Details of the Target
General Information of Target
| Target ID | LDTP03284 | |||||
|---|---|---|---|---|---|---|
| Target Name | ATP synthase F(0) complex subunit B1, mitochondrial (ATP5PB) | |||||
| Gene Name | ATP5PB | |||||
| Gene ID | 515 | |||||
| Synonyms |
ATP5F1; ATP synthase F(0) complex subunit B1, mitochondrial; ATP synthase peripheral stalk-membrane subunit b; ATP synthase proton-transporting mitochondrial F(0) complex subunit B1; ATP synthase subunit b; ATPase subunit b
|
|||||
| 3D Structure | ||||||
| Sequence |
MLSRVVLSAAATAAPSLKNAAFLGPGVLQATRTFHTGQPHLVPVPPLPEYGGKVRYGLIP
EEFFQFLYPKTGVTGPYVLGTGLILYALSKEIYVISAETFTALSVLGVMVYGIKKYGPFV ADFADKLNEQKLAQLEEAKQASIQHIQNAIDTEKSQQALVQKRHYLFDVQRNNIAMALEV TYRERLYRVYKEVKNRLDYHISVQNMMRRKEQEHMINWVEKHVVQSISTQQEKETIAKCI ADLKLLAKKAQAQPVM |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
Eukaryotic ATPase B chain family
|
|||||
| Subcellular location |
Mitochondrion
|
|||||
| Function |
Mitochondrial membrane ATP synthase (F(1)F(0) ATP synthase or Complex V) produces ATP from ADP in the presence of a proton gradient across the membrane which is generated by electron transport complexes of the respiratory chain. F-type ATPases consist of two structural domains, F(1) - containing the extramembraneous catalytic core, and F(0) - containing the membrane proton channel, linked together by a central stalk and a peripheral stalk. During catalysis, ATP synthesis in the catalytic domain of F(1) is coupled via a rotary mechanism of the central stalk subunits to proton translocation. Part of the complex F(0) domain and the peripheric stalk, which acts as a stator to hold the catalytic alpha(3)beta(3) subcomplex and subunit a/ATP6 static relative to the rotary elements.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
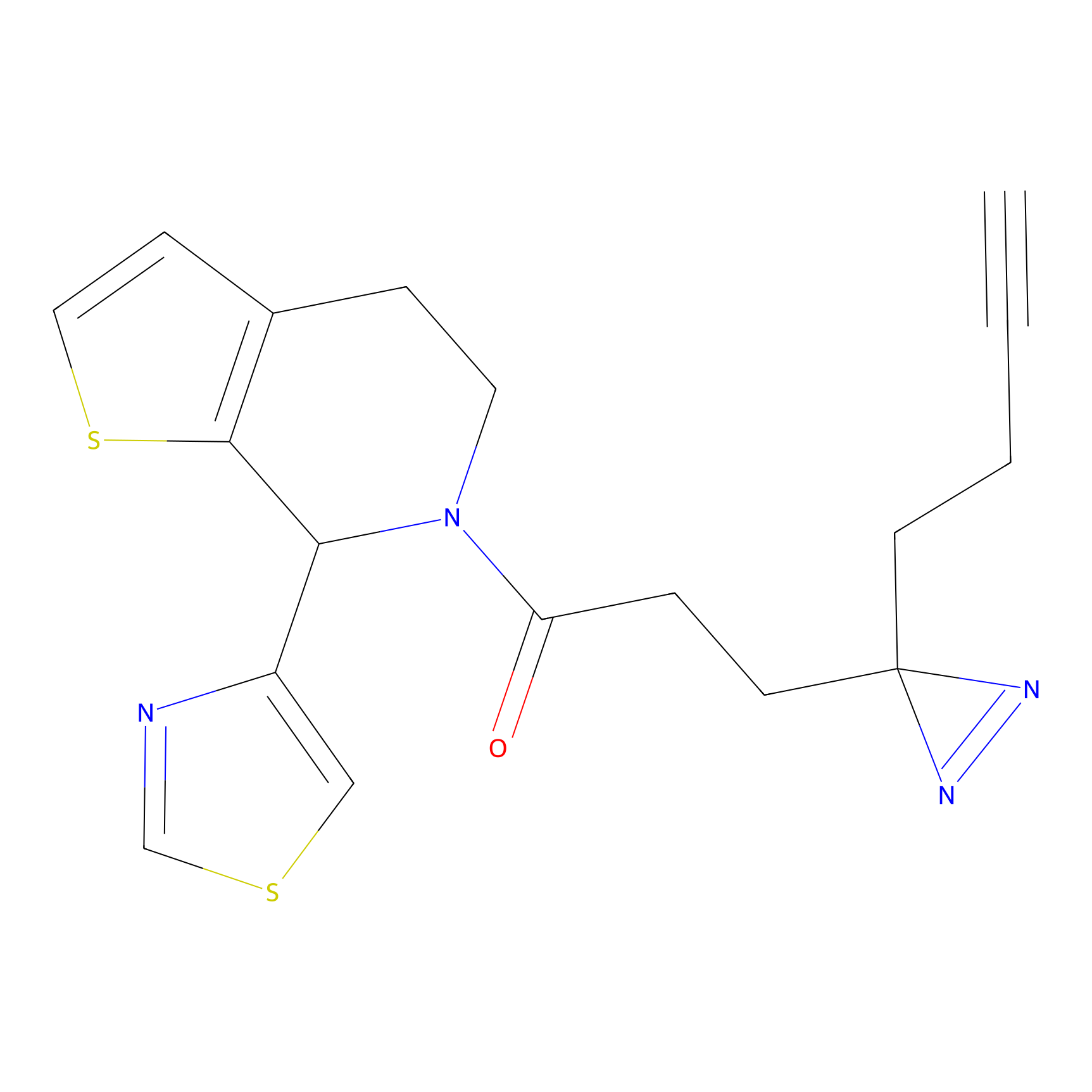
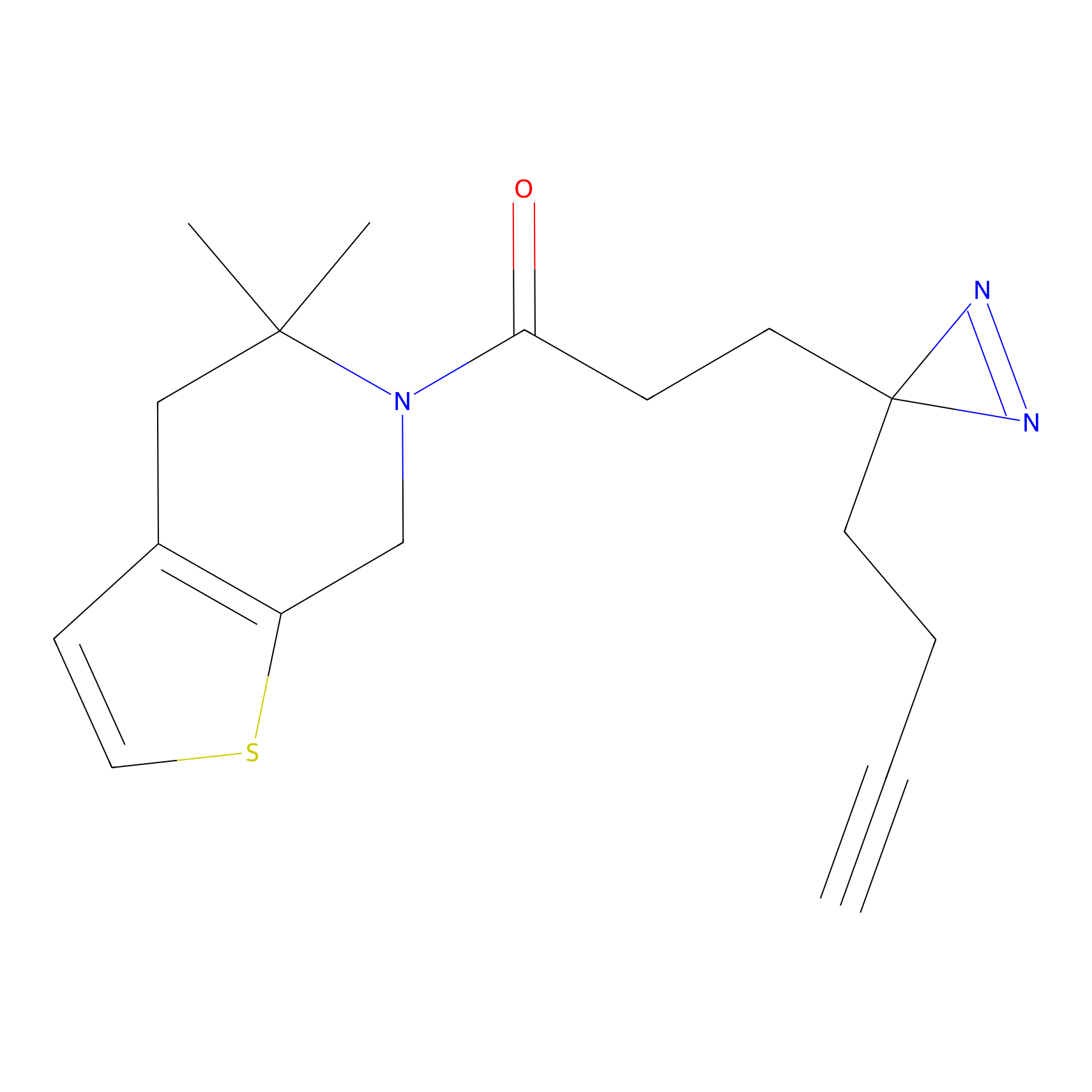
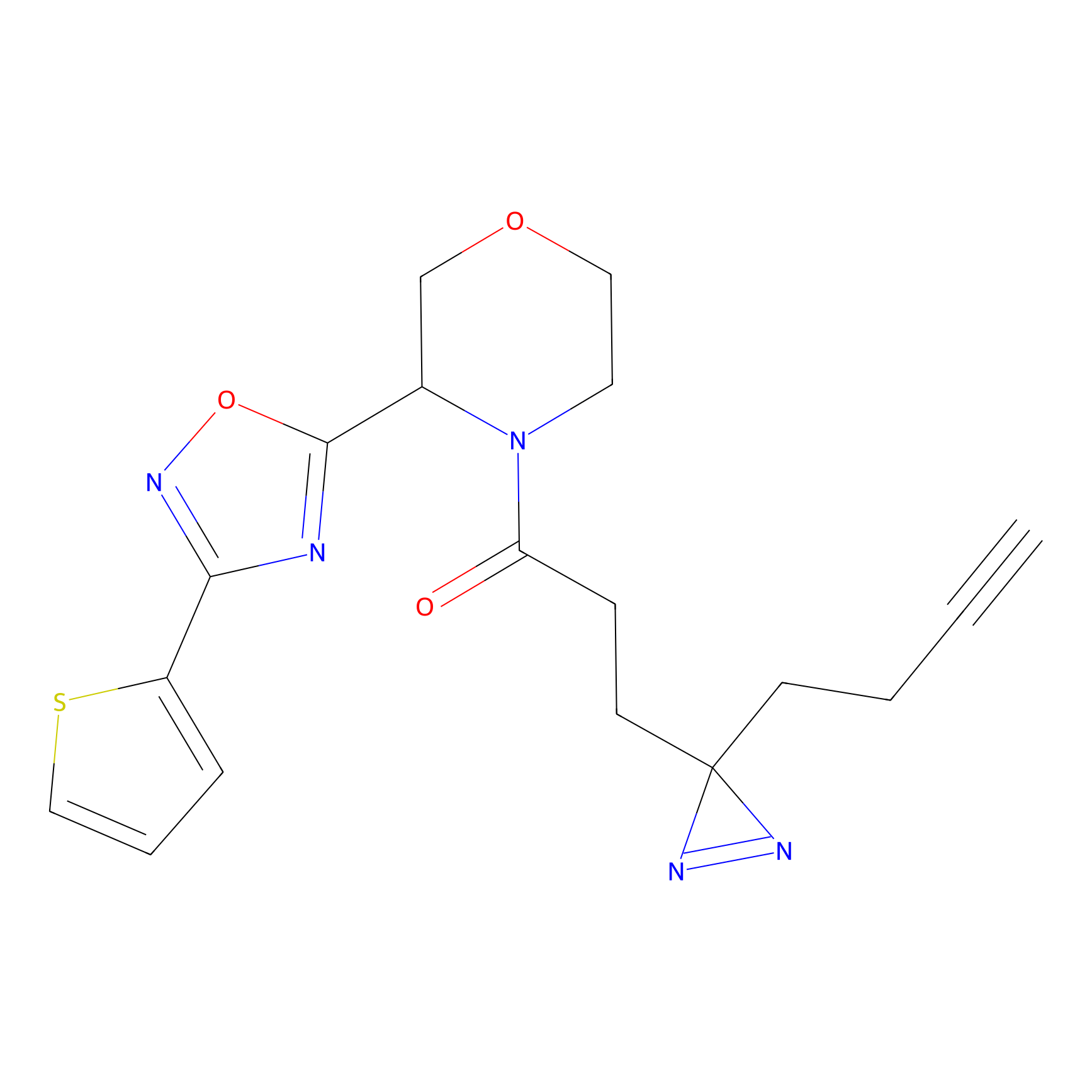
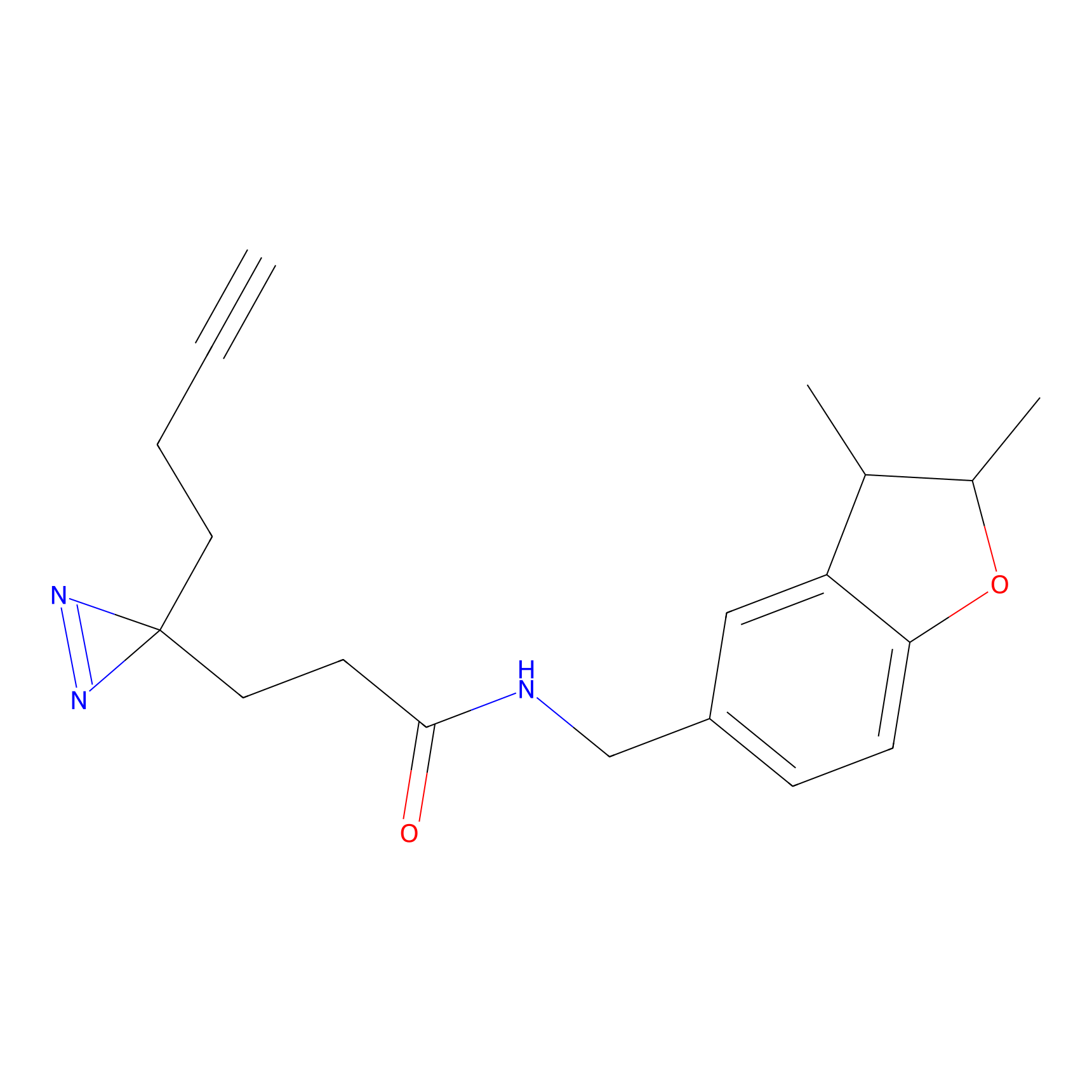
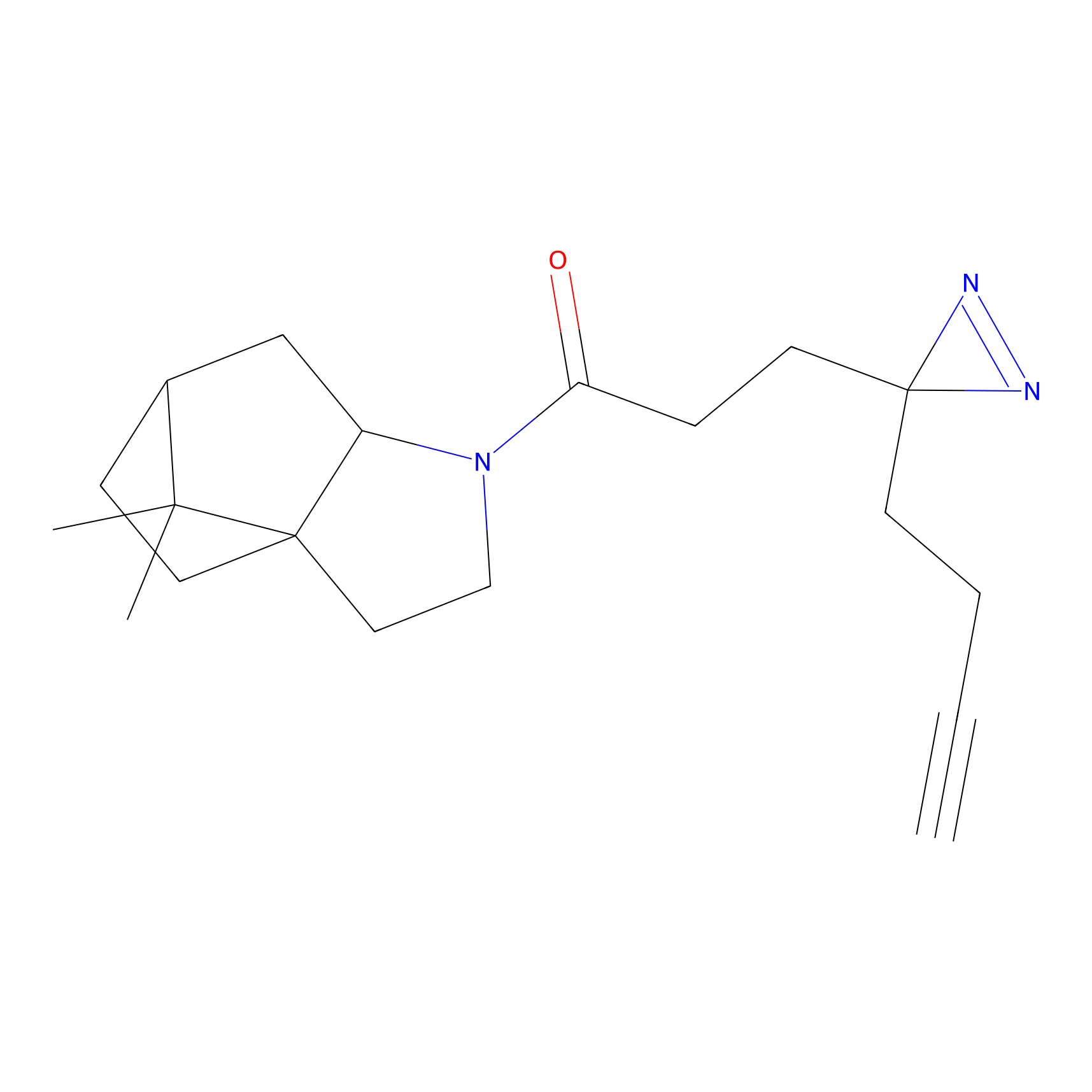
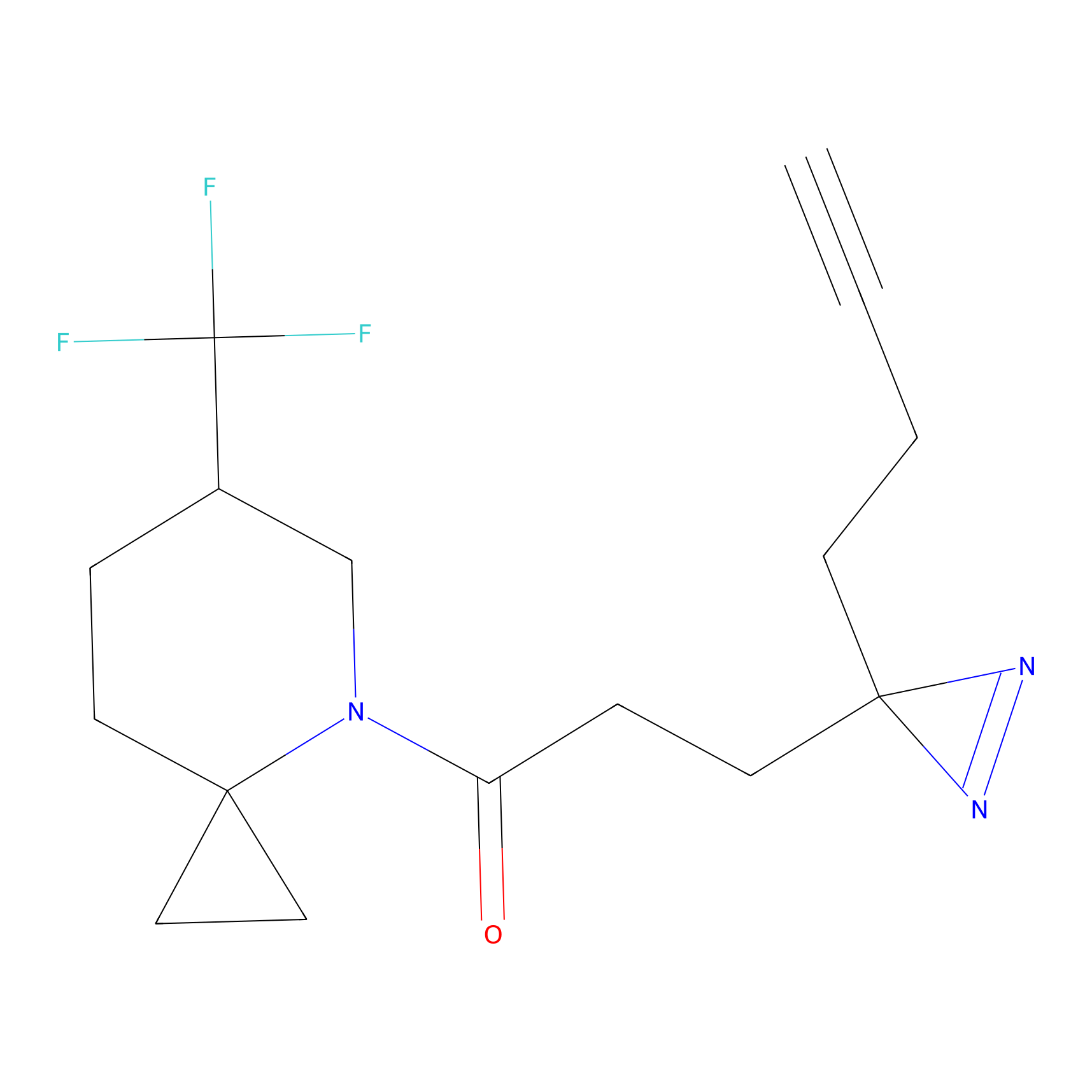
m-APA Probe Info |
 |
9.03 | LDD0402 | [2] | |
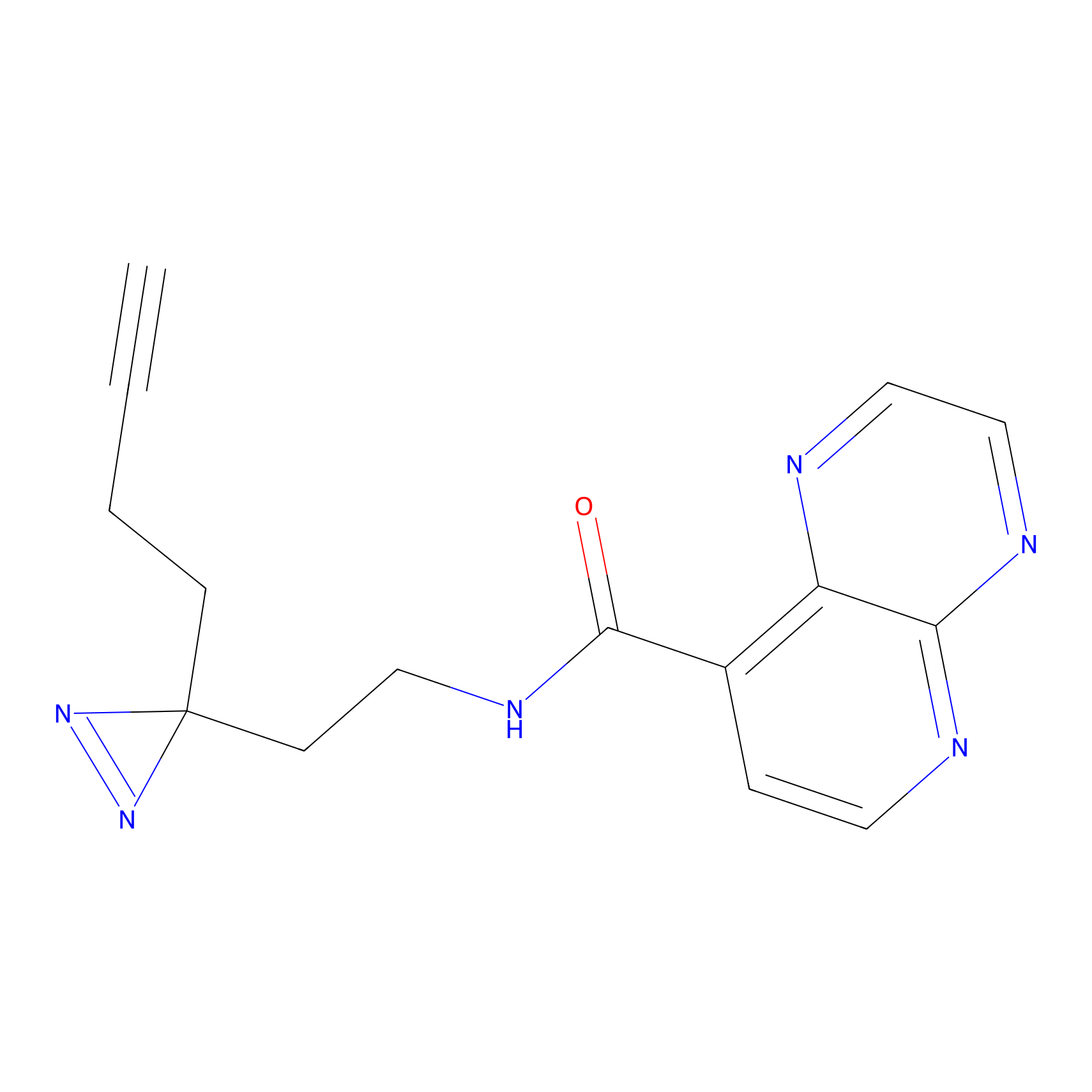
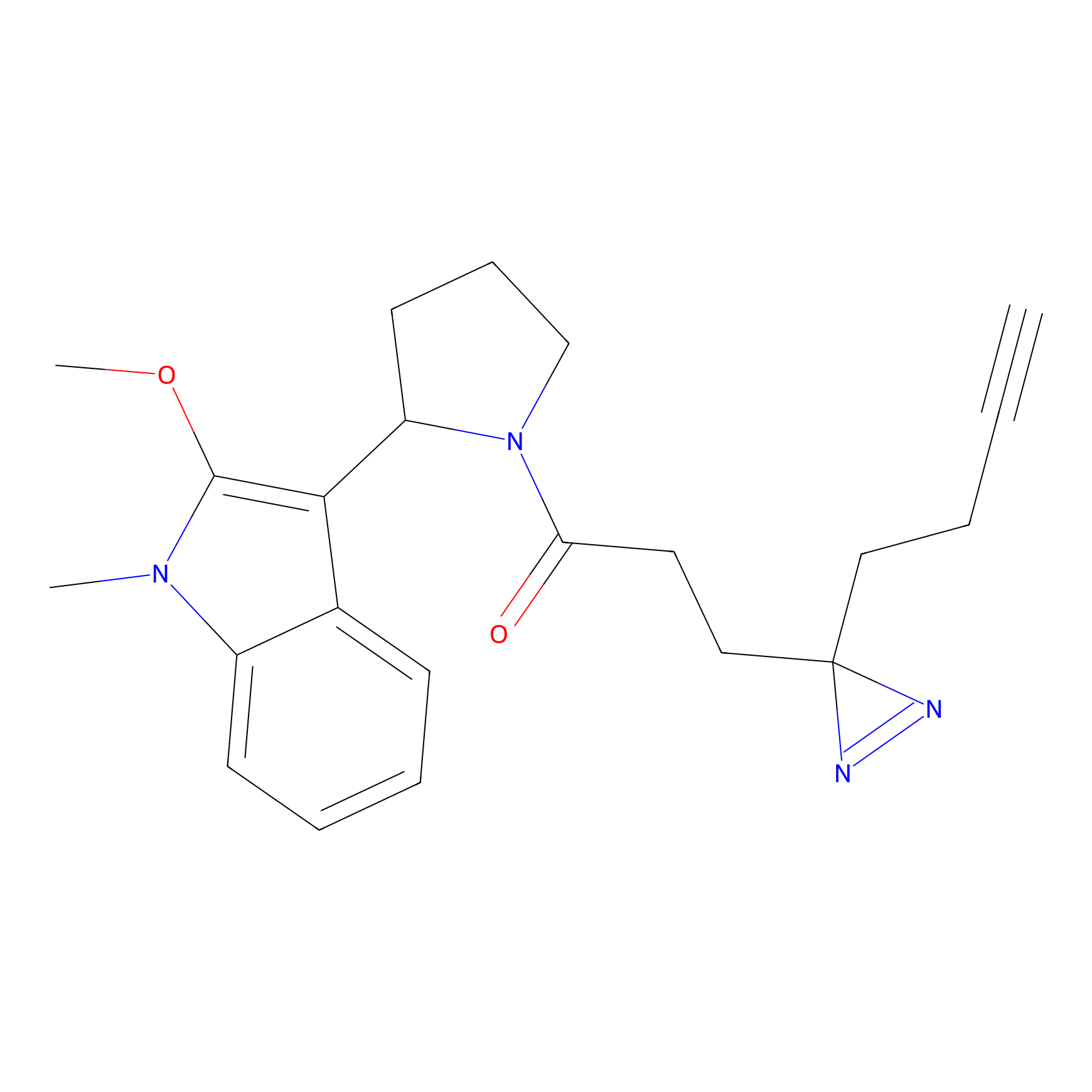
|
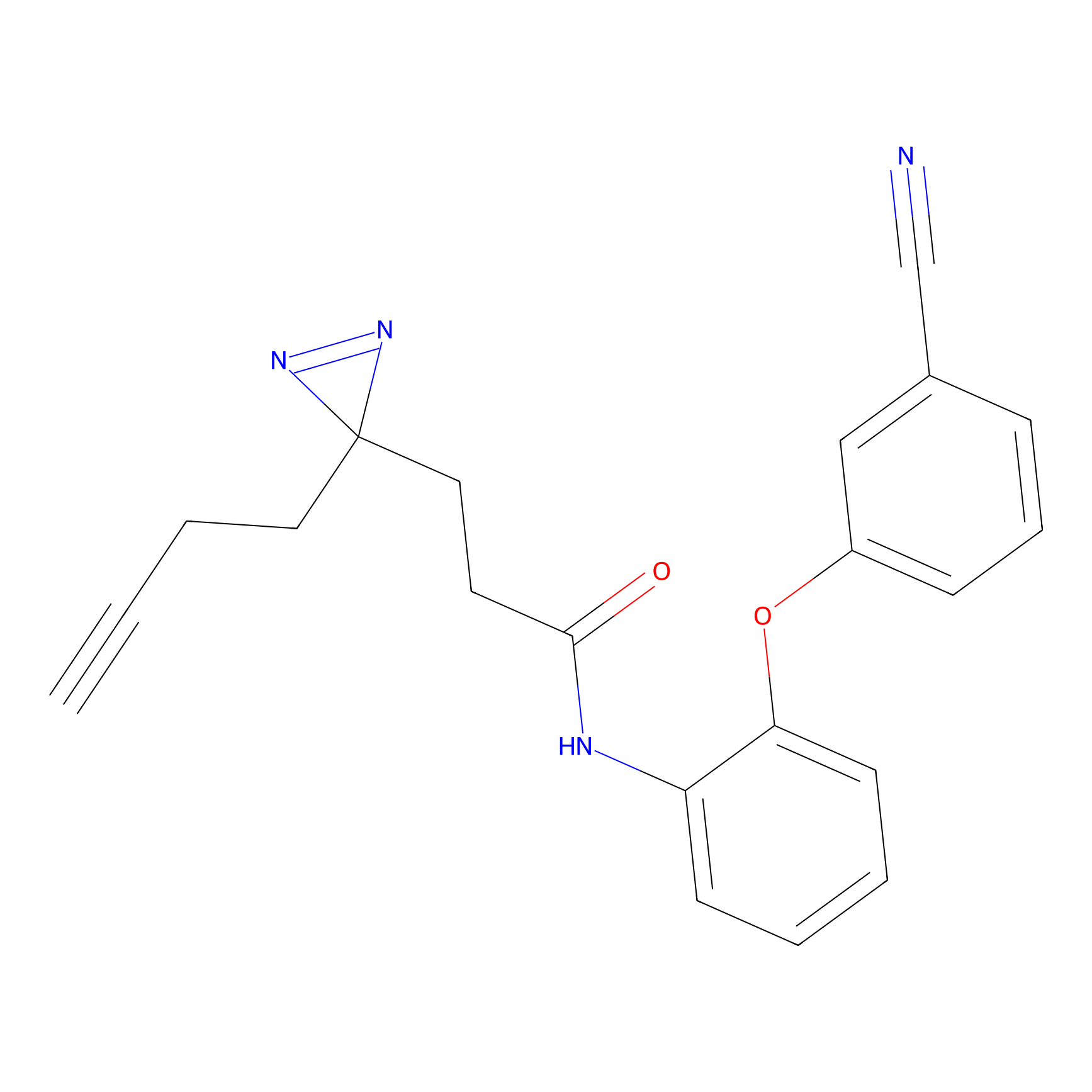
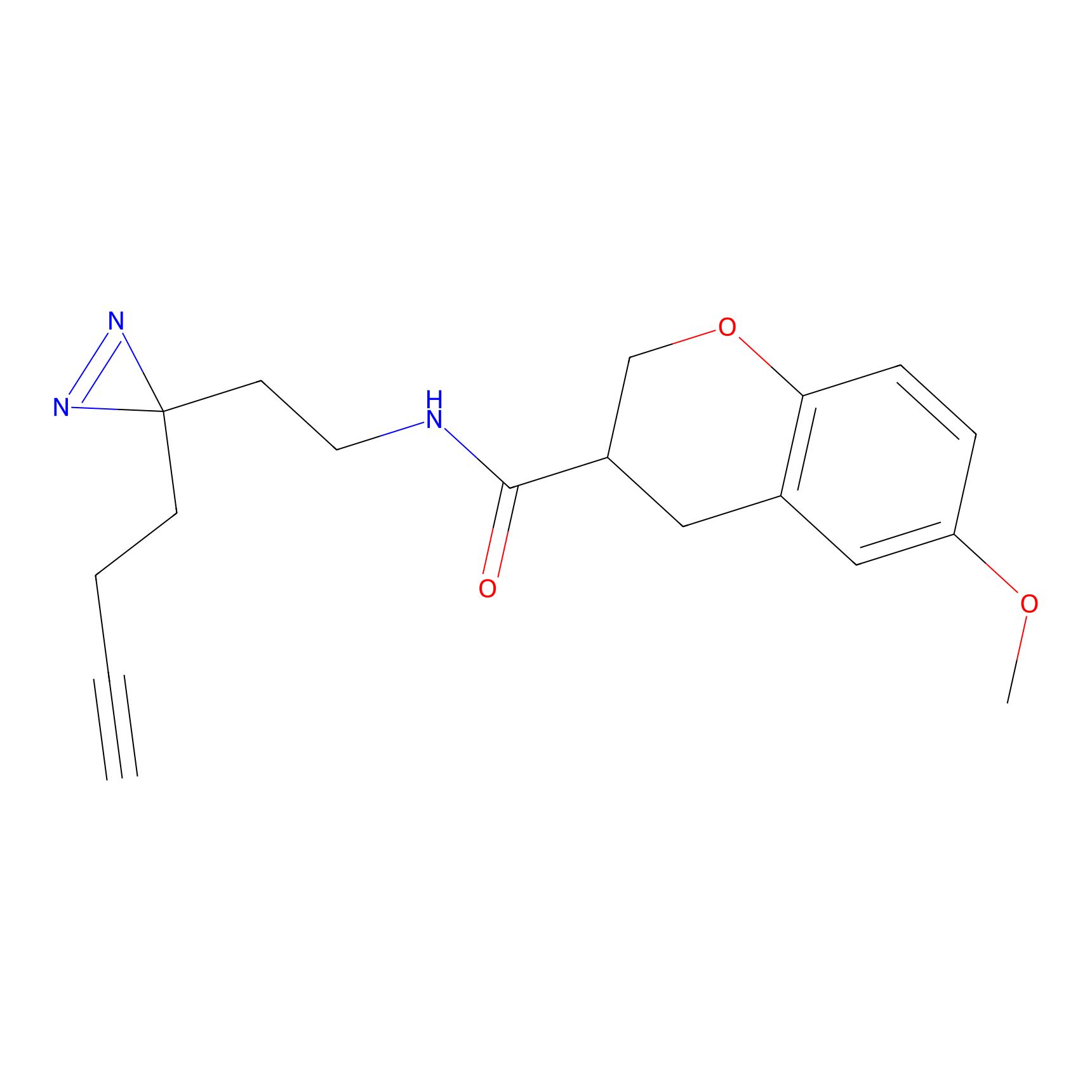
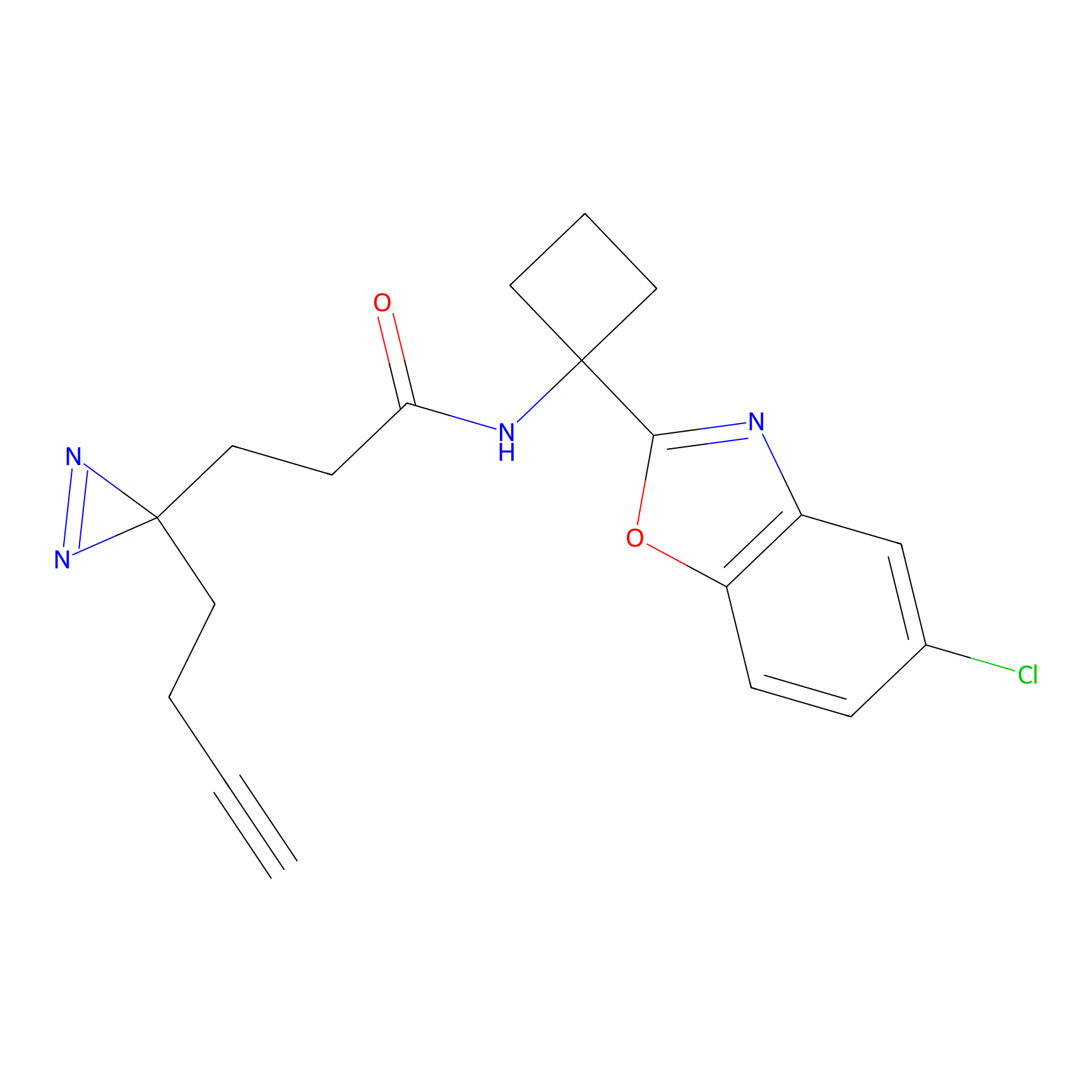
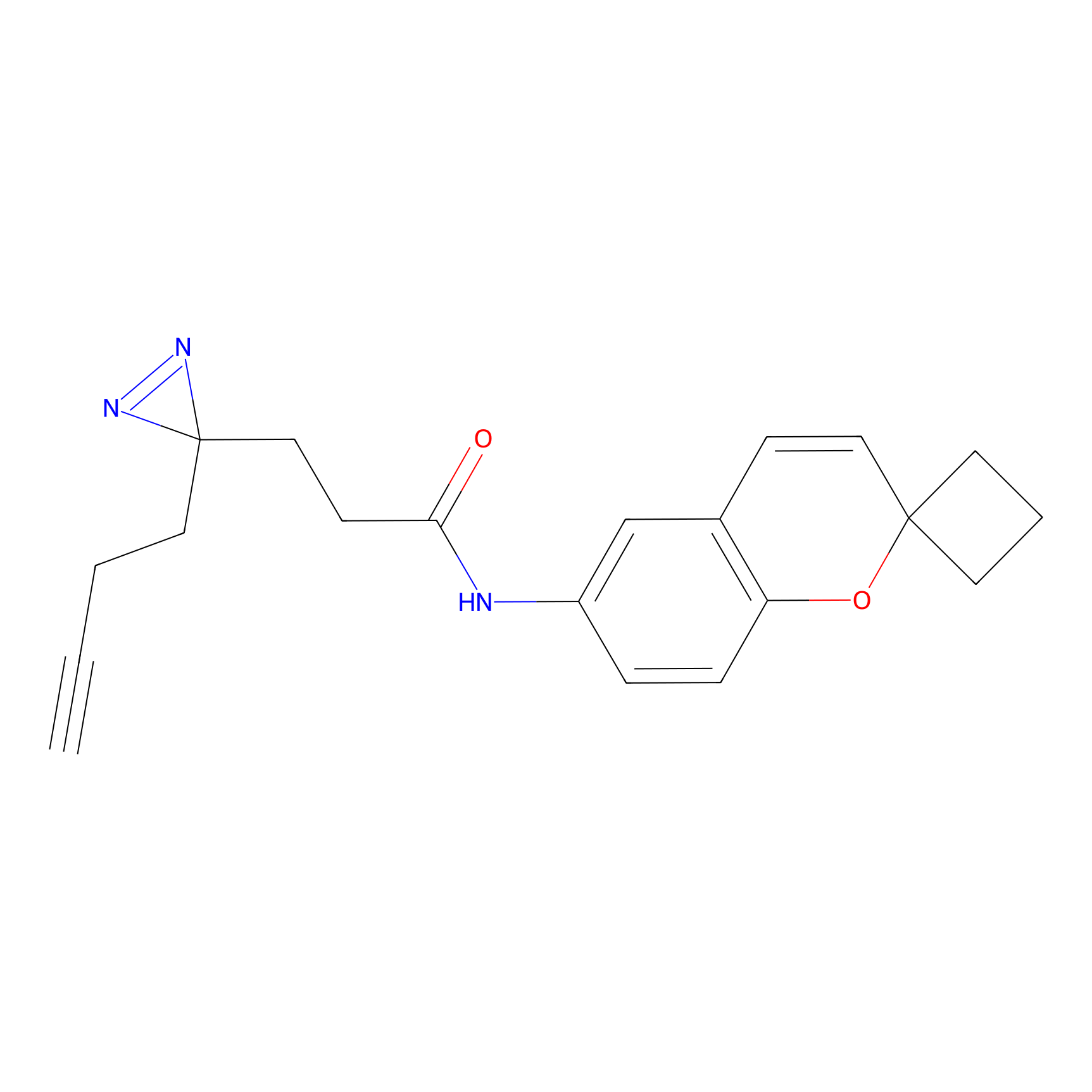
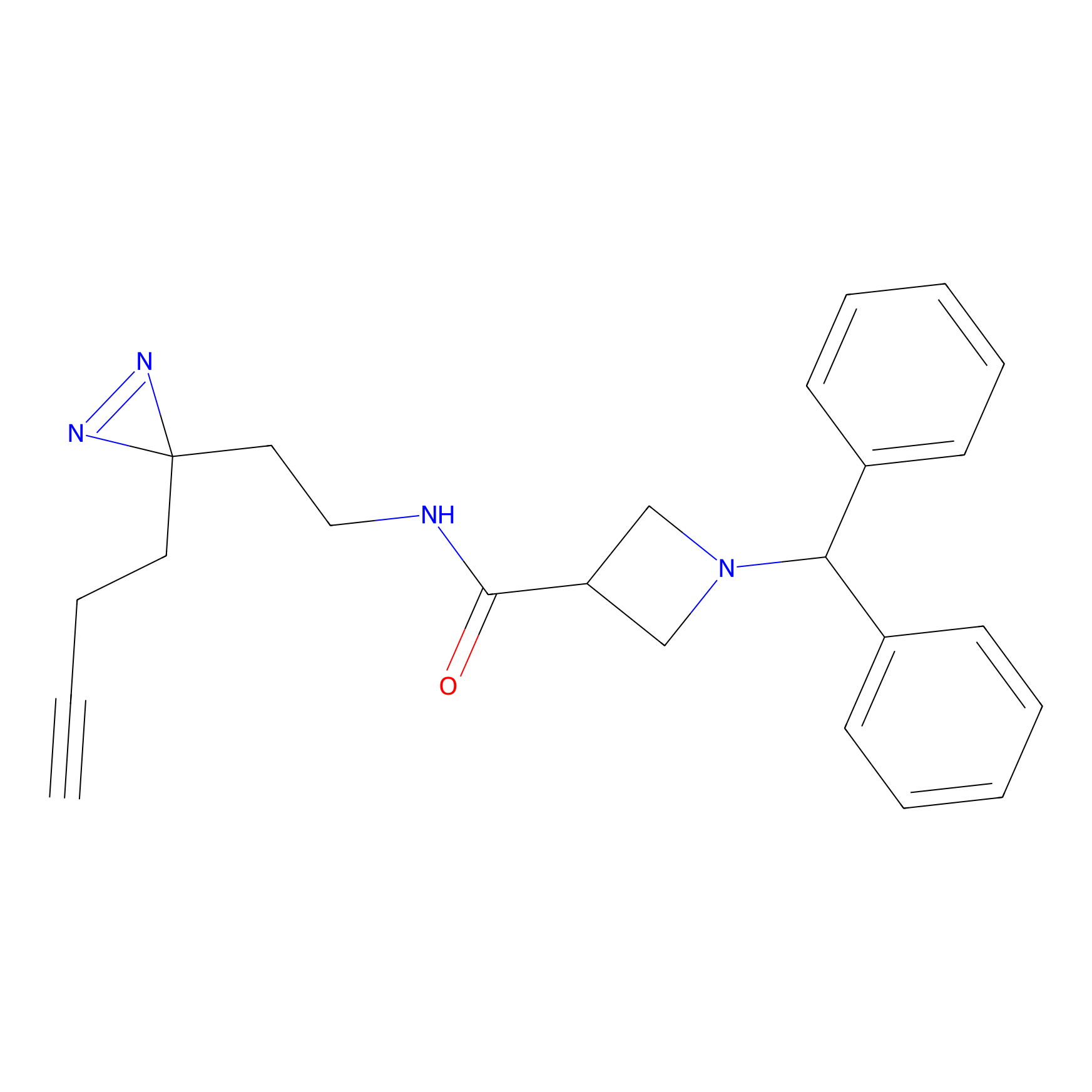
HDSF-alk Probe Info |
 |
3.29 | LDD0197 | [3] | |
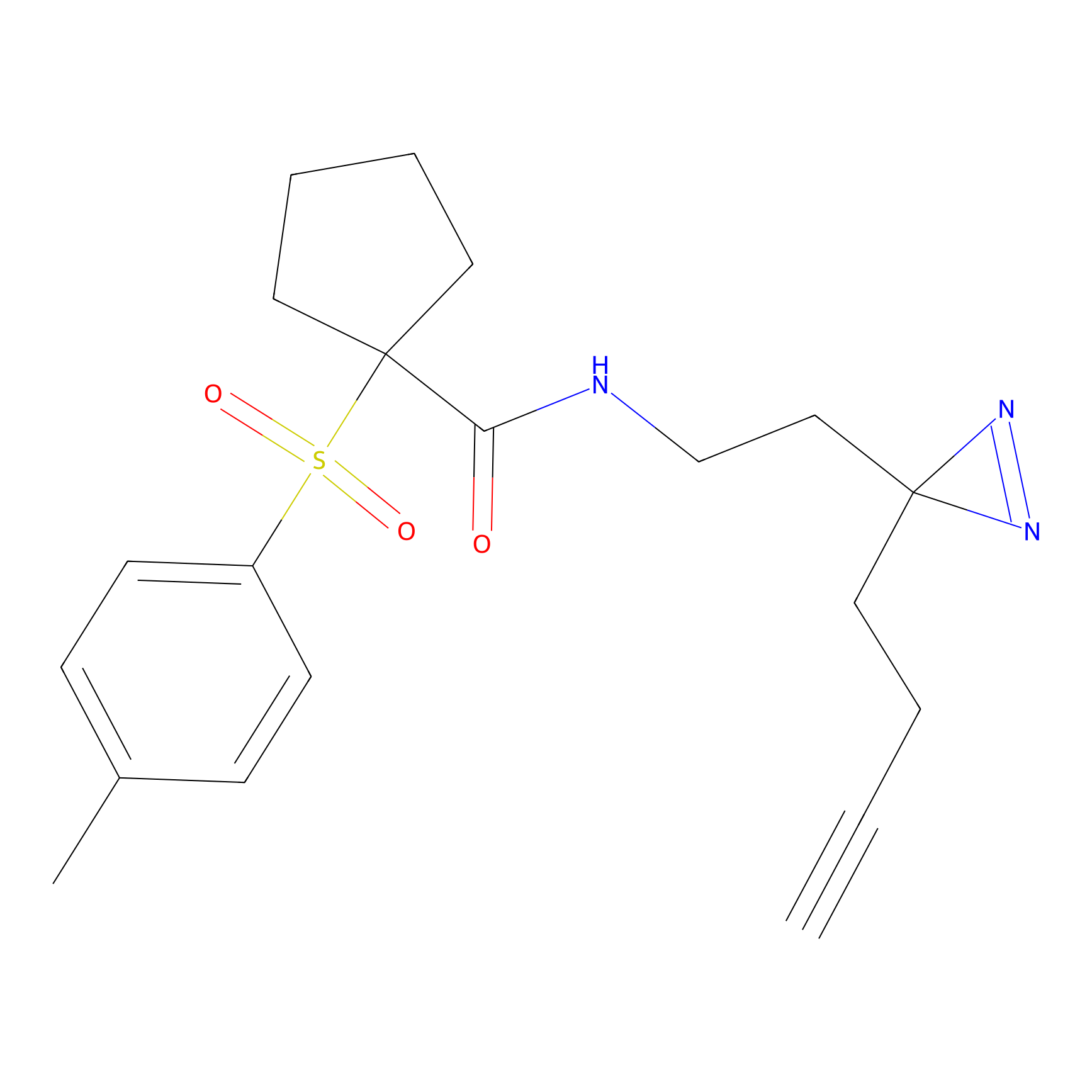
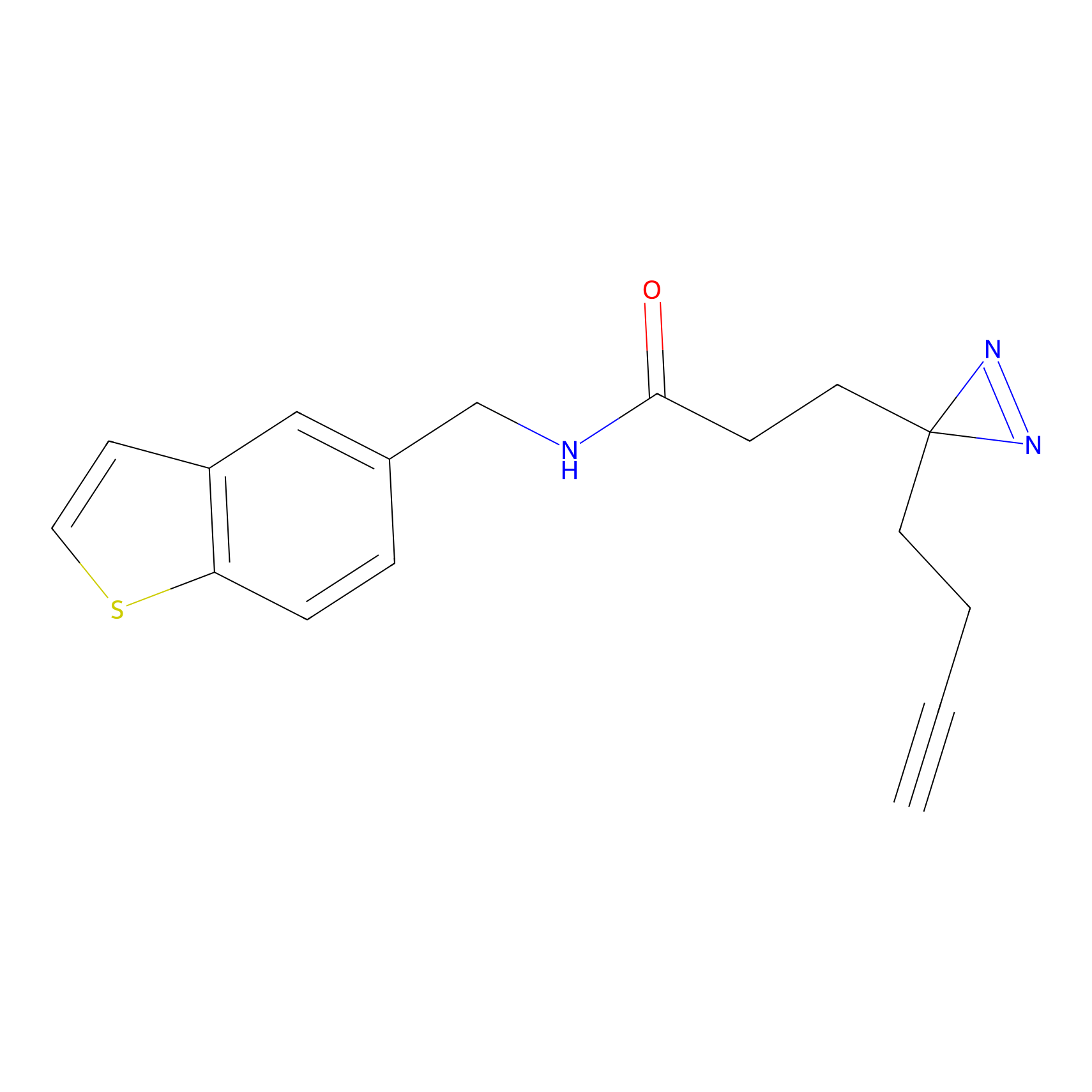
|
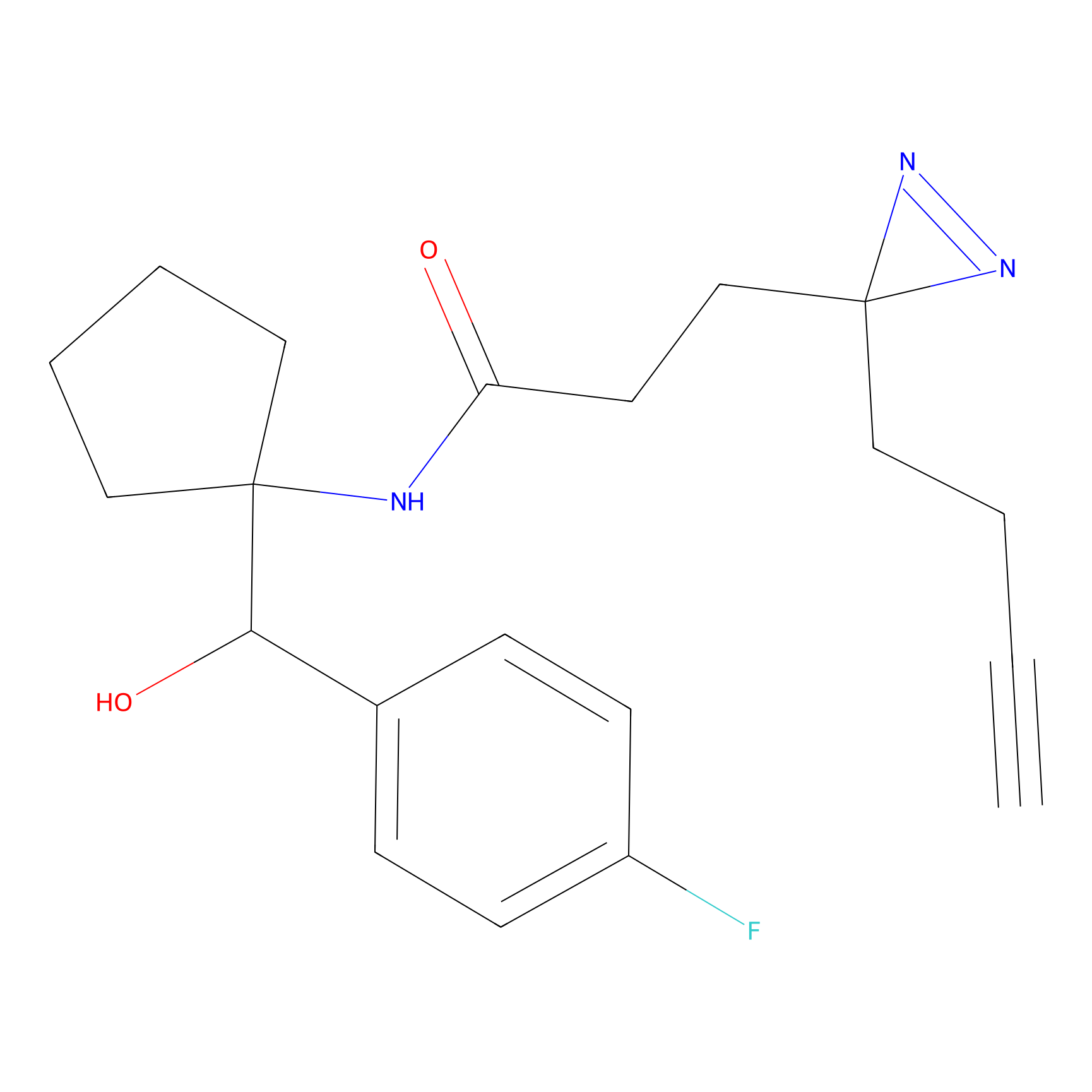
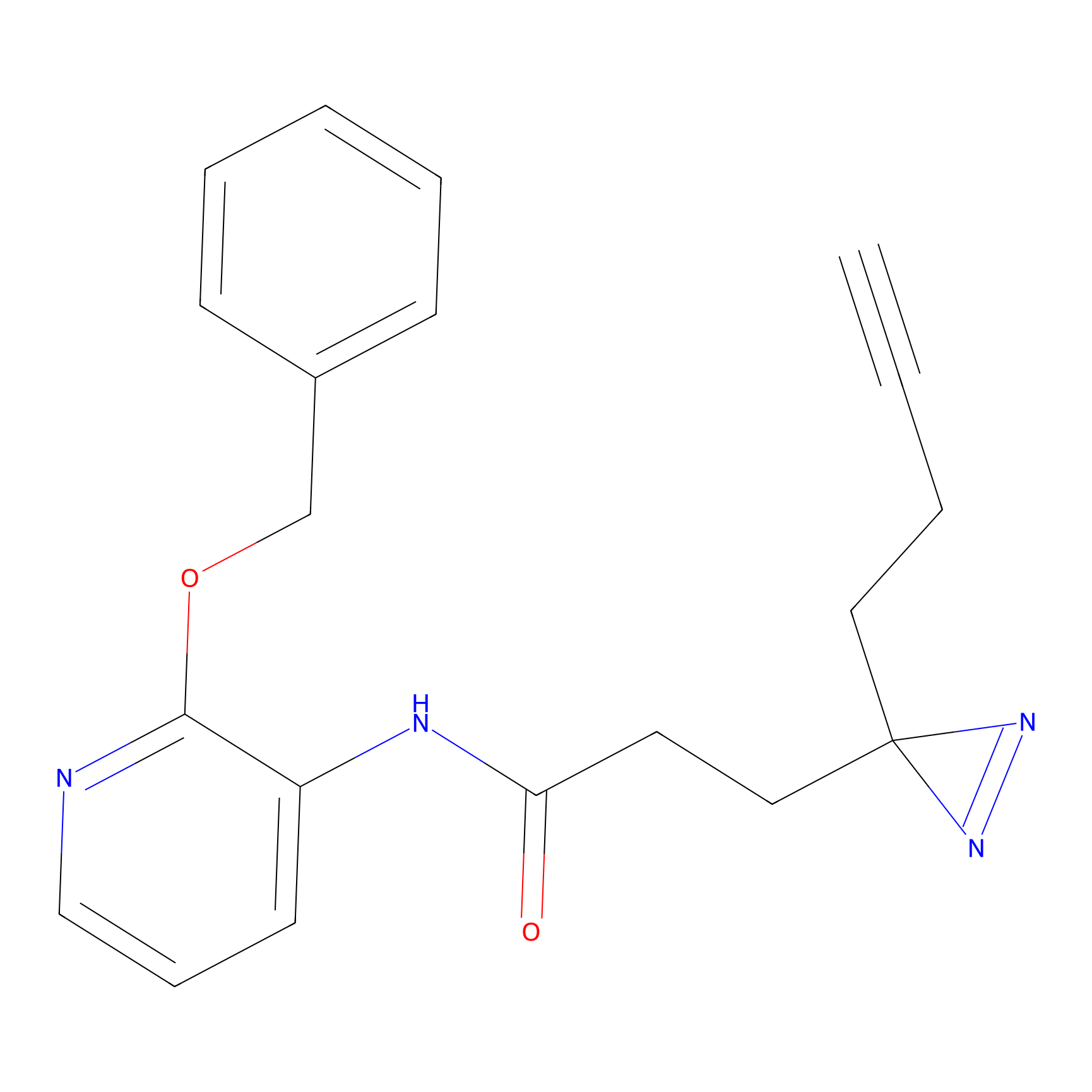
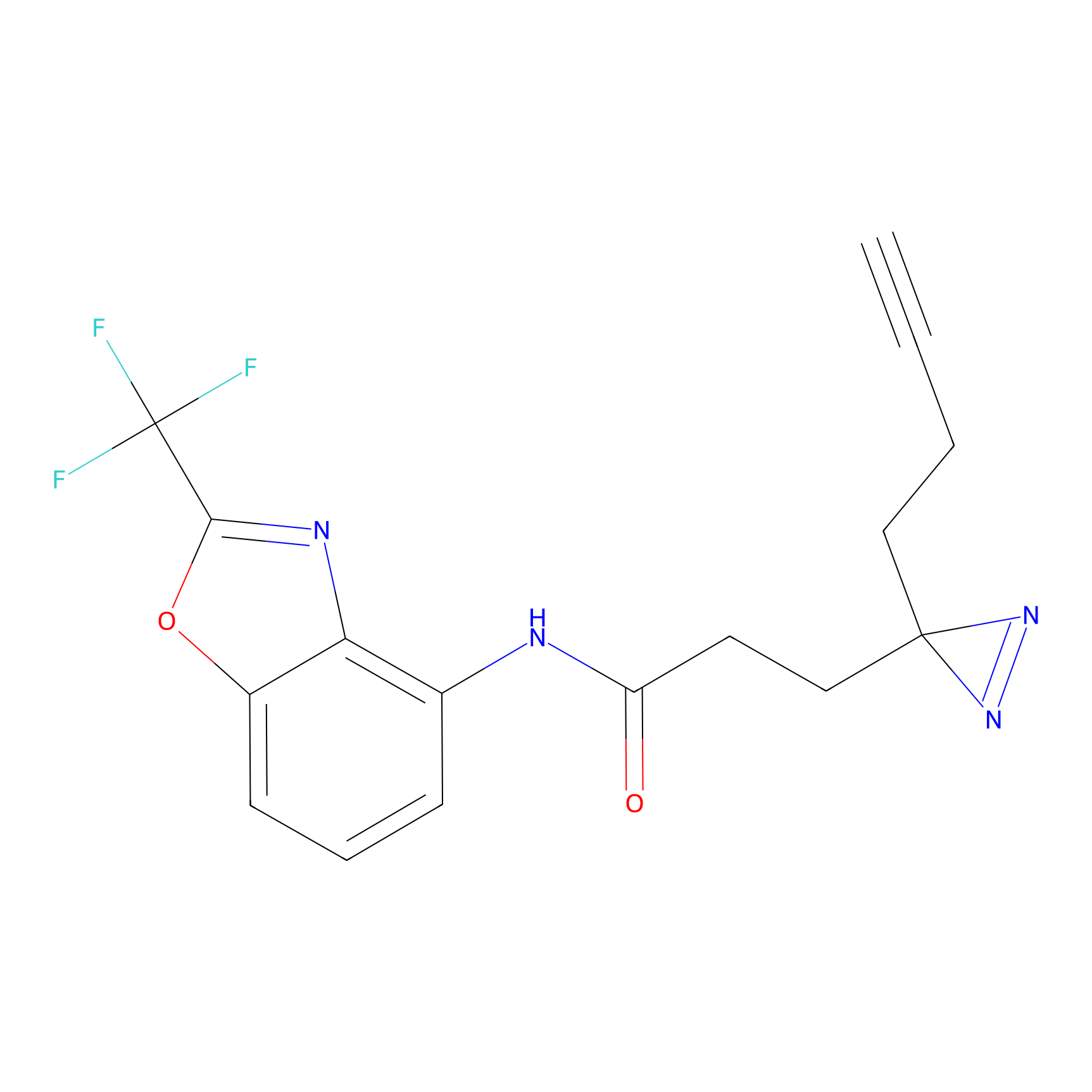
CY4 Probe Info |
 |
100.00 | LDD0244 | [4] | |
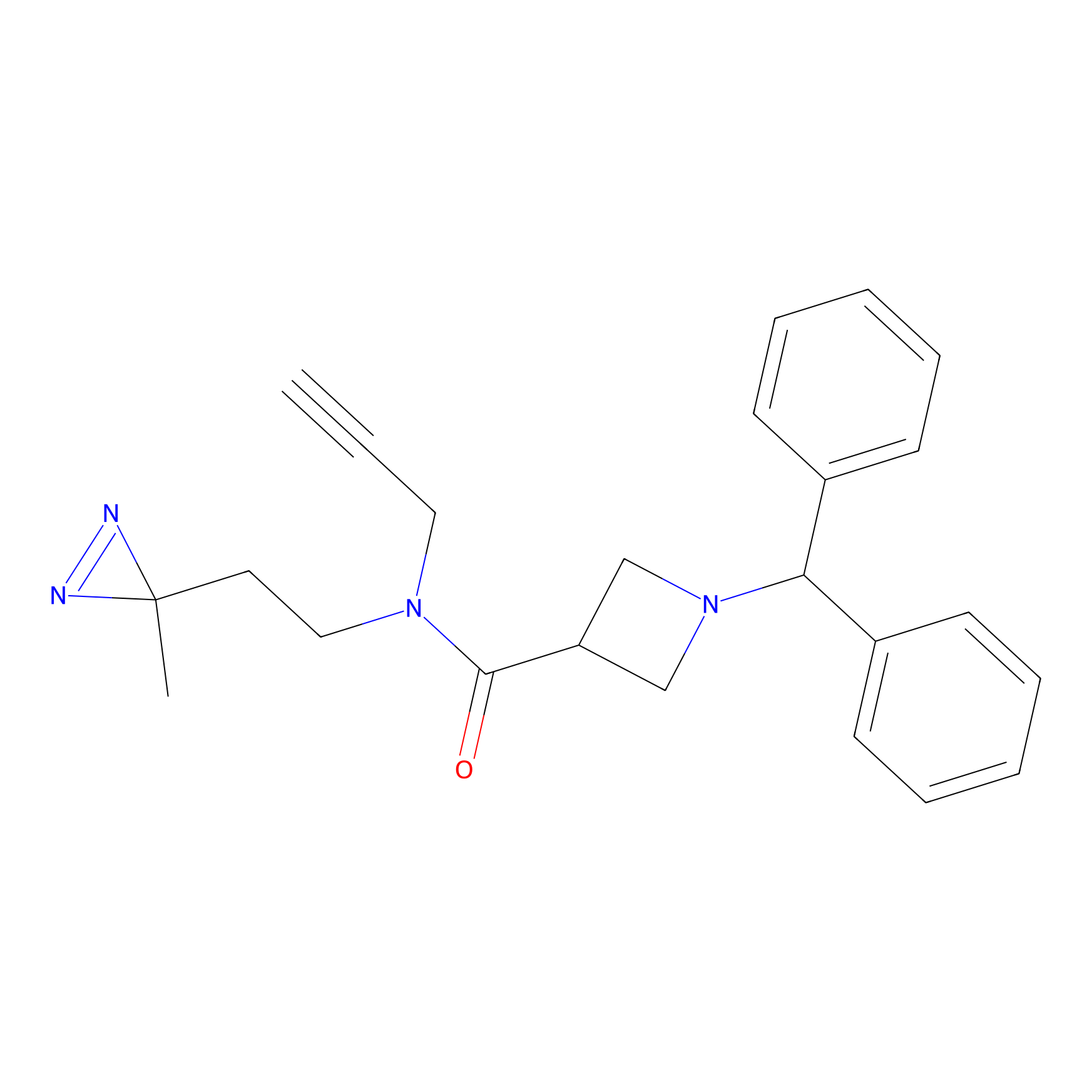
|
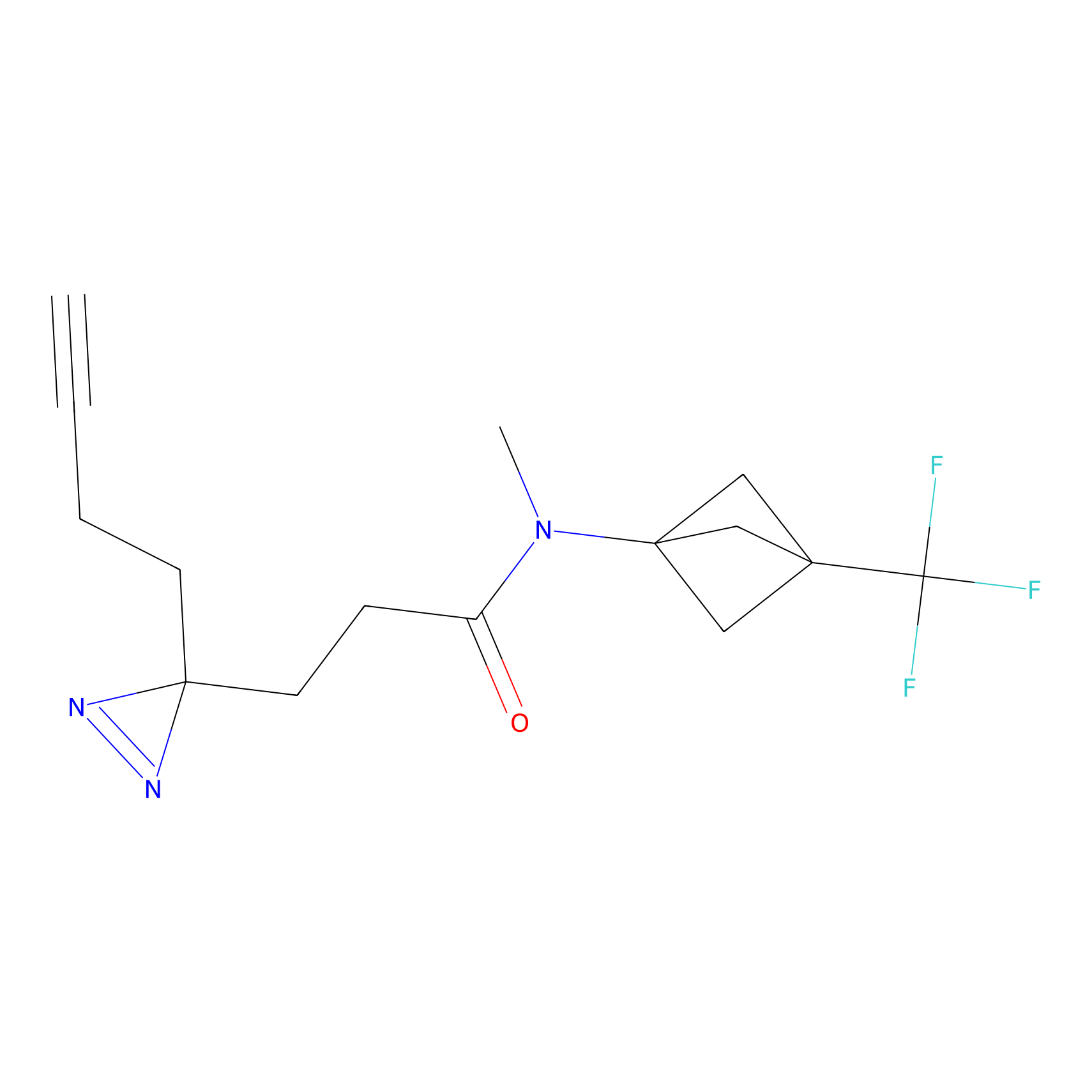
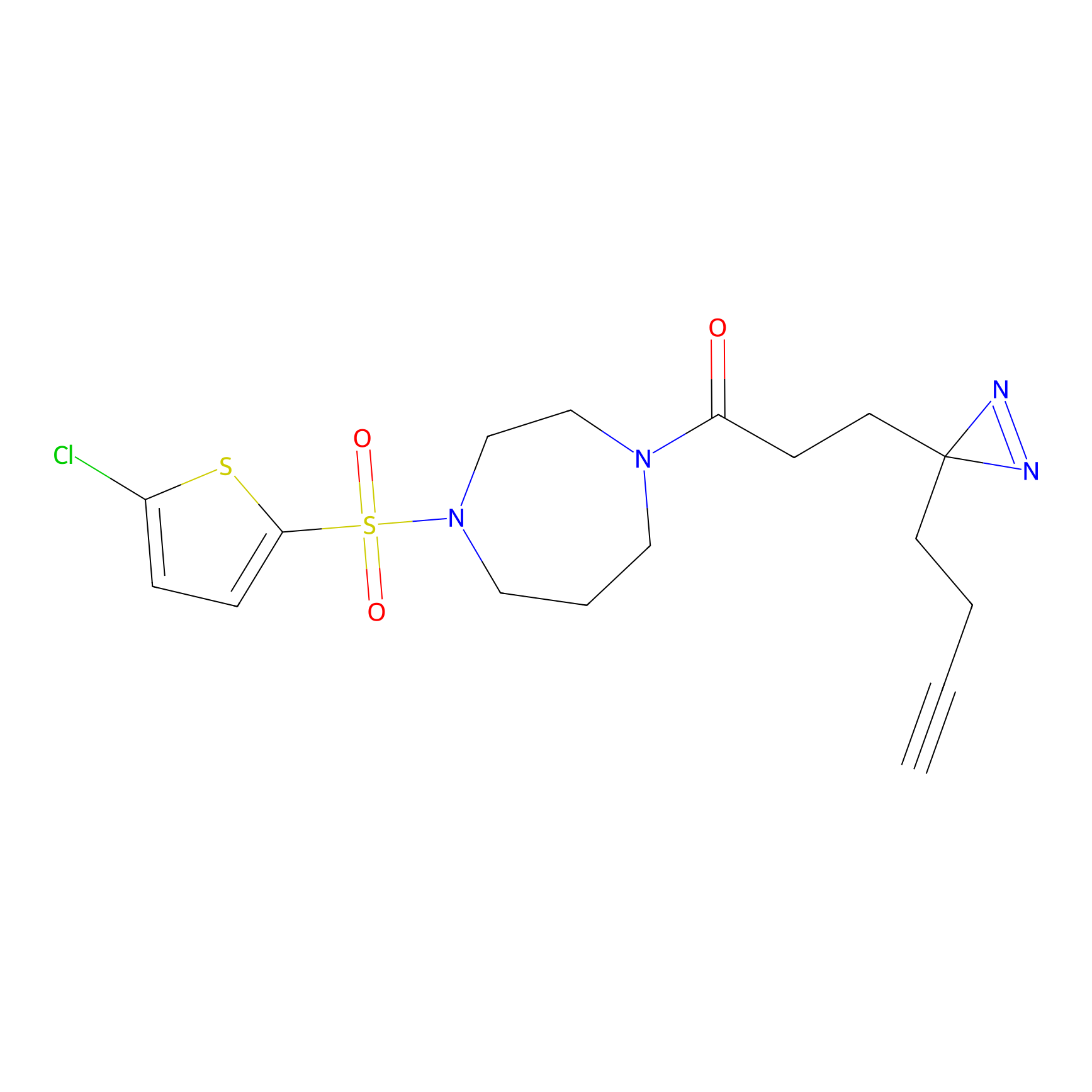
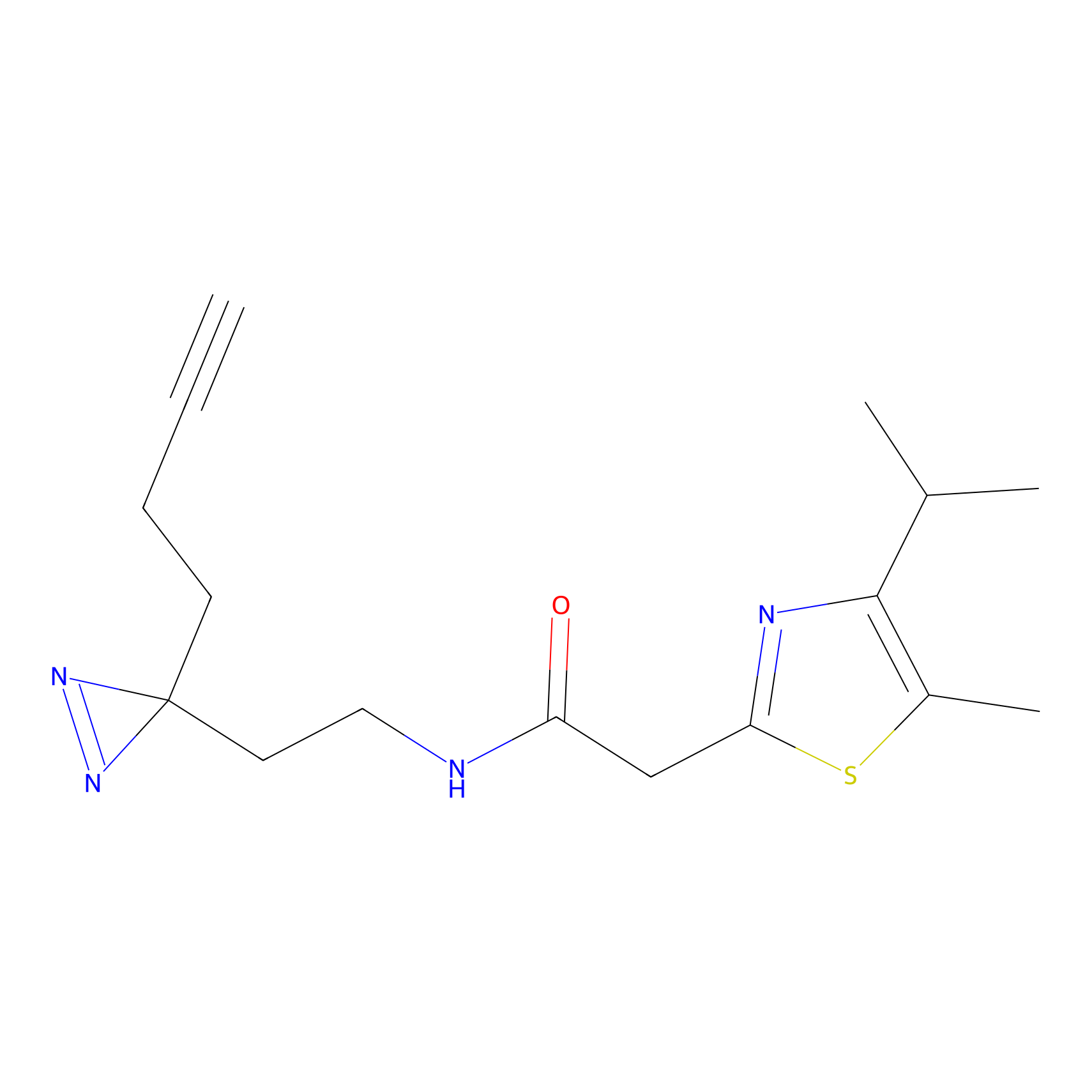
N1 Probe Info |
 |
100.00 | LDD0242 | [4] | |
|
TH211 Probe Info |
 |
Y56(12.30); Y116(11.28) | LDD0260 | [5] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [6] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [6] | |
|
ONAyne Probe Info |
 |
K162(0.00); K53(0.00) | LDD0273 | [7] | |
|
DBIA Probe Info |
 |
C239(15.53) | LDD3319 | [8] | |
|
BTD Probe Info |
 |
C239(0.09) | LDD2116 | [9] | |
|
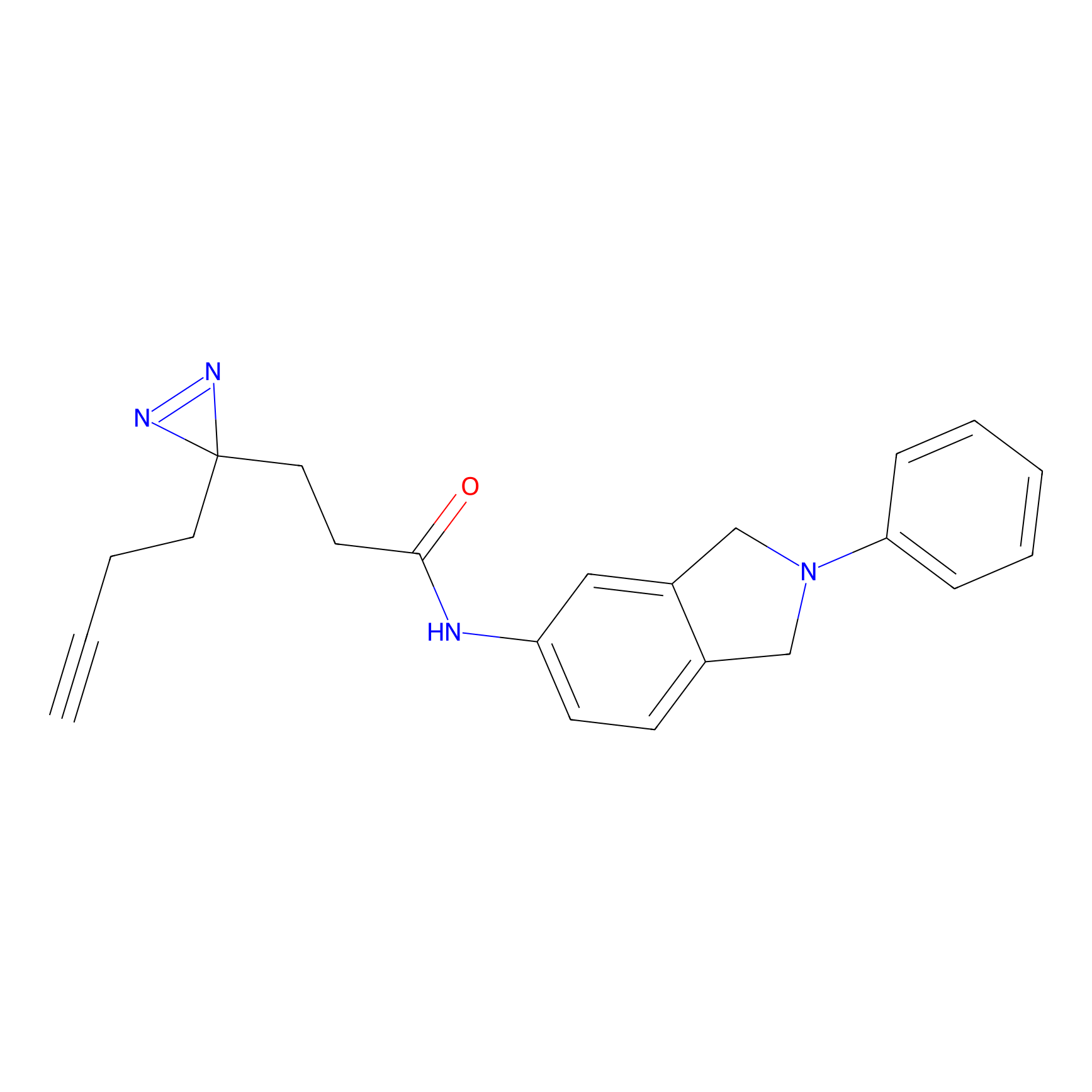
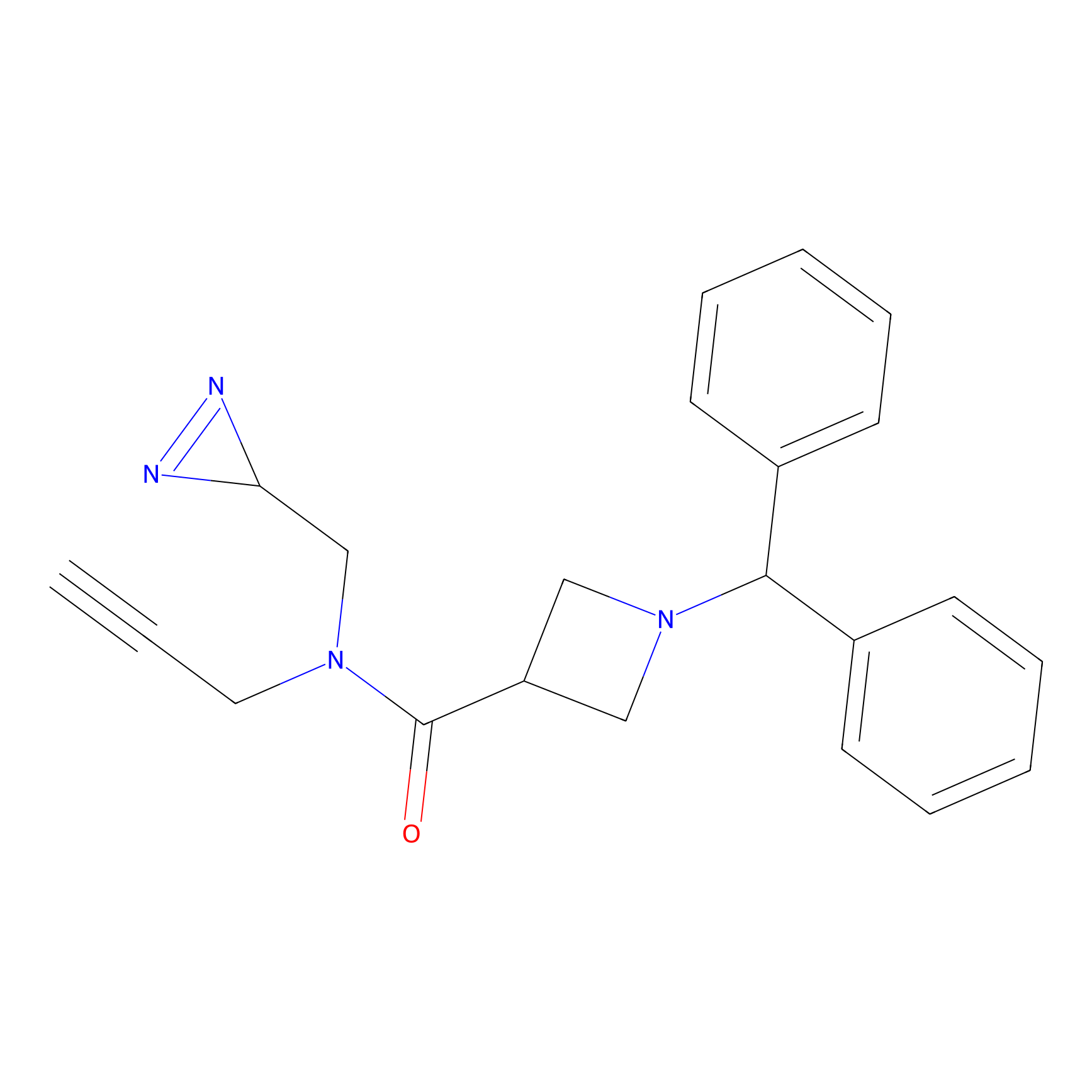
Jackson_14 Probe Info |
 |
2.50 | LDD0123 | [10] | |
|
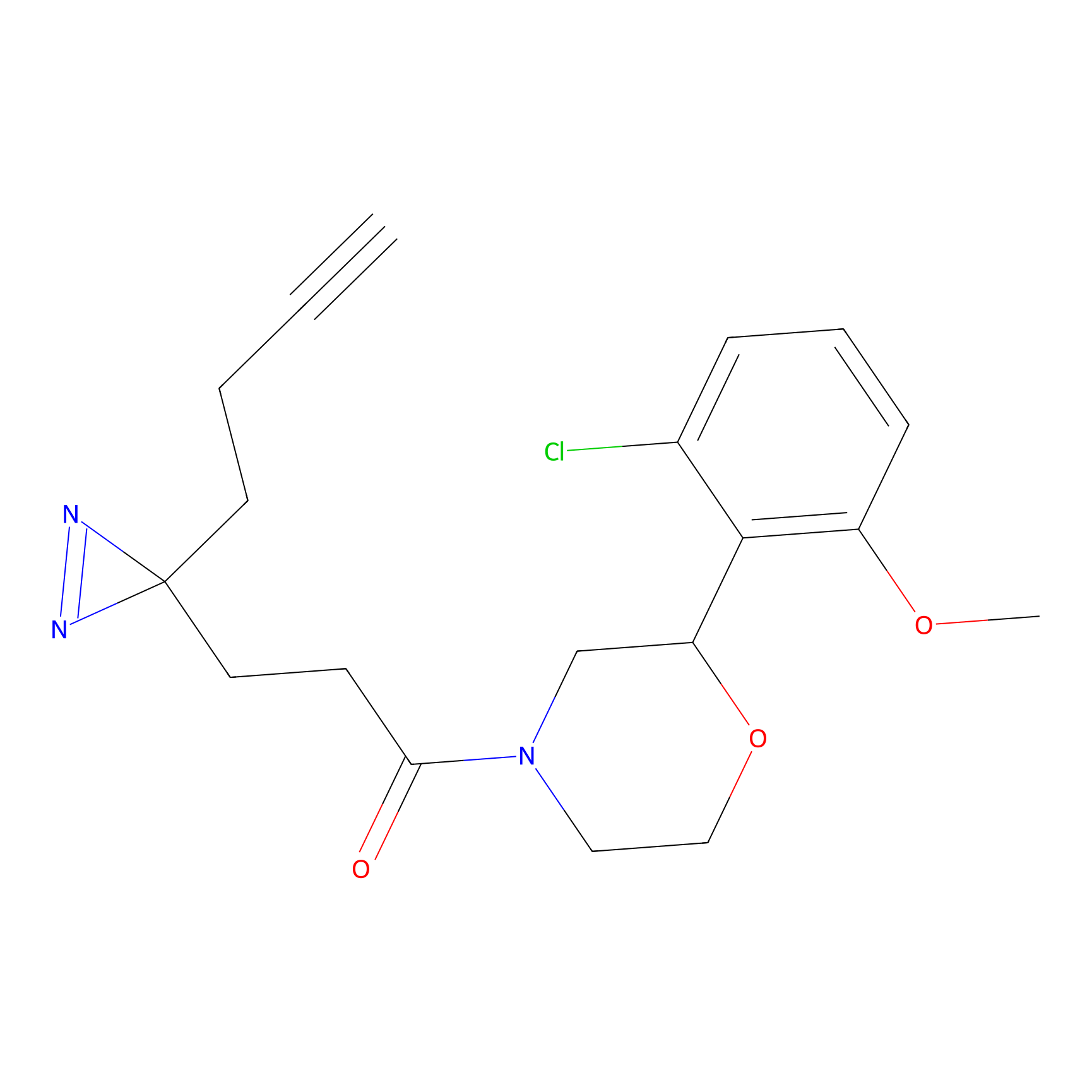
W1 Probe Info |
 |
D198(2.44) | LDD0238 | [11] | |
|
5E-2FA Probe Info |
 |
H222(0.00); H200(0.00); H145(0.00) | LDD2235 | [12] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [13] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [14] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [15] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [16] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [17] | |
|
IPM Probe Info |
 |
N.A. | LDD0147 | [16] | |
|
NHS Probe Info |
 |
K162(0.00); K131(0.00) | LDD0010 | [17] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [18] | |
|
STPyne Probe Info |
 |
K162(0.00); K131(0.00) | LDD0009 | [17] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [19] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [20] | |
|
Acrolein Probe Info |
 |
H222(0.00); H164(0.00); H145(0.00) | LDD0217 | [21] | |
|
AOyne Probe Info |
 |
8.90 | LDD0443 | [22] | |
|
MPP-AC Probe Info |
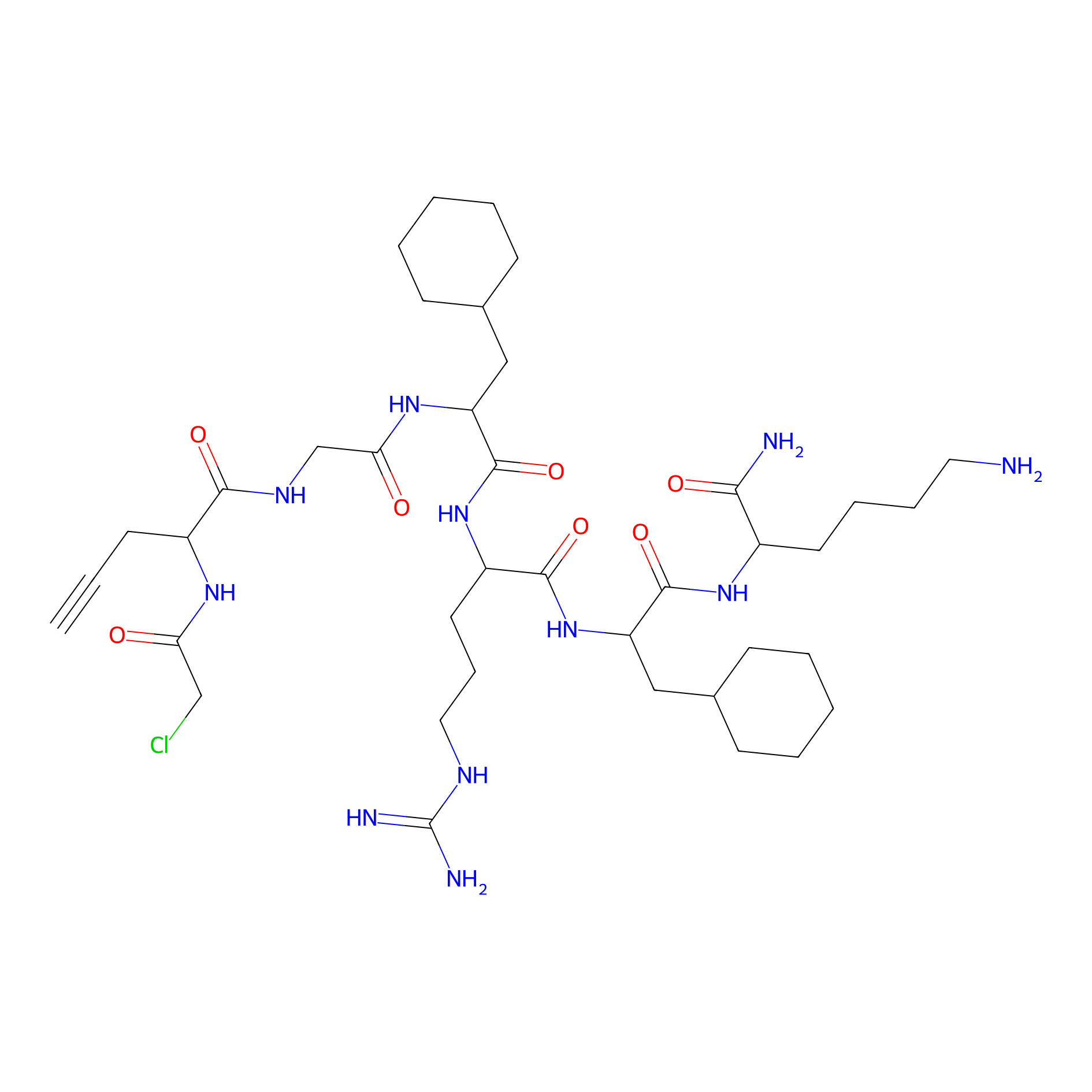 |
N.A. | LDD0428 | [23] | |
|
TER-AC Probe Info |
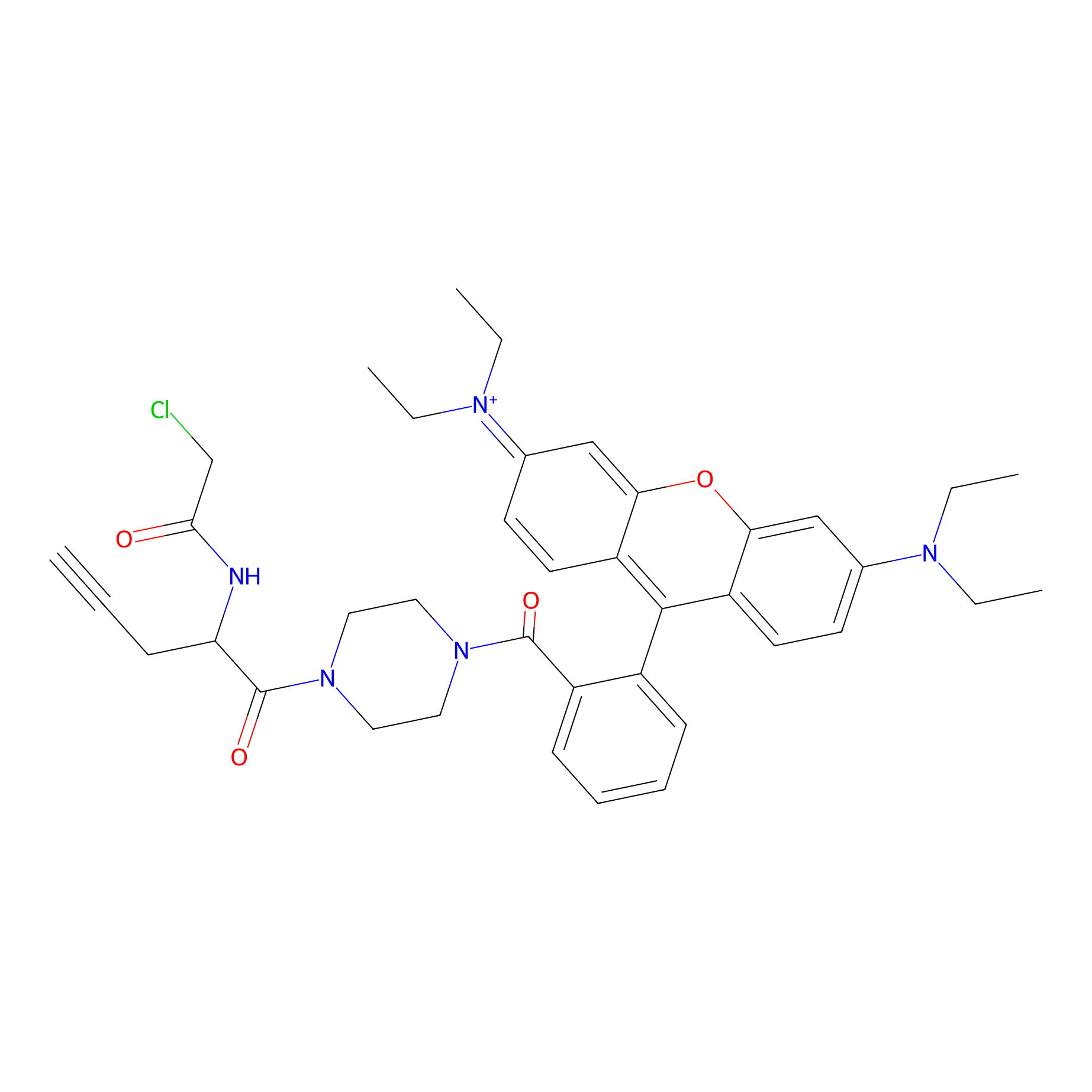 |
N.A. | LDD0426 | [23] | |
|
TPP-AC Probe Info |
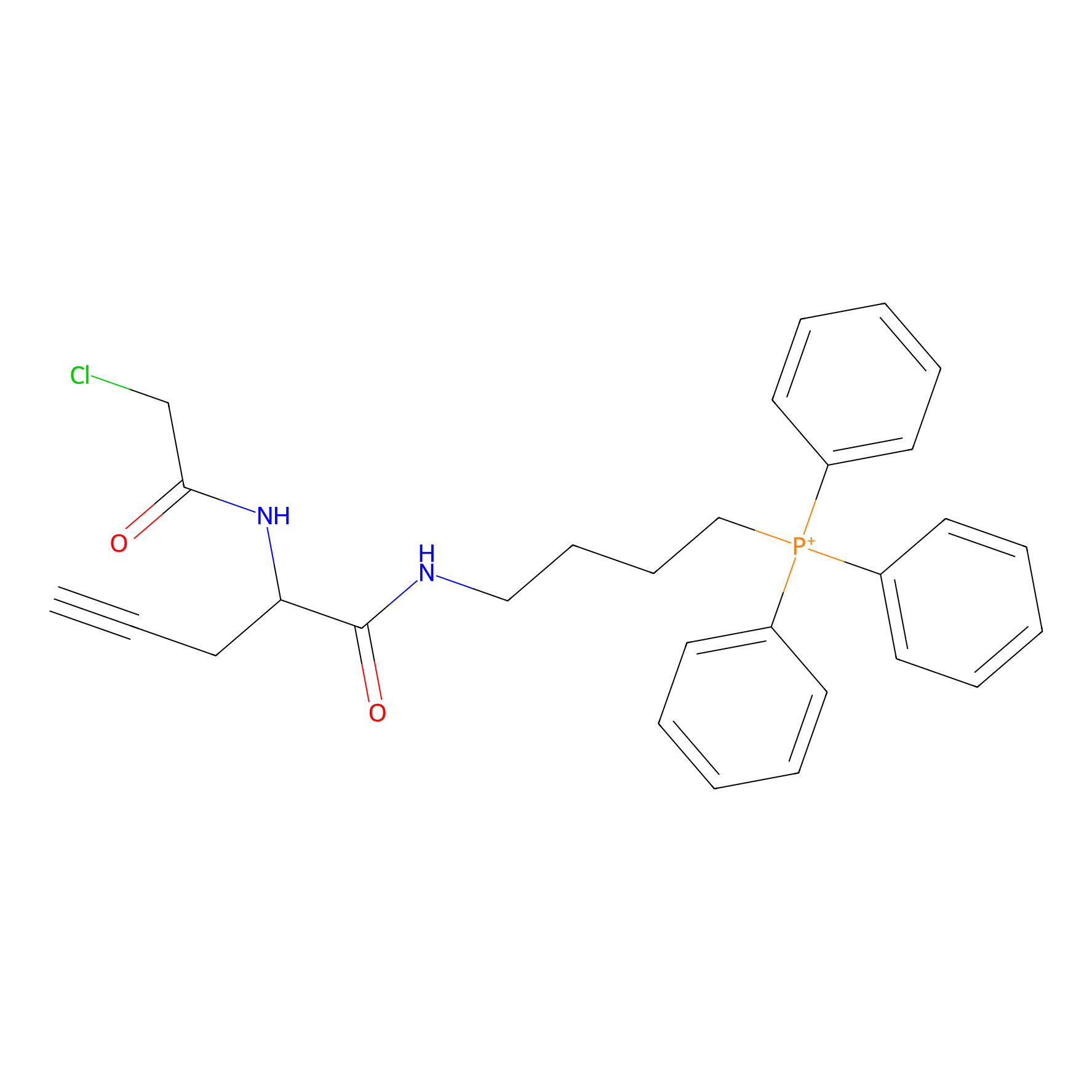 |
N.A. | LDD0427 | [23] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0108 | Chloroacetamide | HeLa | H222(0.00); H164(0.00) | LDD0222 | [21] |
| LDCM0107 | IAA | HeLa | H222(0.00); H164(0.00); H145(0.00); H214(0.00) | LDD0221 | [21] |
| LDCM0022 | KB02 | 42-MG-BA | C239(1.84) | LDD2244 | [8] |
| LDCM0023 | KB03 | 42-MG-BA | C239(6.55) | LDD2661 | [8] |
| LDCM0024 | KB05 | MEWO | C239(15.53) | LDD3319 | [8] |
| LDCM0109 | NEM | HeLa | H222(0.00); H145(0.00); H200(0.00); H164(0.00) | LDD0223 | [21] |
| LDCM0523 | Nucleophilic fragment 24b | MDA-MB-231 | C239(0.09) | LDD2116 | [9] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C239(0.09) | LDD2118 | [9] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C239(0.79) | LDD2120 | [9] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C239(0.04) | LDD2122 | [9] |
| LDCM0531 | Nucleophilic fragment 28b | MDA-MB-231 | C239(0.05) | LDD2124 | [9] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C239(0.05) | LDD2126 | [9] |
| LDCM0555 | Nucleophilic fragment 7b | MDA-MB-231 | C239(0.03) | LDD2149 | [9] |
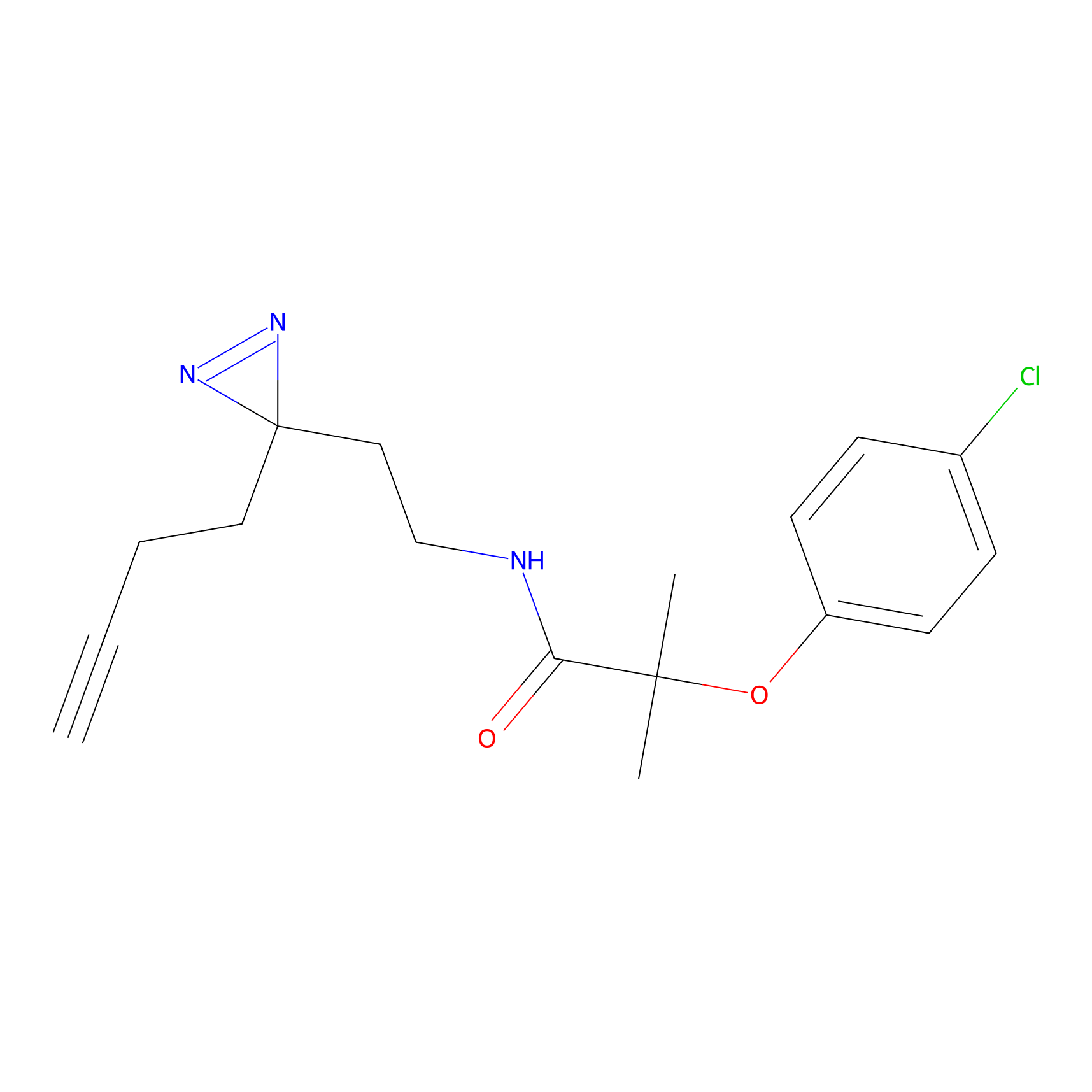
| LDCM0016 | Ranjitkar_cp1 | MDA-MB-231 | 2.50 | LDD0123 | [10] |
| LDCM0111 | W14 | Hep-G2 | D198(2.44) | LDD0238 | [11] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Protein-arginine deiminase type-3 (PADI3) | Protein arginine deiminase family | Q9ULW8 | |||
| Methyltransferase-like protein 27 (METTL27) | . | Q8N6F8 | |||
Transporter and channel
Other
References