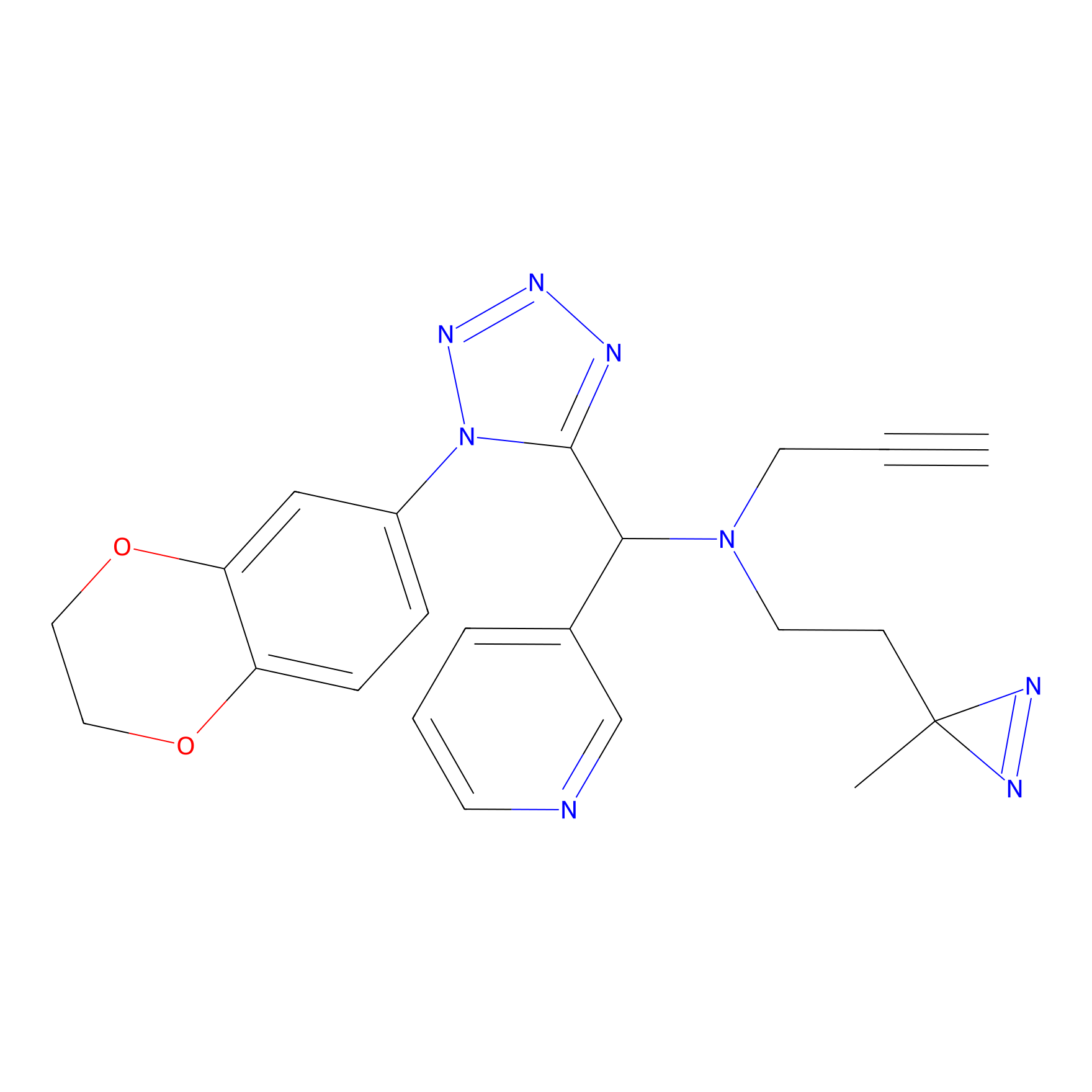
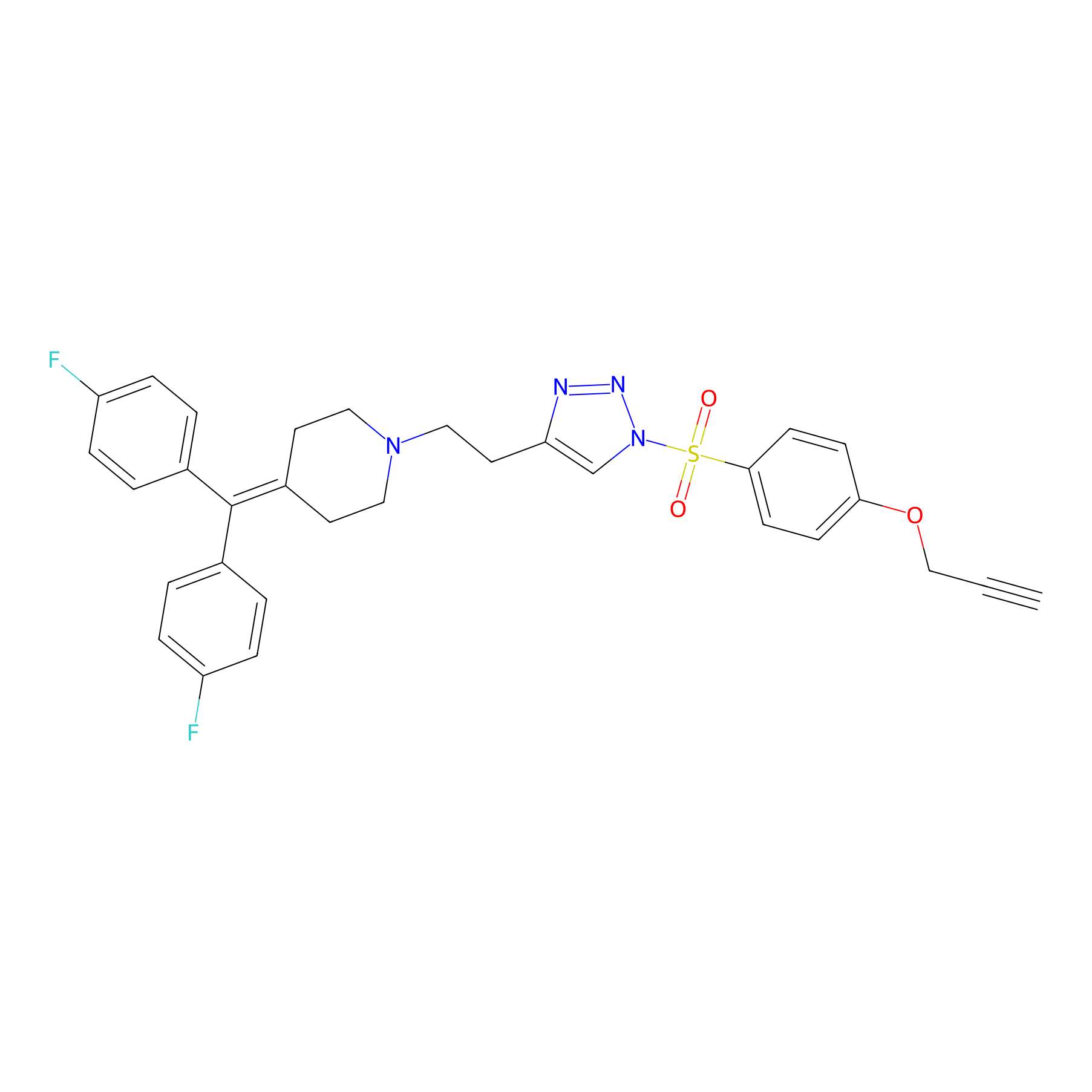
Details of the Target
General Information of Target
| Target ID | LDTP11277 | |||||
|---|---|---|---|---|---|---|
| Target Name | Tubulin beta-2B chain (TUBB2B) | |||||
| Gene Name | TUBB2B | |||||
| Gene ID | 347733 | |||||
| Synonyms |
Tubulin beta-2B chain |
|||||
| 3D Structure | ||||||
| Sequence |
MATPVVTKTAWKLQEIVAHASNVSSLVLGKASGRLLATGGDDCRVNLWSINKPNCIMSLT
GHTSPVESVRLNTPEELIVAGSQSGSIRVWDLEAAKILRTLMGHKANICSLDFHPYGEFV ASGSQDTNIKLWDIRRKGCVFRYRGHSQAVRCLRFSPDGKWLASAADDHTVKLWDLTAGK MMSEFPGHTGPVNVVEFHPNEYLLASGSSDRTIRFWDLEKFQVVSCIEGEPGPVRSVLFN PDGCCLYSGCQDSLRVYGWEPERCFDVVLVNWGKVADLAICNDQLIGVAFSQSNVSSYVV DLTRVTRTGTVARDPVQDHRPLAQPLPNPSAPLRRIYERPSTTCSKPQRVKQNSESERRS PSSEDDRDERESRAEIQNAEDYNEIFQPKNSISRTPPRRSEPFPAPPEDDAATAKEAAKP SPAMDVQFPVPNLEVLPRPPVVASTPAPKAEPAIIPATRNEPIGLKASDFLPAVKIPQQA ELVDEDAMSQIRKGHDTMCVVLTSRHKNLDTVRAVWTMGDIKTSVDSAVAINDLSVVVDL LNIVNQKASLWKLDLCTTVLPQIEKLLQSKYESYVQTGCTSLKLILQRFLPLITDMLAAP PSVGVDISREERLHKCRLCYKQLKSISGLVKSKSGLSGRHGSTFRELHLLMASLD |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Tubulin family
|
|||||
| Subcellular location |
Cytoplasm, cytoskeleton
|
|||||
| Function |
Tubulin is the major constituent of microtubules, a cylinder consisting of laterally associated linear protofilaments composed of alpha- and beta-tubulin heterodimers. Microtubules grow by the addition of GTP-tubulin dimers to the microtubule end, where a stabilizing cap forms. Below the cap, tubulin dimers are in GDP-bound state, owing to GTPase activity of alpha-tubulin. Plays a critical role in proper axon guidance in both central and peripheral axon tracts. Implicated in neuronal migration.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
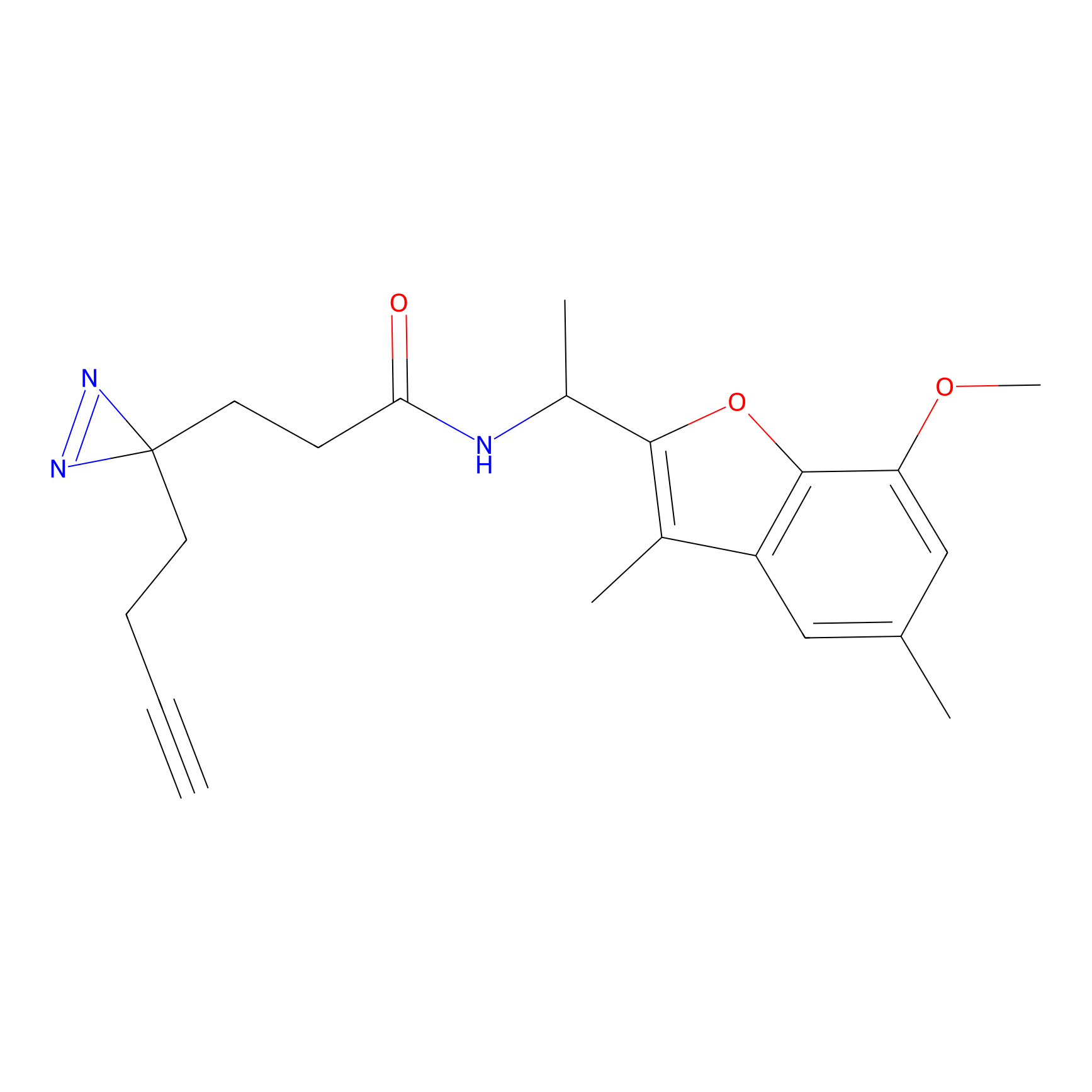
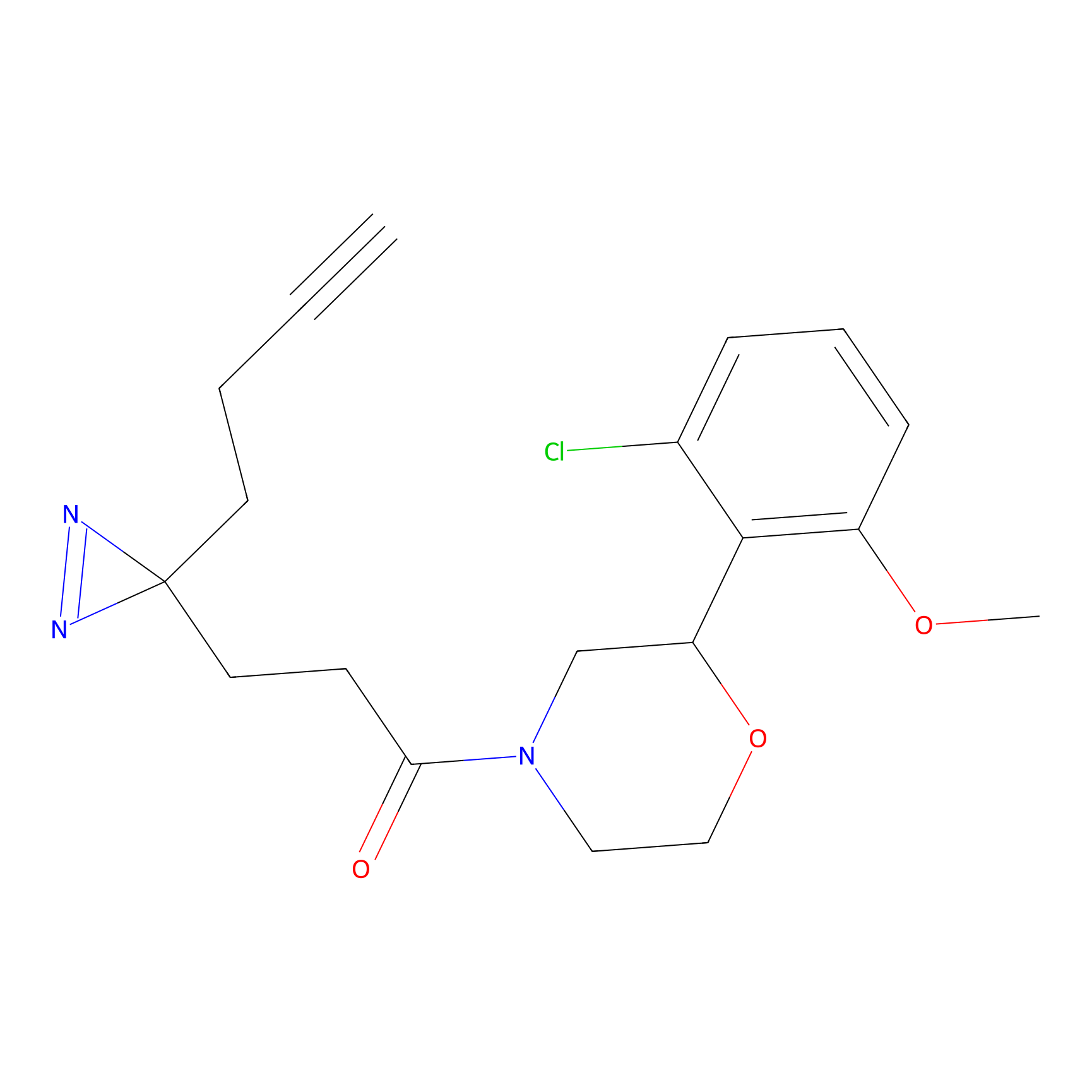
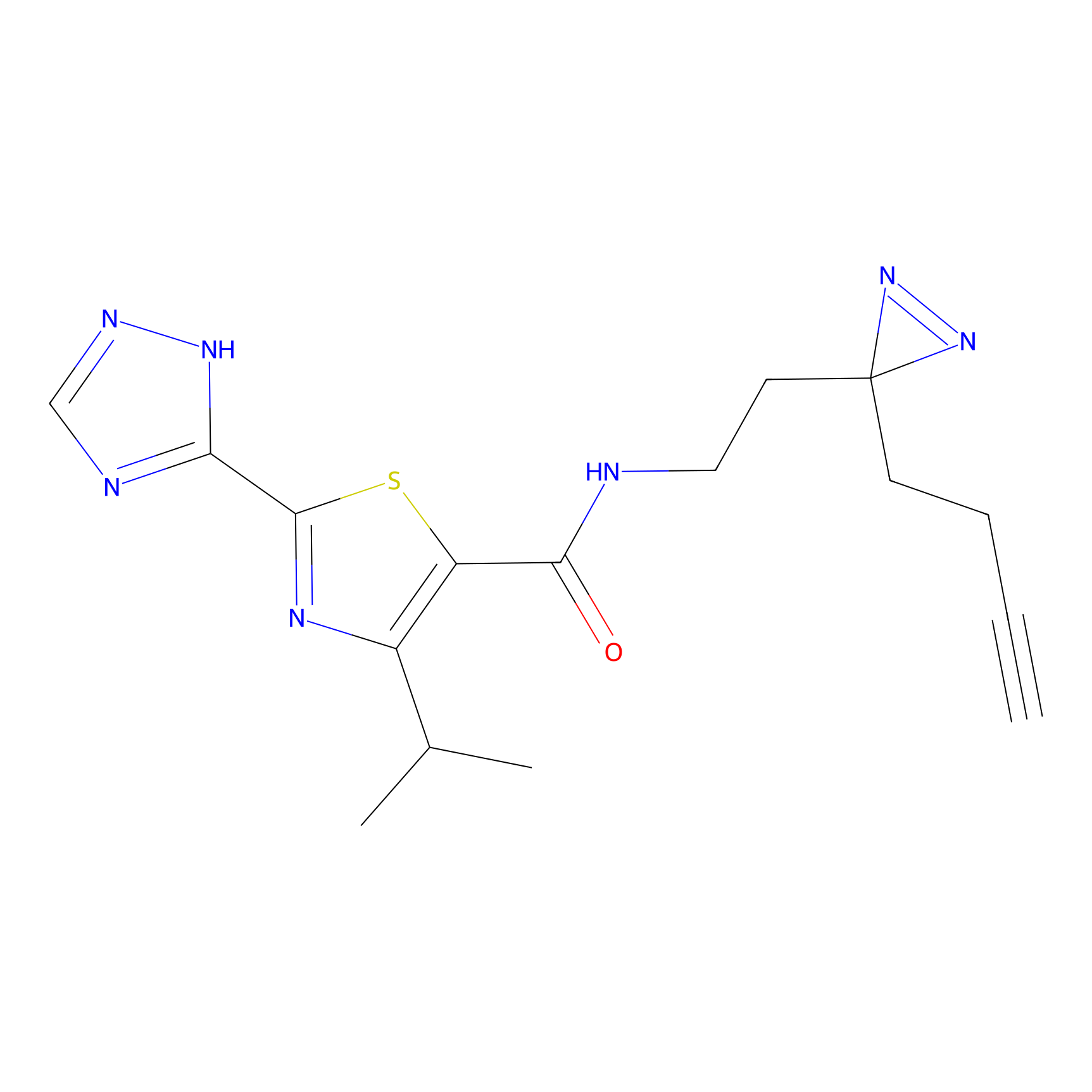
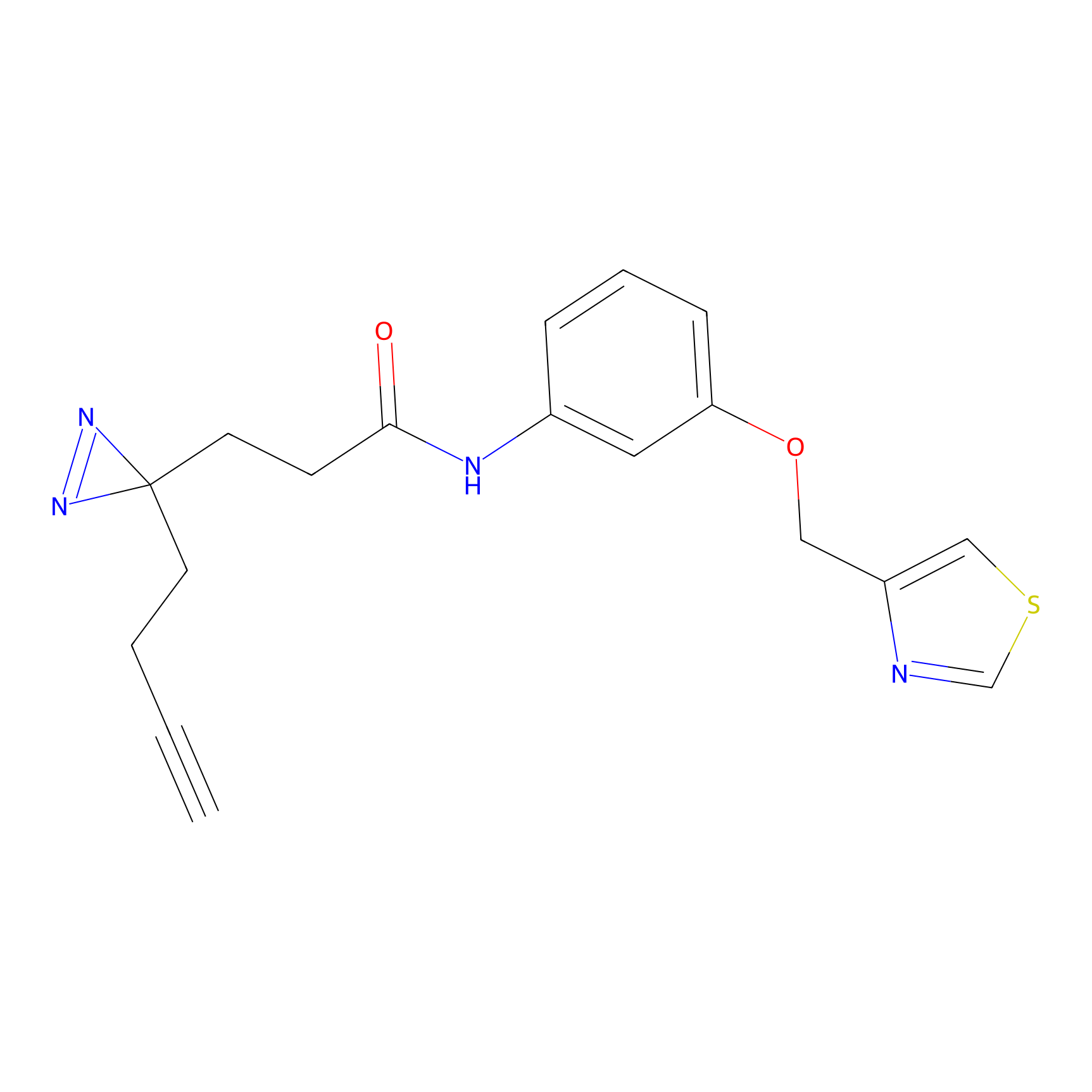
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
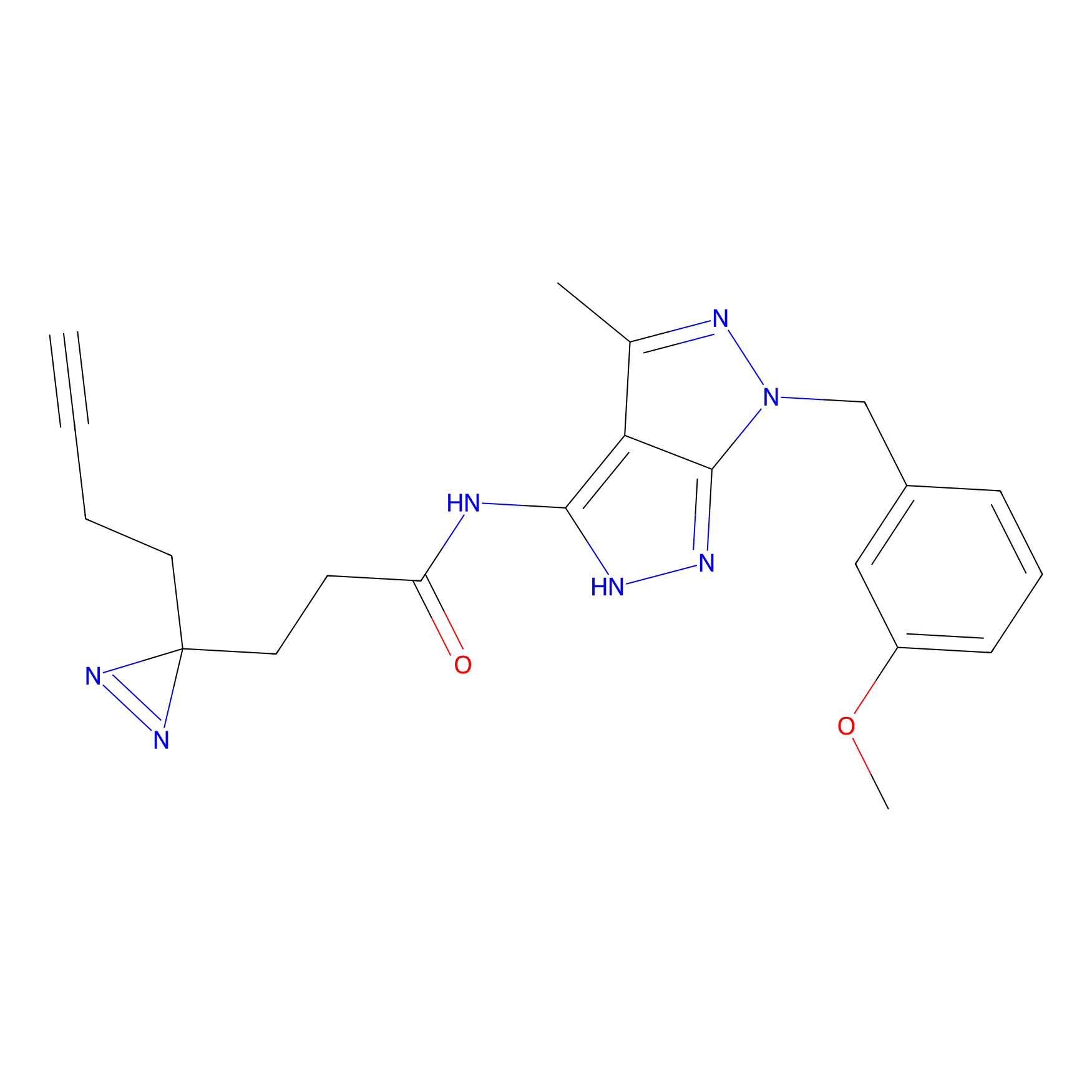
|
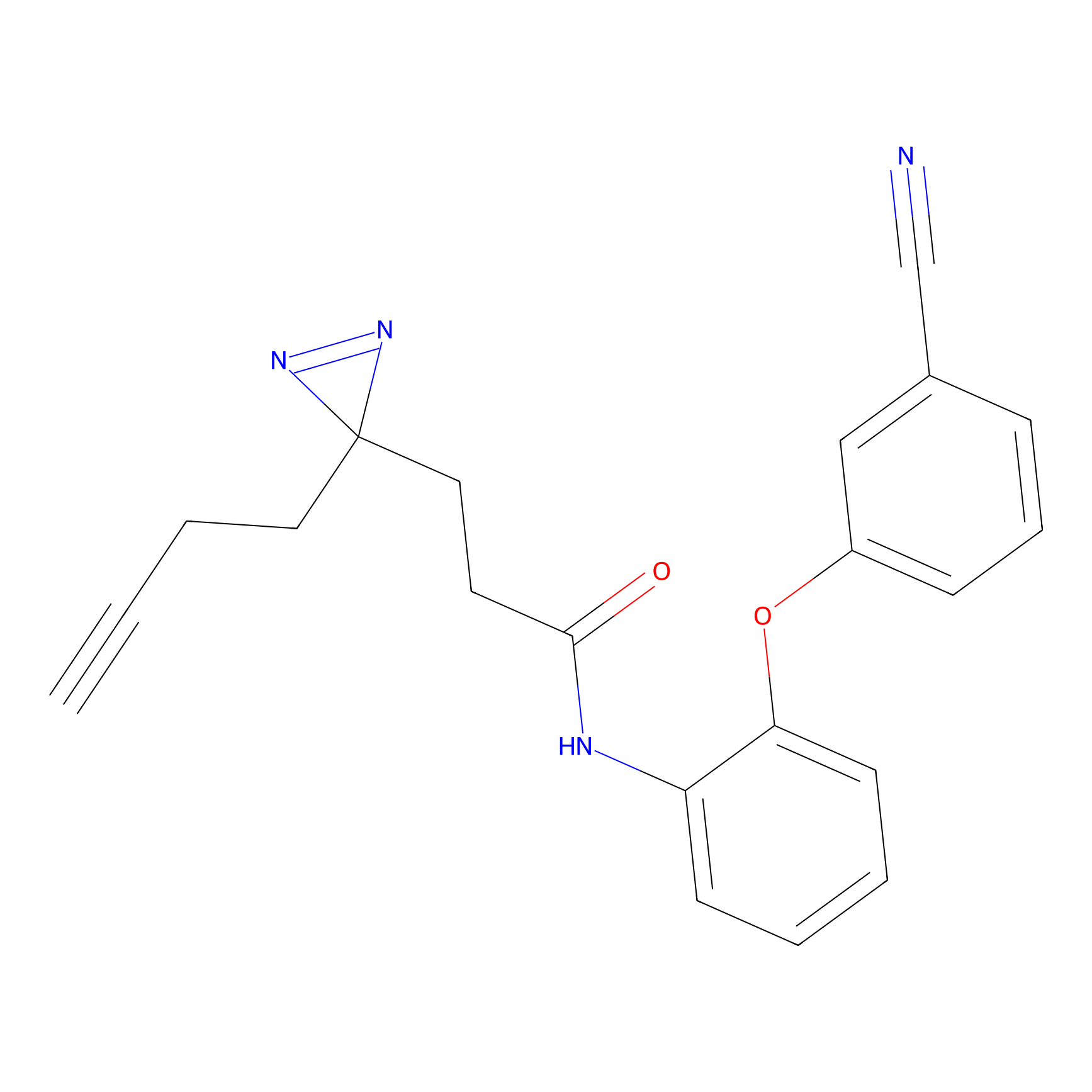
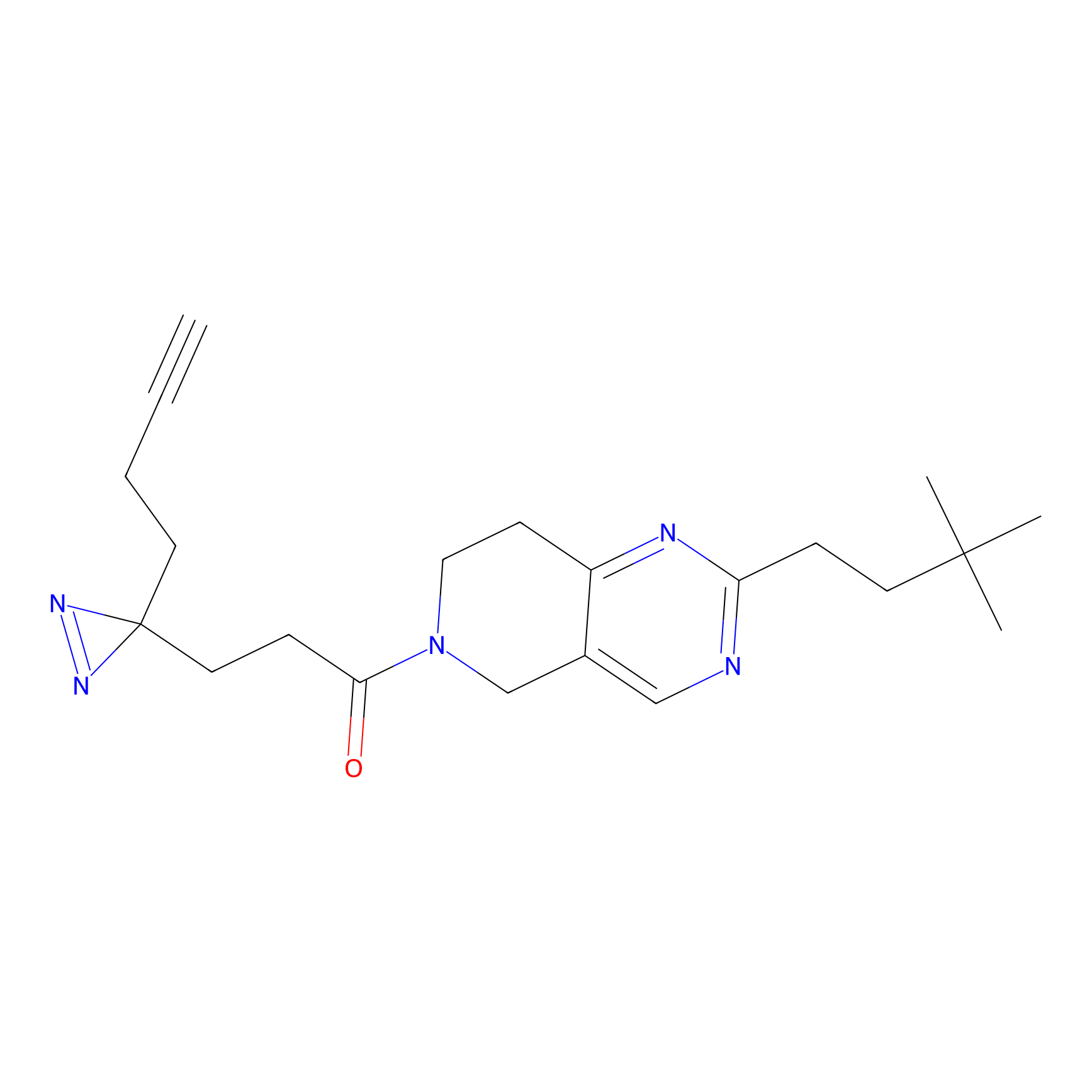
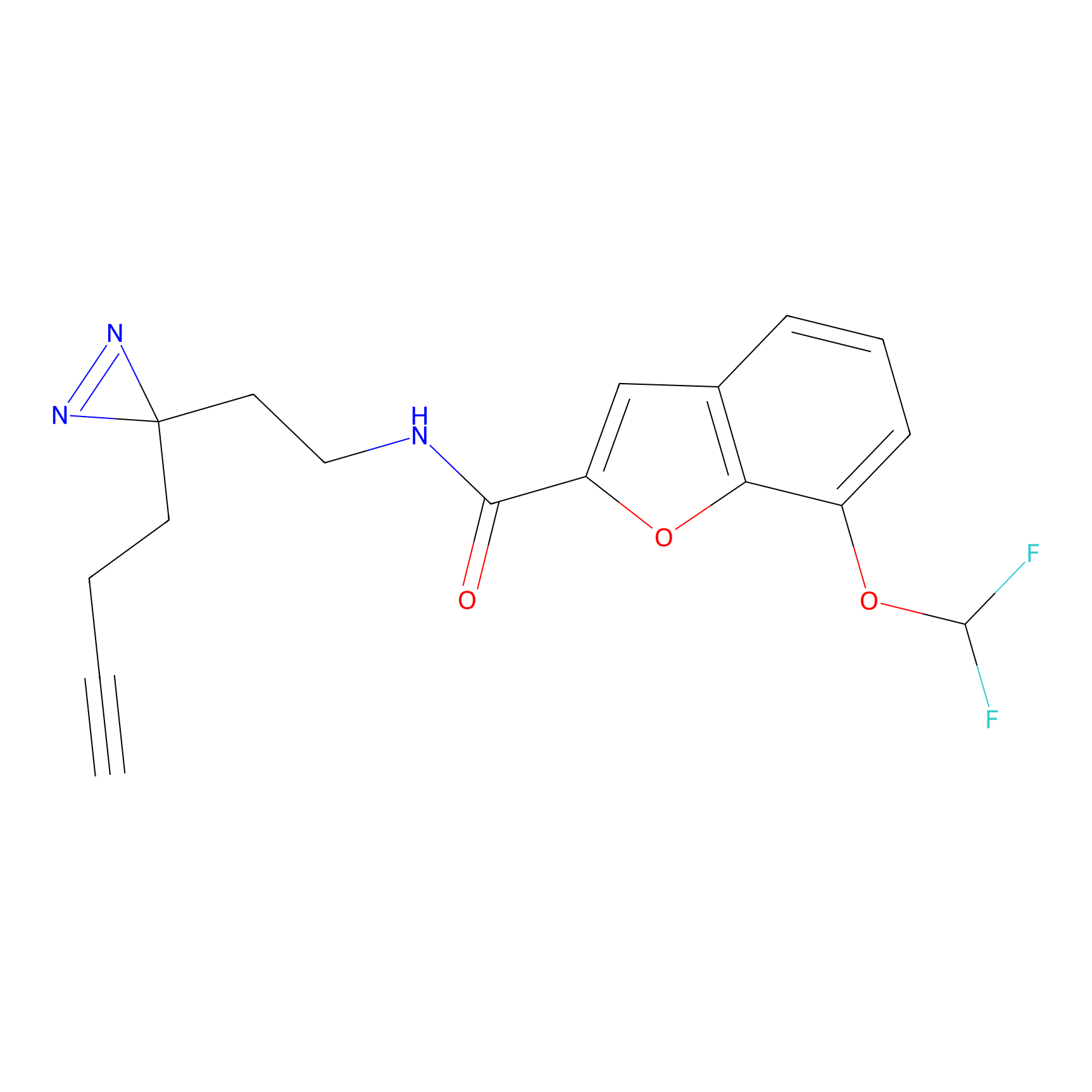
TH211 Probe Info |
 |
Y36(20.00); Y51(20.00); Y59(20.00); Y310(15.87) | LDD0257 | [2] | |
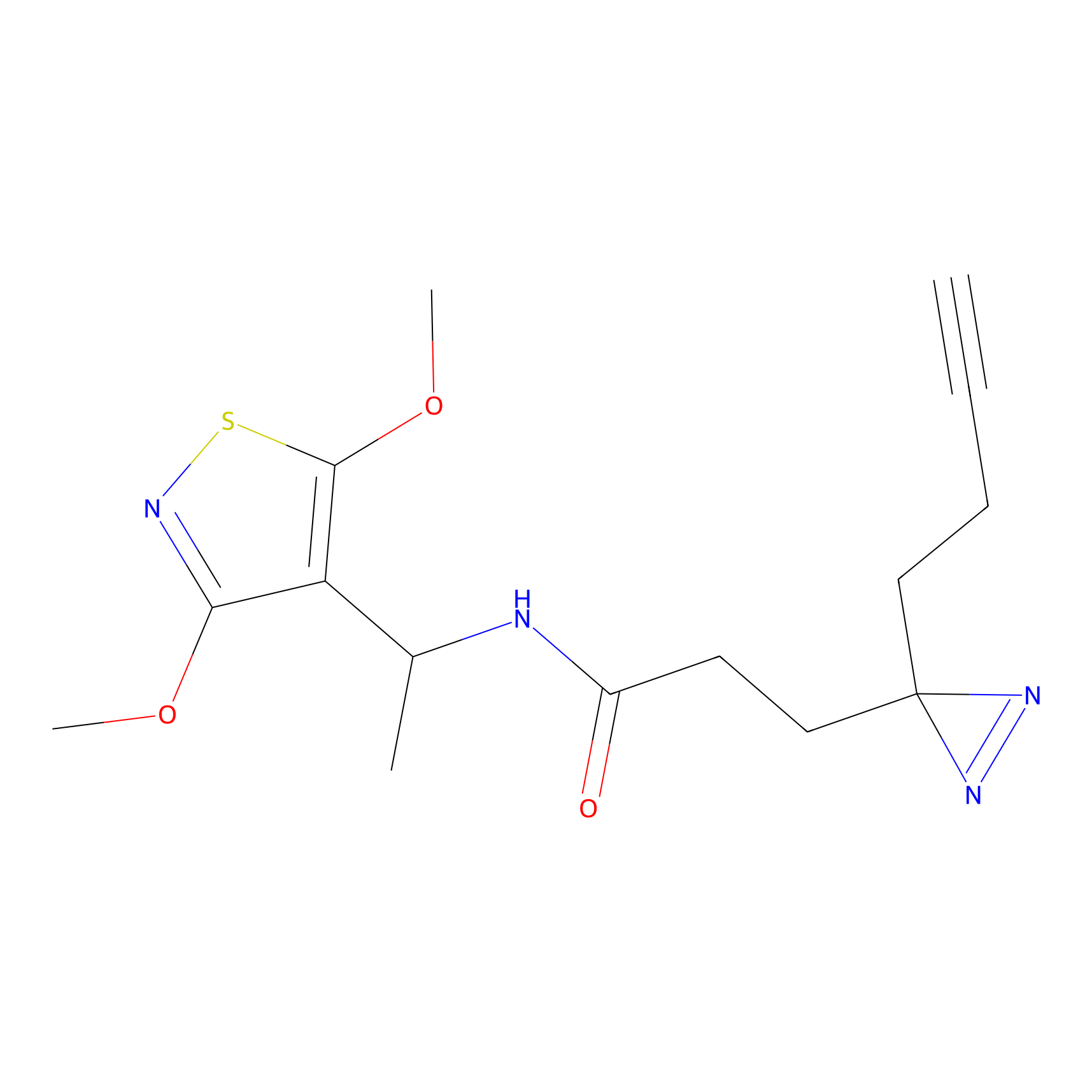
|
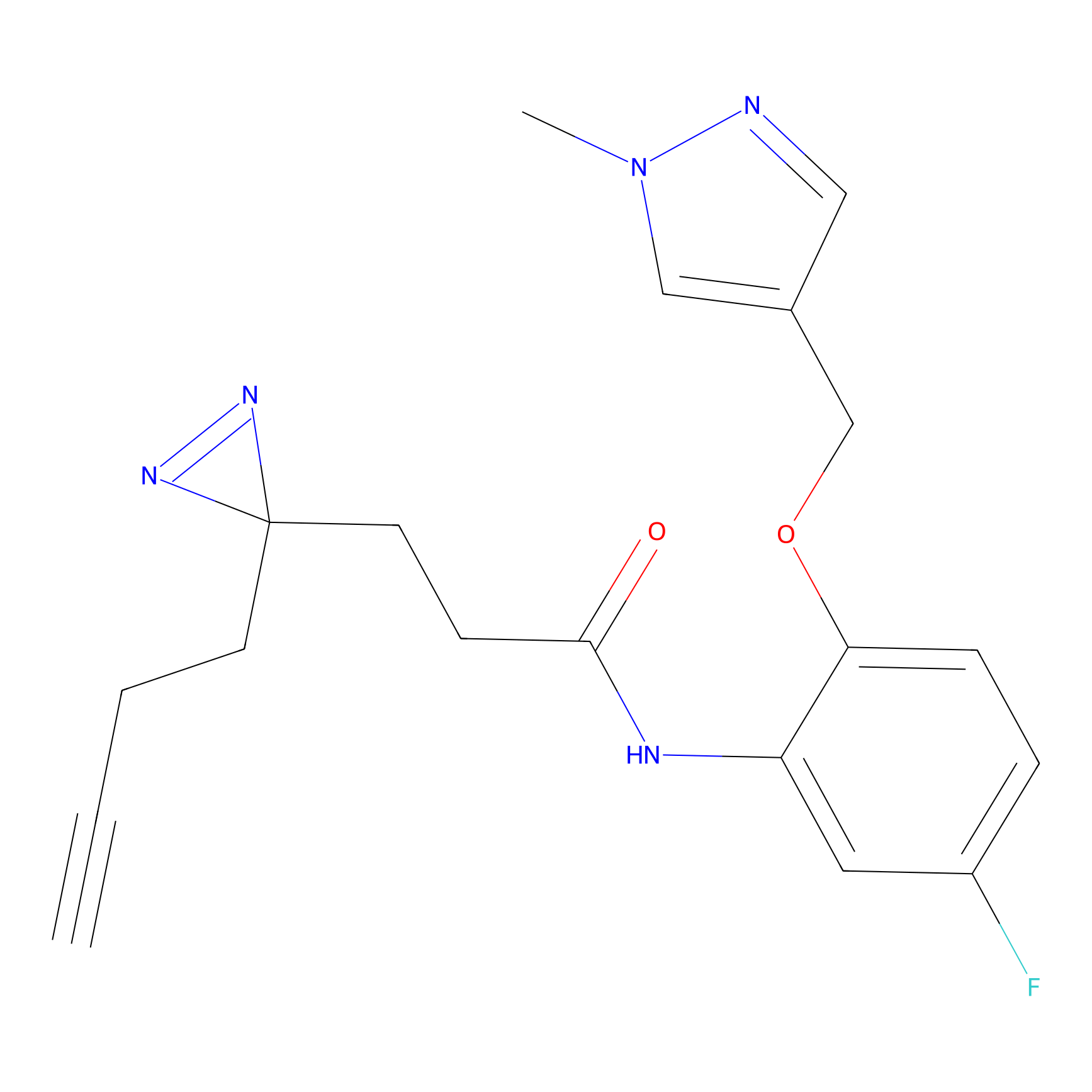
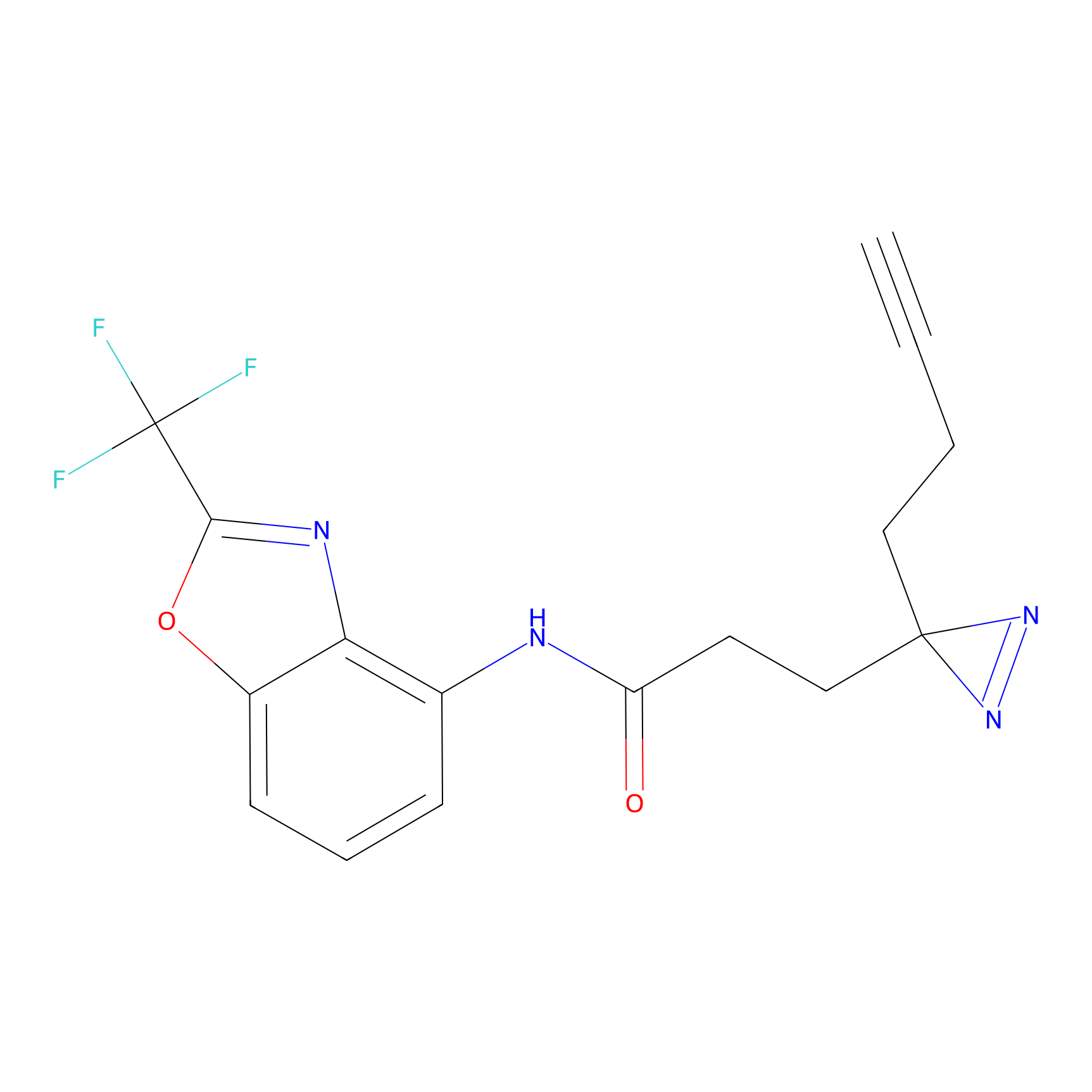
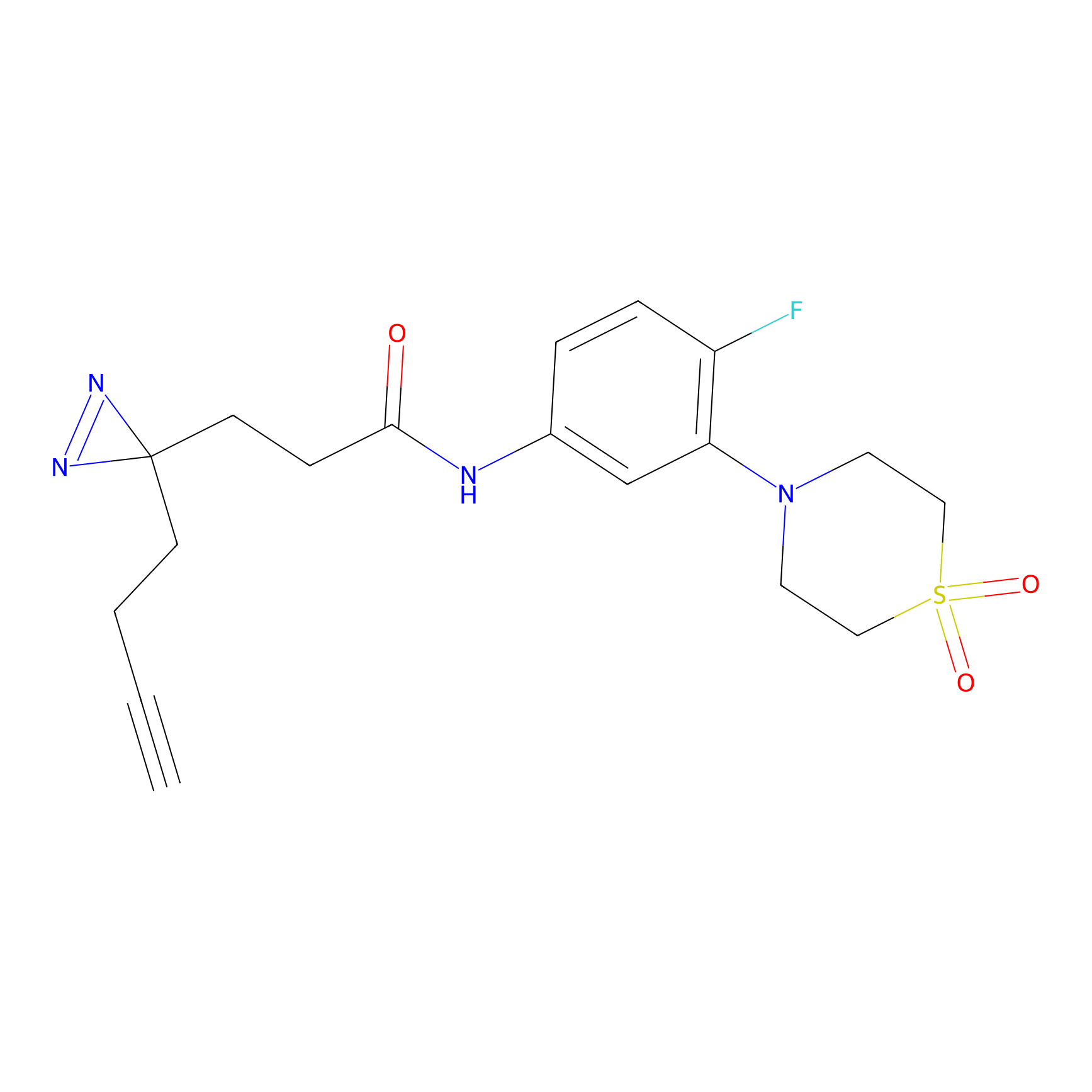
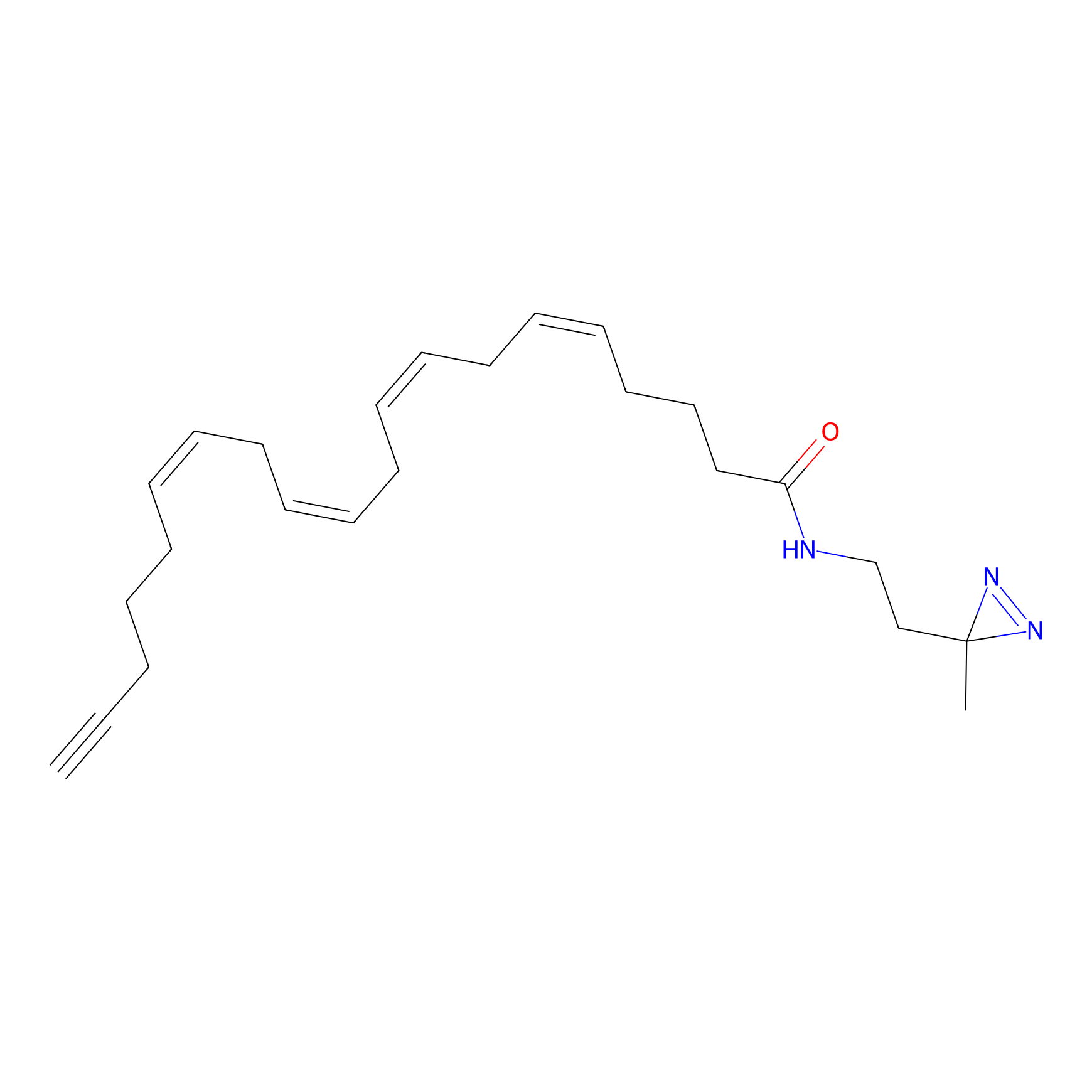
TH214 Probe Info |
 |
Y51(9.48); Y36(9.08); Y310(8.46) | LDD0258 | [2] | |
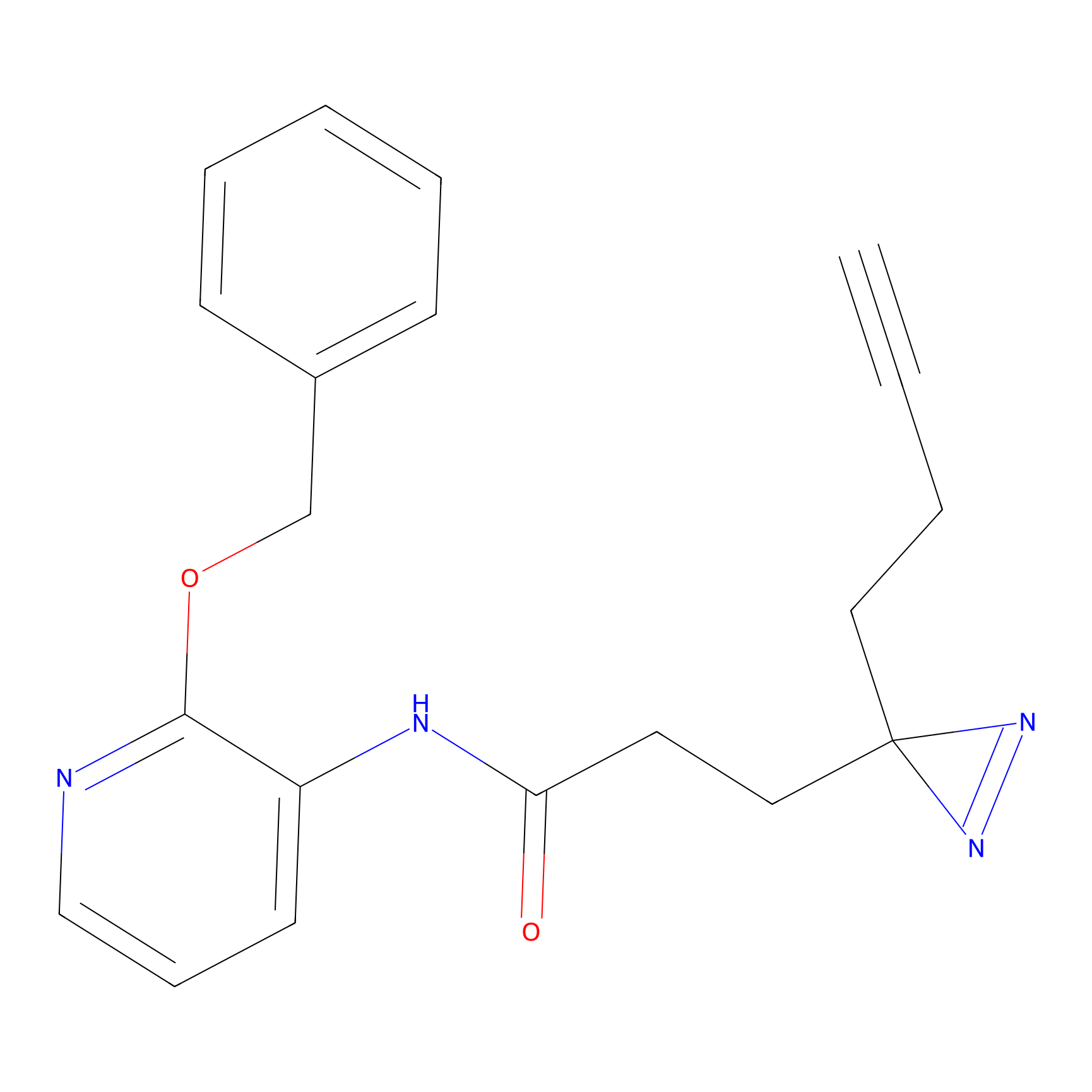
|
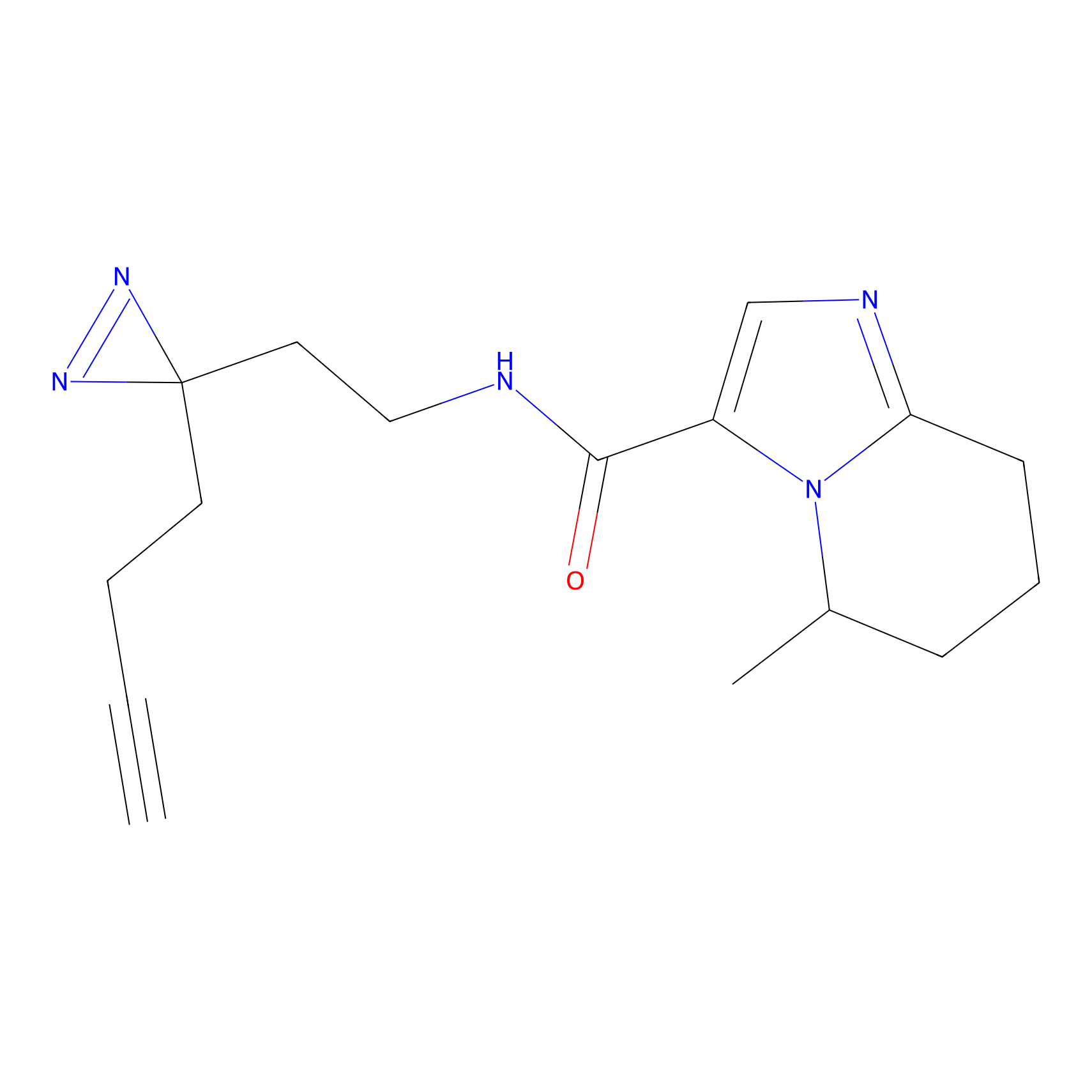
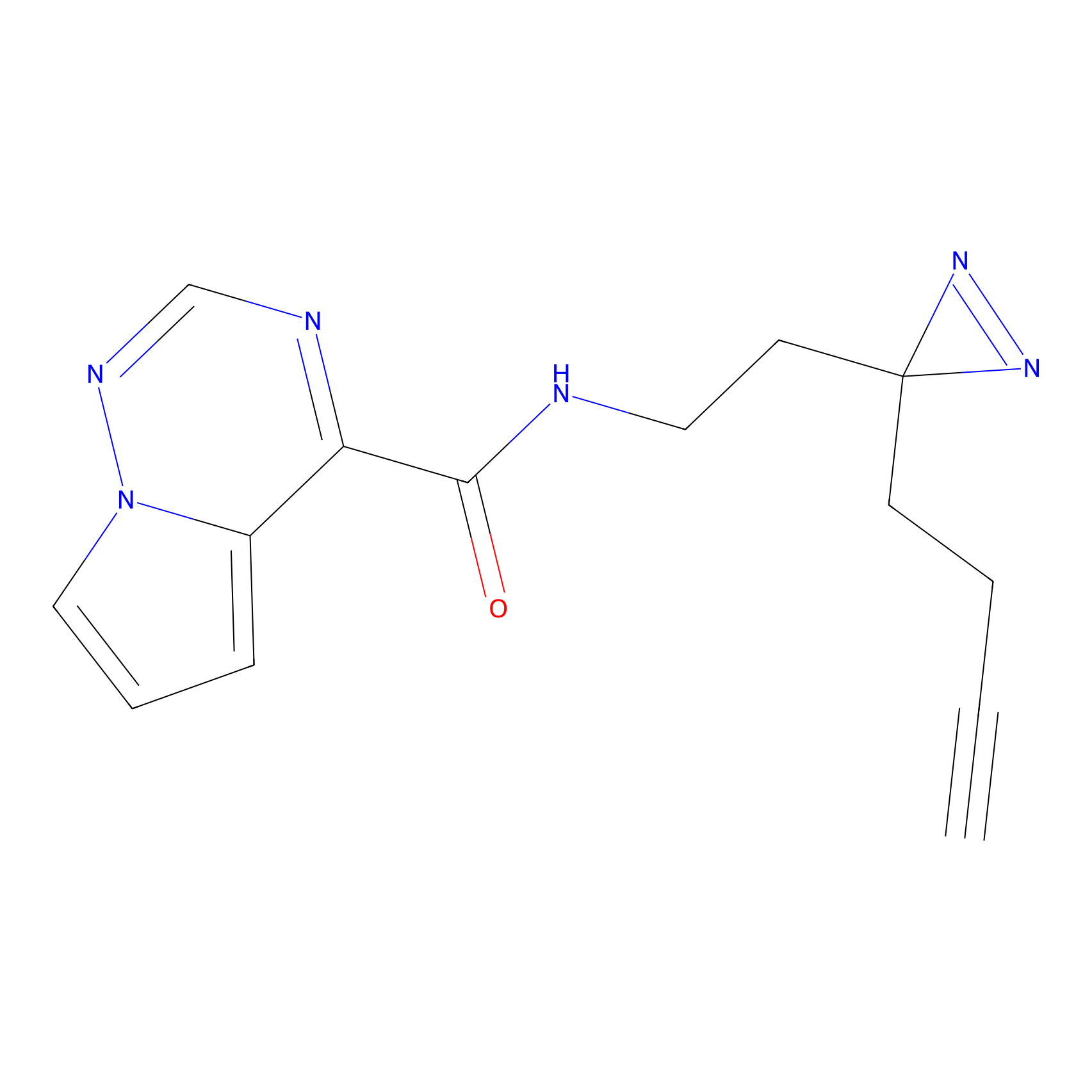
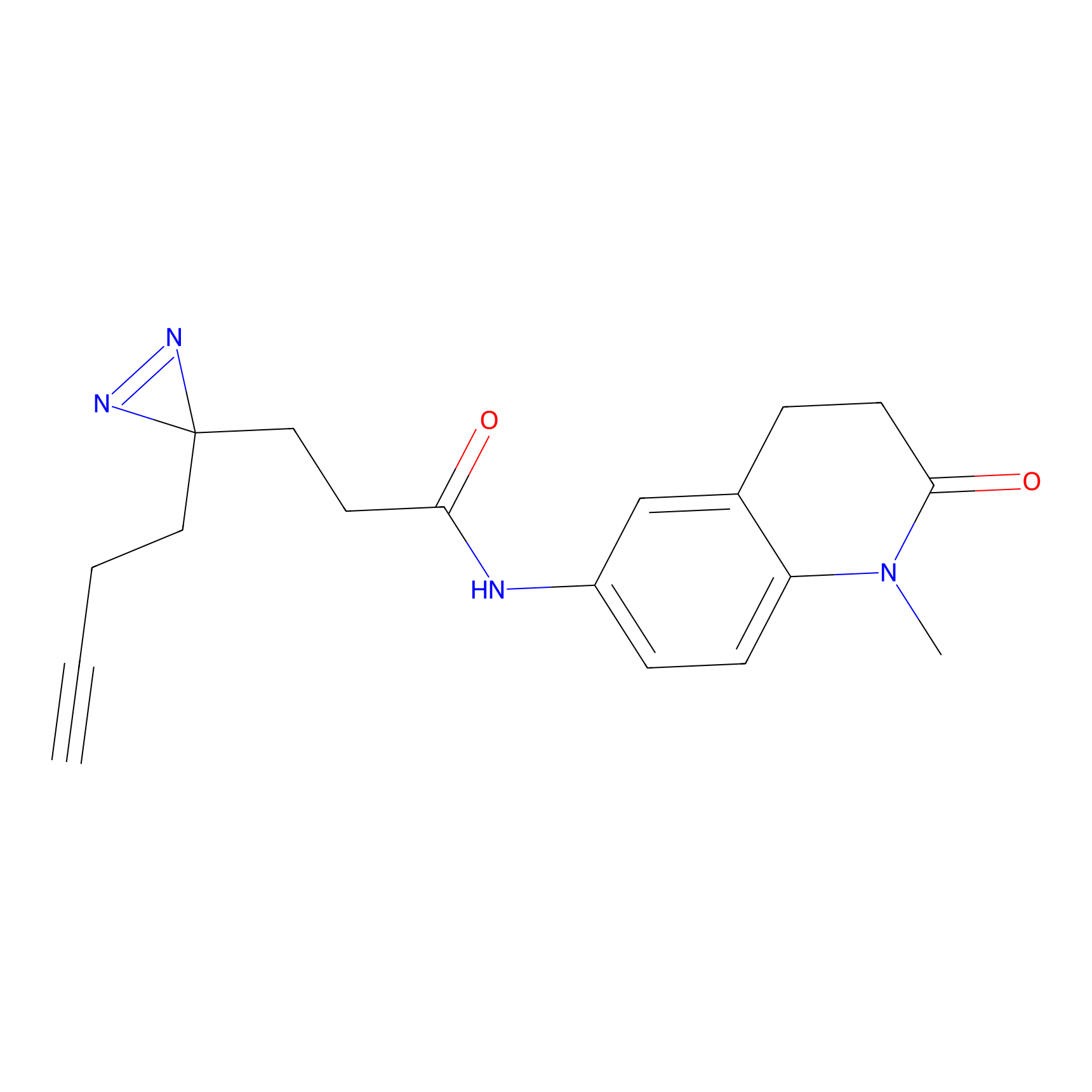
TH216 Probe Info |
 |
Y36(20.00); Y51(20.00); Y159(14.36); Y310(10.30) | LDD0259 | [2] | |
|
AZ-9 Probe Info |
 |
D74(0.84); E383(0.90) | LDD2208 | [3] | |
|
AHL-Pu-1 Probe Info |
 |
C303(2.15) | LDD0168 | [4] | |
|
HHS-482 Probe Info |
 |
Y159(0.80); Y310(0.94); Y36(0.89); Y50(1.06) | LDD0285 | [5] | |
|
HHS-475 Probe Info |
 |
Y222(0.69); Y106(0.78); Y281(0.79); Y340(0.90) | LDD0264 | [6] | |
|
HHS-465 Probe Info |
 |
Y106(10.00); Y159(8.69); Y183(10.00); Y200(3.29) | LDD2237 | [7] | |
|
DBIA Probe Info |
 |
C127(1.42); C129(1.42) | LDD0078 | [8] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C127(0.00); C129(0.00) | LDD0038 | [9] | |
|
IA-alkyne Probe Info |
 |
C12(0.00); C201(0.00); C354(0.00); C239(0.00) | LDD0032 | [10] | |
|
Lodoacetamide azide Probe Info |
 |
C127(0.00); C129(0.00) | LDD0037 | [9] | |
|
JW-RF-010 Probe Info |
 |
C239(0.00); C12(0.00); C129(0.00); C303(0.00) | LDD0026 | [11] | |
|
1d-yne Probe Info |
 |
K216(0.00); K362(0.00); K252(0.00); K103(0.00) | LDD0356 | [12] | |
|
IPM Probe Info |
 |
C127(0.00); C129(0.00) | LDD2156 | [13] | |
|
SF Probe Info |
 |
Y59(0.00); Y222(0.00); Y50(0.00); Y281(0.00) | LDD0028 | [14] | |
|
VSF Probe Info |
 |
C239(0.00); C12(0.00); C129(0.00) | LDD0007 | [15] | |
|
Phosphinate-6 Probe Info |
 |
C201(0.00); C12(0.00); C211(0.00); C354(0.00) | LDD0018 | [16] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [17] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | HEK-293T | C303(2.15) | LDD0168 | [4] |
| LDCM0156 | Aniline | NCI-H1299 | 11.71 | LDD0403 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C127(1.42); C129(1.42) | LDD0078 | [8] |
| LDCM0083 | Avasimibe | A-549 | 2.31 | LDD0143 | [20] |
| LDCM0189 | Compound 16 | HEK-293T | 10.67 | LDD0492 | [19] |
| LDCM0182 | Compound 18 | HEK-293T | 7.06 | LDD0501 | [19] |
| LDCM0191 | Compound 21 | HEK-293T | 7.60 | LDD0493 | [19] |
| LDCM0190 | Compound 34 | HEK-293T | 11.92 | LDD0497 | [19] |
| LDCM0192 | Compound 35 | HEK-293T | 6.64 | LDD0491 | [19] |
| LDCM0193 | Compound 36 | HEK-293T | 5.33 | LDD0494 | [19] |
| LDCM0194 | Compound 37 | HEK-293T | 9.14 | LDD0498 | [19] |
| LDCM0195 | Compound 38 | HEK-293T | 7.03 | LDD0499 | [19] |
| LDCM0196 | Compound 39 | HEK-293T | 6.54 | LDD0496 | [19] |
| LDCM0197 | Compound 40 | HEK-293T | 6.05 | LDD0495 | [19] |
| LDCM0181 | Compound 41 | HEK-293T | 8.06 | LDD0502 | [19] |
| LDCM0183 | Compound 42 | HEK-293T | 4.58 | LDD0500 | [19] |
| LDCM0625 | F8 | Ramos | 0.86 | LDD2187 | [24] |
| LDCM0572 | Fragment10 | Ramos | 0.26 | LDD2189 | [24] |
| LDCM0573 | Fragment11 | Ramos | 0.16 | LDD2190 | [24] |
| LDCM0574 | Fragment12 | Ramos | 0.32 | LDD2191 | [24] |
| LDCM0575 | Fragment13 | Ramos | 0.49 | LDD2192 | [24] |
| LDCM0576 | Fragment14 | Ramos | 1.61 | LDD2193 | [24] |
| LDCM0579 | Fragment20 | Ramos | 0.73 | LDD2194 | [24] |
| LDCM0580 | Fragment21 | Ramos | 0.47 | LDD2195 | [24] |
| LDCM0582 | Fragment23 | Ramos | 0.69 | LDD2196 | [24] |
| LDCM0578 | Fragment27 | Ramos | 0.77 | LDD2197 | [24] |
| LDCM0586 | Fragment28 | Ramos | 1.20 | LDD2198 | [24] |
| LDCM0588 | Fragment30 | Ramos | 0.53 | LDD2199 | [24] |
| LDCM0589 | Fragment31 | Ramos | 0.48 | LDD2200 | [24] |
| LDCM0590 | Fragment32 | Ramos | 0.48 | LDD2201 | [24] |
| LDCM0468 | Fragment33 | Ramos | 0.39 | LDD2202 | [24] |
| LDCM0596 | Fragment38 | Ramos | 0.27 | LDD2203 | [24] |
| LDCM0566 | Fragment4 | Ramos | 0.72 | LDD2184 | [24] |
| LDCM0610 | Fragment52 | Ramos | 0.44 | LDD2204 | [24] |
| LDCM0614 | Fragment56 | Ramos | 0.67 | LDD2205 | [24] |
| LDCM0569 | Fragment7 | Ramos | 0.68 | LDD2186 | [24] |
| LDCM0571 | Fragment9 | Ramos | 0.51 | LDD2188 | [24] |
| LDCM0116 | HHS-0101 | DM93 | Y222(0.69); Y106(0.78); Y281(0.79); Y340(0.90) | LDD0264 | [6] |
| LDCM0117 | HHS-0201 | DM93 | Y222(0.59); Y106(0.68); Y281(0.80); Y340(0.92) | LDD0265 | [6] |
| LDCM0118 | HHS-0301 | DM93 | Y222(0.62); Y106(0.66); Y281(0.85); Y340(0.89) | LDD0266 | [6] |
| LDCM0119 | HHS-0401 | DM93 | Y222(0.63); Y106(0.63); Y281(0.91); Y340(1.26) | LDD0267 | [6] |
| LDCM0120 | HHS-0701 | DM93 | Y106(0.62); Y222(0.63); Y281(0.76); Y340(1.21) | LDD0268 | [6] |
| LDCM0123 | JWB131 | DM93 | Y159(0.80); Y310(0.94); Y36(0.89); Y50(1.06) | LDD0285 | [5] |
| LDCM0124 | JWB142 | DM93 | Y159(0.94); Y310(0.75); Y36(0.63); Y50(0.96) | LDD0286 | [5] |
| LDCM0125 | JWB146 | DM93 | Y159(0.67); Y310(1.49); Y36(1.11); Y50(1.26) | LDD0287 | [5] |
| LDCM0126 | JWB150 | DM93 | Y159(3.60); Y310(2.86); Y36(2.34); Y50(1.54) | LDD0288 | [5] |
| LDCM0127 | JWB152 | DM93 | Y159(1.27); Y310(1.95); Y36(1.64); Y50(1.90) | LDD0289 | [5] |
| LDCM0128 | JWB198 | DM93 | Y159(0.40); Y310(1.16); Y36(1.12); Y50(1.75) | LDD0290 | [5] |
| LDCM0129 | JWB202 | DM93 | Y159(0.61); Y310(0.46); Y36(0.44); Y51(0.50) | LDD0291 | [5] |
| LDCM0130 | JWB211 | DM93 | Y159(0.62); Y310(1.05); Y36(0.94); Y50(1.06) | LDD0292 | [5] |
| LDCM0073 | Kambe_cp3 | PC-3 | 3.04 | LDD0129 | [21] |
| LDCM0022 | KB02 | HEK-293T | C354(0.93); C12(0.93); C239(0.95); C127(0.94) | LDD1492 | [25] |
| LDCM0023 | KB03 | Jurkat | C127,C129(5.42) | LDD0315 | [10] |
| LDCM0024 | KB05 | HEK-293T | C354(0.88); C12(1.00); C239(0.95); C127(1.03) | LDD1502 | [25] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C127(15.00); C129(15.00); C239(1.43); C12(1.05) | LDD2206 | [26] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C129(14.14); C239(1.14); C201(1.01); C211(1.01) | LDD2207 | [26] |
| LDCM0131 | RA190 | MM1.R | C129(1.28) | LDD0304 | [27] |
The Interaction Atlas With This Target
References