Details of the Target
General Information of Target
| Target ID | LDTP06268 | |||||
|---|---|---|---|---|---|---|
| Target Name | Condensin complex subunit 2 (NCAPH) | |||||
| Gene Name | NCAPH | |||||
| Gene ID | 23397 | |||||
| Synonyms |
BRRN; BRRN1; CAPH; KIAA0074; Condensin complex subunit 2; Barren homolog protein 1; Chromosome-associated protein H; hCAP-H; Non-SMC condensin I complex subunit H; XCAP-H homolog |
|||||
| 3D Structure | ||||||
| Sequence |
MGPPGPALPATMNNSSSETRGHPHSASSPSERVFPMPLPRKAPLNIPGTPVLEDFPQNDD
EKERLQRRRSRVFDLQFSTDSPRLLASPSSRSIDISATIPKFTNTQITEHYSTCIKLSTE NKITTKNAFGLHLIDFMSEILKQKDTEPTNFKVAAGTLDASTKIYAVRVDAVHADVYRVL GGLGKDAPSLEEVEGHVADGSATEMGTTKKAVKPKKKHLHRTIEQNINNLNVSEADRKCE IDPMFQKTAASFDECSTAGVFLSTLHCQDYRSELLFPSDVQTLSTGEPLELPELGCVEMT DLKAPLQQCAEDRQICPSLAGFQFTQWDSETHNESVSALVDKFKKNDQVFDINAEVDESD CGDFPDGSLGDDFDANDEPDHTAVGDHEEFRSWKEPCQVQSCQEEMISLGDGDIRTMCPL LSMKPGEYSYFSPRTMSMWAGPDHWRFRPRRKQDAPSQSENKKKSTKKDFEIDFEDDIDF DVYFRKTKAATILTKSTLENQNWRATTLPTDFNYNVDTLVQLHLKPGTRLLKMAQGHRVE TEHYEEIEDYDYNNPNDTSNFCPGLQAADSDDEDLDDLFVGPVGNSDLSPYPCHPPKTAQ QNGDTPEAQGLDITTYGESNLVAEPQKVNKIEIHYAKTAKKMDMKKLKQSMWSLLTALSG KEADAEANHREAGKEAALAEVADEKMLSGLTKDLQRSLPPVMAQNLSIPLAFACLLHLAN EKNLKLEGTEDLSDVLVRQGD |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
CND2 (condensin subunit 2) family
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Regulatory subunit of the condensin complex, a complex required for conversion of interphase chromatin into mitotic-like condense chromosomes. The condensin complex probably introduces positive supercoils into relaxed DNA in the presence of type I topoisomerases and converts nicked DNA into positive knotted forms in the presence of type II topoisomerases. Early in neurogenesis, may play an essential role to ensure accurate mitotic chromosome condensation in neuron stem cells, ultimately affecting neuron pool and cortex size.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| JHH4 | SNV: p.M642V | DBIA Probe Info | |||
| MOLT4 | SNV: p.A166V | IA-alkyne Probe Info | |||
| NCIH1155 | SNV: p.A249T | DBIA Probe Info | |||
| NCIH23 | SNV: p.N14K | DBIA Probe Info | |||
| OVK18 | SNV: p.H196Q | DBIA Probe Info | |||
| RL952 | Insertion: p.H24PfsTer36 | DBIA Probe Info | |||
| SW1271 | SNV: p.C114Y | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [2] | |
|
TH211 Probe Info |
 |
Y111(20.00); Y635(8.53); Y428(8.51) | LDD0257 | [3] | |
|
TH216 Probe Info |
 |
Y635(12.89) | LDD0259 | [3] | |
|
STPyne Probe Info |
 |
K495(10.00); K532(10.00); K630(1.15); K648(6.15) | LDD0277 | [4] | |
|
BTD Probe Info |
 |
C309(0.52) | LDD2089 | [5] | |
|
Sulforaphane-probe2 Probe Info |
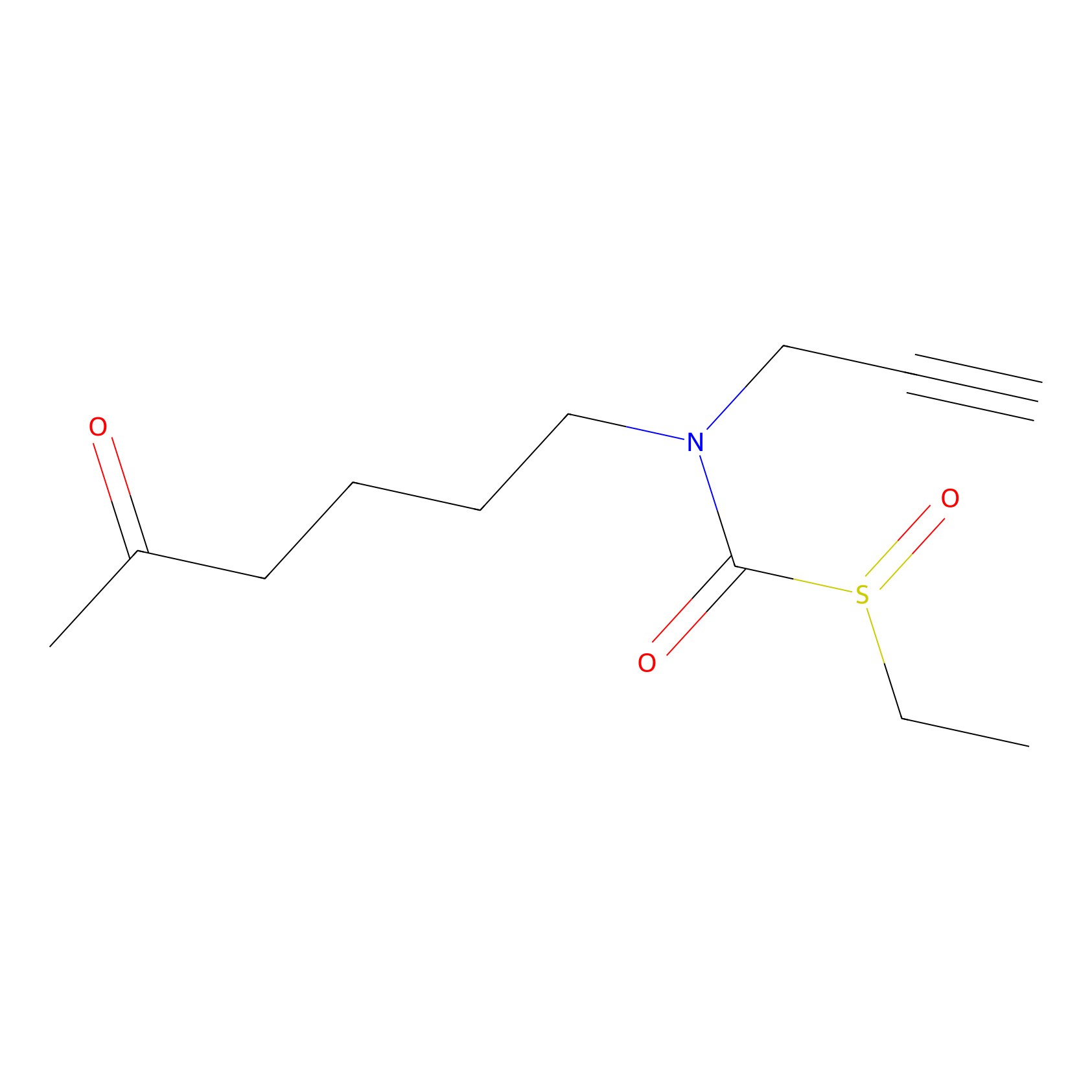 |
2.02 | LDD0160 | [6] | |
|
DA-P3 Probe Info |
 |
4.11 | LDD0179 | [7] | |
|
AHL-Pu-1 Probe Info |
 |
C397(2.21); C296(5.82) | LDD0168 | [8] | |
|
HHS-482 Probe Info |
 |
Y635(0.97) | LDD0285 | [9] | |
|
HHS-475 Probe Info |
 |
Y430(0.86); Y428(0.88); Y635(0.88) | LDD0264 | [10] | |
|
HHS-465 Probe Info |
 |
Y635(8.38) | LDD2237 | [11] | |
|
DBIA Probe Info |
 |
C397(1.02); C402(1.02) | LDD0078 | [12] | |
|
5E-2FA Probe Info |
 |
H669(0.00); H110(0.00); H24(0.00); H173(0.00) | LDD2235 | [13] | |
|
ATP probe Probe Info |
 |
K116(0.00); K637(0.00); K640(0.00); K692(0.00) | LDD0199 | [14] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C309(0.00); C114(0.00); C296(0.00); C714(0.00) | LDD0038 | [15] | |
|
IA-alkyne Probe Info |
 |
C309(0.00); C239(0.00); C114(0.00); C316(0.00) | LDD0036 | [15] | |
|
Lodoacetamide azide Probe Info |
 |
C309(0.00); C114(0.00); C316(0.00); C296(0.00) | LDD0037 | [15] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [16] | |
|
NAIA_4 Probe Info |
 |
C114(0.00); C239(0.00); C255(0.00); C296(0.00) | LDD2226 | [17] | |
|
TFBX Probe Info |
 |
C114(0.00); C418(0.00) | LDD0027 | [16] | |
|
WYneN Probe Info |
 |
C239(0.00); C309(0.00) | LDD0021 | [18] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [18] | |
|
1d-yne Probe Info |
 |
K41(0.00); K692(0.00); K394(0.00) | LDD0356 | [19] | |
|
Compound 10 Probe Info |
 |
C114(0.00); C239(0.00); C255(0.00); C418(0.00) | LDD2216 | [20] | |
|
Compound 11 Probe Info |
 |
C239(0.00); C418(0.00) | LDD2213 | [20] | |
|
IPM Probe Info |
 |
C418(0.00); C239(0.00) | LDD0005 | [18] | |
|
NPM Probe Info |
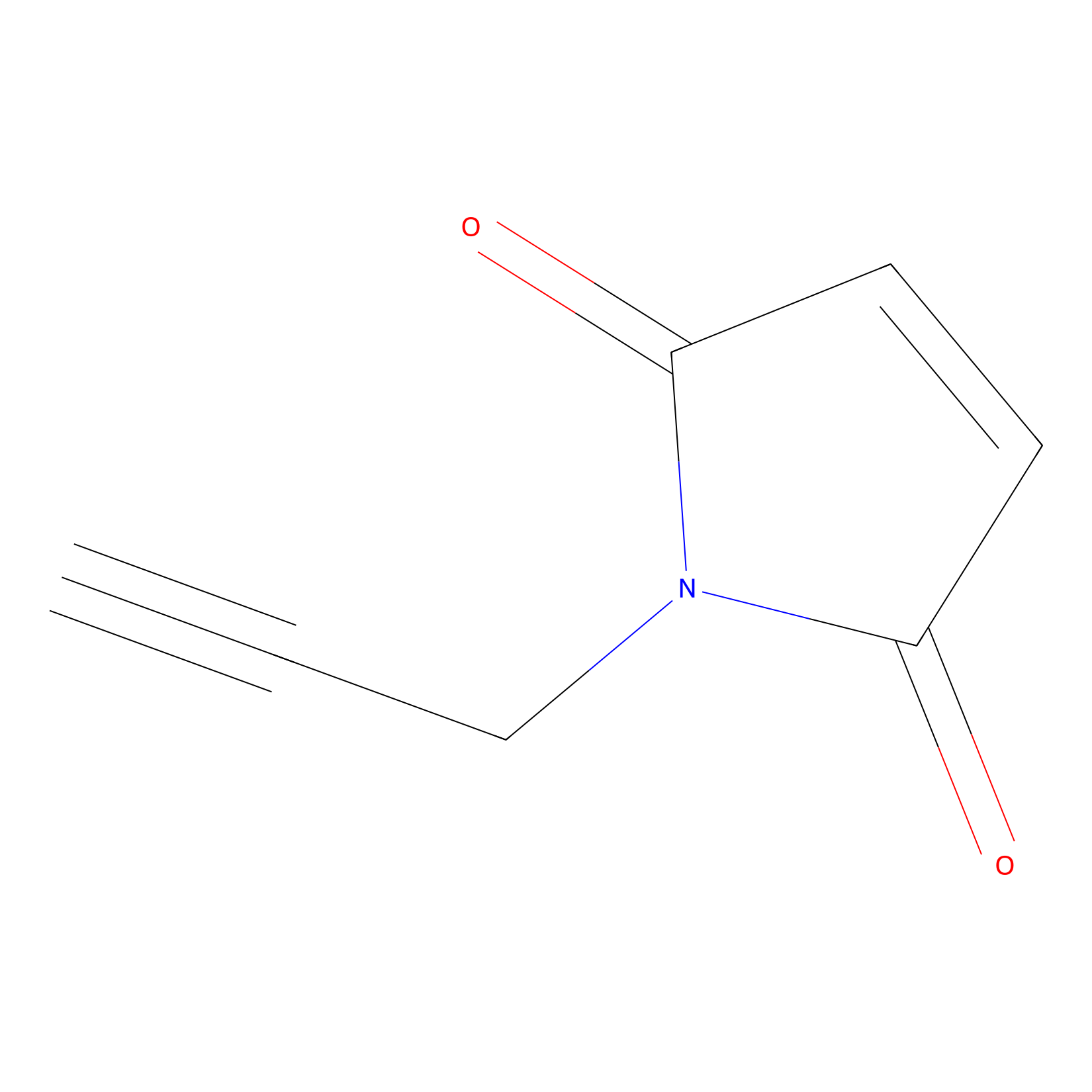 |
N.A. | LDD0016 | [18] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [21] | |
|
VSF Probe Info |
 |
C296(0.00); C239(0.00); C397(0.00) | LDD0007 | [18] | |
|
Phosphinate-6 Probe Info |
 |
C418(0.00); C239(0.00); C402(0.00); C255(0.00) | LDD0018 | [22] | |
|
1c-yne Probe Info |
 |
K495(0.00); K725(0.00); K424(0.00); K661(0.00) | LDD0228 | [19] | |
|
Acrolein Probe Info |
 |
C114(0.00); C309(0.00) | LDD0217 | [23] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [23] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [24] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [25] | |
|
NAIA_5 Probe Info |
 |
C239(0.00); C714(0.00); C114(0.00); C418(0.00) | LDD2223 | [17] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C198 Probe Info |
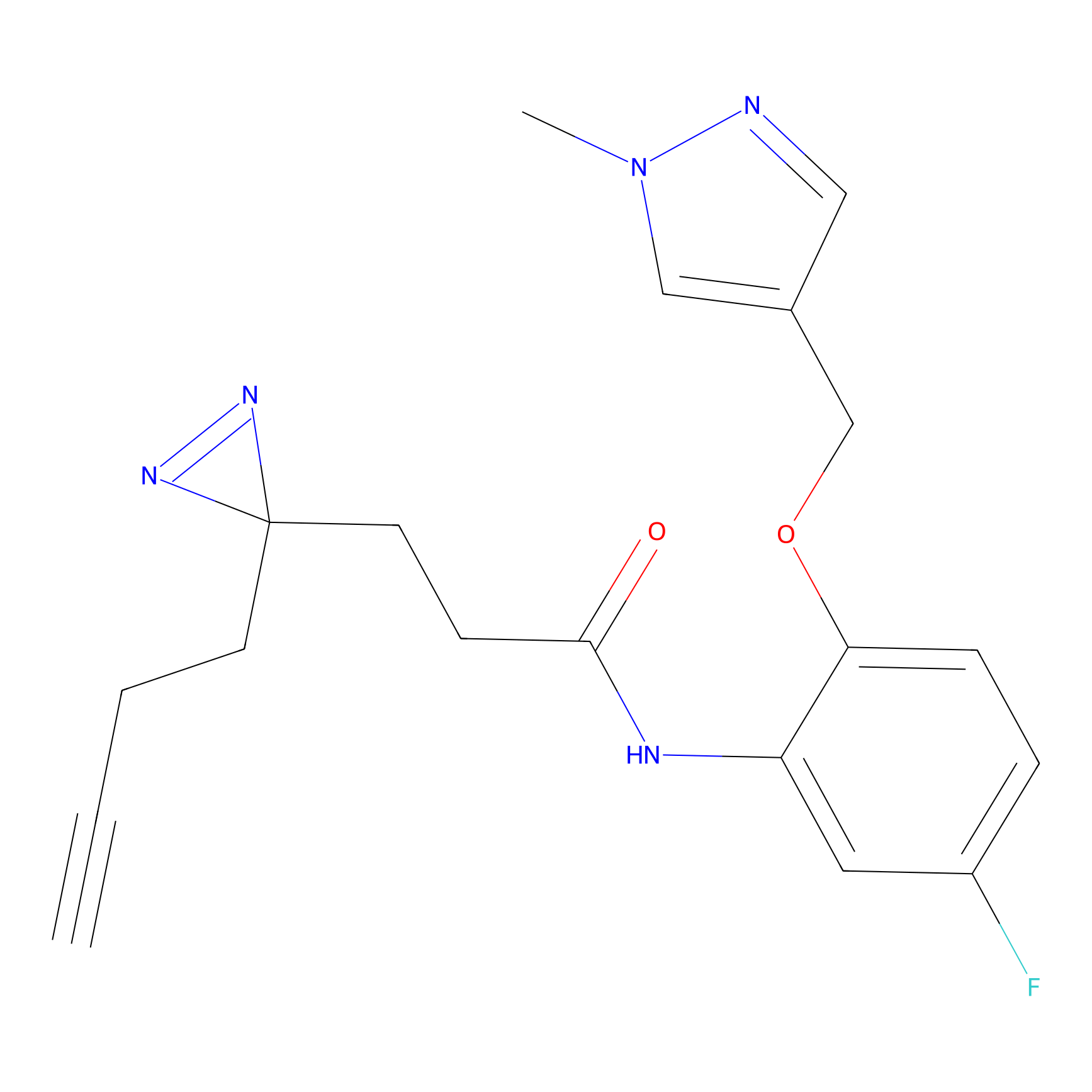 |
11.96 | LDD1874 | [26] | |
|
C264 Probe Info |
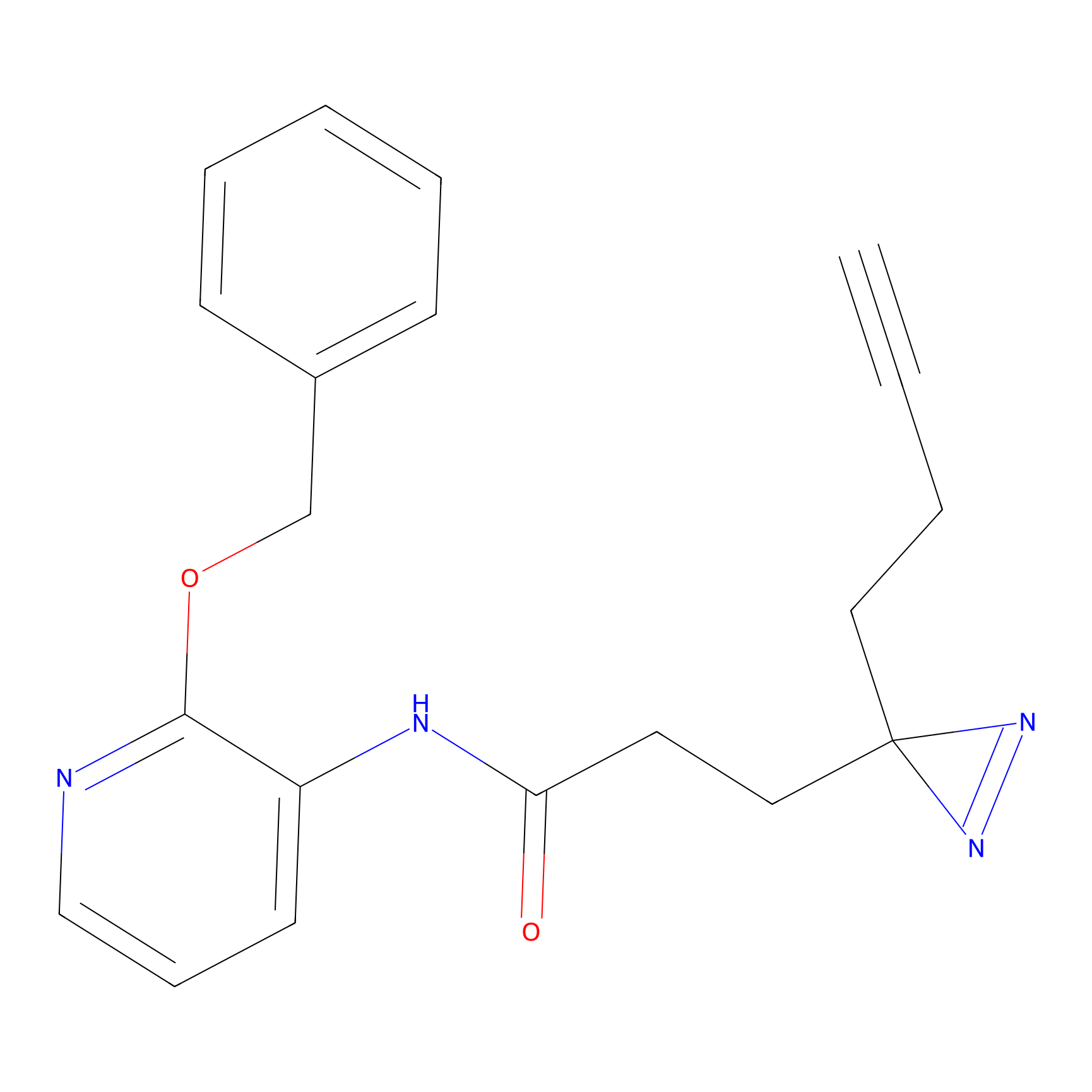 |
16.56 | LDD1935 | [26] | |
|
FFF probe13 Probe Info |
 |
6.53 | LDD0475 | [27] | |
|
FFF probe14 Probe Info |
 |
12.95 | LDD0477 | [27] | |
|
FFF probe2 Probe Info |
 |
6.42 | LDD0463 | [27] | |
|
FFF probe3 Probe Info |
 |
20.00 | LDD0464 | [27] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C418(0.95) | LDD2112 | [5] |
| LDCM0537 | 2-Cyano-N,N-dimethylacetamide | MDA-MB-231 | C239(0.95) | LDD2130 | [5] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C309(1.18) | LDD2117 | [5] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C309(1.81) | LDD2152 | [5] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C418(0.99) | LDD2103 | [5] |
| LDCM0025 | 4SU-RNA | HEK-293T | C397(2.21); C296(5.82) | LDD0168 | [8] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C402(4.55); C397(4.89); C296(3.29) | LDD0169 | [8] |
| LDCM0214 | AC1 | HCT 116 | C418(0.99); C239(1.04); C309(0.34); C397(1.05) | LDD0531 | [12] |
| LDCM0215 | AC10 | HCT 116 | C418(1.12); C239(0.92); C309(0.76); C397(1.26) | LDD0532 | [12] |
| LDCM0216 | AC100 | HCT 116 | C418(1.07); C239(1.33); C255(1.08); C267(1.08) | LDD0533 | [12] |
| LDCM0217 | AC101 | HCT 116 | C418(1.01); C239(1.31); C255(1.05); C267(1.05) | LDD0534 | [12] |
| LDCM0218 | AC102 | HCT 116 | C418(1.03); C239(1.18); C255(1.13); C267(1.13) | LDD0535 | [12] |
| LDCM0219 | AC103 | HCT 116 | C418(1.15); C239(1.35); C255(0.70); C267(0.70) | LDD0536 | [12] |
| LDCM0220 | AC104 | HCT 116 | C418(1.00); C239(1.11); C255(1.01); C267(1.01) | LDD0537 | [12] |
| LDCM0221 | AC105 | HCT 116 | C418(1.01); C239(1.35); C255(1.19); C267(1.19) | LDD0538 | [12] |
| LDCM0222 | AC106 | HCT 116 | C418(1.17); C239(1.52); C255(0.57); C267(0.57) | LDD0539 | [12] |
| LDCM0223 | AC107 | HCT 116 | C418(0.96); C239(1.36); C255(0.79); C267(0.79) | LDD0540 | [12] |
| LDCM0224 | AC108 | HCT 116 | C418(1.05); C239(1.27); C255(1.09); C267(1.09) | LDD0541 | [12] |
| LDCM0225 | AC109 | HCT 116 | C418(0.95); C239(1.05); C255(0.88); C267(0.88) | LDD0542 | [12] |
| LDCM0226 | AC11 | HCT 116 | C418(1.07); C239(0.91); C309(0.76); C397(1.39) | LDD0543 | [12] |
| LDCM0227 | AC110 | HCT 116 | C418(1.01); C239(1.43); C255(0.76); C267(0.76) | LDD0544 | [12] |
| LDCM0228 | AC111 | HCT 116 | C418(1.04); C239(1.39); C255(1.19); C267(1.19) | LDD0545 | [12] |
| LDCM0229 | AC112 | HCT 116 | C418(1.00); C239(1.39); C255(0.50); C267(0.50) | LDD0546 | [12] |
| LDCM0230 | AC113 | HCT 116 | C418(0.99); C239(1.01); C255(0.57); C267(0.57) | LDD0547 | [12] |
| LDCM0231 | AC114 | HCT 116 | C418(1.02); C239(0.98); C255(0.50); C267(0.50) | LDD0548 | [12] |
| LDCM0232 | AC115 | HCT 116 | C418(1.02); C239(1.03); C255(0.50); C267(0.50) | LDD0549 | [12] |
| LDCM0233 | AC116 | HCT 116 | C418(1.03); C239(1.11); C255(0.52); C267(0.52) | LDD0550 | [12] |
| LDCM0234 | AC117 | HCT 116 | C418(1.06); C239(1.11); C255(0.46); C267(0.46) | LDD0551 | [12] |
| LDCM0235 | AC118 | HCT 116 | C418(0.96); C239(0.98); C255(0.56); C267(0.56) | LDD0552 | [12] |
| LDCM0236 | AC119 | HCT 116 | C418(1.08); C239(1.26); C255(0.54); C267(0.54) | LDD0553 | [12] |
| LDCM0237 | AC12 | HCT 116 | C418(1.06); C239(0.85); C309(0.82); C397(1.08) | LDD0554 | [12] |
| LDCM0238 | AC120 | HCT 116 | C418(1.01); C239(1.07); C255(0.48); C267(0.48) | LDD0555 | [12] |
| LDCM0239 | AC121 | HCT 116 | C418(0.85); C239(0.81); C255(0.50); C267(0.50) | LDD0556 | [12] |
| LDCM0240 | AC122 | HCT 116 | C418(1.09); C239(1.06); C255(0.57); C267(0.57) | LDD0557 | [12] |
| LDCM0241 | AC123 | HCT 116 | C418(0.96); C239(0.81); C255(0.70); C267(0.70) | LDD0558 | [12] |
| LDCM0242 | AC124 | HCT 116 | C418(0.89); C239(0.85); C255(0.49); C267(0.49) | LDD0559 | [12] |
| LDCM0243 | AC125 | HCT 116 | C418(0.90); C239(0.91); C255(0.58); C267(0.58) | LDD0560 | [12] |
| LDCM0244 | AC126 | HCT 116 | C418(0.88); C239(0.82); C255(0.57); C267(0.57) | LDD0561 | [12] |
| LDCM0245 | AC127 | HCT 116 | C418(1.02); C239(0.92); C255(0.65); C267(0.65) | LDD0562 | [12] |
| LDCM0246 | AC128 | HCT 116 | C239(0.66); C255(1.29); C267(1.29); C309(0.57) | LDD0563 | [12] |
| LDCM0247 | AC129 | HCT 116 | C239(0.74); C255(1.26); C267(1.26); C309(1.12) | LDD0564 | [12] |
| LDCM0249 | AC130 | HCT 116 | C239(0.82); C255(1.33); C267(1.33); C309(0.71) | LDD0566 | [12] |
| LDCM0250 | AC131 | HCT 116 | C239(0.67); C255(1.48); C267(1.48); C309(0.78) | LDD0567 | [12] |
| LDCM0251 | AC132 | HCT 116 | C239(0.82); C255(1.32); C267(1.32); C309(0.56) | LDD0568 | [12] |
| LDCM0252 | AC133 | HCT 116 | C239(0.79); C255(1.29); C267(1.29); C309(0.67) | LDD0569 | [12] |
| LDCM0253 | AC134 | HCT 116 | C239(0.79); C255(1.30); C267(1.30); C309(0.63) | LDD0570 | [12] |
| LDCM0254 | AC135 | HCT 116 | C239(0.77); C255(1.32); C267(1.32); C309(0.64) | LDD0571 | [12] |
| LDCM0255 | AC136 | HCT 116 | C239(0.80); C255(1.25); C267(1.25); C309(0.46) | LDD0572 | [12] |
| LDCM0256 | AC137 | HCT 116 | C239(0.79); C255(1.12); C267(1.12); C309(0.54) | LDD0573 | [12] |
| LDCM0257 | AC138 | HCT 116 | C239(0.79); C255(1.33); C267(1.33); C309(0.62) | LDD0574 | [12] |
| LDCM0258 | AC139 | HCT 116 | C239(0.95); C255(1.31); C267(1.31); C309(0.75) | LDD0575 | [12] |
| LDCM0259 | AC14 | HCT 116 | C418(0.90); C239(0.81); C309(0.87); C397(1.24) | LDD0576 | [12] |
| LDCM0260 | AC140 | HCT 116 | C239(0.93); C255(1.29); C267(1.29); C309(0.63) | LDD0577 | [12] |
| LDCM0261 | AC141 | HCT 116 | C239(0.85); C255(1.33); C267(1.33); C309(0.66) | LDD0578 | [12] |
| LDCM0262 | AC142 | HCT 116 | C239(0.82); C255(1.33); C267(1.33); C309(0.49) | LDD0579 | [12] |
| LDCM0263 | AC143 | HCT 116 | C418(1.22); C309(0.76); C397(0.99); C402(0.99) | LDD0580 | [12] |
| LDCM0264 | AC144 | HCT 116 | C309(0.62); C418(0.80); C239(1.01); C397(1.09) | LDD0581 | [12] |
| LDCM0265 | AC145 | HCT 116 | C418(0.89); C309(1.07); C239(1.18); C397(1.24) | LDD0582 | [12] |
| LDCM0266 | AC146 | HCT 116 | C309(0.69); C418(0.89); C239(1.19); C397(1.33) | LDD0583 | [12] |
| LDCM0267 | AC147 | HCT 116 | C309(0.74); C418(0.84); C239(1.02); C397(1.25) | LDD0584 | [12] |
| LDCM0268 | AC148 | HCT 116 | C418(0.84); C309(1.14); C397(1.52); C402(1.54) | LDD0585 | [12] |
| LDCM0269 | AC149 | HCT 116 | C309(0.71); C418(0.95); C239(1.37); C397(1.48) | LDD0586 | [12] |
| LDCM0270 | AC15 | HCT 116 | C418(0.88); C309(0.89); C239(0.90); C397(1.23) | LDD0587 | [12] |
| LDCM0271 | AC150 | HCT 116 | C309(0.90); C418(0.96); C239(1.16); C397(1.28) | LDD0588 | [12] |
| LDCM0272 | AC151 | HCT 116 | C309(0.60); C397(0.97); C418(1.00); C402(1.05) | LDD0589 | [12] |
| LDCM0273 | AC152 | HCT 116 | C418(0.76); C309(0.94); C239(1.15); C397(1.45) | LDD0590 | [12] |
| LDCM0274 | AC153 | HCT 116 | C418(0.70); C309(1.10); C397(1.55); C402(1.58) | LDD0591 | [12] |
| LDCM0621 | AC154 | HCT 116 | C418(0.72); C239(1.25); C309(0.69); C397(1.15) | LDD2158 | [12] |
| LDCM0622 | AC155 | HCT 116 | C418(0.85); C239(1.30); C309(0.83); C397(1.25) | LDD2159 | [12] |
| LDCM0623 | AC156 | HCT 116 | C418(0.81); C239(1.28); C309(1.24); C397(1.28) | LDD2160 | [12] |
| LDCM0624 | AC157 | HCT 116 | C418(0.73); C239(1.18); C309(0.79); C397(1.33) | LDD2161 | [12] |
| LDCM0276 | AC17 | HCT 116 | C309(0.68); C418(0.90); C402(1.02); C397(1.02) | LDD0593 | [12] |
| LDCM0277 | AC18 | HCT 116 | C309(0.67); C418(1.07); C397(1.25); C402(1.25) | LDD0594 | [12] |
| LDCM0278 | AC19 | HCT 116 | C309(0.78); C418(0.94); C397(1.08); C402(1.10) | LDD0595 | [12] |
| LDCM0279 | AC2 | HCT 116 | C309(0.76); C397(0.95); C402(0.95); C418(1.01) | LDD0596 | [12] |
| LDCM0280 | AC20 | HCT 116 | C309(0.67); C418(0.89); C397(1.04); C239(1.08) | LDD0597 | [12] |
| LDCM0281 | AC21 | HCT 116 | C309(0.62); C418(0.89); C239(1.24); C397(1.28) | LDD0598 | [12] |
| LDCM0282 | AC22 | HCT 116 | C309(0.73); C418(0.90); C397(0.99); C402(1.01) | LDD0599 | [12] |
| LDCM0283 | AC23 | HCT 116 | C309(0.60); C418(0.99); C239(1.01); C397(1.31) | LDD0600 | [12] |
| LDCM0284 | AC24 | HCT 116 | C309(0.63); C418(0.74); C239(1.08); C397(1.13) | LDD0601 | [12] |
| LDCM0285 | AC25 | HCT 116 | C418(0.96); C402(1.06); C397(1.14); C239(1.17) | LDD0602 | [12] |
| LDCM0286 | AC26 | HCT 116 | C309(0.93); C418(1.00); C397(1.08); C402(1.11) | LDD0603 | [12] |
| LDCM0287 | AC27 | HCT 116 | C309(0.92); C418(0.97); C402(1.08); C397(1.12) | LDD0604 | [12] |
| LDCM0288 | AC28 | HCT 116 | C309(0.91); C418(0.98); C397(1.16); C402(1.18) | LDD0605 | [12] |
| LDCM0289 | AC29 | HCT 116 | C309(0.78); C418(1.03); C402(1.24); C397(1.33) | LDD0606 | [12] |
| LDCM0290 | AC3 | HCT 116 | C309(0.53); C418(0.94); C397(1.01); C402(1.01) | LDD0607 | [12] |
| LDCM0291 | AC30 | HCT 116 | C309(0.81); C402(1.08); C418(1.08); C397(1.11) | LDD0608 | [12] |
| LDCM0292 | AC31 | HCT 116 | C309(0.83); C418(0.88); C402(1.01); C397(1.04) | LDD0609 | [12] |
| LDCM0293 | AC32 | HCT 116 | C309(0.59); C418(1.00); C397(1.33); C402(1.37) | LDD0610 | [12] |
| LDCM0294 | AC33 | HCT 116 | C309(0.66); C418(0.94); C402(1.25); C397(1.26) | LDD0611 | [12] |
| LDCM0295 | AC34 | HCT 116 | C309(0.63); C418(1.05); C402(1.26); C397(1.44) | LDD0612 | [12] |
| LDCM0296 | AC35 | HCT 116 | C255(0.96); C267(0.96); C239(0.97); C397(1.03) | LDD0613 | [12] |
| LDCM0297 | AC36 | HCT 116 | C397(0.91); C402(0.91); C309(1.05); C239(1.06) | LDD0614 | [12] |
| LDCM0298 | AC37 | HCT 116 | C397(0.84); C402(0.84); C309(0.96); C239(1.02) | LDD0615 | [12] |
| LDCM0299 | AC38 | HCT 116 | C309(0.91); C255(1.06); C267(1.06); C397(1.07) | LDD0616 | [12] |
| LDCM0300 | AC39 | HCT 116 | C255(0.81); C267(0.81); C397(0.97); C402(0.97) | LDD0617 | [12] |
| LDCM0301 | AC4 | HCT 116 | C309(0.39); C418(0.85); C397(1.06); C402(1.06) | LDD0618 | [12] |
| LDCM0302 | AC40 | HCT 116 | C309(0.99); C397(0.99); C402(0.99); C255(1.00) | LDD0619 | [12] |
| LDCM0303 | AC41 | HCT 116 | C397(0.94); C402(0.94); C255(0.94); C267(0.94) | LDD0620 | [12] |
| LDCM0304 | AC42 | HCT 116 | C255(0.82); C267(0.82); C397(0.94); C402(0.94) | LDD0621 | [12] |
| LDCM0305 | AC43 | HCT 116 | C255(0.86); C267(0.86); C309(0.87); C397(1.04) | LDD0622 | [12] |
| LDCM0306 | AC44 | HCT 116 | C255(0.87); C267(0.87); C397(0.96); C402(0.96) | LDD0623 | [12] |
| LDCM0307 | AC45 | HCT 116 | C255(0.87); C267(0.87); C239(0.97); C397(1.07) | LDD0624 | [12] |
| LDCM0308 | AC46 | HCT 116 | C309(0.82); C397(0.96); C402(0.96); C418(1.19) | LDD0625 | [12] |
| LDCM0309 | AC47 | HCT 116 | C397(1.00); C402(1.00); C239(1.07); C418(1.09) | LDD0626 | [12] |
| LDCM0310 | AC48 | HCT 116 | C309(0.79); C397(0.99); C402(0.99); C418(1.02) | LDD0627 | [12] |
| LDCM0311 | AC49 | HCT 116 | C309(0.96); C397(1.20); C402(1.20); C239(1.27) | LDD0628 | [12] |
| LDCM0312 | AC5 | HCT 116 | C309(0.38); C418(0.87); C397(0.87); C402(0.87) | LDD0629 | [12] |
| LDCM0313 | AC50 | HCT 116 | C309(0.84); C418(1.18); C397(1.24); C402(1.24) | LDD0630 | [12] |
| LDCM0314 | AC51 | HCT 116 | C309(0.93); C397(1.05); C402(1.05); C239(1.18) | LDD0631 | [12] |
| LDCM0315 | AC52 | HCT 116 | C309(0.82); C397(0.98); C402(0.98); C239(1.10) | LDD0632 | [12] |
| LDCM0316 | AC53 | HCT 116 | C309(0.66); C239(1.02); C397(1.18); C402(1.18) | LDD0633 | [12] |
| LDCM0317 | AC54 | HCT 116 | C309(0.68); C418(1.07); C397(1.13); C402(1.13) | LDD0634 | [12] |
| LDCM0318 | AC55 | HCT 116 | C309(0.90); C239(1.08); C418(1.26); C397(1.41) | LDD0635 | [12] |
| LDCM0319 | AC56 | HCT 116 | C309(0.94); C239(1.19); C418(1.31); C397(1.70) | LDD0636 | [12] |
| LDCM0320 | AC57 | HCT 116 | C309(0.71); C418(1.01); C397(1.02); C402(1.02) | LDD0637 | [12] |
| LDCM0321 | AC58 | HCT 116 | C309(0.67); C418(1.03); C397(1.18); C402(1.18) | LDD0638 | [12] |
| LDCM0322 | AC59 | HCT 116 | C309(0.66); C418(1.00); C397(1.30); C402(1.30) | LDD0639 | [12] |
| LDCM0323 | AC6 | HCT 116 | C309(0.78); C418(1.04); C239(1.08); C397(1.26) | LDD0640 | [12] |
| LDCM0324 | AC60 | HCT 116 | C309(0.77); C397(1.10); C402(1.10); C418(1.17) | LDD0641 | [12] |
| LDCM0325 | AC61 | HCT 116 | C309(0.53); C397(0.99); C402(0.99); C418(1.02) | LDD0642 | [12] |
| LDCM0326 | AC62 | HCT 116 | C309(0.81); C397(1.20); C402(1.20); C418(1.28) | LDD0643 | [12] |
| LDCM0327 | AC63 | HCT 116 | C309(0.67); C397(1.10); C402(1.10); C418(1.40) | LDD0644 | [12] |
| LDCM0328 | AC64 | HCT 116 | C309(0.68); C418(1.15); C397(1.27); C402(1.27) | LDD0645 | [12] |
| LDCM0329 | AC65 | HCT 116 | C309(0.67); C397(1.09); C402(1.09); C418(1.12) | LDD0646 | [12] |
| LDCM0330 | AC66 | HCT 116 | C309(0.59); C397(0.97); C402(0.97); C418(1.14) | LDD0647 | [12] |
| LDCM0331 | AC67 | HCT 116 | C309(0.75); C418(1.11); C397(1.15); C402(1.15) | LDD0648 | [12] |
| LDCM0332 | AC68 | HCT 116 | C418(0.82); C239(0.88); C397(0.95); C402(0.95) | LDD0649 | [12] |
| LDCM0333 | AC69 | HCT 116 | C239(0.83); C418(0.87); C309(0.90); C397(0.92) | LDD0650 | [12] |
| LDCM0334 | AC7 | HCT 116 | C309(0.80); C239(0.93); C397(1.01); C402(1.01) | LDD0651 | [12] |
| LDCM0335 | AC70 | HCT 116 | C309(0.76); C397(0.83); C402(0.83); C239(0.92) | LDD0652 | [12] |
| LDCM0336 | AC71 | HCT 116 | C397(0.79); C402(0.79); C418(0.82); C309(0.82) | LDD0653 | [12] |
| LDCM0337 | AC72 | HCT 116 | C397(0.83); C402(0.83); C309(0.90); C239(0.99) | LDD0654 | [12] |
| LDCM0338 | AC73 | HCT 116 | C397(1.02); C402(1.02); C418(1.11); C309(1.14) | LDD0655 | [12] |
| LDCM0339 | AC74 | HCT 116 | C418(0.89); C239(0.92); C397(0.98); C402(0.98) | LDD0656 | [12] |
| LDCM0340 | AC75 | HCT 116 | C397(1.05); C402(1.05); C239(1.07); C418(1.08) | LDD0657 | [12] |
| LDCM0341 | AC76 | HCT 116 | C397(0.89); C402(0.89); C309(0.90); C418(0.96) | LDD0658 | [12] |
| LDCM0342 | AC77 | HCT 116 | C309(0.90); C418(0.93); C397(0.93); C402(0.93) | LDD0659 | [12] |
| LDCM0343 | AC78 | HCT 116 | C309(0.84); C418(0.93); C397(0.95); C402(0.95) | LDD0660 | [12] |
| LDCM0344 | AC79 | HCT 116 | C397(0.89); C402(0.89); C309(0.95); C418(0.96) | LDD0661 | [12] |
| LDCM0345 | AC8 | HCT 116 | C309(0.67); C239(0.97); C418(1.10); C397(1.10) | LDD0662 | [12] |
| LDCM0346 | AC80 | HCT 116 | C418(0.89); C397(0.94); C402(0.94); C309(1.20) | LDD0663 | [12] |
| LDCM0347 | AC81 | HCT 116 | C397(0.88); C402(0.88); C418(0.89); C239(0.91) | LDD0664 | [12] |
| LDCM0348 | AC82 | HCT 116 | C418(0.93); C239(1.02); C309(1.06); C397(1.09) | LDD0665 | [12] |
| LDCM0349 | AC83 | HCT 116 | C309(0.74); C239(1.17); C418(1.35); C397(1.55) | LDD0666 | [12] |
| LDCM0350 | AC84 | HCT 116 | C309(1.06); C239(1.11); C397(1.33); C402(1.34) | LDD0667 | [12] |
| LDCM0351 | AC85 | HCT 116 | C309(0.88); C239(1.16); C402(1.35); C418(1.39) | LDD0668 | [12] |
| LDCM0352 | AC86 | HCT 116 | C309(1.08); C239(1.13); C418(1.23); C402(1.25) | LDD0669 | [12] |
| LDCM0353 | AC87 | HCT 116 | C309(0.66); C418(0.98); C239(1.01); C397(1.10) | LDD0670 | [12] |
| LDCM0354 | AC88 | HCT 116 | C418(1.05); C239(1.13); C397(1.19); C402(1.27) | LDD0671 | [12] |
| LDCM0355 | AC89 | HCT 116 | C309(1.05); C418(1.15); C239(1.23); C402(1.36) | LDD0672 | [12] |
| LDCM0357 | AC90 | HCT 116 | C309(0.99); C402(1.04); C397(1.07); C418(1.09) | LDD0674 | [12] |
| LDCM0358 | AC91 | HCT 116 | C309(0.65); C239(1.15); C418(1.43); C397(1.50) | LDD0675 | [12] |
| LDCM0359 | AC92 | HCT 116 | C309(0.82); C239(1.14); C418(1.33); C397(1.49) | LDD0676 | [12] |
| LDCM0360 | AC93 | HCT 116 | C309(0.87); C239(1.12); C397(1.28); C402(1.36) | LDD0677 | [12] |
| LDCM0361 | AC94 | HCT 116 | C309(0.80); C397(1.28); C239(1.29); C402(1.37) | LDD0678 | [12] |
| LDCM0362 | AC95 | HCT 116 | C309(1.00); C239(1.09); C397(1.16); C402(1.19) | LDD0679 | [12] |
| LDCM0363 | AC96 | HCT 116 | C309(0.77); C239(1.11); C397(1.32); C402(1.35) | LDD0680 | [12] |
| LDCM0364 | AC97 | HCT 116 | C309(0.93); C239(1.38); C402(1.52); C397(1.57) | LDD0681 | [12] |
| LDCM0365 | AC98 | HCT 116 | C255(1.05); C267(1.05); C397(1.18); C402(1.18) | LDD0682 | [12] |
| LDCM0366 | AC99 | HCT 116 | C397(0.81); C402(0.81); C255(1.03); C267(1.03) | LDD0683 | [12] |
| LDCM0248 | AKOS034007472 | HCT 116 | C418(0.98); C239(0.99); C309(0.74); C397(1.02) | LDD0565 | [12] |
| LDCM0356 | AKOS034007680 | HCT 116 | C309(0.74); C239(0.92); C418(1.00); C397(1.14) | LDD0673 | [12] |
| LDCM0275 | AKOS034007705 | HCT 116 | C309(0.68); C239(1.02); C418(1.04); C397(1.47) | LDD0592 | [12] |
| LDCM0156 | Aniline | NCI-H1299 | 11.39 | LDD0403 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C397(1.02); C402(1.02) | LDD0078 | [12] |
| LDCM0087 | Capsaicin | HEK-293T | 11.52 | LDD0185 | [7] |
| LDCM0108 | Chloroacetamide | HeLa | C309(0.00); H24(0.00) | LDD0222 | [23] |
| LDCM0632 | CL-Sc | Hep-G2 | C418(20.00); C114(1.48) | LDD2227 | [17] |
| LDCM0367 | CL1 | HCT 116 | C309(0.88); C397(0.90); C402(0.90); C239(0.94) | LDD0684 | [12] |
| LDCM0368 | CL10 | HCT 116 | C309(0.79); C418(1.08); C397(1.16); C402(1.16) | LDD0685 | [12] |
| LDCM0369 | CL100 | HCT 116 | C309(0.52); C397(0.95); C402(0.95); C418(0.98) | LDD0686 | [12] |
| LDCM0370 | CL101 | HCT 116 | C309(0.88); C239(0.94); C418(1.00); C397(1.35) | LDD0687 | [12] |
| LDCM0371 | CL102 | HCT 116 | C239(0.75); C309(0.84); C397(1.11); C402(1.11) | LDD0688 | [12] |
| LDCM0372 | CL103 | HCT 116 | C309(0.70); C239(0.80); C418(0.96); C397(1.19) | LDD0689 | [12] |
| LDCM0373 | CL104 | HCT 116 | C239(0.87); C418(1.03); C309(1.07); C397(1.10) | LDD0690 | [12] |
| LDCM0374 | CL105 | HCT 116 | C309(0.68); C418(1.02); C402(1.04); C397(1.05) | LDD0691 | [12] |
| LDCM0375 | CL106 | HCT 116 | C309(0.84); C418(0.91); C402(1.21); C397(1.22) | LDD0692 | [12] |
| LDCM0376 | CL107 | HCT 116 | C309(0.75); C418(1.00); C402(1.14); C397(1.15) | LDD0693 | [12] |
| LDCM0377 | CL108 | HCT 116 | C309(0.78); C418(0.84); C402(1.06); C397(1.07) | LDD0694 | [12] |
| LDCM0378 | CL109 | HCT 116 | C309(0.87); C402(1.03); C397(1.03); C418(1.28) | LDD0695 | [12] |
| LDCM0379 | CL11 | HCT 116 | C309(0.76); C397(1.23); C402(1.23); C239(1.27) | LDD0696 | [12] |
| LDCM0380 | CL110 | HCT 116 | C309(0.67); C418(0.88); C402(1.13); C397(1.21) | LDD0697 | [12] |
| LDCM0381 | CL111 | HCT 116 | C309(0.89); C418(1.02); C397(1.10); C402(1.10) | LDD0698 | [12] |
| LDCM0382 | CL112 | HCT 116 | C309(0.82); C418(0.99); C402(1.04); C397(1.09) | LDD0699 | [12] |
| LDCM0383 | CL113 | HCT 116 | C309(0.90); C418(0.99); C402(1.19); C397(1.20) | LDD0700 | [12] |
| LDCM0384 | CL114 | HCT 116 | C309(0.88); C397(1.00); C402(1.04); C418(1.24) | LDD0701 | [12] |
| LDCM0385 | CL115 | HCT 116 | C309(0.91); C397(1.07); C418(1.12); C402(1.15) | LDD0702 | [12] |
| LDCM0386 | CL116 | HCT 116 | C309(0.83); C402(1.01); C397(1.02); C418(1.14) | LDD0703 | [12] |
| LDCM0387 | CL117 | HCT 116 | C255(0.91); C267(0.91); C309(1.09); C397(1.20) | LDD0704 | [12] |
| LDCM0388 | CL118 | HCT 116 | C397(0.86); C402(0.86); C255(1.01); C267(1.01) | LDD0705 | [12] |
| LDCM0389 | CL119 | HCT 116 | C397(0.96); C402(0.96); C255(0.97); C267(0.97) | LDD0706 | [12] |
| LDCM0390 | CL12 | HCT 116 | C418(0.99); C309(1.13); C397(1.38); C402(1.38) | LDD0707 | [12] |
| LDCM0391 | CL120 | HCT 116 | C255(0.88); C267(0.88); C397(0.93); C402(0.93) | LDD0708 | [12] |
| LDCM0392 | CL121 | HCT 116 | C309(0.80); C239(1.12); C418(1.15); C397(2.56) | LDD0709 | [12] |
| LDCM0393 | CL122 | HCT 116 | C309(0.79); C397(0.99); C402(0.99); C239(1.12) | LDD0710 | [12] |
| LDCM0394 | CL123 | HCT 116 | C309(0.67); C397(1.18); C402(1.18); C239(1.21) | LDD0711 | [12] |
| LDCM0395 | CL124 | HCT 116 | C309(0.84); C239(1.20); C397(1.21); C402(1.21) | LDD0712 | [12] |
| LDCM0396 | CL125 | HCT 116 | C397(0.89); C402(0.89); C309(0.90); C418(1.22) | LDD0713 | [12] |
| LDCM0397 | CL126 | HCT 116 | C309(0.69); C397(0.91); C402(0.91); C418(1.30) | LDD0714 | [12] |
| LDCM0398 | CL127 | HCT 116 | C309(0.64); C397(0.92); C402(0.92); C418(1.21) | LDD0715 | [12] |
| LDCM0399 | CL128 | HCT 116 | C309(0.65); C418(1.10); C397(1.12); C402(1.12) | LDD0716 | [12] |
| LDCM0400 | CL13 | HCT 116 | C309(0.89); C418(1.00); C397(1.11); C402(1.11) | LDD0717 | [12] |
| LDCM0401 | CL14 | HCT 116 | C309(0.78); C397(0.99); C402(0.99); C239(1.14) | LDD0718 | [12] |
| LDCM0402 | CL15 | HCT 116 | C309(0.84); C397(1.06); C402(1.06); C239(1.16) | LDD0719 | [12] |
| LDCM0403 | CL16 | HCT 116 | C309(0.72); C397(1.04); C402(1.04); C418(1.13) | LDD0720 | [12] |
| LDCM0404 | CL17 | HCT 116 | C309(0.62); C397(0.92); C402(0.92); C239(1.11) | LDD0721 | [12] |
| LDCM0405 | CL18 | HCT 116 | C309(0.67); C397(0.99); C402(0.99); C418(1.05) | LDD0722 | [12] |
| LDCM0406 | CL19 | HCT 116 | C309(0.70); C418(0.97); C397(1.07); C402(1.07) | LDD0723 | [12] |
| LDCM0407 | CL2 | HCT 116 | C239(0.91); C397(0.91); C402(0.91); C418(1.11) | LDD0724 | [12] |
| LDCM0408 | CL20 | HCT 116 | C309(0.74); C418(1.09); C397(1.15); C402(1.15) | LDD0725 | [12] |
| LDCM0409 | CL21 | HCT 116 | C309(0.62); C239(1.33); C397(1.38); C402(1.38) | LDD0726 | [12] |
| LDCM0410 | CL22 | HCT 116 | C309(0.72); C418(1.02); C239(1.80); C397(1.83) | LDD0727 | [12] |
| LDCM0411 | CL23 | HCT 116 | C309(0.67); C418(1.01); C239(1.13); C397(1.24) | LDD0728 | [12] |
| LDCM0412 | CL24 | HCT 116 | C309(0.69); C418(0.98); C397(1.36); C402(1.36) | LDD0729 | [12] |
| LDCM0413 | CL25 | HCT 116 | C418(1.13); C239(1.27); C309(0.57); C397(1.31) | LDD0730 | [12] |
| LDCM0414 | CL26 | HCT 116 | C418(1.18); C239(1.16); C309(0.69); C397(1.07) | LDD0731 | [12] |
| LDCM0415 | CL27 | HCT 116 | C418(0.97); C239(1.17); C309(0.69); C397(1.13) | LDD0732 | [12] |
| LDCM0416 | CL28 | HCT 116 | C418(1.23); C239(1.42); C309(0.63); C397(1.06) | LDD0733 | [12] |
| LDCM0417 | CL29 | HCT 116 | C418(1.04); C239(1.45); C309(0.72); C397(1.20) | LDD0734 | [12] |
| LDCM0418 | CL3 | HCT 116 | C418(1.10); C239(0.97); C309(0.95); C397(0.88) | LDD0735 | [12] |
| LDCM0419 | CL30 | HCT 116 | C418(1.22); C239(1.13); C309(0.67); C397(0.94) | LDD0736 | [12] |
| LDCM0420 | CL31 | HCT 116 | C418(1.02); C239(1.17); C255(0.88); C267(0.88) | LDD0737 | [12] |
| LDCM0421 | CL32 | HCT 116 | C418(1.08); C239(1.16); C255(0.90); C267(0.90) | LDD0738 | [12] |
| LDCM0422 | CL33 | HCT 116 | C418(2.37); C239(1.33); C255(0.91); C267(0.91) | LDD0739 | [12] |
| LDCM0423 | CL34 | HCT 116 | C418(1.18); C239(2.05); C255(1.02); C267(1.02) | LDD0740 | [12] |
| LDCM0424 | CL35 | HCT 116 | C418(1.01); C239(1.66); C255(0.93); C267(0.93) | LDD0741 | [12] |
| LDCM0425 | CL36 | HCT 116 | C418(1.11); C239(1.47); C255(1.19); C267(1.19) | LDD0742 | [12] |
| LDCM0426 | CL37 | HCT 116 | C418(0.91); C239(1.91); C255(0.95); C267(0.95) | LDD0743 | [12] |
| LDCM0428 | CL39 | HCT 116 | C418(1.04); C239(1.52); C255(0.99); C267(0.99) | LDD0745 | [12] |
| LDCM0429 | CL4 | HCT 116 | C418(2.65); C239(1.10); C309(0.77); C397(0.96) | LDD0746 | [12] |
| LDCM0430 | CL40 | HCT 116 | C418(1.08); C239(1.38); C255(0.81); C267(0.81) | LDD0747 | [12] |
| LDCM0431 | CL41 | HCT 116 | C418(1.17); C239(1.39); C255(0.91); C267(0.91) | LDD0748 | [12] |
| LDCM0432 | CL42 | HCT 116 | C418(0.95); C239(1.76); C255(0.73); C267(0.73) | LDD0749 | [12] |
| LDCM0433 | CL43 | HCT 116 | C418(1.08); C239(1.63); C255(0.95); C267(0.95) | LDD0750 | [12] |
| LDCM0434 | CL44 | HCT 116 | C418(1.05); C239(1.52); C255(0.87); C267(0.87) | LDD0751 | [12] |
| LDCM0435 | CL45 | HCT 116 | C418(1.09); C239(1.76); C255(0.94); C267(0.94) | LDD0752 | [12] |
| LDCM0436 | CL46 | HCT 116 | C418(1.60); C239(1.11); C255(0.65); C267(0.65) | LDD0753 | [12] |
| LDCM0437 | CL47 | HCT 116 | C418(1.12); C239(1.05); C255(0.98); C267(0.98) | LDD0754 | [12] |
| LDCM0438 | CL48 | HCT 116 | C418(1.08); C239(1.03); C255(1.60); C267(1.60) | LDD0755 | [12] |
| LDCM0439 | CL49 | HCT 116 | C418(1.08); C239(1.04); C255(1.40); C267(1.40) | LDD0756 | [12] |
| LDCM0440 | CL5 | HCT 116 | C418(0.99); C239(0.99); C309(0.90); C397(1.03) | LDD0757 | [12] |
| LDCM0441 | CL50 | HCT 116 | C418(1.13); C239(0.92); C255(1.70); C267(1.70) | LDD0758 | [12] |
| LDCM0442 | CL51 | HCT 116 | C418(1.36); C239(1.06); C255(2.26); C267(2.26) | LDD0759 | [12] |
| LDCM0443 | CL52 | HCT 116 | C418(1.27); C239(1.14); C255(1.44); C267(1.44) | LDD0760 | [12] |
| LDCM0444 | CL53 | HCT 116 | C418(1.27); C239(0.99); C255(1.52); C267(1.52) | LDD0761 | [12] |
| LDCM0445 | CL54 | HCT 116 | C418(1.12); C239(1.00); C255(1.79); C267(1.79) | LDD0762 | [12] |
| LDCM0446 | CL55 | HCT 116 | C418(1.06); C239(0.95); C255(1.05); C267(1.05) | LDD0763 | [12] |
| LDCM0447 | CL56 | HCT 116 | C418(1.09); C239(0.96); C255(0.87); C267(0.87) | LDD0764 | [12] |
| LDCM0448 | CL57 | HCT 116 | C418(1.44); C239(0.95); C255(1.85); C267(1.85) | LDD0765 | [12] |
| LDCM0449 | CL58 | HCT 116 | C418(1.46); C239(1.04); C255(1.32); C267(1.32) | LDD0766 | [12] |
| LDCM0450 | CL59 | HCT 116 | C418(0.94); C239(1.01); C255(1.20); C267(1.20) | LDD0767 | [12] |
| LDCM0451 | CL6 | HCT 116 | C418(1.16); C239(1.04); C309(0.91); C397(1.06) | LDD0768 | [12] |
| LDCM0452 | CL60 | HCT 116 | C418(1.17); C239(1.08); C255(1.14); C267(1.14) | LDD0769 | [12] |
| LDCM0453 | CL61 | HCT 116 | C418(1.31); C239(1.12); C255(0.60); C267(0.60) | LDD0770 | [12] |
| LDCM0454 | CL62 | HCT 116 | C418(1.33); C239(1.17); C255(0.76); C267(0.76) | LDD0771 | [12] |
| LDCM0455 | CL63 | HCT 116 | C418(1.79); C239(1.16); C255(0.94); C267(0.94) | LDD0772 | [12] |
| LDCM0456 | CL64 | HCT 116 | C418(1.37); C239(1.44); C255(0.62); C267(0.62) | LDD0773 | [12] |
| LDCM0457 | CL65 | HCT 116 | C418(1.11); C239(1.03); C255(0.83); C267(0.83) | LDD0774 | [12] |
| LDCM0458 | CL66 | HCT 116 | C418(1.46); C239(1.27); C255(0.62); C267(0.62) | LDD0775 | [12] |
| LDCM0459 | CL67 | HCT 116 | C418(1.42); C239(1.30); C255(0.90); C267(0.90) | LDD0776 | [12] |
| LDCM0460 | CL68 | HCT 116 | C418(1.10); C239(1.54); C255(0.70); C267(0.70) | LDD0777 | [12] |
| LDCM0461 | CL69 | HCT 116 | C418(1.59); C239(1.32); C255(0.89); C267(0.89) | LDD0778 | [12] |
| LDCM0462 | CL7 | HCT 116 | C418(0.90); C239(1.22); C309(0.90); C397(1.11) | LDD0779 | [12] |
| LDCM0463 | CL70 | HCT 116 | C418(1.03); C239(1.15); C255(0.83); C267(0.83) | LDD0780 | [12] |
| LDCM0464 | CL71 | HCT 116 | C418(1.34); C239(1.28); C255(0.76); C267(0.76) | LDD0781 | [12] |
| LDCM0465 | CL72 | HCT 116 | C418(1.29); C239(1.38); C255(0.88); C267(0.88) | LDD0782 | [12] |
| LDCM0466 | CL73 | HCT 116 | C418(1.46); C239(1.28); C255(0.80); C267(0.80) | LDD0783 | [12] |
| LDCM0467 | CL74 | HCT 116 | C418(1.24); C239(1.30); C255(0.78); C267(0.78) | LDD0784 | [12] |
| LDCM0469 | CL76 | HCT 116 | C418(0.94); C239(0.92); C309(1.24); C397(1.10) | LDD0786 | [12] |
| LDCM0470 | CL77 | HCT 116 | C418(1.13); C239(0.86); C309(1.10); C397(0.88) | LDD0787 | [12] |
| LDCM0471 | CL78 | HCT 116 | C418(1.16); C239(0.92); C309(0.98); C397(0.98) | LDD0788 | [12] |
| LDCM0472 | CL79 | HCT 116 | C418(1.00); C239(0.96); C309(0.93); C397(1.21) | LDD0789 | [12] |
| LDCM0473 | CL8 | HCT 116 | C418(6.38); C239(1.59); C309(0.84); C397(1.47) | LDD0790 | [12] |
| LDCM0474 | CL80 | HCT 116 | C418(0.96); C239(0.88); C309(0.77); C397(1.05) | LDD0791 | [12] |
| LDCM0475 | CL81 | HCT 116 | C418(0.93); C239(0.93); C309(0.92); C397(1.11) | LDD0792 | [12] |
| LDCM0476 | CL82 | HCT 116 | C418(0.96); C239(0.87); C309(0.89); C397(1.39) | LDD0793 | [12] |
| LDCM0477 | CL83 | HCT 116 | C418(1.03); C239(0.96); C309(0.91); C397(1.18) | LDD0794 | [12] |
| LDCM0478 | CL84 | HCT 116 | C418(1.03); C239(0.84); C309(0.98); C397(1.51) | LDD0795 | [12] |
| LDCM0479 | CL85 | HCT 116 | C418(0.97); C239(0.92); C309(0.85); C397(0.97) | LDD0796 | [12] |
| LDCM0480 | CL86 | HCT 116 | C418(0.91); C239(0.84); C309(0.80); C397(0.83) | LDD0797 | [12] |
| LDCM0481 | CL87 | HCT 116 | C418(1.19); C239(0.89); C309(0.92); C397(0.99) | LDD0798 | [12] |
| LDCM0482 | CL88 | HCT 116 | C418(1.03); C239(0.85); C309(0.88); C397(1.37) | LDD0799 | [12] |
| LDCM0483 | CL89 | HCT 116 | C418(0.84); C239(0.80); C309(1.06); C397(1.78) | LDD0800 | [12] |
| LDCM0484 | CL9 | HCT 116 | C418(1.20); C239(1.25); C309(0.67); C397(1.01) | LDD0801 | [12] |
| LDCM0485 | CL90 | HCT 116 | C418(2.20); C239(0.87); C309(0.77); C397(0.93) | LDD0802 | [12] |
| LDCM0486 | CL91 | HCT 116 | C418(1.05); C239(1.24); C309(1.07); C397(1.18) | LDD0803 | [12] |
| LDCM0487 | CL92 | HCT 116 | C418(1.05); C239(1.12); C309(1.03); C397(1.12) | LDD0804 | [12] |
| LDCM0488 | CL93 | HCT 116 | C418(0.98); C239(1.05); C309(0.66); C397(0.99) | LDD0805 | [12] |
| LDCM0489 | CL94 | HCT 116 | C418(0.99); C239(1.33); C309(0.76); C397(1.34) | LDD0806 | [12] |
| LDCM0490 | CL95 | HCT 116 | C418(1.53); C239(1.23); C309(0.54); C397(1.20) | LDD0807 | [12] |
| LDCM0491 | CL96 | HCT 116 | C418(0.99); C239(1.21); C309(0.55); C397(0.93) | LDD0808 | [12] |
| LDCM0492 | CL97 | HCT 116 | C418(1.01); C239(1.15); C309(0.50); C397(1.14) | LDD0809 | [12] |
| LDCM0493 | CL98 | HCT 116 | C418(1.06); C239(1.23); C309(0.75); C397(1.02) | LDD0810 | [12] |
| LDCM0494 | CL99 | HCT 116 | C418(1.07); C239(1.20); C309(0.44); C397(1.03) | LDD0811 | [12] |
| LDCM0191 | Compound 21 | HEK-293T | 11.30 | LDD0508 | [27] |
| LDCM0190 | Compound 34 | HEK-293T | 7.87 | LDD0510 | [27] |
| LDCM0192 | Compound 35 | HEK-293T | 6.58 | LDD0509 | [27] |
| LDCM0193 | Compound 36 | HEK-293T | 8.97 | LDD0511 | [27] |
| LDCM0634 | CY-0357 | Hep-G2 | C296(2.12); C309(1.93); C114(1.66) | LDD2228 | [17] |
| LDCM0028 | Dobutamine | HEK-293T | 6.59 | LDD0180 | [7] |
| LDCM0027 | Dopamine | HEK-293T | 4.11 | LDD0179 | [7] |
| LDCM0495 | E2913 | HEK-293T | C309(1.13); C239(0.93); C418(1.20); C316(1.23) | LDD1698 | [28] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C114(2.31); C239(1.60) | LDD1702 | [5] |
| LDCM0031 | Epigallocatechin gallate | HEK-293T | 77.26 | LDD0183 | [7] |
| LDCM0625 | F8 | Ramos | C309(0.99); C114(1.42); C239(1.15); C418(1.33) | LDD2187 | [29] |
| LDCM0572 | Fragment10 | Ramos | C309(0.58); C114(0.75); C239(1.66); C418(3.47) | LDD2189 | [29] |
| LDCM0573 | Fragment11 | MDA-MB-231 | C418(1.63) | LDD1467 | [30] |
| LDCM0574 | Fragment12 | Ramos | C418(2.60) | LDD1394 | [30] |
| LDCM0575 | Fragment13 | Ramos | C418(0.92) | LDD1396 | [30] |
| LDCM0576 | Fragment14 | MDA-MB-231 | C418(1.39) | LDD1397 | [30] |
| LDCM0579 | Fragment20 | Ramos | C418(2.21) | LDD1403 | [30] |
| LDCM0580 | Fragment21 | MDA-MB-231 | C418(1.20) | LDD1404 | [30] |
| LDCM0582 | Fragment23 | MDA-MB-231 | C418(1.57) | LDD1408 | [30] |
| LDCM0583 | Fragment24 | Ramos | C418(1.42) | LDD1410 | [30] |
| LDCM0584 | Fragment25 | MDA-MB-231 | C418(0.86) | LDD1411 | [30] |
| LDCM0585 | Fragment26 | Ramos | C418(1.12) | LDD1412 | [30] |
| LDCM0578 | Fragment27 | MDA-MB-231 | C418(1.11) | LDD1401 | [30] |
| LDCM0586 | Fragment28 | MDA-MB-231 | C418(1.54) | LDD1415 | [30] |
| LDCM0587 | Fragment29 | MDA-MB-231 | C418(1.35) | LDD1417 | [30] |
| LDCM0588 | Fragment30 | MDA-MB-231 | C418(1.62) | LDD1419 | [30] |
| LDCM0589 | Fragment31 | MDA-MB-231 | C418(1.68) | LDD1421 | [30] |
| LDCM0590 | Fragment32 | MDA-MB-231 | C418(13.08) | LDD1423 | [30] |
| LDCM0468 | Fragment33 | HCT 116 | C418(1.17); C239(1.34); C255(0.79); C267(0.79) | LDD0785 | [12] |
| LDCM0596 | Fragment38 | MDA-MB-231 | C418(1.14) | LDD1433 | [30] |
| LDCM0566 | Fragment4 | MDA-MB-231 | C418(3.86) | LDD1378 | [30] |
| LDCM0598 | Fragment40 | Ramos | C418(1.36) | LDD1437 | [30] |
| LDCM0599 | Fragment41 | MDA-MB-231 | C418(1.23) | LDD1438 | [30] |
| LDCM0600 | Fragment42 | Ramos | C418(0.99) | LDD1440 | [30] |
| LDCM0601 | Fragment43 | MDA-MB-231 | C418(12.96) | LDD1441 | [30] |
| LDCM0602 | Fragment44 | MDA-MB-231 | C418(0.78) | LDD1443 | [30] |
| LDCM0603 | Fragment45 | MDA-MB-231 | C418(20.00) | LDD1482 | [30] |
| LDCM0427 | Fragment51 | HCT 116 | C418(1.05); C239(1.97); C255(0.77); C267(0.77) | LDD0744 | [12] |
| LDCM0610 | Fragment52 | MDA-MB-231 | C418(1.19) | LDD1452 | [30] |
| LDCM0614 | Fragment56 | MDA-MB-231 | C418(1.29) | LDD1484 | [30] |
| LDCM0569 | Fragment7 | MDA-MB-231 | C418(2.38) | LDD1383 | [30] |
| LDCM0570 | Fragment8 | MDA-MB-231 | C418(4.42) | LDD1385 | [30] |
| LDCM0571 | Fragment9 | MDA-MB-231 | C418(1.51) | LDD1387 | [30] |
| LDCM0116 | HHS-0101 | DM93 | Y430(0.86); Y428(0.88); Y635(0.88) | LDD0264 | [10] |
| LDCM0117 | HHS-0201 | DM93 | Y428(0.93); Y635(0.94); Y430(1.31) | LDD0265 | [10] |
| LDCM0118 | HHS-0301 | DM93 | Y428(0.70); Y635(1.03); Y430(1.07) | LDD0266 | [10] |
| LDCM0119 | HHS-0401 | DM93 | Y428(0.86); Y635(0.90); Y430(1.08) | LDD0267 | [10] |
| LDCM0120 | HHS-0701 | DM93 | Y635(0.78); Y428(1.09); Y430(1.58) | LDD0268 | [10] |
| LDCM0107 | IAA | HeLa | H110(0.00); H24(0.00) | LDD0221 | [23] |
| LDCM0123 | JWB131 | DM93 | Y635(0.97) | LDD0285 | [9] |
| LDCM0124 | JWB142 | DM93 | Y635(0.90) | LDD0286 | [9] |
| LDCM0125 | JWB146 | DM93 | Y635(1.12) | LDD0287 | [9] |
| LDCM0126 | JWB150 | DM93 | Y635(3.14) | LDD0288 | [9] |
| LDCM0127 | JWB152 | DM93 | Y635(1.94) | LDD0289 | [9] |
| LDCM0128 | JWB198 | DM93 | Y635(1.28) | LDD0290 | [9] |
| LDCM0129 | JWB202 | DM93 | Y635(0.56) | LDD0291 | [9] |
| LDCM0130 | JWB211 | DM93 | Y635(0.91) | LDD0292 | [9] |
| LDCM0022 | KB02 | HCT 116 | C397(4.80); C402(4.80); C309(2.40) | LDD0080 | [12] |
| LDCM0023 | KB03 | HCT 116 | C397(3.65); C402(3.65); C309(0.73) | LDD0081 | [12] |
| LDCM0024 | KB05 | HCT 116 | C397(4.04); C402(4.04); C309(1.20) | LDD0082 | [12] |
| LDCM0030 | Luteolin | HEK-293T | 30.84 | LDD0182 | [7] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C418(0.54) | LDD2121 | [5] |
| LDCM0109 | NEM | HeLa | H173(0.00); H24(0.00) | LDD0223 | [23] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C309(0.52) | LDD2089 | [5] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C418(0.60) | LDD2092 | [5] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C309(0.88) | LDD2093 | [5] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C309(1.05) | LDD2098 | [5] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C418(1.09) | LDD2099 | [5] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C418(0.93); C114(1.26) | LDD2105 | [5] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C239(1.06); C309(1.22) | LDD2107 | [5] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C239(1.93) | LDD2119 | [5] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C309(0.96) | LDD2123 | [5] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C418(0.97); C309(0.85); C114(1.28) | LDD2125 | [5] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C309(0.83) | LDD2126 | [5] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C309(1.04) | LDD2127 | [5] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C239(1.13); C418(0.91) | LDD2129 | [5] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C309(0.99); C114(0.74) | LDD2135 | [5] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C309(1.17) | LDD2136 | [5] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C309(1.11) | LDD2137 | [5] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C418(0.96); C309(1.01) | LDD2140 | [5] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C418(2.06); C114(1.49) | LDD2144 | [5] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C418(0.97); C309(0.93) | LDD2146 | [5] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C418(0.54); C309(0.63); C114(0.77) | LDD2150 | [5] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C114(1.15) | LDD2153 | [5] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C239(1.33) | LDD2206 | [31] |
| LDCM0032 | Oleacein | HEK-293T | 8.09 | LDD0184 | [7] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C114(0.61) | LDD2207 | [31] |
| LDCM0029 | Quercetin | HEK-293T | 13.62 | LDD0181 | [7] |
| LDCM0003 | Sulforaphane | MDA-MB-231 | 2.02 | LDD0160 | [6] |
| LDCM0021 | THZ1 | HCT 116 | C239(1.09); C255(1.38); C267(1.38); C397(0.95) | LDD2173 | [12] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| ATP-dependent RNA helicase DDX3X (DDX3X) | DEAD box helicase family | O00571 | |||
Other
References
