Details of the Target
General Information of Target
| Target ID | LDTP02552 | |||||
|---|---|---|---|---|---|---|
| Target Name | Glycogen phosphorylase, brain form (PYGB) | |||||
| Gene Name | PYGB | |||||
| Gene ID | 5834 | |||||
| Synonyms |
Glycogen phosphorylase, brain form; EC 2.4.1.1 |
|||||
| 3D Structure | ||||||
| Sequence |
MAKPLTDSEKRKQISVRGLAGLGDVAEVRKSFNRHLHFTLVKDRNVATPRDYFFALAHTV
RDHLVGRWIRTQQHYYERDPKRIYYLSLEFYMGRTLQNTMVNLGLQNACDEAIYQLGLDL EELEEIEEDAGLGNGGLGRLAACFLDSMATLGLAAYGYGIRYEFGIFNQKIVNGWQVEEA DDWLRYGNPWEKARPEYMLPVHFYGRVEHTPDGVKWLDTQVVLAMPYDTPVPGYKNNTVN TMRLWSAKAPNDFKLQDFNVGDYIEAVLDRNLAENISRVLYPNDNFFEGKELRLKQEYFV VAATLQDIIRRFKSSKFGCRDPVRTCFETFPDKVAIQLNDTHPALSIPELMRILVDVEKV DWDKAWEITKKTCAYTNHTVLPEALERWPVSMFEKLLPRHLEIIYAINQRHLDHVAALFP GDVDRLRRMSVIEEGDCKRINMAHLCVIGSHAVNGVARIHSEIVKQSVFKDFYELEPEKF QNKTNGITPRRWLLLCNPGLADTIVEKIGEEFLTDLSQLKKLLPLVSDEVFIRDVAKVKQ ENKLKFSAFLEKEYKVKINPSSMFDVHVKRIHEYKRQLLNCLHVVTLYNRIKRDPAKAFV PRTVMIGGKAAPGYHMAKLIIKLVTSIGDVVNHDPVVGDRLKVIFLENYRVSLAEKVIPA ADLSQQISTAGTEASGTGNMKFMLNGALTIGTMDGANVEMAEEAGAENLFIFGLRVEDVE ALDRKGYNAREYYDHLPELKQAVDQISSGFFSPKEPDCFKDIVNMLMHHDRFKVFADYEA YMQCQAQVDQLYRNPKEWTKKVIRNIACSGKFSSDRTITEYAREIWGVEPSDLQIPPPNI PRD |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Glycogen phosphorylase family
|
|||||
| Function |
Glycogen phosphorylase that regulates glycogen mobilization. Phosphorylase is an important allosteric enzyme in carbohydrate metabolism. Enzymes from different sources differ in their regulatory mechanisms and in their natural substrates. However, all known phosphorylases share catalytic and structural properties.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
11.27 | LDD0402 | [1] | |
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [2] | |
|
CY4 Probe Info |
 |
100.00 | LDD0244 | [2] | |
|
N1 Probe Info |
 |
100.00 | LDD0242 | [2] | |
|
TH211 Probe Info |
 |
Y197(10.00); Y263(9.40); Y281(8.87) | LDD0260 | [3] | |
|
TH216 Probe Info |
 |
Y197(6.61) | LDD0259 | [3] | |
|
ONAyne Probe Info |
 |
K597(6.67) | LDD0274 | [4] | |
|
STPyne Probe Info |
 |
K12(6.15); K192(7.60); K248(6.59); K316(8.37) | LDD0277 | [4] | |
|
Probe 1 Probe Info |
 |
Y197(5.89); Y473(26.82); Y821(12.22) | LDD3495 | [5] | |
|
AF-2 Probe Info |
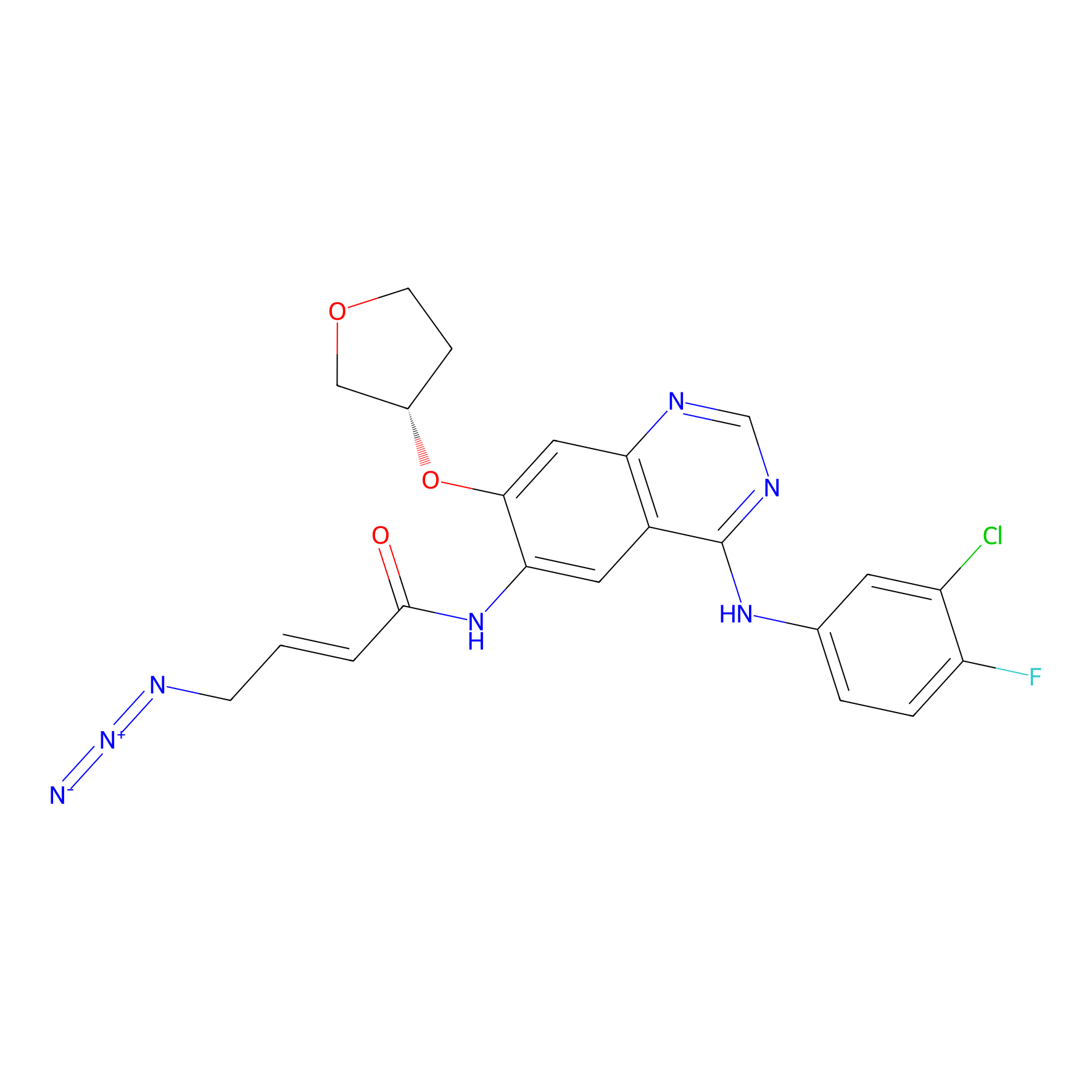 |
1.99 | LDD0422 | [6] | |
|
BTD Probe Info |
 |
C319(1.95) | LDD1700 | [7] | |
|
DA-P3 Probe Info |
 |
4.23 | LDD0185 | [8] | |
|
AHL-Pu-1 Probe Info |
 |
C437(2.05) | LDD0168 | [9] | |
|
EA-probe Probe Info |
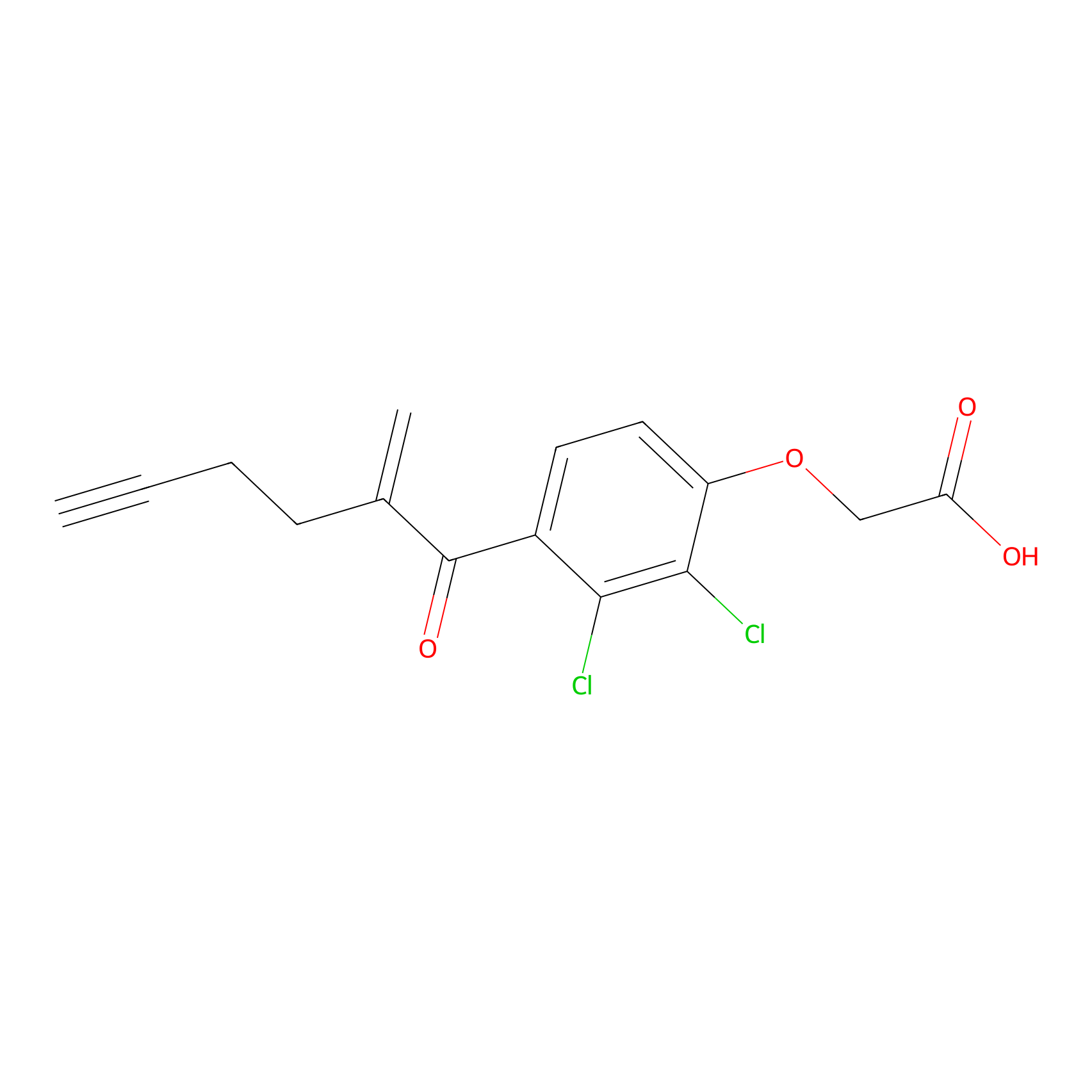 |
C326(1.22); C143(0.98); C496(0.88) | LDD2210 | [10] | |
|
HHS-482 Probe Info |
 |
Y186(0.85); Y197(0.88); Y204(0.97); Y227(1.37) | LDD0285 | [11] | |
|
HHS-475 Probe Info |
 |
Y197(0.72); Y614(0.83); Y76(0.96); Y298(0.99) | LDD0264 | [12] | |
|
HHS-465 Probe Info |
 |
Y186(10.00); Y197(5.25); Y204(10.00); Y234(2.18) | LDD2237 | [13] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0222 | [14] | |
|
DBIA Probe Info |
 |
C437(0.95) | LDD0078 | [15] | |
|
ATP probe Probe Info |
 |
K618(0.00); K622(0.00); K438(0.00); K520(0.00) | LDD0199 | [16] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C446(0.00); C143(0.00); C437(0.00); C808(0.00) | LDD0038 | [17] | |
|
IA-alkyne Probe Info |
 |
C319(0.00); C446(0.00); C437(0.00); C581(0.00) | LDD0036 | [17] | |
|
Lodoacetamide azide Probe Info |
 |
C319(0.00); C446(0.00); C143(0.00); C437(0.00) | LDD0037 | [17] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [18] | |
|
NAIA_4 Probe Info |
 |
C109(0.00); C326(0.00); C373(0.00); C437(0.00) | LDD2226 | [19] | |
|
WYneC Probe Info |
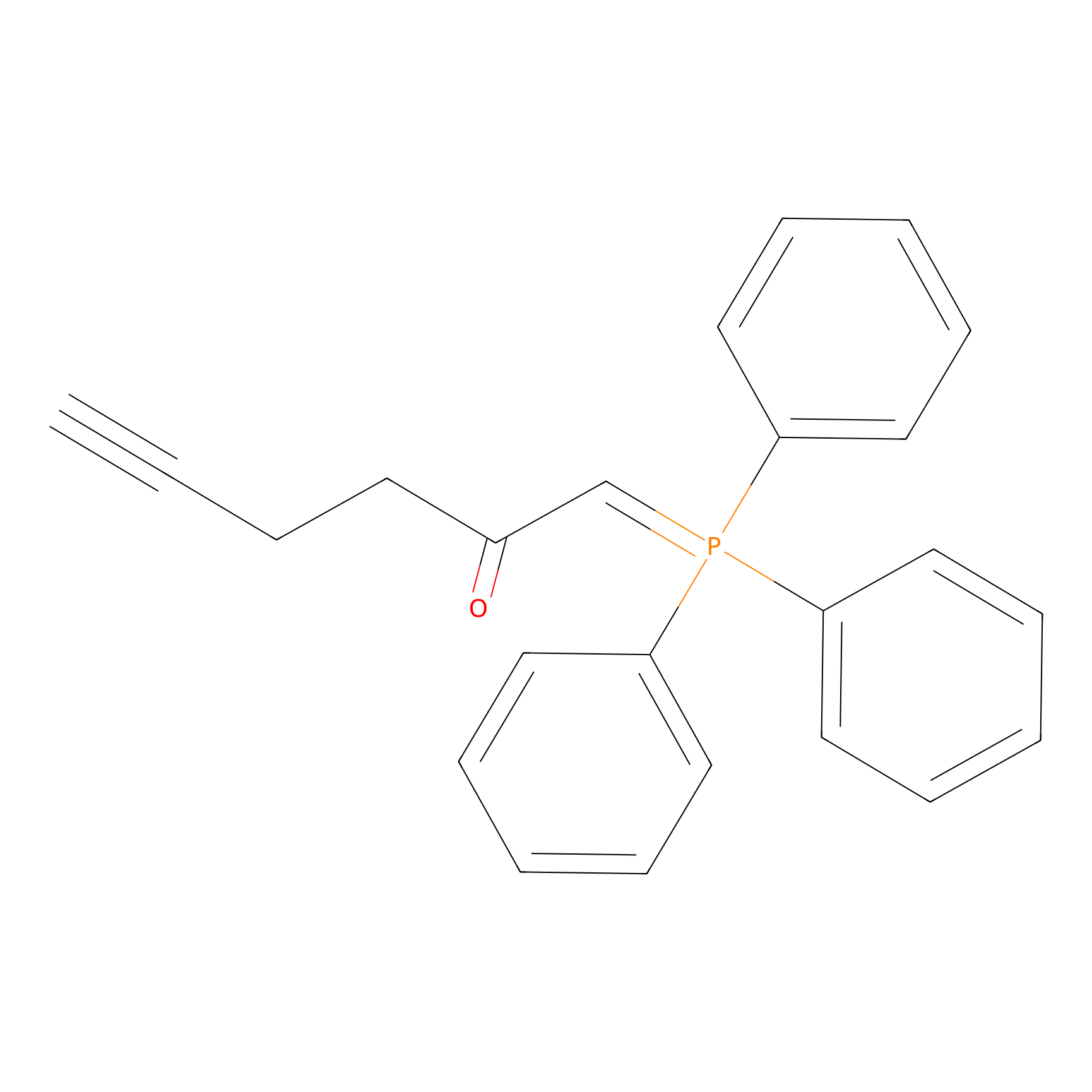 |
N.A. | LDD0014 | [20] | |
|
WYneN Probe Info |
 |
C326(0.00); C496(0.00) | LDD0021 | [20] | |
|
WYneO Probe Info |
 |
C808(0.00); C437(0.00); C326(0.00) | LDD0022 | [20] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [20] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [21] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [22] | |
|
NAIA_5 Probe Info |
 |
C758(0.00); C319(0.00); C446(0.00); C373(0.00) | LDD2223 | [19] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C064 Probe Info |
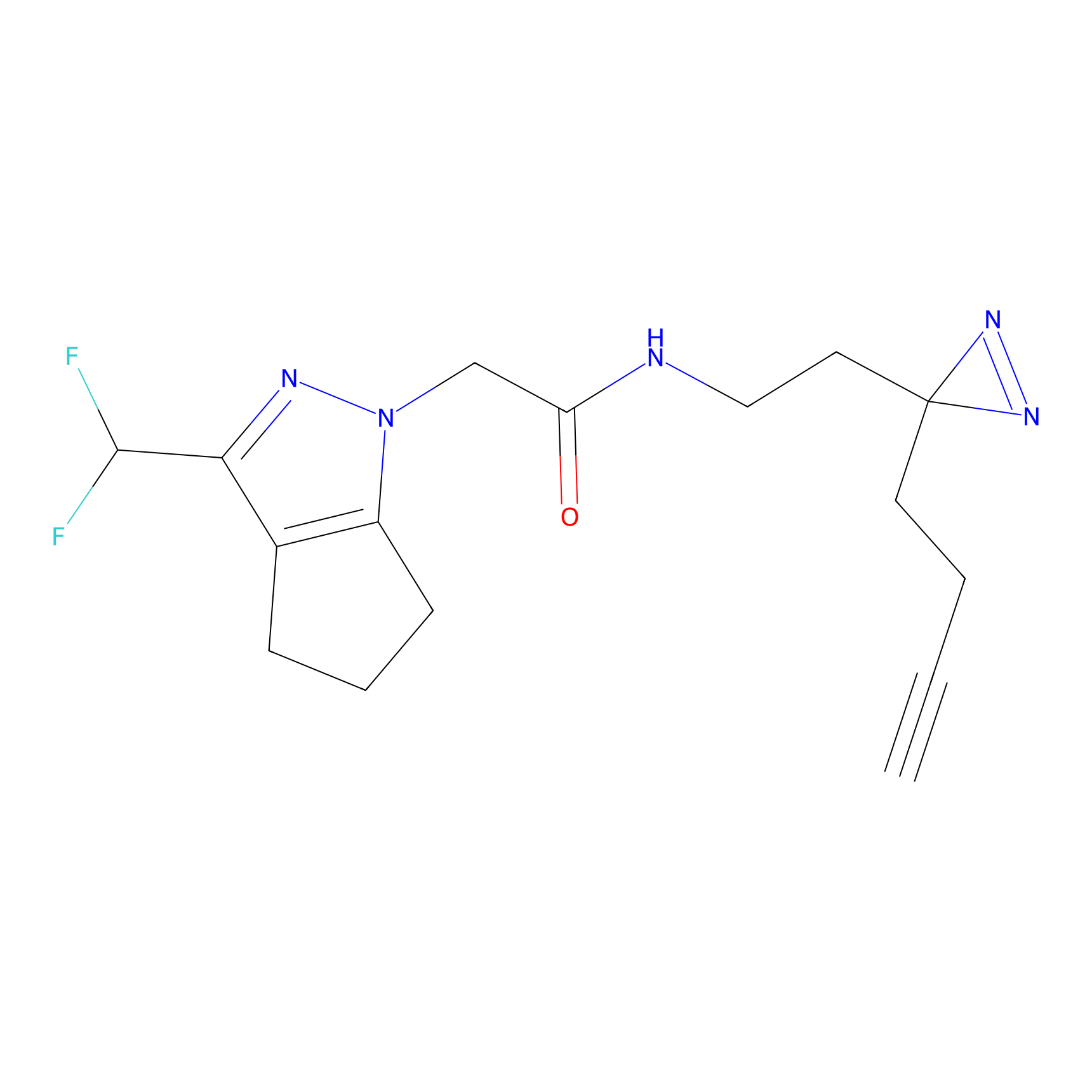 |
6.92 | LDD1761 | [23] | |
|
FFF probe13 Probe Info |
 |
8.07 | LDD0475 | [24] | |
|
FFF probe14 Probe Info |
 |
12.49 | LDD0477 | [24] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [25] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [26] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0068 | [27] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C373(0.75); C319(0.70) | LDD2142 | [7] |
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C373(0.94); C319(0.64) | LDD2112 | [7] |
| LDCM0502 | 1-(Cyanoacetyl)piperidine | MDA-MB-231 | C373(0.57); C319(0.56) | LDD2095 | [7] |
| LDCM0537 | 2-Cyano-N,N-dimethylacetamide | MDA-MB-231 | C373(0.83); C808(0.88); C326(0.69) | LDD2130 | [7] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C373(1.61); C319(0.99); C808(0.99); C326(1.14) | LDD2117 | [7] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C373(1.40); C319(1.02) | LDD2152 | [7] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C319(1.13) | LDD2103 | [7] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C373(0.50); C319(0.54); C808(0.60); C326(0.47) | LDD2132 | [7] |
| LDCM0538 | 4-(Cyanoacetyl)morpholine | MDA-MB-231 | C319(0.59); C808(0.69); C326(0.45) | LDD2131 | [7] |
| LDCM0025 | 4SU-RNA | HEK-293T | C437(2.05) | LDD0168 | [9] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C496(3.05); C437(2.49) | LDD0169 | [9] |
| LDCM0214 | AC1 | HCT 116 | C326(1.32); C319(0.61) | LDD0531 | [15] |
| LDCM0215 | AC10 | HCT 116 | C326(1.55); C319(1.29); C437(1.07) | LDD0532 | [15] |
| LDCM0216 | AC100 | HCT 116 | C326(1.20); C437(1.04) | LDD0533 | [15] |
| LDCM0217 | AC101 | HCT 116 | C326(1.13); C437(0.90) | LDD0534 | [15] |
| LDCM0218 | AC102 | HCT 116 | C326(1.05); C437(0.96) | LDD0535 | [15] |
| LDCM0219 | AC103 | HCT 116 | C326(1.31); C437(1.00) | LDD0536 | [15] |
| LDCM0220 | AC104 | HCT 116 | C326(1.13); C437(0.82) | LDD0537 | [15] |
| LDCM0221 | AC105 | HCT 116 | C326(1.17); C437(0.90) | LDD0538 | [15] |
| LDCM0222 | AC106 | HCT 116 | C326(1.49); C437(0.93) | LDD0539 | [15] |
| LDCM0223 | AC107 | HCT 116 | C326(1.34); C437(0.94) | LDD0540 | [15] |
| LDCM0224 | AC108 | HCT 116 | C326(1.24); C437(0.95) | LDD0541 | [15] |
| LDCM0225 | AC109 | HCT 116 | C326(1.00); C437(0.90) | LDD0542 | [15] |
| LDCM0226 | AC11 | HCT 116 | C326(1.34); C319(2.44); C437(0.95) | LDD0543 | [15] |
| LDCM0227 | AC110 | HCT 116 | C326(1.10); C437(1.00) | LDD0544 | [15] |
| LDCM0228 | AC111 | HCT 116 | C326(1.12); C437(0.92) | LDD0545 | [15] |
| LDCM0229 | AC112 | HCT 116 | C326(1.37); C437(0.90) | LDD0546 | [15] |
| LDCM0230 | AC113 | HCT 116 | C319(0.96) | LDD0547 | [15] |
| LDCM0231 | AC114 | HCT 116 | C319(0.92) | LDD0548 | [15] |
| LDCM0232 | AC115 | HCT 116 | C319(0.83) | LDD0549 | [15] |
| LDCM0233 | AC116 | HCT 116 | C319(0.67) | LDD0550 | [15] |
| LDCM0234 | AC117 | HCT 116 | C319(0.54) | LDD0551 | [15] |
| LDCM0235 | AC118 | HCT 116 | C319(0.57) | LDD0552 | [15] |
| LDCM0236 | AC119 | HCT 116 | C319(0.48) | LDD0553 | [15] |
| LDCM0237 | AC12 | HCT 116 | C326(1.10); C319(0.98); C437(0.91) | LDD0554 | [15] |
| LDCM0238 | AC120 | HCT 116 | C319(1.61) | LDD0555 | [15] |
| LDCM0239 | AC121 | HCT 116 | C319(0.92) | LDD0556 | [15] |
| LDCM0240 | AC122 | HCT 116 | C319(0.50) | LDD0557 | [15] |
| LDCM0241 | AC123 | HCT 116 | C319(0.78) | LDD0558 | [15] |
| LDCM0242 | AC124 | HCT 116 | C319(0.65) | LDD0559 | [15] |
| LDCM0243 | AC125 | HCT 116 | C319(0.76) | LDD0560 | [15] |
| LDCM0244 | AC126 | HCT 116 | C319(0.84) | LDD0561 | [15] |
| LDCM0245 | AC127 | HCT 116 | C319(0.44) | LDD0562 | [15] |
| LDCM0246 | AC128 | HCT 116 | C326(1.30); C319(1.10) | LDD0563 | [15] |
| LDCM0247 | AC129 | HCT 116 | C326(1.10); C319(0.86) | LDD0564 | [15] |
| LDCM0249 | AC130 | HCT 116 | C326(1.31); C319(0.84) | LDD0566 | [15] |
| LDCM0250 | AC131 | HCT 116 | C326(0.93); C319(1.28) | LDD0567 | [15] |
| LDCM0251 | AC132 | HCT 116 | C326(1.11); C319(0.84) | LDD0568 | [15] |
| LDCM0252 | AC133 | HCT 116 | C326(1.15); C319(0.90) | LDD0569 | [15] |
| LDCM0253 | AC134 | HCT 116 | C326(1.35); C319(1.79) | LDD0570 | [15] |
| LDCM0254 | AC135 | HCT 116 | C326(1.30); C319(0.83) | LDD0571 | [15] |
| LDCM0255 | AC136 | HCT 116 | C326(1.18); C319(1.21) | LDD0572 | [15] |
| LDCM0256 | AC137 | HCT 116 | C326(1.17); C319(1.25) | LDD0573 | [15] |
| LDCM0257 | AC138 | HCT 116 | C326(1.59); C319(2.32) | LDD0574 | [15] |
| LDCM0258 | AC139 | HCT 116 | C326(1.28); C319(1.09) | LDD0575 | [15] |
| LDCM0259 | AC14 | HCT 116 | C326(0.74); C319(2.22); C437(1.21) | LDD0576 | [15] |
| LDCM0260 | AC140 | HCT 116 | C326(1.42); C319(1.55) | LDD0577 | [15] |
| LDCM0261 | AC141 | HCT 116 | C326(1.37); C319(1.69) | LDD0578 | [15] |
| LDCM0262 | AC142 | HCT 116 | C326(1.07); C319(1.04) | LDD0579 | [15] |
| LDCM0263 | AC143 | HCT 116 | C326(1.15) | LDD0580 | [15] |
| LDCM0264 | AC144 | HCT 116 | C326(1.38) | LDD0581 | [15] |
| LDCM0265 | AC145 | HCT 116 | C326(1.29) | LDD0582 | [15] |
| LDCM0266 | AC146 | HCT 116 | C326(1.40) | LDD0583 | [15] |
| LDCM0267 | AC147 | HCT 116 | C326(1.30) | LDD0584 | [15] |
| LDCM0268 | AC148 | HCT 116 | C326(1.85) | LDD0585 | [15] |
| LDCM0269 | AC149 | HCT 116 | C326(1.61) | LDD0586 | [15] |
| LDCM0270 | AC15 | HCT 116 | C326(0.83); C437(1.29); C319(1.43) | LDD0587 | [15] |
| LDCM0271 | AC150 | HCT 116 | C326(1.10) | LDD0588 | [15] |
| LDCM0272 | AC151 | HCT 116 | C326(1.07) | LDD0589 | [15] |
| LDCM0273 | AC152 | HCT 116 | C326(1.31) | LDD0590 | [15] |
| LDCM0274 | AC153 | HCT 116 | C326(1.82) | LDD0591 | [15] |
| LDCM0621 | AC154 | HCT 116 | C326(1.11) | LDD2158 | [15] |
| LDCM0622 | AC155 | HCT 116 | C326(1.23) | LDD2159 | [15] |
| LDCM0623 | AC156 | HCT 116 | C326(0.97) | LDD2160 | [15] |
| LDCM0624 | AC157 | HCT 116 | C326(0.96) | LDD2161 | [15] |
| LDCM0276 | AC17 | HCT 116 | C319(0.61); C437(0.77); C326(1.29) | LDD0593 | [15] |
| LDCM0277 | AC18 | HCT 116 | C319(0.58); C437(0.74); C326(1.46) | LDD0594 | [15] |
| LDCM0278 | AC19 | HCT 116 | C319(0.53); C437(0.76); C326(1.21) | LDD0595 | [15] |
| LDCM0279 | AC2 | HCT 116 | C319(0.61); C326(1.17) | LDD0596 | [15] |
| LDCM0280 | AC20 | HCT 116 | C319(0.93); C437(0.95); C326(1.31) | LDD0597 | [15] |
| LDCM0281 | AC21 | HCT 116 | C437(0.68); C319(0.75); C326(1.21) | LDD0598 | [15] |
| LDCM0282 | AC22 | HCT 116 | C319(0.77); C437(0.85); C326(1.21) | LDD0599 | [15] |
| LDCM0283 | AC23 | HCT 116 | C319(0.80); C437(0.84); C326(1.33) | LDD0600 | [15] |
| LDCM0284 | AC24 | HCT 116 | C319(0.82); C437(0.92); C326(1.04) | LDD0601 | [15] |
| LDCM0285 | AC25 | HCT 116 | C319(0.82); C326(1.10); C437(1.18) | LDD0602 | [15] |
| LDCM0286 | AC26 | HCT 116 | C319(0.55); C326(1.37); C437(1.46) | LDD0603 | [15] |
| LDCM0287 | AC27 | HCT 116 | C319(0.77); C326(1.12); C437(1.31) | LDD0604 | [15] |
| LDCM0288 | AC28 | HCT 116 | C319(0.26); C326(1.20); C437(1.29) | LDD0605 | [15] |
| LDCM0289 | AC29 | HCT 116 | C326(1.28); C319(1.37); C437(1.51) | LDD0606 | [15] |
| LDCM0290 | AC3 | HCT 116 | C319(0.49); C326(1.27) | LDD0607 | [15] |
| LDCM0291 | AC30 | HCT 116 | C319(1.16); C326(1.30); C437(1.54) | LDD0608 | [15] |
| LDCM0292 | AC31 | HCT 116 | C319(0.68); C326(1.17); C437(1.39) | LDD0609 | [15] |
| LDCM0293 | AC32 | HCT 116 | C319(0.66); C326(1.52); C437(1.75) | LDD0610 | [15] |
| LDCM0294 | AC33 | HCT 116 | C319(1.15); C326(1.29); C437(1.51) | LDD0611 | [15] |
| LDCM0295 | AC34 | HCT 116 | C437(1.47); C326(1.48); C319(2.22) | LDD0612 | [15] |
| LDCM0296 | AC35 | HCT 116 | C319(0.83); C437(0.91); C808(0.96); C326(1.00) | LDD0613 | [15] |
| LDCM0297 | AC36 | HCT 116 | C808(0.87); C319(0.96); C437(0.96); C326(1.05) | LDD0614 | [15] |
| LDCM0298 | AC37 | HCT 116 | C319(0.70); C437(0.96); C808(1.03); C326(1.19) | LDD0615 | [15] |
| LDCM0299 | AC38 | HCT 116 | C437(0.93); C319(0.94); C808(0.98); C326(1.06) | LDD0616 | [15] |
| LDCM0300 | AC39 | HCT 116 | C319(1.04); C437(1.04); C326(1.15); C808(1.28) | LDD0617 | [15] |
| LDCM0301 | AC4 | HCT 116 | C319(0.52); C326(1.29) | LDD0618 | [15] |
| LDCM0302 | AC40 | HCT 116 | C808(1.05); C437(1.09); C319(1.15); C326(1.29) | LDD0619 | [15] |
| LDCM0303 | AC41 | HCT 116 | C437(0.98); C808(1.02); C319(1.21); C326(1.25) | LDD0620 | [15] |
| LDCM0304 | AC42 | HCT 116 | C808(0.91); C437(1.02); C326(1.17); C319(1.48) | LDD0621 | [15] |
| LDCM0305 | AC43 | HCT 116 | C319(0.91); C437(1.00); C808(1.09); C326(1.15) | LDD0622 | [15] |
| LDCM0306 | AC44 | HCT 116 | C437(0.99); C808(0.99); C326(1.06); C319(1.35) | LDD0623 | [15] |
| LDCM0307 | AC45 | HCT 116 | C437(1.07); C319(1.10); C808(1.17); C326(1.50) | LDD0624 | [15] |
| LDCM0308 | AC46 | HCT 116 | C319(0.80); C808(1.01); C437(1.04); C326(1.11) | LDD0625 | [15] |
| LDCM0309 | AC47 | HCT 116 | C319(0.80); C808(1.01); C326(1.07); C437(1.14) | LDD0626 | [15] |
| LDCM0310 | AC48 | HCT 116 | C319(0.90); C326(0.96); C808(1.13); C437(1.25) | LDD0627 | [15] |
| LDCM0311 | AC49 | HCT 116 | C319(0.84); C808(1.02); C326(1.18); C437(1.40) | LDD0628 | [15] |
| LDCM0312 | AC5 | HCT 116 | C319(0.53); C326(1.33) | LDD0629 | [15] |
| LDCM0313 | AC50 | HCT 116 | C808(0.94); C319(1.05); C326(1.06); C437(1.32) | LDD0630 | [15] |
| LDCM0314 | AC51 | HCT 116 | C808(0.95); C326(1.05); C437(1.06); C319(1.21) | LDD0631 | [15] |
| LDCM0315 | AC52 | HCT 116 | C319(0.82); C326(1.08); C808(1.19); C437(1.22) | LDD0632 | [15] |
| LDCM0316 | AC53 | HCT 116 | C326(0.99); C808(1.06); C319(1.20); C437(1.26) | LDD0633 | [15] |
| LDCM0317 | AC54 | HCT 116 | C319(0.86); C326(0.92); C808(1.12); C437(1.34) | LDD0634 | [15] |
| LDCM0318 | AC55 | HCT 116 | C808(0.87); C319(0.94); C326(1.16); C437(1.32) | LDD0635 | [15] |
| LDCM0319 | AC56 | HCT 116 | C808(0.40); C319(1.14); C326(1.18); C437(1.59) | LDD0636 | [15] |
| LDCM0320 | AC57 | HCT 116 | C319(0.64); C437(1.09); C326(1.12); C808(1.63) | LDD0637 | [15] |
| LDCM0321 | AC58 | HCT 116 | C326(1.03); C808(1.06); C437(1.06); C319(1.30) | LDD0638 | [15] |
| LDCM0322 | AC59 | HCT 116 | C319(0.98); C437(0.98); C326(1.05); C808(1.09) | LDD0639 | [15] |
| LDCM0323 | AC6 | HCT 116 | C437(0.98); C326(1.18); C319(1.50) | LDD0640 | [15] |
| LDCM0324 | AC60 | HCT 116 | C326(1.08); C437(1.10); C319(1.12); C808(1.58) | LDD0641 | [15] |
| LDCM0325 | AC61 | HCT 116 | C319(0.69); C326(1.07); C437(1.10); C808(1.22) | LDD0642 | [15] |
| LDCM0326 | AC62 | HCT 116 | C319(0.82); C326(1.09); C437(1.10); C808(1.23) | LDD0643 | [15] |
| LDCM0327 | AC63 | HCT 116 | C319(0.84); C437(0.99); C326(1.21); C808(1.62) | LDD0644 | [15] |
| LDCM0328 | AC64 | HCT 116 | C319(0.85); C326(1.00); C437(1.03); C808(1.16) | LDD0645 | [15] |
| LDCM0329 | AC65 | HCT 116 | C319(0.84); C326(1.12); C437(1.46); C808(1.53) | LDD0646 | [15] |
| LDCM0330 | AC66 | HCT 116 | C319(1.13); C437(1.18); C808(1.18); C326(1.20) | LDD0647 | [15] |
| LDCM0331 | AC67 | HCT 116 | C808(0.50); C319(0.86); C437(1.11); C326(1.16) | LDD0648 | [15] |
| LDCM0332 | AC68 | HCT 116 | C808(0.89); C326(0.90); C319(1.39) | LDD0649 | [15] |
| LDCM0333 | AC69 | HCT 116 | C326(0.74); C808(1.02); C319(1.17) | LDD0650 | [15] |
| LDCM0334 | AC7 | HCT 116 | C437(0.92); C326(1.02); C319(1.41) | LDD0651 | [15] |
| LDCM0335 | AC70 | HCT 116 | C808(0.83); C319(0.94); C326(1.28) | LDD0652 | [15] |
| LDCM0336 | AC71 | HCT 116 | C326(0.67); C808(1.19); C319(1.22) | LDD0653 | [15] |
| LDCM0337 | AC72 | HCT 116 | C808(0.76); C326(0.98); C319(1.07) | LDD0654 | [15] |
| LDCM0338 | AC73 | HCT 116 | C808(0.56); C319(0.92); C326(1.00) | LDD0655 | [15] |
| LDCM0339 | AC74 | HCT 116 | C808(0.53); C326(0.76); C319(1.14) | LDD0656 | [15] |
| LDCM0340 | AC75 | HCT 116 | C808(0.23); C319(0.85); C326(1.30) | LDD0657 | [15] |
| LDCM0341 | AC76 | HCT 116 | C326(0.71); C319(0.96); C808(1.65) | LDD0658 | [15] |
| LDCM0342 | AC77 | HCT 116 | C326(0.86); C319(1.39); C808(1.40) | LDD0659 | [15] |
| LDCM0343 | AC78 | HCT 116 | C326(0.87); C319(1.10); C808(1.11) | LDD0660 | [15] |
| LDCM0344 | AC79 | HCT 116 | C326(0.84); C808(0.98); C319(1.12) | LDD0661 | [15] |
| LDCM0345 | AC8 | HCT 116 | C437(0.88); C326(1.00); C319(1.49) | LDD0662 | [15] |
| LDCM0346 | AC80 | HCT 116 | C326(0.68); C319(0.90); C808(0.92) | LDD0663 | [15] |
| LDCM0347 | AC81 | HCT 116 | C326(0.76); C808(0.78); C319(1.11) | LDD0664 | [15] |
| LDCM0348 | AC82 | HCT 116 | C808(0.06); C326(0.68); C319(1.09) | LDD0665 | [15] |
| LDCM0349 | AC83 | HCT 116 | C319(0.87); C437(1.17); C326(1.20) | LDD0666 | [15] |
| LDCM0350 | AC84 | HCT 116 | C319(0.88); C437(1.12); C326(1.27) | LDD0667 | [15] |
| LDCM0351 | AC85 | HCT 116 | C319(0.98); C437(1.08); C326(1.28) | LDD0668 | [15] |
| LDCM0352 | AC86 | HCT 116 | C319(0.87); C326(1.18); C437(1.34) | LDD0669 | [15] |
| LDCM0353 | AC87 | HCT 116 | C319(1.14); C326(1.17); C437(1.55) | LDD0670 | [15] |
| LDCM0354 | AC88 | HCT 116 | C326(1.16); C319(1.19); C437(1.20) | LDD0671 | [15] |
| LDCM0355 | AC89 | HCT 116 | C319(0.87); C326(1.14); C437(1.42) | LDD0672 | [15] |
| LDCM0357 | AC90 | HCT 116 | C319(0.96); C326(1.15); C437(1.31) | LDD0674 | [15] |
| LDCM0358 | AC91 | HCT 116 | C319(0.69); C326(1.23); C437(1.27) | LDD0675 | [15] |
| LDCM0359 | AC92 | HCT 116 | C319(0.79); C437(1.08); C326(1.31) | LDD0676 | [15] |
| LDCM0360 | AC93 | HCT 116 | C437(1.07); C319(1.21); C326(1.30) | LDD0677 | [15] |
| LDCM0361 | AC94 | HCT 116 | C437(0.91); C326(1.17); C319(1.22) | LDD0678 | [15] |
| LDCM0362 | AC95 | HCT 116 | C319(0.99); C326(1.25); C437(1.26) | LDD0679 | [15] |
| LDCM0363 | AC96 | HCT 116 | C437(0.93); C326(1.20); C319(1.34) | LDD0680 | [15] |
| LDCM0364 | AC97 | HCT 116 | C319(1.14); C326(1.38); C437(1.66) | LDD0681 | [15] |
| LDCM0365 | AC98 | HCT 116 | C437(1.04); C326(1.63) | LDD0682 | [15] |
| LDCM0366 | AC99 | HCT 116 | C437(1.02); C326(1.30) | LDD0683 | [15] |
| LDCM0545 | Acetamide | MDA-MB-231 | C319(0.42); C808(0.41); C437(0.55) | LDD2138 | [7] |
| LDCM0166 | Afatinib | A431 | 1.99 | LDD0422 | [6] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C373(0.74); C319(0.56); C808(0.81) | LDD2113 | [7] |
| LDCM0248 | AKOS034007472 | HCT 116 | C326(1.15); C319(1.24); C437(0.93) | LDD0565 | [15] |
| LDCM0356 | AKOS034007680 | HCT 116 | C437(1.07); C326(1.24); C319(1.35) | LDD0673 | [15] |
| LDCM0275 | AKOS034007705 | HCT 116 | C326(0.98); C319(1.26); C437(1.52) | LDD0592 | [15] |
| LDCM0156 | Aniline | NCI-H1299 | N.A. | LDD0404 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C437(0.95) | LDD0078 | [15] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C373(0.70); C319(0.48); C808(0.44); C437(0.98) | LDD2091 | [7] |
| LDCM0087 | Capsaicin | HEK-293T | 4.23 | LDD0185 | [8] |
| LDCM0630 | CCW28-3 | 231MFP | C109(1.39); C326(1.17) | LDD2214 | [28] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [14] |
| LDCM0632 | CL-Sc | Hep-G2 | C319(20.00); C437(20.00); C496(1.50); C437(1.16) | LDD2227 | [19] |
| LDCM0367 | CL1 | HCT 116 | C437(0.88); C319(1.32); C326(1.53) | LDD0684 | [15] |
| LDCM0368 | CL10 | HCT 116 | C437(0.93); C326(1.07); C319(1.53) | LDD0685 | [15] |
| LDCM0369 | CL100 | HCT 116 | C319(0.66); C326(1.45) | LDD0686 | [15] |
| LDCM0370 | CL101 | HCT 116 | C326(0.88); C437(1.06); C319(1.71) | LDD0687 | [15] |
| LDCM0371 | CL102 | HCT 116 | C437(1.31); C326(1.37); C319(1.86) | LDD0688 | [15] |
| LDCM0372 | CL103 | HCT 116 | C437(0.75); C326(1.32); C319(2.03) | LDD0689 | [15] |
| LDCM0373 | CL104 | HCT 116 | C437(0.86); C326(1.13); C319(2.10) | LDD0690 | [15] |
| LDCM0374 | CL105 | HCT 116 | C319(0.70); C437(0.76); C326(1.25) | LDD0691 | [15] |
| LDCM0375 | CL106 | HCT 116 | C319(0.58); C437(0.67); C326(1.54) | LDD0692 | [15] |
| LDCM0376 | CL107 | HCT 116 | C319(0.75); C437(0.85); C326(1.67) | LDD0693 | [15] |
| LDCM0377 | CL108 | HCT 116 | C319(0.69); C437(0.72); C326(1.34) | LDD0694 | [15] |
| LDCM0378 | CL109 | HCT 116 | C319(0.69); C437(0.80); C326(1.56) | LDD0695 | [15] |
| LDCM0379 | CL11 | HCT 116 | C437(1.04); C326(1.67); C319(2.47) | LDD0696 | [15] |
| LDCM0380 | CL110 | HCT 116 | C437(0.72); C319(0.78); C326(2.02) | LDD0697 | [15] |
| LDCM0381 | CL111 | HCT 116 | C319(0.75); C437(0.86); C326(1.38) | LDD0698 | [15] |
| LDCM0382 | CL112 | HCT 116 | C437(1.16); C319(1.21); C326(1.45) | LDD0699 | [15] |
| LDCM0383 | CL113 | HCT 116 | C326(1.44); C437(1.55); C319(2.46) | LDD0700 | [15] |
| LDCM0384 | CL114 | HCT 116 | C437(1.35); C326(1.60); C319(3.00) | LDD0701 | [15] |
| LDCM0385 | CL115 | HCT 116 | C437(1.36); C326(1.47); C319(2.07) | LDD0702 | [15] |
| LDCM0386 | CL116 | HCT 116 | C319(1.16); C326(1.29); C437(1.45) | LDD0703 | [15] |
| LDCM0387 | CL117 | HCT 116 | C808(0.80); C319(1.14); C437(1.23); C326(1.68) | LDD0704 | [15] |
| LDCM0388 | CL118 | HCT 116 | C319(0.72); C437(1.08); C808(1.12); C326(1.22) | LDD0705 | [15] |
| LDCM0389 | CL119 | HCT 116 | C319(0.61); C437(1.09); C808(1.12); C326(1.27) | LDD0706 | [15] |
| LDCM0390 | CL12 | HCT 116 | C326(1.02); C437(1.14); C319(1.28) | LDD0707 | [15] |
| LDCM0391 | CL120 | HCT 116 | C319(0.73); C808(1.06); C437(1.07); C326(1.20) | LDD0708 | [15] |
| LDCM0392 | CL121 | HCT 116 | C808(0.79); C437(1.16); C326(1.24); C319(1.26) | LDD0709 | [15] |
| LDCM0393 | CL122 | HCT 116 | C808(0.96); C437(1.08); C319(1.12); C326(1.20) | LDD0710 | [15] |
| LDCM0394 | CL123 | HCT 116 | C319(0.73); C808(1.01); C437(1.10); C326(1.23) | LDD0711 | [15] |
| LDCM0395 | CL124 | HCT 116 | C319(0.70); C808(1.02); C326(1.11); C437(1.27) | LDD0712 | [15] |
| LDCM0396 | CL125 | HCT 116 | C319(0.59); C808(1.03); C326(1.20); C437(1.20) | LDD0713 | [15] |
| LDCM0397 | CL126 | HCT 116 | C437(1.04); C319(1.21); C326(1.22); C808(1.26) | LDD0714 | [15] |
| LDCM0398 | CL127 | HCT 116 | C437(1.05); C319(1.12); C326(1.22); C808(1.62) | LDD0715 | [15] |
| LDCM0399 | CL128 | HCT 116 | C319(0.63); C437(0.99); C326(0.99); C808(1.71) | LDD0716 | [15] |
| LDCM0400 | CL13 | HCT 116 | C437(1.04); C319(1.17); C326(1.47) | LDD0717 | [15] |
| LDCM0401 | CL14 | HCT 116 | C437(0.90); C326(1.09); C319(1.77) | LDD0718 | [15] |
| LDCM0402 | CL15 | HCT 116 | C319(0.76); C437(1.02); C326(3.12) | LDD0719 | [15] |
| LDCM0403 | CL16 | HCT 116 | C437(1.03); C326(1.25) | LDD0720 | [15] |
| LDCM0404 | CL17 | HCT 116 | C319(0.55); C437(0.91); C326(4.91) | LDD0721 | [15] |
| LDCM0405 | CL18 | HCT 116 | C319(0.64); C437(1.03); C326(1.18) | LDD0722 | [15] |
| LDCM0406 | CL19 | HCT 116 | C319(0.78); C326(0.95); C437(1.16) | LDD0723 | [15] |
| LDCM0407 | CL2 | HCT 116 | C437(0.92); C326(1.20); C319(1.29) | LDD0724 | [15] |
| LDCM0408 | CL20 | HCT 116 | C319(0.80); C326(1.09); C437(1.31) | LDD0725 | [15] |
| LDCM0409 | CL21 | HCT 116 | C319(0.61); C437(1.57); C326(2.22) | LDD0726 | [15] |
| LDCM0410 | CL22 | HCT 116 | C326(1.20); C437(1.98); C319(2.00) | LDD0727 | [15] |
| LDCM0411 | CL23 | HCT 116 | C319(0.98); C326(1.10); C437(1.18) | LDD0728 | [15] |
| LDCM0412 | CL24 | HCT 116 | C319(0.83); C326(1.05); C437(1.45) | LDD0729 | [15] |
| LDCM0413 | CL25 | HCT 116 | C326(1.15); C319(0.69); C437(1.36) | LDD0730 | [15] |
| LDCM0414 | CL26 | HCT 116 | C326(1.28); C319(0.78); C437(1.01) | LDD0731 | [15] |
| LDCM0415 | CL27 | HCT 116 | C326(0.98); C319(0.87); C437(1.21) | LDD0732 | [15] |
| LDCM0416 | CL28 | HCT 116 | C326(1.17); C319(0.60); C437(1.26) | LDD0733 | [15] |
| LDCM0417 | CL29 | HCT 116 | C326(1.01); C319(1.42); C437(1.31) | LDD0734 | [15] |
| LDCM0418 | CL3 | HCT 116 | C326(1.21); C319(1.34); C437(0.90) | LDD0735 | [15] |
| LDCM0419 | CL30 | HCT 116 | C326(1.03); C319(0.47); C437(1.04) | LDD0736 | [15] |
| LDCM0420 | CL31 | HCT 116 | C326(1.59); C319(0.63); C437(0.96) | LDD0737 | [15] |
| LDCM0421 | CL32 | HCT 116 | C326(1.37); C437(1.04) | LDD0738 | [15] |
| LDCM0422 | CL33 | HCT 116 | C326(11.33); C437(1.04) | LDD0739 | [15] |
| LDCM0423 | CL34 | HCT 116 | C326(1.72); C437(1.33) | LDD0740 | [15] |
| LDCM0424 | CL35 | HCT 116 | C326(1.38); C437(1.29) | LDD0741 | [15] |
| LDCM0425 | CL36 | HCT 116 | C326(1.49); C437(1.22) | LDD0742 | [15] |
| LDCM0426 | CL37 | HCT 116 | C326(1.67); C437(1.39) | LDD0743 | [15] |
| LDCM0428 | CL39 | HCT 116 | C326(1.65); C437(1.24) | LDD0745 | [15] |
| LDCM0429 | CL4 | HCT 116 | C326(10.44); C319(1.94); C437(0.90) | LDD0746 | [15] |
| LDCM0430 | CL40 | HCT 116 | C326(2.41); C437(1.09) | LDD0747 | [15] |
| LDCM0431 | CL41 | HCT 116 | C326(1.67); C437(1.14) | LDD0748 | [15] |
| LDCM0432 | CL42 | HCT 116 | C326(2.01); C437(1.40) | LDD0749 | [15] |
| LDCM0433 | CL43 | HCT 116 | C326(2.01); C437(1.26) | LDD0750 | [15] |
| LDCM0434 | CL44 | HCT 116 | C326(1.89); C437(1.22) | LDD0751 | [15] |
| LDCM0435 | CL45 | HCT 116 | C326(3.42); C437(1.23) | LDD0752 | [15] |
| LDCM0436 | CL46 | HCT 116 | C326(1.23); C319(0.30); C437(0.81) | LDD0753 | [15] |
| LDCM0437 | CL47 | HCT 116 | C326(0.98); C319(1.06); C437(0.78) | LDD0754 | [15] |
| LDCM0438 | CL48 | HCT 116 | C326(1.06); C319(0.47); C437(0.80) | LDD0755 | [15] |
| LDCM0439 | CL49 | HCT 116 | C326(0.87); C319(0.28); C437(0.87) | LDD0756 | [15] |
| LDCM0440 | CL5 | HCT 116 | C326(0.94); C319(0.76); C437(0.98) | LDD0757 | [15] |
| LDCM0441 | CL50 | HCT 116 | C326(1.02); C319(0.51); C437(0.76) | LDD0758 | [15] |
| LDCM0442 | CL51 | HCT 116 | C326(0.99); C319(0.31); C437(1.02) | LDD0759 | [15] |
| LDCM0443 | CL52 | HCT 116 | C326(0.99); C319(0.36); C437(0.91) | LDD0760 | [15] |
| LDCM0444 | CL53 | HCT 116 | C326(1.00); C319(3.34); C437(0.82) | LDD0761 | [15] |
| LDCM0445 | CL54 | HCT 116 | C326(0.98); C319(0.40); C437(0.85) | LDD0762 | [15] |
| LDCM0446 | CL55 | HCT 116 | C326(0.87); C319(0.67); C437(0.82) | LDD0763 | [15] |
| LDCM0447 | CL56 | HCT 116 | C326(1.08); C319(1.32); C437(0.93) | LDD0764 | [15] |
| LDCM0448 | CL57 | HCT 116 | C326(1.60); C319(1.47); C437(1.02) | LDD0765 | [15] |
| LDCM0449 | CL58 | HCT 116 | C326(1.16); C319(0.41); C437(0.70) | LDD0766 | [15] |
| LDCM0450 | CL59 | HCT 116 | C326(0.92); C319(0.66); C437(0.74) | LDD0767 | [15] |
| LDCM0451 | CL6 | HCT 116 | C326(0.94); C319(0.50); C437(1.04) | LDD0768 | [15] |
| LDCM0452 | CL60 | HCT 116 | C326(0.97); C319(0.28); C437(0.80) | LDD0769 | [15] |
| LDCM0453 | CL61 | HCT 116 | C326(1.01); C319(0.61); C437(1.16) | LDD0770 | [15] |
| LDCM0454 | CL62 | HCT 116 | C326(1.03); C319(0.70); C437(1.15) | LDD0771 | [15] |
| LDCM0455 | CL63 | HCT 116 | C326(1.47); C319(1.48); C437(1.12) | LDD0772 | [15] |
| LDCM0456 | CL64 | HCT 116 | C326(1.08); C319(0.64); C437(1.18) | LDD0773 | [15] |
| LDCM0457 | CL65 | HCT 116 | C326(1.08); C319(1.00); C437(1.11) | LDD0774 | [15] |
| LDCM0458 | CL66 | HCT 116 | C326(1.03); C319(0.75); C437(1.44) | LDD0775 | [15] |
| LDCM0459 | CL67 | HCT 116 | C326(0.93); C319(2.05); C437(1.38) | LDD0776 | [15] |
| LDCM0460 | CL68 | HCT 116 | C326(1.19); C319(8.93); C437(1.40) | LDD0777 | [15] |
| LDCM0461 | CL69 | HCT 116 | C326(1.28); C319(1.05); C437(1.01) | LDD0778 | [15] |
| LDCM0462 | CL7 | HCT 116 | C326(1.00); C319(0.44); C437(1.17) | LDD0779 | [15] |
| LDCM0463 | CL70 | HCT 116 | C326(0.99); C319(1.26); C437(1.20) | LDD0780 | [15] |
| LDCM0464 | CL71 | HCT 116 | C326(1.16); C319(1.16); C437(1.14) | LDD0781 | [15] |
| LDCM0465 | CL72 | HCT 116 | C326(1.02); C319(1.74); C437(1.04) | LDD0782 | [15] |
| LDCM0466 | CL73 | HCT 116 | C326(1.20); C319(1.93); C437(1.23) | LDD0783 | [15] |
| LDCM0467 | CL74 | HCT 116 | C326(1.13); C319(1.82); C437(1.08) | LDD0784 | [15] |
| LDCM0469 | CL76 | HCT 116 | C326(1.06); C808(1.15) | LDD0786 | [15] |
| LDCM0470 | CL77 | HCT 116 | C326(2.97); C808(0.81) | LDD0787 | [15] |
| LDCM0471 | CL78 | HCT 116 | C326(2.50); C808(1.08) | LDD0788 | [15] |
| LDCM0472 | CL79 | HCT 116 | C326(1.01); C808(1.09) | LDD0789 | [15] |
| LDCM0473 | CL8 | HCT 116 | C326(2.85); C319(1.00); C437(1.05) | LDD0790 | [15] |
| LDCM0474 | CL80 | HCT 116 | C326(0.98); C808(1.04) | LDD0791 | [15] |
| LDCM0475 | CL81 | HCT 116 | C326(1.02); C808(0.94) | LDD0792 | [15] |
| LDCM0476 | CL82 | HCT 116 | C326(0.92); C808(1.00) | LDD0793 | [15] |
| LDCM0477 | CL83 | HCT 116 | C326(1.22); C808(1.07) | LDD0794 | [15] |
| LDCM0478 | CL84 | HCT 116 | C326(1.19); C808(0.75) | LDD0795 | [15] |
| LDCM0479 | CL85 | HCT 116 | C326(1.15); C808(0.93) | LDD0796 | [15] |
| LDCM0480 | CL86 | HCT 116 | C326(1.15); C808(0.87) | LDD0797 | [15] |
| LDCM0481 | CL87 | HCT 116 | C326(1.00); C808(0.96) | LDD0798 | [15] |
| LDCM0482 | CL88 | HCT 116 | C326(0.94); C808(0.83) | LDD0799 | [15] |
| LDCM0483 | CL89 | HCT 116 | C326(0.95); C808(0.56) | LDD0800 | [15] |
| LDCM0484 | CL9 | HCT 116 | C326(1.10); C319(0.56); C437(1.11) | LDD0801 | [15] |
| LDCM0485 | CL90 | HCT 116 | C326(14.52); C808(0.79) | LDD0802 | [15] |
| LDCM0486 | CL91 | HCT 116 | C326(1.14); C319(0.85) | LDD0803 | [15] |
| LDCM0487 | CL92 | HCT 116 | C326(1.72); C319(0.83) | LDD0804 | [15] |
| LDCM0488 | CL93 | HCT 116 | C326(1.09); C319(0.43) | LDD0805 | [15] |
| LDCM0489 | CL94 | HCT 116 | C326(1.66); C319(0.61) | LDD0806 | [15] |
| LDCM0490 | CL95 | HCT 116 | C326(1.94); C319(0.54) | LDD0807 | [15] |
| LDCM0491 | CL96 | HCT 116 | C326(1.17); C319(0.69) | LDD0808 | [15] |
| LDCM0492 | CL97 | HCT 116 | C326(1.18); C319(0.60) | LDD0809 | [15] |
| LDCM0493 | CL98 | HCT 116 | C326(1.29); C319(0.66) | LDD0810 | [15] |
| LDCM0494 | CL99 | HCT 116 | C326(1.62); C319(0.72) | LDD0811 | [15] |
| LDCM0634 | CY-0357 | Hep-G2 | C326(2.10); C437(0.82) | LDD2228 | [19] |
| LDCM0198 | Dimethyl Fumarate(DMF) | T cell | C326(4.11) | LDD0526 | [29] |
| LDCM0495 | E2913 | HEK-293T | C319(0.84); C808(1.07); C437(0.92); C446(1.04) | LDD1698 | [30] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C326(0.92) | LDD1702 | [7] |
| LDCM0175 | Ethacrynic acid | HeLa | C326(1.22); C143(0.98); C496(0.88) | LDD2210 | [10] |
| LDCM0625 | F8 | Ramos | C437(0.99); C319(0.96); C326(0.98) | LDD2187 | [31] |
| LDCM0572 | Fragment10 | MDA-MB-231 | C326(1.95) | LDD1389 | [32] |
| LDCM0573 | Fragment11 | MDA-MB-231 | C326(0.76) | LDD1467 | [32] |
| LDCM0574 | Fragment12 | MDA-MB-231 | C326(3.90) | LDD1468 | [32] |
| LDCM0575 | Fragment13 | MDA-MB-231 | C326(1.34) | LDD1469 | [32] |
| LDCM0576 | Fragment14 | MDA-MB-231 | C326(1.13) | LDD1397 | [32] |
| LDCM0579 | Fragment20 | MDA-MB-231 | C326(1.97) | LDD1402 | [32] |
| LDCM0580 | Fragment21 | MDA-MB-231 | C326(1.03) | LDD1404 | [32] |
| LDCM0582 | Fragment23 | MDA-MB-231 | C326(1.02) | LDD1408 | [32] |
| LDCM0584 | Fragment25 | MDA-MB-231 | C326(0.98) | LDD1411 | [32] |
| LDCM0578 | Fragment27 | MDA-MB-231 | C326(0.85) | LDD1401 | [32] |
| LDCM0586 | Fragment28 | MDA-MB-231 | C326(0.98) | LDD1415 | [32] |
| LDCM0587 | Fragment29 | MDA-MB-231 | C326(1.58) | LDD1417 | [32] |
| LDCM0588 | Fragment30 | MDA-MB-231 | C326(1.13) | LDD1419 | [32] |
| LDCM0589 | Fragment31 | MDA-MB-231 | C326(1.18) | LDD1421 | [32] |
| LDCM0590 | Fragment32 | MDA-MB-231 | C326(3.55) | LDD1423 | [32] |
| LDCM0468 | Fragment33 | HCT 116 | C326(1.14); C319(1.20); C437(1.20) | LDD0785 | [15] |
| LDCM0594 | Fragment36 | MDA-MB-231 | C326(1.48) | LDD1431 | [32] |
| LDCM0596 | Fragment38 | MDA-MB-231 | C326(1.15) | LDD1433 | [32] |
| LDCM0597 | Fragment39 | MDA-MB-231 | C326(20.00) | LDD1435 | [32] |
| LDCM0566 | Fragment4 | MDA-MB-231 | C326(1.43) | LDD1378 | [32] |
| LDCM0598 | Fragment40 | MDA-MB-231 | C326(0.62) | LDD1436 | [32] |
| LDCM0599 | Fragment41 | MDA-MB-231 | C326(1.21) | LDD1438 | [32] |
| LDCM0602 | Fragment44 | MDA-MB-231 | C326(0.82) | LDD1443 | [32] |
| LDCM0603 | Fragment45 | MDA-MB-231 | C326(5.26) | LDD1482 | [32] |
| LDCM0604 | Fragment46 | MDA-MB-231 | C326(0.83) | LDD1445 | [32] |
| LDCM0606 | Fragment48 | MDA-MB-231 | C326(1.20) | LDD1447 | [32] |
| LDCM0427 | Fragment51 | HCT 116 | C326(2.54); C437(1.33) | LDD0744 | [15] |
| LDCM0610 | Fragment52 | MDA-MB-231 | C326(1.22) | LDD1452 | [32] |
| LDCM0613 | Fragment55 | MDA-MB-231 | C326(1.13) | LDD1457 | [32] |
| LDCM0614 | Fragment56 | MDA-MB-231 | C326(1.32) | LDD1458 | [32] |
| LDCM0569 | Fragment7 | MDA-MB-231 | C326(1.48) | LDD1383 | [32] |
| LDCM0570 | Fragment8 | MDA-MB-231 | C326(16.59) | LDD1385 | [32] |
| LDCM0571 | Fragment9 | MDA-MB-231 | C326(1.14) | LDD1387 | [32] |
| LDCM0116 | HHS-0101 | DM93 | Y197(0.72); Y614(0.83); Y76(0.96); Y298(0.99) | LDD0264 | [12] |
| LDCM0117 | HHS-0201 | DM93 | Y186(0.80); Y614(0.80); Y298(0.88); Y197(0.90) | LDD0265 | [12] |
| LDCM0118 | HHS-0301 | DM93 | Y76(0.40); Y614(0.70); Y298(0.72); Y204(0.88) | LDD0266 | [12] |
| LDCM0119 | HHS-0401 | DM93 | Y76(0.64); Y197(0.65); Y614(0.78); Y298(0.82) | LDD0267 | [12] |
| LDCM0120 | HHS-0701 | DM93 | Y76(0.32); Y197(0.41); Y614(0.46); Y298(0.55) | LDD0268 | [12] |
| LDCM0123 | JWB131 | DM93 | Y186(0.85); Y197(0.88); Y204(0.97); Y227(1.37) | LDD0285 | [11] |
| LDCM0124 | JWB142 | DM93 | Y186(0.68); Y197(0.73); Y204(0.81); Y227(0.77) | LDD0286 | [11] |
| LDCM0125 | JWB146 | DM93 | Y186(0.83); Y197(1.07); Y204(0.86); Y227(1.33) | LDD0287 | [11] |
| LDCM0126 | JWB150 | DM93 | Y186(2.85); Y197(2.96); Y204(3.11); Y227(3.59) | LDD0288 | [11] |
| LDCM0127 | JWB152 | DM93 | Y186(1.98); Y197(1.48); Y204(2.13); Y227(2.18) | LDD0289 | [11] |
| LDCM0128 | JWB198 | DM93 | Y186(1.15); Y197(1.16); Y204(1.49); Y227(1.46) | LDD0290 | [11] |
| LDCM0129 | JWB202 | DM93 | Y186(0.54); Y197(0.41); Y204(0.45); Y227(1.16) | LDD0291 | [11] |
| LDCM0130 | JWB211 | DM93 | Y186(0.85); Y197(0.90); Y204(1.06); Y227(1.43) | LDD0292 | [11] |
| LDCM0022 | KB02 | MDA-MB-231 | C326(15.32) | LDD1374 | [32] |
| LDCM0023 | KB03 | MDA-MB-231 | C326(1.75) | LDD1376 | [32] |
| LDCM0024 | KB05 | COLO792 | C326(2.06) | LDD3310 | [33] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C373(0.84); C319(0.98) | LDD2102 | [7] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C373(0.85); C319(0.61); C808(0.66); C326(0.82) | LDD2121 | [7] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C373(0.91); C319(0.62); C808(0.46); C326(0.87) | LDD2089 | [7] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C808(1.13) | LDD2090 | [7] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C319(0.95) | LDD2092 | [7] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C373(1.37); C319(1.29); C808(1.36); C326(0.84) | LDD2093 | [7] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C319(1.21) | LDD2094 | [7] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C319(0.60); C808(0.94) | LDD2097 | [7] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C373(1.02); C319(0.94) | LDD2098 | [7] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C373(1.33); C319(0.81); C808(1.00); C326(1.16) | LDD2099 | [7] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C373(1.05); C319(1.02); C808(1.08) | LDD2101 | [7] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C373(0.66) | LDD2104 | [7] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C373(0.97); C319(1.72); C437(1.73) | LDD2105 | [7] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C373(1.33); C319(0.92); C808(1.37); C326(1.17) | LDD2107 | [7] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C373(0.97); C319(0.86); C437(0.89) | LDD2109 | [7] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C373(1.40); C808(0.99); C326(1.26) | LDD2111 | [7] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C373(0.60); C319(0.46) | LDD2114 | [7] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C373(0.59); C319(0.50) | LDD2115 | [7] |
| LDCM0523 | Nucleophilic fragment 24b | MDA-MB-231 | C373(0.54); C319(0.59) | LDD2116 | [7] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C373(0.52); C319(0.56) | LDD2118 | [7] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C373(1.72) | LDD2119 | [7] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C373(0.47); C319(0.66); C808(0.85) | LDD2120 | [7] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C373(0.53); C319(0.54); C437(0.63) | LDD2122 | [7] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C319(0.76); C808(0.78); C326(1.10); C437(0.85) | LDD2123 | [7] |
| LDCM0531 | Nucleophilic fragment 28b | MDA-MB-231 | C319(0.51); C808(0.06) | LDD2124 | [7] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C373(1.29); C319(0.89); C326(0.91); C437(0.75) | LDD2125 | [7] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C319(0.64) | LDD2126 | [7] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C319(0.92); C808(1.07); C437(1.15) | LDD2127 | [7] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C373(0.46); C319(0.57); C808(0.99); C437(0.90) | LDD2128 | [7] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C373(1.34); C319(0.95); C808(1.10) | LDD2129 | [7] |
| LDCM0540 | Nucleophilic fragment 35 | MDA-MB-231 | C808(0.71) | LDD2133 | [7] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C373(0.63); C808(0.48) | LDD2134 | [7] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C373(1.68); C319(1.15); C326(1.10); C437(0.57) | LDD2135 | [7] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C319(1.13); C808(1.24); C326(1.09) | LDD2136 | [7] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C319(0.94); C326(0.92); C437(1.58) | LDD2137 | [7] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C319(1.95) | LDD1700 | [7] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C373(1.23); C319(0.74); C326(0.96); C437(0.76) | LDD2140 | [7] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C373(0.72); C319(0.59) | LDD2141 | [7] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C373(0.64); C319(0.56); C808(1.01) | LDD2143 | [7] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C373(1.48); C319(2.89); C326(1.56) | LDD2144 | [7] |
| LDCM0551 | Nucleophilic fragment 5b | MDA-MB-231 | C373(0.50) | LDD2145 | [7] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C319(0.79); C326(1.09) | LDD2146 | [7] |
| LDCM0553 | Nucleophilic fragment 6b | MDA-MB-231 | C373(0.91); C319(1.05); C808(3.91) | LDD2147 | [7] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C373(0.48); C808(0.42) | LDD2148 | [7] |
| LDCM0555 | Nucleophilic fragment 7b | MDA-MB-231 | C373(0.41); C319(0.65) | LDD2149 | [7] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C373(0.59); C319(0.41); C808(0.36); C326(0.75) | LDD2150 | [7] |
| LDCM0557 | Nucleophilic fragment 8b | MDA-MB-231 | C373(0.39); C319(0.44) | LDD2151 | [7] |
| LDCM0131 | RA190 | MM1.R | C326(1.19) | LDD0304 | [34] |
| LDCM0021 | THZ1 | HCT 116 | C437(1.03) | LDD2173 | [15] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Protein phosphatase 1 regulatory subunit 3B (PPP1R3B) | . | Q86XI6 | |||
References
