Details of the Target
General Information of Target
| Target ID | LDTP07156 | |||||
|---|---|---|---|---|---|---|
| Target Name | Protein wntless homolog (WLS) | |||||
| Gene Name | WLS | |||||
| Gene ID | 79971 | |||||
| Synonyms |
C1orf139; GPR177; Protein wntless homolog; Integral membrane protein GPR177; Protein evenness interrupted homolog; EVI; Putative NF-kappa-B-activating protein 373 |
|||||
| 3D Structure | ||||||
| Sequence |
MAGAIIENMSTKKLCIVGGILLVFQIIAFLVGGLIAPGPTTAVSYMSVKCVDARKNHHKT
KWFVPWGPNHCDKIRDIEEAIPREIEANDIVFSVHIPLPHMEMSPWFQFMLFILQLDIAF KLNNQIRENAEVSMDVSLAYRDDAFAEWTEMAHERVPRKLKCTFTSPKTPEHEGRYYECD VLPFMEIGSVAHKFYLLNIRLPVNEKKKINVGIGEIKDIRLVGIHQNGGFTKVWFAMKTF LTPSIFIIMVWYWRRITMMSRPPVLLEKVIFALGISMTFINIPVEWFSIGFDWTWMLLFG DIRQGIFYAMLLSFWIIFCGEHMMDQHERNHIAGYWKQVGPIAVGSFCLFIFDMCERGVQ LTNPFYSIWTTDIGTELAMAFIIVAGICLCLYFLFLCFMVFQVFRNISGKQSSLPAMSKV RRLHYEGLIFRFKFLMLITLACAAMTVIFFIVSQVTEGHWKWGGVTVQVNSAFFTGIYGM WNLYVFALMFLYAPSHKNYGEDQSNGDLGVHSGEELQLTTTITHVDGPTEIYKLTRKEAQ E |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
Wntless family
|
|||||
| Subcellular location |
Golgi apparatus membrane
|
|||||
| Function |
Regulates Wnt proteins sorting and secretion in a feedback regulatory mechanism. This reciprocal interaction plays a key role in the regulation of expression, subcellular location, binding and organelle-specific association of Wnt proteins. Plays also an important role in establishment of the anterior-posterior body axis formation during development.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| ICC12 | SNV: p.K419N | DBIA Probe Info | |||
| IPC298 | SNV: p.I373T | DBIA Probe Info | |||
| JM1 | SNV: p.L349V | . | |||
| NCIH661 | SNV: p.P263A | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
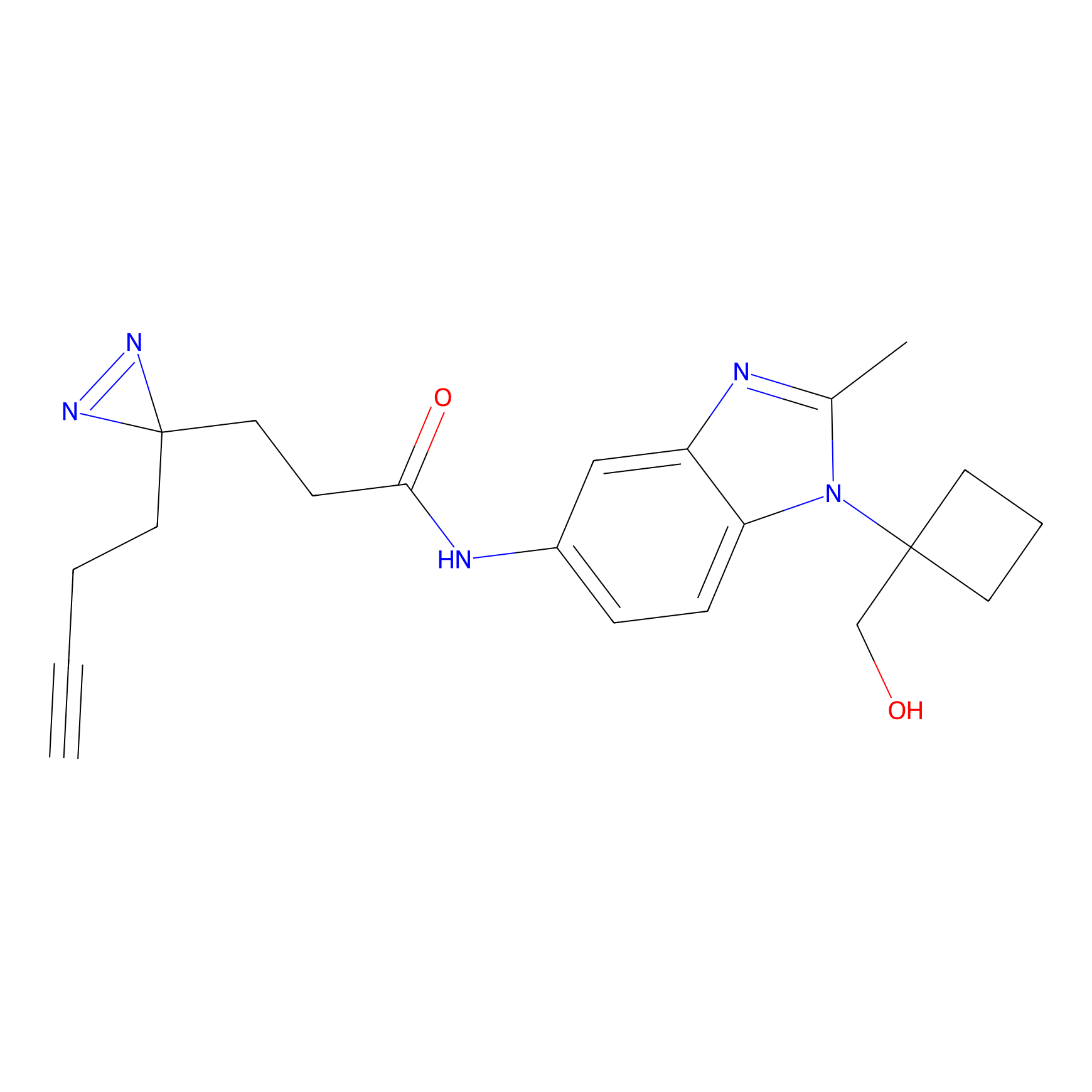
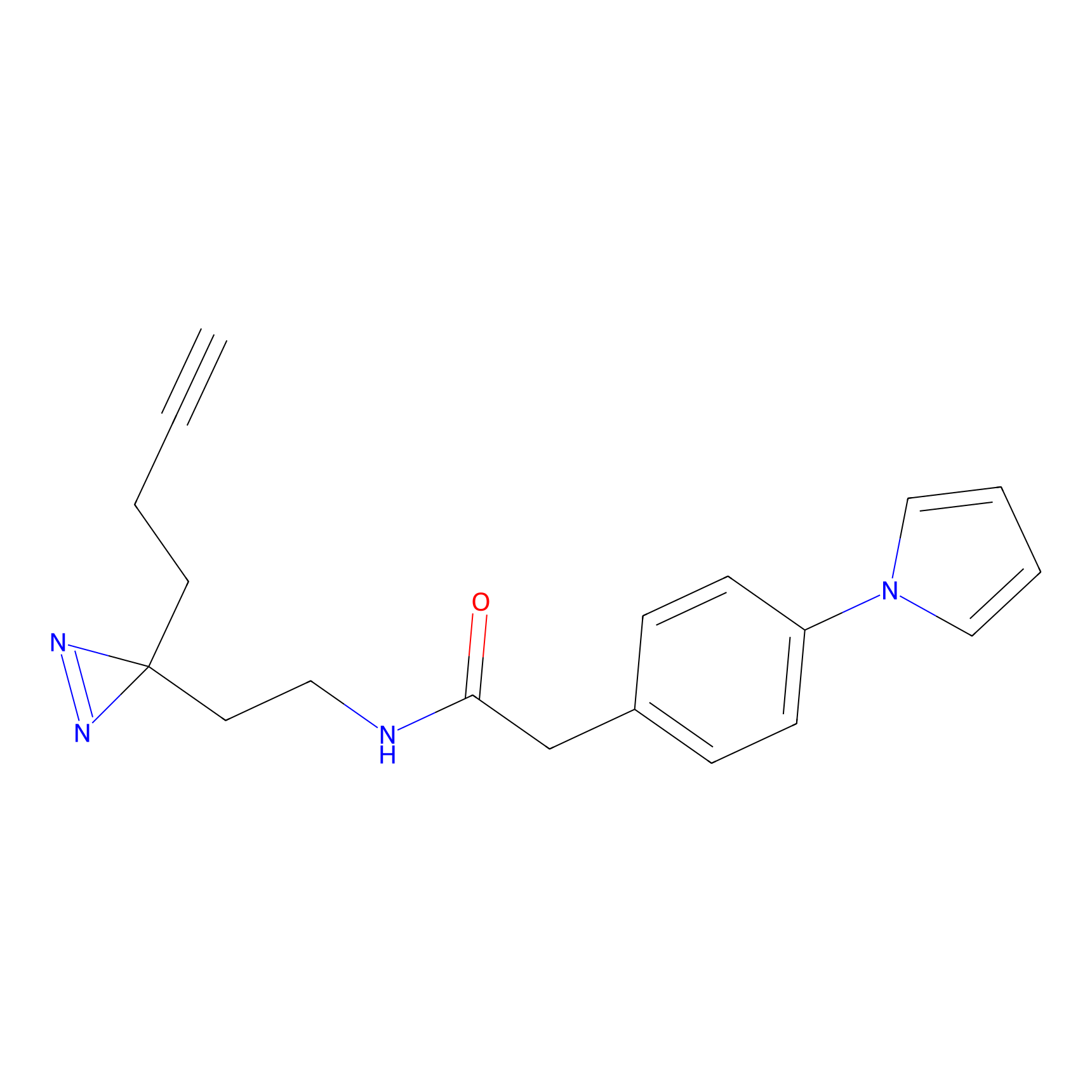
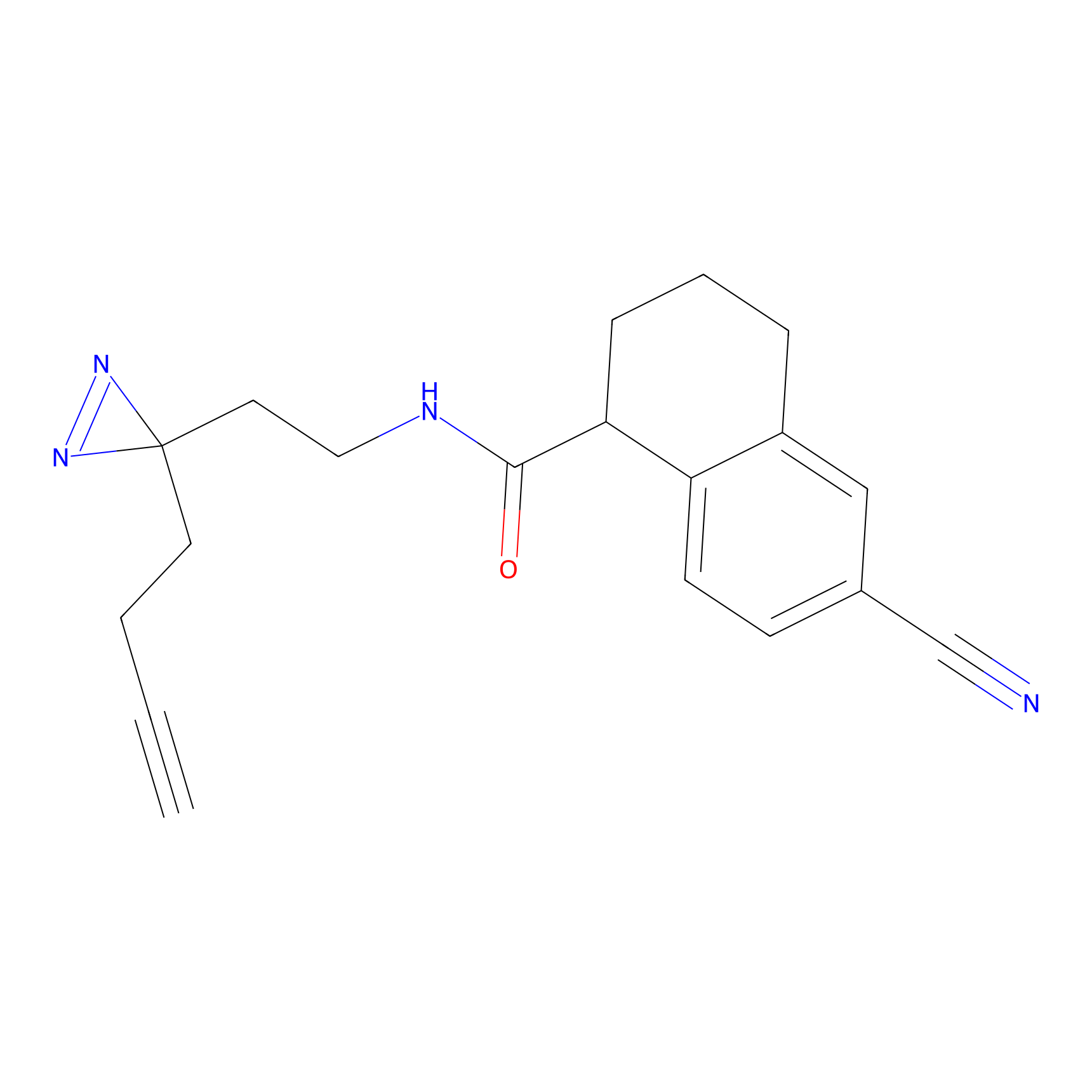
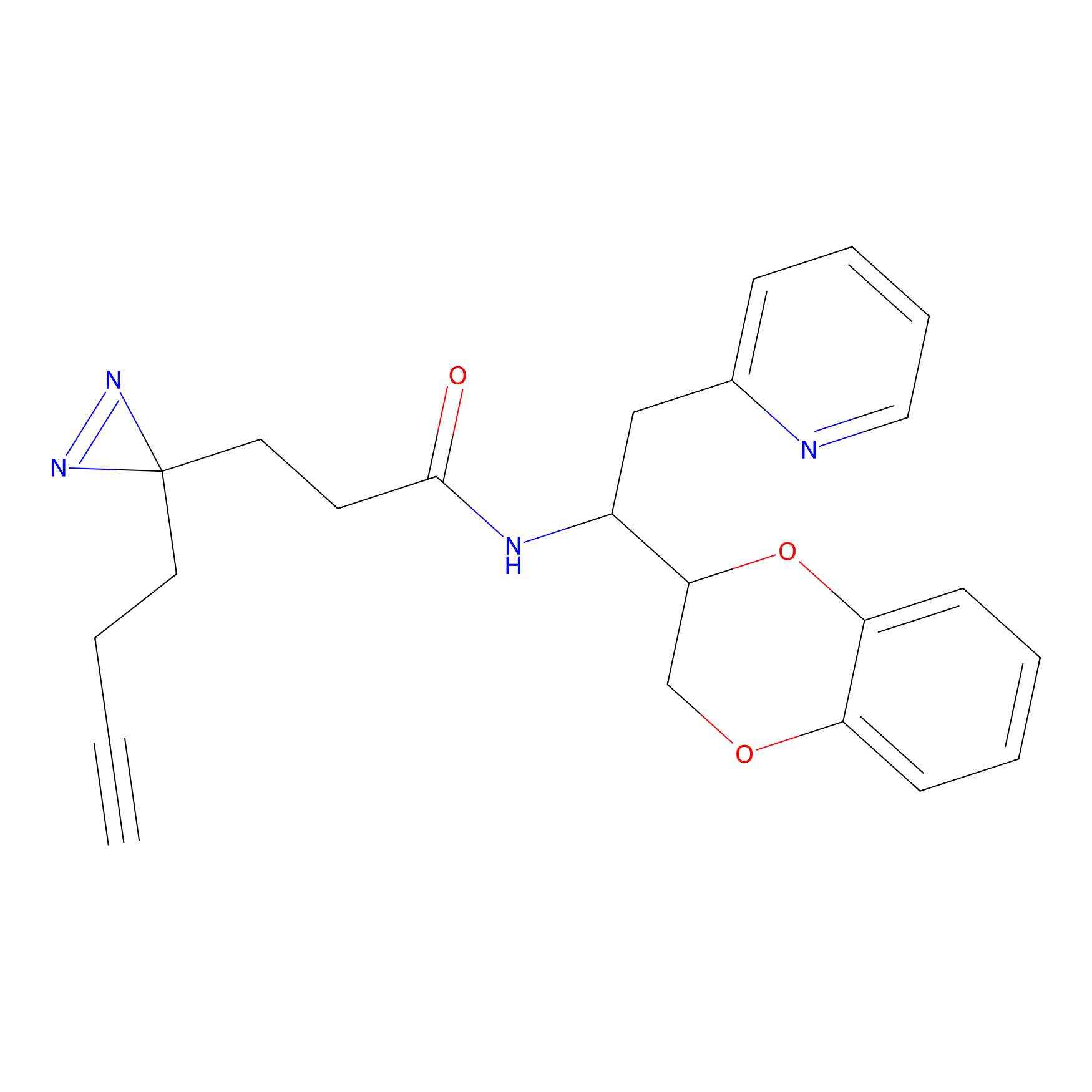
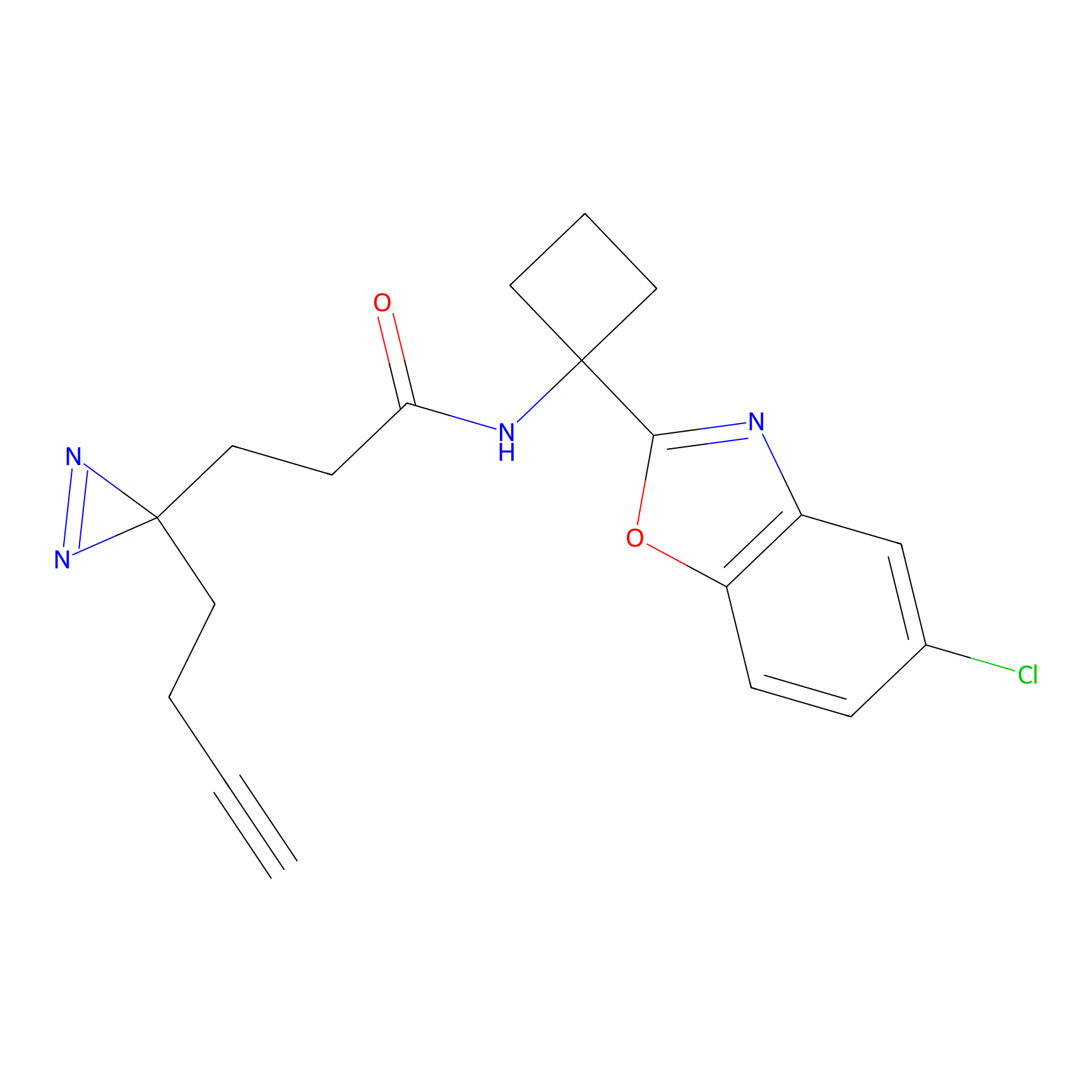
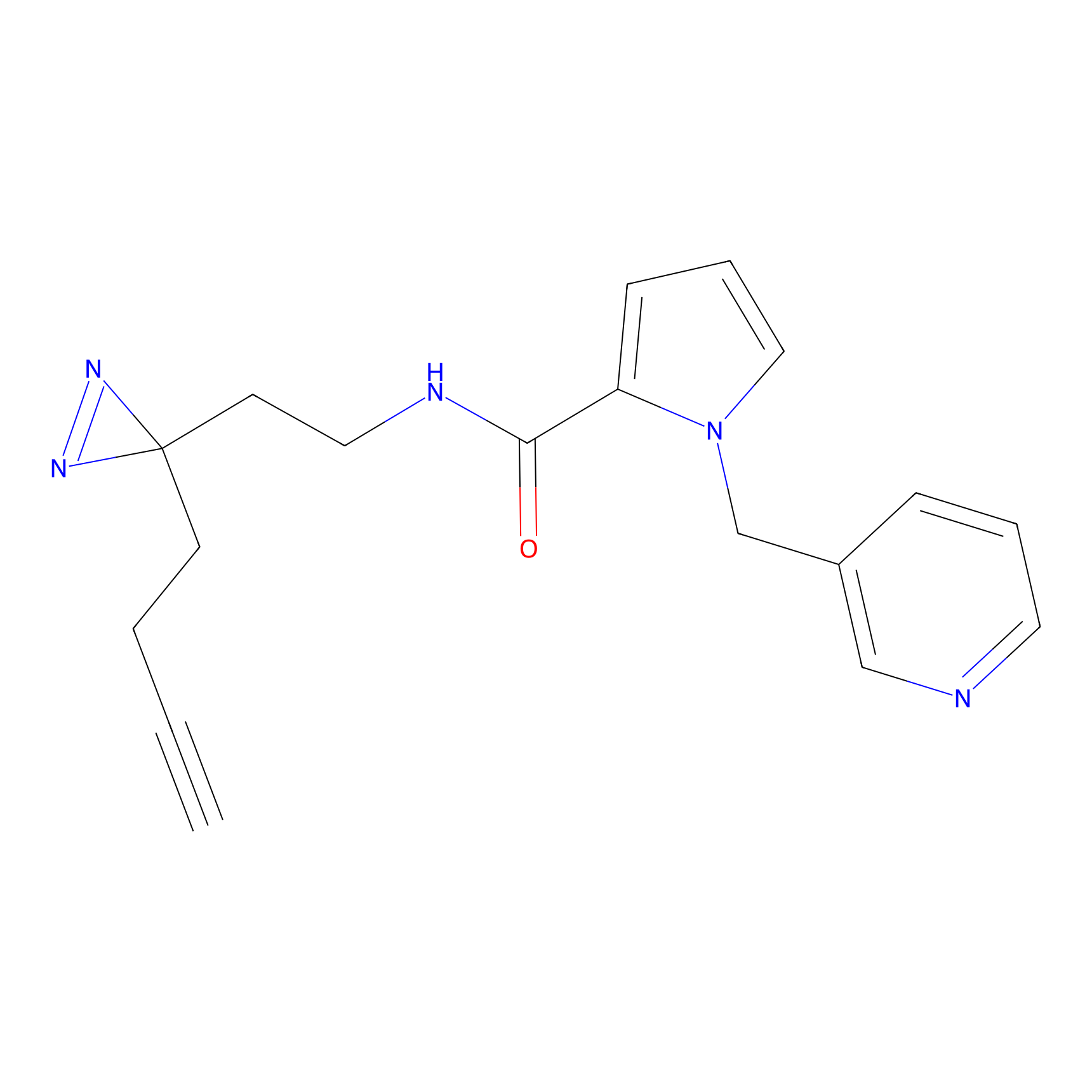
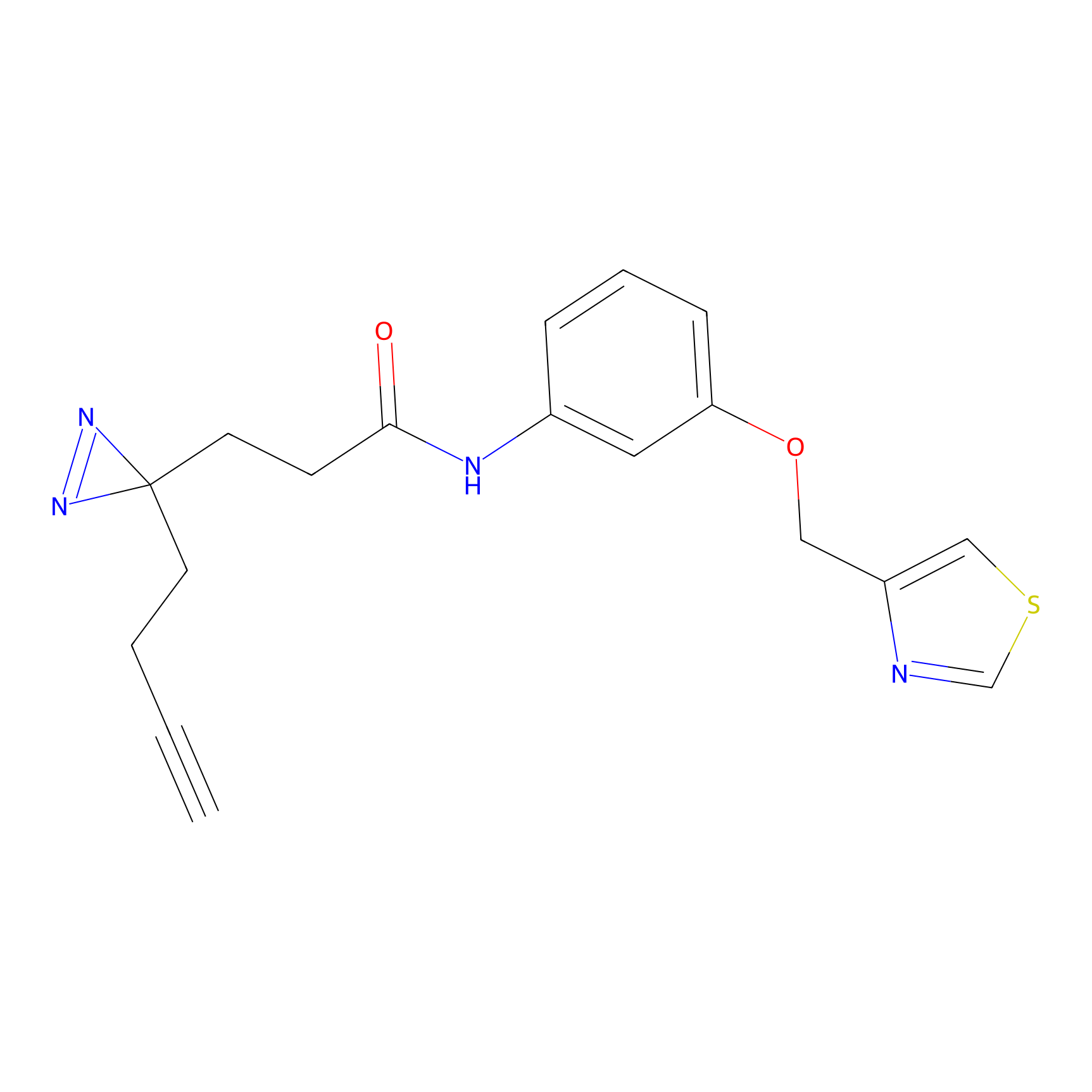
|
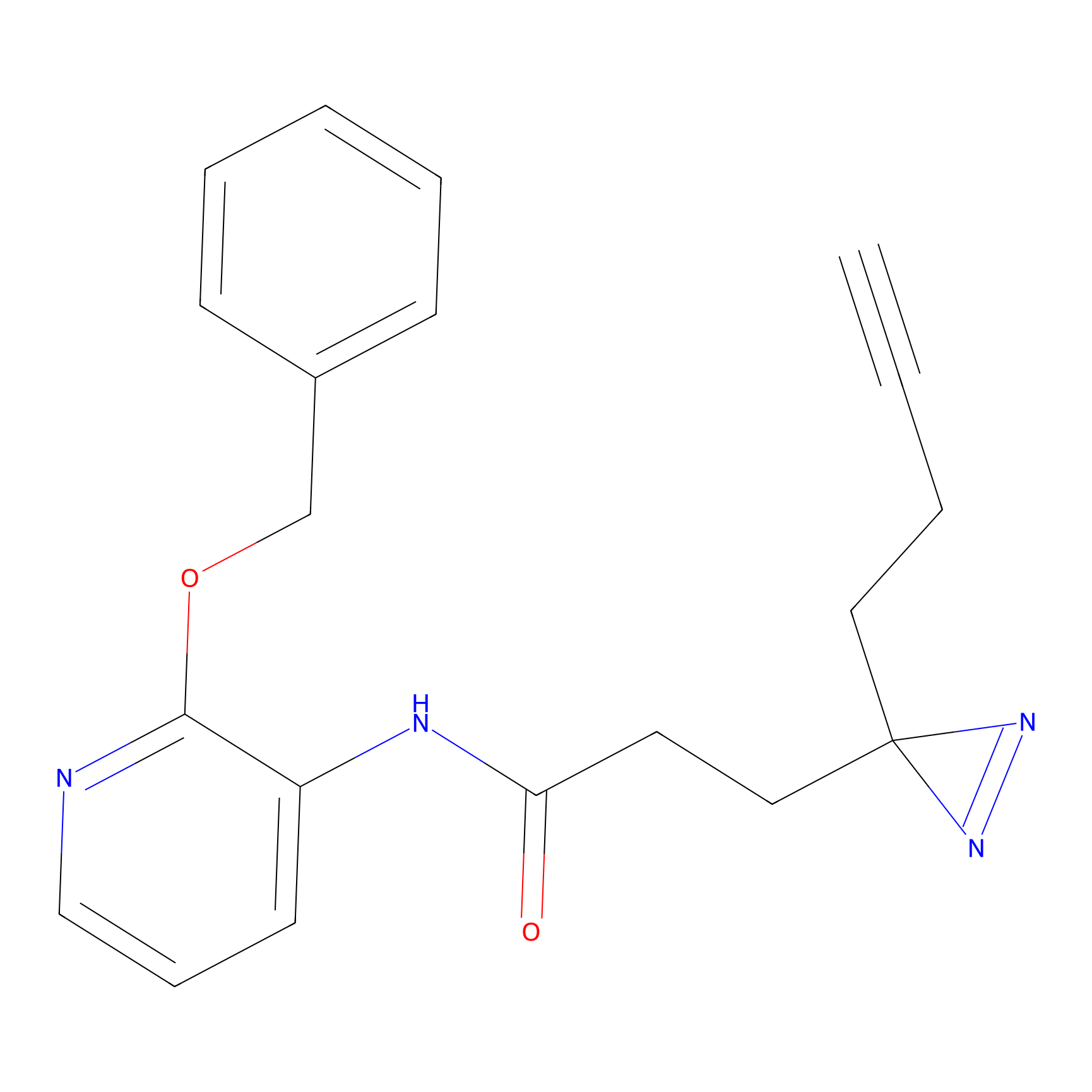
STPyne Probe Info |
 |
K12(6.25) | LDD0277 | [1] | |
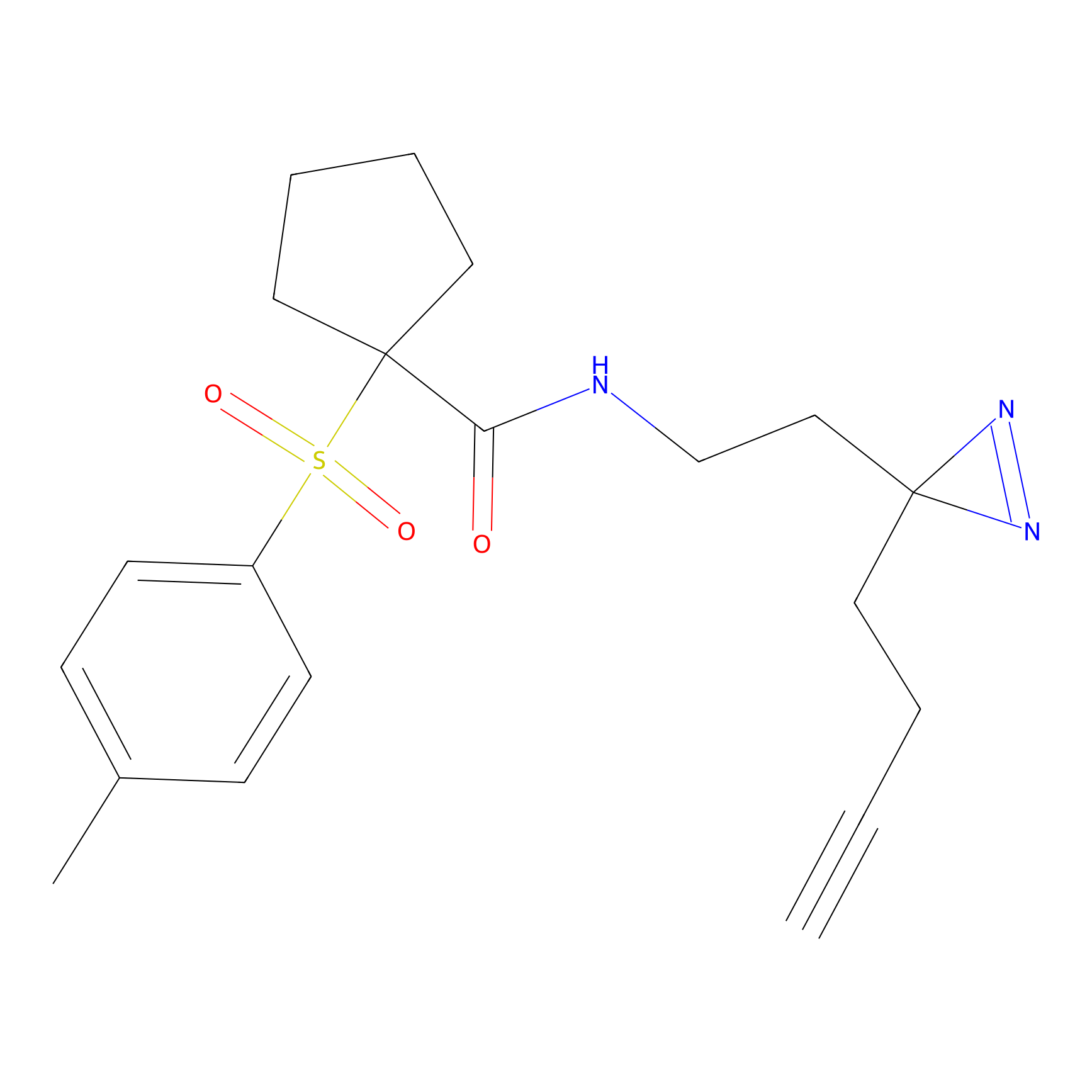
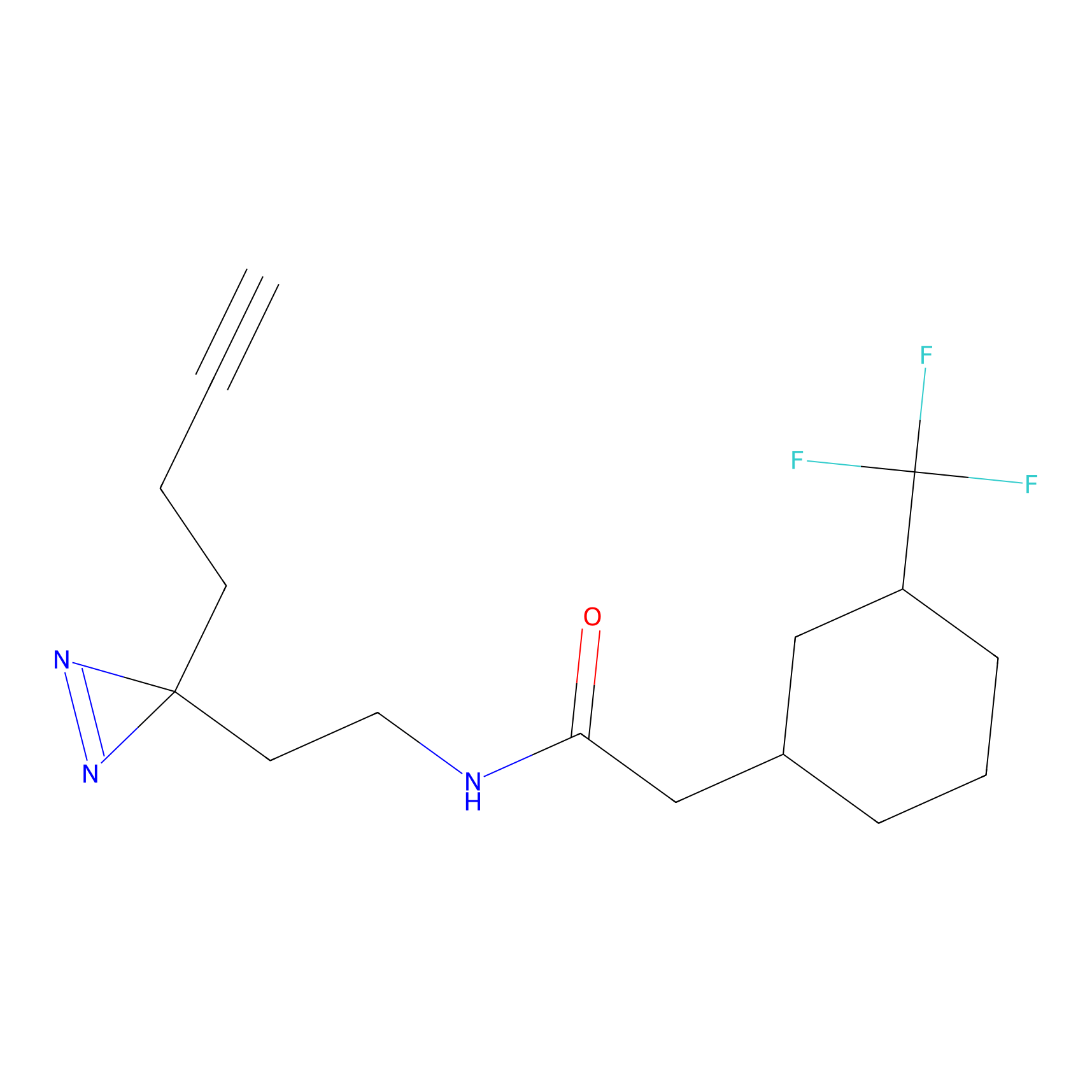
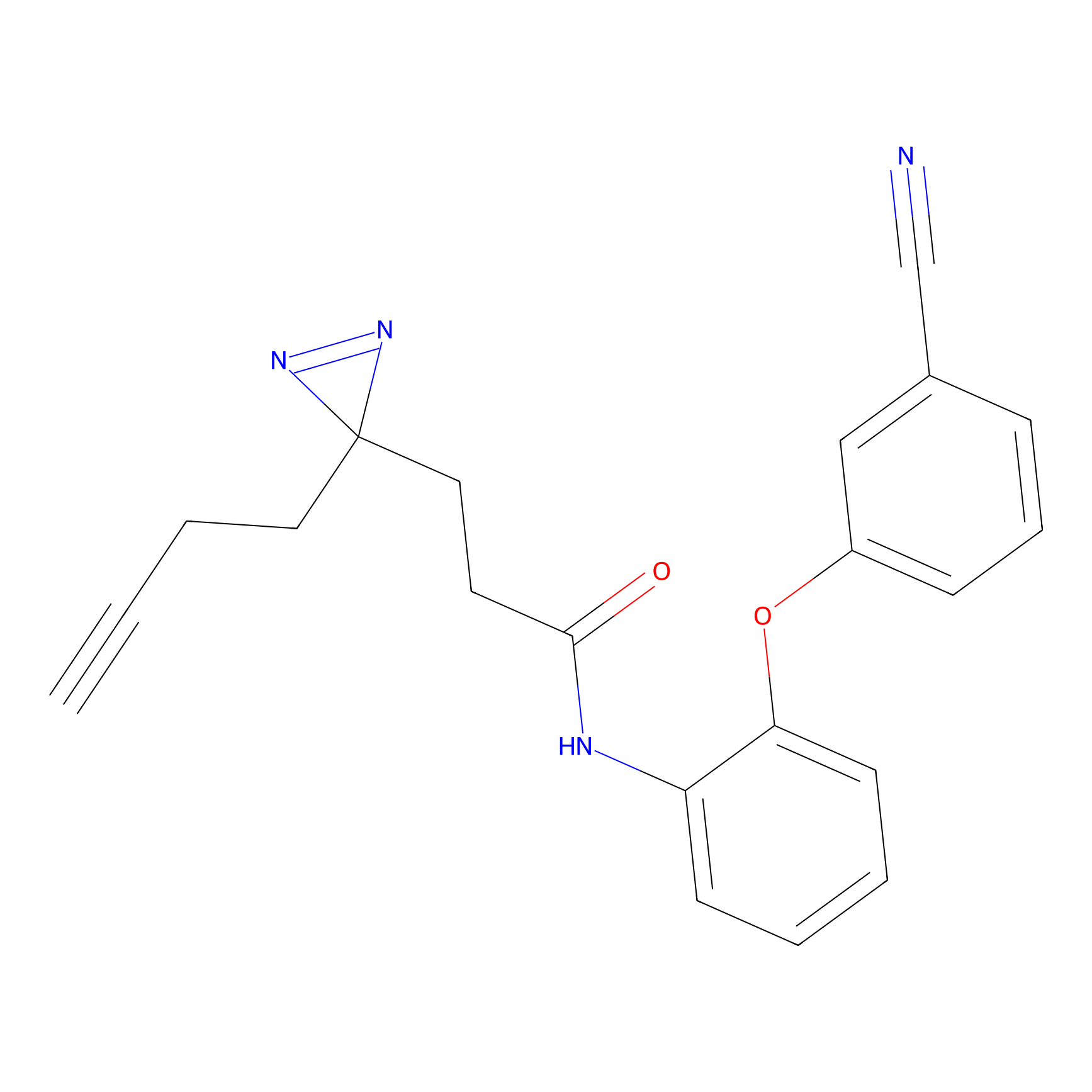
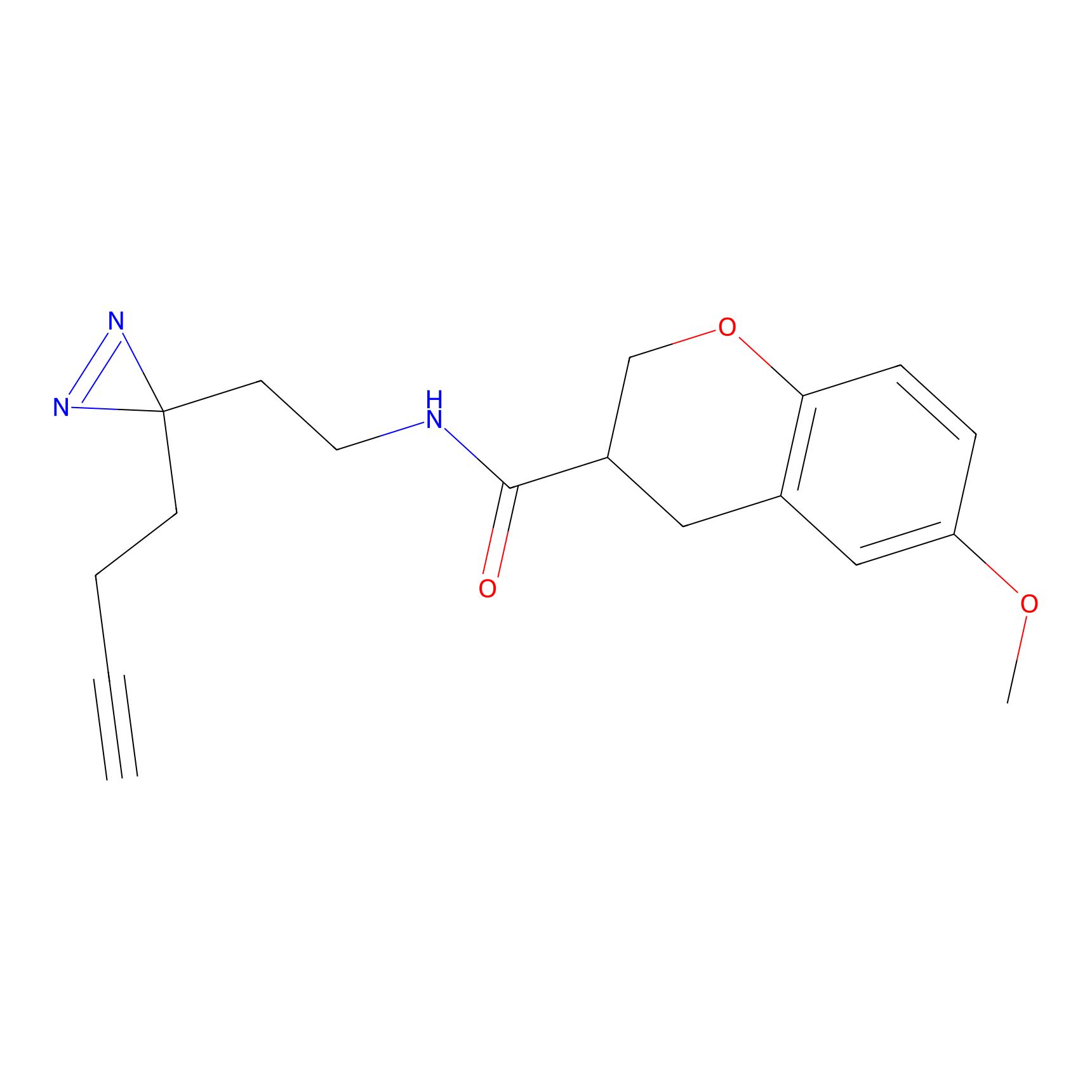
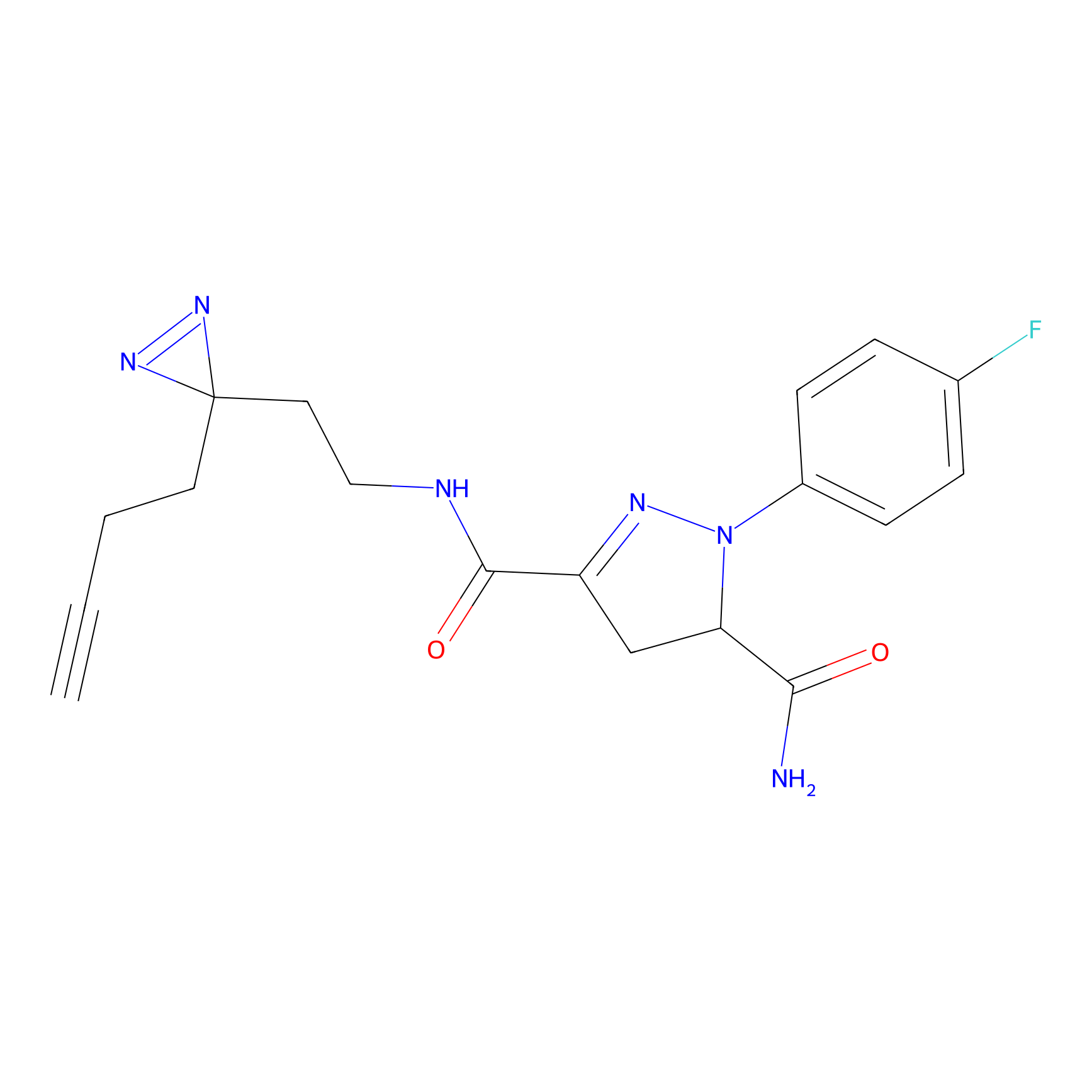
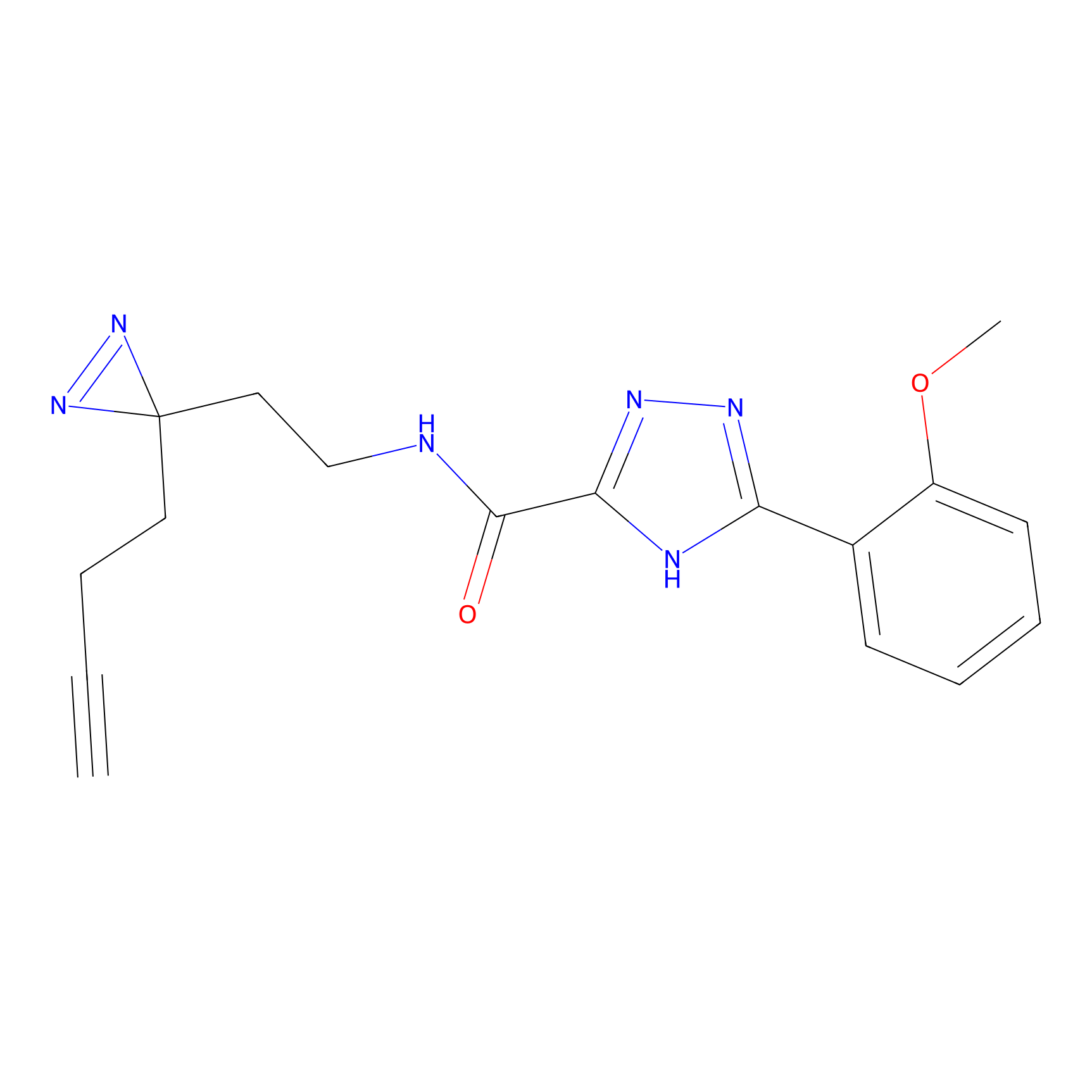
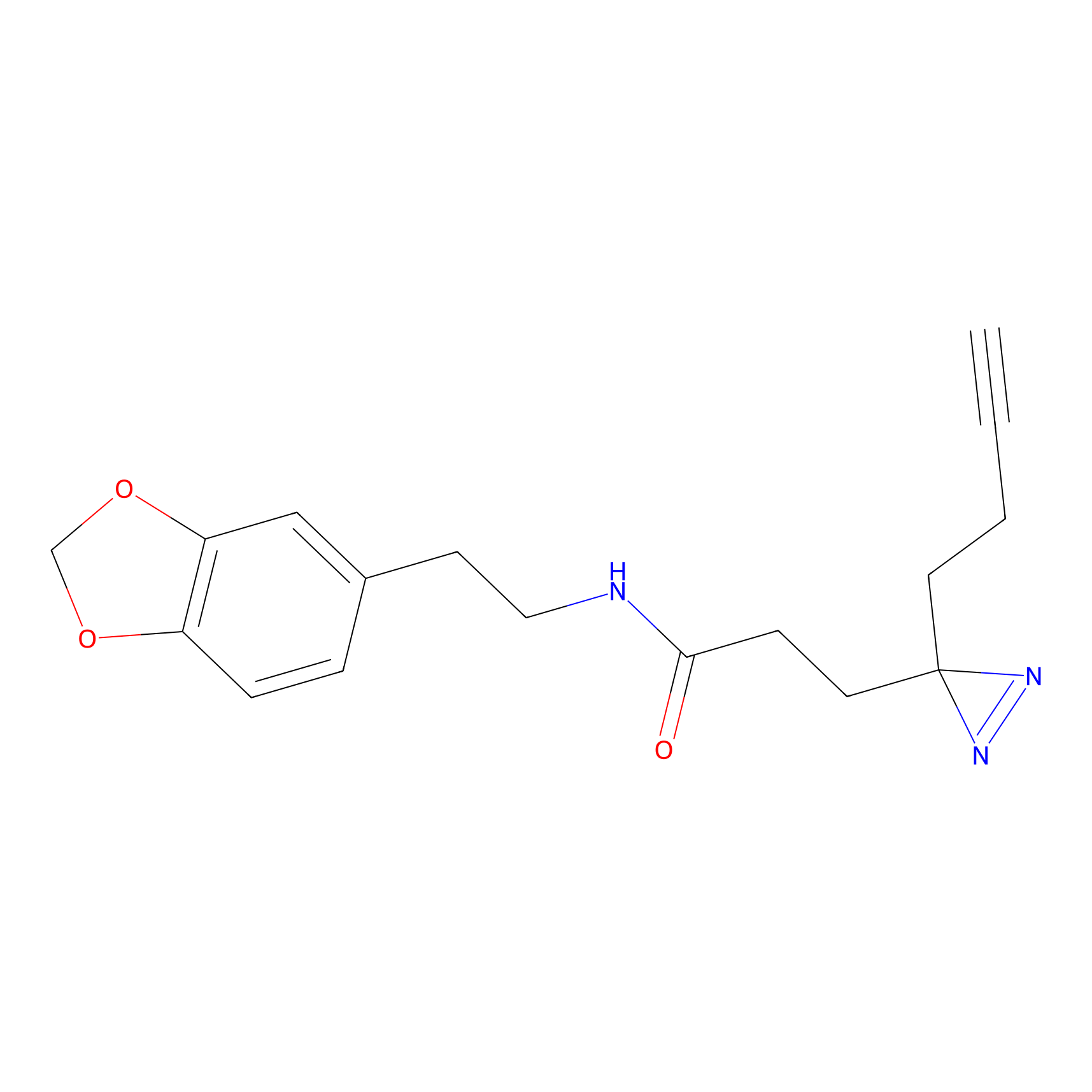
|
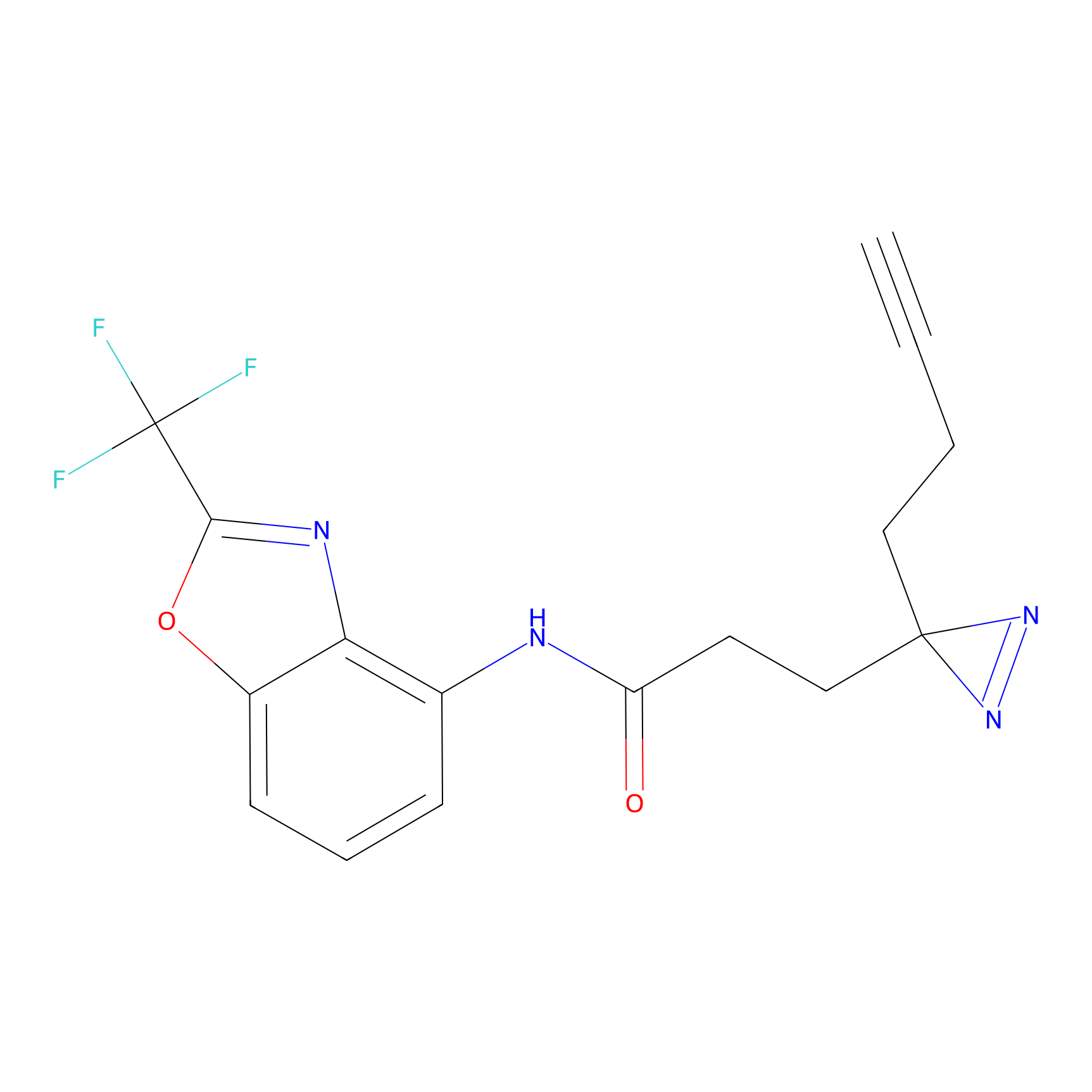
OPA-S-S-alkyne Probe Info |
 |
K217(2.13); K168(5.48); K161(9.24) | LDD3494 | [2] | |
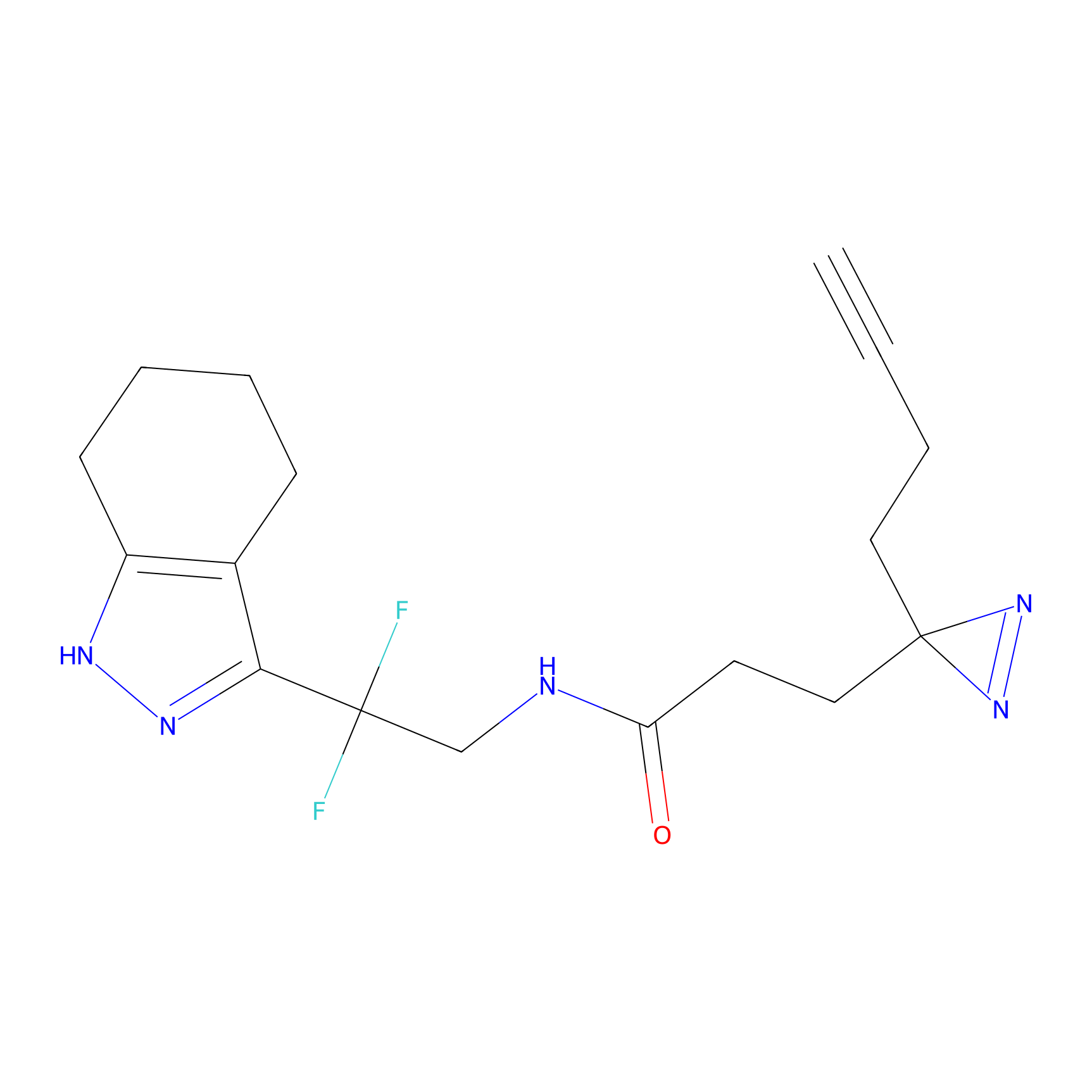
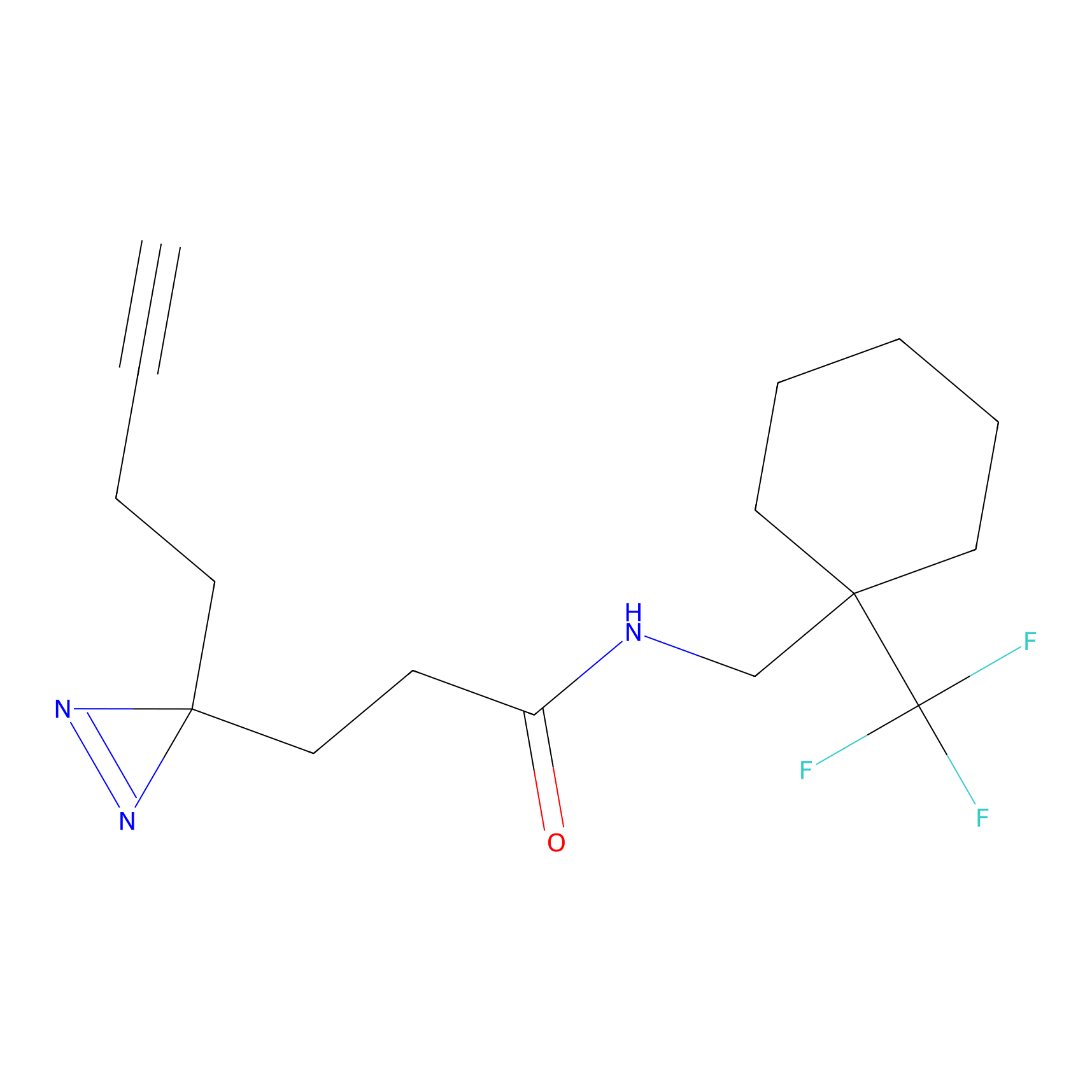
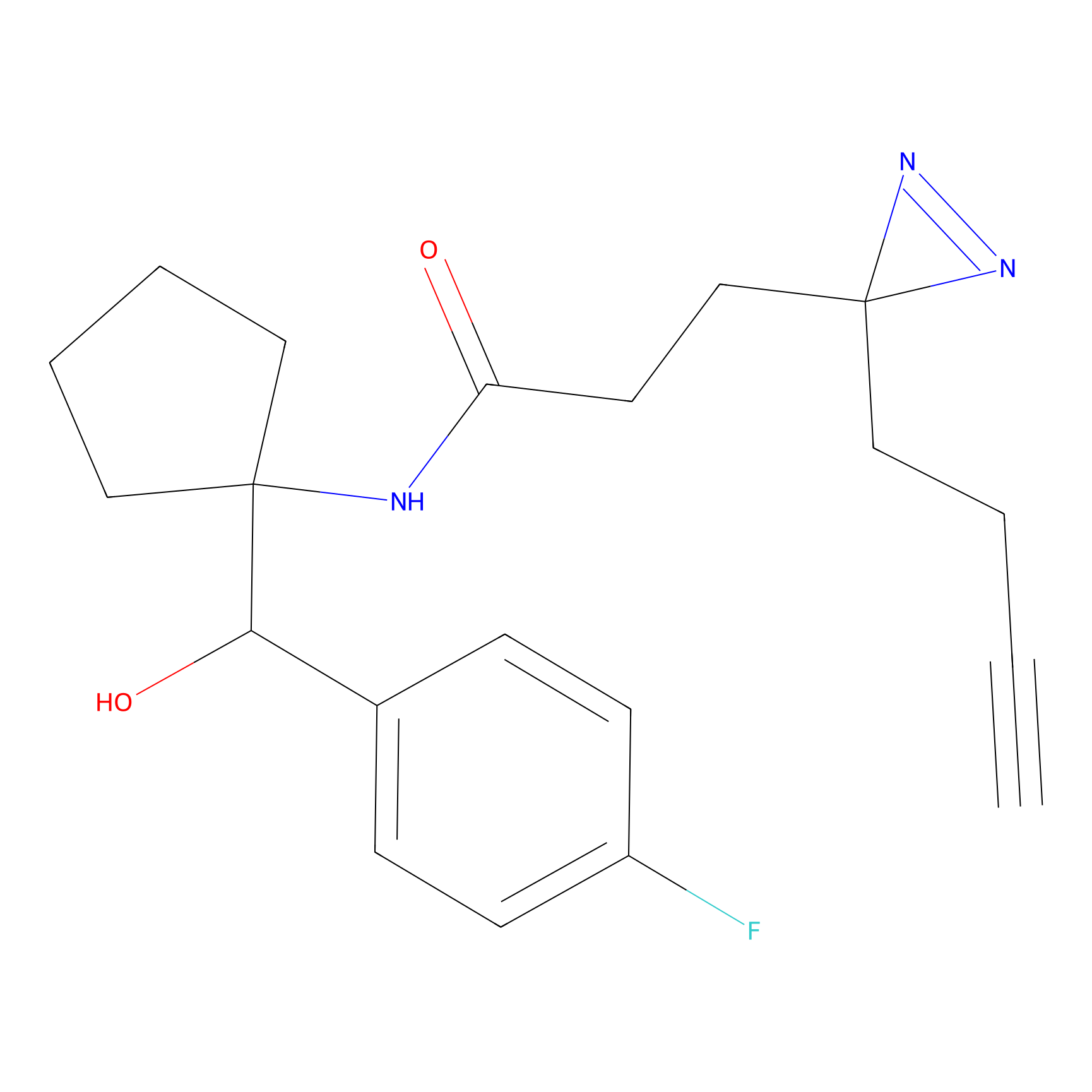
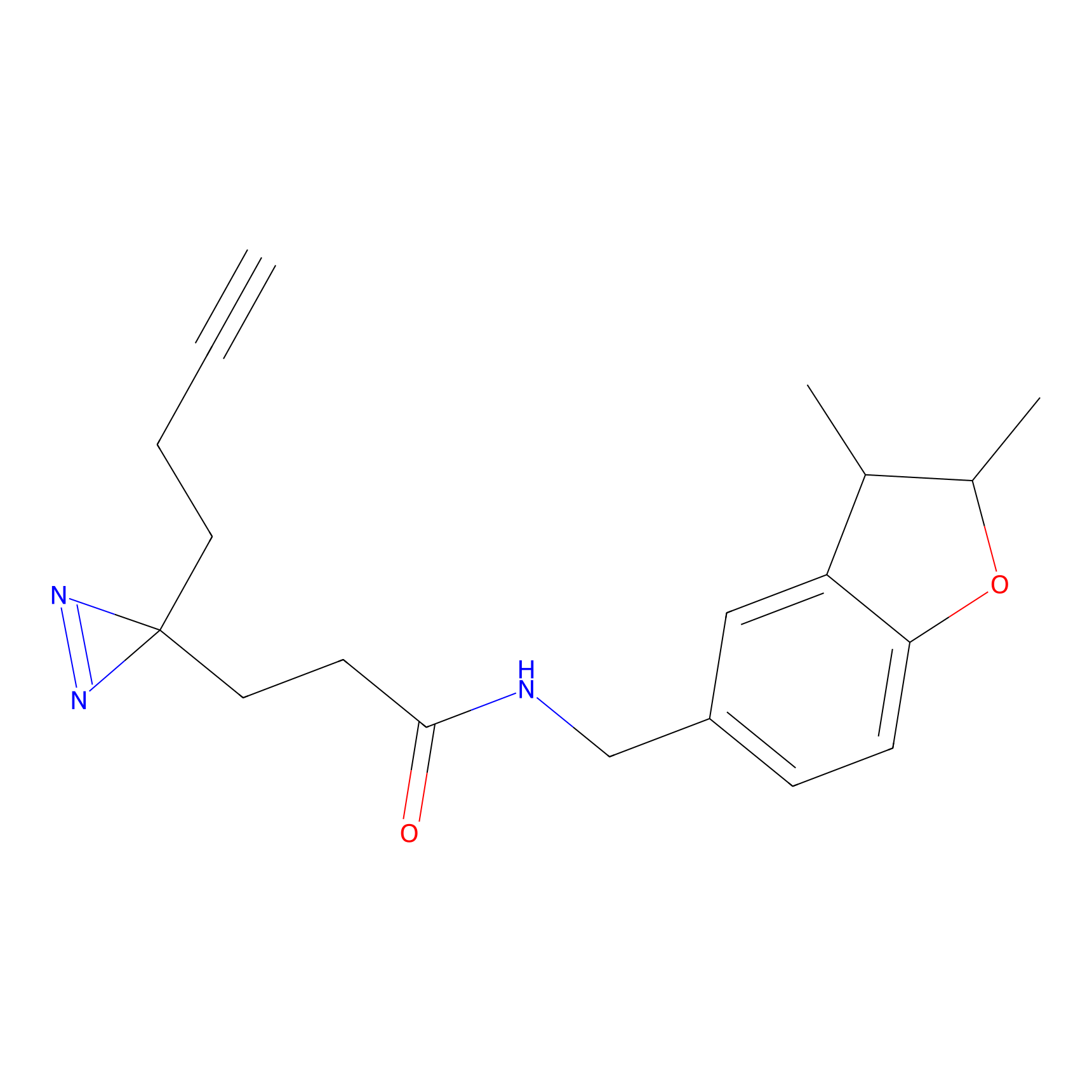
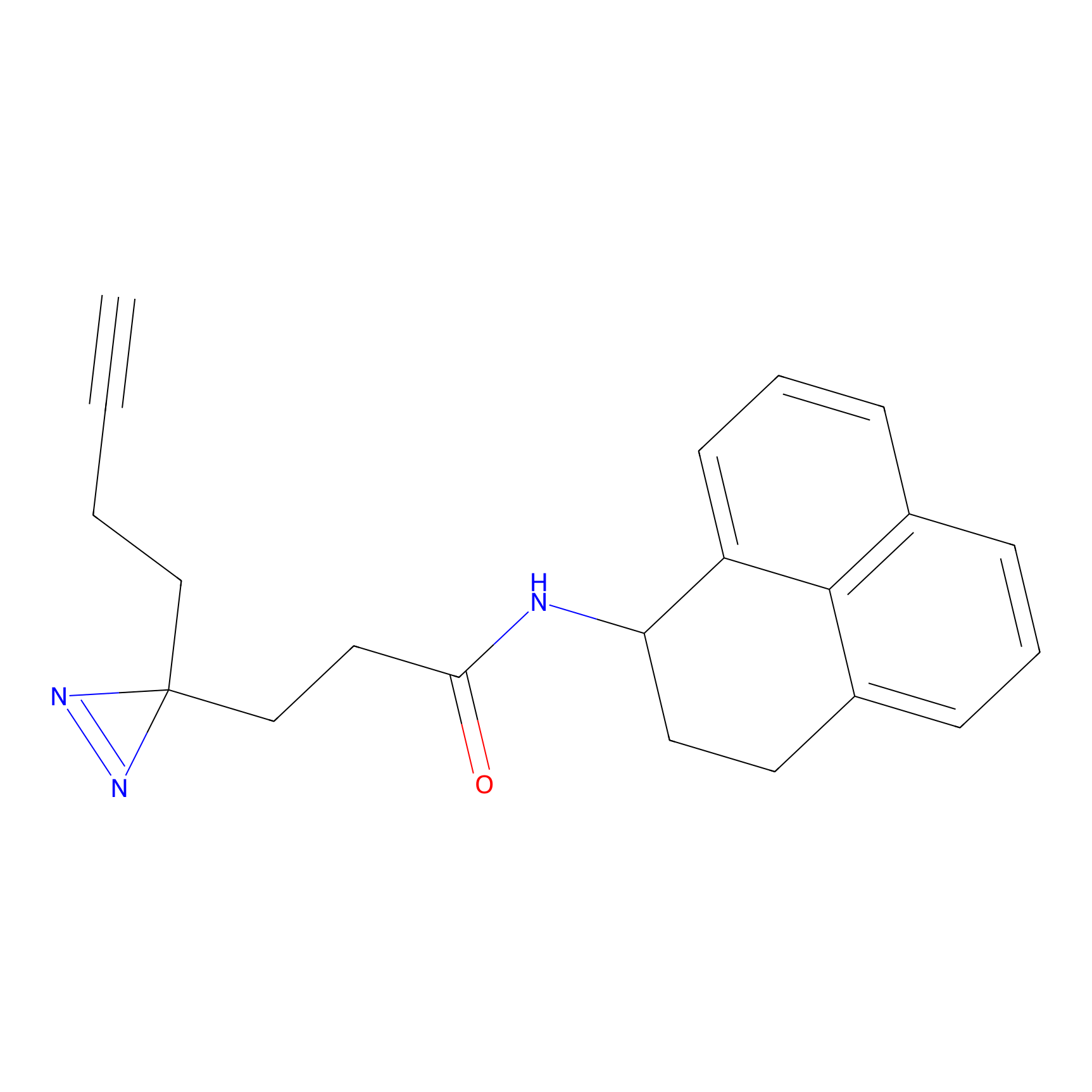
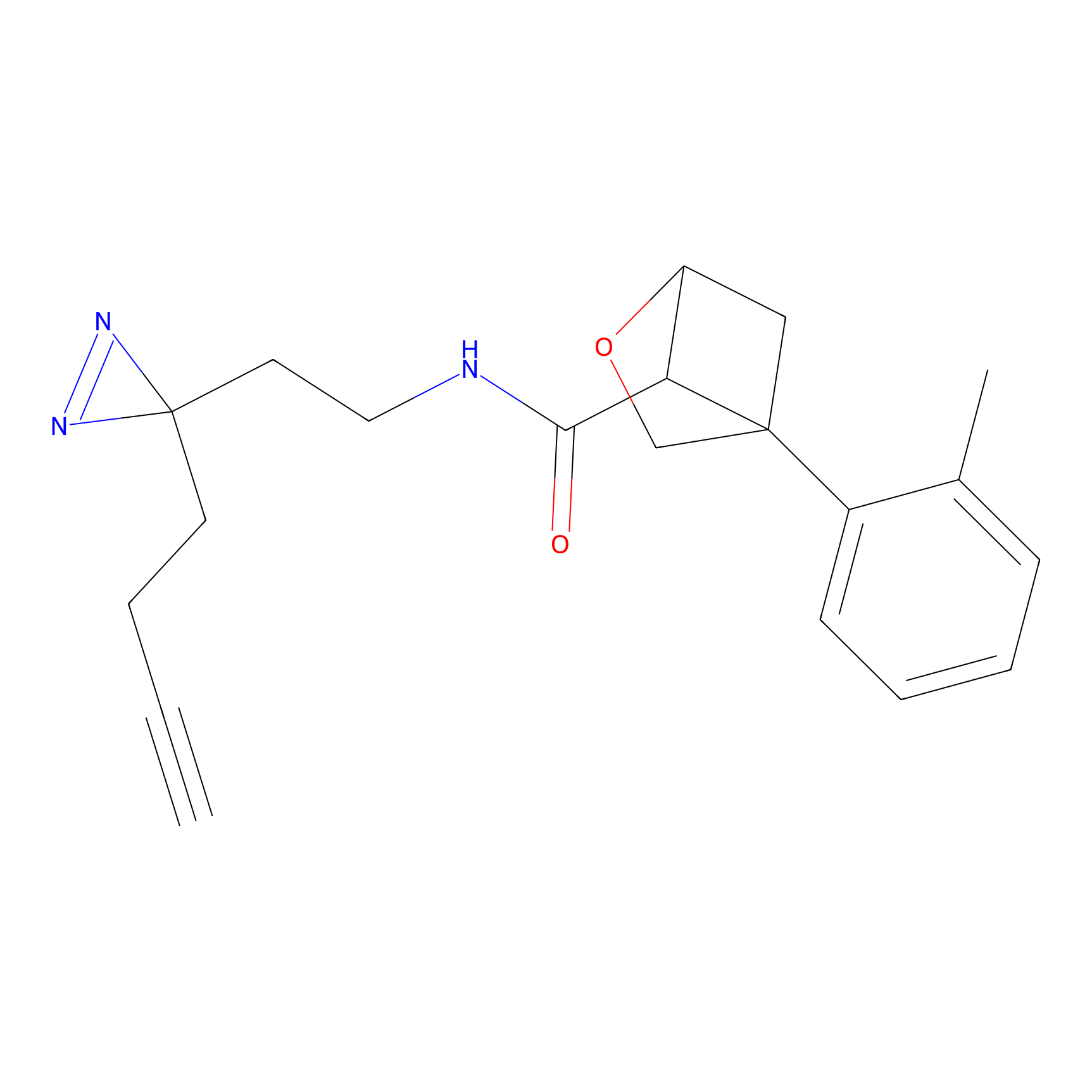
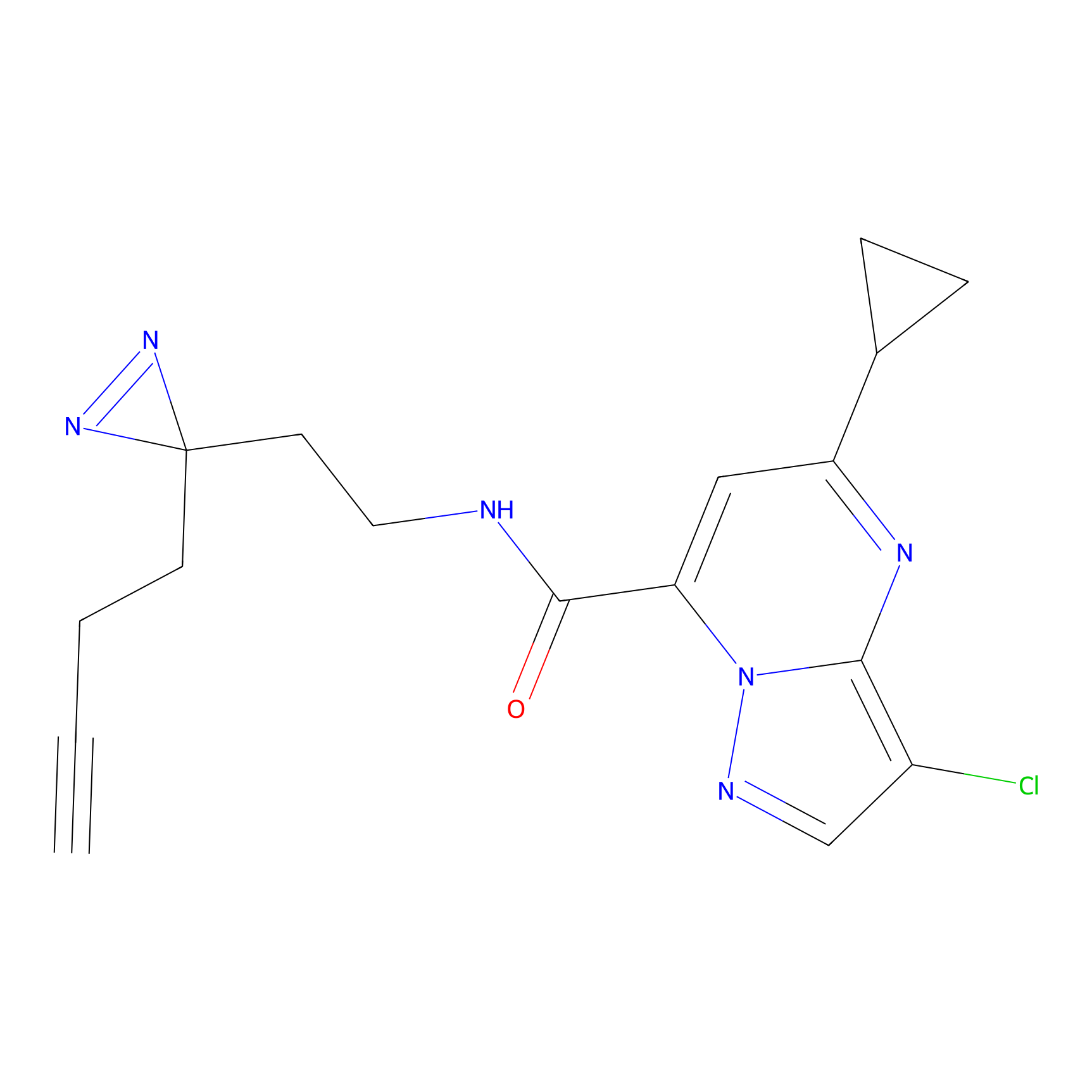
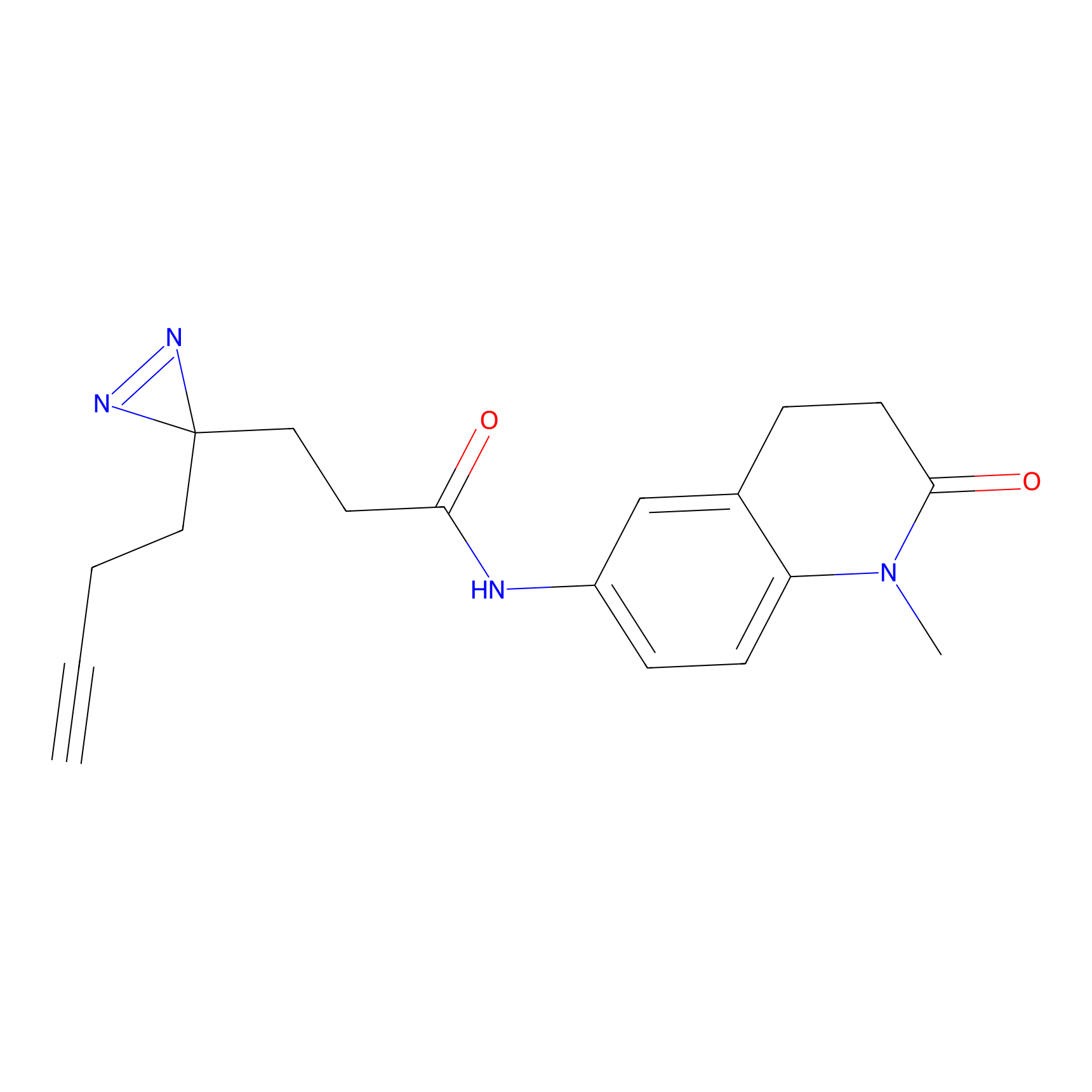
|
DBIA Probe Info |
 |
C71(3.65) | LDD3311 | [3] | |
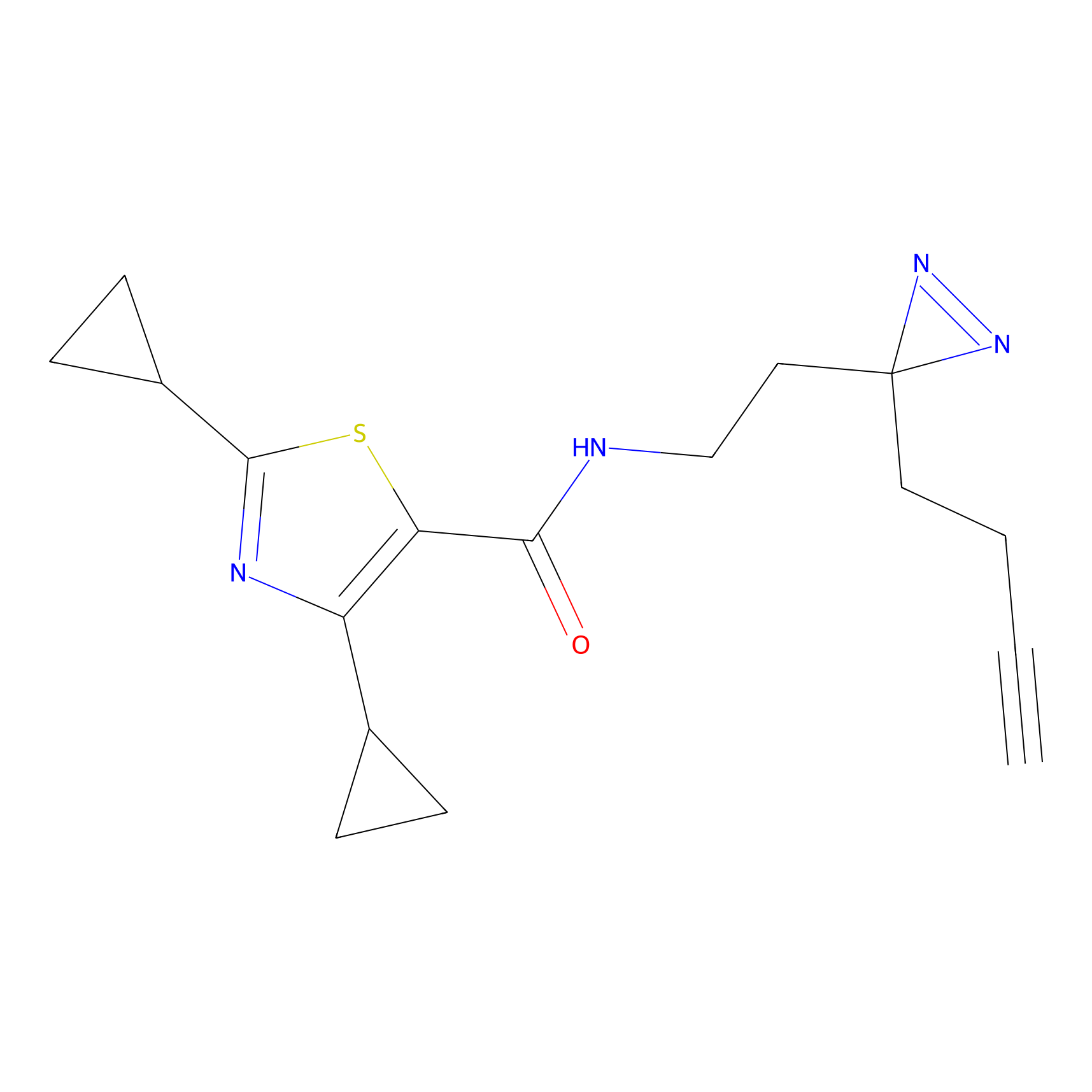
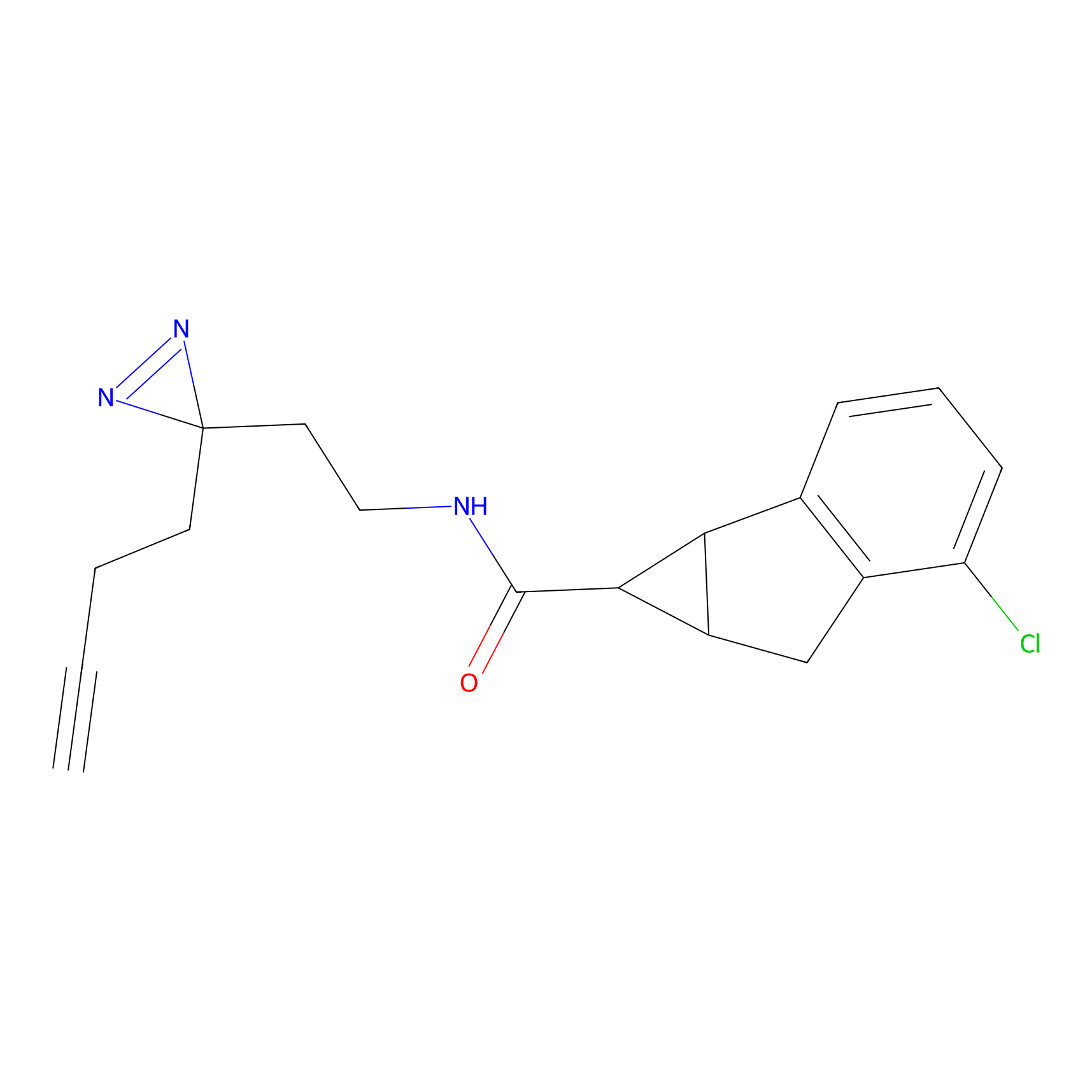
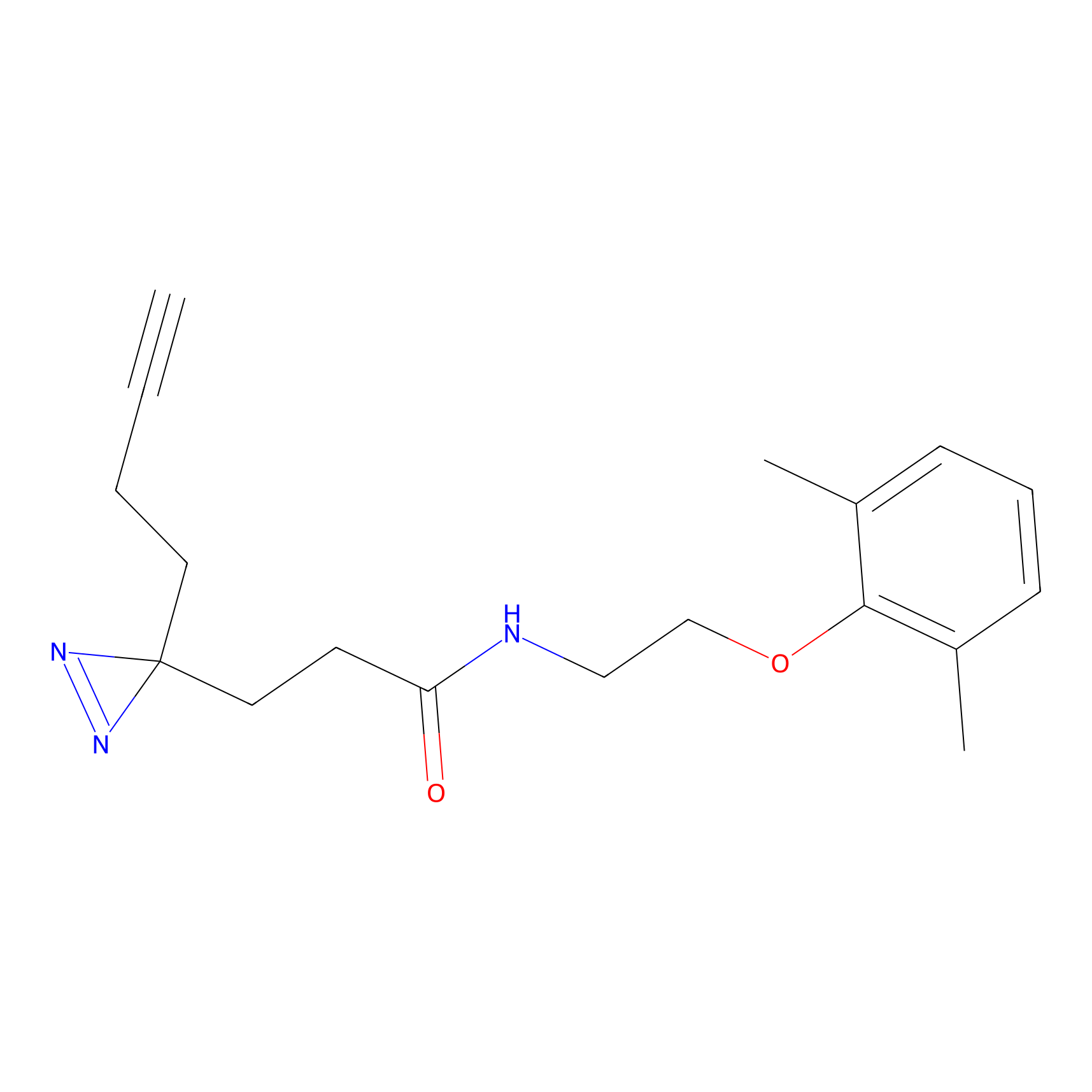
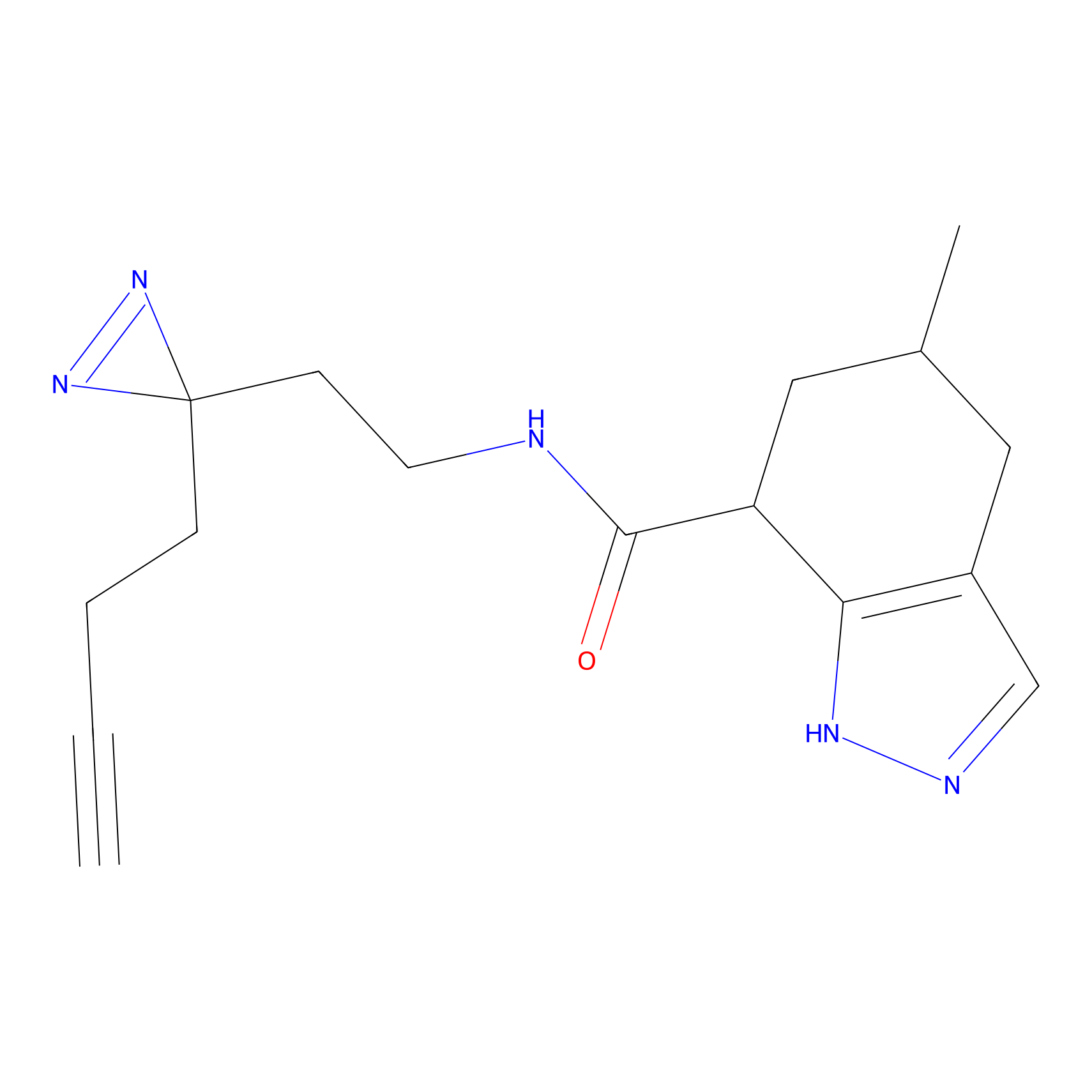
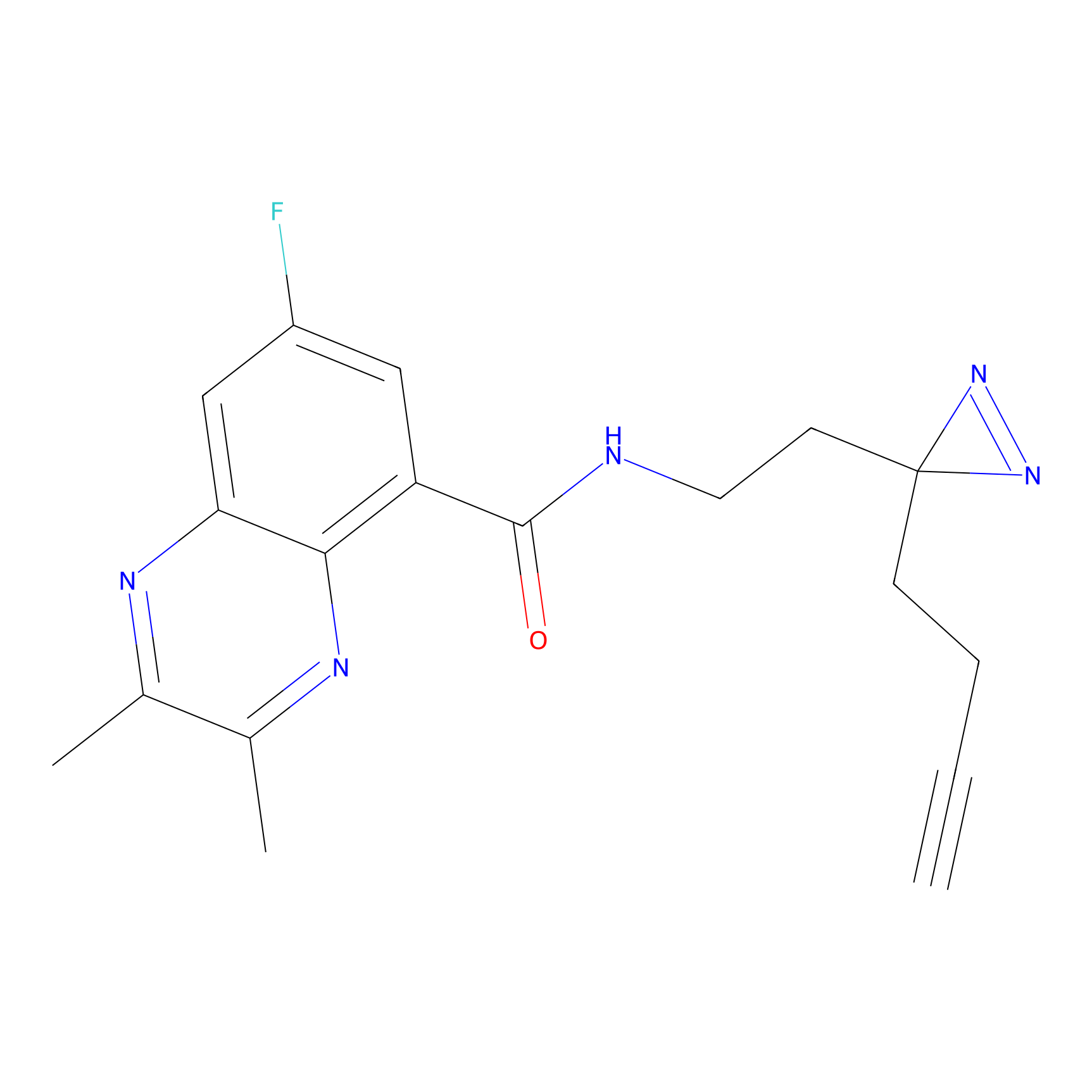
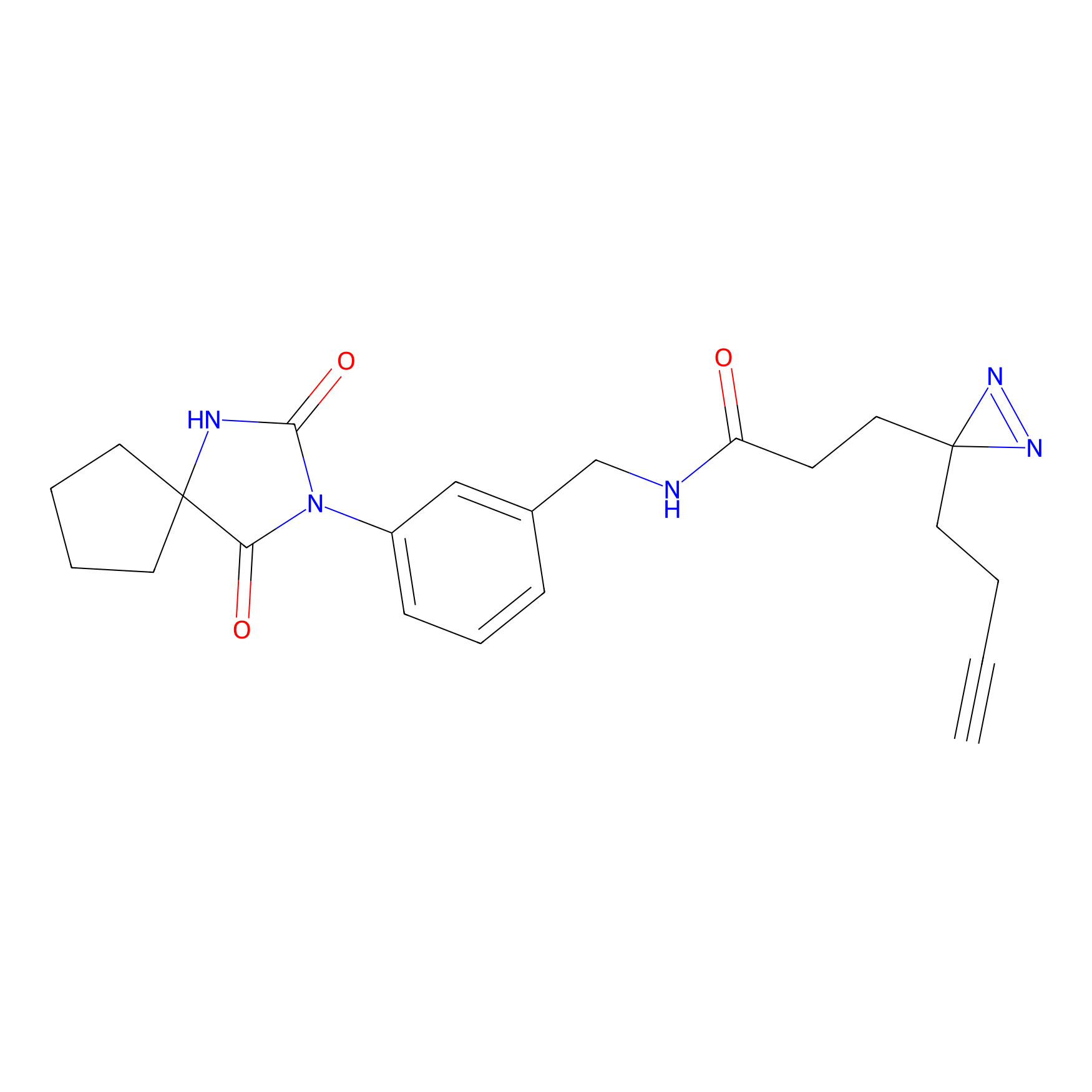
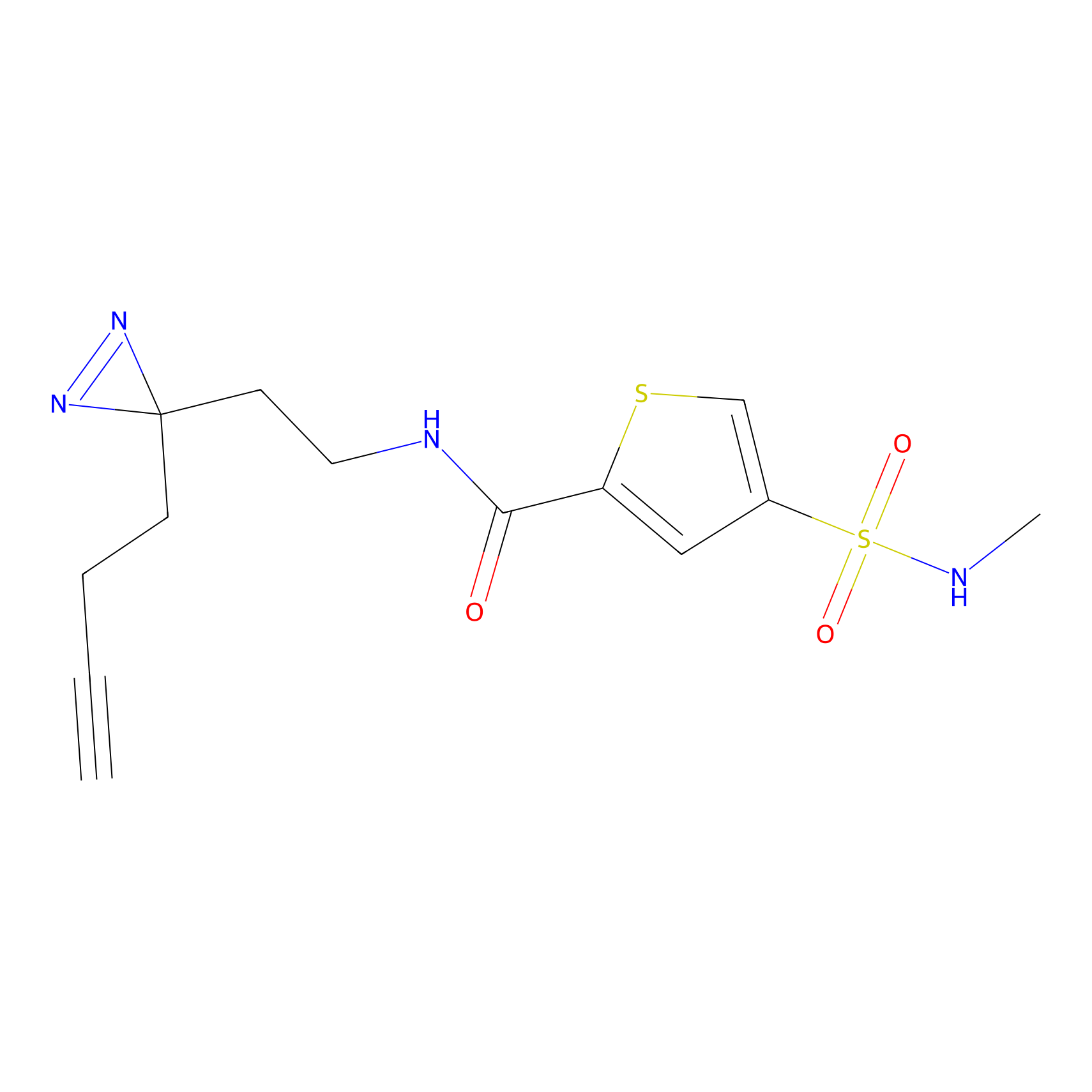
|
BTD Probe Info |
 |
C162(2.19) | LDD1700 | [4] | |
|
ATP probe Probe Info |
 |
K73(0.00); K12(0.00); K13(0.00) | LDD0199 | [5] | |
|
IA-alkyne Probe Info |
 |
C162(0.00); C179(0.00); C71(0.00) | LDD0165 | [6] | |
|
IPM Probe Info |
 |
C162(0.00); C71(0.00) | LDD0005 | [7] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [8] | |
|
AOyne Probe Info |
 |
13.10 | LDD0443 | [9] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0498 | BS-3668 | MDA-MB-231 | C162(0.43) | LDD2091 | [4] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C162(2.80) | LDD1702 | [4] |
| LDCM0107 | IAA | HeLa | H153(0.00); H192(0.00) | LDD0221 | [8] |
| LDCM0022 | KB02 | HEK-293T | C162(0.88); C71(0.95); C179(0.95) | LDD1492 | [13] |
| LDCM0023 | KB03 | HEK-293T | C162(0.95); C71(0.99); C179(0.97) | LDD1497 | [13] |
| LDCM0024 | KB05 | G361 | C71(3.65) | LDD3311 | [3] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0225 | [8] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C162(2.19) | LDD1700 | [4] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C162(1.87) | LDD2144 | [4] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
GPCR
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Adenosine receptor A2a (ADORA2A) | G-protein coupled receptor 1 family | P29274 | |||
| Mu-type opioid receptor (OPRM1) | G-protein coupled receptor 1 family | P35372 | |||
Other
References