Details of the Target
General Information of Target
| Target ID | LDTP07891 | |||||
|---|---|---|---|---|---|---|
| Target Name | Inhibitor of nuclear factor kappa-B kinase-interacting protein (IKBIP) | |||||
| Gene Name | IKBIP | |||||
| Gene ID | 121457 | |||||
| Synonyms |
IKIP; Inhibitor of nuclear factor kappa-B kinase-interacting protein; I kappa-B kinase-interacting protein; IKBKB-interacting protein; IKK-interacting protein |
|||||
| 3D Structure | ||||||
| Sequence |
MSEVKSRKKSGPKGAPAAEPGKRSEGGKTPVARSSGGGGWADPRTCLSLLSLGTCLGLAW
FVFQQSEKFAKVENQYQLLKLETNEFQQLQSKISLISEKWQKSEAIMEQLKSFQIIAHLK RLQEEINEVKTWSNRITEKQDILNNSLTTLSQDITKVDQSTTSMAKDVGLKITSVKTDIR RISGLVTDVISLTDSVQELENKIEKVEKNTVKNIGDLLSSSIDRTATLRKTASENSQRIN SVKKTLTELKSDFDKHTDRFLSLEGDRAKVLKTVTFANDLKPKVYNLKKDFSRLEPLVND LTLRIGRLVTDLLQREKEIAFLSEKISNLTIVQAEIKDIKDEIAHISDMN |
|||||
| Target Bioclass |
Other
|
|||||
| Subcellular location |
Endoplasmic reticulum membrane
|
|||||
| Function | Target of p53/TP53 with pro-apoptotic function. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
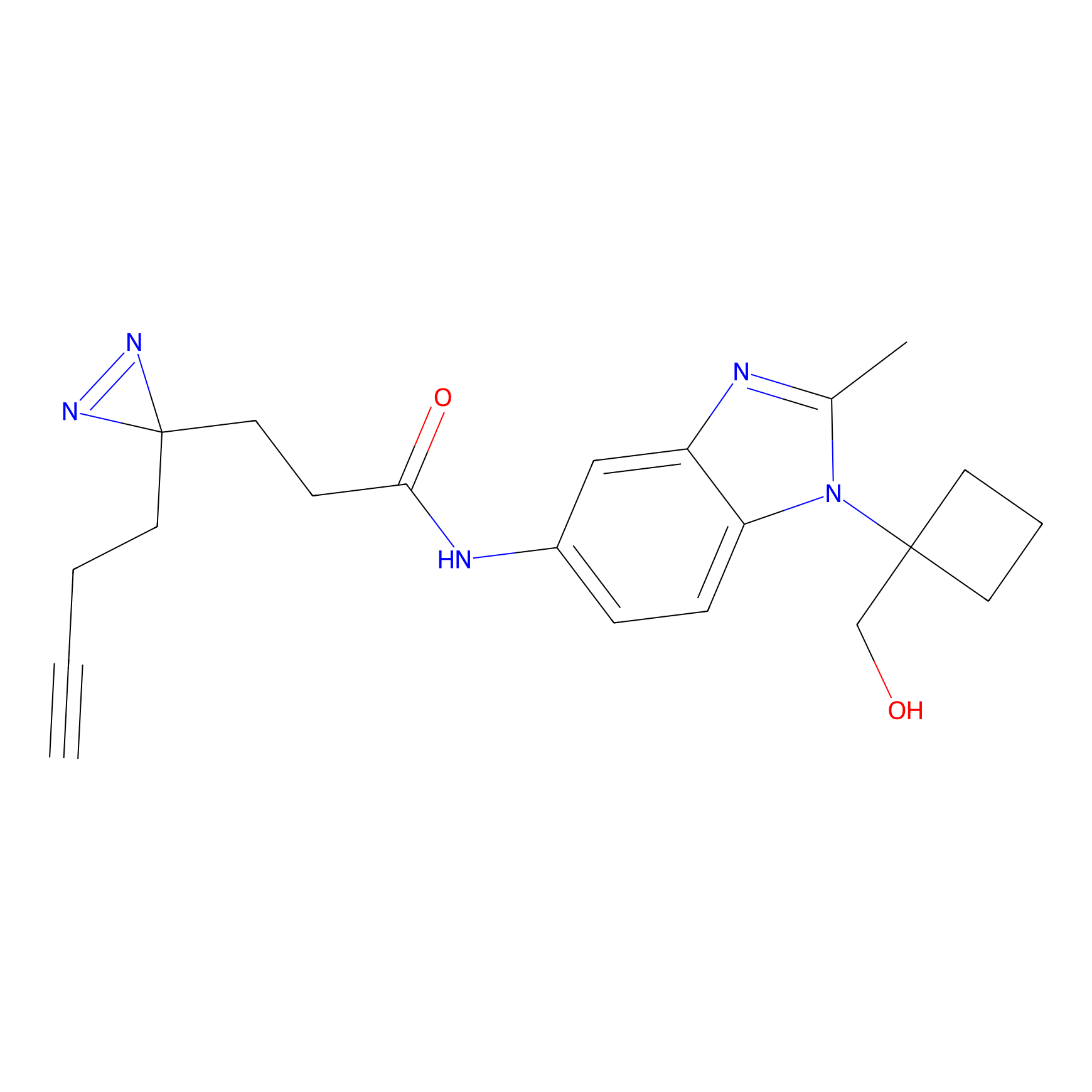
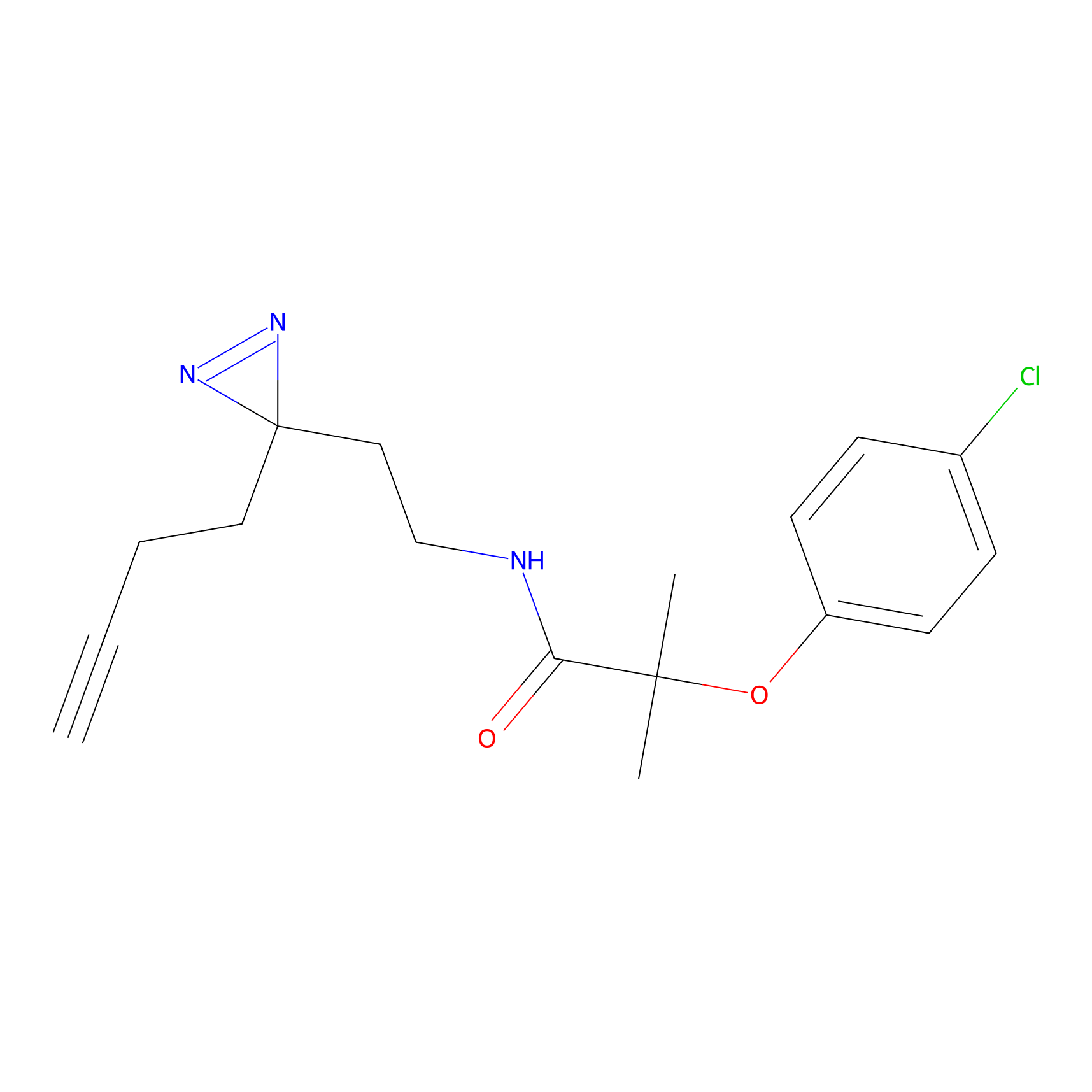
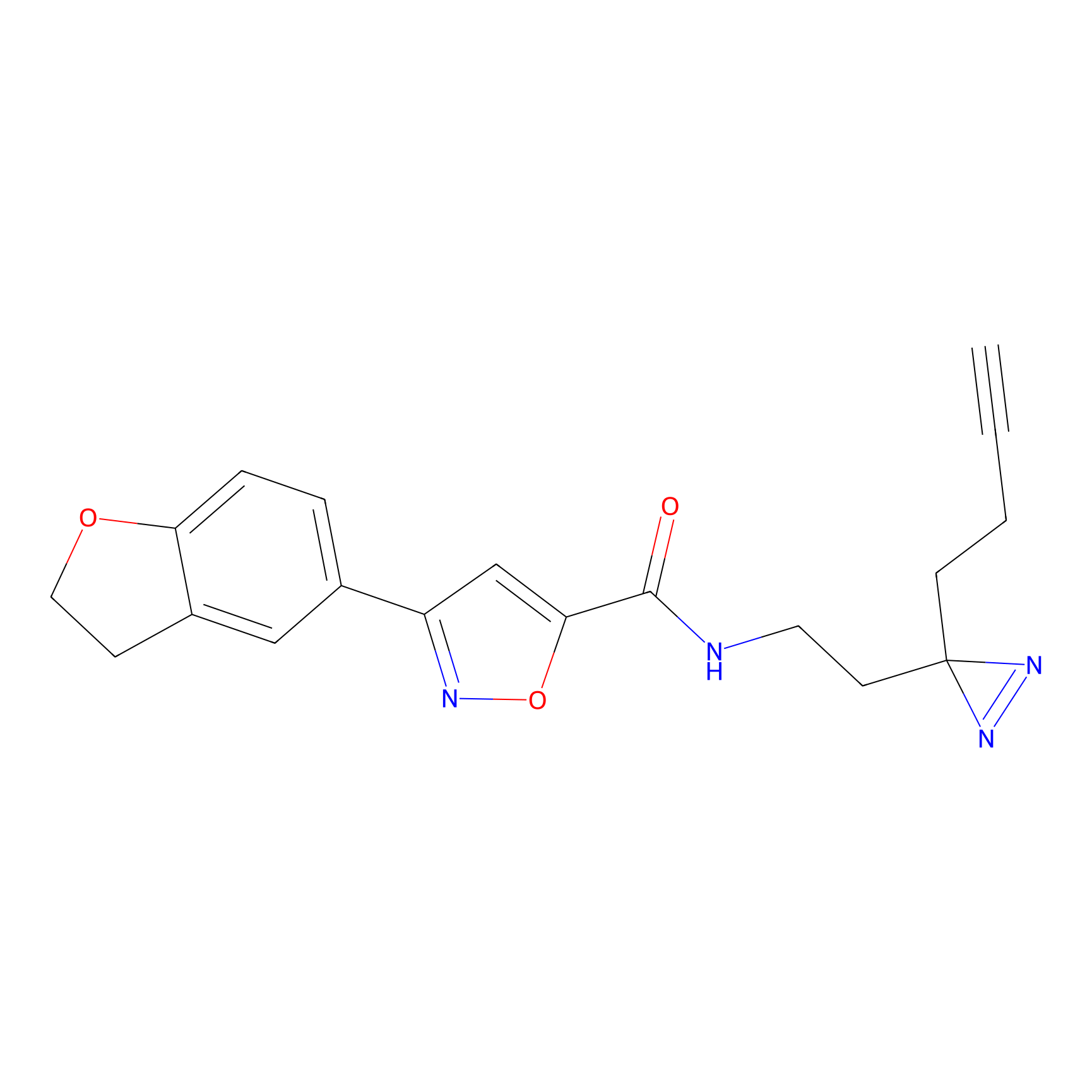
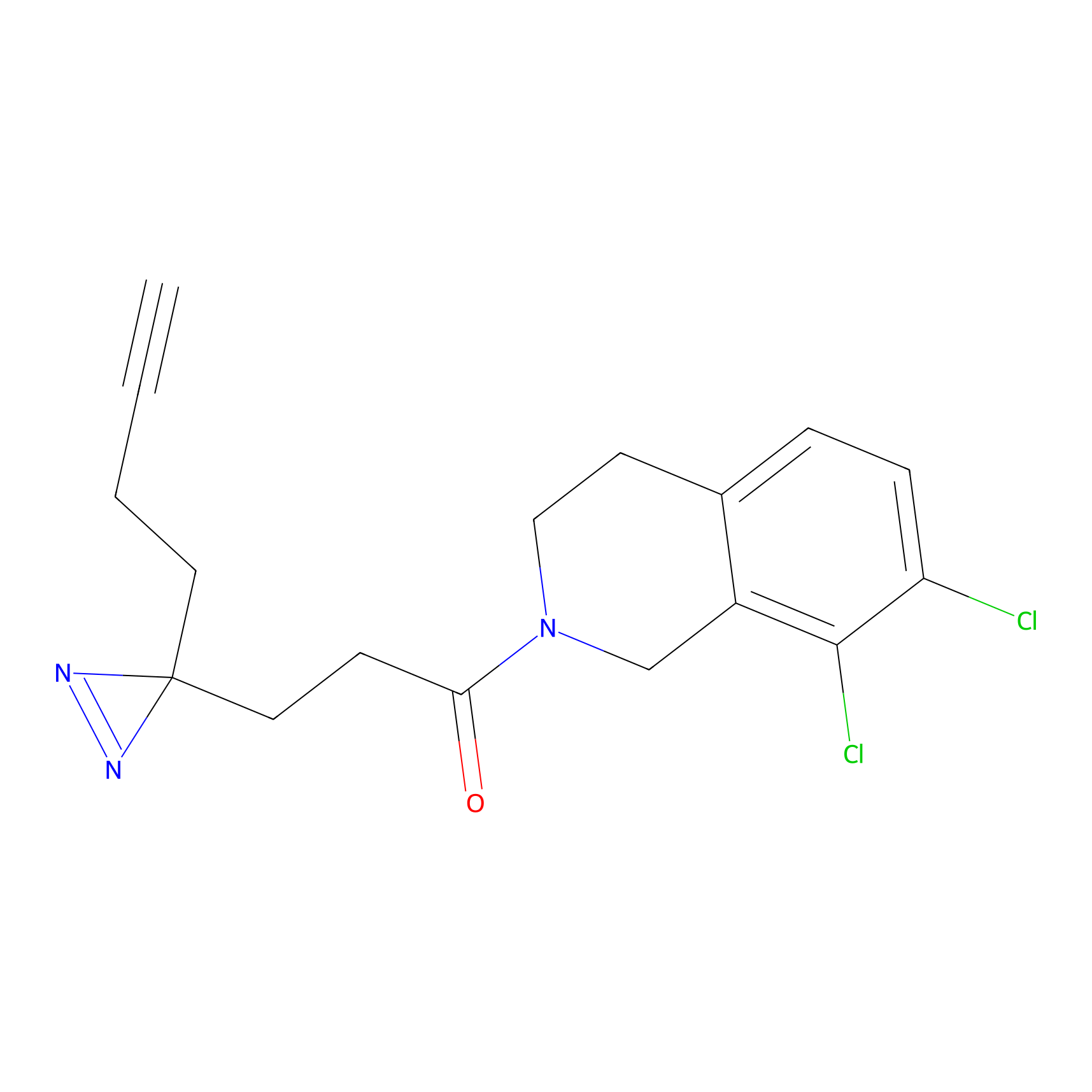
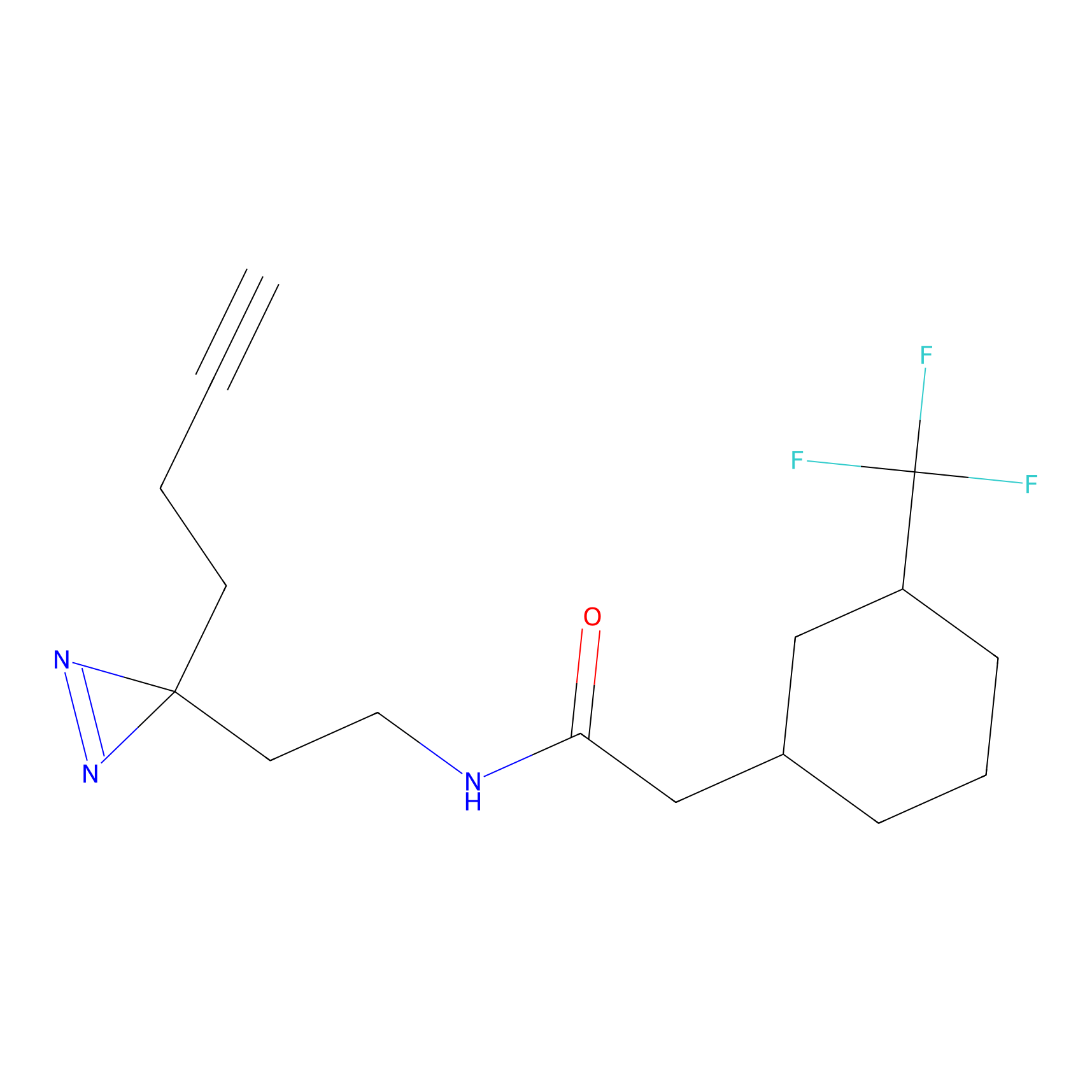
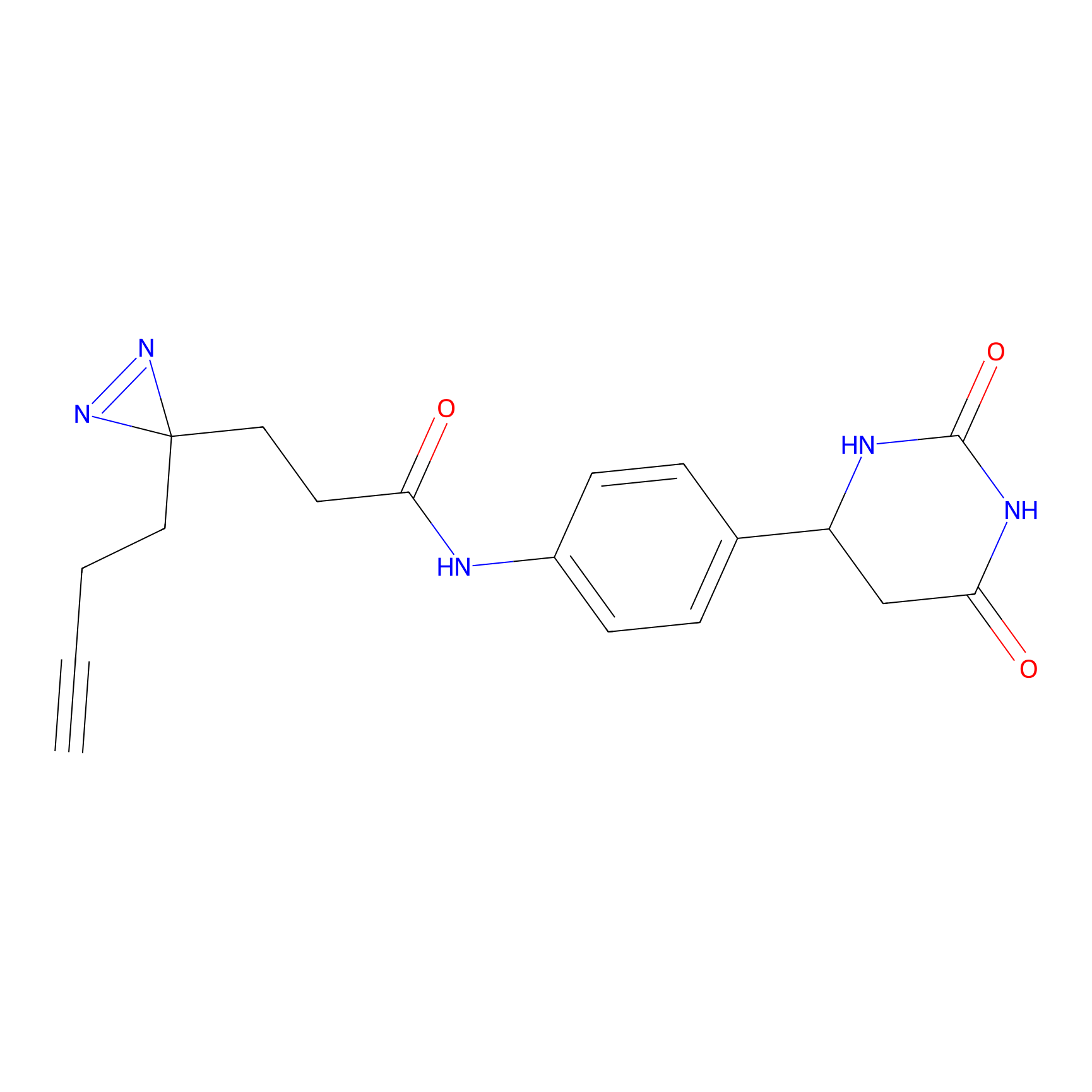
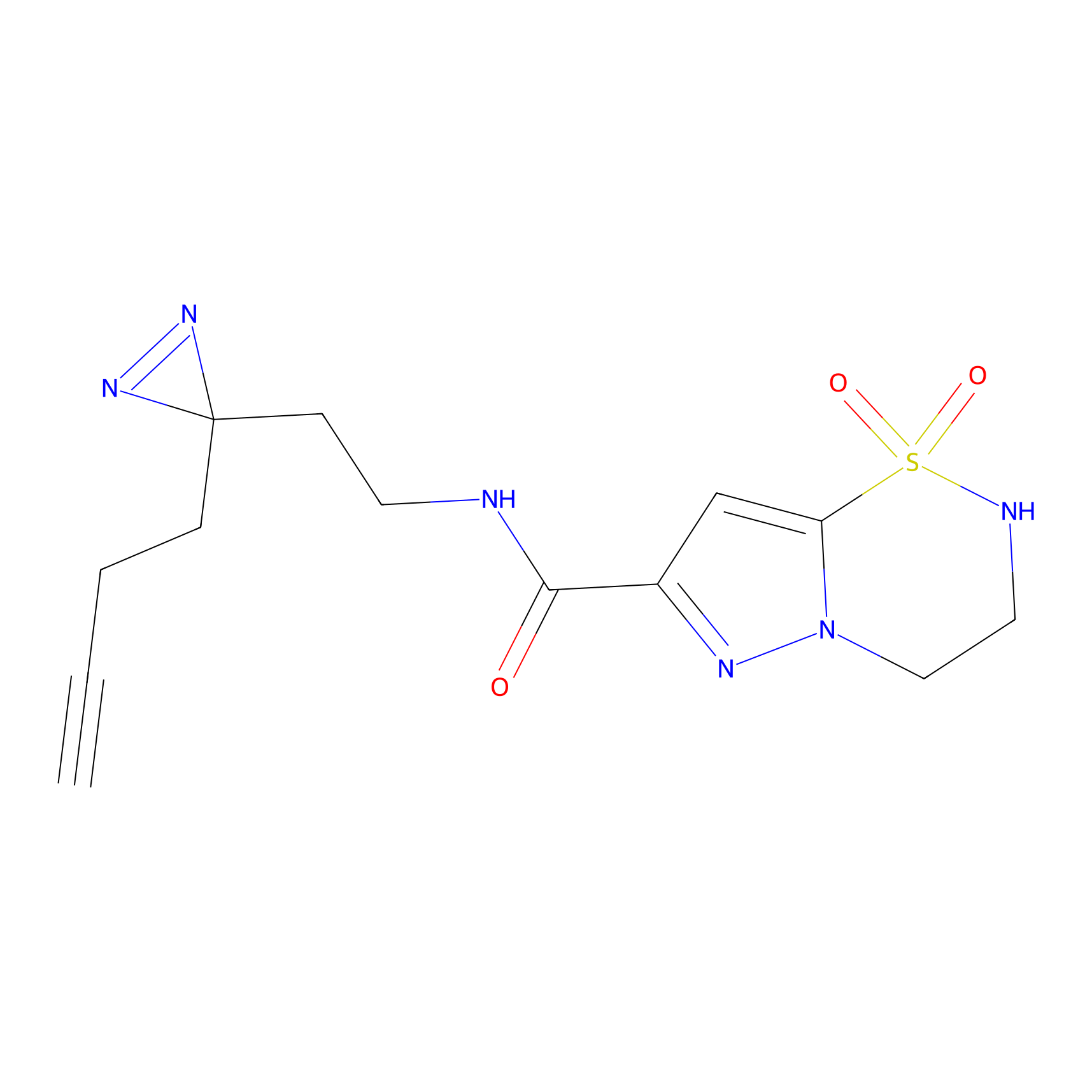
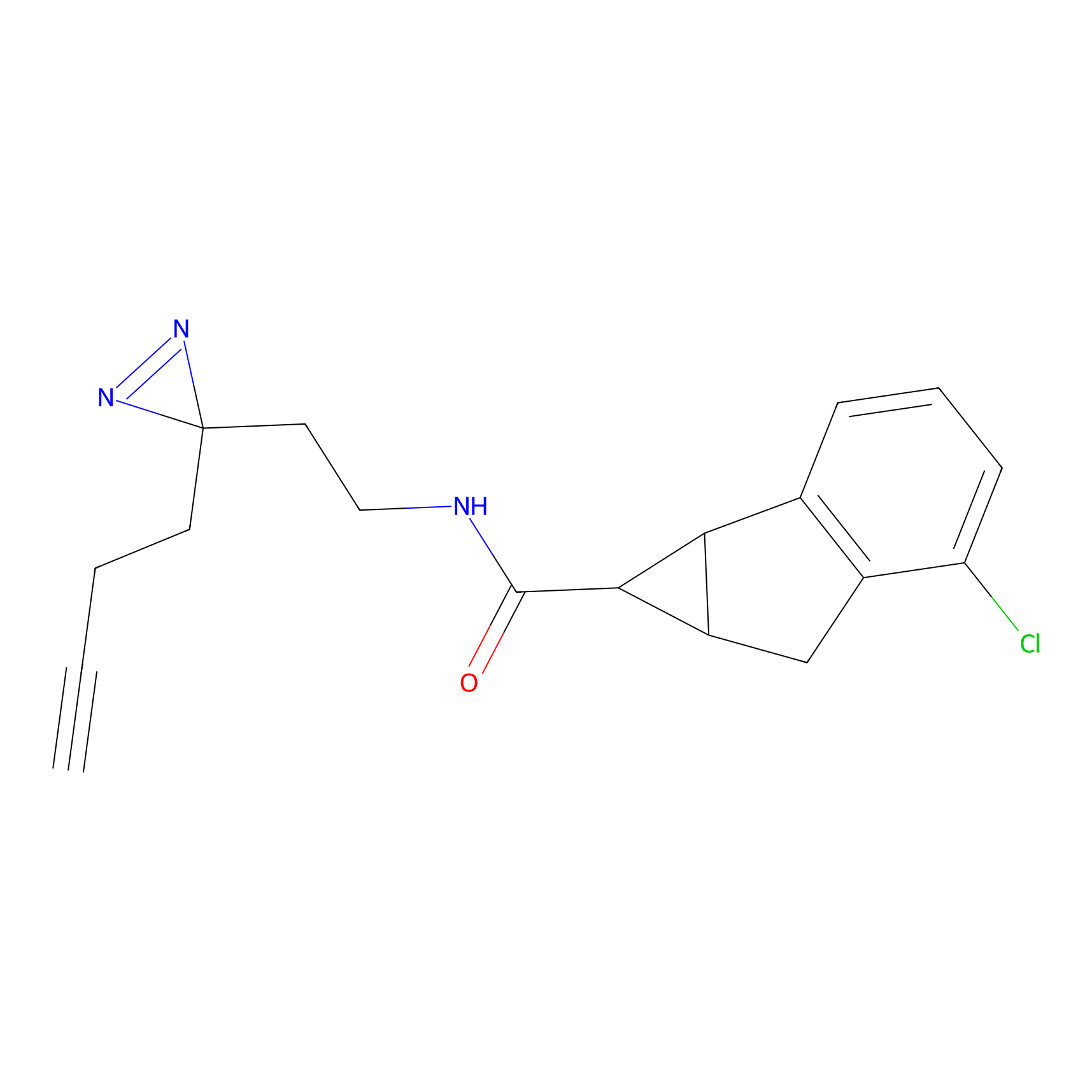
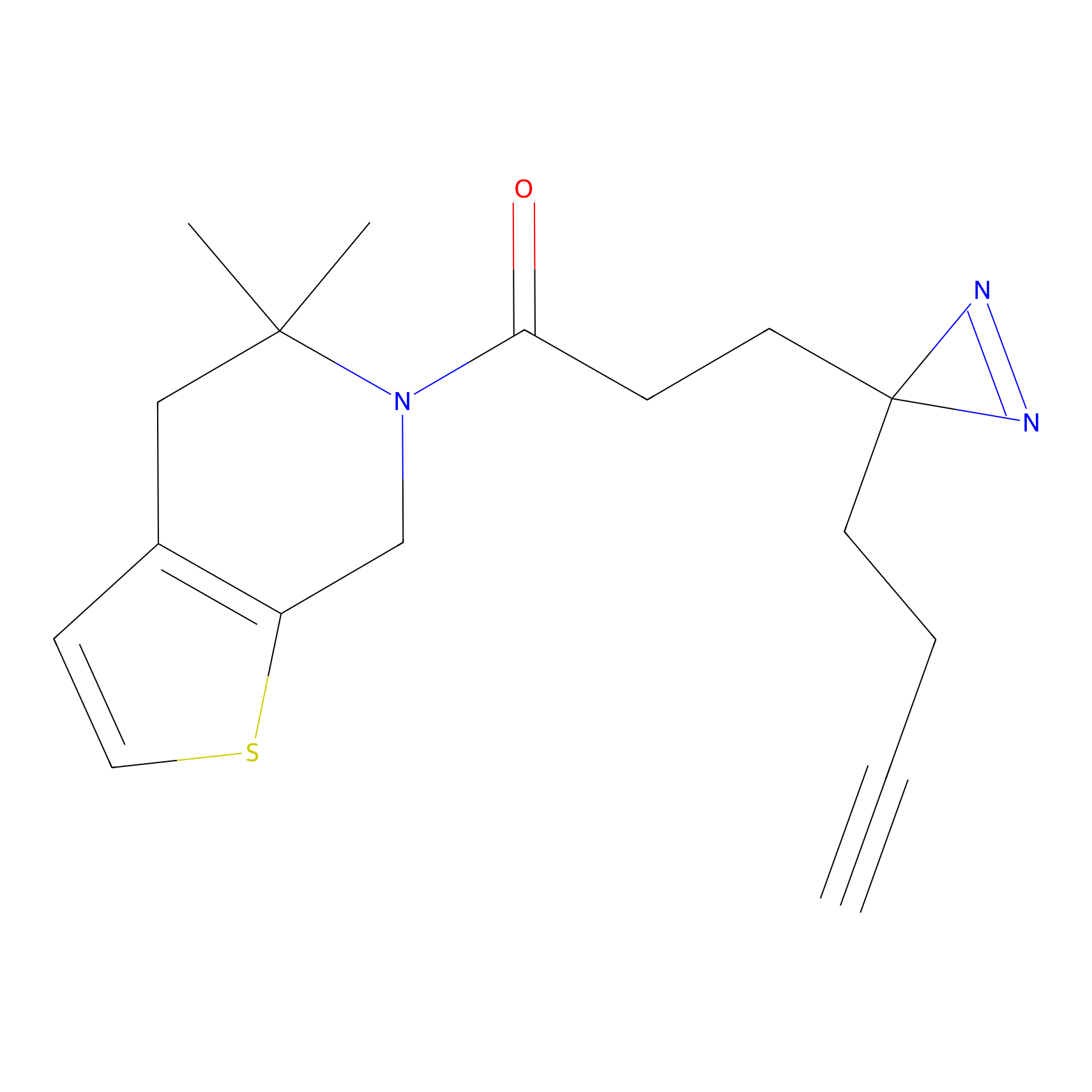
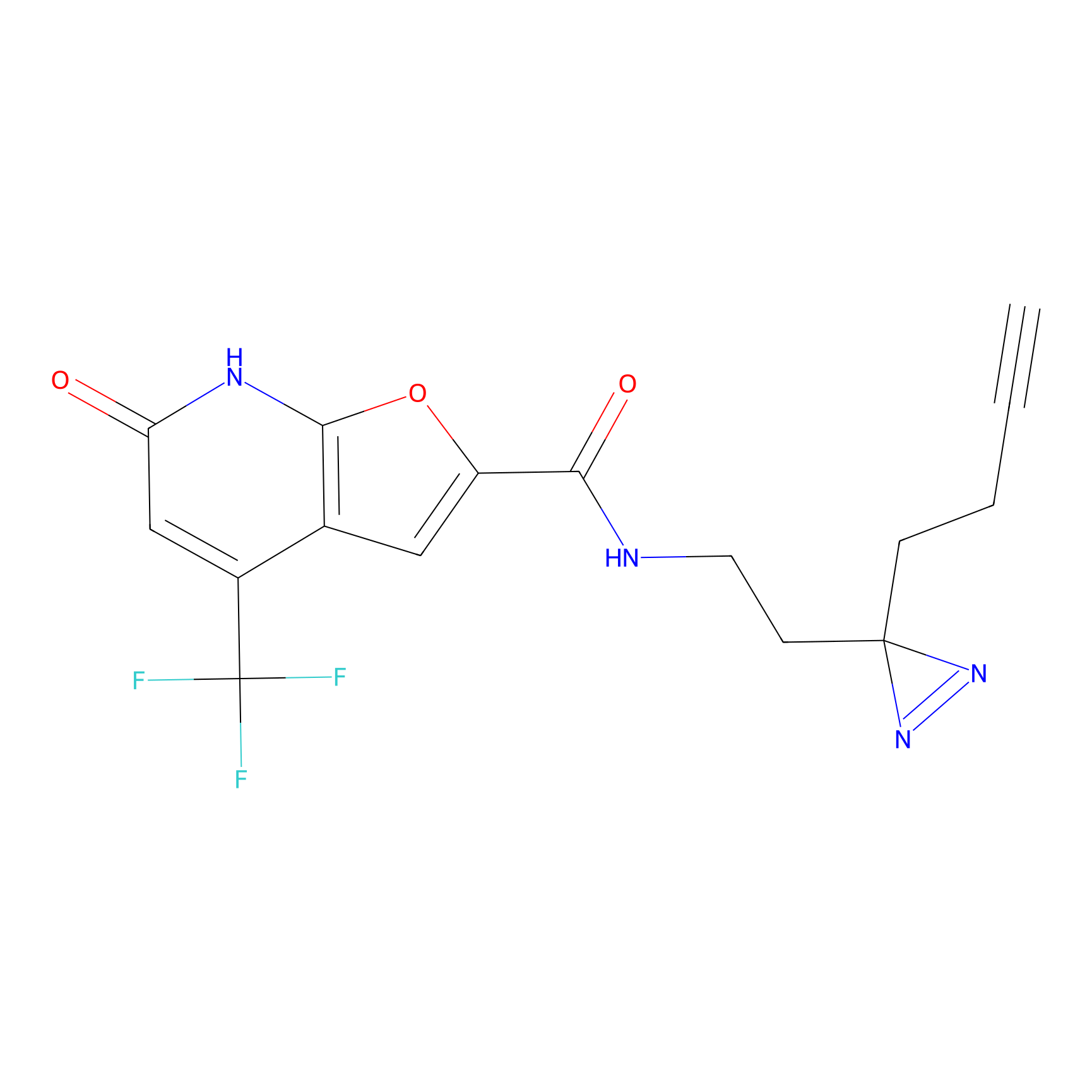
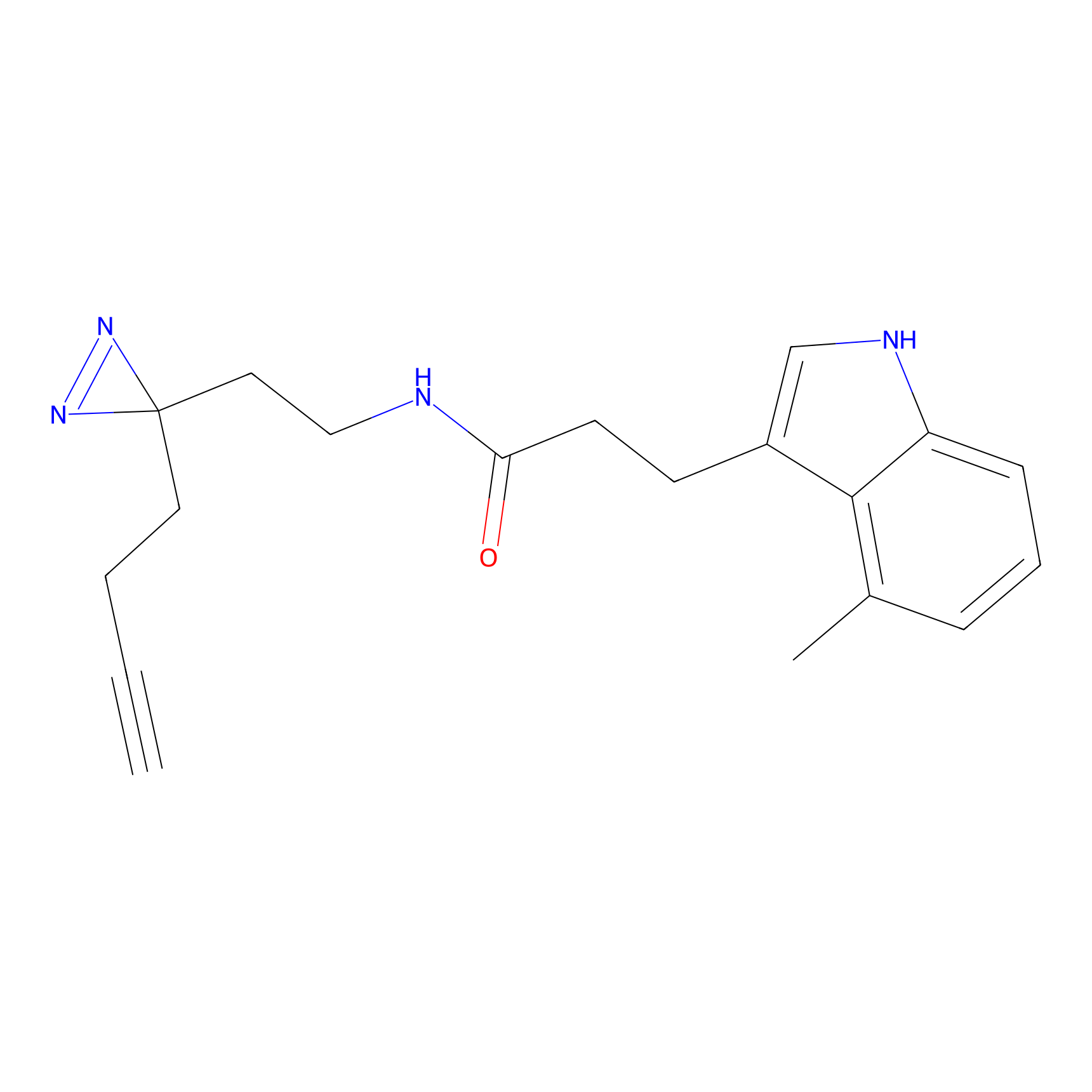
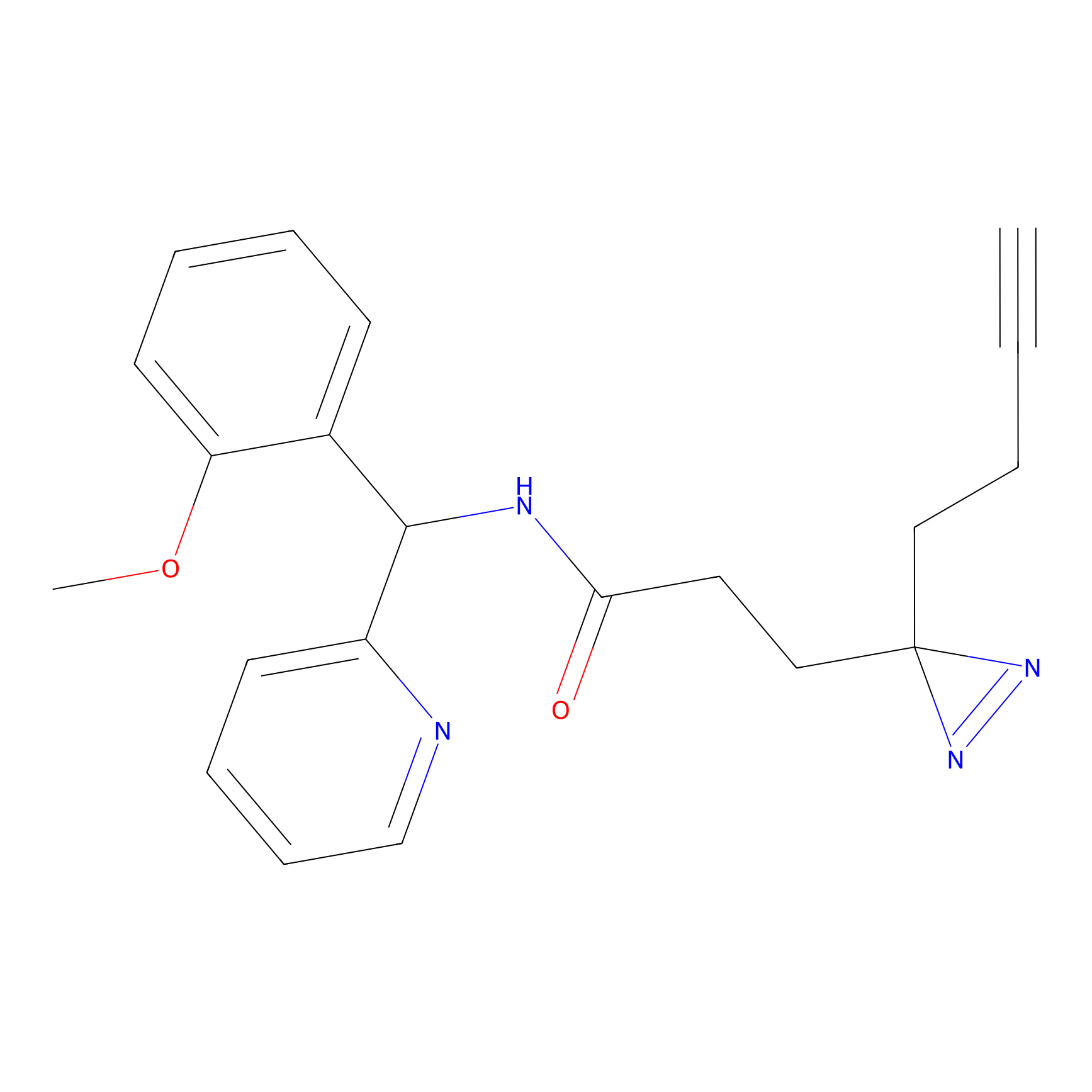
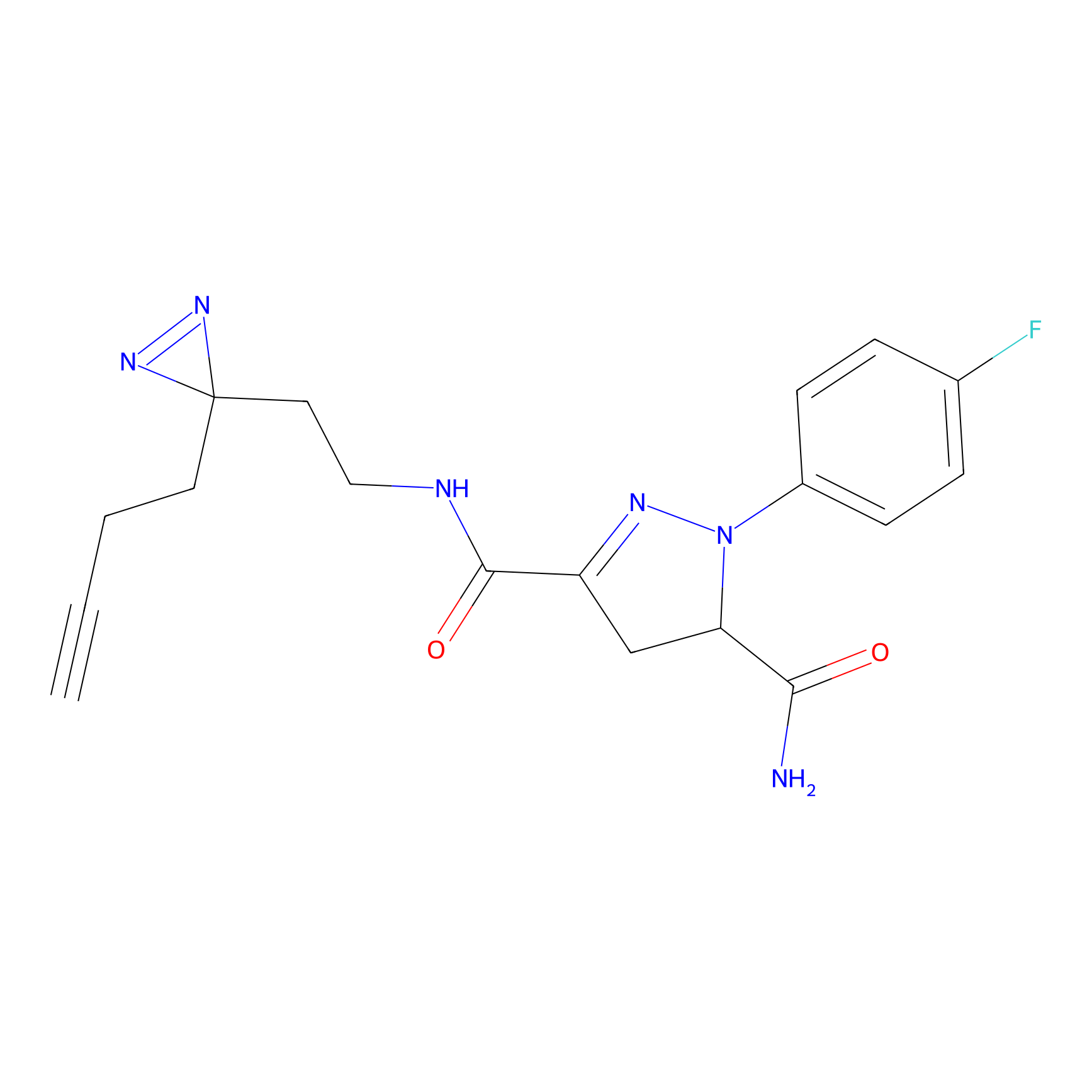
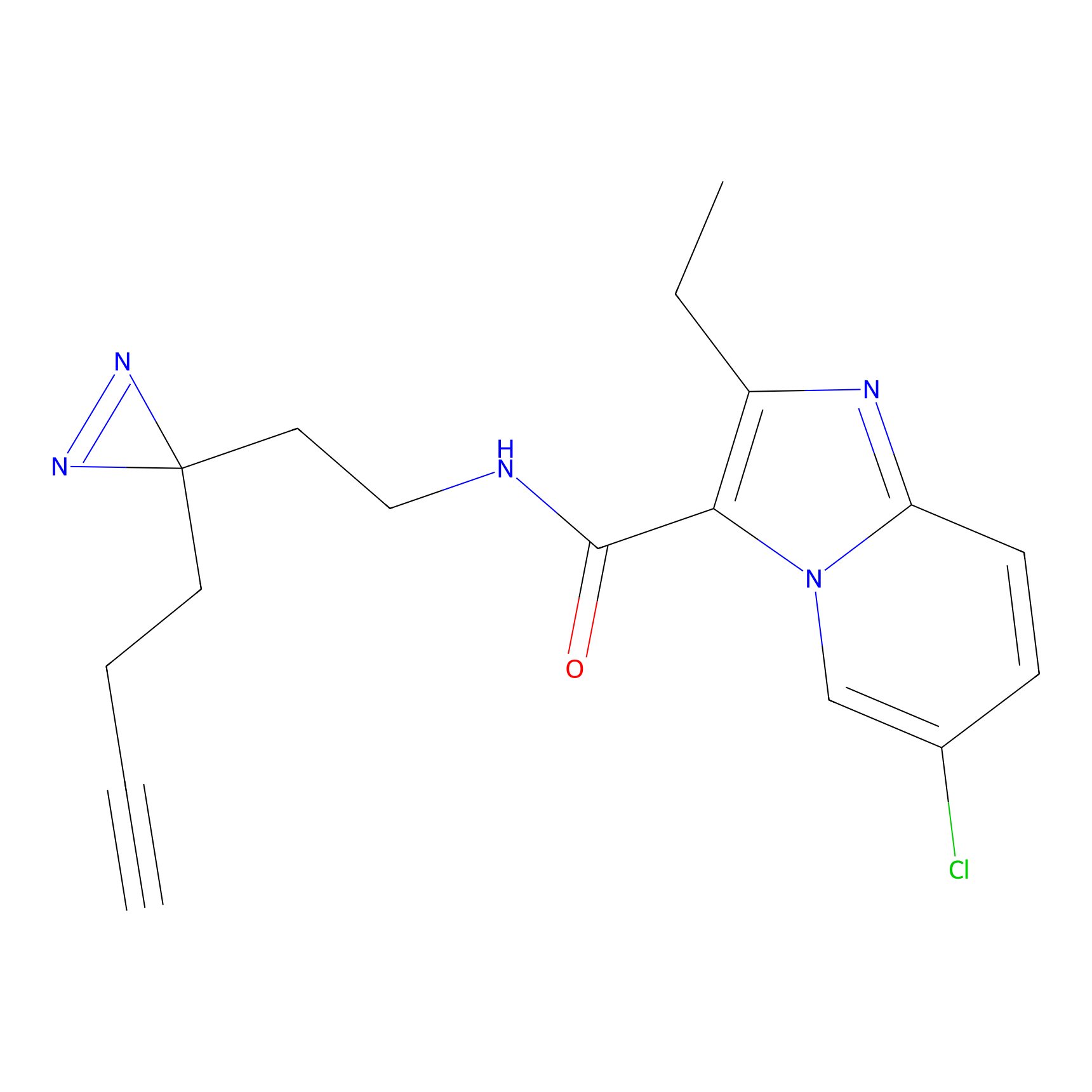
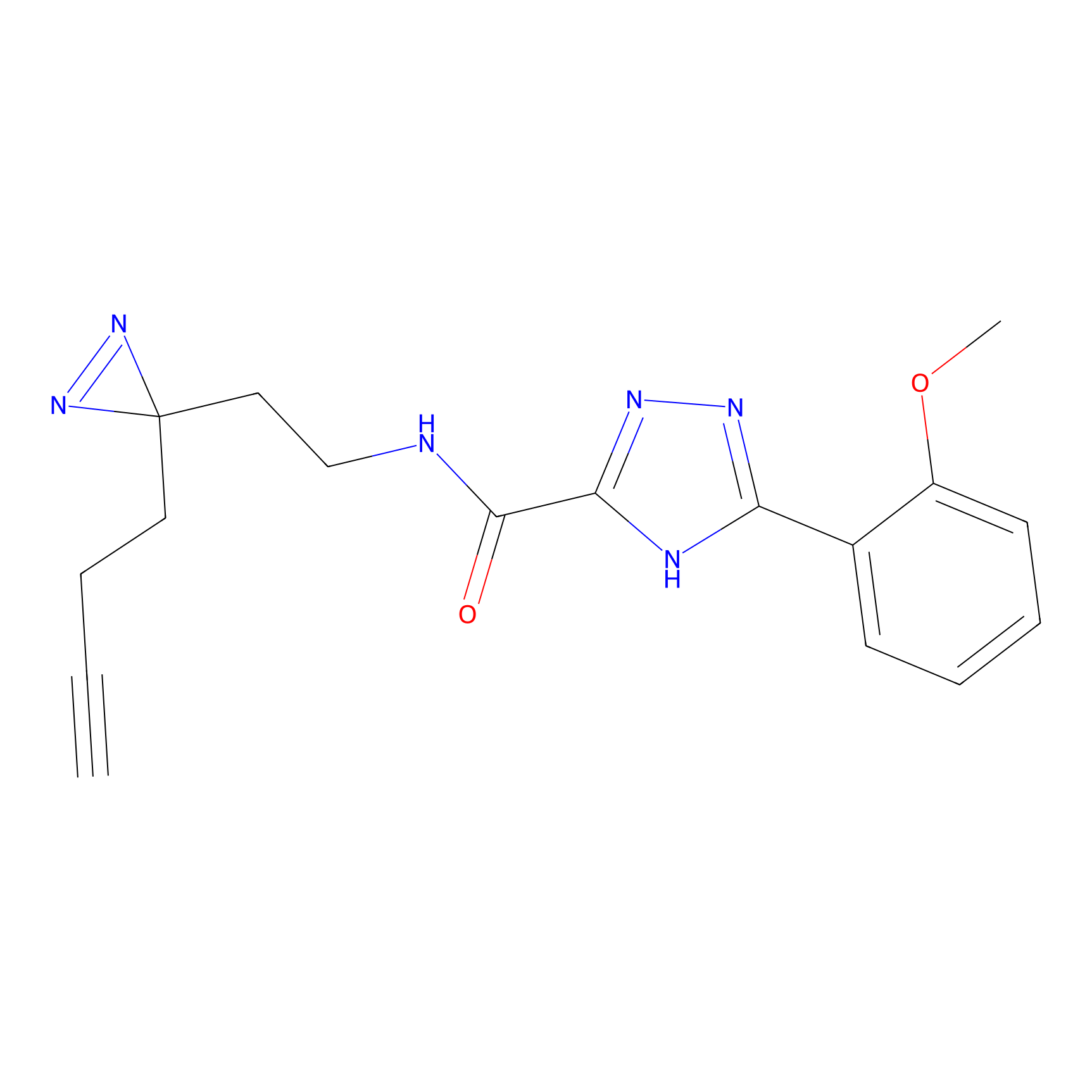
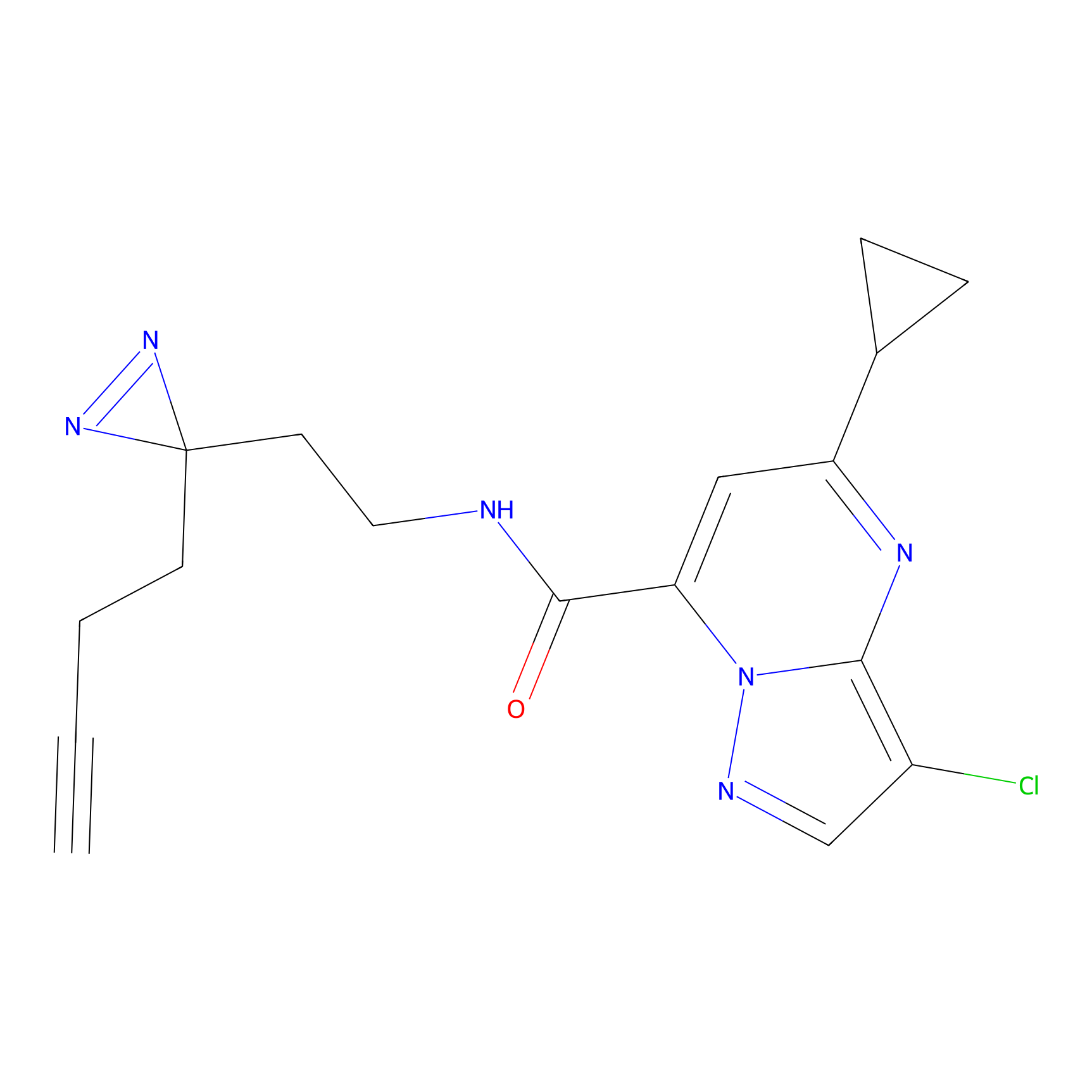
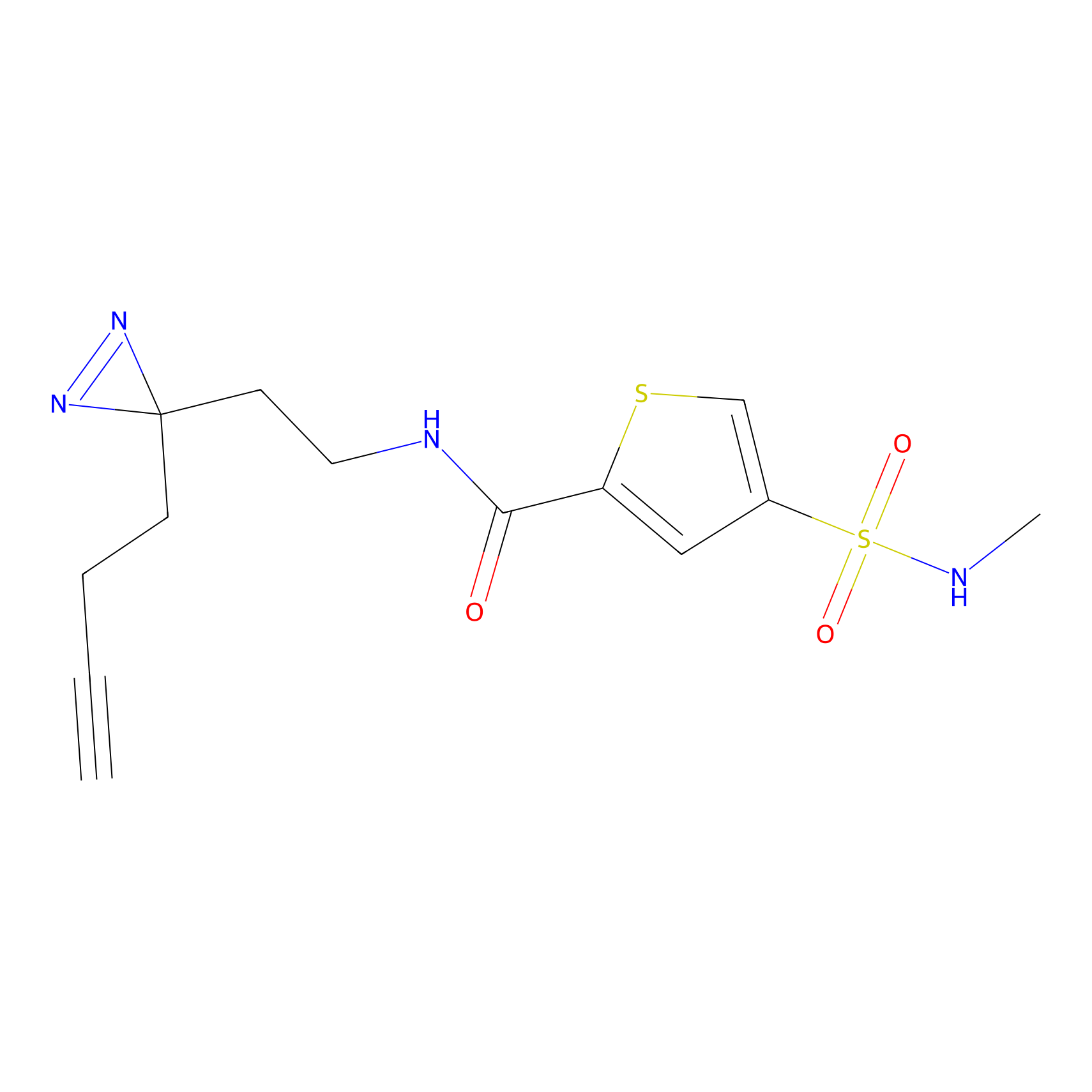
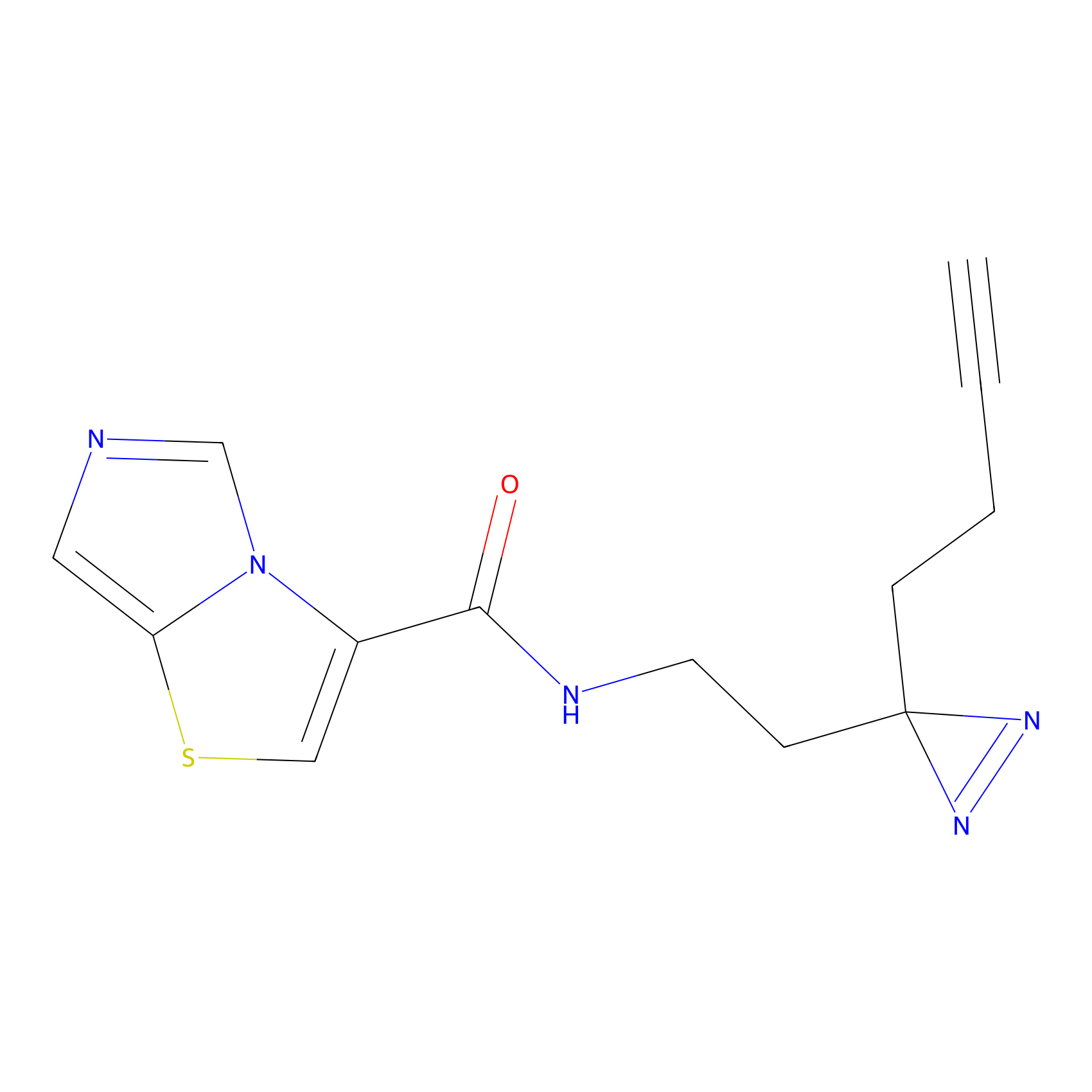
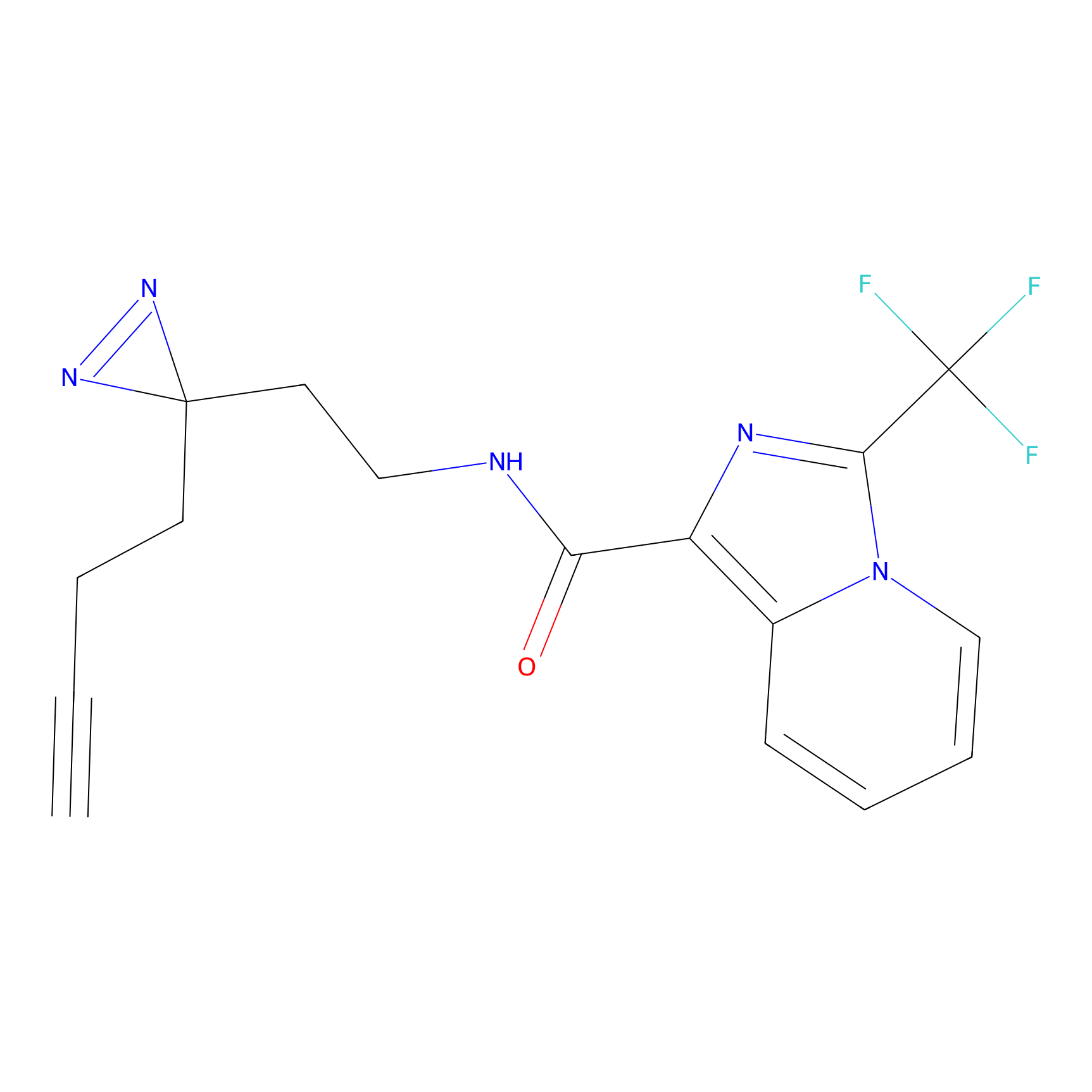
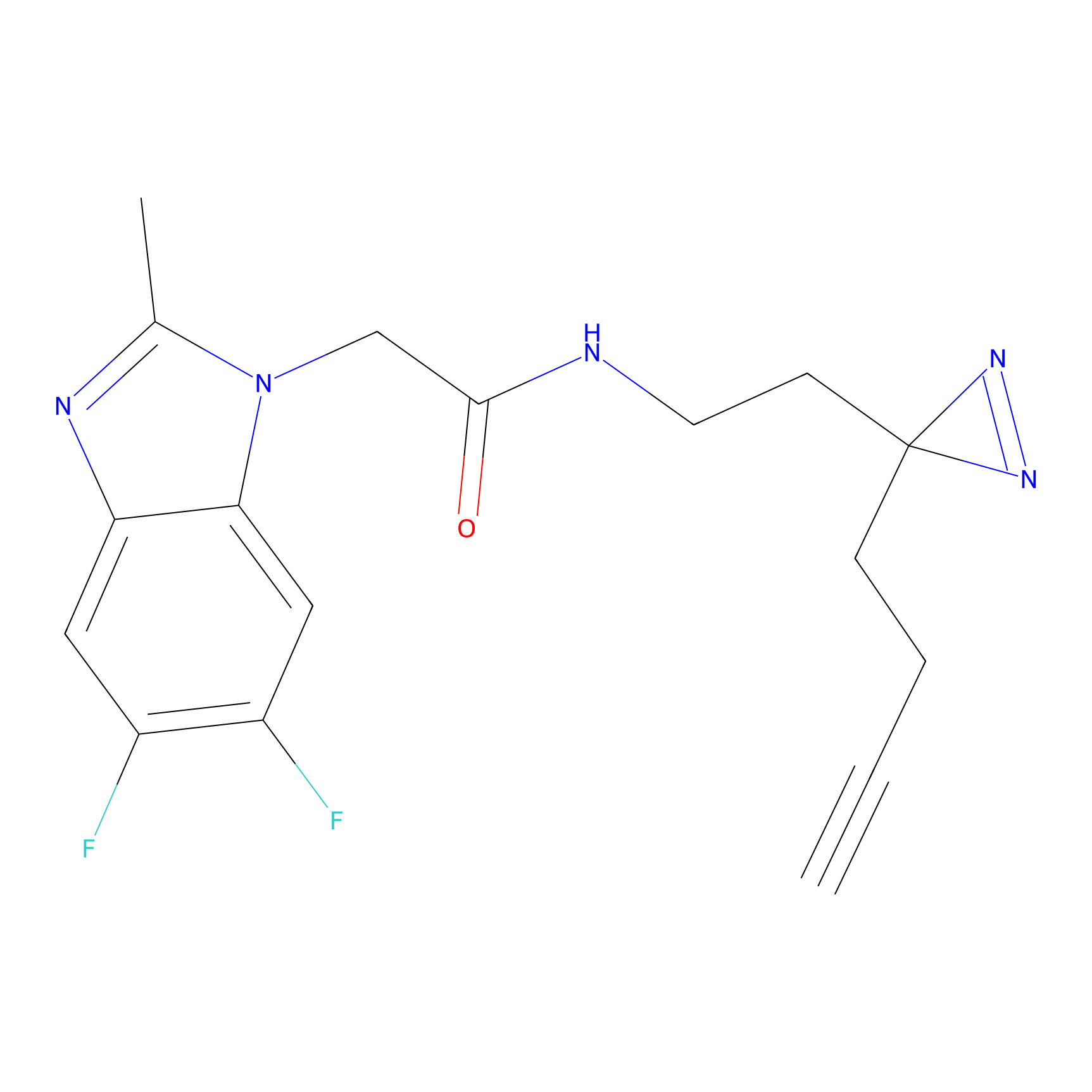
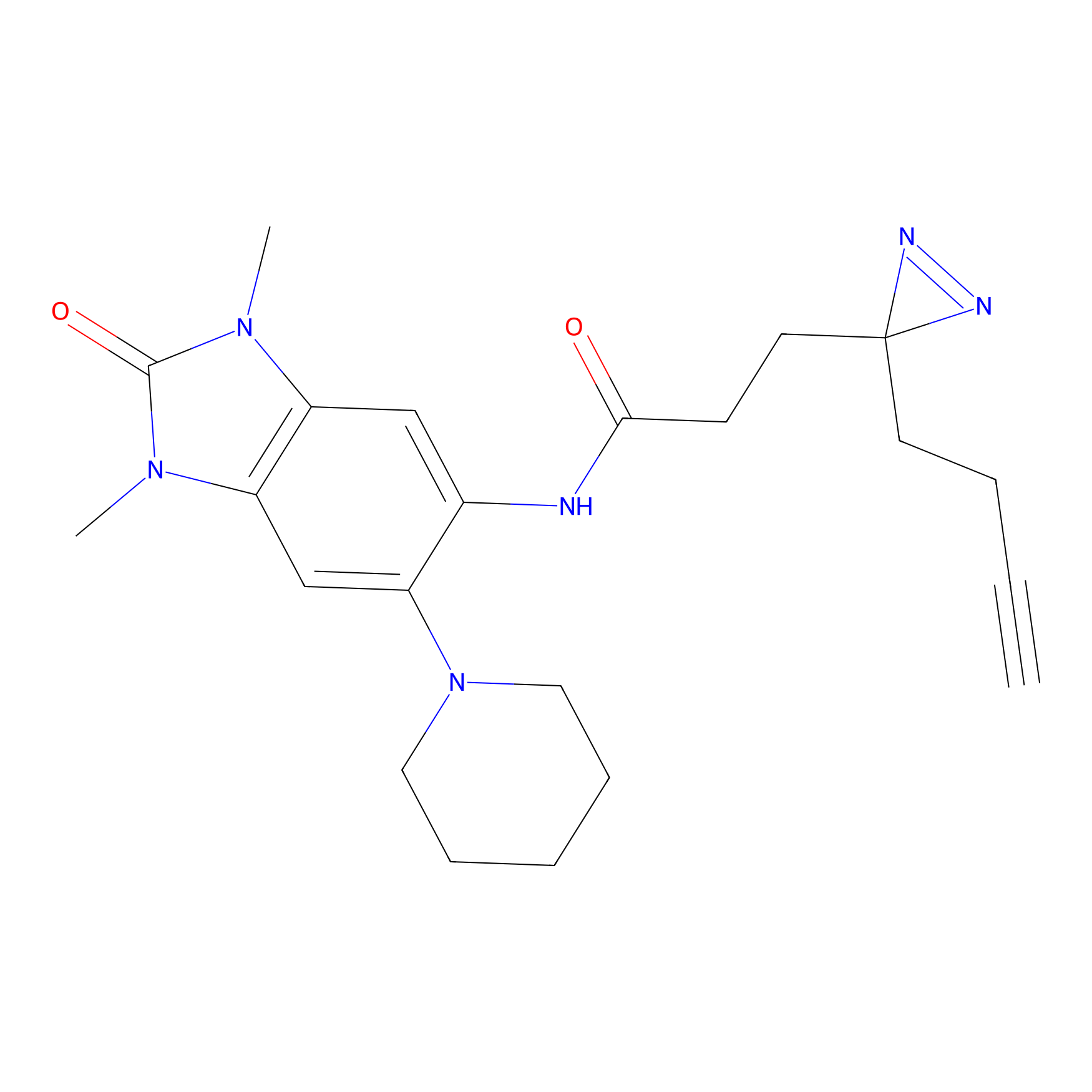
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
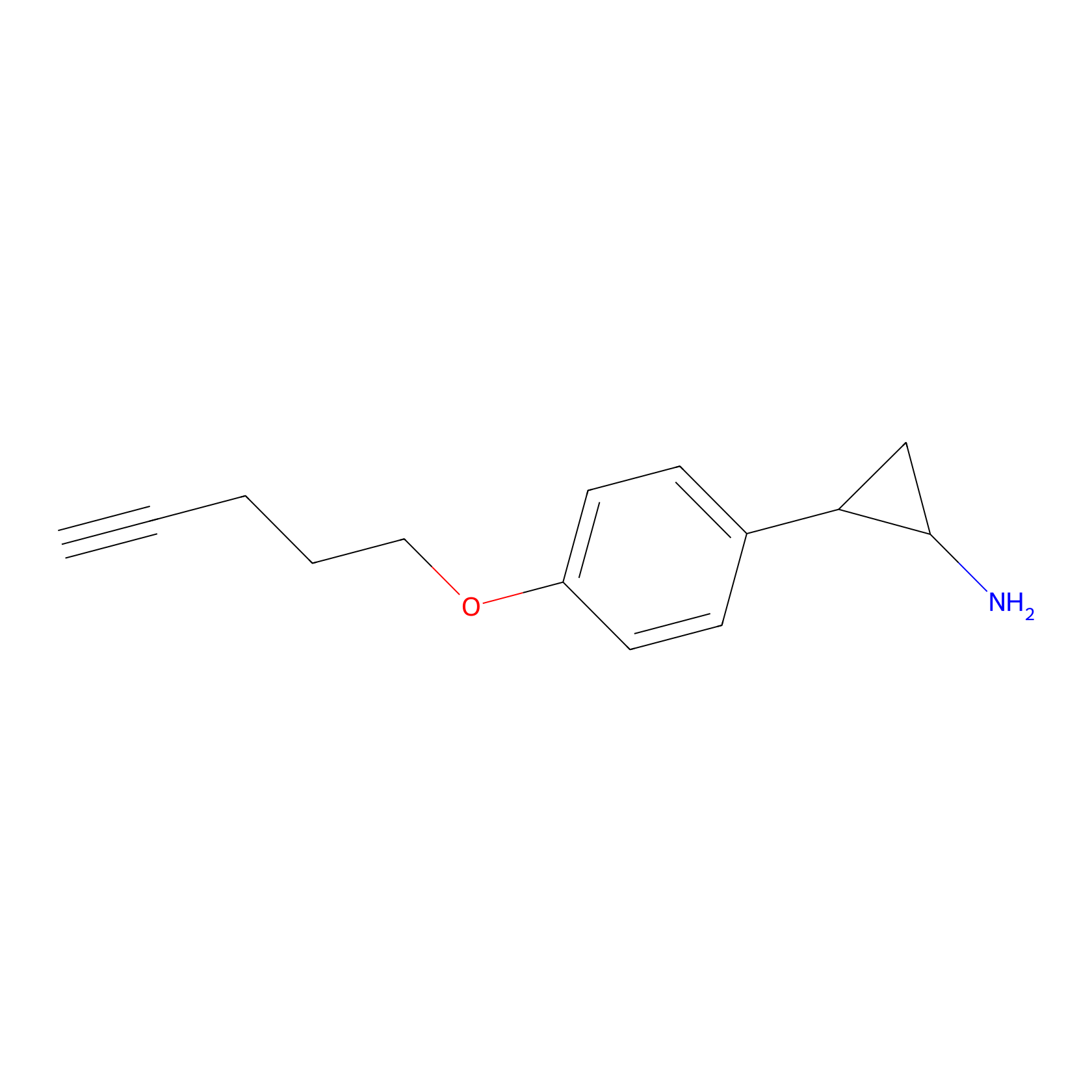
|
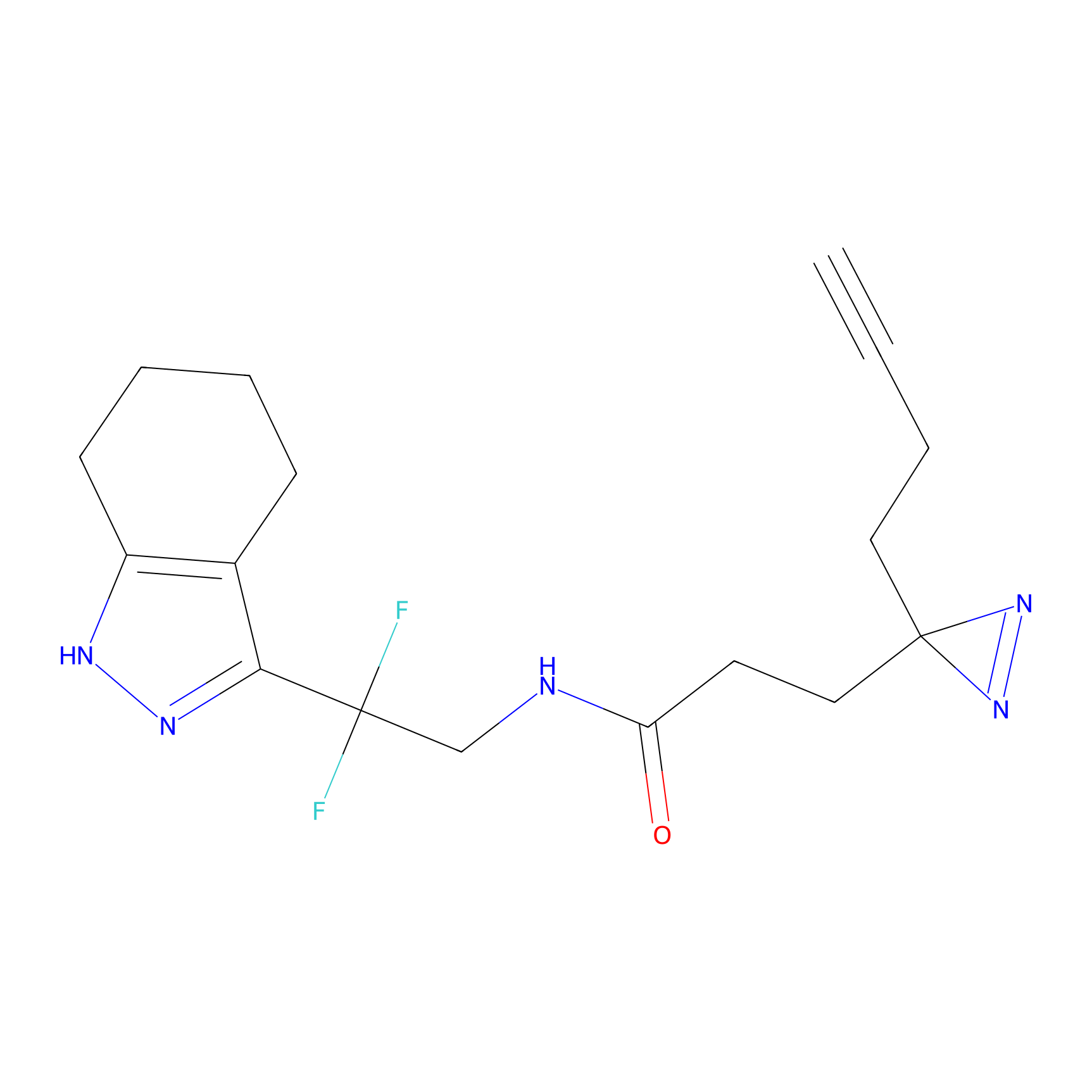
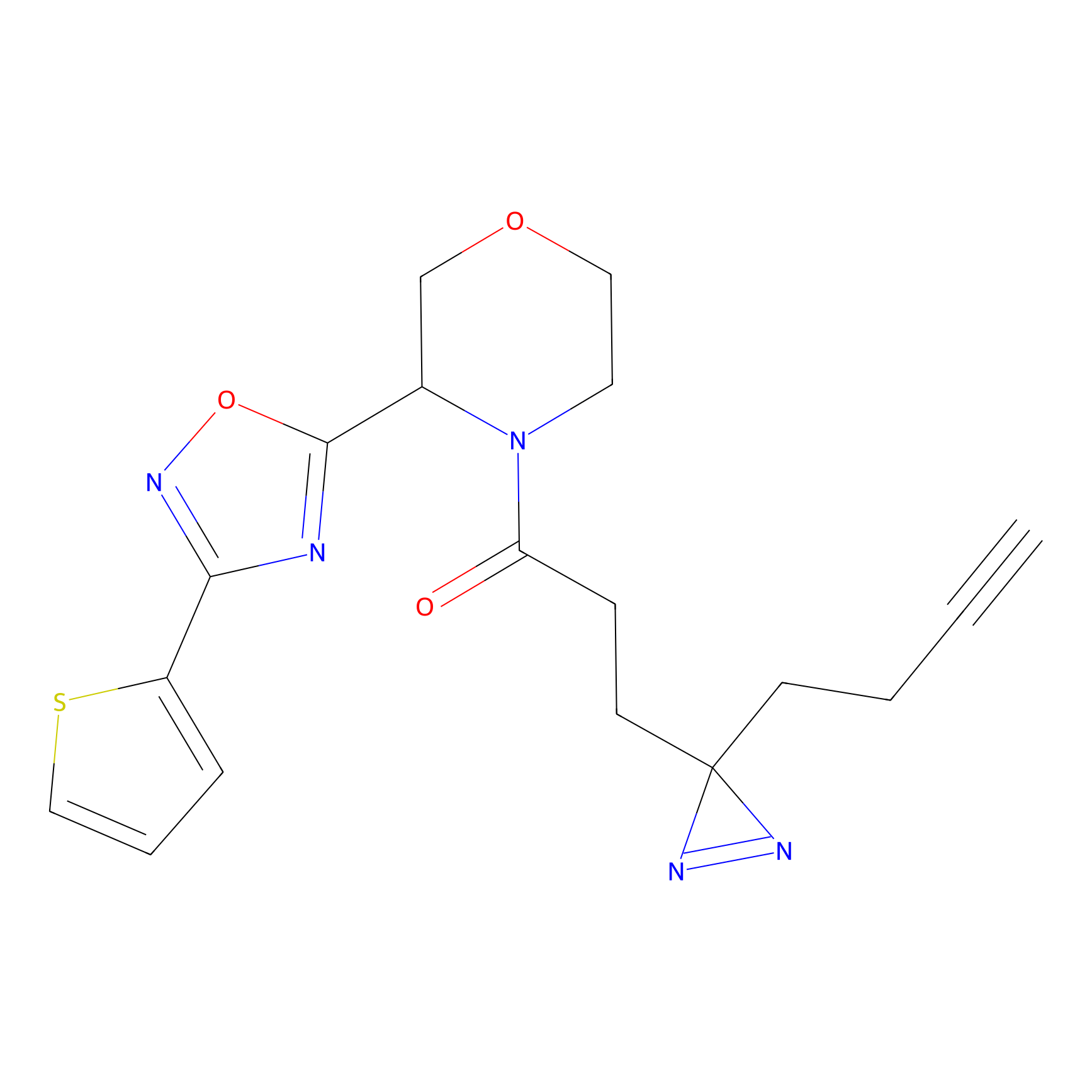
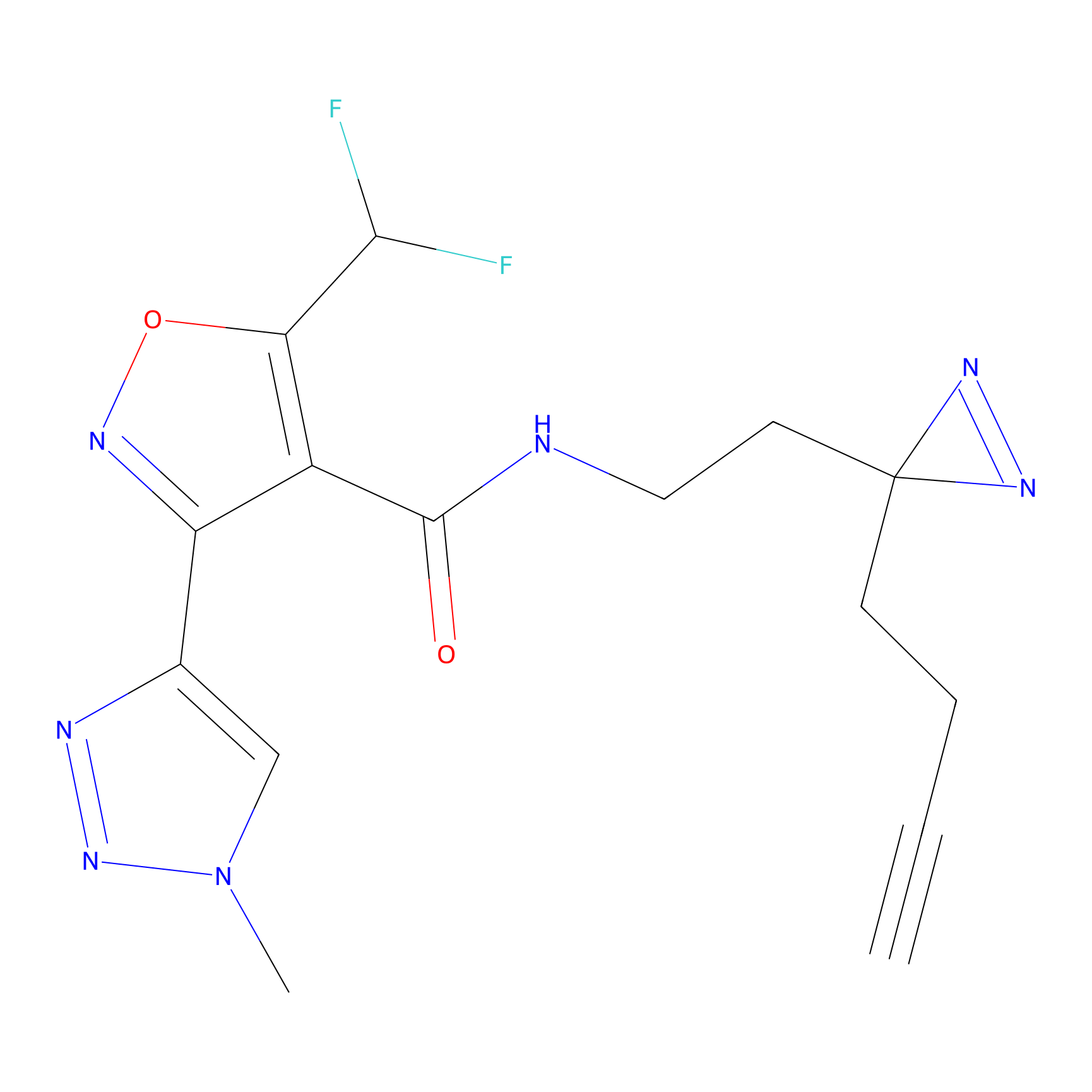
FBPP2 Probe Info |
 |
2.29 | LDD0318 | [1] | |
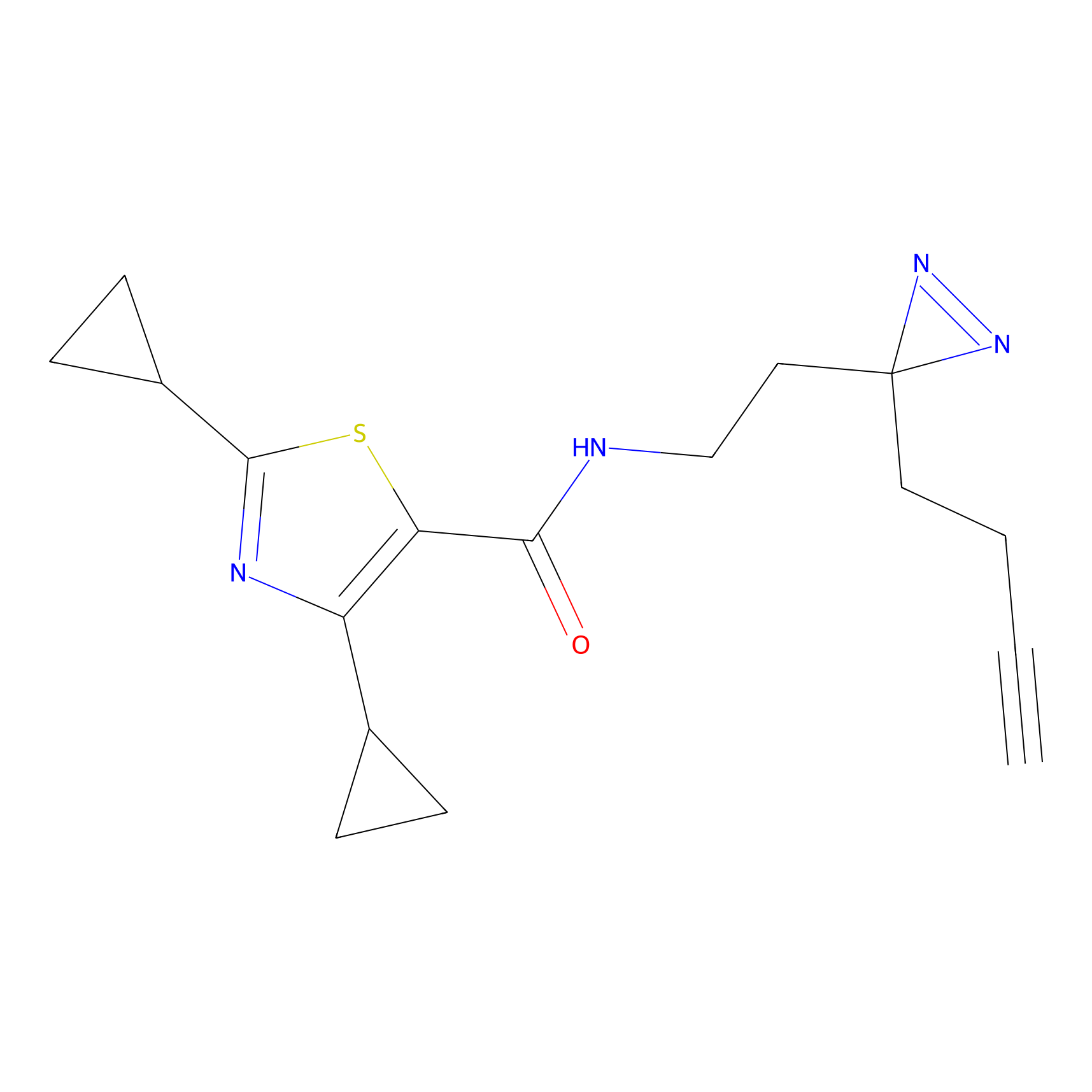
|
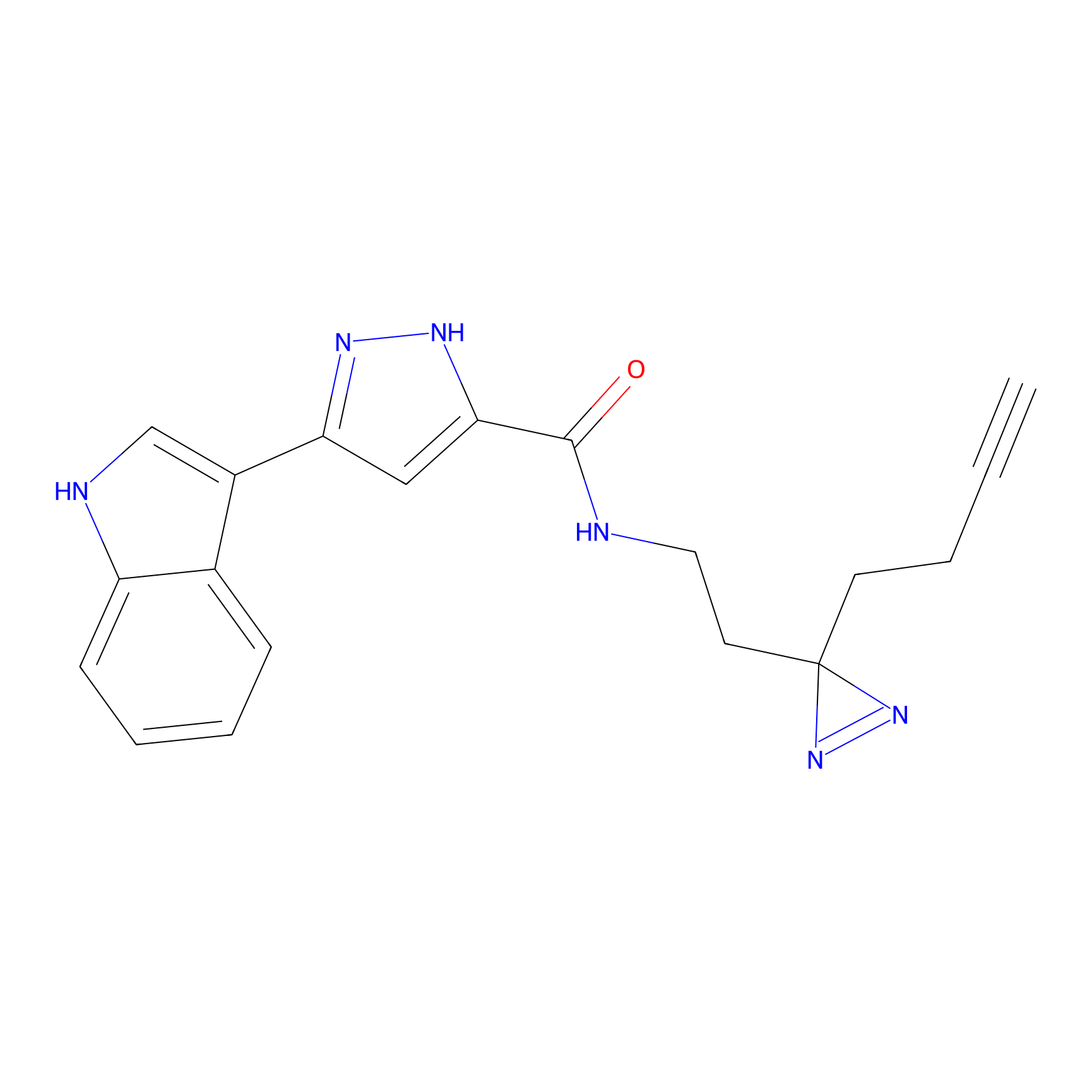
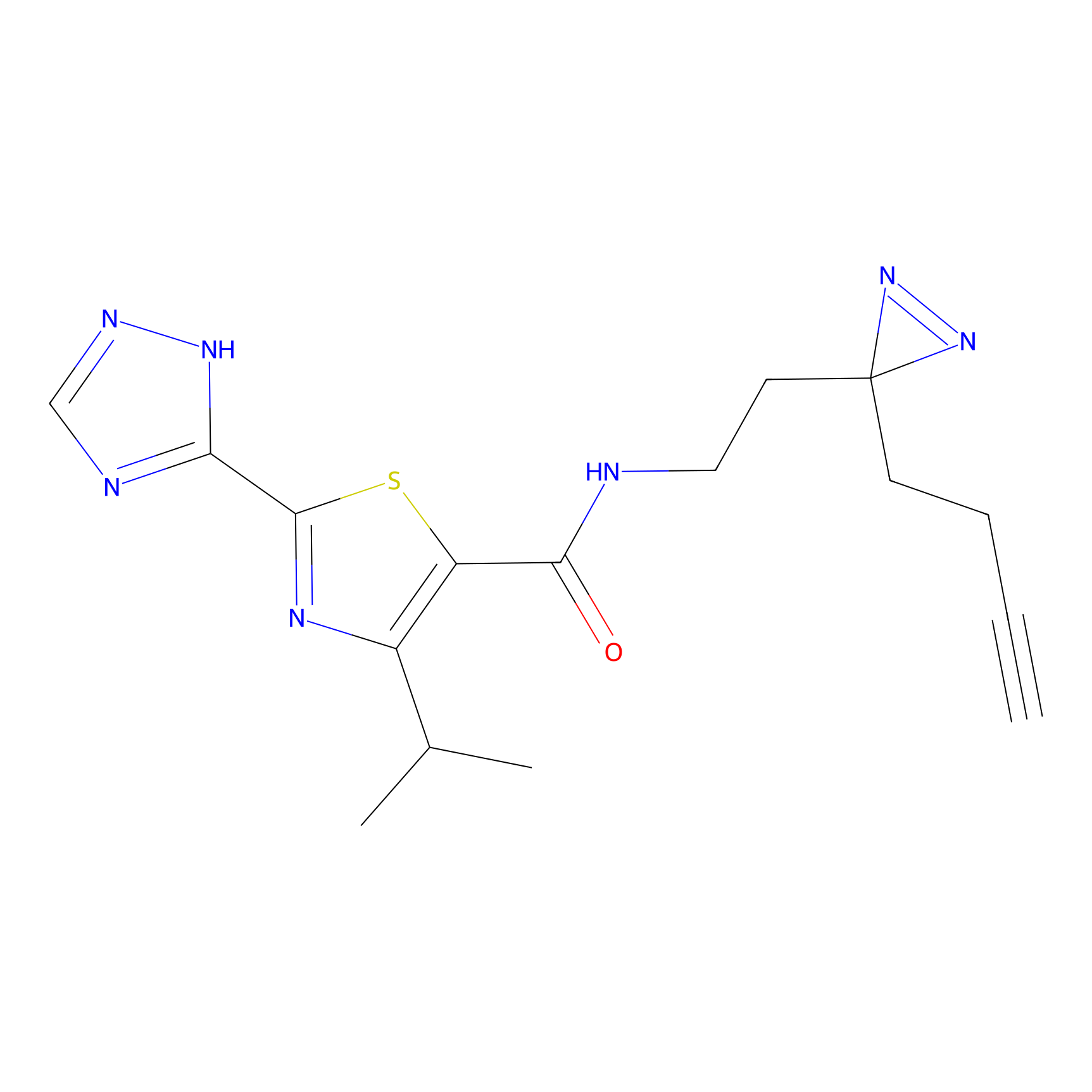
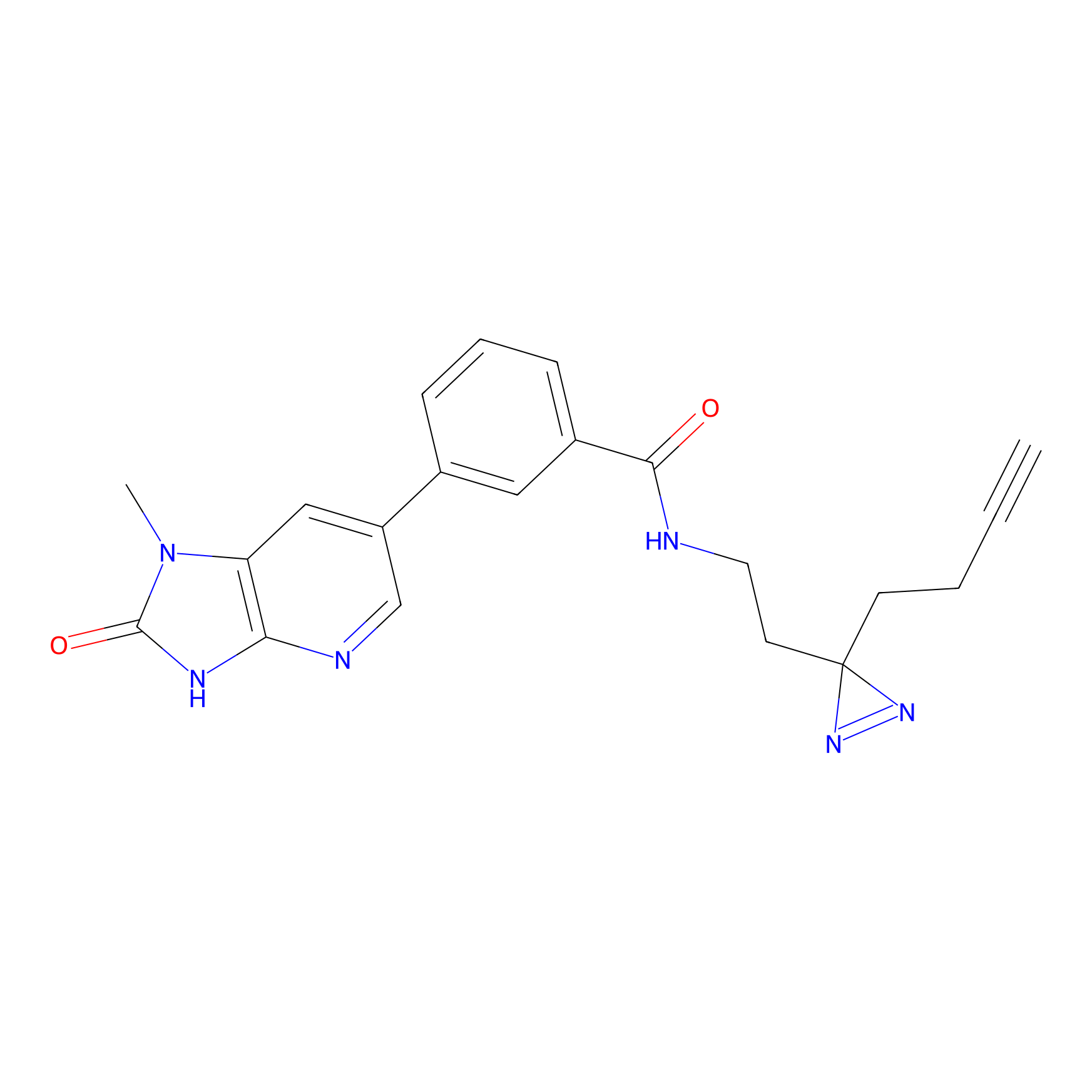
CY4 Probe Info |
 |
100.00 | LDD0244 | [2] | |
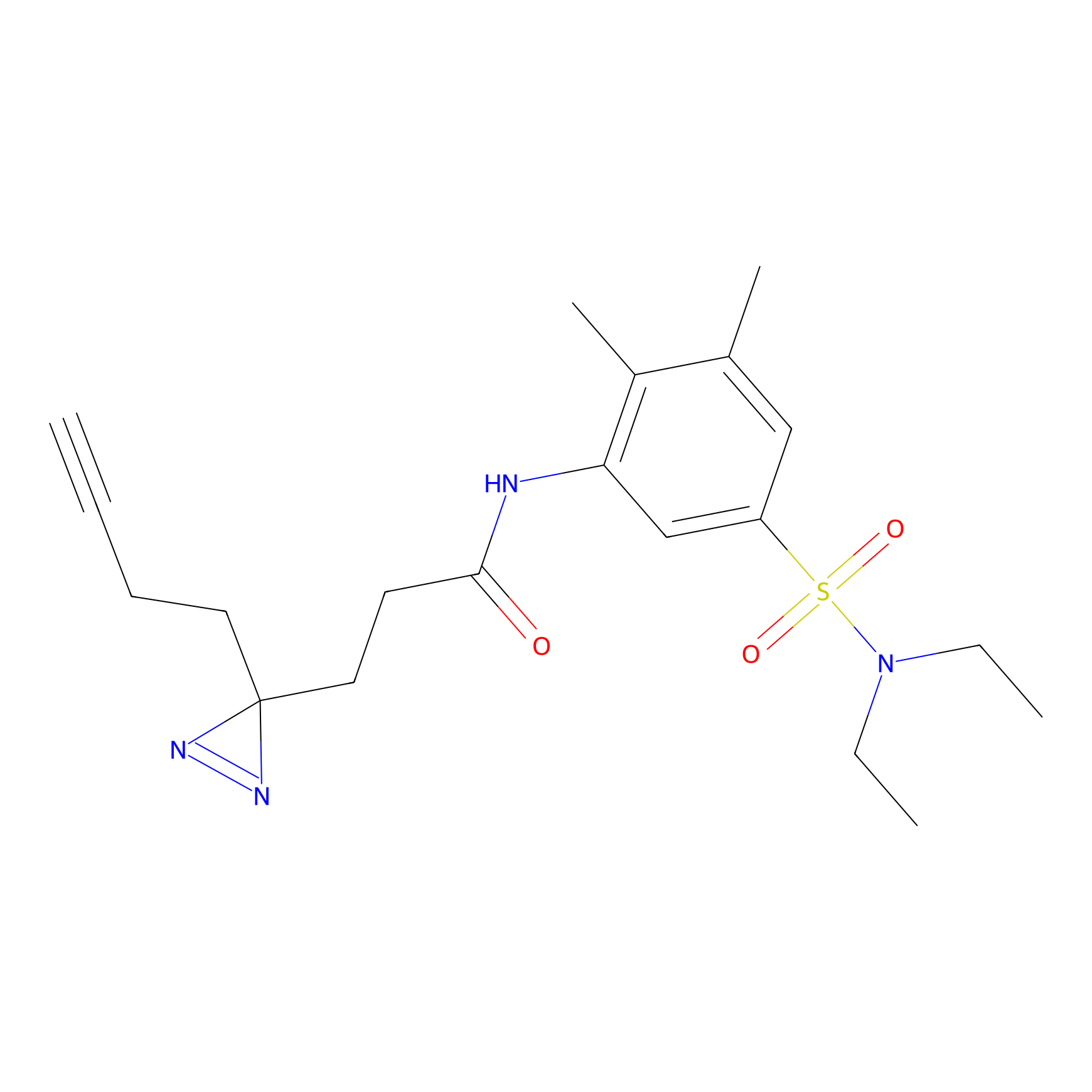
|
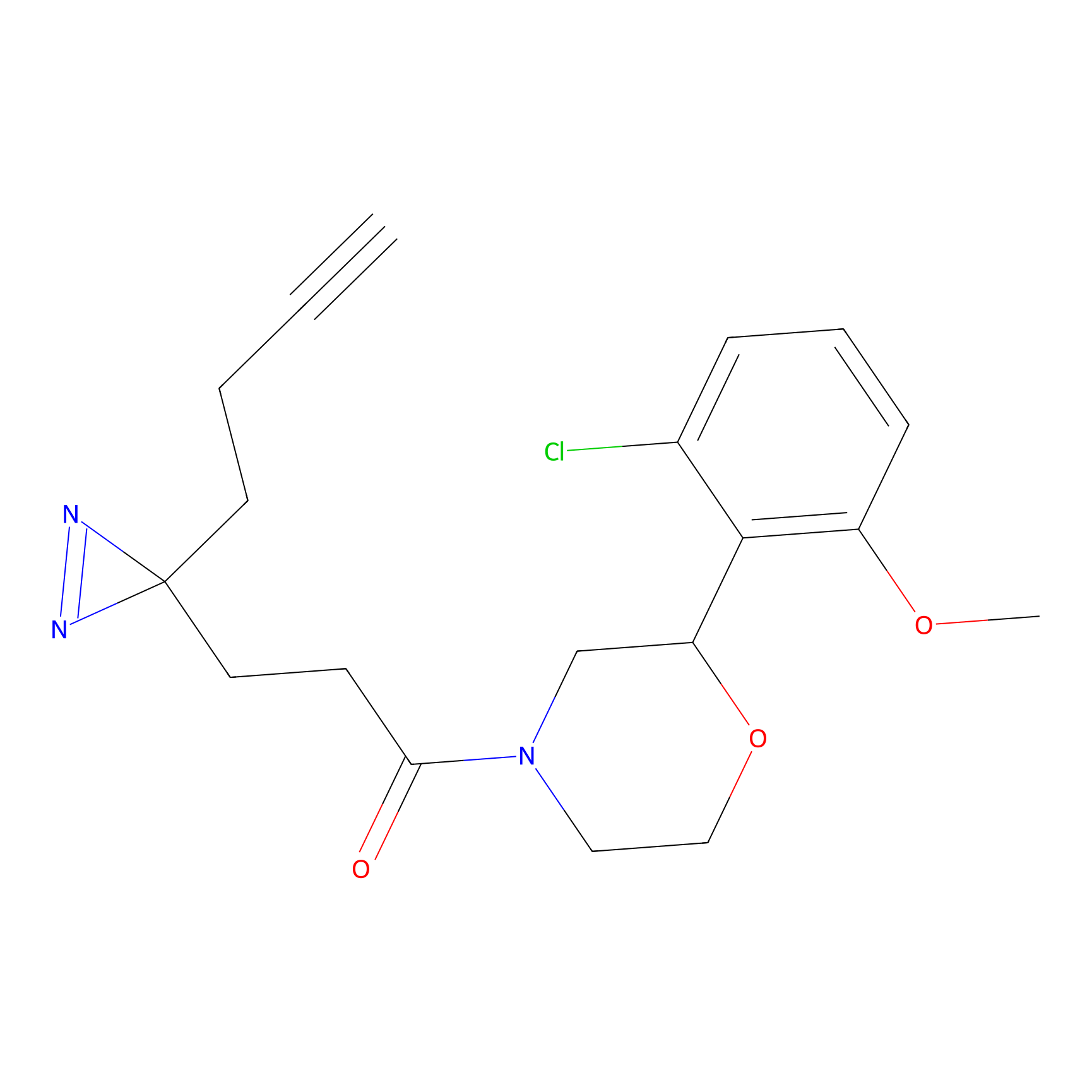
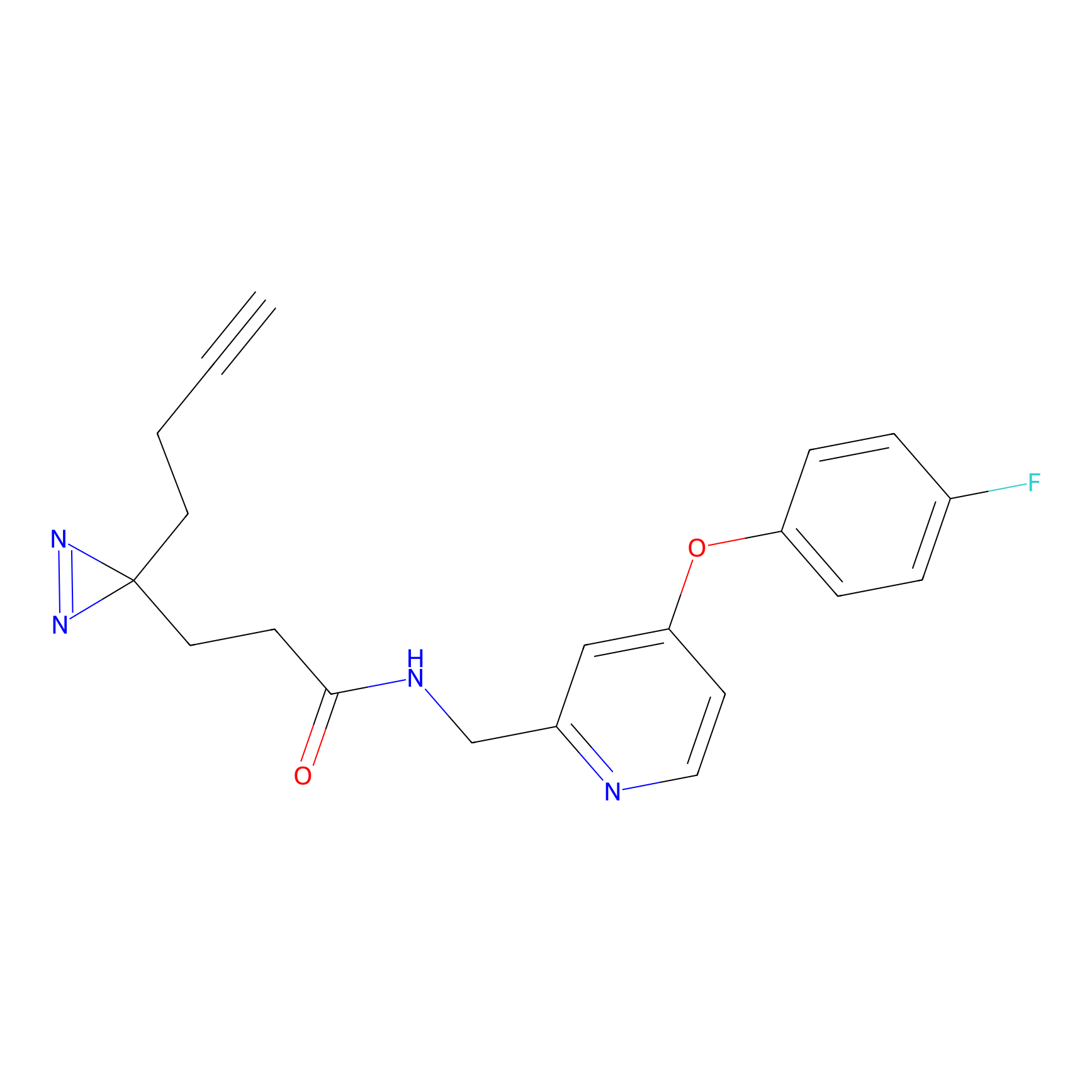
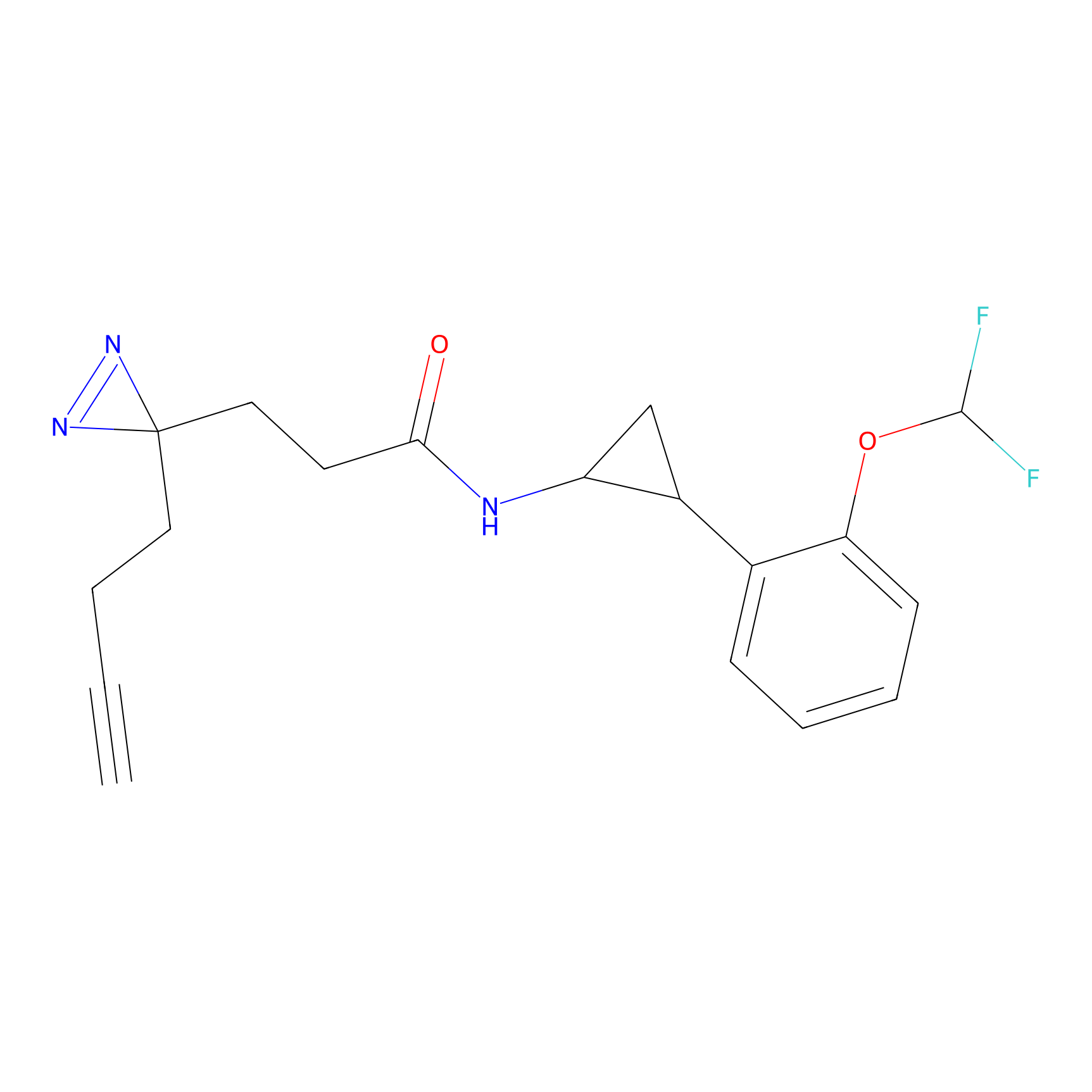
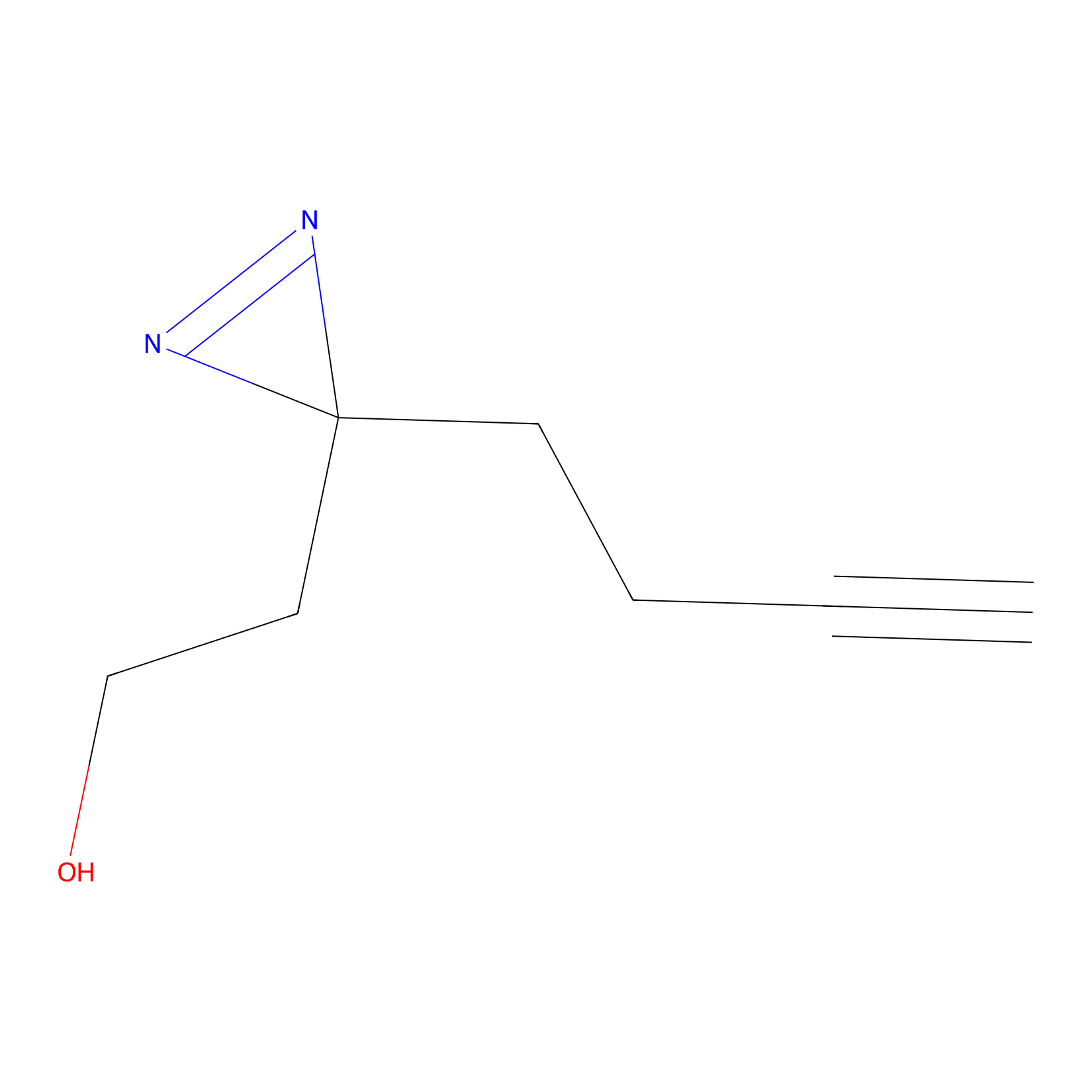
FBP2 Probe Info |
 |
5.73 | LDD0317 | [1] | |
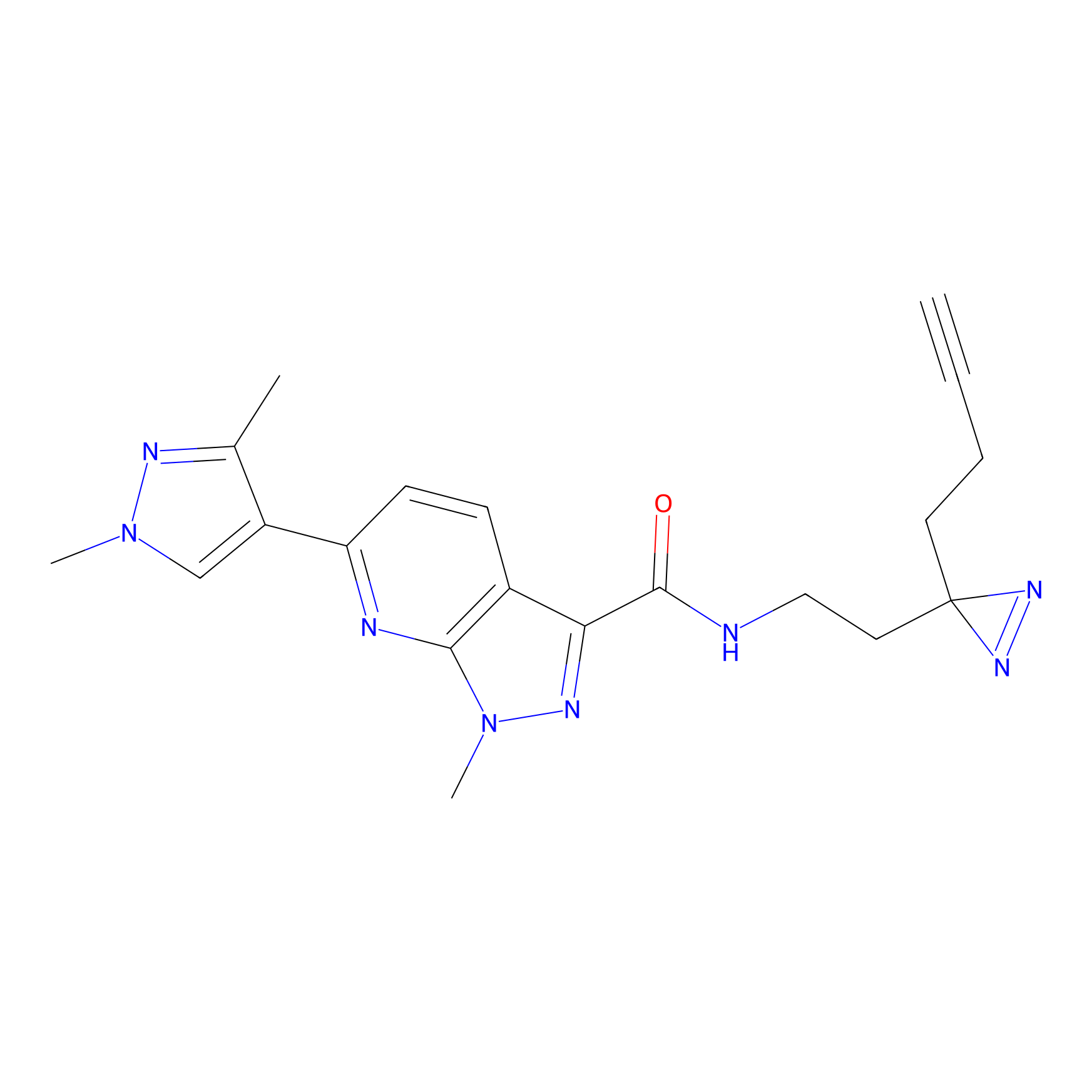
|
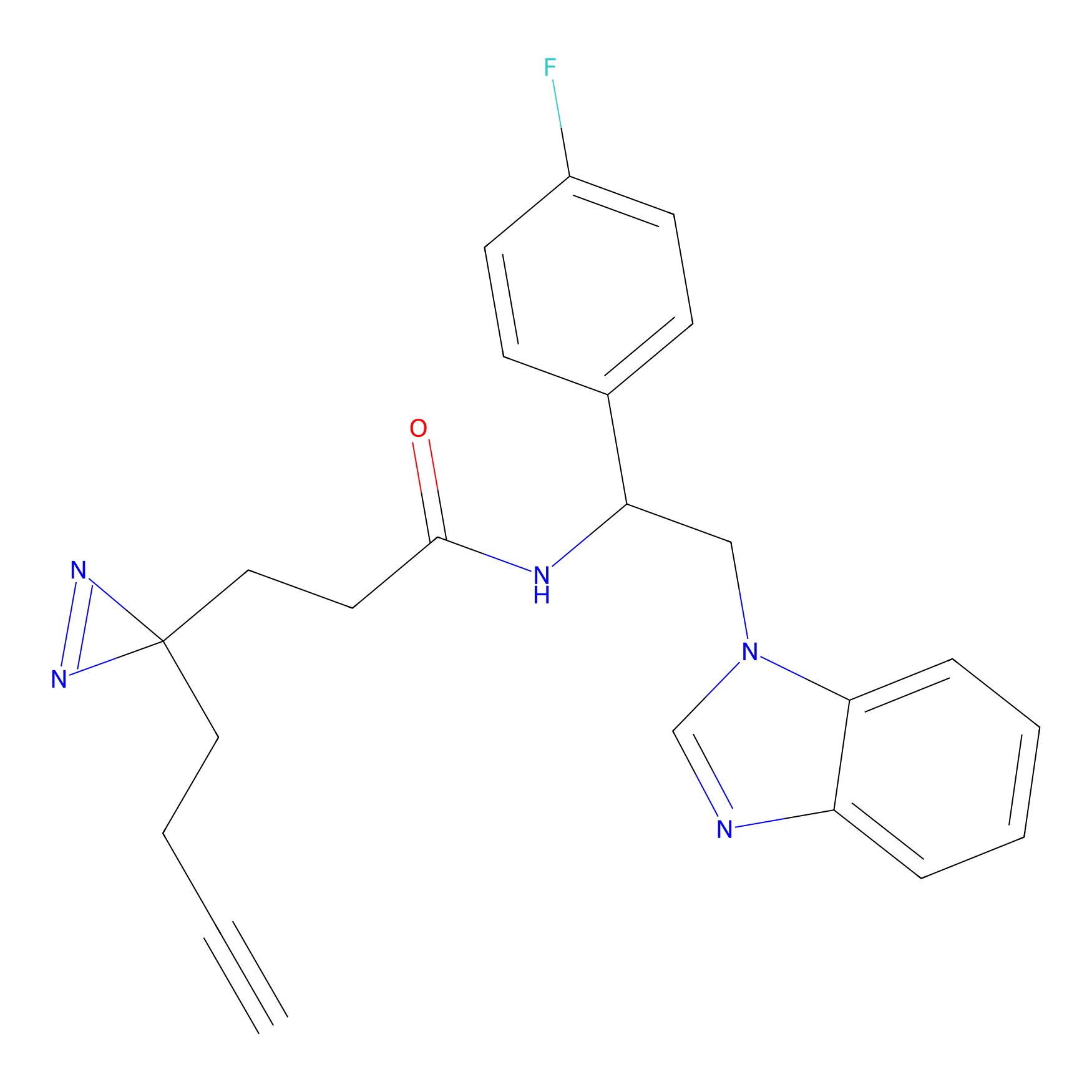
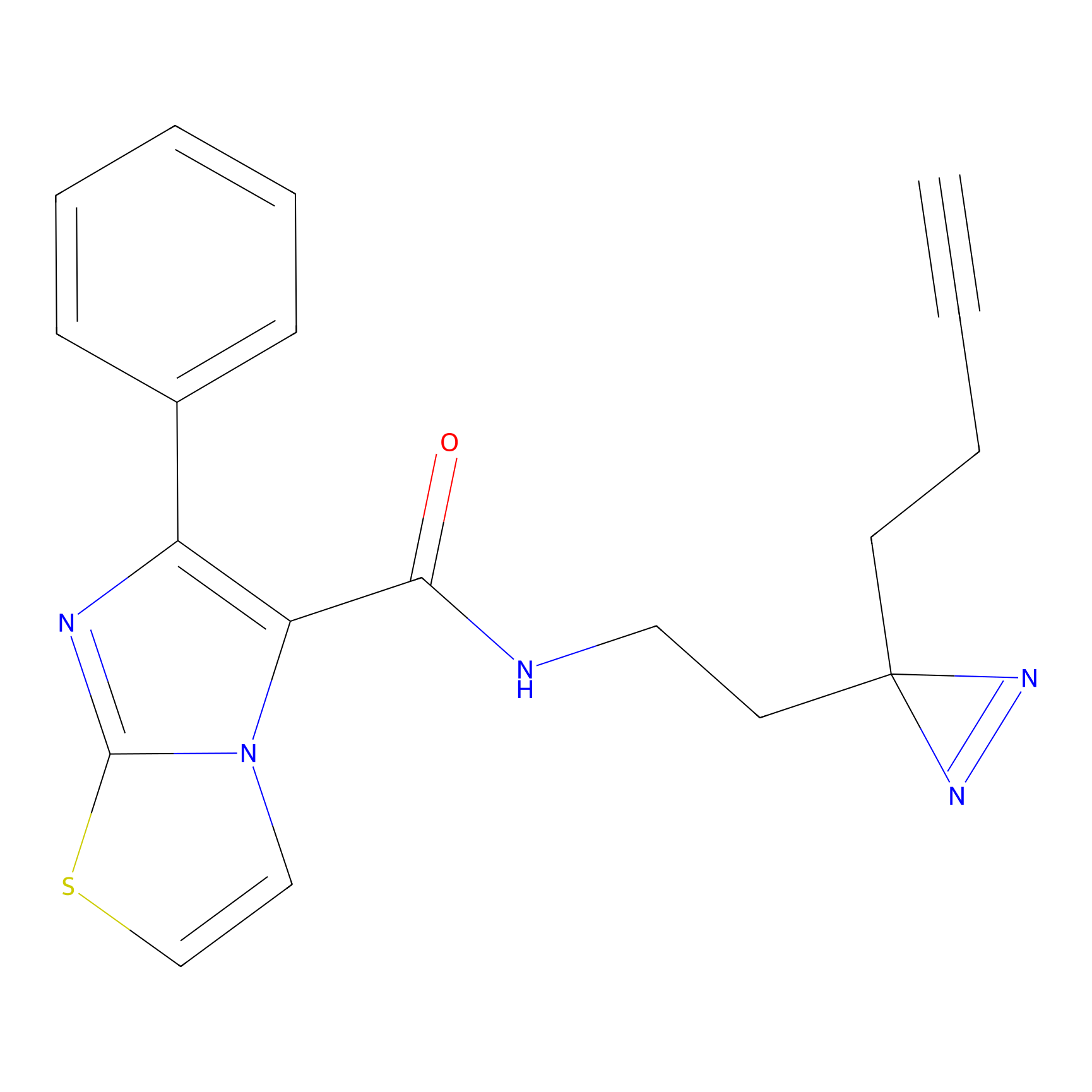
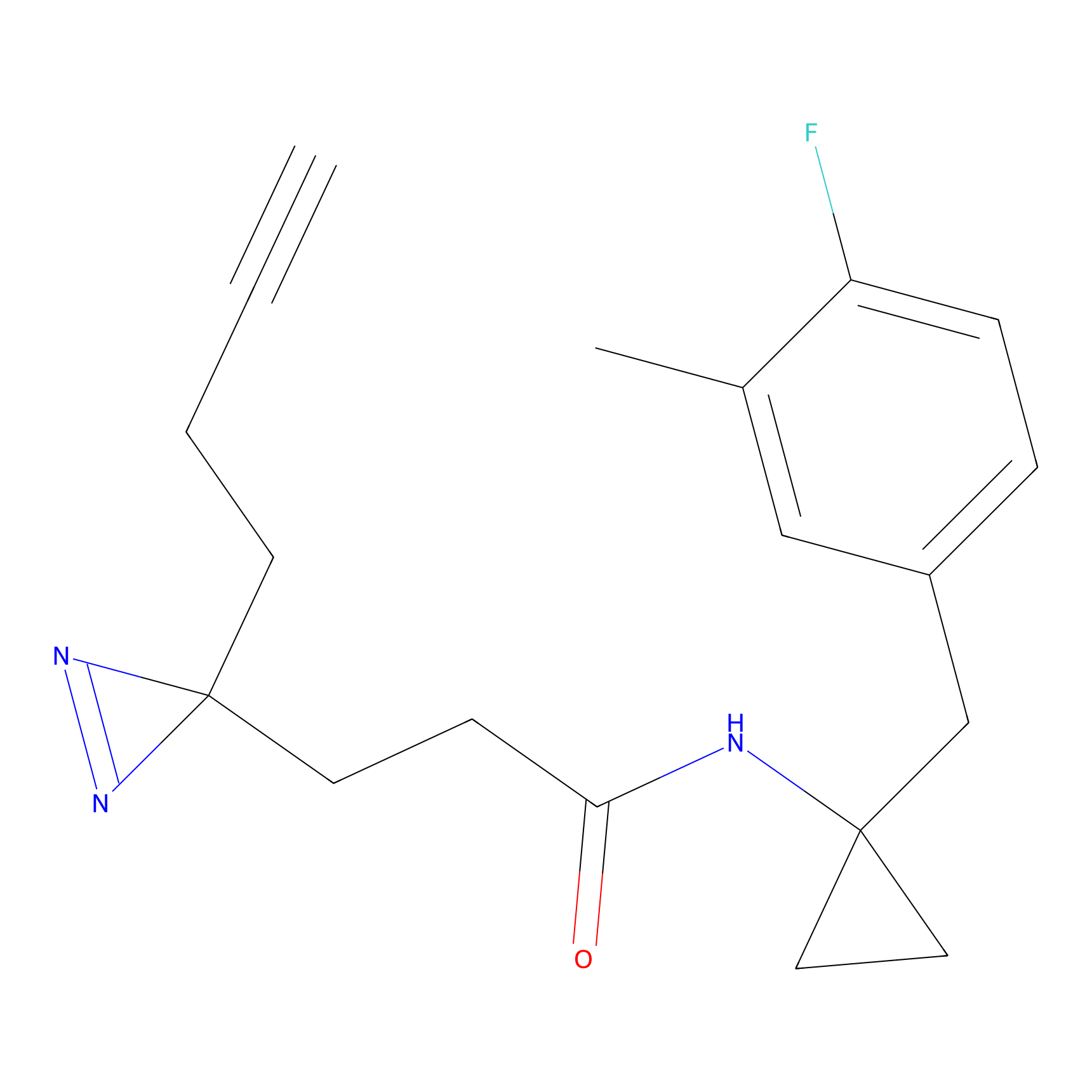
YN-4 Probe Info |
 |
100.00 | LDD0445 | [3] | |
|
ONAyne Probe Info |
 |
K176(1.02) | LDD0274 | [4] | |
|
STPyne Probe Info |
 |
K13(3.33); K176(10.00); K212(5.00); K22(5.96) | LDD0277 | [4] | |
|
m-APA Probe Info |
 |
13.78 | LDD0403 | [5] | |
|
Curcusone 37 Probe Info |
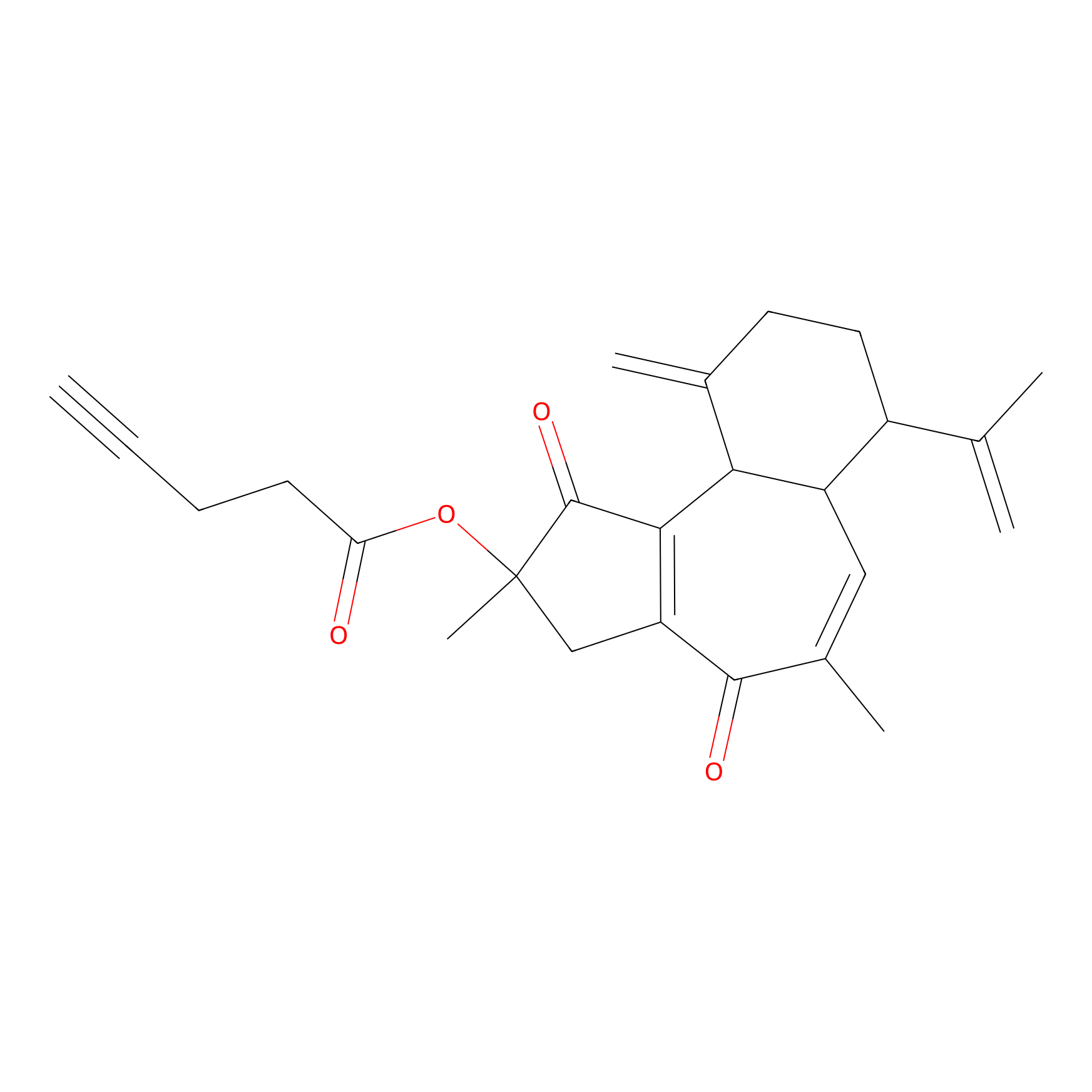 |
2.88 | LDD0188 | [6] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0233 | [7] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [8] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Myogenin (MYOG) | . | P15173 | |||
GPCR
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Probable G-protein coupled receptor 25 (GPR25) | G-protein coupled receptor 1 family | O00155 | |||
Cytokine and receptor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| CKLF-like MARVEL transmembrane domain-containing protein 3 (CMTM3) | Chemokine-like factor family | Q96MX0 | |||
Other
References