Details of the Target
General Information of Target
| Target ID | LDTP10775 | |||||
|---|---|---|---|---|---|---|
| Target Name | Endoplasmic reticulum-Golgi intermediate compartment protein 2 (ERGIC2) | |||||
| Gene Name | ERGIC2 | |||||
| Gene ID | 51290 | |||||
| Synonyms |
ERV41; PTX1; Endoplasmic reticulum-Golgi intermediate compartment protein 2 |
|||||
| 3D Structure | ||||||
| Sequence |
MRLLGAAAVAALGRGRAPASLGWQRKQVNWKACRWSSSGVIPNEKIRNIGISAHIDSGKT
TLTERVLYYTGRIAKMHEVKGKDGVGAVMDSMELERQRGITIQSAATYTMWKDVNINIID TPGHVDFTIEVERALRVLDGAVLVLCAVGGVQCQTMTVNRQMKRYNVPFLTFINKLDRMG SNPARALQQMRSKLNHNAAFMQIPMGLEGNFKGIVDLIEERAIYFDGDFGQIVRYGEIPA ELRAAATDHRQELIECVANSDEQLGEMFLEEKIPSISDLKLAIRRATLKRSFTPVFLGSA LKNKGVQPLLDAVLEYLPNPSEVQNYAILNKEDDSKEKTKILMNSSRDNSHPFVGLAFKL EVGRFGQLTYVRSYQGELKKGDTIYNTRTRKKVRLQRLARMHADMMEDVEEVYAGDICAL FGIDCASGDTFTDKANSGLSMESIHVPDPVISIAMKPSNKNDLEKFSKGIGRFTREDPTF KVYFDTENKETVISGMGELHLEIYAQRLEREYGCPCITGKPKVAFRETITAPVPFDFTHK KQSGGAGQYGKVIGVLEPLDPEDYTKLEFSDETFGSNIPKQFVPAVEKGFLDACEKGPLS GHKLSGLRFVLQDGAHHMVDSNEISFIRAGEGALKQALANATLCILEPIMAVEVVAPNEF QGQVIAGINRRHGVITGQDGVEDYFTLYADVPLNDMFGYSTELRSCTEGKGEYTMEYSRY QPCLPSTQEDVINKYLEATGQLPVKKGKAKN |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
ERGIC family
|
|||||
| Subcellular location |
Endoplasmic reticulum-Golgi intermediate compartment membrane
|
|||||
| Function | Possible role in transport between endoplasmic reticulum and Golgi. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
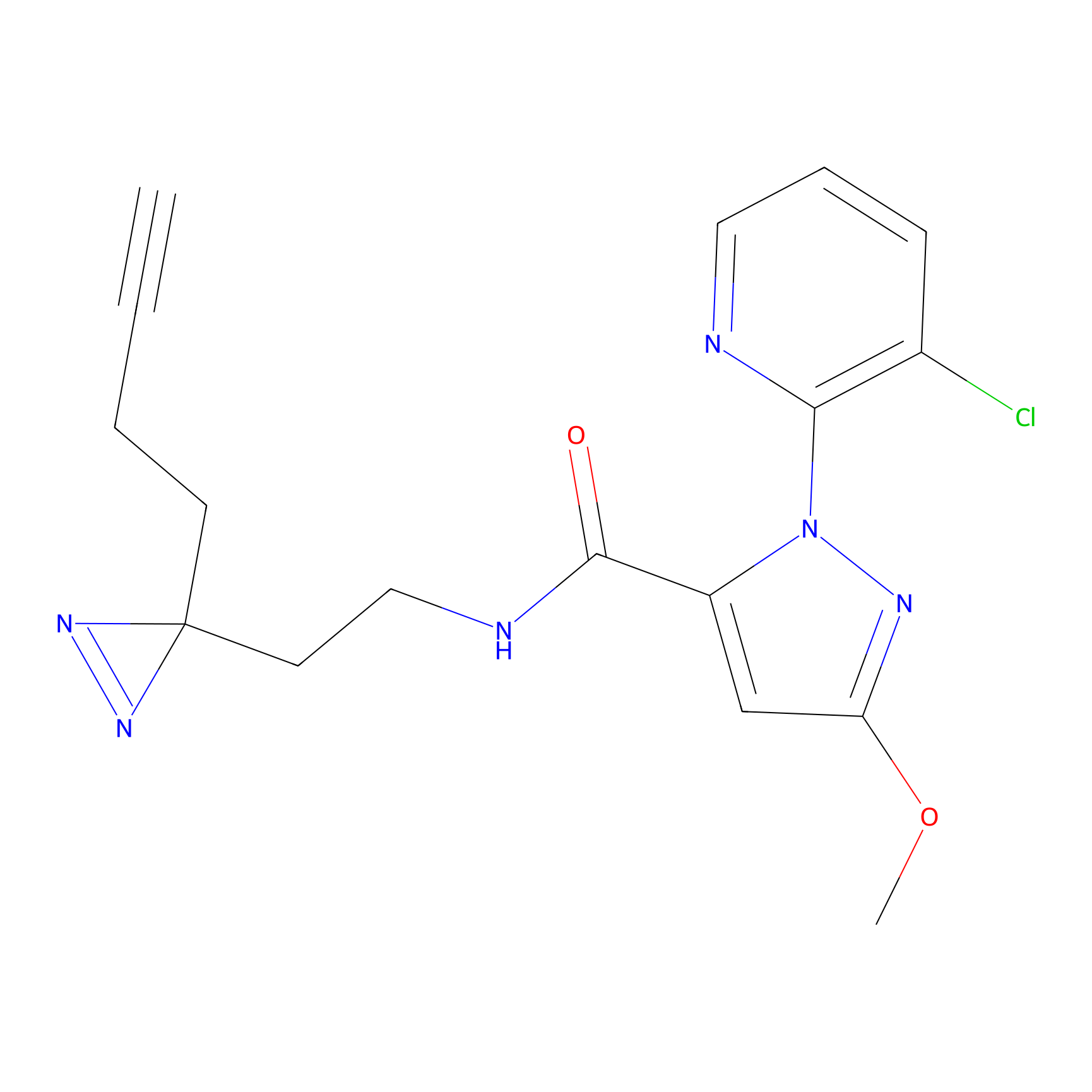
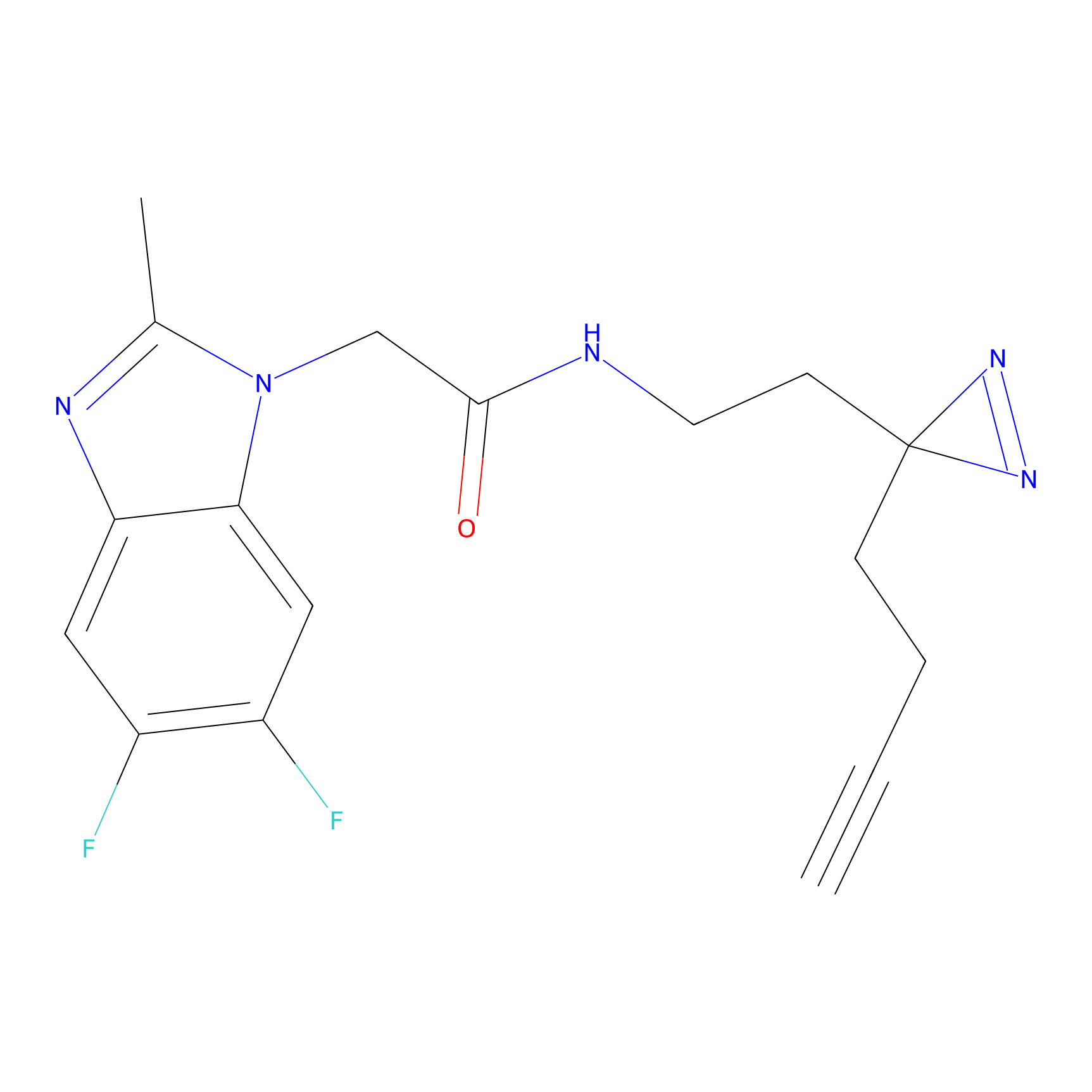
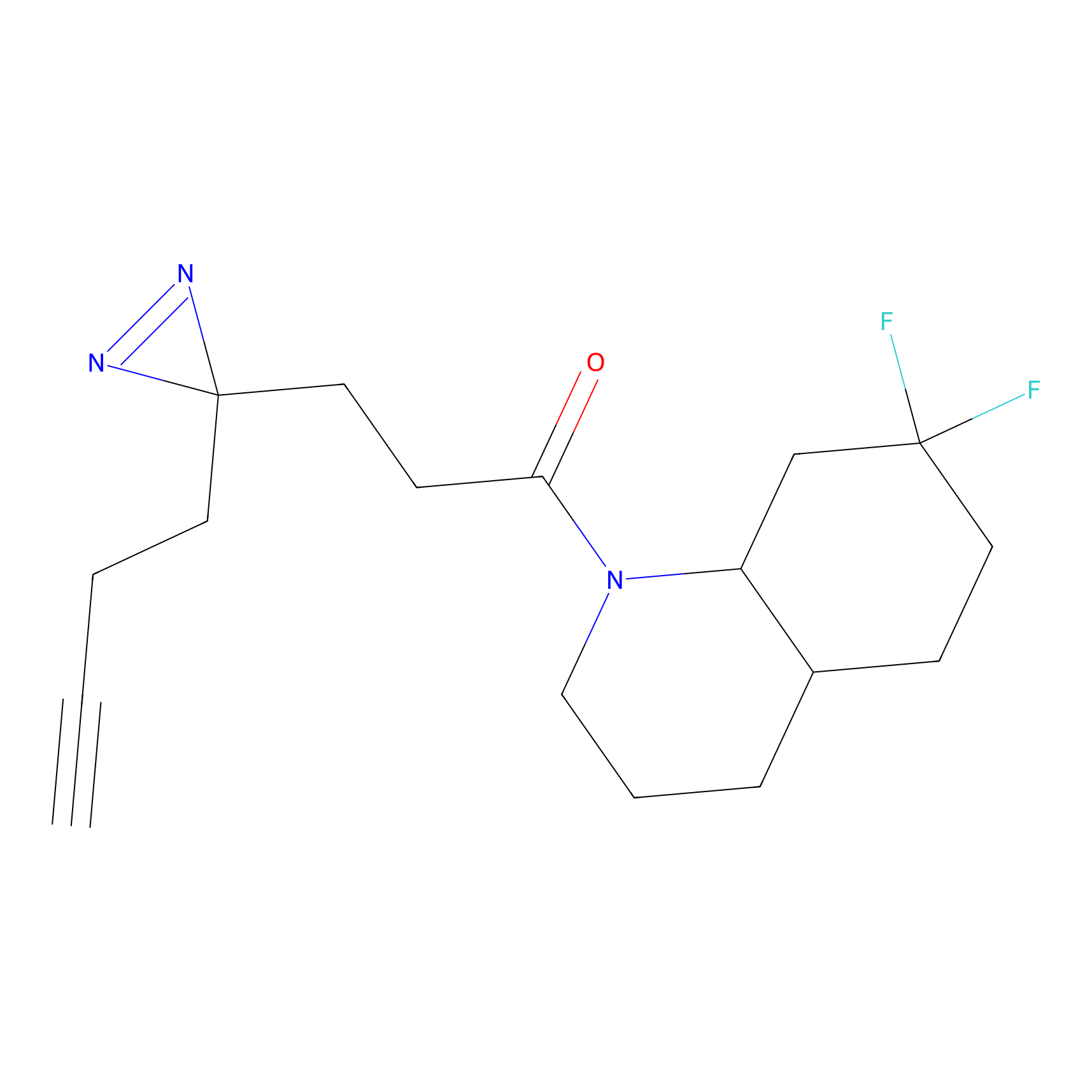
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
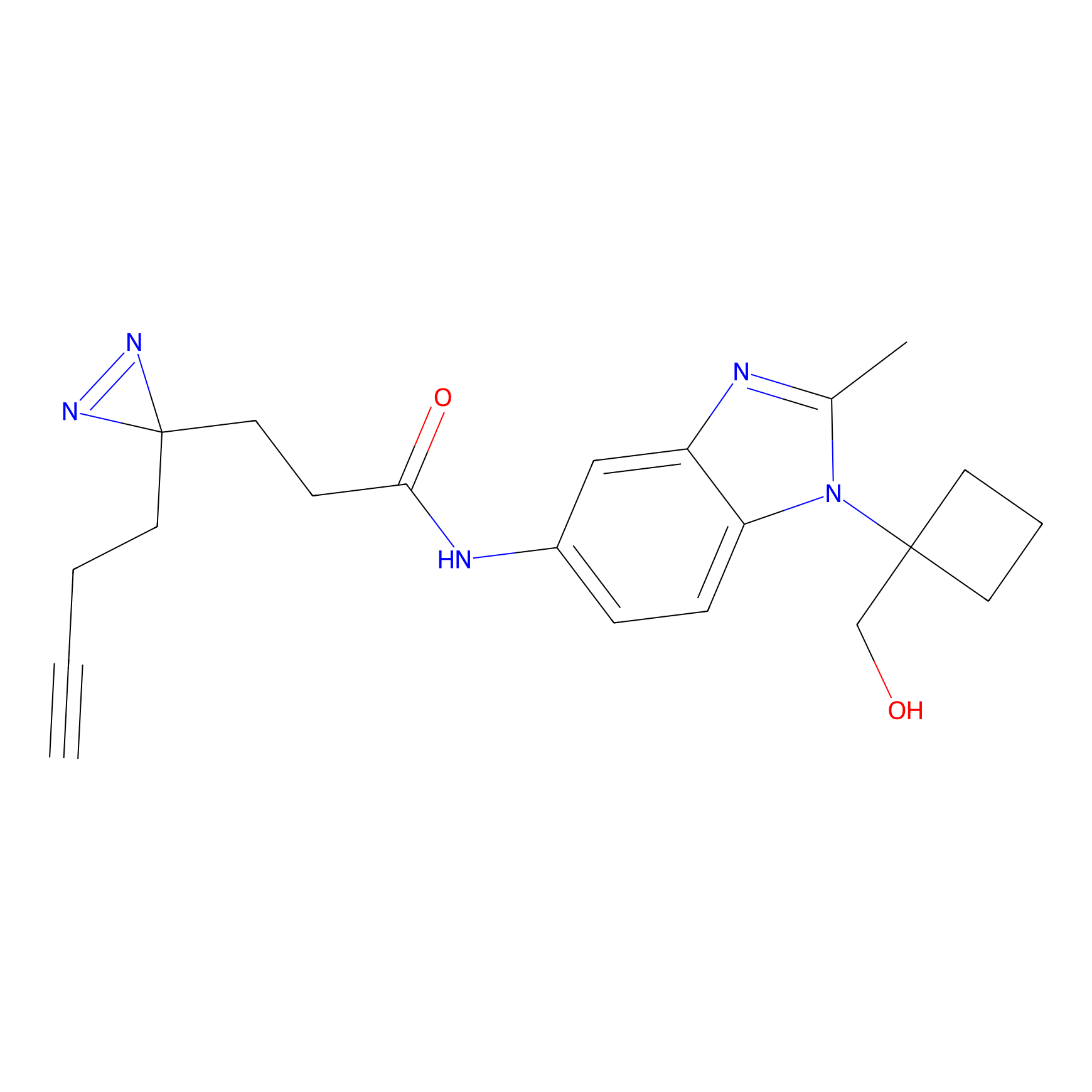
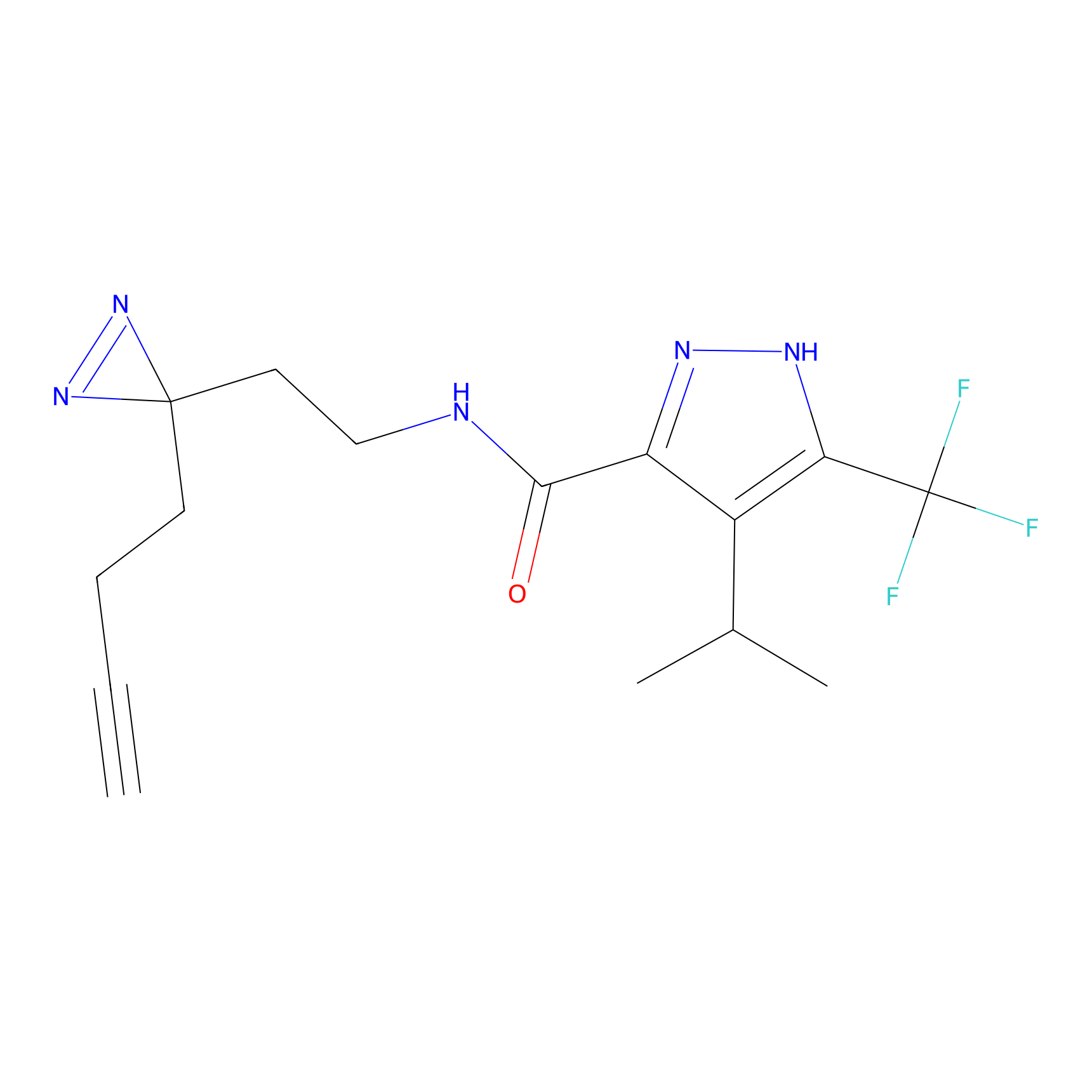
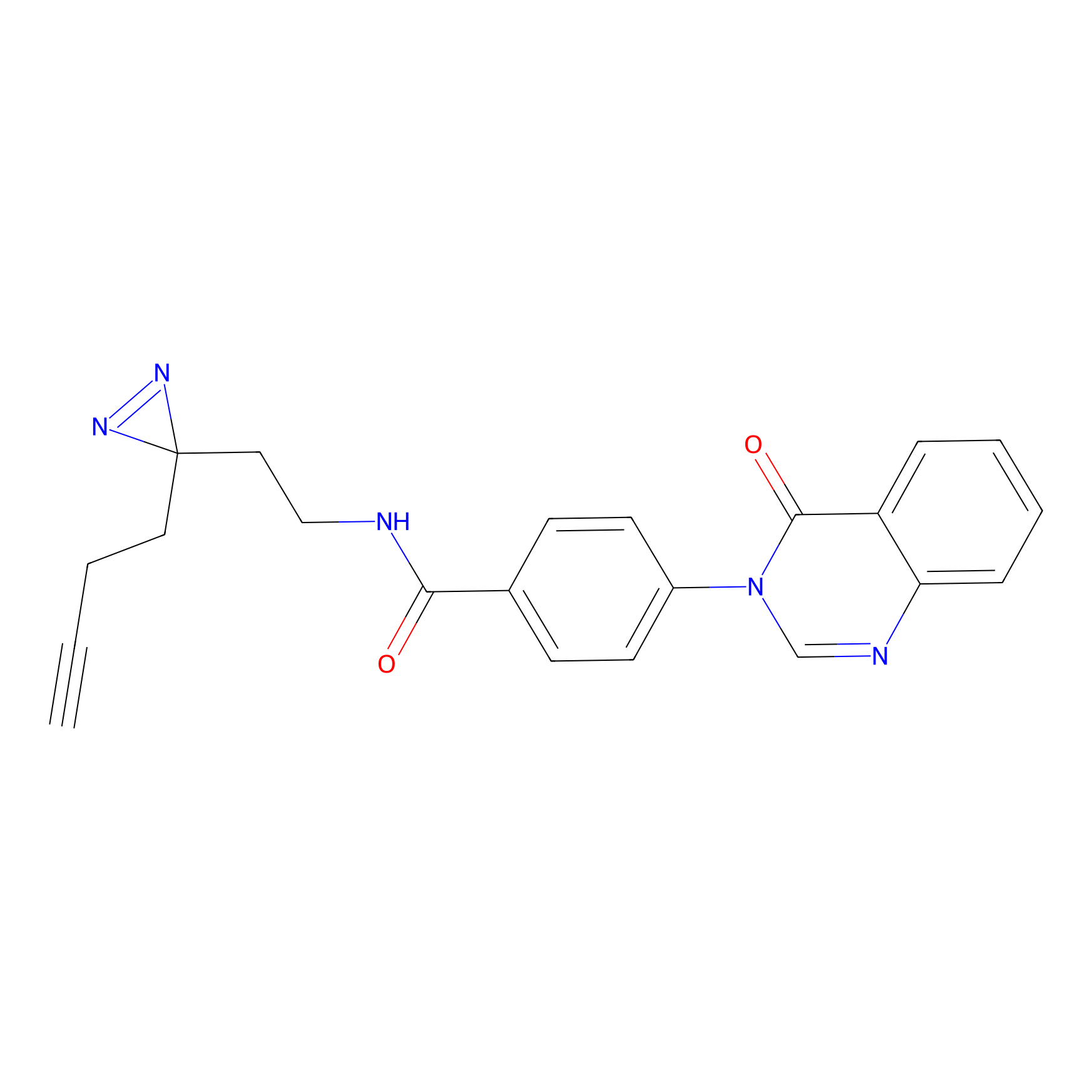
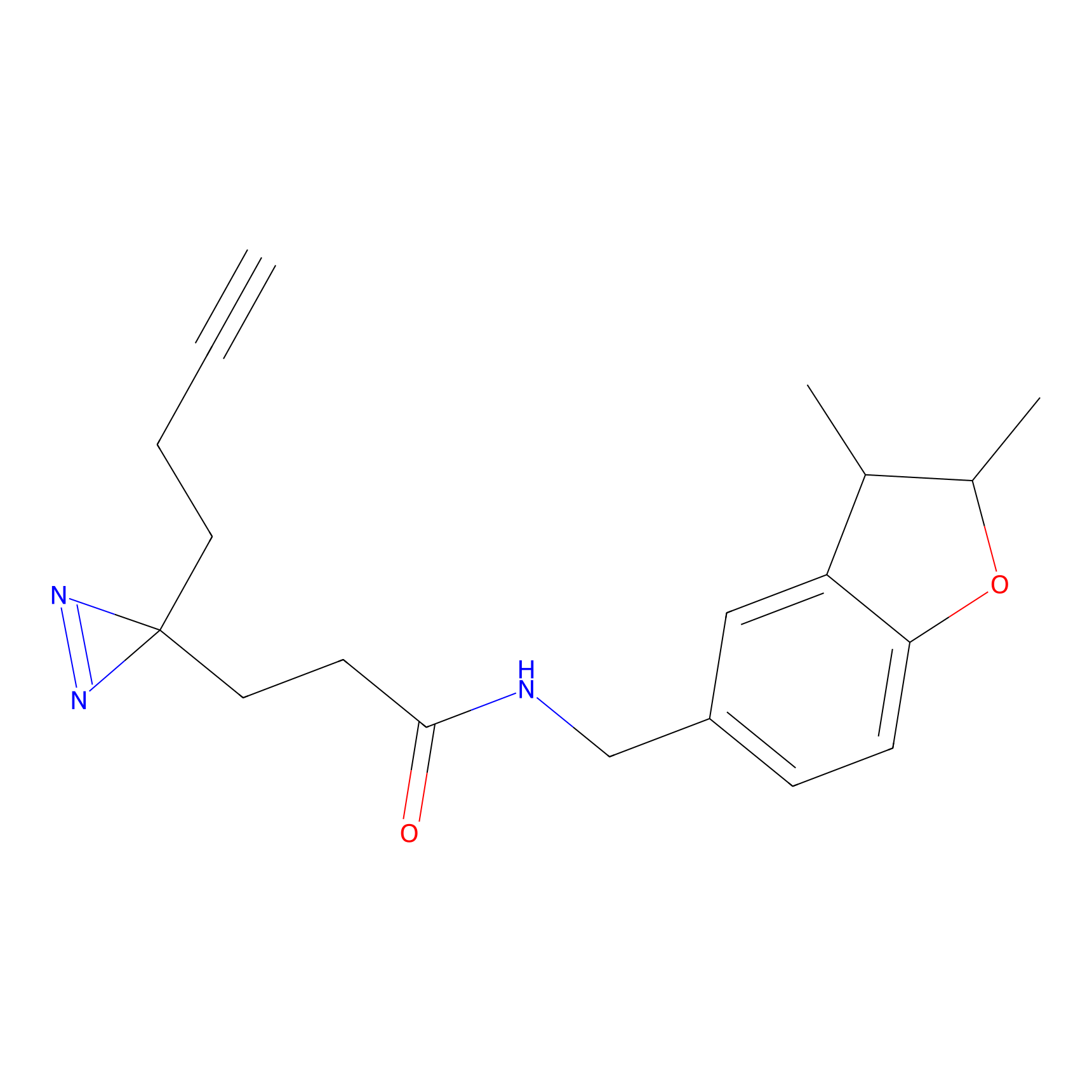
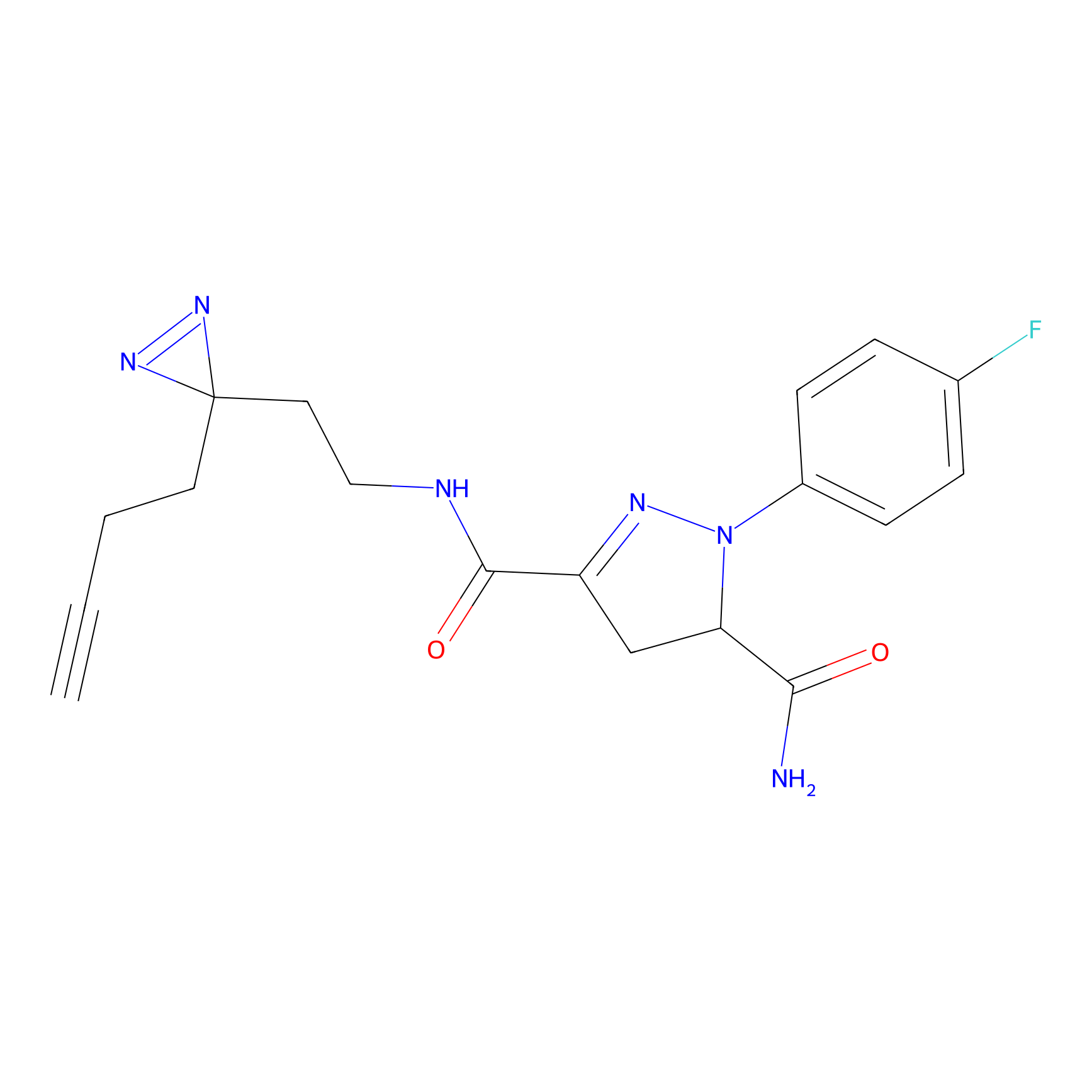
YN-1 Probe Info |
 |
100.00 | LDD0444 | [1] | |
|
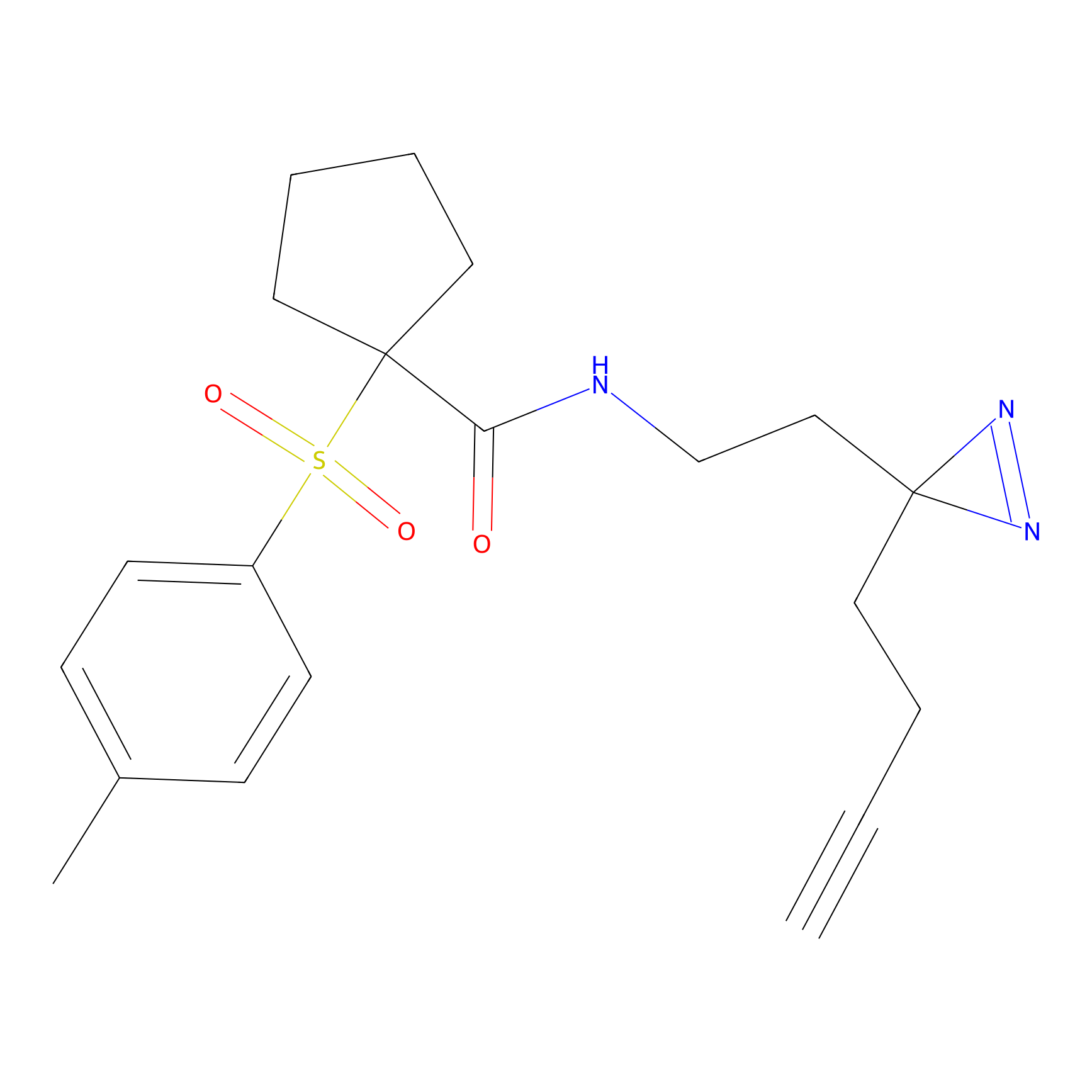
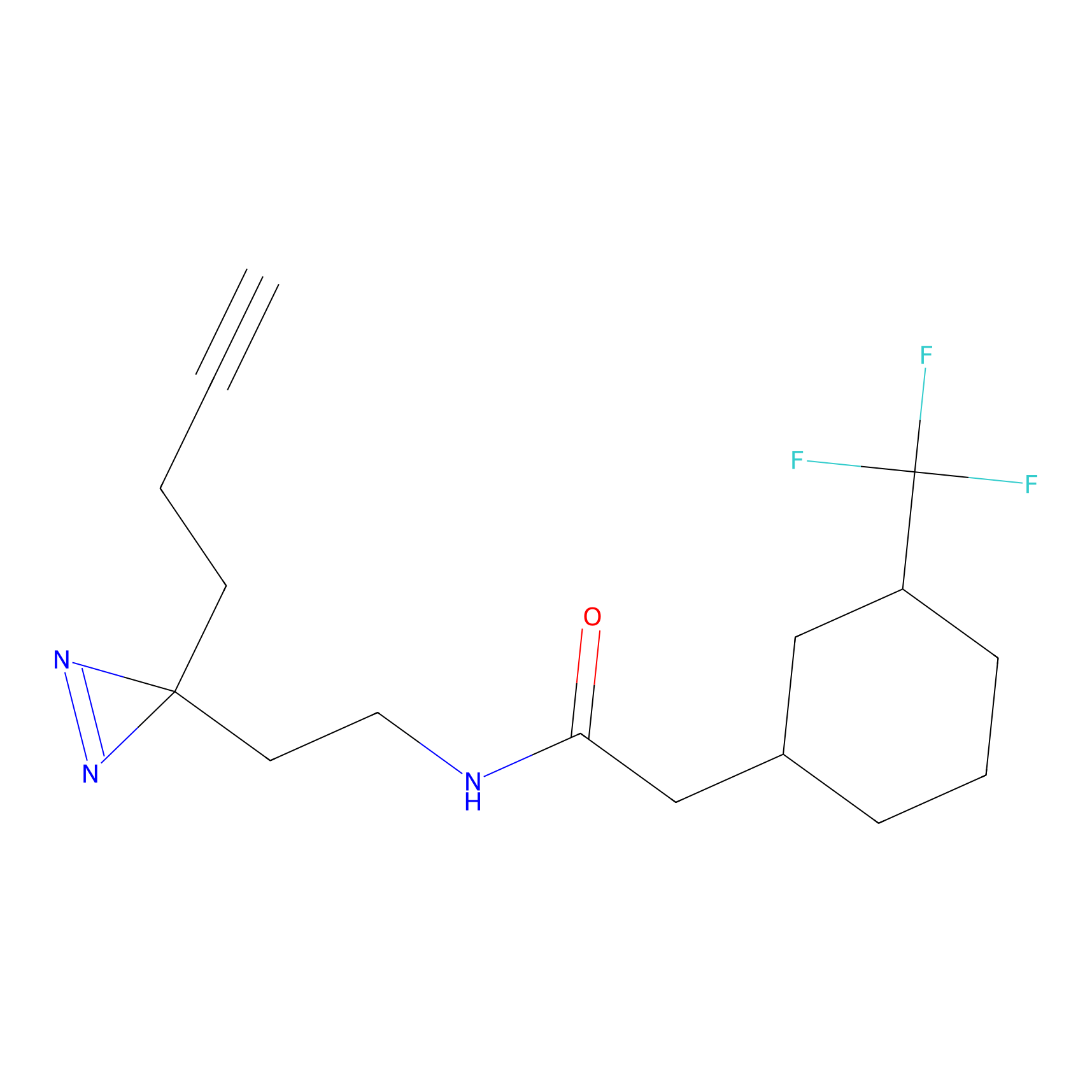
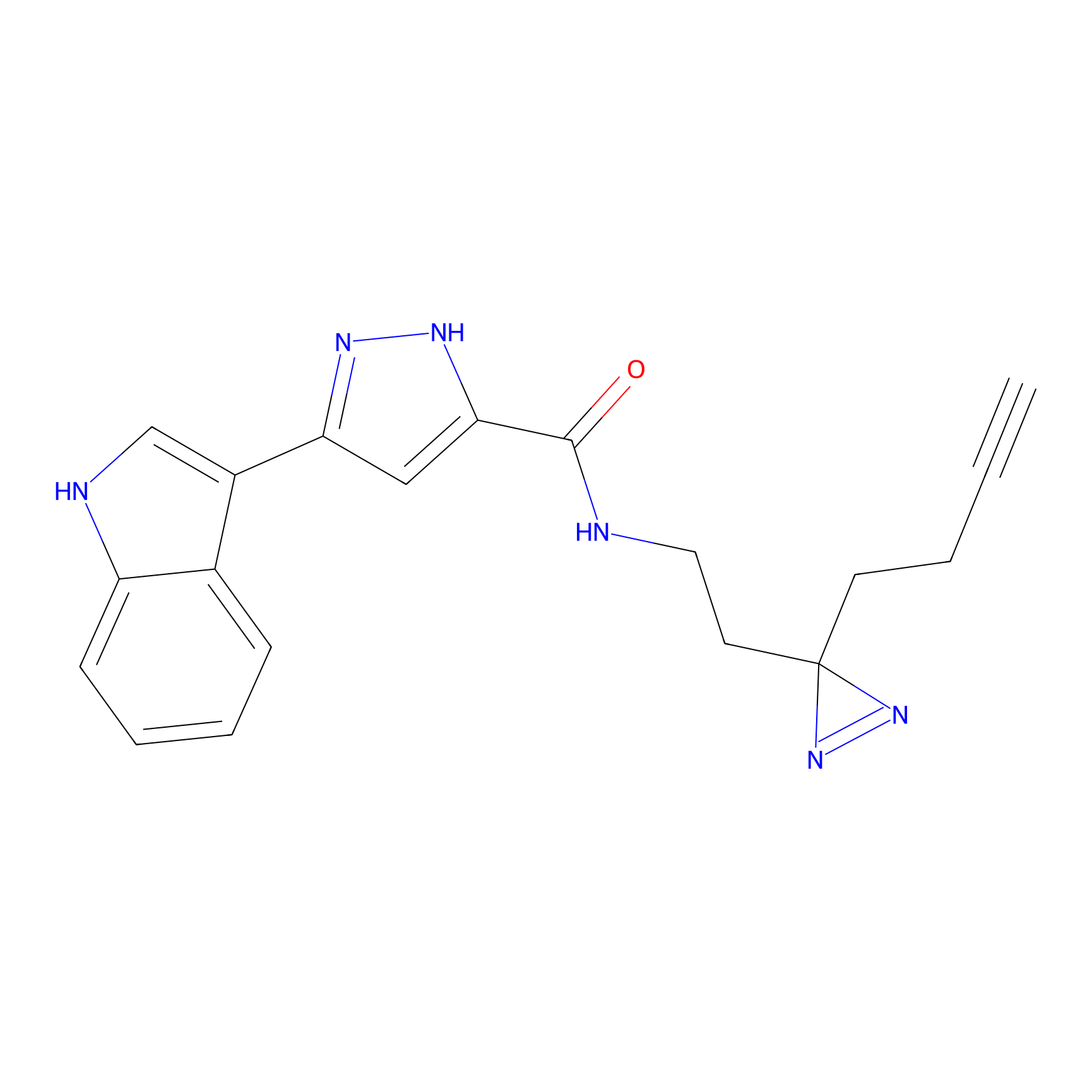
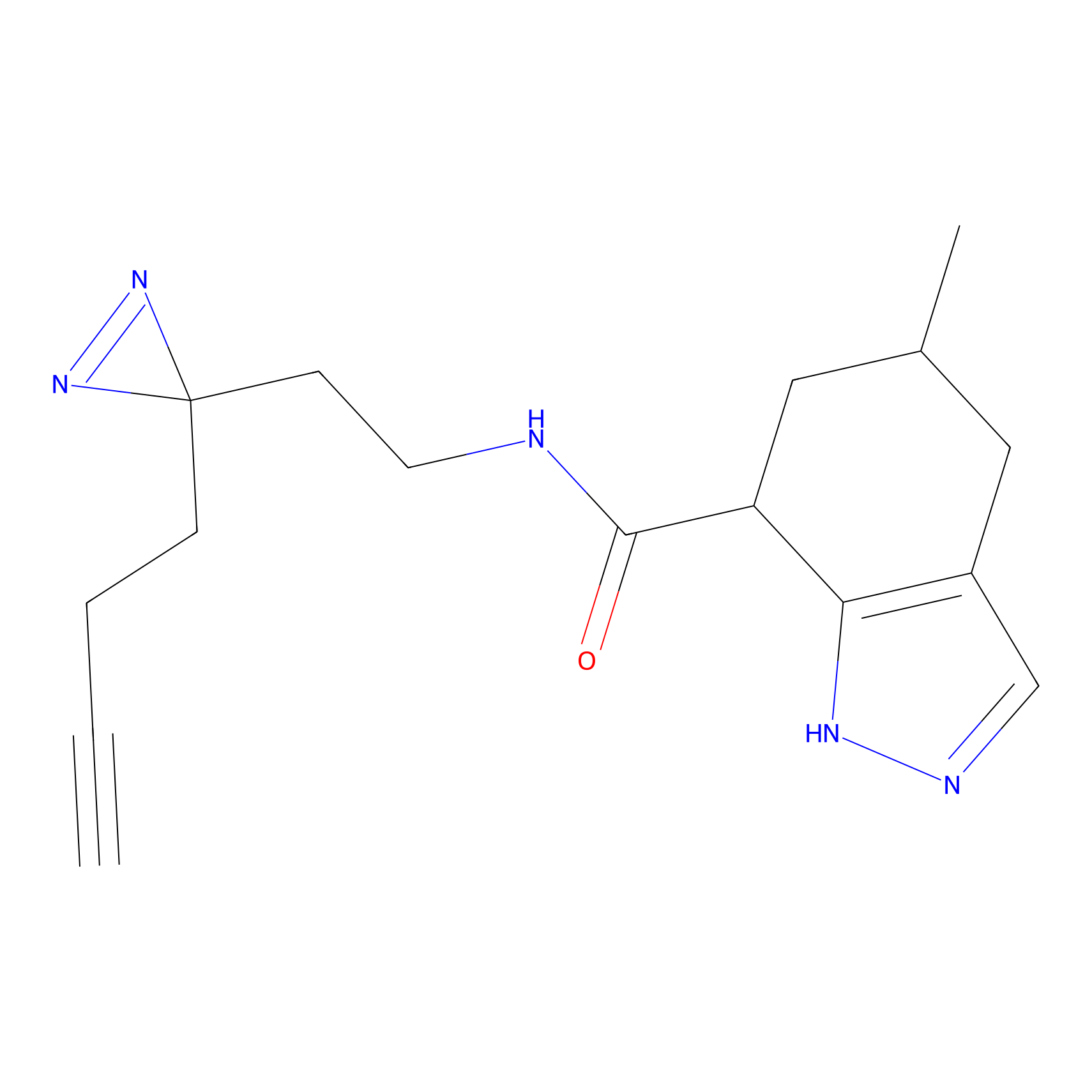
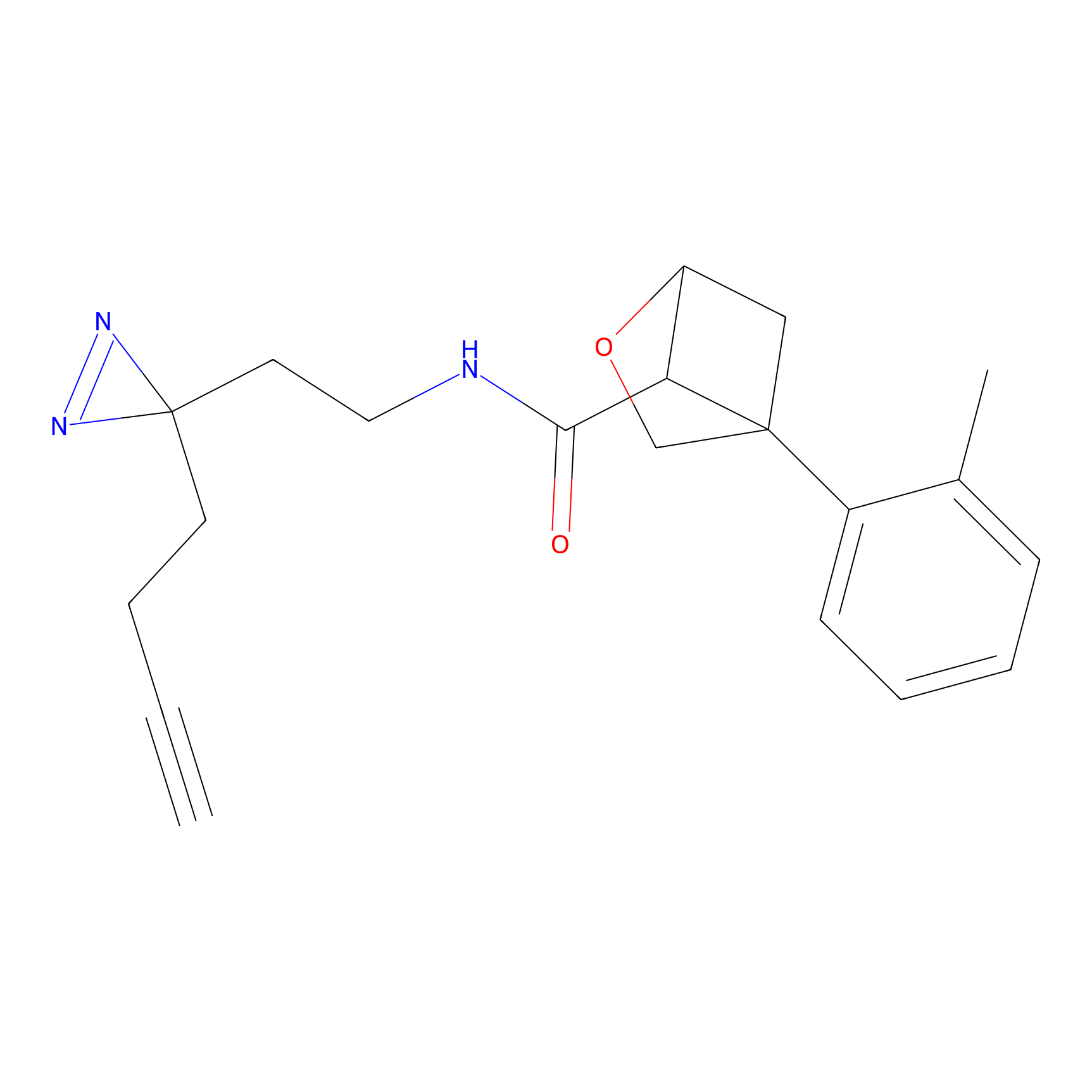
m-APA Probe Info |
 |
11.44 | LDD0403 | [2] | |
|
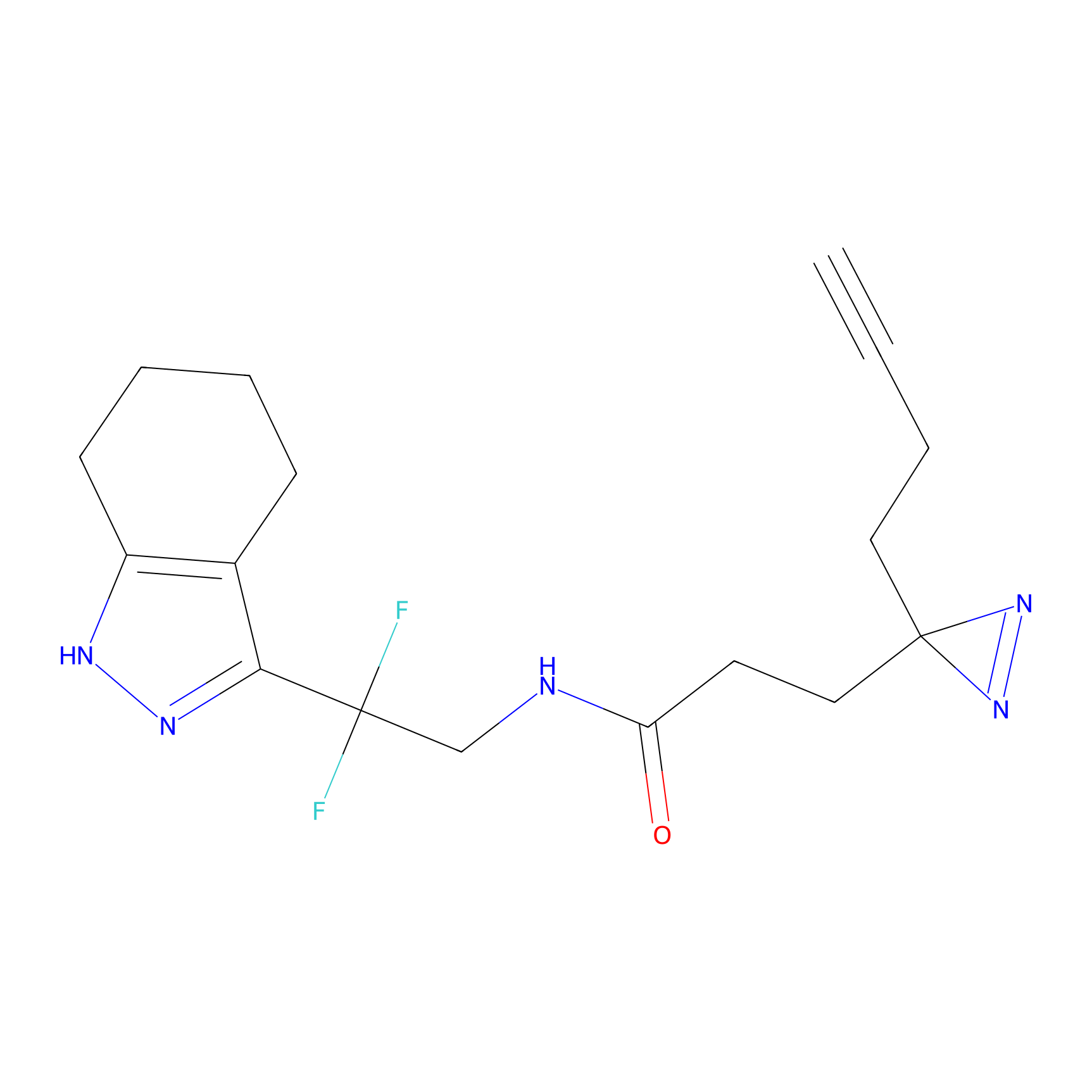
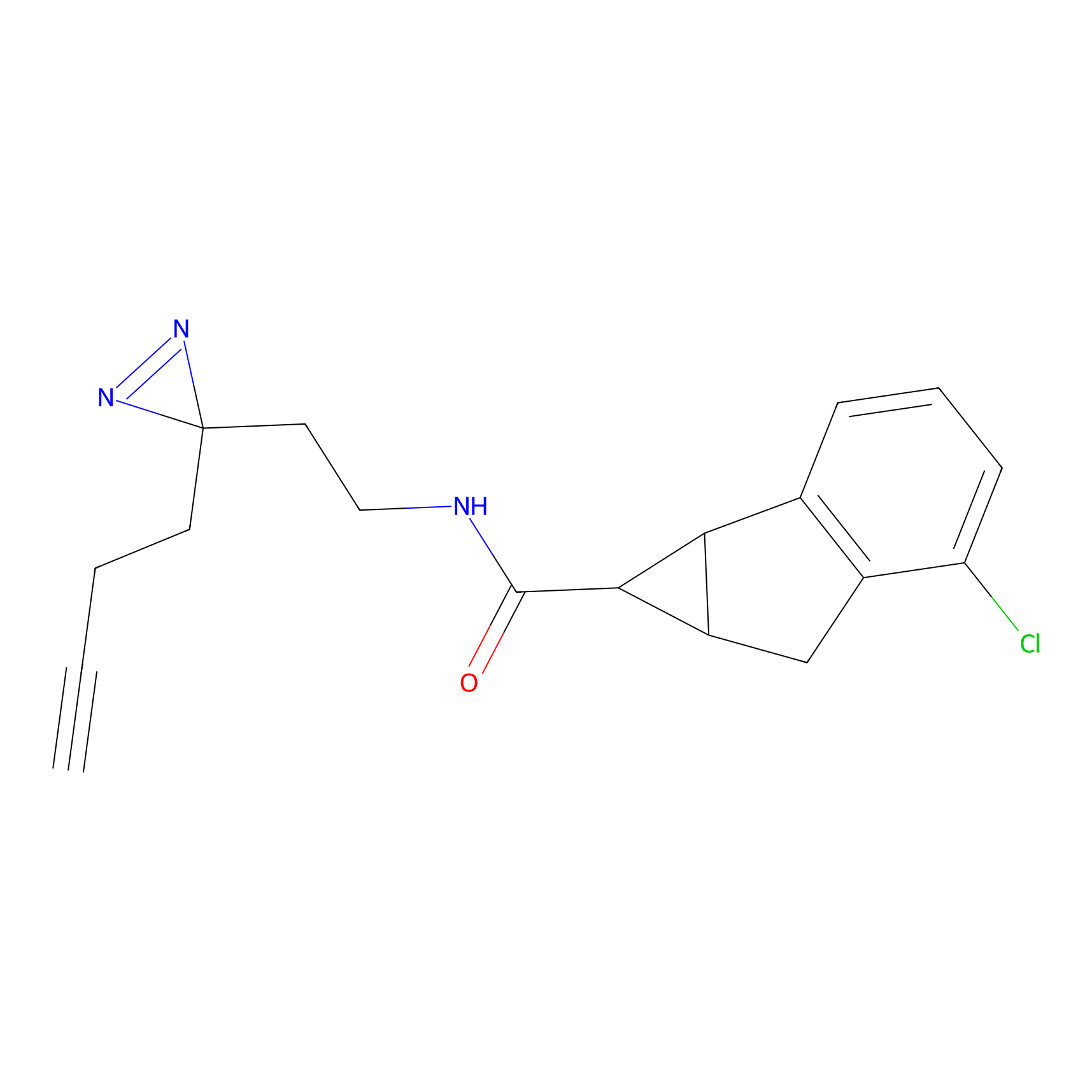
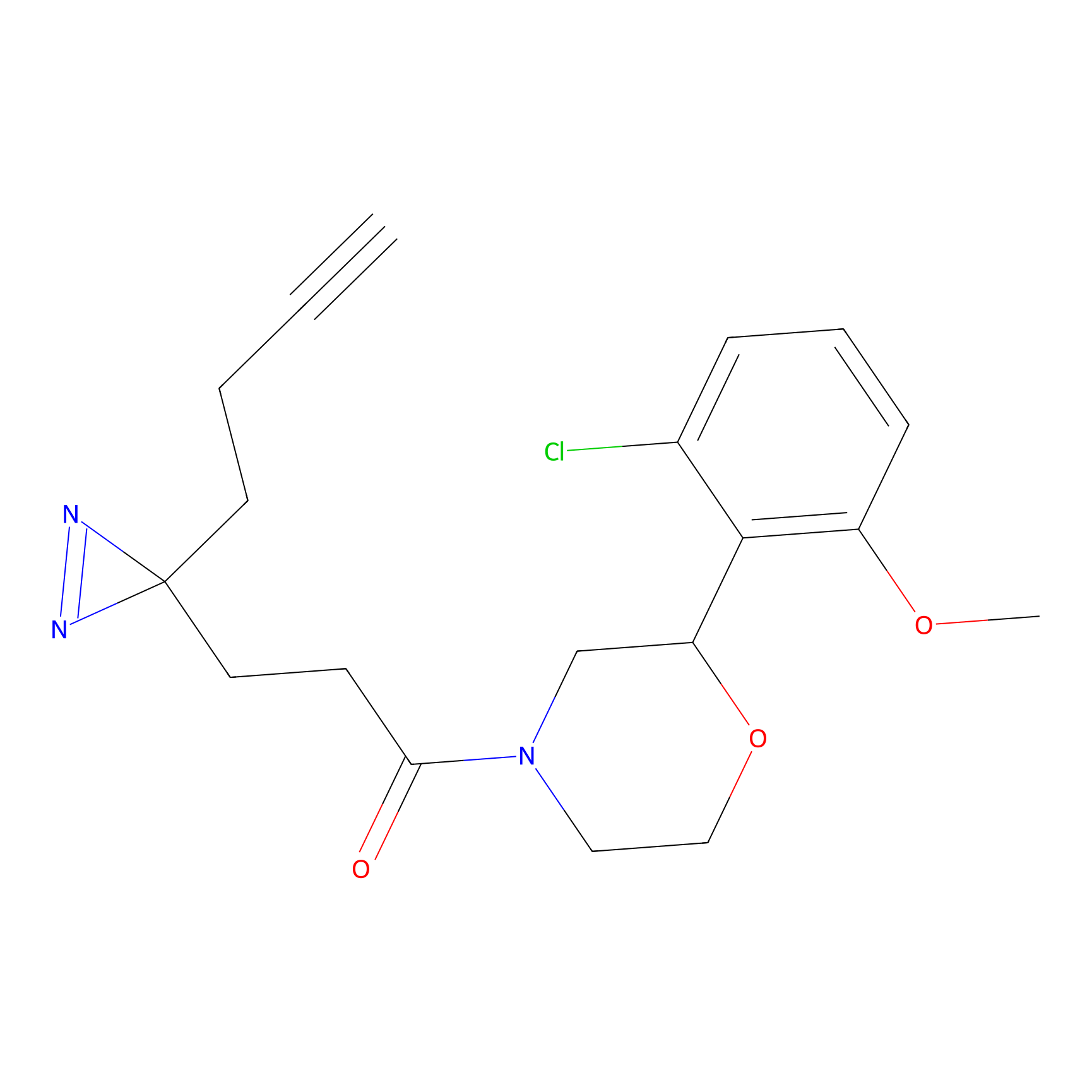
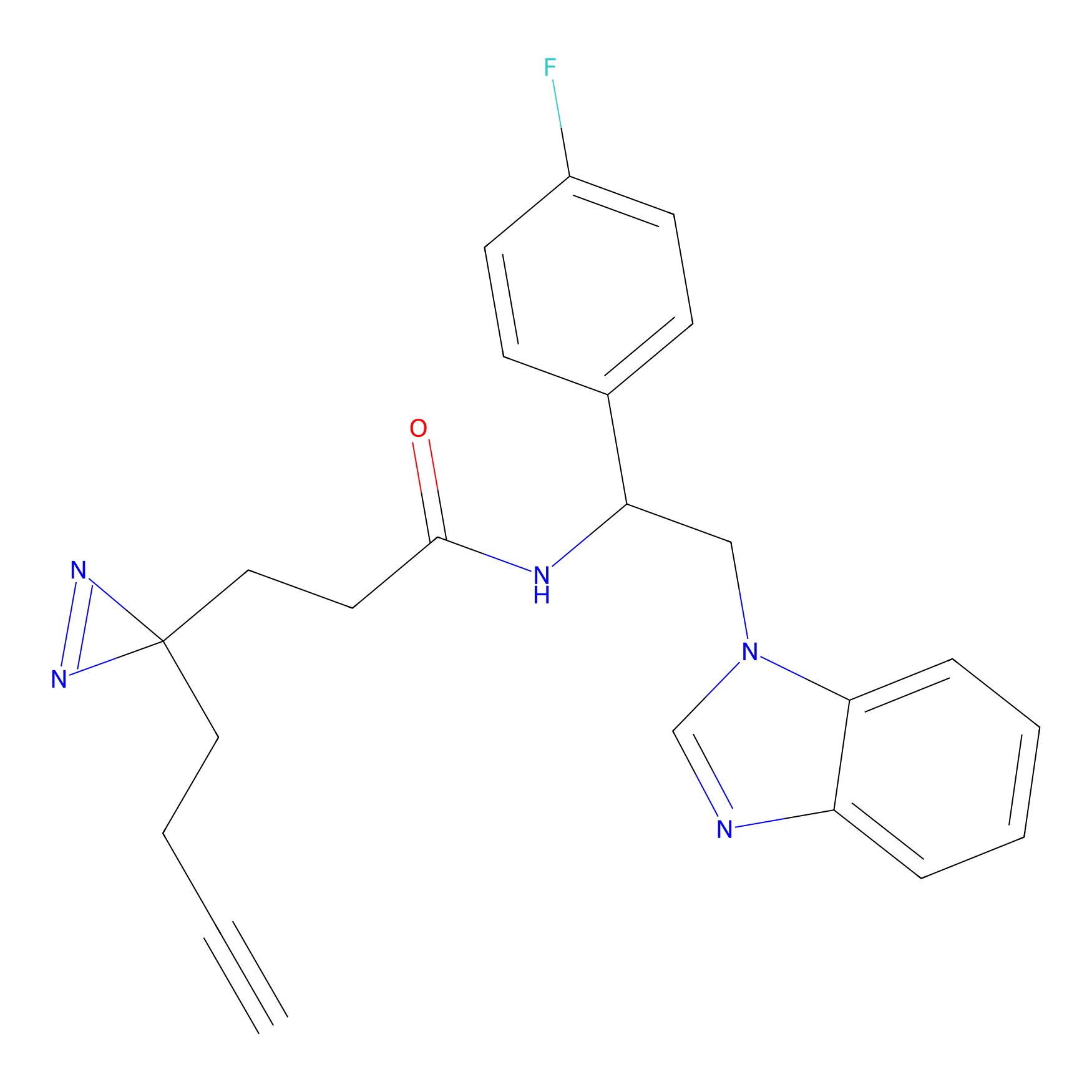
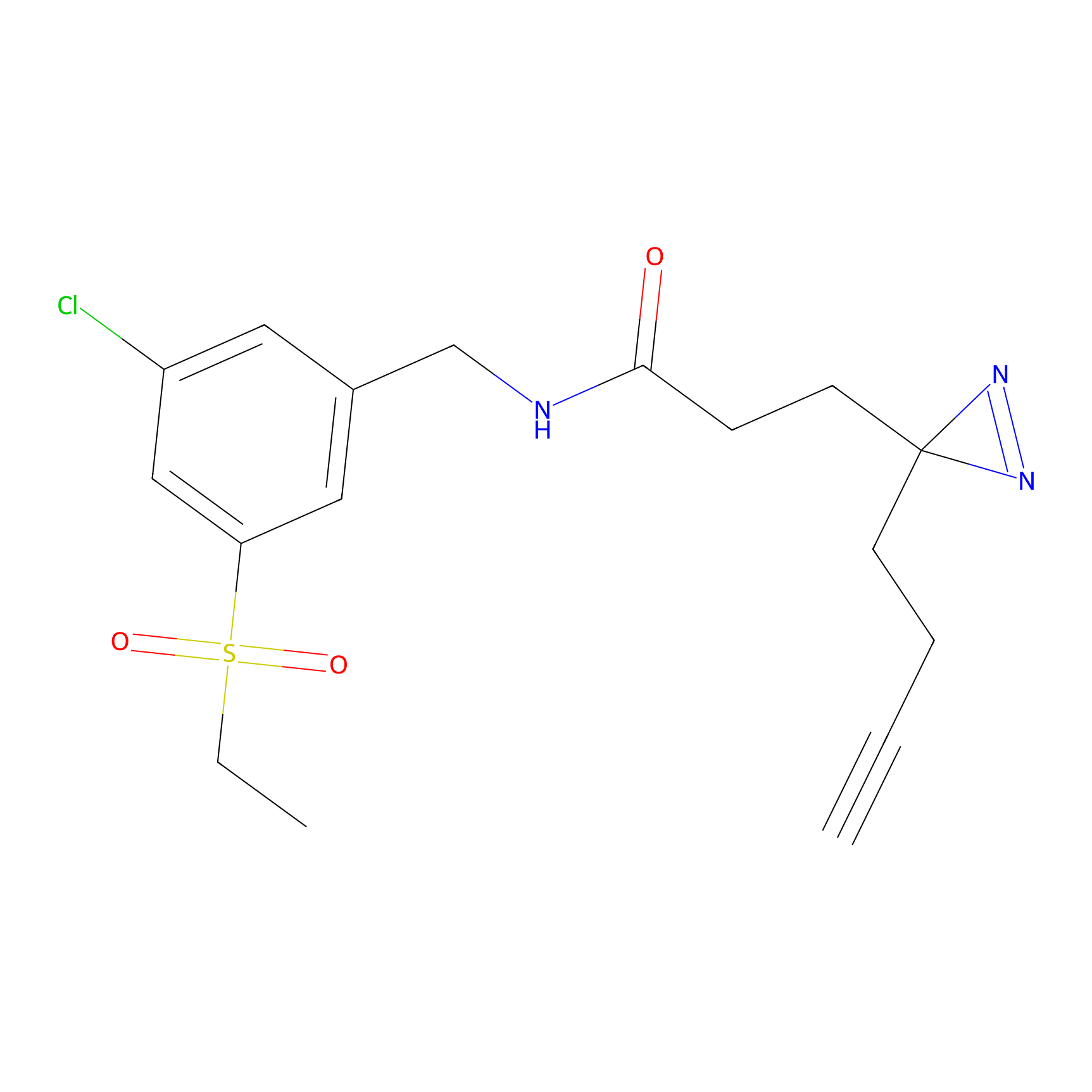
DBIA Probe Info |
 |
C344(0.81); C345(0.81) | LDD1095 | [3] | |
|
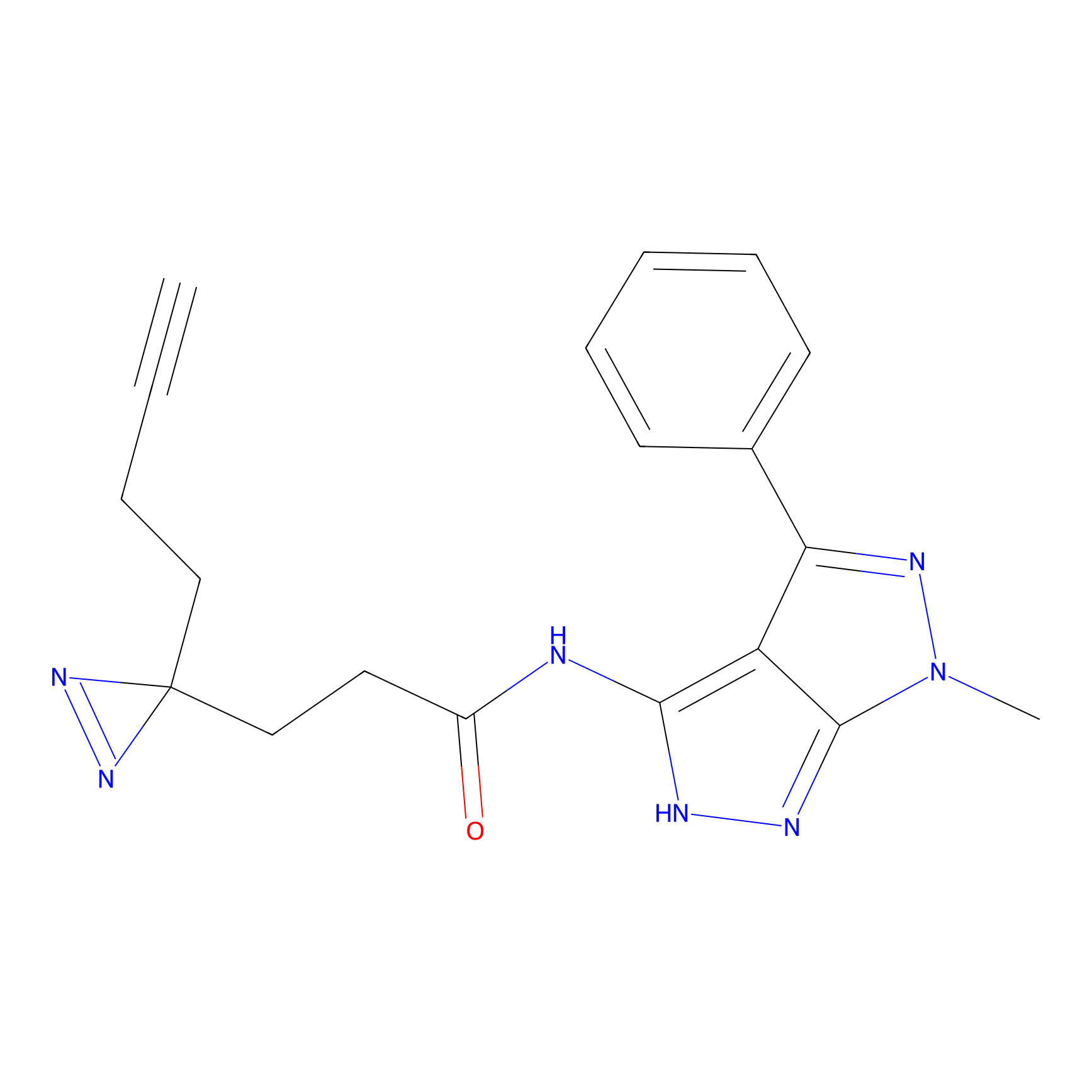
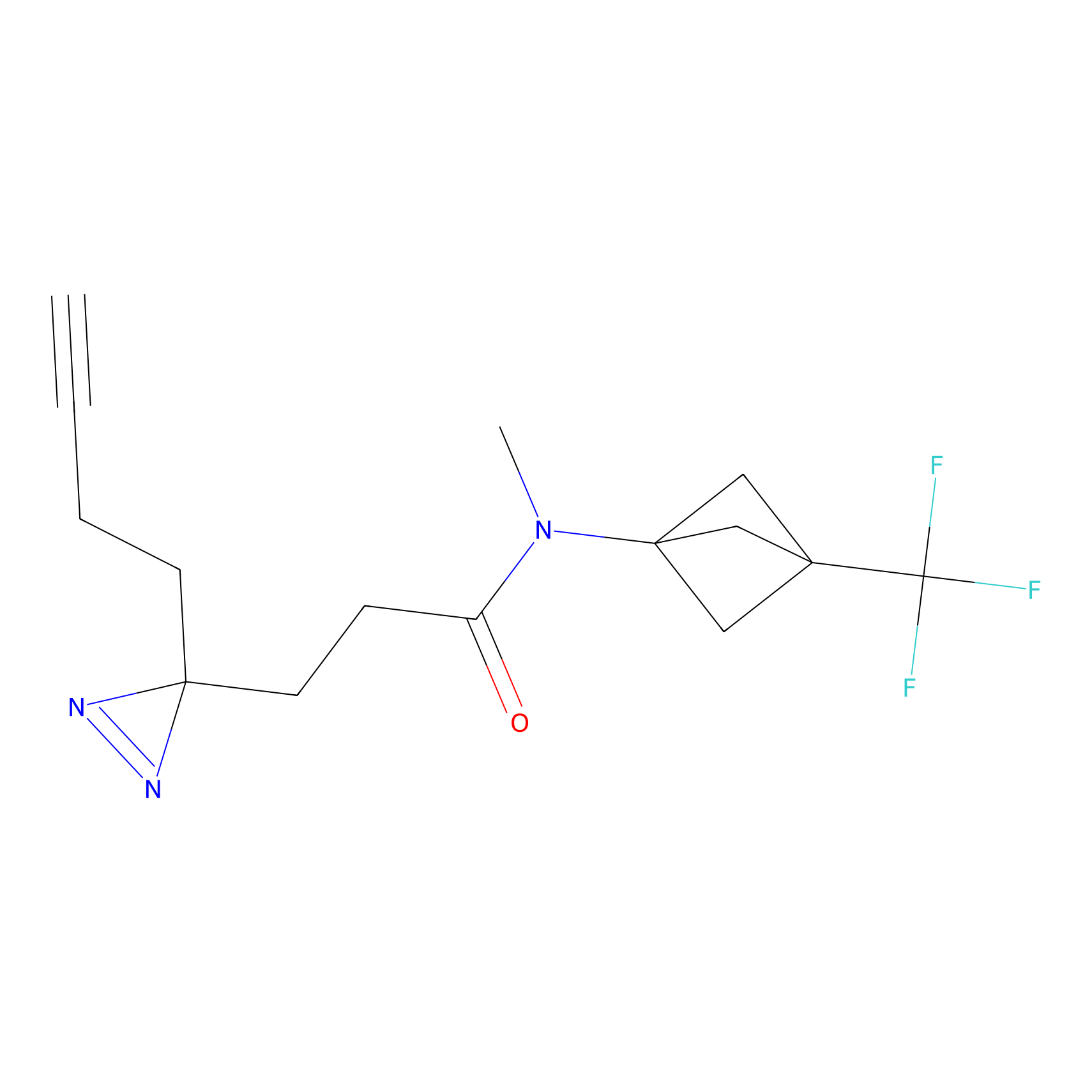
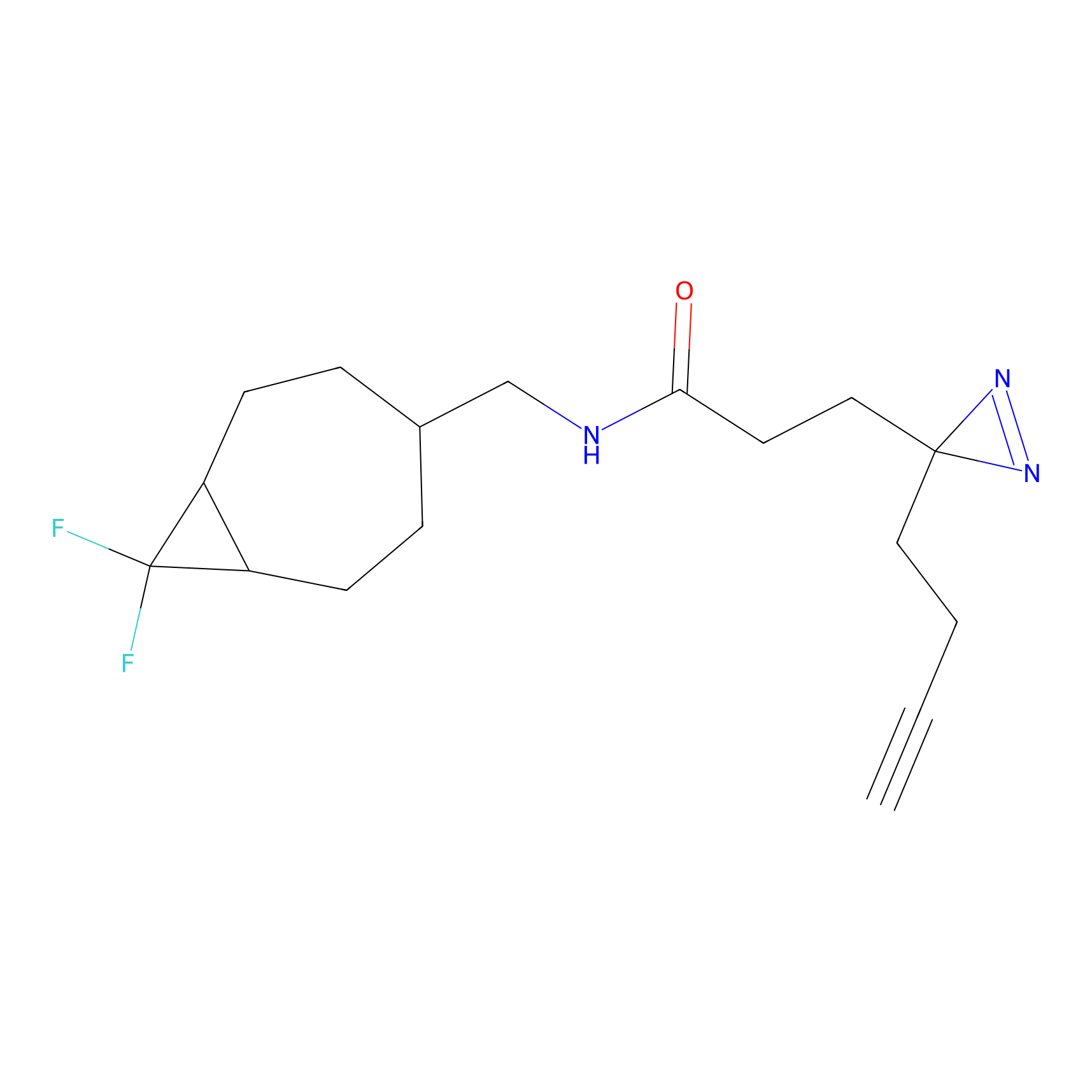
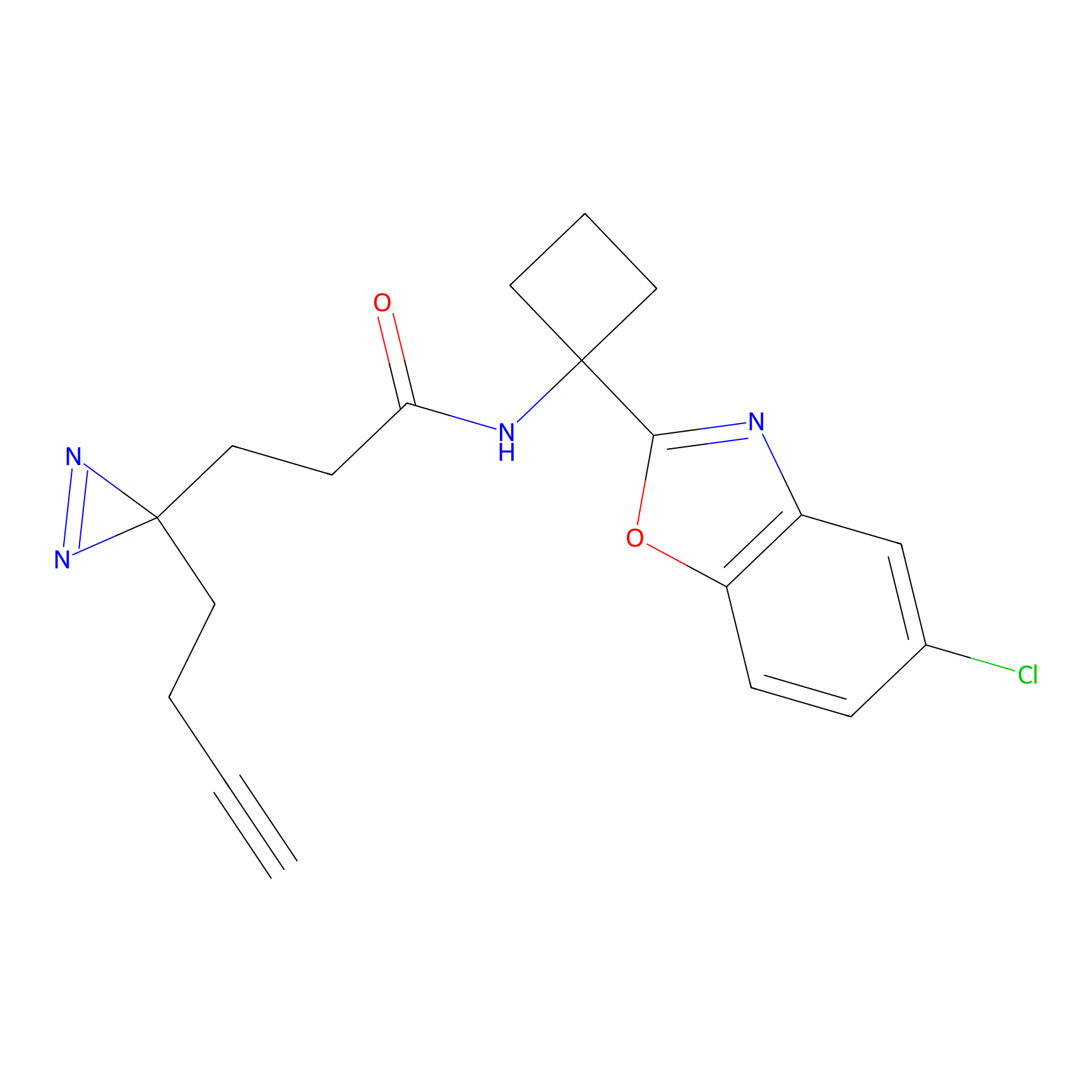
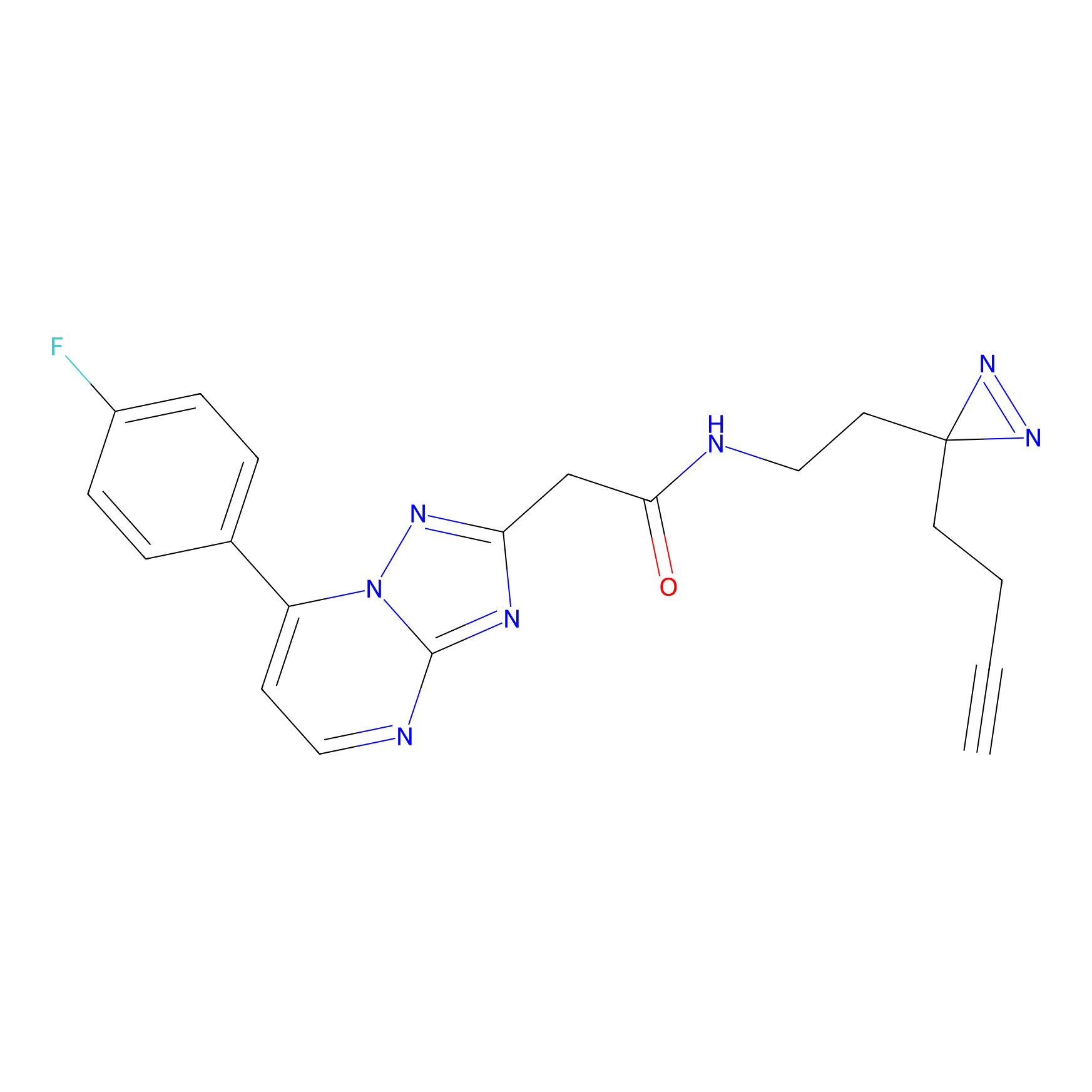
IPM Probe Info |
 |
N.A. | LDD2156 | [4] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [5] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [6] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0216 | AC100 | PaTu 8988t | C344(0.81); C345(0.81) | LDD1095 | [3] |
| LDCM0217 | AC101 | PaTu 8988t | C344(0.83); C345(0.83) | LDD1096 | [3] |
| LDCM0218 | AC102 | PaTu 8988t | C344(0.79); C345(0.79) | LDD1097 | [3] |
| LDCM0219 | AC103 | PaTu 8988t | C344(0.92); C345(0.92) | LDD1098 | [3] |
| LDCM0220 | AC104 | PaTu 8988t | C344(0.78); C345(0.78) | LDD1099 | [3] |
| LDCM0221 | AC105 | PaTu 8988t | C344(0.67); C345(0.67) | LDD1100 | [3] |
| LDCM0222 | AC106 | PaTu 8988t | C344(1.01); C345(1.01) | LDD1101 | [3] |
| LDCM0223 | AC107 | PaTu 8988t | C344(0.91); C345(0.91) | LDD1102 | [3] |
| LDCM0224 | AC108 | PaTu 8988t | C344(1.20); C345(1.20) | LDD1103 | [3] |
| LDCM0225 | AC109 | PaTu 8988t | C344(0.86); C345(0.86) | LDD1104 | [3] |
| LDCM0227 | AC110 | PaTu 8988t | C344(0.83); C345(0.83) | LDD1106 | [3] |
| LDCM0228 | AC111 | PaTu 8988t | C344(0.72); C345(0.72) | LDD1107 | [3] |
| LDCM0229 | AC112 | PaTu 8988t | C344(0.77); C345(0.77) | LDD1108 | [3] |
| LDCM0230 | AC113 | PaTu 8988t | C344(1.20); C345(1.20) | LDD1109 | [3] |
| LDCM0231 | AC114 | PaTu 8988t | C344(1.48); C345(1.48) | LDD1110 | [3] |
| LDCM0232 | AC115 | PaTu 8988t | C344(1.64); C345(1.64) | LDD1111 | [3] |
| LDCM0233 | AC116 | PaTu 8988t | C344(0.99); C345(0.99) | LDD1112 | [3] |
| LDCM0234 | AC117 | PaTu 8988t | C344(1.63); C345(1.63) | LDD1113 | [3] |
| LDCM0235 | AC118 | PaTu 8988t | C344(1.17); C345(1.17) | LDD1114 | [3] |
| LDCM0236 | AC119 | PaTu 8988t | C344(1.24); C345(1.24) | LDD1115 | [3] |
| LDCM0238 | AC120 | PaTu 8988t | C344(1.20); C345(1.20) | LDD1117 | [3] |
| LDCM0239 | AC121 | PaTu 8988t | C344(1.84); C345(1.84) | LDD1118 | [3] |
| LDCM0240 | AC122 | PaTu 8988t | C344(1.16); C345(1.16) | LDD1119 | [3] |
| LDCM0241 | AC123 | PaTu 8988t | C344(1.16); C345(1.16) | LDD1120 | [3] |
| LDCM0242 | AC124 | PaTu 8988t | C344(1.32); C345(1.32) | LDD1121 | [3] |
| LDCM0243 | AC125 | PaTu 8988t | C344(1.10); C345(1.10) | LDD1122 | [3] |
| LDCM0244 | AC126 | PaTu 8988t | C344(0.90); C345(0.90) | LDD1123 | [3] |
| LDCM0245 | AC127 | PaTu 8988t | C344(1.09); C345(1.09) | LDD1124 | [3] |
| LDCM0276 | AC17 | PaTu 8988t | C344(1.07); C345(1.07) | LDD1155 | [3] |
| LDCM0277 | AC18 | PaTu 8988t | C344(0.93); C345(0.93) | LDD1156 | [3] |
| LDCM0278 | AC19 | PaTu 8988t | C344(1.01); C345(1.01) | LDD1157 | [3] |
| LDCM0280 | AC20 | PaTu 8988t | C344(0.95); C345(0.95) | LDD1159 | [3] |
| LDCM0281 | AC21 | PaTu 8988t | C344(0.91); C345(0.91) | LDD1160 | [3] |
| LDCM0282 | AC22 | PaTu 8988t | C344(1.11); C345(1.11) | LDD1161 | [3] |
| LDCM0283 | AC23 | PaTu 8988t | C344(1.12); C345(1.12) | LDD1162 | [3] |
| LDCM0284 | AC24 | PaTu 8988t | C344(0.97); C345(0.97) | LDD1163 | [3] |
| LDCM0320 | AC57 | PaTu 8988t | C344(1.19); C345(1.19) | LDD1199 | [3] |
| LDCM0321 | AC58 | PaTu 8988t | C344(1.39); C345(1.39) | LDD1200 | [3] |
| LDCM0322 | AC59 | PaTu 8988t | C344(1.19); C345(1.19) | LDD1201 | [3] |
| LDCM0324 | AC60 | PaTu 8988t | C344(1.43); C345(1.43) | LDD1203 | [3] |
| LDCM0325 | AC61 | PaTu 8988t | C344(1.35); C345(1.35) | LDD1204 | [3] |
| LDCM0326 | AC62 | PaTu 8988t | C344(1.44); C345(1.44) | LDD1205 | [3] |
| LDCM0327 | AC63 | PaTu 8988t | C344(1.23); C345(1.23) | LDD1206 | [3] |
| LDCM0328 | AC64 | PaTu 8988t | C344(1.60); C345(1.60) | LDD1207 | [3] |
| LDCM0329 | AC65 | PaTu 8988t | C344(1.01); C345(1.01) | LDD1208 | [3] |
| LDCM0330 | AC66 | PaTu 8988t | C344(1.11); C345(1.11) | LDD1209 | [3] |
| LDCM0331 | AC67 | PaTu 8988t | C344(1.52); C345(1.52) | LDD1210 | [3] |
| LDCM0365 | AC98 | PaTu 8988t | C344(0.71); C345(0.71) | LDD1244 | [3] |
| LDCM0366 | AC99 | PaTu 8988t | C344(0.92); C345(0.92) | LDD1245 | [3] |
| LDCM0156 | Aniline | NCI-H1299 | 11.44 | LDD0403 | [2] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [5] |
| LDCM0374 | CL105 | PaTu 8988t | C344(0.68); C345(0.68) | LDD1253 | [3] |
| LDCM0375 | CL106 | PaTu 8988t | C344(0.92); C345(0.92) | LDD1254 | [3] |
| LDCM0376 | CL107 | PaTu 8988t | C344(0.89); C345(0.89) | LDD1255 | [3] |
| LDCM0377 | CL108 | PaTu 8988t | C344(0.78); C345(0.78) | LDD1256 | [3] |
| LDCM0378 | CL109 | PaTu 8988t | C344(0.91); C345(0.91) | LDD1257 | [3] |
| LDCM0380 | CL110 | PaTu 8988t | C344(1.13); C345(1.13) | LDD1259 | [3] |
| LDCM0381 | CL111 | PaTu 8988t | C344(1.55); C345(1.55) | LDD1260 | [3] |
| LDCM0396 | CL125 | PaTu 8988t | C344(1.18); C345(1.18) | LDD1275 | [3] |
| LDCM0397 | CL126 | PaTu 8988t | C344(1.45); C345(1.45) | LDD1276 | [3] |
| LDCM0398 | CL127 | PaTu 8988t | C344(1.21); C345(1.21) | LDD1277 | [3] |
| LDCM0399 | CL128 | PaTu 8988t | C344(1.18); C345(1.18) | LDD1278 | [3] |
| LDCM0420 | CL31 | PaTu 8988t | C344(0.94); C345(0.94) | LDD1299 | [3] |
| LDCM0421 | CL32 | PaTu 8988t | C344(0.90); C345(0.90) | LDD1300 | [3] |
| LDCM0422 | CL33 | PaTu 8988t | C344(1.57); C345(1.57) | LDD1301 | [3] |
| LDCM0423 | CL34 | PaTu 8988t | C344(0.78); C345(0.78) | LDD1302 | [3] |
| LDCM0424 | CL35 | PaTu 8988t | C344(0.99); C345(0.99) | LDD1303 | [3] |
| LDCM0425 | CL36 | PaTu 8988t | C344(1.08); C345(1.08) | LDD1304 | [3] |
| LDCM0426 | CL37 | PaTu 8988t | C344(0.92); C345(0.92) | LDD1305 | [3] |
| LDCM0428 | CL39 | PaTu 8988t | C344(1.03); C345(1.03) | LDD1307 | [3] |
| LDCM0430 | CL40 | PaTu 8988t | C344(0.98); C345(0.98) | LDD1309 | [3] |
| LDCM0431 | CL41 | PaTu 8988t | C344(1.21); C345(1.21) | LDD1310 | [3] |
| LDCM0432 | CL42 | PaTu 8988t | C344(0.84); C345(0.84) | LDD1311 | [3] |
| LDCM0433 | CL43 | PaTu 8988t | C344(1.00); C345(1.00) | LDD1312 | [3] |
| LDCM0434 | CL44 | PaTu 8988t | C344(0.94); C345(0.94) | LDD1313 | [3] |
| LDCM0435 | CL45 | PaTu 8988t | C344(0.97); C345(0.97) | LDD1314 | [3] |
| LDCM0436 | CL46 | PaTu 8988t | C344(0.80); C345(0.80) | LDD1315 | [3] |
| LDCM0437 | CL47 | PaTu 8988t | C344(0.81); C345(0.81) | LDD1316 | [3] |
| LDCM0438 | CL48 | PaTu 8988t | C344(0.82); C345(0.82) | LDD1317 | [3] |
| LDCM0439 | CL49 | PaTu 8988t | C344(0.91); C345(0.91) | LDD1318 | [3] |
| LDCM0441 | CL50 | PaTu 8988t | C344(0.91); C345(0.91) | LDD1320 | [3] |
| LDCM0442 | CL51 | PaTu 8988t | C344(1.00); C345(1.00) | LDD1321 | [3] |
| LDCM0443 | CL52 | PaTu 8988t | C344(0.87); C345(0.87) | LDD1322 | [3] |
| LDCM0444 | CL53 | PaTu 8988t | C344(0.96); C345(0.96) | LDD1323 | [3] |
| LDCM0445 | CL54 | PaTu 8988t | C344(0.98); C345(0.98) | LDD1324 | [3] |
| LDCM0446 | CL55 | PaTu 8988t | C344(0.94); C345(0.94) | LDD1325 | [3] |
| LDCM0447 | CL56 | PaTu 8988t | C344(0.46); C345(0.46) | LDD1326 | [3] |
| LDCM0448 | CL57 | PaTu 8988t | C344(1.73); C345(1.73) | LDD1327 | [3] |
| LDCM0449 | CL58 | PaTu 8988t | C344(1.15); C345(1.15) | LDD1328 | [3] |
| LDCM0450 | CL59 | PaTu 8988t | C344(1.76); C345(1.76) | LDD1329 | [3] |
| LDCM0452 | CL60 | PaTu 8988t | C344(1.22); C345(1.22) | LDD1331 | [3] |
| LDCM0453 | CL61 | PaTu 8988t | C344(1.01); C345(1.01) | LDD1332 | [3] |
| LDCM0454 | CL62 | PaTu 8988t | C344(0.83); C345(0.83) | LDD1333 | [3] |
| LDCM0455 | CL63 | PaTu 8988t | C344(0.90); C345(0.90) | LDD1334 | [3] |
| LDCM0456 | CL64 | PaTu 8988t | C344(1.13); C345(1.13) | LDD1335 | [3] |
| LDCM0457 | CL65 | PaTu 8988t | C344(0.90); C345(0.90) | LDD1336 | [3] |
| LDCM0458 | CL66 | PaTu 8988t | C344(0.99); C345(0.99) | LDD1337 | [3] |
| LDCM0459 | CL67 | PaTu 8988t | C344(1.04); C345(1.04) | LDD1338 | [3] |
| LDCM0460 | CL68 | PaTu 8988t | C344(1.03); C345(1.03) | LDD1339 | [3] |
| LDCM0461 | CL69 | PaTu 8988t | C344(1.16); C345(1.16) | LDD1340 | [3] |
| LDCM0463 | CL70 | PaTu 8988t | C344(0.99); C345(0.99) | LDD1342 | [3] |
| LDCM0464 | CL71 | PaTu 8988t | C344(0.89); C345(0.89) | LDD1343 | [3] |
| LDCM0465 | CL72 | PaTu 8988t | C344(0.94); C345(0.94) | LDD1344 | [3] |
| LDCM0466 | CL73 | PaTu 8988t | C344(1.20); C345(1.20) | LDD1345 | [3] |
| LDCM0467 | CL74 | PaTu 8988t | C344(0.84); C345(0.84) | LDD1346 | [3] |
| LDCM0468 | Fragment33 | PaTu 8988t | C344(1.00); C345(1.00) | LDD1347 | [3] |
| LDCM0427 | Fragment51 | PaTu 8988t | C344(0.88); C345(0.88) | LDD1306 | [3] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [5] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0224 | [5] |
The Interaction Atlas With This Target
References