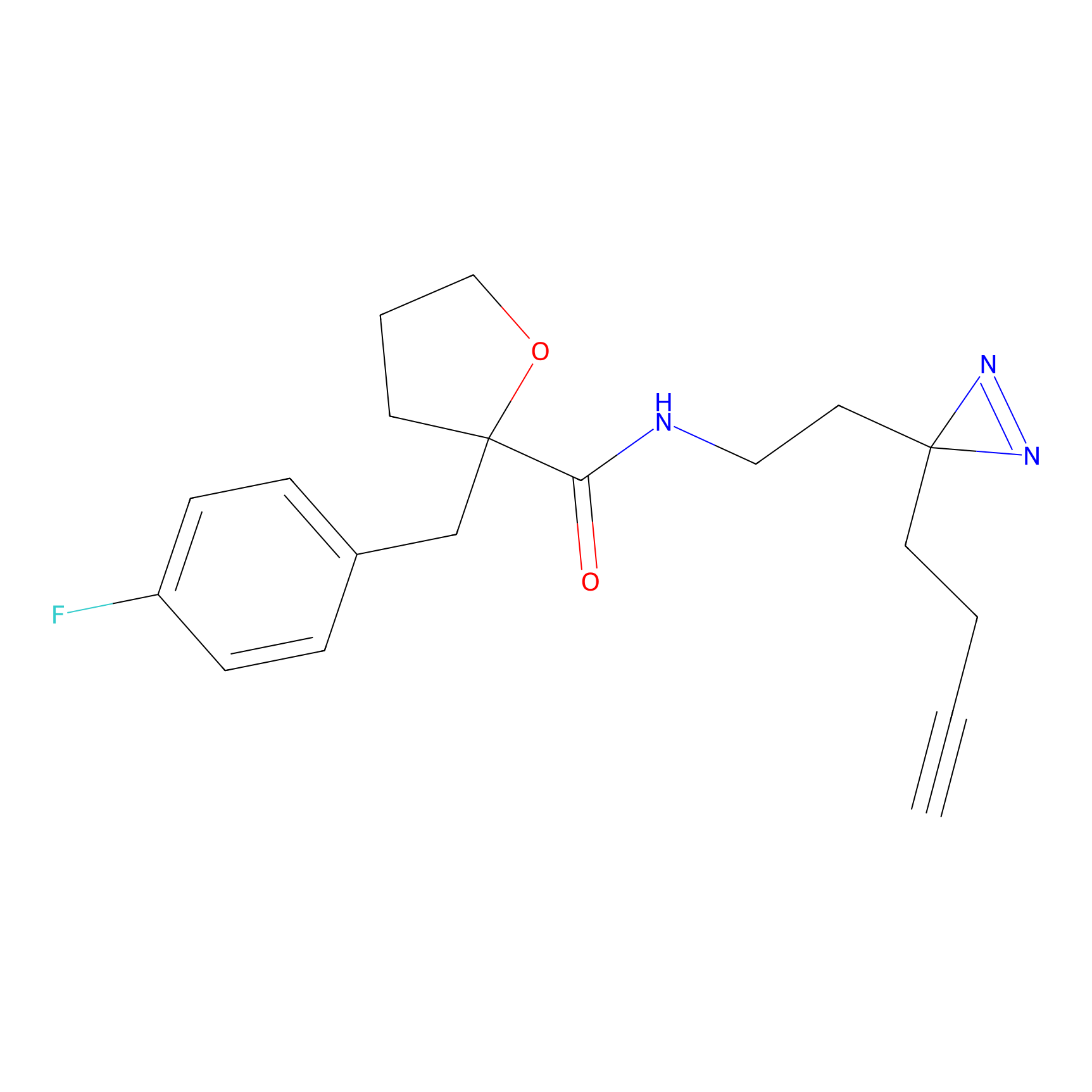
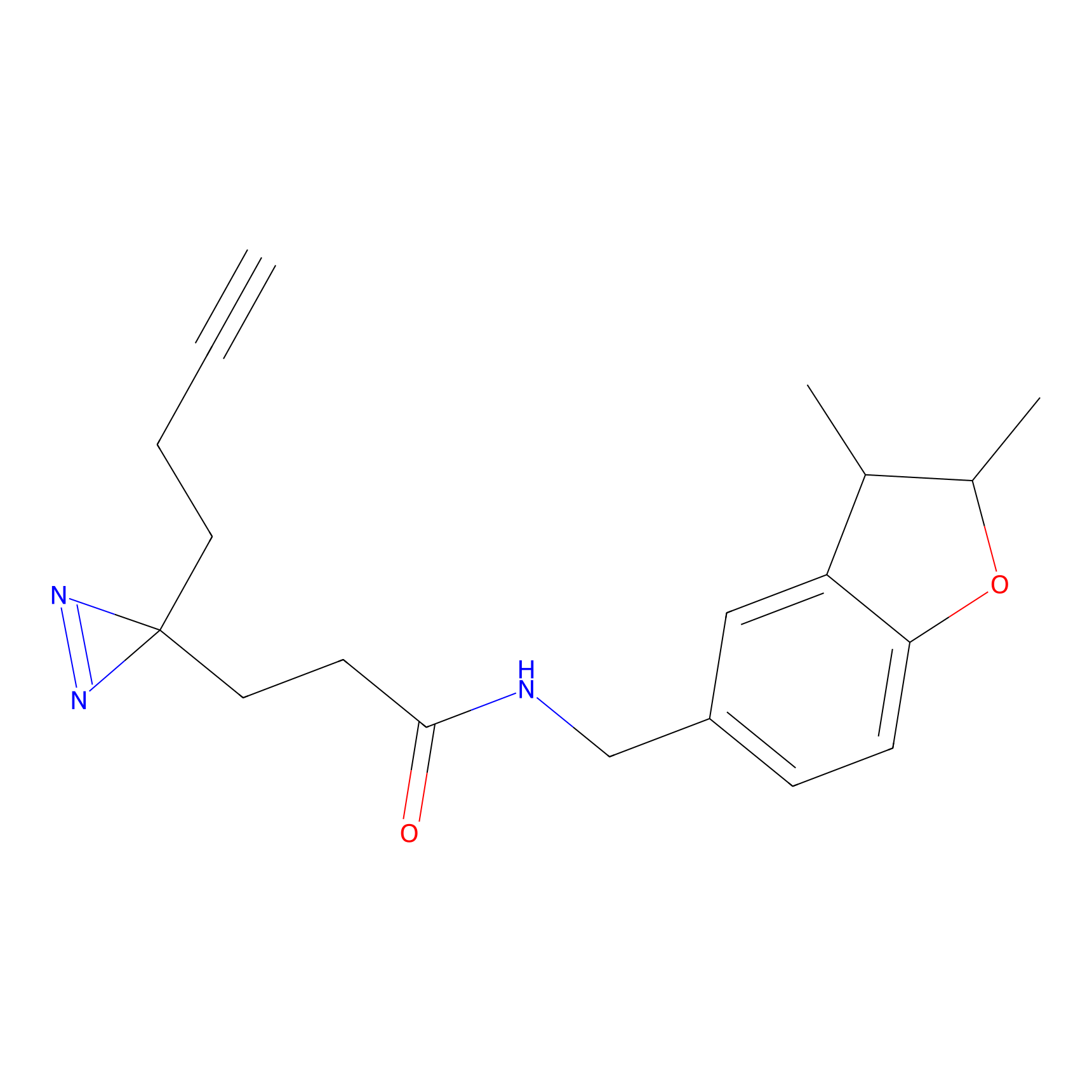
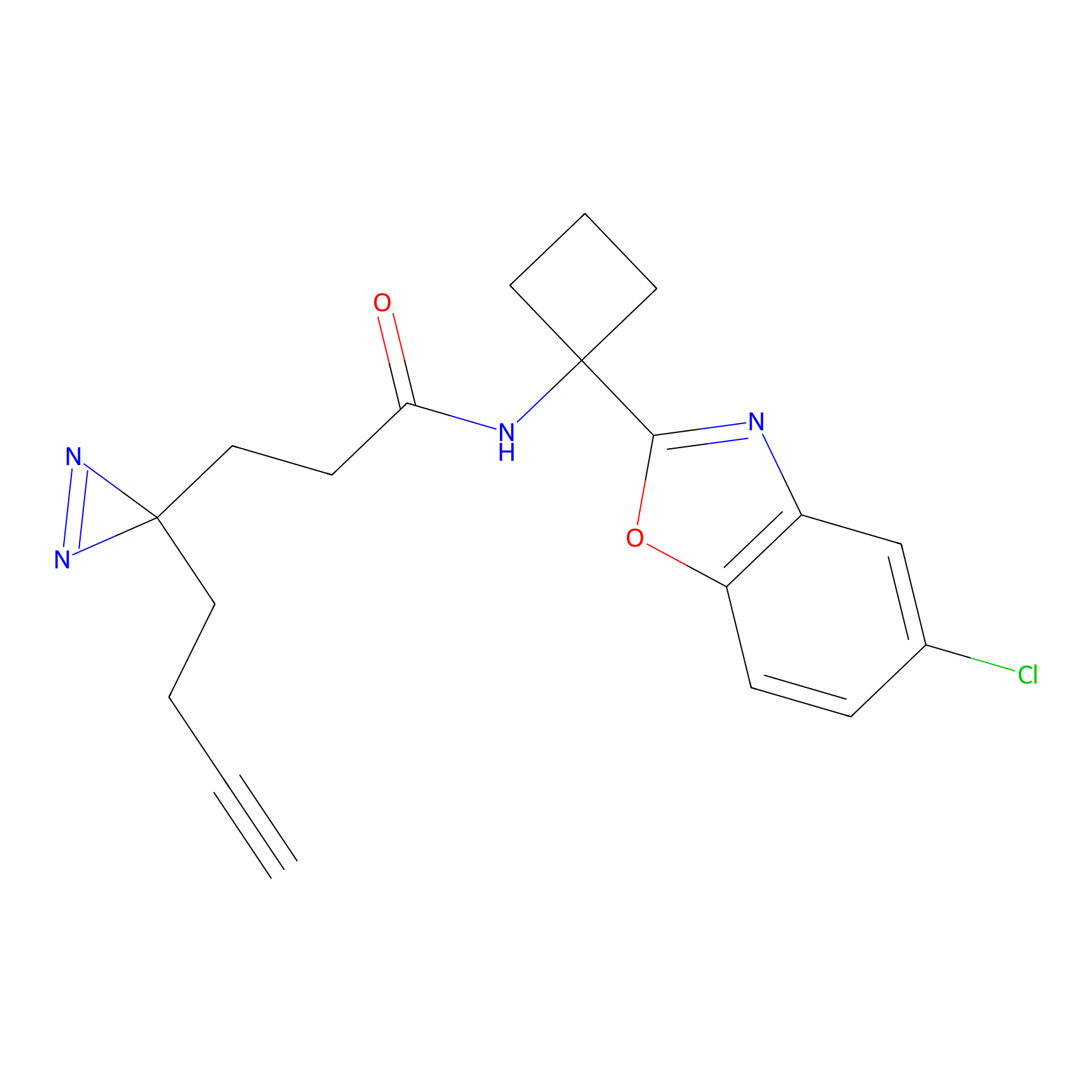
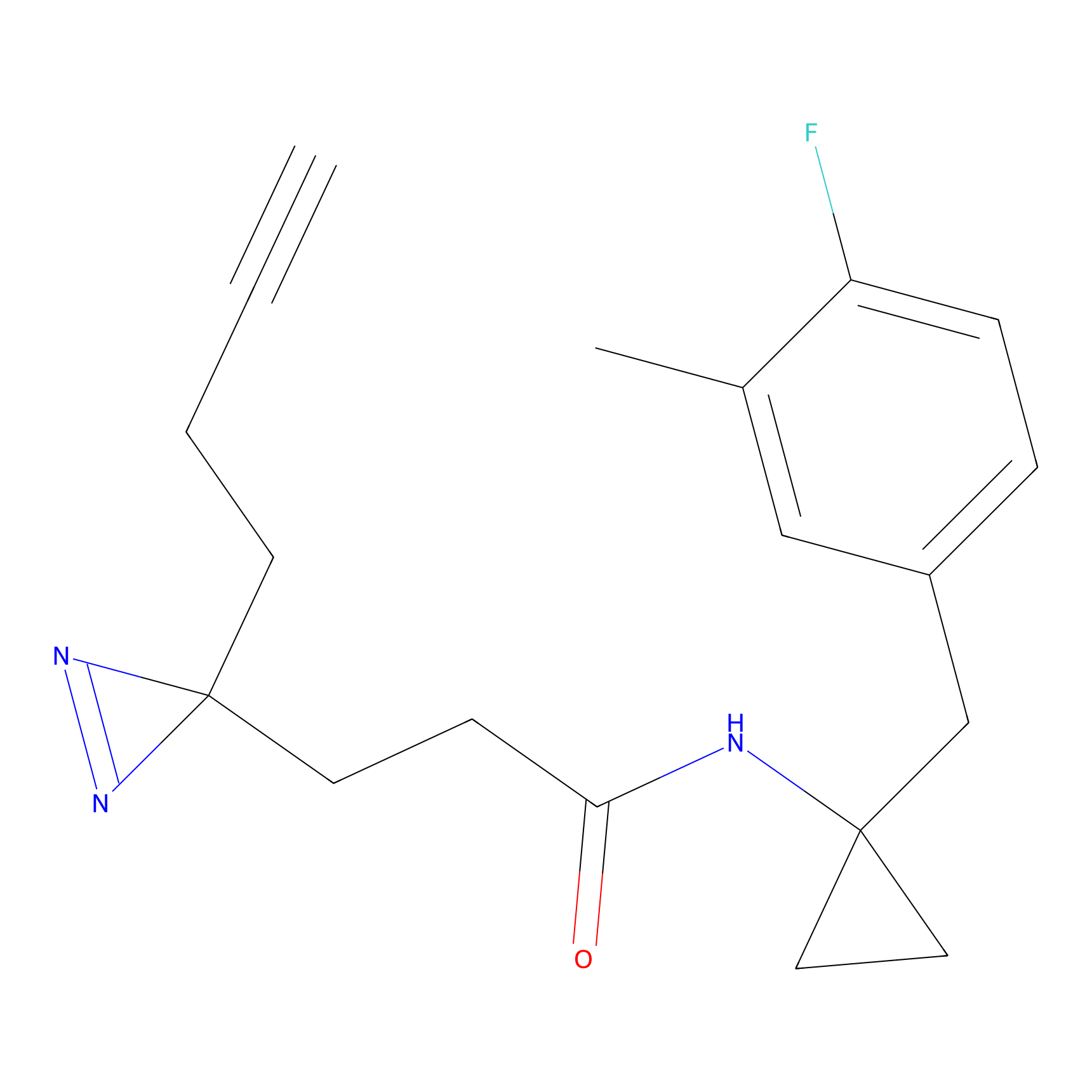
SF Probe Info |
 |
N.A. | LDD0028 | [3] | |
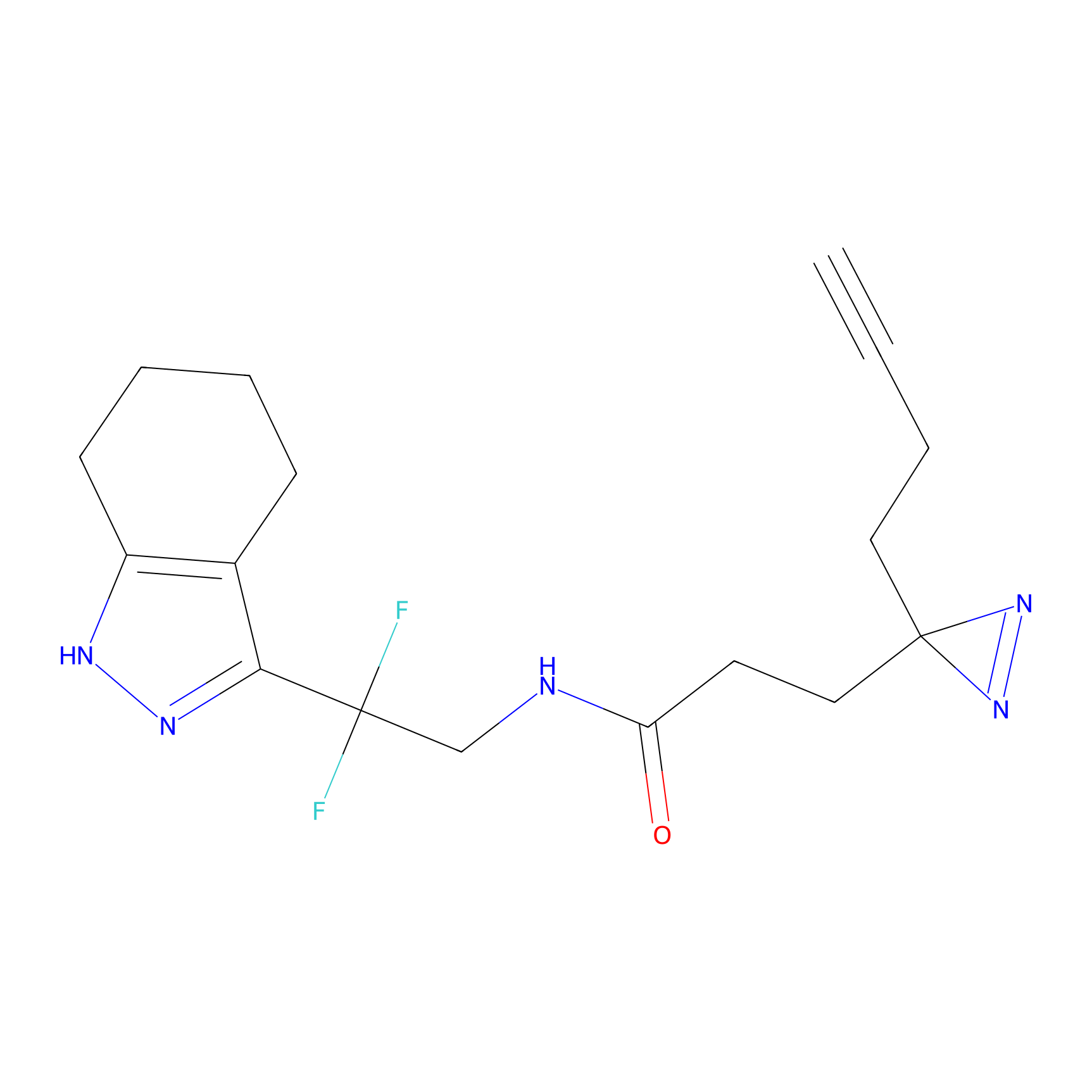
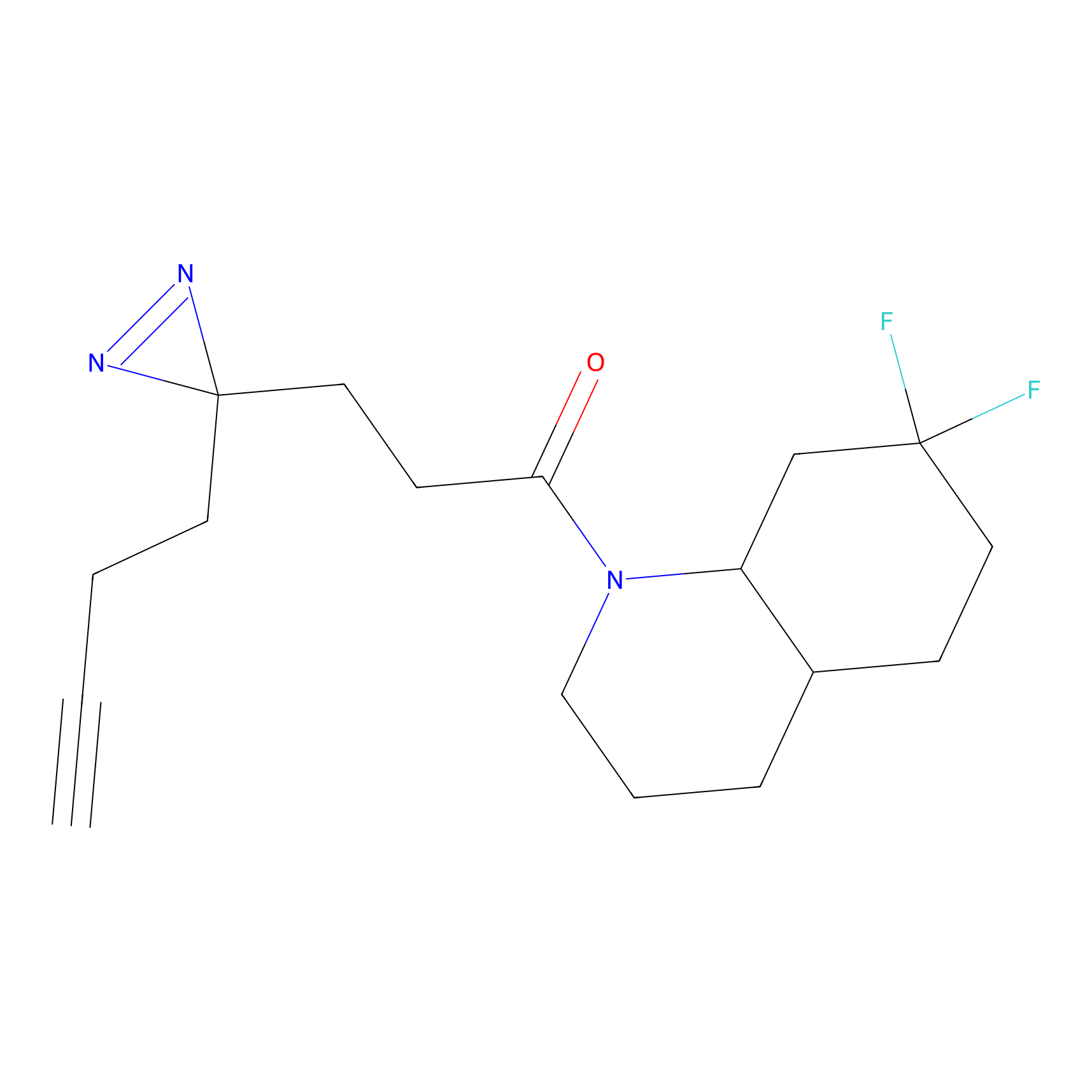
|
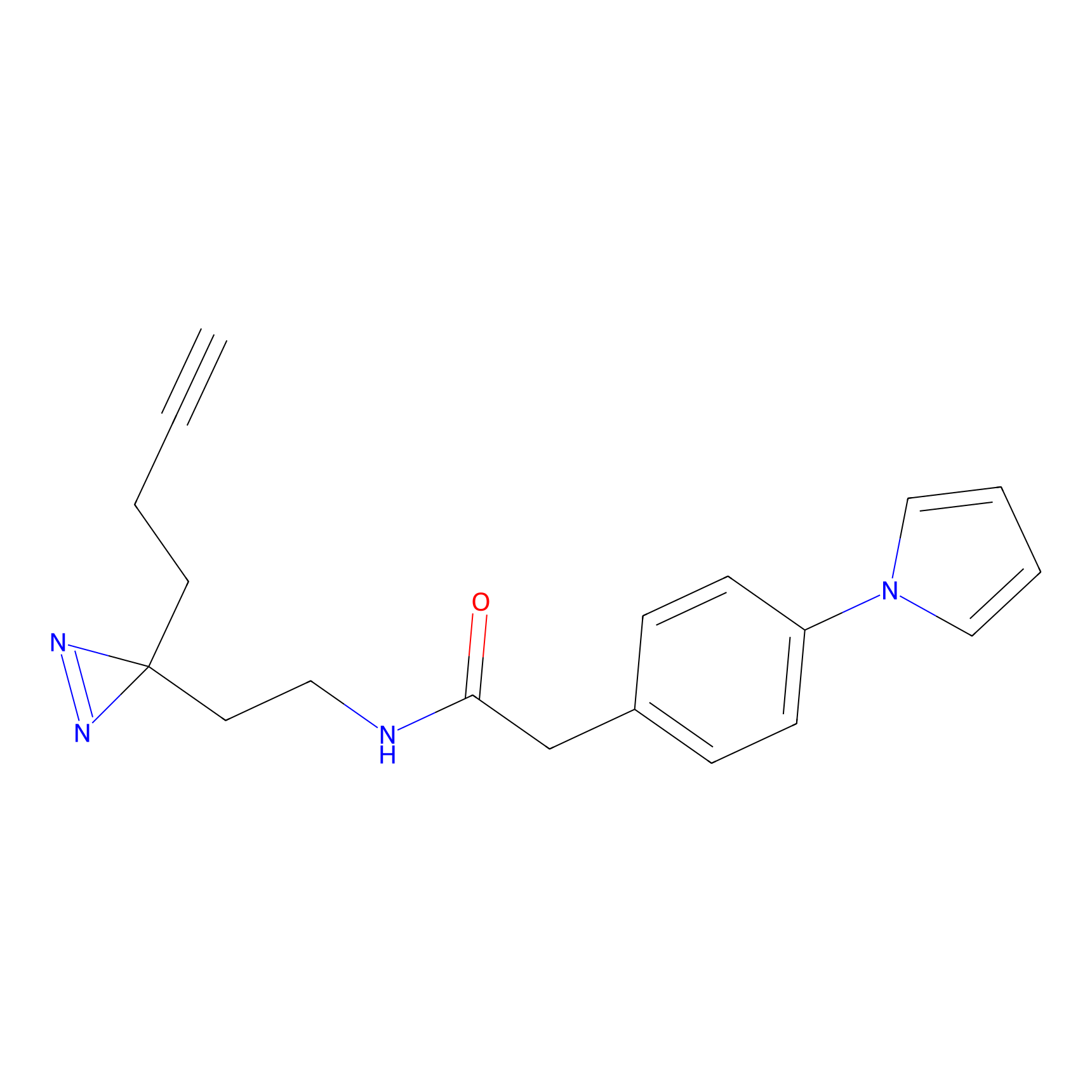
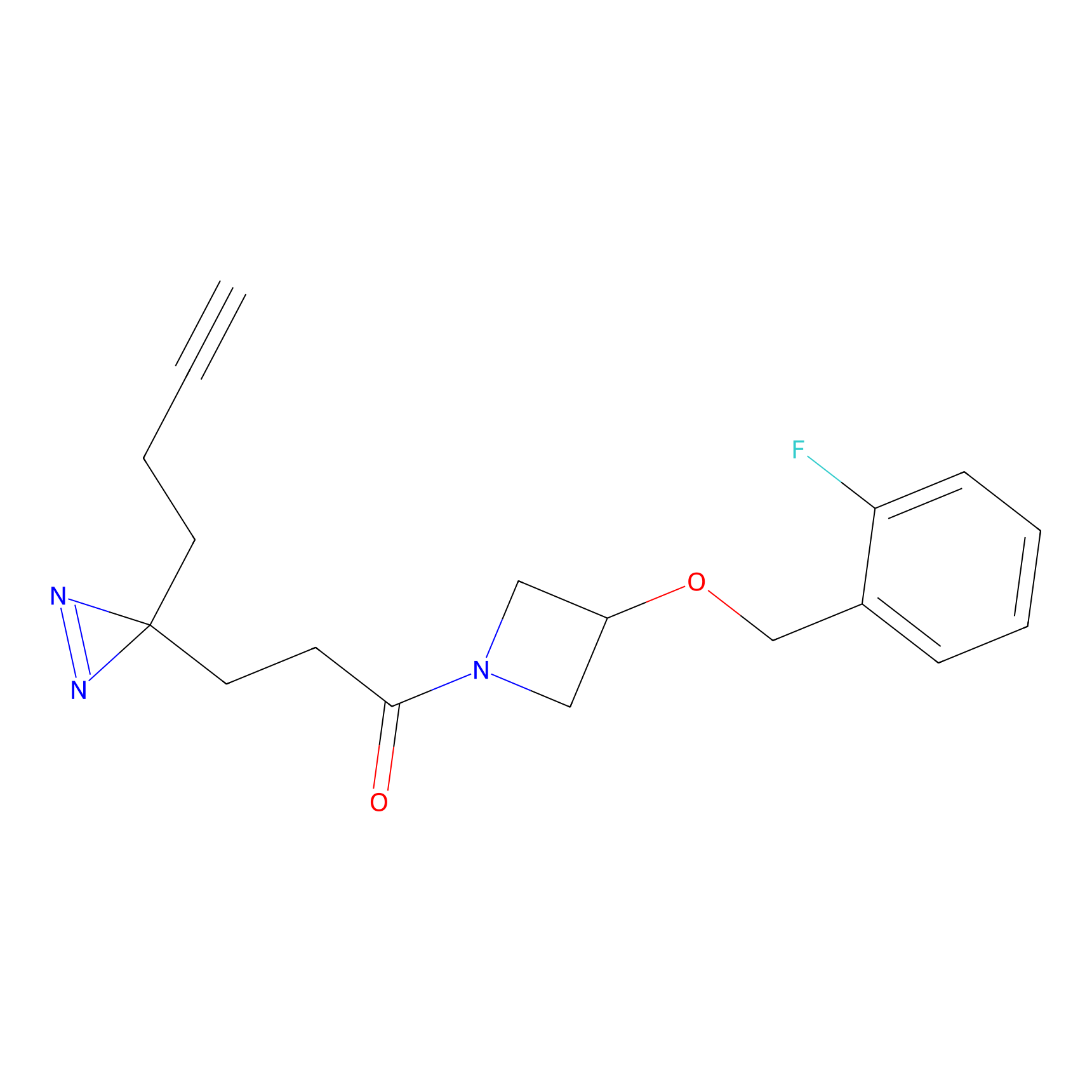
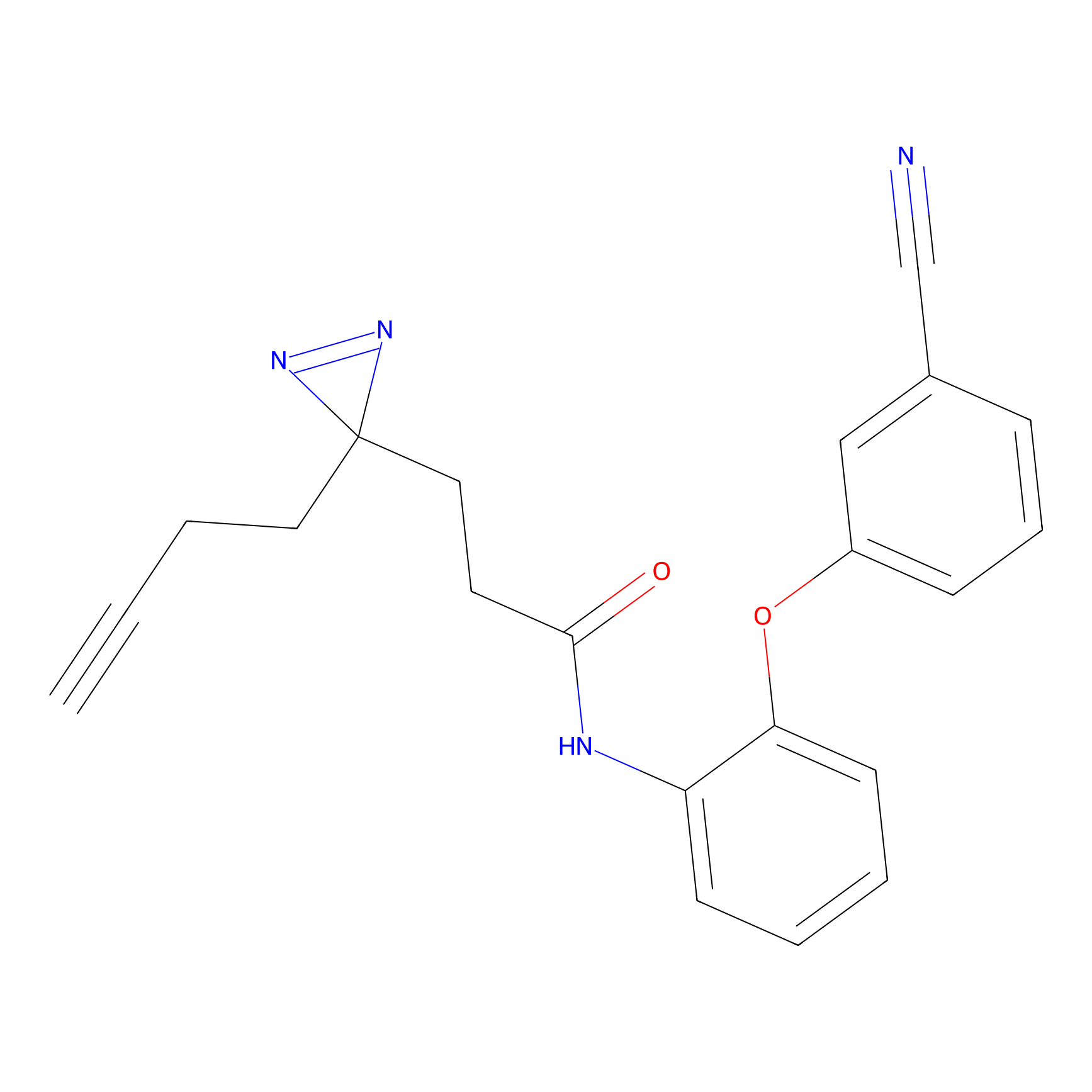
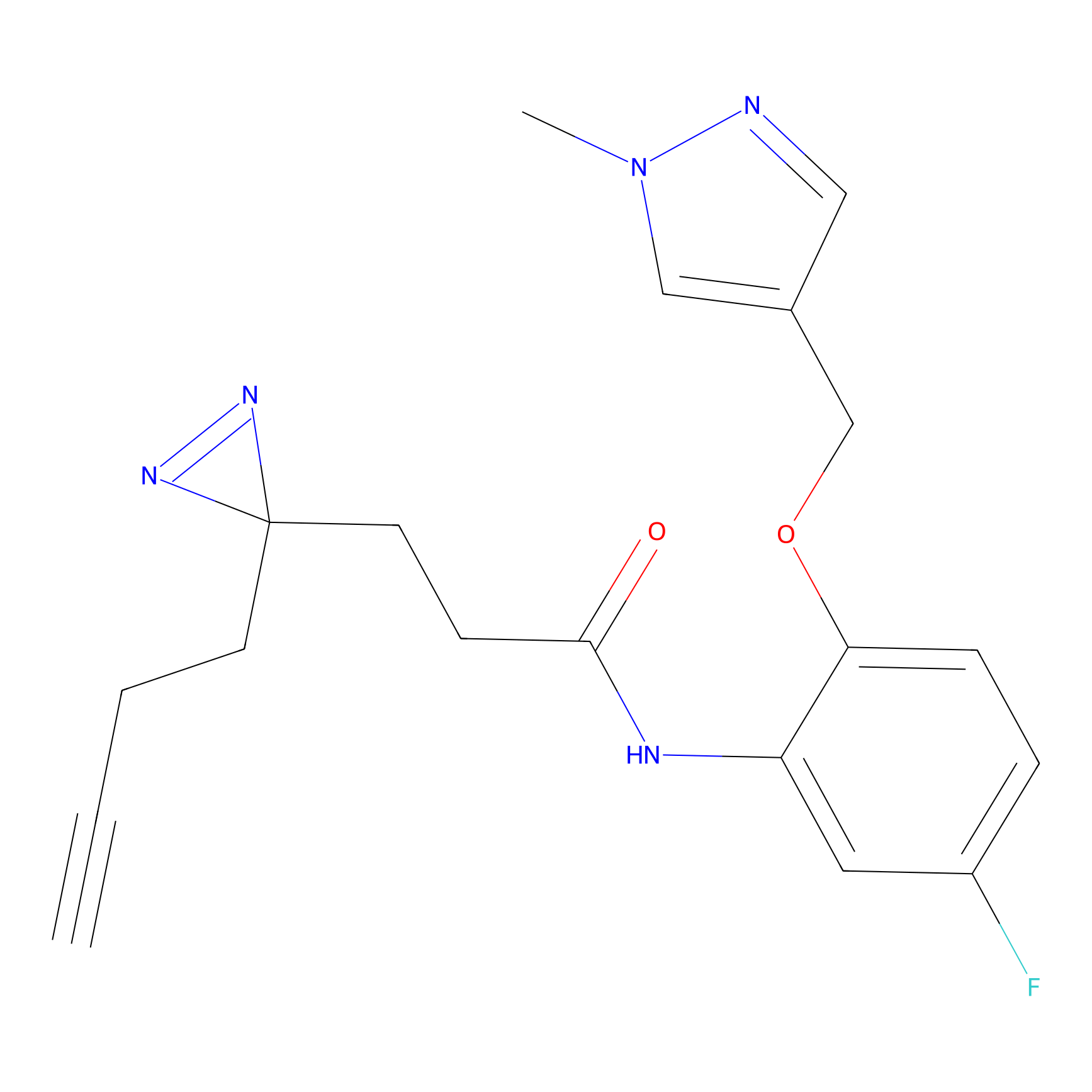
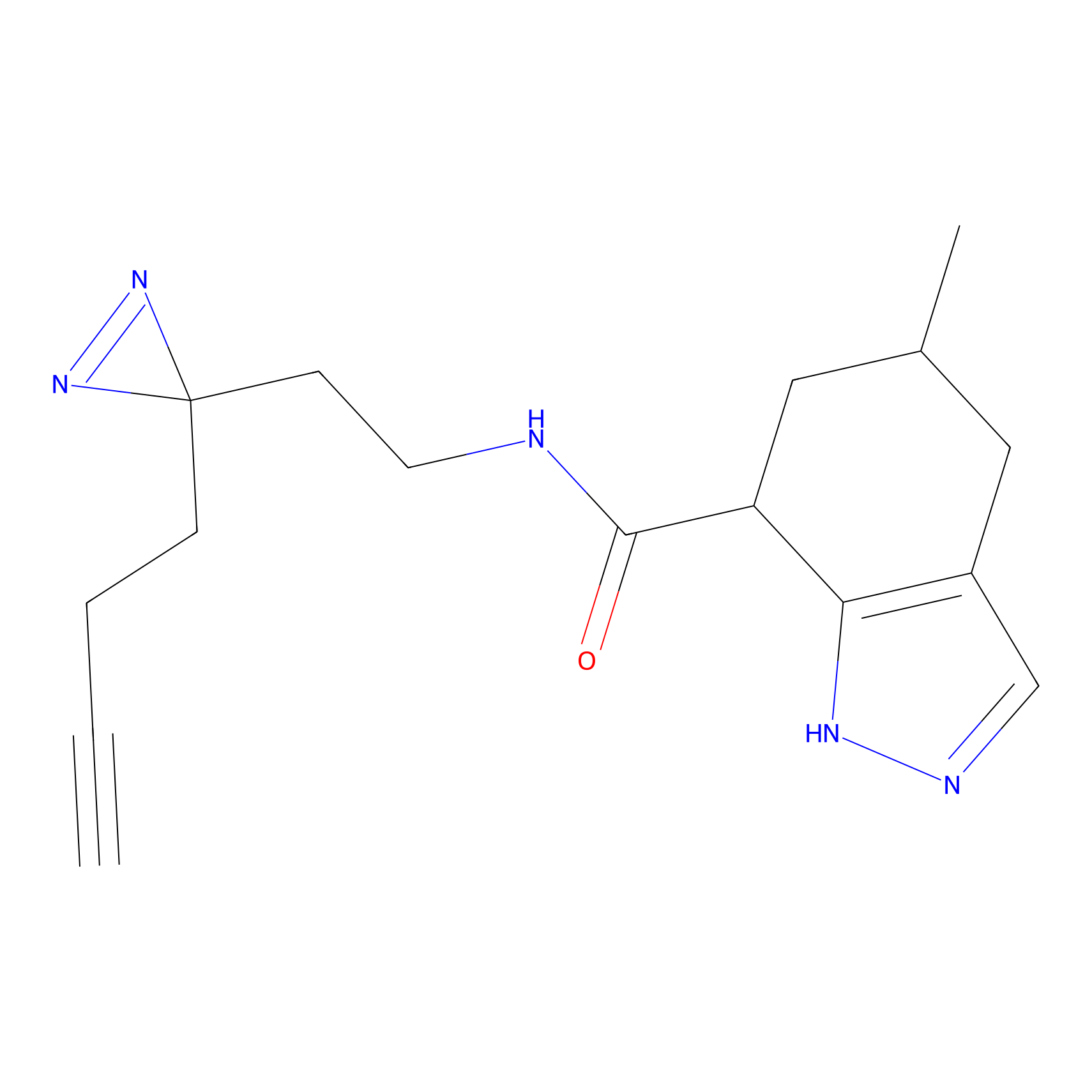
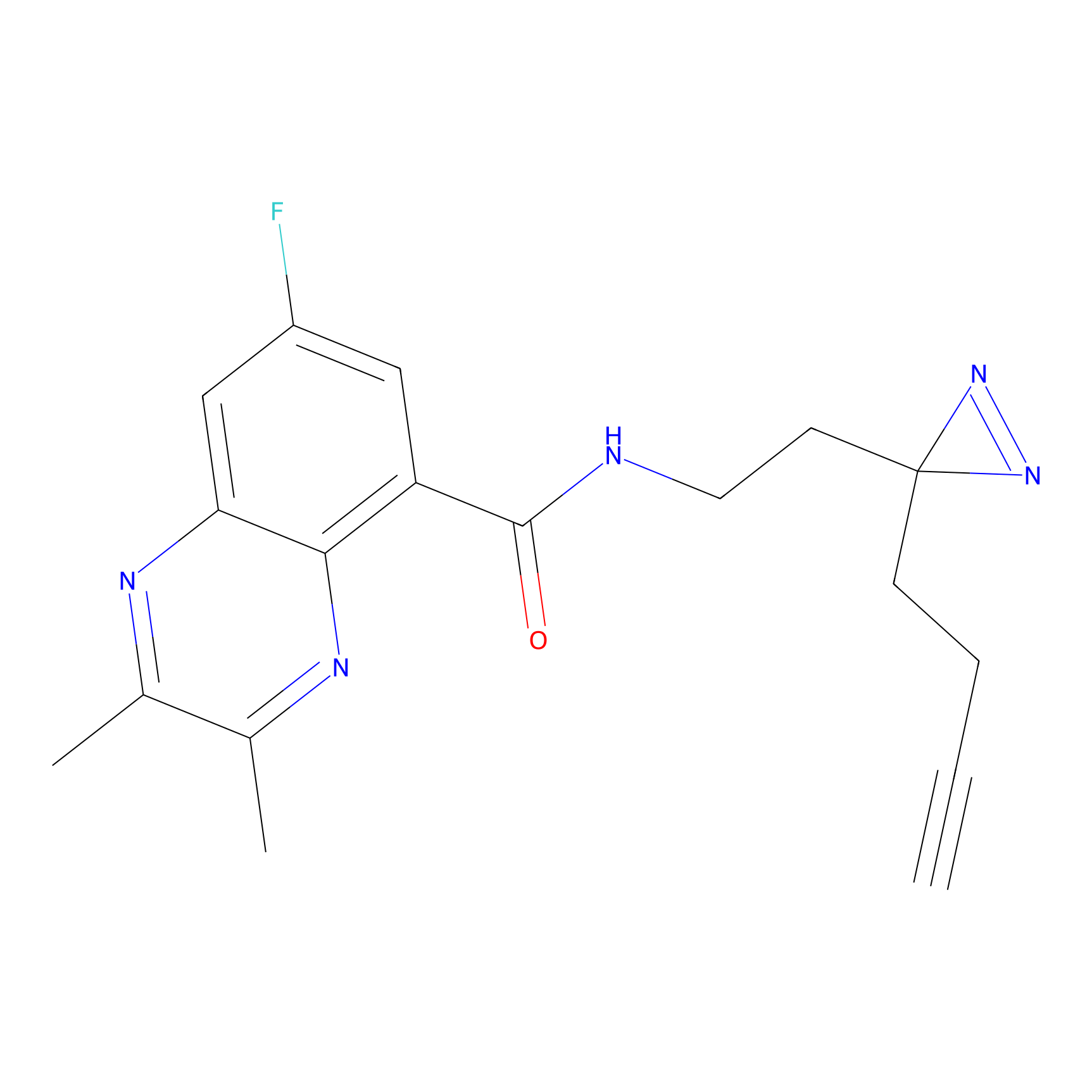
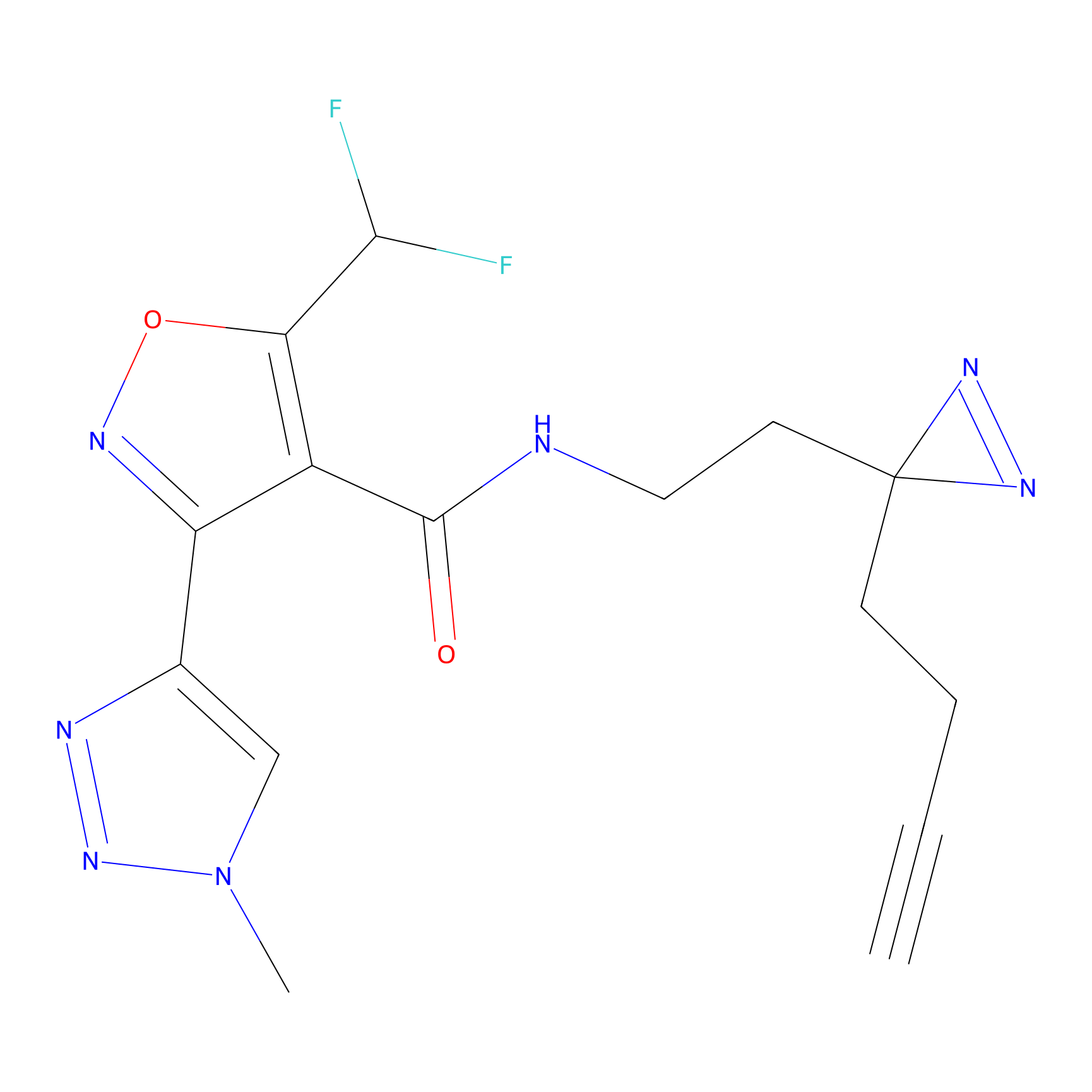
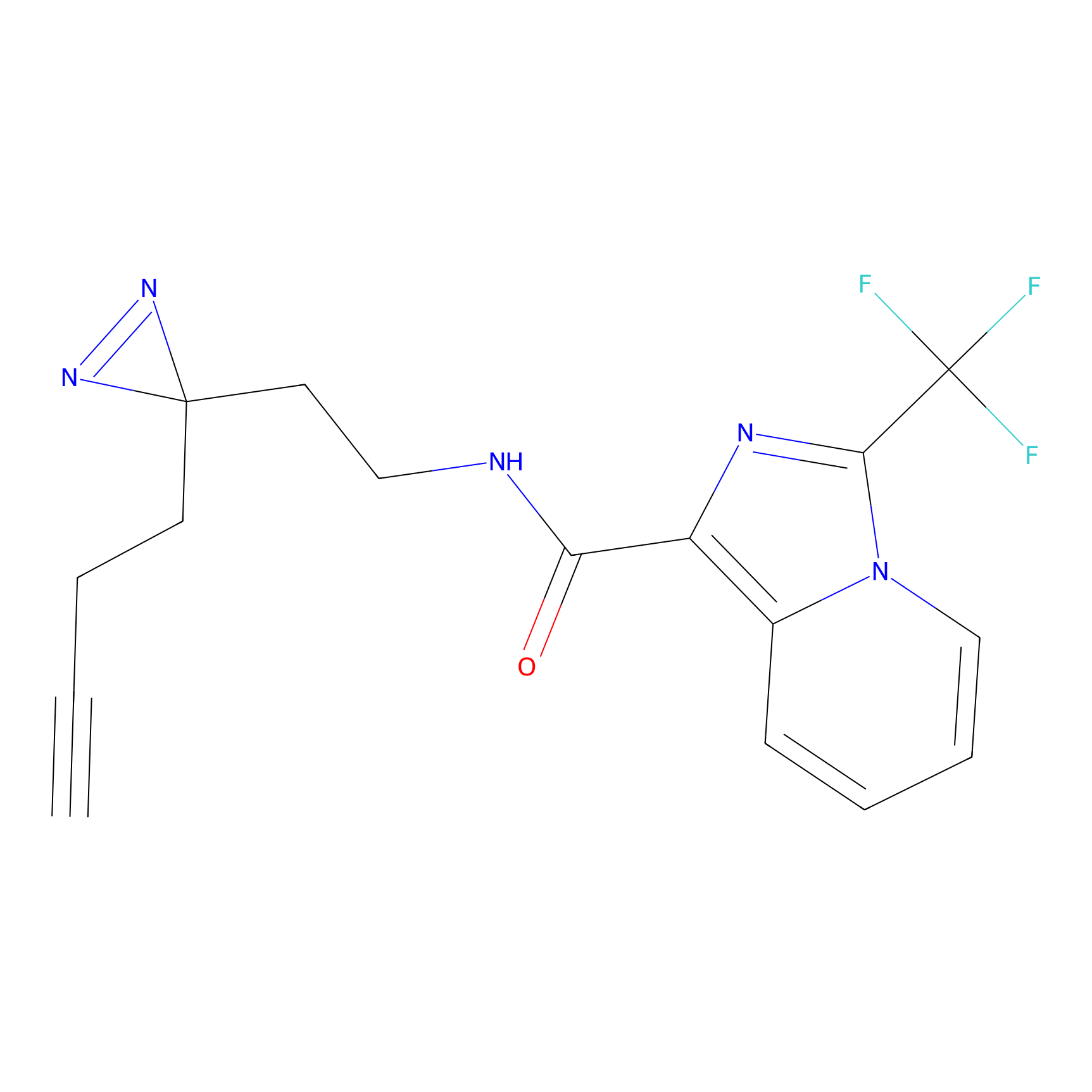
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [4] | |
|
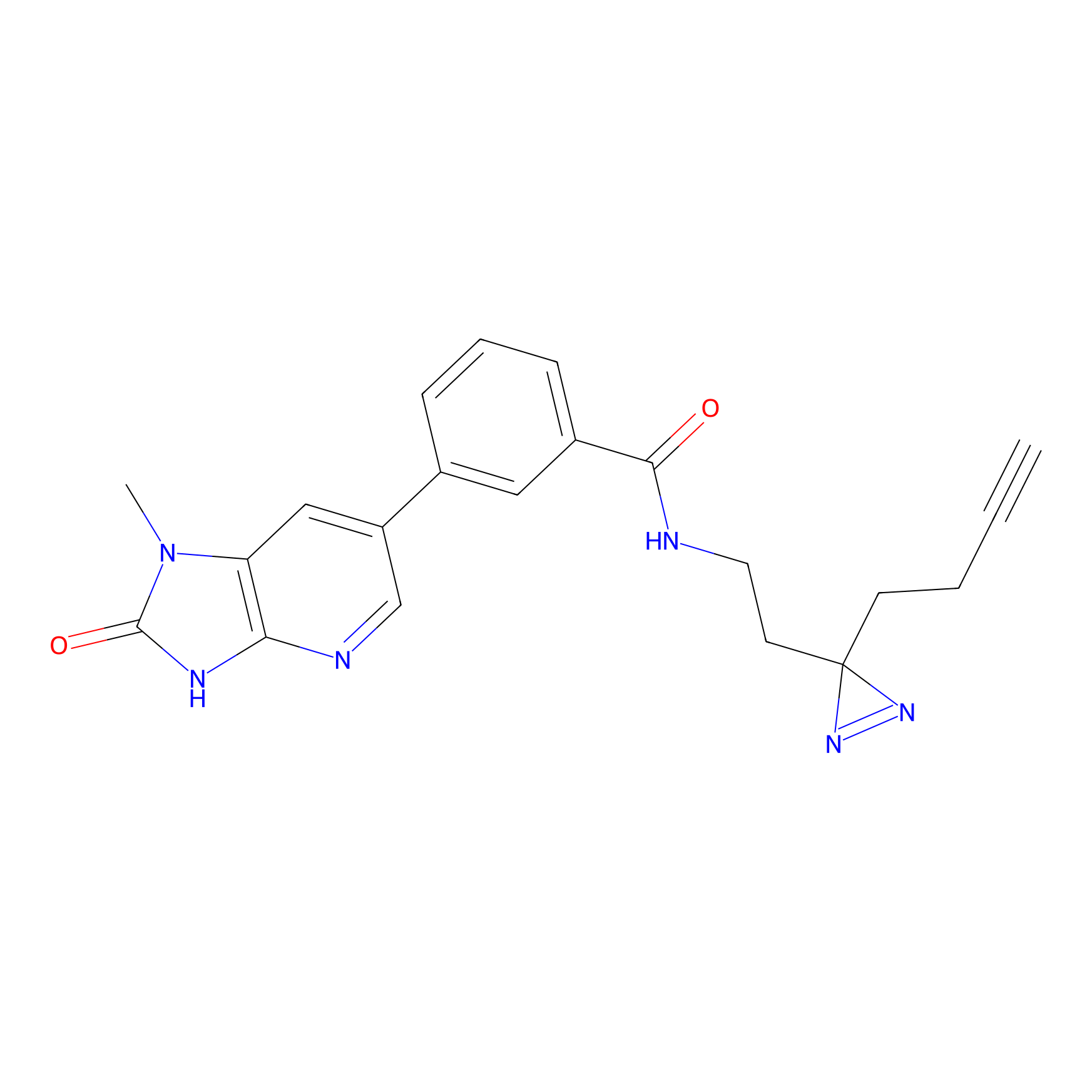
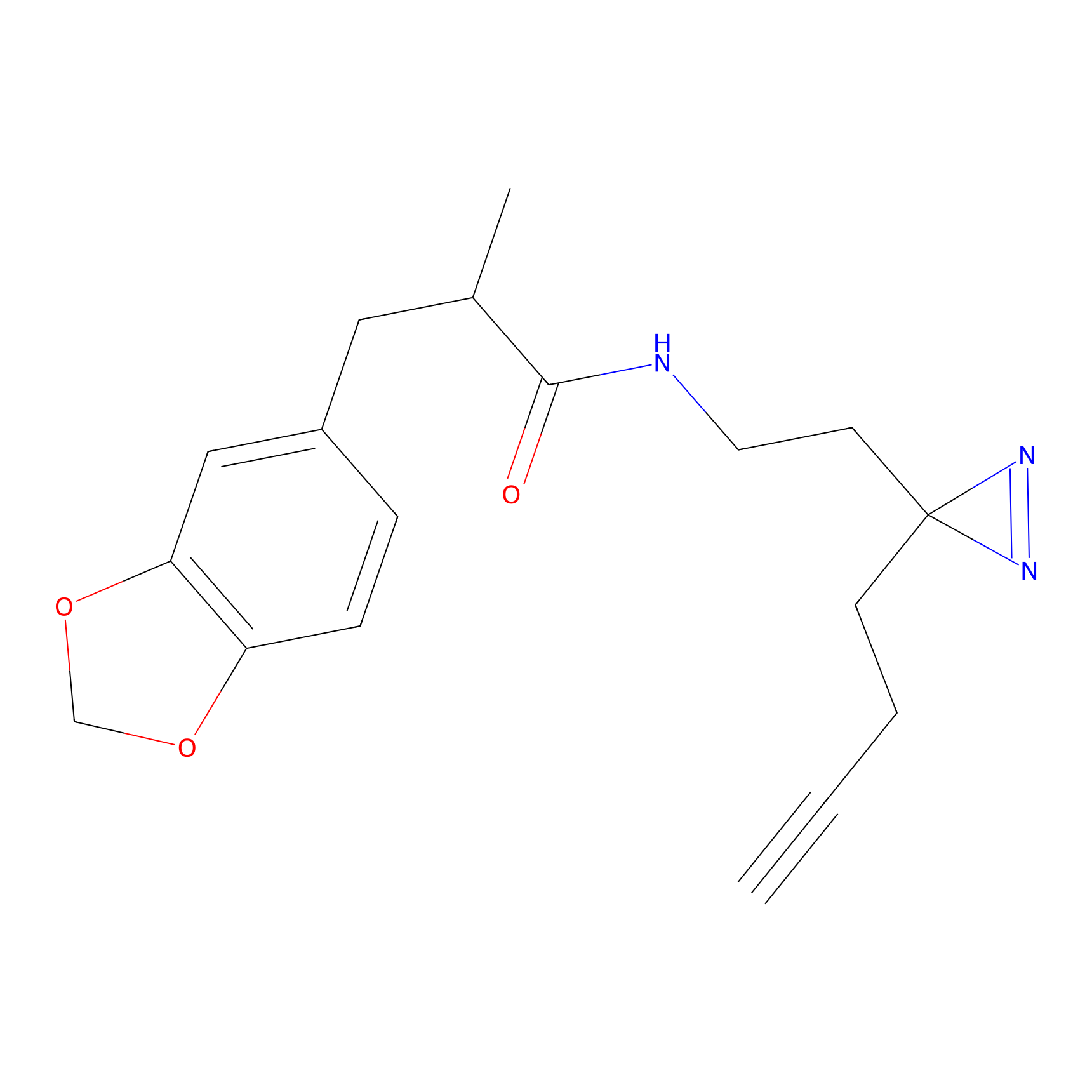
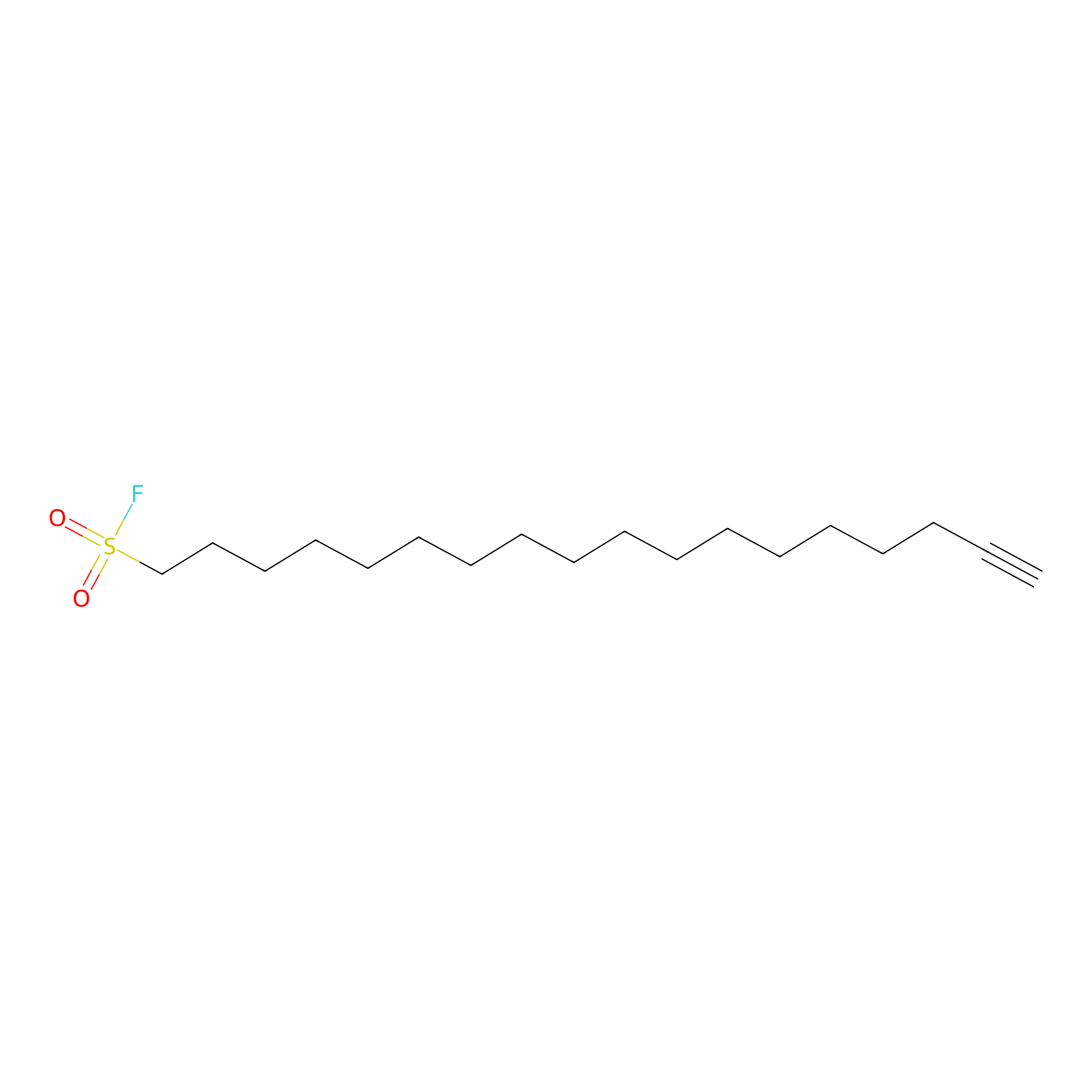
1c-yne Probe Info |
 |
N.A. | LDD0228 | [5] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [6] | |
|
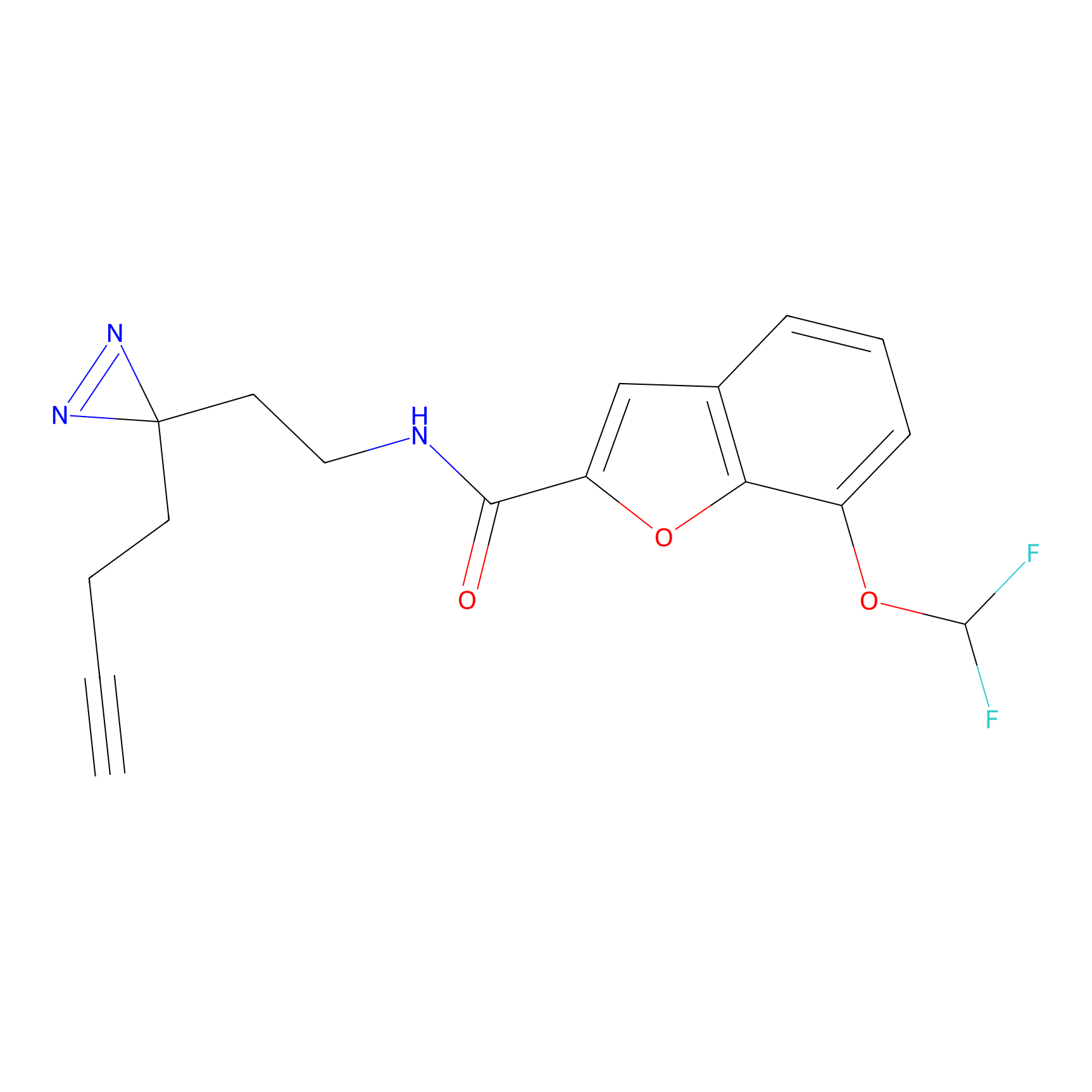
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [7] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
GPCR
Other
References