Details of the Target
General Information of Target
| Target ID | LDTP00417 | |||||
|---|---|---|---|---|---|---|
| Target Name | Programmed cell death protein 5 (PDCD5) | |||||
| Gene Name | PDCD5 | |||||
| Gene ID | 9141 | |||||
| Synonyms |
TFAR19; Programmed cell death protein 5; TF-1 cell apoptosis-related protein 19; Protein TFAR19 |
|||||
| 3D Structure | ||||||
| Sequence |
MADEELEALRRQRLAELQAKHGDPGDAAQQEAKHREAEMRNSILAQVLDQSARARLSNLA
LVKPEKTKAVENYLIQMARYGQLSEKVSEQGLIEILKKVSQQTEKTTTVKFNRRKVMDSD EDDDY |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
PDCD5 family
|
|||||
| Function | May function in the process of apoptosis. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
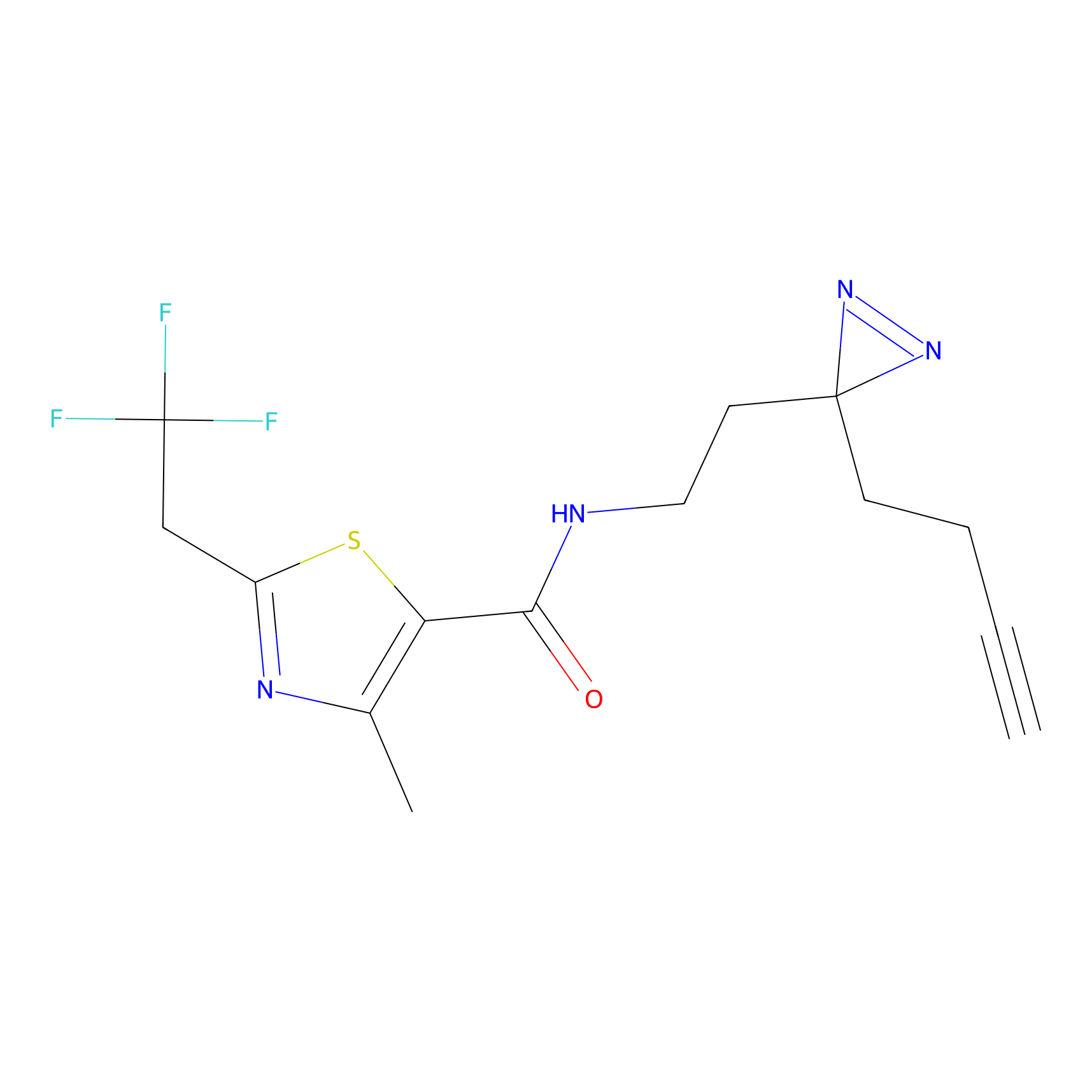
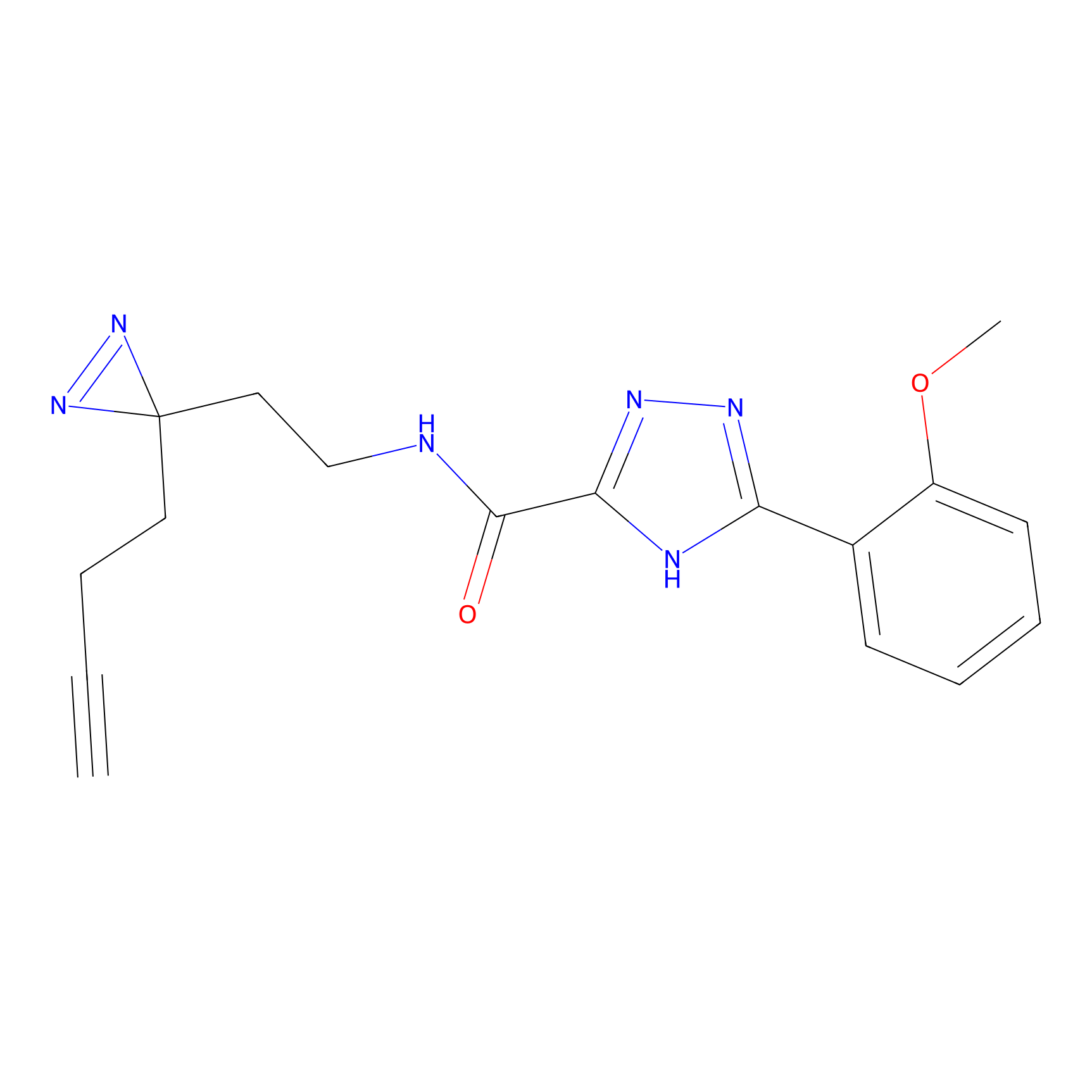
TH211 Probe Info |
 |
Y73(17.42) | LDD0257 | [1] | |
|
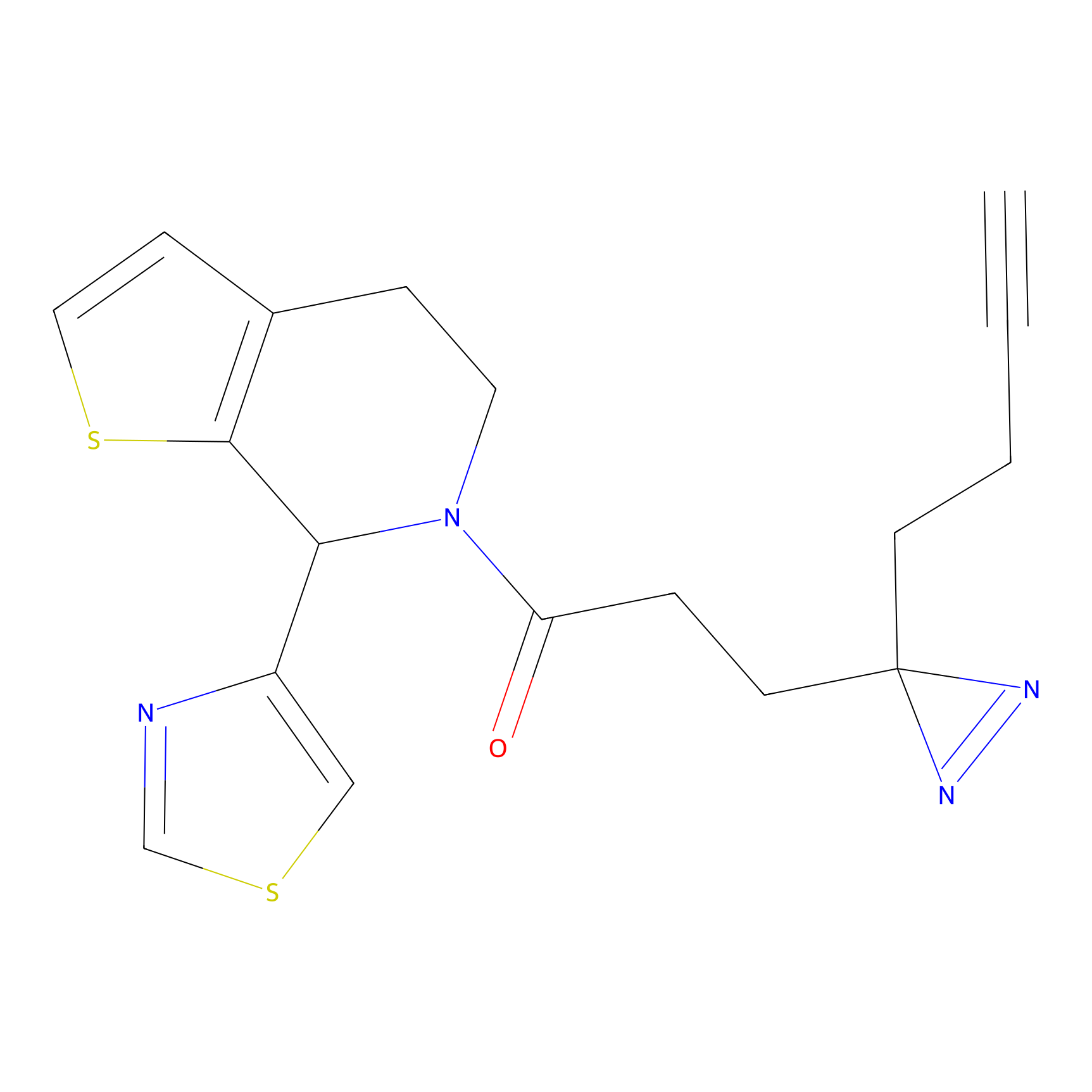
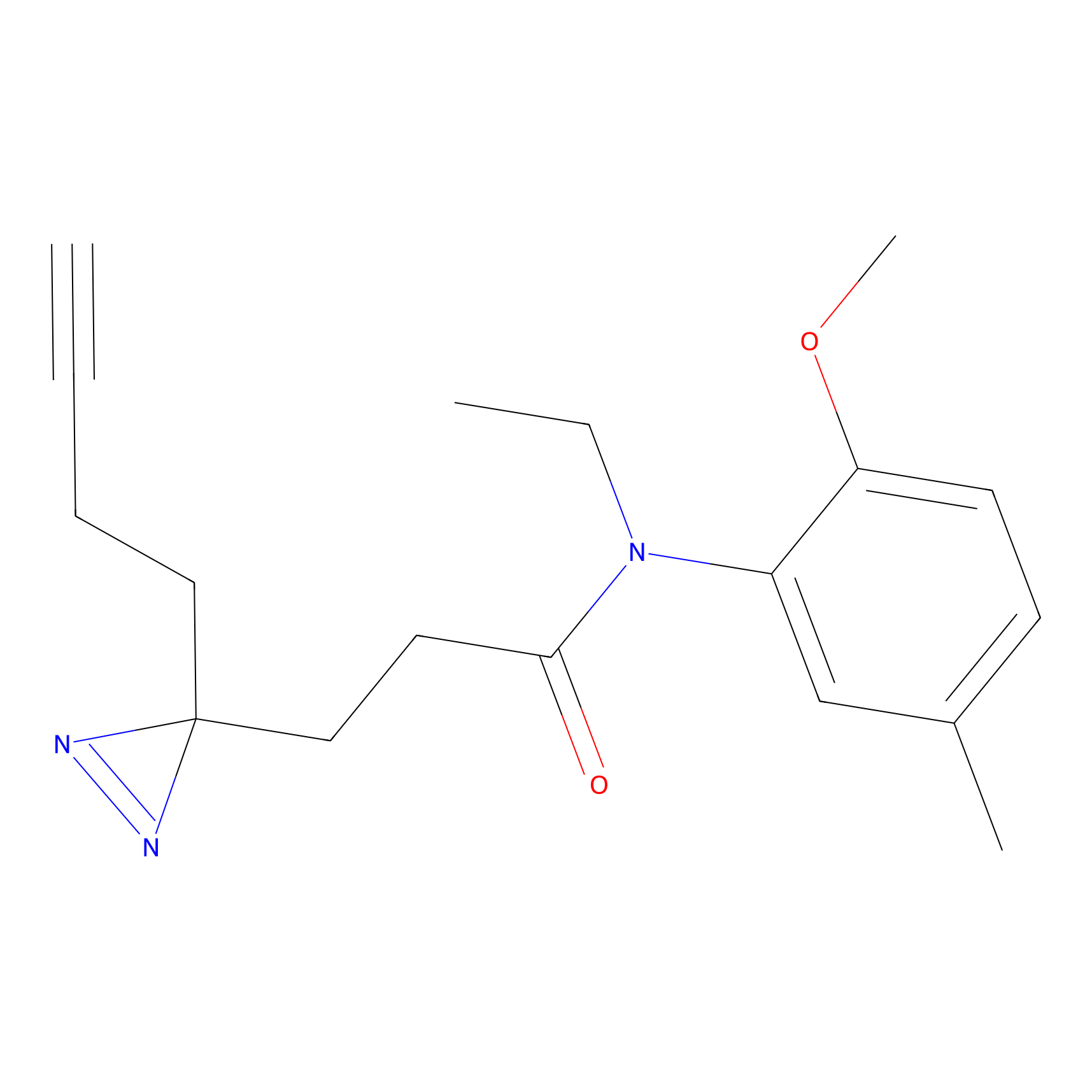
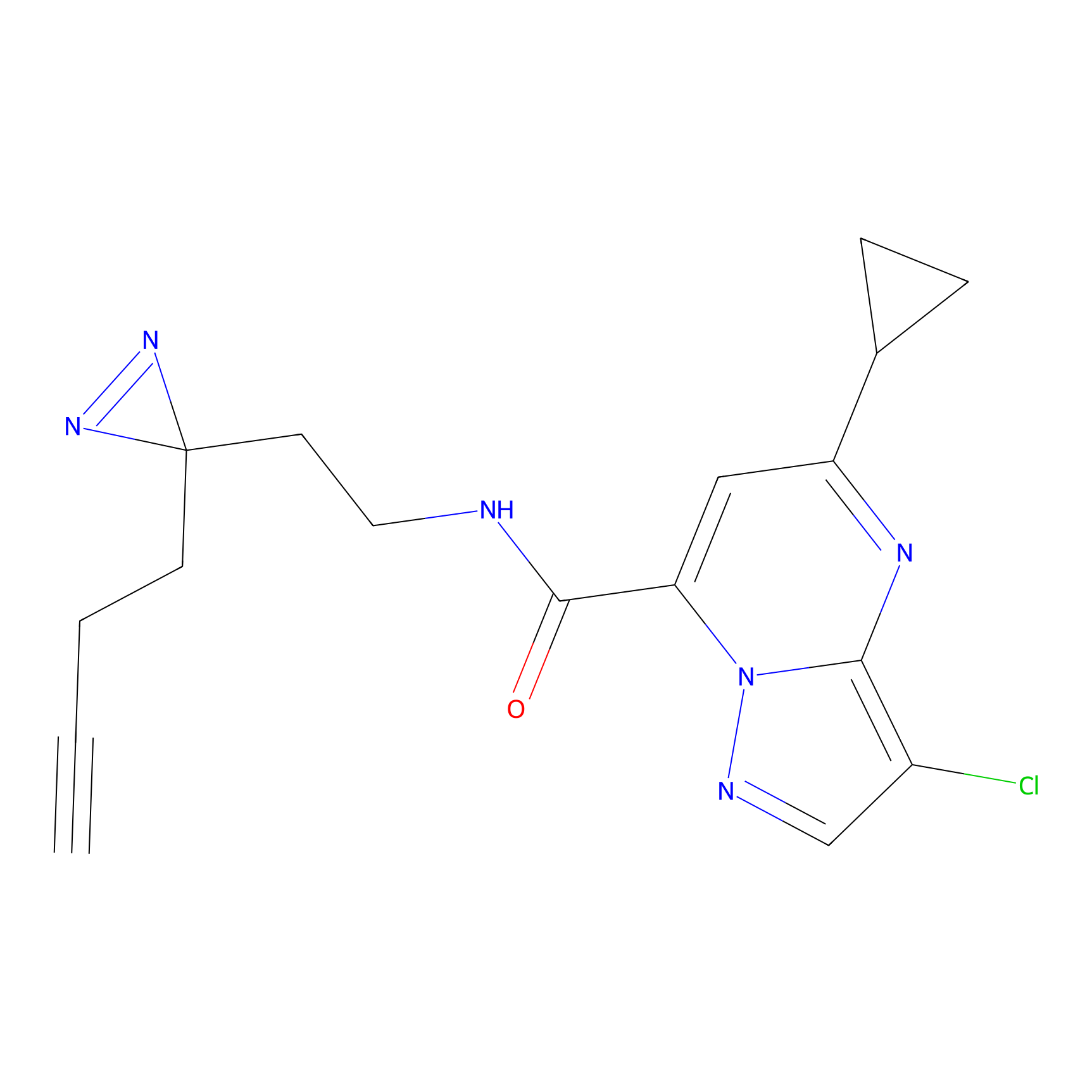
TH214 Probe Info |
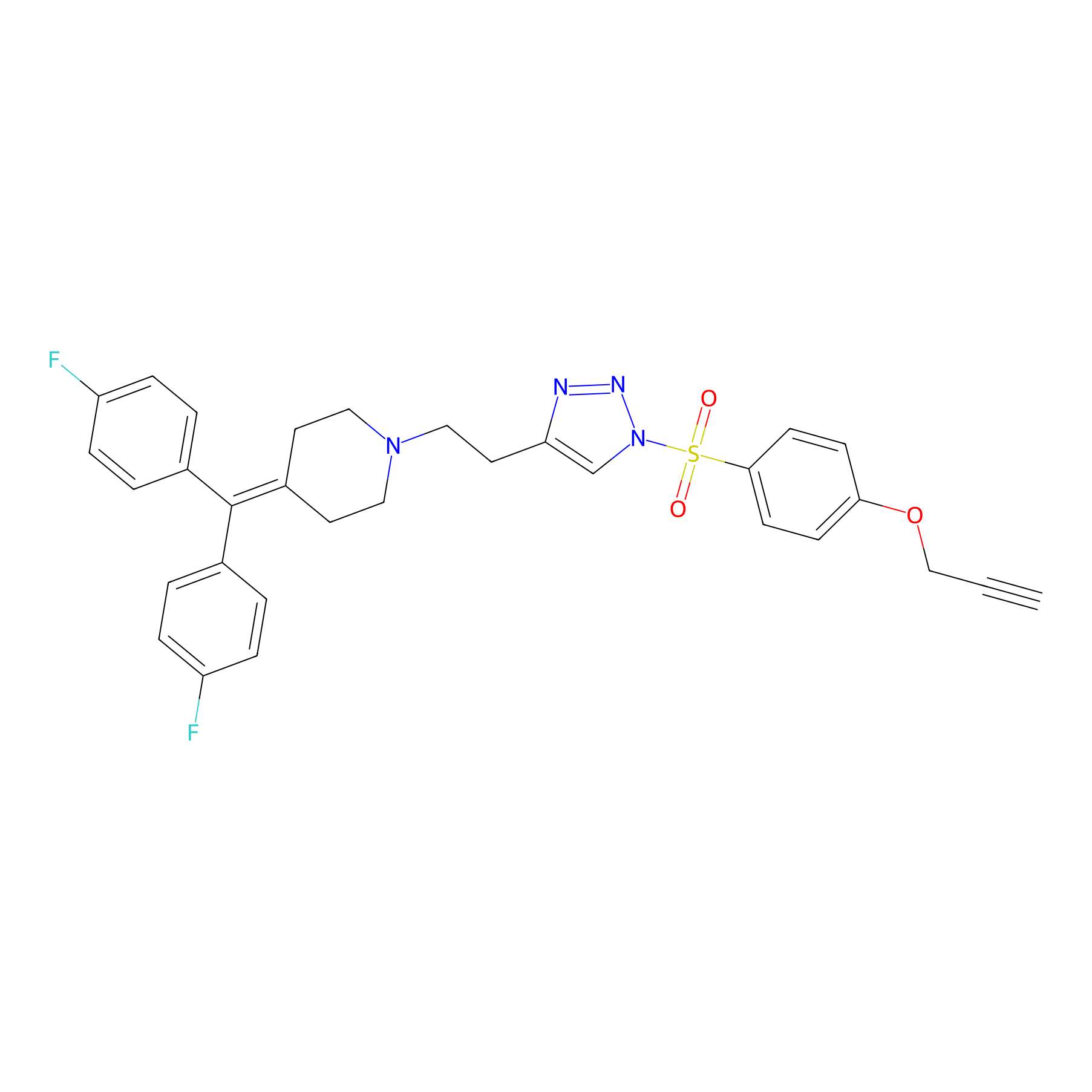 |
Y80(10.23) | LDD0258 | [1] | |
|
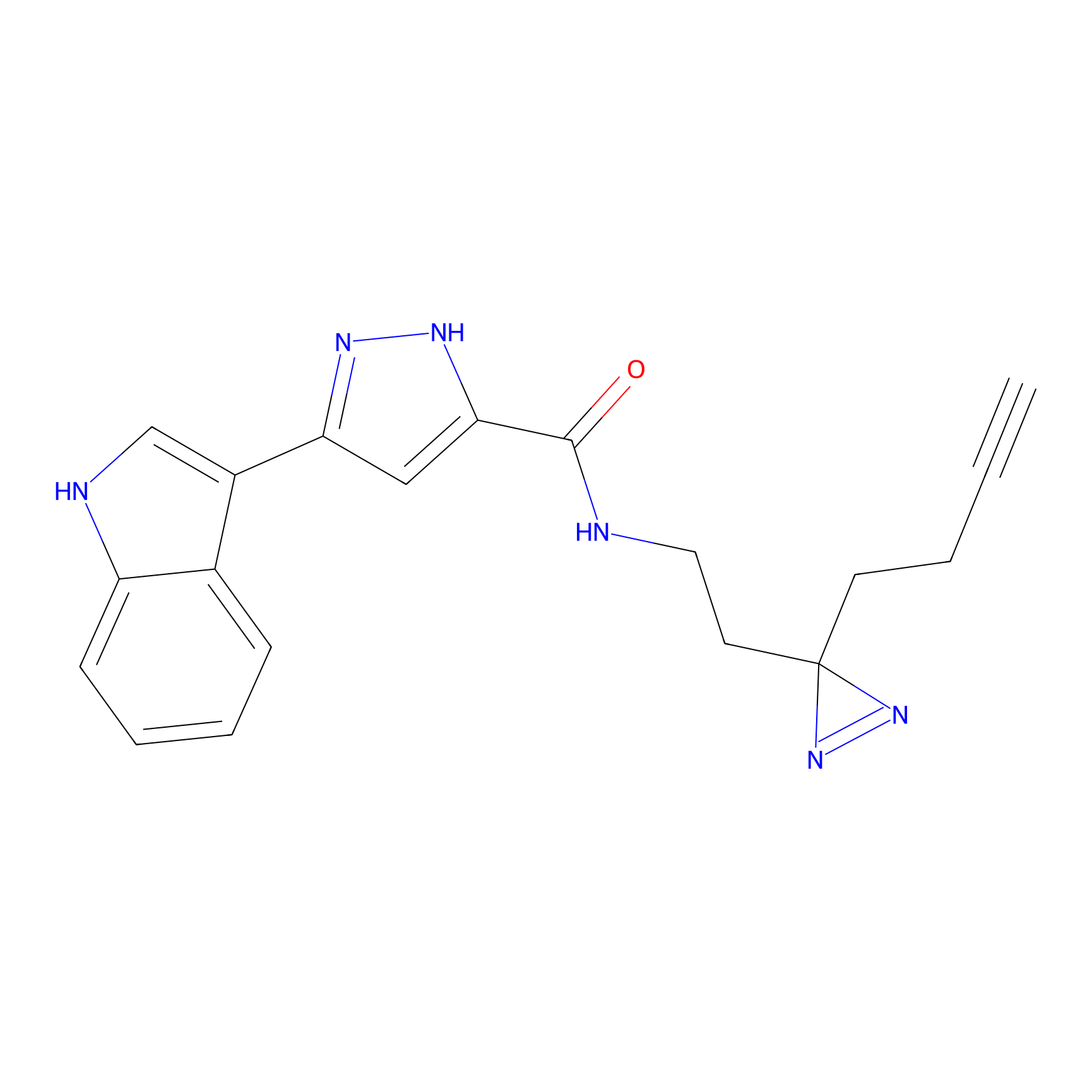
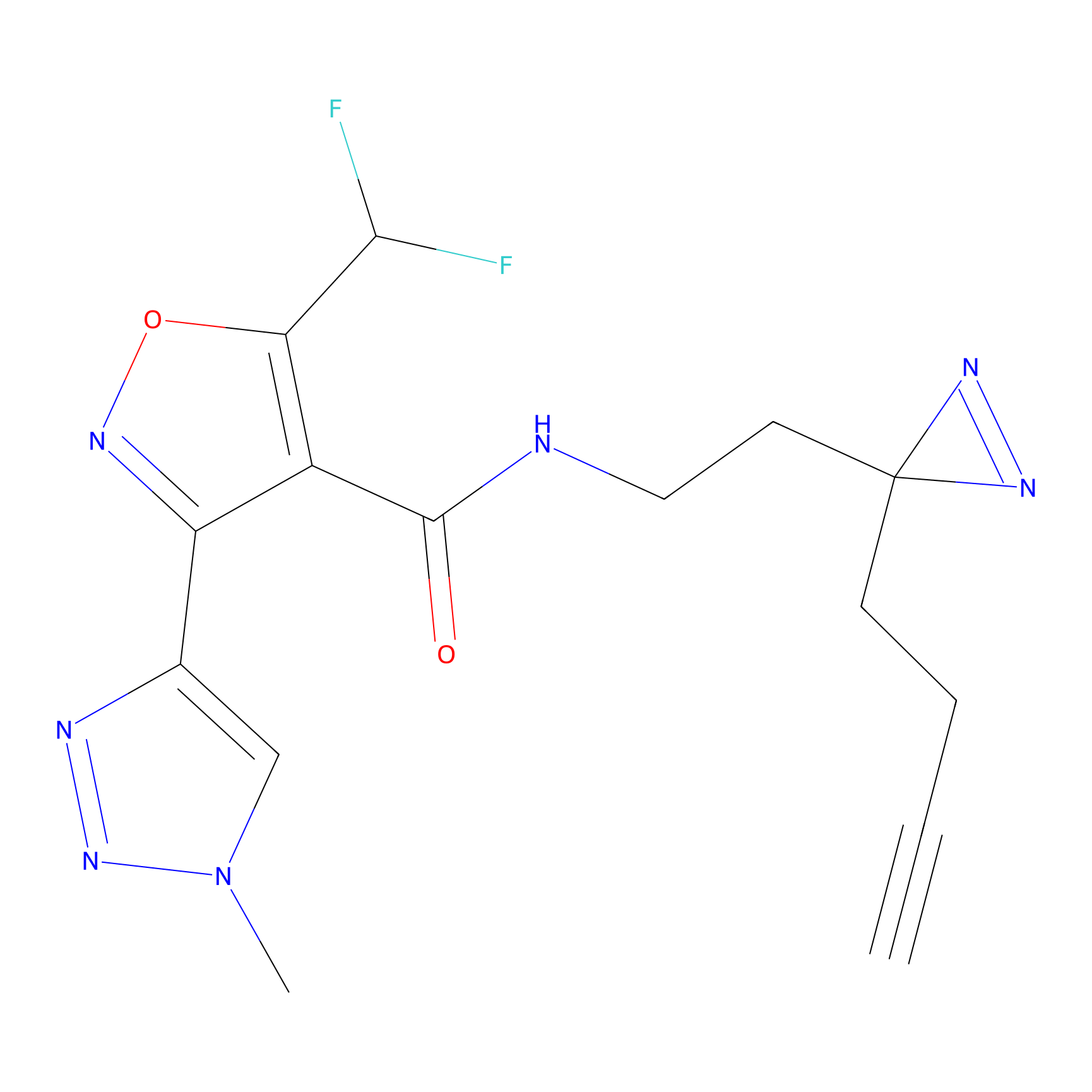
TH216 Probe Info |
 |
Y73(13.09); Y80(10.80) | LDD0259 | [1] | |
|
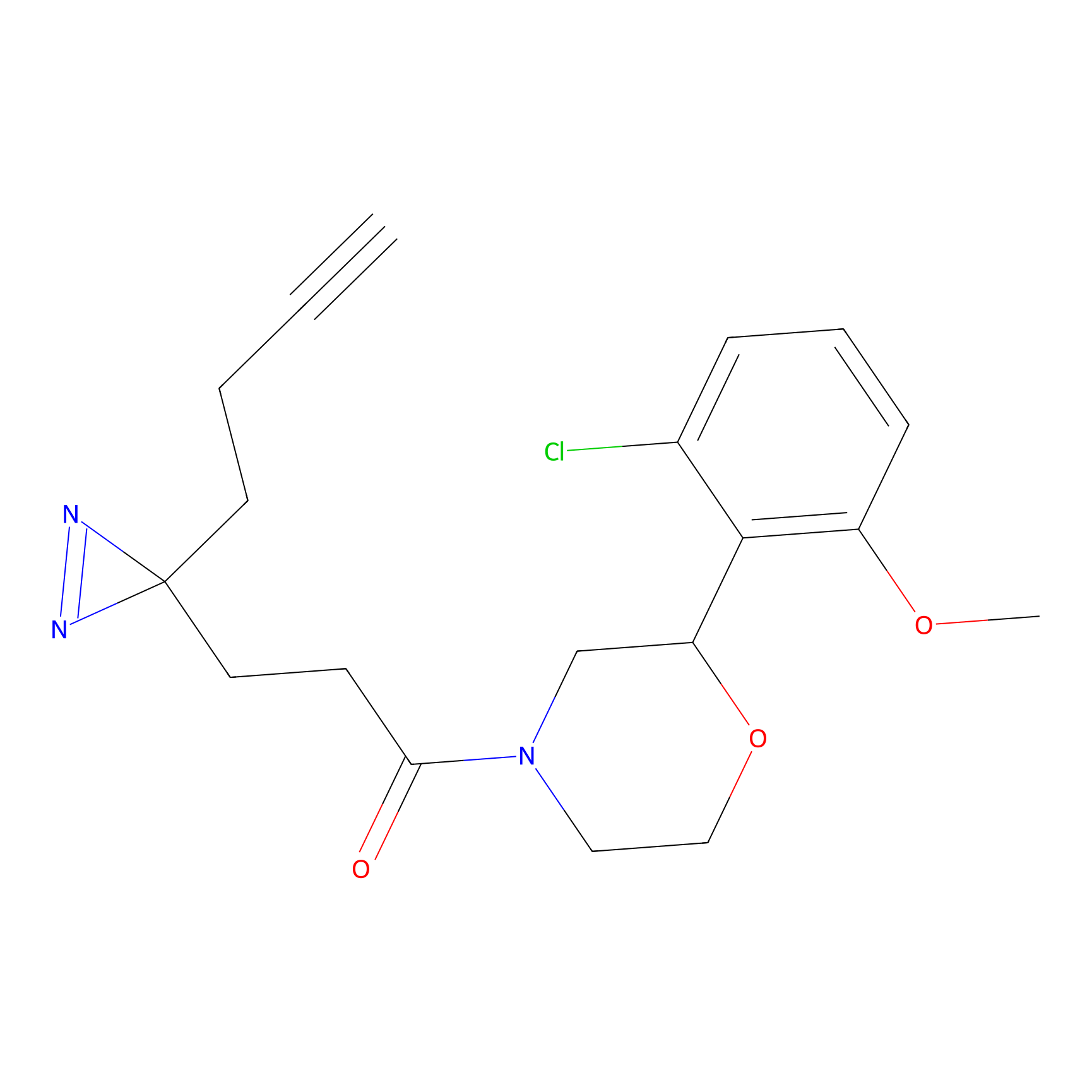
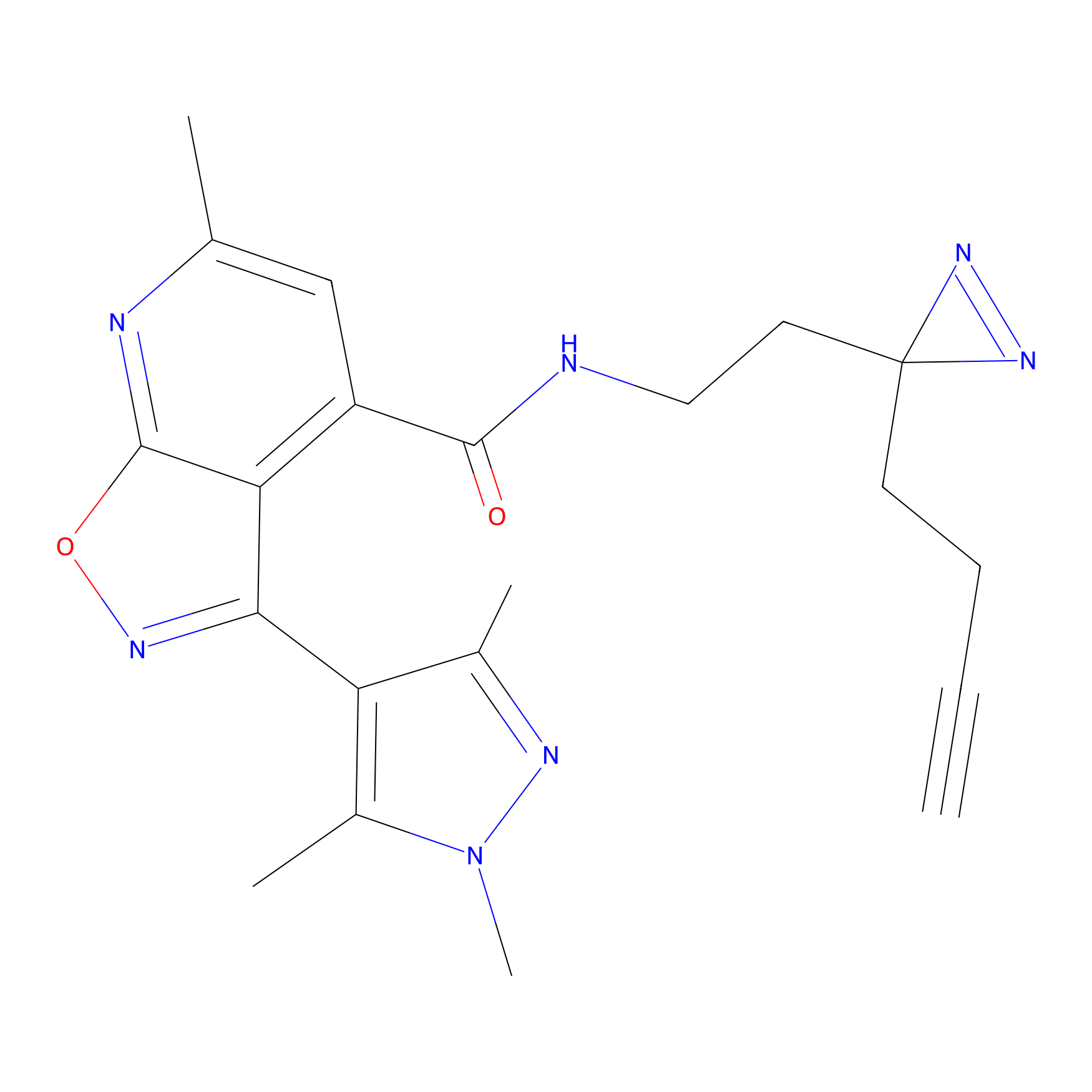
ONAyne Probe Info |
 |
N.A. | LDD0273 | [2] | |
|
OPA-S-S-alkyne Probe Info |
 |
K33(4.82) | LDD3494 | [3] | |
|
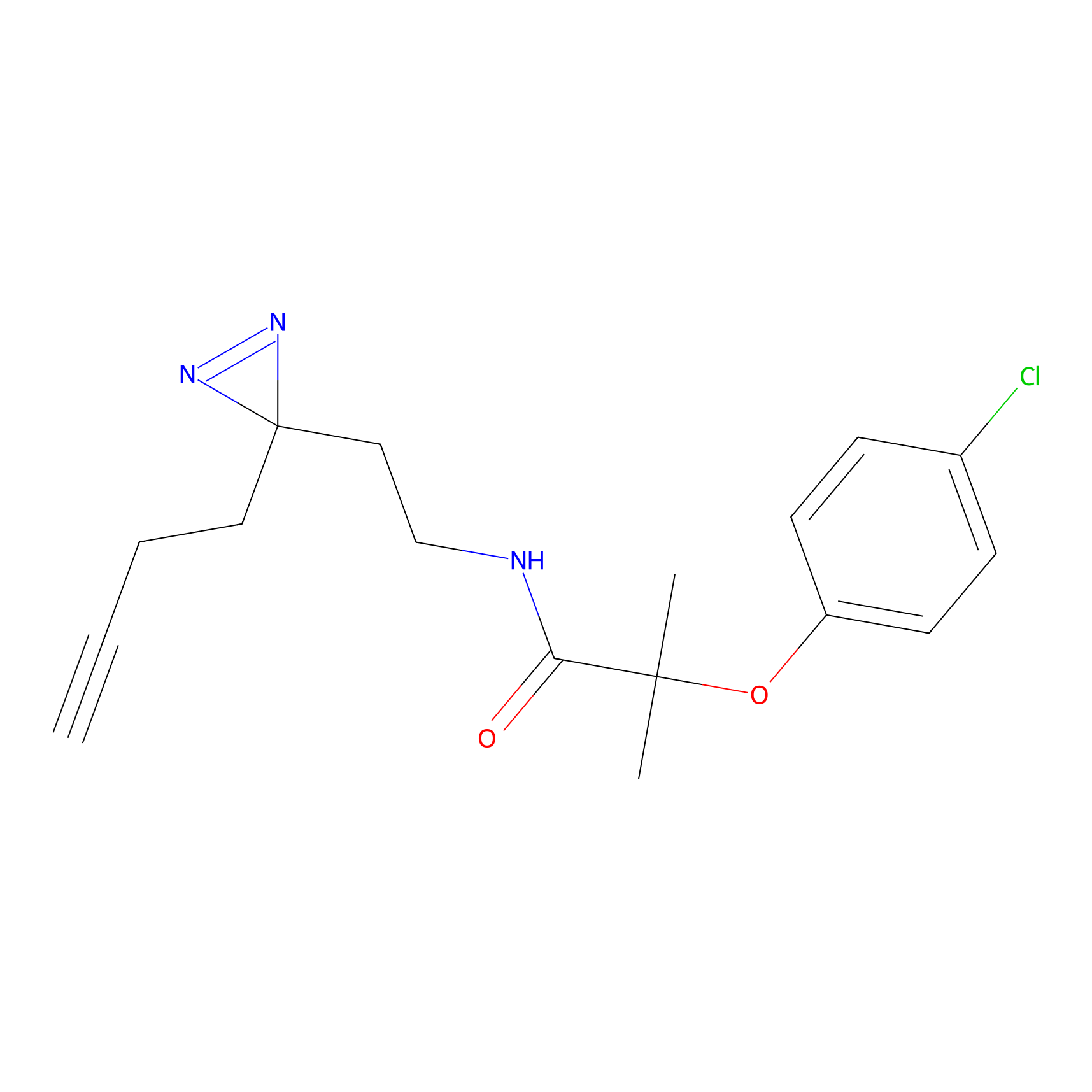
m-APA Probe Info |
 |
10.57 | LDD0403 | [4] | |
|
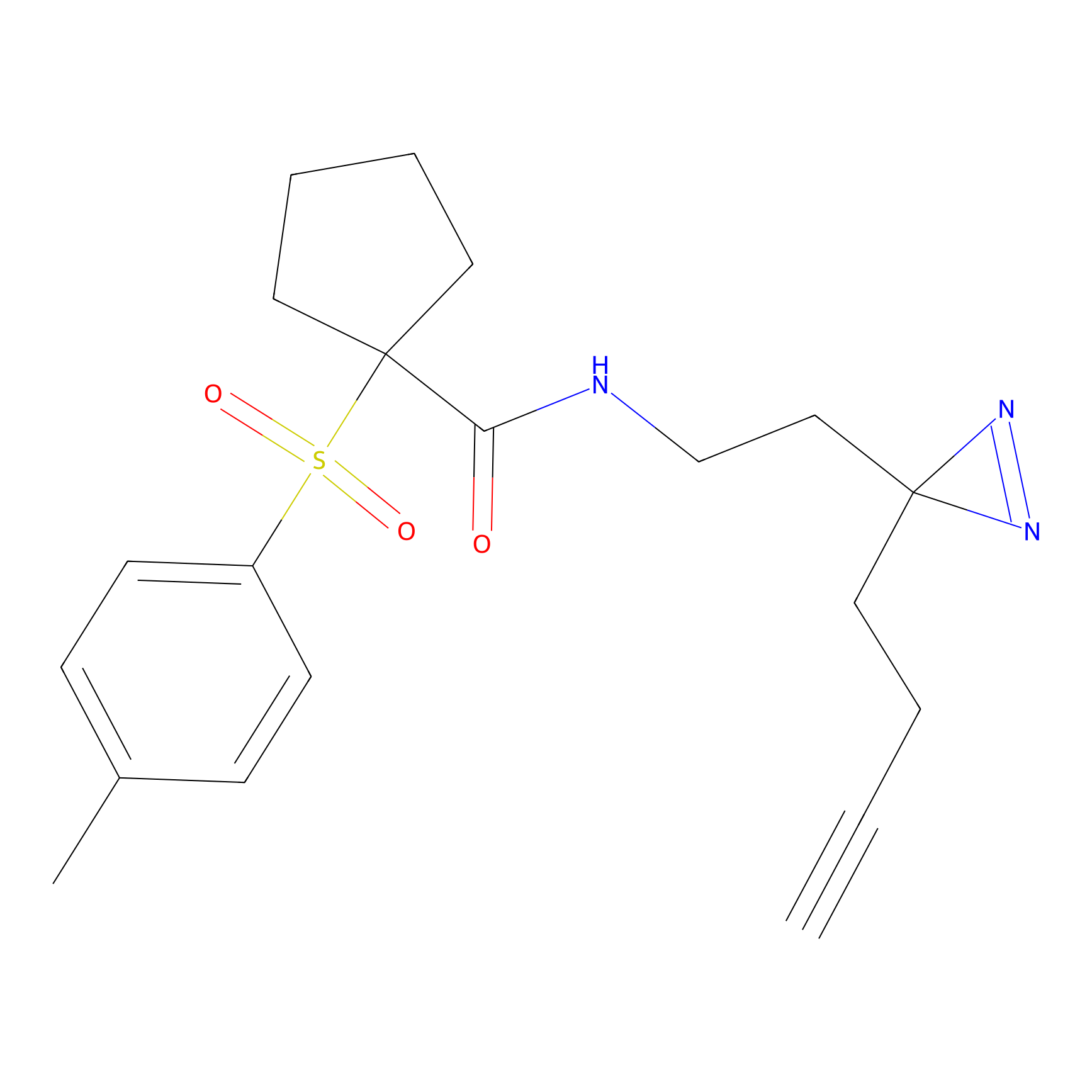
HHS-482 Probe Info |
 |
Y73(1.27); Y80(1.33) | LDD0285 | [5] | |
|
HHS-475 Probe Info |
 |
Y80(0.83); Y73(0.87) | LDD0264 | [6] | |
|
HHS-465 Probe Info |
 |
Y73(9.01); Y80(9.55) | LDD2237 | [7] | |
|
AMP probe Probe Info |
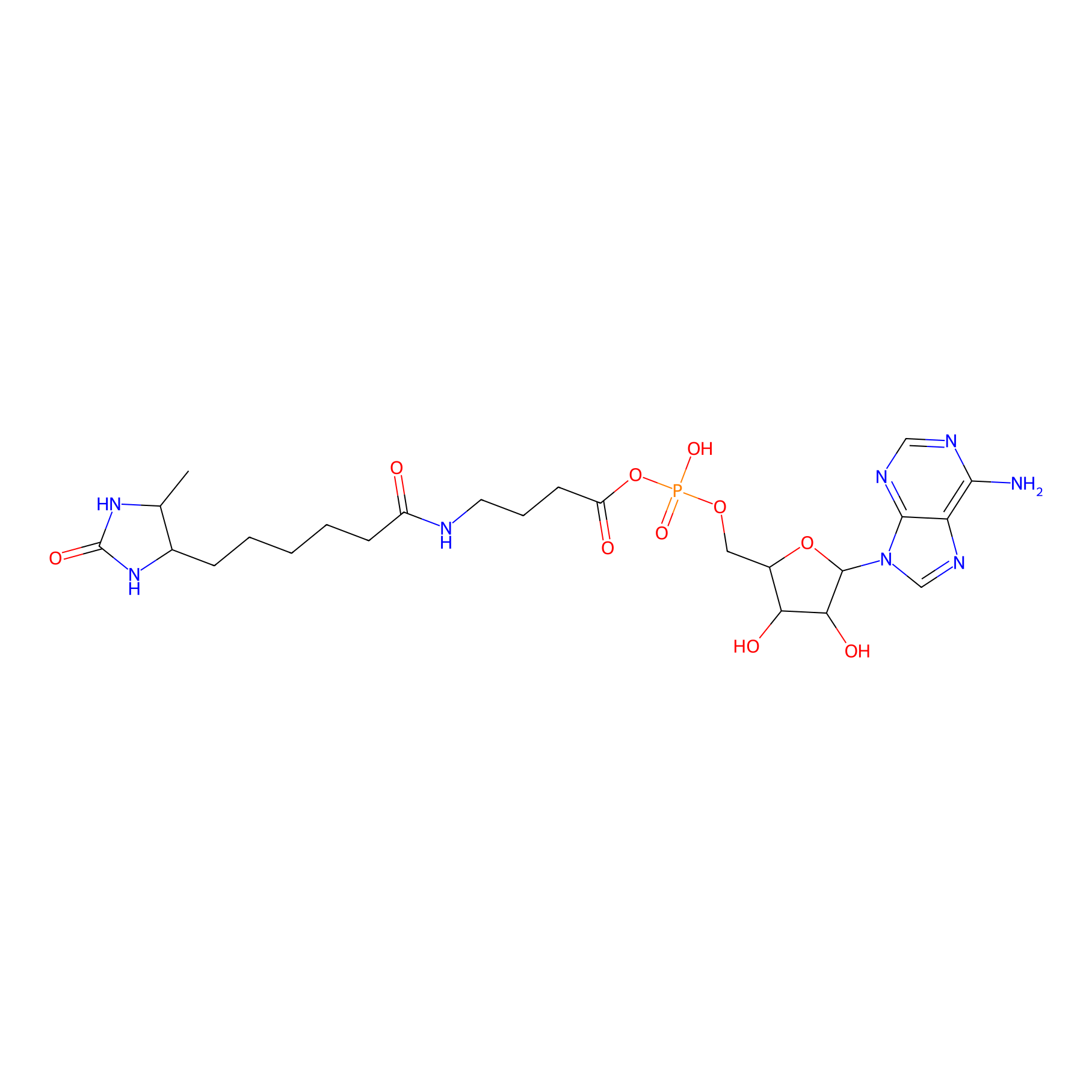 |
K63(0.00); K20(0.00) | LDD0200 | [8] | |
|
ATP probe Probe Info |
 |
K63(0.00); K20(0.00); K66(0.00); K68(0.00) | LDD0199 | [8] | |
|
NHS Probe Info |
 |
K97(0.00); K68(0.00); K33(0.00); K105(0.00) | LDD0010 | [9] | |
|
OSF Probe Info |
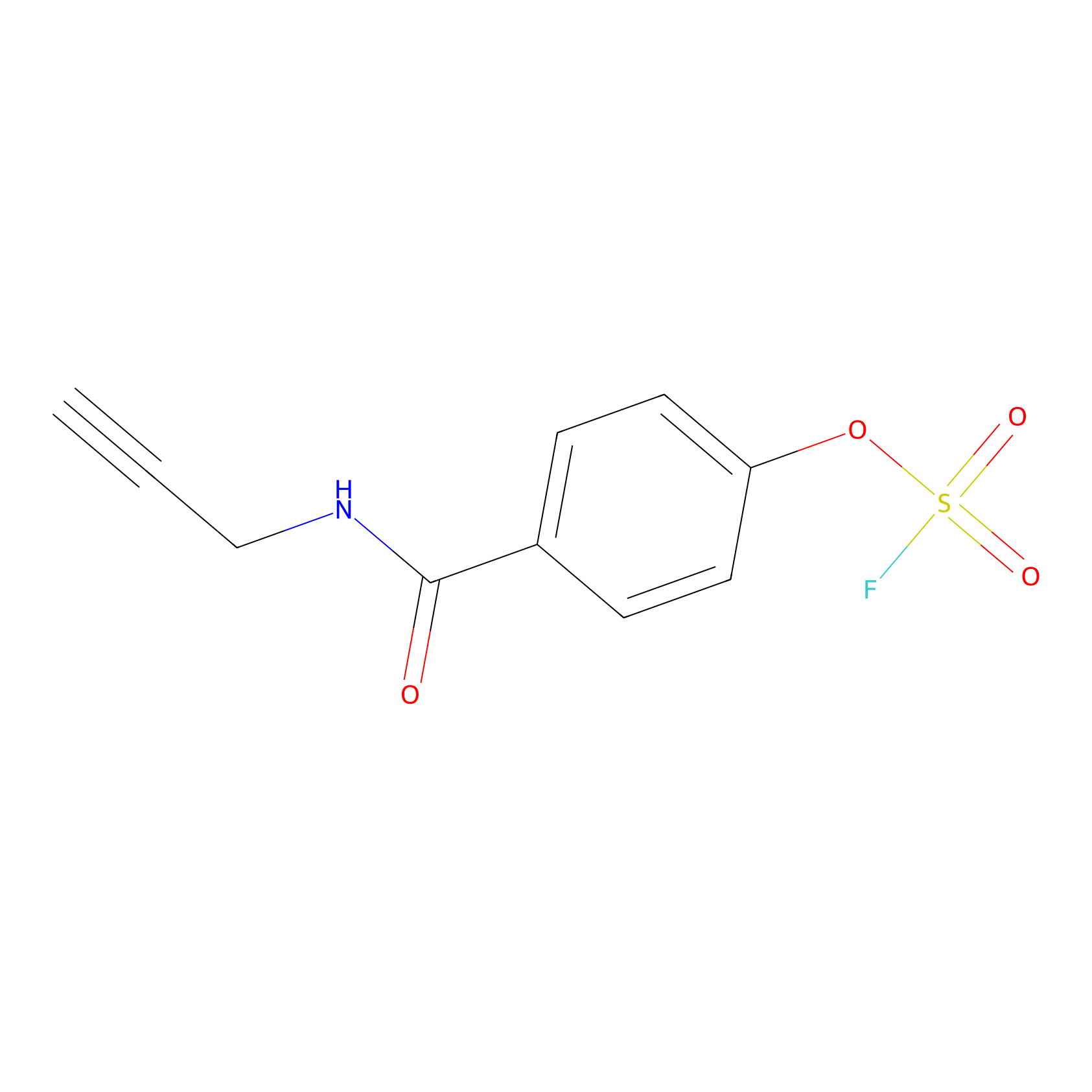 |
N.A. | LDD0029 | [10] | |
|
SF Probe Info |
 |
K63(0.00); K66(0.00); Y80(0.00); K110(0.00) | LDD0028 | [10] | |
|
STPyne Probe Info |
 |
K97(0.00); K63(0.00); K33(0.00); K66(0.00) | LDD0009 | [9] | |
|
1c-yne Probe Info |
 |
K33(0.00); K20(0.00); K68(0.00) | LDD0228 | [11] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [12] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0156 | Aniline | NCI-H1299 | 10.57 | LDD0403 | [4] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [12] |
| LDCM0116 | HHS-0101 | DM93 | Y80(0.83); Y73(0.87) | LDD0264 | [6] |
| LDCM0117 | HHS-0201 | DM93 | Y80(0.74); Y73(0.77) | LDD0265 | [6] |
| LDCM0118 | HHS-0301 | DM93 | Y80(0.87); Y73(1.00) | LDD0266 | [6] |
| LDCM0119 | HHS-0401 | DM93 | Y80(0.69); Y73(0.81) | LDD0267 | [6] |
| LDCM0120 | HHS-0701 | DM93 | Y80(0.80); Y73(0.96) | LDD0268 | [6] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [12] |
| LDCM0123 | JWB131 | DM93 | Y73(1.27); Y80(1.33) | LDD0285 | [5] |
| LDCM0124 | JWB142 | DM93 | Y73(1.54); Y80(1.86) | LDD0286 | [5] |
| LDCM0125 | JWB146 | DM93 | Y73(1.08); Y80(1.89) | LDD0287 | [5] |
| LDCM0126 | JWB150 | DM93 | Y73(3.28); Y80(7.48) | LDD0288 | [5] |
| LDCM0127 | JWB152 | DM93 | Y73(3.11); Y80(5.78) | LDD0289 | [5] |
| LDCM0128 | JWB198 | DM93 | Y80(3.02) | LDD0290 | [5] |
| LDCM0129 | JWB202 | DM93 | Y73(1.01); Y80(1.65) | LDD0291 | [5] |
| LDCM0130 | JWB211 | DM93 | Y73(1.32); Y80(1.97) | LDD0292 | [5] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [12] |
The Interaction Atlas With This Target
References