Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
P8 Probe Info |
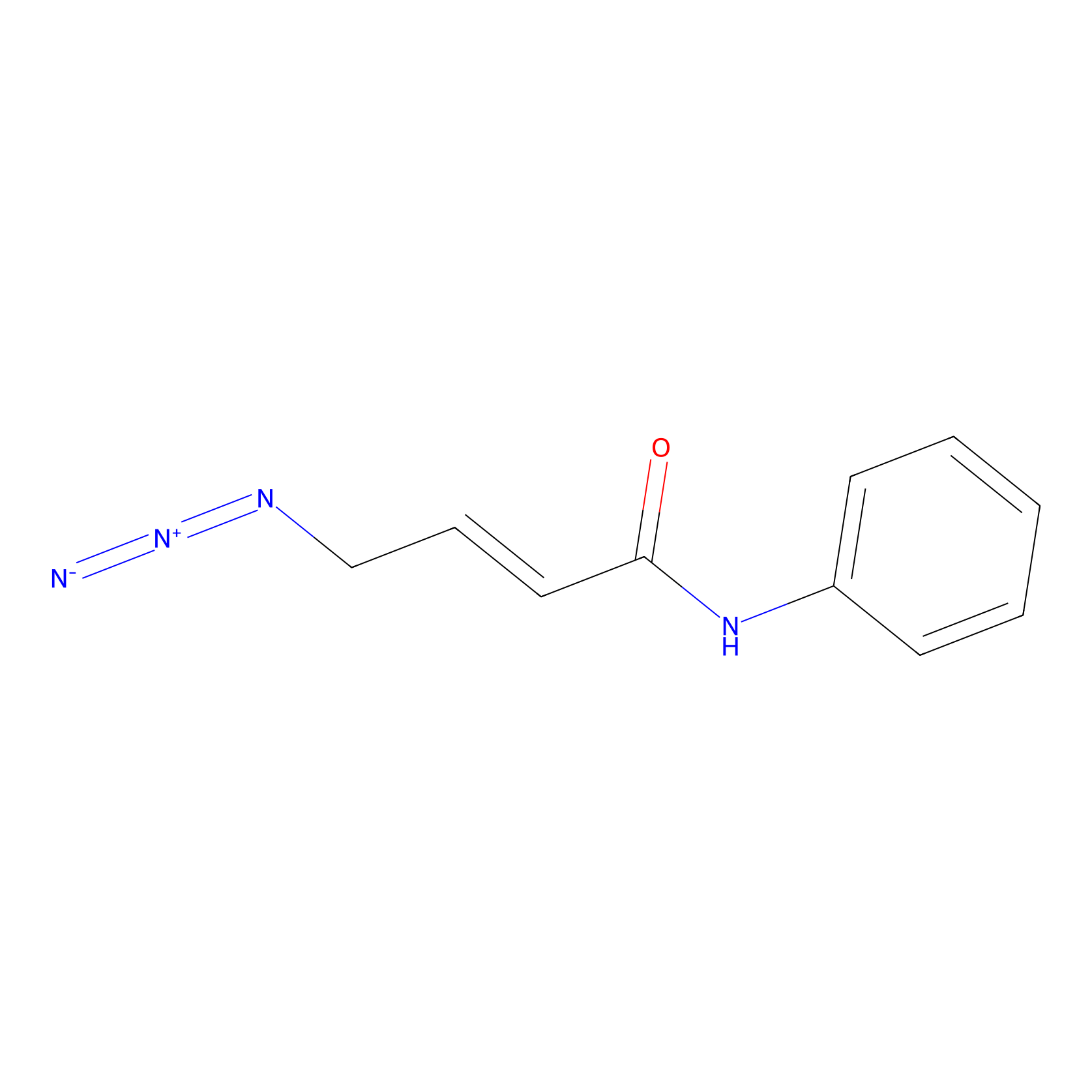 |
10.00 | LDD0451 | [1] | |
|
A-EBA Probe Info |
 |
2.25 | LDD0215 | [2] | |
|
CHEMBL5175495 Probe Info |
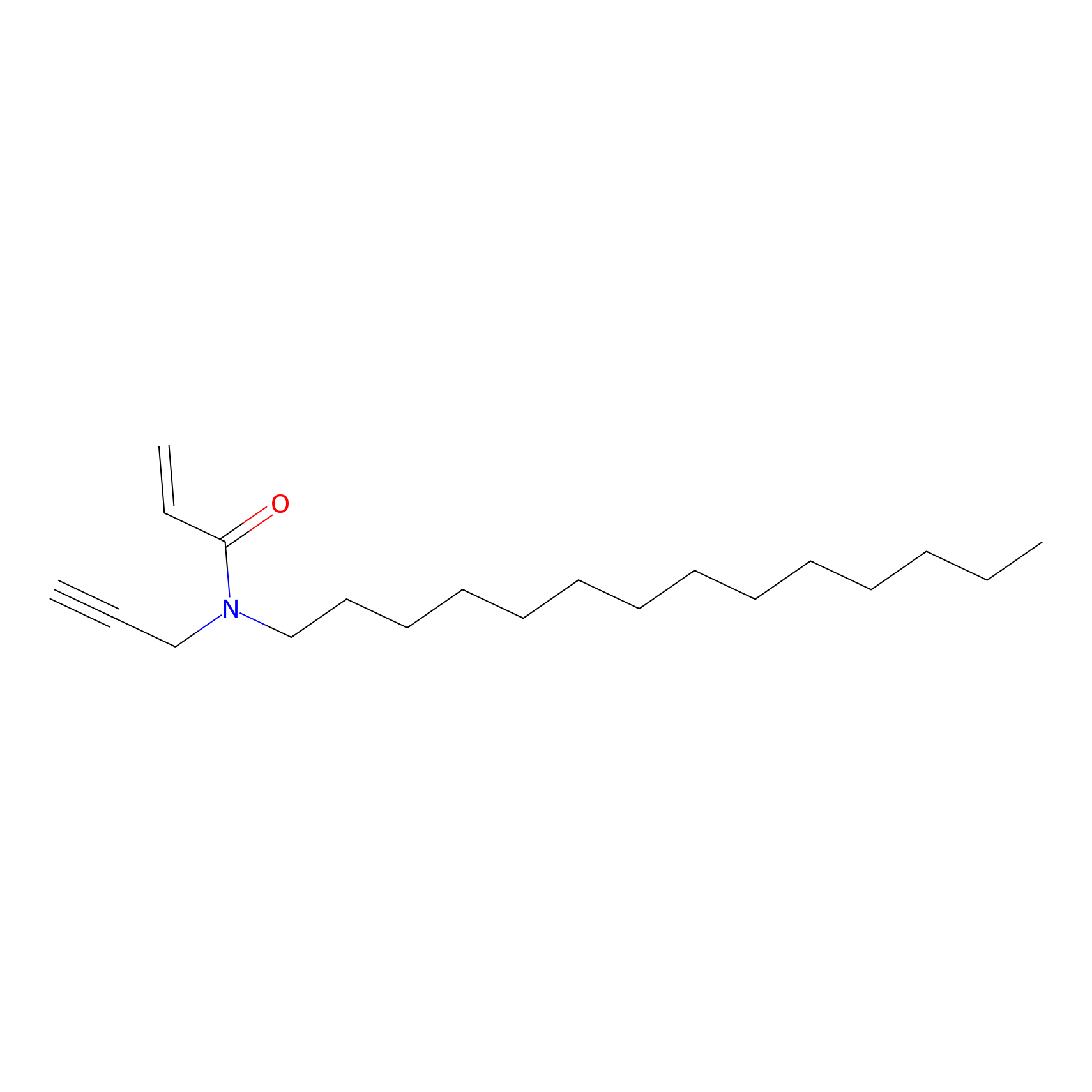 |
6.30 | LDD0196 | [3] | |
|
CY4 Probe Info |
 |
100.00 | LDD0244 | [4] | |
|
N1 Probe Info |
 |
100.00 | LDD0242 | [4] | |
|
C-Sul Probe Info |
 |
5.70 | LDD0066 | [5] | |
|
TH211 Probe Info |
 |
Y331(20.00); Y418(20.00); Y431(20.00); Y230(6.39) | LDD0257 | [6] | |
|
TH214 Probe Info |
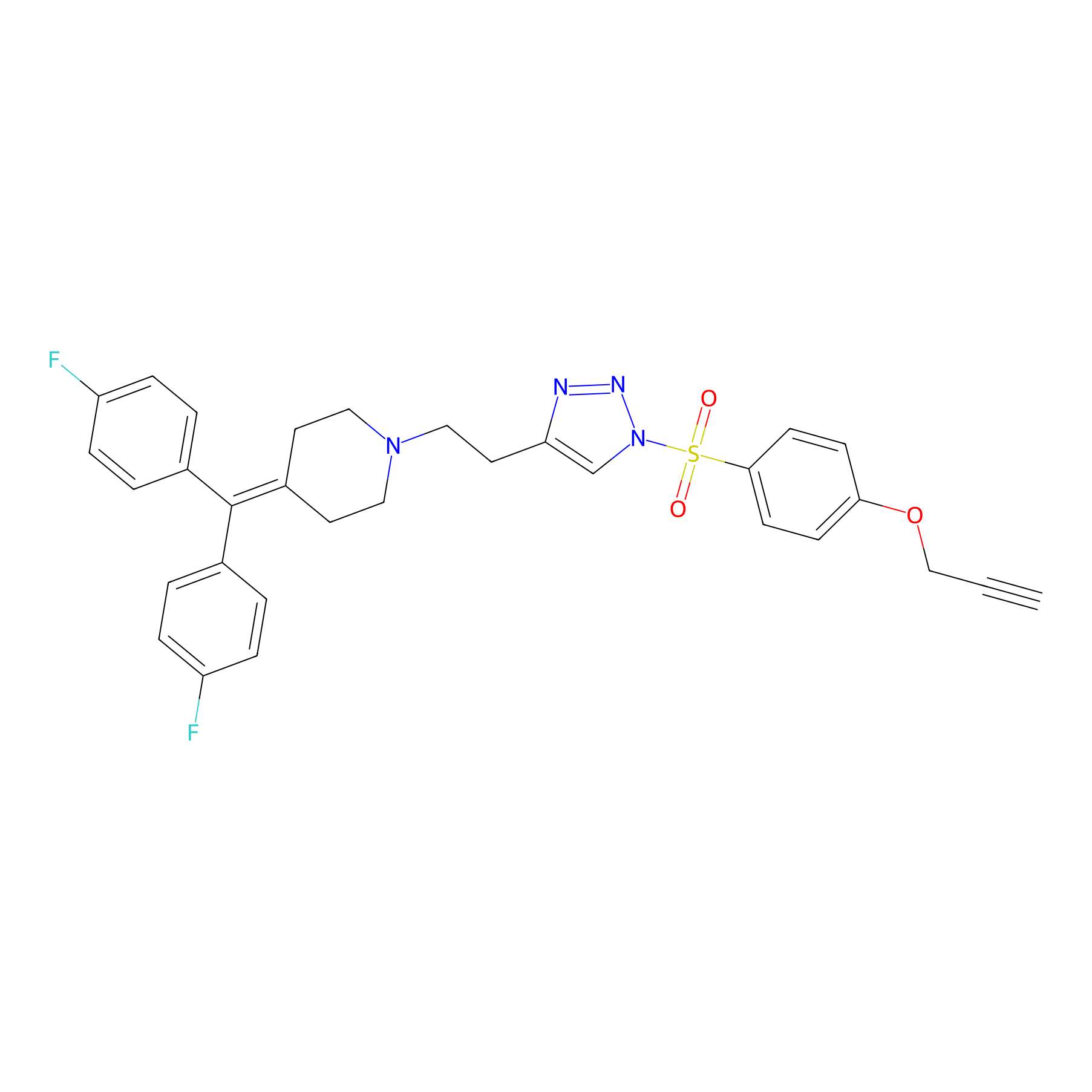 |
Y230(20.00); Y247(20.00) | LDD0258 | [6] | |
|
TH216 Probe Info |
 |
Y196(20.00); Y230(20.00); Y331(20.00); Y395(20.00) | LDD0259 | [6] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [7] | |
|
AZ-9 Probe Info |
 |
D402(1.42) | LDD2208 | [8] | |
|
ONAyne Probe Info |
 |
K426(0.00); K133(0.00); K159(0.00); K485(0.00) | LDD0273 | [9] | |
|
OPA-S-S-alkyne Probe Info |
 |
K522(3.50); K133(3.53); K264(4.08) | LDD3494 | [10] | |
|
Probe 1 Probe Info |
 |
Y230(32.02); Y247(19.59); Y269(12.38); Y395(31.08) | LDD3495 | [11] | |
|
HHS-482 Probe Info |
 |
Y395(0.83); Y418(0.82); Y431(1.06) | LDD0285 | [12] | |
|
HHS-475 Probe Info |
 |
Y395(0.58); Y431(0.86); Y418(0.91) | LDD0264 | [13] | |
|
5E-2FA Probe Info |
 |
H227(0.00); H248(0.00); H477(0.00); H417(0.00) | LDD2235 | [14] | |
|
ATP probe Probe Info |
 |
K522(0.00); K124(0.00); K426(0.00); K259(0.00) | LDD0199 | [15] | |
|
m-APA Probe Info |
 |
H227(0.00); H248(0.00); H477(0.00); H501(0.00) | LDD2231 | [14] | |
|
ATP probe Probe Info |
 |
K124(0.00); K161(0.00); K133(0.00); K480(0.00) | LDD0035 | [16] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0356 | [17] | |
|
NHS Probe Info |
 |
K426(0.00); K259(0.00); K264(0.00); K124(0.00) | LDD0010 | [18] | |
|
OSF Probe Info |
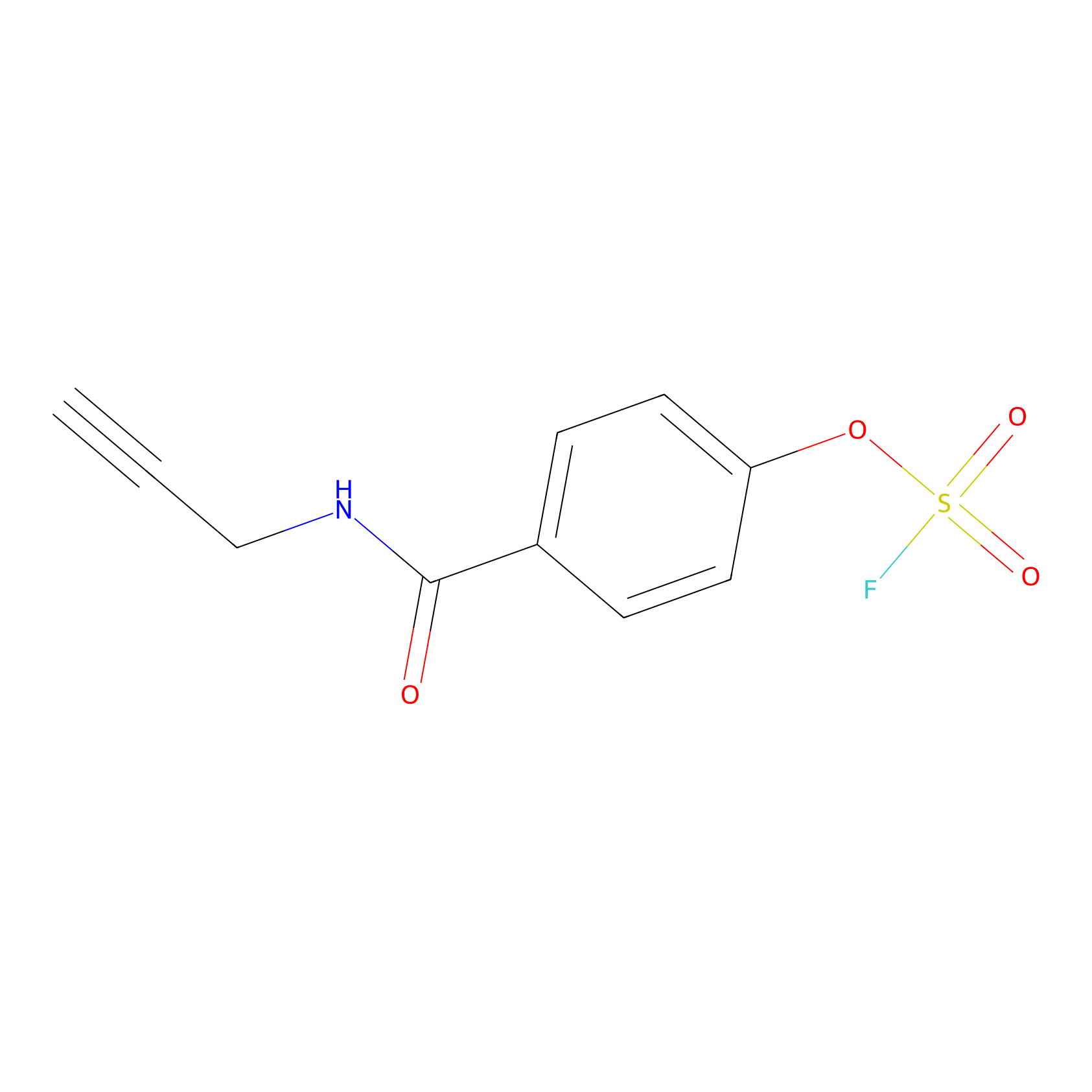 |
H417(0.00); Y418(0.00) | LDD0029 | [19] | |
|
SF Probe Info |
 |
Y431(0.00); Y361(0.00); K485(0.00); K522(0.00) | LDD0028 | [19] | |
|
STPyne Probe Info |
 |
K198(0.00); K201(0.00); K133(0.00) | LDD0009 | [18] | |
|
1c-yne Probe Info |
 |
K133(0.00); K351(0.00); K259(0.00); K161(0.00) | LDD0228 | [17] | |
|
Acrolein Probe Info |
 |
H477(0.00); H248(0.00); H102(0.00); H417(0.00) | LDD0217 | [20] | |
|
Crotonaldehyde Probe Info |
 |
H417(0.00); H102(0.00); H527(0.00) | LDD0219 | [20] | |
|
Methacrolein Probe Info |
 |
H527(0.00); H102(0.00) | LDD0218 | [20] | |
|
W1 Probe Info |
 |
E472(0.00); Q469(0.00); H477(0.00) | LDD0236 | [21] | |
|
MPP-AC Probe Info |
 |
N.A. | LDD0428 | [22] | |
|
TER-AC Probe Info |
 |
N.A. | LDD0426 | [22] | |
|
TPP-AC Probe Info |
 |
N.A. | LDD0427 | [22] | |
|
HHS-465 Probe Info |
 |
K432(0.00); Y418(0.00); Y431(0.00); K133(0.00) | LDD2240 | [23] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C003 Probe Info |
 |
14.42 | LDD1713 | [24] | |
|
C017 Probe Info |
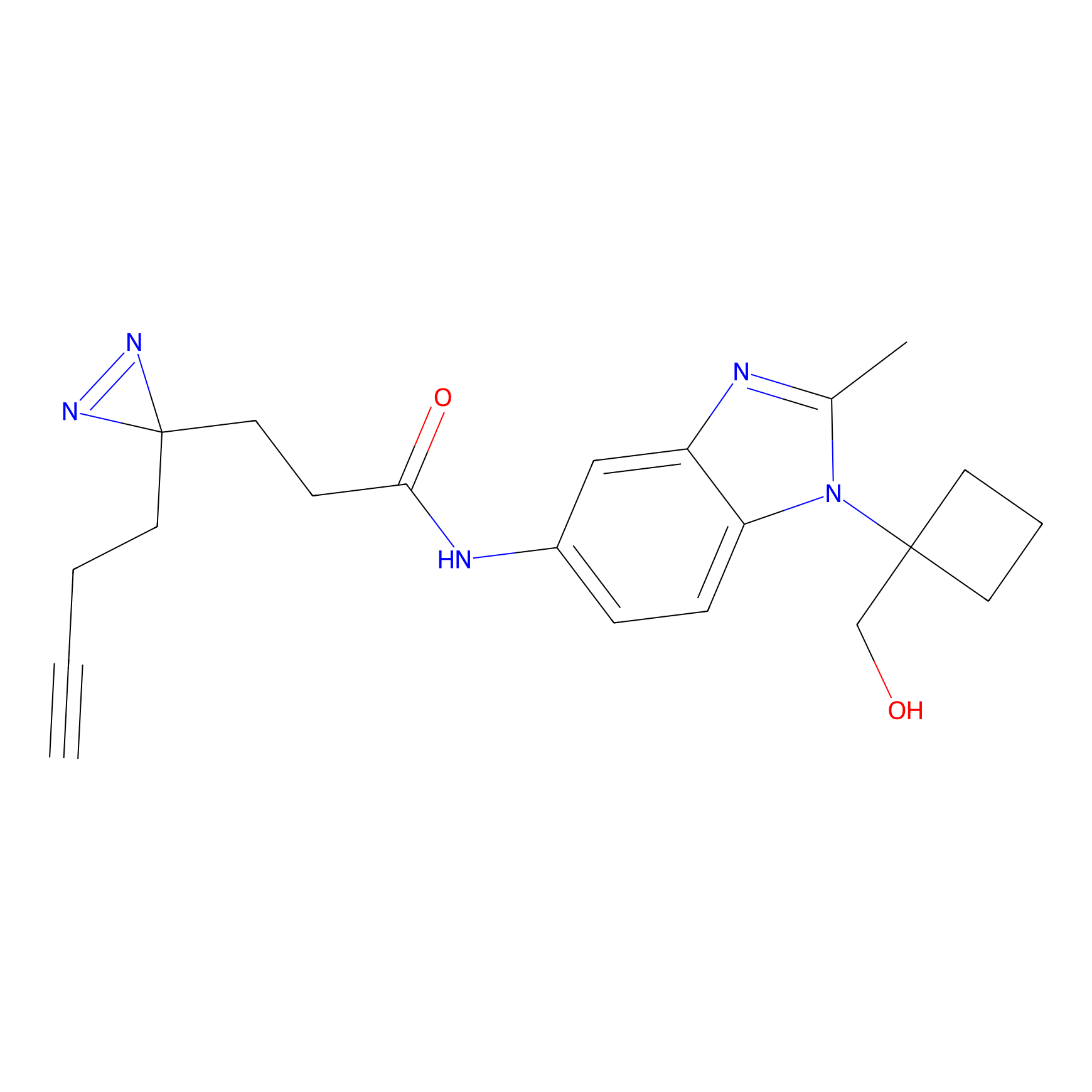 |
7.73 | LDD1725 | [24] | |
|
C040 Probe Info |
 |
7.31 | LDD1740 | [24] | |
|
C041 Probe Info |
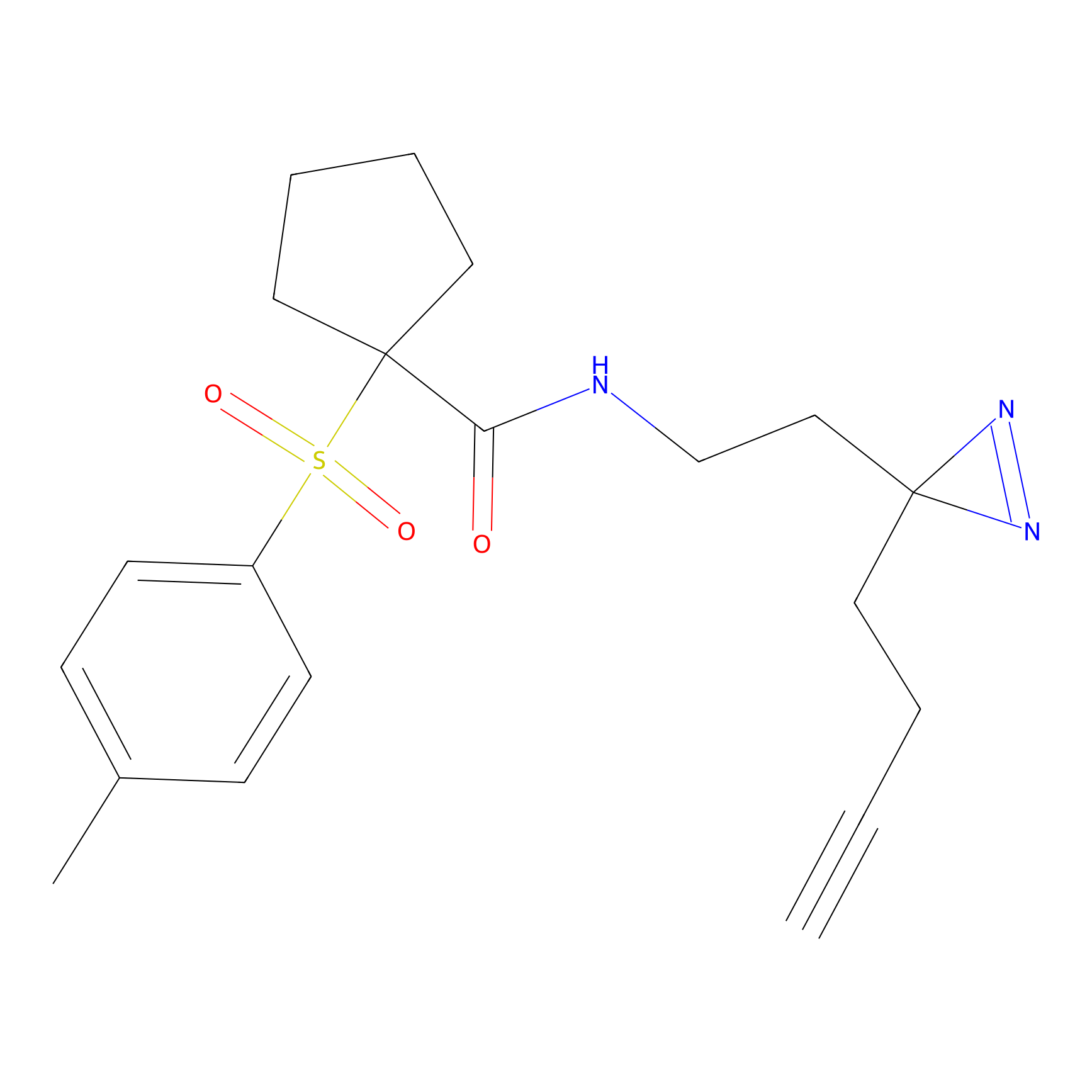 |
6.45 | LDD1741 | [24] | |
|
C044 Probe Info |
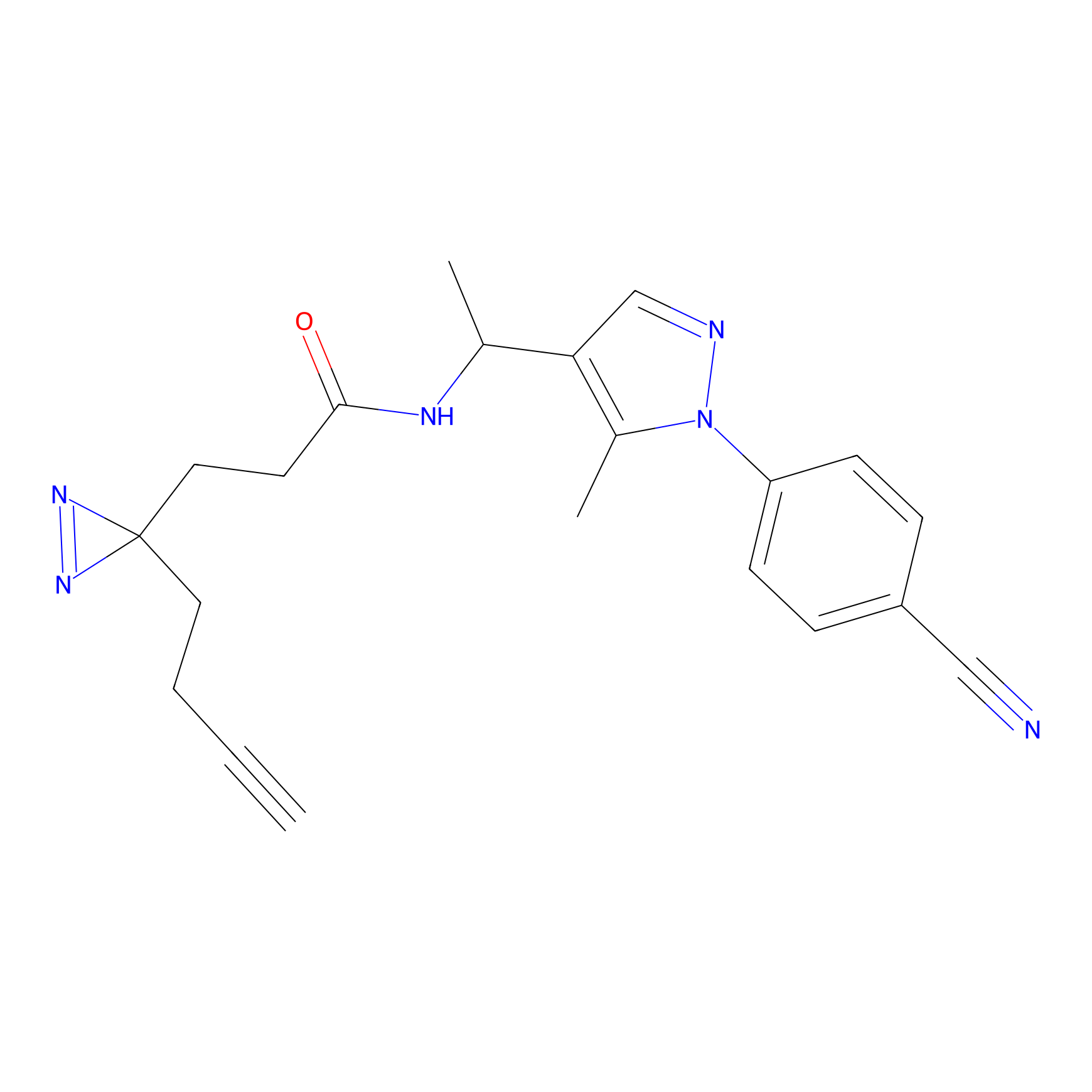 |
5.86 | LDD1743 | [24] | |
|
C056 Probe Info |
 |
18.64 | LDD1753 | [24] | |
|
C063 Probe Info |
 |
9.92 | LDD1760 | [24] | |
|
C082 Probe Info |
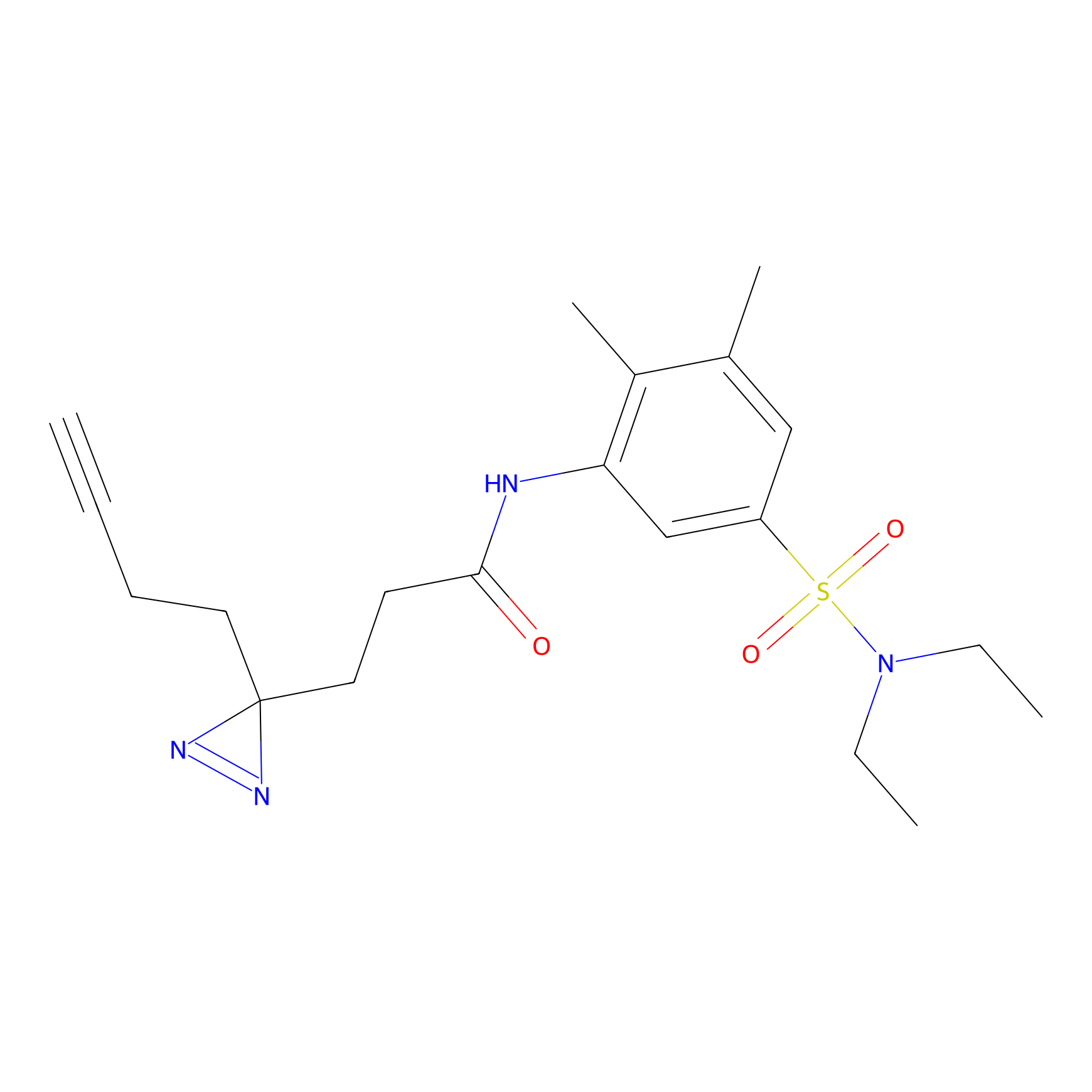 |
5.62 | LDD1774 | [24] | |
|
C087 Probe Info |
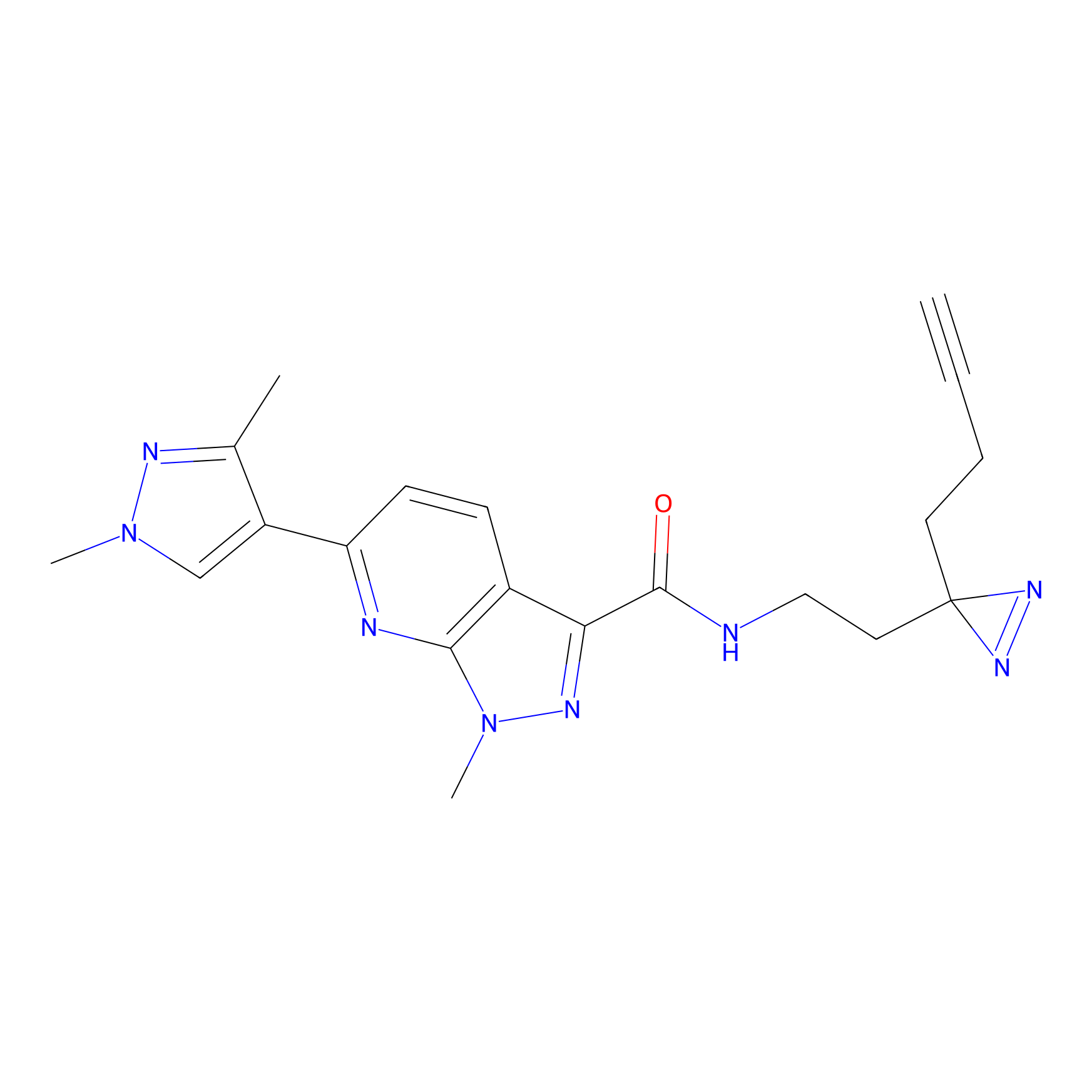 |
7.67 | LDD1779 | [24] | |
|
C102 Probe Info |
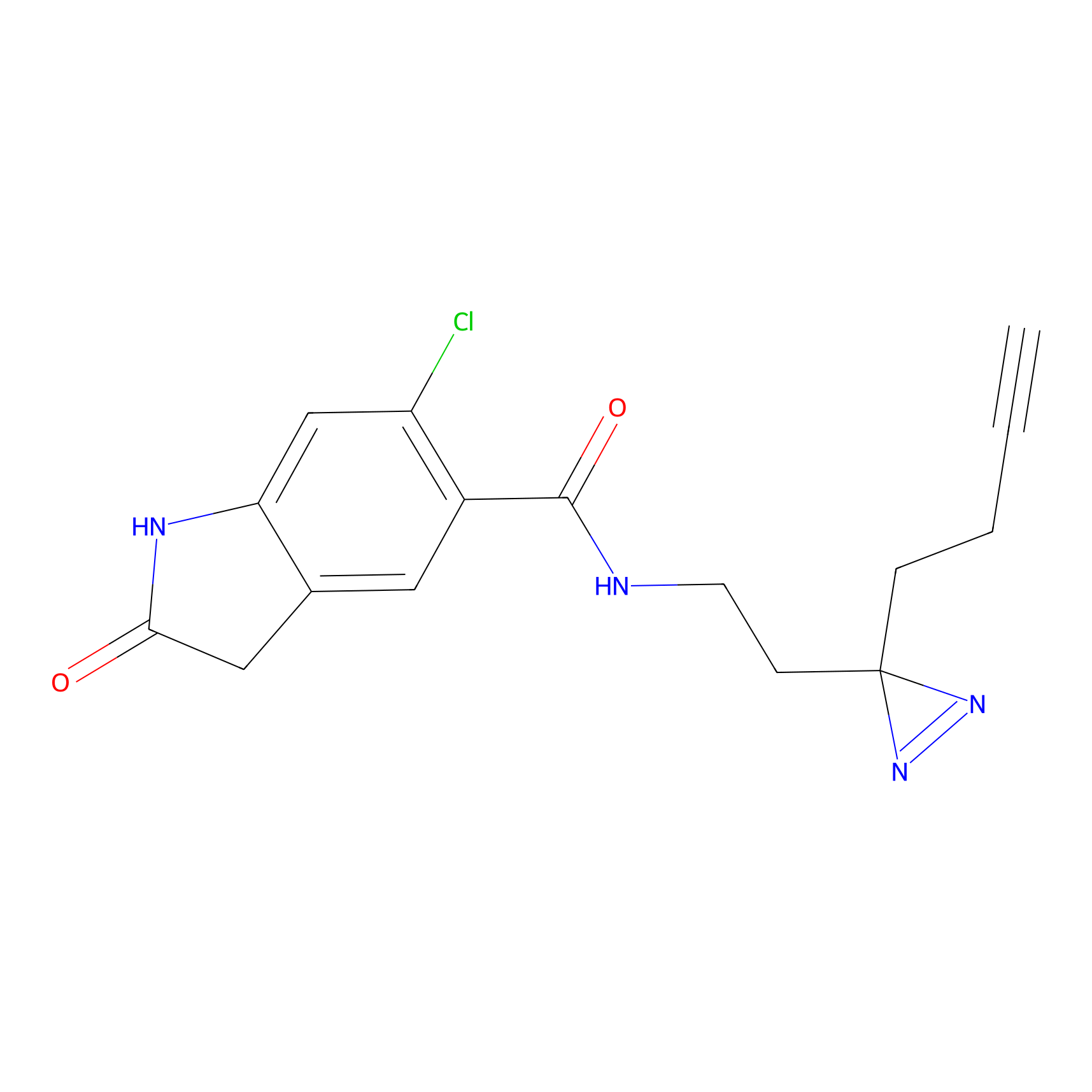 |
5.86 | LDD1790 | [24] | |
|
C106 Probe Info |
 |
16.68 | LDD1793 | [24] | |
|
C112 Probe Info |
 |
39.95 | LDD1799 | [24] | |
|
C158 Probe Info |
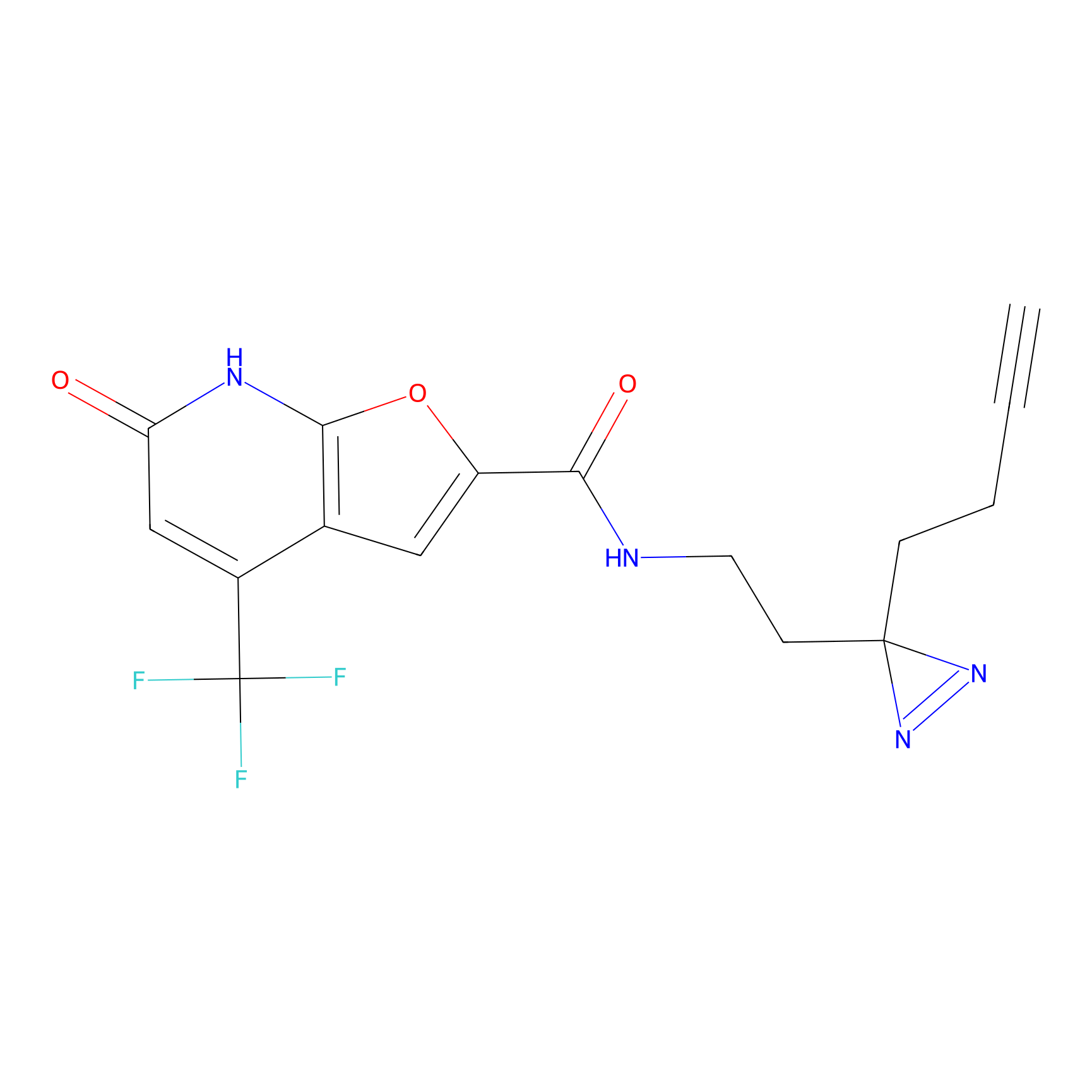 |
15.89 | LDD1838 | [24] | |
|
C161 Probe Info |
 |
11.88 | LDD1841 | [24] | |
|
C201 Probe Info |
 |
25.11 | LDD1877 | [24] | |
|
C218 Probe Info |
 |
12.91 | LDD1892 | [24] | |
|
C232 Probe Info |
 |
34.54 | LDD1905 | [24] | |
|
C269 Probe Info |
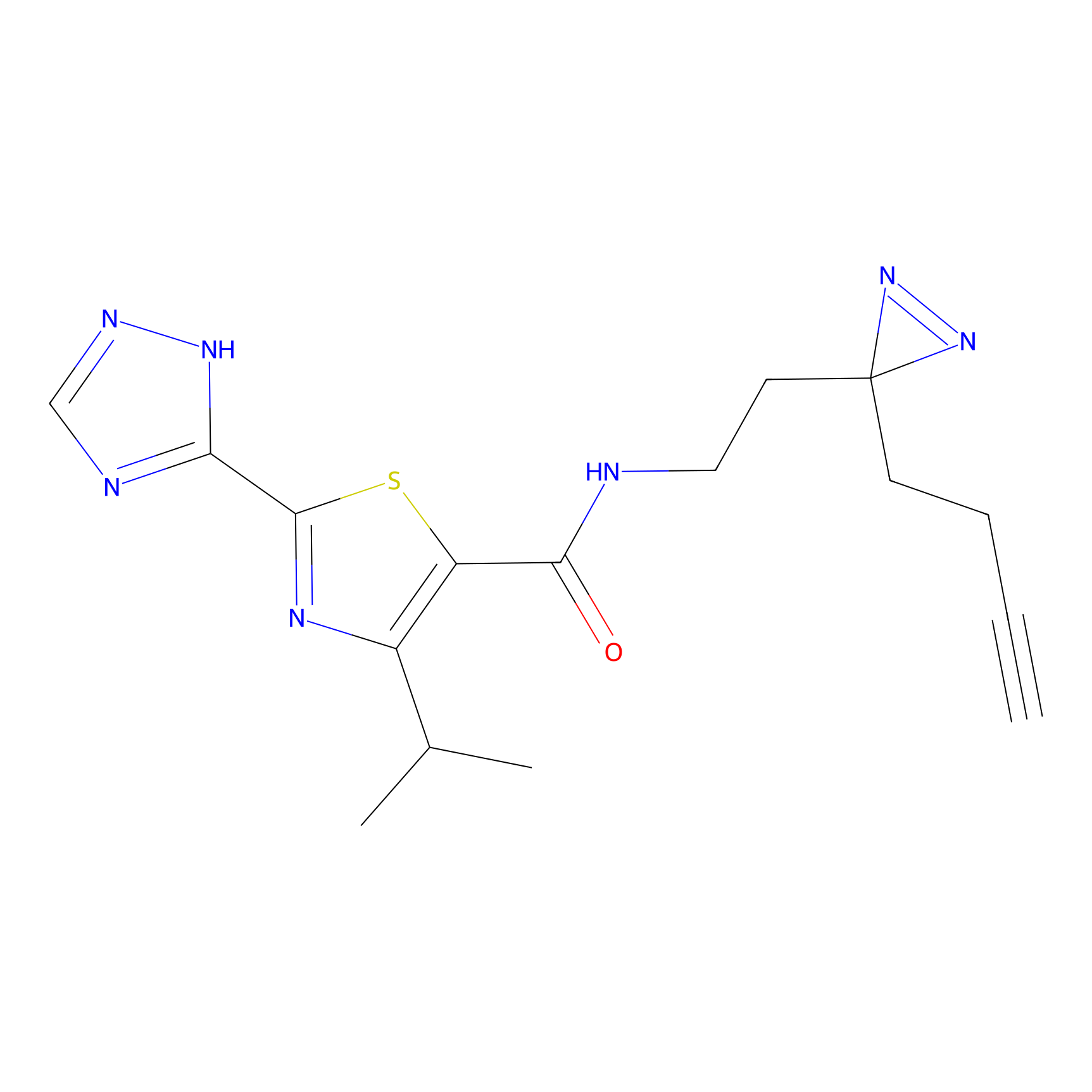 |
9.06 | LDD1939 | [24] | |
|
C274 Probe Info |
 |
11.63 | LDD1944 | [24] | |
|
C284 Probe Info |
 |
22.78 | LDD1954 | [24] | |
|
C305 Probe Info |
 |
11.16 | LDD1974 | [24] | |
|
C338 Probe Info |
 |
14.93 | LDD2001 | [24] | |
|
C355 Probe Info |
 |
22.94 | LDD2016 | [24] | |
|
C362 Probe Info |
 |
39.95 | LDD2023 | [24] | |
|
C388 Probe Info |
 |
51.27 | LDD2047 | [24] | |
|
C391 Probe Info |
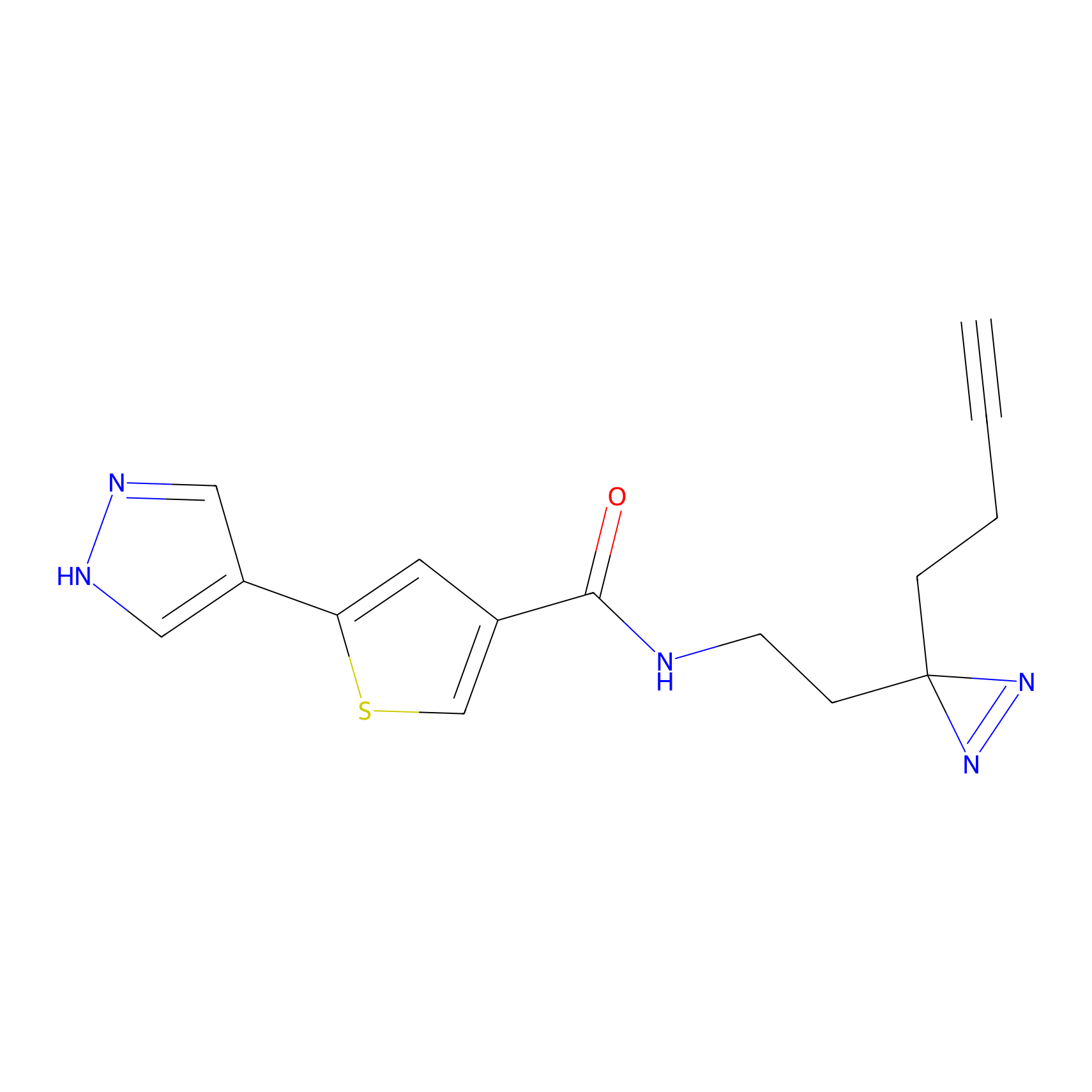 |
15.14 | LDD2050 | [24] | |
|
C407 Probe Info |
 |
19.70 | LDD2064 | [24] | |
|
FFF probe11 Probe Info |
 |
7.22 | LDD0471 | [25] | |
|
FFF probe13 Probe Info |
 |
17.82 | LDD0475 | [25] | |
|
FFF probe14 Probe Info |
 |
13.27 | LDD0477 | [25] | |
|
FFF probe2 Probe Info |
 |
9.64 | LDD0463 | [25] | |
|
FFF probe3 Probe Info |
 |
18.41 | LDD0464 | [25] | |
|
FFF probe6 Probe Info |
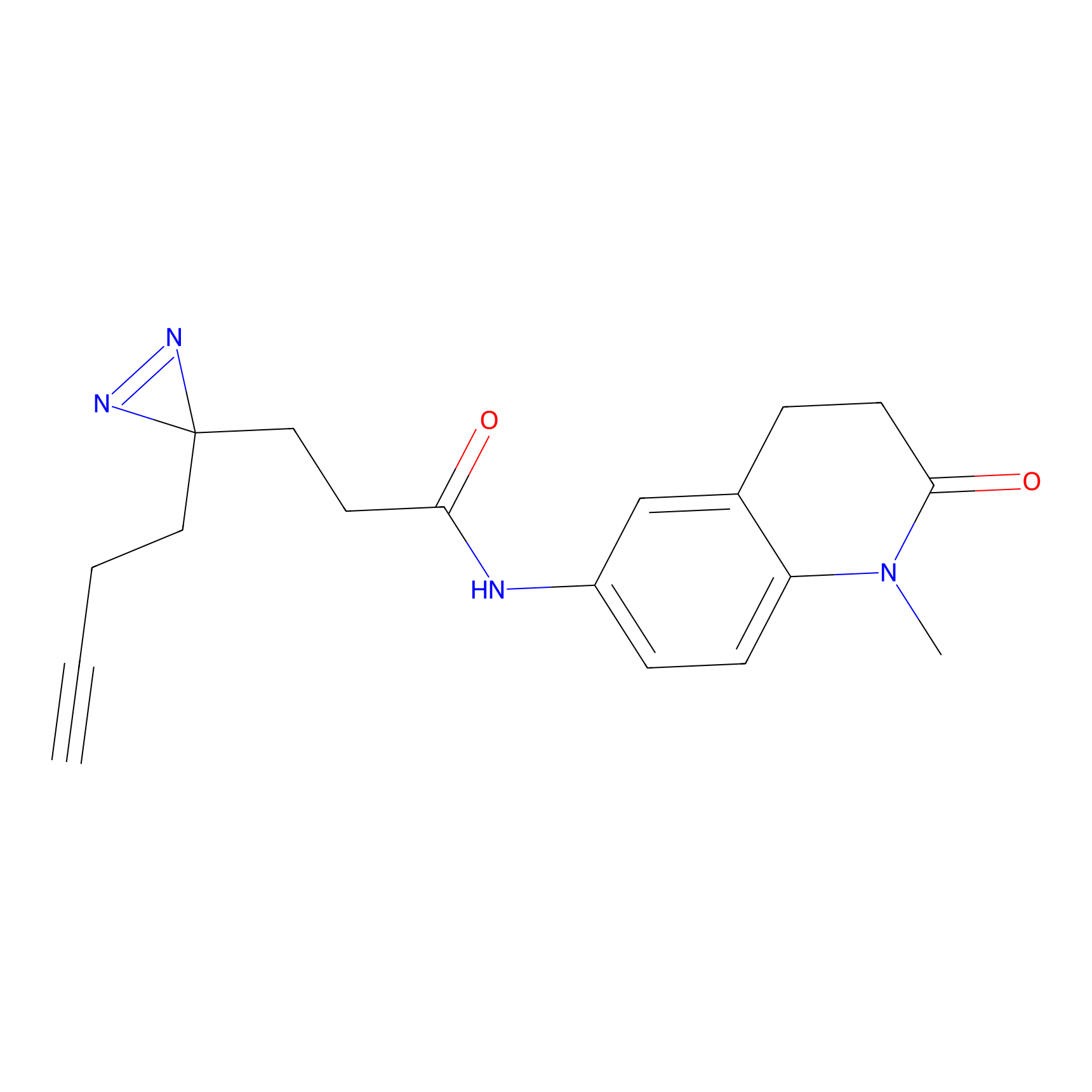 |
8.80 | LDD0467 | [25] | |
|
FFF probe9 Probe Info |
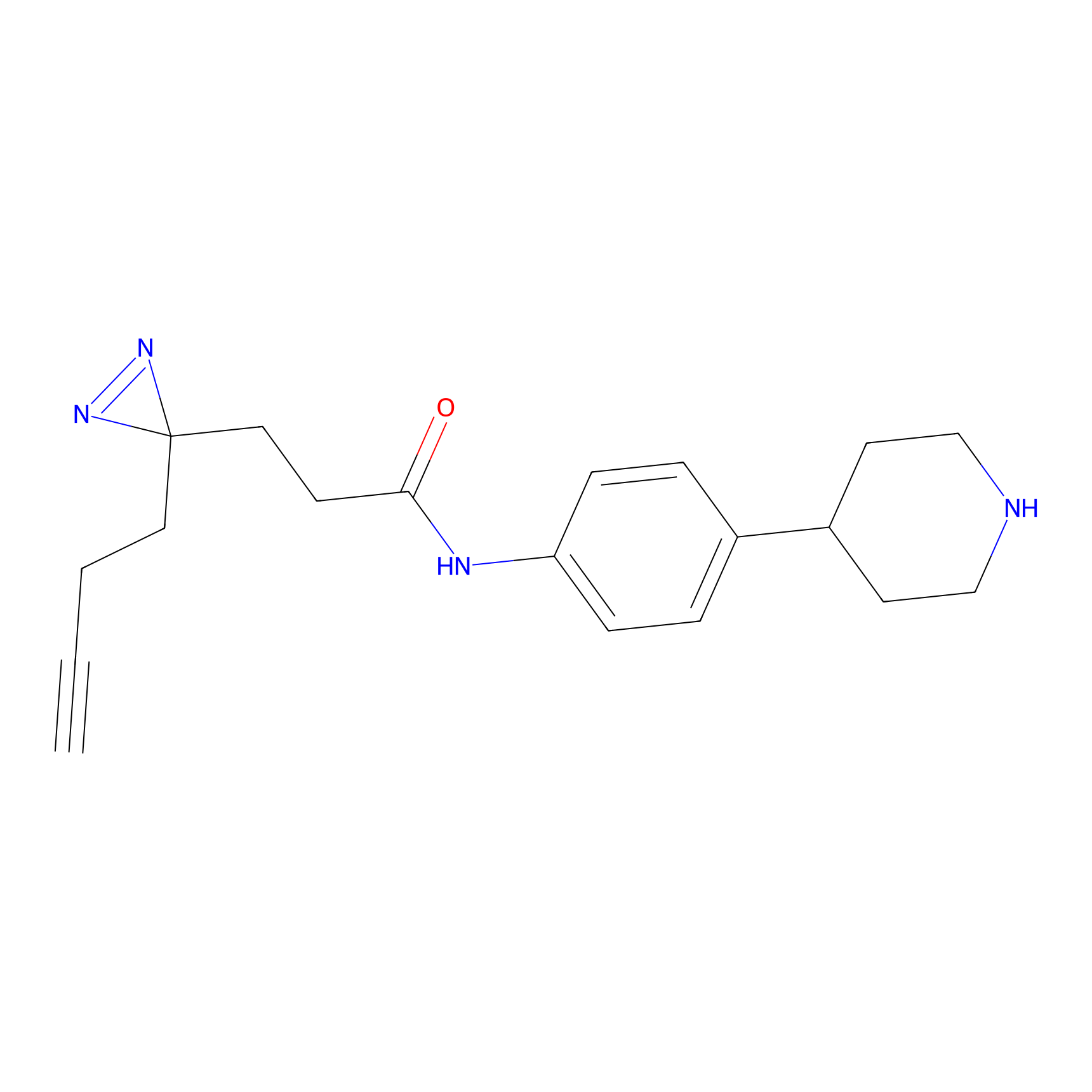 |
5.67 | LDD0470 | [25] | |
|
JN0003 Probe Info |
 |
15.57 | LDD0469 | [25] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [26] | |
|
BD-F Probe Info |
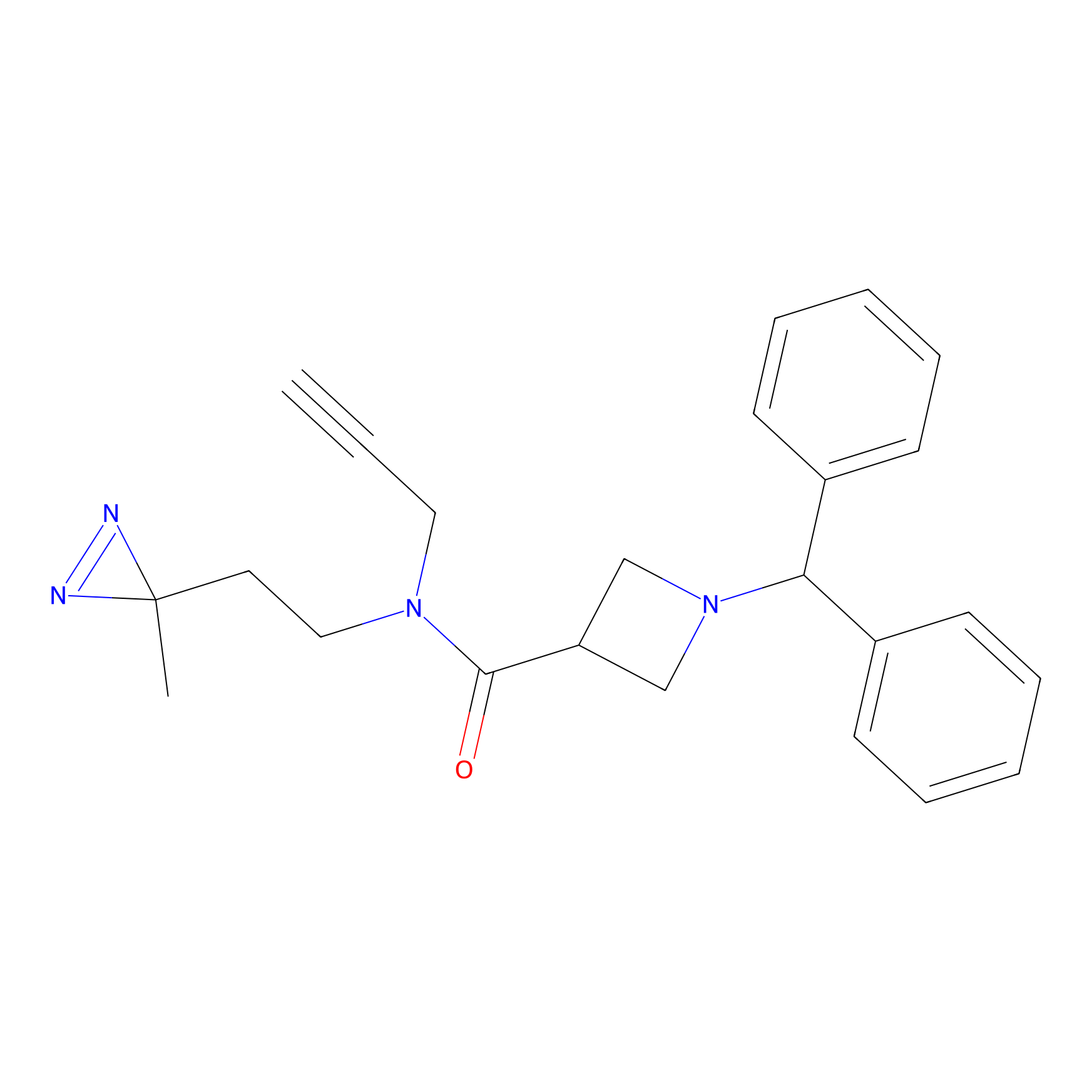 |
E391(0.00); L392(0.00) | LDD0024 | [27] | |
|
TM-F Probe Info |
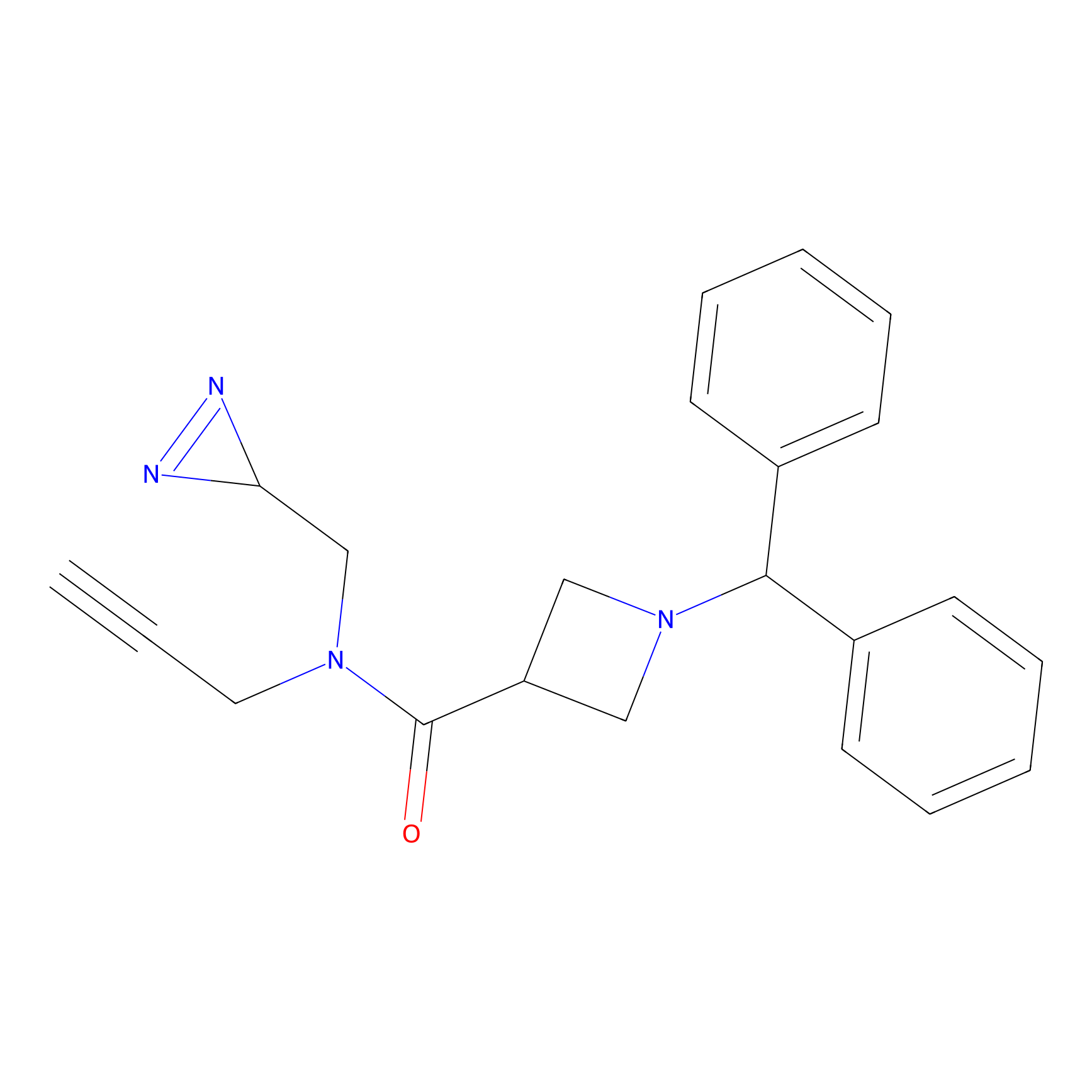 |
A388(0.00); G393(0.00); A397(0.00) | LDD0020 | [27] | |
|
Photocelecoxib Probe Info |
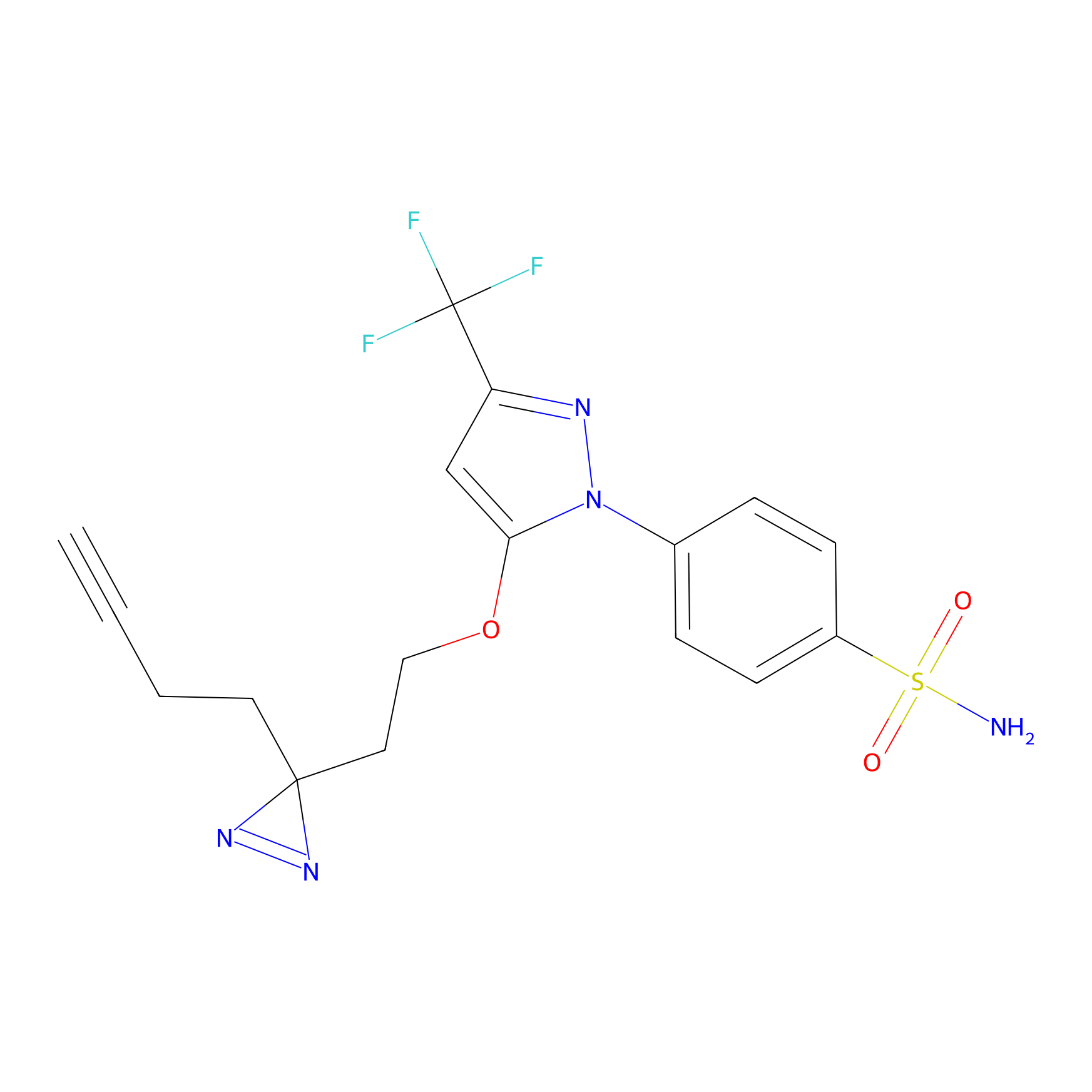 |
I389(0.00); E391(0.00) | LDD0019 | [28] | |
|
Photonaproxen Probe Info |
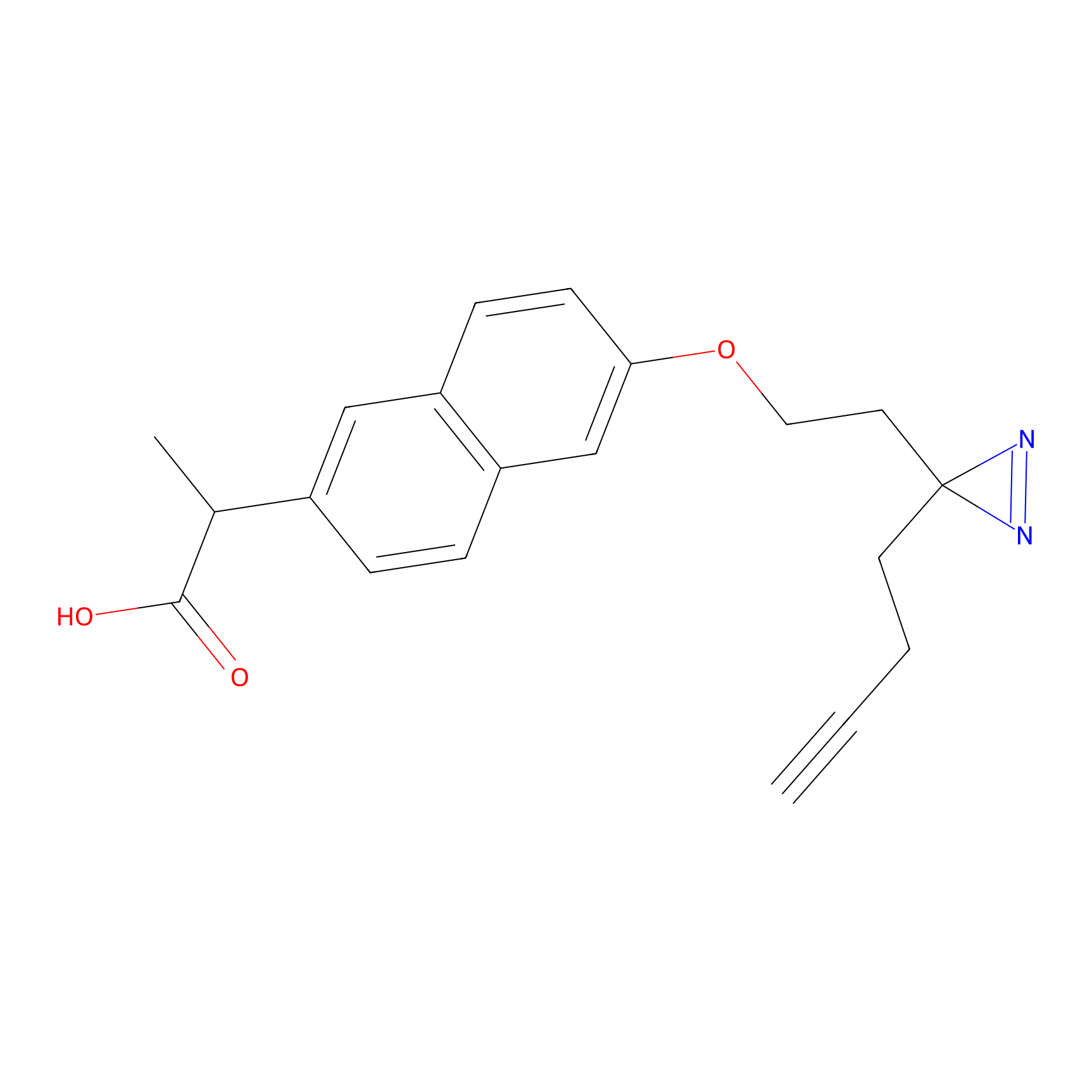 |
Y395(0.00); D402(0.00) | LDD0156 | [28] | |
|
OEA-DA Probe Info |
 |
4.98 | LDD0046 | [29] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0069 | [30] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0108 | Chloroacetamide | HeLa | H477(0.00); H248(0.00); H417(0.00); H102(0.00) | LDD0222 | [20] |
| LDCM0189 | Compound 16 | HEK-293T | 6.31 | LDD0492 | [25] |
| LDCM0191 | Compound 21 | HEK-293T | 3.84 | LDD0493 | [25] |
| LDCM0190 | Compound 34 | HEK-293T | 5.74 | LDD0497 | [25] |
| LDCM0192 | Compound 35 | HEK-293T | 4.81 | LDD0491 | [25] |
| LDCM0193 | Compound 36 | HEK-293T | 3.51 | LDD0494 | [25] |
| LDCM0194 | Compound 37 | HEK-293T | 5.36 | LDD0498 | [25] |
| LDCM0195 | Compound 38 | HEK-293T | 7.10 | LDD0499 | [25] |
| LDCM0197 | Compound 40 | HEK-293T | 3.84 | LDD0495 | [25] |
| LDCM0116 | HHS-0101 | DM93 | Y395(0.58); Y431(0.86); Y418(0.91) | LDD0264 | [13] |
| LDCM0117 | HHS-0201 | DM93 | Y395(0.18); Y418(1.20); Y431(1.21) | LDD0265 | [13] |
| LDCM0118 | HHS-0301 | DM93 | Y395(0.27); Y431(0.78); Y418(0.87) | LDD0266 | [13] |
| LDCM0119 | HHS-0401 | DM93 | Y395(0.23); Y418(1.10); Y431(1.84) | LDD0267 | [13] |
| LDCM0120 | HHS-0701 | DM93 | Y395(0.25); Y431(0.97); Y418(1.62) | LDD0268 | [13] |
| LDCM0107 | IAA | HeLa | H477(0.00); H102(0.00); H417(0.00); H527(0.00) | LDD0221 | [20] |
| LDCM0123 | JWB131 | DM93 | Y395(0.83); Y418(0.82); Y431(1.06) | LDD0285 | [12] |
| LDCM0124 | JWB142 | DM93 | Y395(3.13); Y418(1.07); Y431(1.51) | LDD0286 | [12] |
| LDCM0125 | JWB146 | DM93 | Y395(5.89); Y418(0.89); Y431(1.37) | LDD0287 | [12] |
| LDCM0126 | JWB150 | DM93 | Y395(8.95); Y418(4.08); Y431(3.90) | LDD0288 | [12] |
| LDCM0127 | JWB152 | DM93 | Y395(6.95); Y418(2.11); Y431(2.74) | LDD0289 | [12] |
| LDCM0128 | JWB198 | DM93 | Y395(0.61); Y418(1.19); Y431(1.22) | LDD0290 | [12] |
| LDCM0129 | JWB202 | DM93 | Y395(1.93); Y418(0.76); Y431(0.93) | LDD0291 | [12] |
| LDCM0130 | JWB211 | DM93 | Y395(0.84); Y418(1.02); Y431(1.22) | LDD0292 | [12] |
| LDCM0109 | NEM | HeLa | H477(0.00); H417(0.00); H102(0.00); H527(0.00) | LDD0223 | [20] |
| LDCM0111 | W14 | Hep-G2 | K485(0.52) | LDD0238 | [21] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
Other
The Drug(s) Related To This Target
Approved
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Inositol Nicotinate | . | DB08949 | |||
Investigative
References
