Details of the Target
General Information of Target
| Target ID | LDTP13953 | |||||
|---|---|---|---|---|---|---|
| Target Name | Endophilin-B1 (SH3GLB1) | |||||
| Gene Name | SH3GLB1 | |||||
| Gene ID | 51100 | |||||
| Synonyms |
KIAA0491; Endophilin-B1; Bax-interacting factor 1; Bif-1; SH3 domain-containing GRB2-like protein B1 |
|||||
| 3D Structure | ||||||
| Sequence |
MEKELRSTILFNAYKKEIFTTNNGYKSMQKKLRSNWKIQSLKDEITSEKLNGVKLWITAG
PREKFTAAEFEILKKYLDTGGDVFVMLGEGGESRFDTNINFLLEEYGIMVNNDAVVRNVY HKYFHPKEALVSSGVLNREISRAAGKAVPGIIDEESSGNNAQALTFVYPFGATLSVMKPA VAVLSTGSVCFPLNRPILAFYHSKNQGGKLAVLGSCHMFSDQYLDKEENSKIMDVVFQWL TTGDIHLNQIDAEDPEISDYMMLPYTATLSKRNRECLQESDEIPRDFTTLFDLSIFQLDT TSFHSVIEAHEQLNVKHEPLQLIQPQFETPLPTLQPAVFPPSFRELPPPPLELFDLDETF SSEKARLAQITNKCTEEDLEFYVRKCGDILGVTSKLPKDQQDAKHILEHVFFQVVEFKKL NQEHDIDTSETAFQNNF |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
Endophilin family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
May be required for normal outer mitochondrial membrane dynamics. Required for coatomer-mediated retrograde transport in certain cells. May recruit other proteins to membranes with high curvature. May promote membrane fusion. Involved in activation of caspase-dependent apoptosis by promoting BAX/BAK1 activation. Isoform 1 acts proapoptotic in fibroblasts. Involved in caspase-independent apoptosis during nutrition starvation and involved in the regulation of autophagy. Activates lipid kinase activity of PIK3C3 during autophagy probably by associating with the PI3K complex II (PI3KC3-C2). Associated with PI3KC3-C2 during autophagy may regulate the trafficking of ATG9A from the Golgi complex to the peripheral cytoplasm for the formation of autophagosomes by inducing Golgi membrane tubulation and fragmentation. Involved in regulation of degradative endocytic trafficking and cytokinesis, probably in the context of PI3KC3-C2. Isoform 2 acts antiapoptotic in neuronal cells; involved in maintenance of mitochondrial morphology and promotes neuronal viability.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
FBPP2 Probe Info |
 |
2.75 | LDD0318 | [2] | |
|
TH211 Probe Info |
 |
Y80(9.42); Y155(5.54) | LDD0257 | [3] | |
|
TH216 Probe Info |
 |
Y155(20.00); Y80(12.96) | LDD0259 | [3] | |
|
STPyne Probe Info |
 |
K10(0.21); K156(8.04); K160(5.24); K163(10.00) | LDD0277 | [4] | |
|
DA-P3 Probe Info |
 |
45.31 | LDD0179 | [5] | |
|
DBIA Probe Info |
 |
C304(4.08) | LDD0209 | [6] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [7] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [8] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [9] | |
|
Lodoacetamide azide Probe Info |
 |
C121(0.00); C304(0.00) | LDD0037 | [9] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [10] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [11] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [12] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [13] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [10] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C040 Probe Info |
 |
9.13 | LDD1740 | [14] | |
|
C091 Probe Info |
 |
10.13 | LDD1782 | [14] | |
|
C092 Probe Info |
 |
28.84 | LDD1783 | [14] | |
|
C106 Probe Info |
 |
16.56 | LDD1793 | [14] | |
|
C112 Probe Info |
 |
22.78 | LDD1799 | [14] | |
|
C134 Probe Info |
 |
20.39 | LDD1816 | [14] | |
|
C201 Probe Info |
 |
70.52 | LDD1877 | [14] | |
|
C220 Probe Info |
 |
12.64 | LDD1894 | [14] | |
|
C228 Probe Info |
 |
23.59 | LDD1901 | [14] | |
|
C246 Probe Info |
 |
12.38 | LDD1919 | [14] | |
|
C285 Probe Info |
 |
21.56 | LDD1955 | [14] | |
|
C289 Probe Info |
 |
53.45 | LDD1959 | [14] | |
|
C350 Probe Info |
 |
22.47 | LDD2011 | [14] | |
|
C362 Probe Info |
 |
35.02 | LDD2023 | [14] | |
|
C367 Probe Info |
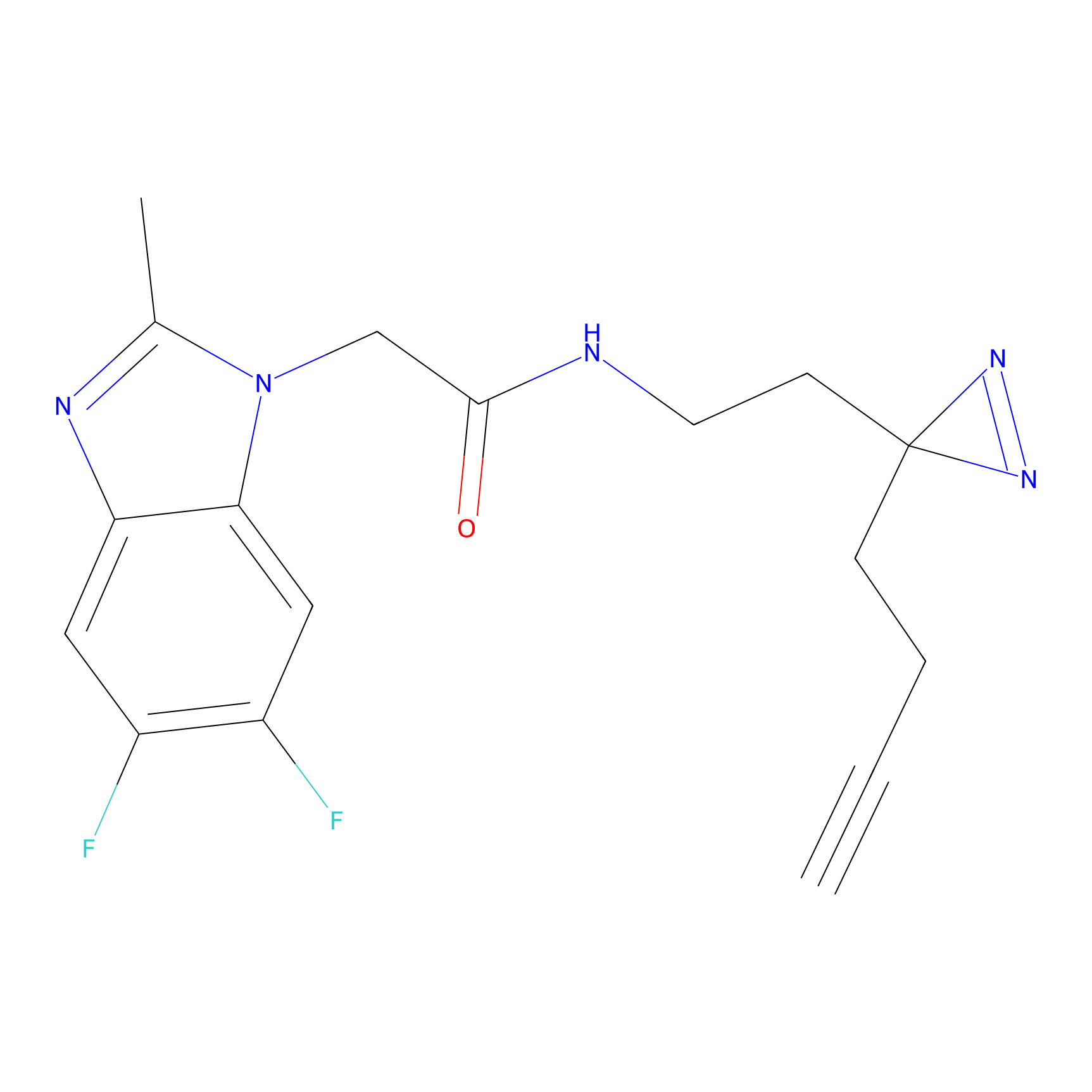 |
5.35 | LDD2028 | [14] | |
|
C388 Probe Info |
 |
50.56 | LDD2047 | [14] | |
|
C390 Probe Info |
 |
22.01 | LDD2049 | [14] | |
|
C407 Probe Info |
 |
12.91 | LDD2064 | [14] | |
|
FFF probe11 Probe Info |
 |
16.14 | LDD0471 | [15] | |
|
FFF probe13 Probe Info |
 |
17.95 | LDD0475 | [15] | |
|
FFF probe14 Probe Info |
 |
19.86 | LDD0477 | [15] | |
|
FFF probe3 Probe Info |
 |
7.39 | LDD0464 | [15] | |
|
FFF probe4 Probe Info |
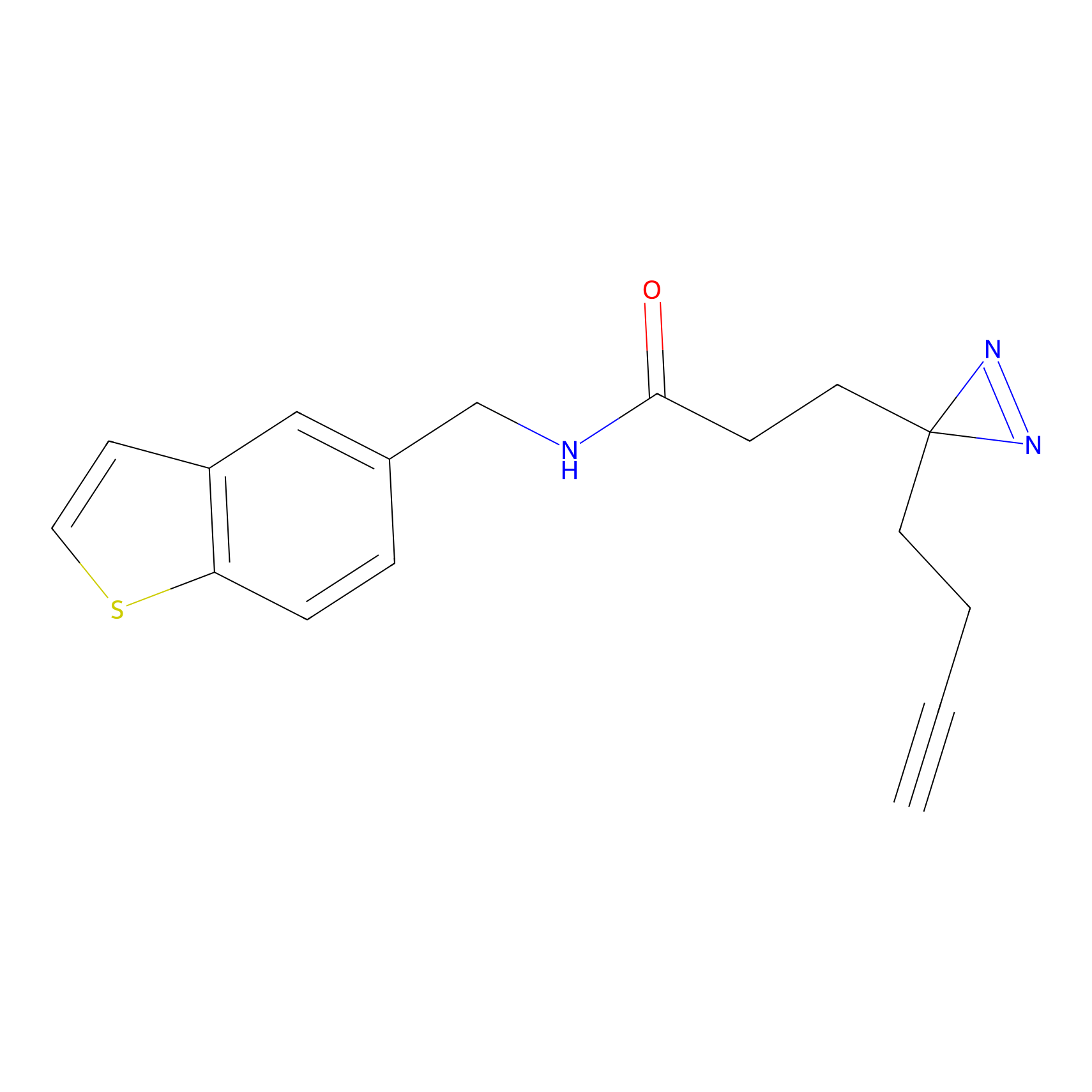 |
7.42 | LDD0466 | [15] | |
|
JN0003 Probe Info |
 |
17.20 | LDD0469 | [15] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [16] | |
|
OEA-DA Probe Info |
 |
17.58 | LDD0046 | [17] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HEK-293T | C226(1.26) | LDD1507 | [18] |
| LDCM0215 | AC10 | HEK-293T | C226(1.02) | LDD1508 | [18] |
| LDCM0226 | AC11 | HEK-293T | C226(0.85) | LDD1509 | [18] |
| LDCM0237 | AC12 | HEK-293T | C226(1.08) | LDD1510 | [18] |
| LDCM0259 | AC14 | HEK-293T | C226(0.97) | LDD1512 | [18] |
| LDCM0270 | AC15 | HEK-293T | C226(1.12) | LDD1513 | [18] |
| LDCM0276 | AC17 | HEK-293T | C226(1.10) | LDD1515 | [18] |
| LDCM0277 | AC18 | HEK-293T | C226(0.92) | LDD1516 | [18] |
| LDCM0278 | AC19 | HEK-293T | C226(0.85) | LDD1517 | [18] |
| LDCM0279 | AC2 | HEK-293T | C226(0.84) | LDD1518 | [18] |
| LDCM0280 | AC20 | HEK-293T | C226(0.99) | LDD1519 | [18] |
| LDCM0281 | AC21 | HEK-293T | C226(1.02) | LDD1520 | [18] |
| LDCM0282 | AC22 | HEK-293T | C226(0.95) | LDD1521 | [18] |
| LDCM0283 | AC23 | HEK-293T | C226(1.07) | LDD1522 | [18] |
| LDCM0284 | AC24 | HEK-293T | C226(0.82) | LDD1523 | [18] |
| LDCM0285 | AC25 | HEK-293T | C226(1.33) | LDD1524 | [18] |
| LDCM0286 | AC26 | HEK-293T | C226(0.90) | LDD1525 | [18] |
| LDCM0287 | AC27 | HEK-293T | C226(0.88) | LDD1526 | [18] |
| LDCM0288 | AC28 | HEK-293T | C226(1.02) | LDD1527 | [18] |
| LDCM0289 | AC29 | HEK-293T | C226(0.96) | LDD1528 | [18] |
| LDCM0290 | AC3 | HEK-293T | C226(1.00) | LDD1529 | [18] |
| LDCM0291 | AC30 | HEK-293T | C226(1.02) | LDD1530 | [18] |
| LDCM0292 | AC31 | HEK-293T | C226(1.22) | LDD1531 | [18] |
| LDCM0293 | AC32 | HEK-293T | C226(0.94) | LDD1532 | [18] |
| LDCM0294 | AC33 | HEK-293T | C226(1.22) | LDD1533 | [18] |
| LDCM0295 | AC34 | HEK-293T | C226(1.06) | LDD1534 | [18] |
| LDCM0296 | AC35 | HEK-293T | C226(0.94) | LDD1535 | [18] |
| LDCM0297 | AC36 | HEK-293T | C226(1.04) | LDD1536 | [18] |
| LDCM0298 | AC37 | HEK-293T | C226(0.89) | LDD1537 | [18] |
| LDCM0299 | AC38 | HEK-293T | C226(0.86) | LDD1538 | [18] |
| LDCM0300 | AC39 | HEK-293T | C226(1.03) | LDD1539 | [18] |
| LDCM0301 | AC4 | HEK-293T | C226(1.01) | LDD1540 | [18] |
| LDCM0302 | AC40 | HEK-293T | C226(0.94) | LDD1541 | [18] |
| LDCM0303 | AC41 | HEK-293T | C226(1.08) | LDD1542 | [18] |
| LDCM0304 | AC42 | HEK-293T | C226(1.01) | LDD1543 | [18] |
| LDCM0305 | AC43 | HEK-293T | C226(0.89) | LDD1544 | [18] |
| LDCM0306 | AC44 | HEK-293T | C226(1.02) | LDD1545 | [18] |
| LDCM0307 | AC45 | HEK-293T | C226(0.95) | LDD1546 | [18] |
| LDCM0308 | AC46 | HEK-293T | C226(0.99) | LDD1547 | [18] |
| LDCM0309 | AC47 | HEK-293T | C226(0.96) | LDD1548 | [18] |
| LDCM0310 | AC48 | HEK-293T | C226(0.73) | LDD1549 | [18] |
| LDCM0311 | AC49 | HEK-293T | C226(1.23) | LDD1550 | [18] |
| LDCM0312 | AC5 | HEK-293T | C226(0.91) | LDD1551 | [18] |
| LDCM0313 | AC50 | HEK-293T | C226(0.89) | LDD1552 | [18] |
| LDCM0314 | AC51 | HEK-293T | C226(0.88) | LDD1553 | [18] |
| LDCM0315 | AC52 | HEK-293T | C226(1.01) | LDD1554 | [18] |
| LDCM0316 | AC53 | HEK-293T | C226(1.10) | LDD1555 | [18] |
| LDCM0317 | AC54 | HEK-293T | C226(0.79) | LDD1556 | [18] |
| LDCM0318 | AC55 | HEK-293T | C226(0.96) | LDD1557 | [18] |
| LDCM0319 | AC56 | HEK-293T | C226(0.99) | LDD1558 | [18] |
| LDCM0320 | AC57 | HEK-293T | C226(1.18) | LDD1559 | [18] |
| LDCM0321 | AC58 | HEK-293T | C226(0.97) | LDD1560 | [18] |
| LDCM0322 | AC59 | HEK-293T | C226(0.98) | LDD1561 | [18] |
| LDCM0323 | AC6 | HEK-293T | C226(1.00) | LDD1562 | [18] |
| LDCM0324 | AC60 | HEK-293T | C226(1.09) | LDD1563 | [18] |
| LDCM0325 | AC61 | HEK-293T | C226(1.10) | LDD1564 | [18] |
| LDCM0326 | AC62 | HEK-293T | C226(1.09) | LDD1565 | [18] |
| LDCM0327 | AC63 | HEK-293T | C226(1.04) | LDD1566 | [18] |
| LDCM0328 | AC64 | HEK-293T | C226(0.87) | LDD1567 | [18] |
| LDCM0334 | AC7 | HEK-293T | C226(0.97) | LDD1568 | [18] |
| LDCM0345 | AC8 | HEK-293T | C226(0.95) | LDD1569 | [18] |
| LDCM0248 | AKOS034007472 | HEK-293T | C226(1.02) | LDD1511 | [18] |
| LDCM0356 | AKOS034007680 | HEK-293T | C226(1.10) | LDD1570 | [18] |
| LDCM0275 | AKOS034007705 | HEK-293T | C226(1.06) | LDD1514 | [18] |
| LDCM0156 | Aniline | NCI-H1299 | 12.00 | LDD0403 | [1] |
| LDCM0632 | CL-Sc | Hep-G2 | C121(1.03) | LDD2227 | [10] |
| LDCM0367 | CL1 | HEK-293T | C226(1.18) | LDD1571 | [18] |
| LDCM0368 | CL10 | HEK-293T | C226(0.50) | LDD1572 | [18] |
| LDCM0369 | CL100 | HEK-293T | C226(1.08) | LDD1573 | [18] |
| LDCM0370 | CL101 | HEK-293T | C226(1.01) | LDD1574 | [18] |
| LDCM0371 | CL102 | HEK-293T | C226(1.13) | LDD1575 | [18] |
| LDCM0372 | CL103 | HEK-293T | C226(0.81) | LDD1576 | [18] |
| LDCM0373 | CL104 | HEK-293T | C226(1.02) | LDD1577 | [18] |
| LDCM0374 | CL105 | HEK-293T | C226(1.05) | LDD1578 | [18] |
| LDCM0375 | CL106 | HEK-293T | C226(0.91) | LDD1579 | [18] |
| LDCM0376 | CL107 | HEK-293T | C226(1.12) | LDD1580 | [18] |
| LDCM0377 | CL108 | HEK-293T | C226(0.95) | LDD1581 | [18] |
| LDCM0378 | CL109 | HEK-293T | C226(1.17) | LDD1582 | [18] |
| LDCM0379 | CL11 | HEK-293T | C226(0.95) | LDD1583 | [18] |
| LDCM0380 | CL110 | HEK-293T | C226(1.20) | LDD1584 | [18] |
| LDCM0381 | CL111 | HEK-293T | C226(0.84) | LDD1585 | [18] |
| LDCM0382 | CL112 | HEK-293T | C226(0.98) | LDD1586 | [18] |
| LDCM0383 | CL113 | HEK-293T | C226(0.98) | LDD1587 | [18] |
| LDCM0384 | CL114 | HEK-293T | C226(1.35) | LDD1588 | [18] |
| LDCM0385 | CL115 | HEK-293T | C226(0.91) | LDD1589 | [18] |
| LDCM0386 | CL116 | HEK-293T | C226(1.05) | LDD1590 | [18] |
| LDCM0387 | CL117 | HEK-293T | C226(1.02) | LDD1591 | [18] |
| LDCM0388 | CL118 | HEK-293T | C226(0.91) | LDD1592 | [18] |
| LDCM0389 | CL119 | HEK-293T | C226(1.04) | LDD1593 | [18] |
| LDCM0390 | CL12 | HEK-293T | C226(0.81) | LDD1594 | [18] |
| LDCM0391 | CL120 | HEK-293T | C226(0.90) | LDD1595 | [18] |
| LDCM0392 | CL121 | HEK-293T | C226(1.01) | LDD1596 | [18] |
| LDCM0393 | CL122 | HEK-293T | C226(1.00) | LDD1597 | [18] |
| LDCM0394 | CL123 | HEK-293T | C226(1.15) | LDD1598 | [18] |
| LDCM0395 | CL124 | HEK-293T | C226(0.91) | LDD1599 | [18] |
| LDCM0396 | CL125 | HEK-293T | C226(1.12) | LDD1600 | [18] |
| LDCM0397 | CL126 | HEK-293T | C226(1.15) | LDD1601 | [18] |
| LDCM0398 | CL127 | HEK-293T | C226(0.96) | LDD1602 | [18] |
| LDCM0399 | CL128 | HEK-293T | C226(1.13) | LDD1603 | [18] |
| LDCM0400 | CL13 | HEK-293T | C226(1.11) | LDD1604 | [18] |
| LDCM0401 | CL14 | HEK-293T | C226(0.85) | LDD1605 | [18] |
| LDCM0402 | CL15 | HEK-293T | C226(1.08) | LDD1606 | [18] |
| LDCM0403 | CL16 | HEK-293T | C226(0.83) | LDD1607 | [18] |
| LDCM0404 | CL17 | HEK-293T | C226(1.13) | LDD1608 | [18] |
| LDCM0405 | CL18 | HEK-293T | C226(0.83) | LDD1609 | [18] |
| LDCM0406 | CL19 | HEK-293T | C226(0.67) | LDD1610 | [18] |
| LDCM0407 | CL2 | HEK-293T | C226(1.07) | LDD1611 | [18] |
| LDCM0408 | CL20 | HEK-293T | C226(0.85) | LDD1612 | [18] |
| LDCM0409 | CL21 | HEK-293T | C226(0.80) | LDD1613 | [18] |
| LDCM0410 | CL22 | HEK-293T | C226(0.62) | LDD1614 | [18] |
| LDCM0411 | CL23 | HEK-293T | C226(1.14) | LDD1615 | [18] |
| LDCM0412 | CL24 | HEK-293T | C226(0.55) | LDD1616 | [18] |
| LDCM0413 | CL25 | HEK-293T | C226(1.22) | LDD1617 | [18] |
| LDCM0414 | CL26 | HEK-293T | C226(1.13) | LDD1618 | [18] |
| LDCM0415 | CL27 | HEK-293T | C226(0.86) | LDD1619 | [18] |
| LDCM0416 | CL28 | HEK-293T | C226(0.84) | LDD1620 | [18] |
| LDCM0417 | CL29 | HEK-293T | C226(0.93) | LDD1621 | [18] |
| LDCM0418 | CL3 | HEK-293T | C226(0.89) | LDD1622 | [18] |
| LDCM0419 | CL30 | HEK-293T | C226(0.79) | LDD1623 | [18] |
| LDCM0420 | CL31 | HEK-293T | C226(0.73) | LDD1624 | [18] |
| LDCM0421 | CL32 | HEK-293T | C226(0.87) | LDD1625 | [18] |
| LDCM0422 | CL33 | HEK-293T | C226(1.19) | LDD1626 | [18] |
| LDCM0423 | CL34 | HEK-293T | C226(0.62) | LDD1627 | [18] |
| LDCM0424 | CL35 | HEK-293T | C226(1.06) | LDD1628 | [18] |
| LDCM0425 | CL36 | HEK-293T | C226(0.64) | LDD1629 | [18] |
| LDCM0426 | CL37 | HEK-293T | C226(0.93) | LDD1630 | [18] |
| LDCM0428 | CL39 | HEK-293T | C226(0.85) | LDD1632 | [18] |
| LDCM0429 | CL4 | HEK-293T | C226(1.00) | LDD1633 | [18] |
| LDCM0430 | CL40 | HEK-293T | C226(0.87) | LDD1634 | [18] |
| LDCM0431 | CL41 | HEK-293T | C226(0.88) | LDD1635 | [18] |
| LDCM0432 | CL42 | HEK-293T | C226(0.88) | LDD1636 | [18] |
| LDCM0433 | CL43 | HEK-293T | C226(0.70) | LDD1637 | [18] |
| LDCM0434 | CL44 | HEK-293T | C226(0.80) | LDD1638 | [18] |
| LDCM0435 | CL45 | HEK-293T | C226(0.73) | LDD1639 | [18] |
| LDCM0436 | CL46 | HEK-293T | C226(0.65) | LDD1640 | [18] |
| LDCM0437 | CL47 | HEK-293T | C226(0.87) | LDD1641 | [18] |
| LDCM0438 | CL48 | HEK-293T | C226(0.74) | LDD1642 | [18] |
| LDCM0439 | CL49 | HEK-293T | C226(1.06) | LDD1643 | [18] |
| LDCM0440 | CL5 | HEK-293T | C226(1.12) | LDD1644 | [18] |
| LDCM0441 | CL50 | HEK-293T | C226(0.99) | LDD1645 | [18] |
| LDCM0443 | CL52 | HEK-293T | C226(0.91) | LDD1646 | [18] |
| LDCM0444 | CL53 | HEK-293T | C226(1.02) | LDD1647 | [18] |
| LDCM0445 | CL54 | HEK-293T | C226(1.25) | LDD1648 | [18] |
| LDCM0446 | CL55 | HEK-293T | C226(0.68) | LDD1649 | [18] |
| LDCM0447 | CL56 | HEK-293T | C226(0.84) | LDD1650 | [18] |
| LDCM0448 | CL57 | HEK-293T | C226(0.74) | LDD1651 | [18] |
| LDCM0449 | CL58 | HEK-293T | C226(0.62) | LDD1652 | [18] |
| LDCM0450 | CL59 | HEK-293T | C226(1.06) | LDD1653 | [18] |
| LDCM0451 | CL6 | HEK-293T | C226(1.03) | LDD1654 | [18] |
| LDCM0452 | CL60 | HEK-293T | C226(0.71) | LDD1655 | [18] |
| LDCM0453 | CL61 | HEK-293T | C226(1.10) | LDD1656 | [18] |
| LDCM0454 | CL62 | HEK-293T | C226(0.95) | LDD1657 | [18] |
| LDCM0455 | CL63 | HEK-293T | C226(0.97) | LDD1658 | [18] |
| LDCM0456 | CL64 | HEK-293T | C226(0.95) | LDD1659 | [18] |
| LDCM0457 | CL65 | HEK-293T | C226(0.95) | LDD1660 | [18] |
| LDCM0458 | CL66 | HEK-293T | C226(0.90) | LDD1661 | [18] |
| LDCM0459 | CL67 | HEK-293T | C226(0.76) | LDD1662 | [18] |
| LDCM0460 | CL68 | HEK-293T | C226(0.83) | LDD1663 | [18] |
| LDCM0461 | CL69 | HEK-293T | C226(0.75) | LDD1664 | [18] |
| LDCM0462 | CL7 | HEK-293T | C226(0.81) | LDD1665 | [18] |
| LDCM0463 | CL70 | HEK-293T | C226(0.70) | LDD1666 | [18] |
| LDCM0464 | CL71 | HEK-293T | C226(0.94) | LDD1667 | [18] |
| LDCM0465 | CL72 | HEK-293T | C226(0.55) | LDD1668 | [18] |
| LDCM0466 | CL73 | HEK-293T | C226(1.20) | LDD1669 | [18] |
| LDCM0467 | CL74 | HEK-293T | C226(1.17) | LDD1670 | [18] |
| LDCM0469 | CL76 | HEK-293T | C226(0.94) | LDD1672 | [18] |
| LDCM0470 | CL77 | HEK-293T | C226(0.96) | LDD1673 | [18] |
| LDCM0471 | CL78 | HEK-293T | C226(0.90) | LDD1674 | [18] |
| LDCM0472 | CL79 | HEK-293T | C226(0.75) | LDD1675 | [18] |
| LDCM0473 | CL8 | HEK-293T | C226(1.19) | LDD1676 | [18] |
| LDCM0474 | CL80 | HEK-293T | C226(0.84) | LDD1677 | [18] |
| LDCM0475 | CL81 | HEK-293T | C226(0.74) | LDD1678 | [18] |
| LDCM0476 | CL82 | HEK-293T | C226(0.62) | LDD1679 | [18] |
| LDCM0477 | CL83 | HEK-293T | C226(1.04) | LDD1680 | [18] |
| LDCM0478 | CL84 | HEK-293T | C226(0.89) | LDD1681 | [18] |
| LDCM0479 | CL85 | HEK-293T | C226(1.13) | LDD1682 | [18] |
| LDCM0480 | CL86 | HEK-293T | C226(1.26) | LDD1683 | [18] |
| LDCM0481 | CL87 | HEK-293T | C226(1.04) | LDD1684 | [18] |
| LDCM0482 | CL88 | HEK-293T | C226(0.93) | LDD1685 | [18] |
| LDCM0483 | CL89 | HEK-293T | C226(1.10) | LDD1686 | [18] |
| LDCM0484 | CL9 | HEK-293T | C226(0.76) | LDD1687 | [18] |
| LDCM0485 | CL90 | HEK-293T | C226(1.58) | LDD1688 | [18] |
| LDCM0486 | CL91 | HEK-293T | C226(0.74) | LDD1689 | [18] |
| LDCM0487 | CL92 | HEK-293T | C226(1.01) | LDD1690 | [18] |
| LDCM0488 | CL93 | HEK-293T | C226(0.80) | LDD1691 | [18] |
| LDCM0489 | CL94 | HEK-293T | C226(0.70) | LDD1692 | [18] |
| LDCM0490 | CL95 | HEK-293T | C226(0.94) | LDD1693 | [18] |
| LDCM0491 | CL96 | HEK-293T | C226(0.94) | LDD1694 | [18] |
| LDCM0492 | CL97 | HEK-293T | C226(1.08) | LDD1695 | [18] |
| LDCM0493 | CL98 | HEK-293T | C226(1.07) | LDD1696 | [18] |
| LDCM0494 | CL99 | HEK-293T | C226(0.85) | LDD1697 | [18] |
| LDCM0027 | Dopamine | HEK-293T | 45.31 | LDD0179 | [5] |
| LDCM0495 | E2913 | HEK-293T | C226(0.85) | LDD1698 | [18] |
| LDCM0031 | Epigallocatechin gallate | HEK-293T | 33.22 | LDD0183 | [5] |
| LDCM0625 | F8 | Ramos | C121(0.54) | LDD2187 | [19] |
| LDCM0572 | Fragment10 | Ramos | C121(1.36) | LDD2189 | [19] |
| LDCM0573 | Fragment11 | Ramos | C121(0.03) | LDD2190 | [19] |
| LDCM0574 | Fragment12 | Ramos | C121(3.06) | LDD2191 | [19] |
| LDCM0575 | Fragment13 | Ramos | C121(0.80) | LDD2192 | [19] |
| LDCM0576 | Fragment14 | Ramos | C121(1.38) | LDD2193 | [19] |
| LDCM0579 | Fragment20 | Ramos | C121(2.85) | LDD2194 | [19] |
| LDCM0580 | Fragment21 | Ramos | C121(0.91) | LDD2195 | [19] |
| LDCM0582 | Fragment23 | Ramos | C121(2.38) | LDD2196 | [19] |
| LDCM0578 | Fragment27 | Ramos | C121(1.00) | LDD2197 | [19] |
| LDCM0586 | Fragment28 | Ramos | C121(0.47) | LDD2198 | [19] |
| LDCM0588 | Fragment30 | Ramos | C121(1.50) | LDD2199 | [19] |
| LDCM0589 | Fragment31 | Ramos | C121(0.84) | LDD2200 | [19] |
| LDCM0590 | Fragment32 | Ramos | C121(1.68) | LDD2201 | [19] |
| LDCM0468 | Fragment33 | HEK-293T | C226(0.94) | LDD1671 | [18] |
| LDCM0596 | Fragment38 | Ramos | C121(1.20) | LDD2203 | [19] |
| LDCM0566 | Fragment4 | Ramos | C121(1.76) | LDD2184 | [19] |
| LDCM0427 | Fragment51 | HEK-293T | C226(1.10) | LDD1631 | [18] |
| LDCM0610 | Fragment52 | Ramos | C121(1.55) | LDD2204 | [19] |
| LDCM0614 | Fragment56 | Ramos | C121(1.24) | LDD2205 | [19] |
| LDCM0569 | Fragment7 | Ramos | C121(1.25) | LDD2186 | [19] |
| LDCM0571 | Fragment9 | Ramos | C121(7.34) | LDD2188 | [19] |
| LDCM0022 | KB02 | A2780 | C21(2.24) | LDD2254 | [20] |
| LDCM0023 | KB03 | Jurkat | C304(4.08) | LDD0209 | [6] |
| LDCM0024 | KB05 | G361 | C21(5.12) | LDD3311 | [20] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C121(0.80) | LDD2207 | [21] |
| LDCM0029 | Quercetin | HEK-293T | 6.70 | LDD0181 | [5] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
Transcription factor
GPCR
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Succinate receptor 1 (SUCNR1) | G-protein coupled receptor 1 family | Q9BXA5 | |||
Immunoglobulin
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| T-cell surface glycoprotein CD4 (CD4) | . | P01730 | |||
Cytokine and receptor
Other
References
