Details of the Target
General Information of Target
| Target ID | LDTP11869 | |||||
|---|---|---|---|---|---|---|
| Target Name | Growth hormone-inducible transmembrane protein (GHITM) | |||||
| Gene Name | GHITM | |||||
| Gene ID | 27069 | |||||
| Synonyms |
DERP2; MICS1; TMBIM5; Growth hormone-inducible transmembrane protein; Dermal papilla-derived protein 2; Mitochondrial morphology and cristae structure 1; MICS1; Transmembrane BAX inhibitor motif-containing protein 5
|
|||||
| 3D Structure | ||||||
| Sequence |
MSTVGLFHFPTPLTRICPAPWGLRLWEKLTLLSPGIAVTPVQMAGKKDYPALLSLDENEL
EEQFVKGHGPGGQATNKTSNCVVLKHIPSGIVVKCHQTRSVDQNRKLARKILQEKVDVFY NGENSPVHKEKREAAKKKQERKKRAKETLEKKKLLKELWESSKKVH |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
BI1 family
|
|||||
| Subcellular location |
Mitochondrion inner membrane
|
|||||
| Function |
Plays an important role in maintenance of mitochondrial morphology and in mediating either calcium or potassium/proton antiport. Mediates proton-dependent calcium efflux from mitochondrion. Functions also as an electroneutral mitochondrial proton/potassium exchanger. Required for the mitochondrial tubular network and cristae organization. Involved in apoptotic release of cytochrome c. Inhibits the proteolytic activity of AFG3L2, stimulating respiration and stabilizing respiratory enzymes in actively respiring mitochondria. However, when mitochondria become hyperpolarized, GHITM loses its inhibitory activity toward AFG3L2 and the now the active AFG3L2 turns first on GHITM and, if hyperpolarization persists, on other proteins of the mitochondria, leading to a broad remodeling of the mitochondrial proteome.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
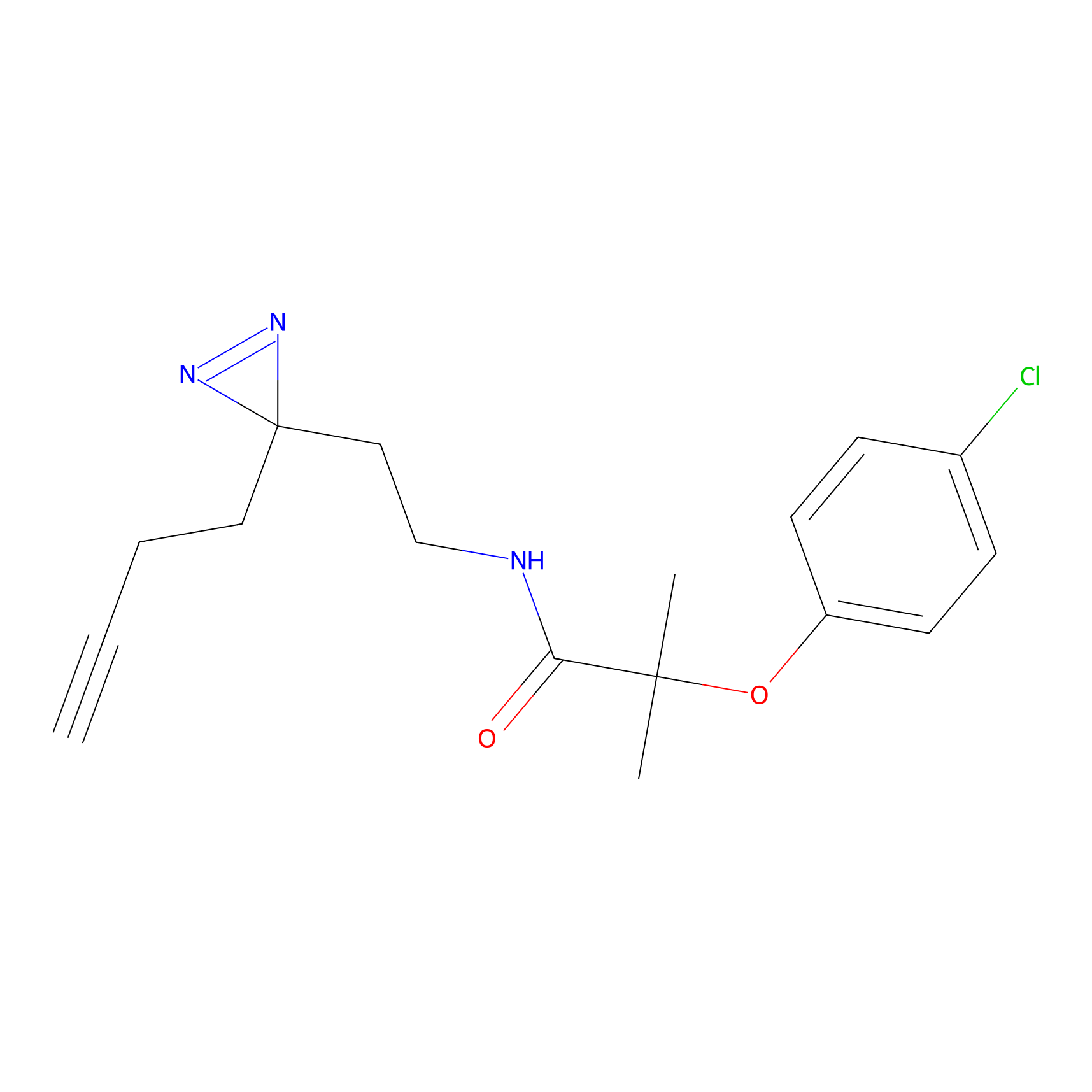
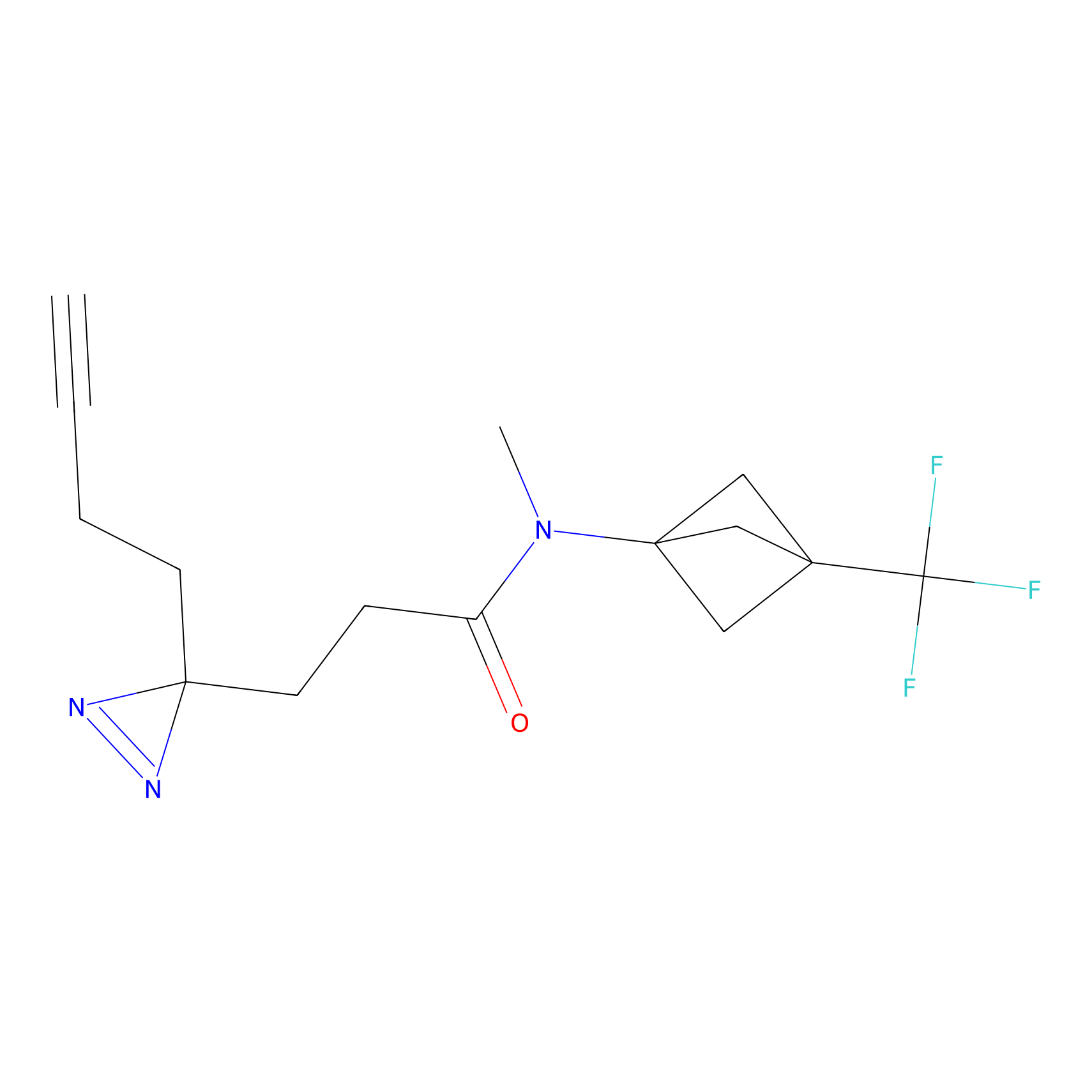
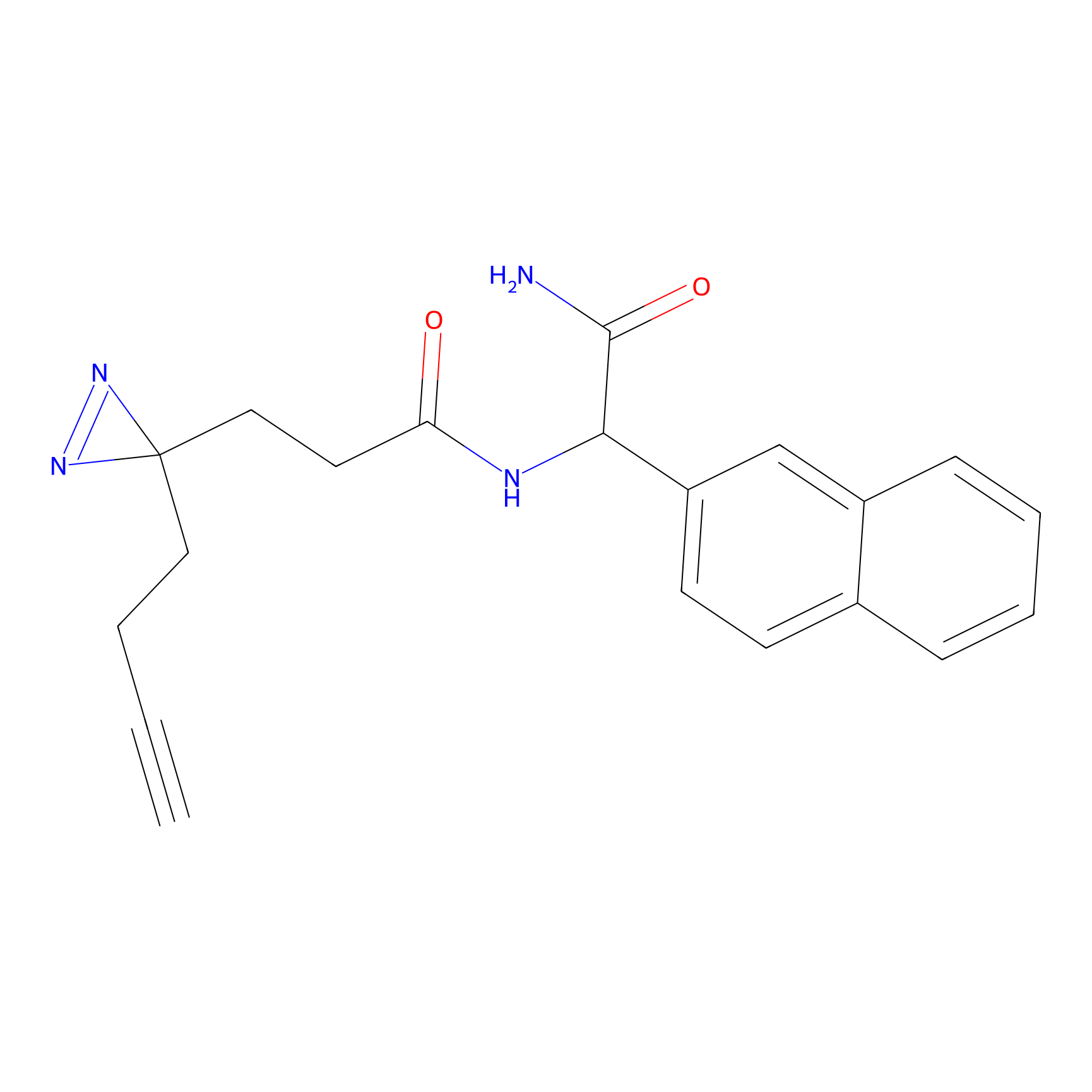
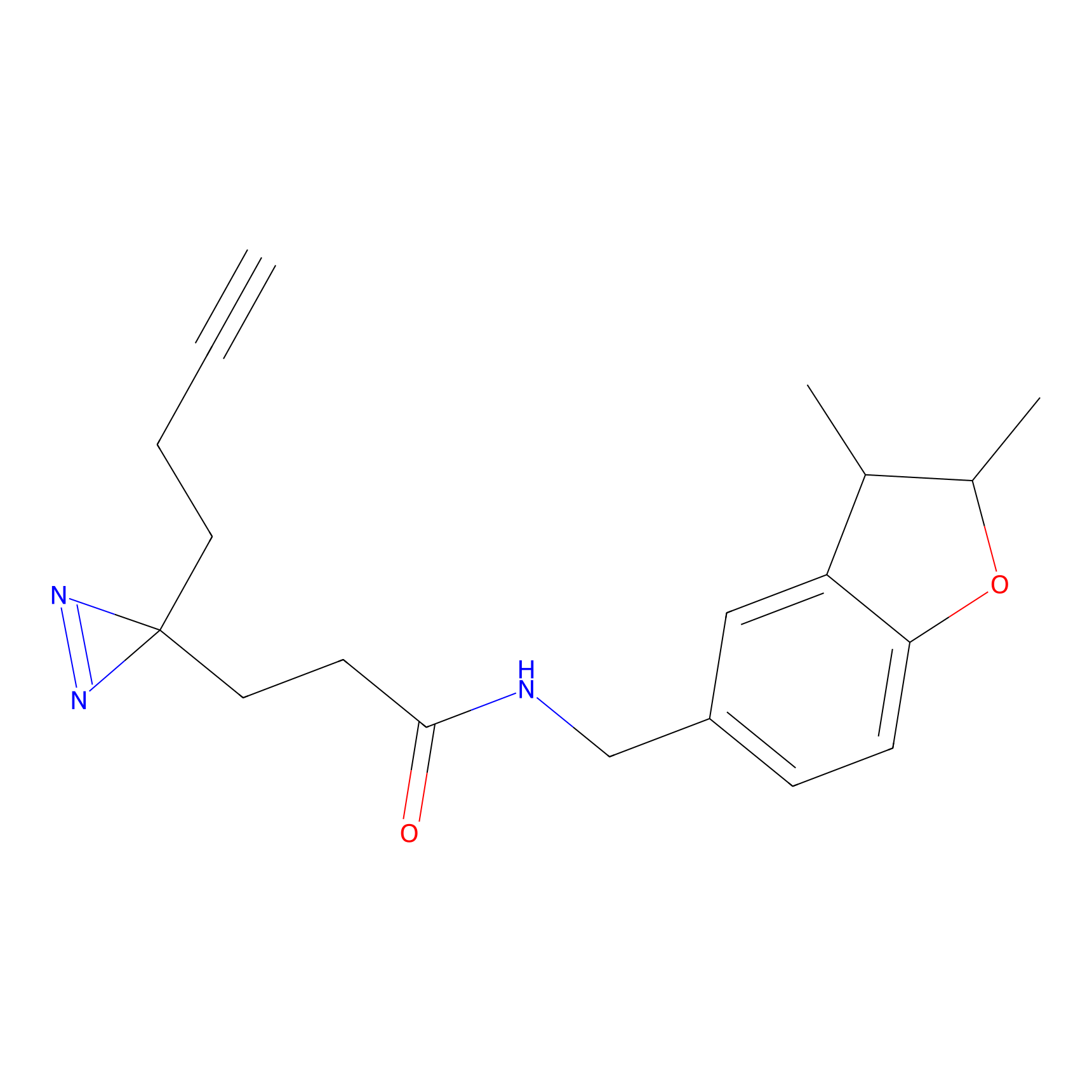
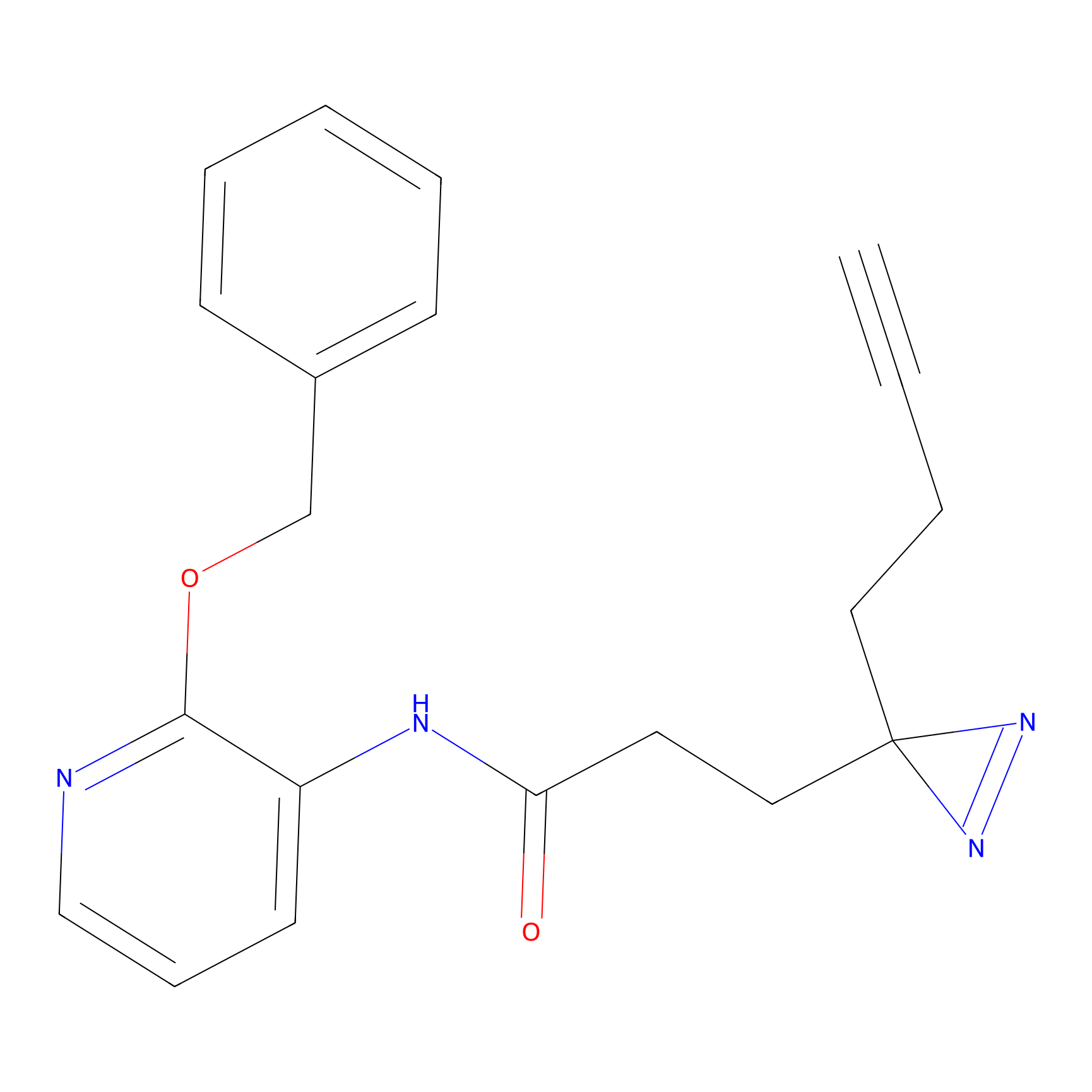
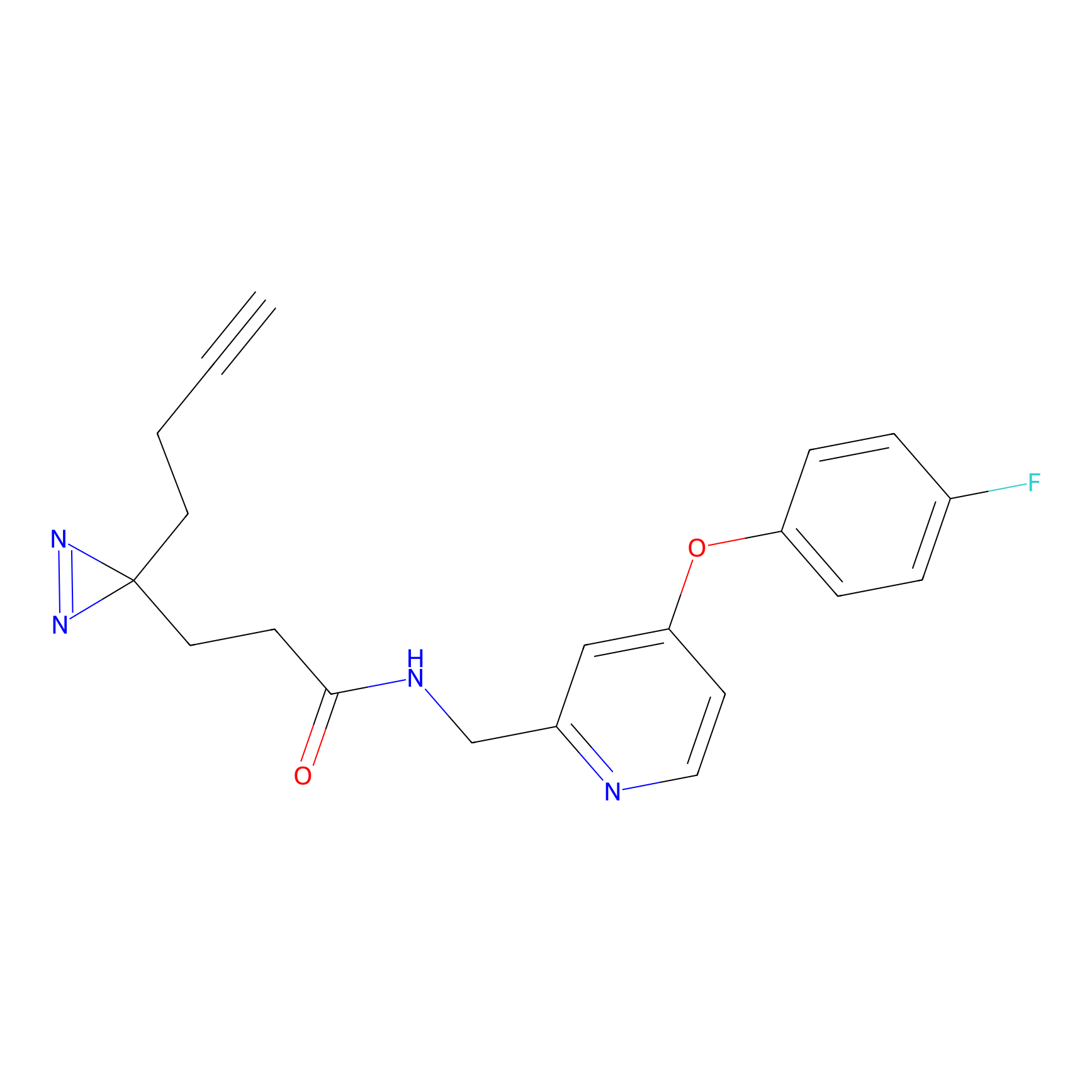
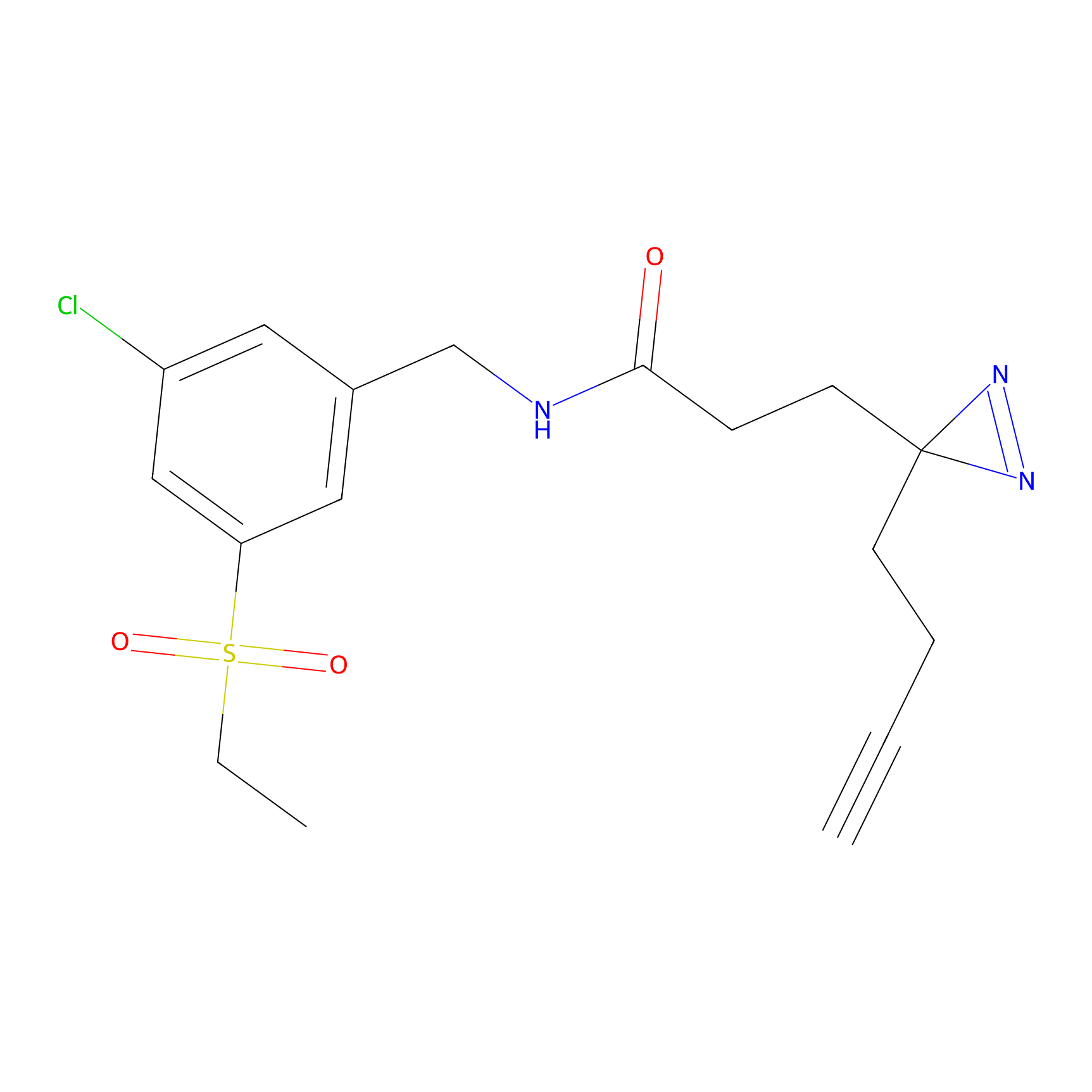
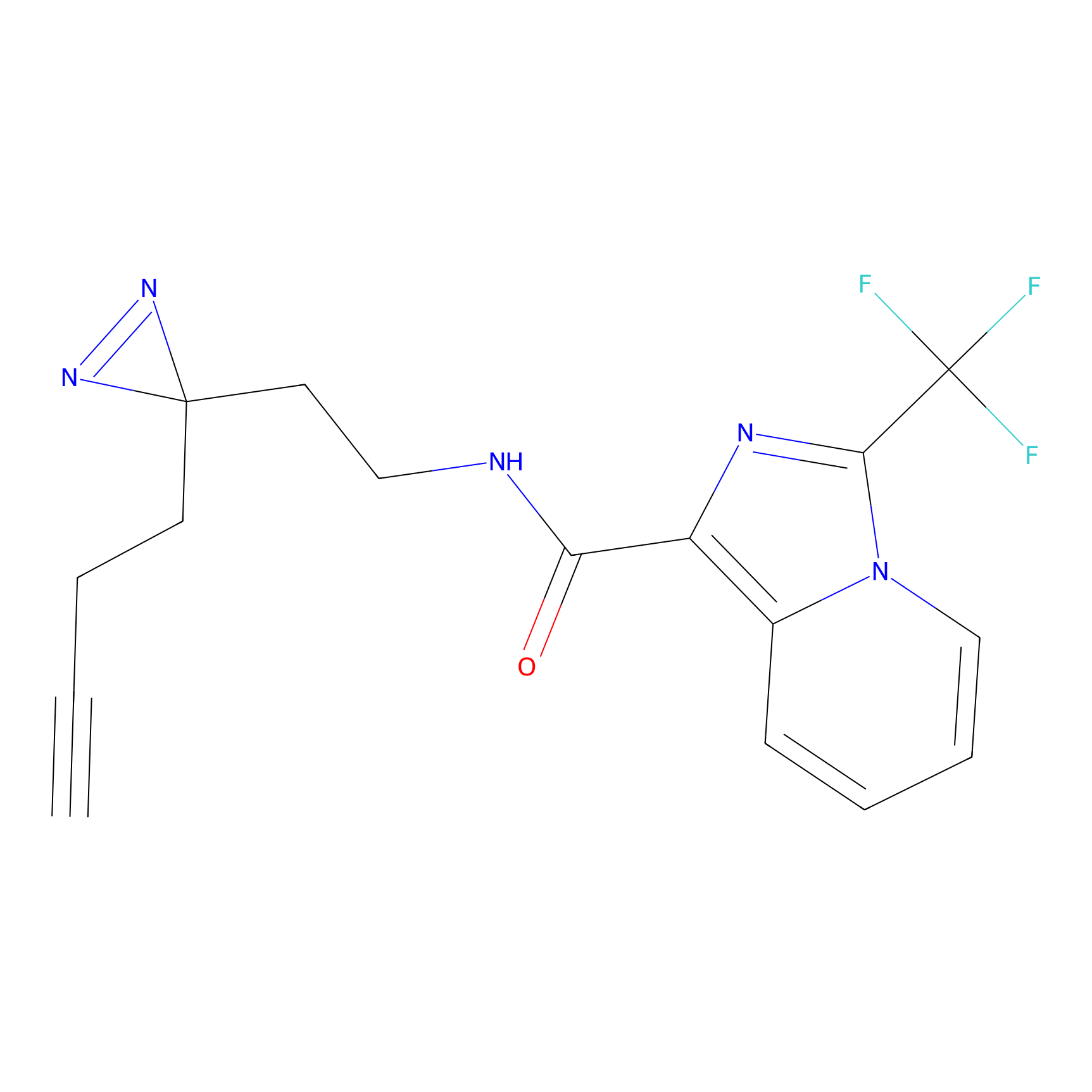
YN-1 Probe Info |
 |
100.00 | LDD0444 | [1] | |
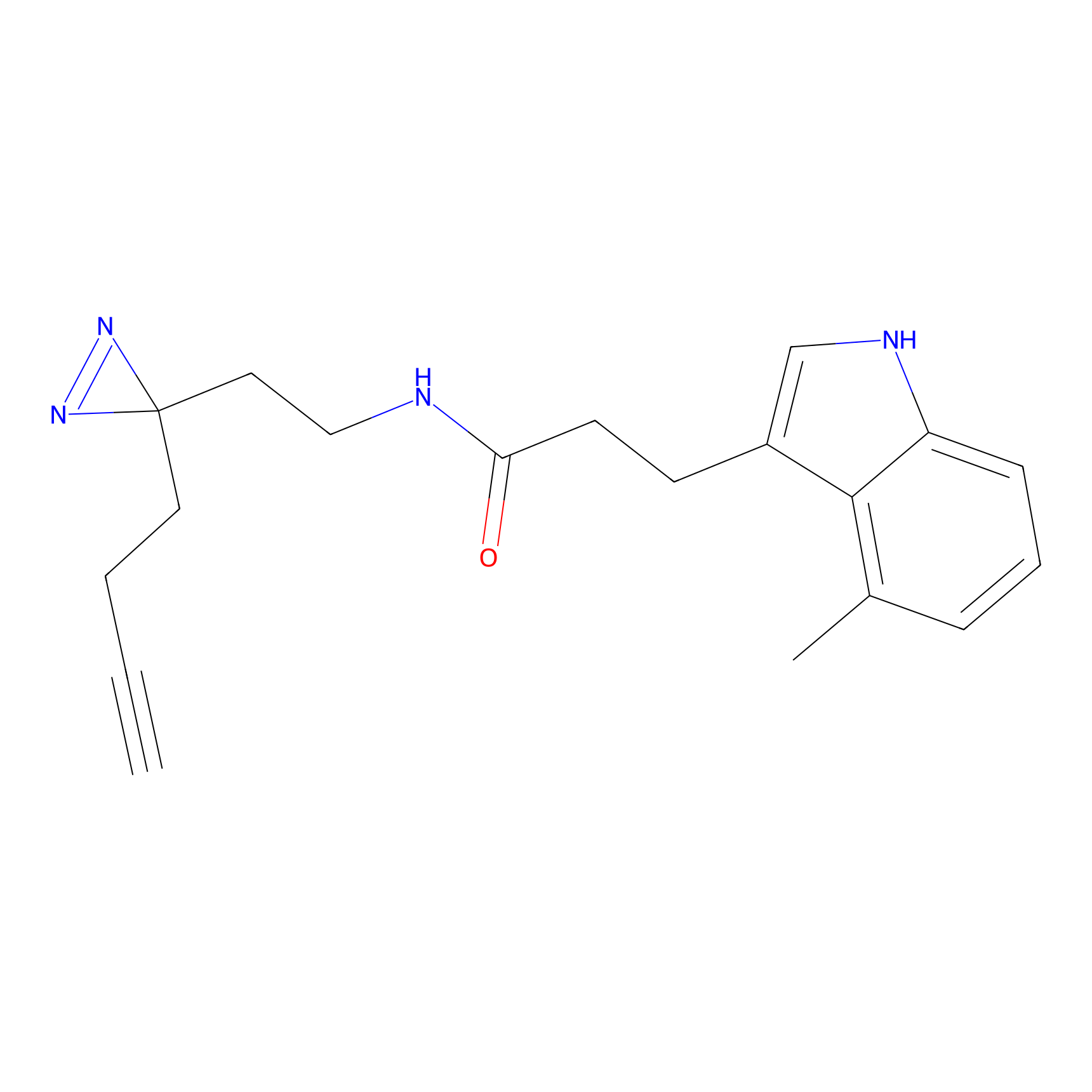
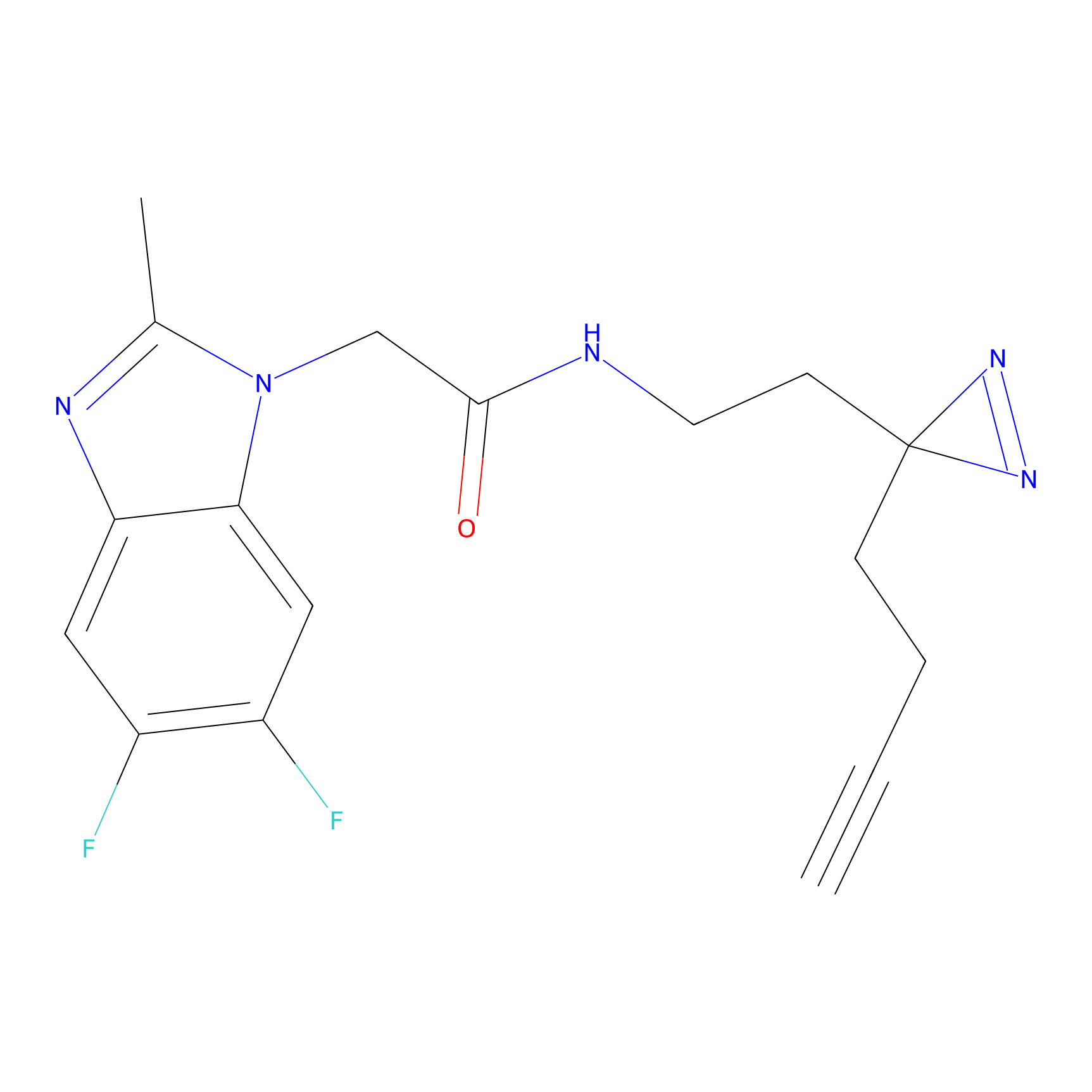
|
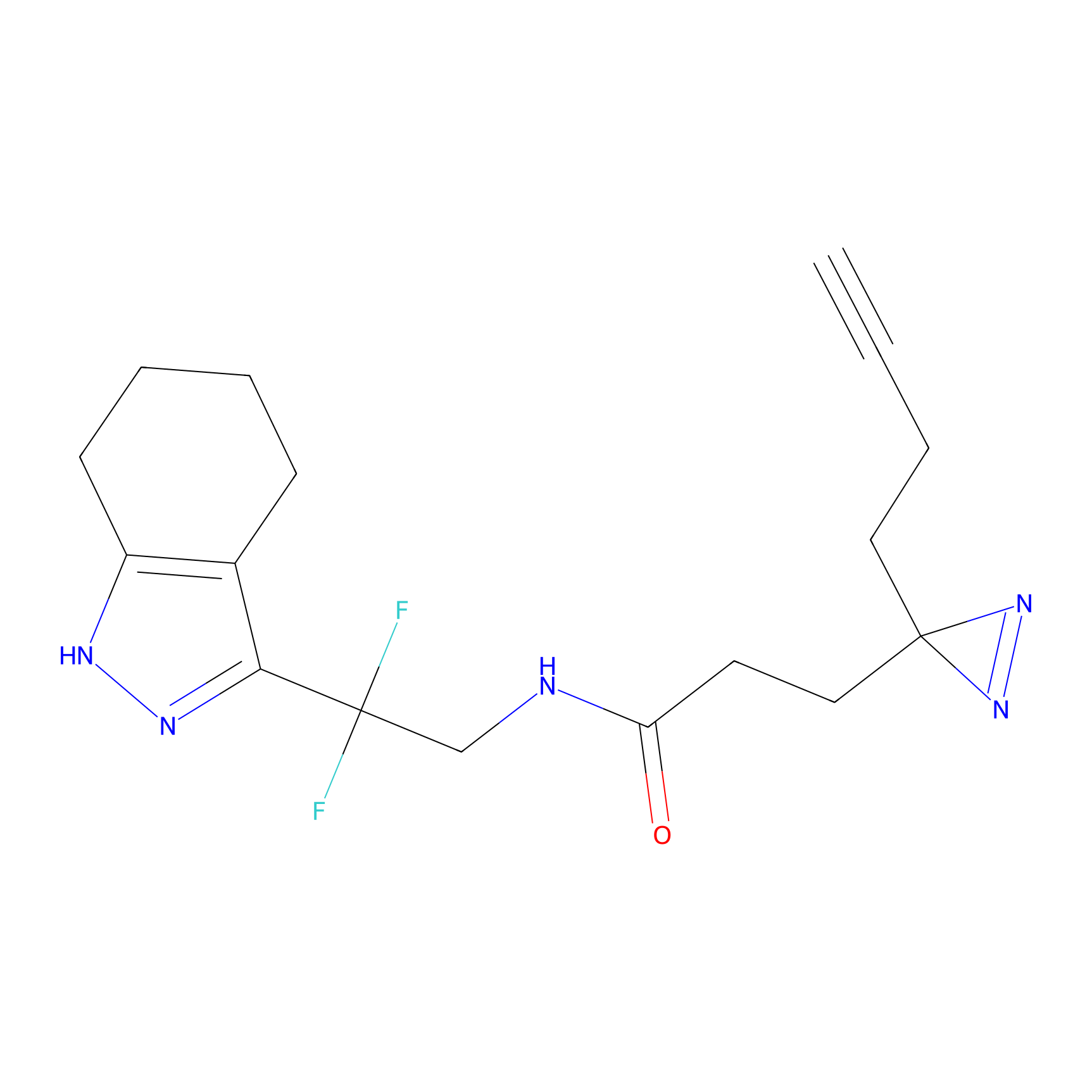
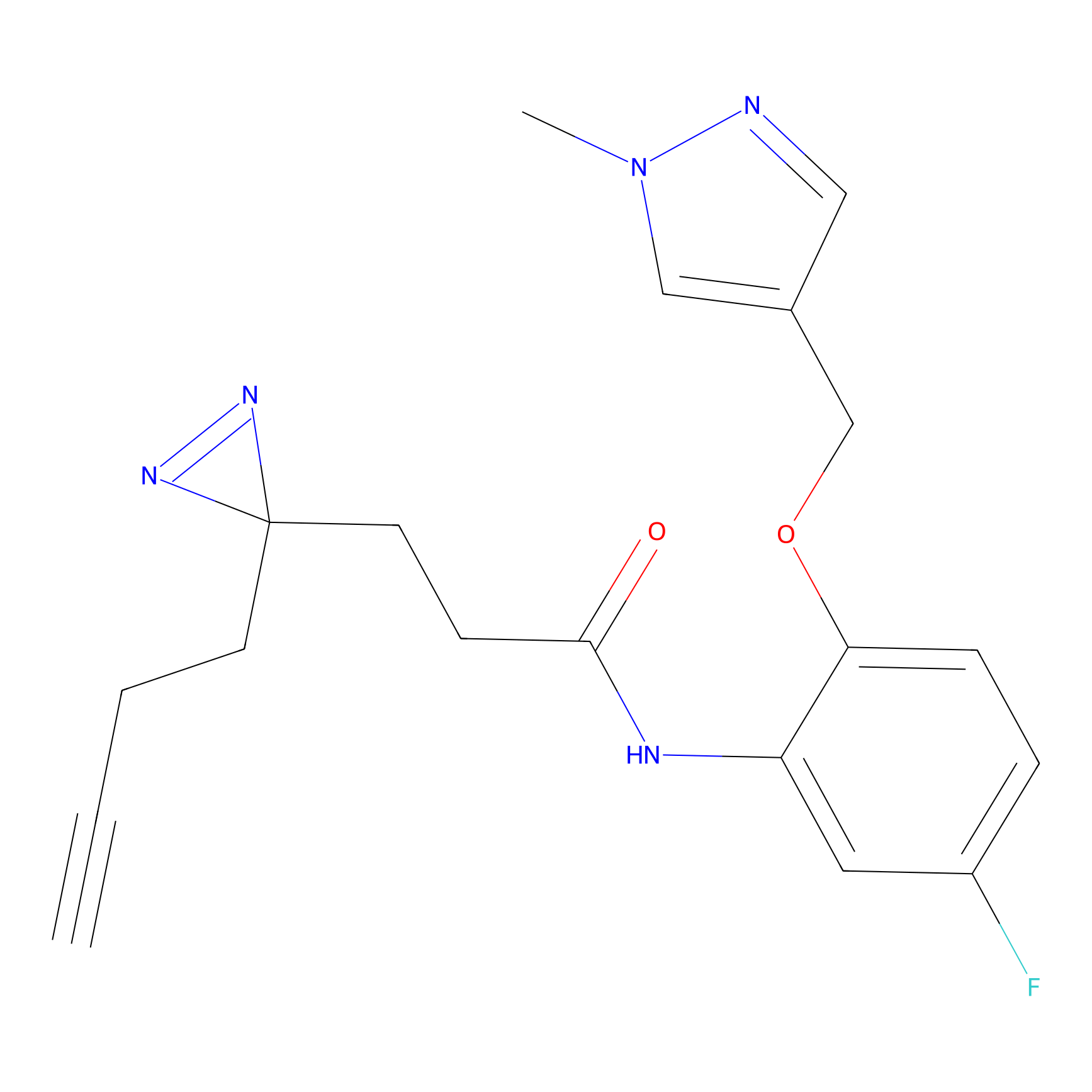
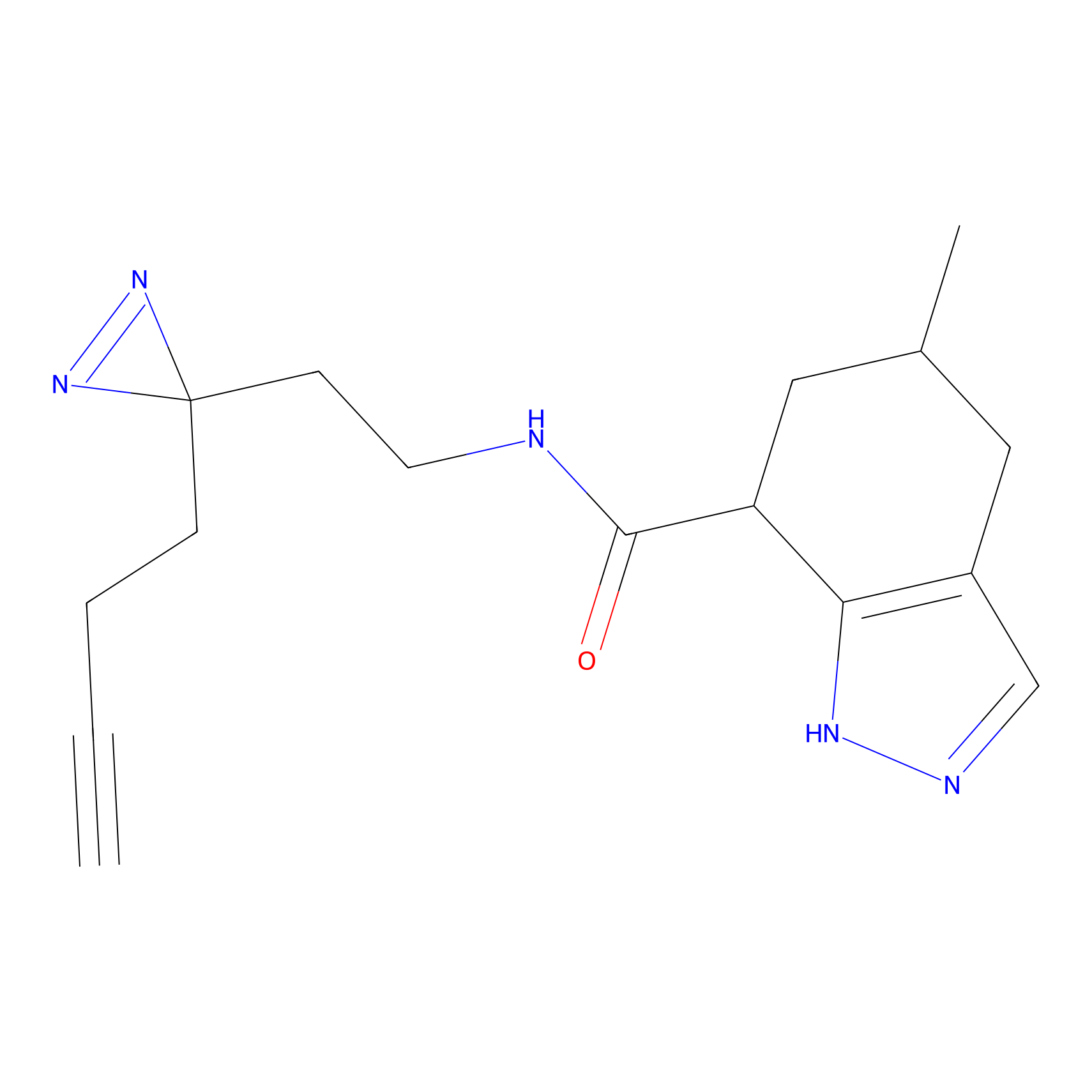
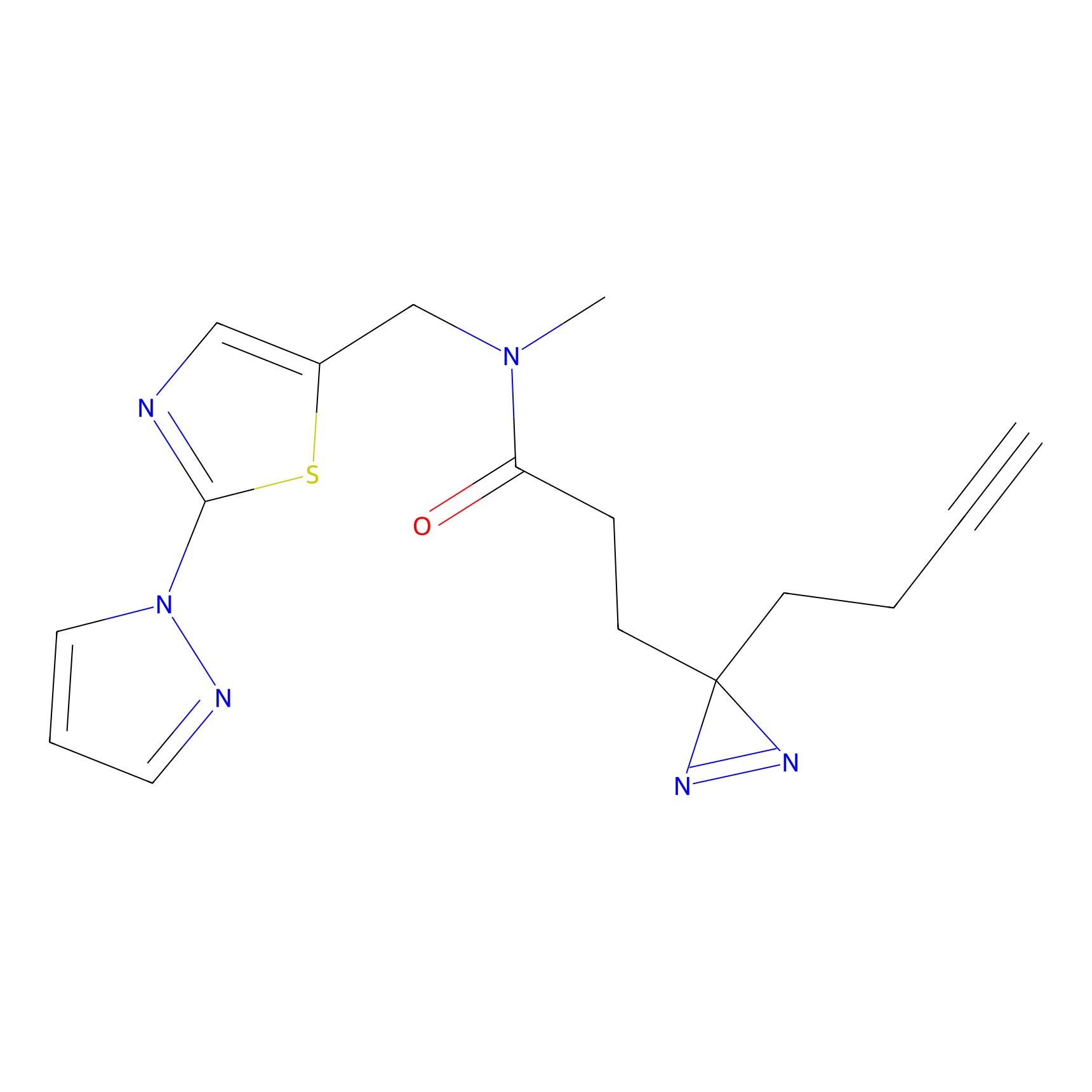
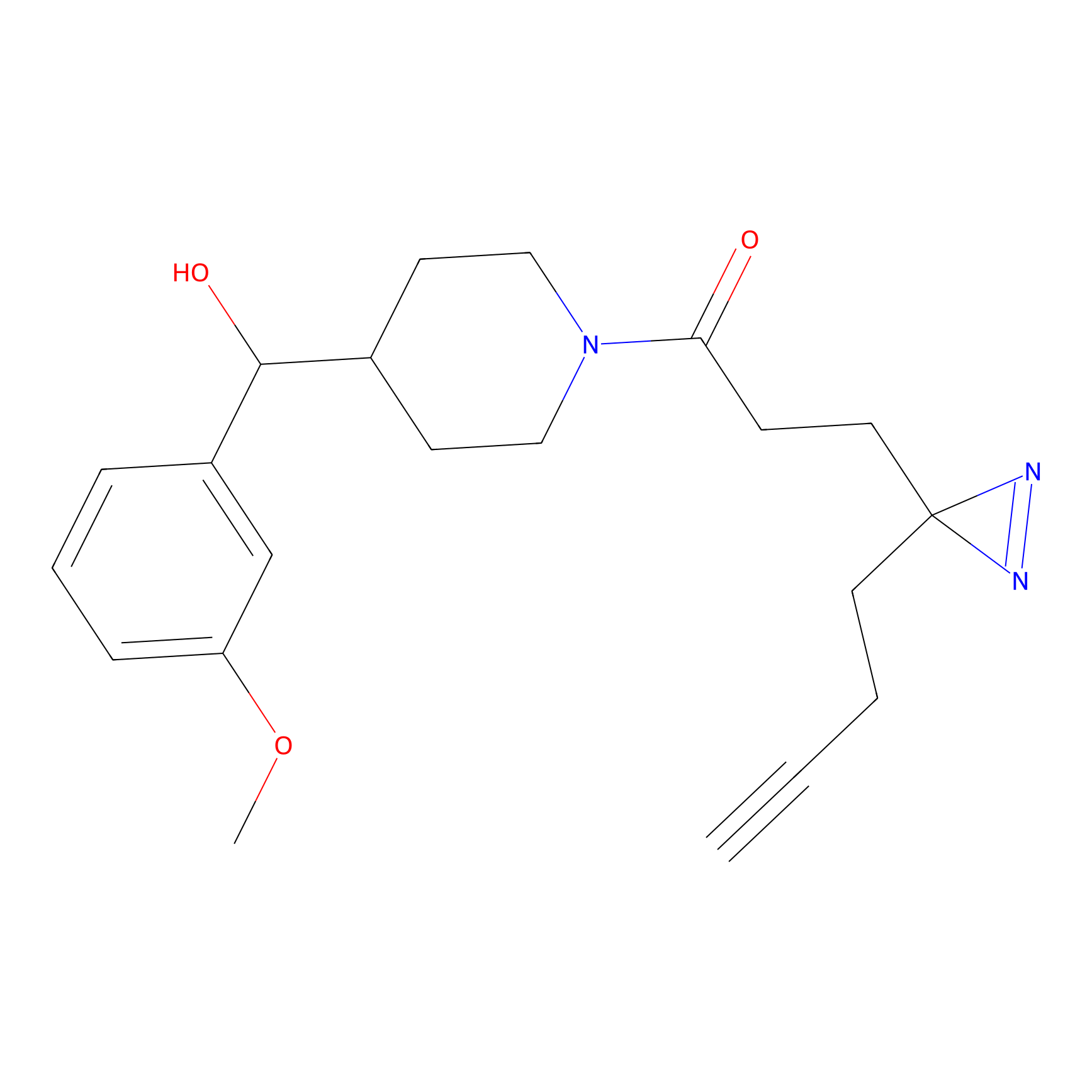
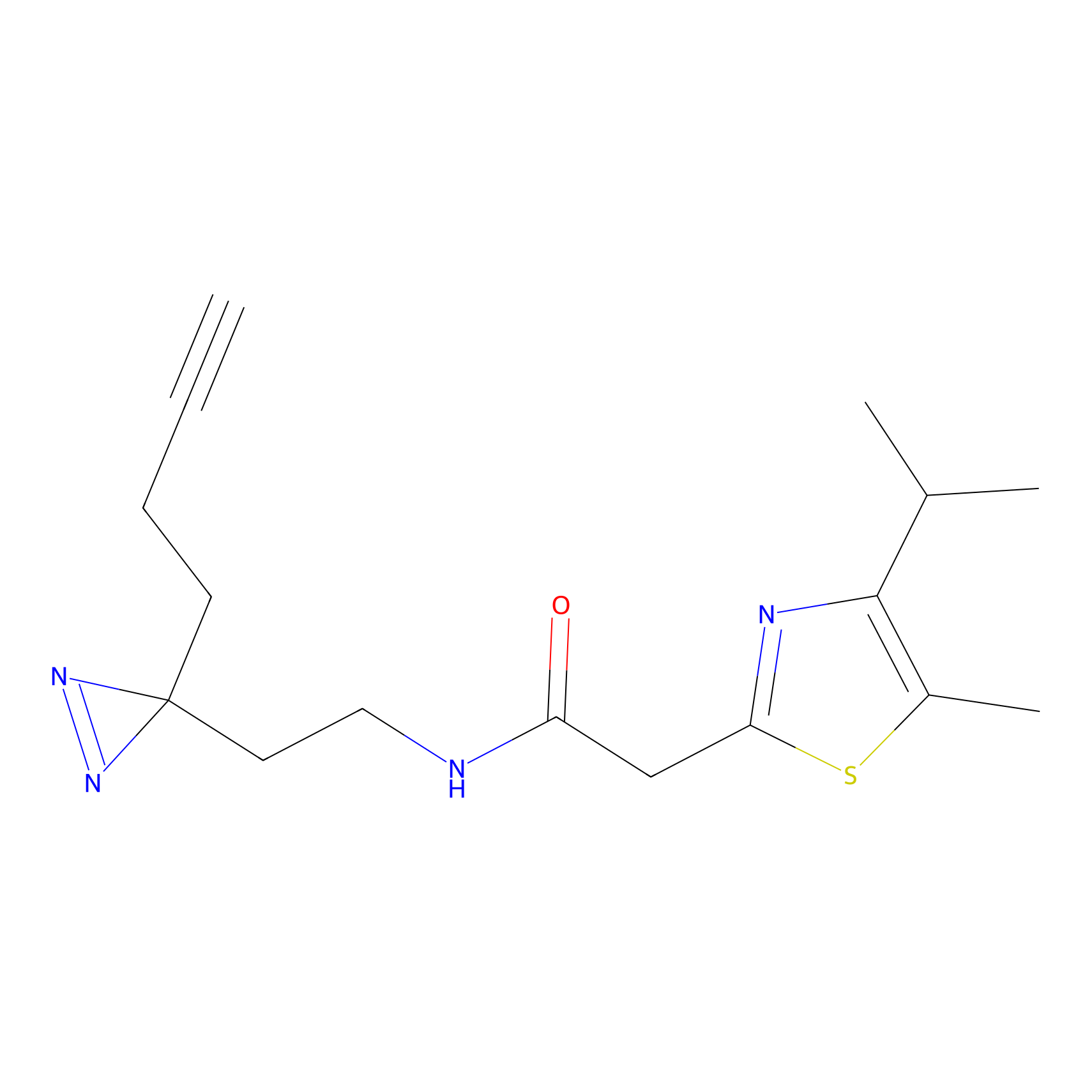
IA-alkyne Probe Info |
 |
C235(2.51) | LDD2157 | [2] | |
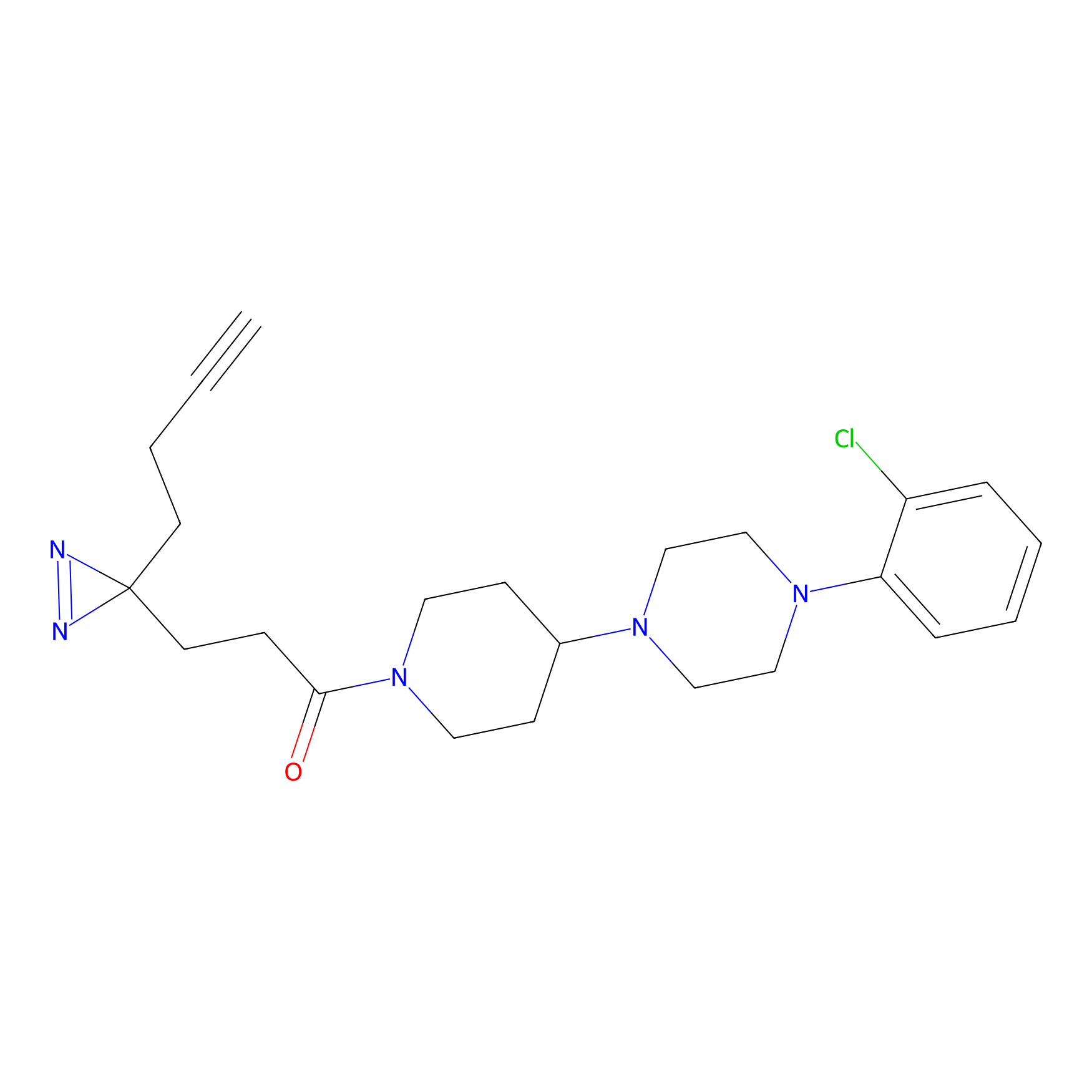
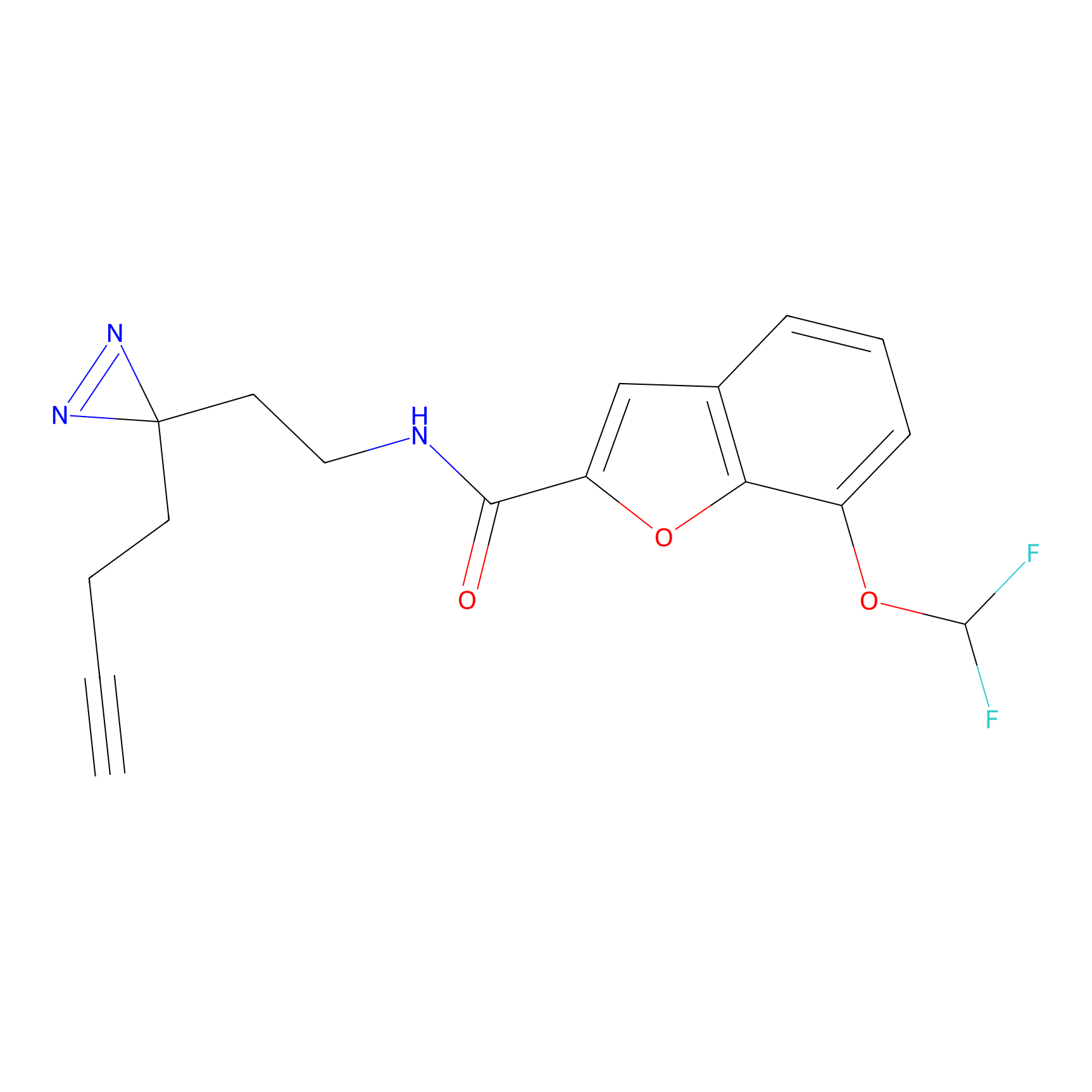
|
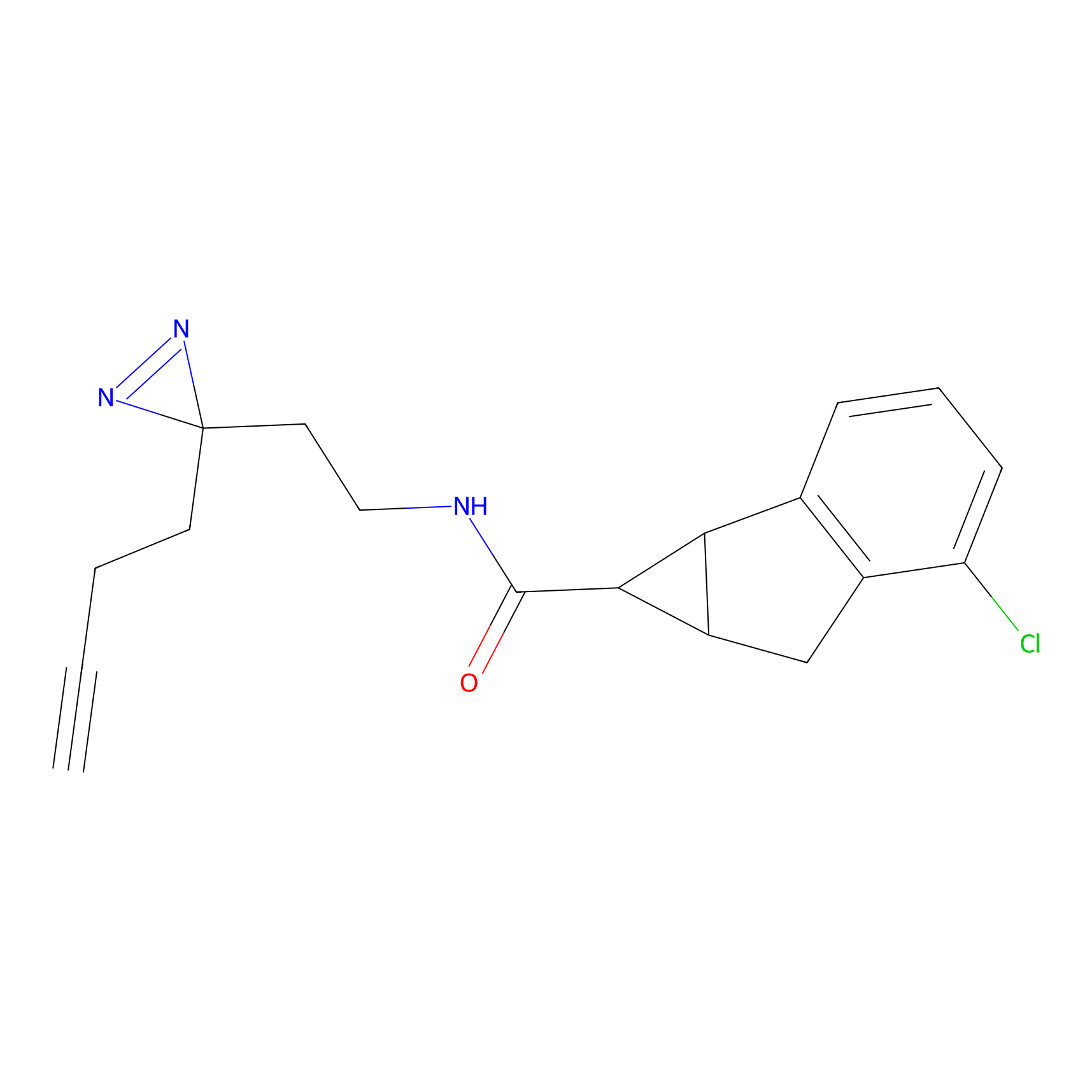
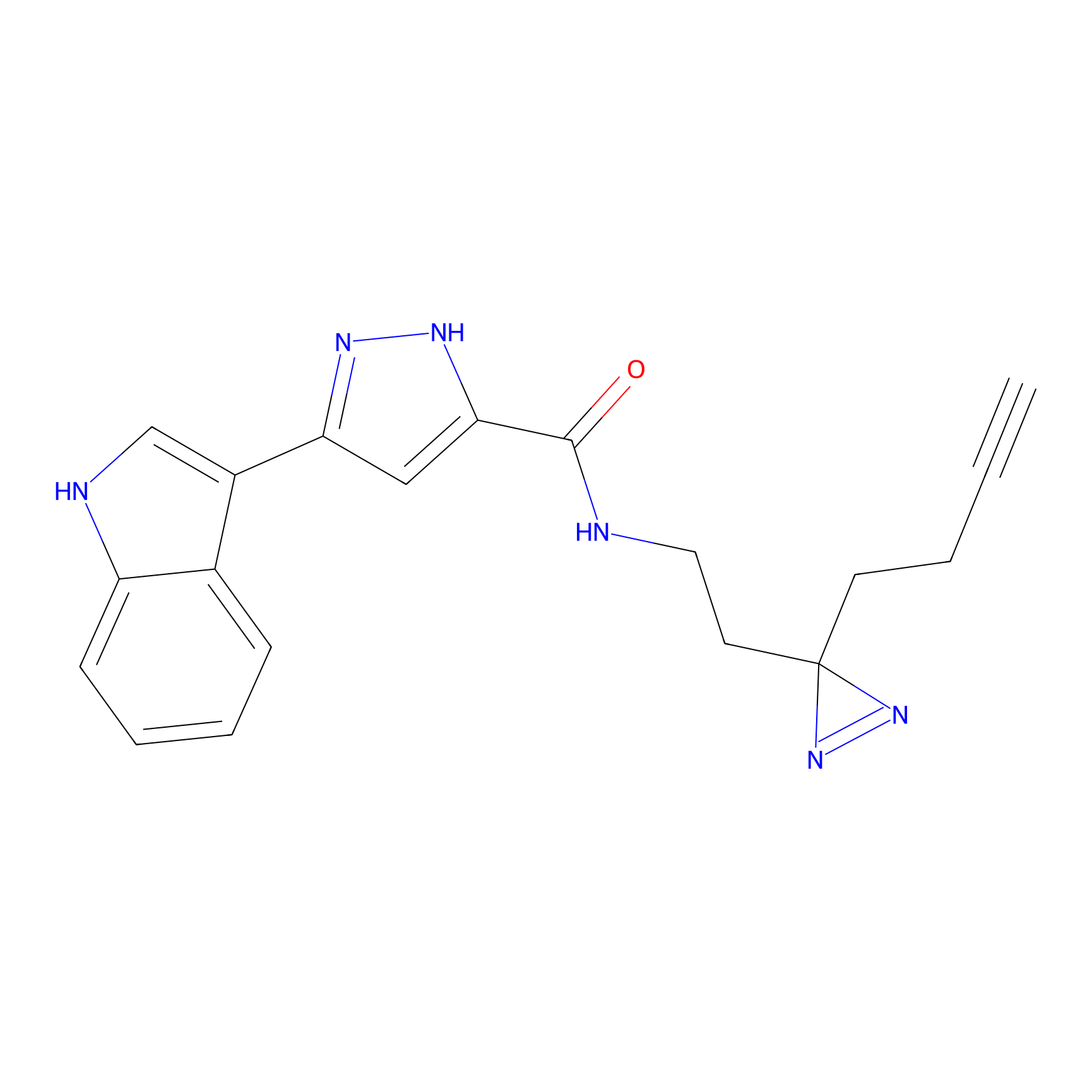
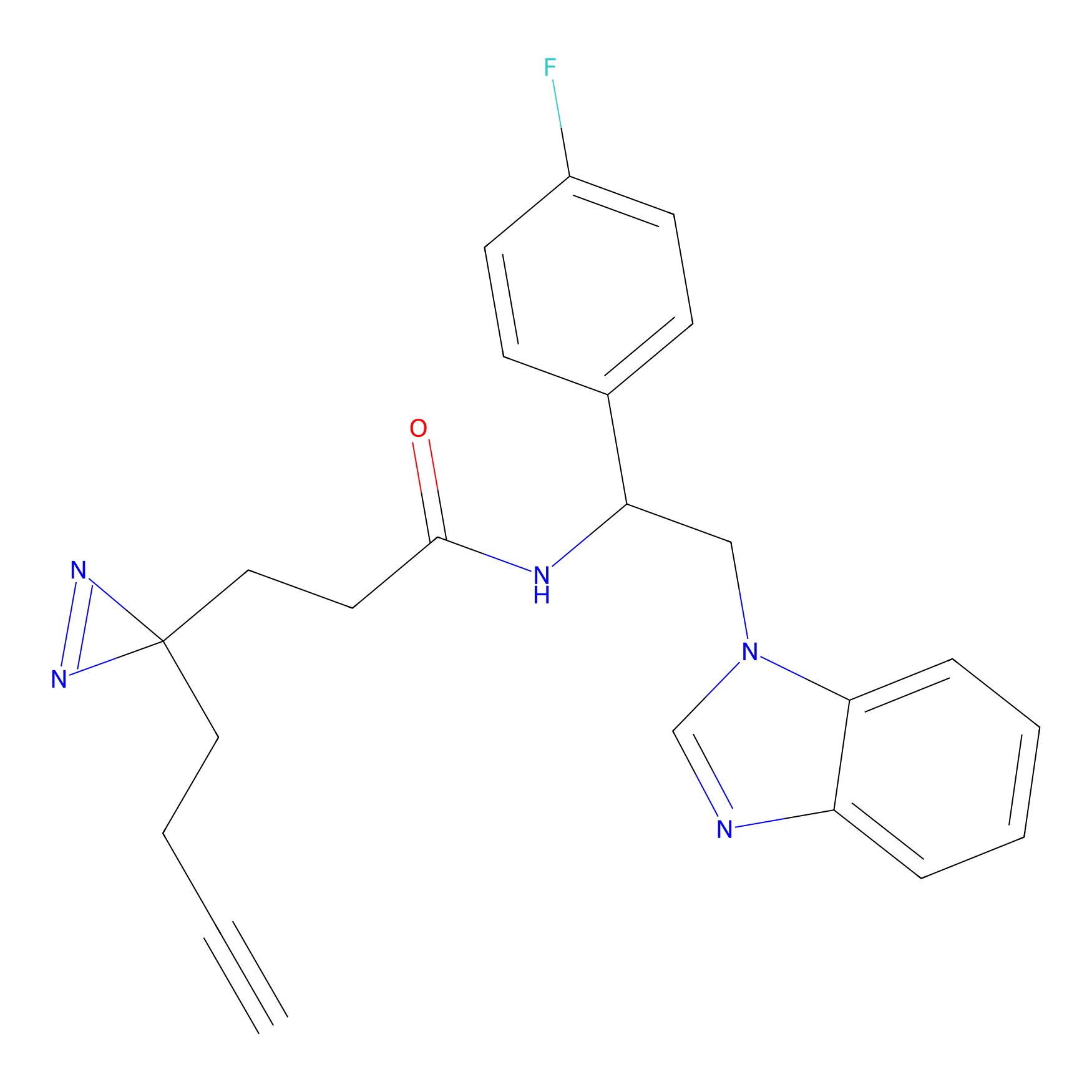
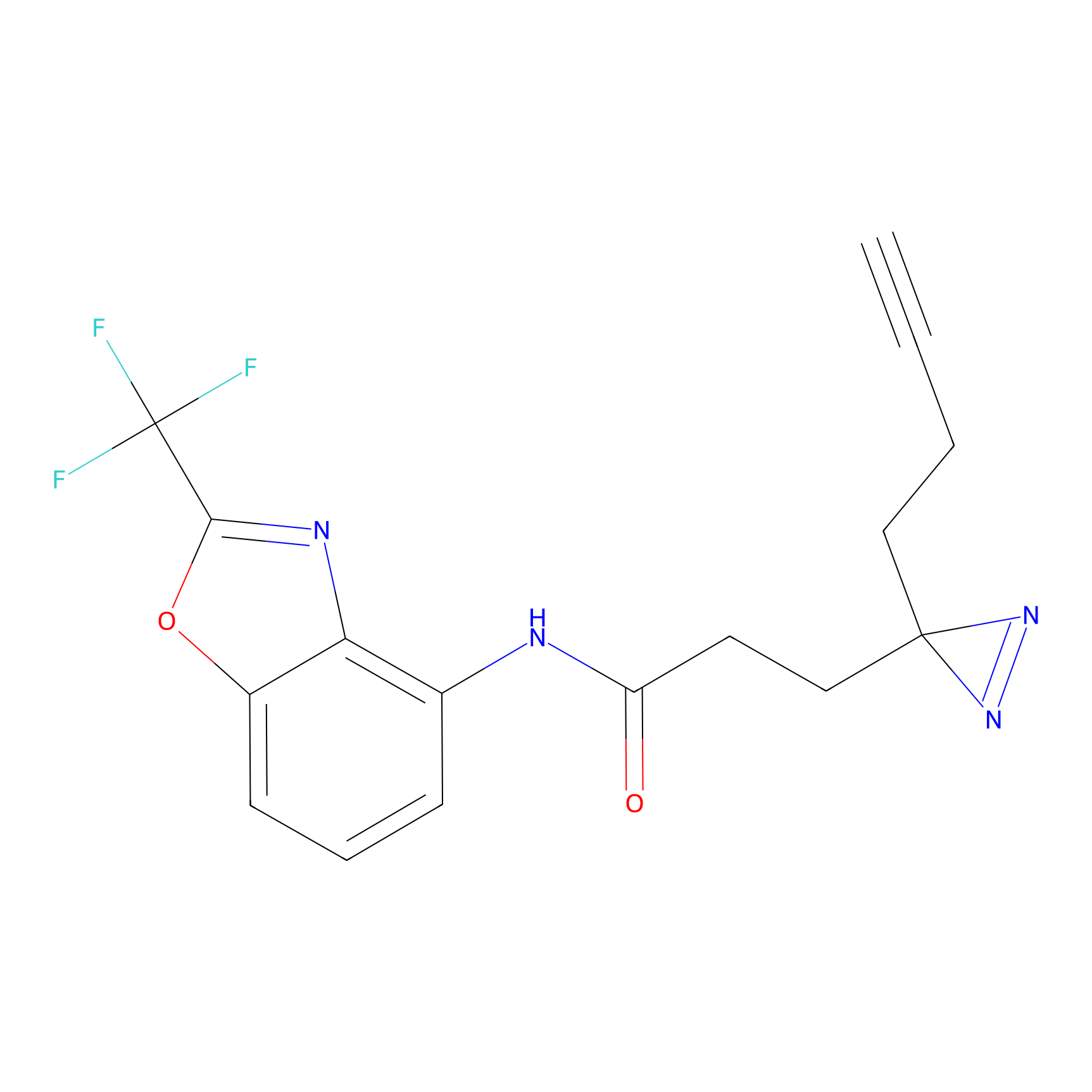
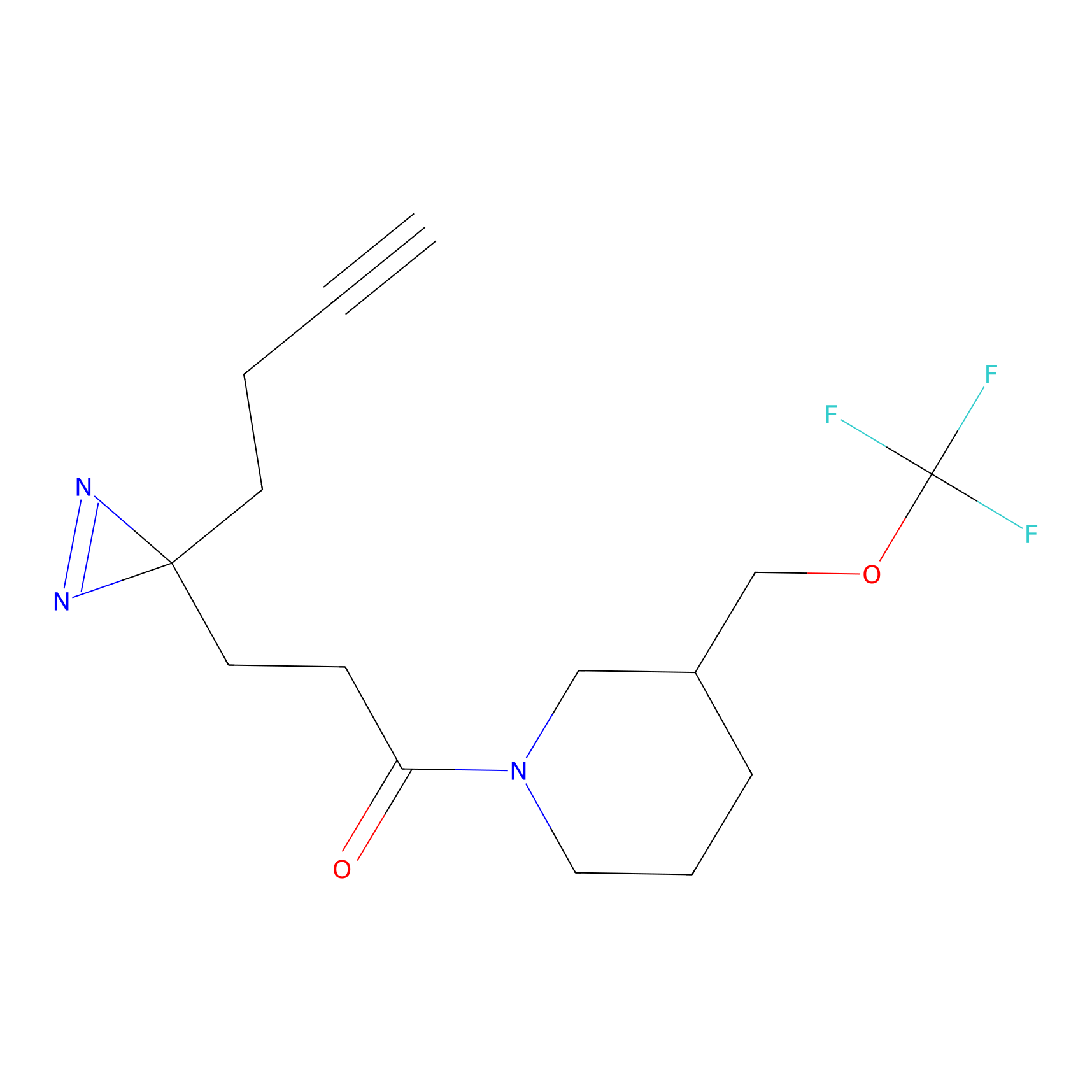
OPA-S-S-alkyne Probe Info |
 |
K76(4.44) | LDD3494 | [3] | |
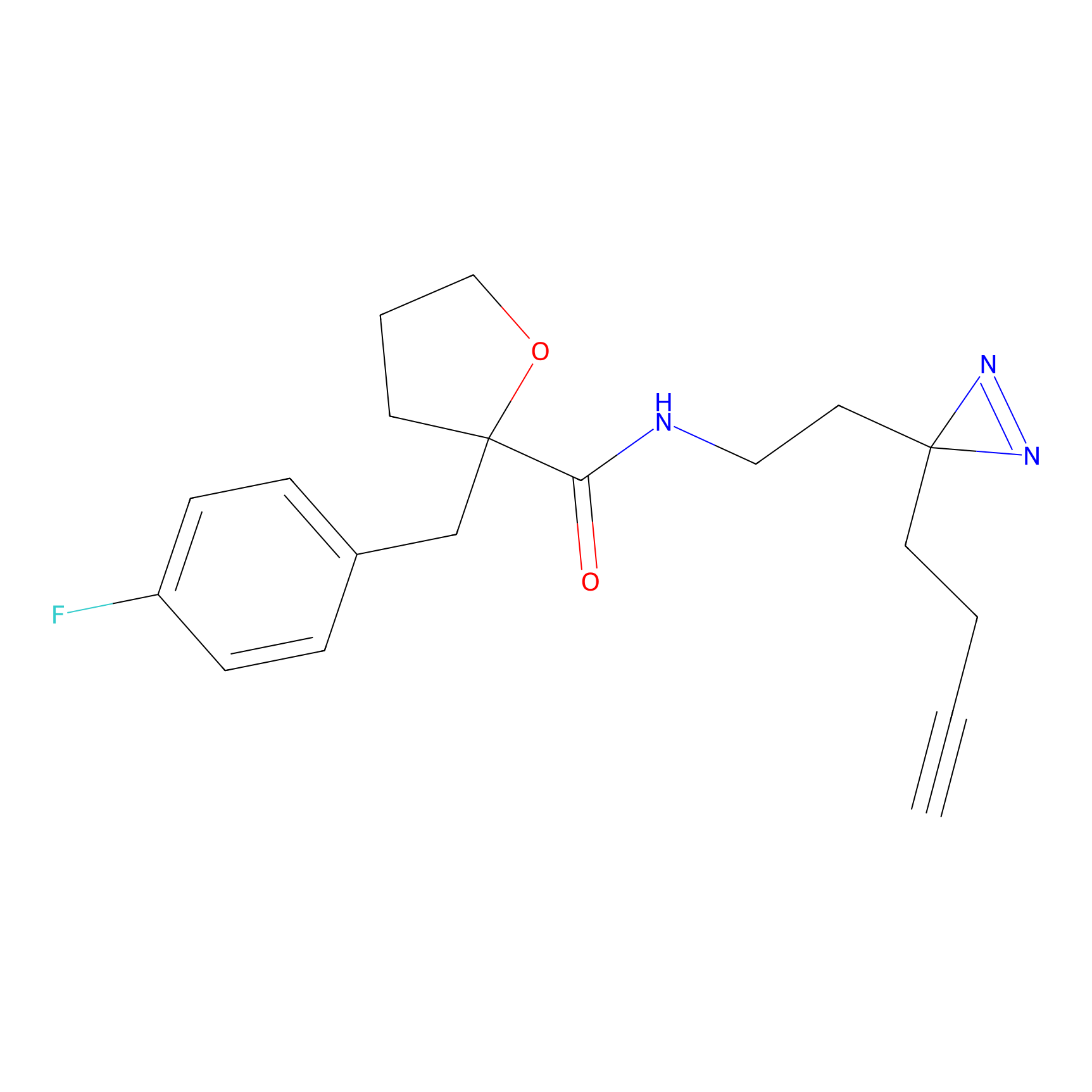
|
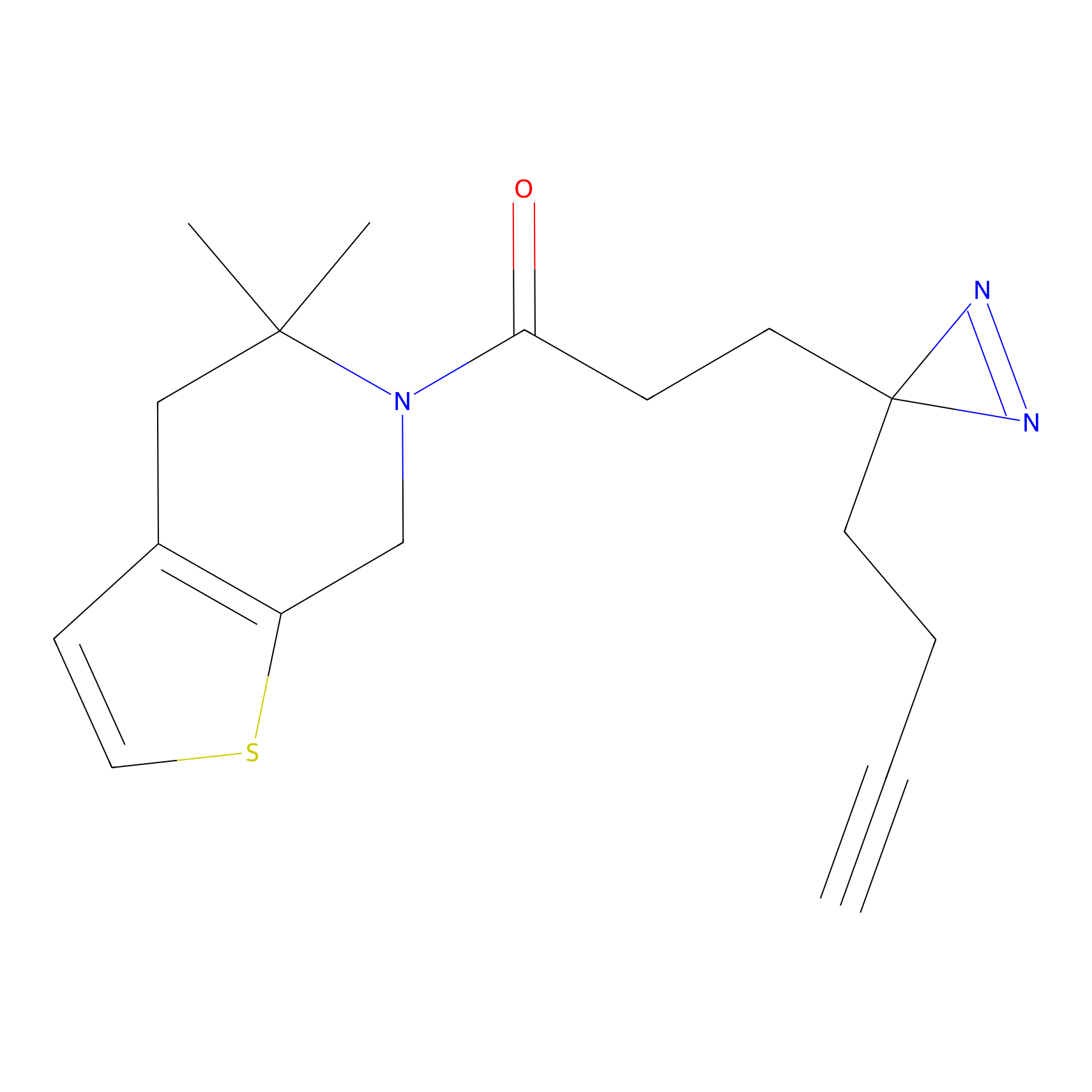
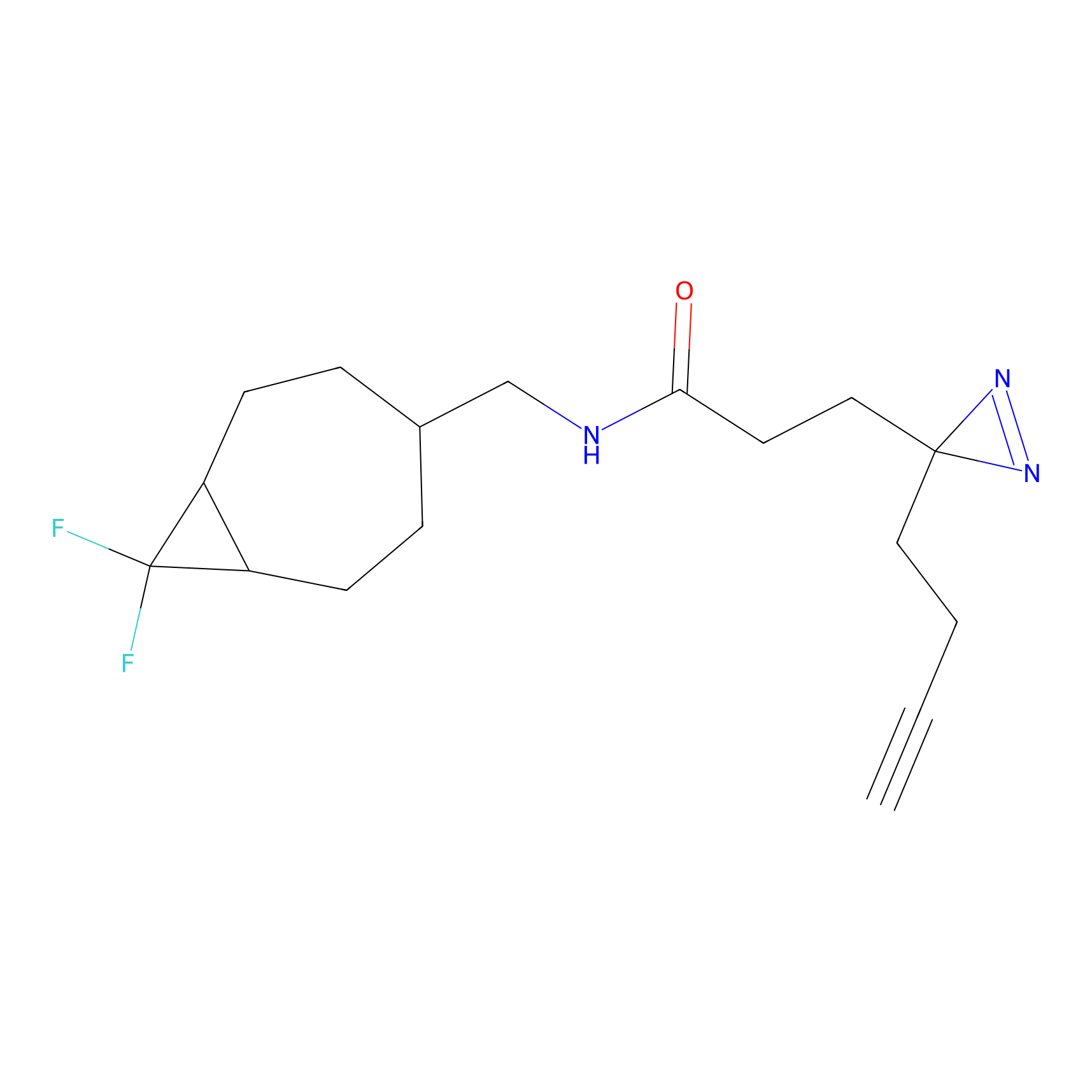
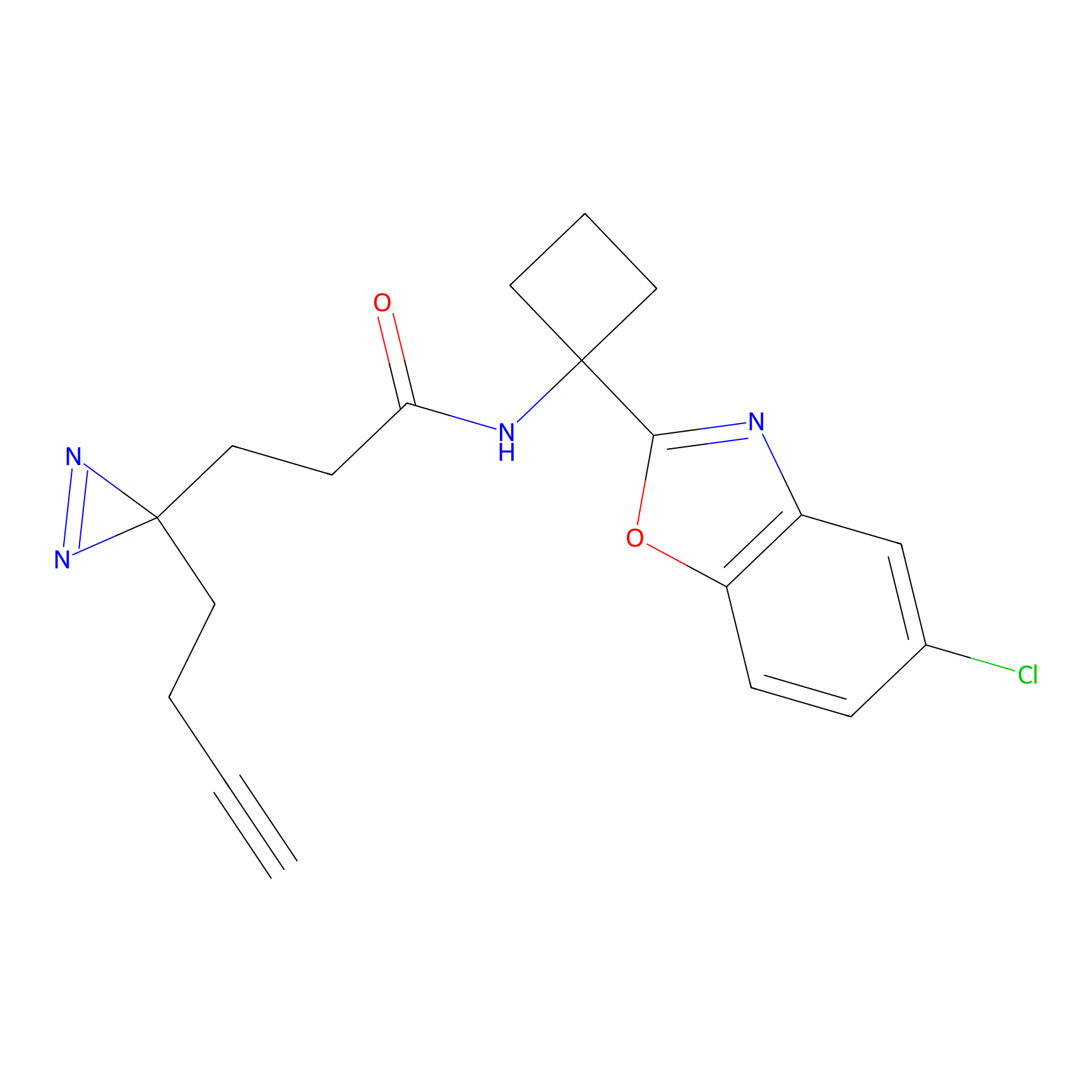
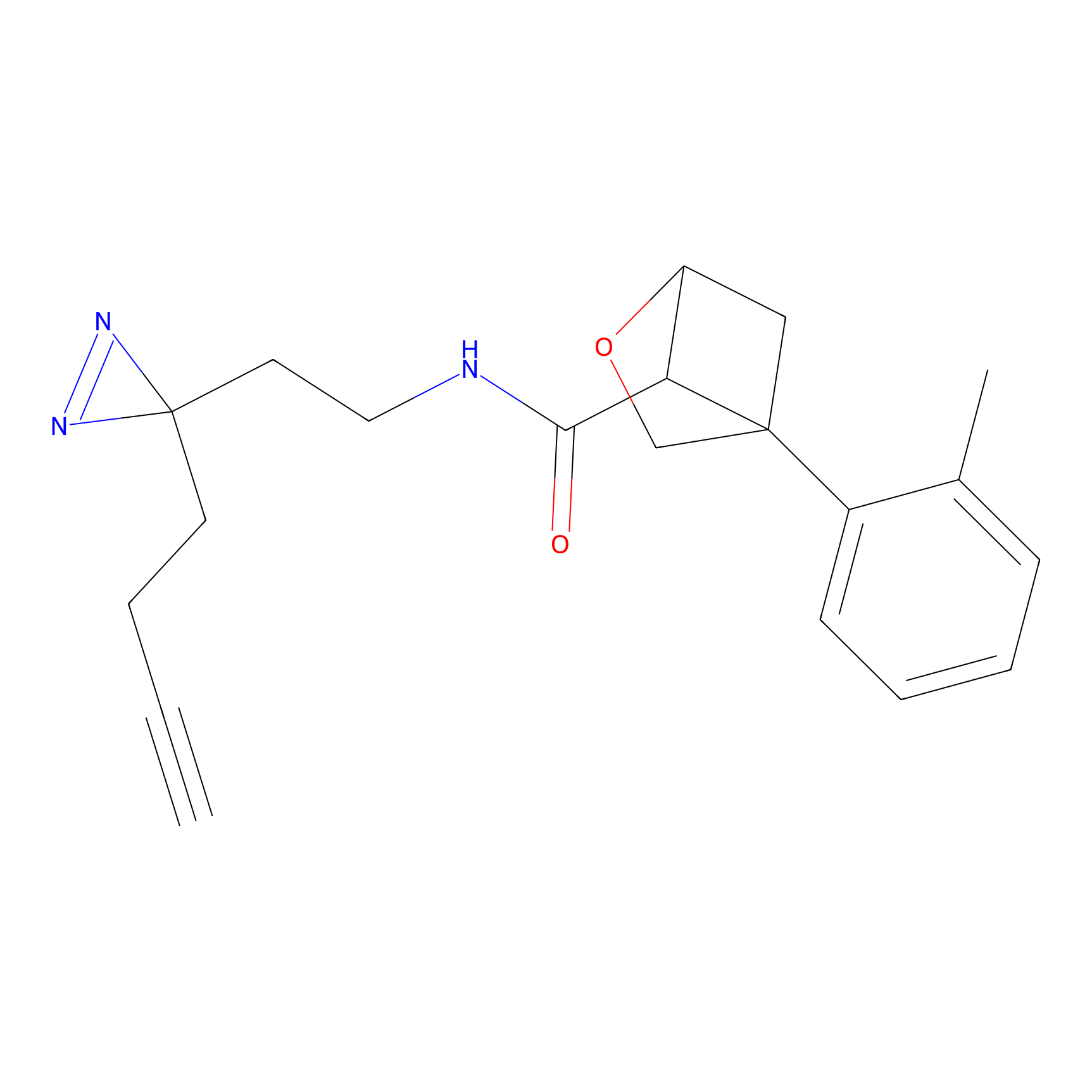
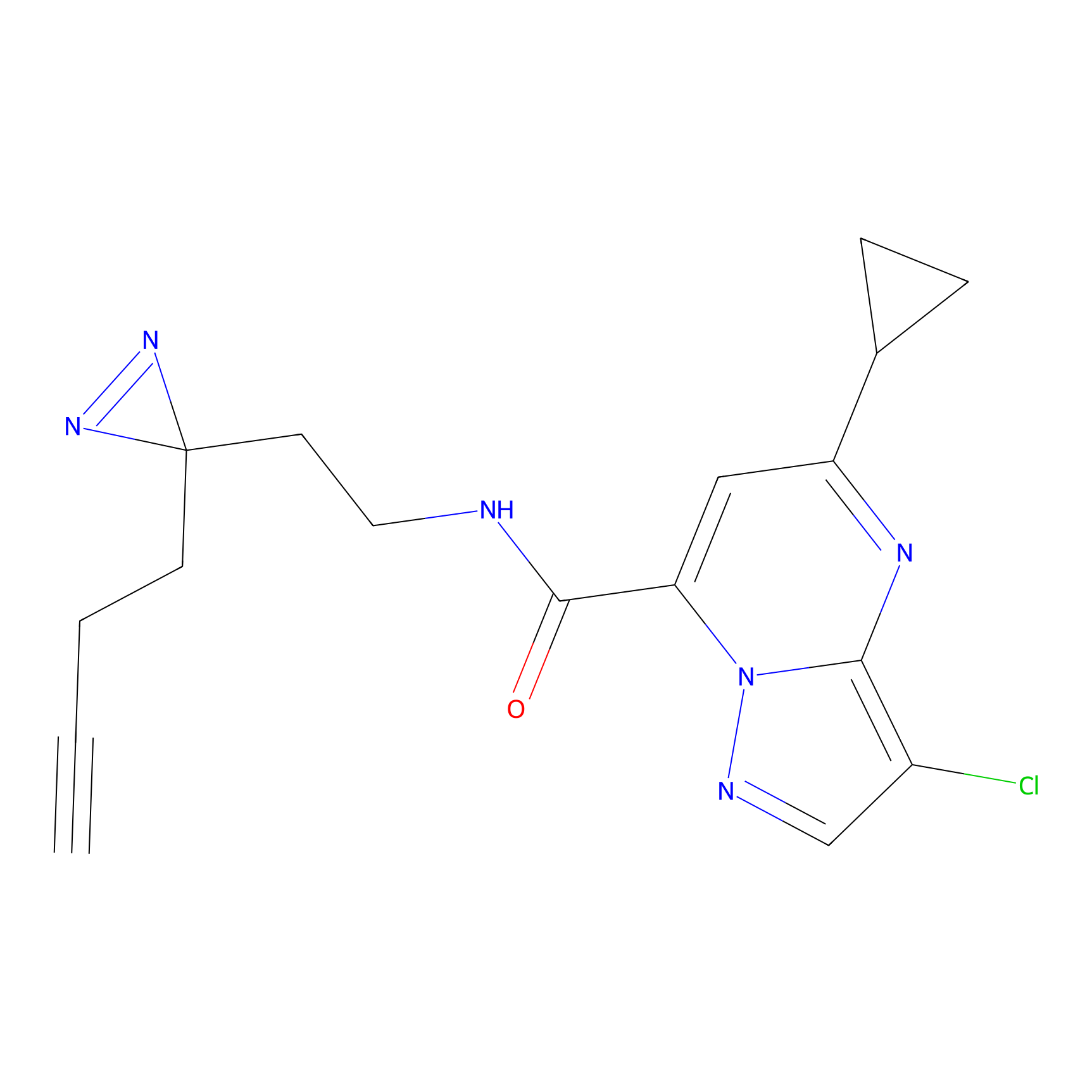
DBIA Probe Info |
 |
C235(2.69) | LDD3338 | [4] | |
|
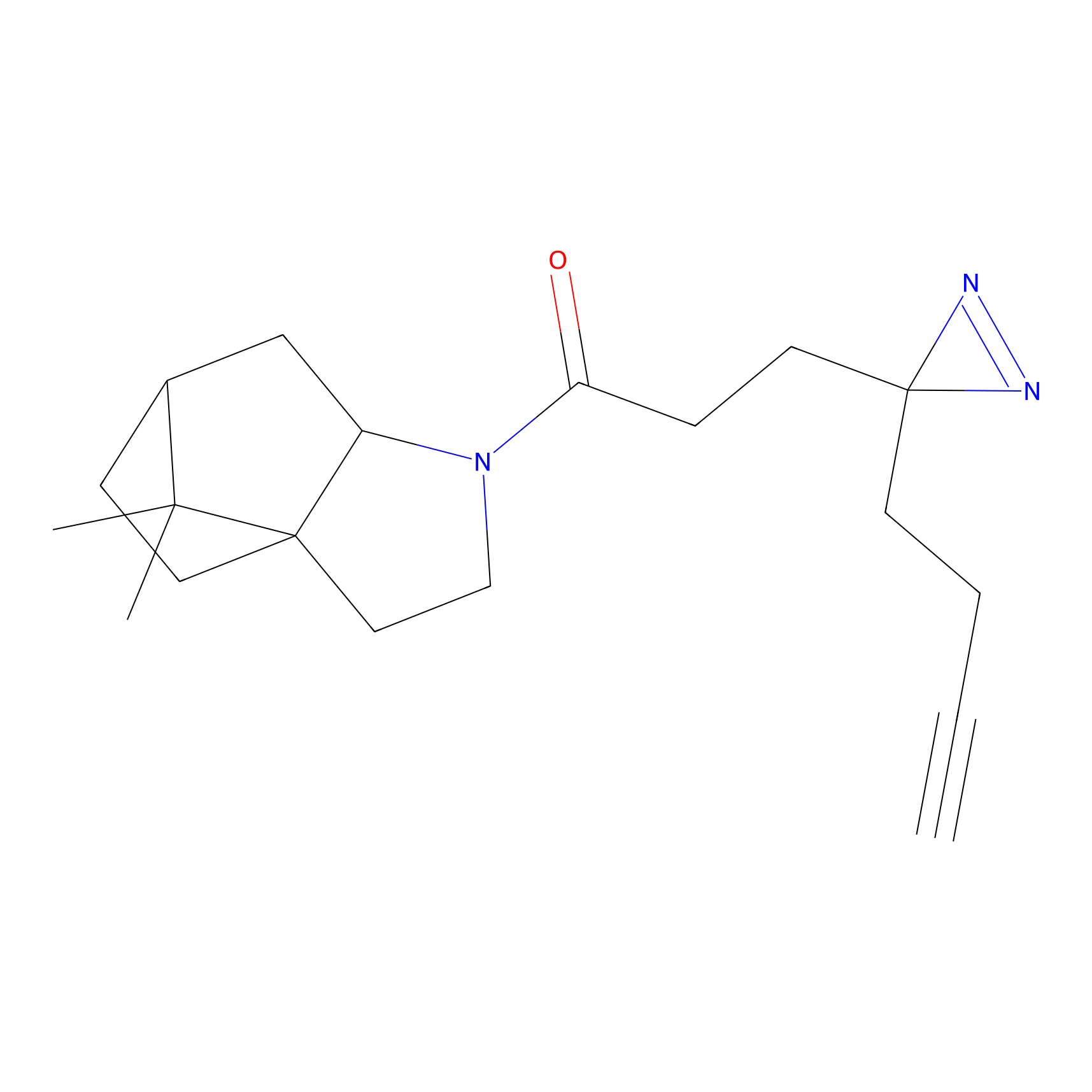
Jackson_14 Probe Info |
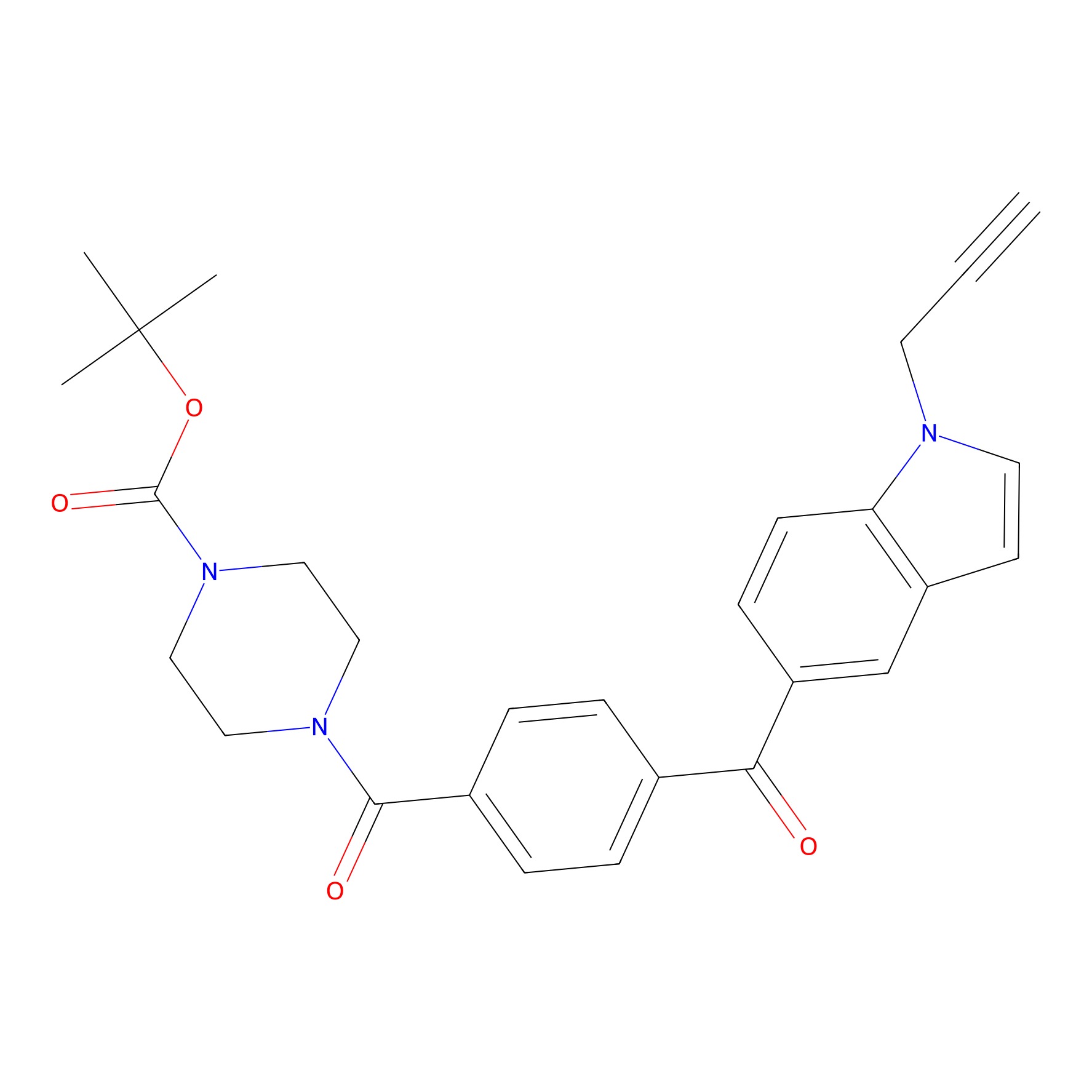 |
2.22 | LDD0123 | [5] | |
|
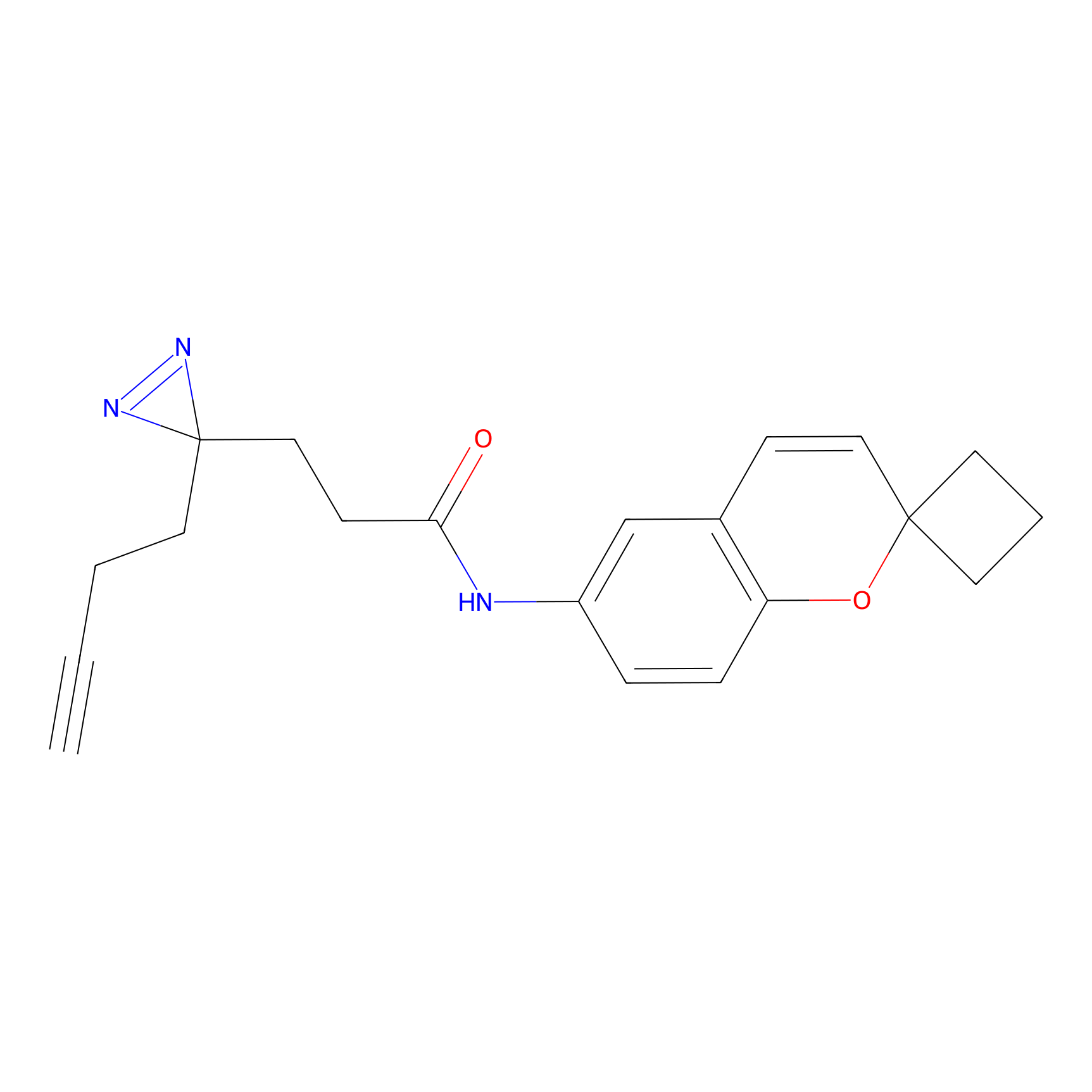
HPAP Probe Info |
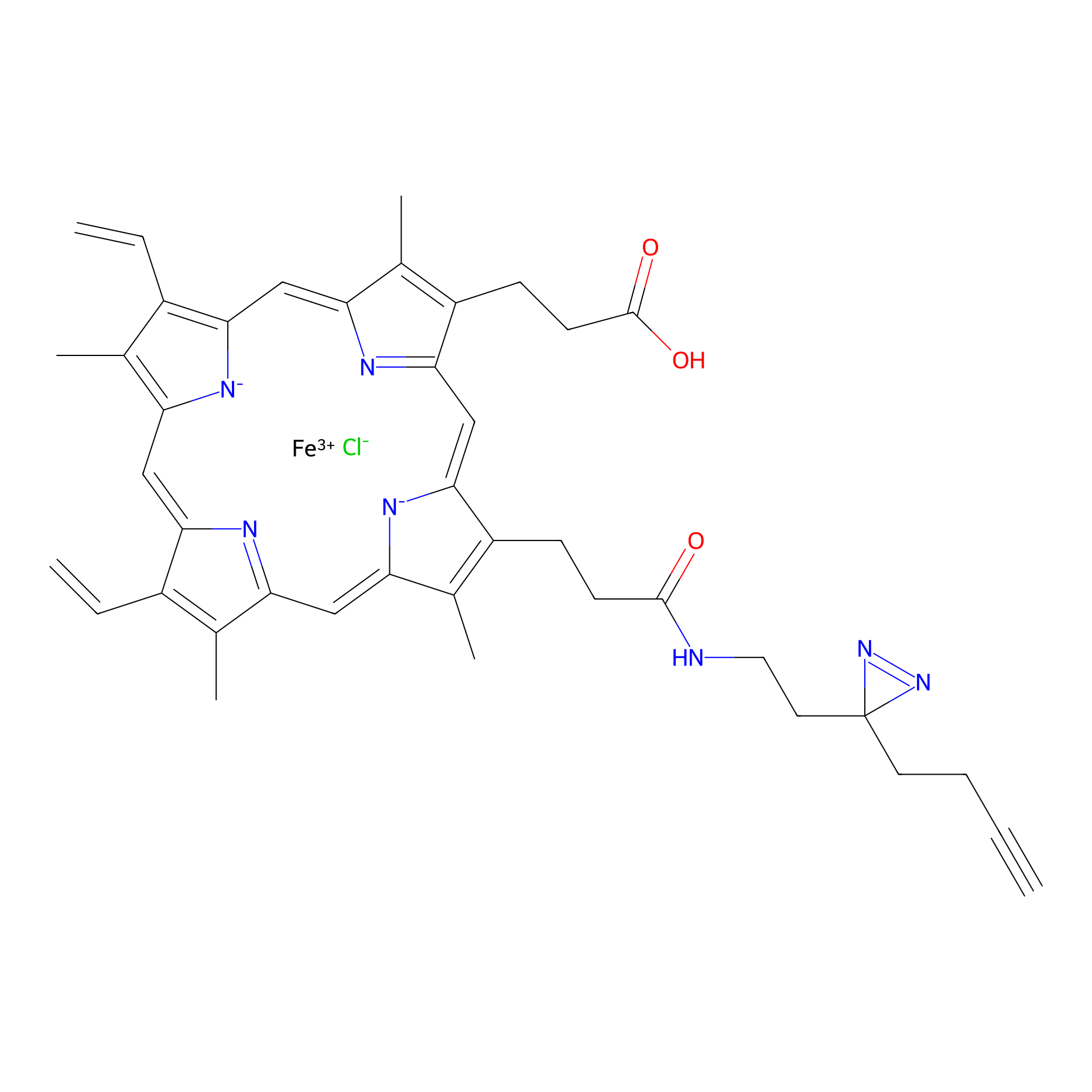 |
3.32 | LDD0062 | [6] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [7] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [8] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [7] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [9] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [10] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0022 | KB02 | CDX: HA1; Afatinib resistant | C235(1.22) | LDD2346 | [4] |
| LDCM0023 | KB03 | CDX: HA1; Afatinib resistant | C235(1.77) | LDD2763 | [4] |
| LDCM0024 | KB05 | MV4-11 | C235(2.69) | LDD3338 | [4] |
| LDCM0014 | Panhematin | HEK-293T | 3.32 | LDD0062 | [6] |
| LDCM0016 | Ranjitkar_cp1 | MDA-MB-231 | 2.22 | LDD0123 | [5] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Tyrosine--tRNA ligase, cytoplasmic (YARS1) | Class-I aminoacyl-tRNA synthetase family | P54577 | |||
Transporter and channel
Other
References