Details of the Target
General Information of Target
| Target ID | LDTP01759 | |||||
|---|---|---|---|---|---|---|
| Target Name | Histone-lysine N-methyltransferase NSD2 (NSD2) | |||||
| Gene Name | NSD2 | |||||
| Gene ID | 7468 | |||||
| Synonyms |
KIAA1090; MMSET; TRX5; WHSC1; Histone-lysine N-methyltransferase NSD2; EC 2.1.1.357; Multiple myeloma SET domain-containing protein; MMSET; Nuclear SET domain-containing protein 2; Protein trithorax-5; Wolf-Hirschhorn syndrome candidate 1 protein
|
|||||
| 3D Structure | ||||||
| Sequence |
MEFSIKQSPLSVQSVVKCIKMKQAPEILGSANGKTPSCEVNRECSVFLSKAQLSSSLQEG
VMQKFNGHDALPFIPADKLKDLTSRVFNGEPGAHDAKLRFESQEMKGIGTPPNTTPIKNG SPEIKLKITKTYMNGKPLFESSICGDSAADVSQSEENGQKPENKARRNRKRSIKYDSLLE QGLVEAALVSKISSPSDKKIPAKKESCPNTGRDKDHLLKYNVGDLVWSKVSGYPWWPCMV SADPLLHSYTKLKGQKKSARQYHVQFFGDAPERAWIFEKSLVAFEGEGQFEKLCQESAKQ APTKAEKIKLLKPISGKLRAQWEMGIVQAEEAASMSVEERKAKFTFLYVGDQLHLNPQVA KEAGIAAESLGEMAESSGVSEEAAENPKSVREECIPMKRRRRAKLCSSAETLESHPDIGK STPQKTAEADPRRGVGSPPGRKKTTVSMPRSRKGDAASQFLVFCQKHRDEVVAEHPDASG EEIEELLRSQWSLLSEKQRARYNTKFALVAPVQAEEDSGNVNGKKRNHTKRIQDPTEDAE AEDTPRKRLRTDKHSLRKRDTITDKTARTSSYKAMEAASSLKSQAATKNLSDACKPLKKR NRASTAASSALGFSKSSSPSASLTENEVSDSPGDEPSESPYESADETQTEVSVSSKKSER GVTAKKEYVCQLCEKPGSLLLCEGPCCGAFHLACLGLSRRPEGRFTCSECASGIHSCFVC KESKTDVKRCVVTQCGKFYHEACVKKYPLTVFESRGFRCPLHSCVSCHASNPSNPRPSKG KMMRCVRCPVAYHSGDACLAAGCSVIASNSIICTAHFTARKGKRHHAHVNVSWCFVCSKG GSLLCCESCPAAFHPDCLNIEMPDGSWFCNDCRAGKKLHFQDIIWVKLGNYRWWPAEVCH PKNVPPNIQKMKHEIGEFPVFFFGSKDYYWTHQARVFPYMEGDRGSRYQGVRGIGRVFKN ALQEAEARFREIKLQREARETQESERKPPPYKHIKVNKPYGKVQIYTADISEIPKCNCKP TDENPCGFDSECLNRMLMFECHPQVCPAGEFCQNQCFTKRQYPETKIIKTDGKGWGLVAK RDIRKGEFVNEYVGELIDEEECMARIKHAHENDITHFYMLTIDKDRIIDAGPKGNYSRFM NHSCQPNCETLKWTVNGDTRVGLFAVCDIPAGTELTFNYNLDCLGNEKTVCRCGASNCSG FLGDRPKTSTTLSSEEKGKKTKKKTRRRRAKGEGKRQSEDECFRCGDGGQLVLCDRKFCT KAYHLSCLGLGKRPFGKWECPWHHCDVCGKPSTSFCHLCPNSFCKEHQDGTAFSCTPDGR SYCCEHDLGAASVRSTKTEKPPPEPGKPKGKRRRRRGWRRVTEGK |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Class V-like SAM-binding methyltransferase superfamily, Histone-lysine methyltransferase family, SET2 subfamily
|
|||||
| Subcellular location |
Cytoplasm; Nucleus
|
|||||
| Function |
Histone methyltransferase which specifically dimethylates nucleosomal histone H3 at 'Lys-36' (H3K36me2). Also monomethylates nucleosomal histone H3 at 'Lys-36' (H3K36me) in vitro. Does not trimethylate nucleosomal histone H3 at 'Lys-36' (H3K36me3). However, specifically trimethylates histone H3 at 'Lys-36' (H3K36me3) at euchromatic regions in embryonic stem (ES) cells. By methylating histone H3 at 'Lys-36', involved in the regulation of gene transcription during various biological processes. In ES cells, associates with developmental transcription factors such as SALL1 and represses inappropriate gene transcription mediated by histone deacetylation. During heart development, associates with transcription factor NKX2-5 to repress transcription of NKX2-5 target genes. Plays an essential role in adipogenesis, by regulating expression of genes involved in pre-adipocyte differentiation. During T-cell receptor (TCR) and CD28-mediated T-cell activation, promotes the transcription of transcription factor BCL6 which is required for follicular helper T (Tfh) cell differentiation. During B-cell development, required for the generation of the B1 lineage. During B2 cell activation, may contribute to the control of isotype class switch recombination (CRS), splenic germinal center formation, and the humoral immune response. Plays a role in class switch recombination of the immunoglobulin heavy chain (IgH) locus during B-cell activation. By regulating the methylation of histone H3 at 'Lys-36' and histone H4 at 'Lys-20' at the IgH locus, involved in TP53BP1 recruitment to the IgH switch region and promotes the transcription of IgA.; [Isoform 1]: Histone methyltransferase which specifically dimethylates nucleosomal histone H3 at 'Lys-36' (H3K36me2).; [Isoform 4]: Histone methyltransferase which specifically dimethylates nucleosomal histone H3 at 'Lys-36' (H3K36me2). Methylation of histone H3 at 'Lys-27' is controversial. Mono-, di- or tri-methylates histone H3 at 'Lys-27' (H3K27me, H3K27me2 and H3K27me3). Does not methylate histone H3 at 'Lys-27'. May act as a transcription regulator that binds DNA and suppresses IL5 transcription through HDAC recruitment.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| 22RV1 | SNV: p.Y502H | DBIA Probe Info | |||
| COLO800 | SNV: p.E1111K | DBIA Probe Info | |||
| EFO27 | Deletion: p.P1343QfsTer49 | DBIA Probe Info | |||
| HCT15 | Deletion: p.P1343QfsTer49 | DBIA Probe Info | |||
| K562 | SNV: p.R600Ter | FFF probe13 Probe Info | |||
| KELLY | SNV: p.W867L | DBIA Probe Info | |||
| KYSE150 | SNV: p.R986H | DBIA Probe Info | |||
| MFE319 | SNV: p.Y641H | DBIA Probe Info | |||
| MM1S | SNV: p.E1099K | . | |||
| RKO | Deletion: . Insertion: p.E1344RfsTer91 |
DBIA Probe Info | |||
| SAOS2 | SNV: p.R501H | . | |||
| SKMEL5 | SNV: p.P36S | DBIA Probe Info | |||
| SKNSH | SNV: p.S1266Y | . | |||
| SUDHL6 | SNV: p.G325D | DBIA Probe Info | |||
| TFK1 | SNV: p.D69G | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
11.35 | LDD0402 | [1] | |
|
Jackson_1 Probe Info |
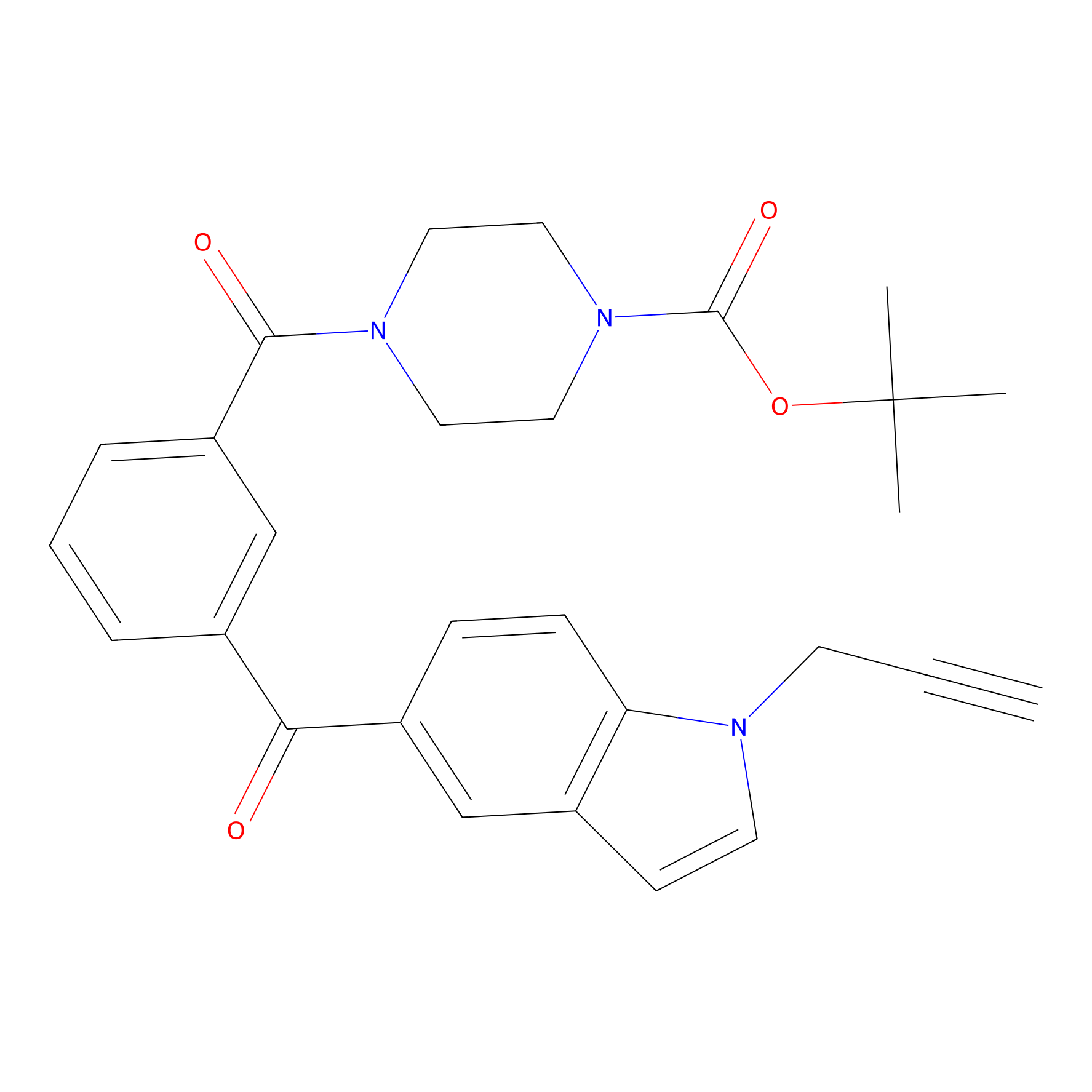 |
20.00 | LDD0122 | [2] | |
|
STPyne Probe Info |
 |
K998(10.00) | LDD0277 | [3] | |
|
P11 Probe Info |
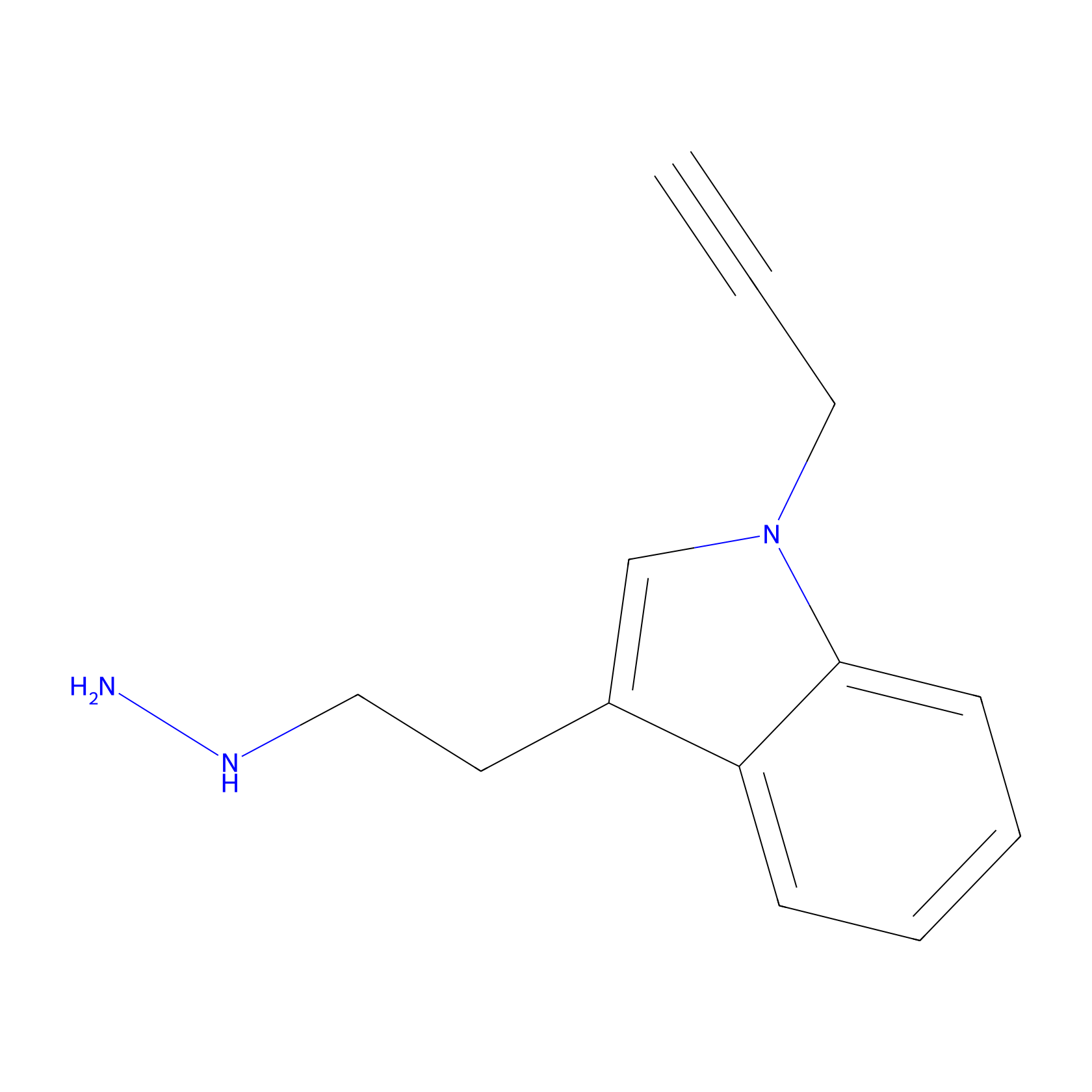 |
20.00 | LDD0201 | [4] | |
|
BTD Probe Info |
 |
C594(1.27) | LDD1700 | [5] | |
|
AHL-Pu-1 Probe Info |
 |
C594(2.58); C406(3.37) | LDD0168 | [6] | |
|
DBIA Probe Info |
 |
C1193(1.13); C1198(1.13); C44(1.01); C730(0.98) | LDD0078 | [7] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C406(0.00); C238(0.00); C1267(0.00); C1102(0.00) | LDD0038 | [8] | |
|
IA-alkyne Probe Info |
 |
C238(0.00); C743(0.00); C464(0.00); C294(0.00) | LDD0036 | [8] | |
|
Lodoacetamide azide Probe Info |
 |
C394(0.00); C743(0.00); C238(0.00); C1267(0.00) | LDD0037 | [8] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [9] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [10] | |
|
WYneO Probe Info |
 |
C406(0.00); C38(0.00) | LDD0022 | [10] | |
|
Compound 10 Probe Info |
 |
C38(0.00); C406(0.00) | LDD2216 | [11] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [11] | |
|
IPM Probe Info |
 |
C406(0.00); C594(0.00); C38(0.00); C207(0.00) | LDD0005 | [10] | |
|
TFBX Probe Info |
 |
C594(0.00); C1193(0.00) | LDD0148 | [12] | |
|
VSF Probe Info |
 |
C44(0.00); C406(0.00); C38(0.00) | LDD0007 | [10] | |
|
Phosphinate-6 Probe Info |
 |
C594(0.00); C406(0.00) | LDD0018 | [13] | |
|
Acrolein Probe Info |
 |
C1323(0.00); C1267(0.00) | LDD0217 | [14] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [14] | |
|
Methacrolein Probe Info |
 |
C406(0.00); C1323(0.00); C207(0.00); C594(0.00) | LDD0218 | [14] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [15] | |
|
NAIA_5 Probe Info |
 |
C406(0.00); C238(0.00) | LDD2223 | [9] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C106 Probe Info |
 |
18.13 | LDD1793 | [16] | |
|
C108 Probe Info |
 |
9.19 | LDD1795 | [16] | |
|
C161 Probe Info |
 |
11.55 | LDD1841 | [16] | |
|
C198 Probe Info |
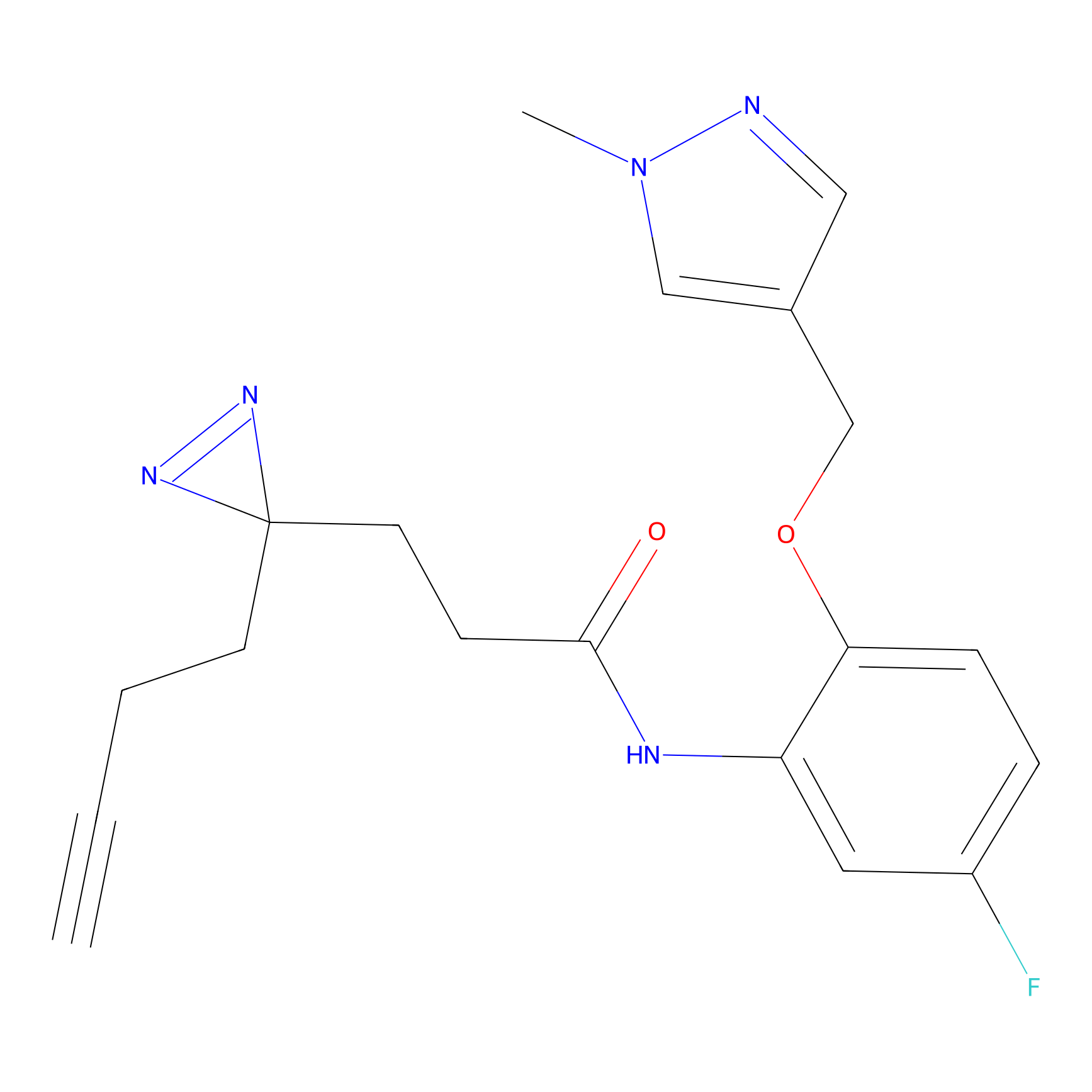 |
10.41 | LDD1874 | [16] | |
|
C201 Probe Info |
 |
28.44 | LDD1877 | [16] | |
|
C277 Probe Info |
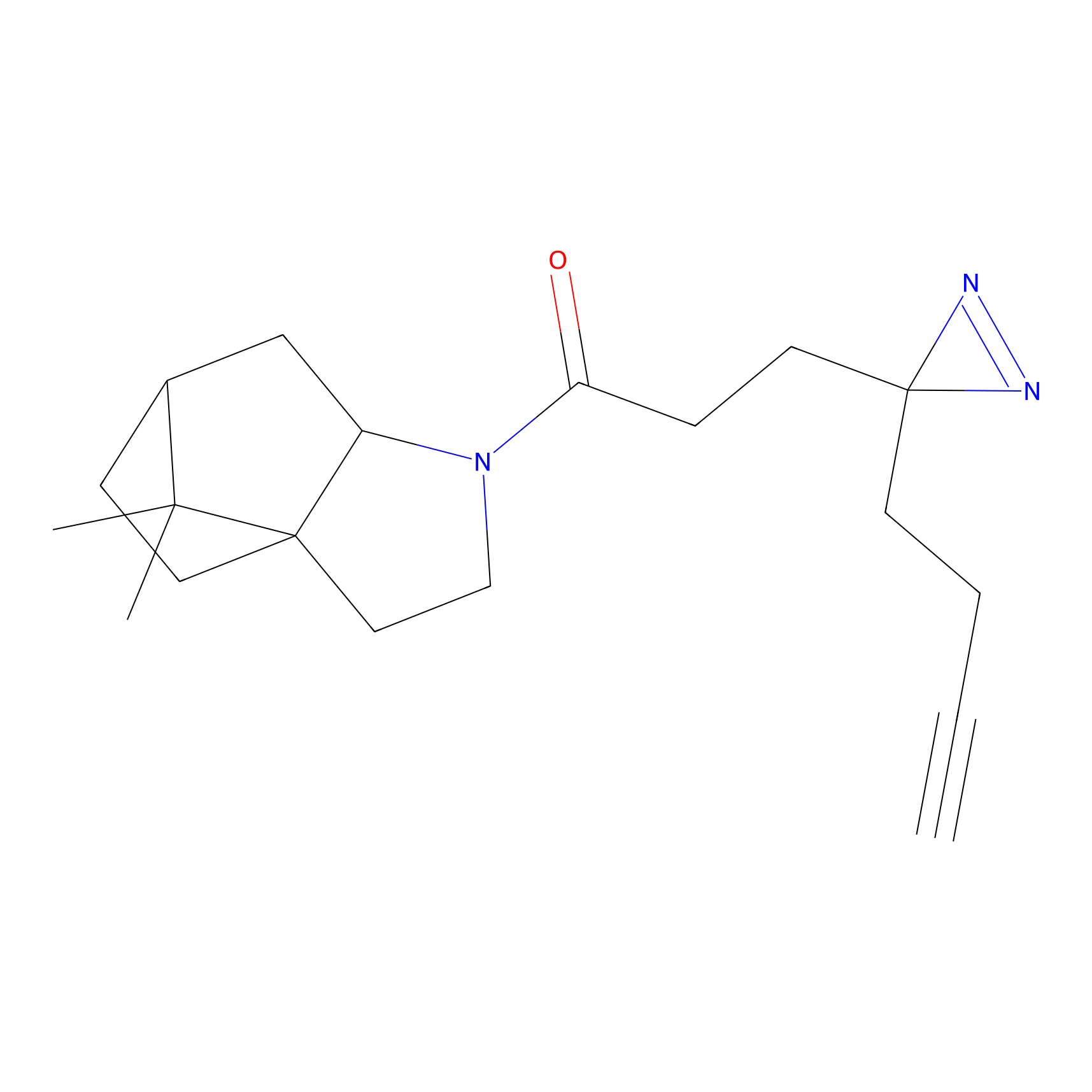 |
10.13 | LDD1947 | [16] | |
|
C284 Probe Info |
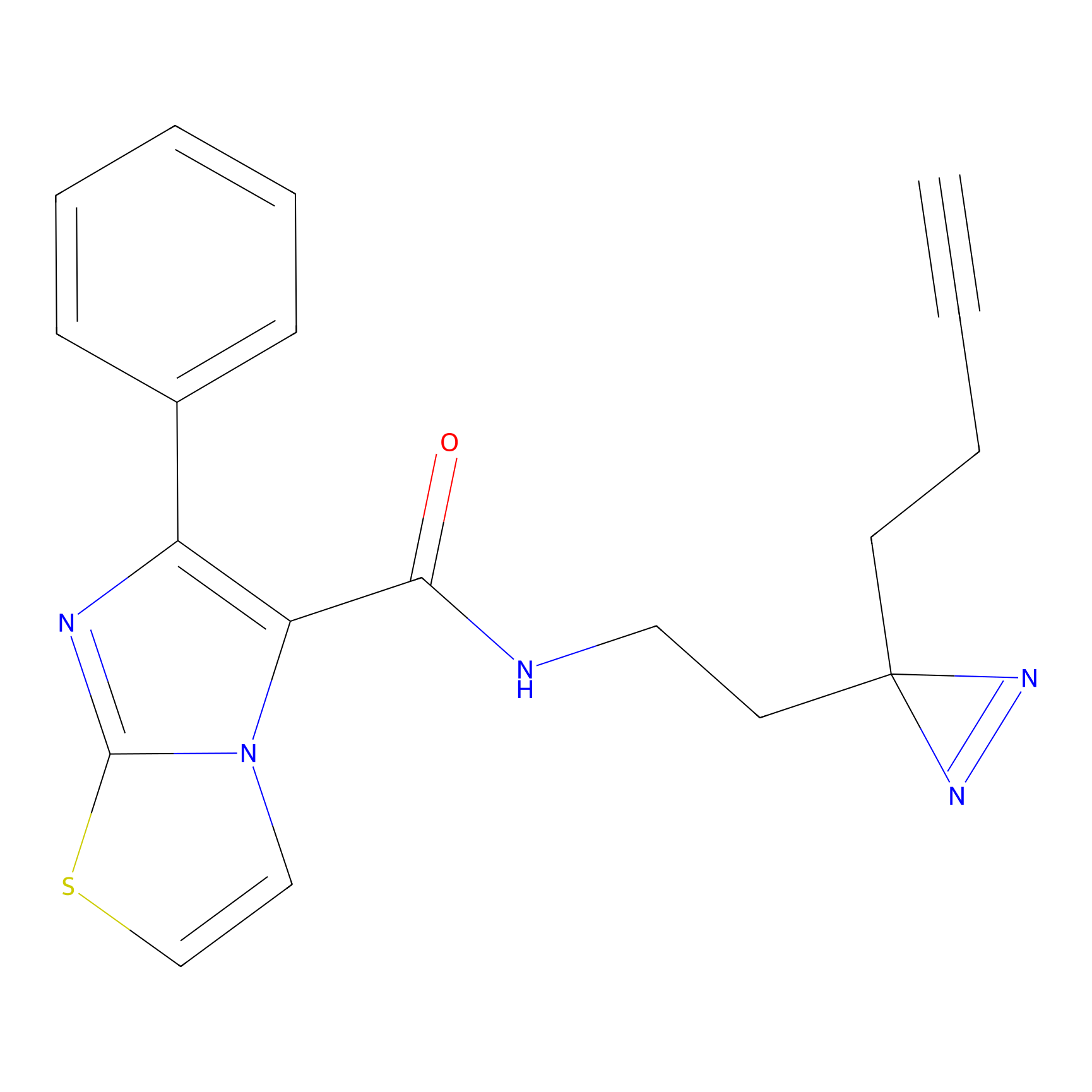 |
22.47 | LDD1954 | [16] | |
|
C343 Probe Info |
 |
11.79 | LDD2005 | [16] | |
|
C344 Probe Info |
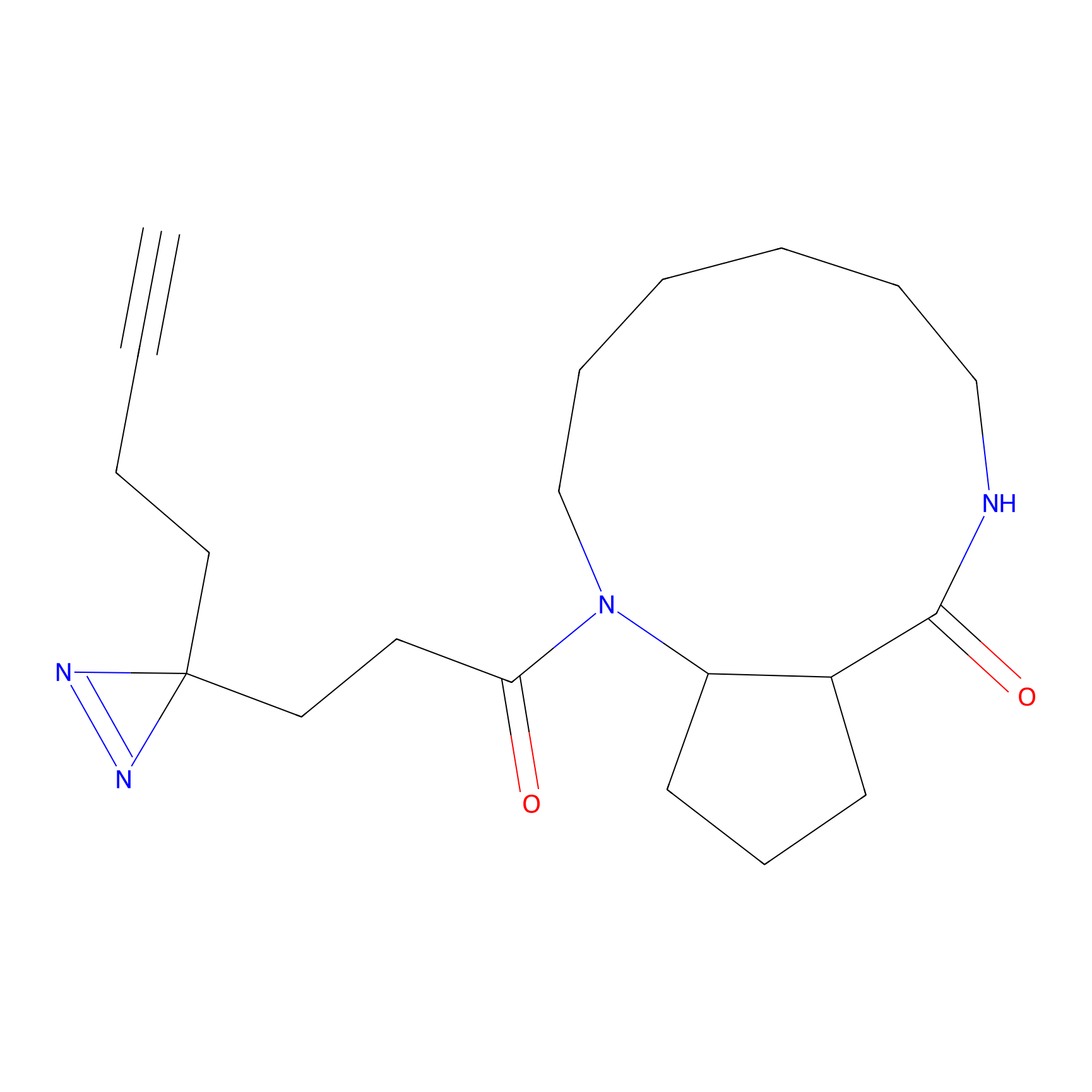 |
6.92 | LDD2006 | [16] | |
|
C346 Probe Info |
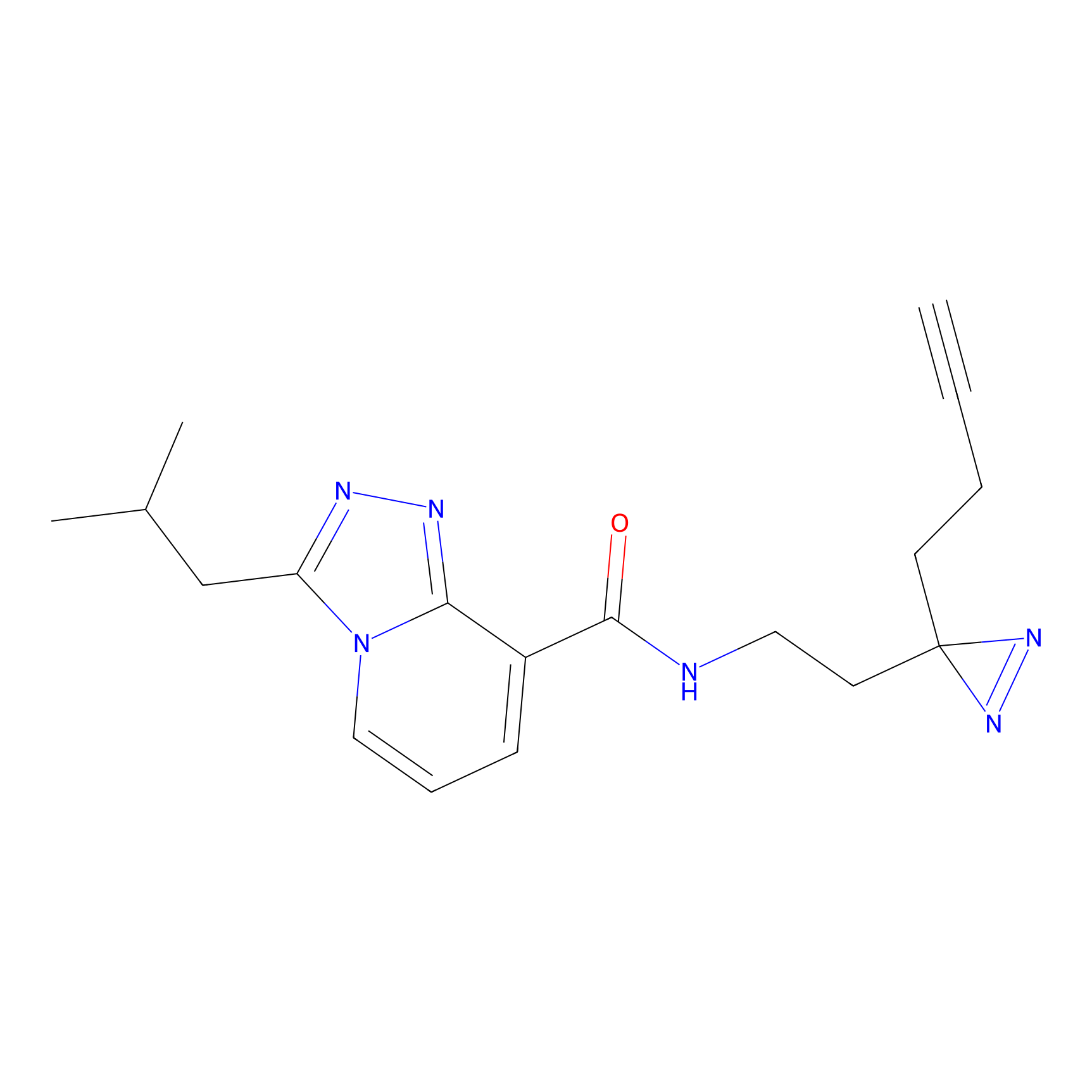 |
18.13 | LDD2007 | [16] | |
|
C363 Probe Info |
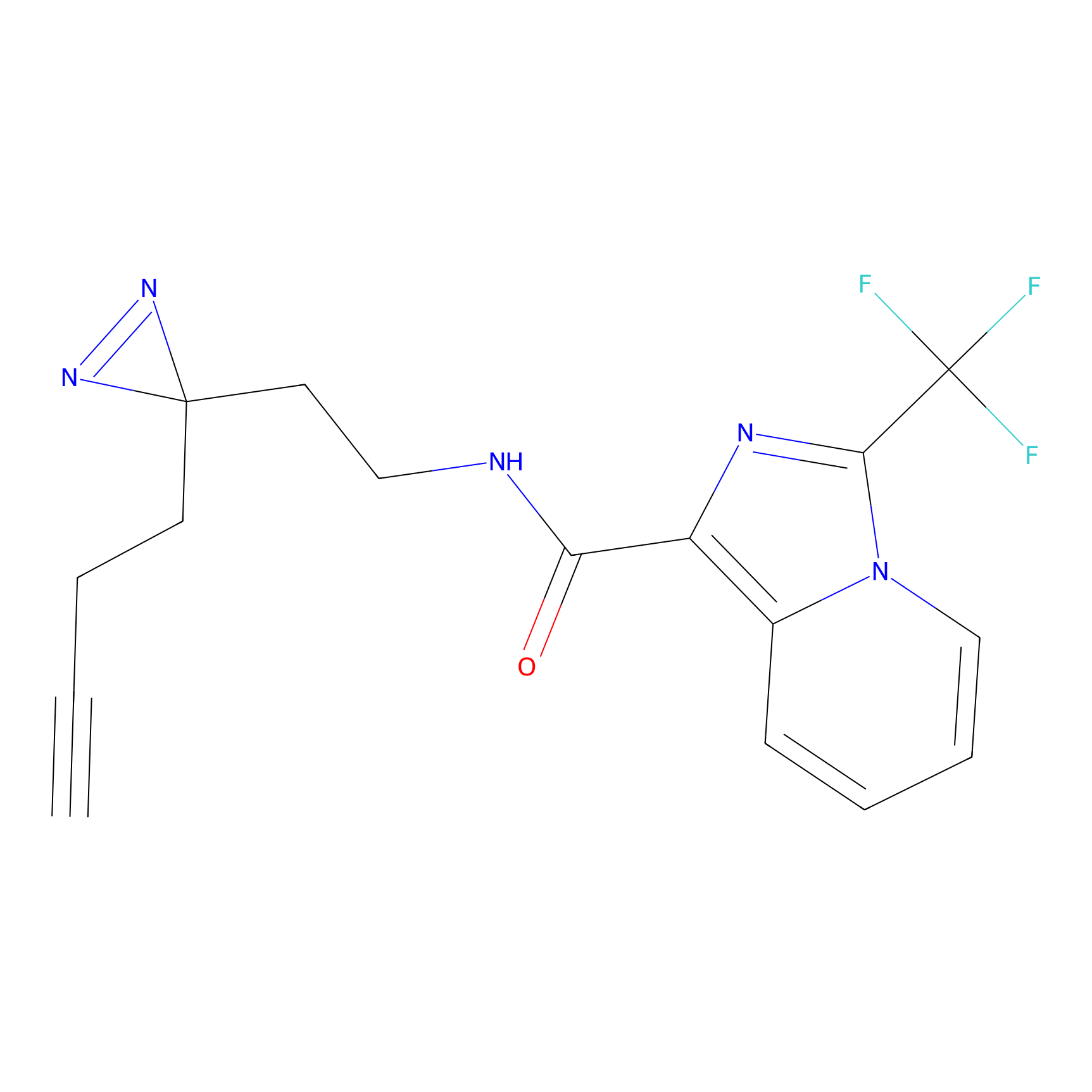 |
16.45 | LDD2024 | [16] | |
|
C376 Probe Info |
 |
7.94 | LDD2036 | [16] | |
|
C386 Probe Info |
 |
8.11 | LDD2045 | [16] | |
|
C399 Probe Info |
 |
7.84 | LDD2058 | [16] | |
|
C419 Probe Info |
 |
11.96 | LDD2074 | [16] | |
|
C420 Probe Info |
 |
11.63 | LDD2075 | [16] | |
|
C431 Probe Info |
 |
15.78 | LDD2086 | [16] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [17] | |
|
FFF probe2 Probe Info |
 |
20.00 | LDD0463 | [17] | |
|
Diazir Probe Info |
 |
N.A. | LDD0011 | [10] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C594(1.16) | LDD2112 | [5] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C594(1.32); C207(0.97) | LDD2117 | [5] |
| LDCM0025 | 4SU-RNA | HEK-293T | C594(2.58); C406(3.37) | LDD0168 | [6] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C406(3.48); C144(3.52) | LDD0169 | [6] |
| LDCM0214 | AC1 | HCT 116 | C1016(0.98); C1018(0.87); C1032(0.87); C1144(1.00) | LDD0531 | [7] |
| LDCM0215 | AC10 | HCT 116 | C1046(0.92); C1056(0.93); C1016(0.94); C1018(0.79) | LDD0532 | [7] |
| LDCM0216 | AC100 | HCT 116 | C1016(0.51); C1018(0.57); C1032(0.51); C1144(0.74) | LDD0533 | [7] |
| LDCM0217 | AC101 | HCT 116 | C1016(0.65); C1018(0.65); C1032(0.65); C1144(0.79) | LDD0534 | [7] |
| LDCM0218 | AC102 | HCT 116 | C1016(0.44); C1018(0.46); C1032(0.44); C1144(0.81) | LDD0535 | [7] |
| LDCM0219 | AC103 | HCT 116 | C1016(0.52); C1018(0.60); C1032(0.52); C1144(0.86) | LDD0536 | [7] |
| LDCM0220 | AC104 | HCT 116 | C1016(0.48); C1018(0.51); C1032(0.48); C1144(0.94) | LDD0537 | [7] |
| LDCM0221 | AC105 | HCT 116 | C1016(0.55); C1018(0.53); C1032(0.55); C1144(0.87) | LDD0538 | [7] |
| LDCM0222 | AC106 | HCT 116 | C1016(0.52); C1018(0.48); C1032(0.52); C1144(0.92) | LDD0539 | [7] |
| LDCM0223 | AC107 | HCT 116 | C1016(0.50); C1018(0.58); C1032(0.38); C1144(0.61) | LDD0540 | [7] |
| LDCM0224 | AC108 | HCT 116 | C1016(0.55); C1018(0.53); C1032(0.84); C1144(0.67) | LDD0541 | [7] |
| LDCM0225 | AC109 | HCT 116 | C1016(0.70); C1018(0.58); C1032(0.81); C1144(0.71) | LDD0542 | [7] |
| LDCM0226 | AC11 | HCT 116 | C1046(0.95); C1056(0.99); C1016(0.81); C1018(0.69) | LDD0543 | [7] |
| LDCM0227 | AC110 | HCT 116 | C1016(0.55); C1018(0.65); C1032(0.61); C1144(0.64) | LDD0544 | [7] |
| LDCM0228 | AC111 | HCT 116 | C1016(0.61); C1018(0.53); C1032(0.82); C1144(0.67) | LDD0545 | [7] |
| LDCM0229 | AC112 | HCT 116 | C1016(0.52); C1018(0.52); C1032(0.54); C1144(0.69) | LDD0546 | [7] |
| LDCM0230 | AC113 | HCT 116 | C1041(0.98); C1046(1.21); C1056(1.15); C1016(1.14) | LDD0547 | [7] |
| LDCM0231 | AC114 | HCT 116 | C1041(0.85); C1046(0.93); C1056(0.93); C1016(1.05) | LDD0548 | [7] |
| LDCM0232 | AC115 | HCT 116 | C1041(0.79); C1046(0.85); C1056(0.83); C1016(0.88) | LDD0549 | [7] |
| LDCM0233 | AC116 | HCT 116 | C1041(0.86); C1046(0.98); C1056(0.91); C1016(0.80) | LDD0550 | [7] |
| LDCM0234 | AC117 | HCT 116 | C1041(0.85); C1046(1.01); C1056(0.92); C1016(0.92) | LDD0551 | [7] |
| LDCM0235 | AC118 | HCT 116 | C1041(0.83); C1046(0.87); C1056(0.89); C1016(0.98) | LDD0552 | [7] |
| LDCM0236 | AC119 | HCT 116 | C1041(0.80); C1046(1.01); C1056(0.91); C1016(0.93) | LDD0553 | [7] |
| LDCM0237 | AC12 | HCT 116 | C1046(0.89); C1056(0.88); C1016(0.75); C1018(0.67) | LDD0554 | [7] |
| LDCM0238 | AC120 | HCT 116 | C1041(0.79); C1046(0.92); C1056(0.90); C1016(0.77) | LDD0555 | [7] |
| LDCM0239 | AC121 | HCT 116 | C1041(0.97); C1046(0.97); C1056(0.96); C1016(1.13) | LDD0556 | [7] |
| LDCM0240 | AC122 | HCT 116 | C1041(0.95); C1046(0.87); C1056(0.94); C1016(0.88) | LDD0557 | [7] |
| LDCM0241 | AC123 | HCT 116 | C1041(0.88); C1046(0.96); C1056(0.92); C1016(1.20) | LDD0558 | [7] |
| LDCM0242 | AC124 | HCT 116 | C1041(0.89); C1046(1.00); C1056(0.97); C1016(1.12) | LDD0559 | [7] |
| LDCM0243 | AC125 | HCT 116 | C1041(0.83); C1046(0.87); C1056(0.89); C1016(0.81) | LDD0560 | [7] |
| LDCM0244 | AC126 | HCT 116 | C1041(0.87); C1046(0.96); C1056(0.93); C1016(0.92) | LDD0561 | [7] |
| LDCM0245 | AC127 | HCT 116 | C1041(0.79); C1046(0.91); C1056(0.91); C1016(1.03) | LDD0562 | [7] |
| LDCM0246 | AC128 | HCT 116 | C1041(1.43); C1046(1.17); C1056(0.90); C1016(1.36) | LDD0563 | [7] |
| LDCM0247 | AC129 | HCT 116 | C1041(2.51); C1046(1.67); C1056(0.84); C1016(1.39) | LDD0564 | [7] |
| LDCM0249 | AC130 | HCT 116 | C1041(1.03); C1046(0.89); C1056(0.76); C1016(1.61) | LDD0566 | [7] |
| LDCM0250 | AC131 | HCT 116 | C1041(1.27); C1046(1.03); C1056(0.78); C1016(1.98) | LDD0567 | [7] |
| LDCM0251 | AC132 | HCT 116 | C1041(1.47); C1046(1.07); C1056(0.66); C1016(1.59) | LDD0568 | [7] |
| LDCM0252 | AC133 | HCT 116 | C1041(1.30); C1046(1.09); C1056(0.89); C1016(1.20) | LDD0569 | [7] |
| LDCM0253 | AC134 | HCT 116 | C1041(1.21); C1046(0.93); C1056(0.65); C1016(1.34) | LDD0570 | [7] |
| LDCM0254 | AC135 | HCT 116 | C1041(0.91); C1046(0.82); C1056(0.73); C1016(2.00) | LDD0571 | [7] |
| LDCM0255 | AC136 | HCT 116 | C1041(1.39); C1046(1.08); C1056(0.77); C1016(1.23) | LDD0572 | [7] |
| LDCM0256 | AC137 | HCT 116 | C1041(1.85); C1046(1.29); C1056(0.73); C1016(1.11) | LDD0573 | [7] |
| LDCM0257 | AC138 | HCT 116 | C1041(1.02); C1046(0.79); C1056(0.56); C1016(1.41) | LDD0574 | [7] |
| LDCM0258 | AC139 | HCT 116 | C1041(1.10); C1046(0.91); C1056(0.71); C1016(2.18) | LDD0575 | [7] |
| LDCM0259 | AC14 | HCT 116 | C1046(0.83); C1056(0.86); C1016(1.04); C1018(0.93) | LDD0576 | [7] |
| LDCM0260 | AC140 | HCT 116 | C1041(1.78); C1046(1.22); C1056(0.66); C1016(1.34) | LDD0577 | [7] |
| LDCM0261 | AC141 | HCT 116 | C1041(1.04); C1046(0.81); C1056(0.58); C1016(1.34) | LDD0578 | [7] |
| LDCM0262 | AC142 | HCT 116 | C1041(1.85); C1046(1.31); C1056(0.78); C1016(1.44) | LDD0579 | [7] |
| LDCM0263 | AC143 | HCT 116 | C1041(1.07); C1046(0.84); C1056(1.03); C207(0.66) | LDD0580 | [7] |
| LDCM0264 | AC144 | HCT 116 | C1280(0.53); C1018(0.63); C1026(0.70); C207(0.78) | LDD0581 | [7] |
| LDCM0265 | AC145 | HCT 116 | C1026(0.60); C1148(0.78); C294(0.81); C1018(0.84) | LDD0582 | [7] |
| LDCM0266 | AC146 | HCT 116 | C1148(0.73); C1280(0.84); C207(0.84); C294(0.91) | LDD0583 | [7] |
| LDCM0267 | AC147 | HCT 116 | C1280(0.54); C1018(0.67); C1148(0.78); C1032(0.83) | LDD0584 | [7] |
| LDCM0268 | AC148 | HCT 116 | C1280(0.53); C207(0.65); C294(0.72); C1148(0.89) | LDD0585 | [7] |
| LDCM0269 | AC149 | HCT 116 | C1280(0.60); C1148(0.78); C1323(0.84); C1324(0.84) | LDD0586 | [7] |
| LDCM0270 | AC15 | HCT 116 | C1016(0.78); C44(0.78); C1026(0.81); C730(0.84) | LDD0587 | [7] |
| LDCM0271 | AC150 | HCT 116 | C1148(0.71); C1144(0.82); C38(0.86); C1245(0.86) | LDD0588 | [7] |
| LDCM0272 | AC151 | HCT 116 | C1026(0.67); C1148(0.80); C1016(0.82); C1144(0.83) | LDD0589 | [7] |
| LDCM0273 | AC152 | HCT 116 | C1280(0.55); C1018(0.64); C1026(0.71); C1032(0.75) | LDD0590 | [7] |
| LDCM0274 | AC153 | HCT 116 | C1280(0.58); C207(0.59); C294(0.91); C1242(0.94) | LDD0591 | [7] |
| LDCM0621 | AC154 | HCT 116 | C1041(1.06); C1046(1.33); C1056(1.06); C1016(0.91) | LDD2158 | [7] |
| LDCM0622 | AC155 | HCT 116 | C1041(0.92); C1046(1.12); C1056(0.90); C1016(0.97) | LDD2159 | [7] |
| LDCM0623 | AC156 | HCT 116 | C1041(1.30); C1046(1.14); C1056(1.23); C1016(0.86) | LDD2160 | [7] |
| LDCM0624 | AC157 | HCT 116 | C1041(1.10); C1046(1.55); C1056(1.14); C1016(0.94) | LDD2161 | [7] |
| LDCM0276 | AC17 | HCT 116 | C44(0.70); C1016(0.75); C1041(0.75); C899(0.79) | LDD0593 | [7] |
| LDCM0277 | AC18 | HCT 116 | C1016(0.62); C1032(0.64); C1041(0.68); C1018(0.76) | LDD0594 | [7] |
| LDCM0278 | AC19 | HCT 116 | C1041(0.62); C1016(0.67); C1032(0.71); C1148(0.74) | LDD0595 | [7] |
| LDCM0279 | AC2 | HCT 116 | C1148(0.68); C1018(0.80); C1032(0.80); C730(0.81) | LDD0596 | [7] |
| LDCM0280 | AC20 | HCT 116 | C1315(0.71); C899(0.78); C44(0.78); C1016(0.81) | LDD0597 | [7] |
| LDCM0281 | AC21 | HCT 116 | C899(0.72); C1148(0.78); C44(0.78); C1016(0.80) | LDD0598 | [7] |
| LDCM0282 | AC22 | HCT 116 | C1315(0.66); C44(0.67); C899(0.72); C207(0.78) | LDD0599 | [7] |
| LDCM0283 | AC23 | HCT 116 | C44(0.66); C899(0.72); C1041(0.75); C1315(0.75) | LDD0600 | [7] |
| LDCM0284 | AC24 | HCT 116 | C44(0.65); C1041(0.69); C1046(0.73); C1056(0.73) | LDD0601 | [7] |
| LDCM0285 | AC25 | HCT 116 | C1324(0.65); C1052(0.69); C1032(0.71); C670(0.75) | LDD0602 | [7] |
| LDCM0286 | AC26 | HCT 116 | C1026(0.68); C1016(0.71); C1052(0.72); C1018(0.73) | LDD0603 | [7] |
| LDCM0287 | AC27 | HCT 116 | C1032(0.75); C1018(0.75); C1016(0.79); C1052(0.82) | LDD0604 | [7] |
| LDCM0288 | AC28 | HCT 116 | C1032(0.62); C1018(0.66); C1052(0.66); C1016(0.67) | LDD0605 | [7] |
| LDCM0289 | AC29 | HCT 116 | C1052(0.65); C1148(0.69); C1046(0.74); C1056(0.74) | LDD0606 | [7] |
| LDCM0290 | AC3 | HCT 116 | C1148(0.73); C730(0.77); C735(0.77); C1016(0.81) | LDD0607 | [7] |
| LDCM0291 | AC30 | HCT 116 | C1148(0.49); C1032(0.66); C1018(0.67); C1016(0.68) | LDD0608 | [7] |
| LDCM0292 | AC31 | HCT 116 | C1018(0.83); C1032(0.84); C670(0.85); C673(0.85) | LDD0609 | [7] |
| LDCM0293 | AC32 | HCT 116 | C1052(0.63); C670(0.74); C673(0.74); C1032(0.75) | LDD0610 | [7] |
| LDCM0294 | AC33 | HCT 116 | C1052(0.62); C1032(0.69); C1016(0.74); C1046(0.75) | LDD0611 | [7] |
| LDCM0295 | AC34 | HCT 116 | C1148(0.65); C1018(0.66); C1032(0.71); C1016(0.73) | LDD0612 | [7] |
| LDCM0296 | AC35 | HCT 116 | C1245(0.54); C1254(0.54); C1324(0.82); C207(0.88) | LDD0613 | [7] |
| LDCM0297 | AC36 | HCT 116 | C207(0.88); C1052(0.89); C1315(0.91); C1144(0.93) | LDD0614 | [7] |
| LDCM0298 | AC37 | HCT 116 | C1052(0.76); C1046(0.81); C1056(0.86); C1041(0.88) | LDD0615 | [7] |
| LDCM0299 | AC38 | HCT 116 | C1315(0.86); C1324(0.87); C1144(0.95); C1148(0.98) | LDD0616 | [7] |
| LDCM0300 | AC39 | HCT 116 | C1245(0.88); C1254(0.88); C1148(0.98); C1041(0.98) | LDD0617 | [7] |
| LDCM0301 | AC4 | HCT 116 | C1148(0.66); C1144(0.75); C730(0.82); C735(0.82) | LDD0618 | [7] |
| LDCM0302 | AC40 | HCT 116 | C1052(0.85); C1046(0.87); C1056(0.90); C1245(0.93) | LDD0619 | [7] |
| LDCM0303 | AC41 | HCT 116 | C1052(0.75); C1324(0.87); C1148(0.89); C1046(0.89) | LDD0620 | [7] |
| LDCM0304 | AC42 | HCT 116 | C1052(0.72); C1046(0.79); C1056(0.86); C1148(0.89) | LDD0621 | [7] |
| LDCM0305 | AC43 | HCT 116 | C1324(0.88); C38(0.88); C1323(0.90); C1315(0.91) | LDD0622 | [7] |
| LDCM0306 | AC44 | HCT 116 | C1324(0.77); C1148(0.87); C1041(0.88); C1056(0.88) | LDD0623 | [7] |
| LDCM0307 | AC45 | HCT 116 | C1046(0.84); C1148(0.87); C1041(0.89); C1056(0.91) | LDD0624 | [7] |
| LDCM0308 | AC46 | HCT 116 | C1288(0.86); C1323(0.89); C1026(0.91); C1242(0.92) | LDD0625 | [7] |
| LDCM0309 | AC47 | HCT 116 | C1041(0.70); C1144(0.94); C1026(0.97); C1056(0.98) | LDD0626 | [7] |
| LDCM0310 | AC48 | HCT 116 | C1323(0.81); C1324(0.83); C44(0.88); C1144(0.89) | LDD0627 | [7] |
| LDCM0311 | AC49 | HCT 116 | C1026(0.78); C1245(0.81); C1254(0.81); C1041(0.81) | LDD0628 | [7] |
| LDCM0312 | AC5 | HCT 116 | C1148(0.70); C38(0.82); C730(0.84); C735(0.84) | LDD0629 | [7] |
| LDCM0313 | AC50 | HCT 116 | C1242(0.90); C1026(0.92); C44(0.93); C1280(0.96) | LDD0630 | [7] |
| LDCM0314 | AC51 | HCT 116 | C1323(0.74); C1324(0.82); C1315(0.86); C1144(0.87) | LDD0631 | [7] |
| LDCM0315 | AC52 | HCT 116 | C1041(0.71); C1288(0.82); C1026(0.82); C1280(0.86) | LDD0632 | [7] |
| LDCM0316 | AC53 | HCT 116 | C1280(0.88); C1026(0.88); C1242(0.89); C1018(0.94) | LDD0633 | [7] |
| LDCM0317 | AC54 | HCT 116 | C1026(0.81); C1016(0.89); C1041(0.89); C1242(0.90) | LDD0634 | [7] |
| LDCM0318 | AC55 | HCT 116 | C1148(0.93); C1041(0.95); C1280(0.97); C1144(1.01) | LDD0635 | [7] |
| LDCM0319 | AC56 | HCT 116 | C1148(0.74); C1041(0.92); C1285(0.94); C1280(0.96) | LDD0636 | [7] |
| LDCM0320 | AC57 | HCT 116 | C1026(0.78); C1148(0.88); C1198(0.90); C1280(0.92) | LDD0637 | [7] |
| LDCM0321 | AC58 | HCT 116 | C1148(0.74); C670(0.84); C673(0.84); C1032(0.87) | LDD0638 | [7] |
| LDCM0322 | AC59 | HCT 116 | C1026(0.70); C294(0.73); C1018(0.80); C1032(0.80) | LDD0639 | [7] |
| LDCM0323 | AC6 | HCT 116 | C1148(0.86); C38(0.86); C1018(0.89); C730(0.90) | LDD0640 | [7] |
| LDCM0324 | AC60 | HCT 116 | C207(0.93); C1148(0.93); C1280(0.96); C1026(0.99) | LDD0641 | [7] |
| LDCM0325 | AC61 | HCT 116 | C294(1.00); C1280(1.03); C44(1.07); C1198(1.09) | LDD0642 | [7] |
| LDCM0326 | AC62 | HCT 116 | C1026(0.80); C1148(0.85); C1280(0.88); C1198(0.95) | LDD0643 | [7] |
| LDCM0327 | AC63 | HCT 116 | C1280(0.77); C1026(0.84); C1148(0.88); C1198(1.03) | LDD0644 | [7] |
| LDCM0328 | AC64 | HCT 116 | C1280(0.82); C294(0.83); C1026(0.91); C1148(0.98) | LDD0645 | [7] |
| LDCM0329 | AC65 | HCT 116 | C1026(0.71); C1148(0.83); C1323(0.86); C1324(0.86) | LDD0646 | [7] |
| LDCM0330 | AC66 | HCT 116 | C1026(0.69); C294(0.70); C1198(0.78); C1148(0.80) | LDD0647 | [7] |
| LDCM0331 | AC67 | HCT 116 | C1026(0.78); C1323(0.78); C1324(0.78); C1148(0.86) | LDD0648 | [7] |
| LDCM0332 | AC68 | HCT 116 | C38(0.61); C207(0.76); C1242(0.80); C730(0.84) | LDD0649 | [7] |
| LDCM0333 | AC69 | HCT 116 | C38(0.43); C730(0.72); C735(0.72); C1242(0.79) | LDD0650 | [7] |
| LDCM0334 | AC7 | HCT 116 | C1018(0.83); C1032(0.86); C730(0.92); C735(0.92) | LDD0651 | [7] |
| LDCM0335 | AC70 | HCT 116 | C38(0.41); C1285(0.76); C1280(0.79); C730(0.79) | LDD0652 | [7] |
| LDCM0336 | AC71 | HCT 116 | C38(0.53); C1242(0.76); C730(0.80); C735(0.80) | LDD0653 | [7] |
| LDCM0337 | AC72 | HCT 116 | C38(0.34); C730(0.79); C735(0.79); C1046(0.87) | LDD0654 | [7] |
| LDCM0338 | AC73 | HCT 116 | C1046(1.11); C1041(1.38); C1056(1.41); C38(0.48) | LDD0655 | [7] |
| LDCM0339 | AC74 | HCT 116 | C38(0.44); C1242(0.87); C1280(0.95); C1245(0.97) | LDD0656 | [7] |
| LDCM0340 | AC75 | HCT 116 | C38(0.46); C1242(0.84); C1280(0.90); C1285(0.90) | LDD0657 | [7] |
| LDCM0341 | AC76 | HCT 116 | C38(0.55); C1280(0.84); C730(0.89); C735(0.89) | LDD0658 | [7] |
| LDCM0342 | AC77 | HCT 116 | C38(0.39); C730(0.84); C735(0.84); C1280(0.95) | LDD0659 | [7] |
| LDCM0343 | AC78 | HCT 116 | C38(0.35); C1280(0.88); C1285(0.90); C730(0.95) | LDD0660 | [7] |
| LDCM0344 | AC79 | HCT 116 | C38(0.45); C730(0.82); C735(0.82); C1285(0.91) | LDD0661 | [7] |
| LDCM0345 | AC8 | HCT 116 | C1018(0.71); C1016(0.73); C1032(0.74); C1026(0.79) | LDD0662 | [7] |
| LDCM0346 | AC80 | HCT 116 | C38(0.37); C730(0.78); C735(0.78); C1242(0.79) | LDD0663 | [7] |
| LDCM0347 | AC81 | HCT 116 | C38(0.45); C1245(0.76); C1254(0.76); C1242(0.97) | LDD0664 | [7] |
| LDCM0348 | AC82 | HCT 116 | C38(0.47); C1280(0.84); C1285(0.84); C730(0.85) | LDD0665 | [7] |
| LDCM0349 | AC83 | HCT 116 | C1315(0.64); C1016(0.83); C1018(0.85); C1148(0.85) | LDD0666 | [7] |
| LDCM0350 | AC84 | HCT 116 | C1315(0.66); C1018(0.72); C1016(0.75); C1032(0.75) | LDD0667 | [7] |
| LDCM0351 | AC85 | HCT 116 | C1315(0.68); C1148(0.75); C1018(0.75); C1245(0.77) | LDD0668 | [7] |
| LDCM0352 | AC86 | HCT 116 | C1315(0.75); C1032(0.77); C1148(0.79); C1018(0.79) | LDD0669 | [7] |
| LDCM0353 | AC87 | HCT 116 | C1026(0.73); C1245(0.77); C1254(0.77); C1018(0.77) | LDD0670 | [7] |
| LDCM0354 | AC88 | HCT 116 | C1026(0.62); C1018(0.64); C1032(0.65); C1016(0.68) | LDD0671 | [7] |
| LDCM0355 | AC89 | HCT 116 | C1026(0.64); C1315(0.65); C1018(0.74); C1148(0.76) | LDD0672 | [7] |
| LDCM0357 | AC90 | HCT 116 | C294(0.65); C1315(0.72); C1018(0.79); C1032(0.80) | LDD0674 | [7] |
| LDCM0358 | AC91 | HCT 116 | C1315(0.58); C1148(0.72); C294(0.77); C1018(0.82) | LDD0675 | [7] |
| LDCM0359 | AC92 | HCT 116 | C1148(0.69); C1315(0.71); C1018(0.74); C1026(0.74) | LDD0676 | [7] |
| LDCM0360 | AC93 | HCT 116 | C1018(0.66); C1032(0.67); C1026(0.68); C1315(0.68) | LDD0677 | [7] |
| LDCM0361 | AC94 | HCT 116 | C1315(0.56); C294(0.60); C1016(0.66); C1018(0.72) | LDD0678 | [7] |
| LDCM0362 | AC95 | HCT 116 | C1315(0.71); C294(0.86); C899(0.95); C1198(1.01) | LDD0679 | [7] |
| LDCM0363 | AC96 | HCT 116 | C1315(0.54); C1148(0.70); C1144(0.78); C1016(0.79) | LDD0680 | [7] |
| LDCM0364 | AC97 | HCT 116 | C1026(0.63); C1148(0.68); C1018(0.69); C1032(0.70) | LDD0681 | [7] |
| LDCM0365 | AC98 | HCT 116 | C1148(0.52); C1016(0.66); C1032(0.66); C1245(0.68) | LDD0682 | [7] |
| LDCM0366 | AC99 | HCT 116 | C1148(0.59); C1016(0.59); C1032(0.59); C1018(0.62) | LDD0683 | [7] |
| LDCM0248 | AKOS034007472 | HCT 116 | C1046(1.06); C1056(1.07); C1016(1.03); C1018(0.84) | LDD0565 | [7] |
| LDCM0356 | AKOS034007680 | HCT 116 | C1018(0.84); C1032(0.88); C1323(0.90); C1324(0.90) | LDD0673 | [7] |
| LDCM0275 | AKOS034007705 | HCT 116 | C1016(0.82); C1026(0.85); C1148(0.87); C1018(0.88) | LDD0592 | [7] |
| LDCM0156 | Aniline | NCI-H1299 | 11.60 | LDD0403 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C1193(1.13); C1198(1.13); C44(1.01); C730(0.98) | LDD0078 | [7] |
| LDCM0088 | C45 | HEK-293T | 20.00 | LDD0201 | [4] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [14] |
| LDCM0632 | CL-Sc | Hep-G2 | C406(0.45) | LDD2227 | [9] |
| LDCM0367 | CL1 | HCT 116 | C1242(0.70); C1245(0.77); C1254(0.77); C1324(0.90) | LDD0684 | [7] |
| LDCM0368 | CL10 | HCT 116 | C44(0.96); C294(0.99); C1242(1.12); C1315(1.17) | LDD0685 | [7] |
| LDCM0369 | CL100 | HCT 116 | C1148(0.68); C1016(0.68); C1018(0.77); C1032(0.77) | LDD0686 | [7] |
| LDCM0370 | CL101 | HCT 116 | C1018(0.78); C1032(0.80); C1016(0.83); C1026(0.87) | LDD0687 | [7] |
| LDCM0371 | CL102 | HCT 116 | C1018(0.75); C730(0.75); C735(0.75); C1032(0.78) | LDD0688 | [7] |
| LDCM0372 | CL103 | HCT 116 | C38(0.85); C899(0.86); C1056(0.91); C1016(0.91) | LDD0689 | [7] |
| LDCM0373 | CL104 | HCT 116 | C1323(0.78); C1324(0.78); C1026(0.83); C38(0.84) | LDD0690 | [7] |
| LDCM0374 | CL105 | HCT 116 | C44(0.72); C1041(0.73); C1016(0.73); C1032(0.74) | LDD0691 | [7] |
| LDCM0375 | CL106 | HCT 116 | C44(0.70); C1041(0.74); C1315(0.77); C1016(0.79) | LDD0692 | [7] |
| LDCM0376 | CL107 | HCT 116 | C1041(0.60); C44(0.67); C1052(0.70); C1016(0.72) | LDD0693 | [7] |
| LDCM0377 | CL108 | HCT 116 | C1041(0.61); C1016(0.66); C1052(0.67); C44(0.68) | LDD0694 | [7] |
| LDCM0378 | CL109 | HCT 116 | C1041(0.58); C1032(0.67); C1016(0.68); C1046(0.69) | LDD0695 | [7] |
| LDCM0379 | CL11 | HCT 116 | C1315(1.01); C294(1.01); C44(1.08); C1242(1.28) | LDD0696 | [7] |
| LDCM0380 | CL110 | HCT 116 | C44(0.66); C1016(0.67); C1026(0.67); C1032(0.70) | LDD0697 | [7] |
| LDCM0381 | CL111 | HCT 116 | C1016(0.62); C1041(0.64); C1032(0.65); C1018(0.72) | LDD0698 | [7] |
| LDCM0382 | CL112 | HCT 116 | C670(0.71); C673(0.71); C1032(0.76); C1148(0.76) | LDD0699 | [7] |
| LDCM0383 | CL113 | HCT 116 | C1052(0.58); C1018(0.63); C1032(0.64); C1016(0.68) | LDD0700 | [7] |
| LDCM0384 | CL114 | HCT 116 | C670(0.61); C673(0.61); C1052(0.66); C1148(0.81) | LDD0701 | [7] |
| LDCM0385 | CL115 | HCT 116 | C1052(0.59); C1032(0.67); C670(0.67); C673(0.67) | LDD0702 | [7] |
| LDCM0386 | CL116 | HCT 116 | C1052(0.67); C670(0.75); C673(0.75); C1026(0.78) | LDD0703 | [7] |
| LDCM0387 | CL117 | HCT 116 | C1052(0.95); C1245(1.02); C1254(1.02); C1026(1.04) | LDD0704 | [7] |
| LDCM0388 | CL118 | HCT 116 | C1046(0.81); C1056(0.84); C1052(0.84); C1041(0.85) | LDD0705 | [7] |
| LDCM0389 | CL119 | HCT 116 | C1052(0.71); C1046(0.72); C1056(0.73); C1245(0.78) | LDD0706 | [7] |
| LDCM0390 | CL12 | HCT 116 | C294(0.96); C1148(0.96); C44(1.01); C1242(1.11) | LDD0707 | [7] |
| LDCM0391 | CL120 | HCT 116 | C1324(0.74); C1046(0.82); C1056(0.84); C1052(0.84) | LDD0708 | [7] |
| LDCM0392 | CL121 | HCT 116 | C1041(0.54); C1056(0.59); C1242(0.67); C1016(0.69) | LDD0709 | [7] |
| LDCM0393 | CL122 | HCT 116 | C1242(0.89); C1288(0.90); C1324(0.91); C1041(0.94) | LDD0710 | [7] |
| LDCM0394 | CL123 | HCT 116 | C1148(0.40); C1288(0.72); C1280(0.78); C1016(0.79) | LDD0711 | [7] |
| LDCM0395 | CL124 | HCT 116 | C1288(0.71); C1041(0.74); C1280(0.89); C1242(0.94) | LDD0712 | [7] |
| LDCM0396 | CL125 | HCT 116 | C294(0.78); C1198(0.79); C1193(0.85); C1018(0.85) | LDD0713 | [7] |
| LDCM0397 | CL126 | HCT 116 | C294(0.67); C1016(0.82); C670(0.83); C673(0.83) | LDD0714 | [7] |
| LDCM0398 | CL127 | HCT 116 | C1026(0.77); C1018(0.86); C1032(0.88); C294(0.92) | LDD0715 | [7] |
| LDCM0399 | CL128 | HCT 116 | C1148(0.74); C1018(0.83); C1026(0.85); C1032(0.85) | LDD0716 | [7] |
| LDCM0400 | CL13 | HCT 116 | C44(0.97); C1315(1.02); C294(1.06); C207(1.10) | LDD0717 | [7] |
| LDCM0401 | CL14 | HCT 116 | C1242(0.78); C1148(0.92); C1144(0.94); C1315(0.98) | LDD0718 | [7] |
| LDCM0402 | CL15 | HCT 116 | C294(0.88); C1315(0.92); C44(1.02); C730(1.02) | LDD0719 | [7] |
| LDCM0403 | CL16 | HCT 116 | C1016(0.67); C1026(0.67); C38(0.83); C1018(0.86) | LDD0720 | [7] |
| LDCM0404 | CL17 | HCT 116 | C1032(0.80); C1016(0.81); C1026(0.82); C1052(0.87) | LDD0721 | [7] |
| LDCM0405 | CL18 | HCT 116 | C899(0.92); C38(1.13); C1032(1.19); C1144(1.21) | LDD0722 | [7] |
| LDCM0406 | CL19 | HCT 116 | C1026(0.74); C1052(0.79); C1056(0.84); C1046(0.89) | LDD0723 | [7] |
| LDCM0407 | CL2 | HCT 116 | C294(0.62); C1315(0.89); C207(0.94); C1026(0.94) | LDD0724 | [7] |
| LDCM0408 | CL20 | HCT 116 | C1046(0.72); C1026(0.74); C1056(0.79); C1052(0.85) | LDD0725 | [7] |
| LDCM0409 | CL21 | HCT 116 | C1026(0.77); C1046(0.78); C899(0.85); C1016(0.89) | LDD0726 | [7] |
| LDCM0410 | CL22 | HCT 116 | C1032(0.82); C1016(0.83); C1026(0.84); C38(0.87) | LDD0727 | [7] |
| LDCM0411 | CL23 | HCT 116 | C1026(0.59); C1046(0.71); C1056(0.75); C1052(0.78) | LDD0728 | [7] |
| LDCM0412 | CL24 | HCT 116 | C1056(0.85); C1046(0.86); C1016(1.02); C1018(1.10) | LDD0729 | [7] |
| LDCM0413 | CL25 | HCT 116 | C1046(0.70); C1056(0.78); C1016(0.73); C1018(0.83) | LDD0730 | [7] |
| LDCM0414 | CL26 | HCT 116 | C1046(0.72); C1056(0.78); C1016(0.78); C1018(0.85) | LDD0731 | [7] |
| LDCM0415 | CL27 | HCT 116 | C1046(0.74); C1056(0.76); C1016(0.78); C1018(0.86) | LDD0732 | [7] |
| LDCM0416 | CL28 | HCT 116 | C1046(0.74); C1056(0.78); C1016(0.84); C1018(0.92) | LDD0733 | [7] |
| LDCM0417 | CL29 | HCT 116 | C1046(0.82); C1056(0.84); C1016(0.99); C1018(1.05) | LDD0734 | [7] |
| LDCM0418 | CL3 | HCT 116 | C1046(1.03); C1056(1.03); C1016(1.00); C1018(0.96) | LDD0735 | [7] |
| LDCM0419 | CL30 | HCT 116 | C1046(0.96); C1056(1.07); C1016(0.78); C1018(0.87) | LDD0736 | [7] |
| LDCM0420 | CL31 | HCT 116 | C1016(0.88); C1018(0.85); C1026(0.84); C1032(0.85) | LDD0737 | [7] |
| LDCM0421 | CL32 | HCT 116 | C1016(1.48); C1018(1.43); C1026(1.53); C1032(1.43) | LDD0738 | [7] |
| LDCM0422 | CL33 | HCT 116 | C1016(1.54); C1018(1.34); C1026(1.74); C1032(1.34) | LDD0739 | [7] |
| LDCM0423 | CL34 | HCT 116 | C1016(1.09); C1018(0.94); C1026(1.25); C1032(0.94) | LDD0740 | [7] |
| LDCM0424 | CL35 | HCT 116 | C1016(0.86); C1018(0.80); C1026(0.91); C1032(0.80) | LDD0741 | [7] |
| LDCM0425 | CL36 | HCT 116 | C1016(0.96); C1018(0.87); C1026(1.05); C1032(0.87) | LDD0742 | [7] |
| LDCM0426 | CL37 | HCT 116 | C1016(0.83); C1018(0.88); C1026(0.78); C1032(0.88) | LDD0743 | [7] |
| LDCM0428 | CL39 | HCT 116 | C1016(0.93); C1018(0.97); C1026(0.90); C1032(0.97) | LDD0745 | [7] |
| LDCM0429 | CL4 | HCT 116 | C1046(0.92); C1056(0.92); C1016(0.97); C1018(1.07) | LDD0746 | [7] |
| LDCM0430 | CL40 | HCT 116 | C1016(0.80); C1018(0.73); C1026(0.86); C1032(0.73) | LDD0747 | [7] |
| LDCM0431 | CL41 | HCT 116 | C1016(1.14); C1018(0.96); C1026(1.33); C1032(0.96) | LDD0748 | [7] |
| LDCM0432 | CL42 | HCT 116 | C1016(1.25); C1018(1.01); C1026(1.49); C1032(1.01) | LDD0749 | [7] |
| LDCM0433 | CL43 | HCT 116 | C1016(1.05); C1018(0.92); C1026(1.19); C1032(0.92) | LDD0750 | [7] |
| LDCM0434 | CL44 | HCT 116 | C1016(1.09); C1018(1.03); C1026(1.16); C1032(1.03) | LDD0751 | [7] |
| LDCM0435 | CL45 | HCT 116 | C1016(0.97); C1018(0.98); C1026(0.96); C1032(0.98) | LDD0752 | [7] |
| LDCM0436 | CL46 | HCT 116 | C1041(0.90); C1046(0.95); C1056(0.94); C1016(0.92) | LDD0753 | [7] |
| LDCM0437 | CL47 | HCT 116 | C1041(0.91); C1046(1.28); C1056(0.94); C1016(0.97) | LDD0754 | [7] |
| LDCM0438 | CL48 | HCT 116 | C1041(0.95); C1046(1.15); C1056(0.97); C1016(1.01) | LDD0755 | [7] |
| LDCM0439 | CL49 | HCT 116 | C1041(0.76); C1046(0.60); C1056(0.78); C1016(0.97) | LDD0756 | [7] |
| LDCM0440 | CL5 | HCT 116 | C1046(1.03); C1056(1.03); C1016(1.06); C1018(1.17) | LDD0757 | [7] |
| LDCM0441 | CL50 | HCT 116 | C1041(0.73); C1046(1.11); C1056(0.81); C1016(0.90) | LDD0758 | [7] |
| LDCM0442 | CL51 | HCT 116 | C1041(0.91); C1046(0.97); C1056(0.89); C1016(0.97) | LDD0759 | [7] |
| LDCM0443 | CL52 | HCT 116 | C1041(0.89); C1046(0.94); C1056(0.88); C1016(1.20) | LDD0760 | [7] |
| LDCM0444 | CL53 | HCT 116 | C1041(0.76); C1046(0.84); C1056(0.81); C1016(1.22) | LDD0761 | [7] |
| LDCM0445 | CL54 | HCT 116 | C1041(0.65); C1046(0.73); C1056(0.65); C1016(0.99) | LDD0762 | [7] |
| LDCM0446 | CL55 | HCT 116 | C1041(0.73); C1046(0.89); C1056(0.84); C1016(0.93) | LDD0763 | [7] |
| LDCM0447 | CL56 | HCT 116 | C1041(0.86); C1046(1.02); C1056(0.91); C1016(0.95) | LDD0764 | [7] |
| LDCM0448 | CL57 | HCT 116 | C1041(0.81); C1046(0.94); C1056(0.88); C1016(1.22) | LDD0765 | [7] |
| LDCM0449 | CL58 | HCT 116 | C1041(0.95); C1046(1.30); C1056(0.94); C1016(0.94) | LDD0766 | [7] |
| LDCM0450 | CL59 | HCT 116 | C1041(0.93); C1046(0.96); C1056(0.96); C1016(1.33) | LDD0767 | [7] |
| LDCM0451 | CL6 | HCT 116 | C1046(1.49); C1056(1.49); C1016(1.52); C1018(1.62) | LDD0768 | [7] |
| LDCM0452 | CL60 | HCT 116 | C1041(0.99); C1046(1.09); C1056(1.00); C1016(0.92) | LDD0769 | [7] |
| LDCM0453 | CL61 | HCT 116 | C1016(1.11); C1018(1.12); C1026(1.13); C1032(1.14) | LDD0770 | [7] |
| LDCM0454 | CL62 | HCT 116 | C1016(0.91); C1018(0.94); C1026(1.02); C1032(0.90) | LDD0771 | [7] |
| LDCM0455 | CL63 | HCT 116 | C1016(1.00); C1018(1.00); C1026(1.08); C1032(1.00) | LDD0772 | [7] |
| LDCM0456 | CL64 | HCT 116 | C1016(1.01); C1018(1.01); C1026(1.05); C1032(1.03) | LDD0773 | [7] |
| LDCM0457 | CL65 | HCT 116 | C1016(0.98); C1018(1.04); C1026(1.05); C1032(1.01) | LDD0774 | [7] |
| LDCM0458 | CL66 | HCT 116 | C1016(0.94); C1018(0.99); C1026(1.00); C1032(0.97) | LDD0775 | [7] |
| LDCM0459 | CL67 | HCT 116 | C1016(0.88); C1018(0.88); C1026(0.91); C1032(0.89) | LDD0776 | [7] |
| LDCM0460 | CL68 | HCT 116 | C1016(1.15); C1018(1.16); C1026(1.21); C1032(1.15) | LDD0777 | [7] |
| LDCM0461 | CL69 | HCT 116 | C1016(1.69); C1018(1.83); C1026(1.35); C1032(1.51) | LDD0778 | [7] |
| LDCM0462 | CL7 | HCT 116 | C1046(1.37); C1056(1.37); C1016(1.33); C1018(1.35) | LDD0779 | [7] |
| LDCM0463 | CL70 | HCT 116 | C1016(1.23); C1018(1.26); C1026(1.33); C1032(1.23) | LDD0780 | [7] |
| LDCM0464 | CL71 | HCT 116 | C1016(0.87); C1018(0.87); C1026(0.88); C1032(0.90) | LDD0781 | [7] |
| LDCM0465 | CL72 | HCT 116 | C1016(0.99); C1018(1.00); C1026(0.97); C1032(1.01) | LDD0782 | [7] |
| LDCM0466 | CL73 | HCT 116 | C1016(0.87); C1018(0.87); C1026(0.94); C1032(0.87) | LDD0783 | [7] |
| LDCM0467 | CL74 | HCT 116 | C1016(0.98); C1018(1.02); C1026(1.07); C1032(1.00) | LDD0784 | [7] |
| LDCM0469 | CL76 | HCT 116 | C1041(20.00); C1046(20.00); C1056(20.00); C1016(1.47) | LDD0786 | [7] |
| LDCM0470 | CL77 | HCT 116 | C1041(0.71); C1046(0.71); C1056(0.71); C1016(0.93) | LDD0787 | [7] |
| LDCM0471 | CL78 | HCT 116 | C1041(0.57); C1046(0.57); C1056(0.57); C1016(0.93) | LDD0788 | [7] |
| LDCM0472 | CL79 | HCT 116 | C1041(0.71); C1046(0.71); C1056(0.71); C1016(1.12) | LDD0789 | [7] |
| LDCM0473 | CL8 | HCT 116 | C1046(1.45); C1056(1.45); C1016(1.70); C1018(2.03) | LDD0790 | [7] |
| LDCM0474 | CL80 | HCT 116 | C1041(3.56); C1046(3.56); C1056(3.56); C1016(1.55) | LDD0791 | [7] |
| LDCM0475 | CL81 | HCT 116 | C1041(0.48); C1046(0.48); C1056(0.48); C1016(1.10) | LDD0792 | [7] |
| LDCM0476 | CL82 | HCT 116 | C1041(0.60); C1046(0.60); C1056(0.60); C1016(1.24) | LDD0793 | [7] |
| LDCM0477 | CL83 | HCT 116 | C1041(0.62); C1046(0.62); C1056(0.62); C1016(1.08) | LDD0794 | [7] |
| LDCM0478 | CL84 | HCT 116 | C1041(0.69); C1046(0.69); C1056(0.69); C1016(1.46) | LDD0795 | [7] |
| LDCM0479 | CL85 | HCT 116 | C1041(0.46); C1046(0.46); C1056(0.46); C1016(0.98) | LDD0796 | [7] |
| LDCM0480 | CL86 | HCT 116 | C1041(0.76); C1046(0.76); C1056(0.76); C1016(1.02) | LDD0797 | [7] |
| LDCM0481 | CL87 | HCT 116 | C1041(0.46); C1046(0.46); C1056(0.46); C1016(1.04) | LDD0798 | [7] |
| LDCM0482 | CL88 | HCT 116 | C1041(0.55); C1046(0.55); C1056(0.55); C1016(1.03) | LDD0799 | [7] |
| LDCM0483 | CL89 | HCT 116 | C1041(0.58); C1046(0.58); C1056(0.58); C1016(1.28) | LDD0800 | [7] |
| LDCM0484 | CL9 | HCT 116 | C1046(1.26); C1056(1.26); C1016(1.22); C1018(1.17) | LDD0801 | [7] |
| LDCM0485 | CL90 | HCT 116 | C1041(0.66); C1046(0.66); C1056(0.66); C1016(1.14) | LDD0802 | [7] |
| LDCM0486 | CL91 | HCT 116 | C1016(1.00); C1018(0.95); C1032(0.95); C1144(0.91) | LDD0803 | [7] |
| LDCM0487 | CL92 | HCT 116 | C1016(0.90); C1018(0.82); C1032(0.82); C1144(1.07) | LDD0804 | [7] |
| LDCM0488 | CL93 | HCT 116 | C1016(1.58); C1018(1.48); C1032(1.48); C1144(0.97) | LDD0805 | [7] |
| LDCM0489 | CL94 | HCT 116 | C1016(0.93); C1018(1.08); C1032(1.08); C1144(0.90) | LDD0806 | [7] |
| LDCM0490 | CL95 | HCT 116 | C1016(0.81); C1018(0.91); C1032(0.91); C1144(0.95) | LDD0807 | [7] |
| LDCM0491 | CL96 | HCT 116 | C1016(0.90); C1018(1.24); C1032(1.24); C1144(1.08) | LDD0808 | [7] |
| LDCM0492 | CL97 | HCT 116 | C1016(0.69); C1018(0.65); C1032(0.65); C1144(0.91) | LDD0809 | [7] |
| LDCM0493 | CL98 | HCT 116 | C1016(0.75); C1018(0.71); C1032(0.71); C1144(0.84) | LDD0810 | [7] |
| LDCM0494 | CL99 | HCT 116 | C1016(0.71); C1018(0.81); C1032(0.81); C1144(0.97) | LDD0811 | [7] |
| LDCM0495 | E2913 | HEK-293T | C207(1.20); C594(1.12); C294(1.08); C394(0.94) | LDD1698 | [18] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C743(1.70); C1267(1.53); C1323(1.19); C1315(1.11) | LDD1702 | [5] |
| LDCM0468 | Fragment33 | HCT 116 | C1016(0.93); C1018(0.96); C1026(0.97); C1032(0.99) | LDD0785 | [7] |
| LDCM0427 | Fragment51 | HCT 116 | C1016(1.27); C1018(0.97); C1026(1.57); C1032(0.97) | LDD0744 | [7] |
| LDCM0022 | KB02 | HCT 116 | C1245(1.96); C1254(1.96); C730(1.81); C735(1.81) | LDD0080 | [7] |
| LDCM0023 | KB03 | HCT 116 | C1245(2.59); C1254(2.59); C730(2.47); C735(2.47) | LDD0081 | [7] |
| LDCM0024 | KB05 | HCT 116 | C1245(1.95); C1254(1.95); C730(1.93); C735(1.93) | LDD0082 | [7] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C207(1.45) | LDD2100 | [5] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C594(1.08) | LDD2104 | [5] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C594(1.24) | LDD2107 | [5] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C594(0.84); C38(0.62); C294(0.95) | LDD2109 | [5] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C38(10.30) | LDD2111 | [5] |
| LDCM0523 | Nucleophilic fragment 24b | MDA-MB-231 | C594(0.58) | LDD2116 | [5] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C594(0.97) | LDD2123 | [5] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C594(0.95); C294(0.92) | LDD2125 | [5] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C594(0.99) | LDD2127 | [5] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C594(0.70) | LDD2129 | [5] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C594(1.22) | LDD2135 | [5] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C594(1.25); C294(1.41) | LDD2136 | [5] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C594(1.02) | LDD2137 | [5] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C594(1.27) | LDD1700 | [5] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C594(0.94) | LDD2140 | [5] |
| LDCM0021 | THZ1 | HCT 116 | C707(1.17); C710(1.17); C720(1.17); C1242(1.46) | LDD2173 | [7] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Androgen receptor (AR) | Nuclear hormone receptor family | P10275 | |||
Other
References
