Details of the Target
General Information of Target
| Target ID | LDTP01562 | |||||
|---|---|---|---|---|---|---|
| Target Name | Peptidyl-prolyl cis-trans isomerase FKBP9 (FKBP9) | |||||
| Gene Name | FKBP9 | |||||
| Gene ID | 11328 | |||||
| Synonyms |
FKBP60; FKBP63; Peptidyl-prolyl cis-trans isomerase FKBP9; PPIase FKBP9; EC 5.2.1.8; 63 kDa FK506-binding protein; 63 kDa FKBP; FKBP-63; FK506-binding protein 9; FKBP-9; Rotamase |
|||||
| 3D Structure | ||||||
| Sequence |
MAFRGWRPPPPPLLLLLLWVTGQAAPVAGLGSDAELQIERRFVPDECPRTVRSGDFVRYH
YVGTFPDGQKFDSSYDRDSTFNVFVGKGQLITGMDQALVGMCVNERRFVKIPPKLAYGNE GVSGVIPPNSVLHFDVLLMDIWNSEDQVQIHTYFKPPSCPRTIQVSDFVRYHYNGTFLDG TLFDSSHNRMKTYDTYVGIGWLIPGMDKGLLGMCVGEKRIITIPPFLAYGEDGDGKDIPG QASLVFDVALLDLHNPKDSISIENKVVPENCERISQSGDFLRYHYNGTLLDGTLFDSSYS RNRTFDTYIGQGYVIPGMDEGLLGVCIGEKRRIVVPPHLGYGEEGRGNIPGSAVLVFDIH VIDFHNPSDSISITSHYKPPDCSVLSKKGDYLKYHYNASLLDGTLLDSTWNLGKTYNIVL GSGQVVLGMDMGLREMCVGEKRTVIIPPHLGYGEAGVDGEVPGSAVLVFDIELLELVAGL PEGYMFIWNGEVSPNLFEEIDKDGNGEVLLEEFSEYIHAQVASGKGKLAPGFDAELIVKN MFTNQDRNGDGKVTAEEFKLKDQEAKHDEL |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Subcellular location |
Endoplasmic reticulum
|
|||||
| Function | PPIases accelerate the folding of proteins during protein synthesis. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
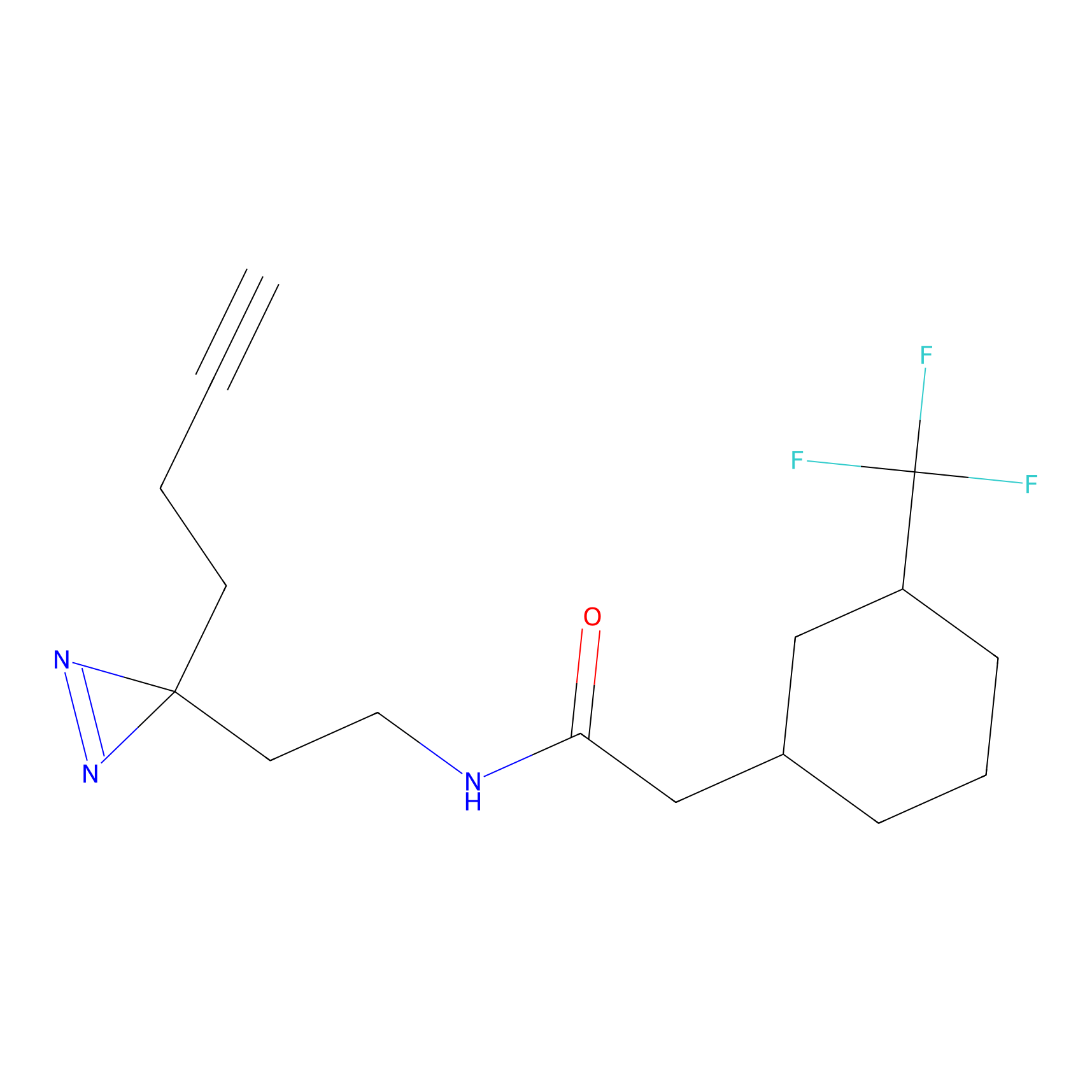
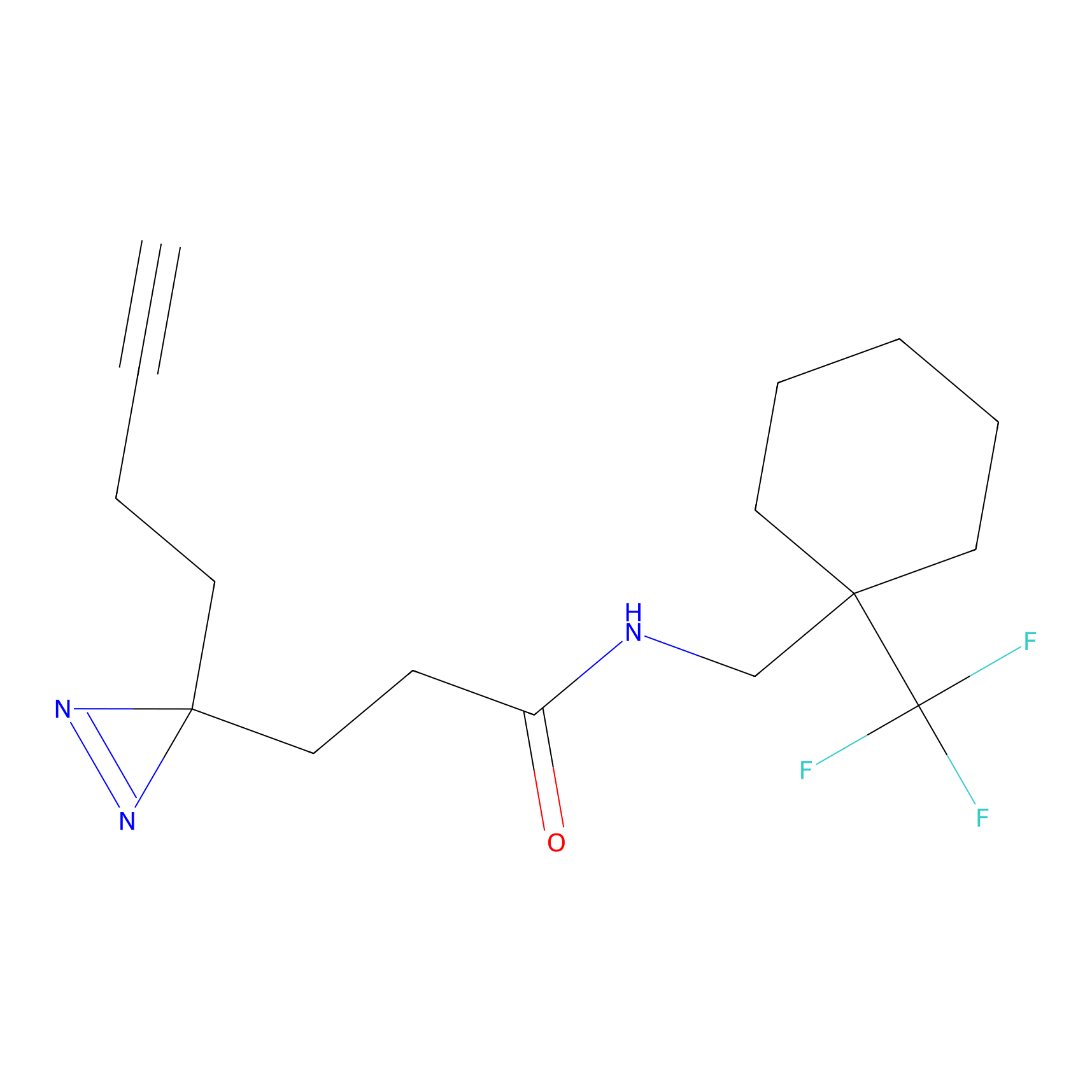
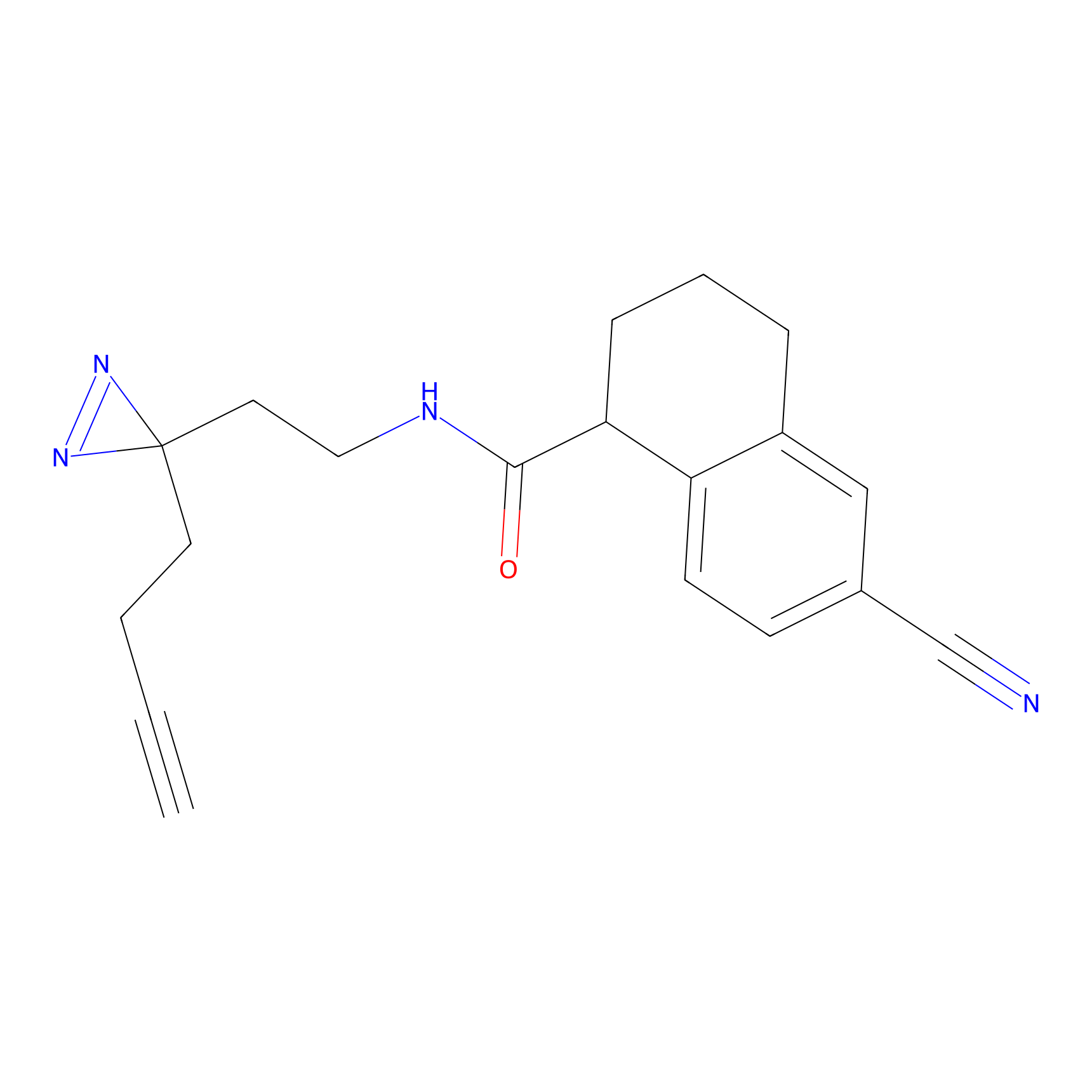
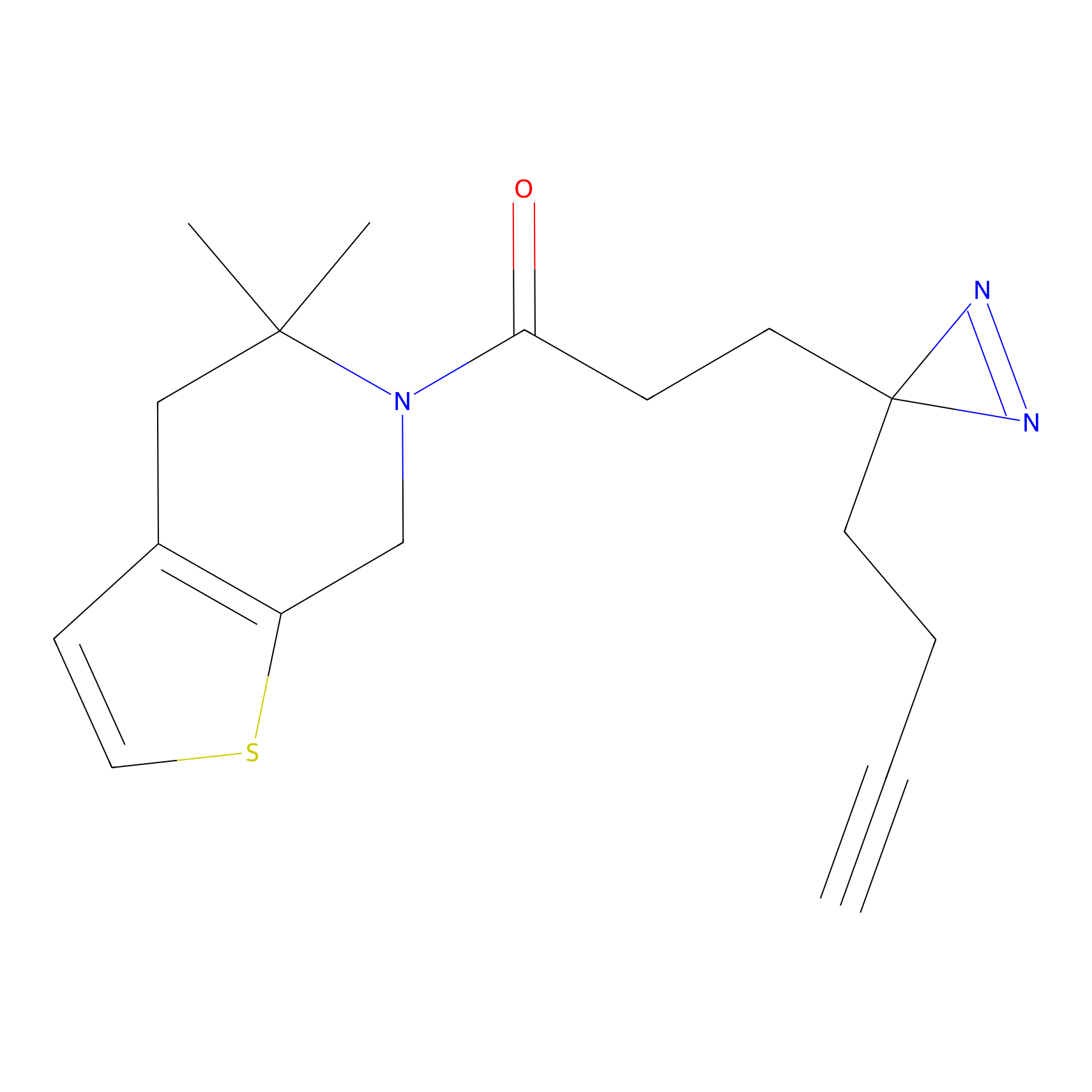
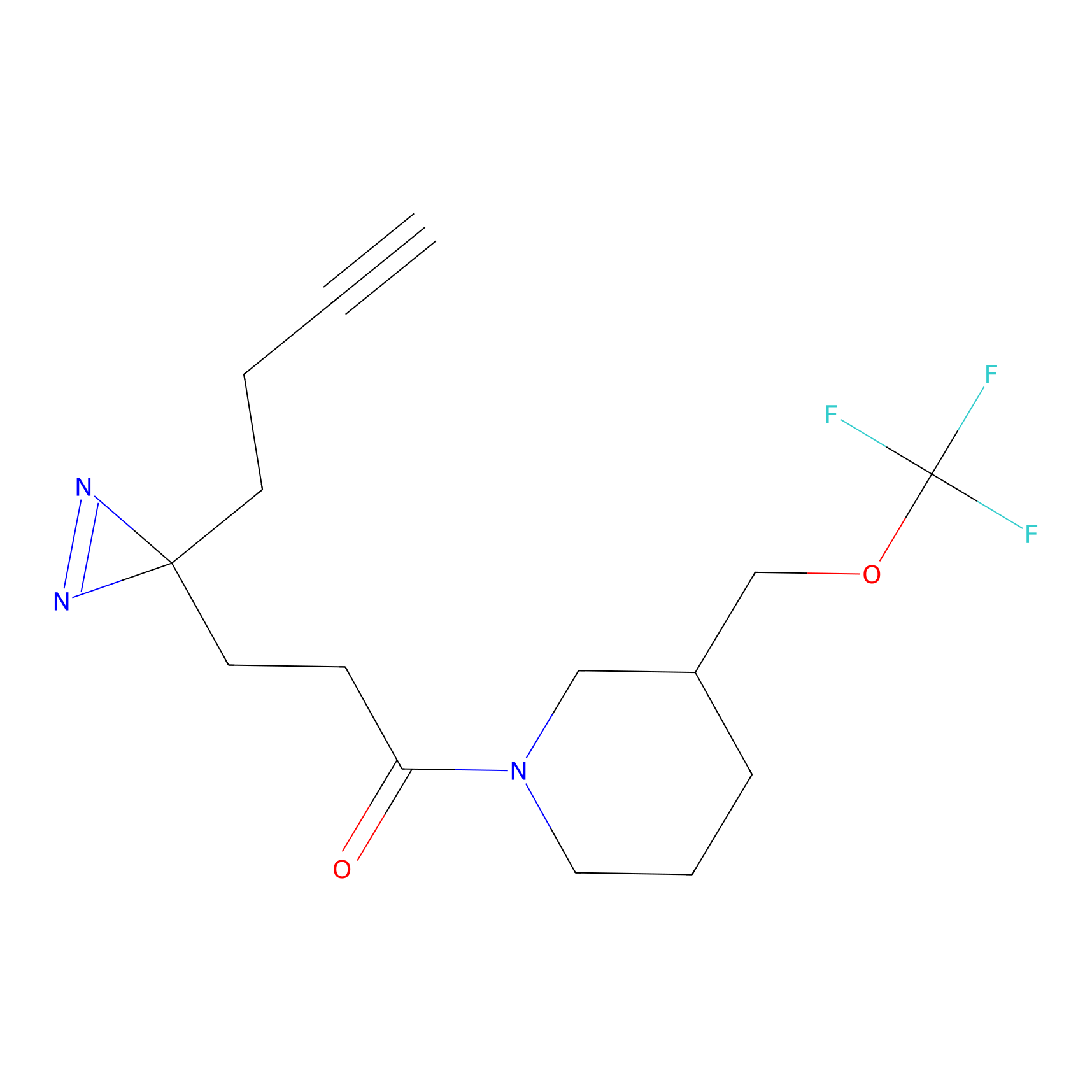
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
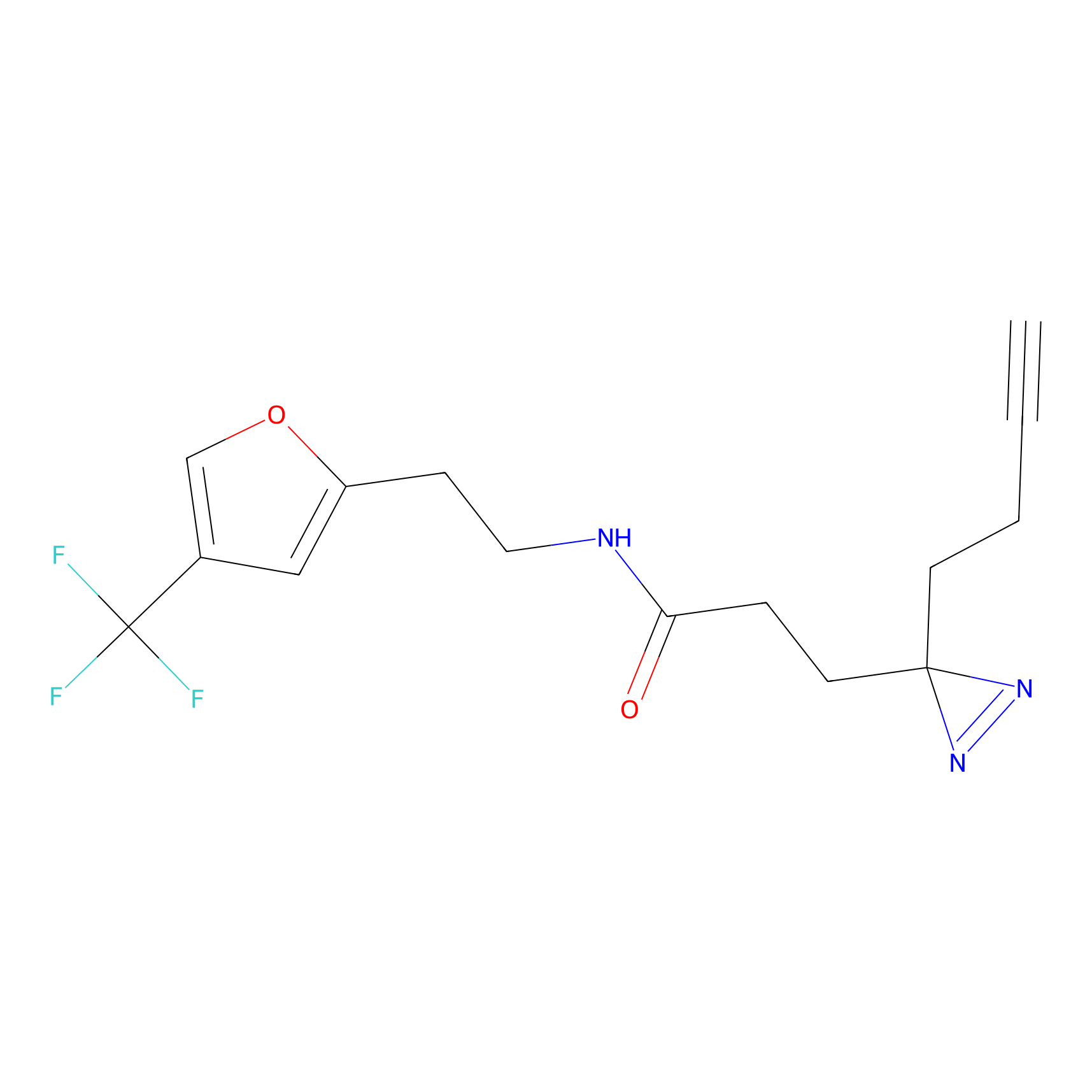
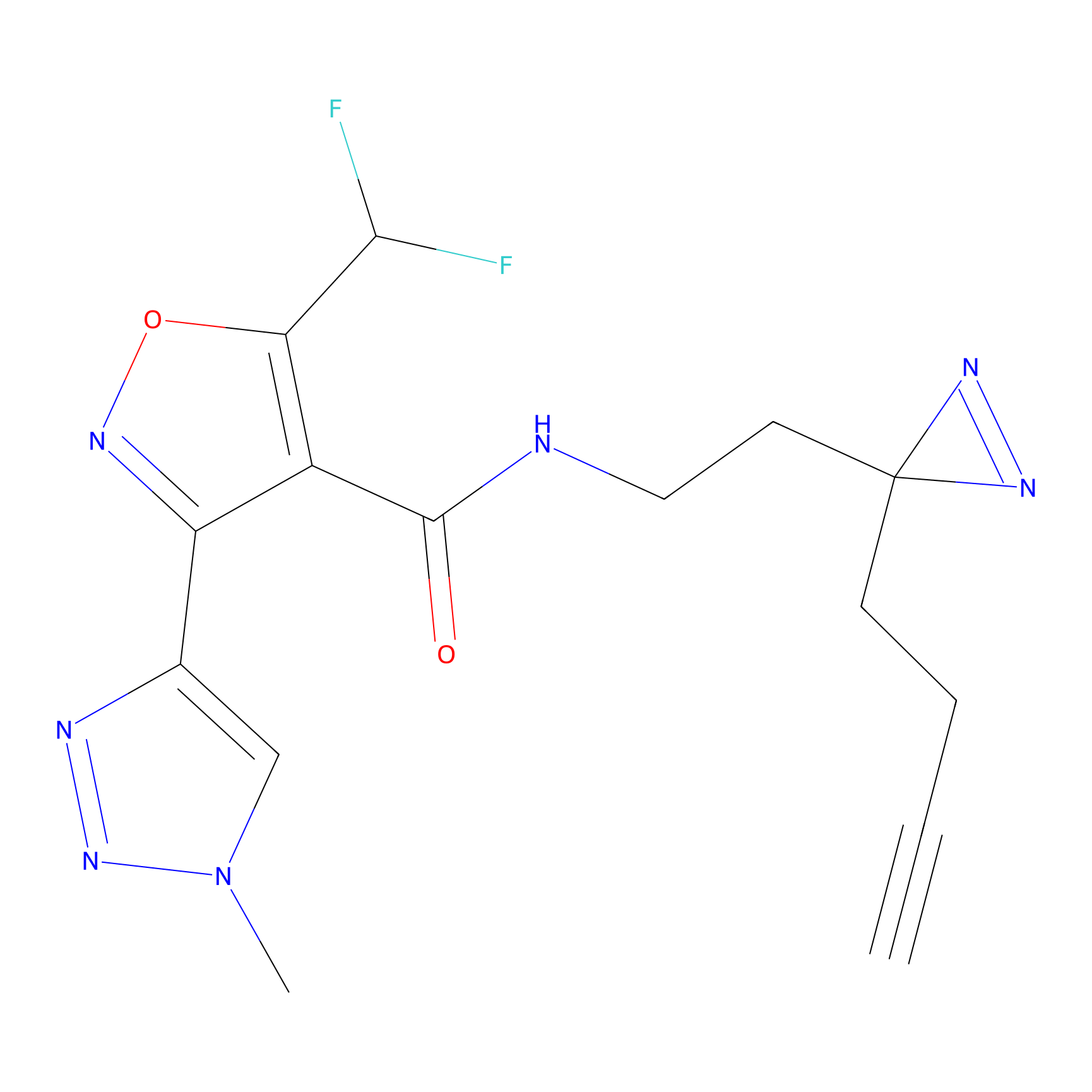
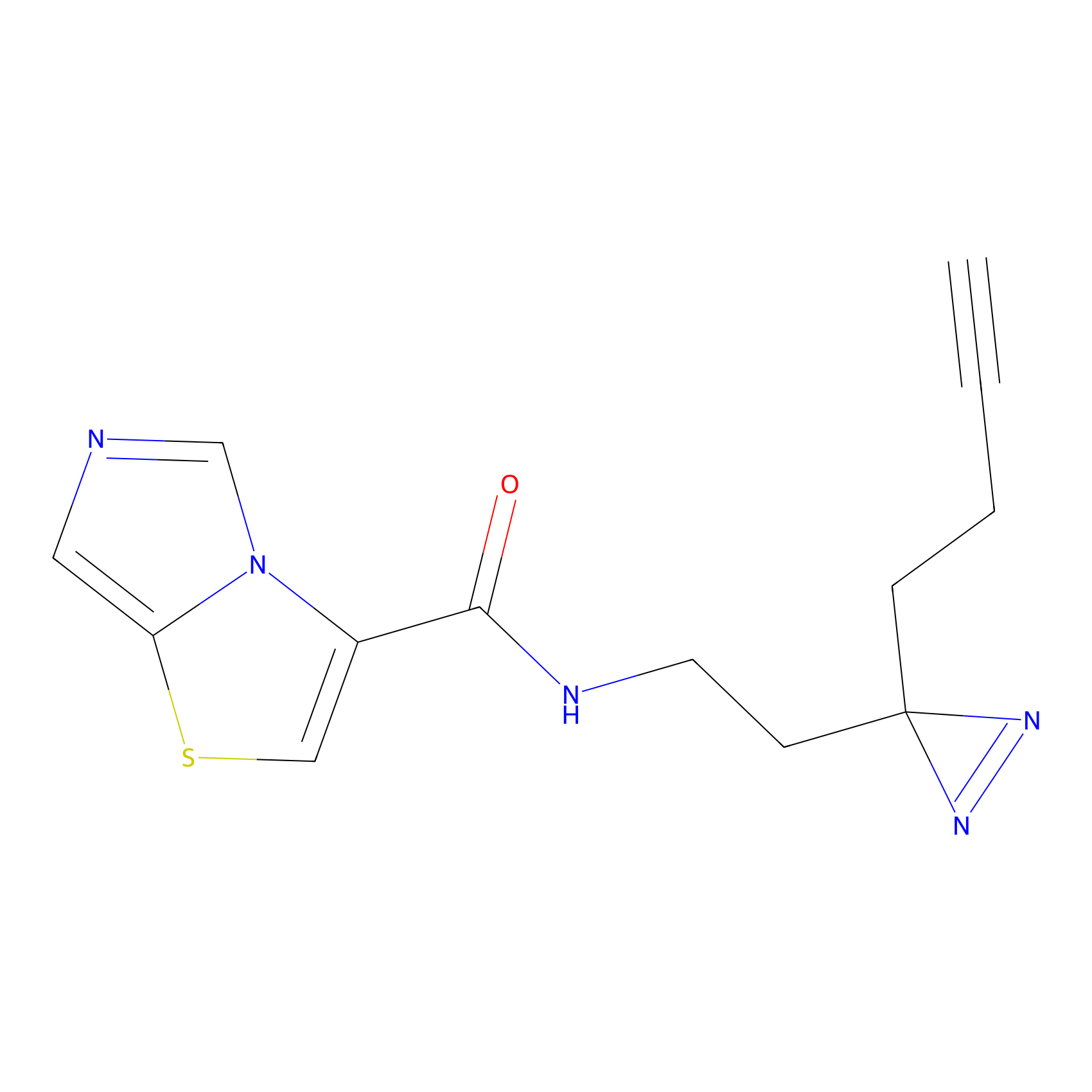
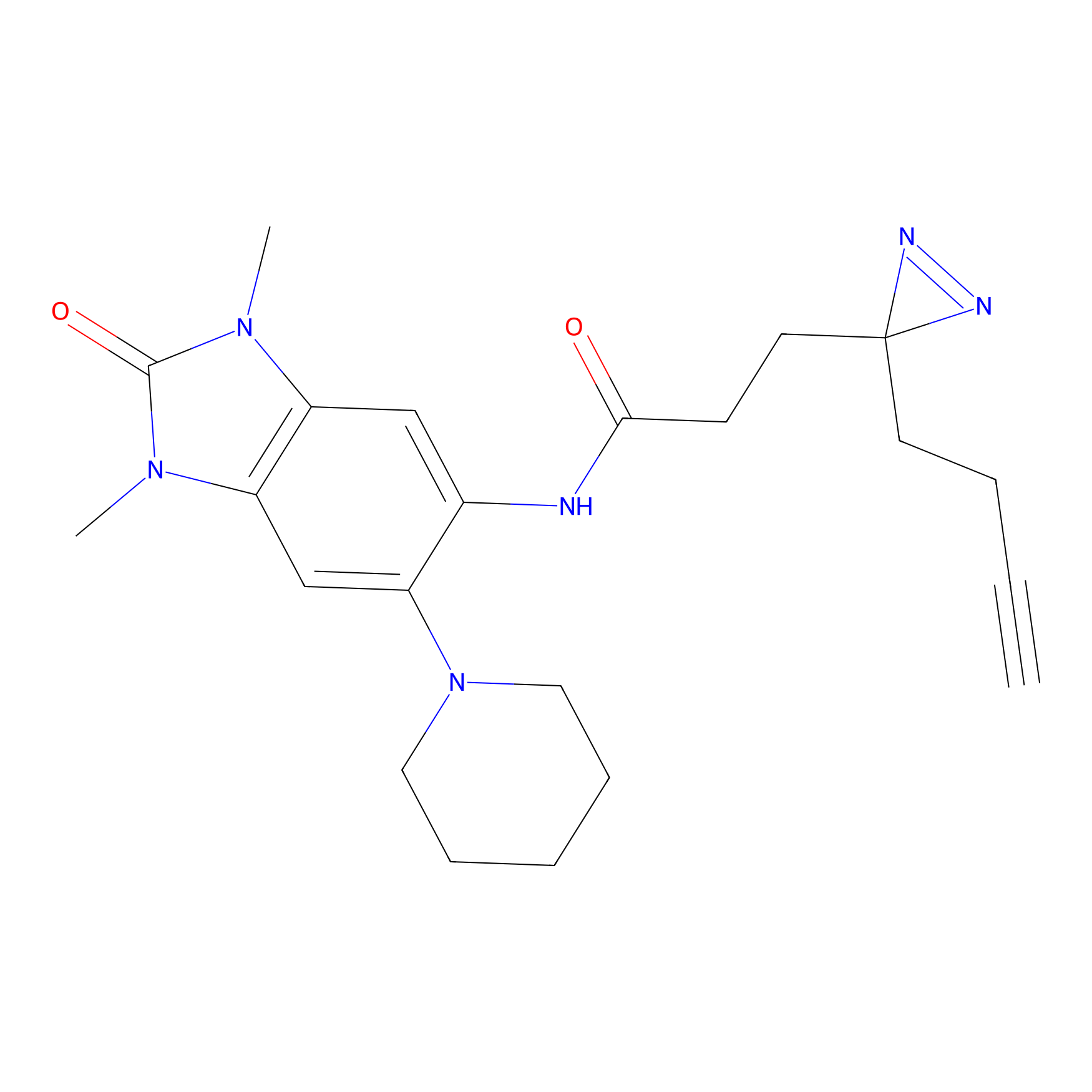
Probe 1 Probe Info |
 |
Y75(29.54) | LDD3495 | [1] | |
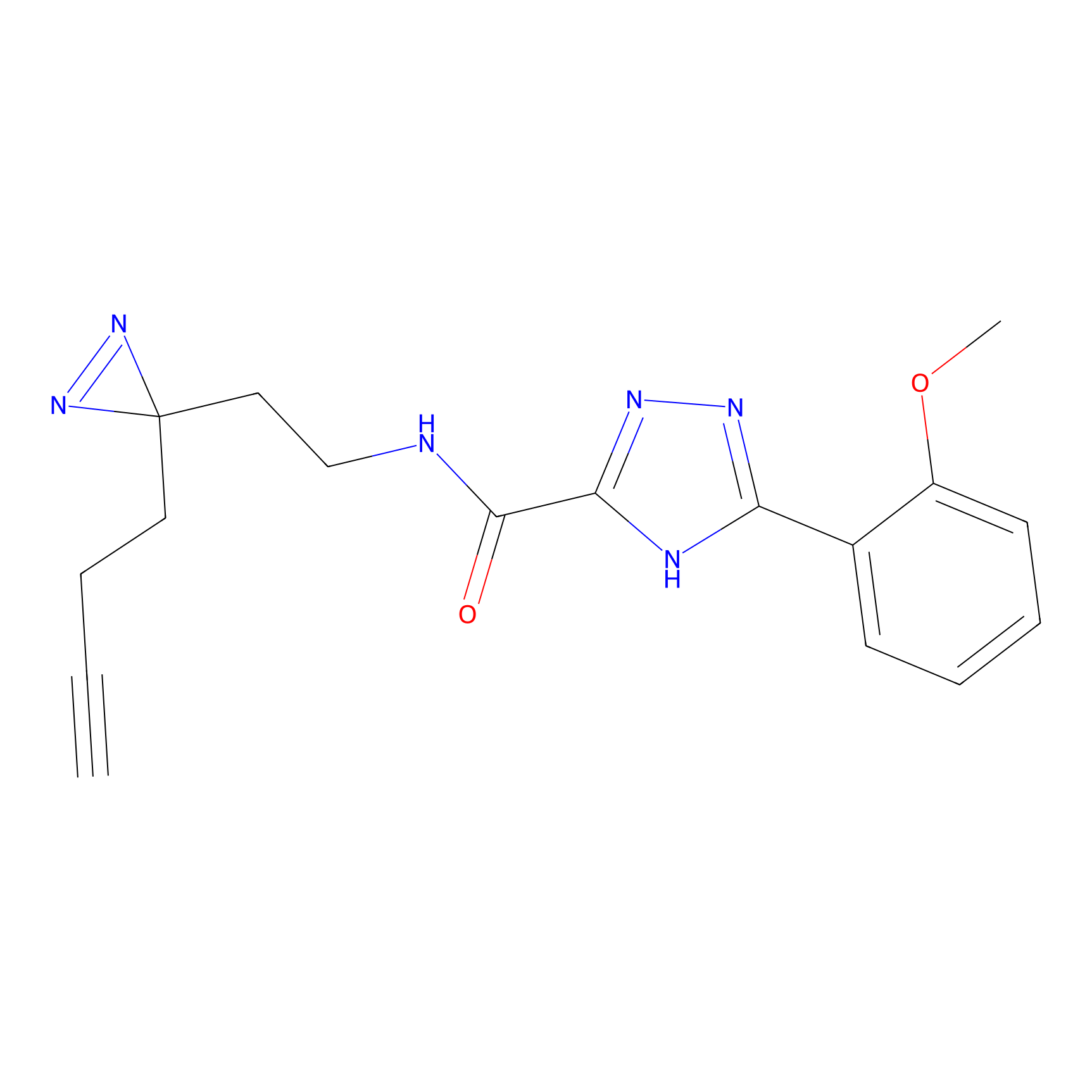
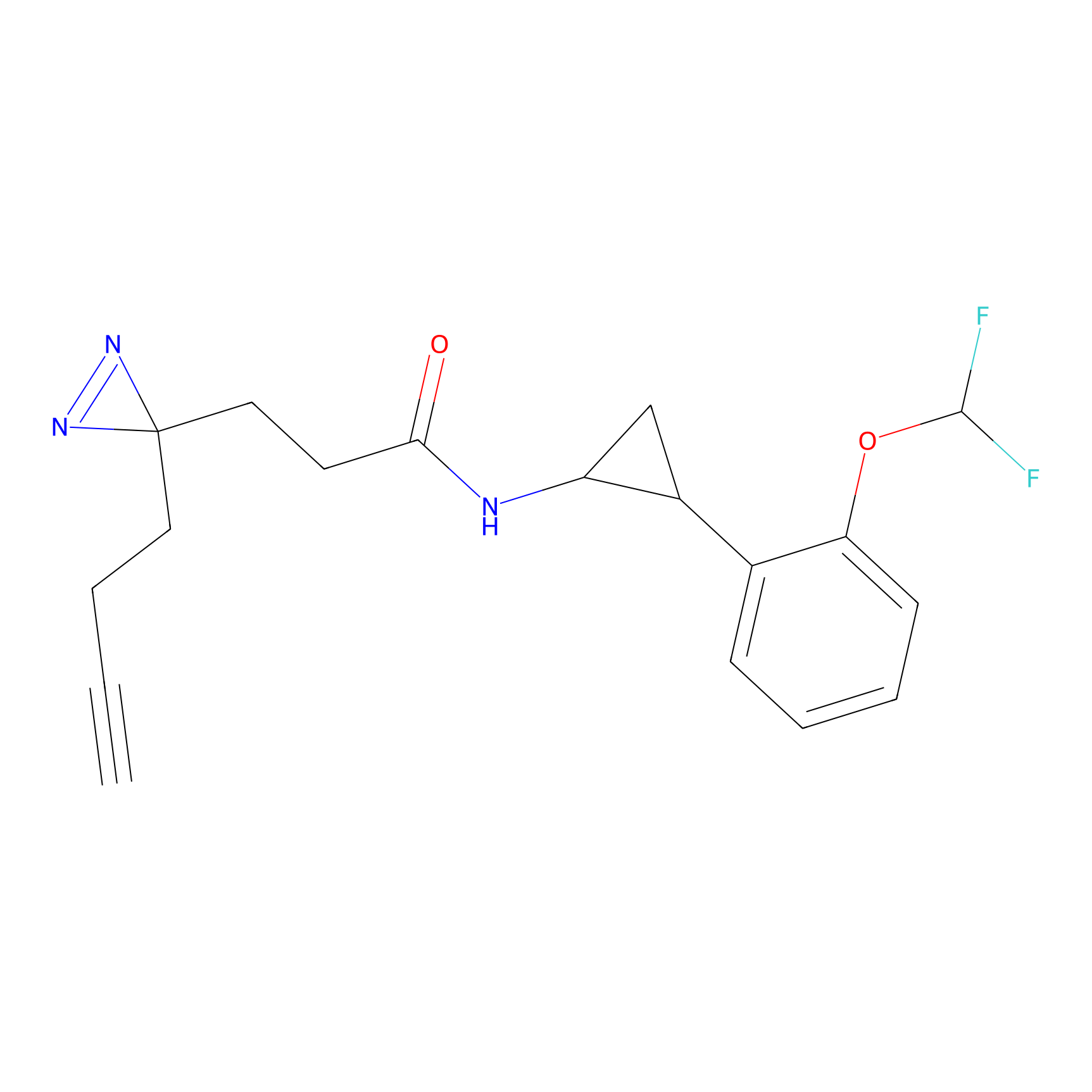
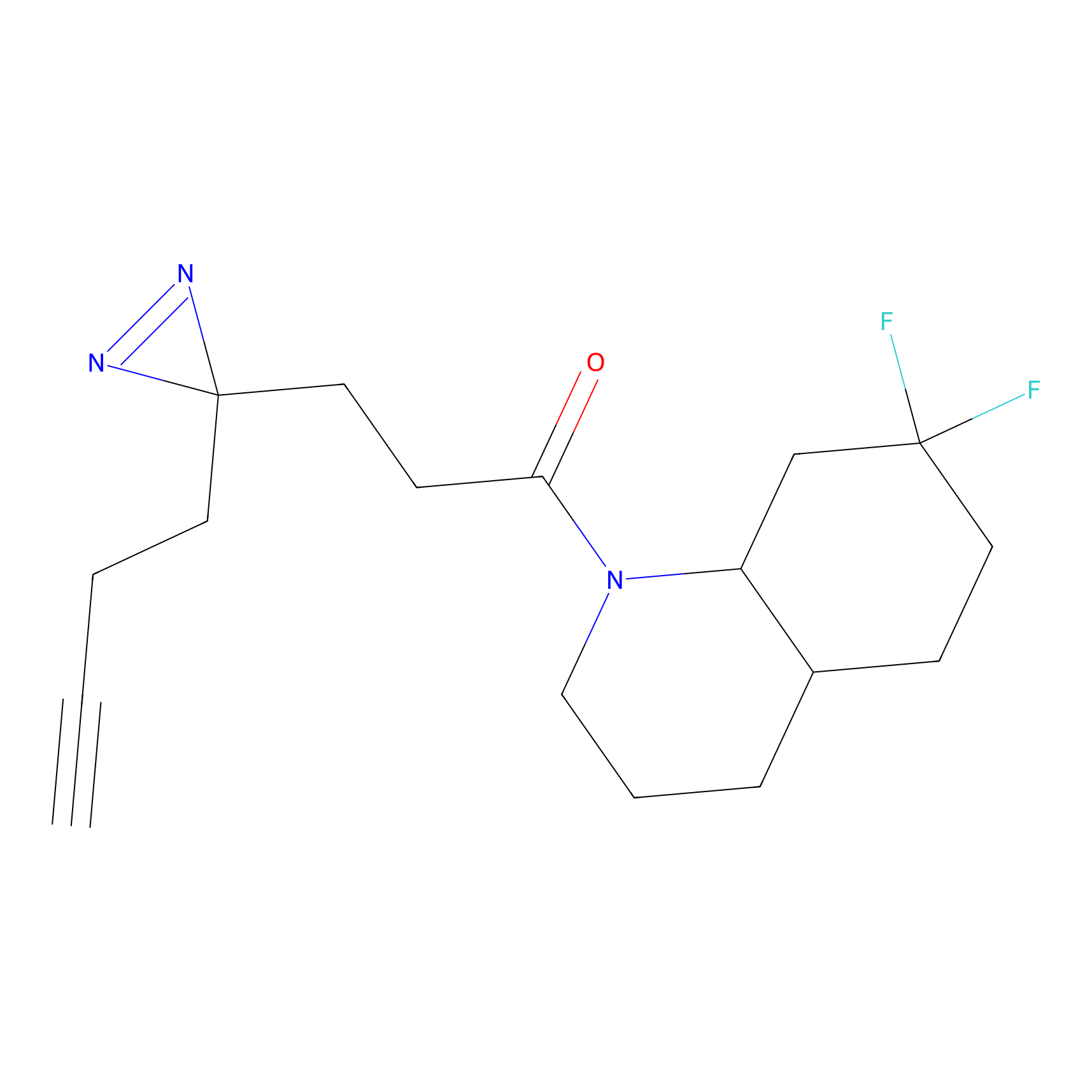
|
DBIA Probe Info |
 |
C47(2.39); C267(3.10) | LDD3310 | [2] | |
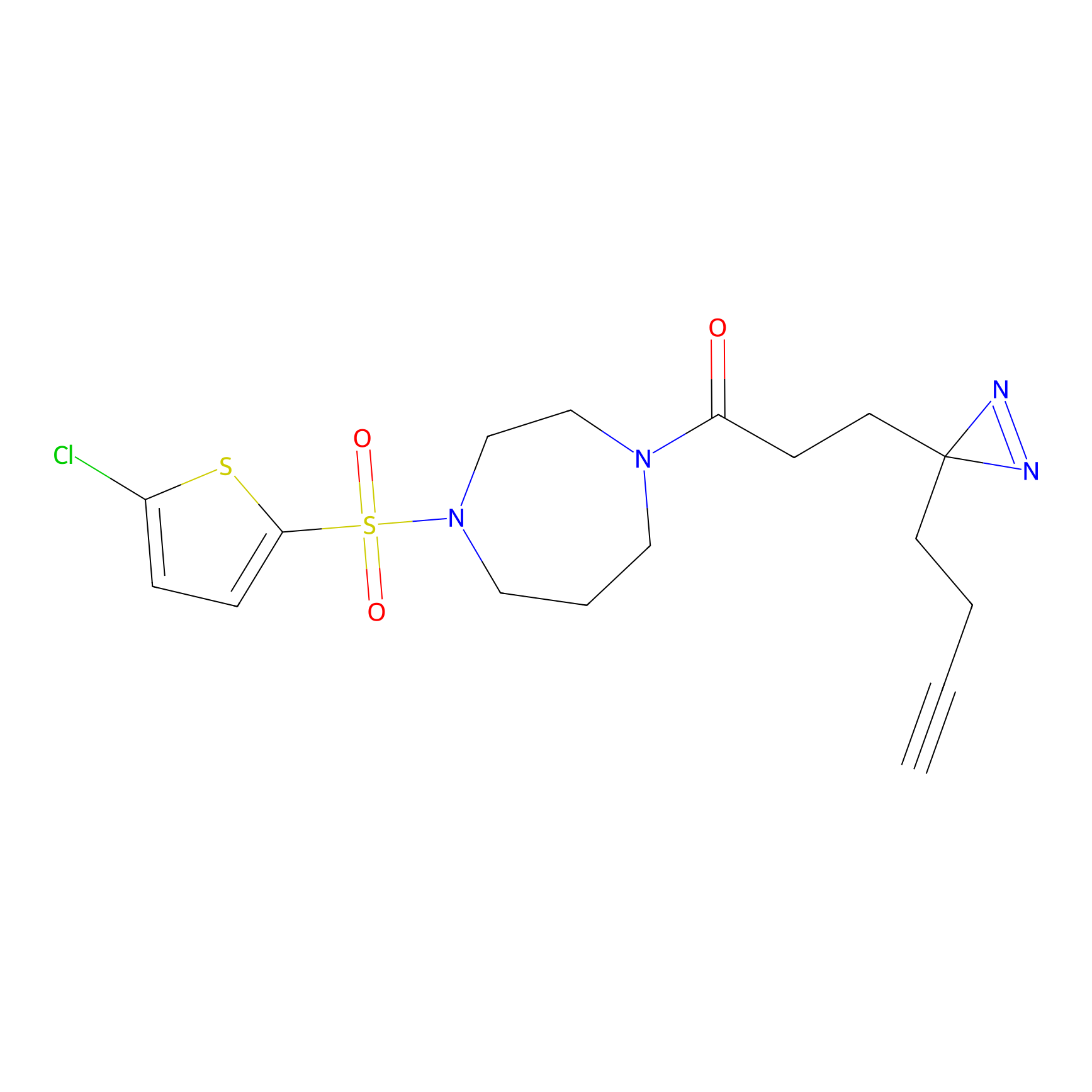
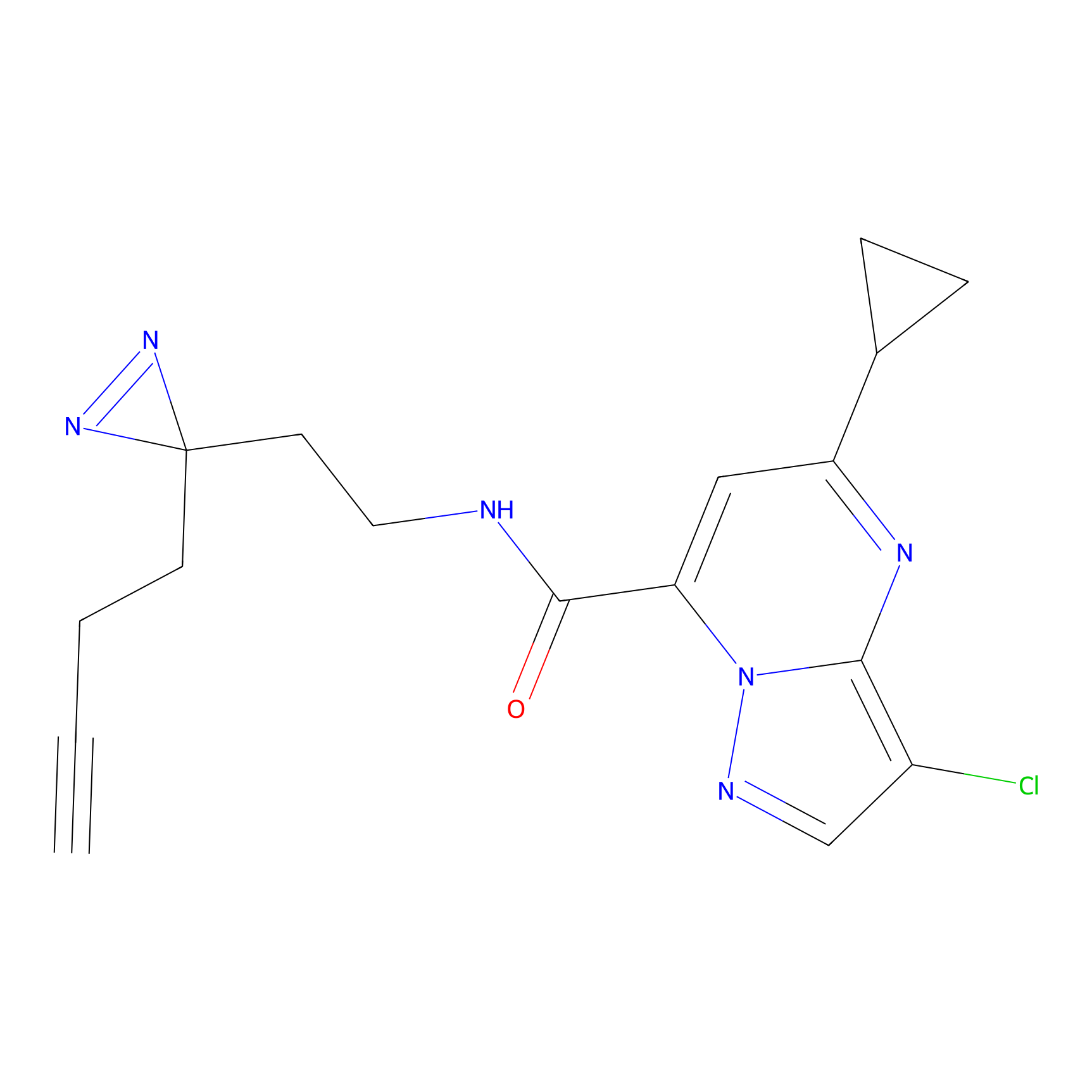
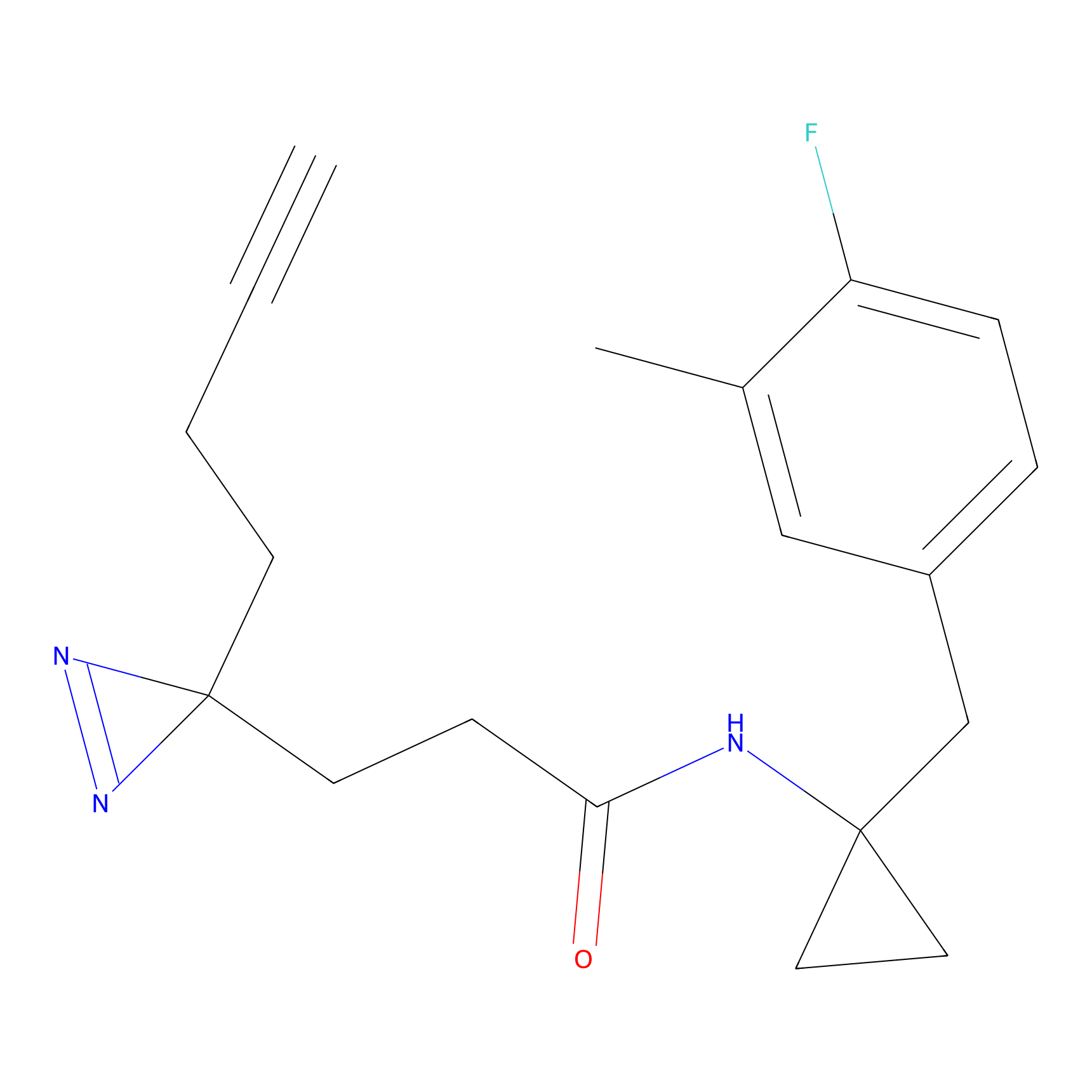
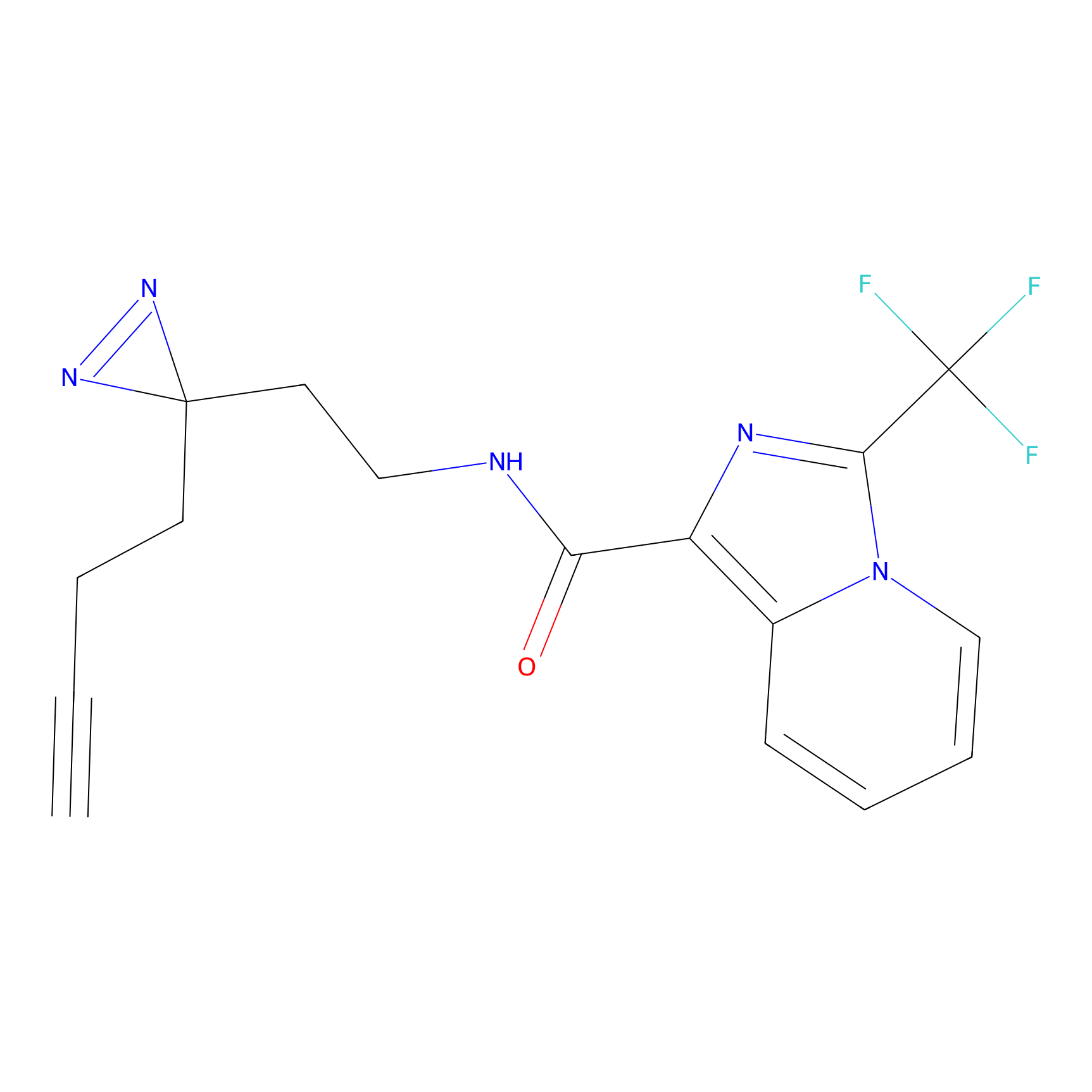
|
HHS-475 Probe Info |
 |
Y196(0.05); Y61(0.77); Y341(1.14) | LDD0264 | [3] | |
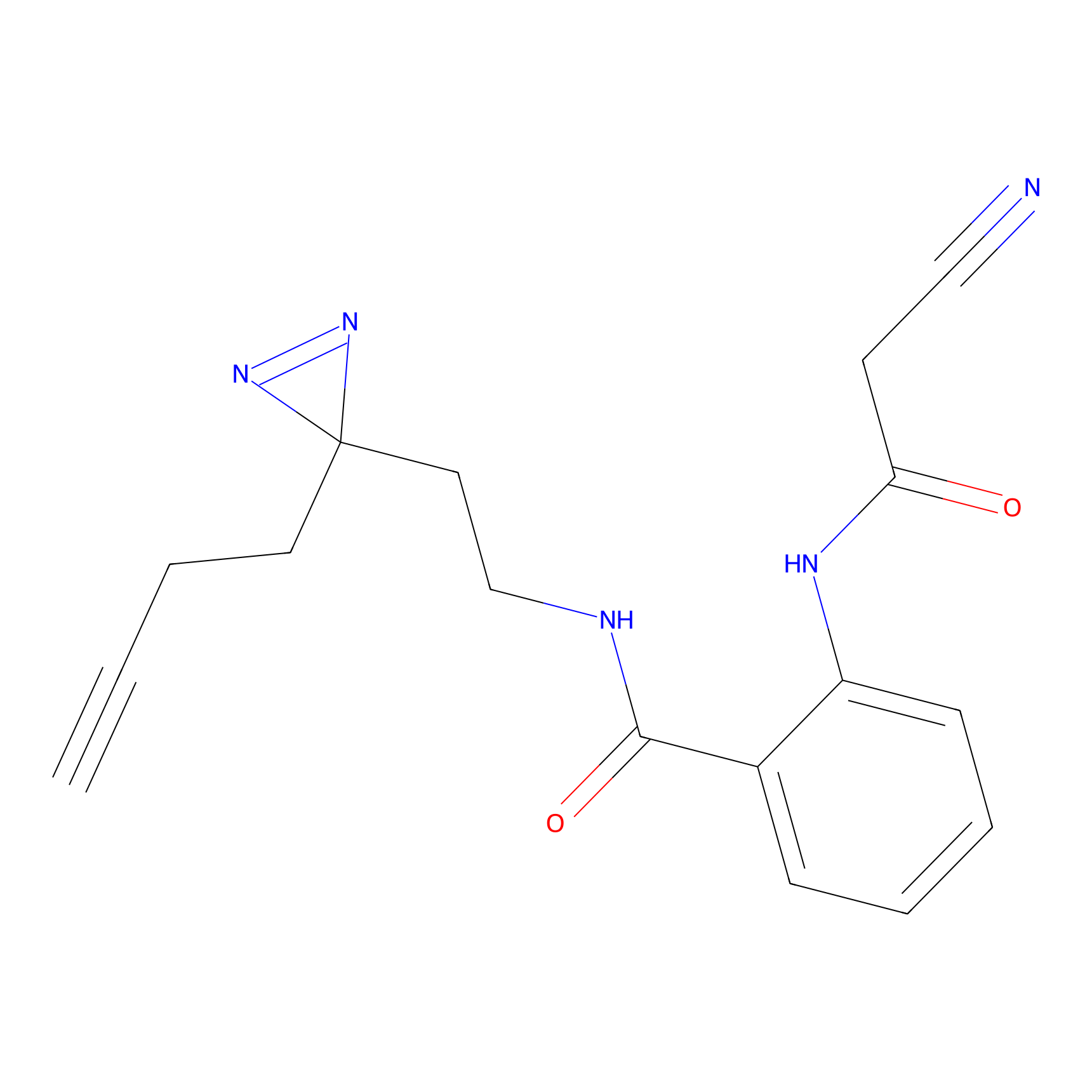
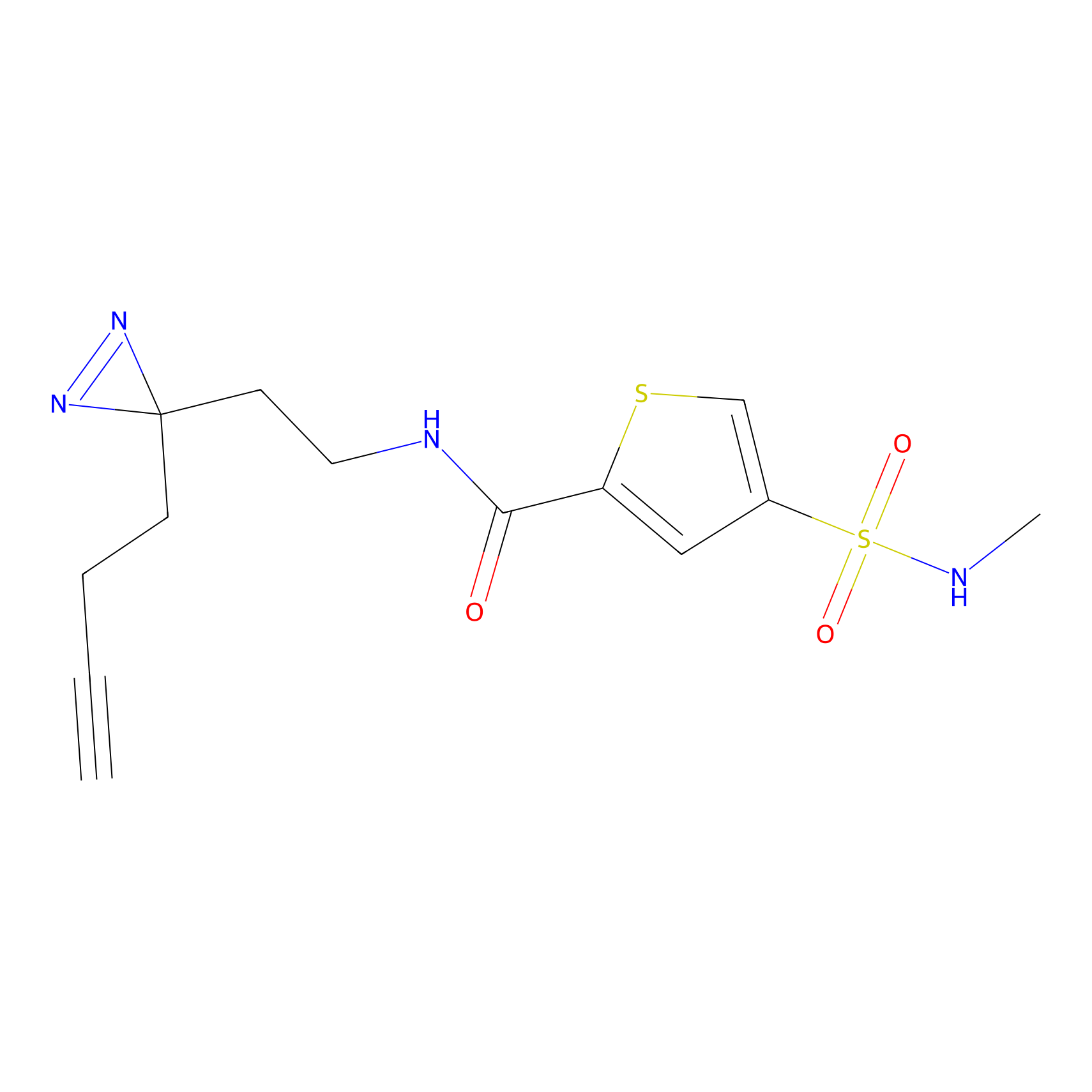
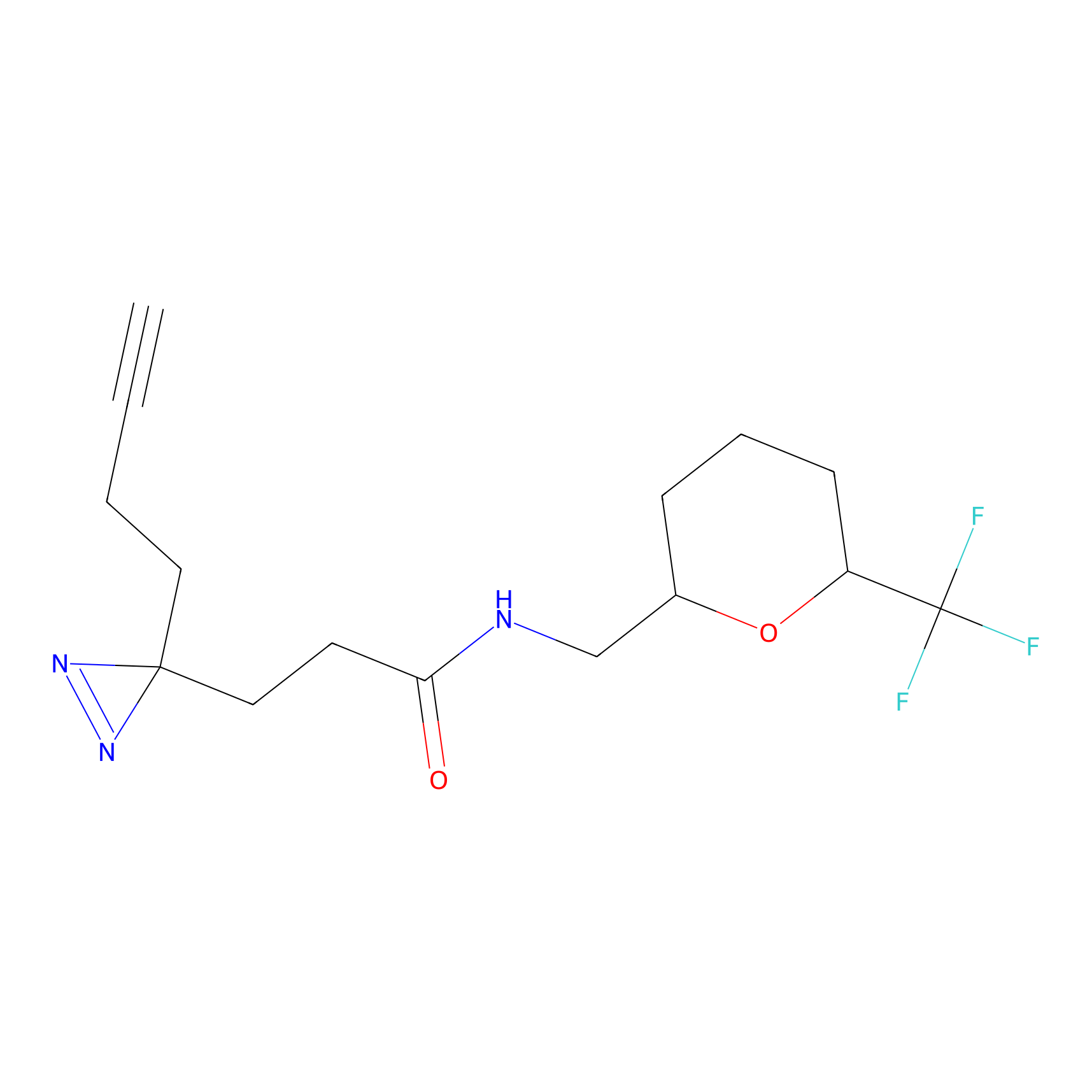
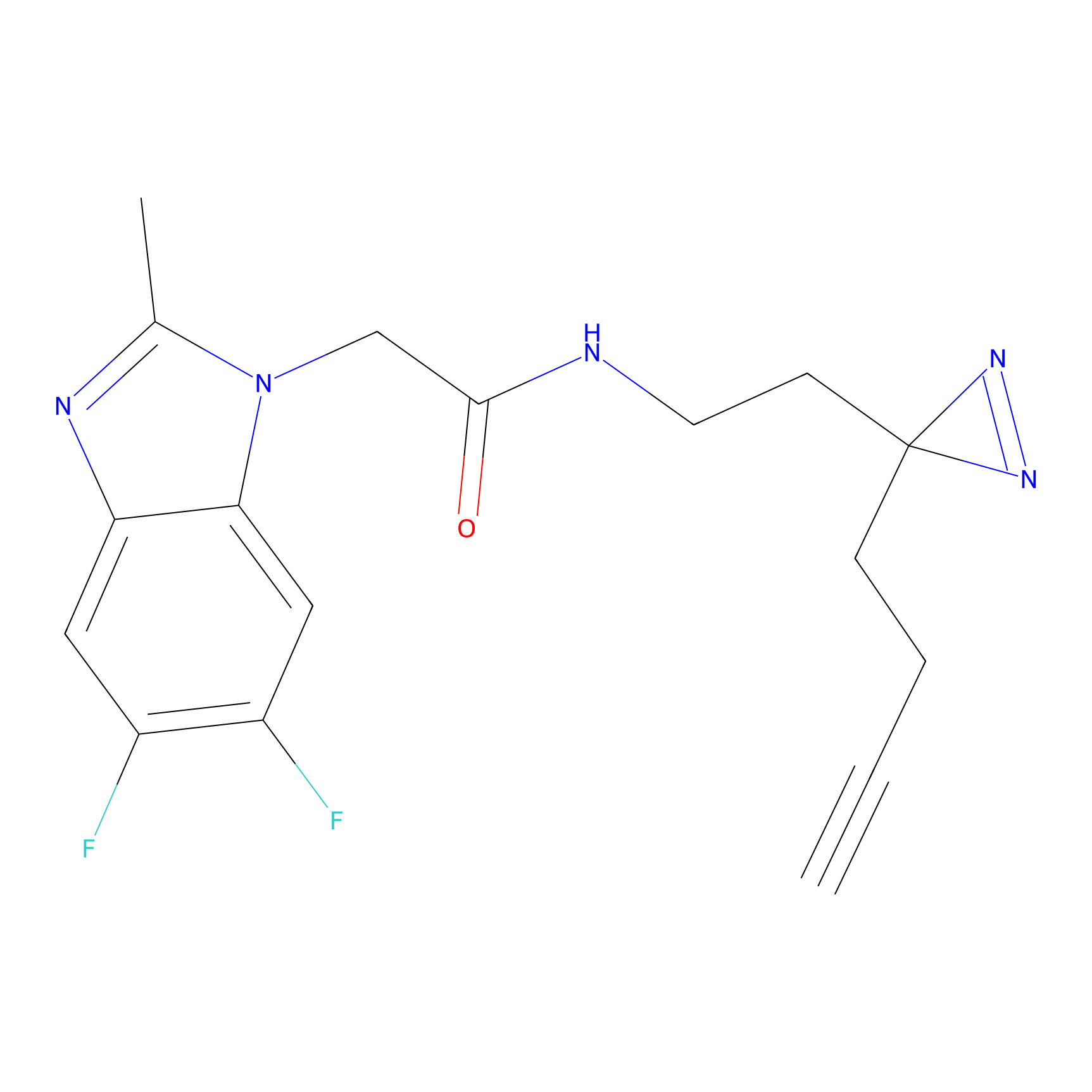
|
IA-alkyne Probe Info |
 |
C47(0.00); C437(0.00); C271(0.00) | LDD0162 | [4] | |
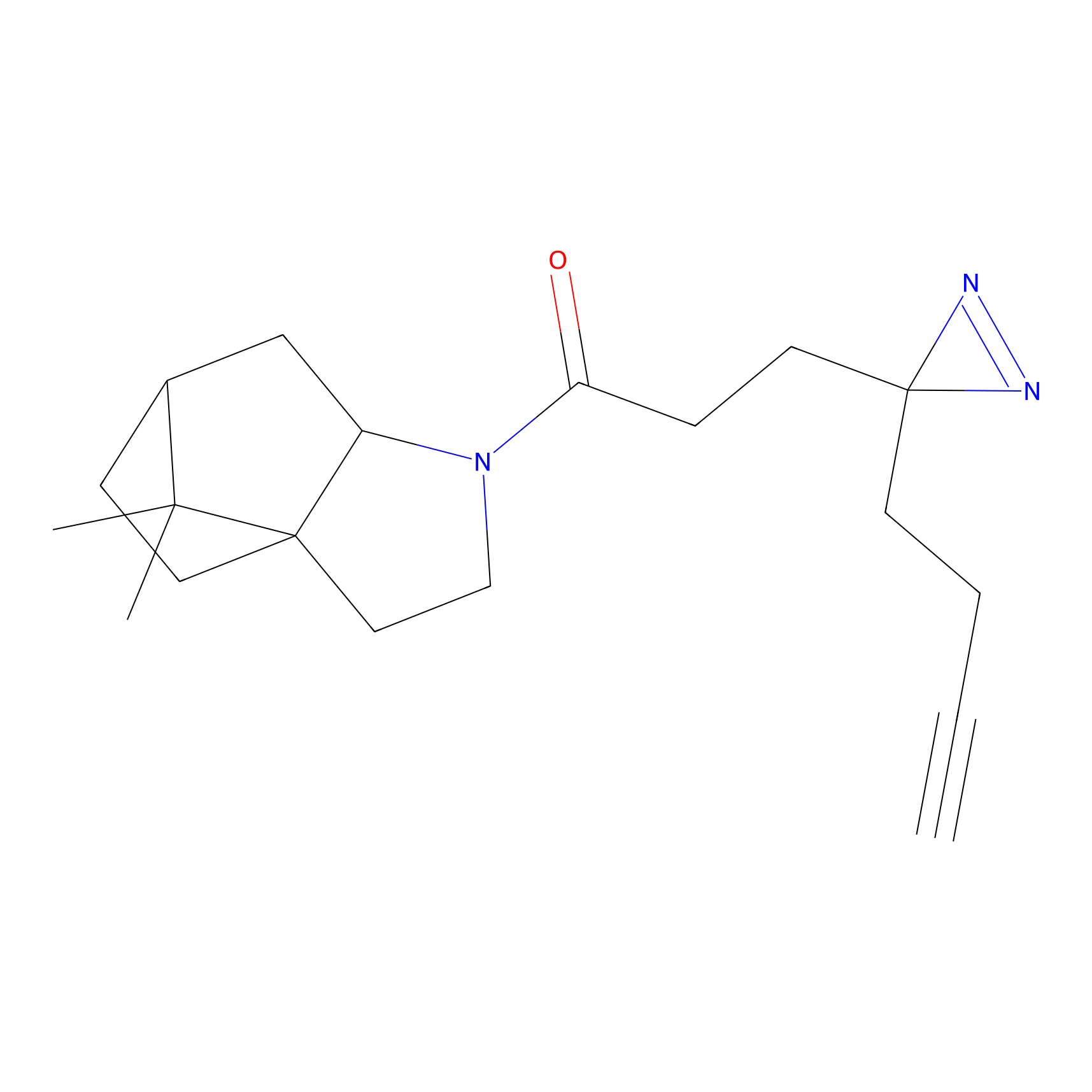
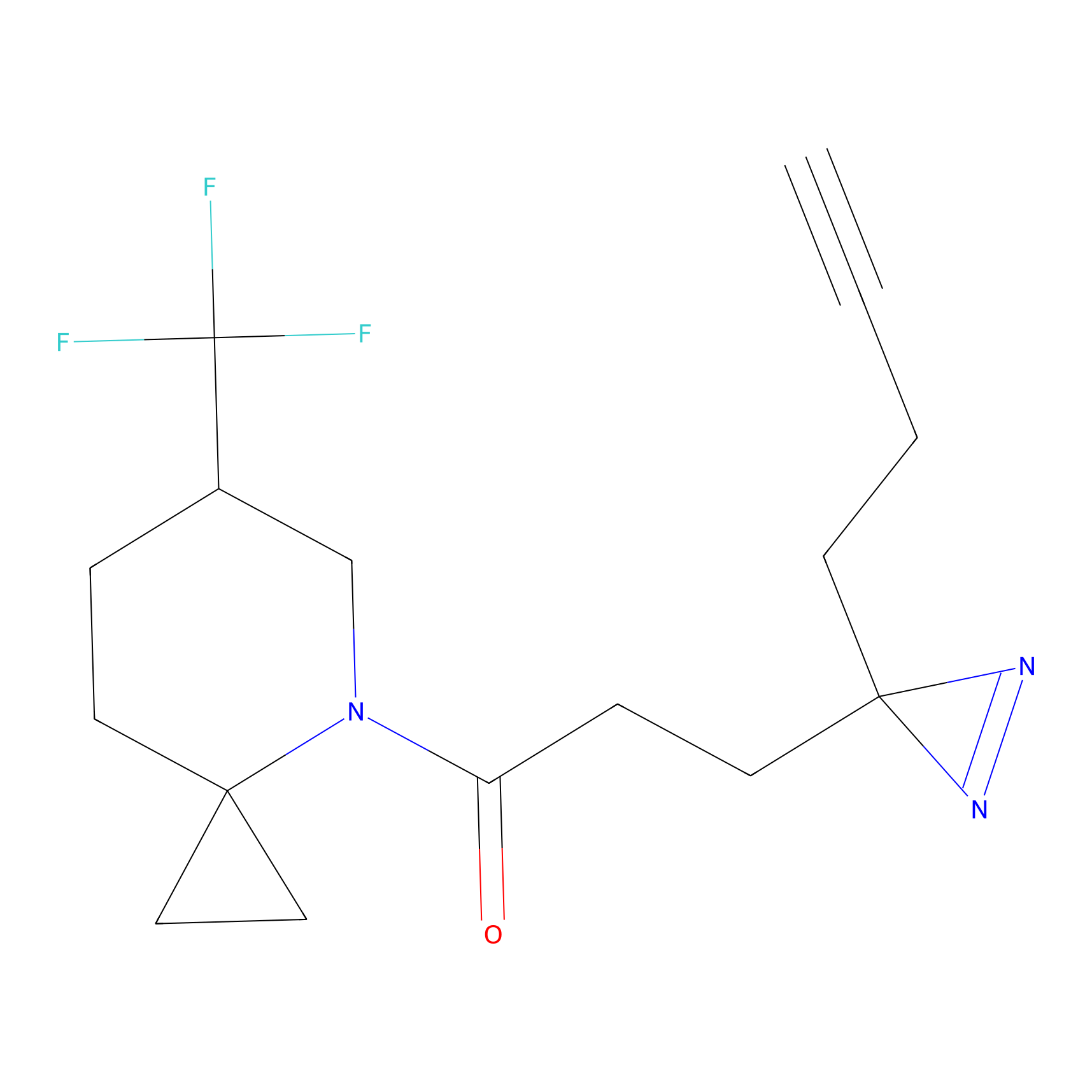
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [5] | |
|
Acrolein Probe Info |
 |
C47(0.00); C437(0.00) | LDD0217 | [6] | |
|
Crotonaldehyde Probe Info |
 |
C47(0.00); C271(0.00) | LDD0219 | [6] | |
|
Methacrolein Probe Info |
 |
C47(0.00); C214(0.00) | LDD0218 | [6] | |
|
NAIA_5 Probe Info |
 |
C437(0.00); C214(0.00); C102(0.00); C47(0.00) | LDD2223 | [7] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0215 | AC10 | PaTu 8988t | C47(0.52) | LDD1094 | [10] |
| LDCM0226 | AC11 | PaTu 8988t | C47(0.60) | LDD1105 | [10] |
| LDCM0237 | AC12 | PaTu 8988t | C47(0.64) | LDD1116 | [10] |
| LDCM0246 | AC128 | PaTu 8988t | C47(0.78) | LDD1125 | [10] |
| LDCM0247 | AC129 | PaTu 8988t | C47(0.70) | LDD1126 | [10] |
| LDCM0249 | AC130 | PaTu 8988t | C47(1.07) | LDD1128 | [10] |
| LDCM0250 | AC131 | PaTu 8988t | C47(0.93) | LDD1129 | [10] |
| LDCM0251 | AC132 | PaTu 8988t | C47(1.16) | LDD1130 | [10] |
| LDCM0252 | AC133 | PaTu 8988t | C47(0.44) | LDD1131 | [10] |
| LDCM0253 | AC134 | PaTu 8988t | C47(0.44) | LDD1132 | [10] |
| LDCM0254 | AC135 | PaTu 8988t | C47(0.43) | LDD1133 | [10] |
| LDCM0255 | AC136 | PaTu 8988t | C47(0.60) | LDD1134 | [10] |
| LDCM0256 | AC137 | PaTu 8988t | C47(0.39) | LDD1135 | [10] |
| LDCM0257 | AC138 | PaTu 8988t | C47(0.71) | LDD1136 | [10] |
| LDCM0258 | AC139 | PaTu 8988t | C47(0.71) | LDD1137 | [10] |
| LDCM0259 | AC14 | PaTu 8988t | C47(0.89) | LDD1138 | [10] |
| LDCM0260 | AC140 | PaTu 8988t | C47(0.93) | LDD1139 | [10] |
| LDCM0261 | AC141 | PaTu 8988t | C47(1.12) | LDD1140 | [10] |
| LDCM0262 | AC142 | PaTu 8988t | C47(0.60) | LDD1141 | [10] |
| LDCM0270 | AC15 | PaTu 8988t | C47(0.69) | LDD1149 | [10] |
| LDCM0276 | AC17 | PaTu 8988t | C47(0.56) | LDD1155 | [10] |
| LDCM0277 | AC18 | PaTu 8988t | C47(0.41) | LDD1156 | [10] |
| LDCM0278 | AC19 | PaTu 8988t | C47(0.79) | LDD1157 | [10] |
| LDCM0279 | AC2 | HEK-293T | C271(1.03) | LDD1518 | [11] |
| LDCM0280 | AC20 | PaTu 8988t | C47(0.49) | LDD1159 | [10] |
| LDCM0281 | AC21 | PaTu 8988t | C47(0.35) | LDD1160 | [10] |
| LDCM0282 | AC22 | PaTu 8988t | C47(0.26) | LDD1161 | [10] |
| LDCM0283 | AC23 | PaTu 8988t | C47(0.38) | LDD1162 | [10] |
| LDCM0284 | AC24 | PaTu 8988t | C47(0.40) | LDD1163 | [10] |
| LDCM0286 | AC26 | HEK-293T | C271(1.07) | LDD1525 | [11] |
| LDCM0295 | AC34 | HEK-293T | C271(1.08) | LDD1534 | [11] |
| LDCM0296 | AC35 | PaTu 8988t | C47(0.52) | LDD1175 | [10] |
| LDCM0297 | AC36 | PaTu 8988t | C47(0.65) | LDD1176 | [10] |
| LDCM0298 | AC37 | PaTu 8988t | C47(0.30) | LDD1177 | [10] |
| LDCM0299 | AC38 | PaTu 8988t | C47(0.46) | LDD1178 | [10] |
| LDCM0300 | AC39 | PaTu 8988t | C47(0.41) | LDD1179 | [10] |
| LDCM0302 | AC40 | PaTu 8988t | C47(0.26) | LDD1181 | [10] |
| LDCM0303 | AC41 | PaTu 8988t | C47(0.35) | LDD1182 | [10] |
| LDCM0304 | AC42 | PaTu 8988t | C47(0.29) | LDD1183 | [10] |
| LDCM0305 | AC43 | PaTu 8988t | C47(0.52) | LDD1184 | [10] |
| LDCM0306 | AC44 | PaTu 8988t | C47(0.12) | LDD1185 | [10] |
| LDCM0307 | AC45 | PaTu 8988t | C47(0.17) | LDD1186 | [10] |
| LDCM0308 | AC46 | PaTu 8988t | C47(0.75) | LDD1187 | [10] |
| LDCM0309 | AC47 | PaTu 8988t | C47(0.77) | LDD1188 | [10] |
| LDCM0310 | AC48 | PaTu 8988t | C47(0.60) | LDD1189 | [10] |
| LDCM0311 | AC49 | PaTu 8988t | C47(0.32) | LDD1190 | [10] |
| LDCM0313 | AC50 | PaTu 8988t | C47(0.26) | LDD1192 | [10] |
| LDCM0314 | AC51 | PaTu 8988t | C47(0.10) | LDD1193 | [10] |
| LDCM0315 | AC52 | PaTu 8988t | C47(0.51) | LDD1194 | [10] |
| LDCM0316 | AC53 | PaTu 8988t | C47(0.25) | LDD1195 | [10] |
| LDCM0317 | AC54 | PaTu 8988t | C47(0.21) | LDD1196 | [10] |
| LDCM0318 | AC55 | PaTu 8988t | C47(0.45) | LDD1197 | [10] |
| LDCM0319 | AC56 | PaTu 8988t | C47(0.24) | LDD1198 | [10] |
| LDCM0321 | AC58 | HEK-293T | C271(1.07) | LDD1560 | [11] |
| LDCM0323 | AC6 | PaTu 8988t | C47(0.70) | LDD1202 | [10] |
| LDCM0334 | AC7 | PaTu 8988t | C47(0.90) | LDD1213 | [10] |
| LDCM0345 | AC8 | PaTu 8988t | C47(1.13) | LDD1224 | [10] |
| LDCM0248 | AKOS034007472 | PaTu 8988t | C47(0.71) | LDD1127 | [10] |
| LDCM0356 | AKOS034007680 | PaTu 8988t | C47(0.72) | LDD1235 | [10] |
| LDCM0275 | AKOS034007705 | PaTu 8988t | C47(0.66) | LDD1154 | [10] |
| LDCM0020 | ARS-1620 | HCC44 | C271(1.16) | LDD2171 | [10] |
| LDCM0108 | Chloroacetamide | HeLa | C102(0.00); C214(0.00); C271(0.00); C47(0.00) | LDD0222 | [6] |
| LDCM0370 | CL101 | PaTu 8988t | C47(0.79) | LDD1249 | [10] |
| LDCM0371 | CL102 | PaTu 8988t | C47(0.72) | LDD1250 | [10] |
| LDCM0372 | CL103 | PaTu 8988t | C47(0.45) | LDD1251 | [10] |
| LDCM0373 | CL104 | PaTu 8988t | C47(0.87) | LDD1252 | [10] |
| LDCM0374 | CL105 | PaTu 8988t | C47(0.87) | LDD1253 | [10] |
| LDCM0375 | CL106 | PaTu 8988t | C47(0.74) | LDD1254 | [10] |
| LDCM0376 | CL107 | PaTu 8988t | C47(0.81) | LDD1255 | [10] |
| LDCM0377 | CL108 | PaTu 8988t | C47(0.62) | LDD1256 | [10] |
| LDCM0378 | CL109 | PaTu 8988t | C47(0.66) | LDD1257 | [10] |
| LDCM0380 | CL110 | PaTu 8988t | C47(0.57) | LDD1259 | [10] |
| LDCM0381 | CL111 | PaTu 8988t | C47(0.28) | LDD1260 | [10] |
| LDCM0387 | CL117 | PaTu 8988t | C47(0.36) | LDD1266 | [10] |
| LDCM0388 | CL118 | PaTu 8988t | C47(0.55) | LDD1267 | [10] |
| LDCM0389 | CL119 | PaTu 8988t | C47(0.41) | LDD1268 | [10] |
| LDCM0391 | CL120 | PaTu 8988t | C47(0.73) | LDD1270 | [10] |
| LDCM0392 | CL121 | PaTu 8988t | C47(0.63) | LDD1271 | [10] |
| LDCM0393 | CL122 | PaTu 8988t | C47(0.54) | LDD1272 | [10] |
| LDCM0394 | CL123 | PaTu 8988t | C47(0.40) | LDD1273 | [10] |
| LDCM0395 | CL124 | PaTu 8988t | C47(0.30) | LDD1274 | [10] |
| LDCM0405 | CL18 | HEK-293T | C271(1.13) | LDD1609 | [11] |
| LDCM0419 | CL30 | HEK-293T | C271(1.01) | LDD1623 | [11] |
| LDCM0420 | CL31 | PaTu 8988t | C47(0.36) | LDD1299 | [10] |
| LDCM0421 | CL32 | PaTu 8988t | C47(0.23) | LDD1300 | [10] |
| LDCM0422 | CL33 | PaTu 8988t | C47(0.14) | LDD1301 | [10] |
| LDCM0423 | CL34 | PaTu 8988t | C47(0.14) | LDD1302 | [10] |
| LDCM0424 | CL35 | PaTu 8988t | C47(0.11) | LDD1303 | [10] |
| LDCM0425 | CL36 | PaTu 8988t | C47(0.11) | LDD1304 | [10] |
| LDCM0426 | CL37 | PaTu 8988t | C47(0.07) | LDD1305 | [10] |
| LDCM0428 | CL39 | PaTu 8988t | C47(0.17) | LDD1307 | [10] |
| LDCM0430 | CL40 | PaTu 8988t | C47(0.28) | LDD1309 | [10] |
| LDCM0431 | CL41 | PaTu 8988t | C47(0.13) | LDD1310 | [10] |
| LDCM0432 | CL42 | PaTu 8988t | C47(0.08) | LDD1311 | [10] |
| LDCM0433 | CL43 | PaTu 8988t | C47(0.06) | LDD1312 | [10] |
| LDCM0434 | CL44 | PaTu 8988t | C47(0.12) | LDD1313 | [10] |
| LDCM0435 | CL45 | PaTu 8988t | C47(0.20) | LDD1314 | [10] |
| LDCM0445 | CL54 | HEK-293T | C271(1.03) | LDD1648 | [11] |
| LDCM0451 | CL6 | HEK-293T | C271(0.96) | LDD1654 | [11] |
| LDCM0458 | CL66 | HEK-293T | C271(1.01) | LDD1661 | [11] |
| LDCM0471 | CL78 | HEK-293T | C271(1.08) | LDD1674 | [11] |
| LDCM0485 | CL90 | HEK-293T | C271(0.96) | LDD1688 | [11] |
| LDCM0427 | Fragment51 | PaTu 8988t | C47(0.05) | LDD1306 | [10] |
| LDCM0116 | HHS-0101 | DM93 | Y196(0.05); Y61(0.77); Y341(1.14) | LDD0264 | [3] |
| LDCM0117 | HHS-0201 | DM93 | Y196(0.52); Y61(0.85); Y341(1.25) | LDD0265 | [3] |
| LDCM0118 | HHS-0301 | DM93 | Y196(0.05); Y61(0.78); Y341(1.33) | LDD0266 | [3] |
| LDCM0119 | HHS-0401 | DM93 | Y196(0.05); Y61(1.21); Y341(2.01) | LDD0267 | [3] |
| LDCM0120 | HHS-0701 | DM93 | Y196(0.05); Y61(0.58); Y341(0.98) | LDD0268 | [3] |
| LDCM0022 | KB02 | 42-MG-BA | C39(2.78) | LDD2244 | [2] |
| LDCM0023 | KB03 | 769-P | C39(2.68) | LDD2663 | [2] |
| LDCM0024 | KB05 | COLO792 | C47(2.39); C267(3.10) | LDD3310 | [2] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [6] |
| LDCM0021 | THZ1 | HCT 116 | C271(1.16) | LDD2173 | [10] |
References