Details of the Target
General Information of Target
| Target ID | LDTP04374 | |||||
|---|---|---|---|---|---|---|
| Target Name | Dynamin-2 (DNM2) | |||||
| Gene Name | DNM2 | |||||
| Gene ID | 1785 | |||||
| Synonyms |
DYN2; Dynamin-2; EC 3.6.5.5; Dynamin 2; Dynamin II |
|||||
| 3D Structure | ||||||
| Sequence |
MGNRGMEELIPLVNKLQDAFSSIGQSCHLDLPQIAVVGGQSAGKSSVLENFVGRDFLPRG
SGIVTRRPLILQLIFSKTEHAEFLHCKSKKFTDFDEVRQEIEAETDRVTGTNKGISPVPI NLRVYSPHVLNLTLIDLPGITKVPVGDQPPDIEYQIKDMILQFISRESSLILAVTPANMD LANSDALKLAKEVDPQGLRTIGVITKLDLMDEGTDARDVLENKLLPLRRGYIGVVNRSQK DIEGKKDIRAALAAERKFFLSHPAYRHMADRMGTPHLQKTLNQQLTNHIRESLPALRSKL QSQLLSLEKEVEEYKNFRPDDPTRKTKALLQMVQQFGVDFEKRIEGSGDQVDTLELSGGA RINRIFHERFPFELVKMEFDEKDLRREISYAIKNIHGVRTGLFTPDLAFEAIVKKQVVKL KEPCLKCVDLVIQELINTVRQCTSKLSSYPRLREETERIVTTYIREREGRTKDQILLLID IEQSYINTNHEDFIGFANAQQRSTQLNKKRAIPNQGEILVIRRGWLTINNISLMKGGSKE YWFVLTAESLSWYKDEEEKEKKYMLPLDNLKIRDVEKGFMSNKHVFAIFNTEQRNVYKDL RQIELACDSQEDVDSWKASFLRAGVYPEKDQAENEDGAQENTFSMDPQLERQVETIRNLV DSYVAIINKSIRDLMPKTIMHLMINNTKAFIHHELLAYLYSSADQSSLMEESADQAQRRD DMLRMYHALKEALNIIGDISTSTVSTPVPPPVDDTWLQSASSHSPTPQRRPVSSIHPPGR PPAVRGPTPGPPLIPVPVGAAASFSAPPIPSRPGPQSVFANSDLFPAPPQIPSRPVRIPP GIPPGVPSRRPPAAPSRPTIIRPAEPSLLD |
|||||
| Target Type |
Clinical trial
|
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
TRAFAC class dynamin-like GTPase superfamily, Dynamin/Fzo/YdjA family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Catalyzes the hydrolysis of GTP and utilizes this energy to mediate vesicle scission at plasma membrane during endocytosis and filament remodeling at many actin structures during organization of the actin cytoskeleton. Plays an important role in vesicular trafficking processes, namely clathrin-mediated endocytosis (CME), exocytic and clathrin-coated vesicle from the trans-Golgi network, and PDGF stimulated macropinocytosis. During vesicular trafficking process, associates to the membrane, through lipid binding, and self-assembles into ring-like structure through oligomerization to form a helical polymer around the vesicle membrane and leading to vesicle scission. Plays a role in organization of the actin cytoskeleton by mediating arrangement of stress fibers and actin bundles in podocytes. During organization of the actin cytoskeleton, self-assembles into ring-like structure that directly bundles actin filaments to form typical membrane tubules decorated with dynamin spiral polymers. Self-assembly increases GTPase activity and the GTP hydrolysis causes the rapid depolymerization of dynamin spiral polymers, and results in dispersion of actin bundles. Remodels, through its interaction with CTTN, bundled actin filaments in a GTPase-dependent manner and plays a role in orchestrating the global actomyosin cytoskeleton. The interaction with CTTN stabilizes the interaction of DNM2 and actin filaments and stimulates the intrinsic GTPase activity that results in actin filament-barbed ends and increases the sensitivity of filaments in bundles to the actin depolymerizing factor, CFL1. Plays a role in the autophagy process, by participating in the formation of ATG9A vesicles destined for the autophagosomes through its interaction with SNX18, by mediating recycling endosome scission leading to autophagosome release through MAP1LC3B interaction. Also regulates maturation of apoptotic cell corpse-containing phagosomes by recruiting PIK3C3 to the phagosome membrane. Also plays a role in cytokinesis. May participate in centrosome cohesion through its interaction with TUBG1. Plays a role in the regulation of neuron morphology, axon growth and formation of neuronal growth cones. Involved in membrane tubulation.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
A-EBA Probe Info |
 |
2.85 | LDD0215 | [2] | |
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [3] | |
|
CY4 Probe Info |
 |
100.00 | LDD0244 | [3] | |
|
N1 Probe Info |
 |
100.00 | LDD0242 | [3] | |
|
TH211 Probe Info |
 |
Y265(20.00); Y726(20.00); Y125(18.46); Y626(5.13) | LDD0257 | [4] | |
|
TH214 Probe Info |
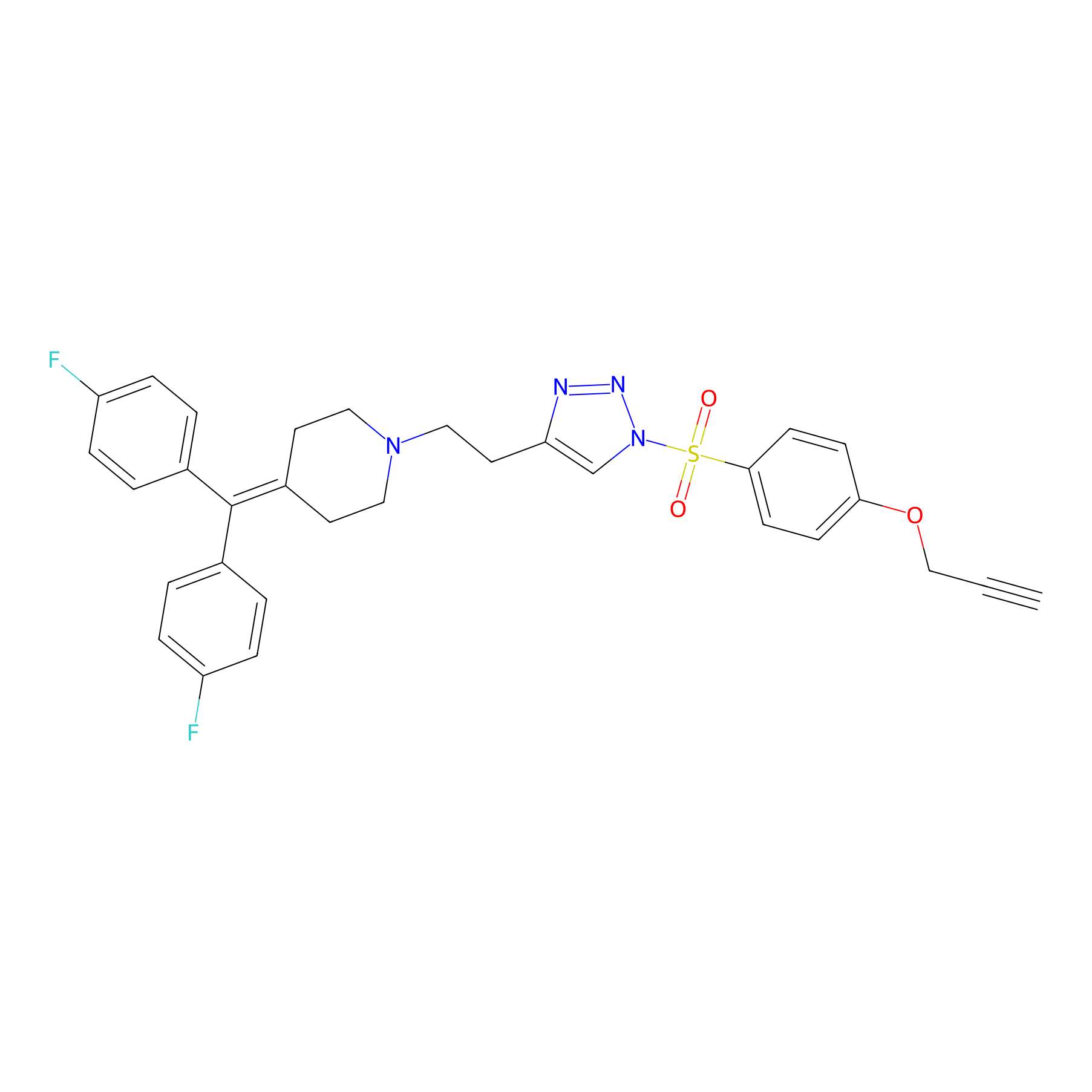 |
Y265(6.42) | LDD0258 | [4] | |
|
TH216 Probe Info |
 |
Y726(20.00); Y231(8.60); Y125(6.33); Y265(5.05) | LDD0259 | [4] | |
|
ONAyne Probe Info |
 |
K299(5.88) | LDD0274 | [5] | |
|
STPyne Probe Info |
 |
K299(9.09); K393(10.00); K508(9.10); K577(1.49) | LDD0277 | [5] | |
|
Probe 1 Probe Info |
 |
Y154(7.14); Y231(27.98); Y314(85.46); Y463(13.83) | LDD3495 | [6] | |
|
DBIA Probe Info |
 |
C86(2.50) | LDD3310 | [7] | |
|
BTD Probe Info |
 |
C86(0.83) | LDD2089 | [8] | |
|
AHL-Pu-1 Probe Info |
 |
C27(2.38) | LDD0171 | [9] | |
|
HHS-475 Probe Info |
 |
Y626(0.80); Y597(0.82); Y390(0.82); Y726(0.87) | LDD0264 | [10] | |
|
HHS-465 Probe Info |
 |
Y125(1.16); Y265(1.26); Y390(9.87); Y597(3.85) | LDD2237 | [11] | |
|
W1 Probe Info |
 |
C27(0.68) | LDD0239 | [12] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [13] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [14] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C427(0.00); C27(0.00); C607(0.00); C86(0.00) | LDD0038 | [15] | |
|
IA-alkyne Probe Info |
 |
C427(0.00); C27(0.00) | LDD0032 | [16] | |
|
Lodoacetamide azide Probe Info |
 |
C427(0.00); C424(0.00); C27(0.00); C607(0.00) | LDD0037 | [15] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [17] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [18] | |
|
NAIA_4 Probe Info |
 |
C27(0.00); C86(0.00) | LDD2226 | [19] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [18] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [20] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [20] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [21] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [21] | |
|
Acrolein Probe Info |
 |
C86(0.00); H85(0.00) | LDD0217 | [22] | |
|
AOyne Probe Info |
 |
6.60 | LDD0443 | [23] | |
|
NAIA_5 Probe Info |
 |
C427(0.00); C27(0.00); C424(0.00); C607(0.00) | LDD2223 | [19] | |
|
HHS-482 Probe Info |
 |
Y390(1.47) | LDD2239 | [11] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C112 Probe Info |
 |
28.64 | LDD1799 | [24] | |
|
C174 Probe Info |
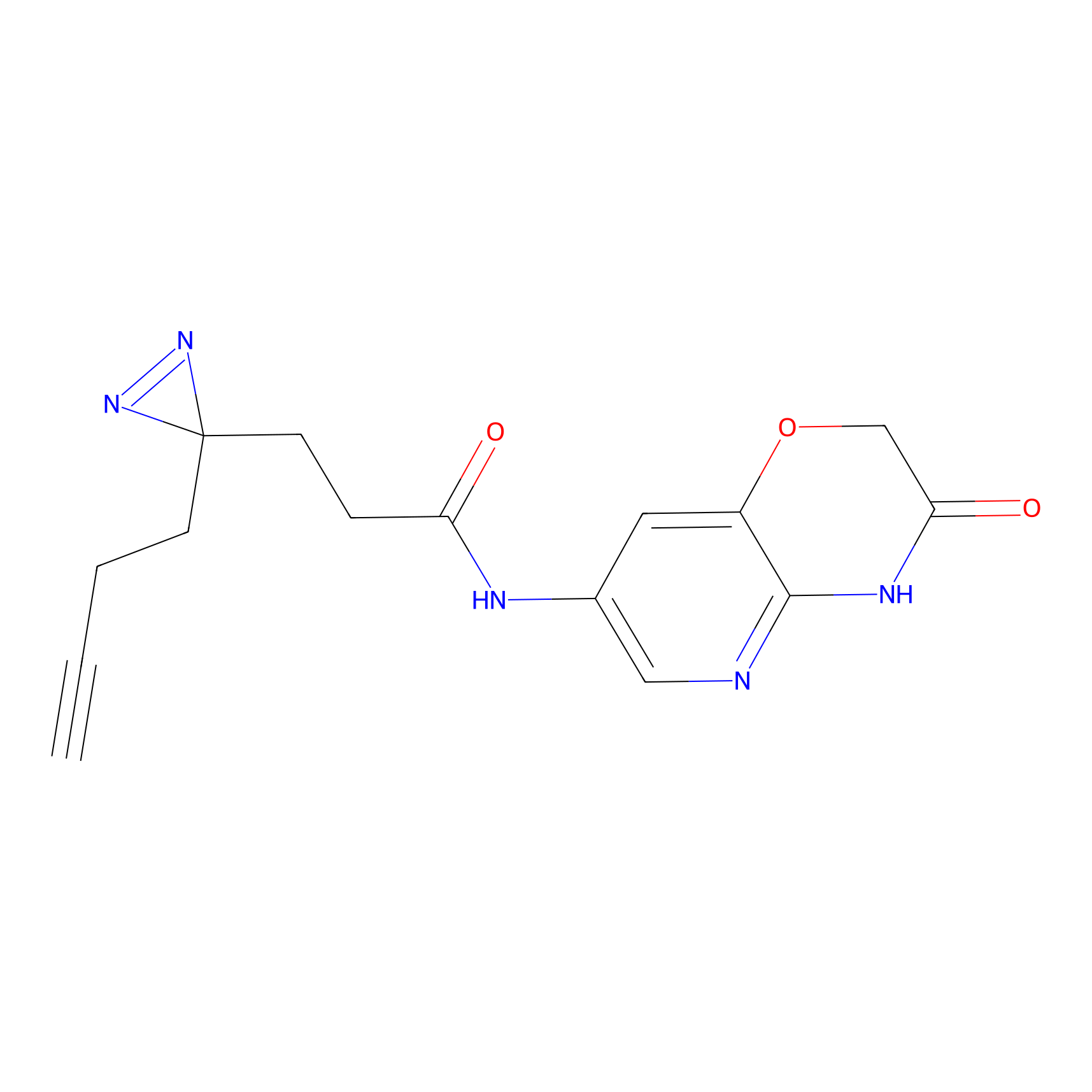 |
6.11 | LDD1854 | [24] | |
|
C244 Probe Info |
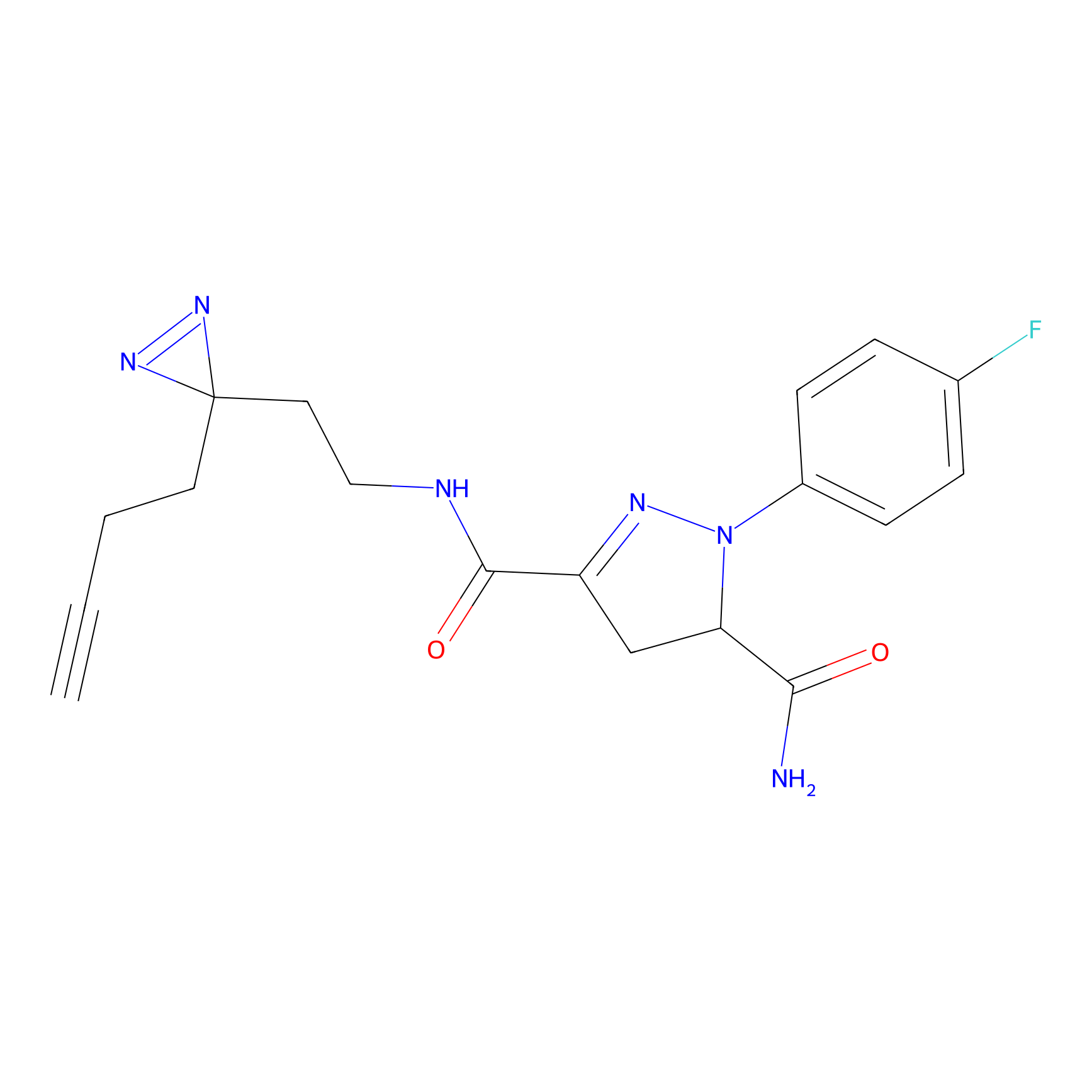 |
16.45 | LDD1917 | [24] | |
|
C264 Probe Info |
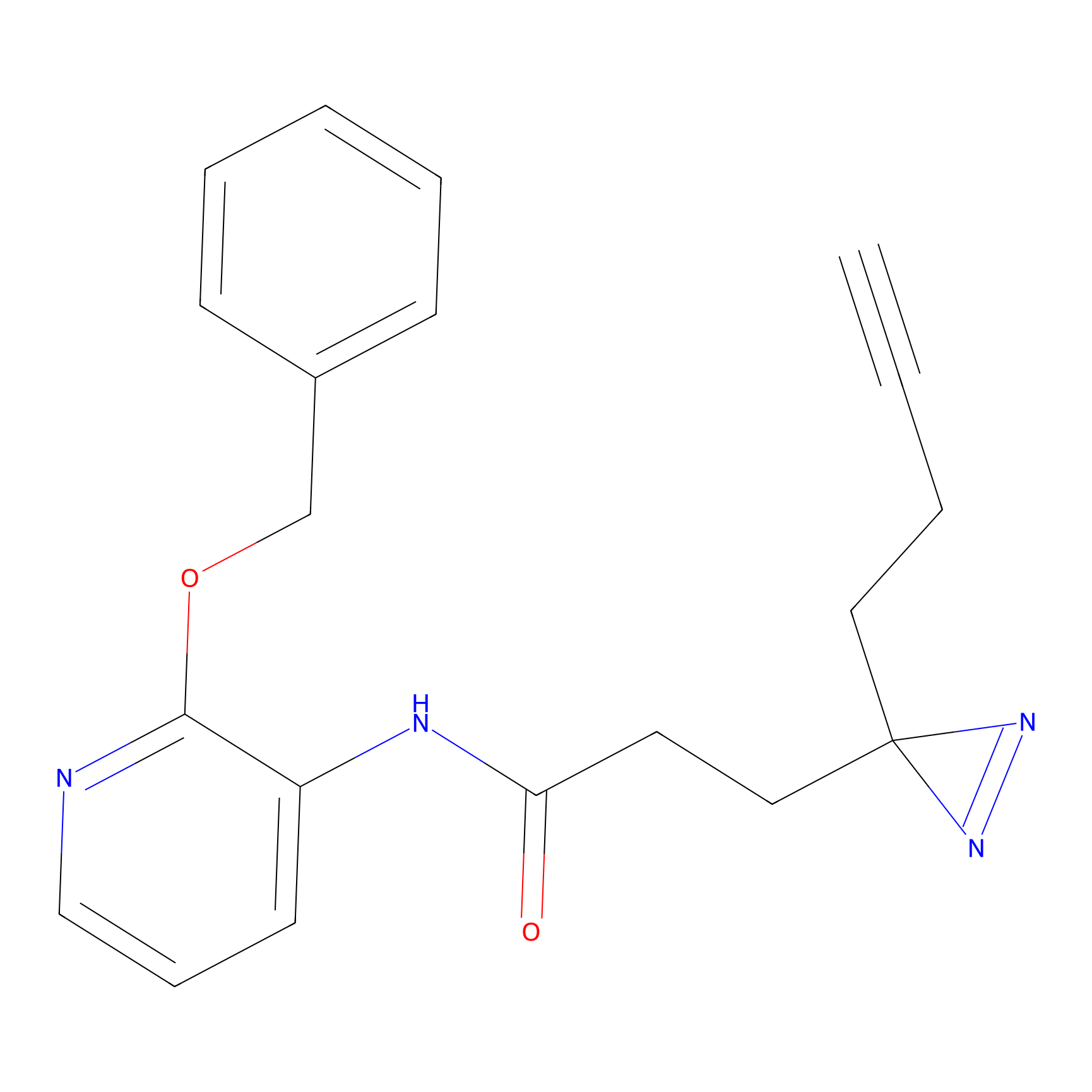 |
18.38 | LDD1935 | [24] | |
|
FFF probe11 Probe Info |
 |
10.88 | LDD0471 | [25] | |
|
FFF probe13 Probe Info |
 |
18.23 | LDD0475 | [25] | |
|
FFF probe14 Probe Info |
 |
13.50 | LDD0477 | [25] | |
|
FFF probe2 Probe Info |
 |
7.59 | LDD0463 | [25] | |
|
FFF probe3 Probe Info |
 |
17.02 | LDD0464 | [25] | |
|
FFF probe6 Probe Info |
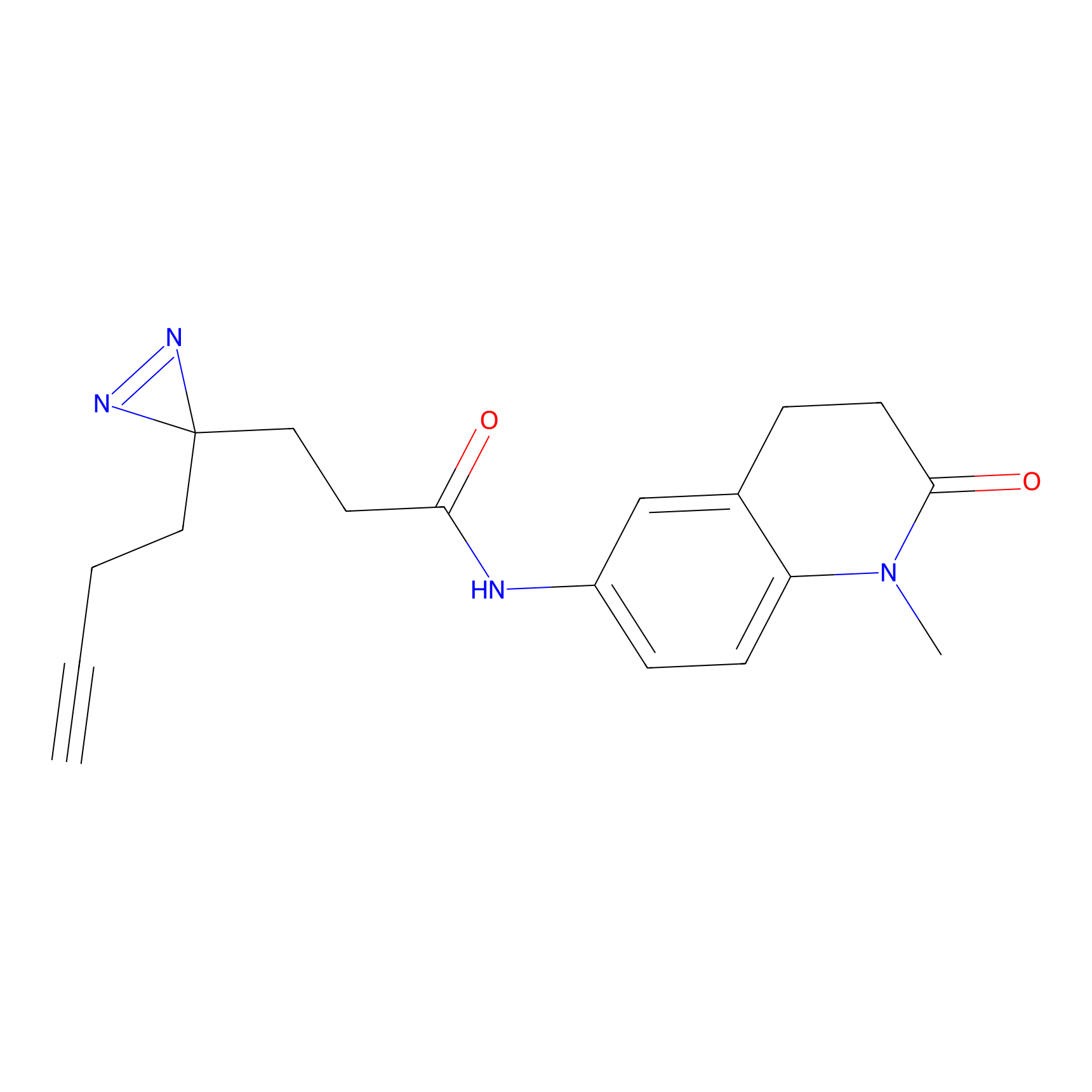 |
10.78 | LDD0467 | [25] | |
|
JN0003 Probe Info |
 |
12.21 | LDD0469 | [25] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [26] | |
|
SAHA-CA-8PAP Probe Info |
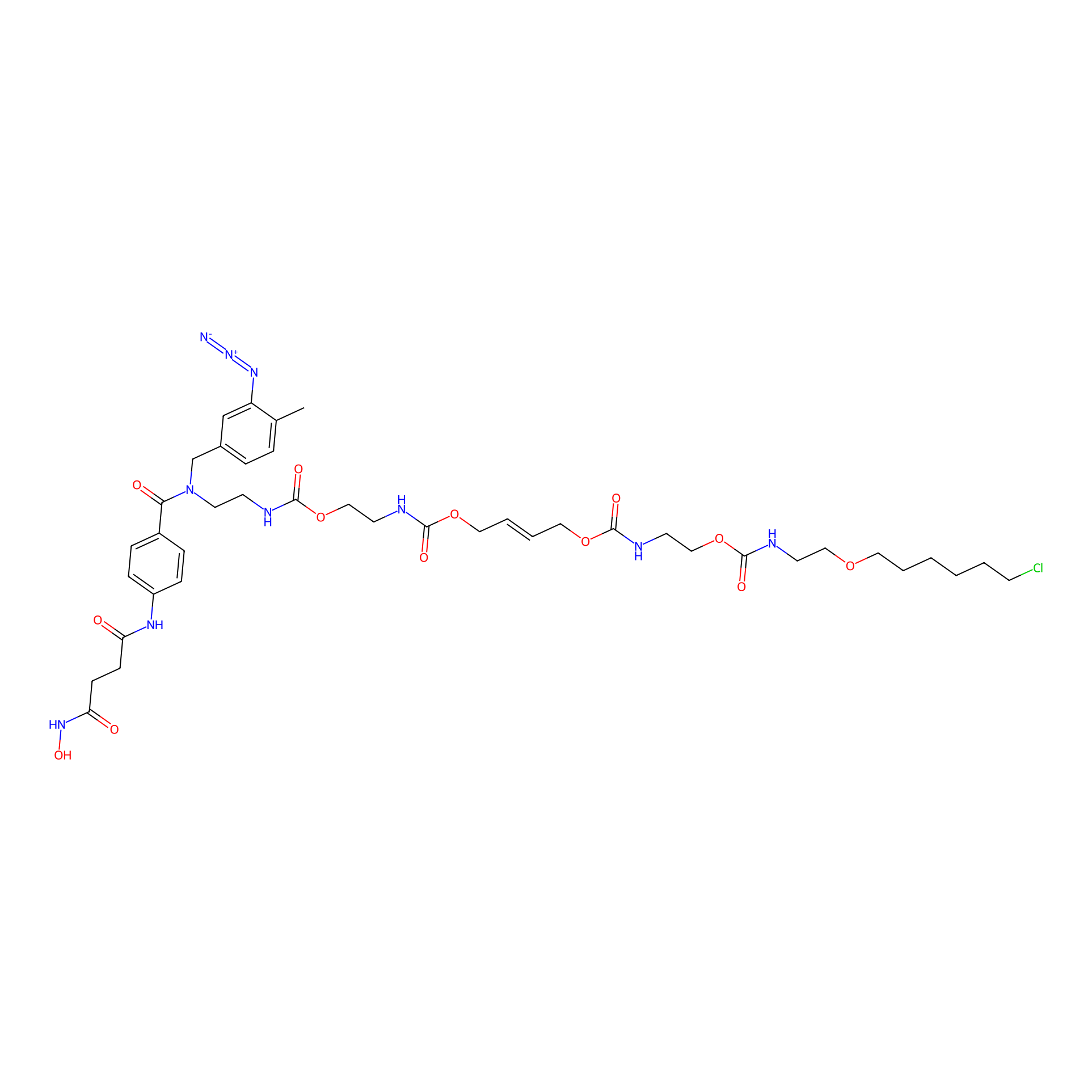 |
6.50 | LDD0362 | [27] | |
|
DA-2 Probe Info |
 |
N.A. | LDD0073 | [28] | |
|
OEA-DA Probe Info |
 |
3.03 | LDD0046 | [29] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C424(0.98); C86(1.32) | LDD2117 | [8] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C424(1.02) | LDD2152 | [8] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C424(0.80) | LDD2103 | [8] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C424(0.95) | LDD2132 | [8] |
| LDCM0026 | 4SU-RNA+native RNA | DM93 | C27(2.38) | LDD0171 | [9] |
| LDCM0214 | AC1 | PaTu 8988t | C427(0.32) | LDD1093 | [30] |
| LDCM0215 | AC10 | PaTu 8988t | C427(0.95) | LDD1094 | [30] |
| LDCM0216 | AC100 | HEK-293T | C427(1.05) | LDD0814 | [30] |
| LDCM0217 | AC101 | HEK-293T | C427(1.33) | LDD0815 | [30] |
| LDCM0218 | AC102 | HEK-293T | C427(1.35) | LDD0816 | [30] |
| LDCM0219 | AC103 | HEK-293T | C427(1.40) | LDD0817 | [30] |
| LDCM0220 | AC104 | HEK-293T | C427(1.45) | LDD0818 | [30] |
| LDCM0221 | AC105 | HEK-293T | C427(0.76) | LDD0819 | [30] |
| LDCM0222 | AC106 | HEK-293T | C427(0.85) | LDD0820 | [30] |
| LDCM0223 | AC107 | HEK-293T | C427(1.07) | LDD0821 | [30] |
| LDCM0224 | AC108 | HEK-293T | C427(0.92) | LDD0822 | [30] |
| LDCM0225 | AC109 | HEK-293T | C427(1.20) | LDD0823 | [30] |
| LDCM0226 | AC11 | PaTu 8988t | C427(0.95) | LDD1105 | [30] |
| LDCM0227 | AC110 | HEK-293T | C427(2.26) | LDD0825 | [30] |
| LDCM0228 | AC111 | HEK-293T | C427(3.06) | LDD0826 | [30] |
| LDCM0229 | AC112 | HEK-293T | C427(2.33) | LDD0827 | [30] |
| LDCM0230 | AC113 | PaTu 8988t | C427(1.35) | LDD1109 | [30] |
| LDCM0231 | AC114 | PaTu 8988t | C427(1.38) | LDD1110 | [30] |
| LDCM0232 | AC115 | PaTu 8988t | C427(1.08) | LDD1111 | [30] |
| LDCM0233 | AC116 | PaTu 8988t | C427(1.32) | LDD1112 | [30] |
| LDCM0234 | AC117 | PaTu 8988t | C427(1.39) | LDD1113 | [30] |
| LDCM0235 | AC118 | PaTu 8988t | C427(1.01) | LDD1114 | [30] |
| LDCM0236 | AC119 | PaTu 8988t | C427(0.93) | LDD1115 | [30] |
| LDCM0237 | AC12 | PaTu 8988t | C427(0.82) | LDD1116 | [30] |
| LDCM0238 | AC120 | PaTu 8988t | C427(1.38) | LDD1117 | [30] |
| LDCM0239 | AC121 | PaTu 8988t | C427(1.36) | LDD1118 | [30] |
| LDCM0240 | AC122 | PaTu 8988t | C427(1.15) | LDD1119 | [30] |
| LDCM0241 | AC123 | PaTu 8988t | C427(1.12) | LDD1120 | [30] |
| LDCM0242 | AC124 | PaTu 8988t | C427(1.09) | LDD1121 | [30] |
| LDCM0243 | AC125 | PaTu 8988t | C427(0.86) | LDD1122 | [30] |
| LDCM0244 | AC126 | PaTu 8988t | C427(1.11) | LDD1123 | [30] |
| LDCM0245 | AC127 | PaTu 8988t | C427(0.95) | LDD1124 | [30] |
| LDCM0259 | AC14 | PaTu 8988t | C427(0.92) | LDD1138 | [30] |
| LDCM0270 | AC15 | PaTu 8988t | C427(0.95) | LDD1149 | [30] |
| LDCM0276 | AC17 | HEK-293T | C424(1.21); C27(1.04); C427(0.93); C86(1.04) | LDD1515 | [31] |
| LDCM0277 | AC18 | HEK-293T | C424(1.08); C27(0.94); C427(1.14); C86(0.99) | LDD1516 | [31] |
| LDCM0278 | AC19 | HEK-293T | C424(1.24); C27(0.87); C427(0.94); C86(1.02) | LDD1517 | [31] |
| LDCM0279 | AC2 | PaTu 8988t | C427(0.33) | LDD1158 | [30] |
| LDCM0280 | AC20 | HEK-293T | C424(1.16); C27(1.00); C427(1.06); C86(0.93) | LDD1519 | [31] |
| LDCM0281 | AC21 | HEK-293T | C424(1.04); C27(1.07); C427(0.93); C86(0.94) | LDD1520 | [31] |
| LDCM0282 | AC22 | HEK-293T | C424(1.22); C27(1.12); C427(1.02); C86(0.93) | LDD1521 | [31] |
| LDCM0283 | AC23 | HEK-293T | C424(1.10); C27(1.09); C427(0.95); C86(1.07) | LDD1522 | [31] |
| LDCM0284 | AC24 | HEK-293T | C424(1.01); C27(1.11); C427(0.82); C86(1.08) | LDD1523 | [31] |
| LDCM0285 | AC25 | HEK-293T | C427(0.99) | LDD0883 | [30] |
| LDCM0286 | AC26 | HEK-293T | C427(0.89) | LDD0884 | [30] |
| LDCM0287 | AC27 | HEK-293T | C427(0.93) | LDD0885 | [30] |
| LDCM0288 | AC28 | HEK-293T | C427(1.27) | LDD0886 | [30] |
| LDCM0289 | AC29 | HEK-293T | C427(0.96) | LDD0887 | [30] |
| LDCM0290 | AC3 | PaTu 8988t | C427(0.31) | LDD1169 | [30] |
| LDCM0291 | AC30 | HEK-293T | C427(1.15) | LDD0889 | [30] |
| LDCM0292 | AC31 | HEK-293T | C427(1.07) | LDD0890 | [30] |
| LDCM0293 | AC32 | HEK-293T | C427(0.80) | LDD0891 | [30] |
| LDCM0294 | AC33 | HEK-293T | C427(1.06) | LDD0892 | [30] |
| LDCM0295 | AC34 | HEK-293T | C427(0.80) | LDD0893 | [30] |
| LDCM0296 | AC35 | HEK-293T | C424(1.08); C27(1.16); C427(1.00); C86(0.91) | LDD1535 | [31] |
| LDCM0297 | AC36 | HEK-293T | C424(1.10); C27(0.99); C427(1.09); C86(0.91) | LDD1536 | [31] |
| LDCM0298 | AC37 | HEK-293T | C424(0.93); C27(1.04); C427(0.92); C86(0.96) | LDD1537 | [31] |
| LDCM0299 | AC38 | HEK-293T | C424(1.11); C27(0.97); C427(1.10); C86(1.02) | LDD1538 | [31] |
| LDCM0300 | AC39 | HEK-293T | C424(1.05); C27(1.13); C427(1.02); C86(1.01) | LDD1539 | [31] |
| LDCM0301 | AC4 | PaTu 8988t | C427(0.28) | LDD1180 | [30] |
| LDCM0302 | AC40 | HEK-293T | C424(1.27); C27(1.06); C427(0.85); C86(1.00) | LDD1541 | [31] |
| LDCM0303 | AC41 | HEK-293T | C424(1.05); C27(1.05); C427(0.80); C86(0.96) | LDD1542 | [31] |
| LDCM0304 | AC42 | HEK-293T | C424(1.02); C27(0.90); C427(1.09); C86(0.90) | LDD1543 | [31] |
| LDCM0305 | AC43 | HEK-293T | C424(0.95); C27(0.96); C427(0.99); C86(0.91) | LDD1544 | [31] |
| LDCM0306 | AC44 | HEK-293T | C424(1.00); C27(0.90); C427(1.02); C86(1.01) | LDD1545 | [31] |
| LDCM0307 | AC45 | HEK-293T | C424(0.98); C27(1.06); C427(1.03); C86(0.90) | LDD1546 | [31] |
| LDCM0308 | AC46 | HEK-293T | C427(1.10) | LDD0906 | [30] |
| LDCM0309 | AC47 | HEK-293T | C427(1.29) | LDD0907 | [30] |
| LDCM0310 | AC48 | HEK-293T | C427(1.18) | LDD0908 | [30] |
| LDCM0311 | AC49 | HEK-293T | C427(1.23) | LDD0909 | [30] |
| LDCM0312 | AC5 | PaTu 8988t | C427(0.22) | LDD1191 | [30] |
| LDCM0313 | AC50 | HEK-293T | C427(1.24) | LDD0911 | [30] |
| LDCM0314 | AC51 | HEK-293T | C427(1.48) | LDD0912 | [30] |
| LDCM0315 | AC52 | HEK-293T | C427(1.39) | LDD0913 | [30] |
| LDCM0316 | AC53 | HEK-293T | C427(1.24) | LDD0914 | [30] |
| LDCM0317 | AC54 | HEK-293T | C427(1.41) | LDD0915 | [30] |
| LDCM0318 | AC55 | HEK-293T | C427(1.33) | LDD0916 | [30] |
| LDCM0319 | AC56 | HEK-293T | C427(1.11) | LDD0917 | [30] |
| LDCM0320 | AC57 | HEK-293T | C427(0.81) | LDD0918 | [30] |
| LDCM0321 | AC58 | HEK-293T | C427(0.71) | LDD0919 | [30] |
| LDCM0322 | AC59 | HEK-293T | C427(0.91) | LDD0920 | [30] |
| LDCM0323 | AC6 | PaTu 8988t | C427(0.87) | LDD1202 | [30] |
| LDCM0324 | AC60 | HEK-293T | C427(1.36) | LDD0922 | [30] |
| LDCM0325 | AC61 | HEK-293T | C427(1.56) | LDD0923 | [30] |
| LDCM0326 | AC62 | HEK-293T | C427(0.90) | LDD0924 | [30] |
| LDCM0327 | AC63 | HEK-293T | C427(0.95) | LDD0925 | [30] |
| LDCM0328 | AC64 | HEK-293T | C427(0.78) | LDD0926 | [30] |
| LDCM0329 | AC65 | HEK-293T | C427(1.19) | LDD0927 | [30] |
| LDCM0330 | AC66 | HEK-293T | C427(0.82) | LDD0928 | [30] |
| LDCM0331 | AC67 | HEK-293T | C427(1.06) | LDD0929 | [30] |
| LDCM0334 | AC7 | PaTu 8988t | C427(0.89) | LDD1213 | [30] |
| LDCM0345 | AC8 | PaTu 8988t | C427(0.85) | LDD1224 | [30] |
| LDCM0349 | AC83 | HEK-293T | C427(0.86) | LDD0947 | [30] |
| LDCM0350 | AC84 | HEK-293T | C427(1.20) | LDD0948 | [30] |
| LDCM0351 | AC85 | HEK-293T | C427(0.90) | LDD0949 | [30] |
| LDCM0352 | AC86 | HEK-293T | C427(1.06) | LDD0950 | [30] |
| LDCM0353 | AC87 | HEK-293T | C427(1.17) | LDD0951 | [30] |
| LDCM0354 | AC88 | HEK-293T | C427(1.27) | LDD0952 | [30] |
| LDCM0355 | AC89 | HEK-293T | C427(1.42) | LDD0953 | [30] |
| LDCM0357 | AC90 | HEK-293T | C427(1.34) | LDD0955 | [30] |
| LDCM0358 | AC91 | HEK-293T | C427(1.17) | LDD0956 | [30] |
| LDCM0359 | AC92 | HEK-293T | C427(1.08) | LDD0957 | [30] |
| LDCM0360 | AC93 | HEK-293T | C427(1.13) | LDD0958 | [30] |
| LDCM0361 | AC94 | HEK-293T | C427(1.35) | LDD0959 | [30] |
| LDCM0362 | AC95 | HEK-293T | C427(1.23) | LDD0960 | [30] |
| LDCM0363 | AC96 | HEK-293T | C427(1.26) | LDD0961 | [30] |
| LDCM0364 | AC97 | HEK-293T | C427(2.80) | LDD0962 | [30] |
| LDCM0365 | AC98 | HEK-293T | C427(0.93) | LDD0963 | [30] |
| LDCM0366 | AC99 | HEK-293T | C427(0.80) | LDD0964 | [30] |
| LDCM0248 | AKOS034007472 | PaTu 8988t | C427(0.92) | LDD1127 | [30] |
| LDCM0356 | AKOS034007680 | PaTu 8988t | C427(1.01) | LDD1235 | [30] |
| LDCM0275 | AKOS034007705 | PaTu 8988t | C427(1.04) | LDD1154 | [30] |
| LDCM0156 | Aniline | NCI-H1299 | 11.70 | LDD0403 | [1] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C424(0.42) | LDD2091 | [8] |
| LDCM0630 | CCW28-3 | 231MFP | C27(1.44) | LDD2214 | [32] |
| LDCM0108 | Chloroacetamide | HeLa | C86(0.00); H85(0.00) | LDD0222 | [22] |
| LDCM0632 | CL-Sc | Hep-G2 | C424(20.00); C86(0.94) | LDD2227 | [19] |
| LDCM0367 | CL1 | HEK-293T | C424(1.15); C27(0.95); C427(0.88); C86(0.88) | LDD1571 | [31] |
| LDCM0368 | CL10 | HEK-293T | C424(0.90); C27(1.23); C427(1.17); C86(1.07) | LDD1572 | [31] |
| LDCM0369 | CL100 | PaTu 8988t | C427(0.22) | LDD1248 | [30] |
| LDCM0370 | CL101 | PaTu 8988t | C427(0.95) | LDD1249 | [30] |
| LDCM0371 | CL102 | PaTu 8988t | C427(0.81) | LDD1250 | [30] |
| LDCM0372 | CL103 | PaTu 8988t | C427(1.00) | LDD1251 | [30] |
| LDCM0373 | CL104 | PaTu 8988t | C427(0.88) | LDD1252 | [30] |
| LDCM0374 | CL105 | HEK-293T | C424(1.12); C27(1.03); C427(1.01); C86(0.83) | LDD1578 | [31] |
| LDCM0375 | CL106 | HEK-293T | C424(1.13); C27(0.99); C427(1.05); C86(0.99) | LDD1579 | [31] |
| LDCM0376 | CL107 | HEK-293T | C424(1.14); C27(1.03); C427(0.99); C86(1.00) | LDD1580 | [31] |
| LDCM0377 | CL108 | HEK-293T | C424(1.17); C27(0.97); C427(1.00); C86(1.04) | LDD1581 | [31] |
| LDCM0378 | CL109 | HEK-293T | C424(1.13); C27(0.95); C427(0.94); C86(0.94) | LDD1582 | [31] |
| LDCM0379 | CL11 | HEK-293T | C424(0.79); C27(1.08); C427(1.26); C86(1.12) | LDD1583 | [31] |
| LDCM0380 | CL110 | HEK-293T | C424(1.16); C27(1.16); C427(1.02); C86(0.94) | LDD1584 | [31] |
| LDCM0381 | CL111 | HEK-293T | C424(1.19); C27(1.10); C427(1.04); C86(0.99) | LDD1585 | [31] |
| LDCM0382 | CL112 | HEK-293T | C427(1.53) | LDD0980 | [30] |
| LDCM0383 | CL113 | HEK-293T | C427(0.95) | LDD0981 | [30] |
| LDCM0384 | CL114 | HEK-293T | C427(1.91) | LDD0982 | [30] |
| LDCM0385 | CL115 | HEK-293T | C427(1.36) | LDD0983 | [30] |
| LDCM0386 | CL116 | HEK-293T | C427(1.04) | LDD0984 | [30] |
| LDCM0387 | CL117 | HEK-293T | C424(1.06); C27(0.95); C427(0.96); C86(0.86) | LDD1591 | [31] |
| LDCM0388 | CL118 | HEK-293T | C424(1.08); C27(0.92); C427(1.07); C86(0.93) | LDD1592 | [31] |
| LDCM0389 | CL119 | HEK-293T | C424(1.11); C27(0.99); C427(0.97); C86(0.84) | LDD1593 | [31] |
| LDCM0390 | CL12 | HEK-293T | C424(0.98); C27(1.01); C427(0.95); C86(1.06) | LDD1594 | [31] |
| LDCM0391 | CL120 | HEK-293T | C424(1.03); C27(0.83); C427(0.91); C86(0.89) | LDD1595 | [31] |
| LDCM0392 | CL121 | HEK-293T | C427(1.41) | LDD0990 | [30] |
| LDCM0393 | CL122 | HEK-293T | C427(1.24) | LDD0991 | [30] |
| LDCM0394 | CL123 | HEK-293T | C427(1.26) | LDD0992 | [30] |
| LDCM0395 | CL124 | HEK-293T | C427(1.31) | LDD0993 | [30] |
| LDCM0396 | CL125 | HEK-293T | C427(0.97) | LDD0994 | [30] |
| LDCM0397 | CL126 | HEK-293T | C427(0.97) | LDD0995 | [30] |
| LDCM0398 | CL127 | HEK-293T | C427(0.78) | LDD0996 | [30] |
| LDCM0399 | CL128 | HEK-293T | C427(0.74) | LDD0997 | [30] |
| LDCM0400 | CL13 | HEK-293T | C424(1.01); C27(0.88); C427(0.89); C86(0.88) | LDD1604 | [31] |
| LDCM0401 | CL14 | HEK-293T | C424(1.10); C27(1.07); C427(1.03); C86(0.93) | LDD1605 | [31] |
| LDCM0402 | CL15 | HEK-293T | C424(1.15); C27(0.93); C427(1.07); C86(0.87) | LDD1606 | [31] |
| LDCM0403 | CL16 | PaTu 8988t | C427(1.20) | LDD1282 | [30] |
| LDCM0404 | CL17 | PaTu 8988t | C427(0.95) | LDD1283 | [30] |
| LDCM0405 | CL18 | PaTu 8988t | C427(1.45) | LDD1284 | [30] |
| LDCM0406 | CL19 | PaTu 8988t | C427(1.24) | LDD1285 | [30] |
| LDCM0407 | CL2 | HEK-293T | C424(1.10); C27(1.12); C427(1.01); C86(0.93) | LDD1611 | [31] |
| LDCM0408 | CL20 | PaTu 8988t | C427(1.44) | LDD1287 | [30] |
| LDCM0409 | CL21 | PaTu 8988t | C427(1.54) | LDD1288 | [30] |
| LDCM0410 | CL22 | PaTu 8988t | C427(1.37) | LDD1289 | [30] |
| LDCM0411 | CL23 | PaTu 8988t | C427(1.26) | LDD1290 | [30] |
| LDCM0412 | CL24 | PaTu 8988t | C427(1.28) | LDD1291 | [30] |
| LDCM0413 | CL25 | PaTu 8988t | C427(1.05) | LDD1292 | [30] |
| LDCM0414 | CL26 | PaTu 8988t | C427(1.36) | LDD1293 | [30] |
| LDCM0415 | CL27 | PaTu 8988t | C427(1.15) | LDD1294 | [30] |
| LDCM0416 | CL28 | PaTu 8988t | C427(1.22) | LDD1295 | [30] |
| LDCM0417 | CL29 | PaTu 8988t | C427(1.40) | LDD1296 | [30] |
| LDCM0418 | CL3 | HEK-293T | C424(1.14); C27(1.00); C427(1.03); C86(0.95) | LDD1622 | [31] |
| LDCM0419 | CL30 | PaTu 8988t | C427(1.15) | LDD1298 | [30] |
| LDCM0420 | CL31 | HEK-293T | C424(0.95); C27(0.87); C427(0.92); C86(1.08) | LDD1624 | [31] |
| LDCM0421 | CL32 | HEK-293T | C424(0.83); C27(0.94); C427(1.09); C86(1.04) | LDD1625 | [31] |
| LDCM0422 | CL33 | HEK-293T | C424(0.87); C27(0.93); C427(1.11); C86(1.05) | LDD1626 | [31] |
| LDCM0423 | CL34 | HEK-293T | C424(0.87); C27(0.95); C427(1.13); C86(1.20) | LDD1627 | [31] |
| LDCM0424 | CL35 | HEK-293T | C424(0.79); C27(1.03); C427(0.98); C86(1.17) | LDD1628 | [31] |
| LDCM0425 | CL36 | HEK-293T | C424(0.88); C27(1.05); C427(0.98); C86(1.11) | LDD1629 | [31] |
| LDCM0426 | CL37 | HEK-293T | C424(1.02); C27(0.54); C427(0.86); C86(0.95) | LDD1630 | [31] |
| LDCM0428 | CL39 | HEK-293T | C424(1.04); C27(1.05); C427(1.06); C86(0.89) | LDD1632 | [31] |
| LDCM0429 | CL4 | HEK-293T | C424(1.10); C27(0.89); C427(1.07); C86(1.06) | LDD1633 | [31] |
| LDCM0430 | CL40 | HEK-293T | C424(1.05); C27(1.08); C427(0.90); C86(1.00) | LDD1634 | [31] |
| LDCM0431 | CL41 | HEK-293T | C424(0.96); C27(1.15); C427(0.82); C86(0.94) | LDD1635 | [31] |
| LDCM0432 | CL42 | HEK-293T | C424(0.90); C27(0.91); C427(1.03); C86(0.99) | LDD1636 | [31] |
| LDCM0433 | CL43 | HEK-293T | C424(0.84); C27(0.92); C427(1.16); C86(1.04) | LDD1637 | [31] |
| LDCM0434 | CL44 | HEK-293T | C424(0.90); C27(0.93); C427(1.02); C86(0.99) | LDD1638 | [31] |
| LDCM0435 | CL45 | HEK-293T | C424(0.76); C27(0.93); C427(0.99); C86(0.99) | LDD1639 | [31] |
| LDCM0436 | CL46 | PaTu 8988t | C427(0.86) | LDD1315 | [30] |
| LDCM0437 | CL47 | PaTu 8988t | C427(0.90) | LDD1316 | [30] |
| LDCM0438 | CL48 | PaTu 8988t | C427(0.81) | LDD1317 | [30] |
| LDCM0439 | CL49 | PaTu 8988t | C427(0.82) | LDD1318 | [30] |
| LDCM0440 | CL5 | HEK-293T | C424(0.96); C27(0.82); C427(0.98); C86(0.98) | LDD1644 | [31] |
| LDCM0441 | CL50 | PaTu 8988t | C427(0.79) | LDD1320 | [30] |
| LDCM0442 | CL51 | PaTu 8988t | C427(0.87) | LDD1321 | [30] |
| LDCM0443 | CL52 | PaTu 8988t | C427(0.83) | LDD1322 | [30] |
| LDCM0444 | CL53 | PaTu 8988t | C427(0.78) | LDD1323 | [30] |
| LDCM0445 | CL54 | PaTu 8988t | C427(0.87) | LDD1324 | [30] |
| LDCM0446 | CL55 | PaTu 8988t | C427(0.80) | LDD1325 | [30] |
| LDCM0447 | CL56 | PaTu 8988t | C427(0.75) | LDD1326 | [30] |
| LDCM0448 | CL57 | PaTu 8988t | C427(0.75) | LDD1327 | [30] |
| LDCM0449 | CL58 | PaTu 8988t | C427(0.75) | LDD1328 | [30] |
| LDCM0450 | CL59 | PaTu 8988t | C427(0.84) | LDD1329 | [30] |
| LDCM0451 | CL6 | HEK-293T | C424(0.96); C27(0.87); C427(0.97); C86(1.03) | LDD1654 | [31] |
| LDCM0452 | CL60 | PaTu 8988t | C427(0.82) | LDD1331 | [30] |
| LDCM0453 | CL61 | HEK-293T | C427(0.92) | LDD1051 | [30] |
| LDCM0454 | CL62 | HEK-293T | C427(0.75) | LDD1052 | [30] |
| LDCM0455 | CL63 | HEK-293T | C427(1.02) | LDD1053 | [30] |
| LDCM0456 | CL64 | HEK-293T | C427(1.15) | LDD1054 | [30] |
| LDCM0457 | CL65 | HEK-293T | C427(1.15) | LDD1055 | [30] |
| LDCM0458 | CL66 | HEK-293T | C427(1.10) | LDD1056 | [30] |
| LDCM0459 | CL67 | HEK-293T | C427(1.23) | LDD1057 | [30] |
| LDCM0460 | CL68 | HEK-293T | C427(1.18) | LDD1058 | [30] |
| LDCM0461 | CL69 | HEK-293T | C427(1.52) | LDD1059 | [30] |
| LDCM0462 | CL7 | HEK-293T | C424(0.88); C27(0.96); C427(0.97); C86(1.08) | LDD1665 | [31] |
| LDCM0463 | CL70 | HEK-293T | C427(1.50) | LDD1061 | [30] |
| LDCM0464 | CL71 | HEK-293T | C427(2.38) | LDD1062 | [30] |
| LDCM0465 | CL72 | HEK-293T | C427(1.34) | LDD1063 | [30] |
| LDCM0466 | CL73 | HEK-293T | C427(1.42) | LDD1064 | [30] |
| LDCM0467 | CL74 | HEK-293T | C427(3.07) | LDD1065 | [30] |
| LDCM0469 | CL76 | HEK-293T | C427(0.92) | LDD1067 | [30] |
| LDCM0470 | CL77 | HEK-293T | C427(0.77) | LDD1068 | [30] |
| LDCM0471 | CL78 | HEK-293T | C427(1.45) | LDD1069 | [30] |
| LDCM0472 | CL79 | HEK-293T | C427(1.16) | LDD1070 | [30] |
| LDCM0473 | CL8 | HEK-293T | C424(1.13); C27(0.87); C427(1.39); C86(0.96) | LDD1676 | [31] |
| LDCM0474 | CL80 | HEK-293T | C427(1.19) | LDD1072 | [30] |
| LDCM0475 | CL81 | HEK-293T | C427(1.08) | LDD1073 | [30] |
| LDCM0476 | CL82 | HEK-293T | C427(0.75) | LDD1074 | [30] |
| LDCM0477 | CL83 | HEK-293T | C427(0.88) | LDD1075 | [30] |
| LDCM0478 | CL84 | HEK-293T | C427(1.15) | LDD1076 | [30] |
| LDCM0479 | CL85 | HEK-293T | C427(1.26) | LDD1077 | [30] |
| LDCM0480 | CL86 | HEK-293T | C427(1.09) | LDD1078 | [30] |
| LDCM0481 | CL87 | HEK-293T | C427(1.08) | LDD1079 | [30] |
| LDCM0482 | CL88 | HEK-293T | C427(1.21) | LDD1080 | [30] |
| LDCM0483 | CL89 | HEK-293T | C427(1.47) | LDD1081 | [30] |
| LDCM0484 | CL9 | HEK-293T | C424(0.73); C27(0.99); C427(0.94); C86(1.03) | LDD1687 | [31] |
| LDCM0485 | CL90 | HEK-293T | C427(1.33) | LDD1083 | [30] |
| LDCM0486 | CL91 | PaTu 8988t | C427(0.25) | LDD1365 | [30] |
| LDCM0487 | CL92 | PaTu 8988t | C427(0.23) | LDD1366 | [30] |
| LDCM0488 | CL93 | PaTu 8988t | C427(0.26) | LDD1367 | [30] |
| LDCM0489 | CL94 | PaTu 8988t | C427(0.36) | LDD1368 | [30] |
| LDCM0490 | CL95 | PaTu 8988t | C427(0.23) | LDD1369 | [30] |
| LDCM0491 | CL96 | PaTu 8988t | C427(0.30) | LDD1370 | [30] |
| LDCM0492 | CL97 | PaTu 8988t | C427(0.62) | LDD1371 | [30] |
| LDCM0493 | CL98 | PaTu 8988t | C427(0.27) | LDD1372 | [30] |
| LDCM0494 | CL99 | PaTu 8988t | C427(0.24) | LDD1373 | [30] |
| LDCM0634 | CY-0357 | Hep-G2 | C27(1.15) | LDD2228 | [19] |
| LDCM0495 | E2913 | HEK-293T | C424(1.12); C27(0.92); C427(0.99); C86(0.90) | LDD1698 | [31] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C27(2.73); C86(1.60) | LDD1702 | [8] |
| LDCM0625 | F8 | Ramos | C27(1.08) | LDD2187 | [33] |
| LDCM0572 | Fragment10 | MDA-MB-231 | C27(1.33) | LDD1389 | [34] |
| LDCM0573 | Fragment11 | MDA-MB-231 | C27(1.00) | LDD1391 | [34] |
| LDCM0574 | Fragment12 | Ramos | C27(0.65) | LDD1394 | [34] |
| LDCM0575 | Fragment13 | MDA-MB-231 | C27(1.00) | LDD1395 | [34] |
| LDCM0576 | Fragment14 | MDA-MB-231 | C27(1.55) | LDD1397 | [34] |
| LDCM0577 | Fragment15 | Ramos | C27(1.09) | LDD1400 | [34] |
| LDCM0579 | Fragment20 | MDA-MB-231 | C27(1.19) | LDD1402 | [34] |
| LDCM0580 | Fragment21 | MDA-MB-231 | C27(0.78) | LDD1404 | [34] |
| LDCM0581 | Fragment22 | MDA-MB-231 | C27(0.98) | LDD1406 | [34] |
| LDCM0582 | Fragment23 | MDA-MB-231 | C27(1.66) | LDD1408 | [34] |
| LDCM0583 | Fragment24 | Ramos | C27(1.64) | LDD1410 | [34] |
| LDCM0584 | Fragment25 | MDA-MB-231 | C27(1.31) | LDD1411 | [34] |
| LDCM0585 | Fragment26 | Ramos | C27(0.91) | LDD1412 | [34] |
| LDCM0578 | Fragment27 | MDA-MB-231 | C27(1.02) | LDD1401 | [34] |
| LDCM0586 | Fragment28 | MDA-MB-231 | C27(0.92) | LDD1415 | [34] |
| LDCM0587 | Fragment29 | MDA-MB-231 | C27(1.27) | LDD1417 | [34] |
| LDCM0588 | Fragment30 | MDA-MB-231 | C27(0.74) | LDD1419 | [34] |
| LDCM0589 | Fragment31 | MDA-MB-231 | C27(2.17) | LDD1421 | [34] |
| LDCM0590 | Fragment32 | MDA-MB-231 | C27(1.12) | LDD1423 | [34] |
| LDCM0468 | Fragment33 | HEK-293T | C427(2.34) | LDD1066 | [30] |
| LDCM0592 | Fragment34 | MDA-MB-231 | C27(0.76) | LDD1427 | [34] |
| LDCM0593 | Fragment35 | MDA-MB-231 | C27(0.80) | LDD1429 | [34] |
| LDCM0594 | Fragment36 | MDA-MB-231 | C27(2.02) | LDD1431 | [34] |
| LDCM0595 | Fragment37 | Ramos | C27(0.92) | LDD1432 | [34] |
| LDCM0596 | Fragment38 | MDA-MB-231 | C27(0.94) | LDD1433 | [34] |
| LDCM0566 | Fragment4 | MDA-MB-231 | C27(1.21) | LDD1378 | [34] |
| LDCM0598 | Fragment40 | MDA-MB-231 | C27(0.81) | LDD1436 | [34] |
| LDCM0599 | Fragment41 | MDA-MB-231 | C27(1.31) | LDD1438 | [34] |
| LDCM0600 | Fragment42 | Ramos | C27(0.97) | LDD1440 | [34] |
| LDCM0601 | Fragment43 | MDA-MB-231 | C27(0.96) | LDD1441 | [34] |
| LDCM0602 | Fragment44 | MDA-MB-231 | C27(0.88) | LDD1443 | [34] |
| LDCM0603 | Fragment45 | MDA-MB-231 | C27(2.53) | LDD1444 | [34] |
| LDCM0604 | Fragment46 | MDA-MB-231 | C27(1.04) | LDD1445 | [34] |
| LDCM0605 | Fragment47 | MDA-MB-231 | C27(1.36) | LDD1446 | [34] |
| LDCM0607 | Fragment49 | MDA-MB-231 | C27(1.16) | LDD1448 | [34] |
| LDCM0608 | Fragment50 | MDA-MB-231 | C27(1.29) | LDD1449 | [34] |
| LDCM0427 | Fragment51 | MDA-MB-231 | C27(0.96) | LDD1450 | [34] |
| LDCM0610 | Fragment52 | MDA-MB-231 | C27(1.05) | LDD1452 | [34] |
| LDCM0611 | Fragment53 | MDA-MB-231 | C27(1.00) | LDD1454 | [34] |
| LDCM0612 | Fragment54 | MDA-MB-231 | C27(1.28) | LDD1456 | [34] |
| LDCM0613 | Fragment55 | MDA-MB-231 | C27(0.89) | LDD1457 | [34] |
| LDCM0614 | Fragment56 | MDA-MB-231 | C27(0.96) | LDD1458 | [34] |
| LDCM0568 | Fragment6 | MDA-MB-231 | C27(1.62) | LDD1382 | [34] |
| LDCM0569 | Fragment7 | MDA-MB-231 | C27(1.14) | LDD1383 | [34] |
| LDCM0570 | Fragment8 | MDA-MB-231 | C27(0.98) | LDD1385 | [34] |
| LDCM0571 | Fragment9 | MDA-MB-231 | C27(0.92) | LDD1387 | [34] |
| LDCM0116 | HHS-0101 | DM93 | Y626(0.80); Y597(0.82); Y390(0.82); Y726(0.87) | LDD0264 | [10] |
| LDCM0117 | HHS-0201 | DM93 | Y563(0.75); Y390(0.76); Y597(0.79); Y626(0.83) | LDD0265 | [10] |
| LDCM0118 | HHS-0301 | DM93 | Y563(0.53); Y597(0.72); Y390(0.79); Y626(0.89) | LDD0266 | [10] |
| LDCM0119 | HHS-0401 | DM93 | Y314(0.52); Y563(0.72); Y125(0.72); Y597(0.85) | LDD0267 | [10] |
| LDCM0120 | HHS-0701 | DM93 | Y314(0.68); Y597(0.77); Y726(0.88); Y390(0.97) | LDD0268 | [10] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [22] |
| LDCM0022 | KB02 | MDA-MB-231 | C27(1.64) | LDD1374 | [34] |
| LDCM0023 | KB03 | MDA-MB-231 | C27(2.21) | LDD1376 | [34] |
| LDCM0024 | KB05 | COLO792 | C86(2.50) | LDD3310 | [7] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C424(1.19) | LDD2102 | [8] |
| LDCM0109 | NEM | HeLa | H288(0.00); H396(0.00); H681(0.00) | LDD0223 | [22] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C86(0.83) | LDD2089 | [8] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C424(1.16) | LDD2090 | [8] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C424(0.89); C86(0.92) | LDD2092 | [8] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C424(1.01) | LDD2093 | [8] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C424(1.24); C86(1.04) | LDD2094 | [8] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C424(1.01) | LDD2098 | [8] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C424(0.91); C86(1.41) | LDD2099 | [8] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C424(1.69) | LDD2105 | [8] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C424(1.05) | LDD2107 | [8] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C424(0.63); C86(0.77) | LDD2109 | [8] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C424(0.90) | LDD2111 | [8] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C424(0.70) | LDD2115 | [8] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C424(2.65); C86(0.96) | LDD2119 | [8] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C424(0.84) | LDD2120 | [8] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C424(0.77); C86(1.03) | LDD2123 | [8] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C424(0.66) | LDD2125 | [8] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C424(0.88) | LDD2127 | [8] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C424(0.97) | LDD2129 | [8] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C424(0.63) | LDD2134 | [8] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C86(1.00) | LDD2135 | [8] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C424(1.30); C86(1.87) | LDD2136 | [8] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C424(0.57); C86(0.99) | LDD2140 | [8] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C424(0.90) | LDD2143 | [8] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C424(2.96); C86(1.85) | LDD2144 | [8] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C86(1.26) | LDD2146 | [8] |
| LDCM0553 | Nucleophilic fragment 6b | MDA-MB-231 | C424(2.28) | LDD2147 | [8] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C424(0.38) | LDD2150 | [8] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C424(2.06) | LDD2153 | [8] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C27(0.69) | LDD2206 | [35] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C27(1.26) | LDD2207 | [35] |
| LDCM0131 | RA190 | MM1.R | C27(1.54) | LDD0304 | [36] |
| LDCM0096 | SAHA | K562 | 6.50 | LDD0362 | [27] |
| LDCM0112 | W16 | Hep-G2 | C27(0.68) | LDD0239 | [12] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
Transcription factor
GPCR
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Frizzled-4 (FZD4) | G-protein coupled receptor Fz/Smo family | Q9ULV1 | |||
Cytokine and receptor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Granulocyte-macrophage colony-stimulating factor (CSF2) | GM-CSF family | P04141 | |||
Other
References
