Details of the Target
General Information of Target
| Target ID | LDTP03608 | |||||
|---|---|---|---|---|---|---|
| Target Name | RAC-beta serine/threonine-protein kinase (AKT2) | |||||
| Gene Name | AKT2 | |||||
| Gene ID | 208 | |||||
| Synonyms |
RAC-beta serine/threonine-protein kinase; EC 2.7.11.1; Protein kinase Akt-2; Protein kinase B beta; PKB beta; RAC-PK-beta |
|||||
| 3D Structure | ||||||
| Sequence |
MNEVSVIKEGWLHKRGEYIKTWRPRYFLLKSDGSFIGYKERPEAPDQTLPPLNNFSVAEC
QLMKTERPRPNTFVIRCLQWTTVIERTFHVDSPDEREEWMRAIQMVANSLKQRAPGEDPM DYKCGSPSDSSTTEEMEVAVSKARAKVTMNDFDYLKLLGKGTFGKVILVREKATGRYYAM KILRKEVIIAKDEVAHTVTESRVLQNTRHPFLTALKYAFQTHDRLCFVMEYANGGELFFH LSRERVFTEERARFYGAEIVSALEYLHSRDVVYRDIKLENLMLDKDGHIKITDFGLCKEG ISDGATMKTFCGTPEYLAPEVLEDNDYGRAVDWWGLGVVMYEMMCGRLPFYNQDHERLFE LILMEEIRFPRTLSPEAKSLLAGLLKKDPKQRLGGGPSDAKEVMEHRFFLSINWQDVVQK KLLPPFKPQVTSEVDTRYFDDEFTAQSITITPPDRYDSLGLLELDQRTHFPQFSYSASIR E |
|||||
| Target Type |
Literature-reported
|
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Protein kinase superfamily, AGC Ser/Thr protein kinase family, RAC subfamily
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
AKT2 is one of 3 closely related serine/threonine-protein kinases (AKT1, AKT2 and AKT3) called the AKT kinase, and which regulate many processes including metabolism, proliferation, cell survival, growth and angiogenesis. This is mediated through serine and/or threonine phosphorylation of a range of downstream substrates. Over 100 substrate candidates have been reported so far, but for most of them, no isoform specificity has been reported. AKT is responsible of the regulation of glucose uptake by mediating insulin-induced translocation of the SLC2A4/GLUT4 glucose transporter to the cell surface. Phosphorylation of PTPN1 at 'Ser-50' negatively modulates its phosphatase activity preventing dephosphorylation of the insulin receptor and the attenuation of insulin signaling. Phosphorylation of TBC1D4 triggers the binding of this effector to inhibitory 14-3-3 proteins, which is required for insulin-stimulated glucose transport. AKT regulates also the storage of glucose in the form of glycogen by phosphorylating GSK3A at 'Ser-21' and GSK3B at 'Ser-9', resulting in inhibition of its kinase activity. Phosphorylation of GSK3 isoforms by AKT is also thought to be one mechanism by which cell proliferation is driven. AKT regulates also cell survival via the phosphorylation of MAP3K5 (apoptosis signal-related kinase). Phosphorylation of 'Ser-83' decreases MAP3K5 kinase activity stimulated by oxidative stress and thereby prevents apoptosis. AKT mediates insulin-stimulated protein synthesis by phosphorylating TSC2 at 'Ser-939' and 'Thr-1462', thereby activating mTORC1 signaling and leading to both phosphorylation of 4E-BP1 and in activation of RPS6KB1. AKT is involved in the phosphorylation of members of the FOXO factors (Forkhead family of transcription factors), leading to binding of 14-3-3 proteins and cytoplasmic localization. In particular, FOXO1 is phosphorylated at 'Thr-24', 'Ser-256' and 'Ser-319'. FOXO3 and FOXO4 are phosphorylated on equivalent sites. AKT has an important role in the regulation of NF-kappa-B-dependent gene transcription and positively regulates the activity of CREB1 (cyclic AMP (cAMP)-response element binding protein). The phosphorylation of CREB1 induces the binding of accessory proteins that are necessary for the transcription of pro-survival genes such as BCL2 and MCL1. AKT phosphorylates 'Ser-454' on ATP citrate lyase (ACLY), thereby potentially regulating ACLY activity and fatty acid synthesis. Activates the 3B isoform of cyclic nucleotide phosphodiesterase (PDE3B) via phosphorylation of 'Ser-273', resulting in reduced cyclic AMP levels and inhibition of lipolysis. Phosphorylates PIKFYVE on 'Ser-318', which results in increased PI(3)P-5 activity. The Rho GTPase-activating protein DLC1 is another substrate and its phosphorylation is implicated in the regulation cell proliferation and cell growth. AKT plays a role as key modulator of the AKT-mTOR signaling pathway controlling the tempo of the process of newborn neurons integration during adult neurogenesis, including correct neuron positioning, dendritic development and synapse formation. Signals downstream of phosphatidylinositol 3-kinase (PI(3)K) to mediate the effects of various growth factors such as platelet-derived growth factor (PDGF), epidermal growth factor (EGF), insulin and insulin-like growth factor I (IGF-I). AKT mediates the antiapoptotic effects of IGF-I. Essential for the SPATA13-mediated regulation of cell migration and adhesion assembly and disassembly. May be involved in the regulation of the placental development. Involved in the inhibition of ciliogenesis associated with RAB8-dependent cilia growth.; One of the few specific substrates of AKT2 identified recently is PITX2. Phosphorylation of PITX2 impairs its association with the CCND1 mRNA-stabilizing complex thus shortening the half-life of CCND1. AKT2 seems also to be the principal isoform responsible of the regulation of glucose uptake. Phosphorylates C2CD5 on 'Ser-197' during insulin-stimulated adipocytes. AKT2 is also specifically involved in skeletal muscle differentiation, one of its substrates in this process being ANKRD2. Down-regulation by RNA interference reduces the expression of the phosphorylated form of BAD, resulting in the induction of caspase-dependent apoptosis. Phosphorylates CLK2 on 'Thr-343'.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| HEC1 | SNV: p.Y177H | DBIA Probe Info | |||
| Ishikawa (Heraklio) 02 ER | SNV: p.Y273H | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
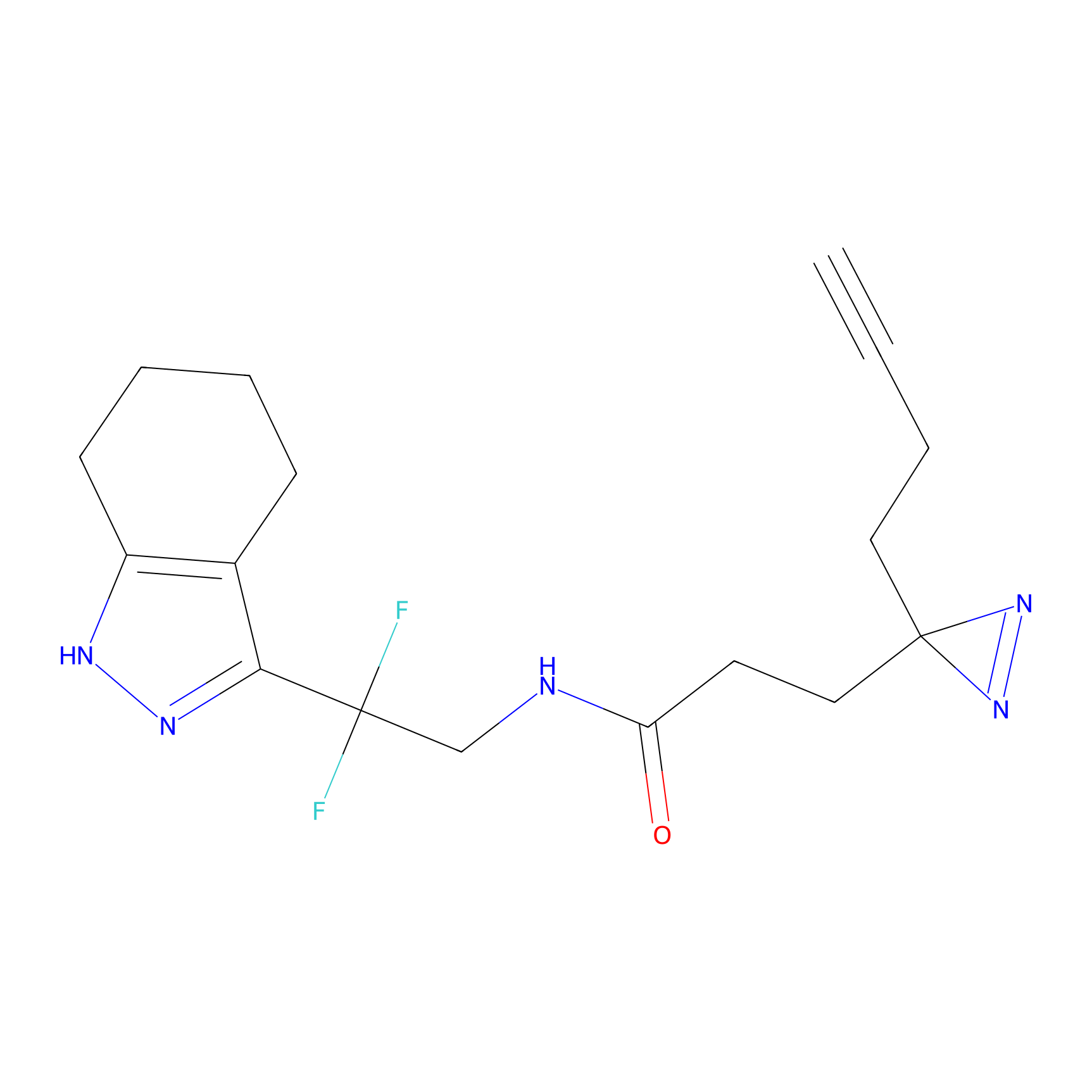
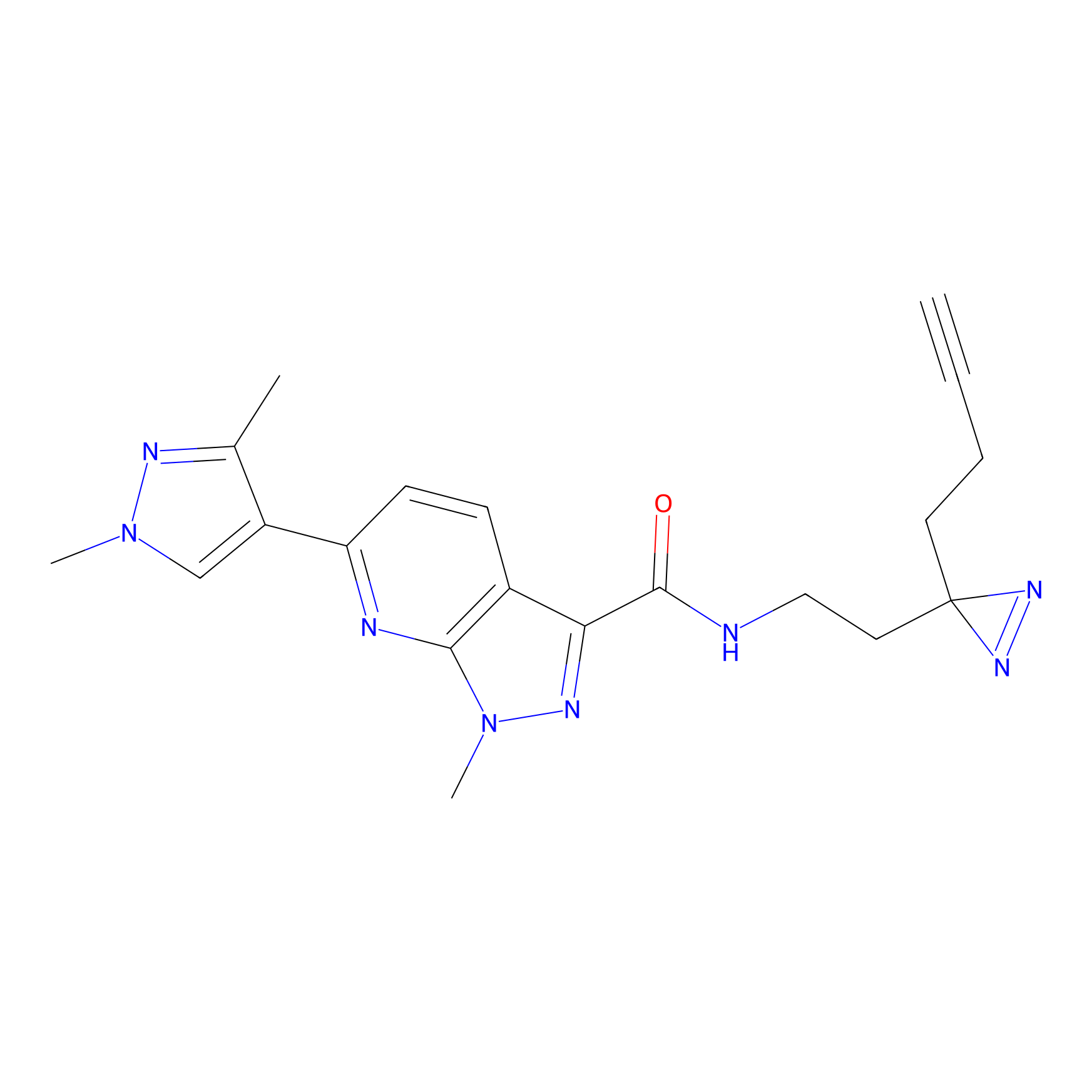
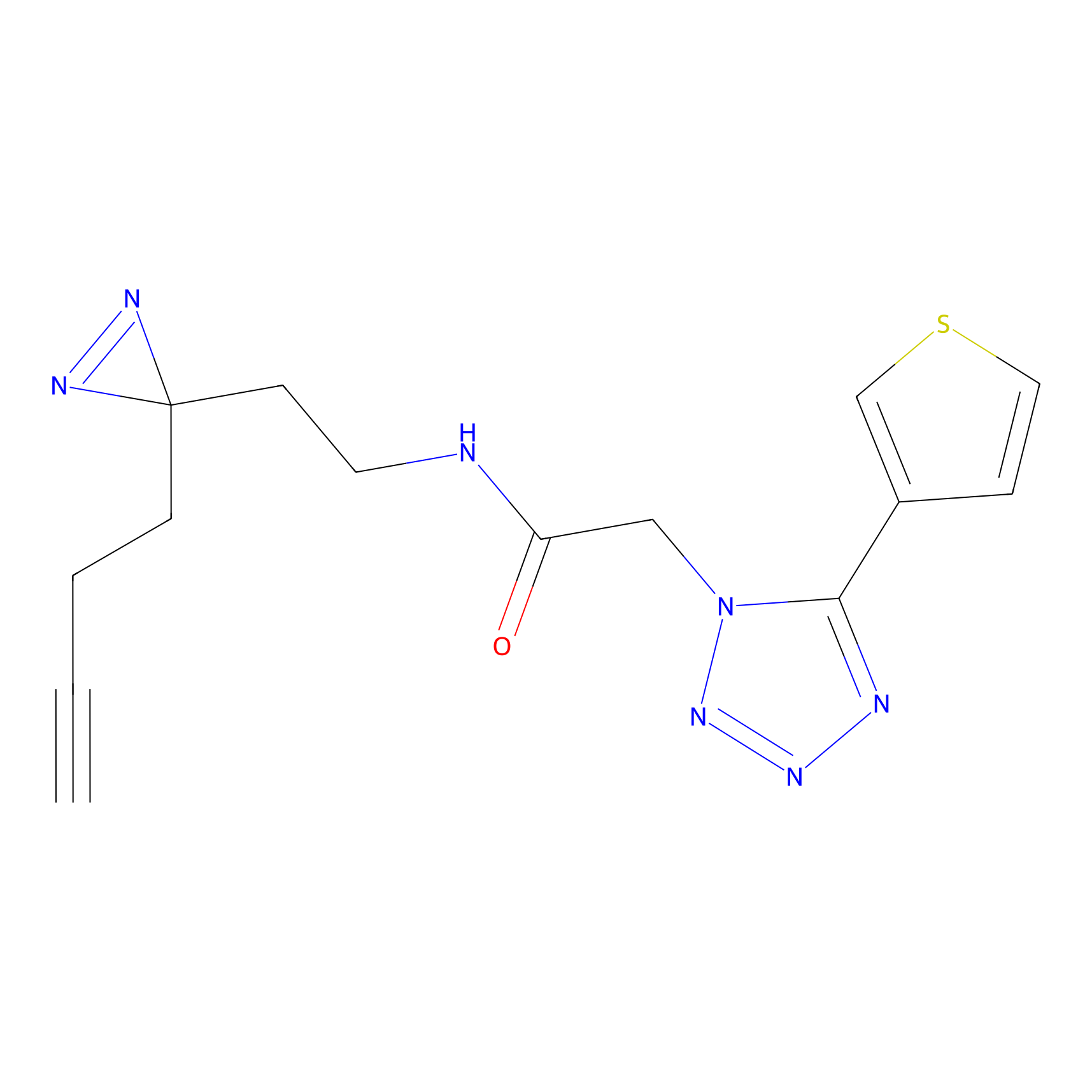
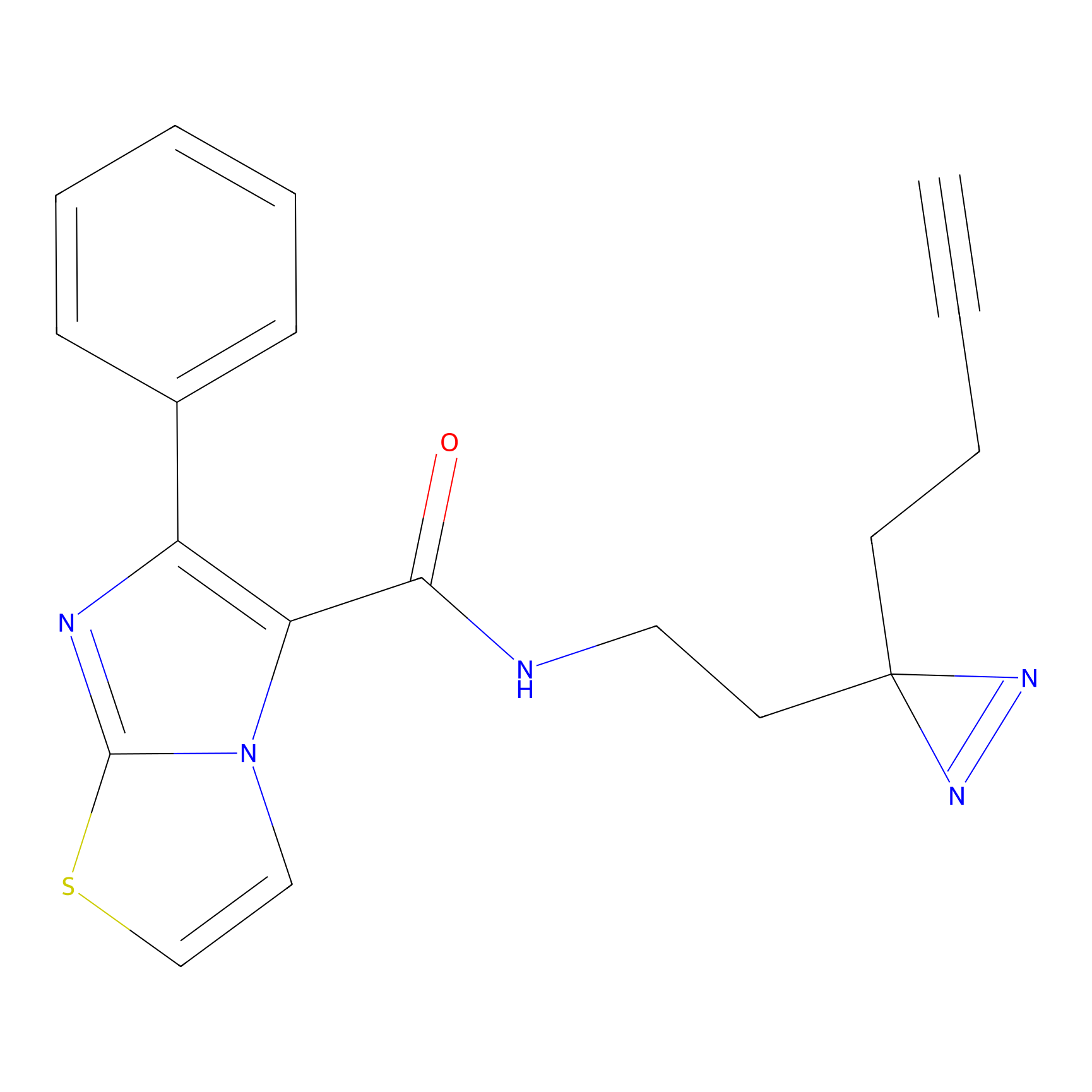
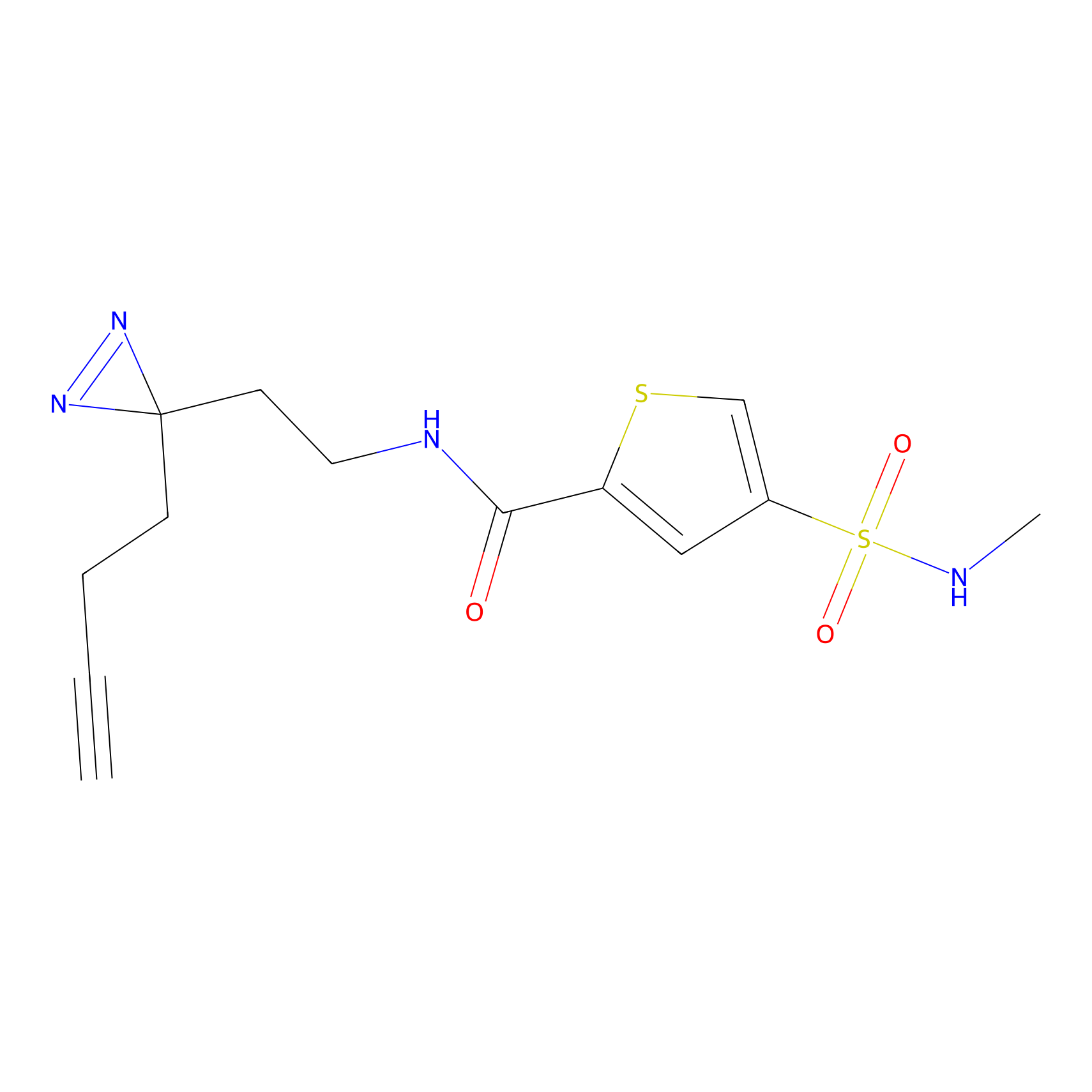
|
m-APA Probe Info |
 |
13.98 | LDD0402 | [1] | |
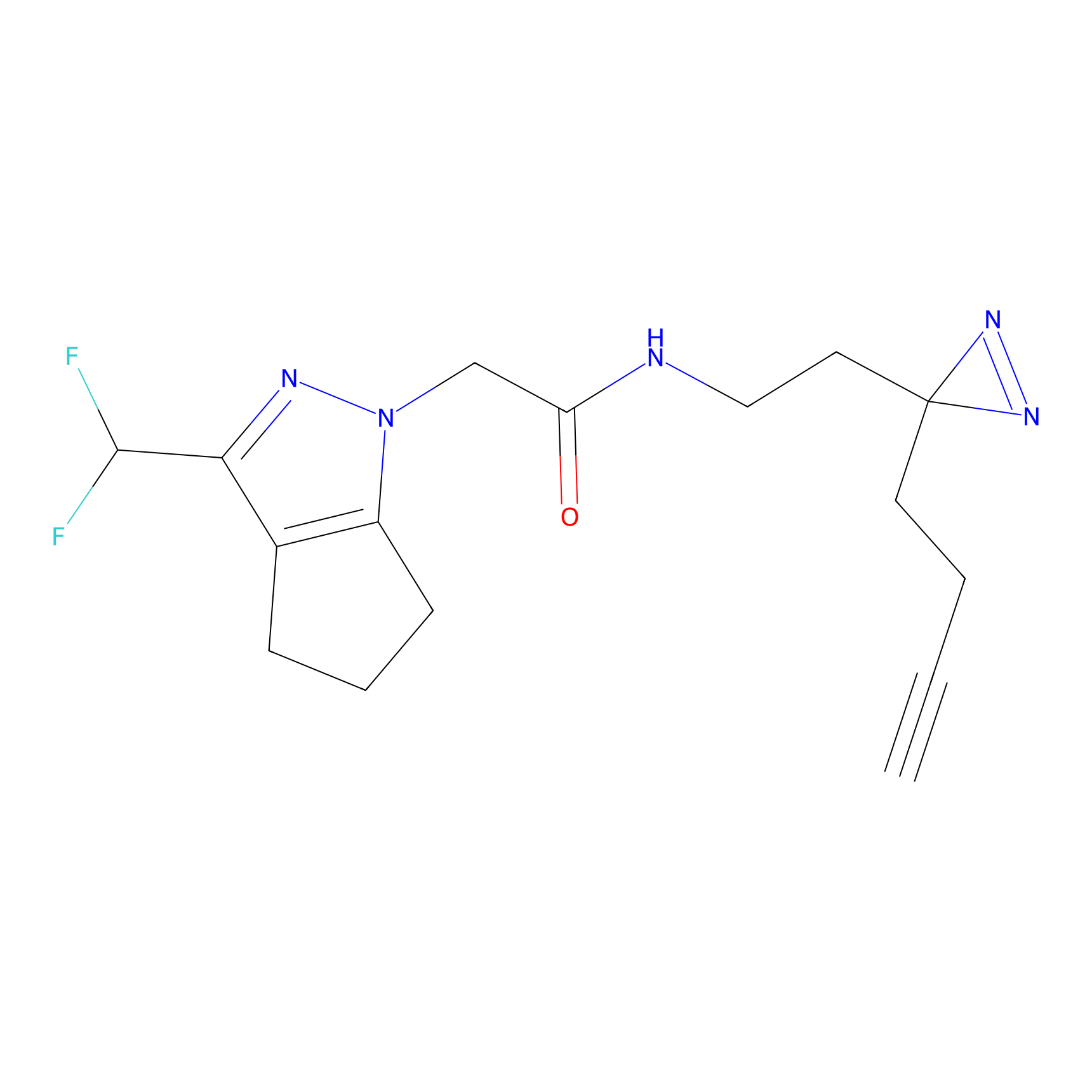
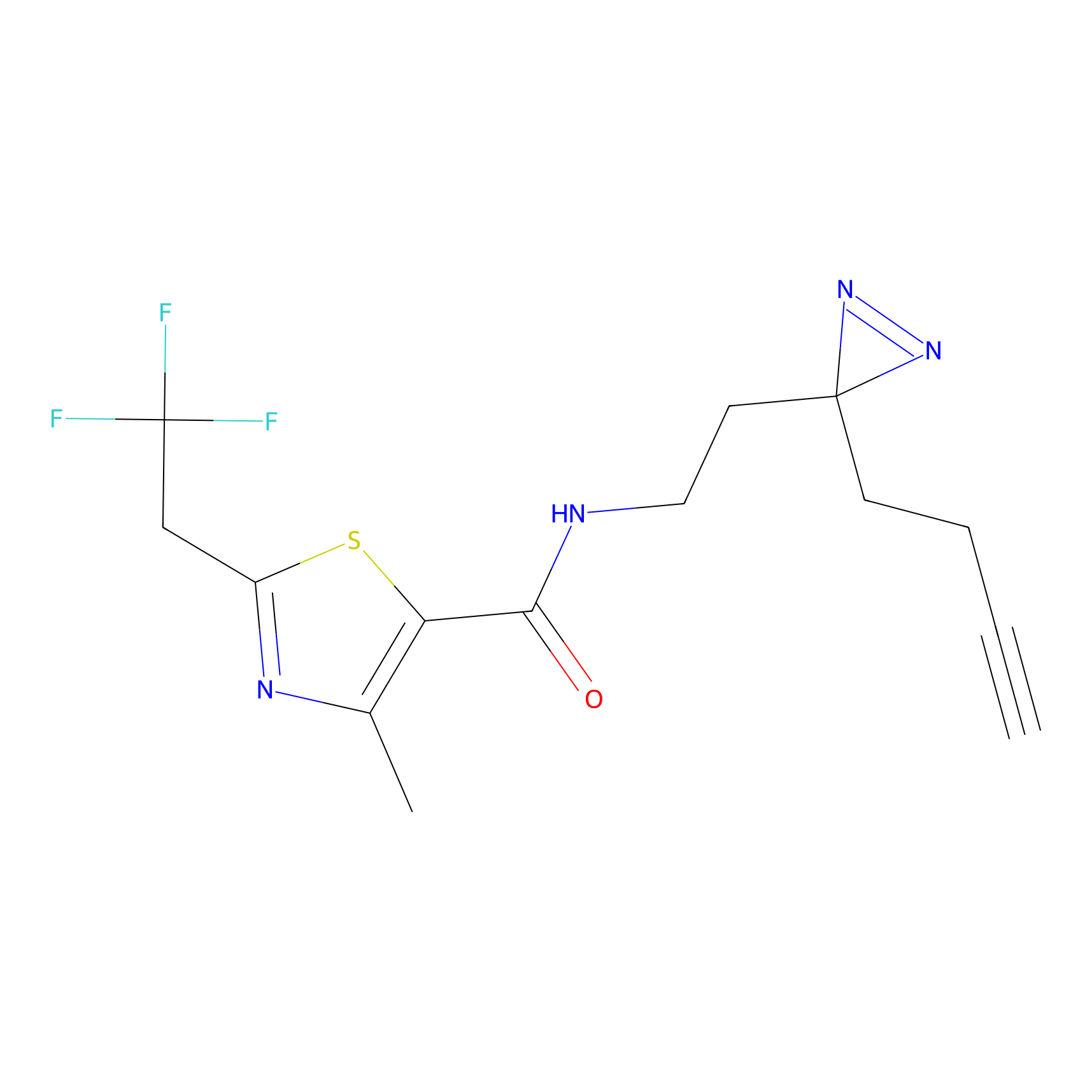
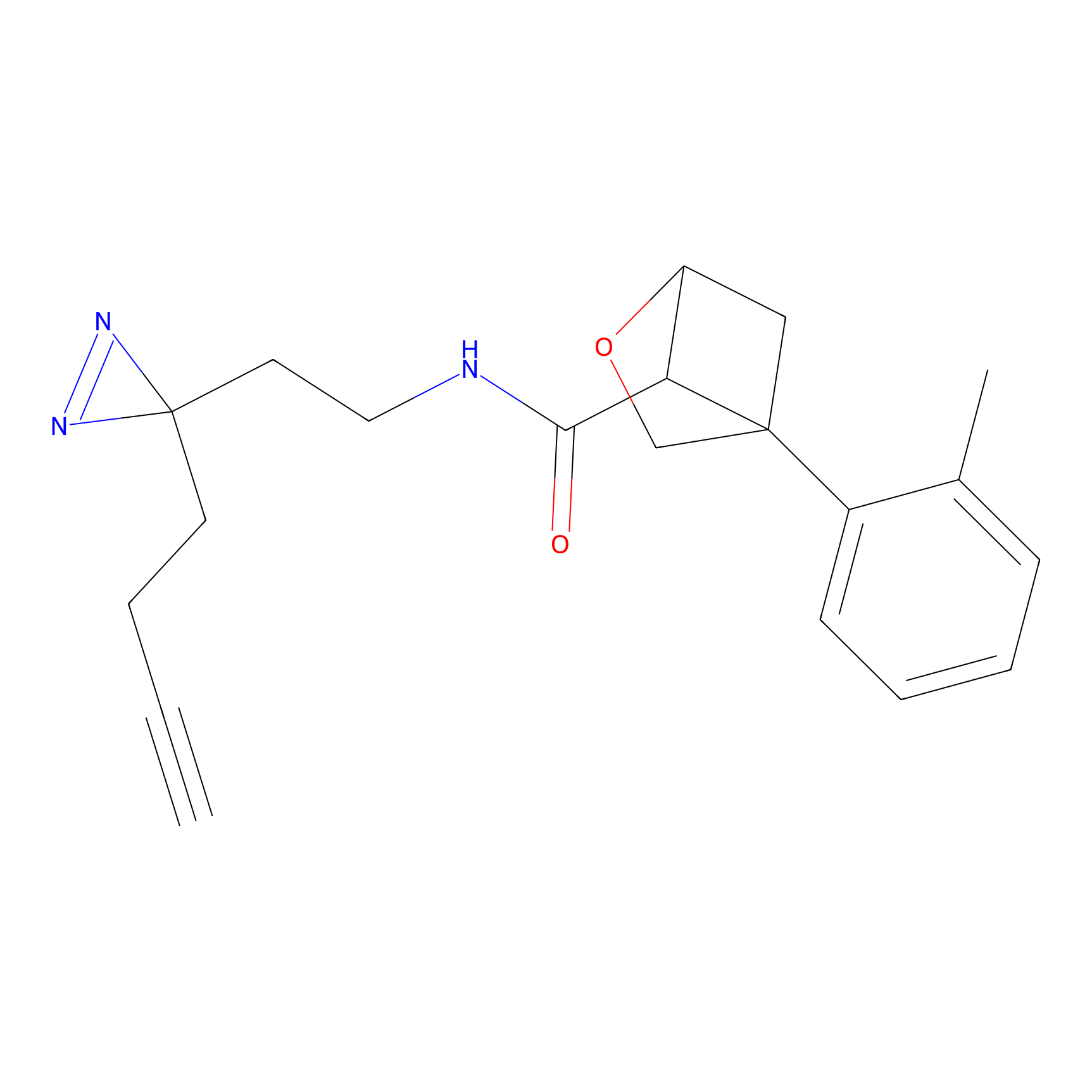
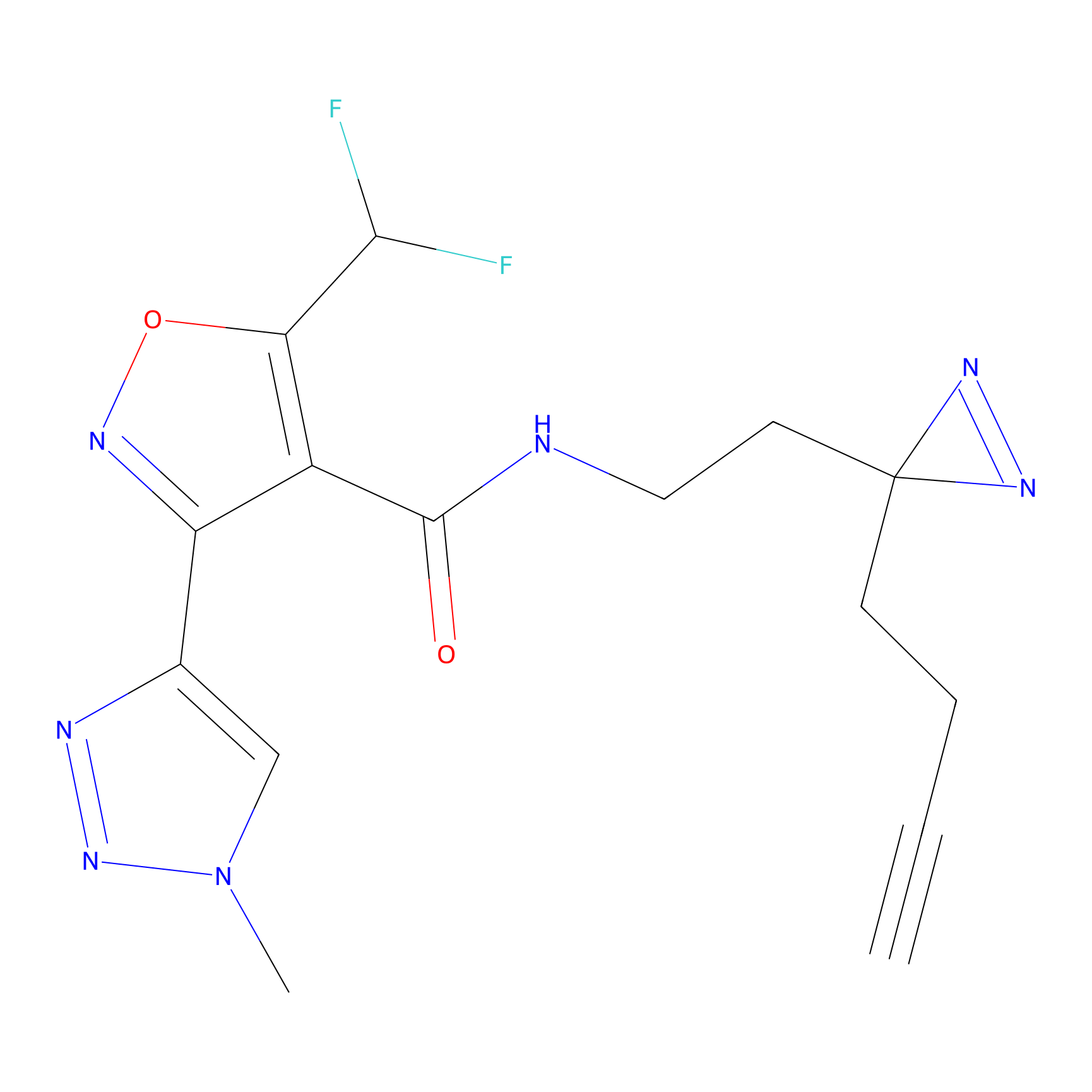
|
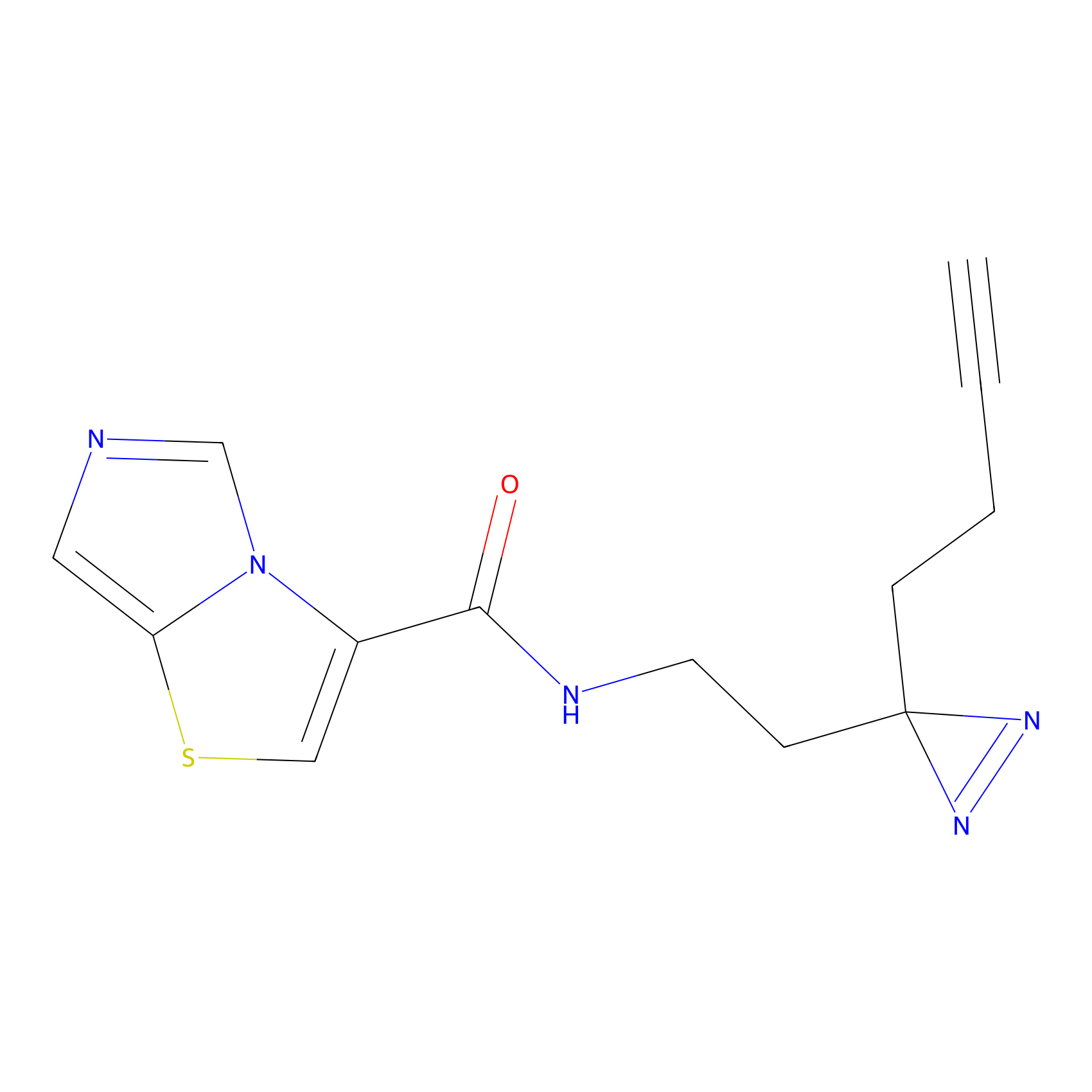
Probe 1 Probe Info |
 |
Y38(18.58); Y122(17.32) | LDD3495 | [2] | |
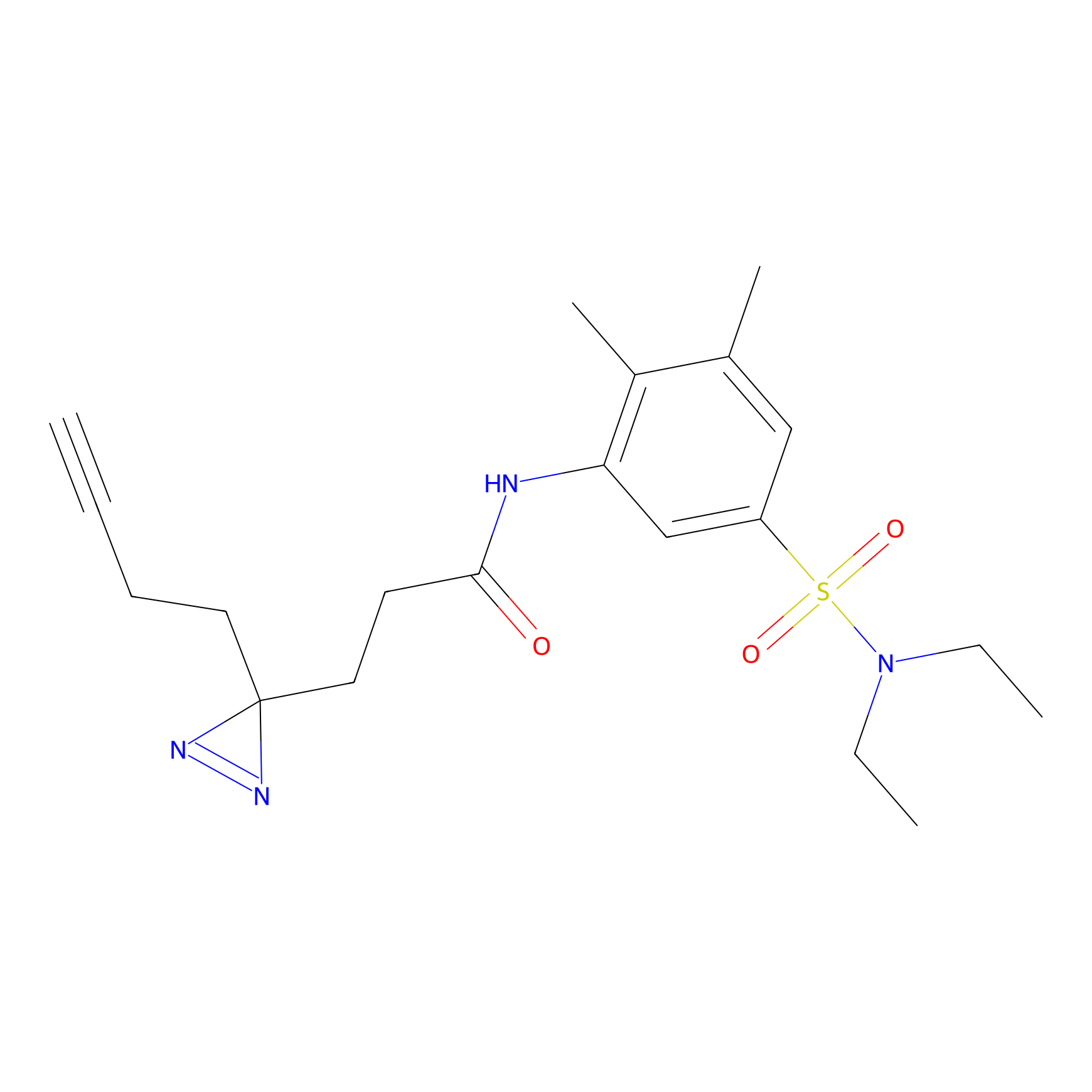
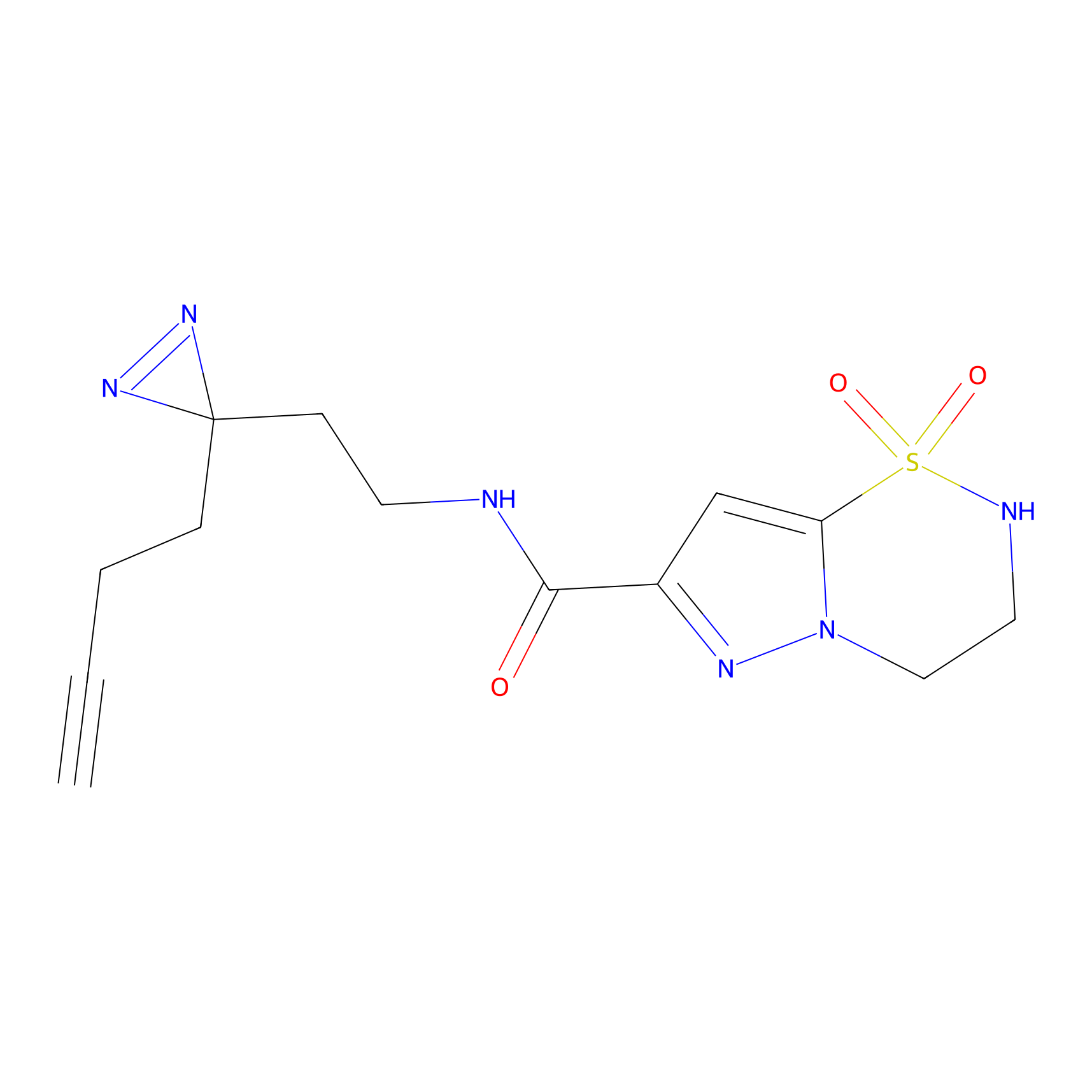
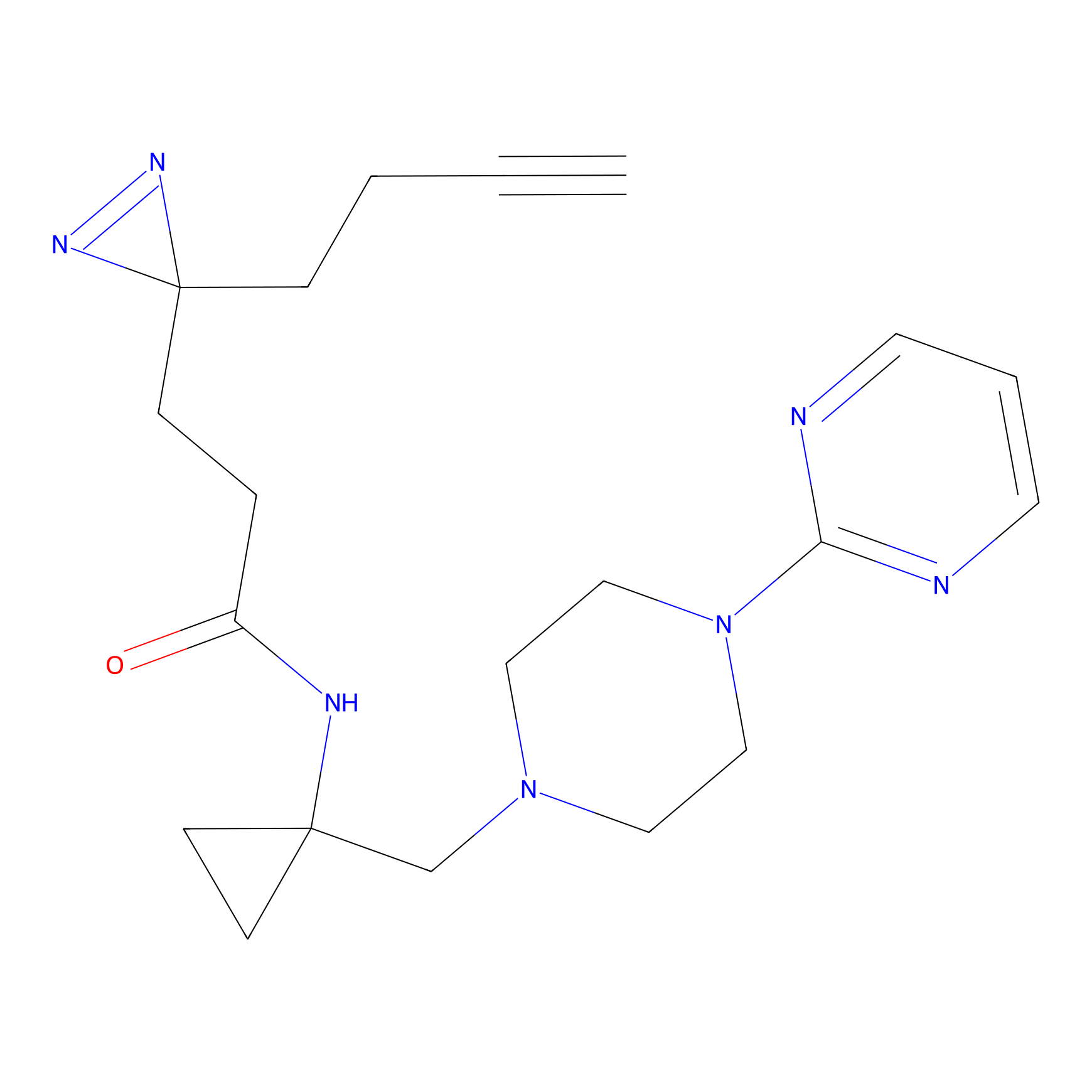
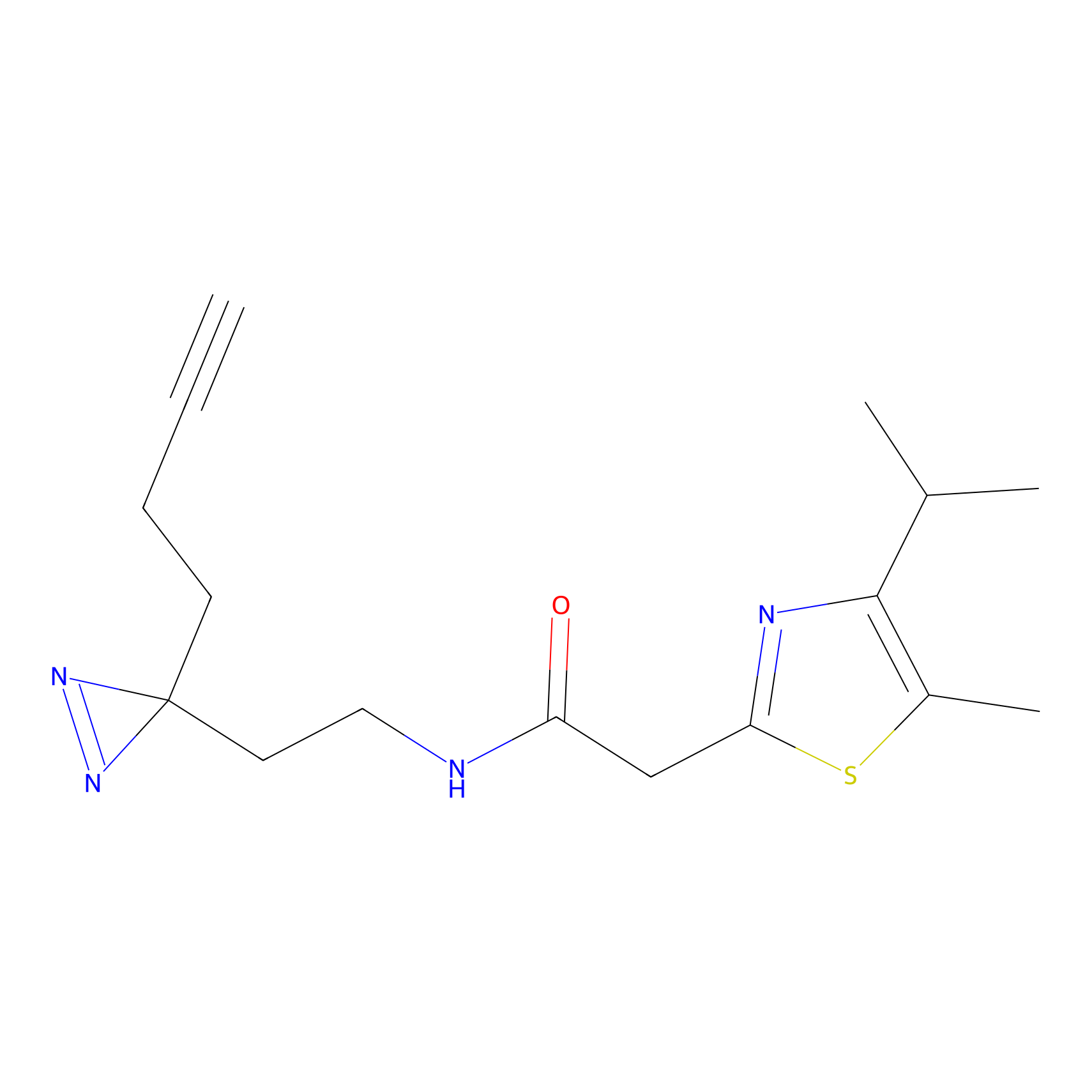
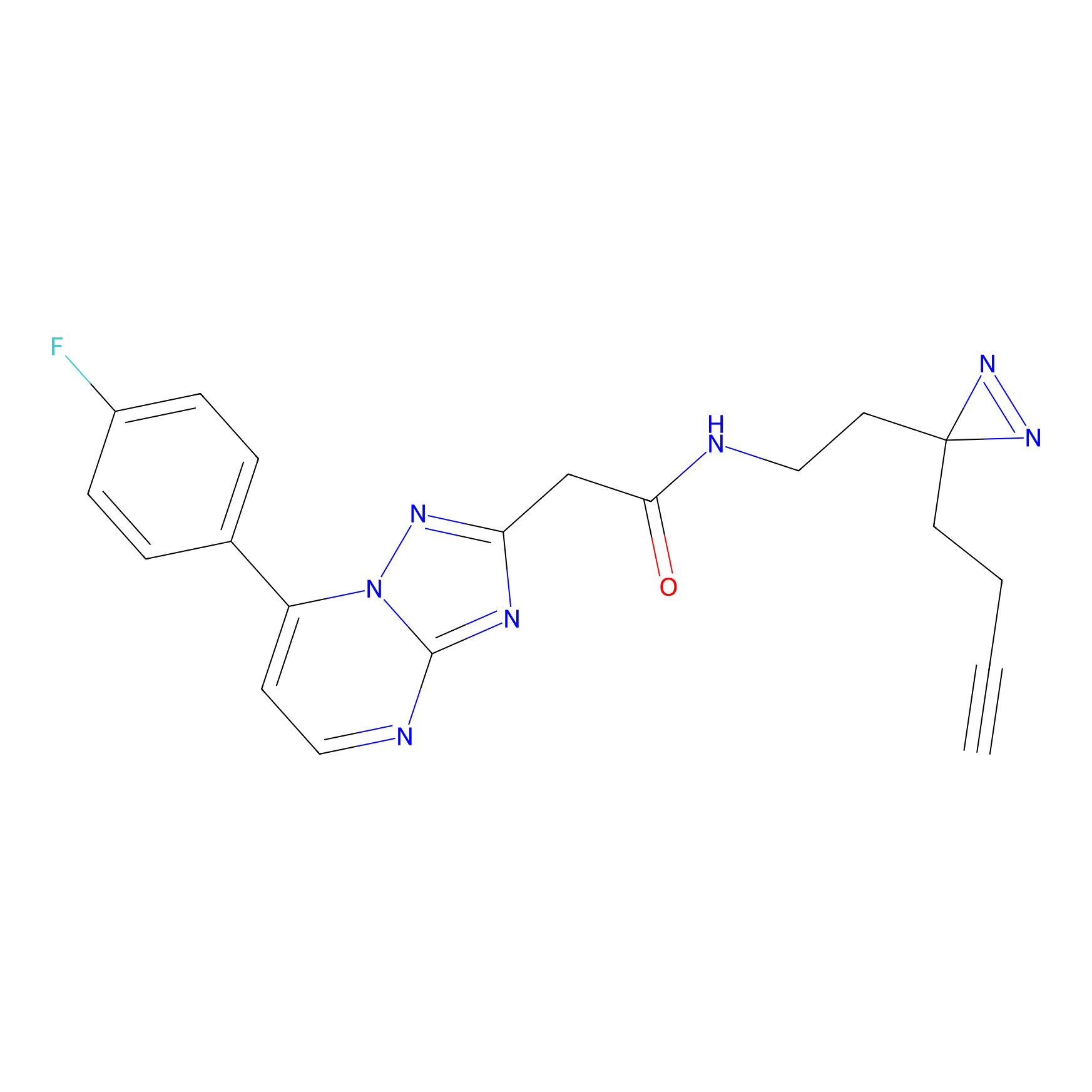
|
AHL-Pu-1 Probe Info |
 |
C311(2.20) | LDD0168 | [3] | |
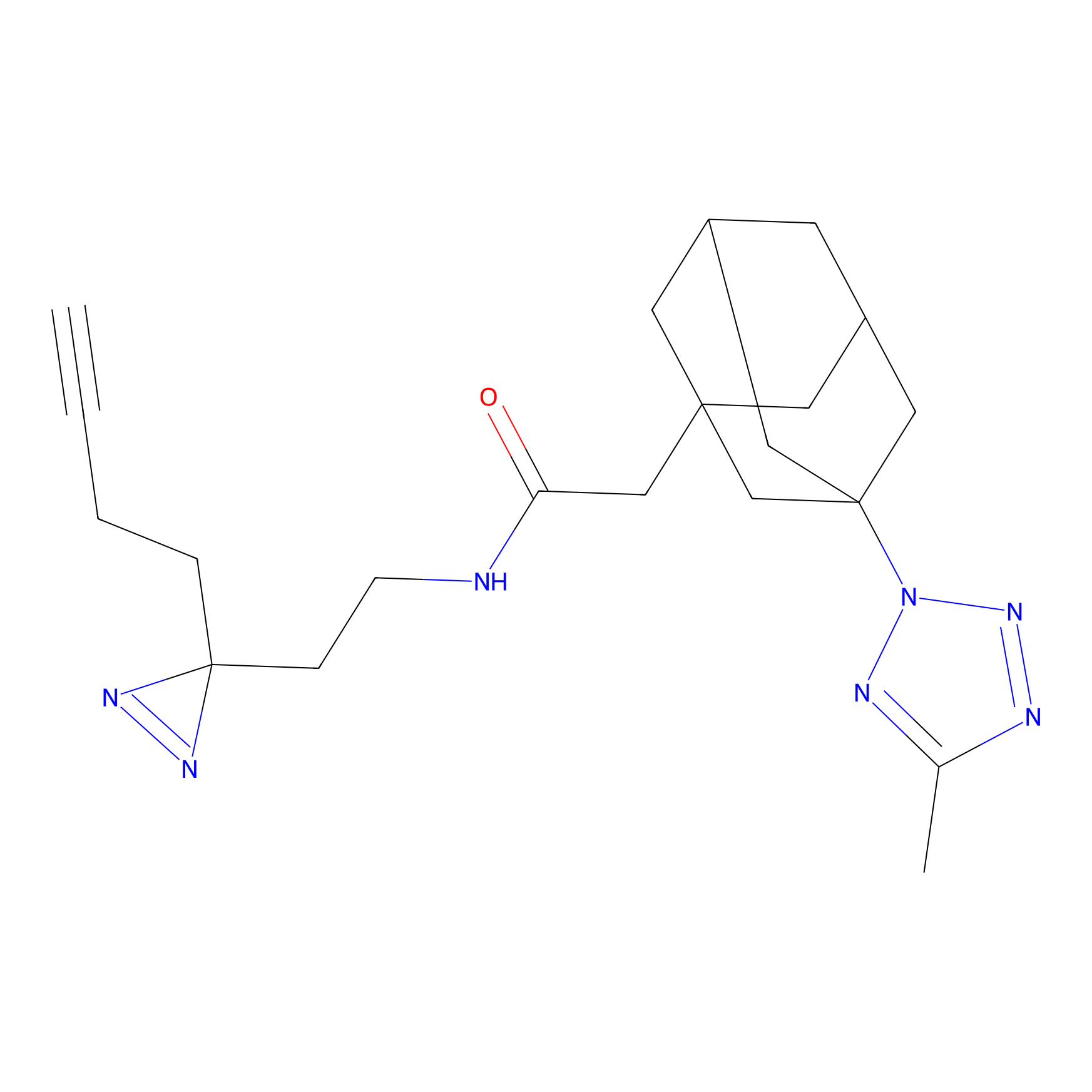
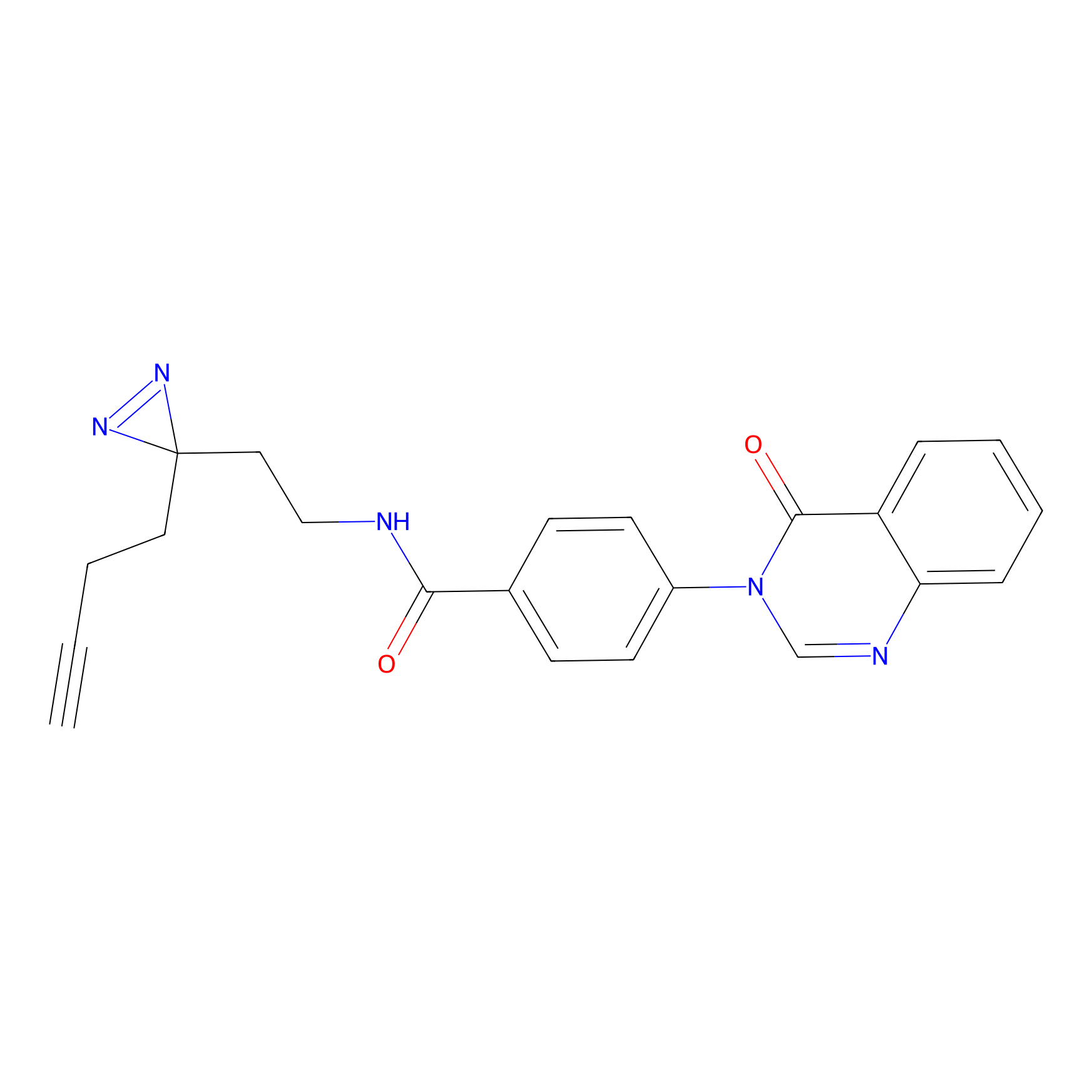
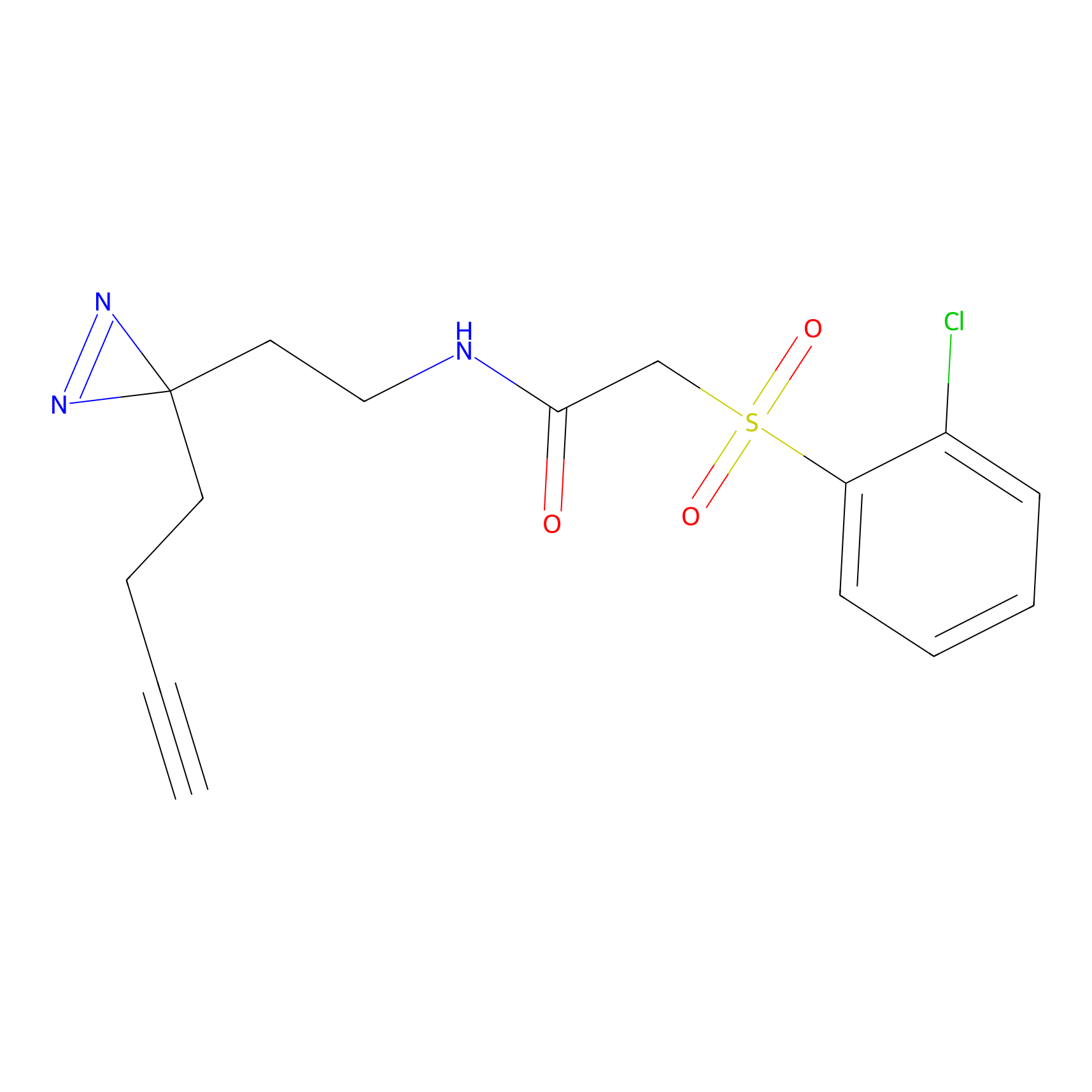
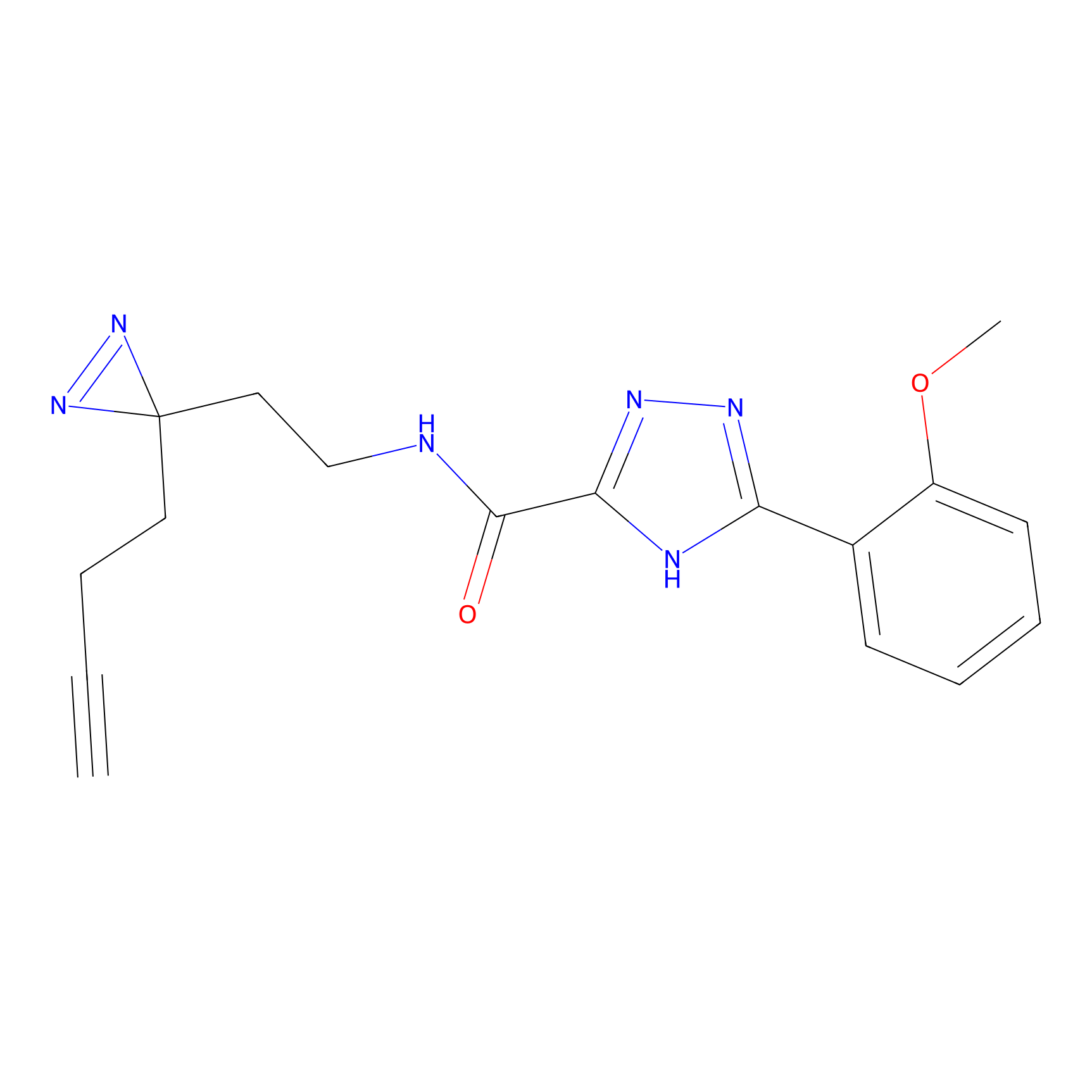
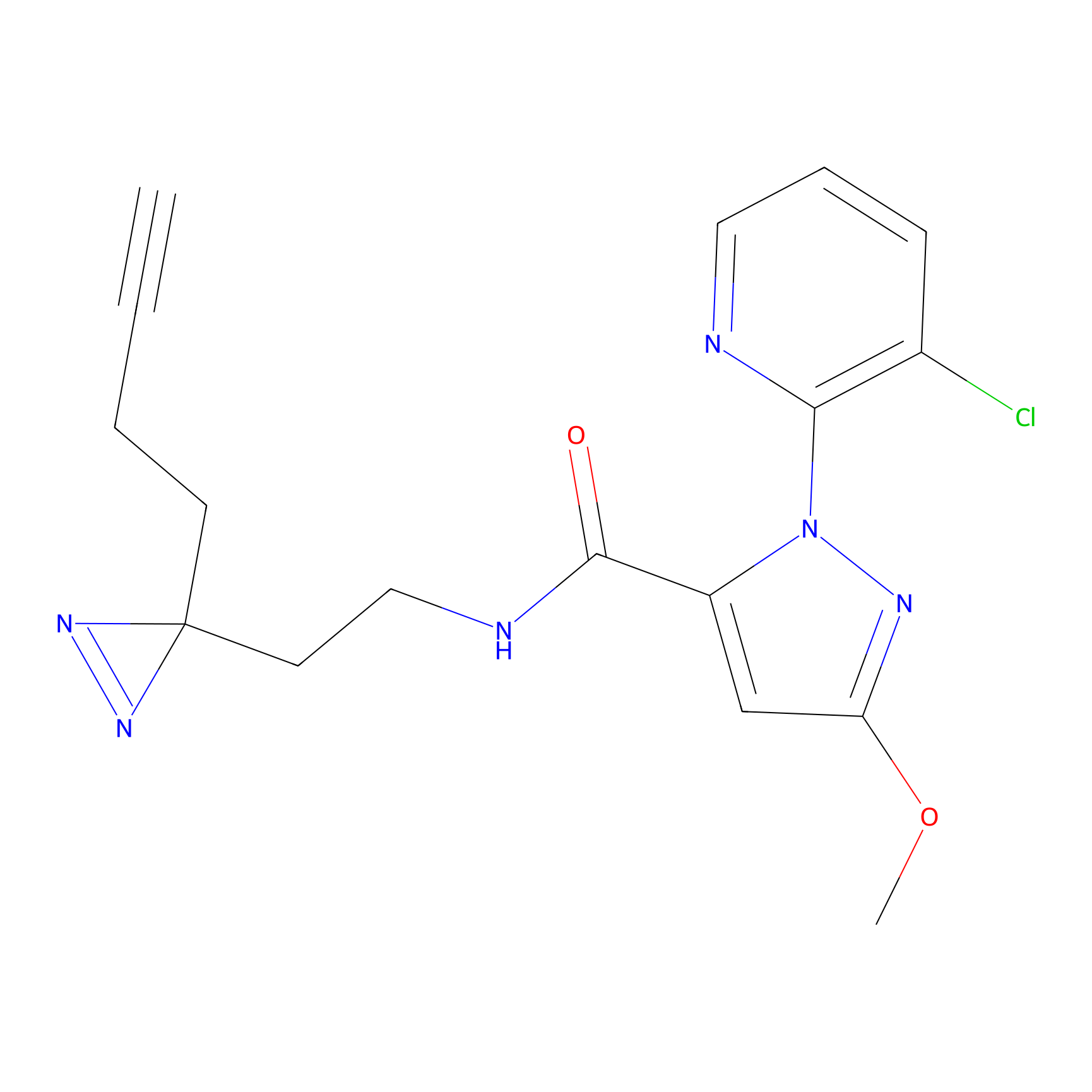
|
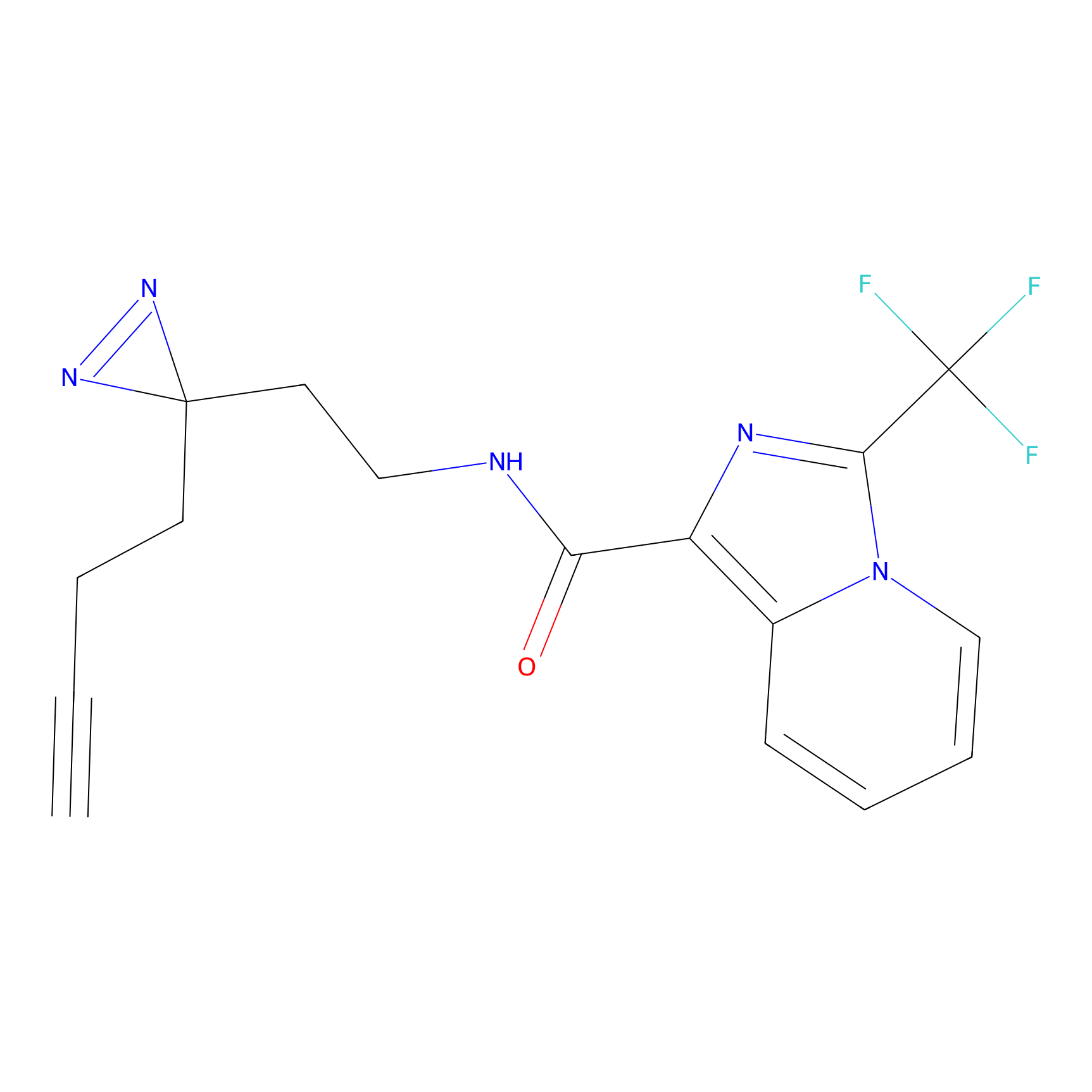
HHS-482 Probe Info |
 |
Y265(0.90); Y327(0.90) | LDD0285 | [4] | |
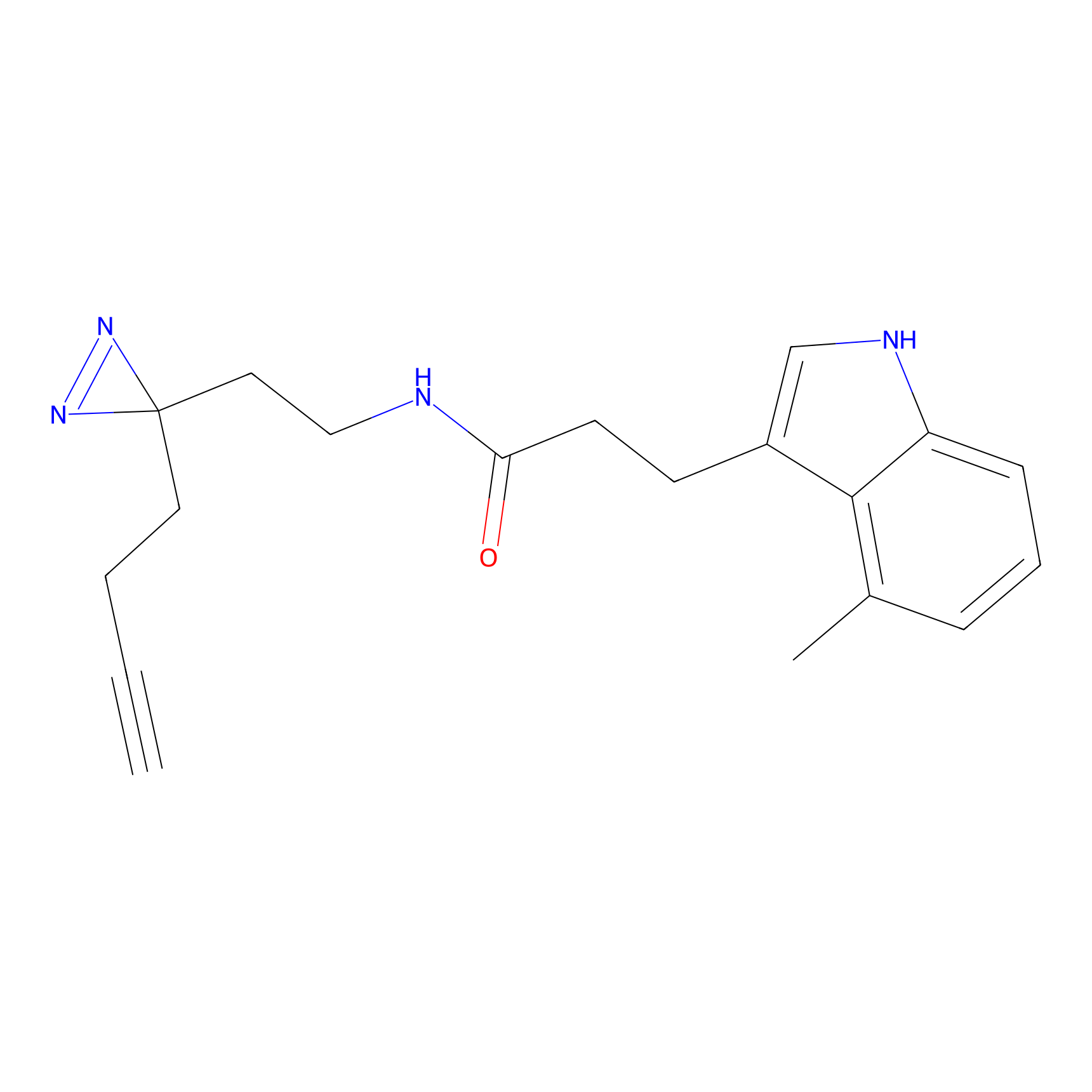
|
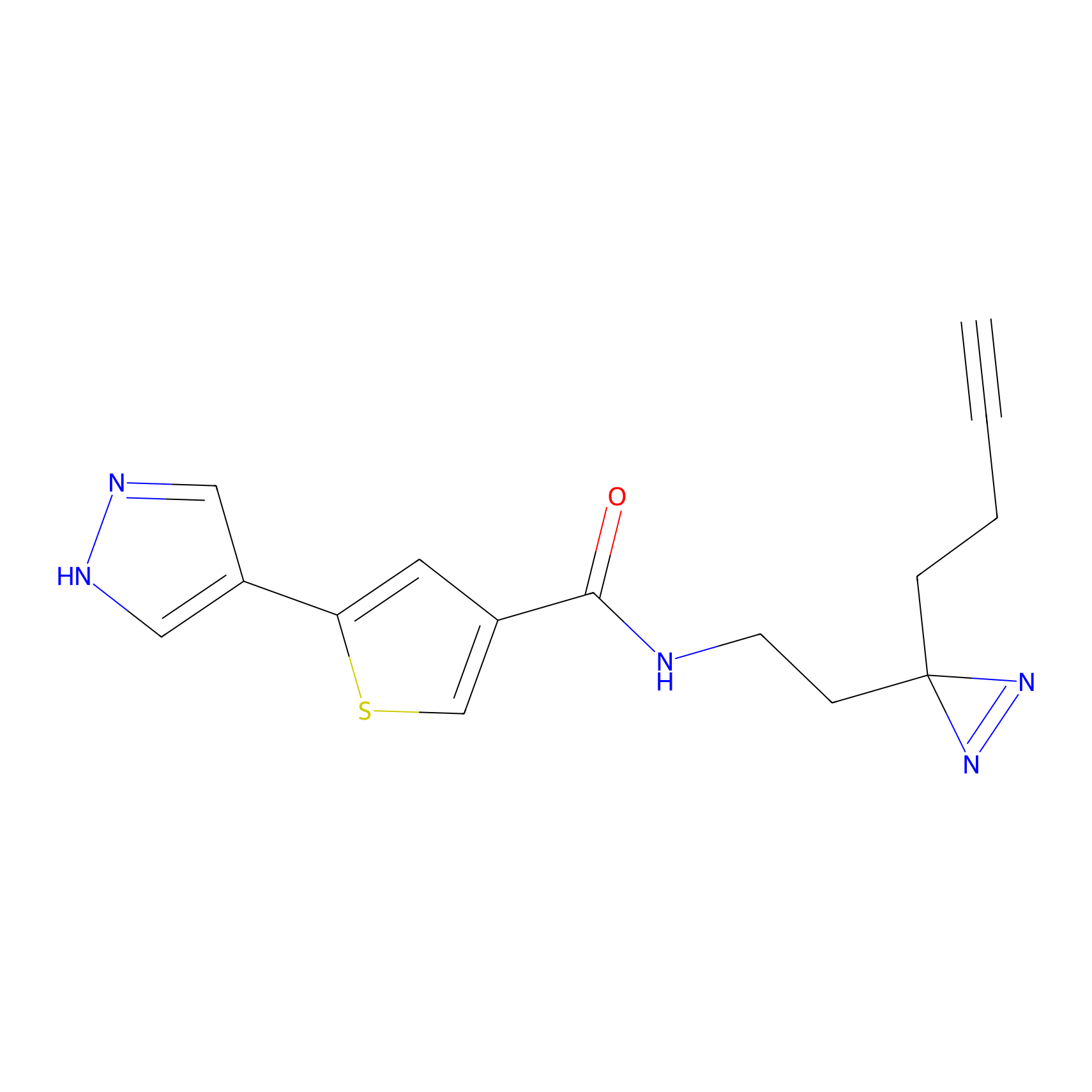
HHS-475 Probe Info |
 |
Y327(0.83); Y265(1.06) | LDD0264 | [5] | |
|
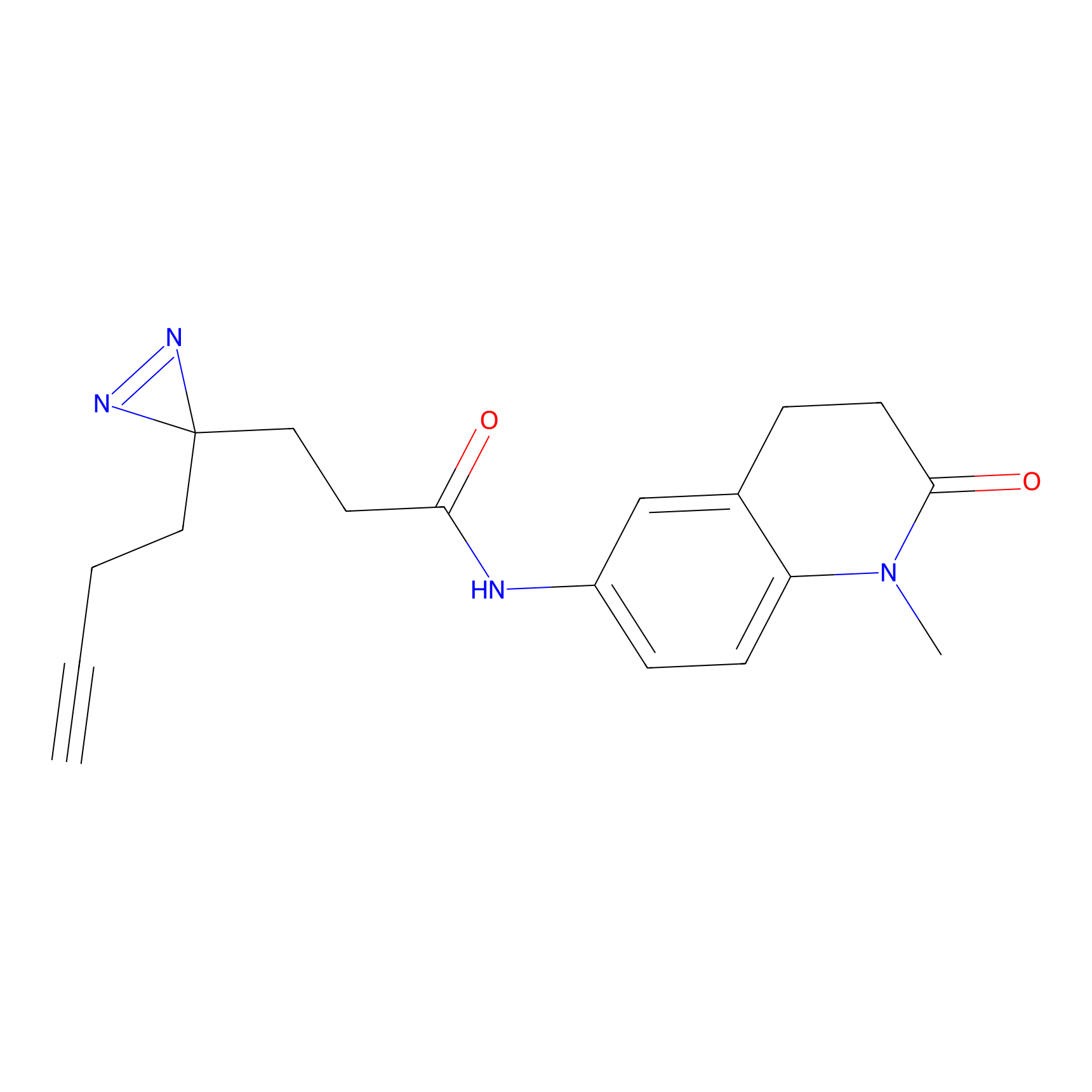
HHS-465 Probe Info |
 |
Y327(10.00) | LDD2237 | [6] | |
|
DBIA Probe Info |
 |
C297(1.01); C311(0.93) | LDD0078 | [7] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [8] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C77(0.00); C60(0.00); C297(0.00); C311(0.00) | LDD0038 | [9] | |
|
IA-alkyne Probe Info |
 |
C77(0.00); C60(0.00); C297(0.00); C311(0.00) | LDD0036 | [9] | |
|
Lodoacetamide azide Probe Info |
 |
C124(0.00); C77(0.00); C60(0.00); C297(0.00) | LDD0037 | [9] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2224 | [10] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [11] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [11] | |
|
IPM Probe Info |
 |
C77(0.00); C297(0.00) | LDD0005 | [11] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [12] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [11] | |
|
TFBX Probe Info |
 |
C297(0.00); C311(0.00) | LDD0148 | [13] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [11] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [14] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | HEK-293T | C311(2.20) | LDD0168 | [3] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C311(2.35) | LDD0169 | [3] |
| LDCM0214 | AC1 | HCT 116 | C124(0.74); C311(1.16); C77(0.99) | LDD0531 | [7] |
| LDCM0215 | AC10 | HCT 116 | C124(1.26); C311(1.12); C77(1.37) | LDD0532 | [7] |
| LDCM0216 | AC100 | HCT 116 | C124(0.74); C311(1.48); C77(1.07) | LDD0533 | [7] |
| LDCM0217 | AC101 | HCT 116 | C124(0.92); C311(1.68); C77(1.04) | LDD0534 | [7] |
| LDCM0218 | AC102 | HCT 116 | C124(0.91); C311(1.40); C77(1.00) | LDD0535 | [7] |
| LDCM0219 | AC103 | HCT 116 | C124(1.15); C311(1.82); C77(1.22) | LDD0536 | [7] |
| LDCM0220 | AC104 | HCT 116 | C124(1.10); C311(1.18); C77(1.00) | LDD0537 | [7] |
| LDCM0221 | AC105 | HCT 116 | C124(1.10); C311(1.66); C77(0.98) | LDD0538 | [7] |
| LDCM0222 | AC106 | HCT 116 | C124(1.23); C311(1.74); C77(1.15) | LDD0539 | [7] |
| LDCM0223 | AC107 | HCT 116 | C124(1.29); C311(1.49); C77(0.94) | LDD0540 | [7] |
| LDCM0224 | AC108 | HCT 116 | C124(0.92); C311(1.92); C77(0.99) | LDD0541 | [7] |
| LDCM0225 | AC109 | HCT 116 | C124(0.84); C311(1.11); C77(1.01) | LDD0542 | [7] |
| LDCM0226 | AC11 | HCT 116 | C124(1.22); C311(1.03); C77(1.78) | LDD0543 | [7] |
| LDCM0227 | AC110 | HCT 116 | C124(0.81); C311(1.70); C77(1.16) | LDD0544 | [7] |
| LDCM0228 | AC111 | HCT 116 | C124(1.10); C311(1.64); C77(1.30) | LDD0545 | [7] |
| LDCM0229 | AC112 | HCT 116 | C124(1.43); C311(1.47); C77(0.89) | LDD0546 | [7] |
| LDCM0230 | AC113 | HCT 116 | C124(0.91); C311(0.78); C77(1.10) | LDD0547 | [7] |
| LDCM0231 | AC114 | HCT 116 | C124(1.37); C311(0.97); C77(1.37) | LDD0548 | [7] |
| LDCM0232 | AC115 | HCT 116 | C124(1.32); C311(0.80); C77(1.38) | LDD0549 | [7] |
| LDCM0233 | AC116 | HCT 116 | C124(1.57); C311(0.90); C77(1.38) | LDD0550 | [7] |
| LDCM0234 | AC117 | HCT 116 | C124(1.14); C311(0.78); C77(1.36) | LDD0551 | [7] |
| LDCM0235 | AC118 | HCT 116 | C124(1.32); C311(0.79); C77(1.33) | LDD0552 | [7] |
| LDCM0236 | AC119 | HCT 116 | C124(1.35); C311(0.75); C77(1.12) | LDD0553 | [7] |
| LDCM0237 | AC12 | HCT 116 | C124(0.94); C311(0.87); C77(1.12) | LDD0554 | [7] |
| LDCM0238 | AC120 | HCT 116 | C124(1.22); C311(0.85); C77(1.11) | LDD0555 | [7] |
| LDCM0239 | AC121 | HCT 116 | C124(1.46); C311(0.72); C77(1.29) | LDD0556 | [7] |
| LDCM0240 | AC122 | HCT 116 | C124(1.34); C311(0.72); C77(1.67) | LDD0557 | [7] |
| LDCM0241 | AC123 | HCT 116 | C124(1.26); C311(0.79); C77(0.96) | LDD0558 | [7] |
| LDCM0242 | AC124 | HCT 116 | C124(1.04); C311(0.96); C77(1.13) | LDD0559 | [7] |
| LDCM0243 | AC125 | HCT 116 | C124(1.03); C311(0.73); C77(1.24) | LDD0560 | [7] |
| LDCM0244 | AC126 | HCT 116 | C124(1.44); C311(0.92); C77(1.56) | LDD0561 | [7] |
| LDCM0245 | AC127 | HCT 116 | C124(1.50); C311(0.86); C77(1.30) | LDD0562 | [7] |
| LDCM0246 | AC128 | HCT 116 | C124(1.50); C311(1.35); C77(1.30) | LDD0563 | [7] |
| LDCM0247 | AC129 | HCT 116 | C124(0.81); C311(0.97); C77(0.86) | LDD0564 | [7] |
| LDCM0249 | AC130 | HCT 116 | C124(1.11); C311(1.16); C77(1.03) | LDD0566 | [7] |
| LDCM0250 | AC131 | HCT 116 | C124(0.83); C311(0.99); C77(1.00) | LDD0567 | [7] |
| LDCM0251 | AC132 | HCT 116 | C124(0.84); C311(1.02); C77(1.09) | LDD0568 | [7] |
| LDCM0252 | AC133 | HCT 116 | C124(1.05); C311(1.13); C77(1.38) | LDD0569 | [7] |
| LDCM0253 | AC134 | HCT 116 | C124(1.05); C311(1.27); C77(1.49) | LDD0570 | [7] |
| LDCM0254 | AC135 | HCT 116 | C124(1.15); C311(1.20); C77(1.46) | LDD0571 | [7] |
| LDCM0255 | AC136 | HCT 116 | C124(1.16); C311(1.19); C77(1.14) | LDD0572 | [7] |
| LDCM0256 | AC137 | HCT 116 | C124(0.93); C311(1.12); C77(1.11) | LDD0573 | [7] |
| LDCM0257 | AC138 | HCT 116 | C124(1.04); C311(1.36); C77(1.36) | LDD0574 | [7] |
| LDCM0258 | AC139 | HCT 116 | C124(1.14); C311(1.20); C77(1.25) | LDD0575 | [7] |
| LDCM0259 | AC14 | HCT 116 | C124(1.29); C311(0.98); C77(1.09) | LDD0576 | [7] |
| LDCM0260 | AC140 | HCT 116 | C124(1.11); C311(1.30); C77(1.22) | LDD0577 | [7] |
| LDCM0261 | AC141 | HCT 116 | C124(1.12); C311(1.33); C77(1.38) | LDD0578 | [7] |
| LDCM0262 | AC142 | HCT 116 | C124(0.92); C311(1.05); C77(1.03) | LDD0579 | [7] |
| LDCM0263 | AC143 | HCT 116 | C124(1.18); C311(1.11); C77(1.06) | LDD0580 | [7] |
| LDCM0264 | AC144 | HCT 116 | C77(1.00); C124(1.26); C311(1.36) | LDD0581 | [7] |
| LDCM0265 | AC145 | HCT 116 | C77(1.00); C311(1.05); C124(1.11) | LDD0582 | [7] |
| LDCM0266 | AC146 | HCT 116 | C77(1.05); C311(1.26); C124(1.33) | LDD0583 | [7] |
| LDCM0267 | AC147 | HCT 116 | C77(1.04); C311(1.37); C124(1.44) | LDD0584 | [7] |
| LDCM0268 | AC148 | HCT 116 | C77(1.01); C311(1.71); C124(1.77) | LDD0585 | [7] |
| LDCM0269 | AC149 | HCT 116 | C77(0.95); C124(1.46); C311(1.69) | LDD0586 | [7] |
| LDCM0270 | AC15 | HCT 116 | C311(0.95); C77(1.15); C124(1.23) | LDD0587 | [7] |
| LDCM0271 | AC150 | HCT 116 | C77(1.00); C124(1.05); C311(1.17) | LDD0588 | [7] |
| LDCM0272 | AC151 | HCT 116 | C124(1.00); C77(1.09); C311(1.14) | LDD0589 | [7] |
| LDCM0273 | AC152 | HCT 116 | C77(0.90); C311(1.28); C124(1.37) | LDD0590 | [7] |
| LDCM0274 | AC153 | HCT 116 | C77(1.03); C124(1.80); C311(2.03) | LDD0591 | [7] |
| LDCM0621 | AC154 | HCT 116 | C124(1.15); C311(1.39); C77(1.11) | LDD2158 | [7] |
| LDCM0622 | AC155 | HCT 116 | C124(1.24); C311(1.38); C77(0.99) | LDD2159 | [7] |
| LDCM0623 | AC156 | HCT 116 | C124(1.04); C311(1.13); C77(1.05) | LDD2160 | [7] |
| LDCM0624 | AC157 | HCT 116 | C124(1.03); C311(1.02); C77(1.10) | LDD2161 | [7] |
| LDCM0276 | AC17 | HCT 116 | C124(0.94); C77(1.07); C311(1.08) | LDD0593 | [7] |
| LDCM0277 | AC18 | HCT 116 | C77(1.18); C311(1.43); C124(1.52) | LDD0594 | [7] |
| LDCM0278 | AC19 | HCT 116 | C77(0.99); C311(1.14); C124(1.21) | LDD0595 | [7] |
| LDCM0279 | AC2 | HCT 116 | C77(1.00); C124(1.11); C311(1.31) | LDD0596 | [7] |
| LDCM0280 | AC20 | HCT 116 | C311(1.11); C77(1.17); C124(1.19) | LDD0597 | [7] |
| LDCM0281 | AC21 | HCT 116 | C77(0.86); C124(1.04); C311(1.15) | LDD0598 | [7] |
| LDCM0282 | AC22 | HCT 116 | C77(0.86); C124(0.96); C311(0.99) | LDD0599 | [7] |
| LDCM0283 | AC23 | HCT 116 | C124(0.93); C77(1.03); C311(1.19) | LDD0600 | [7] |
| LDCM0284 | AC24 | HCT 116 | C311(0.94); C124(0.99); C77(1.09) | LDD0601 | [7] |
| LDCM0285 | AC25 | HCT 116 | C77(0.99); C311(1.03); C124(1.13) | LDD0602 | [7] |
| LDCM0286 | AC26 | HCT 116 | C311(1.14); C77(1.17); C124(1.21) | LDD0603 | [7] |
| LDCM0287 | AC27 | HCT 116 | C77(1.19); C124(1.25); C311(1.30) | LDD0604 | [7] |
| LDCM0288 | AC28 | HCT 116 | C124(1.17); C311(1.23); C77(1.53) | LDD0605 | [7] |
| LDCM0289 | AC29 | HCT 116 | C311(1.15); C77(1.19); C124(1.30) | LDD0606 | [7] |
| LDCM0290 | AC3 | HCT 116 | C124(0.97); C77(1.32); C311(1.36) | LDD0607 | [7] |
| LDCM0291 | AC30 | HCT 116 | C77(1.12); C311(1.24); C124(1.25) | LDD0608 | [7] |
| LDCM0292 | AC31 | HCT 116 | C124(1.22); C311(1.29); C77(1.44) | LDD0609 | [7] |
| LDCM0293 | AC32 | HCT 116 | C311(1.36); C124(1.40); C77(1.49) | LDD0610 | [7] |
| LDCM0294 | AC33 | HCT 116 | C77(1.11); C311(1.20); C124(1.36) | LDD0611 | [7] |
| LDCM0295 | AC34 | HCT 116 | C77(1.09); C124(1.33); C311(1.45) | LDD0612 | [7] |
| LDCM0296 | AC35 | HCT 116 | C124(0.90); C77(0.96); C311(1.14) | LDD0613 | [7] |
| LDCM0297 | AC36 | HCT 116 | C124(0.92); C311(1.01); C77(1.03) | LDD0614 | [7] |
| LDCM0298 | AC37 | HCT 116 | C124(0.97); C311(1.08); C77(1.31) | LDD0615 | [7] |
| LDCM0299 | AC38 | HCT 116 | C124(0.94); C77(1.14); C311(1.25) | LDD0616 | [7] |
| LDCM0300 | AC39 | HCT 116 | C124(1.02); C311(1.34); C77(1.38) | LDD0617 | [7] |
| LDCM0301 | AC4 | HCT 116 | C124(0.96); C77(1.15); C311(1.23) | LDD0618 | [7] |
| LDCM0302 | AC40 | HCT 116 | C124(1.09); C311(1.11); C77(1.30) | LDD0619 | [7] |
| LDCM0303 | AC41 | HCT 116 | C124(0.90); C77(1.14); C311(1.39) | LDD0620 | [7] |
| LDCM0304 | AC42 | HCT 116 | C124(1.10); C77(1.19); C311(1.99) | LDD0621 | [7] |
| LDCM0305 | AC43 | HCT 116 | C124(1.07); C311(1.18); C77(1.25) | LDD0622 | [7] |
| LDCM0306 | AC44 | HCT 116 | C77(1.01); C124(1.05); C311(1.56) | LDD0623 | [7] |
| LDCM0307 | AC45 | HCT 116 | C124(1.18); C311(1.22); C77(1.80) | LDD0624 | [7] |
| LDCM0308 | AC46 | HCT 116 | C77(1.13); C311(1.20); C124(1.22) | LDD0625 | [7] |
| LDCM0309 | AC47 | HCT 116 | C77(0.95); C311(0.98); C124(1.12) | LDD0626 | [7] |
| LDCM0310 | AC48 | HCT 116 | C77(1.06); C311(1.11); C124(1.22) | LDD0627 | [7] |
| LDCM0311 | AC49 | HCT 116 | C77(1.03); C311(1.52); C124(1.62) | LDD0628 | [7] |
| LDCM0312 | AC5 | HCT 116 | C124(0.81); C311(0.98); C77(1.32) | LDD0629 | [7] |
| LDCM0313 | AC50 | HCT 116 | C77(1.01); C311(1.48); C124(1.55) | LDD0630 | [7] |
| LDCM0314 | AC51 | HCT 116 | C77(0.95); C124(1.01); C311(1.01) | LDD0631 | [7] |
| LDCM0315 | AC52 | HCT 116 | C77(1.05); C124(1.10); C311(1.14) | LDD0632 | [7] |
| LDCM0316 | AC53 | HCT 116 | C77(0.98); C311(1.18); C124(1.33) | LDD0633 | [7] |
| LDCM0317 | AC54 | HCT 116 | C77(0.99); C311(1.16); C124(1.33) | LDD0634 | [7] |
| LDCM0318 | AC55 | HCT 116 | C77(1.06); C311(1.19); C124(1.33) | LDD0635 | [7] |
| LDCM0319 | AC56 | HCT 116 | C77(0.96); C311(1.60); C124(1.82) | LDD0636 | [7] |
| LDCM0320 | AC57 | HCT 116 | C77(1.23); C311(1.27) | LDD0637 | [7] |
| LDCM0321 | AC58 | HCT 116 | C77(1.15); C311(1.44) | LDD0638 | [7] |
| LDCM0322 | AC59 | HCT 116 | C77(1.09); C311(1.26) | LDD0639 | [7] |
| LDCM0323 | AC6 | HCT 116 | C311(1.10); C124(1.11); C77(1.33) | LDD0640 | [7] |
| LDCM0324 | AC60 | HCT 116 | C77(1.13); C311(1.35) | LDD0641 | [7] |
| LDCM0325 | AC61 | HCT 116 | C77(1.14); C311(1.29) | LDD0642 | [7] |
| LDCM0326 | AC62 | HCT 116 | C77(1.16); C311(1.34) | LDD0643 | [7] |
| LDCM0327 | AC63 | HCT 116 | C77(1.05); C311(1.23) | LDD0644 | [7] |
| LDCM0328 | AC64 | HCT 116 | C77(1.12); C311(1.40) | LDD0645 | [7] |
| LDCM0329 | AC65 | HCT 116 | C77(1.17); C311(1.48) | LDD0646 | [7] |
| LDCM0330 | AC66 | HCT 116 | C77(1.17); C311(1.58) | LDD0647 | [7] |
| LDCM0331 | AC67 | HCT 116 | C77(1.22); C311(1.57) | LDD0648 | [7] |
| LDCM0332 | AC68 | HCT 116 | C77(0.92); C311(1.15) | LDD0649 | [7] |
| LDCM0333 | AC69 | HCT 116 | C77(0.83); C311(1.24) | LDD0650 | [7] |
| LDCM0334 | AC7 | HCT 116 | C311(0.97); C77(1.05); C124(1.23) | LDD0651 | [7] |
| LDCM0335 | AC70 | HCT 116 | C77(0.96); C311(1.31) | LDD0652 | [7] |
| LDCM0336 | AC71 | HCT 116 | C77(1.00); C311(1.13) | LDD0653 | [7] |
| LDCM0337 | AC72 | HCT 116 | C77(0.87); C311(1.30) | LDD0654 | [7] |
| LDCM0338 | AC73 | HCT 116 | C77(1.10); C311(1.80) | LDD0655 | [7] |
| LDCM0339 | AC74 | HCT 116 | C77(1.00); C311(1.64) | LDD0656 | [7] |
| LDCM0340 | AC75 | HCT 116 | C77(0.90); C311(1.76) | LDD0657 | [7] |
| LDCM0341 | AC76 | HCT 116 | C77(0.87); C311(1.41) | LDD0658 | [7] |
| LDCM0342 | AC77 | HCT 116 | C77(1.01); C311(1.50) | LDD0659 | [7] |
| LDCM0343 | AC78 | HCT 116 | C77(0.78); C311(1.23) | LDD0660 | [7] |
| LDCM0344 | AC79 | HCT 116 | C77(0.92); C311(1.32) | LDD0661 | [7] |
| LDCM0345 | AC8 | HCT 116 | C77(1.01); C311(1.12); C124(1.18) | LDD0662 | [7] |
| LDCM0346 | AC80 | HCT 116 | C77(1.02); C311(1.16) | LDD0663 | [7] |
| LDCM0347 | AC81 | HCT 116 | C77(0.87); C311(1.08) | LDD0664 | [7] |
| LDCM0348 | AC82 | HCT 116 | C77(1.03); C311(1.59) | LDD0665 | [7] |
| LDCM0349 | AC83 | HCT 116 | C77(1.17); C311(1.30); C124(1.50) | LDD0666 | [7] |
| LDCM0350 | AC84 | HCT 116 | C77(1.09); C124(1.36); C311(1.37) | LDD0667 | [7] |
| LDCM0351 | AC85 | HCT 116 | C77(1.18); C124(1.22); C311(1.38) | LDD0668 | [7] |
| LDCM0352 | AC86 | HCT 116 | C124(1.30); C311(1.34); C77(1.39) | LDD0669 | [7] |
| LDCM0353 | AC87 | HCT 116 | C124(1.09); C311(1.27); C77(1.40) | LDD0670 | [7] |
| LDCM0354 | AC88 | HCT 116 | C124(1.26); C311(1.30); C77(1.38) | LDD0671 | [7] |
| LDCM0355 | AC89 | HCT 116 | C77(1.19); C124(1.48); C311(1.48) | LDD0672 | [7] |
| LDCM0357 | AC90 | HCT 116 | C124(1.22); C311(1.22); C77(1.23) | LDD0674 | [7] |
| LDCM0358 | AC91 | HCT 116 | C311(1.19); C124(1.38); C77(1.39) | LDD0675 | [7] |
| LDCM0359 | AC92 | HCT 116 | C77(1.07); C124(1.25); C311(1.32) | LDD0676 | [7] |
| LDCM0360 | AC93 | HCT 116 | C124(1.10); C77(1.14); C311(1.41) | LDD0677 | [7] |
| LDCM0361 | AC94 | HCT 116 | C124(1.03); C311(1.23); C77(1.37) | LDD0678 | [7] |
| LDCM0362 | AC95 | HCT 116 | C77(1.21); C311(1.33); C124(1.45) | LDD0679 | [7] |
| LDCM0363 | AC96 | HCT 116 | C124(1.04); C311(1.21); C77(1.24) | LDD0680 | [7] |
| LDCM0364 | AC97 | HCT 116 | C77(1.44); C124(1.46); C311(1.58) | LDD0681 | [7] |
| LDCM0365 | AC98 | HCT 116 | C77(1.07); C124(1.60); C311(1.70) | LDD0682 | [7] |
| LDCM0366 | AC99 | HCT 116 | C124(0.90); C77(1.07); C311(1.57) | LDD0683 | [7] |
| LDCM0248 | AKOS034007472 | HCT 116 | C124(1.07); C311(0.92); C77(1.29) | LDD0565 | [7] |
| LDCM0356 | AKOS034007680 | HCT 116 | C311(0.96); C77(1.20); C124(1.21) | LDD0673 | [7] |
| LDCM0275 | AKOS034007705 | HCT 116 | C311(1.26); C77(1.41); C124(1.90) | LDD0592 | [7] |
| LDCM0156 | Aniline | NCI-H1299 | 11.80 | LDD0403 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C297(1.01); C311(0.93) | LDD0078 | [7] |
| LDCM0367 | CL1 | HCT 116 | C124(0.74); C311(1.01); C77(1.05) | LDD0684 | [7] |
| LDCM0368 | CL10 | HCT 116 | C77(1.16); C124(1.30); C311(1.34) | LDD0685 | [7] |
| LDCM0369 | CL100 | HCT 116 | C311(1.04); C124(1.17); C77(1.26) | LDD0686 | [7] |
| LDCM0370 | CL101 | HCT 116 | C311(1.05); C77(1.31); C124(1.56) | LDD0687 | [7] |
| LDCM0371 | CL102 | HCT 116 | C311(0.85); C124(1.04); C77(1.57) | LDD0688 | [7] |
| LDCM0372 | CL103 | HCT 116 | C311(0.88); C77(1.02); C124(1.10) | LDD0689 | [7] |
| LDCM0373 | CL104 | HCT 116 | C311(0.89); C124(1.08); C77(1.15) | LDD0690 | [7] |
| LDCM0374 | CL105 | HCT 116 | C311(1.25); C77(1.28); C124(1.34) | LDD0691 | [7] |
| LDCM0375 | CL106 | HCT 116 | C77(1.22); C311(1.53); C124(1.53) | LDD0692 | [7] |
| LDCM0376 | CL107 | HCT 116 | C124(1.20); C77(1.34); C311(1.46) | LDD0693 | [7] |
| LDCM0377 | CL108 | HCT 116 | C77(1.38); C311(1.41); C124(1.72) | LDD0694 | [7] |
| LDCM0378 | CL109 | HCT 116 | C77(1.17); C311(1.22); C124(1.33) | LDD0695 | [7] |
| LDCM0379 | CL11 | HCT 116 | C77(1.05); C124(1.27); C311(1.36) | LDD0696 | [7] |
| LDCM0380 | CL110 | HCT 116 | C77(1.26); C311(1.41); C124(1.53) | LDD0697 | [7] |
| LDCM0381 | CL111 | HCT 116 | C77(1.08); C124(1.13); C311(1.33) | LDD0698 | [7] |
| LDCM0382 | CL112 | HCT 116 | C311(1.01); C124(1.11); C77(1.37) | LDD0699 | [7] |
| LDCM0383 | CL113 | HCT 116 | C77(0.99); C124(1.34); C311(1.34) | LDD0700 | [7] |
| LDCM0384 | CL114 | HCT 116 | C124(1.16); C77(1.36); C311(1.41) | LDD0701 | [7] |
| LDCM0385 | CL115 | HCT 116 | C77(1.10); C311(1.19); C124(1.29) | LDD0702 | [7] |
| LDCM0386 | CL116 | HCT 116 | C124(1.19); C311(1.27); C77(1.35) | LDD0703 | [7] |
| LDCM0387 | CL117 | HCT 116 | C77(1.14); C124(1.41); C311(2.44) | LDD0704 | [7] |
| LDCM0388 | CL118 | HCT 116 | C124(0.99); C77(1.20); C311(1.90) | LDD0705 | [7] |
| LDCM0389 | CL119 | HCT 116 | C77(1.09); C311(1.14); C124(1.15) | LDD0706 | [7] |
| LDCM0390 | CL12 | HCT 116 | C77(1.00); C311(1.30); C124(1.48) | LDD0707 | [7] |
| LDCM0391 | CL120 | HCT 116 | C311(1.01); C124(1.04); C77(1.20) | LDD0708 | [7] |
| LDCM0392 | CL121 | HCT 116 | C311(0.95); C124(1.12); C77(1.12) | LDD0709 | [7] |
| LDCM0393 | CL122 | HCT 116 | C77(0.94); C124(1.20); C311(1.29) | LDD0710 | [7] |
| LDCM0394 | CL123 | HCT 116 | C77(1.03); C124(1.39); C311(1.54) | LDD0711 | [7] |
| LDCM0395 | CL124 | HCT 116 | C77(1.00); C311(1.36); C124(1.48) | LDD0712 | [7] |
| LDCM0396 | CL125 | HCT 116 | C311(1.08); C77(1.19) | LDD0713 | [7] |
| LDCM0397 | CL126 | HCT 116 | C77(1.12); C311(1.69) | LDD0714 | [7] |
| LDCM0398 | CL127 | HCT 116 | C77(1.04); C311(1.14) | LDD0715 | [7] |
| LDCM0399 | CL128 | HCT 116 | C311(1.17); C77(1.18) | LDD0716 | [7] |
| LDCM0400 | CL13 | HCT 116 | C77(0.99); C311(1.13); C124(1.18) | LDD0717 | [7] |
| LDCM0401 | CL14 | HCT 116 | C77(1.05); C124(1.13); C311(1.15) | LDD0718 | [7] |
| LDCM0402 | CL15 | HCT 116 | C77(1.03); C124(1.06); C311(1.24) | LDD0719 | [7] |
| LDCM0403 | CL16 | HCT 116 | C311(0.98); C77(1.13) | LDD0720 | [7] |
| LDCM0404 | CL17 | HCT 116 | C77(1.15); C311(1.19) | LDD0721 | [7] |
| LDCM0405 | CL18 | HCT 116 | C311(1.07); C77(1.23) | LDD0722 | [7] |
| LDCM0406 | CL19 | HCT 116 | C77(1.10); C311(1.19) | LDD0723 | [7] |
| LDCM0407 | CL2 | HCT 116 | C124(0.85); C77(0.85); C311(1.02) | LDD0724 | [7] |
| LDCM0408 | CL20 | HCT 116 | C311(1.19); C77(1.31) | LDD0725 | [7] |
| LDCM0409 | CL21 | HCT 116 | C77(1.38); C311(1.39) | LDD0726 | [7] |
| LDCM0410 | CL22 | HCT 116 | C77(1.61); C311(1.76) | LDD0727 | [7] |
| LDCM0411 | CL23 | HCT 116 | C311(1.09); C77(1.38) | LDD0728 | [7] |
| LDCM0412 | CL24 | HCT 116 | C311(1.29); C77(1.50) | LDD0729 | [7] |
| LDCM0413 | CL25 | HCT 116 | C311(1.63); C77(1.38) | LDD0730 | [7] |
| LDCM0414 | CL26 | HCT 116 | C311(1.12); C77(1.18) | LDD0731 | [7] |
| LDCM0415 | CL27 | HCT 116 | C311(1.13); C77(1.24) | LDD0732 | [7] |
| LDCM0416 | CL28 | HCT 116 | C311(1.21); C77(1.37) | LDD0733 | [7] |
| LDCM0417 | CL29 | HCT 116 | C311(1.12); C77(1.37) | LDD0734 | [7] |
| LDCM0418 | CL3 | HCT 116 | C124(0.87); C311(1.06); C77(0.87) | LDD0735 | [7] |
| LDCM0419 | CL30 | HCT 116 | C311(0.98); C77(1.26) | LDD0736 | [7] |
| LDCM0420 | CL31 | HCT 116 | C311(1.09); C77(1.42) | LDD0737 | [7] |
| LDCM0421 | CL32 | HCT 116 | C311(1.15); C77(1.03) | LDD0738 | [7] |
| LDCM0422 | CL33 | HCT 116 | C311(1.50); C77(1.20) | LDD0739 | [7] |
| LDCM0423 | CL34 | HCT 116 | C311(1.43); C77(1.34) | LDD0740 | [7] |
| LDCM0424 | CL35 | HCT 116 | C311(1.36); C77(1.69) | LDD0741 | [7] |
| LDCM0425 | CL36 | HCT 116 | C311(1.27); C77(1.52) | LDD0742 | [7] |
| LDCM0426 | CL37 | HCT 116 | C311(1.57); C77(1.30) | LDD0743 | [7] |
| LDCM0428 | CL39 | HCT 116 | C311(1.85); C77(1.31) | LDD0745 | [7] |
| LDCM0429 | CL4 | HCT 116 | C124(1.04); C311(0.99); C77(0.98) | LDD0746 | [7] |
| LDCM0430 | CL40 | HCT 116 | C311(1.47); C77(1.46) | LDD0747 | [7] |
| LDCM0431 | CL41 | HCT 116 | C311(1.24); C77(1.11) | LDD0748 | [7] |
| LDCM0432 | CL42 | HCT 116 | C311(1.95); C77(1.34) | LDD0749 | [7] |
| LDCM0433 | CL43 | HCT 116 | C311(1.52); C77(1.44) | LDD0750 | [7] |
| LDCM0434 | CL44 | HCT 116 | C311(1.31); C77(1.38) | LDD0751 | [7] |
| LDCM0435 | CL45 | HCT 116 | C311(1.63); C77(1.52) | LDD0752 | [7] |
| LDCM0436 | CL46 | HCT 116 | C311(1.07); C77(1.29) | LDD0753 | [7] |
| LDCM0437 | CL47 | HCT 116 | C311(1.04); C77(1.22) | LDD0754 | [7] |
| LDCM0438 | CL48 | HCT 116 | C311(0.93); C77(1.31) | LDD0755 | [7] |
| LDCM0439 | CL49 | HCT 116 | C311(0.92); C77(1.19) | LDD0756 | [7] |
| LDCM0440 | CL5 | HCT 116 | C124(1.03); C311(0.92); C77(0.93) | LDD0757 | [7] |
| LDCM0441 | CL50 | HCT 116 | C311(1.18); C77(1.11) | LDD0758 | [7] |
| LDCM0442 | CL51 | HCT 116 | C311(1.16); C77(1.23) | LDD0759 | [7] |
| LDCM0443 | CL52 | HCT 116 | C311(1.11); C77(1.16) | LDD0760 | [7] |
| LDCM0444 | CL53 | HCT 116 | C311(0.99); C77(0.85) | LDD0761 | [7] |
| LDCM0445 | CL54 | HCT 116 | C311(1.11); C77(1.12) | LDD0762 | [7] |
| LDCM0446 | CL55 | HCT 116 | C311(0.98); C77(1.18) | LDD0763 | [7] |
| LDCM0447 | CL56 | HCT 116 | C311(1.10); C77(1.13) | LDD0764 | [7] |
| LDCM0448 | CL57 | HCT 116 | C311(1.02); C77(1.10) | LDD0765 | [7] |
| LDCM0449 | CL58 | HCT 116 | C311(0.99); C77(1.18) | LDD0766 | [7] |
| LDCM0450 | CL59 | HCT 116 | C311(1.14); C77(1.22) | LDD0767 | [7] |
| LDCM0451 | CL6 | HCT 116 | C124(0.95); C311(1.11); C77(0.96) | LDD0768 | [7] |
| LDCM0452 | CL60 | HCT 116 | C311(1.50); C77(1.24) | LDD0769 | [7] |
| LDCM0453 | CL61 | HCT 116 | C311(1.61); C77(0.97) | LDD0770 | [7] |
| LDCM0454 | CL62 | HCT 116 | C311(1.15); C77(0.99) | LDD0771 | [7] |
| LDCM0455 | CL63 | HCT 116 | C311(1.27); C77(1.06) | LDD0772 | [7] |
| LDCM0456 | CL64 | HCT 116 | C311(1.57); C77(0.93) | LDD0773 | [7] |
| LDCM0457 | CL65 | HCT 116 | C311(1.46); C77(0.98) | LDD0774 | [7] |
| LDCM0458 | CL66 | HCT 116 | C311(1.97); C77(1.22) | LDD0775 | [7] |
| LDCM0459 | CL67 | HCT 116 | C311(1.47); C77(0.92) | LDD0776 | [7] |
| LDCM0460 | CL68 | HCT 116 | C311(1.61); C77(1.01) | LDD0777 | [7] |
| LDCM0461 | CL69 | HCT 116 | C311(1.42); C77(1.25) | LDD0778 | [7] |
| LDCM0462 | CL7 | HCT 116 | C124(1.20); C311(1.07); C77(1.27) | LDD0779 | [7] |
| LDCM0463 | CL70 | HCT 116 | C311(1.33); C77(0.98) | LDD0780 | [7] |
| LDCM0464 | CL71 | HCT 116 | C311(1.93); C77(0.99) | LDD0781 | [7] |
| LDCM0465 | CL72 | HCT 116 | C311(1.20); C77(1.04) | LDD0782 | [7] |
| LDCM0466 | CL73 | HCT 116 | C311(1.43); C77(0.92) | LDD0783 | [7] |
| LDCM0467 | CL74 | HCT 116 | C311(1.35); C77(1.02) | LDD0784 | [7] |
| LDCM0469 | CL76 | HCT 116 | C311(1.10); C77(0.98) | LDD0786 | [7] |
| LDCM0470 | CL77 | HCT 116 | C311(1.02); C77(0.92) | LDD0787 | [7] |
| LDCM0471 | CL78 | HCT 116 | C311(1.12); C77(1.09) | LDD0788 | [7] |
| LDCM0472 | CL79 | HCT 116 | C311(1.34); C77(1.06) | LDD0789 | [7] |
| LDCM0473 | CL8 | HCT 116 | C124(1.52); C311(1.54); C77(0.92) | LDD0790 | [7] |
| LDCM0474 | CL80 | HCT 116 | C311(1.06); C77(0.95) | LDD0791 | [7] |
| LDCM0475 | CL81 | HCT 116 | C311(1.15); C77(1.04) | LDD0792 | [7] |
| LDCM0476 | CL82 | HCT 116 | C311(1.56); C77(1.21) | LDD0793 | [7] |
| LDCM0477 | CL83 | HCT 116 | C311(1.36); C77(1.17) | LDD0794 | [7] |
| LDCM0478 | CL84 | HCT 116 | C311(1.81); C77(1.51) | LDD0795 | [7] |
| LDCM0479 | CL85 | HCT 116 | C311(1.13); C77(1.23) | LDD0796 | [7] |
| LDCM0480 | CL86 | HCT 116 | C311(1.06); C77(0.89) | LDD0797 | [7] |
| LDCM0481 | CL87 | HCT 116 | C311(1.16); C77(1.20) | LDD0798 | [7] |
| LDCM0482 | CL88 | HCT 116 | C311(1.41); C77(1.11) | LDD0799 | [7] |
| LDCM0483 | CL89 | HCT 116 | C311(1.93); C77(1.33) | LDD0800 | [7] |
| LDCM0484 | CL9 | HCT 116 | C124(1.24); C311(1.06); C77(0.74) | LDD0801 | [7] |
| LDCM0485 | CL90 | HCT 116 | C311(0.98); C77(0.88) | LDD0802 | [7] |
| LDCM0486 | CL91 | HCT 116 | C124(1.12); C311(1.40); C77(1.14) | LDD0803 | [7] |
| LDCM0487 | CL92 | HCT 116 | C124(1.21); C311(1.03); C77(1.19) | LDD0804 | [7] |
| LDCM0488 | CL93 | HCT 116 | C124(1.00); C311(0.97); C77(1.08) | LDD0805 | [7] |
| LDCM0489 | CL94 | HCT 116 | C124(0.94); C311(1.66); C77(1.29) | LDD0806 | [7] |
| LDCM0490 | CL95 | HCT 116 | C124(1.33); C311(1.36); C77(1.04) | LDD0807 | [7] |
| LDCM0491 | CL96 | HCT 116 | C124(1.04); C311(1.16); C77(1.29) | LDD0808 | [7] |
| LDCM0492 | CL97 | HCT 116 | C124(1.44); C311(1.65); C77(1.21) | LDD0809 | [7] |
| LDCM0493 | CL98 | HCT 116 | C124(1.01); C311(1.36); C77(1.62) | LDD0810 | [7] |
| LDCM0494 | CL99 | HCT 116 | C124(0.92); C311(1.25); C77(1.34) | LDD0811 | [7] |
| LDCM0634 | CY-0357 | Hep-G2 | C124(0.92) | LDD2228 | [10] |
| LDCM0495 | E2913 | HEK-293T | C124(0.95); C311(0.79); C77(1.02) | LDD1698 | [17] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C77(1.23) | LDD1702 | [18] |
| LDCM0468 | Fragment33 | HCT 116 | C311(1.45); C77(0.97) | LDD0785 | [7] |
| LDCM0427 | Fragment51 | HCT 116 | C311(1.64); C77(1.67) | LDD0744 | [7] |
| LDCM0116 | HHS-0101 | DM93 | Y327(0.83); Y265(1.06) | LDD0264 | [5] |
| LDCM0117 | HHS-0201 | DM93 | Y327(0.84); Y265(1.30) | LDD0265 | [5] |
| LDCM0118 | HHS-0301 | DM93 | Y327(0.60); Y265(1.09) | LDD0266 | [5] |
| LDCM0119 | HHS-0401 | DM93 | Y327(0.85); Y265(0.98) | LDD0267 | [5] |
| LDCM0120 | HHS-0701 | DM93 | Y327(0.99); Y265(1.36) | LDD0268 | [5] |
| LDCM0123 | JWB131 | DM93 | Y265(0.90); Y327(0.90) | LDD0285 | [4] |
| LDCM0124 | JWB142 | DM93 | Y265(0.70); Y327(0.51) | LDD0286 | [4] |
| LDCM0125 | JWB146 | DM93 | Y265(3.61); Y327(0.85) | LDD0287 | [4] |
| LDCM0126 | JWB150 | DM93 | Y265(1.24); Y327(4.76) | LDD0288 | [4] |
| LDCM0127 | JWB152 | DM93 | Y265(2.88); Y327(1.31) | LDD0289 | [4] |
| LDCM0128 | JWB198 | DM93 | Y265(0.83); Y327(0.70) | LDD0290 | [4] |
| LDCM0129 | JWB202 | DM93 | Y265(1.03); Y327(0.32) | LDD0291 | [4] |
| LDCM0130 | JWB211 | DM93 | Y265(0.81); Y327(0.75) | LDD0292 | [4] |
| LDCM0022 | KB02 | HCT 116 | C311(2.05); C77(1.05) | LDD0080 | [7] |
| LDCM0023 | KB03 | HCT 116 | C311(1.75); C77(1.26) | LDD0081 | [7] |
| LDCM0024 | KB05 | HCT 116 | C311(3.55); C77(0.89) | LDD0082 | [7] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C297(1.82) | LDD2207 | [19] |
| LDCM0021 | THZ1 | HCT 116 | C297(0.94); C77(1.09) | LDD2173 | [7] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Glycogen synthase kinase-3 beta (GSK3B) | CMGC Ser/Thr protein kinase family | P49841 | |||
| E3 ubiquitin-protein ligase TTC3 (TTC3) | . | P53804 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Heat shock protein HSP 90-beta (HSP90AB1) | Heat shock protein 90 family | P08238 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Vimentin (VIM) | Intermediate filament family | P08670 | |||
| PRKCA-binding protein (PICK1) | . | Q9NRD5 | |||
| Vinexin (SORBS3) | . | O60504 | |||
The Drug(s) Related To This Target
Investigative
References