Details of the Target
General Information of Target
| Target ID | LDTP03152 | |||||
|---|---|---|---|---|---|---|
| Target Name | Protein-L-isoaspartate(D-aspartate) O-methyltransferase (PCMT1) | |||||
| Gene Name | PCMT1 | |||||
| Gene ID | 5110 | |||||
| Synonyms |
Protein-L-isoaspartate(D-aspartate) O-methyltransferase; PIMT; EC 2.1.1.77; L-isoaspartyl protein carboxyl methyltransferase; Protein L-isoaspartyl/D-aspartyl methyltransferase; Protein-beta-aspartate methyltransferase
|
|||||
| 3D Structure | ||||||
| Sequence |
MAWKSGGASHSELIHNLRKNGIIKTDKVFEVMLATDRSHYAKCNPYMDSPQSIGFQATIS
APHMHAYALELLFDQLHEGAKALDVGSGSGILTACFARMVGCTGKVIGIDHIKELVDDSV NNVRKDDPTLLSSGRVQLVVGDGRMGYAEEAPYDAIHVGAAAPVVPQALIDQLKPGGRLI LPVGPAGGNQMLEQYDKLQDGSIKMKPLMGVIYVPLTDKEKQWSRWK |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Methyltransferase superfamily, L-isoaspartyl/D-aspartyl protein methyltransferase family
|
|||||
| Subcellular location |
Cytoplasm, cytosol
|
|||||
| Function |
Initiates the repair of damaged proteins by catalyzing methyl esterification of L-isoaspartyl and D-aspartyl residues produced by spontaneous isomerization and racemization of L-aspartyl and L-asparaginyl residues in aging peptides and proteins. Acts on EIF4EBP2, microtubule-associated protein 2, calreticulin, clathrin light chains a and b, Ubiquitin C-terminal hydrolase isozyme L1, phosphatidylethanolamine-binding protein 1, stathmin, beta-synuclein and alpha-synuclein.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| HCC15 | SNV: p.H157L | DBIA Probe Info | |||
| KMCH1 | SNV: p.A186P | DBIA Probe Info | |||
| TE1 | SNV: p.I109M | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
A-EBA Probe Info |
 |
3.63 | LDD0215 | [2] | |
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [3] | |
|
CY4 Probe Info |
 |
100.00 | LDD0244 | [3] | |
|
N1 Probe Info |
 |
100.00 | LDD0242 | [3] | |
|
TH211 Probe Info |
 |
Y213(6.56) | LDD0257 | [4] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [5] | |
|
BTD Probe Info |
 |
C102(6.69) | LDD1699 | [6] | |
|
OPA-S-S-alkyne Probe Info |
 |
K125(1.38) | LDD3494 | [7] | |
|
DBIA Probe Info |
 |
C102(0.88) | LDD3312 | [8] | |
|
DA-P3 Probe Info |
 |
7.51 | LDD0183 | [9] | |
|
AHL-Pu-1 Probe Info |
 |
C95(2.14) | LDD0169 | [10] | |
|
HHS-475 Probe Info |
 |
Y213(2.35) | LDD0264 | [11] | |
|
HHS-465 Probe Info |
 |
Y213(4.59) | LDD2237 | [12] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [13] | |
|
ATP probe Probe Info |
 |
K4(0.00); K105(0.00); K125(0.00) | LDD0199 | [14] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [15] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [16] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [17] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [16] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [18] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [18] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0151 | [19] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [18] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [18] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [18] | |
|
Phosphinate-6 Probe Info |
 |
C43(0.00); C102(0.00) | LDD0018 | [20] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [21] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [21] | |
|
W1 Probe Info |
 |
C102(0.00); D154(0.00); E150(0.00); Y153(0.00) | LDD0236 | [22] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [23] | |
|
NAIA_5 Probe Info |
 |
C95(0.00); C43(0.00) | LDD2223 | [17] | |
|
TER-AC Probe Info |
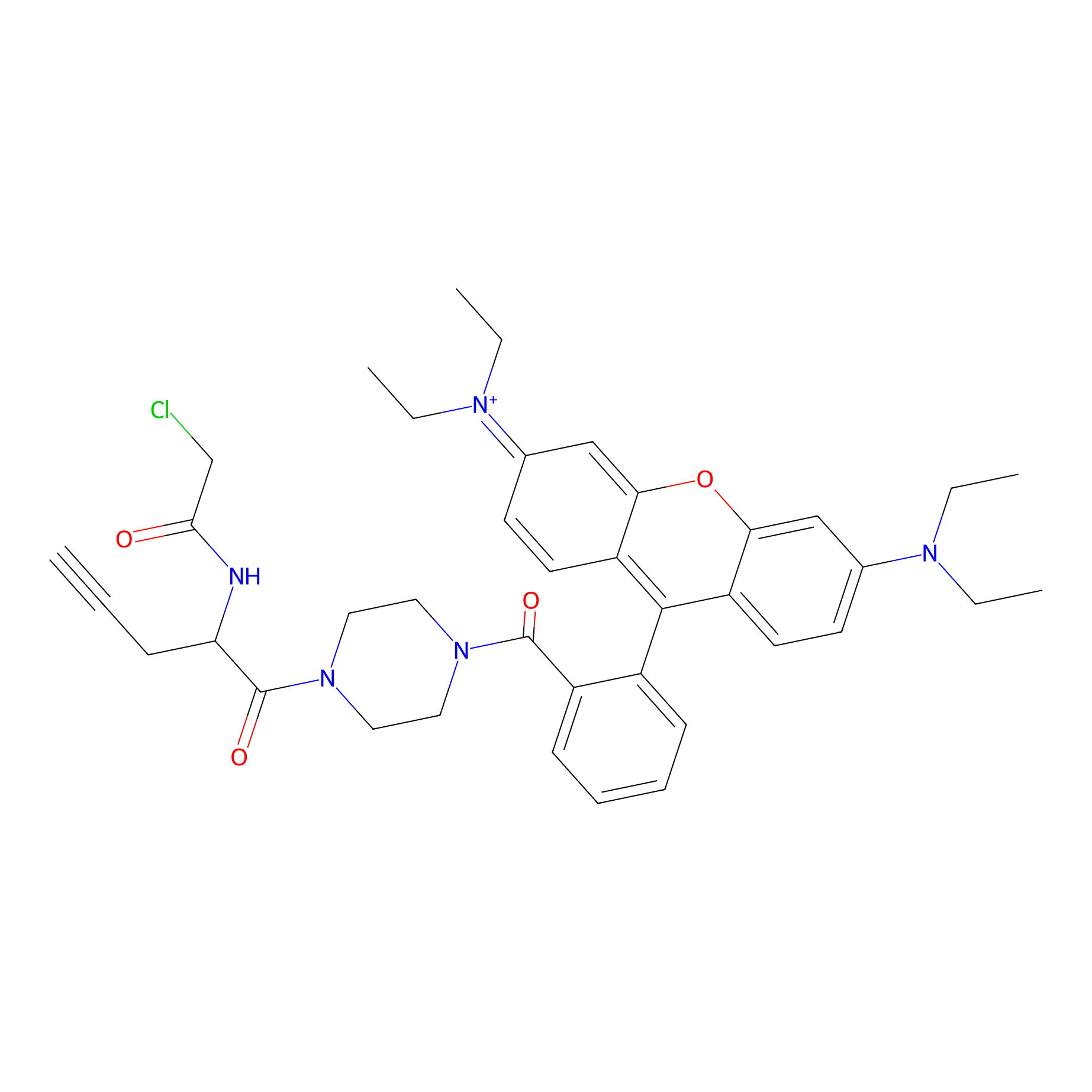 |
N.A. | LDD0426 | [24] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C191 Probe Info |
 |
9.99 | LDD1868 | [25] | |
|
FFF probe13 Probe Info |
 |
12.00 | LDD0475 | [26] | |
|
FFF probe14 Probe Info |
 |
20.00 | LDD0477 | [26] | |
|
FFF probe2 Probe Info |
 |
14.18 | LDD0463 | [26] | |
|
FFF probe3 Probe Info |
 |
7.84 | LDD0465 | [26] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C102(0.76) | LDD2142 | [6] |
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C102(0.90) | LDD2112 | [6] |
| LDCM0502 | 1-(Cyanoacetyl)piperidine | MDA-MB-231 | C102(0.89) | LDD2095 | [6] |
| LDCM0537 | 2-Cyano-N,N-dimethylacetamide | MDA-MB-231 | C102(0.96) | LDD2130 | [6] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C102(0.91) | LDD2117 | [6] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C102(1.11) | LDD2152 | [6] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C102(0.96) | LDD2103 | [6] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C102(0.51) | LDD2132 | [6] |
| LDCM0538 | 4-(Cyanoacetyl)morpholine | MDA-MB-231 | C102(1.68) | LDD2131 | [6] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C95(2.14) | LDD0169 | [10] |
| LDCM0545 | Acetamide | MDA-MB-231 | C102(0.41) | LDD2138 | [6] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C102(0.61) | LDD2113 | [6] |
| LDCM0156 | Aniline | NCI-H1299 | 13.56 | LDD0403 | [1] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C102(0.53) | LDD2091 | [6] |
| LDCM0108 | Chloroacetamide | HeLa | C95(0.00); H157(0.00); C102(0.00) | LDD0222 | [21] |
| LDCM0632 | CL-Sc | Hep-G2 | C95(0.99) | LDD2227 | [17] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C102(0.87) | LDD1702 | [6] |
| LDCM0031 | Epigallocatechin gallate | HEK-293T | 7.51 | LDD0183 | [9] |
| LDCM0625 | F8 | Ramos | C102(1.06); C95(0.69) | LDD2187 | [27] |
| LDCM0572 | Fragment10 | Ramos | C102(1.42) | LDD1390 | [28] |
| LDCM0573 | Fragment11 | Ramos | C102(1.08) | LDD1392 | [28] |
| LDCM0574 | Fragment12 | Ramos | C102(0.53) | LDD1394 | [28] |
| LDCM0575 | Fragment13 | MDA-MB-231 | C102(0.93) | LDD1395 | [28] |
| LDCM0576 | Fragment14 | Ramos | C102(1.35) | LDD1398 | [28] |
| LDCM0577 | Fragment15 | Ramos | C102(1.44) | LDD1400 | [28] |
| LDCM0579 | Fragment20 | Ramos | C102(0.63); C95(0.63) | LDD2194 | [27] |
| LDCM0580 | Fragment21 | MDA-MB-231 | C102(0.86) | LDD1404 | [28] |
| LDCM0581 | Fragment22 | MDA-MB-231 | C102(1.08) | LDD1406 | [28] |
| LDCM0582 | Fragment23 | Ramos | C102(1.16); C95(1.14) | LDD2196 | [27] |
| LDCM0584 | Fragment25 | MDA-MB-231 | C102(0.94) | LDD1411 | [28] |
| LDCM0585 | Fragment26 | Ramos | C102(0.87) | LDD1412 | [28] |
| LDCM0578 | Fragment27 | MDA-MB-231 | C102(1.19) | LDD1401 | [28] |
| LDCM0586 | Fragment28 | MDA-MB-231 | C102(0.81) | LDD1415 | [28] |
| LDCM0587 | Fragment29 | Ramos | C102(1.43) | LDD1418 | [28] |
| LDCM0588 | Fragment30 | MDA-MB-231 | C102(0.68) | LDD1419 | [28] |
| LDCM0589 | Fragment31 | MDA-MB-231 | C102(1.11) | LDD1421 | [28] |
| LDCM0590 | Fragment32 | MDA-MB-231 | C102(1.11) | LDD1423 | [28] |
| LDCM0468 | Fragment33 | MDA-MB-231 | C102(0.86) | LDD1425 | [28] |
| LDCM0592 | Fragment34 | Ramos | C102(1.04) | LDD1428 | [28] |
| LDCM0593 | Fragment35 | Ramos | C102(0.97) | LDD1430 | [28] |
| LDCM0595 | Fragment37 | Ramos | C102(0.84) | LDD1432 | [28] |
| LDCM0596 | Fragment38 | MDA-MB-231 | C102(0.94) | LDD1433 | [28] |
| LDCM0566 | Fragment4 | MDA-MB-231 | C102(0.99) | LDD1378 | [28] |
| LDCM0598 | Fragment40 | MDA-MB-231 | C102(0.83) | LDD1436 | [28] |
| LDCM0599 | Fragment41 | MDA-MB-231 | C102(1.03) | LDD1438 | [28] |
| LDCM0600 | Fragment42 | Ramos | C102(0.85) | LDD1440 | [28] |
| LDCM0601 | Fragment43 | MDA-MB-231 | C102(0.73) | LDD1441 | [28] |
| LDCM0604 | Fragment46 | MDA-MB-231 | C102(0.86) | LDD1445 | [28] |
| LDCM0605 | Fragment47 | MDA-MB-231 | C102(0.95) | LDD1446 | [28] |
| LDCM0607 | Fragment49 | MDA-MB-231 | C102(1.24) | LDD1448 | [28] |
| LDCM0608 | Fragment50 | MDA-MB-231 | C102(20.00) | LDD1449 | [28] |
| LDCM0427 | Fragment51 | Ramos | C102(0.83) | LDD1451 | [28] |
| LDCM0610 | Fragment52 | MDA-MB-231 | C102(0.78) | LDD1452 | [28] |
| LDCM0612 | Fragment54 | MDA-MB-231 | C102(1.03) | LDD1456 | [28] |
| LDCM0613 | Fragment55 | MDA-MB-231 | C102(0.86) | LDD1457 | [28] |
| LDCM0614 | Fragment56 | MDA-MB-231 | C102(0.87) | LDD1458 | [28] |
| LDCM0569 | Fragment7 | Ramos | C102(0.64); C95(0.34) | LDD2186 | [27] |
| LDCM0570 | Fragment8 | MDA-MB-231 | C102(0.94) | LDD1385 | [28] |
| LDCM0571 | Fragment9 | MDA-MB-231 | C102(0.91) | LDD1387 | [28] |
| LDCM0116 | HHS-0101 | DM93 | Y213(2.35) | LDD0264 | [11] |
| LDCM0118 | HHS-0301 | DM93 | Y213(1.18) | LDD0266 | [11] |
| LDCM0119 | HHS-0401 | DM93 | Y213(1.24) | LDD0267 | [11] |
| LDCM0120 | HHS-0701 | DM93 | Y213(1.23) | LDD0268 | [11] |
| LDCM0107 | IAA | HeLa | H15(0.00); H157(0.00) | LDD0221 | [21] |
| LDCM0022 | KB02 | MDA-MB-231 | C102(1.37) | LDD1374 | [28] |
| LDCM0023 | KB03 | MDA-MB-231 | C102(1.22) | LDD1376 | [28] |
| LDCM0024 | KB05 | HMCB | C102(0.88) | LDD3312 | [8] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C102(1.09) | LDD2102 | [6] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C102(0.71) | LDD2121 | [6] |
| LDCM0109 | NEM | HeLa | H15(0.00); K4(0.00) | LDD0223 | [21] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C102(0.65) | LDD2089 | [6] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C102(1.35) | LDD2090 | [6] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C102(1.21) | LDD2092 | [6] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C102(1.28) | LDD2093 | [6] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C102(2.03) | LDD2094 | [6] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C102(0.82) | LDD2097 | [6] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C102(0.80) | LDD2098 | [6] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C102(0.79) | LDD2099 | [6] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C102(1.76) | LDD2100 | [6] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C102(0.63) | LDD2101 | [6] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C102(0.65) | LDD2104 | [6] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C102(1.44) | LDD2105 | [6] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C102(1.01); C95(0.74) | LDD2106 | [6] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C102(0.76) | LDD2107 | [6] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C102(0.99) | LDD2108 | [6] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C102(0.69) | LDD2109 | [6] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C102(0.89) | LDD2111 | [6] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C102(1.07) | LDD2114 | [6] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C102(0.51) | LDD2115 | [6] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C102(2.86) | LDD2119 | [6] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C102(0.86) | LDD2120 | [6] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C102(0.07) | LDD2122 | [6] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C102(0.78) | LDD2123 | [6] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C102(0.73) | LDD2125 | [6] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C102(0.04) | LDD2126 | [6] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C102(0.87) | LDD2127 | [6] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C102(0.96) | LDD2128 | [6] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C102(0.94) | LDD2129 | [6] |
| LDCM0540 | Nucleophilic fragment 35 | MDA-MB-231 | C102(0.49) | LDD2133 | [6] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C102(0.45) | LDD2134 | [6] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C102(0.87) | LDD2135 | [6] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C102(0.92) | LDD2136 | [6] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C102(0.91) | LDD2137 | [6] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C102(1.16) | LDD1700 | [6] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C102(0.80) | LDD2140 | [6] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C102(0.93) | LDD2143 | [6] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C102(3.51) | LDD2144 | [6] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C102(0.84) | LDD2146 | [6] |
| LDCM0553 | Nucleophilic fragment 6b | MDA-MB-231 | C102(2.66) | LDD2147 | [6] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C102(0.44) | LDD2148 | [6] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C102(0.48) | LDD2150 | [6] |
| LDCM0557 | Nucleophilic fragment 8b | MDA-MB-231 | C102(0.08) | LDD2151 | [6] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C102(1.96) | LDD2153 | [6] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C43(0.83) | LDD2206 | [29] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C102(0.72); C95(0.71); C43(0.71) | LDD2207 | [29] |
| LDCM0131 | RA190 | MM1.R | C95(1.35) | LDD0304 | [30] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Wolframin (WFS1) | . | O76024 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Krueppel-like factor 11 (KLF11) | Sp1 C2H2-type zinc-finger protein family | O14901 | |||
Other
References
