Details of the Target
General Information of Target
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| 22RV1 | SNV: p.G139S | . | |||
| KASUMI1 | SNV: p.S357A | . | |||
| KMCH1 | SNV: p.Q88K | . | |||
| MOLT4 | SNV: p.R266H | IA-alkyne Probe Info | |||
| OVCAR8 | SNV: p.Y147C | . | |||
| SKMEL28 | SNV: p.S3F | DBIA Probe Info | |||
| SKNSH | SNV: p.P346H | . | |||
| SW756 | SNV: p.I172V | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
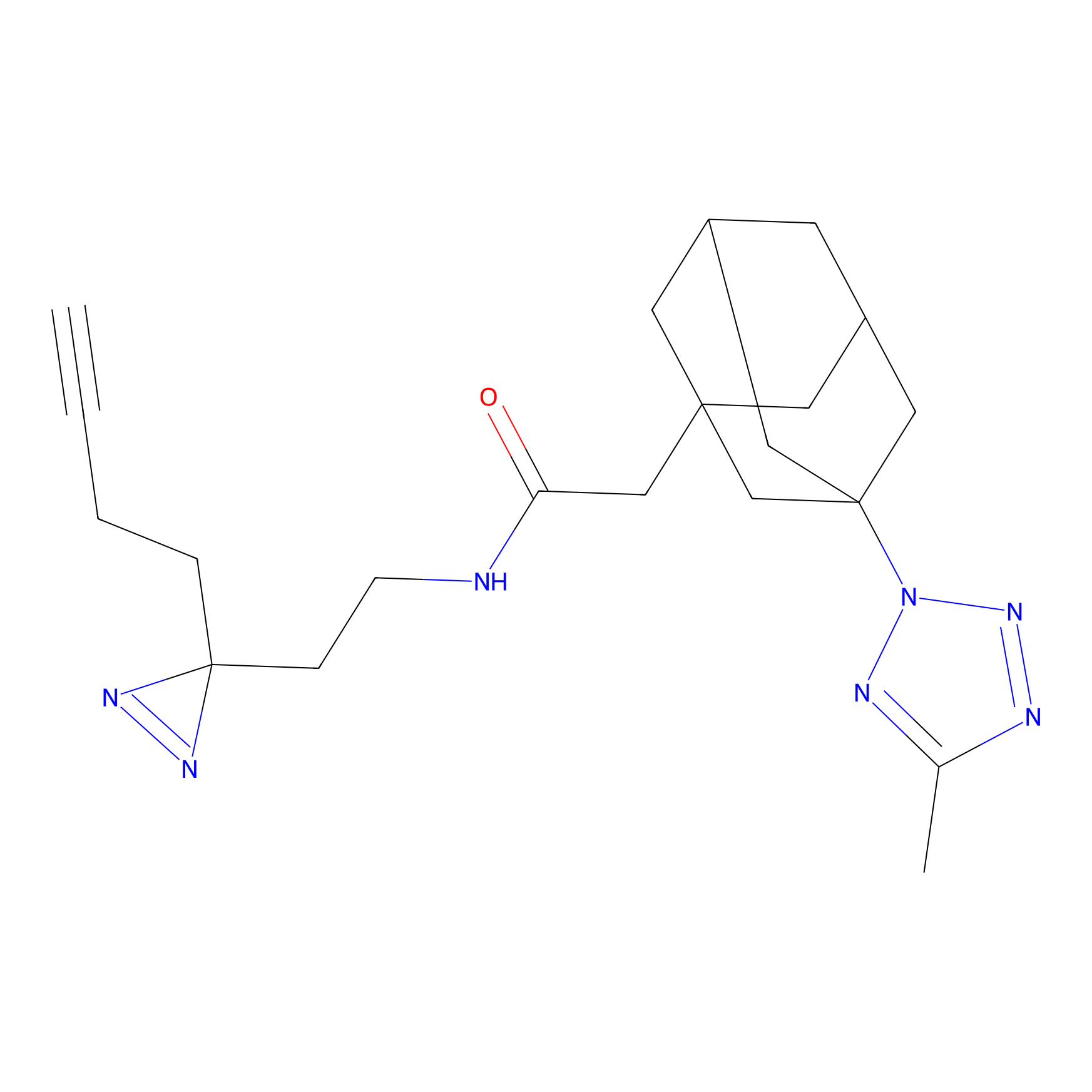
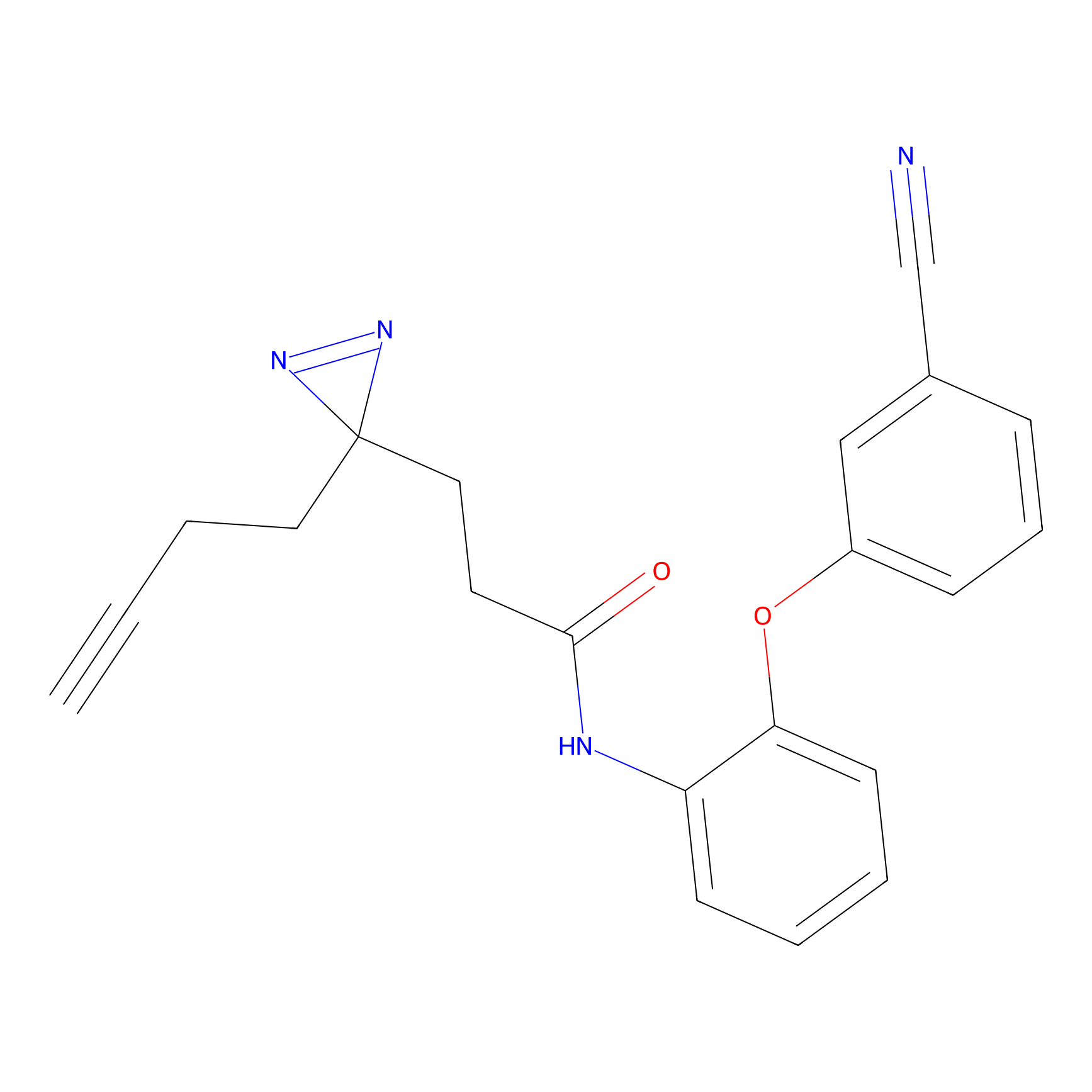
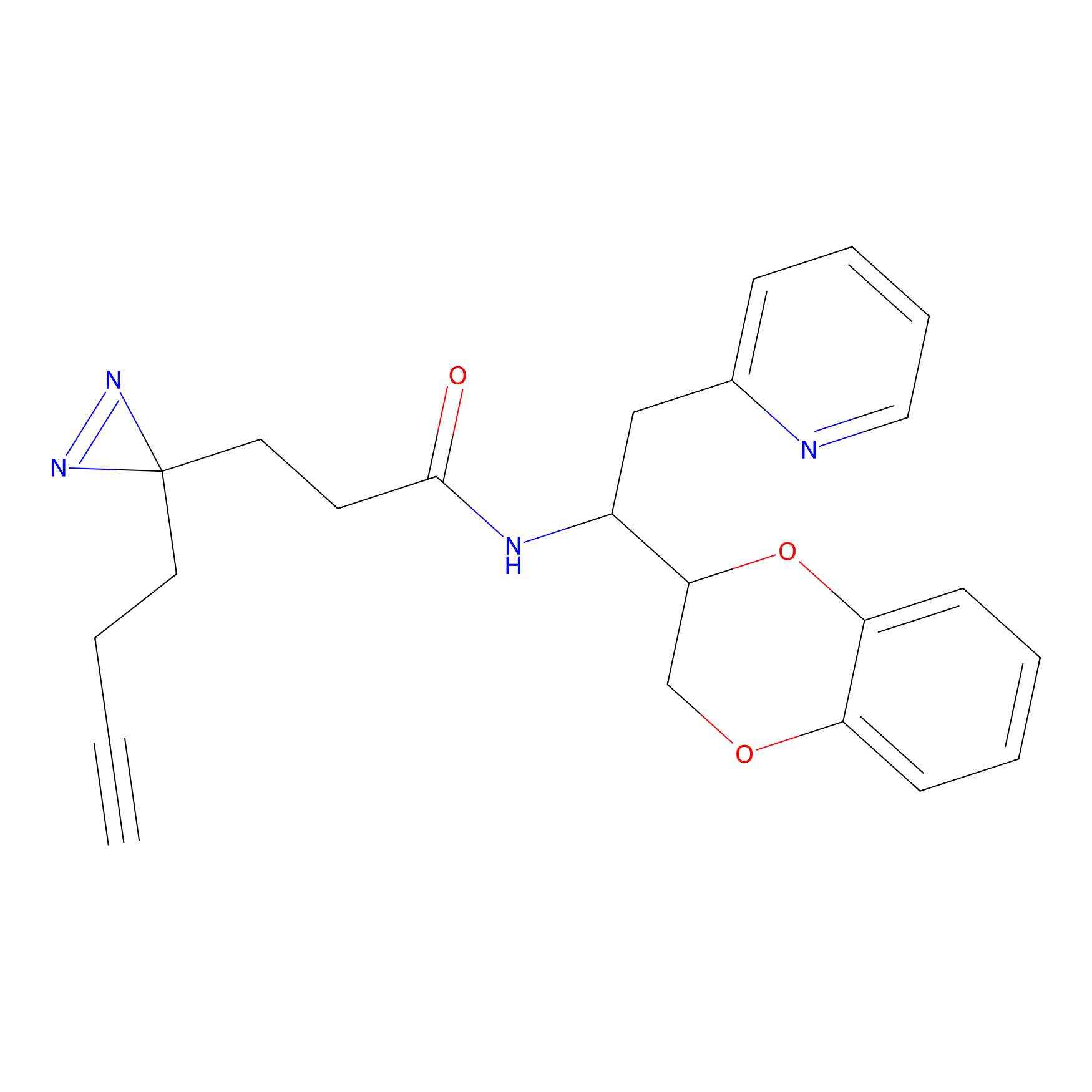
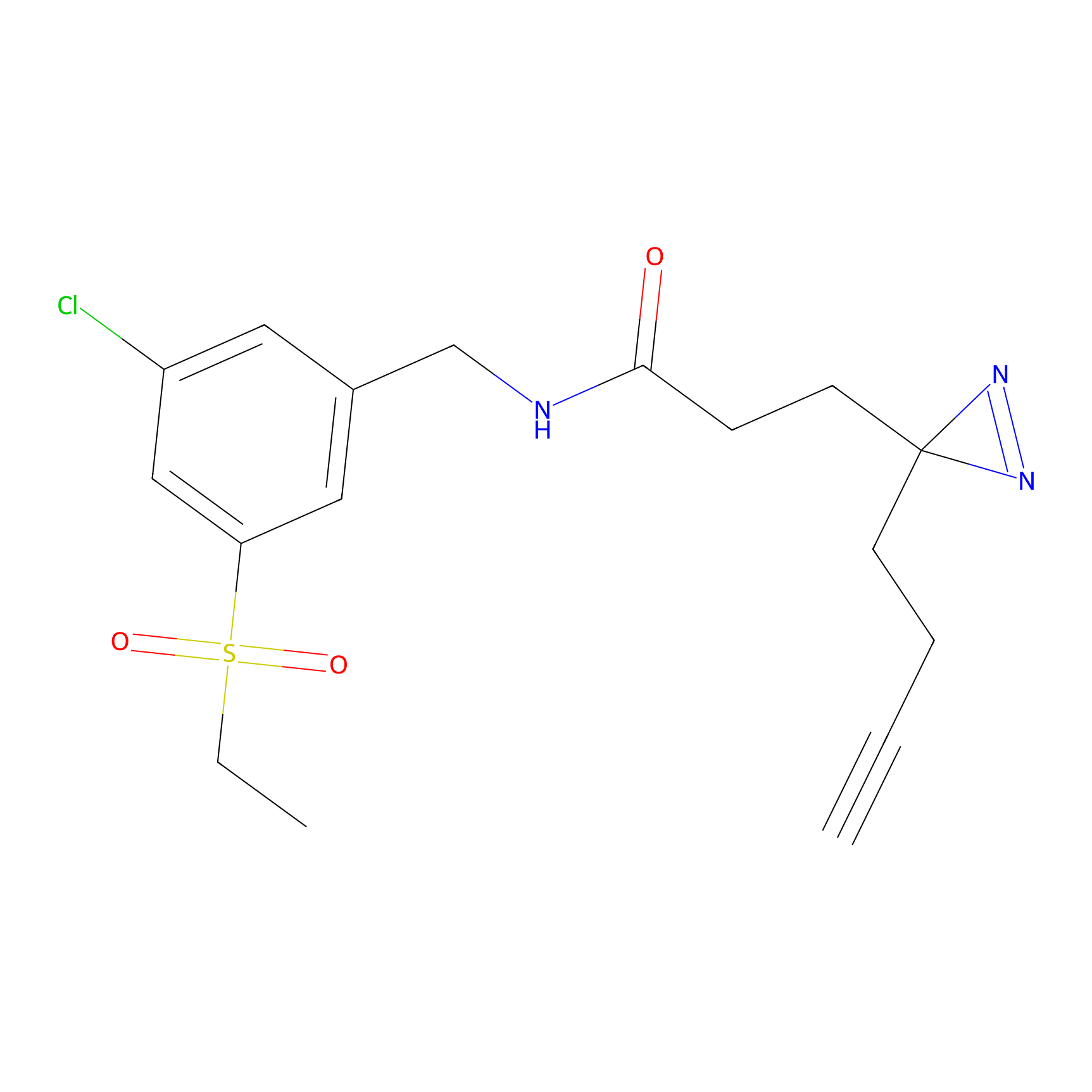
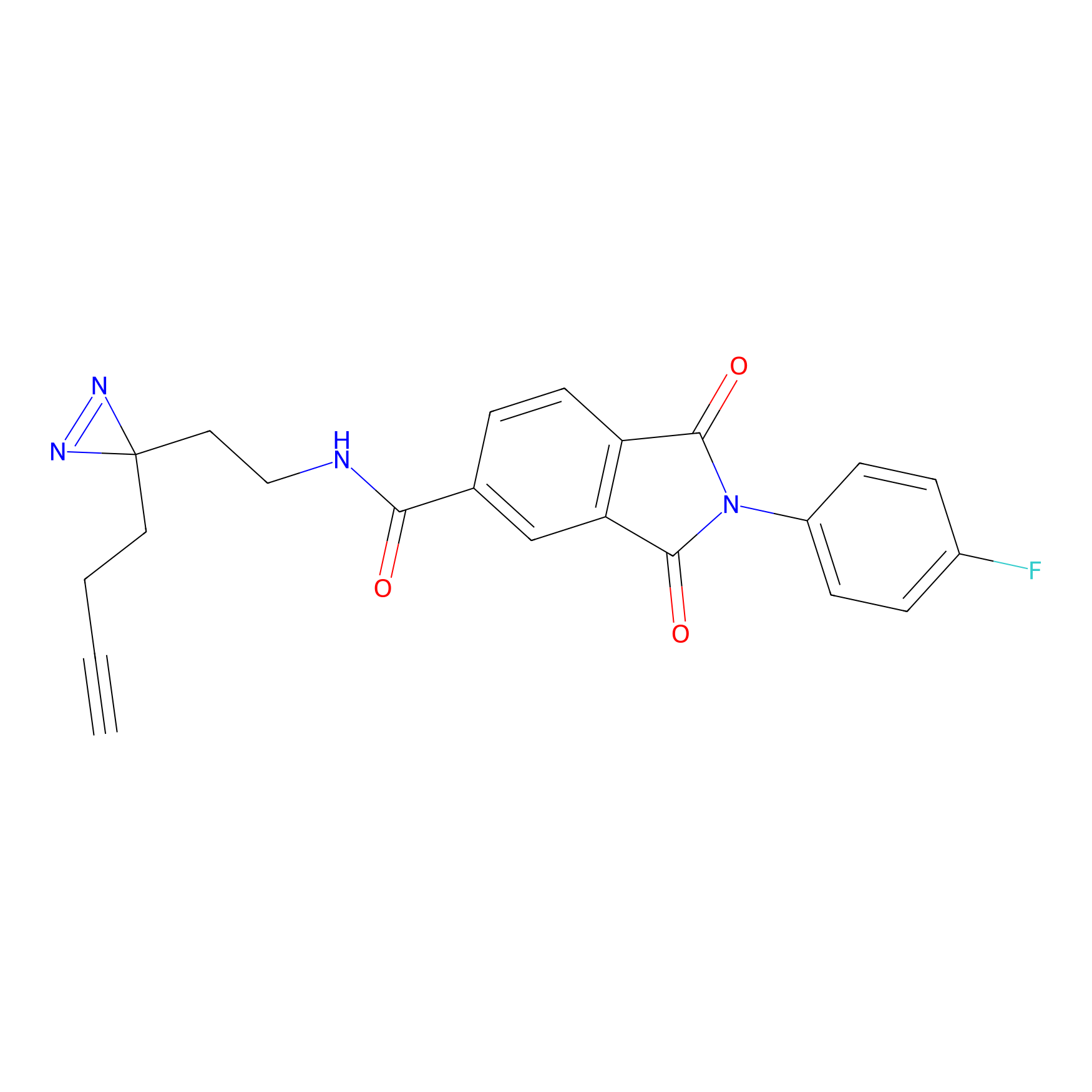
|
TH211 Probe Info |
 |
Y300(9.89) | LDD0257 | [1] | |
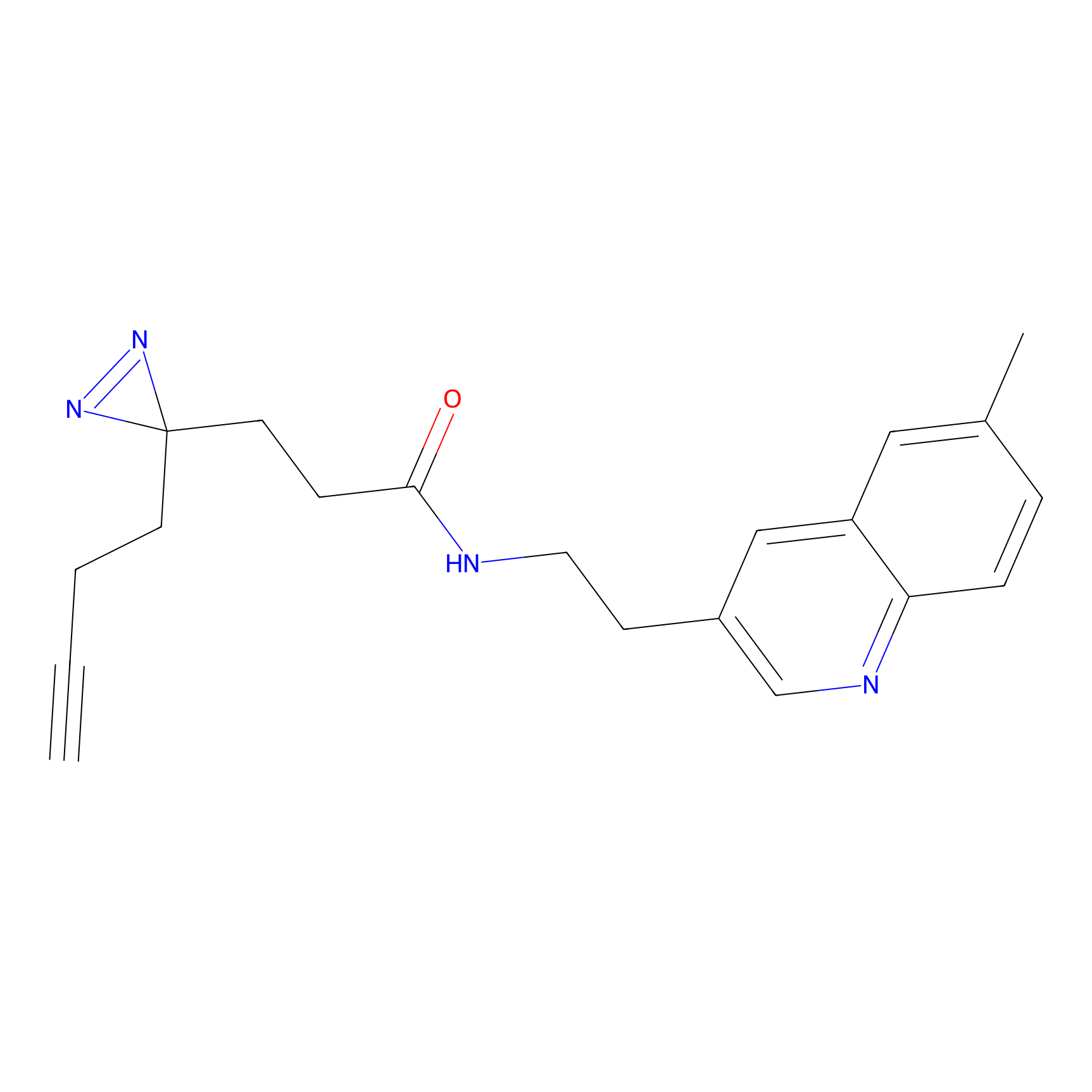
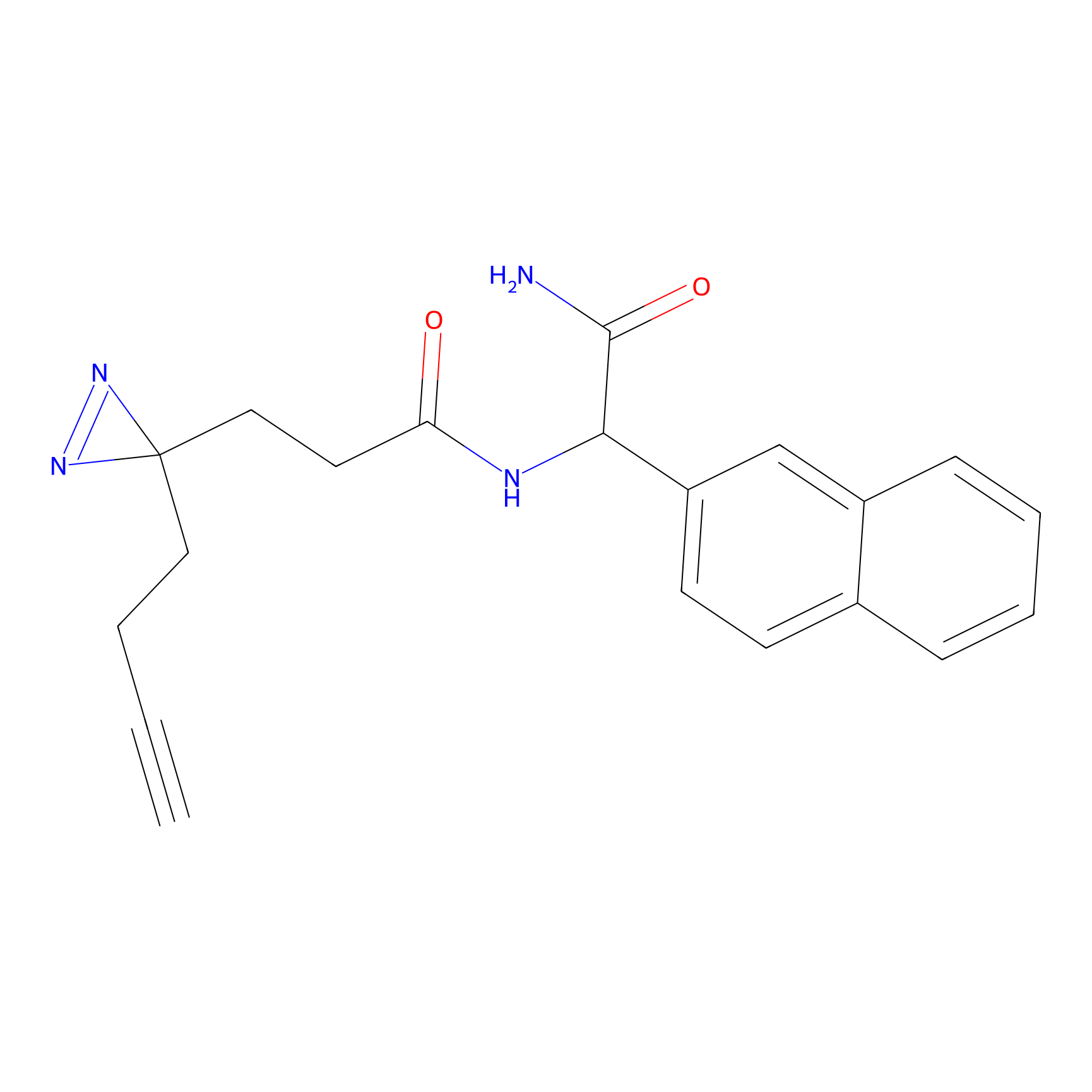
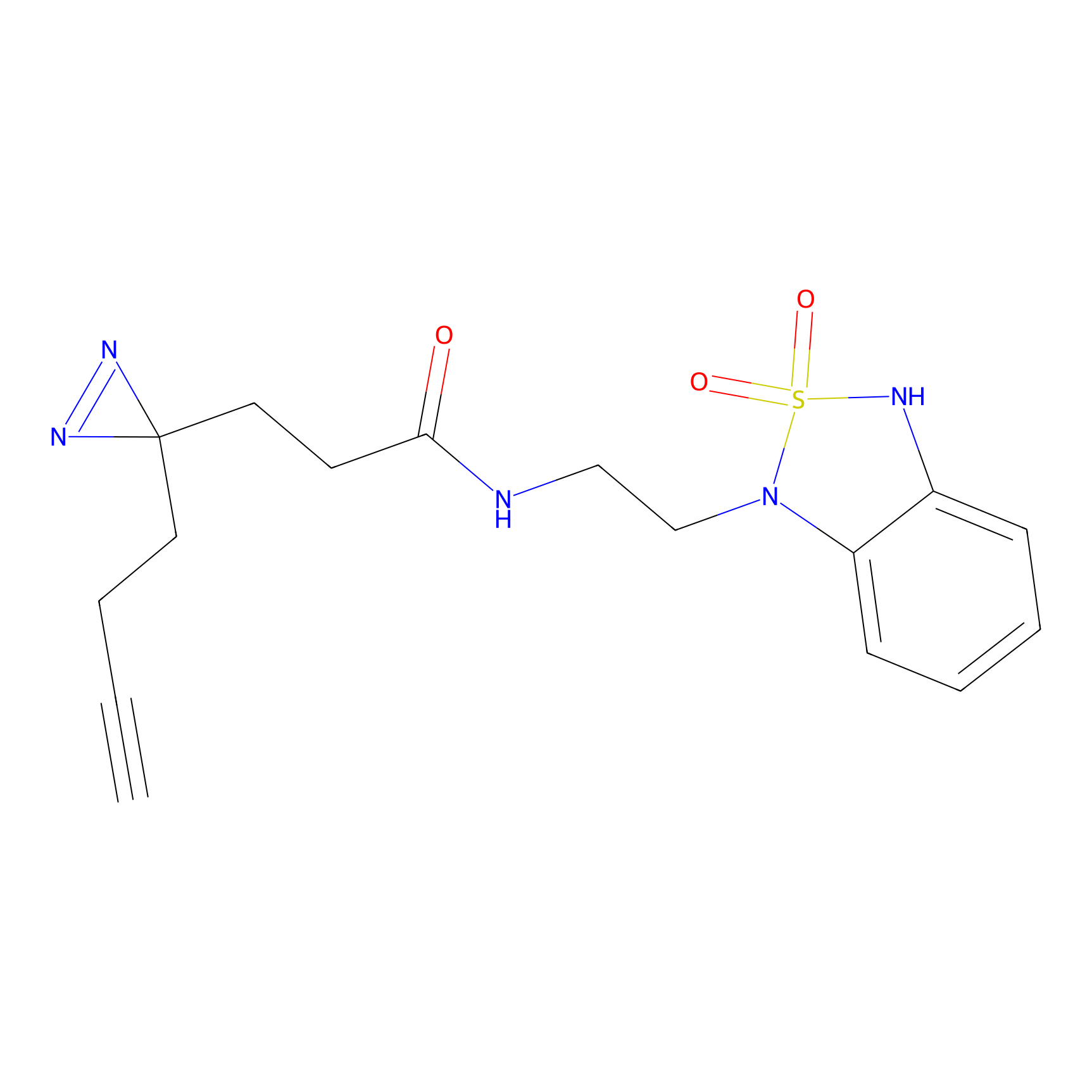
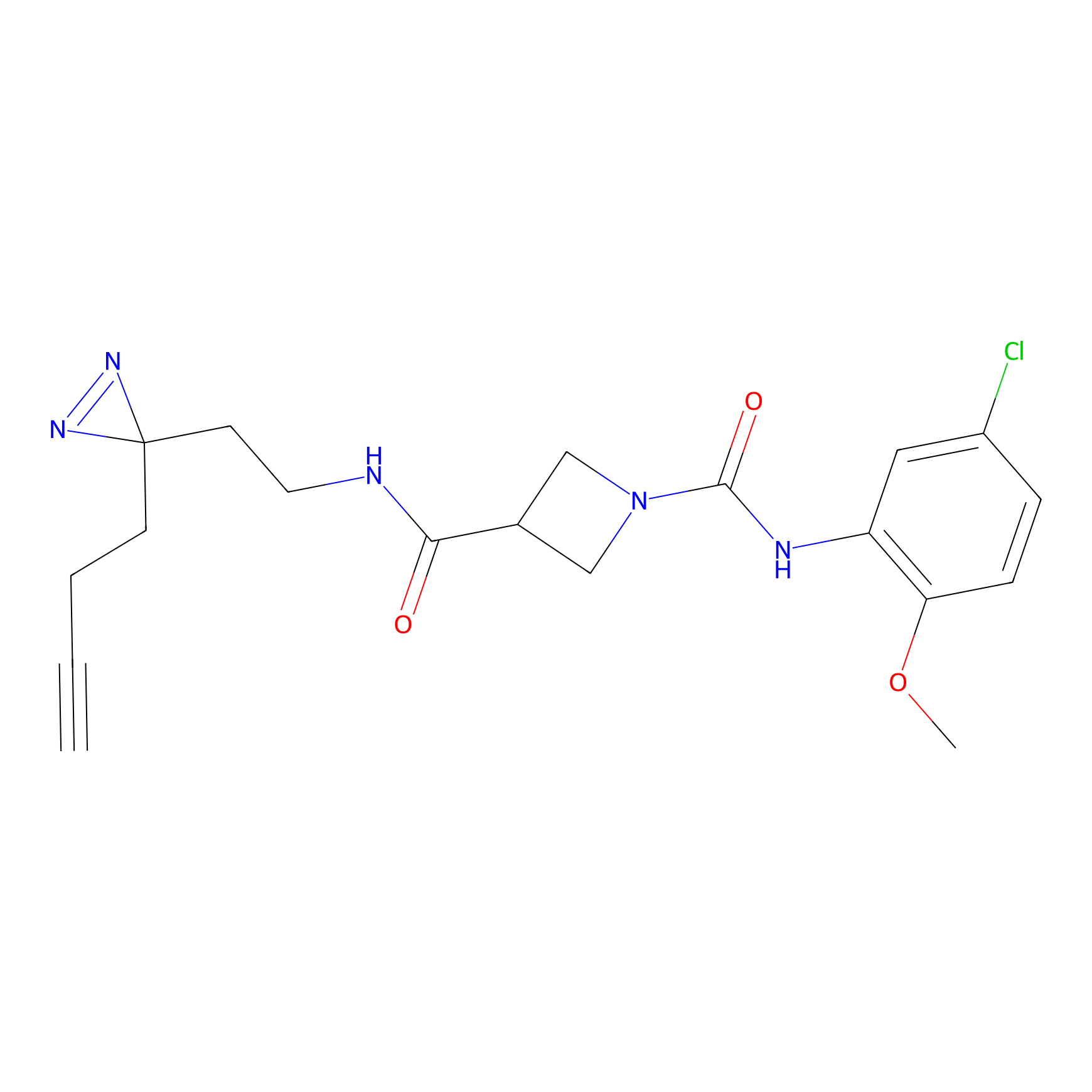
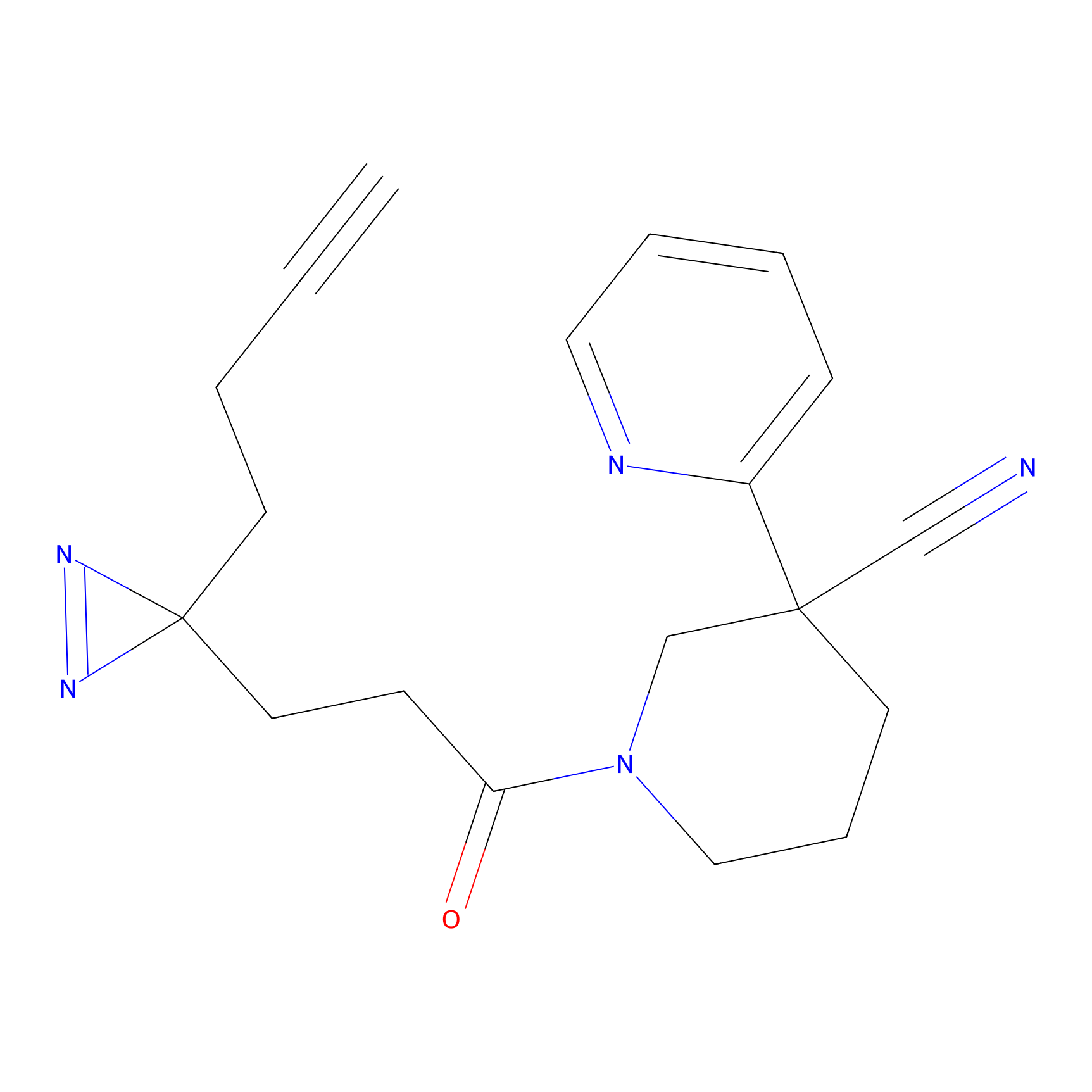
|
STPyne Probe Info |
 |
K83(8.79) | LDD0277 | [2] | |
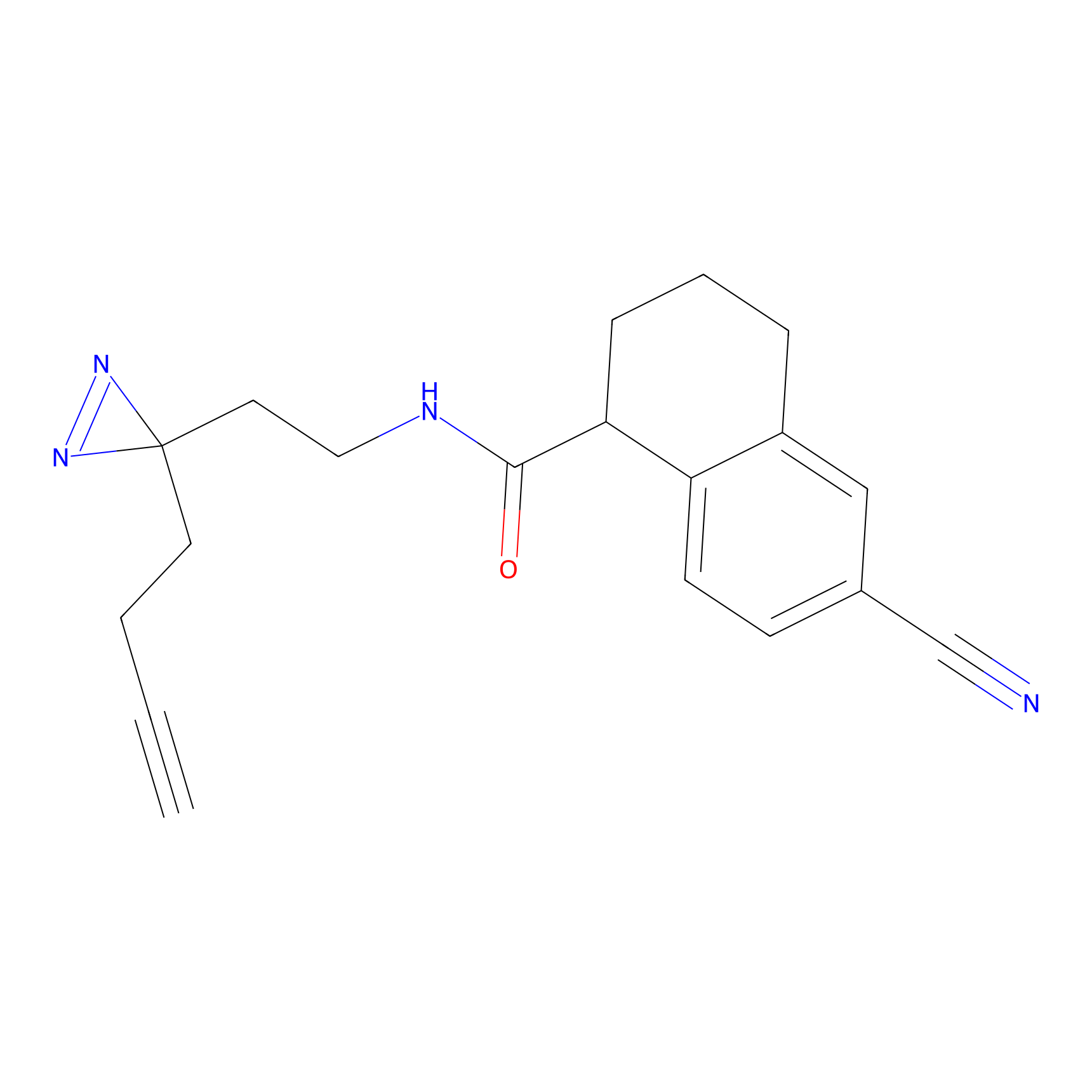
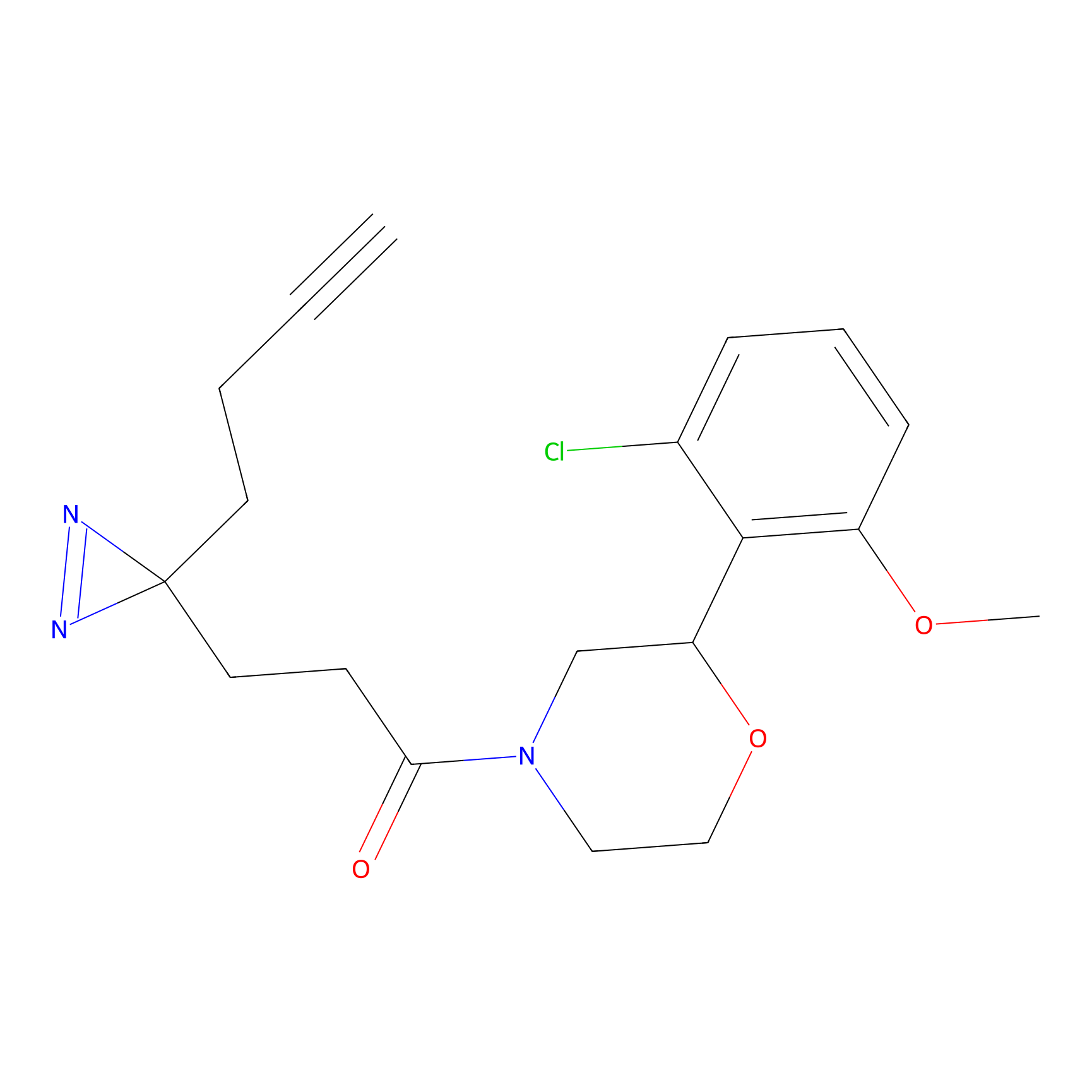
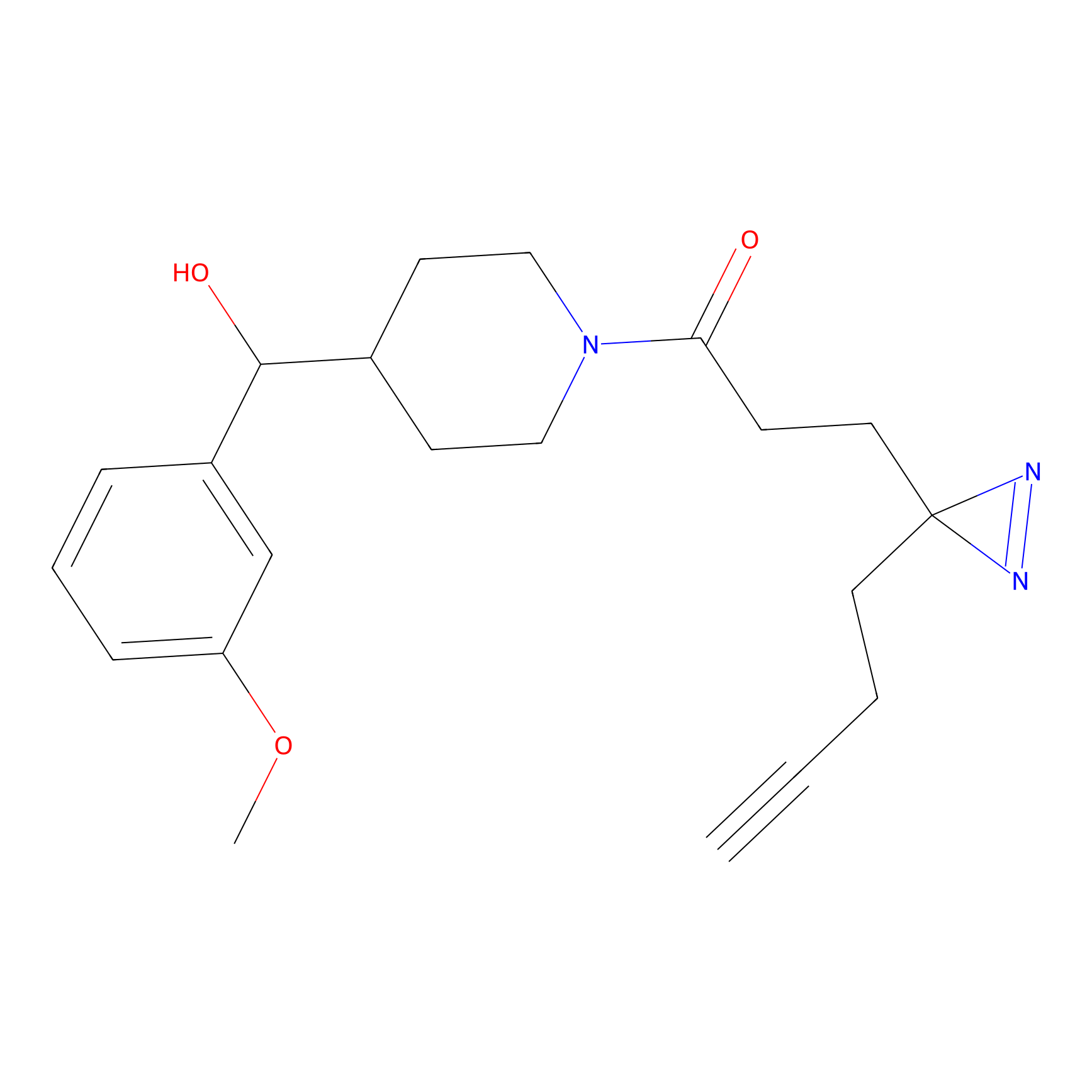
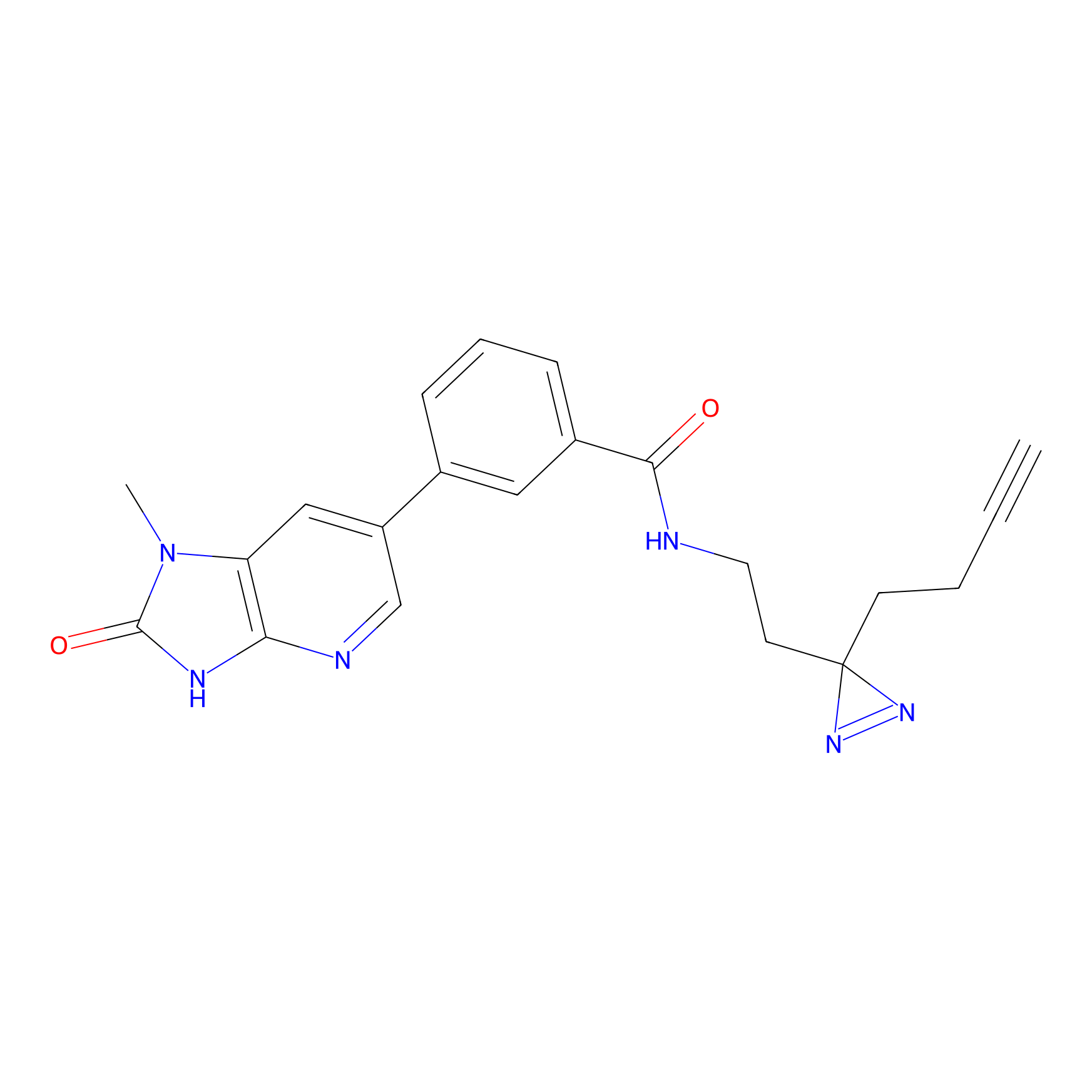
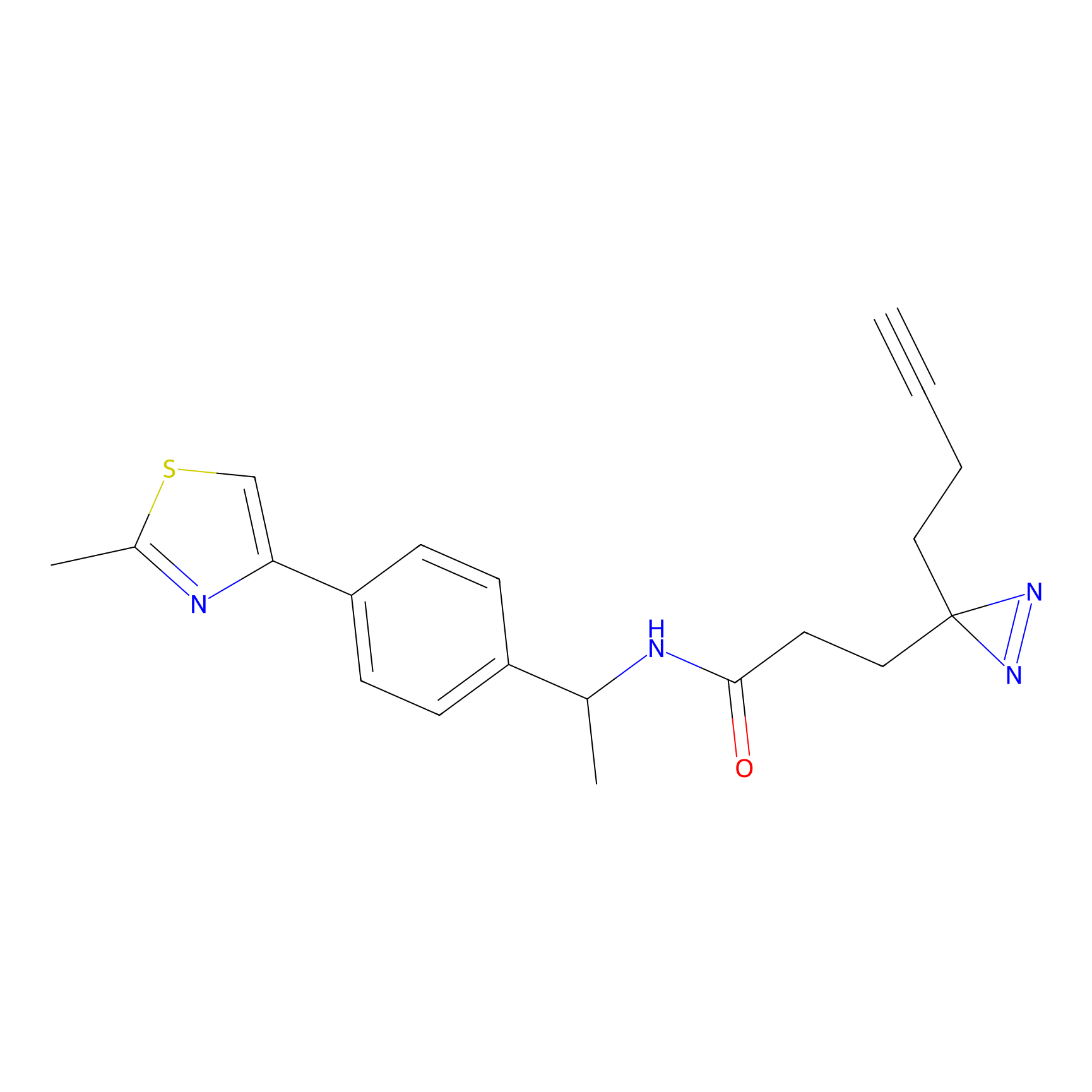
|
m-APA Probe Info |
 |
15.00 | LDD0403 | [3] | |
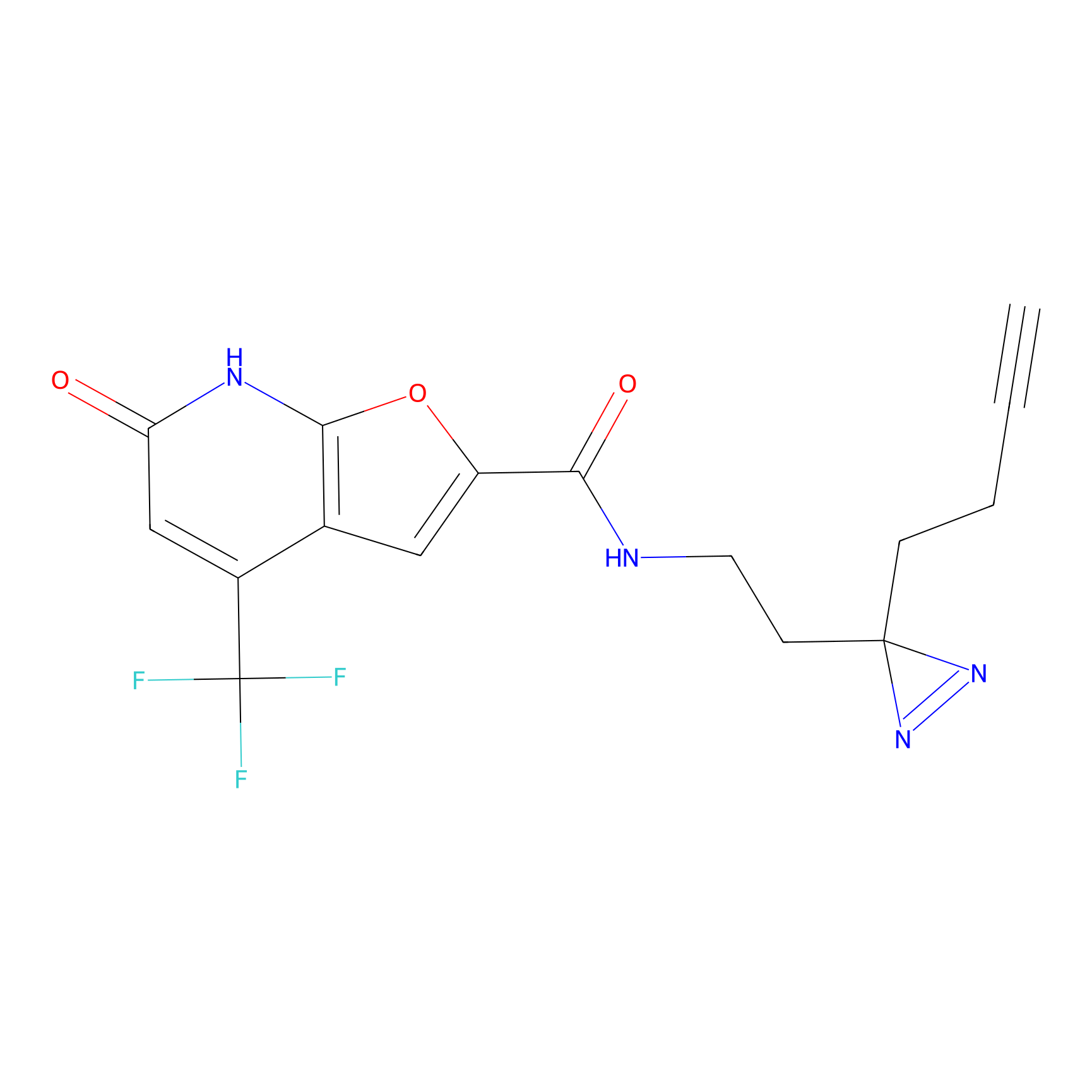
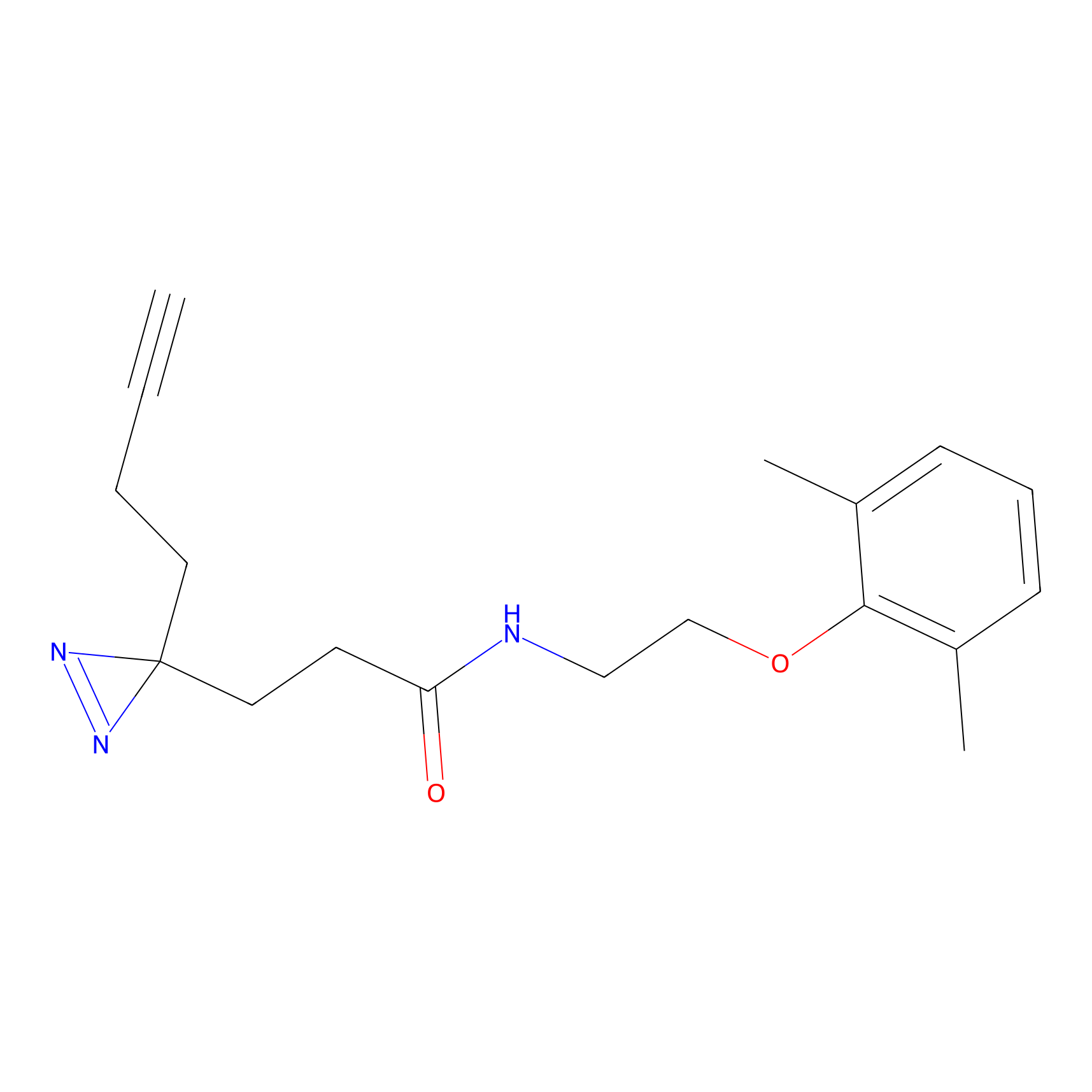
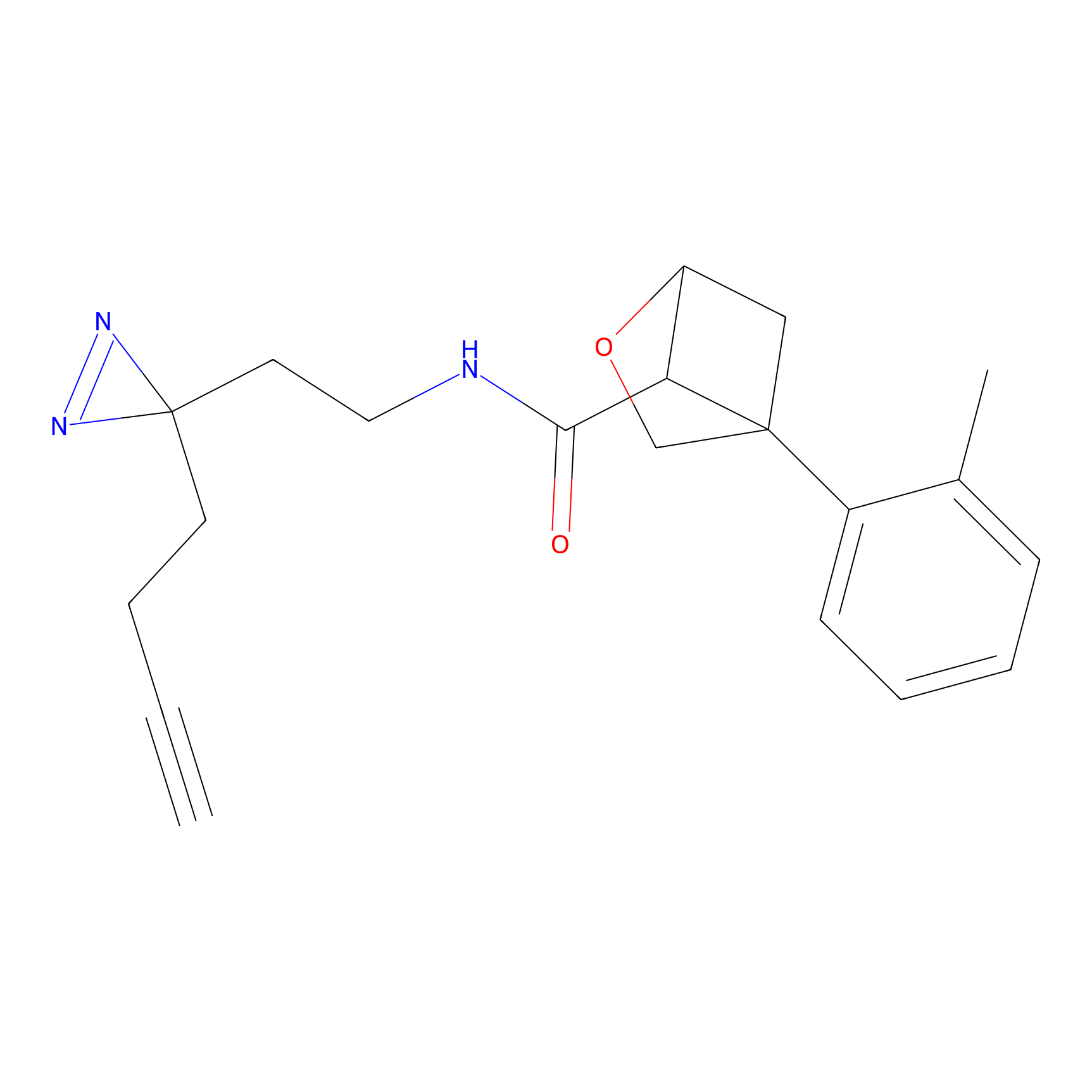
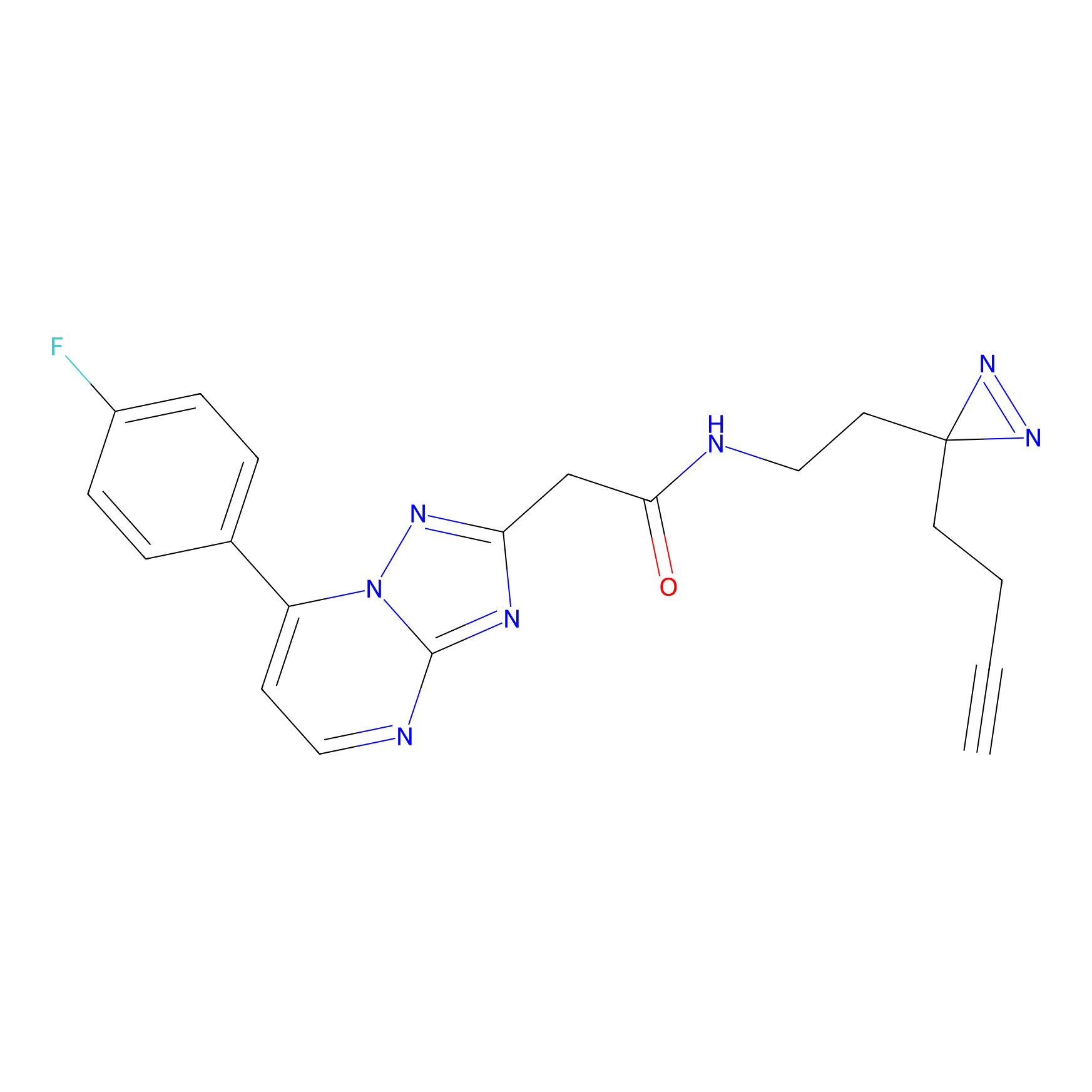
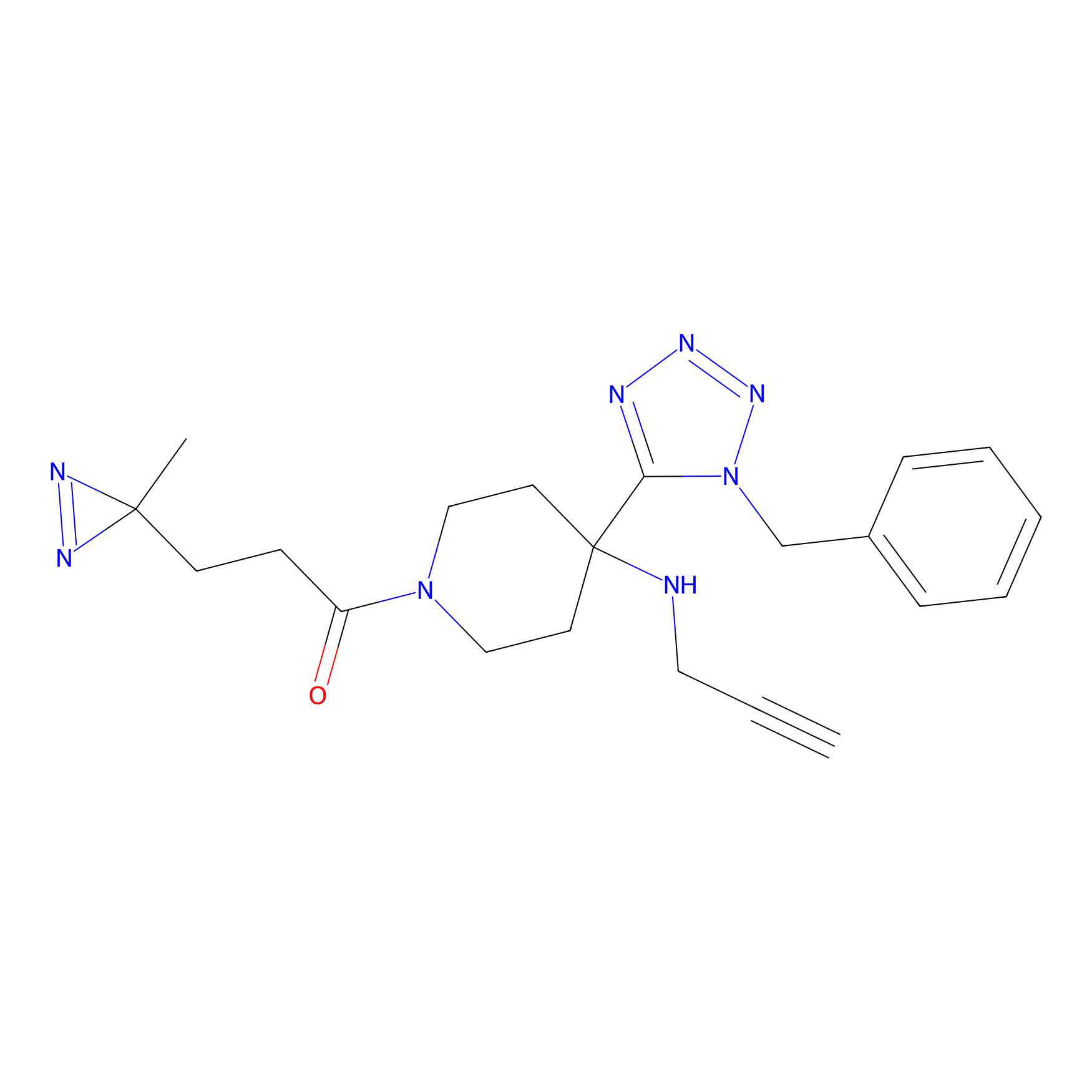
|
AHL-Pu-1 Probe Info |
 |
C299(3.31) | LDD0170 | [4] | |
|
HHS-482 Probe Info |
 |
Y300(0.85) | LDD0286 | [5] | |
|
DBIA Probe Info |
 |
C299(1.09) | LDD0532 | [6] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [7] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [8] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [7] | |
|
IPM Probe Info |
 |
N.A. | LDD2156 | [9] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | DM93 | C299(3.31) | LDD0170 | [4] |
| LDCM0026 | 4SU-RNA+native RNA | DM93 | C299(2.61) | LDD0171 | [4] |
| LDCM0215 | AC10 | HCT 116 | C299(1.09) | LDD0532 | [6] |
| LDCM0226 | AC11 | HCT 116 | C299(0.94) | LDD0543 | [6] |
| LDCM0237 | AC12 | HCT 116 | C299(0.89) | LDD0554 | [6] |
| LDCM0246 | AC128 | HCT 116 | C299(0.69) | LDD0563 | [6] |
| LDCM0247 | AC129 | HCT 116 | C299(0.64) | LDD0564 | [6] |
| LDCM0249 | AC130 | HCT 116 | C299(0.92) | LDD0566 | [6] |
| LDCM0250 | AC131 | HCT 116 | C299(1.15) | LDD0567 | [6] |
| LDCM0251 | AC132 | HCT 116 | C299(0.83) | LDD0568 | [6] |
| LDCM0252 | AC133 | HCT 116 | C299(0.71) | LDD0569 | [6] |
| LDCM0253 | AC134 | HCT 116 | C299(0.77) | LDD0570 | [6] |
| LDCM0254 | AC135 | HCT 116 | C299(0.67) | LDD0571 | [6] |
| LDCM0255 | AC136 | HCT 116 | C299(0.81) | LDD0572 | [6] |
| LDCM0256 | AC137 | HCT 116 | C299(0.91) | LDD0573 | [6] |
| LDCM0257 | AC138 | HCT 116 | C299(0.59) | LDD0574 | [6] |
| LDCM0258 | AC139 | HCT 116 | C299(0.69) | LDD0575 | [6] |
| LDCM0259 | AC14 | HCT 116 | C299(0.87) | LDD0576 | [6] |
| LDCM0260 | AC140 | HCT 116 | C299(0.79) | LDD0577 | [6] |
| LDCM0261 | AC141 | HCT 116 | C299(0.73) | LDD0578 | [6] |
| LDCM0262 | AC142 | HCT 116 | C299(1.00) | LDD0579 | [6] |
| LDCM0270 | AC15 | HCT 116 | C299(1.00) | LDD0587 | [6] |
| LDCM0276 | AC17 | HCT 116 | C299(1.35) | LDD0593 | [6] |
| LDCM0277 | AC18 | HCT 116 | C299(1.55) | LDD0594 | [6] |
| LDCM0278 | AC19 | HCT 116 | C299(1.30) | LDD0595 | [6] |
| LDCM0280 | AC20 | HCT 116 | C299(1.49) | LDD0597 | [6] |
| LDCM0281 | AC21 | HCT 116 | C299(1.27) | LDD0598 | [6] |
| LDCM0282 | AC22 | HCT 116 | C299(1.18) | LDD0599 | [6] |
| LDCM0283 | AC23 | HCT 116 | C299(1.00) | LDD0600 | [6] |
| LDCM0284 | AC24 | HCT 116 | C299(1.40) | LDD0601 | [6] |
| LDCM0285 | AC25 | HCT 116 | C299(0.89) | LDD0602 | [6] |
| LDCM0286 | AC26 | HCT 116 | C299(0.84) | LDD0603 | [6] |
| LDCM0287 | AC27 | HCT 116 | C299(1.08) | LDD0604 | [6] |
| LDCM0288 | AC28 | HCT 116 | C299(0.94) | LDD0605 | [6] |
| LDCM0289 | AC29 | HCT 116 | C299(0.77) | LDD0606 | [6] |
| LDCM0291 | AC30 | HCT 116 | C299(1.12) | LDD0608 | [6] |
| LDCM0292 | AC31 | HCT 116 | C299(1.26) | LDD0609 | [6] |
| LDCM0293 | AC32 | HCT 116 | C299(1.06) | LDD0610 | [6] |
| LDCM0294 | AC33 | HCT 116 | C299(1.11) | LDD0611 | [6] |
| LDCM0295 | AC34 | HCT 116 | C299(1.39) | LDD0612 | [6] |
| LDCM0296 | AC35 | HCT 116 | C299(1.14) | LDD0613 | [6] |
| LDCM0297 | AC36 | HCT 116 | C299(1.29) | LDD0614 | [6] |
| LDCM0298 | AC37 | HCT 116 | C299(1.47) | LDD0615 | [6] |
| LDCM0299 | AC38 | HCT 116 | C299(1.08) | LDD0616 | [6] |
| LDCM0300 | AC39 | HCT 116 | C299(1.28) | LDD0617 | [6] |
| LDCM0302 | AC40 | HCT 116 | C299(1.05) | LDD0619 | [6] |
| LDCM0303 | AC41 | HCT 116 | C299(1.03) | LDD0620 | [6] |
| LDCM0304 | AC42 | HCT 116 | C299(1.24) | LDD0621 | [6] |
| LDCM0305 | AC43 | HCT 116 | C299(0.96) | LDD0622 | [6] |
| LDCM0306 | AC44 | HCT 116 | C299(1.43) | LDD0623 | [6] |
| LDCM0307 | AC45 | HCT 116 | C299(1.38) | LDD0624 | [6] |
| LDCM0323 | AC6 | HCT 116 | C299(1.07) | LDD0640 | [6] |
| LDCM0334 | AC7 | HCT 116 | C299(1.04) | LDD0651 | [6] |
| LDCM0345 | AC8 | HCT 116 | C299(1.10) | LDD0662 | [6] |
| LDCM0248 | AKOS034007472 | HCT 116 | C299(0.98) | LDD0565 | [6] |
| LDCM0356 | AKOS034007680 | HCT 116 | C299(0.99) | LDD0673 | [6] |
| LDCM0275 | AKOS034007705 | HCT 116 | C299(0.83) | LDD0592 | [6] |
| LDCM0156 | Aniline | NCI-H1299 | 15.00 | LDD0403 | [3] |
| LDCM0370 | CL101 | HCT 116 | C299(1.10) | LDD0687 | [6] |
| LDCM0371 | CL102 | HCT 116 | C299(0.79) | LDD0688 | [6] |
| LDCM0372 | CL103 | HCT 116 | C299(0.98) | LDD0689 | [6] |
| LDCM0373 | CL104 | HCT 116 | C299(1.31) | LDD0690 | [6] |
| LDCM0374 | CL105 | HCT 116 | C299(1.10) | LDD0691 | [6] |
| LDCM0375 | CL106 | HCT 116 | C299(1.31) | LDD0692 | [6] |
| LDCM0376 | CL107 | HCT 116 | C299(1.36) | LDD0693 | [6] |
| LDCM0377 | CL108 | HCT 116 | C299(1.24) | LDD0694 | [6] |
| LDCM0378 | CL109 | HCT 116 | C299(1.26) | LDD0695 | [6] |
| LDCM0380 | CL110 | HCT 116 | C299(1.05) | LDD0697 | [6] |
| LDCM0381 | CL111 | HCT 116 | C299(1.17) | LDD0698 | [6] |
| LDCM0382 | CL112 | HCT 116 | C299(0.85) | LDD0699 | [6] |
| LDCM0383 | CL113 | HCT 116 | C299(0.96) | LDD0700 | [6] |
| LDCM0384 | CL114 | HCT 116 | C299(0.80) | LDD0701 | [6] |
| LDCM0385 | CL115 | HCT 116 | C299(0.74) | LDD0702 | [6] |
| LDCM0386 | CL116 | HCT 116 | C299(1.17) | LDD0703 | [6] |
| LDCM0387 | CL117 | HCT 116 | C299(1.29) | LDD0704 | [6] |
| LDCM0388 | CL118 | HCT 116 | C299(0.89) | LDD0705 | [6] |
| LDCM0389 | CL119 | HCT 116 | C299(1.17) | LDD0706 | [6] |
| LDCM0391 | CL120 | HCT 116 | C299(1.05) | LDD0708 | [6] |
| LDCM0469 | CL76 | HCT 116 | C299(0.82) | LDD0786 | [6] |
| LDCM0470 | CL77 | HCT 116 | C299(0.80) | LDD0787 | [6] |
| LDCM0471 | CL78 | HCT 116 | C299(0.50) | LDD0788 | [6] |
| LDCM0472 | CL79 | HCT 116 | C299(0.78) | LDD0789 | [6] |
| LDCM0474 | CL80 | HCT 116 | C299(0.50) | LDD0791 | [6] |
| LDCM0475 | CL81 | HCT 116 | C299(0.54) | LDD0792 | [6] |
| LDCM0476 | CL82 | HCT 116 | C299(1.11) | LDD0793 | [6] |
| LDCM0477 | CL83 | HCT 116 | C299(0.55) | LDD0794 | [6] |
| LDCM0478 | CL84 | HCT 116 | C299(0.53) | LDD0795 | [6] |
| LDCM0479 | CL85 | HCT 116 | C299(0.87) | LDD0796 | [6] |
| LDCM0480 | CL86 | HCT 116 | C299(0.64) | LDD0797 | [6] |
| LDCM0481 | CL87 | HCT 116 | C299(0.54) | LDD0798 | [6] |
| LDCM0482 | CL88 | HCT 116 | C299(0.54) | LDD0799 | [6] |
| LDCM0483 | CL89 | HCT 116 | C299(0.53) | LDD0800 | [6] |
| LDCM0485 | CL90 | HCT 116 | C299(0.62) | LDD0802 | [6] |
| LDCM0124 | JWB142 | DM93 | Y300(0.85) | LDD0286 | [5] |
| LDCM0125 | JWB146 | DM93 | Y300(1.31) | LDD0287 | [5] |
| LDCM0126 | JWB150 | DM93 | Y300(3.43) | LDD0288 | [5] |
| LDCM0127 | JWB152 | DM93 | Y300(1.79) | LDD0289 | [5] |
| LDCM0128 | JWB198 | DM93 | Y300(1.12) | LDD0290 | [5] |
| LDCM0130 | JWB211 | DM93 | Y300(1.22) | LDD0292 | [5] |
| LDCM0076 | kambe_cp71 | PC-3 | 3.45 | LDD0132 | [11] |
| LDCM0022 | KB02 | 42-MG-BA | C299(1.73) | LDD2244 | [12] |
| LDCM0023 | KB03 | 42-MG-BA | C299(2.14) | LDD2661 | [12] |
| LDCM0024 | KB05 | COLO792 | C299(2.03) | LDD3310 | [12] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Homeobox protein Hox-C8 (HOXC8) | Antp homeobox family | P31273 | |||
| T-cell leukemia homeobox protein 3 (TLX3) | . | O43711 | |||
Other
The Drug(s) Related To This Target
Approved
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Lonafarnib | Small molecular drug | DB06448 | |||
Investigative
Discontinued
References