Details of the Target
General Information of Target
| Target ID | LDTP07907 | |||||
|---|---|---|---|---|---|---|
| Target Name | Tubulin alpha-1A chain (TUBA1A) | |||||
| Gene Name | TUBA1A | |||||
| Gene ID | 7846 | |||||
| Synonyms |
TUBA3; Tubulin alpha-1A chain; EC 3.6.5.-; Alpha-tubulin 3; Tubulin B-alpha-1; Tubulin alpha-3 chain) [Cleaved into: Detyrosinated tubulin alpha-1A chain] |
|||||
| 3D Structure | ||||||
| Sequence |
MRECISIHVGQAGVQIGNACWELYCLEHGIQPDGQMPSDKTIGGGDDSFNTFFSETGAGK
HVPRAVFVDLEPTVIDEVRTGTYRQLFHPEQLITGKEDAANNYARGHYTIGKEIIDLVLD RIRKLADQCTGLQGFLVFHSFGGGTGSGFTSLLMERLSVDYGKKSKLEFSIYPAPQVSTA VVEPYNSILTTHTTLEHSDCAFMVDNEAIYDICRRNLDIERPTYTNLNRLIGQIVSSITA SLRFDGALNVDLTEFQTNLVPYPRIHFPLATYAPVISAEKAYHEQLSVAEITNACFEPAN QMVKCDPRHGKYMACCLLYRGDVVPKDVNAAIATIKTKRTIQFVDWCPTGFKVGINYQPP TVVPGGDLAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYAKRAFVHWYVGEGMEEGEFSE AREDMAALEKDYEEVGVDSVEGEGEEEGEEY |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Tubulin family
|
|||||
| Subcellular location |
Cytoplasm, cytoskeleton
|
|||||
| Function |
Tubulin is the major constituent of microtubules, a cylinder consisting of laterally associated linear protofilaments composed of alpha- and beta-tubulin heterodimers. Microtubules grow by the addition of GTP-tubulin dimers to the microtubule end, where a stabilizing cap forms. Below the cap, tubulin dimers are in GDP-bound state, owing to GTPase activity of alpha-tubulin.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| HGC27 | SNV: p.R339H | DBIA Probe Info | |||
| HT3 | SNV: p.Q358Ter | . | |||
| NCIH1155 | SNV: p.R402H | . | |||
| SUPT1 | SNV: p.A99V | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
DAyne Probe Info |
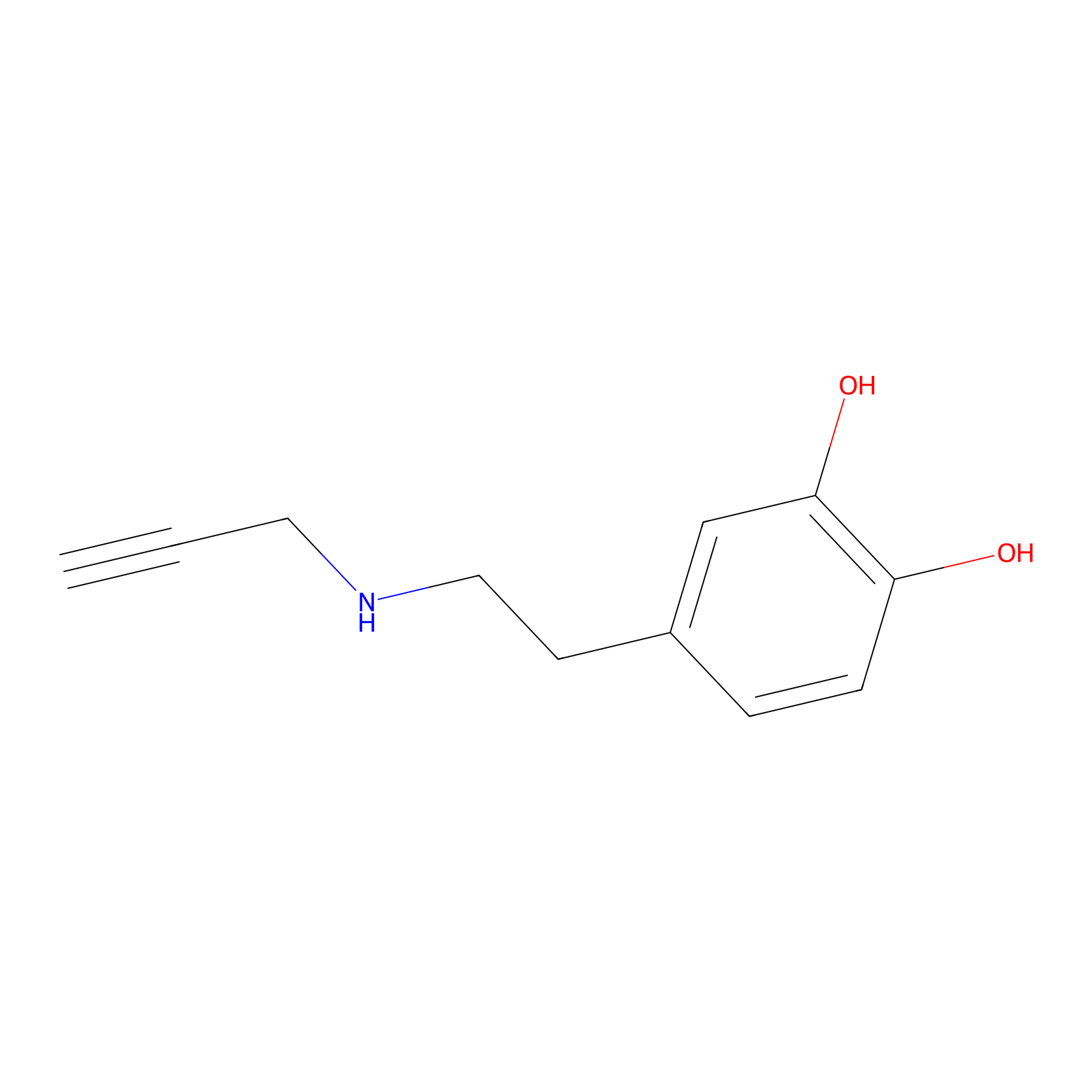 |
2.86 | LDD0261 | [2] | |
|
N1 Probe Info |
 |
4.71 | LDD0242 | [3] | |
|
TH211 Probe Info |
 |
Y357(10.79) | LDD0257 | [4] | |
|
TH216 Probe Info |
 |
Y357(8.28) | LDD0259 | [4] | |
|
Nap-2 Probe Info |
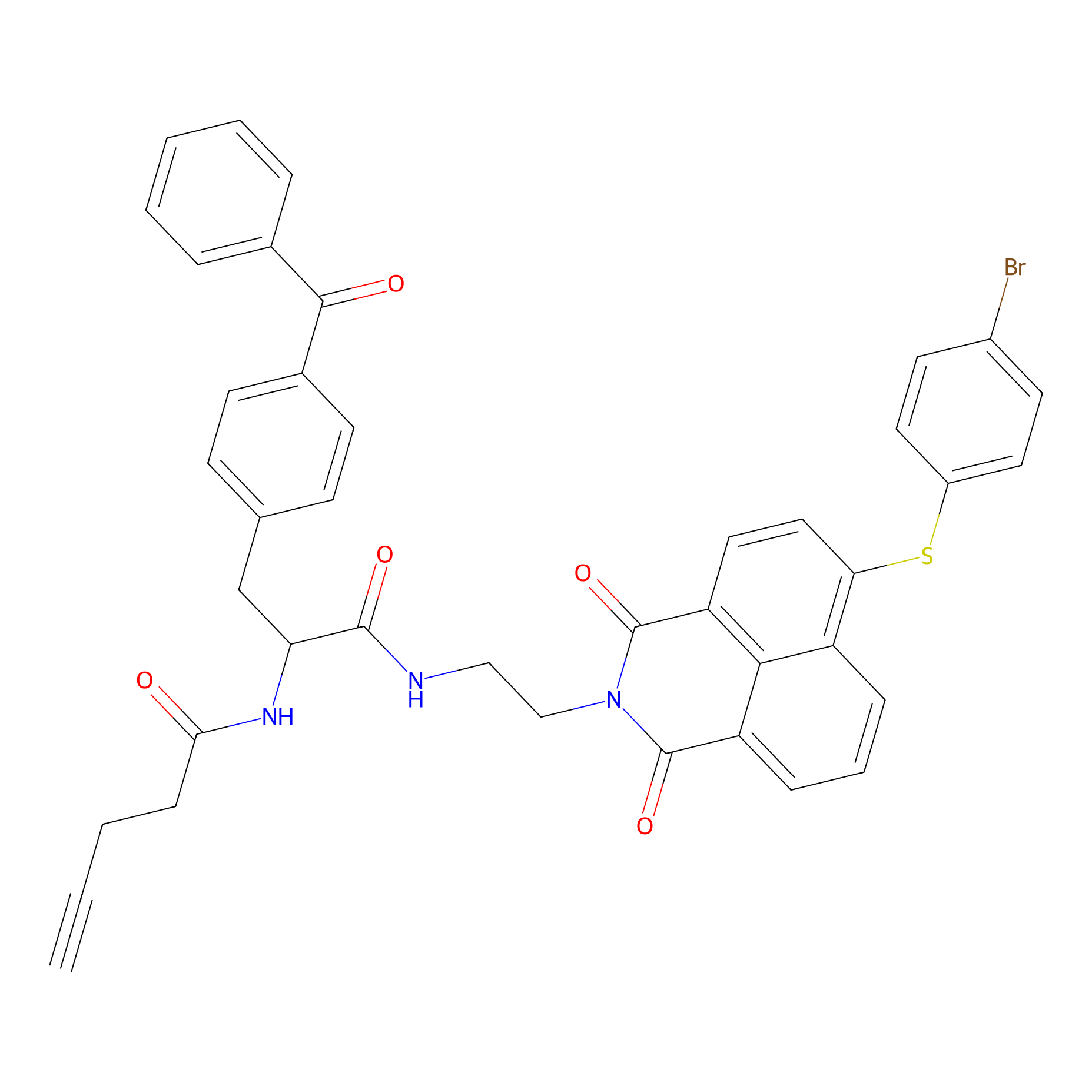 |
N.A. | LDD0435 | [5] | |
|
AZ-9 Probe Info |
 |
D46(1.01); D47(1.01); D345(1.11); E386(1.02) | LDD2208 | [6] | |
|
OPA-S-S-alkyne Probe Info |
 |
K338(2.00); K336(2.00) | LDD3494 | [7] | |
|
P26 Probe Info |
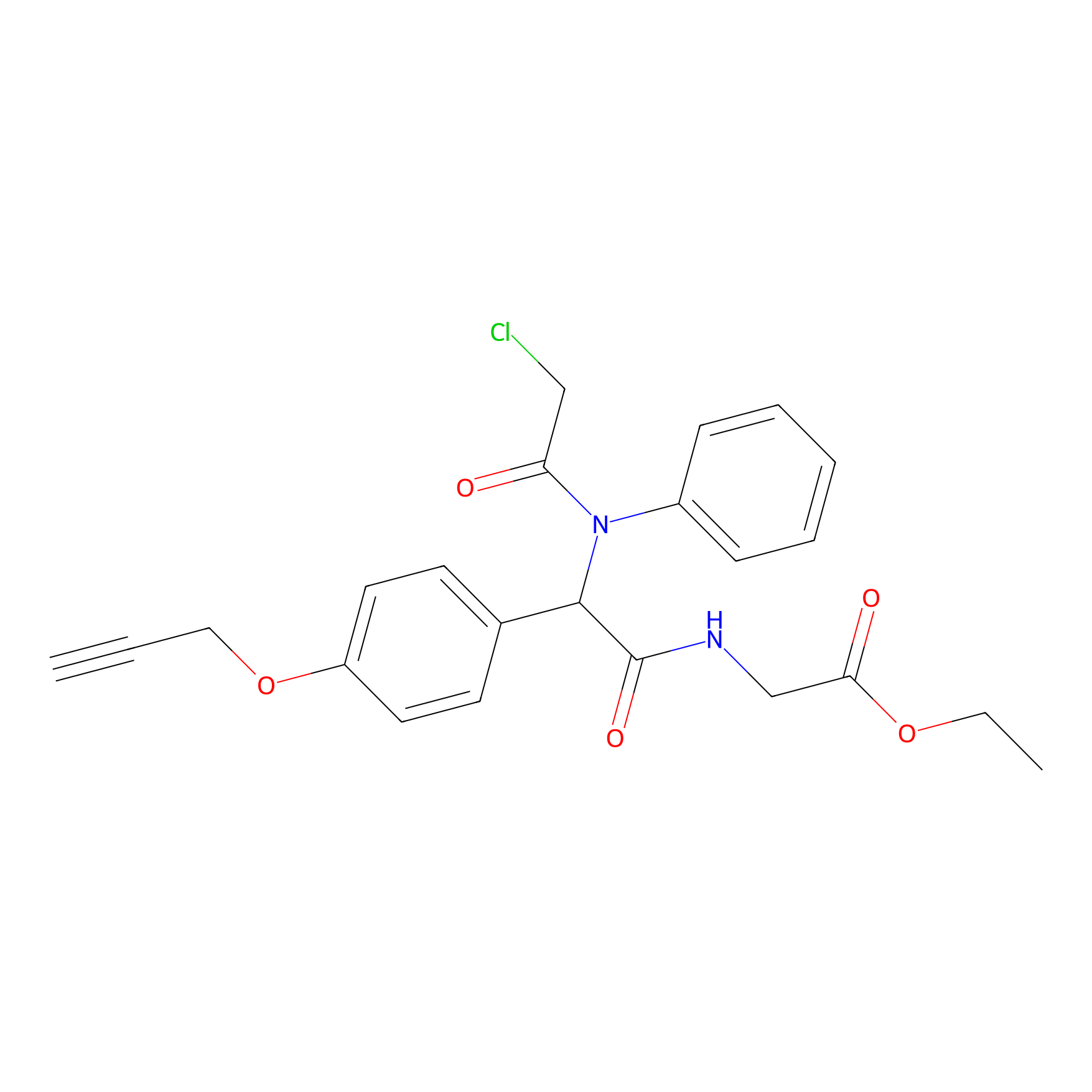 |
1.71 | LDD0409 | [8] | |
|
JZ128-DTB Probe Info |
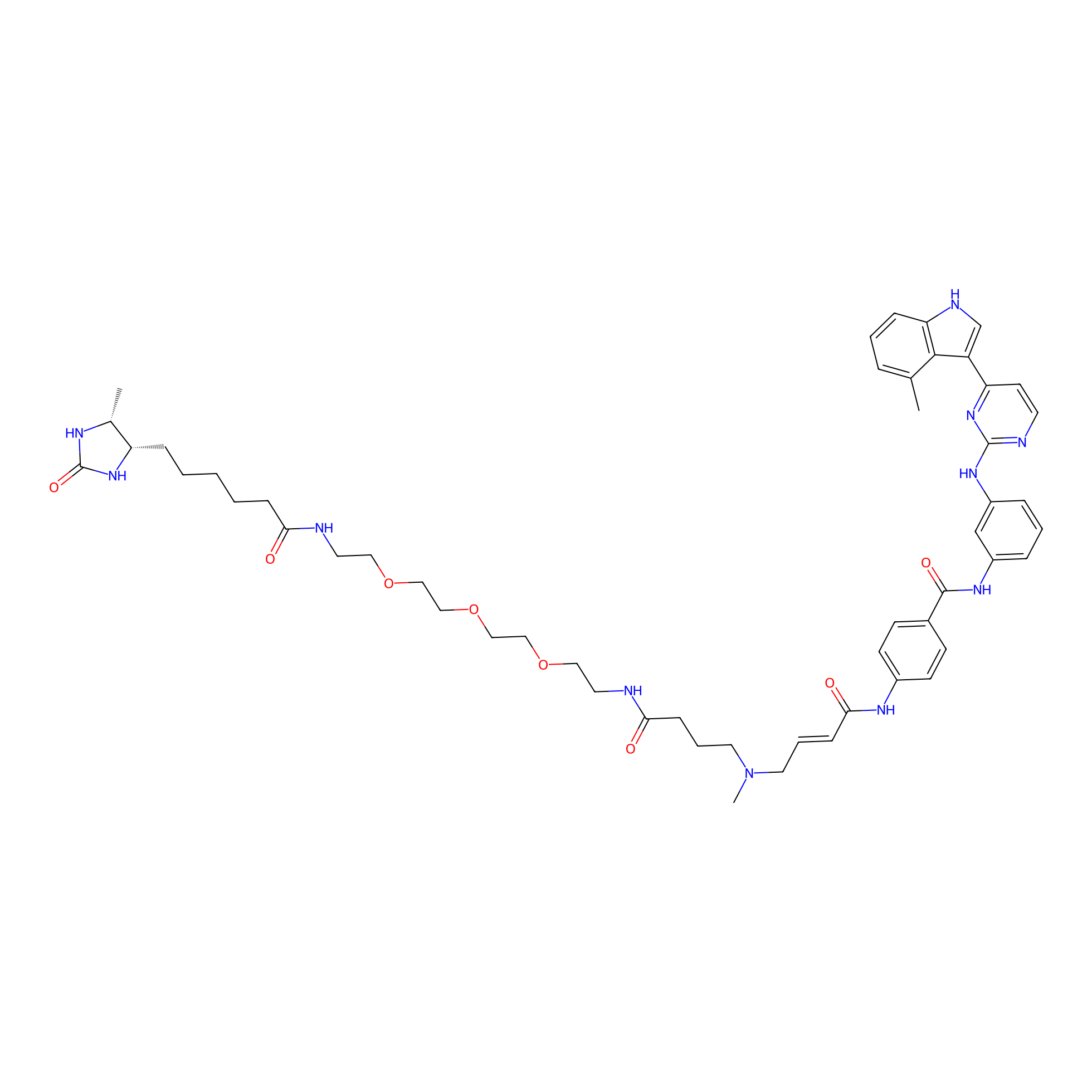 |
C4(0.00); C347(0.00); C200(0.00) | LDD0462 | [9] | |
|
AHL-Pu-1 Probe Info |
 |
C295(7.28) | LDD0170 | [10] | |
|
EA-probe Probe Info |
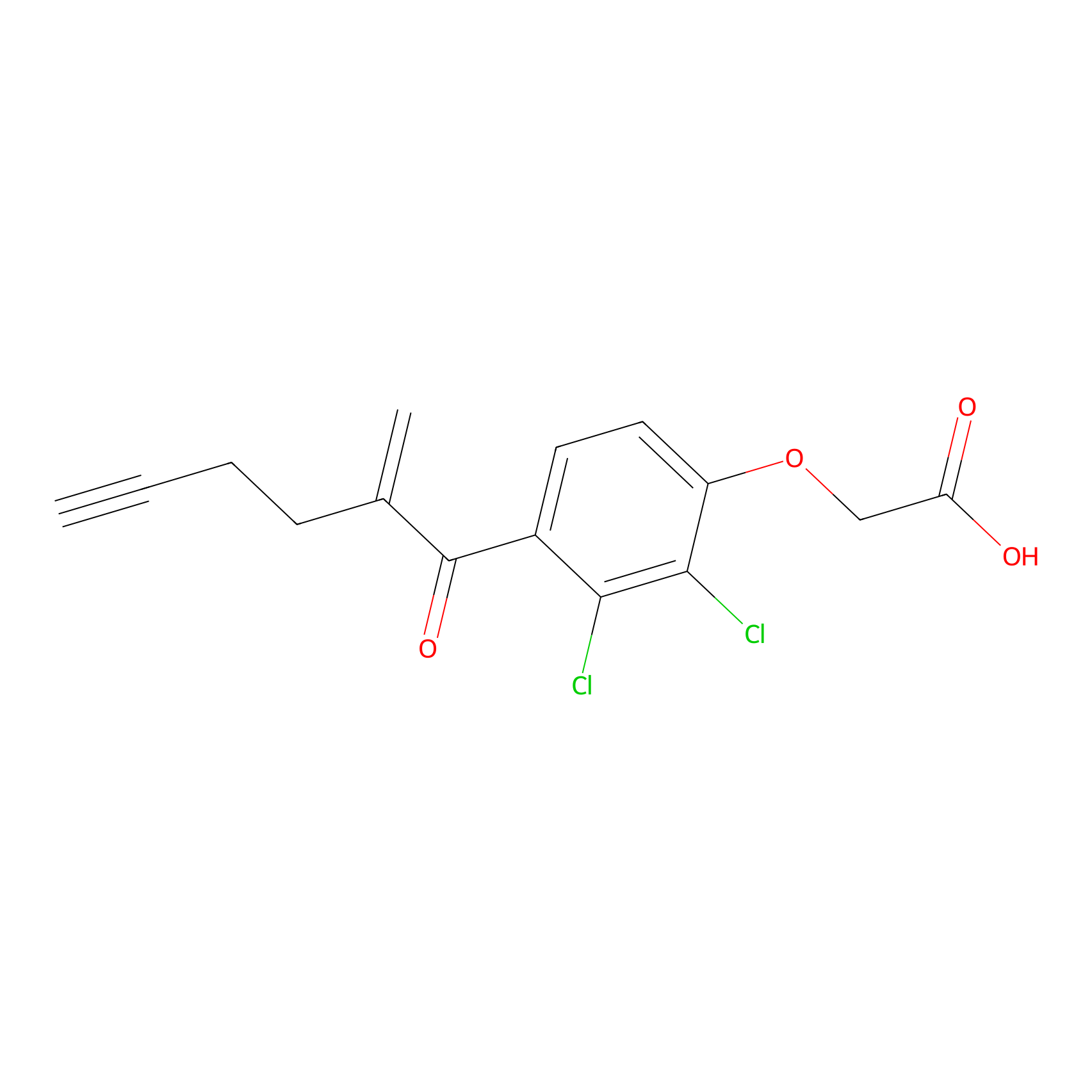 |
C4(1.11); C129(1.04); C347(0.98); C129(1.08) | LDD2210 | [11] | |
|
HHS-482 Probe Info |
 |
Y103(0.88); Y108(0.87); Y161(0.88); Y224(0.87) | LDD0285 | [12] | |
|
HHS-475 Probe Info |
 |
Y408(0.57); Y103(0.64); Y399(0.65); Y262(0.67) | LDD0264 | [13] | |
|
HHS-465 Probe Info |
 |
Y103(10.00); Y108(10.00); Y161(7.71); Y224(10.00) | LDD2237 | [14] | |
|
DBIA Probe Info |
 |
C295(0.98) | LDD0078 | [15] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [16] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [17] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [17] | |
|
IPIAA_L Probe Info |
 |
C376(0.00); C200(0.00); C20(0.00); C295(0.00) | LDD0031 | [18] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [17] | |
|
BTD Probe Info |
 |
N.A. | LDD0004 | [19] | |
|
JW-RF-010 Probe Info |
 |
C315(0.00); C305(0.00); C376(0.00); C295(0.00) | LDD0026 | [20] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [21] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2224 | [21] | |
|
WYneN Probe Info |
 |
C295(0.00); C347(0.00); C376(0.00) | LDD0021 | [19] | |
|
1d-yne Probe Info |
 |
K60(0.00); K96(0.00) | LDD0356 | [22] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [23] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [23] | |
|
ENE Probe Info |
 |
N.A. | LDD0006 | [19] | |
|
IPM Probe Info |
 |
N.A. | LDD2156 | [24] | |
|
OSF Probe Info |
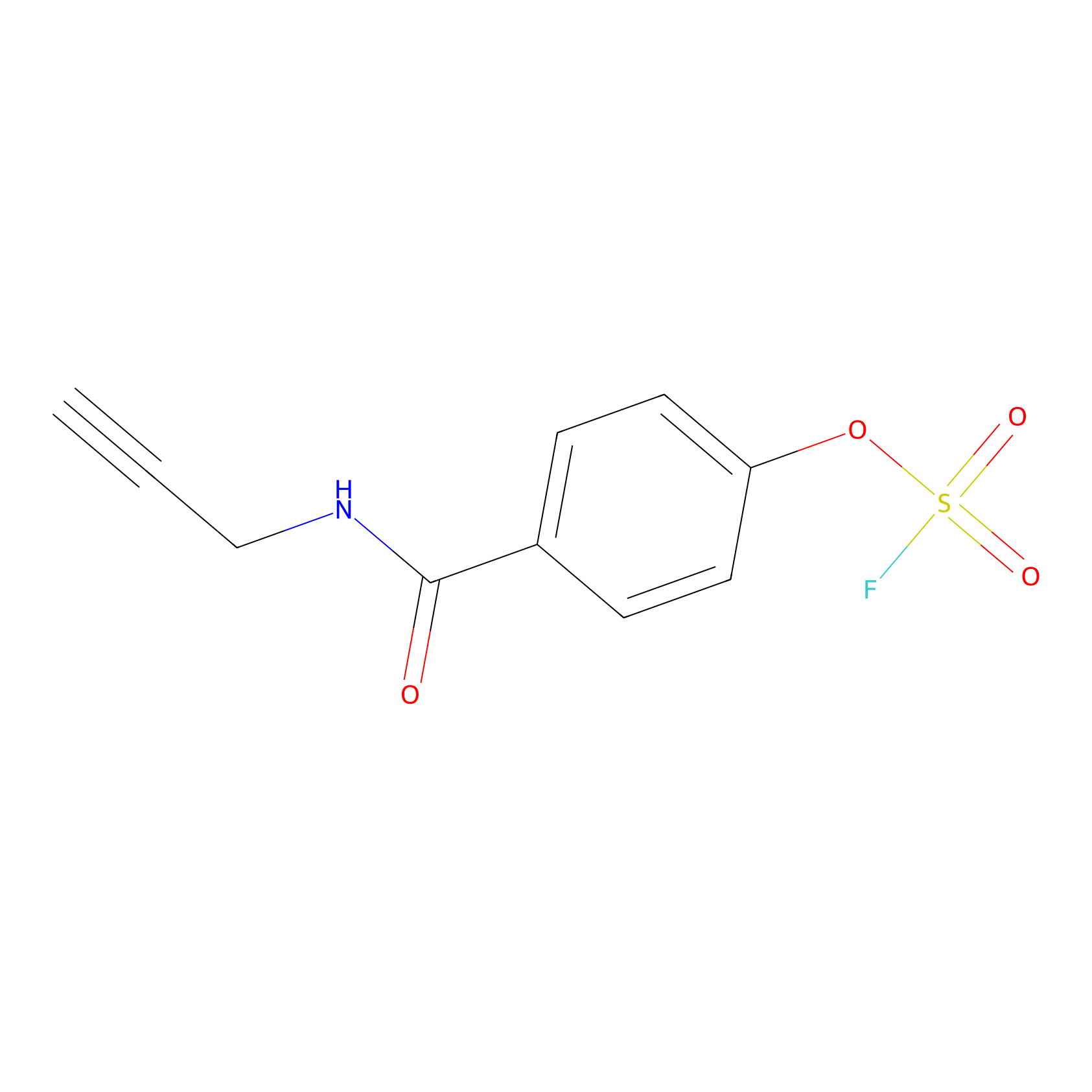 |
Y357(0.00); Y108(0.00); Y224(0.00); H107(0.00) | LDD0029 | [25] | |
|
PF-06672131 Probe Info |
 |
N.A. | LDD0152 | [26] | |
|
SF Probe Info |
 |
Y408(0.00); Y262(0.00); K60(0.00); K338(0.00) | LDD0028 | [25] | |
|
STPyne Probe Info |
 |
K60(0.00); K352(0.00); K401(0.00); K304(0.00) | LDD0009 | [19] | |
|
Phosphinate-6 Probe Info |
 |
C315(0.00); C347(0.00); C25(0.00); C305(0.00) | LDD0018 | [27] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [28] | |
|
1c-yne Probe Info |
 |
K96(0.00); C20(0.00); K352(0.00); K280(0.00) | LDD0228 | [22] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [29] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe12 Probe Info |
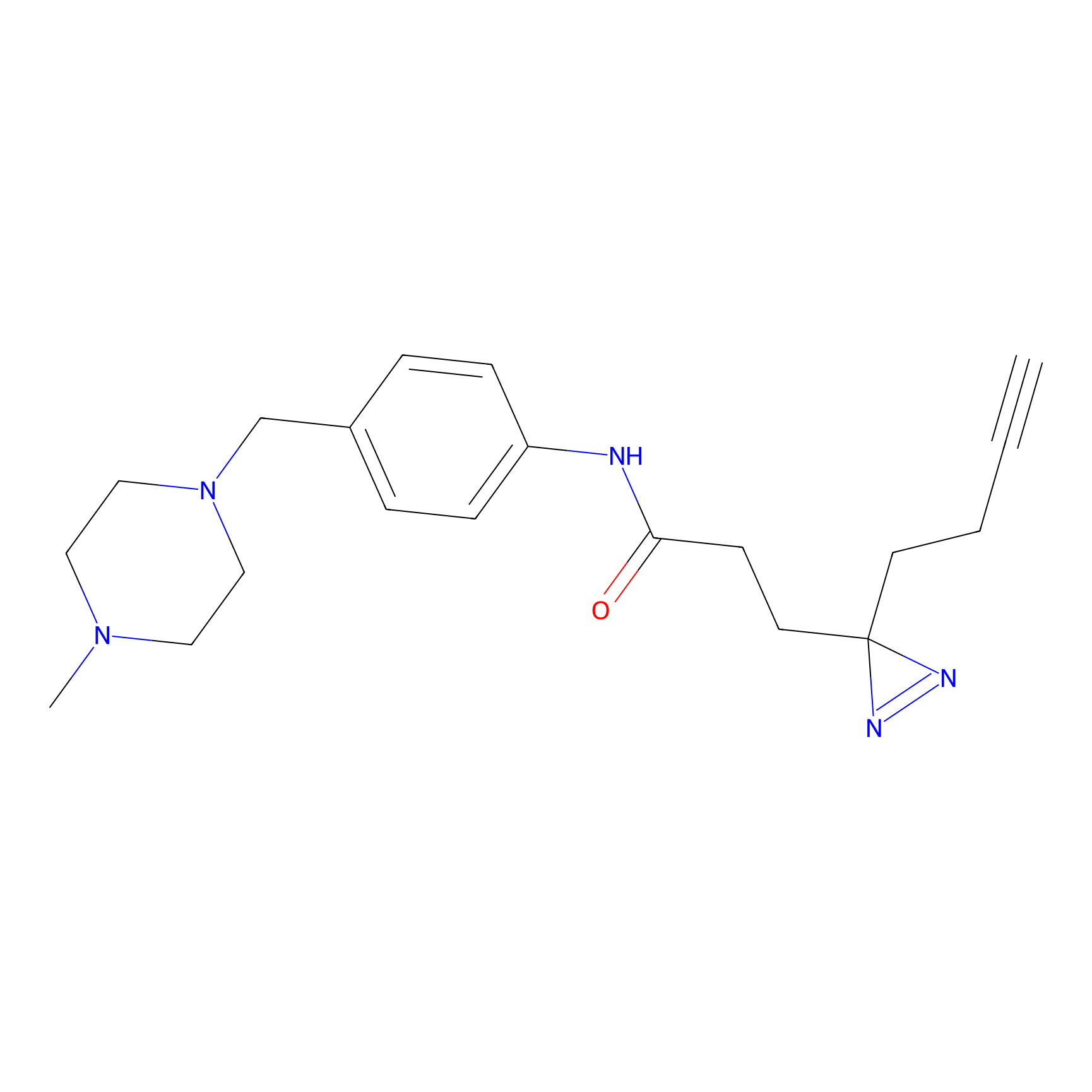 |
6.81 | LDD0473 | [30] | |
|
FFF probe13 Probe Info |
 |
9.78 | LDD0475 | [30] | |
|
FFF probe2 Probe Info |
 |
5.21 | LDD0463 | [30] | |
|
FFF probe3 Probe Info |
 |
5.02 | LDD0464 | [30] | |
|
FFF probe4 Probe Info |
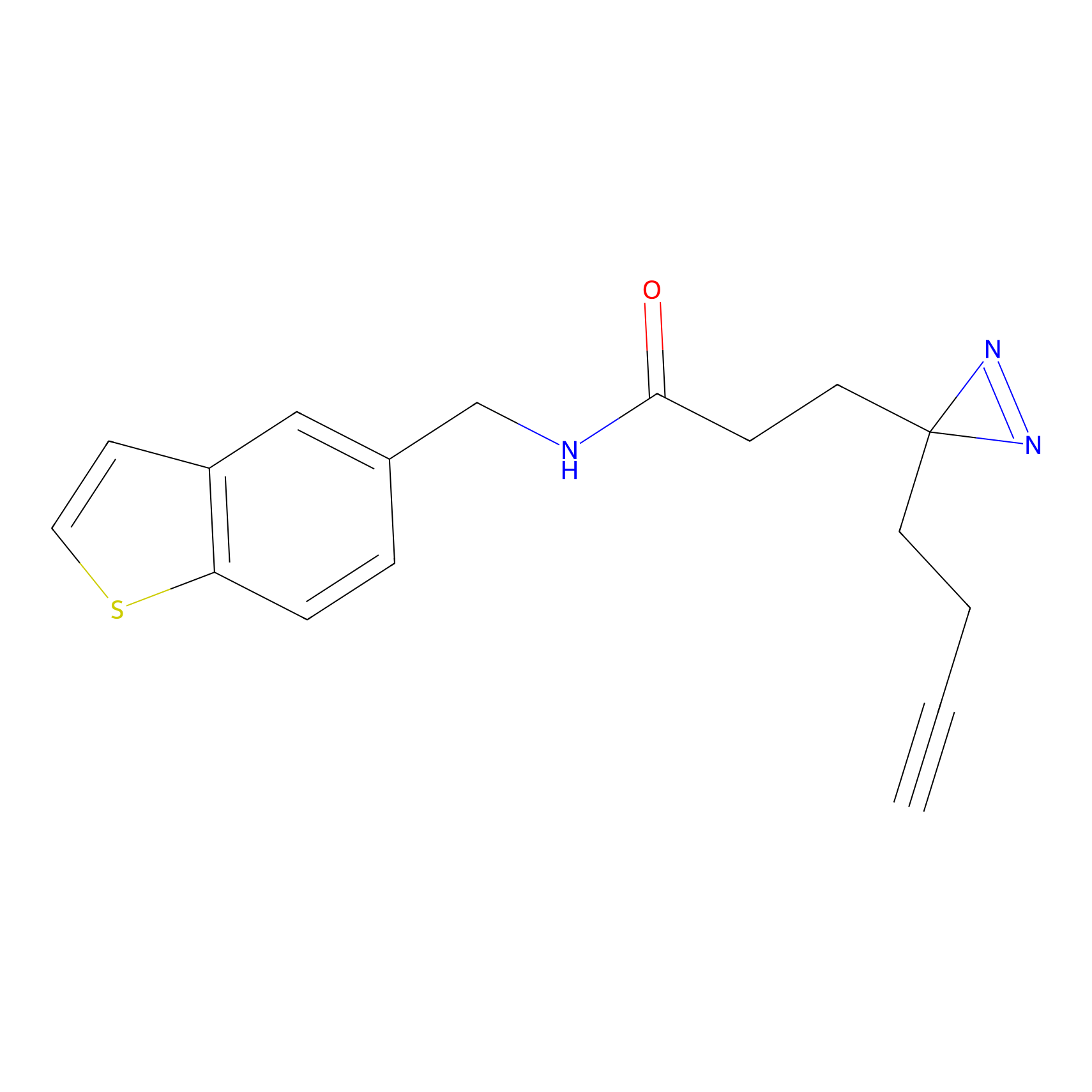 |
6.94 | LDD0466 | [30] | |
|
STS-2 Probe Info |
 |
1.85 | LDD0138 | [31] | |
|
A-DA Probe Info |
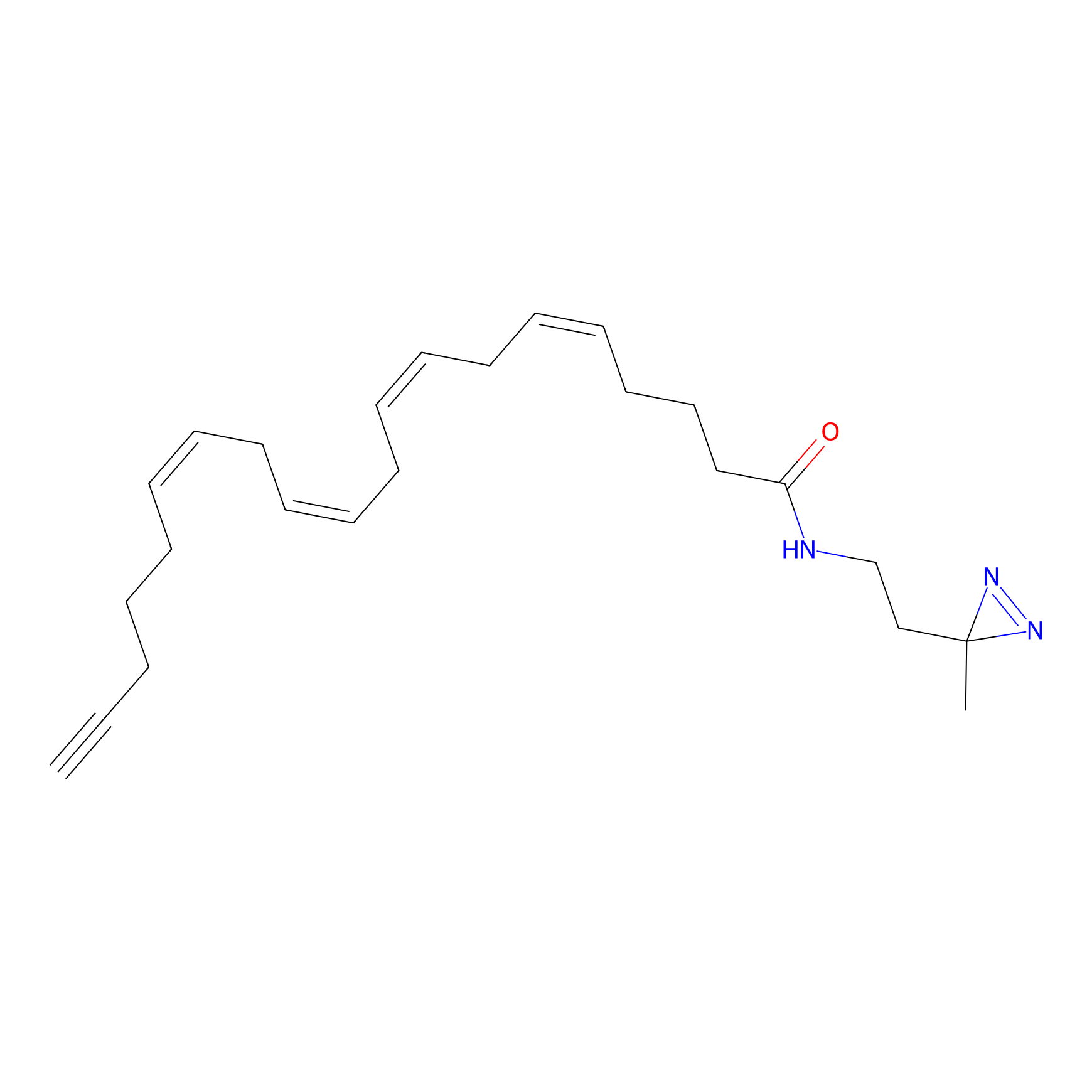 |
2.53 | LDD0143 | [32] | |
|
BD-F Probe Info |
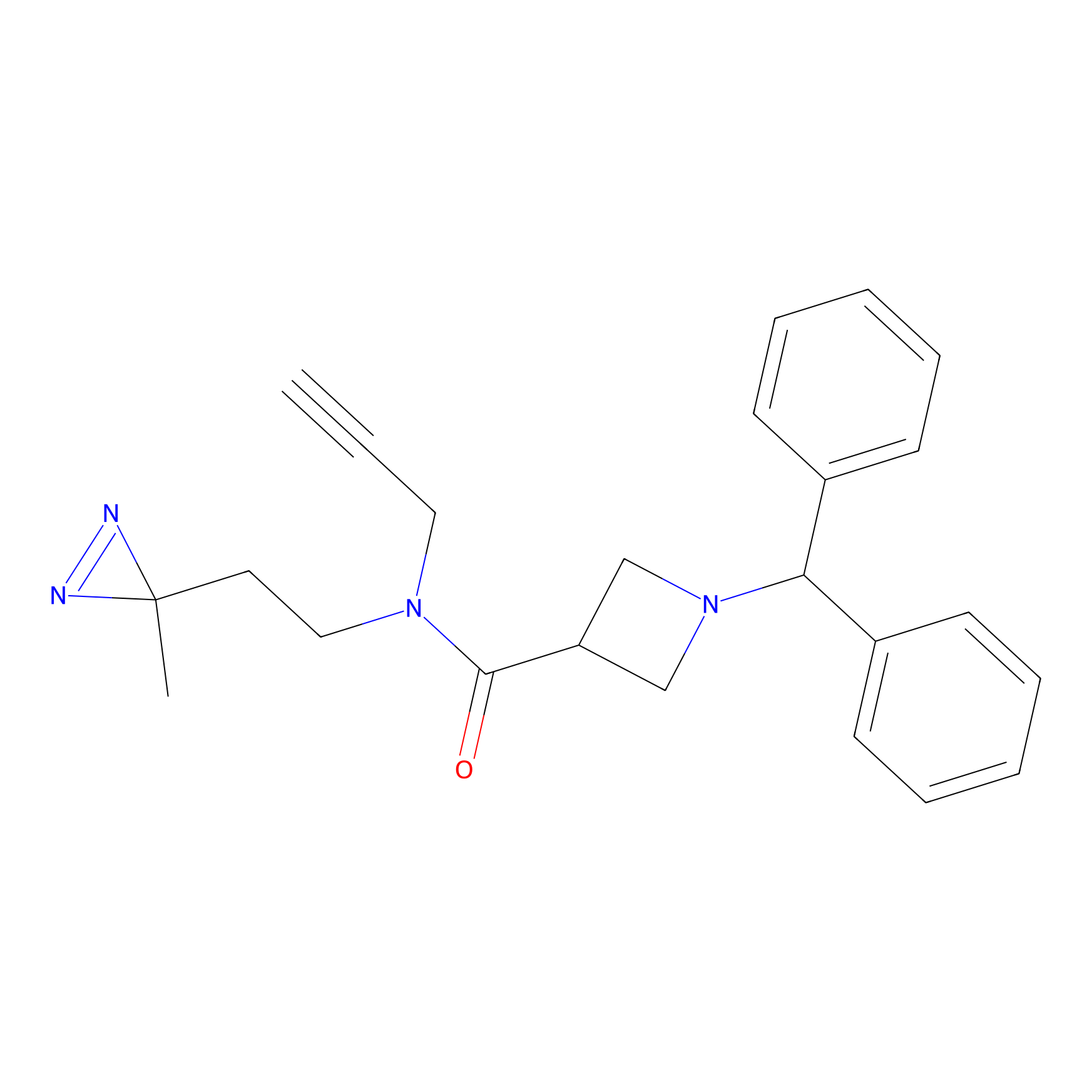 |
E77(0.00); E71(0.00); I75(0.00); R79(0.00) | LDD0024 | [33] | |
|
TM-F Probe Info |
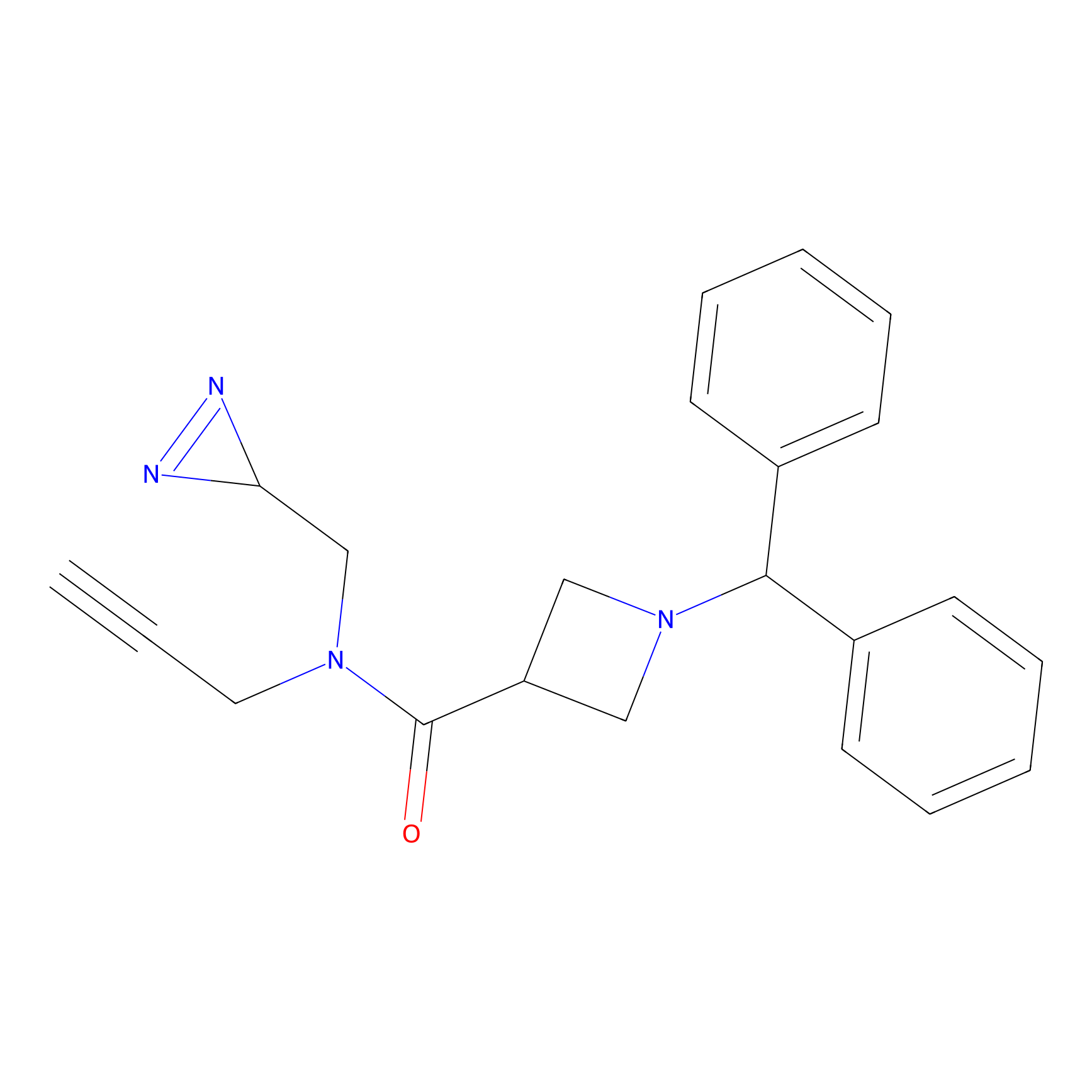 |
E71(0.00); V68(0.00); D76(0.00) | LDD0020 | [33] | |
|
Photocelecoxib Probe Info |
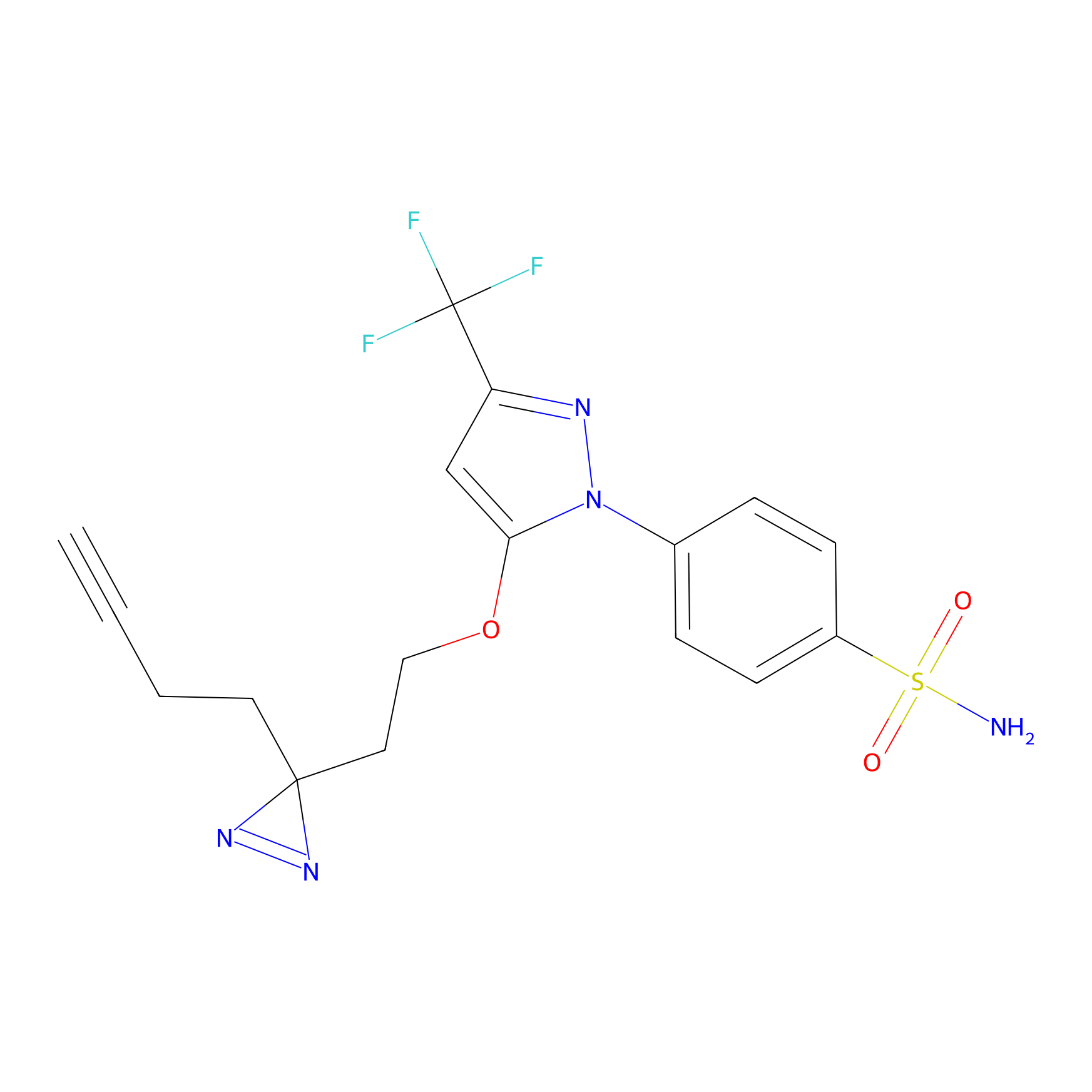 |
F53(0.00); V68(0.00); D69(0.00); P72(0.00) | LDD0019 | [34] | |
|
Photoindomethacin Probe Info |
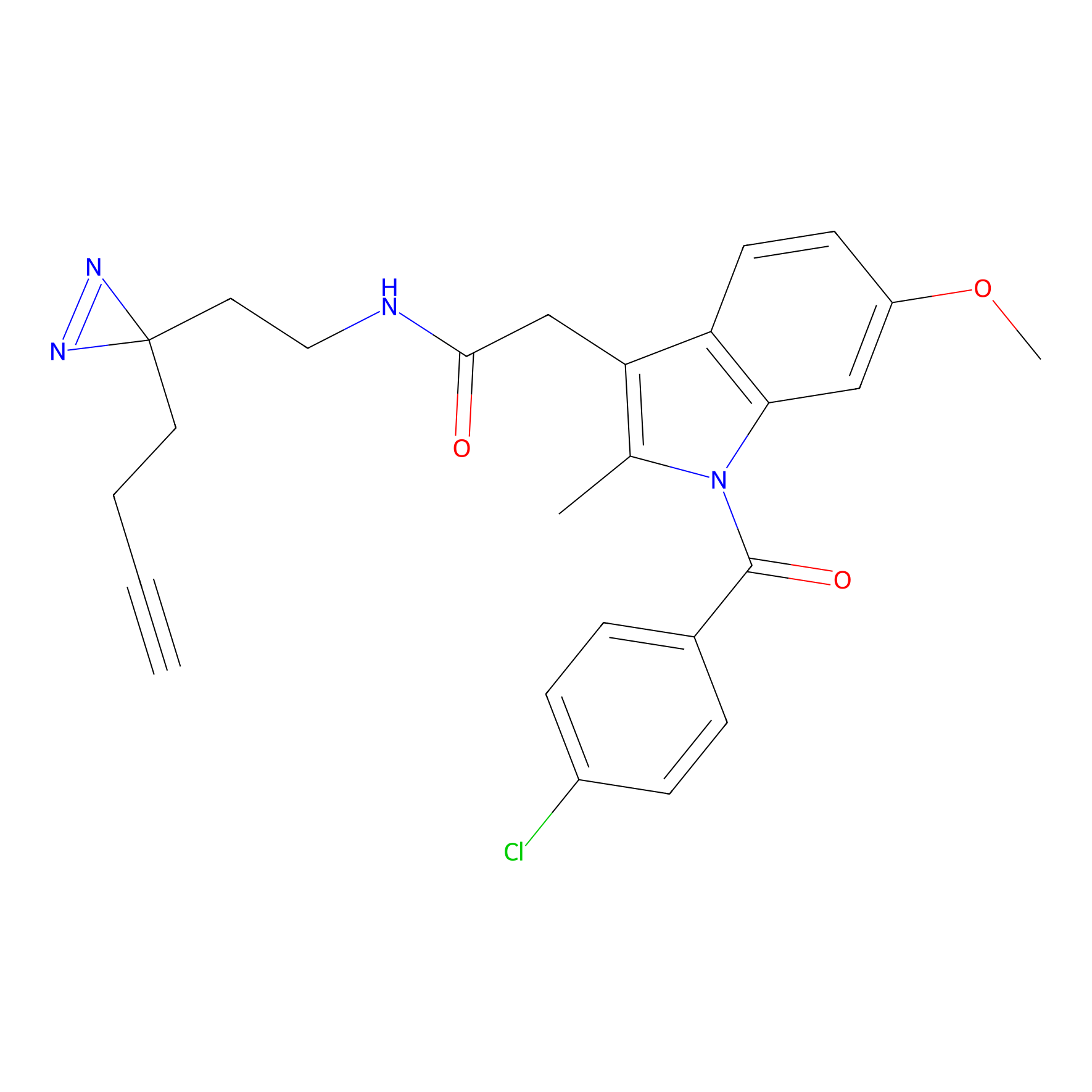 |
E71(0.00); V68(0.00); K370(0.00) | LDD0155 | [34] | |
|
DA-2 Probe Info |
 |
N.A. | LDD0070 | [35] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0068 | [36] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | DM93 | C295(7.28) | LDD0170 | [10] |
| LDCM0156 | Aniline | NCI-H1299 | 11.53 | LDD0403 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C295(0.98) | LDD0078 | [15] |
| LDCM0083 | Avasimibe | A-549 | 2.53 | LDD0143 | [32] |
| LDCM0632 | CL-Sc | Hep-G2 | C347(3.33); C347(0.99) | LDD2227 | [21] |
| LDCM0634 | CY-0357 | Hep-G2 | C347(0.79) | LDD2228 | [21] |
| LDCM0175 | Ethacrynic acid | HeLa | C4(1.11); C129(1.04); C347(0.98); C129(1.08) | LDD2210 | [11] |
| LDCM0625 | F8 | Ramos | C305(2.16); 1.08; C347(1.49); C376(0.35) | LDD2187 | [37] |
| LDCM0572 | Fragment10 | MDA-MB-231 | C347(2.19); C376(2.67); C129(1.65) | LDD1389 | [38] |
| LDCM0573 | Fragment11 | MDA-MB-231 | C347(1.68); C376(1.83); C129(1.15) | LDD1391 | [38] |
| LDCM0574 | Fragment12 | MDA-MB-231 | C347(3.38); C376(3.67); C129(2.80) | LDD1393 | [38] |
| LDCM0575 | Fragment13 | MDA-MB-231 | C347(1.08); C376(1.18) | LDD1395 | [38] |
| LDCM0576 | Fragment14 | MDA-MB-231 | C347(1.93); C376(2.31); C129(1.81) | LDD1397 | [38] |
| LDCM0577 | Fragment15 | MDA-MB-231 | C347(2.19); C376(4.05); C129(2.25) | LDD1399 | [38] |
| LDCM0579 | Fragment20 | MDA-MB-231 | C347(4.61); C376(6.14); C129(3.66) | LDD1402 | [38] |
| LDCM0580 | Fragment21 | MDA-MB-231 | C347(1.22); C376(1.48); C129(1.09) | LDD1404 | [38] |
| LDCM0581 | Fragment22 | MDA-MB-231 | C347(1.27); C376(1.38) | LDD1406 | [38] |
| LDCM0582 | Fragment23 | MDA-MB-231 | C347(2.91); C376(2.75); C129(3.55) | LDD1408 | [38] |
| LDCM0583 | Fragment24 | Ramos | C347(1.76); C376(1.60); C129(1.71) | LDD1410 | [38] |
| LDCM0584 | Fragment25 | MDA-MB-231 | C347(0.88); C376(0.86); C129(0.95) | LDD1411 | [38] |
| LDCM0585 | Fragment26 | Ramos | C347(1.10); C376(0.92); C129(0.88) | LDD1412 | [38] |
| LDCM0578 | Fragment27 | MDA-MB-231 | C347(0.92); C376(0.89); C129(0.98) | LDD1401 | [38] |
| LDCM0586 | Fragment28 | MDA-MB-231 | C347(1.00); C376(1.12); C129(0.99) | LDD1415 | [38] |
| LDCM0587 | Fragment29 | MDA-MB-231 | C347(1.32); C376(1.33); C129(1.13) | LDD1417 | [38] |
| LDCM0588 | Fragment30 | MDA-MB-231 | C347(1.01); C376(1.11); C129(0.88) | LDD1419 | [38] |
| LDCM0589 | Fragment31 | MDA-MB-231 | C347(1.71); C376(2.51); C129(2.22) | LDD1421 | [38] |
| LDCM0590 | Fragment32 | MDA-MB-231 | C347(2.79); C376(3.94); C129(1.65) | LDD1423 | [38] |
| LDCM0468 | Fragment33 | MDA-MB-231 | C347(1.00); C376(1.07) | LDD1425 | [38] |
| LDCM0592 | Fragment34 | MDA-MB-231 | C347(0.88); C376(1.03); C129(1.04) | LDD1427 | [38] |
| LDCM0593 | Fragment35 | MDA-MB-231 | C347(0.84); C376(0.87); C129(1.00) | LDD1429 | [38] |
| LDCM0594 | Fragment36 | MDA-MB-231 | C347(1.66); C376(1.79); C129(1.44) | LDD1431 | [38] |
| LDCM0595 | Fragment37 | Ramos | C347(0.91); C376(0.95) | LDD1432 | [38] |
| LDCM0596 | Fragment38 | MDA-MB-231 | C347(1.13); C376(1.19); C129(1.13) | LDD1433 | [38] |
| LDCM0597 | Fragment39 | MDA-MB-231 | C347(1.69); C376(2.36); C129(1.04) | LDD1435 | [38] |
| LDCM0566 | Fragment4 | MDA-MB-231 | C347(2.11); C376(3.67); C129(2.13) | LDD1378 | [38] |
| LDCM0598 | Fragment40 | MDA-MB-231 | C347(0.84); C376(0.89); C129(0.91) | LDD1436 | [38] |
| LDCM0599 | Fragment41 | MDA-MB-231 | C347(1.44); C376(1.48); C129(1.21) | LDD1438 | [38] |
| LDCM0600 | Fragment42 | Ramos | C347(1.14); C376(0.94); C129(0.94) | LDD1440 | [38] |
| LDCM0601 | Fragment43 | MDA-MB-231 | C347(2.55); C376(2.73); C129(1.36) | LDD1441 | [38] |
| LDCM0602 | Fragment44 | MDA-MB-231 | C347(0.80); C376(0.81); C129(0.90) | LDD1443 | [38] |
| LDCM0603 | Fragment45 | MDA-MB-231 | C347(2.78); C376(3.49); C129(2.45) | LDD1444 | [38] |
| LDCM0604 | Fragment46 | MDA-MB-231 | C347(0.99); C376(1.11); C129(0.93) | LDD1445 | [38] |
| LDCM0605 | Fragment47 | MDA-MB-231 | C347(1.28); C376(1.33); C129(1.25) | LDD1446 | [38] |
| LDCM0606 | Fragment48 | MDA-MB-231 | C347(1.14); C376(1.13); C129(1.13) | LDD1447 | [38] |
| LDCM0607 | Fragment49 | MDA-MB-231 | C347(3.25); C376(4.95) | LDD1448 | [38] |
| LDCM0608 | Fragment50 | MDA-MB-231 | C347(2.44); C376(3.66); C129(2.21) | LDD1449 | [38] |
| LDCM0427 | Fragment51 | MDA-MB-231 | C347(0.84); C376(0.97) | LDD1450 | [38] |
| LDCM0610 | Fragment52 | MDA-MB-231 | C347(1.52); C376(1.54); C129(1.17) | LDD1452 | [38] |
| LDCM0611 | Fragment53 | MDA-MB-231 | C347(0.97); C376(1.19); C129(1.27) | LDD1454 | [38] |
| LDCM0612 | Fragment54 | MDA-MB-231 | C347(1.10); C376(1.15) | LDD1456 | [38] |
| LDCM0613 | Fragment55 | MDA-MB-231 | C347(1.01); C376(1.19) | LDD1457 | [38] |
| LDCM0614 | Fragment56 | MDA-MB-231 | C347(1.21); C376(1.21); C129(1.23) | LDD1458 | [38] |
| LDCM0568 | Fragment6 | MDA-MB-231 | C347(1.27); C376(1.22); C129(1.23) | LDD1382 | [38] |
| LDCM0569 | Fragment7 | MDA-MB-231 | C347(1.96); C376(2.40); C129(1.65) | LDD1383 | [38] |
| LDCM0570 | Fragment8 | MDA-MB-231 | C347(1.95); C376(2.08); C129(1.51) | LDD1385 | [38] |
| LDCM0571 | Fragment9 | MDA-MB-231 | C347(1.73); C376(2.46); C129(1.67) | LDD1387 | [38] |
| LDCM0116 | HHS-0101 | DM93 | Y408(0.57); Y103(0.64); Y399(0.65); Y262(0.67) | LDD0264 | [13] |
| LDCM0117 | HHS-0201 | DM93 | Y399(0.53); Y161(0.53); Y103(0.60); Y312(0.60) | LDD0265 | [13] |
| LDCM0118 | HHS-0301 | DM93 | Y312(0.52); Y262(0.52); Y399(0.61); Y272(0.64) | LDD0266 | [13] |
| LDCM0119 | HHS-0401 | DM93 | Y272(0.39); Y312(0.42); Y262(0.44); Y399(0.49) | LDD0267 | [13] |
| LDCM0120 | HHS-0701 | DM93 | Y103(0.40); Y272(0.47); Y262(0.52); Y312(0.53) | LDD0268 | [13] |
| LDCM0015 | HNE | MDA-MB-231 | C347(0.88) | LDD0346 | [37] |
| LDCM0123 | JWB131 | DM93 | Y103(0.88); Y108(0.87); Y161(0.88); Y224(0.87) | LDD0285 | [12] |
| LDCM0124 | JWB142 | DM93 | Y103(0.60); Y108(0.59); Y161(0.60); Y224(0.61) | LDD0286 | [12] |
| LDCM0125 | JWB146 | DM93 | Y103(1.12); Y108(1.13); Y161(1.08); Y224(1.11) | LDD0287 | [12] |
| LDCM0126 | JWB150 | DM93 | Y103(2.30); Y108(2.21); Y161(2.31); Y224(2.46) | LDD0288 | [12] |
| LDCM0127 | JWB152 | DM93 | Y103(1.74); Y108(1.71); Y161(1.62); Y224(1.83) | LDD0289 | [12] |
| LDCM0128 | JWB198 | DM93 | Y103(1.06); Y108(1.06); Y161(1.04); Y224(1.02) | LDD0290 | [12] |
| LDCM0129 | JWB202 | DM93 | Y103(0.39); Y108(0.39); Y161(0.37); Y224(0.43) | LDD0291 | [12] |
| LDCM0130 | JWB211 | DM93 | Y103(0.91); Y108(0.89); Y161(0.90); Y224(0.89) | LDD0292 | [12] |
| LDCM0179 | JZ128 | PC-3 | C4(0.00); C347(0.00); C200(0.00) | LDD0462 | [9] |
| LDCM0022 | KB02 | HCT 116 | C347(1.63) | LDD0080 | [15] |
| LDCM0023 | KB03 | HCT 116 | C347(3.44) | LDD0081 | [15] |
| LDCM0024 | KB05 | HCT 116 | C347(1.67) | LDD0082 | [15] |
| LDCM0159 | P28 | BxPC-3 | 1.71 | LDD0409 | [8] |
| LDCM0021 | THZ1 | HCT 116 | C315(1.19); C316(1.19); C295(1.23); C305(1.23) | LDD2173 | [15] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Amyloid-beta precursor protein (APP) | APP family | P05067 | |||
| Huntingtin (HTT) | Huntingtin family | P42858 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Transcription factor Jun (JUN) | BZIP family | P05412 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Tubulin beta chain (TUBB) | Tubulin family | P07437 | |||
The Drug(s) Related To This Target
Approved
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Albendazole | Small molecular drug | DB00518 | |||
| Mebendazole | Small molecular drug | DB00643 | |||
| Vinblastine | Small molecular drug | DB00570 | |||
| Artenimol | . | DB11638 | |||
| Colchiceine | . | DB15534 | |||
Investigative
References
