Details of the Target
General Information of Target
| Target ID | LDTP05782 | |||||
|---|---|---|---|---|---|---|
| Target Name | 26S proteasome non-ATPase regulatory subunit 2 (PSMD2) | |||||
| Gene Name | PSMD2 | |||||
| Gene ID | 5708 | |||||
| Synonyms |
TRAP2; 26S proteasome non-ATPase regulatory subunit 2; 26S proteasome regulatory subunit RPN1; 26S proteasome regulatory subunit S2; 26S proteasome subunit p97; Protein 55.11; Tumor necrosis factor type 1 receptor-associated protein 2
|
|||||
| 3D Structure | ||||||
| Sequence |
MEEGGRDKAPVQPQQSPAAAPGGTDEKPSGKERRDAGDKDKEQELSEEDKQLQDELEMLV
ERLGEKDTSLYRPALEELRRQIRSSTTSMTSVPKPLKFLRPHYGKLKEIYENMAPGENKR FAADIISVLAMTMSGERECLKYRLVGSQEELASWGHEYVRHLAGEVAKEWQELDDAEKVQ REPLLTLVKEIVPYNMAHNAEHEACDLLMEIEQVDMLEKDIDENAYAKVCLYLTSCVNYV PEPENSALLRCALGVFRKFSRFPEALRLALMLNDMELVEDIFTSCKDVVVQKQMAFMLGR HGVFLELSEDVEEYEDLTEIMSNVQLNSNFLALARELDIMEPKVPDDIYKTHLENNRFGG SGSQVDSARMNLASSFVNGFVNAAFGQDKLLTDDGNKWLYKNKDHGMLSAAASLGMILLW DVDGGLTQIDKYLYSSEDYIKSGALLACGIVNSGVRNECDPALALLSDYVLHNSNTMRLG SIFGLGLAYAGSNREDVLTLLLPVMGDSKSSMEVAGVTALACGMIAVGSCNGDVTSTILQ TIMEKSETELKDTYARWLPLGLGLNHLGKGEAIEAILAALEVVSEPFRSFANTLVDVCAY AGSGNVLKVQQLLHICSEHFDSKEKEEDKDKKEKKDKDKKEAPADMGAHQGVAVLGIALI AMGEEIGAEMALRTFGHLLRYGEPTLRRAVPLALALISVSNPRLNILDTLSKFSHDADPE VSYNSIFAMGMVGSGTNNARLAAMLRQLAQYHAKDPNNLFMVRLAQGLTHLGKGTLTLCP YHSDRQLMSQVAVAGLLTVLVSFLDVRNIILGKSHYVLYGLVAAMQPRMLVTFDEELRPL PVSVRVGQAVDVVGQAGKPKTITGFQTHTTPVLLAHGERAELATEEFLPVTPILEGFVIL RKNPNYDL |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Proteasome subunit S2 family
|
|||||
| Function |
Component of the 26S proteasome, a multiprotein complex involved in the ATP-dependent degradation of ubiquitinated proteins. This complex plays a key role in the maintenance of protein homeostasis by removing misfolded or damaged proteins, which could impair cellular functions, and by removing proteins whose functions are no longer required. Therefore, the proteasome participates in numerous cellular processes, including cell cycle progression, apoptosis, or DNA damage repair.; Binds to the intracellular domain of tumor necrosis factor type 1 receptor. The binding domain of TRAP1 and TRAP2 resides outside the death domain of TNFR1.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| AU565 | SNV: p.G155C | DBIA Probe Info | |||
| COLO792 | SNV: p.E108K | DBIA Probe Info | |||
| HUH7 | SNV: p.Y71C | DBIA Probe Info | |||
| JURKAT | SNV: p.L317P; p.P342S | Compound 10 Probe Info | |||
| L428 | SNV: p.E574K | DBIA Probe Info | |||
| MOLT4 | SNV: p.R72Q | IA-alkyne Probe Info | |||
| ONS76 | SNV: p.Q291Ter; p.T553I | DBIA Probe Info | |||
| RBE | SNV: p.R763L | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
9.22 | LDD0402 | [1] | |
|
A-EBA Probe Info |
 |
3.66 | LDD0215 | [2] | |
|
CY-1 Probe Info |
 |
24.89 | LDD0243 | [3] | |
|
CY4 Probe Info |
 |
15.65 | LDD0244 | [3] | |
|
TH211 Probe Info |
 |
Y751(7.24) | LDD0257 | [4] | |
|
TH214 Probe Info |
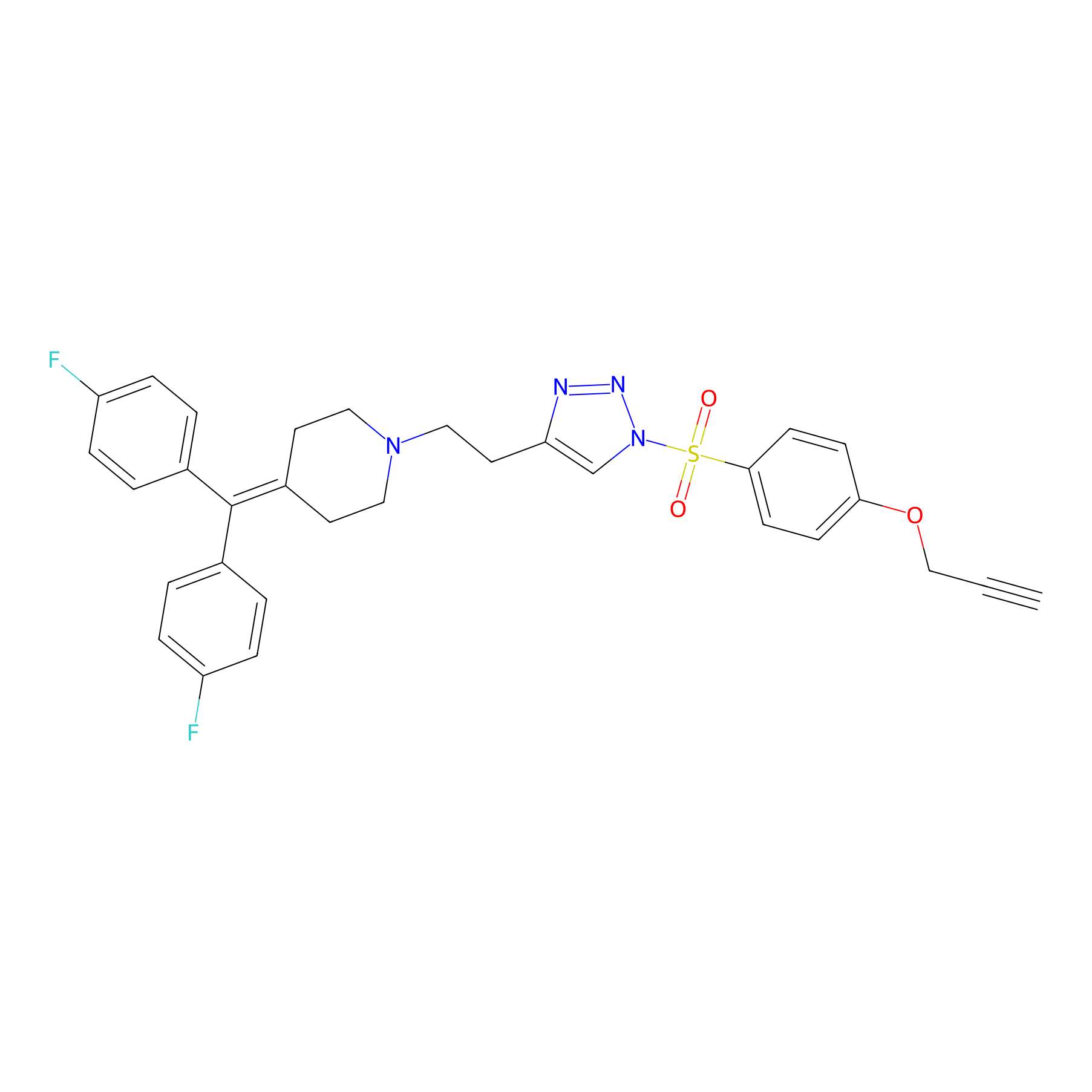 |
Y400(14.13) | LDD0258 | [4] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [5] | |
|
1oxF11yne Probe Info |
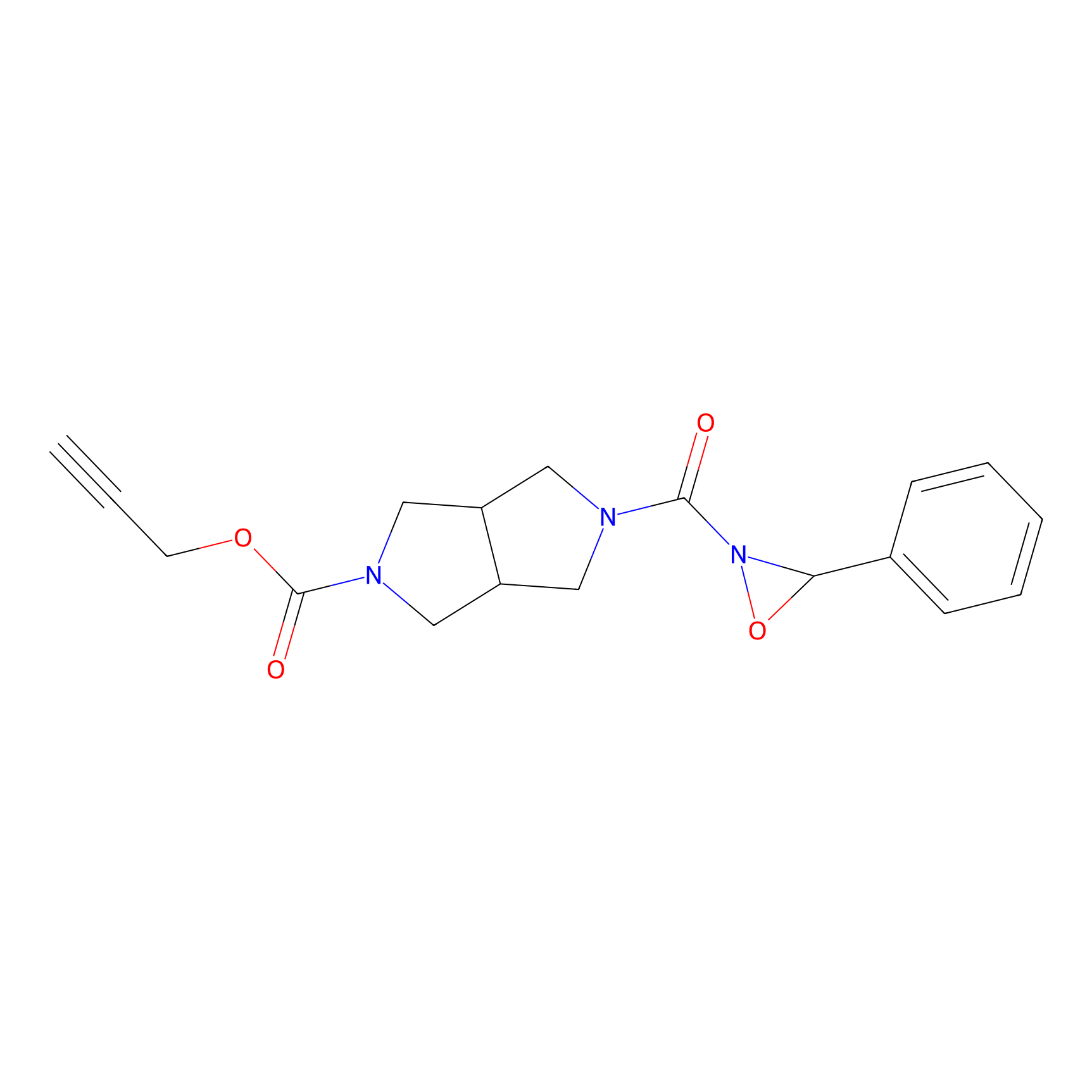 |
N.A. | LDD0193 | [6] | |
|
ONAyne Probe Info |
 |
K66(1.48) | LDD0274 | [7] | |
|
STPyne Probe Info |
 |
K105(10.00); K343(10.00); K350(9.77); K397(0.94) | LDD0277 | [7] | |
|
OPA-S-S-alkyne Probe Info |
 |
K551(2.34) | LDD3494 | [8] | |
|
Probe 1 Probe Info |
 |
Y71(69.10); Y103(38.35); Y110(46.95); Y434(19.19) | LDD3495 | [9] | |
|
BTD Probe Info |
 |
C779(1.81) | LDD1700 | [10] | |
|
AHL-Pu-1 Probe Info |
 |
C459(2.02) | LDD0168 | [11] | |
|
HHS-482 Probe Info |
 |
Y469(1.44) | LDD0285 | [12] | |
|
HHS-475 Probe Info |
 |
Y400(0.71); Y439(0.86); Y751(1.32); Y469(1.59) | LDD0264 | [13] | |
|
HHS-465 Probe Info |
 |
Y400(7.24); Y469(10.00); Y751(10.00) | LDD2237 | [14] | |
|
DBIA Probe Info |
 |
C251(0.89); C448(0.87) | LDD0078 | [15] | |
|
5E-2FA Probe Info |
 |
H868(0.00); H614(0.00) | LDD2235 | [16] | |
|
ATP probe Probe Info |
 |
K350(0.00); K858(0.00); K860(0.00); K178(0.00) | LDD0199 | [17] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C251(0.00); C779(0.00); C285(0.00); C459(0.00) | LDD0038 | [18] | |
|
IA-alkyne Probe Info |
 |
C448(0.00); C139(0.00); C779(0.00); C459(0.00) | LDD0032 | [19] | |
|
IPIAA_H Probe Info |
 |
C616(0.00); C459(0.00) | LDD0030 | [20] | |
|
IPIAA_L Probe Info |
 |
C616(0.00); C459(0.00) | LDD0031 | [20] | |
|
Lodoacetamide azide Probe Info |
 |
C251(0.00); C205(0.00); C139(0.00); C779(0.00) | LDD0037 | [18] | |
|
NAIA_4 Probe Info |
 |
C459(0.00); C779(0.00) | LDD2226 | [21] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [22] | |
|
aHNE Probe Info |
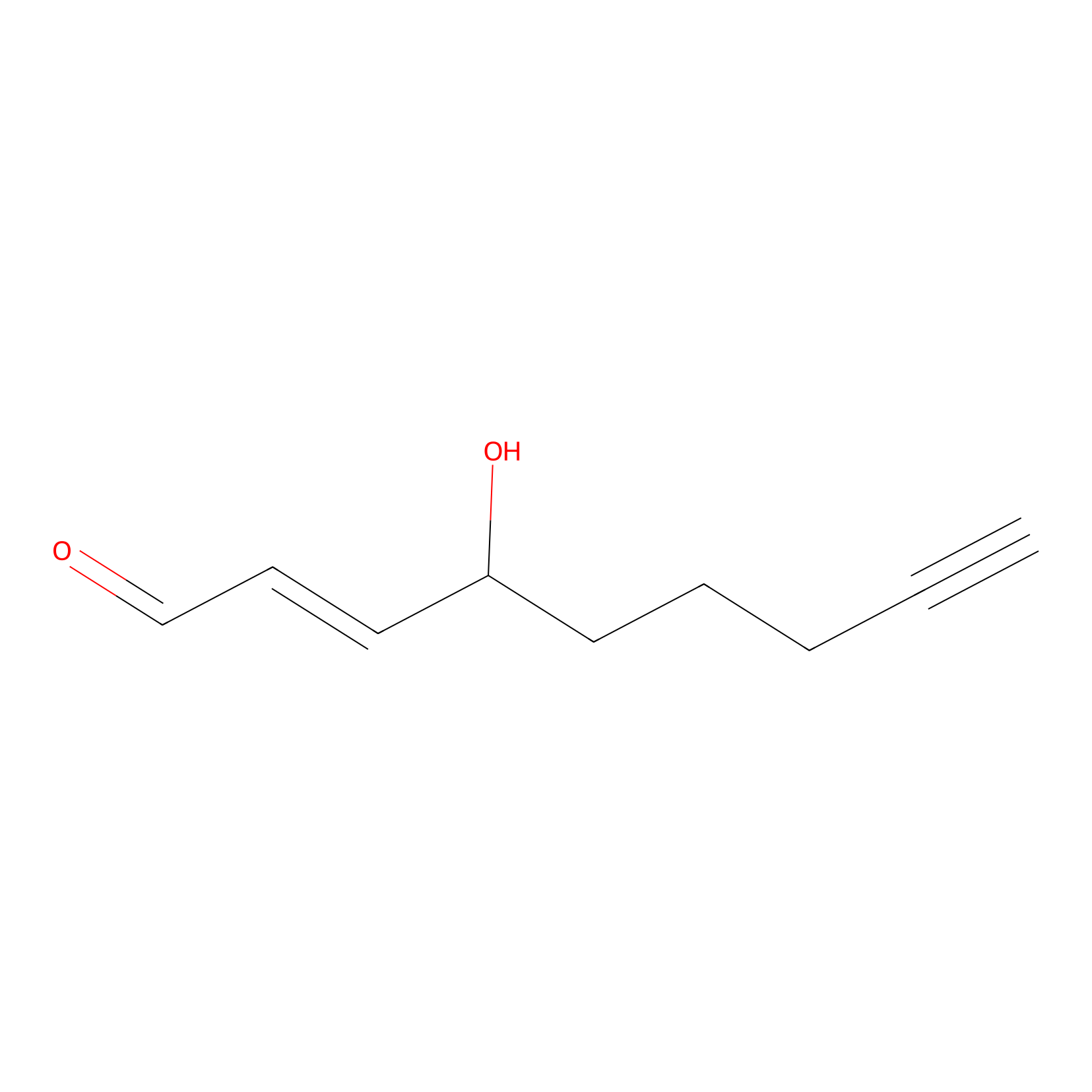 |
N.A. | LDD0001 | [22] | |
|
Compound 10 Probe Info |
 |
C448(0.00); C459(0.00); C779(0.00) | LDD2216 | [23] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [23] | |
|
IPM Probe Info |
 |
C459(0.00); C779(0.00) | LDD0005 | [22] | |
|
NHS Probe Info |
 |
K8(0.00); K350(0.00) | LDD0010 | [22] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [24] | |
|
TFBX Probe Info |
 |
C251(0.00); C448(0.00); C779(0.00); C459(0.00) | LDD0148 | [25] | |
|
Ox-W18 Probe Info |
 |
W154(0.00); W170(0.00) | LDD2175 | [26] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [27] | |
|
Acrolein Probe Info |
 |
C616(0.00); C448(0.00); H156(0.00); H161(0.00) | LDD0217 | [28] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [28] | |
|
Methacrolein Probe Info |
 |
C616(0.00); C448(0.00); C779(0.00) | LDD0218 | [28] | |
|
W1 Probe Info |
 |
C459(0.00); C779(0.00) | LDD0236 | [29] | |
|
NAIA_5 Probe Info |
 |
C251(0.00); C205(0.00); C139(0.00); C779(0.00) | LDD2223 | [21] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe13 Probe Info |
 |
5.69 | LDD0475 | [30] | |
|
FFF probe3 Probe Info |
 |
8.50 | LDD0464 | [30] | |
|
FFF probe4 Probe Info |
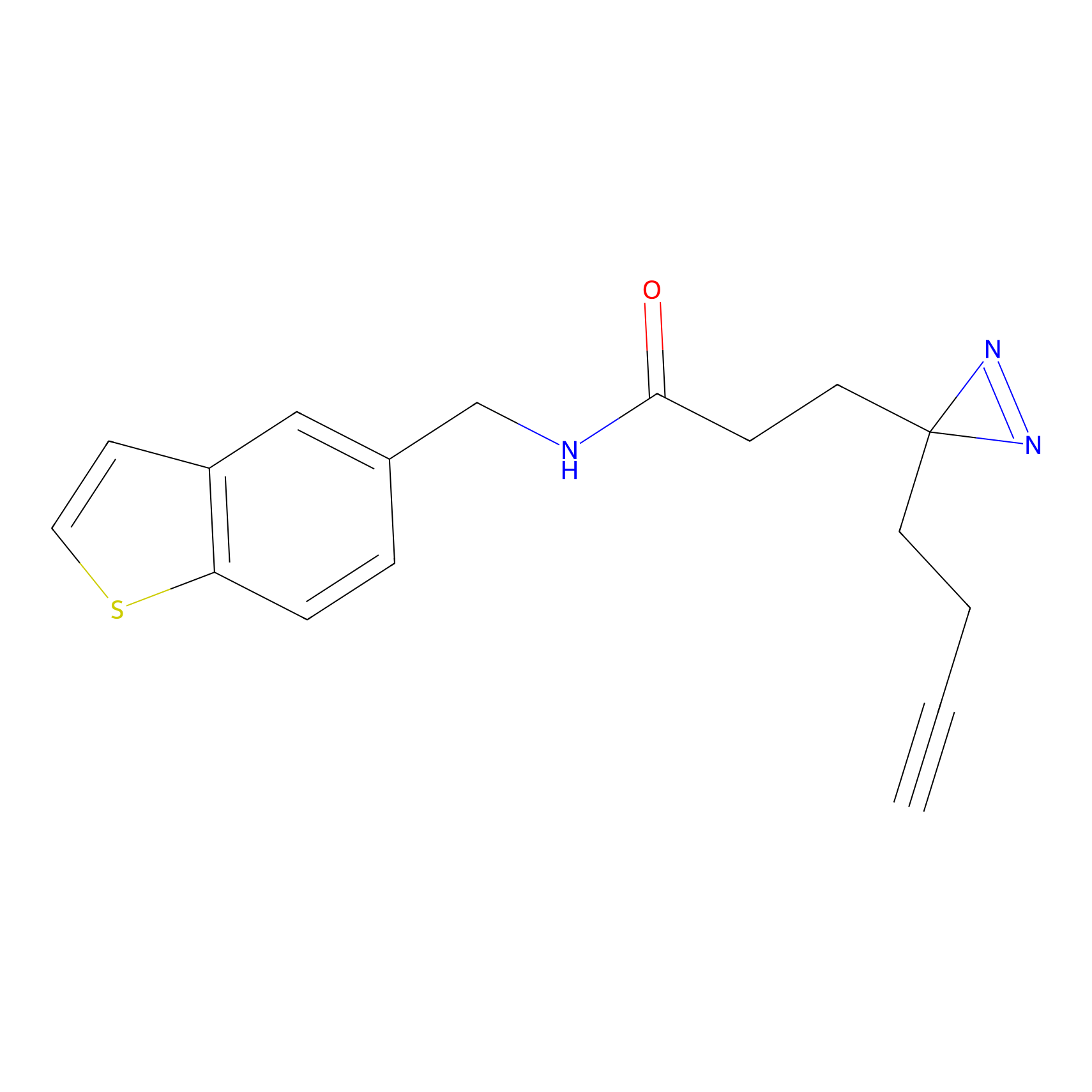 |
5.42 | LDD0466 | [30] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [31] | |
|
DA-2 Probe Info |
 |
N.A. | LDD0070 | [32] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0068 | [33] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C779(0.56); C448(0.42) | LDD2142 | [10] |
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C779(0.96); C251(0.88) | LDD2112 | [10] |
| LDCM0502 | 1-(Cyanoacetyl)piperidine | MDA-MB-231 | C779(0.66); C251(0.59) | LDD2095 | [10] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C779(1.48); C448(1.22); C616(2.51) | LDD2117 | [10] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C779(1.15); C448(1.33) | LDD2152 | [10] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C779(1.68); C448(1.04) | LDD2103 | [10] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C779(0.50); C251(0.54) | LDD2132 | [10] |
| LDCM0538 | 4-(Cyanoacetyl)morpholine | MDA-MB-231 | C779(0.80); C251(0.99) | LDD2131 | [10] |
| LDCM0025 | 4SU-RNA | HEK-293T | C459(2.02) | LDD0168 | [11] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C459(3.01) | LDD0169 | [11] |
| LDCM0562 | Abegg_cp(-)-11 | HeLa | C448(2.01) | LDD0313 | [25] |
| LDCM0214 | AC1 | HCT 116 | C230(0.71); C236(0.71); C251(1.17); C448(1.07) | LDD0531 | [15] |
| LDCM0215 | AC10 | HCT 116 | C230(0.27); C236(0.27); C251(1.00); C448(1.28) | LDD0532 | [15] |
| LDCM0216 | AC100 | HCT 116 | C230(0.46); C236(0.46); C251(1.03); C448(2.19) | LDD0533 | [15] |
| LDCM0217 | AC101 | HCT 116 | C230(0.34); C236(0.34); C251(0.83); C448(1.10) | LDD0534 | [15] |
| LDCM0218 | AC102 | HCT 116 | C230(0.33); C236(0.33); C251(0.83); C448(0.86) | LDD0535 | [15] |
| LDCM0219 | AC103 | HCT 116 | C230(0.20); C236(0.20); C251(0.94); C448(0.73) | LDD0536 | [15] |
| LDCM0220 | AC104 | HCT 116 | C230(0.32); C236(0.32); C251(1.01); C448(1.09) | LDD0537 | [15] |
| LDCM0221 | AC105 | HCT 116 | C230(0.26); C236(0.26); C251(0.81); C448(0.87) | LDD0538 | [15] |
| LDCM0222 | AC106 | HCT 116 | C230(0.28); C236(0.28); C251(0.85); C448(0.79) | LDD0539 | [15] |
| LDCM0223 | AC107 | HCT 116 | C230(0.39); C236(0.51); C251(0.99); C448(0.73) | LDD0540 | [15] |
| LDCM0224 | AC108 | HCT 116 | C230(0.20); C236(0.24); C251(0.90); C448(0.85) | LDD0541 | [15] |
| LDCM0225 | AC109 | HCT 116 | C230(0.26); C236(0.27); C251(1.04); C448(0.79) | LDD0542 | [15] |
| LDCM0226 | AC11 | HCT 116 | C230(0.28); C236(0.28); C251(0.93); C448(1.28) | LDD0543 | [15] |
| LDCM0227 | AC110 | HCT 116 | C230(0.25); C236(0.29); C251(0.91); C448(0.78) | LDD0544 | [15] |
| LDCM0228 | AC111 | HCT 116 | C230(0.25); C236(0.26); C251(0.77); C448(0.67) | LDD0545 | [15] |
| LDCM0229 | AC112 | HCT 116 | C230(0.38); C236(0.25); C251(0.81); C448(0.72) | LDD0546 | [15] |
| LDCM0230 | AC113 | HCT 116 | C251(1.02); C448(1.01); C598(0.88); C779(1.12) | LDD0547 | [15] |
| LDCM0231 | AC114 | HCT 116 | C251(1.21); C448(0.95); C598(0.53); C779(1.22) | LDD0548 | [15] |
| LDCM0232 | AC115 | HCT 116 | C251(0.95); C448(0.89); C598(0.47); C779(1.13) | LDD0549 | [15] |
| LDCM0233 | AC116 | HCT 116 | C251(1.13); C448(0.97); C598(0.53); C779(1.43) | LDD0550 | [15] |
| LDCM0234 | AC117 | HCT 116 | C251(1.26); C448(1.03); C598(0.56); C779(1.07) | LDD0551 | [15] |
| LDCM0235 | AC118 | HCT 116 | C251(1.19); C448(1.04); C598(0.61); C779(1.15) | LDD0552 | [15] |
| LDCM0236 | AC119 | HCT 116 | C251(2.12); C448(1.01); C598(0.58); C779(1.37) | LDD0553 | [15] |
| LDCM0237 | AC12 | HCT 116 | C230(0.35); C236(0.35); C251(0.96); C448(1.40) | LDD0554 | [15] |
| LDCM0238 | AC120 | HCT 116 | C251(1.61); C448(0.85); C598(0.57); C779(1.09) | LDD0555 | [15] |
| LDCM0239 | AC121 | HCT 116 | C251(1.72); C448(1.08); C598(0.70); C779(1.16) | LDD0556 | [15] |
| LDCM0240 | AC122 | HCT 116 | C251(1.14); C448(1.08); C598(0.46); C779(1.01) | LDD0557 | [15] |
| LDCM0241 | AC123 | HCT 116 | C251(1.16); C448(1.32); C598(0.61); C779(0.91) | LDD0558 | [15] |
| LDCM0242 | AC124 | HCT 116 | C251(1.24); C448(1.19); C598(0.56); C779(1.22) | LDD0559 | [15] |
| LDCM0243 | AC125 | HCT 116 | C251(1.57); C448(0.99); C598(0.78); C779(1.12) | LDD0560 | [15] |
| LDCM0244 | AC126 | HCT 116 | C251(1.32); C448(1.01); C598(0.50); C779(0.98) | LDD0561 | [15] |
| LDCM0245 | AC127 | HCT 116 | C251(1.20); C448(1.03); C598(0.55); C779(1.10) | LDD0562 | [15] |
| LDCM0246 | AC128 | HCT 116 | C230(0.76); C236(0.66); C251(1.67); C448(1.57) | LDD0563 | [15] |
| LDCM0247 | AC129 | HCT 116 | C230(1.48); C236(1.41); C251(1.92); C448(1.37) | LDD0564 | [15] |
| LDCM0249 | AC130 | HCT 116 | C230(1.43); C236(1.35); C251(1.55); C448(1.29) | LDD0566 | [15] |
| LDCM0250 | AC131 | HCT 116 | C230(1.52); C236(1.52); C251(1.54); C448(1.14) | LDD0567 | [15] |
| LDCM0251 | AC132 | HCT 116 | C230(0.77); C236(1.12); C251(1.42); C448(1.05) | LDD0568 | [15] |
| LDCM0252 | AC133 | HCT 116 | C230(0.84); C236(0.67); C251(1.52); C448(1.25) | LDD0569 | [15] |
| LDCM0253 | AC134 | HCT 116 | C230(0.87); C236(0.79); C251(1.55); C448(0.81) | LDD0570 | [15] |
| LDCM0254 | AC135 | HCT 116 | C230(0.76); C236(0.70); C251(1.35); C448(1.17) | LDD0571 | [15] |
| LDCM0255 | AC136 | HCT 116 | C230(0.72); C236(0.63); C251(1.38); C448(0.97) | LDD0572 | [15] |
| LDCM0256 | AC137 | HCT 116 | C230(0.87); C236(0.72); C251(1.81); C448(0.95) | LDD0573 | [15] |
| LDCM0257 | AC138 | HCT 116 | C230(1.01); C236(0.82); C251(1.10); C448(0.57) | LDD0574 | [15] |
| LDCM0258 | AC139 | HCT 116 | C230(0.91); C236(1.05); C251(1.15); C448(0.64) | LDD0575 | [15] |
| LDCM0259 | AC14 | HCT 116 | C230(0.29); C236(0.29); C251(1.02); C448(1.21) | LDD0576 | [15] |
| LDCM0260 | AC140 | HCT 116 | C230(0.87); C236(0.75); C251(1.25); C448(0.61) | LDD0577 | [15] |
| LDCM0261 | AC141 | HCT 116 | C230(0.91); C236(0.82); C251(1.20); C448(0.62) | LDD0578 | [15] |
| LDCM0262 | AC142 | HCT 116 | C230(2.04); C236(1.37); C251(1.42); C448(0.86) | LDD0579 | [15] |
| LDCM0263 | AC143 | HCT 116 | C448(0.70); C598(0.85); C251(1.05) | LDD0580 | [15] |
| LDCM0264 | AC144 | HCT 116 | C448(0.62); C598(0.65); C251(1.02) | LDD0581 | [15] |
| LDCM0265 | AC145 | HCT 116 | C448(0.56); C598(0.81); C251(1.04) | LDD0582 | [15] |
| LDCM0266 | AC146 | HCT 116 | C598(0.61); C448(0.70); C251(0.86) | LDD0583 | [15] |
| LDCM0267 | AC147 | HCT 116 | C598(0.64); C448(0.68); C251(1.07) | LDD0584 | [15] |
| LDCM0268 | AC148 | HCT 116 | C598(0.33); C448(0.74); C251(0.78) | LDD0585 | [15] |
| LDCM0269 | AC149 | HCT 116 | C598(0.51); C448(0.52); C251(0.83) | LDD0586 | [15] |
| LDCM0270 | AC15 | HCT 116 | C230(0.32); C236(0.32); C779(1.00); C251(1.05) | LDD0587 | [15] |
| LDCM0271 | AC150 | HCT 116 | C251(0.85); C598(0.89); C448(1.42) | LDD0588 | [15] |
| LDCM0272 | AC151 | HCT 116 | C598(0.78); C251(0.87); C448(1.04) | LDD0589 | [15] |
| LDCM0273 | AC152 | HCT 116 | C598(0.54); C251(0.92); C448(1.11) | LDD0590 | [15] |
| LDCM0274 | AC153 | HCT 116 | C598(0.24); C448(0.45); C251(0.85) | LDD0591 | [15] |
| LDCM0621 | AC154 | HCT 116 | C251(0.95); C448(0.67); C598(0.68) | LDD2158 | [15] |
| LDCM0622 | AC155 | HCT 116 | C251(1.00); C448(0.71); C598(0.70) | LDD2159 | [15] |
| LDCM0623 | AC156 | HCT 116 | C251(0.86); C448(0.57); C598(1.03) | LDD2160 | [15] |
| LDCM0624 | AC157 | HCT 116 | C251(0.99); C448(0.65); C598(0.96) | LDD2161 | [15] |
| LDCM0276 | AC17 | HCT 116 | C779(1.01); C448(1.02); C598(1.03); C230(1.19) | LDD0593 | [15] |
| LDCM0277 | AC18 | HCT 116 | C598(0.82); C448(0.95); C779(1.01); C230(1.10) | LDD0594 | [15] |
| LDCM0278 | AC19 | HCT 116 | C598(0.92); C251(0.95); C779(1.03); C448(1.08) | LDD0595 | [15] |
| LDCM0279 | AC2 | HCT 116 | C779(0.66); C230(0.69); C236(0.69); C598(0.91) | LDD0596 | [15] |
| LDCM0280 | AC20 | HCT 116 | C448(1.01); C779(1.01); C251(1.02); C598(1.04) | LDD0597 | [15] |
| LDCM0281 | AC21 | HCT 116 | C230(0.82); C236(0.82); C598(0.97); C779(1.04) | LDD0598 | [15] |
| LDCM0282 | AC22 | HCT 116 | C598(0.94); C448(0.98); C779(1.04); C251(1.15) | LDD0599 | [15] |
| LDCM0283 | AC23 | HCT 116 | C598(0.77); C251(0.79); C779(0.86); C448(1.02) | LDD0600 | [15] |
| LDCM0284 | AC24 | HCT 116 | C779(1.00); C448(1.00); C251(1.01); C598(1.08) | LDD0601 | [15] |
| LDCM0285 | AC25 | HCT 116 | C779(0.69); C251(1.08); C598(1.10); C448(1.19) | LDD0602 | [15] |
| LDCM0286 | AC26 | HCT 116 | C779(0.66); C448(0.82); C598(0.83); C251(1.26) | LDD0603 | [15] |
| LDCM0287 | AC27 | HCT 116 | C779(0.71); C598(0.78); C448(0.90); C251(1.08) | LDD0604 | [15] |
| LDCM0288 | AC28 | HCT 116 | C779(0.64); C598(0.81); C448(0.95); C251(1.16) | LDD0605 | [15] |
| LDCM0289 | AC29 | HCT 116 | C448(0.64); C598(0.64); C779(0.67); C251(1.05) | LDD0606 | [15] |
| LDCM0290 | AC3 | HCT 116 | C230(0.57); C236(0.57); C779(0.89); C251(1.10) | LDD0607 | [15] |
| LDCM0291 | AC30 | HCT 116 | C448(0.59); C598(0.60); C779(0.64); C251(0.91) | LDD0608 | [15] |
| LDCM0292 | AC31 | HCT 116 | C598(0.69); C448(0.74); C779(0.75); C251(1.30) | LDD0609 | [15] |
| LDCM0293 | AC32 | HCT 116 | C598(0.59); C448(0.63); C779(0.67); C251(1.09) | LDD0610 | [15] |
| LDCM0294 | AC33 | HCT 116 | C598(0.76); C779(0.86); C448(0.93); C251(1.06) | LDD0611 | [15] |
| LDCM0295 | AC34 | HCT 116 | C598(0.72); C448(0.94); C251(0.96); C779(1.04) | LDD0612 | [15] |
| LDCM0296 | AC35 | HCT 116 | C779(0.91); C251(1.01); C448(1.03); C598(1.56) | LDD0613 | [15] |
| LDCM0297 | AC36 | HCT 116 | C779(0.92); C251(0.94); C448(1.10); C598(1.29) | LDD0614 | [15] |
| LDCM0298 | AC37 | HCT 116 | C779(0.82); C448(0.97); C251(1.00); C598(1.28) | LDD0615 | [15] |
| LDCM0299 | AC38 | HCT 116 | C251(0.85); C448(0.97); C779(1.13); C598(1.31) | LDD0616 | [15] |
| LDCM0300 | AC39 | HCT 116 | C598(0.96); C448(0.98); C779(1.00); C251(1.04) | LDD0617 | [15] |
| LDCM0301 | AC4 | HCT 116 | C230(0.53); C236(0.53); C779(0.84); C251(1.22) | LDD0618 | [15] |
| LDCM0302 | AC40 | HCT 116 | C448(0.80); C251(0.93); C779(1.05); C598(1.07) | LDD0619 | [15] |
| LDCM0303 | AC41 | HCT 116 | C448(0.86); C251(0.86); C779(0.94); C598(1.17) | LDD0620 | [15] |
| LDCM0304 | AC42 | HCT 116 | C251(0.81); C448(0.83); C779(1.02); C598(1.12) | LDD0621 | [15] |
| LDCM0305 | AC43 | HCT 116 | C448(0.74); C251(0.81); C598(0.91); C779(0.97) | LDD0622 | [15] |
| LDCM0306 | AC44 | HCT 116 | C448(0.82); C779(0.89); C251(0.98); C598(1.11) | LDD0623 | [15] |
| LDCM0307 | AC45 | HCT 116 | C448(0.69); C779(0.81); C598(0.88); C251(0.93) | LDD0624 | [15] |
| LDCM0308 | AC46 | HCT 116 | C230(0.58); C236(0.58); C251(0.88); C598(0.92) | LDD0625 | [15] |
| LDCM0309 | AC47 | HCT 116 | C230(0.60); C236(0.60); C598(0.82); C779(0.87) | LDD0626 | [15] |
| LDCM0310 | AC48 | HCT 116 | C598(0.64); C230(0.66); C236(0.66); C448(0.91) | LDD0627 | [15] |
| LDCM0311 | AC49 | HCT 116 | C230(0.51); C236(0.51); C598(0.56); C448(0.58) | LDD0628 | [15] |
| LDCM0312 | AC5 | HCT 116 | C230(0.44); C236(0.44); C779(0.87); C448(1.22) | LDD0629 | [15] |
| LDCM0313 | AC50 | HCT 116 | C230(0.45); C236(0.45); C598(0.48); C448(0.67) | LDD0630 | [15] |
| LDCM0314 | AC51 | HCT 116 | C251(0.86); C598(0.99); C779(1.08); C230(1.18) | LDD0631 | [15] |
| LDCM0315 | AC52 | HCT 116 | C230(0.64); C236(0.64); C598(0.84); C251(0.87) | LDD0632 | [15] |
| LDCM0316 | AC53 | HCT 116 | C230(0.65); C236(0.65); C598(0.78); C448(0.82) | LDD0633 | [15] |
| LDCM0317 | AC54 | HCT 116 | C598(0.74); C251(0.86); C779(0.86); C448(0.88) | LDD0634 | [15] |
| LDCM0318 | AC55 | HCT 116 | C230(0.53); C236(0.53); C448(0.70); C598(0.71) | LDD0635 | [15] |
| LDCM0319 | AC56 | HCT 116 | C598(0.35); C448(0.43); C230(0.64); C236(0.64) | LDD0636 | [15] |
| LDCM0320 | AC57 | HCT 116 | C230(0.57); C236(0.57); C779(1.05); C448(1.08) | LDD0637 | [15] |
| LDCM0321 | AC58 | HCT 116 | C230(0.67); C236(0.67); C448(0.87); C779(1.02) | LDD0638 | [15] |
| LDCM0322 | AC59 | HCT 116 | C230(0.50); C236(0.50); C251(1.01); C779(1.04) | LDD0639 | [15] |
| LDCM0323 | AC6 | HCT 116 | C230(0.26); C236(0.26); C598(0.71); C448(0.91) | LDD0640 | [15] |
| LDCM0324 | AC60 | HCT 116 | C230(0.42); C236(0.42); C779(0.96); C448(1.00) | LDD0641 | [15] |
| LDCM0325 | AC61 | HCT 116 | C230(0.63); C236(0.63); C779(1.16); C448(1.18) | LDD0642 | [15] |
| LDCM0326 | AC62 | HCT 116 | C230(0.53); C236(0.53); C251(1.04); C779(1.08) | LDD0643 | [15] |
| LDCM0327 | AC63 | HCT 116 | C230(0.62); C236(0.62); C251(1.12); C779(1.15) | LDD0644 | [15] |
| LDCM0328 | AC64 | HCT 116 | C230(0.51); C236(0.51); C251(1.06); C779(1.09) | LDD0645 | [15] |
| LDCM0329 | AC65 | HCT 116 | C230(0.33); C236(0.33); C448(0.93); C251(0.97) | LDD0646 | [15] |
| LDCM0330 | AC66 | HCT 116 | C230(0.48); C236(0.48); C779(1.02); C251(1.03) | LDD0647 | [15] |
| LDCM0331 | AC67 | HCT 116 | C230(0.30); C236(0.30); C448(0.94); C779(0.97) | LDD0648 | [15] |
| LDCM0332 | AC68 | HCT 116 | C230(0.63); C236(0.63); C598(0.70); C251(0.90) | LDD0649 | [15] |
| LDCM0333 | AC69 | HCT 116 | C230(0.44); C236(0.44); C598(0.68); C779(0.87) | LDD0650 | [15] |
| LDCM0334 | AC7 | HCT 116 | C230(0.28); C236(0.28); C779(0.95); C598(0.99) | LDD0651 | [15] |
| LDCM0335 | AC70 | HCT 116 | C230(0.31); C236(0.31); C598(0.56); C448(0.75) | LDD0652 | [15] |
| LDCM0336 | AC71 | HCT 116 | C230(0.53); C236(0.53); C598(0.89); C779(1.10) | LDD0653 | [15] |
| LDCM0337 | AC72 | HCT 116 | C598(0.78); C779(0.85); C448(0.91); C251(0.92) | LDD0654 | [15] |
| LDCM0338 | AC73 | HCT 116 | C598(0.45); C230(0.46); C236(0.46); C448(0.63) | LDD0655 | [15] |
| LDCM0339 | AC74 | HCT 116 | C230(0.41); C236(0.41); C598(0.49); C448(0.66) | LDD0656 | [15] |
| LDCM0340 | AC75 | HCT 116 | C598(0.39); C230(0.61); C236(0.61); C448(0.62) | LDD0657 | [15] |
| LDCM0341 | AC76 | HCT 116 | C230(0.54); C236(0.54); C598(0.70); C779(0.99) | LDD0658 | [15] |
| LDCM0342 | AC77 | HCT 116 | C230(0.42); C236(0.42); C598(0.65); C779(0.90) | LDD0659 | [15] |
| LDCM0343 | AC78 | HCT 116 | C230(0.67); C236(0.67); C598(0.80); C448(0.91) | LDD0660 | [15] |
| LDCM0344 | AC79 | HCT 116 | C230(0.62); C236(0.62); C598(0.77); C779(0.85) | LDD0661 | [15] |
| LDCM0345 | AC8 | HCT 116 | C230(0.46); C236(0.46); C598(0.84); C251(0.93) | LDD0662 | [15] |
| LDCM0346 | AC80 | HCT 116 | C230(0.71); C236(0.71); C779(0.88); C598(0.88) | LDD0663 | [15] |
| LDCM0347 | AC81 | HCT 116 | C598(0.81); C251(0.88); C779(0.96); C230(0.99) | LDD0664 | [15] |
| LDCM0348 | AC82 | HCT 116 | C598(0.36); C230(0.45); C236(0.45); C448(0.58) | LDD0665 | [15] |
| LDCM0349 | AC83 | HCT 116 | C448(0.58); C598(0.62); C251(0.71); C230(0.73) | LDD0666 | [15] |
| LDCM0350 | AC84 | HCT 116 | C598(0.58); C448(0.61); C251(0.72); C230(0.76) | LDD0667 | [15] |
| LDCM0351 | AC85 | HCT 116 | C598(0.61); C448(0.76); C230(0.79); C236(0.79) | LDD0668 | [15] |
| LDCM0352 | AC86 | HCT 116 | C598(0.65); C448(0.78); C230(0.93); C236(0.93) | LDD0669 | [15] |
| LDCM0353 | AC87 | HCT 116 | C448(0.85); C251(0.90); C598(0.97); C230(1.27) | LDD0670 | [15] |
| LDCM0354 | AC88 | HCT 116 | C598(0.66); C448(0.82); C251(0.90); C230(1.06) | LDD0671 | [15] |
| LDCM0355 | AC89 | HCT 116 | C598(0.72); C448(0.72); C251(0.84); C230(0.99) | LDD0672 | [15] |
| LDCM0357 | AC90 | HCT 116 | C598(0.70); C448(1.05); C230(1.26); C236(1.26) | LDD0674 | [15] |
| LDCM0358 | AC91 | HCT 116 | C598(0.67); C448(0.71); C251(0.86); C230(0.91) | LDD0675 | [15] |
| LDCM0359 | AC92 | HCT 116 | C598(0.61); C448(0.74); C251(0.84); C230(0.93) | LDD0676 | [15] |
| LDCM0360 | AC93 | HCT 116 | C598(0.80); C448(0.83); C251(0.92); C230(1.06) | LDD0677 | [15] |
| LDCM0361 | AC94 | HCT 116 | C598(0.92); C251(0.95); C448(1.03); C230(1.05) | LDD0678 | [15] |
| LDCM0362 | AC95 | HCT 116 | C598(0.69); C448(0.82); C251(1.00); C230(1.16) | LDD0679 | [15] |
| LDCM0363 | AC96 | HCT 116 | C598(0.77); C251(0.92); C448(0.96); C230(1.04) | LDD0680 | [15] |
| LDCM0364 | AC97 | HCT 116 | C598(0.62); C448(0.67); C251(0.82); C230(0.99) | LDD0681 | [15] |
| LDCM0365 | AC98 | HCT 116 | C230(0.14); C236(0.14); C448(0.50); C251(0.98) | LDD0682 | [15] |
| LDCM0366 | AC99 | HCT 116 | C230(0.26); C236(0.26); C448(0.76); C779(0.90) | LDD0683 | [15] |
| LDCM0545 | Acetamide | MDA-MB-231 | C779(0.53) | LDD2138 | [10] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C779(0.85); C448(1.06) | LDD2113 | [10] |
| LDCM0248 | AKOS034007472 | HCT 116 | C230(0.24); C236(0.24); C251(1.19); C448(1.38) | LDD0565 | [15] |
| LDCM0356 | AKOS034007680 | HCT 116 | C230(0.25); C236(0.25); C779(0.96); C251(1.08) | LDD0673 | [15] |
| LDCM0275 | AKOS034007705 | HCT 116 | C230(0.17); C236(0.17); C598(0.77); C251(0.86) | LDD0592 | [15] |
| LDCM0156 | Aniline | NCI-H1299 | N.A. | LDD0404 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C251(0.89); C448(0.87) | LDD0078 | [15] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C779(0.70); C251(1.05); C448(0.80) | LDD2091 | [10] |
| LDCM0630 | CCW28-3 | 231MFP | C779(1.28) | LDD2214 | [34] |
| LDCM0108 | Chloroacetamide | HeLa | C459(0.00); C616(0.00); H161(0.00); C779(0.00) | LDD0222 | [28] |
| LDCM0632 | CL-Sc | Hep-G2 | C448(1.39); C459(1.09) | LDD2227 | [21] |
| LDCM0367 | CL1 | HCT 116 | C251(0.82); C598(0.93); C448(0.96); C779(0.98) | LDD0684 | [15] |
| LDCM0368 | CL10 | HCT 116 | C598(0.46); C230(0.74); C236(0.74); C448(0.85) | LDD0685 | [15] |
| LDCM0369 | CL100 | HCT 116 | C230(0.52); C236(0.52); C779(0.78); C251(1.10) | LDD0686 | [15] |
| LDCM0370 | CL101 | HCT 116 | C230(0.19); C236(0.19); C779(0.93); C251(0.94) | LDD0687 | [15] |
| LDCM0371 | CL102 | HCT 116 | C230(0.32); C236(0.32); C779(0.71); C251(1.03) | LDD0688 | [15] |
| LDCM0372 | CL103 | HCT 116 | C230(0.56); C236(0.56); C779(0.81); C251(1.02) | LDD0689 | [15] |
| LDCM0373 | CL104 | HCT 116 | C230(0.30); C236(0.30); C779(0.89); C598(1.02) | LDD0690 | [15] |
| LDCM0374 | CL105 | HCT 116 | C230(0.78); C236(0.78); C598(0.82); C779(0.91) | LDD0691 | [15] |
| LDCM0375 | CL106 | HCT 116 | C598(0.48); C230(0.51); C236(0.51); C448(0.74) | LDD0692 | [15] |
| LDCM0376 | CL107 | HCT 116 | C230(0.52); C236(0.52); C598(0.68); C448(0.94) | LDD0693 | [15] |
| LDCM0377 | CL108 | HCT 116 | C230(0.62); C236(0.62); C598(0.68); C448(0.81) | LDD0694 | [15] |
| LDCM0378 | CL109 | HCT 116 | C598(0.83); C448(0.87); C251(0.91); C779(1.06) | LDD0695 | [15] |
| LDCM0379 | CL11 | HCT 116 | C598(0.49); C230(0.82); C236(0.82); C448(0.85) | LDD0696 | [15] |
| LDCM0380 | CL110 | HCT 116 | C598(0.61); C230(0.68); C236(0.68); C448(0.79) | LDD0697 | [15] |
| LDCM0381 | CL111 | HCT 116 | C598(0.77); C251(0.78); C448(0.88); C779(1.00) | LDD0698 | [15] |
| LDCM0382 | CL112 | HCT 116 | C779(0.67); C598(0.76); C448(0.91); C251(1.06) | LDD0699 | [15] |
| LDCM0383 | CL113 | HCT 116 | C448(0.63); C598(0.65); C779(0.86); C251(1.00) | LDD0700 | [15] |
| LDCM0384 | CL114 | HCT 116 | C598(0.57); C448(0.58); C251(0.94); C779(0.94) | LDD0701 | [15] |
| LDCM0385 | CL115 | HCT 116 | C598(0.71); C448(0.72); C251(0.94); C779(1.22) | LDD0702 | [15] |
| LDCM0386 | CL116 | HCT 116 | C779(0.81); C448(0.87); C251(0.91); C598(0.94) | LDD0703 | [15] |
| LDCM0387 | CL117 | HCT 116 | C598(0.49); C448(0.62); C251(0.82); C779(1.17) | LDD0704 | [15] |
| LDCM0388 | CL118 | HCT 116 | C448(0.88); C251(0.95); C598(0.96); C779(1.05) | LDD0705 | [15] |
| LDCM0389 | CL119 | HCT 116 | C448(0.84); C598(0.99); C779(1.04); C251(1.08) | LDD0706 | [15] |
| LDCM0390 | CL12 | HCT 116 | C598(0.53); C230(0.70); C236(0.70); C448(0.83) | LDD0707 | [15] |
| LDCM0391 | CL120 | HCT 116 | C251(0.96); C448(0.98); C598(1.05); C779(1.06) | LDD0708 | [15] |
| LDCM0392 | CL121 | HCT 116 | C779(0.88); C251(0.95); C448(1.01); C598(1.19) | LDD0709 | [15] |
| LDCM0393 | CL122 | HCT 116 | C230(0.54); C236(0.54); C598(0.63); C448(0.65) | LDD0710 | [15] |
| LDCM0394 | CL123 | HCT 116 | C230(0.52); C236(0.52); C598(0.71); C448(0.74) | LDD0711 | [15] |
| LDCM0395 | CL124 | HCT 116 | C230(0.59); C236(0.59); C598(0.64); C779(0.69) | LDD0712 | [15] |
| LDCM0396 | CL125 | HCT 116 | C251(0.99); C448(0.99); C779(1.05); C230(1.34) | LDD0713 | [15] |
| LDCM0397 | CL126 | HCT 116 | C230(0.69); C236(0.69); C448(0.90); C779(0.94) | LDD0714 | [15] |
| LDCM0398 | CL127 | HCT 116 | C230(0.75); C236(0.75); C448(0.98); C251(1.08) | LDD0715 | [15] |
| LDCM0399 | CL128 | HCT 116 | C230(0.53); C236(0.53); C448(0.97); C251(1.00) | LDD0716 | [15] |
| LDCM0400 | CL13 | HCT 116 | C598(0.56); C448(0.89); C230(0.90); C236(0.90) | LDD0717 | [15] |
| LDCM0401 | CL14 | HCT 116 | C598(0.75); C448(1.07); C779(1.11); C251(1.23) | LDD0718 | [15] |
| LDCM0402 | CL15 | HCT 116 | C598(0.32); C448(0.83); C230(0.85); C236(0.85) | LDD0719 | [15] |
| LDCM0403 | CL16 | HCT 116 | C448(0.75); C230(0.83); C236(0.83); C251(0.95) | LDD0720 | [15] |
| LDCM0404 | CL17 | HCT 116 | C779(0.60); C598(0.87); C230(0.89); C236(0.89) | LDD0721 | [15] |
| LDCM0405 | CL18 | HCT 116 | C598(0.70); C779(0.80); C230(0.93); C236(0.93) | LDD0722 | [15] |
| LDCM0406 | CL19 | HCT 116 | C779(0.65); C598(0.71); C230(0.73); C236(0.73) | LDD0723 | [15] |
| LDCM0407 | CL2 | HCT 116 | C598(0.90); C448(0.93); C251(0.96); C230(0.98) | LDD0724 | [15] |
| LDCM0408 | CL20 | HCT 116 | C779(0.48); C230(0.70); C236(0.70); C598(0.79) | LDD0725 | [15] |
| LDCM0409 | CL21 | HCT 116 | C598(0.51); C230(0.58); C236(0.58); C779(0.67) | LDD0726 | [15] |
| LDCM0410 | CL22 | HCT 116 | C598(0.33); C230(0.64); C236(0.64); C779(0.72) | LDD0727 | [15] |
| LDCM0411 | CL23 | HCT 116 | C779(0.80); C251(0.87); C448(0.92); C598(0.92) | LDD0728 | [15] |
| LDCM0412 | CL24 | HCT 116 | C230(0.69); C236(0.69); C251(1.25); C448(0.89) | LDD0729 | [15] |
| LDCM0413 | CL25 | HCT 116 | C230(0.61); C236(0.61); C251(0.96); C448(0.90) | LDD0730 | [15] |
| LDCM0414 | CL26 | HCT 116 | C230(1.07); C236(1.07); C251(1.10); C448(0.76) | LDD0731 | [15] |
| LDCM0415 | CL27 | HCT 116 | C230(0.66); C236(0.66); C251(1.21); C448(1.00) | LDD0732 | [15] |
| LDCM0416 | CL28 | HCT 116 | C230(0.67); C236(0.67); C251(1.11); C448(0.97) | LDD0733 | [15] |
| LDCM0417 | CL29 | HCT 116 | C230(0.61); C236(0.61); C251(1.28); C448(0.99) | LDD0734 | [15] |
| LDCM0418 | CL3 | HCT 116 | C230(1.25); C236(1.25); C251(0.83); C448(0.98) | LDD0735 | [15] |
| LDCM0419 | CL30 | HCT 116 | C230(1.50); C236(1.50); C251(1.36); C448(0.99) | LDD0736 | [15] |
| LDCM0420 | CL31 | HCT 116 | C251(1.05); C448(1.01); C598(0.82); C779(0.96) | LDD0737 | [15] |
| LDCM0421 | CL32 | HCT 116 | C251(0.96); C448(0.73); C598(0.87); C779(0.73) | LDD0738 | [15] |
| LDCM0422 | CL33 | HCT 116 | C251(0.92); C448(0.76); C598(0.65); C779(0.63) | LDD0739 | [15] |
| LDCM0423 | CL34 | HCT 116 | C251(0.98); C448(0.66); C598(0.58); C779(0.64) | LDD0740 | [15] |
| LDCM0424 | CL35 | HCT 116 | C251(1.14); C448(0.75); C598(0.58); C779(0.97) | LDD0741 | [15] |
| LDCM0425 | CL36 | HCT 116 | C251(1.07); C448(0.89); C598(0.64); C779(0.89) | LDD0742 | [15] |
| LDCM0426 | CL37 | HCT 116 | C251(0.91); C448(0.75); C598(0.63); C779(1.02) | LDD0743 | [15] |
| LDCM0428 | CL39 | HCT 116 | C251(1.01); C448(0.83); C598(0.65); C779(0.74) | LDD0745 | [15] |
| LDCM0429 | CL4 | HCT 116 | C230(1.46); C236(1.46); C251(0.92); C448(1.01) | LDD0746 | [15] |
| LDCM0430 | CL40 | HCT 116 | C251(1.04); C448(0.79); C598(0.68); C779(0.79) | LDD0747 | [15] |
| LDCM0431 | CL41 | HCT 116 | C251(0.92); C448(0.87); C598(0.81); C779(0.93) | LDD0748 | [15] |
| LDCM0432 | CL42 | HCT 116 | C251(1.15); C448(0.63); C598(0.56); C779(0.84) | LDD0749 | [15] |
| LDCM0433 | CL43 | HCT 116 | C251(0.98); C448(0.69); C598(0.69); C779(0.65) | LDD0750 | [15] |
| LDCM0434 | CL44 | HCT 116 | C251(1.03); C448(0.74); C598(0.64); C779(0.69) | LDD0751 | [15] |
| LDCM0435 | CL45 | HCT 116 | C251(1.14); C448(0.73); C598(0.58); C779(0.64) | LDD0752 | [15] |
| LDCM0436 | CL46 | HCT 116 | C251(0.87); C448(0.88); C598(0.76); C779(1.00) | LDD0753 | [15] |
| LDCM0437 | CL47 | HCT 116 | C251(0.93); C448(0.84); C598(0.72); C779(0.96) | LDD0754 | [15] |
| LDCM0438 | CL48 | HCT 116 | C251(0.95); C448(1.00); C598(0.88); C779(0.91) | LDD0755 | [15] |
| LDCM0439 | CL49 | HCT 116 | C251(1.13); C448(1.14); C598(1.16); C779(0.88) | LDD0756 | [15] |
| LDCM0440 | CL5 | HCT 116 | C230(1.22); C236(1.22); C251(1.17); C448(1.06) | LDD0757 | [15] |
| LDCM0441 | CL50 | HCT 116 | C251(1.09); C448(0.98); C598(0.95); C779(0.90) | LDD0758 | [15] |
| LDCM0442 | CL51 | HCT 116 | C251(1.01); C448(1.13); C598(0.81); C779(0.92) | LDD0759 | [15] |
| LDCM0443 | CL52 | HCT 116 | C251(0.87); C448(0.94); C598(0.81); C779(1.00) | LDD0760 | [15] |
| LDCM0444 | CL53 | HCT 116 | C251(0.88); C448(0.68); C598(0.77); C779(0.98) | LDD0761 | [15] |
| LDCM0445 | CL54 | HCT 116 | C251(0.86); C448(0.89); C598(0.69); C779(0.85) | LDD0762 | [15] |
| LDCM0446 | CL55 | HCT 116 | C251(1.18); C448(1.01); C598(0.86); C779(0.87) | LDD0763 | [15] |
| LDCM0447 | CL56 | HCT 116 | C251(0.90); C448(0.83); C598(0.73); C779(0.90) | LDD0764 | [15] |
| LDCM0448 | CL57 | HCT 116 | C251(1.10); C448(0.94); C598(0.92); C779(0.88) | LDD0765 | [15] |
| LDCM0449 | CL58 | HCT 116 | C251(0.89); C448(1.03); C598(0.98); C779(0.86) | LDD0766 | [15] |
| LDCM0450 | CL59 | HCT 116 | C251(1.09); C448(1.21); C598(0.94); C779(0.95) | LDD0767 | [15] |
| LDCM0451 | CL6 | HCT 116 | C230(1.00); C236(1.00); C251(0.98); C448(0.82) | LDD0768 | [15] |
| LDCM0452 | CL60 | HCT 116 | C251(1.03); C448(0.99); C598(0.81); C779(0.85) | LDD0769 | [15] |
| LDCM0453 | CL61 | HCT 116 | C230(0.72); C236(0.72); C251(1.07); C448(0.84) | LDD0770 | [15] |
| LDCM0454 | CL62 | HCT 116 | C230(0.83); C236(0.83); C251(1.05); C448(0.89) | LDD0771 | [15] |
| LDCM0455 | CL63 | HCT 116 | C230(0.59); C236(0.59); C251(1.18); C448(0.99) | LDD0772 | [15] |
| LDCM0456 | CL64 | HCT 116 | C230(0.60); C236(0.60); C251(1.03); C448(1.08) | LDD0773 | [15] |
| LDCM0457 | CL65 | HCT 116 | C230(0.58); C236(0.58); C251(1.17); C448(1.11) | LDD0774 | [15] |
| LDCM0458 | CL66 | HCT 116 | C230(0.46); C236(0.46); C251(0.90); C448(0.97) | LDD0775 | [15] |
| LDCM0459 | CL67 | HCT 116 | C230(0.53); C236(0.53); C251(0.95); C448(1.09) | LDD0776 | [15] |
| LDCM0460 | CL68 | HCT 116 | C230(0.50); C236(0.50); C251(0.97); C448(0.63) | LDD0777 | [15] |
| LDCM0461 | CL69 | HCT 116 | C230(1.00); C236(1.00); C251(0.90); C448(0.69) | LDD0778 | [15] |
| LDCM0462 | CL7 | HCT 116 | C230(0.69); C236(0.69); C251(1.25); C448(0.97) | LDD0779 | [15] |
| LDCM0463 | CL70 | HCT 116 | C230(0.80); C236(0.80); C251(1.02); C448(0.95) | LDD0780 | [15] |
| LDCM0464 | CL71 | HCT 116 | C230(0.57); C236(0.57); C251(1.11); C448(0.89) | LDD0781 | [15] |
| LDCM0465 | CL72 | HCT 116 | C230(0.67); C236(0.67); C251(1.05); C448(0.67) | LDD0782 | [15] |
| LDCM0466 | CL73 | HCT 116 | C230(0.93); C236(0.93); C251(1.19); C448(0.89) | LDD0783 | [15] |
| LDCM0467 | CL74 | HCT 116 | C230(0.79); C236(0.79); C251(0.98); C448(0.95) | LDD0784 | [15] |
| LDCM0469 | CL76 | HCT 116 | C230(1.13); C236(1.13); C251(1.11); C448(0.91) | LDD0786 | [15] |
| LDCM0470 | CL77 | HCT 116 | C230(3.35); C236(3.35); C251(0.97); C448(0.76) | LDD0787 | [15] |
| LDCM0471 | CL78 | HCT 116 | C230(1.04); C236(1.04); C251(1.04); C448(0.86) | LDD0788 | [15] |
| LDCM0472 | CL79 | HCT 116 | C230(0.76); C236(0.76); C251(1.28); C448(0.91) | LDD0789 | [15] |
| LDCM0473 | CL8 | HCT 116 | C230(0.79); C236(0.79); C251(0.83); C448(0.73) | LDD0790 | [15] |
| LDCM0474 | CL80 | HCT 116 | C230(1.33); C236(1.33); C251(1.15); C448(0.89) | LDD0791 | [15] |
| LDCM0475 | CL81 | HCT 116 | C230(0.95); C236(0.95); C251(1.21); C448(0.96) | LDD0792 | [15] |
| LDCM0476 | CL82 | HCT 116 | C230(0.67); C236(0.67); C251(0.98); C448(0.91) | LDD0793 | [15] |
| LDCM0477 | CL83 | HCT 116 | C230(0.69); C236(0.69); C251(1.16); C448(0.86) | LDD0794 | [15] |
| LDCM0478 | CL84 | HCT 116 | C230(0.64); C236(0.64); C251(0.90); C448(0.67) | LDD0795 | [15] |
| LDCM0479 | CL85 | HCT 116 | C230(0.85); C236(0.85); C251(1.20); C448(0.88) | LDD0796 | [15] |
| LDCM0480 | CL86 | HCT 116 | C230(0.95); C236(0.95); C251(1.43); C448(0.96) | LDD0797 | [15] |
| LDCM0481 | CL87 | HCT 116 | C230(0.68); C236(0.68); C251(1.29); C448(1.15) | LDD0798 | [15] |
| LDCM0482 | CL88 | HCT 116 | C230(0.73); C236(0.73); C251(1.21); C448(0.97) | LDD0799 | [15] |
| LDCM0483 | CL89 | HCT 116 | C230(0.57); C236(0.57); C251(0.97); C448(0.69) | LDD0800 | [15] |
| LDCM0484 | CL9 | HCT 116 | C230(1.10); C236(1.10); C251(1.25); C448(0.90) | LDD0801 | [15] |
| LDCM0485 | CL90 | HCT 116 | C230(1.32); C236(1.32); C251(1.34); C448(1.03) | LDD0802 | [15] |
| LDCM0486 | CL91 | HCT 116 | C230(0.71); C236(0.71); C251(0.86); C448(0.79) | LDD0803 | [15] |
| LDCM0487 | CL92 | HCT 116 | C230(0.67); C236(0.67); C251(0.94); C448(0.96) | LDD0804 | [15] |
| LDCM0488 | CL93 | HCT 116 | C230(0.76); C236(0.76); C251(1.22); C448(1.36) | LDD0805 | [15] |
| LDCM0489 | CL94 | HCT 116 | C230(0.62); C236(0.62); C251(1.04); C448(1.25) | LDD0806 | [15] |
| LDCM0490 | CL95 | HCT 116 | C230(0.57); C236(0.57); C251(1.03); C448(1.01) | LDD0807 | [15] |
| LDCM0491 | CL96 | HCT 116 | C230(0.66); C236(0.66); C251(1.12); C448(1.05) | LDD0808 | [15] |
| LDCM0492 | CL97 | HCT 116 | C230(0.72); C236(0.72); C251(1.01); C448(0.94) | LDD0809 | [15] |
| LDCM0493 | CL98 | HCT 116 | C230(0.65); C236(0.65); C251(1.02); C448(0.97) | LDD0810 | [15] |
| LDCM0494 | CL99 | HCT 116 | C230(0.59); C236(0.59); C251(0.94); C448(1.06) | LDD0811 | [15] |
| LDCM0495 | E2913 | HEK-293T | C779(1.04); C448(0.94); C459(1.08); C598(0.96) | LDD1698 | [35] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C251(4.93); C448(2.89); C459(1.42); C779(1.11) | LDD1702 | [10] |
| LDCM0625 | F8 | Ramos | C251(0.56); C779(0.65); C459(1.44) | LDD2187 | [36] |
| LDCM0572 | Fragment10 | Ramos | C251(0.96); C779(1.50); C459(1.00) | LDD2189 | [36] |
| LDCM0573 | Fragment11 | Ramos | C251(0.71); C779(0.03); C459(0.19) | LDD2190 | [36] |
| LDCM0574 | Fragment12 | Ramos | C251(0.94); C779(0.81); C459(1.36) | LDD2191 | [36] |
| LDCM0575 | Fragment13 | Ramos | C251(1.05); C779(1.04); C459(1.09) | LDD2192 | [36] |
| LDCM0576 | Fragment14 | Ramos | C251(0.51); C779(0.59); C459(1.60) | LDD2193 | [36] |
| LDCM0579 | Fragment20 | Ramos | C251(1.19); C779(0.56); C459(0.69) | LDD2194 | [36] |
| LDCM0580 | Fragment21 | Ramos | C251(0.81); C779(0.99); C459(0.96) | LDD2195 | [36] |
| LDCM0582 | Fragment23 | Ramos | C251(1.53); C779(1.50); C459(2.92) | LDD2196 | [36] |
| LDCM0578 | Fragment27 | Ramos | C251(0.83); C779(0.95); C459(1.49) | LDD2197 | [36] |
| LDCM0586 | Fragment28 | Ramos | C251(0.86); C779(1.22); C459(0.49) | LDD2198 | [36] |
| LDCM0588 | Fragment30 | Ramos | C251(1.26); C779(0.97); C459(1.06) | LDD2199 | [36] |
| LDCM0589 | Fragment31 | Ramos | C251(1.48); C779(0.66) | LDD2200 | [36] |
| LDCM0590 | Fragment32 | Ramos | C251(0.63); C779(0.55); C459(0.71) | LDD2201 | [36] |
| LDCM0468 | Fragment33 | HCT 116 | C230(0.60); C236(0.60); C251(1.02); C448(0.95) | LDD0785 | [15] |
| LDCM0596 | Fragment38 | Ramos | C251(1.28); C779(0.78); C459(0.83) | LDD2203 | [36] |
| LDCM0566 | Fragment4 | Ramos | C251(0.71); C779(0.47) | LDD2184 | [36] |
| LDCM0427 | Fragment51 | HCT 116 | C251(0.94); C448(0.66); C598(0.58); C779(0.67) | LDD0744 | [15] |
| LDCM0610 | Fragment52 | Ramos | C251(1.88); C779(1.71); C459(1.46) | LDD2204 | [36] |
| LDCM0614 | Fragment56 | Ramos | C251(1.28); C779(1.01); C459(0.68) | LDD2205 | [36] |
| LDCM0569 | Fragment7 | Ramos | C251(0.60); C779(1.03) | LDD2186 | [36] |
| LDCM0571 | Fragment9 | Ramos | C251(0.85); C779(2.24); C459(0.88) | LDD2188 | [36] |
| LDCM0116 | HHS-0101 | DM93 | Y400(0.71); Y439(0.86); Y751(1.32); Y469(1.59) | LDD0264 | [13] |
| LDCM0117 | HHS-0201 | DM93 | Y400(0.78); Y469(1.07); Y751(1.26); Y439(1.45) | LDD0265 | [13] |
| LDCM0118 | HHS-0301 | DM93 | Y400(0.87); Y469(1.11); Y751(2.37) | LDD0266 | [13] |
| LDCM0119 | HHS-0401 | DM93 | Y400(0.79); Y439(0.92); Y469(0.97); Y751(1.41) | LDD0267 | [13] |
| LDCM0120 | HHS-0701 | DM93 | Y400(0.95); Y439(0.98); Y469(1.04); Y751(1.10) | LDD0268 | [13] |
| LDCM0107 | IAA | HeLa | C616(0.00); H161(0.00); C448(0.00); H677(0.00) | LDD0221 | [28] |
| LDCM0123 | JWB131 | DM93 | Y469(1.44) | LDD0285 | [12] |
| LDCM0124 | JWB142 | DM93 | Y469(0.65) | LDD0286 | [12] |
| LDCM0125 | JWB146 | DM93 | Y469(0.97) | LDD0287 | [12] |
| LDCM0126 | JWB150 | DM93 | Y469(3.84) | LDD0288 | [12] |
| LDCM0127 | JWB152 | DM93 | Y469(3.21) | LDD0289 | [12] |
| LDCM0128 | JWB198 | DM93 | Y469(2.58) | LDD0290 | [12] |
| LDCM0129 | JWB202 | DM93 | Y469(0.36) | LDD0291 | [12] |
| LDCM0130 | JWB211 | DM93 | Y469(0.94) | LDD0292 | [12] |
| LDCM0022 | KB02 | HCT 116 | C251(1.80) | LDD0080 | [15] |
| LDCM0023 | KB03 | HCT 116 | C251(2.31) | LDD0081 | [15] |
| LDCM0024 | KB05 | HCT 116 | C251(2.46) | LDD0082 | [15] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C779(1.56); C448(0.79) | LDD2102 | [10] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C779(0.68); C448(1.36) | LDD2121 | [10] |
| LDCM0109 | NEM | HeLa | H156(0.00); H876(0.00); H161(0.00); H677(0.00) | LDD0223 | [28] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C779(0.75); C251(0.58); C448(0.84) | LDD2089 | [10] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C779(1.15); C448(1.12) | LDD2090 | [10] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C779(1.01) | LDD2092 | [10] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C779(1.62); C448(1.13); C616(1.60) | LDD2093 | [10] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C779(1.44); C616(2.17) | LDD2094 | [10] |
| LDCM0503 | Nucleophilic fragment 14b | MDA-MB-231 | C779(0.88); C448(0.18) | LDD2096 | [10] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C779(0.98); C448(1.01); C616(1.52) | LDD2097 | [10] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C779(0.72) | LDD2098 | [10] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C779(1.06); C448(1.31); C616(1.53) | LDD2099 | [10] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C779(0.70) | LDD2100 | [10] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C448(1.27); C616(1.02) | LDD2101 | [10] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C779(0.76); C251(0.43) | LDD2104 | [10] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C779(1.24) | LDD2105 | [10] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C779(0.71); C251(0.36); C448(0.53) | LDD2106 | [10] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C779(0.95); C448(1.26); C616(1.52) | LDD2107 | [10] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C448(0.41) | LDD2108 | [10] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C779(1.08); C616(1.12) | LDD2109 | [10] |
| LDCM0517 | Nucleophilic fragment 21b | MDA-MB-231 | C779(2.10) | LDD2110 | [10] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C779(1.17); C448(1.20); C616(1.89) | LDD2111 | [10] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C779(0.99); C616(0.64) | LDD2114 | [10] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C779(0.91); C448(0.52); C616(0.43) | LDD2115 | [10] |
| LDCM0523 | Nucleophilic fragment 24b | MDA-MB-231 | C779(1.09); C616(0.26) | LDD2116 | [10] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C779(1.10); C448(0.42); C616(0.28) | LDD2118 | [10] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C779(2.27); C448(2.60); C616(1.42) | LDD2119 | [10] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C779(0.79); C448(0.53) | LDD2120 | [10] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C779(1.10); C448(0.22); C616(0.22) | LDD2122 | [10] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C779(1.31); C251(1.07); C448(0.86); C616(1.36) | LDD2123 | [10] |
| LDCM0531 | Nucleophilic fragment 28b | MDA-MB-231 | C779(1.37); C448(0.24); C616(0.28) | LDD2124 | [10] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C779(1.11); C251(1.11); C448(0.90); C616(1.45) | LDD2125 | [10] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C779(1.78); C448(0.27); C616(0.19) | LDD2126 | [10] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C779(1.26); C251(0.93); C448(1.07); C616(2.44) | LDD2127 | [10] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C779(0.99) | LDD2128 | [10] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C779(1.12); C448(1.47); C616(0.98) | LDD2129 | [10] |
| LDCM0540 | Nucleophilic fragment 35 | MDA-MB-231 | C779(0.99); C448(0.78); C616(0.54) | LDD2133 | [10] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C779(0.78); C448(0.61); C616(0.59) | LDD2134 | [10] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C779(1.11); C448(1.59); C616(0.62) | LDD2135 | [10] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C779(1.41); C448(1.59) | LDD2136 | [10] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C779(1.16); C251(0.60); C616(1.16) | LDD2137 | [10] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C779(1.81) | LDD1700 | [10] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C779(0.95); C448(0.96); C616(1.10) | LDD2140 | [10] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C779(0.64); C448(0.66) | LDD2141 | [10] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C779(0.99); C448(0.53) | LDD2143 | [10] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C779(2.39); C448(2.04); C616(1.25) | LDD2144 | [10] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C779(1.11); C448(1.18); C616(1.54) | LDD2146 | [10] |
| LDCM0553 | Nucleophilic fragment 6b | MDA-MB-231 | C779(1.82) | LDD2147 | [10] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C448(0.48); C616(0.41) | LDD2148 | [10] |
| LDCM0555 | Nucleophilic fragment 7b | MDA-MB-231 | C779(1.30); C448(0.33); C616(0.37) | LDD2149 | [10] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C779(0.56); C448(0.57) | LDD2150 | [10] |
| LDCM0557 | Nucleophilic fragment 8b | MDA-MB-231 | C779(1.33); C448(0.32) | LDD2151 | [10] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C779(1.90); C448(1.35) | LDD2153 | [10] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C779(0.91) | LDD2206 | [37] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C779(0.86) | LDD2207 | [37] |
| LDCM0131 | RA190 | MM1.R | C251(3.58); C779(1.21); C459(1.19) | LDD0304 | [38] |
| LDCM0021 | THZ1 | HCT 116 | C251(1.24); C448(1.10); C230(0.95); C236(0.95) | LDD2173 | [15] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Calreticulin (CALR) | Calreticulin family | P27797 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Zinc finger and BTB domain-containing protein 39 (ZBTB39) | Krueppel C2H2-type zinc-finger protein family | O15060 | |||
Other
References
