Details of the Target
General Information of Target
| Target ID | LDTP04956 | |||||
|---|---|---|---|---|---|---|
| Target Name | Mitochondrial import inner membrane translocase subunit Tim10 (TIMM10) | |||||
| Gene Name | TIMM10 | |||||
| Gene ID | 26519 | |||||
| Synonyms |
TIM10; Mitochondrial import inner membrane translocase subunit Tim10 |
|||||
| 3D Structure | ||||||
| Sequence |
MDPLRAQQLAAELEVEMMADMYNRMTSACHRKCVPPHYKEAELSKGESVCLDRCVSKYLD
IHERMGKKLTELSMQDEELMKRVQQSSGPA |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
Small Tim family
|
|||||
| Subcellular location |
Mitochondrion inner membrane
|
|||||
| Function |
Mitochondrial intermembrane chaperone that participates in the import and insertion of multi-pass transmembrane proteins into the mitochondrial inner membrane. May also be required for the transfer of beta-barrel precursors from the TOM complex to the sorting and assembly machinery (SAM complex) of the outer membrane. Acts as a chaperone-like protein that protects the hydrophobic precursors from aggregation and guide them through the mitochondrial intermembrane space.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
DBIA Probe Info |
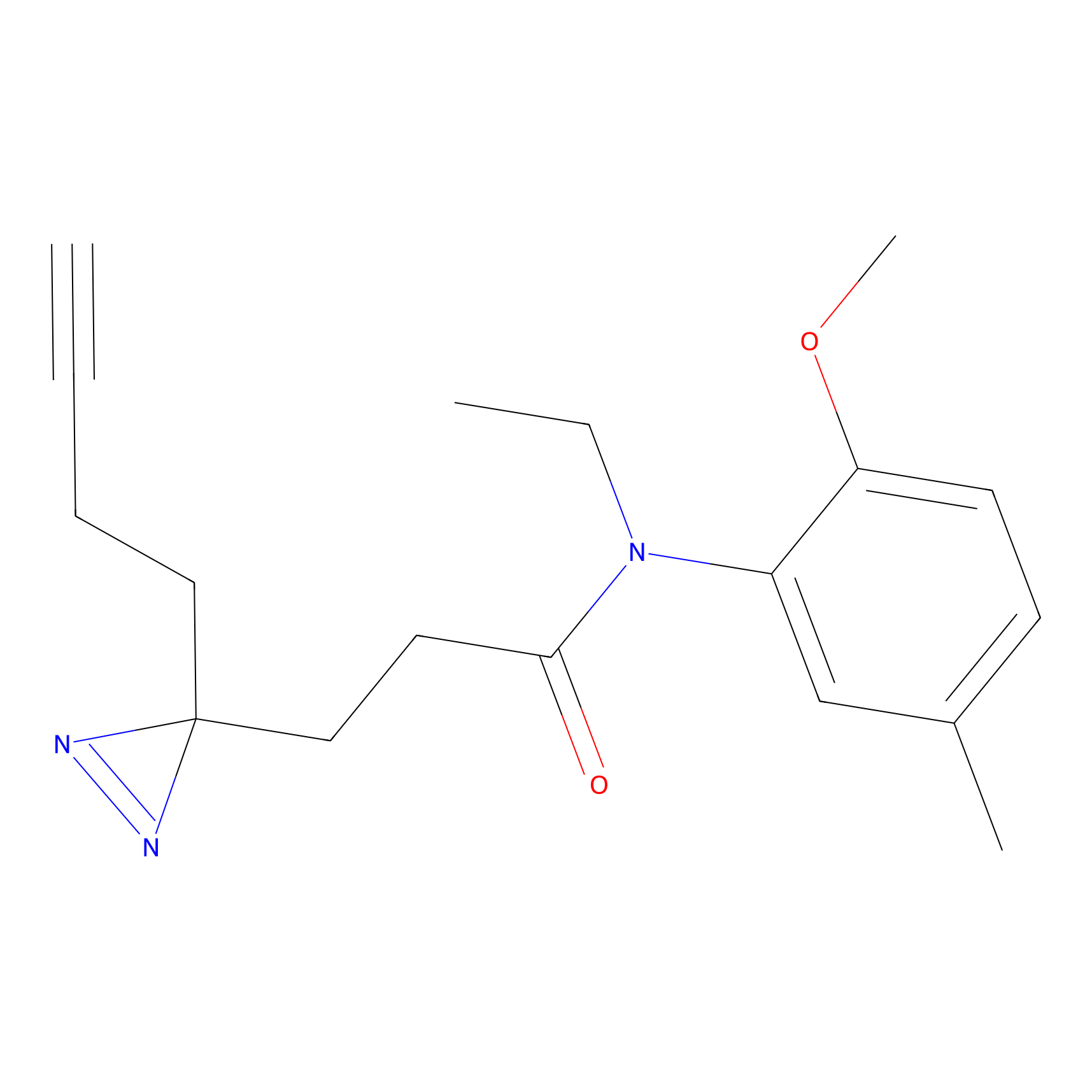
 |
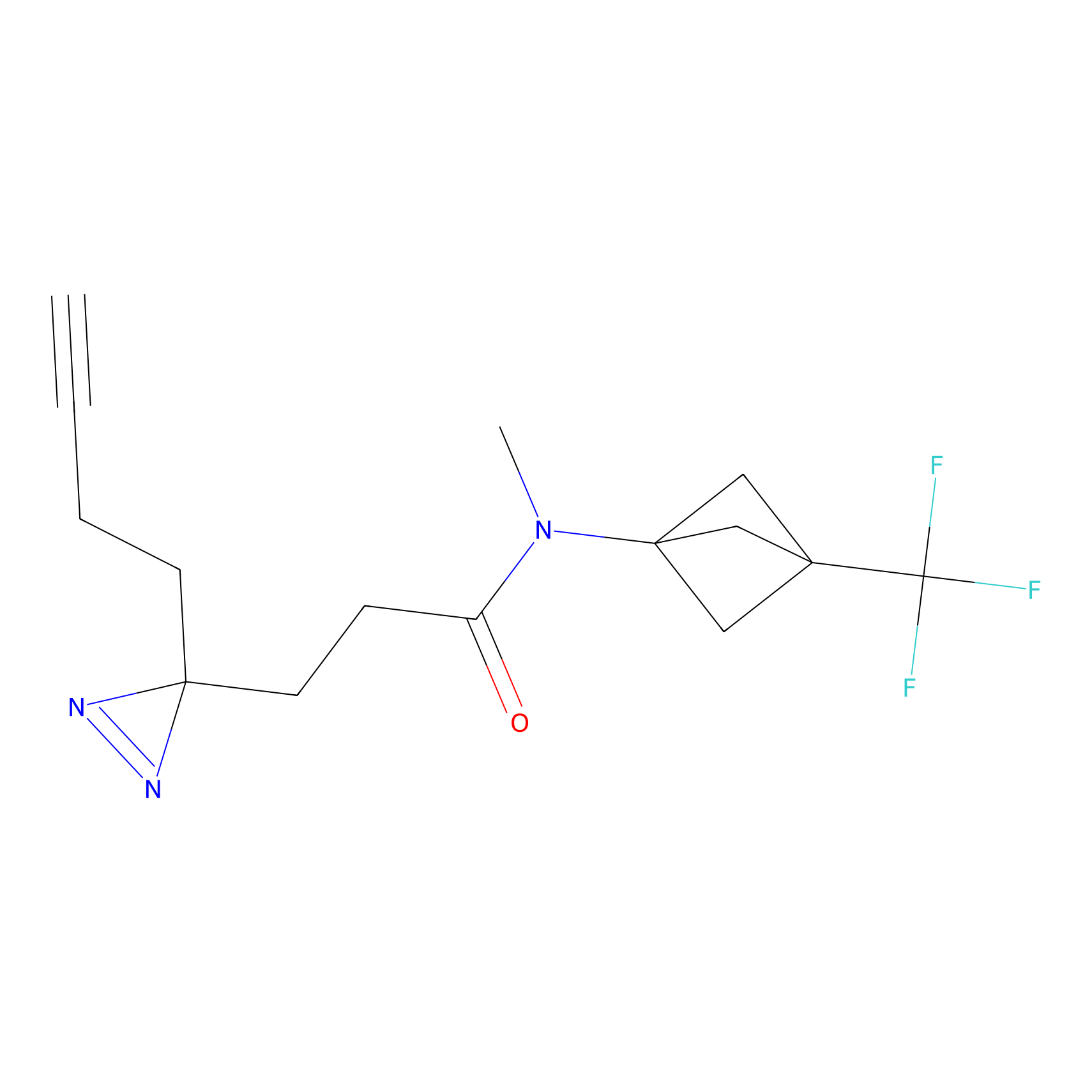
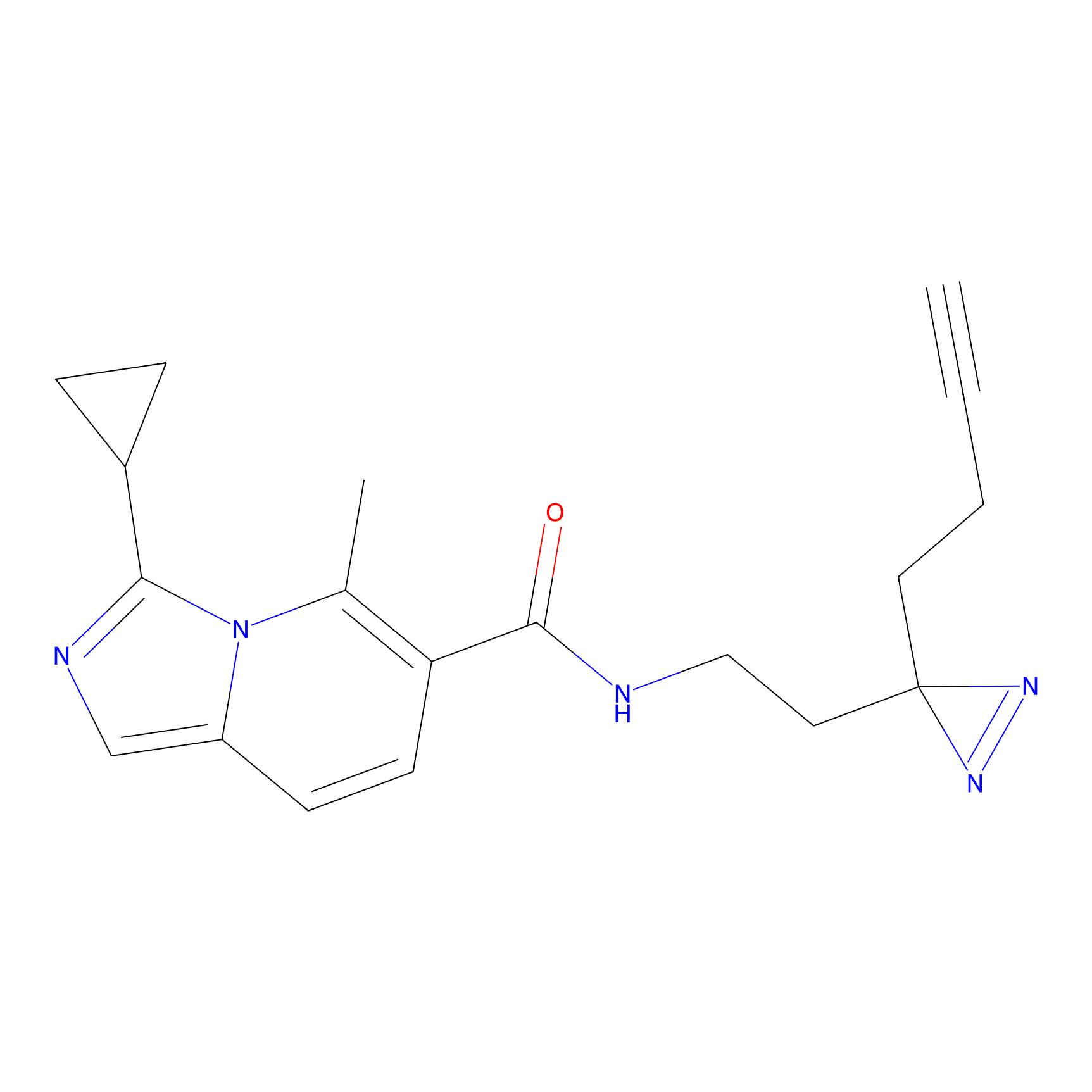
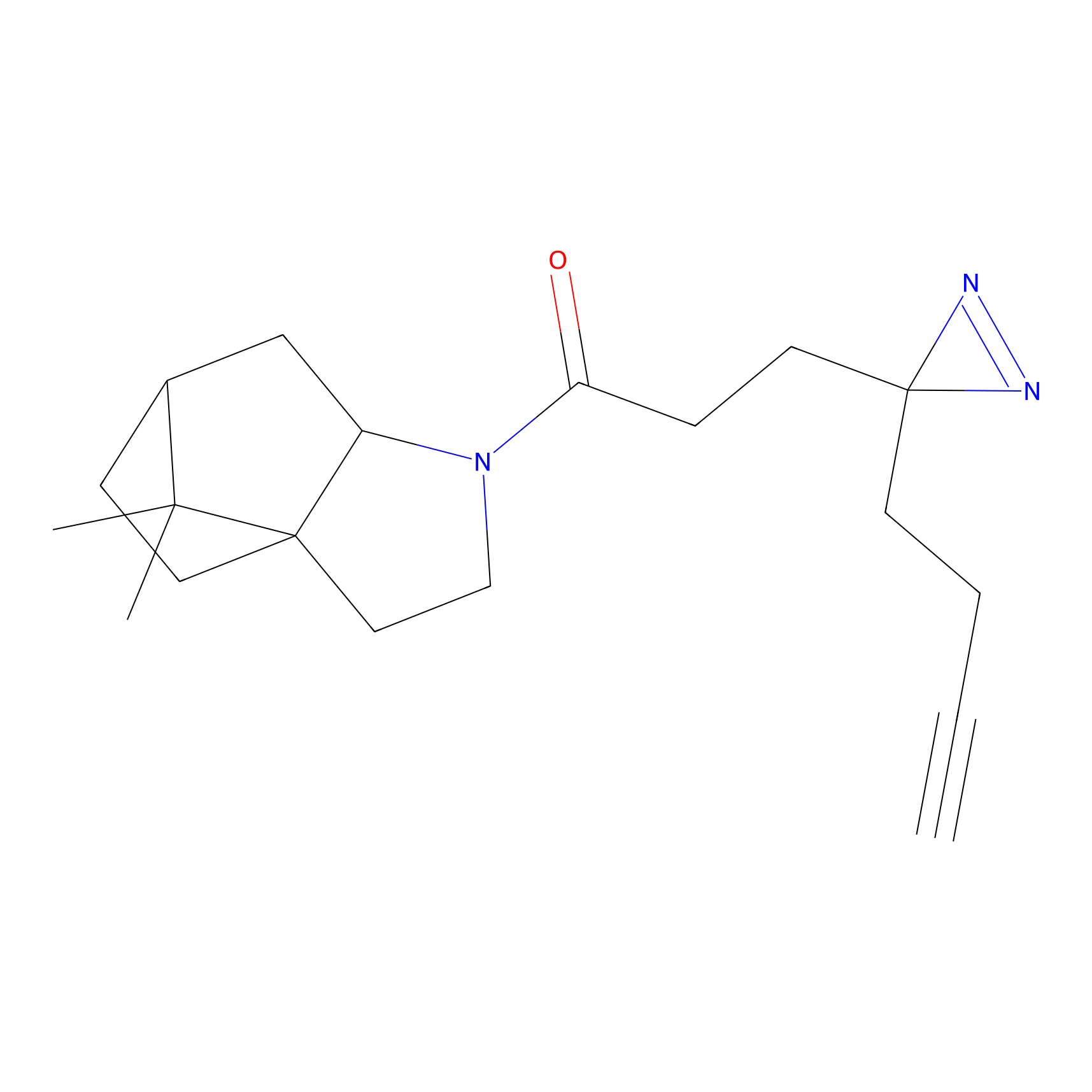
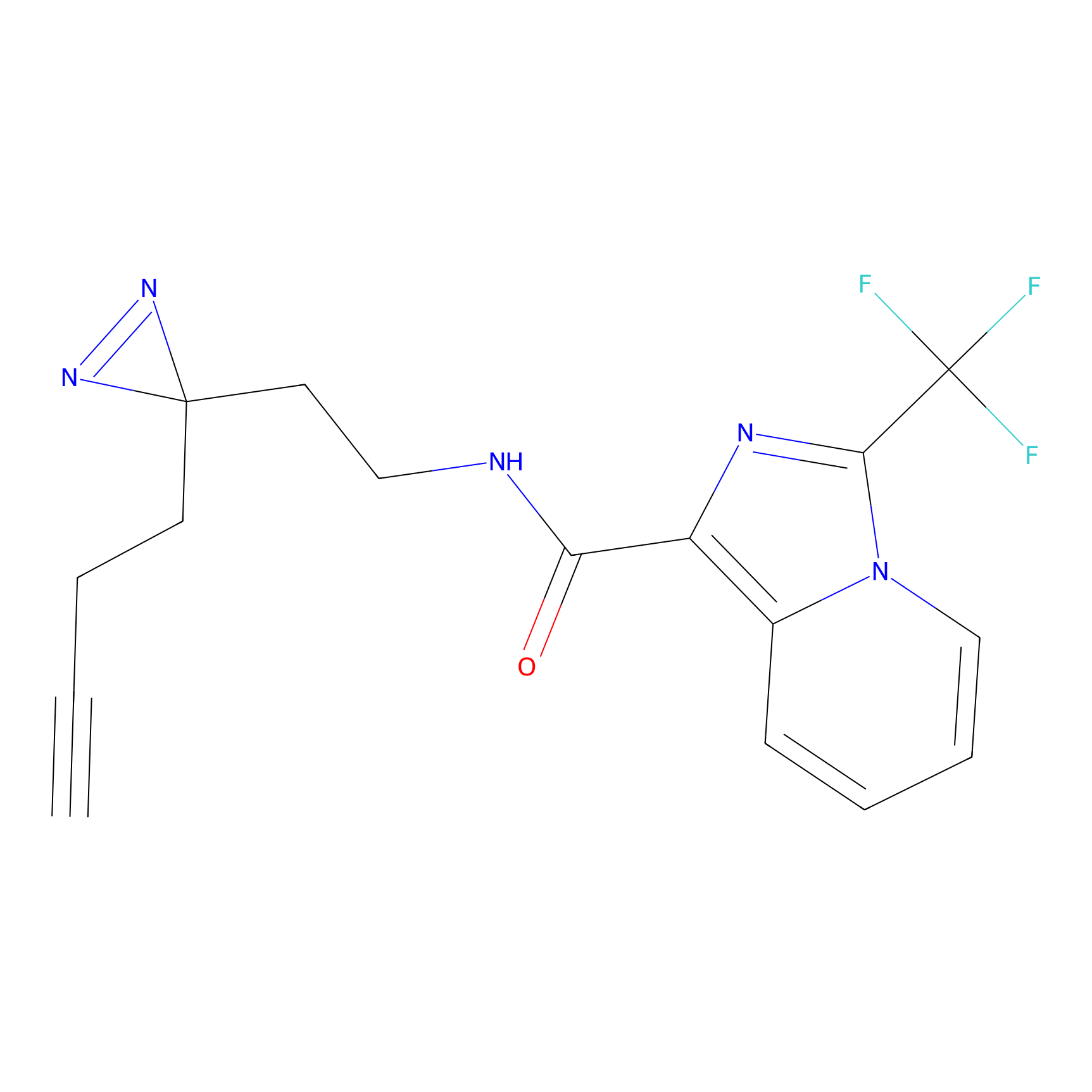
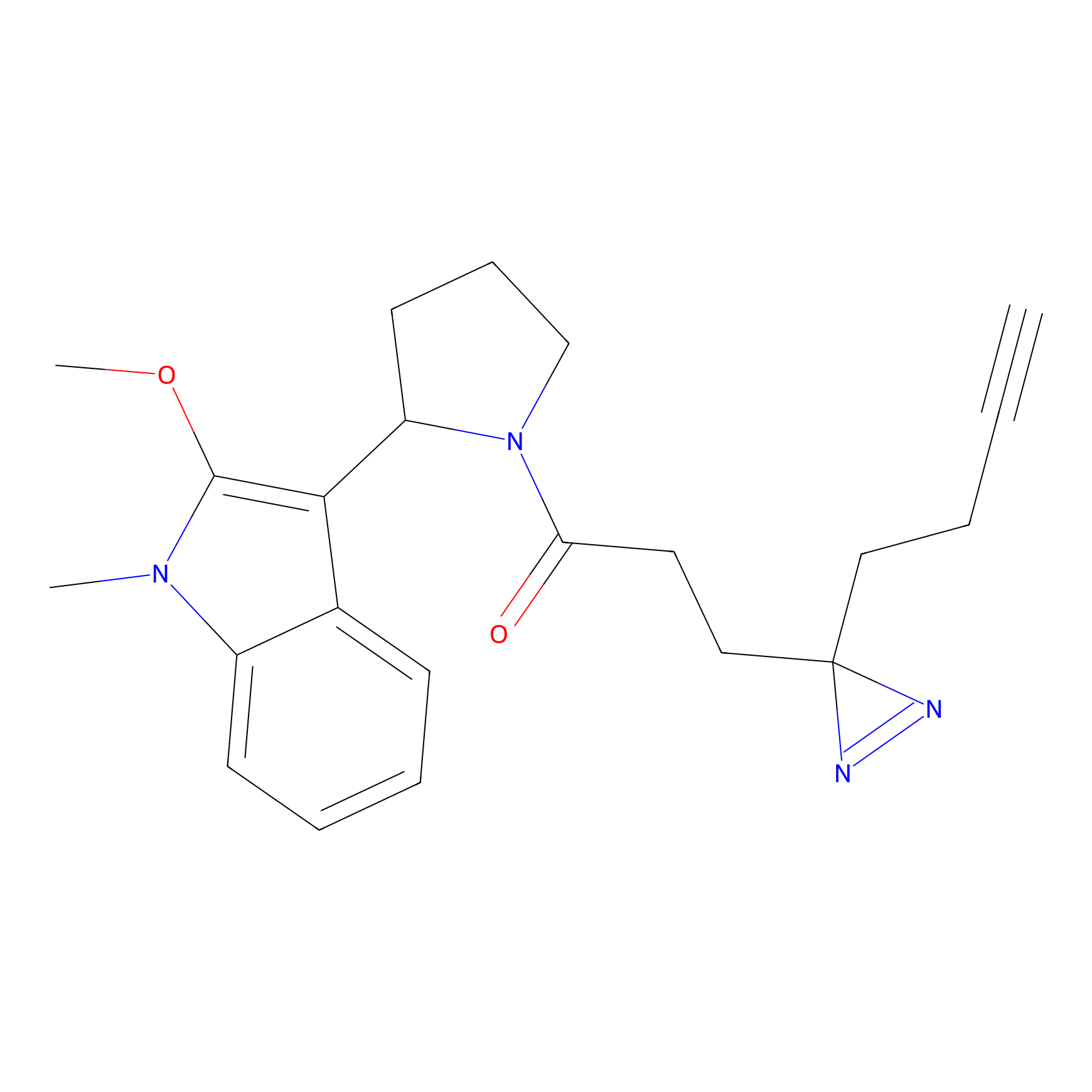
C50(2.68) | LDD3310 | [1] | |
|
BTD Probe Info |
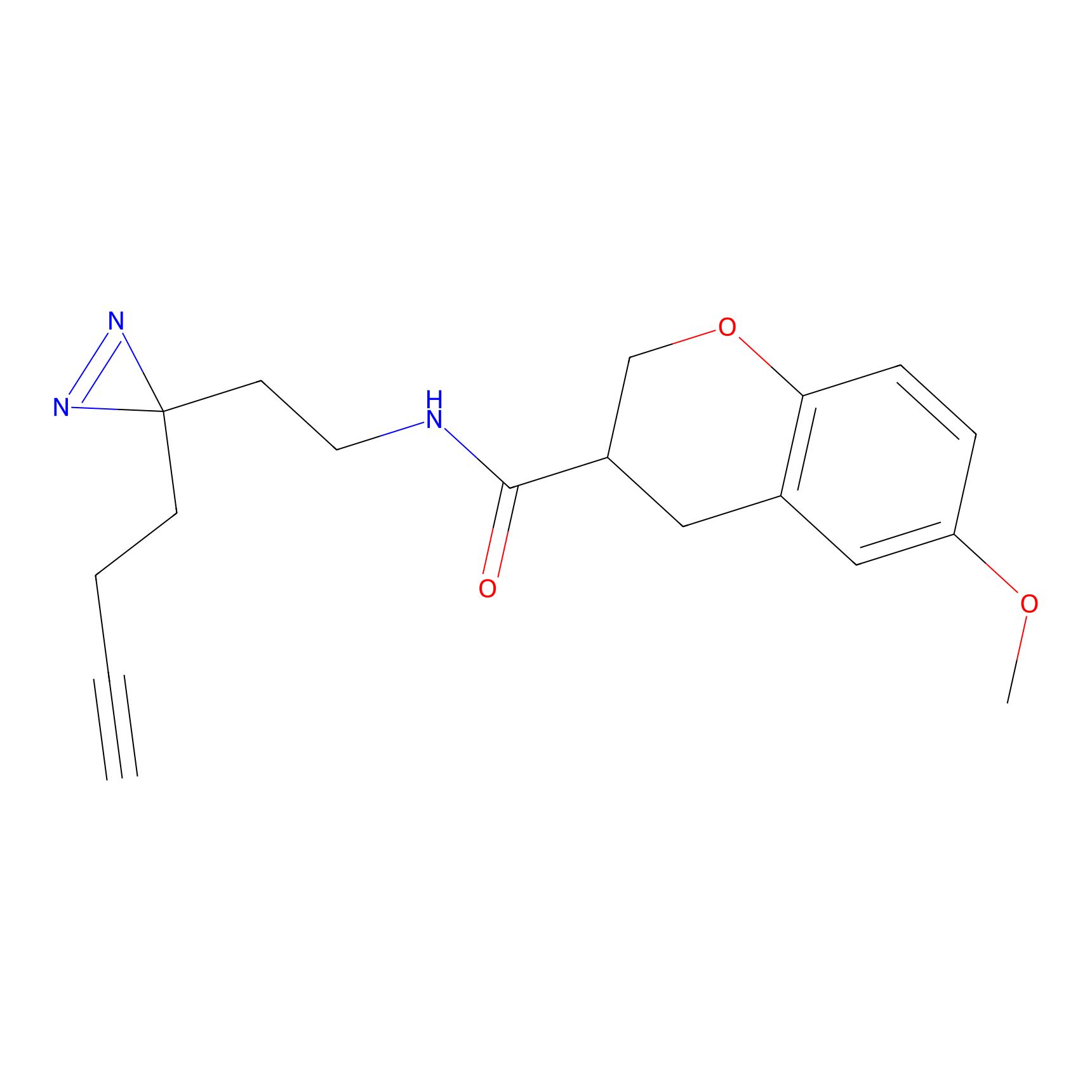
 |
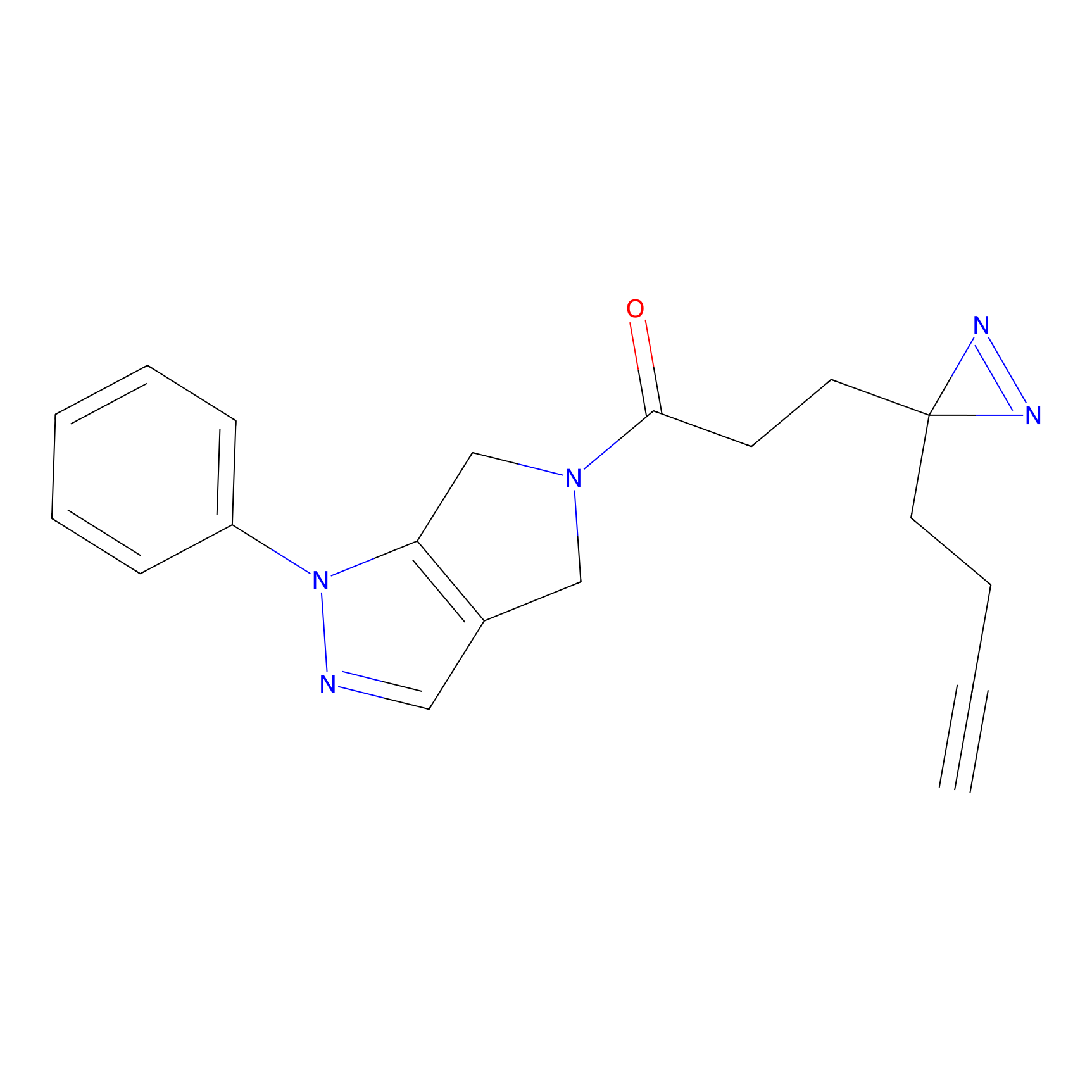
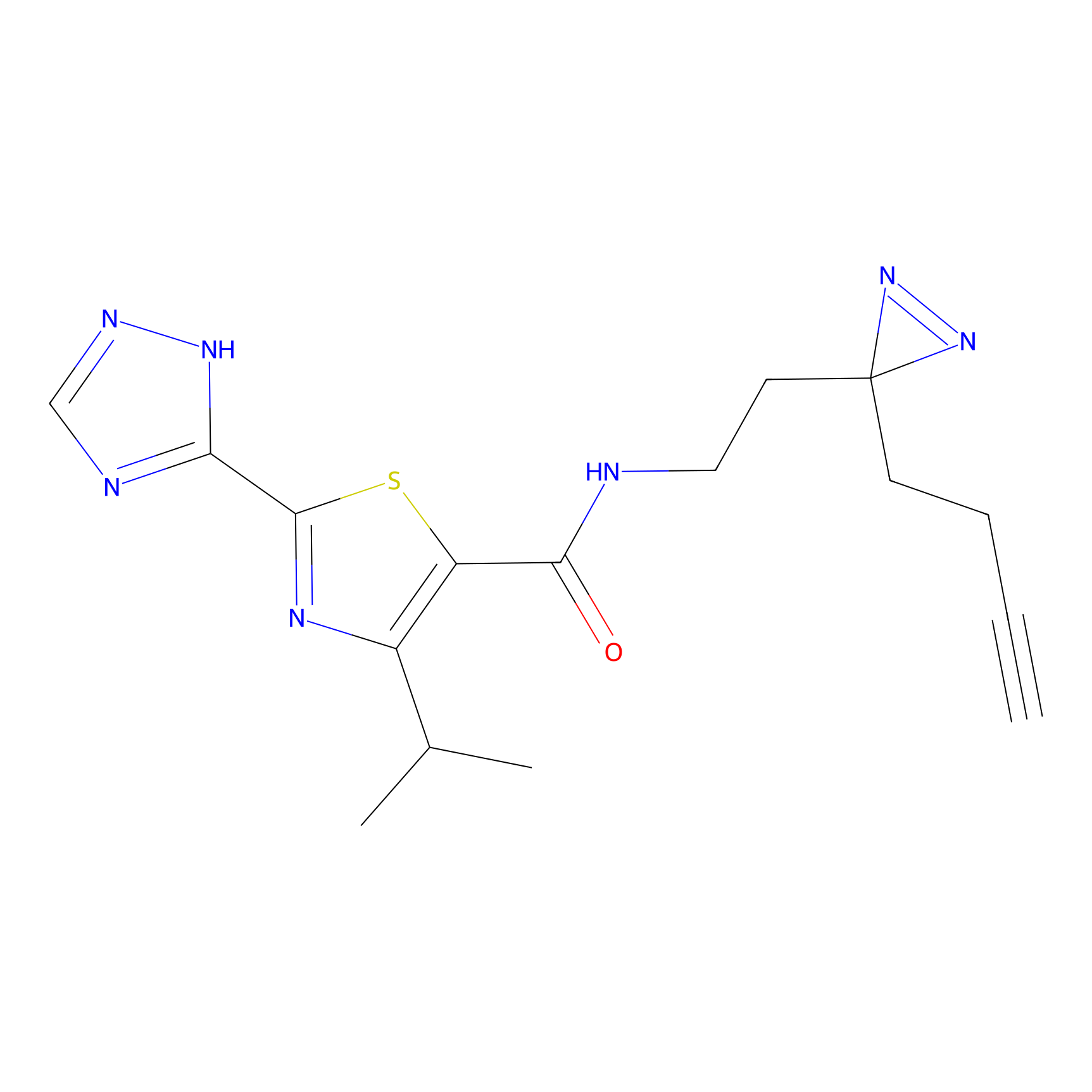
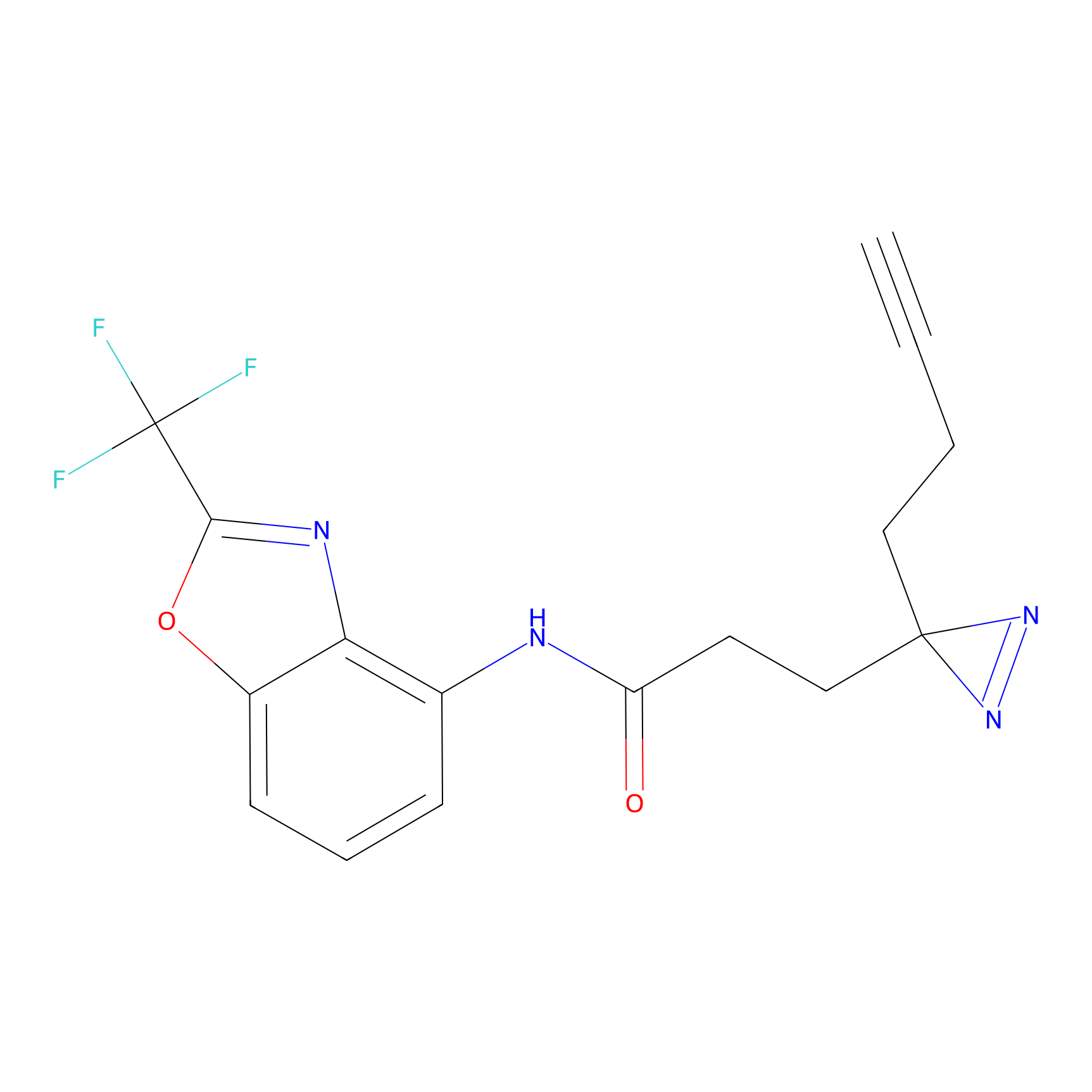
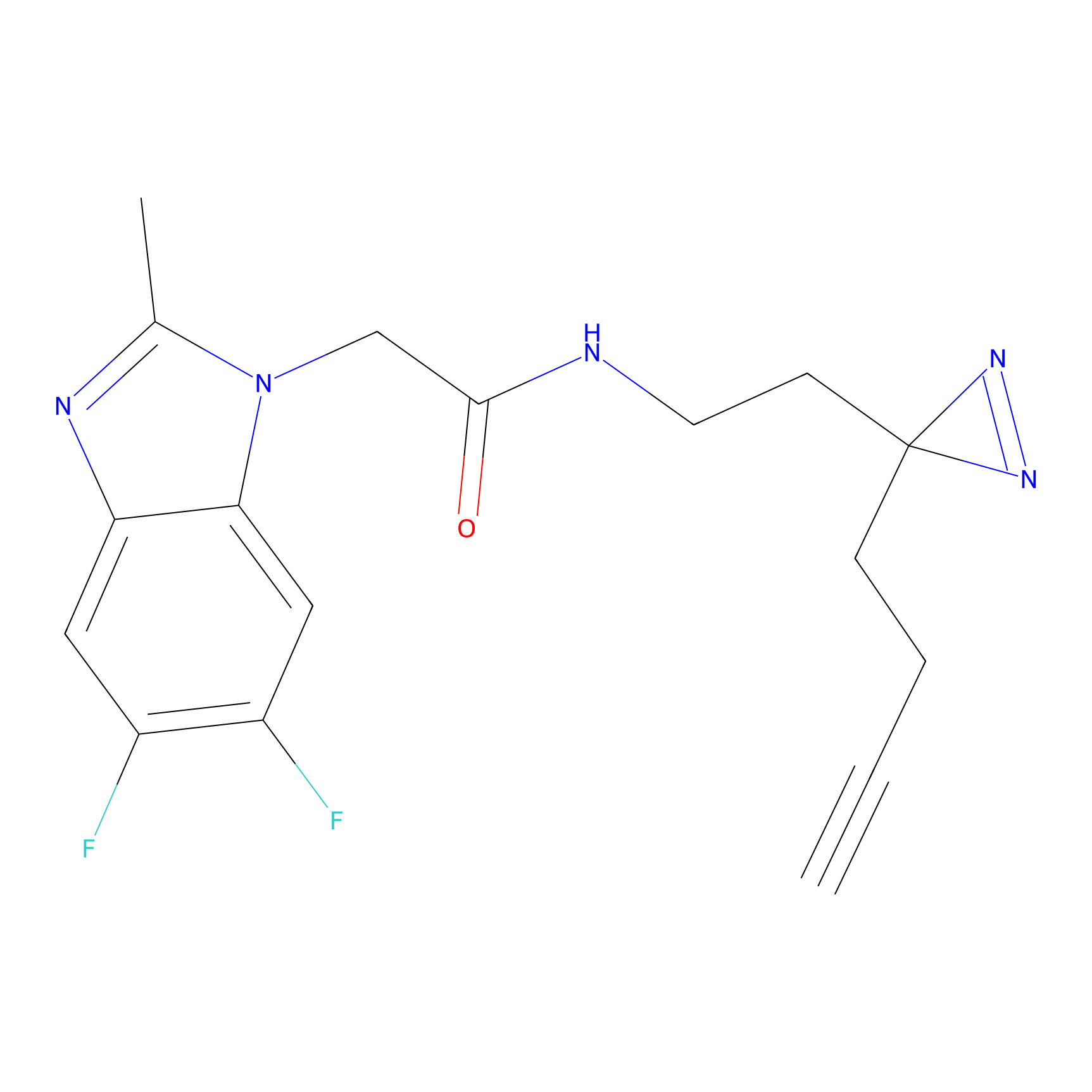
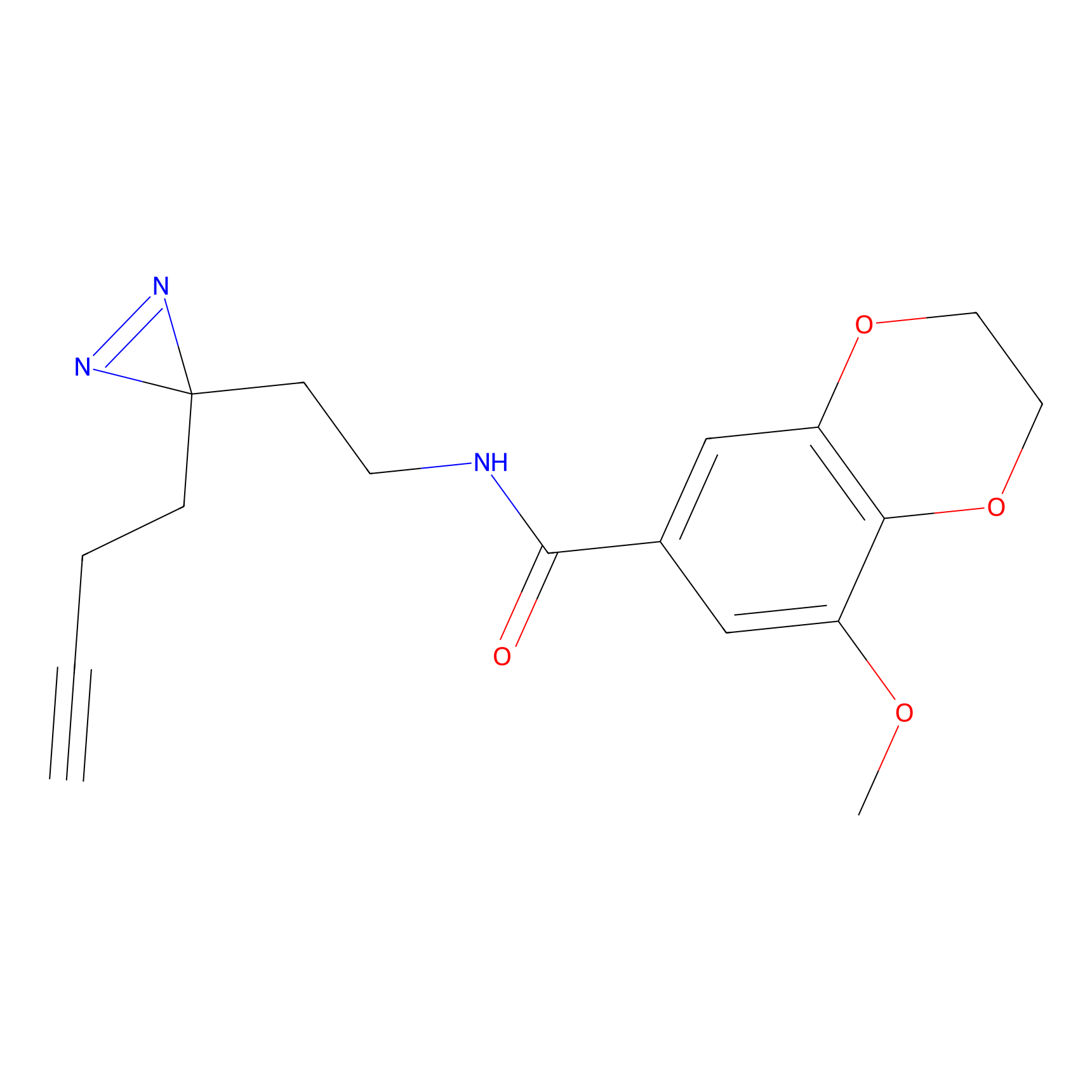
C29(1.13) | LDD1700 | [2] | |
|
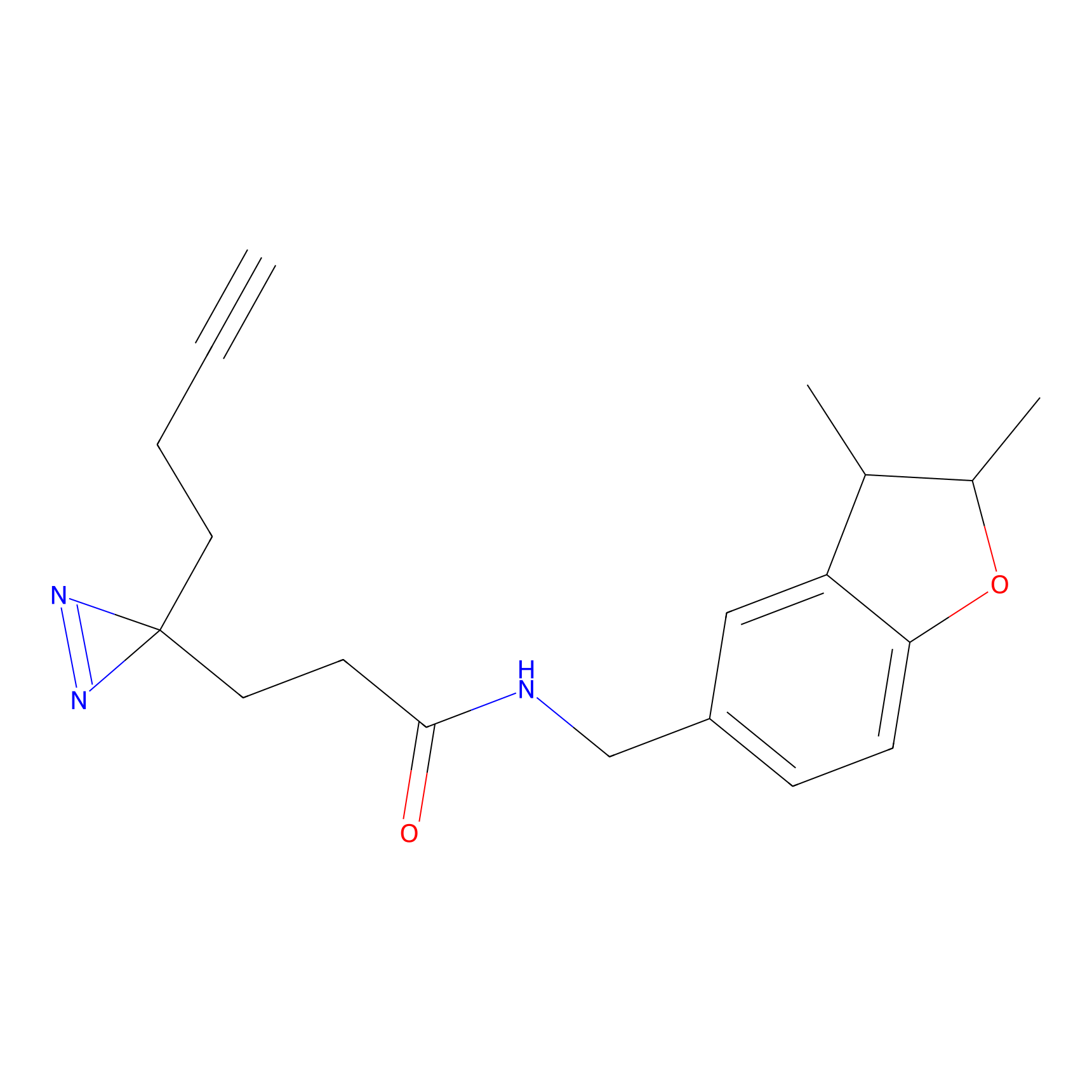
IPM Probe Info |
 |
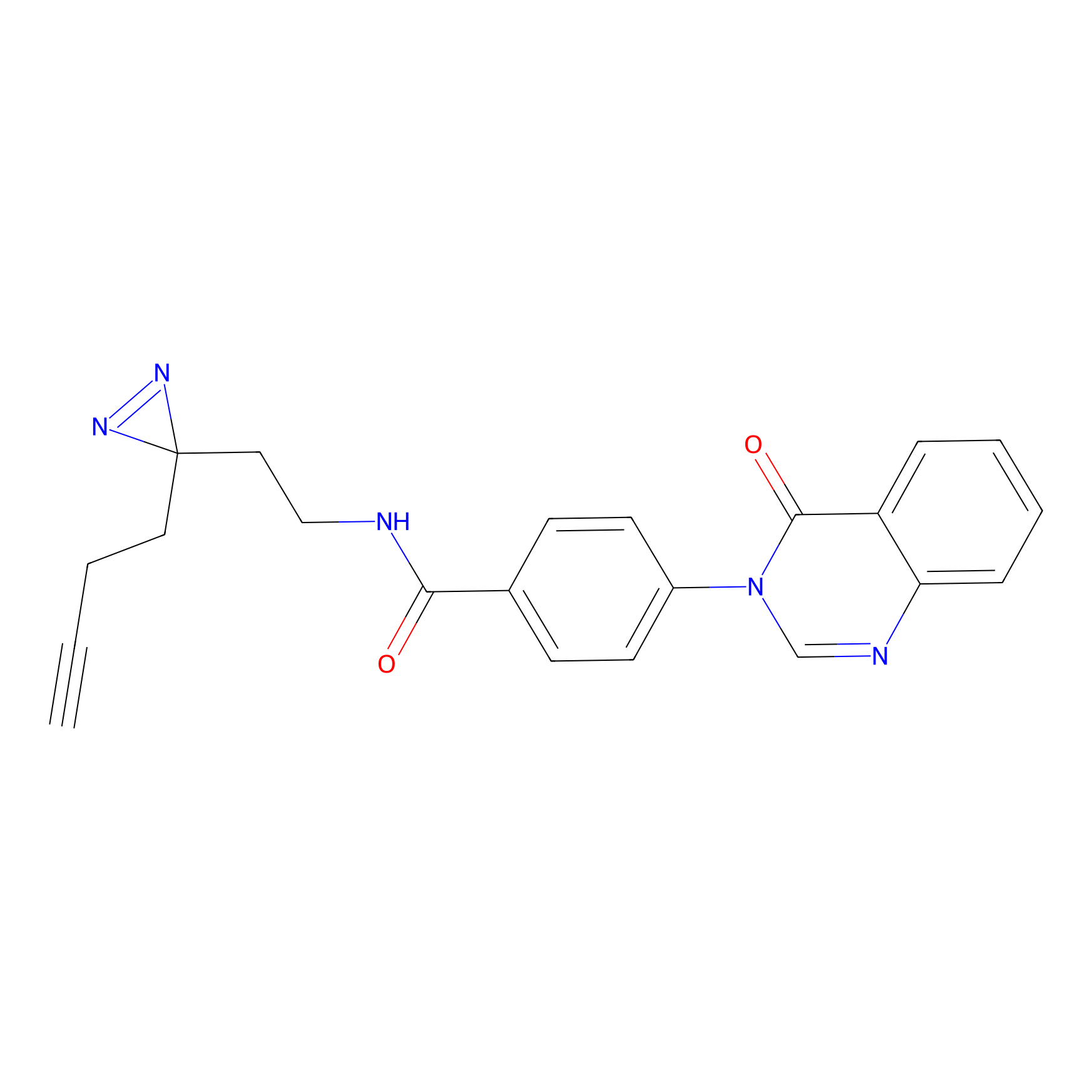
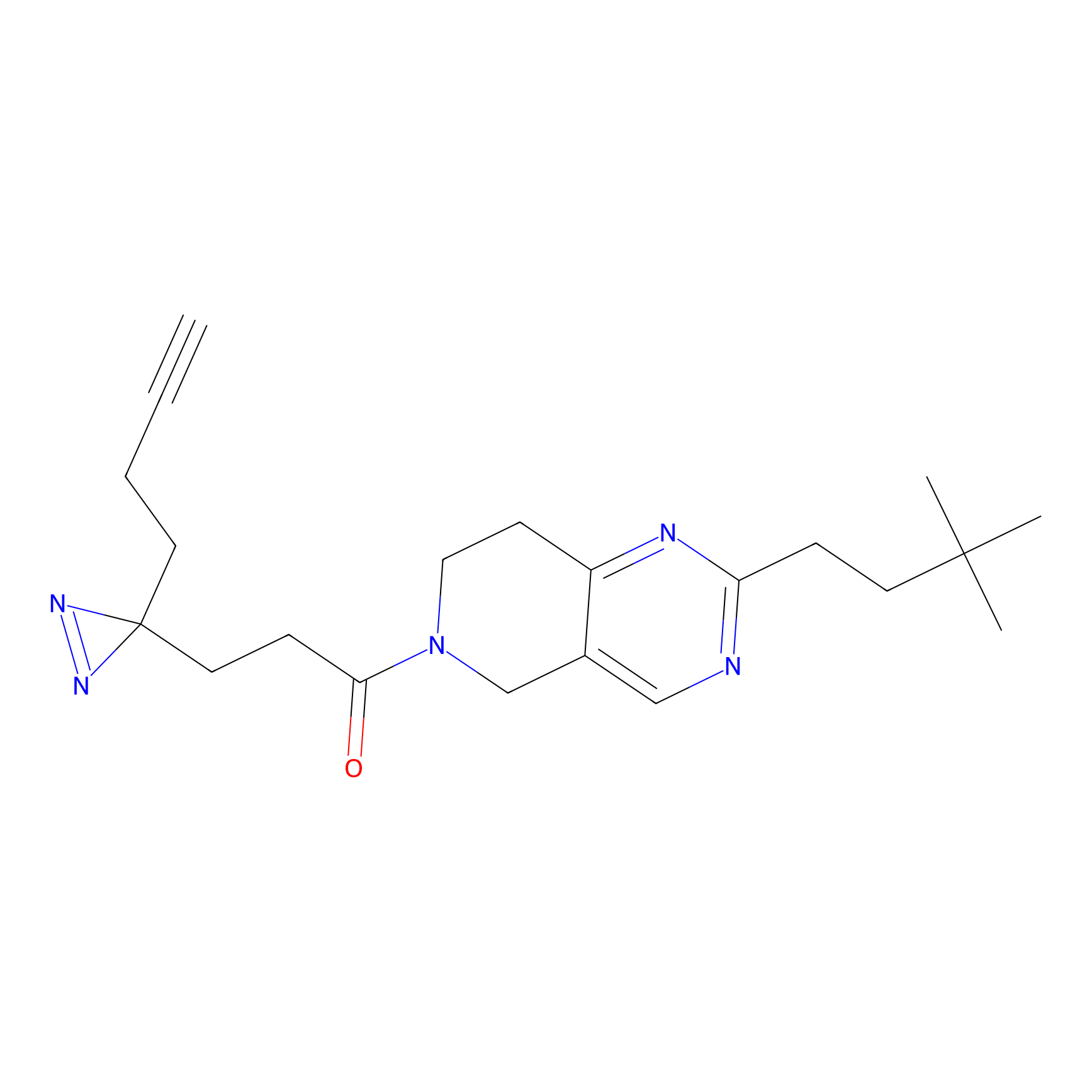
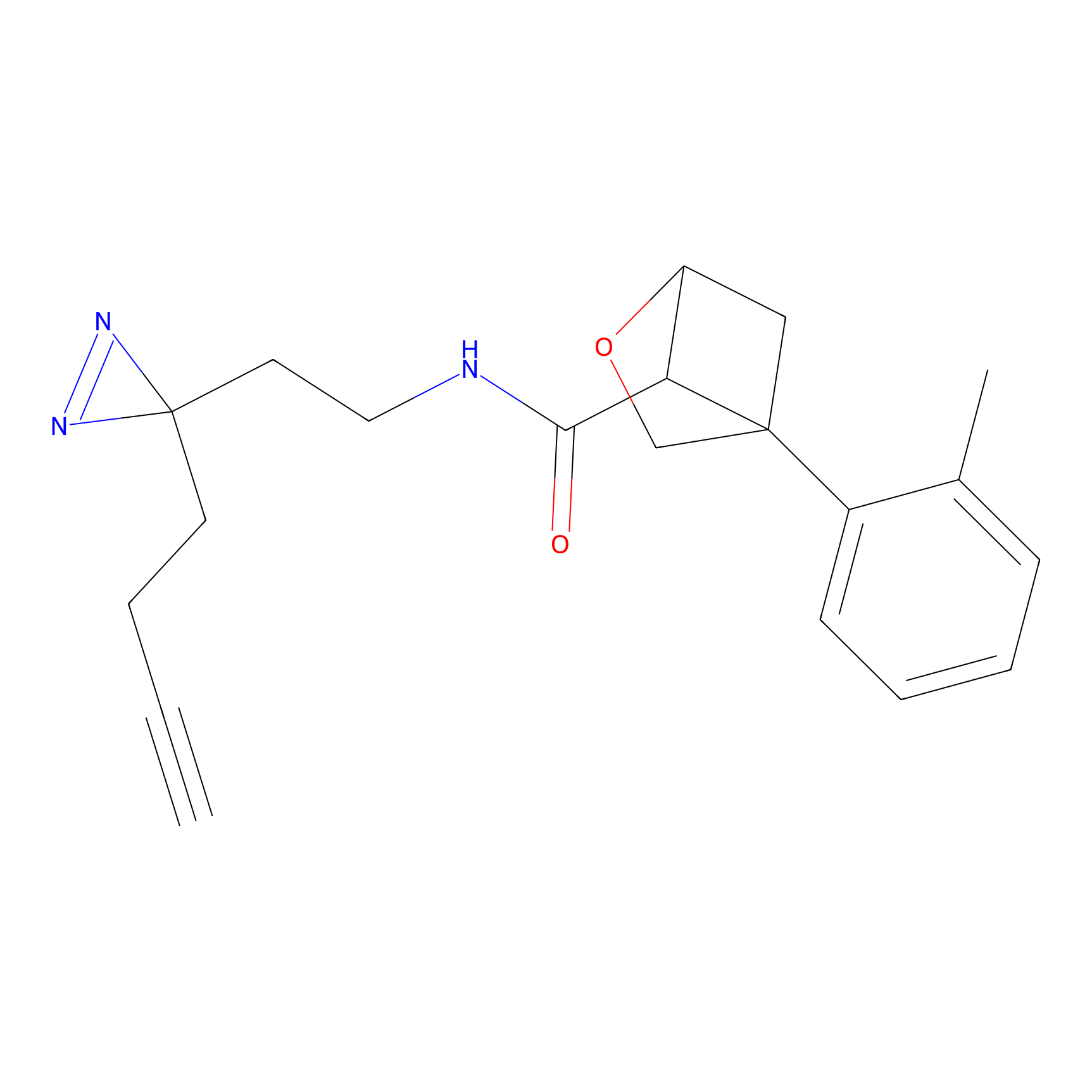
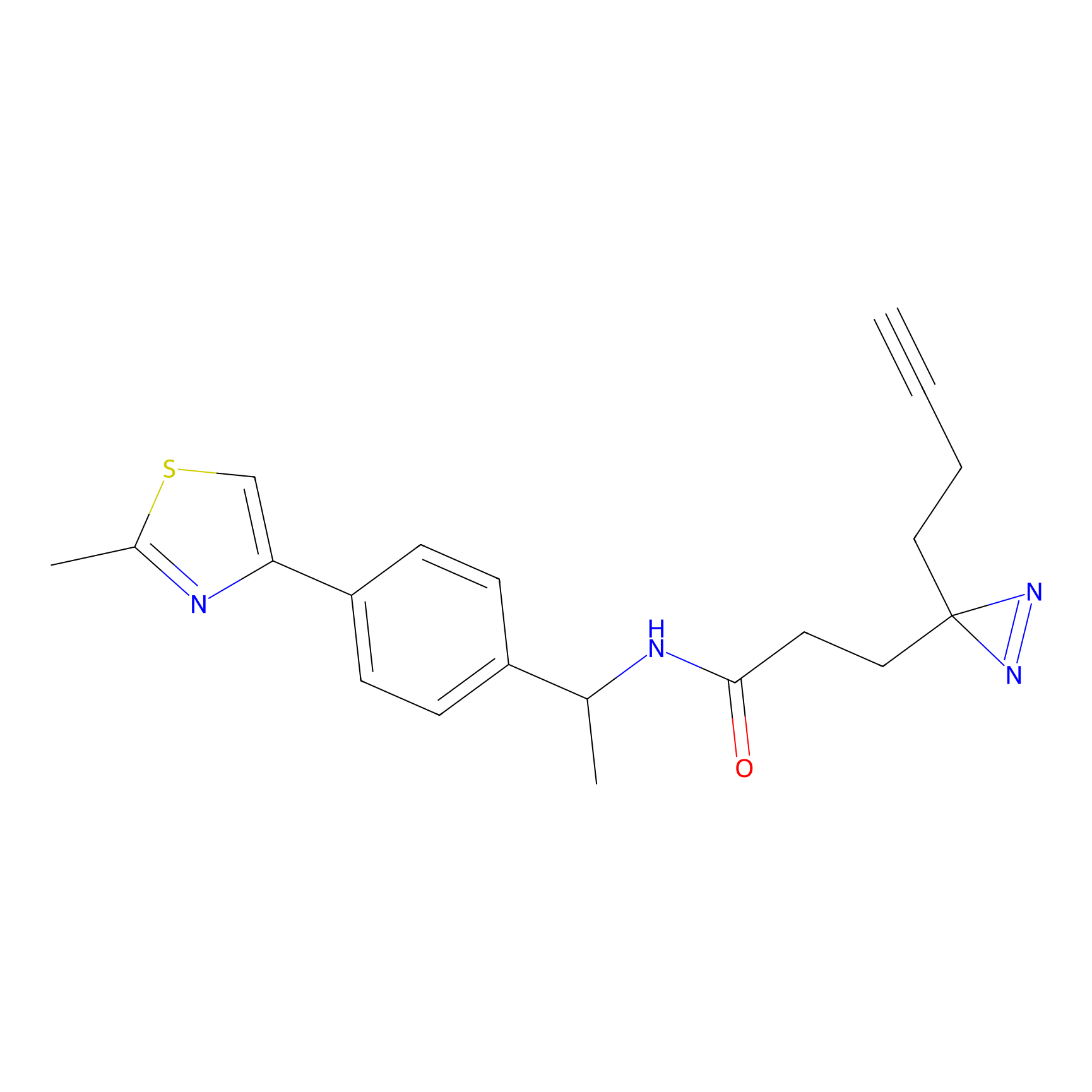
C29(1.40) | LDD1701 | [2] | |
|
Acrolein Probe Info |
 |
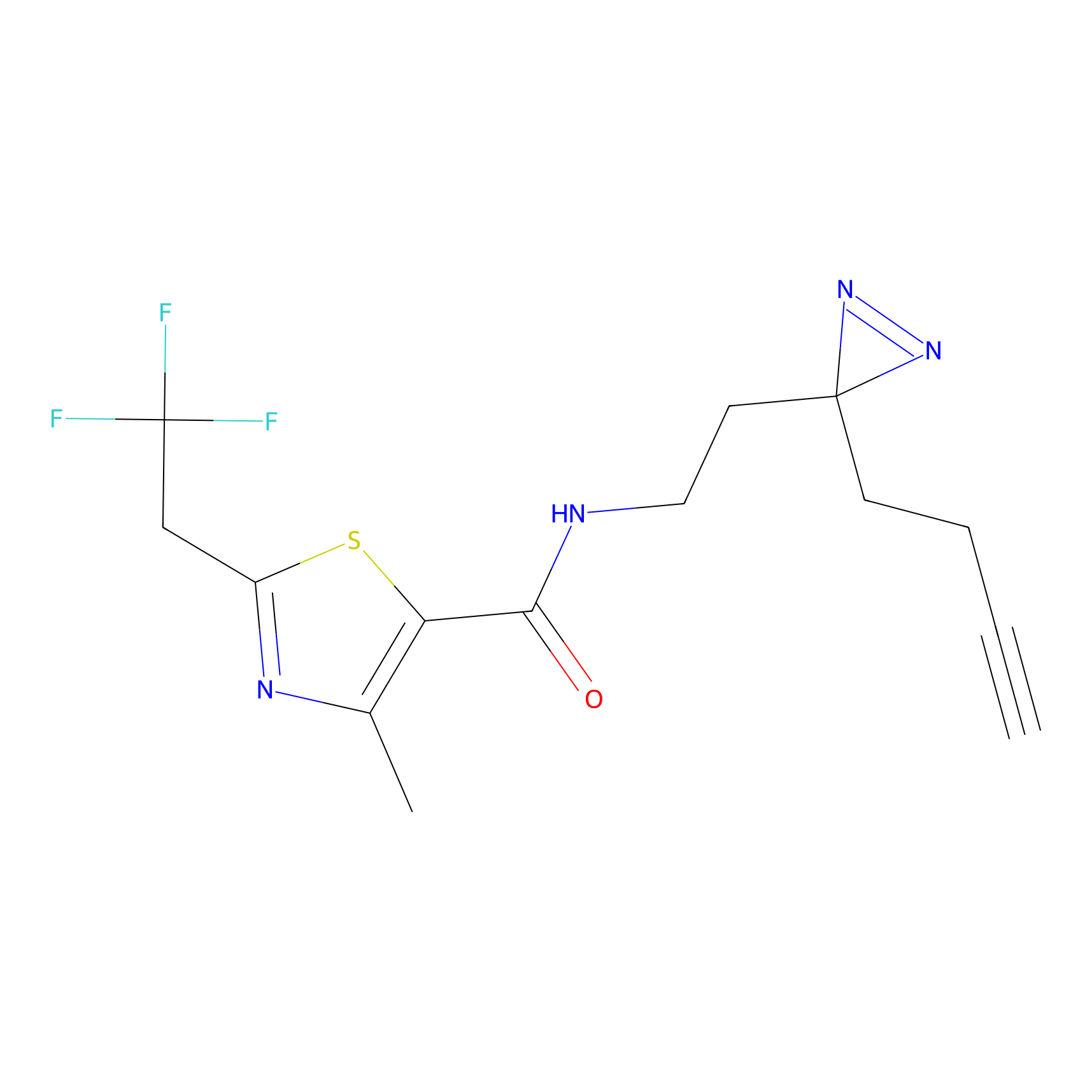
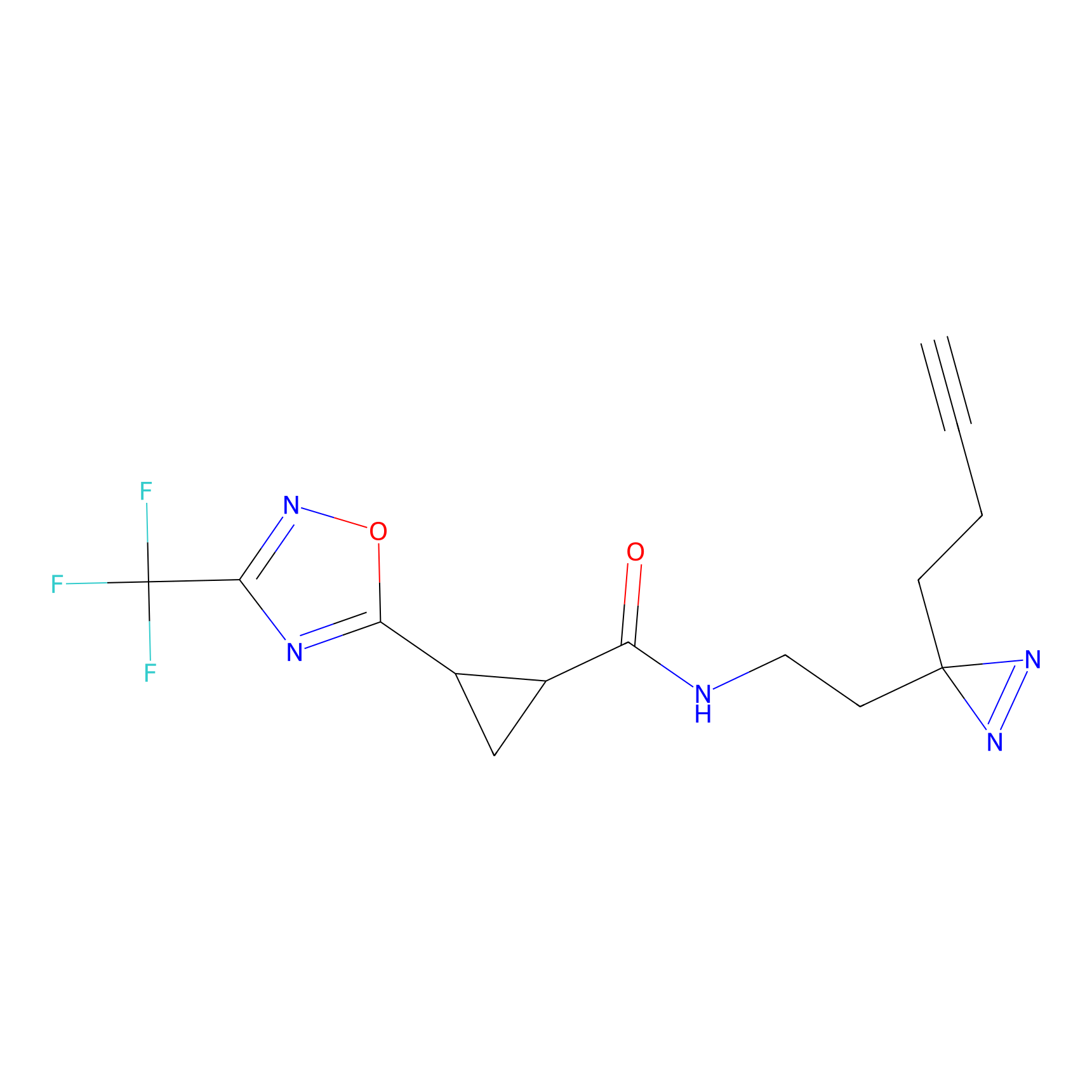
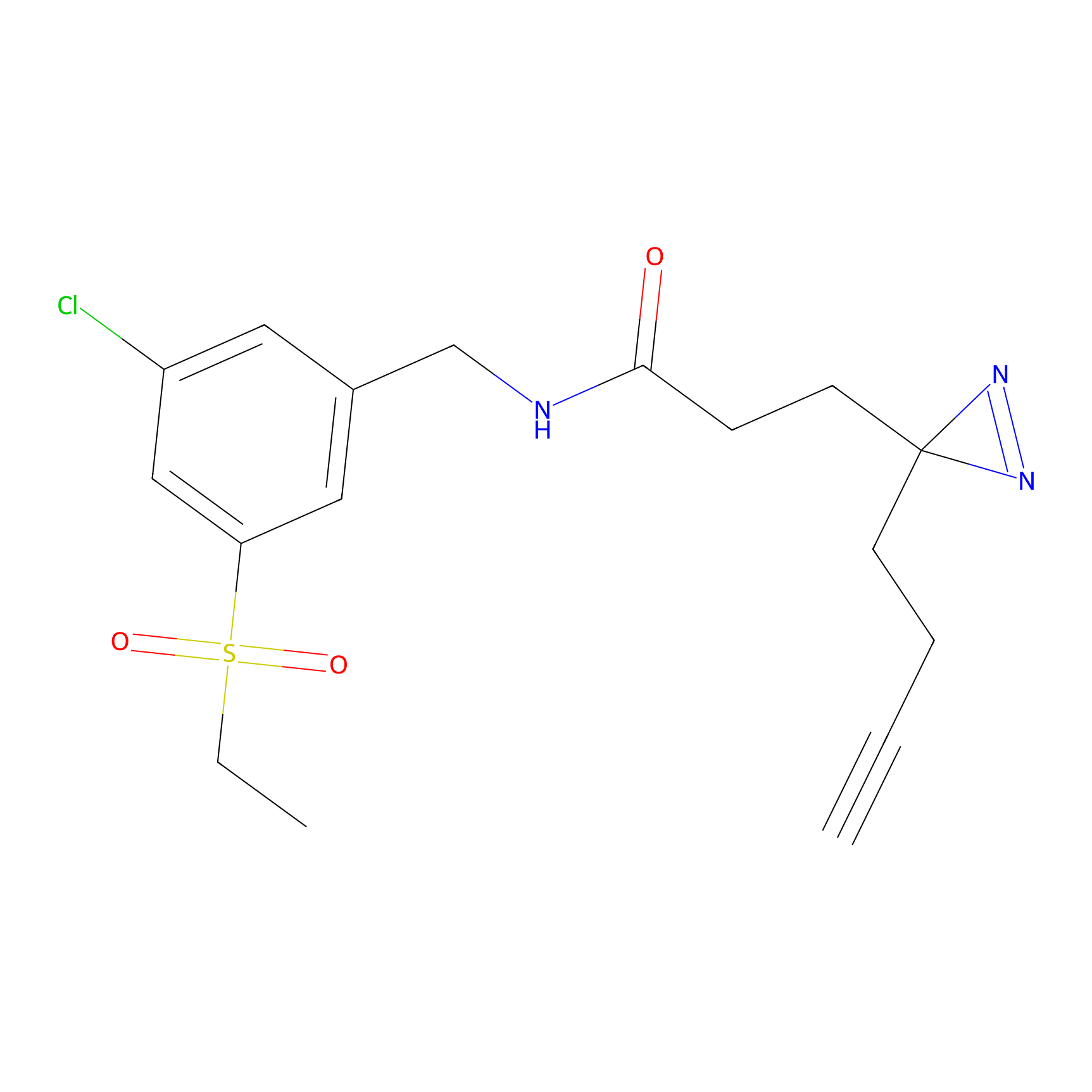
N.A. | LDD0222 | [3] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [4] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [5] | |
|
m-APA Probe Info |
 |
N.A. | LDD2231 | [4] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [6] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [6] | |
|
TFBX Probe Info |
 |
C33(0.00); C29(0.00) | LDD0148 | [7] | |
|
Crotonaldehyde Probe Info |
 |
H62(0.00); H30(0.00) | LDD0219 | [3] | |
|
Methacrolein Probe Info |
 |
C29(0.00); C50(0.00) | LDD0218 | [3] | |
|
NAIA_5 Probe Info |
 |
C33(0.00); C54(0.00); C50(0.00); C29(0.00) | LDD2223 | [8] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C54(0.39) | LDD2142 | [2] |
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C54(0.31) | LDD2112 | [2] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C29(0.45) | LDD2117 | [2] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C29(0.88) | LDD2152 | [2] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C29(0.85); C50(1.13); C54(0.56) | LDD2113 | [2] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [3] |
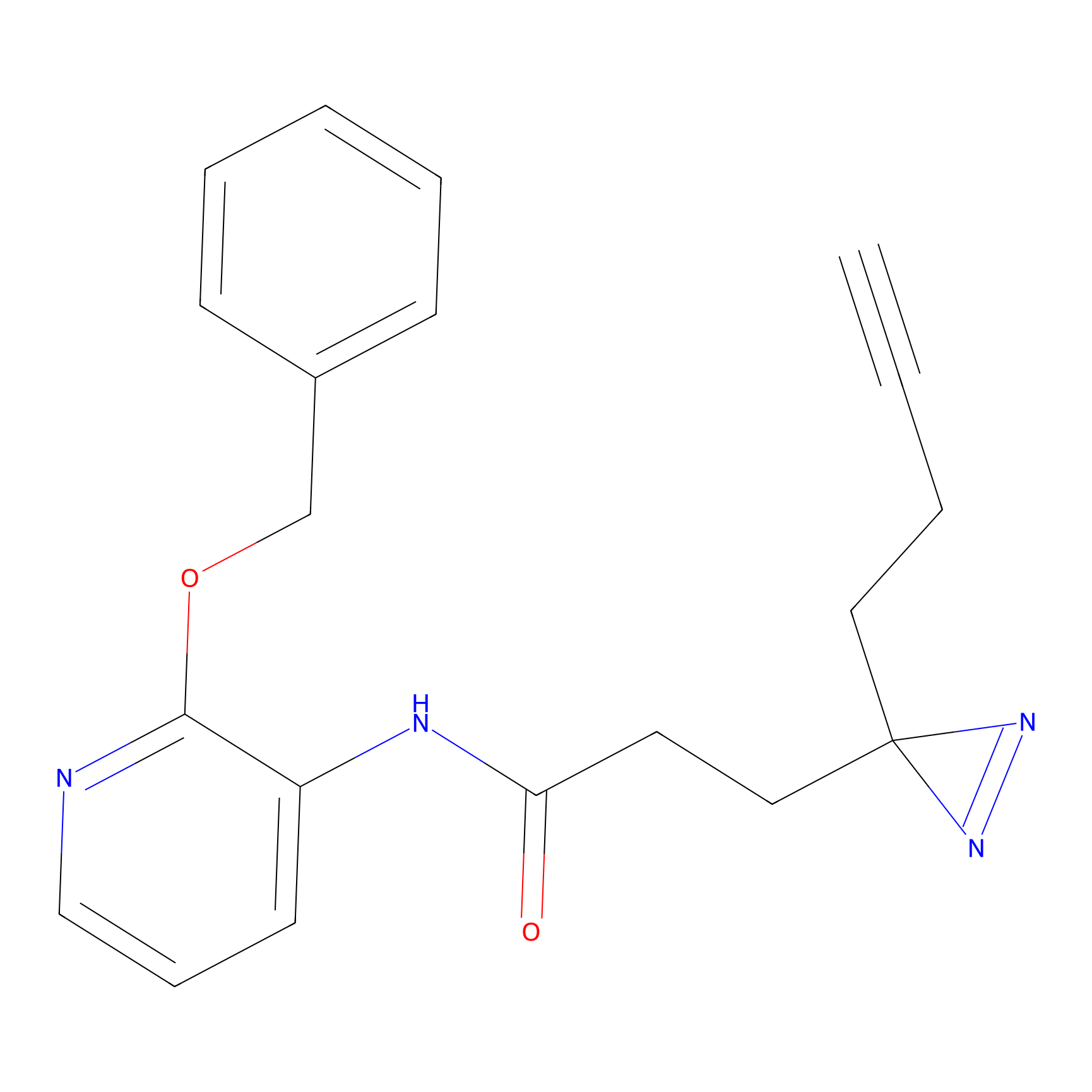
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C29(0.97) | LDD1702 | [2] |
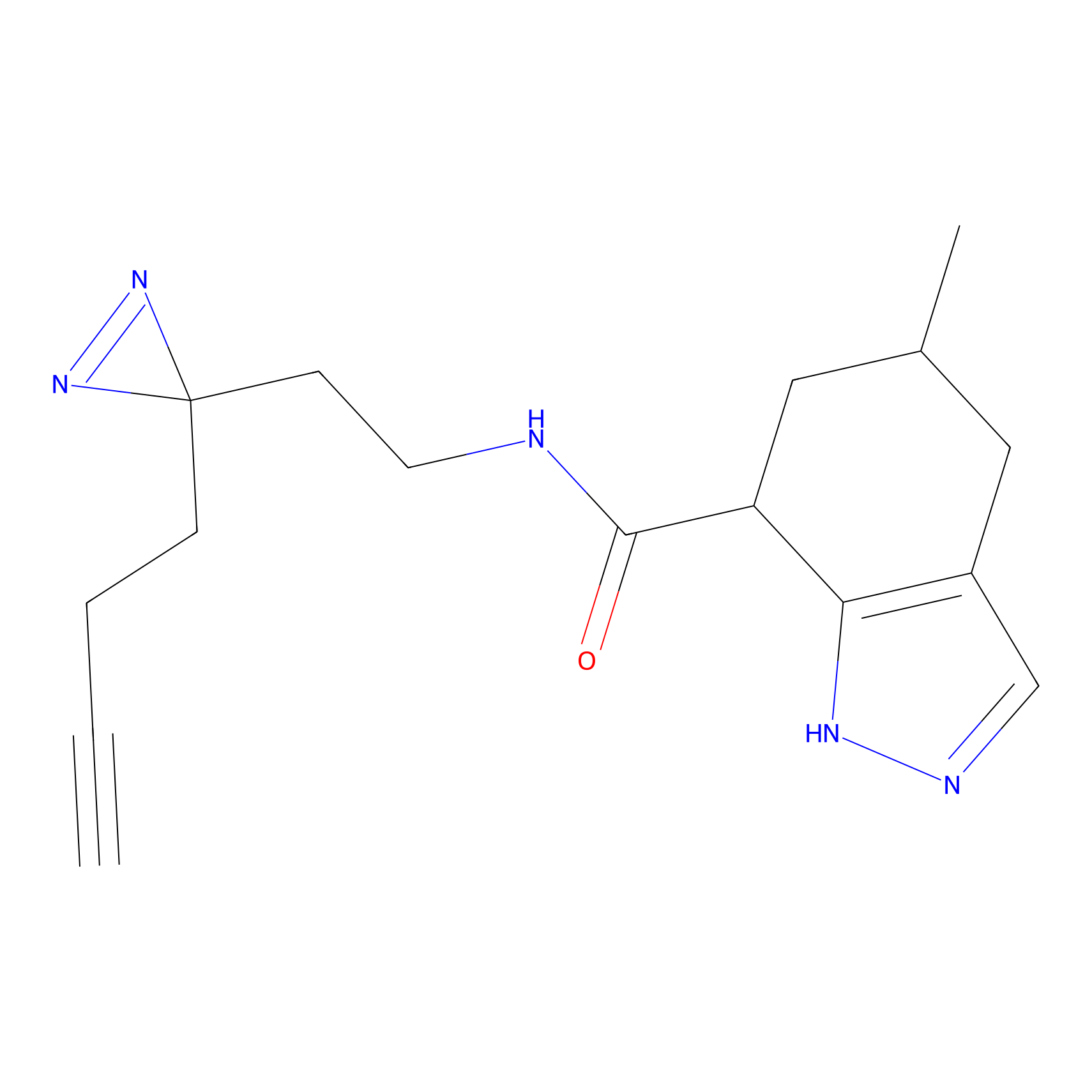
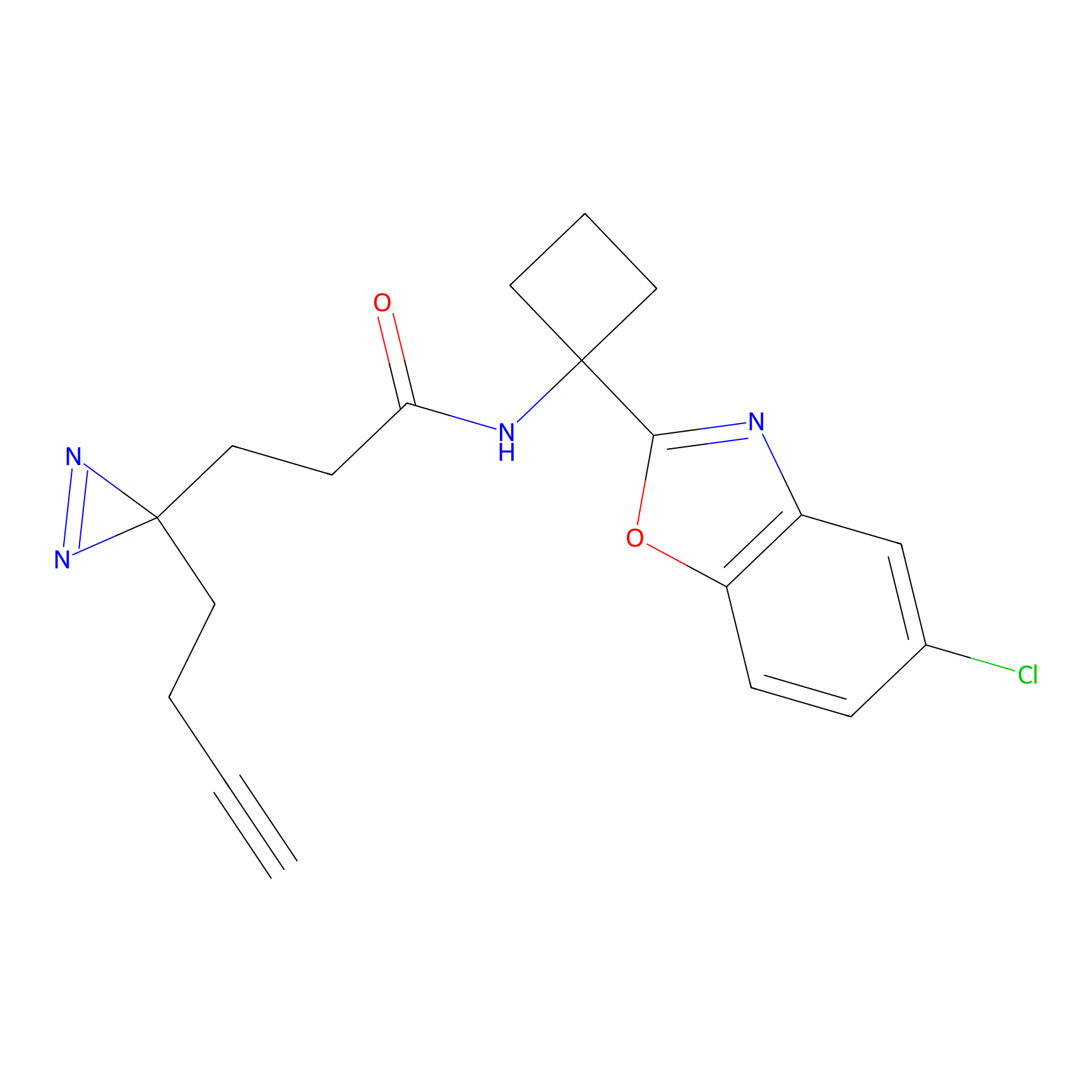
| LDCM0022 | KB02 | A101D | C50(1.17) | LDD2250 | [1] |
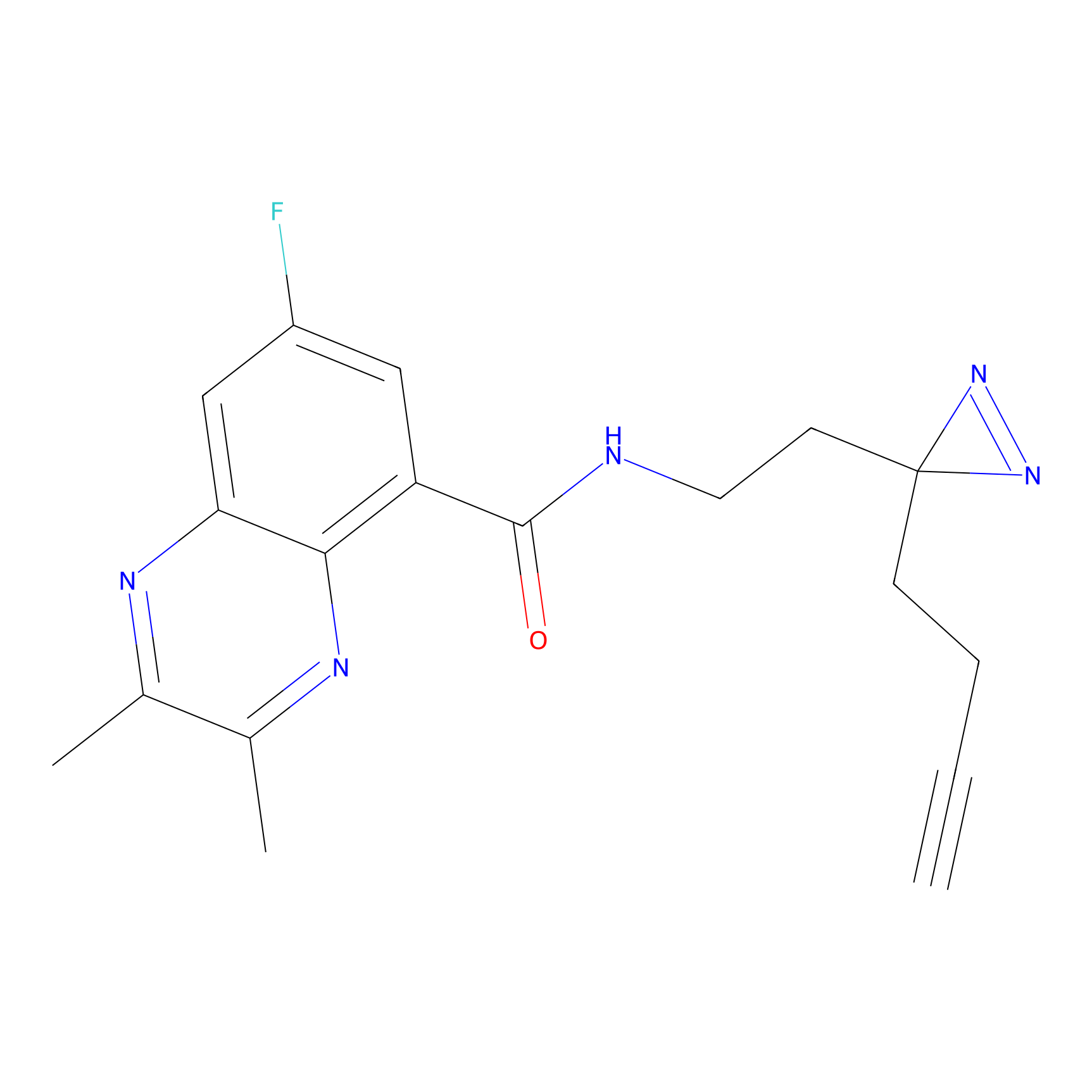
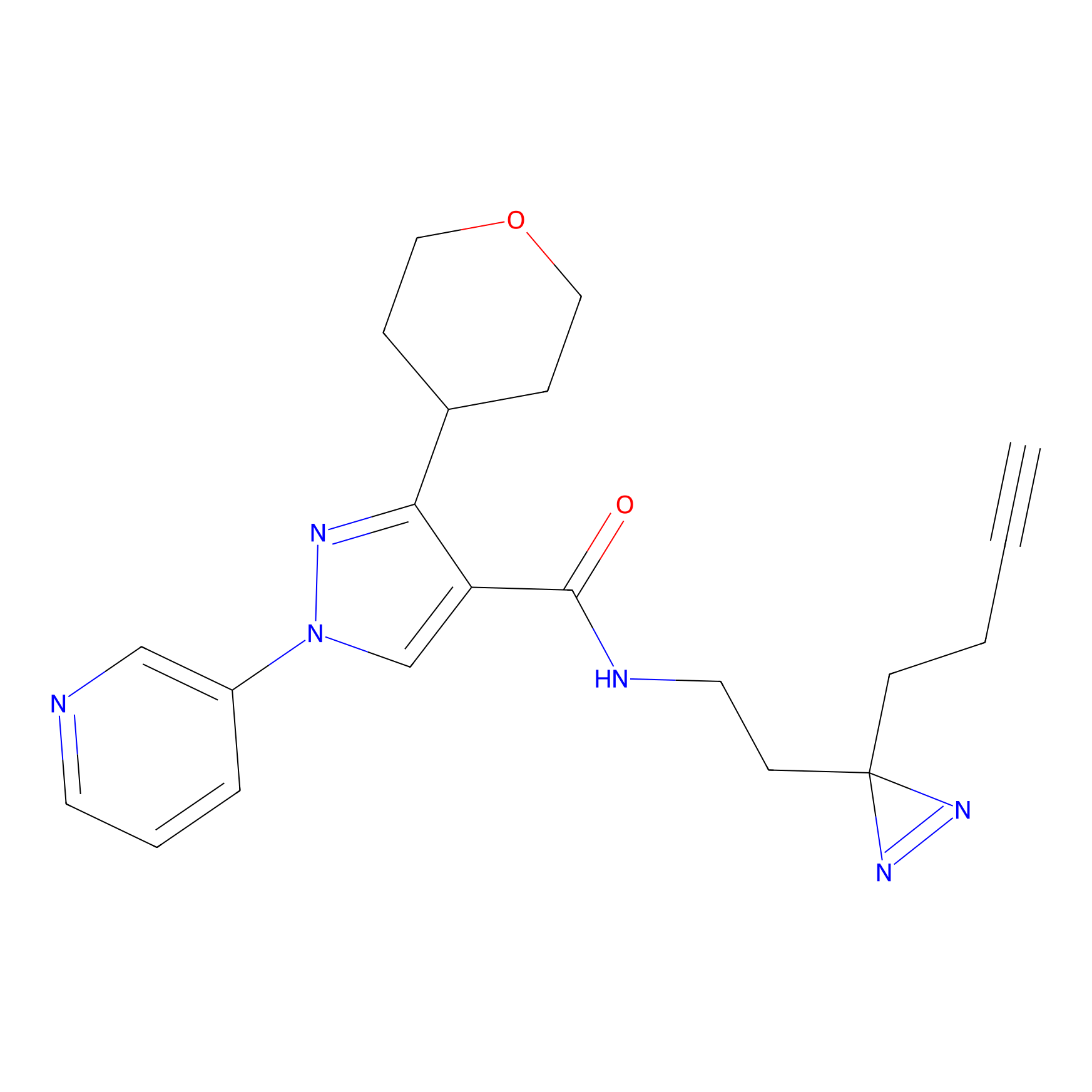
| LDCM0023 | KB03 | MDA-MB-231 | C29(1.40) | LDD1701 | [2] |
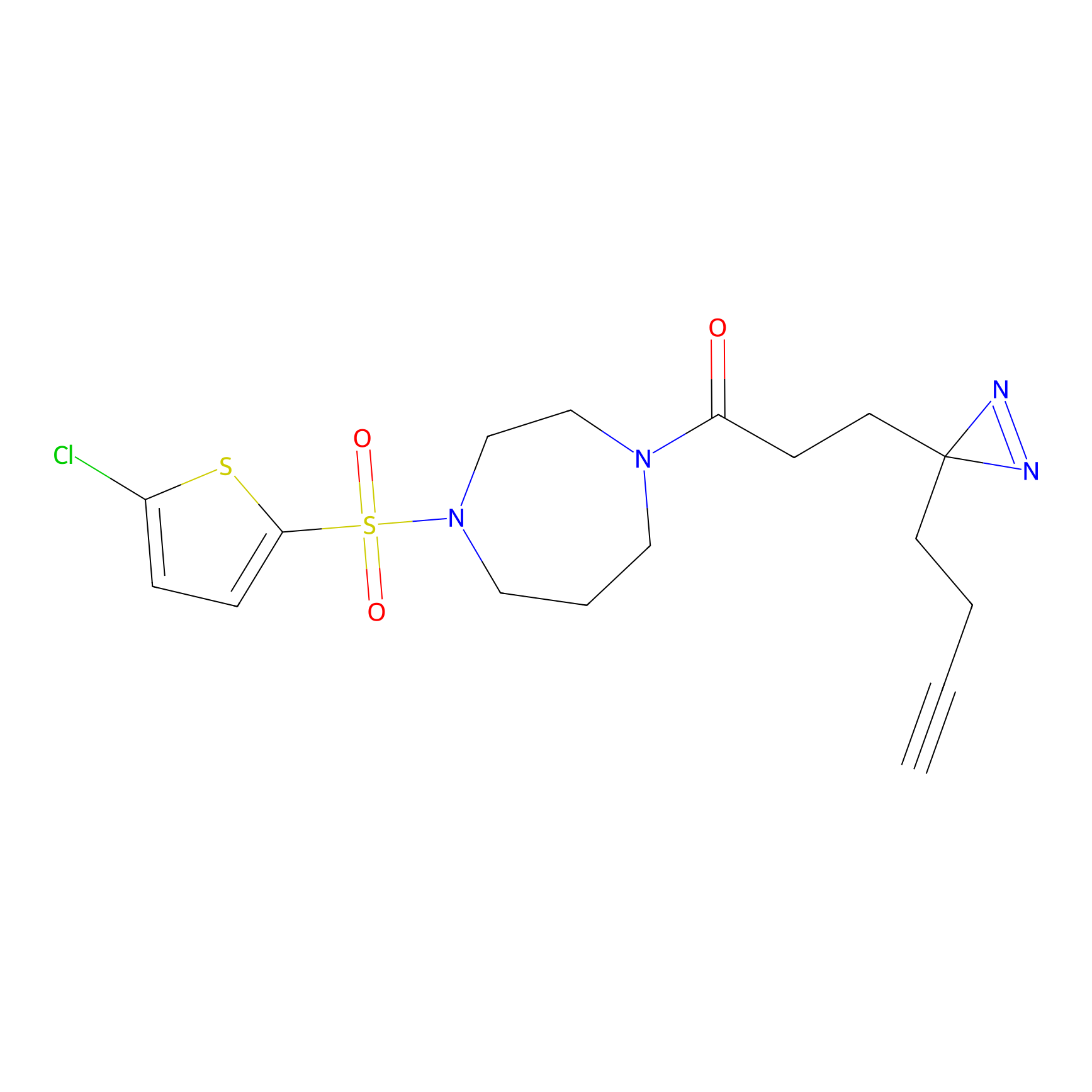
| LDCM0024 | KB05 | COLO792 | C50(2.68) | LDD3310 | [1] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0226 | [3] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C54(3.72) | LDD2092 | [2] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C29(0.72); C54(0.60) | LDD2097 | [2] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C54(0.15) | LDD2100 | [2] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C29(1.49); C54(1.04) | LDD2101 | [2] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C54(0.29) | LDD2104 | [2] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C54(0.34) | LDD2106 | [2] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C29(0.72) | LDD2107 | [2] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C54(0.34) | LDD2108 | [2] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C29(0.72) | LDD2109 | [2] |
| LDCM0517 | Nucleophilic fragment 21b | MDA-MB-231 | C54(0.30) | LDD2110 | [2] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C29(1.45) | LDD2111 | [2] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C29(0.52); C54(0.64) | LDD2115 | [2] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C29(0.36) | LDD2120 | [2] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C29(0.94) | LDD2125 | [2] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C29(0.41) | LDD2128 | [2] |
| LDCM0540 | Nucleophilic fragment 35 | MDA-MB-231 | C29(0.68); C54(0.57) | LDD2133 | [2] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C29(0.48); C50(0.91); C54(0.55) | LDD2134 | [2] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C29(0.79) | LDD2136 | [2] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C29(1.13) | LDD1700 | [2] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C29(0.82) | LDD2140 | [2] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C54(0.67) | LDD2141 | [2] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C29(0.36) | LDD2143 | [2] |
| LDCM0551 | Nucleophilic fragment 5b | MDA-MB-231 | C54(2.85) | LDD2145 | [2] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C29(0.96) | LDD2146 | [2] |
| LDCM0553 | Nucleophilic fragment 6b | MDA-MB-231 | C29(0.60) | LDD2147 | [2] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C29(0.56); C50(0.79); C54(0.39) | LDD2148 | [2] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Putative serine/threonine-protein kinase SIK1B (SIK1B) | CAMK Ser/Thr protein kinase family | A0A0B4J2F2 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Zinc finger protein 655 (ZNF655) | Krueppel C2H2-type zinc-finger protein family | Q8N720 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Mitochondrial import inner membrane translocase subunit Tim9 (TIMM9) | Small Tim family | Q9Y5J7 | |||
| TBC1 domain family member 21 (TBC1D21) | . | Q8IYX1 | |||
References