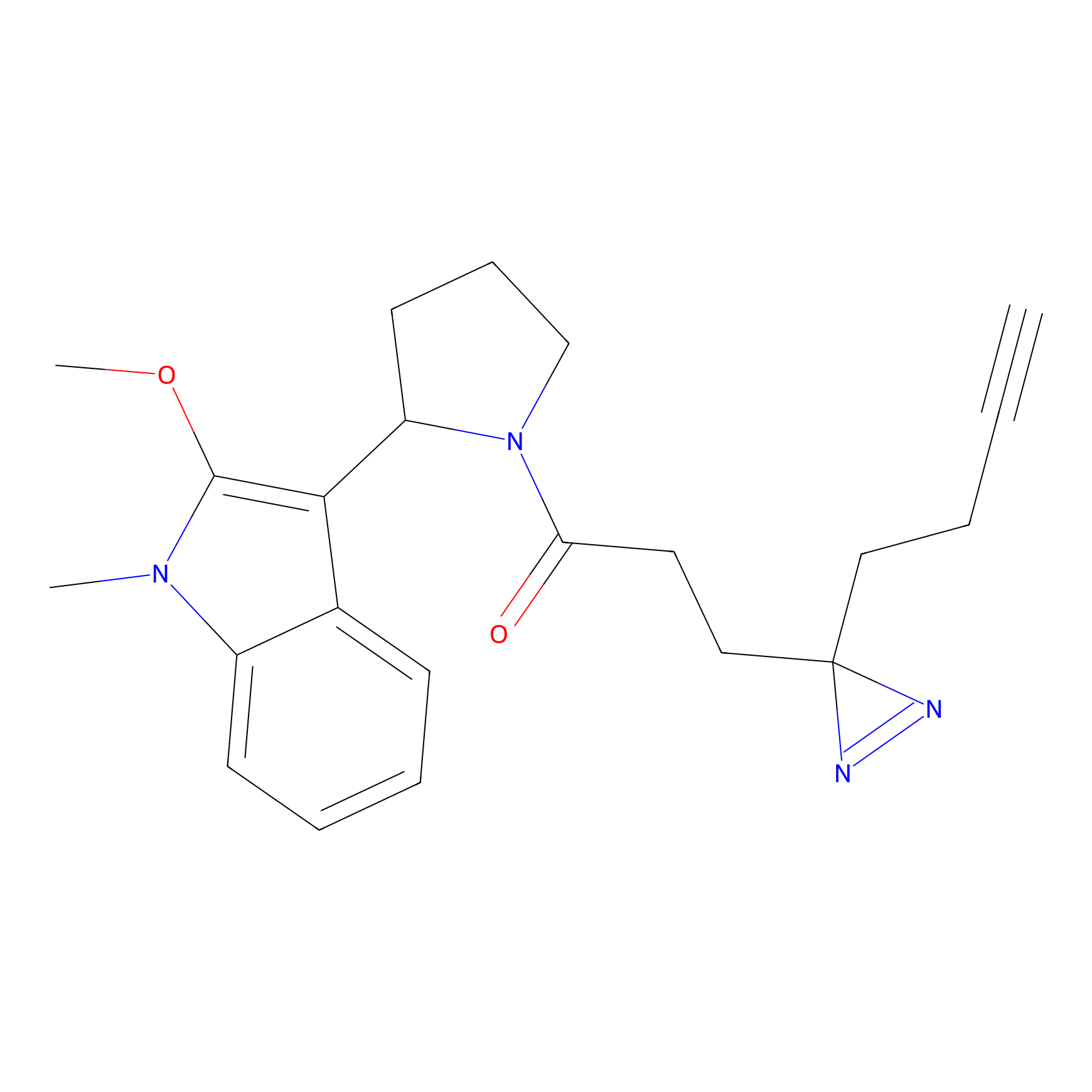
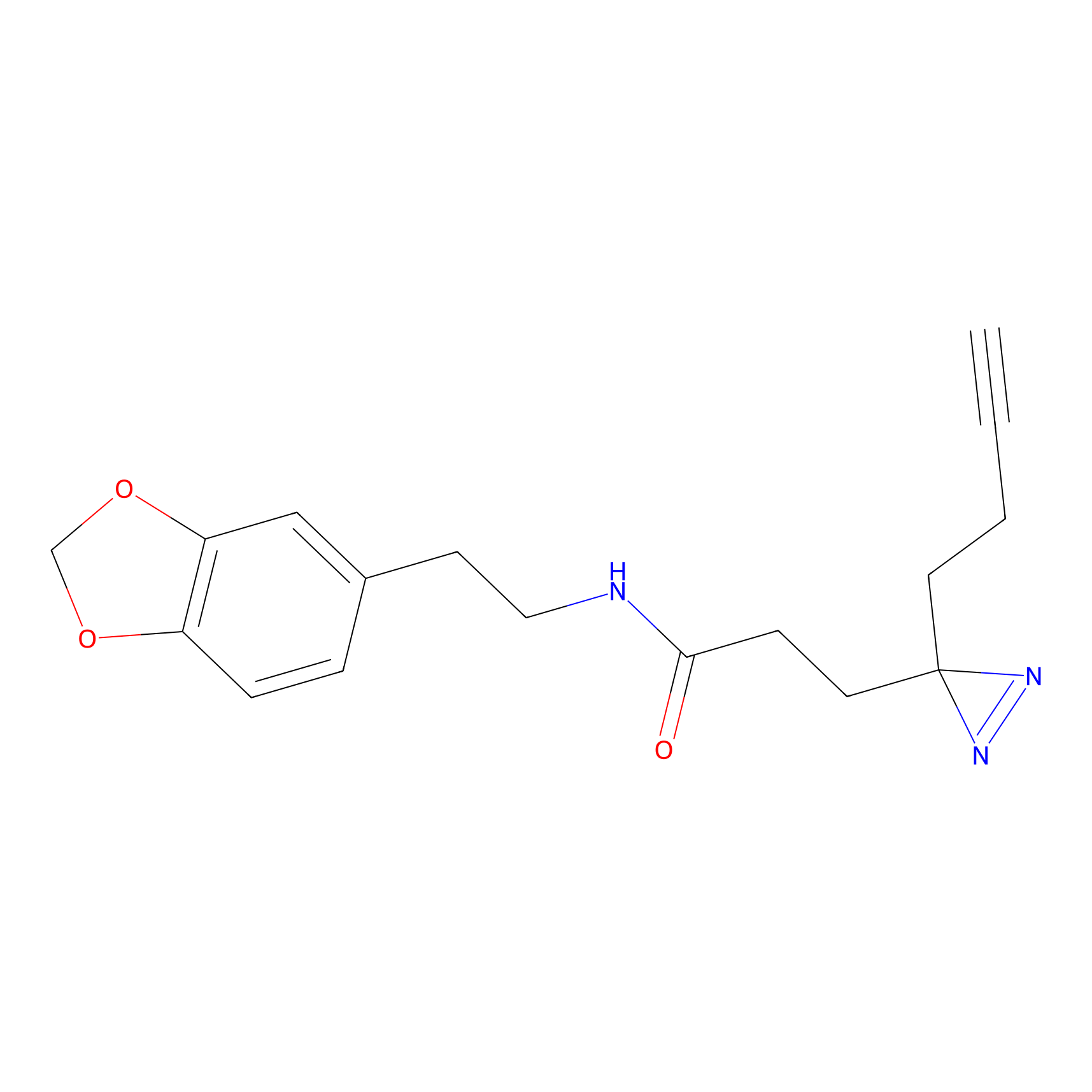
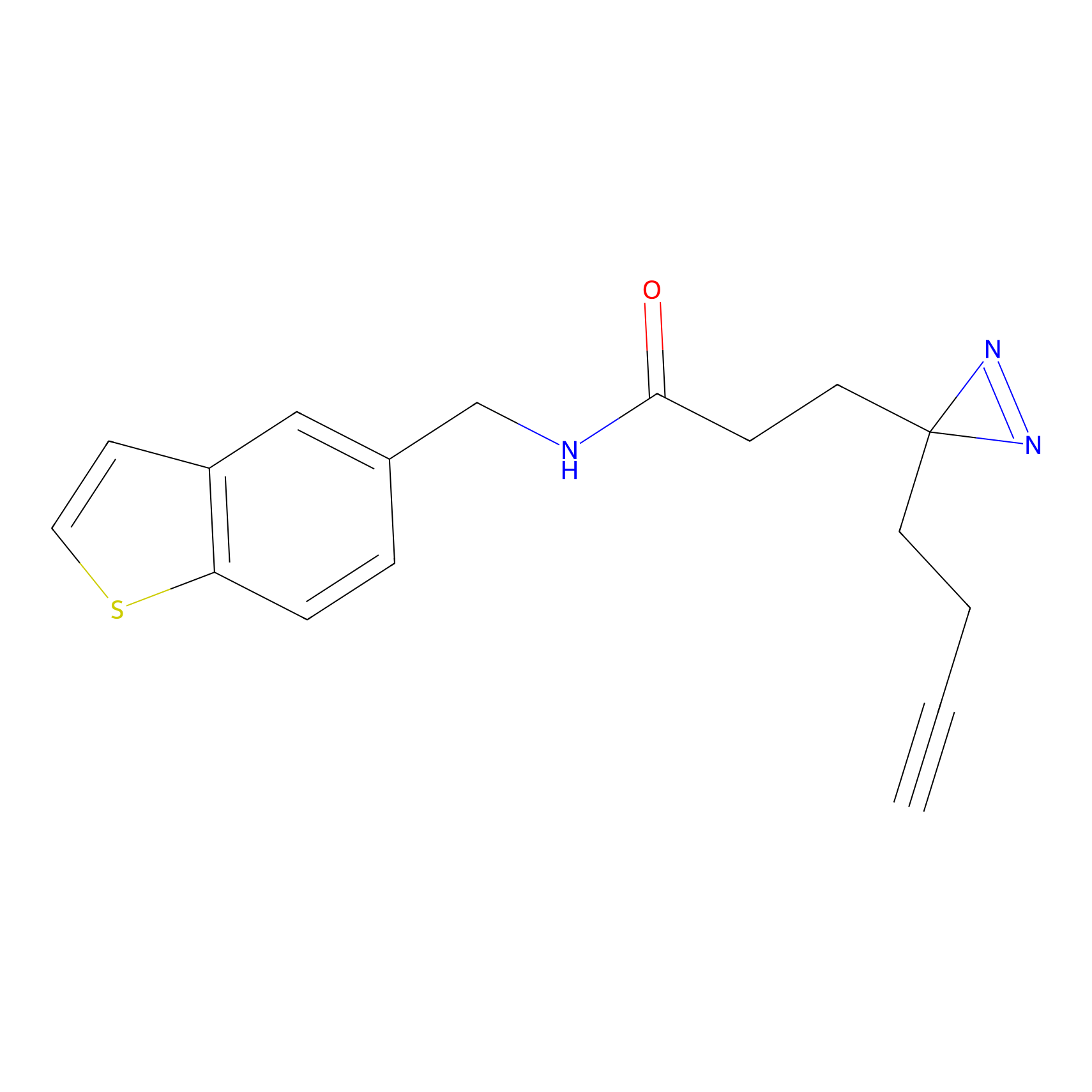
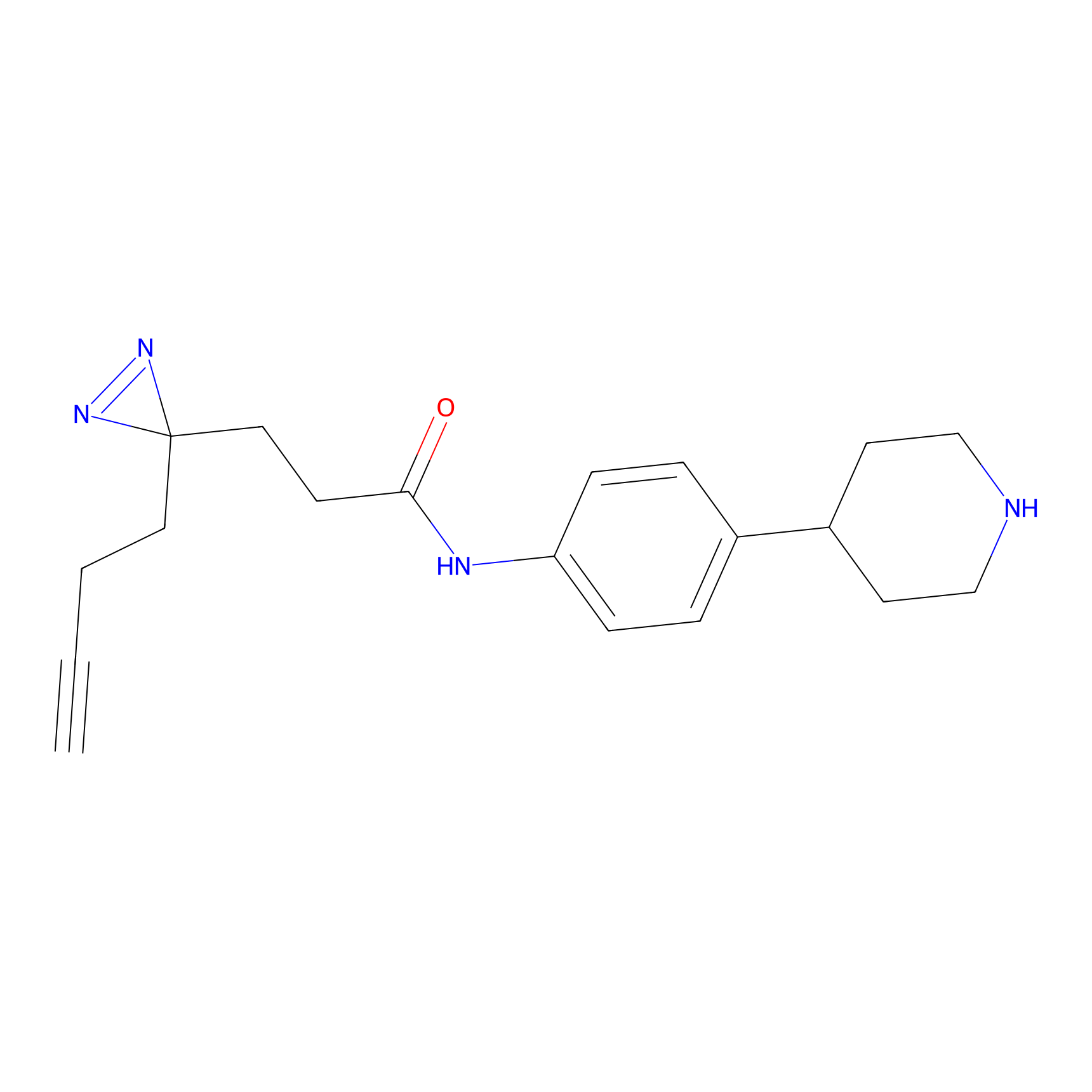
Details of the Target
General Information of Target
| Target ID | LDTP12954 | |||||
|---|---|---|---|---|---|---|
| Target Name | Very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase 3 (HACD3) | |||||
| Gene Name | HACD3 | |||||
| Gene ID | 51495 | |||||
| Synonyms |
BIND1; PTPLAD1; Very-long-chain; 3R)-3-hydroxyacyl-CoA dehydratase 3; EC 4.2.1.134; 3-hydroxyacyl-CoA dehydratase 3; HACD3; Butyrate-induced protein 1; B-ind1; hB-ind1; Protein-tyrosine phosphatase-like A domain-containing protein 1
|
|||||
| 3D Structure | ||||||
| Sequence |
MGALVIRGIRNFNLENRAEREISKMKPSVAPRHPSTNSLLREQISLYPEVKGEIARKDEK
LLSFLKDVYVDSKDPVSSLQVKAAETCQEPKEFRLPKDHHFDMINIKSIPKGKISIVEAL TLLNNHKLFPETWTAEKIMQEYQLEQKDVNSLLKYFVTFEVEIFPPEDKKAIRSK |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Very long-chain fatty acids dehydratase HACD family
|
|||||
| Subcellular location |
Endoplasmic reticulum membrane
|
|||||
| Function |
Catalyzes the third of the four reactions of the long-chain fatty acids elongation cycle. This endoplasmic reticulum-bound enzymatic process, allows the addition of two carbons to the chain of long- and very long-chain fatty acids/VLCFAs per cycle. This enzyme catalyzes the dehydration of the 3-hydroxyacyl-CoA intermediate into trans-2,3-enoyl-CoA, within each cycle of fatty acid elongation. Thereby, it participates in the production of VLCFAs of different chain lengths that are involved in multiple biological processes as precursors of membrane lipids and lipid mediators. May be involved in Rac1-signaling pathways leading to the modulation of gene expression. Promotes insulin receptor/INSR autophosphorylation and is involved in INSR internalization.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
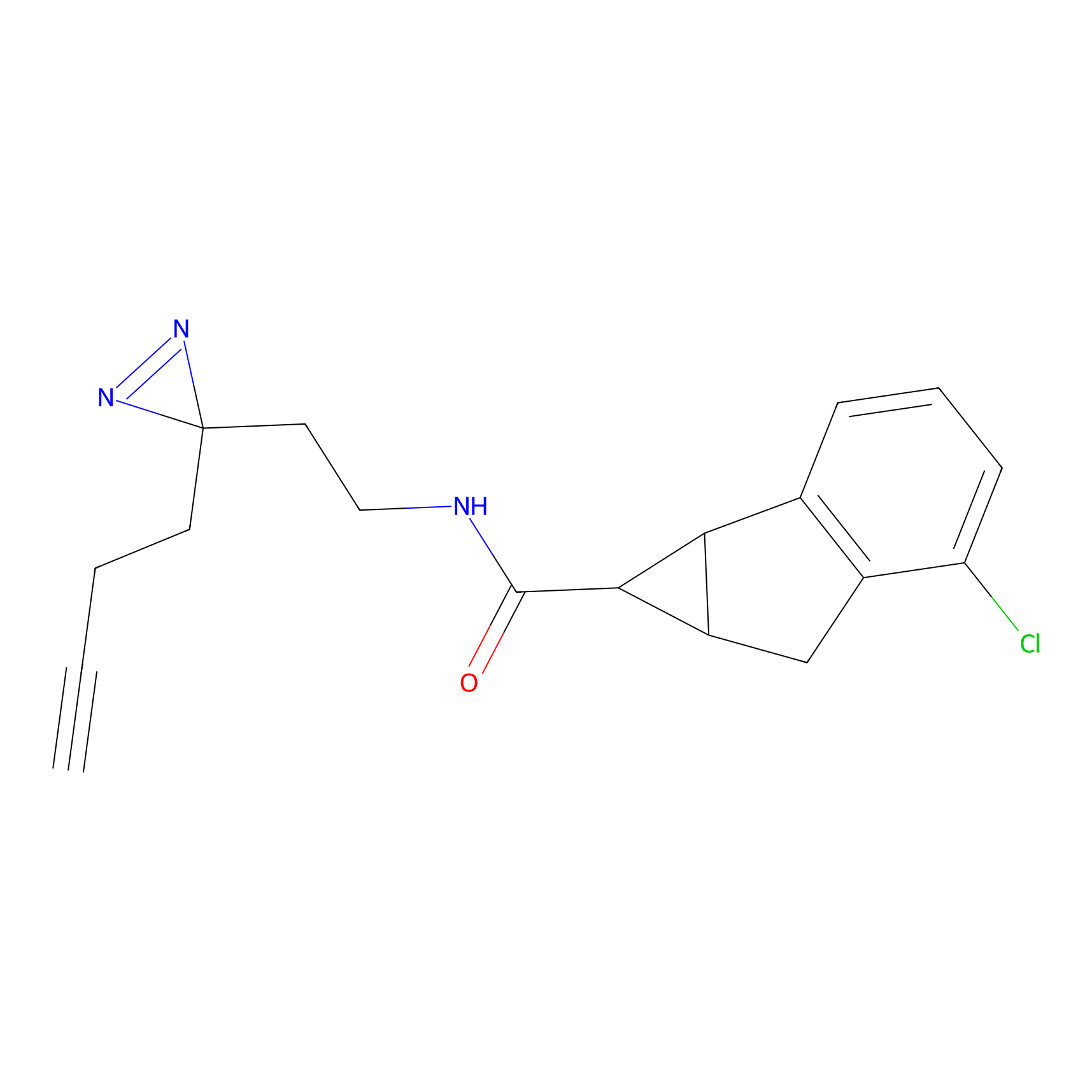
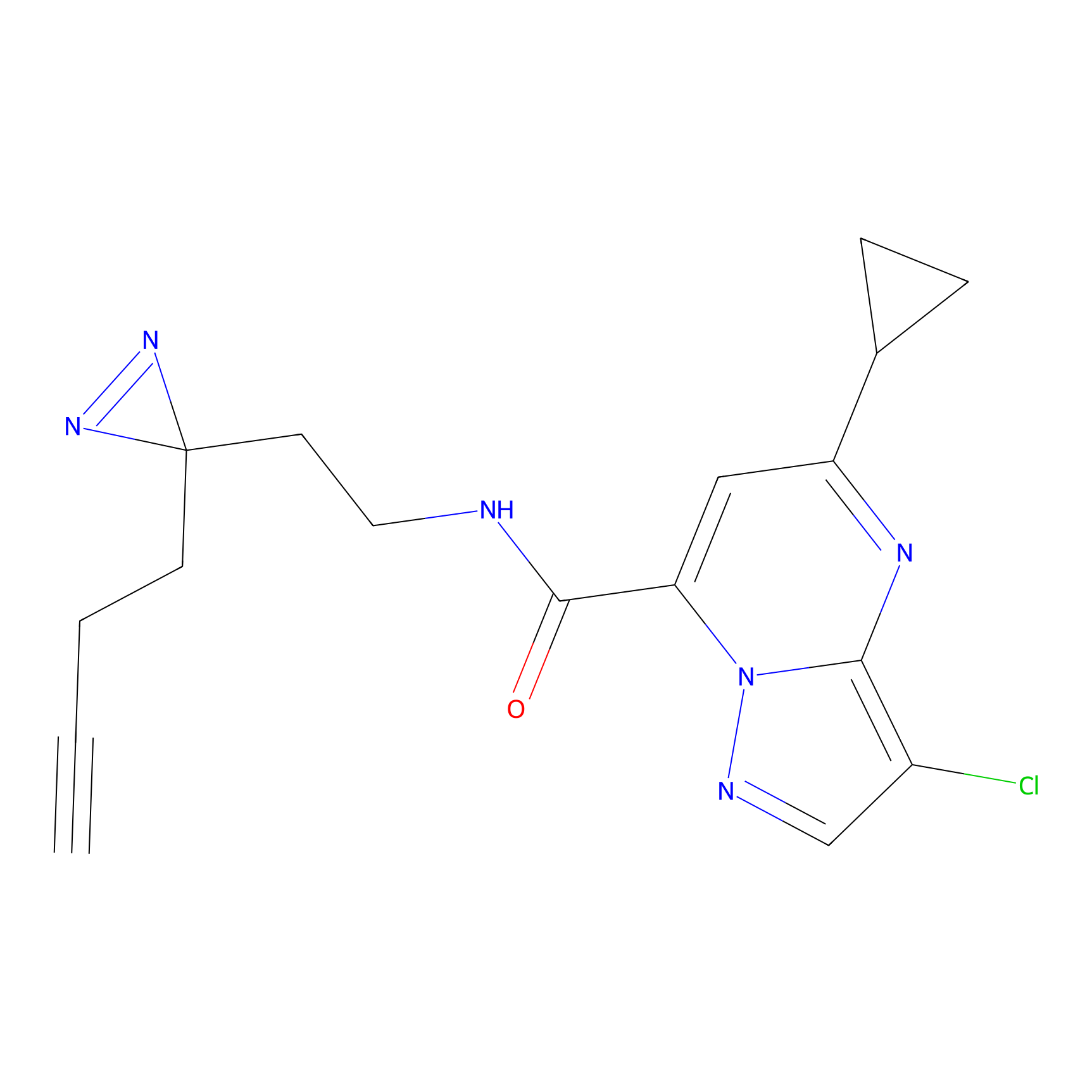
7.45 | LDD0402 | [1] | |
|
HDSF-alk Probe Info |
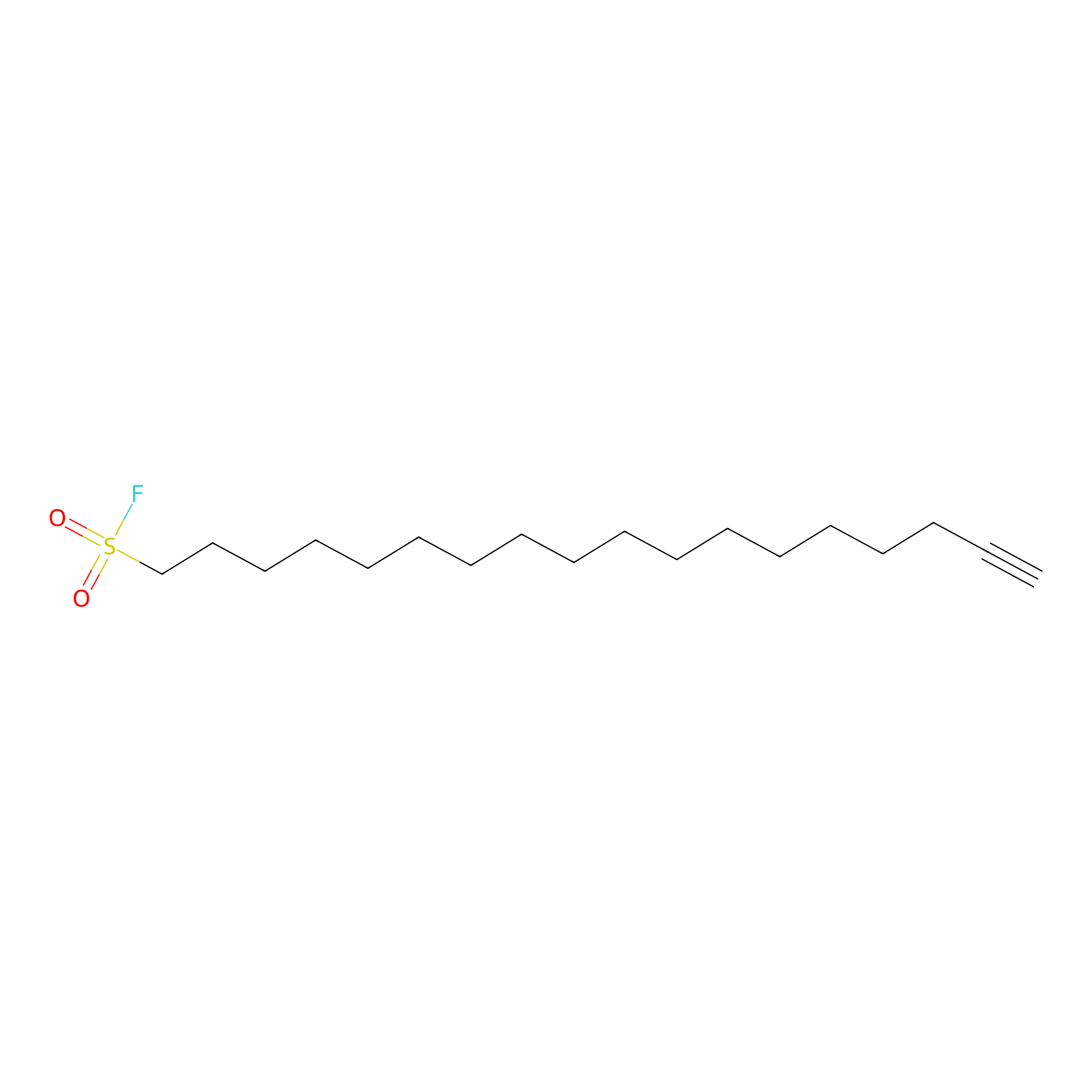 |
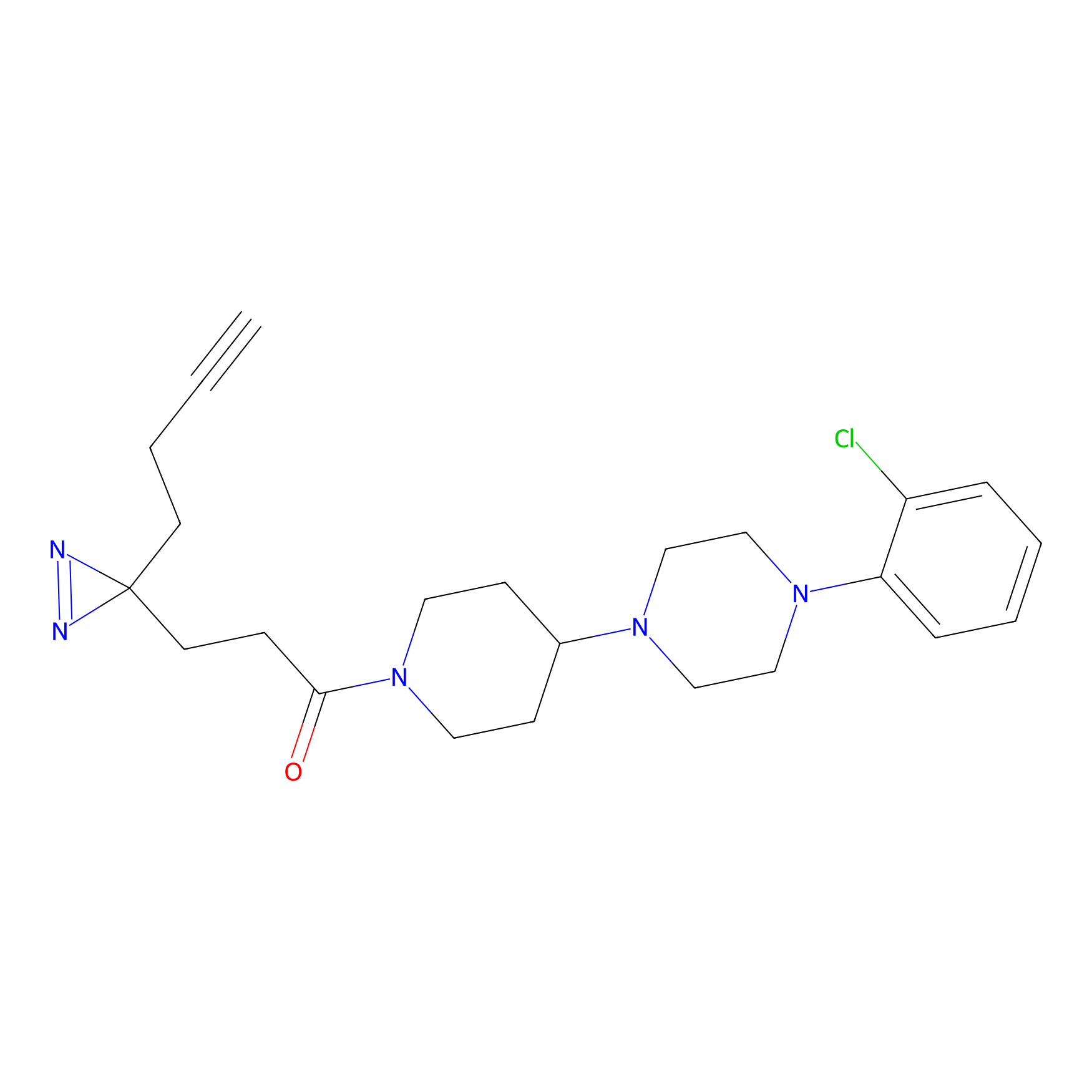
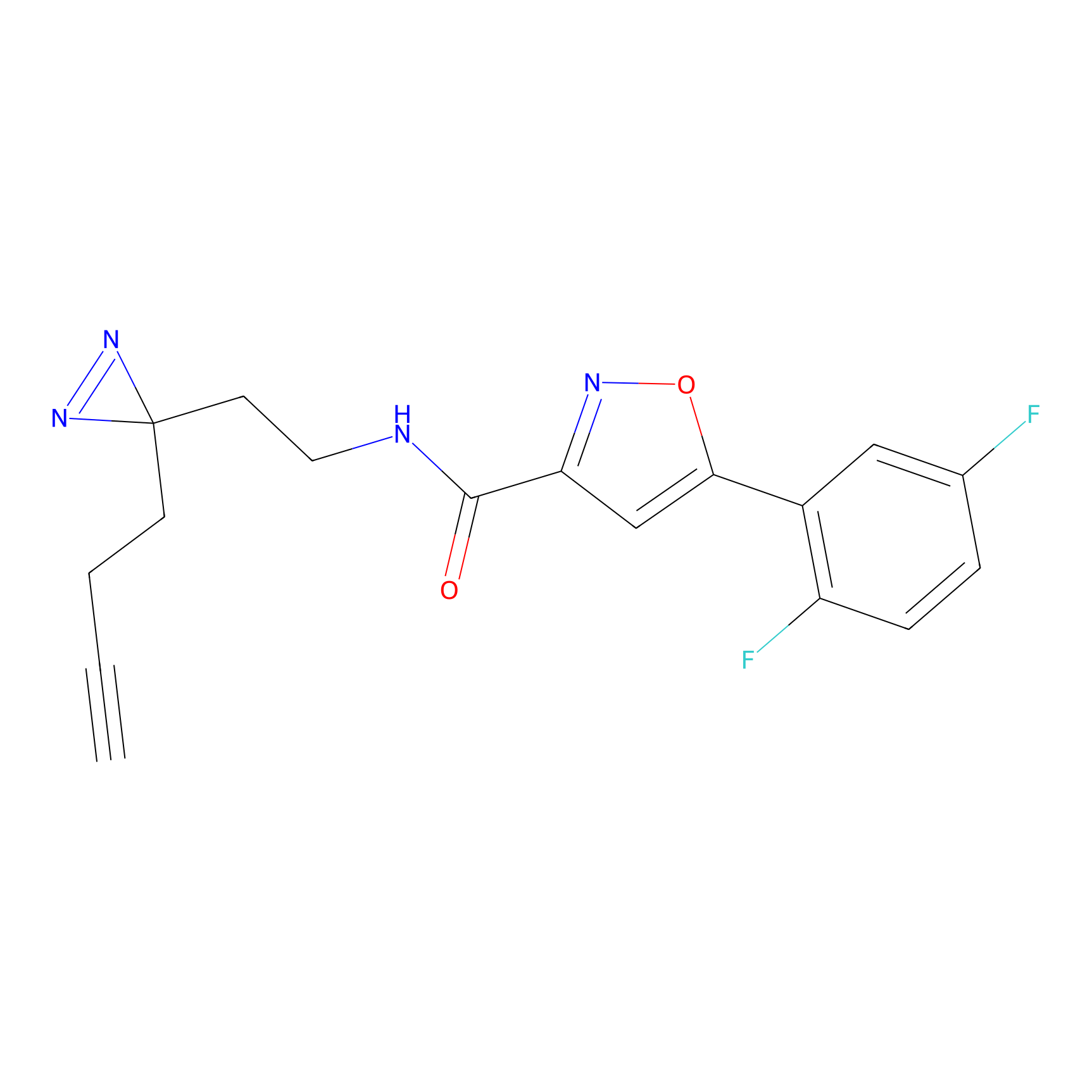
1.76 | LDD0197 | [2] | |
|
CHEMBL5175495 Probe Info |
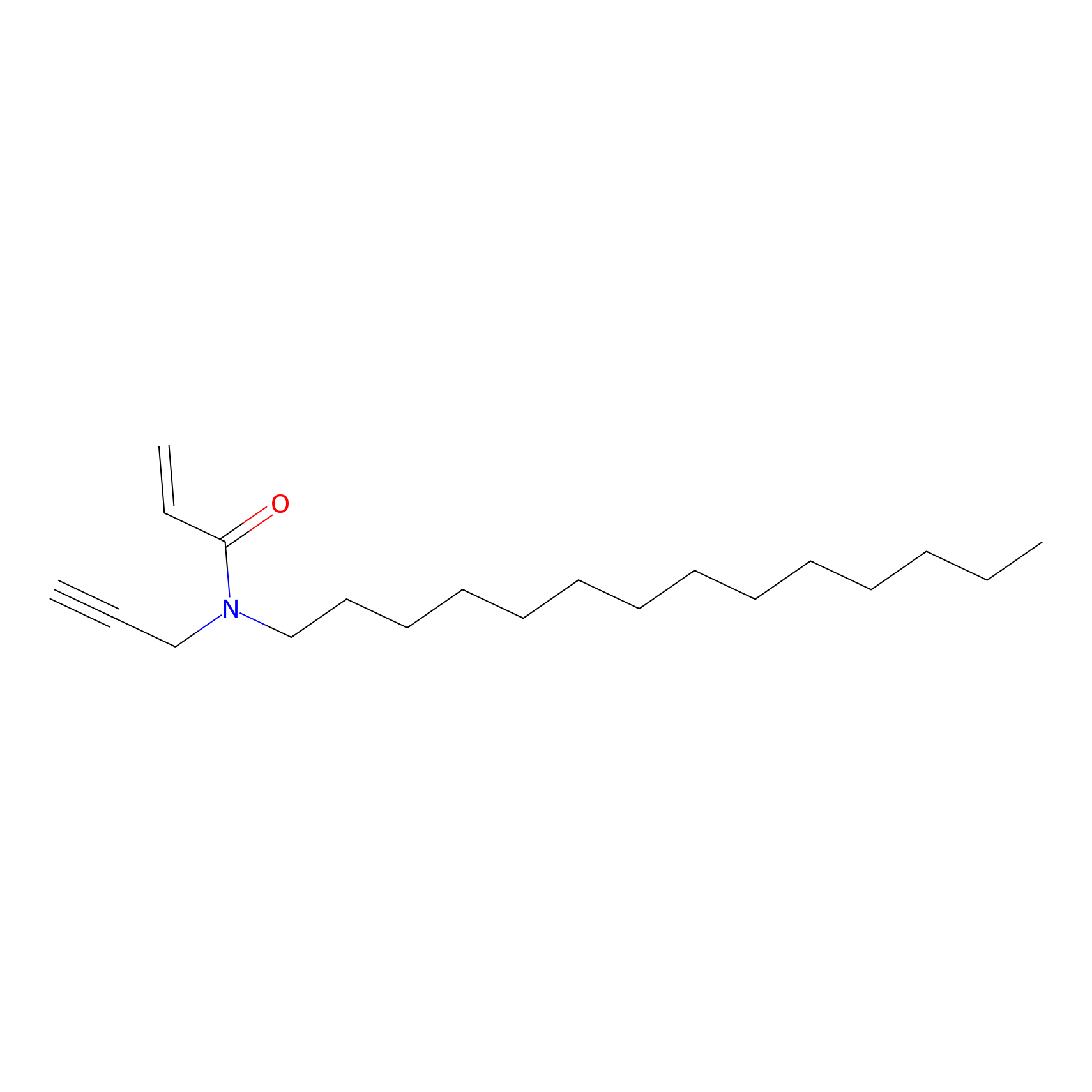 |
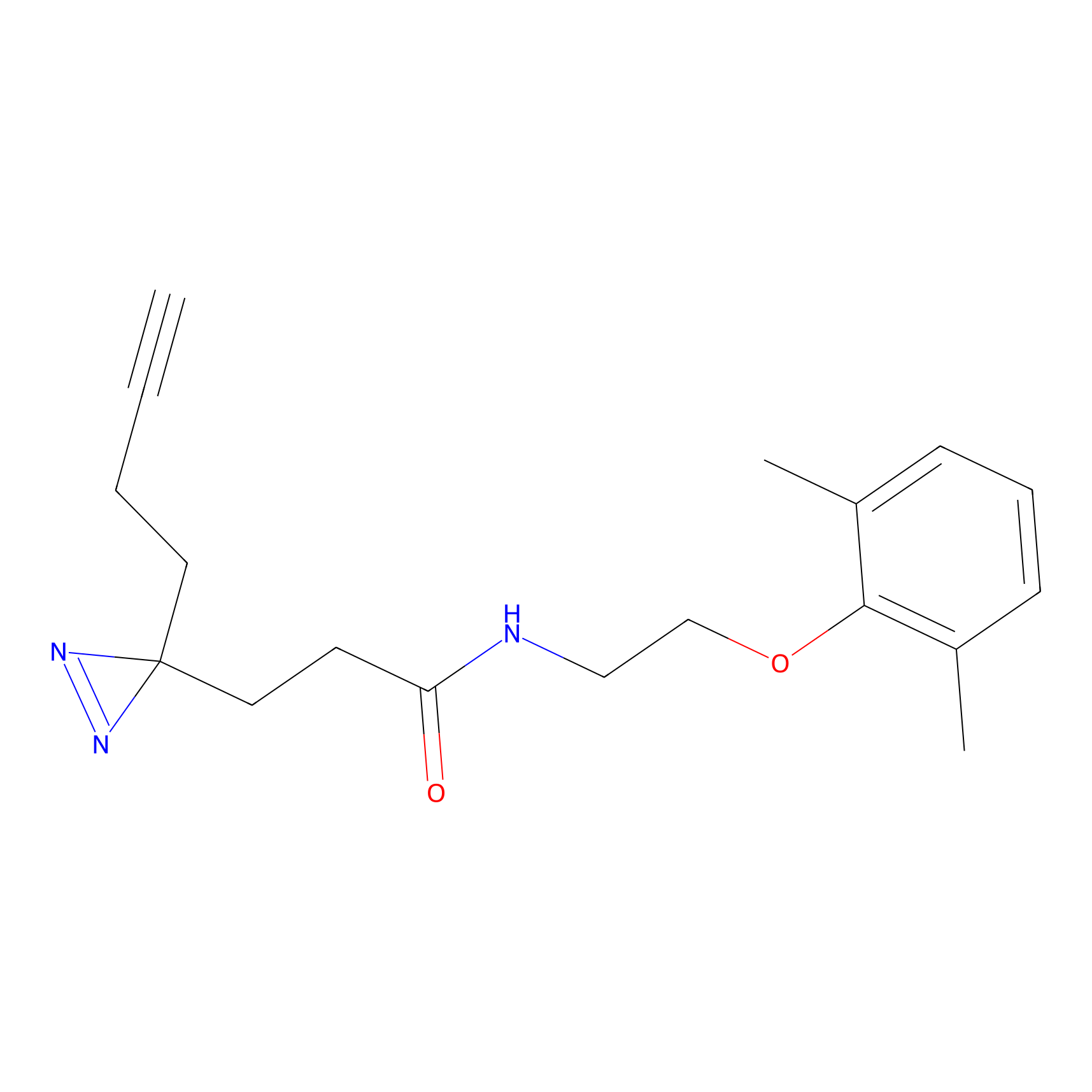
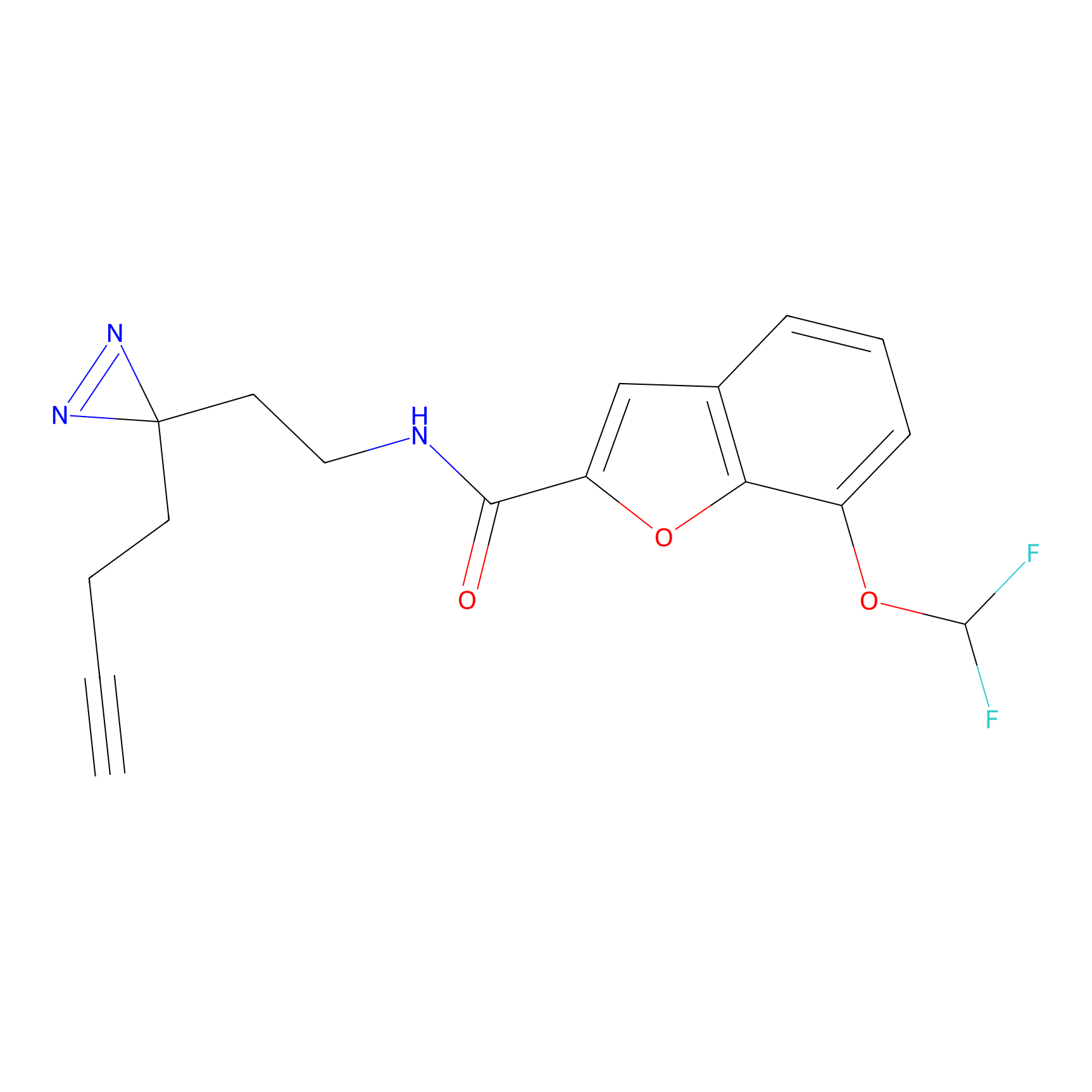
8.07 | LDD0196 | [3] | |
|
CY-1 Probe Info |
 |
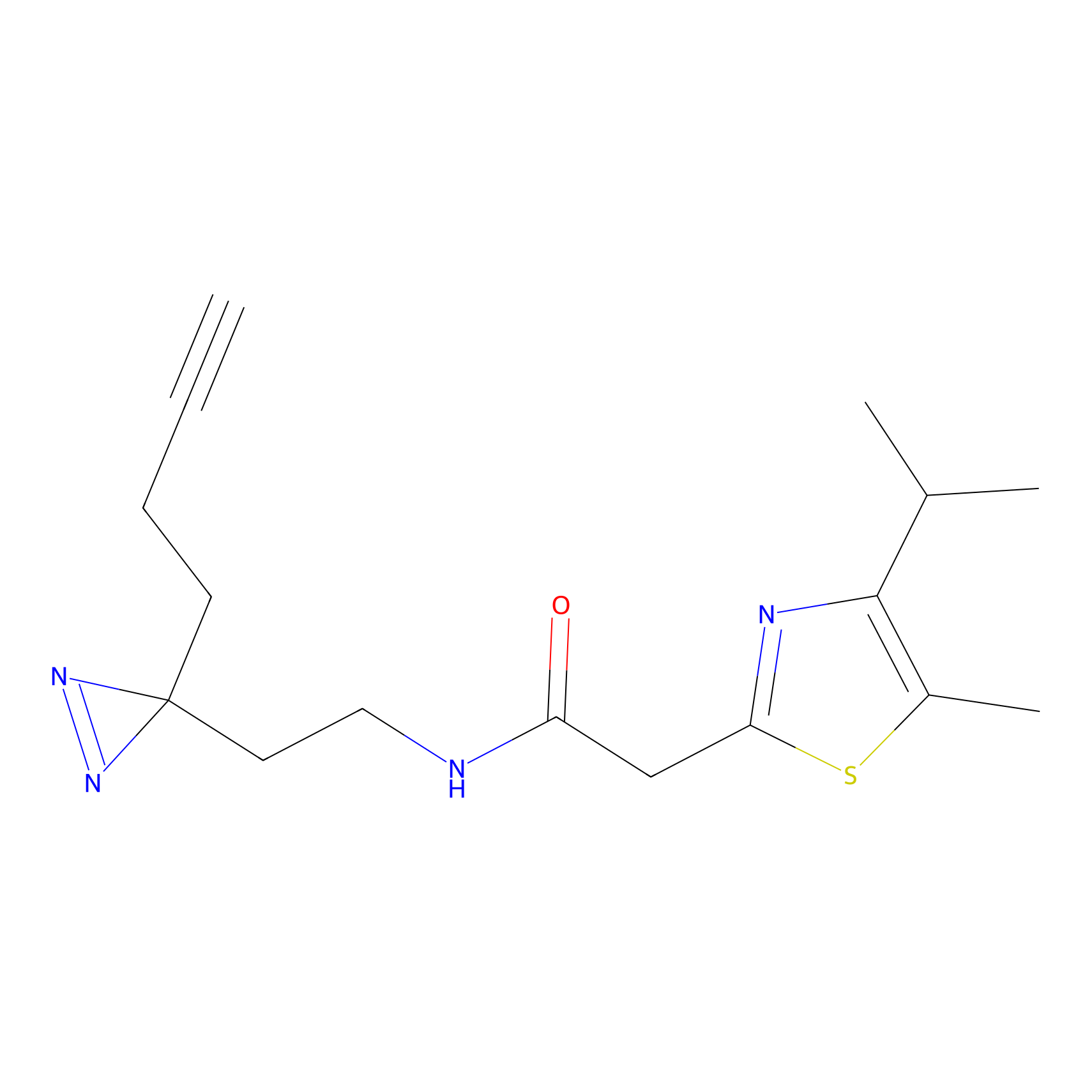
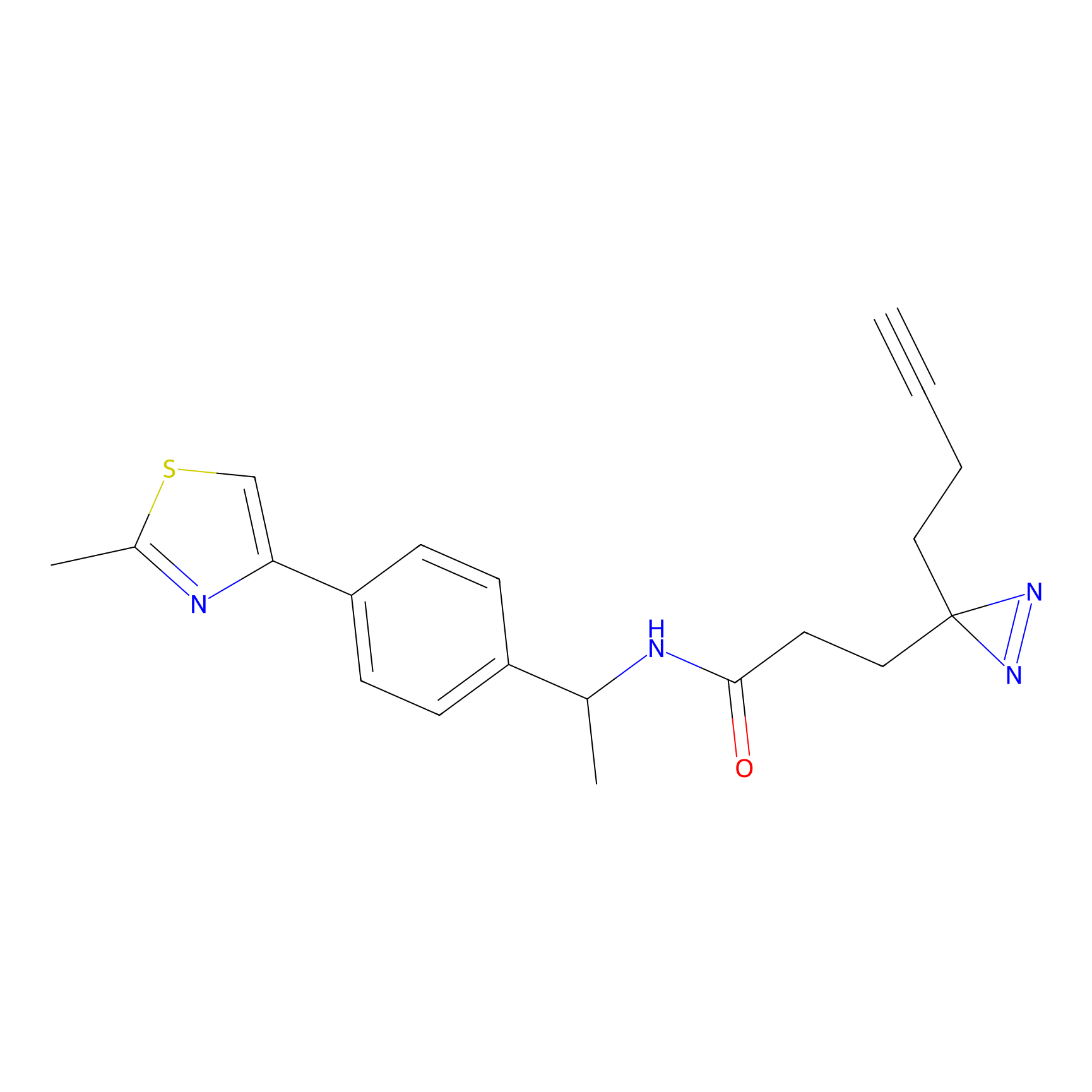
100.00 | LDD0243 | [4] | |
|
CY4 Probe Info |
 |
100.00 | LDD0244 | [4] | |
|
N1 Probe Info |
 |
100.00 | LDD0242 | [4] | |
|
C-Sul Probe Info |
 |
6.55 | LDD0066 | [5] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [6] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [6] | |
|
STPyne Probe Info |
 |
K85(5.40); K98(5.56) | LDD0277 | [7] | |
|
P11 Probe Info |
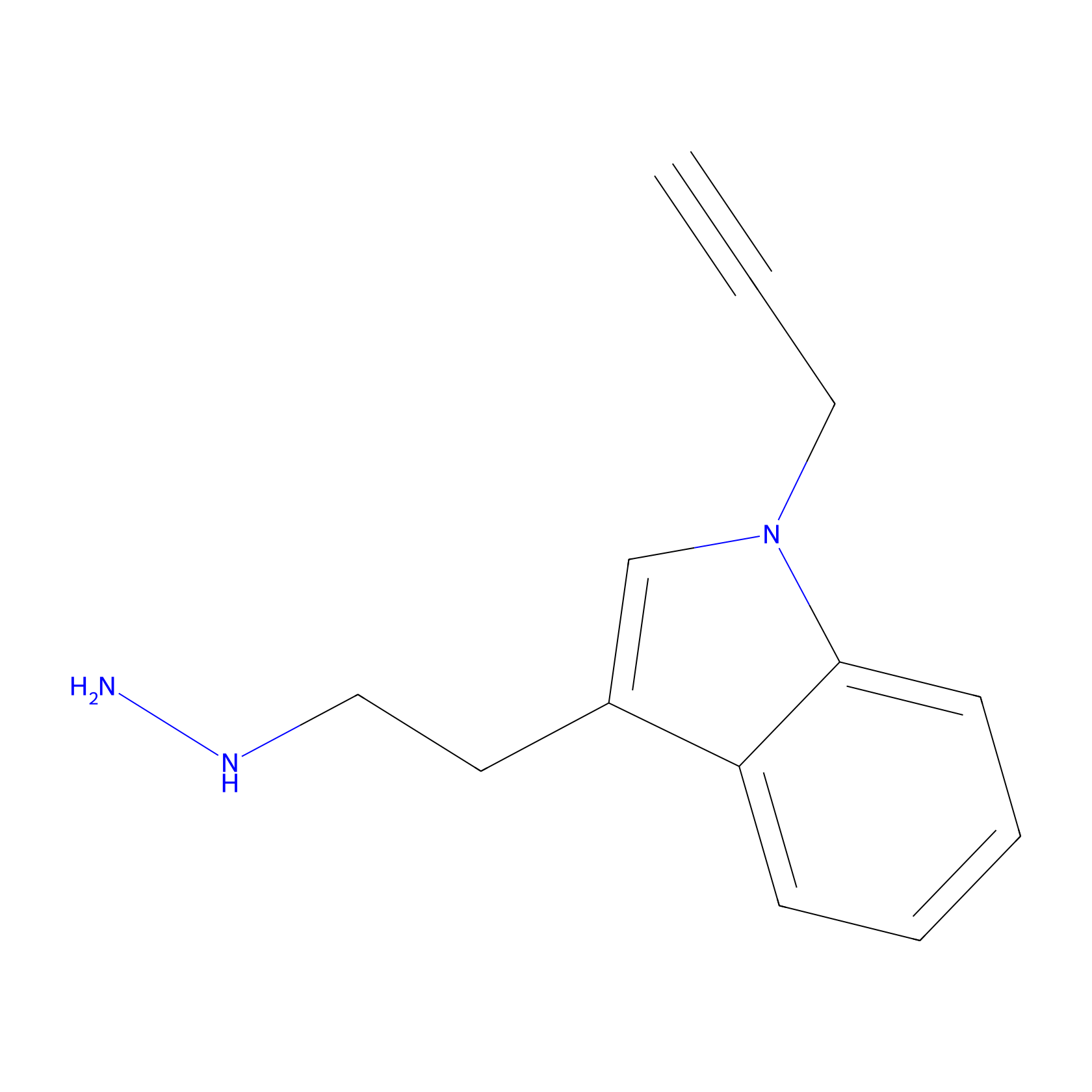 |
1.06 | LDD0210 | [8] | |
|
BTD Probe Info |
 |
C173(1.14) | LDD2100 | [9] | |
|
Jackson_14 Probe Info |
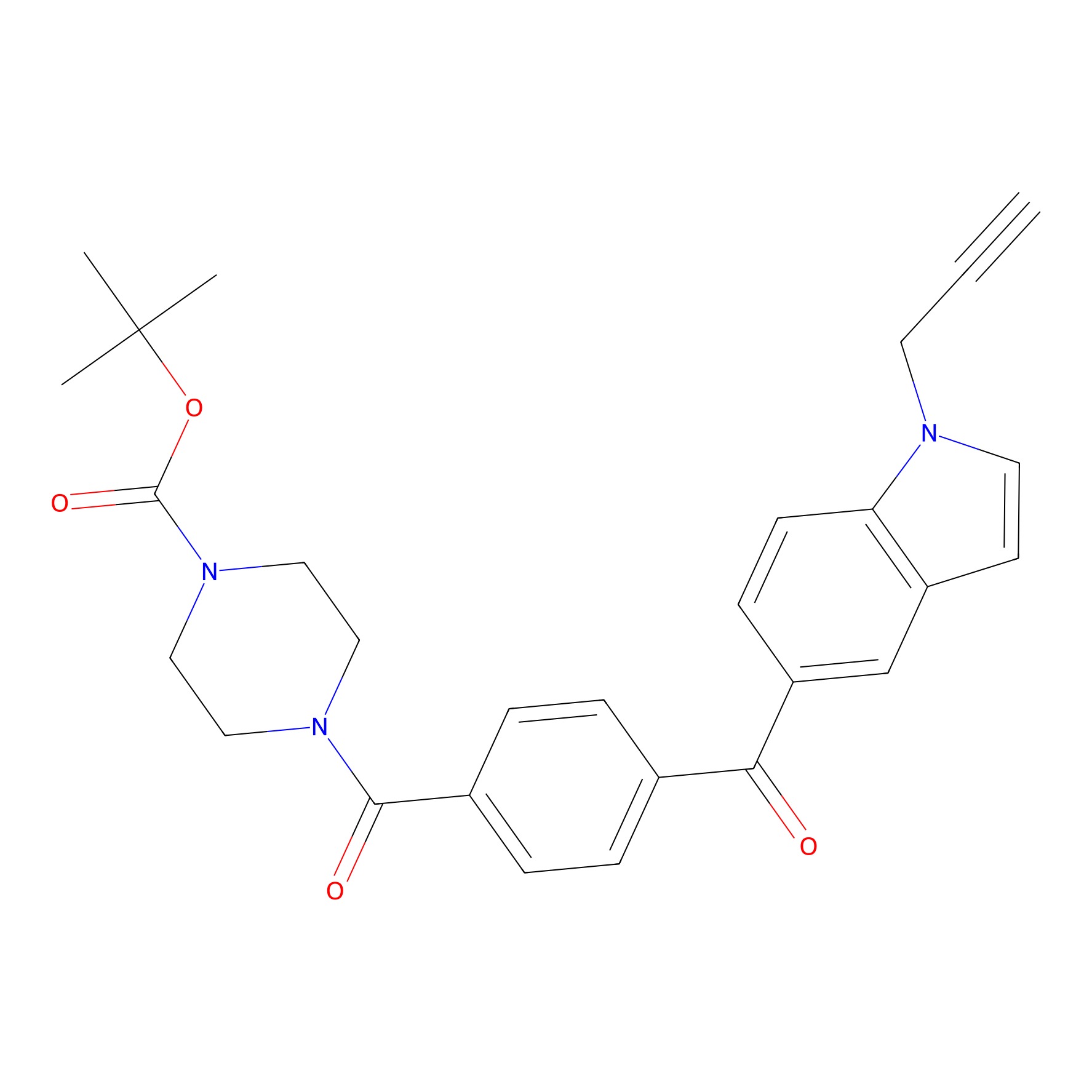 |
2.33 | LDD0123 | [10] | |
|
EA-probe Probe Info |
 |
N.A. | LDD0440 | [11] | |
|
DBIA Probe Info |
 |
C173(32.95) | LDD0209 | [12] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [13] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [14] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [14] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [15] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [16] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HEK-293T | C290(0.88) | LDD1507 | [23] |
| LDCM0215 | AC10 | HEK-293T | C290(0.90) | LDD1508 | [23] |
| LDCM0226 | AC11 | HEK-293T | C290(0.83) | LDD1509 | [23] |
| LDCM0237 | AC12 | HEK-293T | C290(0.91) | LDD1510 | [23] |
| LDCM0259 | AC14 | HEK-293T | C290(1.22) | LDD1512 | [23] |
| LDCM0270 | AC15 | HEK-293T | C290(1.06) | LDD1513 | [23] |
| LDCM0276 | AC17 | HEK-293T | C290(1.07) | LDD1515 | [23] |
| LDCM0277 | AC18 | HEK-293T | C290(0.76) | LDD1516 | [23] |
| LDCM0278 | AC19 | HEK-293T | C290(0.59) | LDD1517 | [23] |
| LDCM0279 | AC2 | HEK-293T | C290(0.83) | LDD1518 | [23] |
| LDCM0280 | AC20 | HEK-293T | C290(0.85) | LDD1519 | [23] |
| LDCM0281 | AC21 | HEK-293T | C290(0.85) | LDD1520 | [23] |
| LDCM0282 | AC22 | HEK-293T | C290(1.04) | LDD1521 | [23] |
| LDCM0283 | AC23 | HEK-293T | C290(0.96) | LDD1522 | [23] |
| LDCM0284 | AC24 | HEK-293T | C290(0.79) | LDD1523 | [23] |
| LDCM0285 | AC25 | HEK-293T | C290(0.91) | LDD1524 | [23] |
| LDCM0286 | AC26 | HEK-293T | C290(0.80) | LDD1525 | [23] |
| LDCM0287 | AC27 | HEK-293T | C290(0.68) | LDD1526 | [23] |
| LDCM0288 | AC28 | HEK-293T | C290(0.81) | LDD1527 | [23] |
| LDCM0289 | AC29 | HEK-293T | C290(0.78) | LDD1528 | [23] |
| LDCM0290 | AC3 | HEK-293T | C290(0.73) | LDD1529 | [23] |
| LDCM0291 | AC30 | HEK-293T | C290(0.82) | LDD1530 | [23] |
| LDCM0292 | AC31 | HEK-293T | C290(0.98) | LDD1531 | [23] |
| LDCM0293 | AC32 | HEK-293T | C290(0.76) | LDD1532 | [23] |
| LDCM0294 | AC33 | HEK-293T | C290(1.03) | LDD1533 | [23] |
| LDCM0295 | AC34 | HEK-293T | C290(0.93) | LDD1534 | [23] |
| LDCM0296 | AC35 | HEK-293T | C290(0.81) | LDD1535 | [23] |
| LDCM0297 | AC36 | HEK-293T | C290(0.85) | LDD1536 | [23] |
| LDCM0298 | AC37 | HEK-293T | C290(0.70) | LDD1537 | [23] |
| LDCM0299 | AC38 | HEK-293T | C290(0.88) | LDD1538 | [23] |
| LDCM0300 | AC39 | HEK-293T | C290(1.14) | LDD1539 | [23] |
| LDCM0301 | AC4 | HEK-293T | C290(0.89) | LDD1540 | [23] |
| LDCM0302 | AC40 | HEK-293T | C290(0.85) | LDD1541 | [23] |
| LDCM0303 | AC41 | HEK-293T | C290(1.17) | LDD1542 | [23] |
| LDCM0304 | AC42 | HEK-293T | C290(0.78) | LDD1543 | [23] |
| LDCM0305 | AC43 | HEK-293T | C290(0.69) | LDD1544 | [23] |
| LDCM0306 | AC44 | HEK-293T | C290(0.96) | LDD1545 | [23] |
| LDCM0307 | AC45 | HEK-293T | C290(0.87) | LDD1546 | [23] |
| LDCM0308 | AC46 | HEK-293T | C290(0.97) | LDD1547 | [23] |
| LDCM0309 | AC47 | HEK-293T | C290(0.99) | LDD1548 | [23] |
| LDCM0310 | AC48 | HEK-293T | C290(0.86) | LDD1549 | [23] |
| LDCM0311 | AC49 | HEK-293T | C290(1.01) | LDD1550 | [23] |
| LDCM0312 | AC5 | HEK-293T | C290(0.78) | LDD1551 | [23] |
| LDCM0313 | AC50 | HEK-293T | C290(0.85) | LDD1552 | [23] |
| LDCM0314 | AC51 | HEK-293T | C290(0.68) | LDD1553 | [23] |
| LDCM0315 | AC52 | HEK-293T | C290(0.85) | LDD1554 | [23] |
| LDCM0316 | AC53 | HEK-293T | C290(0.86) | LDD1555 | [23] |
| LDCM0317 | AC54 | HEK-293T | C290(0.87) | LDD1556 | [23] |
| LDCM0318 | AC55 | HEK-293T | C290(1.01) | LDD1557 | [23] |
| LDCM0319 | AC56 | HEK-293T | C290(0.93) | LDD1558 | [23] |
| LDCM0320 | AC57 | HEK-293T | C290(1.15) | LDD1559 | [23] |
| LDCM0321 | AC58 | HEK-293T | C290(1.15) | LDD1560 | [23] |
| LDCM0322 | AC59 | HEK-293T | C290(0.84) | LDD1561 | [23] |
| LDCM0323 | AC6 | HEK-293T | C290(0.83) | LDD1562 | [23] |
| LDCM0324 | AC60 | HEK-293T | C290(1.12) | LDD1563 | [23] |
| LDCM0325 | AC61 | HEK-293T | C290(1.00) | LDD1564 | [23] |
| LDCM0326 | AC62 | HEK-293T | C290(0.90) | LDD1565 | [23] |
| LDCM0327 | AC63 | HEK-293T | C290(1.21) | LDD1566 | [23] |
| LDCM0328 | AC64 | HEK-293T | C290(0.92) | LDD1567 | [23] |
| LDCM0334 | AC7 | HEK-293T | C290(1.05) | LDD1568 | [23] |
| LDCM0345 | AC8 | HEK-293T | C290(0.91) | LDD1569 | [23] |
| LDCM0248 | AKOS034007472 | HEK-293T | C290(0.83) | LDD1511 | [23] |
| LDCM0356 | AKOS034007680 | HEK-293T | C290(0.96) | LDD1570 | [23] |
| LDCM0275 | AKOS034007705 | HEK-293T | C290(0.83) | LDD1514 | [23] |
| LDCM0089 | C30 | HEK-293T | 1.06 | LDD0210 | [8] |
| LDCM0367 | CL1 | HEK-293T | C290(1.14) | LDD1571 | [23] |
| LDCM0368 | CL10 | HEK-293T | C290(1.69) | LDD1572 | [23] |
| LDCM0369 | CL100 | HEK-293T | C290(0.86) | LDD1573 | [23] |
| LDCM0370 | CL101 | HEK-293T | C290(0.92) | LDD1574 | [23] |
| LDCM0371 | CL102 | HEK-293T | C290(1.19) | LDD1575 | [23] |
| LDCM0372 | CL103 | HEK-293T | C290(0.88) | LDD1576 | [23] |
| LDCM0373 | CL104 | HEK-293T | C290(0.87) | LDD1577 | [23] |
| LDCM0374 | CL105 | HEK-293T | C290(0.85) | LDD1578 | [23] |
| LDCM0375 | CL106 | HEK-293T | C290(0.99) | LDD1579 | [23] |
| LDCM0376 | CL107 | HEK-293T | C290(0.81) | LDD1580 | [23] |
| LDCM0377 | CL108 | HEK-293T | C290(0.77) | LDD1581 | [23] |
| LDCM0378 | CL109 | HEK-293T | C290(0.95) | LDD1582 | [23] |
| LDCM0379 | CL11 | HEK-293T | C290(1.63) | LDD1583 | [23] |
| LDCM0380 | CL110 | HEK-293T | C290(1.24) | LDD1584 | [23] |
| LDCM0381 | CL111 | HEK-293T | C290(1.18) | LDD1585 | [23] |
| LDCM0382 | CL112 | HEK-293T | C290(0.80) | LDD1586 | [23] |
| LDCM0383 | CL113 | HEK-293T | C290(0.92) | LDD1587 | [23] |
| LDCM0384 | CL114 | HEK-293T | C290(1.32) | LDD1588 | [23] |
| LDCM0385 | CL115 | HEK-293T | C290(0.75) | LDD1589 | [23] |
| LDCM0386 | CL116 | HEK-293T | C290(0.91) | LDD1590 | [23] |
| LDCM0387 | CL117 | HEK-293T | C290(0.80) | LDD1591 | [23] |
| LDCM0388 | CL118 | HEK-293T | C290(0.94) | LDD1592 | [23] |
| LDCM0389 | CL119 | HEK-293T | C290(0.81) | LDD1593 | [23] |
| LDCM0390 | CL12 | HEK-293T | C290(1.66) | LDD1594 | [23] |
| LDCM0391 | CL120 | HEK-293T | C290(0.74) | LDD1595 | [23] |
| LDCM0392 | CL121 | HEK-293T | C290(0.91) | LDD1596 | [23] |
| LDCM0393 | CL122 | HEK-293T | C290(1.11) | LDD1597 | [23] |
| LDCM0394 | CL123 | HEK-293T | C290(1.52) | LDD1598 | [23] |
| LDCM0395 | CL124 | HEK-293T | C290(0.86) | LDD1599 | [23] |
| LDCM0396 | CL125 | HEK-293T | C290(1.10) | LDD1600 | [23] |
| LDCM0397 | CL126 | HEK-293T | C290(1.29) | LDD1601 | [23] |
| LDCM0398 | CL127 | HEK-293T | C290(1.01) | LDD1602 | [23] |
| LDCM0399 | CL128 | HEK-293T | C290(1.00) | LDD1603 | [23] |
| LDCM0400 | CL13 | HEK-293T | C290(1.08) | LDD1604 | [23] |
| LDCM0401 | CL14 | HEK-293T | C290(1.00) | LDD1605 | [23] |
| LDCM0402 | CL15 | HEK-293T | C290(1.45) | LDD1606 | [23] |
| LDCM0403 | CL16 | HEK-293T | C290(1.28) | LDD1607 | [23] |
| LDCM0404 | CL17 | HEK-293T | C290(1.75) | LDD1608 | [23] |
| LDCM0405 | CL18 | HEK-293T | C290(1.02) | LDD1609 | [23] |
| LDCM0406 | CL19 | HEK-293T | C290(1.12) | LDD1610 | [23] |
| LDCM0407 | CL2 | HEK-293T | C290(1.22) | LDD1611 | [23] |
| LDCM0408 | CL20 | HEK-293T | C290(1.24) | LDD1612 | [23] |
| LDCM0409 | CL21 | HEK-293T | C290(1.36) | LDD1613 | [23] |
| LDCM0410 | CL22 | HEK-293T | C290(1.33) | LDD1614 | [23] |
| LDCM0411 | CL23 | HEK-293T | C290(1.46) | LDD1615 | [23] |
| LDCM0412 | CL24 | HEK-293T | C290(1.29) | LDD1616 | [23] |
| LDCM0413 | CL25 | HEK-293T | C290(1.88) | LDD1617 | [23] |
| LDCM0414 | CL26 | HEK-293T | C290(0.95) | LDD1618 | [23] |
| LDCM0415 | CL27 | HEK-293T | C290(1.01) | LDD1619 | [23] |
| LDCM0416 | CL28 | HEK-293T | C290(0.90) | LDD1620 | [23] |
| LDCM0417 | CL29 | HEK-293T | C290(1.10) | LDD1621 | [23] |
| LDCM0418 | CL3 | HEK-293T | C290(1.19) | LDD1622 | [23] |
| LDCM0419 | CL30 | HEK-293T | C290(0.85) | LDD1623 | [23] |
| LDCM0420 | CL31 | HEK-293T | C290(0.75) | LDD1624 | [23] |
| LDCM0421 | CL32 | HEK-293T | C290(1.08) | LDD1625 | [23] |
| LDCM0422 | CL33 | HEK-293T | C290(1.83) | LDD1626 | [23] |
| LDCM0423 | CL34 | HEK-293T | C290(1.28) | LDD1627 | [23] |
| LDCM0424 | CL35 | HEK-293T | C290(1.38) | LDD1628 | [23] |
| LDCM0425 | CL36 | HEK-293T | C290(1.26) | LDD1629 | [23] |
| LDCM0426 | CL37 | HEK-293T | C290(1.26) | LDD1630 | [23] |
| LDCM0428 | CL39 | HEK-293T | C290(0.98) | LDD1632 | [23] |
| LDCM0429 | CL4 | HEK-293T | C290(1.12) | LDD1633 | [23] |
| LDCM0430 | CL40 | HEK-293T | C290(0.87) | LDD1634 | [23] |
| LDCM0431 | CL41 | HEK-293T | C290(1.02) | LDD1635 | [23] |
| LDCM0432 | CL42 | HEK-293T | C290(0.91) | LDD1636 | [23] |
| LDCM0433 | CL43 | HEK-293T | C290(0.73) | LDD1637 | [23] |
| LDCM0434 | CL44 | HEK-293T | C290(1.08) | LDD1638 | [23] |
| LDCM0435 | CL45 | HEK-293T | C290(1.20) | LDD1639 | [23] |
| LDCM0436 | CL46 | HEK-293T | C290(1.24) | LDD1640 | [23] |
| LDCM0437 | CL47 | HEK-293T | C290(1.32) | LDD1641 | [23] |
| LDCM0438 | CL48 | HEK-293T | C290(1.19) | LDD1642 | [23] |
| LDCM0439 | CL49 | HEK-293T | C290(1.08) | LDD1643 | [23] |
| LDCM0440 | CL5 | HEK-293T | C290(1.46) | LDD1644 | [23] |
| LDCM0441 | CL50 | HEK-293T | C290(1.23) | LDD1645 | [23] |
| LDCM0443 | CL52 | HEK-293T | C290(0.99) | LDD1646 | [23] |
| LDCM0444 | CL53 | HEK-293T | C290(1.24) | LDD1647 | [23] |
| LDCM0445 | CL54 | HEK-293T | C290(1.79) | LDD1648 | [23] |
| LDCM0446 | CL55 | HEK-293T | C290(0.97) | LDD1649 | [23] |
| LDCM0447 | CL56 | HEK-293T | C290(1.00) | LDD1650 | [23] |
| LDCM0448 | CL57 | HEK-293T | C290(1.32) | LDD1651 | [23] |
| LDCM0449 | CL58 | HEK-293T | C290(1.56) | LDD1652 | [23] |
| LDCM0450 | CL59 | HEK-293T | C290(1.35) | LDD1653 | [23] |
| LDCM0451 | CL6 | HEK-293T | C290(1.39) | LDD1654 | [23] |
| LDCM0452 | CL60 | HEK-293T | C290(1.29) | LDD1655 | [23] |
| LDCM0453 | CL61 | HEK-293T | C290(1.12) | LDD1656 | [23] |
| LDCM0454 | CL62 | HEK-293T | C290(1.00) | LDD1657 | [23] |
| LDCM0455 | CL63 | HEK-293T | C290(0.95) | LDD1658 | [23] |
| LDCM0456 | CL64 | HEK-293T | C290(1.10) | LDD1659 | [23] |
| LDCM0457 | CL65 | HEK-293T | C290(0.92) | LDD1660 | [23] |
| LDCM0458 | CL66 | HEK-293T | C290(1.01) | LDD1661 | [23] |
| LDCM0459 | CL67 | HEK-293T | C290(0.77) | LDD1662 | [23] |
| LDCM0460 | CL68 | HEK-293T | C290(1.12) | LDD1663 | [23] |
| LDCM0461 | CL69 | HEK-293T | C290(1.05) | LDD1664 | [23] |
| LDCM0462 | CL7 | HEK-293T | C290(1.31) | LDD1665 | [23] |
| LDCM0463 | CL70 | HEK-293T | C290(1.31) | LDD1666 | [23] |
| LDCM0464 | CL71 | HEK-293T | C290(1.15) | LDD1667 | [23] |
| LDCM0465 | CL72 | HEK-293T | C290(1.31) | LDD1668 | [23] |
| LDCM0466 | CL73 | HEK-293T | C290(1.39) | LDD1669 | [23] |
| LDCM0467 | CL74 | HEK-293T | C290(0.91) | LDD1670 | [23] |
| LDCM0469 | CL76 | HEK-293T | C290(0.90) | LDD1672 | [23] |
| LDCM0470 | CL77 | HEK-293T | C290(1.06) | LDD1673 | [23] |
| LDCM0471 | CL78 | HEK-293T | C290(0.93) | LDD1674 | [23] |
| LDCM0472 | CL79 | HEK-293T | C290(1.02) | LDD1675 | [23] |
| LDCM0473 | CL8 | HEK-293T | C290(2.78) | LDD1676 | [23] |
| LDCM0474 | CL80 | HEK-293T | C290(0.93) | LDD1677 | [23] |
| LDCM0475 | CL81 | HEK-293T | C290(1.04) | LDD1678 | [23] |
| LDCM0476 | CL82 | HEK-293T | C290(0.98) | LDD1679 | [23] |
| LDCM0477 | CL83 | HEK-293T | C290(1.16) | LDD1680 | [23] |
| LDCM0478 | CL84 | HEK-293T | C290(1.30) | LDD1681 | [23] |
| LDCM0479 | CL85 | HEK-293T | C290(1.33) | LDD1682 | [23] |
| LDCM0480 | CL86 | HEK-293T | C290(1.28) | LDD1683 | [23] |
| LDCM0481 | CL87 | HEK-293T | C290(1.25) | LDD1684 | [23] |
| LDCM0482 | CL88 | HEK-293T | C290(0.81) | LDD1685 | [23] |
| LDCM0483 | CL89 | HEK-293T | C290(1.12) | LDD1686 | [23] |
| LDCM0484 | CL9 | HEK-293T | C290(1.22) | LDD1687 | [23] |
| LDCM0485 | CL90 | HEK-293T | C290(2.28) | LDD1688 | [23] |
| LDCM0486 | CL91 | HEK-293T | C290(0.84) | LDD1689 | [23] |
| LDCM0487 | CL92 | HEK-293T | C290(1.16) | LDD1690 | [23] |
| LDCM0488 | CL93 | HEK-293T | C290(1.45) | LDD1691 | [23] |
| LDCM0489 | CL94 | HEK-293T | C290(1.01) | LDD1692 | [23] |
| LDCM0490 | CL95 | HEK-293T | C290(1.83) | LDD1693 | [23] |
| LDCM0491 | CL96 | HEK-293T | C290(1.34) | LDD1694 | [23] |
| LDCM0492 | CL97 | HEK-293T | C290(1.03) | LDD1695 | [23] |
| LDCM0493 | CL98 | HEK-293T | C290(0.95) | LDD1696 | [23] |
| LDCM0494 | CL99 | HEK-293T | C290(0.91) | LDD1697 | [23] |
| LDCM0495 | E2913 | HEK-293T | C290(0.92) | LDD1698 | [23] |
| LDCM0175 | Ethacrynic acid | HeLa | N.A. | LDD0440 | [11] |
| LDCM0468 | Fragment33 | HEK-293T | C290(0.82) | LDD1671 | [23] |
| LDCM0427 | Fragment51 | HEK-293T | C290(1.02) | LDD1631 | [23] |
| LDCM0022 | KB02 | HEK-293T | C290(0.84) | LDD1492 | [23] |
| LDCM0023 | KB03 | Jurkat | C173(32.95) | LDD0209 | [12] |
| LDCM0024 | KB05 | OAW-28 | C173(2.87) | LDD3368 | [24] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C173(1.14) | LDD2100 | [9] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C173(0.77) | LDD2141 | [9] |
| LDCM0016 | Ranjitkar_cp1 | MDA-MB-231 | 2.33 | LDD0123 | [10] |
References