Details of the Target
General Information of Target
| Target ID | LDTP04459 | |||||
|---|---|---|---|---|---|---|
| Target Name | H(+)/Cl(-) exchange transporter 3 (CLCN3) | |||||
| Gene Name | CLCN3 | |||||
| Gene ID | 1182 | |||||
| Synonyms |
H(+)/Cl(-) exchange transporter 3; Chloride channel protein 3; ClC-3; Chloride transporter ClC-3 |
|||||
| 3D Structure | ||||||
| Sequence |
MESEQLFHRGYYRNSYNSITSASSDEELLDGAGVIMDFQTSEDDNLLDGDTAVGTHYTMT
NGGSINSSTHLLDLLDEPIPGVGTYDDFHTIDWVREKCKDRERHRRINSKKKESAWEMTK SLYDAWSGWLVVTLTGLASGALAGLIDIAADWMTDLKEGICLSALWYNHEQCCWGSNETT FEERDKCPQWKTWAELIIGQAEGPGSYIMNYIMYIFWALSFAFLAVSLVKVFAPYACGSG IPEIKTILSGFIIRGYLGKWTLMIKTITLVLAVASGLSLGKEGPLVHVACCCGNIFSYLF PKYSTNEAKKREVLSAASAAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFAALVAA FVLRSINPFGNSRLVLFYVEYHTPWYLFELFPFILLGVFGGLWGAFFIRANIAWCRRRKS TKFGKYPVLEVIIVAAITAVIAFPNPYTRLNTSELIKELFTDCGPLESSSLCDYRNDMNA SKIVDDIPDRPAGIGVYSAIWQLCLALIFKIIMTVFTFGIKVPSGLFIPSMAIGAIAGRI VGIAVEQLAYYHHDWFIFKEWCEVGADCITPGLYAMVGAAACLGGVTRMTVSLVVIVFEL TGGLEYIVPLMAAVMTSKWVGDAFGREGIYEAHIRLNGYPFLDAKEEFTHTTLAADVMRP RRNDPPLAVLTQDNMTVDDIENMINETSYNGFPVIMSKESQRLVGFALRRDLTIAIESAR KKQEGIVGSSRVCFAQHTPSLPAESPRPLKLRSILDMSPFTVTDHTPMEIVVDIFRKLGL RQCLVTHNGRLLGIITKKDILRHMAQTANQDPASIMFN |
|||||
| Target Type |
Literature-reported
|
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
Chloride channel (TC 2.A.49) family, ClC-3/CLCN3 subfamily
|
|||||
| Subcellular location |
Early endosome membrane; Golgi apparatus membrane
|
|||||
| Function |
[Isoform 1]: Strongly outwardly rectifying, electrogenic H(+)/Cl(-)exchanger which mediates the exchange of chloride ions against protons. The CLC channel family contains both chloride channels and proton-coupled anion transporters that exchange chloride or another anion for protons. The presence of conserved gating glutamate residues is typical for family members that function as antiporters.; [Isoform 2]: Strongly outwardly rectifying, electrogenic H(+)/Cl(-)exchanger which mediates the exchange of chloride ions against protons.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| CAL120 | SNV: p.R539S | DBIA Probe Info | |||
| CHL1 | SNV: p.P491S | DBIA Probe Info | |||
| HCT116 | Deletion: p.D799IfsTer17 | . | |||
| HEC1 | SNV: p.V428I | DBIA Probe Info | |||
| HEC1B | SNV: p.V428I | . | |||
| Ishikawa (Heraklio) 02 ER | SNV: p.I508M | DBIA Probe Info | |||
| KPL1 | SNV: p.W555Ter | DBIA Probe Info | |||
| MCF7 | SNV: p.W555Ter | . | |||
| MDAMB453 | SNV: p.L376I | DBIA Probe Info | |||
| MOLT4 | SNV: p.T192I | . | |||
| NCIH1155 | SNV: p.T514I | DBIA Probe Info | |||
| NCIH2172 | SNV: p.Y256F | . | |||
| NCIH3122 | SNV: p.I695V | DBIA Probe Info | |||
| OVK18 | SNV: p.E26D | DBIA Probe Info | |||
| SG231 | SNV: p.D485N | DBIA Probe Info | |||
| SKN | SNV: p.V379M | DBIA Probe Info | |||
| SNGM | SNV: p.A579V | DBIA Probe Info | |||
| SNU1 | Deletion: p.R311GfsTer14 | DBIA Probe Info | |||
| SUPT1 | SNV: p.G565R | DBIA Probe Info | |||
| SW1271 | SNV: p.R184S | . | |||
| SW620 | SNV: p.E26K | DBIA Probe Info | |||
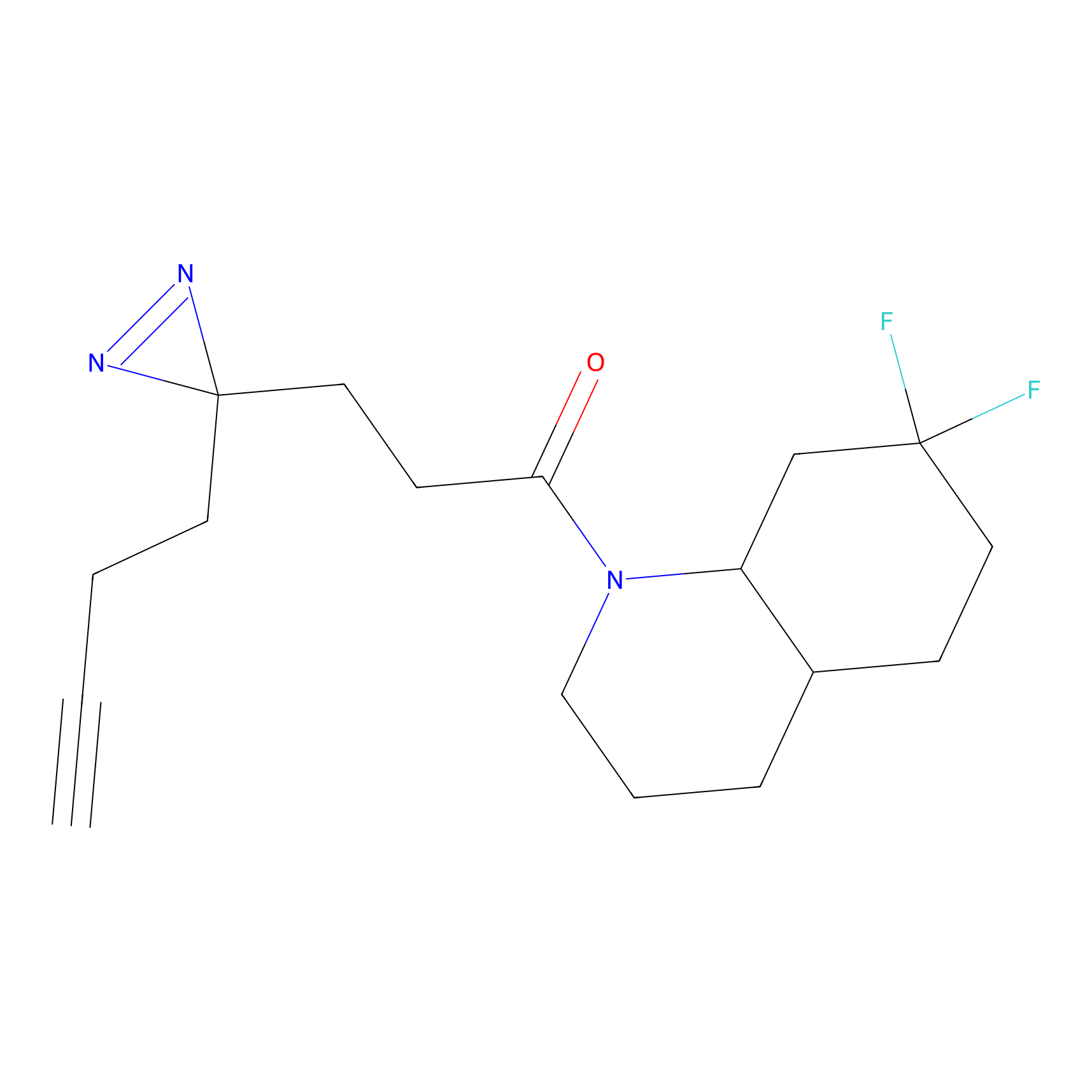
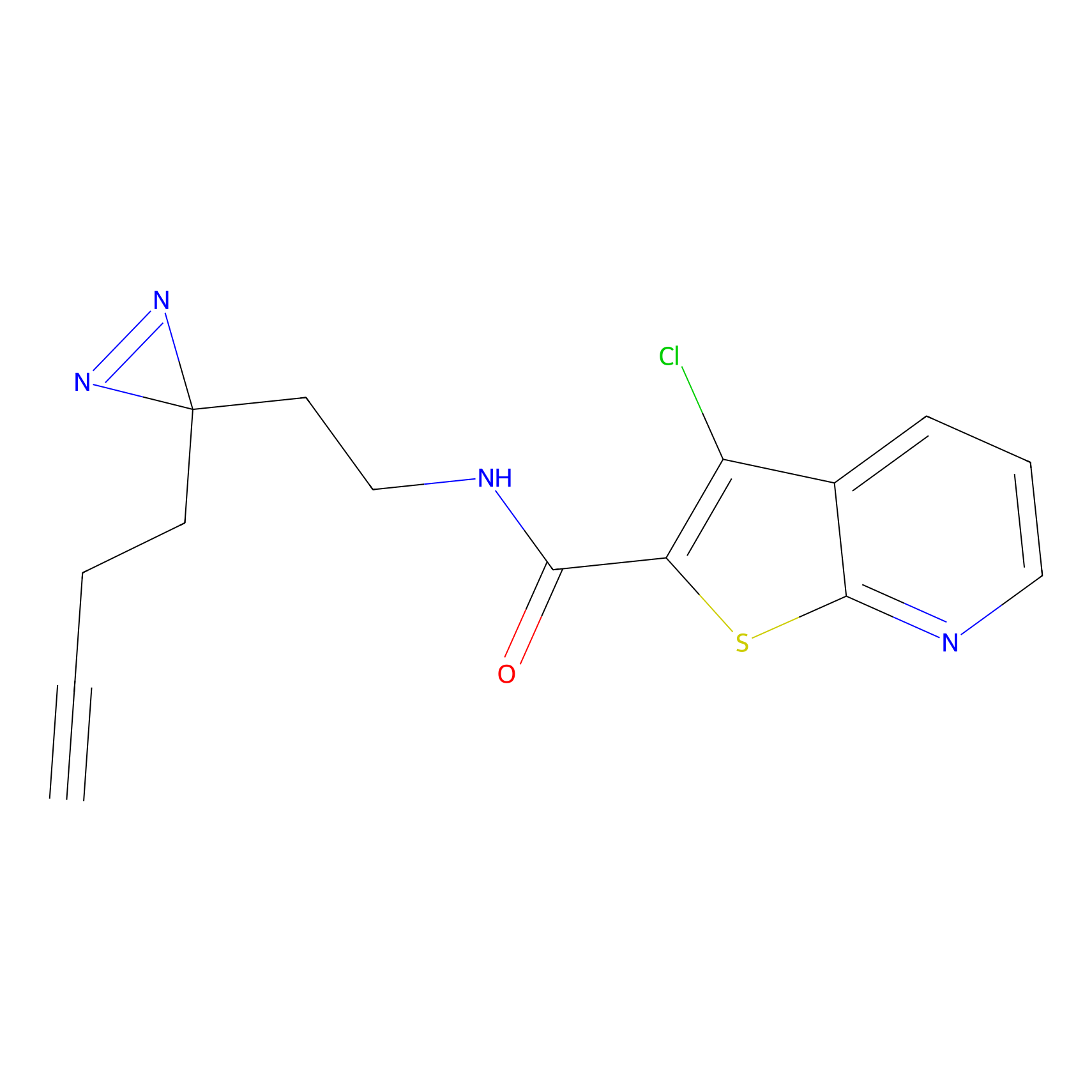
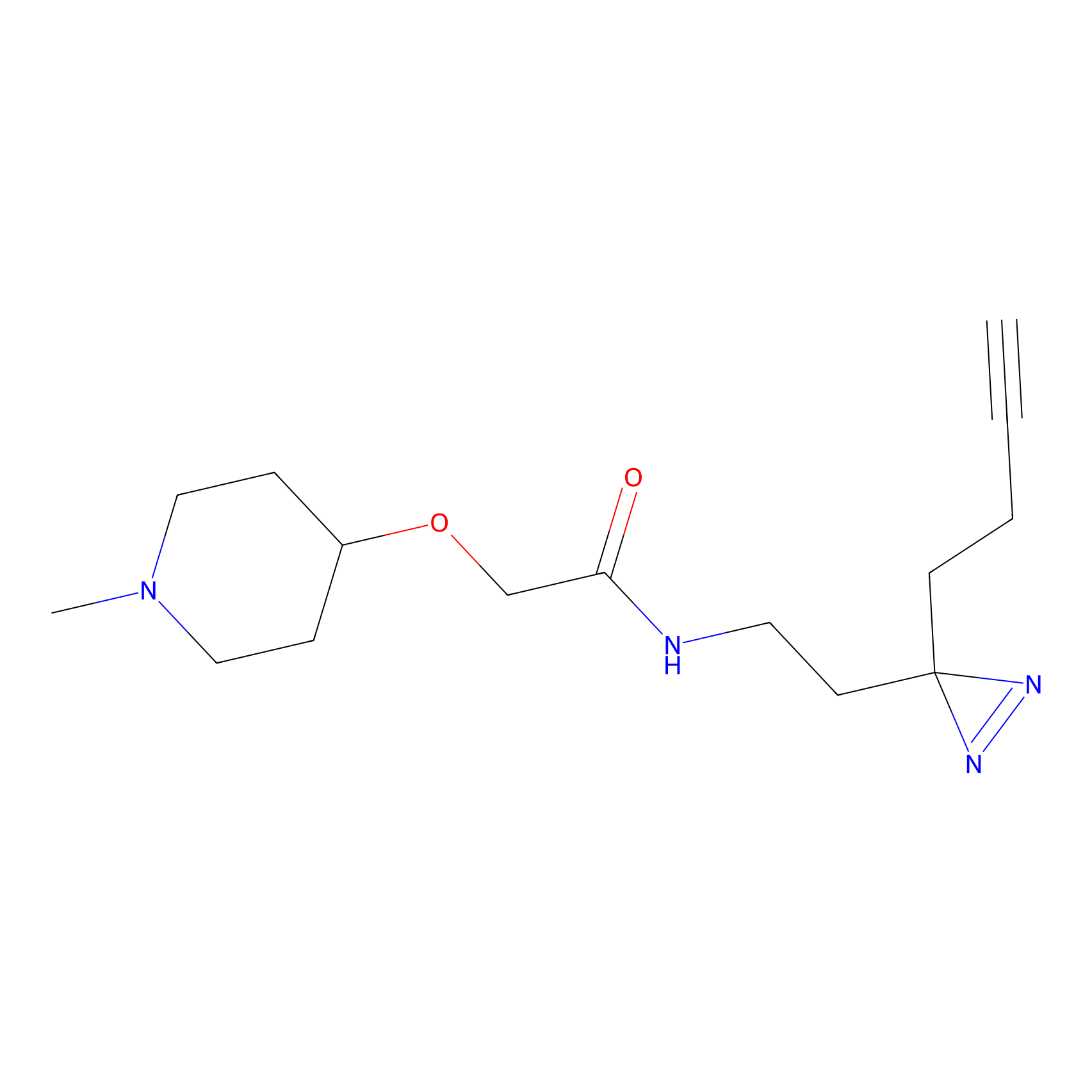
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
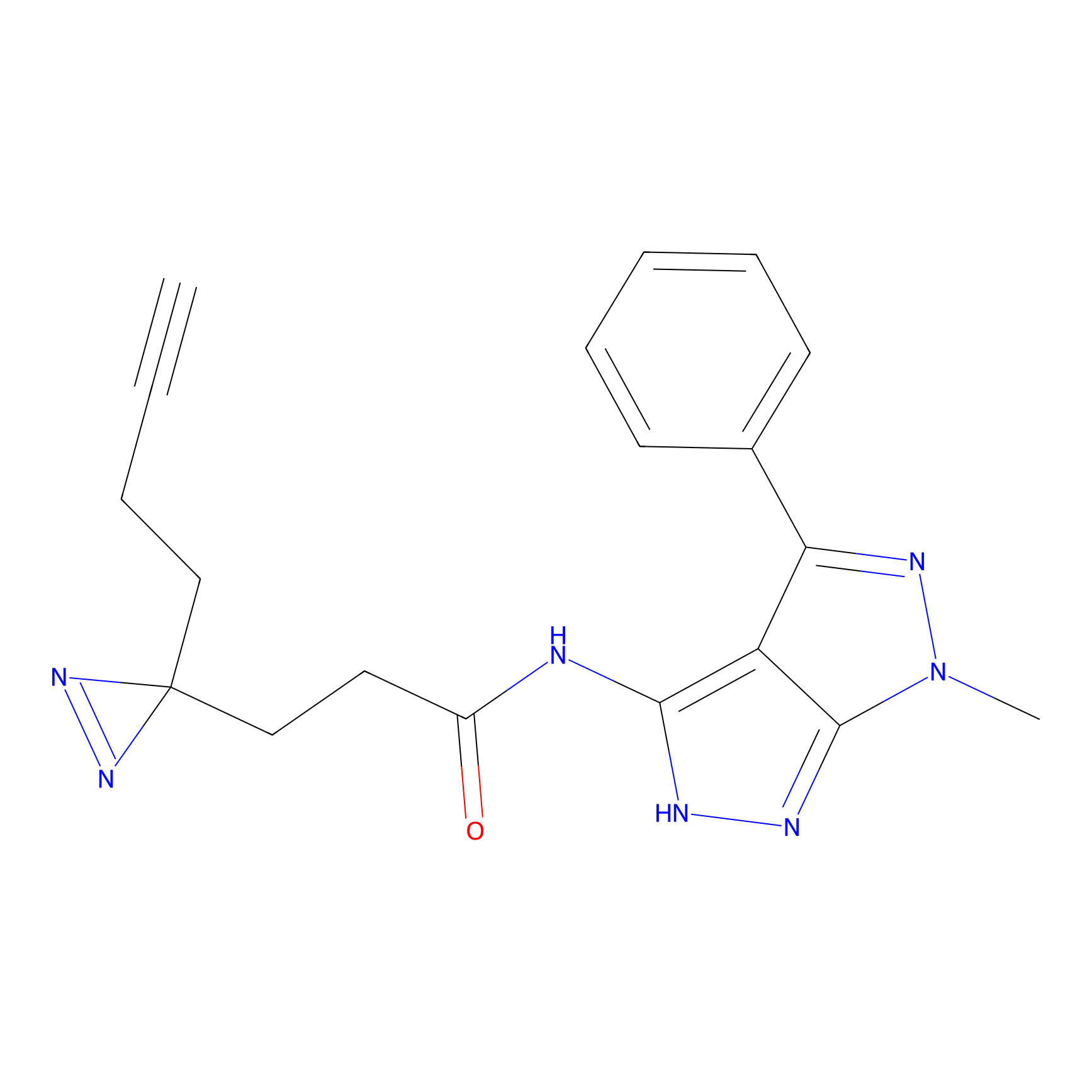
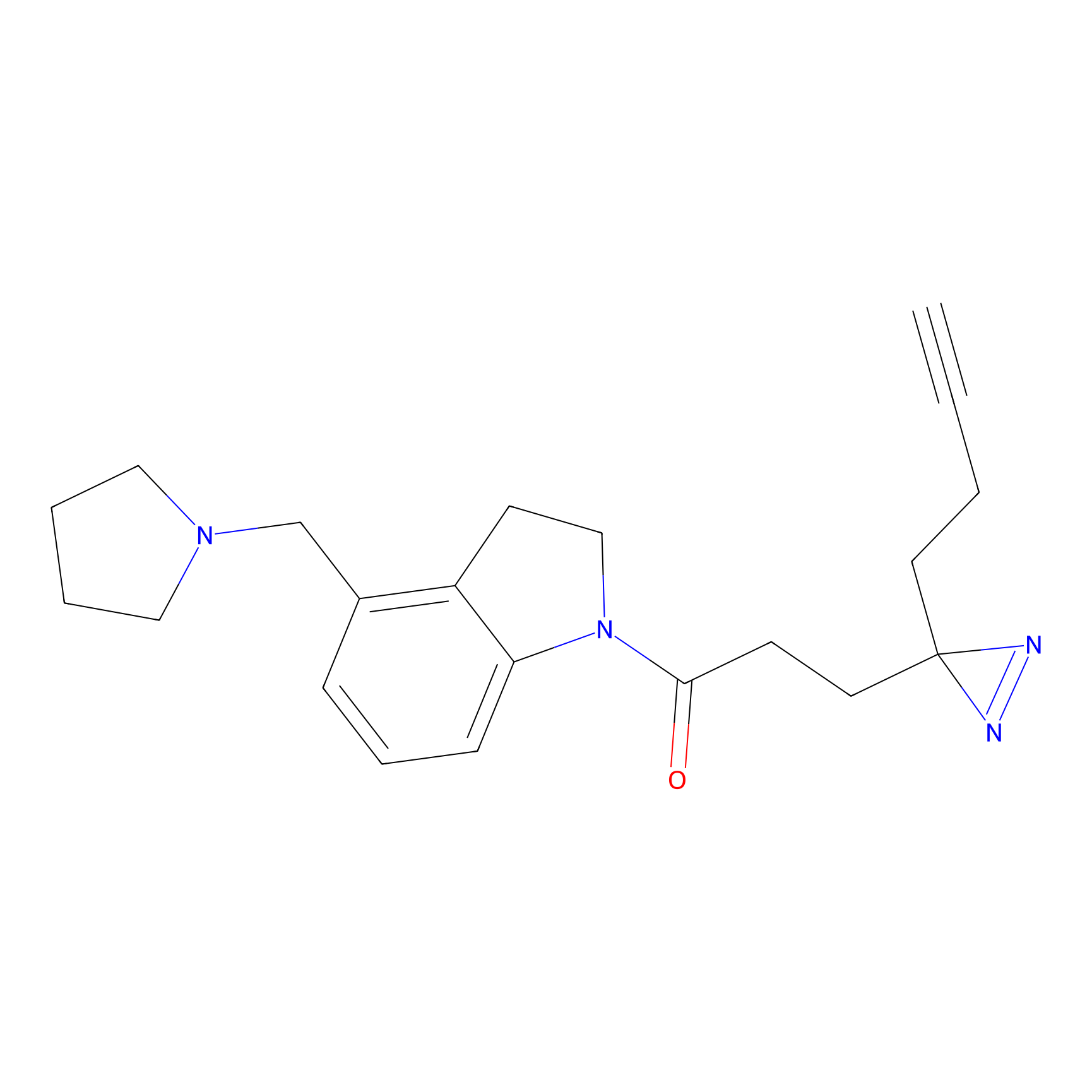
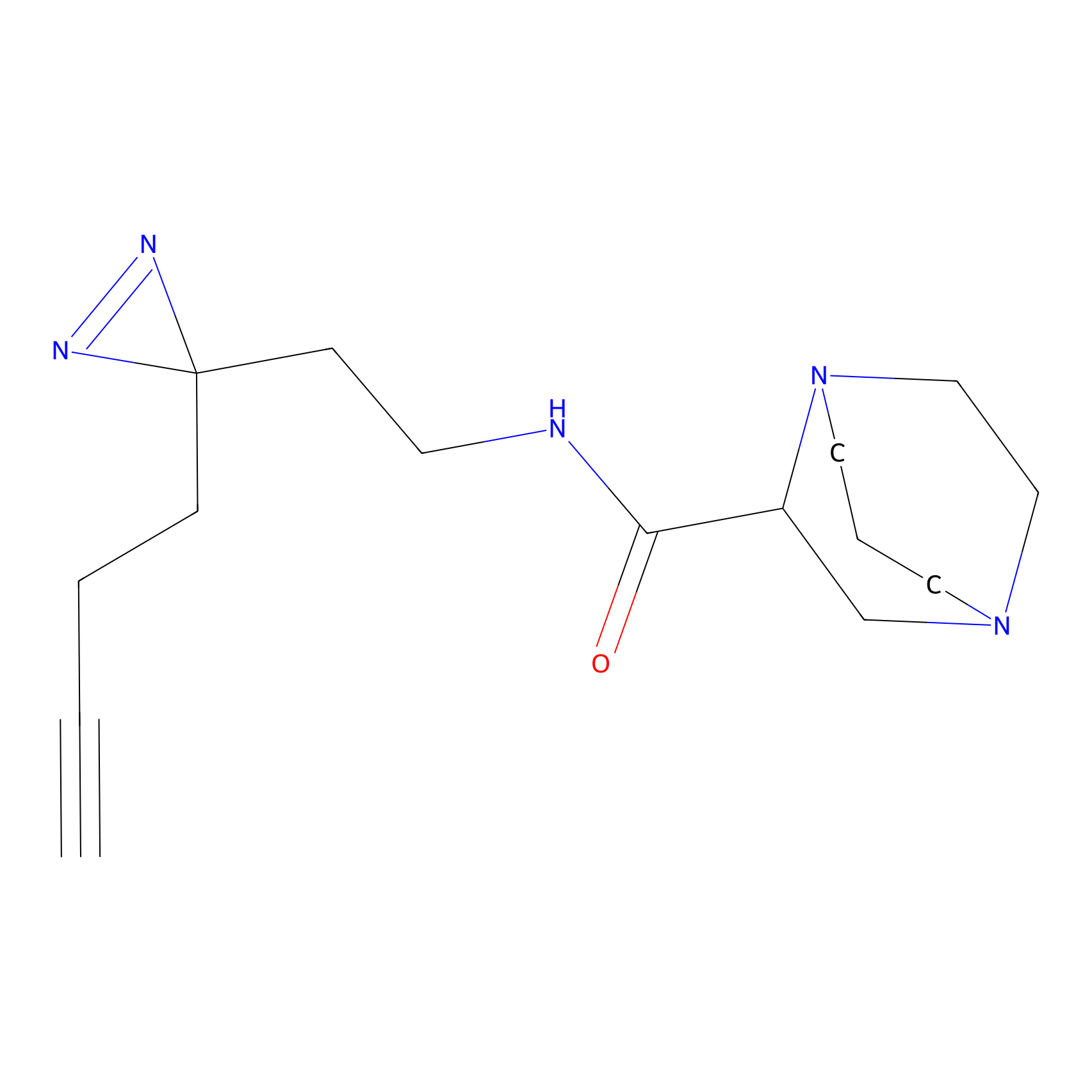
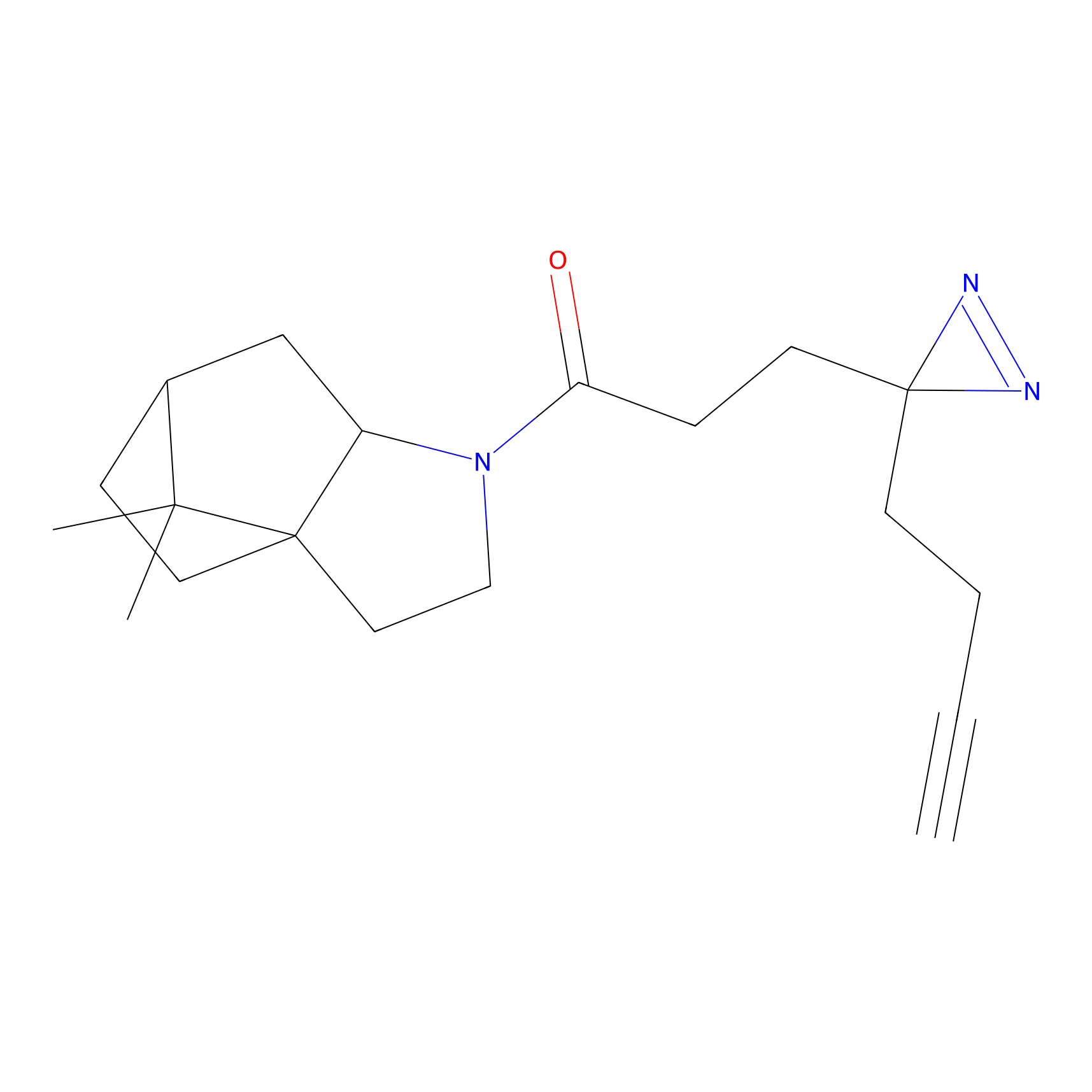
|
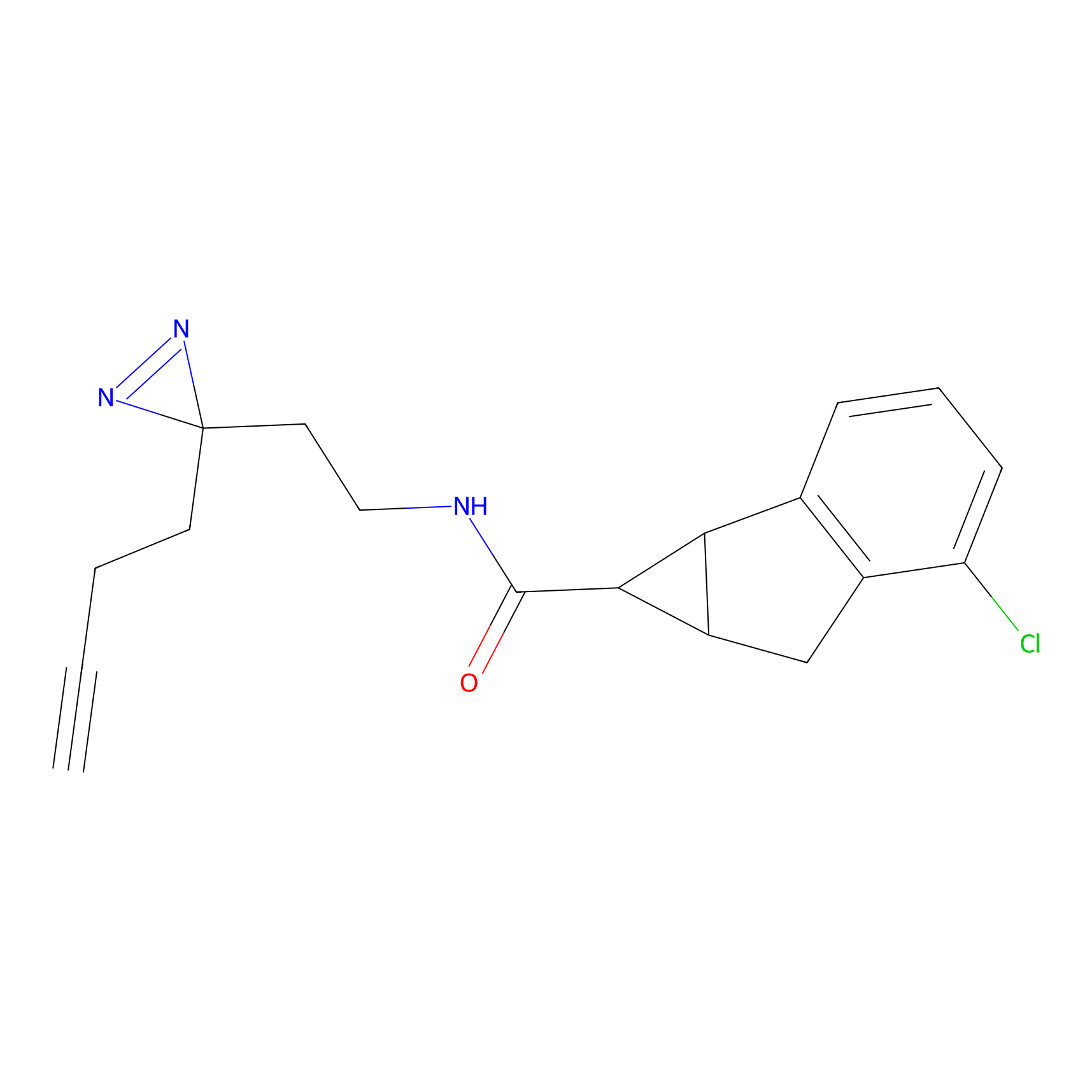
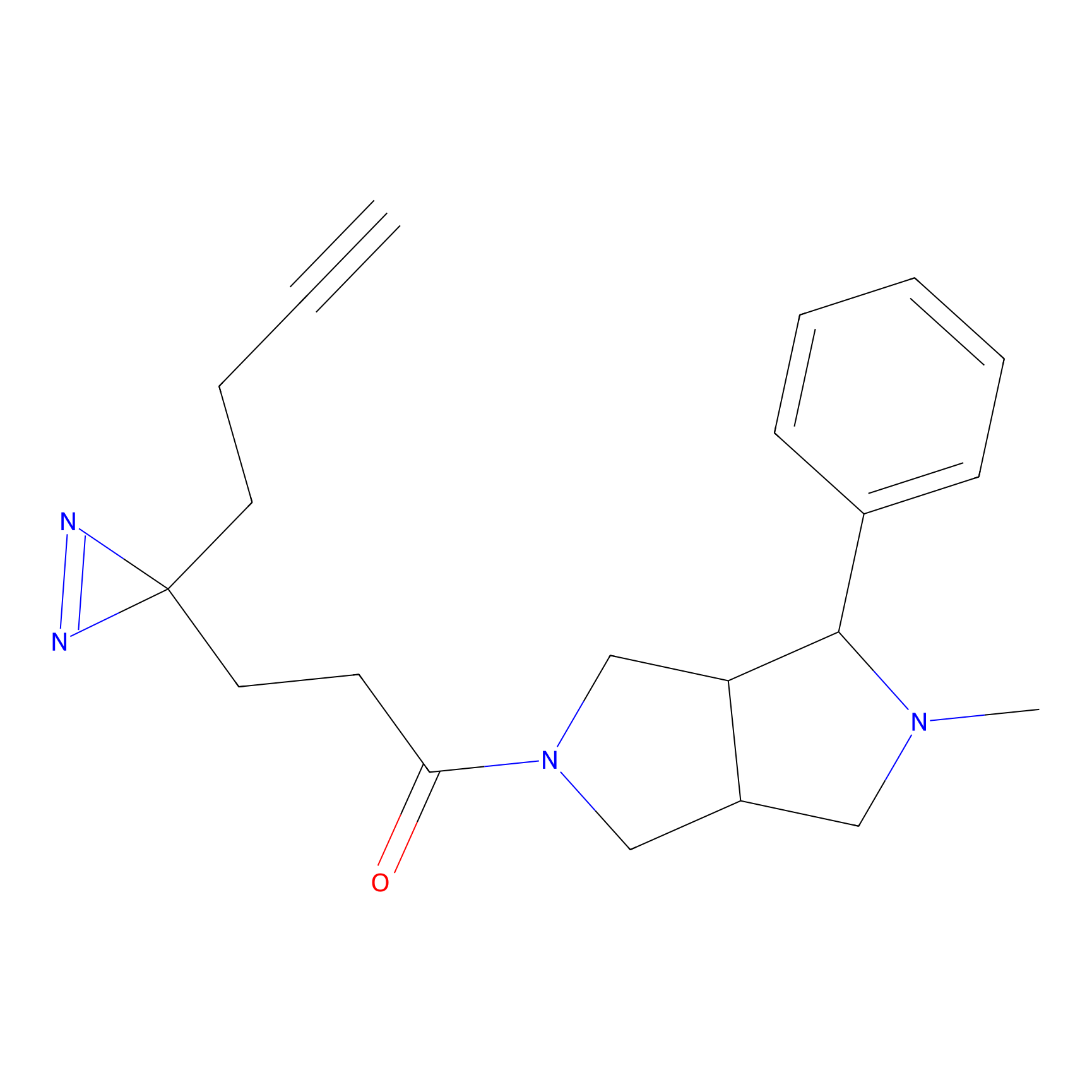
DBIA Probe Info |
 |
C733(1.64) | LDD3310 | [1] | |
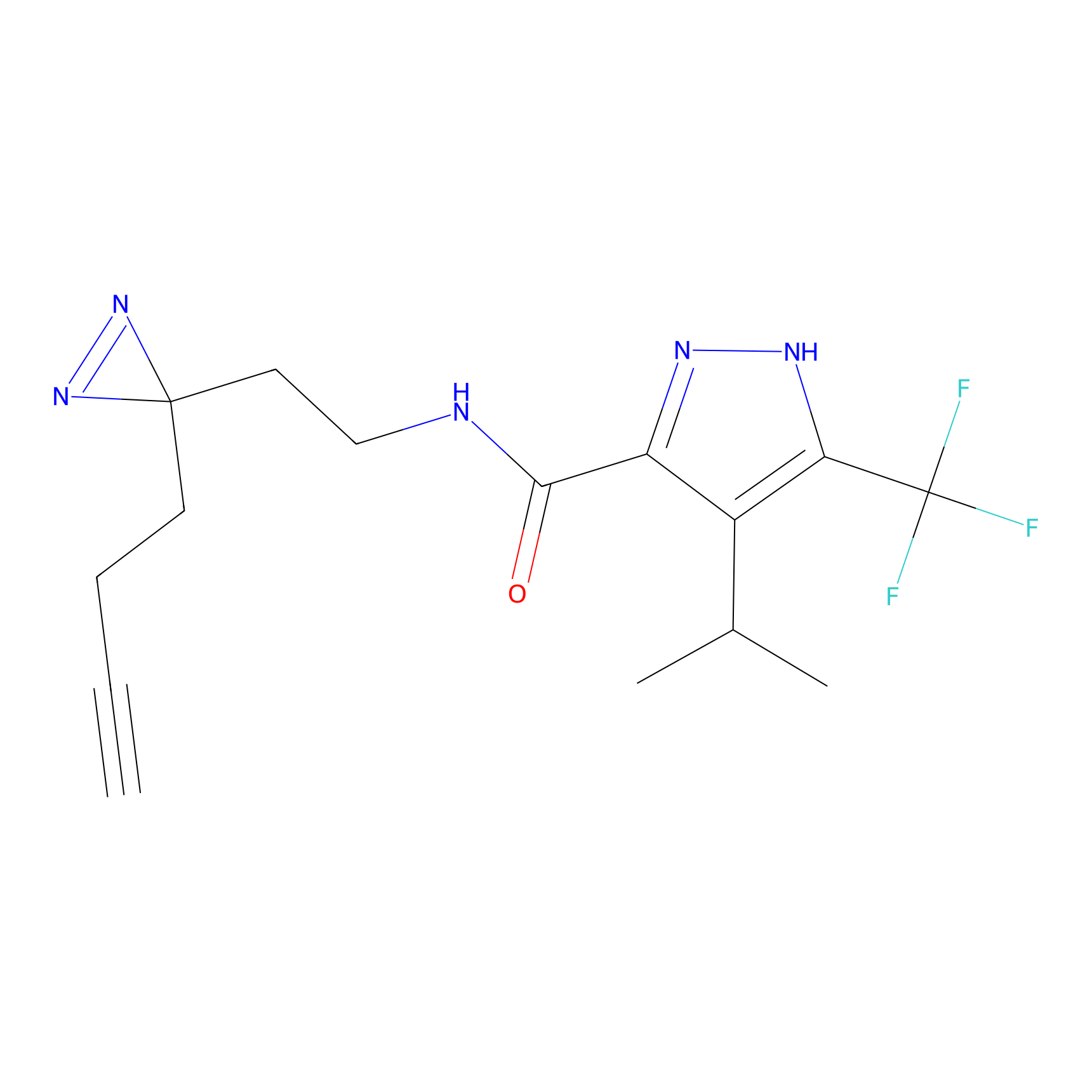
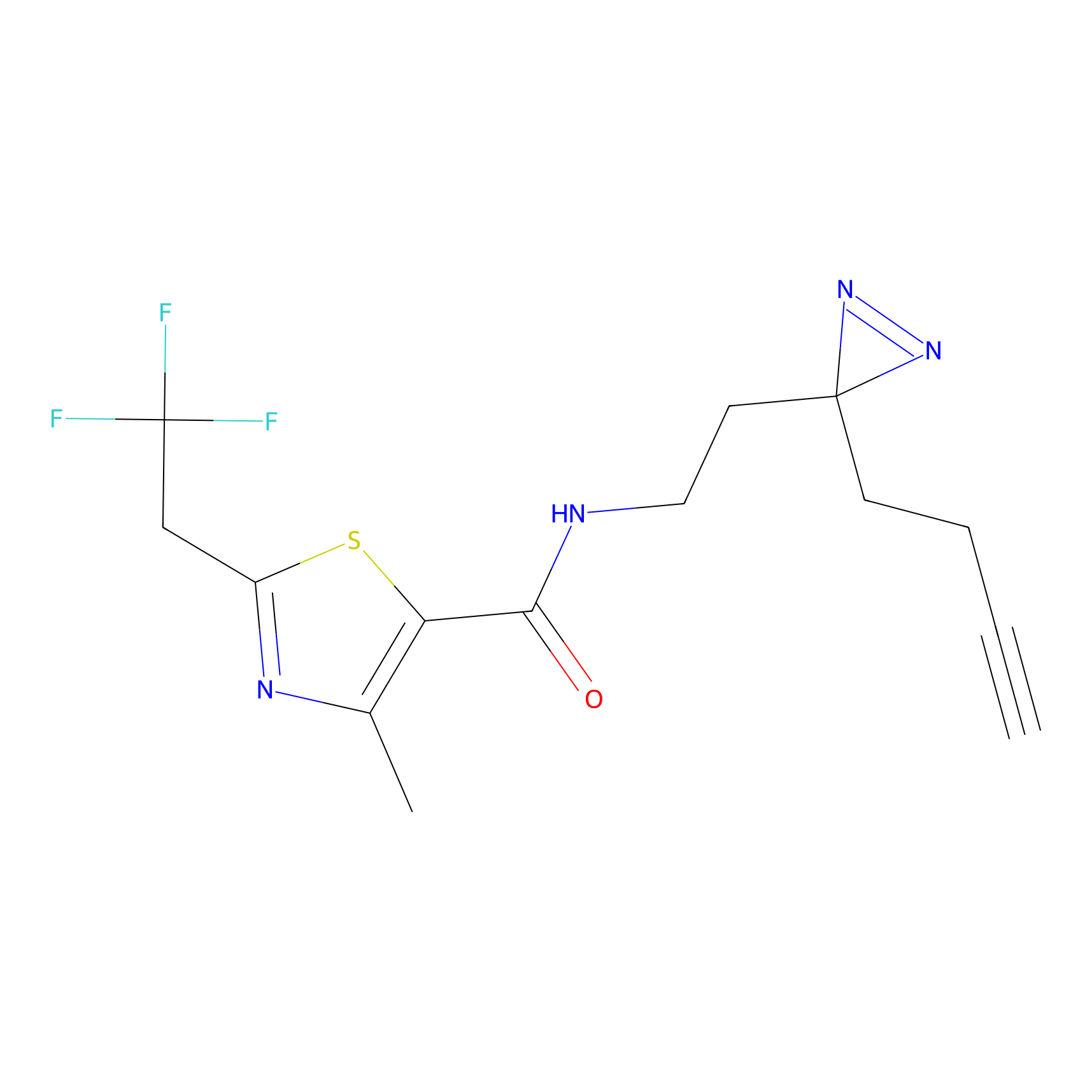
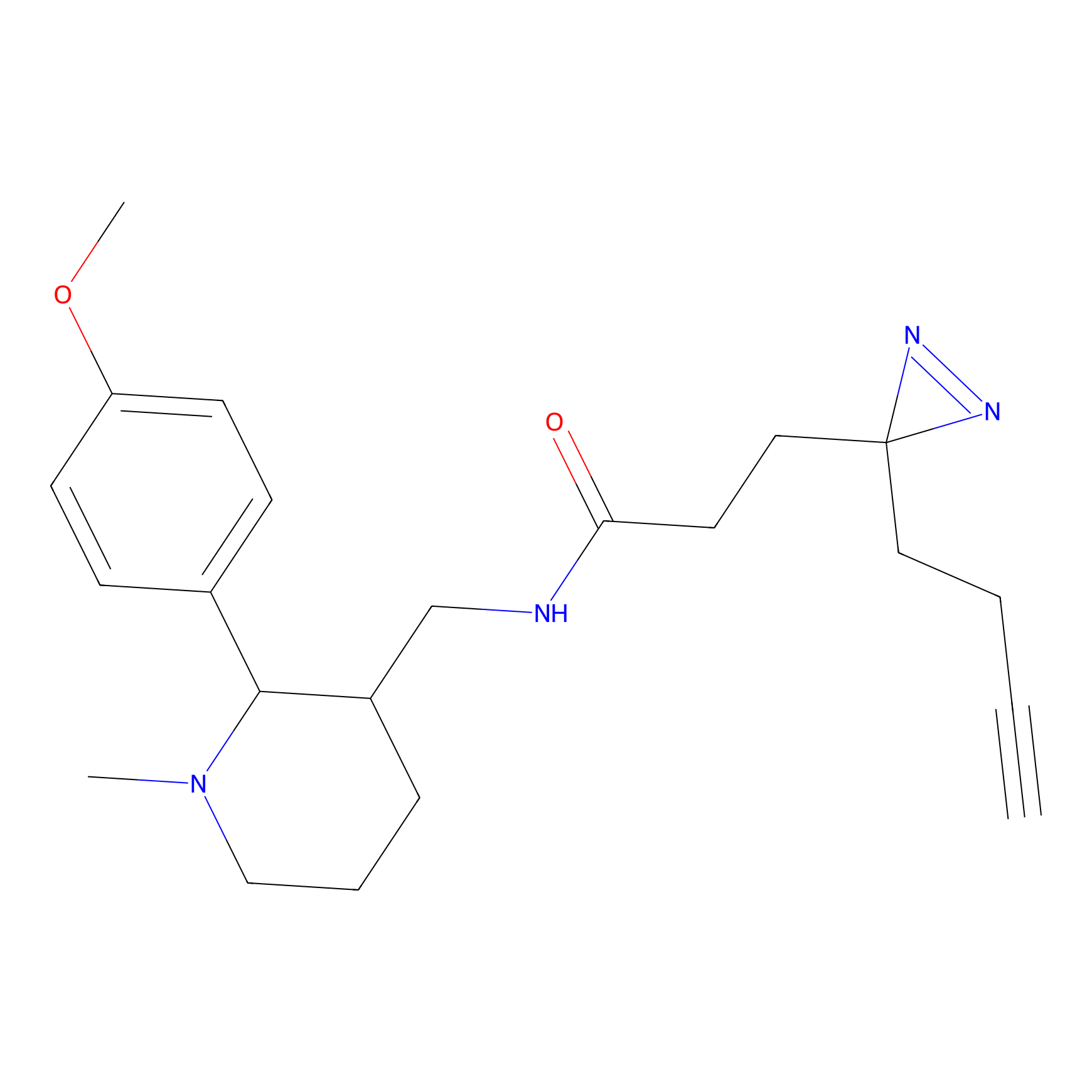
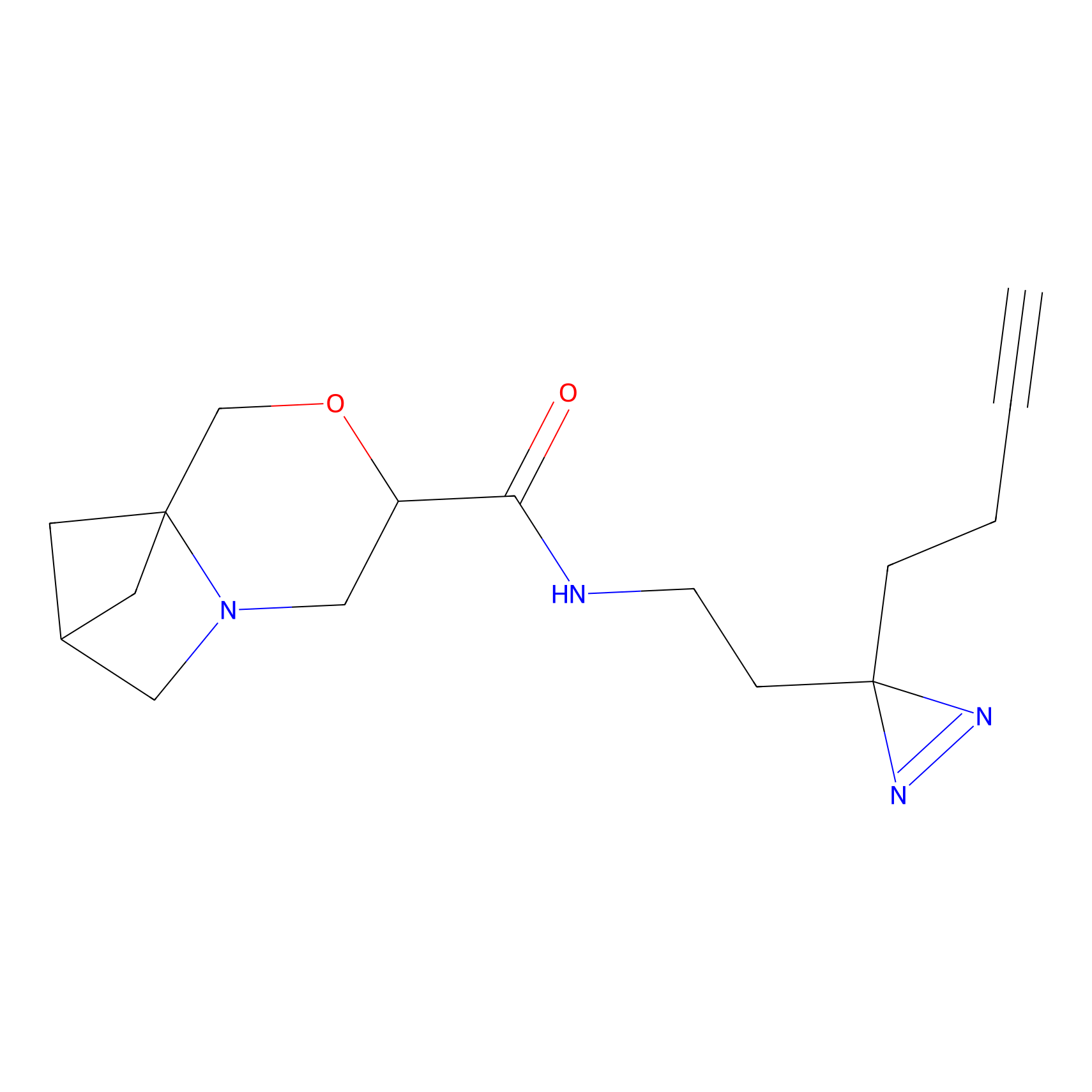
|
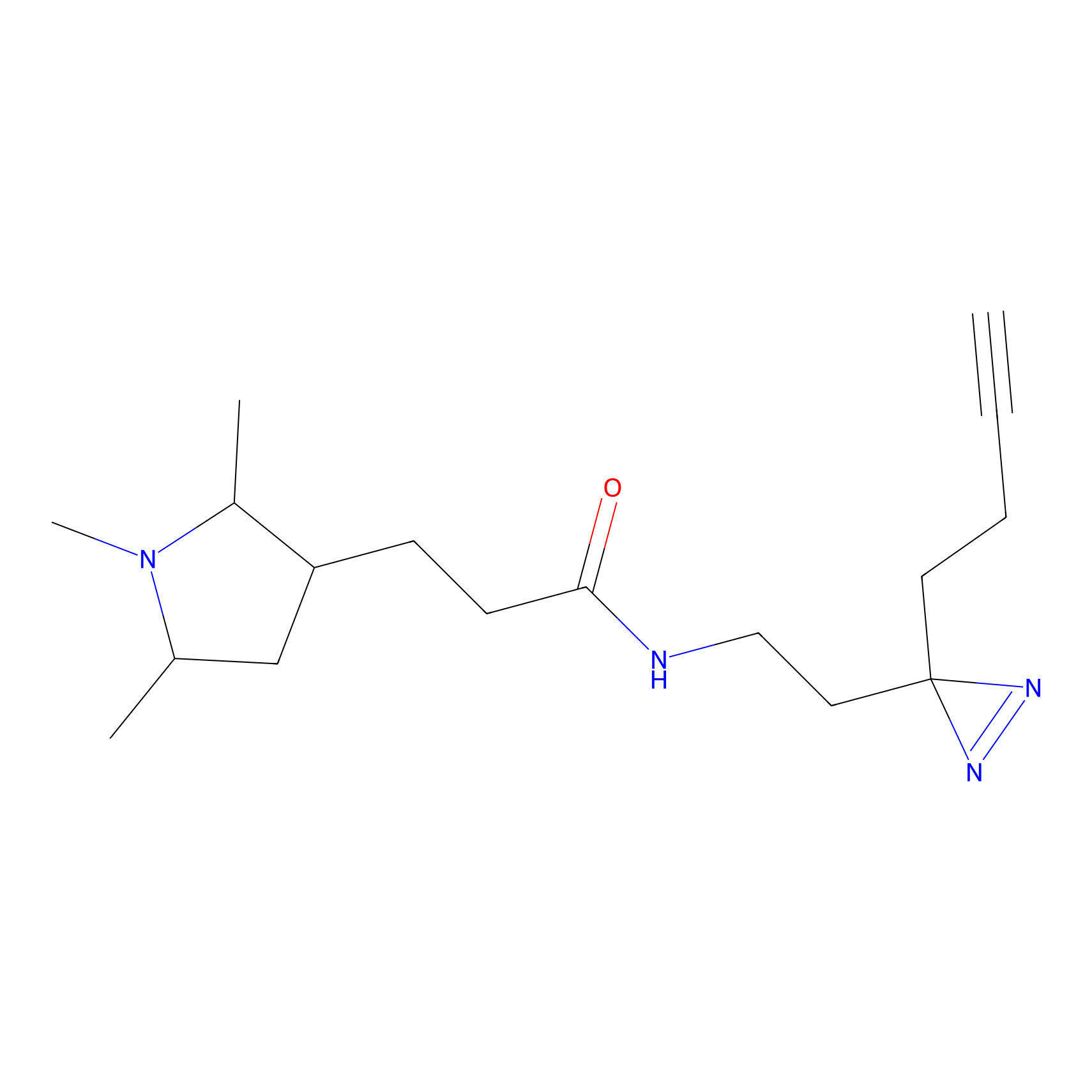
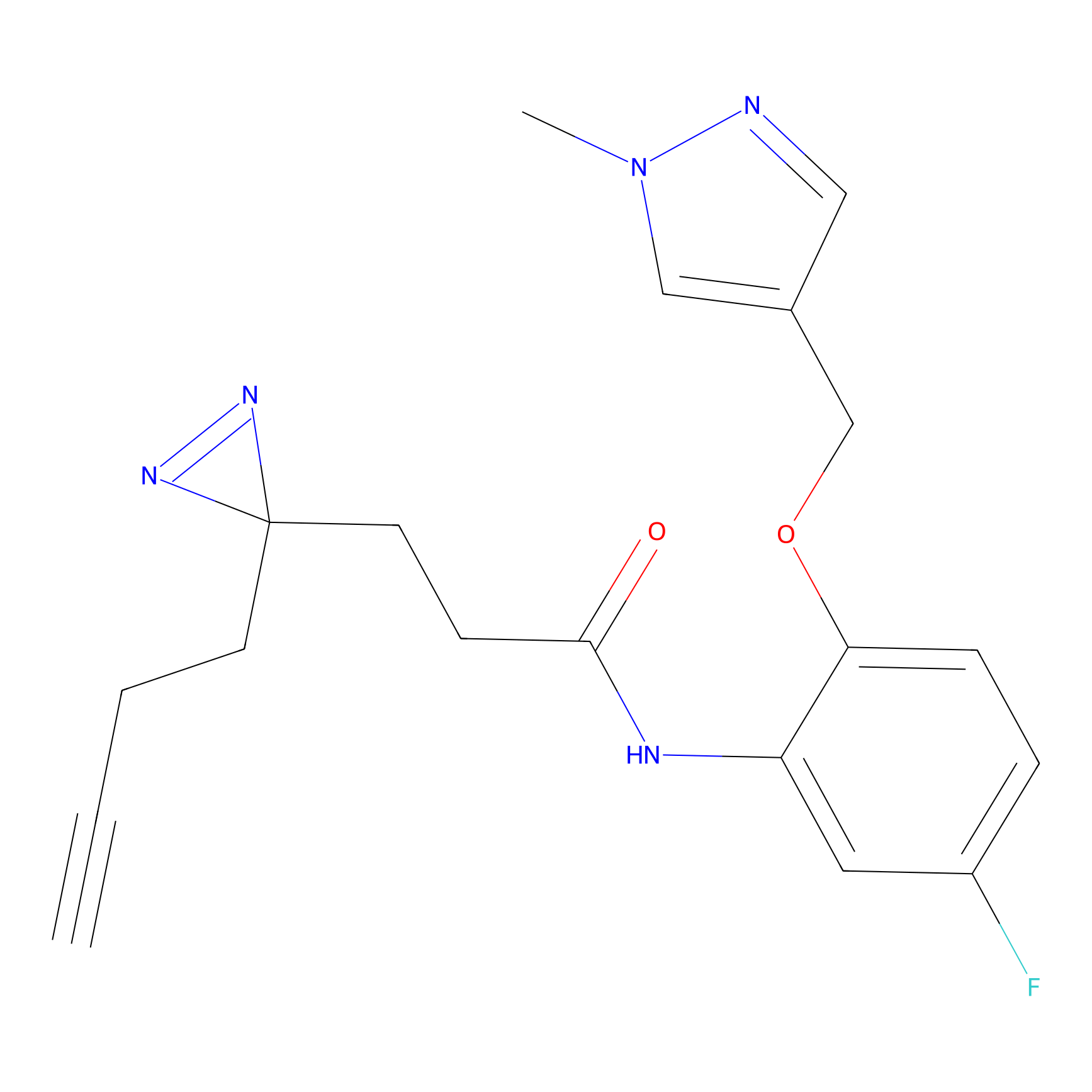
BTD Probe Info |
 |
C783(0.65) | LDD2091 | [2] | |
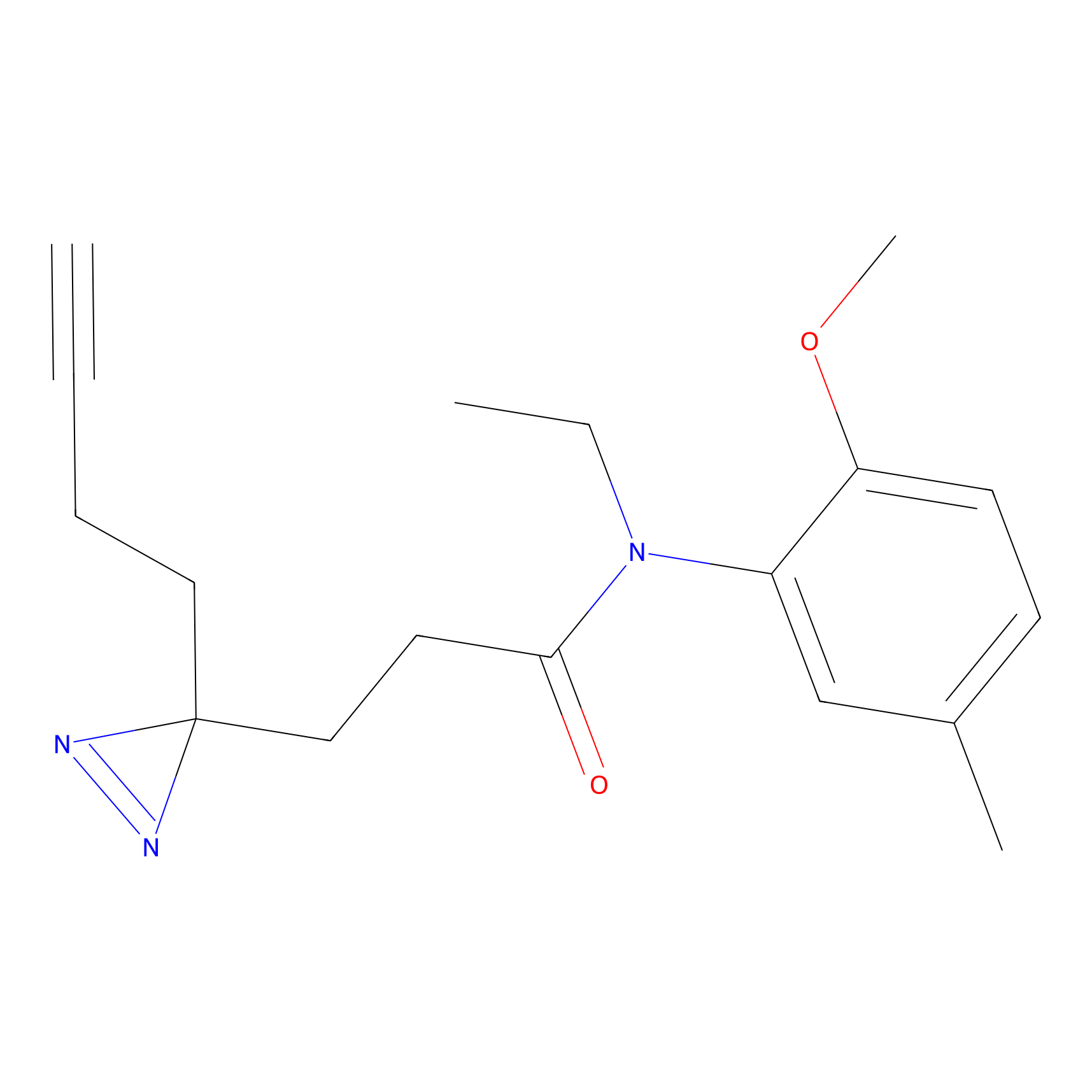
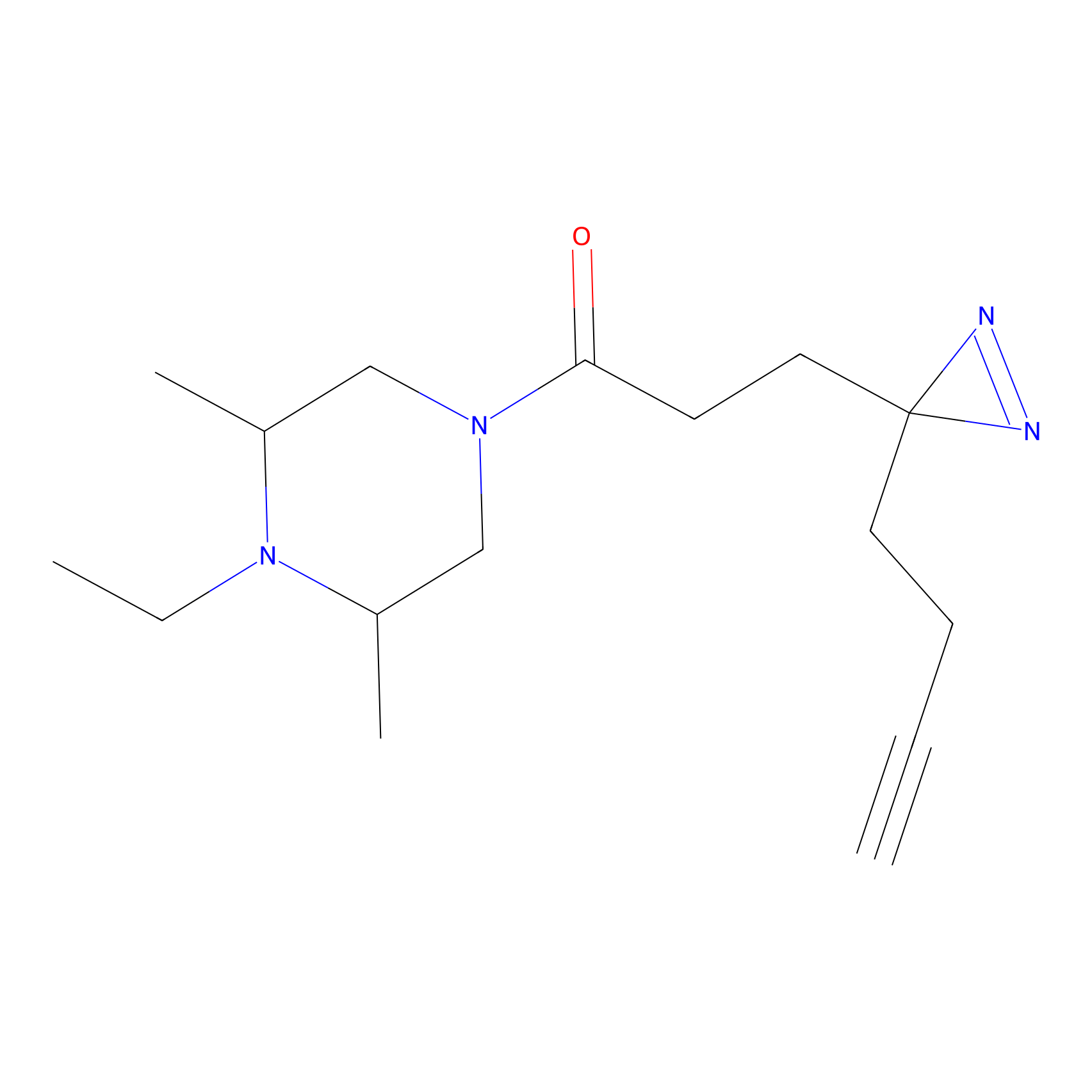
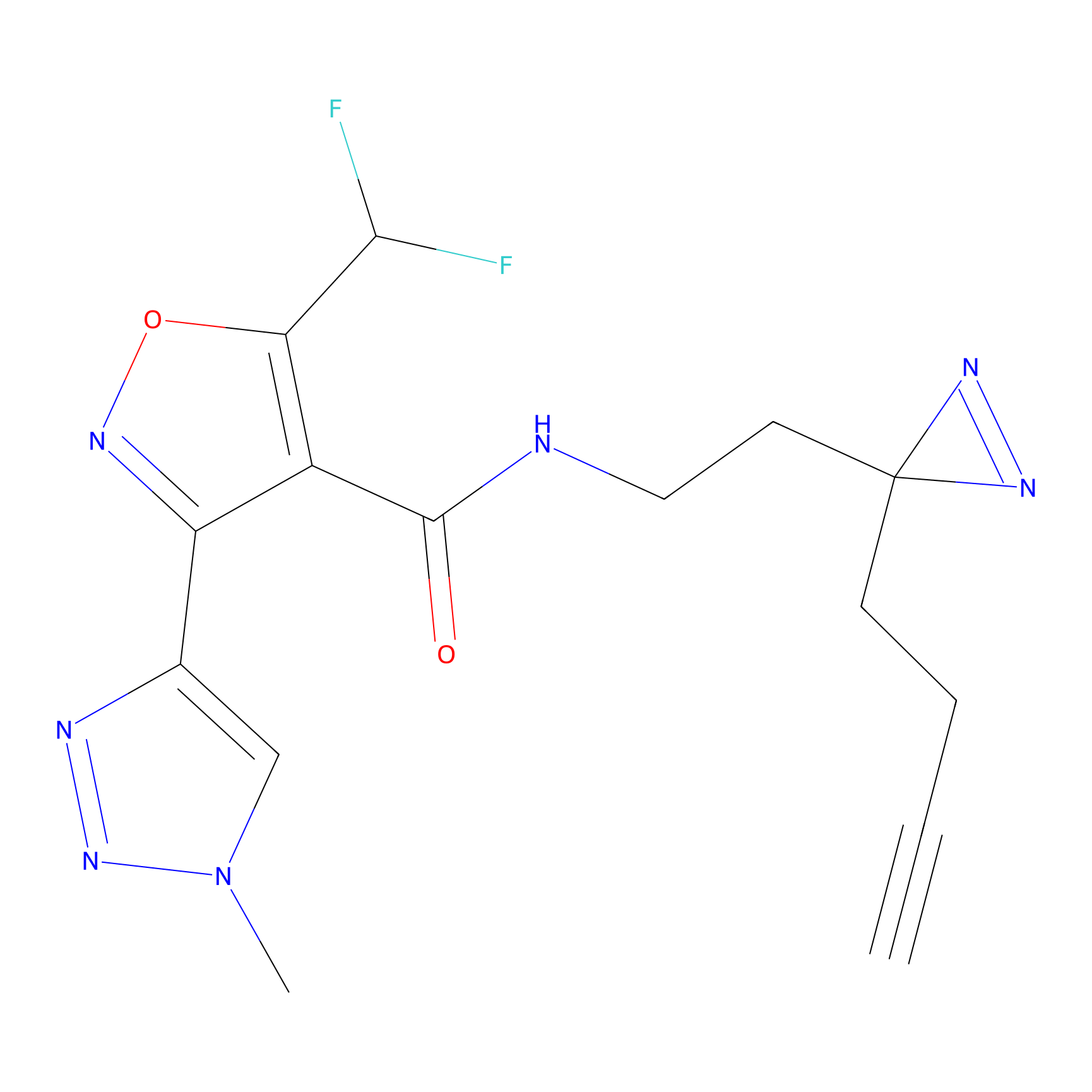
|
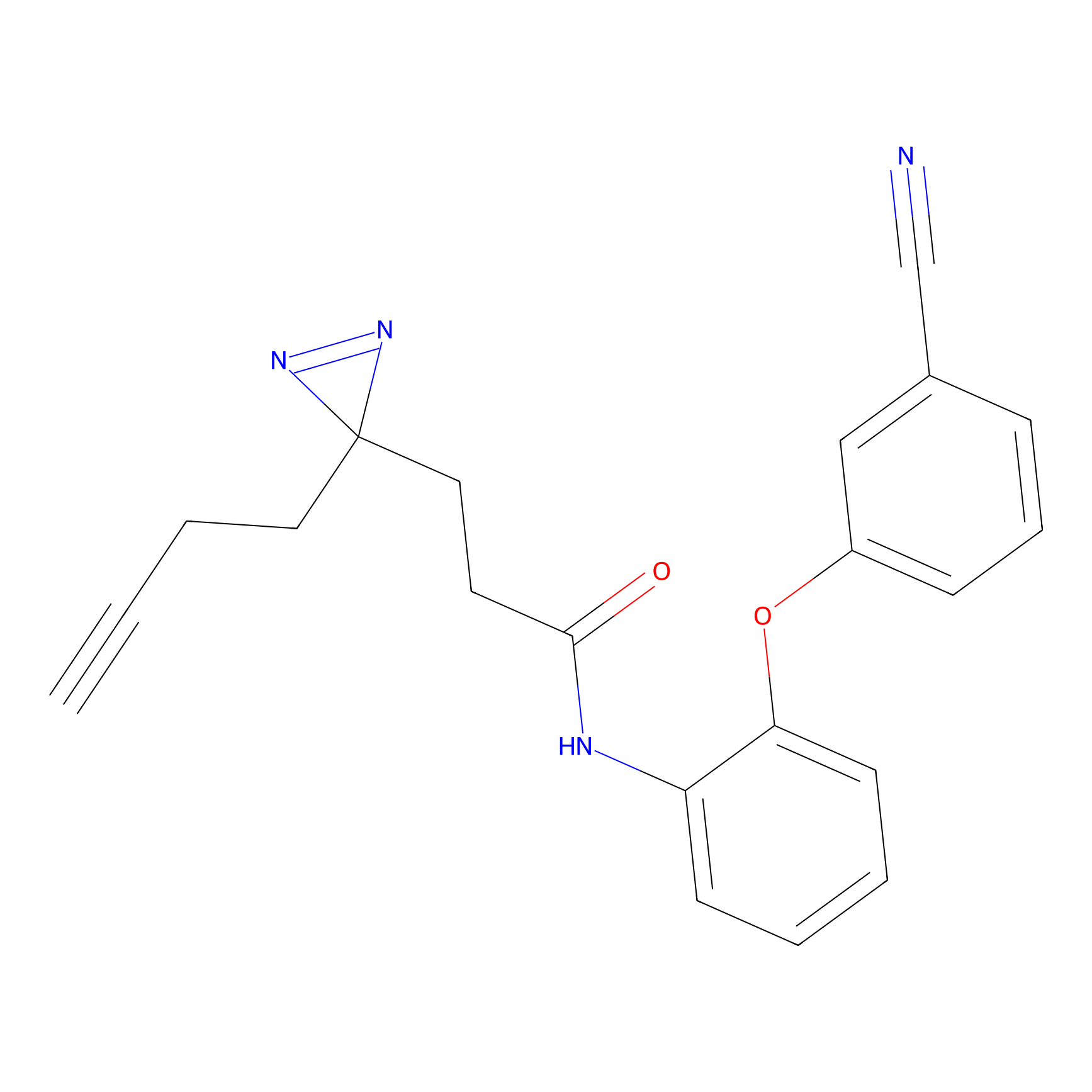
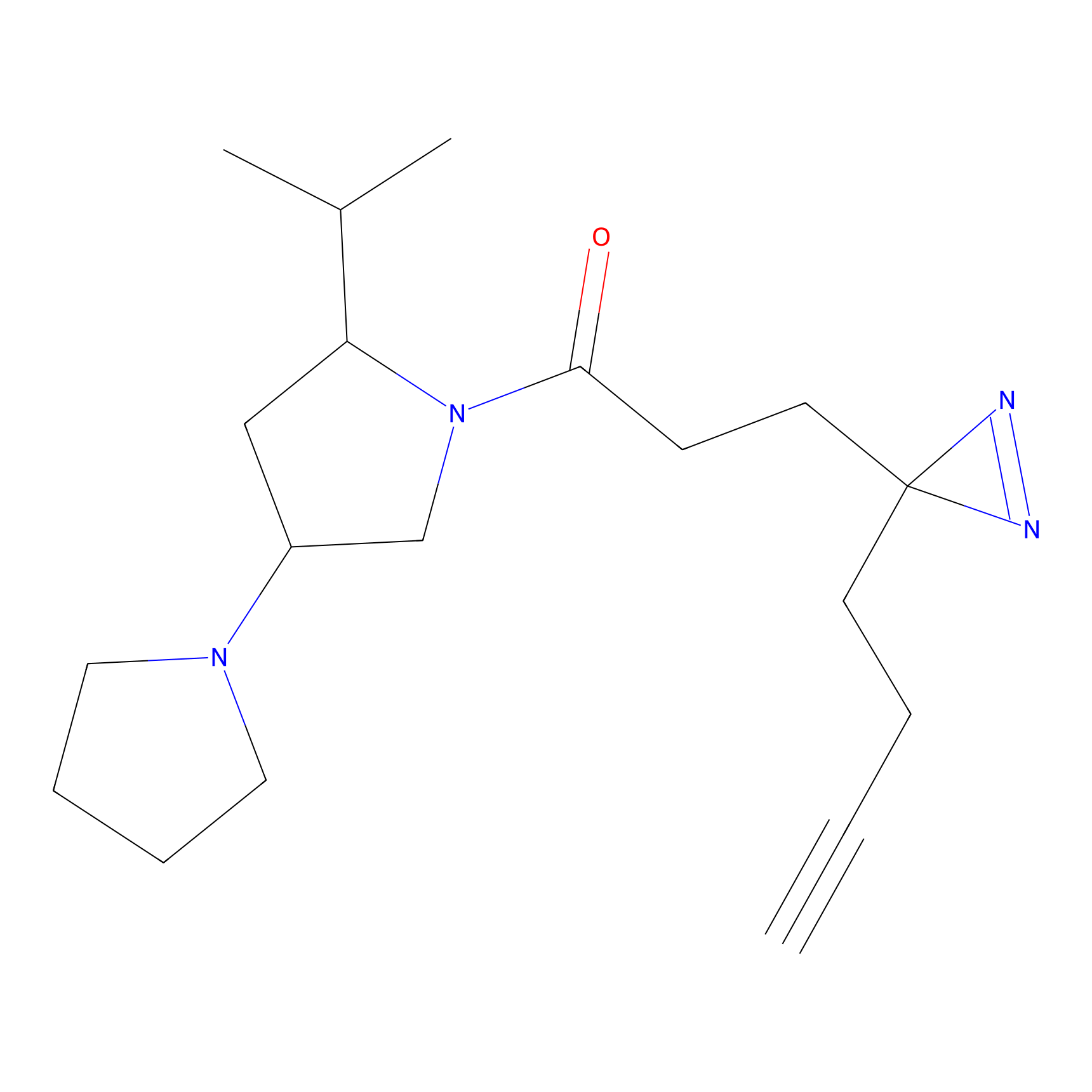
FBPP2 Probe Info |
 |
2.36 | LDD0054 | [3] | |
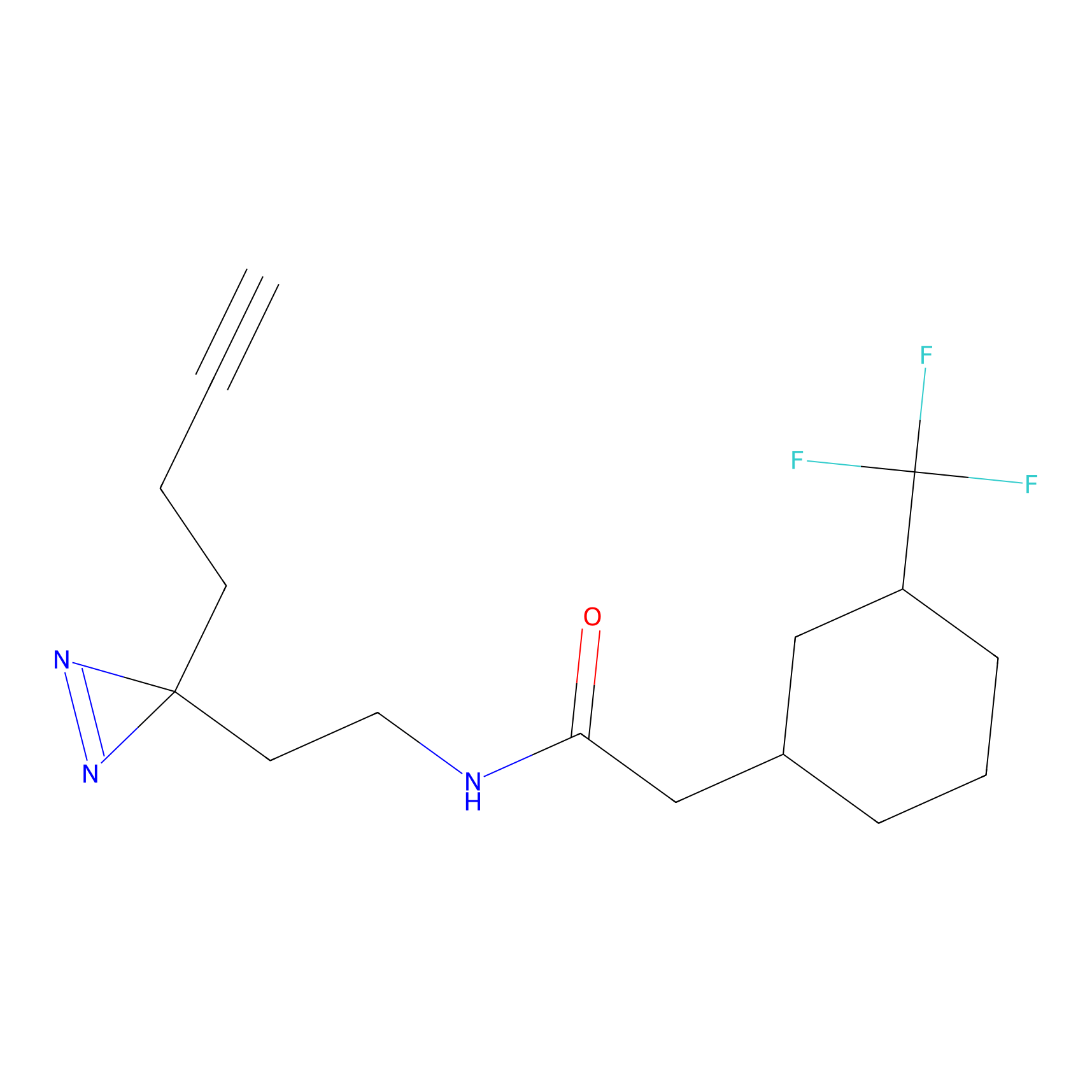
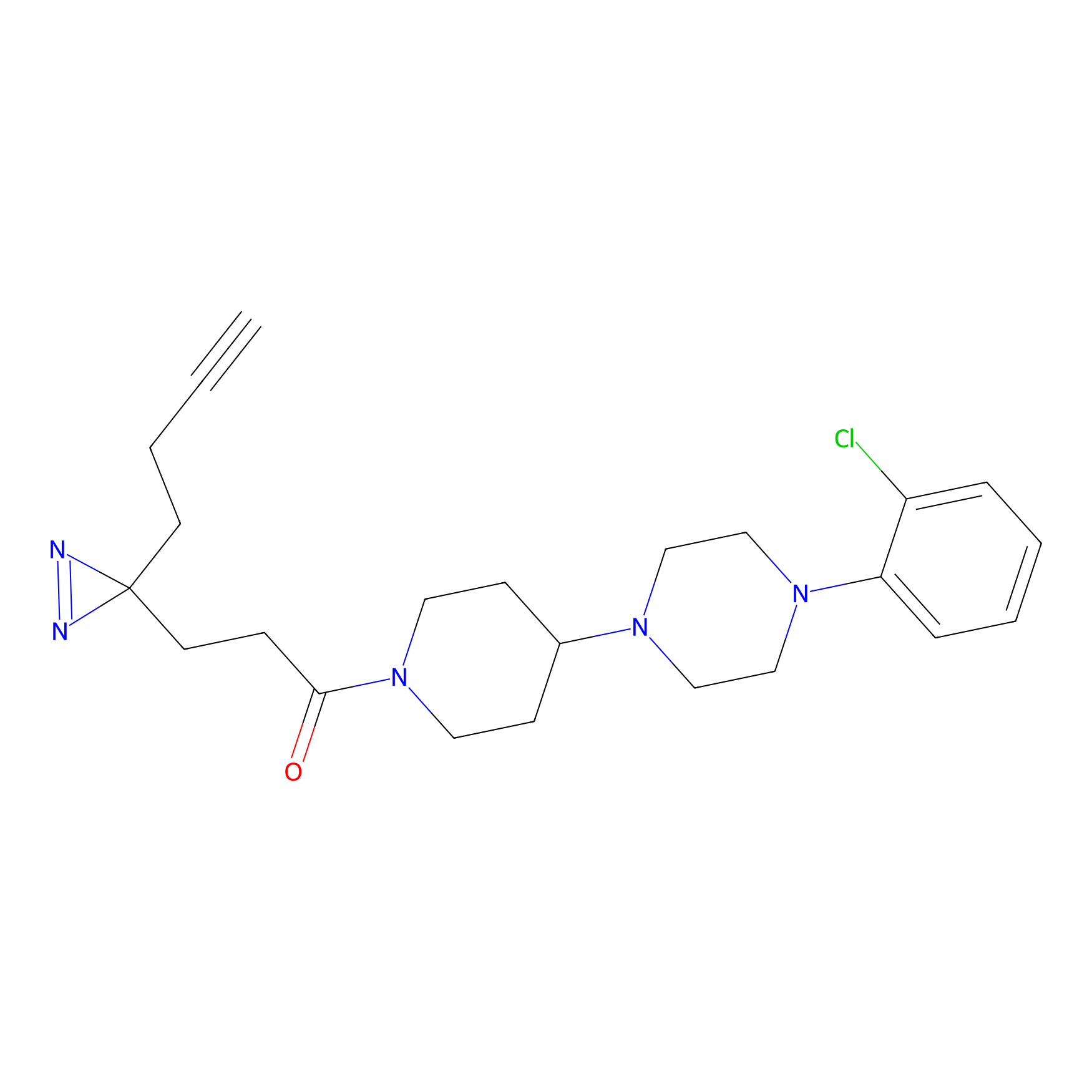
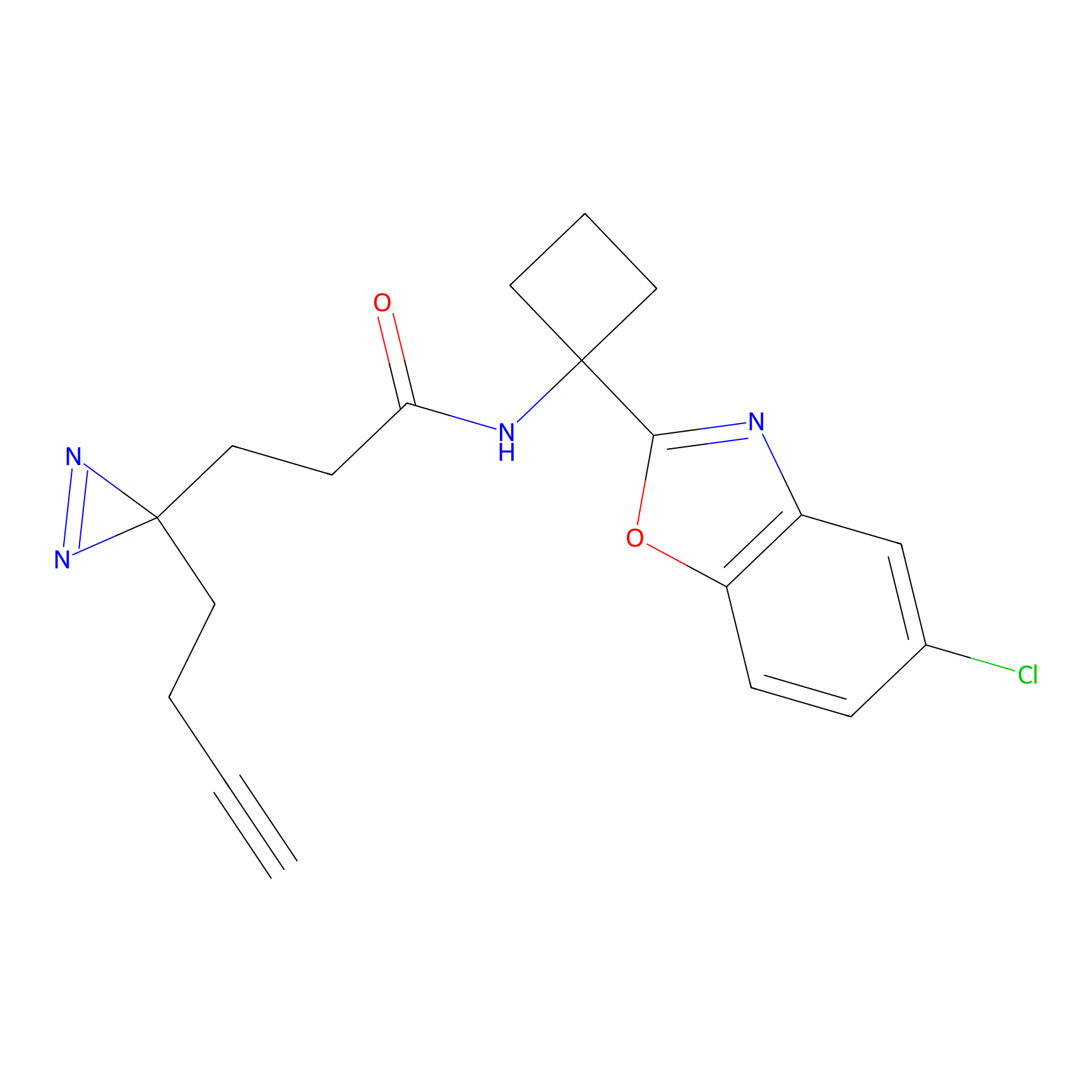
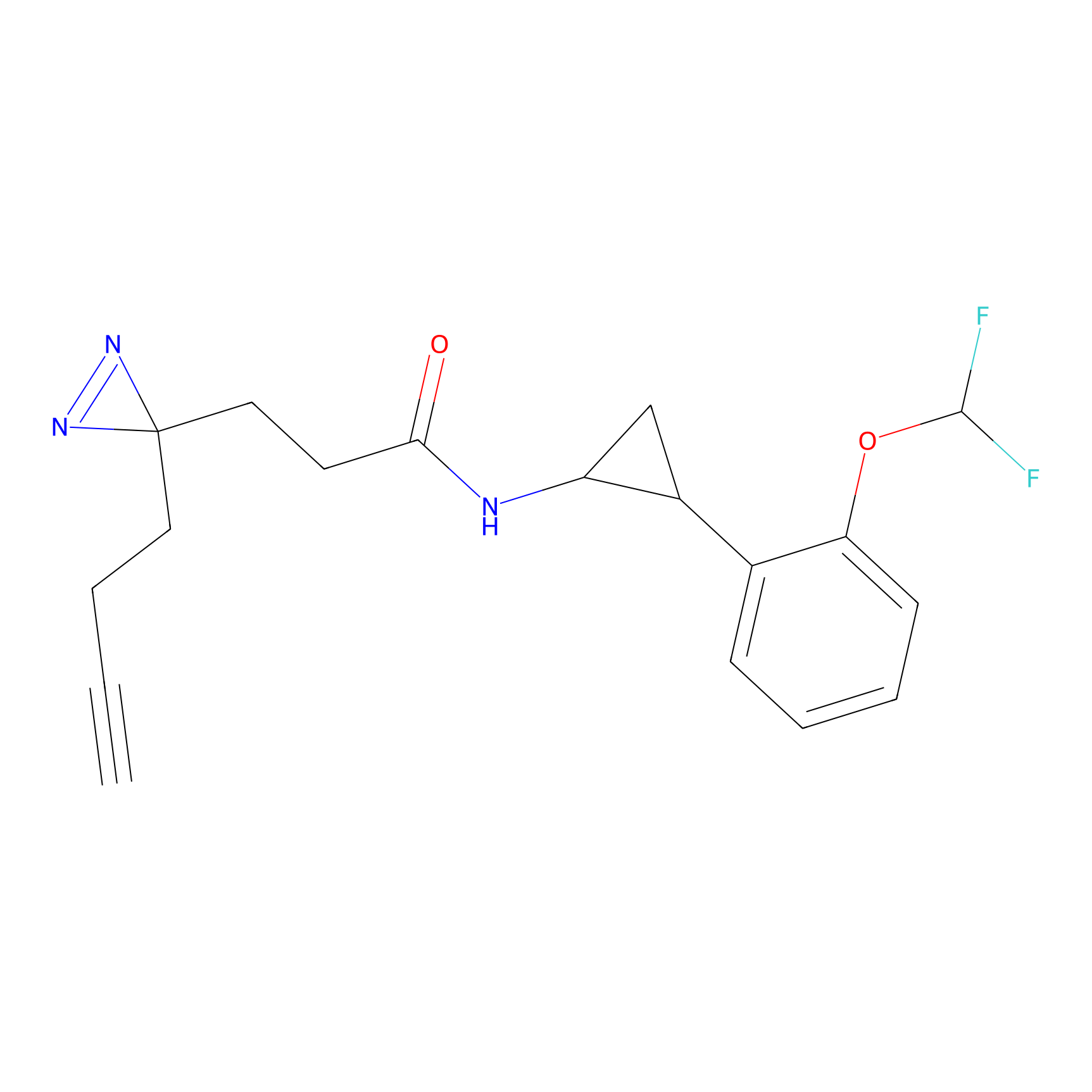
|
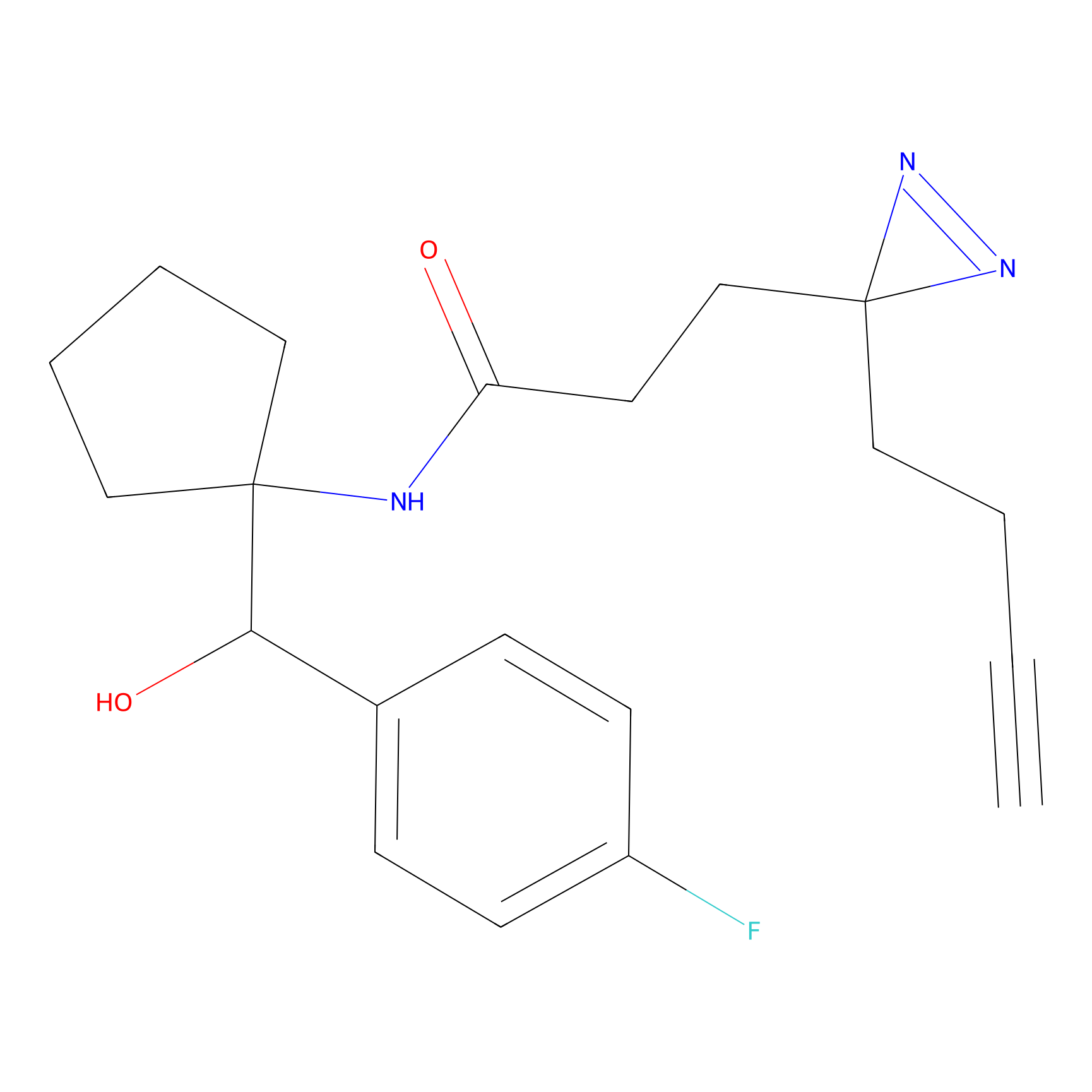
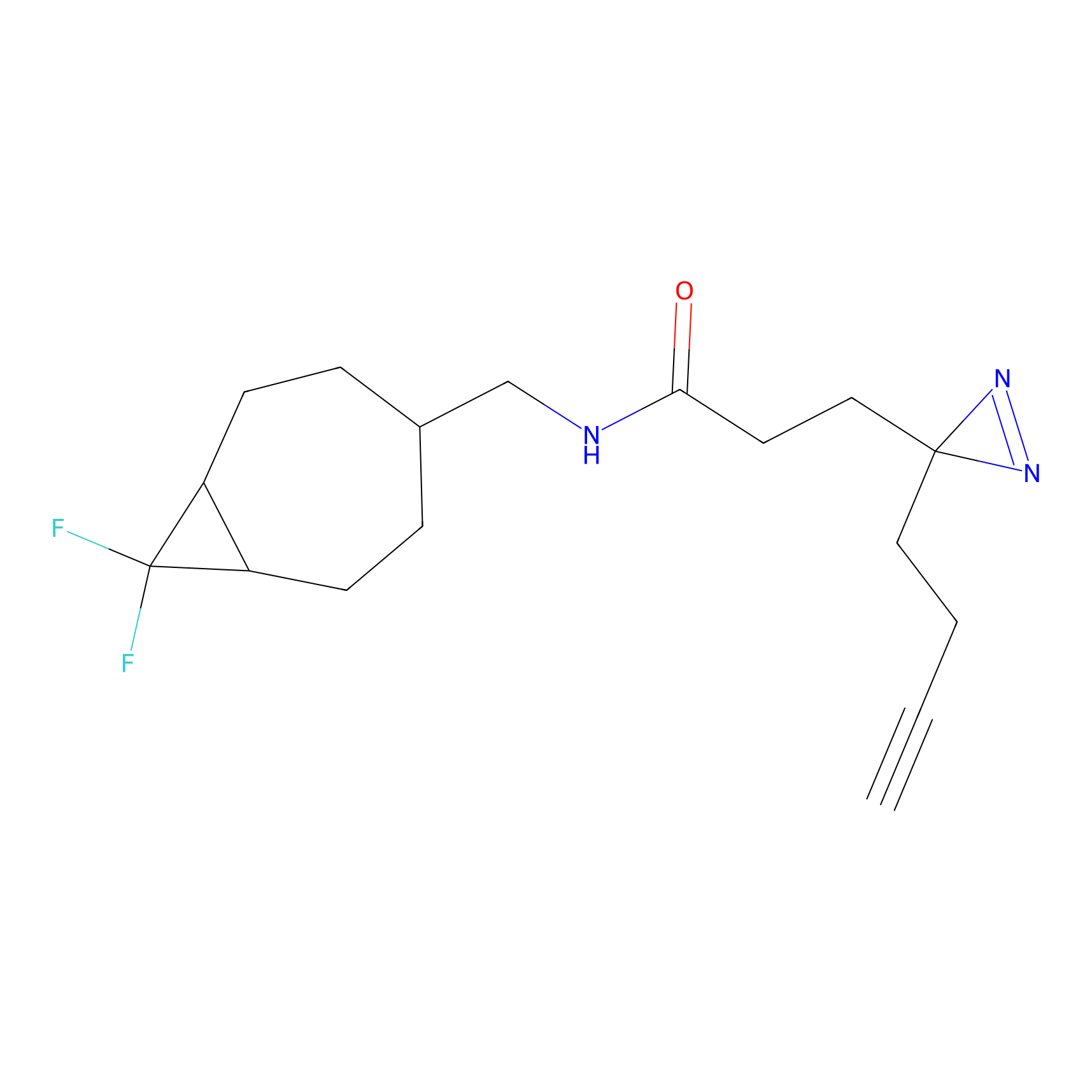
IA-alkyne Probe Info |
 |
C783(0.00); C733(0.00) | LDD0036 | [4] | |
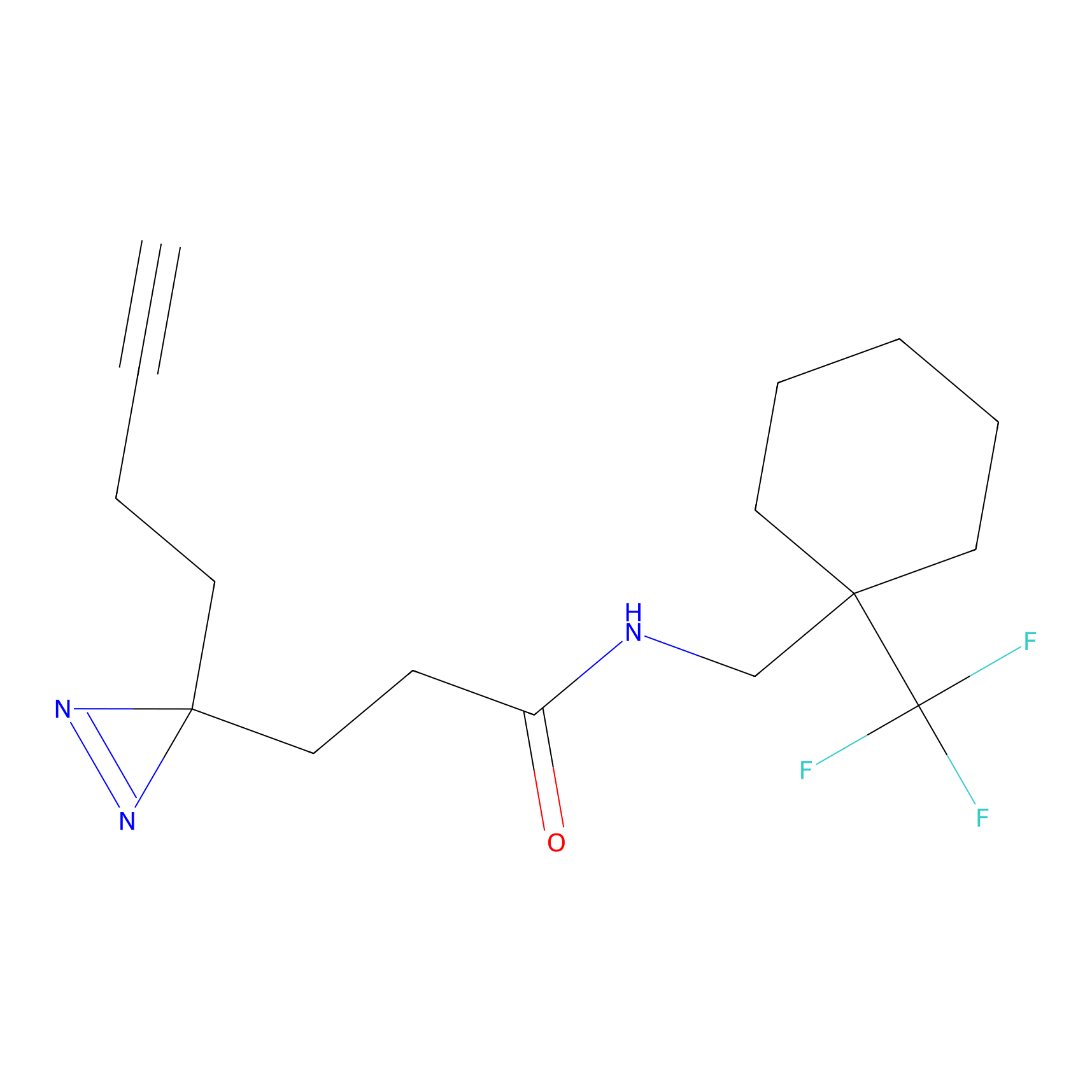
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [4] | |
|
IPM Probe Info |
 |
N.A. | LDD0025 | [5] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2224 | [6] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C733(0.50) | LDD2152 | [2] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C783(0.65) | LDD2091 | [2] |
| LDCM0625 | F8 | Ramos | C237(1.36); C733(1.41) | LDD2187 | [8] |
| LDCM0572 | Fragment10 | Ramos | C237(0.82); C733(1.04) | LDD2189 | [8] |
| LDCM0573 | Fragment11 | Ramos | C237(2.82); C733(3.25) | LDD2190 | [8] |
| LDCM0574 | Fragment12 | Ramos | C237(1.13); C733(0.66) | LDD2191 | [8] |
| LDCM0575 | Fragment13 | Ramos | C237(0.97); C733(1.00) | LDD2192 | [8] |
| LDCM0576 | Fragment14 | Ramos | C237(1.45); C733(0.80) | LDD2193 | [8] |
| LDCM0579 | Fragment20 | Ramos | C733(0.54) | LDD2194 | [8] |
| LDCM0580 | Fragment21 | Ramos | C237(0.96); C733(0.64) | LDD2195 | [8] |
| LDCM0582 | Fragment23 | Ramos | C237(1.11); C733(0.80) | LDD2196 | [8] |
| LDCM0578 | Fragment27 | Ramos | C237(0.60); C733(0.53) | LDD2197 | [8] |
| LDCM0586 | Fragment28 | Ramos | C237(0.71); C733(0.23) | LDD2198 | [8] |
| LDCM0588 | Fragment30 | Ramos | C237(0.81); C733(0.64) | LDD2199 | [8] |
| LDCM0589 | Fragment31 | Ramos | C237(1.01); C733(0.58) | LDD2200 | [8] |
| LDCM0590 | Fragment32 | Ramos | C237(0.94); C733(3.37) | LDD2201 | [8] |
| LDCM0468 | Fragment33 | Ramos | C237(1.58); C733(0.78) | LDD2202 | [8] |
| LDCM0596 | Fragment38 | Ramos | C237(0.80); C733(0.65) | LDD2203 | [8] |
| LDCM0566 | Fragment4 | Ramos | C237(1.11); C733(1.11) | LDD2184 | [8] |
| LDCM0610 | Fragment52 | Ramos | C237(1.34); C733(0.39) | LDD2204 | [8] |
| LDCM0614 | Fragment56 | Ramos | C237(0.99); C733(0.77) | LDD2205 | [8] |
| LDCM0569 | Fragment7 | Ramos | C237(0.91); C733(0.98) | LDD2186 | [8] |
| LDCM0571 | Fragment9 | Ramos | C733(0.85) | LDD2188 | [8] |
| LDCM0022 | KB02 | HEK-293T | C733(1.02); C237(1.03) | LDD1492 | [9] |
| LDCM0023 | KB03 | HEK-293T | C733(0.98); C237(0.99) | LDD1497 | [9] |
| LDCM0024 | KB05 | COLO792 | C733(1.64) | LDD3310 | [1] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C733(1.01) | LDD2121 | [2] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C783(1.74) | LDD2094 | [2] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C733(0.91) | LDD2104 | [2] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C783(1.66) | LDD2107 | [2] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C733(0.51) | LDD2120 | [2] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C733(1.10) | LDD2135 | [2] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C733(0.66) | LDD2143 | [2] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C733(0.66); C783(0.63) | LDD2150 | [2] |
| LDCM0008 | Tranylcypromine | SH-SY5Y | 2.36 | LDD0054 | [3] |
The Interaction Atlas With This Target
References