Details of the Target
General Information of Target
| Target ID | LDTP00449 | |||||
|---|---|---|---|---|---|---|
| Target Name | Proteasome subunit alpha type-7 (PSMA7) | |||||
| Gene Name | PSMA7 | |||||
| Gene ID | 5688 | |||||
| Synonyms |
HSPC; Proteasome subunit alpha type-7; Proteasome subunit RC6-1; Proteasome subunit XAPC7 |
|||||
| 3D Structure | ||||||
| Sequence |
MSYDRAITVFSPDGHLFQVEYAQEAVKKGSTAVGVRGRDIVVLGVEKKSVAKLQDERTVR
KICALDDNVCMAFAGLTADARIVINRARVECQSHRLTVEDPVTVEYITRYIASLKQRYTQ SNGRRPFGISALIVGFDFDGTPRLYQTDPSGTYHAWKANAIGRGAKSVREFLEKNYTDEA IETDDLTIKLVIKALLEVVQSGGKNIELAVMRRDQSLKILNPEEIEKYVAEIEKEKEENE KKKQKKAS |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Peptidase T1A family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Component of the 20S core proteasome complex involved in the proteolytic degradation of most intracellular proteins. This complex plays numerous essential roles within the cell by associating with different regulatory particles. Associated with two 19S regulatory particles, forms the 26S proteasome and thus participates in the ATP-dependent degradation of ubiquitinated proteins. The 26S proteasome plays a key role in the maintenance of protein homeostasis by removing misfolded or damaged proteins that could impair cellular functions, and by removing proteins whose functions are no longer required. Associated with the PA200 or PA28, the 20S proteasome mediates ubiquitin-independent protein degradation. This type of proteolysis is required in several pathways including spermatogenesis (20S-PA200 complex) or generation of a subset of MHC class I-presented antigenic peptides (20S-PA28 complex). Inhibits the transactivation function of HIF-1A under both normoxic and hypoxia-mimicking conditions. The interaction with EMAP2 increases the proteasome-mediated HIF-1A degradation under the hypoxic conditions. Plays a role in hepatitis C virus internal ribosome entry site-mediated translation. Mediates nuclear translocation of the androgen receptor (AR) and thereby enhances androgen-mediated transactivation. Promotes MAVS degradation and thereby negatively regulates MAVS-mediated innate immune response.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| MDST8 | SNV: p.G37E | DBIA Probe Info | |||
| MFE280 | SNV: p.Ter249WextTer1 | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
10.39 | LDD0402 | [1] | |
|
A-EBA Probe Info |
 |
3.35 | LDD0215 | [2] | |
|
N1 Probe Info |
 |
100.00 | LDD0242 | [3] | |
|
TH211 Probe Info |
 |
Y118(20.00); Y153(20.00); Y21(19.27); Y110(13.40) | LDD0257 | [4] | |
|
TH214 Probe Info |
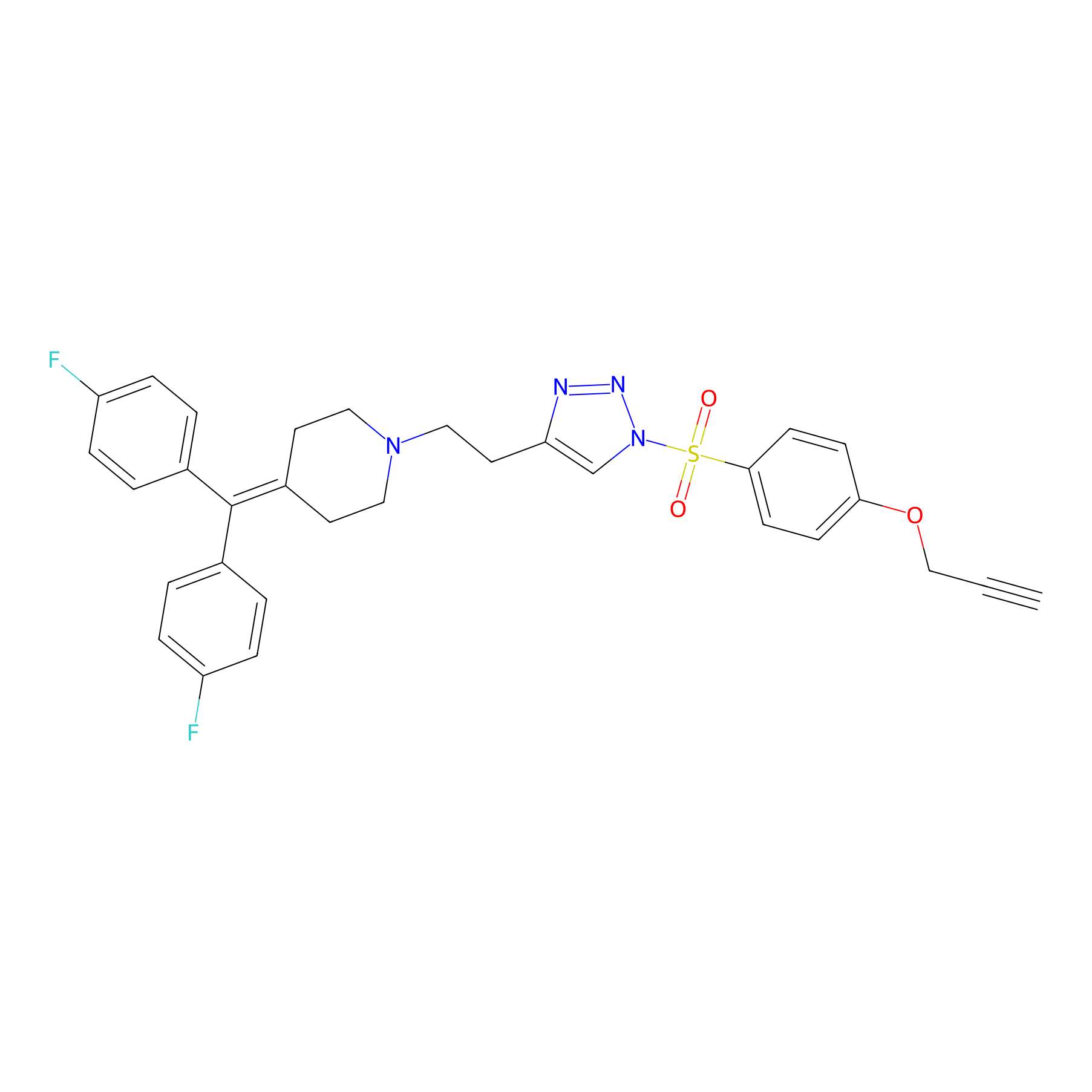 |
Y153(10.65) | LDD0258 | [4] | |
|
TH216 Probe Info |
 |
Y110(20.00); Y118(17.85); Y106(11.93); Y145(11.26) | LDD0259 | [4] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [5] | |
|
STPyne Probe Info |
 |
K115(2.70); K157(8.85); K166(4.67); K218(0.38) | LDD0277 | [6] | |
|
BTD Probe Info |
 |
C91(3.01) | LDD1699 | [7] | |
|
AZ-9 Probe Info |
 |
E182(10.00) | LDD2208 | [8] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [6] | |
|
OPA-S-S-alkyne Probe Info |
 |
K28(2.85) | LDD3494 | [9] | |
|
Probe 1 Probe Info |
 |
Y106(28.95); Y110(51.40); Y176(12.58); Y228(24.01) | LDD3495 | [10] | |
|
AHL-Pu-1 Probe Info |
 |
C63(2.08) | LDD0170 | [11] | |
|
EA-probe Probe Info |
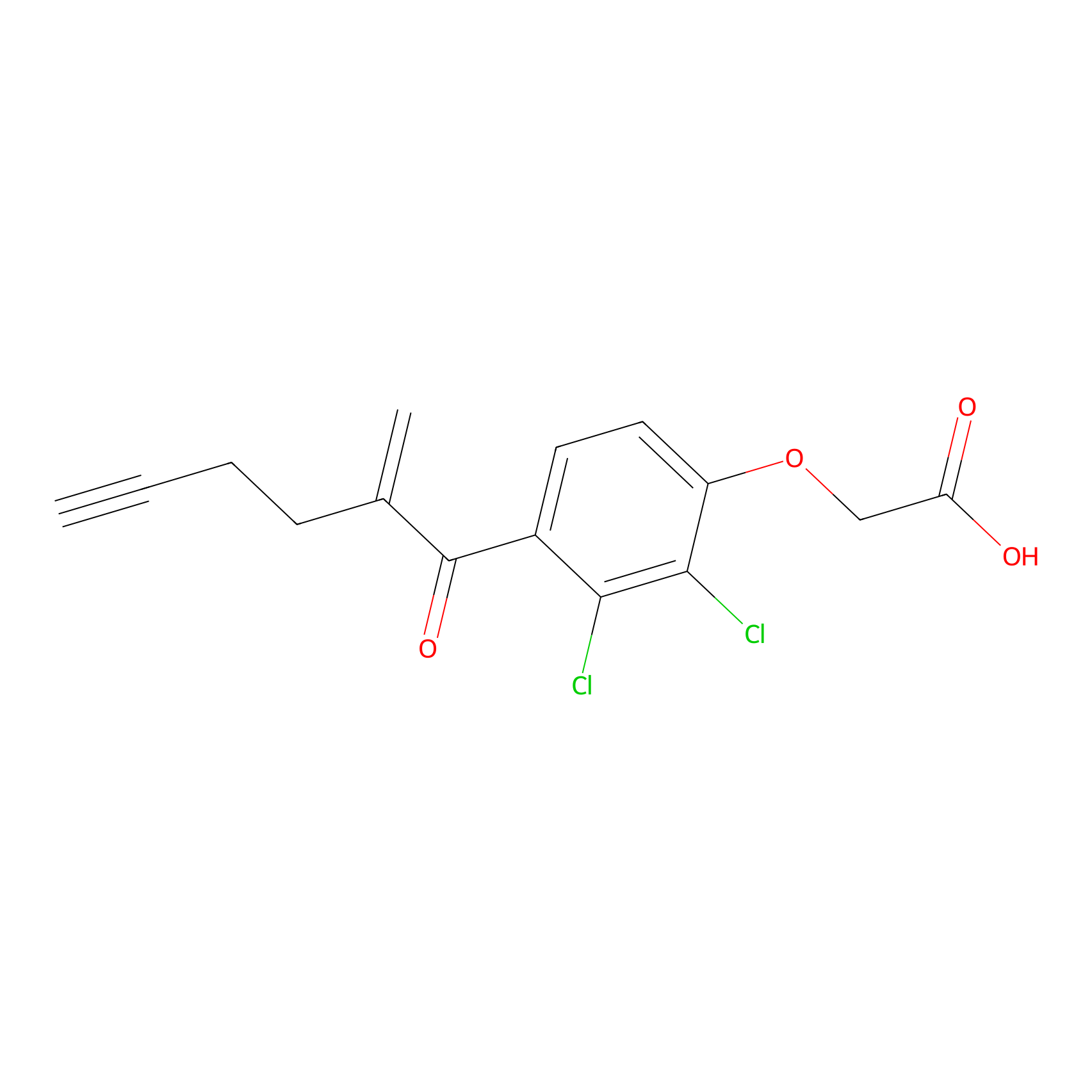 |
N.A. | LDD0440 | [12] | |
|
HHS-482 Probe Info |
 |
Y110(0.74); Y118(0.91); Y145(0.50); Y153(0.85) | LDD0285 | [13] | |
|
HHS-475 Probe Info |
 |
Y110(0.97); Y118(1.03); Y21(3.06) | LDD0264 | [14] | |
|
HHS-465 Probe Info |
 |
Y106(8.78); Y110(8.65); Y118(9.00); Y153(10.00) | LDD2237 | [15] | |
|
DBIA Probe Info |
 |
C63(1.38); C70(1.38) | LDD0080 | [16] | |
|
5E-2FA Probe Info |
 |
H15(0.00); H154(0.00) | LDD2235 | [17] | |
|
ATP probe Probe Info |
 |
K28(0.00); K27(0.00); K234(0.00); K236(0.00) | LDD0199 | [18] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C63(0.00); C70(0.00); C91(0.00) | LDD0038 | [19] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [20] | |
|
IPIAA_L Probe Info |
 |
N.A. | LDD0031 | [21] | |
|
Lodoacetamide azide Probe Info |
 |
C63(0.00); C70(0.00); C91(0.00) | LDD0037 | [19] | |
|
JW-RF-010 Probe Info |
 |
C70(0.00); C63(0.00) | LDD0026 | [22] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [23] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [22] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [24] | |
|
KY-26 Probe Info |
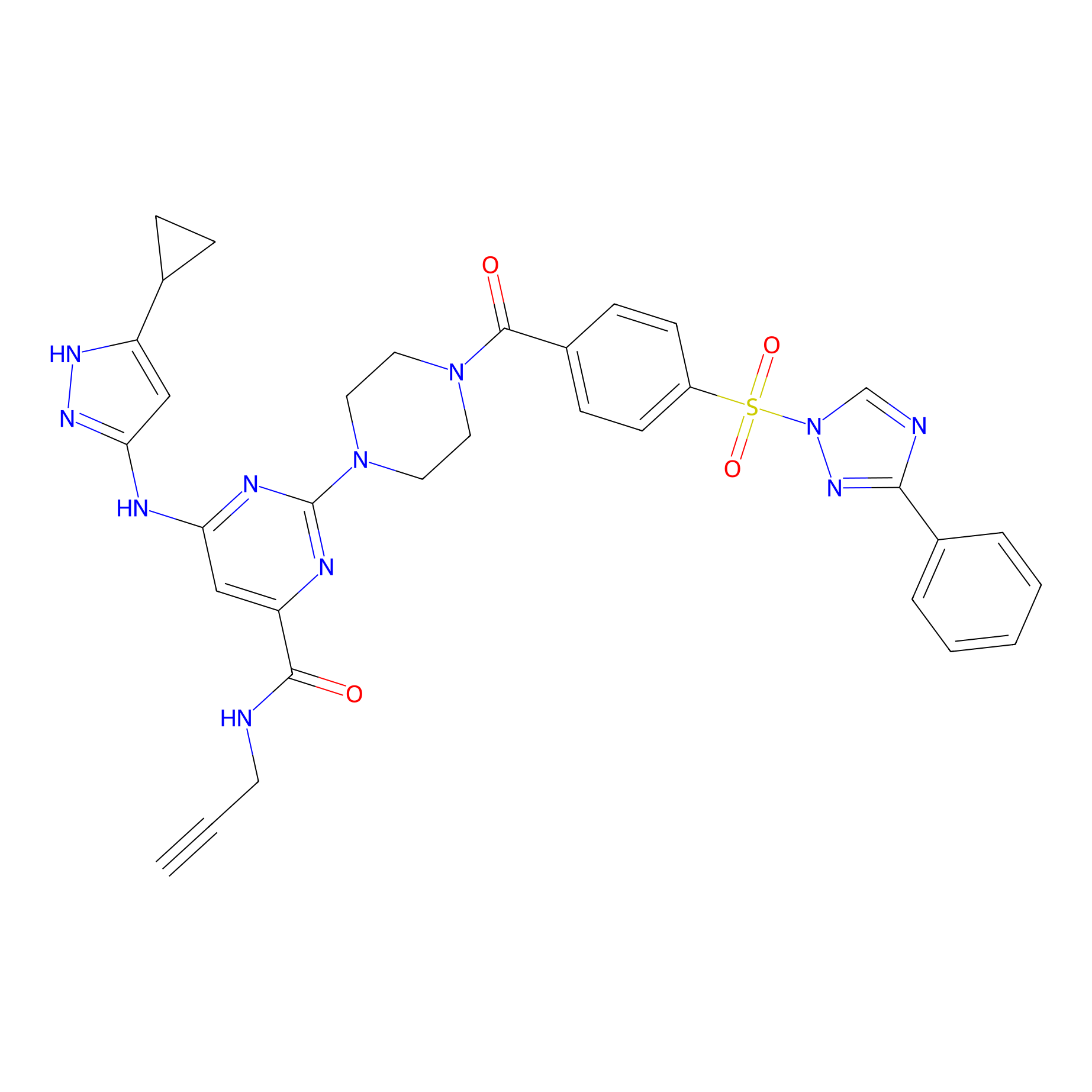 |
N.A. | LDD0301 | [25] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [26] | |
|
IPM Probe Info |
 |
N.A. | LDD0147 | [22] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [24] | |
|
OSF Probe Info |
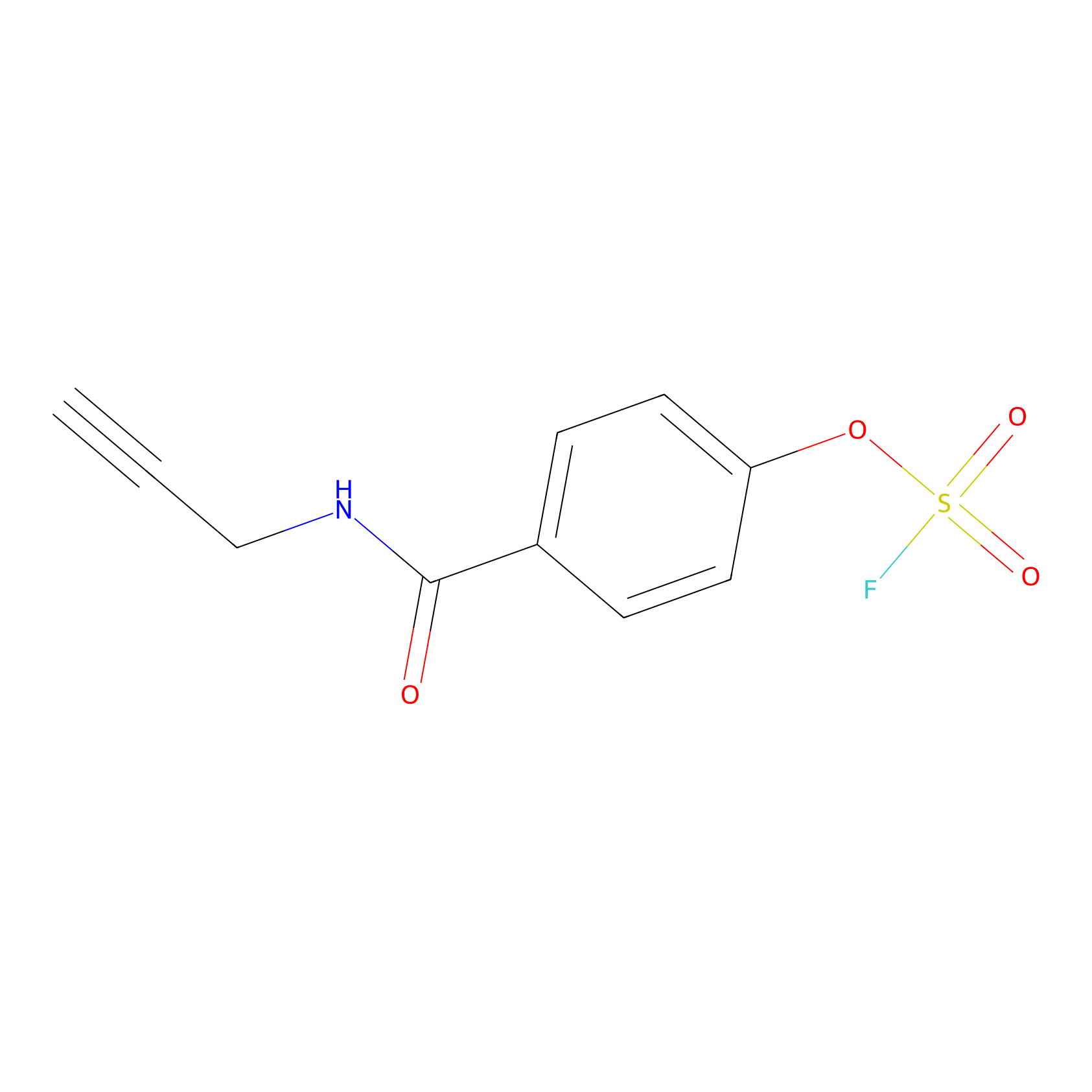 |
Y118(0.00); Y110(0.00) | LDD0029 | [27] | |
|
SF Probe Info |
 |
Y118(0.00); Y106(0.00); Y145(0.00); Y110(0.00) | LDD0028 | [27] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [28] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [29] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [30] | |
|
Acrolein Probe Info |
 |
H154(0.00); C70(0.00) | LDD0217 | [31] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [31] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [31] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [32] | |
|
AOyne Probe Info |
 |
14.90 | LDD0443 | [33] | |
|
MPP-AC Probe Info |
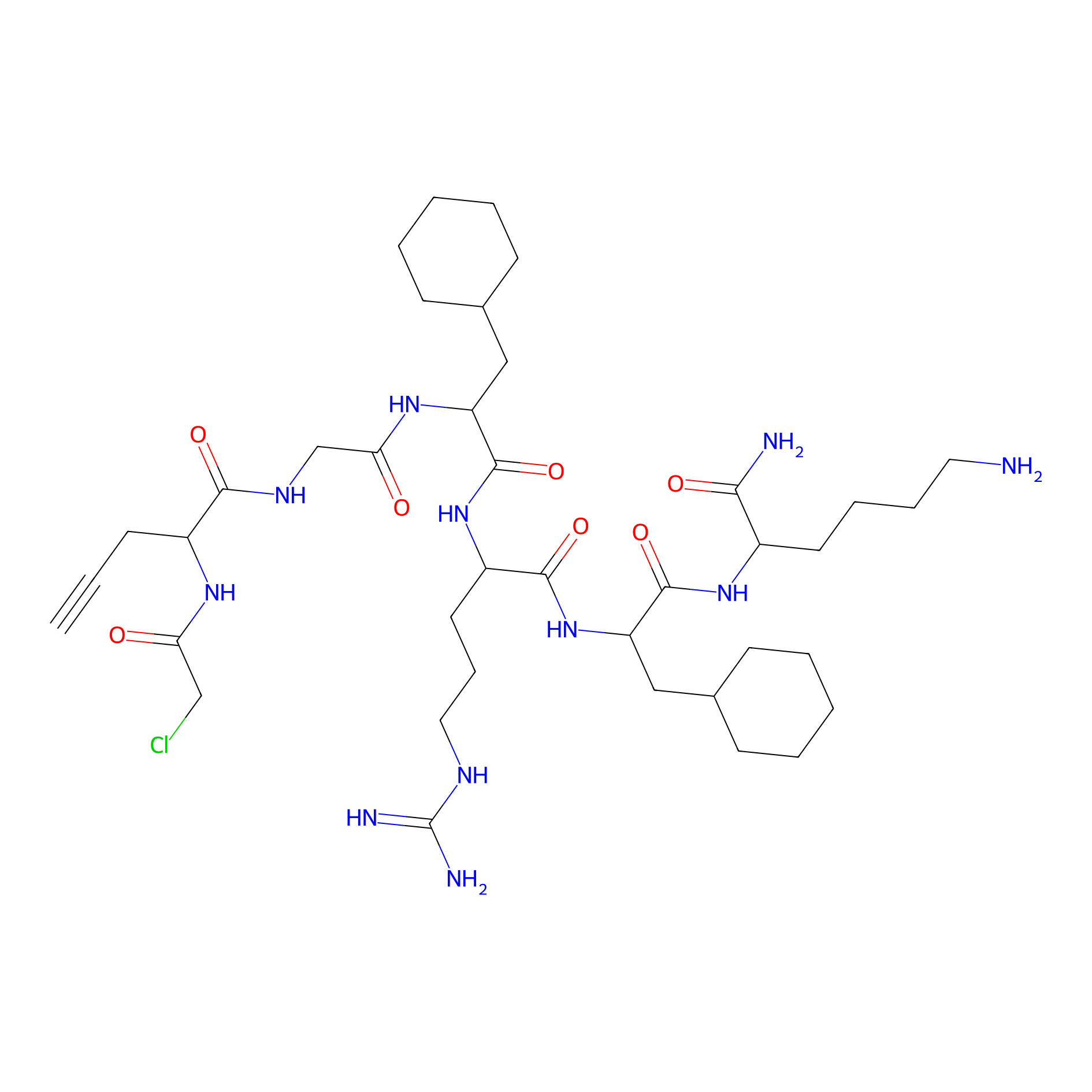 |
N.A. | LDD0428 | [34] | |
|
NAIA_5 Probe Info |
 |
C91(0.00); C70(0.00); C63(0.00) | LDD2223 | [23] | |
|
TER-AC Probe Info |
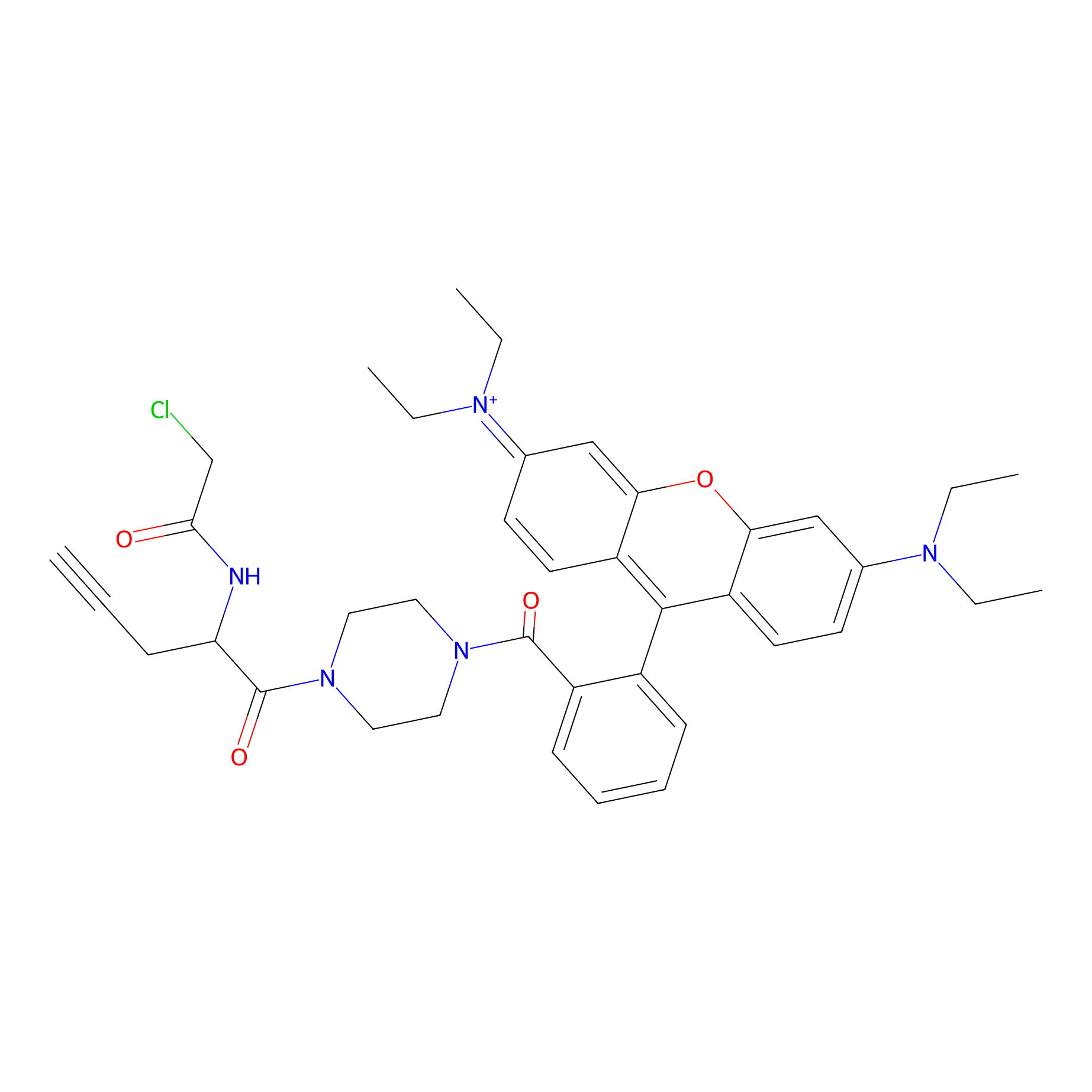 |
N.A. | LDD0426 | [34] | |
|
TPP-AC Probe Info |
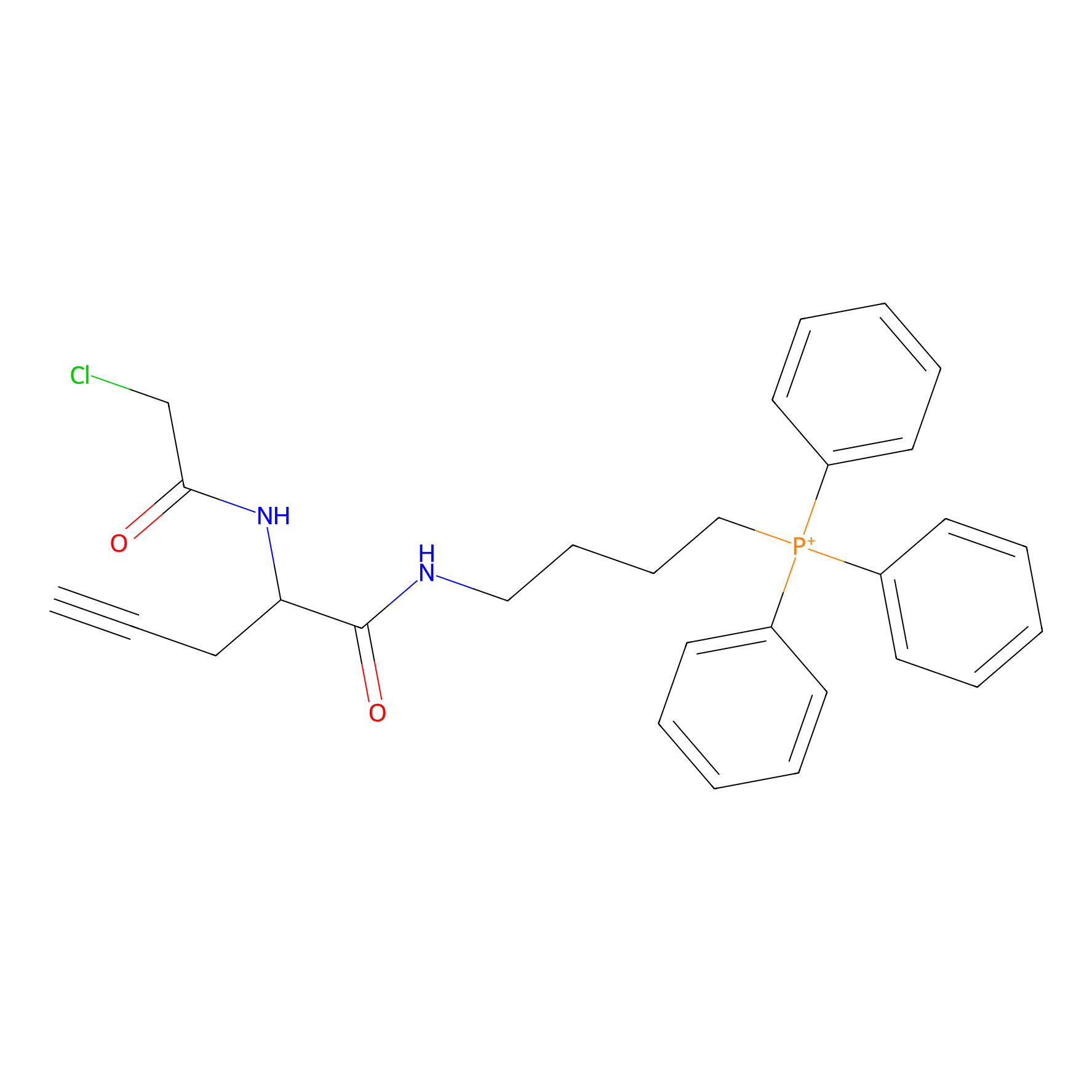 |
N.A. | LDD0427 | [34] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe14 Probe Info |
 |
20.00 | LDD0477 | [35] | |
|
DA-2 Probe Info |
 |
N.A. | LDD0070 | [36] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0068 | [37] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C91(0.32) | LDD2142 | [7] |
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C91(0.51) | LDD2112 | [7] |
| LDCM0502 | 1-(Cyanoacetyl)piperidine | MDA-MB-231 | C91(0.58) | LDD2095 | [7] |
| LDCM0537 | 2-Cyano-N,N-dimethylacetamide | MDA-MB-231 | C91(1.14) | LDD2130 | [7] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C91(0.70) | LDD2117 | [7] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C91(1.34) | LDD2152 | [7] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C91(1.19) | LDD2103 | [7] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C91(0.47) | LDD2132 | [7] |
| LDCM0538 | 4-(Cyanoacetyl)morpholine | MDA-MB-231 | C91(0.79) | LDD2131 | [7] |
| LDCM0025 | 4SU-RNA | DM93 | C63(2.08) | LDD0170 | [11] |
| LDCM0026 | 4SU-RNA+native RNA | DM93 | C63(3.19) | LDD0171 | [11] |
| LDCM0214 | AC1 | HCT 116 | C70(0.75); C63(0.75); C91(1.08) | LDD0531 | [16] |
| LDCM0215 | AC10 | HCT 116 | C70(0.67); C63(0.67); C91(1.17) | LDD0532 | [16] |
| LDCM0216 | AC100 | HCT 116 | C70(0.81); C63(0.81); C91(1.00) | LDD0533 | [16] |
| LDCM0217 | AC101 | HCT 116 | C70(0.87); C63(0.87); C91(1.08) | LDD0534 | [16] |
| LDCM0218 | AC102 | HCT 116 | C70(1.15); C63(1.15); C91(0.98) | LDD0535 | [16] |
| LDCM0219 | AC103 | HCT 116 | C70(0.71); C63(0.71); C91(1.18) | LDD0536 | [16] |
| LDCM0220 | AC104 | HCT 116 | C70(0.61); C63(0.61); C91(1.15) | LDD0537 | [16] |
| LDCM0221 | AC105 | HCT 116 | C70(0.52); C63(0.52); C91(1.21) | LDD0538 | [16] |
| LDCM0222 | AC106 | HCT 116 | C70(0.62); C63(0.62); C91(1.20) | LDD0539 | [16] |
| LDCM0223 | AC107 | HCT 116 | C70(0.68); C63(0.68); C91(0.68) | LDD0540 | [16] |
| LDCM0224 | AC108 | HCT 116 | C70(0.76); C63(0.76); C91(1.04) | LDD0541 | [16] |
| LDCM0225 | AC109 | HCT 116 | C70(1.03); C63(1.03); C91(0.83) | LDD0542 | [16] |
| LDCM0226 | AC11 | HCT 116 | C70(0.56); C63(0.56); C91(1.08) | LDD0543 | [16] |
| LDCM0227 | AC110 | HCT 116 | C70(0.99); C63(0.99); C91(0.78) | LDD0544 | [16] |
| LDCM0228 | AC111 | HCT 116 | C70(0.90); C63(0.90); C91(0.89) | LDD0545 | [16] |
| LDCM0229 | AC112 | HCT 116 | C70(0.69); C63(0.69); C91(0.77) | LDD0546 | [16] |
| LDCM0230 | AC113 | HCT 116 | C91(0.93) | LDD0547 | [16] |
| LDCM0231 | AC114 | HCT 116 | C91(1.12) | LDD0548 | [16] |
| LDCM0232 | AC115 | HCT 116 | C91(1.16) | LDD0549 | [16] |
| LDCM0233 | AC116 | HCT 116 | C91(0.93) | LDD0550 | [16] |
| LDCM0234 | AC117 | HCT 116 | C91(0.87) | LDD0551 | [16] |
| LDCM0235 | AC118 | HCT 116 | C91(0.92) | LDD0552 | [16] |
| LDCM0236 | AC119 | HCT 116 | C91(0.99) | LDD0553 | [16] |
| LDCM0237 | AC12 | HCT 116 | C70(0.79); C63(0.79); C91(1.24) | LDD0554 | [16] |
| LDCM0238 | AC120 | HCT 116 | C91(1.12) | LDD0555 | [16] |
| LDCM0239 | AC121 | HCT 116 | C91(0.86) | LDD0556 | [16] |
| LDCM0240 | AC122 | HCT 116 | C91(0.97) | LDD0557 | [16] |
| LDCM0241 | AC123 | HCT 116 | C91(0.74) | LDD0558 | [16] |
| LDCM0242 | AC124 | HCT 116 | C91(1.08) | LDD0559 | [16] |
| LDCM0243 | AC125 | HCT 116 | C91(1.10) | LDD0560 | [16] |
| LDCM0244 | AC126 | HCT 116 | C91(1.01) | LDD0561 | [16] |
| LDCM0245 | AC127 | HCT 116 | C91(1.09) | LDD0562 | [16] |
| LDCM0246 | AC128 | HCT 116 | C91(0.84) | LDD0563 | [16] |
| LDCM0247 | AC129 | HCT 116 | C91(1.07) | LDD0564 | [16] |
| LDCM0249 | AC130 | HCT 116 | C91(1.06) | LDD0566 | [16] |
| LDCM0250 | AC131 | HCT 116 | C91(0.96) | LDD0567 | [16] |
| LDCM0251 | AC132 | HCT 116 | C91(1.30) | LDD0568 | [16] |
| LDCM0252 | AC133 | HCT 116 | C91(1.22) | LDD0569 | [16] |
| LDCM0253 | AC134 | HCT 116 | C91(1.17) | LDD0570 | [16] |
| LDCM0254 | AC135 | HCT 116 | C91(1.06) | LDD0571 | [16] |
| LDCM0255 | AC136 | HCT 116 | C91(1.20) | LDD0572 | [16] |
| LDCM0256 | AC137 | HCT 116 | C91(1.46) | LDD0573 | [16] |
| LDCM0257 | AC138 | HCT 116 | C91(1.33) | LDD0574 | [16] |
| LDCM0258 | AC139 | HCT 116 | C91(1.24) | LDD0575 | [16] |
| LDCM0259 | AC14 | HCT 116 | C70(0.49); C63(0.49); C91(1.08) | LDD0576 | [16] |
| LDCM0260 | AC140 | HCT 116 | C91(1.14) | LDD0577 | [16] |
| LDCM0261 | AC141 | HCT 116 | C91(1.21) | LDD0578 | [16] |
| LDCM0262 | AC142 | HCT 116 | C91(1.33) | LDD0579 | [16] |
| LDCM0263 | AC143 | HEK-293T | C63(1.00); C70(1.00); C91(0.80) | LDD0861 | [16] |
| LDCM0264 | AC144 | HEK-293T | C63(0.63); C70(0.63); C91(0.69) | LDD0862 | [16] |
| LDCM0265 | AC145 | HEK-293T | C63(0.67); C70(0.67); C91(0.72) | LDD0863 | [16] |
| LDCM0266 | AC146 | HEK-293T | C63(0.68); C70(0.68); C91(0.72) | LDD0864 | [16] |
| LDCM0267 | AC147 | HEK-293T | C63(0.81); C70(0.81); C91(0.73) | LDD0865 | [16] |
| LDCM0268 | AC148 | HEK-293T | C63(0.76); C70(0.76); C91(0.77) | LDD0866 | [16] |
| LDCM0269 | AC149 | HEK-293T | C63(0.99); C70(0.99); C91(0.74) | LDD0867 | [16] |
| LDCM0270 | AC15 | HCT 116 | C70(0.49); C63(0.49); C91(1.16) | LDD0587 | [16] |
| LDCM0271 | AC150 | HEK-293T | C63(0.74); C70(0.74); C91(0.73) | LDD0869 | [16] |
| LDCM0272 | AC151 | HEK-293T | C63(0.71); C70(0.71); C91(0.67) | LDD0870 | [16] |
| LDCM0273 | AC152 | HEK-293T | C63(0.82); C70(0.82); C91(0.83) | LDD0871 | [16] |
| LDCM0274 | AC153 | HEK-293T | C63(0.69); C70(0.69); C91(0.76) | LDD0872 | [16] |
| LDCM0621 | AC154 | HEK-293T | C63(0.80); C70(0.80); C91(0.81) | LDD2162 | [16] |
| LDCM0622 | AC155 | HEK-293T | C63(0.81); C70(0.81); C91(0.77) | LDD2163 | [16] |
| LDCM0623 | AC156 | HEK-293T | C63(0.76); C70(0.76); C91(0.75) | LDD2164 | [16] |
| LDCM0624 | AC157 | HEK-293T | C63(0.96); C70(0.96); C91(0.95) | LDD2165 | [16] |
| LDCM0276 | AC17 | HCT 116 | C91(0.96); C70(1.82); C63(1.82) | LDD0593 | [16] |
| LDCM0277 | AC18 | HCT 116 | C70(0.65); C63(0.65); C91(0.95) | LDD0594 | [16] |
| LDCM0278 | AC19 | HCT 116 | C91(0.93); C70(1.62); C63(1.62) | LDD0595 | [16] |
| LDCM0279 | AC2 | HCT 116 | C70(0.68); C63(0.68); C91(1.20) | LDD0596 | [16] |
| LDCM0280 | AC20 | HCT 116 | C91(0.94); C70(1.77); C63(1.77) | LDD0597 | [16] |
| LDCM0281 | AC21 | HCT 116 | C91(0.78); C70(1.05); C63(1.05) | LDD0598 | [16] |
| LDCM0282 | AC22 | HCT 116 | C91(1.03); C70(1.55); C63(1.55) | LDD0599 | [16] |
| LDCM0283 | AC23 | HCT 116 | C91(0.94); C70(1.15); C63(1.15) | LDD0600 | [16] |
| LDCM0284 | AC24 | HCT 116 | C91(0.91); C70(1.52); C63(1.52) | LDD0601 | [16] |
| LDCM0285 | AC25 | HCT 116 | C70(0.80); C63(0.80); C91(1.05) | LDD0602 | [16] |
| LDCM0286 | AC26 | HCT 116 | C70(0.73); C63(0.73); C91(1.07) | LDD0603 | [16] |
| LDCM0287 | AC27 | HCT 116 | C70(0.69); C63(0.69); C91(1.13) | LDD0604 | [16] |
| LDCM0288 | AC28 | HCT 116 | C70(0.71); C63(0.71); C91(1.14) | LDD0605 | [16] |
| LDCM0289 | AC29 | HCT 116 | C70(0.94); C63(0.94); C91(1.14) | LDD0606 | [16] |
| LDCM0290 | AC3 | HCT 116 | C70(0.84); C63(0.84); C91(1.15) | LDD0607 | [16] |
| LDCM0291 | AC30 | HCT 116 | C70(0.89); C63(0.89); C91(1.06) | LDD0608 | [16] |
| LDCM0292 | AC31 | HCT 116 | C70(0.72); C63(0.72); C91(1.10) | LDD0609 | [16] |
| LDCM0293 | AC32 | HCT 116 | C70(0.54); C63(0.54); C91(1.07) | LDD0610 | [16] |
| LDCM0294 | AC33 | HCT 116 | C70(0.82); C63(0.82); C91(1.04) | LDD0611 | [16] |
| LDCM0295 | AC34 | HCT 116 | C70(0.59); C63(0.59); C91(1.02) | LDD0612 | [16] |
| LDCM0296 | AC35 | HCT 116 | C91(1.03); C70(1.20); C63(1.20) | LDD0613 | [16] |
| LDCM0297 | AC36 | HCT 116 | C70(1.00); C63(1.00); C91(1.17) | LDD0614 | [16] |
| LDCM0298 | AC37 | HCT 116 | C70(1.10); C63(1.10); C91(1.11) | LDD0615 | [16] |
| LDCM0299 | AC38 | HCT 116 | C91(1.14); C70(1.27); C63(1.27) | LDD0616 | [16] |
| LDCM0300 | AC39 | HCT 116 | C70(0.82); C63(0.82); C91(1.14) | LDD0617 | [16] |
| LDCM0301 | AC4 | HCT 116 | C70(0.51); C63(0.51); C91(1.24) | LDD0618 | [16] |
| LDCM0302 | AC40 | HCT 116 | C70(0.65); C63(0.65); C91(0.95) | LDD0619 | [16] |
| LDCM0303 | AC41 | HCT 116 | C70(0.99); C63(0.99); C91(1.08) | LDD0620 | [16] |
| LDCM0304 | AC42 | HCT 116 | C70(0.96); C63(0.96); C91(1.03) | LDD0621 | [16] |
| LDCM0305 | AC43 | HCT 116 | C91(1.01); C70(1.03); C63(1.03) | LDD0622 | [16] |
| LDCM0306 | AC44 | HCT 116 | C70(0.78); C63(0.78); C91(0.89) | LDD0623 | [16] |
| LDCM0307 | AC45 | HCT 116 | C70(0.70); C63(0.70); C91(0.92) | LDD0624 | [16] |
| LDCM0308 | AC46 | HCT 116 | C91(0.96); C70(0.97); C63(0.97) | LDD0625 | [16] |
| LDCM0309 | AC47 | HCT 116 | C70(0.95); C63(0.95); C91(1.10) | LDD0626 | [16] |
| LDCM0310 | AC48 | HCT 116 | C91(1.09); C70(1.40); C63(1.40) | LDD0627 | [16] |
| LDCM0311 | AC49 | HCT 116 | C70(0.41); C63(0.41); C91(0.94) | LDD0628 | [16] |
| LDCM0312 | AC5 | HCT 116 | C70(0.56); C63(0.56); C91(1.27) | LDD0629 | [16] |
| LDCM0313 | AC50 | HCT 116 | C70(0.69); C63(0.69); C91(1.02) | LDD0630 | [16] |
| LDCM0314 | AC51 | HCT 116 | C91(1.08); C70(2.56); C63(2.56) | LDD0631 | [16] |
| LDCM0315 | AC52 | HCT 116 | C70(0.91); C63(0.91); C91(1.05) | LDD0632 | [16] |
| LDCM0316 | AC53 | HCT 116 | C70(0.89); C63(0.89); C91(1.05) | LDD0633 | [16] |
| LDCM0317 | AC54 | HCT 116 | C70(0.74); C63(0.74); C91(1.30) | LDD0634 | [16] |
| LDCM0318 | AC55 | HCT 116 | C70(0.56); C63(0.56); C91(1.19) | LDD0635 | [16] |
| LDCM0319 | AC56 | HCT 116 | C70(0.39); C63(0.39); C91(1.11) | LDD0636 | [16] |
| LDCM0320 | AC57 | HCT 116 | C91(1.07) | LDD0637 | [16] |
| LDCM0321 | AC58 | HCT 116 | C91(1.06) | LDD0638 | [16] |
| LDCM0322 | AC59 | HCT 116 | C91(0.98) | LDD0639 | [16] |
| LDCM0323 | AC6 | HCT 116 | C70(0.56); C63(0.56); C91(1.02) | LDD0640 | [16] |
| LDCM0324 | AC60 | HCT 116 | C91(0.96) | LDD0641 | [16] |
| LDCM0325 | AC61 | HCT 116 | C91(1.15) | LDD0642 | [16] |
| LDCM0326 | AC62 | HCT 116 | C91(1.03) | LDD0643 | [16] |
| LDCM0327 | AC63 | HCT 116 | C91(1.01) | LDD0644 | [16] |
| LDCM0328 | AC64 | HCT 116 | C91(1.02) | LDD0645 | [16] |
| LDCM0329 | AC65 | HCT 116 | C91(1.09) | LDD0646 | [16] |
| LDCM0330 | AC66 | HCT 116 | C91(1.10) | LDD0647 | [16] |
| LDCM0331 | AC67 | HCT 116 | C91(0.91) | LDD0648 | [16] |
| LDCM0332 | AC68 | HCT 116 | C70(0.65); C63(0.65); C91(1.00) | LDD0649 | [16] |
| LDCM0333 | AC69 | HCT 116 | C70(0.64); C63(0.64); C91(1.07) | LDD0650 | [16] |
| LDCM0334 | AC7 | HCT 116 | C70(0.46); C63(0.46); C91(1.08) | LDD0651 | [16] |
| LDCM0335 | AC70 | HCT 116 | C70(0.40); C63(0.40); C91(0.88) | LDD0652 | [16] |
| LDCM0336 | AC71 | HCT 116 | C70(0.56); C63(0.56); C91(1.15) | LDD0653 | [16] |
| LDCM0337 | AC72 | HCT 116 | C70(0.66); C63(0.66); C91(0.98) | LDD0654 | [16] |
| LDCM0338 | AC73 | HCT 116 | C70(0.36); C63(0.36); C91(0.89) | LDD0655 | [16] |
| LDCM0339 | AC74 | HCT 116 | C70(0.34); C63(0.34); C91(0.87) | LDD0656 | [16] |
| LDCM0340 | AC75 | HCT 116 | C70(0.30); C63(0.30); C91(0.84) | LDD0657 | [16] |
| LDCM0341 | AC76 | HCT 116 | C70(0.71); C63(0.71); C91(0.96) | LDD0658 | [16] |
| LDCM0342 | AC77 | HCT 116 | C70(0.48); C63(0.48); C91(0.95) | LDD0659 | [16] |
| LDCM0343 | AC78 | HCT 116 | C70(0.66); C63(0.66); C91(1.38) | LDD0660 | [16] |
| LDCM0344 | AC79 | HCT 116 | C70(0.59); C63(0.59); C91(1.09) | LDD0661 | [16] |
| LDCM0345 | AC8 | HCT 116 | C70(0.50); C63(0.50); C91(1.16) | LDD0662 | [16] |
| LDCM0346 | AC80 | HCT 116 | C70(0.66); C63(0.66); C91(1.13) | LDD0663 | [16] |
| LDCM0347 | AC81 | HCT 116 | C70(0.73); C63(0.73); C91(1.31) | LDD0664 | [16] |
| LDCM0348 | AC82 | HCT 116 | C70(0.21); C63(0.21); C91(0.70) | LDD0665 | [16] |
| LDCM0349 | AC83 | HCT 116 | C70(0.68); C63(0.68) | LDD0666 | [16] |
| LDCM0350 | AC84 | HCT 116 | C70(0.90); C63(0.90) | LDD0667 | [16] |
| LDCM0351 | AC85 | HCT 116 | C70(1.13); C63(1.13) | LDD0668 | [16] |
| LDCM0352 | AC86 | HCT 116 | C70(0.91); C63(0.91) | LDD0669 | [16] |
| LDCM0353 | AC87 | HCT 116 | C70(0.88); C63(0.88) | LDD0670 | [16] |
| LDCM0354 | AC88 | HCT 116 | C70(0.99); C63(0.99) | LDD0671 | [16] |
| LDCM0355 | AC89 | HCT 116 | C70(1.00); C63(1.00) | LDD0672 | [16] |
| LDCM0357 | AC90 | HCT 116 | C70(0.97); C63(0.97) | LDD0674 | [16] |
| LDCM0358 | AC91 | HCT 116 | C70(0.83); C63(0.83) | LDD0675 | [16] |
| LDCM0359 | AC92 | HCT 116 | C70(0.99); C63(0.99) | LDD0676 | [16] |
| LDCM0360 | AC93 | HCT 116 | C70(0.88); C63(0.88) | LDD0677 | [16] |
| LDCM0361 | AC94 | HCT 116 | C70(0.89); C63(0.89) | LDD0678 | [16] |
| LDCM0362 | AC95 | HCT 116 | C70(1.26); C63(1.26) | LDD0679 | [16] |
| LDCM0363 | AC96 | HCT 116 | C70(1.08); C63(1.08) | LDD0680 | [16] |
| LDCM0364 | AC97 | HCT 116 | C70(0.93); C63(0.93) | LDD0681 | [16] |
| LDCM0365 | AC98 | HCT 116 | C70(0.26); C63(0.26); C91(1.38) | LDD0682 | [16] |
| LDCM0366 | AC99 | HCT 116 | C91(1.23); C70(1.27); C63(1.27) | LDD0683 | [16] |
| LDCM0545 | Acetamide | MDA-MB-231 | C91(0.32) | LDD2138 | [7] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C91(0.60) | LDD2113 | [7] |
| LDCM0248 | AKOS034007472 | HCT 116 | C70(0.59); C63(0.59); C91(1.22) | LDD0565 | [16] |
| LDCM0356 | AKOS034007680 | HCT 116 | C70(0.55); C63(0.55); C91(1.14) | LDD0673 | [16] |
| LDCM0275 | AKOS034007705 | HCT 116 | C70(0.33); C63(0.33); C91(0.97) | LDD0592 | [16] |
| LDCM0156 | Aniline | NCI-H1299 | 11.90 | LDD0403 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C63(1.13); C70(1.13) | LDD2171 | [16] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C91(0.34) | LDD2091 | [7] |
| LDCM0108 | Chloroacetamide | HeLa | H154(0.00); C91(0.00) | LDD0222 | [31] |
| LDCM0632 | CL-Sc | Hep-G2 | C91(1.12) | LDD2227 | [23] |
| LDCM0367 | CL1 | HCT 116 | C91(1.02); C70(1.93); C63(1.93) | LDD0684 | [16] |
| LDCM0368 | CL10 | HCT 116 | C70(0.94); C63(0.94); C91(1.02) | LDD0685 | [16] |
| LDCM0369 | CL100 | HCT 116 | C70(0.64); C63(0.64); C91(1.17) | LDD0686 | [16] |
| LDCM0370 | CL101 | HCT 116 | C70(0.54); C63(0.54); C91(1.01) | LDD0687 | [16] |
| LDCM0371 | CL102 | HCT 116 | C70(0.81); C63(0.81); C91(0.99) | LDD0688 | [16] |
| LDCM0372 | CL103 | HCT 116 | C70(0.60); C63(0.60); C91(0.97) | LDD0689 | [16] |
| LDCM0373 | CL104 | HCT 116 | C70(1.01); C63(1.01); C91(1.03) | LDD0690 | [16] |
| LDCM0374 | CL105 | HCT 116 | C70(0.92); C63(0.92); C91(0.97) | LDD0691 | [16] |
| LDCM0375 | CL106 | HCT 116 | C70(0.73); C63(0.73); C91(0.89) | LDD0692 | [16] |
| LDCM0376 | CL107 | HCT 116 | C70(0.77); C63(0.77); C91(1.01) | LDD0693 | [16] |
| LDCM0377 | CL108 | HCT 116 | C70(0.67); C63(0.67); C91(1.05) | LDD0694 | [16] |
| LDCM0378 | CL109 | HCT 116 | C91(0.88); C70(1.20); C63(1.20) | LDD0695 | [16] |
| LDCM0379 | CL11 | HCT 116 | C70(0.62); C63(0.62); C91(0.94) | LDD0696 | [16] |
| LDCM0380 | CL110 | HCT 116 | C70(1.06); C63(1.06); C91(1.10) | LDD0697 | [16] |
| LDCM0381 | CL111 | HCT 116 | C91(0.95); C70(1.26); C63(1.26) | LDD0698 | [16] |
| LDCM0382 | CL112 | HCT 116 | C70(0.80); C63(0.80); C91(0.97) | LDD0699 | [16] |
| LDCM0383 | CL113 | HCT 116 | C70(0.84); C63(0.84); C91(1.07) | LDD0700 | [16] |
| LDCM0384 | CL114 | HCT 116 | C70(0.77); C63(0.77); C91(1.01) | LDD0701 | [16] |
| LDCM0385 | CL115 | HCT 116 | C70(0.63); C63(0.63); C91(1.03) | LDD0702 | [16] |
| LDCM0386 | CL116 | HCT 116 | C70(0.60); C63(0.60); C91(1.16) | LDD0703 | [16] |
| LDCM0387 | CL117 | HCT 116 | C70(0.43); C63(0.43); C91(0.87) | LDD0704 | [16] |
| LDCM0388 | CL118 | HCT 116 | C70(0.95); C63(0.95); C91(1.11) | LDD0705 | [16] |
| LDCM0389 | CL119 | HCT 116 | C70(0.77); C63(0.77); C91(1.01) | LDD0706 | [16] |
| LDCM0390 | CL12 | HCT 116 | C70(0.67); C63(0.67); C91(0.97) | LDD0707 | [16] |
| LDCM0391 | CL120 | HCT 116 | C70(0.82); C63(0.82); C91(1.05) | LDD0708 | [16] |
| LDCM0392 | CL121 | HCT 116 | C91(1.08); C70(1.20); C63(1.20) | LDD0709 | [16] |
| LDCM0393 | CL122 | HCT 116 | C70(0.76); C63(0.76); C91(1.14) | LDD0710 | [16] |
| LDCM0394 | CL123 | HCT 116 | C70(0.87); C63(0.87); C91(1.01) | LDD0711 | [16] |
| LDCM0395 | CL124 | HCT 116 | C70(0.62); C63(0.62); C91(1.14) | LDD0712 | [16] |
| LDCM0396 | CL125 | HCT 116 | C91(1.11) | LDD0713 | [16] |
| LDCM0397 | CL126 | HCT 116 | C91(1.03) | LDD0714 | [16] |
| LDCM0398 | CL127 | HCT 116 | C91(1.06) | LDD0715 | [16] |
| LDCM0399 | CL128 | HCT 116 | C91(1.02) | LDD0716 | [16] |
| LDCM0400 | CL13 | HCT 116 | C70(0.95); C63(0.95); C91(1.13) | LDD0717 | [16] |
| LDCM0401 | CL14 | HCT 116 | C91(0.97); C70(1.14); C63(1.14) | LDD0718 | [16] |
| LDCM0402 | CL15 | HCT 116 | C91(0.88); C70(1.03); C63(1.03) | LDD0719 | [16] |
| LDCM0403 | CL16 | HCT 116 | C70(0.81); C63(0.81); C91(0.93) | LDD0720 | [16] |
| LDCM0404 | CL17 | HCT 116 | C70(0.56); C63(0.56); C91(0.84) | LDD0721 | [16] |
| LDCM0405 | CL18 | HCT 116 | C70(0.69); C63(0.69); C91(1.02) | LDD0722 | [16] |
| LDCM0406 | CL19 | HCT 116 | C91(0.98); C70(1.08); C63(1.08) | LDD0723 | [16] |
| LDCM0407 | CL2 | HCT 116 | C91(1.15); C70(1.31); C63(1.31) | LDD0724 | [16] |
| LDCM0408 | CL20 | HCT 116 | C70(0.59); C63(0.59); C91(0.91) | LDD0725 | [16] |
| LDCM0409 | CL21 | HCT 116 | C70(0.56); C63(0.56); C91(0.81) | LDD0726 | [16] |
| LDCM0410 | CL22 | HCT 116 | C70(0.39); C63(0.39); C91(0.85) | LDD0727 | [16] |
| LDCM0411 | CL23 | HCT 116 | C70(0.69); C63(0.69); C91(0.95) | LDD0728 | [16] |
| LDCM0412 | CL24 | HCT 116 | C70(0.40); C63(0.40); C91(0.89) | LDD0729 | [16] |
| LDCM0413 | CL25 | HCT 116 | C70(0.60); C63(0.60); C91(1.00) | LDD0730 | [16] |
| LDCM0414 | CL26 | HCT 116 | C70(0.64); C63(0.64); C91(0.83) | LDD0731 | [16] |
| LDCM0415 | CL27 | HCT 116 | C70(0.75); C63(0.75); C91(1.12) | LDD0732 | [16] |
| LDCM0416 | CL28 | HCT 116 | C70(0.55); C63(0.55); C91(0.92) | LDD0733 | [16] |
| LDCM0417 | CL29 | HCT 116 | C70(0.54); C63(0.54); C91(0.95) | LDD0734 | [16] |
| LDCM0418 | CL3 | HCT 116 | C70(1.16); C63(1.16); C91(1.00) | LDD0735 | [16] |
| LDCM0419 | CL30 | HCT 116 | C70(1.01); C63(1.01); C91(1.10) | LDD0736 | [16] |
| LDCM0420 | CL31 | HCT 116 | C91(1.34) | LDD0737 | [16] |
| LDCM0421 | CL32 | HCT 116 | C91(0.90) | LDD0738 | [16] |
| LDCM0422 | CL33 | HCT 116 | C91(0.67) | LDD0739 | [16] |
| LDCM0423 | CL34 | HCT 116 | C91(0.96) | LDD0740 | [16] |
| LDCM0424 | CL35 | HCT 116 | C91(1.05) | LDD0741 | [16] |
| LDCM0425 | CL36 | HCT 116 | C91(0.85) | LDD0742 | [16] |
| LDCM0426 | CL37 | HCT 116 | C91(0.95) | LDD0743 | [16] |
| LDCM0428 | CL39 | HCT 116 | C91(0.90) | LDD0745 | [16] |
| LDCM0429 | CL4 | HCT 116 | C70(1.22); C63(1.22); C91(0.68) | LDD0746 | [16] |
| LDCM0430 | CL40 | HCT 116 | C91(0.78) | LDD0747 | [16] |
| LDCM0431 | CL41 | HCT 116 | C91(0.83) | LDD0748 | [16] |
| LDCM0432 | CL42 | HCT 116 | C91(0.95) | LDD0749 | [16] |
| LDCM0433 | CL43 | HCT 116 | C91(0.99) | LDD0750 | [16] |
| LDCM0434 | CL44 | HCT 116 | C91(1.06) | LDD0751 | [16] |
| LDCM0435 | CL45 | HCT 116 | C91(1.03) | LDD0752 | [16] |
| LDCM0436 | CL46 | HCT 116 | C70(2.03); C63(2.03); C91(0.81) | LDD0753 | [16] |
| LDCM0437 | CL47 | HCT 116 | C70(2.02); C63(2.02); C91(0.89) | LDD0754 | [16] |
| LDCM0438 | CL48 | HCT 116 | C70(2.70); C63(2.70); C91(0.90) | LDD0755 | [16] |
| LDCM0439 | CL49 | HCT 116 | C70(1.97); C63(1.97); C91(1.12) | LDD0756 | [16] |
| LDCM0440 | CL5 | HCT 116 | C70(1.30); C63(1.30); C91(1.21) | LDD0757 | [16] |
| LDCM0441 | CL50 | HCT 116 | C70(2.22); C63(2.22); C91(0.91) | LDD0758 | [16] |
| LDCM0442 | CL51 | HCT 116 | C70(3.09); C63(3.09); C91(0.95) | LDD0759 | [16] |
| LDCM0443 | CL52 | HCT 116 | C70(2.92); C63(2.92); C91(0.77) | LDD0760 | [16] |
| LDCM0444 | CL53 | HCT 116 | C70(1.98); C63(1.98); C91(0.79) | LDD0761 | [16] |
| LDCM0445 | CL54 | HCT 116 | C70(2.22); C63(2.22); C91(0.82) | LDD0762 | [16] |
| LDCM0446 | CL55 | HCT 116 | C70(3.13); C63(3.13); C91(0.89) | LDD0763 | [16] |
| LDCM0447 | CL56 | HCT 116 | C70(1.60); C63(1.60); C91(0.86) | LDD0764 | [16] |
| LDCM0448 | CL57 | HCT 116 | C70(1.80); C63(1.80); C91(0.95) | LDD0765 | [16] |
| LDCM0449 | CL58 | HCT 116 | C70(2.18); C63(2.18); C91(0.83) | LDD0766 | [16] |
| LDCM0450 | CL59 | HCT 116 | C70(1.74); C63(1.74); C91(0.95) | LDD0767 | [16] |
| LDCM0451 | CL6 | HCT 116 | C70(0.91); C63(0.91); C91(1.00) | LDD0768 | [16] |
| LDCM0452 | CL60 | HCT 116 | C70(1.57); C63(1.57); C91(0.87) | LDD0769 | [16] |
| LDCM0453 | CL61 | HCT 116 | C70(0.87); C63(0.87); C91(1.02) | LDD0770 | [16] |
| LDCM0454 | CL62 | HCT 116 | C70(0.71); C63(0.71); C91(0.94) | LDD0771 | [16] |
| LDCM0455 | CL63 | HCT 116 | C70(0.62); C63(0.62); C91(1.10) | LDD0772 | [16] |
| LDCM0456 | CL64 | HCT 116 | C70(0.61); C63(0.61); C91(1.05) | LDD0773 | [16] |
| LDCM0457 | CL65 | HCT 116 | C70(0.89); C63(0.89); C91(0.98) | LDD0774 | [16] |
| LDCM0458 | CL66 | HCT 116 | C70(0.31); C63(0.31); C91(0.94) | LDD0775 | [16] |
| LDCM0459 | CL67 | HCT 116 | C70(0.50); C63(0.50); C91(1.00) | LDD0776 | [16] |
| LDCM0460 | CL68 | HCT 116 | C70(0.45); C63(0.45); C91(0.97) | LDD0777 | [16] |
| LDCM0461 | CL69 | HCT 116 | C70(0.70); C63(0.70); C91(0.85) | LDD0778 | [16] |
| LDCM0462 | CL7 | HCT 116 | C70(0.73); C63(0.73); C91(1.02) | LDD0779 | [16] |
| LDCM0463 | CL70 | HCT 116 | C70(0.67); C63(0.67); C91(1.08) | LDD0780 | [16] |
| LDCM0464 | CL71 | HCT 116 | C70(0.54); C63(0.54); C91(1.01) | LDD0781 | [16] |
| LDCM0465 | CL72 | HCT 116 | C70(0.92); C63(0.92); C91(1.04) | LDD0782 | [16] |
| LDCM0466 | CL73 | HCT 116 | C70(0.63); C63(0.63); C91(0.93) | LDD0783 | [16] |
| LDCM0467 | CL74 | HCT 116 | C70(0.66); C63(0.66); C91(0.92) | LDD0784 | [16] |
| LDCM0469 | CL76 | HCT 116 | C70(0.90); C63(0.61); C91(0.91) | LDD0786 | [16] |
| LDCM0470 | CL77 | HCT 116 | C70(0.98); C63(0.93); C91(1.08) | LDD0787 | [16] |
| LDCM0471 | CL78 | HCT 116 | C70(0.92); C63(0.76); C91(0.86) | LDD0788 | [16] |
| LDCM0472 | CL79 | HCT 116 | C70(1.44); C63(0.59); C91(0.93) | LDD0789 | [16] |
| LDCM0473 | CL8 | HCT 116 | C70(0.65); C63(0.65); C91(0.98) | LDD0790 | [16] |
| LDCM0474 | CL80 | HCT 116 | C70(1.26); C63(1.70); C91(1.09) | LDD0791 | [16] |
| LDCM0475 | CL81 | HCT 116 | C70(0.88); C63(0.80); C91(0.96) | LDD0792 | [16] |
| LDCM0476 | CL82 | HCT 116 | C70(0.68); C63(0.45); C91(1.07) | LDD0793 | [16] |
| LDCM0477 | CL83 | HCT 116 | C70(0.49); C63(0.39); C91(1.14) | LDD0794 | [16] |
| LDCM0478 | CL84 | HCT 116 | C70(0.74); C63(0.44); C91(0.97) | LDD0795 | [16] |
| LDCM0479 | CL85 | HCT 116 | C70(0.97); C63(0.72); C91(1.07) | LDD0796 | [16] |
| LDCM0480 | CL86 | HCT 116 | C70(1.34); C63(0.93); C91(0.94) | LDD0797 | [16] |
| LDCM0481 | CL87 | HCT 116 | C70(0.98); C63(1.02); C91(0.91) | LDD0798 | [16] |
| LDCM0482 | CL88 | HCT 116 | C70(0.76); C63(0.52); C91(1.02) | LDD0799 | [16] |
| LDCM0483 | CL89 | HCT 116 | C70(0.65); C63(0.35); C91(1.06) | LDD0800 | [16] |
| LDCM0484 | CL9 | HCT 116 | C70(1.02); C63(1.02); C91(0.96) | LDD0801 | [16] |
| LDCM0485 | CL90 | HCT 116 | C70(10.37); C63(0.75); C91(0.76) | LDD0802 | [16] |
| LDCM0486 | CL91 | HCT 116 | C70(0.54); C63(0.54); C91(1.01) | LDD0803 | [16] |
| LDCM0487 | CL92 | HCT 116 | C70(0.54); C63(0.54); C91(0.99) | LDD0804 | [16] |
| LDCM0488 | CL93 | HCT 116 | C70(0.82); C63(0.82); C91(0.99) | LDD0805 | [16] |
| LDCM0489 | CL94 | HCT 116 | C70(0.65); C63(0.65); C91(1.02) | LDD0806 | [16] |
| LDCM0490 | CL95 | HCT 116 | C70(0.67); C63(0.67); C91(1.03) | LDD0807 | [16] |
| LDCM0491 | CL96 | HCT 116 | C70(0.76); C63(0.76); C91(1.07) | LDD0808 | [16] |
| LDCM0492 | CL97 | HCT 116 | C70(0.84); C63(0.84); C91(1.05) | LDD0809 | [16] |
| LDCM0493 | CL98 | HCT 116 | C70(0.58); C63(0.58); C91(1.05) | LDD0810 | [16] |
| LDCM0494 | CL99 | HCT 116 | C70(0.61); C63(0.61); C91(1.15) | LDD0811 | [16] |
| LDCM0495 | E2913 | HEK-293T | C91(0.98); C63(1.17) | LDD1698 | [38] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C91(5.01) | LDD1702 | [7] |
| LDCM0175 | Ethacrynic acid | HeLa | N.A. | LDD0440 | [12] |
| LDCM0625 | F8 | Ramos | C91(1.48) | LDD2187 | [39] |
| LDCM0572 | Fragment10 | Ramos | C91(0.87) | LDD2189 | [39] |
| LDCM0573 | Fragment11 | Ramos | C91(0.09) | LDD2190 | [39] |
| LDCM0574 | Fragment12 | Ramos | C91(0.79) | LDD2191 | [39] |
| LDCM0575 | Fragment13 | Ramos | C91(0.91) | LDD2192 | [39] |
| LDCM0576 | Fragment14 | Ramos | C91(0.79) | LDD2193 | [39] |
| LDCM0579 | Fragment20 | Ramos | C91(0.85) | LDD2194 | [39] |
| LDCM0580 | Fragment21 | Ramos | C91(0.90) | LDD2195 | [39] |
| LDCM0582 | Fragment23 | Ramos | C91(1.59) | LDD2196 | [39] |
| LDCM0578 | Fragment27 | Ramos | C91(1.25) | LDD2197 | [39] |
| LDCM0586 | Fragment28 | Ramos | C91(0.50) | LDD2198 | [39] |
| LDCM0588 | Fragment30 | Ramos | C91(1.26) | LDD2199 | [39] |
| LDCM0589 | Fragment31 | Ramos | C91(1.04) | LDD2200 | [39] |
| LDCM0590 | Fragment32 | Ramos | C91(1.32) | LDD2201 | [39] |
| LDCM0468 | Fragment33 | HCT 116 | C70(0.46); C63(0.46); C91(0.97) | LDD0785 | [16] |
| LDCM0596 | Fragment38 | Ramos | C91(1.20) | LDD2203 | [39] |
| LDCM0566 | Fragment4 | Ramos | C91(0.70) | LDD2184 | [39] |
| LDCM0427 | Fragment51 | HCT 116 | C91(1.06) | LDD0744 | [16] |
| LDCM0610 | Fragment52 | Ramos | C91(1.22) | LDD2204 | [39] |
| LDCM0614 | Fragment56 | Ramos | C91(1.16) | LDD2205 | [39] |
| LDCM0569 | Fragment7 | Ramos | C91(0.61) | LDD2186 | [39] |
| LDCM0571 | Fragment9 | Ramos | C91(0.58) | LDD2188 | [39] |
| LDCM0116 | HHS-0101 | DM93 | Y110(0.97); Y118(1.03); Y21(3.06) | LDD0264 | [14] |
| LDCM0117 | HHS-0201 | DM93 | Y110(0.84); Y118(0.96); Y21(1.77) | LDD0265 | [14] |
| LDCM0118 | HHS-0301 | DM93 | Y110(0.90); Y118(1.01); Y21(2.89) | LDD0266 | [14] |
| LDCM0119 | HHS-0401 | DM93 | Y110(0.99); Y118(1.24); Y21(2.10) | LDD0267 | [14] |
| LDCM0120 | HHS-0701 | DM93 | Y118(1.11); Y110(1.13); Y21(1.48) | LDD0268 | [14] |
| LDCM0107 | IAA | HeLa | H154(0.00); C91(0.00); H94(0.00) | LDD0221 | [31] |
| LDCM0123 | JWB131 | DM93 | Y110(0.74); Y118(0.91); Y145(0.50); Y153(0.85) | LDD0285 | [13] |
| LDCM0124 | JWB142 | DM93 | Y110(0.59); Y118(0.78); Y145(0.42); Y153(0.48) | LDD0286 | [13] |
| LDCM0125 | JWB146 | DM93 | Y110(1.05); Y118(1.03); Y145(0.60); Y153(0.96) | LDD0287 | [13] |
| LDCM0126 | JWB150 | DM93 | Y110(2.26); Y118(2.67); Y145(3.81); Y153(7.24) | LDD0288 | [13] |
| LDCM0127 | JWB152 | DM93 | Y110(1.54); Y118(1.82); Y145(1.54); Y153(2.00) | LDD0289 | [13] |
| LDCM0128 | JWB198 | DM93 | Y110(0.88); Y118(1.26); Y145(0.75); Y153(1.78) | LDD0290 | [13] |
| LDCM0129 | JWB202 | DM93 | Y110(0.38); Y118(0.53); Y145(0.46); Y153(0.51) | LDD0291 | [13] |
| LDCM0130 | JWB211 | DM93 | Y110(0.91); Y118(1.00); Y145(0.56); Y153(0.60) | LDD0292 | [13] |
| LDCM0022 | KB02 | HCT 116 | C63(1.38); C70(1.38) | LDD0080 | [16] |
| LDCM0023 | KB03 | HCT 116 | C63(2.88); C70(2.88) | LDD0081 | [16] |
| LDCM0024 | KB05 | HCT 116 | C63(1.69); C70(1.69) | LDD0082 | [16] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C91(1.14) | LDD2102 | [7] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C91(0.42) | LDD2121 | [7] |
| LDCM0109 | NEM | HeLa | H154(0.00); C70(0.00) | LDD0223 | [31] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C91(0.41) | LDD2089 | [7] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C91(0.91) | LDD2090 | [7] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C91(1.21) | LDD2092 | [7] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C91(0.83) | LDD2093 | [7] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C91(2.29) | LDD2094 | [7] |
| LDCM0503 | Nucleophilic fragment 14b | MDA-MB-231 | C91(0.06) | LDD2096 | [7] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C91(0.68) | LDD2097 | [7] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C91(0.58) | LDD2098 | [7] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C91(0.74) | LDD2099 | [7] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C91(0.40) | LDD2100 | [7] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C91(0.85) | LDD2101 | [7] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C91(0.41) | LDD2104 | [7] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C91(1.73) | LDD2105 | [7] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C91(0.76) | LDD2107 | [7] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C91(0.46) | LDD2108 | [7] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C91(0.42) | LDD2109 | [7] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C91(0.69) | LDD2111 | [7] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C91(0.46) | LDD2114 | [7] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C91(0.42) | LDD2115 | [7] |
| LDCM0523 | Nucleophilic fragment 24b | MDA-MB-231 | C91(0.18) | LDD2116 | [7] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C91(0.22) | LDD2118 | [7] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C91(2.57) | LDD2119 | [7] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C91(0.69) | LDD2120 | [7] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C91(0.09) | LDD2122 | [7] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C91(0.56) | LDD2123 | [7] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C91(0.59) | LDD2125 | [7] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C91(0.07) | LDD2126 | [7] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C91(0.81) | LDD2127 | [7] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C91(0.85) | LDD2128 | [7] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C91(0.76) | LDD2129 | [7] |
| LDCM0540 | Nucleophilic fragment 35 | MDA-MB-231 | C91(0.50) | LDD2133 | [7] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C91(0.51) | LDD2134 | [7] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C91(0.75) | LDD2135 | [7] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C91(0.90) | LDD2136 | [7] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C91(0.85) | LDD2137 | [7] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C91(1.56) | LDD1700 | [7] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C91(0.55) | LDD2140 | [7] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C91(0.51) | LDD2141 | [7] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C91(0.79) | LDD2143 | [7] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C91(3.89) | LDD2144 | [7] |
| LDCM0551 | Nucleophilic fragment 5b | MDA-MB-231 | C91(0.12) | LDD2145 | [7] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C91(0.65) | LDD2146 | [7] |
| LDCM0553 | Nucleophilic fragment 6b | MDA-MB-231 | C91(3.97) | LDD2147 | [7] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C91(0.42) | LDD2148 | [7] |
| LDCM0555 | Nucleophilic fragment 7b | MDA-MB-231 | C91(0.09) | LDD2149 | [7] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C91(0.35) | LDD2150 | [7] |
| LDCM0557 | Nucleophilic fragment 8b | MDA-MB-231 | C91(0.09) | LDD2151 | [7] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C70(0.72) | LDD2206 | [40] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C63(0.71); C91(0.71) | LDD2207 | [40] |
| LDCM0131 | RA190 | MM1.R | C63(1.28); C70(1.28) | LDD0304 | [41] |
| LDCM0021 | THZ1 | HCT 116 | C63(1.13); C70(1.13) | LDD2173 | [16] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Huntingtin (HTT) | Huntingtin family | P42858 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Endothelial cell-specific chemotaxis regulator (ECSCR) | ECSCR family | Q19T08 | |||
| EPM2A-interacting protein 1 (EPM2AIP1) | . | Q7L775 | |||
References
