Details of the Target
General Information of Target
| Target ID | LDTP00338 | |||||
|---|---|---|---|---|---|---|
| Target Name | Lysosomal alpha-mannosidase (MAN2B1) | |||||
| Gene Name | MAN2B1 | |||||
| Gene ID | 4125 | |||||
| Synonyms |
LAMAN; MANB; Lysosomal alpha-mannosidase; Laman; EC 3.2.1.24; Lysosomal acid alpha-mannosidase; Mannosidase alpha class 2B member 1; Mannosidase alpha-B) [Cleaved into: Lysosomal alpha-mannosidase A peptide; Lysosomal alpha-mannosidase B peptide; Lysosomal alpha-mannosidase C peptide; Lysosomal alpha-mannosidase D peptide; Lysosomal alpha-mannosidase E peptide]
|
|||||
| 3D Structure | ||||||
| Sequence |
MGAYARASGVCARGCLDSAGPWTMSRALRPPLPPLCFFLLLLAAAGARAGGYETCPTVQP
NMLNVHLLPHTHDDVGWLKTVDQYFYGIKNDIQHAGVQYILDSVISALLADPTRRFIYVE IAFFSRWWHQQTNATQEVVRDLVRQGRLEFANGGWVMNDEAATHYGAIVDQMTLGLRFLE DTFGNDGRPRVAWHIDPFGHSREQASLFAQMGFDGFFFGRLDYQDKWVRMQKLEMEQVWR ASTSLKPPTADLFTGVLPNGYNPPRNLCWDVLCVDQPLVEDPRSPEYNAKELVDYFLNVA TAQGRYYRTNHTVMTMGSDFQYENANMWFKNLDKLIRLVNAQQAKGSSVHVLYSTPACYL WELNKANLTWSVKHDDFFPYADGPHQFWTGYFSSRPALKRYERLSYNFLQVCNQLEALVG LAANVGPYGSGDSAPLNEAMAVLQHHDAVSGTSRQHVANDYARQLAAGWGPCEVLLSNAL ARLRGFKDHFTFCQQLNISICPLSQTAARFQVIVYNPLGRKVNWMVRLPVSEGVFVVKDP NGRTVPSDVVIFPSSDSQAHPPELLFSASLPALGFSTYSVAQVPRWKPQARAPQPIPRRS WSPALTIENEHIRATFDPDTGLLMEIMNMNQQLLLPVRQTFFWYNASIGDNESDQASGAY IFRPNQQKPLPVSRWAQIHLVKTPLVQEVHQNFSAWCSQVVRLYPGQRHLELEWSVGPIP VGDTWGKEVISRFDTPLETKGRFYTDSNGREILERRRDYRPTWKLNQTEPVAGNYYPVNT RIYITDGNMQLTVLTDRSQGGSSLRDGSLELMVHRRLLKDDGRGVSEPLMENGSGAWVRG RHLVLLDTAQAAAAGHRLLAEQEVLAPQVVLAPGGGAAYNLGAPPRTQFSGLRRDLPPSV HLLTLASWGPEMVLLRLEHQFAVGEDSGRNLSAPVTLNLRDLFSTFTITRLQETTLVANQ LREAASRLKWTTNTGPTPHQTPYQLDPANITLEPMEIRTFLASVQWKEVDG |
|||||
| Target Type |
Successful
|
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Glycosyl hydrolase 38 family
|
|||||
| Subcellular location |
Lysosome
|
|||||
| Function | Necessary for the catabolism of N-linked carbohydrates released during glycoprotein turnover. Cleaves all known types of alpha-mannosidic linkages. | |||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
Alkylaryl probe 1 Probe Info |
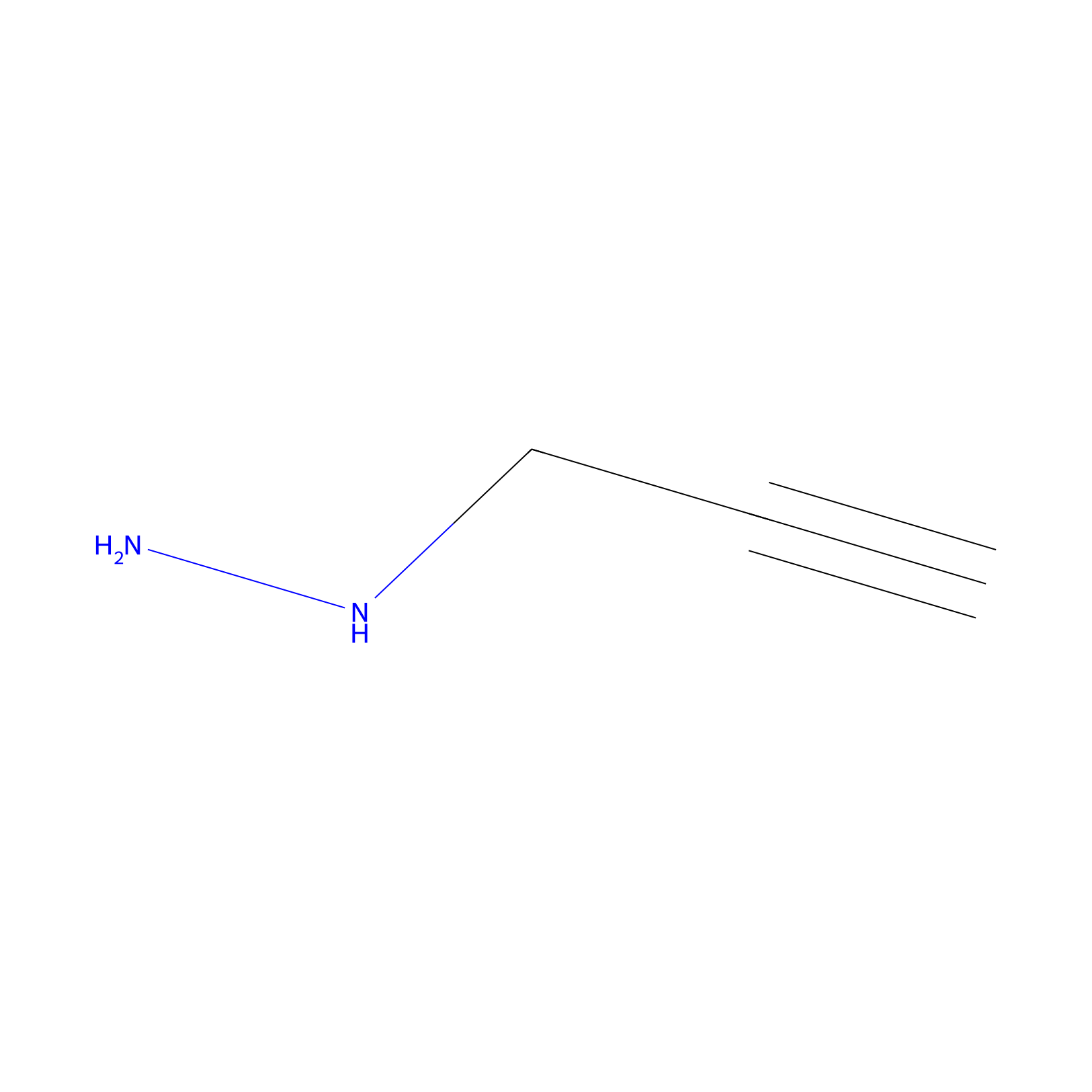 |
20.00 | LDD0387 | [1] | |
|
Alkylaryl probe 2 Probe Info |
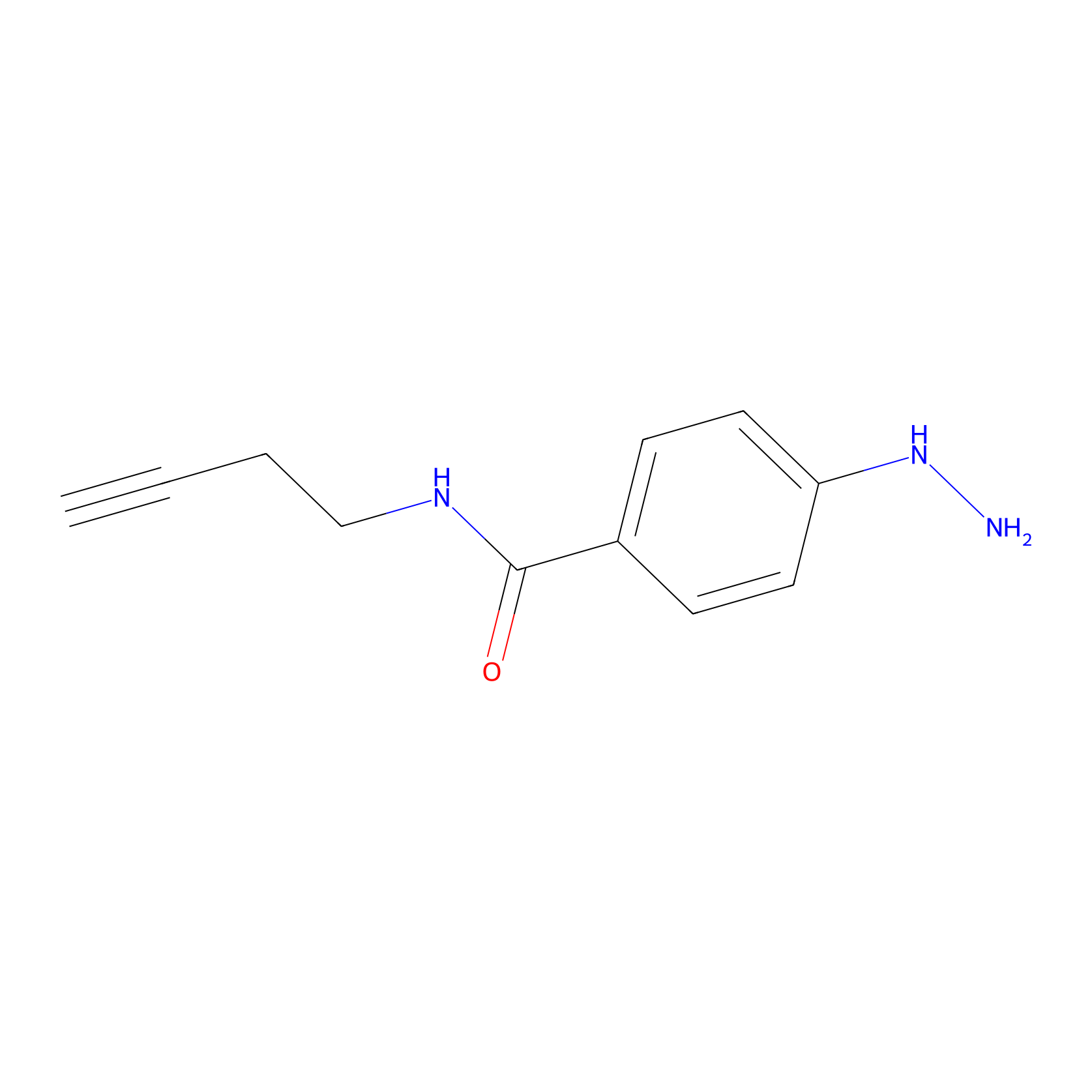 |
20.00 | LDD0391 | [1] | |
|
SAA-alkyne Probe Info |
 |
1.08 | LDD0252 | [2] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [3] | |
|
FBPP2 Probe Info |
 |
7.62 | LDD0054 | [4] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C001 Probe Info |
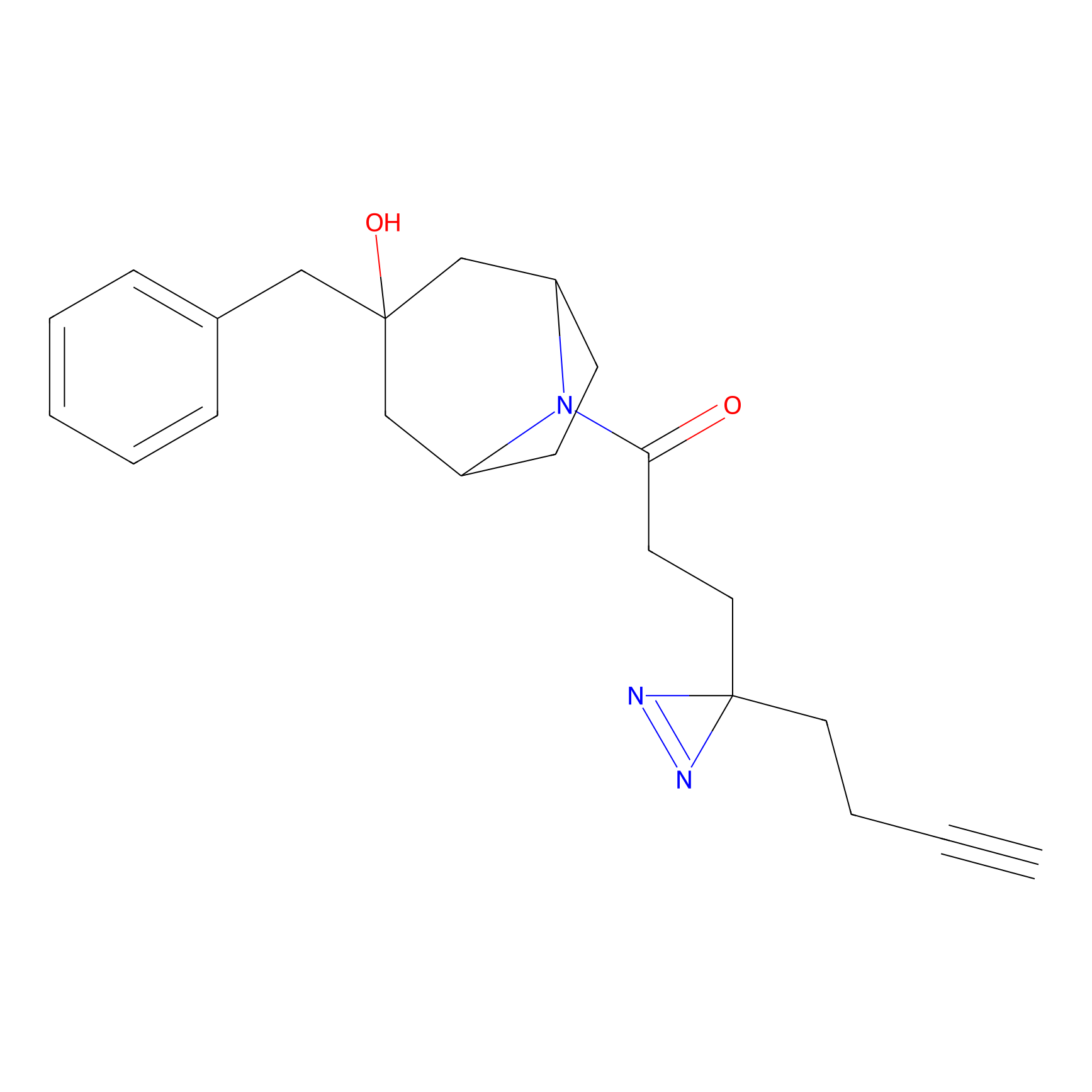 |
5.62 | LDD1711 | [5] | |
|
C045 Probe Info |
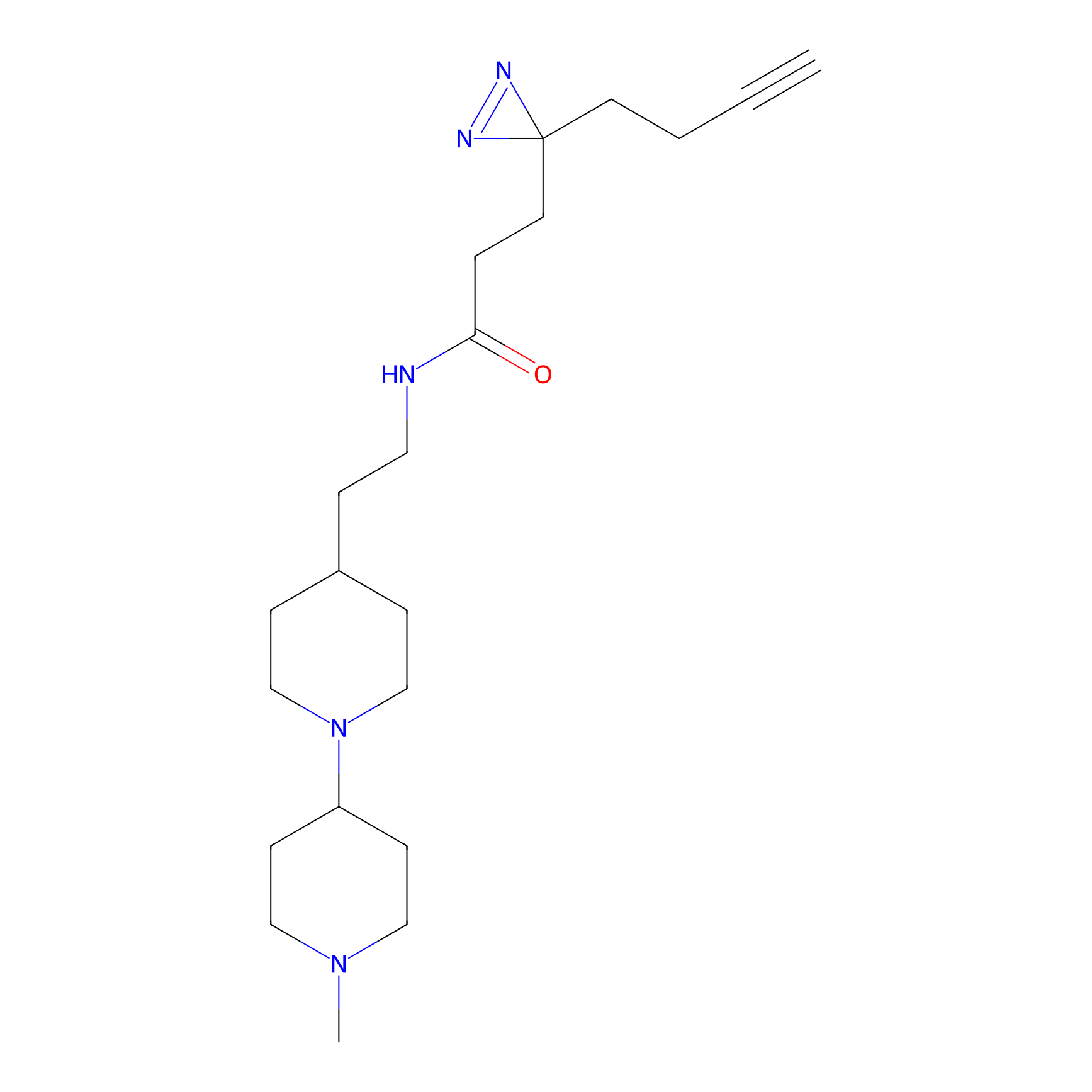 |
7.46 | LDD1744 | [5] | |
|
C106 Probe Info |
 |
16.56 | LDD1793 | [5] | |
|
C108 Probe Info |
 |
8.22 | LDD1795 | [5] | |
|
C115 Probe Info |
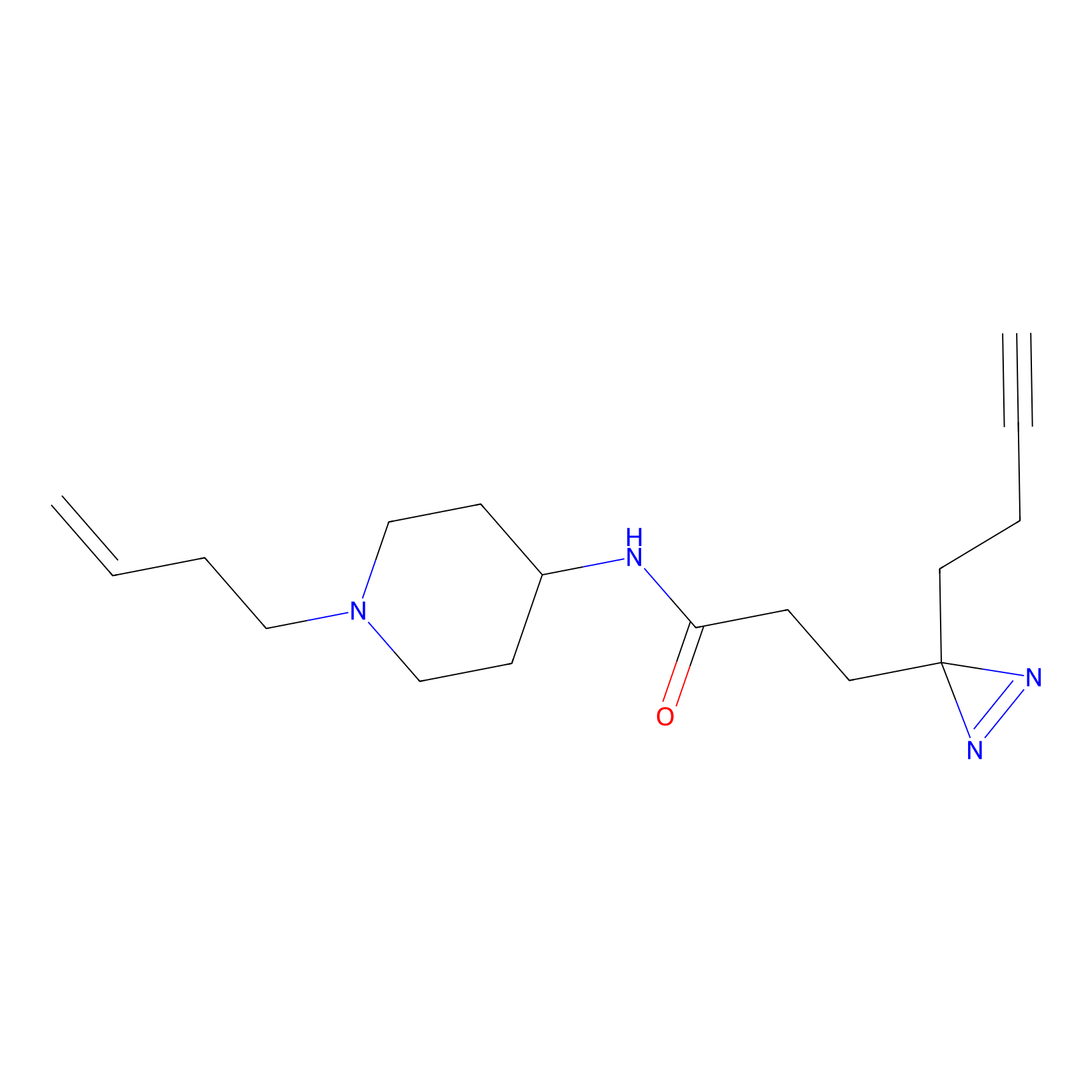 |
5.90 | LDD1802 | [5] | |
|
C130 Probe Info |
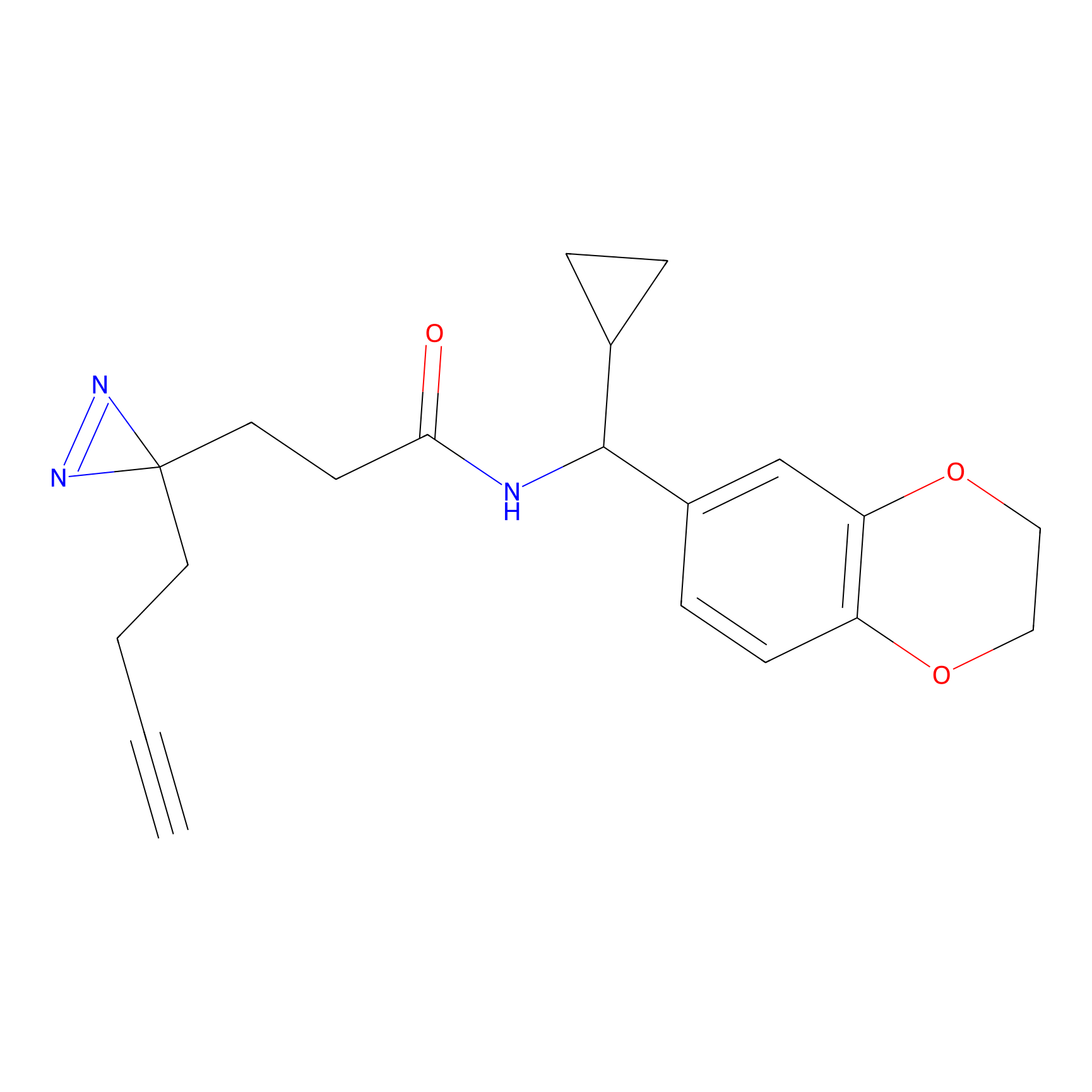 |
6.50 | LDD1812 | [5] | |
|
C141 Probe Info |
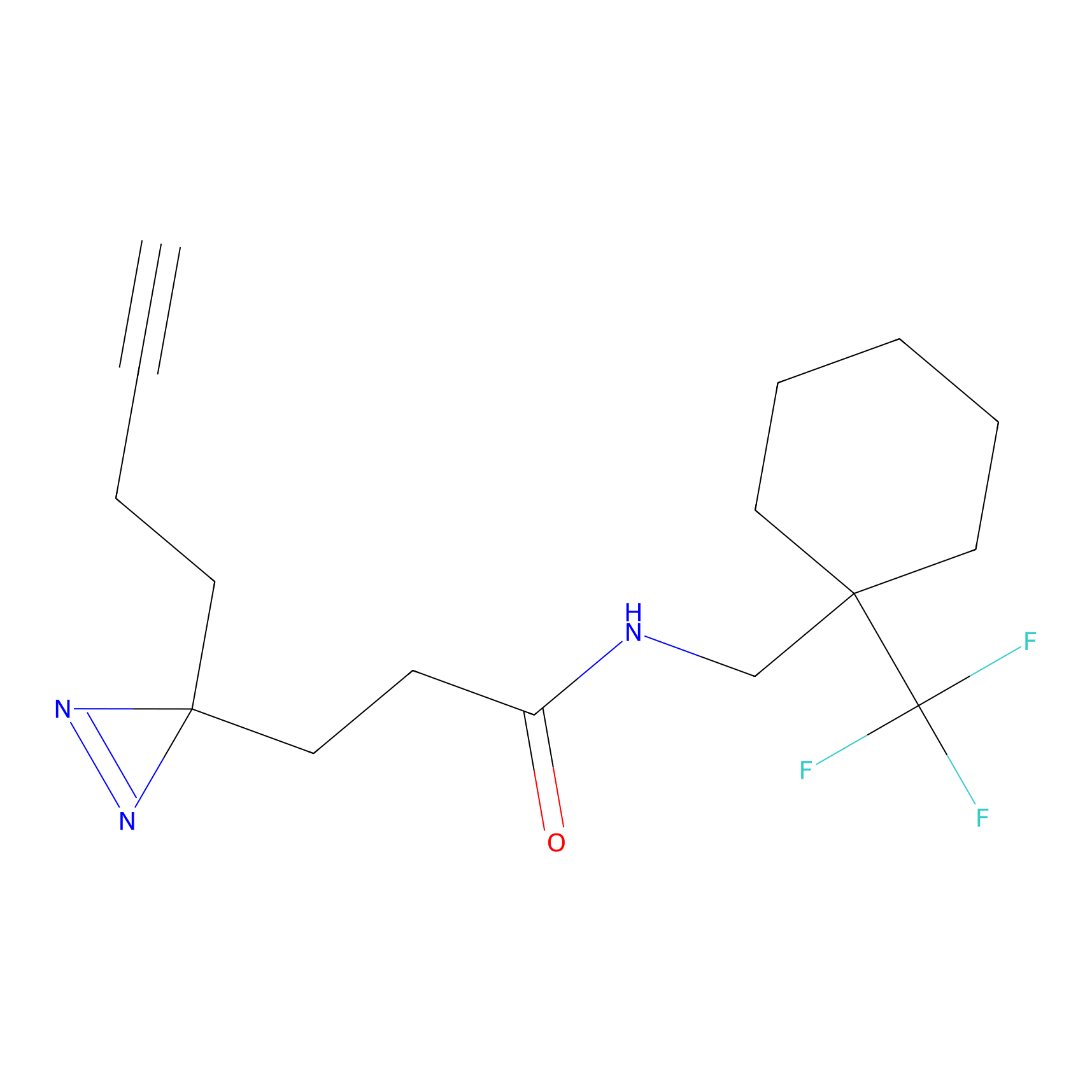 |
10.13 | LDD1823 | [5] | |
|
C149 Probe Info |
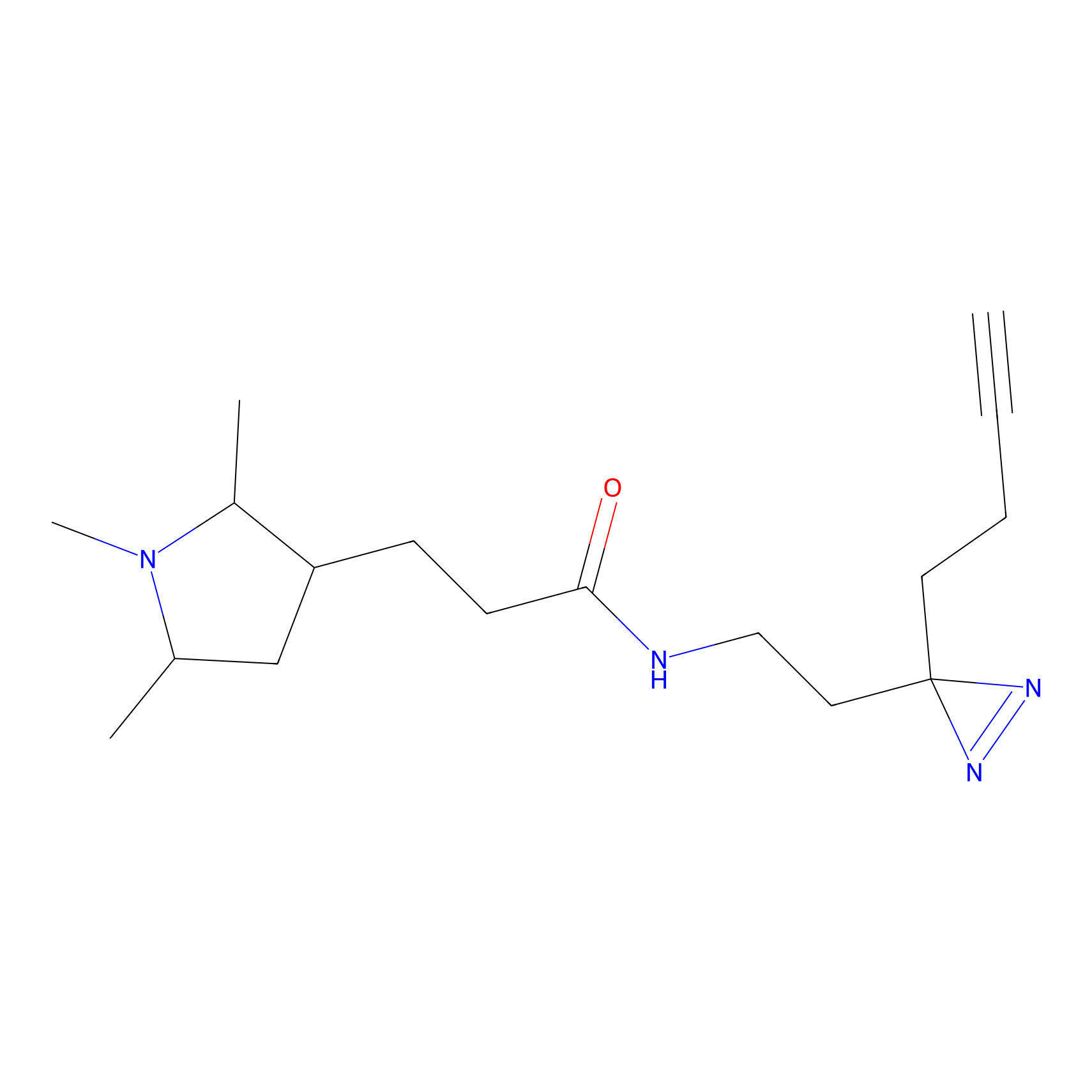 |
6.87 | LDD1830 | [5] | |
|
C170 Probe Info |
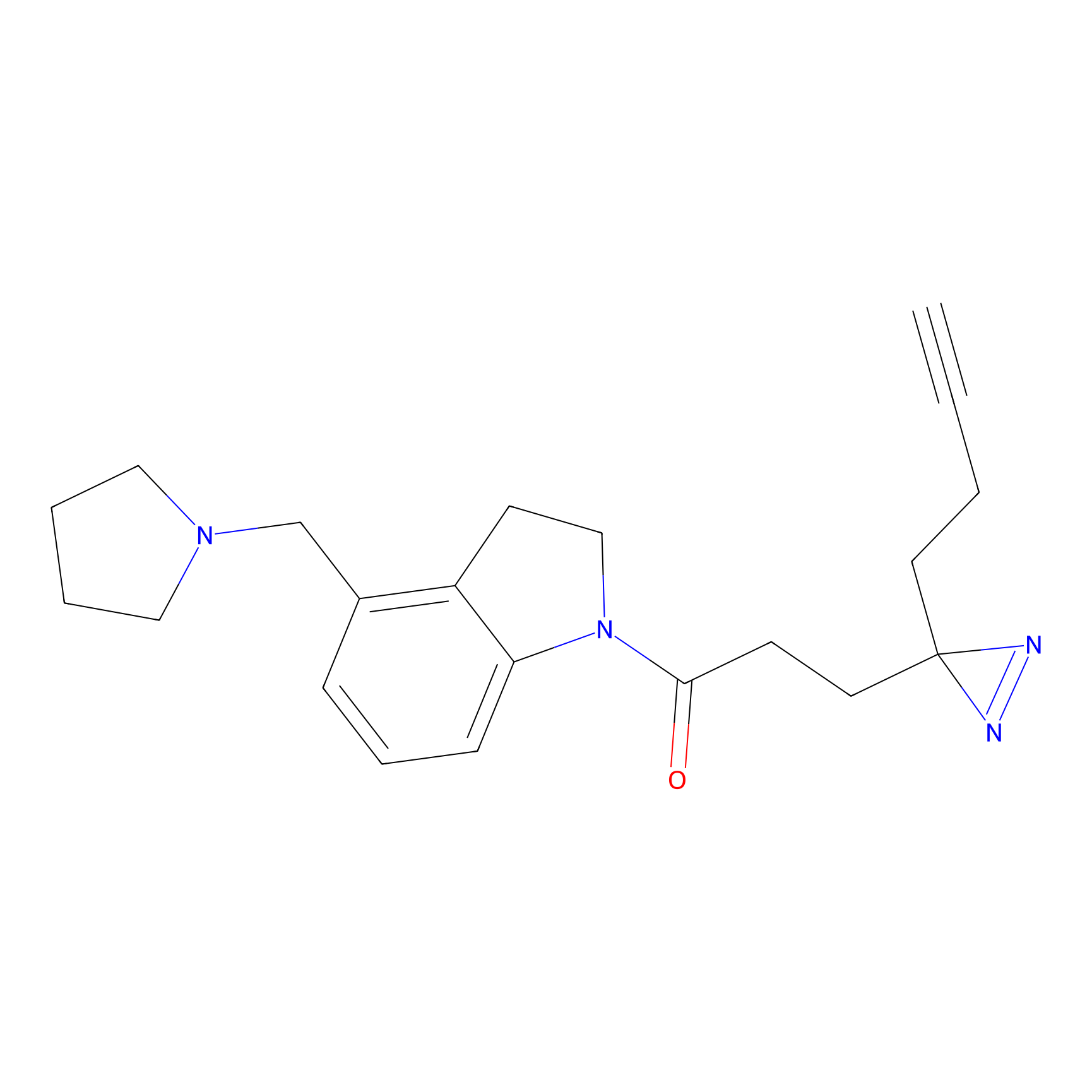 |
18.64 | LDD1850 | [5] | |
|
C171 Probe Info |
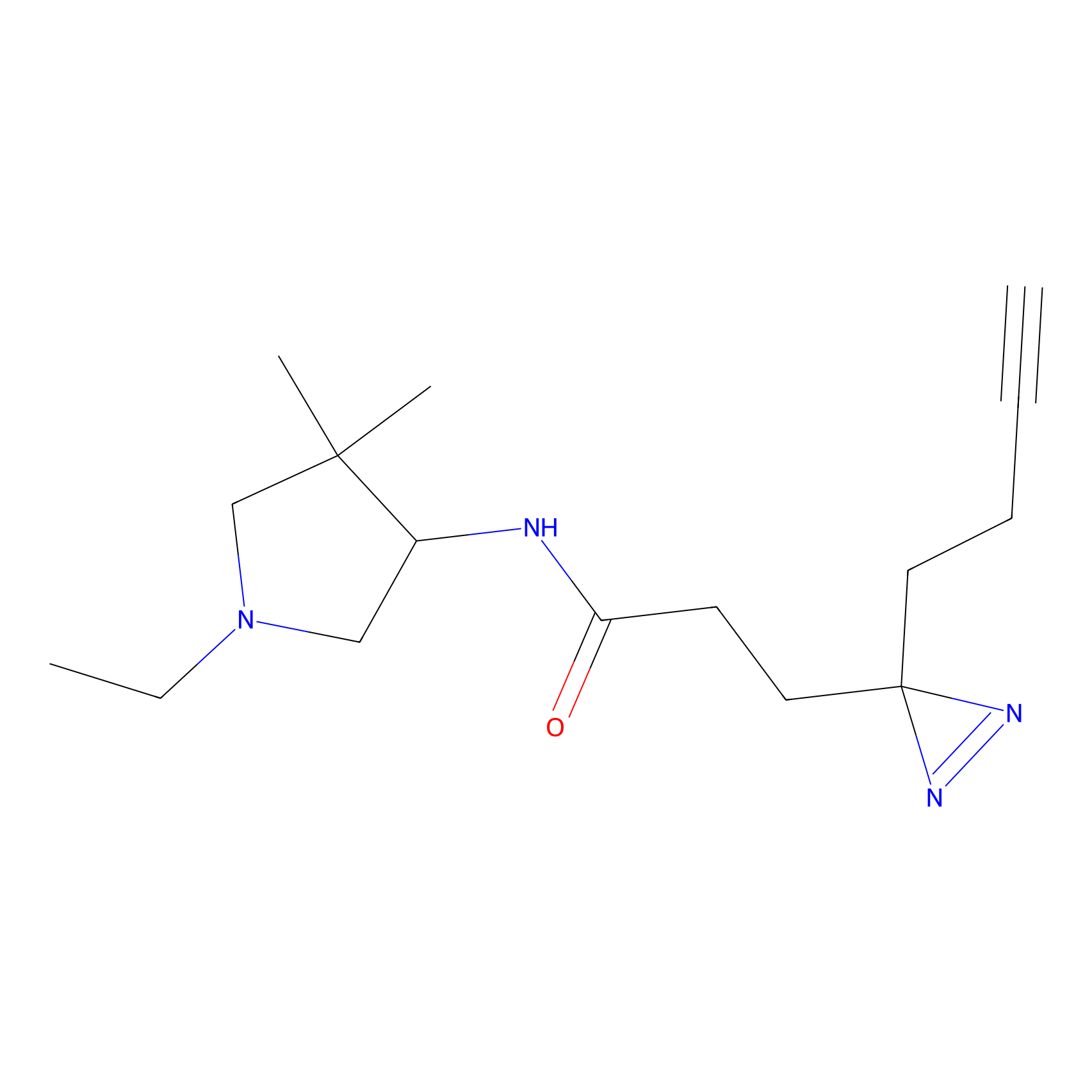 |
7.41 | LDD1851 | [5] | |
|
C186 Probe Info |
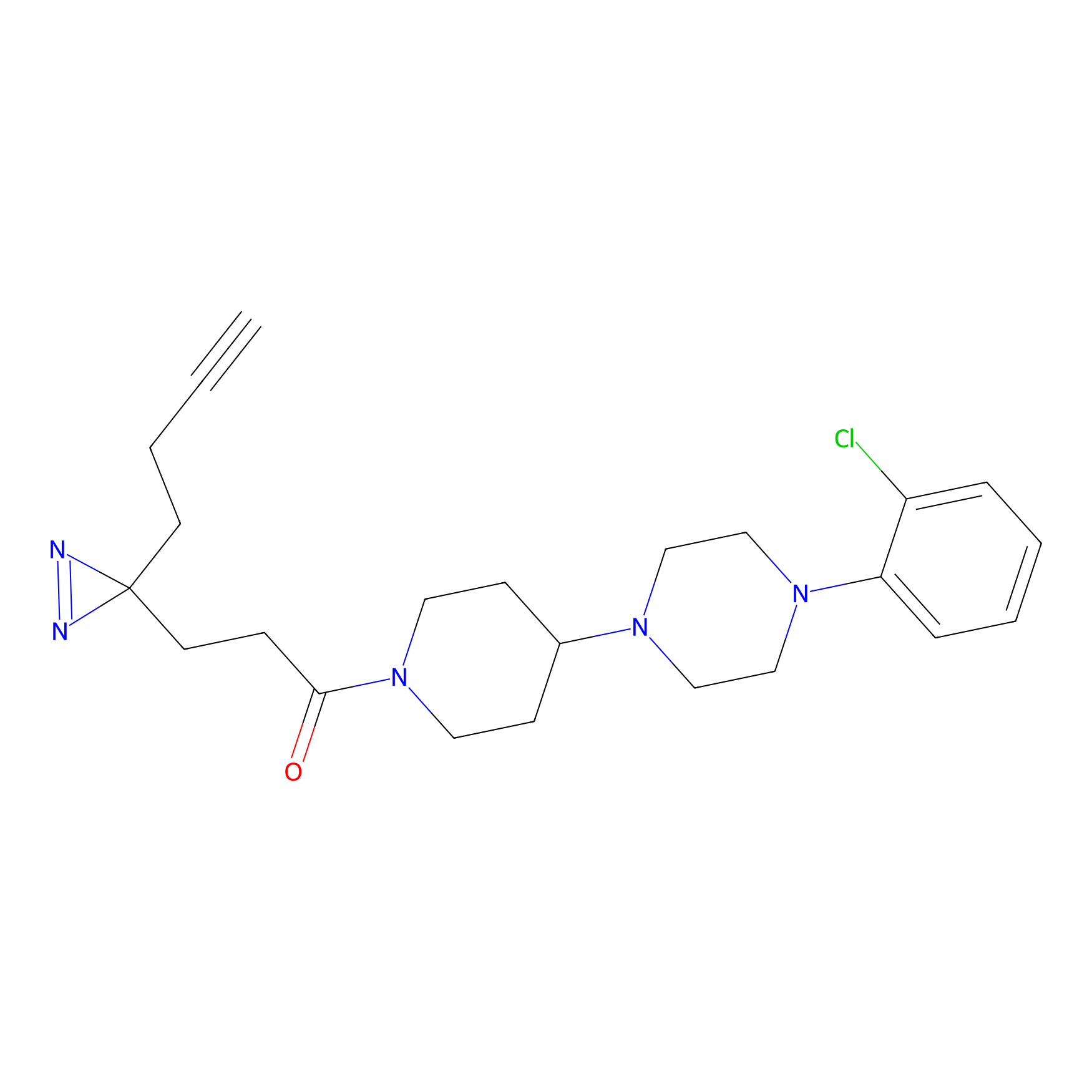 |
10.20 | LDD1864 | [5] | |
|
C208 Probe Info |
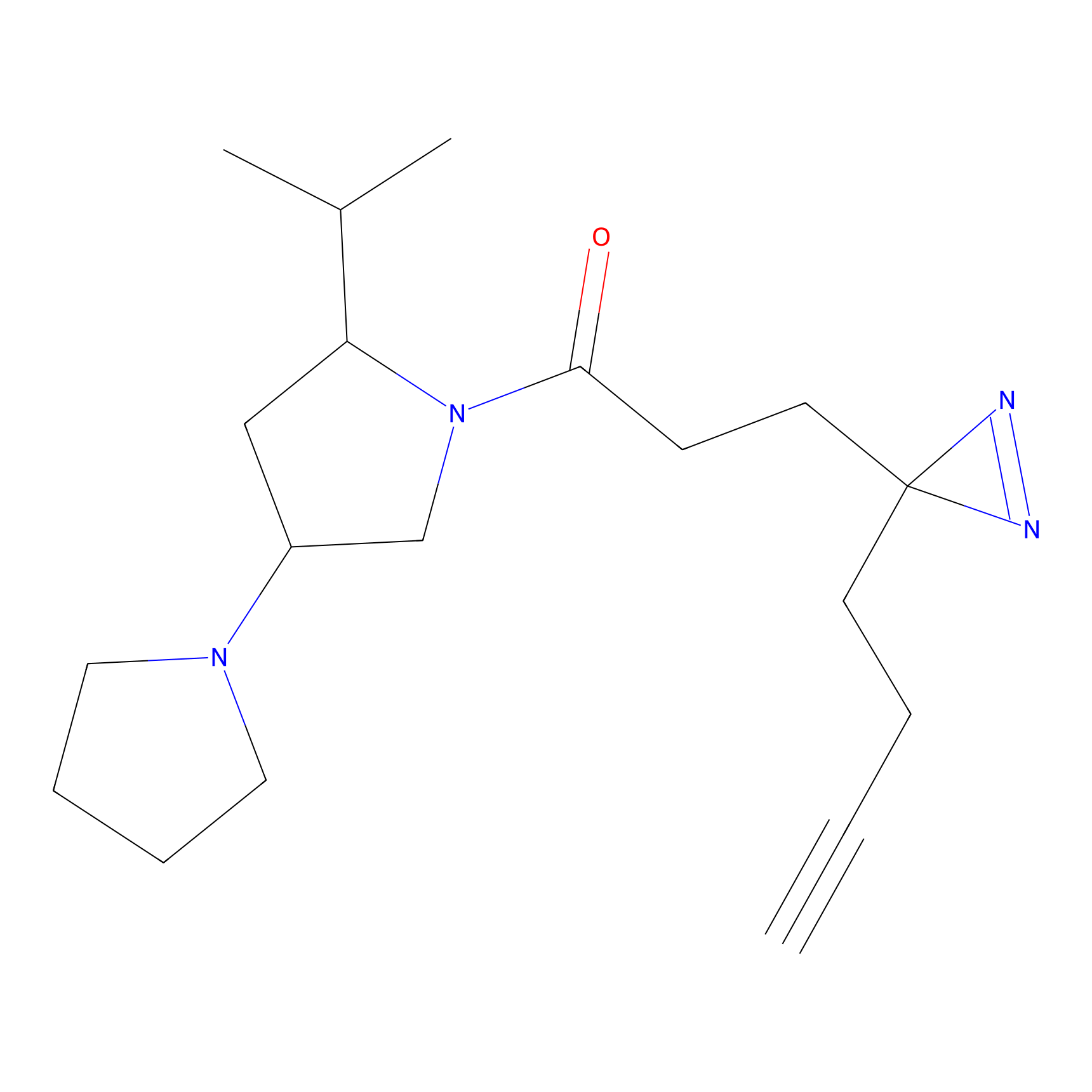 |
6.02 | LDD1883 | [5] | |
|
C223 Probe Info |
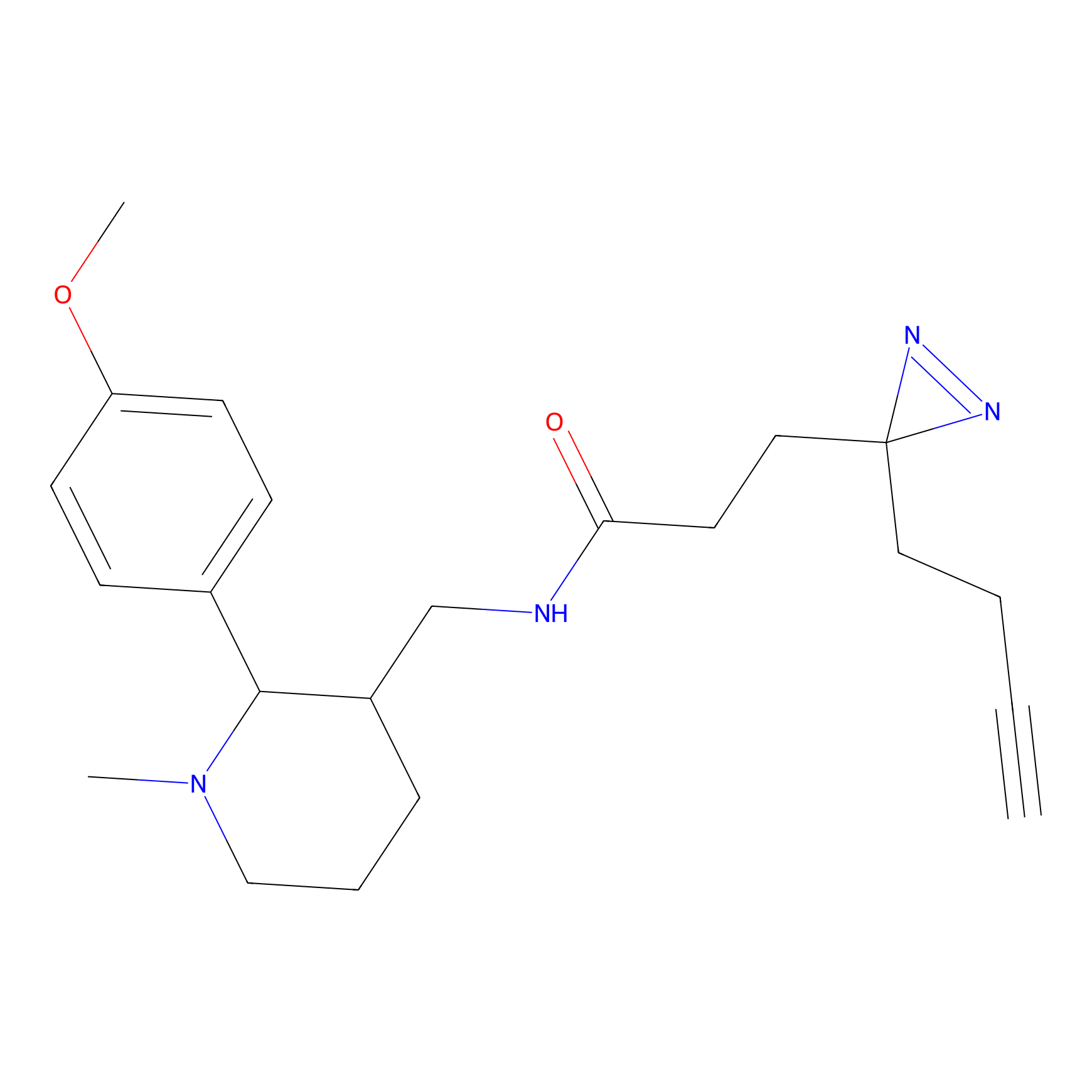 |
5.54 | LDD1897 | [5] | |
|
C237 Probe Info |
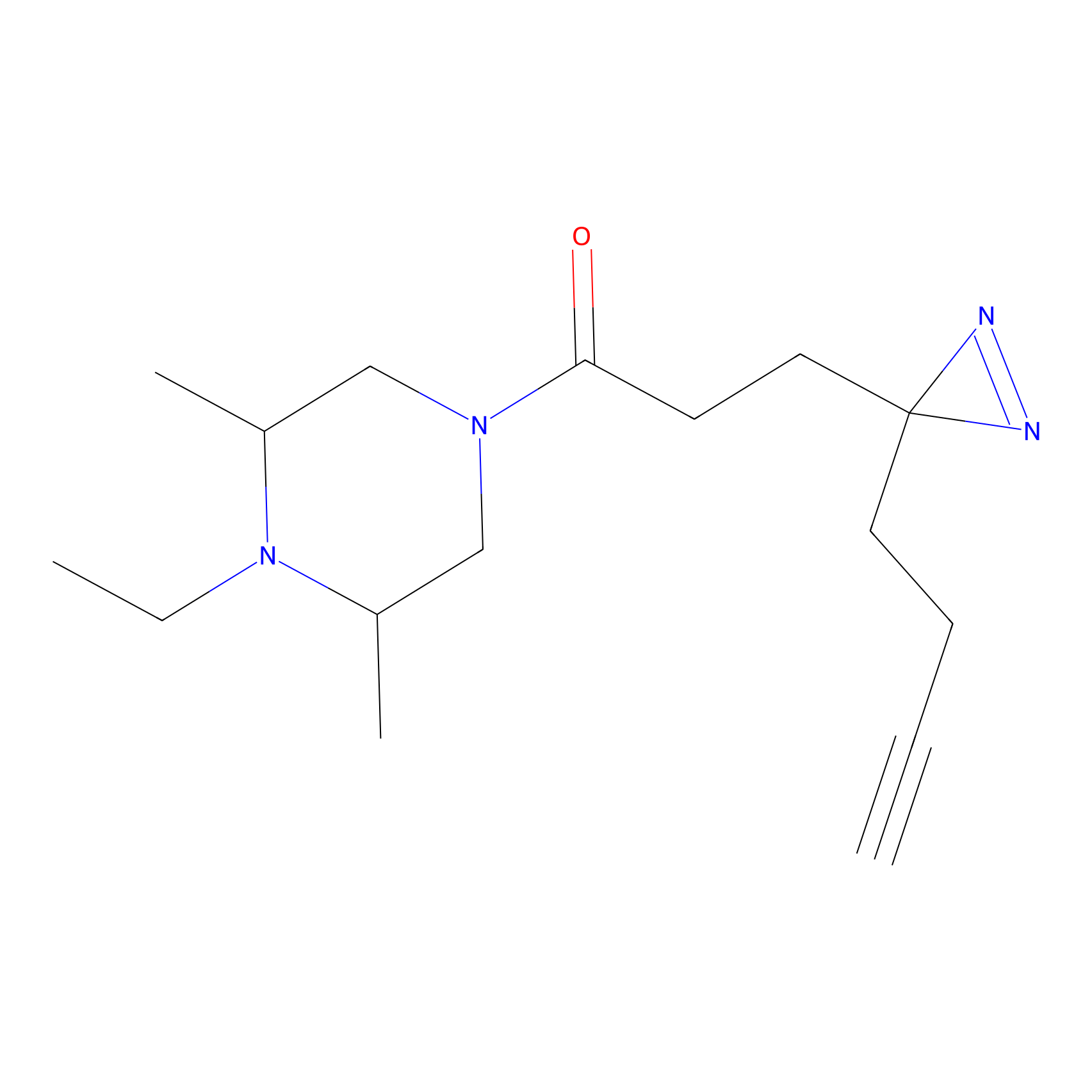 |
6.63 | LDD1910 | [5] | |
|
C302 Probe Info |
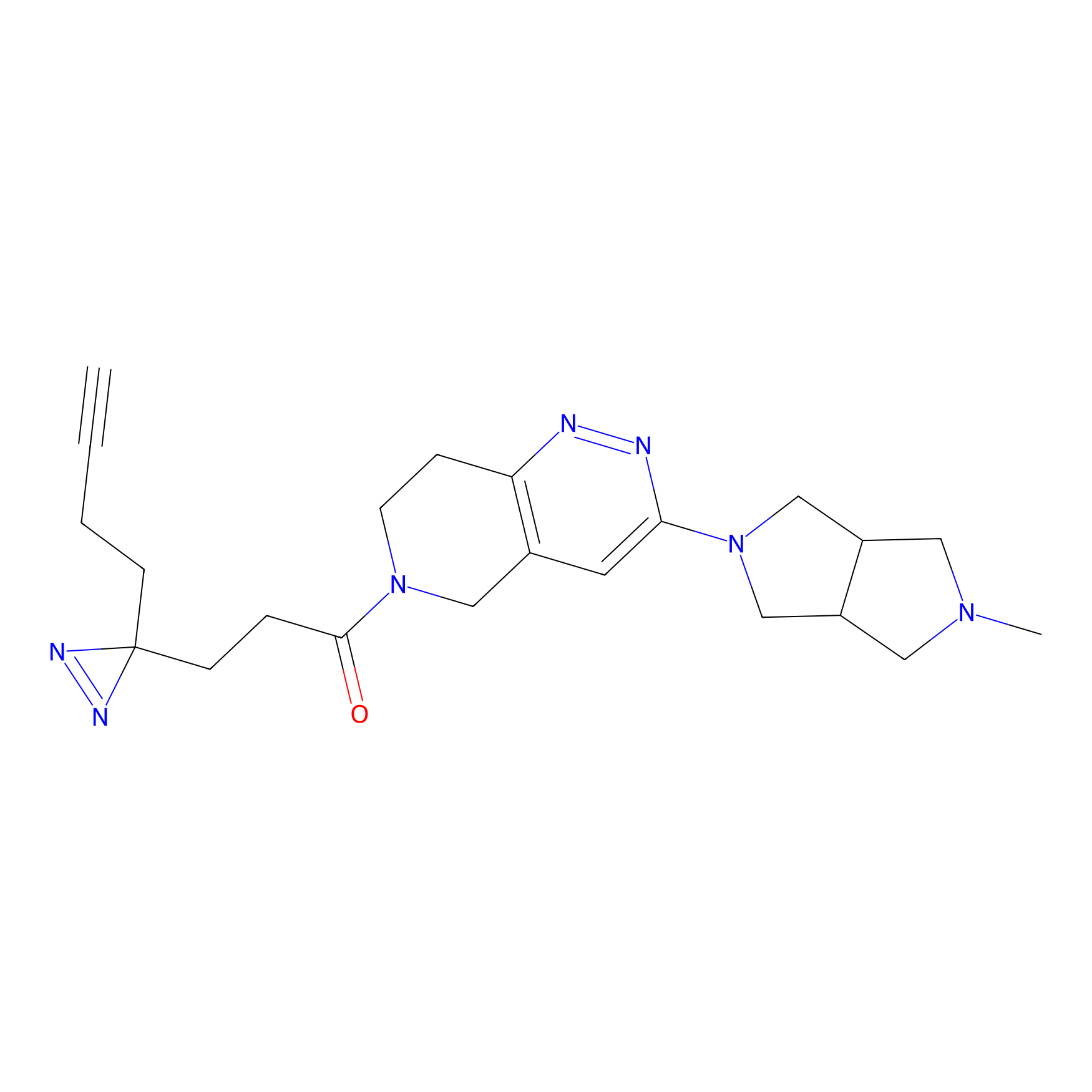 |
5.28 | LDD1971 | [5] | |
|
C310 Probe Info |
 |
14.12 | LDD1977 | [5] | |
|
C343 Probe Info |
 |
11.16 | LDD2005 | [5] | |
|
C346 Probe Info |
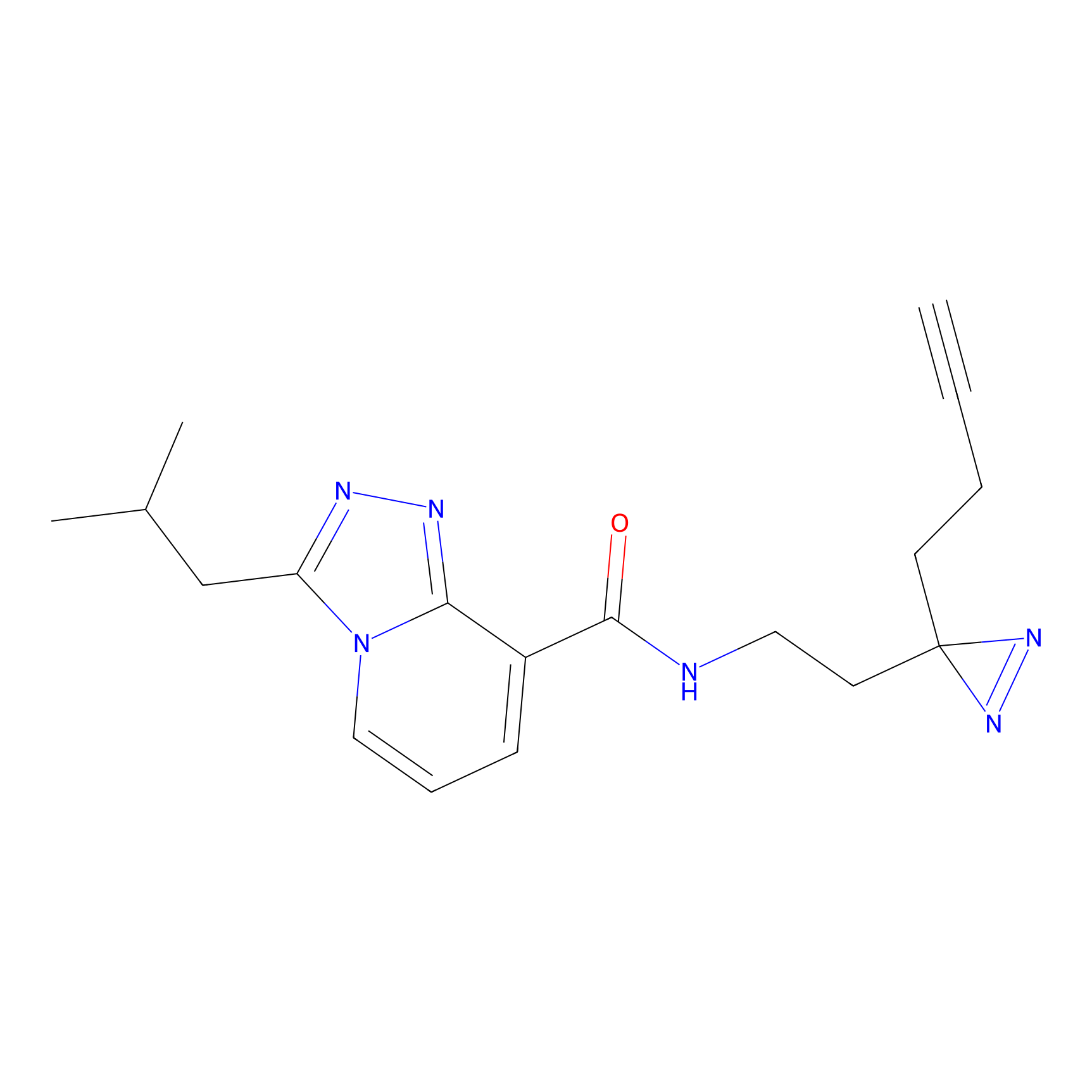 |
11.16 | LDD2007 | [5] | |
|
C376 Probe Info |
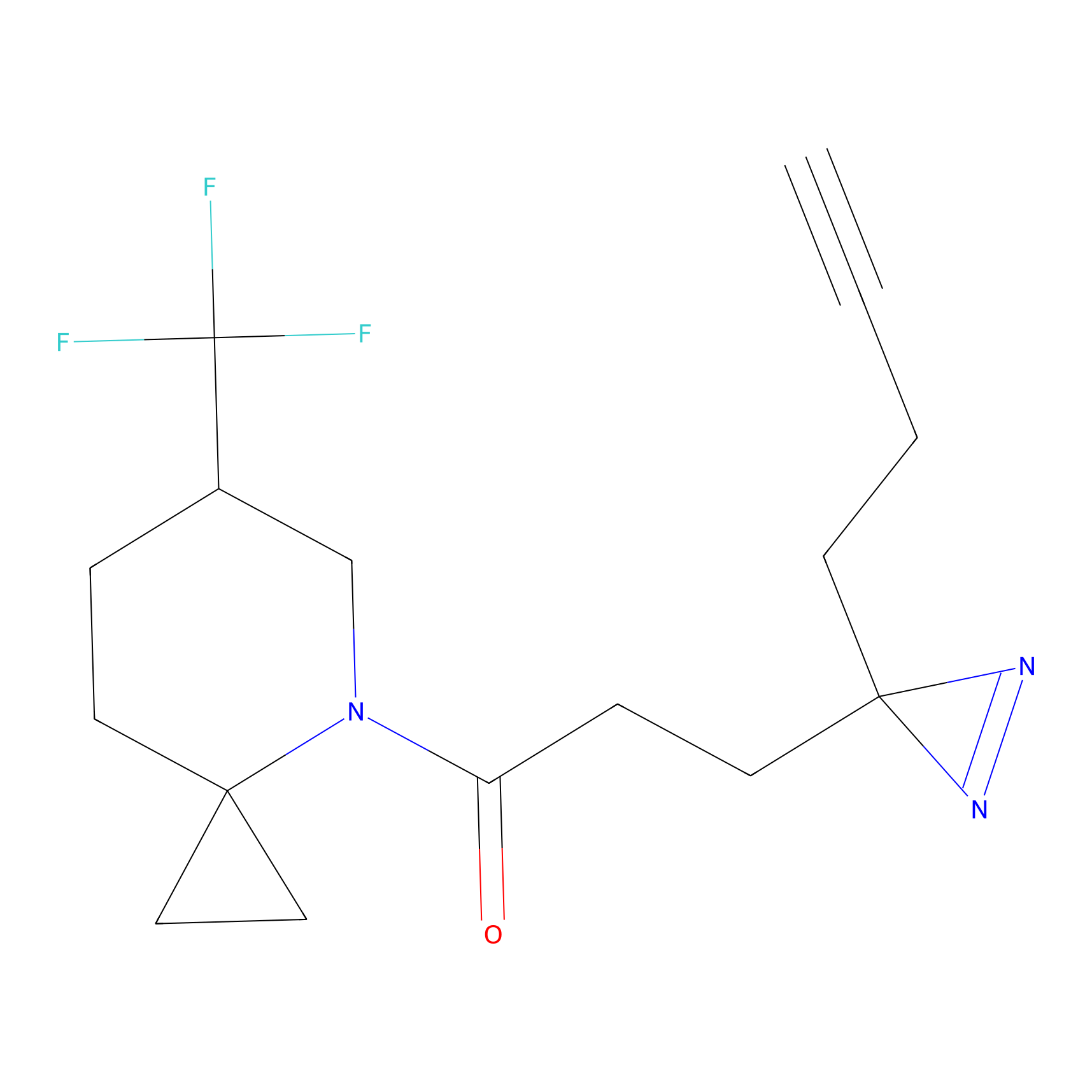 |
8.57 | LDD2036 | [5] | |
|
FFF probe12 Probe Info |
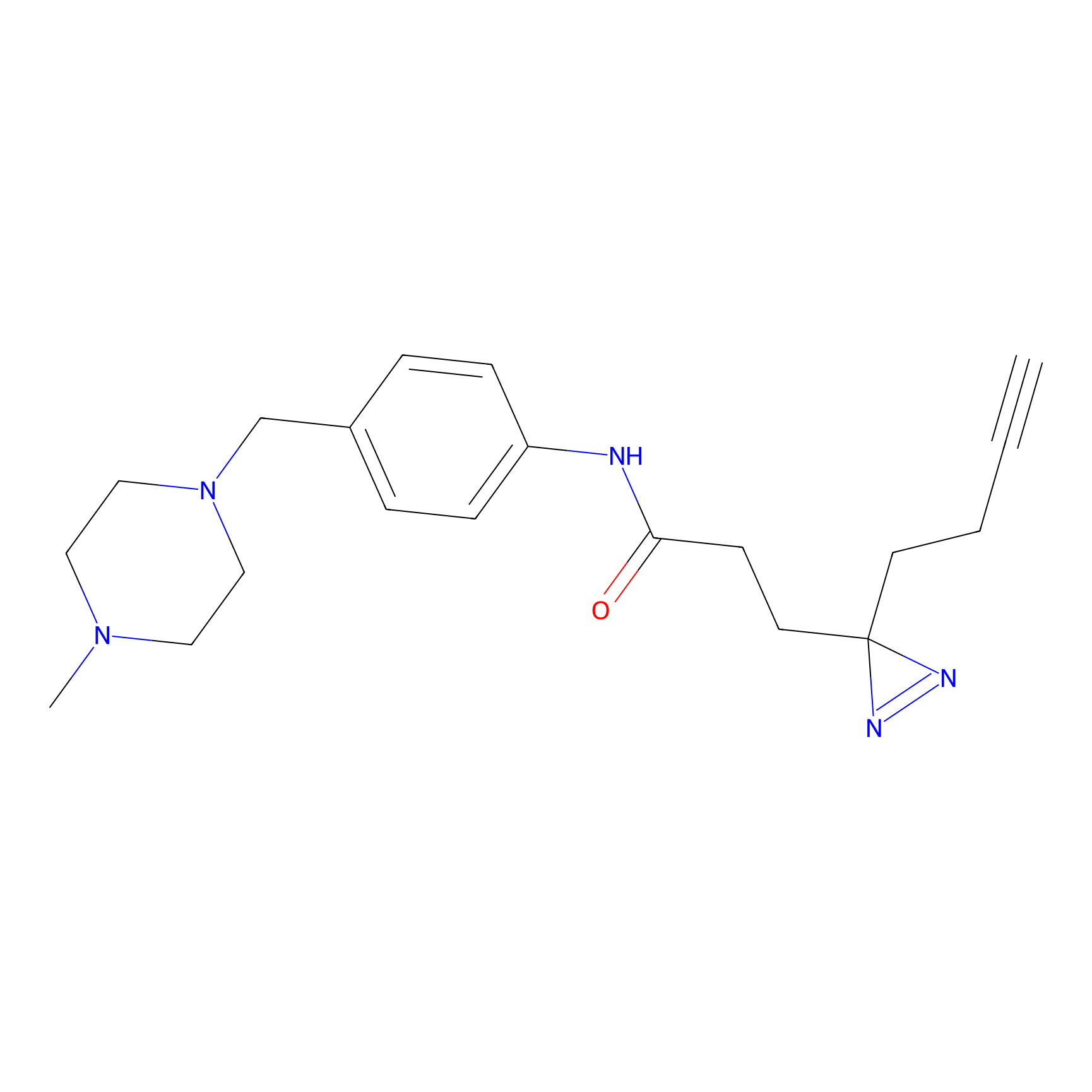 |
20.00 | LDD0473 | [6] | |
|
FFF probe14 Probe Info |
 |
20.00 | LDD0477 | [6] | |
|
FFF probe9 Probe Info |
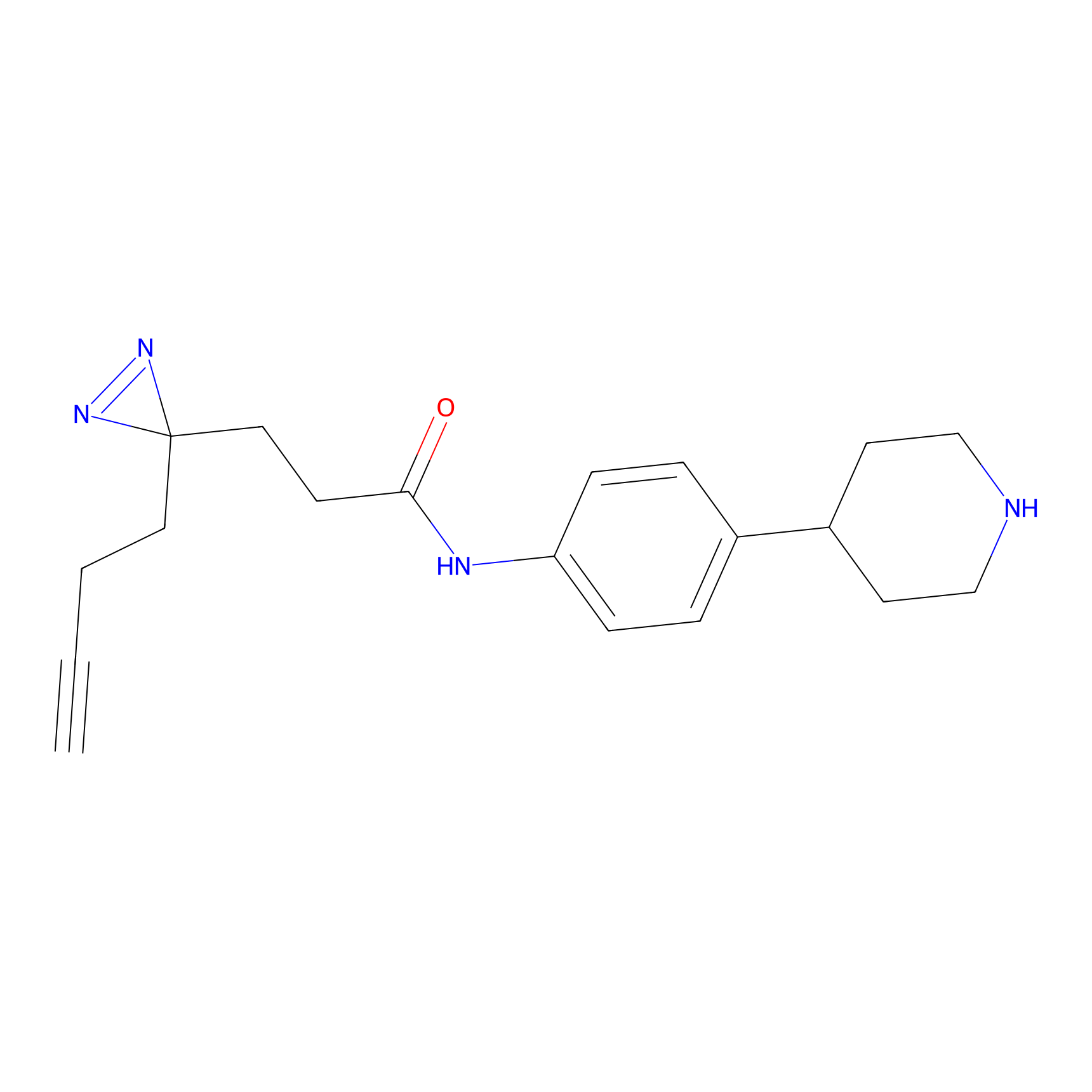 |
20.00 | LDD0470 | [6] | |
|
JN0003 Probe Info |
 |
20.00 | LDD0469 | [6] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [7] | |
Competitor(s) Related to This Target
References
