Details of the Target
General Information of Target
| Target ID | LDTP10471 | |||||
|---|---|---|---|---|---|---|
| Target Name | Protein disulfide-isomerase TMX3 (TMX3) | |||||
| Gene Name | TMX3 | |||||
| Gene ID | 54495 | |||||
| Synonyms |
KIAA1830; TXNDC10; Protein disulfide-isomerase TMX3; EC 5.3.4.1; Thioredoxin domain-containing protein 10; Thioredoxin-related transmembrane protein 3 |
|||||
| 3D Structure | ||||||
| Sequence |
MSPGGKFDFDDGGCYVGGWEAGRAHGYGVCTGPGAQGEYSGCWAHGFESLGVFTGPGGHS
YQGHWQQGKREGLGVERKSRWTYRGEWLGGLKGRSGVWESVSGLRYAGLWKDGFQDGYGT ETYSDGGTYQGQWQAGKRHGYGVRQSVPYHQAALLRSPRRTSLDSGHSDPPTPPPPLPLP GDEGGSPASGSRGGFVLAGPGDADGASSRKRTPAAGGFFRRSLLLSGLRAGGRRSSLGSK RGSLRSEVSSEVGSTGPPGSEASGPPAAAPPALIEGSATEVYAGEWRADRRSGFGVSQRS NGLRYEGEWLGNRRHGYGRTTRPDGSREEGKYKRNRLVHGGRVRSLLPLALRRGKVKEKV DRAVEGARRAVSAARQRQEIAAARAADALLKAVAASSVAEKAVEAARMAKLIAQDLQPML EAPGRRPRQDSEGSDTEPLDEDSPGVYENGLTPSEGSPELPSSPASSRQPWRPPACRSPL PPGGDQGPFSSPKAWPEEWGGAGAQAEELAGYEAEDEAGMQGPGPRDGSPLLGGCSDSSG SLREEEGEDEEPLPPLRAPAGTEPEPIAMLVLRGSSSRGPDAGCLTEELGEPAATERPAQ PGAANPLVVGAVALLDLSLAFLFSQLLT |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Protein disulfide isomerase family
|
|||||
| Subcellular location |
Endoplasmic reticulum membrane
|
|||||
| Function |
Probable disulfide isomerase, which participates in the folding of proteins containing disulfide bonds. May act as a dithiol oxidase. Acts as a regulator of endoplasmic reticulum-mitochondria contact sites via its ability to regulate redox signals.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
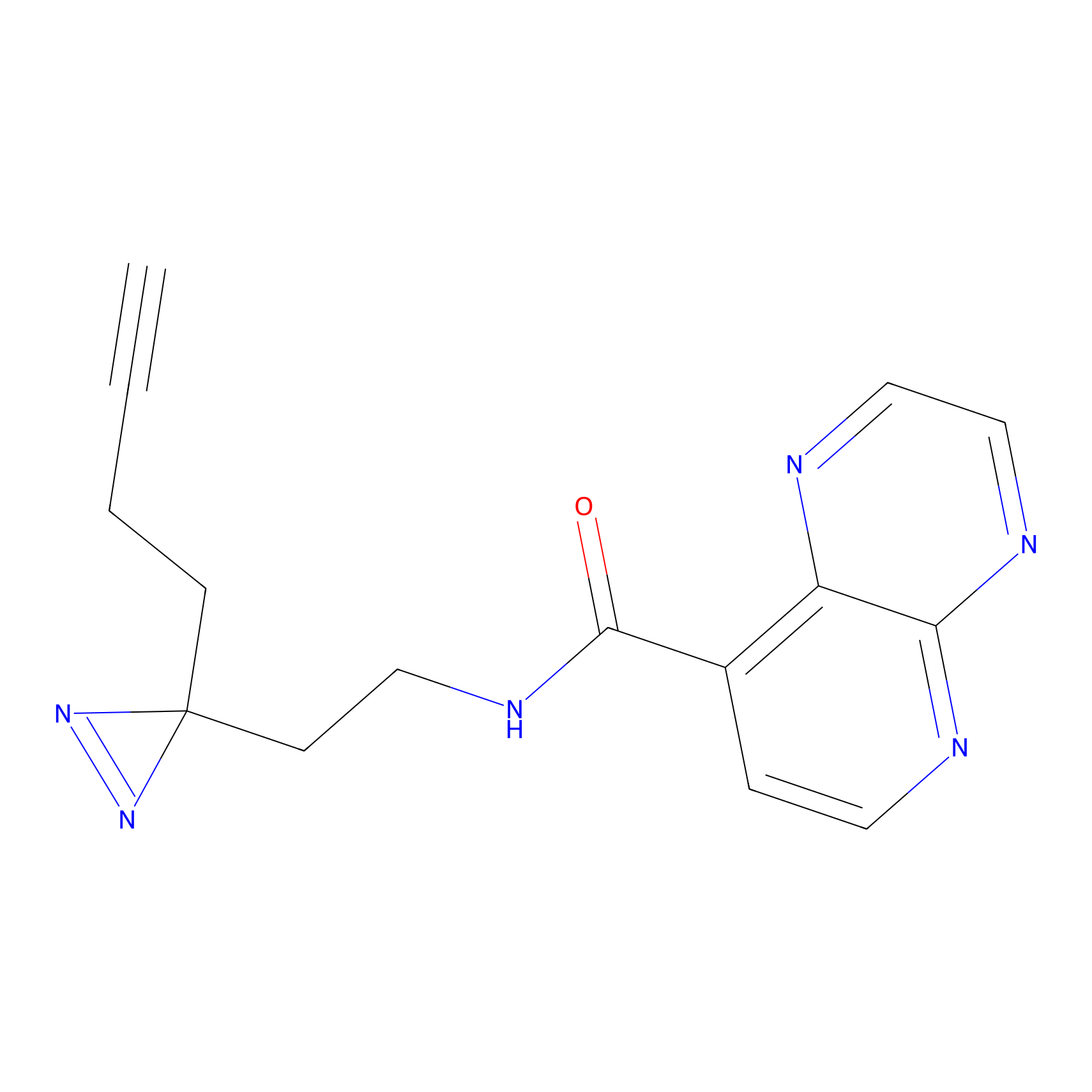
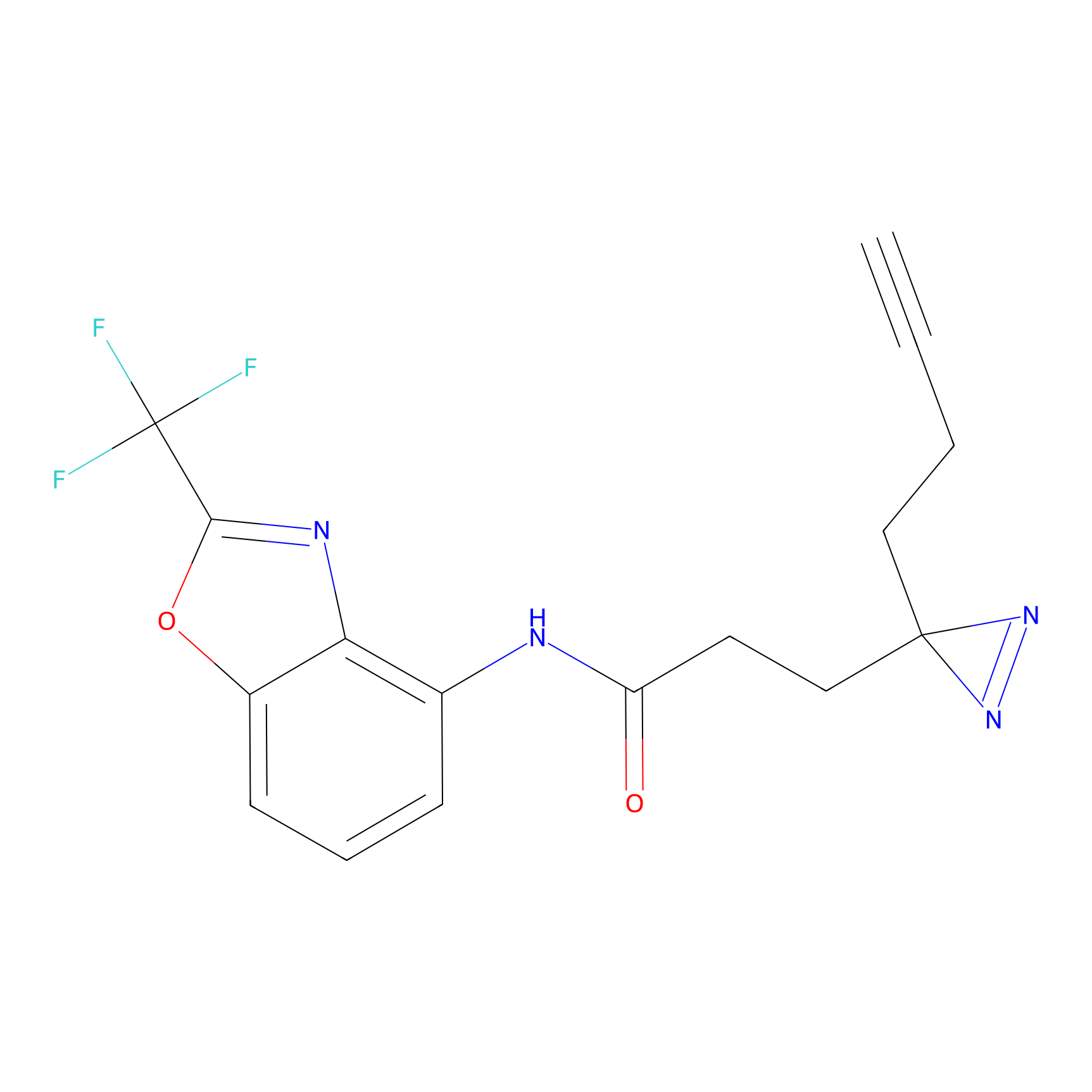
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
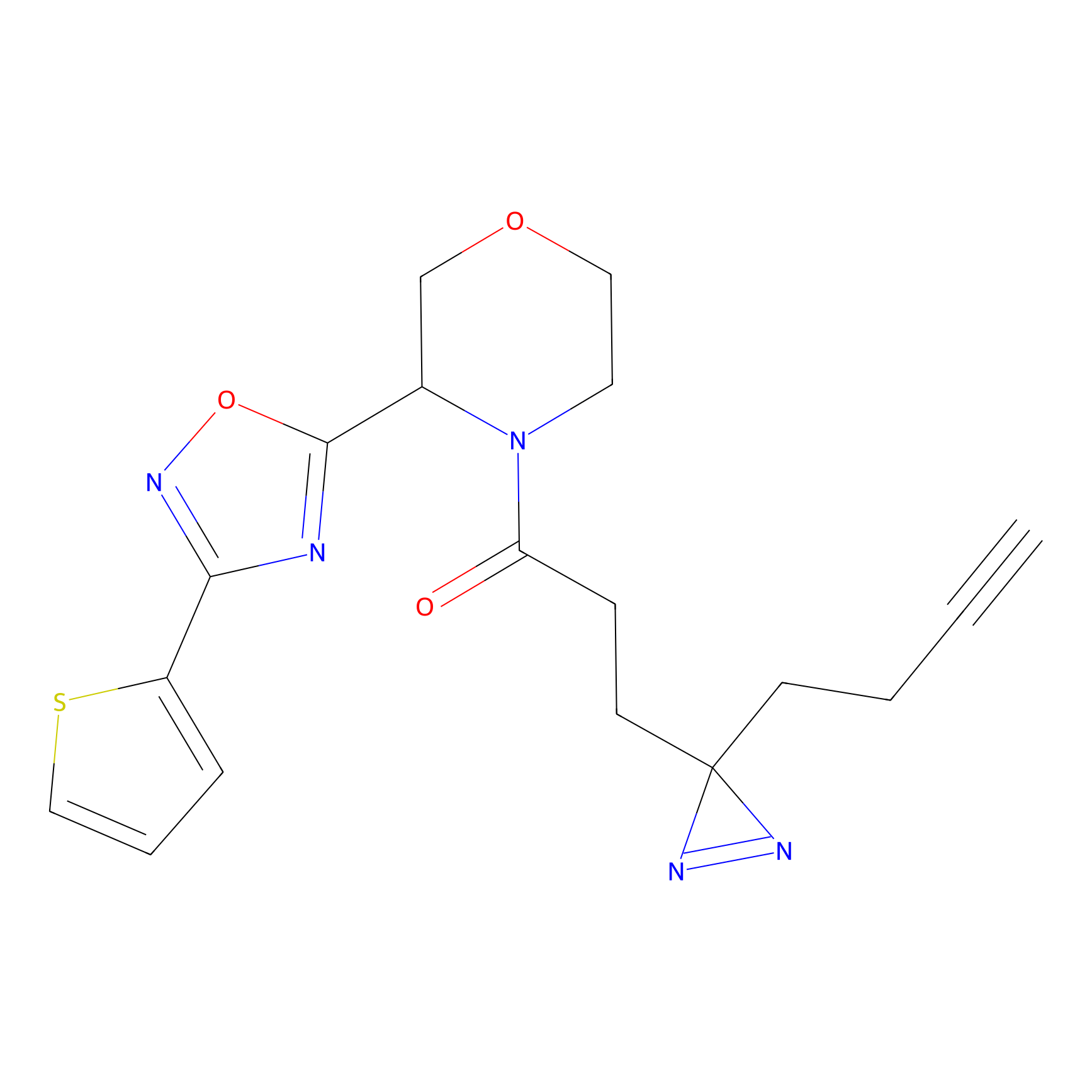
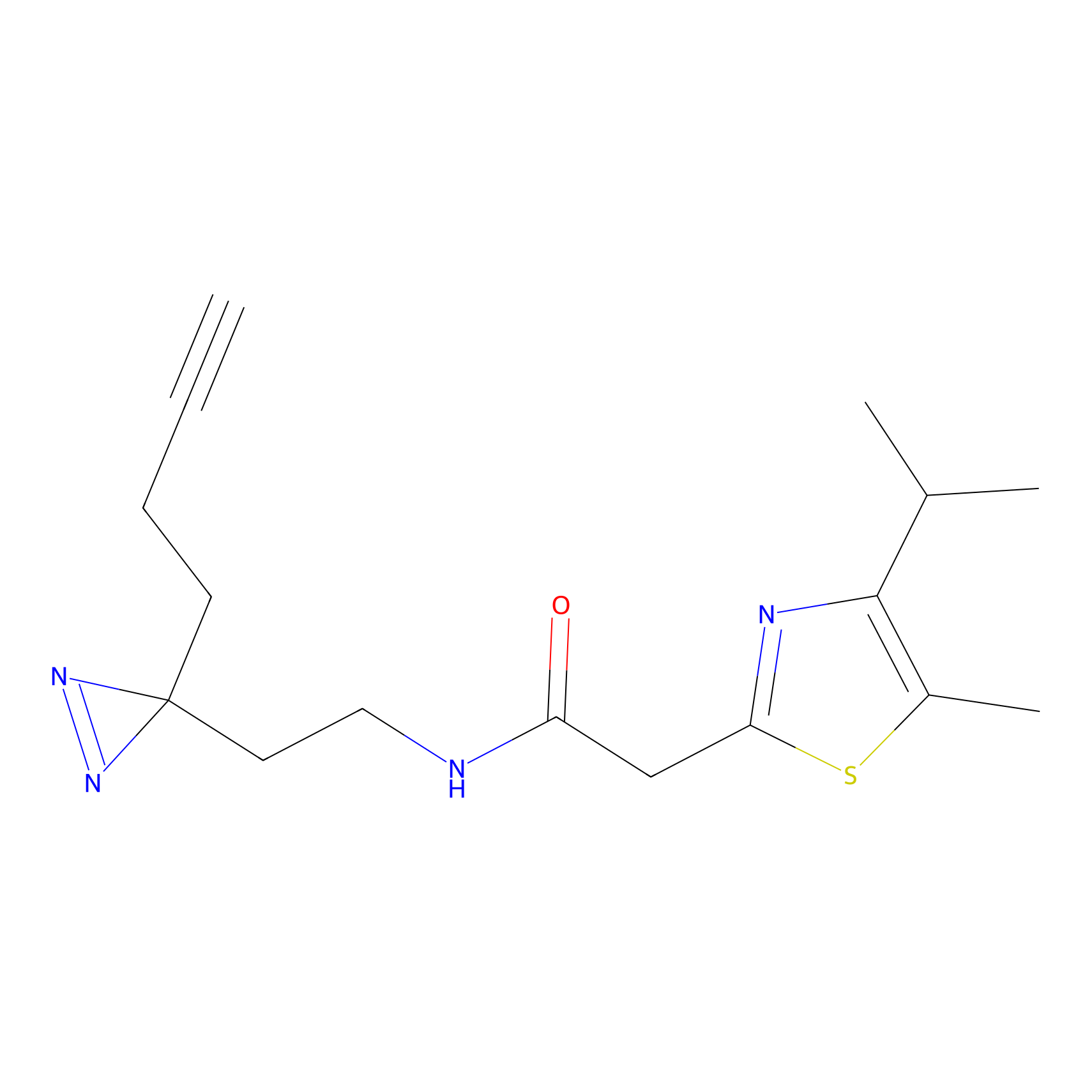
TH211 Probe Info |
 |
Y99(12.61) | LDD0260 | [2] | |
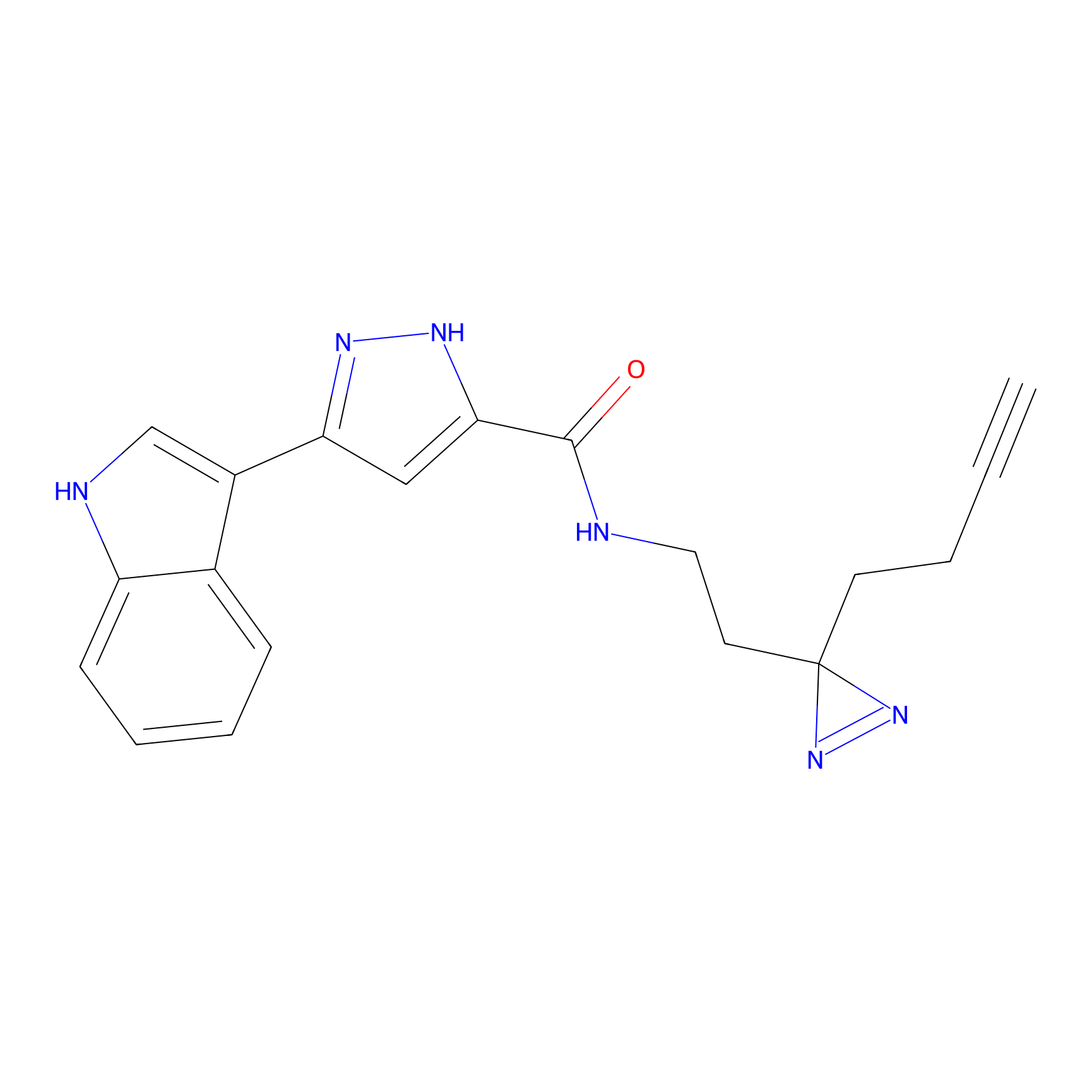
|
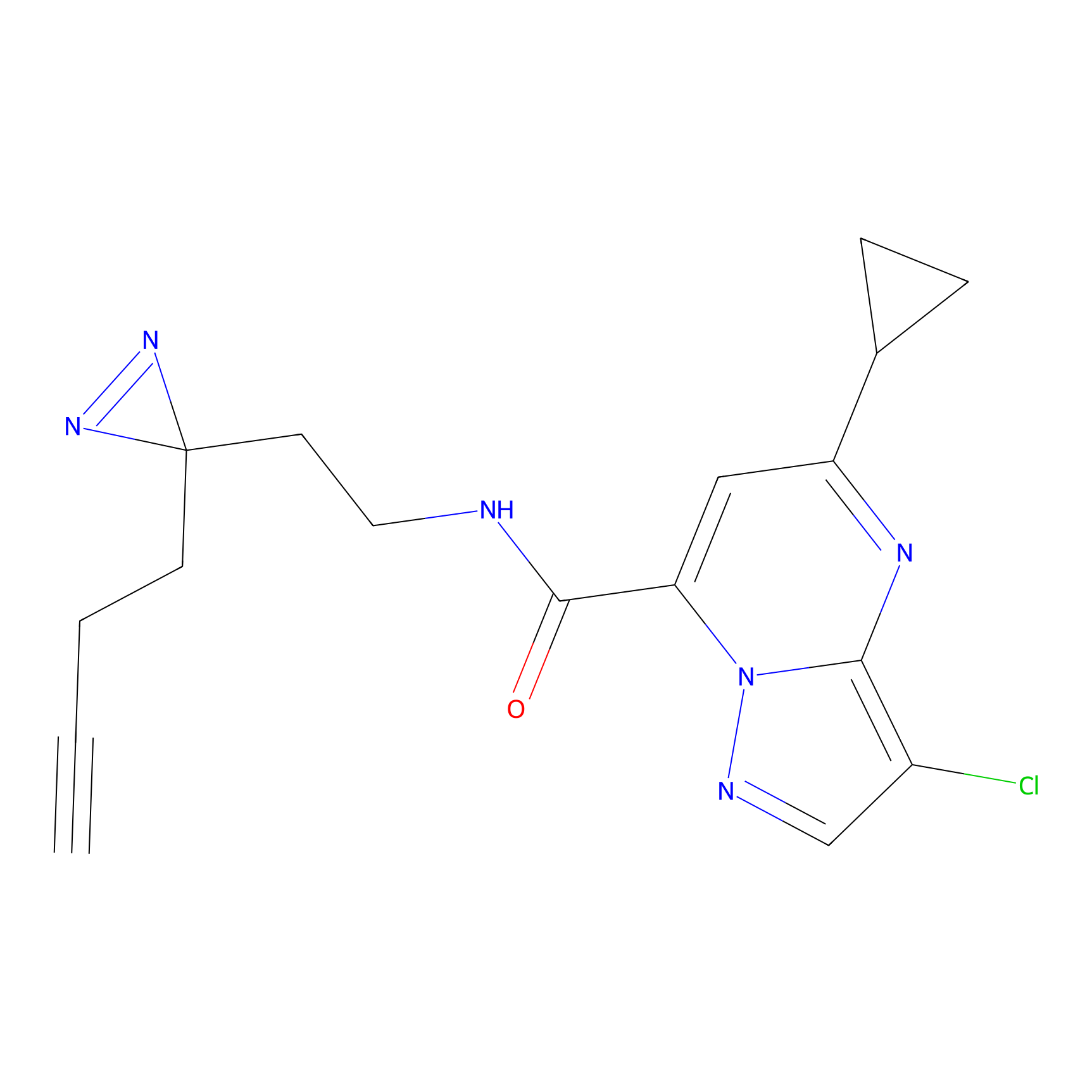
STPyne Probe Info |
 |
K106(3.94); K119(9.32); K267(6.78); K58(8.07) | LDD0277 | [3] | |
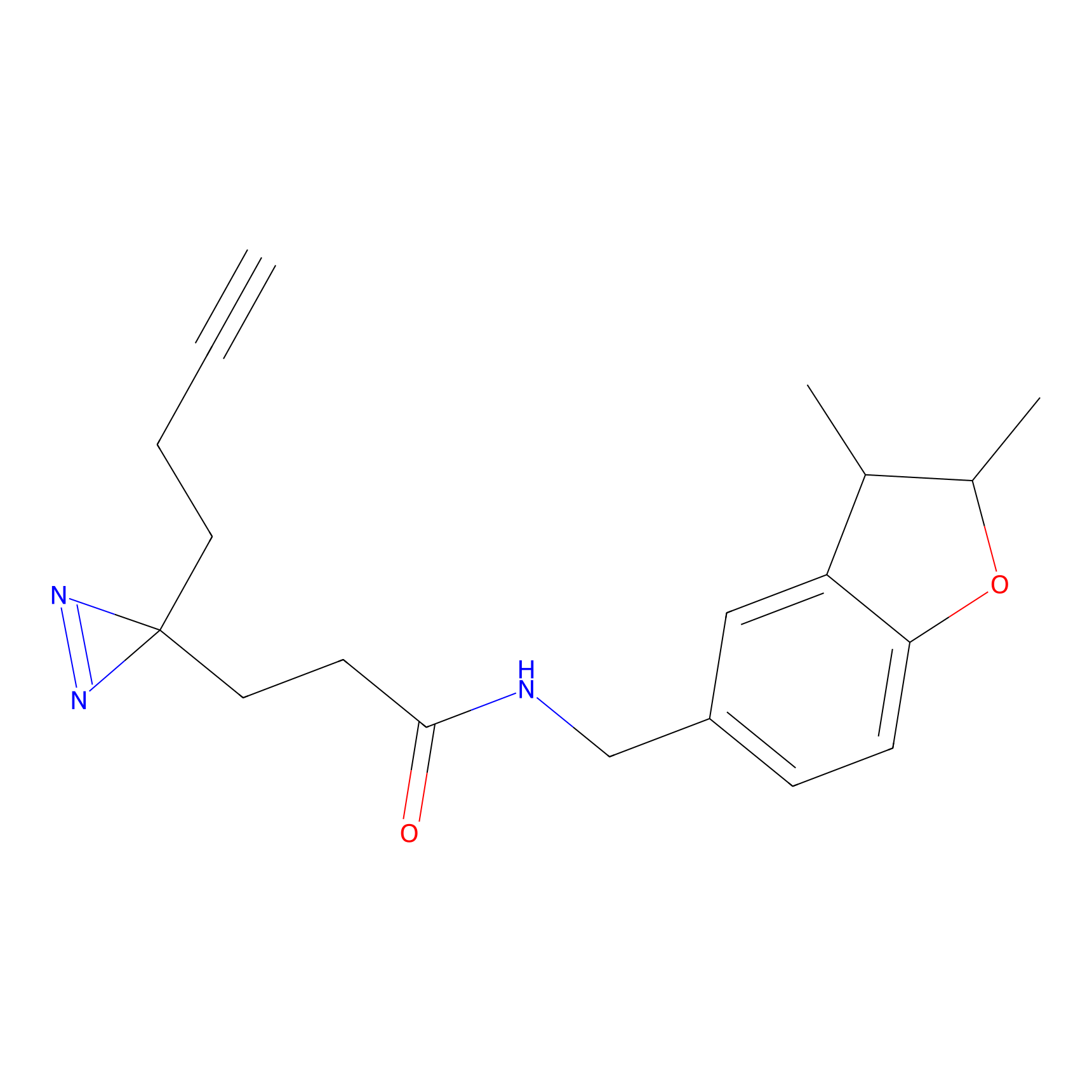
|
OPA-S-S-alkyne Probe Info |
 |
K164(1.61); K119(3.06) | LDD3494 | [4] | |
|
IA-alkyne Probe Info |
 |
C53(1.29); C56(1.29) | LDD0304 | [5] | |
|
HPAP Probe Info |
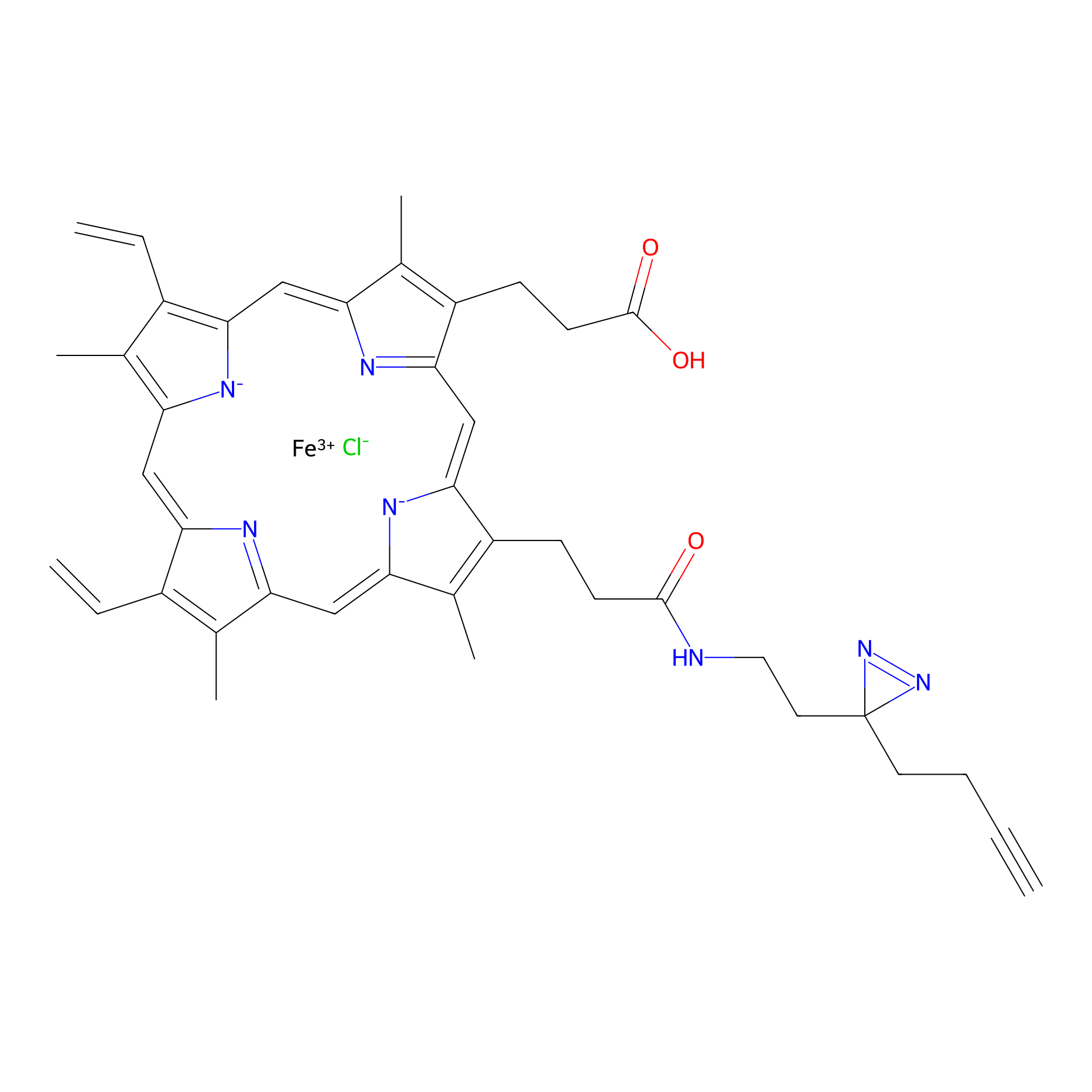 |
3.15 | LDD0064 | [6] | |
|
Alkyne-RA190 Probe Info |
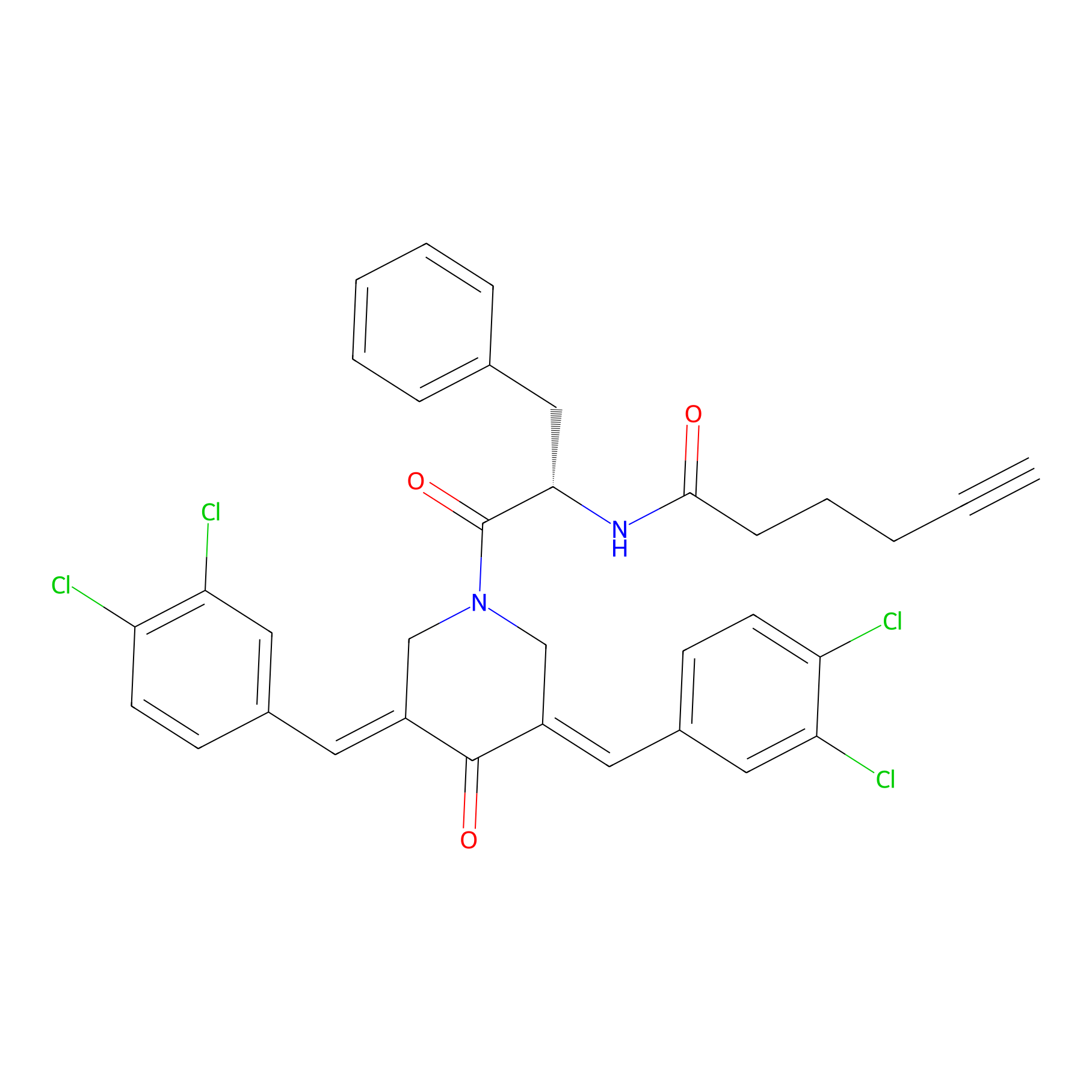 |
2.22 | LDD0299 | [5] | |
|
HHS-475 Probe Info |
 |
Y99(0.86) | LDD0264 | [7] | |
|
HHS-465 Probe Info |
 |
Y99(10.00) | LDD2237 | [8] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0227 | [9] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [10] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [11] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [12] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0156 | Aniline | NCI-H1299 | 11.50 | LDD0403 | [1] |
| LDCM0116 | HHS-0101 | DM93 | Y99(0.86) | LDD0264 | [7] |
| LDCM0117 | HHS-0201 | DM93 | Y99(0.83) | LDD0265 | [7] |
| LDCM0118 | HHS-0301 | DM93 | Y99(0.58) | LDD0266 | [7] |
| LDCM0119 | HHS-0401 | DM93 | Y99(0.78) | LDD0267 | [7] |
| LDCM0120 | HHS-0701 | DM93 | Y99(0.52) | LDD0268 | [7] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0227 | [9] |
| LDCM0014 | Panhematin | hPBMC | 3.15 | LDD0064 | [6] |
| LDCM0131 | RA190 | MM1.R | 2.22 | LDD0299 | [5] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Calreticulin (CALR) | Calreticulin family | P27797 | |||
Immunoglobulin
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Platelet endothelial cell adhesion molecule (PECAM1) | . | P16284 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Four and a half LIM domains protein 3 (FHL3) | . | Q13643 | |||
References