Details of the Target
General Information of Target
| Target ID | LDTP00850 | |||||
|---|---|---|---|---|---|---|
| Target Name | ER lumen protein-retaining receptor 3 (KDELR3) | |||||
| Gene Name | KDELR3 | |||||
| Gene ID | 11015 | |||||
| Synonyms |
ERD23; ER lumen protein-retaining receptor 3; KDEL endoplasmic reticulum protein retention receptor 3; KDEL receptor 3 |
|||||
| 3D Structure | ||||||
| Sequence |
MNVFRILGDLSHLLAMILLLGKIWRSKCCKGISGKSQILFALVFTTRYLDLFTNFISIYN
TVMKVVFLLCAYVTVYMIYGKFRKTFDSENDTFRLEFLLVPVIGLSFLENYSFTLLEILW TFSIYLESVAILPQLFMISKTGEAETITTHYLFFLGLYRALYLANWIRRYQTENFYDQIA VVSGVVQTIFYCDFFYLYVTKVLKGKKLSLPMPI |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
ERD2 family
|
|||||
| Subcellular location |
Endoplasmic reticulum membrane
|
|||||
| Function | Receptor for the C-terminal sequence motif K-D-E-L that is present on endoplasmic reticulum resident proteins and that mediates their recycling from the Golgi back to the endoplasmic reticulum. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
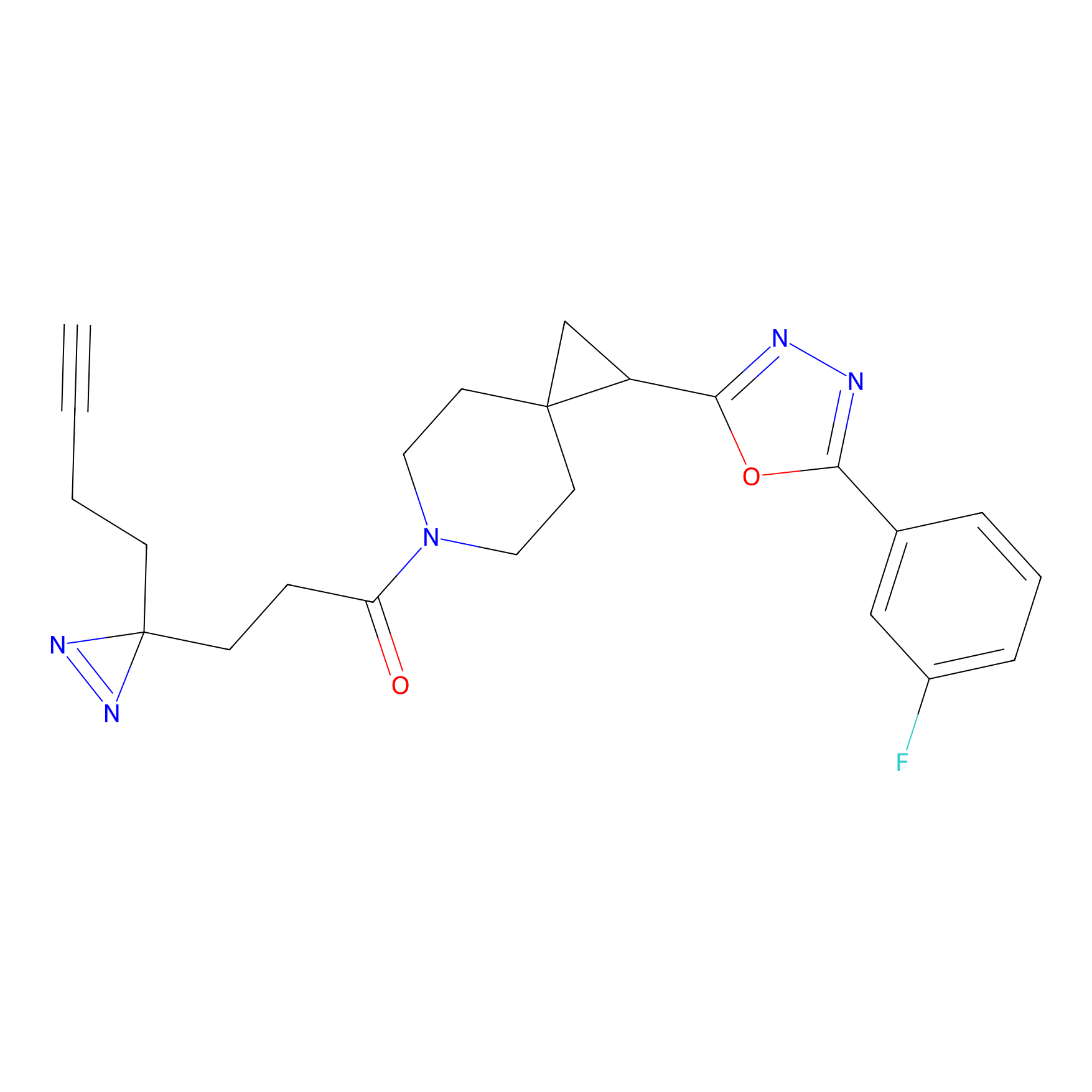
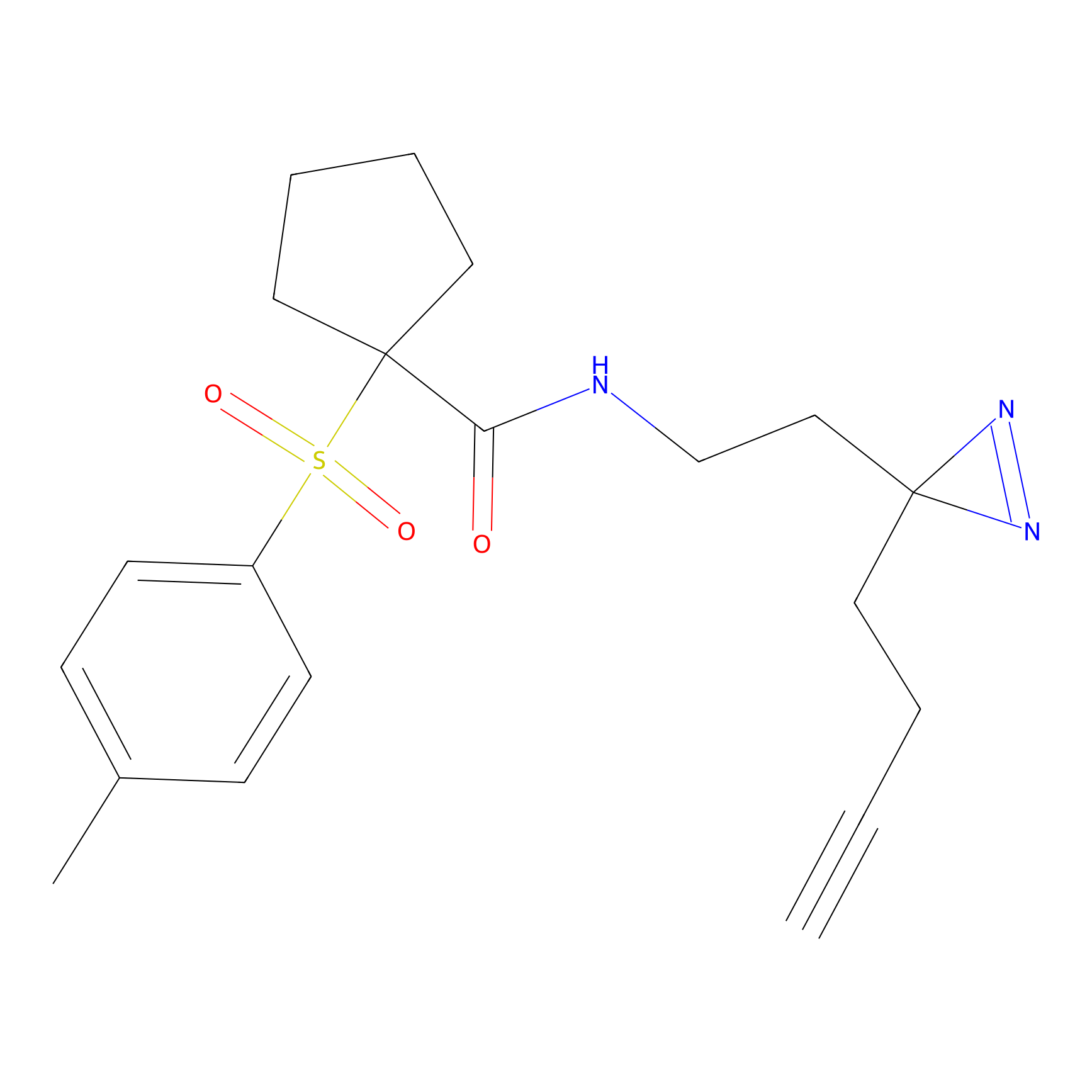
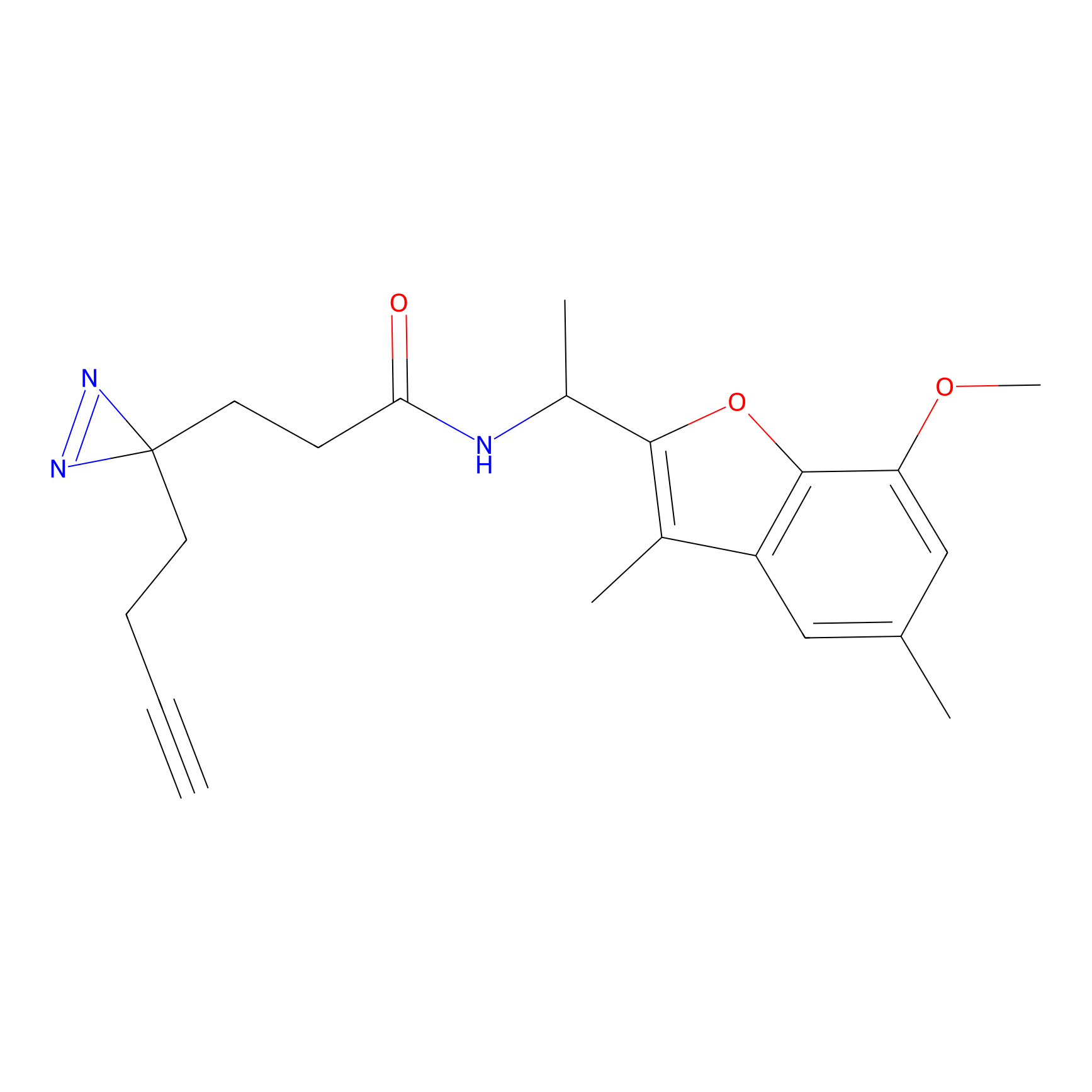
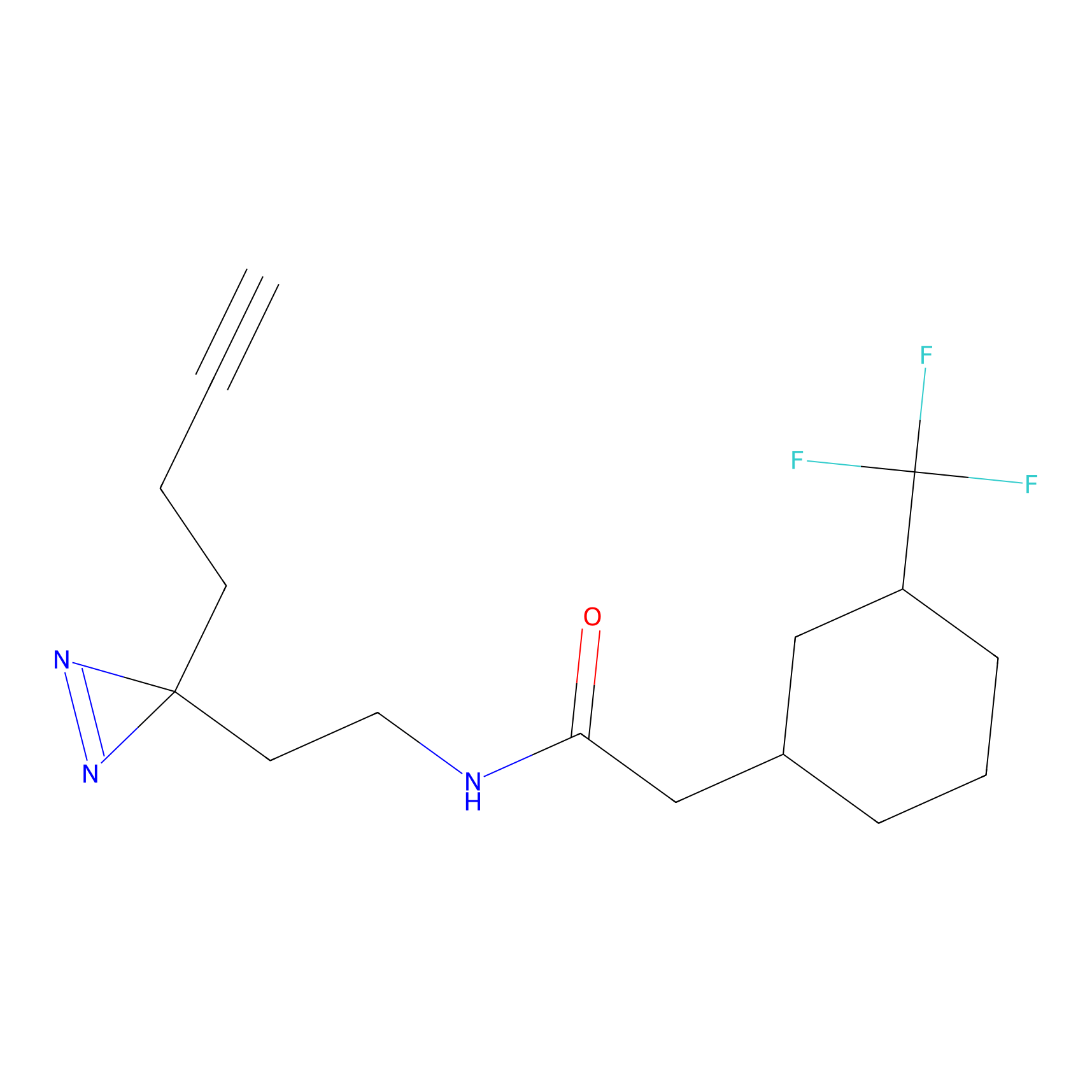
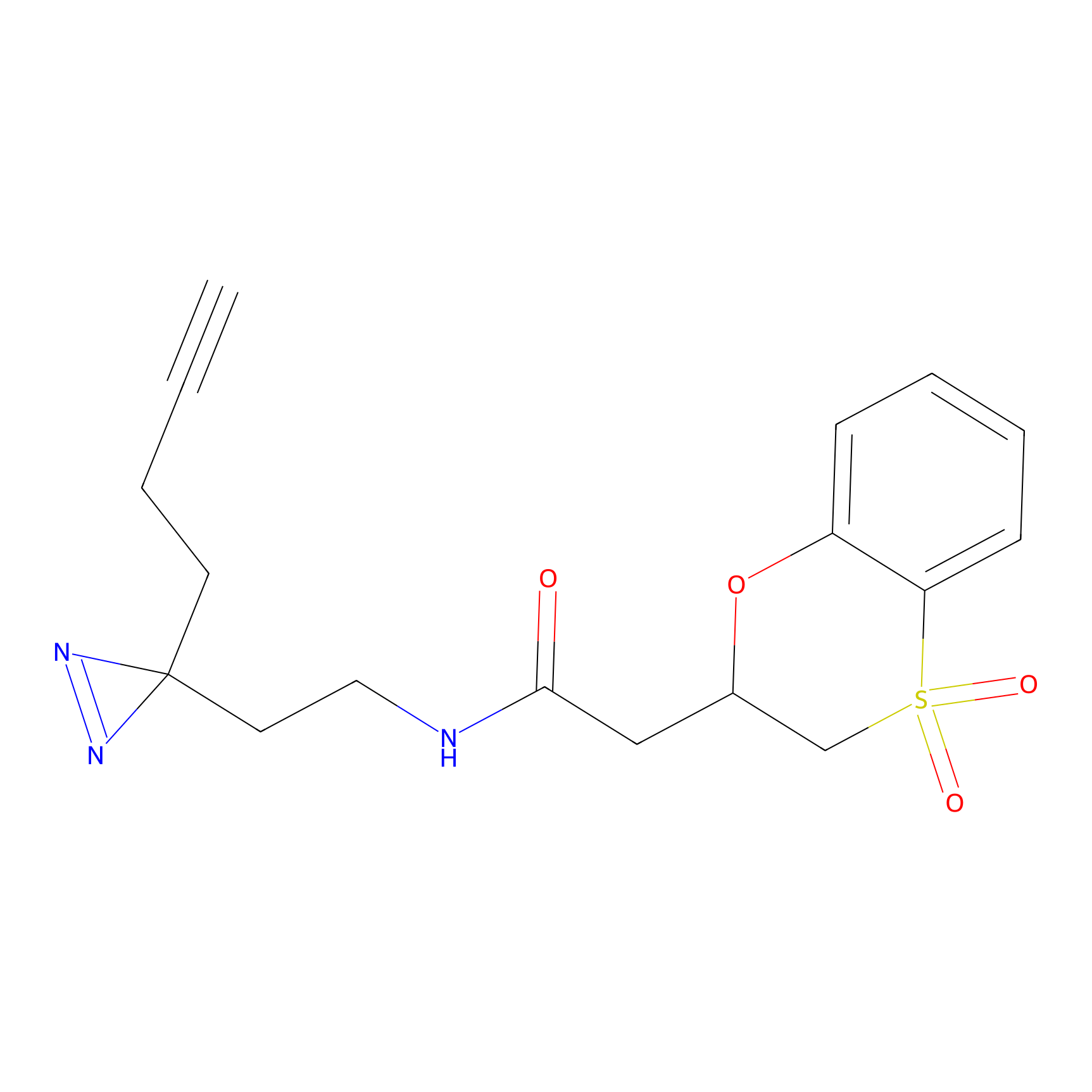
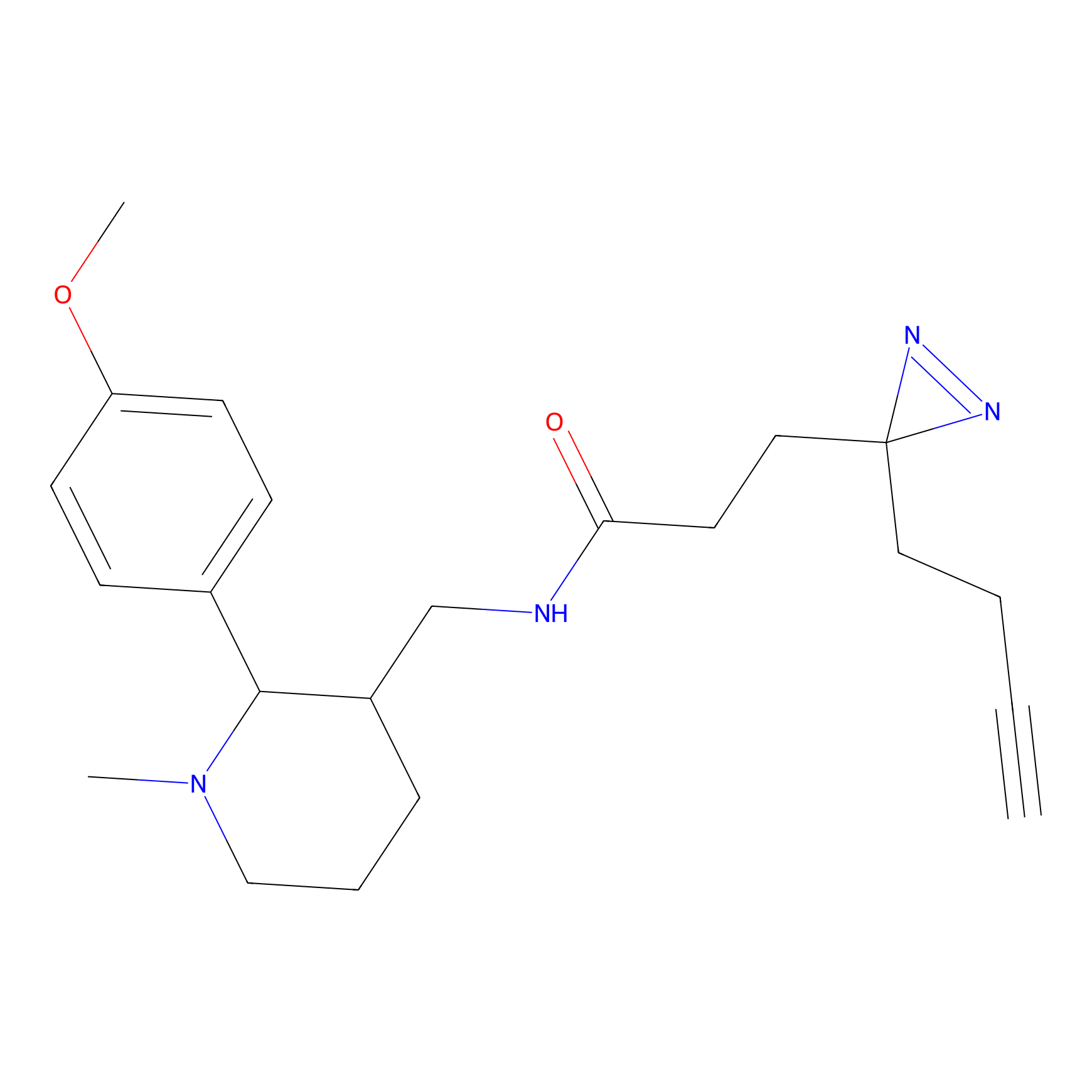
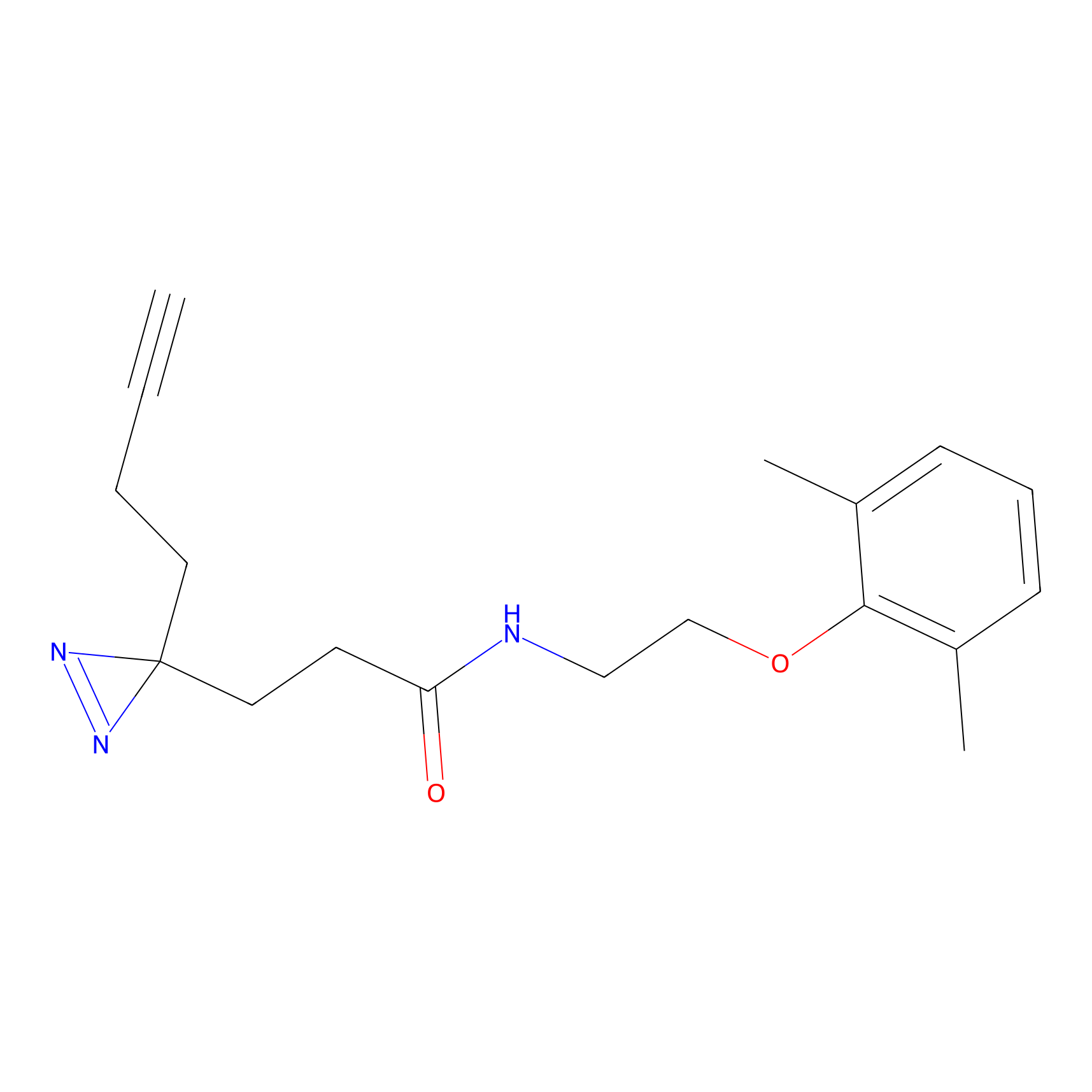
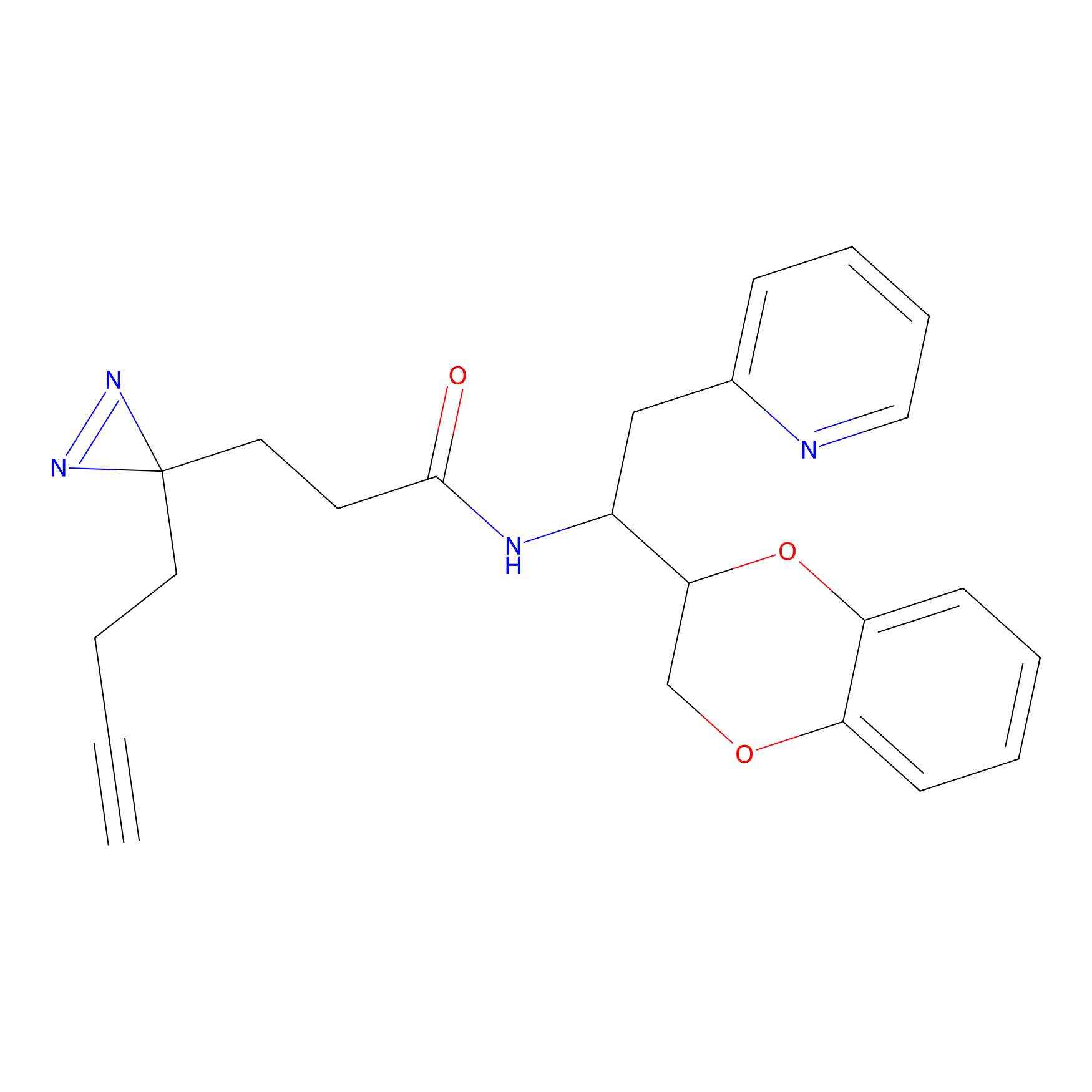
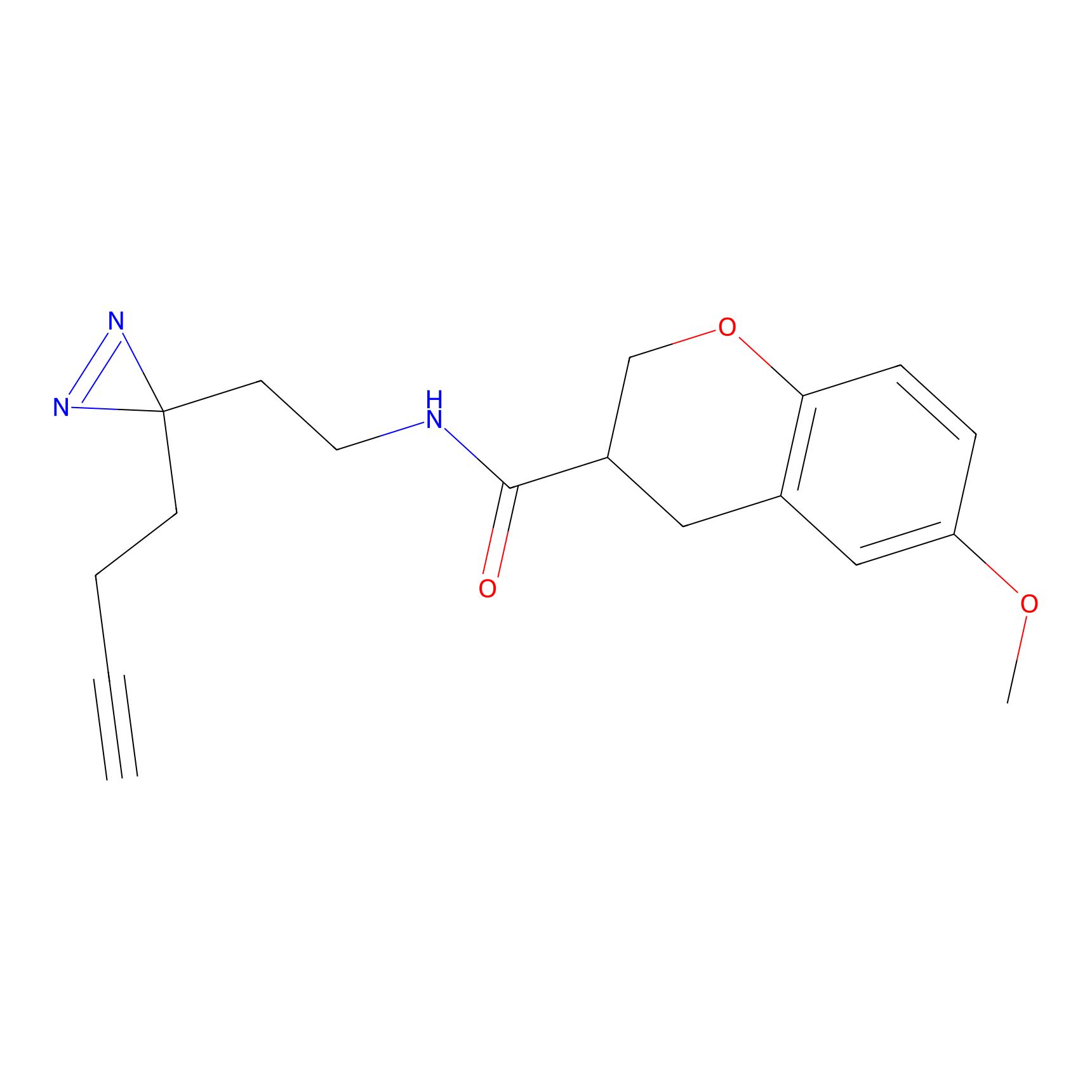
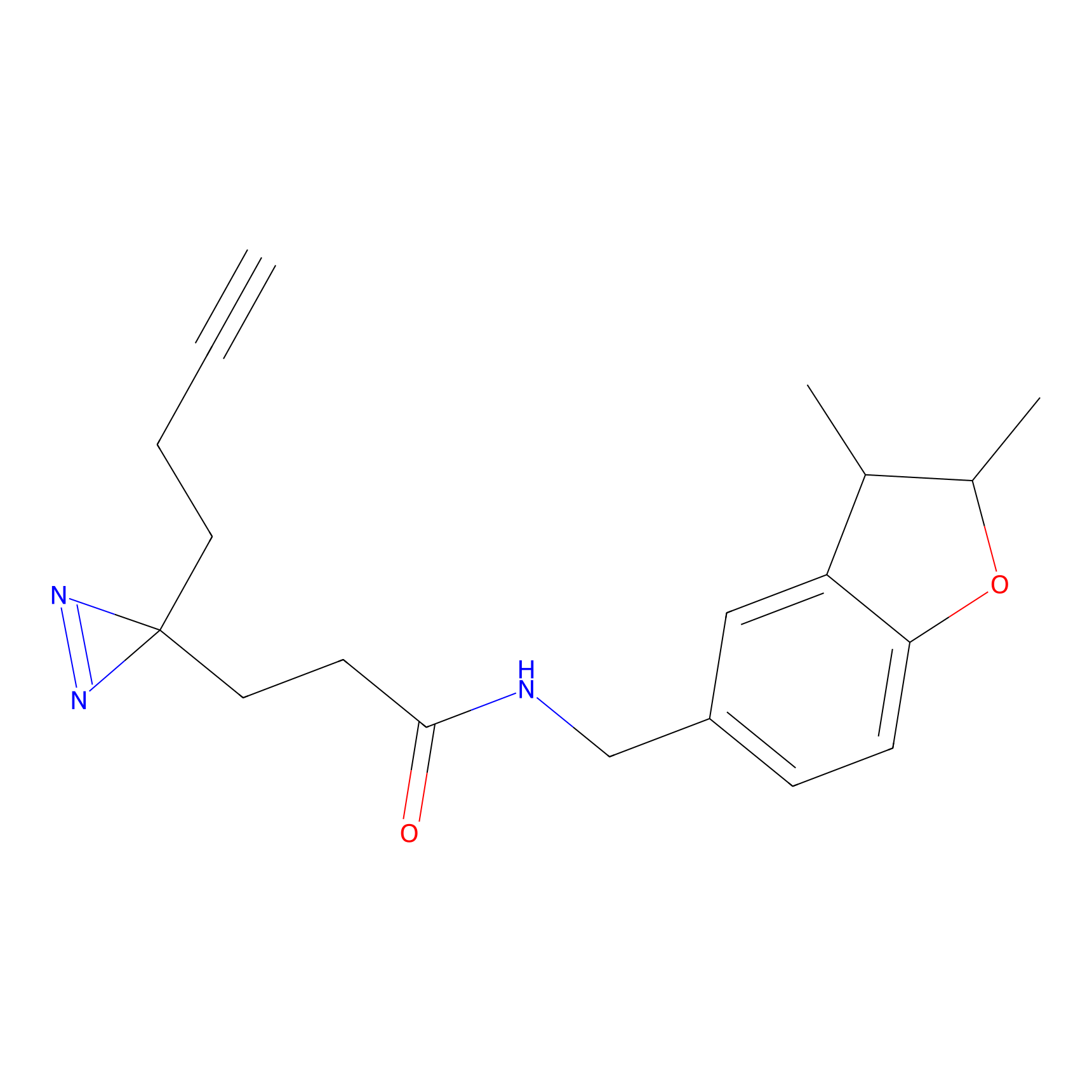
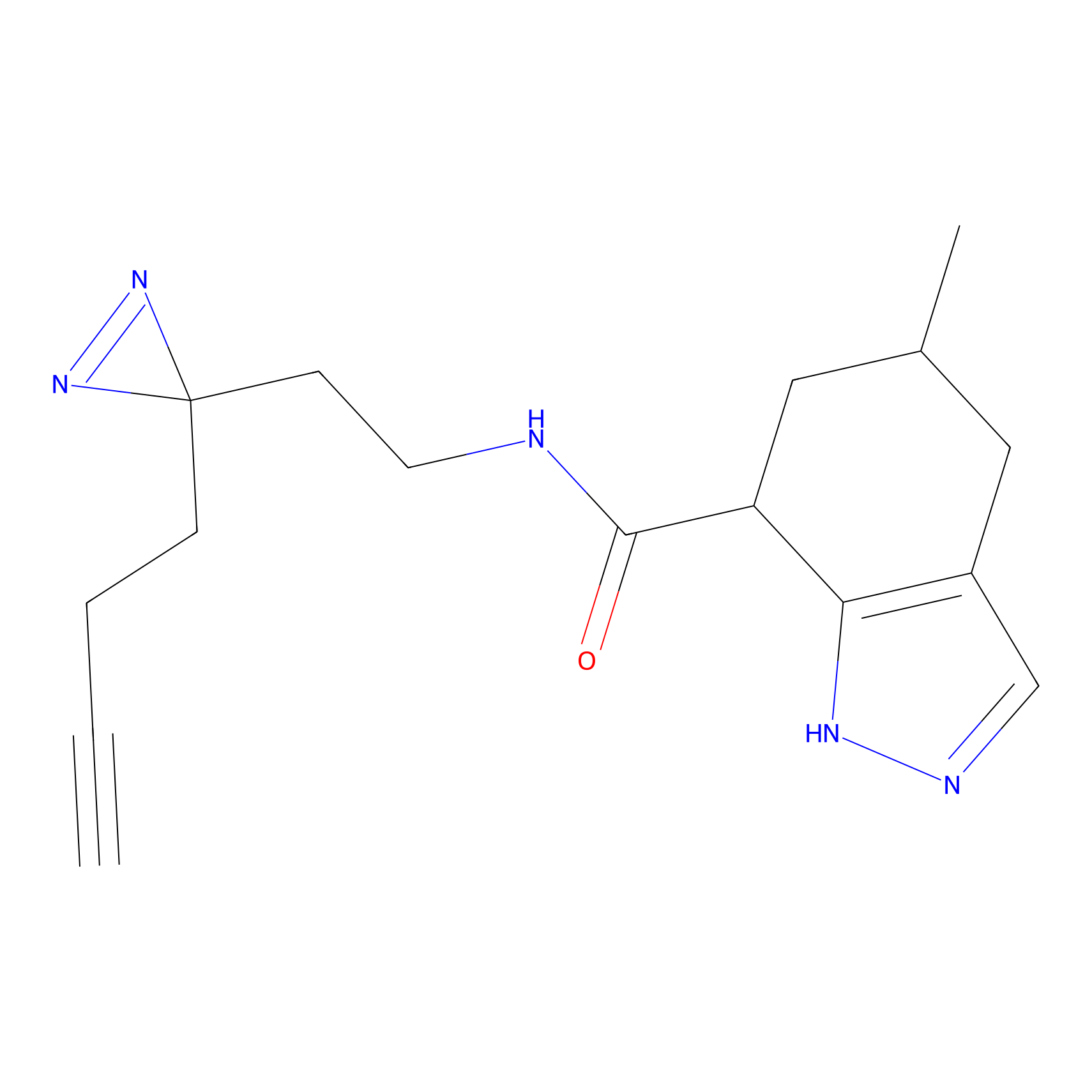
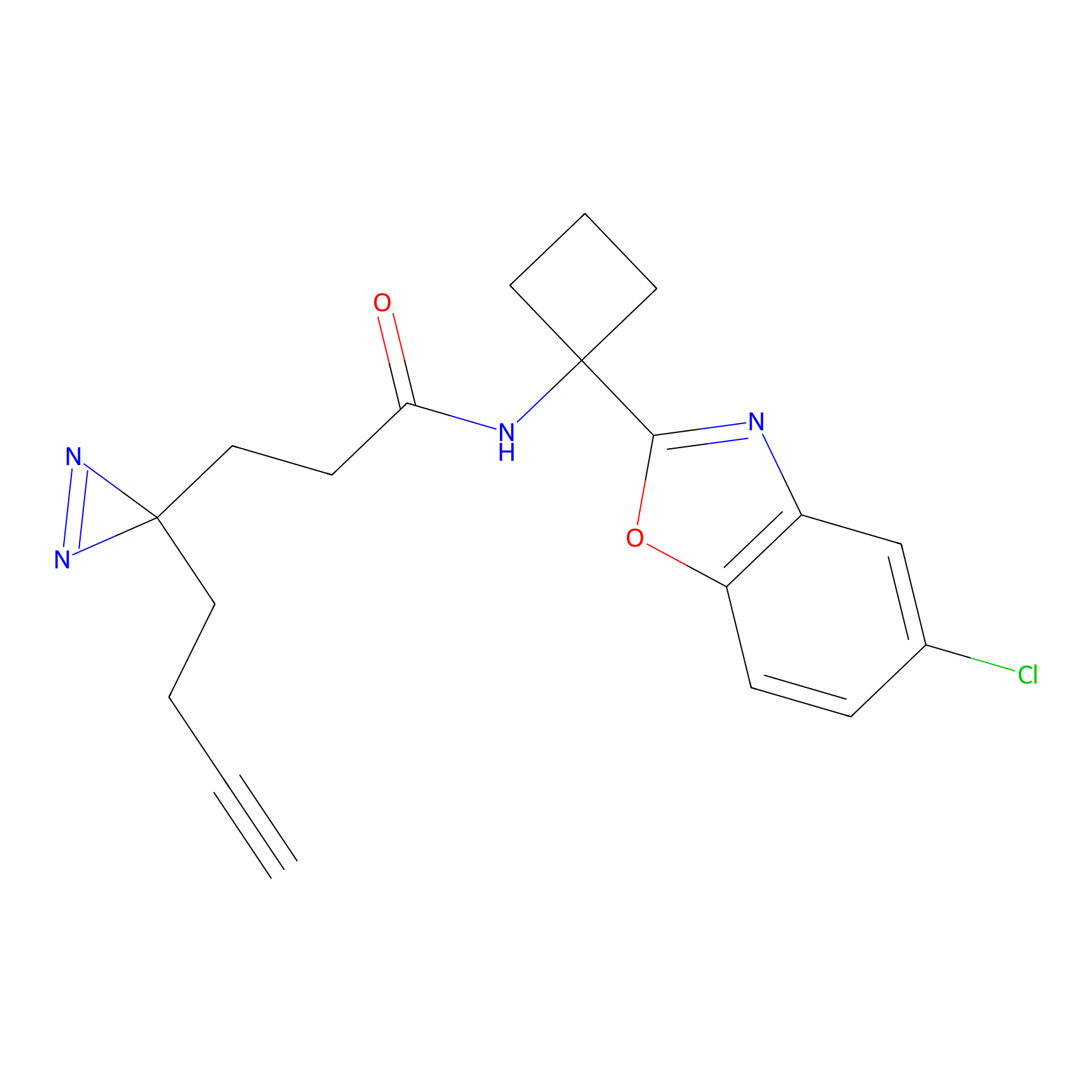
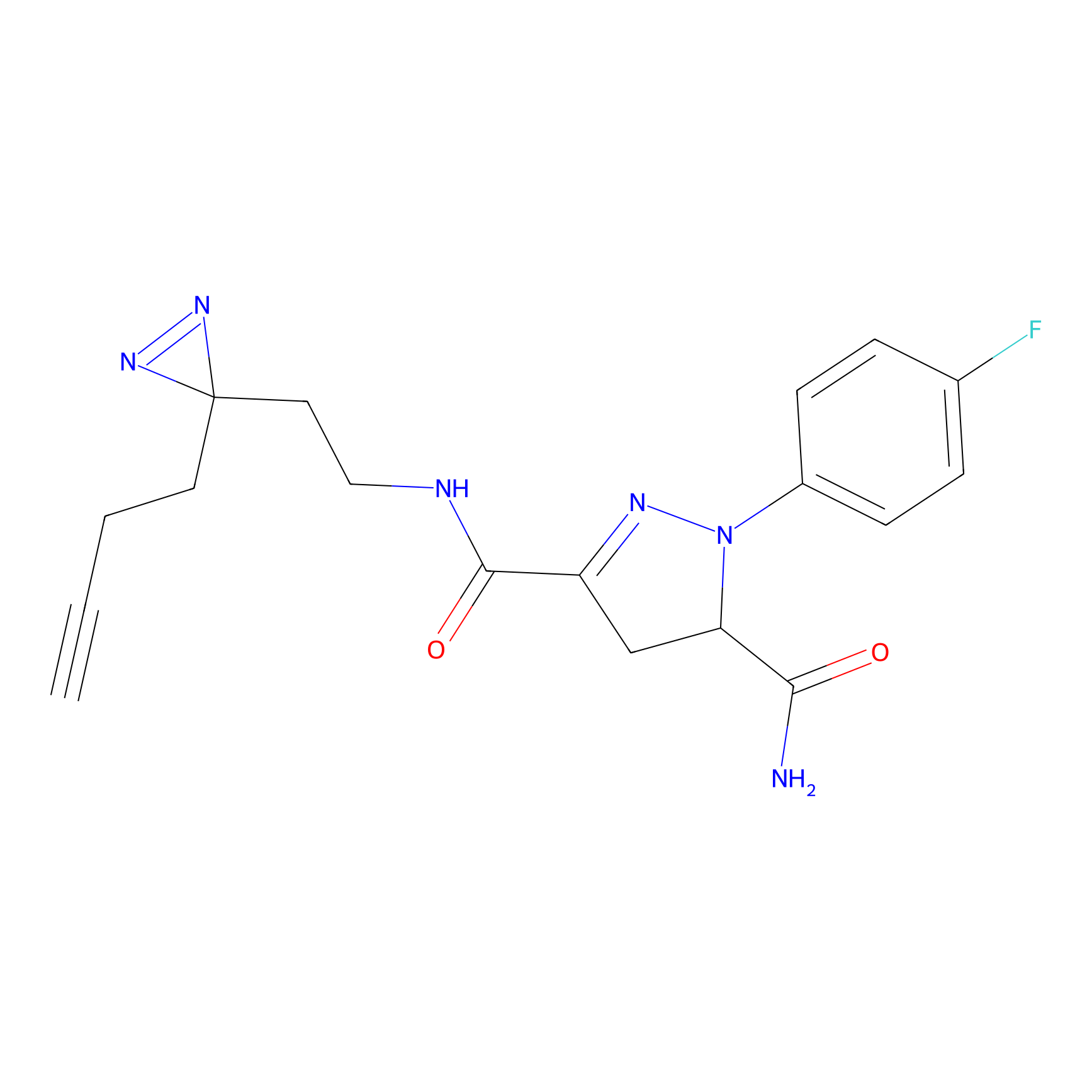
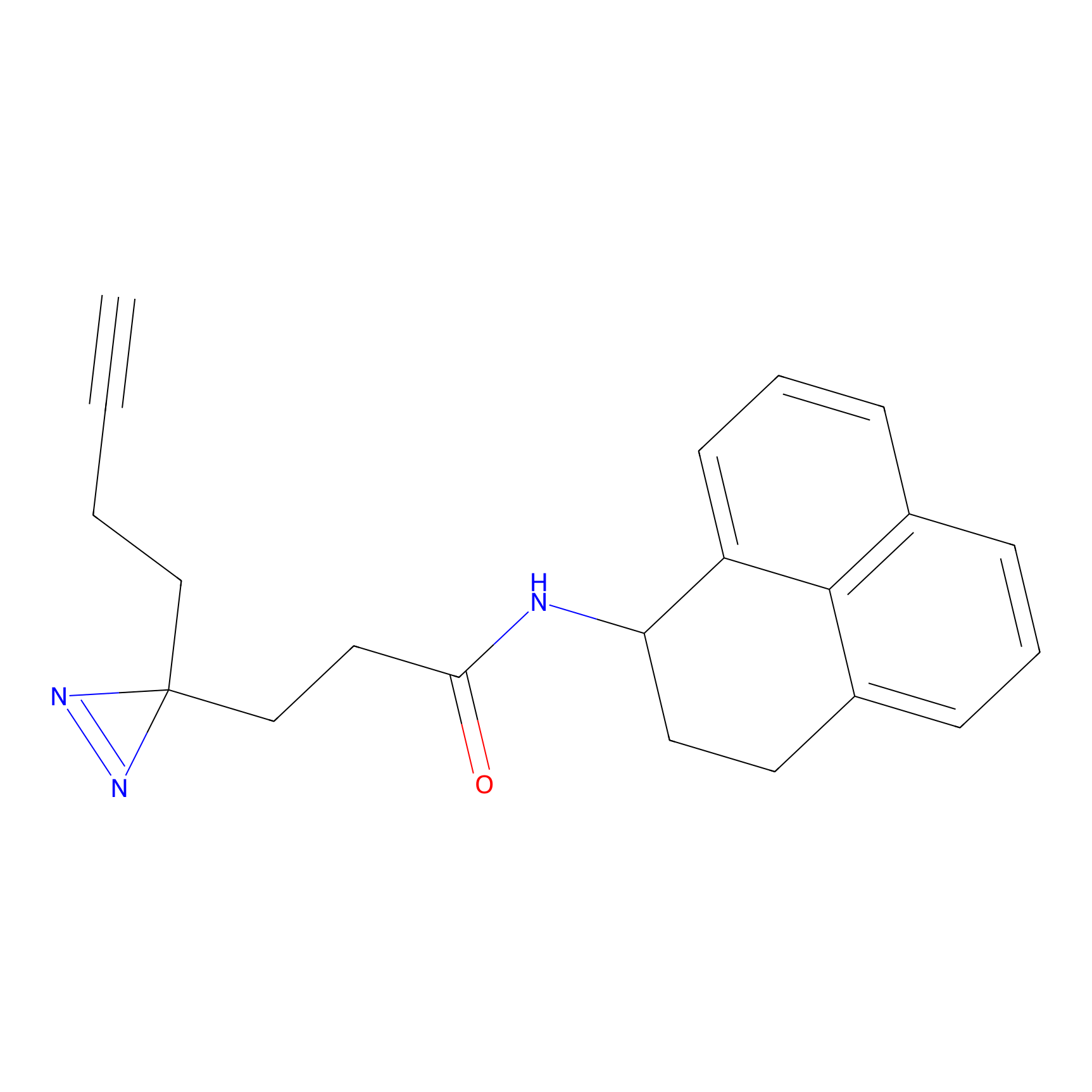
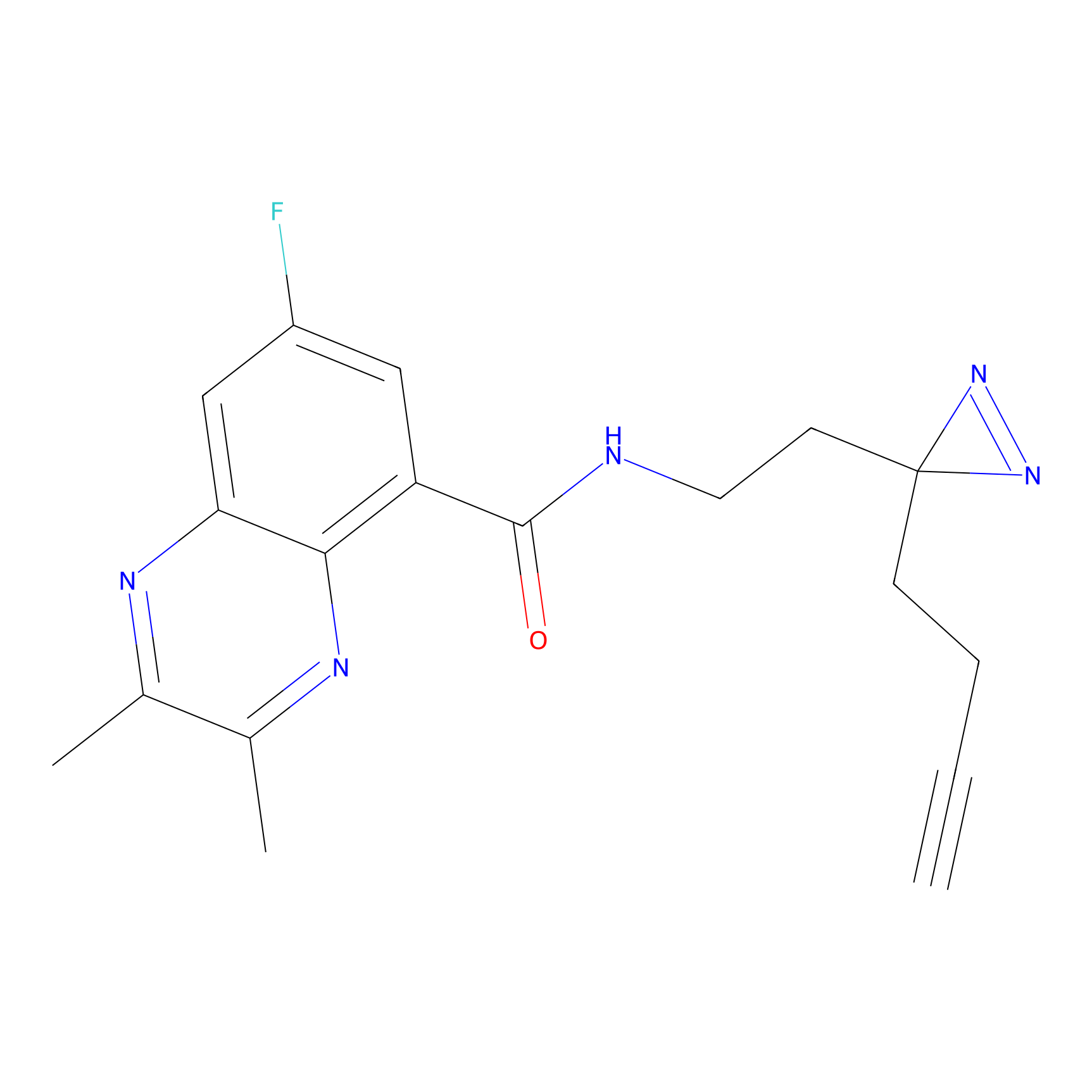
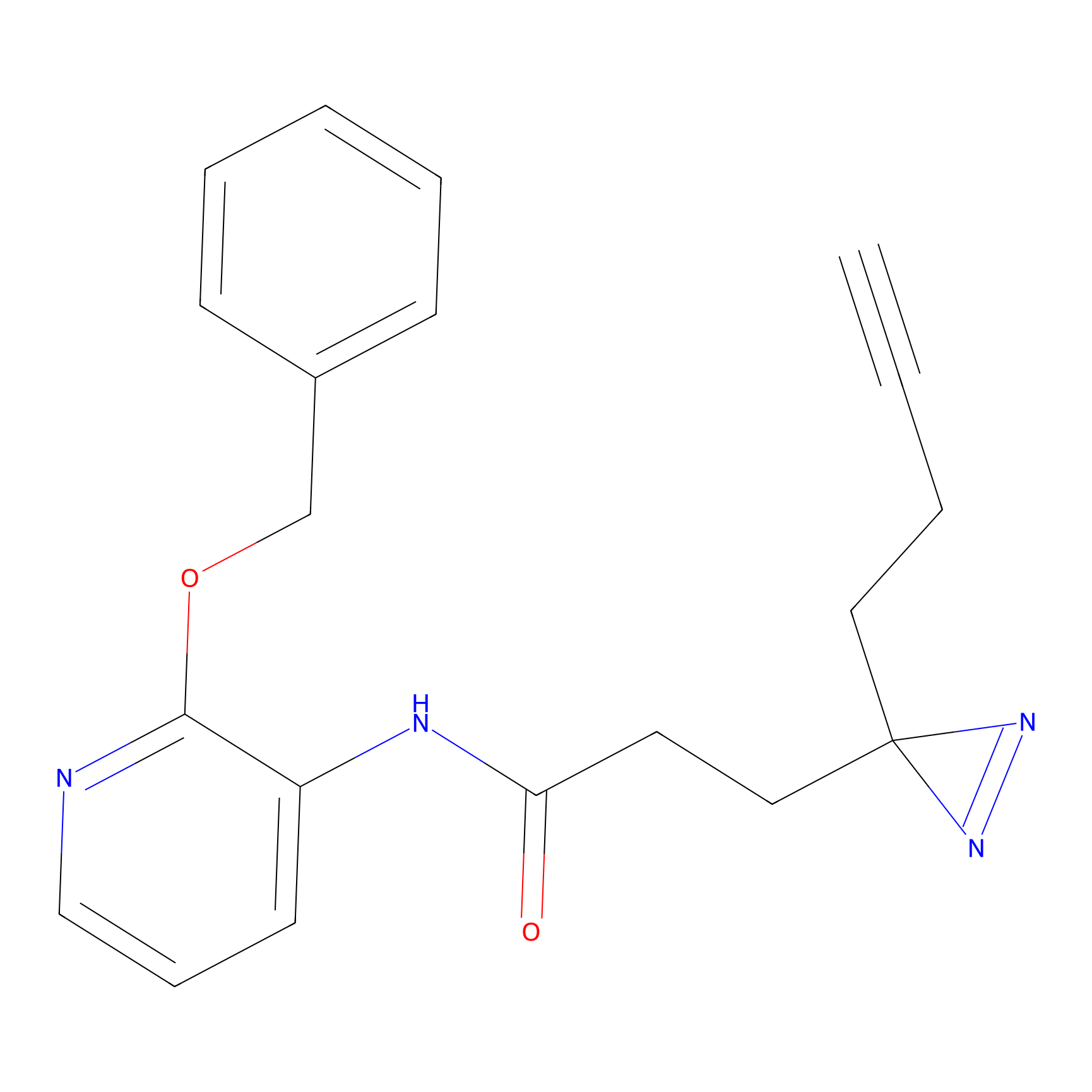
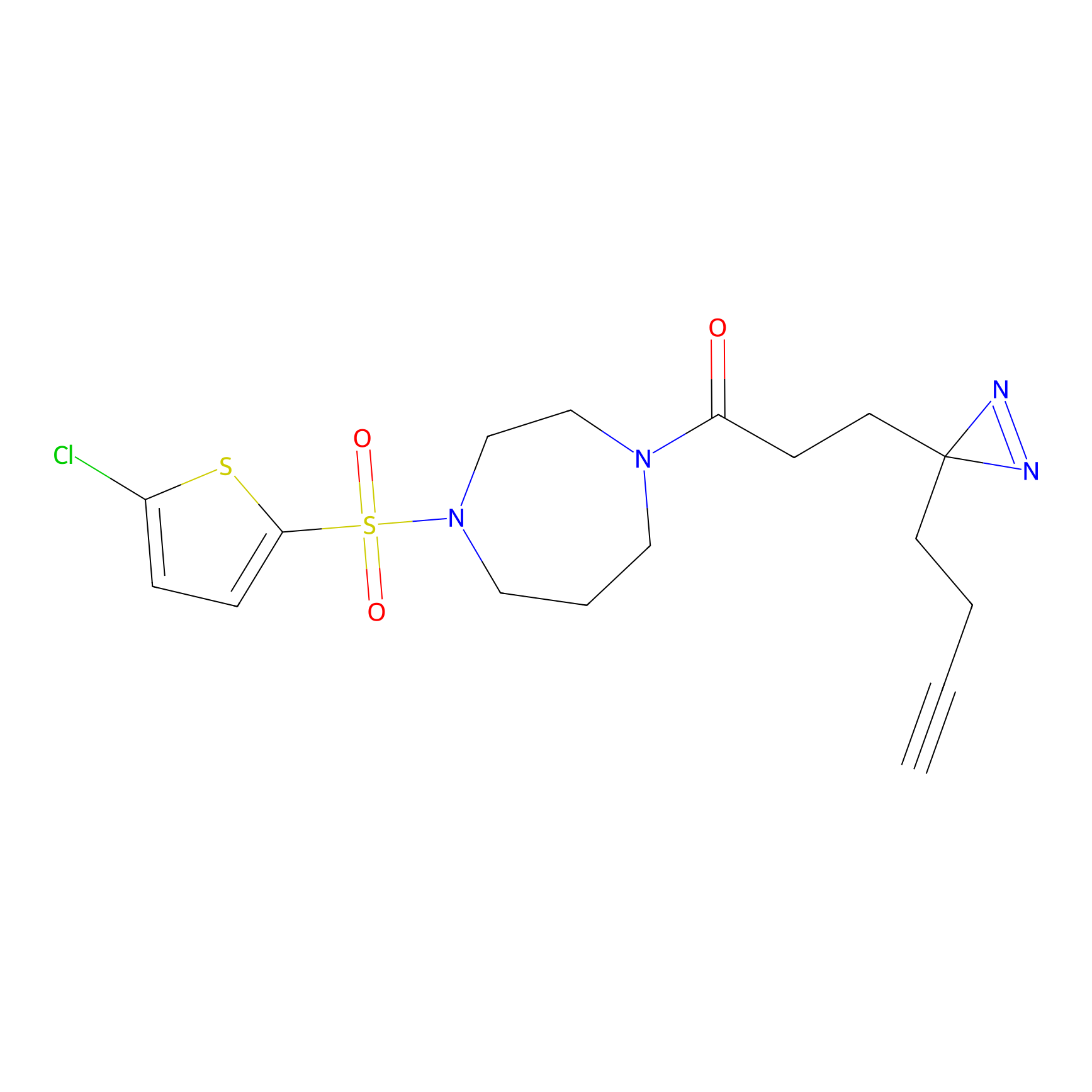
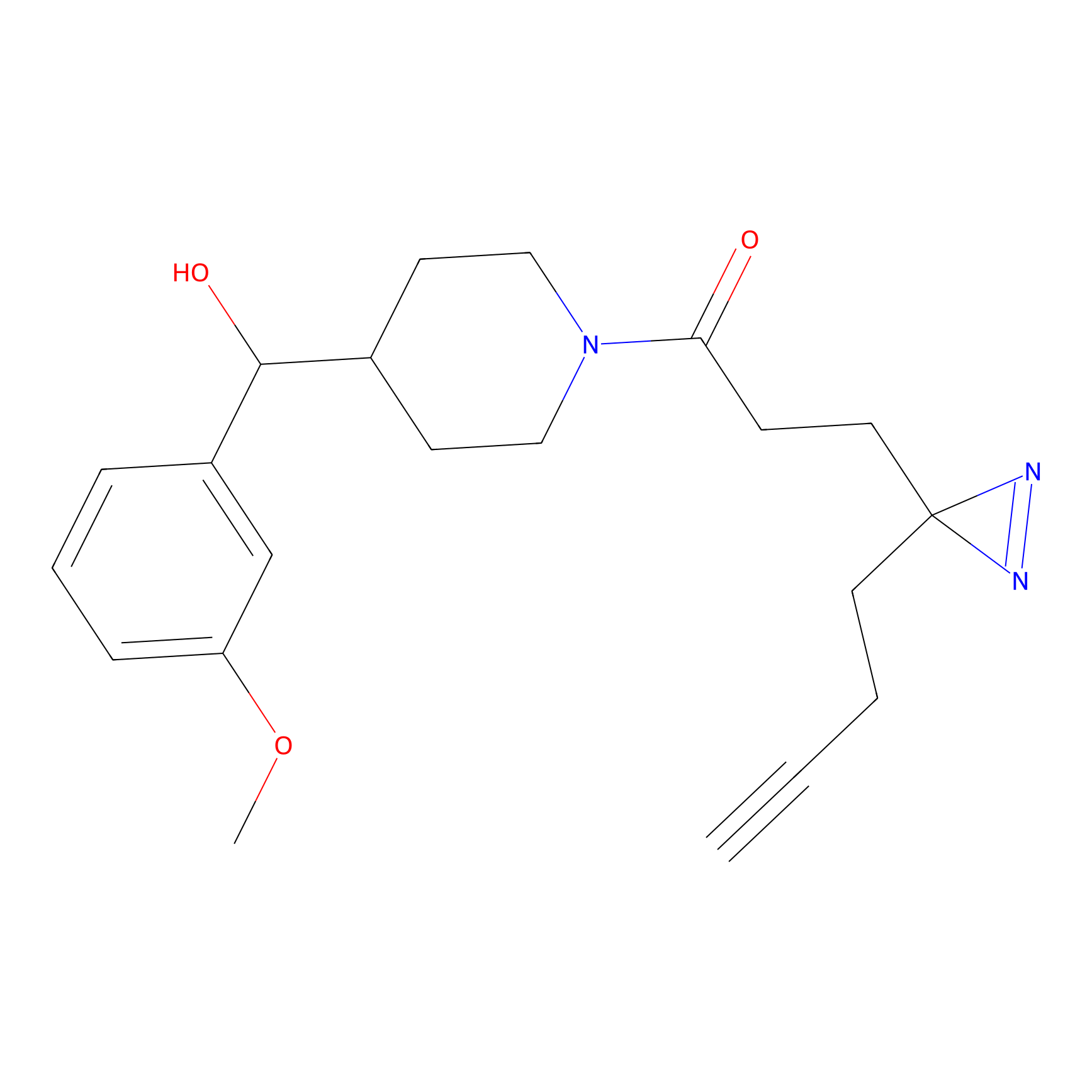
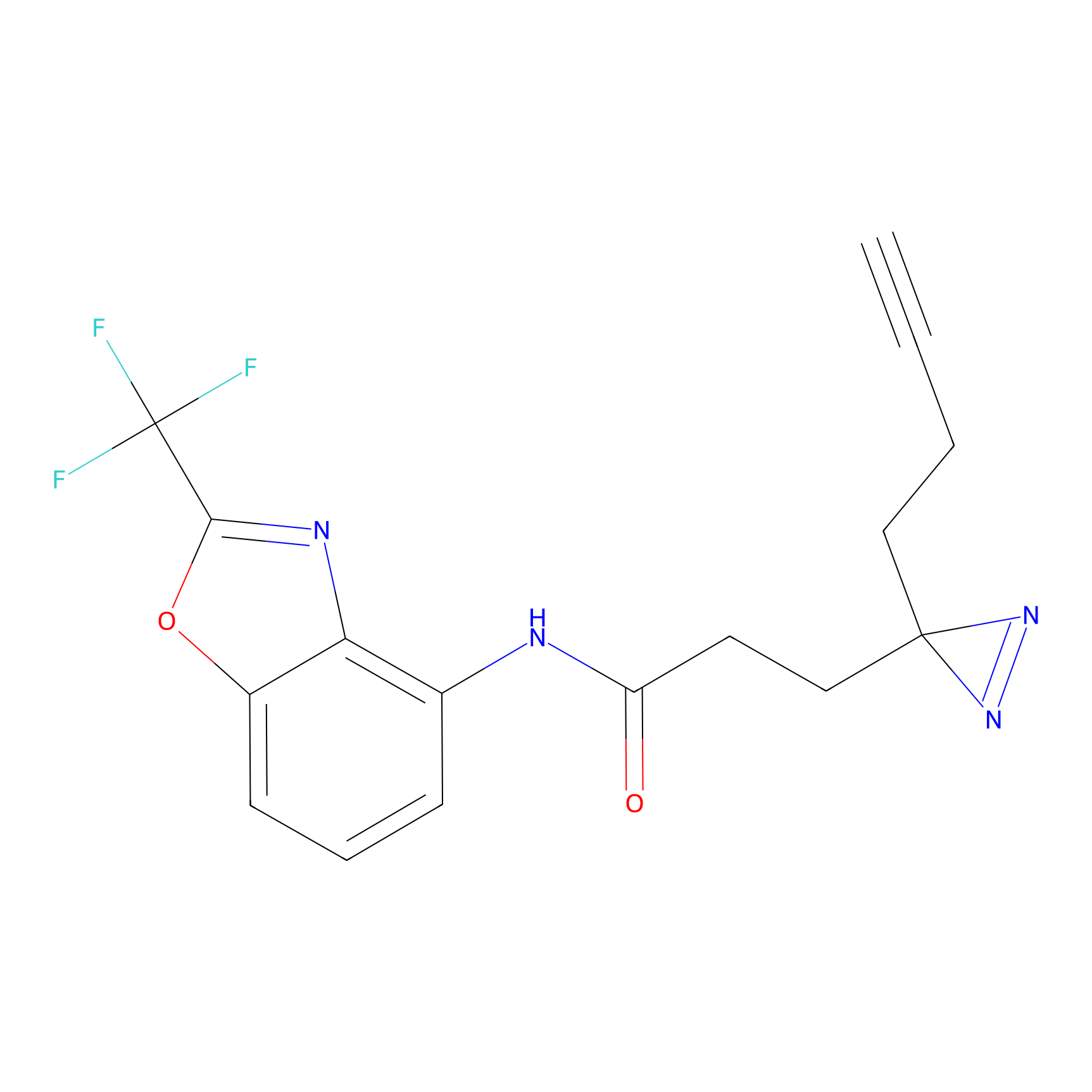
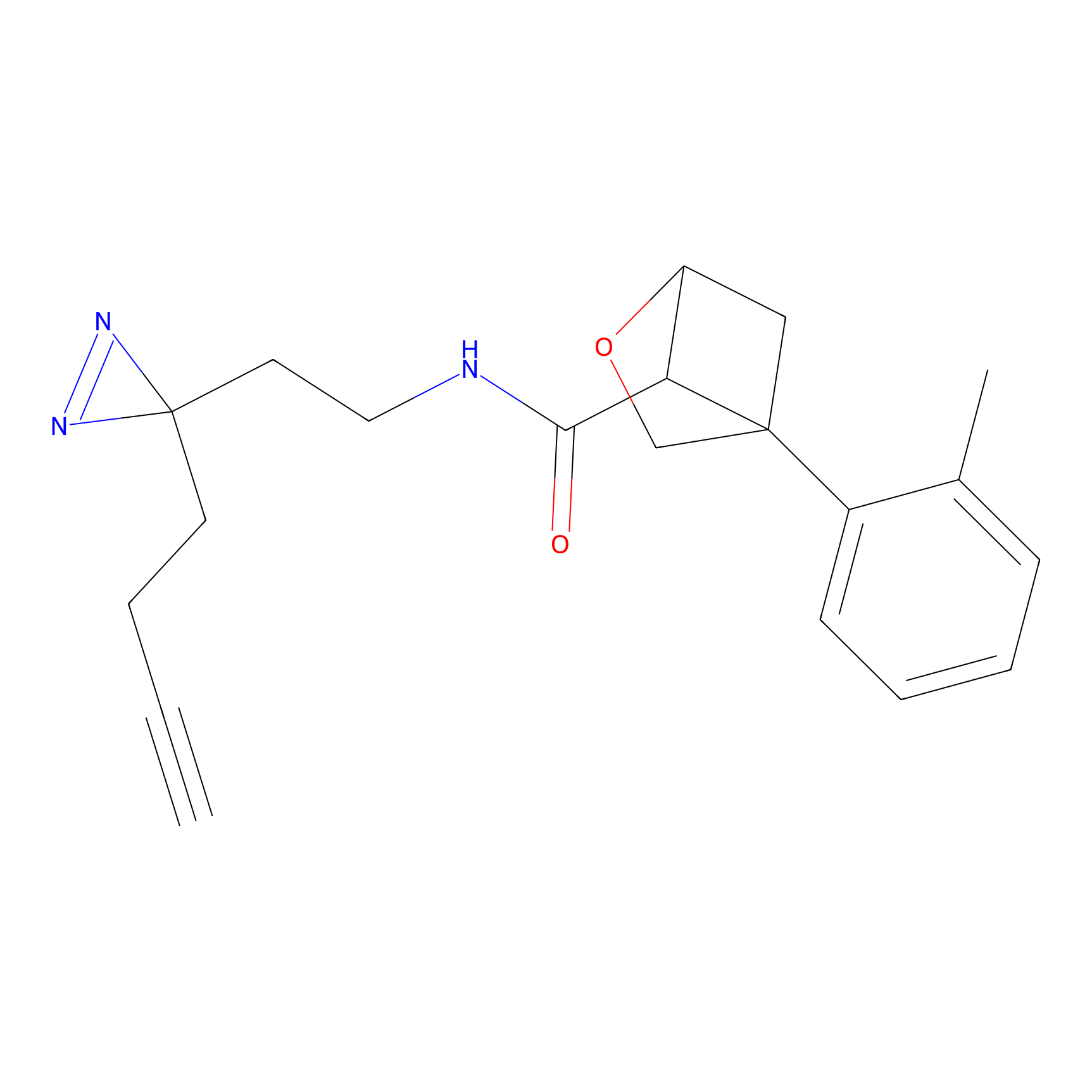
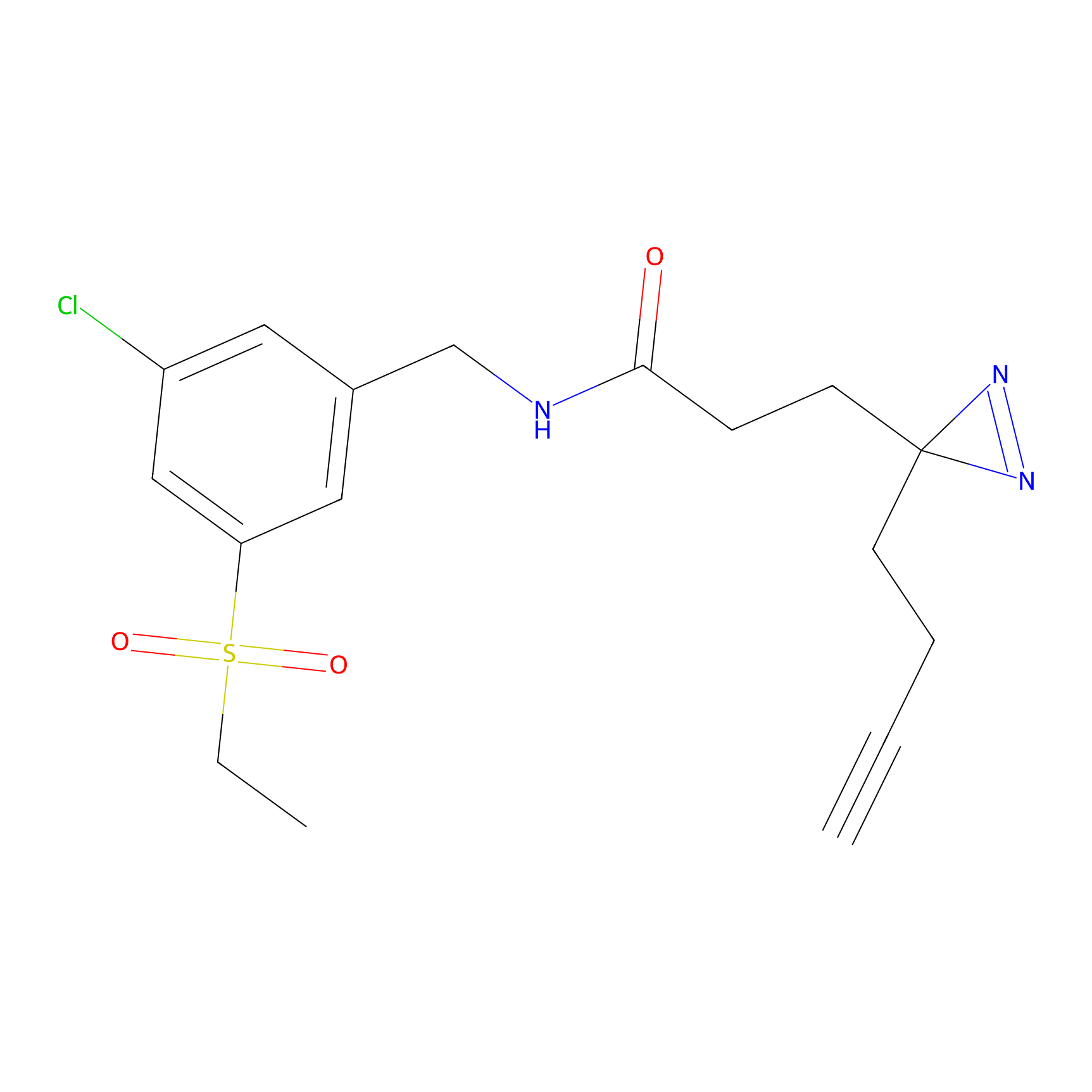
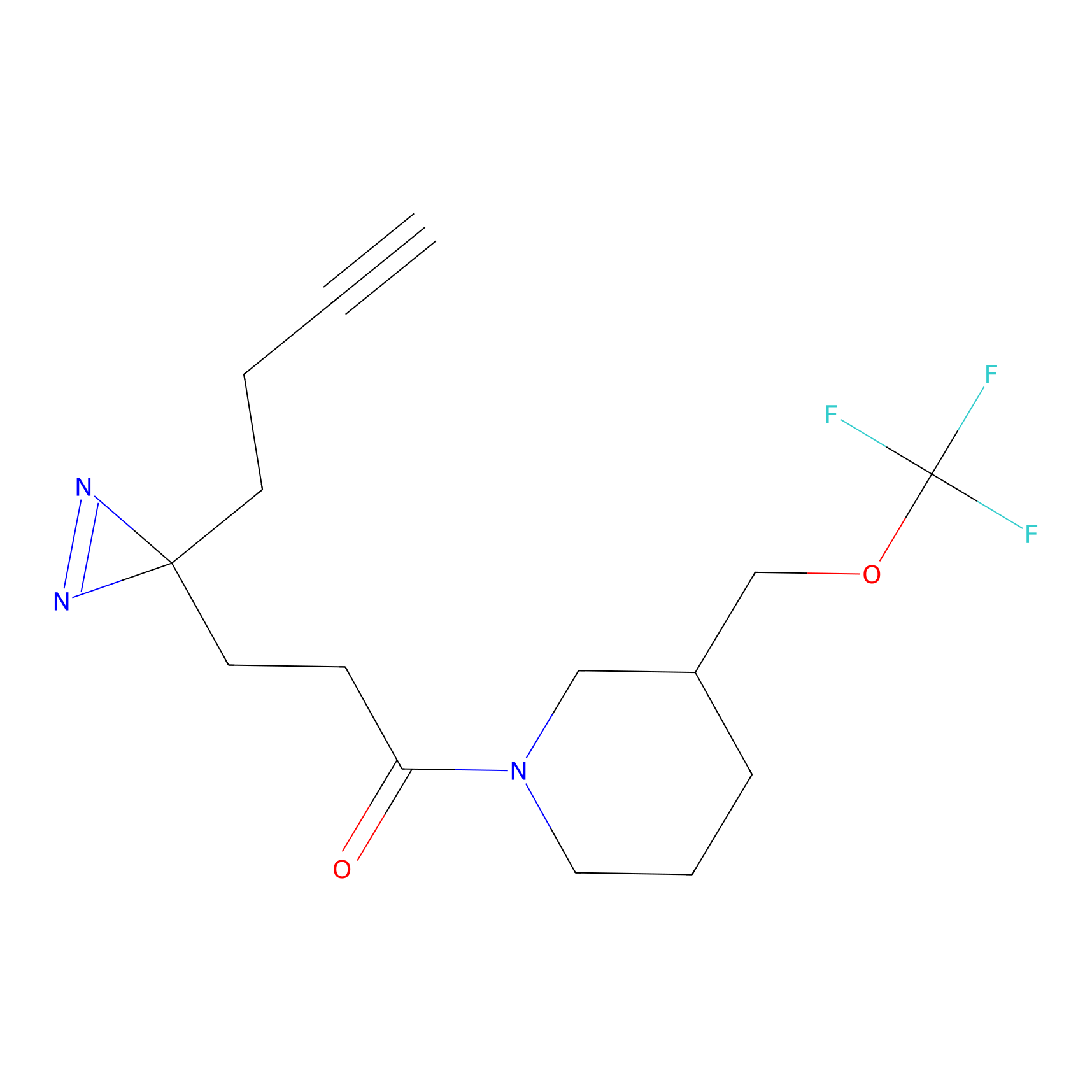
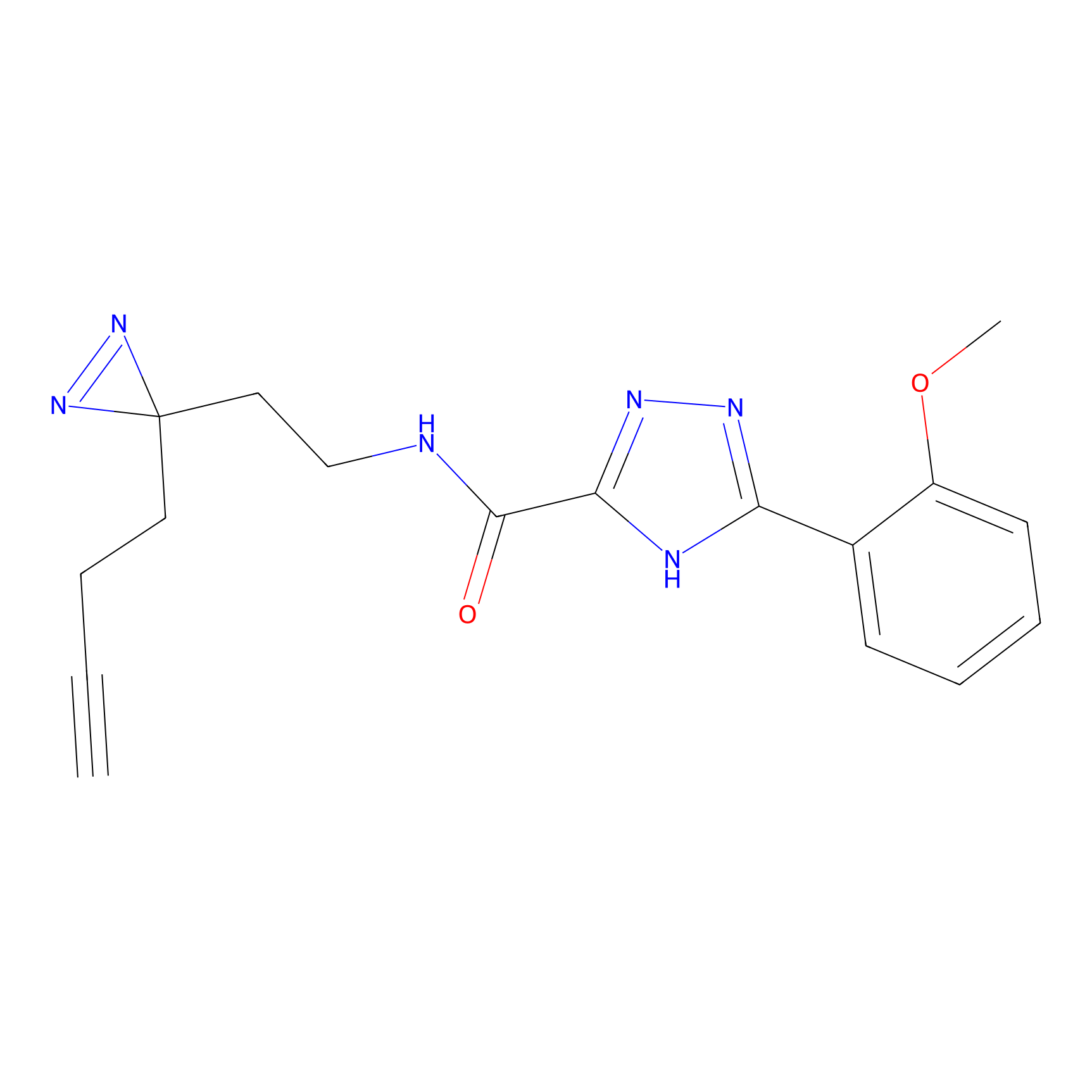
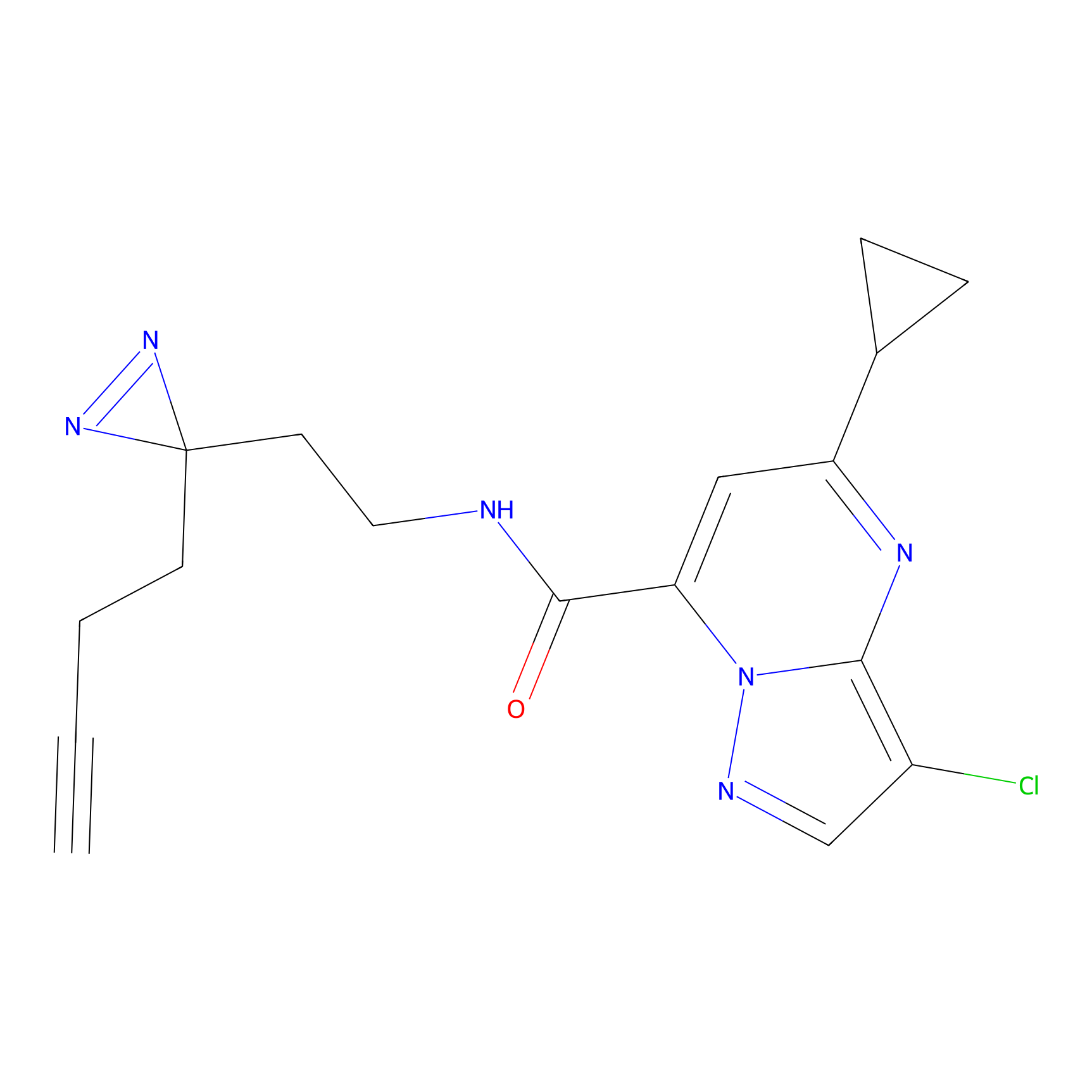
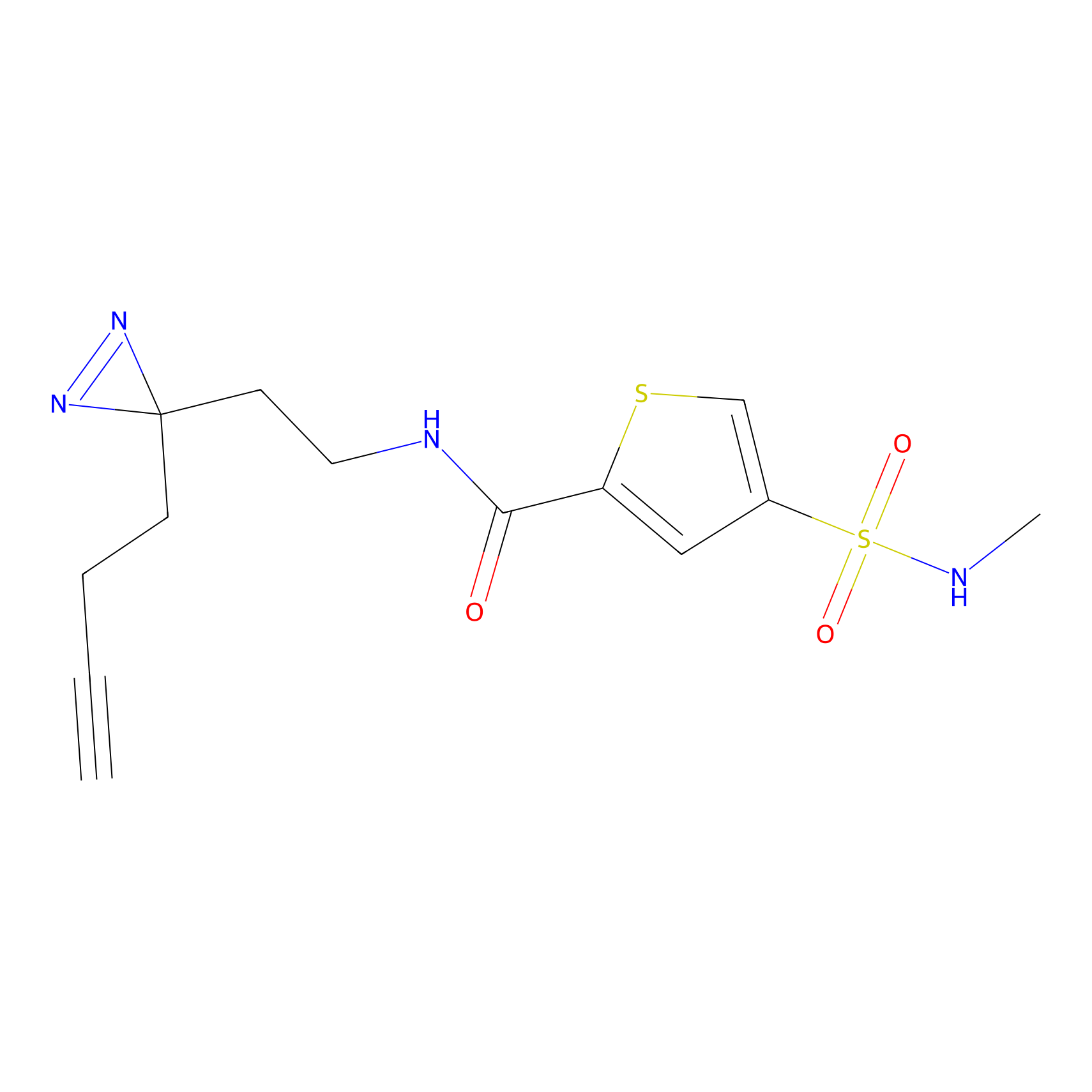
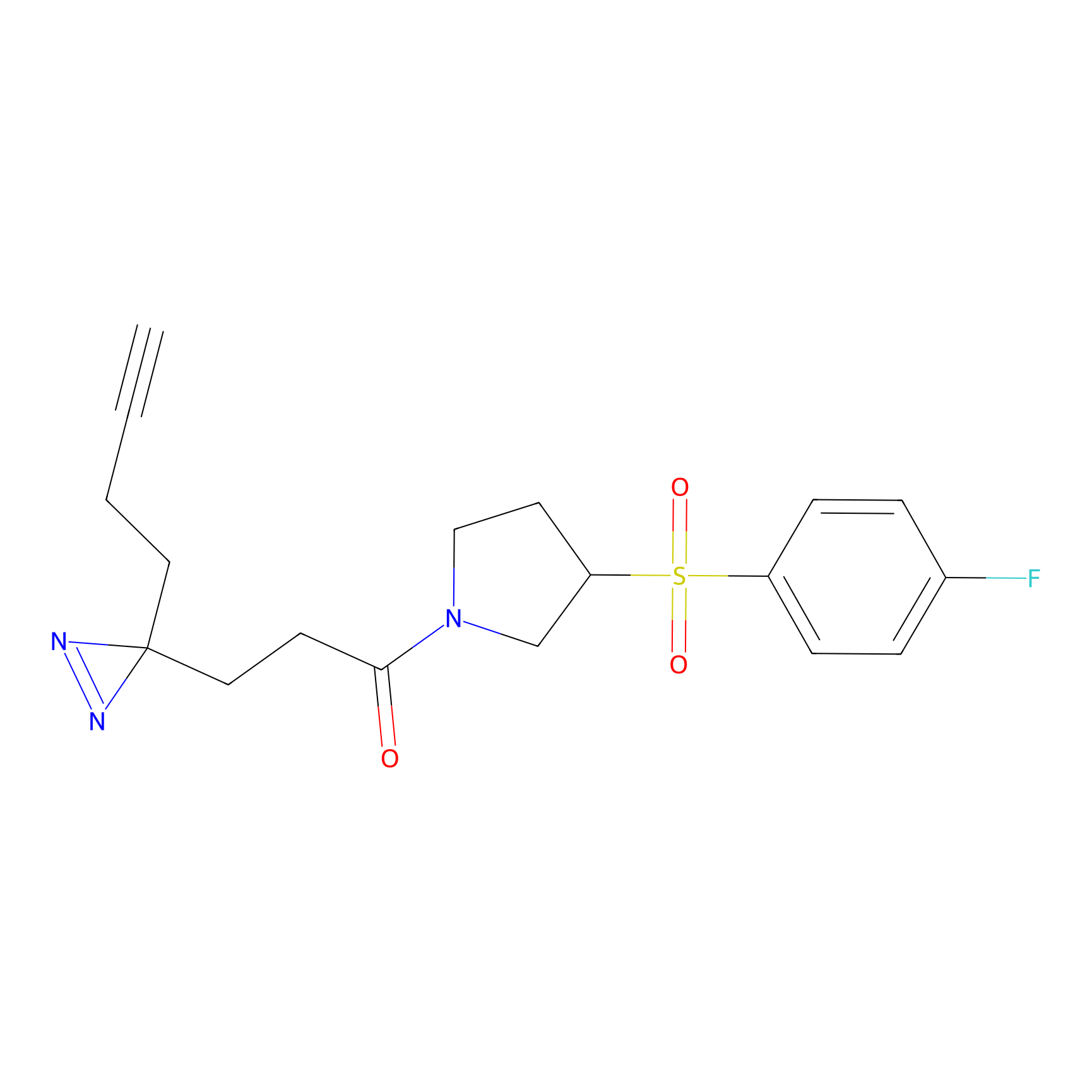
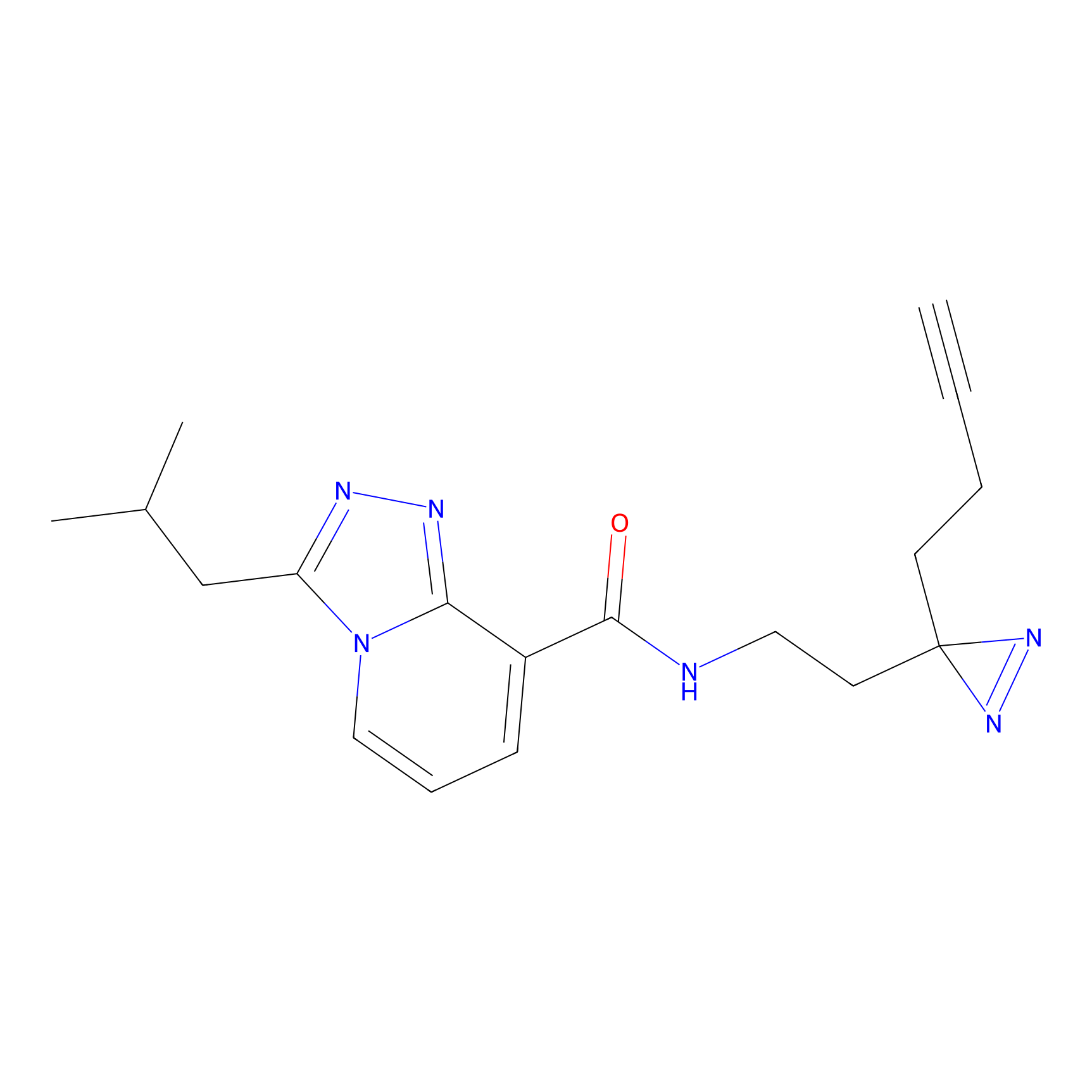
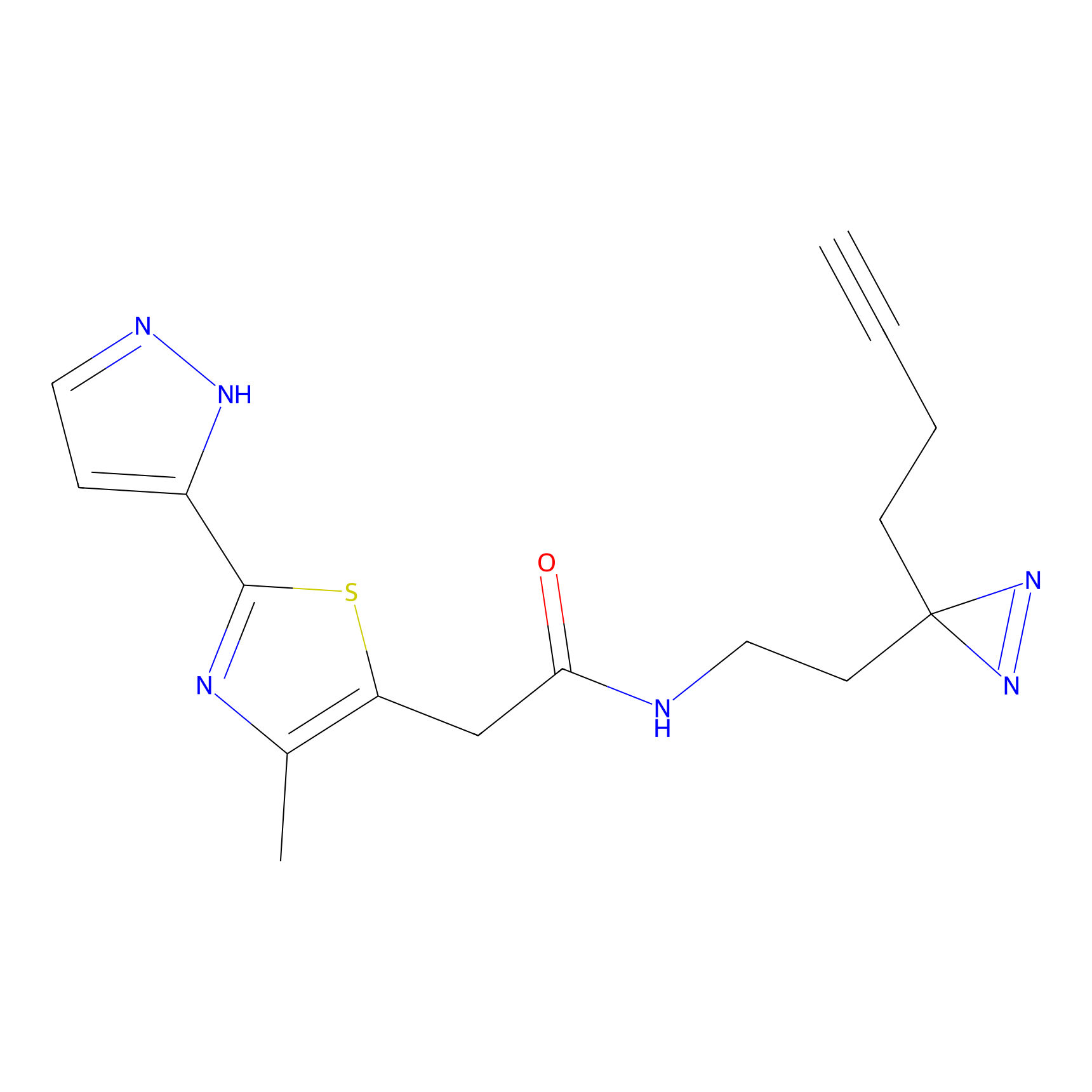
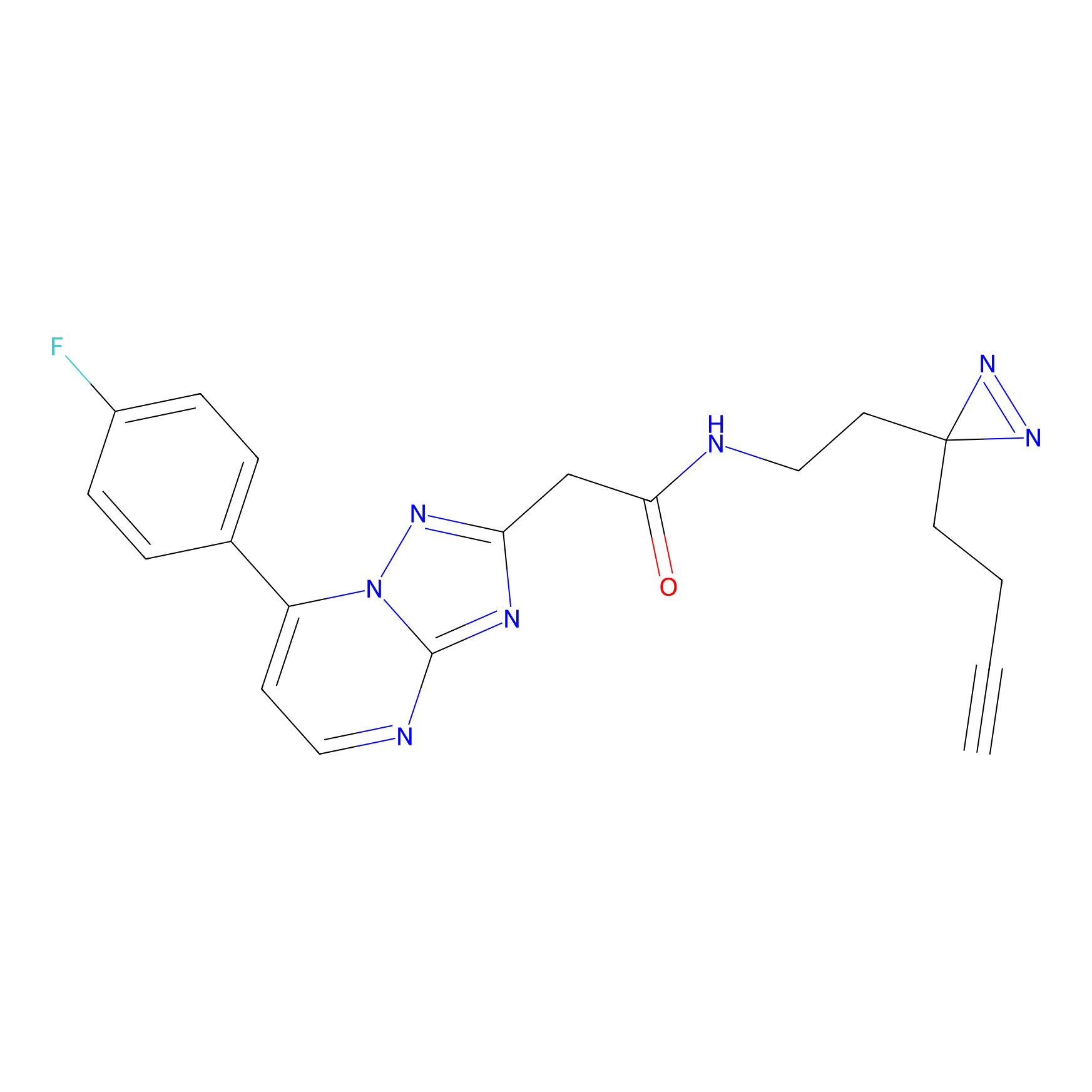
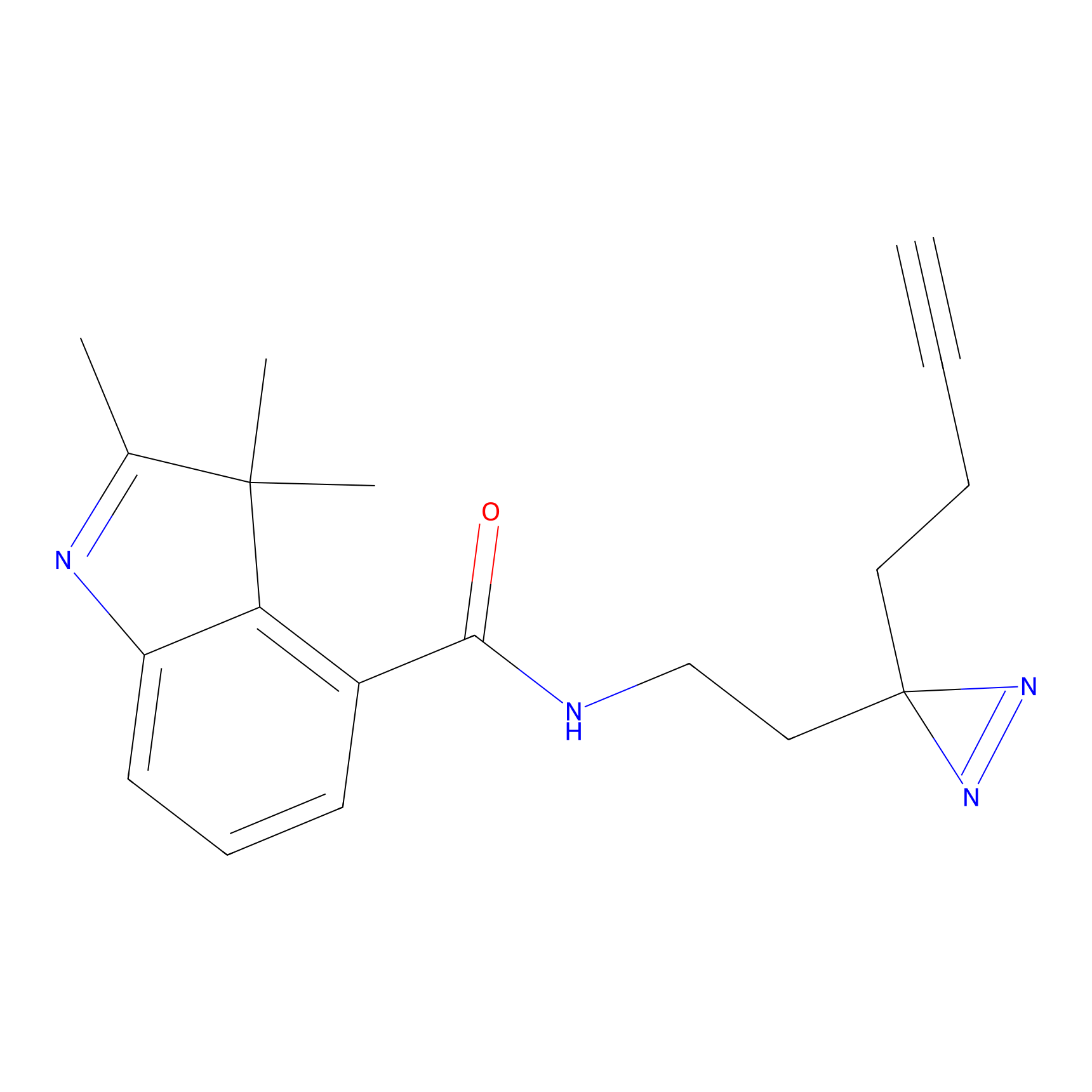
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
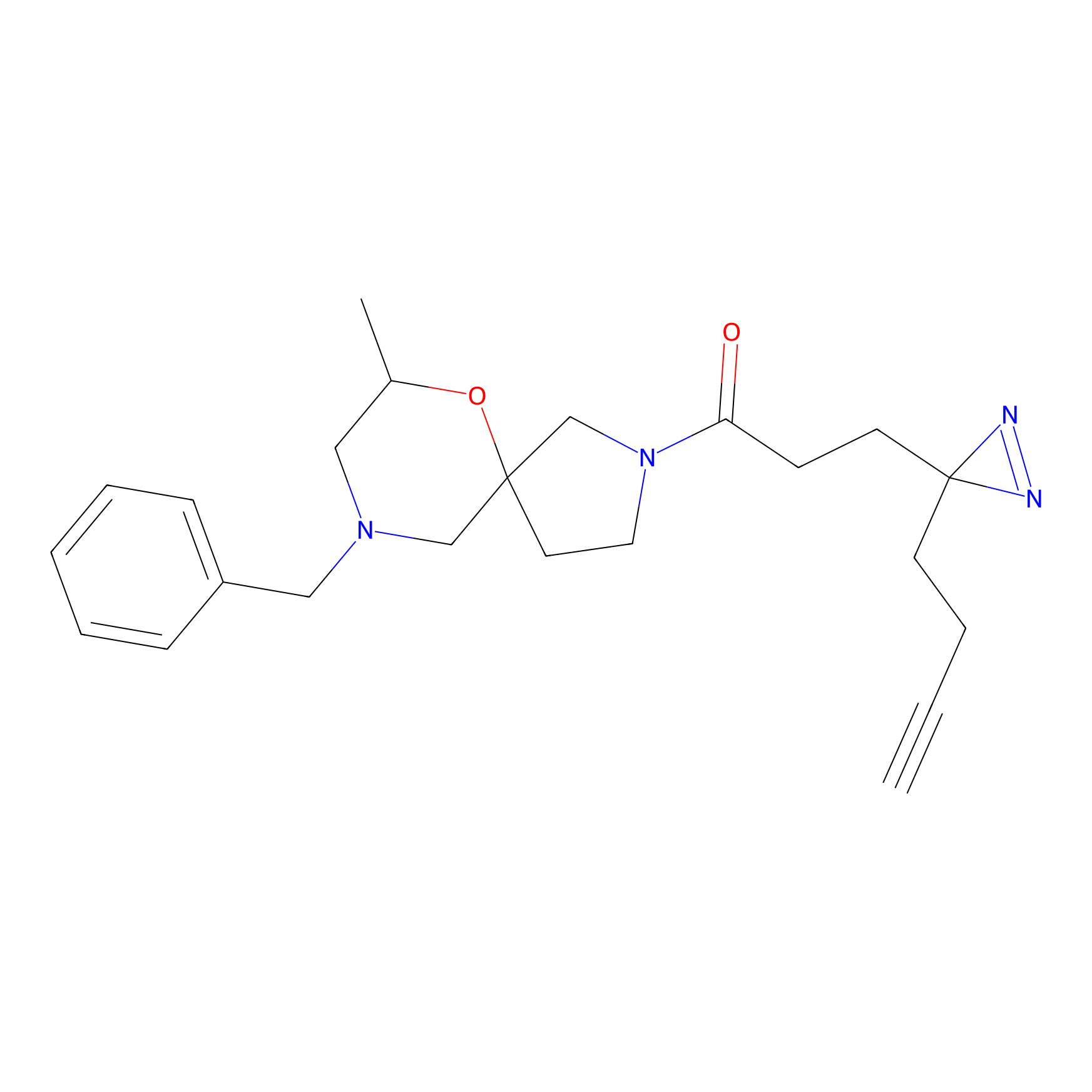
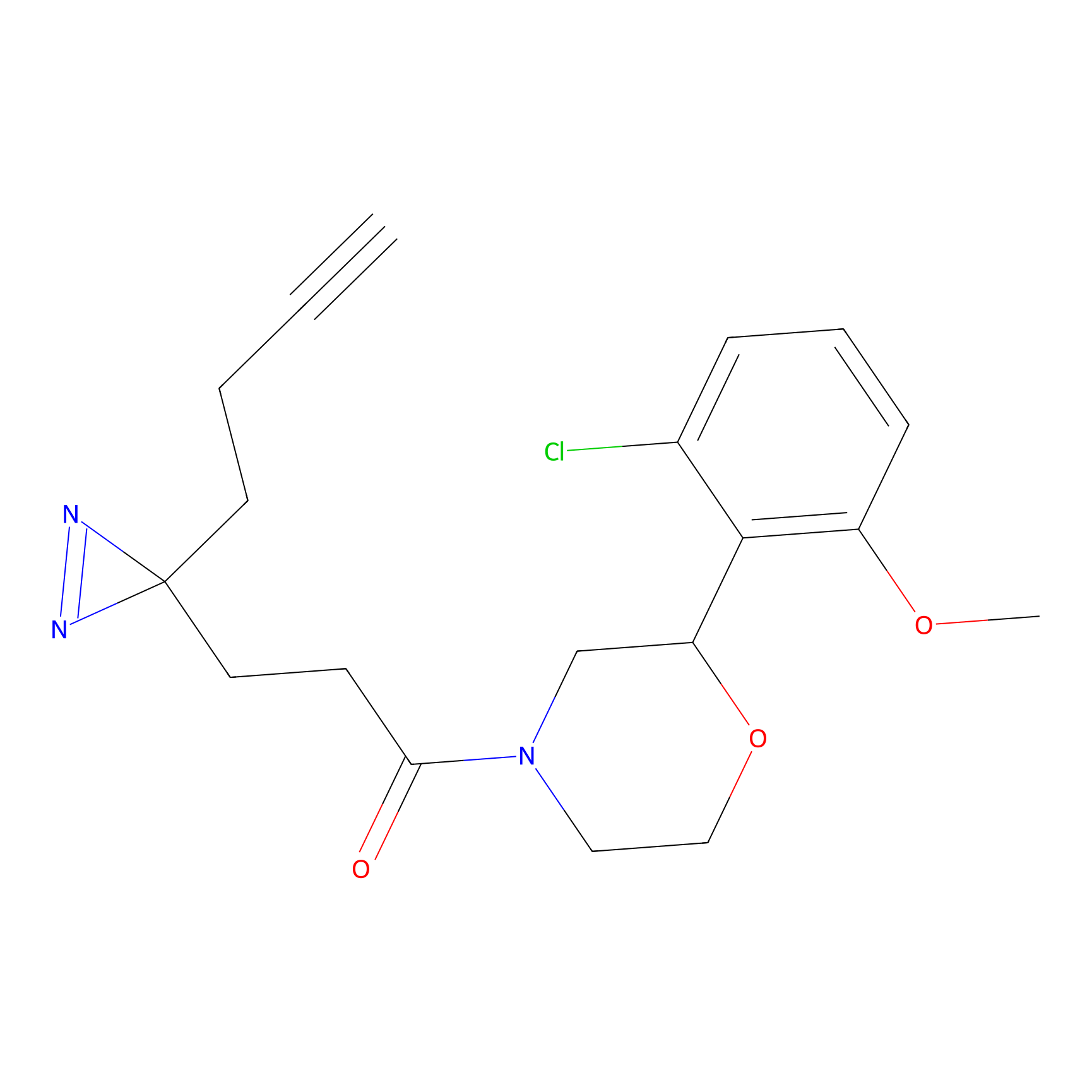
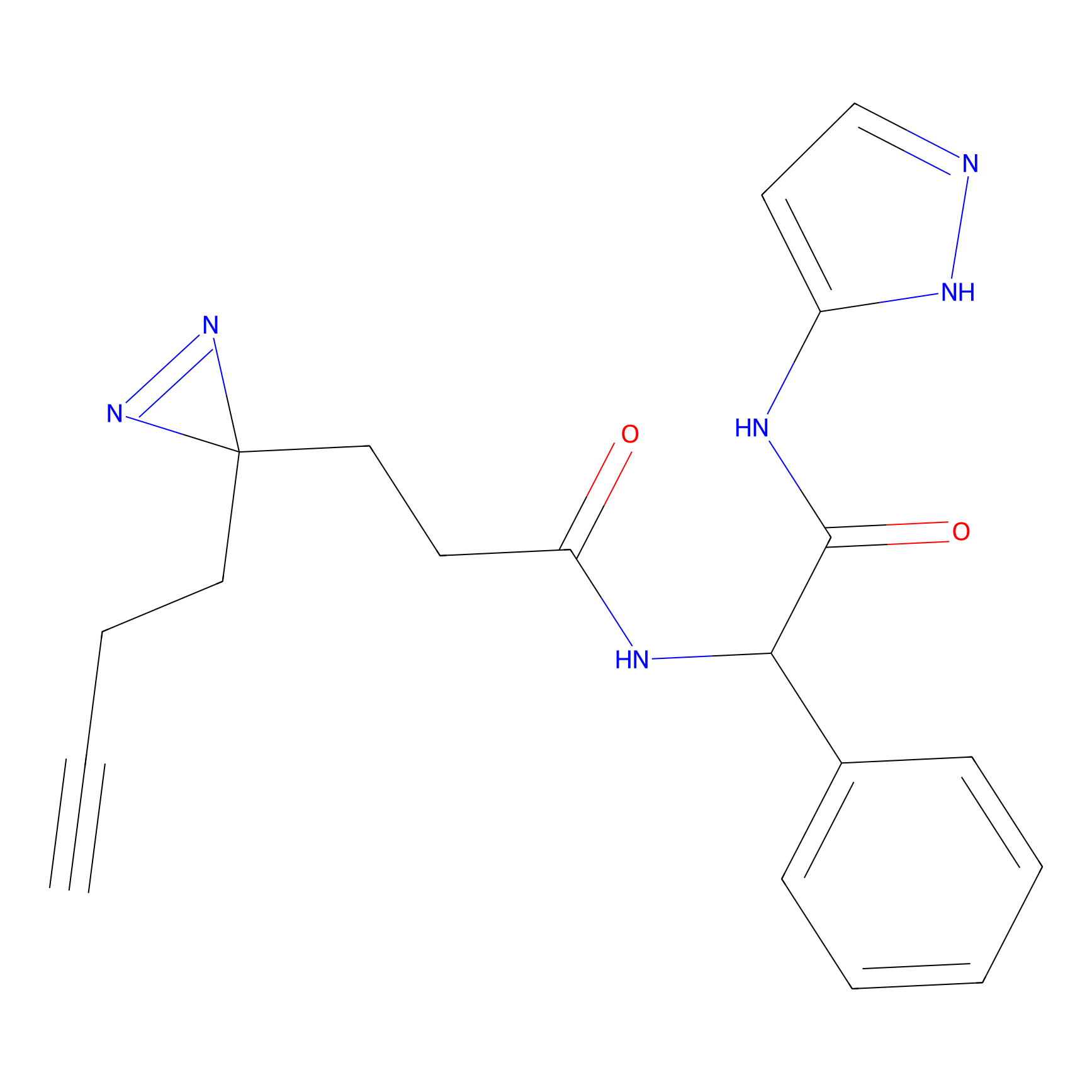
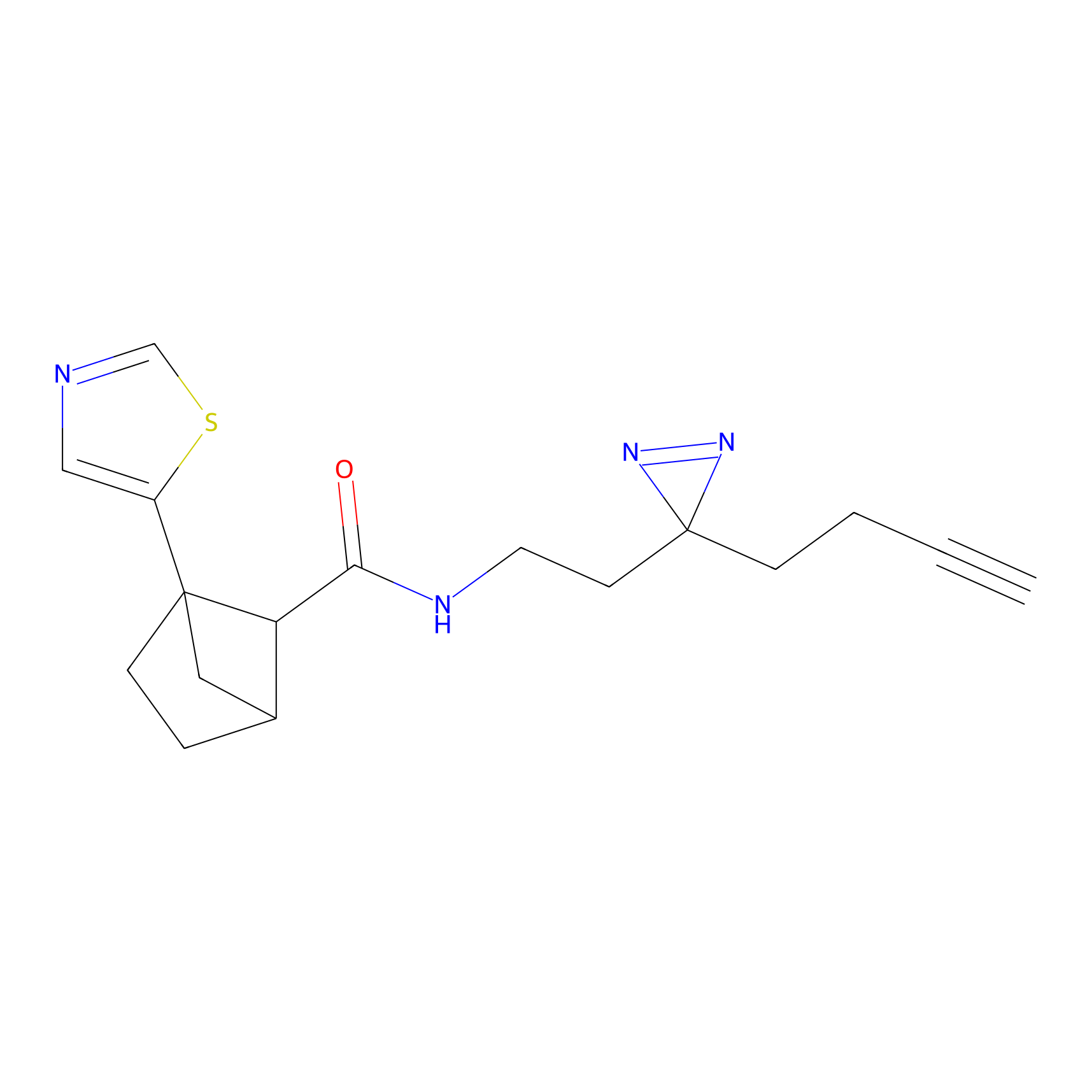
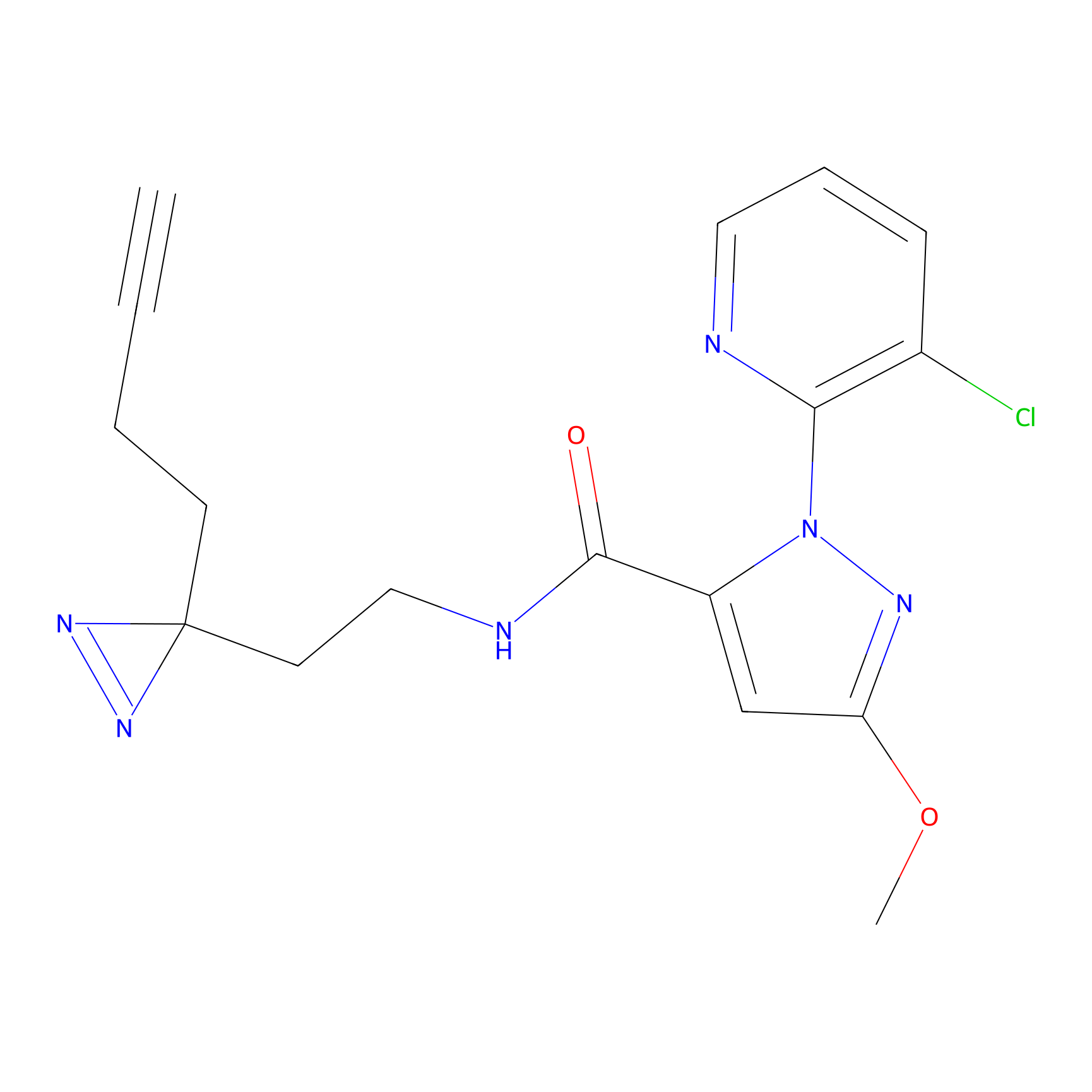
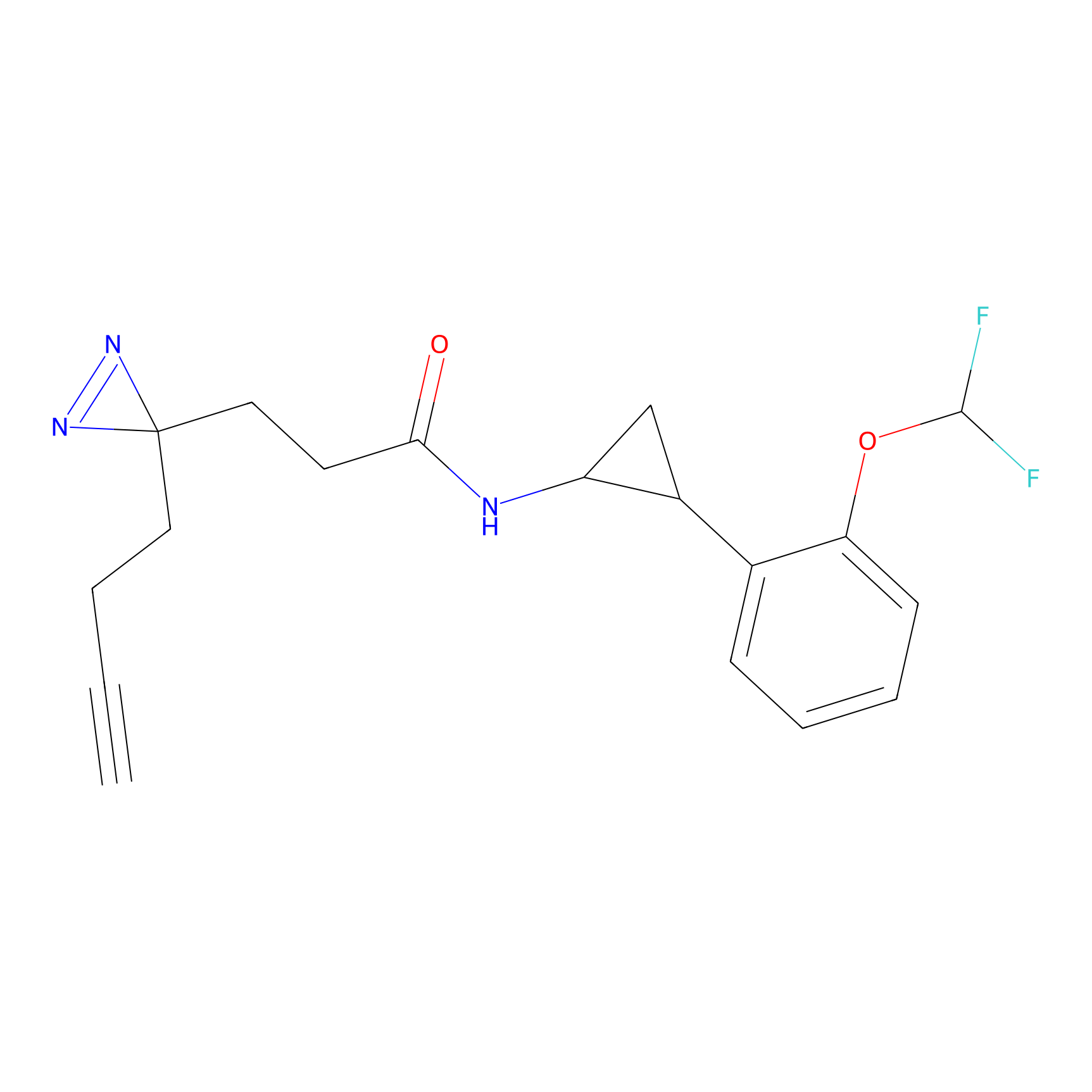
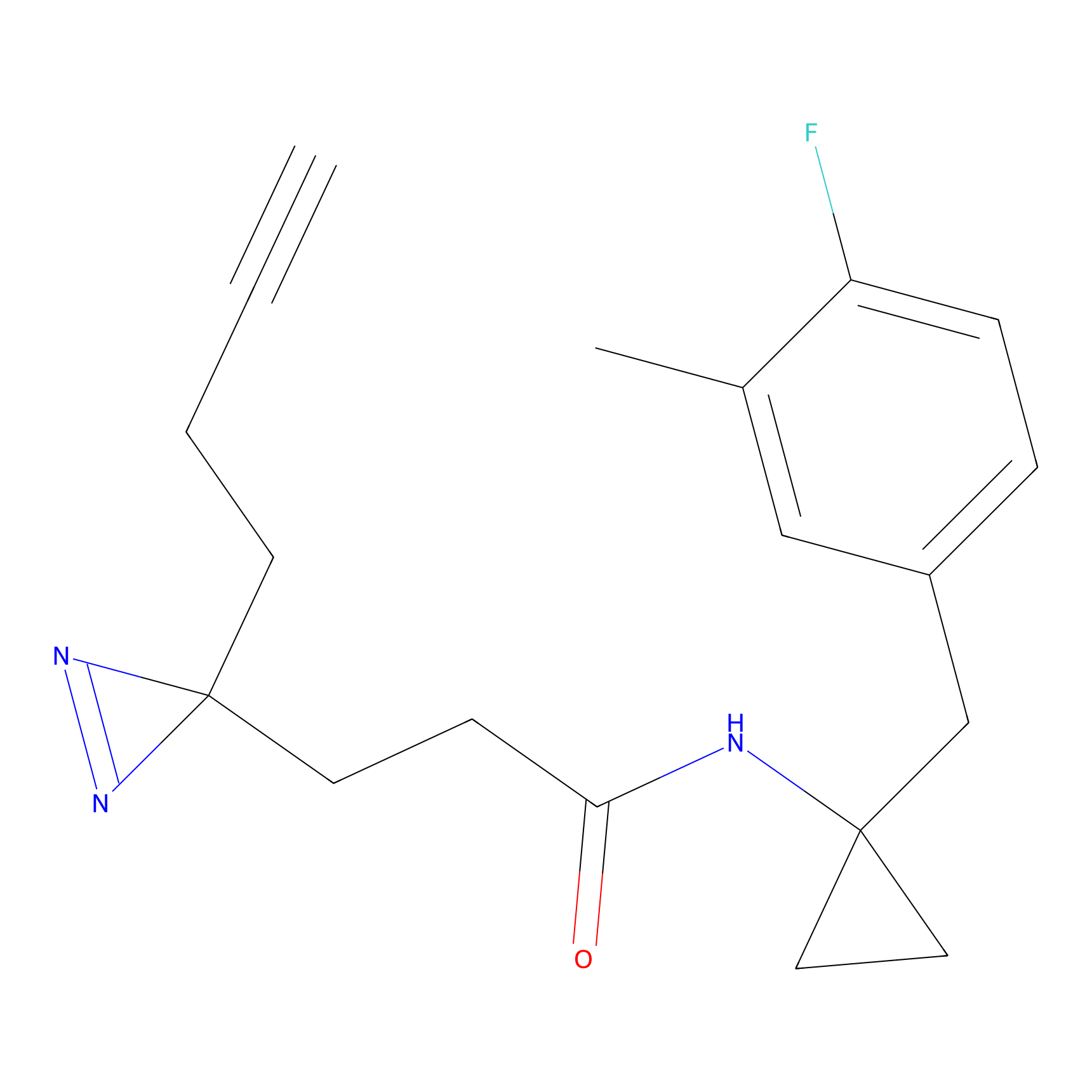
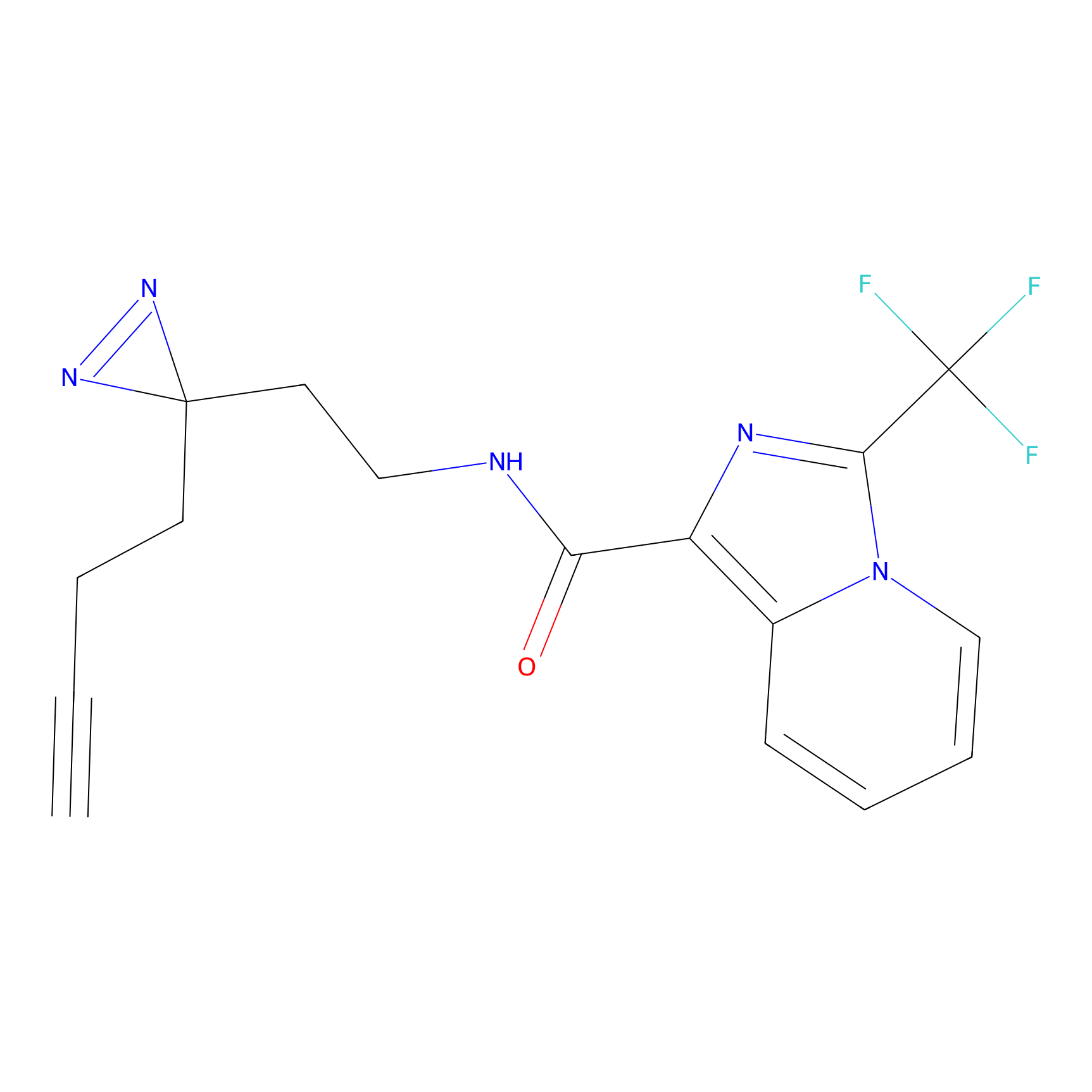
|
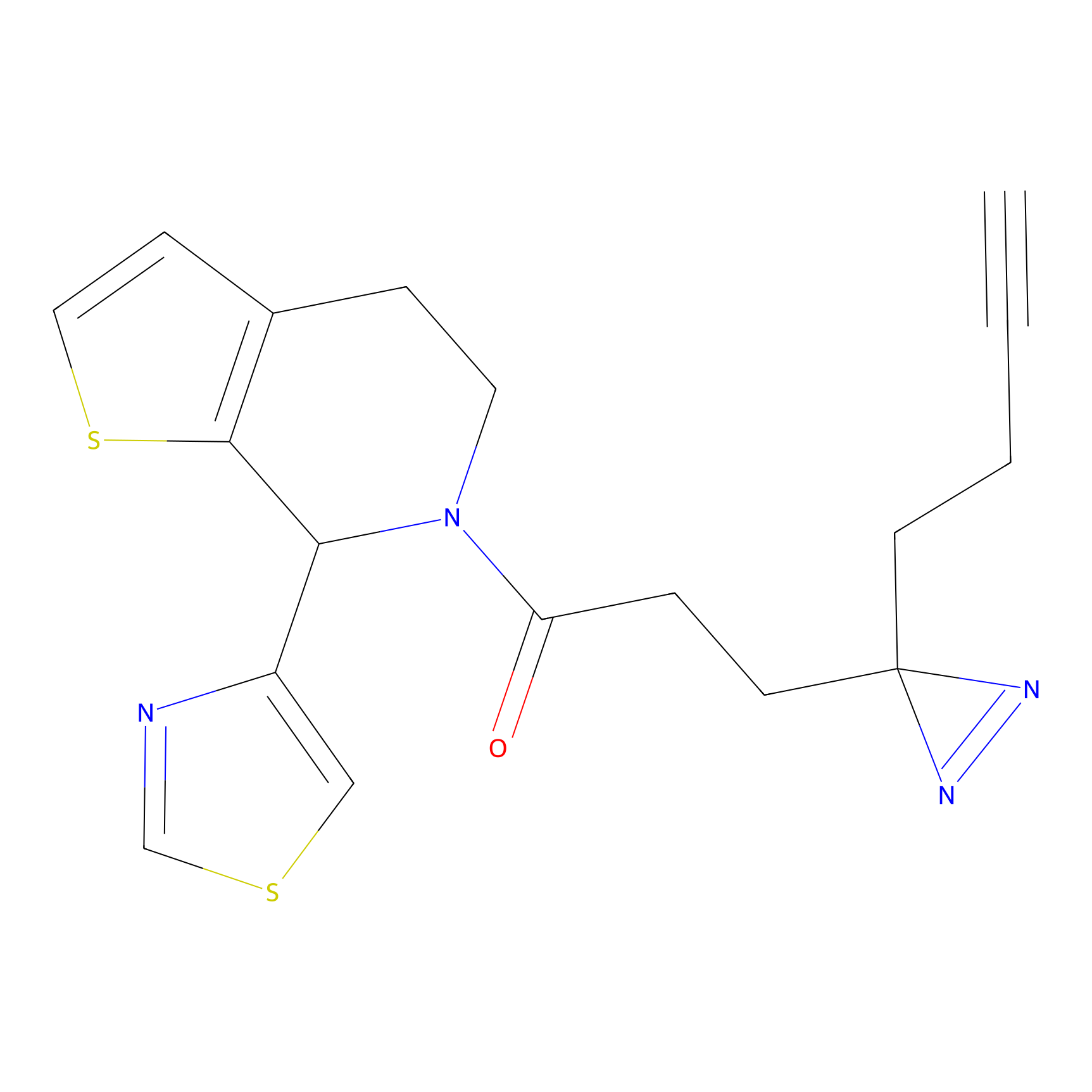
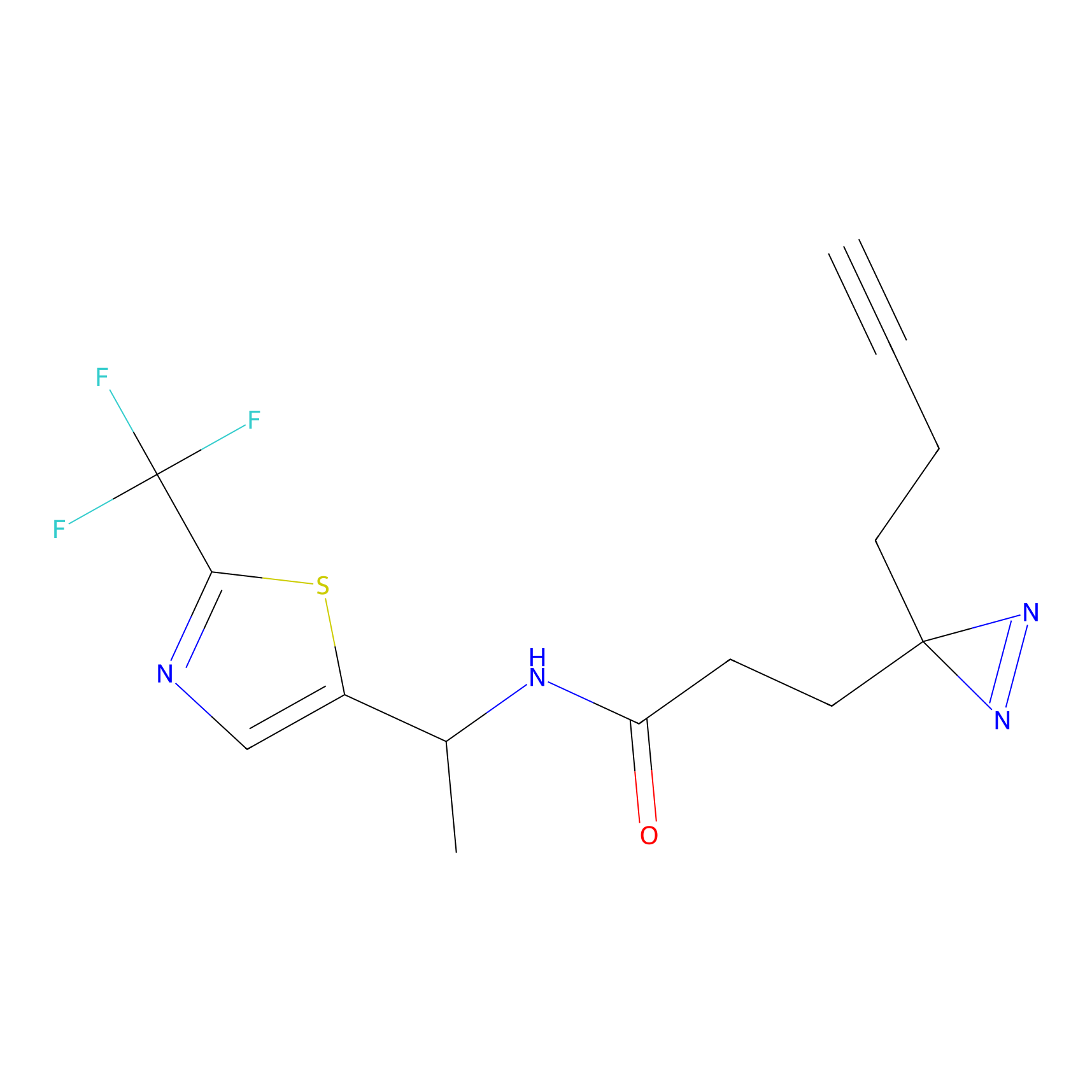
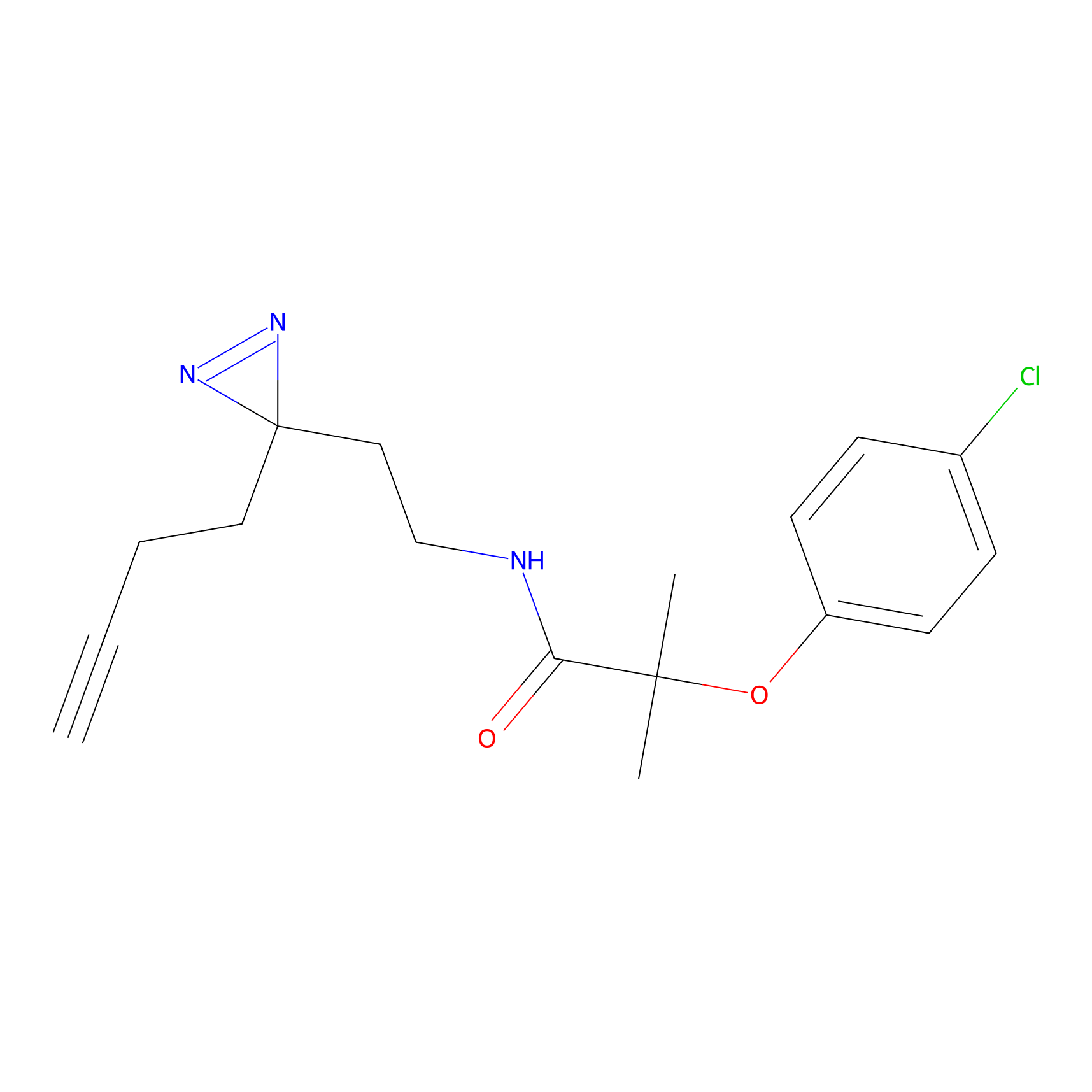
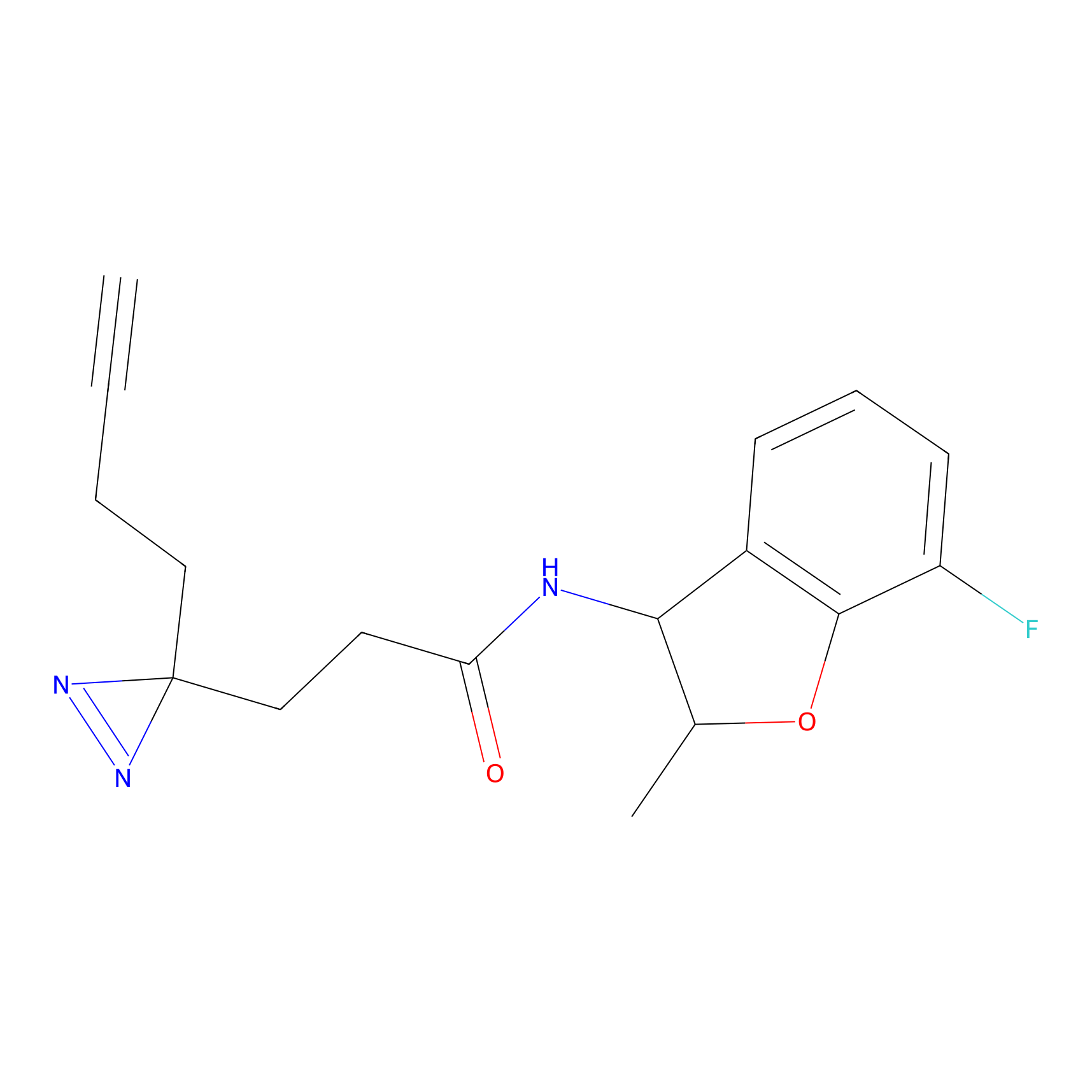
STPyne Probe Info |
 |
K207(10.00) | LDD0277 | [1] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
The Interaction Atlas With This Target
References