Details of the Target
General Information of Target
| Target ID | LDTP04720 | |||||
|---|---|---|---|---|---|---|
| Target Name | IgG receptor FcRn large subunit p51 (FCGRT) | |||||
| Gene Name | FCGRT | |||||
| Gene ID | 2217 | |||||
| Synonyms |
FCRN; IgG receptor FcRn large subunit p51; FcRn; IgG Fc fragment receptor transporter alpha chain; Neonatal Fc receptor |
|||||
| 3D Structure | ||||||
| Sequence |
MGVPRPQPWALGLLLFLLPGSLGAESHLSLLYHLTAVSSPAPGTPAFWVSGWLGPQQYLS
YNSLRGEAEPCGAWVWENQVSWYWEKETTDLRIKEKLFLEAFKALGGKGPYTLQGLLGCE LGPDNTSVPTAKFALNGEEFMNFDLKQGTWGGDWPEALAISQRWQQQDKAANKELTFLLF SCPHRLREHLERGRGNLEWKEPPSMRLKARPSSPGFSVLTCSAFSFYPPELQLRFLRNGL AAGTGQGDFGPNSDGSFHASSSLTVKSGDEHHYCCIVQHAGLAQPLRVELESPAKSSVLV VGIVIGVLLLTAAAVGGALLWRRMRSGLPAPWISLRGDDTGVLLPTPGEAQDADLKDVNV IPATA |
|||||
| Target Type |
Successful
|
|||||
| Target Bioclass |
Immunoglobulin
|
|||||
| Family |
Immunoglobulin superfamily
|
|||||
| Subcellular location |
Cell membrane
|
|||||
| Function |
Cell surface receptor that transfers passive humoral immunity from the mother to the newborn. Binds to the Fc region of monomeric immunoglobulin gamma and mediates its selective uptake from milk. IgG in the milk is bound at the apical surface of the intestinal epithelium. The resultant FcRn-IgG complexes are transcytosed across the intestinal epithelium and IgG is released from FcRn into blood or tissue fluids. Throughout life, contributes to effective humoral immunity by recycling IgG and extending its half-life in the circulation. Mechanistically, monomeric IgG binding to FcRn in acidic endosomes of endothelial and hematopoietic cells recycles IgG to the cell surface where it is released into the circulation. In addition of IgG, regulates homeostasis of the other most abundant circulating protein albumin/ALB.; (Microbial infection) Acts as an uncoating receptor for a panel of echoviruses including Echovirus 5, 6, 7, 9, 11, 13, 25 and 29.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
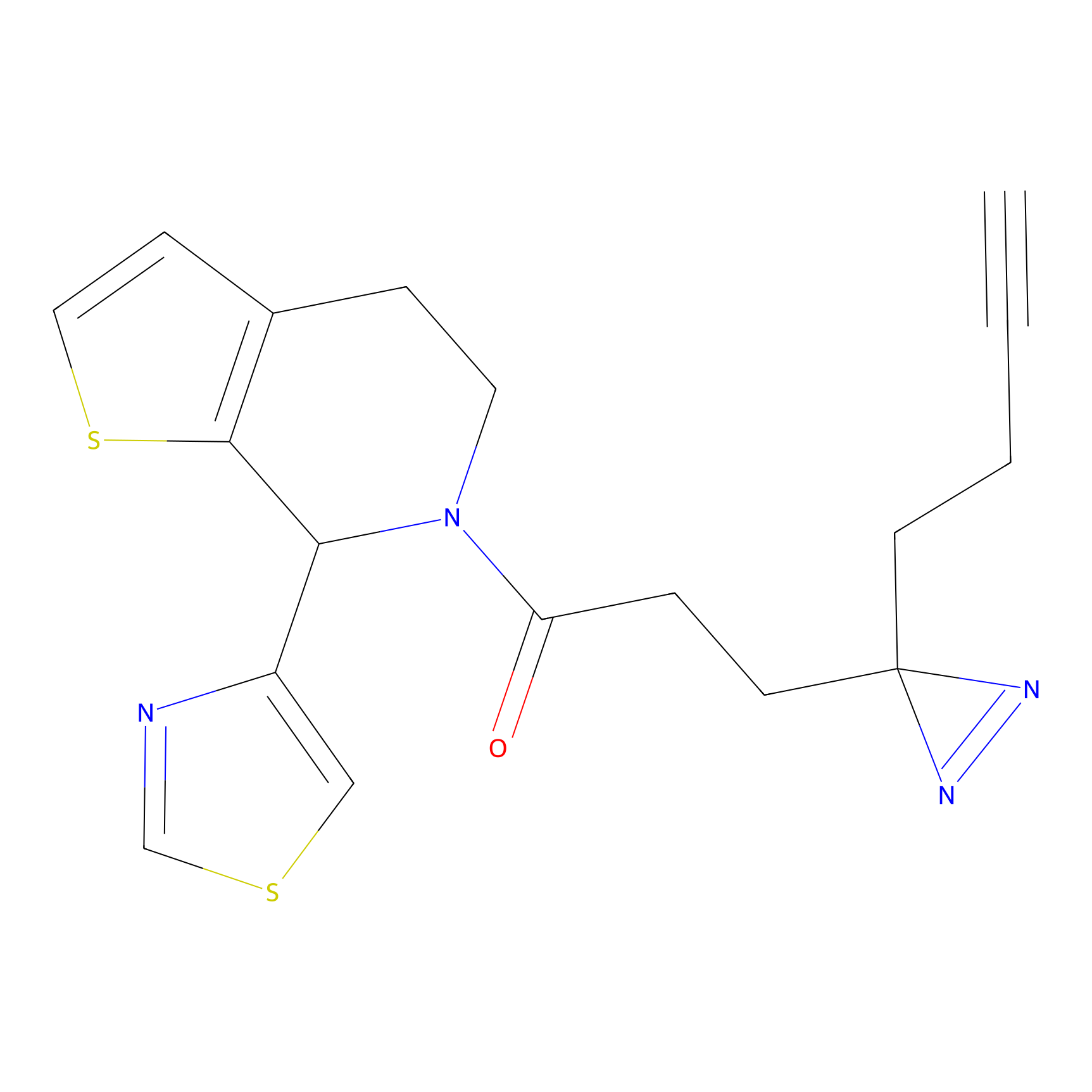
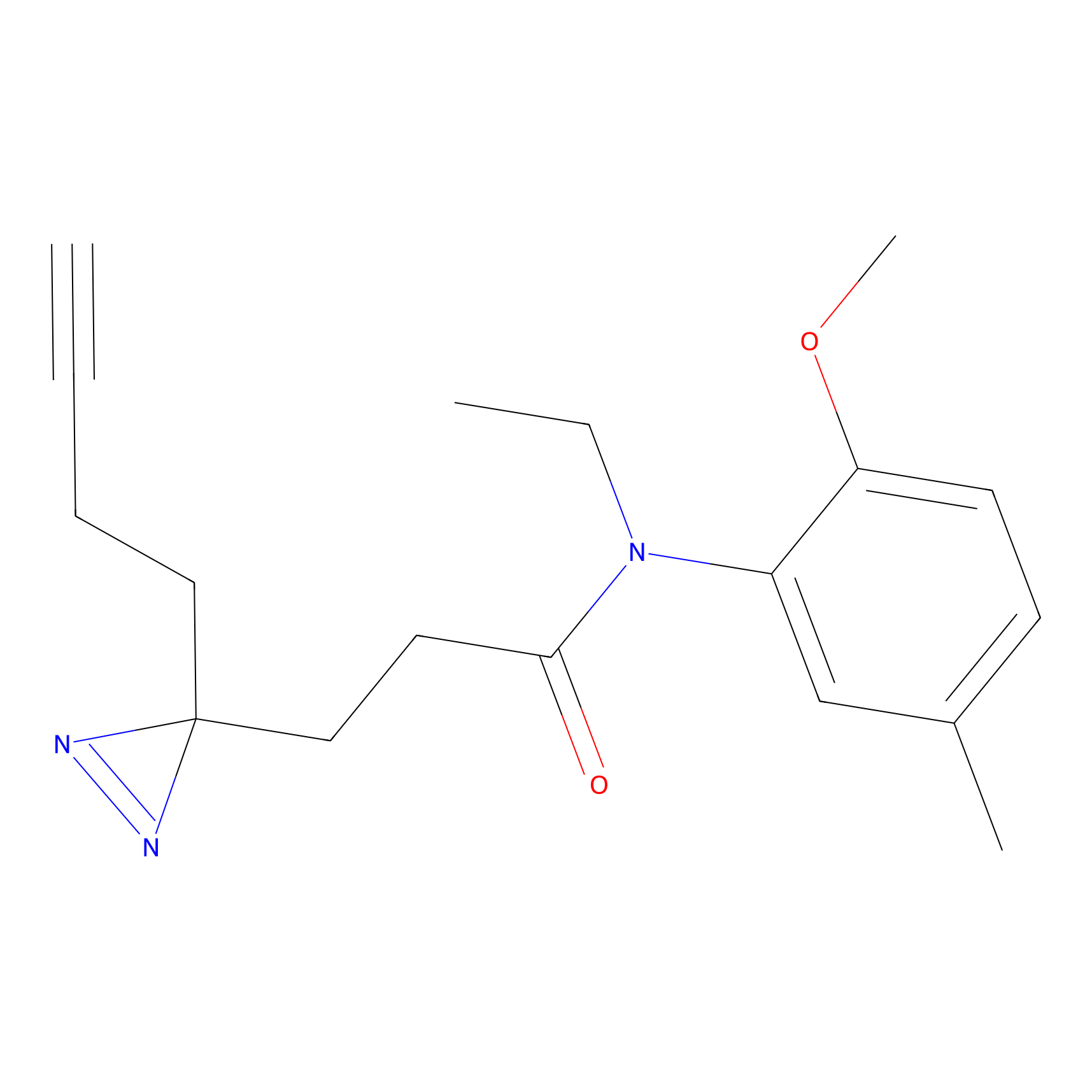
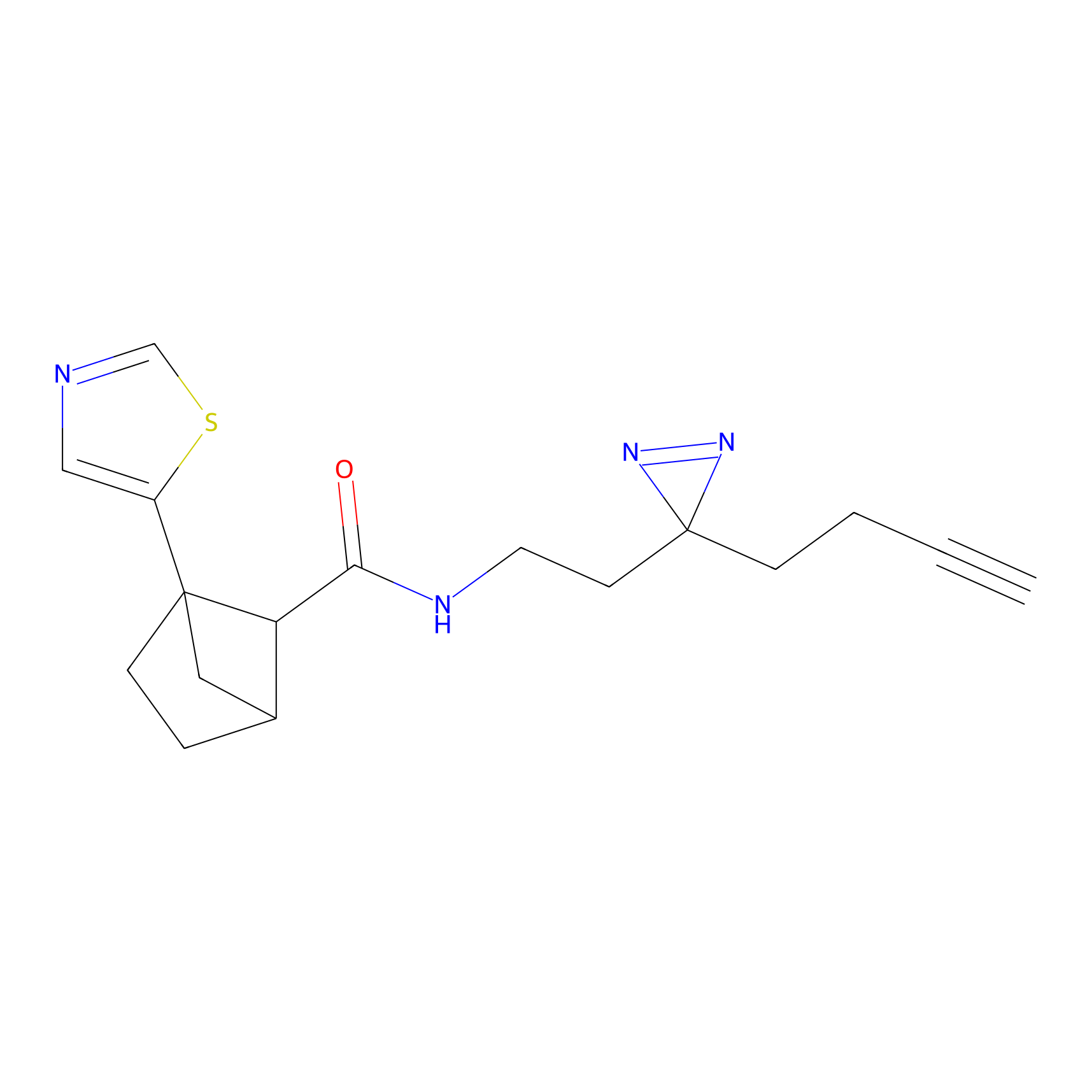
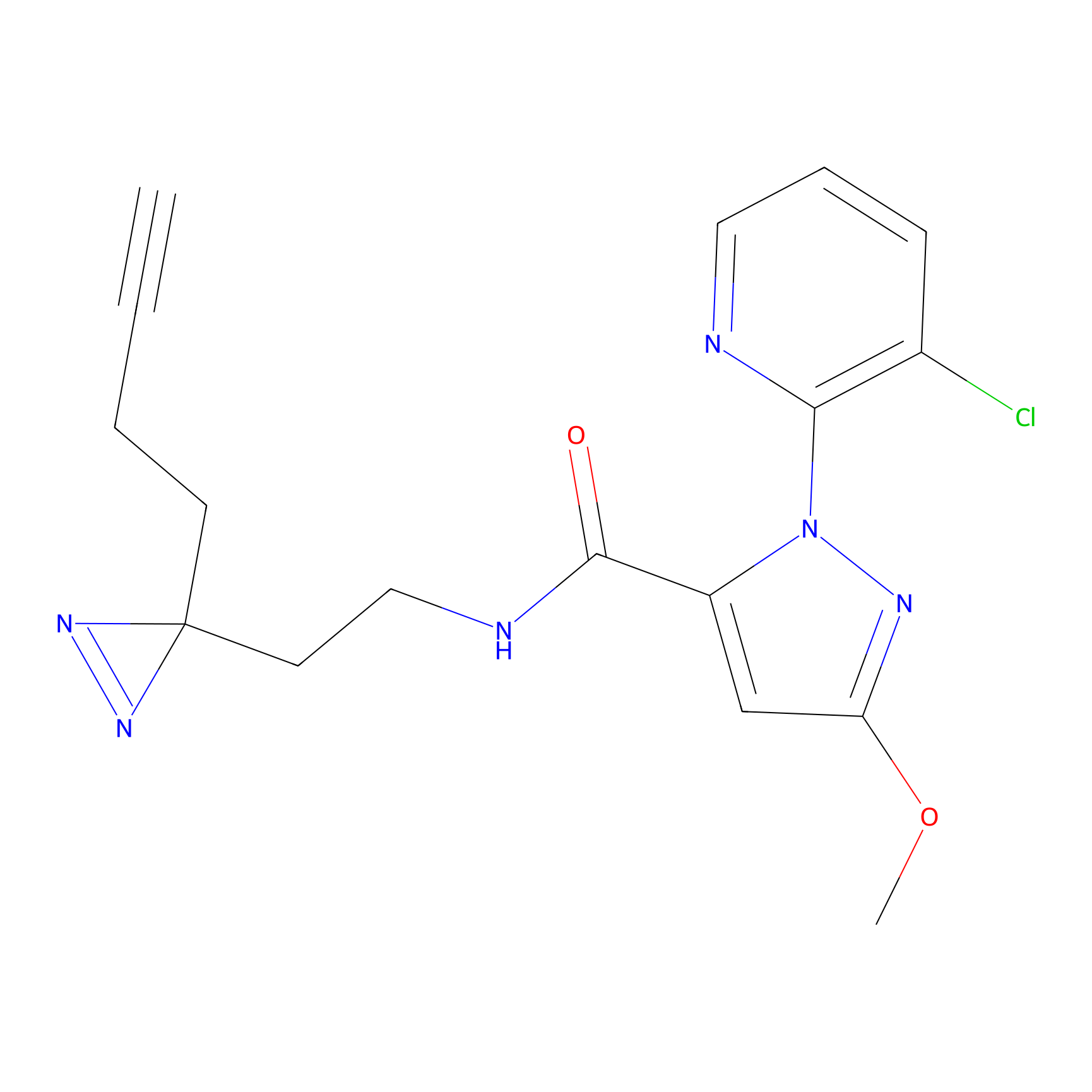
|
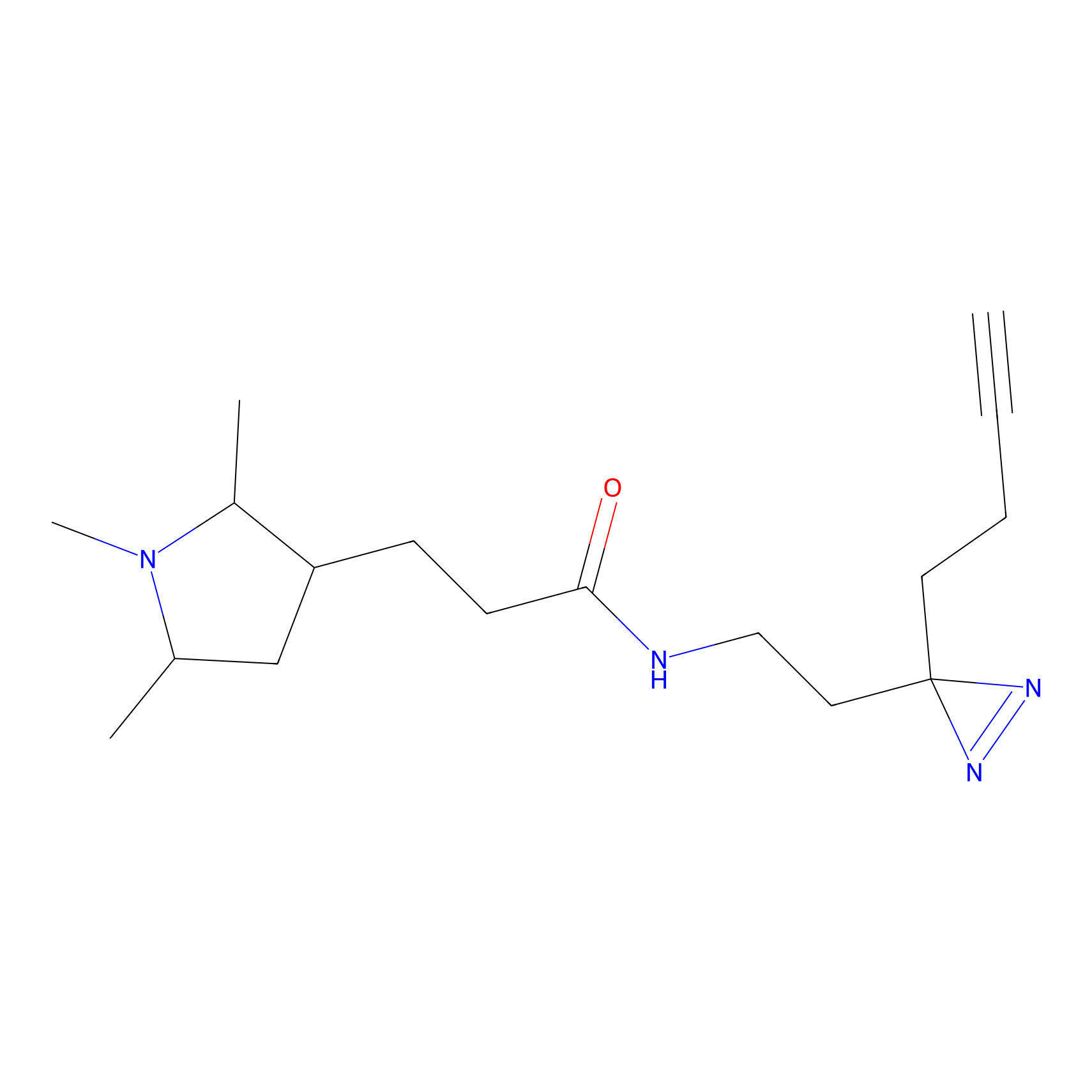
DBIA Probe Info |
 |
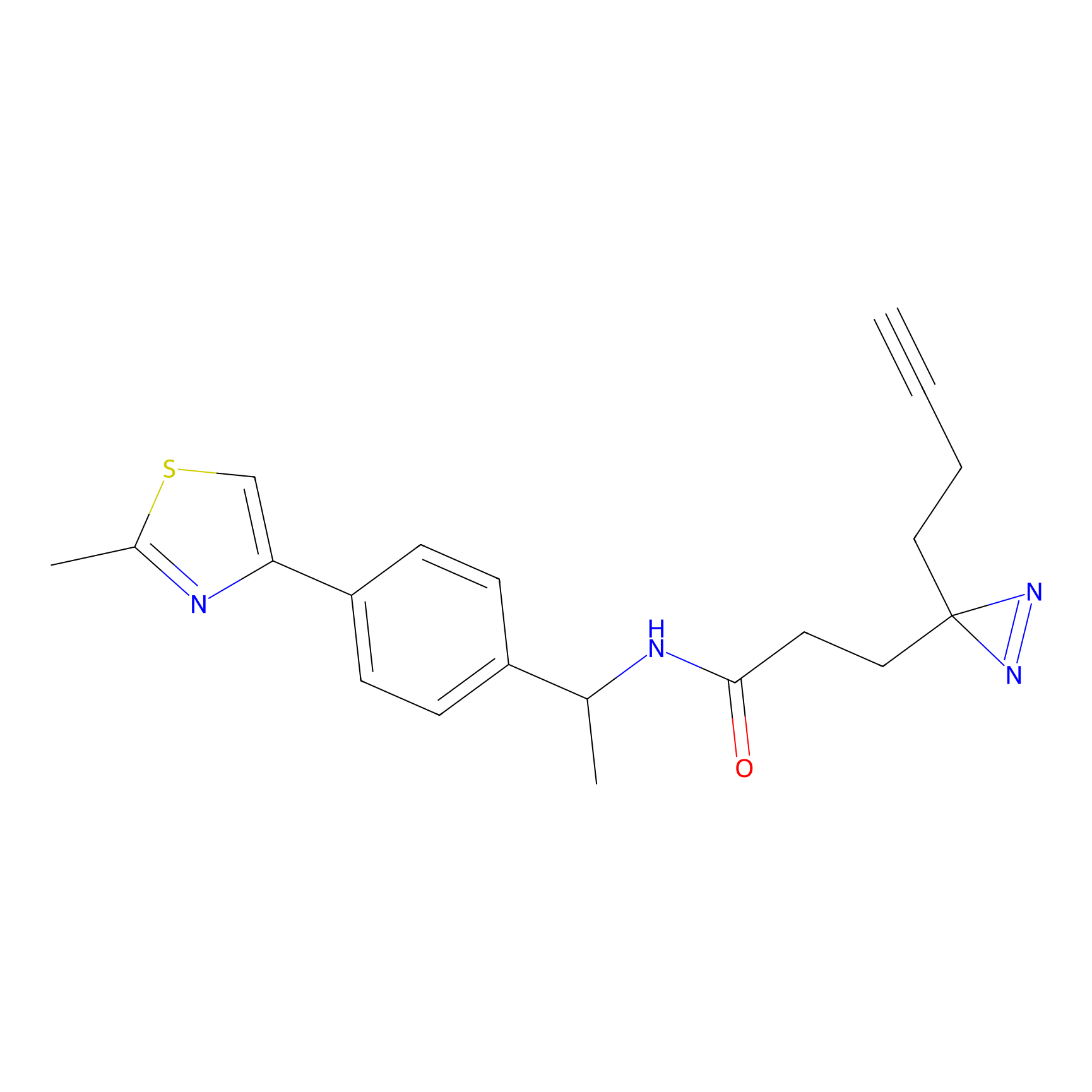
C182(2.17) | LDD3368 | [1] | |
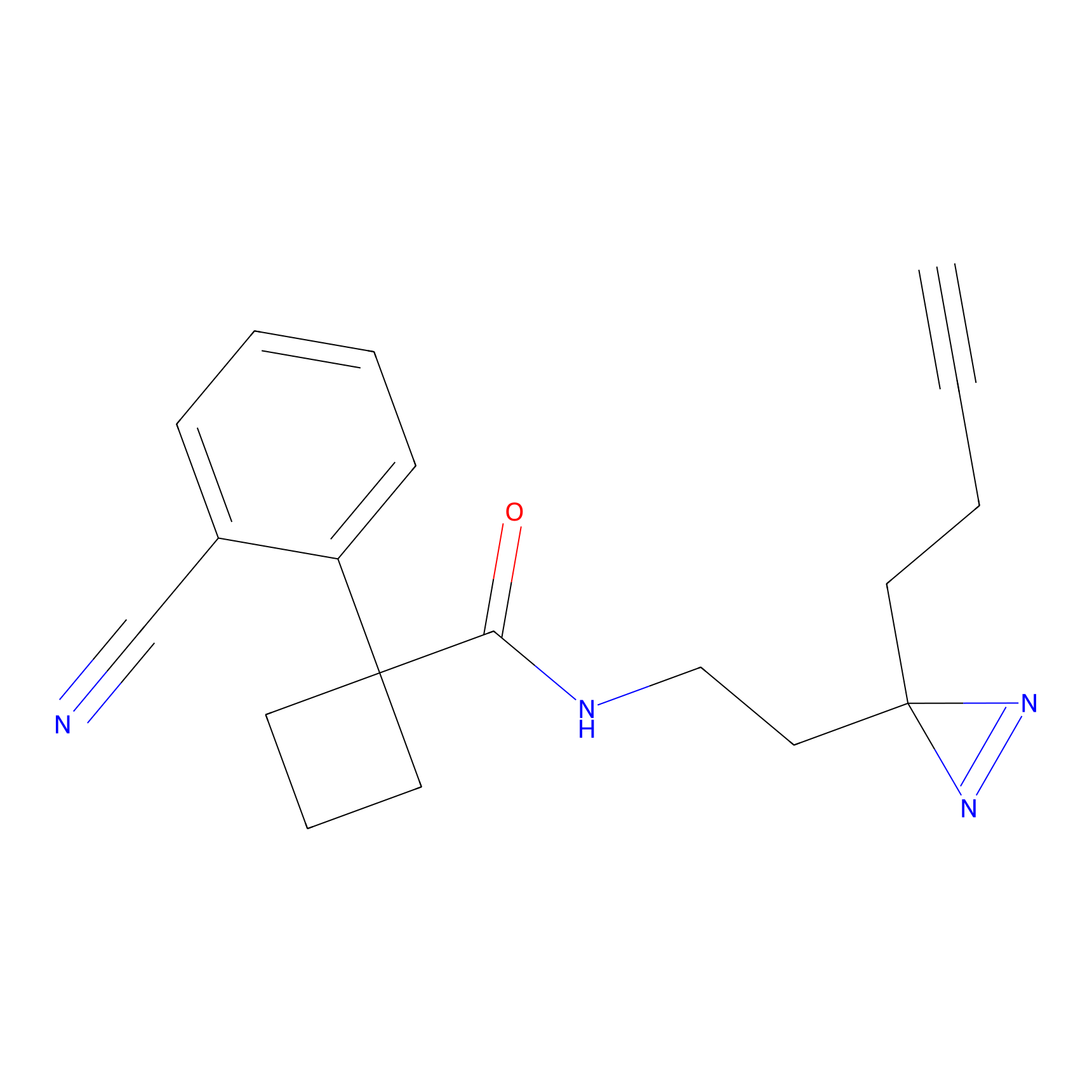
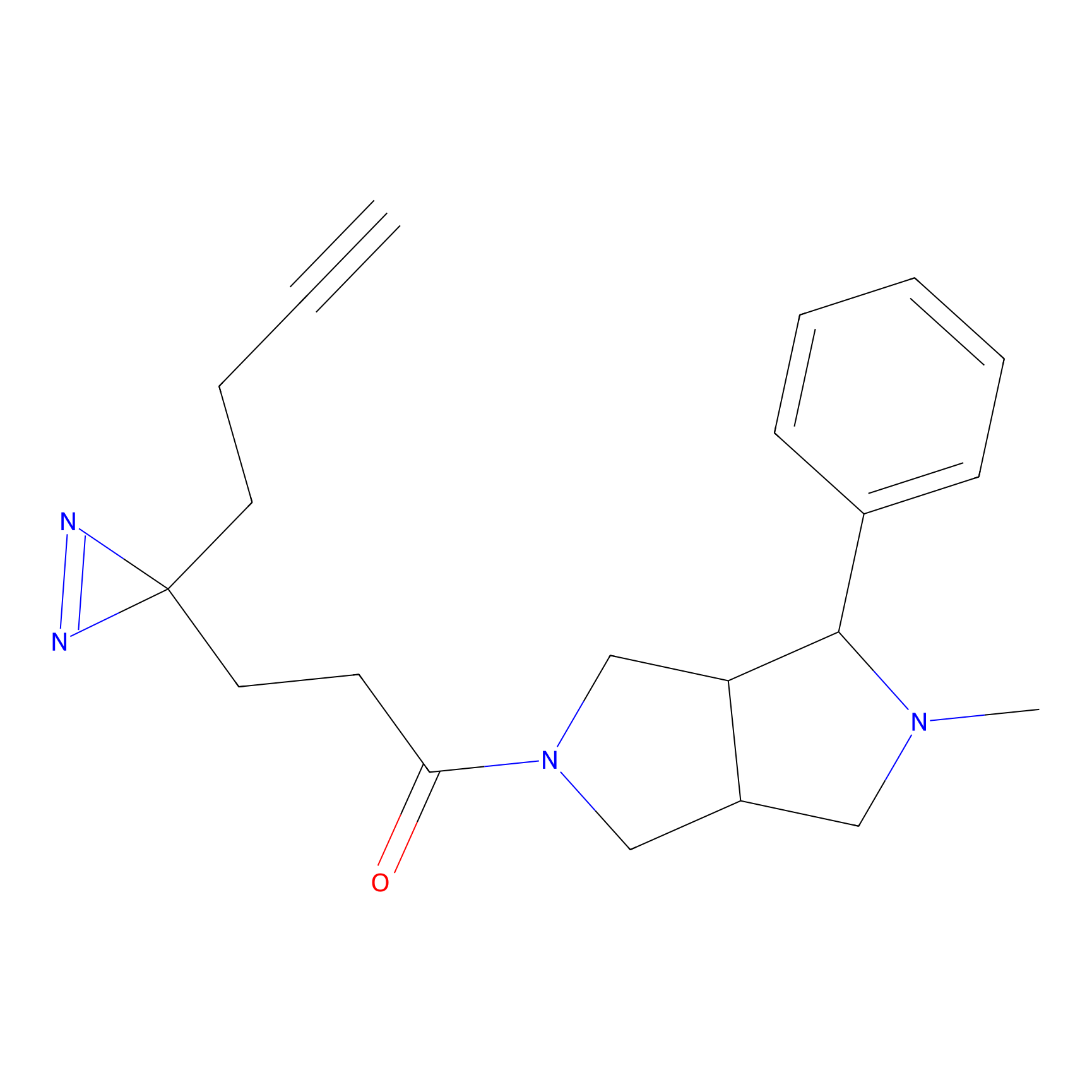
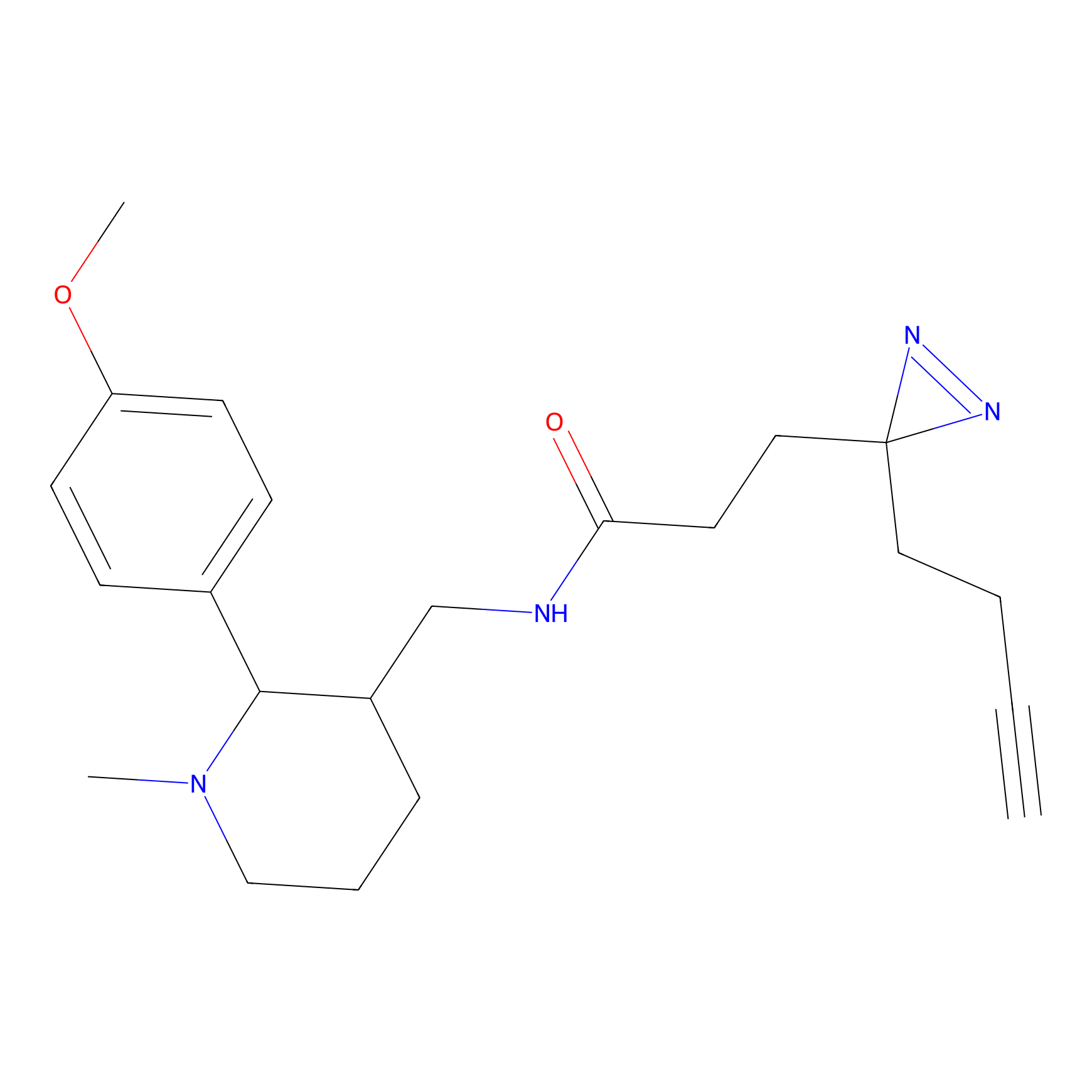
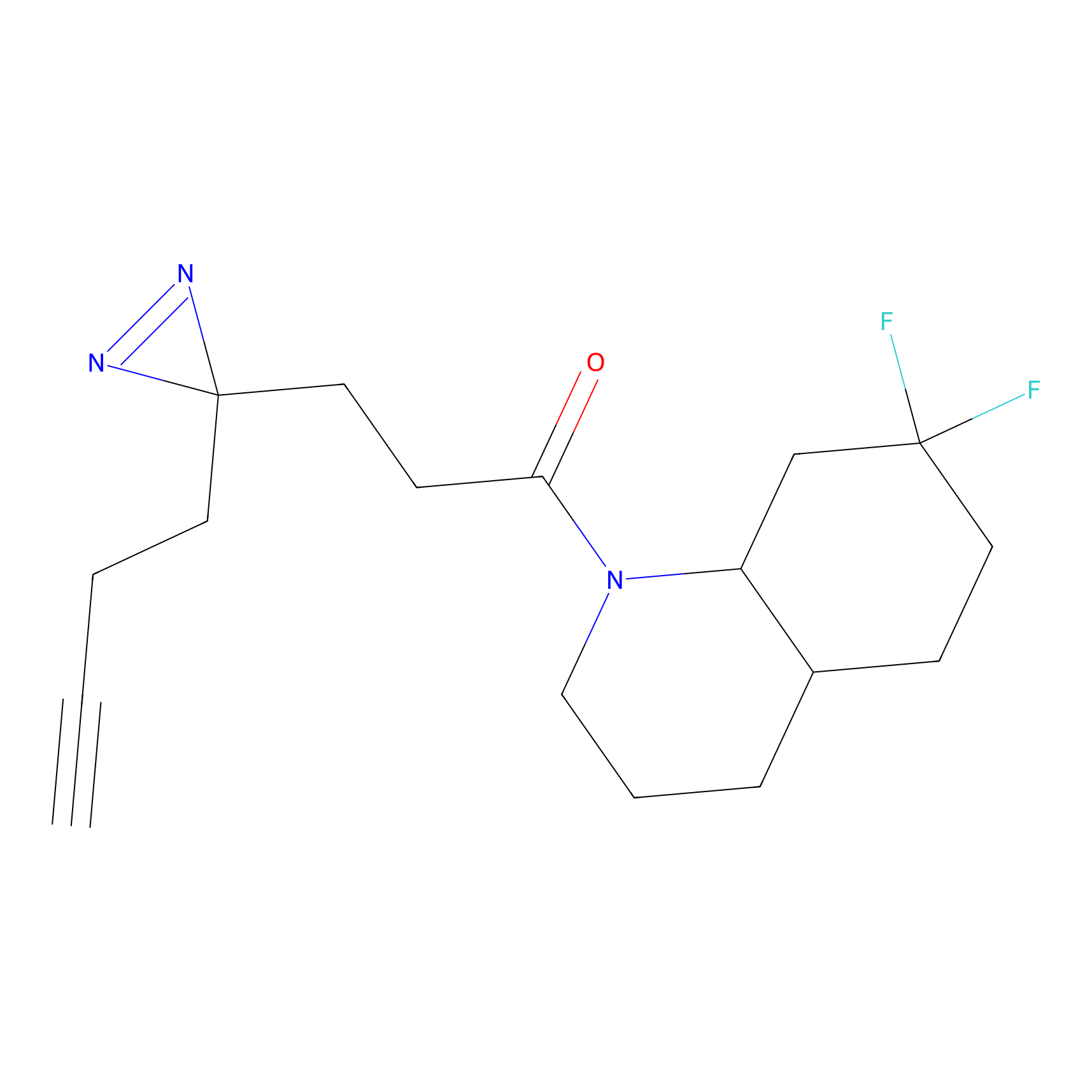
|
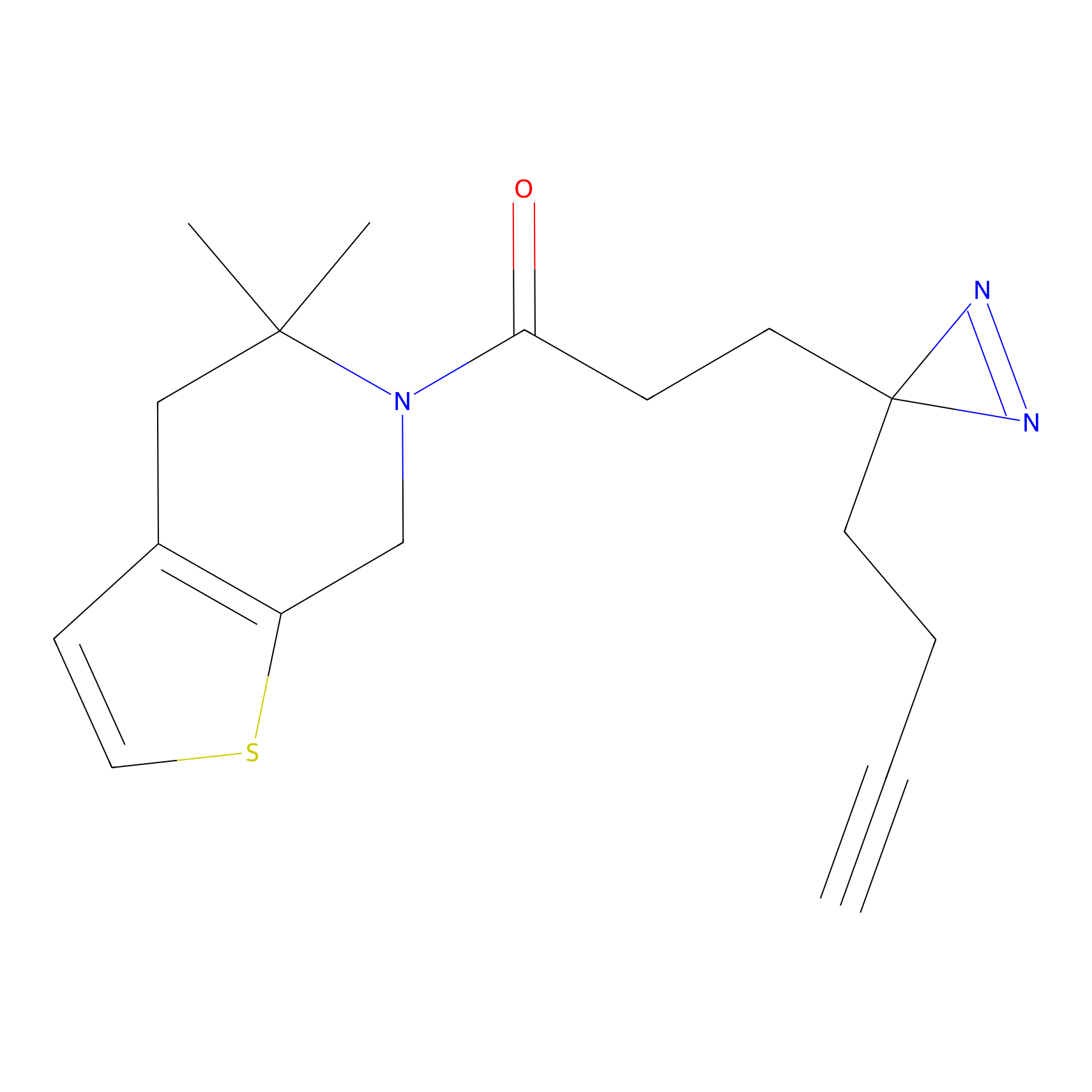
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [2] | |
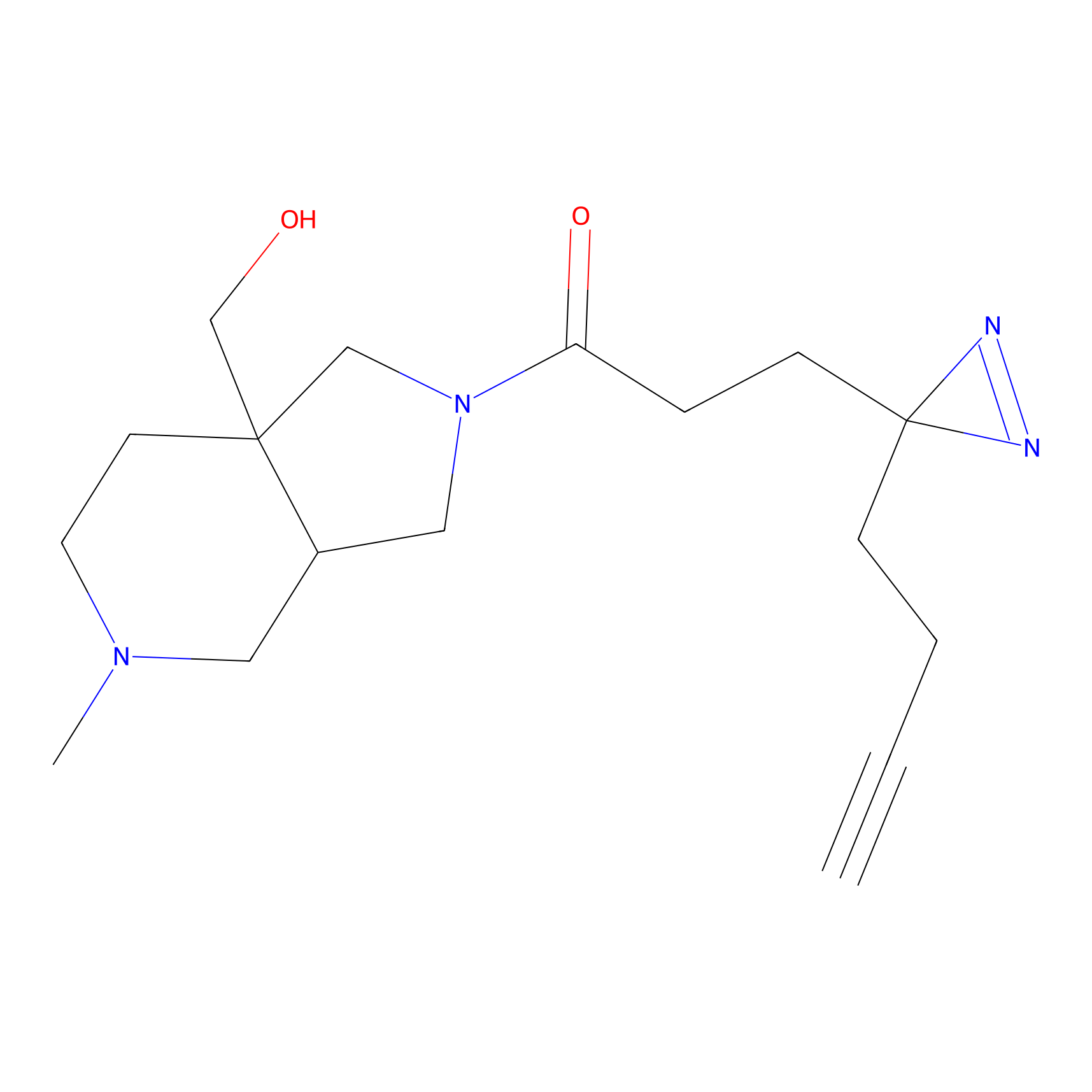
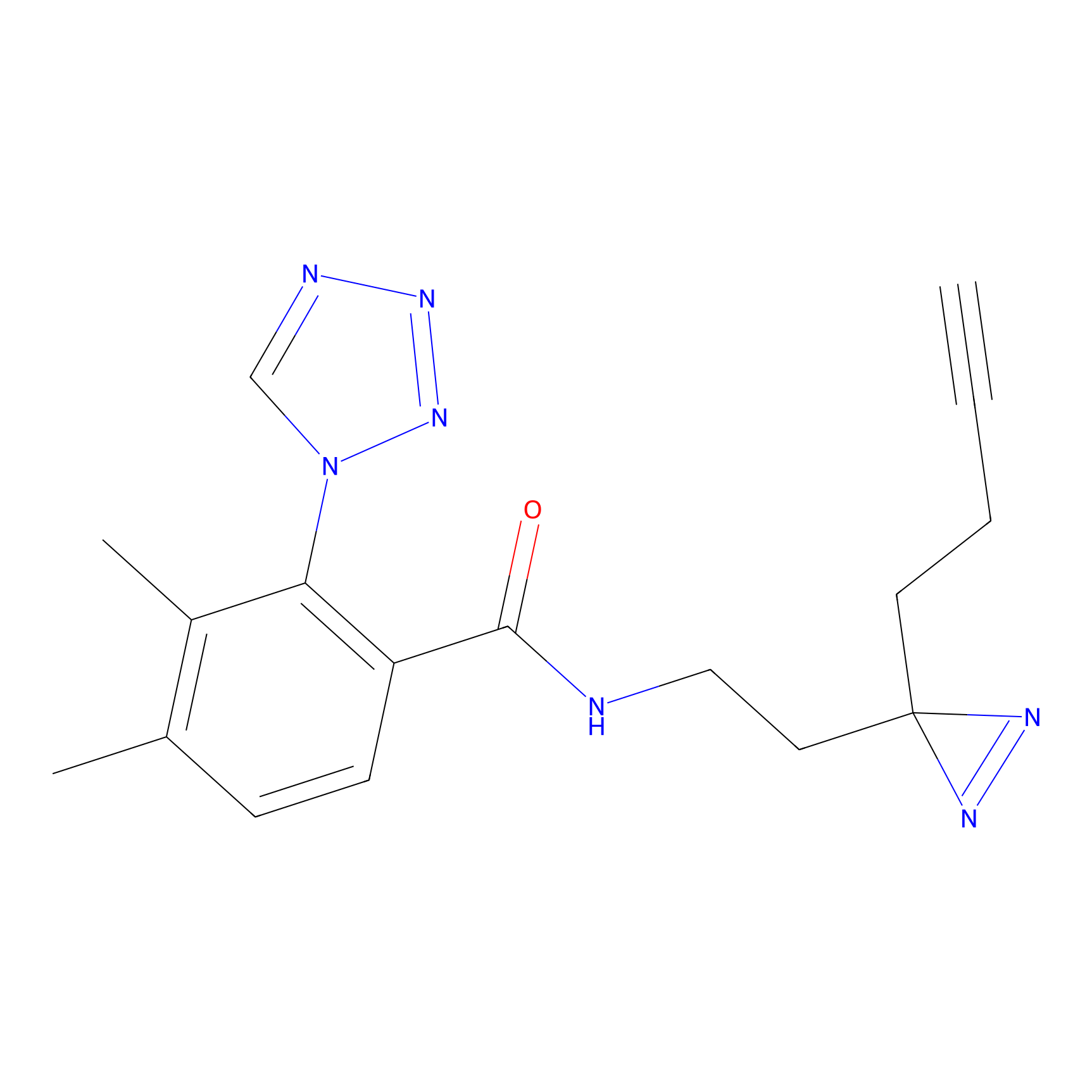
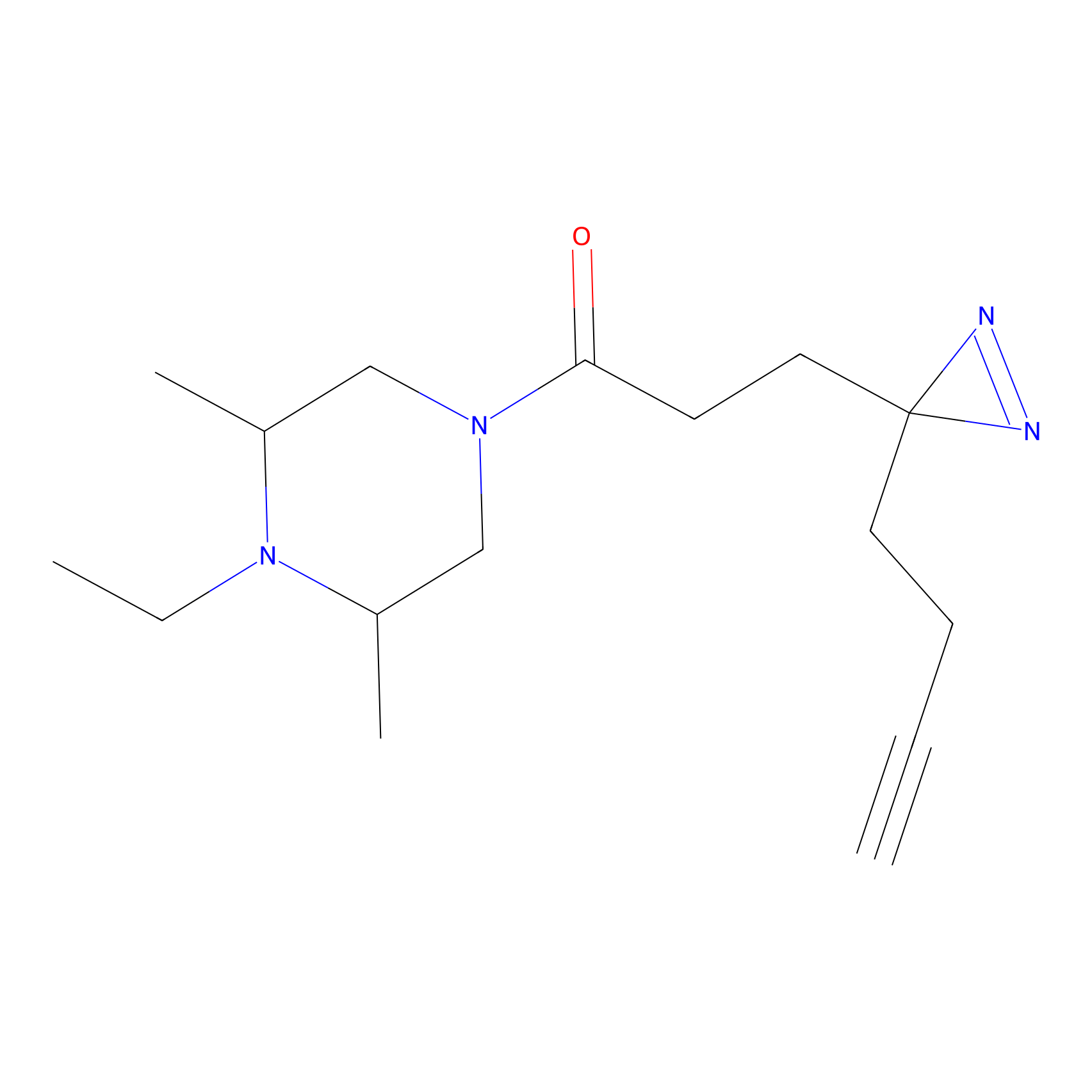
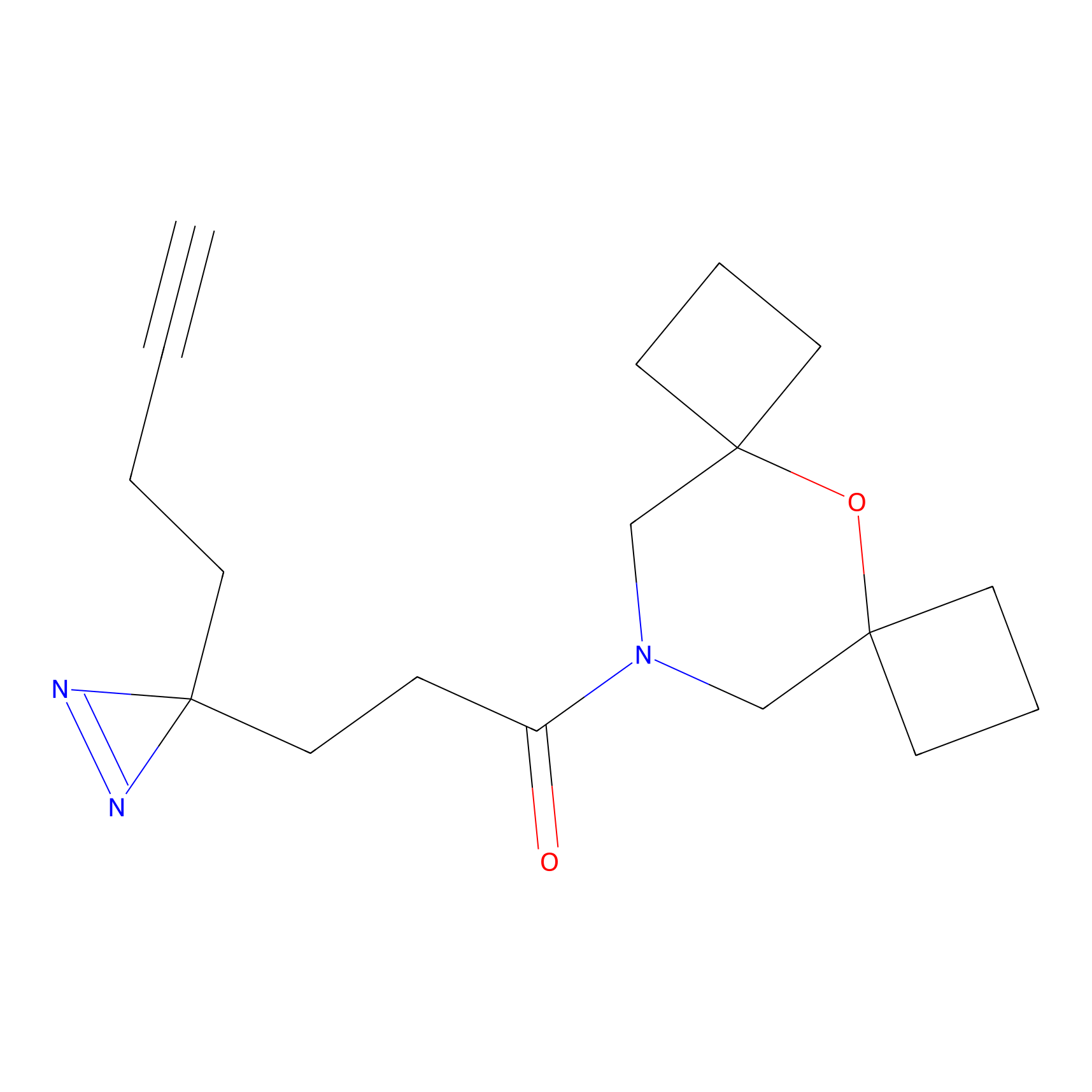
|
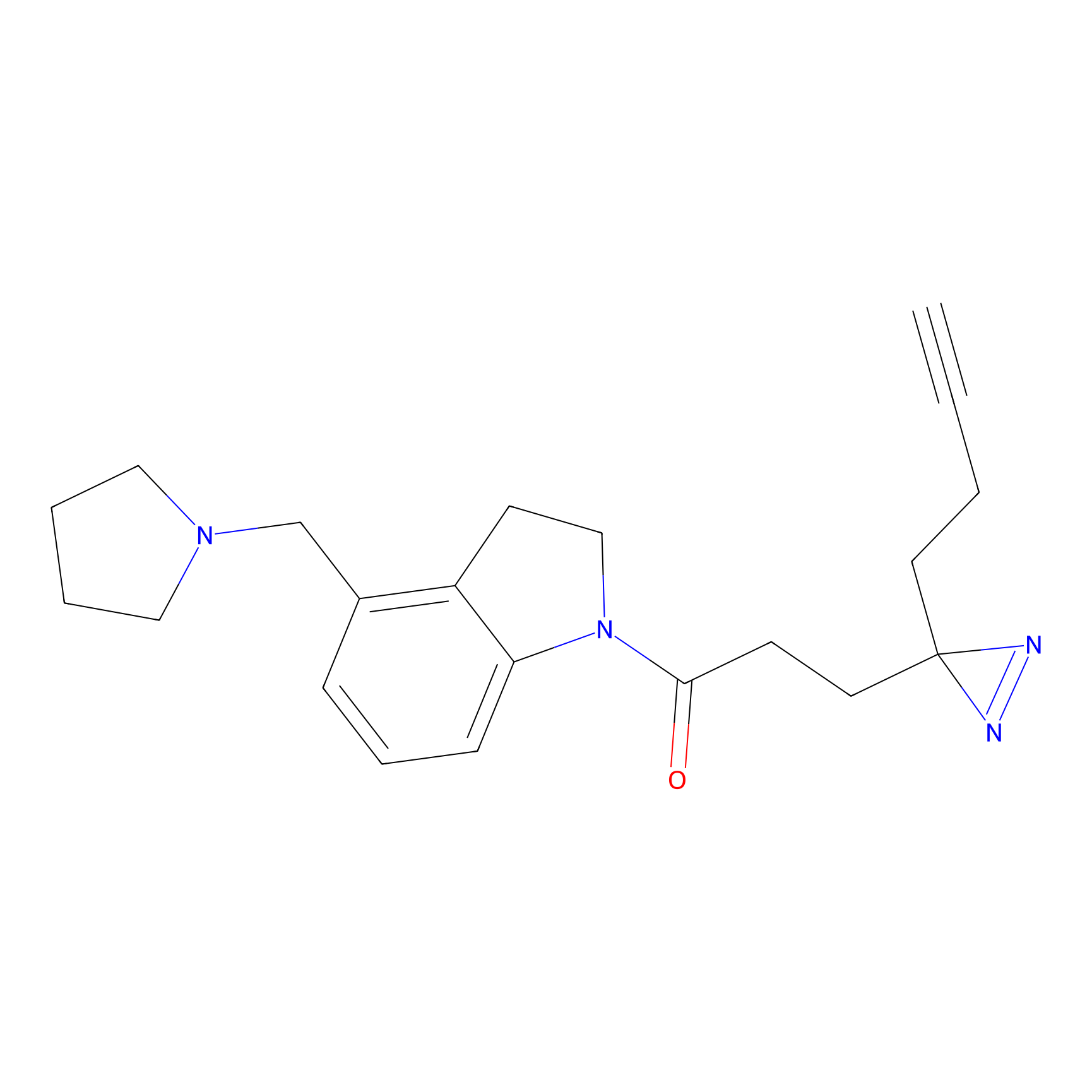
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [2] | |
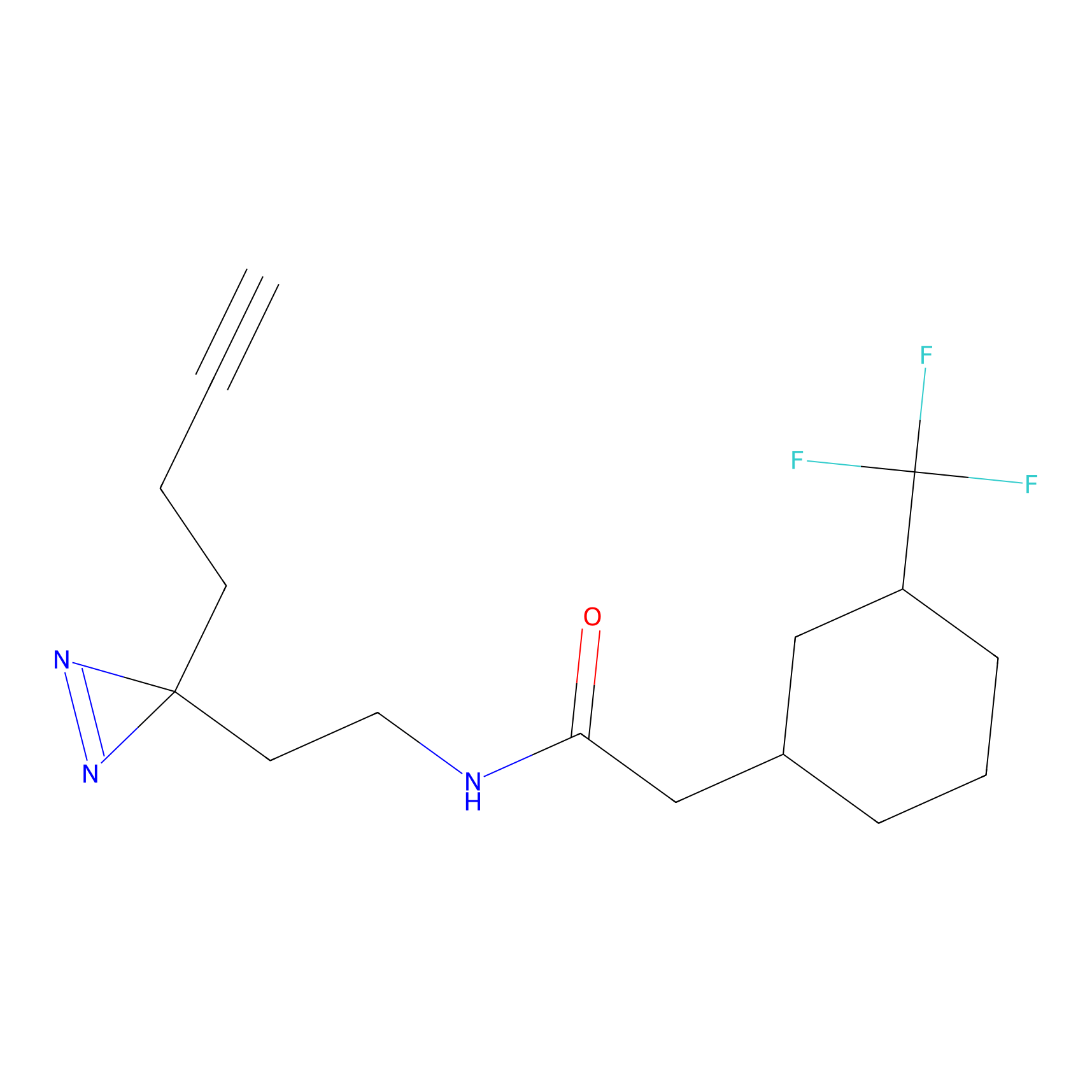
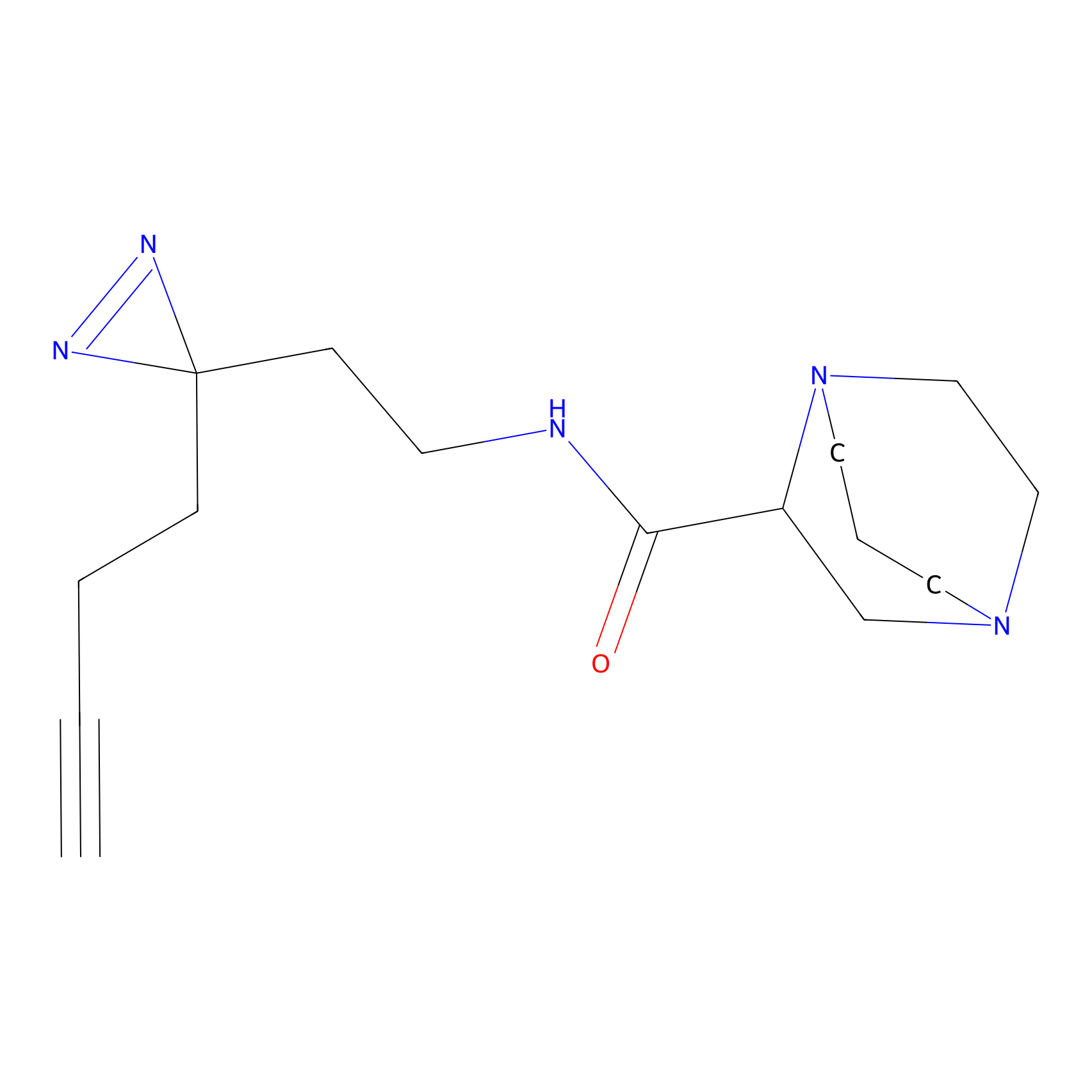
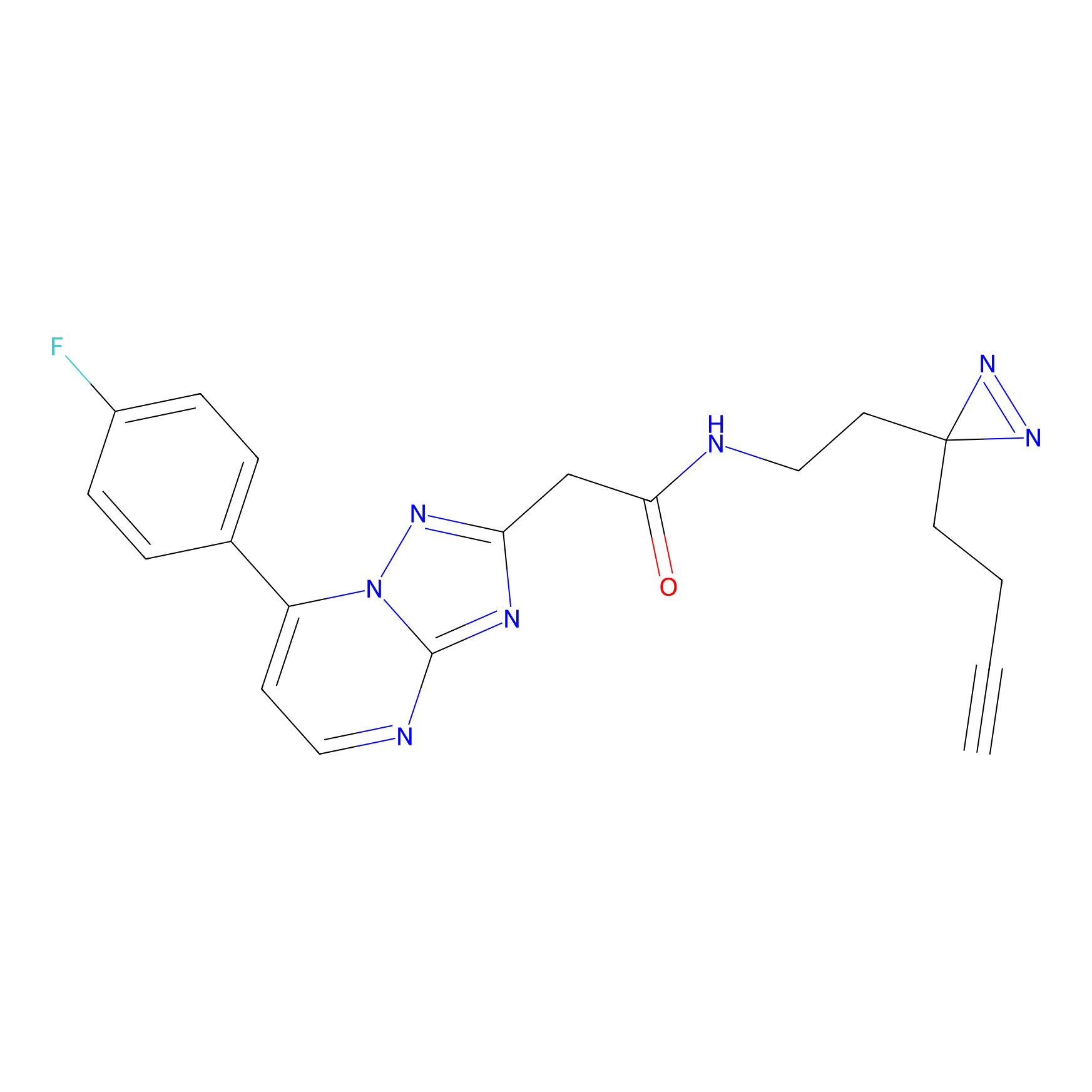
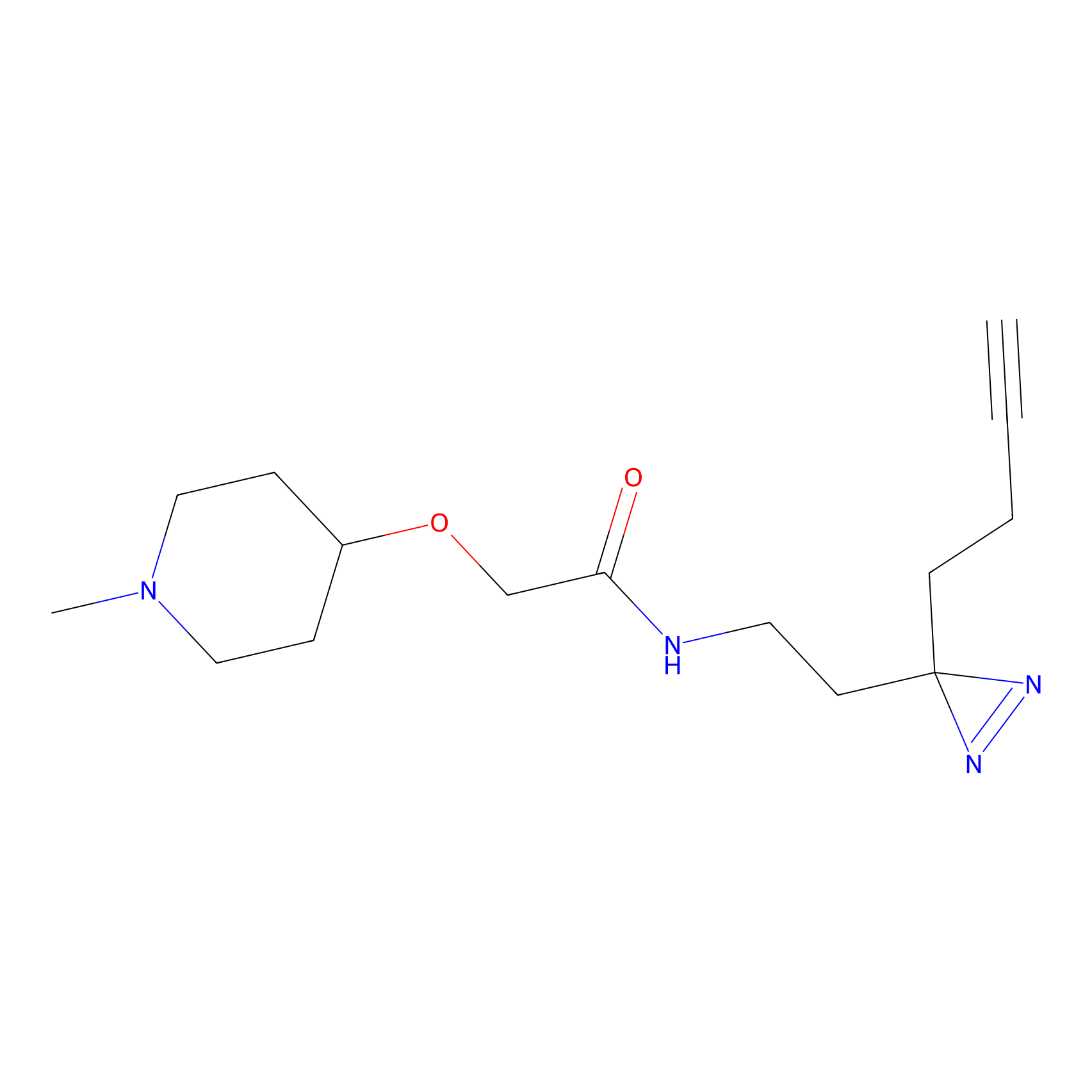
|
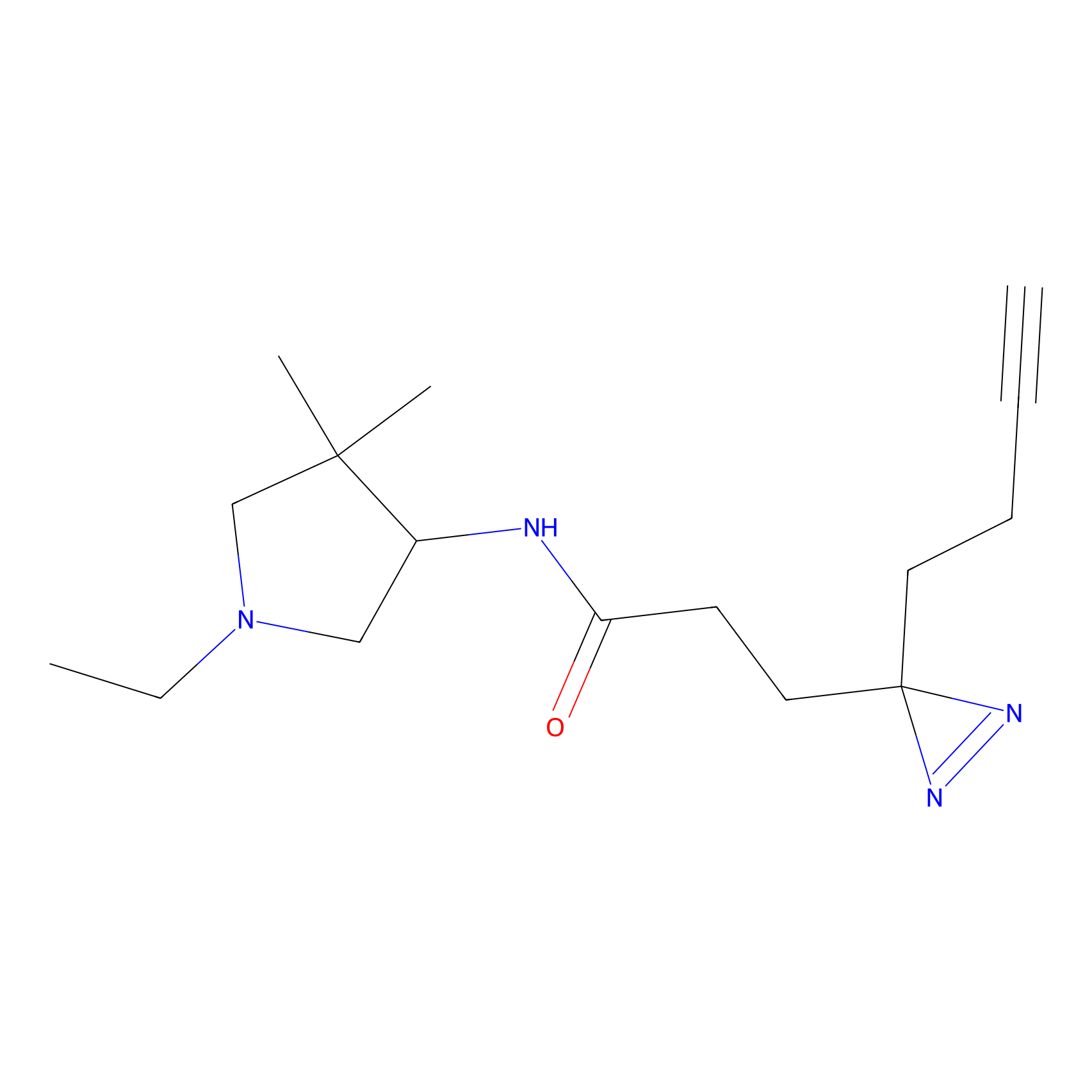
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [2] | |
PAL-AfBPP Probe
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0215 | AC10 | PaTu 8988t | C71(0.67) | LDD1094 | [4] |
| LDCM0226 | AC11 | PaTu 8988t | C71(0.55) | LDD1105 | [4] |
| LDCM0237 | AC12 | PaTu 8988t | C71(0.41) | LDD1116 | [4] |
| LDCM0259 | AC14 | PaTu 8988t | C71(0.70) | LDD1138 | [4] |
| LDCM0270 | AC15 | PaTu 8988t | C71(0.77) | LDD1149 | [4] |
| LDCM0276 | AC17 | PaTu 8988t | C71(0.96) | LDD1155 | [4] |
| LDCM0277 | AC18 | PaTu 8988t | C71(0.88) | LDD1156 | [4] |
| LDCM0278 | AC19 | PaTu 8988t | C71(1.22) | LDD1157 | [4] |
| LDCM0280 | AC20 | PaTu 8988t | C71(1.70) | LDD1159 | [4] |
| LDCM0281 | AC21 | PaTu 8988t | C71(0.85) | LDD1160 | [4] |
| LDCM0282 | AC22 | PaTu 8988t | C71(1.12) | LDD1161 | [4] |
| LDCM0283 | AC23 | PaTu 8988t | C71(0.91) | LDD1162 | [4] |
| LDCM0284 | AC24 | PaTu 8988t | C71(0.65) | LDD1163 | [4] |
| LDCM0289 | AC29 | HEK-293T | C71(0.97) | LDD1528 | [5] |
| LDCM0291 | AC30 | HEK-293T | C119(0.93) | LDD1530 | [5] |
| LDCM0292 | AC31 | HEK-293T | C71(0.89) | LDD1531 | [5] |
| LDCM0296 | AC35 | PaTu 8988t | C71(1.53) | LDD1175 | [4] |
| LDCM0297 | AC36 | PaTu 8988t | C71(0.88) | LDD1176 | [4] |
| LDCM0298 | AC37 | PaTu 8988t | C71(0.65) | LDD1177 | [4] |
| LDCM0299 | AC38 | PaTu 8988t | C71(0.70) | LDD1178 | [4] |
| LDCM0300 | AC39 | PaTu 8988t | C71(0.68) | LDD1179 | [4] |
| LDCM0302 | AC40 | PaTu 8988t | C71(0.91) | LDD1181 | [4] |
| LDCM0303 | AC41 | PaTu 8988t | C71(1.00) | LDD1182 | [4] |
| LDCM0304 | AC42 | PaTu 8988t | C71(0.65) | LDD1183 | [4] |
| LDCM0305 | AC43 | PaTu 8988t | C71(0.64) | LDD1184 | [4] |
| LDCM0306 | AC44 | PaTu 8988t | C71(0.60) | LDD1185 | [4] |
| LDCM0307 | AC45 | PaTu 8988t | C71(0.69) | LDD1186 | [4] |
| LDCM0308 | AC46 | PaTu 8988t | C71(1.11) | LDD1187 | [4] |
| LDCM0309 | AC47 | PaTu 8988t | C71(1.20) | LDD1188 | [4] |
| LDCM0310 | AC48 | PaTu 8988t | C71(0.81) | LDD1189 | [4] |
| LDCM0311 | AC49 | PaTu 8988t | C71(0.89) | LDD1190 | [4] |
| LDCM0312 | AC5 | HEK-293T | C71(1.04) | LDD1551 | [5] |
| LDCM0313 | AC50 | PaTu 8988t | C71(0.98) | LDD1192 | [4] |
| LDCM0314 | AC51 | PaTu 8988t | C71(0.36) | LDD1193 | [4] |
| LDCM0315 | AC52 | PaTu 8988t | C71(0.81) | LDD1194 | [4] |
| LDCM0316 | AC53 | PaTu 8988t | C71(1.07) | LDD1195 | [4] |
| LDCM0317 | AC54 | PaTu 8988t | C71(1.05) | LDD1196 | [4] |
| LDCM0318 | AC55 | PaTu 8988t | C71(0.75) | LDD1197 | [4] |
| LDCM0319 | AC56 | PaTu 8988t | C71(1.14) | LDD1198 | [4] |
| LDCM0323 | AC6 | PaTu 8988t | C71(0.52) | LDD1202 | [4] |
| LDCM0325 | AC61 | HEK-293T | C71(1.01) | LDD1564 | [5] |
| LDCM0326 | AC62 | HEK-293T | C119(0.94) | LDD1565 | [5] |
| LDCM0327 | AC63 | HEK-293T | C71(0.98) | LDD1566 | [5] |
| LDCM0334 | AC7 | PaTu 8988t | C71(0.59) | LDD1213 | [4] |
| LDCM0345 | AC8 | PaTu 8988t | C71(0.88) | LDD1224 | [4] |
| LDCM0248 | AKOS034007472 | PaTu 8988t | C71(0.58) | LDD1127 | [4] |
| LDCM0356 | AKOS034007680 | PaTu 8988t | C71(0.55) | LDD1235 | [4] |
| LDCM0275 | AKOS034007705 | PaTu 8988t | C71(0.64) | LDD1154 | [4] |
| LDCM0367 | CL1 | PaTu 8988t | C71(0.94) | LDD1246 | [4] |
| LDCM0368 | CL10 | PaTu 8988t | C71(0.61) | LDD1247 | [4] |
| LDCM0370 | CL101 | PaTu 8988t | C71(0.60) | LDD1249 | [4] |
| LDCM0371 | CL102 | PaTu 8988t | C71(0.61) | LDD1250 | [4] |
| LDCM0372 | CL103 | PaTu 8988t | C71(0.50) | LDD1251 | [4] |
| LDCM0373 | CL104 | PaTu 8988t | C71(0.84) | LDD1252 | [4] |
| LDCM0374 | CL105 | PaTu 8988t | C71(1.35) | LDD1253 | [4] |
| LDCM0375 | CL106 | PaTu 8988t | C71(1.77) | LDD1254 | [4] |
| LDCM0376 | CL107 | PaTu 8988t | C71(1.08) | LDD1255 | [4] |
| LDCM0377 | CL108 | PaTu 8988t | C71(1.41) | LDD1256 | [4] |
| LDCM0378 | CL109 | PaTu 8988t | C71(1.99) | LDD1257 | [4] |
| LDCM0379 | CL11 | PaTu 8988t | C71(0.85) | LDD1258 | [4] |
| LDCM0380 | CL110 | PaTu 8988t | C71(2.14) | LDD1259 | [4] |
| LDCM0381 | CL111 | PaTu 8988t | C71(0.58) | LDD1260 | [4] |
| LDCM0385 | CL115 | HEK-293T | C71(1.07) | LDD1589 | [5] |
| LDCM0387 | CL117 | PaTu 8988t | C71(1.51) | LDD1266 | [4] |
| LDCM0388 | CL118 | PaTu 8988t | C71(0.75) | LDD1267 | [4] |
| LDCM0389 | CL119 | PaTu 8988t | C71(0.59) | LDD1268 | [4] |
| LDCM0390 | CL12 | PaTu 8988t | C71(0.93) | LDD1269 | [4] |
| LDCM0391 | CL120 | PaTu 8988t | C71(1.35) | LDD1270 | [4] |
| LDCM0392 | CL121 | PaTu 8988t | C71(2.01) | LDD1271 | [4] |
| LDCM0393 | CL122 | PaTu 8988t | C71(1.00) | LDD1272 | [4] |
| LDCM0394 | CL123 | PaTu 8988t | C71(1.24) | LDD1273 | [4] |
| LDCM0395 | CL124 | PaTu 8988t | C71(0.89) | LDD1274 | [4] |
| LDCM0398 | CL127 | HEK-293T | C71(1.13) | LDD1602 | [5] |
| LDCM0400 | CL13 | PaTu 8988t | C71(0.63) | LDD1279 | [4] |
| LDCM0401 | CL14 | PaTu 8988t | C71(0.60) | LDD1280 | [4] |
| LDCM0402 | CL15 | PaTu 8988t | C71(0.83) | LDD1281 | [4] |
| LDCM0407 | CL2 | PaTu 8988t | C71(1.02) | LDD1286 | [4] |
| LDCM0409 | CL21 | HEK-293T | C71(1.02) | LDD1613 | [5] |
| LDCM0410 | CL22 | HEK-293T | C119(0.86) | LDD1614 | [5] |
| LDCM0411 | CL23 | HEK-293T | C71(1.16) | LDD1615 | [5] |
| LDCM0415 | CL27 | HEK-293T | C71(0.98) | LDD1619 | [5] |
| LDCM0418 | CL3 | PaTu 8988t | C71(0.54) | LDD1297 | [4] |
| LDCM0420 | CL31 | PaTu 8988t | C71(0.58) | LDD1299 | [4] |
| LDCM0421 | CL32 | PaTu 8988t | C71(0.48) | LDD1300 | [4] |
| LDCM0422 | CL33 | PaTu 8988t | C71(0.42) | LDD1301 | [4] |
| LDCM0423 | CL34 | PaTu 8988t | C71(0.44) | LDD1302 | [4] |
| LDCM0424 | CL35 | PaTu 8988t | C71(0.56) | LDD1303 | [4] |
| LDCM0425 | CL36 | PaTu 8988t | C71(0.40) | LDD1304 | [4] |
| LDCM0426 | CL37 | PaTu 8988t | C71(0.41) | LDD1305 | [4] |
| LDCM0428 | CL39 | PaTu 8988t | C71(0.42) | LDD1307 | [4] |
| LDCM0429 | CL4 | PaTu 8988t | C71(0.70) | LDD1308 | [4] |
| LDCM0430 | CL40 | PaTu 8988t | C71(0.46) | LDD1309 | [4] |
| LDCM0431 | CL41 | PaTu 8988t | C71(0.43) | LDD1310 | [4] |
| LDCM0432 | CL42 | PaTu 8988t | C71(0.53) | LDD1311 | [4] |
| LDCM0433 | CL43 | PaTu 8988t | C71(0.36) | LDD1312 | [4] |
| LDCM0434 | CL44 | PaTu 8988t | C71(0.54) | LDD1313 | [4] |
| LDCM0435 | CL45 | PaTu 8988t | C71(0.47) | LDD1314 | [4] |
| LDCM0436 | CL46 | PaTu 8988t | C71(0.81) | LDD1315 | [4] |
| LDCM0437 | CL47 | PaTu 8988t | C71(0.72) | LDD1316 | [4] |
| LDCM0438 | CL48 | PaTu 8988t | C71(0.93) | LDD1317 | [4] |
| LDCM0439 | CL49 | PaTu 8988t | C71(0.83) | LDD1318 | [4] |
| LDCM0440 | CL5 | PaTu 8988t | C71(0.71) | LDD1319 | [4] |
| LDCM0441 | CL50 | PaTu 8988t | C71(0.65) | LDD1320 | [4] |
| LDCM0442 | CL51 | PaTu 8988t | C71(0.74) | LDD1321 | [4] |
| LDCM0443 | CL52 | PaTu 8988t | C71(0.95) | LDD1322 | [4] |
| LDCM0444 | CL53 | PaTu 8988t | C71(0.75) | LDD1323 | [4] |
| LDCM0445 | CL54 | PaTu 8988t | C71(0.49) | LDD1324 | [4] |
| LDCM0446 | CL55 | PaTu 8988t | C71(0.51) | LDD1325 | [4] |
| LDCM0447 | CL56 | PaTu 8988t | C71(0.29) | LDD1326 | [4] |
| LDCM0448 | CL57 | PaTu 8988t | C71(0.40) | LDD1327 | [4] |
| LDCM0449 | CL58 | PaTu 8988t | C71(0.73) | LDD1328 | [4] |
| LDCM0450 | CL59 | PaTu 8988t | C71(0.41) | LDD1329 | [4] |
| LDCM0451 | CL6 | PaTu 8988t | C71(0.81) | LDD1330 | [4] |
| LDCM0452 | CL60 | PaTu 8988t | C71(0.52) | LDD1331 | [4] |
| LDCM0453 | CL61 | PaTu 8988t | C71(0.99) | LDD1332 | [4] |
| LDCM0454 | CL62 | PaTu 8988t | C71(0.63) | LDD1333 | [4] |
| LDCM0455 | CL63 | PaTu 8988t | C71(0.79) | LDD1334 | [4] |
| LDCM0456 | CL64 | PaTu 8988t | C71(0.81) | LDD1335 | [4] |
| LDCM0457 | CL65 | PaTu 8988t | C71(0.82) | LDD1336 | [4] |
| LDCM0458 | CL66 | PaTu 8988t | C71(0.92) | LDD1337 | [4] |
| LDCM0459 | CL67 | PaTu 8988t | C71(0.95) | LDD1338 | [4] |
| LDCM0460 | CL68 | PaTu 8988t | C71(0.82) | LDD1339 | [4] |
| LDCM0461 | CL69 | PaTu 8988t | C71(0.81) | LDD1340 | [4] |
| LDCM0462 | CL7 | PaTu 8988t | C71(0.70) | LDD1341 | [4] |
| LDCM0463 | CL70 | PaTu 8988t | C71(0.89) | LDD1342 | [4] |
| LDCM0464 | CL71 | PaTu 8988t | C71(0.68) | LDD1343 | [4] |
| LDCM0465 | CL72 | PaTu 8988t | C71(1.09) | LDD1344 | [4] |
| LDCM0466 | CL73 | PaTu 8988t | C71(0.65) | LDD1345 | [4] |
| LDCM0467 | CL74 | PaTu 8988t | C71(0.73) | LDD1346 | [4] |
| LDCM0473 | CL8 | PaTu 8988t | C71(0.64) | LDD1352 | [4] |
| LDCM0475 | CL81 | HEK-293T | C71(1.05) | LDD1678 | [5] |
| LDCM0476 | CL82 | HEK-293T | C119(0.91) | LDD1679 | [5] |
| LDCM0477 | CL83 | HEK-293T | C71(0.84) | LDD1680 | [5] |
| LDCM0481 | CL87 | HEK-293T | C71(1.45) | LDD1684 | [5] |
| LDCM0484 | CL9 | PaTu 8988t | C71(0.75) | LDD1363 | [4] |
| LDCM0488 | CL93 | HEK-293T | C71(0.23) | LDD1691 | [5] |
| LDCM0489 | CL94 | HEK-293T | C119(0.82) | LDD1692 | [5] |
| LDCM0490 | CL95 | HEK-293T | C71(0.86) | LDD1693 | [5] |
| LDCM0494 | CL99 | HEK-293T | C71(1.11) | LDD1697 | [5] |
| LDCM0495 | E2913 | HEK-293T | C71(1.30) | LDD1698 | [5] |
| LDCM0468 | Fragment33 | PaTu 8988t | C71(0.82) | LDD1347 | [4] |
| LDCM0427 | Fragment51 | PaTu 8988t | C71(0.38) | LDD1306 | [4] |
| LDCM0022 | KB02 | HEK-293T | C182(0.95) | LDD1492 | [5] |
| LDCM0023 | KB03 | HEK-293T | C182(1.02) | LDD1497 | [5] |
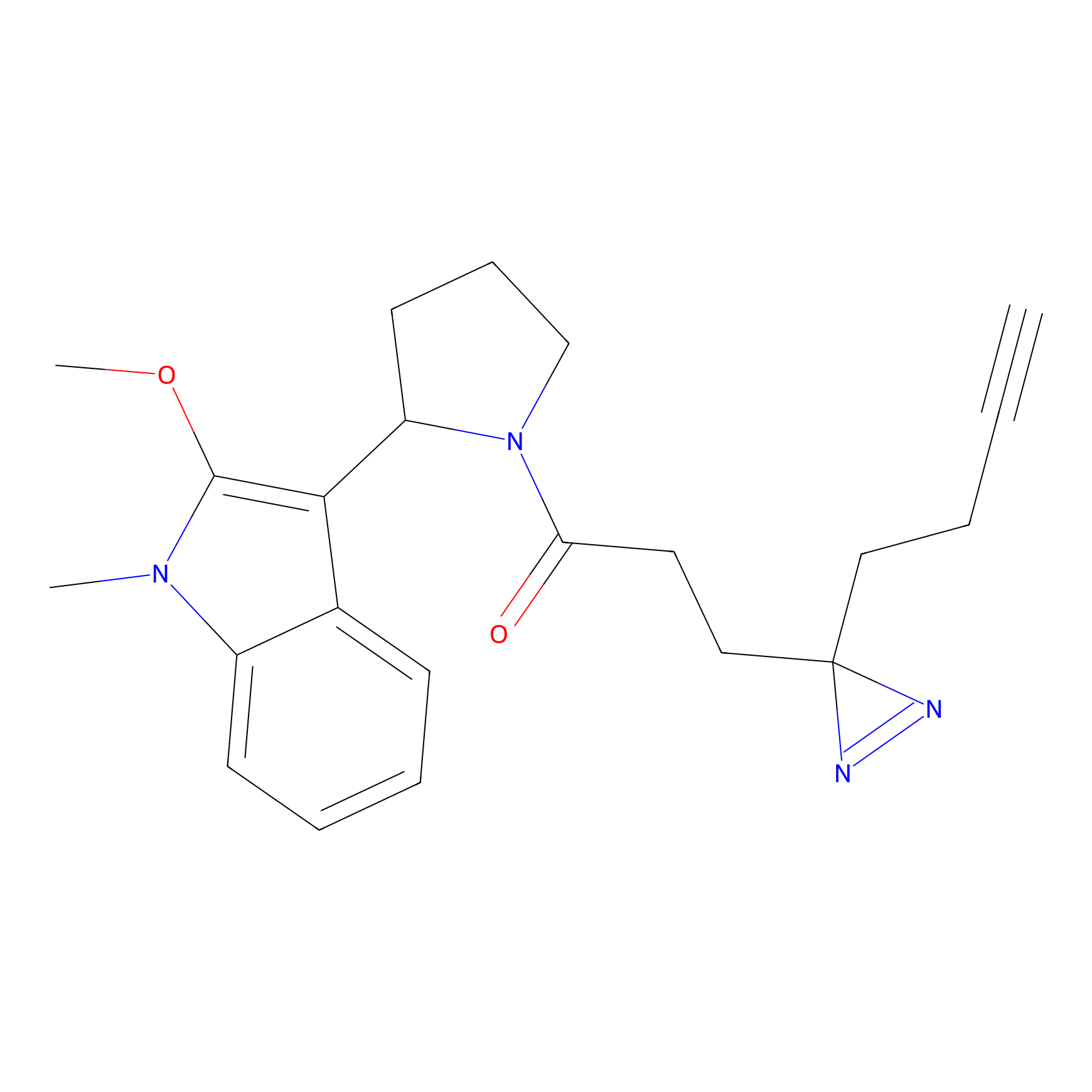
| LDCM0024 | KB05 | OAW-28 | C182(2.17) | LDD3368 | [1] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Peripheral myelin protein 22 (PMP22) | PMP-22/EMP/MP20 family | Q01453 | |||
| Uroplakin-2 (UPK2) | Uroplakin-2 family | O00526 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Protein FAM3C (FAM3C) | FAM3 family | Q92520 | |||
| Ninjurin-2 (NINJ2) | Ninjurin family | Q9NZG7 | |||
References