Details of the Target
General Information of Target
| Target ID | LDTP16078 | |||||
|---|---|---|---|---|---|---|
| Target Name | Heme-binding protein 1 (HEBP1) | |||||
| Gene Name | HEBP1 | |||||
| Gene ID | 50865 | |||||
| Synonyms |
HBP; Heme-binding protein 1; p22HBP |
|||||
| 3D Structure | ||||||
| Sequence |
MAAAAAAAAAVGVRLRDCCSRGAVLLLFFSLSPRPPAAAAWLLGLRPEDTAGGRVSLEGG
TLRAAEGTSFLLRVYFQPGPPATAAPVPSPTLNSGENGTGDWAPRLVFIEEPPGGGGVAP SAVPTRPPGPQRCREQSDWASDVEVLGPLRPGGVAGSALVQVRVRELRKGEAERGGAGGG GKLFSLCAWDGRAWHHHGAAGGFLLRVRPRLYGPGGDLLPPAWLRALGALLLLALSALFS GLRLSLLSLDPVELRVLRNSGSAAEQEQARRVQAVRGRGTHLLCTLLLGQAGANAALAGW LYTSLPPGFGGTGEDYSEEGIHFPWLPALVCTGAVFLGAEICPYSVCSRHGLAIASHSVC LTRLLMAAAFPVCYPLGRLLDWALRQEISTFYTREKLLETLRAADPYSDLVKEELNIIQG ALELRTKVVEEVLTPLGDCFMLRSDAVLDFATVSEILRSGYTRIPVYEGDQRHNIVDILF VKDLAFVDPDDCTPLLTVTRFYNRPLHCVFNDTRLDTVLEEFKKGKSHLAIVQRVNNEGE GDPFYEVMGIVTLEDIIEEIIKSEILDETDLYTDNRKKQRVPQRERKRHDFSLFKLSDTE MRVKISPQLLLATHRFMATEVEPFKSLYLSEKILLRLLKHPNVIQELKFDEKNKKAPEHY LYQRNRPVDYFVLLLQGKVEVEVGKEGLRFENGAFTYYGVPAIMTTACSDNDVRKVGSLA GSSVFLNRSPSRCSGLNRSESPNRERSDFGGSNTQLYSSSNNLYMPDYSVHILSDVQFVK ITRQQYQNALTACHMDSSPQSPDMEAFTDGDSTKAPTTRGTPQTPKDDPAITLLNNRNSL PCSRSDGLRSPSEVVYLRMEELAFTQEEMTDFEEHSTQQLTLSPAAVPTRAASDSECCNI NLDTETSPCSSDFEENVGKKLLRTLSGQKRKRSPEGERTSEDNSNLTPLIT |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
HEBP family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
May bind free porphyrinogens that may be present in the cell and thus facilitate removal of these potentially toxic compound. Binds with a high affinity to one molecule of heme or porphyrins. It binds metalloporphyrins, free porphyrins and N-methylprotoporphyrin with similar affinities.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C-Sul Probe Info |
 |
6.90 | LDD0066 | [1] | |
|
TH211 Probe Info |
 |
Y65(10.72) | LDD0257 | [2] | |
|
TH216 Probe Info |
 |
Y138(11.87) | LDD0259 | [2] | |
|
STPyne Probe Info |
 |
K118(8.33); K64(2.54) | LDD0277 | [3] | |
|
ONAyne Probe Info |
 |
K121(10.00) | LDD0275 | [3] | |
|
Probe 1 Probe Info |
 |
Y31(18.38); Y144(12.42) | LDD3495 | [4] | |
|
DBIA Probe Info |
 |
C36(1.88) | LDD3314 | [5] | |
|
P13 Probe Info |
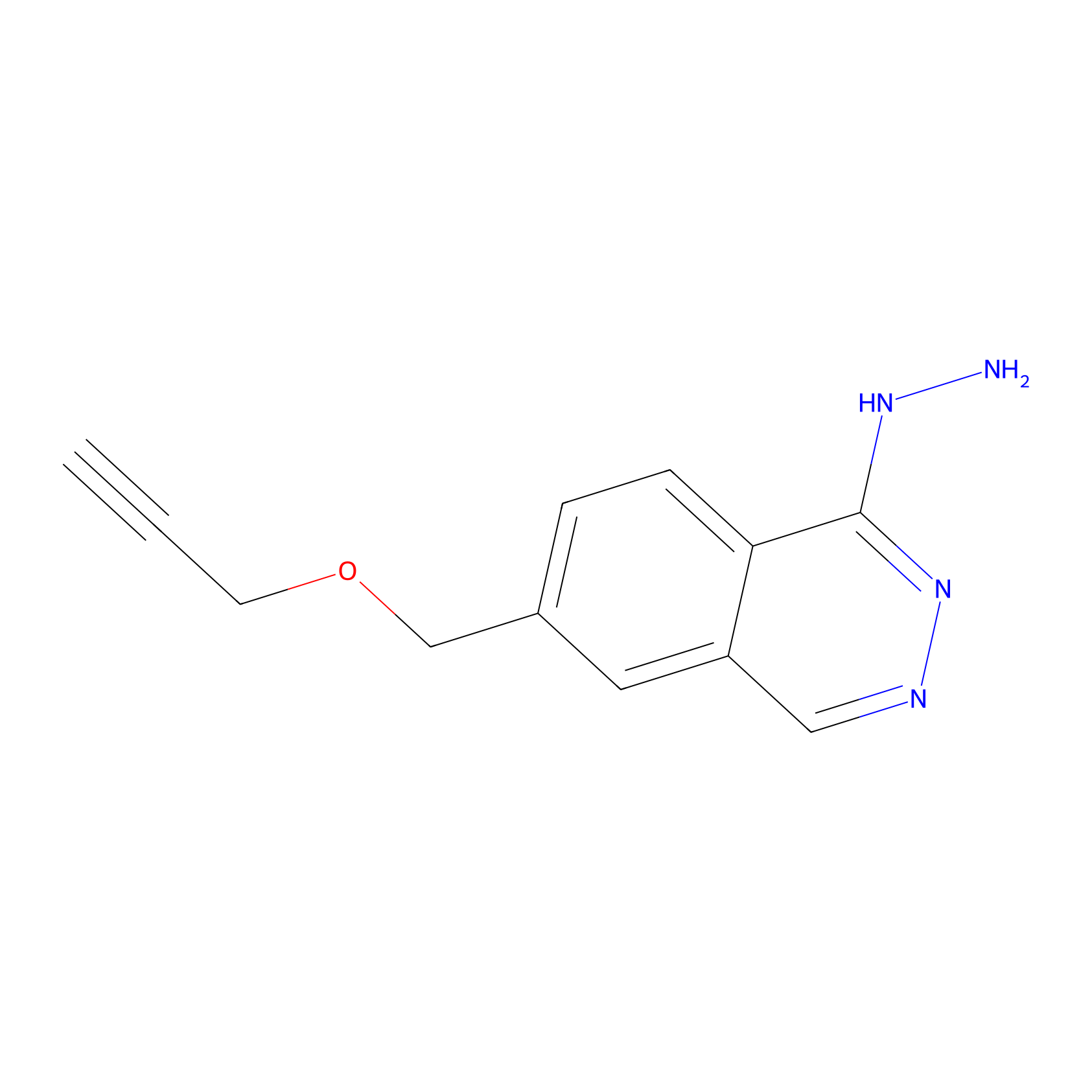 |
13.94 | LDD0203 | [6] | |
|
Jackson_1 Probe Info |
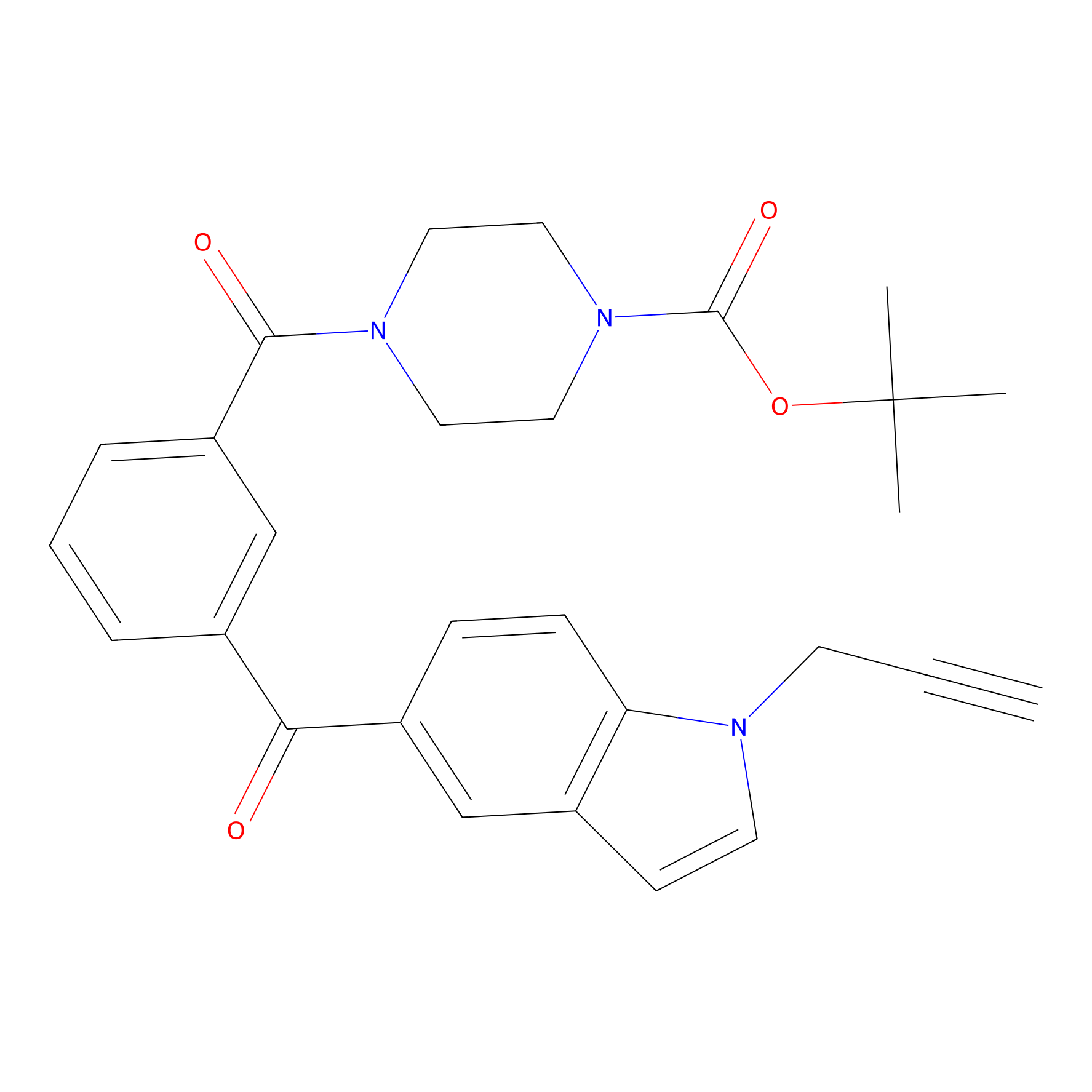 |
2.14 | LDD0120 | [7] | |
|
HPAP Probe Info |
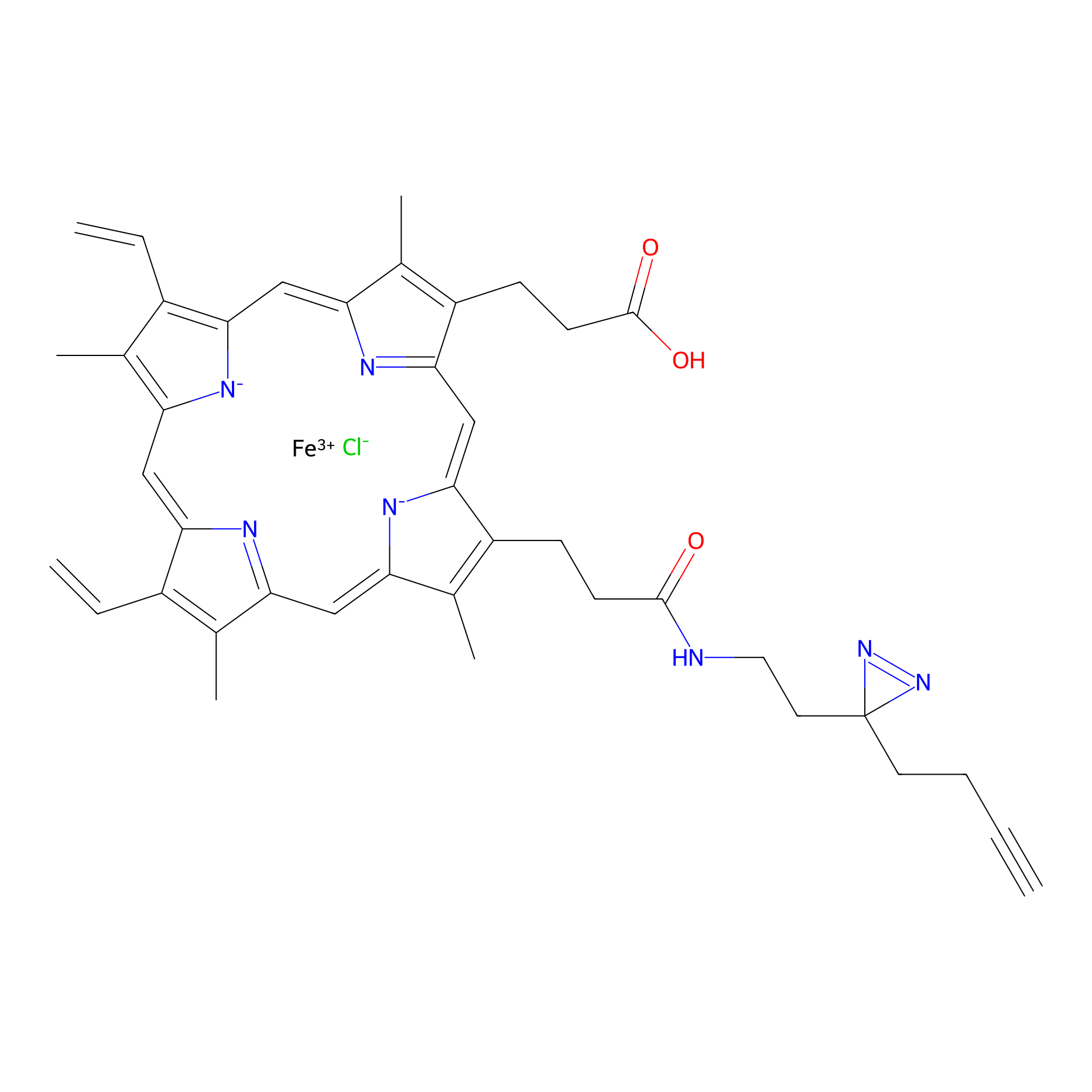 |
3.35 | LDD0062 | [8] | |
|
HHS-482 Probe Info |
 |
Y138(0.83); Y178(0.68) | LDD0285 | [9] | |
|
HHS-475 Probe Info |
 |
Y138(0.45) | LDD0264 | [10] | |
|
HHS-465 Probe Info |
 |
Y178(3.54) | LDD2237 | [11] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0358 | [12] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0162 | [13] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [14] | |
|
NAIA_5 Probe Info |
 |
C36(0.00); C168(0.00) | LDD2223 | [15] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C027 Probe Info |
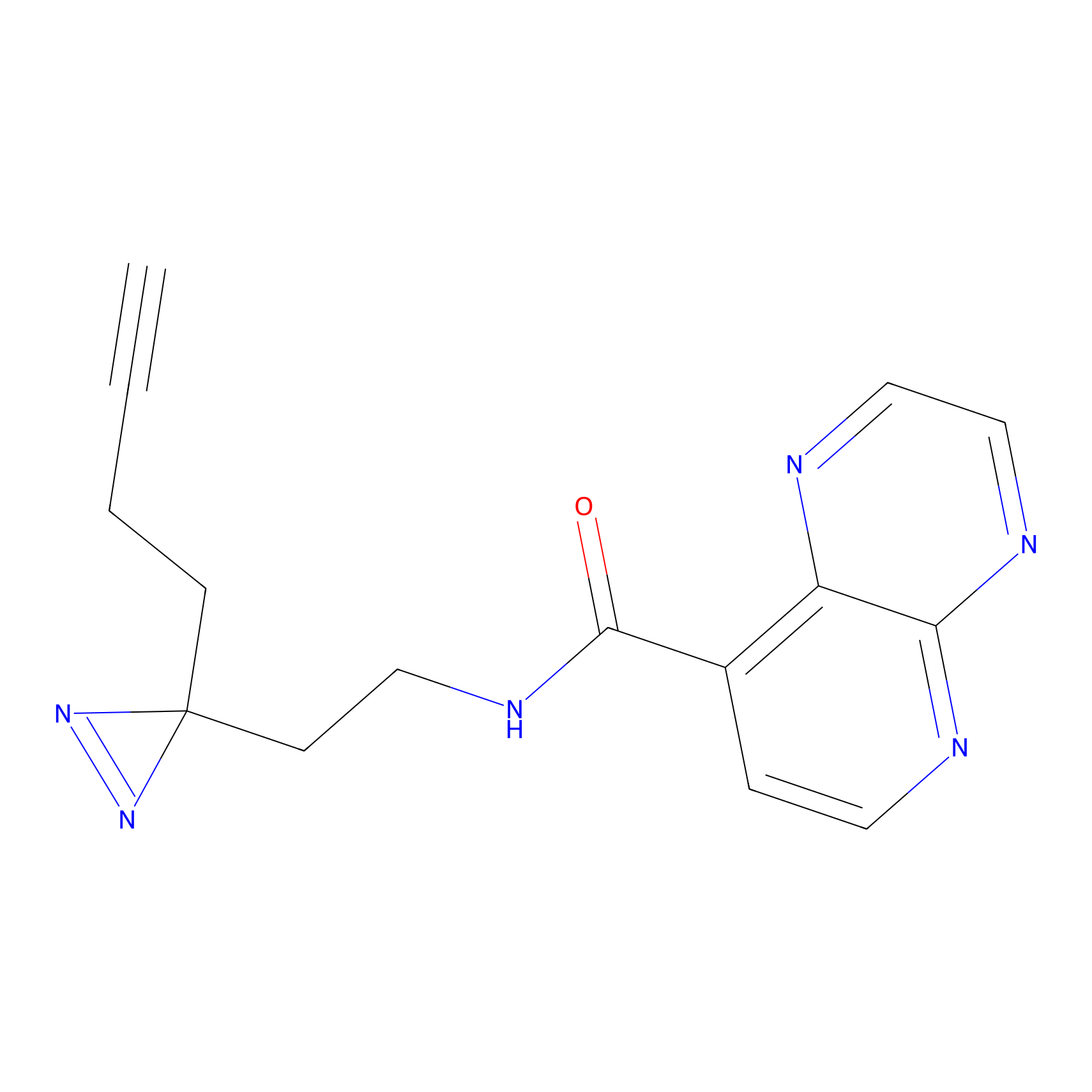 |
6.28 | LDD1733 | [16] | |
|
C056 Probe Info |
 |
18.38 | LDD1753 | [16] | |
|
C094 Probe Info |
 |
39.95 | LDD1785 | [16] | |
|
C145 Probe Info |
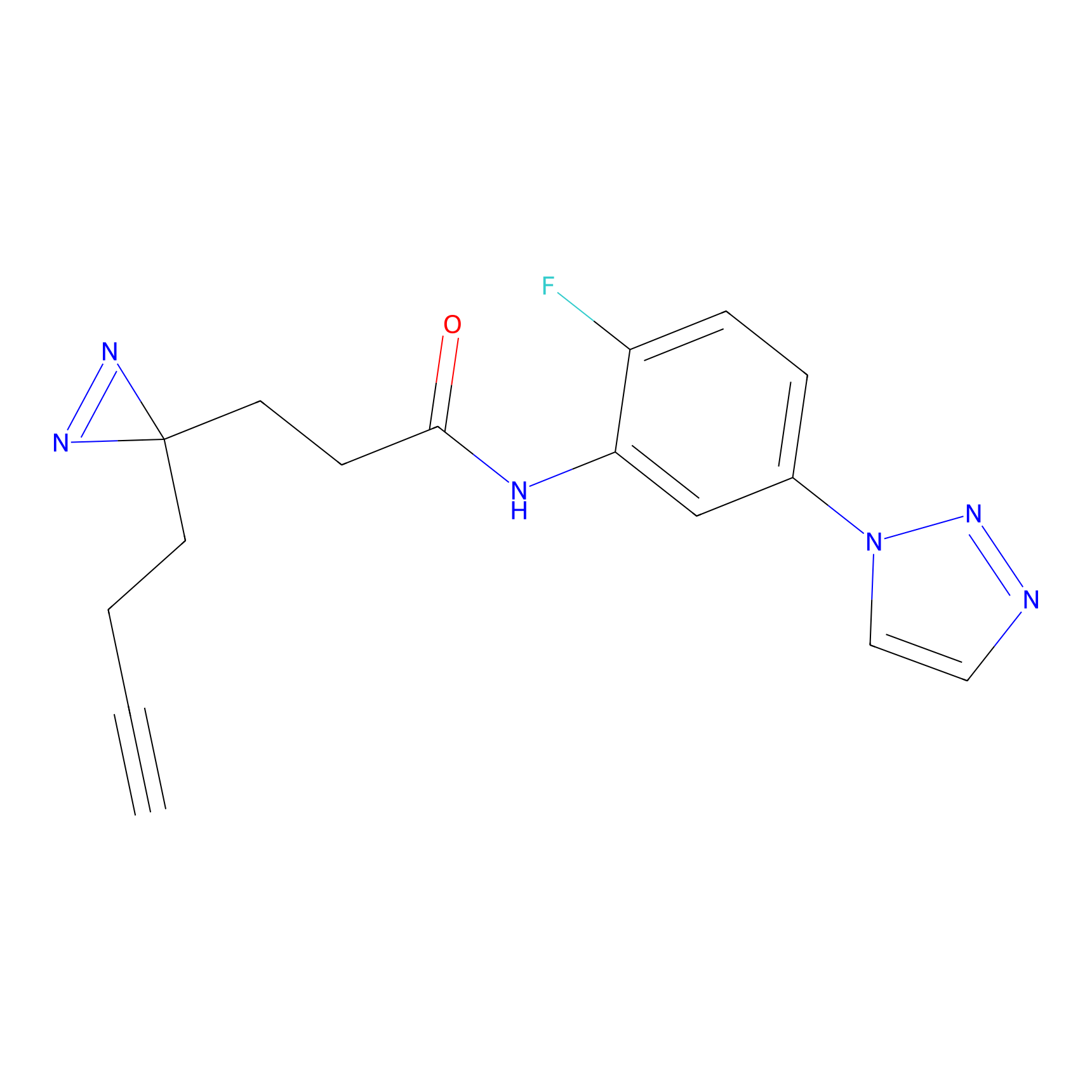 |
8.28 | LDD1827 | [16] | |
|
C158 Probe Info |
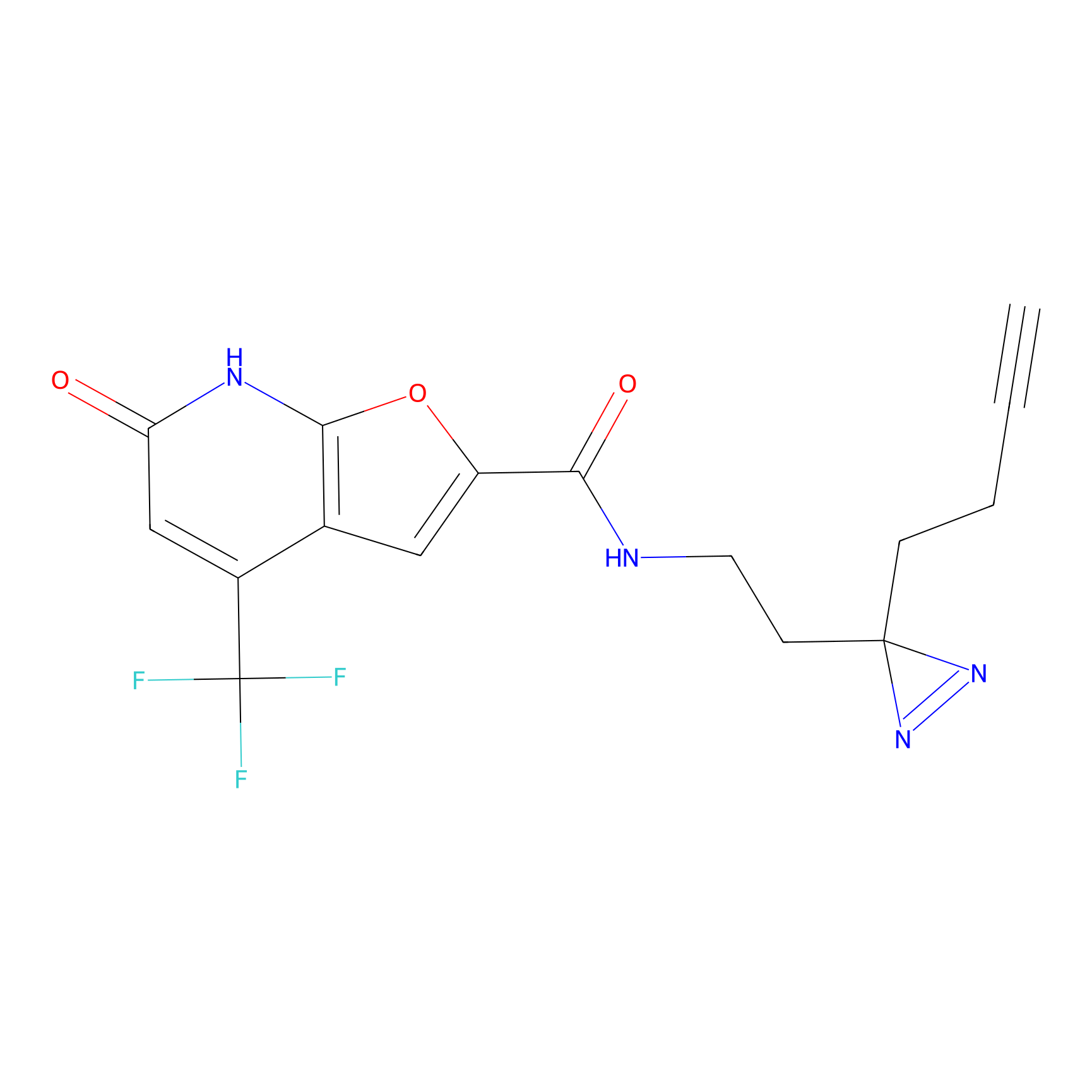 |
12.30 | LDD1838 | [16] | |
|
C206 Probe Info |
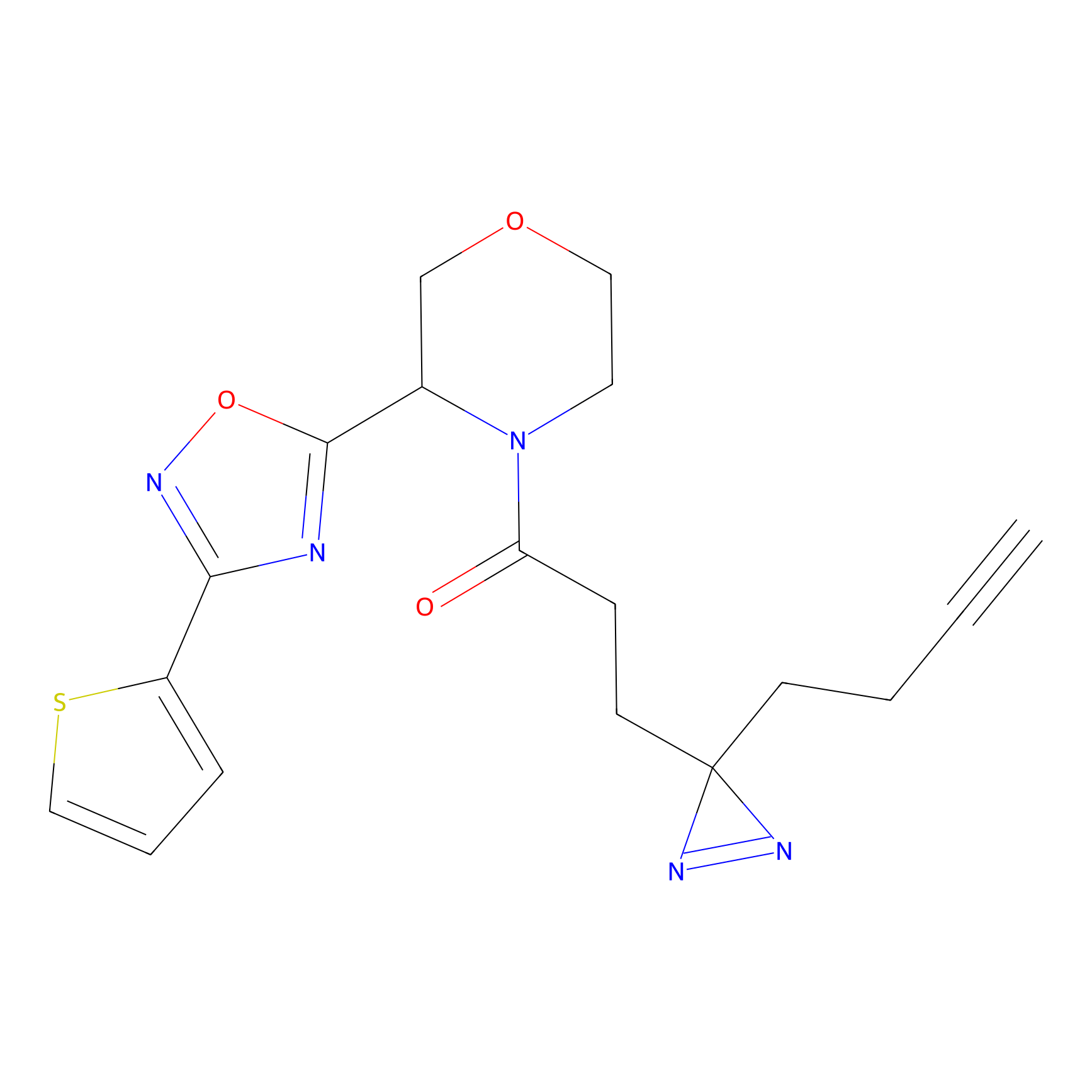 |
15.67 | LDD1881 | [16] | |
|
C234 Probe Info |
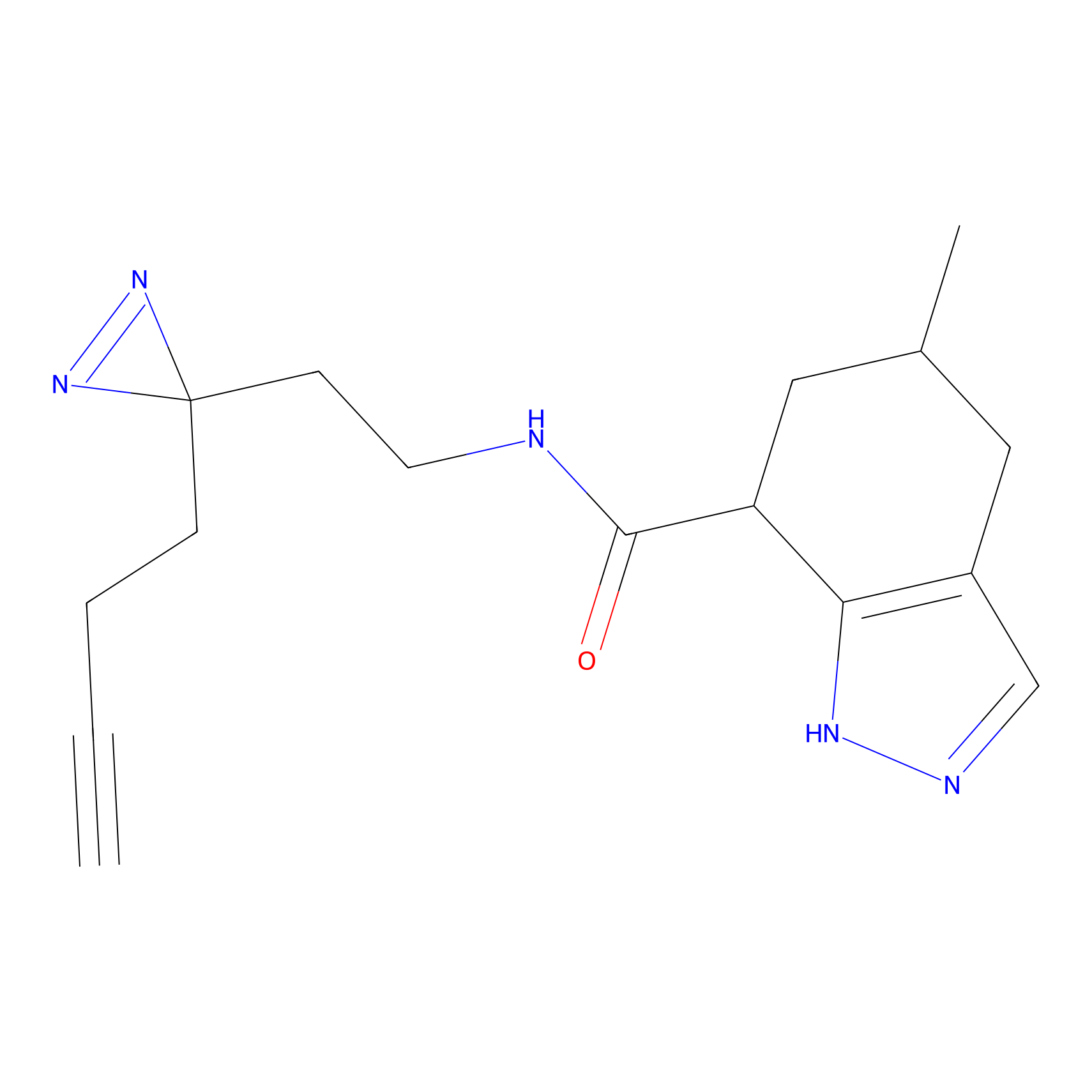 |
6.02 | LDD1907 | [16] | |
|
C235 Probe Info |
 |
99.73 | LDD1908 | [16] | |
|
C244 Probe Info |
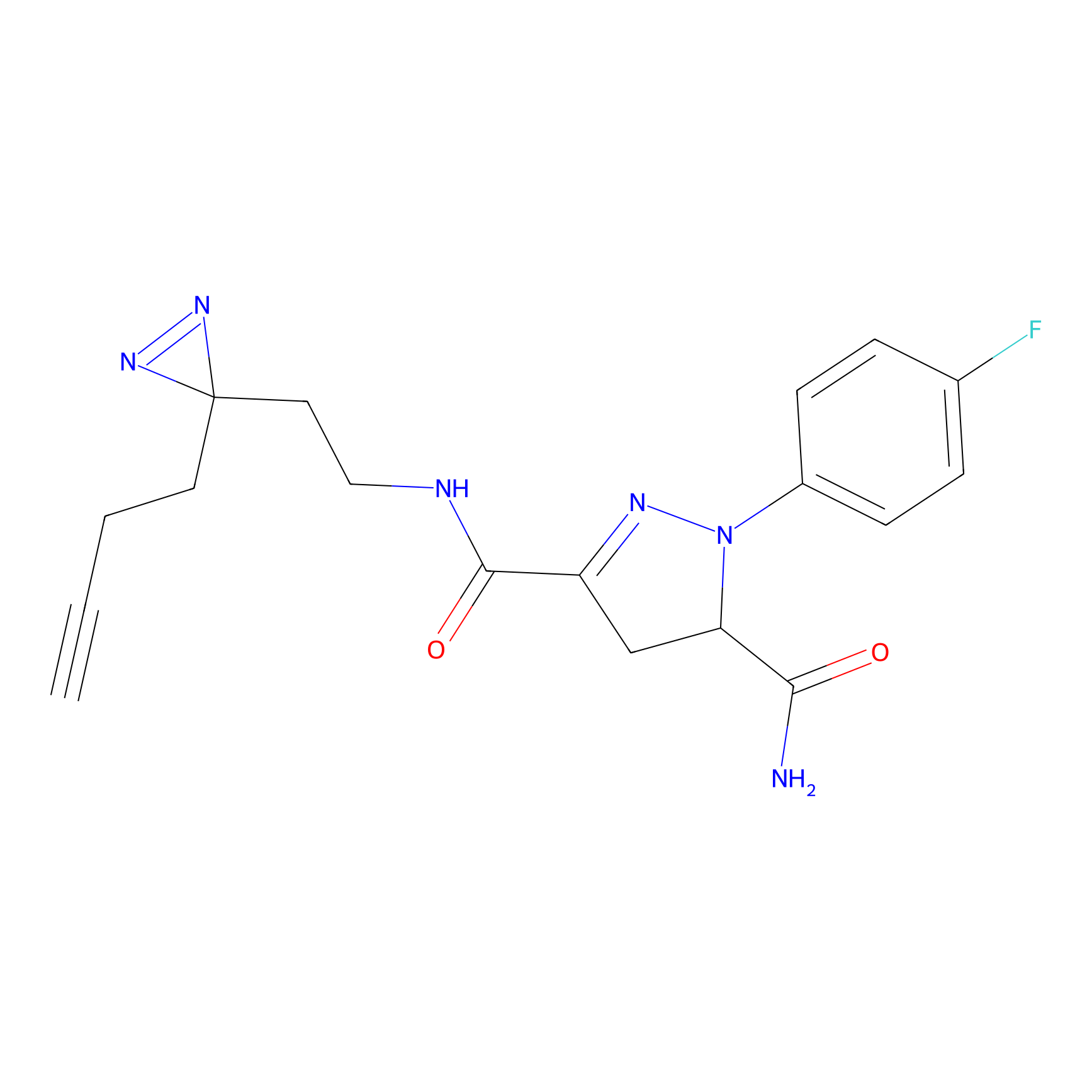 |
27.47 | LDD1917 | [16] | |
|
C249 Probe Info |
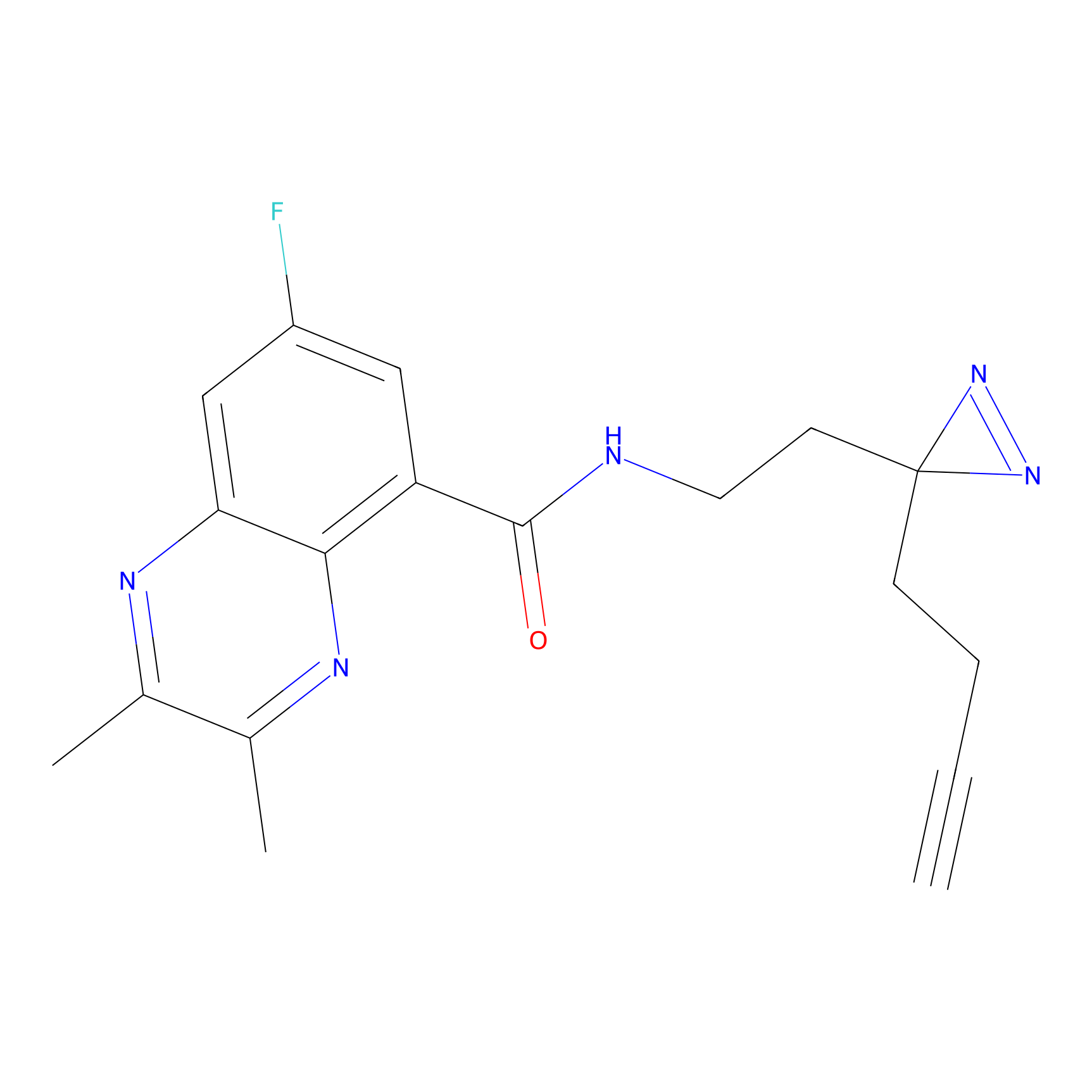 |
14.62 | LDD1922 | [16] | |
|
C251 Probe Info |
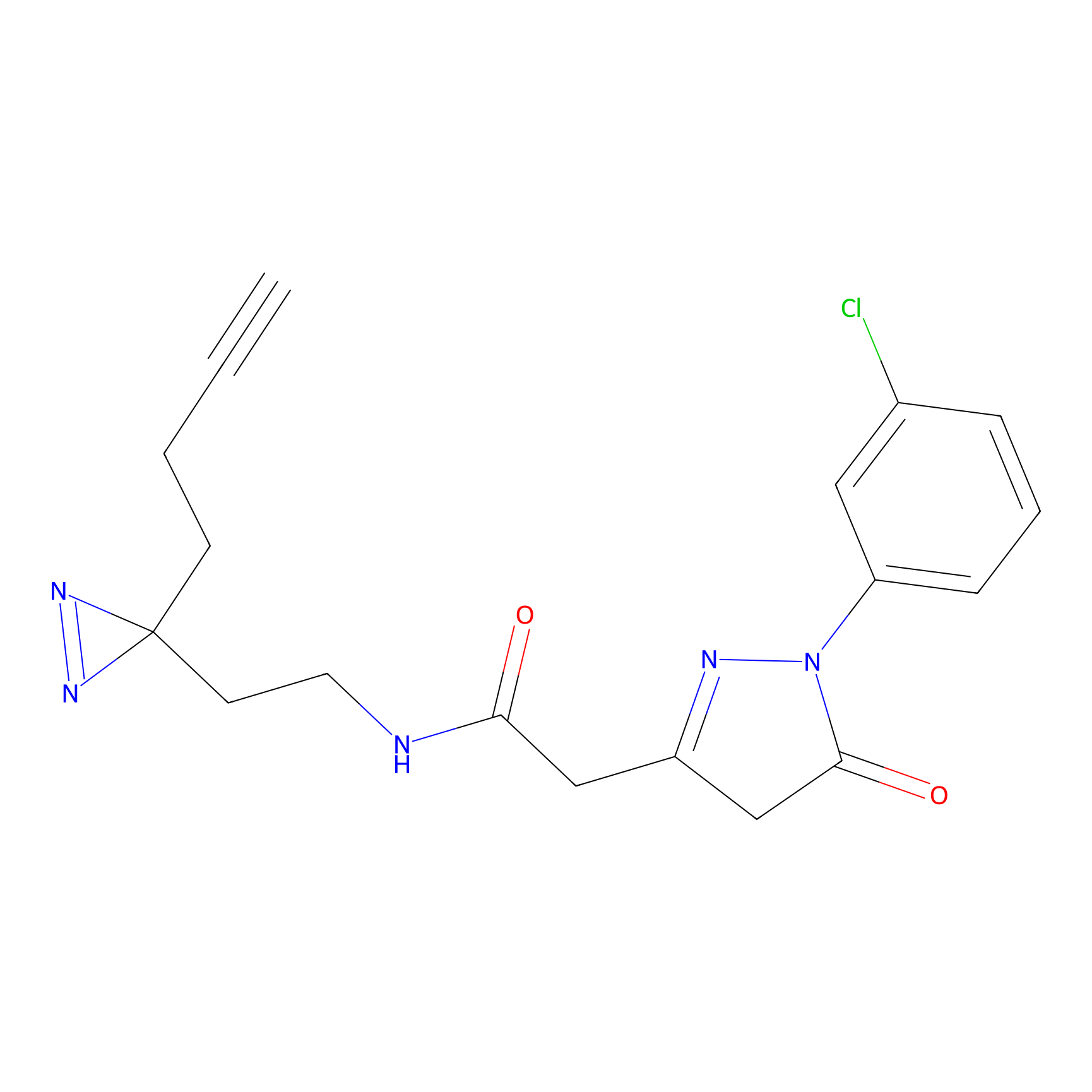 |
27.86 | LDD1924 | [16] | |
|
C252 Probe Info |
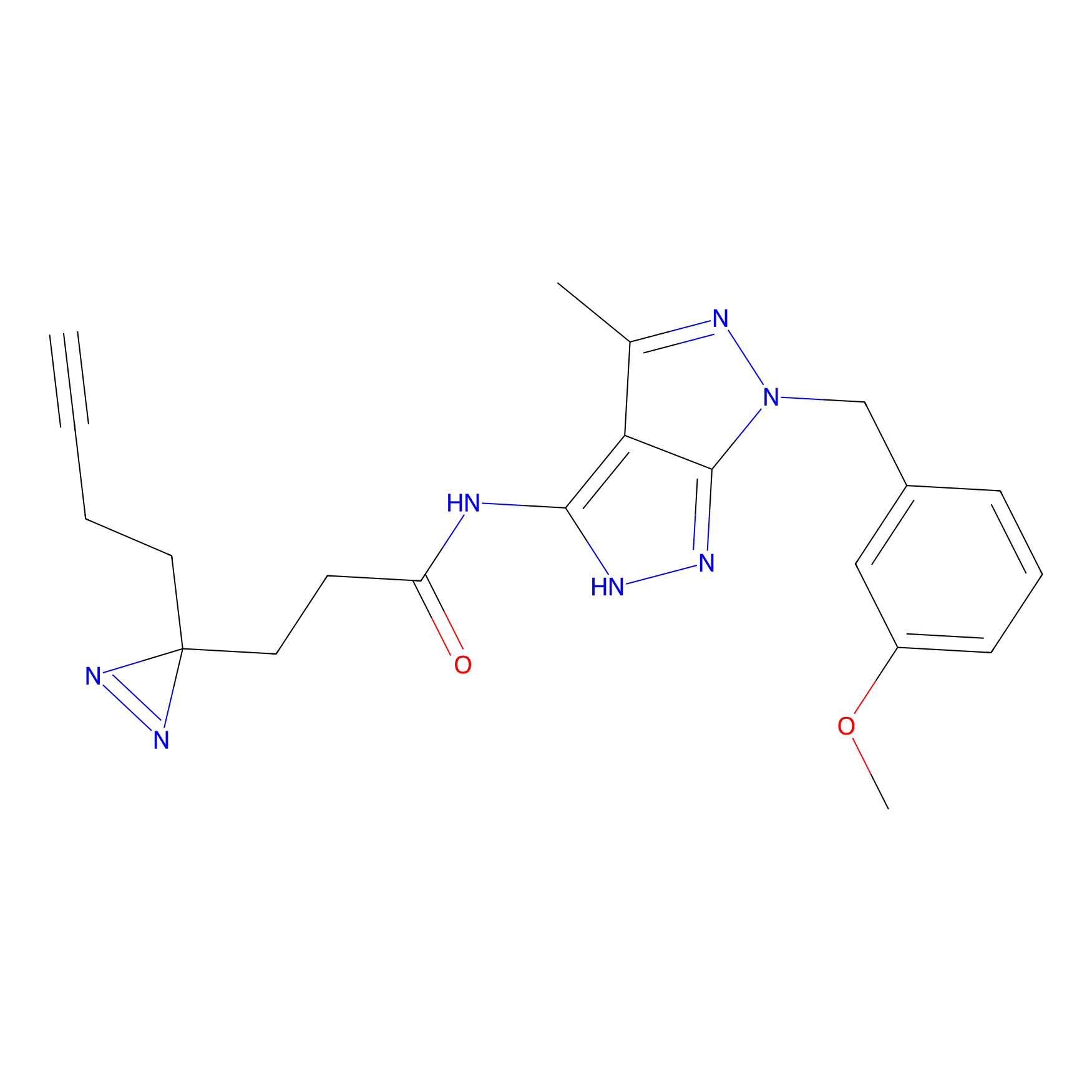 |
9.78 | LDD1925 | [16] | |
|
C266 Probe Info |
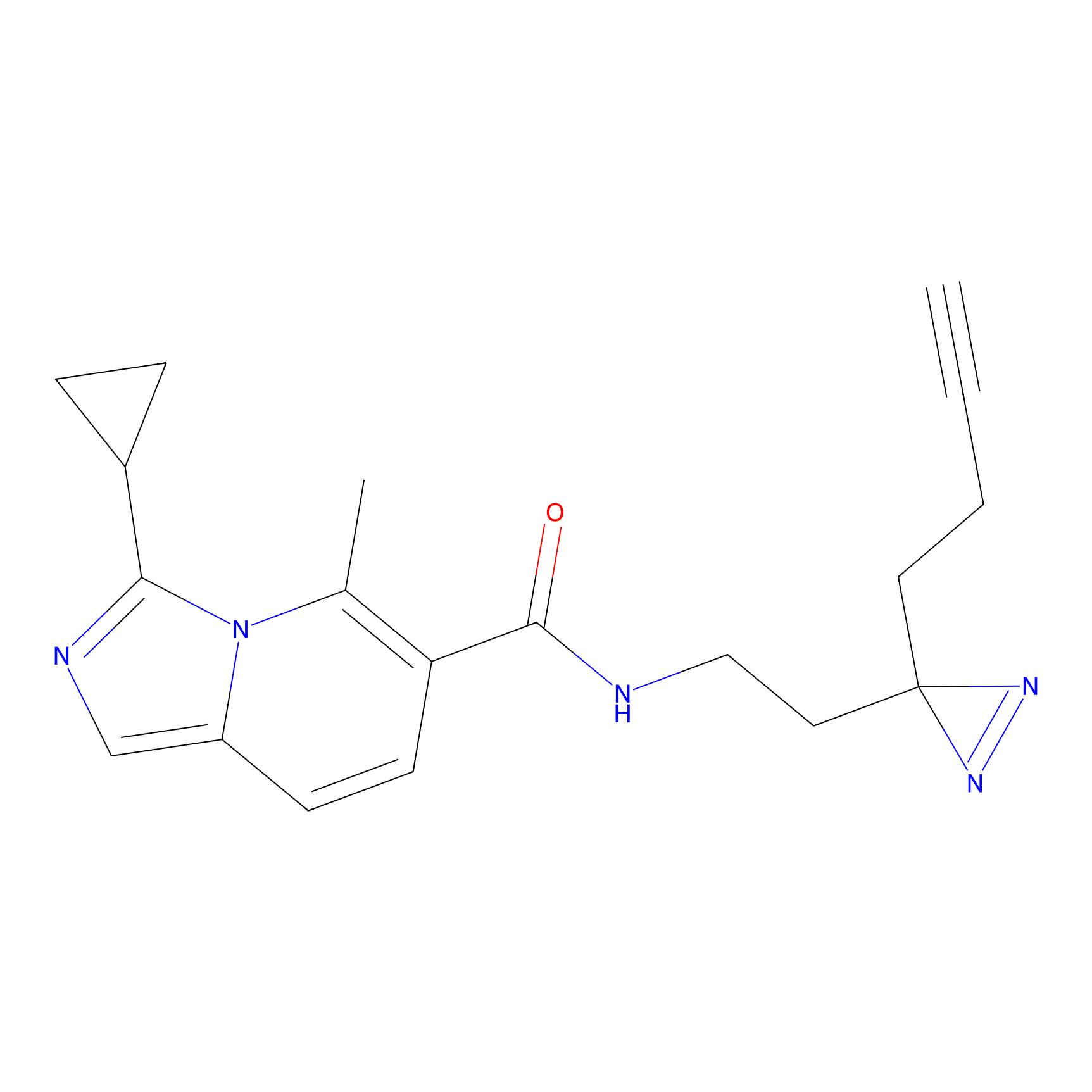 |
7.94 | LDD1937 | [16] | |
|
C285 Probe Info |
 |
22.01 | LDD1955 | [16] | |
|
C287 Probe Info |
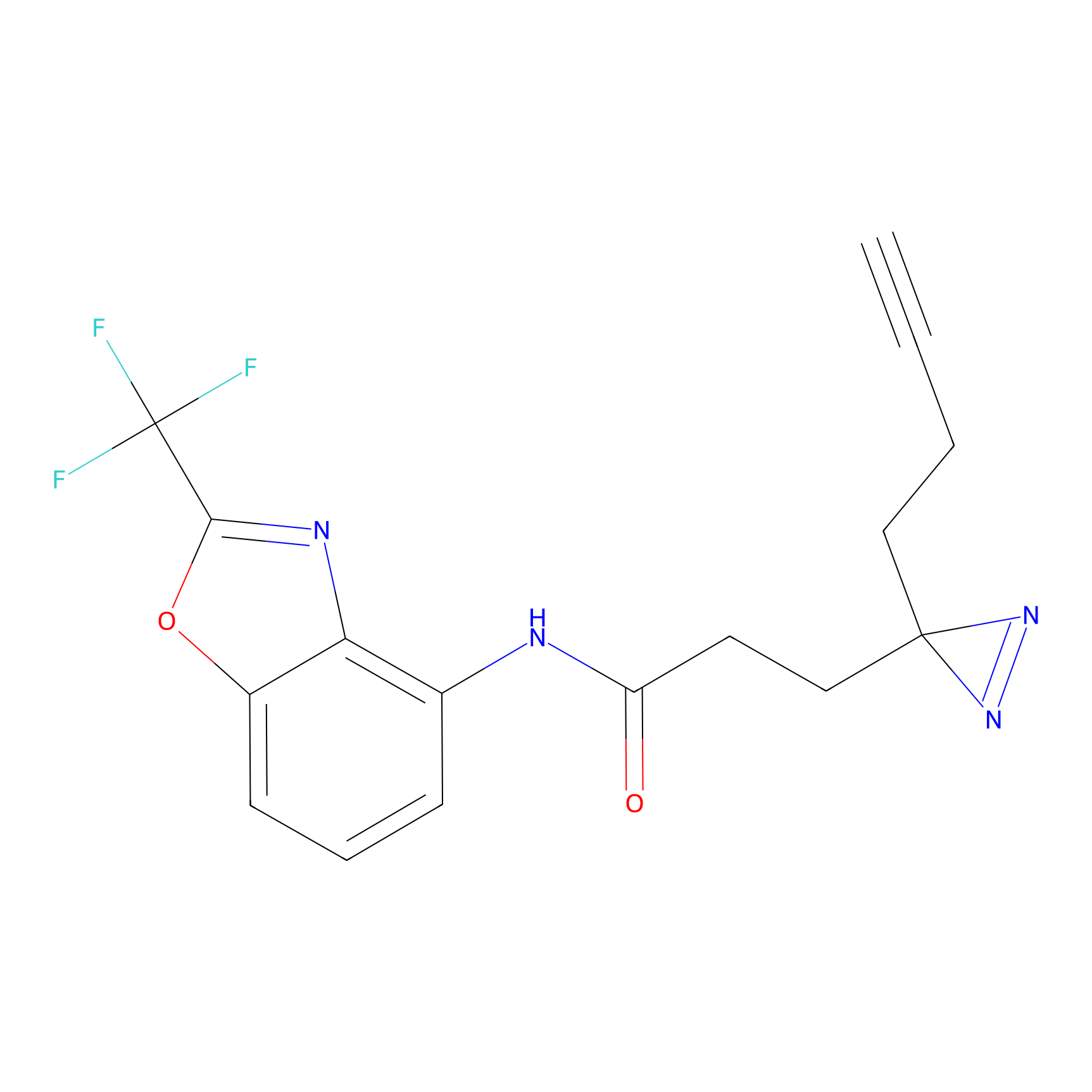 |
15.35 | LDD1957 | [16] | |
|
C289 Probe Info |
 |
55.33 | LDD1959 | [16] | |
|
C299 Probe Info |
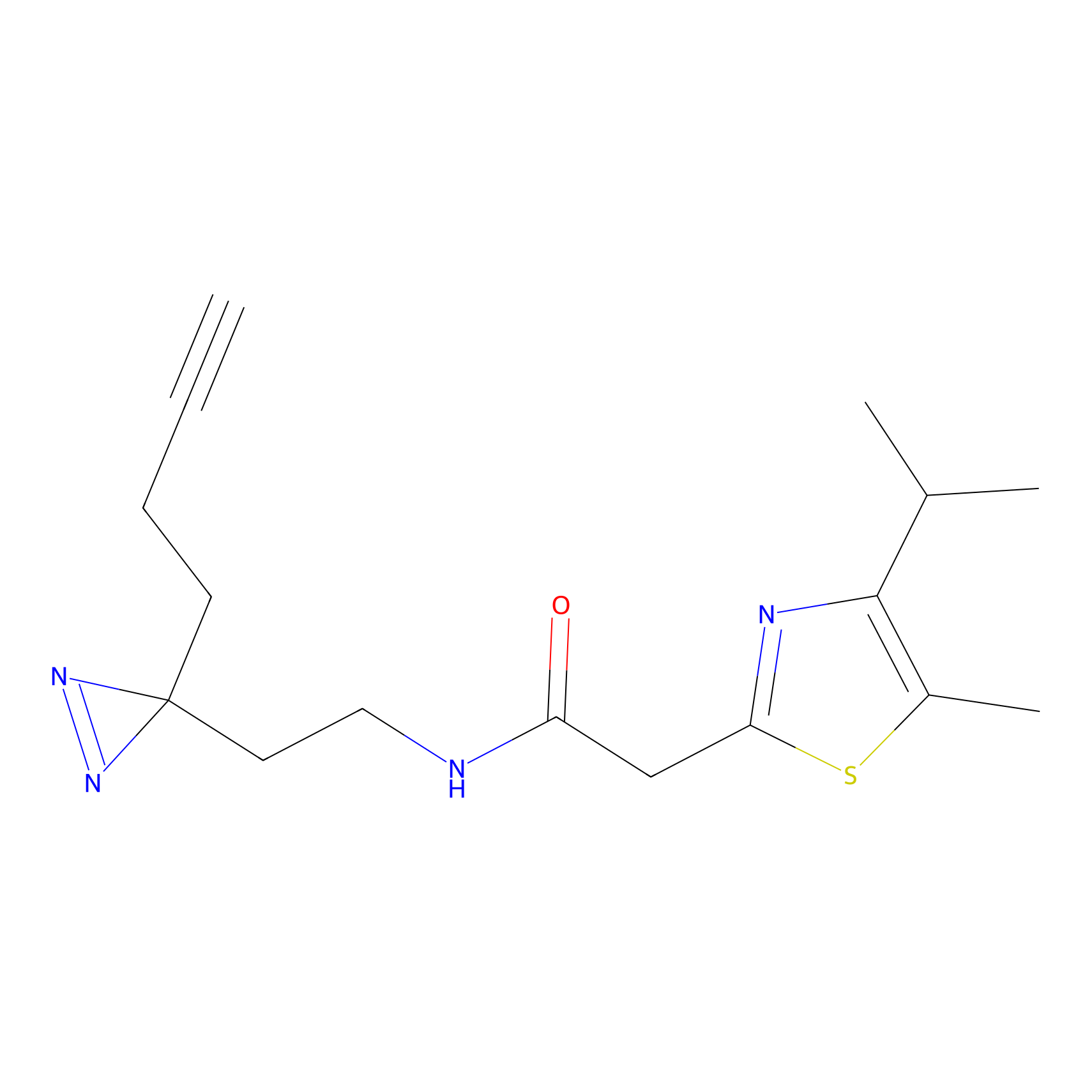 |
81.01 | LDD1968 | [16] | |
|
C346 Probe Info |
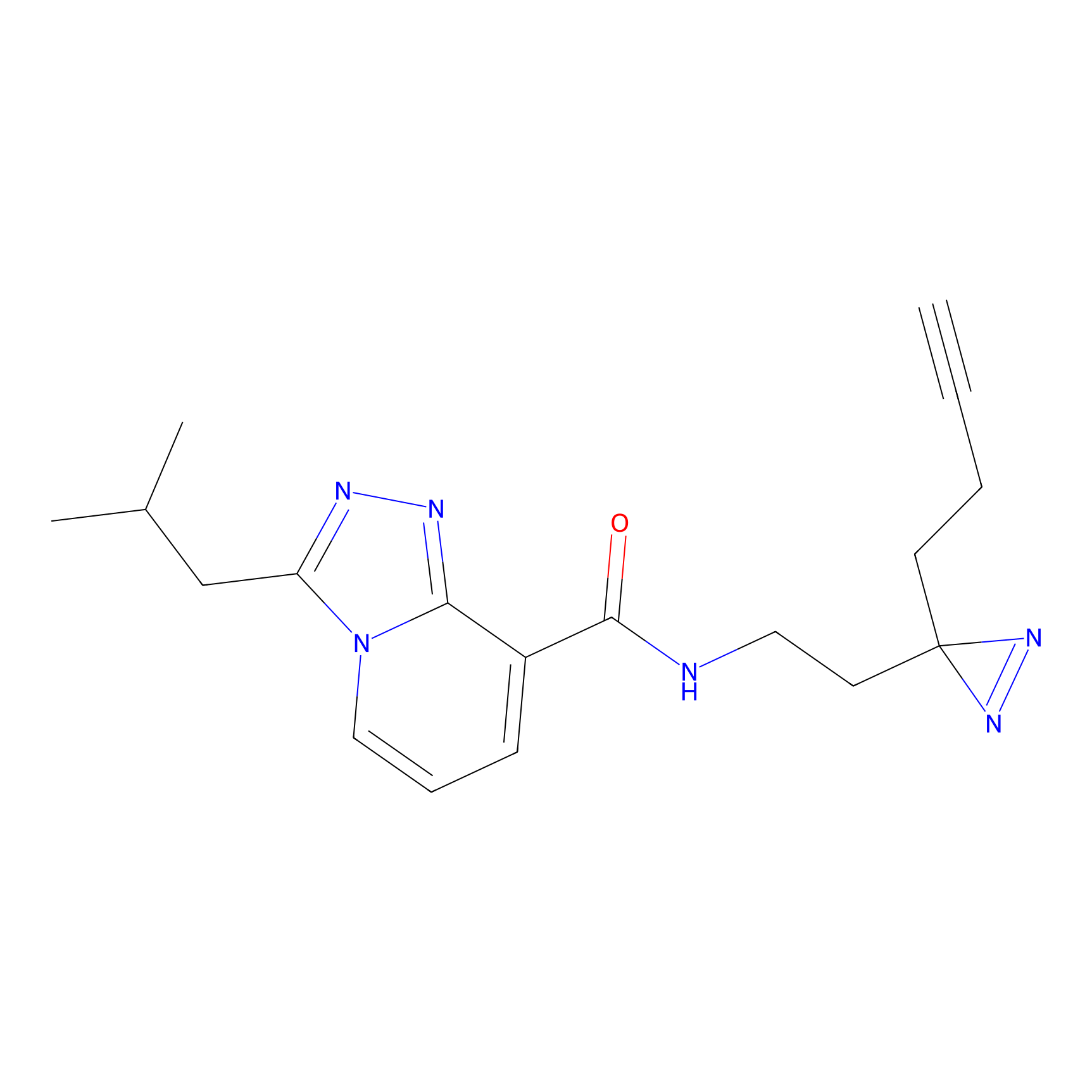 |
34.30 | LDD2007 | [16] | |
|
C347 Probe Info |
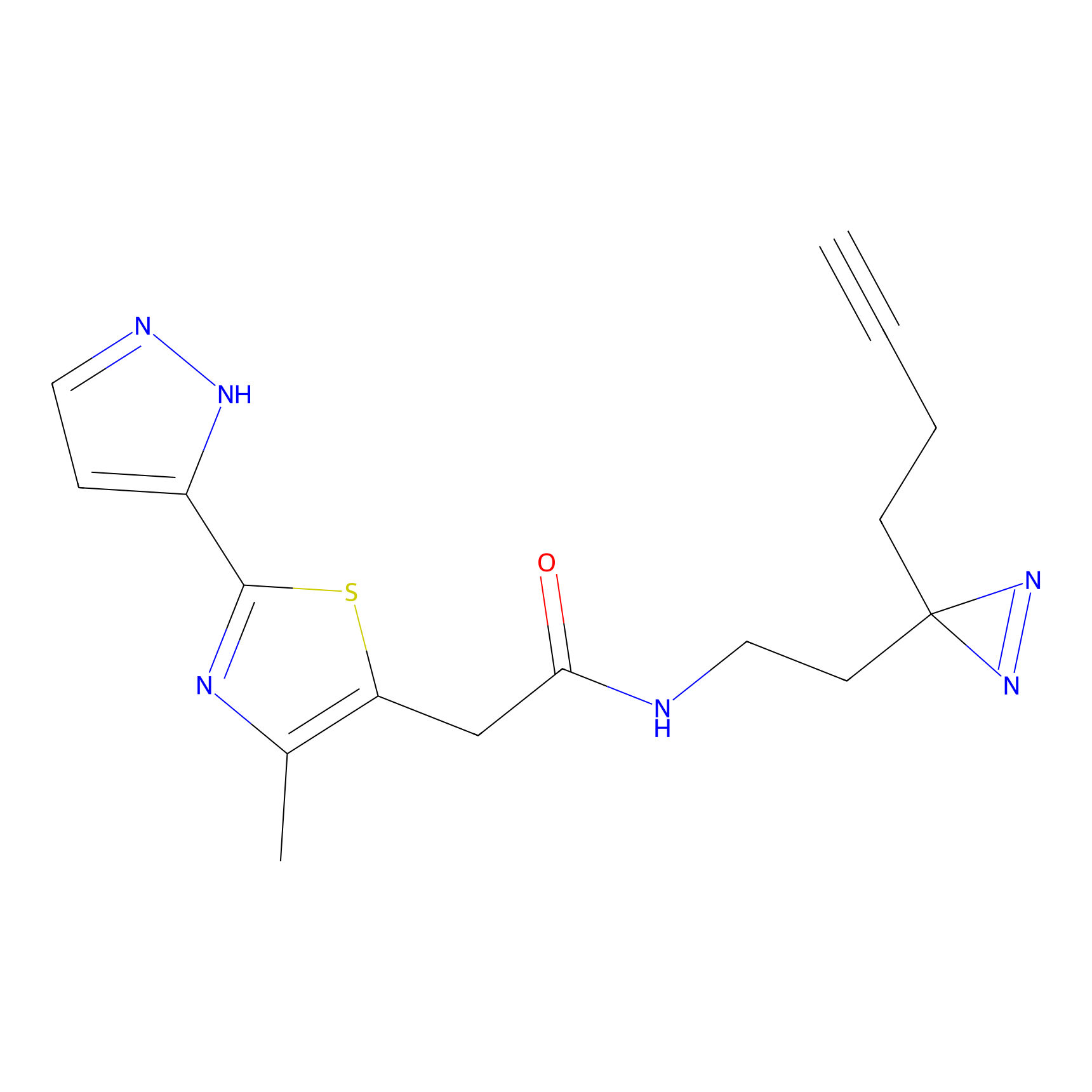 |
11.55 | LDD2008 | [16] | |
|
C355 Probe Info |
 |
29.24 | LDD2016 | [16] | |
|
C361 Probe Info |
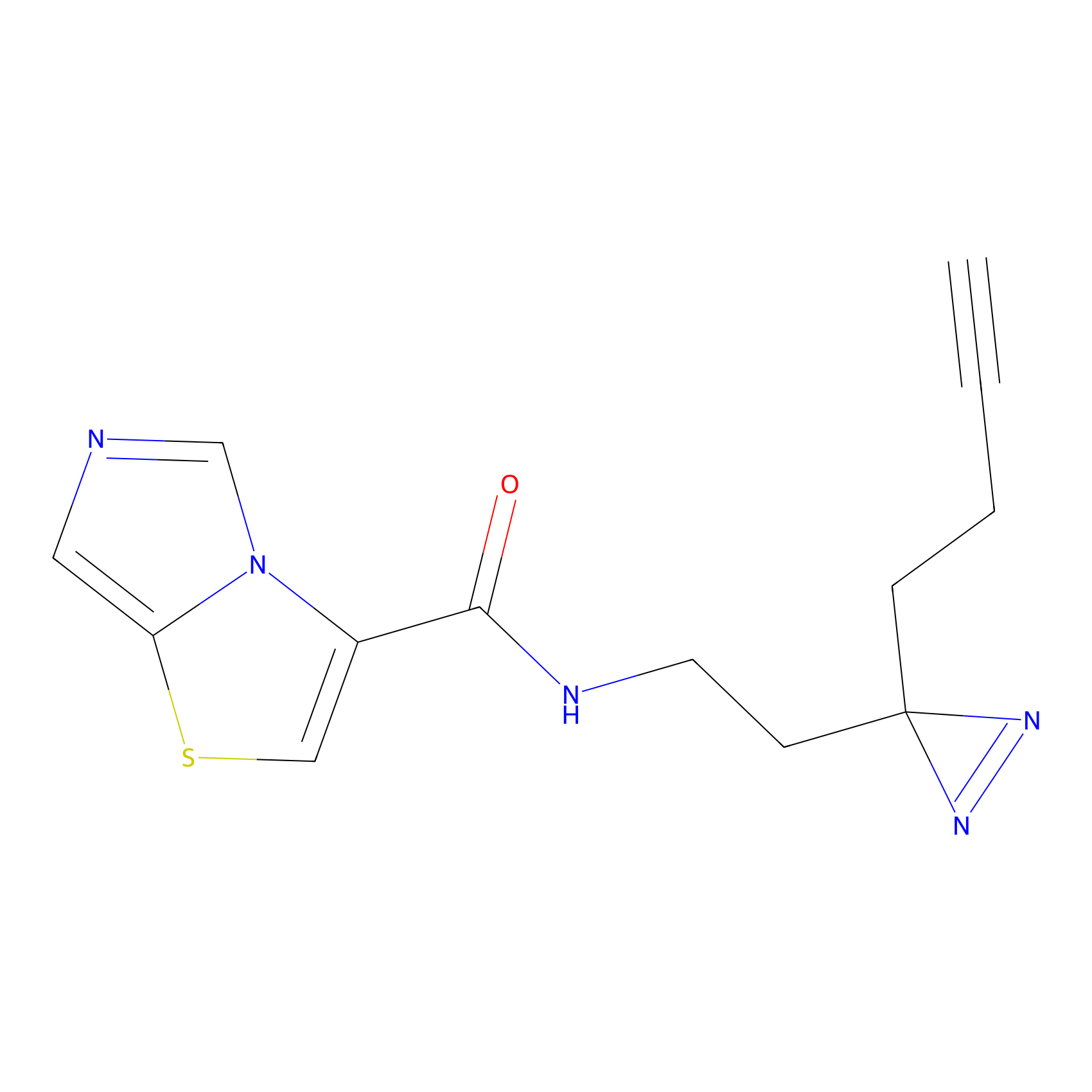 |
39.40 | LDD2022 | [16] | |
|
C373 Probe Info |
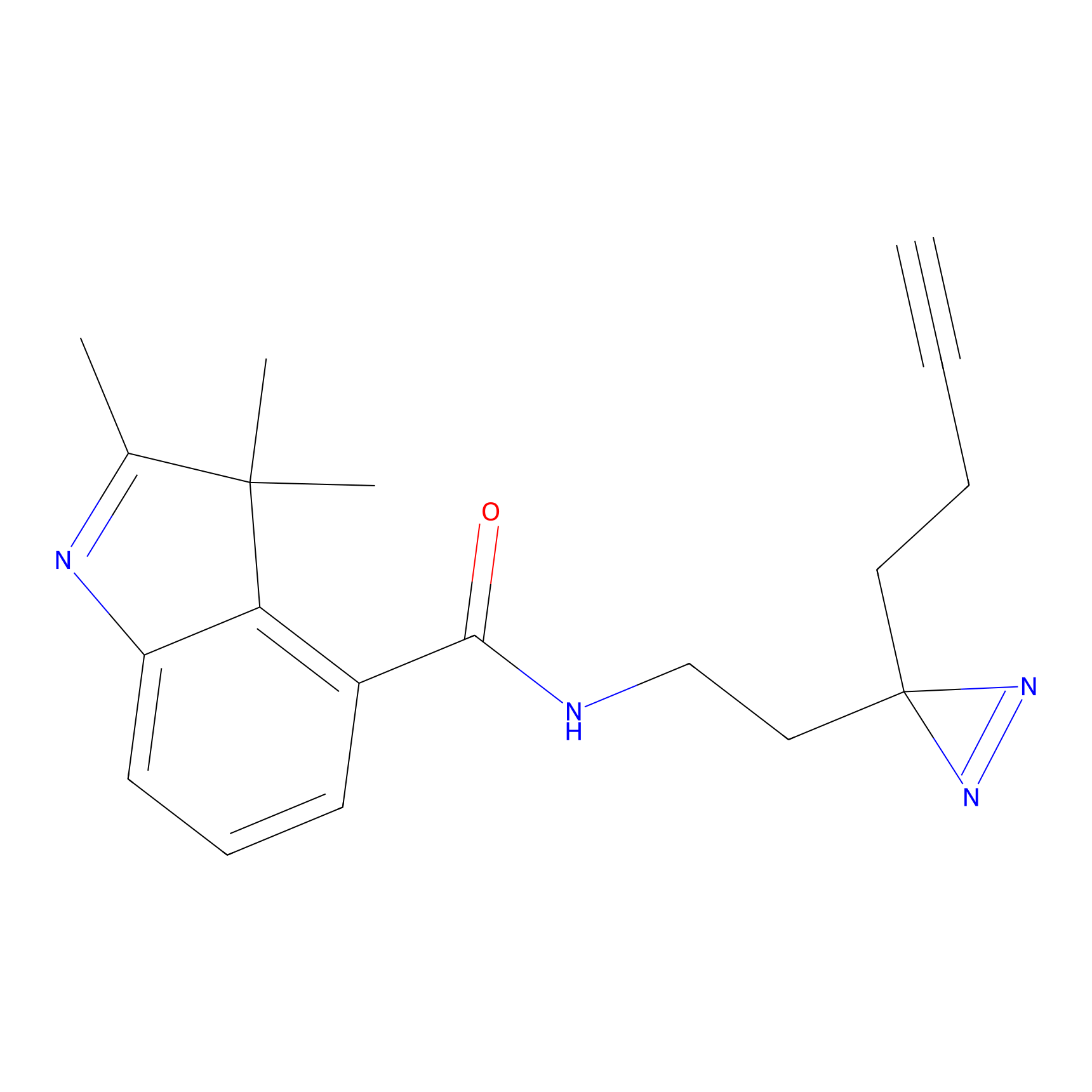 |
44.94 | LDD2033 | [16] | |
|
C399 Probe Info |
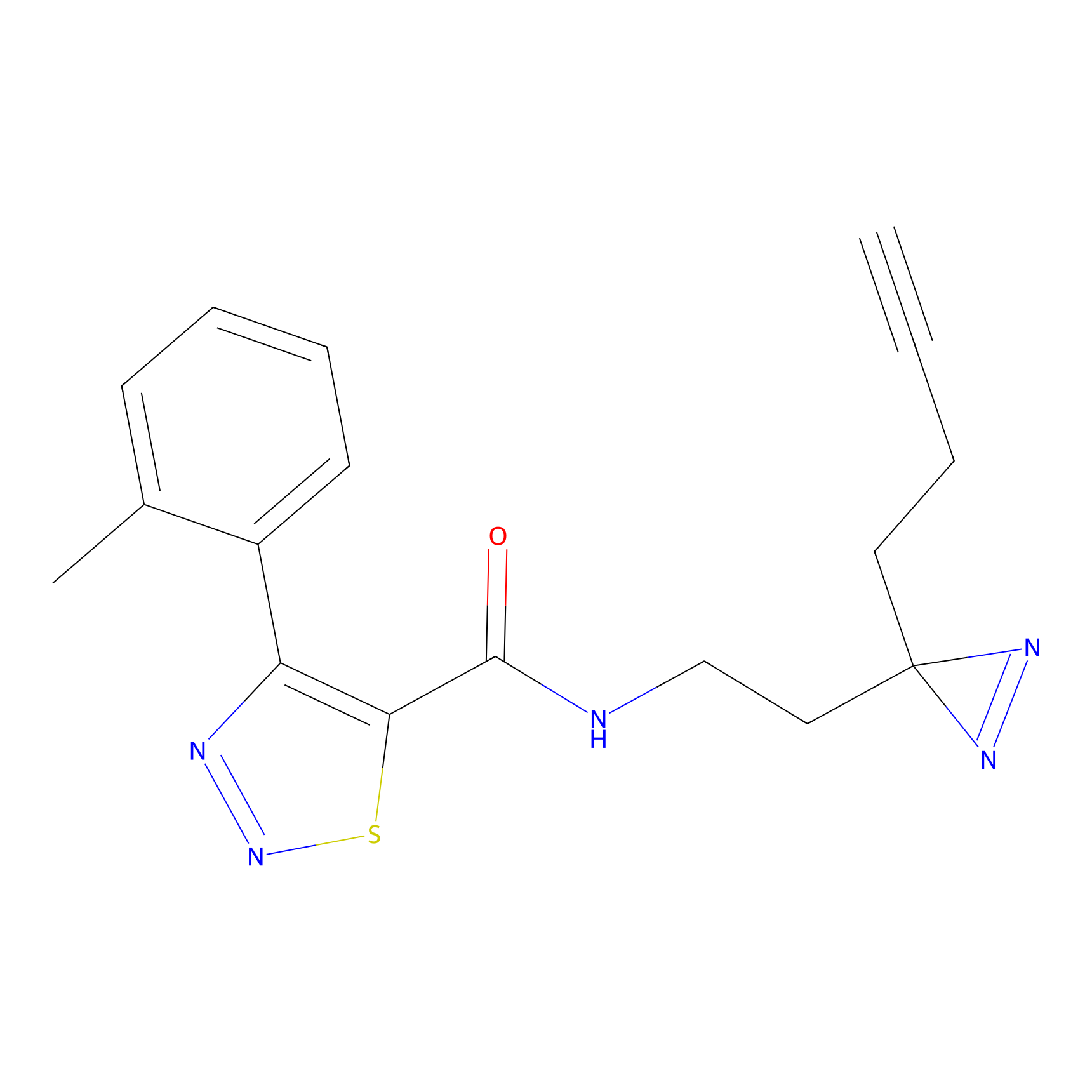 |
8.11 | LDD2058 | [16] | |
|
C403 Probe Info |
 |
16.34 | LDD2061 | [16] | |
|
DA-2 Probe Info |
 |
N.A. | LDD0072 | [17] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0088 | C45 | HEK-293T | 13.94 | LDD0203 | [6] |
| LDCM0070 | Cisar_cp37 | MDA-MB-231 | 2.14 | LDD0120 | [7] |
| LDCM0116 | HHS-0101 | DM93 | Y138(0.45) | LDD0264 | [10] |
| LDCM0117 | HHS-0201 | DM93 | Y138(1.13) | LDD0265 | [10] |
| LDCM0118 | HHS-0301 | DM93 | Y138(1.07) | LDD0266 | [10] |
| LDCM0119 | HHS-0401 | DM93 | Y138(1.58) | LDD0267 | [10] |
| LDCM0120 | HHS-0701 | DM93 | Y138(0.76) | LDD0268 | [10] |
| LDCM0123 | JWB131 | DM93 | Y138(0.83); Y178(0.68) | LDD0285 | [9] |
| LDCM0124 | JWB142 | DM93 | Y138(1.03); Y178(0.41) | LDD0286 | [9] |
| LDCM0125 | JWB146 | DM93 | Y138(1.16); Y178(0.82) | LDD0287 | [9] |
| LDCM0126 | JWB150 | DM93 | Y138(2.26); Y178(5.99) | LDD0288 | [9] |
| LDCM0127 | JWB152 | DM93 | Y138(4.45); Y178(0.19) | LDD0289 | [9] |
| LDCM0128 | JWB198 | DM93 | Y138(0.15); Y178(1.77) | LDD0290 | [9] |
| LDCM0129 | JWB202 | DM93 | Y138(0.71); Y178(0.38) | LDD0291 | [9] |
| LDCM0130 | JWB211 | DM93 | Y138(1.18); Y178(1.05) | LDD0292 | [9] |
| LDCM0022 | KB02 | 769-P | C36(1.81) | LDD2246 | [5] |
| LDCM0023 | KB03 | 769-P | C36(2.01) | LDD2663 | [5] |
| LDCM0024 | KB05 | IGR37 | C36(1.88) | LDD3314 | [5] |
| LDCM0014 | Panhematin | HEK-293T | 3.35 | LDD0062 | [8] |
| LDCM0131 | RA190 | MM1.R | C168(1.55) | LDD0304 | [18] |
References
