Details of the Target
General Information of Target
| Target ID | LDTP06501 | |||||
|---|---|---|---|---|---|---|
| Target Name | Ras-related protein Rab-11B (RAB11B) | |||||
| Gene Name | RAB11B | |||||
| Gene ID | 9230 | |||||
| Synonyms |
YPT3; Ras-related protein Rab-11B; EC 3.6.5.2; GTP-binding protein YPT3 |
|||||
| 3D Structure | ||||||
| Sequence |
MGTRDDEYDYLFKVVLIGDSGVGKSNLLSRFTRNEFNLESKSTIGVEFATRSIQVDGKTI
KAQIWDTAGQERYRAITSAYYRGAVGALLVYDIAKHLTYENVERWLKELRDHADSNIVIM LVGNKSDLRHLRAVPTDEARAFAEKNNLSFIETSALDSTNVEEAFKNILTEIYRIVSQKQ IADRAAHDESPGNNVVDISVPPTTDGQKPNKLQCCQNL |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Small GTPase superfamily, Rab family
|
|||||
| Subcellular location |
Recycling endosome membrane
|
|||||
| Function |
The small GTPases Rab are key regulators of intracellular membrane trafficking, from the formation of transport vesicles to their fusion with membranes. Rabs cycle between an inactive GDP-bound form and an active GTP-bound form that is able to recruit to membranes different set of downstream effectors directly responsible for vesicle formation, movement, tethering and fusion. The small Rab GTPase RAB11B plays a role in endocytic recycling, regulating apical recycling of several transmembrane proteins including cystic fibrosis transmembrane conductance regulator/CFTR, epithelial sodium channel/ENaC, potassium voltage-gated channel, and voltage-dependent L-type calcium channel. May also regulate constitutive and regulated secretion, like insulin granule exocytosis. Required for melanosome transport and release from melanocytes. Also regulates V-ATPase intracellular transport in response to extracellular acidosis. Promotes Rabin8/RAB3IP preciliary vesicular trafficking to mother centriole by forming a ciliary targeting complex containing Rab11, ASAP1, Rabin8/RAB3IP, RAB11FIP3 and ARF4, thereby regulating ciliogenesis initiation (Probable). On the contrary, upon LPAR1 receptor signaling pathway activation, interaction with phosphorylated WDR44 prevents Rab11-RAB3IP-RAB11FIP3 complex formation and cilia growth (Probable).
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
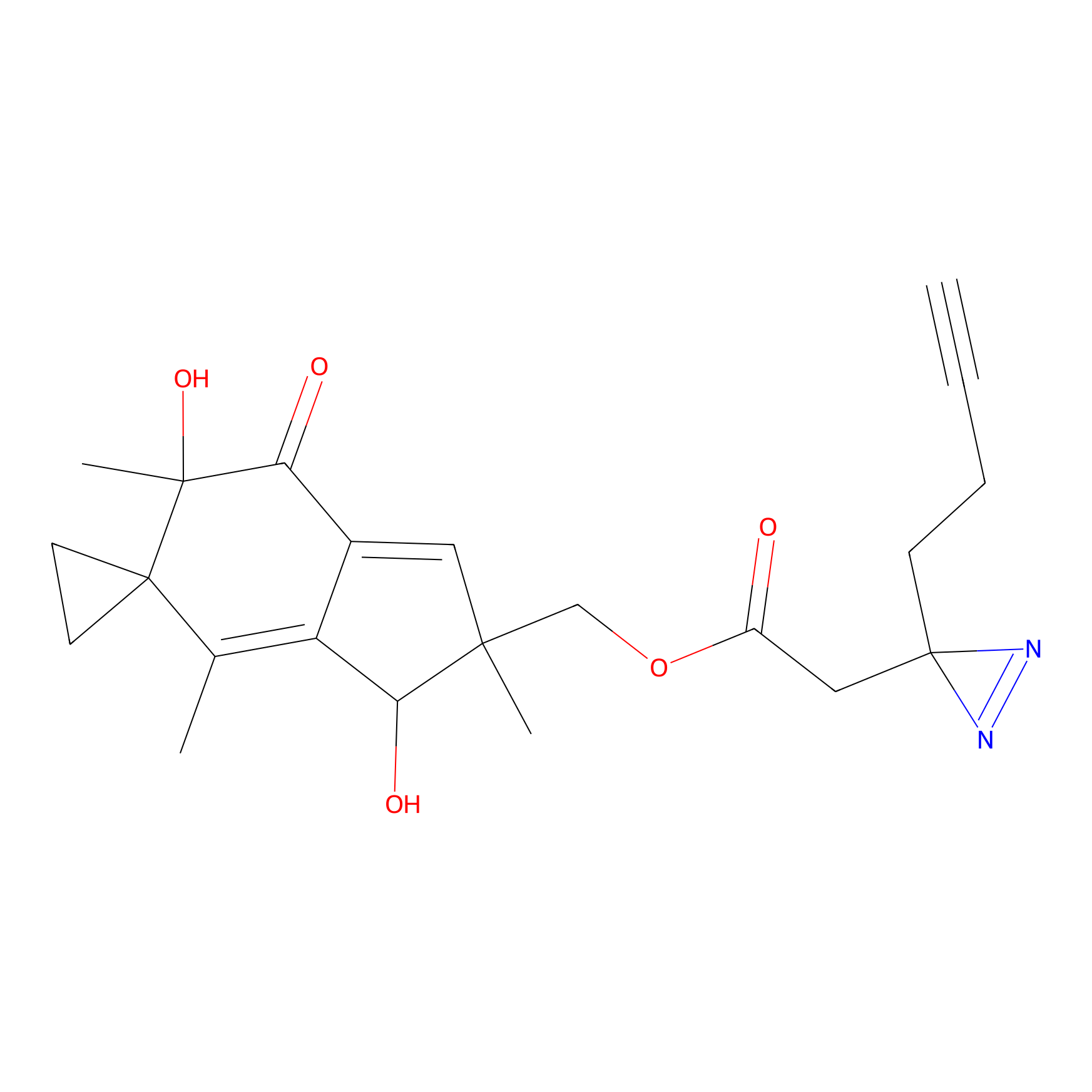
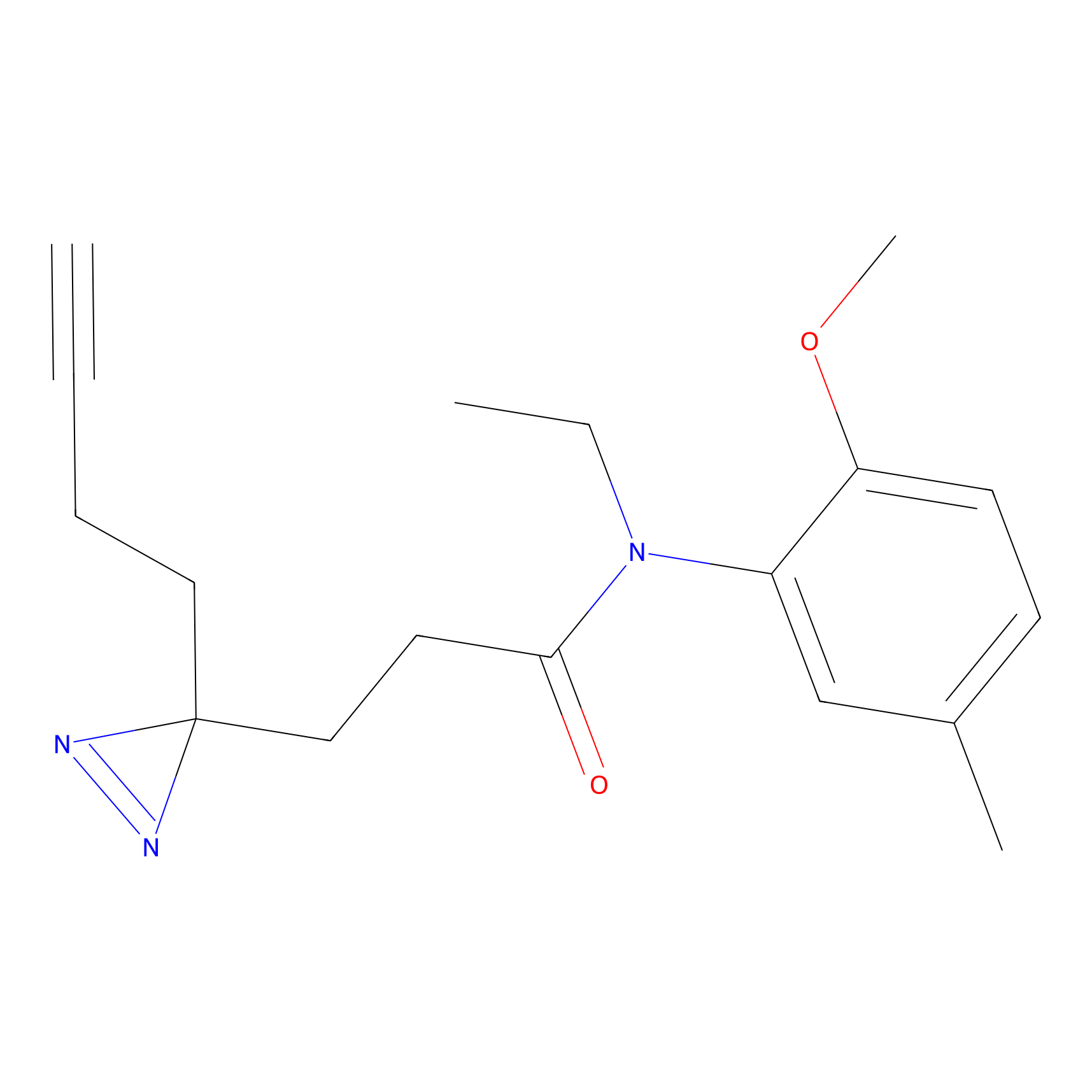
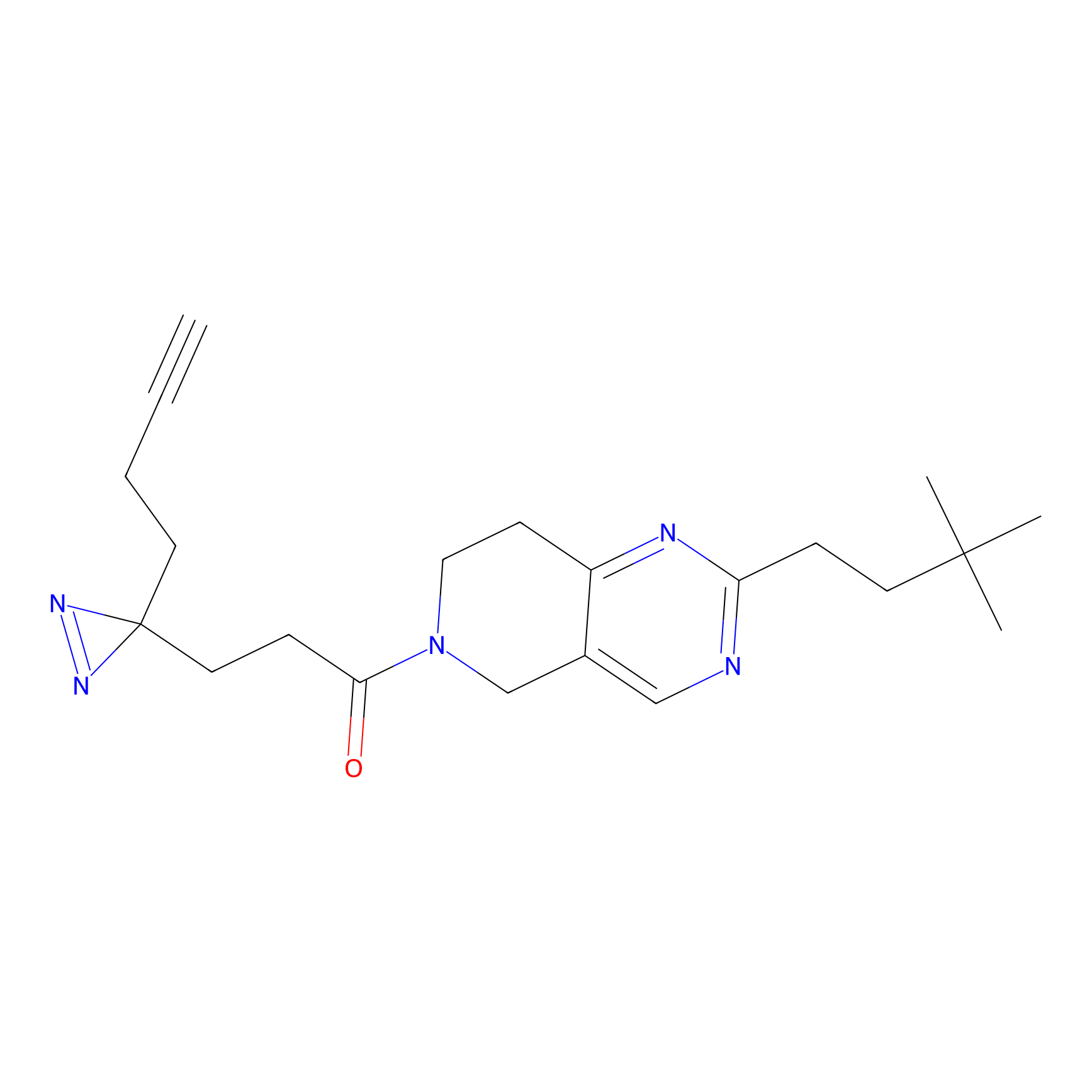
FBPP2 Probe Info |
 |
4.73 | LDD0318 | [1] | |
|
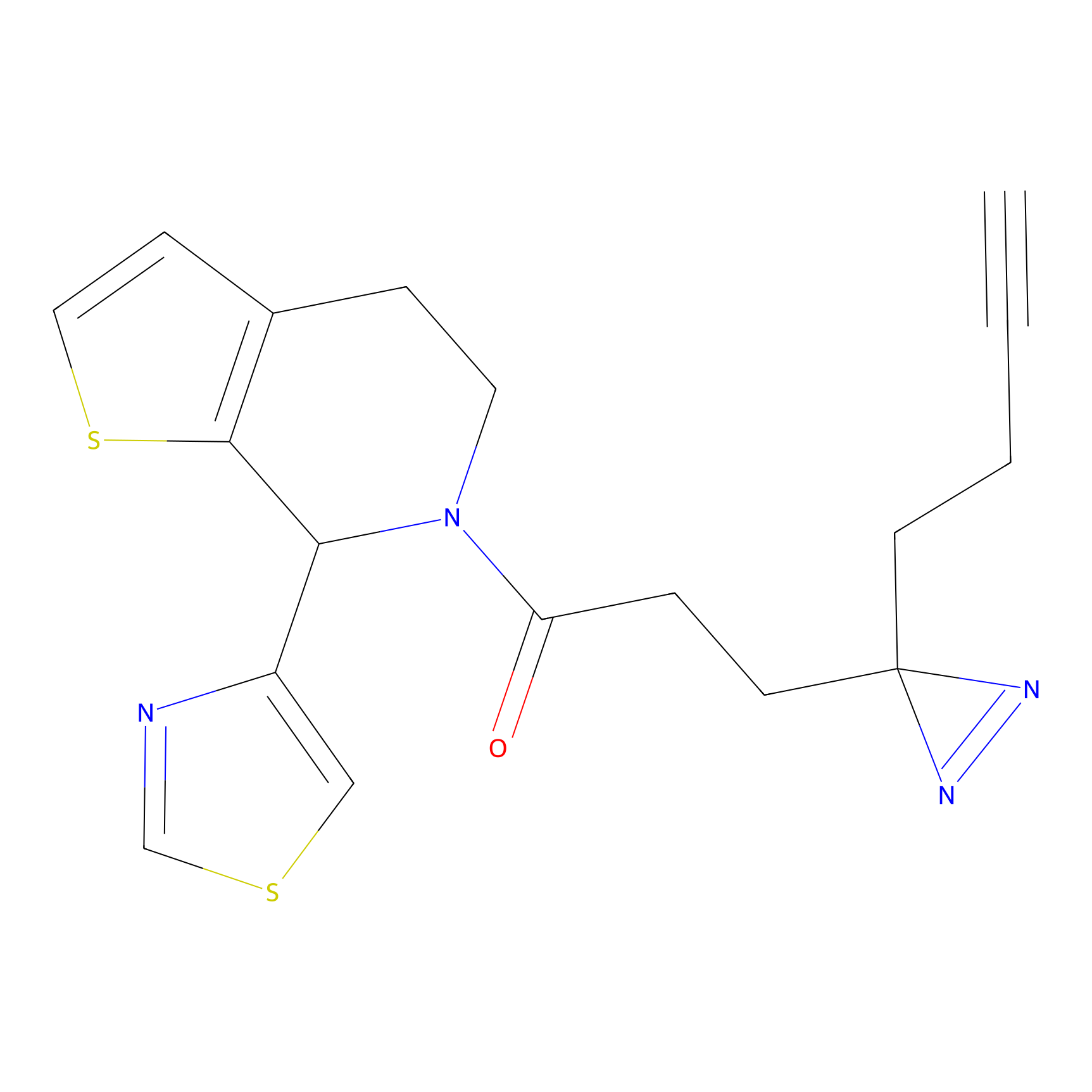
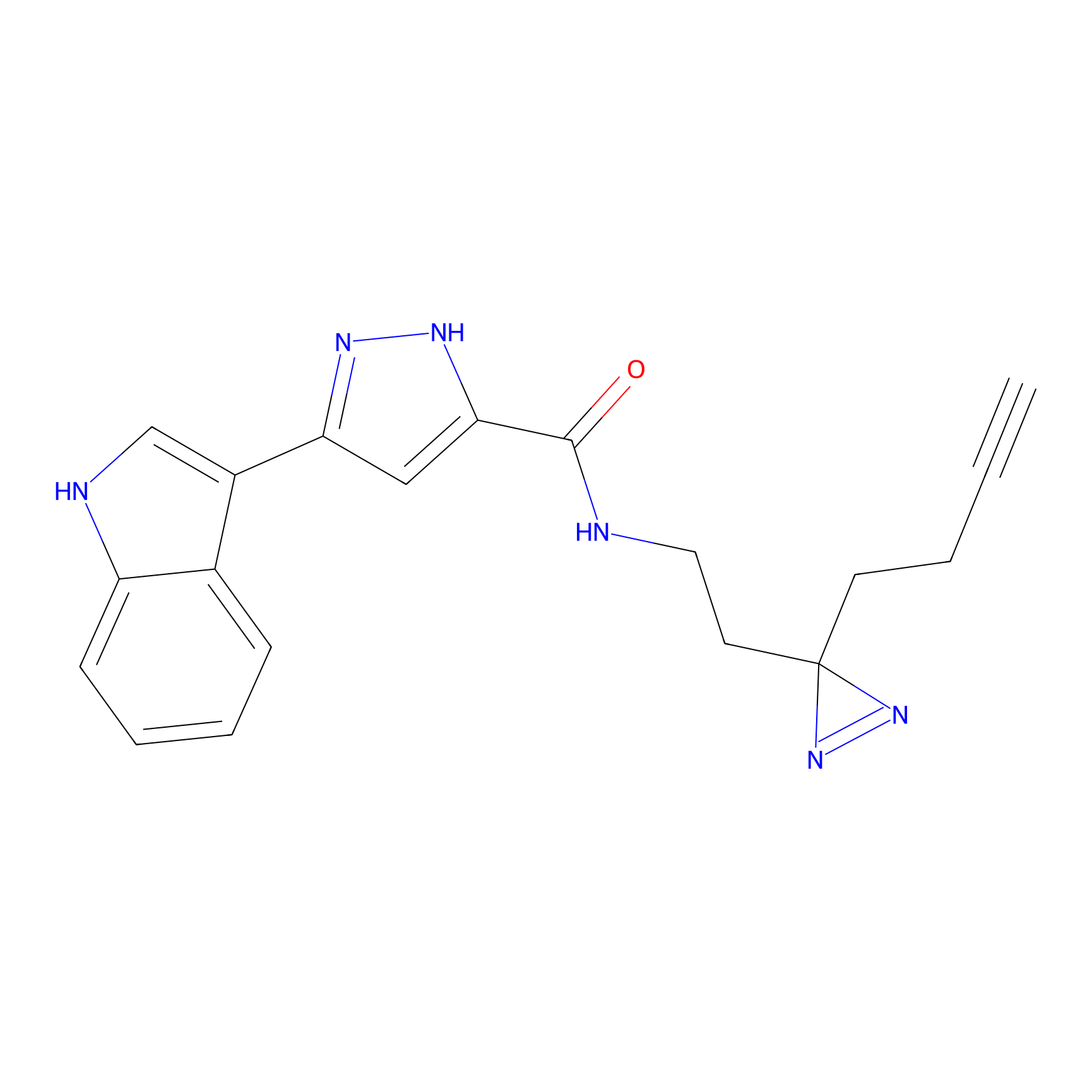
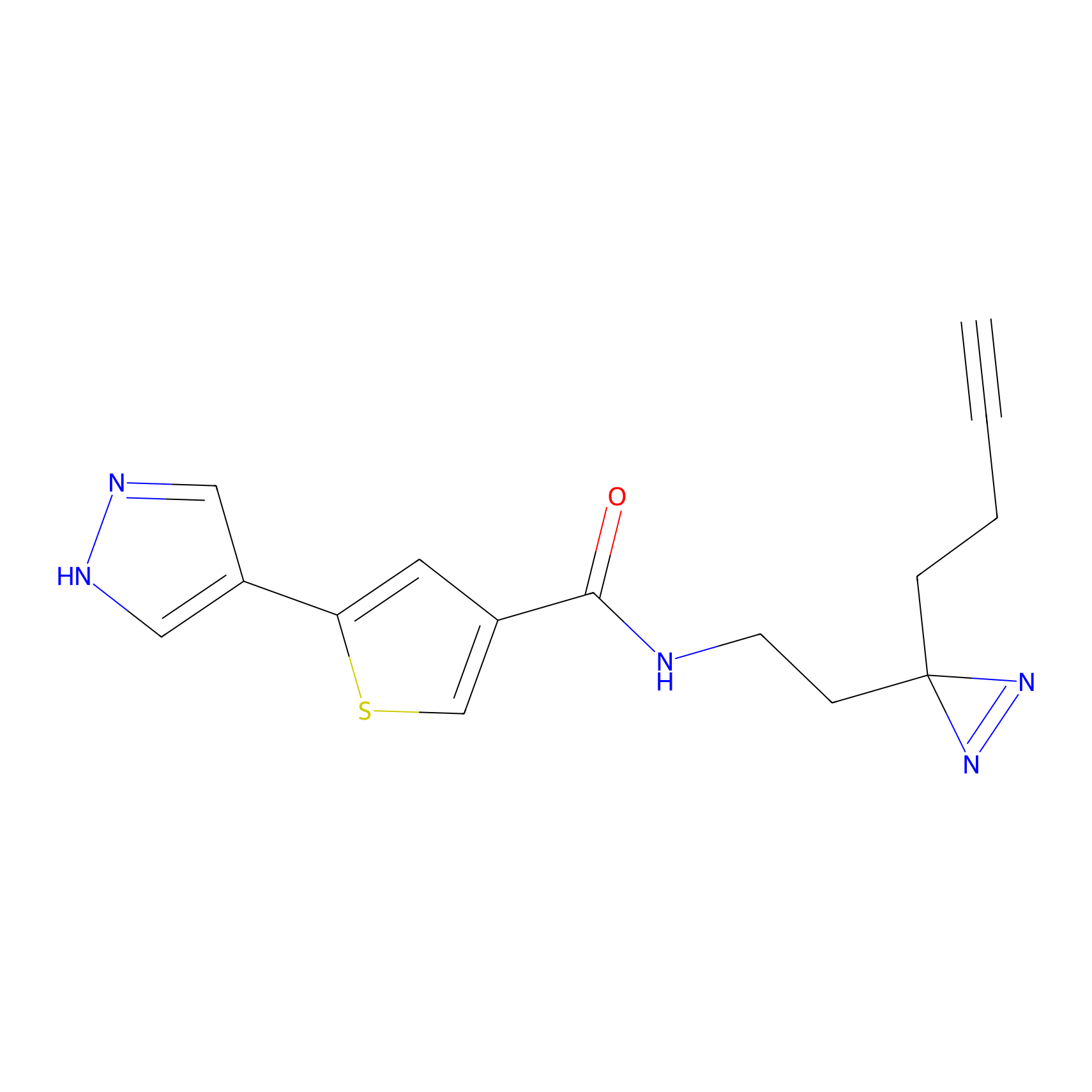
TH211 Probe Info |
 |
Y91(7.45) | LDD0260 | [2] | |
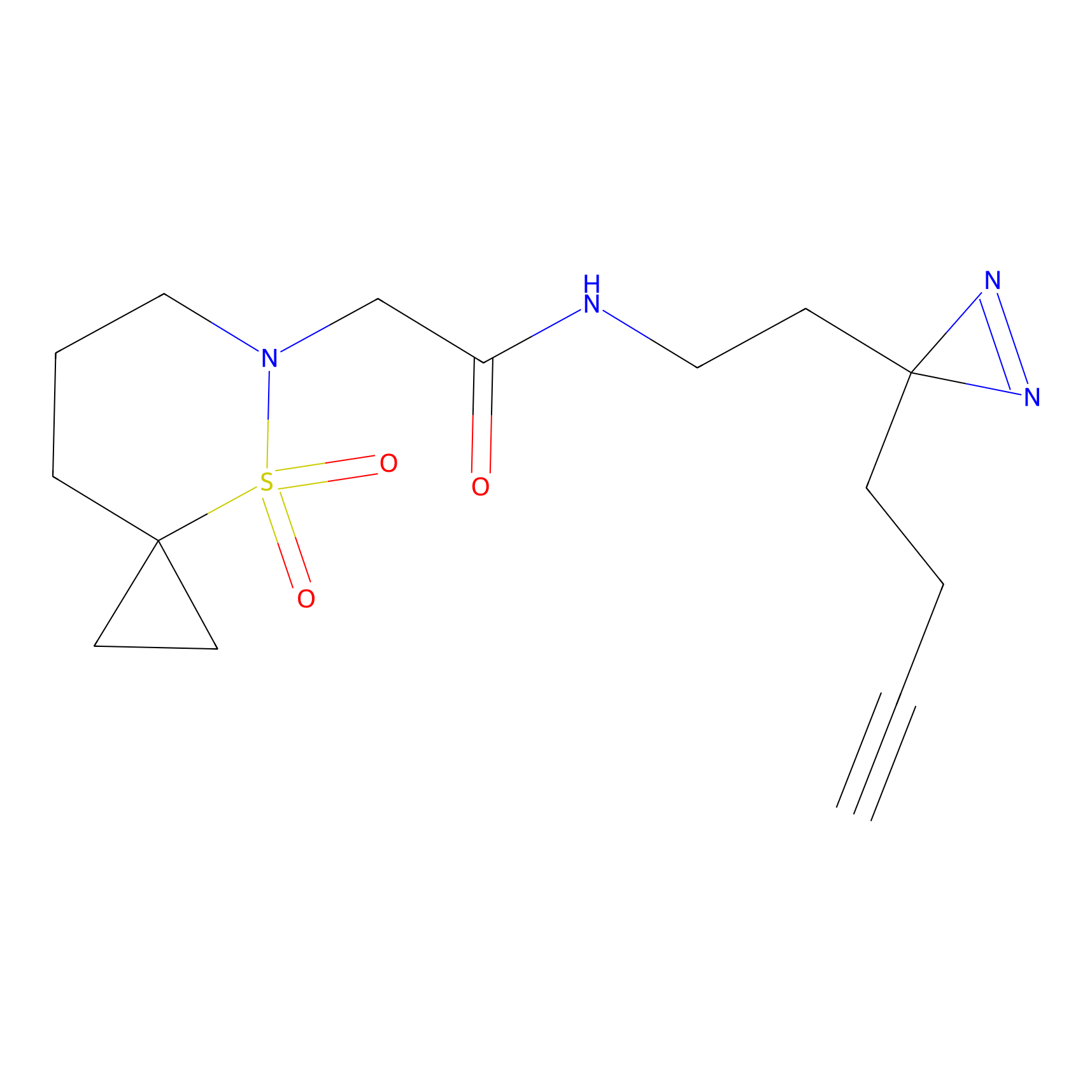
|
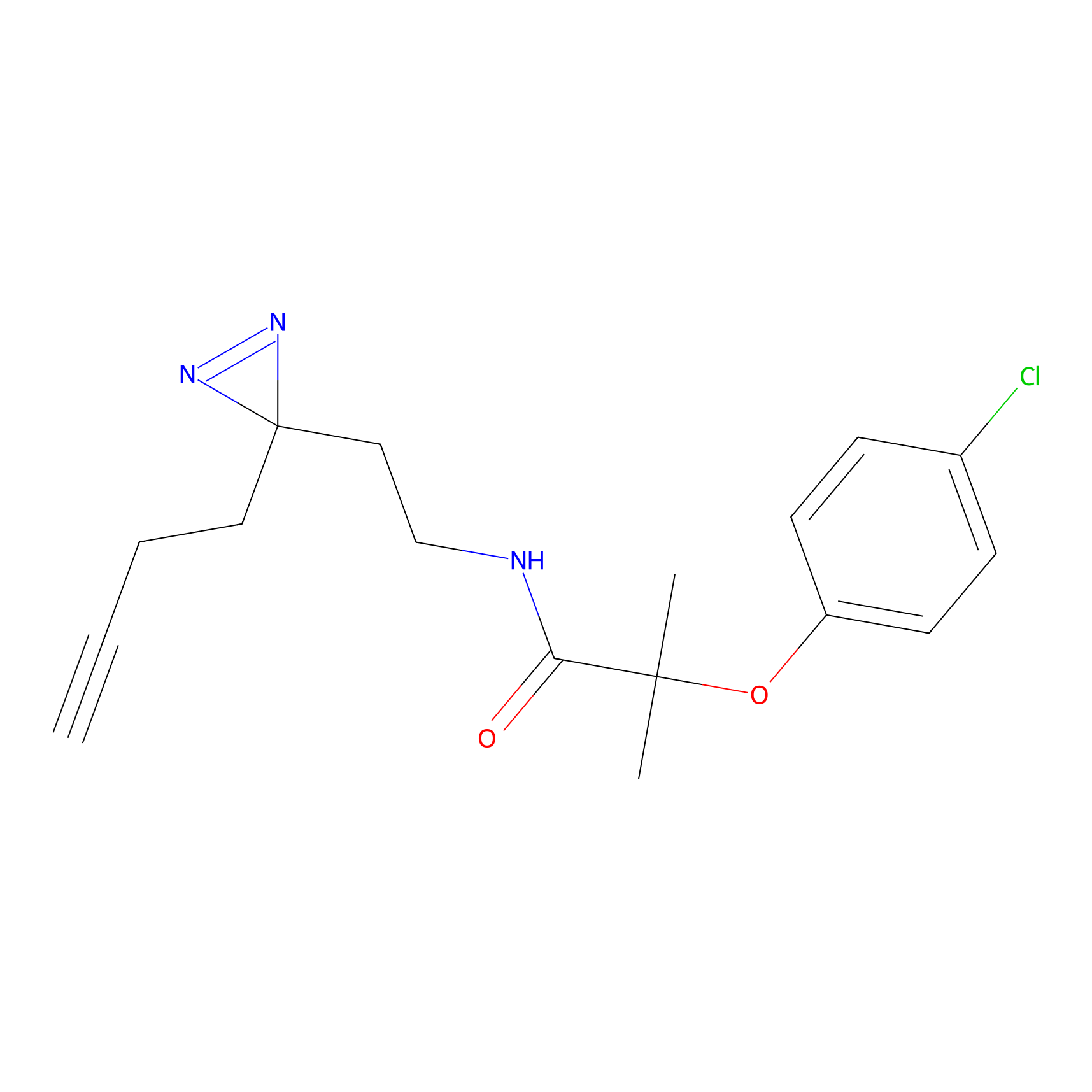
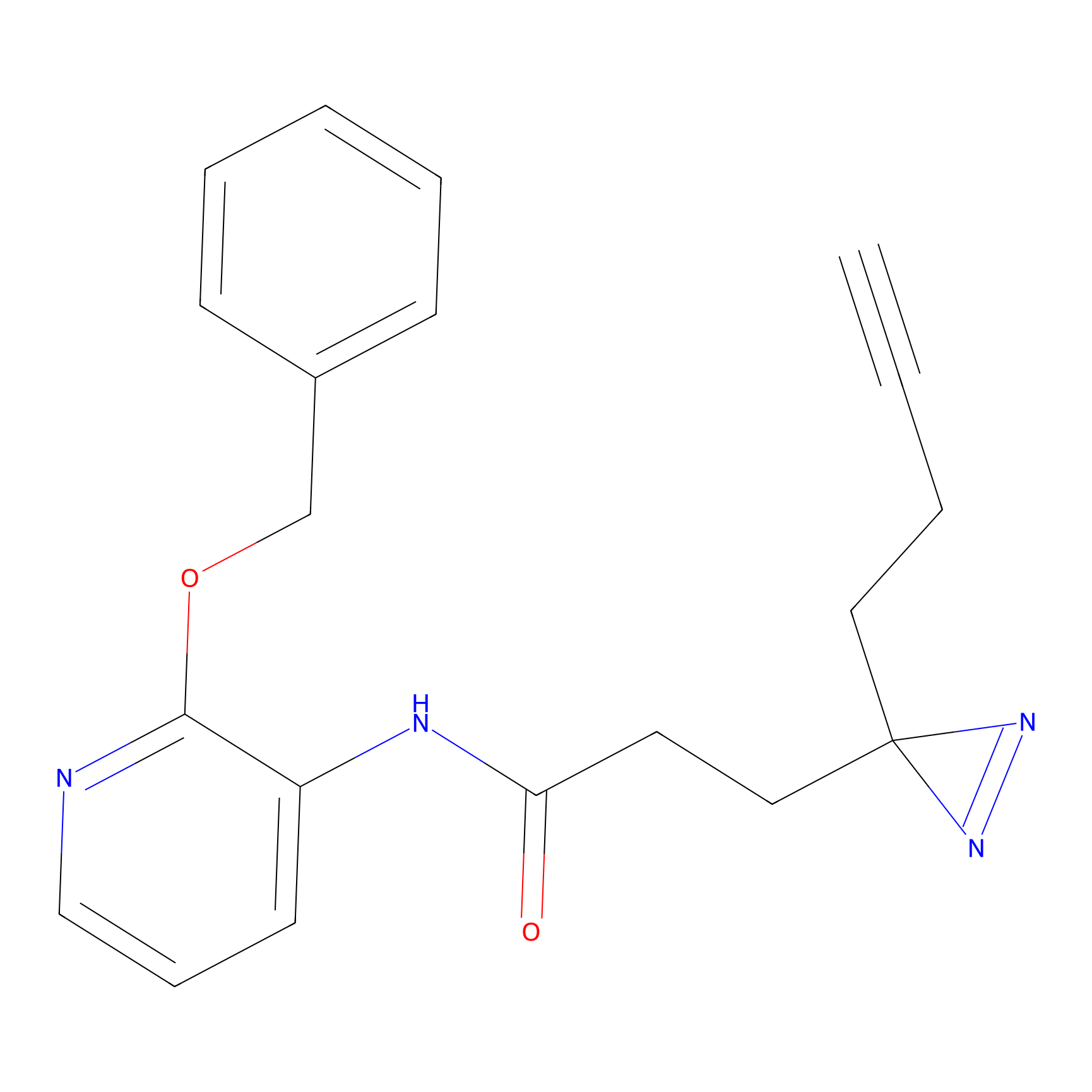
ONAyne Probe Info |
 |
K179(3.12); K58(1.08) | LDD0274 | [3] | |
|
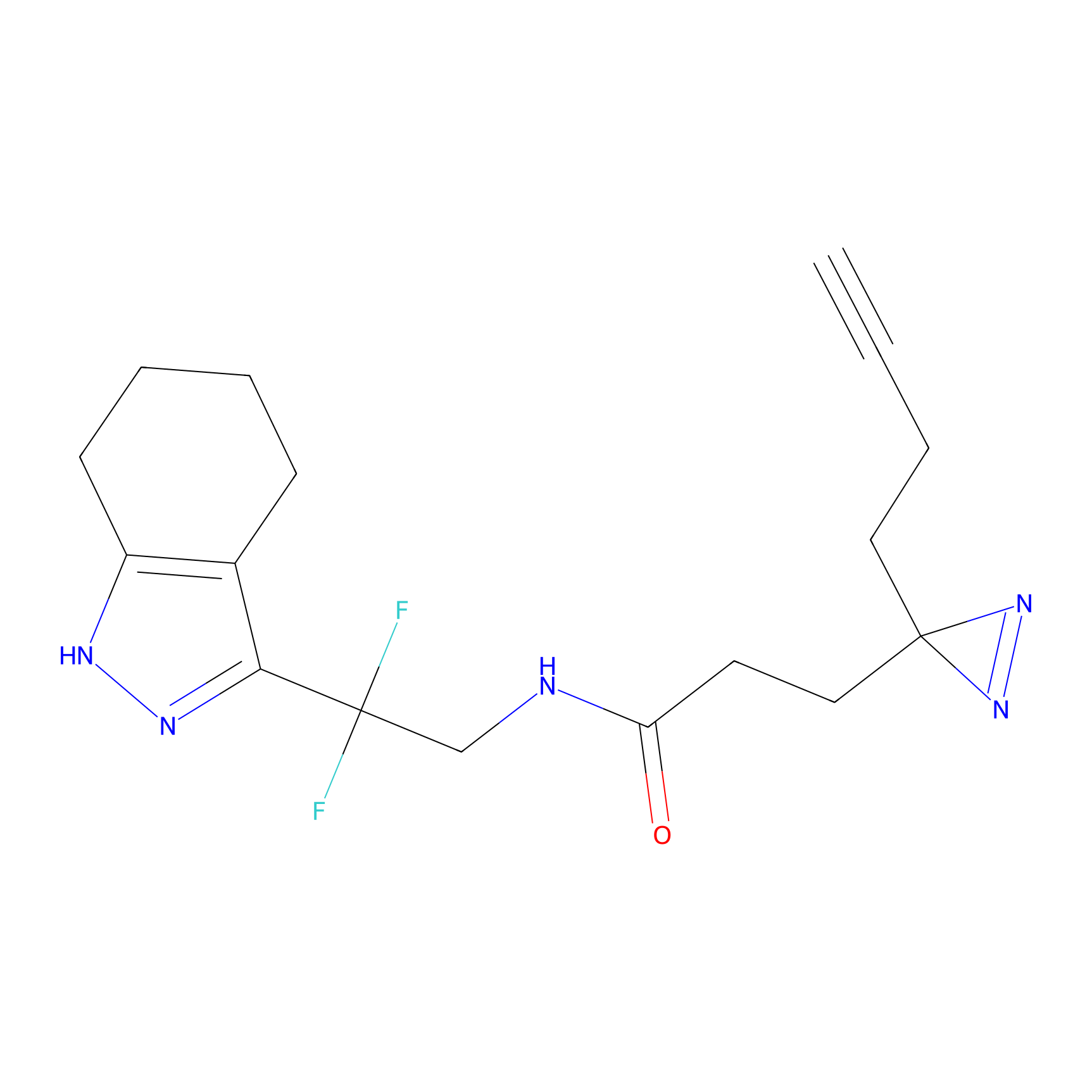
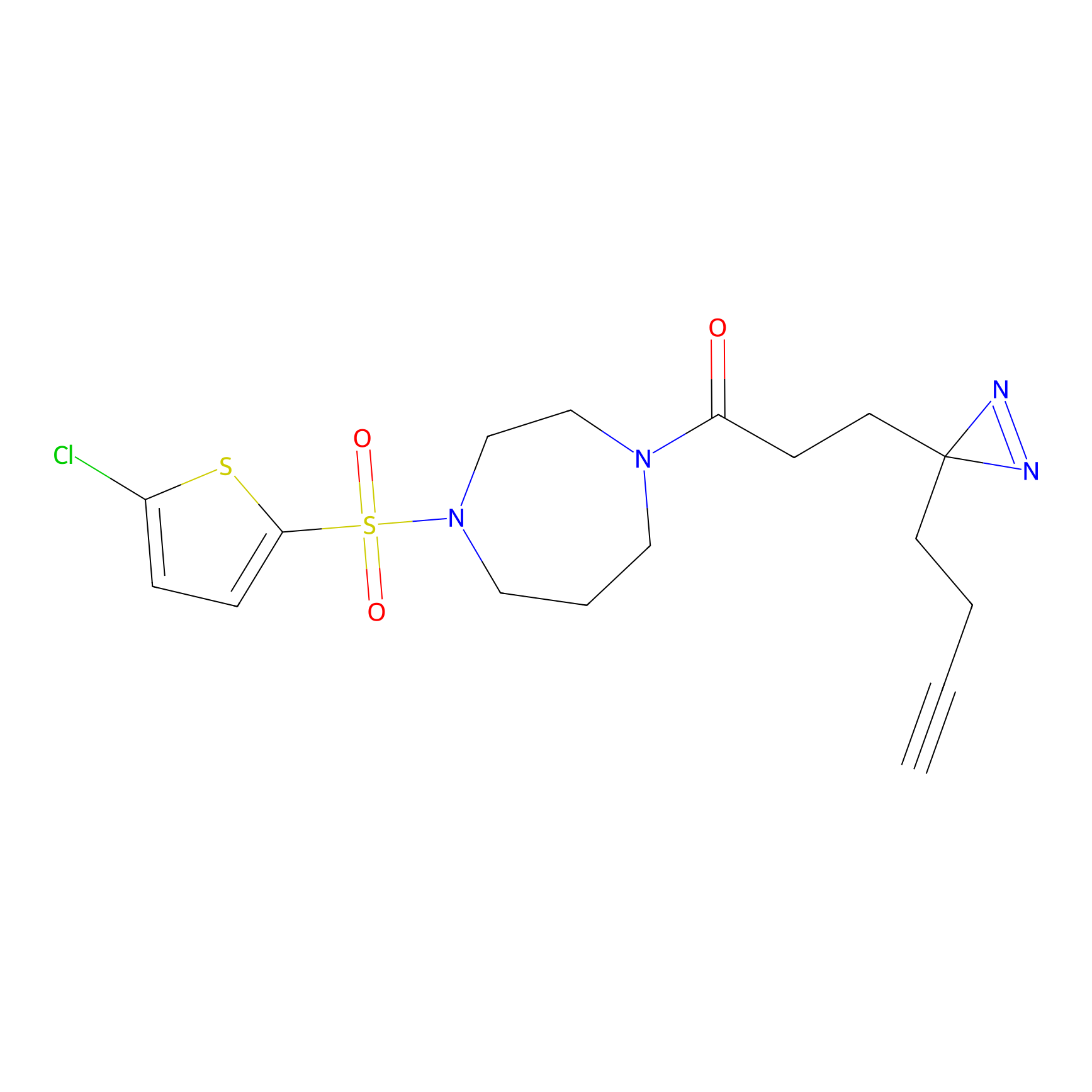
STPyne Probe Info |
 |
K179(9.65) | LDD0277 | [3] | |
|
AZ-9 Probe Info |
 |
E39(1.05) | LDD2208 | [4] | |
|
OPA-S-S-alkyne Probe Info |
 |
K179(3.25) | LDD3494 | [5] | |
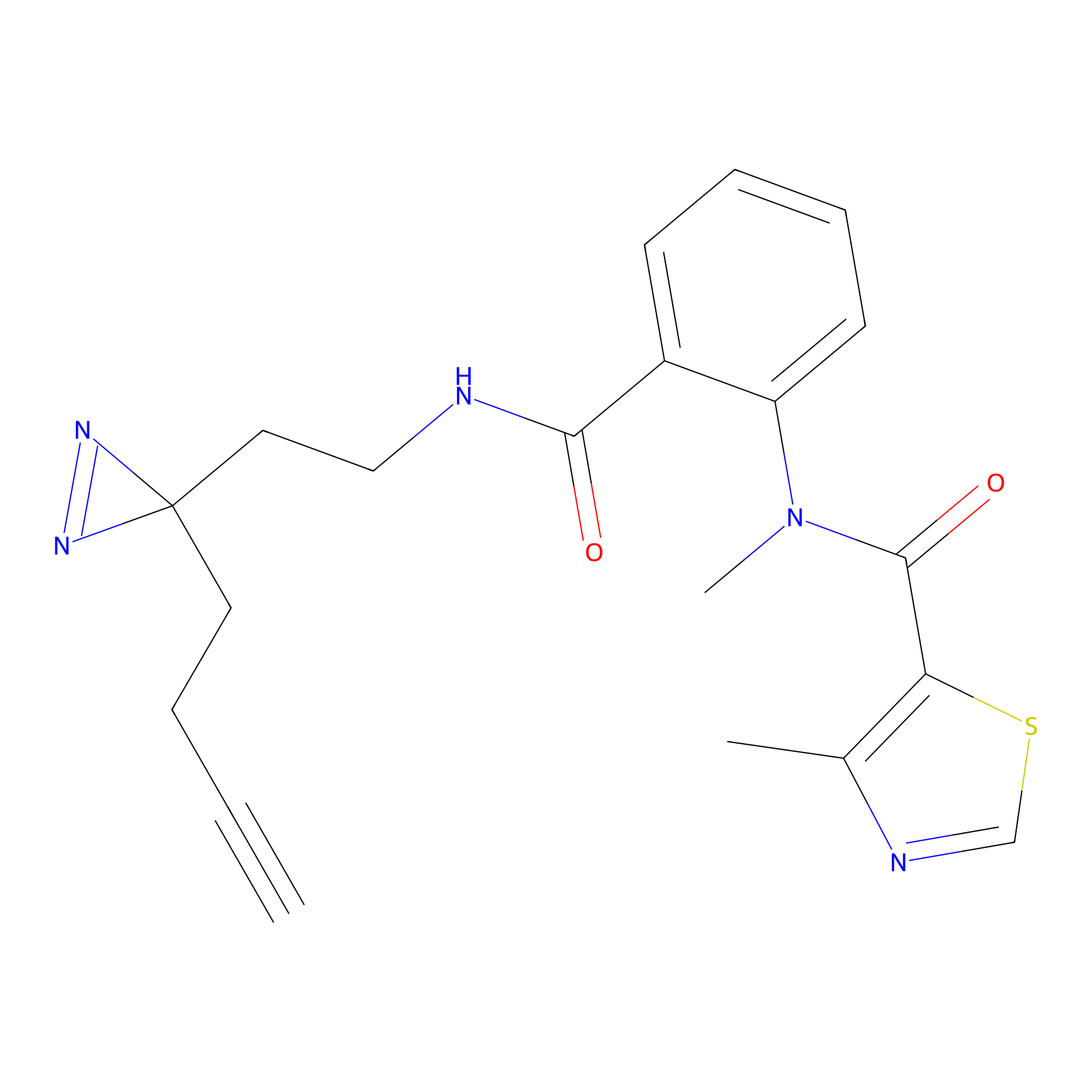
|
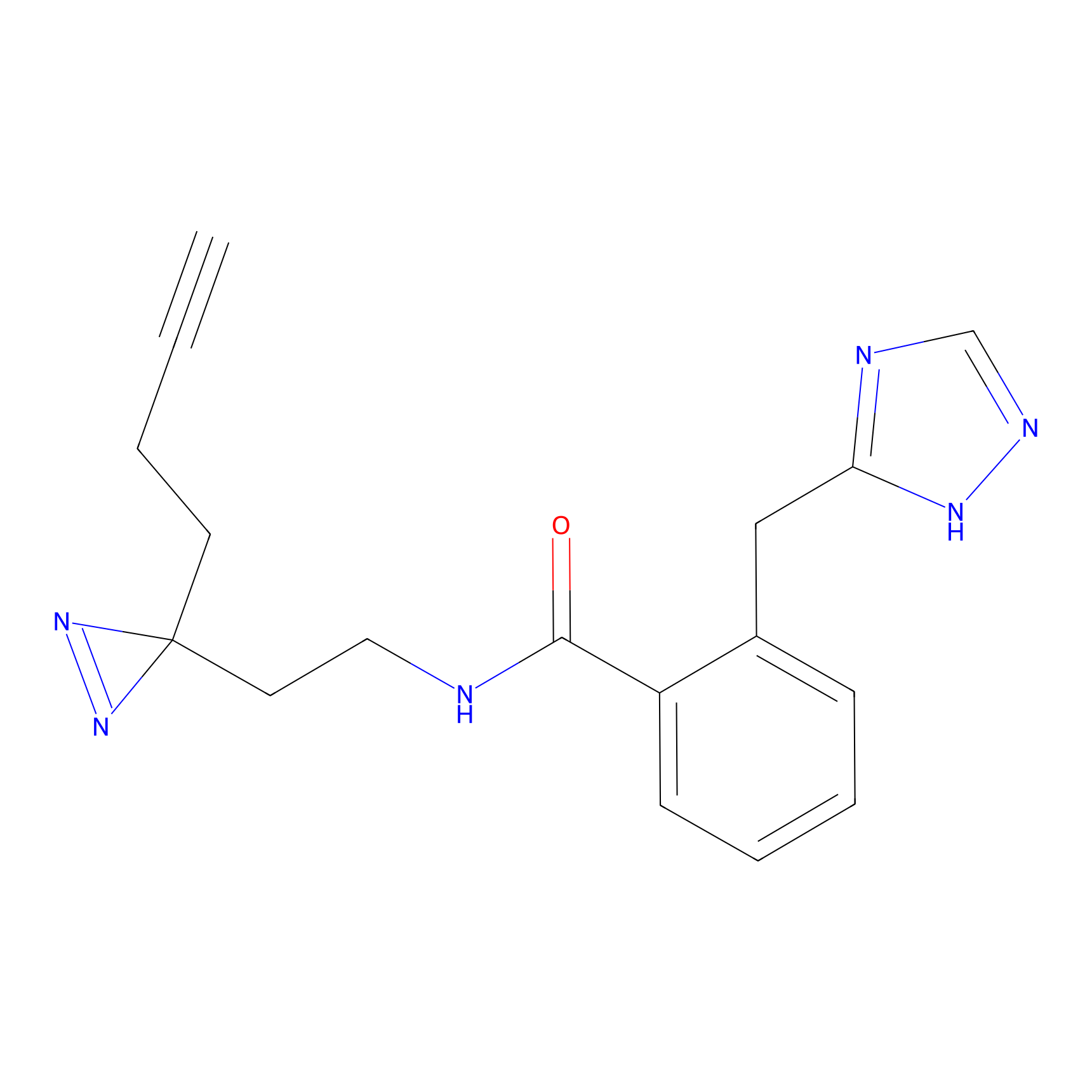
Alkylaryl probe 2 Probe Info |
 |
4.00 | LDD0392 | [6] | |
|
Jackson_14 Probe Info |
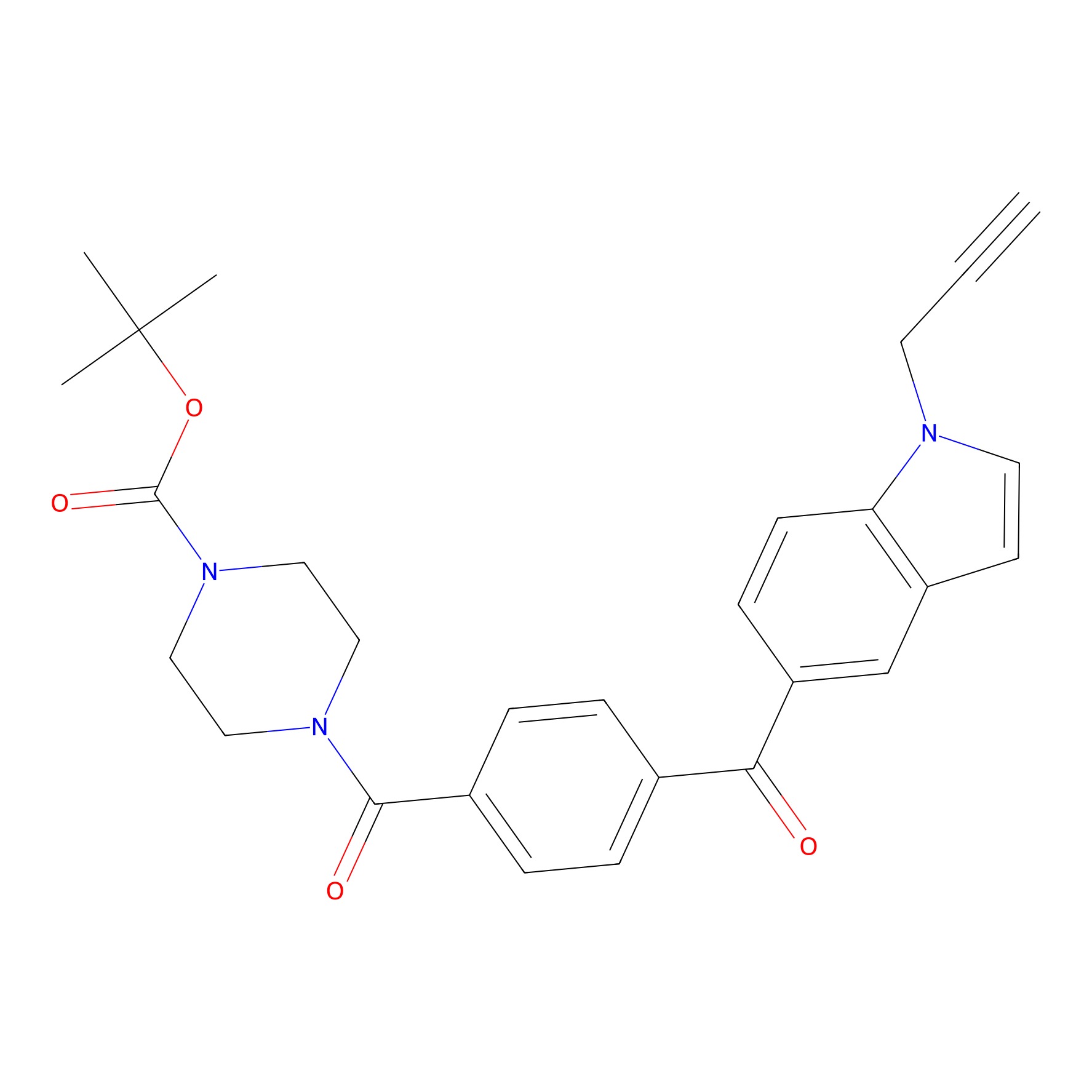 |
2.08 | LDD0123 | [7] | |
|
Acrolein Probe Info |
 |
H96(0.00); H112(0.00) | LDD0217 | [8] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [8] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0108 | Chloroacetamide | HeLa | H96(0.00); H112(0.00) | LDD0222 | [8] |
| LDCM0107 | IAA | HeLa | H112(0.00); H96(0.00) | LDD0221 | [8] |
| LDCM0109 | NEM | HeLa | H112(0.00); H96(0.00) | LDD0223 | [8] |
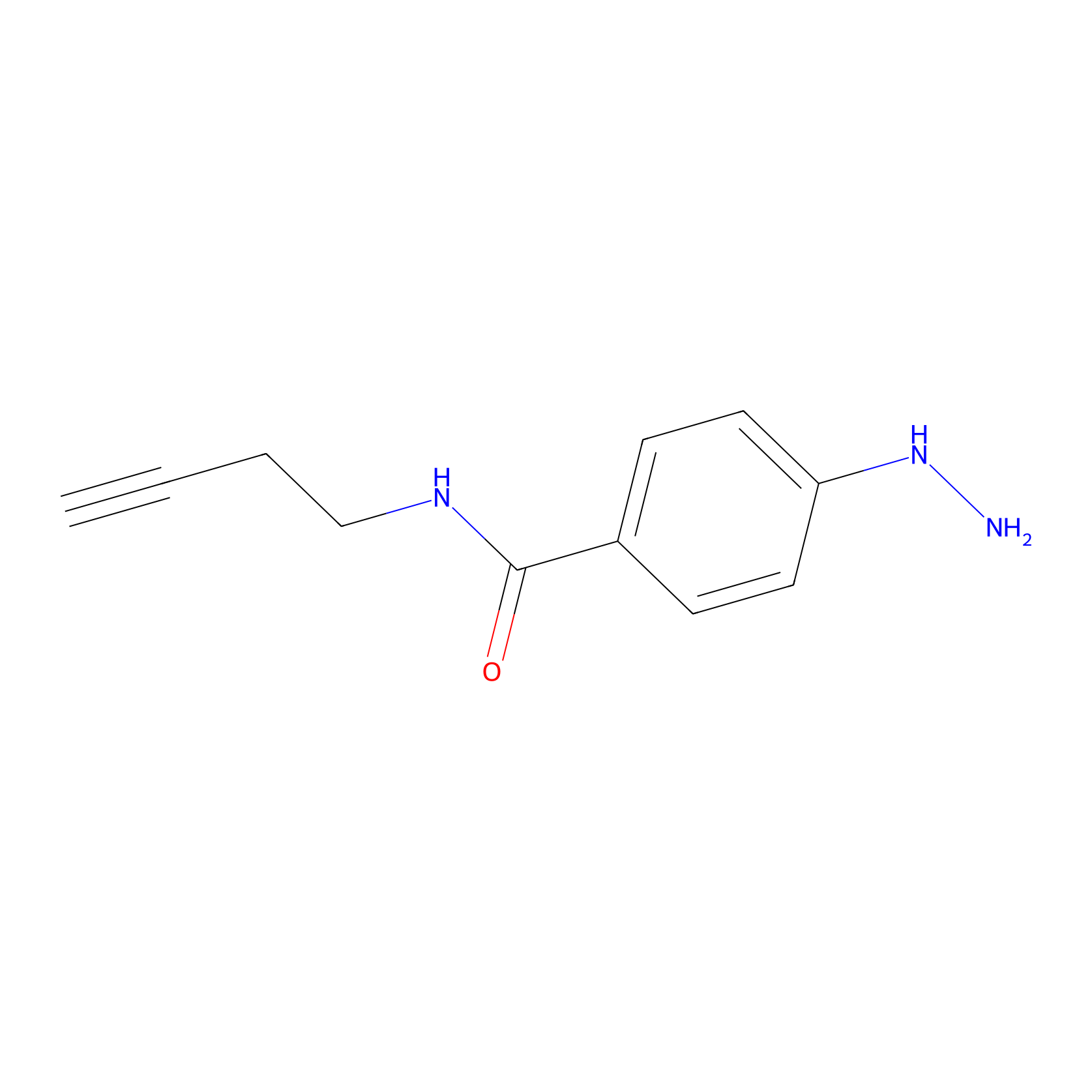
| LDCM0099 | Phenelzine | MDA-MB-231 | 4.00 | LDD0392 | [6] |
| LDCM0016 | Ranjitkar_cp1 | MDA-MB-231 | 2.08 | LDD0123 | [7] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Proto-oncogene tyrosine-protein kinase Src (SRC) | Tyr protein kinase family | P12931 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Glutamate receptor 1 (GRIA1) | Glutamate-gated ion channel family | P42261 | |||
| Wolframin (WFS1) | . | O76024 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Protein c-Fos (FOS) | BZIP family | P01100 | |||
Other
References