Details of the Target
General Information of Target
| Target ID | LDTP01335 | |||||
|---|---|---|---|---|---|---|
| Target Name | Protein SCO1 homolog, mitochondrial (SCO1) | |||||
| Gene Name | SCO1 | |||||
| Gene ID | 6341 | |||||
| Synonyms |
SCOD1; Protein SCO1 homolog, mitochondrial |
|||||
| 3D Structure | ||||||
| Sequence |
MAMLVLVPGRVMRPLGGQLWRFLPRGLEFWGPAEGTARVLLRQFCARQAEAWRASGRPGY
CLGTRPLSTARPPPPWSQKGPGDSTRPSKPGPVSWKSLAITFAIGGALLAGMKHVKKEKA EKLEKERQRHIGKPLLGGPFSLTTHTGERKTDKDYLGQWLLIYFGFTHCPDVCPEELEKM IQVVDEIDSITTLPDLTPLFISIDPERDTKEAIANYVKEFSPKLVGLTGTREEVDQVARA YRVYYSPGPKDEDEDYIVDHTIIMYLIGPDGEFLDYFGQNKRKGEIAASIATHMRPYRKK S |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
SCO1/2 family
|
|||||
| Subcellular location |
Mitochondrion
|
|||||
| Function |
Copper metallochaperone essential for the maturation of cytochrome c oxidase subunit II (MT-CO2/COX2). Not required for the synthesis of MT-CO2/COX2 but plays a crucial role in stabilizing MT-CO2/COX2 during its subsequent maturation. Involved in transporting copper to the Cu(A) site on MT-CO2/COX2. Plays an important role in the regulation of copper homeostasis by controlling the abundance and cell membrane localization of copper transporter CTR1.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
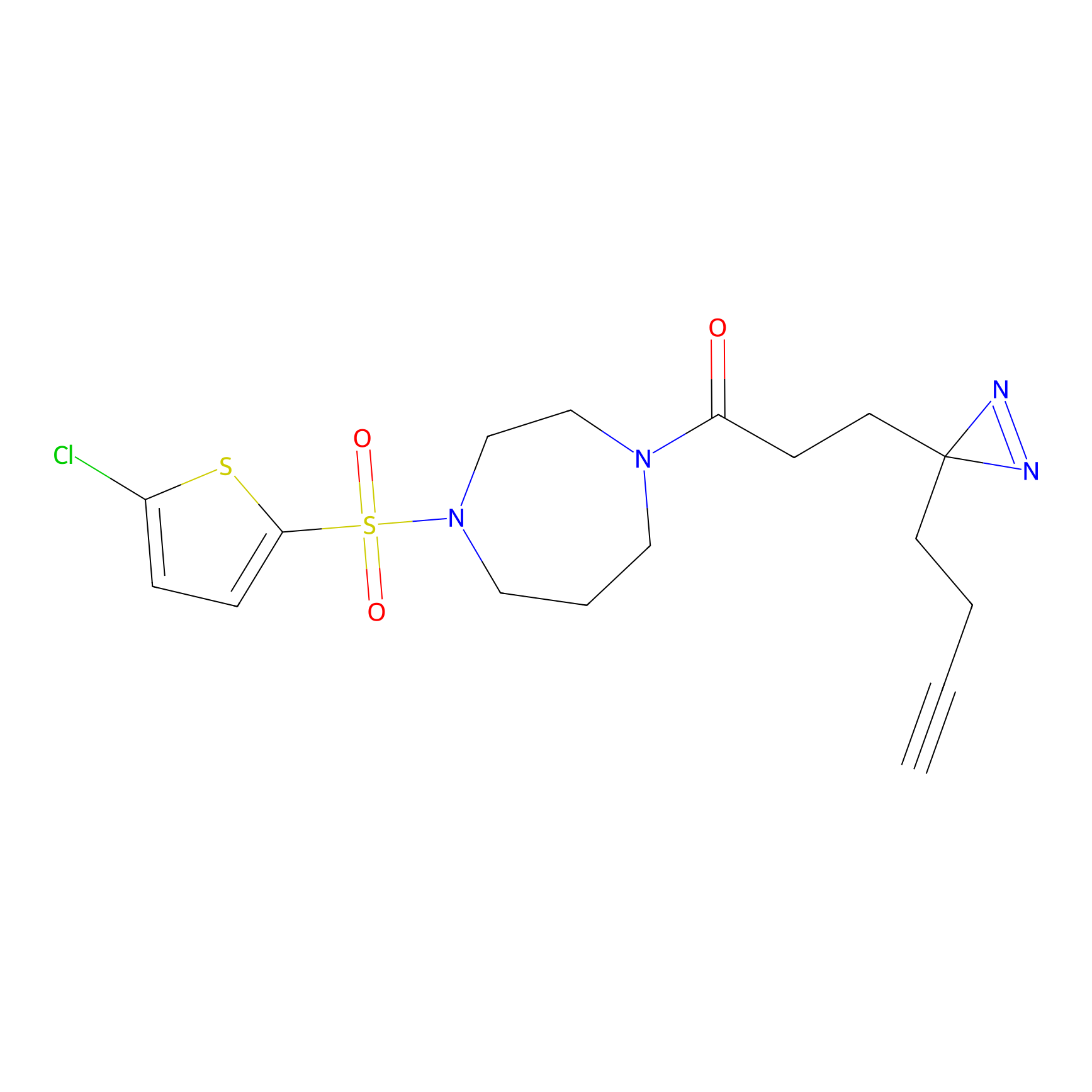
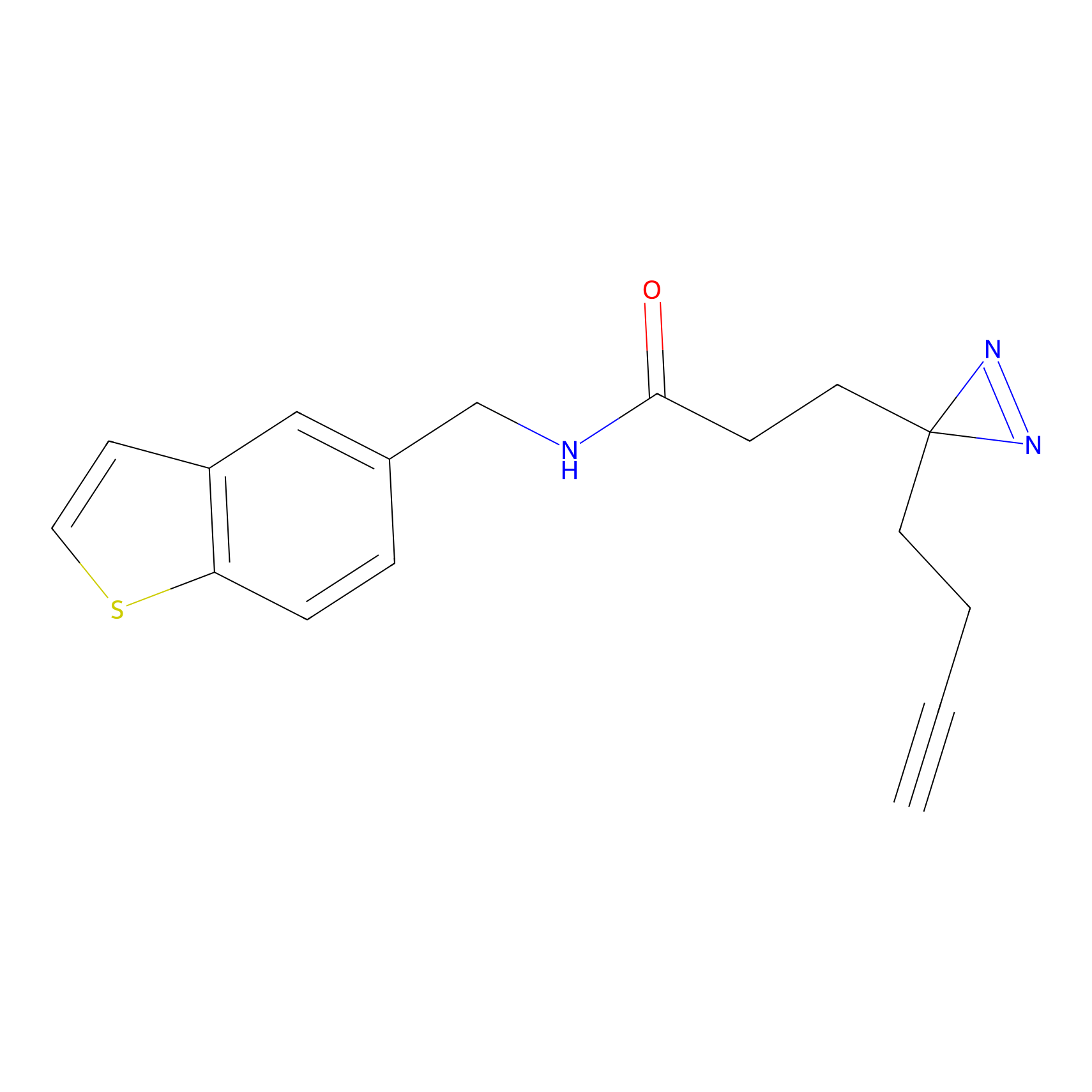
|
m-APA Probe Info |
 |
7.80 | LDD0402 | [1] | |
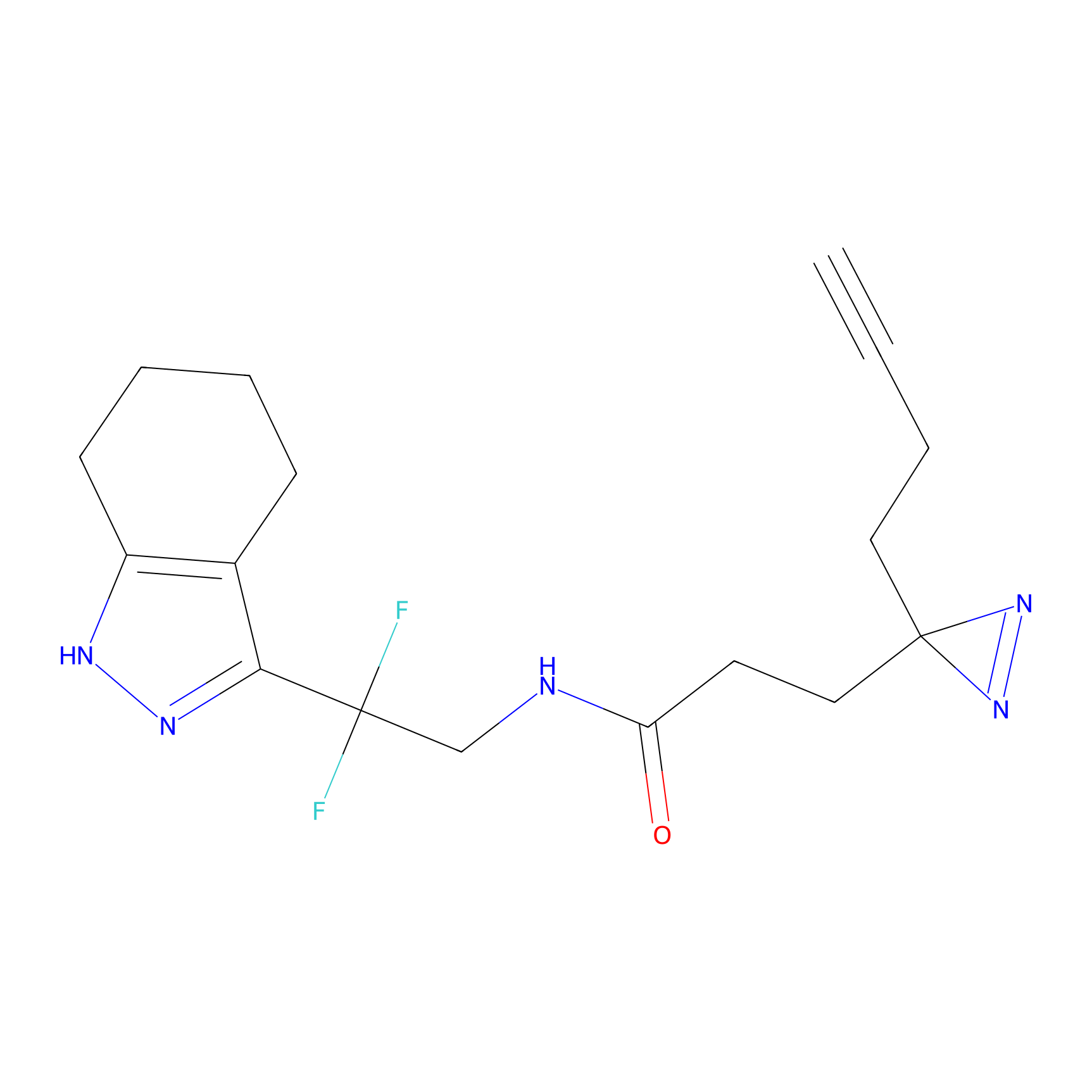
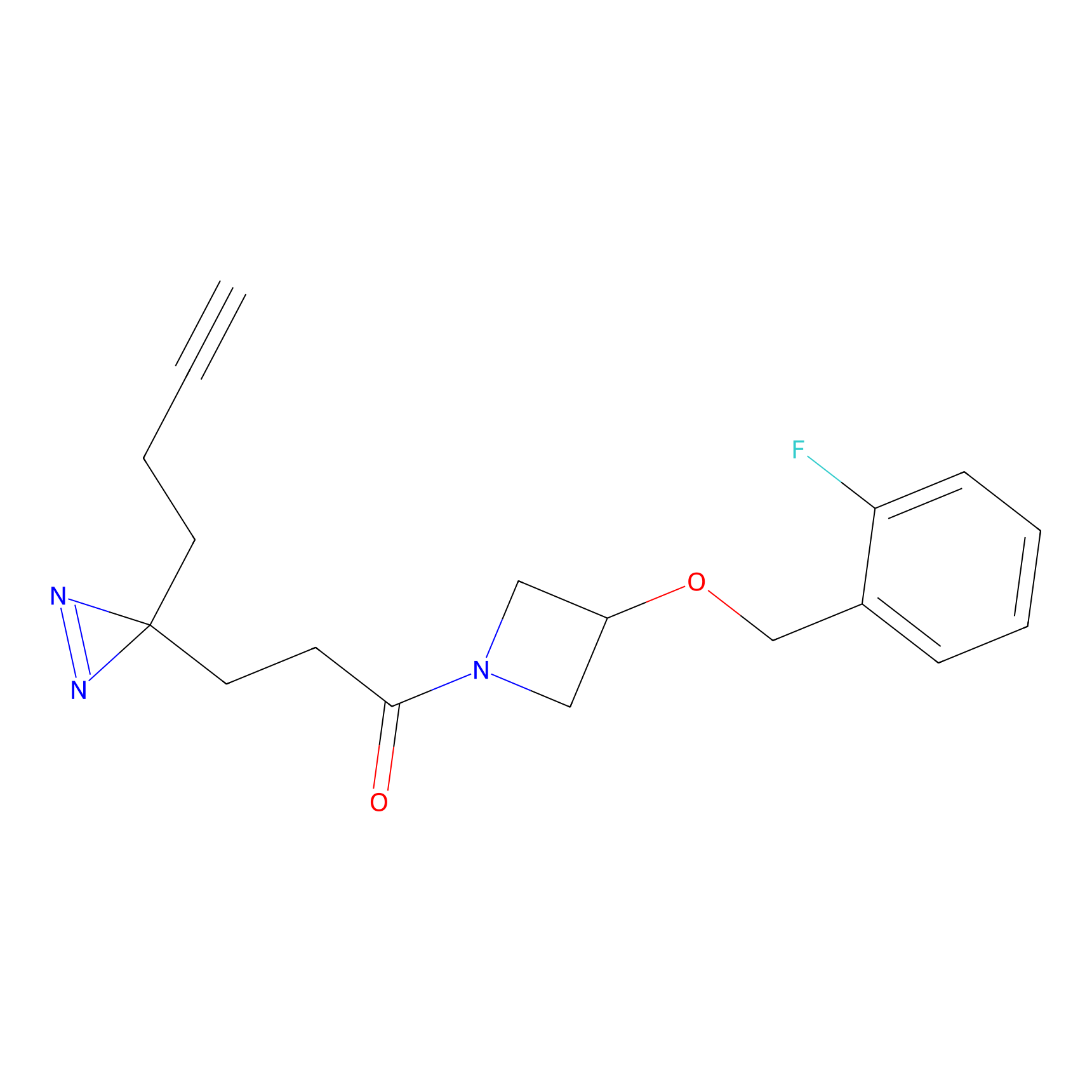
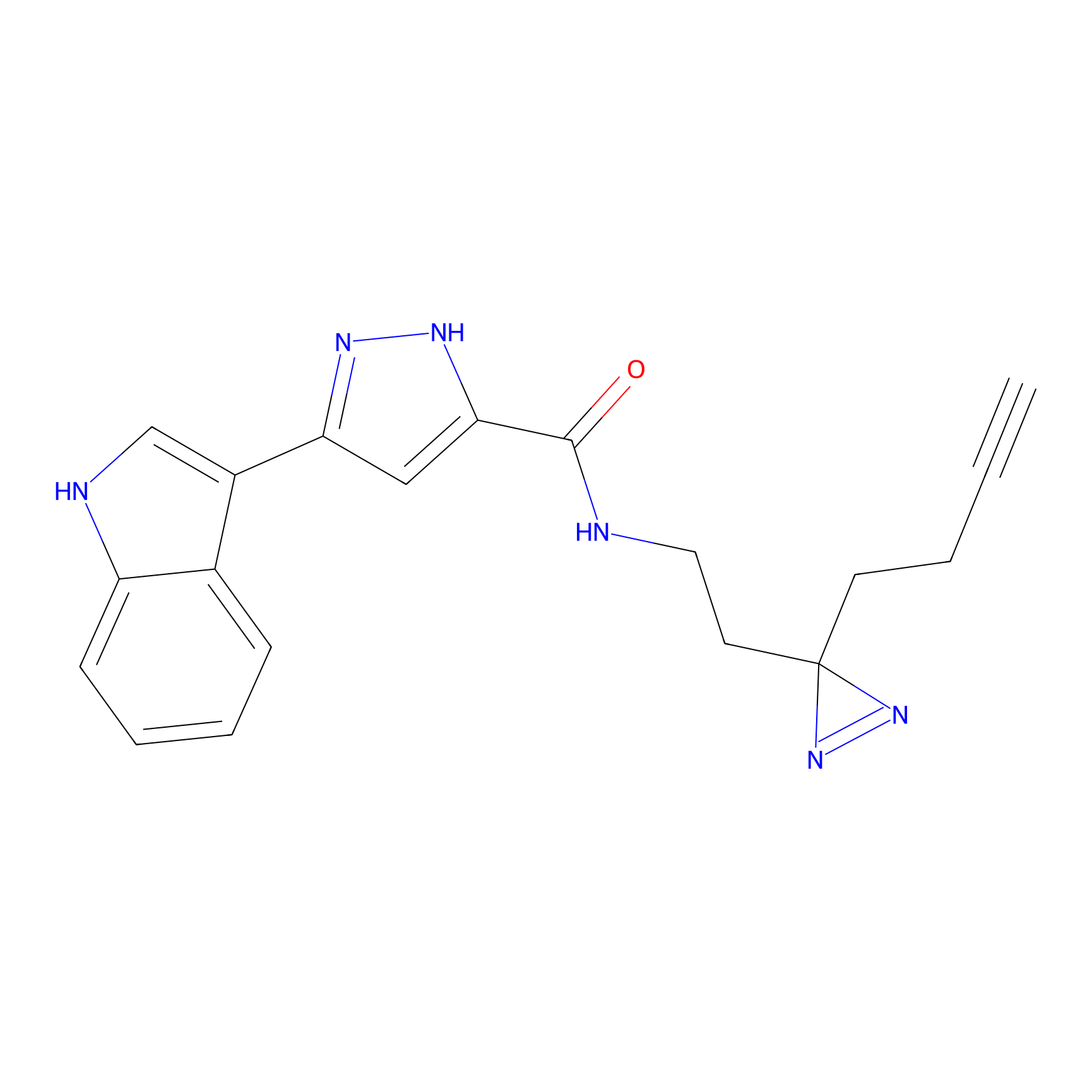
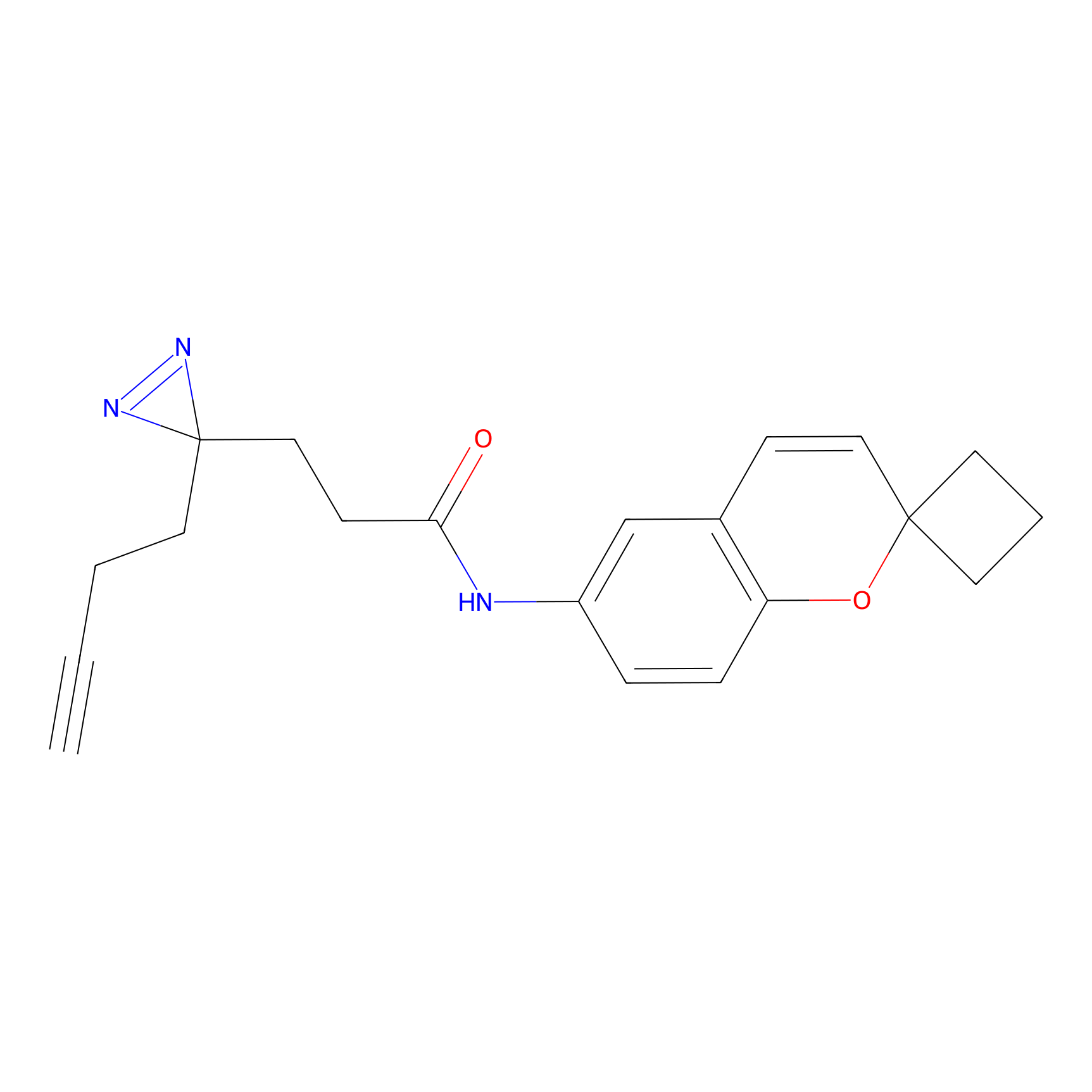
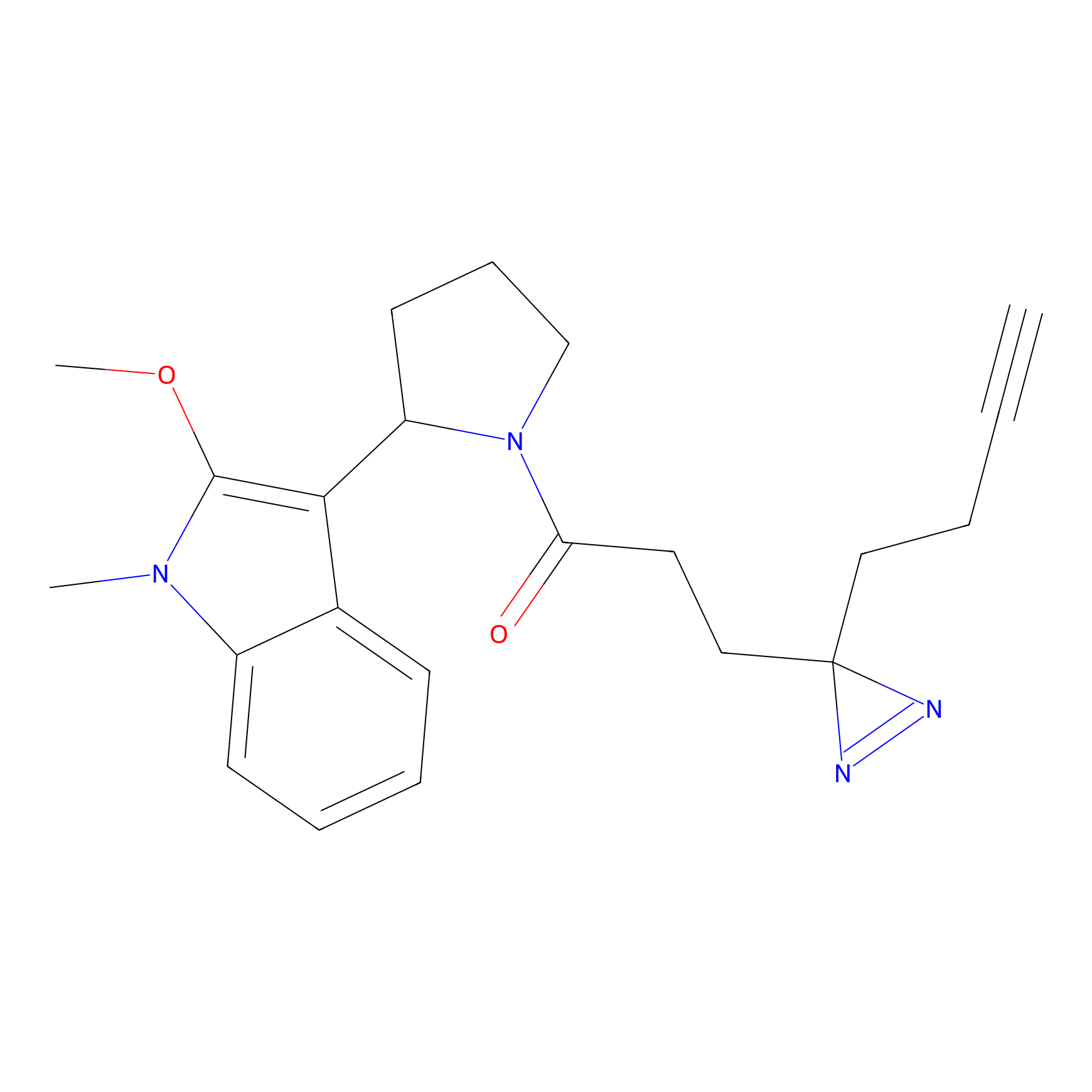
|
Probe 1 Probe Info |
 |
Y244(25.27); Y245(27.46) | LDD3495 | [2] | |
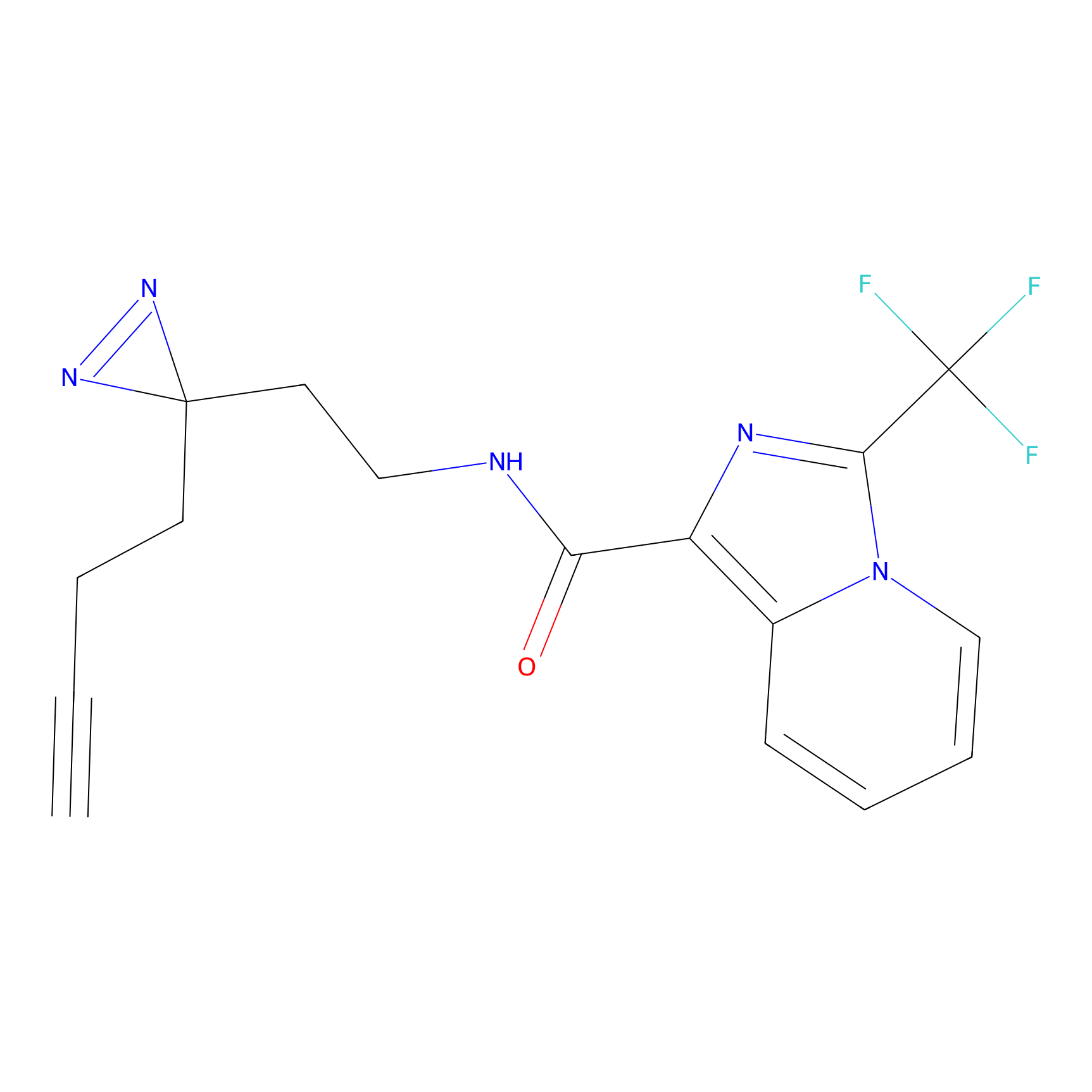
|
HPAP Probe Info |
 |
4.16 | LDD0062 | [3] | |
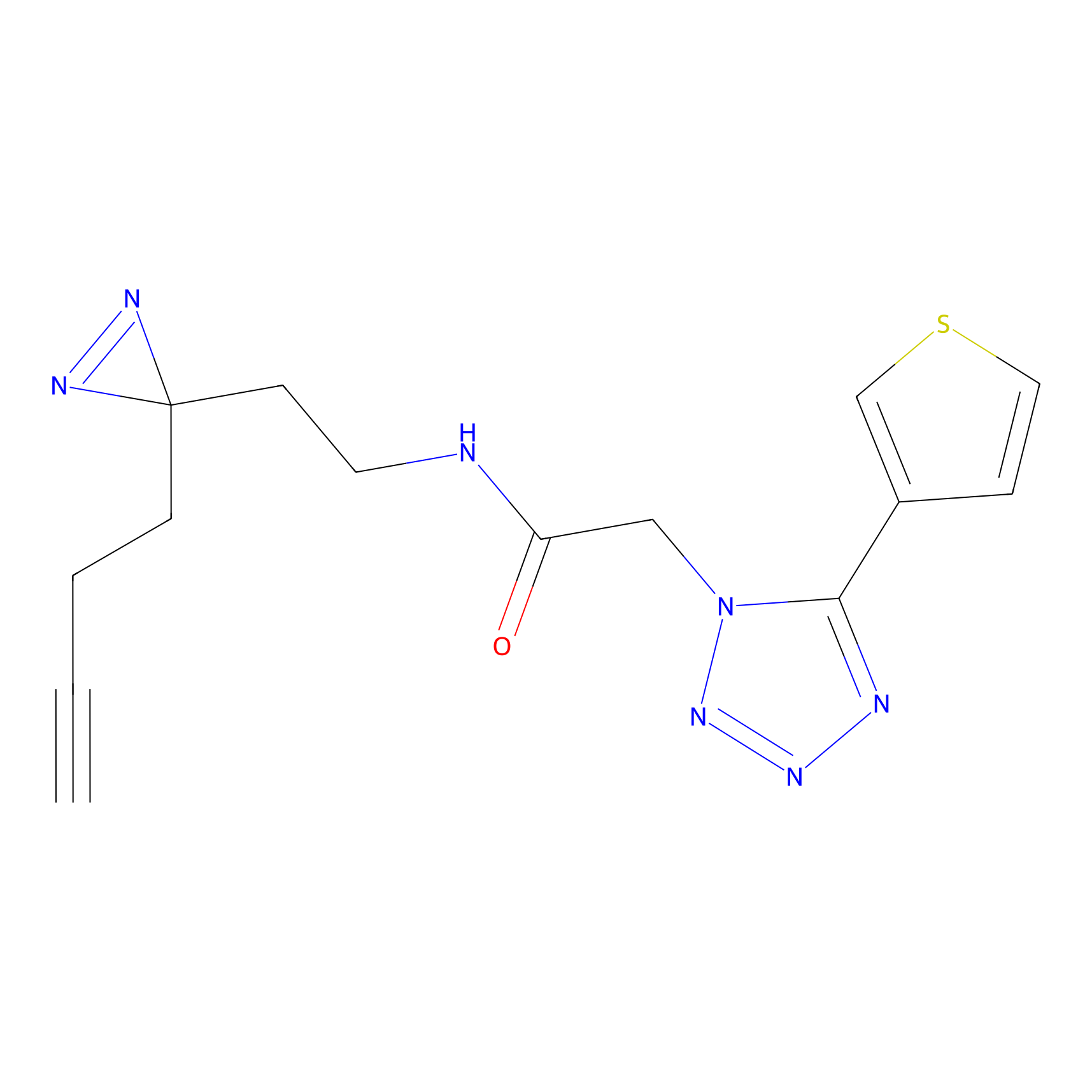
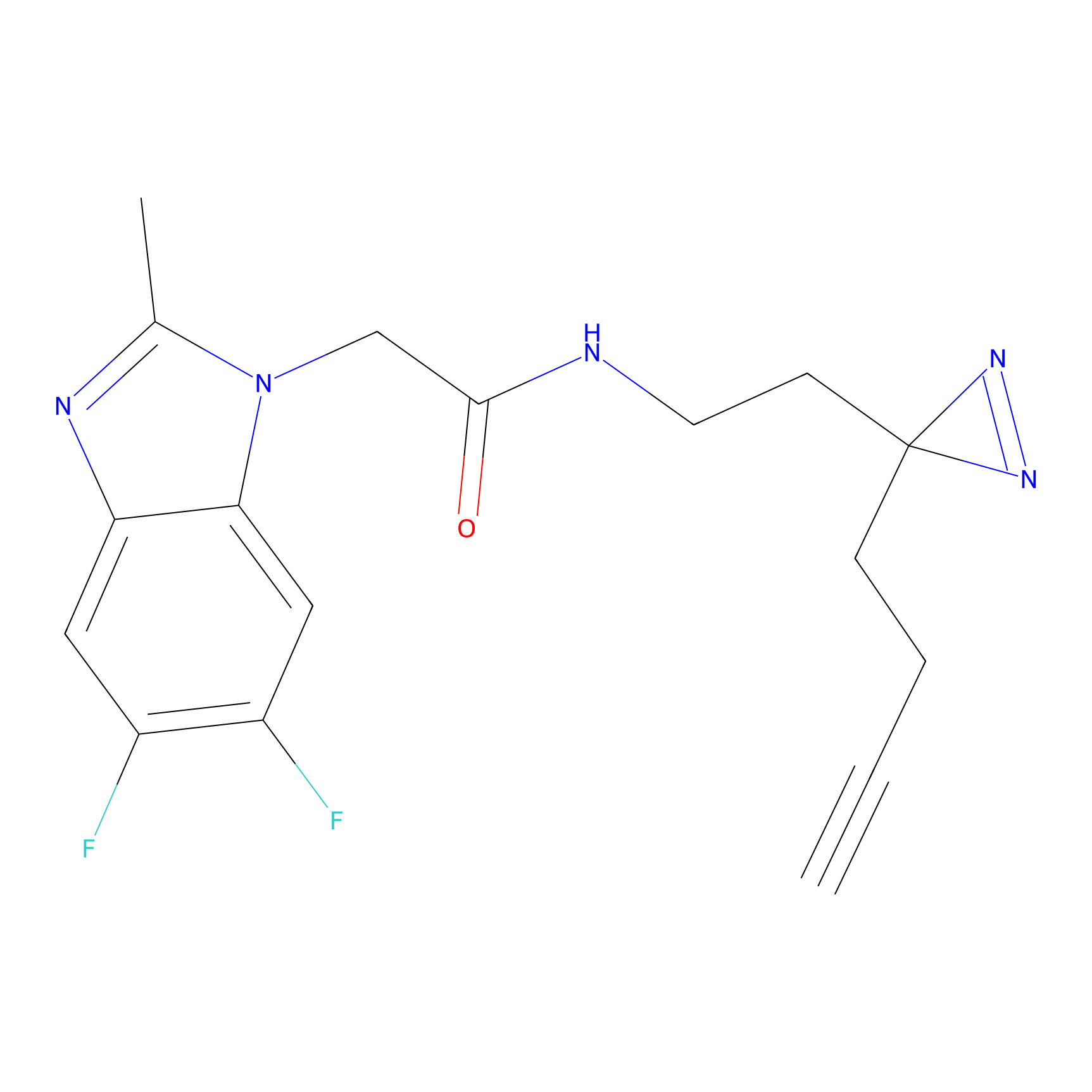
|
DBIA Probe Info |
 |
C173(0.92) | LDD1575 | [4] | |
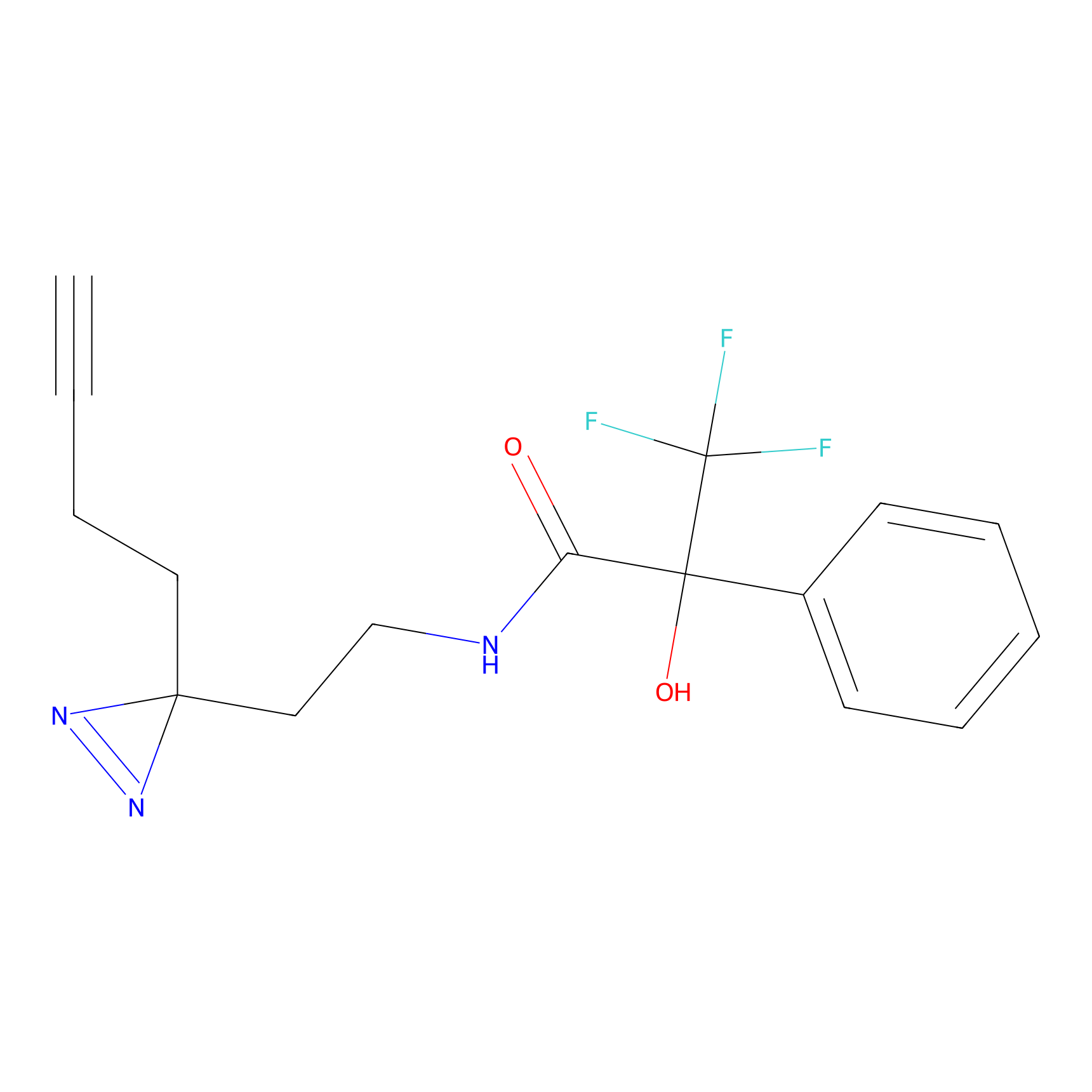
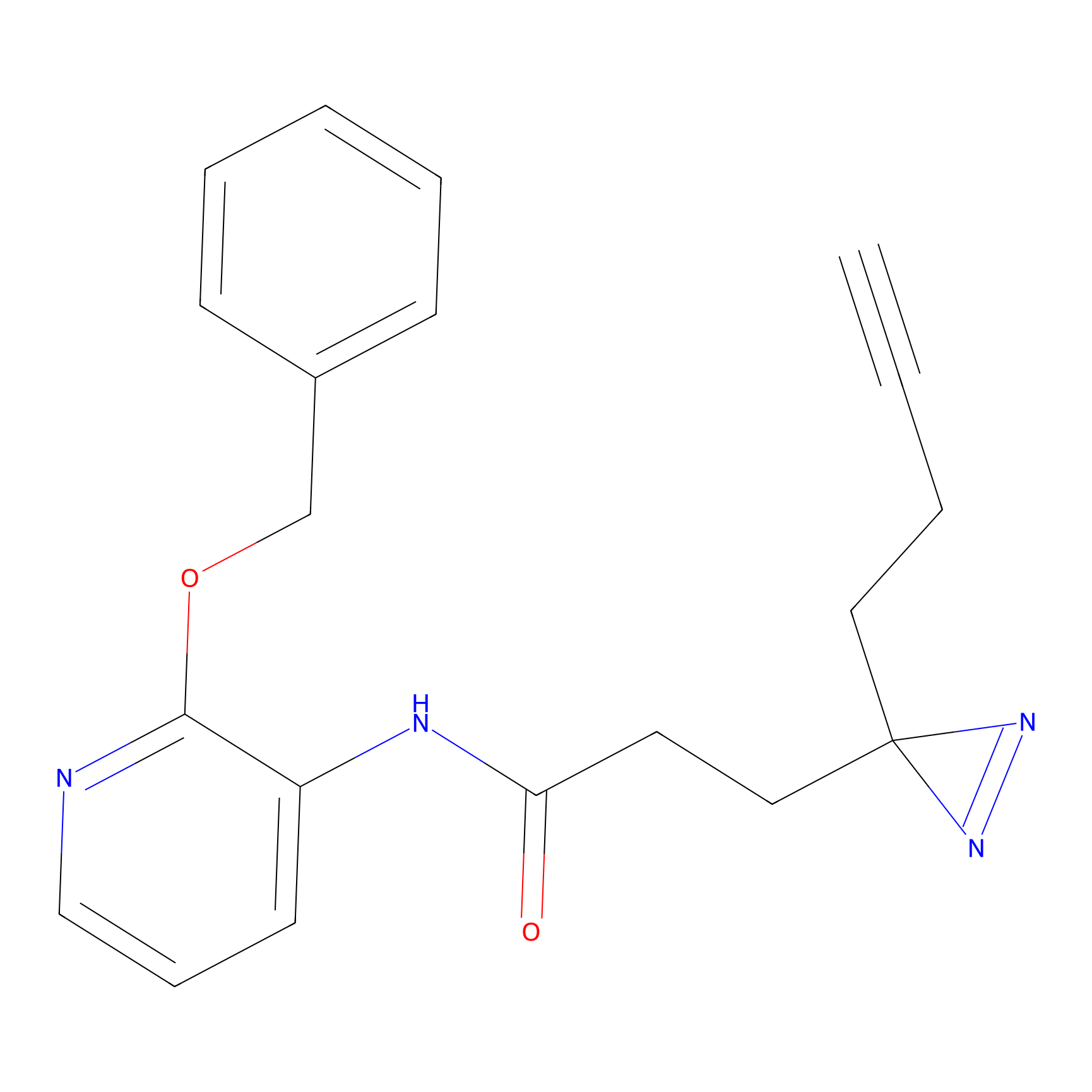
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [5] | |
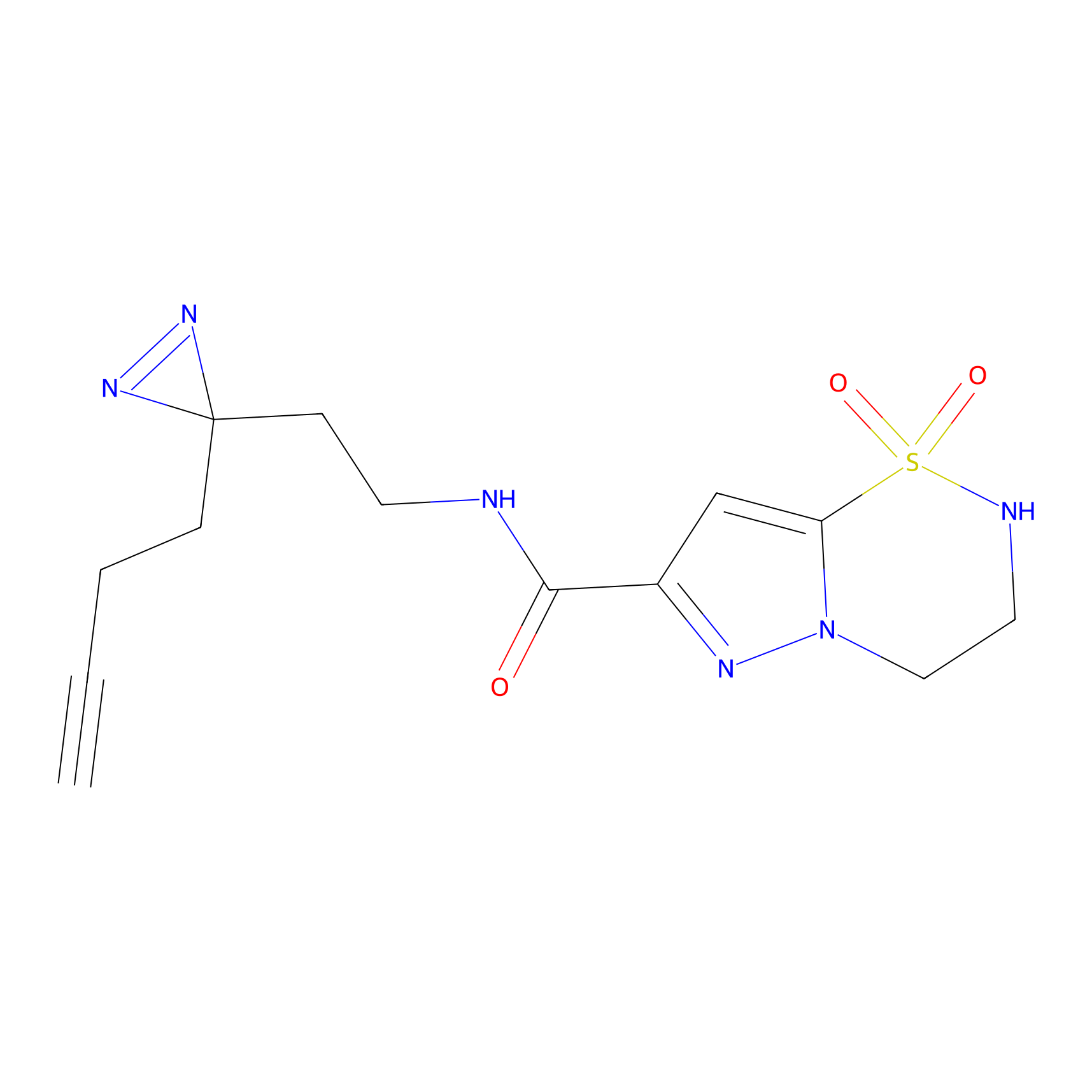
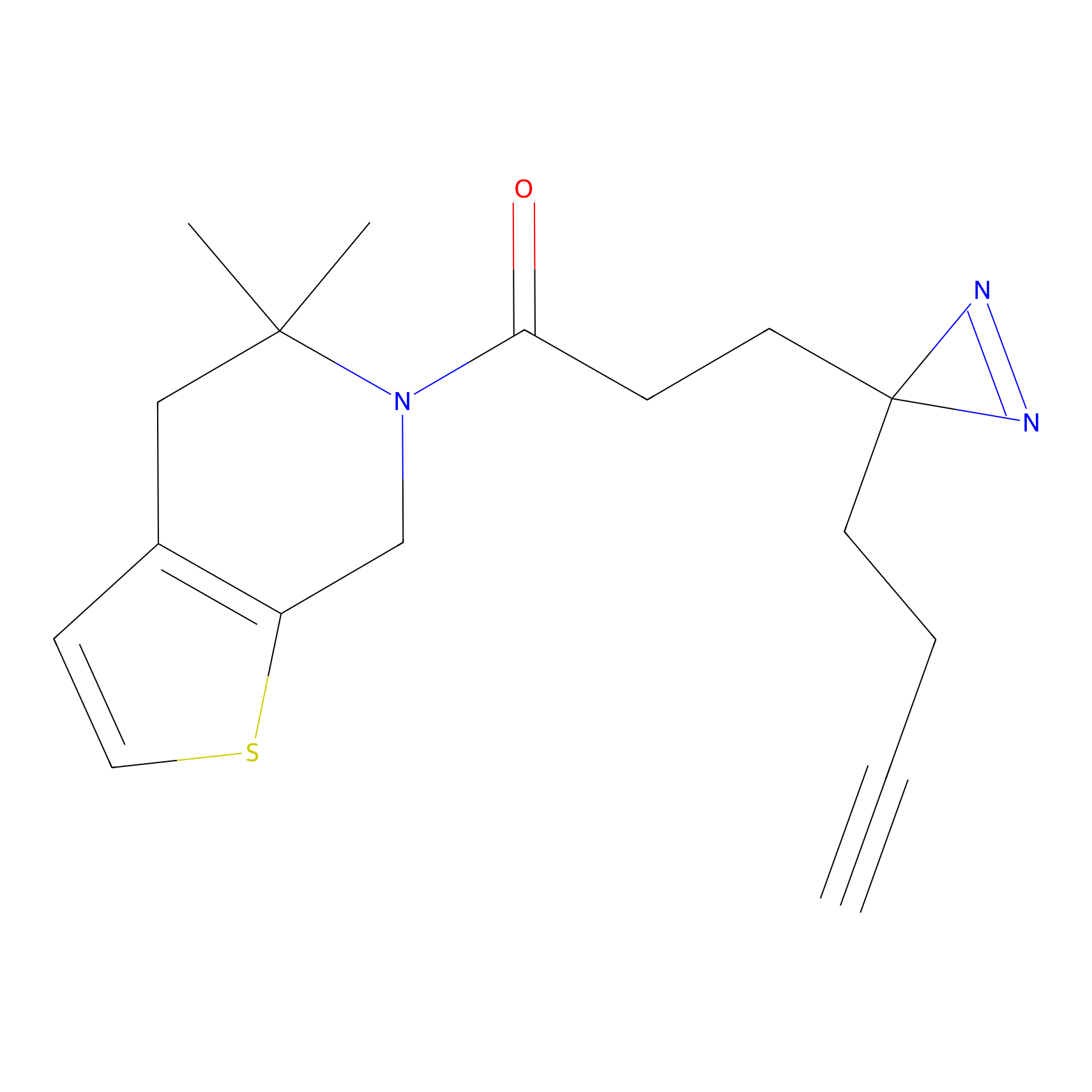
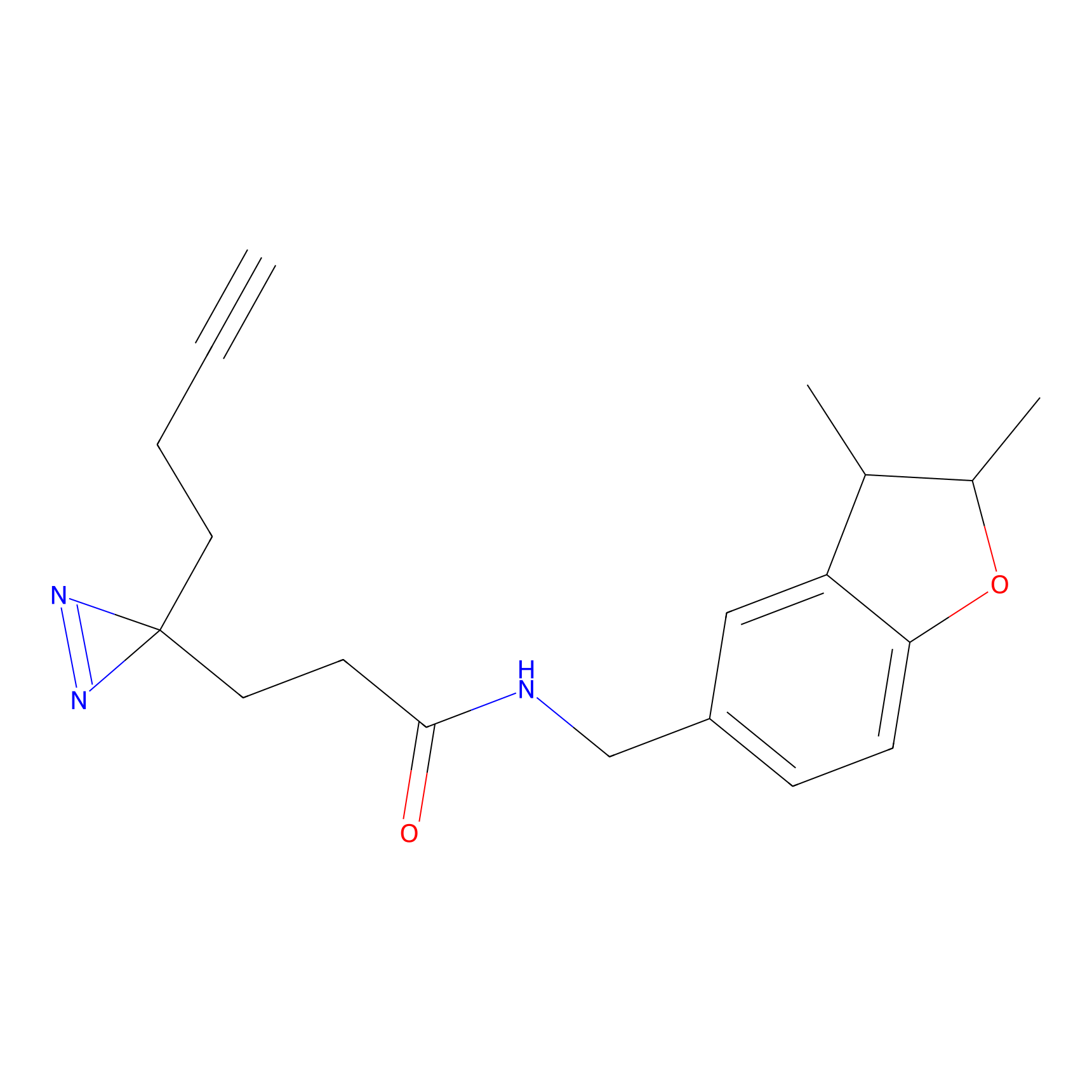
|
ATP probe Probe Info |
 |
K133(0.00); K283(0.00) | LDD0199 | [6] | |
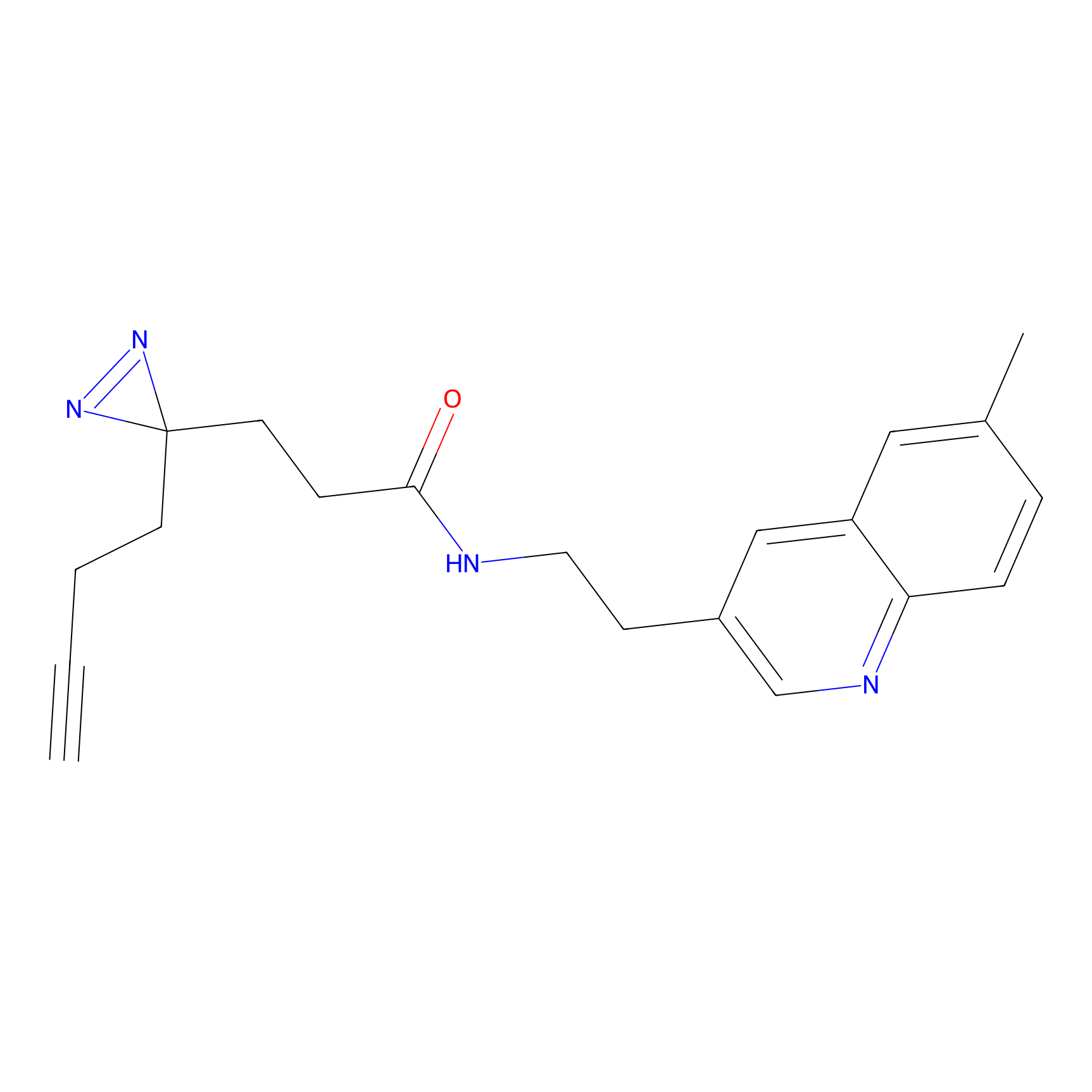
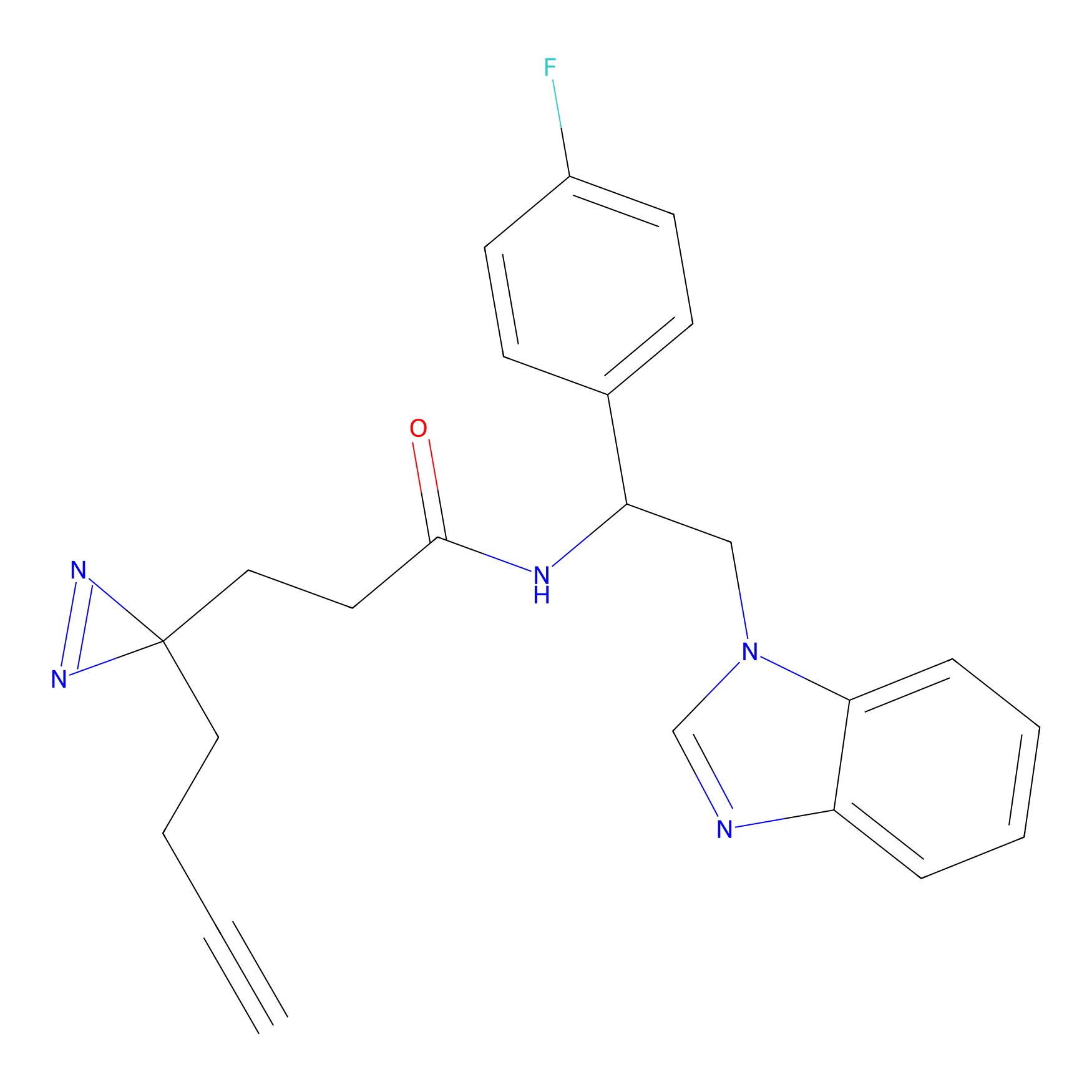
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C169(0.00); C173(0.00) | LDD0038 | [7] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0165 | [8] | |
|
Lodoacetamide azide Probe Info |
 |
C169(0.00); C173(0.00) | LDD0037 | [7] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [9] | |
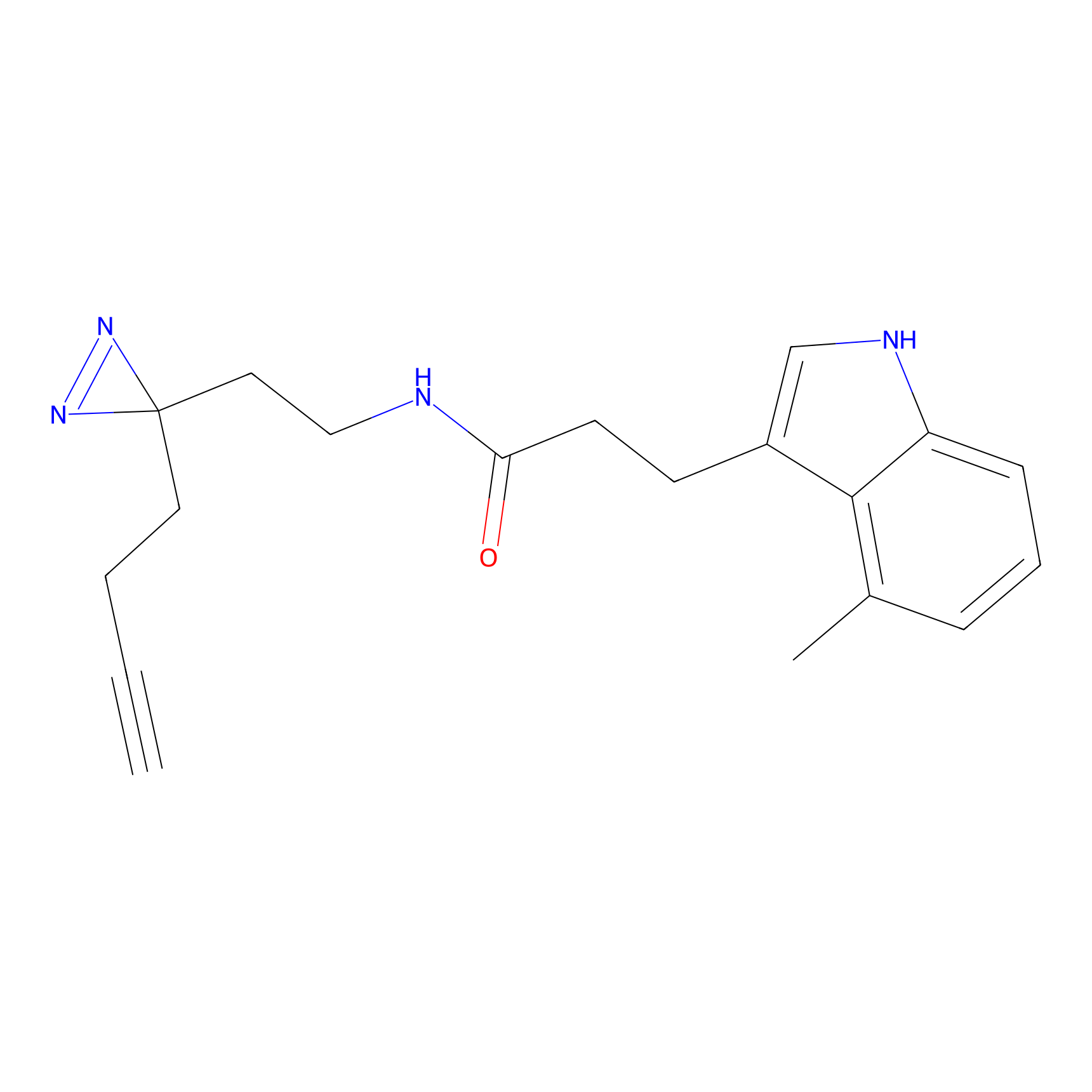
|
1d-yne Probe Info |
 |
N.A. | LDD0357 | [10] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [11] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0156 | Aniline | NCI-H1299 | 11.73 | LDD0403 | [1] |
| LDCM0371 | CL102 | HEK-293T | C173(0.92) | LDD1575 | [4] |
| LDCM0375 | CL106 | HEK-293T | C173(1.01) | LDD1579 | [4] |
| LDCM0380 | CL110 | HEK-293T | C173(0.86) | LDD1584 | [4] |
| LDCM0384 | CL114 | HEK-293T | C173(0.89) | LDD1588 | [4] |
| LDCM0388 | CL118 | HEK-293T | C173(0.85) | LDD1592 | [4] |
| LDCM0393 | CL122 | HEK-293T | C173(1.05) | LDD1597 | [4] |
| LDCM0397 | CL126 | HEK-293T | C173(1.00) | LDD1601 | [4] |
| LDCM0401 | CL14 | HEK-293T | C173(1.15) | LDD1605 | [4] |
| LDCM0407 | CL2 | HEK-293T | C173(0.95) | LDD1611 | [4] |
| LDCM0414 | CL26 | HEK-293T | C173(0.96) | LDD1618 | [4] |
| LDCM0441 | CL50 | HEK-293T | C173(0.95) | LDD1645 | [4] |
| LDCM0454 | CL62 | HEK-293T | C173(1.07) | LDD1657 | [4] |
| LDCM0467 | CL74 | HEK-293T | C173(1.00) | LDD1670 | [4] |
| LDCM0480 | CL86 | HEK-293T | C173(0.96) | LDD1683 | [4] |
| LDCM0493 | CL98 | HEK-293T | C173(0.99) | LDD1696 | [4] |
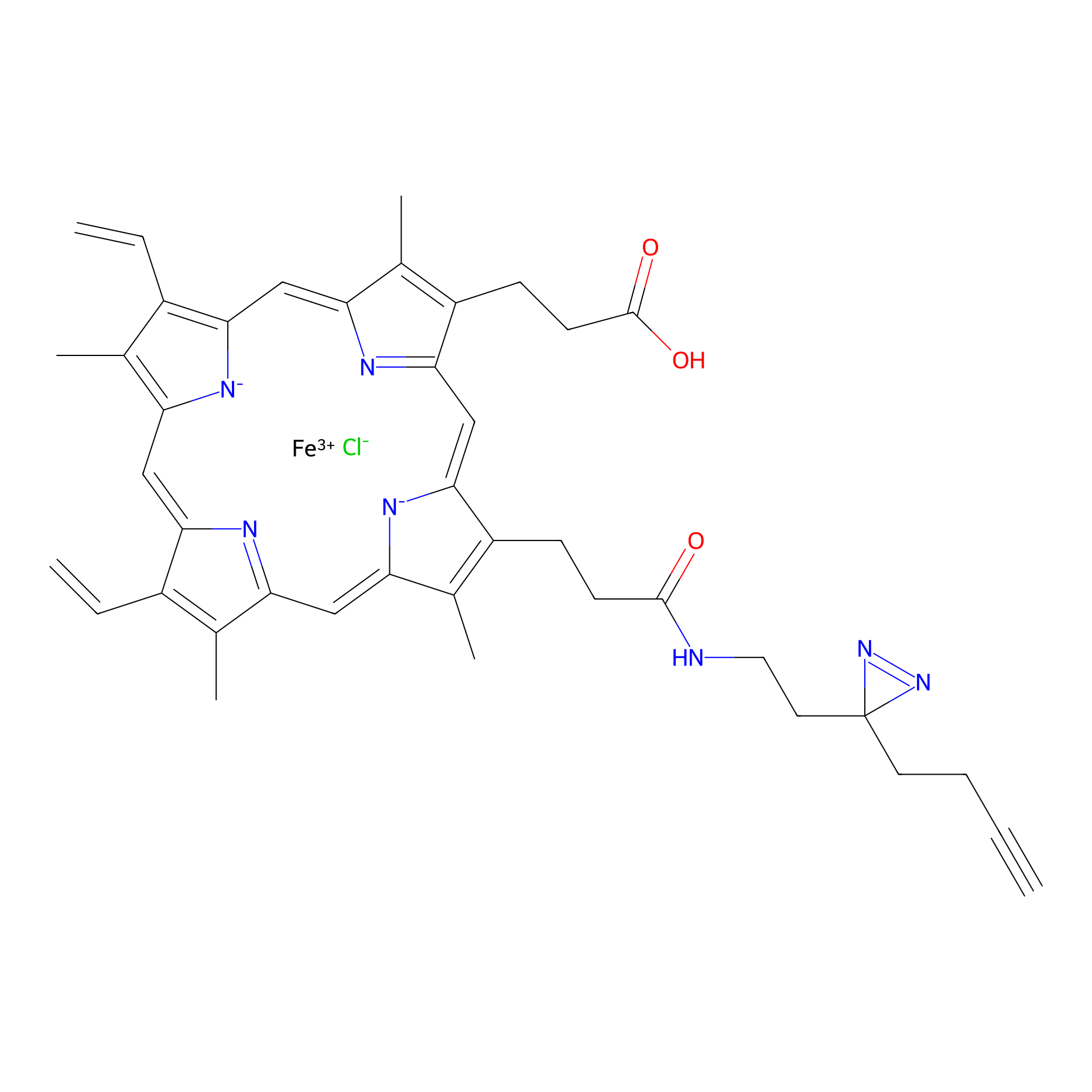
| LDCM0427 | Fragment51 | HEK-293T | C173(0.97) | LDD1631 | [4] |
| LDCM0014 | Panhematin | HEK-293T | 4.16 | LDD0062 | [3] |
The Interaction Atlas With This Target
References