Details of the Target
General Information of Target
| Target ID | LDTP06684 | |||||
|---|---|---|---|---|---|---|
| Target Name | Sister chromatid cohesion protein PDS5 homolog A (PDS5A) | |||||
| Gene Name | PDS5A | |||||
| Gene ID | 23244 | |||||
| Synonyms |
KIAA0648; PDS5; Sister chromatid cohesion protein PDS5 homolog A; Cell proliferation-inducing gene 54 protein; Sister chromatid cohesion protein 112; SCC-112 |
|||||
| 3D Structure | ||||||
| Sequence |
MDFTAQPKPATALCGVVSADGKIAYPPGVKEITDKITTDEMIKRLKMVVKTFMDMDQDSE
DEKQQYLPLALHLASEFFLRNPNKDVRLLVACCLADIFRIYAPEAPYTSHDKLKDIFLFI TRQLKGLEDTKSPQFNRYFYLLENLAWVKSYNICFELEDCNEIFIQLFRTLFSVINNSHN KKVQMHMLDLMSSIIMEGDGVTQELLDSILINLIPAHKNLNKQSFDLAKVLLKRTVQTIE ACIANFFNQVLVLGRSSVSDLSEHVFDLIQELFAIDPHLLLSVMPQLEFKLKSNDGEERL AVVRLLAKLFGSKDSDLATQNRPLWQCFLGRFNDIHVPVRLESVKFASHCLMNHPDLAKD LTEYLKVRSHDPEEAIRHDVIVTIITAAKRDLALVNDQLLGFVRERTLDKRWRVRKEAMM GLAQLYKKYCLHGEAGKEAAEKVSWIKDKLLHIYYQNSIDDKLLVEKIFAQYLVPHNLET EERMKCLYYLYASLDPNAVKALNEMWKCQNMLRSHVRELLDLHKQPTSEANCSAMFGKLM TIAKNLPDPGKAQDFVKKFNQVLGDDEKLRSQLELLISPTCSCKQADICVREIARKLANP KQPTNPFLEMVKFLLERIAPVHIDSEAISALVKLMNKSIEGTADDEEEGVSPDTAIRSGL ELLKVLSFTHPTSFHSAETYESLLQCLRMEDDKVAEAAIQIFRNTGHKIETDLPQIRSTL IPILHQKAKRGTPHQAKQAVHCIHAIFTNKEVQLAQIFEPLSRSLNADVPEQLITPLVSL GHISMLAPDQFASPMKSVVANFIVKDLLMNDRSTGEKNGKLWSPDEEVSPEVLAKVQAIK LLVRWLLGMKNNQSKSANSTLRLLSAMLVSEGDLTEQKRISKSDMSRLRLAAGSAIMKLA QEPCYHEIITPEQFQLCALVINDECYQVRQIFAQKLHKALVKLLLPLEYMAIFALCAKDP VKERRAHARQCLLKNISIRREYIKQNPMATEKLLSLLPEYVVPYMIHLLAHDPDFTRSQD VDQLRDIKECLWFMLEVLMTKNENNSHAFMKKMAENIKLTRDAQSPDESKTNEKLYTVCD VALCVINSKSALCNADSPKDPVLPMKFFTQPEKDFCNDKSYISEETRVLLLTGKPKPAGV LGAVNKPLSATGRKPYVRSTGTETGSNINVNSELNPSTGNRSREQSSEAAETGVSENEEN PVRIISVTPVKNIDPVKNKEINSDQATQGNISSDRGKKRTVTAAGAENIQQKTDEKVDES GPPAPSKPRRGRRPKSESQGNATKNDDLNKPINKGRKRAAVGQESPGGLEAGNAKAPKLQ DLAKKAAPAERQIDLQR |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
PDS5 family
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Probable regulator of sister chromatid cohesion in mitosis which may stabilize cohesin complex association with chromatin. May couple sister chromatid cohesion during mitosis to DNA replication. Cohesion ensures that chromosome partitioning is accurate in both meiotic and mitotic cells and plays an important role in DNA repair.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| CHL1 | SNV: p.R80W | DBIA Probe Info | |||
| GB2 | SNV: p.E481Q | DBIA Probe Info | |||
| HEC1 | SNV: p.R377C | DBIA Probe Info | |||
| K562 | SNV: p.D923N | FFF probe13 Probe Info | |||
| LS180 | SNV: p.R889Ter | DBIA Probe Info | |||
| MEWO | SNV: p.P606S | DBIA Probe Info | |||
| MOLT4 | SNV: p.R979C | IA-alkyne Probe Info | |||
| NCIH358 | SNV: p.Q553Ter | DBIA Probe Info | |||
| RKO | Insertion: p.T1253NfsTer3 SNV: p.A989V |
DBIA Probe Info | |||
| SF295 | SNV: p.D20N | DBIA Probe Info | |||
| TOV21G | SNV: p.E609Ter | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
12.38 | LDD0402 | [1] | |
|
A-EBA Probe Info |
 |
3.36 | LDD0215 | [2] | |
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [3] | |
|
CY4 Probe Info |
 |
100.00 | LDD0244 | [3] | |
|
N1 Probe Info |
 |
100.00 | LDD0242 | [3] | |
|
TH211 Probe Info |
 |
Y472(20.00); Y1121(8.10); Y680(7.57); Y107(5.86) | LDD0257 | [4] | |
|
TH216 Probe Info |
 |
Y472(17.23); Y429(15.65); Y1121(7.40) | LDD0259 | [4] | |
|
STPyne Probe Info |
 |
K229(10.00); K308(10.00); K557(2.30); K558(4.94) | LDD0277 | [5] | |
|
ONAyne Probe Info |
 |
K962(5.00) | LDD0275 | [5] | |
|
Probe 1 Probe Info |
 |
Y1121(26.73) | LDD3495 | [6] | |
|
JZ128-DTB Probe Info |
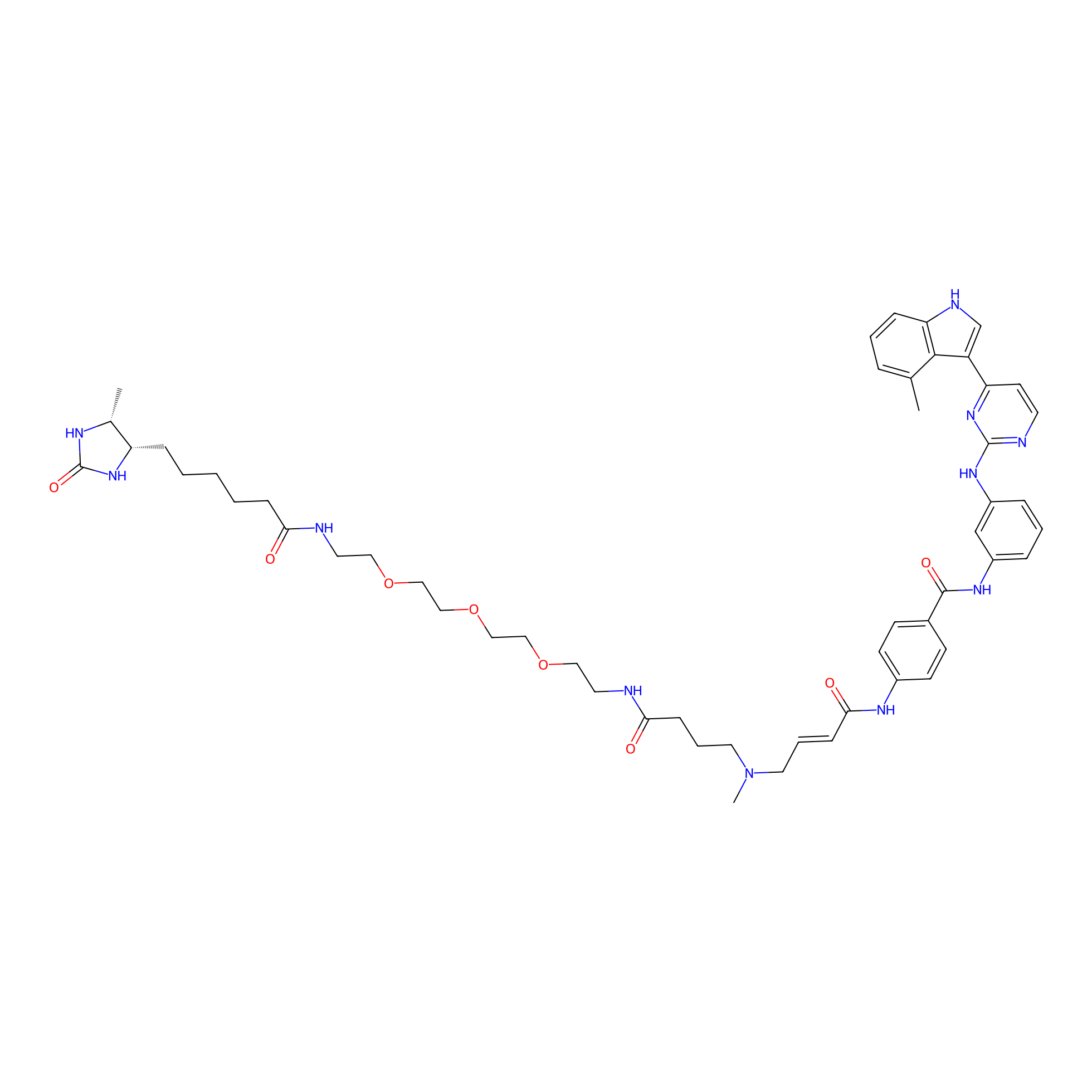 |
C581(0.00); C532(0.00) | LDD0462 | [7] | |
|
BTD Probe Info |
 |
C1093(1.21) | LDD1700 | [8] | |
|
AHL-Pu-1 Probe Info |
 |
C532(2.13); C327(2.48) | LDD0168 | [9] | |
|
Alkyne-RA190 Probe Info |
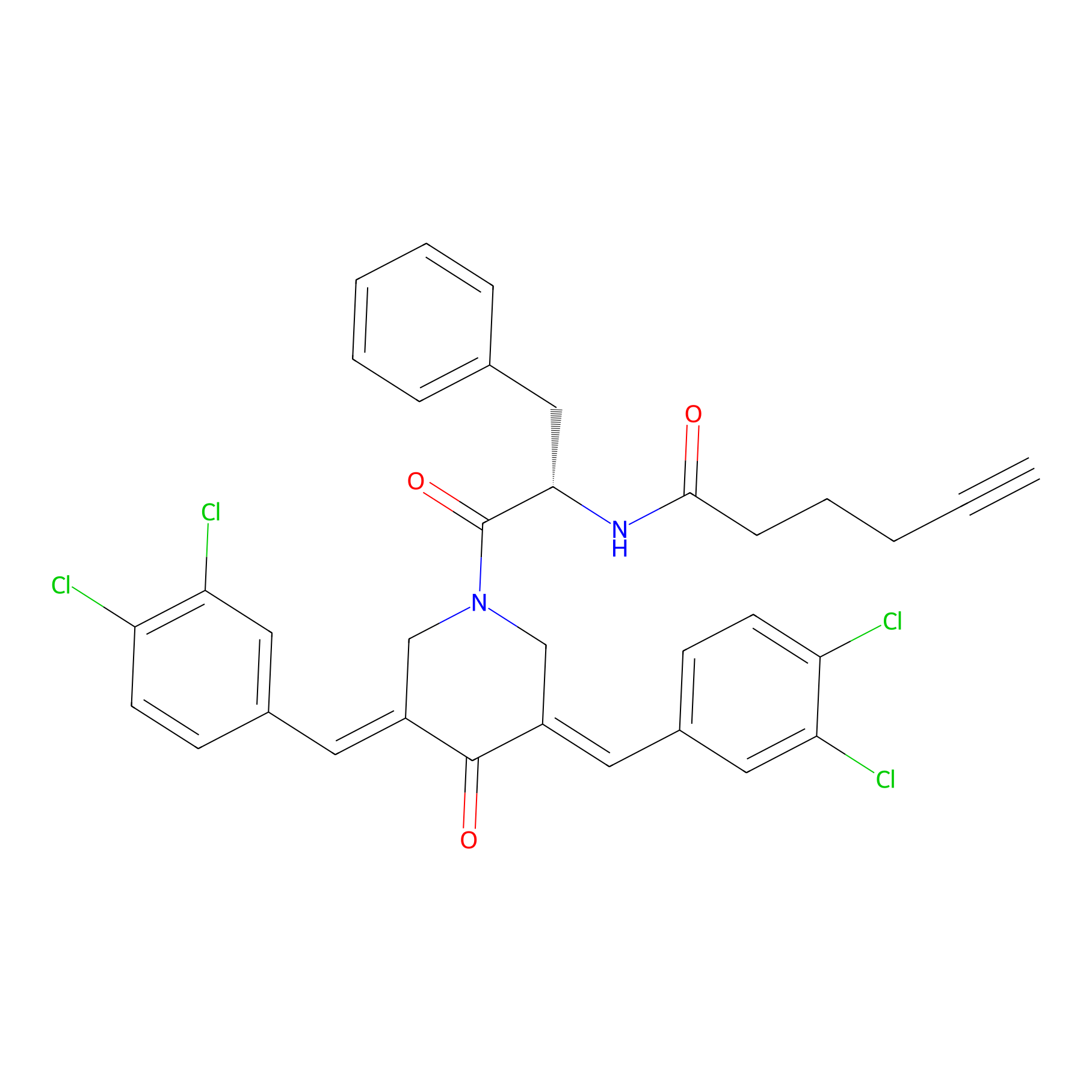 |
2.61 | LDD0300 | [10] | |
|
EA-probe Probe Info |
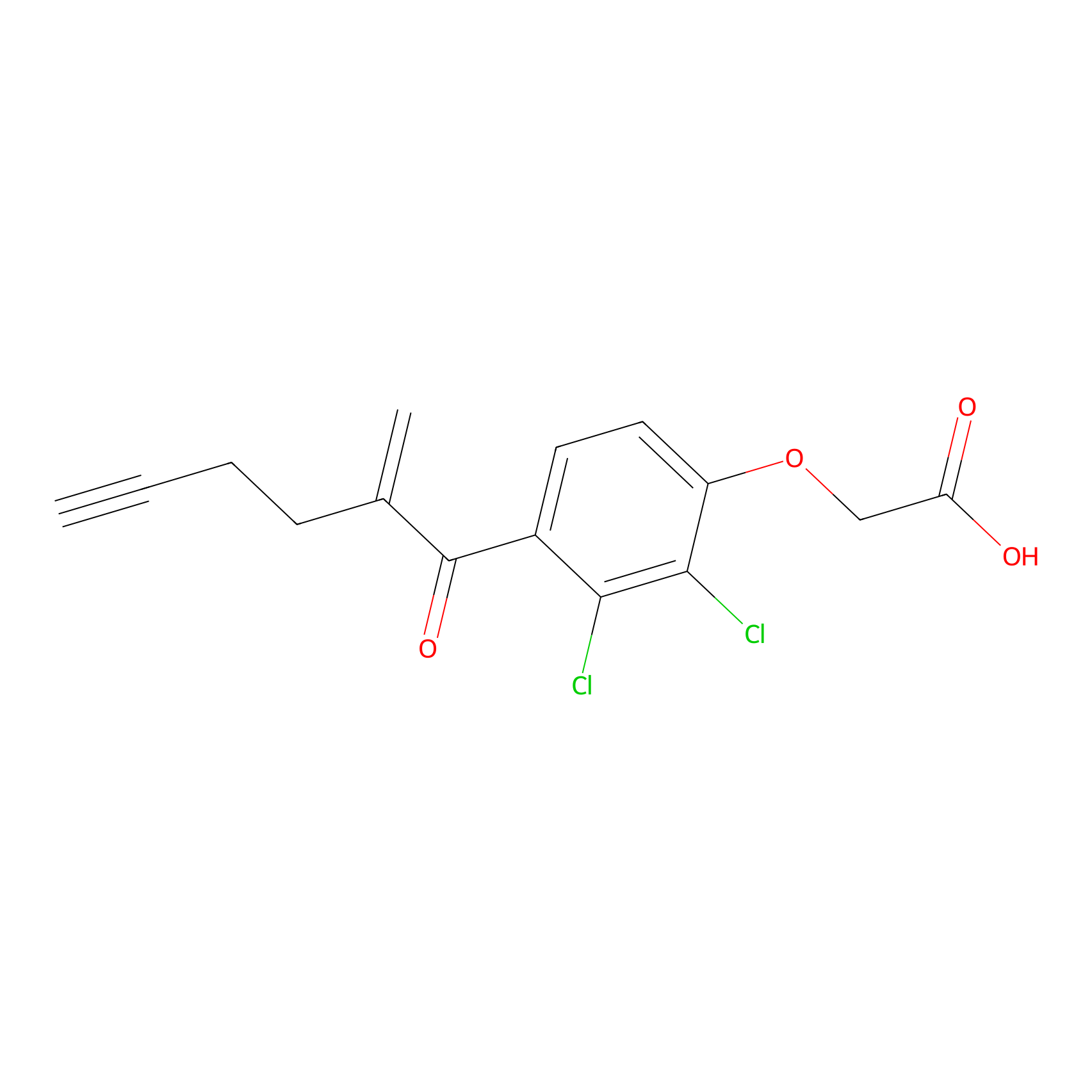 |
N.A. | LDD0440 | [11] | |
|
DBIA Probe Info |
 |
C532(1.23); C430(0.86) | LDD0078 | [12] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C327(0.00); C532(0.00); C350(0.00); C1116(0.00) | LDD0038 | [13] | |
|
IA-alkyne Probe Info |
 |
C1116(0.00); C350(0.00); C532(0.00); C92(0.00) | LDD0032 | [14] | |
|
IPIAA_H Probe Info |
 |
N.A. | LDD0030 | [15] | |
|
IPIAA_L Probe Info |
 |
C327(0.00); C686(0.00) | LDD0031 | [15] | |
|
Lodoacetamide azide Probe Info |
 |
C486(0.00); C327(0.00); C532(0.00); C350(0.00) | LDD0037 | [13] | |
|
NAIA_4 Probe Info |
 |
C327(0.00); C532(0.00) | LDD2226 | [16] | |
|
TFBX Probe Info |
 |
C583(0.00); C581(0.00) | LDD0027 | [17] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [18] | |
|
Compound 10 Probe Info |
 |
C14(0.00); C327(0.00); C532(0.00) | LDD2216 | [19] | |
|
Compound 11 Probe Info |
 |
C14(0.00); C327(0.00); C532(0.00) | LDD2213 | [19] | |
|
ENE Probe Info |
 |
N.A. | LDD0006 | [18] | |
|
IPM Probe Info |
 |
C532(0.00); C327(0.00); C430(0.00); C583(0.00) | LDD0005 | [18] | |
|
NHS Probe Info |
 |
K558(0.00); K366(0.00) | LDD0010 | [18] | |
|
PF-06672131 Probe Info |
 |
N.A. | LDD0152 | [20] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [18] | |
|
Acrolein Probe Info |
 |
C14(0.00); H336(0.00); H622(0.00); C430(0.00) | LDD0217 | [21] | |
|
Methacrolein Probe Info |
 |
C430(0.00); C14(0.00) | LDD0218 | [21] | |
|
W1 Probe Info |
 |
C327(0.00); C581(0.00); C583(0.00); C1093(0.00) | LDD0236 | [22] | |
|
AOyne Probe Info |
 |
13.80 | LDD0443 | [23] | |
|
NAIA_5 Probe Info |
 |
C486(0.00); C327(0.00); C350(0.00); C1116(0.00) | LDD2223 | [16] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C193 Probe Info |
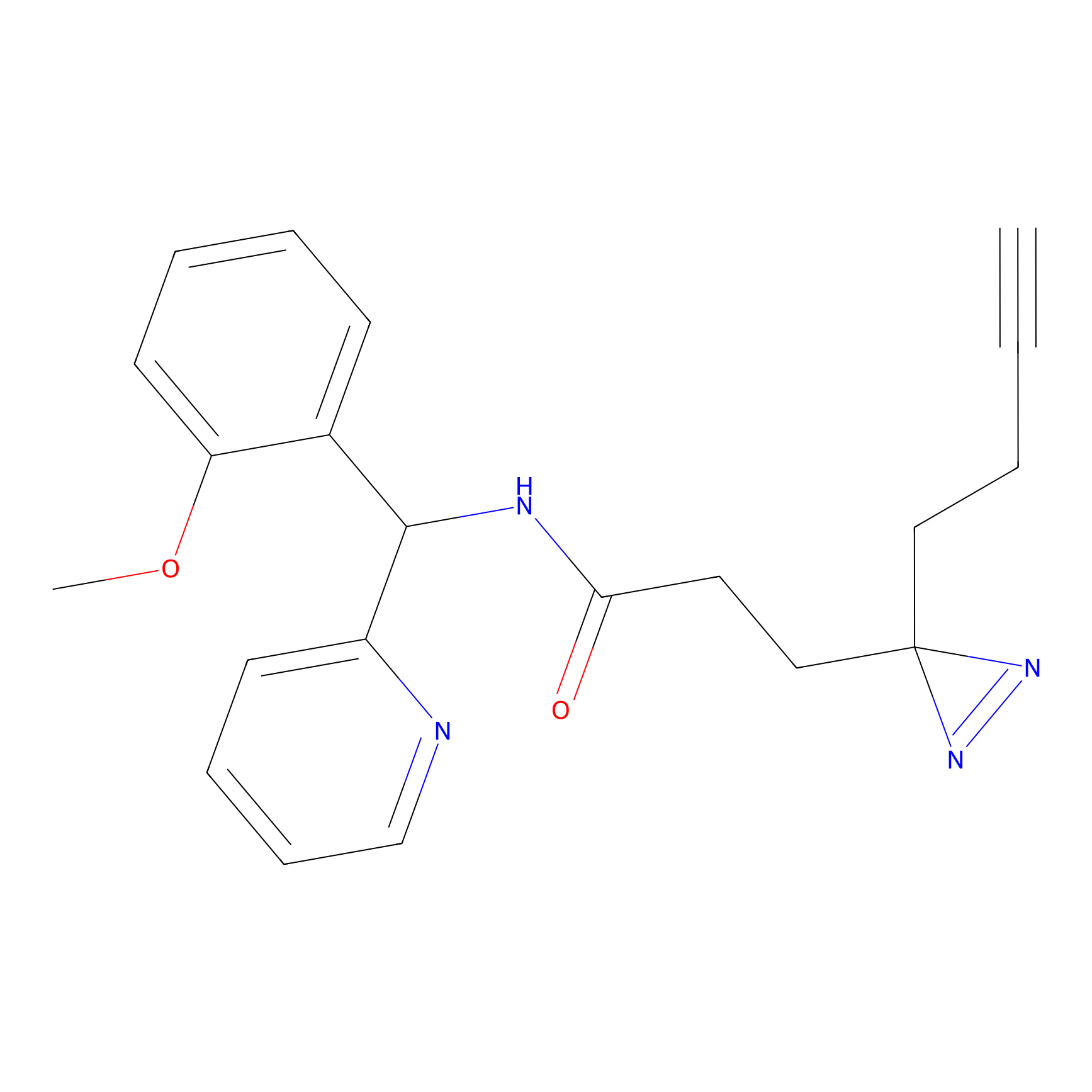 |
5.54 | LDD1869 | [24] | |
|
C380 Probe Info |
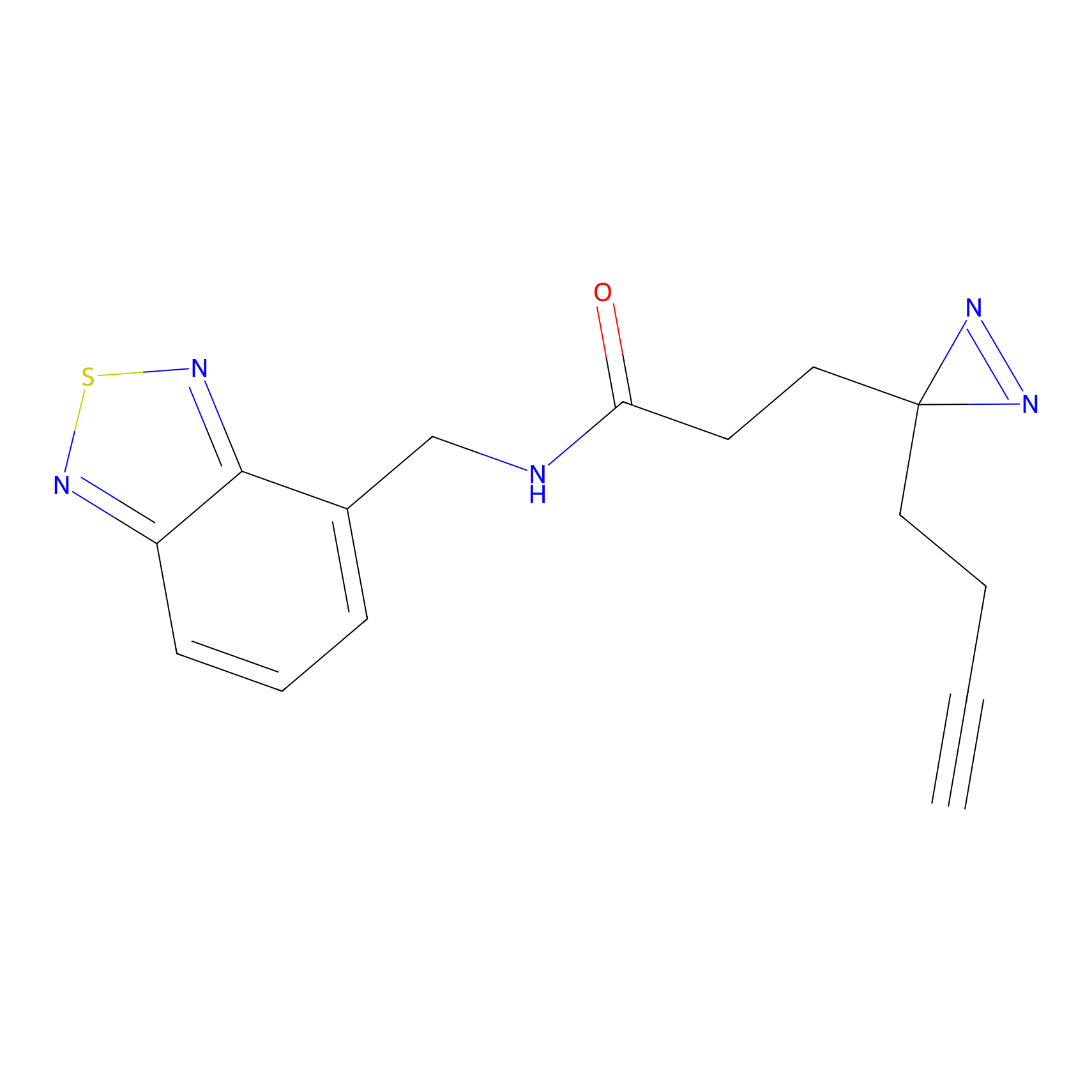 |
4.99 | LDD2039 | [24] | |
|
C420 Probe Info |
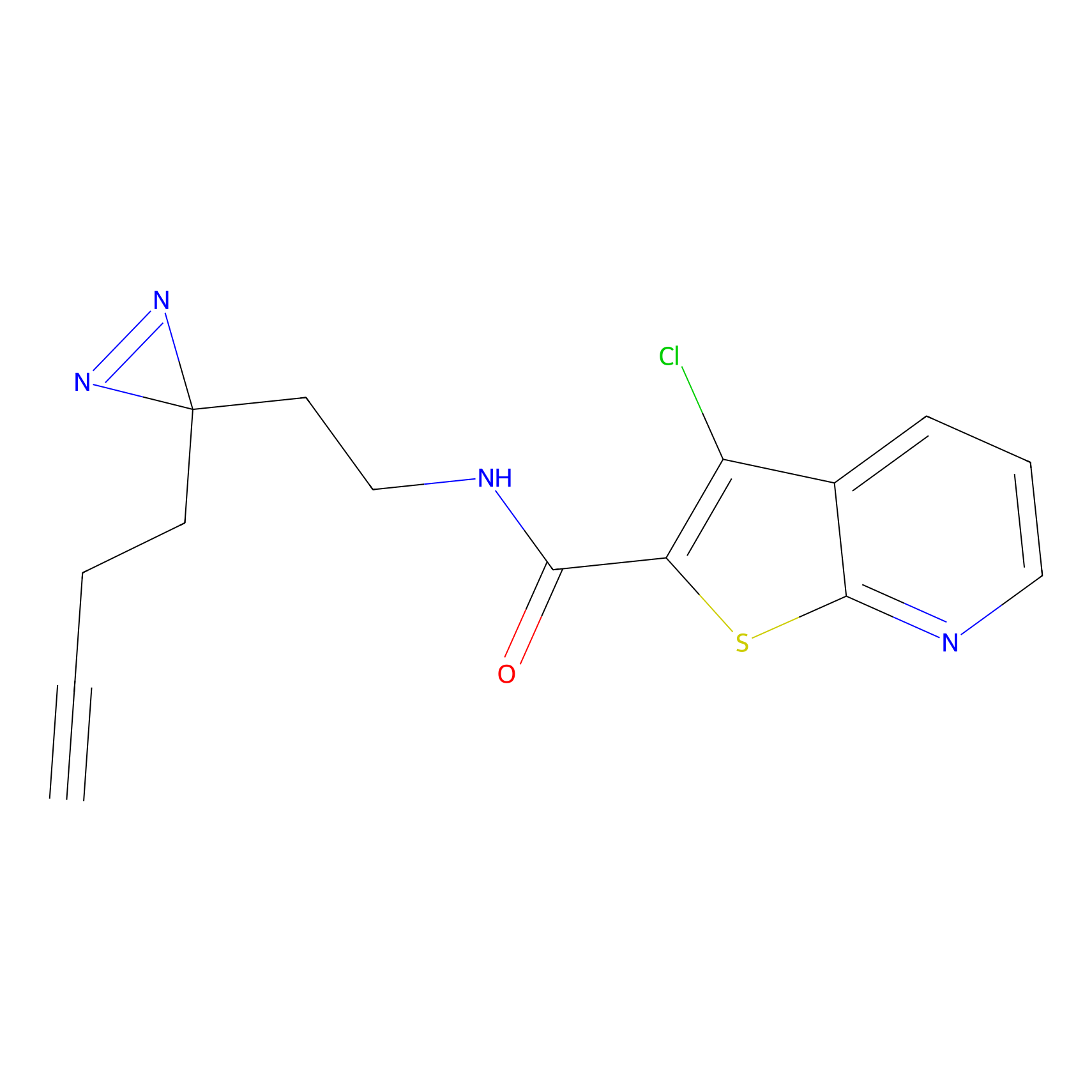 |
12.38 | LDD2075 | [24] | |
|
FFF probe11 Probe Info |
 |
20.00 | LDD0471 | [25] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [25] | |
|
FFF probe3 Probe Info |
 |
20.00 | LDD0465 | [25] | |
|
FFF probe9 Probe Info |
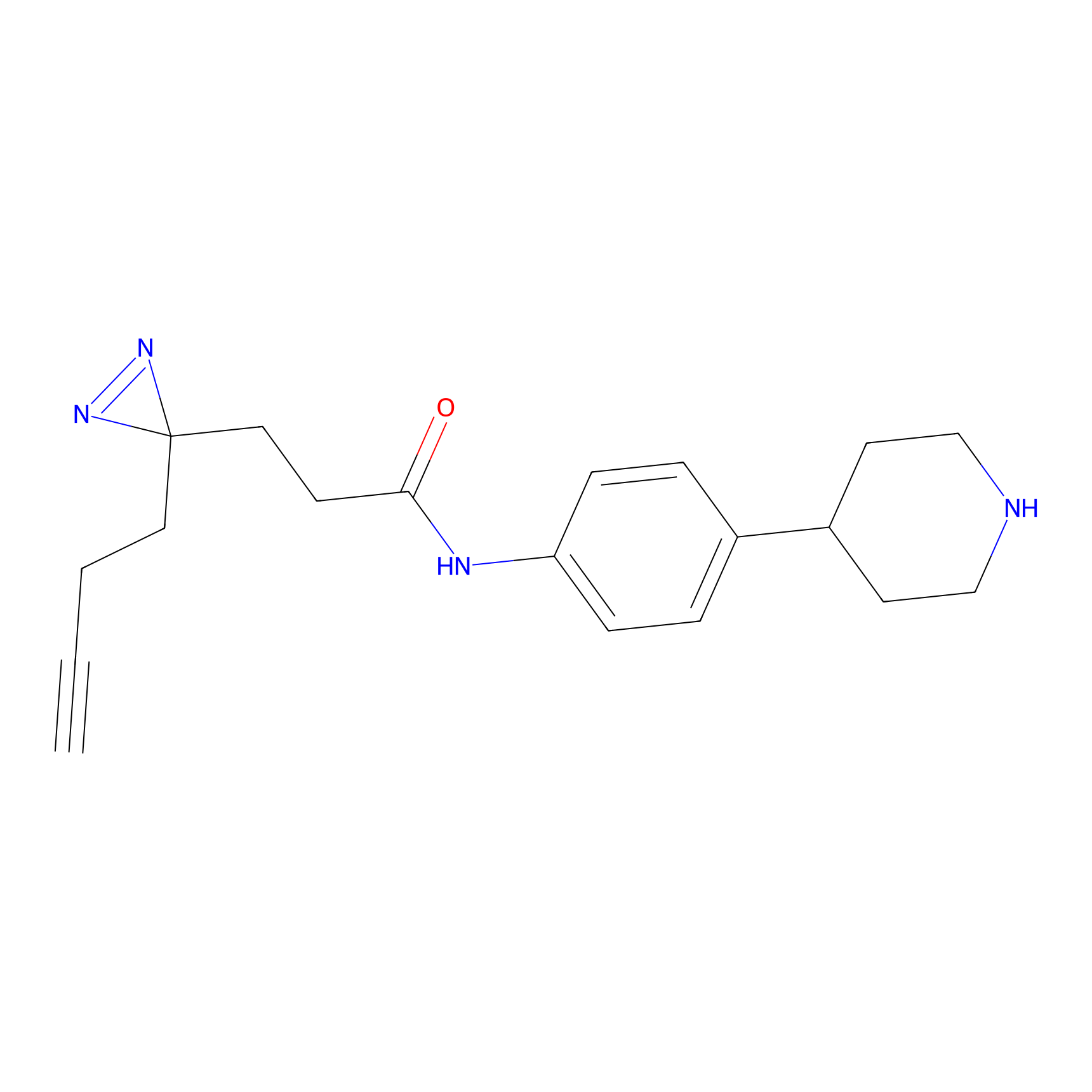 |
7.14 | LDD0470 | [25] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [26] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [27] | |
|
OEA-DA Probe Info |
 |
3.26 | LDD0046 | [28] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C430(0.79); C14(0.75) | LDD2142 | [8] |
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C430(1.22); C14(0.67) | LDD2112 | [8] |
| LDCM0502 | 1-(Cyanoacetyl)piperidine | MDA-MB-231 | C430(0.79); C14(0.64) | LDD2095 | [8] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C430(1.67); C589(1.00) | LDD2117 | [8] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C430(1.61); C589(1.42); C350(0.88) | LDD2152 | [8] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C14(0.82) | LDD2103 | [8] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C430(0.83); C14(0.50) | LDD2132 | [8] |
| LDCM0538 | 4-(Cyanoacetyl)morpholine | MDA-MB-231 | C14(0.75) | LDD2131 | [8] |
| LDCM0025 | 4SU-RNA | HEK-293T | C532(2.13); C327(2.48) | LDD0168 | [9] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C532(2.05); C327(2.79); C1093(2.01) | LDD0169 | [9] |
| LDCM0214 | AC1 | HCT 116 | C327(0.86); C1079(0.78); C1084(0.78); C1093(1.28) | LDD0531 | [12] |
| LDCM0215 | AC10 | HCT 116 | C327(1.01); C1079(0.95); C1084(0.95); C1093(1.12) | LDD0532 | [12] |
| LDCM0216 | AC100 | HCT 116 | C327(1.06); C1079(0.62); C1084(0.62); C1093(1.13) | LDD0533 | [12] |
| LDCM0217 | AC101 | HCT 116 | C327(1.05); C1079(0.67); C1084(0.67); C1093(1.02) | LDD0534 | [12] |
| LDCM0218 | AC102 | HCT 116 | C327(0.86); C1079(0.72); C1084(0.72); C1093(0.98) | LDD0535 | [12] |
| LDCM0219 | AC103 | HCT 116 | C327(1.11); C1079(0.80); C1084(0.80); C1093(1.24) | LDD0536 | [12] |
| LDCM0220 | AC104 | HCT 116 | C327(1.18); C1079(0.82); C1084(0.82); C1093(1.14) | LDD0537 | [12] |
| LDCM0221 | AC105 | HCT 116 | C327(1.05); C1079(0.72); C1084(0.72); C1093(1.14) | LDD0538 | [12] |
| LDCM0222 | AC106 | HCT 116 | C327(1.13); C1079(0.71); C1084(0.71); C1093(1.12) | LDD0539 | [12] |
| LDCM0223 | AC107 | HCT 116 | C327(1.16); C1079(0.80); C1084(0.80); C1093(1.19) | LDD0540 | [12] |
| LDCM0224 | AC108 | HCT 116 | C327(1.23); C1079(0.63); C1084(0.63); C1093(1.28) | LDD0541 | [12] |
| LDCM0225 | AC109 | HCT 116 | C327(0.90); C1079(0.65); C1084(0.65); C1093(1.21) | LDD0542 | [12] |
| LDCM0226 | AC11 | HCT 116 | C327(1.02); C1079(0.87); C1084(0.87); C1093(1.13) | LDD0543 | [12] |
| LDCM0227 | AC110 | HCT 116 | C327(0.99); C1079(0.68); C1084(0.68); C1093(1.15) | LDD0544 | [12] |
| LDCM0228 | AC111 | HCT 116 | C327(1.11); C1079(0.62); C1084(0.62); C1093(1.12) | LDD0545 | [12] |
| LDCM0229 | AC112 | HCT 116 | C327(1.03); C1079(0.71); C1084(0.71); C1093(1.02) | LDD0546 | [12] |
| LDCM0230 | AC113 | HCT 116 | C327(0.92); C1079(1.11); C1084(1.11); C1093(0.99) | LDD0547 | [12] |
| LDCM0231 | AC114 | HCT 116 | C327(0.91); C1079(1.04); C1084(1.04); C1093(1.02) | LDD0548 | [12] |
| LDCM0232 | AC115 | HCT 116 | C327(0.89); C1079(1.20); C1084(1.20); C1093(1.13) | LDD0549 | [12] |
| LDCM0233 | AC116 | HCT 116 | C327(0.90); C1079(1.15); C1084(1.15); C1093(1.40) | LDD0550 | [12] |
| LDCM0234 | AC117 | HCT 116 | C327(1.06); C1079(1.08); C1084(1.08); C1093(1.17) | LDD0551 | [12] |
| LDCM0235 | AC118 | HCT 116 | C327(0.89); C1079(1.05); C1084(1.05); C1093(1.13) | LDD0552 | [12] |
| LDCM0236 | AC119 | HCT 116 | C327(0.82); C1079(1.08); C1084(1.08); C1093(1.06) | LDD0553 | [12] |
| LDCM0237 | AC12 | HCT 116 | C327(1.00); C1079(1.01); C1084(1.01); C1093(1.02) | LDD0554 | [12] |
| LDCM0238 | AC120 | HCT 116 | C327(0.85); C1079(1.26); C1084(1.26); C1093(1.04) | LDD0555 | [12] |
| LDCM0239 | AC121 | HCT 116 | C327(0.81); C1079(0.94); C1084(0.94); C1093(0.93) | LDD0556 | [12] |
| LDCM0240 | AC122 | HCT 116 | C327(0.93); C1079(1.04); C1084(1.04); C1093(1.02) | LDD0557 | [12] |
| LDCM0241 | AC123 | HCT 116 | C327(0.88); C1079(1.05); C1084(1.05); C1093(0.88) | LDD0558 | [12] |
| LDCM0242 | AC124 | HCT 116 | C327(0.85); C1079(1.00); C1084(1.00); C1093(0.96) | LDD0559 | [12] |
| LDCM0243 | AC125 | HCT 116 | C327(1.22); C1079(0.96); C1084(0.96); C1093(1.03) | LDD0560 | [12] |
| LDCM0244 | AC126 | HCT 116 | C327(0.87); C1079(1.33); C1084(1.33); C1093(1.04) | LDD0561 | [12] |
| LDCM0245 | AC127 | HCT 116 | C327(1.05); C1079(1.07); C1084(1.07); C1093(1.21) | LDD0562 | [12] |
| LDCM0246 | AC128 | HCT 116 | C327(1.28); C1079(1.40); C1084(1.40); C1093(1.05) | LDD0563 | [12] |
| LDCM0247 | AC129 | HCT 116 | C327(1.27); C1079(0.84); C1084(0.84); C1093(0.95) | LDD0564 | [12] |
| LDCM0249 | AC130 | HCT 116 | C327(1.28); C1079(1.06); C1084(1.06); C1093(1.23) | LDD0566 | [12] |
| LDCM0250 | AC131 | HCT 116 | C327(1.19); C1079(0.97); C1084(0.97); C1093(1.05) | LDD0567 | [12] |
| LDCM0251 | AC132 | HCT 116 | C327(1.50); C1079(0.83); C1084(0.83); C1093(1.09) | LDD0568 | [12] |
| LDCM0252 | AC133 | HCT 116 | C327(1.36); C1079(0.94); C1084(0.94); C1093(1.07) | LDD0569 | [12] |
| LDCM0253 | AC134 | HCT 116 | C327(1.11); C1079(0.98); C1084(0.98); C1093(1.16) | LDD0570 | [12] |
| LDCM0254 | AC135 | HCT 116 | C327(1.24); C1079(0.99); C1084(0.99); C1093(1.23) | LDD0571 | [12] |
| LDCM0255 | AC136 | HCT 116 | C327(1.11); C1079(0.91); C1084(0.91); C1093(1.09) | LDD0572 | [12] |
| LDCM0256 | AC137 | HCT 116 | C327(1.41); C1079(0.82); C1084(0.82); C1093(0.99) | LDD0573 | [12] |
| LDCM0257 | AC138 | HCT 116 | C327(1.87); C1079(0.89); C1084(0.89); C1093(1.14) | LDD0574 | [12] |
| LDCM0258 | AC139 | HCT 116 | C327(1.44); C1079(0.81); C1084(0.81); C1093(1.41) | LDD0575 | [12] |
| LDCM0259 | AC14 | HCT 116 | C327(1.02); C1079(0.99); C1084(0.99); C1093(0.99) | LDD0576 | [12] |
| LDCM0260 | AC140 | HCT 116 | C327(1.50); C1079(0.88); C1084(0.88); C1093(1.19) | LDD0577 | [12] |
| LDCM0261 | AC141 | HCT 116 | C327(1.72); C1079(0.80); C1084(0.80); C1093(1.21) | LDD0578 | [12] |
| LDCM0262 | AC142 | HCT 116 | C327(1.18); C1079(0.76); C1084(0.76); C1093(1.04) | LDD0579 | [12] |
| LDCM0263 | AC143 | HCT 116 | C327(0.95); C1079(0.88); C1084(0.88); C581(0.88) | LDD0580 | [12] |
| LDCM0264 | AC144 | HCT 116 | C430(0.71); C583(0.85); C581(0.85); C742(0.93) | LDD0581 | [12] |
| LDCM0265 | AC145 | HCT 116 | C430(0.87); C1079(0.89); C1084(0.89); C583(0.93) | LDD0582 | [12] |
| LDCM0266 | AC146 | HCT 116 | C583(0.83); C581(0.85); C430(0.90); C1079(0.95) | LDD0583 | [12] |
| LDCM0267 | AC147 | HCT 116 | C583(0.93); C430(0.97); C581(0.97); C14(1.01) | LDD0584 | [12] |
| LDCM0268 | AC148 | HCT 116 | C742(0.97); C583(1.02); C581(1.04); C589(1.17) | LDD0585 | [12] |
| LDCM0269 | AC149 | HCT 116 | C581(0.84); C583(0.84); C589(1.03); C742(1.06) | LDD0586 | [12] |
| LDCM0270 | AC15 | HCT 116 | C742(0.79); C581(0.88); C430(0.88); C583(0.93) | LDD0587 | [12] |
| LDCM0271 | AC150 | HCT 116 | C1079(0.84); C1084(0.84); C583(0.89); C581(0.90) | LDD0588 | [12] |
| LDCM0272 | AC151 | HCT 116 | C583(0.84); C581(0.85); C742(0.93); C589(0.97) | LDD0589 | [12] |
| LDCM0273 | AC152 | HCT 116 | C742(0.89); C583(0.97); C430(0.98); C581(0.98) | LDD0590 | [12] |
| LDCM0274 | AC153 | HCT 116 | C742(0.94); C583(0.97); C581(0.99); C589(1.11) | LDD0591 | [12] |
| LDCM0621 | AC154 | HCT 116 | C327(1.13); C1079(1.01); C1084(1.01); C1093(1.05) | LDD2158 | [12] |
| LDCM0622 | AC155 | HCT 116 | C327(1.10); C1079(1.08); C1084(1.08); C1093(1.10) | LDD2159 | [12] |
| LDCM0623 | AC156 | HCT 116 | C327(0.99); C1079(0.96); C1084(0.96); C1093(1.08) | LDD2160 | [12] |
| LDCM0624 | AC157 | HCT 116 | C327(1.05); C1079(1.08); C1084(1.08); C1093(0.92) | LDD2161 | [12] |
| LDCM0276 | AC17 | HCT 116 | C904(0.79); C917(0.79); C925(0.79); C430(0.80) | LDD0593 | [12] |
| LDCM0277 | AC18 | HCT 116 | C904(0.73); C917(0.73); C925(0.73); C1079(0.84) | LDD0594 | [12] |
| LDCM0278 | AC19 | HCT 116 | C430(0.81); C583(0.91); C1079(0.96); C1084(0.96) | LDD0595 | [12] |
| LDCM0279 | AC2 | HCT 116 | C327(0.78); C1079(0.80); C1084(0.80); C904(0.84) | LDD0596 | [12] |
| LDCM0280 | AC20 | HCT 116 | C430(0.78); C1079(0.84); C1084(0.84); C583(1.08) | LDD0597 | [12] |
| LDCM0281 | AC21 | HCT 116 | C904(0.69); C917(0.69); C925(0.69); C430(0.81) | LDD0598 | [12] |
| LDCM0282 | AC22 | HCT 116 | C904(0.72); C917(0.72); C925(0.72); C430(0.83) | LDD0599 | [12] |
| LDCM0283 | AC23 | HCT 116 | C430(0.78); C904(0.79); C917(0.79); C925(0.79) | LDD0600 | [12] |
| LDCM0284 | AC24 | HCT 116 | C430(0.77); C904(0.98); C917(0.98); C925(0.98) | LDD0601 | [12] |
| LDCM0285 | AC25 | HCT 116 | C327(0.73); C14(0.86); C583(0.94); C581(0.94) | LDD0602 | [12] |
| LDCM0286 | AC26 | HCT 116 | C742(0.71); C581(0.82); C589(0.87); C583(0.88) | LDD0603 | [12] |
| LDCM0287 | AC27 | HCT 116 | C581(0.78); C742(0.83); C589(0.83); C327(0.86) | LDD0604 | [12] |
| LDCM0288 | AC28 | HCT 116 | C742(0.57); C581(0.82); C583(0.88); C589(0.97) | LDD0605 | [12] |
| LDCM0289 | AC29 | HCT 116 | C327(0.74); C742(0.76); C581(0.76); C583(0.88) | LDD0606 | [12] |
| LDCM0290 | AC3 | HCT 116 | C327(0.73); C486(0.91); C1079(0.91); C1084(0.91) | LDD0607 | [12] |
| LDCM0291 | AC30 | HCT 116 | C581(0.78); C742(0.78); C583(0.83); C589(0.89) | LDD0608 | [12] |
| LDCM0292 | AC31 | HCT 116 | C742(0.68); C581(0.76); C583(0.86); C327(0.88) | LDD0609 | [12] |
| LDCM0293 | AC32 | HCT 116 | C742(0.67); C581(0.78); C589(0.87); C583(0.90) | LDD0610 | [12] |
| LDCM0294 | AC33 | HCT 116 | C581(0.75); C583(0.79); C327(0.86); C589(0.89) | LDD0611 | [12] |
| LDCM0295 | AC34 | HCT 116 | C581(0.82); C14(0.87); C327(0.88); C583(0.95) | LDD0612 | [12] |
| LDCM0296 | AC35 | HCT 116 | C532(0.92); C581(0.96); C1093(0.97); C14(1.03) | LDD0613 | [12] |
| LDCM0297 | AC36 | HCT 116 | C581(0.86); C532(0.93); C430(0.97); C583(0.99) | LDD0614 | [12] |
| LDCM0298 | AC37 | HCT 116 | C581(0.85); C532(0.90); C14(0.94); C583(0.95) | LDD0615 | [12] |
| LDCM0299 | AC38 | HCT 116 | C430(0.89); C532(0.94); C581(0.96); C1093(0.99) | LDD0616 | [12] |
| LDCM0300 | AC39 | HCT 116 | C327(0.77); C14(0.96); C1079(1.03); C1084(1.03) | LDD0617 | [12] |
| LDCM0301 | AC4 | HCT 116 | C904(0.73); C917(0.73); C925(0.73); C1079(0.87) | LDD0618 | [12] |
| LDCM0302 | AC40 | HCT 116 | C581(0.92); C327(0.93); C430(0.98); C14(1.00) | LDD0619 | [12] |
| LDCM0303 | AC41 | HCT 116 | C581(0.85); C1079(0.94); C1084(0.94); C583(0.95) | LDD0620 | [12] |
| LDCM0304 | AC42 | HCT 116 | C581(0.93); C589(0.95); C583(0.98); C430(0.98) | LDD0621 | [12] |
| LDCM0305 | AC43 | HCT 116 | C581(0.88); C583(0.93); C589(0.97); C430(1.00) | LDD0622 | [12] |
| LDCM0306 | AC44 | HCT 116 | C583(0.97); C581(0.98); C532(1.00); C430(1.03) | LDD0623 | [12] |
| LDCM0307 | AC45 | HCT 116 | C581(0.88); C583(0.92); C14(1.00); C430(1.13) | LDD0624 | [12] |
| LDCM0308 | AC46 | HCT 116 | C589(0.88); C1079(0.90); C1084(0.90); C583(0.95) | LDD0625 | [12] |
| LDCM0309 | AC47 | HCT 116 | C327(0.79); C589(0.92); C486(0.92); C583(0.95) | LDD0626 | [12] |
| LDCM0310 | AC48 | HCT 116 | C486(0.73); C1079(0.81); C1084(0.81); C583(0.97) | LDD0627 | [12] |
| LDCM0311 | AC49 | HCT 116 | C589(0.89); C581(1.09); C583(1.13); C486(1.27) | LDD0628 | [12] |
| LDCM0312 | AC5 | HCT 116 | C1079(0.77); C1084(0.77); C904(0.77); C917(0.77) | LDD0629 | [12] |
| LDCM0313 | AC50 | HCT 116 | C589(0.96); C581(1.02); C583(1.08); C327(1.12) | LDD0630 | [12] |
| LDCM0314 | AC51 | HCT 116 | C1079(0.89); C1084(0.89); C486(0.93); C589(0.94) | LDD0631 | [12] |
| LDCM0315 | AC52 | HCT 116 | C589(0.85); C583(0.90); C581(0.93); C486(0.96) | LDD0632 | [12] |
| LDCM0316 | AC53 | HCT 116 | C327(0.79); C581(0.95); C583(0.98); C589(0.99) | LDD0633 | [12] |
| LDCM0317 | AC54 | HCT 116 | C581(0.98); C583(1.00); C589(1.08); C486(1.12) | LDD0634 | [12] |
| LDCM0318 | AC55 | HCT 116 | C327(0.79); C589(1.05); C430(1.10); C486(1.17) | LDD0635 | [12] |
| LDCM0319 | AC56 | HCT 116 | C589(1.02); C581(1.26); C583(1.29); C430(1.40) | LDD0636 | [12] |
| LDCM0320 | AC57 | HCT 116 | C486(0.70); C14(0.90); C581(0.92); C742(0.96) | LDD0637 | [12] |
| LDCM0321 | AC58 | HCT 116 | C486(0.81); C581(0.85); C583(0.89); C589(0.94) | LDD0638 | [12] |
| LDCM0322 | AC59 | HCT 116 | C486(0.65); C581(0.92); C589(0.93); C583(0.94) | LDD0639 | [12] |
| LDCM0323 | AC6 | HCT 116 | C742(0.81); C430(0.85); C581(0.91); C1079(0.92) | LDD0640 | [12] |
| LDCM0324 | AC60 | HCT 116 | C486(0.58); C589(0.90); C581(0.94); C742(0.95) | LDD0641 | [12] |
| LDCM0325 | AC61 | HCT 116 | C486(0.58); C742(0.84); C14(0.89); C589(0.90) | LDD0642 | [12] |
| LDCM0326 | AC62 | HCT 116 | C486(0.83); C430(0.97); C742(0.98); C589(1.04) | LDD0643 | [12] |
| LDCM0327 | AC63 | HCT 116 | C486(0.66); C581(0.98); C583(1.01); C14(1.01) | LDD0644 | [12] |
| LDCM0328 | AC64 | HCT 116 | C486(0.74); C742(1.02); C14(1.04); C589(1.08) | LDD0645 | [12] |
| LDCM0329 | AC65 | HCT 116 | C486(0.51); C589(0.87); C14(0.91); C742(0.93) | LDD0646 | [12] |
| LDCM0330 | AC66 | HCT 116 | C486(0.58); C742(0.86); C589(0.88); C581(0.88) | LDD0647 | [12] |
| LDCM0331 | AC67 | HCT 116 | C486(0.63); C589(0.93); C14(0.93); C581(0.99) | LDD0648 | [12] |
| LDCM0332 | AC68 | HCT 116 | C1079(0.85); C1084(0.85); C1093(0.96); C327(0.99) | LDD0649 | [12] |
| LDCM0333 | AC69 | HCT 116 | C742(1.07); C1093(1.09); C327(1.14); C430(1.15) | LDD0650 | [12] |
| LDCM0334 | AC7 | HCT 116 | C742(0.73); C904(0.84); C917(0.84); C925(0.84) | LDD0651 | [12] |
| LDCM0335 | AC70 | HCT 116 | C1079(0.61); C1084(0.61); C430(0.92); C742(1.11) | LDD0652 | [12] |
| LDCM0336 | AC71 | HCT 116 | C1079(0.68); C1084(0.68); C1093(0.93); C327(1.04) | LDD0653 | [12] |
| LDCM0337 | AC72 | HCT 116 | C430(0.95); C581(0.95); C1079(1.02); C1084(1.02) | LDD0654 | [12] |
| LDCM0338 | AC73 | HCT 116 | C589(1.03); C742(1.13); C581(1.28); C1093(1.28) | LDD0655 | [12] |
| LDCM0339 | AC74 | HCT 116 | C1093(0.97); C742(1.13); C589(1.14); C581(1.25) | LDD0656 | [12] |
| LDCM0340 | AC75 | HCT 116 | C589(1.02); C1093(1.09); C742(1.16); C581(1.27) | LDD0657 | [12] |
| LDCM0341 | AC76 | HCT 116 | C1079(0.83); C1084(0.83); C1093(1.09); C430(1.11) | LDD0658 | [12] |
| LDCM0342 | AC77 | HCT 116 | C589(0.93); C430(1.03); C1093(1.08); C1079(1.09) | LDD0659 | [12] |
| LDCM0343 | AC78 | HCT 116 | C1079(0.62); C1084(0.62); C327(0.95); C589(0.97) | LDD0660 | [12] |
| LDCM0344 | AC79 | HCT 116 | C1079(0.95); C1084(0.95); C581(0.99); C430(1.02) | LDD0661 | [12] |
| LDCM0345 | AC8 | HCT 116 | C742(0.73); C581(0.91); C327(0.93); C583(0.94) | LDD0662 | [12] |
| LDCM0346 | AC80 | HCT 116 | C589(1.01); C1093(1.03); C430(1.04); C581(1.06) | LDD0663 | [12] |
| LDCM0347 | AC81 | HCT 116 | C1079(0.65); C1084(0.65); C589(1.00); C327(1.03) | LDD0664 | [12] |
| LDCM0348 | AC82 | HCT 116 | C1093(1.04); C589(1.08); C742(1.13); C581(1.44) | LDD0665 | [12] |
| LDCM0349 | AC83 | HCT 116 | C742(0.70); C327(0.83); C589(0.90); C581(0.93) | LDD0666 | [12] |
| LDCM0350 | AC84 | HCT 116 | C742(0.74); C327(0.87); C430(0.97); C581(1.02) | LDD0667 | [12] |
| LDCM0351 | AC85 | HCT 116 | C742(0.66); C327(0.69); C430(0.84); C581(0.91) | LDD0668 | [12] |
| LDCM0352 | AC86 | HCT 116 | C486(0.72); C742(0.73); C327(0.78); C430(0.86) | LDD0669 | [12] |
| LDCM0353 | AC87 | HCT 116 | C327(0.65); C742(0.77); C430(0.90); C486(0.90) | LDD0670 | [12] |
| LDCM0354 | AC88 | HCT 116 | C742(0.74); C430(0.87); C327(0.89); C581(0.91) | LDD0671 | [12] |
| LDCM0355 | AC89 | HCT 116 | C742(0.70); C327(0.75); C430(0.86); C589(0.93) | LDD0672 | [12] |
| LDCM0357 | AC90 | HCT 116 | C742(0.68); C486(0.80); C583(0.87); C430(0.88) | LDD0674 | [12] |
| LDCM0358 | AC91 | HCT 116 | C742(0.62); C486(0.89); C327(0.92); C581(0.99) | LDD0675 | [12] |
| LDCM0359 | AC92 | HCT 116 | C742(0.69); C581(0.90); C430(0.95); C583(1.00) | LDD0676 | [12] |
| LDCM0360 | AC93 | HCT 116 | C327(0.80); C742(0.81); C581(0.85); C486(0.88) | LDD0677 | [12] |
| LDCM0361 | AC94 | HCT 116 | C742(0.69); C904(0.90); C917(0.90); C925(0.90) | LDD0678 | [12] |
| LDCM0362 | AC95 | HCT 116 | C742(0.73); C904(0.90); C917(0.90); C925(0.90) | LDD0679 | [12] |
| LDCM0363 | AC96 | HCT 116 | C742(0.75); C486(0.96); C583(1.00); C1079(1.10) | LDD0680 | [12] |
| LDCM0364 | AC97 | HCT 116 | C742(0.71); C581(0.88); C589(1.02); C583(1.03) | LDD0681 | [12] |
| LDCM0365 | AC98 | HCT 116 | C486(0.57); C581(0.68); C742(0.74); C583(0.87) | LDD0682 | [12] |
| LDCM0366 | AC99 | HCT 116 | C486(0.57); C742(0.62); C1079(0.67); C1084(0.67) | LDD0683 | [12] |
| LDCM0545 | Acetamide | MDA-MB-231 | C430(0.79) | LDD2138 | [8] |
| LDCM0248 | AKOS034007472 | HCT 116 | C327(0.96); C1079(0.87); C1084(0.87); C1093(0.87) | LDD0565 | [12] |
| LDCM0356 | AKOS034007680 | HCT 116 | C742(0.75); C581(0.91); C430(0.92); C583(0.94) | LDD0673 | [12] |
| LDCM0275 | AKOS034007705 | HCT 116 | C742(0.77); C581(0.97); C430(0.99); C589(1.00) | LDD0592 | [12] |
| LDCM0156 | Aniline | NCI-H1299 | N.A. | LDD0404 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C532(1.23); C430(0.86) | LDD0078 | [12] |
| LDCM0104 | BDHI 13 | Jurkat | 5.15 | LDD0206 | [29] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C430(0.93) | LDD2091 | [8] |
| LDCM0108 | Chloroacetamide | HeLa | C686(0.00); C14(0.00); H622(0.00); C1093(0.00) | LDD0222 | [21] |
| LDCM0632 | CL-Sc | Hep-G2 | C1116(20.00); C14(20.00); C1093(1.81); C430(1.68) | LDD2227 | [16] |
| LDCM0367 | CL1 | HCT 116 | C589(0.78); C1093(0.95); C583(0.98); C581(0.99) | LDD0684 | [12] |
| LDCM0368 | CL10 | HCT 116 | C589(0.96); C583(0.99); C581(1.00); C14(1.05) | LDD0685 | [12] |
| LDCM0369 | CL100 | HCT 116 | C904(0.82); C917(0.82); C925(0.82); C1079(0.87) | LDD0686 | [12] |
| LDCM0370 | CL101 | HCT 116 | C742(0.72); C904(0.81); C917(0.81); C925(0.81) | LDD0687 | [12] |
| LDCM0371 | CL102 | HCT 116 | C430(0.75); C581(0.80); C742(0.81); C904(0.82) | LDD0688 | [12] |
| LDCM0372 | CL103 | HCT 116 | C742(0.77); C430(0.82); C581(0.84); C583(0.89) | LDD0689 | [12] |
| LDCM0373 | CL104 | HCT 116 | C742(0.70); C430(0.79); C581(0.88); C327(0.94) | LDD0690 | [12] |
| LDCM0374 | CL105 | HCT 116 | C430(0.81); C904(0.86); C917(0.86); C925(0.86) | LDD0691 | [12] |
| LDCM0375 | CL106 | HCT 116 | C430(0.89); C589(1.03); C904(1.03); C917(1.03) | LDD0692 | [12] |
| LDCM0376 | CL107 | HCT 116 | C430(0.88); C1079(0.95); C1084(0.95); C904(0.98) | LDD0693 | [12] |
| LDCM0377 | CL108 | HCT 116 | C430(0.85); C904(0.87); C917(0.87); C925(0.87) | LDD0694 | [12] |
| LDCM0378 | CL109 | HCT 116 | C430(0.65); C583(0.88); C581(0.96); C589(0.97) | LDD0695 | [12] |
| LDCM0379 | CL11 | HCT 116 | C14(0.81); C589(0.89); C581(1.06); C583(1.07) | LDD0696 | [12] |
| LDCM0380 | CL110 | HCT 116 | C430(0.67); C486(0.88); C589(0.98); C583(1.00) | LDD0697 | [12] |
| LDCM0381 | CL111 | HCT 116 | C430(0.81); C1079(0.88); C1084(0.88); C327(0.94) | LDD0698 | [12] |
| LDCM0382 | CL112 | HCT 116 | C327(0.80); C742(0.92); C14(0.96); C1093(0.96) | LDD0699 | [12] |
| LDCM0383 | CL113 | HCT 116 | C581(0.79); C583(0.85); C327(0.86); C589(0.86) | LDD0700 | [12] |
| LDCM0384 | CL114 | HCT 116 | C581(0.71); C327(0.74); C583(0.83); C589(0.84) | LDD0701 | [12] |
| LDCM0385 | CL115 | HCT 116 | C581(0.75); C327(0.81); C583(0.86); C589(0.92) | LDD0702 | [12] |
| LDCM0386 | CL116 | HCT 116 | C327(0.83); C581(0.85); C1079(0.89); C1084(0.89) | LDD0703 | [12] |
| LDCM0387 | CL117 | HCT 116 | C14(0.99); C589(1.05); C581(1.18); C430(1.26) | LDD0704 | [12] |
| LDCM0388 | CL118 | HCT 116 | C581(0.95); C583(0.96); C532(1.01); C430(1.06) | LDD0705 | [12] |
| LDCM0389 | CL119 | HCT 116 | C581(0.92); C589(0.94); C327(0.95); C583(0.99) | LDD0706 | [12] |
| LDCM0390 | CL12 | HCT 116 | C589(0.92); C14(0.95); C581(1.04); C583(1.17) | LDD0707 | [12] |
| LDCM0391 | CL120 | HCT 116 | C581(0.84); C583(0.91); C1079(0.98); C1084(0.98) | LDD0708 | [12] |
| LDCM0392 | CL121 | HCT 116 | C486(0.76); C430(0.77); C327(0.84); C532(0.91) | LDD0709 | [12] |
| LDCM0393 | CL122 | HCT 116 | C1079(0.92); C1084(0.92); C486(0.97); C581(0.97) | LDD0710 | [12] |
| LDCM0394 | CL123 | HCT 116 | C327(0.76); C581(0.89); C583(0.93); C589(0.98) | LDD0711 | [12] |
| LDCM0395 | CL124 | HCT 116 | C589(0.90); C581(0.99); C583(1.04); C327(1.06) | LDD0712 | [12] |
| LDCM0396 | CL125 | HCT 116 | C486(0.54); C430(0.91); C581(0.95); C583(0.95) | LDD0713 | [12] |
| LDCM0397 | CL126 | HCT 116 | C486(0.64); C430(0.74); C589(0.82); C742(0.89) | LDD0714 | [12] |
| LDCM0398 | CL127 | HCT 116 | C486(0.66); C430(0.77); C742(0.81); C14(0.82) | LDD0715 | [12] |
| LDCM0399 | CL128 | HCT 116 | C486(0.64); C581(0.93); C583(0.94); C430(0.96) | LDD0716 | [12] |
| LDCM0400 | CL13 | HCT 116 | C14(0.84); C589(1.02); C581(1.02); C1079(1.04) | LDD0717 | [12] |
| LDCM0401 | CL14 | HCT 116 | C581(0.89); C583(0.90); C589(0.91); C1079(0.97) | LDD0718 | [12] |
| LDCM0402 | CL15 | HCT 116 | C14(0.74); C589(0.83); C581(0.94); C583(1.01) | LDD0719 | [12] |
| LDCM0403 | CL16 | HCT 116 | C581(0.76); C583(0.85); C430(0.96); C589(0.96) | LDD0720 | [12] |
| LDCM0404 | CL17 | HCT 116 | C581(0.83); C14(0.86); C583(0.90); C430(0.94) | LDD0721 | [12] |
| LDCM0405 | CL18 | HCT 116 | C581(0.89); C583(0.92); C430(0.95); C14(0.96) | LDD0722 | [12] |
| LDCM0406 | CL19 | HCT 116 | C581(0.87); C14(0.88); C583(0.89); C327(0.94) | LDD0723 | [12] |
| LDCM0407 | CL2 | HCT 116 | C327(0.92); C589(0.93); C1093(0.95); C583(0.97) | LDD0724 | [12] |
| LDCM0408 | CL20 | HCT 116 | C581(0.78); C583(0.82); C589(0.84); C14(0.92) | LDD0725 | [12] |
| LDCM0409 | CL21 | HCT 116 | C581(0.75); C583(0.84); C14(0.93); C589(0.99) | LDD0726 | [12] |
| LDCM0410 | CL22 | HCT 116 | C14(0.89); C589(0.90); C581(1.03); C583(1.07) | LDD0727 | [12] |
| LDCM0411 | CL23 | HCT 116 | C589(0.80); C430(0.85); C583(1.04); C14(1.07) | LDD0728 | [12] |
| LDCM0412 | CL24 | HCT 116 | C589(0.89); C583(0.96); C581(0.97); C430(1.07) | LDD0729 | [12] |
| LDCM0413 | CL25 | HCT 116 | C327(0.92); C1079(1.58); C1084(1.58); C1093(1.18) | LDD0730 | [12] |
| LDCM0414 | CL26 | HCT 116 | C327(1.06); C1079(1.45); C1084(1.45); C1093(1.11) | LDD0731 | [12] |
| LDCM0415 | CL27 | HCT 116 | C327(0.85); C1079(1.06); C1084(1.06); C1093(1.15) | LDD0732 | [12] |
| LDCM0416 | CL28 | HCT 116 | C327(1.14); C1079(1.60); C1084(1.60); C1093(1.11) | LDD0733 | [12] |
| LDCM0417 | CL29 | HCT 116 | C327(1.22); C1079(1.35); C1084(1.35); C1093(1.19) | LDD0734 | [12] |
| LDCM0418 | CL3 | HCT 116 | C327(0.93); C1079(0.98); C1084(0.98); C1093(0.94) | LDD0735 | [12] |
| LDCM0419 | CL30 | HCT 116 | C327(1.13); C1079(1.16); C1084(1.16); C1093(1.14) | LDD0736 | [12] |
| LDCM0420 | CL31 | HCT 116 | C327(0.97); C1079(0.87); C1084(0.87); C1093(1.09) | LDD0737 | [12] |
| LDCM0421 | CL32 | HCT 116 | C327(1.41); C1079(0.82); C1084(0.82); C1093(1.14) | LDD0738 | [12] |
| LDCM0422 | CL33 | HCT 116 | C327(1.66); C1079(0.75); C1084(0.75); C1093(1.14) | LDD0739 | [12] |
| LDCM0423 | CL34 | HCT 116 | C327(1.74); C1079(1.18); C1084(1.18); C1093(1.31) | LDD0740 | [12] |
| LDCM0424 | CL35 | HCT 116 | C327(1.88); C1079(1.43); C1084(1.43); C1093(1.21) | LDD0741 | [12] |
| LDCM0425 | CL36 | HCT 116 | C327(2.11); C1079(1.08); C1084(1.08); C1093(1.24) | LDD0742 | [12] |
| LDCM0426 | CL37 | HCT 116 | C327(1.75); C1079(1.28); C1084(1.28); C1093(1.31) | LDD0743 | [12] |
| LDCM0428 | CL39 | HCT 116 | C327(1.66); C1079(1.04); C1084(1.04); C1093(1.26) | LDD0745 | [12] |
| LDCM0429 | CL4 | HCT 116 | C327(1.03); C1079(1.11); C1084(1.11); C1093(0.97) | LDD0746 | [12] |
| LDCM0430 | CL40 | HCT 116 | C327(1.43); C1079(1.08); C1084(1.08); C1093(1.21) | LDD0747 | [12] |
| LDCM0431 | CL41 | HCT 116 | C327(1.74); C1079(0.97); C1084(0.97); C1093(1.30) | LDD0748 | [12] |
| LDCM0432 | CL42 | HCT 116 | C327(1.91); C1079(1.62); C1084(1.62); C1093(1.75) | LDD0749 | [12] |
| LDCM0433 | CL43 | HCT 116 | C327(1.94); C1079(1.47); C1084(1.47); C1093(1.39) | LDD0750 | [12] |
| LDCM0434 | CL44 | HCT 116 | C327(1.78); C1079(1.06); C1084(1.06); C1093(1.31) | LDD0751 | [12] |
| LDCM0435 | CL45 | HCT 116 | C327(1.69); C1079(1.26); C1084(1.26); C1093(1.40) | LDD0752 | [12] |
| LDCM0436 | CL46 | HCT 116 | C327(0.90); C1093(1.00); C14(1.18); C532(0.93) | LDD0753 | [12] |
| LDCM0437 | CL47 | HCT 116 | C327(0.89); C1093(1.02); C14(1.21); C532(0.89) | LDD0754 | [12] |
| LDCM0438 | CL48 | HCT 116 | C327(0.84); C1093(0.93); C14(1.06); C532(0.91) | LDD0755 | [12] |
| LDCM0439 | CL49 | HCT 116 | C327(0.94); C1093(1.12); C14(0.99); C532(1.02) | LDD0756 | [12] |
| LDCM0440 | CL5 | HCT 116 | C327(1.03); C1079(1.07); C1084(1.07); C1093(1.01) | LDD0757 | [12] |
| LDCM0441 | CL50 | HCT 116 | C327(0.97); C1093(0.97); C14(0.94); C532(1.27) | LDD0758 | [12] |
| LDCM0442 | CL51 | HCT 116 | C327(0.94); C1093(1.09); C14(1.01); C532(1.00) | LDD0759 | [12] |
| LDCM0443 | CL52 | HCT 116 | C327(0.89); C1093(1.01); C14(1.18); C532(0.97) | LDD0760 | [12] |
| LDCM0444 | CL53 | HCT 116 | C327(1.15); C1093(0.81); C14(0.98); C532(1.42) | LDD0761 | [12] |
| LDCM0445 | CL54 | HCT 116 | C327(0.95); C1093(0.97); C14(0.92); C532(1.00) | LDD0762 | [12] |
| LDCM0446 | CL55 | HCT 116 | C327(0.97); C1093(1.04); C14(0.95); C532(0.80) | LDD0763 | [12] |
| LDCM0447 | CL56 | HCT 116 | C327(0.96); C1093(0.92); C14(0.83); C532(1.00) | LDD0764 | [12] |
| LDCM0448 | CL57 | HCT 116 | C327(0.97); C1093(0.93); C14(0.91); C532(0.87) | LDD0765 | [12] |
| LDCM0449 | CL58 | HCT 116 | C327(0.98); C1093(1.07); C14(0.98); C532(0.86) | LDD0766 | [12] |
| LDCM0450 | CL59 | HCT 116 | C327(0.85); C1093(1.07); C14(0.90); C532(0.89) | LDD0767 | [12] |
| LDCM0451 | CL6 | HCT 116 | C327(0.94); C1079(1.08); C1084(1.08); C1093(1.07) | LDD0768 | [12] |
| LDCM0452 | CL60 | HCT 116 | C327(0.96); C1093(1.14); C14(0.98); C532(1.06) | LDD0769 | [12] |
| LDCM0453 | CL61 | HCT 116 | C327(1.08); C1079(0.81); C1084(0.81); C1093(1.07) | LDD0770 | [12] |
| LDCM0454 | CL62 | HCT 116 | C327(1.12); C1079(0.82); C1084(0.82); C1093(1.10) | LDD0771 | [12] |
| LDCM0455 | CL63 | HCT 116 | C327(0.98); C1079(0.94); C1084(0.94); C1093(1.24) | LDD0772 | [12] |
| LDCM0456 | CL64 | HCT 116 | C327(1.02); C1079(1.14); C1084(1.14); C1093(1.29) | LDD0773 | [12] |
| LDCM0457 | CL65 | HCT 116 | C327(1.09); C1079(0.90); C1084(0.90); C1093(1.11) | LDD0774 | [12] |
| LDCM0458 | CL66 | HCT 116 | C327(1.03); C1079(1.33); C1084(1.33); C1093(1.31) | LDD0775 | [12] |
| LDCM0459 | CL67 | HCT 116 | C327(0.93); C1079(1.05); C1084(1.05); C1093(1.29) | LDD0776 | [12] |
| LDCM0460 | CL68 | HCT 116 | C327(1.36); C1079(0.89); C1084(0.89); C1093(1.27) | LDD0777 | [12] |
| LDCM0461 | CL69 | HCT 116 | C327(0.94); C1079(0.93); C1084(0.93); C1093(1.16) | LDD0778 | [12] |
| LDCM0462 | CL7 | HCT 116 | C327(1.08); C1079(1.09); C1084(1.09); C1093(1.20) | LDD0779 | [12] |
| LDCM0463 | CL70 | HCT 116 | C327(1.00); C1079(0.85); C1084(0.85); C1093(1.51) | LDD0780 | [12] |
| LDCM0464 | CL71 | HCT 116 | C327(1.01); C1079(0.91); C1084(0.91); C1093(1.25) | LDD0781 | [12] |
| LDCM0465 | CL72 | HCT 116 | C327(0.98); C1079(0.95); C1084(0.95); C1093(1.12) | LDD0782 | [12] |
| LDCM0466 | CL73 | HCT 116 | C327(1.03); C1079(0.89); C1084(0.89); C1093(1.16) | LDD0783 | [12] |
| LDCM0467 | CL74 | HCT 116 | C327(0.92); C1079(0.92); C1084(0.92); C1093(1.14) | LDD0784 | [12] |
| LDCM0469 | CL76 | HCT 116 | C327(1.25); C1093(1.11); C14(1.04); C430(0.94) | LDD0786 | [12] |
| LDCM0470 | CL77 | HCT 116 | C327(4.84); C1093(0.98); C14(1.28); C430(0.84) | LDD0787 | [12] |
| LDCM0471 | CL78 | HCT 116 | C327(1.21); C1093(1.10); C14(1.20); C430(0.89) | LDD0788 | [12] |
| LDCM0472 | CL79 | HCT 116 | C327(1.42); C1093(1.28); C14(1.09); C430(0.98) | LDD0789 | [12] |
| LDCM0473 | CL8 | HCT 116 | C327(1.35); C1079(1.42); C1084(1.42); C1093(1.33) | LDD0790 | [12] |
| LDCM0474 | CL80 | HCT 116 | C327(1.32); C1093(0.98); C14(1.05); C430(0.99) | LDD0791 | [12] |
| LDCM0475 | CL81 | HCT 116 | C327(1.30); C1093(1.13); C14(1.09); C430(0.91) | LDD0792 | [12] |
| LDCM0476 | CL82 | HCT 116 | C327(1.52); C1093(1.51); C14(1.02); C430(1.18) | LDD0793 | [12] |
| LDCM0477 | CL83 | HCT 116 | C327(1.68); C1093(1.48); C14(0.96); C430(1.18) | LDD0794 | [12] |
| LDCM0478 | CL84 | HCT 116 | C327(2.24); C1093(1.72); C14(0.99); C430(1.30) | LDD0795 | [12] |
| LDCM0479 | CL85 | HCT 116 | C327(1.16); C1093(1.23); C14(1.17); C430(0.99) | LDD0796 | [12] |
| LDCM0480 | CL86 | HCT 116 | C327(1.08); C1093(1.04); C14(1.11); C430(0.99) | LDD0797 | [12] |
| LDCM0481 | CL87 | HCT 116 | C327(1.14); C1093(1.29); C14(0.90); C430(0.99) | LDD0798 | [12] |
| LDCM0482 | CL88 | HCT 116 | C327(1.69); C1093(1.38); C14(0.94); C430(1.16) | LDD0799 | [12] |
| LDCM0483 | CL89 | HCT 116 | C327(1.67); C1093(2.02); C14(1.00); C430(1.53) | LDD0800 | [12] |
| LDCM0484 | CL9 | HCT 116 | C327(1.25); C1079(1.08); C1084(1.08); C1093(1.10) | LDD0801 | [12] |
| LDCM0485 | CL90 | HCT 116 | C327(1.06); C1093(0.91); C14(1.04); C430(0.97) | LDD0802 | [12] |
| LDCM0486 | CL91 | HCT 116 | C327(0.72); C1079(0.88); C1084(0.88); C1093(1.18) | LDD0803 | [12] |
| LDCM0487 | CL92 | HCT 116 | C327(0.83); C1079(0.93); C1084(0.93); C1093(1.12) | LDD0804 | [12] |
| LDCM0488 | CL93 | HCT 116 | C327(1.07); C1079(0.94); C1084(0.94); C1093(1.19) | LDD0805 | [12] |
| LDCM0489 | CL94 | HCT 116 | C327(1.02); C1079(0.89); C1084(0.89); C1093(1.31) | LDD0806 | [12] |
| LDCM0490 | CL95 | HCT 116 | C327(0.85); C1079(0.90); C1084(0.90); C1093(1.23) | LDD0807 | [12] |
| LDCM0491 | CL96 | HCT 116 | C327(0.84); C1079(0.94); C1084(0.94); C1093(1.31) | LDD0808 | [12] |
| LDCM0492 | CL97 | HCT 116 | C327(0.93); C1079(0.85); C1084(0.85); C1093(1.36) | LDD0809 | [12] |
| LDCM0493 | CL98 | HCT 116 | C327(0.85); C1079(0.84); C1084(0.84); C1093(1.19) | LDD0810 | [12] |
| LDCM0494 | CL99 | HCT 116 | C327(0.72); C1079(0.92); C1084(0.92); C1093(1.26) | LDD0811 | [12] |
| LDCM0634 | CY-0357 | Hep-G2 | C14(0.64) | LDD2228 | [16] |
| LDCM0495 | E2913 | HEK-293T | C430(0.91); C1093(0.84); C532(0.87); C14(1.08) | LDD1698 | [30] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C589(22.87); C327(19.40); C583(2.00) | LDD1702 | [8] |
| LDCM0175 | Ethacrynic acid | HeLa | N.A. | LDD0440 | [11] |
| LDCM0204 | EV-97 | T cell | C532(8.80) | LDD0524 | [31] |
| LDCM0625 | F8 | Ramos | C1116(1.97); C350(3.69); 0.32; C430(2.00) | LDD2187 | [32] |
| LDCM0572 | Fragment10 | MDA-MB-231 | C242(0.99) | LDD1389 | [33] |
| LDCM0573 | Fragment11 | MDA-MB-231 | C242(1.41) | LDD1391 | [33] |
| LDCM0574 | Fragment12 | MDA-MB-231 | C242(1.06) | LDD1393 | [33] |
| LDCM0575 | Fragment13 | Ramos | C242(0.68) | LDD1396 | [33] |
| LDCM0576 | Fragment14 | MDA-MB-231 | C242(1.63) | LDD1397 | [33] |
| LDCM0577 | Fragment15 | MDA-MB-231 | C242(1.17) | LDD1399 | [33] |
| LDCM0579 | Fragment20 | MDA-MB-231 | C242(1.04) | LDD1402 | [33] |
| LDCM0580 | Fragment21 | MDA-MB-231 | C242(0.67) | LDD1404 | [33] |
| LDCM0581 | Fragment22 | Ramos | C242(0.83) | LDD1407 | [33] |
| LDCM0582 | Fragment23 | Ramos | C1116(0.58); C350(0.89); 2.15; C430(2.21) | LDD2196 | [32] |
| LDCM0584 | Fragment25 | MDA-MB-231 | C242(1.41) | LDD1411 | [33] |
| LDCM0585 | Fragment26 | Ramos | C242(1.11) | LDD1412 | [33] |
| LDCM0578 | Fragment27 | MDA-MB-231 | C242(0.64) | LDD1401 | [33] |
| LDCM0586 | Fragment28 | MDA-MB-231 | C242(0.76) | LDD1415 | [33] |
| LDCM0587 | Fragment29 | MDA-MB-231 | C242(1.25) | LDD1417 | [33] |
| LDCM0588 | Fragment30 | Ramos | C1116(0.50); C350(1.02); 1.29; C430(0.97) | LDD2199 | [32] |
| LDCM0589 | Fragment31 | MDA-MB-231 | C242(1.18) | LDD1421 | [33] |
| LDCM0590 | Fragment32 | MDA-MB-231 | C242(1.00) | LDD1423 | [33] |
| LDCM0468 | Fragment33 | HCT 116 | C327(1.13); C1079(1.17); C1084(1.17); C1093(1.05) | LDD0785 | [12] |
| LDCM0594 | Fragment36 | MDA-MB-231 | C242(1.56) | LDD1431 | [33] |
| LDCM0596 | Fragment38 | MDA-MB-231 | C242(1.10) | LDD1433 | [33] |
| LDCM0566 | Fragment4 | MDA-MB-231 | C242(0.92) | LDD1378 | [33] |
| LDCM0598 | Fragment40 | Ramos | C242(1.42) | LDD1437 | [33] |
| LDCM0599 | Fragment41 | MDA-MB-231 | C242(1.27) | LDD1438 | [33] |
| LDCM0600 | Fragment42 | Ramos | C242(0.88) | LDD1440 | [33] |
| LDCM0601 | Fragment43 | MDA-MB-231 | C242(1.07) | LDD1441 | [33] |
| LDCM0604 | Fragment46 | MDA-MB-231 | C242(1.19) | LDD1445 | [33] |
| LDCM0605 | Fragment47 | MDA-MB-231 | C242(1.17) | LDD1446 | [33] |
| LDCM0606 | Fragment48 | MDA-MB-231 | C242(1.10) | LDD1447 | [33] |
| LDCM0607 | Fragment49 | MDA-MB-231 | C242(5.03) | LDD1448 | [33] |
| LDCM0427 | Fragment51 | HCT 116 | C327(1.69); C1079(1.74); C1084(1.74); C1093(1.64) | LDD0744 | [12] |
| LDCM0610 | Fragment52 | MDA-MB-231 | C242(0.88) | LDD1452 | [33] |
| LDCM0611 | Fragment53 | MDA-MB-231 | C242(0.86) | LDD1454 | [33] |
| LDCM0614 | Fragment56 | MDA-MB-231 | C242(0.80) | LDD1458 | [33] |
| LDCM0568 | Fragment6 | MDA-MB-231 | C242(0.88) | LDD1382 | [33] |
| LDCM0569 | Fragment7 | Ramos | C1116(1.50); C430(1.47); C1093(2.46); C327(7.70) | LDD2186 | [32] |
| LDCM0570 | Fragment8 | MDA-MB-231 | C242(0.86) | LDD1385 | [33] |
| LDCM0571 | Fragment9 | MDA-MB-231 | C242(0.73) | LDD1387 | [33] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [21] |
| LDCM0179 | JZ128 | PC-3 | C581(0.00); C532(0.00) | LDD0462 | [7] |
| LDCM0022 | KB02 | HCT 116 | C532(16.71); C486(2.73); C1093(1.40); C430(1.40) | LDD0080 | [12] |
| LDCM0023 | KB03 | HCT 116 | C532(2.07); C486(1.45); C1093(1.68); C430(0.91) | LDD0081 | [12] |
| LDCM0024 | KB05 | HCT 116 | C532(17.10); C486(1.77); C1093(1.55); C430(1.55) | LDD0082 | [12] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C14(0.88) | LDD2102 | [8] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C14(0.58) | LDD2121 | [8] |
| LDCM0109 | NEM | HeLa | H622(0.00); H725(0.00) | LDD0223 | [21] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C430(1.03); C14(0.60) | LDD2089 | [8] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C14(1.29) | LDD2090 | [8] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C430(0.92); C14(0.94) | LDD2092 | [8] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C430(1.25); C14(1.07); C589(1.04); C350(2.10) | LDD2093 | [8] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C430(1.01); C14(0.75) | LDD2094 | [8] |
| LDCM0503 | Nucleophilic fragment 14b | MDA-MB-231 | C532(0.80) | LDD2096 | [8] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C430(1.05) | LDD2097 | [8] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C430(1.61); C14(1.19); C589(1.09) | LDD2099 | [8] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C430(1.11); C14(0.91) | LDD2100 | [8] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C589(0.64) | LDD2104 | [8] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C14(0.60) | LDD2105 | [8] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C14(0.68) | LDD2106 | [8] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C350(1.54); C1093(0.91) | LDD2107 | [8] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C430(1.09); C14(0.73) | LDD2108 | [8] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C430(0.82); C350(1.05) | LDD2109 | [8] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C589(1.16); C350(1.91); C1093(1.26) | LDD2111 | [8] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C430(0.82); C14(0.85); C589(0.90) | LDD2114 | [8] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C430(1.99); C589(2.48) | LDD2119 | [8] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C14(0.98) | LDD2120 | [8] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C430(1.38) | LDD2125 | [8] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C589(0.89) | LDD2127 | [8] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C1093(1.17) | LDD2128 | [8] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C430(1.04); C14(1.35); C589(1.11) | LDD2129 | [8] |
| LDCM0540 | Nucleophilic fragment 35 | MDA-MB-231 | C430(0.96); C589(0.84) | LDD2133 | [8] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C430(0.96) | LDD2134 | [8] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C430(1.30); C14(0.99); C589(1.18); C350(1.49) | LDD2135 | [8] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C430(2.15); C14(1.39); C532(1.72) | LDD2136 | [8] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C430(1.04) | LDD2137 | [8] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C1093(1.21) | LDD1700 | [8] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C430(0.98); C14(0.85) | LDD2140 | [8] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C14(0.76) | LDD2141 | [8] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C14(0.87) | LDD2143 | [8] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C430(1.65); C14(0.90) | LDD2144 | [8] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C430(1.02); C589(1.04) | LDD2146 | [8] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C430(0.76); C14(0.74); C589(0.73); C350(0.72) | LDD2150 | [8] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C14(1.55) | LDD2153 | [8] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C350(0.79); C1093(0.56); C14(0.56); C532(0.36) | LDD2206 | [34] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C589(1.15); C1116(1.10); C14(0.63); C92(0.49) | LDD2207 | [34] |
| LDCM0131 | RA190 | MM1.R | 2.61 | LDD0300 | [10] |
| LDCM0021 | THZ1 | HCT 116 | C532(1.09); C327(1.10); C1093(0.92) | LDD2173 | [12] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| mRNA export factor GLE1 (GLE1) | GLE1 family | Q53GS7 | |||
| MyoD family inhibitor (MDFI) | MDFI family | Q99750 | |||
Other
References
