Details of the Target
General Information of Target
| Target ID | LDTP06281 | |||||
|---|---|---|---|---|---|---|
| Target Name | Condensin complex subunit 1 (NCAPD2) | |||||
| Gene Name | NCAPD2 | |||||
| Gene ID | 9918 | |||||
| Synonyms |
CAPD2; CNAP1; KIAA0159; Condensin complex subunit 1; Chromosome condensation-related SMC-associated protein 1; Chromosome-associated protein D2; hCAP-D2; Non-SMC condensin I complex subunit D2; XCAP-D2 homolog
|
|||||
| 3D Structure | ||||||
| Sequence |
MAPQMYEFHLPLSPEELLKSGGVNQYVVQEVLSIKHLPPQLRAFQAAFRAQGPLAMLQHF
DTIYSILHHFRSIDPGLKEDTLQFLIKVVSRHSQELPAILDDTTLSGSDRNAHLNALKMN CYALIRLLESFETMASQTNLVDLDLGGKGKKARTKAAHGFDWEEERQPILQLLTQLLQLD IRHLWNHSIIEEEFVSLVTGCCYRLLENPTINHQKNRPTREAITHLLGVALTRYNHMLSA TVKIIQMLQHFEHLAPVLVAAVSLWATDYGMKSIVGEIVREIGQKCPQELSRDPSGTKGF AAFLTELAERVPAILMSSMCILLDHLDGENYMMRNAVLAAMAEMVLQVLSGDQLEAAARD TRDQFLDTLQAHGHDVNSFVRSRVLQLFTRIVQQKALPLTRFQAVVALAVGRLADKSVLV CKNAIQLLASFLANNPFSCKLSDADLAGPLQKETQKLQEMRAQRRTAAASAVLDPEEEWE AMLPELKSTLQQLLQLPQGEEEIPEQIANTETTEDVKGRIYQLLAKASYKKAIILTREAT GHFQESEPFSHIDPEESEETRLLNILGLIFKGPAASTQEKNPRESTGNMVTGQTVCKNKP NMSDPEESRGNDELVKQEMLVQYLQDAYSFSRKITEAIGIISKMMYENTTTVVQEVIEFF VMVFQFGVPQALFGVRRMLPLIWSKEPGVREAVLNAYRQLYLNPKGDSARAKAQALIQNL SLLLVDASVGTIQCLEEILCEFVQKDELKPAVTQLLWERATEKVACCPLERCSSVMLLGM MARGKPEIVGSNLDTLVSIGLDEKFPQDYRLAQQVCHAIANISDRRKPSLGKRHPPFRLP QEHRLFERLRETVTKGFVHPDPLWIPFKEVAVTLIYQLAEGPEVICAQILQGCAKQALEK LEEKRTSQEDPKESPAMLPTFLLMNLLSLAGDVALQQLVHLEQAVSGELCRRRVLREEQE HKTKDPKEKNTSSETTMEEELGLVGATADDTEAELIRGICEMELLDGKQTLAAFVPLLLK VCNNPGLYSNPDLSAAASLALGKFCMISATFCDSQLRLLFTMLEKSPLPIVRSNLMVATG DLAIRFPNLVDPWTPHLYARLRDPAQQVRKTAGLVMTHLILKDMVKVKGQVSEMAVLLID PEPQIAALAKNFFNELSHKGNAIYNLLPDIISRLSDPELGVEEEPFHTIMKQLLSYITKD KQTESLVEKLCQRFRTSRTERQQRDLAYCVSQLPLTERGLRKMLDNFDCFGDKLSDESIF SAFLSVVGKLRRGAKPEGKAIIDEFEQKLRACHTRGLDGIKELEIGQAGSQRAPSAKKPS TGSRYQPLASTASDNDFVTPEPRRTTRRHPNTQQRASKKKPKVVFSSDESSEEDLSAEMT EDETPKKTTPILRASARRHRS |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
CND1 (condensin subunit 1) family
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Regulatory subunit of the condensin complex, a complex required for conversion of interphase chromatin into mitotic-like condense chromosomes. The condensin complex probably introduces positive supercoils into relaxed DNA in the presence of type I topoisomerases and converts nicked DNA into positive knotted forms in the presence of type II topoisomerases. May target the condensin complex to DNA via its C-terminal domain. May promote the resolution of double-strand DNA catenanes (intertwines) between sister chromatids. Condensin-mediated compaction likely increases tension in catenated sister chromatids, providing directionality for type II topoisomerase-mediated strand exchanges toward chromatid decatenation. Required for decatenation of non-centromeric ultrafine DNA bridges during anaphase. Early in neurogenesis, may play an essential role to ensure accurate mitotic chromosome condensation in neuron stem cells, ultimately affecting neuron pool and cortex size.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
11.46 | LDD0402 | [1] | |
|
A-EBA Probe Info |
 |
4.52 | LDD0215 | [2] | |
|
AZ-9 Probe Info |
 |
5.62 | LDD0393 | [3] | |
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [4] | |
|
CY4 Probe Info |
 |
100.00 | LDD0244 | [4] | |
|
TH211 Probe Info |
 |
Y122(10.04) | LDD0257 | [5] | |
|
ONAyne Probe Info |
 |
K416(0.85) | LDD0274 | [6] | |
|
Probe 1 Probe Info |
 |
Y697(14.63); Y701(7.80) | LDD3495 | [7] | |
|
BTD Probe Info |
 |
C286(1.40); C596(0.70); C767(0.49) | LDD1700 | [8] | |
|
Sulforaphane-probe2 Probe Info |
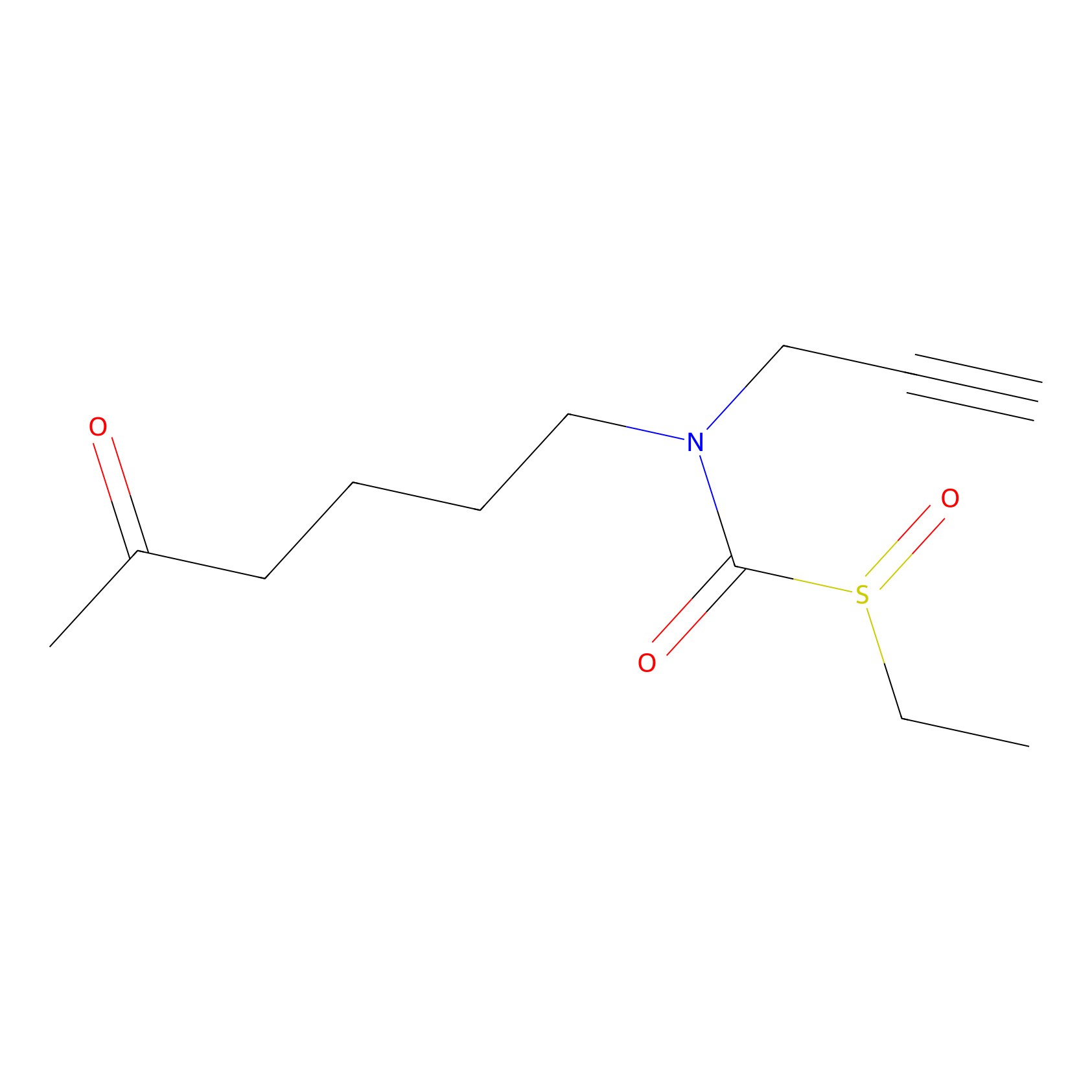 |
3.60 | LDD0042 | [9] | |
|
DA-P3 Probe Info |
 |
4.89 | LDD0182 | [10] | |
|
AHL-Pu-1 Probe Info |
 |
C421(2.04); C596(3.31) | LDD0169 | [11] | |
|
HHS-475 Probe Info |
 |
Y1325(1.25) | LDD0264 | [12] | |
|
HHS-465 Probe Info |
 |
Y1164(1.27); Y1325(8.86) | LDD2237 | [13] | |
|
DBIA Probe Info |
 |
C439(1.25); C766(1.07); C767(1.07); C1045(1.03) | LDD0078 | [14] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [15] | |
|
ATP probe Probe Info |
 |
K1209(0.00); K1288(0.00); K452(0.00); K456(0.00) | LDD0199 | [16] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C286(0.00); C1229(0.00); C596(0.00); C1000(0.00) | LDD0038 | [17] | |
|
IA-alkyne Probe Info |
 |
C596(0.00); C1229(0.00); C772(0.00) | LDD0032 | [18] | |
|
IPIAA_L Probe Info |
 |
N.A. | LDD0031 | [19] | |
|
Lodoacetamide azide Probe Info |
 |
C286(0.00); C1229(0.00); C596(0.00); C1000(0.00) | LDD0037 | [17] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [20] | |
|
NAIA_4 Probe Info |
 |
C439(0.00); C596(0.00); C767(0.00) | LDD2226 | [21] | |
|
TFBX Probe Info |
 |
C121(0.00); C286(0.00) | LDD0027 | [20] | |
|
WYneN Probe Info |
 |
C596(0.00); C767(0.00); C286(0.00) | LDD0021 | [22] | |
|
WYneO Probe Info |
 |
C286(0.00); C596(0.00) | LDD0022 | [22] | |
|
Compound 10 Probe Info |
 |
C596(0.00); C767(0.00) | LDD2216 | [23] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [23] | |
|
ENE Probe Info |
 |
C596(0.00); C767(0.00); C421(0.00) | LDD0006 | [22] | |
|
IPM Probe Info |
 |
C421(0.00); C286(0.00); C596(0.00); C767(0.00) | LDD0005 | [22] | |
|
NPM Probe Info |
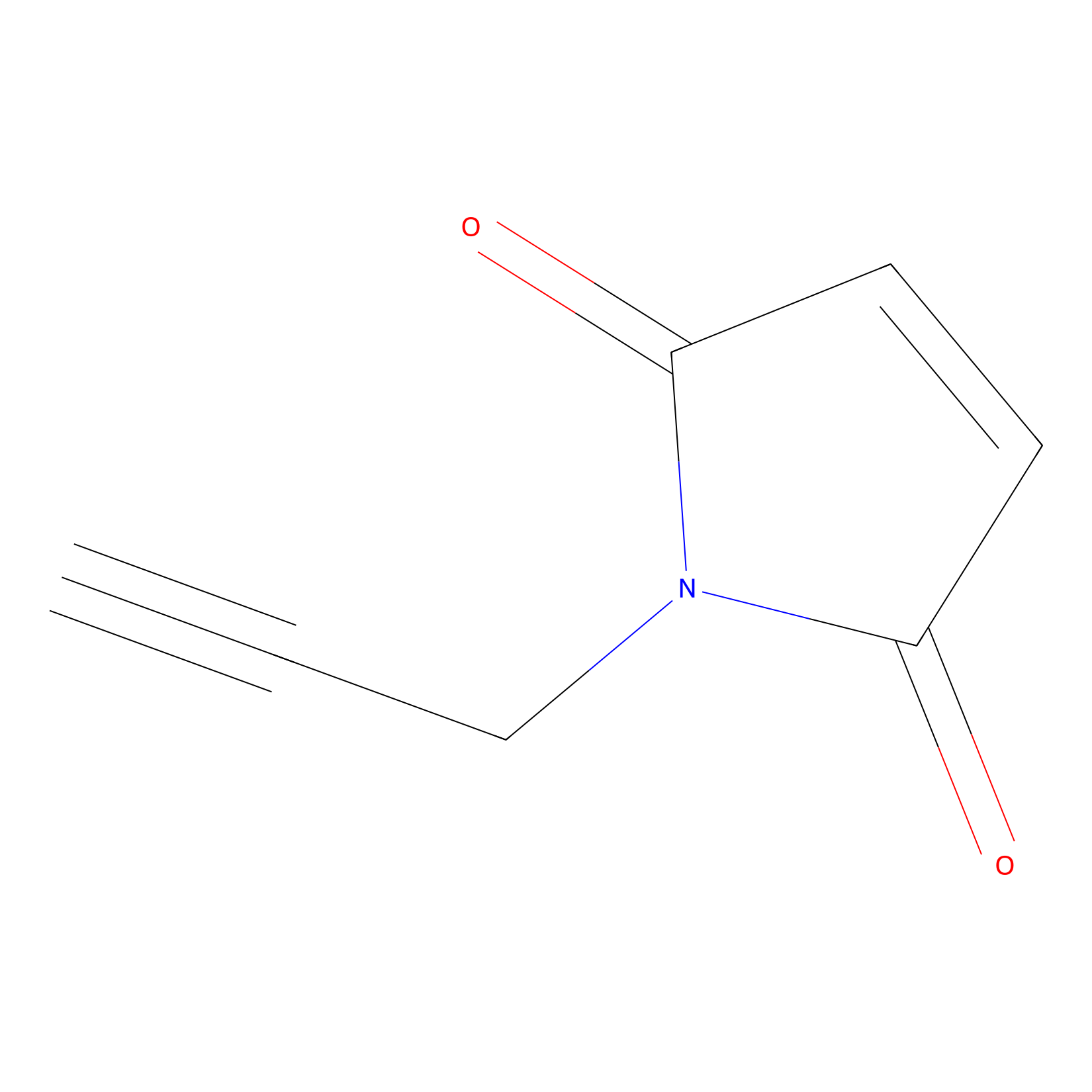 |
N.A. | LDD0016 | [22] | |
|
PF-06672131 Probe Info |
 |
N.A. | LDD0017 | [24] | |
|
PPMS Probe Info |
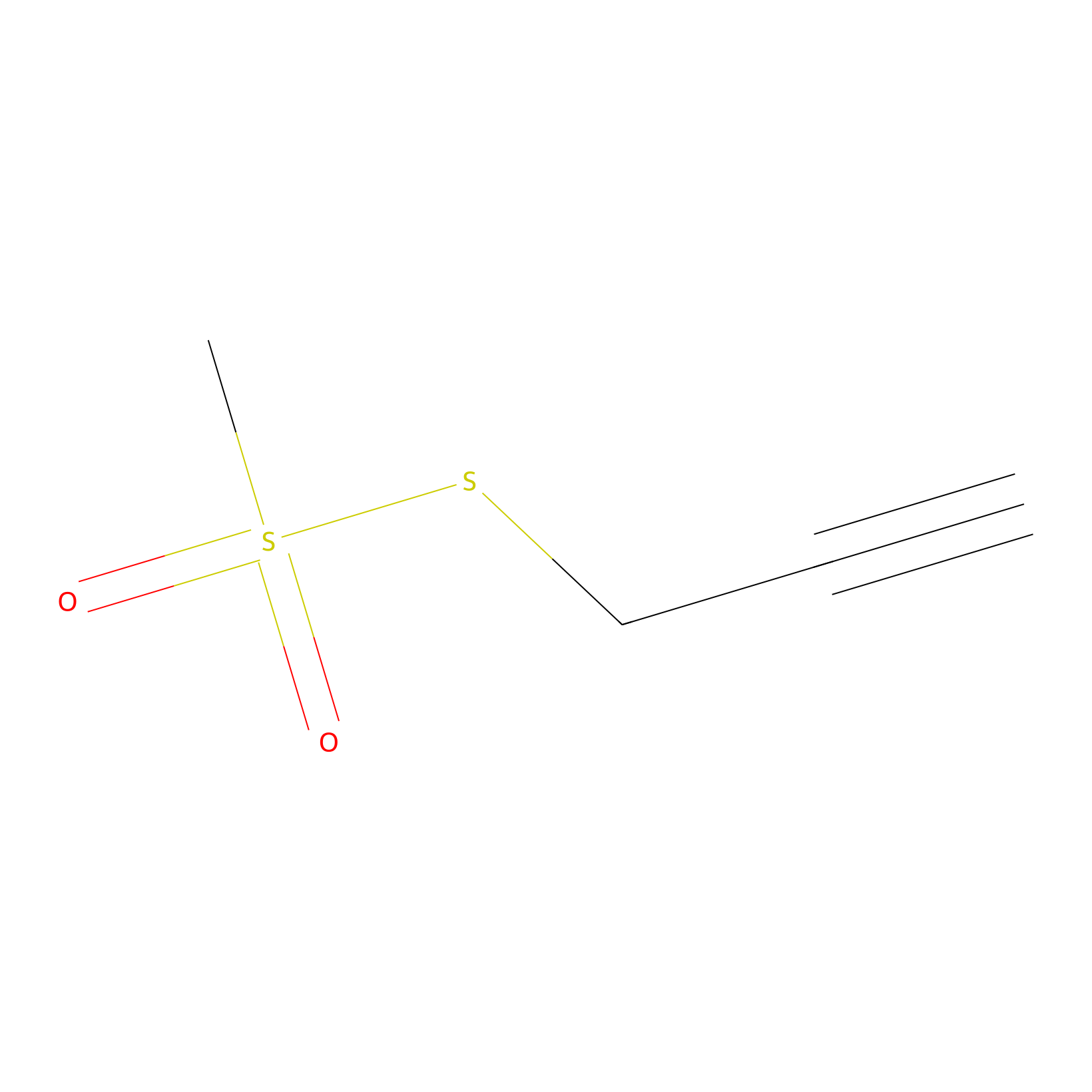 |
N.A. | LDD0008 | [22] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [25] | |
|
STPyne Probe Info |
 |
K395(0.00); K633(0.00); K763(0.00) | LDD0009 | [22] | |
|
VSF Probe Info |
 |
C286(0.00); C596(0.00) | LDD0007 | [22] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [26] | |
|
1c-yne Probe Info |
 |
K1122(0.00); K616(0.00); K456(0.00); K571(0.00) | LDD0228 | [27] | |
|
Acrolein Probe Info |
 |
C816(0.00); C596(0.00); C1229(0.00); H92(0.00) | LDD0217 | [28] | |
|
Crotonaldehyde Probe Info |
 |
C596(0.00); C1229(0.00); C766(0.00) | LDD0219 | [28] | |
|
Methacrolein Probe Info |
 |
C1229(0.00); C816(0.00); C766(0.00); C596(0.00) | LDD0218 | [28] | |
|
W1 Probe Info |
 |
C1229(0.00); C766(0.00); C767(0.00); C596(0.00) | LDD0236 | [29] | |
|
AOyne Probe Info |
 |
9.90 | LDD0443 | [30] | |
|
NAIA_5 Probe Info |
 |
C772(0.00); C1229(0.00); C1000(0.00); C121(0.00) | LDD2223 | [21] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe11 Probe Info |
 |
5.02 | LDD0472 | [31] | |
|
FFF probe13 Probe Info |
 |
13.32 | LDD0475 | [31] | |
|
FFF probe2 Probe Info |
 |
7.19 | LDD0463 | [31] | |
|
FFF probe3 Probe Info |
 |
8.32 | LDD0464 | [31] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0137 | [32] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [32] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [33] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C286(1.00) | LDD2112 | [8] |
| LDCM0537 | 2-Cyano-N,N-dimethylacetamide | MDA-MB-231 | C421(1.01); C286(1.01) | LDD2130 | [8] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C421(1.19); C286(0.94); C596(0.92); C767(1.02) | LDD2117 | [8] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C421(1.41); C766(1.32) | LDD2152 | [8] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C286(1.00); C816(0.92) | LDD2103 | [8] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C286(0.81) | LDD2132 | [8] |
| LDCM0538 | 4-(Cyanoacetyl)morpholine | MDA-MB-231 | C421(0.94); C286(0.87); C596(0.71) | LDD2131 | [8] |
| LDCM0025 | 4SU-RNA | DM93 | C421(3.65) | LDD0170 | [11] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C421(2.04); C596(3.31) | LDD0169 | [11] |
| LDCM0214 | AC1 | HCT 116 | C766(0.94); C767(1.02); C1229(1.23); C1000(1.06) | LDD0531 | [14] |
| LDCM0215 | AC10 | HCT 116 | C766(0.93); C767(1.22); C1000(0.97); C1022(1.11) | LDD0532 | [14] |
| LDCM0216 | AC100 | HCT 116 | C766(1.17); C767(1.07); C1229(0.36); C1000(0.79) | LDD0533 | [14] |
| LDCM0217 | AC101 | HCT 116 | C766(0.74); C767(1.25); C1229(0.63); C1000(0.81) | LDD0534 | [14] |
| LDCM0218 | AC102 | HCT 116 | C766(0.69); C767(0.79); C1229(0.35); C1000(0.74) | LDD0535 | [14] |
| LDCM0219 | AC103 | HCT 116 | C766(0.55); C767(1.60); C1229(0.26); C1000(0.80) | LDD0536 | [14] |
| LDCM0220 | AC104 | HCT 116 | C766(0.86); C767(1.54); C1229(0.30); C1000(0.81) | LDD0537 | [14] |
| LDCM0221 | AC105 | HCT 116 | C766(0.95); C767(1.46); C1229(0.35); C1000(0.73) | LDD0538 | [14] |
| LDCM0222 | AC106 | HCT 116 | C766(0.50); C767(1.37); C1229(0.25); C1000(0.77) | LDD0539 | [14] |
| LDCM0223 | AC107 | HCT 116 | C766(0.71); C767(1.18); C1229(0.30); C1000(1.18) | LDD0540 | [14] |
| LDCM0224 | AC108 | HCT 116 | C766(0.64); C767(0.81); C1229(0.46); C1000(0.95) | LDD0541 | [14] |
| LDCM0225 | AC109 | HCT 116 | C766(0.80); C767(1.16); C1229(0.81); C1000(0.93) | LDD0542 | [14] |
| LDCM0226 | AC11 | HCT 116 | C766(0.94); C767(1.27); C1000(1.00); C1022(0.90) | LDD0543 | [14] |
| LDCM0227 | AC110 | HCT 116 | C766(0.58); C767(0.84); C1229(0.39); C1000(0.96) | LDD0544 | [14] |
| LDCM0228 | AC111 | HCT 116 | C766(0.51); C767(0.87); C1229(0.38); C1000(0.92) | LDD0545 | [14] |
| LDCM0229 | AC112 | HCT 116 | C766(0.72); C767(1.89); C1229(0.42); C1000(1.06) | LDD0546 | [14] |
| LDCM0230 | AC113 | HCT 116 | C766(0.89); C767(1.04); C1229(0.97); C1000(0.92) | LDD0547 | [14] |
| LDCM0231 | AC114 | HCT 116 | C766(0.94); C767(0.95); C1229(1.01); C1000(0.80) | LDD0548 | [14] |
| LDCM0232 | AC115 | HCT 116 | C766(1.03); C767(1.00); C1229(0.84); C1000(0.79) | LDD0549 | [14] |
| LDCM0233 | AC116 | HCT 116 | C766(2.58); C767(1.52); C1229(0.91); C1000(0.82) | LDD0550 | [14] |
| LDCM0234 | AC117 | HCT 116 | C766(1.09); C767(1.08); C1229(1.11); C1000(0.80) | LDD0551 | [14] |
| LDCM0235 | AC118 | HCT 116 | C766(1.02); C767(1.08); C1229(0.99); C1000(0.86) | LDD0552 | [14] |
| LDCM0236 | AC119 | HCT 116 | C766(1.16); C767(1.16); C1229(1.00); C1000(0.87) | LDD0553 | [14] |
| LDCM0237 | AC12 | HCT 116 | C766(1.10); C767(1.06); C1000(1.01); C1022(0.95) | LDD0554 | [14] |
| LDCM0238 | AC120 | HCT 116 | C766(0.85); C767(1.03); C1229(0.94); C1000(0.83) | LDD0555 | [14] |
| LDCM0239 | AC121 | HCT 116 | C766(1.01); C767(0.93); C1229(1.15); C1000(0.93) | LDD0556 | [14] |
| LDCM0240 | AC122 | HCT 116 | C766(1.19); C767(1.00); C1229(0.97); C1000(0.94) | LDD0557 | [14] |
| LDCM0241 | AC123 | HCT 116 | C766(1.09); C767(0.92); C1229(1.17); C1000(0.90) | LDD0558 | [14] |
| LDCM0242 | AC124 | HCT 116 | C766(1.13); C767(0.99); C1229(1.19); C1000(0.85) | LDD0559 | [14] |
| LDCM0243 | AC125 | HCT 116 | C766(1.04); C767(0.99); C1229(1.05); C1000(0.99) | LDD0560 | [14] |
| LDCM0244 | AC126 | HCT 116 | C766(1.10); C767(1.05); C1229(1.18); C1000(1.10) | LDD0561 | [14] |
| LDCM0245 | AC127 | HCT 116 | C766(1.18); C767(1.03); C1229(0.96); C1000(0.88) | LDD0562 | [14] |
| LDCM0246 | AC128 | HCT 116 | C766(1.29); C767(1.29); C1229(1.07); C1045(1.23) | LDD0563 | [14] |
| LDCM0247 | AC129 | HCT 116 | C766(1.25); C767(1.25); C1229(0.88); C1045(1.21) | LDD0564 | [14] |
| LDCM0249 | AC130 | HCT 116 | C766(1.47); C767(1.47); C1229(0.91); C1045(1.20) | LDD0566 | [14] |
| LDCM0250 | AC131 | HCT 116 | C766(1.23); C767(1.23); C1229(1.23); C1045(1.43) | LDD0567 | [14] |
| LDCM0251 | AC132 | HCT 116 | C766(1.59); C767(1.59); C1229(1.32); C1045(1.35) | LDD0568 | [14] |
| LDCM0252 | AC133 | HCT 116 | C766(1.31); C767(1.31); C1229(1.23); C1045(1.23) | LDD0569 | [14] |
| LDCM0253 | AC134 | HCT 116 | C766(1.41); C767(1.41); C1229(1.41); C1045(1.06) | LDD0570 | [14] |
| LDCM0254 | AC135 | HCT 116 | C766(1.29); C767(1.29); C1229(0.91); C1045(1.26) | LDD0571 | [14] |
| LDCM0255 | AC136 | HCT 116 | C766(1.20); C767(1.20); C1229(0.96); C1045(1.15) | LDD0572 | [14] |
| LDCM0256 | AC137 | HCT 116 | C766(1.40); C767(1.40); C1229(1.15); C1045(1.24) | LDD0573 | [14] |
| LDCM0257 | AC138 | HCT 116 | C766(2.06); C767(2.06); C1229(1.09); C1045(1.44) | LDD0574 | [14] |
| LDCM0258 | AC139 | HCT 116 | C766(1.49); C767(1.49); C1229(0.90); C1045(1.32) | LDD0575 | [14] |
| LDCM0259 | AC14 | HCT 116 | C766(0.77); C767(1.15); C1000(1.02); C1022(1.16) | LDD0576 | [14] |
| LDCM0260 | AC140 | HCT 116 | C766(1.81); C767(1.81); C1229(0.91); C1045(1.28) | LDD0577 | [14] |
| LDCM0261 | AC141 | HCT 116 | C766(1.56); C767(1.56); C1229(0.92); C1045(1.19) | LDD0578 | [14] |
| LDCM0262 | AC142 | HCT 116 | C766(1.48); C767(1.48); C1229(1.19); C1045(1.14) | LDD0579 | [14] |
| LDCM0263 | AC143 | HCT 116 | C766(1.13); C767(1.14); C1229(0.83); C1000(0.71) | LDD0580 | [14] |
| LDCM0264 | AC144 | HCT 116 | C1229(0.41); C1249(0.74); C121(0.76); C1000(0.85) | LDD0581 | [14] |
| LDCM0265 | AC145 | HCT 116 | C1229(0.58); C1249(0.87); C121(0.92); C1000(0.93) | LDD0582 | [14] |
| LDCM0266 | AC146 | HCT 116 | C1229(0.51); C1249(0.82); C286(0.89); C121(0.92) | LDD0583 | [14] |
| LDCM0267 | AC147 | HCT 116 | C1229(0.43); C1249(0.74); C1045(0.87); C1052(0.90) | LDD0584 | [14] |
| LDCM0268 | AC148 | HCT 116 | C1229(0.47); C1249(0.84); C1000(0.97); C286(1.04) | LDD0585 | [14] |
| LDCM0269 | AC149 | HCT 116 | C1229(0.48); C1249(0.82); C1045(0.92); C1052(0.97) | LDD0586 | [14] |
| LDCM0270 | AC15 | HCT 116 | C1022(0.88); C816(0.92); C1000(0.95); C766(0.96) | LDD0587 | [14] |
| LDCM0271 | AC150 | HCT 116 | C1229(0.66); C1249(0.86); C772(0.91); C1000(0.98) | LDD0588 | [14] |
| LDCM0272 | AC151 | HCT 116 | C1229(0.74); C1249(0.85); C767(1.00); C1045(1.03) | LDD0589 | [14] |
| LDCM0273 | AC152 | HCT 116 | C1229(0.42); C1249(0.70); C772(0.94); C1052(1.00) | LDD0590 | [14] |
| LDCM0274 | AC153 | HCT 116 | C1229(0.51); C1249(0.96); C286(0.98); C772(0.99) | LDD0591 | [14] |
| LDCM0621 | AC154 | HCT 116 | C766(1.10); C767(0.98); C1229(0.50); C1000(1.01) | LDD2158 | [14] |
| LDCM0622 | AC155 | HCT 116 | C766(1.02); C767(0.86); C1229(0.50); C1000(1.08) | LDD2159 | [14] |
| LDCM0623 | AC156 | HCT 116 | C766(0.91); C767(0.84); C1229(0.93); C1000(1.11) | LDD2160 | [14] |
| LDCM0624 | AC157 | HCT 116 | C766(0.87); C767(0.76); C1229(0.80); C1000(1.13) | LDD2161 | [14] |
| LDCM0276 | AC17 | HCT 116 | C121(0.62); C1229(0.81); C1249(0.83); C596(0.88) | LDD0593 | [14] |
| LDCM0277 | AC18 | HCT 116 | C121(0.66); C1249(0.72); C1022(0.73); C1229(0.82) | LDD0594 | [14] |
| LDCM0278 | AC19 | HCT 116 | C1229(0.79); C1249(0.79); C596(0.82); C286(0.85) | LDD0595 | [14] |
| LDCM0279 | AC2 | HCT 116 | C121(0.63); C1022(0.89); C1045(0.90); C1052(0.90) | LDD0596 | [14] |
| LDCM0280 | AC20 | HCT 116 | C1229(0.81); C1249(0.82); C1000(0.84); C596(0.89) | LDD0597 | [14] |
| LDCM0281 | AC21 | HCT 116 | C1022(0.70); C1249(0.82); C121(0.86); C1045(0.86) | LDD0598 | [14] |
| LDCM0282 | AC22 | HCT 116 | C121(0.81); C596(0.81); C1249(0.86); C1000(0.92) | LDD0599 | [14] |
| LDCM0283 | AC23 | HCT 116 | C1249(0.85); C1045(0.92); C1229(0.92); C772(0.92) | LDD0600 | [14] |
| LDCM0284 | AC24 | HCT 116 | C596(0.73); C121(0.88); C1249(0.88); C772(0.93) | LDD0601 | [14] |
| LDCM0285 | AC25 | HCT 116 | C766(0.84); C1229(0.86); C1000(0.87); C1052(0.91) | LDD0602 | [14] |
| LDCM0286 | AC26 | HCT 116 | C1229(0.81); C1249(0.83); C766(0.84); C1022(0.89) | LDD0603 | [14] |
| LDCM0287 | AC27 | HCT 116 | C766(0.80); C1052(0.81); C286(0.81); C1022(0.83) | LDD0604 | [14] |
| LDCM0288 | AC28 | HCT 116 | C286(0.79); C1022(0.79); C1229(0.87); C1052(0.87) | LDD0605 | [14] |
| LDCM0289 | AC29 | HCT 116 | C1229(0.71); C1249(0.81); C1022(0.85); C816(0.88) | LDD0606 | [14] |
| LDCM0290 | AC3 | HCT 116 | C121(0.60); C1022(0.89); C1045(0.91); C1052(0.91) | LDD0607 | [14] |
| LDCM0291 | AC30 | HCT 116 | C1229(0.70); C1249(0.80); C766(0.82); C1022(0.84) | LDD0608 | [14] |
| LDCM0292 | AC31 | HCT 116 | C1022(0.72); C1249(0.79); C1229(0.81); C766(0.86) | LDD0609 | [14] |
| LDCM0293 | AC32 | HCT 116 | C766(0.73); C1022(0.74); C1229(0.79); C1249(0.80) | LDD0610 | [14] |
| LDCM0294 | AC33 | HCT 116 | C1229(0.71); C1022(0.73); C1249(0.74); C766(0.82) | LDD0611 | [14] |
| LDCM0295 | AC34 | HCT 116 | C1229(0.63); C1249(0.75); C766(0.81); C1022(0.83) | LDD0612 | [14] |
| LDCM0296 | AC35 | HCT 116 | C1000(0.80); C767(0.87); C286(0.90); C596(0.90) | LDD0613 | [14] |
| LDCM0297 | AC36 | HCT 116 | C121(0.84); C1000(0.90); C596(0.93); C1022(0.93) | LDD0614 | [14] |
| LDCM0298 | AC37 | HCT 116 | C1229(0.75); C1000(0.78); C767(0.84); C439(0.85) | LDD0615 | [14] |
| LDCM0299 | AC38 | HCT 116 | C1000(0.92); C121(0.94); C596(0.94); C767(0.95) | LDD0616 | [14] |
| LDCM0300 | AC39 | HCT 116 | C1022(0.89); C1229(0.91); C1000(0.91); C121(0.94) | LDD0617 | [14] |
| LDCM0301 | AC4 | HCT 116 | C121(0.63); C1022(0.74); C1045(0.80); C1052(0.80) | LDD0618 | [14] |
| LDCM0302 | AC40 | HCT 116 | C1229(0.72); C1000(0.82); C121(0.85); C1022(0.91) | LDD0619 | [14] |
| LDCM0303 | AC41 | HCT 116 | C1229(0.81); C1022(0.86); C121(0.86); C439(0.88) | LDD0620 | [14] |
| LDCM0304 | AC42 | HCT 116 | C1022(0.92); C1229(0.94); C1249(0.97); C121(1.01) | LDD0621 | [14] |
| LDCM0305 | AC43 | HCT 116 | C1000(0.89); C121(0.98); C1022(0.99); C1045(1.00) | LDD0622 | [14] |
| LDCM0306 | AC44 | HCT 116 | C1229(0.73); C1000(0.86); C121(0.88); C1022(0.93) | LDD0623 | [14] |
| LDCM0307 | AC45 | HCT 116 | C1229(0.79); C121(0.89); C1022(0.91); C1249(0.95) | LDD0624 | [14] |
| LDCM0308 | AC46 | HCT 116 | C286(0.95); C121(0.95); C1000(0.95); C816(0.97) | LDD0625 | [14] |
| LDCM0309 | AC47 | HCT 116 | C1229(0.82); C286(0.91); C121(0.92); C1022(0.93) | LDD0626 | [14] |
| LDCM0310 | AC48 | HCT 116 | C439(0.77); C286(0.89); C596(0.96); C816(0.96) | LDD0627 | [14] |
| LDCM0311 | AC49 | HCT 116 | C1229(0.94); C1249(0.95); C286(0.97); C772(1.01) | LDD0628 | [14] |
| LDCM0312 | AC5 | HCT 116 | C121(0.70); C1022(0.73); C1045(0.88); C1052(0.88) | LDD0629 | [14] |
| LDCM0313 | AC50 | HCT 116 | C286(0.92); C816(0.97); C439(1.01); C1249(1.01) | LDD0630 | [14] |
| LDCM0314 | AC51 | HCT 116 | C439(0.88); C286(0.88); C596(0.90); C1229(0.95) | LDD0631 | [14] |
| LDCM0315 | AC52 | HCT 116 | C439(0.74); C1229(0.77); C121(0.87); C1000(0.97) | LDD0632 | [14] |
| LDCM0316 | AC53 | HCT 116 | C439(0.86); C816(0.89); C1249(0.90); C1229(0.94) | LDD0633 | [14] |
| LDCM0317 | AC54 | HCT 116 | C439(0.70); C1229(0.94); C286(0.94); C1249(0.95) | LDD0634 | [14] |
| LDCM0318 | AC55 | HCT 116 | C1229(0.85); C439(0.86); C1249(0.94); C816(0.98) | LDD0635 | [14] |
| LDCM0319 | AC56 | HCT 116 | C439(0.93); C1229(0.98); C816(0.98); C121(1.02) | LDD0636 | [14] |
| LDCM0320 | AC57 | HCT 116 | C1229(0.48); C1000(0.77); C1249(0.82); C286(0.91) | LDD0637 | [14] |
| LDCM0321 | AC58 | HCT 116 | C1229(0.51); C1249(0.79); C286(0.81); C766(0.92) | LDD0638 | [14] |
| LDCM0322 | AC59 | HCT 116 | C1229(0.46); C1249(0.78); C286(0.85); C766(0.86) | LDD0639 | [14] |
| LDCM0323 | AC6 | HCT 116 | C1045(0.86); C1052(0.87); C1000(0.88); C1249(0.92) | LDD0640 | [14] |
| LDCM0324 | AC60 | HCT 116 | C1229(0.46); C1249(0.77); C286(0.81); C1000(0.89) | LDD0641 | [14] |
| LDCM0325 | AC61 | HCT 116 | C1229(0.61); C286(0.86); C1052(0.95); C1249(0.95) | LDD0642 | [14] |
| LDCM0326 | AC62 | HCT 116 | C1229(0.52); C1249(0.81); C286(0.86); C766(0.96) | LDD0643 | [14] |
| LDCM0327 | AC63 | HCT 116 | C1229(0.50); C1249(0.78); C1000(0.82); C1052(0.90) | LDD0644 | [14] |
| LDCM0328 | AC64 | HCT 116 | C1229(0.52); C1249(0.81); C286(0.83); C766(0.95) | LDD0645 | [14] |
| LDCM0329 | AC65 | HCT 116 | C1229(0.49); C1249(0.82); C766(0.88); C286(0.94) | LDD0646 | [14] |
| LDCM0330 | AC66 | HCT 116 | C1229(0.48); C1249(0.83); C286(0.85); C766(0.98) | LDD0647 | [14] |
| LDCM0331 | AC67 | HCT 116 | C1229(0.45); C1249(0.77); C286(0.86); C766(0.97) | LDD0648 | [14] |
| LDCM0332 | AC68 | HCT 116 | C121(0.81); C1249(0.85); C286(0.92); C767(0.94) | LDD0649 | [14] |
| LDCM0333 | AC69 | HCT 116 | C286(0.85); C1249(0.86); C1229(0.87); C767(0.88) | LDD0650 | [14] |
| LDCM0334 | AC7 | HCT 116 | C816(0.82); C1052(0.93); C1000(0.96); C1045(0.97) | LDD0651 | [14] |
| LDCM0335 | AC70 | HCT 116 | C1229(0.66); C816(0.76); C1249(0.89); C121(0.91) | LDD0652 | [14] |
| LDCM0336 | AC71 | HCT 116 | C286(0.76); C121(0.87); C1052(0.97); C767(0.97) | LDD0653 | [14] |
| LDCM0337 | AC72 | HCT 116 | C1229(0.75); C1249(0.85); C121(0.87); C286(0.87) | LDD0654 | [14] |
| LDCM0338 | AC73 | HCT 116 | C1229(0.80); C286(0.89); C1249(0.90); C121(1.03) | LDD0655 | [14] |
| LDCM0339 | AC74 | HCT 116 | C286(0.70); C816(0.82); C1249(0.87); C767(1.00) | LDD0656 | [14] |
| LDCM0340 | AC75 | HCT 116 | C816(0.83); C286(0.85); C1229(0.89); C121(0.97) | LDD0657 | [14] |
| LDCM0341 | AC76 | HCT 116 | C286(0.67); C1249(0.86); C121(0.86); C816(0.91) | LDD0658 | [14] |
| LDCM0342 | AC77 | HCT 116 | C286(0.71); C1229(0.82); C816(0.86); C121(0.86) | LDD0659 | [14] |
| LDCM0343 | AC78 | HCT 116 | C286(0.79); C439(0.91); C1052(0.92); C121(0.95) | LDD0660 | [14] |
| LDCM0344 | AC79 | HCT 116 | C286(0.73); C1249(0.84); C766(0.86); C1229(0.94) | LDD0661 | [14] |
| LDCM0345 | AC8 | HCT 116 | C766(0.81); C1022(0.87); C1045(0.88); C1052(0.91) | LDD0662 | [14] |
| LDCM0346 | AC80 | HCT 116 | C286(0.72); C766(0.80); C1229(0.87); C816(0.88) | LDD0663 | [14] |
| LDCM0347 | AC81 | HCT 116 | C286(0.78); C439(0.79); C772(0.85); C816(0.89) | LDD0664 | [14] |
| LDCM0348 | AC82 | HCT 116 | C286(0.78); C121(0.87); C1229(0.92); C816(0.96) | LDD0665 | [14] |
| LDCM0349 | AC83 | HCT 116 | C1229(0.57); C1249(0.69); C1045(0.93); C772(0.94) | LDD0666 | [14] |
| LDCM0350 | AC84 | HCT 116 | C1229(0.53); C1249(0.68); C1045(0.97); C766(1.05) | LDD0667 | [14] |
| LDCM0351 | AC85 | HCT 116 | C1229(0.66); C1249(0.81); C1045(0.81); C1052(0.84) | LDD0668 | [14] |
| LDCM0352 | AC86 | HCT 116 | C1229(0.64); C1249(0.78); C1045(0.90); C1052(0.94) | LDD0669 | [14] |
| LDCM0353 | AC87 | HCT 116 | C1045(0.79); C1052(0.88); C1229(0.96); C772(1.00) | LDD0670 | [14] |
| LDCM0354 | AC88 | HCT 116 | C1229(0.69); C1045(0.78); C1052(0.83); C1249(0.85) | LDD0671 | [14] |
| LDCM0355 | AC89 | HCT 116 | C1229(0.58); C1249(0.73); C772(0.97); C1045(0.97) | LDD0672 | [14] |
| LDCM0357 | AC90 | HCT 116 | C1045(0.86); C1052(0.88); C772(0.89); C596(0.98) | LDD0674 | [14] |
| LDCM0358 | AC91 | HCT 116 | C1229(0.50); C1249(0.73); C1045(0.82); C286(0.92) | LDD0675 | [14] |
| LDCM0359 | AC92 | HCT 116 | C1229(0.48); C1249(0.74); C1045(0.82); C772(0.83) | LDD0676 | [14] |
| LDCM0360 | AC93 | HCT 116 | C1229(0.62); C1249(0.71); C1045(0.85); C1000(0.86) | LDD0677 | [14] |
| LDCM0361 | AC94 | HCT 116 | C1229(0.69); C772(0.78); C1045(0.81); C1249(0.82) | LDD0678 | [14] |
| LDCM0362 | AC95 | HCT 116 | C1229(0.75); C1045(0.80); C1052(0.86); C1249(0.88) | LDD0679 | [14] |
| LDCM0363 | AC96 | HCT 116 | C1229(0.50); C1249(0.75); C1045(0.81); C286(0.85) | LDD0680 | [14] |
| LDCM0364 | AC97 | HCT 116 | C1229(0.49); C1249(0.71); C772(0.81); C1045(0.84) | LDD0681 | [14] |
| LDCM0365 | AC98 | HCT 116 | C1229(0.26); C1249(0.61); C816(0.67); C1052(0.68) | LDD0682 | [14] |
| LDCM0366 | AC99 | HCT 116 | C1229(0.47); C766(0.67); C816(0.68); C1045(0.72) | LDD0683 | [14] |
| LDCM0545 | Acetamide | MDA-MB-231 | C421(0.48); C286(0.37) | LDD2138 | [8] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C286(0.48); C766(1.00) | LDD2113 | [8] |
| LDCM0248 | AKOS034007472 | HCT 116 | C766(0.96); C767(1.16); C1000(0.98); C1022(1.55) | LDD0565 | [14] |
| LDCM0356 | AKOS034007680 | HCT 116 | C816(0.87); C1045(0.90); C1000(0.93); C1052(0.95) | LDD0673 | [14] |
| LDCM0275 | AKOS034007705 | HCT 116 | C1022(0.74); C766(0.90); C1249(0.92); C816(0.93) | LDD0592 | [14] |
| LDCM0156 | Aniline | NCI-H1299 | N.A. | LDD0404 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C439(1.25); C766(1.07); C767(1.07); C1045(1.03) | LDD0078 | [14] |
| LDCM0102 | BDHI 8 | Jurkat | C121(37.59) | LDD0204 | [34] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C286(0.62) | LDD2091 | [8] |
| LDCM0630 | CCW28-3 | 231MFP | C439(1.39) | LDD2214 | [35] |
| LDCM0108 | Chloroacetamide | HeLa | C816(0.00); C1229(0.00); C596(0.00); C1249(0.00) | LDD0222 | [28] |
| LDCM0632 | CL-Sc | Hep-G2 | C596(20.00); C772(0.73); C816(0.62) | LDD2227 | [21] |
| LDCM0367 | CL1 | HCT 116 | C1022(0.72); C816(1.00); C1000(1.03); C772(1.05) | LDD0684 | [14] |
| LDCM0368 | CL10 | HCT 116 | C1022(0.81); C772(0.92); C816(0.92); C1249(1.08) | LDD0685 | [14] |
| LDCM0369 | CL100 | HCT 116 | C121(0.78); C1022(0.80); C1045(0.86); C1052(0.86) | LDD0686 | [14] |
| LDCM0370 | CL101 | HCT 116 | C286(0.86); C816(0.90); C1045(0.91); C1022(0.92) | LDD0687 | [14] |
| LDCM0371 | CL102 | HCT 116 | C816(0.84); C286(0.93); C439(0.94); C1249(1.00) | LDD0688 | [14] |
| LDCM0372 | CL103 | HCT 116 | C286(0.81); C816(0.81); C766(0.87); C1045(1.04) | LDD0689 | [14] |
| LDCM0373 | CL104 | HCT 116 | C286(0.81); C1022(0.85); C816(0.85); C766(0.94) | LDD0690 | [14] |
| LDCM0374 | CL105 | HCT 116 | C1022(0.67); C121(0.80); C1249(0.80); C772(0.83) | LDD0691 | [14] |
| LDCM0375 | CL106 | HCT 116 | C1022(0.72); C1249(0.77); C772(0.85); C1045(0.87) | LDD0692 | [14] |
| LDCM0376 | CL107 | HCT 116 | C1249(0.73); C1022(0.79); C772(0.84); C1229(0.85) | LDD0693 | [14] |
| LDCM0377 | CL108 | HCT 116 | C1249(0.72); C772(0.83); C121(0.83); C1229(0.87) | LDD0694 | [14] |
| LDCM0378 | CL109 | HCT 116 | C1229(0.71); C1249(0.78); C1022(0.80); C1045(0.93) | LDD0695 | [14] |
| LDCM0379 | CL11 | HCT 116 | C1022(0.79); C816(0.90); C772(0.93); C1249(1.04) | LDD0696 | [14] |
| LDCM0380 | CL110 | HCT 116 | C1249(0.77); C1022(0.79); C1045(0.96); C286(1.00) | LDD0697 | [14] |
| LDCM0381 | CL111 | HCT 116 | C1229(0.79); C1249(0.80); C1022(0.88); C772(0.92) | LDD0698 | [14] |
| LDCM0382 | CL112 | HCT 116 | C772(0.81); C1229(0.88); C1249(0.93); C1022(0.94) | LDD0699 | [14] |
| LDCM0383 | CL113 | HCT 116 | C1249(0.77); C1229(0.80); C1022(0.84); C766(0.85) | LDD0700 | [14] |
| LDCM0384 | CL114 | HCT 116 | C1229(0.86); C1000(0.91); C1249(0.92); C816(0.95) | LDD0701 | [14] |
| LDCM0385 | CL115 | HCT 116 | C1022(0.79); C1229(0.79); C1249(0.80); C816(0.84) | LDD0702 | [14] |
| LDCM0386 | CL116 | HCT 116 | C1022(0.78); C766(0.78); C1052(0.85); C1229(0.86) | LDD0703 | [14] |
| LDCM0387 | CL117 | HCT 116 | C1000(1.01); C1249(1.05); C772(1.12); C1022(1.13) | LDD0704 | [14] |
| LDCM0388 | CL118 | HCT 116 | C1229(0.86); C121(0.87); C1000(0.91); C1052(0.95) | LDD0705 | [14] |
| LDCM0389 | CL119 | HCT 116 | C121(0.74); C1229(0.79); C1000(0.87); C1249(0.94) | LDD0706 | [14] |
| LDCM0390 | CL12 | HCT 116 | C816(0.88); C1022(0.88); C772(0.90); C1249(0.96) | LDD0707 | [14] |
| LDCM0391 | CL120 | HCT 116 | C1229(0.83); C1022(0.88); C121(0.89); C1000(0.95) | LDD0708 | [14] |
| LDCM0392 | CL121 | HCT 116 | C439(0.67); C286(0.85); C596(0.91); C772(0.94) | LDD0709 | [14] |
| LDCM0393 | CL122 | HCT 116 | C439(0.69); C121(0.83); C286(0.89); C816(0.93) | LDD0710 | [14] |
| LDCM0394 | CL123 | HCT 116 | C439(0.59); C816(0.83); C1229(0.88); C121(0.98) | LDD0711 | [14] |
| LDCM0395 | CL124 | HCT 116 | C439(0.69); C1229(0.82); C286(0.92); C1249(0.95) | LDD0712 | [14] |
| LDCM0396 | CL125 | HCT 116 | C1229(0.70); C1249(0.96); C286(0.96); C772(1.06) | LDD0713 | [14] |
| LDCM0397 | CL126 | HCT 116 | C1229(0.65); C1249(0.87); C286(0.93); C772(1.01) | LDD0714 | [14] |
| LDCM0398 | CL127 | HCT 116 | C1229(0.76); C286(0.86); C1249(0.90); C1000(0.91) | LDD0715 | [14] |
| LDCM0399 | CL128 | HCT 116 | C1229(0.48); C1249(0.80); C286(0.85); C1000(0.95) | LDD0716 | [14] |
| LDCM0400 | CL13 | HCT 116 | C1022(0.66); C816(0.85); C1249(0.97); C1000(1.01) | LDD0717 | [14] |
| LDCM0401 | CL14 | HCT 116 | C286(0.97); C1022(1.01); C1000(1.02); C1249(1.06) | LDD0718 | [14] |
| LDCM0402 | CL15 | HCT 116 | C1022(0.78); C816(0.78); C1249(1.11); C1052(1.21) | LDD0719 | [14] |
| LDCM0403 | CL16 | HCT 116 | C1249(0.87); C1000(0.92); C772(0.97); C1045(1.01) | LDD0720 | [14] |
| LDCM0404 | CL17 | HCT 116 | C816(0.73); C1249(0.83); C1000(0.94); C1045(1.06) | LDD0721 | [14] |
| LDCM0405 | CL18 | HCT 116 | C1045(0.82); C1052(0.85); C816(0.86); C1249(0.86) | LDD0722 | [14] |
| LDCM0406 | CL19 | HCT 116 | C766(0.80); C816(0.82); C1249(0.83); C1045(0.90) | LDD0723 | [14] |
| LDCM0407 | CL2 | HCT 116 | C1022(0.80); C816(0.98); C596(0.99); C1000(1.00) | LDD0724 | [14] |
| LDCM0408 | CL20 | HCT 116 | C766(0.70); C816(0.79); C1249(0.83); C1229(0.86) | LDD0725 | [14] |
| LDCM0409 | CL21 | HCT 116 | C816(0.92); C1249(0.95); C286(1.06); C1229(1.13) | LDD0726 | [14] |
| LDCM0410 | CL22 | HCT 116 | C816(0.76); C1249(0.93); C766(0.96); C286(1.03) | LDD0727 | [14] |
| LDCM0411 | CL23 | HCT 116 | C816(0.41); C766(0.79); C1249(0.79); C1000(0.83) | LDD0728 | [14] |
| LDCM0412 | CL24 | HCT 116 | C766(0.68); C816(0.79); C1249(0.87); C1045(1.02) | LDD0729 | [14] |
| LDCM0413 | CL25 | HCT 116 | C766(1.97); C767(2.41); C1229(1.36); C1000(1.01) | LDD0730 | [14] |
| LDCM0414 | CL26 | HCT 116 | C766(0.80); C767(1.14); C1229(1.14); C1000(0.91) | LDD0731 | [14] |
| LDCM0415 | CL27 | HCT 116 | C766(0.92); C767(1.24); C1229(1.29); C1000(1.12) | LDD0732 | [14] |
| LDCM0416 | CL28 | HCT 116 | C766(2.32); C767(2.09); C1229(0.97); C1000(0.87) | LDD0733 | [14] |
| LDCM0417 | CL29 | HCT 116 | C766(0.74); C767(1.07); C1229(0.97); C1000(0.94) | LDD0734 | [14] |
| LDCM0418 | CL3 | HCT 116 | C766(3.16); C767(2.01); C1229(1.30); C1000(1.04) | LDD0735 | [14] |
| LDCM0419 | CL30 | HCT 116 | C766(0.83); C767(1.15); C1229(1.15); C1000(0.96) | LDD0736 | [14] |
| LDCM0420 | CL31 | HCT 116 | C766(1.89); C767(1.42); C1229(1.04); C1000(0.95) | LDD0737 | [14] |
| LDCM0421 | CL32 | HCT 116 | C766(1.02); C767(0.98); C1229(0.85); C1000(0.83) | LDD0738 | [14] |
| LDCM0422 | CL33 | HCT 116 | C766(0.87); C767(1.06); C1229(0.90); C1000(1.05) | LDD0739 | [14] |
| LDCM0423 | CL34 | HCT 116 | C766(0.81); C767(1.05); C1229(0.86); C1000(1.09) | LDD0740 | [14] |
| LDCM0424 | CL35 | HCT 116 | C766(0.86); C767(0.99); C1229(0.79); C1000(0.86) | LDD0741 | [14] |
| LDCM0425 | CL36 | HCT 116 | C766(0.68); C767(1.01); C1229(0.90); C1000(0.93) | LDD0742 | [14] |
| LDCM0426 | CL37 | HCT 116 | C766(0.76); C767(0.94); C1229(0.90); C1000(1.11) | LDD0743 | [14] |
| LDCM0428 | CL39 | HCT 116 | C766(0.87); C767(1.01); C1229(1.08); C1000(0.97) | LDD0745 | [14] |
| LDCM0429 | CL4 | HCT 116 | C766(2.89); C767(1.53); C1229(1.00); C1000(1.03) | LDD0746 | [14] |
| LDCM0430 | CL40 | HCT 116 | C766(0.76); C767(1.22); C1229(0.95); C1000(0.79) | LDD0747 | [14] |
| LDCM0431 | CL41 | HCT 116 | C766(0.91); C767(1.11); C1229(0.90); C1000(0.80) | LDD0748 | [14] |
| LDCM0432 | CL42 | HCT 116 | C766(0.83); C767(1.05); C1229(0.87); C1000(1.34) | LDD0749 | [14] |
| LDCM0433 | CL43 | HCT 116 | C766(0.83); C767(0.94); C1229(0.78); C1000(1.00) | LDD0750 | [14] |
| LDCM0434 | CL44 | HCT 116 | C766(1.02); C767(0.94); C1229(1.07); C1000(1.08) | LDD0751 | [14] |
| LDCM0435 | CL45 | HCT 116 | C766(0.76); C767(1.01); C1229(0.92); C1000(1.18) | LDD0752 | [14] |
| LDCM0436 | CL46 | HCT 116 | C766(1.61); C767(1.44); C1229(1.81); C1000(1.10) | LDD0753 | [14] |
| LDCM0437 | CL47 | HCT 116 | C766(1.23); C767(1.04); C1229(1.39); C1000(1.01) | LDD0754 | [14] |
| LDCM0438 | CL48 | HCT 116 | C766(1.65); C767(1.43); C1229(1.11); C1000(0.94) | LDD0755 | [14] |
| LDCM0439 | CL49 | HCT 116 | C766(2.48); C767(1.79); C1229(1.05); C1000(1.01) | LDD0756 | [14] |
| LDCM0440 | CL5 | HCT 116 | C766(1.04); C767(0.96); C1229(1.21); C1000(0.98) | LDD0757 | [14] |
| LDCM0441 | CL50 | HCT 116 | C766(1.52); C767(1.31); C1229(1.85); C1000(0.98) | LDD0758 | [14] |
| LDCM0442 | CL51 | HCT 116 | C766(0.94); C767(1.23); C1229(1.51); C1000(1.07) | LDD0759 | [14] |
| LDCM0443 | CL52 | HCT 116 | C766(1.76); C767(1.56); C1229(1.39); C1000(1.00) | LDD0760 | [14] |
| LDCM0444 | CL53 | HCT 116 | C766(3.47); C767(2.39); C1229(0.95); C1000(1.02) | LDD0761 | [14] |
| LDCM0445 | CL54 | HCT 116 | C766(2.49); C767(1.76); C1229(2.72); C1000(1.06) | LDD0762 | [14] |
| LDCM0446 | CL55 | HCT 116 | C766(0.94); C767(0.92); C1229(2.24); C1000(1.05) | LDD0763 | [14] |
| LDCM0447 | CL56 | HCT 116 | C766(1.12); C767(1.16); C1229(1.08); C1000(0.98) | LDD0764 | [14] |
| LDCM0448 | CL57 | HCT 116 | C766(2.46); C767(1.80); C1229(3.24); C1000(1.05) | LDD0765 | [14] |
| LDCM0449 | CL58 | HCT 116 | C766(1.03); C767(1.02); C1229(1.84); C1000(0.92) | LDD0766 | [14] |
| LDCM0450 | CL59 | HCT 116 | C766(2.46); C767(1.78); C1229(1.24); C1000(1.01) | LDD0767 | [14] |
| LDCM0451 | CL6 | HCT 116 | C766(1.81); C767(1.78); C1229(1.28); C1000(1.06) | LDD0768 | [14] |
| LDCM0452 | CL60 | HCT 116 | C766(1.74); C767(1.46); C1229(0.94); C1000(0.97) | LDD0769 | [14] |
| LDCM0453 | CL61 | HCT 116 | C766(1.28); C767(1.19); C1229(0.99); C1000(0.96) | LDD0770 | [14] |
| LDCM0454 | CL62 | HCT 116 | C766(1.04); C767(1.22); C1229(0.88); C1000(0.89) | LDD0771 | [14] |
| LDCM0455 | CL63 | HCT 116 | C766(1.15); C767(1.83); C1229(0.82); C1000(0.94) | LDD0772 | [14] |
| LDCM0456 | CL64 | HCT 116 | C766(1.28); C767(4.62); C1229(0.78); C1000(1.04) | LDD0773 | [14] |
| LDCM0457 | CL65 | HCT 116 | C766(2.65); C767(1.47); C1229(0.84); C1000(0.92) | LDD0774 | [14] |
| LDCM0458 | CL66 | HCT 116 | C766(0.97); C767(2.72); C1229(0.81); C1000(1.21) | LDD0775 | [14] |
| LDCM0459 | CL67 | HCT 116 | C766(1.46); C767(1.44); C1229(0.75); C1000(0.95) | LDD0776 | [14] |
| LDCM0460 | CL68 | HCT 116 | C766(1.01); C767(3.36); C1229(1.03); C1000(0.95) | LDD0777 | [14] |
| LDCM0461 | CL69 | HCT 116 | C766(1.72); C767(2.78); C1229(0.94); C1000(0.85) | LDD0778 | [14] |
| LDCM0462 | CL7 | HCT 116 | C766(1.44); C767(1.57); C1229(1.01); C1000(1.08) | LDD0779 | [14] |
| LDCM0463 | CL70 | HCT 116 | C766(1.56); C767(1.36); C1229(0.88); C1000(0.97) | LDD0780 | [14] |
| LDCM0464 | CL71 | HCT 116 | C766(1.05); C767(1.66); C1229(0.79); C1000(0.88) | LDD0781 | [14] |
| LDCM0465 | CL72 | HCT 116 | C766(1.31); C767(1.28); C1229(0.82); C1000(0.95) | LDD0782 | [14] |
| LDCM0466 | CL73 | HCT 116 | C766(0.99); C767(2.22); C1229(0.77); C1000(0.87) | LDD0783 | [14] |
| LDCM0467 | CL74 | HCT 116 | C766(1.54); C767(1.60); C1229(0.90); C1000(0.92) | LDD0784 | [14] |
| LDCM0469 | CL76 | HCT 116 | C766(1.02); C767(1.08); C1000(0.98); C1022(0.96) | LDD0786 | [14] |
| LDCM0470 | CL77 | HCT 116 | C766(0.86); C767(1.51); C1000(0.94); C1022(1.37) | LDD0787 | [14] |
| LDCM0471 | CL78 | HCT 116 | C766(2.54); C767(1.47); C1000(0.98); C1022(0.76) | LDD0788 | [14] |
| LDCM0472 | CL79 | HCT 116 | C766(0.79); C767(1.16); C1000(0.95); C1022(0.92) | LDD0789 | [14] |
| LDCM0473 | CL8 | HCT 116 | C766(9.14); C767(6.47); C1229(1.19); C1000(1.28) | LDD0790 | [14] |
| LDCM0474 | CL80 | HCT 116 | C766(0.88); C767(1.09); C1000(0.98); C1022(1.17) | LDD0791 | [14] |
| LDCM0475 | CL81 | HCT 116 | C766(0.89); C767(1.24); C1000(0.98); C1022(0.84) | LDD0792 | [14] |
| LDCM0476 | CL82 | HCT 116 | C766(0.87); C767(1.11); C1000(1.11); C1022(0.90) | LDD0793 | [14] |
| LDCM0477 | CL83 | HCT 116 | C766(1.92); C767(1.88); C1000(1.17); C1022(0.96) | LDD0794 | [14] |
| LDCM0478 | CL84 | HCT 116 | C766(1.63); C767(1.54); C1000(1.48); C1022(1.18) | LDD0795 | [14] |
| LDCM0479 | CL85 | HCT 116 | C766(1.07); C767(1.08); C1000(0.98); C1022(0.88) | LDD0796 | [14] |
| LDCM0480 | CL86 | HCT 116 | C766(1.58); C767(1.20); C1000(0.94); C1022(0.89) | LDD0797 | [14] |
| LDCM0481 | CL87 | HCT 116 | C766(1.08); C767(1.16); C1000(1.04); C1022(0.89) | LDD0798 | [14] |
| LDCM0482 | CL88 | HCT 116 | C766(0.88); C767(0.89); C1000(1.16); C1022(0.91) | LDD0799 | [14] |
| LDCM0483 | CL89 | HCT 116 | C766(1.17); C767(1.01); C1000(1.58); C1022(1.33) | LDD0800 | [14] |
| LDCM0484 | CL9 | HCT 116 | C766(1.89); C767(2.05); C1229(1.08); C1000(0.88) | LDD0801 | [14] |
| LDCM0485 | CL90 | HCT 116 | C766(2.41); C767(2.01); C1000(1.30); C1022(1.42) | LDD0802 | [14] |
| LDCM0486 | CL91 | HCT 116 | C766(1.19); C767(1.34); C1229(0.93); C1000(1.02) | LDD0803 | [14] |
| LDCM0487 | CL92 | HCT 116 | C766(4.91); C767(3.01); C1229(0.96); C1000(1.28) | LDD0804 | [14] |
| LDCM0488 | CL93 | HCT 116 | C766(3.05); C767(2.04); C1229(1.04); C1000(1.07) | LDD0805 | [14] |
| LDCM0489 | CL94 | HCT 116 | C766(1.45); C767(1.11); C1229(1.03); C1000(1.04) | LDD0806 | [14] |
| LDCM0490 | CL95 | HCT 116 | C766(8.98); C767(5.07); C1229(1.07); C1000(1.18) | LDD0807 | [14] |
| LDCM0491 | CL96 | HCT 116 | C766(1.63); C767(1.42); C1229(1.02); C1000(1.29) | LDD0808 | [14] |
| LDCM0492 | CL97 | HCT 116 | C766(2.71); C767(1.99); C1229(0.95); C1000(1.49) | LDD0809 | [14] |
| LDCM0493 | CL98 | HCT 116 | C766(1.93); C767(1.98); C1229(0.99); C1000(1.18) | LDD0810 | [14] |
| LDCM0494 | CL99 | HCT 116 | C766(1.25); C767(1.24); C1229(1.05); C1000(1.18) | LDD0811 | [14] |
| LDCM0191 | Compound 21 | HEK-293T | 4.70 | LDD0508 | [31] |
| LDCM0190 | Compound 34 | HEK-293T | 4.60 | LDD0510 | [31] |
| LDCM0193 | Compound 36 | HEK-293T | 4.04 | LDD0511 | [31] |
| LDCM0634 | CY-0357 | Hep-G2 | C439(2.91); C596(0.87); C286(0.60) | LDD2228 | [21] |
| LDCM0495 | E2913 | HEK-293T | C286(0.95); C596(1.08); C121(0.90); C1000(1.01) | LDD1698 | [36] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C421(42.68); C596(1.78); C1229(1.31) | LDD1702 | [8] |
| LDCM0202 | EV-93 | T cell | C767(4.41) | LDD0514 | [37] |
| LDCM0204 | EV-97 | T cell | C767(4.83) | LDD0524 | [37] |
| LDCM0625 | F8 | Ramos | C121(0.83); C1229(0.60); C596(1.36); C286(1.67) | LDD2187 | [38] |
| LDCM0572 | Fragment10 | Ramos | C767(6.39) | LDD1390 | [39] |
| LDCM0573 | Fragment11 | MDA-MB-231 | C439(0.88) | LDD1467 | [39] |
| LDCM0574 | Fragment12 | MDA-MB-231 | C439(6.71) | LDD1468 | [39] |
| LDCM0575 | Fragment13 | Ramos | C767(1.44) | LDD1396 | [39] |
| LDCM0576 | Fragment14 | MDA-MB-231 | C767(3.64) | LDD1397 | [39] |
| LDCM0577 | Fragment15 | Ramos | C767(9.89) | LDD1400 | [39] |
| LDCM0579 | Fragment20 | Ramos | C121(3.96); C1229(1.46); C596(1.64); C816(4.61) | LDD2194 | [38] |
| LDCM0580 | Fragment21 | MDA-MB-231 | C439(0.67) | LDD1473 | [39] |
| LDCM0582 | Fragment23 | Ramos | C767(1.52) | LDD1409 | [39] |
| LDCM0584 | Fragment25 | MDA-MB-231 | C767(0.97) | LDD1411 | [39] |
| LDCM0578 | Fragment27 | MDA-MB-231 | C767(0.83) | LDD1401 | [39] |
| LDCM0586 | Fragment28 | MDA-MB-231 | C439(0.87) | LDD1475 | [39] |
| LDCM0587 | Fragment29 | MDA-MB-231 | C767(1.02) | LDD1417 | [39] |
| LDCM0588 | Fragment30 | Ramos | C121(2.17); C1229(0.90); C596(1.35); C286(2.05) | LDD2199 | [38] |
| LDCM0589 | Fragment31 | MDA-MB-231 | C439(20.00) | LDD1477 | [39] |
| LDCM0590 | Fragment32 | Ramos | C121(1.16); C1229(0.64); C596(2.03); C816(0.42) | LDD2201 | [38] |
| LDCM0468 | Fragment33 | HCT 116 | C766(1.13); C767(1.63); C1229(1.00); C1000(1.07) | LDD0785 | [14] |
| LDCM0593 | Fragment35 | Ramos | C767(1.41) | LDD1430 | [39] |
| LDCM0595 | Fragment37 | Ramos | C767(0.95) | LDD1432 | [39] |
| LDCM0596 | Fragment38 | MDA-MB-231 | C767(1.21) | LDD1433 | [39] |
| LDCM0566 | Fragment4 | MDA-MB-231 | C767(20.00) | LDD1378 | [39] |
| LDCM0598 | Fragment40 | Ramos | C767(1.43) | LDD1437 | [39] |
| LDCM0599 | Fragment41 | MDA-MB-231 | C767(1.19) | LDD1438 | [39] |
| LDCM0600 | Fragment42 | Ramos | C767(0.93) | LDD1440 | [39] |
| LDCM0427 | Fragment51 | HCT 116 | C766(0.78); C767(1.12); C1229(0.79); C1000(1.01) | LDD0744 | [14] |
| LDCM0610 | Fragment52 | MDA-MB-231 | C767(1.07) | LDD1452 | [39] |
| LDCM0614 | Fragment56 | MDA-MB-231 | C439(1.24) | LDD1484 | [39] |
| LDCM0569 | Fragment7 | Ramos | C767(20.00) | LDD1384 | [39] |
| LDCM0570 | Fragment8 | MDA-MB-231 | C767(15.32) | LDD1385 | [39] |
| LDCM0571 | Fragment9 | Ramos | C767(2.06) | LDD1388 | [39] |
| LDCM0116 | HHS-0101 | DM93 | Y1325(1.25) | LDD0264 | [12] |
| LDCM0117 | HHS-0201 | DM93 | Y1325(1.82) | LDD0265 | [12] |
| LDCM0118 | HHS-0301 | DM93 | Y1325(2.56) | LDD0266 | [12] |
| LDCM0119 | HHS-0401 | DM93 | Y1325(2.48) | LDD0267 | [12] |
| LDCM0120 | HHS-0701 | DM93 | Y1325(2.19) | LDD0268 | [12] |
| LDCM0107 | IAA | HeLa | C1229(0.00); H859(0.00); H92(0.00) | LDD0221 | [28] |
| LDCM0022 | KB02 | HCT 116 | C439(3.02); C596(2.56); C772(1.03) | LDD0080 | [14] |
| LDCM0023 | KB03 | HCT 116 | C439(1.82); C596(1.22); C772(1.66) | LDD0081 | [14] |
| LDCM0024 | KB05 | HCT 116 | C439(14.92); C596(2.04); C772(1.02) | LDD0082 | [14] |
| LDCM0030 | Luteolin | HEK-293T | 4.89 | LDD0182 | [10] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C286(1.18) | LDD2102 | [8] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C421(0.65); C286(0.39); C596(0.52) | LDD2121 | [8] |
| LDCM0109 | NEM | HeLa | H372(0.00); H843(0.00); H92(0.00); H859(0.00) | LDD0223 | [28] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C421(0.76); C596(0.49) | LDD2089 | [8] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C286(2.07); C596(1.05) | LDD2090 | [8] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C286(0.96) | LDD2092 | [8] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C421(1.37); C767(1.11) | LDD2093 | [8] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C286(1.47) | LDD2094 | [8] |
| LDCM0503 | Nucleophilic fragment 14b | MDA-MB-231 | C286(0.27); C767(1.24) | LDD2096 | [8] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C421(0.99) | LDD2097 | [8] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C286(1.89) | LDD2098 | [8] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C286(0.56); C596(0.78); C816(1.11) | LDD2099 | [8] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C286(1.16) | LDD2100 | [8] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C767(0.96) | LDD2101 | [8] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C286(1.01) | LDD2104 | [8] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C286(2.55); C816(1.74) | LDD2105 | [8] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C421(1.04); C286(0.94); C596(0.99); C816(0.72) | LDD2107 | [8] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C596(0.80); C816(0.64) | LDD2109 | [8] |
| LDCM0517 | Nucleophilic fragment 21b | MDA-MB-231 | C286(3.46) | LDD2110 | [8] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C596(0.81) | LDD2111 | [8] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C286(0.66) | LDD2114 | [8] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C286(0.63) | LDD2115 | [8] |
| LDCM0523 | Nucleophilic fragment 24b | MDA-MB-231 | C286(0.75) | LDD2116 | [8] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C421(0.73); C286(0.63); C596(1.01) | LDD2118 | [8] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C816(1.07) | LDD2119 | [8] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C286(0.88); C596(1.08); C816(0.58) | LDD2120 | [8] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C421(0.42); C286(0.59); C596(0.45); C816(0.16) | LDD2122 | [8] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C286(0.61); C596(0.84); C816(0.54) | LDD2123 | [8] |
| LDCM0531 | Nucleophilic fragment 28b | MDA-MB-231 | C421(0.35); C286(0.63); C816(0.24) | LDD2124 | [8] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C286(0.59); C596(0.72); C766(0.68); C816(0.70) | LDD2125 | [8] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C286(0.68) | LDD2126 | [8] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C421(1.23); C286(1.03); C596(0.91); C766(0.84) | LDD2127 | [8] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C286(0.72); C816(0.58) | LDD2128 | [8] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C421(1.07) | LDD2129 | [8] |
| LDCM0540 | Nucleophilic fragment 35 | MDA-MB-231 | C286(0.80); C766(0.83) | LDD2133 | [8] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C286(1.09); C767(0.66); C766(0.51) | LDD2134 | [8] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C286(0.53); C767(1.12); C766(0.76) | LDD2135 | [8] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C421(1.41); C286(1.40); C596(1.14) | LDD2136 | [8] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C286(0.79); C596(0.98); C767(0.60); C816(0.63) | LDD2137 | [8] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C286(1.40); C596(0.70); C767(0.49) | LDD1700 | [8] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C421(1.35); C596(0.65); C766(1.27); C816(0.49) | LDD2140 | [8] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C286(0.77) | LDD2141 | [8] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C421(0.96); C286(1.16) | LDD2143 | [8] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C421(2.10); C286(4.15); C766(2.01) | LDD2144 | [8] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C421(1.04); C286(0.49) | LDD2146 | [8] |
| LDCM0553 | Nucleophilic fragment 6b | MDA-MB-231 | C286(4.25); C766(1.64) | LDD2147 | [8] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C767(1.07) | LDD2148 | [8] |
| LDCM0555 | Nucleophilic fragment 7b | MDA-MB-231 | C421(0.67); C286(0.61); C816(1.05) | LDD2149 | [8] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C421(0.49); C286(0.53); C596(0.42) | LDD2150 | [8] |
| LDCM0557 | Nucleophilic fragment 8b | MDA-MB-231 | C286(0.84) | LDD2151 | [8] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C286(4.48); C596(1.42) | LDD2153 | [8] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C439(1.09); C421(0.70) | LDD2206 | [40] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C1229(0.86); C596(0.85); C421(0.76); C767(0.53) | LDD2207 | [40] |
| LDCM0003 | Sulforaphane | MCF-7 | 3.60 | LDD0042 | [9] |
| LDCM0021 | THZ1 | HCT 116 | C767(1.06); C439(1.14); C766(0.96); C596(1.02) | LDD2173 | [14] |
| LDCM0112 | W16 | Hep-G2 | C767(2.25) | LDD0239 | [29] |
The Interaction Atlas With This Target
References
