Details of the Target
General Information of Target
| Target ID | LDTP02608 | |||||
|---|---|---|---|---|---|---|
| Target Name | ADP/ATP translocase 1 (SLC25A4) | |||||
| Gene Name | SLC25A4 | |||||
| Gene ID | 291 | |||||
| Synonyms |
AAC1; ANT1; ADP/ATP translocase 1; ADP,ATP carrier protein 1; ADP,ATP carrier protein, heart/skeletal muscle isoform T1; Adenine nucleotide translocator 1; ANT 1; Solute carrier family 25 member 4 |
|||||
| 3D Structure | ||||||
| Sequence |
MGDHAWSFLKDFLAGGVAAAVSKTAVAPIERVKLLLQVQHASKQISAEKQYKGIIDCVVR
IPKEQGFLSFWRGNLANVIRYFPTQALNFAFKDKYKQLFLGGVDRHKQFWRYFAGNLASG GAAGATSLCFVYPLDFARTRLAADVGKGAAQREFHGLGDCIIKIFKSDGLRGLYQGFNVS VQGIIIYRAAYFGVYDTAKGMLPDPKNVHIFVSWMIAQSVTAVAGLVSYPFDTVRRRMMM QSGRKGADIMYTGTVDCWRKIAKDEGAKAFFKGAWSNVLRGMGGAFVLVLYDEIKKYV |
|||||
| Target Type |
Successful
|
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
Mitochondrial carrier (TC 2.A.29) family
|
|||||
| Subcellular location |
Mitochondrion inner membrane
|
|||||
| Function |
ADP:ATP antiporter that mediates import of ADP into the mitochondrial matrix for ATP synthesis, and export of ATP out to fuel the cell. Cycles between the cytoplasmic-open state (c-state) and the matrix-open state (m-state): operates by the alternating access mechanism with a single substrate-binding site intermittently exposed to either the cytosolic (c-state) or matrix (m-state) side of the inner mitochondrial membrane. In addition to its ADP:ATP antiporter activity, also involved in mitochondrial uncoupling and mitochondrial permeability transition pore (mPTP) activity. Plays a role in mitochondrial uncoupling by acting as a proton transporter: proton transport uncouples the proton flows via the electron transport chain and ATP synthase to reduce the efficiency of ATP production and cause mitochondrial thermogenesis. Proton transporter activity is inhibited by ADP:ATP antiporter activity, suggesting that SLC25A4/ANT1 acts as a master regulator of mitochondrial energy output by maintaining a delicate balance between ATP production (ADP:ATP antiporter activity) and thermogenesis (proton transporter activity). Proton transporter activity requires free fatty acids as cofactor, but does not transport it. Also plays a key role in mPTP opening, a non-specific pore that enables free passage of the mitochondrial membranes to solutes of up to 1.5 kDa, and which contributes to cell death. It is however unclear if SLC25A4/ANT1 constitutes a pore-forming component of mPTP or regulates it. Acts as a regulator of mitophagy independently of ADP:ATP antiporter activity: promotes mitophagy via interaction with TIMM44, leading to inhibit the presequence translocase TIMM23, thereby promoting stabilization of PINK1.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
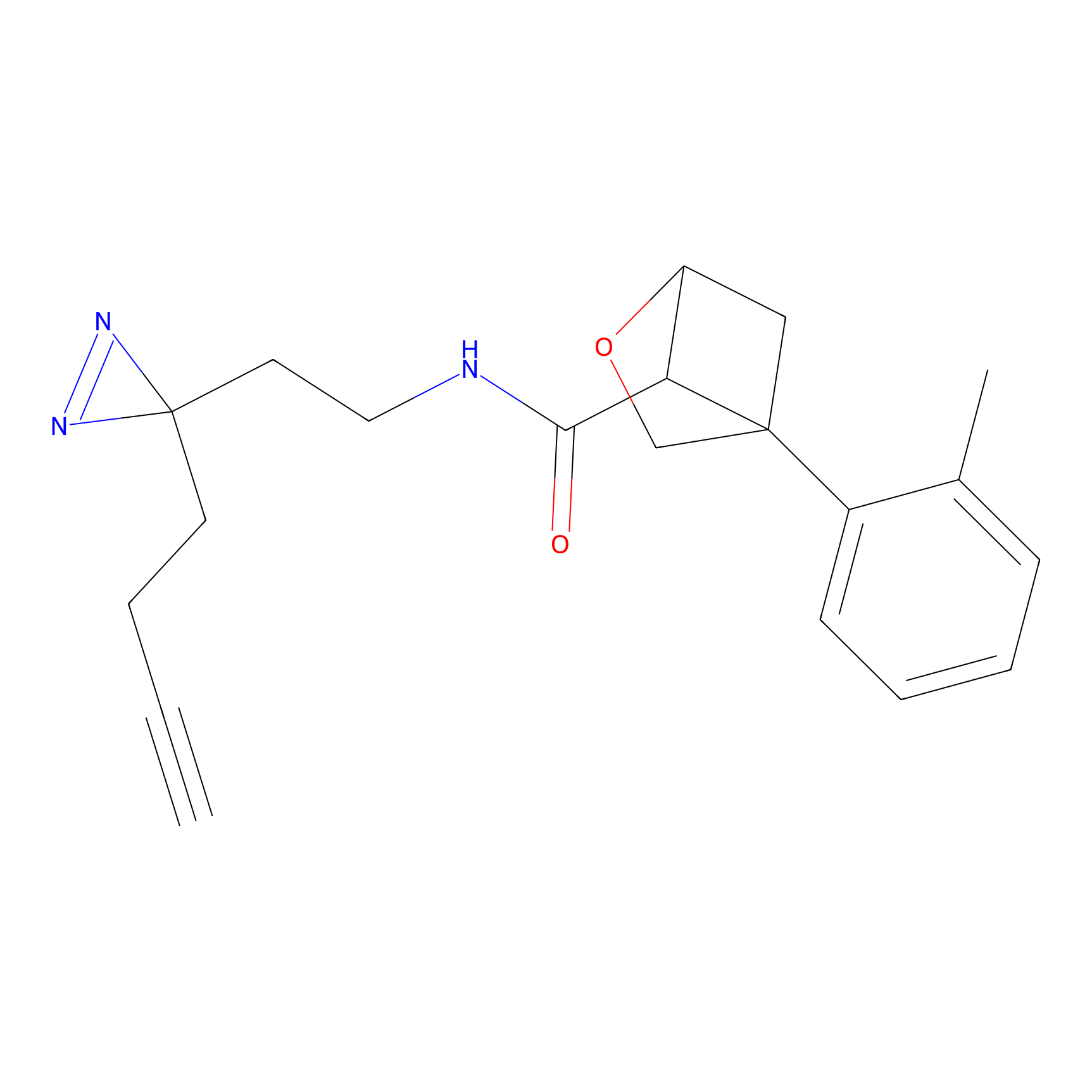
|
m-APA Probe Info |
 |
13.36 | LDD0402 | [1] | |
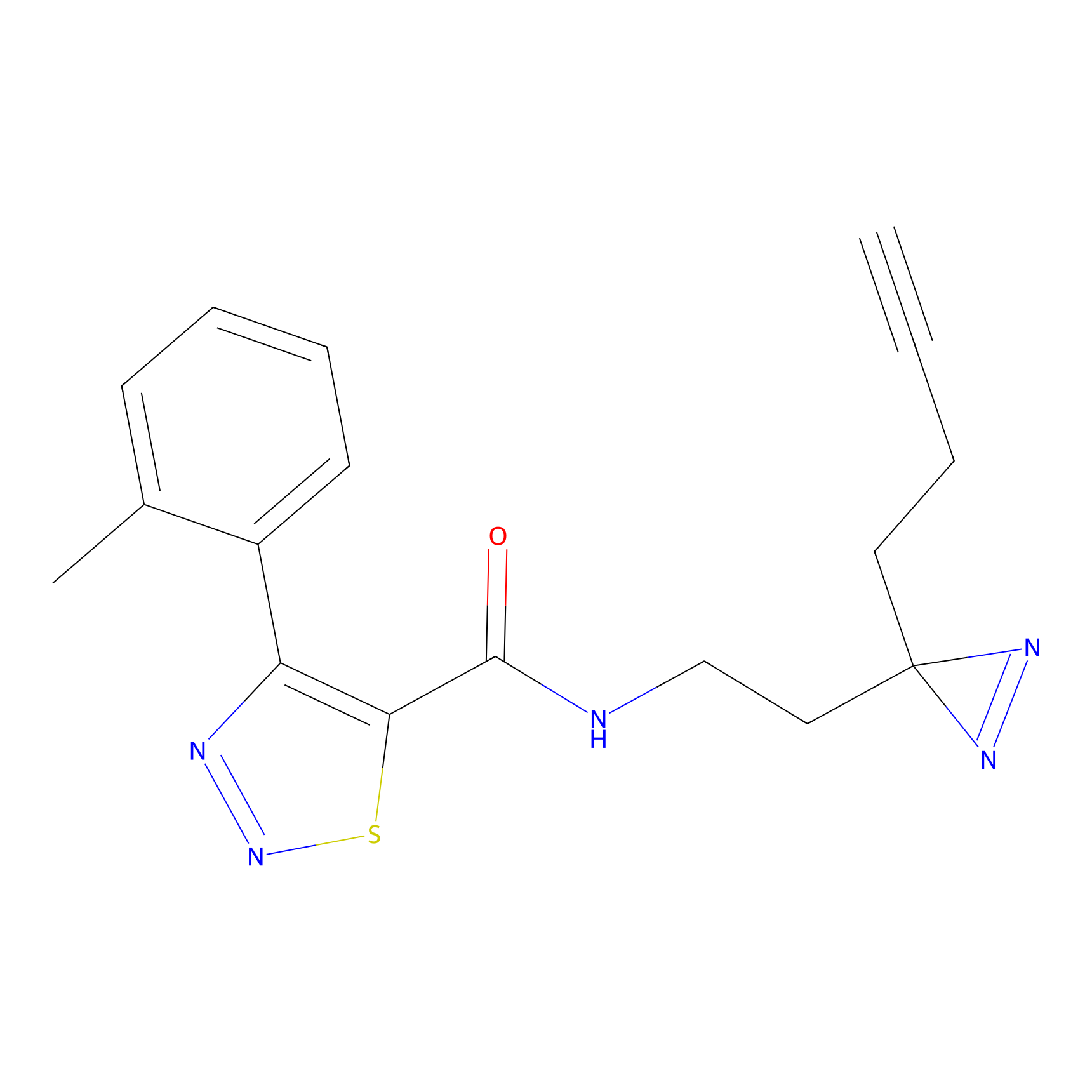
|
TH211 Probe Info |
 |
Y81(5.37) | LDD0257 | [2] | |
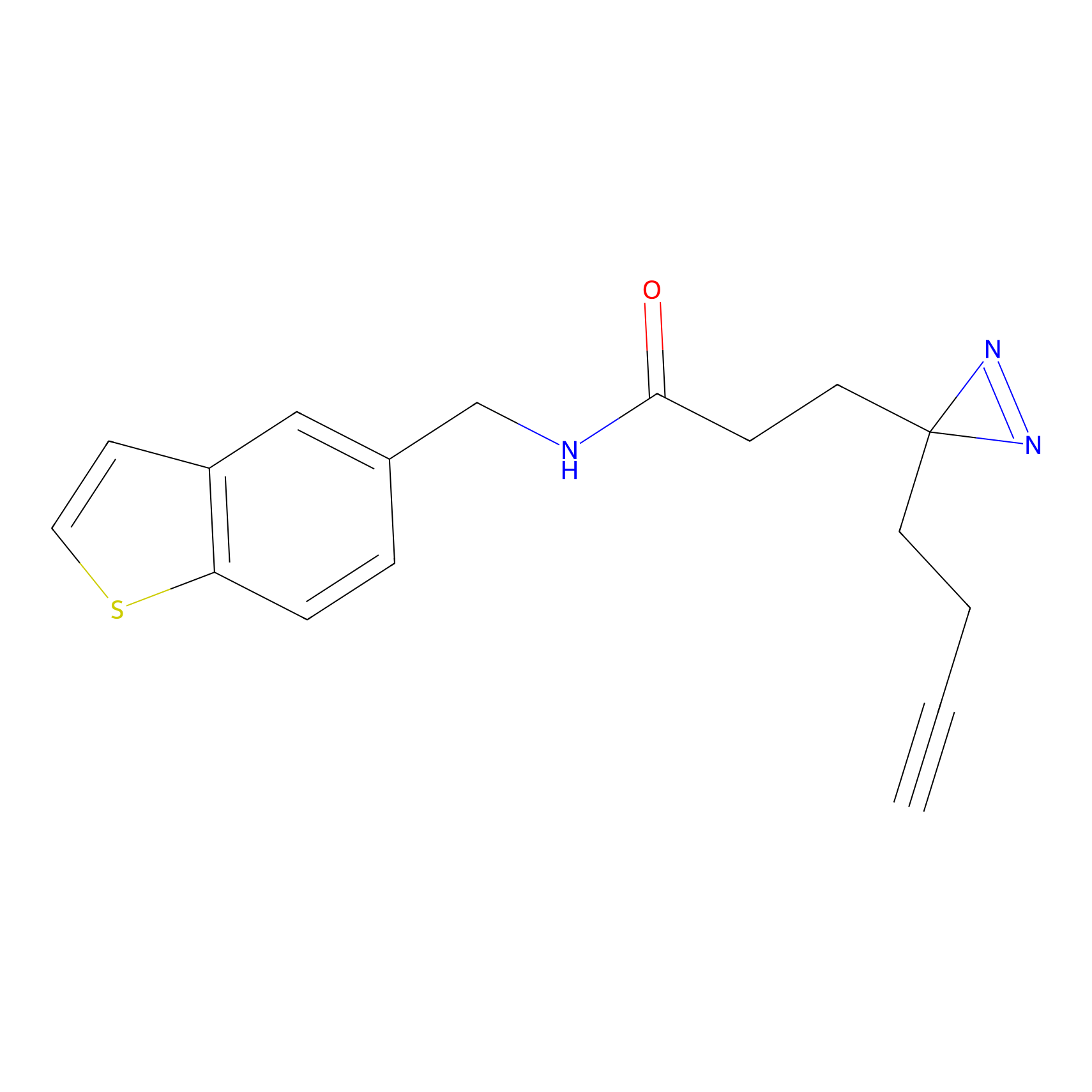
|
ONAyne Probe Info |
 |
K147(1.01) | LDD0274 | [3] | |
|
OPA-S-S-alkyne Probe Info |
 |
K245(1.72) | LDD3494 | [4] | |
|
JZ128-DTB Probe Info |
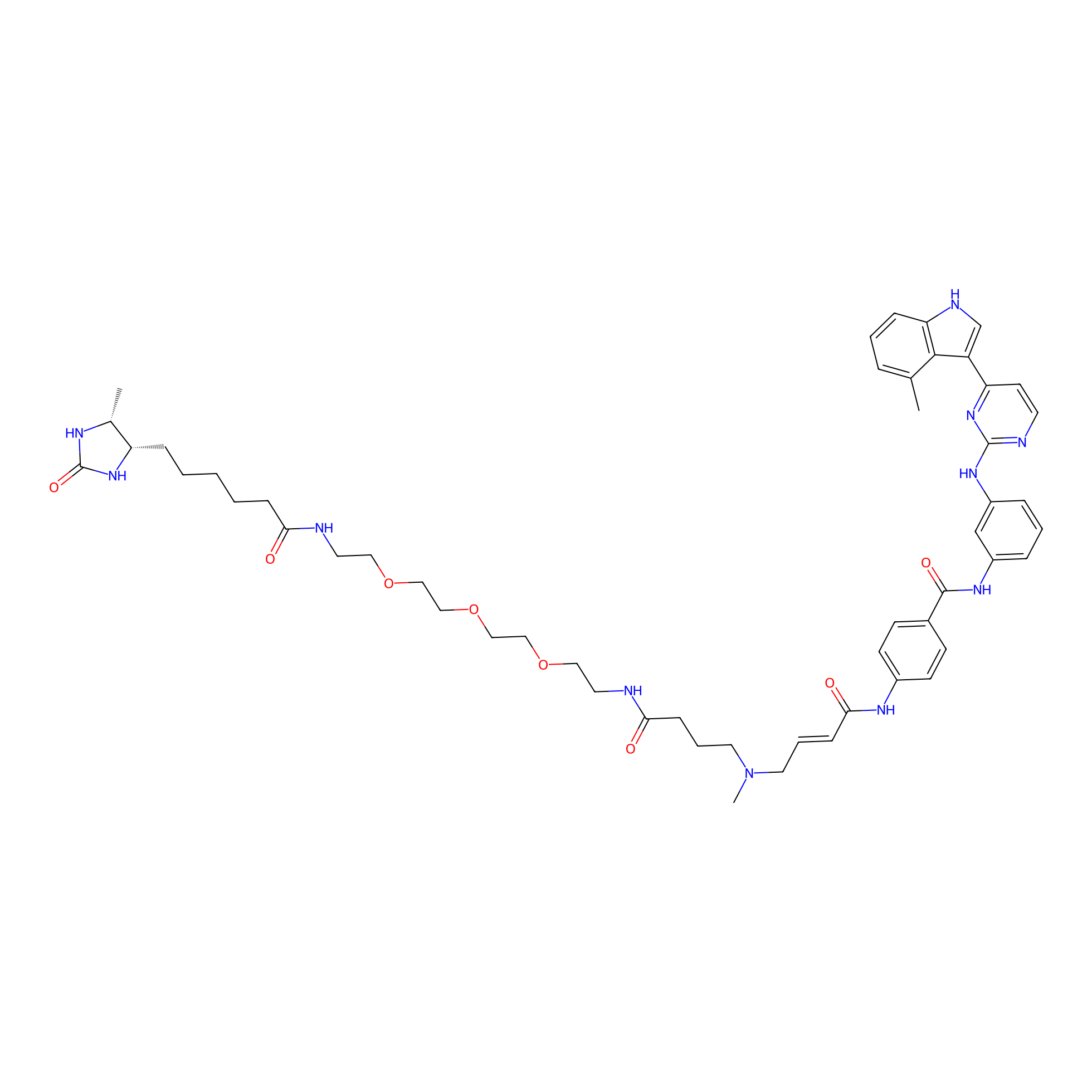 |
C257(0.00); C129(0.00) | LDD0462 | [5] | |
|
BTD Probe Info |
 |
C160(1.03) | LDD2099 | [6] | |
|
EA-probe Probe Info |
 |
N.A. | LDD0440 | [7] | |
|
DBIA Probe Info |
 |
C160(0.97); C257(0.90) | LDD0078 | [8] | |
|
ATP probe Probe Info |
 |
K92(0.00); K94(0.00); K43(0.00); K272(0.00) | LDD0199 | [9] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C160(0.00); C257(0.00) | LDD0038 | [10] | |
|
IA-alkyne Probe Info |
 |
C257(0.00); C129(0.00) | LDD0032 | [11] | |
|
Lodoacetamide azide Probe Info |
 |
C257(0.00); C160(0.00) | LDD0037 | [10] | |
|
ENE Probe Info |
 |
C57(0.00); C257(0.00); C160(0.00) | LDD0006 | [12] | |
|
IPM Probe Info |
 |
C160(0.00); C257(0.00) | LDD0005 | [12] | |
|
NHS Probe Info |
 |
K96(0.00); K33(0.00); K43(0.00); K272(0.00) | LDD0010 | [12] | |
|
STPyne Probe Info |
 |
K94(0.00); K23(0.00); K96(0.00); K43(0.00) | LDD0009 | [12] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [12] | |
|
Phosphinate-6 Probe Info |
 |
C257(0.00); C57(0.00); C160(0.00); C129(0.00) | LDD0018 | [13] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [14] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [15] | |
|
Acrolein Probe Info |
 |
C257(0.00); C160(0.00) | LDD0217 | [16] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [16] | |
|
Methacrolein Probe Info |
 |
C257(0.00); C160(0.00) | LDD0218 | [16] | |
|
W1 Probe Info |
 |
C160(0.00); C257(0.00) | LDD0236 | [17] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [18] | |
|
NAIA_5 Probe Info |
 |
C160(0.00); C257(0.00); C129(0.00) | LDD2223 | [19] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C160(1.11) | LDD2117 | [6] |
| LDCM0214 | AC1 | HCT 116 | C129(0.61); C160(0.94); C257(0.93); C57(0.93) | LDD0531 | [8] |
| LDCM0215 | AC10 | HCT 116 | C129(0.64); C160(0.92); C257(0.87) | LDD0532 | [8] |
| LDCM0216 | AC100 | HCT 116 | C129(0.59); C160(1.27); C257(0.67); C57(1.28) | LDD0533 | [8] |
| LDCM0217 | AC101 | HCT 116 | C129(0.72); C160(1.00); C257(0.67); C57(1.23) | LDD0534 | [8] |
| LDCM0218 | AC102 | HCT 116 | C129(0.76); C160(1.05); C257(0.66); C57(0.91) | LDD0535 | [8] |
| LDCM0219 | AC103 | HCT 116 | C129(0.61); C160(0.96); C257(0.70); C57(1.09) | LDD0536 | [8] |
| LDCM0220 | AC104 | HCT 116 | C129(0.75); C160(1.06); C257(0.63); C57(0.99) | LDD0537 | [8] |
| LDCM0221 | AC105 | HCT 116 | C129(0.77); C160(1.09); C257(0.68); C57(1.05) | LDD0538 | [8] |
| LDCM0222 | AC106 | HCT 116 | C129(0.55); C160(1.11); C257(0.55); C57(1.12) | LDD0539 | [8] |
| LDCM0223 | AC107 | HCT 116 | C129(0.48); C160(1.00); C257(1.13); C57(0.94) | LDD0540 | [8] |
| LDCM0224 | AC108 | HCT 116 | C129(1.02); C160(1.14); C257(0.80); C57(0.88) | LDD0541 | [8] |
| LDCM0225 | AC109 | HCT 116 | C129(0.90); C160(1.07); C257(0.87); C57(1.01) | LDD0542 | [8] |
| LDCM0226 | AC11 | HCT 116 | C129(0.57); C160(0.93); C257(0.87) | LDD0543 | [8] |
| LDCM0227 | AC110 | HCT 116 | C129(0.77); C160(0.98); C257(0.86); C57(1.18) | LDD0544 | [8] |
| LDCM0228 | AC111 | HCT 116 | C129(0.95); C160(1.03); C257(0.87); C57(1.08) | LDD0545 | [8] |
| LDCM0229 | AC112 | HCT 116 | C129(0.61); C160(0.98); C257(0.99); C57(0.83) | LDD0546 | [8] |
| LDCM0230 | AC113 | HCT 116 | C129(0.98); C160(0.93); C257(1.06); C57(0.88) | LDD0547 | [8] |
| LDCM0231 | AC114 | HCT 116 | C129(1.25); C160(0.76); C257(0.85); C57(0.92) | LDD0548 | [8] |
| LDCM0232 | AC115 | HCT 116 | C129(1.58); C160(0.79); C257(0.90); C57(1.01) | LDD0549 | [8] |
| LDCM0233 | AC116 | HCT 116 | C129(1.39); C160(0.85); C257(0.85); C57(0.97) | LDD0550 | [8] |
| LDCM0234 | AC117 | HCT 116 | C129(1.30); C160(0.85); C257(0.84); C57(1.06) | LDD0551 | [8] |
| LDCM0235 | AC118 | HCT 116 | C129(1.28); C160(0.86); C257(0.88); C57(0.86) | LDD0552 | [8] |
| LDCM0236 | AC119 | HCT 116 | C129(1.61); C160(0.88); C257(0.85); C57(1.01) | LDD0553 | [8] |
| LDCM0237 | AC12 | HCT 116 | C129(0.75); C160(1.00); C257(1.02) | LDD0554 | [8] |
| LDCM0238 | AC120 | HCT 116 | C129(1.34); C160(0.81); C257(0.95); C57(0.67) | LDD0555 | [8] |
| LDCM0239 | AC121 | HCT 116 | C129(1.05); C160(0.89); C257(1.02); C57(1.03) | LDD0556 | [8] |
| LDCM0240 | AC122 | HCT 116 | C129(1.07); C160(0.84); C257(0.76); C57(0.91) | LDD0557 | [8] |
| LDCM0241 | AC123 | HCT 116 | C129(1.50); C160(0.87); C257(0.88); C57(0.90) | LDD0558 | [8] |
| LDCM0242 | AC124 | HCT 116 | C129(1.22); C160(0.83); C257(0.84); C57(0.92) | LDD0559 | [8] |
| LDCM0243 | AC125 | HCT 116 | C129(1.34); C160(0.81); C257(0.99); C57(0.92) | LDD0560 | [8] |
| LDCM0244 | AC126 | HCT 116 | C129(1.05); C160(0.77); C257(0.83); C57(1.17) | LDD0561 | [8] |
| LDCM0245 | AC127 | HCT 116 | C129(1.36); C160(0.76); C257(0.79); C57(0.88) | LDD0562 | [8] |
| LDCM0246 | AC128 | HCT 116 | C129(0.82); C160(1.12); C257(1.08) | LDD0563 | [8] |
| LDCM0247 | AC129 | HCT 116 | C129(1.01); C160(0.96); C257(1.10) | LDD0564 | [8] |
| LDCM0249 | AC130 | HCT 116 | C129(0.84); C160(0.94); C257(0.98) | LDD0566 | [8] |
| LDCM0250 | AC131 | HCT 116 | C129(1.13); C160(0.96); C257(1.10) | LDD0567 | [8] |
| LDCM0251 | AC132 | HCT 116 | C129(0.76); C160(1.00); C257(1.04) | LDD0568 | [8] |
| LDCM0252 | AC133 | HCT 116 | C129(0.78); C160(1.02); C257(1.03) | LDD0569 | [8] |
| LDCM0253 | AC134 | HCT 116 | C129(0.91); C160(1.01); C257(1.05) | LDD0570 | [8] |
| LDCM0254 | AC135 | HCT 116 | C129(0.86); C160(1.00); C257(1.08) | LDD0571 | [8] |
| LDCM0255 | AC136 | HCT 116 | C129(0.86); C160(1.01); C257(1.04) | LDD0572 | [8] |
| LDCM0256 | AC137 | HCT 116 | C129(0.76); C160(0.95); C257(1.09) | LDD0573 | [8] |
| LDCM0257 | AC138 | HCT 116 | C129(0.72); C160(1.03); C257(1.19) | LDD0574 | [8] |
| LDCM0258 | AC139 | HCT 116 | C129(1.03); C160(1.10); C257(1.10) | LDD0575 | [8] |
| LDCM0259 | AC14 | HCT 116 | C129(0.57); C160(0.89); C257(0.85) | LDD0576 | [8] |
| LDCM0260 | AC140 | HCT 116 | C129(0.95); C160(1.15); C257(1.10) | LDD0577 | [8] |
| LDCM0261 | AC141 | HCT 116 | C129(0.82); C160(1.01); C257(1.12) | LDD0578 | [8] |
| LDCM0262 | AC142 | HCT 116 | C129(0.74); C160(1.02); C257(0.96) | LDD0579 | [8] |
| LDCM0263 | AC143 | HCT 116 | C129(0.54); C257(0.77); C160(0.98) | LDD0580 | [8] |
| LDCM0264 | AC144 | HCT 116 | C129(0.68); C257(0.68); C160(1.08) | LDD0581 | [8] |
| LDCM0265 | AC145 | HCT 116 | C257(0.78); C129(0.86); C160(1.34) | LDD0582 | [8] |
| LDCM0266 | AC146 | HCT 116 | C257(0.68); C129(1.13); C160(1.15) | LDD0583 | [8] |
| LDCM0267 | AC147 | HCT 116 | C257(0.74); C129(0.83); C160(1.06) | LDD0584 | [8] |
| LDCM0268 | AC148 | HCT 116 | C257(0.72); C129(0.88); C160(1.13) | LDD0585 | [8] |
| LDCM0269 | AC149 | HCT 116 | C257(0.68); C160(1.06); C129(1.55) | LDD0586 | [8] |
| LDCM0270 | AC15 | HCT 116 | C129(0.61); C160(0.83); C257(0.89) | LDD0587 | [8] |
| LDCM0271 | AC150 | HCT 116 | C257(0.78); C129(0.84); C160(1.24) | LDD0588 | [8] |
| LDCM0272 | AC151 | HCT 116 | C129(0.67); C257(0.76); C160(1.21) | LDD0589 | [8] |
| LDCM0273 | AC152 | HCT 116 | C257(0.66); C129(1.03); C160(1.18) | LDD0590 | [8] |
| LDCM0274 | AC153 | HCT 116 | C257(0.67); C129(0.82); C160(1.21) | LDD0591 | [8] |
| LDCM0621 | AC154 | HCT 116 | C129(0.71); C160(1.32); C257(0.66) | LDD2158 | [8] |
| LDCM0622 | AC155 | HCT 116 | C129(1.11); C160(1.35); C257(0.66) | LDD2159 | [8] |
| LDCM0623 | AC156 | HCT 116 | C129(0.84); C160(1.11); C257(0.79) | LDD2160 | [8] |
| LDCM0624 | AC157 | HCT 116 | C129(0.90); C160(1.18); C257(0.76) | LDD2161 | [8] |
| LDCM0276 | AC17 | HCT 116 | C257(0.86); C160(0.87); C129(1.04); C57(1.08) | LDD0593 | [8] |
| LDCM0277 | AC18 | HCT 116 | C160(0.81); C257(0.84); C129(0.86); C57(1.54) | LDD0594 | [8] |
| LDCM0278 | AC19 | HCT 116 | C257(0.84); C160(0.87); C129(0.88); C57(0.97) | LDD0595 | [8] |
| LDCM0279 | AC2 | HCT 116 | C129(0.78); C160(0.99); C257(1.08); C57(1.24) | LDD0596 | [8] |
| LDCM0280 | AC20 | HCT 116 | C160(0.84); C129(0.90); C257(0.94); C57(1.23) | LDD0597 | [8] |
| LDCM0281 | AC21 | HCT 116 | C160(0.81); C257(0.84); C129(0.90); C57(0.96) | LDD0598 | [8] |
| LDCM0282 | AC22 | HCT 116 | C160(0.86); C129(1.00); C257(1.01); C57(1.14) | LDD0599 | [8] |
| LDCM0283 | AC23 | HCT 116 | C160(0.89); C257(0.97); C129(0.98); C57(1.05) | LDD0600 | [8] |
| LDCM0284 | AC24 | HCT 116 | C257(0.90); C57(1.01); C160(1.02); C129(1.08) | LDD0601 | [8] |
| LDCM0285 | AC25 | HCT 116 | C129(0.73); C57(0.99); C257(1.00); C160(1.12) | LDD0602 | [8] |
| LDCM0286 | AC26 | HCT 116 | C129(0.57); C257(0.84); C57(1.00); C160(1.13) | LDD0603 | [8] |
| LDCM0287 | AC27 | HCT 116 | C129(0.62); C257(0.87); C57(1.03); C160(1.09) | LDD0604 | [8] |
| LDCM0288 | AC28 | HCT 116 | C129(0.68); C257(0.82); C57(0.86); C160(1.35) | LDD0605 | [8] |
| LDCM0289 | AC29 | HCT 116 | C129(0.64); C257(0.91); C57(1.03); C160(1.04) | LDD0606 | [8] |
| LDCM0290 | AC3 | HCT 116 | C129(0.79); C160(1.02); C257(1.14); C57(1.40) | LDD0607 | [8] |
| LDCM0291 | AC30 | HCT 116 | C129(0.68); C257(0.82); C57(0.97); C160(1.00) | LDD0608 | [8] |
| LDCM0292 | AC31 | HCT 116 | C129(0.51); C257(0.82); C57(0.94); C160(1.13) | LDD0609 | [8] |
| LDCM0293 | AC32 | HCT 116 | C129(0.51); C257(0.83); C160(1.05); C57(1.05) | LDD0610 | [8] |
| LDCM0294 | AC33 | HCT 116 | C129(0.59); C257(0.84); C160(1.00); C57(1.04) | LDD0611 | [8] |
| LDCM0295 | AC34 | HCT 116 | C129(0.59); C257(0.85); C160(0.88); C57(1.03) | LDD0612 | [8] |
| LDCM0296 | AC35 | HCT 116 | C160(0.92); C129(0.94); C57(0.95); C257(1.16) | LDD0613 | [8] |
| LDCM0297 | AC36 | HCT 116 | C129(0.90); C57(0.90); C160(0.91); C257(1.06) | LDD0614 | [8] |
| LDCM0298 | AC37 | HCT 116 | C129(0.96); C57(0.99); C257(1.00); C160(1.02) | LDD0615 | [8] |
| LDCM0299 | AC38 | HCT 116 | C160(0.89); C129(0.91); C57(0.93); C257(1.05) | LDD0616 | [8] |
| LDCM0300 | AC39 | HCT 116 | C129(0.77); C160(0.89); C57(0.94); C257(1.09) | LDD0617 | [8] |
| LDCM0301 | AC4 | HCT 116 | C129(0.62); C160(1.00); C257(1.06); C57(1.37) | LDD0618 | [8] |
| LDCM0302 | AC40 | HCT 116 | C57(0.89); C257(0.99); C160(1.01); C129(1.08) | LDD0619 | [8] |
| LDCM0303 | AC41 | HCT 116 | C129(0.63); C57(0.84); C160(0.90); C257(0.97) | LDD0620 | [8] |
| LDCM0304 | AC42 | HCT 116 | C57(0.85); C160(0.94); C257(1.03); C129(1.05) | LDD0621 | [8] |
| LDCM0305 | AC43 | HCT 116 | C160(0.88); C57(0.90); C257(0.97); C129(1.10) | LDD0622 | [8] |
| LDCM0306 | AC44 | HCT 116 | C57(0.96); C160(0.96); C129(1.01); C257(1.09) | LDD0623 | [8] |
| LDCM0307 | AC45 | HCT 116 | C129(0.86); C57(0.90); C160(0.92); C257(1.03) | LDD0624 | [8] |
| LDCM0308 | AC46 | HCT 116 | C129(0.88); C160(0.92); C57(0.93); C257(1.00) | LDD0625 | [8] |
| LDCM0309 | AC47 | HCT 116 | C57(0.86); C129(0.94); C160(0.95); C257(0.97) | LDD0626 | [8] |
| LDCM0310 | AC48 | HCT 116 | C129(0.63); C57(0.90); C160(0.95); C257(0.96) | LDD0627 | [8] |
| LDCM0311 | AC49 | HCT 116 | C57(0.87); C257(1.02); C129(1.02); C160(1.07) | LDD0628 | [8] |
| LDCM0312 | AC5 | HCT 116 | C129(0.64); C160(1.05); C257(1.16); C57(1.36) | LDD0629 | [8] |
| LDCM0313 | AC50 | HCT 116 | C160(0.91); C129(0.93); C257(0.95); C57(1.01) | LDD0630 | [8] |
| LDCM0314 | AC51 | HCT 116 | C57(0.82); C129(0.91); C160(0.96); C257(1.00) | LDD0631 | [8] |
| LDCM0315 | AC52 | HCT 116 | C57(0.79); C160(0.92); C257(0.96); C129(0.97) | LDD0632 | [8] |
| LDCM0316 | AC53 | HCT 116 | C57(0.78); C129(0.86); C160(0.93); C257(0.98) | LDD0633 | [8] |
| LDCM0317 | AC54 | HCT 116 | C57(0.74); C257(0.96); C160(1.03); C129(1.05) | LDD0634 | [8] |
| LDCM0318 | AC55 | HCT 116 | C129(0.94); C160(0.99); C57(1.04); C257(1.09) | LDD0635 | [8] |
| LDCM0319 | AC56 | HCT 116 | C57(0.86); C129(0.93); C160(1.00); C257(1.06) | LDD0636 | [8] |
| LDCM0320 | AC57 | HCT 116 | C257(0.89); C160(0.96); C57(1.16) | LDD0637 | [8] |
| LDCM0321 | AC58 | HCT 116 | C160(0.86); C257(0.88); C57(1.02) | LDD0638 | [8] |
| LDCM0322 | AC59 | HCT 116 | C160(0.89); C257(0.95); C57(1.10) | LDD0639 | [8] |
| LDCM0323 | AC6 | HCT 116 | C129(0.64); C160(0.88); C257(0.89) | LDD0640 | [8] |
| LDCM0324 | AC60 | HCT 116 | C160(0.84); C257(0.89); C57(0.97) | LDD0641 | [8] |
| LDCM0325 | AC61 | HCT 116 | C257(0.79); C160(0.89); C57(0.98) | LDD0642 | [8] |
| LDCM0326 | AC62 | HCT 116 | C257(0.76); C160(0.91); C57(1.10) | LDD0643 | [8] |
| LDCM0327 | AC63 | HCT 116 | C160(0.87); C57(0.91); C257(0.99) | LDD0644 | [8] |
| LDCM0328 | AC64 | HCT 116 | C257(0.93); C160(0.93); C57(1.05) | LDD0645 | [8] |
| LDCM0329 | AC65 | HCT 116 | C257(1.01); C160(1.01); C57(1.38) | LDD0646 | [8] |
| LDCM0330 | AC66 | HCT 116 | C160(0.95); C257(0.96); C57(1.29) | LDD0647 | [8] |
| LDCM0331 | AC67 | HCT 116 | C160(0.86); C257(0.92); C57(1.22) | LDD0648 | [8] |
| LDCM0332 | AC68 | HCT 116 | C129(0.84); C257(0.89); C160(0.96); C57(0.97) | LDD0649 | [8] |
| LDCM0333 | AC69 | HCT 116 | C129(0.80); C257(0.88); C160(0.94); C57(0.99) | LDD0650 | [8] |
| LDCM0334 | AC7 | HCT 116 | C129(0.66); C257(0.95); C160(0.98) | LDD0651 | [8] |
| LDCM0335 | AC70 | HCT 116 | C129(0.83); C257(0.95); C160(1.15); C57(1.47) | LDD0652 | [8] |
| LDCM0336 | AC71 | HCT 116 | C257(0.94); C129(0.95); C160(1.01); C57(1.04) | LDD0653 | [8] |
| LDCM0337 | AC72 | HCT 116 | C160(0.95); C257(0.96); C129(1.03); C57(1.09) | LDD0654 | [8] |
| LDCM0338 | AC73 | HCT 116 | C129(0.92); C160(0.99); C257(1.06); C57(1.20) | LDD0655 | [8] |
| LDCM0339 | AC74 | HCT 116 | C129(0.84); C257(1.01); C160(1.04); C57(1.15) | LDD0656 | [8] |
| LDCM0340 | AC75 | HCT 116 | C129(0.94); C160(1.04); C257(1.07); C57(1.18) | LDD0657 | [8] |
| LDCM0341 | AC76 | HCT 116 | C129(0.90); C257(0.97); C160(1.01); C57(1.17) | LDD0658 | [8] |
| LDCM0342 | AC77 | HCT 116 | C129(0.80); C257(0.88); C160(0.89); C57(1.03) | LDD0659 | [8] |
| LDCM0343 | AC78 | HCT 116 | C129(0.76); C160(0.86); C257(0.87); C57(0.96) | LDD0660 | [8] |
| LDCM0344 | AC79 | HCT 116 | C129(0.89); C160(0.99); C257(1.01); C57(1.07) | LDD0661 | [8] |
| LDCM0345 | AC8 | HCT 116 | C129(0.85); C160(0.90); C257(0.97) | LDD0662 | [8] |
| LDCM0346 | AC80 | HCT 116 | C129(0.87); C257(0.94); C160(0.99); C57(0.99) | LDD0663 | [8] |
| LDCM0347 | AC81 | HCT 116 | C257(0.98); C129(1.00); C57(1.02); C160(1.07) | LDD0664 | [8] |
| LDCM0348 | AC82 | HCT 116 | C129(0.96); C160(1.03); C257(1.05); C57(1.22) | LDD0665 | [8] |
| LDCM0349 | AC83 | HCT 116 | C257(0.91); C57(0.98); C160(1.02) | LDD0666 | [8] |
| LDCM0350 | AC84 | HCT 116 | C257(0.84); C160(0.97); C57(1.16) | LDD0667 | [8] |
| LDCM0351 | AC85 | HCT 116 | C257(0.78); C57(1.01); C160(1.04) | LDD0668 | [8] |
| LDCM0352 | AC86 | HCT 116 | C257(0.85); C160(1.04); C57(1.10) | LDD0669 | [8] |
| LDCM0353 | AC87 | HCT 116 | C257(0.75); C160(0.97); C57(1.33) | LDD0670 | [8] |
| LDCM0354 | AC88 | HCT 116 | C257(0.85); C160(1.00); C57(1.24) | LDD0671 | [8] |
| LDCM0355 | AC89 | HCT 116 | C257(0.77); C57(1.00); C160(1.23) | LDD0672 | [8] |
| LDCM0357 | AC90 | HCT 116 | C57(1.03); C257(1.07); C160(1.16) | LDD0674 | [8] |
| LDCM0358 | AC91 | HCT 116 | C257(0.88); C160(1.11); C57(1.17) | LDD0675 | [8] |
| LDCM0359 | AC92 | HCT 116 | C257(0.84); C160(1.17); C57(1.29) | LDD0676 | [8] |
| LDCM0360 | AC93 | HCT 116 | C257(0.82); C57(1.00); C160(1.06) | LDD0677 | [8] |
| LDCM0361 | AC94 | HCT 116 | C257(0.78); C57(1.08); C160(1.20) | LDD0678 | [8] |
| LDCM0362 | AC95 | HCT 116 | C257(0.69); C57(1.04); C160(1.17) | LDD0679 | [8] |
| LDCM0363 | AC96 | HCT 116 | C257(0.79); C57(1.07); C160(1.22) | LDD0680 | [8] |
| LDCM0364 | AC97 | HCT 116 | C257(0.74); C160(1.14); C57(1.43) | LDD0681 | [8] |
| LDCM0365 | AC98 | HCT 116 | C129(0.44); C257(0.65); C57(0.95); C160(0.99) | LDD0682 | [8] |
| LDCM0366 | AC99 | HCT 116 | C257(0.75); C129(0.77); C57(0.92); C160(1.20) | LDD0683 | [8] |
| LDCM0248 | AKOS034007472 | HCT 116 | C129(0.75); C160(0.91); C257(0.97) | LDD0565 | [8] |
| LDCM0356 | AKOS034007680 | HCT 116 | C129(0.61); C160(0.93); C257(0.96) | LDD0673 | [8] |
| LDCM0275 | AKOS034007705 | HCT 116 | C129(0.59); C257(0.87); C160(0.88) | LDD0592 | [8] |
| LDCM0156 | Aniline | NCI-H1299 | 12.06 | LDD0403 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C160(0.97); C257(0.90) | LDD0078 | [8] |
| LDCM0630 | CCW28-3 | 231MFP | C129(1.23) | LDD2214 | [25] |
| LDCM0108 | Chloroacetamide | HeLa | C257(0.00); C160(0.00) | LDD0222 | [16] |
| LDCM0367 | CL1 | HCT 116 | C129(0.73); C160(0.83); C257(1.18); C57(1.57) | LDD0684 | [8] |
| LDCM0368 | CL10 | HCT 116 | C160(0.77); C257(0.91); C129(0.96); C57(1.34) | LDD0685 | [8] |
| LDCM0369 | CL100 | HCT 116 | C129(0.68); C160(0.99); C257(1.21); C57(1.22) | LDD0686 | [8] |
| LDCM0370 | CL101 | HCT 116 | C129(0.57); C160(0.84); C257(0.88) | LDD0687 | [8] |
| LDCM0371 | CL102 | HCT 116 | C129(0.65); C160(0.90); C257(0.93) | LDD0688 | [8] |
| LDCM0372 | CL103 | HCT 116 | C129(0.75); C257(0.97); C160(1.01) | LDD0689 | [8] |
| LDCM0373 | CL104 | HCT 116 | C129(0.75); C160(0.89); C257(0.98) | LDD0690 | [8] |
| LDCM0374 | CL105 | HCT 116 | C129(0.65); C160(0.84); C257(0.85); C57(1.15) | LDD0691 | [8] |
| LDCM0375 | CL106 | HCT 116 | C129(0.63); C160(0.80); C257(0.92); C57(1.45) | LDD0692 | [8] |
| LDCM0376 | CL107 | HCT 116 | C129(0.73); C160(0.83); C257(0.98); C57(1.21) | LDD0693 | [8] |
| LDCM0377 | CL108 | HCT 116 | C129(0.77); C257(0.80); C160(1.06); C57(1.12) | LDD0694 | [8] |
| LDCM0378 | CL109 | HCT 116 | C129(0.73); C160(0.78); C257(0.87); C57(1.07) | LDD0695 | [8] |
| LDCM0379 | CL11 | HCT 116 | C160(0.92); C129(0.96); C257(1.18); C57(1.90) | LDD0696 | [8] |
| LDCM0380 | CL110 | HCT 116 | C129(0.68); C160(0.80); C257(0.84); C57(1.29) | LDD0697 | [8] |
| LDCM0381 | CL111 | HCT 116 | C129(0.81); C160(0.82); C257(0.86); C57(1.24) | LDD0698 | [8] |
| LDCM0382 | CL112 | HCT 116 | C129(0.85); C57(0.96); C160(1.01); C257(1.02) | LDD0699 | [8] |
| LDCM0383 | CL113 | HCT 116 | C129(0.60); C257(0.81); C160(0.95); C57(0.95) | LDD0700 | [8] |
| LDCM0384 | CL114 | HCT 116 | C129(0.73); C257(0.93); C160(0.96); C57(0.98) | LDD0701 | [8] |
| LDCM0385 | CL115 | HCT 116 | C129(0.63); C257(0.89); C160(0.97); C57(1.14) | LDD0702 | [8] |
| LDCM0386 | CL116 | HCT 116 | C129(0.57); C257(0.93); C160(0.97); C57(1.19) | LDD0703 | [8] |
| LDCM0387 | CL117 | HCT 116 | C129(0.89); C57(0.90); C160(0.99); C257(1.10) | LDD0704 | [8] |
| LDCM0388 | CL118 | HCT 116 | C129(0.71); C160(0.90); C57(0.95); C257(1.03) | LDD0705 | [8] |
| LDCM0389 | CL119 | HCT 116 | C160(0.91); C129(0.99); C257(1.03); C57(1.09) | LDD0706 | [8] |
| LDCM0390 | CL12 | HCT 116 | C129(0.87); C160(0.91); C257(1.33); C57(1.36) | LDD0707 | [8] |
| LDCM0391 | CL120 | HCT 116 | C129(0.77); C160(0.93); C57(0.98); C257(1.07) | LDD0708 | [8] |
| LDCM0392 | CL121 | HCT 116 | C257(0.70); C57(0.85); C129(0.92); C160(0.93) | LDD0709 | [8] |
| LDCM0393 | CL122 | HCT 116 | C57(0.74); C129(0.77); C160(0.84); C257(0.95) | LDD0710 | [8] |
| LDCM0394 | CL123 | HCT 116 | C57(0.69); C129(0.84); C257(0.89); C160(0.93) | LDD0711 | [8] |
| LDCM0395 | CL124 | HCT 116 | C57(0.80); C160(0.92); C129(0.94); C257(1.01) | LDD0712 | [8] |
| LDCM0396 | CL125 | HCT 116 | C57(0.99); C160(1.04); C257(1.10) | LDD0713 | [8] |
| LDCM0397 | CL126 | HCT 116 | C160(0.96); C257(1.00); C57(1.33) | LDD0714 | [8] |
| LDCM0398 | CL127 | HCT 116 | C57(0.84); C160(0.88); C257(1.03) | LDD0715 | [8] |
| LDCM0399 | CL128 | HCT 116 | C257(0.91); C160(1.02); C57(1.22) | LDD0716 | [8] |
| LDCM0400 | CL13 | HCT 116 | C160(0.90); C257(1.05); C129(1.30); C57(1.39) | LDD0717 | [8] |
| LDCM0401 | CL14 | HCT 116 | C160(0.85); C129(0.87); C257(1.18); C57(1.35) | LDD0718 | [8] |
| LDCM0402 | CL15 | HCT 116 | C160(0.78); C129(1.05); C257(1.09); C57(1.87) | LDD0719 | [8] |
| LDCM0403 | CL16 | HCT 116 | C257(0.67); C160(0.84); C129(1.00); C57(1.59) | LDD0720 | [8] |
| LDCM0404 | CL17 | HCT 116 | C257(0.60); C160(0.68); C129(0.78); C57(1.02) | LDD0721 | [8] |
| LDCM0405 | CL18 | HCT 116 | C257(0.51); C160(0.64); C129(0.72); C57(1.43) | LDD0722 | [8] |
| LDCM0406 | CL19 | HCT 116 | C257(0.55); C129(0.69); C160(0.79); C57(0.83) | LDD0723 | [8] |
| LDCM0407 | CL2 | HCT 116 | C160(0.79); C129(0.81); C57(1.05); C257(1.37) | LDD0724 | [8] |
| LDCM0408 | CL20 | HCT 116 | C257(0.65); C160(0.71); C129(0.75); C57(1.00) | LDD0725 | [8] |
| LDCM0409 | CL21 | HCT 116 | C257(0.61); C129(0.65); C160(0.69); C57(1.02) | LDD0726 | [8] |
| LDCM0410 | CL22 | HCT 116 | C257(0.76); C129(0.77); C160(0.82); C57(0.92) | LDD0727 | [8] |
| LDCM0411 | CL23 | HCT 116 | C257(0.72); C160(0.79); C129(0.89); C57(2.08) | LDD0728 | [8] |
| LDCM0412 | CL24 | HCT 116 | C129(0.78); C160(0.79); C257(0.65); C57(0.77) | LDD0729 | [8] |
| LDCM0413 | CL25 | HCT 116 | C129(0.67); C160(0.82); C257(0.68); C57(1.63) | LDD0730 | [8] |
| LDCM0414 | CL26 | HCT 116 | C129(1.05); C160(0.75); C257(0.71); C57(0.87) | LDD0731 | [8] |
| LDCM0415 | CL27 | HCT 116 | C129(0.64); C160(0.83); C257(0.72); C57(0.73) | LDD0732 | [8] |
| LDCM0416 | CL28 | HCT 116 | C129(0.80); C160(0.71); C257(0.56); C57(1.12) | LDD0733 | [8] |
| LDCM0417 | CL29 | HCT 116 | C129(0.68); C160(0.69); C257(0.61); C57(0.87) | LDD0734 | [8] |
| LDCM0418 | CL3 | HCT 116 | C129(1.84); C160(0.68); C257(1.39); C57(1.23) | LDD0735 | [8] |
| LDCM0419 | CL30 | HCT 116 | C129(0.82); C160(0.90); C257(0.60); C57(1.11) | LDD0736 | [8] |
| LDCM0420 | CL31 | HCT 116 | C129(0.76); C160(1.00); C257(0.78); C57(1.25) | LDD0737 | [8] |
| LDCM0421 | CL32 | HCT 116 | C129(0.79); C160(0.88); C257(0.74); C57(1.12) | LDD0738 | [8] |
| LDCM0422 | CL33 | HCT 116 | C129(0.92); C160(0.82); C257(0.72); C57(1.26) | LDD0739 | [8] |
| LDCM0423 | CL34 | HCT 116 | C129(0.91); C160(1.20); C257(0.69); C57(1.66) | LDD0740 | [8] |
| LDCM0424 | CL35 | HCT 116 | C129(1.04); C160(0.86); C257(0.71); C57(1.62) | LDD0741 | [8] |
| LDCM0425 | CL36 | HCT 116 | C129(1.07); C160(0.92); C257(0.82); C57(1.32) | LDD0742 | [8] |
| LDCM0426 | CL37 | HCT 116 | C129(1.73); C160(0.94); C257(0.82); C57(1.36) | LDD0743 | [8] |
| LDCM0428 | CL39 | HCT 116 | C129(1.29); C160(0.91); C257(0.88); C57(1.78) | LDD0745 | [8] |
| LDCM0429 | CL4 | HCT 116 | C129(0.62); C160(0.97); C257(1.02); C57(1.58) | LDD0746 | [8] |
| LDCM0430 | CL40 | HCT 116 | C129(0.76); C160(0.92); C257(0.81); C57(1.28) | LDD0747 | [8] |
| LDCM0431 | CL41 | HCT 116 | C129(0.79); C160(0.87); C257(0.75); C57(1.20) | LDD0748 | [8] |
| LDCM0432 | CL42 | HCT 116 | C129(1.29); C160(0.95); C257(0.91); C57(1.53) | LDD0749 | [8] |
| LDCM0433 | CL43 | HCT 116 | C129(1.27); C160(0.88); C257(0.82); C57(1.26) | LDD0750 | [8] |
| LDCM0434 | CL44 | HCT 116 | C129(0.98); C160(0.85); C257(0.70); C57(1.74) | LDD0751 | [8] |
| LDCM0435 | CL45 | HCT 116 | C129(1.03); C160(0.91); C257(0.72); C57(1.82) | LDD0752 | [8] |
| LDCM0436 | CL46 | HCT 116 | C160(1.15); C257(1.47); C57(1.62) | LDD0753 | [8] |
| LDCM0437 | CL47 | HCT 116 | C160(0.97); C257(1.33); C57(1.12) | LDD0754 | [8] |
| LDCM0438 | CL48 | HCT 116 | C160(0.95); C257(1.30); C57(0.86) | LDD0755 | [8] |
| LDCM0439 | CL49 | HCT 116 | C160(1.06); C257(1.22); C57(1.15) | LDD0756 | [8] |
| LDCM0440 | CL5 | HCT 116 | C129(0.75); C160(0.78); C257(1.21); C57(1.16) | LDD0757 | [8] |
| LDCM0441 | CL50 | HCT 116 | C160(1.11); C257(1.39); C57(0.86) | LDD0758 | [8] |
| LDCM0442 | CL51 | HCT 116 | C160(1.04); C257(1.25); C57(0.97) | LDD0759 | [8] |
| LDCM0443 | CL52 | HCT 116 | C160(0.99); C257(1.23); C57(1.21) | LDD0760 | [8] |
| LDCM0444 | CL53 | HCT 116 | C160(0.91); C257(1.29); C57(0.73) | LDD0761 | [8] |
| LDCM0445 | CL54 | HCT 116 | C160(0.96); C257(1.35); C57(0.74) | LDD0762 | [8] |
| LDCM0446 | CL55 | HCT 116 | C160(1.07); C257(1.35); C57(0.89) | LDD0763 | [8] |
| LDCM0447 | CL56 | HCT 116 | C160(0.89); C257(1.21); C57(0.85) | LDD0764 | [8] |
| LDCM0448 | CL57 | HCT 116 | C160(0.91); C257(1.18); C57(0.75) | LDD0765 | [8] |
| LDCM0449 | CL58 | HCT 116 | C160(0.99); C257(1.34); C57(0.89) | LDD0766 | [8] |
| LDCM0450 | CL59 | HCT 116 | C160(1.02); C257(1.26); C57(0.89) | LDD0767 | [8] |
| LDCM0451 | CL6 | HCT 116 | C129(0.95); C160(0.75); C257(1.00); C57(1.50) | LDD0768 | [8] |
| LDCM0452 | CL60 | HCT 116 | C160(0.96); C257(1.27); C57(0.91) | LDD0769 | [8] |
| LDCM0453 | CL61 | HCT 116 | C129(0.88); C160(0.89); C257(0.95); C57(1.07) | LDD0770 | [8] |
| LDCM0454 | CL62 | HCT 116 | C129(0.92); C160(0.93); C257(0.97); C57(1.11) | LDD0771 | [8] |
| LDCM0455 | CL63 | HCT 116 | C129(0.67); C160(0.85); C257(0.85); C57(1.18) | LDD0772 | [8] |
| LDCM0456 | CL64 | HCT 116 | C129(0.72); C160(0.83); C257(0.79); C57(1.77) | LDD0773 | [8] |
| LDCM0457 | CL65 | HCT 116 | C129(0.82); C160(0.96); C257(1.02); C57(1.04) | LDD0774 | [8] |
| LDCM0458 | CL66 | HCT 116 | C129(0.75); C160(0.87); C257(0.82); C57(1.10) | LDD0775 | [8] |
| LDCM0459 | CL67 | HCT 116 | C129(0.80); C160(0.81); C257(0.81); C57(1.30) | LDD0776 | [8] |
| LDCM0460 | CL68 | HCT 116 | C129(0.73); C160(0.76); C257(0.78); C57(1.16) | LDD0777 | [8] |
| LDCM0461 | CL69 | HCT 116 | C129(0.85); C160(0.72); C257(0.76); C57(1.29) | LDD0778 | [8] |
| LDCM0462 | CL7 | HCT 116 | C129(1.21); C160(0.92); C257(1.06); C57(1.18) | LDD0779 | [8] |
| LDCM0463 | CL70 | HCT 116 | C129(0.71); C160(0.81); C257(0.76); C57(1.15) | LDD0780 | [8] |
| LDCM0464 | CL71 | HCT 116 | C129(0.74); C160(0.82); C257(0.88); C57(1.10) | LDD0781 | [8] |
| LDCM0465 | CL72 | HCT 116 | C129(0.88); C160(0.92); C257(0.88); C57(1.20) | LDD0782 | [8] |
| LDCM0466 | CL73 | HCT 116 | C129(0.80); C160(0.80); C257(0.94); C57(1.23) | LDD0783 | [8] |
| LDCM0467 | CL74 | HCT 116 | C129(0.87); C160(0.81); C257(0.90); C57(1.10) | LDD0784 | [8] |
| LDCM0469 | CL76 | HCT 116 | C129(0.65); C160(0.83); C257(0.86); C57(1.21) | LDD0786 | [8] |
| LDCM0470 | CL77 | HCT 116 | C129(0.96); C160(0.88); C257(1.17); C57(1.02) | LDD0787 | [8] |
| LDCM0471 | CL78 | HCT 116 | C129(0.73); C160(0.95); C257(0.99); C57(1.11) | LDD0788 | [8] |
| LDCM0472 | CL79 | HCT 116 | C129(0.81); C160(0.83); C257(0.90); C57(1.21) | LDD0789 | [8] |
| LDCM0473 | CL8 | HCT 116 | C129(0.86); C160(0.73); C257(1.23); C57(1.20) | LDD0790 | [8] |
| LDCM0474 | CL80 | HCT 116 | C129(0.82); C160(0.86); C257(0.76); C57(1.16) | LDD0791 | [8] |
| LDCM0475 | CL81 | HCT 116 | C129(0.75); C160(0.89); C257(0.99); C57(1.48) | LDD0792 | [8] |
| LDCM0476 | CL82 | HCT 116 | C129(0.66); C160(0.96); C257(0.90); C57(1.43) | LDD0793 | [8] |
| LDCM0477 | CL83 | HCT 116 | C129(0.63); C160(0.88); C257(0.92); C57(1.35) | LDD0794 | [8] |
| LDCM0478 | CL84 | HCT 116 | C129(0.69); C160(1.06); C257(0.93); C57(1.30) | LDD0795 | [8] |
| LDCM0479 | CL85 | HCT 116 | C129(0.76); C160(0.91); C257(0.90); C57(1.40) | LDD0796 | [8] |
| LDCM0480 | CL86 | HCT 116 | C129(0.84); C160(0.96); C257(1.01); C57(1.78) | LDD0797 | [8] |
| LDCM0481 | CL87 | HCT 116 | C129(0.67); C160(0.85); C257(0.96); C57(1.24) | LDD0798 | [8] |
| LDCM0482 | CL88 | HCT 116 | C129(0.60); C160(0.84); C257(0.91); C57(1.20) | LDD0799 | [8] |
| LDCM0483 | CL89 | HCT 116 | C129(0.72); C160(1.10); C257(1.02); C57(1.49) | LDD0800 | [8] |
| LDCM0484 | CL9 | HCT 116 | C129(0.57); C160(0.69); C257(1.18); C57(1.31) | LDD0801 | [8] |
| LDCM0485 | CL90 | HCT 116 | C129(1.07); C160(1.05); C257(1.04); C57(1.11) | LDD0802 | [8] |
| LDCM0486 | CL91 | HCT 116 | C129(0.77); C160(1.06); C257(1.14); C57(1.03) | LDD0803 | [8] |
| LDCM0487 | CL92 | HCT 116 | C129(0.89); C160(1.03); C257(1.27); C57(1.05) | LDD0804 | [8] |
| LDCM0488 | CL93 | HCT 116 | C129(0.82); C160(1.10); C257(1.28); C57(1.59) | LDD0805 | [8] |
| LDCM0489 | CL94 | HCT 116 | C129(0.62); C160(0.88); C257(1.03); C57(1.19) | LDD0806 | [8] |
| LDCM0490 | CL95 | HCT 116 | C129(0.57); C160(0.97); C257(1.08); C57(1.16) | LDD0807 | [8] |
| LDCM0491 | CL96 | HCT 116 | C129(0.71); C160(0.97); C257(0.99); C57(1.26) | LDD0808 | [8] |
| LDCM0492 | CL97 | HCT 116 | C129(0.76); C160(0.91); C257(0.90); C57(1.29) | LDD0809 | [8] |
| LDCM0493 | CL98 | HCT 116 | C129(0.67); C160(0.99); C257(1.05); C57(1.03) | LDD0810 | [8] |
| LDCM0494 | CL99 | HCT 116 | C129(0.77); C160(0.96); C257(1.06); C57(1.15) | LDD0811 | [8] |
| LDCM0495 | E2913 | HEK-293T | C160(0.87) | LDD1698 | [26] |
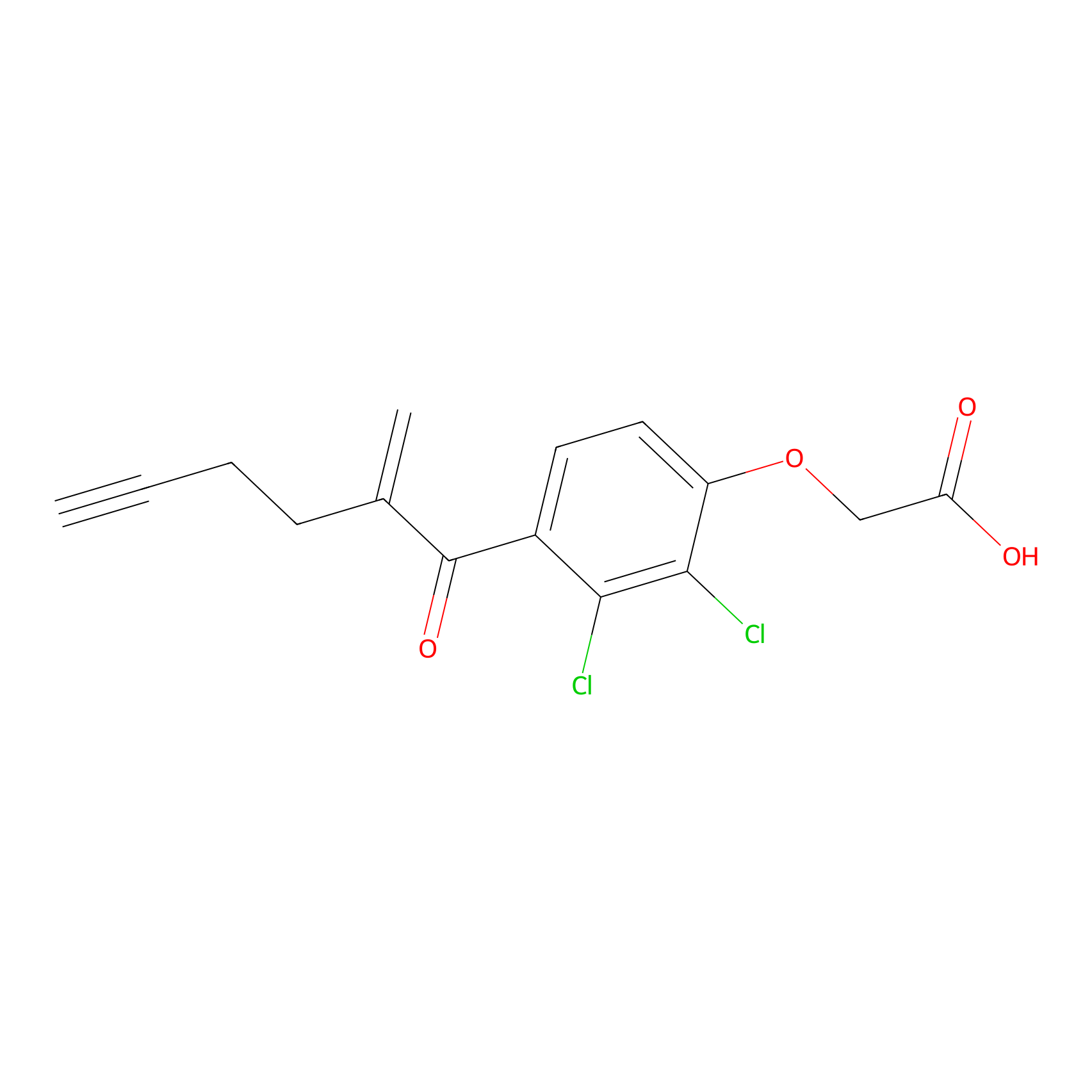
| LDCM0175 | Ethacrynic acid | HeLa | N.A. | LDD0440 | [7] |
| LDCM0625 | F8 | Ramos | C257(1.56); C160(1.33); C129(0.73) | LDD2187 | [27] |
| LDCM0572 | Fragment10 | Ramos | C257(2.97); C160(5.42); C129(0.79) | LDD2189 | [27] |
| LDCM0573 | Fragment11 | Ramos | C257(0.28); C129(0.08) | LDD2190 | [27] |
| LDCM0574 | Fragment12 | Ramos | C257(2.35); C160(11.15); C129(0.40) | LDD2191 | [27] |
| LDCM0575 | Fragment13 | Ramos | C257(1.23); C160(1.43); C129(0.53) | LDD2192 | [27] |
| LDCM0576 | Fragment14 | Ramos | C257(9.09); C160(20.00); C129(3.02) | LDD2193 | [27] |
| LDCM0579 | Fragment20 | Ramos | C257(2.63); C160(10.92) | LDD2194 | [27] |
| LDCM0580 | Fragment21 | Ramos | C257(1.78); C160(3.72); C129(0.87) | LDD2195 | [27] |
| LDCM0582 | Fragment23 | Ramos | C257(0.98); C160(0.65); C129(0.53) | LDD2196 | [27] |
| LDCM0578 | Fragment27 | Ramos | C257(1.18); C160(0.89); C129(0.70) | LDD2197 | [27] |
| LDCM0586 | Fragment28 | Ramos | C257(1.69); C160(0.86); C129(0.49) | LDD2198 | [27] |
| LDCM0588 | Fragment30 | Ramos | C257(1.25); C160(1.62); C129(0.45) | LDD2199 | [27] |
| LDCM0589 | Fragment31 | Ramos | C257(1.48); C160(1.56); C129(0.74) | LDD2200 | [27] |
| LDCM0590 | Fragment32 | Ramos | C257(3.32); C160(4.20); C129(0.68) | LDD2201 | [27] |
| LDCM0468 | Fragment33 | HCT 116 | C129(0.80); C160(0.83); C257(0.72); C57(1.43) | LDD0785 | [8] |
| LDCM0596 | Fragment38 | Ramos | C257(2.06); C160(2.39); C129(0.74) | LDD2203 | [27] |
| LDCM0566 | Fragment4 | Ramos | C257(4.31); C160(7.40); C129(1.02) | LDD2184 | [27] |
| LDCM0427 | Fragment51 | HCT 116 | C129(1.10); C160(1.05); C257(0.73); C57(1.57) | LDD0744 | [8] |
| LDCM0610 | Fragment52 | Ramos | C257(1.21); C160(2.66); C129(0.46) | LDD2204 | [27] |
| LDCM0614 | Fragment56 | Ramos | C257(0.81); C160(1.01); C129(0.21) | LDD2205 | [27] |
| LDCM0569 | Fragment7 | Ramos | C257(3.95); C160(5.85); C129(1.61) | LDD2186 | [27] |
| LDCM0571 | Fragment9 | Ramos | C257(2.93); C160(17.20); C129(0.98) | LDD2188 | [27] |
| LDCM0015 | HNE | MDA-MB-231 | C257(1.25); C129(0.93) | LDD0346 | [27] |
| LDCM0107 | IAA | HeLa | C257(0.00); C160(0.00) | LDD0221 | [16] |
| LDCM0179 | JZ128 | PC-3 | C257(0.00); C129(0.00) | LDD0462 | [5] |
| LDCM0022 | KB02 | HEK-293T | C257(0.92); C160(0.70); C129(0.98) | LDD1492 | [26] |
| LDCM0023 | KB03 | HEK-293T | C257(0.96); C160(0.77); C129(1.02) | LDD1497 | [26] |
| LDCM0024 | KB05 | SKMEL24 | C160(2.83) | LDD3323 | [28] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [16] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C160(1.03) | LDD2099 | [6] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C160(0.83) | LDD2107 | [6] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C160(1.00) | LDD2109 | [6] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C160(0.97) | LDD2125 | [6] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C160(0.96) | LDD2140 | [6] |
| LDCM0551 | Nucleophilic fragment 5b | MDA-MB-231 | C160(0.85) | LDD2145 | [6] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C257(1.19); C129(1.10); C257(1.07); C257(1.02) | LDD2206 | [29] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C129(1.37); C57(1.16); C257(0.98); C257(0.79) | LDD2207 | [29] |
| LDCM0131 | RA190 | MM1.R | C160(1.09) | LDD0304 | [30] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Leucine-rich repeat serine/threonine-protein kinase 2 (LRRK2) | TKL Ser/Thr protein kinase family | Q5S007 | |||
The Drug(s) Related To This Target
Approved
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Clodronate | Small molecular drug | D00HNB | |||
| Clodronic Acid | Small molecular drug | DB00720 | |||
| Etidronic Acid | Small molecular drug | DB01077 | |||
Investigative
References