Details of the Target
General Information of Target
| Target ID | LDTP09949 | |||||
|---|---|---|---|---|---|---|
| Target Name | Transportin-1 (TNPO1) | |||||
| Gene Name | TNPO1 | |||||
| Gene ID | 3842 | |||||
| Synonyms |
KPNB2; MIP1; TRN; Transportin-1; Importin beta-2; Karyopherin beta-2; M9 region interaction protein; MIP |
|||||
| 3D Structure | ||||||
| Sequence |
MASQPPPPPKPWETRRIPGAGPGPGPGPTFQSADLGPTLMTRPGQPALTRVPPPILPRPS
QQTGSSSVNTFRPAYSSFSSGYGAYGNSFYGGYSPYSYGYNGLGYNRLRVDDLPPSRFVQ QAEESSRGAFQSIESIVHAFASVSMMMDATFSAVYNSFRAVLDVANHFSRLKIHFTKVFS AFALVRTIRYLYRRLQRMLGLRRGSENEDLWAESEGTVACLGAEDRAATSAKSWPIFLFF AVILGGPYLIWKLLSTHSDEVTDSINWASGEDDHVVARAEYDFAAVSEEEISFRAGDMLN LALKEQQPKVRGWLLASLDGQTTGLIPANYVKILGKRKGRKTVESSKVSKQQQSFTNPTL TKGATVADSLDEQEAAFESVFVETNKVPVAPDSIGKDGEKQDL |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
Importin beta family, Importin beta-2 subfamily
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Functions in nuclear protein import as nuclear transport receptor. Serves as receptor for nuclear localization signals (NLS) in cargo substrates. May mediate docking of the importin/substrate complex to the nuclear pore complex (NPC) through binding to nucleoporin and the complex is subsequently translocated through the pore by an energy requiring, Ran-dependent mechanism. At the nucleoplasmic side of the NPC, Ran binds to the importin, the importin/substrate complex dissociates and importin is re-exported from the nucleus to the cytoplasm where GTP hydrolysis releases Ran. The directionality of nuclear import is thought to be conferred by an asymmetric distribution of the GTP- and GDP-bound forms of Ran between the cytoplasm and nucleus. Involved in nuclear import of M9-containing proteins. In vitro, binds directly to the M9 region of the heterogeneous nuclear ribonucleoproteins (hnRNP), A1 and A2 and mediates their nuclear import. Involved in hnRNP A1/A2 nuclear export. Mediates the nuclear import of ribosomal proteins RPL23A, RPS7 and RPL5. In vitro, mediates nuclear import of H2A, H2B, H3 and H4 histones. In vitro, mediates nuclear import of SRP19. Mediates nuclear import of ADAR/ADAR1 isoform 1 and isoform 5 in a RanGTP-dependent manner.; (Microbial infection) In case of HIV-1 infection, binds and mediates the nuclear import of HIV-1 Rev.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| COLO678 | SNV: p.L809V | DBIA Probe Info | |||
| COLO829 | SNV: p.Q38H | DBIA Probe Info | |||
| HCC70 | SNV: p.L679V | DBIA Probe Info | |||
| HEC1 | SNV: p.C597Y | DBIA Probe Info | |||
| HEC1B | SNV: p.C597Y | . | |||
| HT | SNV: p.T514A | . | |||
| J82 | SNV: p.W805Ter | DBIA Probe Info | |||
| JURKAT | Deletion: p.C844VfsTer45 | Compound 10 Probe Info | |||
| OVCAR8 | SNV: p.R351S | DBIA Probe Info | |||
| SKMEL2 | SNV: p.P196S | DBIA Probe Info | |||
| SNU1196 | Deletion: p.T555FfsTer29 | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
11.30 | LDD0402 | [1] | |
|
A-EBA Probe Info |
 |
2.74 | LDD0215 | [2] | |
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [3] | |
|
N1 Probe Info |
 |
100.00 | LDD0242 | [3] | |
|
TH211 Probe Info |
 |
Y51(11.03) | LDD0257 | [4] | |
|
TH216 Probe Info |
 |
Y51(20.00) | LDD0259 | [4] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [5] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [5] | |
|
BTD Probe Info |
 |
C806(5.07) | LDD1699 | [6] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [7] | |
|
OPA-S-S-alkyne Probe Info |
 |
K889(1.10) | LDD3494 | [8] | |
|
JZ128-DTB Probe Info |
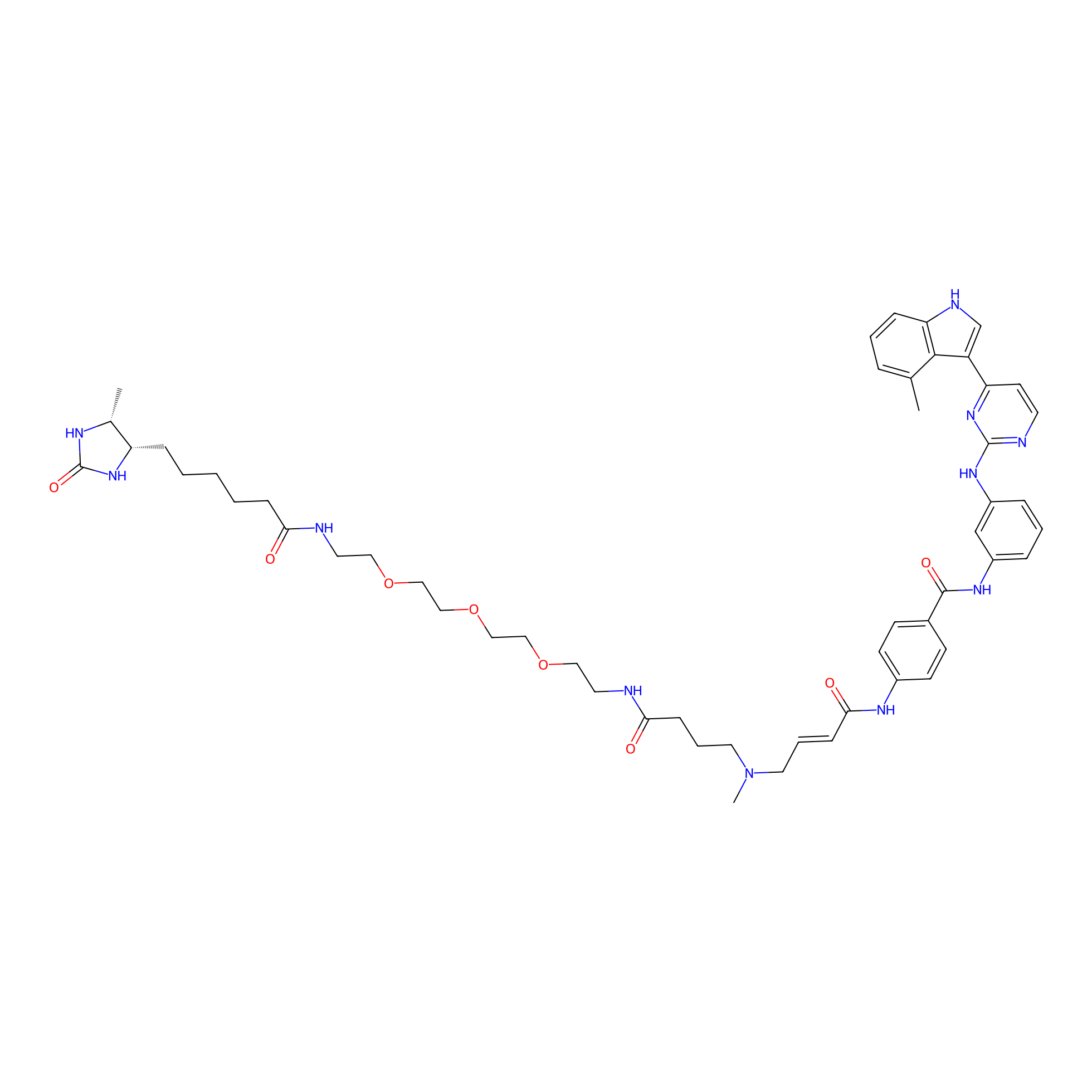 |
C285(0.00); C790(0.00); C142(0.00); C103(0.00) | LDD0462 | [9] | |
|
THZ1-DTB Probe Info |
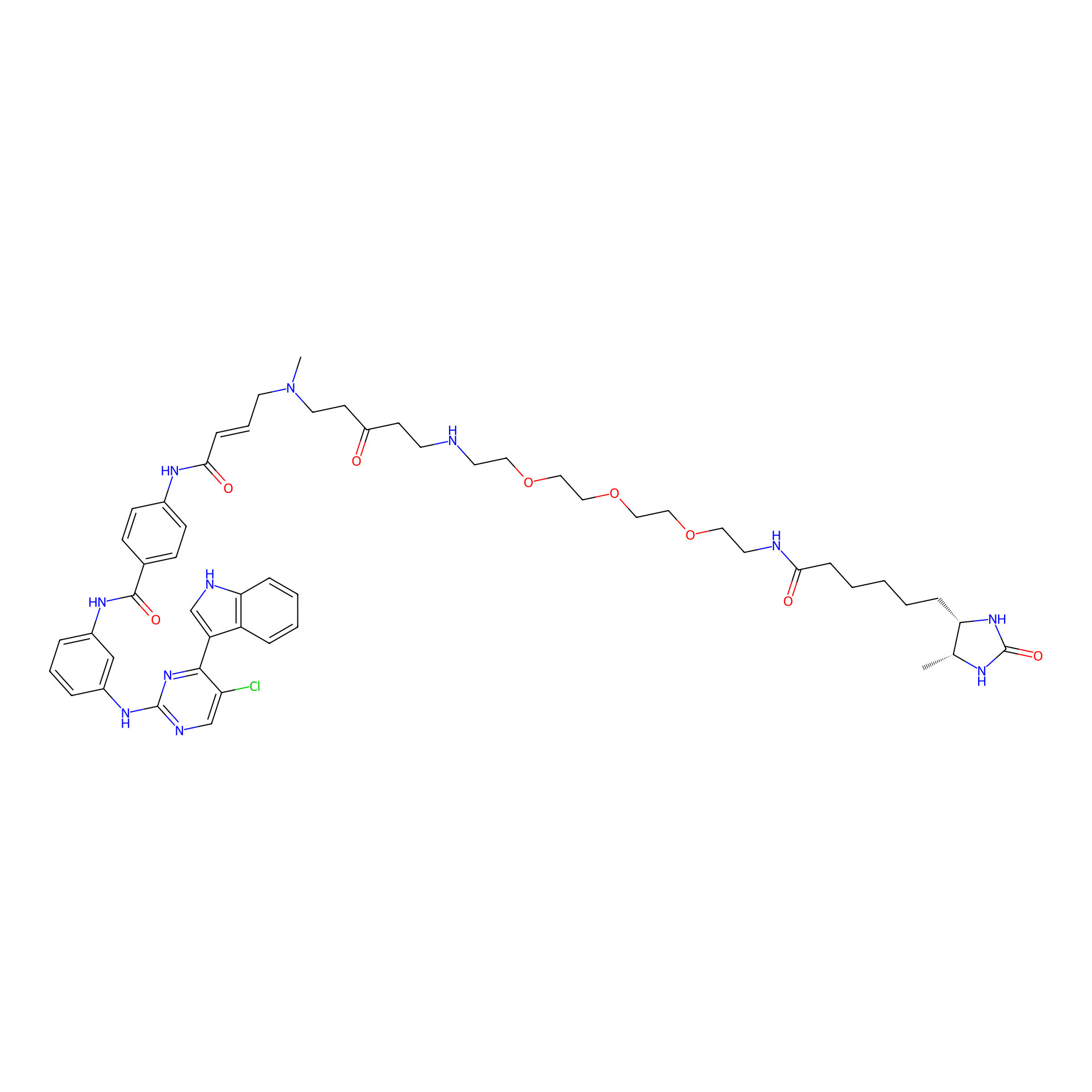 |
C103(1.10); C285(1.08); C790(1.06); C142(1.02) | LDD0460 | [9] | |
|
AZ-9 Probe Info |
 |
10.00 | LDD2154 | [10] | |
|
AHL-Pu-1 Probe Info |
 |
C164(3.13) | LDD0169 | [11] | |
|
EA-probe Probe Info |
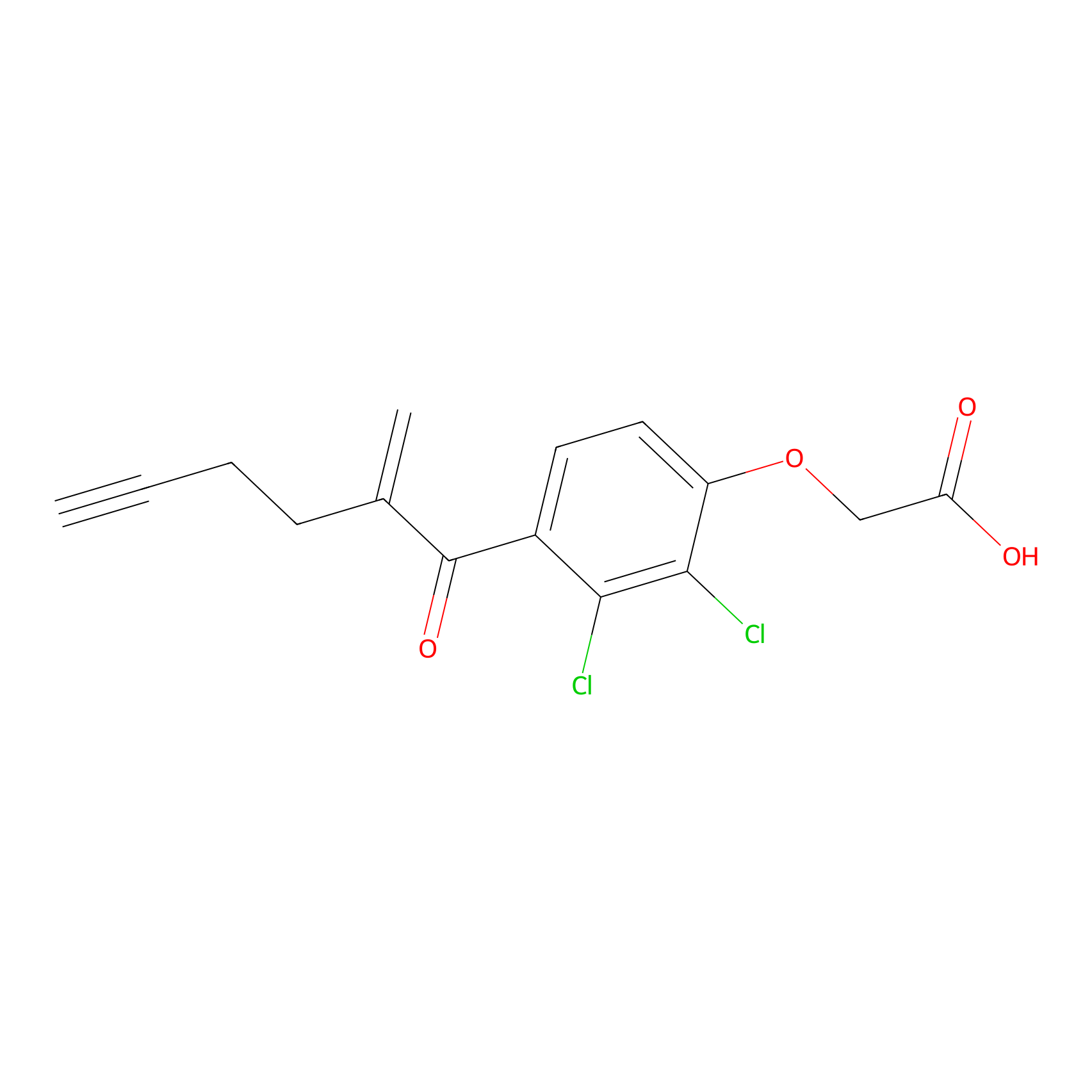 |
N.A. | LDD0440 | [12] | |
|
HHS-475 Probe Info |
 |
Y51(0.74); Y788(0.75) | LDD0264 | [13] | |
|
DBIA Probe Info |
 |
C467(1.53); C205(0.89) | LDD0078 | [14] | |
|
ATP probe Probe Info |
 |
K85(0.00); K869(0.00); K889(0.00) | LDD0199 | [15] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C386(0.00); C620(0.00); C164(0.00); C806(0.00) | LDD0038 | [16] | |
|
IA-alkyne Probe Info |
 |
C142(0.00); C103(0.00); C683(0.00) | LDD0032 | [17] | |
|
IPIAA_L Probe Info |
 |
C164(0.00); C103(0.00) | LDD0031 | [18] | |
|
Lodoacetamide azide Probe Info |
 |
C386(0.00); C620(0.00); C164(0.00); C806(0.00) | LDD0037 | [16] | |
|
JW-RF-010 Probe Info |
 |
C103(0.00); C164(0.00); C205(0.00) | LDD0026 | [19] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [20] | |
|
TFBX Probe Info |
 |
C467(0.00); C205(0.00); C620(0.00); C103(0.00) | LDD0027 | [19] | |
|
WYneN Probe Info |
 |
C164(0.00); C620(0.00); C386(0.00) | LDD0021 | [21] | |
|
WYneO Probe Info |
 |
C620(0.00); C103(0.00) | LDD0022 | [21] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0356 | [22] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [23] | |
|
ENE Probe Info |
 |
N.A. | LDD0006 | [21] | |
|
IPM Probe Info |
 |
C386(0.00); C683(0.00); C142(0.00); C205(0.00) | LDD0005 | [21] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [21] | |
|
PPMS Probe Info |
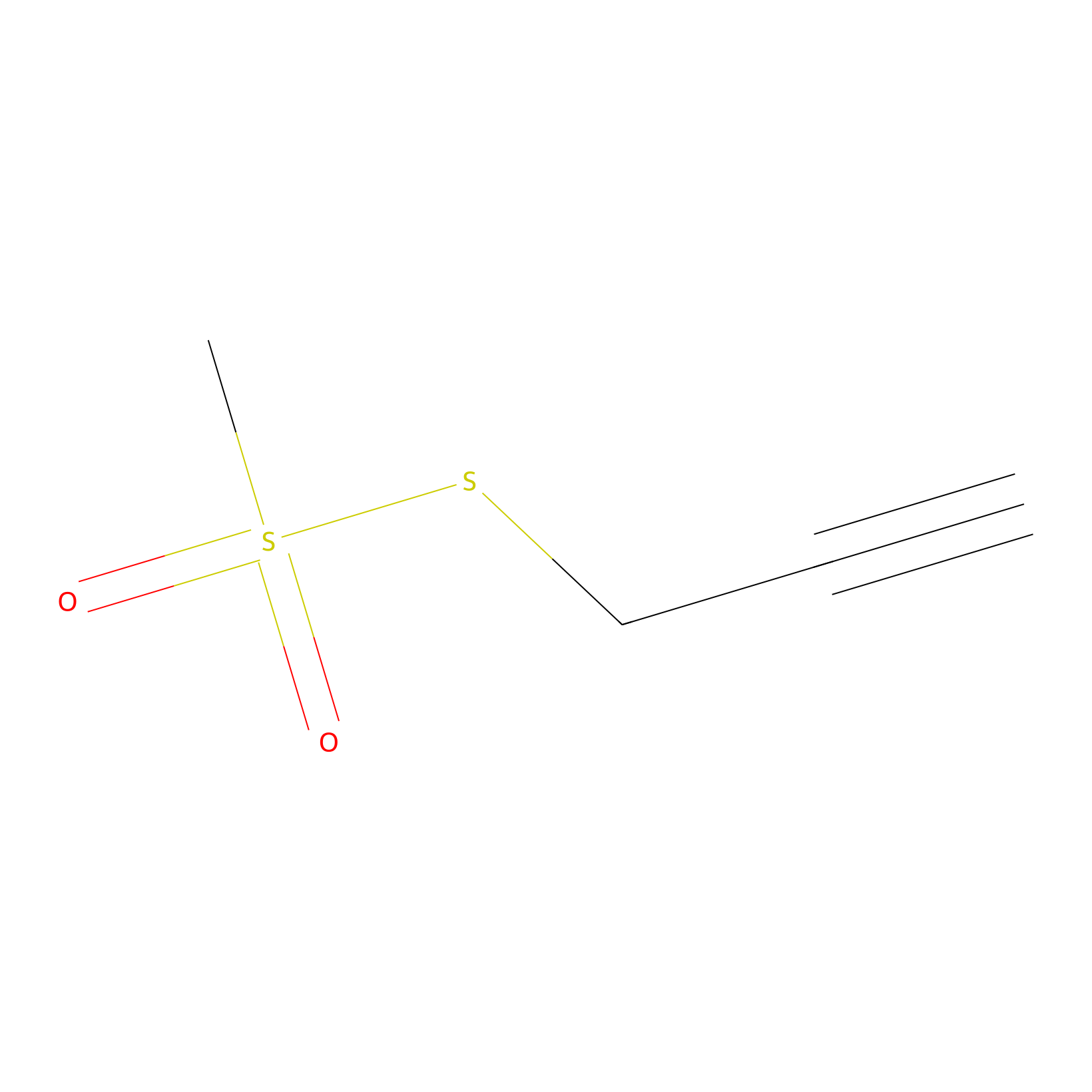 |
N.A. | LDD0008 | [21] | |
|
STPyne Probe Info |
 |
K889(0.00); K81(0.00); K869(0.00) | LDD0009 | [21] | |
|
VSF Probe Info |
 |
C164(0.00); C153(0.00); C103(0.00); C142(0.00) | LDD0007 | [21] | |
|
Phosphinate-6 Probe Info |
 |
C862(0.00); C790(0.00); C103(0.00) | LDD0018 | [24] | |
|
Ox-W18 Probe Info |
 |
W134(0.00); W878(0.00) | LDD2175 | [25] | |
|
1c-yne Probe Info |
 |
K487(0.00); K687(0.00) | LDD0228 | [22] | |
|
Acrolein Probe Info |
 |
C103(0.00); C164(0.00); C153(0.00); C142(0.00) | LDD0217 | [26] | |
|
Cinnamaldehyde Probe Info |
 |
N.A. | LDD0220 | [26] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [26] | |
|
Methacrolein Probe Info |
 |
C103(0.00); C142(0.00); C153(0.00) | LDD0218 | [26] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [27] | |
|
AOyne Probe Info |
 |
11.00 | LDD0443 | [28] | |
|
NAIA_5 Probe Info |
 |
C386(0.00); C862(0.00); C164(0.00); C205(0.00) | LDD2223 | [20] | |
|
HHS-465 Probe Info |
 |
Y51(0.00); Y788(0.00); K64(0.00) | LDD2240 | [29] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C004 Probe Info |
 |
6.41 | LDD1714 | [30] | |
|
C017 Probe Info |
 |
7.01 | LDD1725 | [30] | |
|
C106 Probe Info |
 |
23.26 | LDD1793 | [30] | |
|
C108 Probe Info |
 |
9.06 | LDD1795 | [30] | |
|
C140 Probe Info |
 |
7.78 | LDD1822 | [30] | |
|
C161 Probe Info |
 |
11.00 | LDD1841 | [30] | |
|
C163 Probe Info |
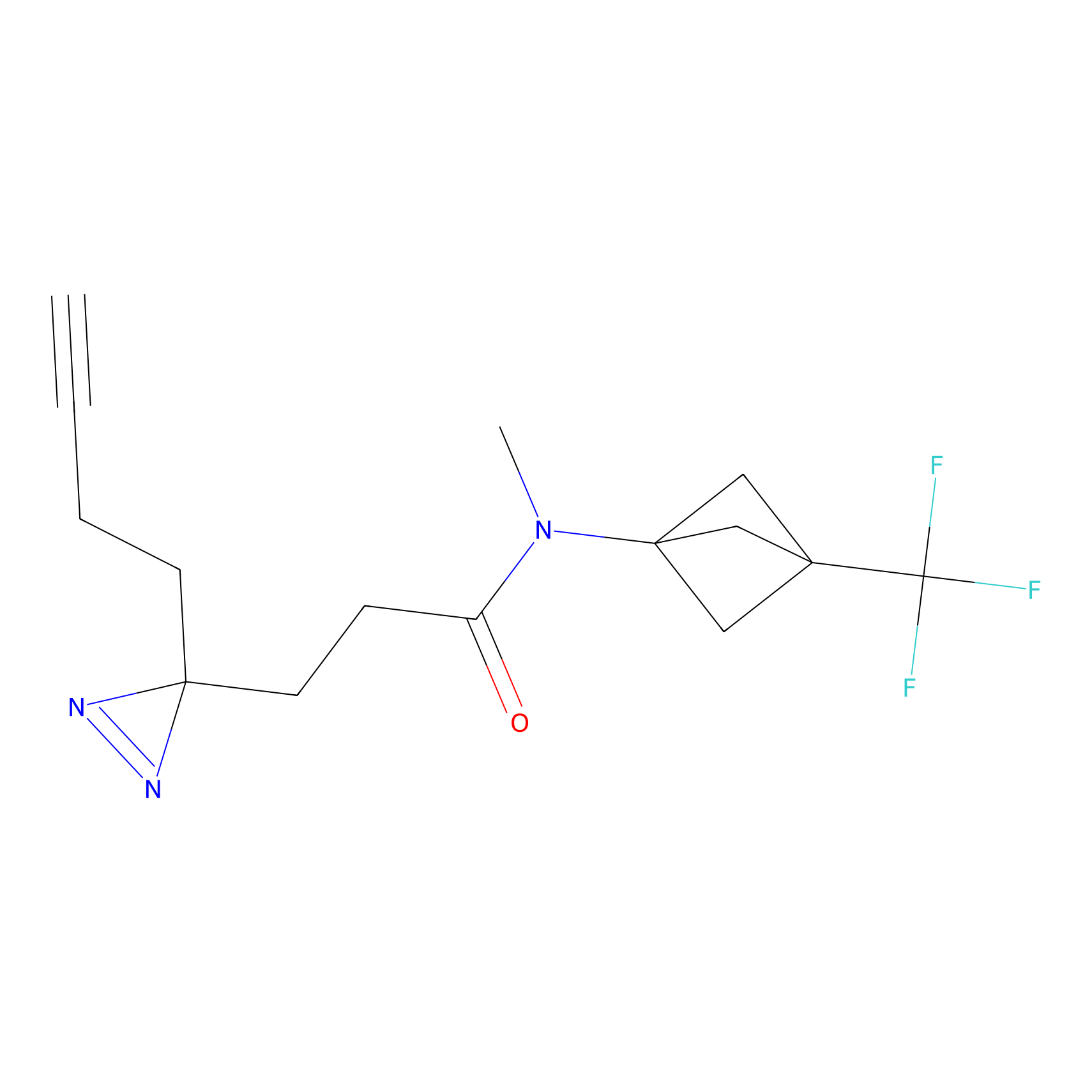 |
8.75 | LDD1843 | [30] | |
|
C178 Probe Info |
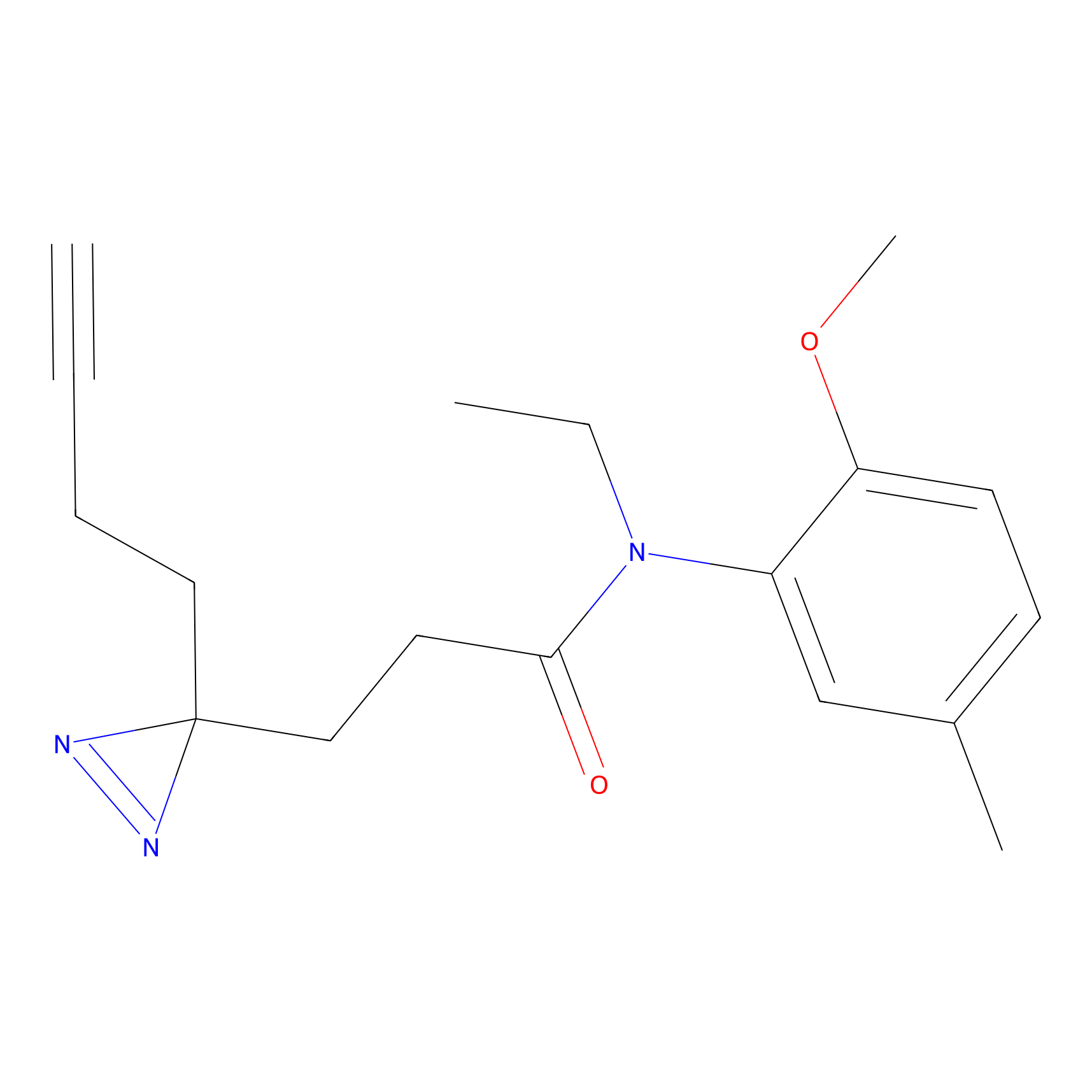 |
15.24 | LDD1857 | [30] | |
|
C191 Probe Info |
 |
23.43 | LDD1868 | [30] | |
|
C220 Probe Info |
 |
17.51 | LDD1894 | [30] | |
|
C228 Probe Info |
 |
15.45 | LDD1901 | [30] | |
|
C277 Probe Info |
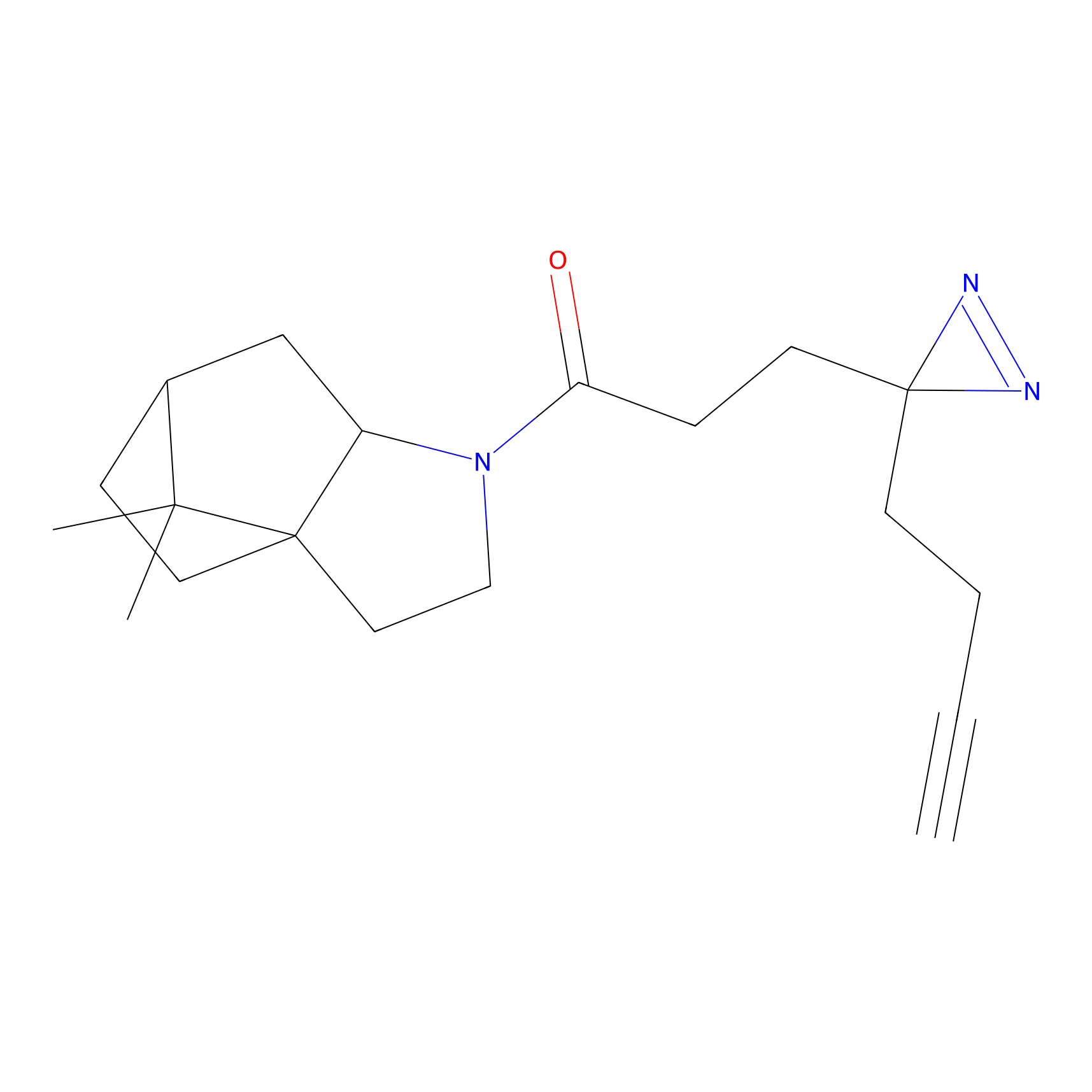 |
14.83 | LDD1947 | [30] | |
|
C305 Probe Info |
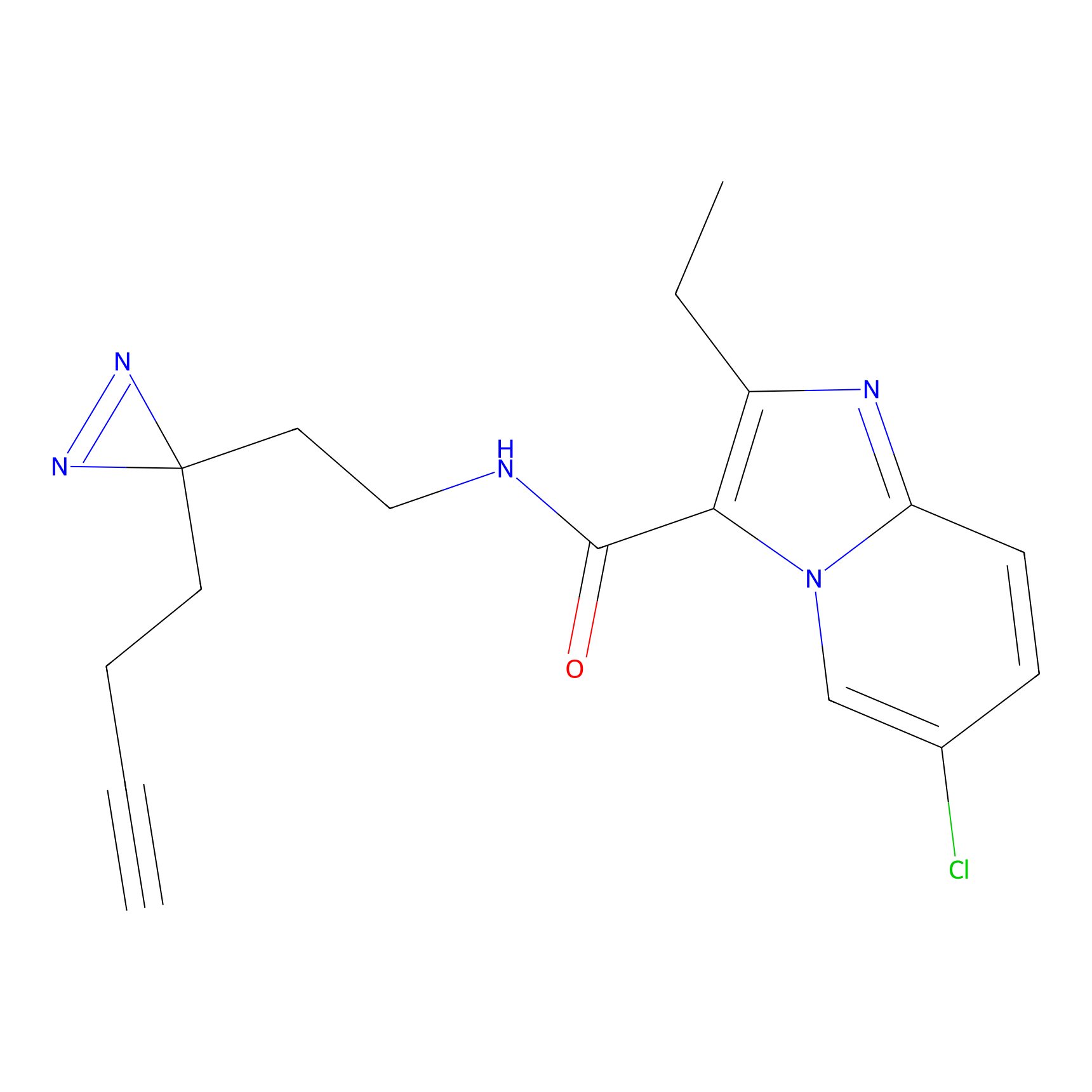 |
17.75 | LDD1974 | [30] | |
|
C310 Probe Info |
 |
8.57 | LDD1977 | [30] | |
|
C326 Probe Info |
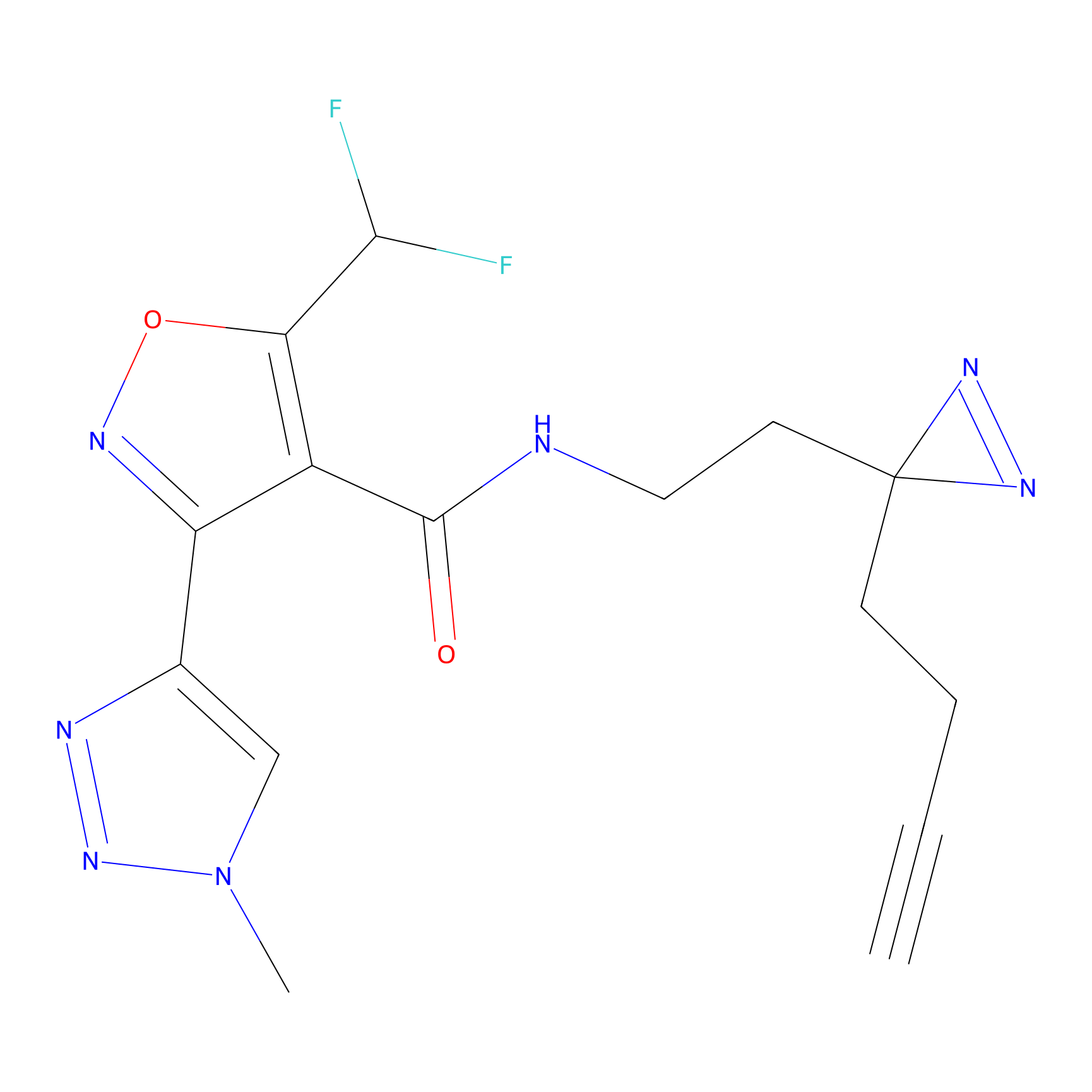 |
8.28 | LDD1990 | [30] | |
|
C337 Probe Info |
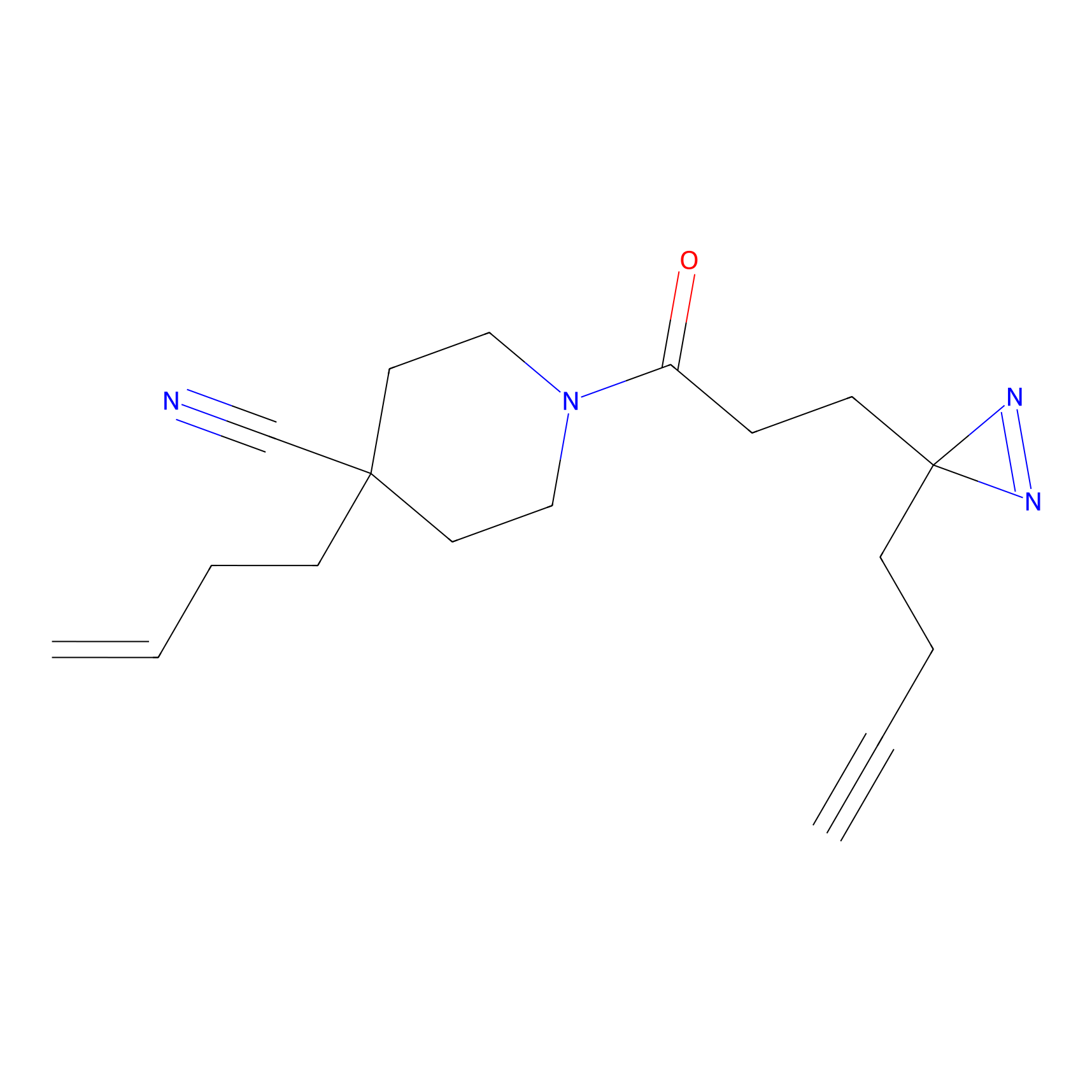 |
5.24 | LDD2000 | [30] | |
|
C343 Probe Info |
 |
14.93 | LDD2005 | [30] | |
|
C376 Probe Info |
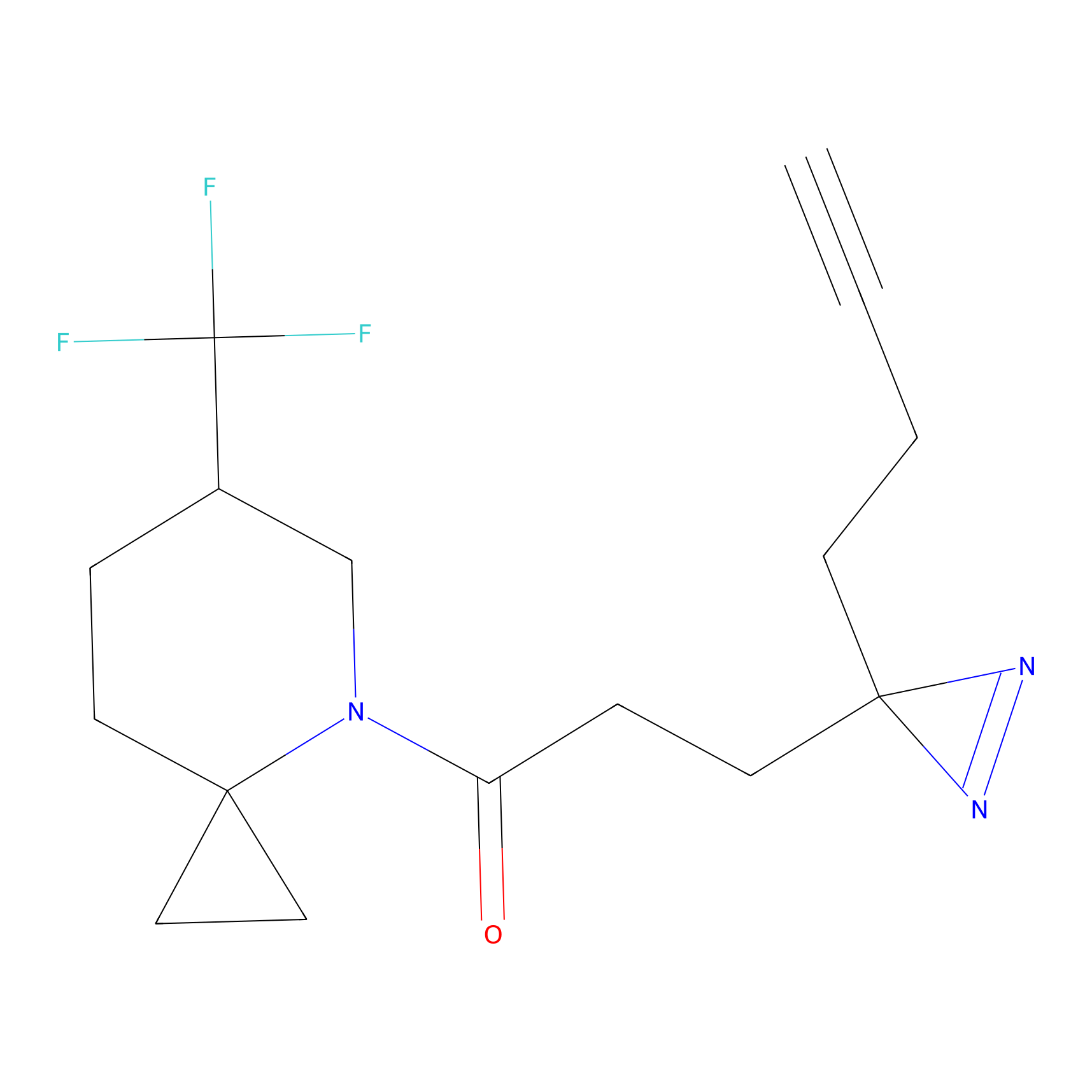 |
8.17 | LDD2036 | [30] | |
|
FFF probe11 Probe Info |
 |
5.84 | LDD0471 | [31] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [31] | |
|
FFF probe14 Probe Info |
 |
20.00 | LDD0477 | [31] | |
|
FFF probe15 Probe Info |
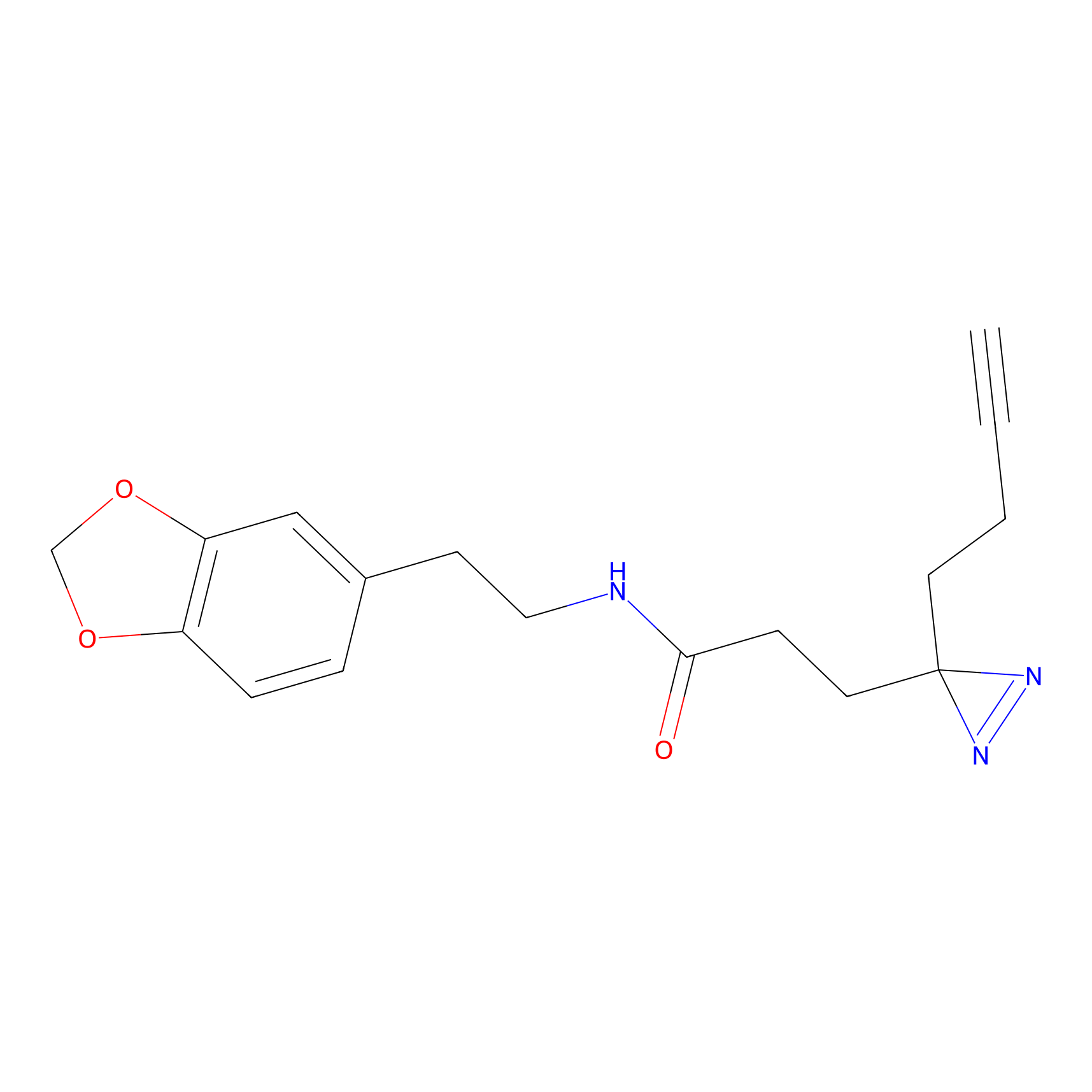 |
12.50 | LDD0490 | [31] | |
|
FFF probe2 Probe Info |
 |
10.06 | LDD0463 | [31] | |
|
FFF probe3 Probe Info |
 |
7.33 | LDD0464 | [31] | |
|
JN0003 Probe Info |
 |
13.29 | LDD0469 | [31] | |
|
STS-1 Probe Info |
 |
7.33 | LDD0136 | [32] | |
|
STS-2 Probe Info |
 |
13.33 | LDD0138 | [32] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [33] | |
|
BD-F Probe Info |
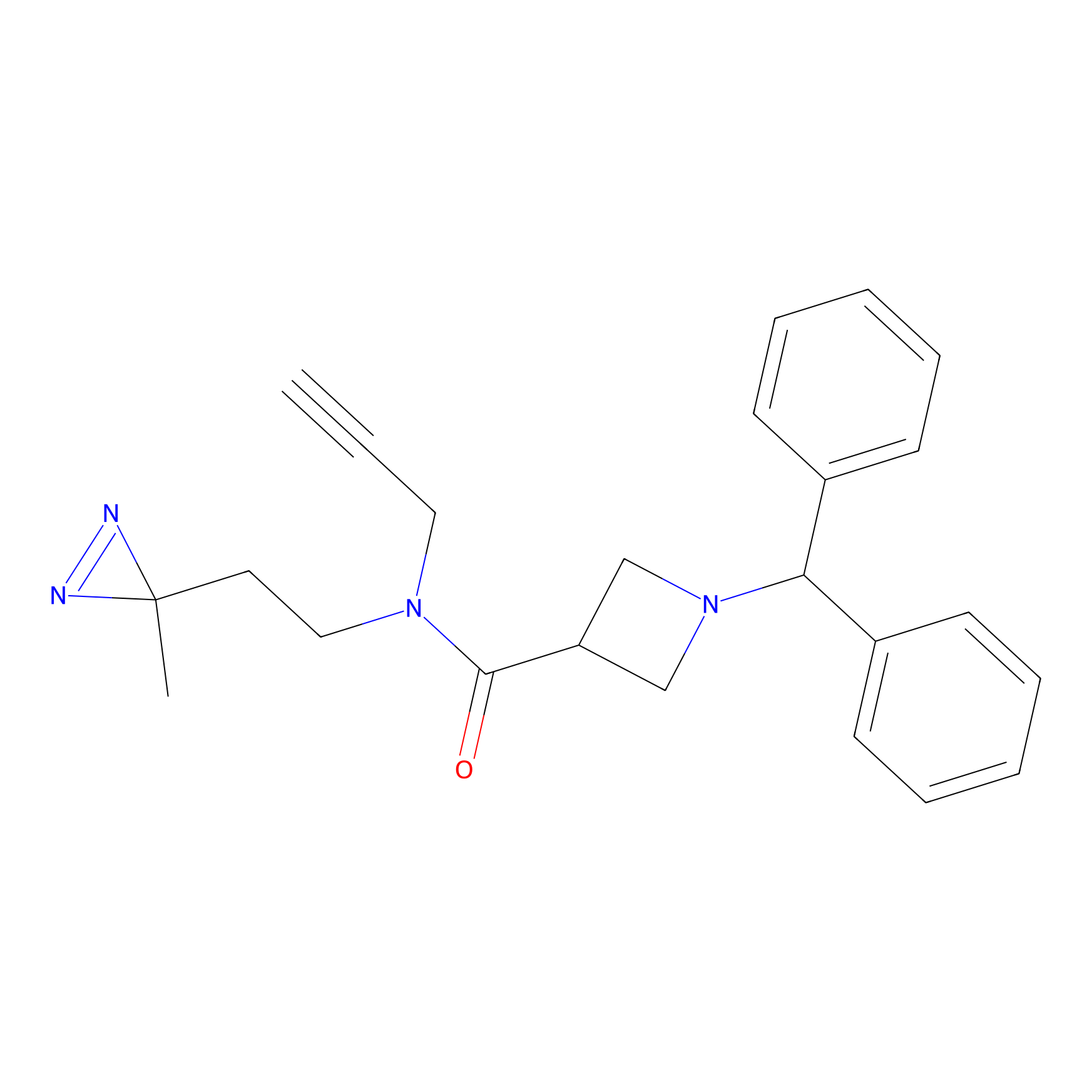 |
N55(0.00); Q50(0.00); N49(0.00) | LDD0024 | [34] | |
|
OEA-DA Probe Info |
 |
6.75 | LDD0046 | [35] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C620(0.81) | LDD2112 | [6] |
| LDCM0537 | 2-Cyano-N,N-dimethylacetamide | MDA-MB-231 | C620(1.02); C103(0.91) | LDD2130 | [6] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C467(1.01); C103(0.88) | LDD2117 | [6] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C467(0.85) | LDD2152 | [6] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C806(1.07) | LDD2103 | [6] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C103(0.34) | LDD2132 | [6] |
| LDCM0538 | 4-(Cyanoacetyl)morpholine | MDA-MB-231 | C620(0.80); C103(0.60) | LDD2131 | [6] |
| LDCM0025 | 4SU-RNA | DM93 | C164(2.75); C103(2.59) | LDD0170 | [11] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C164(3.13) | LDD0169 | [11] |
| LDCM0214 | AC1 | HCT 116 | C142(0.79); C103(0.93); C153(0.88); C205(0.83) | LDD0531 | [14] |
| LDCM0215 | AC10 | HCT 116 | C142(0.72); C103(0.76); C153(0.83); C205(0.80) | LDD0532 | [14] |
| LDCM0216 | AC100 | HCT 116 | C142(0.38); C862(1.28); C103(0.98); C153(0.75) | LDD0533 | [14] |
| LDCM0217 | AC101 | HCT 116 | C142(0.38); C862(2.19); C103(0.81); C153(0.76) | LDD0534 | [14] |
| LDCM0218 | AC102 | HCT 116 | C142(0.46); C862(1.56); C103(0.86); C153(0.69) | LDD0535 | [14] |
| LDCM0219 | AC103 | HCT 116 | C142(0.39); C862(2.38); C103(0.74); C153(0.58) | LDD0536 | [14] |
| LDCM0220 | AC104 | HCT 116 | C142(0.35); C862(3.33); C103(0.81); C153(0.82) | LDD0537 | [14] |
| LDCM0221 | AC105 | HCT 116 | C142(0.38); C862(1.41); C103(0.79); C153(0.74) | LDD0538 | [14] |
| LDCM0222 | AC106 | HCT 116 | C142(0.38); C862(2.13); C103(0.88); C153(0.77) | LDD0539 | [14] |
| LDCM0223 | AC107 | HCT 116 | C142(0.36); C862(2.70); C103(0.83); C153(0.70) | LDD0540 | [14] |
| LDCM0224 | AC108 | HCT 116 | C142(0.45); C862(1.20); C103(0.86); C153(0.76) | LDD0541 | [14] |
| LDCM0225 | AC109 | HCT 116 | C142(0.46); C862(2.11); C103(0.94); C153(0.92) | LDD0542 | [14] |
| LDCM0226 | AC11 | HCT 116 | C142(0.72); C103(0.91); C153(0.82); C205(0.78) | LDD0543 | [14] |
| LDCM0227 | AC110 | HCT 116 | C142(0.44); C862(1.57); C103(0.84); C153(0.62) | LDD0544 | [14] |
| LDCM0228 | AC111 | HCT 116 | C142(0.40); C862(4.98); C103(0.72); C153(0.63) | LDD0545 | [14] |
| LDCM0229 | AC112 | HCT 116 | C142(0.46); C862(2.68); C103(0.79); C153(0.72) | LDD0546 | [14] |
| LDCM0230 | AC113 | HCT 116 | C862(0.93); C103(1.59); C205(1.03); C386(0.87) | LDD0547 | [14] |
| LDCM0231 | AC114 | HCT 116 | C862(0.93); C103(0.85); C205(1.02); C386(1.25) | LDD0548 | [14] |
| LDCM0232 | AC115 | HCT 116 | C862(0.91); C103(0.87); C205(1.00); C386(0.77) | LDD0549 | [14] |
| LDCM0233 | AC116 | HCT 116 | C862(0.48); C103(0.92); C205(0.88); C386(0.74) | LDD0550 | [14] |
| LDCM0234 | AC117 | HCT 116 | C862(0.30); C103(0.80); C205(0.97); C386(0.87) | LDD0551 | [14] |
| LDCM0235 | AC118 | HCT 116 | C862(0.29); C103(1.03); C205(0.95); C386(0.97) | LDD0552 | [14] |
| LDCM0236 | AC119 | HCT 116 | C862(0.19); C103(0.97); C205(0.79); C386(0.96) | LDD0553 | [14] |
| LDCM0237 | AC12 | HCT 116 | C142(0.84); C103(0.94); C153(0.89); C205(0.81) | LDD0554 | [14] |
| LDCM0238 | AC120 | HCT 116 | C862(1.14); C103(0.74); C205(0.93); C386(0.51) | LDD0555 | [14] |
| LDCM0239 | AC121 | HCT 116 | C862(0.72); C103(0.97); C205(1.01); C386(0.92) | LDD0556 | [14] |
| LDCM0240 | AC122 | HCT 116 | C862(0.21); C103(0.92); C205(0.92); C386(0.90) | LDD0557 | [14] |
| LDCM0241 | AC123 | HCT 116 | C862(0.65); C103(0.84); C205(0.92); C386(0.85) | LDD0558 | [14] |
| LDCM0242 | AC124 | HCT 116 | C862(0.46); C103(1.02); C205(0.94); C386(1.16) | LDD0559 | [14] |
| LDCM0243 | AC125 | HCT 116 | C862(0.79); C103(0.82); C205(0.84); C386(1.26) | LDD0560 | [14] |
| LDCM0244 | AC126 | HCT 116 | C862(0.77); C103(0.82); C205(0.89); C386(1.18) | LDD0561 | [14] |
| LDCM0245 | AC127 | HCT 116 | C862(0.24); C103(0.94); C205(0.85); C386(1.19) | LDD0562 | [14] |
| LDCM0246 | AC128 | HCT 116 | C103(1.15); C205(0.96); C386(1.66); C467(1.45) | LDD0563 | [14] |
| LDCM0247 | AC129 | HCT 116 | C103(1.47); C205(1.52); C386(1.58); C467(1.47) | LDD0564 | [14] |
| LDCM0249 | AC130 | HCT 116 | C103(1.23); C205(1.02); C386(1.63); C467(1.21) | LDD0566 | [14] |
| LDCM0250 | AC131 | HCT 116 | C103(1.23); C205(1.76); C386(1.27); C467(1.63) | LDD0567 | [14] |
| LDCM0251 | AC132 | HCT 116 | C103(1.42); C205(0.91); C386(1.46); C467(0.96) | LDD0568 | [14] |
| LDCM0252 | AC133 | HCT 116 | C103(1.21); C205(0.90); C386(1.60); C467(1.02) | LDD0569 | [14] |
| LDCM0253 | AC134 | HCT 116 | C103(1.32); C205(0.97); C386(1.63); C467(0.99) | LDD0570 | [14] |
| LDCM0254 | AC135 | HCT 116 | C103(1.41); C205(0.86); C386(1.65); C467(0.96) | LDD0571 | [14] |
| LDCM0255 | AC136 | HCT 116 | C103(1.21); C205(0.89); C386(1.18); C467(0.97) | LDD0572 | [14] |
| LDCM0256 | AC137 | HCT 116 | C103(1.32); C205(1.03); C386(1.28); C467(1.05) | LDD0573 | [14] |
| LDCM0257 | AC138 | HCT 116 | C103(1.42); C205(0.84); C386(1.51); C467(0.96) | LDD0574 | [14] |
| LDCM0258 | AC139 | HCT 116 | C103(1.45); C205(0.83); C386(1.46); C467(1.09) | LDD0575 | [14] |
| LDCM0259 | AC14 | HCT 116 | C142(0.78); C103(0.72); C153(0.80); C205(0.86) | LDD0576 | [14] |
| LDCM0260 | AC140 | HCT 116 | C103(1.32); C205(0.78); C386(1.85); C467(1.03) | LDD0577 | [14] |
| LDCM0261 | AC141 | HCT 116 | C103(1.42); C205(0.88); C386(1.46); C467(1.01) | LDD0578 | [14] |
| LDCM0262 | AC142 | HCT 116 | C103(1.41); C205(0.86); C386(1.39); C467(0.90) | LDD0579 | [14] |
| LDCM0263 | AC143 | HCT 116 | C706(0.65); C153(0.68); C683(0.73); C620(0.76) | LDD0580 | [14] |
| LDCM0264 | AC144 | HCT 116 | C153(0.52); C683(0.58); C620(0.69); C205(0.83) | LDD0581 | [14] |
| LDCM0265 | AC145 | HCT 116 | C683(0.60); C153(0.70); C790(0.74); C620(0.74) | LDD0582 | [14] |
| LDCM0266 | AC146 | HCT 116 | C683(0.53); C620(0.60); C790(0.61); C205(0.71) | LDD0583 | [14] |
| LDCM0267 | AC147 | HCT 116 | C683(0.56); C153(0.58); C790(0.67); C620(0.76) | LDD0584 | [14] |
| LDCM0268 | AC148 | HCT 116 | C620(0.61); C683(0.66); C153(0.84); C205(0.85) | LDD0585 | [14] |
| LDCM0269 | AC149 | HCT 116 | C683(0.56); C620(0.58); C153(0.66); C205(0.74) | LDD0586 | [14] |
| LDCM0270 | AC15 | HCT 116 | C706(0.68); C103(0.69); C142(0.69); C153(0.77) | LDD0587 | [14] |
| LDCM0271 | AC150 | HCT 116 | C683(0.64); C153(0.67); C790(0.67); C620(0.69) | LDD0588 | [14] |
| LDCM0272 | AC151 | HCT 116 | C683(0.67); C153(0.68); C620(0.79); C467(0.86) | LDD0589 | [14] |
| LDCM0273 | AC152 | HCT 116 | C620(0.61); C683(0.64); C153(0.79); C806(0.85) | LDD0590 | [14] |
| LDCM0274 | AC153 | HCT 116 | C620(0.50); C683(0.67); C205(0.89); C103(0.91) | LDD0591 | [14] |
| LDCM0621 | AC154 | HCT 116 | C103(0.97); C153(0.58); C205(0.82); C386(0.98) | LDD2158 | [14] |
| LDCM0622 | AC155 | HCT 116 | C103(0.89); C153(0.58); C205(0.77); C386(0.86) | LDD2159 | [14] |
| LDCM0623 | AC156 | HCT 116 | C103(0.96); C153(0.72); C205(0.85); C386(0.93) | LDD2160 | [14] |
| LDCM0624 | AC157 | HCT 116 | C103(0.78); C153(0.99); C205(1.30); C386(1.02) | LDD2161 | [14] |
| LDCM0276 | AC17 | HCT 116 | C620(0.88); C683(0.92); C103(0.94); C706(0.95) | LDD0593 | [14] |
| LDCM0277 | AC18 | HCT 116 | C467(0.75); C103(0.84); C620(0.85); C683(0.86) | LDD0594 | [14] |
| LDCM0278 | AC19 | HCT 116 | C153(0.86); C683(0.89); C103(0.91); C467(0.97) | LDD0595 | [14] |
| LDCM0279 | AC2 | HCT 116 | C706(0.75); C142(0.80); C153(0.80); C205(0.81) | LDD0596 | [14] |
| LDCM0280 | AC20 | HCT 116 | C683(0.80); C386(0.82); C806(0.91); C620(0.93) | LDD0597 | [14] |
| LDCM0281 | AC21 | HCT 116 | C683(0.77); C620(0.84); C103(0.88); C205(0.89) | LDD0598 | [14] |
| LDCM0282 | AC22 | HCT 116 | C706(0.82); C467(0.84); C620(0.84); C683(0.89) | LDD0599 | [14] |
| LDCM0283 | AC23 | HCT 116 | C683(0.76); C806(0.81); C620(0.81); C386(0.84) | LDD0600 | [14] |
| LDCM0284 | AC24 | HCT 116 | C706(0.74); C386(0.87); C683(0.92); C153(1.02) | LDD0601 | [14] |
| LDCM0285 | AC25 | HCT 116 | C683(0.74); C467(0.75); C205(0.80); C620(0.84) | LDD0602 | [14] |
| LDCM0286 | AC26 | HCT 116 | C683(0.65); C205(0.68); C790(0.70); C467(0.74) | LDD0603 | [14] |
| LDCM0287 | AC27 | HCT 116 | C683(0.65); C205(0.65); C467(0.65); C790(0.84) | LDD0604 | [14] |
| LDCM0288 | AC28 | HCT 116 | C683(0.66); C467(0.67); C205(0.69); C790(0.80) | LDD0605 | [14] |
| LDCM0289 | AC29 | HCT 116 | C683(0.61); C205(0.63); C467(0.72); C790(0.75) | LDD0606 | [14] |
| LDCM0290 | AC3 | HCT 116 | C706(0.76); C205(0.82); C285(0.86); C297(0.86) | LDD0607 | [14] |
| LDCM0291 | AC30 | HCT 116 | C467(0.68); C205(0.72); C683(0.75); C790(0.76) | LDD0608 | [14] |
| LDCM0292 | AC31 | HCT 116 | C683(0.62); C467(0.70); C205(0.71); C790(0.75) | LDD0609 | [14] |
| LDCM0293 | AC32 | HCT 116 | C683(0.55); C205(0.64); C706(0.75); C103(0.79) | LDD0610 | [14] |
| LDCM0294 | AC33 | HCT 116 | C683(0.64); C706(0.66); C205(0.70); C790(0.76) | LDD0611 | [14] |
| LDCM0295 | AC34 | HCT 116 | C683(0.64); C706(0.67); C103(0.76); C205(0.77) | LDD0612 | [14] |
| LDCM0296 | AC35 | HCT 116 | C790(0.64); C706(0.84); C806(0.87); C142(0.98) | LDD0613 | [14] |
| LDCM0297 | AC36 | HCT 116 | C790(0.76); C142(0.82); C153(0.82); C706(0.84) | LDD0614 | [14] |
| LDCM0298 | AC37 | HCT 116 | C790(0.61); C706(0.85); C806(0.88); C205(0.94) | LDD0615 | [14] |
| LDCM0299 | AC38 | HCT 116 | C790(0.52); C806(0.64); C706(0.65); C683(0.97) | LDD0616 | [14] |
| LDCM0300 | AC39 | HCT 116 | C790(0.54); C205(0.73); C706(0.84); C806(0.86) | LDD0617 | [14] |
| LDCM0301 | AC4 | HCT 116 | C142(0.78); C706(0.79); C285(0.81); C297(0.81) | LDD0618 | [14] |
| LDCM0302 | AC40 | HCT 116 | C386(0.61); C790(0.65); C706(0.80); C806(0.81) | LDD0619 | [14] |
| LDCM0303 | AC41 | HCT 116 | C790(0.53); C386(0.59); C706(0.59); C806(0.74) | LDD0620 | [14] |
| LDCM0304 | AC42 | HCT 116 | C790(0.50); C706(0.59); C806(0.77); C142(0.86) | LDD0621 | [14] |
| LDCM0305 | AC43 | HCT 116 | C790(0.54); C706(0.67); C386(0.68); C142(0.81) | LDD0622 | [14] |
| LDCM0306 | AC44 | HCT 116 | C790(0.53); C706(0.57); C386(0.67); C806(0.77) | LDD0623 | [14] |
| LDCM0307 | AC45 | HCT 116 | C386(0.53); C790(0.62); C706(0.68); C806(0.73) | LDD0624 | [14] |
| LDCM0308 | AC46 | HCT 116 | C103(0.92); C790(0.93); C205(1.00); C620(1.01) | LDD0625 | [14] |
| LDCM0309 | AC47 | HCT 116 | C790(0.82); C205(0.84); C806(0.88); C103(1.03) | LDD0626 | [14] |
| LDCM0310 | AC48 | HCT 116 | C790(0.34); C205(0.67); C467(0.77); C620(0.83) | LDD0627 | [14] |
| LDCM0311 | AC49 | HCT 116 | C103(0.85); C706(0.89); C620(0.96); C806(0.97) | LDD0628 | [14] |
| LDCM0312 | AC5 | HCT 116 | C706(0.68); C205(0.74); C285(0.83); C297(0.83) | LDD0629 | [14] |
| LDCM0313 | AC50 | HCT 116 | C706(0.77); C103(0.85); C620(0.96); C806(1.00) | LDD0630 | [14] |
| LDCM0314 | AC51 | HCT 116 | C790(0.68); C706(0.72); C205(0.89); C103(0.92) | LDD0631 | [14] |
| LDCM0315 | AC52 | HCT 116 | C790(0.80); C706(0.87); C205(0.90); C103(0.90) | LDD0632 | [14] |
| LDCM0316 | AC53 | HCT 116 | C806(0.81); C706(0.88); C790(0.90); C103(0.92) | LDD0633 | [14] |
| LDCM0317 | AC54 | HCT 116 | C706(0.73); C790(0.91); C806(0.93); C467(0.95) | LDD0634 | [14] |
| LDCM0318 | AC55 | HCT 116 | C706(0.82); C467(0.94); C806(0.98); C103(1.01) | LDD0635 | [14] |
| LDCM0319 | AC56 | HCT 116 | C806(0.80); C706(0.80); C103(0.89); C620(0.96) | LDD0636 | [14] |
| LDCM0320 | AC57 | HCT 116 | C142(0.69); C153(0.74); C620(0.81); C205(0.87) | LDD0637 | [14] |
| LDCM0321 | AC58 | HCT 116 | C142(0.65); C620(0.77); C153(0.79); C683(0.80) | LDD0638 | [14] |
| LDCM0322 | AC59 | HCT 116 | C142(0.74); C706(0.77); C153(0.83); C205(0.84) | LDD0639 | [14] |
| LDCM0323 | AC6 | HCT 116 | C142(0.65); C103(0.66); C205(0.77); C620(0.78) | LDD0640 | [14] |
| LDCM0324 | AC60 | HCT 116 | C142(0.69); C620(0.70); C706(0.78); C153(0.80) | LDD0641 | [14] |
| LDCM0325 | AC61 | HCT 116 | C142(0.65); C153(0.72); C706(0.74); C620(0.76) | LDD0642 | [14] |
| LDCM0326 | AC62 | HCT 116 | C706(0.73); C142(0.76); C620(0.77); C153(0.79) | LDD0643 | [14] |
| LDCM0327 | AC63 | HCT 116 | C142(0.69); C620(0.73); C153(0.78); C706(0.82) | LDD0644 | [14] |
| LDCM0328 | AC64 | HCT 116 | C620(0.72); C142(0.73); C153(0.81); C205(0.85) | LDD0645 | [14] |
| LDCM0329 | AC65 | HCT 116 | C790(0.58); C205(0.73); C806(0.73); C142(0.73) | LDD0646 | [14] |
| LDCM0330 | AC66 | HCT 116 | C790(0.54); C142(0.78); C706(0.81); C205(0.81) | LDD0647 | [14] |
| LDCM0331 | AC67 | HCT 116 | C790(0.58); C620(0.74); C706(0.75); C683(0.76) | LDD0648 | [14] |
| LDCM0332 | AC68 | HCT 116 | C620(0.79); C683(0.79); C205(0.80); C386(0.80) | LDD0649 | [14] |
| LDCM0333 | AC69 | HCT 116 | C142(0.76); C386(0.81); C683(0.83); C620(0.86) | LDD0650 | [14] |
| LDCM0334 | AC7 | HCT 116 | C103(0.73); C142(0.74); C790(0.75); C153(0.78) | LDD0651 | [14] |
| LDCM0335 | AC70 | HCT 116 | C862(0.26); C683(0.63); C205(0.80); C467(0.85) | LDD0652 | [14] |
| LDCM0336 | AC71 | HCT 116 | C142(0.81); C683(0.81); C620(0.93); C153(0.97) | LDD0653 | [14] |
| LDCM0337 | AC72 | HCT 116 | C862(0.35); C142(0.84); C386(0.87); C683(0.91) | LDD0654 | [14] |
| LDCM0338 | AC73 | HCT 116 | C862(0.48); C142(0.92); C706(0.83); C620(0.95) | LDD0655 | [14] |
| LDCM0339 | AC74 | HCT 116 | C706(0.79); C683(0.87); C620(0.91); C386(0.96) | LDD0656 | [14] |
| LDCM0340 | AC75 | HCT 116 | C862(0.61); C103(0.96); C706(0.97); C205(1.06) | LDD0657 | [14] |
| LDCM0341 | AC76 | HCT 116 | C862(0.76); C706(0.84); C142(0.84); C386(0.88) | LDD0658 | [14] |
| LDCM0342 | AC77 | HCT 116 | C706(0.74); C683(0.83); C103(0.89); C142(0.91) | LDD0659 | [14] |
| LDCM0343 | AC78 | HCT 116 | C142(0.69); C706(0.71); C683(0.75); C386(0.78) | LDD0660 | [14] |
| LDCM0344 | AC79 | HCT 116 | C862(0.69); C706(0.74); C142(0.78); C683(0.84) | LDD0661 | [14] |
| LDCM0345 | AC8 | HCT 116 | C103(0.75); C142(0.76); C205(0.78); C706(0.80) | LDD0662 | [14] |
| LDCM0346 | AC80 | HCT 116 | C386(0.78); C706(0.79); C620(0.82); C683(0.84) | LDD0663 | [14] |
| LDCM0347 | AC81 | HCT 116 | C862(0.63); C386(0.66); C806(0.77); C706(0.79) | LDD0664 | [14] |
| LDCM0348 | AC82 | HCT 116 | C862(0.40); C706(0.77); C683(0.88); C103(0.96) | LDD0665 | [14] |
| LDCM0349 | AC83 | HCT 116 | C706(0.70); C620(0.74); C205(0.80); C806(0.81) | LDD0666 | [14] |
| LDCM0350 | AC84 | HCT 116 | C706(0.63); C620(0.76); C205(0.79); C806(0.81) | LDD0667 | [14] |
| LDCM0351 | AC85 | HCT 116 | C706(0.73); C620(0.73); C205(0.75); C285(0.75) | LDD0668 | [14] |
| LDCM0352 | AC86 | HCT 116 | C706(0.73); C285(0.73); C297(0.73); C103(0.74) | LDD0669 | [14] |
| LDCM0353 | AC87 | HCT 116 | C706(0.83); C285(0.89); C297(0.89); C205(0.91) | LDD0670 | [14] |
| LDCM0354 | AC88 | HCT 116 | C706(0.68); C205(0.90); C285(0.91); C297(0.91) | LDD0671 | [14] |
| LDCM0355 | AC89 | HCT 116 | C706(0.57); C620(0.78); C205(0.82); C806(0.88) | LDD0672 | [14] |
| LDCM0357 | AC90 | HCT 116 | C205(0.87); C706(0.92); C285(1.04); C297(1.04) | LDD0674 | [14] |
| LDCM0358 | AC91 | HCT 116 | C706(0.66); C205(0.85); C103(0.87); C620(0.89) | LDD0675 | [14] |
| LDCM0359 | AC92 | HCT 116 | C706(0.59); C620(0.80); C205(0.81); C683(0.83) | LDD0676 | [14] |
| LDCM0360 | AC93 | HCT 116 | C706(0.58); C285(0.76); C297(0.76); C205(0.81) | LDD0677 | [14] |
| LDCM0361 | AC94 | HCT 116 | C706(0.62); C620(0.82); C103(0.83); C683(0.88) | LDD0678 | [14] |
| LDCM0362 | AC95 | HCT 116 | C706(0.64); C285(0.67); C297(0.67); C620(0.68) | LDD0679 | [14] |
| LDCM0363 | AC96 | HCT 116 | C706(0.65); C467(0.79); C620(0.80); C285(0.81) | LDD0680 | [14] |
| LDCM0364 | AC97 | HCT 116 | C706(0.55); C285(0.70); C297(0.70); C620(0.75) | LDD0681 | [14] |
| LDCM0365 | AC98 | HCT 116 | C683(0.33); C142(0.42); C285(0.50); C297(0.50) | LDD0682 | [14] |
| LDCM0366 | AC99 | HCT 116 | C285(0.38); C297(0.38); C683(0.44); C142(0.51) | LDD0683 | [14] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C706(1.17) | LDD2113 | [6] |
| LDCM0248 | AKOS034007472 | HCT 116 | C142(0.83); C103(0.81); C153(0.88); C205(0.91) | LDD0565 | [14] |
| LDCM0356 | AKOS034007680 | HCT 116 | C706(0.68); C142(0.75); C205(0.76); C103(0.82) | LDD0673 | [14] |
| LDCM0275 | AKOS034007705 | HCT 116 | C706(0.63); C142(0.70); C153(0.76); C103(0.76) | LDD0592 | [14] |
| LDCM0156 | Aniline | NCI-H1299 | N.A. | LDD0405 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C467(1.53); C205(0.89) | LDD0078 | [14] |
| LDCM0151 | AZ-11 | HeLa | 10.00 | LDD2154 | [10] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C467(0.96); C103(0.74) | LDD2091 | [6] |
| LDCM0108 | Chloroacetamide | HeLa | C103(0.00); C164(0.00); C620(0.00); H87(0.00) | LDD0222 | [26] |
| LDCM0632 | CL-Sc | Hep-G2 | C467(20.00); C205(1.76); C683(1.06); C862(0.30) | LDD2227 | [20] |
| LDCM0367 | CL1 | HCT 116 | C620(0.77); C386(0.91); C706(0.92); C103(0.94) | LDD0684 | [14] |
| LDCM0368 | CL10 | HCT 116 | C620(0.69); C706(0.72); C683(0.92); C142(0.98) | LDD0685 | [14] |
| LDCM0369 | CL100 | HCT 116 | C706(0.78); C790(0.79); C285(0.80); C297(0.80) | LDD0686 | [14] |
| LDCM0370 | CL101 | HCT 116 | C142(0.66); C103(0.69); C153(0.71); C285(0.78) | LDD0687 | [14] |
| LDCM0371 | CL102 | HCT 116 | C103(0.74); C806(0.77); C142(0.85); C285(0.88) | LDD0688 | [14] |
| LDCM0372 | CL103 | HCT 116 | C806(0.71); C103(0.86); C153(0.92); C142(0.93) | LDD0689 | [14] |
| LDCM0373 | CL104 | HCT 116 | C706(0.75); C142(0.80); C103(0.81); C153(0.83) | LDD0690 | [14] |
| LDCM0374 | CL105 | HCT 116 | C467(0.74); C683(0.75); C205(0.83); C620(0.86) | LDD0691 | [14] |
| LDCM0375 | CL106 | HCT 116 | C683(0.67); C620(0.78); C467(0.88); C153(0.93) | LDD0692 | [14] |
| LDCM0376 | CL107 | HCT 116 | C683(0.70); C205(0.78); C620(0.79); C467(0.88) | LDD0693 | [14] |
| LDCM0377 | CL108 | HCT 116 | C467(0.73); C683(0.79); C205(0.87); C620(0.89) | LDD0694 | [14] |
| LDCM0378 | CL109 | HCT 116 | C683(0.72); C706(0.85); C386(0.85); C153(0.88) | LDD0695 | [14] |
| LDCM0379 | CL11 | HCT 116 | C620(0.79); C706(0.80); C683(1.06); C103(1.08) | LDD0696 | [14] |
| LDCM0380 | CL110 | HCT 116 | C683(0.66); C467(0.74); C620(0.84); C706(0.85) | LDD0697 | [14] |
| LDCM0381 | CL111 | HCT 116 | C806(0.64); C683(0.81); C153(0.83); C706(0.85) | LDD0698 | [14] |
| LDCM0382 | CL112 | HCT 116 | C205(0.73); C683(0.75); C467(0.78); C790(0.87) | LDD0699 | [14] |
| LDCM0383 | CL113 | HCT 116 | C683(0.62); C467(0.65); C706(0.77); C205(0.78) | LDD0700 | [14] |
| LDCM0384 | CL114 | HCT 116 | C467(0.54); C706(0.64); C683(0.66); C386(0.72) | LDD0701 | [14] |
| LDCM0385 | CL115 | HCT 116 | C683(0.60); C467(0.61); C706(0.62); C205(0.70) | LDD0702 | [14] |
| LDCM0386 | CL116 | HCT 116 | C706(0.61); C683(0.64); C205(0.67); C467(0.78) | LDD0703 | [14] |
| LDCM0387 | CL117 | HCT 116 | C790(0.68); C806(0.77); C706(0.82); C386(0.84) | LDD0704 | [14] |
| LDCM0388 | CL118 | HCT 116 | C790(0.57); C706(0.74); C142(0.82); C153(0.82) | LDD0705 | [14] |
| LDCM0389 | CL119 | HCT 116 | C790(0.70); C706(0.77); C205(0.85); C806(0.88) | LDD0706 | [14] |
| LDCM0390 | CL12 | HCT 116 | C620(0.80); C706(0.83); C142(0.97); C153(0.97) | LDD0707 | [14] |
| LDCM0391 | CL120 | HCT 116 | C790(0.72); C706(0.76); C205(0.77); C467(0.85) | LDD0708 | [14] |
| LDCM0392 | CL121 | HCT 116 | C467(0.71); C806(0.76); C790(0.77); C153(0.83) | LDD0709 | [14] |
| LDCM0393 | CL122 | HCT 116 | C467(0.81); C806(0.81); C706(0.82); C103(0.84) | LDD0710 | [14] |
| LDCM0394 | CL123 | HCT 116 | C806(0.79); C620(0.88); C467(0.90); C706(0.92) | LDD0711 | [14] |
| LDCM0395 | CL124 | HCT 116 | C706(0.82); C806(0.85); C103(0.85); C620(0.96) | LDD0712 | [14] |
| LDCM0396 | CL125 | HCT 116 | C467(0.90); C153(0.95); C142(0.95); C205(0.96) | LDD0713 | [14] |
| LDCM0397 | CL126 | HCT 116 | C142(0.79); C153(0.85); C620(0.89); C706(0.95) | LDD0714 | [14] |
| LDCM0398 | CL127 | HCT 116 | C620(0.80); C683(0.81); C142(0.82); C153(0.85) | LDD0715 | [14] |
| LDCM0399 | CL128 | HCT 116 | C142(0.79); C683(0.79); C205(0.83); C153(0.85) | LDD0716 | [14] |
| LDCM0400 | CL13 | HCT 116 | C706(0.67); C620(0.71); C142(0.92); C153(0.92) | LDD0717 | [14] |
| LDCM0401 | CL14 | HCT 116 | C706(0.62); C620(0.63); C142(0.85); C153(0.85) | LDD0718 | [14] |
| LDCM0402 | CL15 | HCT 116 | C620(0.71); C706(0.73); C386(0.95); C285(1.00) | LDD0719 | [14] |
| LDCM0403 | CL16 | HCT 116 | C153(0.79); C142(0.87); C467(0.88); C706(0.92) | LDD0720 | [14] |
| LDCM0404 | CL17 | HCT 116 | C862(0.44); C706(0.67); C142(0.89); C620(0.89) | LDD0721 | [14] |
| LDCM0405 | CL18 | HCT 116 | C862(0.51); C706(0.53); C153(0.72); C620(0.75) | LDD0722 | [14] |
| LDCM0406 | CL19 | HCT 116 | C706(0.68); C862(0.76); C620(0.80); C683(0.88) | LDD0723 | [14] |
| LDCM0407 | CL2 | HCT 116 | C386(0.80); C620(0.81); C706(0.85); C103(1.01) | LDD0724 | [14] |
| LDCM0408 | CL20 | HCT 116 | C706(0.56); C862(0.70); C620(0.84); C142(0.85) | LDD0725 | [14] |
| LDCM0409 | CL21 | HCT 116 | C706(0.59); C862(0.65); C620(0.75); C153(0.83) | LDD0726 | [14] |
| LDCM0410 | CL22 | HCT 116 | C706(0.63); C103(0.90); C205(0.98); C620(0.98) | LDD0727 | [14] |
| LDCM0411 | CL23 | HCT 116 | C706(0.54); C153(0.71); C142(0.81); C620(0.84) | LDD0728 | [14] |
| LDCM0412 | CL24 | HCT 116 | C142(0.95); C862(0.95); C103(1.22); C153(0.85) | LDD0729 | [14] |
| LDCM0413 | CL25 | HCT 116 | C142(0.95); C862(0.79); C103(1.23); C153(0.79) | LDD0730 | [14] |
| LDCM0414 | CL26 | HCT 116 | C142(1.01); C862(0.65); C103(1.12); C153(0.82) | LDD0731 | [14] |
| LDCM0415 | CL27 | HCT 116 | C142(0.84); C862(0.93); C103(1.24); C153(0.77) | LDD0732 | [14] |
| LDCM0416 | CL28 | HCT 116 | C142(0.86); C862(0.54); C103(1.21); C153(0.74) | LDD0733 | [14] |
| LDCM0417 | CL29 | HCT 116 | C142(0.89); C862(0.82); C103(0.97); C153(0.84) | LDD0734 | [14] |
| LDCM0418 | CL3 | HCT 116 | C142(0.95); C103(0.99); C153(0.95); C205(0.92) | LDD0735 | [14] |
| LDCM0419 | CL30 | HCT 116 | C142(0.91); C862(0.34); C103(0.72); C153(0.78) | LDD0736 | [14] |
| LDCM0420 | CL31 | HCT 116 | C142(0.74); C862(0.56); C103(1.24); C153(0.74) | LDD0737 | [14] |
| LDCM0421 | CL32 | HCT 116 | C142(0.62); C103(1.04); C153(0.62); C205(1.05) | LDD0738 | [14] |
| LDCM0422 | CL33 | HCT 116 | C142(0.63); C103(1.20); C153(0.63); C205(0.93) | LDD0739 | [14] |
| LDCM0423 | CL34 | HCT 116 | C142(0.78); C103(1.09); C153(0.78); C205(1.17) | LDD0740 | [14] |
| LDCM0424 | CL35 | HCT 116 | C142(0.81); C103(1.05); C153(0.81); C205(1.27) | LDD0741 | [14] |
| LDCM0425 | CL36 | HCT 116 | C142(0.72); C103(1.02); C153(0.72); C205(1.17) | LDD0742 | [14] |
| LDCM0426 | CL37 | HCT 116 | C142(0.72); C103(1.15); C153(0.72); C205(1.22) | LDD0743 | [14] |
| LDCM0428 | CL39 | HCT 116 | C142(0.74); C103(1.26); C153(0.74); C205(1.00) | LDD0745 | [14] |
| LDCM0429 | CL4 | HCT 116 | C142(1.04); C103(0.94); C153(1.04); C205(1.15) | LDD0746 | [14] |
| LDCM0430 | CL40 | HCT 116 | C142(0.76); C103(1.01); C153(0.76); C205(1.14) | LDD0747 | [14] |
| LDCM0431 | CL41 | HCT 116 | C142(0.66); C103(0.95); C153(0.66); C205(1.07) | LDD0748 | [14] |
| LDCM0432 | CL42 | HCT 116 | C142(0.89); C103(1.03); C153(0.89); C205(1.19) | LDD0749 | [14] |
| LDCM0433 | CL43 | HCT 116 | C142(0.72); C103(1.07); C153(0.72); C205(1.20) | LDD0750 | [14] |
| LDCM0434 | CL44 | HCT 116 | C142(0.63); C103(1.06); C153(0.63); C205(1.12) | LDD0751 | [14] |
| LDCM0435 | CL45 | HCT 116 | C142(0.75); C103(1.11); C153(0.75); C205(0.94) | LDD0752 | [14] |
| LDCM0436 | CL46 | HCT 116 | C142(1.12); C103(0.98); C153(0.98); C205(1.11) | LDD0753 | [14] |
| LDCM0437 | CL47 | HCT 116 | C142(1.00); C103(0.88); C153(0.95); C205(1.07) | LDD0754 | [14] |
| LDCM0438 | CL48 | HCT 116 | C142(1.02); C103(0.96); C153(1.07); C205(1.23) | LDD0755 | [14] |
| LDCM0439 | CL49 | HCT 116 | C142(0.99); C103(1.11); C153(0.93); C205(1.19) | LDD0756 | [14] |
| LDCM0440 | CL5 | HCT 116 | C142(1.03); C103(1.00); C153(1.03); C205(0.95) | LDD0757 | [14] |
| LDCM0441 | CL50 | HCT 116 | C142(0.91); C103(1.05); C153(0.92); C205(1.17) | LDD0758 | [14] |
| LDCM0442 | CL51 | HCT 116 | C142(0.98); C103(1.32); C153(1.00); C205(1.13) | LDD0759 | [14] |
| LDCM0443 | CL52 | HCT 116 | C142(1.02); C103(1.04); C153(0.92); C205(1.06) | LDD0760 | [14] |
| LDCM0444 | CL53 | HCT 116 | C142(1.01); C103(1.11); C153(0.95); C205(1.33) | LDD0761 | [14] |
| LDCM0445 | CL54 | HCT 116 | C142(1.05); C103(1.02); C153(0.84); C205(0.93) | LDD0762 | [14] |
| LDCM0446 | CL55 | HCT 116 | C142(1.09); C103(0.97); C153(0.89); C205(1.35) | LDD0763 | [14] |
| LDCM0447 | CL56 | HCT 116 | C142(0.93); C103(1.24); C153(0.85); C205(1.06) | LDD0764 | [14] |
| LDCM0448 | CL57 | HCT 116 | C142(0.89); C103(0.83); C153(0.76); C205(1.11) | LDD0765 | [14] |
| LDCM0449 | CL58 | HCT 116 | C142(1.04); C103(0.87); C153(1.01); C205(1.13) | LDD0766 | [14] |
| LDCM0450 | CL59 | HCT 116 | C142(0.97); C103(1.10); C153(0.98); C205(0.99) | LDD0767 | [14] |
| LDCM0451 | CL6 | HCT 116 | C142(0.94); C103(0.92); C153(0.94); C205(0.82) | LDD0768 | [14] |
| LDCM0452 | CL60 | HCT 116 | C142(1.00); C103(1.02); C153(0.95); C205(1.20) | LDD0769 | [14] |
| LDCM0453 | CL61 | HCT 116 | C142(1.13); C103(0.80); C153(1.04); C205(0.89) | LDD0770 | [14] |
| LDCM0454 | CL62 | HCT 116 | C142(1.18); C103(0.81); C153(1.02); C205(0.82) | LDD0771 | [14] |
| LDCM0455 | CL63 | HCT 116 | C142(0.93); C103(0.67); C153(0.85); C205(0.76) | LDD0772 | [14] |
| LDCM0456 | CL64 | HCT 116 | C142(1.23); C103(0.71); C153(1.17); C205(0.71) | LDD0773 | [14] |
| LDCM0457 | CL65 | HCT 116 | C142(1.03); C103(0.86); C153(0.98); C205(0.84) | LDD0774 | [14] |
| LDCM0458 | CL66 | HCT 116 | C142(1.24); C103(0.80); C153(1.04); C205(0.80) | LDD0775 | [14] |
| LDCM0459 | CL67 | HCT 116 | C142(0.94); C103(0.71); C153(1.09); C205(0.82) | LDD0776 | [14] |
| LDCM0460 | CL68 | HCT 116 | C142(0.77); C103(0.73); C153(0.83); C205(0.81) | LDD0777 | [14] |
| LDCM0461 | CL69 | HCT 116 | C142(0.78); C103(0.66); C153(0.83); C205(0.80) | LDD0778 | [14] |
| LDCM0462 | CL7 | HCT 116 | C142(1.03); C103(1.05); C153(1.03); C205(1.00) | LDD0779 | [14] |
| LDCM0463 | CL70 | HCT 116 | C142(0.98); C103(0.79); C153(0.91); C205(0.88) | LDD0780 | [14] |
| LDCM0464 | CL71 | HCT 116 | C142(0.86); C103(0.84); C153(0.88); C205(0.81) | LDD0781 | [14] |
| LDCM0465 | CL72 | HCT 116 | C142(1.19); C103(0.75); C153(1.24); C205(0.82) | LDD0782 | [14] |
| LDCM0466 | CL73 | HCT 116 | C142(0.96); C103(0.79); C153(1.04); C205(0.78) | LDD0783 | [14] |
| LDCM0467 | CL74 | HCT 116 | C142(0.84); C103(0.76); C153(0.99); C205(0.84) | LDD0784 | [14] |
| LDCM0469 | CL76 | HCT 116 | C142(0.82); C103(0.73); C153(0.86); C205(0.95) | LDD0786 | [14] |
| LDCM0470 | CL77 | HCT 116 | C142(0.86); C103(0.75); C153(0.90); C205(1.22) | LDD0787 | [14] |
| LDCM0471 | CL78 | HCT 116 | C142(0.93); C103(1.23); C153(0.97); C205(0.91) | LDD0788 | [14] |
| LDCM0472 | CL79 | HCT 116 | C142(0.88); C103(0.90); C153(0.98); C205(1.04) | LDD0789 | [14] |
| LDCM0473 | CL8 | HCT 116 | C142(0.97); C103(0.93); C153(0.97); C205(1.20) | LDD0790 | [14] |
| LDCM0474 | CL80 | HCT 116 | C142(0.87); C103(0.80); C153(0.89); C205(0.97) | LDD0791 | [14] |
| LDCM0475 | CL81 | HCT 116 | C142(0.85); C103(0.90); C153(0.95); C205(1.04) | LDD0792 | [14] |
| LDCM0476 | CL82 | HCT 116 | C142(0.94); C103(0.90); C153(0.98); C205(1.12) | LDD0793 | [14] |
| LDCM0477 | CL83 | HCT 116 | C142(0.93); C103(0.82); C153(0.95); C205(1.03) | LDD0794 | [14] |
| LDCM0478 | CL84 | HCT 116 | C142(1.04); C103(0.89); C153(1.05); C205(0.97) | LDD0795 | [14] |
| LDCM0479 | CL85 | HCT 116 | C142(0.85); C103(0.75); C153(0.89); C205(0.99) | LDD0796 | [14] |
| LDCM0480 | CL86 | HCT 116 | C142(0.89); C103(0.98); C153(0.97); C205(1.03) | LDD0797 | [14] |
| LDCM0481 | CL87 | HCT 116 | C142(0.95); C103(1.02); C153(0.95); C205(0.97) | LDD0798 | [14] |
| LDCM0482 | CL88 | HCT 116 | C142(0.91); C103(0.89); C153(0.92); C205(1.09) | LDD0799 | [14] |
| LDCM0483 | CL89 | HCT 116 | C142(1.26); C103(1.16); C153(1.17); C205(1.21) | LDD0800 | [14] |
| LDCM0484 | CL9 | HCT 116 | C142(0.96); C103(0.95); C153(0.96); C205(1.04) | LDD0801 | [14] |
| LDCM0485 | CL90 | HCT 116 | C142(1.03); C103(1.16); C153(0.96); C205(1.19) | LDD0802 | [14] |
| LDCM0486 | CL91 | HCT 116 | C142(0.80); C103(0.92); C153(0.83); C205(0.76) | LDD0803 | [14] |
| LDCM0487 | CL92 | HCT 116 | C142(0.85); C103(0.93); C153(0.87); C205(0.85) | LDD0804 | [14] |
| LDCM0488 | CL93 | HCT 116 | C142(0.95); C103(0.99); C153(0.90); C205(1.02) | LDD0805 | [14] |
| LDCM0489 | CL94 | HCT 116 | C142(0.80); C103(0.85); C153(0.87); C205(0.81) | LDD0806 | [14] |
| LDCM0490 | CL95 | HCT 116 | C142(0.78); C103(0.87); C153(0.79); C205(0.83) | LDD0807 | [14] |
| LDCM0491 | CL96 | HCT 116 | C142(0.71); C103(0.94); C153(0.84); C205(0.82) | LDD0808 | [14] |
| LDCM0492 | CL97 | HCT 116 | C142(0.81); C103(0.84); C153(0.87); C205(0.84) | LDD0809 | [14] |
| LDCM0493 | CL98 | HCT 116 | C142(0.76); C103(0.89); C153(0.83); C205(0.79) | LDD0810 | [14] |
| LDCM0494 | CL99 | HCT 116 | C142(0.81); C103(0.94); C153(0.82); C205(0.78) | LDD0811 | [14] |
| LDCM0185 | Compound 17 | HEK-293T | 4.14 | LDD0512 | [31] |
| LDCM0191 | Compound 21 | HEK-293T | 4.05 | LDD0508 | [31] |
| LDCM0190 | Compound 34 | HEK-293T | 3.86 | LDD0510 | [31] |
| LDCM0192 | Compound 35 | HEK-293T | 3.08 | LDD0509 | [31] |
| LDCM0193 | Compound 36 | HEK-293T | 3.63 | LDD0511 | [31] |
| LDCM0495 | E2913 | HEK-293T | C706(0.94); C620(1.01); C806(0.78); C205(0.92) | LDD1698 | [36] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C467(12.35); C205(8.33); C386(7.26); C103(3.56) | LDD1702 | [6] |
| LDCM0175 | Ethacrynic acid | HeLa | N.A. | LDD0440 | [12] |
| LDCM0625 | F8 | Ramos | C862(1.15); C467(1.34); C620(1.08); C205(1.42) | LDD2187 | [37] |
| LDCM0572 | Fragment10 | Ramos | C862(9.40); C467(8.34); C620(1.28); C205(1.77) | LDD2189 | [37] |
| LDCM0573 | Fragment11 | Ramos | C862(0.20); C467(0.55); C205(0.63); C683(20.00) | LDD2190 | [37] |
| LDCM0574 | Fragment12 | Ramos | C862(5.63); C467(7.54); C620(1.34); C683(1.81) | LDD2191 | [37] |
| LDCM0575 | Fragment13 | Ramos | C862(1.00); C467(1.39); C620(1.05); C205(0.57) | LDD2192 | [37] |
| LDCM0576 | Fragment14 | Ramos | C862(6.49); C467(8.66); C620(3.44); C205(11.30) | LDD2193 | [37] |
| LDCM0579 | Fragment20 | Ramos | C862(20.00); C467(14.01); C620(2.85); C103(20.00) | LDD2194 | [37] |
| LDCM0580 | Fragment21 | Ramos | C862(1.18); C467(1.93); C620(0.86); C205(1.23) | LDD2195 | [37] |
| LDCM0582 | Fragment23 | Ramos | C862(2.52); C467(1.11); C620(3.08); C103(2.78) | LDD2196 | [37] |
| LDCM0578 | Fragment27 | Ramos | C862(0.85); C467(1.01); C620(0.97); C205(0.61) | LDD2197 | [37] |
| LDCM0586 | Fragment28 | Ramos | C862(0.81); C467(1.05); C620(0.77); C205(1.93) | LDD2198 | [37] |
| LDCM0588 | Fragment30 | Ramos | C862(1.35); C467(1.30); C620(1.39); C205(0.19) | LDD2199 | [37] |
| LDCM0589 | Fragment31 | Ramos | C862(1.36); C467(2.01); C620(1.21); C205(1.86) | LDD2200 | [37] |
| LDCM0590 | Fragment32 | Ramos | C862(2.89); C467(3.64); C620(0.76); C205(1.26) | LDD2201 | [37] |
| LDCM0468 | Fragment33 | HCT 116 | C142(0.90); C103(0.68); C153(0.98); C205(0.76) | LDD0785 | [14] |
| LDCM0596 | Fragment38 | Ramos | C862(3.18); C467(3.91); C620(1.20); C205(0.51) | LDD2203 | [37] |
| LDCM0566 | Fragment4 | Ramos | C862(5.50); C467(5.25); C620(1.07); C205(3.29) | LDD2184 | [37] |
| LDCM0427 | Fragment51 | HCT 116 | C142(0.91); C103(1.12); C153(0.91); C205(1.17) | LDD0744 | [14] |
| LDCM0610 | Fragment52 | Ramos | C862(1.31); C467(2.11); C620(1.41); C205(0.24) | LDD2204 | [37] |
| LDCM0614 | Fragment56 | Ramos | C862(0.97); C467(1.22); C620(1.78); C205(0.32) | LDD2205 | [37] |
| LDCM0569 | Fragment7 | Ramos | C862(3.82); C467(4.83); C620(1.26); C205(2.66) | LDD2186 | [37] |
| LDCM0571 | Fragment9 | Ramos | C862(17.46); C467(20.00); C620(2.70); C103(20.00) | LDD2188 | [37] |
| LDCM0116 | HHS-0101 | DM93 | Y51(0.74); Y788(0.75) | LDD0264 | [13] |
| LDCM0117 | HHS-0201 | DM93 | Y51(0.76); Y788(0.88) | LDD0265 | [13] |
| LDCM0118 | HHS-0301 | DM93 | Y51(0.61); Y788(0.88) | LDD0266 | [13] |
| LDCM0119 | HHS-0401 | DM93 | Y51(0.77); Y788(0.99) | LDD0267 | [13] |
| LDCM0120 | HHS-0701 | DM93 | Y788(0.67); Y51(0.83) | LDD0268 | [13] |
| LDCM0015 | HNE | MDA-MB-231 | C103(1.24); C862(1.66); C164(0.88) | LDD0346 | [37] |
| LDCM0107 | IAA | HeLa | C103(0.00); H87(0.00); C153(0.00) | LDD0221 | [26] |
| LDCM0179 | JZ128 | PC-3 | C285(0.00); C790(0.00); C142(0.00); C103(0.00) | LDD0462 | [9] |
| LDCM0022 | KB02 | HCT 116 | C142(9.92); C153(9.92); C683(2.25); C467(2.79) | LDD0080 | [14] |
| LDCM0023 | KB03 | HCT 116 | C142(14.44); C153(14.44); C683(2.86); C467(3.49) | LDD0081 | [14] |
| LDCM0024 | KB05 | HCT 116 | C142(20.00); C153(20.00); C683(3.49); C467(2.56) | LDD0082 | [14] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C103(0.71) | LDD2121 | [6] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [26] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C620(1.13); C103(0.44) | LDD2089 | [6] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C806(1.23) | LDD2090 | [6] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C806(0.85) | LDD2092 | [6] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C467(1.36) | LDD2093 | [6] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C806(0.79) | LDD2094 | [6] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C205(1.04); C806(1.30) | LDD2098 | [6] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C467(1.13); C103(1.02) | LDD2099 | [6] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C806(2.25) | LDD2100 | [6] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C706(1.32) | LDD2101 | [6] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C620(0.58) | LDD2104 | [6] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C205(0.92) | LDD2105 | [6] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C467(0.97); C620(1.01); C103(1.01); C205(1.01) | LDD2107 | [6] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C806(1.05) | LDD2108 | [6] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C467(1.14); C103(1.01); C205(1.28) | LDD2109 | [6] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C467(1.45); C103(1.02) | LDD2111 | [6] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C620(0.51); C806(0.90) | LDD2114 | [6] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C103(0.47); C706(0.76) | LDD2115 | [6] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C205(0.60); C862(0.29) | LDD2118 | [6] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C467(1.39); C103(2.19) | LDD2119 | [6] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C706(0.83) | LDD2122 | [6] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C205(0.74); C806(0.75); C386(1.22) | LDD2123 | [6] |
| LDCM0531 | Nucleophilic fragment 28b | MDA-MB-231 | C205(1.05) | LDD2124 | [6] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C467(0.91); C103(0.86); C205(0.79); C706(0.79) | LDD2125 | [6] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C467(1.12); C205(1.21); C706(0.97) | LDD2127 | [6] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C467(1.21) | LDD2129 | [6] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C467(1.34); C103(1.01) | LDD2135 | [6] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C467(1.71); C103(0.99); C706(1.14) | LDD2136 | [6] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C467(0.95); C103(0.90); C205(0.98); C706(0.74) | LDD2137 | [6] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C467(0.96); C103(0.68) | LDD2140 | [6] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C806(0.95) | LDD2141 | [6] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C467(2.00) | LDD2144 | [6] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C467(1.01); C103(1.24); C205(0.96) | LDD2146 | [6] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C467(1.12); C806(0.58); C706(0.61) | LDD2148 | [6] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C620(0.95); C103(0.49) | LDD2150 | [6] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C683(0.99); C683(0.88); C467(0.77); C153(0.55) | LDD2206 | [38] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C862(1.33); C205(1.10); C103(1.04); C164(1.00) | LDD2207 | [38] |
| LDCM0131 | RA190 | MM1.R | C142(1.42); C153(1.42) | LDD0304 | [39] |
| LDCM0021 | THZ1 | HeLa S3 | C103(1.10); C285(1.08); C790(1.06); C142(1.02) | LDD0460 | [9] |
The Interaction Atlas With This Target
References
